Abstract
The E3 strain of E. coli was isolated in an outbreak of respiratory disease in broiler chickens, and experimental aerosol exposure of chickens to this strain induced disease similar to that seen in the field. In order to establish whether the virulent phenotype of this strain was associated with carriage of particular plasmids, four plasmid-cured derivatives, each lacking two or more of the plasmids carried by the wild-type strain, were assessed for virulence. Virulence was found to be associated with one large plasmid, pVM01. Plasmid pVM01 was marked by introduction of the transposon TnphoA, carrying kanamycin resistance, and was then cloned by transformation of E. coli strain DH5α. The cloned plasmid was then reintroduced by conjugation into an avirulent plasmid-cured derivative of strain E3 which lacked pVM01. The conjugant was shown to be as virulent as the wild-type strain E3, establishing that this plasmid is required for virulence following aerosol exposure. This virulence plasmid conferred expression of a hydroxamate siderophore, but not colicins, on both strain E3 and strain DH5α. Carriage of this plasmid was required for strain E3 to colonize the respiratory tracts of chickens but was not necessary for colonization of the gastrointestinal tract. However, the virulence plasmid did not confer virulence, or the capacity to colonize the respiratory tract, on strain DH5α. Thus, these studies have established that infection of chickens with E. coli strain E3 by the respiratory route is dependent on carriage of a conjugative virulence plasmid, which confers the capacity to colonize specifically the respiratory tract and which also carries genes for expression of a hydroxymate siderophore. These findings will facilitate identification of the specific genes required for virulence in these pathogens.
Lower-respiratory-tract infections are the most common disease syndrome associated with Escherichia coli in poultry. Although in extreme cases mortality can be over 20%, it is the high morbidity and associated loss in productivity which is responsible for the greatest economic loss (31).
There is evidence to suggest that virulent strains of avian E. coli belong to a limited number of clone complexes (60, 61) and that particular clones may be specific to particular manifestations of E. coli infection (47). A number of characteristics have been associated with virulence in avian E. coli, including colicin V production (22, 23, 50, 59), adhesins (17, 18, 20, 32, 46, 67), serum resistance (21, 37, 47, 48, 59, 66), and iron sequestering (37, 41, 43, 47, 59, 63), but specific attempts to establish the requirements of these factors for virulence are limited. While initial studies of avian E. coli led to the conclusion that certain serogroups, O1, O2, and O78 in particular, were more commonly associated with colibacillosis (27, 28, 29, 33, 35, 36), the most prevalent serotypes vary with geographic location and many isolates are untypeable (3, 6, 12, 19, 38, 45).
Although plasmid-encoded virulence genes have been well investigated and described for human E. coli, their role in the pathogenicity of avian E. coli is less well understood. It has been observed that the presence of high-molecular-weight plasmids is a feature of E. coli isolated from chickens with colibacillosis (23, 59), and other studies have implicated plasmid-encoded genes as possible virulence determinants in avian E. coli (37, 49, 56, 64). In addition, some unique chromosomal regions have been associated with virulence in a pathogenic strain of avian E. coli (13), and a strain in which the chromosomally encoded fim cluster had been deleted has been shown to have some reduction in virulence (44).
Avian E. coli is thought to gain entry to the chicken by inhalation of coliform-contaminated dust (30) or by colonization of the upper respiratory tract through ingestion of food and water contaminated with feces (34). The virulence determinants required to successfully colonize and replicate in the respiratory tract can be assessed experimentally only if the route of inoculation mimics that of the natural infection. No studies have examined the genetic basis for virulence following aerosol exposure. Studies which have examined the role of plasmid-encoded genes have assessed virulence using air sac and intravenous inoculation (56) or mortality following intratracheal instillation of a bolus of E. coli (37).
The aim of this study was to assess whether plasmid-encoded genes were associated with virulence of E. coli when chickens were exposed by the aerosol route. The association of virulence with phenotypic markers such as colicin V and siderophore production and the correlation between virulence and the ability to colonize the respiratory and intestinal tracts were also assessed.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study are described in Table 1. The wild-type strain E3 was isolated in pure culture from the pericardium of a 40-day-old broiler bird with colibacillosis. Isolates with the same antimicrobial sensitivity pattern and plasmid profile as E3 were also isolated in pure culture from the liver, air sacs, and joints of the bird and from other diseased birds in the same flock. DH5α was provided by H. S. Nagesha (School of Veterinary Science, The University of Melbourne). E. coli SM10 λpir carries a kanamycin resistance-encoding transposon (TnphoA) on the suicide plasmid pRT733 (57). Strains derived in this study are named on the basis of origin (first part of name) and colony pick number (last part of name).
TABLE 1.
Plasmid profiles and phenotypic characteristics of E. coli strains used in this study
| Strain | Plasmid(s) | Phenotypic characteristicsa |
|---|---|---|
| SM10 λpir | pRT733::TnphoAb | Ampr Knr |
| E3 | pVM01, pVM02, pVM03, pVM04, pVM05, pVM06 | Ampr SFr Wr Cs Ter Col+ Hyd+ |
| E3/1.1 | pVM02, pVM05, pVM06 | Amps SFs Ws Cs Ter Col+ Hyd− |
| E3/2 | pVM02 | Amps SFs Ws Cs Ter Col+ Hyd− |
| E3/2.4 | pVM02, pVM03, pVM05, pVM06 | Ampr SFr Wr Cs Ter Col+ Hyd− |
| E3/3.4 | pVM01, pVM02, pVM05, pVM06 | Amps SFs Ws Cs Ter Col+ Hyd+ |
| E3/3.4/28 | pVM01::TnphoA, pVM02, pVM05, pVM06 | Amps SFs Ws Cs Ter Knr Col+ Hyd+ |
| E3/3.4/32 | pVM01::TnphoA, pVM02, pVM05, pVM06 | Amps SFs Ws Cs Ter Knr Col+ Hyd+ |
| E3/2.4/1 | pVM01::TnphoA, pVM02, pVM03, pVM05, pVM06 | Ampr SFr Wr Cs Ter Knr Col+ Hyd+ |
| E3/2.4/6 | pVM01::TnphoA, pVM02, pVM03, pVM05, pVM06 | Ampr SFr Wr Cs Ter Knr Col+ Hyd+ |
| E3/2.4/9 | pVM01::TnphoA, pVM02, pVM03, pVM05, pVM06 | Ampr SFr Wr Cs Ter Knr Col+ Hyd+ |
| E3/2.4/10 | pVM01::TnphoA, pVM02, pVM03, pVM05, pVM06 | Ampr SFr Wr Cs Ter Knr Col+ Hyd+ |
| DH5α | Amps Knr Hyd− | |
| DH5α/4 | pVM01::TnphoA | Amps Knr Hyd+ |
| DH5α/5 | pVM01::TnphoA | Amps Knr Hyd+ |
Abbreviations: Amp, ampicillin; SF, sulfafurazole; W, trimethoprim; C, chloramphenicol; Te, tetracycline; Kn, kanamycin; r, resistant; s, sensitive; Col+, colicin production; Hyd+, hydroxamate production.
Described by Taylor et al. (57).
Determination of phenotypic characteristics.
O serotypes were determined using standard methods (9, 58). H serotyping was carried out using the method of Chandler and Bettelheim (14). The full range of O and H antisera in the International scheme were used.
The production of heat-labile enterotoxin was determined in tissue culture using a modification of the method of Sack and Sack (10, 54). The immunoassay for heat-labile enterotoxin was carried out as described previously (8). Production of Shiga-like toxins I and II was determined in cell culture (24, 40) and by immunoassay (1). Enterohemolysin production was determined using the two-plate method (7). Alpha hemolysin was detected by growth on sheep blood agar. The antimicrobial sensitivities of E. coli isolates were assessed using the calibrated dichotomous sensitivity (CDS) test (6a) on Sensitest agar (Oxoid, Heidelberg West, Victoria, Australia). Antimicrobial discs (Oxoid) used were ampicillin (AMP), 25 μg; sulfafurazole (SF), 300 μg; trimethoprim (W), 2.5 μg; chloramphenicol (C), 30 μg; and tetracycline (TE), 30 μg. Colicin production was detected using the double-layer technique (42). Detection of hydroxamate in culture supernatant was carried out using the method of Csaky (15) with modifications in hydrolysis conditions (25).
Plasmid curing.
E. coli strain E3 was grown in SOC broth (55) or brain heart infusion (BHI) broth (Oxoid) at 45 to 46°C and subcultured into fresh broth every 24 h. The culture was plated onto nutrient agar and MacConkey agar, and single colonies were selected for plasmid analysis.
Preparation of plasmid DNA.
Small-scale preparation of plasmid DNA was carried out using a modification of the method of Kado and Liu (39). E. coli organisms were inoculated into 3 ml of BHI broth and incubated overnight in an orbital shaker at 37°C. A 1.5-ml aliquot of the overnight culture was centrifuged at 16,250 × g for 45 s, and the pellet was resuspended in 200 μl of E buffer (40 mM Tris–2 mM EDTA [pH 7.9]). The cells were lysed by the addition of 400 μl of lysing solution (3% sodium dodecyl sulfate [SDS]–50 mM Tris [pH 12.6]), made from stock solutions just prior to use. The solution was mixed by gentle inversion and then heated at 55°C for 20 min before the addition of 1 volume of unbuffered phenol-chloroform (50:50, vol/vol). After mixing by gentle inversion, the solution was centrifuged (at 16,250 × g for 20 min). The aqueous phase was extracted with 1 volume of buffered phenol (pH 8)–chloroform–isoamyl alcohol (50:25:24:1, vol/vol), and the DNA was precipitated from the aqueous phase with ethanol. The DNA pellet was resuspended in 18 μl of E buffer. Large-scale preparation of plasmid DNA from avian E. coli was carried out using the Qiagen (Clifton Hill, Victoria, Australia) Plasmid Midi Kit (Qiagen-tip 100) as recommended by the manufacturer, except that E. coli was grown in 50 ml of BHI broth. The modifications recommended by the manufacturer to obtain higher yields of low-copy-number plasmids were also used.
Agarose gel electrophoresis.
For standard agarose gel electrophoresis (SAGE), plasmids were separated in 0.7% (wt/vol) gels containing E buffer in a field of 2.5 V/cm for 4 to 5 h. Gels were stained in E buffer containing 0.5 mg of ethidium bromide/ml for 10 min and were then washed briefly in water, and DNA was visualized by UV transillumination. Pulsed-field gel electrophoresis (PFGE) was carried out in a CHEF-DR III System (Bio-Rad, Regents Park, New South Wales, Australia). Plasmids were separated in 1.0% (wt/vol) DNA-grade agarose (Progen, Darra, Queensland, Australia) containing TBE (89 mM Tris, 89 mM boric acid, and 2 mM EDTA) for 18 to 22 h at 6 V/cm with a switch time of 1 to 20 s and an included angle of 120°. Gels were stained in TBE containing 0.5 mg of ethidium bromide/ml for 30 min and then destained in TBE for 30 min, and DNA was visualized by UV transillumination.
Insertion of a kanamycin resistance marker into pVM01.
TnphoA mutants were derived by conjugating E. coli SM10 λpir with E. coli strain E3/3.4. Strain E3/3.4 was used because it contained pVM01 but, unlike strain E3, was sensitive to ampicillin, so that conjugants which cointegrated the suicide plasmid could be eliminated. Suspensions of SM10 λpir and E3/3.4 were made using scrapings of colonies from fresh Luria-Bertani (LB) agar in sterile saline as previously described (2). Mating was carried out by mixing the suspensions on LB agar at a ratio of 2:1 (donor to recipient) and incubating at 37°C for 5 to 6 h. The mating mix was scraped from the agar and resuspended in 1 ml of sterile saline. Conjugants were selected on LB agar containing glucose (0.2%, wt/vol), 5-bromo-4-chloro-3-indolyl phosphate (50 μg/ml), kanamycin (100 μg/ml), and tetracycline (60 μg/ml). In order to eliminate those conjugants which had cointegrated the suicide plasmid, selected isolates were screened for sensitivity to ampicillin. Insertion of TnphoA into pVM01 was confirmed by probing Southern blots of plasmid DNA with the 2.8-kb BglII fragment of pRT733.
Transformation of DH5α with pVM01::TnphoA.
Preparation and electrotransformation of E. coli strain DH5α were carried out as recommended by the manufacturer (Gene Pulser; Bio-Rad). DH5α was transformed with plasmid DNA from strains E3/3.4/28 and E3/3.4/32. Transformants were selected on LB agar containing kanamycin (100 μg/ml). Transformation was confirmed by plasmid screening and by probing Southern blots of plasmid DNA with the 2.8-kb BglII fragment of pRT733.
Reintroduction of pVM01 into E3/2.4.
Conjugation was carried out using strain DH5α/4 or DH5α/5 as the donor and strain E3/2.4 as the recipient. Suspensions of each strain were made by scraping colonies from LB agar plates and resuspending them in phosphate-buffered saline (PBS). Mating was carried out by mixing the suspensions on LB agar at a ratio of 1:1 (donor to recipient) and incubating at 37°C for 5 to 6 h. The mating mix was scraped from the agar and resuspended in PBS. Conjugants were selected on LB agar containing ampicillin (50 μg/ml) and kanamycin (100 μg/ml). Successful conjugation was confirmed by plasmid profile analysis.
Southern blot hybridization.
Plasmid DNA was transferred to a nylon membrane (Hybond N+; Amersham, Baulkham Hills, New South Wales, Australia) using a modification of a previously published method (55). After electrophoresis, the gel was exposed to UV irradiation (C-63 Mineralight Transilluminator; UVP International, Inc.) for 10 min and then was wet with 0.4 M NaOH. The gel was then inverted and placed on five to six sheets of Whatman 3MM paper which had been wet with 0.4 M NaOH. The membrane and a further two to three sheets of 0.4 M NaOH-wetted Whatman 3MM paper were placed on top of the gel, and dry paper towels were placed on top of this. Transfer was allowed to proceed for 4 h, after which the membrane was washed briefly in 2 × SSC (1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate). DNA probes were labeled with [α-32P]dCTP using a random primed DNA labeling kit (Boehringer Mannheim). Prehybridization and hybridization were carried out in Denhardt's buffer overnight at 68°C. Membranes were rinsed in 2 × SSC at room temperature, washed first in 2 × SSC and then in 0.2 × SSC at 68°C for 30 min each, and exposed to Kodak X-Omat AR film at −70°C.
Pathogenicity testing.
To prepare E. coli cultures for infecting birds by the aerosol route, 250 ml of BHI broth (Oxoid CM471) was inoculated with a loopful of the stock culture and incubated in an orbital shaker at 37°C for 22 to 24 h. When E3/2.4 conjugants containing pVM01::TnphoA were assessed, kanamycin (100 μg/ml) and ampicillin (50 μg/ml) were added to the BHI broth prior to inoculation; when DH5α/4 and DH5α/5 were assessed, kanamycin alone (100 μg/ml) was added. The optical densities at 600 nm (OD600) of the 10−1 and 10−2 dilutions of this culture were determined. An OD600 of approximately 1.4 for the 10−1 dilution and approximately 0.2 for the 10−2 dilution corresponded to about 1.5 × 1010 CFU in the original culture. After incubation, cultures were centrifuged in a Sorvall GSA rotor (at 3,687 × g for 10 min) and the cell pellet was resuspended in BHI broth to give a concentration of 1010 to 1010.5 CFU/ml. The estimated colony count was confirmed by plating 100 μl of a 10−7 and a 10−8 dilution of the final culture onto separate nutrient agar plates (Oxoid CM3) and six samples of 20 μl of the 10−8 dilution onto a single divided nutrient agar plate. The virulence of E. coli strains was assessed by an aerosol exposure method described previously (26). Briefly, 1-day-old specific-pathogen-free White Leghorn Hybrid chicks were given an intranasal inoculation of infectious bronchitis virus (IBV) vaccine (Webster's VicS) at 10 times the immunizing dose and were then exposed to an aerosol of E. coli for 20 min. Each group of chickens was housed on litter in separate plastic bubble isolators. Further aerosols of E. coli were administered to the birds on day 4 and day 7 of the experiment, and surviving birds were killed on day 11 with an intravenous injection of barbiturate. Birds were examined for airsacculitis, pericarditis, perihepatitis, yolk sac infection, and peritonitis. Swabs collected during postmortem examinations were plated directly onto MacConkey agar no. 3. MacConkey agar plates were incubated at 37°C for 16 to 18 h. A single representative colony was selected and subcultured onto nutrient agar, then incubated at 37°C for 16 to 18 h. Further tests were carried out on a single colony selected from the nutrient agar plate. When E3/2.4 conjugants containing pVM01::TnphoA were assessed, kanamycin (100 μg/ml) and ampicillin (50 μg/ml) were added to both the MacConkey and nutrient agar plates, and when DH5α/4 and DH5α/5 were assessed, kanamycin alone (100 μg/ml) was added to both the MacConkey and nutrient agar plates. Organisms which were gram-negative rods, oxidase negative, catalase positive, indole positive, methyl red positive, Voges-Proskauer negative, and citrate negative were considered to be E. coli. Antimicrobial sensitivity was used to confirm that the E. coli reisolates were the same as the original inoculate.
Statistical analysis.
A two-tailed Fisher's exact test was used to analyze mortality, lesion, and reisolation rates.
RESULTS
Phenotypic characterization of strain E3.
E3 was serotype O nontypeable:H28 and did not produce alpha-hemolysin or enterohemolysin, heat-labile enterotoxin, or Shiga-like toxin I or II. A colicin(s) not belonging to the ColV immunity group and a hydroxamate siderophore were produced by E3. E3 was resistant to ampicillin, sulfafurazole, trimethoprim, and tetracycline and sensitive to chloramphenicol.
Pathogenicity of E3.
The virulence of E3 has been assessed repeatedly as part of the development of the pathogenicity testing method, and challenge with E3 was used as the positive control for all experiments in this study. The results shown in Tables 2 and 3 detail the lesion and mortality rates following aerosol exposure to E3 in the positive control groups for the six separate experiments. The average mortality rate for E3 was 10%, and the average lesion rate was 79%. Except for experiment 4, in which the lesion rate for challenge with E3 was 100% and significantly different from the lesion rates for challenge with E3 in experiments 2 and 3 (P < 0.05 by Fisher's exact test), the lesion rates for challenge with E3 were not significantly different in different experiments.
TABLE 2.
Pathogenicities of avian E. coli strain E3 and its plasmid-cured derivatives following aerosol exposure
| Strain | Expt no. | Mortality ratea | Lesion rateb | Rate of reisolationc from:
|
||
|---|---|---|---|---|---|---|
| Cloaca | Trachea | Air sac | ||||
| E3 | 1 | 2/15* | 13/15* | 15/15* | 14/15* | 14/15*,† |
| E3 | 2 | 2/14* | 9/14* | 12/14* | 12/14* | 14/14*,† |
| E3/1.1 | 1 | 1/15* | 0/15† | 15/15* | 4/15†,‡ | 1/15‡ |
| E3/2.4 | 1 | 0/13* | 1/13† | 12/13* | 4/13† | 0/13‡ |
| E3/2.4 | 2 | 2/13* | 0/13† | 3/13† | 3/13†,‡ | 1/13‡ |
| E3/2 | 1 | 0/14* | 0/14† | 14/14* | 0/14‡ | 0/14‡ |
| E3/3.4 | 2 | 2/13* | 9/13* | 13/13* | 13/13* | 8/13* |
No. of birds which died or were euthanized/total no. of birds. In each column, proportions with the same superscript symbols are not significantly different (P > 0.05 by Fisher's exact test).
No. of birds with lesions/total no. of birds.
No. of birds from which the inoculated strain of E. coli was isolated from each of the three sites swabbed/total no. of birds.
TABLE 3.
Pathogenicities of E. coli strain DH5α carrying pVM01 and strain E3/2.4 after reintroduction of pVM01 compared to that of the virulent parent strain E3
| Strain | Expt no. | Mortality ratea | Lesion rateb | Rate of reisolationc from:
|
||
|---|---|---|---|---|---|---|
| Cloaca | Trachea | Air sac | ||||
| E3 | 3 | 0/16*,† | 11/16* | 4/16*,†,∥ | 13/16*,†,§ | 9/10* |
| E3 | 4 | 4/16* | 16/16† | 8/16*,†,‡ | 15/16*,† | 14/16* |
| E3 | 5 | 0/16*,† | 12/16*,† | 10/16†,‡ | 15/16*,† | 11/16*,† |
| E3 | 6 | 1/15*,† | 12/15*,† | 15/15§ | 15/15† | 13/15* |
| DH5α/4 | 6 | 0/17† | 3/17‡,§ | 1/17∥,# | 0/15‡ | 0/17‡ |
| DH5α/5 | 6 | 0/17† | 4/17‡,§ | 0/17# | 0/15‡ | 0/17‡ |
| E3/2.4 | 5 | 0/17† | 2/17‡ | 3/17*,∥,# | 8/17§ | 1/17‡ |
| E3/2.4/1 | 3 | 0/14*,† | 7/14*,§ | 10/14‡,** | 10/14*,§ | 3/14‡,§ |
| E3/2.4/6 | 4 | 0/16*,† | 13/16*,† | 10/16†,‡ | 14/16*,† | 9/16*,†,§ |
| E3/2.4/9 | 4 | 1/17*,† | 13/17*,† | 16/17§,** | 15/17*,† | 10/17*,†,§ |
| E3/2.4/9 | 5 | 1/17*,† | 12/17* | 17/17§ | 16/17*,† | 11/17*,† |
| E3/2.4/10 | 3 | 1/16*,† | 7/16*,‡,§ | 14/16‡,§,** | 13/16*,†,§ | 7/16† |
No. of birds which died or were euthanized/total no. of birds. In each column, proportions with the same superscript symbols are not significantly different (P > 0.05 by Fisher's exact test).
No. of birds with lesions/total no. of birds.
No. of birds from which the inoculated strain of E. coli was isolated from each of the three sites swabbed/total no. of birds.
Plasmid curing.
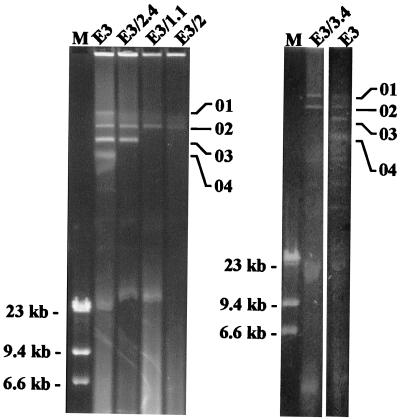
The plasmid profile of E3 is shown in Fig. 1. Four different plasmid-cured strains (E3/1.1, E3/2, E3/2.4, and E3/3.4) were obtained (Fig. 1 and Table 1).
FIG. 1.
Comparison of the plasmid profiles of avian E. coli strain E3 and its plasmid-cured derivatives. Positions of plasmids pVM01 (01), pVM02 (02), pVM03 (03), and pVM04 (04) are indicated. Other bands of greater mobility are either smaller plasmids or different forms of the higher-molecular-weight plasmids. The positions of fragments of HindIII-digested phage lambda DNA, which were used as molecular size markers, are shown (M). This image was obtained using a Nikon Scantouch scanner and Adobe Photoshop.
A colicin(s), not belonging to the ColV immunity group, was produced by each of the plasmid-cured strains. A hydroxamate siderophore was produced by E3/3.4 but not by any of the other plasmid-cured derivatives of E3. Loss of pVM03 in cured strains E3/1.1, E3/2, and E3/3.4 was associated with loss of resistance to ampicillin, sulfafurazole, and trimethoprim. Despite the loss of two plasmids (pVM01 and pVM04), E3/2.4 had the same antimicrobial resistance pattern as the wild-type strain, E3. Plasmids pVM02 and pVM03 were introduced into DH5α by electrotransformation with plasmid DNA from E3, and the resultant transformants were selected using ampicillin or tetracycline. Plasmid profiles of selected DH5α containing plasmids pVM02 and pVM03 confirmed that these plasmids mediate tetracycline and ampicillin resistance, respectively. The plasmid profiles and phenotypic characteristics of E3 and its plasmid-cured derivatives, as well as those of the other bacterial strains used in this study are described in Table 1.
Pathogenicity of plasmid-cured derivatives of E3.
In experiments 1 and 2, the virulence of E3 was compared with that of its cured derivatives, E3/1.1, E3/2, E3/2.4, and E3/3.4 (Table 2). Lesions caused by E3 included severe peritonitis, severe pericarditis, perihepatitis, airsacculitis, and yolk sac infection. E3 was isolated from the lesions in all of the affected birds. Aerosol exposure to E3/1.1 produced a single mortality, and although there were no lesions, E3/1.1 was isolated from the cloaca, trachea, and left abdominal air sac of the dead bird. E3/1.1 could not be isolated from the air sacs of any of the other birds in the group and was isolated in very low numbers (1 colony/agar plate) from the tracheas of 4 of 15 birds.
E3/2 was isolated from the cloacae of all the birds but could not be isolated from the tracheas or air sacs of any of the birds.
In the first experiment, one bird which was exposed to E3/2.4 had flecks of caseous material distributed throughout its intestinal mesentery, and cultures of this material were positive for E3/2.4. In experiment 2, there were two mortalities following administration of E3/2.4. Neither bird had any lesions, but E3/2.4 was isolated from the cloaca, trachea, pericardium, and air sac of one bird and from the trachea of the other bird.
There were two mortalities following the administration of E3/3.4. One of the birds had pericarditis, perihepatitis, and airsacculitis. The other bird was killed because it showed signs of respiratory distress, but other than an accumulation of mucus in the trachea, there were no lesions. Except for experiment 2, where E3/2.4 did not successfully colonize the intestinal tract, there was no significant difference between the rates of reisolation of E3, E3/1.1, E3/2, and E3/2.4 from the cloacae of infected birds. However, the reisolation rates of E3 from tracheas and air sacs were significantly higher than the reisolation rates of E3/1.1, E3/2, and E3/2.4 from these sites in birds which were infected with these strains (P < 0.05 by Fisher's exact test). There was no significant difference between the rates of reisolation of E3 and E3/3.4 from tracheas (P = 1) or air sacs (P > 0.3).
Insertion of a kanamycin resistance gene into pVM01.
Two kanamycin-resistant, tetracycline-resistant, ampicillin-sensitive conjugants were obtained. Southern blot analysis showed that TnphoA had inserted exclusively into pVM01 in conjugants E3/3.4/28 and E3/3.4/32.
Transformation of DH5α with pVM01::TnphoA.
Plasmid pVM01::TnphoA was introduced into E. coli strain DH5α by electrotransformation with plasmid DNA isolated from strains E3/3.4/28 and E3/3.4/32. Transformants were selected on LB agar containing kanamycin (100 μg/ml), and their plasmid profiles were analyzed by PFGE and by Southern blot analysis. DH5α/4 and DH5α/5 both contained a single plasmid which corresponded in size to pVM01 and hybridized with the 2.8-kb BglII fragment of TnphoA.
Reintroduction of pVM01 into E3/2.4.
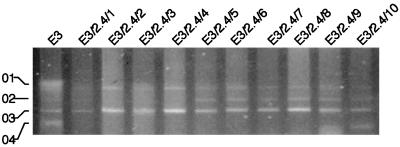
Transformation of E3/2.4 with plasmid DNA from strains E3/3.4/28 and E3/3.4/32 was unsuccessful, so conjugation was carried out using strain DH5α/4 or DH5α/5 as the donor and strain E3/2.4 as the recipient. Conjugants were selected on LB agar containing ampicillin (50 μg/ml) and kanamycin (100 μg/ml), and acquisition of plasmid pVM01::TnphoA was confirmed by plasmid profile analysis. Figure 2 shows the plasmid profiles of conjugants. Plasmid pVM01::TnphoA was present in all the isolates. Four of the conjugants, E3/2.4/1, E3/2.4/6, E3/2.4/9, and E3/2.4/10, were selected for pathogenicity testing.
FIG. 2.
Plasmid profiles of selected E. coli strain E3/2.4 progeny following conjugation with DH5α/4 or DH5α/5 and selection in the presence of ampicillin and kanamycin. Positions of plasmids pVM01 (01), pVM02 (02), pVM03 (03), and pVM04 (04) are indicated in the parental E3 strain. Strains E3/2.4/1 and E3/2.4/6 (derived from DH5α/4) and strains E3/2.4/9 and E3/2.4/10 (derived from DH5α/5), all of which carried pVM01, pVM02, and pVM03, were chosen for further characterization. This image was obtained using a Nikon Scantouch scanner and Adobe Photoshop.
Phenotypic characteristics of strains carrying pVM01::TnphoA.
A colicin(s) not belonging to the ColV immunity group was produced by strains E3/3.4/28, E3/3.4/32, E3/2.4/1, E3/2.4/6, E3/2.4/9, and E3/2.4/10. Colicin was not produced by DH5α/4 or DH5α/5. A hydroxamate siderophore was produced by E3/3.4/28, E3/3.4/32, E3/2.4/1, E3/2.4/6, E3/2.4/9, and E3/2.4/10, as well as by DH5α/4 and DH5α/5. All strains retained their original antimicrobial resistances, and all were also resistant to kanamycin. The plasmid profiles and phenotypic characteristics of these strains are described in Table 1.
Pathogenicity of DH5α containing pVM01::TnphoA.
Three of 17 birds exposed to DH5α/4 and 4 of 17 birds exposed to DH5α/5 developed very mild airsacculitis (Table 3), but E. coli was not isolated from the air sacs of any of the birds in either group. E. coli with the same antimicrobial resistance pattern as the inoculated strain was isolated from the cloaca of one bird in the DH5α/4 group.
Pathogenicity of E3/2.4 containing pVM01::TnphoA.
Three separate experiments were carried out to determine the pathogenicities of strains E3/2.4/1, E3/2.4/6, E3/2.4/9, and E3/2.4/10. Experiment 3 compared E3/2.4/1 and E3/2.4/10 with E3, experiment 4 compared E3/2.4/6 and E3/2.4/9 with E3, and experiment 5 compared E3/2.4/9 and E3/2.4 with E3. The results of these experiments are combined in Table 3. Exposure to E3 caused mild to severe airsacculitis in 11 of 16, 16 of 16, and 12 of 16 birds and pericarditis and perihepatitis in 4 of 16, 5 of 16, and 8 of 16 birds, respectively. Exposure to E3/2.4/1 or E3/2.4/6 resulted in mild to moderate airsacculitis in 7 of 14 and 13 of 16 birds, respectively. Neither of these strains caused pericarditis or perihepatitis. Exposure to E3/2.4/9 produced mild to moderate airsacculitis in 13 of 17 and 12 of 17 birds and pericarditis and perihepatitis in 3 of 17 and 2 of 17 birds. Exposure to E3/2.4/10 produced mild to moderate airsacculitis in 7 of 16 birds and pericarditis and perihepatitis in 1 of 16 birds. Exposure to E3/2.4 produced very mild airsacculitis in 2 of 17 birds and no pericarditis or perihepatitis.
The lesion rates caused by exposure to E3 were not significantly different from those caused by exposure to E3/2.4/1, E3/2.4/6, E3/2.4/9, and E3/2.4/10 when they were compared in the same experiment (P > 0.1). There was no significant difference in reisolation rates from the tracheas (P > 0.3) and air sacs (P > 0.06) of birds exposed to E3, E3/2.4/1, E3/2.4/6, E3/2.4/9, and E3/2.4/10.
In experiment 5, both the lesion rates and the reisolation rates from the tracheas and air sacs in birds exposed to E3 and E3/2.4/9 were significantly higher than those in birds exposed to E3/2.4 (P < 0.05).
DISCUSSION
E3 was shown to be a virulent strain of avian E. coli which contained six plasmids, and loss of virulence was correlated with the loss of pVM01, one of these plasmids. E3 was resistant to a number of antimicrobial agents and produced a colicin(s) and a hydroxamate siderophore. All of the plasmid-cured derivatives of E3 produced a colicin(s), but only E3/3.4, which was the only derivative strain which still possessed pVM01, also retained production of aerobactin, and only E3/3.4 was virulent. Notably, the loss of virulence did not appear to be correlated with a decreased ability to colonize the intestines of exposed birds, as assessed by isolation from the cloacae, but it did appear to be correlated with an inability to colonize the respiratory tract.
The association between pVM01 and virulence was further established by cloning the plasmid in E. coli strain DH5α and then reintroducing this cloned plasmid into the avirulent cured strain E3/2.4. This cloning was facilitated by introduction of a kanamycin resistance gene on the transposon TnphoA into pVM01, enabling positive selection for transformants and conjugants carrying the plasmid. All four E3/2.4-derived conjugants carrying pVM01::TnphoA were virulent, establishing that virulence could be regained by the avirulent derivatives upon reintroduction of the virulence plasmid pVM01. The reintroduction of pVM01 also restored the capacity of E3/2.4 to colonize the respiratory tracts of exposed birds.
Examination of the phenotype of DH5α carrying pVM01, and of the conjugants into which pVM01 had been reintroduced, established that this plasmid carried genes for the expression of a hydroxamate siderophore, but not for colicin production.
A number of putative virulence-determining genes have been found on ColV plasmids of E. coli from humans and other animals, but it has been shown that colicin V itself is not a virulence determinant (11, 51, 62). The production of colicin V has been associated with aerobactin production and complement resistance in avian E. coli (43, 63, 65), but there are a significant number of E. coli strains which produce aerobactin in the absence of colicin V (43).
E3 and its plasmid-cured derivatives produced a colicin(s) which did not belong to the colicin V immunity group. Eighteen other colicins have been described in E. coli, and the genes for colicin production are found exclusively on plasmids (53). The genes which encode the colicin(s) produced by E3 and its cured derivatives must be located on pVM02, as that is the only plasmid present in strain E3/2. This putative colicin plasmid did not confer virulence, as all the plasmid-cured strains retained pVM02 but only E3/3.4 was virulent.
Although our study did not support an association between colicin production and virulence, it did support the association between the production of siderophores and virulence. It has previously been shown that the ability to grow under iron-limited conditions is a feature of pathogenic avian E. coli (17). The loss of a 95-kb plasmid from a virulent avian E. coli strain has been shown to reduce virulence, but this difference in virulence was obvious only when the E. coli bacteria were given by air sac injection (56). The wild-type and plasmid-cured strains in this study both appeared to be virulent when given by intravenous injection, and although both strains produced aerobactin, the plasmid-cured strain grew slightly less well under iron-restricted conditions. Plasmid curing and reintroduction experiments have shown that a 100-MDa plasmid of a virulent avian E. coli strain encoding serum resistance and aerobactin production was required for virulence (37), but virulence in this study was assessed by determining the 50% lethal dose (LD50) of E. coli administered by intratracheal inoculation, rather than by aerosol exposure and assessment of colonization and respiratory tract lesions as in our study. Others have shown possible links between plasmid-encoded genes and virulence (49, 64), but they used an embryo lethality test to determine virulence, the results of which should be interpreted with caution, since they correlate only moderately well with a chicken lethality test (48).
Despite evidence that aerobactin production (41) and other putative virulence determinants like serum resistance are important for the virulence of avian E. coli, it is likely that colonization of the upper airways is paramount. Virulent avian E. coli strains colonize the tracheas of experimentally infected birds in greater numbers than avirulent strains (16), are cleared less quickly from the respiratory tract, and can produce a bacteremia after aerosol exposure (4). Piliated E. coli strains are more virulent for chickens than nonpiliated strains (5, 17, 46). Both type 1 and P pili can be expressed by avian E. coli, and both may have roles in virulence (19, 20). Deletion of the fim cluster, which encodes type 1 pili, has been shown to reduce colonization under some conditions (44). However, both type 1 pili and P pili are encoded on the E. coli chromosome, so although it is possible that some other adherence mechanism is responsible for the virulence characteristics expressed from pVM01, it is also possible that the aerobactin system encoded by pVM01 may enhance the ability of E. coli organisms to colonize the respiratory tract after they have adhered to the respiratory epithelium. Furthermore, virulence cannot be conferred in the absence of a suitable chromosomal background, as was shown in experiment 6, where carriage of pVM01 did not confer virulence on the laboratory strain DH5α.
It has been postulated for some time that colonization may be a prerequisite for expression of other virulence factors (52), and recently it has been shown that the binding of P pili results in the production of a sensor-regulatory protein which is essential for the activation of the iron starvation response in uropathogenic E. coli (68). The association of hydroxamate siderophore production and the capacity to colonize the respiratory tract suggests that there may be an association between these properties in pathogenic avian E. coli as well.
Initial examination of the adhesive properties of E3 and its plasmid-cured derivatives using HeLa cells was unrevealing (data not shown), but further examination using an avian cell line would be more appropriate. Nevertheless, data presented here would tend to indicate that efficient colonization of the respiratory tract is a prerequisite for disease production. E3 colonized the tracheas and air sacs of infected birds to a significantly greater extent than its avirulent plasmid-cured derivatives, and this loss of colonizing ability could be restored by reintroduction of pVM01. Although restoration of this putative virulence plasmid to a cured derivative of E3 restored its virulence, the introduction of pVM01 into DH5α did not confer virulence, indicating that a suitable genetic background is required for plasmid-mediated virulence determinants to be fully expressed. Thus, our findings are likely to complement those which have identified chromosomal regions of virulent avian E. coli associated with virulence (13, 44). Our studies have laid the basis for further examination of the virulence determinants of avian E. coli in the respiratory tract, using a model of infection that closely mimics the naturally occurring disease, and will enable attention to be focused on the role and function of genes carried on the conjugative virulence plasmid pVM01 in successful colonization of the avian respiratory tract.
ACKNOWLEDGMENTS
We thank Simon Stuart for determining colicin and siderophore production. This work was supported by a grant from the Rural Industries Research and Development Corporation.
REFERENCES
- 1.Acheson D W, Keusch G T, Lightowlers M, Donohue R A. Enzyme linked immunosorbent assay for Shiga toxin and Shiga-like toxin II using P1 glycoprotein from hydatid cysts. J Infect Dis. 1990;161:134–137. doi: 10.1093/infdis/161.1.134. [DOI] [PubMed] [Google Scholar]
- 2.Adams L M, Simmons C P, Rezmann L, Strugnell R A, Robins-Browne R M. Identification and characterization of a K88- and Cs31A-like operon of a rabbit enteropathogenic Escherichia coli strain which encodes fimbriae involved in the colonization of rabbit intestine. Infect Immun. 1997;65:5222–5230. doi: 10.1128/iai.65.12.5222-5230.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allan B J, van den Hurk J V, Potter A A. Characterization of Escherichia coli isolated from cases of avian colibacillosis. Can J Vet Res. 1993;57:146–151. [PMC free article] [PubMed] [Google Scholar]
- 4.Arp L H, Graham C, Cheville N F. Comparison of clearance rates of virulent and avirulent Escherichia coli in turkeys after aerosol exposure. Avian Dis. 1979;23:386–391. [Google Scholar]
- 5.Arp L H, Jensen A E. Piliation, hemagglutination, motility, and generation time of Escherichia coli that are virulent or avirulent for turkeys. Avian Dis. 1980;24:153–161. [Google Scholar]
- 6.Barbour E K, Nabbut N H, Al N H. Use of epidemiologic markers to identify the source of Escherichia coli infections in poultry. Am J Vet Res. 1985;46:989–991. [PubMed] [Google Scholar]
- 6a.Bell S M. Antibiotic sensitivity testing by the CDS method. In: Hartwig N, editor. Proceedings of the Clinical Microbiology Update Program, no. 21, 1984. Sydney, Australia: New South Wales Branch of the Australian Society for Microbiology; 1984. [Google Scholar]
- 7.Bettelheim K A. Identification of enterohaemorrhagic Escherichia coli by means of their production of enterohaemolysin. J Appl Bacteriol. 1995;79:178–180. doi: 10.1111/j.1365-2672.1995.tb00932.x. [DOI] [PubMed] [Google Scholar]
- 8.Bettelheim K A, Gracey M, Wadstrom T. The use of the coagulation test to determine whether Australian and New Zealand isolates of Escherichia coli produce the heat-labile enterotoxin. Zentbl Bakteriol Mikrobiol Hyg Abt 1 Orig A. 1985;260:293–299. doi: 10.1016/s0176-6724(85)80017-1. [DOI] [PubMed] [Google Scholar]
- 9.Bettelheim K A, Thompson C J. New method of serotyping Escherichia coli: implementation and verification. J Clin Microbiol. 1987;25:781–786. doi: 10.1128/jcm.25.5.781-786.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bettelheim K A, Wilson M W, Shooter R A, O'Farrell S M. Studies on enterotoxicity of environmental Escherichia coli belonging to serotypes normally considered enterotoxigenic. J Hyg. 1980;84:411–424. doi: 10.1017/s0022172400026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Binns M M, Davies D L, Hardy K G. Cloned fragments of the plasmid ColV, I-K94 specifying virulence and serum resistance. Nature. 1979;279:778–781. doi: 10.1038/279778a0. [DOI] [PubMed] [Google Scholar]
- 12.Blanco J E, Blanco M, Mora A, Jansen W H, Garcia V, Vazquez M L, Blanco J. Serotypes of Escherichia coli isolated from septicaemic chickens in Galicia (northwest Spain) Vet Microbiol. 1998;61:229–235. doi: 10.1016/s0378-1135(98)00182-5. [DOI] [PubMed] [Google Scholar]
- 13.Brown P K, Curtiss R. Unique chromosomal regions associated with virulence of an avian pathogenic Escherichia coli strain. Proc Natl Acad Sci USA. 1996;93:11149–11154. doi: 10.1073/pnas.93.20.11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandler M E, Bettelheim K A. A rapid method of identifying Escherichia coli H antigens. Zentbl Bakteriol Mikrobiol Hyg Abt 1 Orig A. 1974;229:74–79. [PubMed] [Google Scholar]
- 15.Csaky T Z. On the estimation of bound hydroxylamine in biological materials. Acta Chem Scand. 1948;2:450–454. [Google Scholar]
- 16.Dho M, Lafont J P. Escherichia coli colonization of the trachea in poultry: comparison of virulent and avirulent strains in gnotoxenic strains. Avian Dis. 1982;26:787–797. [PubMed] [Google Scholar]
- 17.Dho M, Lafont J P. Adhesive properties and iron uptake ability in Escherichia coli lethal and nonlethal for chicks. Avian Dis. 1984;28:1016–1025. [PubMed] [Google Scholar]
- 18.Dozois C M, Chanteloup N, Dho M M, Bree A, Desautels C, Fairbrother J M. Bacterial colonization and in vivo expression of F1 (type 1) fimbrial antigens in chickens experimentally infected with pathogenic Escherichia coli. Avian Dis. 1994;38:231–239. [PubMed] [Google Scholar]
- 19.Dozois C M, Fairbrother J M, Harel J, Bosse M. pap- and pil-related DNA sequences and other virulence determinants associated with Escherichia coli isolated from septicemic chickens and turkeys. Infect Immun. 1992;60:2648–2656. doi: 10.1128/iai.60.7.2648-2656.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dozois C M, Pourbakhsh S A, Fairbrother J M. Expression of P and type 1 (F1) fimbriae in pathogenic Escherichia coli from poultry. Vet Microbiol. 1995;45:297–309. doi: 10.1016/0378-1135(94)00127-i. [DOI] [PubMed] [Google Scholar]
- 21.Ellis M G, Arp L H, Lamont S J. Serum resistance and virulence of Escherichia coli isolated from turkeys. Am J Vet Res. 1988;49:2034–2047. [PubMed] [Google Scholar]
- 22.Emery D A, Nagaraja K V, Shaw D P, Newman J A, White D G. Virulence factors of Escherichia coli associated with colisepticemia in chickens and turkeys. Avian Dis. 1992;36:504–511. [PubMed] [Google Scholar]
- 23.Fantinatti F, Silveira W D, Castro A. Characteristics associated with pathogenicity of avian septicaemic Escherichia coli strains. Vet Microbiol. 1994;41:75–86. doi: 10.1016/0378-1135(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 24.Gentry M K, Dalrymple J M. Quantitative microtiter cytotoxicity assay for Shigella toxins. J Clin Microbiol. 1980;12:361–366. doi: 10.1128/jcm.12.3.361-366.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson F, Magrath D I. The isolation and characterization of hydroxamic acid (aerobactin) formed by Aerobacter aerogenes 62-1. Biochim Biophys Acta. 1969;192:175–184. doi: 10.1016/0304-4165(69)90353-5. [DOI] [PubMed] [Google Scholar]
- 26.Ginns C A, Browning G F, Benham M L, Whithear K G. Development and application of an aerosol challenge method for reproduction of avian colibacillosis. Avian Pathol. 1998;27:505–511. doi: 10.1080/03079459808419375. [DOI] [PubMed] [Google Scholar]
- 27.Glantz P J, Narotsky S, Bubash G. Escherichia coli serotypes isolated from salpingitis and chronic respiratory disease of poultry. Avian Dis. 1962;6:322–328. [Google Scholar]
- 28.Gross W B. Escherichia coli as a complicating factor in chronic respiratory disease of chickens and infectious sinusitis of turkeys. Poultry Sci. 1956;35:765–771. [Google Scholar]
- 29.Gross W B. Symposium on chronic respiratory diseases of poultry. II. The role of Escherichia coli in the cause of chronic respiratory disease and certain other respiratory diseases. Am J Vet Res. 1958;19:448–452. [PubMed] [Google Scholar]
- 30.Gross W B. Colibacillosis. In: Calnek B W, Beard C W, Reid W M, Yoder H W J, editors. Diseases of poultry—1991. Ames: Iowa State University Press; 1991. pp. 138–144. [Google Scholar]
- 31.Gross W G. Diseases due to Escherichia coli in poultry. In: Gyles C L, editor. Escherichia coli in domestic animals and humans—1994. Wallingford, United Kingdom: CAB International; 1994. pp. 237–259. [Google Scholar]
- 32.Gyimah J E, Panigrahy B. Adhesin-receptor interactions mediating the attachment of pathogenic Escherichia coli to chicken tracheal epithelium. Avian Dis. 1988;32:74–78. [PubMed] [Google Scholar]
- 33.Harry E G. A study of 119 outbreaks of coli-septicaemia in broiler flocks. Vet Rec. 1964;76:443–449. [Google Scholar]
- 34.Harry E G, Hemsley L A. The relationship between environmental contamination with septicaemia strains of Escherichia coli and their incidence in chickens. Vet Rec. 1965;77:241–245. [PubMed] [Google Scholar]
- 35.Harry E G, Hemsley L A. The association between the presence of septicaemia strains of Escherichia coli in the respiratory and intestinal tracts of chickens and the occurrence of Coli septicaemia. Vet Rec. 1965;77:35–40. [PubMed] [Google Scholar]
- 36.Hemsley L A, Harry E G. Coliform pericarditis (Coli septicaemia) in broiler chickens: a three year study on one farm. Vet Rec. 1965;77:103–107. [PubMed] [Google Scholar]
- 37.Ike K, Kawahara K, Danbara H, Kume K. Serum resistance and aerobactin iron uptake in avian Escherichia coli mediated by conjugative 100-megadalton plasmid. J Vet Med Sci. 1992;54:1091–1098. doi: 10.1292/jvms.54.1091. [DOI] [PubMed] [Google Scholar]
- 38.Ike K, Kume K, Kawahara K, Danbara H. Serotyping of O and pilus antigens of Escherichia coli strains isolated from chickens with coli-septicemia. Jpn J Vet Sci. 1990;52:1023–1027. doi: 10.1292/jvms1939.52.1023. [DOI] [PubMed] [Google Scholar]
- 39.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konowalchuk J, Spiers J I, Stavric S. Vero response of a cytotoxin of Escherichia coli. Infect Immun. 1977;18:775–779. doi: 10.1128/iai.18.3.775-779.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lafont J P, Dho M, D'Hauteville H M, Bree A, Sansonetti P J. Presence and expression of aerobactin genes in virulent avian strains of Escherichia coli. Infect Immun. 1987;55:193–197. doi: 10.1128/iai.55.1.193-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis M J. Transferable drug resistance and other transferable agents in strains of Escherichia coli from two human populations. Lancet. 1968;i:1389–1393. doi: 10.1016/s0140-6736(68)91973-9. [DOI] [PubMed] [Google Scholar]
- 43.Linggood M A, Roberts M, Ford S, Parry S H, Williams P H. Incidence of the aerobactin iron uptake system among Escherichia coli isolates from infections of farm animals. J Gen Microbiol. 1987;133:835–842. doi: 10.1099/00221287-133-4-835. [DOI] [PubMed] [Google Scholar]
- 44.Marc D, Arne P, Bree A, Dho M M. Colonization ability and pathogenic properties of a Fim− mutant of an avian strain of Escherichia coli. Res Microbiol. 1998;149:473–485. doi: 10.1016/s0923-2508(98)80002-8. [DOI] [PubMed] [Google Scholar]
- 45.Murray C J. Salmonella and Escherichia coli from veterinary and human sources in Australia during 1985 and 1986. Aust Vet J. 1987;64:256–257. doi: 10.1111/j.1751-0813.1987.tb09698.x. [DOI] [PubMed] [Google Scholar]
- 46.Naveh M W, Zusman T, Skutelsky E, Ron E Z. Adherence pili in avian strains of Escherichia coli: effect on pathogenicity. Avian Dis. 1984;28:651–661. [PubMed] [Google Scholar]
- 47.Ngeleka M, Kwaga J, White D G, Whittam T S, Riddell C, Goodhope R, Potter A A, Allan B. Escherichia coli cellulitis in broiler chickens—clonal relationships among strains and analysis of virulence-associated factors of isolates from diseased birds. Infect Immun. 1996;64:3118–3126. doi: 10.1128/iai.64.8.3118-3126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nolan L K, Wooley R E, Brown J, Spears K R, Dickerson H W, Dekich M. Comparison of a complement resistance test, a chicken embryo lethality test, and the chicken lethality test for determining virulence of avian Escherichia coli. Avian Dis. 1992;36:395–397. [PubMed] [Google Scholar]
- 49.Nolan L K, Wooley R E, Cooper R K. Transposon mutagenesis used to study the role of complement resistance in the virulence of an avian Escherichia coli isolate. Avian Dis. 1992;36:398–402. [PubMed] [Google Scholar]
- 50.Peighambari S M, Vaillancourt J P, Wilson R A, Gyles C L. Characteristics of Escherichia coli isolates from avian cellulitis. Avian Dis. 1995;39:116–124. [PubMed] [Google Scholar]
- 51.Quackenbush R L, Falkow S. Relationships between colicin V activity and virulence in Escherichia coli. Infect Immun. 1979;24:562–564. doi: 10.1128/iai.24.2.562-564.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reed W P, Williams R C., Jr Bacterial adherence: first step in pathogenesis of certain infections. J Chronic Dis. 1978;31:67–72. doi: 10.1016/0021-9681(78)90091-7. [DOI] [PubMed] [Google Scholar]
- 53.Riley M A. Molecular mechanisms of colicin evolution. Mol Biol Evol. 1993;10:1380–1395. doi: 10.1093/oxfordjournals.molbev.a040081. [DOI] [PubMed] [Google Scholar]
- 54.Sack D A, Sack R B. Test for enterotoxigenic Escherichia coli using Y-1 adrenal cells in mini culture. Infect Immun. 1975;11:334–336. doi: 10.1128/iai.11.2.334-336.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N. Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 56.Sekizaki T, Nonomura I, Imada Y. Loss of virulence associated with plasmid curing of chicken pathogenic Escherichia coli. Jpn J Vet Sci. 1989;51:659–661. doi: 10.1292/jvms1939.51.659. [DOI] [PubMed] [Google Scholar]
- 57.Taylor R K, Manoil C, Mekalanos J J. Broad-host-range vector for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J Bacteriol. 1989;171:1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson C J. New method of serotyping Escherichia coli: mathematical development. J Clin Microbiol. 1987;25:774–780. doi: 10.1128/jcm.25.5.774-780.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vidotto M C, Muller E E, de Freitas J C, Alfieri A A, Guimaraes I G, Santos D S. Virulence factors of avian Escherichia coli. Avian Dis. 1990;34:531–538. [PubMed] [Google Scholar]
- 60.White D G, Dho M M, Wilson R A, Whittam T S. Clonal relationships and variation in virulence among Escherichia coli strains of avian origin. Microb Pathog. 1993;14:399–409. doi: 10.1006/mpat.1993.1039. [DOI] [PubMed] [Google Scholar]
- 61.Whittam T S, Wilson R A. Genetic relationships among pathogenic strains of avian Escherichia coli. Infect Immun. 1988;56:2458–2466. doi: 10.1128/iai.56.9.2458-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williams P H, Warner P J. ColV plasmid-mediated, colicin V-independent iron uptake system of invasive strains of Escherichia coli. Infect Immun. 1980;29:411–416. doi: 10.1128/iai.29.2.411-416.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wittig W, Prager R, Tietze E, Seltmann G, Tschape H. Arch. Exp. Veterinaermed. 42:221–229. 1988. Aerobactin-positive Escherichia coli as causative agents of extra-intestinal infections among animals. [PubMed] [Google Scholar]
- 64.Wooley R E, Gibbs P S, Dickerson H W, Brown J, Nolan L K. Analysis of plasmids cloned from a virulent avian Escherichia coli and transformed into Escherichia coli DH5-alpha. Avian Dis. 1996;40:533–539. [PubMed] [Google Scholar]
- 65.Wooley R E, Nolan L K, Brown J, Gibbs P S, Giddings C W, Turner K S. Association of K-1 capsule, smooth lipopolysaccharides, traT gene, and colicin V production with complement resistance and virulence of avian Escherichia coli. Avian Dis. 1993;37:1092–1096. [PubMed] [Google Scholar]
- 66.Wooley R E, Spears K R, Brown J, Nolan L K, Fletcher O J. Relationship of complement resistance and selected virulence factors in pathogenic avian Escherichia coli. Avian Dis. 1992;36:679–684. [PubMed] [Google Scholar]
- 67.Yerushalmi Z, Smorodinsky N I, Naveh M W, Ron E Z. Adherence pili of avian strains of Escherichia coli O78. Infect Immun. 1990;58:1129–1131. doi: 10.1128/iai.58.4.1129-1131.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang J P, Normark S. Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science. 1996;273:1234–1236. doi: 10.1126/science.273.5279.1234. [DOI] [PubMed] [Google Scholar]