Abstract
Early-life stress (ELS) leads to increased vulnerability to psychiatric disorders including depression later in life. Neuroinflammatory processes have been implicated in ELS-induced negative health outcomes, but how ELS impacts microglia, the main tissue-resident macrophages of the central nervous system, is unknown. Here, we determined the effects of ELS-induced by limited bedding and nesting material during the first week of life (postnatal days [P]2–9) on microglial (i) morphology; (ii) hippocampal gene expression; and (iii) synaptosome phagocytic capacity in male pups (P9) and adult (P200) mice. The hippocampus of ELS-exposed adult mice displayed altered proportions of morphological subtypes of microglia, as well as microglial transcriptomic changes related to the tumor necrosis factor response and protein ubiquitination. ELS exposure leads to distinct gene expression profiles during microglial development from P9 to P200 and in response to an LPS challenge at P200. Functionally, synaptosomes from ELS-exposed mice were phagocytosed less by age-matched microglia. At P200, but not P9, ELS microglia showed reduced synaptosome phagocytic capacity when compared to control microglia. Lastly, we confirmed the ELS-induced increased expression of the phagocytosis-related gene GAS6 that we observed in mice, in the dentate gyrus of individuals with a history of child abuse using in situ hybridization. These findings reveal persistent effects of ELS on microglial function and suggest that altered microglial phagocytic capacity is a key contributor to ELS-induced phenotypes.
Subject terms: Molecular neuroscience, Depression
Introduction
Exposure to early-life stress (ELS) has long-lasting effects on brain structure and function and increases the risk for psychiatric illness later in life [1–3]. Human and rodent studies have demonstrated that stress during sensitive developmental periods impacts mood [2–5], cognition [6–10], and the neuroimmune system [11–16].
While the mechanisms for early-life programming of later-life health remain poorly understood, there is increasing evidence that long-term impact on the neuroimmune systems and microglia in particular might contribute to these effects [17, 18]. Microglia are innate immune cells in the brain parenchyma that can respond to environmental cues such as stress by means of cytokine release and phagocytosis [19–22] and are crucial for proper brain development and function by, e.g., synaptic pruning [23–25].
There is ample evidence from maternal inflammation and early-life infection studies in rodents that early experiences can enduringly change microglial phenotypes [26]. This is thought to be mediated via epigenetic mechanisms that reinforce microglial training or desensitization, i.e. hyper- or hyposensitivity, towards secondary inflammatory challenges in later life [15, 27–32]. Considering the well-documented interactions between stress and inflammation [33–35], ELS might, similarly, program microglia [36, 37]. In fact, we and others have previously shown age-dependent effects of ELS on microglia that are largely based on morphological characterization at basal [14, 38, 39] and challenged states, e.g., in response to amyloid-β pathology [14]. While transcriptomic studies of microglia in the context of ELS are rare, microglial gene expression profiling shortly after postnatal stress revealed ELS-induced alterations in chemotactic and phagocytic processes [13]. However, a thorough understanding of ELS’ short and long-term impact on microglial gene expression and function, and whether such changes also occur in the human brain is currently lacking. Therefore, we studied (1) the immediate effects of ELS on microglial gene expression; (2) the long-term effects of ELS on microglial morphology and gene expression profile in mice, in basal and immune-challenged states, in order to unmask potentially latent impacts of ELS; (3) the implications of these alterations for microglial phagocytic capacity; and (4) whether one of our target genes is similarly altered in the human condition.
We here demonstrate for the first time that ELS leads to long-term changes in the microglial transcriptome at P200, modifies the trajectory of gene expression changes during development and in response to LPS, and reduces microglial phagocytosis of synapses at P9 and P200. Finally, we validate in a post-mortem human cohort that GAS6, a phagocytosis-related gene found upregulated in ELS mice, is also increased in the hippocampal microglia of individuals with a history of child abuse.
Materials and methods
Experimental design, breeding, and early-life stress model
To determine the acute and long-term effects of ELS on microglia, we exposed seven different cohorts of mice to ELS or control (CTR) conditions from postnatal days (P)2 to P9 via the limited bedding and nesting model (see Supplementary Methods). We confirmed across the generated cohorts for this study the reduction in body weight gain between P2 and P9 in ELS-exposed mice (Fig. S1A) [38, 40]. Male mice were used in all experiments.
Cohort 1 was used for morphological characterization of microglia 24 hours after PBS or LPS (5 mg/kg) injection at the age of 3–5 months (CTR-PBS: n = 5, ELS-PBS: n = 4, CTR-LPS: n = 10, ELS-LPS: n = 6). Cohort 2 was sacrificed at P9, while cohort 3 was allowed to mature until P200 and injected with either PBS or LPS (1 mg/kg). Microglia from cohorts 2 and 3 were isolated for transcriptomic characterization (P9-CTR: n = 12, P9-ELS: n = 12, P200-CTR-PBS: n = 7, P200-ELS-PBS: n = 6, P200-CTR-LPS: n = 7, P200-ELS-LPS: n = 5, Fig. 1A).
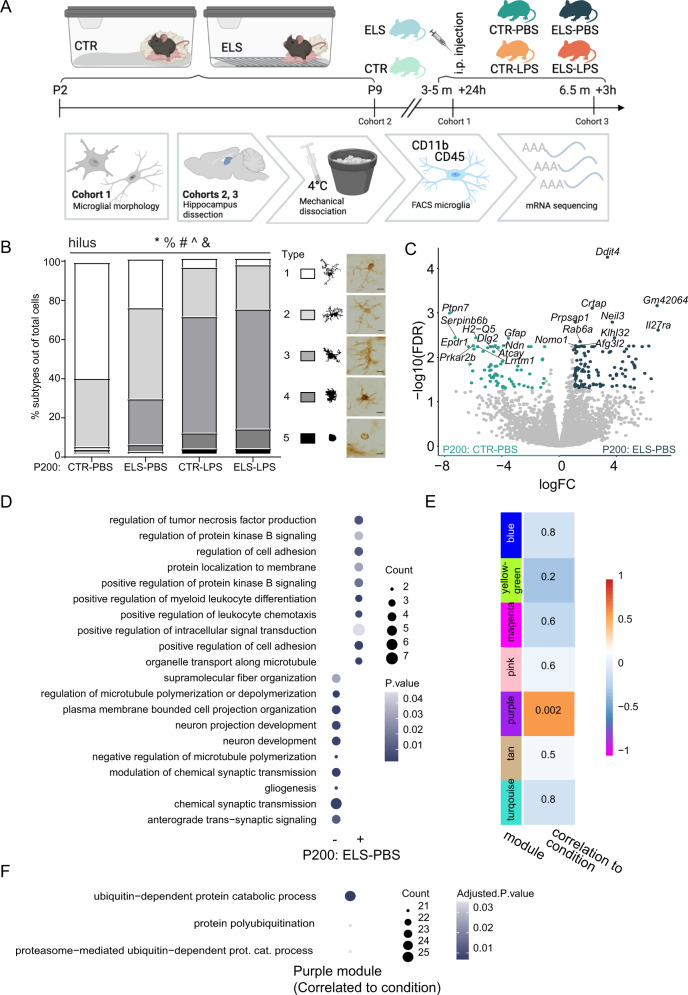
Fig. 1. ELS exerts long-term, but no immediate effects on the microglia transcriptome.
A Overview of the experimental design for the microglia morphometric and transcriptomic analysis (cohorts 1, 2, and 3): from P2 to P9 mice were exposed to limited bedding and nesting material resulting in early-life stress. Control mice were left undisturbed. For the morphological analysis of microglia (cohort 1), mice received an i.p. injection with LPS (5 mg/kg) at 3–5 months (CTR-PBS (n = 5), ELS-PBS (n = 4), CTR-LPS (n = 10), ELS-LPS (n = 6) and were sacrificed 24 h later. Two other cohorts were created for transcriptomic analysis of microglia. Cohort 2 was sacrificed immediately after ELS at P9 (P9-CTR (n = 12), P9-ELS (n = 13)) and cohort 3 was left undisturbed until P200, when they received an i.p. injection with PBS or LPS (1 mg/kg) (P200-CTR-PBS (n = 7), P200-ELS-PBS (n = 6), P200-CTR-LPS (n = 7), P200-ELS-LPS (n = 5) and were sacrificed 3 hours later. From cohort 2 and 3, hippocampi were dissected, microglia were FACS-purified, and expression profiled. Created with BioRender.com. B Effects of condition (CTR/ELS) and treatment (PBS/LPS) on the proportion of morphological microglia subtypes in the hilus of 3–5 months mice, including representative images of morphological Iba1+ subtypes. Objective ×40, scale bar = 10 µm. General Linear Model Multivariate test, *: main effect treatment, %: interaction effect treatment*condition, #: treatment effect for subtype 1, 3, and 4, ^: condition effect for subtype 1, &: interaction treatment*condition for subtype 1. p < 0.05. Stacked bar plot depicts the average proportion of each cell type per group. C Volcano plot depicting differentially expressed genes between P200: ELS-PBS and P200: CTR-PBS (logFC < />1, FDR < 0.05). Each dot represents a gene. Significantly more abundant genes in P200: CTR-PBS are marked in turquoise and significantly more abundant genes in P200: ELS-PBS are marked in dark turquoise. The 10 most enriched genes in each condition are labeled. D Gene ontology (GO) analysis of relatively lower (−) and abundant (+) significant genes in P200: ELS-PBS when compared to P200: CTR-PBS. Top 10 significantly enriched GO terms (p < 0.05) based on gene count are depicted (Table S5). E Pearson correlation of modules detected with weighted gene co-expression network analysis and condition (ELS/CTR). P value is indicated as number and R2 as color for each correlation. F Significantly enriched GO terms associated with the pink module genes (adjusted p value < 0.05, Table S7). CTR control, ELS early-life stress, GO gene ontology, h hours, LPS lipopolysaccharide, m months, PBS phosphate-buffered saline.
For a functional readout of microglial function, we extracted microglia from cohorts 4 and 5, which were kept until P9 or P200, respectively (P9-CTR: n = 8, P9-ELS: n = 12, P200-CTR: n = 8, P200-ELS: n = 3). The extracted microglia were incubated with age-matched synapses isolated from cohorts 6 and 7, which were sacrificed at P9 and P150, respectively (P9-CTR-synapses: n = 12, P9-ELS-synapses: n = 11, P150-CTR-synapses: n = 8, P150-ELS-synapses: n = 7).
Power calculations were based on previously reported experiments [14, 41, 42]. All experimental procedures were conducted according to the Dutch national law and European Union directives on animal experiments and were approved by the Animal Welfare Body of the University of Amsterdam.
Lipopolysaccharide injection
Adult mice were intraperitoneally (i.p.) injected with either Dulbecco’s Phosphate-Buffered Saline (PBS, Sigma-Aldrich D8537) or lipopolysaccharide (LPS, Sigma-Aldrich, E. coli, O111:B4, L4391) dissolved in PBS at a dose of 5 mg/kg (cohort 1) or 1 mg/kg (cohort 3) body weight. Twenty-four (cohort 1) or three hours (cohort 3) after injection, blood was collected via tail cuts, and mice were sacrificed with i.p. euthasol injection and transcardial perfusion.
Cytokine cytometric bead assay
Plasma protein concentrations of IFNγ, IL-6, IL-10, IL12p70, MCP-1, and TNF were measured in PBS- and LPS-injected mice at P200 (cohort 4), using a mouse inflammation kit, BD Cytometric Bead Assay (BS Bioscience, Vianen, the Netherlands), according to manufacturer’s instructions. Samples were acquired on the flow cytometer, LSR Fortessa. Data were analyzed using a two-tailed two-way analysis of variance (ANOVA) after checking for homogeneity of variance and normality.
Immunohistochemistry
Microglia were detected in free-floating paraformaldehyde-perfused brain tissue by targeting ionized calcium-binding adaptor molecule 1 (rabbit anti-IBA1, 019–19741, Wako, see Supplementary Methods for detailed immunohistochemistry protocol).
Morphological analysis was done at ×10 magnification on a Nikon Eclipse Ni-E microscope. Cell density was obtained by counting the number of IBA1+ cell bodies in the dentate gyrus and cornu ammonis and normalizing to total area, while coverage was measured by dividing the thresholded IBA1 signal by the total area. Microglia in the hilus of the dentate gyrus and the stratum lacunosum-moleculare of the cornu ammonis were classified into five morphological phenotypes as previously described [14, 16, 43].
After checking for homogeneity of variance and normality, coverage and density data were analyzed using a two-tailed two-way ANOVA, while the morphological subtypes were analyzed with a general linear multivariate model, all using SPSS 25 (IBM software) and graphed using ggplot2 (v3.3.3.9000) [44] in R. Data were considered statistically significant when p < 0.05.
Microglia isolation and mRNA sequencing
Microglia were isolated from left and right hippocampi of saline-perfused mice as described [45]. Briefly, after mechanical dissociation and myelin removal in adult (but not P9) brains using Percoll (Cytiva, 17–5445–02), cells were incubated with a blocker for the mouse Fc Receptor (5 µg/ml, eBioscience, 14–0161) for 15 min. Afterwards, cells were stained with anti-mouse CD11b-PE (1.2 µg/ml, eBioscience, 12–0112) and anti-mouse CD45-FITC (2.5 µg/ml, eBioscience, 11–0451) for 30 min. Shortly before cell sorting, DAPI (0.15 µg/ml, Biolegend, 422801) and DRAQ5 (2 µM, Thermo Scientific, 62251) were added to the cell suspension. Single, viable (DAPI-, DRAQ5+) microglia (CD45int, CD11b+) were sorted with the Beckman Coulter MoFlo XDP and were collected in 350 µl RNA lysis buffer (Qiagen, 1053393), and stored at −80 °C until further use. Following RNA isolation, mRNA sequencing, and sequence alignment after quality check, bioinformatic analyses were performed in RStudio (v4.0.2). Differential gene expression was determined by a log-fold change of 0.1 and FDR < 0.05. Gene ontology analysis was performed using enrichR (v3.0) [46] based on the “GO_Biological_Process_2021” database. Of the cited GO terms distinctly overrepresented in CTR or ELS groups, we also highlighted their associated genes; for terms with >4 associated genes, the top 5 are listed based on absolute logFC (see Supplementary Methods for further details on microglia isolation, mRNA sequencing, and downstream analyses).
Ex vivo synaptosome phagocytosis assay
We adapted a flow cytometry-based ex vivo phagocytosis assay [42]. After sacrifice via rapid decapitation, microglia were enriched from whole brains (P9) or half brains from the cortex until the midbrain (P200) using an isotonic Percoll gradient (Supplementary Methods). We incubated 50,000 (P9) or 80,000 (P200) cells with age-matched hippocampal synaptosomes from P9 (1.2 µg) or P150 (2 µg) mice in 300 µl DMEM-F12. We used one tube per mouse as negative control to ensure signal specificity. Synaptosomes were extracted based on a published protocol [47] (Supplementary Methods), and were conjugated to pHrodo-red (P36600, Invitrogen) according to manufacturer instructions. Staining was performed by first blocking the mouse Fc Receptor (5 µg/ml, eBioscience, 14–0161) for 15 min and then by incubating with anti-mouse CD11b-APC (1 µg/ml, eBioscience, 17–0112–82) for 30 min. DAPI (0.15 µg/ml, Biolegend, 422801) was added before flow cytometry analysis using the BD FACS Diva (BD Biosciences).
Approximately 1500 DAPI−/CD11b+ cells were recorded per tube, and phagocytosis was defined as the proportion of CD11b+pHrodo+ cells divided by the total number of DAPI−/CD11b+ cells. Data analysis was done using a mixed linear model using the nlme package in R [48], correcting for the seeding of multiple tubes from each animal, as well as nest effects in P9 samples. The normality of the residuals was inspected by generating a quantile-quantile plot in R.
Human cohort and fluorescent in situ hybridization in post-mortem human hippocampus
Fresh-frozen hippocampal tissue, from well-characterized age-matched adult males, (depressed suicides with a history of child abuse, n = 7, and matched sudden-death controls, n = 6) were obtained from the Douglas-Bell Canada Brain Bank (Montreal, Canada). Characterization of early-life histories was based on adapted Childhood Experience of Care and Abuse interviews assessing experiences of sexual and physical abuse (see Supplementary Methods for further details on human cohort). Group characteristics can be found in Table S1, together with correlations between covariates (age, post-mortem interval (PMI), pH, substance dependence, and medication) and the variables measured in this study.
Hippocampal tissues were cut into 10 µm-thick sections with a cryostat and collected on Superfrost charged slides. In situ hybridization was performed for Hs-TMEM119 and Hs-GAS6 using Advanced Cell Diagnostics RNAscope® probes and reagents following the manufacturer’s instructions (see Supplementary Methods for further details). Sections were imaged using Olympus VS120 virtual slide microscope at ×20 magnification. Dentate gyrus area was demarcated manually and QuPath (v0.3.2) [49] was employed for automated cell detection based on DAPI (Vector Laboratories) staining and RNAscope signal quantification. For each probe, cells bearing three or more fluorescence puncta were counted as positive. Data were analyzed using a two-sided T test after testing for homogeneity of variance and normality.
Results
Morphological characterization of hippocampal microglia from ELS-exposed mice under basal and immune-challenged conditions
Microglia can adapt a range of morphologies in response to homeostatic disbalance [16, 50, 51]. We determined the effect of ELS on microglia density, coverage, and morphology in subregions of the hippocampus of adult mice under basal conditions and in response to LPS (Fig. 1A). ELS exposure did not affect IBA1+ cell numbers or coverage in the dentate gyrus and the cornus ammonis subregions (Fig. S1B). However, LPS increased IBA1+ cell density in both areas (DG: F = 27.179, p < 0.001; CA: F = 23.821, p < 0.001,) and reduced IBA1+ coverage especially in the dentate gyrus (F = 5.528, p = 0.028; cornus ammonis: F = 4.223, p = 0.053, Fig. S1C).
To further investigate microglial morphology, we characterized microglial subtypes [16, 43, 52] in the hilus (Fig. 1B) and stratum lacunosum-moleculare (SLM, Fig. S1D). We identified two main morphological subtypes in PBS-injected mice, characterized by either a small cell soma and long-branched ramifications (type 1) or a larger cell soma and thicker, branched ramifications (type 2). In PBS-injected animals, ELS decreased the proportion of type 1 microglia in the hilus (interaction treatment*condition: F = 9.621, p = 0.006; main effect condition: F = 11.135, p = 0.003) and SLM (main effect condition: F = 8.606, p = 0.008). Additionally, two other subtypes (type 3 and 4) were observed mostly in ELS mice, characterized by fewer ramifications and larger cell bodies than subtypes 1 and 2. Number of Type 3 microglia was increased by ELS in PBS-injected mice in both regions (Figs. 1B, S1D).
LPS significantly affected the proportions of morphological subtypes in the hilus (GLM main effect treatment, F = 8.386, p < 0.001, Fig. 1B) and SLM (GLM main effect treatment, F = 8.386, p < 0.001, Fig. S1D), with modulation by ELS in the hilus (GLM interaction effect treatment*condition, F = 3.181, p = 0.035, Fig. 1B). LPS reduced type 1 microglia in the hilus (F = 49.442, p < 0.001) and SLM (F = 50.609, p < 0.001) and increased type 3 (hilus: F = 20.789, p < 0.001; SLM: F = 33.000, p < 0.001) and type 4 (hilus: F = 8.537, p = 0.008; SLM: F = 5.719, p = 0.027) microglia regardless of early-life condition. A fifth subtype, characterized by an ameboid morphology, was also detected in some LPS-injected mice (F = 3.171, P = 0.090) (Figs. 1B, S1D).
In summary, under basal conditions ELS altered the proportion of morphological subtypes associated with immune reactivity. Expectedly, LPS treatment also induced a morphological profile associated with inflammation, independent of early-life condition. In this experiment, a relatively high LPS dose was used (5 mg/kg, i.p.), which might have overruled the potentially subtler effects of ELS on microglia. To better detect these modulatory effects of ELS on LPS responses, we used a dose of 1 mg/kg LPS for the transcriptomic experiment.
ELS impacts the microglia transcriptome on the long-term at P200 but not immediately after stress exposure at P9
To determine the acute (P9) and long-term (P200) effects of ELS and LPS on microglial gene expression, we performed mRNA sequencing on purified hippocampal microglia (Figs. 1A, S2A). We confirmed the purity of sorted microglia by the high expression of microglial signature genes, but not of other brain cell types (Fig. S2B, Table S2). Correlation analysis of the first six principal components (PC) with the experimental variables revealed “age” as the main source of variability in the dataset (with PC1, R² = 0.97, FDR < 0.001), followed by “treatment” (with PC2, R² = 0.23, FDR < 0.01; with PC3, R² = 0.67, FDR < 0.001), and “condition” (with PC6, R² = 0.15, FDR < 0.05; Fig. S2C).
When comparing microglial transcriptomes between CTR and ELS-exposed animals at P9, almost no transcriptional changes were found. Only one differentially expressed gene (DEG) was detected, triggering receptor expressed on myeloid cells (Trem1) (logFC </>1, FDR < 0.05, Fig. S2D, Table S3). This gene was however differentially expressed in only 3 out of a total of 13 ELS-exposed mice (Fig. S2E) and was therefore not considered biologically relevant for ELS.
In adulthood, we detected 186 DEGs when comparing gene expression profiles of CTR and ELS-exposed animals at basal state, injected with PBS (P200: ELS-PBS versus P200: CTR-PBS), (Fig. 1C, Table S4). Gene ontology (GO) analysis revealed that the genes downregulated by ELS were involved in “regulation of microtubule (de)polymerization/plasma membrane bounded cell projection organization/neuron development” (Map, Stmn2, Stmn3) and “gliogenesis” (Cdh2, Metrn). Genes upregulated by ELS were associated with inflammatory pathways and processes, such as “regulation of tumor necrosis factor production” (Ripk, Gas6, Trim27, Pf4), “regulation of protein kinase B signaling” (C1qbp, Gas6, Fermt2, Myorg), “positive regulation of leukocyte chemotaxis” (Akirin1, C1qbp, Gas6), and “positive regulation of cell adhesion” (Frmd5, C1qbp, Rell2, Dusp26) (Fig. 1D, Table S5).
To identify modules of genes with similar expression patterns in an unbiased manner, Weighted Gene Co-expression Network Analysis (WGCNA) was performed on all P200 samples (P200: CTR-PBS, P200: ELS-PBS, P200: CTR-LPS, P200: ELS-LPS). Thirteen co-expression modules were identified (Fig. S2F), of which one (purple, Table S6) significantly correlated with early-life condition (R2 = 0.6, p = 0.002, Fig. 1E). GO analysis of genes in this module suggests a role in protein ubiquitination (Fig. 1F, Table S7).
In brief, ELS does not impact the microglial transcriptome at P9, but at P200 we detected an upregulation of genes associated with inflammatory processes and protein ubiquitination and a downregulation of genes linked to morphological reconstruction.
Shared and unique transcriptional changes between P9 and P200 CTR and ELS microglia
To investigate how ELS impacts microglial transcriptional changes over development from P9 to P200, we compared the transcriptomes of P9 and P200 microglia from CTR and ELS-exposed animals, revealing DEGs shared between (gray dots, 2899) and unique for CTR and ELS microglia (P9: ELS, light blue dots, 617; P200: ELS-PBS, dark turquoise dots, 473; P9: CTR, light green dots, 797; P200: CTR-PBS, turquoise dots, 353) (Fig. 2A, Tables S8, S9). These developmental DEGs (P200 compared to P9) in CTR and ELS animals mostly did not overlap with the transcriptional changes between CTR-PBS and ELS-PBS animals at P200, pointing to an altered maturation gene expression program of mouse microglia in response to ELS (Fig. 2B).
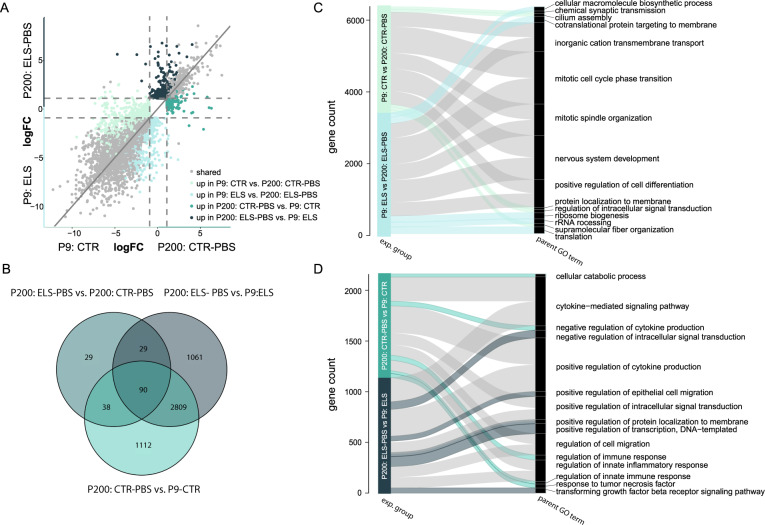
Fig. 2. ELS alters microglia development from P9 to P200.
A Four-way plot depicting expression changes in developmental (P200 vs. P9)-associated genes in ELS and CTR microglia (logFC < />1, FDR < 0.05, Tables S8, S9). Each dot represents a gene. Light green dots mark DEGs uniquely enriched in CTR microglia of P9 compared to P200 animals. Light blue dots mark DEGs uniquely enriched in ELS microglia of P9 compared to P200 animals. Turquoise dots mark DEGs uniquely enriched in CTR microglia of P200 compared to P9 animals. Dark turquoise dots mark DEGs uniquely enriched in ELS microglia of P200 compared to P9 animals. Overlapping genes in the development from P9 to P200 of ELS and CTR microglia are marked in gray. B Venn diagram depicting the gene expression overlap of all DEGs between adult ELS effects (ELS vs. CTR, Table S3) and developmental effects (P200 vs. P9) in ELS and CTR microglia. C, D Alluvial plots illustrating the top 5 enriched parent GO terms P9- (C) and P200- associated (D) genes in CTR and ELS microglia. Significant GO terms (p < 0.05) for each experimental group were reduced into parent GO terms (Tables S10, S11), which were ranked based on the total gene count belonging to that parent GO term. Color indicates the experimental group and ribbon thickness depicts the number of genes overlapping with parent GO term-specific genes. CTR control, ELS early-life stress, exp. experimental, GO gene ontology, PBS phosphate-buffered saline.
GO analysis was performed on the shared and unique DEGs and redundant GO terms were reduced into parent GO terms (Fig. 2C, D, Tables S10, S11). The shared DEGs in CTR and ELS microglia revealed that P9 microglia had relatively enriched expression of genes associated with mitosis and neurodevelopmental processes (e.g., “mitotic cell cycle phase transition”, “positive regulation of cell differentiation”, “nervous system development”, “chemical synapse organization”; Fig. 2C, gray ribbons), whereas, at P200, microglia upregulated genes associated with inflammation (e.g., “positive regulation of cytokine production”, “regulation of cell migration”, “regulation of innate inflammatory response”, Fig. 2D, gray ribbons).
In CTR animals specifically, P9 microglia were uniquely enriched in genes associated with the “regulation of intracellular signal transduction” (e.g., Gnai1, Mapk11, Arhgef17, Bst1, Rhov), “supramolecular fiber organization” (e.g., Shroom2, Col9a3, Tspan15, Myo5b, Ccdc13) and “chemical synaptic transmission” (e.g., Chrna4, Crhbp, Hrh1, Pcdhb16, Pcdhb5) (Fig. 2C, light green ribbons), while P200 microglia were uniquely enriched in genes controlling the immune-response (e.g., “response to tumor necrosis factor (TNF)”:, e.g., Acod1, Hyal3, Ccl3, Nfe2l2, Ccl25; “negative regulation of cytokine production”:, e.g., Acod1, Mefv, Ptpn22, Ppp1r11, Fcgr2b; “regulation of innate immune response”:, e.g., Acod1, Ptpn22, Trim21, Birc3, Polr3f; Fig. 2D, turquoise ribbons).
In ELS-exposed animals, P9 microglia were uniquely enriched in genes associated with biological processes such as “cellular macromolecule biosynthetic process” (e.g., Rps16, Rps15a, Rpl39, Rps27a, Rps12), “cotranslational protein targeting to membrane” (e.g., Rps16, Rps15a, Rpl39, Rps27a, Rps12), “ribosome biogenesis” (e.g., Rps16, Rpl39, Rpl17, Rps24, Rpl9) and “rRNA processing“ (e.g., Rps16, Rpl39, Rpl17, Rps24, Rpl9) (Fig. 2C, light blue ribbons), whereas genes uniquely enriched at P200 were related to the “negative regulation of intracellular signal transduction” (e.g., Ddit4, Gper1, Pik3cb, Prkaa2, Per1), “positive regulation of transcription, DNA-templated” (e.g., Gper1, Thrb, Ciita, Nr4a1, Zbtb16), and “transforming growth factor beta (TGFβ) signaling pathway” (e.g., Gdf9, Src, Smurf1, Zfyve9, Arhgef18) (Fig. 2D, dark turquoise ribbons).
These observations show that independent of early-life condition, P9 microglia are involved in processes related to cell division, cell differentiation and neurodevelopment, while adult microglia perform more inflammation-related. Additionally, ELS at P9 specifically induced processes related to protein translation and biosynthesis of other macromolecules, and at P200 induced genes related to TGFβ signaling.
ELS impacts the microglia gene expression response to an LPS challenge in adulthood
To determine if ELS alters the transcriptional response of microglia to a systemic immune challenge, we compared microglial transcriptomes of P200 CTR and ELS mice i.p. injected with either PBS or LPS. We identified shared (gray dots, 810), CTR-specific (CTR-PBS, turquoise dots, 299; CTR-LPS, orange dots, 423), and ELS-specific (ELS-PBS, dark turquoise dots, 253; ELS-LPS, dark orange dots, 236) genes dysregulated in response to LPS (Fig. 3A, Tables S12, S13).
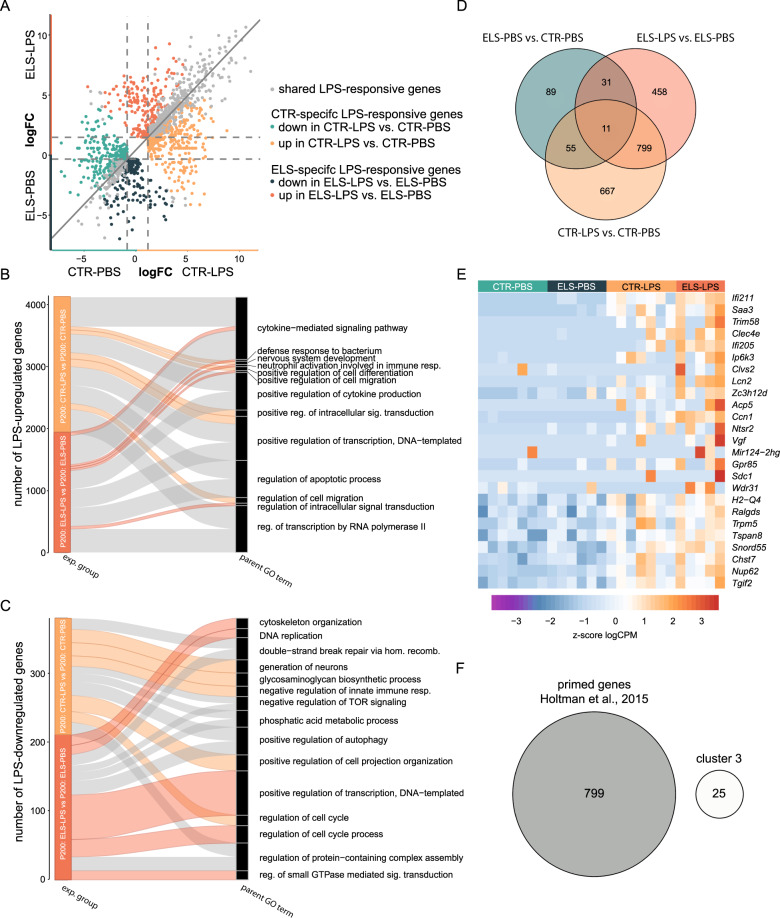
Fig. 3. ELS alters the microglia immune response to LPS.
A Four-way plot depicting gene expression changes in response to LPS (LPS vs. PBS) in ELS and CTR microglia (logFC < />1, FDR < 0.05, Tables S12, S13). Each dot represents a gene. CTR-specific LPS-responsive genes are marked in turquoise and orange, whereby the turquoise dots mark DEGs relatively lowest and orange dots mark DEGs relatively abundant expressed in CTR-LPS compared to CTR-PBS. ELS-specific LPS-responsive genes are marked in dark turquoise and dark orange, whereby the dark turquoise dots mark DEGs relatively lowest and the dark orange dots mark DEGs relatively abundant expressed in CTR-LPS compared to CTR-PBS. Overlapping genes in the LPS response of ELS and CTR microglia are marked in gray. B, C Alluvial plots illustrating the top 5 enriched parent GO terms, for upregulated (B) and downregulated (C) genes in LPS- compared to PBS-treated CTR and ELS microglia. Significant GO terms (p < 0.05) for each experimental group were reduced into parent GO terms (Tables S14, S15), which were ranked based on the total gene count belonging to that parent GO term. Color indicates the experimental group and ribbon thickness depicts the number of genes overlapping with parent GO term-specific genes. D Venn diagram depicting the gene expression overlap between ELS- (P200: ELS-PBS vs. P200: CTR-PBS, Table S3) and LPS- (P200: ELS-LPS vs. P200: ELS-PBS and P200: CTR-LPS vs P200: CTR-PBS) introduced transcriptomic differences in P200 ELS and CTR microglia. E Heatmap depicting z scores of logCPM of primed/trained genes (cluster 3, Fig. S3, Table S16). F Venn diagram showing gene overlap between genes detected to be primed/trained by ELS and LPS (cluster 3, Table S16) and trained genes detected by Holtman et al. [54] (Table S2). CTR control, ELS early-life stress, LPS lipopolysaccharide, PBS phosphate-buffered saline.
Measurement of plasma levels of select cytokines (IFNɣ, IL-6, IL-10, MCP-1 and TNF) confirmed the effectiveness of LPS injections (Fig. S3A), without further modulation by ELS. LPS-induced upregulated genes in both conditions, as expected, were associated with inflammatory response GO terms, such as “cytokine-mediated signaling pathway”, “positive regulation of cytokine production”, “regulation of apoptotic process” (Fig. 3B, gray ribbons, Table S14). Shared downregulated genes in LPS-exposed CTR and ELS mice were related to GO terms such as “double-strand break repair via homologous recombination”, “positive regulation of autophagy”, and “regulation of protein-containing complex assembly” (Fig. 3C, gray ribbons, Table S15).
The majority of LPS-induced DEGs in ELS (ELS-LPS) compared to CTR (CTR-LPS) microglia did not overlap with the gene expression changes induced by ELS itself (ELS-PBS vs CTR-PBS) (Fig. 3D), indicating that the differential response to LPS in ELS microglia were not simply due to the differential expression profile caused by ELS.
Cluster analysis of LPS-responsive genes shared between CTR and ELS microglia identified six gene clusters (Fig. S3B, Table S16). Genes in cluster 3 are upregulated by LPS in CTR microglia and even more so in ELS microglia (Fig. 3E), indicative of microglia training [29, 53] by ELS. The list of trained genes in ELS microglia is distinct from a common training gene set detected in (accelerated) aging, and mouse models of Alzheimer’s disease and amyotrophic lateral sclerosis (Table S2, Fig. 3F) [54].
Next, GO analysis was performed on the unique transcriptional changes in CTR and ELS microglia in response to LPS, respectively. LPS-induced genes in CTR microglia were related to “positive regulation of cell differentiation” (e.g., Snai1, Mapk11, Bmpr1b, Zbtb16, Ctnna2), “positive regulation of intracellular signal transduction” (e.g., Nedd4, Adra1a, Fn1, Lck, Fermt2) and “regulation of cell migration” (e.g., Plet1, Snai1, Ctnna2, Fermt2, Sema3c) (Fig. 3B, orange ribbons), whereas LPS-downregulated genes in CTR microglia were associated with the “glycosaminoglycan biosynthetic process” (Pxylp1, Ndst2, Ndst1, Hs2st1, B4galt4), “negative regulation of innate immune response” (Susd4, Dcst1, Trim21) and “positive regulation of cell projection organization” (e.g., Map3k13, Reln, Grip1, Fut9, Ptprd) (Fig. 3C, orange ribbons).
Genes induced in LPS-treated ELS microglia were involved in inflammatory processes such as “defense response to bacterium” (e.g., Nos2, Chga, Slpi, Isg15, Optn), “neutrophil activation involved in immune response” (e.g., Hp, Tnfaip6, Tarm1, Rab37, Sell) and “positive regulation of cell migration” (e.g., Sema3e, Pdpn, Edn1, Sod2, Rhoc) (Fig. 3B, dark orange ribbons), whereas LPS-downregulated genes were associated with “cytoskeleton organization” (Sema6a, Cecr2, Zmym6, Mast1, Arap3), “DNA replication” (e.g., Cdc6, Dna2, Chek1, Dbf4, Polg2) and “regulation of cell cycle process” (Chek1, Sbf4, Cul9, Zfyve26) (Fig. 3C, dark orange ribbons).
Summarizing, while we observed shared regulation of genes in response to LPS in both adult CTR and ELS microglia, ELS appears to prime a distinct set of LPS-responsive genes in microglia, resulting in a distinct transcriptional response to LPS.
ELS microglia phagocytes less synaptosomes ex vivo at P200, but not at P9
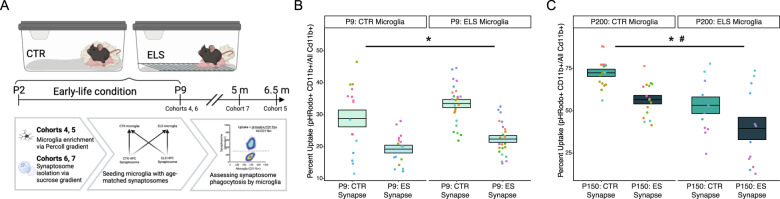
To further our understanding of the functional consequences of ELS on microglia and to complement our transcriptomic data, we incubated whole brain microglia from P9 and P200 CTR and ELS mice with labeled age-matched hippocampal synaptosomes from CTR and ELS mice (Fig. S4).
While at P9 microglia phagocytosis of synaptosomes did not depend on the origin of microglia (Fig. 4B, t(4) = 0.910, p = 0.414), P200 microglia from ELS mice exhibited decreased phagocytosis of synaptosomes (Fig. 4C, microglia-condition: t(9) = −2.226, p = 0.050). At both ages, ELS synaptosomes were phagocytosed less than CTR synaptosomes, with no interaction between synaptosome source and microglia source (P9 – Fig. 4B, synapse condition: t(58) = −5.720, p < 0.001; interaction: t(58) = −0.695, p = 0.490; P200–Fig. 4C, synapse condition: t(15) = −4.779, p < 0.001; interaction: t(15) = 0.425, p = 0.677).
Fig. 4. Assessing ELS effects on microglial phagocytosis ex vivo.
A Experimental design for phagocytosis assay (cohorts 4–7). Whole brain (P9, cohort 4) or hemi-cortex brains (P200, cohort 5) were enriched for microglia using Percoll and incubated with age-matched pHrodo-labeled hippocampal synaptosomes (isolated from cohorts 6–7) for 30 min before flow cytometry analysis. Data were analyzed using a mixed model to correct for animal source and nest for P9 microglia, and animal source for P200 microglia. Created with Biorender.com. B Reduced uptake of labeled hippocampal synaptosomes isolated from P9-ELS mice as compared to those from CTR mice, independent of early-life condition of microglia source. C At P200, ELS synapses are phagocytosed less than CTR synapses, with a trend towards decreased uptake by ELS microglia. Mixed effect linear model, *: synaptosome-condition effect, p < 0.05, #: microglia-condition effect, p = 0.05. CTR control, ELS early-life stress, HPC hippocampus. Center values represent the mean, and boundaries of the box plots represent the SEM.
Increased microglial GAS6 expression in the hippocampus of post-mortem samples of depressed individuals with a history of childhood abuse
Although animal models are essential to understand the short- and long-term neurobiological consequences of ELS, they cannot fully mirror the complexity of the human brain and experience [55]. It is therefore important to study the cellular and molecular consequences of ELS, as experienced through child abuse, in post-mortem human brain samples.
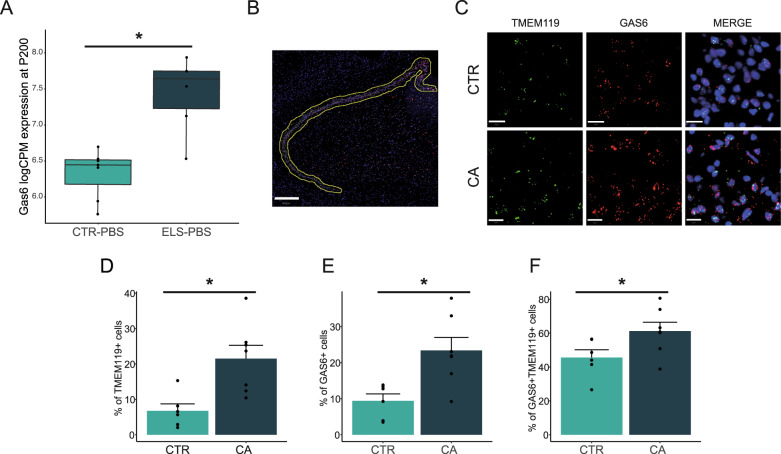
To validate the ELS-induced alterations observed in mice, we selected Gas6 as a target gene due to its role in phagocytosis [56], its strong upregulation in ELS-PBS vs. CTR-PBS mice (Fig. 5A), and its reported expression also in human microglia [57]. We studied GAS6 in the dentate gyrus (Fig. 5B) of post-mortem samples from depressed suicides with a history of childhood abuse (CA, n = 7) and matched healthy controls (CTR, n = 6) using RNAScope in situ hybridization (Fig. 5C, D, Supplementary Table). Using a TMEM119 probe to label microglia, we found that CA subjects displayed increased numbers of microglia (Fig. 5E, t(11) = 3.308, p = 0.007), GAS6+ cells (Fig. 5F, t(11) = 3.238, p = 0.008), and GAS6+ microglia (Fig. 5G, t(11) = 2.208, p = 0.049) in the dentate gyrus. These results highlight the translational value of our ELS mouse model.
Fig. 5. GAS6 expression is increased in mouse and human hippocampal microglia following ELS.
A Increased expression of microglial Gas6 in mice following early-life stress was validated in B human post-mortem dentate gyrus (DG) using RNAscope fluorescent in situ hybridization. Hippocampi from male depressed suicide subjects with a history of childhood abuse (CA, n = 7) were compared with age-matched control samples (CTR, n = 6). TMEM119 was used to probe microglia. Scale bar: 500 µm. C Representative image depicting CA-associated changes in the number of cells expressing GAS6, TMEM19, and double positive cells. The number of subpopulations expressing the mRNA of interest were counted and shown as percentage to total cells detected using DAPI staining. Scale bar: 20 µm. CA is associated with a significant increase in D microglia counts as identified by TMEM19+ cells, E total GAS6+ cells, and F microglia expressing GAS6+, detected by cells positive for both TMEM19 and GAS6. Data were analyzed using a two-tailed t test. *, condition effect, p < 0.05. CTR control, CA childhood abuse. Center values in the box plots represent the mean, borders represent the first and third quartiles, and whiskers represent 1.5*IQR value. Bar graphs represent mean and SEM.
Discussion
We demonstrate in this study that ELS exposure in mice, induced by limiting bedding and nesting material, changed the proportion of morphological microglia subtypes and microglial transcriptome in the adult hippocampus, without impacting the transcriptome immediately after stress exposure. Additionally, ELS modulated age-related changes in the microglial gene expression profile between P9 and P200, and the microglia transcriptional response to an LPS challenge in adulthood. The impact of ELS on microglia was also evident at the functional level in the ex vivo phagocytosis of synaptosomes, where P200 (but not P9) ELS microglia exhibited reduced phagocytic capacity. At both ages, ELS synapses were phagocytosed less by both CTR and ELS microglia. Lastly, in order to provide evidence for the translational value of our findings, we demonstrate that one of the identified targets altered by ELS exposure in mouse microglia (i.e., increased GAS6 expression) is also increased in the hippocampal microglia of individuals that experienced childhood adversity.
ELS affects the microglial transcriptome in adulthood but not immediately after stress exposure
Concerning the immediate effects of ELS on microglia, we did not detect any change in hippocampal microglial transcriptomes after ELS at P9. This is in contrast with the short-term effects on microglial transcriptomes after exposure to brief daily maternal separation [13], as well as with our reported ELS-induced reduction of IBA1 coverage in the dentate gyrus and IBA1 + cell complexity in the hilus at P9 [14]. The discrepancy with the findings by Delpech et al. might be due to differences between the ELS model used (brief daily maternal separation versus limited bedding and nesting) and specific ages studied (P14 versus P9). The morphological effects we described previously were subregion-specific [14], while the generated transcriptomic profiles in the present study were from whole hippocampi, possibly diluting out subregion-specific changes. Alternatively, such transcriptional changes might be latent and emerge only later in life. In fact, there is accumulating evidence for mediation of ELS-associated phenotypes by epigenetic alterations [30, 58–60]. It is thus possible that ELS-induced effects on microglia at P9 manifest at the epigenetic level, e.g., via stressor- and brain-region-dependent alterations to DNA methylation as described by others [61], that lead to later-life alterations in gene expression and function.
Moreover, the effects of ELS on other cell types in the brain, such as neurons [40], oligodendrocytes [62] and astrocytes [63, 64] also raise the possibility that transcriptional changes in microglia occur only at later time points through interactions with other brain cells. Notably, in our transcriptomic data, despite the high overlap between our detected genes with other previously published microglia datasets (Fig. S2B) [65], we also detected the presence of genes canonically defined as expressed by other cell types (e.g., Gfap for astrocytes, Serpinb6b for endothelial cells, and Dlg2 for neurons). Beyond any potential contamination, the detection of these genes could be interpreted in various ways. These could be present in microglia due to phagocytosis of these other cell types, or atypical microglial expression of these genes, which has been previously described [66, 67]. For instance, microglia have been documented to express Gfap under disease or injury states [66, 67]. It will be valuable to investigate direct effects of ELS on microglia and their interactions with the mentioned cell types via, e.g., RNAscope, single-cell mRNA sequencing or genetic manipulations, in future experiments.
Concerning the long-term effects of ELS on P200 microglia, ELS induced a shift in microglial morphological subtypes, which are indicative for immune reactive cells [52]. These alterations were accompanied by increased expression of genes related to inflammatory response, in line with the previously reported ‘immune-activated’ microglial phenotype following ELS [36, 38, 39], and downregulation of genes involved in microtubule (de)polymerization, typically involved in morphological modulations [68]. The motility and dynamics of the microglial cytoskeleton are important for core functions such as chemotaxis [69] and phagocytosis [70], thus potentially contributing to the observed reduction in phagocytic capacity of adult ELS-derived microglia. Furthermore, genes involved in neuronal development, gliogenesis, and microtubule regulation were downregulated by ELS at P200, which could contribute to earlier reported ELS-induced deficits in various forms of cellular plasticity [38, 40, 71].
With an unbiased weighted gene co-expression network analysis (WGCNA) analysis, we identified an ELS-associated gene module in P200 microglia specifically related to protein ubiquitination and degradation of ubiquitinated proteins, consistent with reported alterations in the ubiquitin-proteasome system in the hippocampus and cortex of adult rats exposed to maternal separation [72]. Ubiquitin is crucial for protein degradation processes and is also involved in inflammatory pathways [73]. This could contribute to ELS-induced increased risk for diseases with a neuroimmune component such as Alzheimer’s disease, a link that has been proposed in pre-clinical models [74–77] and epidemiological studies [78–80].
In summary, while at P9 we did not detect effects of ELS on microglial gene expression, latent changes nonetheless occur, and resulted in the observed differential microglial gene expression in adult ELS-exposed mice.
ELS modulates microglia development between P9 and P200
In line with the hypothesis of ELS-associated alterations to developmental trajectories, we observed both shared and unique shifts in gene expression profiles between P9 and P200 in CTR and ELS mice. Microglia upregulated inflammatory pathways across developmental patterns independent of early-life condition, consistent with earlier reported developmental pattern of microglial gene expression, transitioning from processes related to the cell cycle and pruning to those related to immune surveillance [24, 81–84]. We detected several differences in the transcriptional changes from P9 to P200 between CTR and ELS animals. For instance, development of CTR microglia specifically involved the Tnf pathway, whereas the Tgfβ pathway was specifically induced during development of ELS microglia. Tgfβ signaling has been reported to drive microglial survival [85], and Tgfβ serum levels have been linked to ELS, as it positively correlated with plasma cortisol levels after an acute stressor in 2-year old primates that experienced ELS [86].
These results indicate that ELS and CTR microglia follow different developmental trajectories, specifically in immune-response related genes. The exact implications of this differential trajectory for ELS microglia remain to be determined, but it might underlie how ELS modulates the response to later-life challenges, such as other forms of stress and infection reported by others and us [27, 31, 32, 82, 87].
ELS modulates microglia immune reactivity to LPS challenge in adulthood
We were able to confirm an LPS-associated increased of various inflammatory cytokines, without interaction with ELS. This is in contrast with our recently reported ELS-induced exacerbation of LPS-induced increased plasma levels of IL-6, CXCL1, and CCL2 [88]. This difference in cytokine levels is likely due to different LPS doses used (1 versus 5 mg/kg) and the time post injection before cytokine levels were determined (3 versus 24 hours). Beyond confirming an LPS-induced upregulation of inflammatory genes [22, 27, 28] and downregulation of autophagy genes [89, 90] in both CTR and ELS microglia, we also uncovered a differential LPS response dependent on early-life exposure. These results are in line with the two-hit hypothesis, which postulates that a ‘first hit’ increases sensitivity to later-life challenges that are unmasked by a ‘second hit’ [91]. We previously reported that a first hit in the form of an LPS challenge or deficiency of the DNA repair enzyme Ercc1 interacts with a second LPS hit to unmask long-lasting epigenetic changes leading to either reduced or exaggerated LPS responses as compared to naïve mice injected with LPS [41, 53, 92]. Here, we demonstrate that ELS can similarly serve as a first hit that affects the transcriptional response of microglia to an LPS challenge in adulthood. Interestingly, beyond exacerbating the expression of a group of LPS-responsive genes, suggesting microglia priming [53], ELS also led to a distinct transcriptional profile in response to LPS.
ELS impacts on microglial phagocytosis of synaptosomes
In line with our transcriptomic data, we demonstrated deficits in the ex vivo uptake of synaptosomes in P200, but not P9, microglia isolated from ELS mice. This is important considering the key role of microglial phagocytosis (i.e., pruning) in brain development and function [25, 93], and the role of neuron-microglia crosstalk in mediating the hippocampal response to stress [94, 95].
At P9, we found reduced microglial phagocytosis, which was driven by the origin of the synaptosomes (i.e., the early-life condition of the mice we extracted synaptosomes from) rather than the origin of the microglia. This suggests that ELS leads to an altered molecular signature in the developmental synapses in the hippocampus. Such reduction of the phagocytosis of ELS synapses, is in line with ELS-induced impaired pruning of excitatory synapses in the hypothalamus at P9 [96]. These synaptic signatures could drive the lasting microglial (mal-)adaptations, as we found in our adult data, where the early-life condition of both synaptosomes and microglia contributed to reduced synaptic uptake, possibly ultimately contributing to the well-established ELS effects on hippocampal plasticity [40, 97, 98]. The downregulation of synaptic phagocytosis by P200 ELS microglia might seem counterintuitive considering the literature on ELS-induced reduction of dendritic and synaptic complexity [99–101], which would imply increased phagocytosis. In fact, we previously demonstrated increased CD68 immunoreactivity in two separate cohorts of adult ELS-exposed mice [14, 38], and observed here the upregulation of genes regulating phagocytosis (e.g., C1qbp and Gas6) in our adult transcriptomic data. These are in line with the increased phagocytosis of bacterial particles observed in mice exposed to maternal separation [13], despite the difference in age. Our data, together with the reported findings by Delpech et al. [13] and Bolton et al. [96], suggest that ELS effects of phagocytosis, rather than being generalized, are complex and dependent on, e.g., specific substrates and possible eat-me signals. The identity of these different pathways, their regulators, and how ELS modulates them, is still unexplored and awaits future studies.
One specific gene that we explored further is the opsonin Gas6. We demonstrated Gas6 to be increased not only in ELS-exposed microglia in mice, but also, importantly, in the post-mortem hippocampi of patients with a history of childhood abuse, both globally and specifically in microglia. GAS6 is a ligand for the tyrosine kinase receptors TYRO3, AXL, and MERTK (TAM) that stimulate microglial phagocytosis [102]. Its signaling is also known to dampen the LPS inflammatory response of primary cultured microglia [103], mediated by TGF-β expression [104], which was increased in ELS microglia. GAS6 is present at high levels in the brain throughout development, continues to be expressed in adulthood in rodents and humans [57, 105], and may act as a neurotrophic factor for hippocampal neurons [106]. The modulation of this pathway by ELS is in line with the findings by Bolton et al., where the impaired synaptic phagocytosis of hypothalamic excitatory neurons was mediated via the AXL and MERTK receptors [96]. While the increase in GAS6 might raise expectations towards increased microglial phagocytic capacity, it is important to note that GAS6 both activates and is secreted by microglia [56, 104]. Because activation of the GAS6 receptor MERTK has been shown to stimulate synaptic phagocytosis in astrocytes [107], the observed increase in microglial GAS6 might be a mechanism to recruit other phagocytes to compensate for their deficient functioning.
Moreover, the induction of the TAM pathway by secreted ligands such as GAS6 inhibits prolonged and unrestricted inherent immune responses in macrophages/microglia. Activation of TAM receptors triggers the expression of suppressors of cytokine signaling proteins, which either terminate cytokine receptor-mediated signaling or inhibit nuclear factor kappa B (NF-κB) transcriptional activity [108]. In line with this, we found relative downregulation of NF-κB-related GO terms in ELS-PBS microglia versus CTR-PBS microglia. According to this mechanism, overexpression of Gas6 in dentate gyrus microglia after ELS might represent a compensatory mechanism to prevent microglia from becoming hyperresponsive to activation due to stressors or other stimuli in adulthood. This upregulation would imply a non-inflammatory phenotype of microglia in major depression, consistent with recent post-mortem investigations [109] that revealed, e.g., increased homeostatic marker expression in the microglia of depressed individuals [110, 111].
An additional significance of the hippocampal GAS6-TAM pathway is its dual role in regulating neurogenesis both directly by supporting neural stem cells, and indirectly by inhibiting microglia and astrocytes [103]. Given the ample evidence of long-term modulation of adult hippocampal neurogenesis by ELS [112], there might be clinical implications for targeting TAM receptor-mediated signaling pathways to treat conditions accompanied by neurogenesis loss [108, 113], such as depression [109], for which ELS is a major risk factor [3]. Importantly, especially in the context of deriving translational insights from our data, we were not able to include female mice in our study. This is pertinent given the documented sex differences in microglia [114], phenotype after ELS exposure [40, 115], and LPS response [116]. These effects should be followed up in future studies.
Overall, we report here that ELS has long-term effects on hippocampal microglia. ELS altered the distribution of morphological subtypes of microglia, the adult microglia transcriptome at basal state, the microglial developmental trajectory, and their response to an acute immune challenge during adulthood. We provide evidence that these changes have functional consequences for phagocytosis, and that microglia are lastingly impacted in the human brain after childhood abuse. These data highlight the key role of microglia in the lasting effects of ELS exposure, thereby possibly mediating the ELS-induced increased vulnerability to psychopathologies with a neuroinflammatory component such as Alzheimer’s disease and depression.
Supplementary information
Acknowledgements
The authors would like to thank E. Wesseling for assistance during microglia and RNA isolation, and C. Verseijden for carrying out the plasma cytokine measurements. The authors would like to acknowledge the expert advice on cell sorting of J. Teunis and G. Mesander from the UMCG Central Flow Cytometry Unit.
Author contributions
AK, BJLE conceptualized and supervised this study and reviewed and edited the manuscript. KR, LK, and JK conceptualized the study and wrote the manuscript. KR and JK performed all mouse-related experimental work. LK analyzed the sequencing data and prepared the related figures. NM and RR planned the human post-mortem experiments and analyses, which were conducted by RR. GT characterized the post-mortem samples, and SS helped in tissue sectioning. LV assisted with the morphological characterization of microglia. JK isolated the synaptosomes used for phagocytosis. JK, AvI, and GCS set up and ran the phagocytosis assay. GCS assisted with the operation of the flow cytometer. NB and LK were instrumental in the isolation of microglia for the sequencing experiment; SMK was instrumental with analyses and interpretation of RNAseq. All authors contributed to editing the manuscript.
Data availability
The mRNA sequencing data generated in this study is available at the Gene Expression Omnibus database, under accession number GSE207067. All other data will be made available upon request.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kitty Reemst, Laura Kracht, Janssen M. Kotah, Reza Rahimian.
These authors jointly supervised this work: Bart J.L. Eggen, Aniko Korosi.
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-022-02265-6.
References
- 1.Short AK, Baram TZ. Early-life adversity and neurological disease: age-old questions and novel answers. Nat Rev Neurol. 2019;15:657–69. doi: 10.1038/s41582-019-0246-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson CA, Scott RD, Bhutta ZA, Harris NB, Danese A, Samara M. Adversity in childhood is linked to mental and physical health throughout life. BMJ. 2020;371:m3048. doi: 10.1136/bmj.m3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saleh A, Potter GG, McQuoid DR, Boyd B, Turner R, MacFall JR, et al. Effects of early life stress on depression, cognitive performance and brain morphology. Psychol Med. 2017;47:171–81. doi: 10.1017/S0033291716002403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green JG, McLaughlin KA, Berglund PA, Gruber MJ, Sampson NA, Zaslavsky AM, et al. Childhood adversities and adult psychiatric disorders in the national comorbidity survey replication I: associations with first onset of DSM-IV disorders. Arch Gen Psychiatry. 2010;67:113–23. doi: 10.1001/archgenpsychiatry.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacMillan HL, Jan Fleming FE, David Streiner FL, Lin E, Boyle MH, Jamieson E, et al. Article childhood abuse and lifetime psychopathology in a community sample. Am J Psychiatry. 2001;158:1878–83. doi: 10.1176/appi.ajp.158.11.1878. [DOI] [PubMed] [Google Scholar]
- 6.Grainger SA, Crawford JD, Kochan NA, Mather KA, Chander RJ, Draper B, et al. An investigation into early-life stress and cognitive function in older age. Int Psychogeriatr. 2020;32:1325–9. doi: 10.1017/S1041610219001583. [DOI] [PubMed] [Google Scholar]
- 7.Mueller SC, Maheu FS, Dozier M, Peloso E, Mandell D, Leibenluft E, et al. Early-life stress is associated with impairment in cognitive control in adolescence: an fMRI study. Neuropsychologia. 2010;48:3037–44. doi: 10.1016/j.neuropsychologia.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McIlwrick S, Pohl T, Chen A, Touma C. Late-onset cognitive impairments after early-life stress are shaped by inherited differences in stress reactivity. Front Cell Neurosci. 2017;11:9. doi: 10.3389/fncel.2017.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reemst K, Ruigrok SR, Bleker L, Naninck E, Ernst T, Kotah J, et al. Sex-dependence and comorbidities of the early-life adversity induced mental and metabolic disease risks: where are we at? Neurosci Biobehav Rev. 2022;104627. [DOI] [PubMed]
- 10.Bonapersona V, Kentrop J, Van Lissa CJ, van der Veen R, Joëls M, Sarabdjitsingh RA. The behavioral phenotype of early life adversity: a 3-level meta-analysis of rodent studies. Neurosci Biobehav Rev. 2019;102:299–307. doi: 10.1016/j.neubiorev.2019.04.021. [DOI] [PubMed] [Google Scholar]
- 11.Fagundes CP, Glaser R, Kiecolt-Glaser JK. Stressful early life experiences and immune dysregulation across the lifespan. Brain Behav Immun. 2013;27:8–12. doi: 10.1016/j.bbi.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumeister D, Akhtar R, Ciufolini S, Pariante CM, Mondelli V. Childhood trauma and adulthood inflammation: a meta-analysis of peripheral C-reactive protein, interleukin-6 and tumour necrosis factor-α. Mol Psychiatry. 2016;21:642–9. doi: 10.1038/mp.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delpech JCC, Wei L, Hao J, Yu X, Madore C, Butovsky O, et al. Early life stress perturbs the maturation of microglia in the developing hippocampus. Brain Behav Immun. 2016;57:79–93. doi: 10.1016/j.bbi.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoeijmakers L, Ruigrok SR, Amelianchik A, Ivan D, van Dam AMM, Lucassen PJ, et al. Early-life stress lastingly alters the neuroinflammatory response to amyloid pathology in an Alzheimer’s disease mouse model. Brain Behav Immun. 2017;63:160–75. doi: 10.1016/j.bbi.2016.12.023. [DOI] [PubMed] [Google Scholar]
- 15.Ferle V, Repouskou A, Aspiotis G, Raftogianni A, Chrousos G, Stylianopoulou F, et al. Synergistic effects of early life mild adversity and chronic social defeat on rat brain microglia and cytokines. Physiol Behav. 2020;215:112791. doi: 10.1016/j.physbeh.2019.112791. [DOI] [PubMed] [Google Scholar]
- 16.Diz-Chaves Y, Pernía O, Carrero P, Garcia-Segura LM. Prenatal stress causes alterations in the morphology of microglia and the inflammatory response of the hippocampus of adult female mice. J Neuroinflamm. 2012;9:580. doi: 10.1186/1742-2094-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson FK, Kaffman A. Early life stress perturbs the function of microglia in the developing rodent brain: new insights and future challenges. Brain Behav Immun. 2018;69:18–27. doi: 10.1016/j.bbi.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bollinger JL, Wohleb ES. The formative role of microglia in stress-induced synaptic deficits and associated behavioral consequences. Neurosci Lett. 2019;711:134369. doi: 10.1016/j.neulet.2019.134369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butovsky O, Weiner HL. Microglial signatures and their role in health and disease. Nat. Rev. Neurosci. 2018;19:622–35. doi: 10.1038/s41583-018-0057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kierdorf K, Prinz M. Microglia in steady state. J. Clin. Investig. 2017;127:201–9. doi: 10.1172/JCI90602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf SA, Boddeke HWGM, Kettenmann H. Microglia in physiology and disease. Annu Rev Physiol. 2017;79:619–43. doi: 10.1146/annurev-physiol-022516-034406. [DOI] [PubMed] [Google Scholar]
- 22.Kettenmann H, Hanisch UK, Noda M, Verkhratsky A. Physiology of microglia. Physiol Rev. 2011;91:461–553. doi: 10.1152/physrev.00011.2010. [DOI] [PubMed] [Google Scholar]
- 23.Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74:691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thion MS, Garel S. Microglial ontogeny, diversity and neurodevelopmental functions. Curr Opin Genet Dev. 2020;65:186–94. doi: 10.1016/j.gde.2020.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Reemst K, Noctor SCSC, Lucassen PJPJ, Hol EMEM. The indispensable roles of microglia and astrocytes during brain development. Front Hum Neurosci. 2016;10:566. doi: 10.3389/fnhum.2016.00566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kentner AC, Bilbo SD, Brown AS, Hsiao EY, McAllister AK, Meyer U, et al. Maternal immune activation: reporting guidelines to improve the rigor, reproducibility, and transparency of the model. Neuropsychopharmacology. 2019;44:245–58. doi: 10.1038/s41386-018-0185-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bilbo SD, Schwarz JM. Early-life programming of later-life brain and behavior: a critical role for the immune system. Front Behav Neurosci. 2009;3:14. doi: 10.3389/neuro.08.014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diz-Chaves Y, Astiz M, Bellini MJJ, Garcia-Segura LM. Prenatal stress increases the expression of proinflammatory cytokines and exacerbates the inflammatory response to LPS in the hippocampal formation of adult male mice. Brain Behav Immun. 2013;28:196–206. doi: 10.1016/j.bbi.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Neher JJ, Cunningham C. Priming microglia for innate immune memory in the brain. Trends Immunol. 2019;40:358–74. doi: 10.1016/j.it.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 30.Schaafsma W, Basterra LB, Jacobs S, Brouwer N, Meerlo P, Schaafsma A, et al. Maternal inflammation induces immune activation of fetal microglia and leads to disrupted microglia immune responses, behavior, and learning performance in adulthood. Neurobiol Dis. 2017;106:291–300. doi: 10.1016/j.nbd.2017.07.017. [DOI] [PubMed] [Google Scholar]
- 31.Mattei D, Ivanov A, Ferrai C, Jordan P, Guneykaya D, Buonfiglioli A, et al. Maternal immune activation results in complex microglial transcriptome signature in the adult offspring that is reversed by minocycline treatment. Transl Psychiatry. 2017;7:e1120. doi: 10.1038/tp.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikezu S, Yeh H, Delpech JC, Woodbury ME, Van Enoo AA, Ruan Z, et al. Inhibition of colony stimulating factor 1 receptor corrects maternal inflammation-induced microglial and synaptic dysfunction and behavioral abnormalities. Mol Psychiatry. 2020;26:1808–31. doi: 10.1038/s41380-020-0671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chrousos GP. Stress and disorders of the stress system. Nat Rev Endocrinol. 2009;5:374–81. doi: 10.1038/nrendo.2009.106. [DOI] [PubMed] [Google Scholar]
- 34.Petrillo MG, Bortner CD, Cidlowski JA. Glucocorticoids: inflammation and immunity. In: The Hypothalamic-Pituitary-Adrenal Axis in Health and Disease: Cushing’s Syndrome and Beyond. 43–63 (Springer International Publishing; 2016).
- 35.De Pablos RM, Herrera AJ, Espinosa-Oliva AM, Sarmiento M, Muñoz MF, Machado A, et al. Chronic stress enhances microglia activation and exacerbates death of nigral dopaminergic neurons under conditions of inflammation. J Neuroinflamm. 2014;11:34. doi: 10.1186/1742-2094-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Catale C, Gironda S, Lo Iacono L, Carola V. Microglial function in the effects of early-life stress on brain and behavioral development. J Clin Med. 2020;9:468. doi: 10.3390/jcm9020468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korosi A, Hoeijmakers L, Lucassen PJ, Korosi A. The interplay of early-life stress, nutrition, and immune activation programs adult hippocampal structure and function. Front Mol Neurosci. 2015;7:1–16. doi: 10.3389/fnmol.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yam KY, Schipper L, Reemst K, Ruigrok SR, Abbink MR, Hoeijmakers L, et al. Increasing availability of ω−3 fatty acid in the early-life diet prevents the early-life stress-induced cognitive impairments without affecting metabolic alterations. FASEB J. 2019;33:5729–40. doi: 10.1096/fj.201802297R. [DOI] [PubMed] [Google Scholar]
- 39.Roque A, Ochoa-Zarzosa A, Torner L. Maternal separation activates microglial cells and induces an inflammatory response in the hippocampus of male rat pups, independently of hypothalamic and peripheral cytokine levels. Brain Behav Immun. 2016;55:39–48. doi: 10.1016/j.bbi.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 40.Naninck EFG, Hoeijmakers L, Kakava-Georgiadou N, Meesters A, Lazic SE, Lucassen PJ, et al. Chronic early life stress alters developmental and adult neurogenesis and impairs cognitive function in mice. Hippocampus. 2015;25:309–28. doi: 10.1002/hipo.22374. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Kracht L, Lerario AM, Dubbelaar ML, Brouwer N, Wesseling EM, et al. Epigenetic regulation of innate immune memory in microglia. J Neuroinflamm. 2022;19:1–19. doi: 10.1186/s12974-022-02463-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madore C, Leyrolle Q, Morel L, Rossitto M, Greenhalgh AD, Delpech JC, et al. Essential omega-3 fatty acids tune microglial phagocytosis of synaptic elements in the mouse developing brain. Nat Commun. 2020;11:6133. doi: 10.1038/s41467-020-19861-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwarz JM, Sholar PW, Bilbo SD. Sex differences in microglial colonization of the developing rat brain. J Neurochem. 2012;120:948–63. doi: 10.1111/j.1471-4159.2011.07630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wickham H ggplot2: elegant graphics for data analysis. 2016.
- 45.Galatro TF, Vainchtein ID, Brouwer N, Boddeke EWGM, Eggen BJL. Isolation of microglia and immune infiltrates from mouse and primate central nervous system. Methods Mol Biol. 2017;1559:333–42. [DOI] [PubMed]
- 46.Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, et al. Gene set knowledge discovery with enrichr. Curr Protoc. 2021;1:e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzalez-Lozano MA, Koopmans F, Sullivan PF, Protze J, Krause G, Verhage M, et al. Stitching the synapse: cross-linking mass spectrometry into resolving synaptic protein interactions. Sci Adv. 2020;6:16878. doi: 10.1126/sciadv.aax5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team. nlme: Linear and Nonlinear Mixed Effects Models. 2019.
- 49.Bankhead P, Loughrey MB, Fern ndez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker FR, Beynon SB, Jones KA, Zhao Z, Kongsui R, Cairns M, et al. Dynamic structural remodelling of microglia in health and disease: a review of the models, the signals and the mechanisms. Brain Behav Immun. 2014;37:1–14. doi: 10.1016/j.bbi.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 51.Heng Y, Zhang X, Borggrewe M, van Weering HRJ, Brummer ML, Nijboer TW, et al. Systemic administration of β-glucan induces immune training in microglia. J Neuroinflammation. 2021;18:57. doi: 10.1186/s12974-021-02103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karperien A, Ahammer H, Jelinek HF. Quantitating the subtleties of microglial morphology with fractal analysis. Front Cell Neurosci. 2013;7:3. doi: 10.3389/fncel.2013.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raj DDA, Jaarsma D, Holtman IR, Olah M, Ferreira FM, Schaafsma W, et al. Priming of microglia in a DNA-repair deficient model of accelerated aging. Neurobiol Aging. 2014;35:2147–60. doi: 10.1016/j.neurobiolaging.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 54.Holtman IR, Raj DD, Miller JA, Schaafsma W, Yin Z, Brouwer N, et al. Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: a co-expression meta-analysis. Acta Neuropathol Commun. 2015;3:31. doi: 10.1186/s40478-015-0203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang D, Levine JLS, Avila-Quintero V, Bloch M, Kaffman A. Systematic review and meta-analysis: effects of maternal separation on anxiety-like behavior in rodents. Transl Psychiatry. 2020;10:174. doi: 10.1038/s41398-020-0856-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bennett ML, Bennett FC, Liddelow SA, Ajami B, Zamanian JL, Fernhoff NB, et al. New tools for studying microglia in the mouse and human CNS. Proc Natl Acad Sci. 2016;113:E1738–46. doi: 10.1073/pnas.1525528113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olah M, Patrick E, Villani AC, Xu J, White CC, Ryan KJ, et al. A transcriptomic atlas of aged human microglia. Nat Commun. 2018;9:539. doi: 10.1038/s41467-018-02926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barnett Burns S, Almeida D, Turecki G. The epigenetics of early life adversity: current limitations and possible solutions. Prog Mol BiolTransl Sci. 2018;157:343–425. doi: 10.1016/bs.pmbts.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 59.Weaver ICG, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–54. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Szyf M. DNA methylation, behavior and early life adversity. J Genet Genomics. 2013;40:331–8. doi: 10.1016/j.jgg.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 61.Catale C, Bussone S, Lo Iacono L, Viscomi MT, Palacios D, Troisi A, et al. Exposure to different early-life stress experiences results in differentially altered DNA methylation in the brain and immune system. Neurobiol Stress. 2020;13:100249. doi: 10.1016/j.ynstr.2020.100249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teissier A, Le Magueresse C, Olusakin J, Andrade da Costa BLS, De Stasi AM, Bacci A, et al. Early-life stress impairs postnatal oligodendrogenesis and adult emotional behaviour through activity-dependent mechanisms. Mol Psychiatry. 2020;25:1159–74. doi: 10.1038/s41380-019-0493-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abbink MR, van Deijk ALF, Heine VM, Verheijen MH, Korosi A. The involvement of astrocytes in early-life adversity induced programming of the brain. Glia. 2019;67:1637–53. doi: 10.1002/glia.23625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abbink MR, Kotah JM, Hoeijmakers L, Mak A, Yvon-Durocher G, Van Der Gaag B, et al. Characterization of astrocytes throughout life in wildtype and APP/PS1 mice after early-life stress exposure. J Neuroinflamm. 2020;17:91. doi: 10.1186/s12974-020-01762-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McKenzie AT, Wang M, Hauberg ME, Fullard JF, Kozlenkov A, Keenan A, et al. Brain cell type specific gene expression and co-expression network architectures. Sci Rep. 2018;8:8868. doi: 10.1038/s41598-018-27293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno-García Á, Bernal-Chico A, Colomer T, Rodríguez-Antigüedad A, Matute C, Mato S. Gene expression analysis of astrocyte and microglia endocannabinoid signaling during autoimmune demyelination. Biomolecules. 2020;10:1–19. doi: 10.3390/biom10091228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noristani HN, Gerber YN, Sabourin JC, Le Corre M, Lonjon N, Mestre-Frances N, et al. RNA-Seq analysis of microglia reveals time-dependent activation of specific genetic programs following spinal cord injury. Front Mol Neurosci. 2017;10:90. doi: 10.3389/fnmol.2017.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park JY, Kim HY, Jou I, Park SM. GM1 induces p38 and microtubule dependent ramification of rat primary microglia in vitro. Brain Res. 2008;1244:13–23. doi: 10.1016/j.brainres.2008.09.072. [DOI] [PubMed] [Google Scholar]
- 69.Das R, Chinnathambi S. Actin-mediated microglial chemotaxis via g-protein coupled purinergic receptor in Alzheimer’s Disease. Neuroscience. 2020;448:325–336. doi: 10.1016/j.neuroscience.2020.09.024. [DOI] [PubMed] [Google Scholar]
- 70.Okazaki T, Saito D, Inden M, Kawaguchi K, Wakimoto S, Nakahari T, et al. Moesin is involved in microglial activation accompanying morphological changes and reorganization of the actin cytoskeleton. J Physiol Sci. 2020;70:52. doi: 10.1186/s12576-020-00779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Derks NAV, Krugers HJ, Hoogenraad CC, Joels M, Sarabdjitsingh RA, Joëls M, et al. Effects of early life stress on synaptic plasticity in the developing hippocampus of male and female rats. PLoS One. 2016;11:e0164551–17. doi: 10.1371/journal.pone.0164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sierra-Fonseca JA, Hamdan JN, Cohen AA, Cardenas SM, Saucedo S, Lodoza GA, et al. Neonatal maternal separation modifies proteostasis marker expression in the adult hippocampus. Front Mol Neurosci. 2021;14:155. doi: 10.3389/fnmol.2021.661993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmidt MF, Gan ZY, Komander D, Dewson G. Ubiquitin signalling in neurodegeneration: mechanisms and therapeutic opportunities. Cell Death Differ. 2021;28:570–590. doi: 10.1038/s41418-020-00706-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Knuesel I, Chicha L, Britschgi M, Schobel SA, Bodmer M, Hellings JA, et al. Maternal immune activation and abnormal brain development across CNS disorders. Nat Rev Neurol. 2014;10:643–60. doi: 10.1038/nrneurol.2014.187. [DOI] [PubMed] [Google Scholar]
- 75.Hoeijmakers L, Lesuis SL, Krugers H, Lucassen PJ, Korosi A. A preclinical perspective on the enhanced vulnerability to Alzheimer’s disease after early-life stress. Neurobiol Stress. 2018;8:172–85. doi: 10.1016/j.ynstr.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Madore C, Yin Z, Leibowitz J, Butovsky O. Microglia, lifestyle stress, and neurodegeneration. Immunity. 2020;52:222–40. [DOI] [PMC free article] [PubMed]
- 77.Lesuis SL, Hoeijmakers L, Korosi A, De Rooij SR, Swaab DF, Kessels HW, et al. Vulnerability and resilience to Alzheimer’s disease: early life conditions modulate neuropathology and determine cognitive reserve. Alzheimer’s Res Ther. 2018;10:1–20. doi: 10.1186/s13195-018-0422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Donley GAR, Lönnroos E, Tuomainen TP, Kauhanen J. Association of childhood stress with late-life dementia and Alzheimer’s disease: the KIHD study. Eur J Public Health. 2018;28:1069–73. doi: 10.1093/eurpub/cky134. [DOI] [PubMed] [Google Scholar]
- 79.Ravona-Springer R, Beeri MS, Goldbourt U. Younger age at crisis following parental death in male children and adolescents is associated with higher risk for dementia at old age. Alzheimer Dis Assoc Disord. 2012;26:68–73. doi: 10.1097/WAD.0b013e3182191f86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Radford K, Delbaere K, Draper B, Mack HA, Daylight G, Cumming R, et al. Childhood stress and adversity is associated with late-life dementia in aboriginal Australians. Am J Geriatr Psychiatry. 2017;25:1097–106. doi: 10.1016/j.jagp.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 81.Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron. 2019;101:207–23.e10. doi: 10.1016/j.neuron.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 2016;353:aad8670. doi: 10.1126/science.aad8670. [DOI] [PubMed] [Google Scholar]
- 83.Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019;50:253–71.e6. doi: 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kracht L, Borggrewe M, Eskandar S, Brouwer N, Chuva de Sousa Lopes SM, Laman JD, et al. Human fetal microglia acquire homeostatic immune-sensing properties early in development. Science. 2020;369:530–7. doi: 10.1126/science.aba5906. [DOI] [PubMed] [Google Scholar]
- 85.Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17:131–43. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Smith ELP, Batuman OA, Trost RC, Coplan JD, Rosenblum LA. Transforming growth factor-β1 and cortisol in differentially reared primates. Brain Behav Immun. 2002;16:140–9. doi: 10.1006/brbi.2001.0629. [DOI] [PubMed] [Google Scholar]
- 87.Ben-Yehuda H, Matcovitch-Natan O, Kertser A, Spinrad A, Prinz M, Amit I, et al. Maternal Type-I interferon signaling adversely affects the microglia and the behavior of the offspring accompanied by increased sensitivity to stress. Mol Psychiatry. 2020;25:1050–67. doi: 10.1038/s41380-019-0604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reemst K, Broos JY, Abbink MR, Cimetti C, Giera M, Kooij G, et al. Early-life stress and dietary fatty acids impact the brain lipid/oxylipin profile into adulthood, basally and in response to LPS. Front Immunol. 2022;13:967437. doi: 10.3389/fimmu.2022.967437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee JW, Nam H, Kim LE, Jeon Y, Min H, Ha S, et al. TLR4 (toll-like receptor 4) activation suppresses autophagy through inhibition of FOXO3 and impairs phagocytic capacity of microglia. Autophagy. 2019;15:753–70. doi: 10.1080/15548627.2018.1556946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ye X, Zhu M, Che X, Wang H, Liang XJ, Wu C, et al. Lipopolysaccharide induces neuroinflammation in microglia by activating the MTOR pathway and downregulating Vps34 to inhibit autophagosome formation. J Neuroinflamm. 2020;17:18. doi: 10.1186/s12974-019-1644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nederhof E, Schmidt MV. Mismatch or cumulative stress: toward an integrated hypothesis of programming effects. Physiol Behav. 2012;106:691–700. doi: 10.1016/j.physbeh.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 92.Schaafsma W, Zhang X, van Zomeren KC, Jacobs S, Georgieva PB, Wolf SA, et al. Long-lasting pro-inflammatory suppression of microglia by LPS-preconditioning is mediated by RelB-dependent epigenetic silencing. Brain Behav Immun. 2015;48:205–21. doi: 10.1016/j.bbi.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 93.Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–8. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 94.Milior G, Lecours C, Samson L, Bisht K, Poggini S, Pagani F, et al. Fractalkine receptor deficiency impairs microglial and neuronal responsiveness to chronic stress. Brain Behav Immun. 2016;55:114–25. doi: 10.1016/j.bbi.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 95.Wohleb ES, Terwilliger R, Duman CH, Duman RS. Stress-induced neuronal colony stimulating factor 1 provokes microglia-mediated neuronal remodeling and depressive-like behavior. Biol Psychiatry. 2018;83:38–49. doi: 10.1016/j.biopsych.2017.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bolton JL, Short AK, Othy S, Kooiker CL, Shao M, Gunn BG, et al. Early stress-induced impaired microglial pruning of excitatory synapses on immature CRH-expressing neurons provokes aberrant adult stress responses. Cell Rep. 2022;38:110600. doi: 10.1016/j.celrep.2022.110600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brunson KL, Eghbal-Ahmadi M, Bender R, Chen Y, Baram TZ. Long-term, progressive hippocampal cell loss and dysfunction induced by early-life administration of corticotropin-releasing hormone reproduce the effects of early-life stress. Proc Natl Acad Sci USA. 2001;98:8856–61. doi: 10.1073/pnas.151224898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kanatsou S, Karst H, Kortesidou D, Van Den Akker RA, den Blaauwen J, Harris AP, et al. Overexpression of mineralocorticoid receptors in the mouse forebrain partly alleviates the effects of chronic early life stress on spatial memory, neurogenesis and synaptic function in the dentate gyrus. Front Cell Neurosci. 2017;11:132. doi: 10.3389/fncel.2017.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liao XM, Yang XD, Jia J, Li JT, Xie XM, Su YA, et al. Blockade of corticotropin-releasing hormone receptor 1 attenuates early-life stress-induced synaptic abnormalities in the neonatal hippocampus. Hippocampus. 2014;24:528–40. doi: 10.1002/hipo.22254. [DOI] [PubMed] [Google Scholar]
- 100.Xu B, Zhang X, He Y, Liu C, Li L, Liu Q, et al. The impacts of early-life adversity on striatal and hippocampal memory functions. Neuroscience. 2022;490:11–24. doi: 10.1016/j.neuroscience.2022.02.029. [DOI] [PubMed] [Google Scholar]
- 101.Ivy AS, Rex CS, Chen Y, Dubé C, Maras PM, Grigoriadis DE, et al. Hippocampal dysfunction and cognitive impairments provoked by chronic early-life stress involve excessive activation of CRH receptors. J Neurosci. 2010;30:13005. doi: 10.1523/JNEUROSCI.1784-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Grommes C, Lee CYD, Wilkinson BL, Jiang Q, Koenigsknecht-Talboo JL, Varnum B, et al. Regulation of microglial phagocytosis and inflammatory gene expression by Gas6 acting on the Axl/Mer family of tyrosine kinases. J Neuroimmune Pharm. 2008;3:130–40. doi: 10.1007/s11481-007-9090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gilchrist SE, Pennelli GM, Hafizi S. Gas6/tam signalling negatively regulates inflammatory induction of gm-csf in mouse brain microglia. Cells. 2021;10:3281. doi: 10.3390/cells10123281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Goudarzi S, Gilchrist SE, Hafizi S. Gas6 induces myelination through anti-inflammatory IL-10 and TGF-β upregulation in white matter and glia. Cells. 2020;9:1779. doi: 10.3390/cells9081779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Prieto AL, Weber JL, Tracy S, Heeb MJ, Lai C. Gas6, a ligand for the receptor protein-tyrosine kinase Tyro-3, is widely expressed in the central nervous system. Brain Res. 1999;816:646–61. doi: 10.1016/S0006-8993(98)01159-7. [DOI] [PubMed] [Google Scholar]
- 106.Funakoshi H, Yonemasu T, Nakano T, Matumoto K, Nakamura T. Identification of Gas6, a putative ligand for Sky and Axl receptor tyrosine kinases, as a novel neurotrophic factor for hippocampal neurons. J Neurosci Res. 2002;68:150–60. doi: 10.1002/jnr.10211. [DOI] [PubMed] [Google Scholar]
- 107.Chung WS, Clarke LE, Wang GX, Stafford BK, Sher A, Chakraborty C, et al. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013;504:394–400. doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Johnson K, Ji R. TAM receptors: Two pathways to regulate adult neurogenesis. Neural Regen Res. 2015;10:344–5. doi: 10.4103/1673-5374.153671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rahimian R, Wakid M, O’Leary LA, Mechawar N. The emerging tale of microglia in psychiatric disorders. Neurosci Biobehav Rev. 2021;131:1–29. doi: 10.1016/j.neubiorev.2021.09.023. [DOI] [PubMed] [Google Scholar]
- 110.Böttcher C, Fernández-Zapata C, Snijders GJL, Schlickeiser S, Sneeboer MAM, Kunkel D, et al. Single-cell mass cytometry of microglia in major depressive disorder reveals a non-inflammatory phenotype with increased homeostatic marker expression. Transl Psychiatry. 2020;10:310. doi: 10.1038/s41398-020-00992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Snijders GJLJ, Sneeboer MAM, Fernández-Andreu A, Udine E, Boks MP, Ormel PR, et al. Distinct non-inflammatory signature of microglia in post-mortem brain tissue of patients with major depressive disorder. Mol Psychiatry. 2021;26:3336–49. doi: 10.1038/s41380-020-00896-z. [DOI] [PubMed] [Google Scholar]
- 112.Korosi A, Naninck EFG, Oomen CA, Schouten M, Krugers H, Fitzsimons C, et al. Early-life stress mediated modulation of adult neurogenesis and behavior. Behav Brain Res. 2012;227:400–9. doi: 10.1016/j.bbr.2011.07.037. [DOI] [PubMed] [Google Scholar]
- 113.Ji R, Tian S, Lu HJ, Lu Q, Zheng Y, Wang X, et al. TAM receptors affect adult brain neurogenesis by negative regulation of microglial cell activation. J Immunol. 2013;191:6165–77. doi: 10.4049/jimmunol.1302229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Han J, Fan Y, Zhou K, Blomgren K, Harris RA. Uncovering sex differences of rodent microglia. J Neuroinflamm. 2021;18:74. doi: 10.1186/s12974-021-02124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Abbink MR, Naninck EFG, Lucassen PJ, Korosi A. Early-life stress diminishes the increase in neurogenesis after exercise in adult female mice. Hippocampus. 2017;27:839–44. doi: 10.1002/hipo.22745. [DOI] [PubMed] [Google Scholar]
- 116.Dockman RL, Carpenter JM, Diaz AN, Benbow RA, Filipov NM. Sex differences in behavior, response to LPS, and glucose homeostasis in middle-aged mice. Behav Brain Res. 2022;418:113628. doi: 10.1016/j.bbr.2021.113628. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mRNA sequencing data generated in this study is available at the Gene Expression Omnibus database, under accession number GSE207067. All other data will be made available upon request.