Abstract
Bacteria are a suspected pathogenic factor in inflammatory bowel disease, but the identity of the relevant microbial species remains unresolved. The pANCA autoantibody is associated with most cases of ulcerative colitis (UC) and hence reflects an immune response associated with the disease process. This study addresses the hypothesis that pANCA identifies an antigen(s) expressed by bacteria resident in the human colonic mucosa. Libraries of colonic bacteria were generated using aerobic and anaerobic microbiologic culture conditions, and bacterial pools and clonal isolates were evaluated for cross-reactive antigens by immunoblot analysis using the pANCA monoclonal antibody Fab 5-3. Two major species of proteins immunoreactive to pANCA monoclonal antibodies were detected in bacteria from the anaerobic libraries. Colony isolates of the expressing bacteria were identified as Bacteroides caccae and Escherichia coli. Isolation and partial sequencing of the B. caccae antigen identified a 100-kDa protein without database homologous sequences. The E. coli protein was biochemically and genetically identified as the outer membrane porin OmpC. Enzyme-linked immunosorbent assay with human sera demonstrated elevated immunoglobulin G anti-OmpC in UC patients compared to healthy controls. These findings demonstrate that a pANCA monoclonal antibody detects a recurrent protein epitope expressed by colonic bacteria and implicates colonic bacterial proteins as a target of the disease-associated immune response.
Ulcerative colitis (UC) is a chronic inflammatory mucosal disease associated with a strong familial pattern and a number of genetic loci implicated in disease susceptibility (5, 29, 36, 37, 49, 53, 54). Variation in penetrance, as well as demographic and epidemiologic features, indicates an important role for environmental factors in the inflammatory process of UC (38, 48). Gut-colonizing microorganisms are strategically situated for such an epidemiologic role. A number of studies have implicated enteric bacteria in human inflammatory bowel disease (IBD), particularly Crohn's disease (reviewed in reference 20). Analyses of several rodent IBD model systems have revealed a pathologic role for enteric bacteria. Rodents rendered germfree were protected from disease onset (25, 27, 31–33). Antimicrobial immunity has been pathogenetically implicated by Elson and colleagues, who identified disease-related humoral and T-cell responses to colonic bacteria in C3H/HeJBir mice (6, 8).
Sixty to 70% of UC patients and 25% of Crohn's disease patients produce disease-specific autoantibodies to a neutrophil protein with a perinuclear distribution, pANCA (14, 34, 39, 40, 43, 52). Using human monoclonal pANCA antibodies, we have recently characterized the neutrophil autoantigen and epitope specificity (a PKKAK motif of histone H1) (15, 19a). In other immune-mediated diseases, marker antibodies have been useful in identifying disease-relevant antigenic targets, even in those disease with T-cell-mediated effector mechanisms (2, 12, 35). pANCA and IBD-associated antibacterial serum antibodies were recently reported to cross-compete for bacterial and pANCA antigen binding (42). Here, we address the hypothesis that UC-associated pANCA reflects the response to a microbial antigenic target expressed by disease-associated gut colonists. This search identified anaerobic bacterial species (Escherichia coli and Bacteroides strains) bearing proteins with a pANCA cross-reactive epitope.
MATERIALS AND METHODS
Antibodies and detection reagents.
Fab 5-2, Fab 5-3, and P313 recombinant Fabs were produced and purified as previously described (15). The P313 expression vector was a generous gift from Carlos Barbas III (3). Alkaline phosphatase-conjugated goat anti-human Fab and goat anti-human Fcγ were purchased from Pierce (Rockford, Ill.) and Sigma Chemical Co. (St. Louis, Mo.), respectively.
Human specimens.
Endoscopic colon pinch biopsies were obtained from three Crohn's disease patients undergoing diagnostic procedures at the Cedars-Sinai Medical Center. All patients had active colonic disease and were not under antibiotic treatment. Endoscopic biopsy samples were directly dropped from the pinching claw into anaerobic transport medium (Anaerobe Systems, San Jose, Calif.) and transported on ice within 0.5 to 1 h for to UCLA (University of California, Los Angeles) for microbiologic processing. Serum aliquots were obtained from the IBD serum research archive at Cedars-Sinai Medical Center; the patient demography of this archive and method of selecting probands and concurrent healthy controls from the archive have been previously reported (49, 52). Forty human sera from UC patients and healthy controls were studied; quantitation of UC-pANCA binding activity was previously performed on all archival specimens as previously described (40). Procedures for subject recruitment, informed consent, and specimen procurement were in accordance with protocols approved by the Institutional Human Subject Protection Committees of UCLA and Cedars-Sinai Medical Center.
Bacterial strains.
Bacteroides isolates were clinical isolates from the UCLA Clinical Laboratories (3955-3, 4579, 4552, 4578, 4536, 4562, 4556, and 4570), University of North Carolina at Chapel Hill (UNC, LG1, LG1-33, and CPT-6; gift from R. B. Sartor), and the American Type Culture Collection (43185). E. coli OmpF− (RAM725) and OmpC− (RAM726) mutants and an OmpC− OmpF− double mutant were generated by Rajeev Misra. The genotype of RAM725 is [MC4100 Φ(ompF′-lacZ+)16-13] F′ araD139 Δ(argF-lac)U169 rpsL150 rel-1 flb-5301 ptsF25 deoC1 thi-1 Φ(ompF-lacZ+)16-13. The genotype of RAM7256 is [MC4100 Φ(ompC′-lacZ+)10-15] F′ araD139 Δ(argF-lac)U169 rpsL150 rel-1 flb-5301 ptsF25 deoC1 thi-1 Φ(ompC-lacZ+)10-15.
Bacteria were inoculated into 15 ml of Luria-Bertani broth and cultured vigorously in a 37°C shaker for 16 h. Cultures were harvested by centrifugation, resuspended in lysis buffer (50 mM Tris-Cl [pH 7.5], 300 mM NaCl, 10 mM EDTA, 0.1% sodium dodecyl sulfate [SDS]), and subjected to two periods of 1-min sonication at 50% intensity in a Misonix (Farmingdale, N.Y.) ultrasonic processor sonicator. The soluble fraction of each lysate was isolated by centrifugation (12,000 × g for 18 min) and subjected to Western analysis.
Bacterial culture.
Reagents are from Becton Dickinson BBL (Franklin Lakes, N.J.) except where noted. In anaerobic chambers, colonic biopsies for each patient were pooled in 2 ml of thioglycolate broth (TGL) and manually homogenized with a mortar and pestle. Then 100 μl of homogenate was added to 100 μl of brucella broth (Difco, Detroit, Mich.) in 40% glycerol and frozen at −80°C; 100 μl of this sample was cultured in liquid medium or agar at 37°C under various oxygen availability conditions (Table 1). Microaerophilic cultures were incubated unshaken in a commercial chamber (CampyPak). Anaerobic cultures were incubated unshaken in an anaerobic hood with 10% CO2–90% N2 atmosphere. Mixed plate cultures cultured on brucella blood agar (BBA) plates were harvested by cotton swabbing and resuspended in saline for Western analysis and subculture. Laked-kanamycin-vancomycin (LKV) and phenylethyl alcohol (PEA) blood agar plates were used for selective bacterial culture under anaerobic conditions. Cultures were restreaked to isolate single colonies, which were then expanded in brain heart infusion (BHI) broth under anaerobic conditions for 48 h at 37°C. For large-scale cultures, the p2Lc3 Bacteroides caccae isolate was inoculated from glycerol frozen stocks onto five LKV plates and incubated anaerobically for 72 h. Plate cultures were harvested with a cotton swab and inoculated into 10 liters of BHI broth for a 48-h fermentation using 10% CO2–90% N2 atmosphere at 37°C in a MicroFerm fermentor (New Brunswick, New Jersey, N.J.).
TABLE 1.
Bacterial isolation conditions
| Medium | O2 | Growth | Isolate(s) | Species |
|---|---|---|---|---|
| Trypticase soy broth | Aerobic | No | None | |
| Sheep blood agar | Aerobic | No | None | |
| BBA | Microaerophilic | No | None | |
| TGL | Anaerobic | Yes | p2c2, p2c5 | E. coli |
| BBA | Anaerobic | Yes | p1Bc5, p1Bc9 | E. coli |
| BBA-PEA | Anaerobic | Yes | p3 | B. caccae |
| BBA-LKV | Anaerobic | Yes | p2Lc2, p2Lc3, p2Lc5 | B. caccae |
16S rRNA gene polymorphism analysis.
Bacterial isolates were identified by 16S rRNA sequence analysis (41, 50). BHI broth cultures for each of seven isolates were centrifuged, and the cell pellet was washed twice with Dulbecco's phosphate-buffered saline (PBS). Cell pellets of ∼104 to 105 were used as a template for PCR amplification of the 1.5-kb fragment polymorphic region of the 16S rRNA gene. Oligonucleotides 5′-AGA GTT TGA T(C/T)(A/C) TGG C-3′ (forward) and 5′-G(C/T)T ACC TTG TTA CGA CTT-3′ (reverse) were used for amplification by PCR using 150 mM MgCl2 and elongation temperature of 60°C. Bacteroides-specific reverse primer 5′-CCT TGT TAC GAC TTA GCC-3′ was used for a more efficient amplification and sequencing of DNA from the Bacteroides isolates. Amplified 16S segments were analyzed with the National Center for Biotechnology Information BLASTN program and National Institutes of Health prokaryote database (version 1.4.11, November 1997) (1).
Preparation of subcellular fractions and protein purification.
Ten-liter fermentor cultures were harvested by centrifugation at 3,000 × g for 30 min at 4°C and resuspended in 300 ml of lysis buffer (50 mM Tris-Cl [pH 7.5], 300 mM NaCl, 10 mM EDTA, 0.1% SDS, protease inhibitors). Cells were lysed by two periods of 1-min pulse sonication (1 s on-0.2 s off) at 50% intensity in a Misonix ultrasonic processor sonicator. The soluble fraction of each lysate was isolated by centrifugation (12,000 × g for 18 min). Soluble proteins were precipitated with ammonium sulfate at 40% saturation with continuous mixing for a minimum of 24 h at 4°C, followed by centrifugation at 10,000 × g for 30 min, and dialyzed against PBS (50 mM sodium phosphate, 150 mM sodium chloride [pH 7.2]) at 4°C. Resuspended proteins were then precipitated with 50% acetone for 2 h at 4°C, centrifuged at 14,000 × g for 10 min, and resuspended in Dulbecco's PBS.
Western immunoblot analysis.
Bacterial cell lysates of mixed cultures were quantified by Bradford analysis, and equivalent protein amounts (10 μg/well) were separated on polyacrylamide gels under reducing conditions in Laemmli buffer; 10, 13, and 8% acrylamide were used for the mixed culture, E. coli, and Bacteroides isolates, respectively. Proteins were transferred overnight to nitrocellulose membranes (Amersham Life Sciences, Buckinghamshire, England) in Tris glycine buffer (National Diagnostics, Atlanta, Ga.) and verified by Ponceau S red staining (Sigma). Membranes were blocked in 5% nonfat milk (Carnation, Glendale, Calif.) in PBS with 0.1% Tween 20 (PBS-Tween) for 1 h. Primary and secondary antibodies diluted in 1% milk–PBS–Tween were incubated with membranes for 1 h. Fab 5-3 and P313 anti-tetanus toxoid were used at 2 μg/ml, while human serum was used at 1:100 dilution. Immunoblots were developed with goat anti-human Fab-alkaline phosphatase or goat anti-human Fcγ-alkaline phosphatase, with 5-bromo-4-chloro-3-indolylphosphate–nitroblue tetrazolium as the chromagenic substrate.
Preparative gel electrophoresis.
E. coli isolate samples were electrophoresed on a 13% full-size polyacrylamide gel (Bio-Rad, Richmond, Calif.). Proteins were electrophoretically transferred overnight to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad) in 10 mM CAPS [3-(cyclohexylamino)-1-propanesulfonic acid] 20% methanol buffer at pH 11.0. Membranes were immunoblotted, and reactive bands were excised from the membrane and subjected to solid-phase NH2-terminal microsequencing using a Beckman-Porton 2090E sequencer (Beckman Instruments, Anaheim, Calif.). Partially purified p2Lc3 B. caccae isolate proteins were identically processed, except for use of 8% polyacrylamide (due to the larger size of the immunoreactive protein species).
Tryptic digests, peptide analysis, and amino acid sequencing.
Protein samples purified by SDS-polyacrylamide gel electrophoresis (PAGE) were processed at the Keck Institute, Yale University. Gel-embedded proteins were washed six times with 0.1% trifluoroacetic acid (TFA)–60% CH3CN followed by one wash with 50% H2O–50% acetonitrile, one wash with 50 mM NH4HCO3–50% CH3CN, and one wash with 10 mM NH4HCO3–50% CH3CN. Samples were dried in a SpeedVac and digested with 1 μg of modified trypsin (Promega, Madison, Wis.) in 15 μl of 10 mM NH4HCO3 per 15 mm3 of gel. Five percent of tryptic digests were subject to matrix-assisted laser desorption ionization—mass Spectroscopy (MALDI-MS) for peptide mass searching (10, 21). The remaining portion of each sample was extracted with 0.1% TFA–60% CH3CN, dried, and dissolved in 0.05% TFA–5% acetonitrile. Peptides were separated by size exclusion high-pressure liquid chromatography (HPLC) on a Sephadex 200HR 10/30 column, and candidate HPLC peaks were evaluated for purity by MALDI-MS. Several fractions were subjected to Edman degradation, and the resultant amino acid sequences were characterized using the National Center for Biotechnology Information BLASTP program (version 1.4.11, November 1997) (1) and National Institutes of Health nonredundant database. Alignments were performed using the CLUSTAL W multiple-sequence alignment program (version 1.7, June 1997) (47).
ELISA analysis.
E. coli porins were purified from the OmpF− mutant strain according to standard methods (23), yielding a porin preparation of >80% purity by SDS-PAGE (data not shown), comprised predominantly of OmpC but including low levels of the minor porins OmpG and PhoE (16, 26). Microtiter plates (Costar 3069; Costar, Cambridge, Mass.) were coated with this OmpC-enriched porin at 1 μg/well in 50 μl of Dulbecco's PBS for 15 h at 4°C. Wells were washed with PBS–0.05% Tween 20 (Sigma), blocked with 1% bovine serum albumin in PBS-0.05% Tween 20 for 1 h, and washed again prior to incubation with sera. Human sera were reacted with duplicate wells at various dilutions (1:100 to 1:1,000) for 2 h at room temperature. The wells were washed four times with PBS–0.05% Tween 20 and then reacted for 1 h with 1:1,000 dilution of alkaline phosphatase-labeled goat anti-human Fcγ. Plates were washed three times in PBS–0.05% Tween 20 and twice with Tris-buffered saline (50 mM Tris [pH 7.5, 0.9% NaCl) and then developed for 15 min with Sigma 104 phosphatase substrate. Absorbances were measured at 405 nm with a Bio-Rad enzyme-linked immunosorbent assay (ELISA) reader and Macintosh analytic software. Data was statistically analyzed by Student's t test and chi-squared test.
RESULTS
Analysis of colonic bacterial cultures.
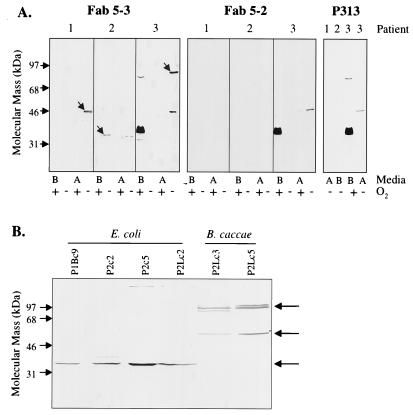
Colonic bacterial cultures were established under various oxygen availability conditions using endoscopic biopsies from three patients (Table 1). Equivalent amounts of protein from each mixed culture cell lysate were resolved by SDS-PAGE and transferred to nitrocellulose membranes for immunoblotting. Membranes were probed with monoclonal Fab 5-3, as a representative of pANCA, or with anti-tetanus toxoid Fab P313 as a negative control (Fig. 1A). Specific bands were detected by Fab 5-3 only under anaerobic conditions. These included prominent proteins migrating at ∼35, ∼48, and ∼80 kDa. Proteins detected in aerobic bacterial pools reacted with Fab P313 and thus represented nonspecific bands. No specific reactivity was detected for the second pANCA monoclonal antibody, Fab 5-2.
FIG. 1.
Immunoblot analysis of colonic bacterial cultures. (A) Mixed bacterial cultures were produced from colonic endoscopic biopsies of three patients under various medium and oxygen availabilities. Equivalent amounts (10 μg/lane) of mixed bacterial cultures were separated on a 10% polyacrylamide gel, transferred to nitrocellulose membranes, and probed with recombinant Fab monoclonal antibodies: Fab 5-3 pANCA (left panel), Fab 5-2 pANCA (middle panel), and P313 anti-tetanus toxoid (right panel). Medium A, sheep blood agar (aerobic) or BBA (anaerobic); medium B, Trypticase soy broth (aerobic) or TGL (anaerobic). Arrows indicate proteins specifically detected by Fab 5-3. (B) Equivalent amounts (10 μg/lane) of anaerobic colonic isolates were separated on a 10% polyacrylamide gel, transferred to nitrocellulose membranes, and probed with 5-3 pANCA monoclonal Fab. Isolates originated from the following anaerobic cultures: p1Bc5 and p1Bc9 were from the patient 1 specimen after BBA selection; p2c2 and p2c5 were from the patient 2 specimen after BBA selection; p2Lc2, p2Lc3, and p2Lc5 were from the patient 2 specimen after LKV selection. Two major immunoreactive protein species were identified: ∼35-kDa protein expressed by E. coli isolates and ∼100-kDa protein doublet expressed by B. caccae isolates. A ∼48-kDa species variably accompanied the ∼100-kDa B. caccae protein.
Bacterial cultures expressing immunoreactive proteins were restreaked on selective agar and colony purified (Table 1). Isolated colonies were expanded by anaerobic culture in BHI broth and tested for expression of Fab 5-3 immunoreactive proteins by Western blot analysis (Fig. 1B). Seven clonal isolates expressing two different immunoreactive protein profiles were obtained. The first was a single ∼35-kDa protein exemplified by the anaerobic culture of the specimen from patient 2. The second was exemplified by LKV cultures from patient 2, expressing a ∼100-kDa doublet and variably a ∼48-kDa species. The ∼48-kDa species was variably present in preparations, depending on culture time or sample handling conditions, suggesting that it was a proteolytic product of the ∼100-kDa protein.
The immunoreactive bacterial isolates were identified by DNA sequence analysis of PCR-amplified segments from a polymorphic region of the 16S rRNA gene. The two isolates expressing the ∼100-kDa immunoreactive protein were identified as members of the genus Bacteroides, with the highest similarity to B. caccae (P < 5.1 × 10−115). Identification of the Bacteroides isolates was supported by microscopic examination (short gram-negative rods) and by growth on LKV but not PEA plates. The isolates expressing the ∼35-kDa reactive protein were identified by 16S rRNA phylogeny as strains of E. coli (P < 2.4 × 10−160). This identification was supported by microscopic analysis (gram-negative rods) and by sensitivity to LKV and PEA. One E. coli clone, presumably representing a kanamycin-resistant strain, was isolated following selection on LKV plates.
Characterization of the Bacteroides ∼100-kDa protein.
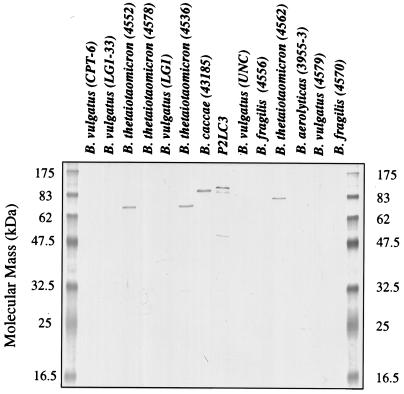
Expression of the ∼100-kDa immunoreactive protein was further evaluated for its species prevalence in a panel of Bacteroides clinical isolates (Fig. 2). Expression of a closely comigrating protein was observed in an independent B. caccae isolate (43185), but no expression was detected in seven isolates representing B. vulgatus, B. fragilis, and B. aerolyticus. An ∼80-kDa protein was detected in three of four isolates of B. thetaiotaomicron. Since our original mixed anaerobic cultures expressed an immunoreactive protein of that size (Fig. 1A), it is conceivable that it represented the product of a population of B. thetaiotaomicron in those cultures.
FIG. 2.
Expression of Fab 5-3 immunoreactive protein in Bacteroides strains. Equivalent amounts (10 μg/lane) of Bacteroides clinical isolates were separated on a 13% polyacrylamide gel, transferred to nitrocellulose membranes, and probed with 5-3 pANCA monoclonal Fab. p2Lc3 is included as a positive control for the reactive ∼100-kDa protein.
The immunoreactive B. caccae protein from p2Lc3 was characterized by a biochemical approach. Cell lysates were fractionated by SDS-PAGE and transferred to PVDF membranes, and the reactive protein doublet was subjected to N-terminal sequencing. This analysis revealed that the protein was N-terminally blocked. In-gel tryptic digests were performed with the SDS-PAGE-purified protein, and the resulting peptides were HPLC purified and analyzed for peptide mass by MALDI-MS (10). No matching digest profiles of these peptide masses were identified in the available databases. Amino acid sequences were obtained by Edman degradation for several tryptic peptides; the longest sequenced peptides are listed in Table 2. Searches for these peptides in available databases yielded only modestly similar homologues. Therefore, the 100-kDa immunoreactive protein has not been previously reported by these methods and is probably a novel species.
TABLE 2.
Tryptic peptides of the B. caccae 100-kDa proteina
| Peptide | Peak | Purity (%) | Size | Amino acid sequence |
|---|---|---|---|---|
| 1 | 71 | 100 | 1,162.9 | Asp-Pro-Ser-Ser-Leu-Ala-Ile-Phe-Gly-Val-Arg |
| 3 | 35 | 100 | 1,199.1 | Gly-Pro-Ser-Glu-Ala-Asp-Ala-Phe-Tyr-Asn-Cys |
| Tyr | ||||
| 2 | 45 | 87 | 1,007.7 | Tyr-Phe-Leu-Ser-Ala-Ser-Tyr-Arg-X |
| Ala Gly-Asn-Gly-Glu | ||||
| Ser |
The protein was isolate by SDS-PAGE from p2Lc3, and tryptic peptides were produced and separated by size exclusion HPLC. The tryptic peptides were analyzed by MALDI-MS for size determination and subjected to Edman degradation for NH2-terminal peptide sequence.
Identification of the E. coli immunoreactive protein.
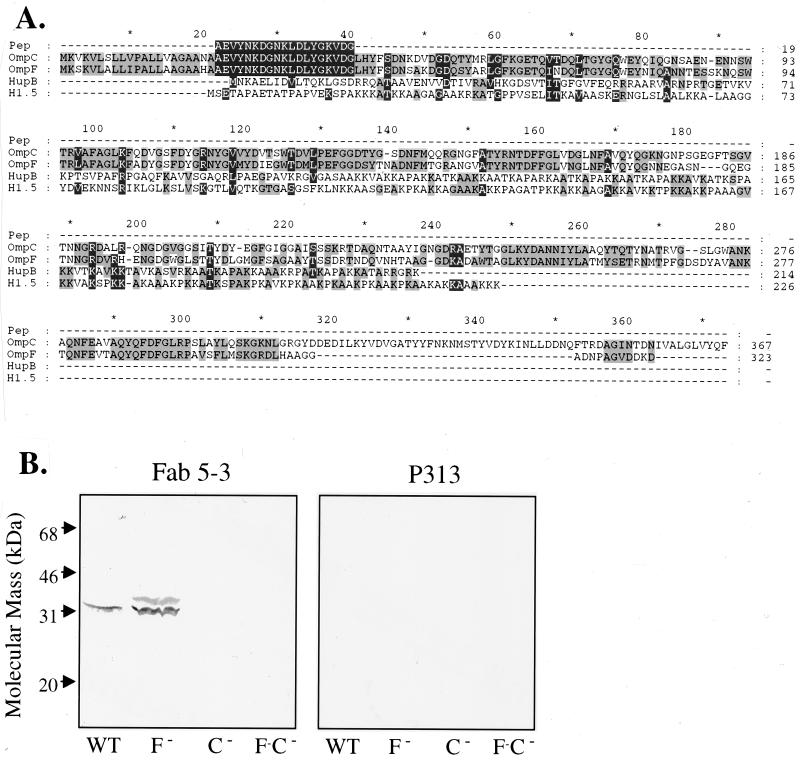
The ∼35-kDa reactive protein species was gel purified by SDS-PAGE, transferred to PVDF membranes, and excised for solid-phase Edman degradation sequencing. A single 19-amino-acid NH2-terminal peptide sequence was obtained for E. coli isolates p2c2, p1Bc9, and p1Bc5 and was found to be identical to amino acids 22 to 40 of the outer membrane porin precursors encoded by the ompC and ompF genes of E. coli (Fig. 3A). To confirm this identification and distinguish between OmpC and OmpF, a genetic analysis was employed. E. coli mutants bearing disruptive insertions of OmpC, OmpF, or both were obtained and compared for immunoreactivity with Fab 5-3 (Fig. 3B). Strains bearing mutant ompC lacked an immunoreactive protein, whereas strains bearing a mutant ompF gene expressed retained the ∼35-kDa species. This analysis established that the immunoreactive protein was OmpC and indicated that the relevant epitope resided in the one of the polymorphic regions distinguishing these two porins. OmpC lacked linear sequence homology with human histone H1 and mycobacterial HupB, two proteins also bearing the Fab 5-3 epitope (Fig. 3A).
FIG. 3.
Identification of the E. coli immunoreactive protein. (A) The 19-amino-acid NH2-terminal peptide sequence obtained from the E. coli ∼35-kDa immunoreactive protein was aligned with the peptide sequences of the E. coli OmpC and OmpF genes. The peptide initiates at position 22, corresponding to the beginning of the mature peptide (the first 21 amino acids are the leader peptide of the precursor protein). Sequences for the human histone H1.5 and Mycobacterium tuberculosis HupB gene products are also aligned, using the CLUSTAL W multiple sequence alignment program. (B) Equivalent number of cells (∼107 per lane) of E. coli XL1 (wild type) and OmpC−, OmpF−, and OmpC−OmpF− mutants were separated on a 12% polyacrylamide gel, transferred to nitrocellulose membranes, and probed with Fab 5-3 or P313 monoclonal antibody.
Seroreactivity of UC patients to OmpC.
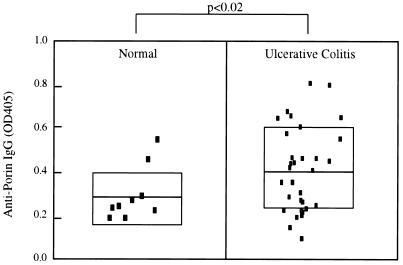
Human sera (31 UC patients and 10 healthy controls) were evaluated by ELISA for binding to E. coli porin (Fig. 4). The mean absorbance of the UC group (0.42 ± 0.19, mean ± standard deviation [SD]) was significantly higher than for healthy controls (0.29 ± 0.11); P < 0.02). However, for sera with high pANCA titers (>1,000 U, n = 8), an elevated mean anti-porin immunoglobulin G (IgG) absorbance was observed (0.55 ± 0.19), and this was significant compared to the normal group (P < 0.005). The frequencies of seropositive individuals (>0.51, the mean + 2 SD of the control population) were 1 of 9, 9 of 31, and 4 of 8 for the control, UC, and high-pANCA UC groups. These frequencies were not significantly different (P < 0.05 by chi-squared test). Due to the small size of the test population, the UC group was not stratified with regard to clinical parameters (extent of disease, response to therapy, extraintestinal manifestations).
FIG. 4.
Human seroreactivity to E. coli porin. ELISA wells were coated with OmpC-enriched porin (purified from OmpF− E. coli) and reacted with serum samples at 1:200 (qualitatively similar results were obtained with 1:100 to 1:1,000 dilutions). Wells were developed with phosphatase–anti-IgG and chromogenic substrate, and absorbances were tabulated for 10 healthy control and 30 UC patient samples. Bars indicate arithmetic means for each group, and shaded areas indicate ± 1 SD. P values were calculated between the two groups by Student's t test. OD405, optical density at 405 nm.
DISCUSSION
This study addresses the hypothesis that colonic bacteria bear proteins cross-reactive to a pANCA epitope. Using the Fab 5-3 pANCA monoclonal antibody, immunoreactive bacteria were detected in the anaerobic population and identified as two species of Bacteroides (caccae and thetaiotaomicron) and E. coli. Purification and partial sequencing of the B. caccae protein indicated that it is a previously unidentified protein. The E. coli protein was identified biochemically and genetically as the outer membrane porin, OmpC.
Structural relationship of the cross-reactive bacterial proteins and the pANCA autoantigen.
The identity of the perinuclear neutrophil autoantigen detected by UC-associated pANCA is emerging as epitopes of the non-core histone chromosomal family. Studies with human sera have implicated the HMG (high mobility group)-1 and -2 proteins as a pANCA antigen in a UC patient subset and in most individuals with autoimmune hepatitis (45, 46). Histone H1 and the HMG proteins are members of the linker nucleoprotein family and are closely related in sequence and subcellular localization. Fab 5-3 strongly reacts with HupB, a novel protein of Mycobacterium globally similar in primary amino acid sequence to human histone H1, particularly in the COOH-terminal random coil. All of these proteins share recurrent KKAK peptides which constitute part of the recurrent Fab 5-3 epitope. However, seroreactivities for HupB, histone H1, and neutrophils were uncorrelated by direct binding and absorption studies. It therefore appears that pANCA-positive sera detect additional antigenic specificities distinct from the epitope identified by the pANCA monoclonal antibodies (7a, 15a).
In contrast to HupB, E. coli OmpC lacked significant linear sequence similarity to these proteins, suggesting that the pANCA epitope expressed by OmpC is conformational. OmpC porin is formed as a trimer with each subunit structured as a highly conserved 16-stranded beta barrel with short turns on the inside and long divergent loops exposed at the exterior (11, 51). A recent immunoreactivity study of porins established that cross-reactivity between family members is characteristic of antibodies targeting the conserved transmembrane beta sheets, whereas antiporin antibodies detect divergent epitopes in the external loops of the porin protein (44). OmpC expression is a virulence trait for certain coliforms (4), and its external loop epitopes are strongly immunogenic (7, 24, 28). The specificity of Fab 5-3 binding for OmpC versus OmpF suggests that it detects an epitope present in one of the external loop structures which distinguish these two porin proteins. Moreover, bacterial expression of OmpC and that of OmpF are inversely regulated by environmental stimuli, with OmpC favored under anaerobic conditions (9, 19). An immune response in the anaerobic environment of the gut is therefore likely to preferentially target OmpC.
The present study identified higher anti-OmpC IgG levels in UC patients than in healthy controls, particularly in those with high pANCA titers. This latter group is notable because of evidence that they represent a distinct patient subset with more severe clinical course, postcolectomy pouchitis, and resistance to anti-tumor necrosis factor therapy. Since OmpC is a major outer membrane protein of E. coli, the previously reported cross-reactivity of serum UC pANCA with E. coli membranes further suggests a relationship of OmpC with pANCA in IBD (18, 22, 39, 42). However, the present study did not significantly correlate levels of serum pANCA and anti-OmpC IgG. A comprehensive population-based serum study will be required to resolve the correlation of anti-OmpC IgG with pANCA cross-reactivity, disease specificity, extent and activity of disease, and response to treatment. It will also be informative to assess the secreted IgA compartment, which has not been reported for either OmpC and pANCA specificities.
pANCA immunoreactive bacteria and IBD.
Bacteroides species and E. coli are common colonic bacteria, typically displaying a commensal, nonvirulent phenotype (17). However, the abundance of E. coli ranges several orders of magnitude even in healthy adults, and the level of expression of OmpC is under environmental control (9). Similar complexity is likely to pertain the Bacteroides species and their immunoreactive protein. It will be informative to assess the abundance of these bacteria and levels of immunoreactive protein expression in stool specimens, comparing populations of control subjects and clinically stratified UC patients.
Innate or specific immune recognition processes can result in destructive inflammatory responses triggered by such commensals, probably through proinflammatory bacterial products which incite or amplify chronic mucosal injury (38). For example, monoassociation studies have implicated Bacteroides vulgatus as a pathological factor in a rat colitis model (30, 33). With regard to this organism, Fig. 2 revealed that these B. vulgatus isolates lacked expression of the Fab 5-3 immunoreactive protein. However, the most important prediction from the present study is that the OmpC or 100-kDa proteins are targets of the pathogenic T-cell response in human or animal models of colitis. Bacteroides and E. coli proteins have been identified as antigenic targets for colitis-inducing T-cell lines in the C3H/HeJBir mouse (8). Recently, IBD-associated T-cell clones were observed to recognize extracts from B. thetaiotaomicron, Bifidobacterium bifidum, and E. coli, in several cases recognizing a bacterial antigen cross-reactive between these species (13). The similar pattern of cross-reactive bacterial recognition by these T-cell clones and the UC-associated monoclonal antibody in the present study is provocative. It will now be important to directly test whether a component of such T-cell responses are indeed specific to the OmpC or 100-kDa proteins.
ACKNOWLEDGMENTS
We thank Audrey Fowler for advice and direction of protein microsequencing, Gunter Harth for guidance in biochemical protein purification, Sydney Finegold and Balfour Sartor for advice and bacterial clinical isolates, Carlos Barbas III for the P313 Fab vector, and Abigail Salyers for advice on 16S rRNA taxonomy. We are especially grateful for the technical assistance of Karin Reimann, Kathleen Lechowitzc, and Kevin Ward.
This work was supported by NIH grants DK46763, EY00360, CA1200, AI38545, DK43026, and AI07126, the Crohn's and Colitis Foundation of America, the Jonnson Comprehensive Cancer Center, and the Feintech Family Chair of Inflammatory Bowel disease.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Meyers E W, Lippman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arden S D, Roep B O, Neophytou P I, Usac E F, Duinkerken G, De Vries R R P, Hutton J C. Imogen 38: a novel 38-kD islet mitochondrial autoantigen recognized by T cells from a newly diagnosed type 1 diabetic patient. J Clin Investig. 1996;97:551–561. doi: 10.1172/JCI118448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbas C F, Kang A S, Lerner R, Benkovic S J. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernardini M L, Sanna M G, Fontaine A, Sansonetti P J. OmpC is involved in invasion of epithelial cells by Shigella flexneri. Infect Immun. 1993;61:3625–3635. doi: 10.1128/iai.61.9.3625-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouma G, Oudkerk P, Crusius J B, Schreuder G M, Hellemans H P, Meijer B U, Kostense P J, Giphart M J, Meuwissen S G, Pena A S. Evidence for genetic heterogeneity in inflammatory bowel disease (IBD); HLA genes in the predisposition to suffer from ulcerative colitis (UC) and Crohn's disease (CD) Clin Exp Immunol. 1997;109:175–179. doi: 10.1046/j.1365-2249.1997.4121510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandwein S L, McCabe R P, Cong Y, Waites K B, Ridwan B U, Dean P A, Ohkusa T, Birkenmeier E H, Sundberg J P, Elson C O. Spontaneously colitic C3H/HeJBir mice demonstrate selective antibody reactivity to antigens of the enteric bacterial flora. J Immunol. 1997;159:44–52. [PubMed] [Google Scholar]
- 7.Calandra G B, Lukacs L J, Jonas L C, Santosham M, Ward J I, Greenberg D P, Daum R S, Matthews H, Vella P P, Ryan J L. Anti-PRP antibody levels after a primary series of PRP-OMPC and persistence of antibody titres following primary and booster doses. Vaccine. 1993;11(Suppl. 1):S58–S62. doi: 10.1016/0264-410x(93)90162-q. [DOI] [PubMed] [Google Scholar]
- 7a.Cohavy O, Harth G, Horwitz M, Eggena M, Landers C, Sutton C, Targan S R, Braun J. Identification of a novel mycobacterial histone H1 homologue (HupB) as an antigenic target of pANCA monoclonal antibody and serum immunoglobulin A from patients with Crohn's disease. Infect Immun. 1999;67:6510–6517. doi: 10.1128/iai.67.12.6510-6517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cong Y, Brandwein S L, McCabe R P, Lazenby A, Birkenmeier E H, Sundberg J P, Elson C O. CD4+ T cells reactive to enteric bacterial antigens in spontaneously colitic C3H/HeJBir mice; increased T helper cell type 1 response and ability to transfer disease. J Exp Med. 1998;187:855–864. doi: 10.1084/jem.187.6.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Contreras I, Munoz L, Toro C S, Mora G C. Heterologous expression of Escherichia coli porin genes in Salmonella typhi Ty2: regulation by medium osmolarity, temperature and oxygen availability. FEMS Microbiol Lett. 1995;133:105–111. doi: 10.1111/j.1574-6968.1995.tb07869.x. [DOI] [PubMed] [Google Scholar]
- 10.Cottrell J S. Protein identification by peptide mass fingerprinting. Pepl Res. 1994;7:115–124. [PubMed] [Google Scholar]
- 11.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J N, Rosenbusch J P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 12.Crameri R, Faith A, Hemmann S, Jaussi R, Ismail C, Menz G, Blaser K. Humoral and cell-mediated autoimmunity in allergy to Aspergillus fumigatus. J Exp Med. 1996;184:265–270. doi: 10.1084/jem.184.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duchmann R, May E, Heike M, Knolle P, Neurath M, Zum B K. T cell specificity and cross reactivity towards enterobacteria, Bacteroides, Bifidobacterium, and antigens from resident intestinal flora in humans. Gut. 1999;44:812–818. doi: 10.1136/gut.44.6.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duerr R H, Targan S R, Landers C J, Sutherland L R, Shanahan F. Anti-neutrophil cytoplasmic antibodies in ulcerative colitis. Comparison with other colitides/diarrheal illnesses. Gastroenterology. 1991;100:1590–1596. doi: 10.1016/0016-5085(91)90657-7. [DOI] [PubMed] [Google Scholar]
- 15.Eggena M, Targan S R, Iwanczyk L, Vidrich A, Gordon L K, Braun J. Phage display cloning and characterization of an immunogenetic marker (perinuclear anti-neutrophil cytoplasmic antibody) in ulcerative colitis. J Immunol. 1996;156:4005–4011. [PubMed] [Google Scholar]
- 16.Fajardo D A, Cheung J, Ito C, Sugawara E, Nikaido H, Misra R. Biochemistry and regulation of a novel Escherichia coli K-12 porin protein, OmpG, which produces unusually large channels. J Bacteriol. 1998;180:4452–4459. doi: 10.1128/jb.180.17.4452-4459.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finegold S M, Sutter V L. Fecal flora in different populations, with special reference to diet. Am J Clin Nutr. 1978;1:S116–S122. doi: 10.1093/ajcn/31.10.S116. [DOI] [PubMed] [Google Scholar]
- 18.Fleshner P R, Vasiliauskas E A, Kam L Y, Abreu-Martin M T, Targan S R. High level perinuclear antineutrophil cytoplasmic antibody (pANCA) in ulcerative colitis patients before colectomy predicts the development of chronic pouchitis after ileal pouch anal anastomosis. Gastroenterology. 1999;116:A716. doi: 10.1136/gut.49.5.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forst S, Delgado J, Inouye M. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Gordon L K, Eggena M, Targan S R, Braun J. Mast cell and neuroendocrine cytoplasmic autoantigen(s) detected by monoclonal pANCA antibodies. Clin Immunol. 2000;94:42–50. doi: 10.1006/clim.1999.4805. [DOI] [PubMed] [Google Scholar]
- 20.Janowitz H D, Croen E C, Sachar D B. The role of the fecal stream in Crohn's disease: an historical and analytic review. Inflamm Bowel Dis. 1998;4:29–39. doi: 10.1097/00054725-199802000-00006. [DOI] [PubMed] [Google Scholar]
- 21.Jimenez C R, Burlingame A L. Ultramicroanalysis of peptide profiles in biological samples using MALDI mass spectrometry. Exp Nephrol. 1998;6:421–428. doi: 10.1159/000020551. [DOI] [PubMed] [Google Scholar]
- 22.Kam L Y, Vasiliauskas E A, Landers C, Targan S R. Magnitude of response to Remicade correlates with marker antibody expression. Gastroenterology. 1999;116:A744. [Google Scholar]
- 23.Le Dain A C, Hase C C, Tommassen J, Martinac B. Porins of Escherichia coli: unidirectional gating by pressure. EMBO J. 1996;15:3524–3528. [PMC free article] [PubMed] [Google Scholar]
- 24.Lutwyche P, Exner M M, Hancock R E, Trust T J. A conserved Aeromonas salmonicida porin provides protective immunity to rainbow trout. Infect Immun. 1995;63:3137–3142. doi: 10.1128/iai.63.8.3137-3142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller M J, Sadowska-Krowicka H, Chotinaruemol S, Kakkis J L, Clark D A. Amelioration of chronic ileitis by nitric oxide synthase inhibition. J Pharmacol Exp Ther. 1993;264:11–16. [PubMed] [Google Scholar]
- 26.Mizuno T, Chou M Y, Inouye M. A comparative study on the genes for three porins of the Escherichia coli outer membrane. DNA sequence of the osmoregulated ompC gene. J Biol Chem. 1983;258:6932–6940. [PubMed] [Google Scholar]
- 27.Morrissey P J, Charrier K. Induction of wasting disease in SCID mice by the transfer of normal CD4+/CD45RBhi T cells and the regulation of this autoreactivity by CD4+/CD45RBlo T cells. Res Immunol. 1994;145:357–362. doi: 10.1016/s0923-2494(94)80200-9. [DOI] [PubMed] [Google Scholar]
- 28.Mulholland E K, Todd J, Rowe M, Campbell H, Byass P, Vella P P, Ahonkhai V I, Greenwood B M. Persistence of antibody at 18 months following vaccination of young Gambian infants with PRP-OMPC Haemophilus influenzae type b conjugate vaccine. Ann Trop Paediatr. 1993;13:153–158. doi: 10.1080/02724936.1993.11747639. [DOI] [PubMed] [Google Scholar]
- 29.Naom I, Lee J, Ford D, Bowman S J, Lanchbury J S, Haris I, Hodgson S V, Easton D, Lennard-Jones J, Mathew C G. Analysis of the contribution of HLA genes to genetic predisposition in inflammatory bowel disease. Am J Hum Genet. 1996;59:226–233. [PMC free article] [PubMed] [Google Scholar]
- 30.Onderdonk A B, Franklin M L, Cisneros R L. Production of experimental ulcerative colitis in gnotobiotic guinea pigs with simplified microflora. Infect Immun. 1981;32:225–231. doi: 10.1128/iai.32.1.225-231.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Podolsky D K. Inflammatory bowel disease. N Engl J Med. 1991;325:1008–1016. doi: 10.1056/NEJM199110033251406. [DOI] [PubMed] [Google Scholar]
- 32.Rachmilewitz D, Stamler J S, Karmeli F, Mullins M E, Singel D J, Loscalzo J, Xavier R J, Podolsky D K. Peroxynitrite-induced rat colitis—a new model of colonic inflammation. Gastroenterology. 1993;105:1681–1688. doi: 10.1016/0016-5085(93)91063-n. [DOI] [PubMed] [Google Scholar]
- 33.Rath H C, Herfarth H H, Ikeda J S, Grenther W B, Hamm T E, Jr, Balish E, Taurog J D, Hammer R E, Wilson K H, Sartor R B. Normal luminal bacteria, especially bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J Clin Investig. 1996;98:945–953. doi: 10.1172/JCI118878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reumaux D, Meziere C, Colombel J-F, Duthilleul P, Muller S. Distinct production of autoantibodies to nuclear components in ulcerative colitis and in Crohn's disease. Clin Immunol Immunopathol. 1995;77:349–357. doi: 10.1006/clin.1995.1162. [DOI] [PubMed] [Google Scholar]
- 35.Rosenberg S A. A new era for cancer immunotherapy based on the genes that encode cancer antigens. Immunity. 1999;10:281–287. doi: 10.1016/s1074-7613(00)80028-x. [DOI] [PubMed] [Google Scholar]
- 36.Roth M-P, Petersen G M, McElree C, Vadheim C M, Panish J F, Rotter J I. Familial empiric risk estimates of inflammatory bowel disease in Ashkenazi Jews. Gastroenterology. 1989;96:1016–1020. doi: 10.1016/0016-5085(89)91618-1. [DOI] [PubMed] [Google Scholar]
- 37.Roussomoustakaki M, Satsangi J, Welsh K, Louis E, Fanning G, Targan S R, Landers C, Jewell D P. Genetic markers may predict disease behavior in patients with ulcerative colitis. Gastroenterology. 1997;112:1845–1853. doi: 10.1053/gast.1997.v112.pm9178675. [DOI] [PubMed] [Google Scholar]
- 38.Sartor R B. The influence of normal microbial flora on the development of chronic mucosal inflammation. Res Immunol. 1997;148:567–576. doi: 10.1016/s0923-2494(98)80151-x. [DOI] [PubMed] [Google Scholar]
- 39.Satsangi J, Landers C J, Welsh K I, Koss K, Targan S R, Jewell D P. The presence of anti-neutrophil antibodies reflects clinical and genetic heterogeneity within inflammatory bowel disease. Inflamm Bowel Dis. 1998;4:18–26. doi: 10.1097/00054725-199802000-00004. [DOI] [PubMed] [Google Scholar]
- 40.Saxon A, Shanahan F, Landers C, Ganz T, Targan S R. A distinct subset of antineutrophil cytoplasmic antibodies is associated with inflammatory bowel disease. J Allergy Clin Immunol. 1990;86:202–210. doi: 10.1016/s0091-6749(05)80067-3. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt T M, Relman D A. Phylogenetic identification of uncultured pathogens using ribosomal RNA sequences. Methods Enzymol. 1994;235:205–222. doi: 10.1016/0076-6879(94)35142-2. [DOI] [PubMed] [Google Scholar]
- 42.Seibold F, Brandwein S, Simpson S, Terhorst C, Elson C O. pANCA represents a cross-reactivity to enteric bacterial antigens. J Clin Immunol. 1998;18:153–160. doi: 10.1023/a:1023203118100. [DOI] [PubMed] [Google Scholar]
- 43.Shanahan F, Duerr R H, Rotter J I, Yang H-Y, Sutherland L R, McElree C, Landers C J, Targan S R. Neutrophil autoantibodies in ulcerative colitis: familial aggregation and genetic heterogeneity. Gastroenterology. 1992;103:456–461. doi: 10.1016/0016-5085(92)90834-l. [DOI] [PubMed] [Google Scholar]
- 44.Singh S P, Upshaw Y, Abdullah T, Singh S R, Klebba P E. Structural relatedness of enteric bacterial porins assessed with monoclonal antibodies to Salmonella typhimurium OmpD and OmpC. J Bacteriol. 1992;174:1965–1973. doi: 10.1128/jb.174.6.1965-1973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sobajima J, Ozaki S, Uesugi H, Osakada F, Inoue M, Fukuda Y, Shirakawa H, Yoshida M, Rokuhara A, Imai H, Kiyosawa K, Nakao K. High mobility group (HMG) non-histone chromosomal proteins HMG1 and HMG2 are significant target antigens of perinuclear anti-neutrophil cytoplasmic antibodies in autoimmune hepatitis. Gut. 1999;44:867–873. doi: 10.1136/gut.44.6.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sobajima J, Ozaki S, Uesugi H, Osakada F, Shirakawa H, Yoshida M, Nakao K. Prevalence and characterization of perinuclear anti-neutrophil cytoplasmic antibodies (P-ANCA) directed against HMG1 and HMG2 in ulcerative colitis (UC) Clin Exp Immunol. 1998;111:402–407. doi: 10.1046/j.1365-2249.1998.00491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson N P, Driscoll R, Pounder R E, Wakefield A J. Genetics versus environment in inflammatory bowel disease: results of a British twin study. Bmj (Clin Res Ed) 1996;312:95–96. doi: 10.1136/bmj.312.7023.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toyoda H, Wang S-J, Yang H-Y, Redford A, Magalong D, Tyan D, McElree C, Pressman S, Shanahan F, Targan S R, Rotter J I. Distinct associations of HLA class II genes with inflammatory bowel disease. Gastroenterology. 1993;104:741–748. doi: 10.1016/0016-5085(93)91009-7. [DOI] [PubMed] [Google Scholar]
- 50.von Wintzingerode F, Gobel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 51.Watanabe M, Rosenbusch J, Schirmer T, Karplus M. Computer simulations of the OmpF porin from the outer membrane of Escherichia coli. Biophys J. 1997;72:2094–2102. doi: 10.1016/S0006-3495(97)78852-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang H-Y, Rotter J I, Toyoda H, Landers C, Tyan D, McElree C K, Targan S R. Ulcerative colitis: a genetically heterogeneous disorder defined by genetic (HLA class II) and subclinical (antineutrophil cytoplasmic antibodies) markers. J Clin Investig. 1993;92:1080–1084. doi: 10.1172/JCI116613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang H-Y, Vora K A, Targan S R, Toyoda H, Beaudet A, Rotter J I. Intercellular adhesion molecule 1 gene association with immunologic subsets of inflammatory bowel disease. Gastroenterology. 1995;109:440–446. doi: 10.1016/0016-5085(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 54.Yang P, Jarnerot G, Danielsson D, Tysk C, Lindberg E. P-ANCA in monozygotic twins with inflammatory bowel disease. Gut. 1995;36:887–890. doi: 10.1136/gut.36.6.887. [DOI] [PMC free article] [PubMed] [Google Scholar]