Abstract
Background: Atrial fibrillation (AF) is the leading cause of ischemic stroke and treatment has focused on reducing this risk through anticoagulation. Direct Oral Anticoagulants (DOACs) are the first-line guideline-recommended therapy since they are as effective and overall safer than warfarin in preventing AF-related stroke. Although patients bleed less from DOACs compared to warfarin, bleeding remains the primary safety concern with this therapy.
Hypothesis: Genetic variants known to modify the function of metabolic enzymes or transporters involved in the pharmacokinetics (PK) of DOACs could increase the risk of bleeding.
Aim: To assess the association of eight, functional PK-related single nucleotide variants (SNVs) in five genes (ABCB1, ABCG2, CYP2J2, CYP3A4, CYP3A5) with the risk of bleeding from DOACs in non-valvular AF patients.
Methods: A retrospective cohort study was carried out with 2,364 self-identified white non-valvular AF patients treated with either rivaroxaban or apixaban. Genotyping was performed with Illumina Infinium CoreExome v12.1 bead arrays by the Michigan Genomics Initiative biobank. The primary endpoint was a composite of major and clinically relevant non-major bleeding. Cox proportional hazards regression with time-varying analysis assessed the association of the eight PK-related SNVs with the risk of bleeding from DOACs in unadjusted and covariate-adjusted models. The pre-specified primary analysis was the covariate-adjusted, additive genetic models. Six tests were performed in the primary analysis as three SNVs are in the same haplotype, and thus p-values below the Bonferroni-corrected level of 8.33e-3 were considered statistically significant.
Results: In the primary analysis, none of the SNVs met the Bonferroni-corrected level of statistical significance (all p > 0.1). In exploratory analyses with other genetic models, the ABCB1 (rs4148732) GG genotype tended to be associated with the risk of bleeding from rivaroxaban [HR: 1.391 (95%CI: 1.019–1.900); p = 0.038] but not from apixaban (p = 0.487).
Conclusion: Eight functional PK-related genetic variants were not significantly associated with bleeding from either rivaroxaban or apixaban in more than 2,000 AF self-identified white outpatients.
Keywords: atrial fibrillation, bleeding, anticoagulation, pharmacogenetics, DOAC
Graphical Abstract
Introduction
Atrial fibrillation (AF) is the most common cardiac arrhythmia worldwide and the leading cause of ischemic stroke (Virani et al., 2020). In the US, AF causes about 465,000 hospitalizations (Virani et al., 2020) and 158,000 deaths yearly (Centers for Disease Control and Prevention, 2020), and accounts for up to 20.4% of all ischemic strokes (Alkhouli et al., 2018). Treatment of AF is largely focused on preventing cerebral ischemia through the use of anticoagulant drugs, which reduces the risk of stroke by 66% (Hart et al., 2007). Currently, direct oral anticoagulants (DOACs) are the guideline-recommended first-line therapy (January et al., 2019) since these drugs have proven to be at least equally effective as warfarin in preventing AF-related stroke (Van Ganse et al., 2020). Although DOACs are more convenient and considered safer than warfarin overall (Wang et al., 2020), bleeding still is the main safety concern of this long-term oral therapy. Over 6 years, emergency department (ED) visits for DOAC-related bleeding jumped from 2.3% to 37.9% with the two most prescribed DOACs in the US: rivaroxaban and apixaban (Geller et al., 2020).
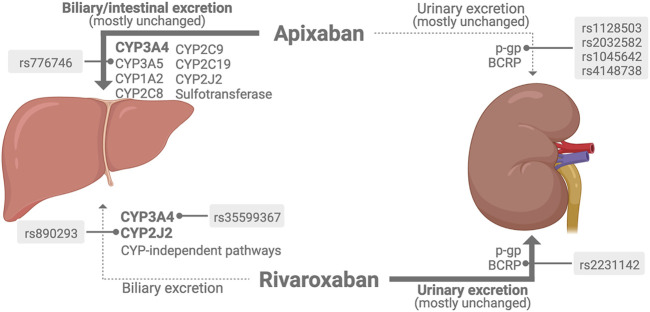
Rivaroxaban and apixaban have in common the direct and highly selective inhibition of factor Xa. However, there are subtle pharmacokinetic (PK) differences between these two DOACs (see Graphical Abstract). Two-thirds of rivaroxaban is metabolized by the liver through CYP3A4 (18%), CYP2J2 (14%), and CYP-independent mechanisms (Mueck et al., 2013, 2014). A third of non-metabolized rivaroxaban is renally excreted via the efflux transporters p-glycoprotein (p-gp) – encoded by ABCB1 gene – and breast cancer resistance protein (BCRP) – encoded by ABCG2 gene (Gnoth et al., 2011; Mueck et al., 2013). In contrast, apixaban is primarily metabolized by CYP3A4/5 with minor contributions from CYP1A2, CYP2C8, CYP2C9, CYP2C19, and CYP2J2 isoenzymes (Wang et al., 2010), and secondly by sulfotransferase (Wang et al., 2009). Apixaban is also a substrate of both p-gp and BCRP efflux transporters (Zhang et al., 2013) and is mainly excreted in feces (47%) and urine (29%) (Byon et al., 2019).
Genetic variants known to modify the function of metabolic enzymes or transporters involved in the pharmacokinetics (PK) of DOACs could hypothetically increase the risk of bleeding from these anticoagulants (Kuehl et al., 2001; Salama et al., 2006; Tomlinson et al., 2010; Wang et al., 2011). For example, the 6981A>G (rs776746) single nucleotide variant (SNV) in CYP3A5 (CYP3A5*3) causes a splicing defect that results in complete loss of enzyme function (Kuehl et al., 2001; Pharmacogene Variation Consortium (PharmVar)). The effect of genetic variants on the PK of DOACs is being increasingly investigated in various pharmacogenetic studies (a summary of previous pharmacogenetic studies of DOACs is displayed in Supplementary Table S1), but the evidence is still unclear. Most of the previous pharmacogenetic studies in this area only focused on changes in plasma concentrations of DOACs (Dimatteo et al., 2016; Ueshima et al., 2017, 2018; Cosmi et al., 2019), and thus it is still unknown whether that translates into differences in the clinical bleeding outcomes.
The few pharmacogenetic studies that used bleeding as an outcome have small sample sizes (typically n < 400), which may have led to their inconclusive results (Ing Lorenzini et al., 2016; Sennesael et al., 2018; Roşian et al., 2020b). The largest pharmacogenetic study performed to date with 1,806 Finnish patients found the ABCB1 (rs4148738) GG genotype as a protective factor for bleeding from apixaban if compared to A carriers (Lähteenmäki et al., 2021). However, those results have not yet been reproduced in other clinical studies. Therefore, this study aims to assess the association of eight functional PK-related genetic variants and the risk of bleeding from DOACs in a large sample of patients diagnosed with non-valvular AF treated with either rivaroxaban or apixaban.
Materials and methods
Study design and cohort identification
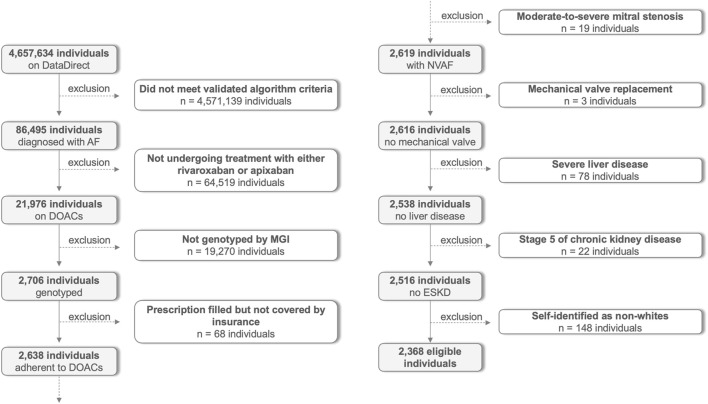
This single-center retrospective cohort study was carried out with 2,364 white outpatients diagnosed with non-valvular AF undergoing treatment with either rivaroxaban or apixaban. Outpatients at Michigan Medicine – a large academic healthcare system affiliated with the University of Michigan in the United States – with atrial fibrillation (n = 86,495) were initially identified according to a validated algorithm (Vanderbilt University PheKB, 2012) through a self-serve tool (DataDirect) that enables the automated data collection from the Electronic Health Record (EHR).
After automated identification based on the diagnosis of AF with subsequent prescription of rivaroxaban or apixaban, each individual was screened for inclusion and exclusion criteria. The complete flow chart showing the participants’ selection is found in Figure 1. Eligible self-identified white and genotyped patients aged 18 or older undergoing treatment for non-valvular AF either with rivaroxaban or apixaban were carefully selected between 1 October 2012 and 31 August 2022. Participants were excluded if they: 1) were diagnosed with moderate-to-severe mitral stenosis; 2) had a history of mechanical valve replacement; 3) were in stage 5 of chronic kidney disease defined as a creatinine clearance of less than 15 mL per minute estimated by the Cockcroft-Gault equation (Cockcroft and Gault, 1976); 4) required renal replacement therapy; 5) were diagnosed with liver disease including those with non-alcoholic fatty liver disease, cirrhosis, total bilirubin >2× normal with AST/ALT/AP > 3× normal, or other severe liver impairment as noted by the physician; 6) were not routinely followed-up by Michigan Medicine; and 7) did not have genotype data available through the Michigan Genomics Initiative (MGI) biobank (University of Michigan, 2022). The study was carried out in accordance with the Declaration of Helsinki and was approved by the local Institutional Review Board with a waiver of informed consent.
FIGURE 1.
Flow chart with participants’ selection. AF: Atrial fibrillation; DOACs: Direct oral anticoagulants; MGI: Michigan Genomics Initiative; NVAF: Non-valvular atrial fibrillation; ESKD: End-stage kidney disease.
Clinical and biochemical evaluation
All clinical and biochemical data for eligible participants were obtained from the University of Michigan data warehouse and completed by trained investigators, as previously described (Hanigan et al., 2020). The first day of the study (index date) was set as the start date of DOAC therapy. Briefly, complete biochemical and clinical summaries were performed through comorbidities identified from the participant’s active problem list or keyword note search at the baseline – defined as the period up to 1 year before the index date. The duration of DOAC treatment was determined through prescription history and keyword search using EMERSE (Hanauer et al., 2015). Study data were collected through a standardized method using a customized form using REDCap (Harris et al., 2009, 2019), and electronic data capture tools are hosted at the University of Michigan. CHA2DS2-VASc, HAS-BLED, and ATRIA scores were calculated as previously described (Lip et al., 2010; Pisters et al., 2010; Fang et al., 2011). Elixhauser comorbidities scores were computed using the “icd” package for R (Wasey JO, 2018).
Study endpoints and adjudication of events
The primary endpoint of this study was a composite of major and clinically relevant non-major (CRNM) bleeding according to the International Society on Thrombosis and Haemostasis (ISTH) criteria (Schulman et al., 2010; Kaatz et al., 2015). The bleeding event must have occurred within the start and stop dates of the DOAC prescription. Briefly, major bleeding was defined as clinically overt non-surgical bleeding with the symptomatic presentation and 1) fatal outcome; and/or 2) involvement of critical anatomical area or site such as intracranial, spinal, intraocular followed by vision changes, pericardial, articular, retroperitoneal, intramuscular with compartment syndrome; and/or 3) hemoglobin fall fall of 2 g/dl or more, or leading to transfusion of two or more units of whole blood or red cells. CRNM bleeding was defined as overt bleeding that does not meet major bleeding criteria but requires at least one of the following criteria: 1) medical intervention by a healthcare professional; and/or 2) hospitalization or increased level of care; and/or 3) a face-to-face evaluation. Since the ISTH are strictly clinical criteria and there is no validated algorithm to help electronically identify bleeding cases according to these criteria, bleeding events were adjudicated and classified by trained clinicians who reviewed all participants’ medical notes using EMERSE (Hanauer et al., 2015). Participants were censored at the time of the first CRNM or major bleeding event or at the end of the study (the date when medical notes were last reviewed). Follow-up time was calculated as the difference between censoring and index dates.
Exposure to and drug interaction with DOACs
Normalized daily doses of rivaroxaban and apixaban were calculated by multiplying the prescribed strength by the number of daily doses (usually one for rivaroxaban and two for apixaban) and then divided by the daily maintenance dose of each drug (20 mg for rivaroxaban and 10 mg for apixaban). Time-varying exposure to DOACs was calculated as the normalized dose multiplied by the duration of treatment within each treatment period (i.e., prescription start and stop dates). Cumulative exposure was the sum of all time-varying exposure over the follow-up period. Drug-drug interactions with DOACs were defined as the concomitant and systemic use of either CYP/p-gp inhibitors or inducers with good or excellent documentation of evidence on Micromedex (IBM Micromedex Solutions, 2022). Twelve drugs were considered as CYP/p-gp inhibitors 1) amiodarone, 2) clarithromycin, 3) conivaptan, 4) cyclosporine, 5) diltiazem, 6) dronedarone, 7) fluconazole, 8) itraconazole, 9) ritonavir, 10) erythromycin, 11) ketoconazole, and 12) verapamil; and seven drugs were considered as CYP/p-gp inducers 1) apalutamide, 2) carbamazepine, 3) fosphenytoin, 4) nevirapine, 5) oxcarbazepine, 6) phenytoin, and 7) rifampin for drug interactions with DOACs.
Candidate variant selection and genotyping
Genes involved in the PK properties of rivaroxaban and apixaban were considered potential candidates. PubMed and the Pharmacogenomics Knowledgebase (PharmGKB) were searched in December 2021 using the following terms [(polymorphism OR genetic variant) AND (drug response OR pharmacogenetic effect) AND (gene)]. Candidate genetic variants were selected according to the following criteria: 1) minor allele frequency (MAF) ≥5% in European ancestry; 2) variants with clinical and/or functional effects; and 3) genetic variants that have been imputed or genotyped by Michigan Genomics Initiative and whose information is part of our genomic bank.
Genomic DNA was extracted from peripheral blood leukocytes and genotyping was performed as part of the MGI, as previously described elsewhere (Fritsche et al., 2018, 2019). Briefly, samples were genotyped at the University of Michigan Advanced Genomics Core lab using Illumina Infinium CoreExome v12.1 bead arrays® (Illumina, San Diego, CA) with standard quality checks (Zajac et al., 2019). Eight functional single nucleotide variants (SNVs) distributed in five different genes encoding either metabolic enzymes or transporters related to the PK of DOACs were chosen for analysis. The PK properties of DOACs mapping the eight SNVs were illustrated using BioRender (BioRender, 2022) and are found in the Graphical Abstract. The eight selected SNVs are as follows: three ABCB1 SNVs located in exons 13 (rs1128503, c.1236 T>C, p. Gly412Gly), 22 (rs2032582, c.2677T>G/A, p. Ser893Thr/Ala) and 26 (rs1045642, c.3435T>C, p. Ile1145Ile); one ABCB1 SNV in intron 18 (rs4148738, c.2482–2236G>A); one ABCG2 SNV in exon 5 (rs22131142, c.421C>A, p. Glu141Lys); one CYP3A4 SNV in intron 6 (rs35599367, c.15389C>T); one CYP3A5 SNV in intron 3 (rs776746, c.6981A>G); and one CYP2J2 SNV in the gene promoter (rs890293, c.-76G>T). All eight SNVs were imputed. Genotype imputation was performed by the MGI using the Michigan Imputation Served and Trans-Omics for Precision Medicine (TOPMed) reference panel, resulting in ∼52 million variants (post quality control filtering) with imputation r 2 > 0.3 (Taliun et al., 2021).
Since the three closely positioned ABCB1 SNVs – 1236T>C (rs1128503), 2677T>G/A (rs2032582), and 3435T>C (rs1045642) – are in high linkage disequilibrium, these three SNVs were analyzed as diplotypes. As 1236C-2677G-3435C is considered the wild-type, three diplotype subgroups encoding an additive 0/1/2 modeling were created as follows: 1) homozygous: 1236C-2677G-3435C/1236C-2677G-3435C; 2) heterozygous: 1236C-2677G-3435C/haplotype other than 1236C-2677G-3435C; and 3) other: haplotype other than 1236C-2677G-3435C/haplotype other than 1236C-2677G-3435C. Haplotypes were estimated from the genotype data by using the PHASE (version 2.1.1) software package.
Statistical analysis
Data distribution was assessed for normality using the Kolmogorov-Smirnov test and visual inspection of plots. Normally distributed baseline data are presented as mean ± SD and non-normally distributed data are presented as the median and interquartile range (IQR). Categorical and continuous baseline data were tested using chi-square and Student t-tests, respectively. Hardy-Weinberg Equilibrium was assessed using the chi-square test. Participants were stratified into two subgroups according to their bleeding status. Multivariable Cox proportional hazards regression models with time-varying analysis were used to assess independent clinical predictors for bleeding risk from either rivaroxaban or apixaban (those with p < 0.05). Then, Cox proportional hazards regression models with time-varying analysis were used again to assess the association of each SNV with the risk of bleeding from DOACs with the significant clinical predictors used as covariates in model adjustment for the SNVs. Hazard ratios (HR) and respective 95% confidence intervals (95%CI) were calculated for unadjusted (Model 1) and adjusted models (Model 2). The primary analysis was pre-specified as the clinical covariate-adjusted models (Model 2) in the additive genetic model for each SNV (i.e., six statistical tests for the primary SNV analysis because three of the eight SNVs are in the same haplotype). Exploratory analyses were performed to assess the association of other genetic models (i.e., dominant and recessive), each ABCB1 haplotype, and for rivaroxaban and apixaban separately. p-values below the Bonferroni-corrected level of 8.33 × 10−3 were considered statistically significant for the primary SNV analyses. This primary analysis had an estimated power of 80% to detect an HR of ≥1.26 for each variant. RGui was used for all statistical analyses.
Results
Baseline characteristics
The clinical and biochemical baseline characteristics of participants stratified by composite endpoint are shown in Table 1. Participants who bled with DOACs were older, had higher rates of previous surgery or trauma, transcatheter aortic valve replacement, previous bleeds, prior thrombosis, former smokers, and higher CHA2DS2-VASc, HAS-BLED, ATRIA, and Elixhauser comorbidities scores than the participants who did not bleed. There was no significant difference in any of the other variables assessed, such as sex, ethnicity, previous stroke, transient ischemic attack, body mass index, blood pressure, creatinine clearance, platelet count, hemoglobin, or follow-up time.
TABLE 1.
Clinical and biochemical baseline characteristics of participants.
| Characteristics | Overall | CRNM + major bleeding | p-value | |
|---|---|---|---|---|
| No | Yes | |||
| Self-identified white participants, n (%) | 2364 (100.0) | 1952 (82.6) | 412 (17.4) | — |
| Age, years | 68.3 (13.6) | 67.7 (13.8) | 71.0 (12.4) | <0.001 |
| Sex, n (%) | 0.553 | |||
| Female | 758 (32.1) | 631 (32.3) | 127 (30.8) | |
| Male | 1606 (67.9) | 1321 (67.7) | 285 (69.2) | |
| Ethnicity, n (%) | 0.553 | |||
| Non-Hispanic or Latino | 2312 (97.8) | 1911 (97.9) | 401 (97.3) | |
| Hispanic or Latino | 10 (0.4) | 7 (0.4) | 3 (0.7) | |
| Unknown | 42 (1.8) | 34 (1.7) | 8 (1.9) | |
| DOAC, n (%) | <0.001 | |||
| Rivaroxaban | 802 (33.9) | 654 (33.5) | 148 (35.9) | |
| Apixaban | 1324 (56.0) | 1124 (57.6) | 200 (48.6) | |
| Both DOACs not simultaneously | 238 (10.1) | 174 (8.9) | 64 (15.5) | |
| Daily dose of DOAC, mg | ||||
| Rivaroxaban | 19.3 ± 3.5 | 19.3 ± 3.6 | 19.3 ± 2.3 | 0.897 |
| Apixaban | 7.8 ± 4.1 | 7.6 ± 4.2 | 9.1 ± 2.6 | <0.001 |
| Cumulative dose of DOAC, g | ||||
| Rivaroxaban | 17.2 ± 15.5 | 16.4 ± 15.2 | 21.0 ± 17.0 | <0.001 |
| Apixaban | 7.9 ± 6.5 | 7.5 ± 6.4 | 9.7 ± 7.0 | <0.001 |
| Drug-drug interactions with DOACs, n (%) | ||||
| CYP/p-gp inhibitors | 1046 (44.5) | 807 (41.3) | 239 (58.0) | <0.001 |
| CYP/p-gp inducers | 33 (1.4) | 24 (1.2) | 9 (2.2) | 0.133 |
| Previous surgery or trauma, n (%) | 1449 (61.3) | 1162 (59.5) | 287 (69.7) | <0.001 |
| Transcatheter aortic valve replacement, n (%) | 22 (0.9) | 14 (0.7) | 8 (1.9) | 0.019 |
| Previous bleeding, n (%) | 491 (20.8) | 384 (19.7) | 107 (26.0) | 0.005 |
| Previous stroke, n (%) | 50 (2.1) | 37 (1.9) | 13 (3.2) | 0.106 |
| Previous transient ischemic attack, n (%) | 37 (1.6) | 30 (1.5) | 7 (1.7) | 0.810 |
| Previous thromboembolism, n (%) | 90 (3.8) | 65 (3.3) | 25 (6.1) | 0.008 |
| CHA2DS2-VASc score | 2.4 ± 1.5 | 2.4 ± 1.5 | 2.8 ± 1.6 | <0.001 |
| Elixhauser comorbidities score | 11.0 ± 11.3 | 10.7 ± 11.3 | 12.4 ± 11.1 | <0.001 |
| HAS-BLED score | 1.3 ± 1.1 | 1.2 ± 1.0 | 1.5 ± 1.1 | <0.001 |
| ATRIA score | 2.3 ± 2.0 | 2.2 ± 2.0 | 2.7 ± 2.0 | 0.002 |
| Smoking habit, n (%) | <0.001 | |||
| Never | 1052 (44.5) | 896 (45.9) | 156 (37.9) | |
| Current | 67 (2.8) | 61 (3.1) | 6 (1.5) | |
| Former | 1222 (51.7) | 972 (49.8) | 250 (60.7) | |
| Unknown | 23 (1.0) | 23 (1.2) | 0 (0.0) | |
| Body mass index, kg/m2 | 29.9 (8.4) | 30.0 (8.4) | 29.8 (8.8) | 0.715 |
| Systolic blood pressure, mmHg | 127.5 (21.5) | 127.5 (22.0) | 128.0 (19.5) | 0.350 |
| Diastolic blood pressure, mmHg | 71.0 (11.5) | 71.0 (11.5) | 70.0 (10.3) | 0.174 |
| Creatinine clearance, mL/min | 92.2 (49.0) | 92.6 (48.8) | 89.7 (50.6) | 0.248 |
| Platelet count, 109/L | 209.8 (74.0) | 209.0 (71.0) | 212.5 (89.3) | 0.069 |
| Hemoglobin, g/dL | 13.5 (2.5) | 13.5 (2.4) | 13.3 (2.6) | 0.388 |
| Follow-up time, (days) | 828.2 ± 739.8 | 834.2 ± 740.9 | 800.2 ± 734.5 | 0.397 |
CRNM: clinically relevant non-major; DOAC: direct oral anticoagulants; CYP: Cytochrome P450; p-gp: p-glycoprotein.
Additionally, the clinical and biochemical baseline characteristics of participants stratified by major bleeding and by DOAC treatment are shown in Supplementary Tables S2, S3, respectively. Briefly, patients who had major bleeding were older, changed from one to another DOAC more frequently, and were significantly more exposed to CYP/p-gp inhibitors while on DOACs than their counterparts. Also, they underwent more significant transcatheter aortic valve replacement and had higher CHA2DS2-VASc, HAS-BLED, ATRIA, and Elixhauser comorbidities scores than those who did not bleed. A significantly higher frequency of former smokers and lower diastolic blood pressure, creatinine clearance, and hemoglobin levels at baseline was observed among patients who majorly bled than their counterparts (Supplementary Table S2).
Regarding patients on rivaroxaban, they were significantly healthier than patients on apixaban. Although patients on rivaroxaban bled more frequently than their counterparts, they were significantly younger, had less frequency of surgery or trauma, transcatheter aortic valve replacement, previous bleeding, and stroke, had significantly lower CHA2DS2-VASc, HAS-BLED, ATRIA and Elixhauser comorbidities scores and higher creatinine clearance than those on apixaban. In contrast, patients on rivaroxaban had significantly higher body mass index, were more frequently exposed to CYP/p-gp inhibitors while anticoagulated and were followed up for a longer time than their counterparts (Supplementary Table S3).
DOAC treatment and prevalence and incidence of bleeding
A total of 802 participants (33.9%) were exclusively treated with rivaroxaban, 1,324 participants (56.0%) were exclusively treated with apixaban, and 238 participants (10.1%) started anticoagulation with one of the two DOACs and then switched to the other DOAC. The two main causes observed for this change during the medical chart reviews were: 1) non-coverage of the DOAC initially prescribed by the health insurance (affordability), and 2) bleeding.
A total of 412 CRNM and major bleeding events were observed in unique patients with a prevalence of 17.4% at a mean follow-up time of 2.3 ± 2.0 years (828.2 ± 739.8 days). Of these 412 patients who bled, 35 of them (8.5%) recurred either major or CRNM bleeding. The prevalence of bleeding was significantly higher among the 1,040 participants treated with rivaroxaban than among those 1,562 participants treated with apixaban (17.69% versus 14.59%, p = 0.034, respectively). The overall incidence was 7.25 bleeding events per 100 person-years, being significantly higher among participants treated with rivaroxaban than apixaban [8.32 (95%CI: 7.16–9.61) vs. 6.57 (95%CI: 5.74–7.48) bleeding events per 100 person-years; p-value = 0.018, respectively], and resulting in an incidence rate ratio of 1.27 (95%CI: 1.04–1.54) bleeding events per 100 person-years for rivaroxaban versus apixaban.
Although there was no difference in the follow-up time between the bleeding groups, the cumulative dose of both rivaroxaban and apixaban were significantly higher among participants who bled in comparison to those who did not bleed (Rivaroxaban: 21.02 ± 16.95 versus 16.38 ± 15.18 g, p < 0.001. Apixaban: 9.70 ± 7.01 versus 7.48 ± 6.39 g, p < 0.001). The daily dose of apixaban was significantly higher in those who bled than their counterparts (9.09 ± 2.62 versus 7.60 ± 4.20 mg, p < 0.001), and there was no significant difference in the daily doses of rivaroxaban between bleeding groups (19.30 ± 2.35 versus 19.27 ± 3.61 mg, p = 0.897). Regarding drug-drug interactions with DOACs, concomitant use of CYP/p-gp inhibitors was significantly higher among participants who bled compared to those who did not bleed (239 (58.0%) versus 807 (41.3%), p < 0.001, respectively). However, there was no significant difference in the concomitant use of CYP/p-gp inducers between the study groups (24 (1.2%) versus 9 (2.2%), p = 0.133).
Independent clinical predictors of bleeding from DOACs
Clinical and biochemical variables that either differed at baseline or varied over the follow-up time were tested using multivariable Cox proportional hazards regression with time-varying analysis to identify independent clinical predictors of major + CRNM bleeding from DOACs in our patient sample. The results are shown in Table 2. Age, previous bleeding, Elixhauser comorbidities score, previous thromboembolism, former and current smoking, use of rivaroxaban (vs. apixaban), and normalized dose of DOAC were independently associated with the study endpoint and were selected as covariates for the SNV analyses. After adjustment for clinical predictors, compared to apixaban users, rivaroxaban users were at a 33% higher risk of bleeding (p = 0.003).
TABLE 2.
Multivariable Cox proportional hazards regression with time-varying analysis assessing clinical characteristics as clinical predictors of major + CRNM bleeding from DOACs.
| β | SE (β) | P | eβ (HR) | 95% CI for HR | ||
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| Age, years | 0.024 | 0.006 | <0.001 | 1.025 | 1.012 | 1.037 |
| History of surgery or trauma | 0.067 | 0.116 | 0.585 | 1.069 | 0.842 | 1.359 |
| Previous bleeding | 0.346 | 0.122 | 0.005 | 1.413 | 1.110 | 1.799 |
| CHA2DS2-VASc score | 0.064 | 0.042 | 0.133 | 1.066 | 0.981 | 1.158 |
| Elixhauser comorbidities score | 0.016 | 0.005 | 0.002 | 1.016 | 1.006 | 1.026 |
| Previous thromboembolism | 0.484 | 0.230 | 0.042 | 1.623 | 1.019 | 2.586 |
| Former and current smoking | 0.122 | 0.049 | 0.016 | 1.130 | 1.023 | 1.248 |
| DOAC (apixaban = 0; rivaroxaban = 1) | 0.289 | 0.097 | 0.003 | 1.335 | 1.105 | 1.613 |
| Normalized dose of DOAC, % | 0.007 | 0.002 | 0.006 | 1.007 | 1.002 | 1.012 |
| Concomitant use of CYP/p-gp inhibitors | 0.099 | 0.124 | 0.447 | 1.105 | 0.855 | 1.428 |
| Creatinine clearance, mL/min | 1.768e−05 | 0.002 | 0.991 | 1.000 | 0.997 | 1.003 |
DOAC: direct oral anticoagulants; CYP: Cytochrome P450; p-gp: p-glycoprotein; HR: hazard ratio; 95%CI: 95% confidence interval.
PK-related genetic variants and the risk of bleeding from DOACs
The genotype and allele frequencies of the eight genetic variants are shown in Table 3. All genotype frequencies were in Hardy-Weinberg equilibrium with p-values >0.05, and all of the allele frequencies were similar to those previously reported for Europeans in publicly available databases (i.e., Ensembl, gnomAD) or previous publications. In the pre-specified primary analysis (i.e., clinical covariate-adjusted Model 2 with the additive genetic model), none of the SNVs met the Bonferroni-corrected level of statistical significance (Table 4). In exploratory analyses with the dominant and recessive genetic models, some of the tests tended to show an association for SNVs in CYP3A5 (rs776746) and ABCB1 (rs4148732) (Supplementary Table S4). Also, another exploratory analysis was performed to scrutinize the association of PK-related genetic variants with bleeding from rivaroxaban and apixaban separately (Supplementary Table S5). Similar to the results for the two DOACs combined, none of the variants were statistically significant in the additive genetic model, and the associations with CYP3A5 (rs776746) and ABCB1 (rs4148732) in other genetic models seemed to be stronger for rivaroxaban than apixaban. The ABCB1 (rs4148732) GG genotype tended to be associated with a higher risk of bleeding from rivaroxaban [HR: 1.39 (95%CI: 1.02–1.90; p = 0.038] but not from apixaban (p = 0.487). Since the ABCB1 diplotypes were not significantly associated with bleeding from DOACs, an exploratory analysis scrutinizing the association of each ABCB1 haplotype with the risk of bleeding was performed, and the results are shown in Table 5. The 1236C-2677G-3435T ABCB1 haplotype tended to be protective against bleeding (p = 0.036).
TABLE 3.
Genotype and allele frequency, and Hardy-Weinberg equilibrium (HWE) assessment of the eight PK-related genetic variants.
| Allele frequency | Statistical comparison of allele frequency (χ2; p-value) | Genotypes and frequencies, n (%) | HWE (χ2; p-value) | |
|---|---|---|---|---|
| European ancestry (reference) | Our cohort | |||
| G: 95.00% | G: 95.37% | 0.000; p = 1.000 | CYP3A4 (rs35599367) | 0.187; p = 0.666 |
| A: 5.00% | A: 4.63% | GG: 2,151 (91.0) | ||
| Cunningham et al. (2022) | GA: 207 (8.8) | |||
| AA: 6 (0.2) | ||||
| A: 5.70% | A: 7.15% | 0.082; p = 0.774 | CYP3A5 (rs776746) | 0.985; p = 0.321 |
| G: 94.30% | G: 92.85% | AA: 8 (0.4) | ||
| Cunningham et al. (2022) | AG: 322 (13.6) | |||
| GG: 2,034 (86.0) | ||||
| T: 5.70% | T: 6.79% | 0.082; p = 0.774 | CYP2J2 (rs890293) | 1.778; p = 0.182 |
| G: 94.30% | G: 93.21% | TT: 15 (0.6) | ||
| Cunningham et al. (2022) | GT: 291 (12.3) | |||
| GG: 2,058 (87.1) | ||||
| C: 90.60% | C: 89.57% | 0.058; p = 0.809 | ABCG2 (rs2231142) | 1.922; p = 0.165 |
| A: 9.40% | A: 10.43% | CC: 1,903 (80.5) | ||
| Cunningham et al. (2022) | CA: 429 (18.1) | |||
| AA: 32 (1.4) | ||||
| T: 41.60% | T: 43.51% | 0.082; p = 0.775 | ABCB1 (rs1128503) | 0.777; p = 0.378 |
| C: 58.40% | C: 56.49% | TT: 458 (19.4) | ||
| Cunningham et al. (2022) | TC: 1,141 (48.3) | |||
| CC: 765 (32.3) | ||||
| T: 41.00% | T: 45.05% | 0.202; p = 0.653 | ABCB1 (rs2032582) | 0.465; p = 0.495 |
| G: 57.30% | G: 54.95% | TT: 488 (20.6) | ||
| Cunningham et al. (2022) | TG: 1,154 (48.8) | |||
| GG: 722 (30.6) | ||||
| T: 51.80% | T: 53.74% | 0.080; p = 0.777 | ABCB1 (rs1045642) | 0.262; p = 0.609 |
| C: 48.20% | C: 46.26% | TT: 689 (29.1) | ||
| Cunningham et al. (2022) | TC: 1,163 (49.2) | |||
| CC: 512 (21.7) | ||||
| G: 49.41% | G: 44.97% | 0.321; p = 0.571 | ABCB1 (rs4148732) | 1.770; p = 0.183 |
| A: 50.59% | A: 55.03% | GG: 494 (20.9) | ||
| Lähteenmäki et al. (2021) | GA: 1,138 (48.1) | |||
| AA: 732 (31.0) | ||||
TABLE 4.
Cox proportional hazards regression with time-varying analysis assessing the association of additive PK-related genetic variant models with DOACs bleeding risk.
| CYP3A4 (rs35599367) | HR (95%CI; p-value) |
| AA vs. AG vs. GG genotypes | |
| Model 1 | 0.876 (0.691–1.110); 0.274 |
| Model 2 | 0.891 (0.708–1.122); 0.327 |
| CYP3A5 (rs776746) | HR (95%CI; p-value) |
| GG vs. GA vs. AA genotypes | |
| Model 1 | 0.960 (0.685–1.347); 0.814 |
| Model 2 | 0.943 (0.687–1.294); 0.716 |
| CYP2J2 (rs890293) | HR (95%CI; p-value) |
| GG vs. GT vs. TT genotypes | |
| Model 1 | 1.133 (0.873–1.471); 0.349 |
| Model 2 | 1.131 (0.871–1.468); 0.357 |
| ABCG2 (rs2231142) | HR (95%CI; p-value) |
| CC vs. CA vs. AA genotypes | |
| Model 1 | 1.076 (0.882–1.314); 0.469 |
| Model 2 | 1.055 (0.863–1.289); 0.602 |
| ABCB1 (rs4148732) | HR (95%CI; p-value) |
| GG vs. GA vs. AA genotypes | |
| Model 1 | 1.113 (0.969–1.277); 0.129 |
| Model 2 | 1.096 (0.956–1.256); 0.188 |
| ABCB1 C-G-C diplotypes | HR (95%CI; p-value) |
| Homozygous vs. hetero vs. other | |
| Model 1 | 0.999 (0.868–1.148); 0.983 |
| Model 2 | 1.027 (0.895–1.179); 0.707 |
Model 1: Unadjusted model. Model 2: Fully adjusted for age, previous bleeding, Elixhauser comorbidities score, previous thromboembolism, smoking, normalized dose, and DOAC. The underlined genotypes were encoded as the risk genotype in the additive genetic model, and thus the HR for each variant was hypothesized to be > 1.
TABLE 5.
Cox proportional hazards regression with time-varying analysis assessing the independent association of 1236C>T (rs1128503), 2677G>T/A (rs2032582), and 3435C>T (rs1045642) ABCB1 haplotypes with bleeding from DOACs.
| Haplotypes | Overall n (%) | Major + CRNM bleeding a | χ2; p-value a | HR (95%CI; p-value a | ||
|---|---|---|---|---|---|---|
| No | Yes | Unadjusted | Fully adjusted b | |||
| C-G-C | 1,961 (41.5) | 1,621 (65.7) | 340 (63.1) | 3.117; 0.210 | 0.999 (0.868–1.148); 0.983 | 1.027 (0.895–1.179); 0.707 |
| T-T-T | 1,888 (39.9) | 1,546 (63.6) | 342 (64.6) | 2.012; 0.366 | 1.127 (0.976–1.301); 0.102 | 1.092 (0.948–1.258); 0.223 |
| C-G-T | 558 (11.8) | 470 (22.7) | 88 (20.1) | 1.278; 0.528 | 0.826 (0.672–1.017); 0.071 | 0.794 (0.640–0.985); 0.036 |
| T-T-C | 90 (1.9) | 71 (3.4) | 19 (4.6) | 2.772; 0.302 | 0.971 (0.641–1.471); 0.890 | 1.071 (0.699–1.642); 0.753 |
| C-T-T | 90 (1.9) | 75 (3.8) | 15 (3.6) | 0.003; 0.958 | 0.959 (0.607–1.514); 0.856 | 0.970 (0.617–1.525); 0.893 |
| T-G-C | 74 (1.6) | 65 (3.3) | 9 (2.2) | 1.118; 0.290 | 0.601 (0.292–1.236); 0.166 | 0.613 (0.290–1.298); 0.201 |
| C-T-C | 62 (1.3) | 51 (2.6) | 11 (2.7) | 0.246; 0.884 | 0.896 (0.513–1.566); 0.700 | 0.970 (0.592–1.590); 0.905 |
| T-G-T | 5 (0.1) | 5 (0.3) | 0 (0.0) | 0.192; 0.594 | 2.2542e−06 (8.035e−07–6.325e−06); <0.001 | 4.996e−06 (1.773e−06–1.408e−05); <0.001 |
CRNM: Clinically relavant non-major bleeding; χ2: chi-square; OR: odds ratio; 95%CI: 95% confidence interval.
Fully adjusted for age, previous bleeding, Elixhauser comorbidities score, previous thromboembolism, smoking, normalized dose, and DOAC.
Discussion
To the best of our knowledge, this is the largest pharmacogenetic study to assess the association of PK-related genetic variants with the risk of bleeding in non-valvular AF white outpatients undergoing treatment with either rivaroxaban or apixaban. Eight SNVs known to be functionally involved with either the metabolism or excretion of DOACs were selected for analysis (Kuehl et al., 2001; Salama et al., 2006; Tomlinson et al., 2010; Wang et al., 2011). None of the eight SNVs met the pre-specified Bonferroni-corrected level of statistical significance for the risk of bleeding from DOACs. Our findings are in line with recent evidence from a candidate gene study that tested virtually the same genetic variants without any of them being associated with bleeding risk from apixaban (Attelind et al., 2022). In exploratory analyses, a tendency of association with a p-value <0.05 was observed between the CYP3A5 rs776746 G carriers and reduced risk of bleeding from DOACs. However, the number of patients with the CYP3A5 rs776746 AA genotype in this sample was extremally small (n = 8), and thus this result is most likely due to chance. Future pharmacogenetic studies should be conducted with this variant in AF patients of African ancestry, in whom the A allele is much more frequent (in more than 70%) (Cunningham et al., 2022). The ABCB1 rs4148732 GG genotype also tended to be associated with an increased risk of bleeding on DOACs in exploratory analyses, and there was a large number of patients with the GG genotype (n = 494). However, this finding is also likely due to chance because it is not supported by previous pharmacogenetic studies of bleeding. None of the ABCB1 rs4148732 genotypes was significantly associated with bleeding from apixaban in a previous study (Roşian et al., 2020a), and our results contradict another study finding that the ABCB1 rs4148732GG was a protective genotype against bleeding from apixaban (Lähteenmäki et al., 2021). This previous study did not correct the level of significance for multiple comparisons (Bromley et al., 2009), and thus their previous finding for the ABCB1 rs4148732GG genotype, like in our exploratory analysis, is most likely due to chance.
There are three main reasons that most likely explain the lack of significant association between the PK-related genetic variants and bleeding risk from DOACs in our findings. First, although genetic variants affecting the functionality of enzymes involved in the main metabolic pathways of rivaroxaban (CYP3A4 and CYP2J2) and apixaban (CYP3A4) have been tested, several other enzymes also play a role in the metabolism of these drugs (e.g., CYP1A2, CYP2C8, CYP2C9, CYP2C19, sulfotransferase). Therefore, these other enzymes may compensate for the genetic effects on the main pathway and preserve the overall metabolism of DOACs. These more complex PK pathways of rivaroxaban and apixaban differ from other well-known pharmacogenetic examples, in which the drug’s metabolism or transport is primarily dependent on a single enzyme or transporter (e.g., clopidogrel and CYP2C19 or simvastatin and SLCO1B1) (Cooper-DeHoff et al., 2022; Lee et al., 2022). Furthermore, as opposed to a purely PK outcome, we assessed the clinical outcome of bleeding, and PK is just one side of this pharmacological double-edged sword. The coagulation cascade is highly complex and works from the interaction of multiple factors and cofactors, and genetic variants affecting pharmacodynamic (PD) changes of DOACs were not assessed in this or any other pharmacogenetic study to date.
Second, the effects of these genetic variants on DOAC exposure may not be strong enough to translate into differences in clinical outcomes. Bleeding complications during DOAC treatment have been shown to be more frequent when peak DOAC plasma levels reached 266% for rivaroxaban and 156% for apixaban compared to baseline (Testa et al., 2019). However, the only two pharmacogenetic studies with significant results for peak plasma levels of DOACs showed smaller effect sizes. Increases of 106% and 26% for peak plasma concentrations of rivaroxaban and apixaban were respectively reported in individuals with the ABCB1 rs4148732 GG (Cosmi et al., 2019) and AA genotypes compared to the homozygous counterparts (Dimatteo et al., 2016). The notion that genetic variants would have small effects on DOAC exposure that would not translate into clinical outcomes is supported by recent evidence that carriers of T allele ABCG2 (rs2231142) have a 5% and 17% higher exposure to apixaban in heterozygous and homozygous, respectively, without any association to bleeding risk from this DOAC (Attelind et al., 2022). Of course, future studies need to confirm these findings, but this preliminary evidence suggests that these genetic variants would not have an effect size large enough on the PK to translate into clinical hemorrhagic outcomes.
The third reason that could explain the lack of significant association in our study is the candidate gene study design. Candidate gene studies are hypothesis-driven with the advantage of having high statistical power in detecting gene-drug associations. However, these studies miss the thousands of other genes and millions of other variants present in the genome–such as the PD-related genetic variants mentioned above or other yet unknown PK variants. Indeed, recent evidence has shown that the vast majority of candidate variants selected for pharmacogenetic studies (98%) are not the best candidates (Linskey et al., 2021). Hence, genome-wide association studies (GWAS) are crucial to discover the most important genetic variants underlying bleeding from DOACs. Although recent evidence from a large GWAS has shown that the genetic variant in the drug transporter gene ABCG2 is associated with the pharmacokinetics of apixaban, this genetic variant had a small influence on drug exposure (Attelind et al., 2022). Future GWAS continues to be needed to identify and uncover the genetic mechanisms underlying the risk of bleeding from DOACs. Another weakness of candidate gene studies lies in the fact that its design also misses polygenic effects (i.e., the effects of multiple genetic variants combined). The most recent evidence shows that interindividual variability in drug response is explained by polygenic inheritance (Lanfear et al., 2020; Johnson et al., 2022). Therefore, it may be that each of the genetic variants has a small effect size on bleeding that is not statistically significant if analyzed individually through a candidate gene study, but perhaps the small effect sizes need to be aggregated into a polygenic score weighted so that its clinical significance can then be determined.
Although our study has some major strengths, such as having clinical outcomes rather than blood levels of DOACs and testing the largest patient sample to date, some limitations must be taken into consideration. This was an observational study, and our findings are limited to a single health system that primarily serves southeastern Michigan. All data were obtained from the EHR, which limits the research to certain variables. Although our data collection has followed standardized protocols, there is no standardization for the clinical care routine and, as a limitation of observational studies, unmeasured residual confounding may have been incorporated. The drug prescribing was defined from EHR data, and clinician reasoning could not be obtained to determine the thought process of prescribing these drugs. We did not have access to pharmacy prescription fill records, so adherence to the DOAC therapy could not be assessed. Finally, not all potential candidates (e.g., rs1042028 in SULT1A1) were covered by the genotyping performed by Michigan Genomics Initiative.
Conclusion
In conclusion, eight functional PK-related genetic variants were not significantly associated with the bleeding risk in more than 2,000 white Americans diagnosed with non-valvular atrial fibrillation and treated with either rivaroxaban or apixaban in a large health system.
Data availability statement
The datasets presented in this article are not readily available because of patient privacy and confidentiality restrictions. Requests to access the datasets should be directed to https://precisionhealth.umich.edu/tools-resources/data-access-tools/how-to-access-data-tools-analytic-environments/
Ethics statement
The studies involving human participants were reviewed and approved by University of Michigan Institutional Review Board. The ethics committee waived the requirement of written informed consent for participation.
Author contributions
Conceptualization: AC-S, JL, H-JZ, GB, and NL. Design: AC-S, JL, H-JZ, GB, and NL. Data analysis: AC-S. Writing: AC-S and JL. Critical review: MD, H-JZ, GB, NL, and JL.
Funding
AC-S is funded by the Michigan Institute for Clinical & Health Research (U074551), and the National Heart, Lung, and Blood Institute of the NIH (1F32HL162231-01A1). GB is funded by Boston Scientific, NHLBI, AHRQ, and Blue Cross Blue Shield of Michigan. JL is funded by the National Heart, Lung, and Blood Institute of NIH (K08 HL146990).
Conflict of interest
MD has received honoraria from Jansen and research funding from BMS/Pfizer, Amgen, Agency for Healthcare Research and Quality, NIH/National Institute of Aging, and the American Heart Association in the past 2 years. GB is a consultant for Pfizer, Bristol-Myers Squib, Janssen, Boston Scientific, and Abbott Vascular.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.1007113/full#supplementary-material.
References
- Alkhouli M., Alqahtani F., Aljohani S., Alvi M., Holmes D. R. (2018). Burden of atrial fibrillation–associated ischemic stroke in the United States. JACC. Clin. Electrophysiol. 4, 618–625. 10.1016/j.jacep.2018.02.021 [DOI] [PubMed] [Google Scholar]
- Attelind S., Hallberg P., Wadelius M., Hamberg A.-K., Siegbahn A., Granger C. B., et al. (2022). Genetic determinants of apixaban plasma levels and their relationship to bleeding and thromboembolic events. Front. Genet. 13, 982955. Available at:. 10.3389/fgene.2022.982955 https://www.frontiersin.org/articles/10.3389/fgene.2022.982955 (Accessed November 2, 2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- BioRender (2022). BioRender. Available at: https://biorender.com/.
- Bromley C. M., Close S., Cohen N., Favis R., Fijal B., Gheyas F., et al. (2009). Designing pharmacogenetic projects in industry: Practical design perspectives from the industry pharmacogenomics working group. Pharmacogenomics J. 9, 14–22. 10.1038/tpj.2008.11 [DOI] [PubMed] [Google Scholar]
- Byon W., Garonzik S., Boyd R. A., Frost C. E. (2019). Apixaban: A clinical pharmacokinetic and pharmacodynamic review. Clin. Pharmacokinet. 58, 1265–1279. 10.1007/s40262-019-00775-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (2020). Underlying cause of death, 1999-2018. Available at: https://wonder.cdc.gov/ucd-icd10.html. [Google Scholar]
- Cockcroft D. W., Gault M. H. (1976). Prediction of creatinine clearance from serum creatinine. Nephron 16, 31–41. 10.1159/000180580 [DOI] [PubMed] [Google Scholar]
- Cooper-DeHoff R. M., Niemi M., Ramsey L. B., Luzum J. A., Tarkiainen E. K., Straka R. J., et al. (2022). The clinical pharmacogenetics implementation consortium guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and statin-associated Musculoskeletal Symptoms. Clin. Pharmacol. Ther. 111, 1007–1021. 10.1002/cpt.2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosmi B., Salomone L., Cini M., Guazzaloca G., Legnani C. (2019). Observational study of the inter-individual variability of the plasma concentrations of direct oral anticoagulants (dabigatran, rivaroxaban, apixaban) and the effect of rs4148738 polymorphism of ABCB1. J. Cardiol. Ther. 7, 8–14. 10.12970/2311-052x.2019.07.02 [DOI] [Google Scholar]
- Cunningham F., Allen J. E., Allen J., Alvarez-Jarreta J., Amode M. R., Armean I. M., et al. (2022). Ensembl 2022. Nucleic Acids Res. 50, D988–D995. 10.1093/nar/gkab1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimatteo C., D’Andrea G., Vecchione G., Paoletti O., Tiscia G. L., Santacroce R., et al. (2016). ABCB1 SNP rs4148738 modulation of apixaban interindividual variability. Thromb. Res. 145, 24–26. 10.1016/j.thromres.2016.07.005 [DOI] [PubMed] [Google Scholar]
- Fang M. C., Go A. S., Chang Y., Borowsky L. H., Pomernacki N. K., Udaltsova N., et al. (2011). A new risk scheme to predict warfarin-associated hemorrhage: The ATRIA (Anticoagulation and Risk Factors in Atrial Fibrillation) Study. J. Am. Coll. Cardiol. 58, 395–401. 10.1016/j.jacc.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche L. G., Beesley L. J., VandeHaar P., Peng R. B., Salvatore M., Zawistowski M., et al. (2019). Exploring various polygenic risk scores for skin cancer in the phenomes of the Michigan genomics initiative and the UK biobank with a visual catalog: PRSWeb. PLoS Genet. 15, e1008202. 10.1371/journal.pgen.1008202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche L. G., Gruber S. B., Wu Z., Schmidt E. M., Zawistowski M., Moser S. E., et al. (2018). Association of polygenic risk scores for multiple cancers in a Phenome-wide study: Results from the Michigan genomics initiative. Am. J. Hum. Genet. 102, 1048–1061. 10.1016/j.ajhg.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller A. I., Shehab N., Lovegrove M. C., Rose K. O., Weidle N. J., Goring S. K., et al. (2020). Emergency visits for oral anticoagulant bleeding. J. Gen. Intern. Med. 35, 371–373. 10.1007/s11606-019-05391-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnoth M. J., Buetehorn U., Muenster U., Schwarz T., Sandmann S. (2011). In vitro and in vivo P-glycoprotein transport characteristics of rivaroxaban. J. Pharmacol. Exp. Ther. 338, 372–380. 10.1124/jpet.111.180240 [DOI] [PubMed] [Google Scholar]
- Hanauer D. A., Mei Q., Law J., Khanna R., Zheng K. (2015). Supporting information retrieval from electronic health records: A report of University of Michigan’s nine-year experience in developing and using the electronic medical record search Engine (EMERSE). J. Biomed. Inf. 55, 290–300. 10.1016/j.jbi.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanigan S., Das J., Pogue K., Barnes G. D., Dorsch M. P. (2020). The real world use of combined P-glycoprotein and moderate CYP3A4 inhibitors with rivaroxaban or apixaban increases bleeding. J. Thromb. Thrombolysis 49, 636–643. 10.1007/s11239-020-02037-3 [DOI] [PubMed] [Google Scholar]
- Harris P. A., Taylor R., Minor B. L., Elliott V., Fernandez M., O’Neal L., et al. (2019). The REDCap consortium: Building an international community of software platform partners. J. Biomed. Inf. 95, 103208. 10.1016/j.jbi.2019.103208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris P. A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J. G. (2009). Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J. Biomed. Inf. 42, 377–381. 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart R. G., Pearce L. A., Aguilar M. I. (2007). Meta-analysis: Antithrombotic therapy to prevent stroke in patients who have nonvalvular atrial fibrillation. Ann. Intern. Med. 146, 857–867. 10.7326/0003-4819-146-12-200706190-00007 [DOI] [PubMed] [Google Scholar]
- IBM Micromedex Solutions (2022). IBM Micromedex Solutions. Available at: https://www.micromedexsolutions.com/home/dispatch (Accessed February 2, 2022). [Google Scholar]
- Ing Lorenzini K., Daali Y., Fontana P., Desmeules J., Samer C. (2016). Rivaroxaban-induced hemorrhage associated with ABCB1 genetic defect. Front. Pharmacol. 7, 494. 10.3389/fphar.2016.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- January C. T., Wann L. S., Calkins H., Chen L. Y., Cigarroa J. E., Cleveland J. C., et al. (2019). 2019 AHA/ACC/HRS focused Update of the 2014 AHA/ACC/HRS guideline for the Management of patients with atrial fibrillation: A report of the American College of Cardiology/American heart association Task Force on clinical Practice guidelines and the heart Rhythm Society. J. Am. Coll. Cardiol. 74, 104–132. 10.1016/j.jacc.2019.01.011 [DOI] [PubMed] [Google Scholar]
- Johnson D., Wilke M. A. P., Lyle S. M., Kowalec K., Jorgensen A., Wright G. E. B., et al. (2022). A Systematic review and analysis of the Use of polygenic scores in pharmacogenomics. Clin. Pharmacol. Ther. 111, 919–930. 10.1002/cpt.2520 [DOI] [PubMed] [Google Scholar]
- Kaatz S., Ahmad D., Spyropoulos A. C., Schulman S. (2015). Definition of clinically relevant non-major bleeding in studies of anticoagulants in atrial fibrillation and venous thromboembolic disease in non-surgical patients: Communication from the SSC of the ISTH. J. Thromb. Haemost. 13, 2119–2126. 10.1111/jth.13140 [DOI] [PubMed] [Google Scholar]
- Kuehl P., Zhang J., Lin Y., Lamba J., Assem M., Schuetz J., et al. (2001). Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat. Genet. 27, 383–391. 10.1038/86882 [DOI] [PubMed] [Google Scholar]
- Lähteenmäki J., Vuorinen A.-L., Pajula J., Harno K., Lehto M., Niemi M., et al. (2021). Pharmacogenetics of bleeding and thromboembolic events in direct oral anticoagulant users. Clin. Pharmacol. Ther. 110, 768–776. 10.1002/cpt.2316 [DOI] [PubMed] [Google Scholar]
- Lanfear D. E., Luzum J. A., She R., Gui H., Donahue M. P., O’Connor C. M., et al. (2020). Polygenic score for Beta-Blocker Survival Benefit in European ancestry patients with reduced Ejection Fraction heart Failure. Circ. Heart Fail. 13, 007012. 10.1161/circheartfailure.119.007012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. R., Luzum J. A., Sangkuhl K., Gammal R. S., Sabatine M. S., Stein C. M., et al. (2022). Clinical pharmacogenetics implementation consortium guideline for CYP2C19 genotype and clopidogrel therapy: 2022 Update. Clin. Pharmacol. Ther. 112, 959–967. 10.1002/cpt.2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskey D. W., Linskey D. C., McLeod H. L., Luzum J. A. (2021). The need to shift pharmacogenetic research from candidate gene to genome-wide association studies. Pharmacogenomics 22, 1143–1150. 10.2217/pgs-2021-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lip G. Y. H., Nieuwlaat R., Pisters R., Lane D. A., Crijns H. J. G. M. (2010). Refining clinical risk stratification for predicting stroke and thromboembolism in atrial fibrillation using a novel risk factor-based approach: The euro heart survey on atrial fibrillation. Chest 137, 263–272. 10.1378/chest.09-1584 [DOI] [PubMed] [Google Scholar]
- Mueck W., Kubitza D., Becka M. (2013). Co-administration of rivaroxaban with drugs that share its elimination pathways: Pharmacokinetic effects in healthy subjects. Br. J. Clin. Pharmacol. 76, 455–466. 10.1111/bcp.12075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueck W., Stampfuss J., Kubitza D., Becka M. (2014). Clinical pharmacokinetic and pharmacodynamic profile of rivaroxaban. Clin. Pharmacokinet. 53, 1–16. 10.1007/s40262-013-0100-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharmacogene Variation Consortium (PharmVar) (2003). Available at: https://www.pharmvar.org/gene/CYP3A5 (Accessed June 25, 2022).
- Pisters R., Lane D. A., Nieuwlaat R., de Vos C. B., Crijns H. J. G. M., Lip G. Y. H. (2010). A novel user-friendly score (HAS-BLED) to assess 1-year risk of major bleeding in patients with atrial fibrillation: The euro heart survey. Chest 138, 1093–1100. 10.1378/chest.10-0134 [DOI] [PubMed] [Google Scholar]
- Roşian A. N., Iancu M., Trifa A. P., Roşian Ş. H., Mada C., Gocan C. P., et al. (2020a). An exploratory association analysis of ABCB1 rs1045642 and ABCB1 rs4148738 with non-major bleeding risk in atrial fibrillation patients treated with dabigatran or apixaban. J. Pers. Med. 10, 1333–E214. 10.3390/jpm10030133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roşian A. N., Roşian Ş. H., Kiss B., Ştefan M. G., Trifa A. P., Ober C. D., et al. (2020b). Interindividual variability of apixaban plasma concentrations: Influence of clinical and genetic factors in a real-life cohort of atrial fibrillation patients. Genes 11, E438. 10.3390/genes11040438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama N. N., Yang Z., Bui T., Ho R. J. Y. (2006). MDR1 haplotypes significantly minimize intracellular uptake and transcellular P-gp substrate transport in recombinant LLC-PK1 cells. J. Pharm. Sci. 95, 2293–2308. 10.1002/jps.20717 [DOI] [PubMed] [Google Scholar]
- Schulman S., Anger S. U., Bergqvist D., Eriksson B., Lassen M. R., Fisher W., et al. (2010). Definition of major bleeding in clinical investigations of antihemostatic medicinal products in surgical patients. J. Thromb. Haemost. 8, 202–204. 10.1111/j.1538-7836.2009.03678.x [DOI] [PubMed] [Google Scholar]
- Sennesael A.-L., Larock A.-S., Douxfils J., Elens L., Stillemans G., Wiesen M., et al. (2018). Rivaroxaban plasma levels in patients admitted for bleeding events: Insights from a prospective study. Thromb. J. 16, 28. 10.1186/s12959-018-0183-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliun D., Harris D. N., Kessler M. D., Carlson J., Szpiech Z. A., Torres R., et al. (2021). Sequencing of 53, 831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299. 10.1038/s41586-021-03205-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa S., Legnani C., Antonucci E., Paoletti O., Dellanoce C., Cosmi B., et al. (2019). Drug levels and bleeding complications in atrial fibrillation patients treated with direct oral anticoagulants. J. Thromb. Haemost. 17, 1064–1072. 10.1111/jth.14457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson B., Hu M., Lee V. W. Y., Lui S. S. H., Chu T. T. W., Poon E. W. M., et al. (2010). ABCG2 polymorphism is associated with the low-density lipoprotein cholesterol response to rosuvastatin. Clin. Pharmacol. Ther. 87, 558–562. 10.1038/clpt.2009.232 [DOI] [PubMed] [Google Scholar]
- Ueshima S., Hira D., Fujii R., Kimura Y., Tomitsuka C., Yamane T., et al. (2017). Impact of ABCB1, ABCG2, and CYP3A5 polymorphisms on plasma trough concentrations of apixaban in Japanese patients with atrial fibrillation. Pharmacogenet. Genomics 27, 329–336. 10.1097/FPC.0000000000000294 [DOI] [PubMed] [Google Scholar]
- Ueshima S., Hira D., Kimura Y., Fujii R., Tomitsuka C., Yamane T., et al. (2018). Population pharmacokinetics and pharmacogenomics of apixaban in Japanese adult patients with atrial fibrillation. Br. J. Clin. Pharmacol. 84, 1301–1312. 10.1111/bcp.13561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- University of Michigan (2022). Michigan genomics Initiative. University of Michigan. Available at: https://precisionhealth.umich.edu/our-research/michigangenomics/. [Google Scholar]
- Van Ganse E., Van Ganse E., Van Ganse E., Danchin N., Mahé I., Mahé I., et al. (2020). Comparative safety and Effectiveness of oral anticoagulants in nonvalvular atrial fibrillation: The NAXOS study. Stroke 51, 2066–2075. 10.1161/STROKEAHA.120.028825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderbilt University PheKB (2012). A knowledgebase for discovering phenotypes from electronic medical records. Vanderbilt University PheKB. Available at: https://phekb.org/phenotype/atrial-fibrillation-demonstration-project. [Google Scholar]
- Virani S. S., Alonso A., Benjamin E. J., Bittencourt M. S., Callaway C. W., Carson A. P., et al. (2020). Heart disease and stroke Statistics-2020 Update: A report from the American heart association. Circulation 141, e139–e596. 10.1161/CIR.0000000000000757 [DOI] [PubMed] [Google Scholar]
- Wang D., Guo Y., Wrighton S., Cooke G., Sadee W. (2011). Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 11, 274–286. 10.1038/tpj.2010.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Raghavan N., He K., Luettgen J. M., Humphreys W. G., Knabb R. M., et al. (2009). Sulfation of o-demethyl apixaban: Enzyme identification and species comparison. Drug Metab. Dispos. 37, 802–808. 10.1124/dmd.108.025593 [DOI] [PubMed] [Google Scholar]
- Wang L., Zhang D., Raghavan N., Yao M., Ma L., Frost C. E., et al. (2010). In vitro assessment of metabolic drug-drug interaction potential of apixaban through cytochrome P450 phenotyping, inhibition, and induction studies. Drug Metab. Dispos. 38, 448–458. 10.1124/dmd.109.029694 [DOI] [PubMed] [Google Scholar]
- Wang Y. P., Kehar R., Iansavitchene A., Lazo-Langner A. (2020). Bleeding risk in nonvalvular atrial fibrillation patients receiving direct oral anticoagulants and warfarin: A Systematic review and Meta-analysis of observational studies. TH Open 4, e145–e152. 10.1055/s-0040-1714918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasey J. O. (2018). Package “icd” version 3.3. Comorbidities from ICD-9 and ICD-10 codes, manipulation and validation. Available at: https://www.rdocumentation.org/packages/icd/versions/3.3 (Accessed May 8, 2022).
- Zajac G. J. M., Fritsche L. G., Weinstock J. S., Dagenais S. L., Lyons R. H., Brummett C. M., et al. (2019). Estimation of DNA contamination and its sources in genotyped samples. Genet. Epidemiol. 43, 980–995. 10.1002/gepi.22257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., He K., Herbst J. J., Kolb J., Shou W., Wang L., et al. (2013). Characterization of efflux transporters involved in distribution and disposition of apixaban. Drug Metab. Dispos. 41, 827–835. 10.1124/dmd.112.050260 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this article are not readily available because of patient privacy and confidentiality restrictions. Requests to access the datasets should be directed to https://precisionhealth.umich.edu/tools-resources/data-access-tools/how-to-access-data-tools-analytic-environments/