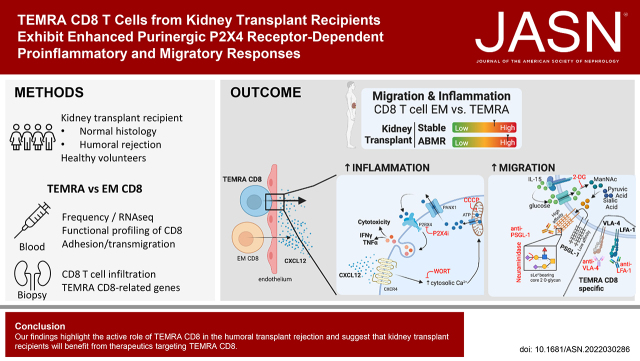
Significance Statement
The pathogenic role of terminally differentiated effector memory (TEMRA) CD8+ T cells has been implicated in kidney transplant failure. The authors showed that humoral rejection of kidney allografts is associated with an accumulation of cytolytic TEMRA CD8+ T cells in blood and in kidney graft biopsies. They demonstrated that TEMRA CD8+ T cells from kidney transplant recipients exhibit enhanced migratory properties compared with effector memory CD8+ T cells and that the chemokine CXCL12 not only promotes migration of TEMRA CD8+ T cells toward nonlymphoid organs but also triggers a purinergic P2X4 receptor–dependent proinflammatory response. They also found that agents aimed at potential TEMRA CD8+ T cell–specific targets inhibited the migration of TEMRA CD8+ T cells from kidney transplant recipients, suggesting a possible strategy in treating kidney transplant failure.
Keywords: cell adhesion, chemokine, lymphocytes, kidney transplantation, immunology, endothelium, chronic allograft rejection, cell activation, adhesion molecule, purinergic P2X4 receptors
Visual Abstract
Abstract
Background
The mechanisms regulating CD8+ T cell migration to nonlymphoid tissue during inflammation have not been fully elucidated, and the migratory properties of effector memory CD8+ T cells that re-express CD45RA (TEMRA CD8+ T cells) remain unclear, despite their roles in autoimmune diseases and allotransplant rejection.
Methods
We used single-cell proteomic profiling and functional testing of CD8+ T cell subsets to characterize their effector functions and migratory properties in healthy volunteers and kidney transplant recipients with stable or humoral rejection.
Results
We showed that humoral rejection of a kidney allograft is associated with an accumulation of cytolytic TEMRA CD8+ T cells in blood and kidney graft biopsies. TEMRA CD8+ T cells from kidney transplant recipients exhibited enhanced migratory properties compared with effector memory (EM) CD8+ T cells, with enhanced adhesion to activated endothelium and transmigration in response to the chemokine CXCL12. CXCL12 directly triggers a purinergic P2×4 receptor–dependent proinflammatory response of TEMRA CD8+ T cells from transplant recipients. The stimulation with IL-15 promotes the CXCL12-induced migration of TEMRA and EM CD8+ T cells and promotes the generation of functional PSGL1, which interacts with the cell adhesion molecule P-selectin and adhesion of these cells to activated endothelium. Although disruption of the interaction between functional PSGL1 and P-selectin prevents the adhesion and transmigration of both TEMRA and EM CD8+ T cells, targeting VLA-4 or LFA-1 (integrins involved in T cell migration) specifically inhibited the migration of TEMRA CD8+ T cells from kidney transplant recipients.
Conclusions
Our findings highlight the active role of TEMRA CD8+ T cells in humoral transplant rejection and suggest that kidney transplant recipients may benefit from therapeutics targeting these cells.
The recirculation of memory T cells is a central process in the establishment of efficient immunosurveillance involving the localization of memory CD8 T cells in secondary lymphoid organs and nonlymphoid tissues. Alteration of their migratory properties has been shown to lead to inefficient or exacerbated responses. Therefore, interference with recirculation signals is an appealing strategy in inflammatory diseases, and various treatments, such as anti-α4 integrin mAb (natalizumab) for multiple sclerosis,1 anti–LFA-1 (efalizumab) for psoriasis,2 or anti-α4/β7 mAb (vedolizumab) for patients with ulcerative colitis3 or active Crohn disease, have been assessed.4 Although many reports have been published, the mechanisms regulating the migration of CD8 T cells are only partially understood,5,6 and the migratory properties of effector memory (EM) re-expressing CD45RA (namely, terminally differentiated effector memory [TEMRA]) CD8 T cells have not been elucidated. The recruitment of memory CD8 T cells into nonlymphoid tissues is tightly regulated by modification of the vascular endothelium that, upon activation, expresses chemokines and adhesion molecules, such as P- and E-selectins, ICAM-1, VCAM-1, CXCL12, and CXCL8,5,7 and by the generation of functional ligands by memory CD8 T cells upon post-translational modifications, such as the transition of the inactive to active state of LFA-1 by inside-out/outside-in signals8 or core 2 O-linked glycosylation and the subsequent addition of sialyl Lewis x (sLex) onto core 2 O-glycan structures.9 Chemokines, such as CXCL12, have been shown to not only regulate T cell recruitment but also trigger the activation of human CD4 T cells through the autocrine stimulation of the purinergic receptor.10 Whether CXCL12 could also activate human memory CD8 T cell subsets remains unclear.
The pathogenic role of TEMRA CD8 T cells has been indicated in many pathologic settings, such as in lupus,11,12 ANCA-associated vasculitis,12,13 Sjogren syndrome,14 scleroderma,15 Susac syndrome,16 and kidney transplant failure.17,18 For example, an increase in TEMRA CD8 T cells in the periphery of kidney transplant recipients (KTs) is associated with a higher risk of kidney dysfunction, despite immunosuppression.17,18 We and others have shown that TEMRA CD8 T cells activation and proliferation are tightly regulated by IL-15.19,20 Upon activation, TEMRA CD8 T cells amplify the immune response by secreting high levels of IFN-γ and TNF-α, which activate the endothelium and upregulate CX3CL1.20 Direct comparative studies of the trafficking potential of human TEMRA and EM CD8 T cell subsets in the resting state (healthy volunteers [HVs]) or in the context of strong allostimulation (KTs) and analysis of the subsequent mechanisms that regulate their trafficking into nonlymphoid tissues have not been performed. Addressing this issue could help identify novel therapeutic strategies to prevent the migration of TEMRA CD8 T cells into grafts.
Here, by combining single-cell proteome profiling and functional assay of CD8 subsets, we found that humoral rejection was associated with an accumulation in the periphery and in the kidney graft of cytolytic TEMRA CD8 T cells. We found that the TEMRA CD8 T cells from the KTs exhibited enhanced migratory properties compared with the EM CD8 T cells, whereas no difference was found between the TEMRA and EM CD8 T cells from the HVs. Notably, the selective targeting of LFA-1 or VLA-4 was effective in preventing the migration of TEMRA CD8 T cells but not of EM CD8 T cells from the KTs. IL-15 potentiates the CXCL12-induced migration of TEMRA CD8 T cells and promotes the generation of functional PSGL1 by enhancing glycolysis-dependent sialic acid biosynthesis in these cells. Finally, we demonstrated that CXCL12 triggers a purinergic P2 × 4 receptor–dependent proinflammatory response of TEMRA CD8 T cells. These data show the potential of immunotherapy regulating the migration of TEMRA CD8 T cells to improve kidney allograft survival.
Methods
Blood Samples and Study Approval
PBMCs were separated from blood samples collected in EDTA tubes on a Ficoll gradient layer according to the manufacturer’s recommendations and were frozen in autologous serum with 10% DMSO. PBMCs were prospectively collected from HVs and KTs (Table 1) in the DIVAT biocollection (http://www.divat.fr/) and stored in the Biologic Resource Center of the Nantes University Hospital, France (BRIF: BB-0033–00040) and from patients with multiple sclerosis (Table 2). All donors were informed of the final use of their blood and signed a written informed consent form. The HVs were enrolled by the Etablissement Français du Sang (Nantes, France) within the context of a research contract. A convention was signed between our laboratory (CR2TI–Institut National de la Santé et de la Recherche Médicale UMR 1064) and the blood bank (Etablissement Français du Sang Pays de La Loire), and the approval of an ethical committee was thus not necessary. The University Hospital Ethical Committee and the Committee for the Protection of Patients from Biologic Risks approved the studies involving patients. The overall study design is shown in Supplemental Figure 1 with the distribution of the different clinical groups.
Table 1.
Description of the quantitative and qualitative characteristics of KTs (mean ± SD or n [percentage])
| Characteristics | Stable Patients, n=27 | mTOR Section, n=47 | ABMR Section, n=64 | ||
|---|---|---|---|---|---|
| With mTOR, n=9 | Without mTOR, n=38 | ABMR Biopsy, n=12 | Normal Biopsy, n=52 | ||
| At blood sampling | |||||
| Recipient age, yr | 54.20±12.74 | 57.44±15.00 | 56.87±13.39 | 58.33±11.72 | 58.44±10.82 |
| Donor age, yr | 49.20±13.44 | 58.00±15.72 | 58.95±15.21 | 58.33±12.47 | 58.83±12.50 |
| Time after-transplantation, yr | 5.21±3.40 | 0.97±0.11 | 1.03±0.06 | 1.03±0.08 | 1.03±0.07 |
| Creatinemia, μmol/L | 141±48 | 170±74 | 161±69 | 149±47 | 143±50 |
| Men recipients | 20 (80) | 7 (78) | 30 (79) | 7 (58) | 33 (63) |
| Men donors | 12 (48) | 5 (56) | 15 (39) | 2 (17) | 29 (56) |
| Incompatibility HLA-A, B, DR >4 | 16 (64) | 2 (22) | 14 (37) | 3 (25) | 11 (21) |
| Induction therapy | |||||
| Polyclonal Ab | 10 (40) | 1 (11) | 10 (26) | 5 (42) | 10 (19) |
| mAb | 15 (60) | 8 (89) | 28 (74) | 7 (58) | 40 (77) |
| Maintenance therapy | |||||
| MMF | 19 (76) | 4 (44) | 37 (97) | 12 (100) | 52 (100) |
| CNI | 23 (92) | 7 (78) | 38 (100) | 12 (100) | 52 (100) |
| mTOR inhibitor | 3 (12) | 9 (100) | 0 (0) | 0 (0) | 0 (0) |
| Corticotherapy | 6 (24) | 9 (100) | 27 (71) | 10 (83) | 40 (77) |
| CMV serology (donor/recipient) | |||||
| D−/R− | 6 (22) | 3 (33) | 31 (82) | 3 (25) | 34 (65) |
| D−/R+ | 5 (19) | 1 (11) | 7 (18) | 2 (17) | 18 (35) |
| D+/R− | 8 (30) | 1 (11) | 0 (0) | 4 (33) | 0 (0) |
| D+/R+ | 8 (30) | 4 (44) | 0 (0) | 3 (25) | 0 (0) |
ABMR, antibody-mediated rejection; MMF, mycophenolate mofetil; CNI, calcineurin inhibitors; CMV, cytomegalovirus; D, donor; R, recipient.
Table 2.
Description of the quantitative and qualitative characteristics of patients with multiple sclerosis (mean ± SD or n [percentage])
| Characteristics | n (%) or mean ± SD |
|---|---|
| Men/women | 2/9 |
| Age at MS diagnosis, yr | 31.09±7.48 |
| Age at blood sampling (M00), yr | 39.00±7.07 |
| Previous treatment | |
| IFN-β | 7 (64) |
| Glatiramer acetate | 6 (55) |
| Immunosuppressor | 2 (18) |
| Response to natalizumab at 1 yr | 11 (100) |
MS, multiple sclerosis; M00, month 0 (inclusion time).
Reagents
The antibodies used for cytometric analyses and key reagents are listed in Supplemental Tables 1 and 2.
CD8 T Cell Isolation and Subset Purification
PBMCs were thawed and rested overnight (O/N) in TexMacs medium. CD8 T cells were purified using the human REAlease CD8 MicroBead Kit. Naïve (CD45RA+CCR7+ or CD45RA+CD28+), EM (CD45RA−CCR7− or CD45RA−CD28+), and TEMRA (CD45RA+CCR7− or CD45RA+CD28−) CD3+CD8+ cells were sorted by FACS (FACSAria; BD Biosciences, Le Pont de Claix, France; purity >95%).
Expression of Cell Adhesion Molecules by Human CD8 T Cell Subsets
Purified CD8 T cells were cultured for 1 or 3 days in 96-well U-bottom plates at 37°C in 5% CO2. At the indicated times, CD8 T cell phenotyping was performed using cell surface markers (CD3, CD8, CD45RA, CD28, CD162, CD11a, and CD49d). Cells were acquired with a BD Celesta flow cytometer as for all of the flow cytometry analyses, and the analyses were performed using FlowJo, version 10.8.1 (BD Biosciences).
Expression of Cytotoxic Molecules by Human CD8 T Cell Subsets
A total of 2 × 106 frozen PBMCs were surface stained with specific antibodies for CD3, CD8, CD45RA, and CCR7, and after fixation and permeabilization, intracellular staining was performed using antibodies against Granzyme B (GZMb) and Perforin-1 (PERF-1). The Fixable Viability Dye 440UV was used to exclude dead cells from the analysis.
Redirected Killing Assay
P815 mastocytoma target cells were labeled with eFluor V450 Cell Proliferation Dye and Fixable Viability Dye 440UV, washed twice, and incubated with anti-CD3 (1 μg/ml) for 30 minutes at room temperature. After a wash step, P1815 was incubated with Fc block. Purified CD8 T cells were rested for 45 minutes in completed medium at 37°C and 5% CO2 and then incubated with anti-CD3–coated P815 cells at different effector-target ratios (1:1 and 1:5) in V-bottom plates for 4 hours at 37°C and 5% CO2. The cells were then extracellularly stained with anti-CD3 FITC mAb and intracellularly with anticaspase 3 PE. Cytolytic activity was assessed as the percentage of active caspase 3 among CD3− Cell Proliferation Dye eFluor V450+ Fixable Viability Dye 440UV− P815 cells.
CXCL12-Induced Activation of Human CD8 T Cell Subsets
Purified CD8 T cells were cultured for 4 hours in 96-well flat-bottom plates at 37°C in 5% CO2 and when indicated, treated with plate-bound anti-CD3 (2 μg/ml; produced in house), CXCL12 (100 nM), CCCP (10 μM), wortmannin (10 μM), and selective P2 × 1 (NF023; 10 μM), P2 × 4 (5-BDBD; 20 μM), or P2 × 7 (A438079; 20 μM) receptor inhibitors. Anti-CD107 mAb PE was added at culture initiation. After 4 hours of stimulation, the cells were stained for cell surface markers (CD3, CD8, CD45RA, CCR7, and CD69).
Calcium Flux
Purified CD8 T cells were stained with the cytosolic Ca2+ indicator Fura-2 AM probe (5 μM for 30 minutes at room temperature) and then resuspended in HBSS + CaCl2 (1 mM) + HEPES (10 mM) + BSA (1%). After an initial recording of 30 seconds, the cells were stimulated with CXCL12 (1 μg/ml), and acquisition was performed for an additional 90 seconds using 605/12 (404-nm laser) and 695/40 (488-nm laser) LSR II flow cytometry. Stimulation with ionomycin (500 ng/ml) was used as a positive control. The results are expressed as the ratio of fluorescence collected by the 605/12 and 695/40 detectors.
P-Selectin Binding
Purified CD8 T cells were incubated with IL-15 (10 ng/ml) at 37°C O/N as indicated. Recombinant human P-selectin IgG Fc chimeric protein (2 μg/ml) was incubated with cells diluted in 1% FBS/Dulbecco PBS containing Ca2+ and Mg2+ for 30 minutes at room temperature. The cells were then washed, and selectin binding was detected using FITC anti-human IgG Fc. CD8 T cell subsets were then identified according to the expression of CD45RA and CD28. When indicated, the cells were pretreated with neuraminidase (0.5 U/ml; 37°C for 20 minutes) or with 2-deoxy-d-glucose (2-DG; 50 μM; 4 hours at 37°C) before the detection of P-selectin binding.
Detection of Mannosamine Incorporation into the Sialic Acid Biosynthetic Pathway with “Click-IT” Chemistry
Purified CD8 T cells were incubated with 25 μM Click-IT ManNAz and when indicated, with IL-15 (10 ng/ml) or pretreated for 4 hours with 2-DG (50 μM). After 36 hours, the cells were fixed with 4% PFA or permeabilized using the BD Cytofix/Cytoperm Kit. Incorporation of ManNAz into the sialic acid biosynthetic pathway was quantified by flow cytometry using alkyne Alexa Fluor 488 and the Click-iT Cell Reaction Buffer Kit according to the manufacturer's instructions.
ATP Measurements
Freshly isolated CD8 T cells (3 × 105 cells) were stimulated for 20 minutes with CXCL12 (100 ng/ml) and immediately centrifuged at 4°C to stop the reaction. When indicated, the CD8 T cells were preincubated with a pannexin-1 (Panx1) inhibitor (10Panx, 100 μM) for 20 minutes. ATP concentrations in the cell supernatant were determined using the ApoSENSOR ATP Cell Viability Bioluminescence Assay Kit and a Tecan microplate reader.
Adhesion Model
Human dermal microvascular endothelial cells (HDMECs; 15 × 104 cells) activated for 24 hours with TNF-α (100 U/ml) were seeded O/N in 24-well flat-bottom plates at 37°C in endothelial cell growth medium–2 microvascular medium. Purified CD8 T cell subsets (1 × 105 cells) were added to the HDMECs. After 2 hours, nonadherent cells were discarded by two washes with PBS. The cells were then collected after trypsin-EDTA digestion, and the number of adherent CD8 T cell subsets was quantified using 123count eBeads Counting Beads and FACS analysis. When indicated, the CD8 T cell subsets were stimulated O/N with IL-15 (10 ng/ml) and then treated with neuraminidase (2 U/ml; 37°C for 30 minutes), anti-PSGL1 mAb (50 μg/ml; 37°C for 1 hour), or 2-DG (50 μM; 4 hours).
Transmigration Model
HDMECs (25 × 104 cells) activated for 24 hours with TNF-α (100 U/ml) were seeded O/N onto 2% gelatin-coated Transwell membrane inserts (3-μm pore polycarbonate membrane; Corning Life Science) in endothelial cell growth medium at 37°C. On the day of the assay, purified CD8 T cell subsets (5 × 105) were added to the top Transwell migration chamber, and various chemokines (CXCL12, 50 ng/ml; IL-8, 250 ng/ml; CCL5, CX3CL1, CXCL9, and CXCL10, 100 ng/ml) were added to the bottom Transwell migration chamber. Migration was evaluated after 4 hours by quantification of the number of migrated cells in the bottom chamber using 123count eBeads Counting Beads and FACS analysis. When indicated, the CD8 T cell subsets were stimulated O/N with IL-15 (10 ng/ml) and then treated with neuraminidase (2 U/ml; 37°C for 30 minutes), anti-PSGL1 mAb (50 μg/ml; 37°C for 1 hour), or 2-DG (50 μM; 4 hours).
Cell Migration and Live-Cell Imaging of Calcium by Time-Lapse Microscopy
TEMRA (CD45RA+CCR7−) and EM (CD45RA−CCR7−) CD3+CD8+ cells from the HVs and the KTs were sorted by FACS and rested O/N in complete RPMI medium supplemented with IL-15 (2 ng/ml). The CD8 T cell subsets stained with the cytosolic Ca2+ indicator Fura-2 AM probe (1 μg/ml for 45 minutes at room temperature) were seeded either on a confluent monolayer of HDMECs activated for 24 hours with TNF-α (100 U/ml) in μ-Slide eight wells or on poly-l-lysine precoated μ-Slide eight wells before the addition of CXCL12 (100 ng/ml). Cell polarization, cell migration, and calcium influx were tracked by time-lapse microscopy with a Leica DMI 6000B microscope through a ×20 objective equipped with MetaMorph software (Molecular Devices), capturing 120 sequential images at 20-second intervals. The migration paths and the calcium influx of individual cells were determined with ImageJ software, the TrackMate extras plug-in, and in-house scripts (R Studio) and used to calculate the average velocity during the observation period. The cell shape (roundness and circularity) of all cells during the recording period was calculated using ImageJ software and an in-house script (MATLAB).
RNA Sequencing
Naïve (CD45RA+CD28+), EM (CD45RA−CD28−), and TEMRA (CD45RA+CD28−) CD3+CD8+ T cells were FACS sorted (FACSAria; BD Biosciences; purity >95%) from freshly isolated human PBMCs obtained from six KTs. Cell pellets were resuspended in RLT buffer (Qiagen) containing 1% β-mercaptoethanol before subsequent RNA extraction using a RNeasy Micro Kit according to the manufacturer’s instructions (Qiagen). Quality and quantity of the RNA were assessed by spectrometry (Nanodrop) and electrophoresis (Agilent RNA 6000 Pico Kit). Smart-Seq2 libraries were prepared by the Broad Technology Labs and sequenced by the Broad Genomics Platform according to the SmartSeq2 protocol with some modifications.21 Briefly, total RNA was purified using RNA-SPRI beads, polyA+ mRNA was reverse transcribed to cDNA, and amplified cDNA was subject to transposon-based fragmentation that used dual indexing to bar code each fragment of each converted transcript with a combination of sample-specific barcodes. Sequencing was carried out as paired-end 2 × 25-bp reads with an additional eight cycles per index. Data were separated by barcode and aligned using TopHat version 2.0.10 with the default settings. Transcripts were quantified by the Broad Technology Labs computational pipeline using Cuffquant version 2.2.1.22 Briefly, data were processed through Cuffnorm if 50% of the reads aligned and if at least 100,000 pairs were aligned per sample. The default settings, including geometric normalization, were used for normalization, and expression-level data in the form of log2-transformed fragments per kilobase of transcript per million mapped fragments values were used for subsequent analyses. Differentially expressed genes between EM and TEMRA were identified with the linear modeling and empirical Bayes method from the limma R package with estimation of the mean-variance relationship (limma trend23). Genes with a Benjamini–Hochberg false discovery rate inferior to 5% and a fold change superior to 1.5 were considered significantly differentially expressed. The biologic significance of differential genes was assessed using the clusterProfiler24 R package.
Gene ontology (GO) categories enriched from the list of expressed genes with a false discovery rate <5% and with at least five genes were selected. RNA sequencing (RNA-seq) were deposited in the Gene Expression Omnibus under the accession number GSE198296.
Gene Set Enrichment Analysis of TEMRA CD8 Signature in Kidney Transplant Biopsies
Raw data were extracted from the Gene Expression Omnibus under the Gene Expression Omnibus accession number GSE147089. Affymetrix microarrays were processed with the R affy package using the robust multichip average normalization. After filtrating out low-expressed transcripts, defined as transcripts with median expression lower than the average expression of all measurements, 25,547 transcripts, corresponding to 14,085 unique genes, were analyzed using gene set enrichment analysis (http://www.broad.mit.edu/gsea/).25 Two sets of TEMRA-related genes were defined on the basis of our previous identification of TEMRA-restricted genes18 or a review of the literature (gene lists are in Supplemental Table 3). Default parameters and 10,000 random permutations of sample labels were used.
Polyfunctional CD8 T Cell Evaluation by the Single-Cell 32-Plex Proteomics
Cryopreserved CD8 T cells were thawed in RPMI 1640 medium supplemented with 10% FBS, 1× glutamax, and 1× penicillin-streptomycin-neomycin solution. Cells were then recovered in complete RPMI medium with IL-2 (10 ng/ml) at a density of 106 cells per milliliter in a 37°C and 5% CO2 incubator. After O/N recovery, the percentage of the viable cells was confirmed by trypan blue. Viable cells were enriched using Ficoll-Paque Plus if viability was lower than 80%. CD8 T cells were stimulated with plate-bound anti-human CD3 (1 μg/ml) and soluble anti-human CD28 (5 μg/ml) at a density of 106 cells per milliliter for 24 hours at 37°C and 5% CO2. The stimulated cells were then labeled with membrane stain (1:500 dilution; IsoPlexis) on chip cell detection and resuspended in complete RPMI medium at a density of 106 cells per milliliter. Approximately 30 μl of cell suspension (30,000 cells) was loaded into a human adaptive IsoCode Chip (IsoPlexis) containing approximately 12,000 cellular microchambers, each of which was prepatterned with a complete copy of the 32-plex antibody array for single-cell secretomic evaluation. Cells on the chip were incubated at 37°C and 5% CO2 for an additional 13.5 hours on the IsoLight automation system (IsoPlexis). Following this final incubation, subsequently secreted proteins from approximately 1000 single cells were captured by the 32-plex antibody bar-coded chip and analyzed by a back-end fluorescence ELISA-based assay. Polyfunctionality of T cells defined as a cell cosecreting two or more cytokines was analyzed by the IsoSpeak software across the seven functional groups: Th1 proinflammatory (GM-CSF, IFN-γ, IL-2, IL-12, TNF-α, and TNF-β); Th2 proinflammatory (IL-4, IL-5, IL-7, IL-9, and IL-13); chemoattractive (CCL11, IL-8, IP-10, MCP-1, MCP-4, MIP-1α, MIP-1β, and RANTES); regulatory (IL-10, IL-15, IL-22, and TGF-β1); Th17 proinflammatory (IL-1β, IL-6, IL-17A, IL-17F, and IL-21); cytolytic (GZMb and PERF); and other (sCD40L and sCD137). Protein signals from zero-cell microchambers were used to assess the cytokine-specific background. Cutoffs for any given cytokine were computed on the basis of background levels from wells not containing cells plus three SDs. In addition, signals with a signal-noise ratio of at least two (relative to the background threshold) and from at least 20 single cells or 2% of all single cells (whichever quantity was larger) were considered as significantly secreted. The polyfunctional strength index (PSI) of T cells was computed using a prespecified formula26–30 defined as the percentage of polyfunctional cells multiplied by the sum of the mean fluorescence intensity (MFI) of the proteins secreted by those cells:
Immunofluorescence Detection of CD8 on FFPE Kidney Biopsy
Slices (4 μm) of FFPE kidney biopsies were deparaffinized, and antigen retrieval was done with high- or low-pH solutions (TR1 or TR2). All samples were stained simultaneously. Endogenous peroxidases were first inhibited with H2O2 for 10 minutes. Anti-CD8 antibody (1:75 dilution) was added for 1 hour at room temperature and revealed with polymers (Poly-E and HRP-2-Step) followed by incubation with the fluorochrome Opal520 (10 minutes). The nuclei were finally counterstained with DAPI (1:1000). The slides were mounted with Vectashield Vibrance Antifade mounting medium. Large images were acquired with the Nikon A1RSi confocal resonant microscope using the Nikon denoise.ai algorithm. The images were analyzed with the Qupath software.31 The cell detection tool was used on the DAPI channel to detect nuclei and then estimate the cell membrane. In each cell, the intensities of each channel were calculated and then exported in Excel to count the number of positive cells for one or more channels. The figure was made under Fiji32 to adjust the contrast and set the scale bar.
Statistical Analyses
All statistical analyses were performed using GraphPad Prism. Mann–Whitney U tests, Kruskal–Wallis tests followed by Dunn post hoc tests, and paired Wilcoxon tests were used as appropriate, and the type of test used is included in the figures. All P values are given as exact values or using asterisks as follows: *P=0.05, **P=0.01, ***P<0.001, and ****P<0.001.
Results
Expansion of TEMRA CD8 in KTs with Biopsy-Proven ABMR
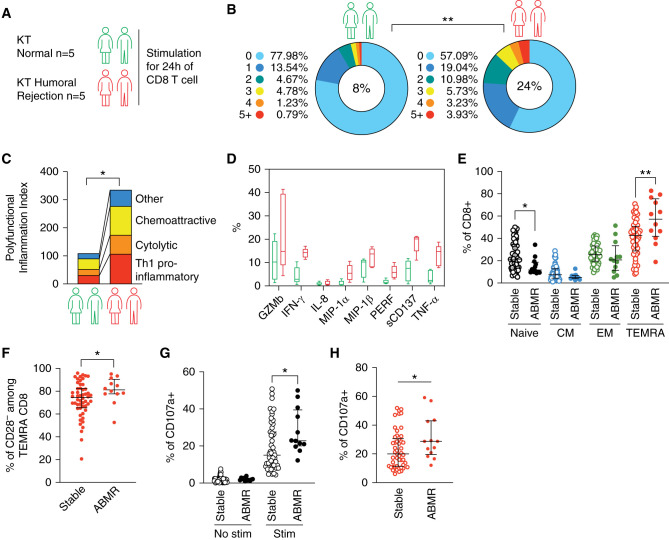
Purified CD8 T cells isolated from KTs with either normal kidney biopsy or biopsy-proven humoral rejection were ex vivo restimulated prior to single-cell cytokines analysis using single-cell 32-plex profiling (Figure 1A).29 The frequency of polyfunctional cells (i.e., producing two or more cytokines) was significantly higher in CD8 T cells from KTs with humoral rejection as compared with those from KTs with normal biopsy (24% versus 8%, respectively; P=0.008) (Figure 1B). The analysis of the strength index demonstrated not only that the percentage of polyfunctional cells was higher for CD8 T cells from KTs with humoral rejection but also that the intensity of all profiled secreted cytokines was higher (Figure 1C) (P=0.02), including a higher production of proinflammatory cytokines (IFN-γ, TNF-α, MIP-1α, and MIP-1β) and higher expression of cytotoxic molecules (GZMb and PERF) (Figure 1D). The rapid and combined production of proinflammatory cytokines and cytotoxic molecules suggests that TEMRA CD8 T cells account preferentially for this response and are dysregulated in KTs with humoral rejection. To test this hypothesis, we recruited 12 KTs with biopsy-proven ABMR and 52 age- and sex-matched KTs with normal histologic kidney biopsy 12 months after transplantation. The profiling of CD8 T cells revealed that KTs with ABMR exhibit an attrition of naïve CD8 T cells associated with an expansion of TEMRA CD8 T cells compared with KTs with normal kidney biopsy (Figure 1E). Considering the absolute number, we were able to demonstrate that KTs with ABMR have a significant increase in the number of TEMRA CD8 per milliliter compared with KTs with stable graft function (P=0.01; 412±199 versus 109±16, respectively) (Supplemental Figure 2A). In KTs with ABMR, the TEMRA CD8 T cells were more differentiated as shown by a higher frequency of CD28– among TEMRA CD8 T cells (80.9±3.3 versus 72.7±2.1, respectively; P=0.05) (Figure 1F). A higher cytotoxic response by CD8 T cells was evidenced in KTs with ABMR (Figure 1G) (P=0.05), the magnitude of the cytotoxic response was positively correlated with the frequency of TEMRA CD8 (r=0.62; P<0.001) (Supplemental Figure 2B), and the cytotoxic response of TEMRA CD8 was exacerbated in KTs with ABMR compared with KTs with stable graft function (31.1±4.2 versus 23.1±1.9, respectively; P=0.05) (Figure 1H). Collectively, these findings support an active role of the CD8 T cell compartment in the process of kidney graft rejection and especially, an overactivation of TEMRA CD8 T cells.
Figure 1.
KTs with biopsy-proven ABMR showed an expansion of TEMRA CD8. (A) CD8 T cells from KTs with humoral rejection (n=5) or with normal histologic biopsy (n=5) stimulated O/N with anti-CD3 and anti-CD28 mAbs prior to single-cell cytokines analysis using single-cell 32-plex profiling. (B) The frequency of polyfunctional CD8 T cells is shown per number of cytokines produced as well as the mean overall polyfunctional CD8 T cells (i.e., producing two or more cytokines) inside the pie chart. (C and D) PSI defined as the percentage of polyfunctional T cells (two or more cytokines secreted) in the sample multiplied by the mean fluorescence signal intensity of the proteins secreted by those cells, and the contribution of each analyte to the PSI is shown. (E–H) KTs were recruited 1 year after their transplantation and with either biopsy-proven ABMR (n=12) or normal histologic biopsy (n=52; recipient age and sex matched). (E) Frequency of functional subsets among CD8 T cells. (F) Percentage of CD28– among TEMRA CD8. (G and H) Cytotoxic response of (G) CD8 T cells or (H) TEMRA CD8 after 4 hours of ex vivo stimulation with CD3 (2 μg/ml) and IL-15 (10 ng/ml). Data represent the median ± interquartile range (25%–75%) and the value (point) of a single patient and one representative patient. The P values were calculated using the Mann–Whitney U test. *P=0.05; **P=0.01. CM, central memory; Stim, stimulation; ABMR, antibody-mediated rejection.
The TEMRA CD8 T Cells from the KTs Exhibit an Enhanced Cytotoxic and Migratory Response Compared with the EM CD8 T Cells
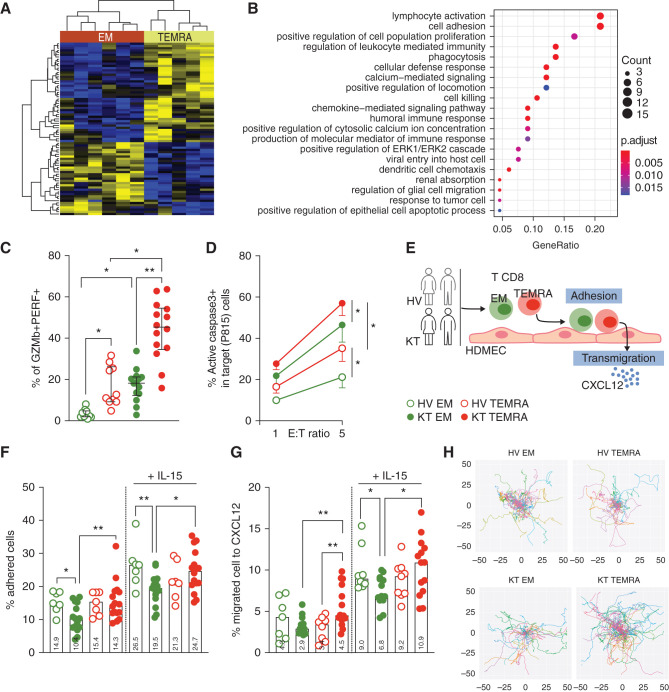
We then perform transcriptomic analysis of EM CD8 T cells and TEMRA CD8 T cells purified from five KTs with stable graft function. Unsupervised analysis revealed a CD8 subset-specific signature of 133 genes (Figure 2A, Supplemental Table 4), and an enrichment in genes related to lymphocyte activation (GO 0046649) and cell adhesion (GO 0007155) in TEMRA CD8 T cells was identified (Figure 2B). The RNA-seq analysis suggested that despite their stable graft function, TEMRA CD8 T cells from KTs are more prone to mediate inflammatory response and to migrate toward lymphoid and nonlymphoid organs compared with EM CD8 T cells. The analysis of the coexpression of the cytotoxic molecules GZMb and PERF confirmed that the reservoir of preformed cytotoxic molecules was higher in TEMRA CD8 T cells from KTs that readily expressed GZMb and PERF as compared with EM CD8 T cells from KTs (Figure 2C) (P=0.01) and also with TEMRA CD8 T cells from HVs (Figure 2C) (P=0.05). In line with these results, TEMRA CD8 T cells from KTs exhibited the highest cytolytic activity compared with EM CD8 T cells from KTs and with TEMRA CD8 T cells from HVs (Figure 2D) (P=0.05).
Figure 2.
TEMRA CD8 T cells from KTs exhibited an enhanced cytotoxic and migratory response compared with EM CD8 T cells. (A) Heat map showing scaled expression values of differentially expressed genes for CD8 T subsets (EM and TEMRA) purified from five KTs. (B) Gene set enrichment analysis of differentially expressed genes for CD8 T subsets (EM and TEMRA) purified from five KTs. (C) Coexpression of GZMb and PERF by TEMRA and EM CD8 T cell subsets (EM, green; TEMRA, red) isolated from KTs (filled circles) and HVs (open circles). (D) Cytolytic activity of TEMRA and EM CD8 T cell subsets (EM, green; TEMRA, red) isolated from KTs (filled circles) and HVs (open circles; n=7 individuals). Redirected killing was quantified at different effector-target (E/T) ratios against sensitized mastocytoma cells (P815). (F–H) TEMRA and EM CD8 T cell subsets (EM, green; TEMRA, red) were isolated from KTs (filled circles) and HVs (open circles) and stimulated, when indicated, O/N with IL-15 (10 ng/ml) before the assessment of their migratory properties. (F) Adhesion of CD8 T cell subsets from KTs and HVs before and after IL-15 stimulation to TNF-α–treated HDMECs. (G) Transmigration of CD8 T cell subsets from KTs and HVs before and after IL-15 stimulation across TNF-α–treated HDMEC monolayers in response to CXCL12 (50 ng/ml). (H) Migration of Fura-2–labeled CD8 T cell subsets from KTs and HVs onto TNF-α–treated HDMEC monolayers. The path of individual cells aligned with their origins at x=y=0 is shown. Data in (C, D, and F–H) represent the median (gray histograms) or mean±SEM and the value (point) of each single individual. The data in (H) represent one experiment representative of five independent experiments. The P values were calculated using the Wilcoxon matched pairs signed rank test. *P=0.05; **P=0.01. P815, cell line identity.
The enrichment for gene signatures associated with cell adhesion by TEMRA CD8 T cells from KTs supported the functional characterization of the migratory properties of TEMRA CD8 T cells (Figure 2E). We show that the TEMRA CD8 T cells from the KTs exhibited an enhanced adhesion to TNF-α–activated endothelial cells (HDMECs) compared with the EM CD8 T cells from the KTs (Figure 2F). By contrast, the EM and TEMRA CD8 T cells from HVs exhibited similar adhesion to activated HDMECs (Figure 2F). O/N stimulation of CD8 T cell subsets with IL-15, a key cytokine regulating the activation and proliferation of TEMRA CD8 T cells,19,20 results in increased adhesion of memory CD8 T cells; with KTs, TEMRA CD8 T cells showed stronger adherence to the endothelium in comparison with KT EM CD8 T cells (Figure 2F).
We then screened chemokines to identify those that induce the transmigration of TEMRA CD8 T cells through the endothelium using published RNA-seq14 and proteomic33 data and functional transmigration assays. The EM and TEMRA CD8 T cells from the HVs expressed similar levels of receptors of CXCL8, CXCL9, CXCL10, CXCL12, and CCL5 (Supplemental Figures 1 and 3A), with CX3CR1 the only receptor that was preferentially expressed by the TEMRA CD8 T cells (Supplemental Figure 3B), in line with recent reports.34,35 The screening of chemokines (CCL5, CXCL9, CXL10, CXCL12, CX3CL1, and CCL8) showed that the memory CD8 T cells (TEMRA or EM) from HVs transmigrate across a monolayer of activated HDMECs in response to CXCL12 added to the lower chamber (Supplemental Figure 3C). The susceptibility of the TEMRA and EM CD8 T cells to the chemoattractant CXCL12 was validated using time-lapse imaging, and we showed that CXCL12 promotes the deformation of both the TEMRA and EM CD8 T cells from both the KTs and HVs (Supplemental Figure 3D). In line with their enhanced adhesion to endothelium, the TEMRA CD8 T cells from the KTs exhibited an enhanced ability to transmigrate across the endothelium in response to CXCL12 compared with the EM CD8 T cells from the KTs and with TEMRA CD8 T cells from the HVs (Figure 2G) (P=0.01). Finally, tracking the cell movement of the TEMRA or EM CD8 T cells cultured with activated HDMECs using live-cell imaging showed that the TEMRA CD8 T cells from the KTs exhibited the strongest patrolling behavior as compared with EM CD8 T cells from the KTs and with TEMRA CD8 T cells from HVs (Figure 2H, Supplemental Figure 3E), characterized by a higher average velocity (Figure 2H). Collectively, these findings demonstrated that the TEMRA CD8 T cells from the KTs have enhanced cytolytic and migratory properties compared with TEMRA CD8 T cells from the HVs as well as their EM CD8 T cell counterparts, with a sensitivity to the chemoattractant CXCL12 and enhanced migratory features induced by IL-15.
Accumulation of TEMRA CD8 T Cells in the Kidney Graft of KTs with ABMR
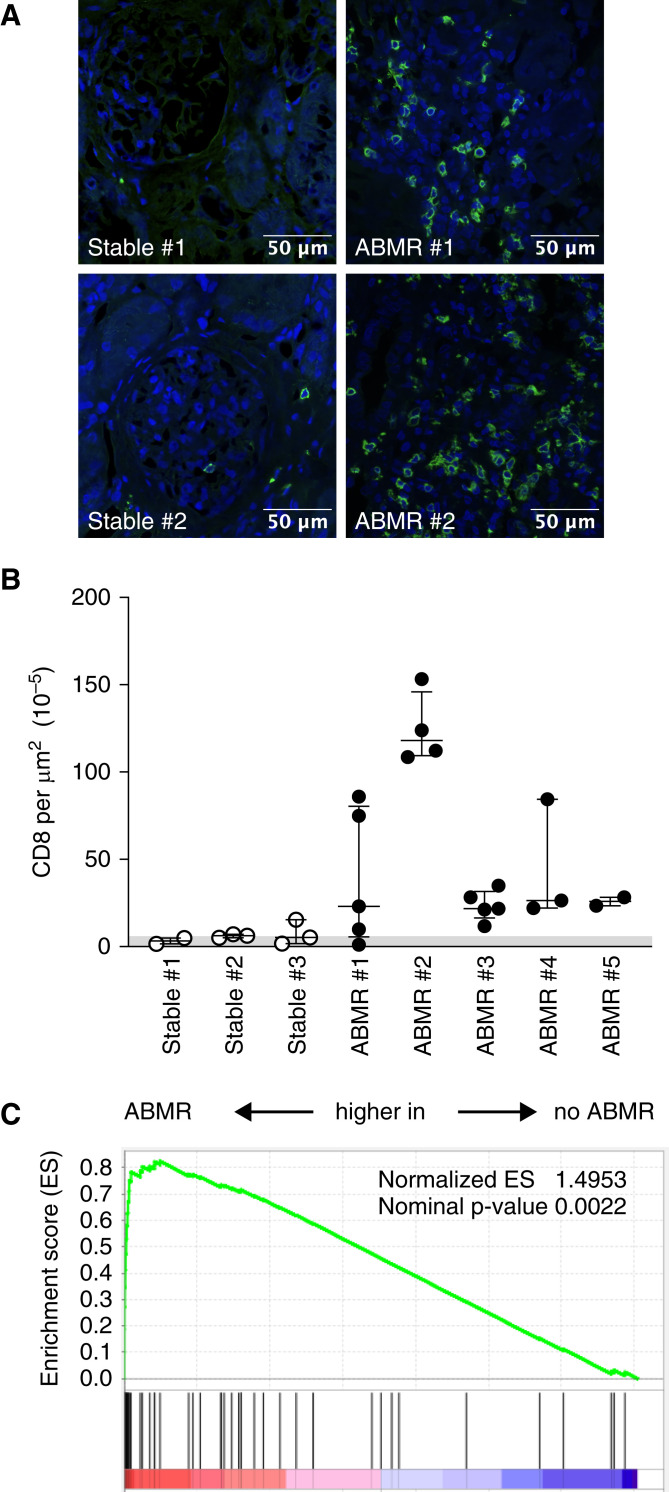
We then analyze in situ the presence of CD8 T cells in kidney biopsies from KTs with normal histology (n=3) or with ABMR (n=5). Only a few CD8 T cells could be detected in KTs with normal histology (Figure 3, A and B). By contrast, a massive CD8 T cell infiltrate was evidenced in biopsies from KTs with ABMR (Figure 3, A and B). To specifically analyze the presence of TEMRA CD8 in the kidneys from KTs with ABMR, two gene signatures of TEMRA CD8 T cells were defined on the basis of either our previous TEMRA-specific transcriptomic analysis18 or the literature (Supplemental Table 3). Gene set enrichment analysis was used to identify enrichment of TEMRA-related genes in gene expression data of kidney biopsies from KTs with and without ABMR (n=56 and n=168, respectively; accession no. GSE147089).36 Both lists of TEMRA-related genes were highly enriched in patients with ABMR lesions (normalized enrichment scores of 1.4953 and 1.2942 and nominal P values of 0.002 and 0.05 using our TEMRA-specific signature or literature-based TEMRA signature, respectively) (Figure 3C, Supplemental Figure 4A). A strong positive correlation between expression of CD8 and TEMRA-associated genes CX3CR1, cytotoxic molecules (GNLY, GZMb, GZMh, and PERF-1), and KLRD1 was observed, and ABMR KTs had the highest coexpression of CD8 and TEMRA-associated genes compared with patients with normal histology (Supplemental Figure 4B). Collectively, our results showed an accumulation of TEMRA CD8 within the kidney graft of patients with ABMR.
Figure 3.
TEMRA CD8 T cells are accumulated in the kidney graft of KTs with ABMR. (A and B) Representative examples of (A) infiltration by CD8 T cells (green cells) and (B) quantification of CD8 T cells (10−5 cells per micrometer squared) in kidney biopsies from KTs with normal histology (stable; n=3) or ABMR (n=5). Each point represents the absolute number of CD8 T cells per region of interest (n=2–5 per patient). (C) Gene set enrichment analysis of TEMRA gene signatures derived from the literature (Supplemental Table 3) in gene expression data of kidney biopsies from KTs with or without ABMR (n=56 and 168, respectively; GSE147089).36 The normalized enrichment score (ES) is the degree to which a gene set is over-represented normalized across analyzed gene sets. The nominal P value is the statistical significance of the enrichment score. Correlation between expression of CD8 and TEMRA-associated genes in patients with ABMR (pink) and normal biopsy (shaded gray). Correlation was calculated using nonparametric Spearman correlation. ABMR, Antibody-mediated rejection.
T Cell Receptor and CXCL12 Stimulation Triggered a Potent Purinergic P2 × 4 Receptor–Dependent Proinflammatory Response of the TEMRA CD8 T Cells from the KTs
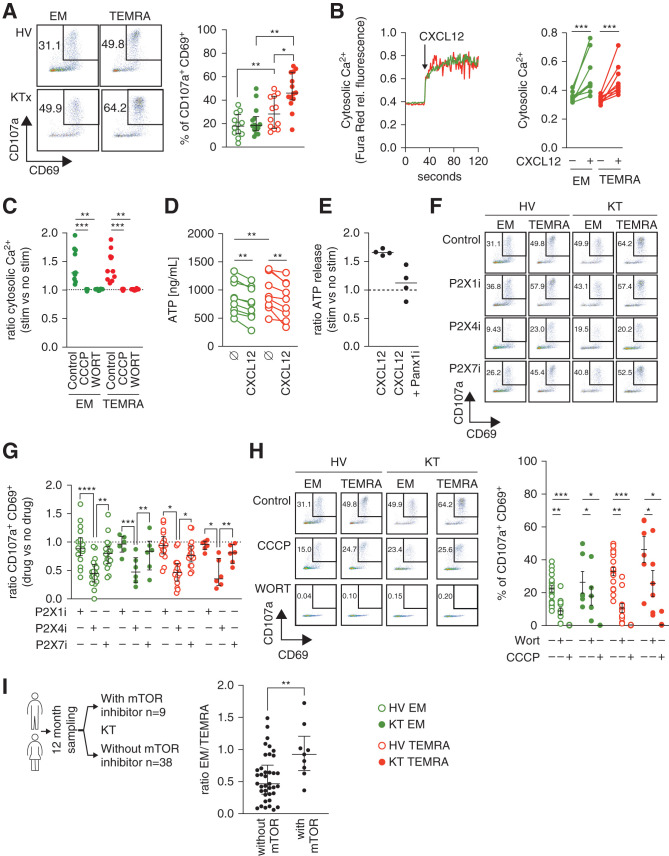
In addition to its role in the regulation of T cell migration,37–39 CXCL12 was recently shown to elicit the activation and proliferation of human CD4 T cells when combined with T cell receptor (TCR) stimulation.10 The susceptibility of human CD8 T cells to CXCL12 and the effect of the differentiation status of human CD8 T cell subsets (naïve versus TEMRA versus EM) remain unclear. The stronger migratory properties of the TEMRA CD8 T cells from the KTs compared with their EM counterparts, especially in the context of CXCL12-driven transmigration, prompted us to determine whether CXCL12 differentially induces the immune activation of the TEMRA and EM CD8 T cells from the KTs and the HVs. Combined stimulation of CXCL12 and TCR led to the upregulation of CD69 and CD107a, with a higher magnitude for the TEMRA CD8 T cells than for the EM CD8 T cells from the KTs (47.9±4.1 versus 22.0±3.2, respectively; P=0.01) (Figure 4A) and also a higher response of TEMRA CD8 T cells from the KTs than those from HVs (47.9±4.1 versus 29.4±3.9, respectively; P=0.05) (Figure 4A). Stimulation with CXCL12 alone failed to activate memory CD8 T cells (TEMRA and EM) (Supplemental Figure 5A), and naïve CD8 cells were not susceptible to CXCL12 and TCR stimulation (Supplemental Figure 5B).
Figure 4.
TCR and CXCL12 stimulation triggered a potent purinergic P2 × 4 receptor–dependent proinflammatory response of the TEMRA CD8 T cells from the KTs. (A) Frequency of CD69+CD107a+ cells among the EM (green circles) and TEMRA (red circles) CD8 T cells from KTs (filled circles) and HVs (open circles) after 4 hours of stimulation with plate-bound anti-CD3 mAb (2 μg/ml) + CXCL12 (50 ng/ml). (B and C) Cytosolic Ca2+ levels in the CD8 T cell subsets before and after the addition of CXCL12 (50 ng/ml) were recorded by flow cytometry for 140 seconds. When indicated, CD8 T cell subsets were pretreated with CCCP (10 μM) or wortmannin (10 μM). The results are expressed as (B) the ratio of Fura Red fluorescence collected by the 605/12 and 695/40 detectors (i.e., cytosolic Ca2+) or (C) the ratio of cytosolic Ca2+ between CXCL12 ± drug versus the basal level. (D) Intracellular ATP reservoir of EM (green circles) and TEMRA (red circles) CD8 T cells from HVs treated or not for 20 minutes with CXCL12 (100 ng/ml). (E) ATP release by purified CD8 T cells from HVs treated with 10Panx1 (100 μM) and stimulated for 20 minutes with CXCL12 (100 ng/ml). The results are expressed as the ratio of ATP after versus before stimulation. (F–H) Frequency of CD69+CD107a+ cells among the EM (green circles) and TEMRA (red circles) CD8 T cells from KTs (filled circles) and HVs (open circles) after 4 hours of stimulation with plate-bound anti-CD3 mAb (2 μg/ml) or anti-CD3 + CXCL12 (50 ng/ml). When indicated, CCCP (10 μM), wortmannin (10 μM), and selective P2 × 1 (10 μM), P2 × 4 (20 μM), or P2 × 7 (20 μM) receptor inhibitors were added simultaneously to anti-CD3 + CXCL12 cells. (I) Frequency of EM and TEMRA among CD8 T cells in KTs recruited 1 year after their transplantation and treated with mTOR inhibitor (n=9). The KTs of the control group (n=38) did not receive mTOR inhibitor and were matched for the recipient age and sex. Data represent the median (black lines) and the value (point) of each single individual. The P values were calculated using (A, C, and F) the nonparametric ANOVA (Kruskal–Wallis) with Dunn multiple comparisons test, (B) the Wilcoxon matched pairs signed rank test, or (H) the Mann–Whitney signed rank U test. *P=0.05; **P=0.01; ***P<0.001; ****P<0.001. KTx, kidney transplant recipients; CCCP, carbonyl cyanide m-chlorophenyl hydrazone; WORT, wortmannin; rel., relative; stim, stimulation.
One essential component of the signal transduction cascade of CXCR4 is an increase in intracellular Ca2+ concentration via the activation of phosphatidylinositol 3-kinase (PI3K),40 and mitochondria play a key role in the control of Ca2+ release into the cytosol via the mitochondrial Na+/Ca2+/Li+ exchanger.41 We showed that CXCL12 alone is sufficient to induce an immediate rise in intracellular Ca2+ concentration with a similar magnitude in the EM and TEMRA CD8 T cells (Figure 4B). Selective inhibition of PI3K and mitochondrial oxidative phosphorylation prevented the CXCL12-induced increase in Ca2+ concentration (Figure 4C), demonstrating that the CXCL12-CXCR4 interaction signal is mediated through PI3K and mitochondrial activity in the EM and TEMRA CD8 T cells. We also showed that the stimulation with CXCL12 alone results in a significant and rapid decrease of intracellular ATP (Figure 4D) in both TEMRA and EM CD8 T cells. These data suggest that the CXCL12-CXCR4 interaction signal is sufficient to initiate the rapid consumption of the initial ATP pool and thus, a rapid immune response. The mammalian P2× receptor family (P2 × 1–7) of ATP-gated ion channels has been shown to contribute to calcium influx, especially in response to extracellular ATP (eATP).42 eATP is released by T cells upon stimulation via the gap junction hemichannel Panx143,44 and can regulate CD4 T cell activation45 through the purinergic receptors P2 × 1 and P2 × 4.10,44 We demonstrated that CXCL12 induces a rapid Panx1-dependent rise in eATP by the EM and TEMRA CD8 T cells (Figure 4E). The inhibition of P2 × 1 and P2 × 7 using NF023 and A438079, respectively, did not prevent TEMRA and EM CD8 activation, whereas the specific and selective P2 × 4 receptor inhibitor 5-BDBD inhibited their activation by >50% (Figure 4, F and G). A similar effect was also achieved by the inhibition of mitochondrial oxidative phosphorylation (Figure 4H), and inhibition of PI3K completely prevented the activation of the TEMRA and EM CD8 T cells (Figure 4H). Collectively, our results showed that CXCL12 not only chemoattracts human TEMRA and EM CD8 T cells but also activates their cytotoxic effector program through the release of eATP followed by its ligation to the purinergic P2 × 4 receptor. Moreover, we demonstrated that TEMRA CD8 T cells from the KTs have an enhanced susceptibility to CXCL12 stimulation, resulting in a very potent induction of their proinflammatory response.
Finally, given the susceptibility of TEMRA CD8 T cells from KTs to PI3K inhibitor, we investigated whether the treatment of KTs with mammalian target of rapamycin (mTOR) inhibitor (sirolimus) affected the frequency of memory CD8 T cells. We obtained samples from nine KTs treated with an mTOR inhibitor and 38 age- and sex-matched KTs not treated with an mTOR inhibitor at 12 months after transplantation (Figure 4I). Interestingly, the frequency of TEMRA CD8 was significantly lower in KTs treated with an mTOR inhibitor than in KTs not receiving an mTOR inhibitor (ratio of EM to TEMRA: 0.56±0.05 versus 0.95±0.13, respectively; P=0.05) (Figure 4I). These observations suggested that the migration toward the kidney graft and the expansion of cytolytic TEMRA CD8 T cells could be restrained by an mTOR inhibitor.
IL-15–Induced Conversion of Functional PSGL1 Is Similar in TEMRA CD8 and EM CD8 T Cells from KTs
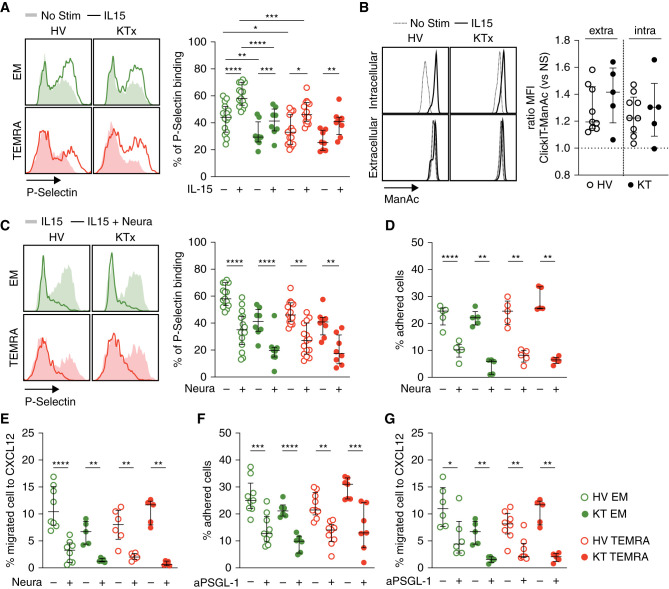
The enhanced migratory properties of the TEMRA CD8 T cells from the KTs could result from an enhanced ability of PSGL1 to interact with E/P-selectin, a key regulator in the rolling of T cells onto the endothelium. To test this hypothesis, we measured the ability of purified CD8 T cell subsets to bind to recombinant P-selectin before and after IL-15 stimulation (Figure 5A). The memory CD8 T cells (TEMRA and EM) showed stronger binding to P-selectin than naïve CD8 T cells (Supplemental Figure 6A). An increase in P-selectin binding was observed for both EM and TEMRA CD8 and for KTs and HVs after stimulation with IL-15 (Figure 5A). The ability of TEMRA CD8 T cells to bind to P-selectin was not different between KTs and HVs (Figure 5A). In contrast, EM CD8 T cells from KTs showed a weaker binding to P-selectin compared with HVs both in the steady state and after stimulation with IL-15. Finally, we observed that EM from HVs has a greater ability to bind to P-selectin than TEMRA. This enhanced P-selectin binding of EM is limited to HVs as no difference between EM and TEMRA from KTs was observed (Figure 5A). Appropriate glycosylation is needed for selectin binding, with a sequential synthesis of core 2 O-glycans, including the sialic acid precursor N-acetyl-d-mannosamine (ManNAc) and the addition of sLeX to generate the functional form of PSGL1.46 We show that IL-15 stimulation results in a direct increased incorporation of the ManNAc in the intracytoplasmic compartment as well as on proteins at the cellular surface (Figure 5B, Supplemental Figure 6B). Removal of sialic acid by neuraminidase after IL-15 stimulation prevented the binding of P-selectin to the TEMRA and EM CD8 T cells (Figure 5C), the adhesion of the TEMRA and EM CD8 T cells to activated endothelium (Figure 5D), and their transmigration (Figure 5E), with similar efficacy between the KTs and the HVs. Finally, blocking the interaction between PSGL1 and P-selectin using the anti-PSGL1 mAb clone CHO131 that specifically recognizes the sLeX-bearing core 2 O-glycan structures prevented the adhesion of the TEMRA and EM CD8 T cells to the activated endothelium and transmigration in response to CXCL12 (Figure 5, F and G). These results demonstrated that IL-15 directly promotes the generation of sLeX-bearing core 2 O-glycan structures and the formation of functional PSGL1 that controls the ability of TEMRA and EM CD8 T cells to interact with the activated endothelium. Furthermore, these results showed that the migration of the TEMRA CD8 T cells from the KTs can be altered by interfering with the interaction between P-selectin and functional PSGL1.
Figure 5.
IL-15 promotes the generation of functional PSGL1 by enhancing sialic acid biosynthesis. (A) CD8 T cell subsets (EM, green; TEMRA, red) were purified from HVs and KTs and stimulated, when indicated, O/N with IL-15 (10 ng/ml). Binding of CD8 T cell subsets to P-selectin chimeric proteins was quantified by flow cytometry. (B) Purified CD8 T cells from HVs and KTs were incubated O/N with Click-IT ManNAz (ManNAc) and when indicated, IL-15 (10 ng/ml). Incorporation of ManNAc was quantified after cell permeabilization (intra) or cell fixation (extra), and the results are expressed as the ratio between IL-15–treated and nonstimulated cells. (C) CD8 T cell subsets (EM, green; TEMRA, red) purified from HVs and KTs were stimulated O/N with IL-15 (10 ng/ml) and, when indicated, treated for 30 minutes with neuraminidase (2 U/ml). Binding of CD8 T cell subsets to P-selectin chimeric proteins was quantified. (D) Adhesion to TNF-α–treated HDMEC monolayers and (E) CXCL12-induced transmigration of CD8 T cell subsets treated O/N with IL-15 (10 ng/ml) followed by pretreatment with neuraminidase (2 U/ml; 30 minutes). (F) Adhesion to TNF-α–treated HDMEC monolayers and (G) CXCL12-induced transmigration of CD8 T cell subsets from HVs and KTs treated O/N with IL-15 (10 ng/ml) followed by preincubation with blocking anti-PSGL1 mAb (50 μg/ml; 60 minutes). Data represent the mean±SEM, the value (point) of each single individual, and (A–C) one representative histogram. The P values were calculated using the Wilcoxon matched pairs signed rank test. *P=0.05; **P=0.01; ***P<0.001; ****P<0.001. KTx, kidney transplant recipients; Stim, stimulation; Neura, neuraminidase; NS, no stimulation; aPSGL-1, anti–P-selectin glycoprotein ligand-1 mAb.
Selective Inhibition of Glycolysis Controls the Migration of the TEMRA CD8 T Cells by Impairing Sialic Acid Biosynthesis and the Transition to the Functional Conformational Structure of PSGL1
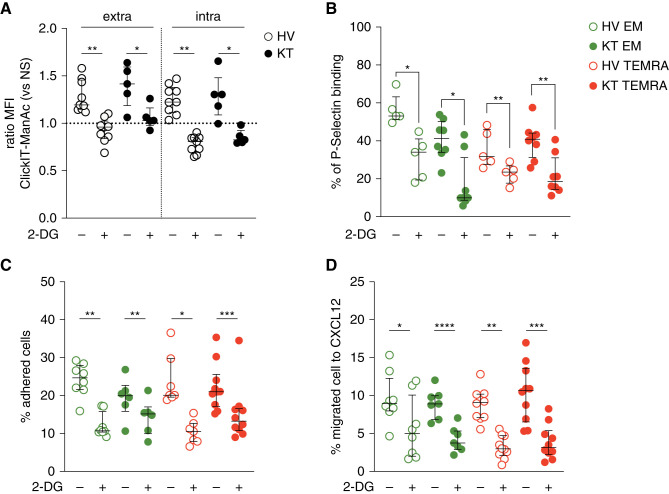
Stimulation with IL-15 enhanced glycolysis in murine CD8 memory T cells47 and in the TEMRA CD8 T cells from HVs or immune-challenged patients (KTs),20 and 2-DG, an inhibitor of glycolysis, efficiently prevents the IL-15–induced activation of TEMRA CD8 T cells.20 In addition to inhibition of glycolysis, 2-DG could directly alter protein glycosylation in natural killer (NK) cells,48 and mannose metabolism was essential for CD4 T1H differentiation.49 We thus hypothesized that the selective targeting of glycolysis could prevent not only CD8 T cell activation but also migration by limiting sialic acid biosynthesis and the transition to a functional conformation of PSGL1. Short-term treatment of the IL-15–stimulated TEMRA or EM CD8 T cells from HVs or the KTs with 2-DG resulted in a decrease in the incorporation of ManNAc both in the cytoplasmic compartment and the protein cell membrane (Figure 6A) and in their binding to P-selectin (Figure 6B). Inhibition of glycolysis resulted in a decrease of functional PSGL1, whereas total PSGL1 expression remained unchanged in TEMRA and EM CD8 T cells (Supplemental Figure 7). This inhibition of sialic acid biosynthesis resulted in a decrease in the adhesion of the TEMRA and EM CD8 T cells to the activated endothelium (Figure 6C) and in their CXCL12-induced transmigration (Figure 6D). Collectively, these data demonstrated that interfering with glycolysis alters the trafficking potential of TEMRA and EM CD8 T cells from the KTs and the HVs by limiting sialic acid production.
Figure 6.
Selective inhibition of glycolysis controls the migration of TEMRA and EM CD8 T cells by impairing sialic acid biosynthesis and the transition to the functional conformational structure of PSGL1. (A) Purified CD8 T cells from HVs and KTs were incubated O/N with Click-IT ManNAz (ManNAc) and IL-15 (10 ng/ml). When indicated, CD8 T cells were pretreated with 2-DG (50 μM; 4 hours at 37°C) before the quantification of the incorporation of ManNAc after cell permeabilization (intra) or cell fixation (extra). The results are expressed as the ratio between 2-DG + IL-15 versus IL-15. (B–D) CD8 T cell subsets (EM, green; TEMRA, red) purified from HVs and KTs were incubated O/N with IL-15 (10 ng/ml). CD8 T cells were then treated with 2-DG (50 μM; 4 hours at 37°C). (B) Binding of CD8 T cell subsets to P-selectin chimeric proteins, (C) their adhesion to TNF-α–treated HDMEC monolayers, and (D) their CXCL12-induced transmigration were quantified. Data represent the mean±SEM and the value (point) of each single individual. The P values were calculated using the Wilcoxon matched pairs signed rank test. *P=0.05; **P=0.01; ***P<0.001; ****P<0.001. NS, no stimulation.
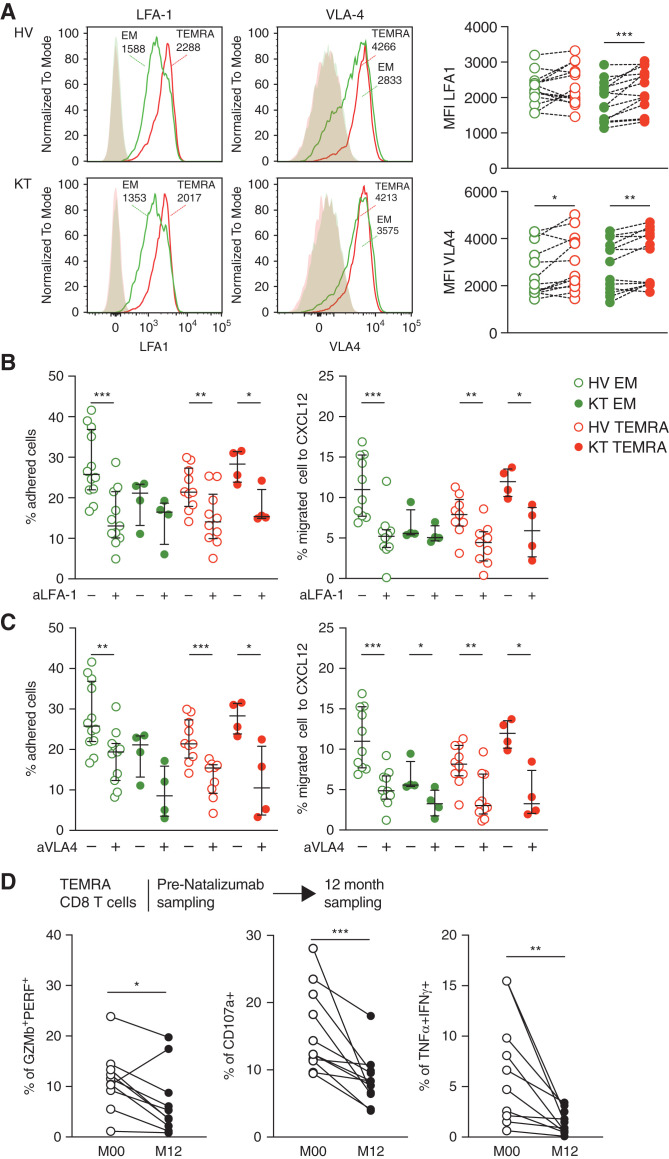
Selective Inhibition of the Migration TEMRA CD8 from the KTs Can Be Achieved by Targeting LFA-1 and VLA-4
Although selectins and addressins control the initial adhesion of cell to the endothelium,50,51 the firm adhesion to the endothelium is induced by high-affinity integrin activation and especially, LFA-1/ICAM-152,53 and VLA-4/VCAM-18 interactions. Therefore, we characterized the involvement of LFA-1 and VLA-4 pathways in the regulation of the enhanced migration of TEMRA CD8 T cells from KTs. TEMRA CD8 T cells from KTs exhibited an enhanced expression of LFA-1 and VLA-4 compared with their EM CD8 counterparts (Figure 7A). Moreover, the adhesion and the transmigration of TEMRA CD8 T cells from KTs can be impaired using blocking anti–LFA-1 and VLA-4 antibodies, whereas the migration of EM CD8 T cells from KTs was marginally altered (Figure 7, B and C). We gathered longitudinal samples from patients with multiple sclerosis treated with natalizumab, a treatment that blocks the action of VLA-4, to explore the involvement of VLA-4 in the function of TEMRA CD8 T cells in more direct clinical settings (Figure 7D). The frequency of TEMRA CD8 T cells expressing cytotoxic molecules PERF and GZMb fell with continuous treatment over a period of 12 months (Figure 7D) (P=0.01). These changes were reflected in the activation-induced functional profiles of TEMRA CD8 T cells, which produce fewer proinflammatory cytokines TNF-α and IFN-γ and exhibit lower cytotoxic function (Figure 7D) (P=0.01). Collectively, these data suggest that the migration and the effector function of TEMRA CD8 T cells from KTs could be selectively controlled by blocking the VLA-4 pathway.
Figure 7.
Blocking LFA-1 or VLA-4 results in the selective inhibition of the migration of TEMRA CD8 from the KTs. (A) Representative example and quantification of the expression of LFA-1 and VLA-4 by CD8 T cell subsets (EM, green; TEMRA, red) purified from HVs and KTs. (B and C) Adhesion to TNF-α–treated HDMEC monolayers and CXCL12-induced transmigration of CD8 T cell subsets from HVs and KTs treated O/N with IL-15 (10 ng/ml) followed by preincubation with (B) blocking anti–LFA-1 mAb (50 μg/ml; 60 minutes) or (C) blocking anti–VLA-4 mAb (50 μg/ml; 60 minutes). (D) Coexpression of GZMb and PERF by TEMRA CD8 T cells isolated from MS patients before (open circles) and after (filled circles) 1 year of treatment with natalizumab. The production of inflammatory cytokines TNF-α and IFN-γ and the cytotoxic function (CD107a) by TEMRA CD8 T cells were analyzed after 4 hours ex vivo polyclonal restimulation. Data represent the mean±SEM and the value (point) of each single individual. The P values were calculated using the Wilcoxon matched pairs signed rank test. *P=0.05; **P=0.01; ***P<0.001. MS, multiple sclerosis; M00/M12, month 0/month12; aLFA-1, anti–lymphocyte function-associated antigen 1 mAb; aVLA-4, anti–very late antigen-4 mAb.
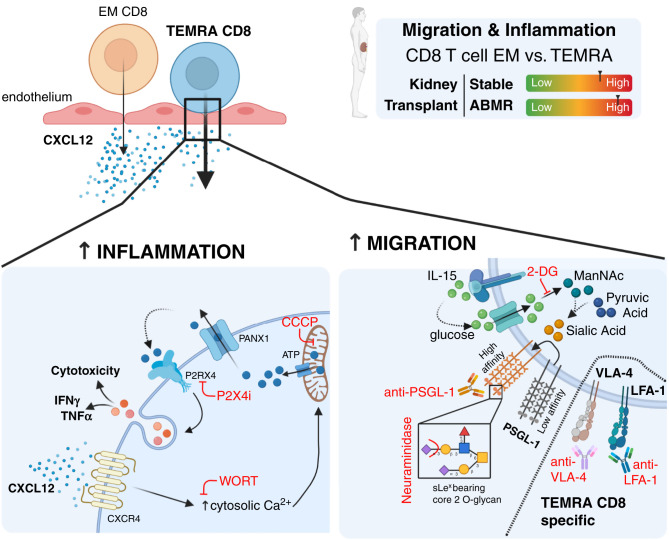
Taken together, these results demonstrated that the TEMRA CD8 T cells from the KTs exhibited enhanced migratory fitness compared with the EM CD8 T cells. CXCL12 not only chemoattracts TEMRA CD8 T cells but also triggers P2 × 4-dependent activation of their immune program, favoring the establishment of a local inflamed environment and promoting kidney graft failure.
Discussion
Using static and dynamic approaches, we showed that the strong challenge resulting from kidney allogeneic transplantation enhanced the migratory fitness of the TEMRA CD8 T cells compared with the EM CD8 T cells, with an enhanced adhesion to activated endothelium, a stronger ability to transmigrate across a monolayer of activated HDMECs in response to CXCL12, and stronger patrolling behavior (higher velocity and CXCL12-induced morphologic deformation). Importantly, KTs with ABMR showed an accumulation of cytolytic TEMRA CD8 T cells in the blood and in the kidney graft. We showed that these enhanced migratory features were restricted to the KTs as those of the TEMRA CD8 and EM CD8 T cells from the HVs did not differ; additionally, the transmigration rate was higher for the TEMRA CD8 T cells from the KTs than the TEMRA CD8 T cells from the HVs. We also demonstrated that IL-15 enhances the generation of sLex from glucose and the addition of sLex onto core 2 O-glycans of PSGL and thereby, favors the migration of TEMRA and EM CD8 T cells. Our results also identified new targets to control the migration of TEMRA CD8 directly by targeting functional PSGL1, VLA-4, or LFA-1 or indirectly by interfering with glycolysis. Finally, we found that CXCL12 not only chemoattracts TEMRA and EM CD8 T cells but is also a potent activator of their immune function with a higher susceptibility of TEMRA CD8 from KTs. CXCL12 directly triggers a stimulatory signal with an increase in cytosolic Ca2+, a Panx1-dependent release of eATP that selectively activates the purinergic receptor P2 × 4. Combined with TCR-derived signals, CXCL12 activates the cytotoxic response of TEMRA and EM CD8 T cells, demonstrating that CXCL12 acts as a costimulatory signal for TEMRA and EM CD8 T cells.
The TEMRA CD8 T cells from the KTs exhibited enhanced migratory properties compared with the EM CD8 T cells and suggest that TEMRA CD8 T cells are preferentially recruited into the kidney graft (Figure 8). The results from single-cell transcriptomic analysis of kidneys54,55 also indicated the in situ recruitment of TEMRA CD8 T cells and their contribution to the inflammatory response. First, single-cell transcriptomic analysis of kidney transplant biopsies with mixed cellular and humoral rejection showed that CXCL12 is expressed by pericytes, fibroblasts, and myofibroblasts and that the cognate receptor CXCR4 is expressed by T cells and monocytes.55 T cells infiltrating the kidney biopsy were preferentially annotated as CD8 memory, CD8 effector GZMH, and CD8 effector GZMK T cells.55 Then, the single-cell transcriptomic analysis of kidney biopsies from patients diagnosed with lupus nephritis demonstrated that CXCL12 was detected in CD16+ monocytes and epithelial cells that were also the main source of CX3CL1.54 CXCR4 was detected in almost all infiltrating cell clusters, whereas CX3CR1 expression was restricted to CD56dimCD16+ NK cells, myeloid cells, and cytotoxic CD8 T cells. Strong infiltration of cytotoxic CD8 T cells and NK cells was observed.54 As we showed that protein expression of CX3CR1 is restricted to TEMRA CD8 T cells (Supplemental Figure 3B), these CD8 T cells could be TEMRA CD8 T cells that migrated in the kidney. This hypothesis is supported by the strong enrichment of two signatures of TEMRA-related genes in gene expression data of kidney biopsies from KTs with ABMR (Figure 3C) and the accumulation of CD8 T cells in the kidneys from KTs with ABMR (Figure 3, A and B). Similar observations were recently reported by Lamarthée et al.56 (preprint) using single-cell RNA-seq of kidney graft biopsies. Interestingly, CD8 TEMRA was equally recipient and donor derived, suggesting that TEMRA CD8 exhibits tissue residency after transplantation.
Figure 8.
TEMRA CD8 T cells from KTs exhibit enhanced purinergic P2 × 4 receptor–dependent proinflammatory and migratory responses. The potential therapeutic targets to control the migration of CD8 T cells are shown in red. ABMR, antibody mediated rejection; WORT, wortmannin; CCCP, carbonyl cyanide m-chlorophenyl hydrazone.
Collectively, these results suggested that TEMRA CD8 T cells from KTs may preferentially migrate to the kidney graft compared with EM CD8 T cells in response to CXCL12 secreted by nonimmune and immune cells, leading to the generation and exacerbation of the local inflammatory response that will ultimately contribute to kidney graft failure (Figure 8). Activation of TEMRA CD8 could result either from stimulation of the donor-specific TCR or from the binding of antigen-antibody complexes to CD16 on TEMRA CD8.18 Interestingly, patients with severe coronavirus disease 2019 accumulate highly activated CD16+ TEMRA.57 CD16 engagement leads to activation of TEMRA CD8 from patients with severe coronavirus disease 2019 and promotes the release of monocyte and neutrophil chemoattractants (CCL2 and XCL8) from endothelial cells, and the complement cleavage product C3a enhances the formation of CD16+ T cells.
IL-15 is an essential cytokine known to regulate many functions of CD8 memory T cells, including the homeostatic renewal, proliferation, survival, effector function, migration, and maintenance of various CD8 memory T cell subsets.58 For instance, the formation of tissue-specific effector and memory CD8 T cell can be inhibited by blocking of IL-15. One of the main characteristics of IL-15 is that it functions as both a homeostatic cytokine and an inflammatory cytokine. Infections and various forms of inflammatory challenges result in increased IL-15 in the plasma of patients. Among hematopoietic cells, IL-15 is mainly expressed by monocytes, inflammatory monocytes, macrophages, neutrophils, and dendritic cells. Unlike naive CD8 T cells, memory CD8 T cells acquire the ability to migrate from the circulation to nonlymphoid tissues without the need for reactivation by antigen-presenting cells. Generation of ligands for adhesion molecules P- and E-selectin is essential in the process of migration to nonlymphoid tissues, and IL-15 signaling promotes the generation of functional ligands for P- and E-selectin molecules in murine memory CD8 T cells.59 In the context of kidney transplantation, elevated levels of IL-15 are detected in allograft-rejected kidneys,60,61 and many cells that memory CD8 T cells might encounter in the renal graft could produce IL-15, including renal tubular epithelial cells,62,63 endothelial cells,64 monocytes, and dendritic cells. Post-transplant renal tubulitis was linked to IL-15 production by renal tubular epithelial cells.63 We have also shown that IL-15–stimulated TEMRA from KTs promotes inflammation by inducing CX3CL1 expression by endothelial cells in an IFN-γ– and TNF-α–dependent manner.20 We now provide evidence that IL-15 promotes the migratory ability of CD8 TEMRA and that CD8 TEMRA in KTs shows a greater susceptibility to migrate to nonlymphoid tissues. Increased production of IL-15 in the circulation and kidney could, therefore, promote the activation and local recruitment of CD8 TEMRA in patients undergoing rejection and stimulate the inflammatory process.
The rise in eATP is an archaic danger signal used by eukaryotes to detect cellular damage65 and results in the activation of innate and adaptive immunity during inflammation. eATP is preferentially detected by the purinergic P2× receptor family (P2 × 1–7) with different sensitivities to eATP concentrations. P2 × 4 is the most sensitive purinergic receptor, whereas P2 × 7 activation requires a much higher concentration of ATP.43 Lymphocyte heterogeneity must be considered in the analysis of eATP and the role of P2×. For instance, stimulation of P2 × 7 enhances the generation of Th166 and Th1767 CD4 T cells, whereas it leads to the pore-induced cell death of regulatory CD4 T cells.68 P2 × 7 is dispensable for the generation of murine short-lived effector CD8 T cells, whereas it is required for the establishment of functional long-lived central and tissue-resident memory CD8 T cells through fine tuning of the metabolic fitness of CD8 memory T cells.69 Inhibition of P2 × 4 prevented CD4 T cell migration in vitro and in vivo in response to CXCL12 stimulation10 and delayed murine lung allograft rejection.70 We demonstrated that the CXCL12-induced activation of TEMRA CD8 T cells relies on P2 × 4 and not on P2 × 1 or P2 × 7 receptors. Specific inhibition of P2 × 4 efficiently prevented the cytotoxic function of TEMRA CD8 T cells and was observed in resting situations (i.e., HVs) as well as in the context of strong alloimmune challenge (i.e., KTs). eATP can be released either by a passive mechanism (by dying and/or inflamed cells) or by active release via the Panx1 cell membrane transporter.71 Resident kidney endothelial and epithelial cells have been shown to be sources of eATP,72 and we demonstrated here that human CD8 memory T cells release eATP in Panx1-dependent fashion upon stimulation by CXCL12. Interestingly, we previously showed that TEMRA and EM CD8 T cells exhibit high levels of preformed ATP compared with naïve CD8 T cells.20 Given the enhanced migratory properties of the TEMRA CD8 T cells from the KTs compared with the EM CD8 T cells from the KTs and the TEMRA CD8 T cells from the HVs, we propose that the TEMRA CD8 T cells from the KTs favor and enhance the generation of an inflammatory context, including the generation of a local eATP cloud by passive and active releases (Figure 8).
Our findings suggest that current immunosuppressive drugs are more effective at controlling EM than TEMRA CD8 T cells and strengthen the need to specifically control TEMRA CD8 T cells by preventing their activation and/or their migration into inflamed tissues. This study identified a number of possible therapeutic targets that can be used to prevent TEMRA CD8 T cells migration and their activation in some cases (Figure 8). Preventing the interactions between PSGL1/P-selectin or VLA-4/VCAM-1 and interfering with CXCL12-derived signals represent two methods to prevent the migration of TEMRA CD8 T cells. Anti-PSGL1 mAb and recombinant PSGL1 Ig have been tested successfully in various preclinical models, but the translation from convincing experimental models to clinical efficacy was shown to be difficult.73 An alternative approach would be to use glycomimetics: synthetic compounds with similar structures to those of native carbohydrates.73 Panselectin inhibitors, such as bimosiamose tested in psoriasis74 or targeted small inhibitors targeting GSnP-6,75 could be used to prevent TEMRA CD8 T cell infiltration. We show that KT TEMRA CD8 T cells but not KT EM CD8 T cells were susceptible to the disruption of VLA-4/VCAM-1, and we provide evidence that 1-year treatment with natalizumab resulted in the decrease of the effector function of TEMRA CD8 in patients with multiple sclerosis. Therefore, KTs identified as high risk of kidney graft failure on the basis of both clinical metrics and a high frequency of TEMRA CD8 T cells18 may benefit from a natalizumab repositioning. Finally, we demonstrated that P2 × 4 inhibitors could efficiently control the activation of both TEMRA and EM CD8 T cells induced by TCR and CXCL12, a potential benefit in controlling the diversity of CD8 memory T cells compared with standard immunosuppressive drugs. Targeting TEMRA CD8 T cells with P2 × 4 inhibitors could, therefore, become a novel approach for controlling their migration and activation. Additional experiments are needed to evaluate the costs and benefits of targeting metabolic processes or purinergic receptors to inhibit the inflammatory response triggered by TEMRA CD8 T cells.
Disclosures
M. Cadoux reports employment with BioMAdvanced Diagnostics. M. Cyr reports employment with IsoPlexis, ownership interest in IsoPlexis, and research funding from IsoPlexis. R. Danger reports ownership interest in BioMAdvanced Diagnostics Company and is a cofounder of BioMAdvanced Diagnostics Company. D. Laplaud reports consultancy agreements with Alexion, Biogen, BMS-Celgene, Merck, MSD, Novartis, Roche, and Sanofi; research funding from Roche; and an advisory or leadership role with Biogen, BMS, Merck, and Roche. R. Liblau reports consultancy agreements with Biogen, GSK, Merck-Serono, Novartis, Orion, and Sanofi-Genzyme; research funding from Nolan and Roche; and speakers bureau for Biogen, Merck-Serono, Novartis, and Sanofi-Genzyme. W. Ni reports employment with IsoPlexis, ownership interest in IsoPlexis, and research funding from IsoPlexis. All remaining authors have nothing to disclose.
Funding
This work was funded by a grant from the LabEX IGO program supported by Agence Nationale de la Recherche (ANR) grant ANR-11-LABX-0016-01 via the “Investment into the Future” program and supported in part by Fondation Centaure. This work was performed as a part of the IHU-Cesti project, which received financial support from the French government managed by ANR grant ANR-10-IBHU-005 via the “Investment into the Future” program. Funding was also received from Agence Nationale de la Recherche grant ANR-10-INBS-04, the ANR project BIKET (ANR-17-CE17-0008), and the ANR project KTD-innov (ANR-17-RHUS-0010). The IHU-Cesti project is also supported by Nantes Metropole and the Pays de la Loire Region. Additionally, this work was supported by Fondation pour l'Aide à la Recherche sur la Sclérose en Plaques, Fondation de Coopération Scientifique Campus Paris-Saclay, and the FP7 VISICORT project, which has received funding from the European Union’s Seventh Framework Programme for research, technologic development, and demonstration under FP7 Health grant 602470.
Supplementary Material
Acknowledgments
The authors thank the HVs and the patients for their participation in the study; the DTC core facility and the IBISA MicroPICell facility (Biogenouest), a member of the national infrastructure France-Bioimaging supported by Agence Nationale de la Recherche grant ANR-10-INBS-04, for their excellent technical support; and B. Pignolet for the MS patient’s biobank.
We thank the biologic resource center for biobanking (CHU Nantes, Hôtel Dieu, Tumorothèque, Nantes, France). The Biological Resource Center of the Nantes University Hospital, France (BRIF: BB-0033-00040) guarantees the quality of the biologic samples.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Contributor Information
Collaborators: DIVAT Consortium, Gilles Blancho, Julien Branchereau, Diego Cantarovich, Agnès Chapelet, Jacques Dantal, Clément Deltombe, Lucile Figueres, Claire Garandeau, Magali Giral, Caroline Gourraud-Vercel, Maryvonne Hourmant, Georges Karam, Clarisse Kerleau, Christophe Masset, Delphine Kervela, Sabine Lebot, Aurélie Meurette, Simon Ville, Christine Kandell, Anne Moreau, Karine Renaudin, Anne Cesbron, Florent Delbos, Alexandre Walencik, and Anne Devis
Author Contributions
S. Brouard, S. Bruneau, M. Cyr, R. Danger, N. Degauque, M. Giral, J. Harb, C. Masset, W. Ni, C. Pecqueur, and A. Vivet conceptualized the study; O. Bonizec, M. Giral, P. Guérif, R. Liblau, H.L. Mai, and C. Masset were responsible for data curation; O. Bonizec, S. Bruneau, M. Cadoux, M. Cyr, N. Degauque, T.-M. Doan Ngoc, A. Garcia, A. Glemain, C. Masset, W. Ni, C. Pecqueur, G. Tilly, and A. Vivet were responsible for investigation; S. Blandin, O. Bonizec, S. Bruneau, M. Cadoux, M. Cyr, R. Danger, N. Degauque, T.-M. Doan Ngoc, L. Dubreil, M. Feyeux, A. Garcia, J. Harb, H.L. Mai, C. Masset, W. Ni, G. Tilly, and A. Vivet were responsible for formal analysis; S. Blandin, M. Cyr, R. Danger, N. Degauque, T.-M. Doan Ngoc, L. Dubreil, M. Feyeux, A. Garcia, J. Harb, C. Masset, W. Ni, and G. Tilly were responsible for methodology; N. Degauque and G. Tilly were responsible for project administration; S. Bruneau, M. Cyr, A. Garcia, M. Giral, A. Glemain, P. Guérif, D. Laplaud, R. Liblau, W. Ni, and C. Pecqueur were responsible for resources; R. Danger and M. Feyeux were responsible for software; N. Degauque was responsible for validation; M. Cyr, R. Danger, N. Degauque, L. Dubreil, and M. Feyeux were responsible for visualization; S. Brouard, N. Degauque, and C. Pecqueur were responsible for funding acquisition; N. Degauque, C. Pecqueur, and G. Tilly provided supervision; N. Degauque wrote the original draft; and S. Brouard, R. Liblau, and A. Vivet reviewed and edited the manuscript.
Data Sharing Statement
Original data reported in this paper of the type of experimental data have been deposited in the Gene Expression Omnibus (accession no. GSE198296).
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2022030286/-/DCSupplemental.
Supplementary Table 1. List of mAbs used.
Supplementary Table 2. List of main reagents used.
Supplementary Table 3. Gene signature of TEMRA CD8 on the basis of a review of the literature.
Supplementary Table 4. Differential expressed genes in TEMRA CD8 versus EM CD8 in KTs.
Supplementary Figure 1. Overall study design.
Supplementary Figure 2. Expansion of TEMRA CD8 in KTs with biopsy-proven ABMR.
Supplementary Figure 3. Migratory properties of memory CD8 T cells (TEMRA and EM): adhesion to activated HDMECs and chemokine screening by transmigration assays.
Supplementary Figure 4. Migratory properties of memory CD8 T cells (TEMRA and EM): adhesion to activated HDMECs and chemokine screening by transmigration assays.
Supplementary Figure 5. CXCL12 acts as a costimulatory signal for memory (TEMRA and EM) CD8 T cells.
Supplementary Figure 6. Short-term IL-15 stimulation enhances the selectivity of memory (TEMRA and EM) CD8 T cells to P-selectin.
Supplementary Figure 7. Inhibition of glycolysis impairs the generation of functional PSGL1.
References
- 1.Kapoor R, Ho P-R, Campbell N, Chang I, Deykin A, Forrestal F, et al. ; ASCEND investigators : Effect of natalizumab on disease progression in secondary progressive multiple sclerosis (ASCEND): A phase 3, randomised, double-blind, placebo-controlled trial with an open-label extension. Lancet Neurol 17: 405–415, 2018 [DOI] [PubMed] [Google Scholar]
- 2.Lebwohl M, Tyring SK, Hamilton TK, Toth D, Glazer S, Tawfik NH, et al. ; Efalizumab Study Group : A novel targeted T-cell modulator, efalizumab, for plaque psoriasis. N Engl J Med 349: 2004–2013, 2003 [DOI] [PubMed] [Google Scholar]
- 3.Feagan BG, Rutgeerts P, Sands BE, Hanauer S, Colombel J-F, Sandborn WJ, et al. ; GEMINI 1 Study Group : Vedolizumab as induction and maintenance therapy for ulcerative colitis. N Engl J Med 369: 699–710, 2013 [DOI] [PubMed] [Google Scholar]
- 4.Sandborn WJ, Feagan BG, Rutgeerts P, Hanauer S, Colombel J-F, Sands BE, et al. ; GEMINI 2 Study Group : Vedolizumab as induction and maintenance therapy for Crohn’s disease. N Engl J Med 369: 711–721, 2013 [DOI] [PubMed] [Google Scholar]
- 5.Schulz O, Hammerschmidt SI, Moschovakis GL, Förster R: Chemokines and chemokine receptors in lymphoid tissue dynamics. Annu Rev Immunol 34: 203–242, 2016 [DOI] [PubMed] [Google Scholar]
- 6.Randolph GJ, Ivanov S, Zinselmeyer BH, Scallan JP: The lymphatic system: Integral roles in immunity. Annu Rev Immunol 35: 31–52, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mueller SN, Gebhardt T, Carbone FR, Heath WR: Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol 31: 137–161, 2013 [DOI] [PubMed] [Google Scholar]
- 8.Abram CL, Lowell CA: The ins and outs of leukocyte integrin signaling. Annu Rev Immunol 27: 339–362, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowe JB: Glycosylation in the control of selectin counter-receptor structure and function. Immunol Rev 186: 19–36, 2002 [DOI] [PubMed] [Google Scholar]
- 10.Ledderose C, Liu K, Kondo Y, Slubowski CJ, Dertnig T, Denicoló S, et al. : Purinergic P2X4 receptors and mitochondrial ATP production regulate T cell migration. J Clin Invest 128: 3583–3594, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKinney EF, Lyons PA, Carr EJ, Hollis JL, Jayne DRW, Willcocks LC, et al. : A CD8+ T cell transcription signature predicts prognosis in autoimmune disease. Nat Med 16: 586–591, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKinney EF, Lee JC, Jayne DRW, Lyons PA, Smith KGC: T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 523: 612–616, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Néel A, Bucchia M, Néel M, Tilly G, Caristan A, Yap M, et al. : Dampening of CD8+ T cell response by B cell depletion therapy in antineutrophil cytoplasmic antibody-associated vasculitis. Arthritis Rheumatol 71: 641–650, 2019 [DOI] [PubMed] [Google Scholar]
- 14.Tasaki S, Suzuki K, Nishikawa A, Kassai Y, Takiguchi M, Kurisu R, et al. : Multiomic disease signatures converge to cytotoxic CD8 T cells in primary Sjögren’s syndrome. Ann Rheum Dis 76: 1458–1466, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuschiotti P, Medsger TA Jr., Morel PA: Effector CD8+ T cells in systemic sclerosis patients produce abnormally high levels of interleukin-13 associated with increased skin fibrosis. Arthritis Rheum 60: 1119–1128, 2009 [DOI] [PubMed] [Google Scholar]
- 16.Gross CC, Meyer C, Bhatia U, Yshii L, Kleffner I, Bauer J, et al. : CD8+ T cell-mediated endotheliopathy is a targetable mechanism of neuro-inflammation in Susac syndrome. Nat Commun 10: 5779, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yap M, Boeffard F, Clave E, Pallier A, Danger R, Giral M, et al. : Expansion of highly differentiated cytotoxic terminally differentiated effector memory CD8+ T cells in a subset of clinically stable kidney transplant recipients: A potential marker for late graft dysfunction. J Am Soc Nephrol 25: 1856–1868, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacquemont L, Tilly G, Yap M, Doan-Ngoc T-M, Danger R, Guérif P, et al. : Terminally differentiated effector memory CD8+ T cells identify kidney transplant recipients at high risk of graft failure. J Am Soc Nephrol 31: 876–891, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Setoguchi R: IL-15 boosts the function and migration of human terminally differentiated CD8+ T cells by inducing a unique gene signature. Int Immunol 28: 293–305, 2016 [DOI] [PubMed] [Google Scholar]
- 20.Tilly G, Doan-Ngoc T-M, Yap M, Caristan A, Jacquemont L, Danger R, et al. : IL-15 harnesses pro-inflammatory function of TEMRA CD8 in kidney-transplant recipients. Front Immunol 8: 778, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trombetta JJ, Gennert D, Lu D, Satija R, Shalek AK, Regev A: Preparation of single-cell RNA-seq libraries for next generation sequencing. Curr Protoc Mol Biol 107: 4.22.1–4.22.17, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. : Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Law CW, Chen Y, Shi W, Smyth GK: voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15: R29, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu G, Wang L-G, Han Y, He Q-Y: clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16: 284–287, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. : Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102: 15545–15550, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Creelan BC, Wang C, Teer JK, Toloza EM, Yao J, Kim S, et al. : Tumor-infiltrating lymphocyte treatment for anti-PD-1-resistant metastatic lung cancer: A phase 1 trial. Nat Med 27: 1410–1418, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spiegel JY, Patel S, Muffly L, Hossain NM, Oak J, Baird JH, et al. : CAR T cells with dual targeting of CD19 and CD22 in adult patients with recurrent or refractory B cell malignancies: A phase 1 trial. Nat Med 27: 1419–1431, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JB, Khan DH, Hurren R, Xu M, Na Y, Kang H, et al. : Venetoclax enhances T cell-mediated antileukemic activity by increasing ROS production. Blood 138: 234–245, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossi J, Paczkowski P, Shen Y-W, Morse K, Flynn B, Kaiser A, et al. : Preinfusion polyfunctional anti-CD19 chimeric antigen receptor T cells are associated with clinical outcomes in NHL. Blood 132: 804–814, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue Q, Bettini E, Paczkowski P, Ng C, Kaiser A, McConnell T, et al. : Single-cell multiplexed cytokine profiling of CD19 CAR-T cells reveals a diverse landscape of polyfunctional antigen-specific response. J Immunother Cancer 5: 85, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. : QuPath: Open source software for digital pathology image analysis. Sci Rep 7: 16878, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. : Fiji: An open-source platform for biological-image analysis. Nat Methods 9: 676–682, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rieckmann JC, Geiger R, Hornburg D, Wolf T, Kveler K, Jarrossay D, et al. : Social network architecture of human immune cells unveiled by quantitative proteomics. Nat Immunol 18: 583–593, 2017 [DOI] [PubMed] [Google Scholar]
- 34.Buggert M, Vella LA, Nguyen S, Wu VH, Chen Z, Sekine T, et al. : The identity of human tissue-emigrant CD8+ T cells. Cell 183: 1946–1961, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bengsch B, Ohtani T, Khan O, Setty M, Manne S, O’Brien S, et al. : Epigenomic-guided mass cytometry profiling reveals disease-specific features of exhausted CD8 T cells. Immunity 48: 1029–1045, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Callemeyn J, Lerut E, de Loor H, Arijs I, Thaunat O, Koenig A, et al. : Transcriptional changes in kidney allografts with histology of antibody-mediated rejection without anti-HLA donor-specific antibodies. J Am Soc Nephrol 31: 2168–2183, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffith JW, Sokol CL, Luster AD: Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu Rev Immunol 32: 659–702, 2014 [DOI] [PubMed] [Google Scholar]
- 38.Xu J, Torres E, Mora AL, Shim H, Ramirez A, Neujahr D, et al. : Attenuation of obliterative bronchiolitis by a CXCR4 antagonist in the murine heterotopic tracheal transplant model. J Heart Lung Transplant 27: 1302–1310, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scimone ML, Felbinger TW, Mazo IB, Stein JV, Von Andrian UH, Weninger W: CXCL12 mediates CCR7-independent homing of central memory cells, but not naive T cells, in peripheral lymph nodes. J Exp Med 199: 1113–1120, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Busillo JM, Benovic JL: Regulation of CXCR4 signaling. Biochim Biophys Acta 1768: 952–963, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trebak M, Kinet J-P: Calcium signalling in T cells. Nat Rev Immunol 19: 154–169, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.North RA: Molecular physiology of P2X receptors. Physiol Rev 82: 1013–1067, 2002 [DOI] [PubMed] [Google Scholar]
- 43.Schenk U, Westendorf AM, Radaelli E, Casati A, Ferro M, Fumagalli M, et al. : Purinergic control of T cell activation by ATP released through pannexin-1 hemichannels. Sci Signal 1: ra6, 2008 [DOI] [PubMed] [Google Scholar]
- 44.Woehrle T, Yip L, Elkhal A, Sumi Y, Chen Y, Yao Y, et al. : Pannexin-1 hemichannel-mediated ATP release together with P2X1 and P2X4 receptors regulate T-cell activation at the immune synapse. Blood 116: 3475–3484, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Filippini A, Taffs RE, Sitkovsky MV: Extracellular ATP in T-lymphocyte activation: Possible role in effector functions. Proc Natl Acad Sci U S A 87: 8267–8271, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ley K, Kansas GS: Selectins in T-cell recruitment to non-lymphoid tissues and sites of inflammation. Nat Rev Immunol 4: 325–335, 2004 [DOI] [PubMed] [Google Scholar]
- 47.van der Windt GJW, O’Sullivan D, Everts B, Huang SC-C, Buck MD, Curtis JD, et al. : CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc Natl Acad Sci U S A 110: 14336–14341, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andresen L, Skovbakke SL, Persson G, Hagemann-Jensen M, Hansen KA, Jensen H, et al. : 2-deoxy D-glucose prevents cell surface expression of NKG2D ligands through inhibition of N-linked glycosylation. J Immunol 188: 1847–1855, 2012 [DOI] [PubMed] [Google Scholar]
- 49.Zygmunt BM, Węgrzyn A, Gajska W, Yevsa T, Chodaczek G, Guzmán CA: Mannose metabolism is essential for Th1 cell differentiation and IFN-γ production. J Immunol 201: 1400–1411, 2018 [DOI] [PubMed] [Google Scholar]
- 50.Chen S, Springer TA: An automatic braking system that stabilizes leukocyte rolling by an increase in selectin bond number with shear. J Cell Biol 144: 185–200, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McEver RP: Selectins: Initiators of leucocyte adhesion and signalling at the vascular wall. Cardiovasc Res 107: 331–339, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Constantin G, Majeed M, Giagulli C, Piccio L, Kim JY, Butcher EC, et al. : Chemokines trigger immediate β2 integrin affinity and mobility changes: Differential regulation and roles in lymphocyte arrest under flow. Immunity 13: 759–769, 2000 [DOI] [PubMed] [Google Scholar]
- 53.Walling BL, Kim M: LFA-1 in T cell migration and differentiation. Front Immunol 9: 952, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arazi A, Rao DA, Berthier CC, Davidson A, Liu Y, Hoover PJ, et al. ; Accelerating Medicines Partnership in SLE network : The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol 20: 902–914, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu H, Malone AF, Donnelly EL, Kirita Y, Uchimura K, Ramakrishnan SM, et al. : Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J Am Soc Nephrol 29: 2069–2080, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamarthée B, Callemeyn J, Herck YV, Antoranz A, Anglicheau D, Becker JU, et al. : Transcriptional and spatial profiling of the kidney allograft unravels a central role for FcyRIII+ innate immune cells in rejection. Medrxiv. (Preprint posted July 10, 2022) [DOI] [PMC free article] [PubMed]
- 57.Georg P, Astaburuaga-García R, Bonaguro L, Brumhard S, Michalick L, Lippert LJ, et al. ; PA-COVID-19 Study Group : Complement activation induces excessive T cell cytotoxicity in severe COVID-19. Cell 185: 493–512, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nolz JC, Richer MJ: Control of memory CD8+ T cell longevity and effector functions by IL-15. Mol Immunol 117: 180–188, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nolz JC, Harty JT: IL-15 regulates memory CD8+ T cell O-glycan synthesis and affects trafficking. J Clin Invest 124: 1013–1026, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pavlakis M, Strehlau J, Lipman M, Shapiro M, Maslinski W, Strom TB: Intragraft IL-15 transcripts are increased in human renal allograft rejection. Transplantation 62: 543–545, 1996 [DOI] [PubMed] [Google Scholar]
- 61.Strehlau J, Pavlakis M, Lipman M, Shapiro M, Vasconcellos L, Harmon W, et al. : Quantitative detection of immune activation transcripts as a diagnostic tool in kidney transplantation. Proc Natl Acad Sci U S A 94: 695–700, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shinozaki M, Hirahashi J, Lebedeva T, Liew FY, Salant DJ, Maron R, et al. : IL-15, a survival factor for kidney epithelial cells, counteracts apoptosis and inflammation during nephritis. J Clin Invest 109: 951–960, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weiler M, Rogashev B, Einbinder T, Hausmann MJ, Kaneti J, Chaimovitz C, et al. : Interleukin-15, a leukocyte activator and growth factor, is produced by cortical tubular epithelial cells. J Am Soc Nephrol 9: 1194–1201, 1998 [DOI] [PubMed] [Google Scholar]
- 64.Oppenheimer-Marks N, Brezinschek RI, Mohamadzadeh M, Vita R, Lipsky PE: Interleukin 15 is produced by endothelial cells and increases the transendothelial migration of T cells In vitro and in the SCID mouse-human rheumatoid arthritis model In vivo. J Clin Invest 101: 1261–1272, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heil M, Land WG: Danger signals - damaged-self recognition across the tree of life. Front Plant Sci 5: 578, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salles ÉM, Menezes MN, Siqueira R, Borges da Silva H, Amaral EP, Castillo-Méndez SI, et al. : P2X7 receptor drives Th1 cell differentiation and controls the follicular helper T cell population to protect against Plasmodium chabaudi malaria. PLoS Pathog 13: e1006595, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, et al. : ATP drives lamina propria T(H)17 cell differentiation. Nature 455: 808–812, 2008 [DOI] [PubMed] [Google Scholar]
- 68.Schenk U, Frascoli M, Proietti M, Geffers R, Traggiai E, Buer J, et al. : ATP inhibits the generation and function of regulatory T cells through the activation of purinergic P2X receptors. Sci Signal 4: ra12, 2011 [DOI] [PubMed] [Google Scholar]
- 69.Borges da Silva H, Beura LK, Wang H, Hanse EA, Gore R, Scott MC, et al. : The purinergic receptor P2RX7 directs metabolic fitness of long-lived memory CD8+ T cells. Nature 559: 264–268, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu K, Vergani A, Zhao P, Ben Nasr M, Wu X, Iken K, et al. : Inhibition of the purinergic pathway prolongs mouse lung allograft survival. Am J Respir Cell Mol Biol 51: 300–310, 2014 [DOI] [PubMed] [Google Scholar]
- 71.Wanhainen KM, Jameson SC, da Silva HB: Self-regulation of memory CD8 T cell metabolism through extracellular ATP signaling. Immunometabolism 1: e190009, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Solini A, Usuelli V, Fiorina P: The dark side of extracellular ATP in kidney diseases. J Am Soc Nephrol 26: 1007–1016, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patel MS, Miranda-Nieves D, Chen J, Haller CA, Chaikof EL: Targeting P-selectin glycoprotein ligand-1/P-selectin interactions as a novel therapy for metabolic syndrome. Transl Res 183: 1–13, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Friedrich M, Bock D, Philipp S, Ludwig N, Sabat R, Wolk K, et al. : Pan-selectin antagonism improves psoriasis manifestation in mice and man. Arch Dermatol Res 297: 345–351, 2006 [DOI] [PubMed] [Google Scholar]
- 75.Krishnamurthy VR, Sardar MYR, Ying Y, Song X, Haller C, Dai E, et al. : Glycopeptide analogues of PSGL-1 inhibit P-selectin in vitro and in vivo. Nat Commun 6: 6387, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.