Abstract
In this study, we characterized seven members of the cp32/18 family of supercoiled plasmids in Borrelia burgdorferi 297. Complete sequence analysis of a 21-kb plasmid (cp18-2) confirmed that the strain 297 plasmids are similar in overall content and organization to their B31 counterparts. Of the 31 open reading frames (ORFs) in cp18-2, only three showed sequence relatedness to proteins with known functions, and only one, a ParA/SopA ortholog, was related to nonborrelial polypeptides. Besides the lipoproteins, none of the ORFs appeared likely to encode a surface-exposed protein. Comparison with the B31 genomic sequence indicated that paralogs for most of the ORFs in cp18-2 can be identified on other genetic elements. cp18-2 was found to lack a 9- to 10-kb fragment present in the 32-kb homologs which, by extrapolation from the B31 cp32 sequences, contains at least 15 genes presumed to be unnecessary for plasmid maintenance. Sequence analysis of the lipoprotein-encoding variable loci provided evidence that recombinatorial processes within these regions may result in the acquisition of exogenous DNA. Pairwise analysis with random shuffling revealed that the multiple lipoproteins (Mlp; formerly designated 2.9 LPs) fall into two distinct homology groups which appear to have arisen by gene fusion events similar to those recently proposed to have generated the three OspE, OspF, and Elp lipoprotein families (D. R. Akins, M. J. Caimano, X. Yang, F. Cerna, M. V. Norgard, and J. D. Radolf, Infect. Immun. 67:1526–1532, 1999). Comparative analysis of the variable regions also indicated that recombination within the loci of each plasmid may occur independently. Last, comparison of variable loci revealed that the cp32/18 plasmid complements of the B31 and 297 isolates differ substantially, indicating that the two strains have been subject to divergent adaptive pressures. In addition to providing evidence for two different types of recombinatorial events involving cp32/18 plasmids, these findings underscore the need for genetic analysis of diverse borrelial isolates in order to elucidate the Lyme disease spirochete's complex parasitic strategies.
Borrelia burgdorferi, the most common arthropod vector-borne pathogen in North America, is maintained in a complex enzootic cycle involving Ixodes scapularis and a mammalian host, typically rodents (28, 40). B. burgdorferi is distinguished from other prokaryotes by the extraordinary complexity of its genome, which is composed of a linear chromosome and a variable plasmid complement consisting of as many as 21 linear and circular plasmids (6, 18, 20). Two lines of evidence implicate plasmid-encoded proteins in the physiological and antigenic changes which underlie the Lyme disease spirochete's adaptation to different host environments as well as virulence expression within the mammalian host. First, most of the differentially expressed borrelial genes identified to date are plasmid encoded (2, 3, 11, 12, 19, 26, 39, 43, 48, 51, 53, 58). Second, loss of plasmids during continuous in vitro cultivation results in diminished infectivity (35, 42, 61, 63). B. burgdorferi plasmids are also of interest because they contain multiple paralogous genes which appear to have undergone extensive recombination during their evolution (2, 5, 8, 10, 15, 20, 39, 45, 49, 64).
The publication of the near-complete genome of a B. burgdorferi B31 clone (20) was a major advance in our understanding of this pathogen's complex molecular and evolutionary biology. The entire genome of this strain was not published, however, due to the difficulties associated with the sequencing and assembly of a family of highly homologous 32-kb circular plasmids (10, 52). Recently, complete sequences for seven of these plasmids have become available from the B. burgdorferi genome database (BBGD; http://www.tigr.org). Each contains a large number of well-conserved open reading frames (ORFs) as well as two variable loci with genes encoding differentially expressed, polymorphic lipoproteins, some of which have been shown to be surface exposed (2, 3, 13, 19, 27, 32, 39, 49, 51, 53–55). Truncated forms of these supercoiled plasmids (designated cp18s) have been described for the N40 and 297 strains but not fully characterized (2, 50).
B. burgdorferi initially was thought to be a single species. In recent years, however, it has become apparent that B. burgdorferi sensu lato is composed of multiple distinct species and genomic groups (4, 24, 31, 59) and that this heterogeneity may relate to differences in disease manifestations (57). Although less marked, genetic diversity also has been detected among the B. burgdorferi sensu stricto isolates which cause Lyme disease in North America (30, 33), and recent findings suggest that this heterogeneity also translates into differences in invasive potential (29, 44, 60). An important implication of these findings is that genetic characterization of multiple B. burgdorferi isolates may be needed in order to understand the relationship(s) between genomic content, borrelial virulence, and disease pathogenesis. To this end, we have extended our previous studies of cp32/18-encoded, differentially expressed lipoproteins (1–3, 39, 62) by comprehensively analyzing the cp32/18 family of plasmids in the 297 strain, a human cerebrospinal fluid isolate (46). In addition to providing evidence for two different types of recombination events involving cp32/18 plasmids, these findings underscore the need for genetic analysis of diverse borrelial isolates in order to elucidate the Lyme disease spirochete's complex parasitic strategies.
MATERIALS AND METHODS
Bacterial strains and media.
Virulent low-passage B. burgdorferi 297, the source of DNA template for all PCR experiments, was passaged and maintained as previously described (1). Electrocompetent Escherichia coli DH5α and DH10B (Gibco/BRL Life Technologies Inc., Gaithersburg, Md.) cells were used for all transformations. Strains and transformants were grown on tryptone-yeast agar or broth supplemented with the appropriate antibiotic.
Nucleotide sequencing.
Greater than 85% of the cp18-2 sequence was derived by analyzing overlapping genomic clones from a Lambda Zap II library (Stratagene, La Jolla, Calif.) (3, 39). Oligonucleotides used as probes for library screening or as primers for PCR amplification of duplex probes are shown in Table 1. The region of cp18-2 extending from mlp9 to p21 (Fig. 1) was amplified by long-distance PCR (GeneAmp XL-PCR kit; Perkin-Elmer Corp., Branchburg, N.J.) using the forward and reverse primers mlp 5′-cons and p21-R+C, respectively (2). The PCR parameters consisted of a preincubation at 94°C for 3 min to fully denature the template DNA, 35 cycles of 94°C for 1 min and 58°C for 8 min, followed by a final incubation at 72°C for 10 min; PCR products were gel purified using the Geneclean glass milk procedure according to the instructions of the manufacturer (Bio101 Inc., Vista, Calif.). Nucleotide sequencing was performed with an Applied Biosystems Inc. model 373A automated DNA sequencer and PRISM ready reaction DyeDeoxy terminator cycle sequencing kits as instructed by the manufacturer (Applied Biosystems, Inc., Foster City, Calif.).
TABLE 1.
Oligonucleotide primers and probes used in this study
| Designation | Sequence | Purpose |
|---|---|---|
| p21-spec | 5′-GCAAGTAATAATGAGTTAAAAGTTAAGCAAAG-3′ | Library screening |
| p21-specR+C | 5′-CTTTGCTTAACTTTTAACTCATTATTACTTTGC-3′ | Long-distance PCRd |
| elpB2 5′ | 5′-GAGATCAATGGAGATATTGATG-3′ | Library screening |
| elpB2 3′ | 5′-CACCACCCCAAGTTTTGCGTATAG-3′ | Library screening |
| ospF-LD-F | 5′-CATCTATAACTTTGTTTGCCAATTCATTGC-3′ | Long-distance PCRbc |
| 2.10-LD-F | 5′-GGCCAACAACCAGTCTTGTTGTTTATTCGC-3′ | Long-distance PCRbc |
| 2.11-LD-F | 5′-GCCCCCCGTTGGTTTTGTTCTCTTATTTGC-3′ | Long-distance PCRb,c |
| elpA1-LD-F | 5′-GAGCTTTGAGCTTTAATTATTTTGAGCAGCACAAGTAGA-3′ | Long-distance PCRbc |
| elpA2-LD-F | 5′-CGGAATTTTTAATATATTTTTACGCTGTC-3′ | Long-distance PCRbc |
| elpB1-LD-F | 5′-CGTGAAGTGAAAGTGAAAAAAGTAGAAGTATCCG-3′ | Long-distance PCRbc |
| elpB2-LD-F | 5′-CGTGAAGTGAAAGTGAAAAAAGTAGAAGTATCCG-′ | Long-distance PCRbc |
| mlp 5′-cons | 5′-AAAGGAGTAACAATGAAAATYATCAAC-3a | Long-distance PCRd |
| mlp1-spec | 5′-GAGACGGTAGTGGCAACTTAATAGAGC-3′ | Long-distance PCRd |
| mlp3-spec | 5′-GTTACTACCTGCGGCAATGGTGGTTCC-3′ | Long-distance PCRd |
| mlp5-spec | 5′-AAGCTAATACTCCACAAGATTGTAAAAATAA-3′ | Long-distance PCRd |
| mlp7A-spec | 5′-CGGTATAGATACTTTCACGGCTCAAAGC-3′ | Long-distance PCRd |
| mlp7B-spec | 5′-CAATAGTGTCCAGCAAGCAAAGCAG-3′ | Long-distance PCRd |
| mlp8-spec | 5′-ACAGAGCAGTCAGATAGTACATGTGGTG-3′ | Long-distance PCRd |
| mlp9-spec | 5′-AATCCAAACAAATGTAATGATAATAAACATGG-3′ | Long-distance PCRd |
| mlp10-spec | 5′-CCACATACAAAACTTTAGTTAAAGAATCTCT-3′ | Long-distance PCRd |
| cp32-1 BBP41cons(R+C) | 5′-TCGCTTCTAACTTTCCTAGCGGTTAACTTCTG-3′ | Long-distance PCRc |
| cp32cons (4306)R+C | 5′-CTCGCTCCATAACACATAATGCAGTGTTA-3′ | Long-distance PCRb |
| cp32cons (8952)R+C | 5′-AAGTCTTACCACTTCAGAACTAAATCCA-3′ | Long-distance PCRb |
| cp32cons (11813)R+C | 5′-ACAACAAAAGTGTGTCTATGTACTTTAAGA-3′ | Long-distance PCRb |
| cp18/32 (8435) 3′ LD | 5′-TTTCTCTTAAGTATTTAACCTCTTAAGCTTTAAAACAGC-3′ | Long-distance PCRb |
Y, nucleotides C or T.
Deletion analysis.
Variable-locus sequence analysis.
mlp and ospE/ospF/elp linkage analysis.
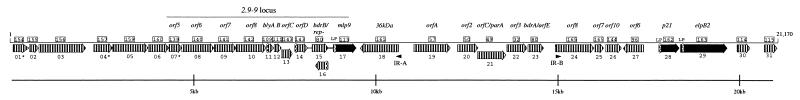
FIG. 1.
Schematic linear representation of cp18-2. ORFs are represented as open arrows. Striped arrows represent ORFs belonging to paralogous families in the B31 strain; the boxed numbers above each arrow indicate the Borrelia paralogous family to which the ORF belongs (BBGD). Black arrows designate the previously characterized, differentially expressed p21 and Mlp9 lipoproteins. Predicted leader peptides (LP) are represented by curved lines; IRs are indicated as solid arrowheads. The names of previously characterized proteins are given above the corresponding ORFs. Asterisks indicate possible pseudogenes. The approximate distances in base pairs are indicated below.
PCR amplification of the truncated portion of cp18-2.
Long-distance PCR to obtain the deleted portion of cp18-2 was performed as described above, using the forward and reverse primers listed in Table 1. The products were gel purified for sequencing as previously described.
Linkage of elpA1, elpA2, bbk2.10, and bbk2.11 with mlp genes and analysis of downstream variable regions.
Long-distance PCR to link elpA1, elpA2, bbk2.10, ospF, and bbk2.11 with mlp genes was performed as described above, using the forward and reverse primers listed in Table 1. The variable regions downstream from elpA1, elpA2, elpB1, ospF, bbk2.10, and bbk2.11 were obtained either as genomic clones or by PCR amplification using the forward and reverse primers listed in Table 1. PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen, Carlsbad, Calif.) and sequenced as described above.
Computer analysis.
Routine sequence analysis and multiple sequence alignments were performed using the MacVector version 6.5 software (Oxford Molecular Group Inc., Campbell, Calif.). ORFs were analyzed for potential signal peptides and cellular localization using PSORT (34). Transmembrane domains were predicted with the TMpred program (23). To generate an unrooted phylogenetic tree, pairwise sequence alignments were first generated using the ClustalW program (56). Sequence alignments were then analyzed using the Protdist and Neighbor programs included in the PHYLIP program suite (17). The resulting neighbor-joining tree was viewed using TreeView, version 1.5 (36). Binary comparisons with randomizations were generated using the GAP program from the Genetics Computer Group (Madison, Wis.) sequence analysis software package, version 10.0. Shuffle scores were calculated by subtracting the average quality score based on 100 randomizations from the quality score of the pairwise alignment; the adjusted quality score of the pairwise alignment then was divided by the standard deviation of the average quality score of randomizations. Scores are expressed as the number of standard deviations, with scores ≥9 indicating statistically significant alignments (41).
Nucleotide sequence accession numbers.
The entire sequence of 297 cp18-2 was submitted to GenBank under accession no. AF169008. Revised sequences for the variable ospE/ospF/elp loci have GenBank accession no. U18292 (bbk2.10), U30617 (bbk2.11), U19754 (ospF), AF023852 (ospE/elpB1), AF023853 (p21/elpB2), AF077602 (elpA1), and AF077603 (elpA2). Accession numbers for genes encoded within the 2.9 loci can be found in references 39 and 62.
RESULTS AND DISCUSSION
Strategy for molecular characterization of seven cp32/18 plasmids in B. burgdorferi 297.
The availability of complete sequences for seven cp32 plasmids from B. burgdorferi B31 (from the BBGD; see above), in concert with our own previous studies (2, 3, 39, 62), enabled us to devise a strategy for a molecular and evolutionary characterization of seven homologous plasmids in B. burgdorferi 297. The strategy was based on the assumption that, as in strain B31, each plasmid consists of large, highly conserved stretches with two interspersed variable regions. We initiated this effort by determining the entire sequence of a truncated family member, using a combination of overlapping genomic library clones and long-distance PCR products. In addition to its smaller size, this plasmid, designated cp18-2 (2), was an attractive candidate for complete sequence analysis for two reasons: (i) its variable loci encode lipoproteins (p21/ElpB2 and Mlp9, respectively) which are selectively expressed during mammalian infection (1, 2, 62) and (ii) loss of this plasmid occurs early during in vitro passage and correlates with attenuation of virulence in the murine model of Lyme disease (D. R. Akins, M. J. Caimano, and J. D. Radolf, unpublished data). Next, to confirm the overall similarity of cp32 plasmids in B. burgdorferi B31 and 297, we amplified from other strain 297 cp32 plasmids the fragment presumed to have been deleted from cp18-2. The two unique, variable lipoprotein-encoding loci in each plasmid were then fully sequenced and linked by long-distance PCR. Last, these results enabled us to assess how recombination has influenced the evolution of individual lipoprotein families as well as the variable regions within the plasmids.
Complete sequence of cp18-2. (i) Content and organization.
A linear representation of cp18-2 is shown in Fig. 1. The plasmid contains a total of 21,170 bp, 86% of which is coding sequence. cp18-2 is very similar in overall gene content and organization to its untruncated B. burgdorferi B31 counterparts. This was expected given the high degree of intra- and interstrain similarity already demonstrated for this family of plasmids. Following the convention previously established by Fraser et al. (20); see also the BBGD website), the 31 ORFs were numbered sequentially; the truncated ORF immediately following the deletion junction (described in detail below) was arbitrarily designated ORF01. Interestingly, all but three ORFs are transcribed in the same direction. The salient features of the 31 genes and corresponding polypeptides are summarized in Table 2. Two genes (ORF04 and ORF07), in addition to ORF01, lack identifiable Shine-Dalgarno sequences and therefore may not be translated. All of the genes except ORF16 (rep- in the 2.9 locus [39] could be assigned to a paralogous family from B. burgdorferi B31 (Fig. 1 and Table 2). Paralogs for ORF16 are present in the B31 cp32s but were not annotated (BBGD). The reason for this omission is unclear given that at least one of the rep- genes in B. burgdorferi 297 has been shown to be transcribed during in vitro growth (39).
TABLE 2.
ORFs encoded on B. burgdorferi 297 cp18-2
| ORFa | Alternate name | Family | bp
|
Orientationb | Potential RBSc | No. of amino acids | Mol wt | pI | PSORT results
|
TMpred results
|
Borrelia paralogous family distribution
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5′ | 3′ | Leader | Leader | Location | No. of transmembrane domains | Position | Score | DNA typed | Paralog locatione | ||||||||
| 1 | 154 | 161 | 415 | 255 | F | 5′ truncated | 85 | 9,880 | 3.98 | N | C | 0 | NA | NA | C/L | A, L, M, N, O, P, Q, R, S | |
| 2 | 155 | 415 | 648 | 234 | F | AACGAA-11 | 78 | 9,343 | 6.59 | N | C | 0 | NA | NA | C/L | A, L, M, N, O, P, Q, R, S | |
| 3 | 156 | 659 | 1945 | 1,287 | F | TGAGG-5 | 429 | 47,854 | 9.5 | N | Ce | 1 | 134–153 | 919f | C | L, M, N, O, P, Q, R, S | |
| 4 | 157 | 2188 | 2649 | 462 | F | None | 154 | 17,557 | 8.93 | N | C | 0 | NA | NA | C/L | A, L, M, N, O, P, Q, R, S | |
| 5 | 159 | 2656 | 3603 | 948 | F | AAAGGA-6 | 316 | 36,059 | 7.96 | N | C | 0 | NA | NA | C/L | A, L, M, N, O, P, Q, R, S | |
| 6 | 160 | 3624 | 4175 | 552 | F | AAAGGG-14 | 184 | 20,764 | 6.23 | N | C | 0 | NA | NA | C/L | A, L, M, N, O, P, Q, R, S | |
| 7 | 2.9 orf5 | 139 | 4208 | 4537 | 330 | F | None | 110 | 12,848 | 5.68 | N | C | 0 | NA | NA | C/L | A, L, M, N, O, P, Q, R, S |
| 8 | 2.9 orf6 | 140 | 4537 | 5412 | 876 | F | ATGGAGTT-6 | 292 | 33,272 | 5.02 | N | C | 0 | NA | NA | C/L | A, L, M, N, O, P, Q, R, S |
| 9 | 2.9 orf7 | 141 | 5421 | 6023 | 602 | F | ATGGATAG-9 | 201 | 22,731 | 9.83 | N | Ce | 1 | 50–67 | 684f | C/L | A, L, M, N, O, P, Q, R, S |
| 10 | 2.9 orf8 | 142 | 6036 | 6845 | 809 | F | AGGGGGG-4 | 270 | 30614 | 5.64 | N | C | 0 | NA | NA | C/L | A, L, M, N, O, P, Q, R, S |
| 11 | blyA | 109 | 6925 | 7125 | 201 | F | AAAGGATT-5 | 67 | 7,477 | 9.71 | Y, uncleaved | IM | 1 | 18–36 | 2876 | C | L, M, N, O, P, Q, R, S |
| 12 | blyB | 111 | 7132 | 7476 | 344 | F | AAAGAGA-7 | 115 | 13,200 | 5.59 | N | C | 0 | NA | NA | C | L, M, N, O, P, Q, R, S |
| 13 | 2.9 orfC | 143 | 7469 | 7801 | 333 | F | AAGGAAAG-8 | 111 | 12,698 | 0.18 | Y, uncleaved | IM | 0 | NA | NA | C/L | A, G, L, M, N, O, P, Q, R, S |
| 14 | 2.9 orfD | 143 | 7797 | 8145 | 349 | F | AAAGAGG-5 | 117 | 13,616 | 9.22 | Y, cleavedf | P/OMf | 1 | 5–22 | 1702 | C/L | A, G, L, M, N, O, P, Q, R, S |
| 15 | bdrB | 80 | 8255 | 8719 | 465 | F | ATAAGGGAA-5 | 155 | 17,853 | 4.94 | N | IM | 1 | 132–154 | 2639 | C/L | F, G, H, L, M, N, O, P, Q, R, S |
| 16 | rep- | NA | 8712 | 8371 | 342 | R | AAGTGAGG-16 | 114 | 10.38 | Y, cleavedg | IM | 3 | 3–23 | 629 | NA | ||
| 32–50 | 2037 | ||||||||||||||||
| 72–90 | 1786 | ||||||||||||||||
| 17 | mlp9 | 113 | 8828 | 9460 | 633 | F | AAAGGAG-5 | 211 | 24,081 | 5.04 | Y, uncleavedh | IMh | 1 | 3–19 | 1336 | C | L, M, N, O, P, Q, R, S |
| 18 | 36kDa | 161 | 10563 | 9547 | 1,016 | R | AACGGAGG-6 | 339 | 39,970 | 7.91 | N | C | 0 | NA | NA | C | C, L, M, N, O, P, Q, R, S |
| 19 | orfA | 57 | 10943 | 12040 | 1,098 | F | AACGGAGA-6 | 366 | 44,309 | 9.65 | N | C | 0 | NA | NA | C/L/X | X, A, C, D, E, F, G, H, I, L, M, N, O, P, Q, R, S, T, U |
| 20 | orf2 | 50 | 12054 | 12617 | 563 | F | TATGAGG-8 | 188 | 22,580 | 9.66 | N | C | 0 | NA | NA | C/ | A, B, C, E, F, G, H, I, J, K, L, M, N, O, P, Q, R, S |
| 21 | orfC/parA | 49 | 12596 | 13348 | 753 | F | CAAGGA-11 | 251 | 29,091 | 8.8 | N | Ce | 1 | 139–154 | 711e | C/L | A, B,C, E, F, G, H, I, J, K, L, M, N, O, P, Q, R, S |
| 22 | orf3 | 32 | 13393 | 13950 | 558 | F | TAAGGAG-5 | 186 | 22,011 | 9.51 | N | C | 0 | NA | NA | C/L/X | X, A, B, C, E, F, G, H, I, J, K, L, M, N, O, P, Q, R, S, U |
| 23 | bdrA | 80 | 13963 | 14484 | 522 | F | TAAGAGGA-4 | 174 | 20,268 | 5.58 | N | IM | 1 | 147–167 | 2324 | C/L | F, G, H, L, M, N, O, P, Q, R, S |
| 24 | orf8 | 165 | 15022 | 16050 | 1,029 | F | AACGGAGG-6 | 343 | 39,666 | 5.15 | N | C | 0 | NA | NA | C | C, L, M, N, O, P, Q, R, S |
| 25 | orf7 | 165 | 16072 | 16350 | 278 | F | AATGGAG-9 (L) | 93 | 10,848 | 9.51 | N | Ce | 1 | 69–89 | 833e | C | C, L, M, N, O, P, Q, R, S |
| 26 | orf10 | 144 | 16405 | 16827 | 423 | F | AGGAGAGG-6 | 141 | 16,745 | 9.61 | N | Ce | 1 | 52–75 | 999e | C/L | A, L, M, N, O, P, Q, R, S |
| 27 | orf6 | 96 | 17612 | 16839 | 774 | R | AAAGGAGAGG-5 | 258 | 30,270 | 9.9 | N | Ce | 1 | 74–90 | 550e | C/L | C, H, L, M, N, O, P, Q, R, S |
| 28 | p21 | 162 | 18043 | 18597 | 555 | F | TATGGAGA-6 | 185 | 20,747 | 7.19 | Y, cleaved | Lipoprotein | 1 | 9–26 | 1632 | C | L, N, P, R |
| 29 | elpB2 | 163 | 18628 | 19845 | 1,218 | F | AGGGGAGA-6 | 406 | 46,539 | 5.02 | Y, uncleaved | IMh | 1 | 6–21 | 1262 | C | F, L, N, O, P, Q |
| 30 | 114 | 19998 | 20387 | 390 | F | AAAAG-12 (L) | 130 | 15,661 | 9.64 | Y, uncleaved | IM | 1 | 1–20 | 803 | C | L, N, O, P, Q, R | |
| 31 | 115 | 20747 | 21169 | 423 | F | TTAGGAGA-6 | 141 | 16,214 | 9.22 | N | C | 0 | NA | NA | C | L, M, N, O, P, Q, R, S | |
Potential pseudogenes are indicated in boldface.
F, forward; R, reverse.
Number following each potential ribosome binding site (RBS) indicates the distance to the AUG start codon. ORFs with putative leucine start codons are indicated by (L).
Refers to the type of genetic element on which ORFs belonging to the designated paralogous family have been described in B31: C, circular plasmid; L, linear plasmid; X, chromosome (BBGD).
Discrepancy between the presence of a predicted transmembrane domain (TMpred) and cytoplasm localization (PSORT) most likely due to relatively low (<1,000) TMpred score.
Experimental studies demonstrated that the 2.9 OrfD leader peptide is not cleaved (39).
Atypical leader peptide.
Mlp leader peptide has been shown to be lipid modified in vitro (39). ElpB2 contains appropriate lipid modification signal sequences.
(ii) Genomic distribution of paralogous ORFs.
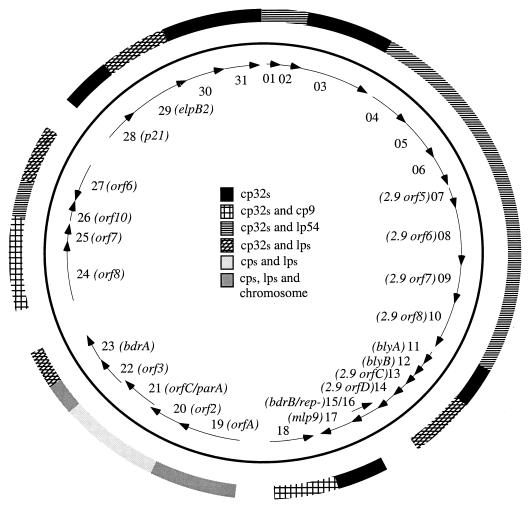
The B. burgdorferi genome, and its plasmid components in particular, has undergone extensive duplications and rearrangements throughout its evolution (5, 8, 15, 20, 39, 45, 49, 64). Lacking the genomic sequence for strain 297, we used the B31 sequence to examine the genomic distribution of cp18-2 paralogos. This analysis revealed that cp18-2 is a mosaic or patchwork consisting of small, plasmid-specific regions and a number of paralogous sequences distributed to various extents throughout the genome (Fig. 2 and Table 2). The widely distributed ORFs can be grouped into five categories. The first contains two genes (ORF19 and ORF22) with chromosomal, linear, and circular plasmid-encoded paralogs. The second contains two genes (ORF20 and ORF21) with paralogs present on nearly all of the B31 plasmids but not the chromosome. The third contains several ORFs with paralogs located on cp9 and the cp32s. The fourth contains members of 10 paralogous families also represented only on lp54, seven of which (ORF04 to ORF10) form a contiguous stretch on both plasmids (Fig. 2 and Table 2). The fifth group is heterogeneous and contains paralogs present on one or more plasmids (linear and/or circular) other than cp9 and lp54.
FIG. 2.
Distribution of cp18-2 paralogous families within the B. burgdorferi B31 genome.
(iii) Homologies with other proteins.
Database searches revealed that only three ORFs matched proteins with known functions. Two of these (ORF11 and ORF12; previously designated blyA and blyB, respectively) are both cp32/18 and B. burgdorferi specific and encode the hemolytic activity described by Guina and Oliver (21). The third gene (ORF21; previously designated orfC [64]) belongs to a paralogous family (family 49) whose members recently were shown by Stevenson et al. (49) to be related to the ParA/SopA protein involved in the partitioning of low-copy-number plasmids between daughter cells following cell division (22). As with the B31 paralogs, ORF21 contains the ATP-1 and ATP-2 motifs required for binding and hydrolysis of ATP as well as two motifs postulated to be involved in the binding of accessory proteins or in membrane attachment during partitioning (data not shown) (49). Stevenson and coworkers (49) also proposed that sequence variation among ParA/SopA homologs prevents incompatibility of cp32 homologs within individual borrelial strains; studies are currently under way to examine the sequence heterogeneity of the strain 297 paralogs. In par-mediated plasmid partitioning, ParA/SopA interacts with a protein, ParB/SopB, which complexes to a cis-acting site, parS/sopC, near the origin of plasmid replication (22). The applicability of this scenario to the partitioning of borrelial plasmids, however, is uncertain given that directed searches failed to identify a homolog for ParB or a site resembling parS/sopC in cp18-2 or, for that matter, in the B. burgdorferi B31 genome. If these functions are encoded on cp18-2, the relevant sequences evidently lack detectable homologies.
(iv) Export signals and transmembrane domains.
The 31 ORFs were analyzed for N-terminal export signals, transmembrane domains, and cellular location, using the PSORT and TMpred algorithms. A summary of these data are included in Table 2. PSORT identified N-terminal export signals on eight polypeptides (ORF11 [blyA], ORF13 [orfC], ORF14 [orfD], ORF16 [rep-], ORF17 [mlp9], ORF28 [p21], ORF29 [elpB2], and ORF30), seven of which have been characterized previously (2, 21, 39, 62). It is noteworthy that PSORT failed to recognize the signal peptidase II (SPase II) cleavage sites for two of the three lipoproteins (Mlp9 and ElpB2). Although lipid modification of ElpB2 has not been confirmed experimentally, an alkaline phosphatase fusion containing the Mlp9 leader peptide is lipidated when expressed in E. coli (39). Of the five nonlipoproteins with predicted leader peptides, only ORF14 and ORF16 were predicted to have SPase I cleavage sites. In both cases, however, these predictions are questionable. The putative leader sequence of ORF16 has three prolines within the hydrophobic core region and a predicted cleavage site 30 amino acids downstream from the end of the hydrophobic stretch. With respect to ORF14, previous analyses using Triton X-114 phase partitioning and in vivo lipid labeling of the recombinant protein expressed in E. coli indicated that the protein's leader peptide also is not cleaved (39). TMpred identified potential internal transmembrane domains, predictive of a cytoplasmic membrane location, in 10 ORFs, while one (ORF16) was predicted to have two internal transmembrane domains in addition to its leader peptide. PSORT, in contrast, failed to identify the putative transmembrane domains for 6 of these 10 polypeptides, placing them instead within the cytoplasmic compartment. These discrepancies probably reflect the relatively low TMpred scores (<1,000) for all seven membrane-spanning regions. The two proteins predicted by both PSORT and TMpred to possess internal transmembrane domains were ORF15 and ORF23. These two polypeptides are members of the family (family 80) of borrelia direct repeat (Bdr) proteins (formerly rep+ within the 2.9 loci [39] and orfE [64]) which characteristically possess an internal, tandemly repeated motif with K-I-D core sequence and C-terminal cytoplasmic membrane anchors (9, 65). The above analyses clearly highlight the need for caution when interpreting computer-based predictions of protein topology and cellular location. Equally important, they indicate that most of the coding capacity of the plasmid is devoted to polypeptides residing within intracellular compartments.
(v) Inverted repeats.
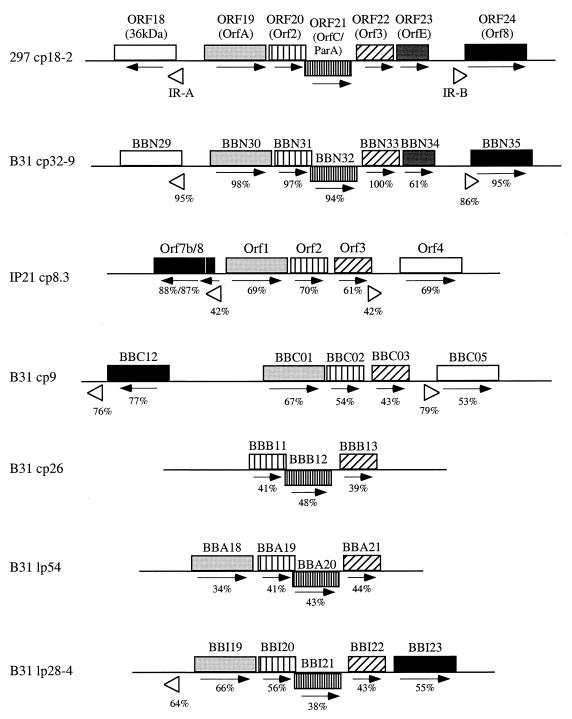
Flanking the stretch from ORF18 to ORF24 are 185- and 191-bp inverted repeats (IRs) which are 42% identical to the 183- and 184-bp IRs on cp8.3 of B. burgdorferi Ip21 (15) (Fig. 1 and 3). The cp18-2 IRs are similar to their cp8.3 counterparts in that they overlap the Shine-Dalgarno sequences and translational starts of the adjacent genes (Fig. 3). They differ, however, in that they are theoretically unable to form stem-loop structures comparable to those which were proposed to interfere with translation of cp8.3 transcripts (15) (data not shown). Three of the genes (ORF19, ORF20, and ORF22) flanked by the IRs in cp18-2 are paralogs for the genes flanked by the IRs in cp8.3; it is noteworthy that the parA/sopA homolog is one of the two genes missing from this region in cp8.3 (Fig. 3). A survey of the B31 genome revealed that similar IRs are found only on the cp32 family and cp9 and that the cp32 IRs flank the same five ORFs as in cp18-2 whereas the cp9 IRs flank the same three ORFs as in cp8.3. (20), BBGD. Variable combinations of these five ORFs are found in similar configurations on B31 linear plasmids which lack the IRs (Fig. 3). One possible interpretation of these observations is that the IR-flanked genes were acquired initially by cp32/18 and/or cp9 plasmids (perhaps via a transpositional insertion) and that various combinations of these genes subsequently recombined into other plasmids. The absence of the ParA/SopA homologs in cp8.3 and cp9 raises the intriguing possibility that the replication and partitioning mechanisms used by these small plasmids differ from those of the larger supercoiled plasmids. In this regard, it should be noted that plasmids which replicate as rolling circles typically are less than 10 kb (25) and that a B. burgdorferi chromosomal gene (BB0607) encodes a protein related to the DNA helicase involved in rolling circle replication in Bacillus subtilis (37).
FIG. 3.
Organization of IRs and/or related flanking ORFs in selected B. burgdorferi plasmids. ORFs are depicted as shaded boxes, with similarly shaded boxes indicating paralogous ORFs; the arrow below each ORF indicates the direction of transcription. Percent similarities between cp18-2 and corresponding paralogs (using amino acid sequences) and IRs are indicated.
Identification of the cp18-2 deletion.
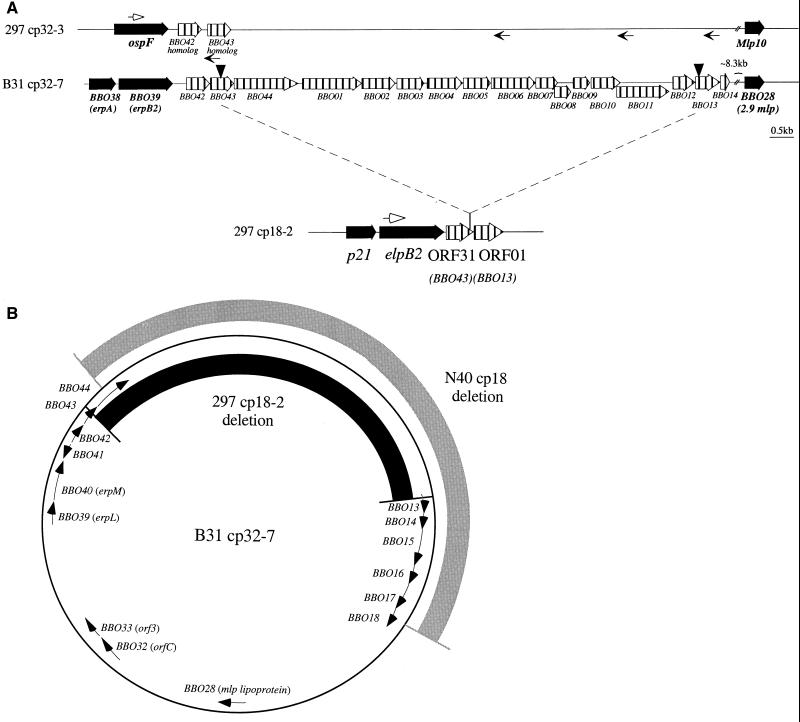
Stevenson and coworkers (50) showed that the truncated cp32 homolog in B. burgdorferi N40 lacks an approximate 12.5-kb fragment. However, because their study predated the availability of the B31 cp32 sequences, they were unable to identify the missing ORFs. Early in our sequencing effort, it became apparent that the N40 and 297 truncations differed substantially. This observation prompted us to define the deletion in cp18-2. Using 5′ primers specific for each of the separately encoded ospE, ospF, and elp genes and a reverse primer located within ORF01 (Table 1), 9- to 10-kb fragments were amplified from strain 297 DNA (Fig. 4A and data not shown). Additional PCRs using the same 5′ primers and nested reverse primers (Table 1) generated smaller fragments of the exact sizes predicted from the 297 and B31 sequences (Fig. 4A and data not shown). Together with the cp18-2 sequence, these results established the high degree of similarity between the strain B31 and 297 cp32s.
FIG. 4.
The cp18-2 deletion. (A) cp32 ORFs predicted to be deleted from cp18-2. Small arrows designate the nested primer pairs used to confirm the similarity between the cp18-2 deletion in a full-length 297 cp32 (represented by cp32-3) and the corresponding segments from the B31 homologs (represented by cp32-7). (B) The deletions in cp18-2 and cp18 of N40 are shown alongside a representative B31 plasmid (cp32-7).
The cp18-2 deletion junction consists of a fusion between ORF31 and ORF01 (Fig. 4A). As a result, ORF31 lacks the 45 amino acids at the C termini of its family 115 paralogs (represented by BBO43), possessing seven unique C-terminal residues instead (data not shown). ORF01, on the other side of the deletion junction, lacks both a Shine-Dalgarno sequence and translational start; from its second residue on, ORF01 is identical to its family 154 paralogs (beginning with amino acid 14) but terminates at amino acid 64 due to an independent frameshift (data not shown). A comparison of the cp18-2 and N40 cp18 deletions (Fig. 4B) shows that the N40 deletion begins within the first cp32 ORF deleted from cp18-2 (represented by BBO44) and extends five ORFs further downstream to the family 160 paralog (represented by BBO18). The simplest explanation for these overlapping deletions is that they occurred separately but involved genes which are nonessential for stable plasmid maintenance and/or which can be complemented in trans by functions encoded elsewhere. The latter possibility is supported by the presence of highly similar paralogs for each of the deleted ORFs on the full-length cp32/18 plasmids in strain B31 (BBGD). Database searches of the 15 B31 ORFs presumed to be encoded by the cp18-2 deletion failed to reveal significant homologies with other prokaryotic proteins. PSORT predicted that all 15 genes encode proteins lacking N-terminal export signals and that 13 of the 15 are cytoplasmic (data not shown), further underscoring the paucity of plasmid coding capacity devoted to surface-exposed proteins. TMpred predicted that three genes (BBO03, BBO08, and BBO44) encode proteins with a single internal transmembrane domain (data not shown). As before, the discrepancies between the two programs probably reflect the relatively low TMpred scores for these three putative membrane-spanning regions.
Evolutionary analysis of the cp32/18-specific, variable loci.
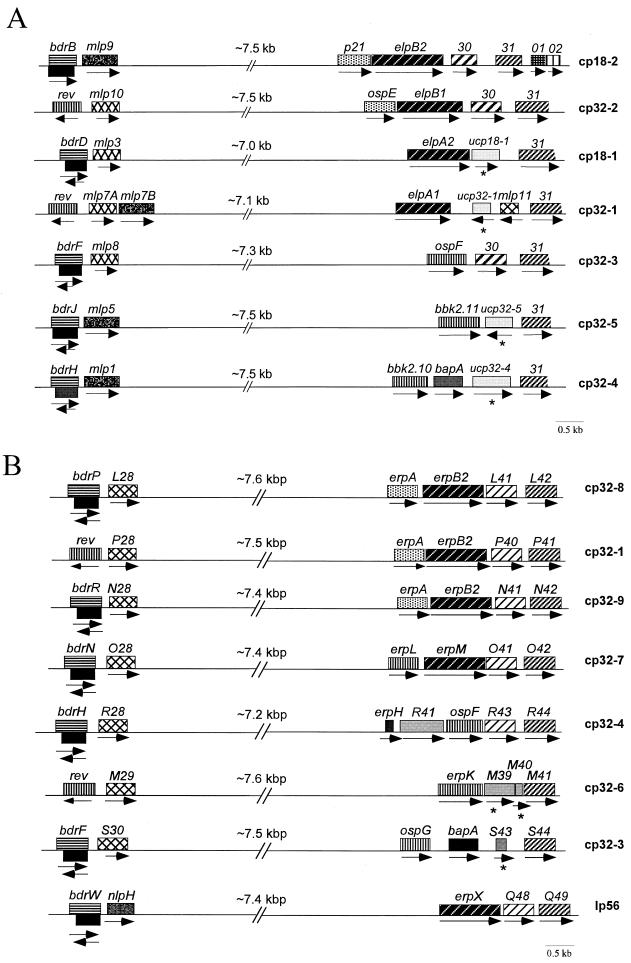
Sequence divergence among cp32/18 plasmids can be attributed primarily to variable regions defined by the genes encoding the Mlp (formerly 2.9-LP [39]) and OspE/OspF/Elp lipoprotein antigens (2). A characterization of these plasmid-specific loci, therefore, was essential for definition of the B. burgdorferi 297 homologs. Sequences for the loci encoding the Mlp lipoproteins were presented in two prior studies (39, 62). In a third report (2), we provided partial sequences for the seven OspE/OspF/Elp-encoding regions and established linkage between the loci containing the ospF, p21/elpB2, and ospE/elpB1 genes and 2.9 loci containing the mlp8, mlp9, and mlp10 genes, respectively (2). To complete this phase of the study, we sequenced all remaining gaps and linked the four unpaired ospE/ospF/elp loci with mlp loci by long-distance PCR.
Schematics for the variable loci of all seven plasmids are shown in Fig. 5A. Though corresponding variable loci are clearly similar in configuration, a variety of rearrangements, insertions, and deletions, believed to be reflective of past recombination events (2, 49), are readily apparent. Two discoveries relating to the ospE/ospF/elp loci are of particular importance because they suggest that these localized recombination events may involve exogenously acquired DNAs. The first is that a mlp paralog (mlp11) resides downstream from elpA1 in cp32-1; to the best of our knowledge, this is the only instance in which a mlp gene has been identified outside of a 2.9 locus. Phylogenetic analysis (Fig. 6B) and sequence alignments (not shown) demonstrate that this lipoprotein is an outlier from the other 297 Mlps, leading us to speculate that it was acquired from another borrelial strain. The second is that cp18-1, cp32-1, cp32-4, and cp32-5 from 297 contain nonparalogous ORFs in place of the cp18-2 ORF30 paralogs present in the other three plasmids. Because these unique circular plasmid-encoded (ucp) genes lack homologies with other sequences within the B31 genome, it seems highly plausible that they were acquired as the result of some form of horizontal gene transfer. In light of the relatively small proportion of total cp32/18 coding capacity devoted to exported proteins, it was of interest to note that three ucp genes (ucp32-1, ucp32-4, and ucp18-1) were predicted by PSORT to encode proteins with N-terminal export signals and that the signal sequences of two (ucp32-1 and ucp32-4) have putative SPase I cleavage sites.
FIG. 5.
Schematic representation of the variable loci in B. burgdorferi 297 (A) and B31 (B). Similarly shaded boxes are used to indicate paralogous ORFs; the arrow below each ORF indicates the direction of transcription. In panel A, paralogs of cp18-2 ORF30 and ORF31 are designated 30 and 31; nonparalogous ORFs are indicated by asterisks.
FIG. 6.
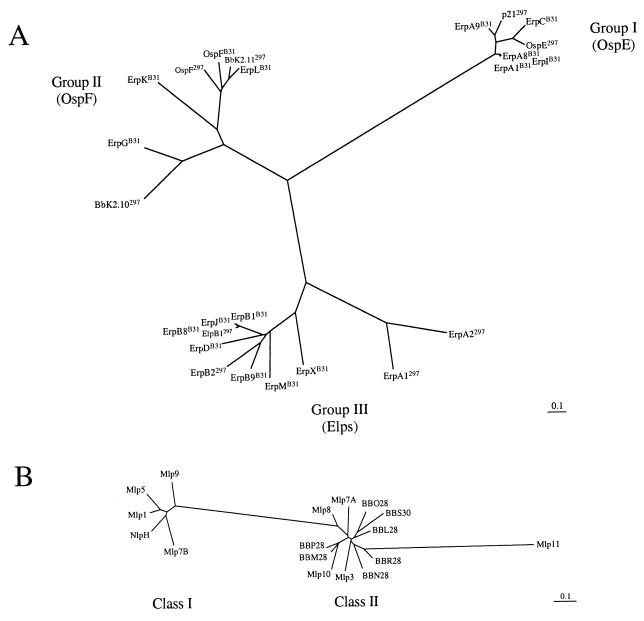
Unrooted neighbor-joining phylograms of OspE, OspF, and Elp (A) and Mlp (B) homologs in B. burgdorferi 297 and B31 cp32/18 plasmids. Scale bars represent the base pair substitutions per site.
Recently, we presented evidence that the OspE, OspF, and Elp homologs within the 297 and B31 strains arose as fusions between a common N terminus and three unrelated ancestral proteins (2). The phylogram in Fig. 6A supports this by demonstrating that these proteins segregate into three distinct homology groups. In light of the evidence for past rearrangements at the 2.9 loci (Fig. 5A), it seemed logical to consider the possibility that analogous fusion events contributed to the evolution of the Mlps. We began this line of inquiry by first assessing the phylogenetic relationships among Mlps; the phylograms in Fig. 6B show that the Mlps fall into two divergent classes. Sequence alignments show that the proteins in each class exhibit extensive sequence similarity across their entire lengths (Fig. 7A and B); sequence relatedness for the entire complement, in contrast, falls off strikingly after position 51 and is highly dependent on the introduction of large gaps in order to accommodate the smaller class II proteins (Fig. 7C). A similar degradation in sequence relatedness among the OspE, OspF, and Elp homologs was an important clue to the potential contribution of gene fusion to the evolutionary histories of these lipoproteins (2). Consequently, we examined the Mlps for common ancestry by calculating binary comparison scores for pairwise alignments with 100 randomizations (41). The resulting score is expressed in standard deviations, with a score equal to or greater than 9 standard deviations being strong evidence that the sequences in question arose from the same ancestral gene. As shown in Table 3, the binary comparison scores for full-length proteins were all significant and extremely so among proteins of the same class. These scores fell off markedly, and in many cases became insignificant, for interclass comparisons of proteins without their leader sequences (data not shown). When these comparisons were repeated with the first 51 amino acids of each protein omitted, significant scores were obtained only for lipoproteins within the same class (Table 3). The most straightforward interpretation of these results is that the two Mlp classes evolved from chimeric progenitors in which shared N termini were joined to unrelated proteins. One implication of this conclusion is that the tandemly arranged genes in cp32-1 (mlp7A/mlp7B) encoding class I and class II Mlp lipoproteins had to have arisen by a different mechanism than the gene duplication events thought to have generated other tandemly arrayed genes encoding lipoproteins (e.g., OspA and OspB) with sequence homology along their entire lengths.
FIG. 7.
Sequence relatedness among the Mlp lipoproteins. The sequences of class I (A), class II (B), and the entire complement of proteins (C) were aligned using the ClustalW algorithm. Shaded residues represent identical or conserved matches.
TABLE 3.
Binary comparison scores for B. burgdorferi 297 Mlp lipoproteinsa
| Protein | Mlp3 | Mlp5 | Mlp7A | Mlp7B | Mlp8 | Mlp9 | Mlp10 |
|---|---|---|---|---|---|---|---|
| Mlp1 | 18 [0] | 154 [113] | 18 [0] | 102 [91] | 26 [0] | 128 [76] | 29 [0] |
| Mlp3 | 12 [0] | 88 [63] | 18 [0] | 75 [58] | 16 [0] | 90 [56] | |
| Mlp5 | 14 [0] | 144 [107] | 28 [0] | 145 [94] | 30 [0] | ||
| Mlp7A | 20 [0] | 87 [61] | 17 [0] | 83 [62] | |||
| Mlp7B | 25 [0] | 129 [121] | 20 [0] | ||||
| Mlp8 | 25 [0] | 86 [65] | |||||
| Mlp9 | 31 [1] |
Binary comparison scores are indicated in standard deviations for pairwise alignments performed using either full-length proteins or proteins without the first 51 amino acids (bracketed). Statistically significant alignments (≥9 standard deviations) are in boldface.
To complete this analysis, we used the lipoprotein genes and other ORFs as physically linked markers to assess the interdependence of recombinatorial processes within the variable loci. The data in Fig. 5A and 6, taken together, argue strongly that recombination events occur independently at each locus. For example, whereas the p21/elpB2 and ospE/elpB1 loci of cp18-2 and cp32-2 are highly similar, they differ significantly at the 2.9 loci with respect to both the class of Mlp and the presence of bdr/rep- (cp18-2) as opposed to rev (cp32-2) genes. Similarly, cp32-3 and cp32-5 encode closely related ospF paralogs but have different downstream ORFs at this locus (an ORF30 paralog versus ucp32-5) as well as mlp genes of different classes at their 2.9 loci. Plasmids cp18-1 and cp32-1 both have elpA genes but otherwise differ markedly at both loci. Last, although the two truncated plasmids cp18-1 and cp18-2 both contain distantly related elp genes, one is bicistronic and paired with a lipoprotein gene (p21) of a different class, indicating a subsequent downstream insertion; at the other locus the two plasmids contain mlp genes of different classes. Interestingly, the importance of recombination as a mechanism for generating sequence diversity at the variable loci is less apparent if one repeats this analysis using the B31 cp32 plasmids. The combined data in Fig. 5B and 6 reveal the two principal reasons for this. The first is that there is less sequence diversity among the B31 variable-locus lipoproteins; this is particularly true for the Mlp-like paralogs which, with a single exception (NlpH on lp56), fall into a single homology group. The other is the greater redundancy of the erp loci, most notably the three virtually identical erpA/erpB2 pairings. A direct comparison of the 297 and B31 variable loci (Fig. 5A and B, respectively) is also noteworthy because it reveals numerous other differences in configuration and lipoprotein gene content, as well as the presence of unrelated nonparalogous genes, which, as a whole, may be indicative of divergent adaptive pressures acting on the two isolates.
Summary and conclusions.
In the present study, we exploited existing sequence information and the highly conserved nature of the cp32/18 plasmids in order to characterize seven homologs from B. burgdorferi 297. Though clearly sequence intensive, a major advantage of our strategy was that it enabled us to delineate the principal features of an entire family of plasmids without having to completely sequence each member. Inherent in this approach, however, is the need to extrapolate from heterologous genomic sequences. Nevertheless, we believe that any potential inaccuracies due to genomic polymorphisms between strains 297 and B31 are unlikely to alter our major findings and are easily counterbalanced by the plethora of new genetic information pertaining to a highly virulent, neurotropic clinical isolate (46). The need for such information is highlighted by our finding, using both needle and tick inoculation in mouse infectivity tests, that the B31 clone used for the genomic sequence is less virulent than either the parental B31 isolate or B. burgdorferi 297 (M. J. Caimano, D. R. Akins, S. K. Wikel, and J. D. Radolf, unpublished data). While these differences could reflect the loss of unrelated sequences during cloning, it is our working hypothesis that the greater virulence of B. burgdorferi 297 reflects, at least in part, the greater sequence diversity of its cp32/18 plasmids. Pending the development of efficient mutagenesis techniques for virulent borrelial isolates, it may not be possible to directly evaluate this conjecture in the near future. Nevertheless, the work reported here sets the stage for a variety of studies ranging from expression mapping and cellular localization of the cp32/18-encoded gene products in spirochetes cultivated under various in vitro and in vivo conditions to molecular epidemiological correlations of spirochetal genetic diversity and disease expression.
It is now well established that recombinatorial processes have conferred a remarkable degree of plasticity upon the B. burgdorferi genome, presumably in the context of enhancing its adaption to diverse ecological niches. Based on the analyses reported here, we now propose that two different types of recombinatorial events have occurred within the cp32/18 family. The first involved exchanges between these plasmids and the bacterium's other genetic elements. This process appears to have been relatively unrestricted in that it utilized diverse donor sequences and recipient sites; its principal constraint was that it was limited to sequences already present within the borrelial genome. One of its primary functions appears to have been plasmid building, that is, generating the scaffolding or framework needed to assemble novel genetic elements. The second type of recombinatorial event, in contrast, was confined to relatively small regions of the cp32/18 plasmids but was less restrictive in that it utilized nonparalogous DNAs, including exogenous genes. A major consequence of this second type of event was to individualize plasmids via the variable regions. While the nonparalogous ORFs present in both the B31 erp and 297 ospE/ospF/elp variable loci are the most tangible evidence for this second process, we believe that it also was involved in the importation of the novel sequences which fused to form the progenitors of the lipoprotein families within the variable loci. Some evidence for this is provided by the mlp outlier in the elpA1 locus; in fact, the proximity of ucp32-1 and mlp11 in cp32-1 (Fig. 5A) suggests that they were introduced on the same recombining DNA fragment. A final question concerns the milieu which serves as the incubator for lateral gene exchange. With the exception of the recently described vls locus (63), it has not been possible to demonstrate genetic rearrangements in Lyme disease spirochetes during the mammalian phase of infection (16, 47). Because of the low spirochetal burdens in B. burgdorferi-infected tissues (7), it seems more likely that such exchanges occur during and shortly after the tick feeding phase(s) of the enzootic cycle, when spirochetes are in close proximity, highly metabolically active, and actively replicating (14, 38) and that novel antigenic phenotypes are subsequently selected within the mammalian host.
ACKNOWLEDGMENTS
We gratefully acknowledge funding for this work provided by grants AI-29735 and AI-45538 from the Lyme disease program of the National Institute of Allergy and Infectious Diseases and by grants from the Arthritis Foundation (via the Egalitcheff Research Fund) and the Centers for Disease Control and Prevention (U50/CCU614875). M.J.C. was supported in part by Molecular Microbiology Training Grant AI-07520 (NIAID).
ADDENDUM IN PROOF
Recent work by Richard Marconi and coworkers has suggested that the bdr paralogs belong to three subfamilies, designated bdrD, bdrE, and bdrF. To clarify the relatedness of these genes both within and among Borrelia isolates, a new nomenclature has been suggested. To that end, we have redesignated the bdr paralogs in strain 297 as follows: bdrA to bdrD1, bdrB to bdrE7 (cp18-2), bdrD to bdrE3 (cp18-1), bdrF to bdrE6 (cp32-3), bdrH to bdrE1 (cp32-4), and bdrJ to bdrE5 (cp32-5).
REFERENCES
- 1.Akins D R, Bourell K W, Caimano M J, Norgard M V, Radolf J D. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J Clin Investig. 1998;101:2240–2250. doi: 10.1172/JCI2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akins D R, Caimano M J, Yang X, Cerna F, Norgard M V, Radolf J D. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect Immun. 1999;67:1526–1532. doi: 10.1128/iai.67.3.1526-1532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akins D R, Porcella S F, Popova T G, Shevchenko D, Baker S I, Li M, Norgard M V, Radolf J D. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homolog. Mol Microbiol. 1995;18:507–520. doi: 10.1111/j.1365-2958.1995.mmi_18030507.x. [DOI] [PubMed] [Google Scholar]
- 4.Baranton G, Postic D, Saint Girons I, Boerlin P, Piffaretti J C, Assous M, Grimont P A. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov. and group VS461 associated with Lyme borreliosis. Int J Syst Bacteriol. 1992;42:378–383. doi: 10.1099/00207713-42-3-378. [DOI] [PubMed] [Google Scholar]
- 5.Barbour A G, Carter C J, Bundoc V, Hinnebusch J. The nucleotide sequence of a linear plasmid of Borrelia burgdorferi reveals similarities to those of circular plasmids of other prokaryotes. J Bacteriol. 1996;178:6635–6639. doi: 10.1128/jb.178.22.6635-6639.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour A G, Garon C F. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987;237:409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- 7.Barthold S W. Lyme borreliosis in the laboratory mouse. J Spirochet Tick-Borne Dis. 1996;3:22–44. [Google Scholar]
- 8.Carlyon J A, LaVoie C, Sung S-Y, Marconi R T. Analysis of the organization of multicopy linear- and circular-plasmid-carried open reading frames in Borrelia burgdorferi sensu lato isolates. Infect Immun. 1998;66:1149–1158. doi: 10.1128/iai.66.3.1149-1158.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlyon J A, Marconi R T. Cloning and molecular characterization of a multicopy, linear plasmid-carried, repeat motif-containing gene from Borrelia turicatae, a causative agent of relapsing fever. J Bacteriol. 1998;180:4974–4981. doi: 10.1128/jb.180.18.4974-4981.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casjens S, van Vugt R, Tilly K, Rosa P A, Stevenson B. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J Bacteriol. 1997;179:217–227. doi: 10.1128/jb.179.1.217-227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cassatt D R, Patel N K, Ulbrandt N D, Hanson M S. DbpA, but not OspA, is expressed by Borrelia burgdorferi during spirochetemia and is a target for protective antibodies. Infect Immun. 1998;66:5379–5387. doi: 10.1128/iai.66.11.5379-5387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Champion C I, Blanco D R, Skare J T, Haake D A, Giladi M, Foley D, Miller J N, Lovett M A. A 9.0 kilobase-pair circular plasmid of Borrelia burgdorferi encodes an exported protein: evidence for expression only during infection. Infect Immun. 1994;62:2653–2661. doi: 10.1128/iai.62.7.2653-2661.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das S, Barthold S W, Giles S S, Montgomery R R, Telford III S R, Fikrig E. Temporal pattern of Borrelia burgdorferi p21 expression in ticks and mammalian host. J Clin Investig. 1997;99:987–995. doi: 10.1172/JCI119264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Silva A M, Fikrig E. Growth and migration of Borrelia burgdorferi in Ixodes ticks during blood feeding. Am J Trop Med Hyg. 1995;53:397–404. doi: 10.4269/ajtmh.1995.53.397. [DOI] [PubMed] [Google Scholar]
- 15.Dunn J J, Buchstein S R, Butler L-L, Fisenne S, Polin D S, Lade B N, Luft B J. Complete nucleotide sequence of a circular plasmid from the Lyme disease spirochete, Borrelia burgdorferi. J Bacteriol. 1994;176:2706–2717. doi: 10.1128/jb.176.9.2706-2717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Hage N, Lieto L D, Stevenson B. Stability of erp loci during Borrelia burgdorferi infection: recombination is not required for chronic infection of immunocompetent mice. Infect Immun. 1999;67:3146–3150. doi: 10.1128/iai.67.6.3146-3150.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 18.Ferdows M S, Barbour A G. Megabase-sized linear DNA in the bacterium Borrelia burgdorferi, the Lyme disease agent. Proc Natl Acad Sci USA. 1989;86:5969–5973. doi: 10.1073/pnas.86.15.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fikrig E, Chen M, Barthold S W, Anguita J, Feng W, Telford III S R, Flavell R A. Borrelia burgdorferi erpT expression in the arthropod vector and murine host. Mol Microbiol. 1999;31:281–280. doi: 10.1046/j.1365-2958.1999.01171.x. [DOI] [PubMed] [Google Scholar]
- 20.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J-F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 21.Guina T, Oliver D B. Cloning and analysis of a Borrelia burgdorferi membrane-interactive protein exhibiting haemolytic activity. Mol Microbiol. 1997;24:1201–1213. doi: 10.1046/j.1365-2958.1997.4291786.x. [DOI] [PubMed] [Google Scholar]
- 22.Helinski D R, Toukdarian A E, Novick R P. Replication control and other stable maintenance mechanisms of plasmids. In: Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2295–2324. [Google Scholar]
- 23.Hofmann K, Stoffel W. Tmbase, a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166–166. [Google Scholar]
- 24.Kawabata H, Masuzawa T, Yanagihara Y. Genomic analysis of Borrelia japonica sp. nov. isolated from Ixodes ovatus in Japan. Microbiol Immunol. 1993;37:843–848. doi: 10.1111/j.1348-0421.1993.tb01714.x. [DOI] [PubMed] [Google Scholar]
- 25.Khan S A. Rolling-circle replication of bacterial plasmids. Microbiol Mol Biol Rev. 1997;61:442–455. doi: 10.1128/mmbr.61.4.442-455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lahdenne P, Porcella S F, Hagman K E, Akins D R, Popova T G, Cox D L, Radolf J D, Norgard M V. Molecular characterization of a 6.6-kilodalton Borrelia burgdorferi outer membrane-associated lipoprotein (lp6.6) which appears to be downregulated during mammalian infection. Infect Immun. 1997;65:412–421. doi: 10.1128/iai.65.2.412-421.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam T T, Nguyen T P K, Montgomery R R, Kantor F S, Fikrig E, Flavell R A. Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect Immun. 1994;62:290–298. doi: 10.1128/iai.62.1.290-298.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lane R S, Piesman J, Burgdorfer W. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu Rev Entomol. 1991;36:587–609. doi: 10.1146/annurev.en.36.010191.003103. [DOI] [PubMed] [Google Scholar]
- 29.Liveris D, Varde S, Iyer R, Koenig S, Bittker S, Cooper D, McKenna D, Nowakowski J, Nadelman R B, Wormser G P, Schwartz I. Genetic diversity of Borrelia burgdorferi in Lyme disease patients as determined by culture versus direct PCR with clinical specimens. J Clin Microbiol. 1999;37:565–569. doi: 10.1128/jcm.37.3.565-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liveris D, Wormser G P, Nowakowski J, Nadelman R, Bittker S, Cooper D, Moy F H, Forseter G, Pavia C S, Schwartz I. Molecular typing of Borrelia burgdorferi from Lyme diseaes patients by PCR-restriction fragment length polymorphism analysis. J Clin Microbiol. 1996;34:1306–1309. doi: 10.1128/jcm.34.5.1306-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marconi R T, Garon C F. Identification of a third genomic group of Borrelia burgdorferi through signature nucleotide analysis and 16S rRNA sequence determination. J Gen Microbiol. 1992;138:533–536. doi: 10.1099/00221287-138-3-533. [DOI] [PubMed] [Google Scholar]
- 32.Marconi R T, Konkel M E, Garon C F. Variability of osp genes and gene products among species of Lyme disease spirochetes. Infect Immun. 1993;61:2611–2617. doi: 10.1128/iai.61.6.2611-2617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathiesen D A, Oliver J H, Jr, Kolbert C P, Tullson E D, Johnson B J B, Campbell G L, Mitchell P D, Reed K D, Telford III S R, Anderson J F, Lane R S, Persing D H. Genetic heterogeneity of Borrelia burgdorferi in the United States. J Infect Dis. 1997;175:98–107. doi: 10.1093/infdis/175.1.98. [DOI] [PubMed] [Google Scholar]
- 34.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 35.Norris S J, Howell J K, Garza S A, Ferdows M S, Barbour A G. High- and low-infectivity phenotypes of clonal populations of in vitro-cultured Borrelia burgdorferi. Infect Immun. 1995;63:2206–2212. doi: 10.1128/iai.63.6.2206-2212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 37.Petit M-A, Dervyn E, Rose M, Entian K-D, McGovern S, Ehrlich S D, Bruand C. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling circle replication. Mol Microbiol. 1998;29:261–273. doi: 10.1046/j.1365-2958.1998.00927.x. [DOI] [PubMed] [Google Scholar]
- 38.Piesman J, Oliver J R, Sinsky R J. Growth kinetics of the Lyme disease spirochete (Borrelia burgdorferi) in vector ticks (Ixodes dammini) Am J Trop Med Hyg. 1990;42:352–357. doi: 10.4269/ajtmh.1990.42.352. [DOI] [PubMed] [Google Scholar]
- 39.Porcella S F, Popova T G, Akins D R, Li M, Radolf J D, Norgard M V. Borrelia burgdorferi supercoiled plasmids encode multiple tandem open reading frames and a lipoprotein gene family. J Bacteriol. 1996;178:3293–3307. doi: 10.1128/jb.178.11.3293-3307.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan J R, Levine J F, Apperson C S, Lubke L, Wirtz R A, Spears P A, Orndorff P E. An experimental chain of infection reveals that distinct Borrelia burgdorferi populations are selected in arthropod and mammalian hosts. Mol Microbiol. 1998;30:365–379. doi: 10.1046/j.1365-2958.1998.01071.x. [DOI] [PubMed] [Google Scholar]
- 41.Saier M H., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58:71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwan T G, Burgdorfer W, Garon C F. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect Immun. 1988;56:1831–1836. doi: 10.1128/iai.56.8.1831-1836.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwan T G, Piesman J, Golde W T, Dolan M C, Rosa P A. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seinost G, Dykhuizen D E, Dattwyler R J, Golde W T, Dunn J J, Wang I-N, Wormser G P, Schriefer M E, Luft B J. Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect Immun. 1999;67:3518–3524. doi: 10.1128/iai.67.7.3518-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simpson W J, Garon C F, Schwan T G. Borrelia burgdorferi contains repeated DNA sequences that are species specific and plasmid associated. Infect Immun. 1990;58:847–853. doi: 10.1128/iai.58.4.847-853.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steere A C, Grodzicki R L, Craft J E, Shrestha M, Kornblatt A N, Malawista S E. Recovery of Lyme disease spirochetes from patients. Yale J Biol Med. 1984;57:557–560. [PMC free article] [PubMed] [Google Scholar]
- 47.Stevenson B, Bockenstedt L K, Barthold S W. Expression and gene sequence of outer surface protein C of Borrelia burgdorferi reisolated from chronically infected mice. Infect Immun. 1994;62:3568–3571. doi: 10.1128/iai.62.8.3568-3571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevenson B, Bono J L, Schwan T G, Rosa P A. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect Immun. 1998;66:2648–2654. doi: 10.1128/iai.66.6.2648-2654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevenson B, Casjens S, Rosa P. Evidence of past recombination events among the genes encoding Erp antigens of Borrelia burgdorferi. Microbiology. 1998;144:1869–1879. doi: 10.1099/00221287-144-7-1869. [DOI] [PubMed] [Google Scholar]
- 50.Stevenson B, Casjens S, van Vugt R, Porcella S F, Tilly K, Bono J L, Rosa P A. Characterization of cp18, a naturally truncated member of the cp32 family of Borrelia burgdorferi plasmids. J Bacteriol. 1997;179:4285–4291. doi: 10.1128/jb.179.13.4285-4291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stevenson B, Schwan T G, Rosa P A. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevenson B, Tilly K, Rosa P A. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi. J Bacteriol. 1996;178:3508–3516. doi: 10.1128/jb.178.12.3508-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suk K, Das S, Sun W, Jwang B, Barthold S W, Flavell R A, Fikrig E. Borrelia burgdorferi genes selectively expressed in the infected host. Proc Natl Acad Sci USA. 1995;92:4269–4273. doi: 10.1073/pnas.92.10.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sung S-Y, Lavoie C P, Carlyon J A, Marconi R T. Genetic divergence and evolutionary instability in ospE-related members of the upstream homology box gene family in Borrelia burgdorferi sensu lato complex isolates. Infect Immun. 1998;66:4656–4668. doi: 10.1128/iai.66.10.4656-4668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Theisen M. Molecular cloning and characterization of nlpH, encoding a novel, surface-exposed, polymorphic, plasmid-encoded 33-kilodalton lipoprotein of Borrelia afzelii. J Bacteriol. 1996;178:6435–6442. doi: 10.1128/jb.178.22.6435-6442.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through weighting positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Dam A P, Kuiper H, Vos K, Widjojokusumo A, de Jongh B M, Spanjaard L, Ramselaar A C P, Kramer M D, Dankert J. Different genospecies of Borrelia burgdorferi are associated with distinct clinical manifestations of Lyme borreliosis. Clin Infect Dis. 1993;17:708–717. doi: 10.1093/clinids/17.4.708. [DOI] [PubMed] [Google Scholar]
- 58.Wallich R, Brenner C, Kramer M D, Simon M M. Molecular cloning and immunological characterization of a novel linear-plasmid-encoded gene, pG, of Borrelia burgdorferi expressed only in vivo. Infect Immun. 1995;63:3327–3335. doi: 10.1128/iai.63.9.3327-3335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Welsh J, Pretzman C, Postic D, Saint Girons I, Baranton G, McClelland M. Genomic fingerprinting by arbitrarily primed polymerase chain reaction resolves Borrelia burgdorferi into three distinct phyletic groups. Int J Syst Bacteriol. 1992;42:370–377. doi: 10.1099/00207713-42-3-370. [DOI] [PubMed] [Google Scholar]
- 60.Wormser G P, Liveris D, Nowakowski J, Nadelman R B, Cavaliere L F, McKenna D, Holmgren D, Schwartz I. Association of specific subtypes of Borrelia burgdorferi with hematogenous dissemination in early Lyme disease. J Infect Dis. 1999;180:720–725. doi: 10.1086/314922. [DOI] [PubMed] [Google Scholar]
- 61.Xu Y, Kodner C, Coleman L, Johnson R C. Correlation of plasmids with infectivity of Borrelia burgdorferi sensu stricto type strain B31. Infect Immun. 1996;64:3870–3876. doi: 10.1128/iai.64.9.3870-3876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang X, Popova T G, Hagman K E, Wikel S K, Schoeler G G, Caimano M J, Radolf J D, Norgard M V. Identification, characterization, and expression of three new members of the Borrelia burgdorferi Mlp (2.9) lipoprotein gene family. Infect Immun. 1999;67:6008–6018. doi: 10.1128/iai.67.11.6008-6018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J R, Hardham J M, Barbour A G, Norris S J. Antigenic variation in Lyme disease Borreliae by promiscuous recombination of Vmp-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 64.Zückert W R, Meyer J. Circular and linear plasmids of Lyme disease spirochetes share extensive homology: characterization of a repeated DNA element. J Bacteriol. 1996;178:2287–2298. doi: 10.1128/jb.178.8.2287-2298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zückert W R, Meyer J, Barbour A G. Comparative analysis and immunological characterization of the Borrelia Bdr protein family. Infect Immun. 1999;67:3257–3266. doi: 10.1128/iai.67.7.3257-3266.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]