Abstract
Vicia faba L. (faba bean) is a legume cultivated worldwide which commonly establishes effective symbiosis with the symbiovar viciae of species from the Rhizobium leguminosarum phylogenetic group. However, on the basis of the rrs, recA, and atpD gene phylogenies, in this work we identified a strain named EFBRI 42 nodulating V. faba as Rhizobium azibense. This is the first report on the nodulation of Vicia by R. azibense which commonly nodulates P. vulgaris and to date encompasses strains harboring the nodC genes typical of the symbiovars gallicum and phaseoli. However, the strain EFBRI 42 carries a nodC gene typical of the symbiovar viciae for which we report here by the first time this symbiovar in R. azibense. This finding showed the existence of symbiotic genes horizontal transfer events during the coevolution of R. azibense with P. vulgaris and V. faba in their respective distribution centers of Mesoamerica and the Middle East.
Keywords: Rhizobium azibense, Symbiovar viciae, Vicia faba, Egypt
Vicia faba L. (faba bean) is a legume probably indigenous to the Near East (Cubero 1974) which is currently cultivated worldwide for the high nutritive value of its seeds, its usefulness as forage and cover crop, and by its ability for nitrogen fixation in symbiosis with rhizobia (Etemadi et al. 2019; Maaluf et al. 2019).
As other legumes, V. faba establishes nitrogen-fixing symbiosis with rhizobial strains whose diversity has been studied in several countries of North Africa where this legume is mostly nodulated by strains phylogenetically related to Rhizobium leguminosarum, R. laguerreae and R. etli (Shamseldin et al. 2009; Youseif et al. 2014; Hassan et al. 2015; Belhadi et al. 2018; Benidire et al. 2018; Missbah El Idrissi et al. 2020).
Several of these studies have been carried out in Egypt where this legume has been cultivated for many centuries (Shamseldin et al., 2009; Youseif et al. 2014; Hassan et al. 2015) showing that the strains nodulating V. faba mostly belong to the symbiovar viciae of Rhizobium leguminosarum and R. etli (Shamseldin et al. 2009; Youseif et al. 2014; Hassan et al. 2015); which is a symbiotic variant able to nodulate specifically legumes of the Vicia cross inoculation group (Rogel et al. 2011; Peix et al. 2015). However, some strains effectively nodulating faba bean have not been assigned to a species and symbiovar until now, as occurs with the strain EFBRI 42 isolated in Egypt (Shamseldin et al., 2009). Therefore, in the present study, we identified this strain through the analysis of the core genes, like rrs, recA, and atpD, and the symbiotic gene nodC, which were not previously analyzed for this strain (Shamseldin et al. 2009), and are commonly used for the rhizobia identification at species and symbiovar levels (Peix et al. 2015).
To obtain sequences of these genes, we extracted DNA of the strain EFBRI 42 grown on TY plates (Triptone Yeast Agar) [(Beringer)] during 24 h at 28 °C. Genomic DNA was obtained using the DNeasy UltraClean Microbial DNA Isolation Kit (Qiagen) following the manufacturer’s protocol. The amplification and sequencing of rrs, recA, atpD and nodC genes were carried out in the conditions and with the primers previously reported (Carro et al. 2012; Gaunt et al. 2001; Laguerre et al. 2001).
The obtained sequences were compared with those from GenBank using the BLASTN program (Altschul et al. 1990) and the sequences of the closely related bacteria were downloaded from GenBank for phylogenetic analyses. The sequences were aligned using the Clustal W program (Thompson et al. 1977). The phylogenetic distances were calculated according to Kimura’s two-parameter model (Kimura 1980). The phylogenetic trees were inferred using the neighbor joining model (Saitou and Nei 1987) MEGA 7.0 (Kumar et al. 2016) was used for all phylogenetic analyses (Figs. 1 and 2).
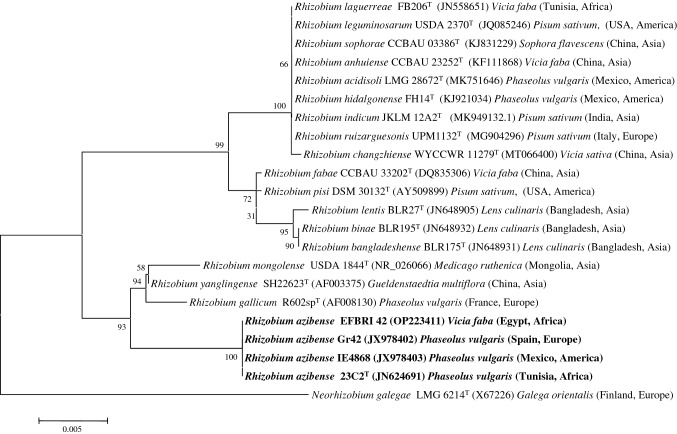
Fig. 1.
Neighbor-joining phylogenetic tree based on rrs gene sequences (1310 nt) showing the taxonomic location of the strain EFBRI 42 within the genus Rhizobium. Bootstrap values calculated for 1000 replications are indicated. Bar: 5 nt substitution per 1000 nt. Accession numbers from Genbank are given in brackets
Fig. 2.
Neighbor-joining phylogenetic tree based on recA and atpD concatenated gene sequences (700 nt) showing the position of the strain EFBRI 42 within the genus Rhizobium. Bootstrap values calculated for 1000 replications are indicated. Bar, 1 nt substitution per 100 nt. Accession numbers from Genbank are given in brackets
The species from the phylogenetic group of R. leguminosarum are the most common endosymbionts of Vicia species and other legumes from its cross inoculation group as Pisum and Lens (Fig. 1). However, the strain EFBRI 42, isolated in Egypt, represented a separate genotype from R. leguminosarum according to the rrs-RFLP pattern analysis (Shamseldin et al. 2009). Accordingly, the sequence of rrs gene of strain EFBRI 42 showed 100% similarity with respect to the species Rhizobium azibense, which encompasses strains nodulating P. vulgaris in different continents and belongs to a group phylogenetically divergent of R. leguminosarum (Fig. 1).
The four species of this phylogenetic group, R. azibense, R. gallicum, R. mongolense, and R. yanglingense, have closely related rrs genes and therefore, the identification of the strain EFBRI 42 was confirmed by the analysis of the recA and atpD housekeeping genes, which allowed the differentiation of Rhizobium species with closely related rrs genes (Peix et al. 2015). The results of the concatenated recA and atpD gene sequences confirmed the affiliation of the strain EFBRI 42 to R. azibense with similarity values higher than 99.0% in both genes (Fig. 2). These results constitute the first report on the nodulation of V. faba by R. azibense, which to date only included strains isolated from P. vulgaris nodules (Mnasri et al. 2014).
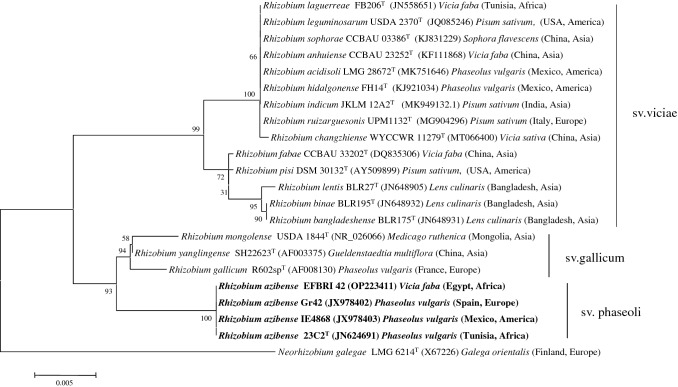
The identification at symbiovar level is mainly based on the nodC gene analysis in the case of the genus Rhizobium (Peix et al. 2015). Based on the results of this analysis the Egyptian strain EFBRI 42 belongs to the symbiovar viciae with its nodC gene being closely related (higher than 98% similarity) to Rhizobium strains isolated from Lens culinaris nodules in Morocco, Syria and Iran (Fig. 3). This is the first report of the symbiovar viciae within the species R. azibense which to date only contains strains nodulating P. vulgaris belonging to the symbiovars gallicum and phaseoli (Fig. 3). The strains of the species R. azibense have been isolated in three different continents, Africa, America and Europe (Mnasri et al. 2014). The strains 23C2T isolated in Tunisia (Africa) and IE4868 isolated in Mexico (America) belong to the symbiovar gallicum, whereas the strain GR42 isolated in Spain (Europe) belongs to the symbiovar phaseoli (Fig. 3). Both symbiovars, phaseoli and gallicum, have been isolated from P. vulgaris nodules in its distribution centers (Silva et al. 2003; Mnasri et al. 2014; Bustos et al. 2017), which are located in Mesoamerica (Bitocchi et al. 2012). Therefore, probably the symbiotic genes typical of these symbiovars arrived to Europe and Africa together with the seeds of P. vulgaris and were transferred to strains of species indigenous to these continents. This seems to be clear in the case of R. leguminosarum strains carrying the symbiovar phaseoli, because an origin outside America has been proposed for this species (Alvarez-Martínez et al. 2009), but still there are not enough data to hypothesize on the geographical origin of the species R. azibense. Nevertheless, the fact of the existence of the symbiovar viciae within this species opens the door to think that this species has coevolved for long times with Vicia species whose geographical origin has been located in a region of the Middle East that include Egypt (Caracuta et al. 2015). In any case, the existence within R. azibense of three symbiovars to date nodulating legumes indigenous to different continents, as occurs with P. vulgaris and V. faba, proved the existence of horizontal transfer events affecting the symbiotic genes during the coevolution of R. azibense with P. vulgaris and V. faba in their respective distribution centers.
Fig. 3.
Neighbor-joining phylogenetic tree based on nodC gene sequences (370 nt) showing the position of the strain EFBRI 42. Bootstrap values calculated for 1000 replications are indicated. Bar, 2 nt substitution per 100 nt. Accession numbers from Genbank are given in brackets
Conclusion
In this study, we report for the first time the nodulation of V. faba by the species R. azibense and the definition of the symbiovar viciae within this species, which to date only contained symbiovars nodulating P. vulgaris, such as gallicum and phaseoli. This finding confirmed the existence of horizontal transfer events affecting the symbiotic genes during the coevolution of R. azibense with different legume hosts.
Acknowledgements
The authors thank the Strategic Research Programs for Units of Excellence from Junta de Castilla y León (CLU-2019-05 and CLU-2O18-04).
Author contributions
AS isolate rhizobial strains, DNA isolation and amplification of 16S rRNA; AP amplify the recA and atpD and nodC genes; AS and AP did the sequences of the genes; EV did the comparisons of the three phylogenetic trees. All the three authors shared the initial writing of the manuscript. EV did revision of the final copy.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Data availability
Sequences of 16S rRNA, recA, atpD, and nodC genes of strain EFBRI are deposited in the gene bank under accession numbers OP223411, OP381820, OP381822, and OP381821, respectively.
Declarations
Conflict of interest
The authors declare that there are no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alvarez-Martínez ER, Valverde A, Ramírez-Bahena MH, García-Fraile P, Tejedor C, Mateos PF, Santillana N, Zúñiga D, Peix A, Velázquez E. The analysis of core and symbiotic genes of rhizobia nodulating Vicia from different continents reveals their common phylogenetic origin and suggests the distribution of Rhizobium leguminosarum strains together with Vicia seeds. Arch Microbiol. 2009;191:659–668. doi: 10.1007/s00203-009-0495-6. [DOI] [PubMed] [Google Scholar]
- Belhadi D, de Lajudie P, Ramdani N, Le Roux C, Boulila F, Tisseyre P, Boulila A, Benguedouar A, Kaci Y, Laguerre G. Vicia faba L. in the Bejaia region of Algeria is nodulated by Rhizobium leguminosarum sv. viciae, Rhizobium laguerreae and two new genospecies. Syst Appl Microbiol. 2018;41:122–130. doi: 10.1016/j.syapm.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Benidire L, Lahrouni M, Daoui K, Fatemi ZEA, Gomez Carmona R, Göttfert M, Oufdou K. Phenotypic and genetic diversity of Moroccan rhizobia isolated from Vicia faba and study of genes that are likely to be involved in their osmotolerance. Syst Appl Microbiol. 2018;41:51–61. doi: 10.1016/j.syapm.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Bitocchi E, Nanni L, Bellucci E, Rossi M, Giardini A, Zeuli PS, Logozzo G, Stougaard J, McClean P, Attene G, Papa R. Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proc Nat Acad Sci USA. 2012;109:E788–E796. doi: 10.1073/pnas.1108973109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos P, Santamaría RI, Pérez-Carrascal OM, Acosta JL, Lozano L, Juárez S, Martínez-Flores I, Martínez-Romero E, Cevallos MÁ, Romero D, Dávila G, Vinuesa P, Miranda F, Ormeño E, González V. Complete genome sequences of three Rhizobium gallicum symbionts associated with common bean (Phaseolus vulgaris) Genome Announc. 2017;5(11):e00030-17. doi: 10.1128/genomeA.00030-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caracuta V, Barzilai O, Khalaily H, Milevski I, Paz Y, Vardi J, Regev L, Boaretto E. The onset of faba bean farming in the Southern Levant. Sci Rep. 2015;5:14370. doi: 10.1038/srep14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro L, Spröer C, Alonso P, Trujillo ME. Diversity of Micromonospora strains isolated from nitrogen fixing nodules and rhizosphere of Pisum sativum analyzed by multilocus sequence analysis. Syst Appl Microbiol. 2012;35:73–80. doi: 10.1016/j.syapm.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Cubero JI. On the evolution of Vicia faba L. Theor Appl Genet. 1974;45:47–51. doi: 10.1007/BF00283475. [DOI] [PubMed] [Google Scholar]
- Etemadi F, Hashemi M, Barker AV, Zandvakili OR, Liu X. Agronomy, nutritional value, and medicinal application of faba bean (Vicia faba L.) Horti Plant J. 2019;5:170–182. doi: 10.1016/j.hpj.2019.04.004. [DOI] [Google Scholar]
- Gaunt MW, Turner SL, Rigottier-Gois L, Lloyd-Macgilp SA, Young JWP. Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int J Syst Evol Microbiol. 2001;51:2037–2048. doi: 10.1099/00207713-51-6-2037. [DOI] [PubMed] [Google Scholar]
- Hassan MM, Fahmi AI, Eissa RA, Nagaty HH. Diversity of rhizobia nodulating faba bean (Vicia faba) growing in Egypt. J Microbiol Biochem Technol. 2015;7(7):152–159. doi: 10.4172/1948-5948.1000197. [DOI] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;3:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N. Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiol. 2001;147:981–993. doi: 10.1099/00221287-147-4-981. [DOI] [PubMed] [Google Scholar]
- Maalouf F, Hu J, O'Sullivan DM, Zong X, Hamwieh A, Kumar S, Baum M. Breeding and genomics status in faba bean (Vicia faba) Plant Breed. 2019;138:465–473. doi: 10.1111/pbr.12644. [DOI] [Google Scholar]
- Missbah El Idrissi M, Lamin H, Bouhnik O, Lamrabet M, Alami S, Jabrone Y, Bennis M, Bedmar EJ, Abdelmoumen H. Characterization of Pisum sativum and Vicia faba microsymbionts in Morocco and definition of symbiovar viciae in Rhizobium acidisoli. Syst Appl Microbiol. 2020;43:126084. doi: 10.1016/j.syapm.2020.126084. [DOI] [PubMed] [Google Scholar]
- Mnasri B, Liu TY, Saidi S, Chen WF, Chen WX, Zhang XX, Mhamdi R. Rhizobium azibense sp. nov., a nitrogen fixing bacterium isolated from root-nodules of Phaseolus vulgaris. Int J Syst Evol Microbiol. 2014;64:1501–1506. doi: 10.1099/ijs.0.058651-0. [DOI] [PubMed] [Google Scholar]
- Peix A, Ramírez-Bahena MH, Velázquez E, Bedmar EJ. Bacterial associations with legumes. Crit Rev Plant Sci. 2015;34:17–42. doi: 10.1080/07352689.2014.897899. [DOI] [Google Scholar]
- Rogel MA, Ormeño-Orrillo E, Martinez Romero E. Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst Appl Microbiol. 2011;34:96–104. doi: 10.1016/j.syapm.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbour-joining method: a new method for reconstructing phylogenetics trees. Mol Biol Evol. 1987;44:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shamseldin A, El-Saadani M, Sadowsky MJ, An CS. Rapid identification and discrimination among Egyptian genotypes of Rhizobium leguminosarum bv. viciae and Sinorhizobium meliloti nodulating faba bean (Vicia faba L.) by analysis of nodC, ARDRA, and rDNA sequence analysis. Soil Biol Biochem. 2009;41:45–53. doi: 10.1016/j.soilbio.2008.09.014. [DOI] [Google Scholar]
- Silva C, Vinuesa P, Eguiarte LE, Martínez-Romero E, Souza V. Rhizobium etli and Rhizobium gallicum nodulate common bean (Phaseolus vulgaris) in a traditionally managed milpa plot in Mexico: population genetics and biogeographic implications. Appl Environ Microbiol. 2003;69:884–893. doi: 10.1128/AEM.69.2.884-893.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The clustalX windows interface: flexible strategies for multiple sequence alignement aided by quality analysis tools. Nucl Acids Res. 1977;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youseif SH, Abd El-Megeed FH, Ageez A, Cocking EC, Saleh SA. Phylogenetic multilocus sequence analysis of native rhizobia nodulating faba bean (Vicia faba L.) in Egypt. Syst Appl Microbiol. 2014;37:560–569. doi: 10.1016/j.syapm.2014.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences of 16S rRNA, recA, atpD, and nodC genes of strain EFBRI are deposited in the gene bank under accession numbers OP223411, OP381820, OP381822, and OP381821, respectively.