Abstract
We investigated the reason for the inability of lipopolysaccharide (LPS)-resistant (Lps-defective [Lpsd]) C57BL/10ScCr mice to produce beta interferon (IFN-β) when stimulated with bacteria. For this purpose, the IFN-β and other macrophage cytokine responses induced by LPS and several killed gram-negative and gram-positive bacteria in LPS-sensitive (Lps-normal [Lpsn]; C57BL/10ScSn and BALB/c) and Lpsd (C57BL/10ScCr and BALB/c/l) mice in vitro and in vivo were investigated on the mRNA and protein levels. In addition, double-stranded RNA (dsRNA) was used as a nonbacterial stimulus. LPS and all gram-negative bacteria employed induced IFN-β in the Lpsn mice but not in the Lpsd mice. All gram-positive bacteria tested failed to induce significant amounts of IFN-β in all four of the mouse strains used. As expected, all other cytokines tested (tumor necrosis factor alpha, interleukin 1α [IL-1α], IL-6, and IL-10) were differentially induced by gram-negative and gram-positive bacteria. Stimulation with dsRNA induced IFN-β and all other cytokines mentioned above in all mouse strains, regardless of their LPS sensitivities. The results suggest strongly that LPS is the only bacterial component capable of inducing IFN-β in significant amounts that are readily detectable under the conditions used in this study. Consequently, in mice, IFN-β is inducible only by gram-negative bacteria, but not in C57BL/10ScCr or other LPS-resistant mice.
In mice, sensitivity to lipopolysaccharide (LPS) is determined by a locus on chromosome 4 which has been designated the Lps gene (36). Mice with defective Lps genes are highly resistant to the biological activity of LPS. The LPS-resistant phenotype has been described in three mouse strains, C57BL/10ScCr (Cr) (5), C3H/HeJ (30), and C57BL/ScN (37). In addition to these naturally occurring mutants, a fourth LPS-resistant strain, BALB/c/l, was produced independently in two laboratories (32, 38) by backcrossing the defective Lps gene from C3H/HeJ into the BALB/c background. The LPS-resistant strains are designated Lpsd (Lps defective), and the LPS-sensitive strains are designated Lpsn (Lps normal). Very recently, evidence has been presented that Lps and Toll-like receptor-4 (Tlr4) genes are identical. Cr mice were shown to be homozygous for a null mutation of the Lps/Tlr4 gene, while C3H/HeJ mice carry a missense mutation of the gene, predicted to replace a proline with a histidine at position 712 of the Tlr4 polypeptide chain (24). Mice of all the Lpsd strains mentioned above are, under normal conditions, highly resistant to all LPS effects. There is, however, an important difference between the Cr and the C3H/HeJ and BALB/c/l strains. This concerns their abilities to produce gamma interferon (IFN-γ) in response to microorganisms; the response is normal in C3H/HeJ and BALB/c/l but highly impaired in Cr mice (11). As demonstrated in an earlier study, IFN-γ is a key mediator of the LPS hypersensitivity induced by infection (10, 17). Consequently, when C3H/HeJ or BALB/c/l mice are infected or treated with killed bacteria, they become partial LPS responders, while Cr mice retain their LPS-resistant phenotype (12).
The defective IFN-γ responses of Cr mice have been demonstrated both in vivo and in vitro by treatment of the mice with live or killed bacteria or following infection with certain parasites (Plasmodium chabaudi chabaudi and Leishmania major). Cr mice, however, do not exhibit a general defect in IFN-γ response, since splenocytes of these mice produce high levels of IFN-γ when stimulated with the T-cell mitogen concanavalin A or with monoclonal antibodies to CD3. The defective IFN-γ response of these mice therefore seems to be confined to IFN-γ induced by microorganisms (11, 21, 40).
Recently, we observed that splenocytes of Cr mice, supplemented with macrophages of the related, IFN-γ-normal C57BL/10ScSn (Sn) mice, acquire the ability to produce IFN-γ in response to gram-negative bacteria. A similar effect was also achieved with supernatants of Sn macrophages that had been stimulated with killed gram-negative bacteria. Such supernatants by themselves do not directly induce IFN-γ in Cr splenocytes; however, they enable these cells to produce IFN-γ in the presence of a bacterial stimulus. The helper factor present in the active supernatants was identified as IFN-β. This provided evidence that IFN-β is a cofactor for IFN-γ production by gram-negative bacteria and that it is missing in Cr mice (40). The reason for the absence of an IFN-β production in Cr mice, however, remained unclear.
In the present study, we used Cr and BALB/c/l mice and the corresponding Lpsn mice of the strains Sn and BALB/c, respectively, to investigate the reason for the absence of IFN-β production in Cr mice stimulated with gram-negative bacteria. We show that IFN-β production induced in mice by bacteria is a function of the LPS component, and therefore the inability of Cr mice to produce IFN-β after stimulation with bacteria is directly related to their LPS-resistant phenotype.
MATERIALS AND METHODS
Animals.
Lpsd C57BL/10ScCr (Cr) and BALB/c/l (32) and Lpsn C57BL/10ScSn (Sn) and BALB/c mouse strains were bred under specific-pathogen-free conditions in the animal facilities of the Max-Planck-Institut für Immunbiologie. Four-week-old mice of either sex were used as donors of bone marrow-derived precursor cells, and 6- to 8-week-old animals were used as donors of splenocytes and for in vivo experiments.
Treatment of mice.
For injection, the agents under test were dissolved or suspended (bacteria) in pyrogen-free phosphate-buffered saline (PBS) and administered to mice (0.2 ml/animal) intravenously in the lateral tail vein. One hour after injection, the mice were sacrificed under anesthesia, and the spleens were removed and treated for RNA extraction as described below. The spleen samples were stored at −80°C until they were used.
Macrophages.
Macrophages were cultured from bone marrow precursor cells of the various mouse strains in the presence of L-cell-conditioned medium, as previously described (9). Cells obtained after 10 days of culture were centrifuged, washed twice, and suspended in serum-free, high-glucose formulations of Dulbecco modified Eagle medium at a concentration of 106/ml. The macrophages (3 × 106/well) were placed in six-well plates (Costar, Cambridge, Mass.) and cultured at 37°C in a humidified atmosphere containing 8% CO2 for 24 h. Thereafter, the macrophage supernatants were replaced by fresh medium and 30 μl of the stimulating agent under test per well was added. Cultivation then continued for different periods of time. Culture supernatants for cytokine measurement were collected and stored in aliquots at −80°C. Total macrophage RNA was extracted as described below.
Materials.
The gram-negative bacteria Salmonella enterica serovar Typhimurium (C5), Escherichia coli (J5), Proteus mirabilis, Pseudomonas aeruginosa, Shigella enteritidis, and Vibrio cholerae and the gram-positive Listeria monocytogenes and Staphylococcus aureus were obtained from overnight cultures. Other gram-positive bacteria employed, Lactobacillus bulgaricus and Streptococcus thermophilus, were a kind gift from C. De Simone, University of L'Aquila, L'Aquila, Italy, and Propionibacterium acnes ATCC 12930 was kindly provided by S. Schlecht, Max-Planck-Institut für Immunbiologie, Freiburg, Germany. All bacteria were washed twice with pyrogen-free PBS (pH 7.2) and killed by heating them at 65°C for 1 h. They were subsequently centrifuged, washed twice with pyrogen-free distilled water, and lyophilized. Endotoxin contamination in the gram-positive bacterial preparations was <0.1 pg/mg as determined by the Limulus amoebocyte lysate test (33). For use, the bacteria were suspended in pyrogen-free PBS, pH 7.2.
LPS of Salmonella enterica serovar Abortus equi in its uniform triethylamine salt form was obtained as described earlier (14). A sterile aqueous stock solution (10 mg/ml) was prepared and stored at 4°C. Before use, the LPS was diluted further with pyrogen-free PBS to the desired concentration.
Double-stranded RNA [dsRNA; poly(I):poly(C)] was purchased from Boehringer (Mannheim, Germany). Recombinant murine IFN-β was a kind gift from M. Moriyama (Toray Industries Inc., Tokyo, Japan). Monoclonal anti-IFN-β (rat immunoglobulin G1) was purchased from Yamasa Shoyu (Tokyo, Japan), and monoclonal anti-IFN-α (rat immunoglobulin G1) was purchased from GIBCO BRL (Gaithersburg, Md.).
ELISAs.
For measurement of cytokines in supernatants of stimulated macrophages, commercial enzyme-linked immunosorbent assay (ELISA) kits were used. For interleukin 1α (IL-1α), an ELISA kit was obtained from Genzyme, Cambridge, Mass.; for IL-6 and IL-10, the kits were from PerSeptive Diagnostics, Cambridge, Mass. The detection limits of the assays were 0.5 pg/ml for IL-1α, 10 pg/ml for IL-6, and 5 pg/ml for IL-10. IFN-γ in supernatants of spleen cell cultures was estimated by a previously described ELISA (29). The limit of detection was 75 pg/ml.
Bioassays.
Tumor necrosis factor alpha (TNF-α) in macrophage supernatants was measured in a cytotoxicity test using a TNF-α-sensitive L 929 cell line as described earlier (1, 8). The detection limit of the test was 4 pg of TNF-α/ml of supernatant.
IFN-β was measured in a bioassay described previously (40). The assay is based on the fact that Cr splenocytes which produce no IFN-γ when stimulated with serovar Typhimurium do so when exogenous IFN-β is added as a cofactor. The amounts of IFN-γ thus induced by a given number of serovar Typhimurium cells is dependent on the dose of IFN-β added. In this assay, the IFN-β present in macrophage supernatants was determined by comparing the potency of such supernatants to support an IFN-γ response to serovar Typhimurium in Cr splenocytes with that of standard recombinant murine IFN-β. Splenocyte suspensions were prepared from the spleens of three or more mice, pooled, and adjusted to a concentration of 2 × 107/ml of Dulbecco modified Eagle medium. The spleen cells (100 μl/well) were placed in 96-well plastic plates (Nunc, Roskilde, Denmark) together with 100 μl of diluted macrophage supernatant or recombinant murine IFN-β (standard)/well and 10 μl of serovar Typhimurium cells (20 μg)/well. Cultures containing supernatant of unstimulated macrophages and cultures without serovar Typhimurium served as controls. After 24 h of culture at 37°C in a humidified atmosphere containing 5% CO2, supernatants for IFN-γ measurement were collected and stored frozen at −80°C until they were used. Inhibition of the IFN-γ response by preincubation of active macrophage supernatants with monoclonal anti-IFN-β (2 μg/100 μl) for 30 min at 4°C was used to verify the IFN-β specificity of the assay. A similar preincubation with monoclonal anti-IFN-α had no effect on the IFN-γ response. The detection limit of the test was 25 U of IFN-β/ml.
The amount of biologically active IL-1 was measured with human dermal fibroblasts, as described previously (20). The detection limit of the assay was 10 pg/ml.
RNA extraction.
Total RNA was isolated from cells and from spleens by a guanidinium isothiocyanate–phenol–chloroform-isoamyl alcohol procedure (3). Briefly, live cells or freshly removed spleens were homogenized in 1.8 ml of solution D (4 M guanidinium isothiocyanate, 25 mM sodium citrate [pH 7], 0.5% sarcosyl, and 0.1 M 2-mercaptoethanol) per organ. Subsequently, 1/10 of the volume of 2 M sodium acetate (pH 4) was added. The homogenates were stored at −80°C until RNA extraction. Total RNA was extracted from homogenates with 1 volume of Tris-EDTA-saturated phenol and 1/5 homogenate volume of chloroform-isoamyl alcohol (49:1). RNA was precipitated from the aqueous phase by the addition of 1 volume of isopropanol chilled to −20°C. The precipitates were washed twice with cold 70% ethanol and resuspended in 15 to 20 μl of RNase-free H2O. The RNA concentration was determined by absorbance at 260 nm.
Northern blot analysis.
RNA samples (2 to 15 μg) were fractionated on 1.2% denaturing agarose-formaldehyde gels and transferred to Nytran filters as described previously (18). The RNAs were hybridized overnight at 65°C with random-primed 32P-labeled cDNA probes as described previously. The IFN-β probe was a 571-bp cDNA fragment spanning the complete open reading frame for murine IFN-β (40). The IFN-α probe was a 581-bp fragment spanning the complete open reading frame for murine IFN-α4. This fragment was generated by PCR using the primers mmIFNA1 (5′-CACCATGGCTAGGCTCTGT-3′) and mmIFNA1R (5′-CACTTTGTCTCAGGACTCCA-3′) and subcloned into Bluescript (Stratagene). The conditions for PCR were 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s for 30 cycles using mouse genomic DNA as a template. The TNF-α probe was a 1,100-bp cDNA fragment spanning the complete open reading frame for murine TNF-α, kindly provided by B. Beutler, Howard Hughes Medical Institute, Dallas, Tex. Under the above-mentioned conditions, in agreement with earlier findings (6, 22) a major TNF-α band (approximately at the position of 18S RNA), and sometimes a second, weaker TNF-α band (below 28S RNA), appeared after hybridization.
The amount of total RNA applied to the gel for each sample was visualized by the intensity of the ethidium bromide-stained band (18S rRNA) on the Nytran filter used for hybridization.
For technical reasons, it was impossible to use Northern blotting to study the induction of TNF-α mRNA in macrophages stimulated with dsRNA. The radiolabeled TNF-α probe hybridized with the exogenously added dsRNA present in the total-RNA extracts of macrophages and obscured the TNF-α mRNA signal (see Fig. 1 and 2).
FIG. 1.
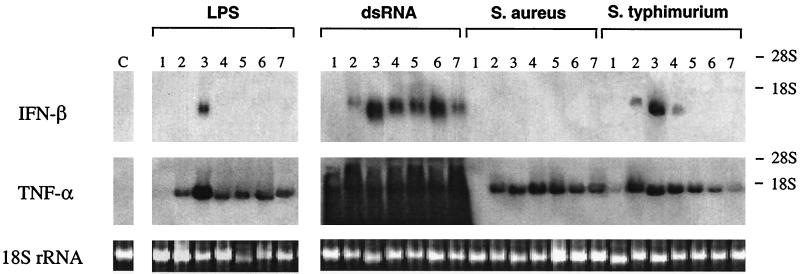
Kinetics of IFN-β and TNF-α mRNA induction in Sn macrophages stimulated with different agents. Macrophages (106/ml) were stimulated with LPS (10 μg/ml), dsRNA (10 μg/ml), serovar Typhimurium (100 μg/ml), and S. aureus (100 μg/ml) for different times. IFN-β and TNF-α mRNAs were detected by Northern blot analysis of total macrophage RNA (pools of duplicates), as described in Materials and Methods. The exposure time for IFN-β was 2 weeks, and that for TNF-α was 12 h. RNA applied to the gel (approximately 2 μg/lane) was visualized for each sample by the intensity of the ethidium bromide-stained 18S rRNA bands. Stimulation times were as follows: lanes 1, 20 min; lanes 2, 1 h; lanes 3, 3 h; lanes 4, 6 h; lanes 5, 9 h; lanes 6, 12 h; lanes 7, 24 h; lane C, unstimulated macrophages.
FIG. 2.
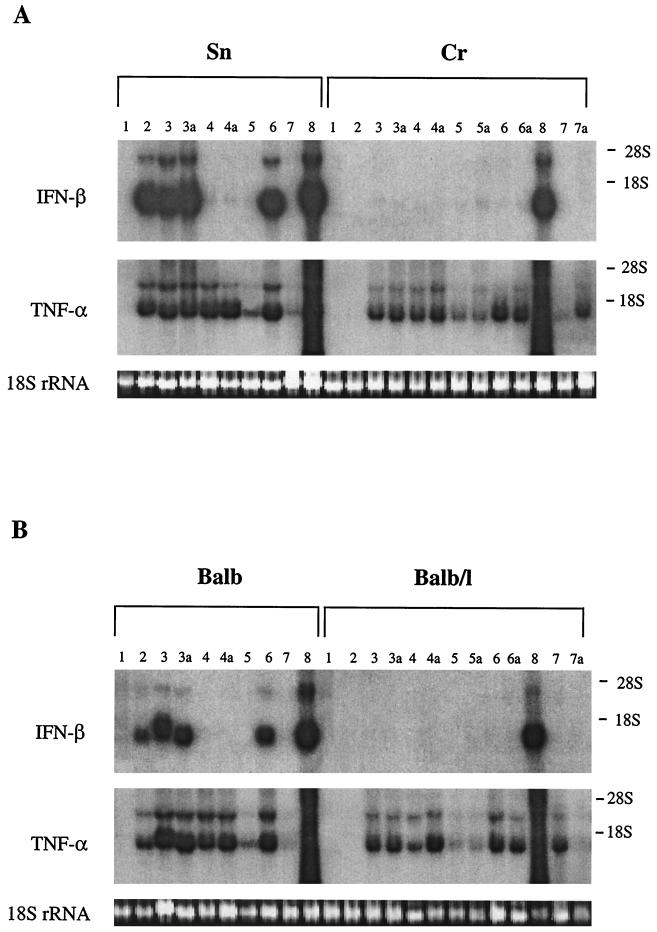
Induction of IFN-β and TNF-α mRNAs in Lpsn and Lpsd macrophages stimulated with LPS and different bacteria. Untreated control macrophages (106/ml [lanes 1]) or macrophages treated with LPS (10 μg/ml [lanes 2]), serovar Typhimurium (100 μg/ml [lanes 3] and 500 μg/ml [lanes 3a]), S. aureus (100 μg/ml [lanes 4] and 500 μg/ml [lanes 4a]), P. acnes (100 μg/ml [lanes 5] and 500 μg/ml [lanes 5a]), E. coli (100 μg/ml [lanes 6] and 500 μg/ml [lanes 6a]), S. thermophilus (100 μg/ml [lanes 7] and 500 μg/ml [lanes 7a]), and dsRNA (10 μg/ml [lanes 8]) were cultured for 3 h. Thereafter, IFN-β and TNF-α mRNAs were detected by Northern blot analysis of total macrophage RNA (pools of duplicates), as described in Materials and Methods. The exposure time for IFN-β was 2 weeks, and that for TNF-α was 3 h. RNAs applied to the gel (approximately 6 μg/lane) were visualized for each sample by the intensity of the ethidium bromide-stained 18S rRNA bands.
RPA.
Cytokine mRNAs were detected by an RNase protection assay (RPA) (15) using a RiboQuant Multi-Probe RPA system (Pharmingen, San Diego, Calif.) as described in the Pharmingen standard protocol. Briefly, the template set (mCK-3b) was used for a T7 RNA polymerase-dependent synthesis of 32P-labeled antisense RNA probes. The RNA samples were hybridized overnight with an excess of labeled probes. After treatment with RNases A and T1 and with proteinase K, the samples were loaded on a 6% polyacrylamide-Tris-borate-EDTA-urea gel and run at 1,900 V with 1× Tris-borate-EDTA electrophoresis buffer, pH 8.3. The gel was dried, and autoradiography film (BIOMAX MS; Kodak, Rochester, N.Y.) was exposed using an intensifying screen (Cronex Lightening Plus; Dupont).
RESULTS
Kinetics of induction of IFN-β and TNF-α mRNAs by different stimuli in macrophages.
To determine the time of maximal expression of IFN-β and TNF-α mRNAs, macrophages of the Lpsn Sn mice were stimulated with LPS, serovar Typhimurium, S. aureus, or dsRNA for different periods of time (up to 24 h). Unstimulated macrophages cultured in parallel served as a control. Total macrophage RNA was then isolated, and expression of IFN-β and TNF-α mRNAs was investigated by Northern blot analysis (Fig. 1). Control macrophages expressed no IFN-β or TNF-α mRNA at any time point. Macrophages stimulated with LPS or either of the two bacteria exhibited a strong expression of TNF-α mRNA after 1 h that remained detectable for up to 24 h. LPS and bacteria, however, differed in their potencies to induce IFN-β mRNA. While LPS and the gram-negative serovar Typhimurium induced a transient expression with a peak at 3 h, the gram-positive S. aureus induced no expression of IFN-β mRNA. dsRNA was the only stimulus used that was capable of a strong, long-lasting induction of IFN-β mRNA in Sn macrophages. Due to technical reasons, the induction of TNF-α mRNA by dsRNA could not be evaluated (see Materials and Methods). Evidence that dsRNA does induce TNF-α in macrophages is presented below. The results show that 3 h of stimulation is a good compromise time point for measuring the induction of all messages, and it was adopted in the following experiments.
Comparative analysis of IFN-β and TNF-α mRNAs induced by gram-negative and gram-positive bacteria in Lpsn and Lpsd macrophages.
Macrophages of Lpsn (Sn and BALB/c) and Lpsd (Cr and BALB/c/l) mice were stimulated with gram-negative serovar Typhimurium and E. coli) and gram-positive (S. aureus, P. acnes, and S. thermophilus) bacteria. In addition, LPS and dsRNA were used as control stimuli. The induction of IFN-β and TNF-α mRNAs after 3 h of stimulation is shown in Fig. 2. As expected, LPS induced IFN-β and TNF-α mRNA in macrophages of Lpsn but not Lpsd mice. Interestingly, while the two gram-negative bacteria induced TNF-α mRNA in all types of macrophages, they induced significant amounts of IFN-β mRNA only in the Lpsn macrophages. Also, the gram-positive bacteria induced varying amounts of TNF-α mRNA in all types of macrophages. However, an induction of IFN-β mRNA by gram-positive bacteria was either undetectable or extremely weak. Thus, in some cases, after longer exposure times of the autoradiography film very faint bands of IFN-β mRNA became visible. These were several orders of magnitude weaker than those induced in Lpsn macrophages by gram-negative bacteria. Since similar weak bands were also seen in some of the untreated control macrophages as well as in Cr macrophages treated with gram-negative bacteria, it cannot be decided whether such bands represent constitutive IFN-β mRNA expression or whether they represent a very weak mRNA induction. In separate experiments in which macrophages of Lpsn and Lpsd mice were stimulated with L. monocytogenes (300 μg/106 cells), similar results were obtained (not shown). Both types of macrophages exhibited strong expression of TNF-α mRNA; however, IFN-β mRNA was not detectable. On long exposure of the autoradiography film (2 weeks), an extremely weak IFN-β signal became discernible. As expected, dsRNA was a potent inducer of IFN-β in all types of macrophages. Again, the inducibility of TNF-α by dsRNA could not be evaluated for the reasons given in Materials and Methods. From these results, we conclude that the induction of IFN-β mRNA by bacteria requires the participation of LPS. The induction is observed, as a rule, only with gram-negative bacteria and consequently proceeds only in Lpsn macrophages.
We also investigated whether IFN-α might be produced by macrophages in response to LPS, bacteria, and dsRNA. All samples investigated in Fig. 2 were also analyzed by Northern blotting for the presence of IFN-α mRNA (not shown). However, it was not detectable in any of the samples, even after prolonged exposure times (up to 6 weeks).
Production of IFN-β and other cytokines by Lpsn and Lpsd macrophages stimulated with gram-negative and gram-positive bacteria.
For the induction of IFN-β, macrophages of the different mouse strains were stimulated with different gram-negative and gram-positive bacteria and, in addition, with LPS for 8 h. As shown in Table 1, IFN-β could not be detected in unstimulated macrophages. LPS and all gram-negative bacteria induced varying amounts of IFN-β in Sn and BALB/c but not in Cr and BALB/c/l macrophages. In contrast, gram-positive bacteria did not induce IFN-β in either the Lpsn or the Lpsd macrophages. Therefore, IFN-β production is induced only by gram-negative bacteria and only in LPS responder macrophages.
TABLE 1.
| Stimulus | IFN-β in supernatants ofb:
|
|||
|---|---|---|---|---|
|
Lpsn macrophages
|
Lpsd macrophages
|
|||
| Sn | BALB/c | Cr | BALB/c/l | |
| None | − | − | − | − |
| LPS | + | + | − | − |
| Gram-negative bacteriac | + | + | − | − |
| Gram-positive bacteriad | − | − | − | − |
Lpsn (Sn and BALB/c) and Lpsd (Cr and BALB/c/l) macrophages (106/ml) were stimulated with 10 μg of LPS/ml or 100 μg of killed bacteria/ml for 8 h. IFN-β in the supernatant was assayed in a bioassay as described in Materials and Methods.
+, 500 to 10,000 U of IFN-β/ml; −, <25 U of IFN-β/ml.
Serovar Typhimurium, E. coli, P. mirabilis, P. aeruginosa, S. enteritidis, and V. cholerae.
S. aureus, S. thermophilus, and L. bulgaricus.
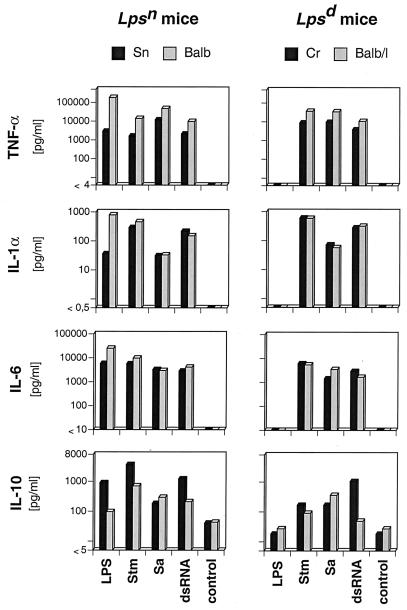
For the induction of TNF-α, IL-1α, IL-6, and IL-10, macrophages from the four mouse strains were stimulated with the different bacteria and LPS, and in addition, with dsRNA for 24 h. Figure 3 shows the results obtained with serovar Typhimurium, S. aureus, LPS, and dsRNA. Unstimulated macrophages of all mouse strains exhibited a very low production (10 to 20 pg/ml) of IL-10, while all other cytokines investigated were not detectable. In general, LPS induced the formation of all of the above-mentioned cytokines only in Lpsn macrophages. In contrast, serovar Typhimurium and S. aureus induced all cytokines tested for in macrophages of all four mouse strains, regardless of their LPS responsiveness. Additional gram-negative bacteria tested (E. coli, P. mirabilis, P. aeruginosa, S. enteritidis, and V. cholerae) induced levels of macrophage cytokines comparable to those induced by serovar Typhimurium (not shown). Other gram-positive bacteria (P. acnes, S. thermophilus, and L. bulgaricus) induced IL-1α, IL-6, and IL-10 at levels approximately 10 times lower than those induced by S. aureus, and in contrast to S. aureus, they elicited only very low levels of TNF-α (not shown). The presence of IL-1 measured in supernatants by ELISA was also confirmed in a bioassay, indicating that this cytokine was present in biologically active form (results not shown).
FIG. 3.
Levels of cytokines in supernatants of Lpsn and Lpsd macrophages stimulated with different agents. Macrophages (106/ml) of Sn, BALB/c (Balb), Cr, and BALB/c/l (Balb/l) mice were cultured with LPS (0.1 μg/ml), serovar Typhimurium (Stm; 100 μg/ml), S. aureus (Sa; 100 μg/ml), and dsRNA (10 μg/ml) for 24 h. The cytokines in cell-free supernatants were measured as described in Materials and Methods. The values are means of duplicates. One representative experiment of four is shown.
dsRNA stimulated the production of all cytokines, including TNF-α, in all types of macrophages under investigation (Fig. 3). Thus, although we could not evaluate the induction of TNF-α mRNA by Northern blot analysis (see above), dsRNA was a potent inducer of TNF-α in both Lpsd and Lpsn macrophages.
The results described above indicate that while a number of macrophage cytokines are inducible by gram-negative and gram-positive bacteria regardless of whether they contain LPS, the induction of IFN-β by bacteria is dependent mainly on LPS.
In vivo induction of IFN-β, TNF-α, and IL-6 mRNAs in Lpsn and Lpsd mice injected with serovar Typhimurium and S. aureus.
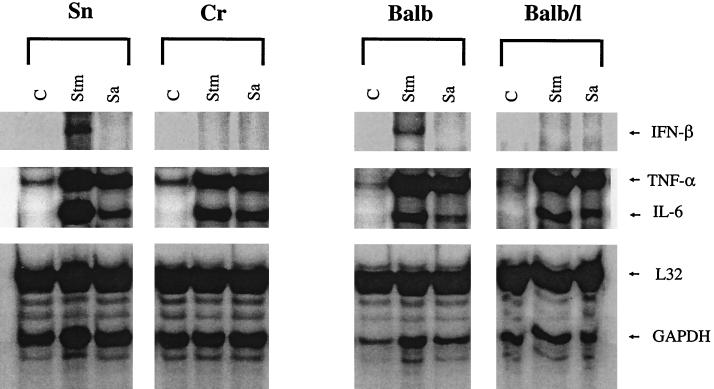
Groups of three mice were injected with heat-killed serovar Typhimurium or S. aureus (15 μg/g of body weight) intravenously, and 1 h later the animals were sacrificed and their spleens were removed for RNA isolation. The presence of IFN-β, TNF-α, and IL-6 mRNA as analyzed by RPA is shown in Fig. 4. All controls showed a weak constitutive expression of TNF-α and no expression of IFN-β or IL-6 mRNA. Both bacteria induced strong TNF-α and IL-6 mRNA responses in all mouse strains used. IFN-β mRNA, however, was inducible only by the gram-negative serovar Typhimurium and only in Lpsn mice. An IFN-β response to S. aureus was absent in all mouse strains. As expected, LPS administered to the mice in vivo induced IFN-β, TNF-α, and IL-6 mRNAs only in Lpsn mice (results not shown).
FIG. 4.
Expression of IFN-β, TNF-α, and IL-6 mRNAs in the spleens of Lpsn and Lpsd mice injected with serovar Typhimurium and S. aureus. Lpsn (Sn and BALB/c [Balb]) and Lpsd (Cr and BALB/c/l [Balb/l]) mice (three animals per strain) were injected intravenously with 15 μg of killed serovar Typhimurium (Stm) or S. aureus (Sa) per g of body weight. Untreated mice served as controls (C). One hour after treatment, the mice were sacrificed and their spleens were removed and homogenized, as described in Materials and Methods. Total spleen RNA was extracted separately for each animal and pooled for the three identically treated animals of each group. Ten micrograms of each RNA pool were used for detection of cytokine mRNA in an RPA with a cytokine template set. Two constitutive gene probes (L32 and GAPDH [glyceraldehyde-3-phosphate dehydrogenase]) for standardization of RNA amounts were also used according to the instructions of the manufacturer.
DISCUSSION
One interesting biological activity of IFN-β that we described earlier (40), and which has subsequently been confirmed by other studies (16, 25), is its property of acting as a cofactor in the induction of IFN-γ. It was also shown that gram-negative bacteria induce IFN-β in the Lpsn Sn mice but not in the related Lpsd Cr mice (40). This finding was unexpected, because bacteria (gram negative and gram positive) generally induce different cytokines in both Lpsn and Lpsd mice (7, 8, 19). We investigated the reason for the above-mentioned inability of Cr mice to produce IFN-β by comparing their IFN-β and other macrophage cytokine responses to those of Lpsd BALB/c/l and Lpsn Sn and BALB/c mice following stimulation with different gram-negative and gram-positive bacteria.
IFN-β mRNA and IFN-β activity were inducible by LPS and by a large number of gram-negative bacteria in macrophages of Lpsn mice (Sn and BALB/c) cultured in vitro. Further, IFN-β mRNA was detectable in both Lpsn strains of mice injected with LPS or serovar Typhimurium. In contrast, neither LPS nor gram-negative bacteria induced IFN-β in Lpsd mice that was in any way comparable to that seen in Lpsn mice. Thus, in BALB/c/l mice IFN-β mRNA was practically undetectable and in Cr mice only extremely weak bands were visible after long exposure. Gram-positive bacteria induced no IFN-β activity in any of the mice, regardless of their LPS sensitivity, and in only some cases a very weak IFN-β mRNA signal was recognizable after prolonged exposure of the autoradiography film. All bacteria, however, induced comparable amounts of TNF-α, IL-1α, IL-6, and IL-10 in vitro or in vivo in Lpsn and Lpsd mice. Thus, while macrophage cytokines known to be induced by LPS are also inducible by all bacteria in Lpsn and Lpsd mice, IFN-β is an exception, being inducible practically only by LPS and therefore generally by gram-negative microorganisms. The inability of Cr and BALB/c/l mice to produce significant amounts of IFN-β when stimulated by gram-negative bacteria is, therefore, related to their lack of LPS responsiveness. A similar absence of IFN-β response to bacteria is predicted for the C3H/HeJ and C57BL/10ScN Lpsd mice (not investigated here). Therefore, LPS emerges as the only notable bacterial component capable of inducing IFN-β, and gram-positive bacteria emerge as a class of microorganisms that are practically incapable of inducing this cytokine.
Recently, evidence has been accumulating that the induction of cytokines by bacterial components is effected via Toll-like receptors (Tlr) 2 and 4 acting as signaling molecules (2, 4, 24, 26, 28, 42, 43). In the present study, the induction of IFN-β and other cytokines by LPS in normal mice and its complete absence in Tlr4/Lps-defective mice (Cr and BALB/c/l) is evidence that LPS signal transduction proceeded solely via Tlr4. Further, the cytokine responses to bacteria obtained in the absence of LPS signaling, i.e., those induced by gram-positive bacteria or by gram-negative bacteria in Cr and BALB/c/l mice, must proceed via Tlr2 (and/or by as-yet-unidentified signaling receptors). Since IFN-β is selectively absent from such responses, it may be concluded that in mice the induction of IFN-β is strictly Tlr4 dependent, while the induction of other cytokines by LPS or other bacterial components can proceed by the Tlr4 or Tlr2 signaling pathway, respectively.
It has been shown that IFN-β is the predominant type I interferon induced in mice by LPS (39). IFN-α and IFN-β share the same cellular receptor, which explains their similar biological activities (23). In this connection, we have shown that both type I IFNs act as cofactors of IFN-γ induction (40). We also investigated the possibility of IFN-α being produced as an alternative to IFN-β but could obtain no evidence for its presence, either on Northern blots or in supernatants from stimulated macrophages, using a bioassay. Interestingly, synthetic oligodeoxynucleotides containing unmethylated CpG motifs from bacterial DNA and nucleic acid fraction from Mycobacterium bovis BCG have been shown to induce IFN-α/β (31, 34, 41). Therefore, the absence of detectable IFN-α/β activity in Lpsn and Lpsd mice stimulated with different gram-positive bacteria and also in Lpsd mice stimulated with gram-negative bacteria observed in our study was surprising. It raises the question of how much of the bacterial DNA is really available for bioactivity in bacterium-treated mice.
As mentioned above, IFN-β acts as a cofactor in the induction of IFN-γ. The present results also allow further conclusions to be made regarding IFN-γ induction by bacteria. Since LPS is the only bacterial component inducing significant amounts of IFN-β, it follows that generally only gram-negative bacteria induce IFN-γ via the IFN-β-dependent pathway. An IFN-β-dependent production of IFN-γ is therefore absent from the LPS-resistant Cr and BALB/c/l mice. BALB/c/l mice, however, produce IFN-γ in response to gram-negative bacteria, indicating that these bacteria induce IFN-γ by additional, IFN-β-independent mechanisms. The latter pathway is also utilized very efficiently by gram-positive bacteria, since many of these bacteria are known to be excellent inducers of IFN-γ. Therefore, the inability of Cr mice to produce IFN-γ in response to any bacteria is related not only to the absence of IFN-β but also to a general inability to respond to the array of IFN-γ-inducing components present in bacteria and, as shown in earlier studies, also in parasites, such as P. chabaudi chabaudi (11) and L. major (21). It is interesting that when exogenous IFN-β is administered to Cr mice, they acquire the ability to produce IFN-γ after stimulation with gram-negative (reference 40 and this study) but not gram-positive (unpublished data) bacteria. This indicates that the IFN-β-dependent pathway of IFN-γ induction is intact in these mice and that this pathway is utilized only by gram-negative bacteria.
Type I interferons are strongly inducible by viruses and play an important role in antiviral defence (23, 27, 35). Consequently, they have been investigated most extensively in this connection. The significance of IFN-α or IFN-β in bacterial infections has been much less studied. It has been shown that mice deficient for the IFN-α/β receptor are indistinguishable from wild-type mice in their susceptibility to L. monocytogenes infection (35). In view of the present results, the above-mentioned investigation allows no conclusions to be made regarding the significance of IFN-α/β in this infection model, since L. monocytogenes induces no IFN-α/β and it would make no difference if the mice are deficient for the IFN-α/β receptor or not. In our hands, administration of exogenous recombinant murine IFN-β to mice infected with serovar Typhimurium had no detectable protective effect (M. Matsuura and C. Galanos, unpublished data). This may be understandable, considering that IFN-β is already induced during serovar Typhimurium infection and additional exogenous IFN-β may have no further discernible effect. For this reason, any antibacterial effect of exogenously administered IFN-β might be best recognized during infections by gram-positive bacteria, since these do not induce this cytokine. In this connection, it is interesting that a protective effect of exogenously administered IFN-β has been reported in the case of infection with L. monocytogenes (13). For future investigations of the role of IFN-α/β in infections by gram-negative bacteria, new models will have to be developed. Of special interest would be the use of IFN-α/β receptor-deficient mice.
ACKNOWLEDGMENTS
We are indebted to N. Goos and H. Stübig for excellent technical assistance and U. Müller for his help in the preparation of the manuscript.
This work was supported in part by BMBF-Gesundheit, projekt O1KI9854/8.
REFERENCES
- 1.Aggarwal B B, Kohr W J, Hass P E, Moffat B, Spencer S A, Henzel W J, Bringman T S, Nedwin G E, Goeddel D V, Harkins R N. Human tumor necrosis factor. Production, purification, and characterization. J Biol Chem. 1985;260:2345–2354. [PubMed] [Google Scholar]
- 2.Brightbill H D, Libraty D H, Krutzik S R, Yang R B, Belisle J T, Bleharski J R, Maitland M, Norgard M V, Plevy S E, Smale S T, Brennan P J, Bloom B R, Godowski P J, Modlin R L. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science. 1999;285:732–736. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 3.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 4.Chow J C, Young D W, Golenbock D T, Christ W J, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 5.Coutinho A, Meo T. Genetic bases for unresponsiveness to lipopolysaccharide in C57BL/10Cr mice. Immunogenetics. 1978;7:17–24. doi: 10.1007/BF01843983. [DOI] [PubMed] [Google Scholar]
- 6.De S K, Chen H L, Pace J L, Hunt J S, Terranova P F, Enders G C. Expression of tumor necrosis factor-alpha in mouse spermatogenic cells. Endocrinology. 1993;133:389–396. doi: 10.1210/endo.133.1.8319585. [DOI] [PubMed] [Google Scholar]
- 7.Dong Z, Qi X, Fidler I J. Tyrosine phosphorylation of mitogen-activated protein kinases is necessary for activation of murine macrophages by natural and synthetic bacterial products. J Exp Med. 1993;177:1071–1077. doi: 10.1084/jem.177.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freudenberg M A, Galanos C. Tumor necrosis factor alpha mediates lethal activity of killed gram-negative and gram-positive bacteria in d-galactosamine-treated mice. Infect Immun. 1991;59:2110–2115. doi: 10.1128/iai.59.6.2110-2115.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freudenberg M A, Keppler D, Galanos C. Requirement for lipopolysaccharide-responsive macrophages in galactosamine-induced sensitization to endotoxin. Infect Immun. 1986;51:891–895. doi: 10.1128/iai.51.3.891-895.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freudenberg M A, Kopf M, Galanos C. Lipopolysaccharide-sensitivity of interferon-g-receptor deficient mice. J Endotox Res. 1996;3:291–295. [Google Scholar]
- 11.Freudenberg M A, Kumazawa Y, Meding S, Langhorne J, Galanos C. Gamma interferon production in endotoxin-responder and -nonresponder mice during infection. Infect Immun. 1991;59:3484–3491. doi: 10.1128/iai.59.10.3484-3491.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freudenberg M A, Salomao R, Sing A, Mitov I, Galanos C. Reconciling the concepts of endotoxin sensitization and tolerance. Proceedings of the 4th Conference of the International Endotoxin Society. Prog Clin Biol Res. 1998;397:261–268. [PubMed] [Google Scholar]
- 13.Fujiki T, Tanaka A. Antibacterial activity of recombinant murine beta interferon. Infect Immun. 1988;56:548–551. doi: 10.1128/iai.56.3.548-551.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galanos C, Lüderitz O, Westphal O. Preparation and properties of a standardized lipopolysaccharide from Salmonella abortus equi (Novo-Pyrexal) Zentbl Bakteriol Orig A. 1979;243:226–244. [PubMed] [Google Scholar]
- 15.Gilman M. Ribonuclease protection assay. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Stuhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley and Sons, Inc.; 1993. pp. 4.7.1–4.7.8. [Google Scholar]
- 16.Hunter C A, Gabriel K E, Radzanowski T, Neyer L E, Remington J S. Type I interferons enhance production of IFN-gamma by NK cells. Immunol Lett. 1997;59:1–5. doi: 10.1016/s0165-2478(97)00091-6. [DOI] [PubMed] [Google Scholar]
- 17.Katschinski T, Galanos C, Coumbos A, Freudenberg M A. Gamma interferon mediates Propionibacterium acnes-induced hypersensitivity to lipopolysaccharide in mice. Infect Immun. 1992;60:1994–2001. doi: 10.1128/iai.60.5.1994-2001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knopf H P, Otto F, Engelhardt R, Freudenberg M A, Galanos C, Herrmann F, Schumann R R. Discordant adaptation of human peritoneal macrophages to stimulation by lipopolysaccharide and the synthetic lipid A analogue SDZ MRL 953. Down-regulation of TNF-alpha and IL-6 is paralleled by an up-regulation of IL-1 beta and granulocyte colony-stimulating factor expression. J Immunol. 1994;153:287–299. [PubMed] [Google Scholar]
- 19.Linder H, Engberg I, Hoschutzky H, Mattsby-Baltzer I, Svanborg C. Adhesion-dependent activation of mucosal interleukin-6 production. Infect Immun. 1991;59:4357–4362. doi: 10.1128/iai.59.12.4357-4362.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loppnow H, Flad H D, Durrbaum I, Musehold J, Fetting R, Ulmer A J, Herzbeck H, Brandt E. Detection of interleukin 1 with human dermal fibroblasts. Immunobiology. 1989;179:283–291. doi: 10.1016/S0171-2985(89)80023-3. [DOI] [PubMed] [Google Scholar]
- 21.Müller I, Freudenberg M, Kropf P, Kiderlen A F, Galanos C. Leishmania major infection in C57BL/10 mice differing at the Lps locus: a new non-healing phenotype. Med Microbiol Immunol. 1997;186:75–81. doi: 10.1007/s004300050048. [DOI] [PubMed] [Google Scholar]
- 22.Pennica D, Hayflick J S, Bringman T S, Palladino M A, Goeddel D V. Cloning and expression in Escherichia coli of the cDNA for murine tumor necrosis factor. Proc Natl Acad Sci USA. 1985;82:6060–6064. doi: 10.1073/pnas.82.18.6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pestka S, Langer J A, Zoon K C, Samuel C E. Interferons and their actions. Annu Rev Biochem. 1987;56:727–777. doi: 10.1146/annurev.bi.56.070187.003455. [DOI] [PubMed] [Google Scholar]
- 24.Poltorak A, He X, Smirnova I, Liu M Y, Huffel C V, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 25.Sareneva T, Matikainen S, Kurimoto M, Julkunen I. Influenza A virus-induced IFN-alpha/beta and IL-18 synergistically enhance IFN-gamma gene expression in human T cells. J Immunol. 1998;160:6032–6038. [PubMed] [Google Scholar]
- 26.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning C J. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J Biol Chem. 1999;274:17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 27.Sen G C, Ransohoff R M. Interferon-induced antiviral actions and their regulation. Adv Virus Res. 1993;42:57–102. doi: 10.1016/s0065-3527(08)60083-4. [DOI] [PubMed] [Google Scholar]
- 28.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slade S J, Langhorne J. Production of interferon-gamma during infection of mice with Plasmodium chabaudi chabaudi. Immunobiology. 1989;179:353–365. doi: 10.1016/S0171-2985(89)80041-5. [DOI] [PubMed] [Google Scholar]
- 30.Sultzer B M. Genetic control of host responses to endotoxin. Infect Immun. 1972;5:107–113. doi: 10.1128/iai.5.1.107-113.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun S, Zhang X, Tough D F, Sprent J. Type I interferon-mediated stimulation of T cells by CpG DNA. J Exp Med. 1998;188:2335–2342. doi: 10.1084/jem.188.12.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takakuwa T, Knopf H P, Sing A, Carsetti R, Galanos C, Freudenberg M A. Induction of CD14 expression in Lpsn, Lpsd and tumor necrosis factor receptor-deficient mice. Eur J Immunol. 1996;26:2686–2692. doi: 10.1002/eji.1830261121. [DOI] [PubMed] [Google Scholar]
- 33.Tanamoto K, Zähringer U, McKenzie G R, Galanos C, Rietschel E T, Lüderitz O, Kusumoto S, Shiba T. Biological activities of synthetic lipid A analogs: pyrogenicity, lethal toxicity, anticomplement activity, and induction of gelation of Limulus amoebocyte lysate. Infect Immun. 1984;44:421–426. doi: 10.1128/iai.44.2.421-426.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tokunaga T, Yano O, Kuramoto E, Kimura Y, Yamamoto T, Kataoka T, Yamamoto S. Synthetic oligonucleotides with particular base sequences from the cDNA encoding proteins of Mycobacterium bovis BCG induce interferons and activate natural killer cells. Microbiol Immunol. 1992;36:55–66. doi: 10.1111/j.1348-0421.1992.tb01642.x. [DOI] [PubMed] [Google Scholar]
- 35.van den Broek M F, Müller U, Huang S, Aguet M, Zinkernagel R M. Antiviral defense in mice lacking both alpha/beta and gamma interferon receptors. J Virol. 1995;69:4792–4796. doi: 10.1128/jvi.69.8.4792-4796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vogel S N. The Lps gene. In: Beutler B, editor. Tumor necrosis factors: the molecules and their emerging role in medicine. New York, N.Y: Raven Press; 1992. pp. 485–513. [Google Scholar]
- 37.Vogel S N, Hansen C T, Rosenstreich D L. Characterization of a congenitally LPS-resistant, athymic mouse strain. J Immunol. 1979;122:619–622. [PubMed] [Google Scholar]
- 38.Vogel S N, Wax J S, Perera P Y, Padlan C, Potter M, Mock B A. Construction of a BALB/c congenic mouse, C.C3H-Lpsd, that expresses the Lpsd allele: analysis of chromosome 4 markers surrounding the Lps gene. Infect Immun. 1994;62:4454–4459. doi: 10.1128/iai.62.10.4454-4459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogel S N, Weedon L L, Wahl L M, Rosenstreich D L. BCG-induced enhancement of endotoxin sensitivity in C3H/HeJ mice. II. T cell modulation of macrophage sensitivity to LPS in vitro. Immunobiology. 1982;160:479–493. doi: 10.1016/S0171-2985(82)80010-7. [DOI] [PubMed] [Google Scholar]
- 40.Yaegashi Y, Nielsen P, Sing A, Galanos C, Freudenberg M A. Interferon beta, a cofactor in the interferon gamma production induced by gram-negative bacteria in mice. J Exp Med. 1995;181:953–960. doi: 10.1084/jem.181.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamamoto S, Kuramoto E, Shimada S, Tokunaga T. In vitro augmentation of natural killer cell activity and production of interferon-alpha/beta and -gamma with deoxyribonucleic acid fraction from Mycobacterium bovis BCG. Jpn J Cancer Res. 1988;79:866–873. doi: 10.1111/j.1349-7006.1988.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang R B, Mark M R, Gray A, Huang A, Xie M H, Zhang M, Goddard A, Wood W I, Gurney A L, Godowski P J. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signalling. Nature. 1998;395:284–288. doi: 10.1038/26239. [DOI] [PubMed] [Google Scholar]
- 43.Yoshimura A, Lien E, Ingalls R R, Tuomanen E, Dziarski R, Golenbock D. Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]