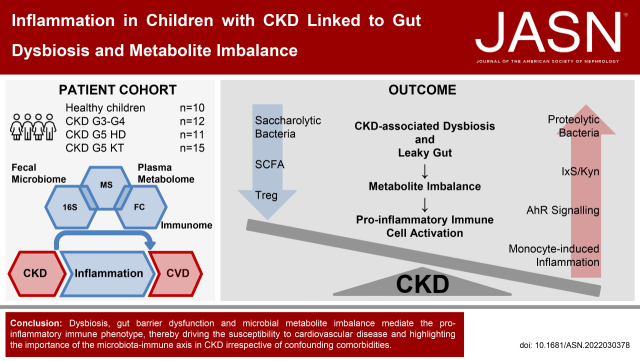
Significance Statement
Controlling chronic inflammatory processes, which are a major risk factor for cardiovascular disease, is of outstanding importance in CKD to reduce the rate of CKD-associated morbidity. This investigation connects microbial dysbiosis and bacterial metabolite imbalance to a proinflammatory immune cell signature. The fact that these dysbiosis-driven immunologic changes are already detectable in children with CKD, in whom comorbidities usually found in adults are absent, highlights the importance and specificity of CKD-related microbiota-immune interaction for chronic inflammation. Personalized dietary interventions and microbiota-targeted therapies may be a promising area of research to improve the prognosis of young and old patients with CKD.
Keywords: cardiovascular disease, children, chronic kidney disease, hypertension, immunology, pediatric nephrology, vascular disease, chronic inflammation, dysbiosis
Visual Abstract
Abstract
Background
CKD is characterized by a sustained proinflammatory response of the immune system, promoting hypertension and cardiovascular disease. The underlying mechanisms are incompletely understood but may be linked to gut dysbiosis. Dysbiosis has been described in adults with CKD; however, comorbidities limit CKD-specific conclusions.
Methods
We analyzed the fecal microbiome, metabolites, and immune phenotypes in 48 children (with normal kidney function, CKD stage G3–G4, G5 treated by hemodialysis [HD], or kidney transplantation) with a mean±SD age of 10.6±3.8 years.
Results
Serum TNF-α and sCD14 were stage-dependently elevated, indicating inflammation, gut barrier dysfunction, and endotoxemia. We observed compositional and functional alterations of the microbiome, including diminished production of short-chain fatty acids. Plasma metabolite analysis revealed a stage-dependent increase of tryptophan metabolites of bacterial origin. Serum from patients on HD activated the aryl hydrocarbon receptor and stimulated TNF-α production in monocytes, corresponding to a proinflammatory shift from classic to nonclassic and intermediate monocytes. Unsupervised analysis of T cells revealed a loss of mucosa-associated invariant T (MAIT) cells and regulatory T cell subtypes in patients on HD.
Conclusions
Gut barrier dysfunction and microbial metabolite imbalance apparently mediate the proinflammatory immune phenotype, thereby driving the susceptibility to cardiovascular disease. The data highlight the importance of the microbiota-immune axis in CKD, irrespective of confounding comorbidities.
Despite ongoing efforts to improve the treatment of patients with CKD, they still suffer from high morbidity and mortality, primarily due to cardiovascular disease (CVD).1 Besides known risk factors, such as arterial hypertension and proteinuria,1 recent studies suggest a crucial role of microbially produced metabolites in promoting inflammation2,3 and, as a consequence, progression of CKD and CVD.1,3,4
Longstanding evidence shows that an imbalance of the intestinal bacterial community with changes in its functional composition, termed dysbiosis, is common in adult patients with CKD.5,6 Beyond the influence of CKD, a variety of other factors, e.g., diet and drugs, are suspected to contribute to dysbiosis in CKD.7 These changes are paralleled by an altered bacterial metabolism of nutrients and a systemic accumulation of uremic toxins of bacterial origin, such as indoxyl sulfate (IxS).8 It is conceivable that dysbiosis and metabolite imbalance aggravate systemic inflammation, which could provide a potential mechanism for the high rate of premature CVD.3
Recently, we demonstrated a positive association between IxS and early CVD in children with CKD.9 In contrast to adults with CKD, children are less affected by risk factors, such as diabetes, obesity, and metabolic syndrome, but mainly suffer from congenital anomalies of the kidney and urinary tract.10 Thus, a pediatric cohort offers the unique opportunity to analyze the effect of CKD on microbiota-host interaction in early CVD development more specifically.
Our cohort includes pediatric patients with CKD, those on hemodialysis (HD), patients after kidney transplantation (KT), and age-matched healthy controls (HCs). We show, for the first time, a CKD-specific dataset of gut microbiome composition, altered microbial metabolism of nutrients, and the corresponding effect on inflammation and immune cell dysregulation in children with CKD. The fact that these dysbiosis-driven immunologic changes are already detectable in children with CKD highlights the potential of microbiota-targeted therapies to improve prognosis of patients with CKD across all ages.
Methods
Study Population
In this cross-sectional study, we recruited patients from the Department of Pediatric Gastroenterology, Nephrology, and Metabolic Diseases at Charité University Hospital in Berlin, Germany, between February 2018 and June 2018. Written informed consent was obtained from all participants and/or their parents before study entry. The study was approved by the local ethical review board (EA2/162/17). All procedures were performed in accordance with the ethical standards of the institutional and national research committees and the 1964 Helsinki Declaration and its later amendments or comparable standards.
Patients (aged 3–18 years) were enrolled in the following groups:
CKD group: patients with CKD stage G3–G4 and an eGFR of 15–60 ml/min per 1.73 m2
HD group: patients with CKD stage G5D, receiving maintenance HD, and enrolled at an earliest of 4 weeks after initiation of HD
KT group: patients after successful KT, at an earliest of 4 weeks after KT, without a history of rejection or chronic graft failure, and an eGFR of >60 ml/min per 1.73 m2
HC group: individuals with normal kidney function, admitted to the hospital or outpatient department for reasons other than kidney disease, who did not meet the exclusion criteria
We excluded all individuals with a body weight <15 kg, acute or chronic inflammatory diseases, fever, diabetes, chronic liver disease, inflammatory bowel disease, or other gastrointestinal disorders (constipation, diarrhea, short bowel syndrome). Patients with antibiotic prophylaxis or treatment within the 4 weeks before recruitment were also excluded.
Clinical Assessment, Biobanking, and Routine Laboratory Measurements
At the time of enrollment, we obtained baseline demographic (age, sex, diagnosis, body weight, and height) and clinical data from all patients. eGFR was calculated according to the bedside Schwartz formula on the basis of serum creatinine11; percentiles of weight and body mass index (BMI) were determined according to national references.12 Office systolic and diastolic BPs were documented as an average of three oscillometric measurements using local devices and normalized to national references.13 Arterial hypertension was defined as BP values above the 95th percentile.
Heparinized blood specimens were collected as part of routine laboratory sampling and used for the measurement of creatinine, urea, uric acid, phosphate, albumin, C-reactive protein, parathyroid hormone, and triglyceride levels. Serum, EDTA, and heparin plasma were stored at −80°C until further use. For the measurement of TNF-α, IL-2, IL-4, IL-6, IL-8, IL-1β, IL-10, IL-13, IL-12p70, and IFN-γ, plasma was analyzed using the Meso Scale Discovery V-PLEX Plus Proinflammatory Panel 1 (human) according to the manufacturer’s protocol. All samples were run on a single plate on a Meso Scale Discovery plate reader (model 1250). Zonulin-1 (Zo-1) and soluble CD14 (sCD14) were analyzed in patients’ serum using the human Zo-1 ELISA kit (Biomatik Corporation, Kitchener, Canada) and sCD14 Quantikine ELISA kit (R&D Systems, Minneapolis, MN) according to the manufacturers’ protocol.
PBMCs were isolated by density gradation using Pancoll (PAN-Biotech, Aidenbach, Germany) using standard protocols and stored in liquid nitrogen until use.
Stool specimens (2–4 g) were collected in tubes (catalog number 80.623.022; Sarstedt, Nümbrecht, Germany), stored for 24 hours at a maximum of 4–8°C, and transferred to the study center for freezing at −80°C until further use. Patients and parents were provided with detailed information about collection, storage, and transportation of stool specimens.
Generation of 16S Ribosomal RNA Amplicon Libraries and Sequencing
Stool DNA was isolated using the QIAamp Fast DNA Stool Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. The extracted DNA was used as a template to amplify the V3–V4 region of the bacterial 16S ribosomal RNA gene. Bacterial 16S amplicon libraries were sequenced using the dual-indexed, paired-end (2 × 300 bp) approach for the Illumina MiSeq platform, as recommended by the manufacturer. For a detailed description, please refer to the information provided in Supplemental Appendix 1.
Targeted Metabolomics
The analysis of plasma metabolites focused on tryptophan (TRP) metabolites, trimethylamine N-oxide (TMAO), and short-chain fatty acids (SCFA). For the TRP and TMAO analysis, liquid chromatography–mass spectrometry analysis was performed; SCFA were analyzed by gas chromatography–mass spectrometry. Details of the mass spectrometric methods are reported in Supplemental Appendix 1.
Monocyte Isolation and Serum Incubation
PBMCs were isolated from a healthy donors and monocytes were enriched using CD14 MicroBeads (Miltenyi Biotec) according to the manufacturer’s protocol. We plated 1 × 106 cells on a 96-well plate and incubated the cells for 24 hours at 37°C in standard culture medium containing 20% patient serum, 50 μM or 125 μM IxS (IxS potassium salt; Sigma-Aldrich, or 10 μM synthetic aryl-hydrocarbon receptor (AhR) antagonist (CH-223191; Sigma-Aldrich). Afterward, we measured TNF-α in the supernatant using a human TNF-α ELISA kit (Thermo Fisher) according to the manufacturer’s protocol.
Flow Cytometry Analysis
PBMCs were analyzed by multicolor flow cytometry. To avoid batch effects, all collected cells were thawed and measured at the same time. After thawing, cells were either stained immediately in FACS buffer or restimulated in a final volume of 200 μl RPMI 1640 (Sigma-Aldrich) supplemented with 10% FBS (Merck), 100 U/ml penicillin (Sigma-Aldrich), 100 mg/ml streptomycin (Sigma-Aldrich), 50 ng/ml PMA (Sigma-Aldrich), 250 ng/ml ionomycin (Sigma-Aldrich), and 1.3 μl/ml Golgistop (BD). Cells were fixed and permeabilized (eBioscience Foxp3/Transcription Factor Staining Buffer Set; Thermo Fisher) and labeled using mAbs (Supplemental Table 1). Cells were analyzed using a LSRFortessa flow cytometer and FACSDiva software (BD). Data analysis was performed with FlowJo.
For FlowSOM analysis, cells were manually gated (live single cells) on T cells (CD3+) and monocytic cells (CD3− CD4+) and exported for the respective analysis. This resulted in 1.1–6.2 × 105 monocytic cells per sample and 1.4–6.6 × 105 T cells per sample for further analysis. Markers used for FlowSOM analysis of the respective panel are shown in the corresponding heatmap. Data were clustered onto a 10 × 10 node square SOM, as implemented in the CATALYST package version 1.14.1 (wrapper for FlowSOM). Lower-resolution ConsensusClusterPlus metaclustering was then performed to reduce the data to eight metaclusters. Phenotypic relationships between individual clusters were explored using the UMAP algorithm for dimension reduction of 105 gated cells from all samples acquired, as implemented in the runDR and runUMAP functions from the CATALYST package (R version 4.0.3). Resource intensive computation was performed on the HPC for Research cluster of the Berlin Institute of Health.
Statistical Analysis
Microbiome and Metabolome Analysis
α-Diversities of microbial communities (Shannon diversity as computed by the RTK [0.93.1] tool, defined at the operational taxonomic unit [OTU] level), were compared between groups using the Kruskal–Wallis test. We assessed β-diversity using the Euclidean (metabolome) and Canberra (microbiome) dissimilarity index between samples, computed using the vegan package version 2.5-7. We also performed principal coordinates analysis using the vegan package version 2.5-7 (employing Euclidean and Canberra distance metrics as above). PERMANOVA was performed using the adonis function from the vegan package (2.5-7). We estimate differential abundance on the phylum and genus level between groups using the package DESeq2 version 1.30.1. The DESeq2 pipeline uses negative binomial distribution models to test for differential abundance between testing conditions. We ran the pipeline with normalized counts under default settings. P-values were adjusted according to a Benjamini–Hochberg (BH) false discovery rate (FDR) correction. A Q-value of <0.1 was considered statistically significant. We performed heatmap visualization in ggplot2 version 3.3.5 and visualized abundance plots using cowplot version 1.1.1.
For each pair of patient groups, features from the TRP analysis were compared using two-sided Mann–Whitney U (MWU) tests with effect sizes calculated as the Cliff δ metric per the R orddom package version 3.1. Effect sizes were taken as Spearman Rho. All significance estimates were adjusted for multiple tests using BH-FDR correction. To assess the effect of patient groups on TRP pathway metabolites (concentrations), multifactor ANOVAs were calculated per metabolite to account for multiple groups and potential confounders (including age, sex, ethnical background, underlying disease category, BMI, and eGFR).
We calculated the coabundance network of host, microbiome, and metabolome features from the dataset as a whole by assessing pairwise Spearman correlations and adjusted for multiple testing using BH-FDR correction, as implemented in the R psych package version 1.9.12. Edges for which absolute Rho was >0.3 and Q was <0.1 were visualized using the iGraph R package. Correlations were also separately assessed by stratifying for group effects using the R coin package version 1.3.1.
Flow Cytometry
For all FlowSOM clusters, we computed log2 fold changes (log2fc). We visualized cluster log2fc using the ggplot2 package version 3.3.5. For all subpopulations of regulatory T cells (Treg cells) and mucosal-associated invariant T (MAIT) cells, we calculated log2fc and assessed statistical significance using two-sided MWU tests between patients on HD and HCs using BH-FDR correction. A Q-value <0.1 was considered statistically significant. Data were visualized using the EnhancedVolcano package version 4.1.2.
Box Plots
We performed analysis and graphical representation using GraphPad Prism 9.3.1 (GraphPad Software, San Diego, CA). Box plots depict the median and interquartile range, with whiskers from the minimum to the maximum. Overlaid data points represent individual patients or measurements. Normality was assessed using Q-Q plots and Shapiro–Wilk test. For more than two groups, we performed one-way ANOVA with Tukey post hoc test or KW test with Dunn post hoc test, as appropriate. For two group comparisons, we performed the two-sided t test or two-sided MWU test, as appropriate.
For all analyses, a P value <0.05 (unadjusted or adjusted, as appropriate) and a Q value <0.1 was considered statistically significant.
Results
Childhood CKD Marked by Arterial Hypertension, Inflammation, and Leaky Gut
Ten healthy individuals and 38 patients were enrolled in the study (Supplemental Figure 1). Baseline characteristics are given in Table 1. The mean±SD age of participants was 10.6±3.8 years. Congenital anomalies of the kidney and urinary tract was the most prevalent disease. Patients treated with HD had a median (range) time on dialysis of 6 (3–29) months, with a median (range) residual diuresis of 50 (0–1800) ml/d and a mean±SD Kt/V of 1.56±0.27, indicating adequacy of HD treatment. Patients after KT had a stable graft function with a mean±SD eGFR of 78.8±19.4 ml/min per 1.73 m2. The median (range) time from transplantation was 48 (10–125) months.
Table 1.
Patients baseline characteristics
| Characteristics | HC | CKD G3–G4 | HD | KT |
|---|---|---|---|---|
| Patients, n | 10 | 12 | 11 | 15 |
| Age (yr), mean±SD | 12.1±4.2 | 8.3±2.3 | 13.6±3.2 | 9.1±2.9 |
| Female, n (%) | 7 (70) | 5 (42) | 3 (27) | 6 (40) |
| Diagnosis, n (%) | ||||
| CAKUT | N/A | 5 (42) | 5 (45) | 8 (53) |
| Tubulointerstitial | N/A | 2 (17) | 1 (9) | 3 (20) |
| Glomerulopathy | N/A | 4 (33) | 4 (36) | 3 (20) |
| Post-AKI | N/A | 1 (8) | 1 (9) | 1 (7) |
| Healthy | 10 (100) | N/A | N/A | N/A |
| Weight (percentile), mean±SD | 61±23 | 38±21 | 8±7 | 52±28 |
| BMI (percentile), mean±SD | 57±21 | 35±19 | 31±19 | 59±25 |
| eGFR (ml/min per 1.73 m2), mean±SD | 100.2±29.3 | 29.6±14.6 | 6.6±1.1 | 78.6±19.4 |
| Urea (mg/dl), mean±SD | 30±7 | 112±67 | 147±39 | 36±14 |
| Uric acid (mg/dl), mean±SD | 4.8±1.3 | 7.6±2.2 | 6.7±1.2 | 5.4±1.4 |
| Phosphate (mmol/L), mean±SD | 1.3±0.1 | 1.6±0.3 | 2.0±0.5 | 1.5±0.3 |
| Albumin (g/L), mean±SD | 47.7±5.3 | 43.8±13.0 | 38.0±4.8 | 40.0±3.4 |
| CrP (mg/L), mean±SD | 0.4±0.16 | 0.8±1.0 | 1.7±1.6 | 2.1±1.8 |
| PTH (pmol/L), mean±SD | N/A | 13.2±8.5 | 38.5±38.4 | 7.7±3.8 |
| Triglycerides (mg/dl), mean±SD | N/A | 166±75 | 186±54 | 112±57 |
| Proteinuria (mg/g), n (%) | N/A | N/A | ||
| <30 | 7 (58) | 15 (100) | ||
| 30–300 | 4 (33) | |||
| >300 | 1 (8) | |||
| Antihypertensive treatment, n (%) | 0 (0) | 10 (83) | 10 (91) | 8 (53) |
| Antihypertensive drugs, n | 0 | 1.8±1.5 | 2.4±1.9 | 0.8±0.9 |
| Arterial hypertension (BP >95th percentile), n (%) | N/A | 2 (17) | 6 (55) | 5 (33) |
Patients (N=48) were grouped into four categories (patients with CKD, those on HD, patients after KT, and HCs). BMI, body mass index; CAKUT, congenital anomalies of the kidney and urinary tract; CrP, C-reactive protein; N/A = not applicable; post-AKI, patients with CKD after AKI; PTH, parathyroid hormone.
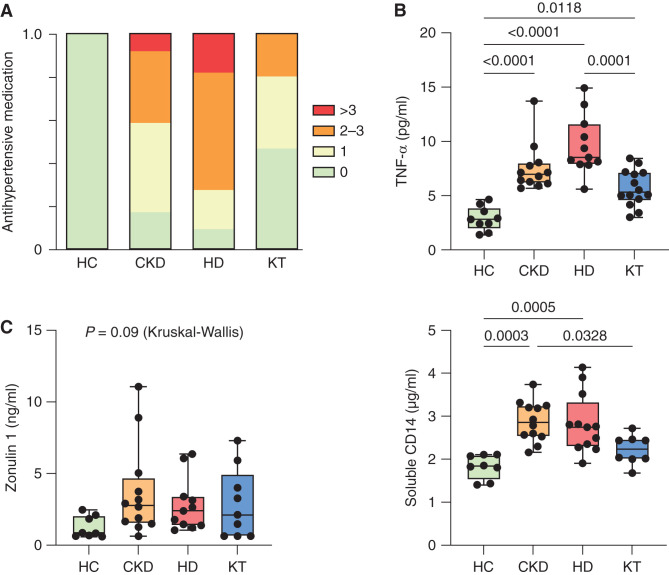
Both the CKD group (CKD stage G3–G4) and the HD group (CKD stage G5D) were hypertensive (83% and 91%, respectively) and received antihypertensive treatment (Figure 1A, Table 1). Six patients on HD (55%) exhibited hypertensive BPs despite treatment (Table 1). In the HD and CKD groups, serum levels of the proinflammatory cytokine TNF-α were increased (Figure 1B), whereas differences in other cytokines did not reach significance (Supplemental Table 2). We investigated serum levels of the tight junction protein Zo-1 and the LPS binding protein sCD14, which were elevated in those with CKD and those on HD compared with HCs and patients after KT (Figure 1C), indicating stage-dependent intestinal barrier dysfunction and CKD-associated endotoxemia. Thus, in the absence of classic cardiovascular risk factors other than hypertension, CKD in children is characterized by elevated serum markers of inflammation and leaky gut.
Figure 1.
Arterial hypertension and systemic inflammation are linked to impaired intestinal barrier function in pediatric CKD. (A) The number of antihypertensive drugs per individual (n=48 patients) is shown in patients with CKD (CKD G3–4), those with HD, patients after KT, and HCs. (B) Plasma TNF-α (n=46 patients) was analyzed by chemiluminescence immunoassay. (C) Gut barrier function was assessed using Zo-1 and sCD14 (n=40 patients) ELISA measurements in plasma. Data are shown as a box (median and interquartile range) and whiskers (minimum–maximum) with overlaid dot plot. P≤0.05 is shown, as measured by ordinary one-way ANOVA or Kruskal–Wallis test followed by Tukey or Dunn post hoc correction for multiple comparisons, as appropriate.
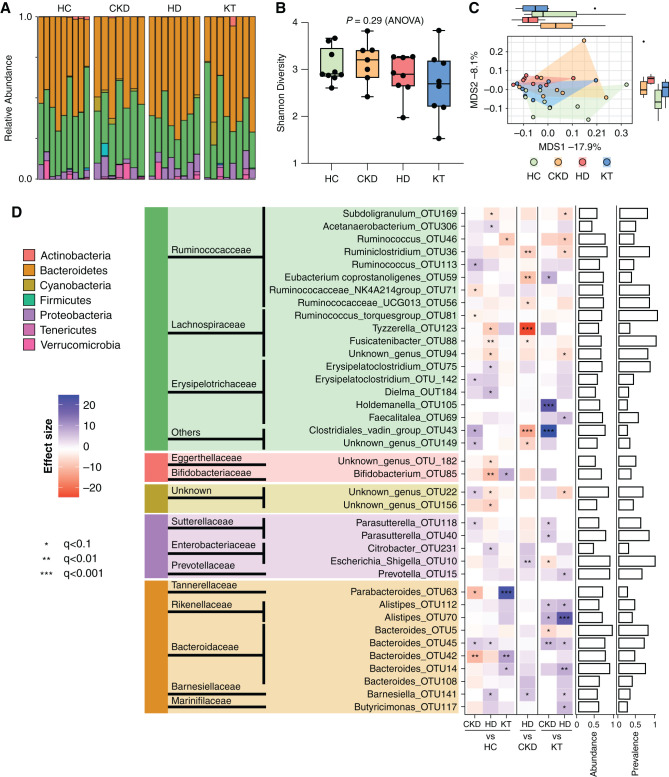
Microbiome Alterations in CKD
Because intestinal barrier dysfunction may be associated with dysbiosis, we next sought to analyze the taxonomic composition of the gut microbiome by 16S sequencing. We observed a high compositional variability at the phylum level (Figure 2A). Microbiome taxonomic richness and diversity showed no statistically significant changes (Figure 2B). However, analysis of β-diversity (Canberra distance) indicated significant differences in microbiome composition between groups, with the CKD and HD groups separating most clearly from the HCs (P=0.01; Figure 2C). Analysis of bacterial composition on the OTU level revealed significant alterations predominantly in patients on HD (Figure 2D). First, relative abundances of Firmicutes and Actinobacteria, such as OTU88 in the genus of Fusicatenibacter (belonging to the Lachnospiraceae family), Subdoligranulum_OTU169 (Ruminococcaceae), and Bifidobacterium_OTU85 (Bifidobacteriaceae) were significantly diminished in patients on HD compared with HCs. Second, we found an increase in relative abundance of Proteobacteria, such as Citrobacter_OTU231 (Enterobacteriaceae) in patients on HD, Parasutterella_OTU118 (Sutterellaceae) in patients with CKD, and several genera of Bacteroides (Bacteroidaceae) in CKD and HD groups compared with HCs and patients after KT.
Figure 2.
Taxonomic changes of the gut microbiome are most pronounced in HD patients. Analysis of gut microbiota from 16S ribosomal RNA sequencing in children (n=32) with CKD (CKD G3–4), patients with HD, patients after KT, and HCs. (A) Relative abundance on the phylum level of individuals according to their respective group. (B) α-Diversity as measured by Shannon diversity. Data are shown as a box (median and interquartile range) and whiskers (minimum–maximum) with overlaid dot plot. (C) β-Diversity assessment by principal coordinate analysis on the basis of Canberra distance (P=0.01 by PERMANOVA). (D) Analyses of group differences on the OTU level are shown as a heatmap, and their phylogenetic origin is visualized on the genus, family, and phylum level. Patient groups were tested against each other (pairwise). The heatmap shows significant changes in abundance using the DESeq2 version 1.30.1 package. Multiple groups of the same genus reported by Lotus due to a lack of coverage in available phylogenetic databases are marked by numbers. Bar charts (right) show abundance and prevalence; abundance is calculated as log(genus count)/log(maximum[genus count]); prevalence for each genus is calculated across the whole dataset. All significance estimates were adjusted for multiple tests using BH-FDR correction. MDS, multidimensional scaling. *q<0.1, **q<0.01, ***q<0.001.
Analysis on the phylum level showed significant changes in Tenericutes, Cyanobacteria, and Actinobacteria, as visualized in Supplemental Figure 2. In addition, patients on HD tend toward a higher abundance of Proteobacteria and lower abundance of Firmicutes. We constructed a phylogenetic tree to better visualize phylogenetic classification and changes among the groups (Supplemental Figure 3), again emphasizing the CKD- and HD-related reduction of OTUs from Lachnospiraceae and Ruminococcaceae and an increase in Enterobacteriaceae in patients on HD.
Taken together, the taxonomic microbiome changes in CKD are stage dependent, most pronounced in the HD group, and less pronounced after KT. The abundance of proteolytic bacteria (such as Citrobacter) increases in those on HD, whereas saccharolytic bacteria (such as Bifidobacterium) decrease.
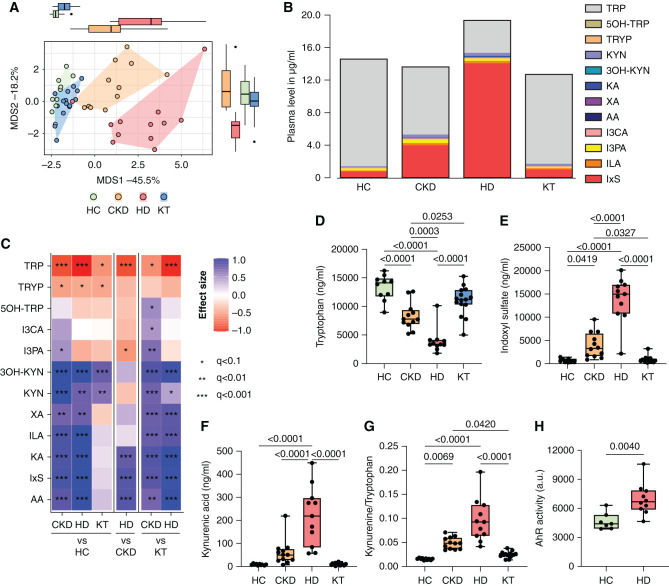
CKD Stage–Dependent Imbalance of Bacterial Metabolites
To investigate the functional effects of the alterations in microbiome composition on host physiology, we focused a plasma metabolite analysis on TRP metabolites and SCFA. Dietary TRP is a substrate for both cellular metabolism and bacterial proteolytic fermentation; the latter is a source of microbially produced uremic toxins. We found significant differences in the abundance of TRP metabolites in the CKD and HD groups compared with the HC and KT groups (Figure 3A). Patients with CKD and those on HD showed a large-scale shift from TRP to its indole and kynurenine (KYN) metabolites, which was predominantly driven by an increase of IxS (Figure 3B). We found a significant decrease in plasma TRP concentrations in patients with CKD and those on HD, whereas IxS and five KYN metabolites (KYN, kynurenic acid [KA], 3-hydroxykynurenine, anthranilic acid, and xanthurenic acid) were significantly elevated (Figure 3C). Individual values of TRP, IxS, and KA are shown across the different groups in Figure 3, D–F. The activation of the cellular KYN pathway is also indicated by the KYN/TRP ratio, which was significantly elevated in patients with CKD and those on HD (Figure 3G). In a multivariate ANOVA, only group and eGFR had an effect on several of the compounds (Supplemental Table 3), confirming their accumulation with declining eGFR (for TRP metabolite transitions please refer to Supplemental Table 4). Because indole and KYN metabolites have been shown to activate the AhR, we measured the AhR-activating potential of serum from the HD and HC groups in vitro. Serum from patients on HD induced significantly increased AhR activity compared with HCs (Figure 3H). Similarly, IxS activates the AhR in a dose-dependent manner (Supplemental Figure 4).
Figure 3.
Stage-dependent activation of plasma TRP metabolism activates the AhR. TRP and its metabolites were measured in plasma of children at different stages of CKD compared with HCs (n=48). (A) Multivariate analysis (principal coordinate analysis) of all measured metabolites discriminates between patients with CKD G3–4, patients on HD, patients after KT, and HCs. (B) Cumulative load of TRP and its metabolites. (C) Univariate analysis, depicted as a heatmap, shows effect sizes (Cliff δ) for each pair of patient groups. Colors denote the effect directions (blue, positive; red, negative) and magnitudes (the darker the color, the stronger the magnitude); asterisks represent the association significance. Statistical significance was assessed by MWU test and BH-FDR correction. Group differences of (D) TRP, (E) IxS, and (F) kynurenic acid (KA) were further visualized in box plots. (G) The KYN/TRP ratio indicates the activity of TRP degradation to KYN metabolites. (H) The activity of the AhR was analyzed using a transfected reporter cell line after 48 hours of incubation with serum of HCs (n=7) and patients on HD (n=10). P≤0.05 is shown, as measured by ordinary one-way ANOVA or Kruskal–Wallis test and adjusted by post hoc Tukey or Dunn correction for multiple testing (D–G) or by t test (H). Data are shown as a box (median and interquartile range) and whiskers (minimum–maximum) with overlaid dot plot. AA, anthranilic acid; I3CA, indole-3-carboxyaldehyde; ILA, indole lactate; MDS, multidimensional scaling; I3PA, indole-3-propionic acid; 3OH-KYN, 3-hydroxykynurenine; 5OH-TRP, 5-hydroxytryptophan; TRYP, tryptamin; XA, xanthurenic acid. *q<0.1, **q<0.01, ***q<0.001.
To functionally validate our findings of a decreased abundance of saccharolytic microbes further, we measured the abundance of a central enzyme for bacterial SCFA production by quantitative PCR in feces. The abundance of butyryl-CoA-dehydrogenase was lower in patients on HD relative to the other patient groups (Supplemental Figure 5A). Thus, the observed stage-dependent taxonomic microbiome changes result in an altered production potential of certain bacterial metabolites. Next, we analyzed serum SCFA levels, which showed a reduction of acetate, propionate, and isobutyrate, but not butyrate, in patients on HD compared with HCs (Supplemental Figure 5B).
In addition, we measured TMAO as a well-described toxin of microbial origin14 that accumulates in CKD and potentially drives CVD progression.14,15 TMAO was stage-dependently increased with the highest levels observed in patients on HD. Again, TMAO levels were almost at normal levels in the KT group (Supplemental Figure 6).
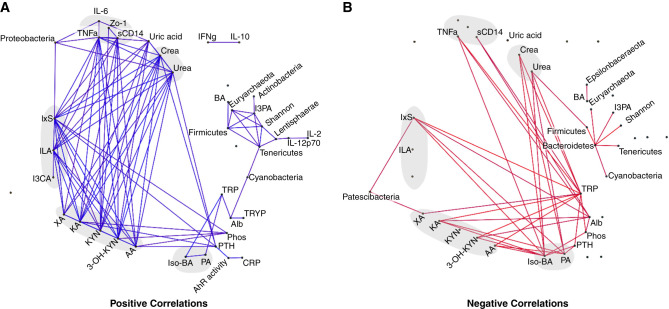
To comprehensively visualize the relationship between clinical, microbial, and metabolomic parameters, we performed a correlation network analysis (Figure 4, Supplemental Appendix 2). Here, TNF-α correlated positively with sCD14, Proteobacteria, IxS, several KYN metabolites, and biomarkers of kidney function (Figure 4A, positive correlations). Conversely, the SCFA propionate and isobutyrate correlated inversely with TNF-α, IxS, KYN metabolites, and kidney function (Figure 4B, negative correlations). Firmicutes correlated positively with microbial diversity, butyrate, and indole-3-propionic acid. These associations support our assumption of kidney function as a catalyst of gut bacteria–driven inflammation.
Figure 4.
Kidney function correlates with markers of gut bacteria-driven inflammation. Laboratory parameters, TRP metabolites (n=48), cytokines (n=46), and taxonomic data (n=32) were associated using pairwise Spearman correlations and adjusted for multiple testing using BH-FDR correction. Edges for which absolute Rho>0.3 and Q<0.1 are visualized. For better visualization, eGFR was removed because creatinine and urea convey similar information. (A) Positive correlations. (B) Negative correlations. AA, anthranilic acid; Alb, albumin; BA, butyric acid; Crea, creatinine; CRP, C-reactive protein; I3CA, indole-3-carboxyaldehyde; ILA, indole lactate; I3PA, indole-3-propionic acid; Iso-BA, isobutyric acid; 3-OH-KYN, 3-hydroxykynurenine; PA, propionic acid; phos, phosphate; TRYP, tryptamin; XA, xanthurenic acid.
Monocyte Subsets Contribute to the Proinflammatory Phenotype in CKD
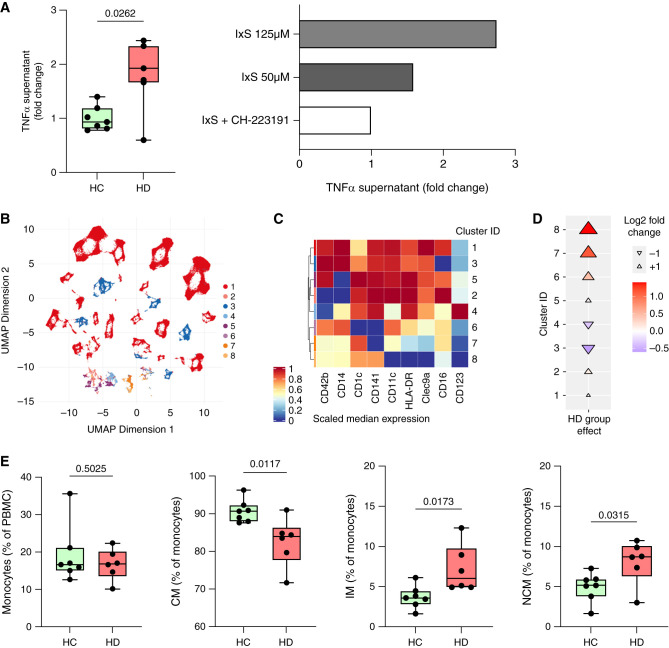
The AhR is expressed in various immune cells, including myeloid cells, and is known to modulate their function.16 The accumulation of AhR ligands in CKD contributes to immune cell activation. Therefore, we isolated monocytes from healthy donors and incubated them with serum from patients on HD or HCs. Monocytes incubated with HD serum showed a significantly increased production of TNF-α (Figure 5A). Similarly, incubation of isolated monocytes with IxS dose-dependently increased TNF-α production, which could be reversed by coincubation with the synthetic AhR antagonist CH-223191, highlighting the importance of AhR-mediated immune activation in CKD (Figure 5A).
Figure 5.
Monocyte subtypes promote inflammation in CKD. Monocytes isolated from healthy donors were incubated with serum from patients on HD (n=7) and HCs (n=7). Monocytes were incubated with IxS in presence or absence of the AhR antagonist CH-223191 (10 µM). (A) TNF-α was measured in the culture supernatant after 24-hour incubation using ELISA. PBMCs were isolated from patients on HD (n=6) and HCs (n=7) for surface staining and multicolor flow cytometry was performed. (B) Unsupervised clustering by FlowSOM revealed eight different cell clusters characterized by (B) the differential expression of nine surface markers describing myeloid and dendritic cells. (D) Cuneiform plots depict the log2fc for these clusters between patients on HD and HCs (indicated by color, size, and directionality of the triangles). (E) Classic hierarchic gating of total, classic (CM; CD16+), nonclassic (NCM; CD14+), and intermediate (IM; CD14+CD16+) monocytes is shown. UMAP, Uniform Manifold Approximation and Projection.
To get a broader overview of changes in relevant immune cell populations in CKD, we recollected PBMCs from seven HCs and six patients on HD for immunophenotyping by flow cytometry. Unsupervised FlowSOM analysis17 of our monocyte and dendritic cell targeting flow panel (Figure 5B; Supplemental Table 1 for antibodies used, Supplemental Figure 7 for gating strategy) showed phenotypic alterations, with monocyte subtypes (cluster 3, Figure 5C) being decreased (Figure 5D) and dendritic cells (cluster 7, Figure 5C) being increased (Figure 5D) in patients on HD. Using classic hierarchic gating, we observed similar abundances of total monocytes (identified by HLA-DR, CD14, and CD1618), but a significant shift from classic (CD14+CD16−) toward nonclassic (CD14−CD16+) and intermediate (CD14+CD16+) monocytes in HD (Figure 5E), the latter two being known for their potent production of TNF-α.19 Hierarchic gating within the dendritic cell population revealed no significant differences of dendritic cell subtypes (Supplemental Figure 8). Thus, patients on HD are characterized by alterations within myeloid cells, showing a shift from classic to proinflammatory intermediate and nonclassic monocytes.
Proinflammatory T Cell Subsets in CKD
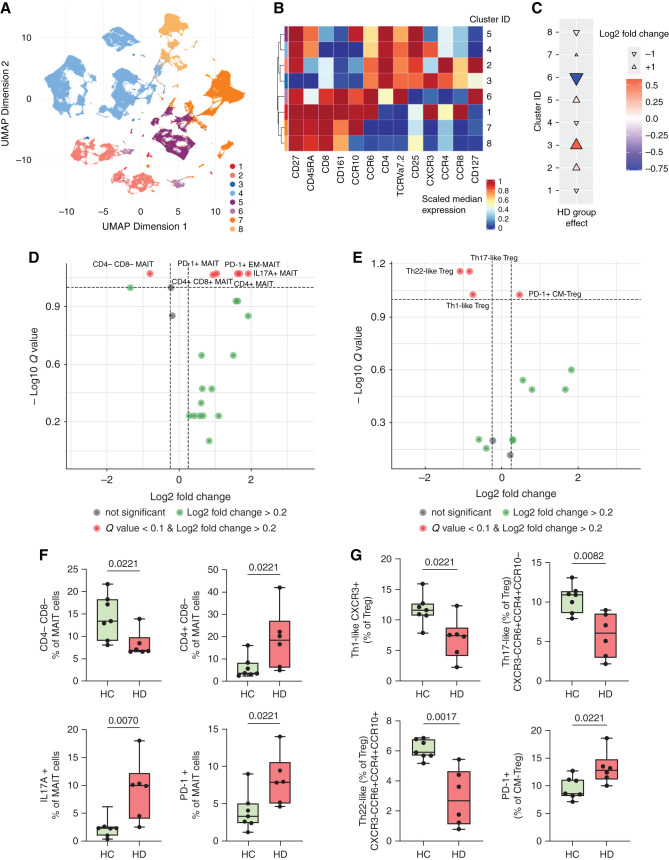
Because SCFA and TRP metabolites are known to modulate T cell differentiation and function, we performed FlowSOM analysis with a T cell targeting panel (Figure 6A, Supplemental Figure 7, Supplemental Table 1, T surface panel). MAIT cells and a subpopulation of Treg cells (clusters 6 and 3, respectively; Figure 6B) exhibited the largest effects between patients on HD and HCs (Figure 6C). We confirmed this finding by classic hierarchic gating, in which circulating MAIT cells (CD3+CD161+TCRVα7.2+) were decreased in those on HD (Supplemental Figure 9A). Next, we performed a more detailed analysis of MAIT cell subpopulations. After adjusting for multiple testing, we found a decrease in CD4−CD8− MAIT cells, whereas CD4+CD8− and CD4+CD8+ cells were enriched in those on HD (Figure 6, D and F). Moreover, MAIT cells of patients on HD displayed an effector memory phenotype (CD45RA−, CD62L−) and expressed more PD-1, indicative of increased activation. Importantly, upon in vitro restimulation, MAIT cells of patients on HD secreted significantly more IL-17A (Figure 6, D and F).
Figure 6.
HD patients display proinflammatory phenotypes in mucosa-associated invariant T cells and regulatory T cells. (A) Unsupervised clustering of patients on HD (n=6) and HCs (n=7) by FlowSOM revealed eight T cell clusters on the basis of (B) the differential expression of surface marker. (C) Cuneiform plots showing the log2fc for these clusters between patients on HD and HCs (indicated by color, size, and directionality of the triangles). Volcano plot of (D) MAIT and (E) Treg subpopulations by hierarchic gating. The y axis indicates Q value by MWU test and BH-FDR correction; x axis indicates log2fc between patients on HD and HCs. Significantly altered subpopulations are depicted as box (median and interquartile range) and whiskers (minimum–maximum) with overlaid dot plots for (F) MAIT cells and (G) Treg cells. For (F) and (G), P≤0.05 is shown, as measured by t test or MWU test as appropriate. UMAP, Uniform Manifold Approximation and Projection.
In addition, we identified changes within a CD4+CD25+CD127− cell cluster (cluster 3, Figure 6, A–C), indicating alterations in Treg cell subpopulations. Total Treg cell proportions were not altered between HCs and patients on HD (Supplemental Figure 9B). However, because different subpopulations with partly different functions are known to exist within the total Treg cell population, we extended the Treg cell characterization (Supplemental Table 1, T surface and T activation).20 Adjusted for multiple testing, several Treg cell subpopulations were significantly changed in patients on HD (Figure 6E). In particular, Treg cells from patients on HD that had a central-memory phenotype (CD45RA−, CD62L+) showed a higher expression of PD-1, indicating an activation state. Th1-like (CXCR3+), Th17-like (CXCR3−CCR6+CCR4+CCR10−), and Th22-like (CXCR3−CCR6+CCR4+CCR10+) Treg cells were significantly diminished in patients on HD compared with HCs (Figure 6E, G). Taken together, we observed significant alterations in T cell subtypes, namely MAIT cells and Treg, which play an important role in mucosal immunity and inflammation.
Discussion
Inflammation is a hallmark of CKD, detectable even at a young age, and is particularly prognosis determining. In children with CKD, we show that intestinal barrier dysfunction and dysbiosis are associated with a systemic bacterial metabolite imbalance that contributes to the proinflammatory phenotype of several immune cells, including monocytes and T cells. We also provide novel insight into the microbiota-immune crosstalk mediated by TRP metabolites and SCFA. Thus, we demonstrate a stage-dependent aberration of the microbiome-immune axis, which is both a contributor to inflammation and a potential target for anti-inflammatory therapeutic strategies.
Early cardiovascular pathologies and complications in children with CKD have been described previously.21,22 The burden of these pathologies accumulates with age and causes a fatal increase of CKD-associated mortality.1 This is remarkable given that CKD occurs in children in the virtual absence of diseases that can cause both CKD and CVD, such as diabetes and metabolic syndrome. This is particularly relevant because traditional comorbidities in adults represent additional influences on the microbiota-immune interaction. Therefore, we can assume the microbiome and immune signatures presented here are influenced to a smaller degree by traditional comorbidities and represent specific signatures of CKD.
The cohort enrolled for this study exhibits a markedly increased risk for CVD, indicated by a high prevalence of arterial hypertension despite antihypertensive treatment. Inflammation is a main driver of CVD in CKD.23 We describe CKD stage–dependent elevated serum levels of TNF-α, whereas C-reactive protein levels were not significantly altered. For the first time, we demonstrate that children with CKD already exhibit considerable stage-dependent intestinal barrier dysfunction, indicated by increased serum levels of Zo-1 and sCD14. Zo-1 regulates intestinal permeability24 and is associated with impaired barrier function in various conditions, such as obesity, diabetes,25 and autoimmune diseases.26 Impaired barrier function permits the translocation of luminal LPS into the circulation. Consequently, we detected a CKD stage–dependent increase in serum levels of the LPS binding protein sCD14, suggesting LPS-induced inflammation.6,27,28 Elevated serum levels of sCD14 have been associated with an increased risk of CVD in two independent cohort studies.29,30 Intestinal barrier dysfunction develops along with, or as a consequence of, dysbiosis, presumably because of an imbalance of microbial metabolites, such as SCFA.31,32 Therefore, we analyzed the composition of the gut microbiome in our cohort. In line with studies of adult patients with CKD,5,8,33,34 we observed CKD stage–dependent compositional changes. This microbial signature was especially present in patients with HD, displaying an increase in proteolytic bacteria and a decrease in saccharolytic bacteria. So far, only one other study investigated the microbiome in children with CKD. Although the authors show partly comparable results on the phylogenetic family level, the increase of Enterobacteriaceae and the decrease of Bifidobacteriaceae in their cohort were most pronounced in patients on peritoneal dialysis.35 Their cohort was characterized by a wider age range and a wider range in terms of time on dialysis, whereas the lower range in our cohort may have had a favorable effect on statistical power. Because adults, unlike children, exhibit a variety of comorbidities whose presence can have an additional obscuring influence on the microbiome (e.g., diabetes,36 obesity,37 and fatty liver disease38), we assume the data presented here are less influenced by such comorbidities and, therefore, are more clearly associated with CKD.
The enrichment of Enterobacteriaceae in patients on HD affects the microbial metabolism of nutrients because these bacteria express tryptophanases, which metabolize TRP to indoles.34 Dietary TRP is metabolized both by somatic cells and by the intestinal microbiota to metabolites with various functions.39,40 We found TRP metabolites in the blood of patients with CKD increased in a stage-dependent manner, which was predominantly driven by the disproportionate increase of metabolites of bacterial origin. Therefore, dysbiosis and intestinal barrier dysfunction, as present in patients with CKD and those on HD in our cohort, are likely contributing to the accumulation of uremic solutes41 in addition to their reduced renal elimination.34,42 In our cohort, microbially derived IxS and indole lactate and host-derived KYN, KA, 3-hydroxykynurenine, xanthurenic acid, and anthranilic acid were significantly elevated in patients with CKD and those on HD, whereas serum levels of TRP were reduced. Both indole and KYN derivates are ligands of the AhR, thereby influencing innate and adaptive immune responses.16,39,40 Consequently, the AhR activation potential of serum from our patients on HD was increased, confirming previous findings in adults.43
Myeloid cells are known to be modulated by the AhR and are thus particularly affected by uremic TRP metabolites.16,39,40 As indicated by unsupervised clustering and confirmed by hierarchic gating, we demonstrated a pathologic shift from classic toward intermediate and nonclassic monocytes in patients on HD. This monocyte signature was previously associated with an increased risk of CVD.44,45 Most notably, we showed that isolated monocytes exhibited a higher production of TNF-α after incubation with serum from patients on HD compared with HCs, which was dependent on AhR function. Thus, the increased TNF-α concentrations in circulation are, at least partly, a consequence of the effect of the increased and mainly microbially produced TRP metabolites on the AhR of monocytes.
In line with the depletion of saccharolytic bacteria,46 we show lower systemic levels of the SCFA acetate, propionate, and isobutyrate. SCFA play a pivotal role in gut homeostasis and in the protection from CVD in vitro47 and in vivo.48 Because SCFA enhance the abundance and suppressive function of Treg cells in mice49 and humans,50 we further analyzed T cells. On the basis of the expression of chemokine receptors,20,51 we found a decrease of Th1-like, Th17-like and Th22-like Treg cells in patients on HD. The lower frequencies of these Treg cell subtypes might be explained by both lower peripheral induction, because of reduced availability of SCFA,49 and increased recruitment of Treg to sites of inflammation, e.g., atherosclerotic plaques.52 Moreover, the frequency of dysfunctional Treg cells with a central-memory phenotype expressing PD-1 was elevated in patients on HD. Circulating Treg cells expressing PD-1 are known to exhibit reduced suppressive function and molecular signatures of exhaustion.53 In light of the importance of chronic inflammation for CVD development, the diminished anti-inflammatory function because of a reduced abundance of Treg cell subtypes and increased abundance of dysfunctional, exhausted Treg is likely to play a role in CVD development.51,52
Interestingly, unsupervised FlowSOM analysis indicated that circulating MAIT cells were reduced in patients on HD; they expressed markers of exhaustion (PD-1) and produced higher amounts of IL-17A. This pattern has been described for autoimmune, inflammatory, and cardiometabolic diseases,54 such as obesity and diabetes.55 It is still a subject of discussion whether decreased MAIT cell abundance is mainly a consequence of migration to inflamed tissue or increased apoptosis.54 In animal models, MAIT cells can promote inflammation and dysbiosis, leading to metabolic dysfunction during obesity.54 A large cross-sectional analysis in patients with cardiometabolic diseases highlighted the positive association between decreased MAIT cell abundances and CVD risk.56 A reduction of MAIT cells has been described in CKD of predominantly diabetic cause, albeit without functional cytokine expression as shown here.57 Taken together, Treg cells and MAIT cells are two important cell populations, which are known to be regulated by the microbiota, that are phenotypically altered in patients on HD, further emphasizing the importance of the microbiota-immune axis in CKD.
Notably, patients after KT still showed elevated serum levels of TNF-α compared with HCs, albeit lower levels than in patients with CKD and those on HD. Patients after KT showed no differences in markers of intestinal barrier function and only slight alterations in the microbiota composition. In contrast, adult patients after KT exhibited a significant decrease in α-diversity and an increase of Proteobacteria, which is partly driven by the use of immunosuppressive drugs.58 In a murine KT model, mice after allograft transplantation exhibited dysbiosis even in the absence of immunosuppression.59 Because gut dysbiosis and impaired microbial production of SCFA seem to play a role in allograft rejection as well,59 the absence of dysbiosis in children after KT might be attributable to a more favorable transplant outcome in children compared with adults.
There are of course limitations to our study. Although our study included approximately 10%–15% of the German pediatric CKD population treated with HD,10 our conclusions are made on the basis of a limited number of patients. Therefore, we cannot meaningfully deconfound for medication effects, but instead provide information on prescribed medications to facilitate the use of our study for meta-analyses in the future (Supplemental Appendix 3). Our study was not designed to investigate clinical end points because a follow-up until adulthood would be necessary. Furthermore, our results indicate a high cardiovascular risk in our pediatric cohort, although cardiovascular phenotyping of the cohort is limited to office BP measurements. Deeper cardiovascular phenotyping and data integration of multiple omics layers should be performed in future studies.
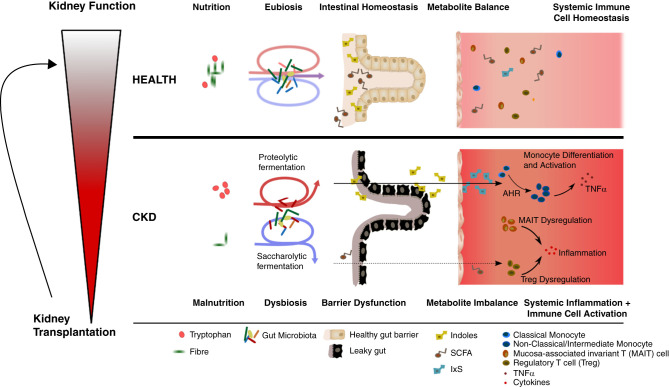
In conclusion, this study is the first to show CKD stage–dependent alterations of the microbiota-metabolite-immune axis in children with CKD (Figure 7). Our data demonstrate alterations at all levels of this pivotal axis. In this context, TRP metabolites act as a mechanistic link between the microbiota and the immune system, contributing to a proinflammatory phenotype in an AhR-dependent manner—even at a young age. SCFA deficiency may further exacerbate CKD-associated chronic inflammation and intestinal barrier dysfunction. These data provide strong evidence that the microbiota is an important stimulus for persistent inflammation. Restoring intestinal eubiosis could favorably influence the proinflammatory sequelae. Therefore, the microbiota appears to be a promising target of future therapeutic strategies aimed at sustained containment of inflammation to prevent chronic sequelae, such as CVD and premature mortality in CKD.
Figure 7.
Gut microbiome - host interaction promotes chronic inflammation in children with CKD. Pediatric patients with CKD stage dependently develop a leaky gut barrier and systemic imbalance of microbiome-derived metabolites. Patients with CKD exhibit a reduction in SCFA and an increase of indole metabolites of bacterial origin due to alterations of the taxonomic composition of the gut microbiome and nutritional alterations. These changes are associated with increased serum TNF-α levels, AhR-dependent secretion of TNF-α from monocytes, and a shift from classic to intermediate and nonclassic monocytes. Additionally, patients with CKD show a dysregulation of MAIT and Treg cell subsets. KT leads to a partial normalization of these dysregulated features.
Disclosures
P. Bufler reports having consultancy agreements with AbbVie, Albireo, Alexion, Amgen, Mirum, Nestlé Nutrition Institute, Nutricia GmbH, Orphalan, and Univar; and serving in an advisory or leadership role for Albireo, Amgen, Mirum, Orphalan, and Promethera. K.-U. Eckardt reports having consultancy agreements with Akebia, AstraZeneca, Bayer, Boehringer Ingelheim, Genzyme, Novartis, Otsuka, Travere, and Vifor; receiving honoraria from Akebia, AstraZeneca, Bayer, Boehringer Ingelheim, Genzyme, Novartis, Otsuka, Travere, and Vifor; receiving research funding from Amgen, AstraZeneca, Bayer, Evotec, Fresenius, Genzyme, Shire, and Vifor; and serving in an advisory or leadership role for Kidney International and The BMJ (on the editorial boards). S.K. Forslund reports receiving research funding from Sanprobi and Vertex. J.A. Kirwan reports having consultancy agreements with Centogene GmbH; serving as a board member of German Society for Metabolomic Research DGMet (unpaid position), a central committee member of Metabolomics Quality Assurance and Quality Control Consortium (unpaid position), and on the NAKO Metabolomics Advisory Group (unpaid position); and serving as a member of the Precision Medicine Working Group of the Metabolomics Society (unpaid position). A. Maifeld reports having consultancy agreements with Charité Research Organisation GmbH, Berlin, Germany. All remaining authors have nothing to disclose.
Funding
This work was supported by the Peter Stiftung für die Nierenwissenschaft grant 125833 and the Corona Foundation in the German Stifterverband grant S199/10080/2019. This work was also supported by local resources of the participating institutions. J. Holle was supported by German Society of Pediatric Nephrology. J. Holle, M.I. Wimmer, S.K. Forslund, and U. Löber were supported by the German Center for Cardiovascular Research, Partner Site Berlin. U. Löber was supported by the German Federal Ministry of Education and Research (EMBARK) grant 01KI1909B under the framework of Joint Programming Initiative on Antimicrobial Resistance (EMBARK) grant JPIAMR2019-109. S. Geisberger was supported by the Bundesministerium für Bildung und Forschung project Multimodal Clinical Mass Spectrometry to Target Treatment Resistance. N. Wilck was supported by the European Research Council under the European Union’s Horizon 2020 research and innovation program grant 852796 and by the Corona Foundation in the German Stifterverband. N. Wilck and S.K. Forslund were supported by the Deutsche Forschungsgemeinschaft (German Research Foundation) for CRC 1365 (project 394046635) and CRC 1470 (project 437531118).
Supplementary Material
Acknowledgments
The authors thank Gudrun Koch, Jana Czychi, Gabriele N’diaye, and Kerstin Sommer for their technical assistance and Nadine Unterwalder for her kind support in analyzing serum cytokine levels.
The sponsors had no involvement in study design, data collection, analysis and interpretation of data, or in the decision to submit this article for publication.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Author Contributions
H. Anandakumar, H. Bartolomaeus, O. Drechsel, J. Holle, M. Kuhring, U. Löber, N. Wilck, and M.I. Wimmer were responsible for visualization; H. Anandakumar, T.U.P. Bartolomaeus, U. Brüning, O. Drechsel, S.K. Forslund, U. Löber, and M.I. Wimmer were responsible for software; H. Bartolomaeus, T.U.P. Bartolomaeus, O. Drechsel, S.K. Forslund, J. Holle, M. Kuhring, U. Löber, and A. Thürmer were responsible for formal analysis; H. Bartolomaeus, F. Behrens, J. Holle, and N. Wilck wrote the original draft; H. Bartolomaeus, U. Brüning, S. Geisberger, J. Holle, S. Kempa, J.A. Kirwan, B. Kleuser, A. Maifeld, V. McParland, F. Schumacher, A. Thürmer, and D.L. Vu were responsible for methodology; H. Bartolomaeus, D. Engler, S. Geisberger, J. Holle, S. Kempa, J.A. Kirwan, S. Kitschke, L.D. Kuhrt, A. Maifeld, V. McParland, A. Thürmer, and D.L. Vu were responsible for investigation; H. Bartolomaeus, D. Engler, J. Holle, S. Kitschke, L.D. Kuhrt, U. Löber, A. Thürmer, and N. Wilck were responsible for data curation; H. Bartolomaeus, J. Holle, D. Mueller, and N. Wilck conceptualized the study; F. Behrens, U. Brüning, O. Drechsel, S.K. Forslund, J.A. Kirwan, B. Kleuser, M. Kuhring, U. Löber, A. Thürmer, and N. Wilck were responsible for validation; P. Bufler, K.-U. Eckardt, J. Holle, J.A. Kirwan, U. Querfeld, and N. Wilck were responsible for resources; K.-U. Eckardt, S.K. Forslund, D. Mueller, U. Querfeld, and N. Wilck provided supervision; J. Holle, D. Mueller, and N. Wilck were responsible for project administration; D. Mueller and N. Wilck were responsible for funding acquisition; and all authors reviewed and edited the manuscript.
Data Sharing Statement
Sequencing data are deposited in the European Nucleotide Archive (PRJEB50493). Lotus 16S export is available at https://github.com/Theda-sys/Pediatric_CKD_Cohort_Holle_2022. Metabolomics data are accessible via Metabolights: www.ebi.ac.uk/metabolights/MTBLS5066.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2022030378/-/DCSupplemental.
Supplemental Appendix 1. Supplemental methods.
Supplemental Appendix 2. Correlations between microbiome, metabolome and clinical features (Q values and Spearman’s rho).
Supplemental Appendix 3. Medication intake of study participants.
Supplemental Table 1. Antibodies used for flow cytometry.
Supplemental Table 2. Markers of inflammation in children with CKD and healthy individuals at study enrollment.
Supplemental Table 3. Multivariable ANOVAs on possible confounders influencing the serum levels of tryptophan metabolites in children with chronic kidney disease.
Supplemental Table 4. Tryptophan metabolite transitions.
Supplemental Figure 1. Study description.
Supplemental Figure 2. Microbiome analysis on phylogenetic phylum level.
Supplemental Figure 3. Phylogenetic tree analysis of 16S amplicon sequencing.
Supplemental Figure 4. Activity of the aryl hydrocarbon receptor in response to different concentrations of indoxyl sulfate.
Supplemental Figure 5. Potential for short chain fatty acid (SCFA) production and its serum levels in children with chronic kidney disease.
Supplemental Figure 6. Systemic levels of TMAO in pediatric CKD.
Supplemental Figure 7. Gating strategy.
Supplemental Figure 8. Dendritic cell differentiation in human peripheral blood mononuclear cells.
Supplemental Figure 9. Hierarchical gating of mucosa-associated invariant T cells and regulatory T cells.
References
- 1.Gansevoort RT, Correa-Rotter R, Hemmelgarn BR, Jafar TH, Heerspink HJ, Mann JF, et al. : Chronic kidney disease and cardiovascular risk: Epidemiology, mechanisms, and prevention. Lancet 382: 339–352, 2013 [DOI] [PubMed] [Google Scholar]
- 2.Claro LM, Moreno-Amaral AN, Gadotti AC, Dolenga CJ, Nakao LS, Azevedo MLV, et al. : The impact of uremic toxicity induced inflammatory response on the cardiovascular burden in chronic kidney disease. Toxins (Basel) 10: 384, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Onal EM, Afsar B, Covic A, Vaziri ND, Kanbay M: Gut microbiota and inflammation in chronic kidney disease and their roles in the development of cardiovascular disease. Hypertens Res 42: 123–140, 2019 [DOI] [PubMed] [Google Scholar]
- 4.Vanholder R, Schepers E, Pletinck A, Nagler EV, Glorieux G: The uremic toxicity of indoxyl sulfate and p-cresyl sulfate: A systematic review. J Am Soc Nephrol 25: 1897–1907, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaziri ND, Wong J, Pahl M, Piceno YM, Yuan J, DeSantis TZ, et al. : Chronic kidney disease alters intestinal microbial flora. Kidney Int 83: 308–315, 2013 [DOI] [PubMed] [Google Scholar]
- 6.Poesen R, Ramezani A, Claes K, Augustijns P, Kuypers D, Barrows IR, et al. : Associations of soluble CD14 and endotoxin with mortality, cardiovascular disease, and progression of kidney disease among patients with CKD. Clin J Am Soc Nephrol 10: 1525–1533, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramezani A, Massy ZA, Meijers B, Evenepoel P, Vanholder R, Raj DS: Role of the gut microbiome in uremia: A potential therapeutic target. Am J Kidney Dis 67: 483–498, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Yang S, Li S, Zhao L, Hao Y, Qin J, et al. : Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents. Gut 69: 2131–2142, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holle J, Querfeld U, Kirchner M, Anninos A, Okun J, Thurn-Valsassina D, et al. : Indoxyl sulfate associates with cardiovascular phenotype in children with chronic kidney disease. Pediatr Nephrol 34: 2571–2582, 2019 [DOI] [PubMed] [Google Scholar]
- 10.Harambat J, van Stralen KJ, Kim JJ, Tizard EJ: Epidemiology of chronic kidney disease in children. Pediatr Nephrol 27: 363–373, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz GJ, Muñoz A, Schneider MF, Mak RH, Kaskel F, Warady BA, et al. : New equations to estimate GFR in children with CKD. J Am Soc Nephrol 20: 629–637, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosario AS, Kurth BM, Stolzenberg H, Ellert U, Neuhauser H: Body mass index percentiles for children and adolescents in Germany based on a nationally representative sample (KiGGS 2003–2006). Eur J Clin Nutr 64: 341–349, 2010 [DOI] [PubMed] [Google Scholar]
- 13.Lurbe E, Agabiti-Rosei E, Cruickshank JK, Dominiczak A, Erdine S, Hirth A, et al. : 2016 European Society of Hypertension guidelines for the management of high blood pressure in children and adolescents. J Hypertens 34: 1887–1920, 2016 [DOI] [PubMed] [Google Scholar]
- 14.Tang WH, Wang Z, Kennedy DJ, Wu Y, Buffa JA, Agatisa-Boyle B, et al. : Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res 116: 448–455, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Witkowski M, Weeks TL, Hazen SL: Gut microbiota and cardiovascular disease. Circ Res 127: 553–570, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothhammer V, Quintana FJ: The aryl hydrocarbon receptor: An environmental sensor integrating immune responses in health and disease. Nat Rev Immunol 19: 184–197, 2019 [DOI] [PubMed] [Google Scholar]
- 17.Quintelier K, Couckuyt A, Emmaneel A, Aerts J, Saeys Y, Van Gassen S: Analyzing high-dimensional cytometry data using FlowSOM. Nat Protoc 16: 3775–3801, 2021 [DOI] [PubMed] [Google Scholar]
- 18.Abeles RD, McPhail MJ, Sowter D, Antoniades CG, Vergis N, Vijay GK, et al. : CD14, CD16 and HLA-DR reliably identifies human monocytes and their subsets in the context of pathologically reduced HLA-DR expression by CD14(hi)/CD16(neg) monocytes: Expansion of CD14(hi)/CD16(pos) and contraction of CD14(lo)/CD16(pos) monocytes in acute liver failure. Cytometry A 81: 823–834, 2012 [DOI] [PubMed] [Google Scholar]
- 19.Wong KL, Tai JJ, Wong WC, Han H, Sem X, Yeap WH, et al. : Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood 118: e16–e31, 2011 [DOI] [PubMed] [Google Scholar]
- 20.Duhen T, Duhen R, Lanzavecchia A, Sallusto F, Campbell DJ: Functionally distinct subsets of human FOXP3+ Treg cells that phenotypically mirror effector Th cells. Blood 119: 4430–4440, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitsnefes MM: Cardiovascular disease in children with chronic kidney disease. J Am Soc Nephrol 23: 578–585, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaefer F, Doyon A, Azukaitis K, Bayazit A, Canpolat N, Duzova A, et al. ; 4C Study Consortium : Cardiovascular phenotypes in children with CKD: The 4C Study. Clin J Am Soc Nephrol 12: 19–28, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jankowski J, Floege J, Fliser D, Böhm M, Marx N: Cardiovascular disease in chronic kidney disease: Pathophysiological insights and therapeutic options. Circulation 143: 1157–1172, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fasano A: Zonulin and its regulation of intestinal barrier function: The biological door to inflammation, autoimmunity, and cancer. Physiol Rev 91: 151–175, 2011 [DOI] [PubMed] [Google Scholar]
- 25.Yuan JH, Xie QS, Chen GC, Huang CL, Yu T, Chen QK, et al. : Impaired intestinal barrier function in type 2 diabetic patients measured by serum LPS, Zonulin, and IFABP. J Diabetes Complications 35: 107766, 2021 [DOI] [PubMed] [Google Scholar]
- 26.Tajik N, Frech M, Schulz O, Schälter F, Lucas S, Azizov V, et al. : Targeting zonulin and intestinal epithelial barrier function to prevent onset of arthritis. Nat Commun 11: 1995, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schütt C, Schilling T, Grunwald U, Schönfeld W, Krüger C: Endotoxin-neutralizing capacity of soluble CD14. Res Immunol 143: 71–78, 1992 [DOI] [PubMed] [Google Scholar]
- 28.Zanoni I, Granucci F: Role of CD14 in host protection against infections and in metabolism regulation. Front Cell Infect Microbiol 3: 32, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stanislawski MA, Lange LA, Raffield LM, Zakai NA, Meyer M, Ferrier K, et al. : Soluble CD14 levels in the jackson heart study: Associations with cardiovascular disease risk and genetic variants. Arterioscler Thromb Vasc Biol 41: e369–e378, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson NC, Koh I, Reiner AP, Judd SE, Irvin MR, Howard G, et al. : Soluble CD14, ischemic stroke, and coronary heart disease risk in a prospective study: The REGARDS Cohort. J Am Heart Assoc 9: e014241, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiippala K, Jouhten H, Ronkainen A, Hartikainen A, Kainulainen V, Jalanka J, et al. : The potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients 10: 988, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geirnaert A, Calatayud M, Grootaert C, Laukens D, Devriese S, Smagghe G, et al. : Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci Rep 7: 11450, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hobby GP, Karaduta O, Dusio GF, Singh M, Zybailov BL, Arthur JM: Chronic kidney disease and the gut microbiome. Am J Physiol Renal Physiol 316: F1211–F1217, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gryp T, Huys GRB, Joossens M, Van Biesen W, Glorieux G, Vaneechoutte M: Isolation and quantification of uremic toxin precursor-generating gut bacteria in chronic kidney disease patients. Int J Mol Sci 21: 1986, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crespo-Salgado J, Vehaskari VM, Stewart T, Ferris M, Zhang Q, Wang G, et al. : Intestinal microbiota in pediatric patients with end stage renal disease: A Midwest Pediatric Nephrology Consortium study. Microbiome 4: 50, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. ; MetaHIT consortium : Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528: 262–266, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouter KE, van Raalte DH, Groen AK, Nieuwdorp M: Role of the gut microbiome in the pathogenesis of obesity and obesity-related metabolic dysfunction. Gastroenterology 152: 1671–1678, 2017 [DOI] [PubMed] [Google Scholar]
- 38.Chu H, Williams B, Schnabl B: Gut microbiota, fatty liver disease, and hepatocellular carcinoma. Liver Res 2: 43–51, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roager HM, Licht TR: Microbial tryptophan catabolites in health and disease. Nat Commun 9: 3294, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, et al. : A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551: 648–652, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaziri ND, Yuan J, Norris K: Role of urea in intestinal barrier dysfunction and disruption of epithelial tight junction in chronic kidney disease. Am J Nephrol 37: 1–6, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poesen R, Viaene L, Verbeke K, Claes K, Bammens B, Sprangers B, et al. : Renal clearance and intestinal generation of p-cresyl sulfate and indoxyl sulfate in CKD. Clin J Am Soc Nephrol 8: 1508–1514, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dou L, Poitevin S, Sallée M, Addi T, Gondouin B, McKay N, et al. : Aryl hydrocarbon receptor is activated in patients and mice with chronic kidney disease. Kidney Int 93: 986–999, 2018 [DOI] [PubMed] [Google Scholar]
- 44.Duni A, Vartholomatos G, Balafa O, Ikonomou M, Tseke P, Lakkas L, et al. : The association of circulating CD14++CD16+ monocytes, natural killer cells and regulatory T cells subpopulations with phenotypes of cardiovascular disease in a cohort of peritoneal dialysis patients. Front Med (Lausanne) 8: 724316, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heine GH, Ortiz A, Massy ZA, Lindholm B, Wiecek A, Martínez-Castelao A, et al. ; European Renal and Cardiovascular Medicine (EURECA-m) working group of the European Renal Association-European Dialysis and Transplant Association (ERA-EDTA) : Monocyte subpopulations and cardiovascular risk in chronic kidney disease. Nat Rev Nephrol 8: 362–369, 2012 [DOI] [PubMed] [Google Scholar]
- 46.Felizardo RJF, Watanabe IKM, Dardi P, Rossoni LV, Câmara NOS: The interplay among gut microbiota, hypertension and kidney diseases: The role of short-chain fatty acids. Pharmacol Res 141: 366–377, 2019 [DOI] [PubMed] [Google Scholar]
- 47.Bartolomaeus H, Balogh A, Yakoub M, Homann S, Markó L, Höges S, et al. : Short-chain fatty acid propionate protects from hypertensive cardiovascular damage. Circulation 139: 1407–1421, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsu CN, Lu PC, Hou CY, Tain YL: Blood pressure abnormalities associated with gut microbiota-derived short chain fatty acids in children with congenital anomalies of the kidney and urinary tract. J Clin Med 8: 1090, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. : Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504: 451–455, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haghikia A, Zimmermann F, Schumann P, Jasina A, Roessler J, Schmidt D, et al. : Propionate attenuates atherosclerosis by immune-dependent regulation of intestinal cholesterol metabolism. Eur Heart J 43: 518–533, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Halim L, Romano M, McGregor R, Correa I, Pavlidis P, Grageda N, et al. : An atlas of human regulatory T helper-like cells reveals features of Th2-like Tregs that support a tumorigenic environment. Cell Rep 20: 757–770, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saigusa R, Winkels H, Ley K: T cell subsets and functions in atherosclerosis. Nat Rev Cardiol 17: 387–401, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lowther DE, Goods BA, Lucca LE, Lerner BA, Raddassi K, van Dijk D, et al. : PD-1 marks dysfunctional regulatory T cells in malignant gliomas. JCI Insight 1: e85935, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toubal A, Kiaf B, Beaudoin L, Cagninacci L, Rhimi M, Fruchet B, et al. : Mucosal-associated invariant T cells promote inflammation and intestinal dysbiosis leading to metabolic dysfunction during obesity. Nat Commun 11: 3755, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magalhaes I, Pingris K, Poitou C, Bessoles S, Venteclef N, Kiaf B, et al. : Mucosal-associated invariant T cell alterations in obese and type 2 diabetic patients. J Clin Invest 125: 1752–1762, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Touch S, Assmann KE, Aron-Wisnewsky J, Marquet F, Rouault C, Fradet M, et al. : Mucosal-associated invariant T (MAIT) cells are depleted and prone to apoptosis in cardiometabolic disorders [published online ahead of print April 27, 2018]. FASEB J 10.1096/fj.201800052RR [DOI] [PubMed] [Google Scholar]
- 57.Juno JA, Waruk JLM, Wragg KM, Mesa C, Lopez C, Bueti J, et al. : Mucosal-associated invariant T cells are depleted and exhibit altered chemokine receptor expression and elevated granulocyte macrophage-colony stimulating factor production during end-stage renal disease. Front Immunol 9: 1076, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swarte JC, Douwes RM, Hu S, Vich Vila A, Eisenga MF, van Londen M, et al. : Characteristics and dysbiosis of the gut microbiome in renal transplant recipients. J Clin Med 9: 386, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu H, Singer J, Kwan TK, Loh YW, Wang C, Tan J, et al. : Gut microbial metabolites induce donor-specific tolerance of kidney allografts through induction of T regulatory cells by short-chain fatty acids. J Am Soc Nephrol 31: 1445–1461, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.