Summary
Background
Neuroblastoma (NB) is the most common solid extracranial paediatric tumour. Genome-wide association studies have driven the discovery of common risk variants, but no large study has investigated the contribution of rare variants to NB susceptibility. Here, we conducted a whole-exome sequencing (WES) of 664 NB cases and 822 controls and used independent validation datasets to identify genes with rare risk variants and involved pathways.
Methods
WES was performed at 50× depth and variants were jointly called in cases and controls. We developed two models to identify mutations with high clinical impact (P/LP model) and to discover less penetrant risk mutations affecting non-canonical cancer pathways (RPV model). We performed a gene-level collapsing test using Firth's logistic regression in 242 selected cancer predisposition genes (CPGs) and a gene-sets burden analysis of biologically-informed pathways.
Findings
Twelve percent of patients carried P/LP variants in CPGs and showed a significant enrichment (P = 2.3 × 10−4) compared to controls (6%). We identified P/LP variants in 45 CPGs enriched in homologous recombination (HR) pathway. The most P/LP enriched genes in NB were BRCA1, ALK and RAD51C. Additionally, we found higher RPV burden in gene-sets of neuron differentiation, neural tube development and synapse assembly, and in gene-sets associated with neurodevelopmental disorders (NDD).
Interpretation
The high fraction of NB patients with P/LP variants indicates the need of genetic counselling. Furthermore, inherited rare variants predispose to NB development by affecting mechanisms related to HR and neurodevelopmental processes, and demonstrate that NDD genes are altered in NB at the germline level.
Funding
Associazione Italiana per la Ricerca sul Cancro, Fondazione Italiana per la Lotta al Neuroblastoma, Associazione Oncologia Pediatrica e Neuroblastoma, Regione Campania, Associazione Giulio Adelfio onlus, and Italian Health Ministry.
Keywords: Neuroblastoma, Genetic susceptibility, Rare variants, Whole-exome sequencing
Research in context.
Evidence before this study
Neuroblastoma is the most common solid extracranial paediatric tumour originating from the sympathetic nervous system during development. It is characterized by diverse clinical manifestations and often associated with a poor survival (40% of the children survive longer than 5 years). Only 1–2% of patients present with a family history of neuroblastoma, mainly explained by mutations in ALK and PHOX2, the vast majority of cases appear to arise sporadically. Genome-wide association studies have identified common polymorphisms that are highly associated with NB susceptibility. Although it is known that 10–20% of paediatric tumours harbour rare pathogenic germline variants, only a few studies have been focused on cases and controls investigating the prevalence of clinically relevant mutations in neuroblastoma.
Added value of this study
In this work, we performed exome sequencing of a large cohort of neuroblastoma cases and controls and used independent validation datasets to investigate the contribution of inherited rare genetic variants to neuroblastoma predisposition. We have designed two analytic approaches to identify mutations with high clinical impact in well-known cancer predisposing genes and to discover non-canonical cancer gene pathways affected by less penetrant risk mutations. We demonstrated that 12% of neuroblastoma patients carry at least one clinically-relevant pathogenic variants occurring preferentially in genes belonging to the homologous recombination pathway. These inherited variants were significantly more frequent in NB cases than in controls (6%) and many genes were validated in two independent sets of neuroblastoma cases. We report mutations in RAD51C associated to NB which advise functional studies to validate its role as candidate susceptibility gene. Using a different approach, our analysis also suggested a key role of genes of neurodevelopmental processes in contributing to neuroblastoma susceptibility. Gene-sets of neural tube, neuron and synapse development along with known causative-genes of neurodevelopmental disorders were enriched in rare damaging variants in neuroblastoma patients compared to controls, supporting the concept that some molecular mechanisms are shared between neuroblastoma and neurodevelopmental disorders. Aggregated variant data from the NB cohort are available at http://nbgen.ceinge.unina.it.
Implications of all the available evidence
Our study highlights a broad spectrum of germline clinically relevant variants in a large cohort of neuroblastoma patients and indicates that inherited alterations in homologous recombination and neurodevelopmental genes contribute to the disease initiation. Many of these variants hold potential for therapeutic actionability and, the detection of these alterations in selected neuroblastoma patients may ultimately facilitate the process of integrating genetic testing in the paediatric oncology clinic for the selection of the appropriate therapy, and expand the knowledge in cases where the genotype–phenotype association is still unclear.
Introduction
Neuroblastoma (NB) is the most common solid extracranial paediatric tumour that originates from the adrenal medulla or paraspinal ganglia (sympathetic nervous system) during development.1 Clinical manifestations may vary broadly, from spontaneous tumour regression without therapeutic interventions to a very poor prognosis in older children despite high intensive chemotherapy.1 Clinical and biological factors are used to define distinct risk strata and to determine treatment plans for patients based upon age at diagnosis, stage, tumour histopathology, ploidy, and genomic aberrations.1 In young children, MYCN amplification and other arm-level chromosomal alterations such as 1p and 11q deletions, unbalanced 17q gain and TERT rearrangements have been reported as poor prognostic features.2,3 Children older than six years present unique structural variants, with 19p loss and 1q gain being the most frequent.4 Somatic coding and non-coding point mutations in ALK and ATRX and in regulatory elements, respectively, have been reported in primary tumours as cancer drivers with the potential to predict patient prognosis.5, 6, 7 Gene expression signatures have been reported to be predictive of clinical outcome in high-risk patients.8,9 However, despite the advances in discovering clinically actionable genomic alterations, NB still causes 15% of all deaths due to cancer in children and less than 40% of the children survive longer than 5 years.10
Only 1–2% of patients present with a family history of disease, mainly explained by mutations in ALK and PHOX2B11,12; the vast majority of cases appear to arise sporadically. In the last decade, the genetic aetiology of NB has increasingly been revealed by genome-wide association studies (GWAS), which led us to identify single nucleotide polymorphisms (SNPs) within or upstream genes (CASC15, CASC14, BARD1, LMO1, DUSP12, DDX4/IL31RA, HACE1, LIN28B, and TP53) that are highly associated with NB susceptibility and/or NB aggressiveness in European and Chinese populations.13,14
We have previously demonstrated that NB shares common disease predisposing variants, regulating SLC16A1, BARD1, MSX1, and SHOX2 genes, with neural crest-derived diseases such as melanoma and congenital heart disease.15,16
About 10–20% of paediatric cancer patients harbours germline pathogenic/likely pathogenic (P/LP) variants in cancer predisposition genes (CPGs).17,18 Although large sequencing studies on cases and controls investigating the prevalence of clinically relevant mutations in specific paediatric tumours have been reported,19 such investigations are still missing in NB. Only two studies have reported an enrichment of rare predicted pathogenic variants in ALK, BARD1, AXIN2, CHEK2, PINK1 in a relatively small cohort of NB cases.5,20
The detection of germline alterations in selected NB patients may ultimately facilitate the process of integrating genetic testing in the paediatric oncology clinic, expand the knowledge in cases where the genotype–phenotype association is unclear, and help in the identification of new candidate CPGs.
Here, we used a cohort of 664 NB patients and 882 controls for a large whole-exome sequencing (WES) study to examine the contribution of rare pathogenic germline variants to NB using two different strategies designed to identify mutations with high clinical impact in well-known CPGs and to discover non-canonical cancer gene pathways altered by less penetrant risk mutations.
Methods
Study samples
A total of 793 NB samples were obtained from the BIT Biobank (Gaslini Institute, Genova, Italy). In addition, 1203 healthy controls were selected from a cohort of Italian individuals used in previous studies.21 Germline data from 179 whole-genome sequencing (WGS) samples from a multi-centre study (EGA-NB, ID: EGAS00001004349, EGAS00001001308) and additional 222 WES samples (TARGET-NB, https://ocg.cancer.gov/programs/target/data-matrix) were used as validation datasets. As additional and independent control set for replication purposes, aggregated variant frequencies were obtained from 2424 independent non-related European controls from the Network for Italian Genomes (NIG, http://www.nig.cineca.it/). All cohorts used in this study and their respective inclusion in different analyses are shown in Table S1.
Ethics
The study protocol was approved by the University of Naples “Federico II” Ethics Committee (N. 76/13). All participants provided written informed consent according to the local Ethics Committee requirements.
Sequencing and data processing
Germline DNA from peripheral blood was extracted using QIAamp DNA mini kit (QIAGEN) according to manufacturer's instructions, quantified with Qubit 4 Fluorometer using the dsDNA BR assay kit (Thermo Fisher Scientific) and assessed by agarose gel (2%) electrophoresis using the DNA 2100 Excel Band DNA ladder 100bp (SMOBIO) for quality-control before WES (50× target depth) on Illumina HiSeq instrument.
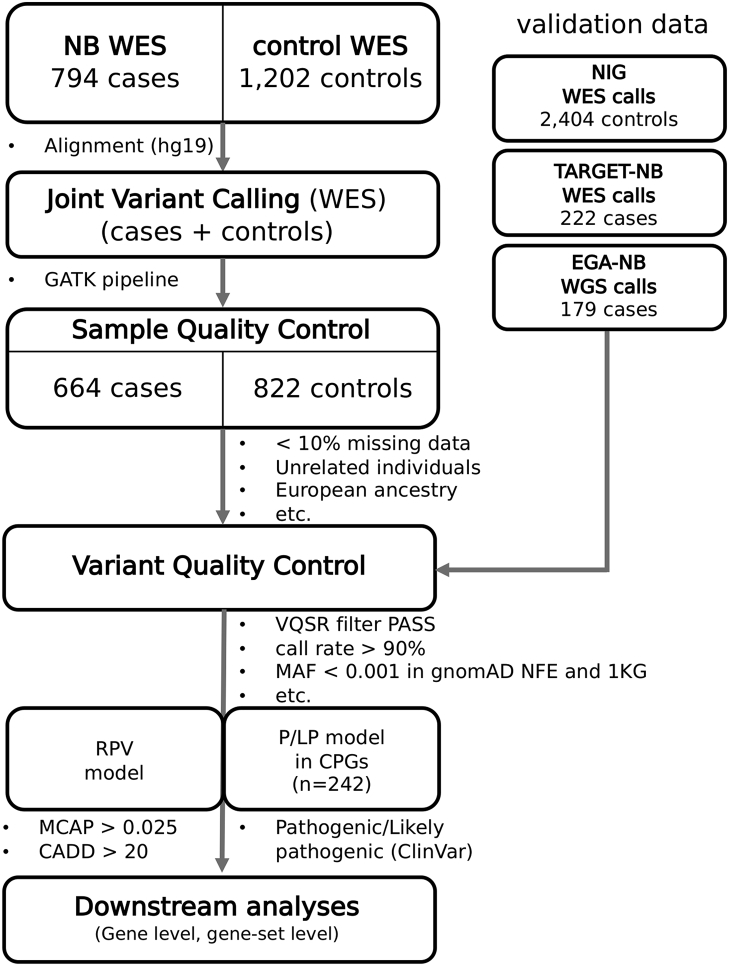
All FASTQ files were uploaded onto a high performance computing system and processed with the standard NGS analysis pipeline based on Genome Analysis Toolkit (GATK) best practice.22 Briefly, adapter and low-quality trimming was performed on the FASTQ files, using the fastp tool (v0.20.1).23 According to the Broad Institute recommendations, sequence reads were aligned to the hg19 reference human genome assembly (GRCh37, including decoy contigs) using the BWA-MEM algorithm (v0.7.17).24 BAM files were sorted by coordinate and duplicates were removed using the GATK MarkDuplicates (v4.2.0.0) and bases were recalibrated according to the GATK guidelines (BaseRecalibrator and ApplyBQSR v4.2.0.0).22 Genomic VCF files (gVCF) were produced for each sample with GATK Haplotype Caller (v4.2.0.0) and pooled into a GenomicsDB datastore for joint genotyping with GATK GenotypeGVCFs (v4.2.0.0) using an interval file with the coordinates of UCSC exonic regions (https://genome.ucsc.edu/).25 A schematic representation of the analysis workflow is depicted in Fig. 1.
Fig. 1.
Workflow of WES and rare variant analysis. WES, whole genome sequencing; RPV, rare pathogenic variant; P/LP, pathogenic/likely pathogenic variant; CPG, cancer predisposition gene; NIG, Network of Italian Genomes.
Germline WES of the parents of probands (trios) with germline P/LP were processed with the same pipeline described above to produce alignment files (BAM), and variants were called through the Strelka2 germline workflow.27 Variants were filtered for those flagged as “PASS” in the resulting VCF according to the embedded scoring method.26
Sample and variant quality control
Samples with genotyping call rate less than 90% and outliers on coverage (20× depth in less than 25% of the exonic regions) were removed. Remaining samples were processed with PLINK v2.027 to select a set of informative SNPs with missingness <0.02, minor allele frequency >0.05, and in Hardy Weinberg Equilibrium (--snps-only --max-alleles 2 --geno 0.02 --maf 0.05 --hwe 1e-5). These markers were then pruned (--indep-pairwise 1500 500 0.2) and used to estimate autosomal heterozygosity, relatedness and population outliers. Samples with a heterozygosity rate deviating more than 3 standard deviations from the mean, duplicates and related samples (pi-hat >0.1875, halfway between a second and third degree relative) were removed. Population outliers (ethnicity different from European) were identified via principal component analysis and projected onto genotypes from 1000 Genomes populations.28 Individuals with an absolute deviation from the median exceeding 6× interquartile range of the first 10 principal components were removed. Sex was imputed from WES data if not available in the medical records. The above filtering steps (summarized in Table S2) were performed with bamdst (https://github.com/shiquan/bamdst) using the coordinates of UCSC exonic regions (https://genome.ucsc.edu/) as target, PLINK v2.0 and bigsnpr v1.10.8.27,29 Variants were recalibrated using GATK Variant Quality Score Recalibration (VQSR) and filtered at the recommended 99% sensitivity tranche. This method, based on a Gaussian mixture model, uses all annotations simultaneously and adjust cutoffs depending on the context to improve the filtering of probable artifacts, rather than setting arbitrary hard thresholds on variant metrics. Variants were normalized, split if multiallelic and left-aligned with bcftools v1.12.30 Variants with missingness higher than 10% were removed. The final multi-sample VCF file contained 1,227,809 variants from 1486 samples (664 NB cases and 822 controls).
Qualifying pathogenic variant models
Ensembl Variant Effect Prediction (VEP) v104.131 was used for the annotation and prioritization of genetic variants, including predictions on splice variants with SpliceAI.32 Functional effect of single amino acid variants in proteins was estimated as the difference in the Gibbs free energy of unfolding, between the wild-type and the variant protein (ΔΔG) with DDGun.33 Data were filtered for coding variants (missense variant, stop lost, stop gained, start lost, splice donor variant, splice acceptor variant, in-frame deletion, in-frame insertion, frame-shift variant) present in less than 0.1% gnomAD and 1000 Genomes populations.28,34 Indels were further filtered to those with a frequency <0.1% in the tested cohort and those mapping within regions of low complexity/mappability (universal masks from the Simons Genome Diversity Project),35 or in homopolymer regions (>5 same nucleotides) were excluded.
Using these variants, we computed two different models of rare pathogenic variants: i) the first, we named P/LP, included only variants clinically annotated as pathogenic (P) or likely pathogenic (LP) in selected CPGs; ii) a second model, named rare pathogenic variants (RPV), included all variants annotated as pathogenic according to M-CAP and CADD predictions.36,37
In the P/LP model, variants were considered qualifying if listed as “pathogenic” or “likely pathogenic” in ClinVar v202012.38 We excluded variants with conflicting clinical significance from different submitters in ClinVar. Details of the filtering steps for classification of variants according to this model are reported in Table S3. The proportions of samples with one or more P/LP variant in CPGs were compared with those from the control group with Fisher's Exact test. Association testing between P/LP carriership (carriers vs non-carriers) and clinical features (MYCN amplification, high-risk NB, stage and age at diagnosis) was performed using logistic regression. Kaplan Meier curves were calculated for overall survival and plotted with the survminer R package (https://cran.r-project.org/web/packages/survminer/index.html). Association with overall survival was evaluated with Cox proportional hazard regression.
Given that the selection of CPGs and P/LP variants in ClinVar database might be biased towards the identification of genes and mutations with high clinical impact and not be suitable to investigate less penetrant pathogenic variants that impact on other molecular mechanisms beyond those related to DNA repair functions, we adopted a burden gene-set analysis using a combination of two predictor scores (RPV model). To compute the RPV model, we used a filtering strategy based on computationally predicted scores rather than selecting variants reported in ClinVar. To select qualifying variants, a sequential data processing with M-CAP and CADD was performed (Table S4).36,37 These computational resources were used because of their documented power to predict pathogenicity of DNA substitutions for clinical utility, assigning priority to M-CAP scores (pathogenicity cutoff >0.025, 5% misclassification rate) over CADD scores (pathogenicity cutoff >0.20, 26% misclassification rate). A control model (null model) with only rare synonymous variants was built and tested accordingly to investigate eventual sources of inflation (Table S5).
Selection of cancer predisposition genes
To compute the P/LP model, annotated data were filtered to capture variants in genes with known association with cancer predisposition (N = 242), defined according to literature and databases. In particular, genes were selected through the harmonization with HUGO Gene Nomenclature Committee (HGNC) identifiers of the CPG lists published in Mirabello et al.,39 Gröbner et al.,40 and genes annotated as germline CPGs in COSMIC. We excluded ribosomal genes (RPL11, RPL15, RPL26, RPL27, RPL31, RPL35A, RPL5, RPS10, RPS19, RPS24, RPS26, RPS27, RPS29, RPS7) and included 9 genes (ATRX, ELP1, FAN1, KIF1B, MSX1, PINK1, RAD50, RIT1, ROS1) as additional CPG candidates because of known implications in NB or other related tumours (Table S6). The gene-set enrichment analysis of mutated CPGs over all tested CPGs (used as background in the hypergeometric test) was performed in WebGestalt using pathways from KEGG.41
Gene collapsing analysis and gene-set burden analysis
To evaluate the cumulative effects of multiple P/LP variants in each CPG we performed a collapsing analysis. For each gene, the proportion of cases was compared to the proportion of controls carrying one or more variants in that gene. To account for eventual case–control imbalances and allow the computation of effect size estimates and standard error which are robust to inflation due to low variant counts, collapsing analysis was modelled based on the Firth's bias-reduced logistic regression implemented in the logistf R package (https://cran.r-project.org/web/packages/logistf/index.html). Only genes with at least two carriers of P/LP variants in the patient group were tested. Confidence intervals and P-values were computed with the profile penalized log likelihood method. Mutations in genes of interest were plotted as lollipop plot and annotated with respective protein domains derived from PFAM database (https://pfam.xfam.org/).
We compared the gene-set RPV burden, that is the number of genes with a RPV variant in selected gene-sets, between cases and controls, implementing a strategy already adopted in previous studies (Figure S1).42 Gene-sets including at least 4 genes were retrieved from Harmonizome (https://maayanlab.cloud/Harmonizome), literature and DisGeNET database (Table S7).43, 44, 45 We describe the gene-set selection process in the results section. The gene-sets related to neuronal development and differentiation were built starting from the GO terms “nervous system development” (GO:0007399) and “neuron differentiation” (GO:0030182) and their downstream classes. Logistic regression was adopted to regress the phenotype on the individual burden score defined as the number of genes with at least one variant count in the tested gene-set. Specifically, for each sample, RPV variants were collapsed by gene and genes summed across each target gene-set to get a burden score, used as a predictor in the regression. To control for background variation, sex and the number of genes with rare synonymous variants were used as covariates in the logistic model as suggested in previous studies.46
Statistics
Analyses were performed using R v4.0.4.47 All statistical tests were 2-sided. Unless otherwise indicated, the cut-off for substantial enrichment was defined as a false discovery rate (FDR) ≤0.05 to account for type I errors. When relevant, further details are available in the method details for the specific analysis.
Role of funders
The funders only provided funding, and had no role in the study design, data collection, data analysis, interpretation, and writing of the report. Authors were not precluded from accessing data in the study, and they accept responsibility to submit for publication.
Results
Prevalence of rare germline pathogenic/likely pathogenic variants in cancer predisposition genes
Our primary analyses were based on patients (N = 793) and controls (N = 1203) with available WES data. To enable case–control analyses and avoid biases potentially introduced by different calling algorithms, germline variants were jointly called from WES data and only positions with a high call rate (>90%) in the whole case–control cohort were brought forward. To reduce confounding due to population stratification, we removed those individuals with an inferred outlier ancestry (non-European) from the principal component analysis (see Methods, Fig. 1 and Figure S2a).
After quality control, 1486 samples (664 NB cases and 822 controls) passed all filtering steps with a mean depth of 55.0 ± 18.4× and 44.7 ± 13.3× before and after duplicate reads removal, respectively, and an average of 78.3 ± 4.6% of target regions were covered with depth higher than 20× (Figure S2b and Table S2). The clinical characteristics of the NB patients (Table 1) were comparable to those reported in previous studies.48 A total of 238 patients (35.8%) were classified as stage 4 according to the International Neuroblastoma Staging System (INSS), 108 (16.3%) had MYCN amplification, and 318 patients (47.9%) were ≥18 months old at the time of diagnosis.
Table 1.
Demographics and clinical characteristics of NB patients included in this study.
| Characteristic | NB patients (percentage) |
|---|---|
| No. of subjects | 664 |
| Age at diagnosis | |
| ≥18 months | 318 (47.9%) |
| < 18 months | 264 (39.8%) |
| unknown | 82 (12.3%) |
| Sex | |
| Male | 342 (51.5%) |
| Female | 292 (44.0%) |
| Unknown | 30 (4.5%) |
| MYCN amplification | |
| Amplified | 108 (16.3%) |
| Non amplified | 370 (55.7%) |
| Gain | 41 (6.2%) |
| Unknown | 145 (21.8%) |
| Stage of cancera | |
| 1/2/3/4s | 254 (38.3%) |
| 4 | 238 (35.8%) |
| Unknown | 172 (25.9%) |
| Risk | |
| Low/intermediate | 270 (40.7%) |
| High | 219 (33.0%) |
| Unknown | 175 (26.3%) |
According to the International Neuroblastoma Staging System (INSS).
We evaluated the frequency of P/LP variants in 242 a priori defined CPGs in the set of patients and control group. These included manually curated genes obtained from previous studies39,40 and genes already annotated as CPG in the COSMIC database (see Methods and Table S6). A total of 80 NB patients (12.05%) carried one or more P/LP variants in a CPG and showed significant enrichment (Fisher's exact test P = 2.3 × 10−4, OR = 1.99) compared to controls (Fig. 2a) encompassing 6.45% of individuals with at least one P/LP variant. Specifically, we identified 84 rare germline P/LP variants in 45 out of 242 CPGs in NB cases (Fig. 2b and Table S8). These P/LP included 36 missense mutations (42.8%), 15 frame-shift variants (17.9%), 9 splice region variants (10.7%) and 24 start-loss/stop-gained variants (28.6%). A total of 12 P/LP variants were annotated as splice variants or missense/splice variants and all of them (except one with a delta score = 0.4) were predicted with high precision (delta score >0.8) to be donor loss (n = 8) or acceptor loss (n = 3) (Table S8). NB patients were enriched in P/LP variant carriers also when focusing only on genes with an autosomal dominant or autosomal recessive mode of inheritance (Fisher's exact test P = 2.2 × 10−4 and P = 0.02, respectively, Fig. 2a). Twelve and thirteen out of the 45 genes were also mutated with P/LP in 31 individuals from two independent sets of 222 and 179 NB cases, respectively (Fig. 2c and Table S8). Integration of variants from these three cohorts of patients showed ten genes mutated in more than four individuals with BRCA1 (N = 8), CHEK2 (N = 6), G6PC (N = 6), ALK (N = 5) BLM (N = 5), BRCA2 (N = 5) and RAD51C (N = 5) being the most frequently mutated among all tested cohorts (Fig. 2c and Table S8). No P/LP variant was detected in the familial NB susceptibility gene PHOXB2, probably due to its lower mutation frequency (5–10%) in familial NB compared to ALK (75–80%).13 Pathway enrichment analysis of the 45 CPGs with P/LP variants against all selected CPGs (N = 242) indicated significant enrichment in homologous recombination (HR) pathway genes (hypergeometric test FDR = 7.6 × 10−3, Table S9).
Fig. 2.
Proportion of NB patients with germline PLP variants detected in CPGs.(a) Proportions of NB patients and controls carrying P/LP variants involving all CPGs and CPGs with autosomal dominant (AD) or autosomal recessive (AR) mode of inheritance. ∗∗∗P = 2.3 × 10−4; ∗∗P = 2.2 × 10−3; ∗P = 0.02 (Fisher's exact test). (b) Percentage of P/LP variant carriers in NB cases and controls. (c) Distribution of P/LP carriers in the tested cohort (NB) and in the validation datasets (TARGET-NB and EGA-NB). Each cell shows the number of carriers for each gene with a P/LP variant in the tested cohort.
The presence of a P/LP germline variant in a CPG was not associated with worse overall survival or other tested clinical features such as INSS stage, age at diagnosis, high-risk and MYCN amplification (Figure S3 and Table S10).
For a subset of five NB with a P/LP variant, germline WES for parents was available and used to analyse inheritance among trios. Identical variants affecting BRCA1, G6PC1, SMARCA4, ALK and FGFR3 were identified in probands and parents. Variants were inherited from the mother in three cases and from the father in two cases (Figure S4).
BRCA1, ALK, and RAD51C enriched for P/LP variants in NB patients compared to controls
We carried out rare variant gene collapsing analysis in the NB cohort versus healthy controls and tested genes with at least two carriers in the NB group (N = 24). Although no CPG showed significant enrichment after FDR adjustment, BRCA1 showed the highest frequency of P/LP carriers in NB patients (N = 7, 1.05%) and yield a nominally significant enrichment (Firth's logistic regression P = 2.7 × 10−3, OR = 18.76) as well as BRCA2 (N = 4, 0.060%, Firth's logistic regression P = 0.032, OR = 11.21) (Fig. 2b, Table 2, Table S11). A P = 0.07 (Firth's logistic regression) was recorded for ALK and RAD51C (OR = 8.70). None of these genes was significantly enriched using a model with synonymous variants only (null model), indicating sufficient control for inflation (Table S11).
Table 2.
Association results for the 24 CPGs with at least 2 carriers in the NB cases cohort.
| Gene | NB freqa | Ctrl freqb | NIG ctrl freqc | NB (N = 664) vs controls (N = 822) |
NB (N = 664) vs NIG controls (N = 2424) |
||||
|---|---|---|---|---|---|---|---|---|---|
| OR | P | FDR | OR | P | FDR | ||||
| BRCA1 | 0.0105 | 0.0000 | 0.0008 | 18.76 | 0.0027 | 0.0648 | 10.95 | 0.0003 | 0.0077 |
| ALK | 0.0045 | 0.0000 | 0.0000 | 8.70 | 0.0726 | 0.3620 | 25.53 | 0.0048 | 0.0384 |
| RAD51C | 0.0045 | 0.0000 | 0.0000 | 8.70 | 0.0726 | 0.3620 | 25.53 | 0.0048 | 0.0384 |
| NF1 | 0.0030 | 0.0000 | 0.0000 | 6.21 | 0.1659 | 0.3620 | 18.24 | 0.0224 | 0.0768 |
| SERPINA1 | 0.0030 | 0.0000 | 0.0000 | 6.21 | 0.1659 | 0.3620 | 18.24 | 0.0224 | 0.0768 |
| TP53 | 0.0030 | 0.0012 | 0.0000 | 2.07 | 0.4740 | 0.5417 | 18.24 | 0.0224 | 0.0768 |
| WRN | 0.0030 | 0.0012 | 0.0000 | 2.07 | 0.4740 | 0.5417 | 18.26 | 0.0223 | 0.0768 |
| MSH2 | 0.0030 | 0.0012 | 0.0004 | 2.07 | 0.4740 | 0.5417 | 6.43 | 0.0694 | 0.2082 |
| ATM | 0.0045 | 0.0012 | 0.0017 | 2.90 | 0.2524 | 0.5048 | 2.86 | 0.1583 | 0.3234 |
| BRCA2 | 0.0060 | 0.0000 | 0.0033 | 11.21 | 0.0319 | 0.3620 | 2.36 | 0.1338 | 0.3234 |
| CHEK2 | 0.0030 | 0.0012 | 0.0008 | 2.07 | 0.4740 | 0.5417 | 3.76 | 0.1530 | 0.3234 |
| NBN | 0.0030 | 0.0012 | 0.0008 | 2.07 | 0.4740 | 0.5417 | 3.65 | 0.1617 | 0.3234 |
| G6PC | 0.0060 | 0.0012 | 0.0041 | 3.73 | 0.1305 | 0.3620 | 1.91 | 0.2388 | 0.4409 |
| COL7A1 | 0.0030 | 0.0061 | 0.0012 | 0.56 | 0.4358 | 0.5417 | 2.61 | 0.2685 | 0.4469 |
| RAD50 | 0.0060 | 0.0012 | 0.0033 | 3.73 | 0.1305 | 0.3620 | 1.93 | 0.2793 | 0.4469 |
| AGL | 0.0030 | 0.0000 | 0.0017 | 6.21 | 0.1659 | 0.3620 | 2.03 | 0.3908 | 0.5211 |
| LZTR1 | 0.0030 | 0.0000 | 0.0017 | 6.21 | 0.1659 | 0.3620 | 2.03 | 0.3897 | 0.5211 |
| TSHR | 0.0030 | 0.0049 | 0.0017 | 0.69 | 0.6289 | 0.6861 | 2.03 | 0.3908 | 0.5211 |
| ERCC3 | 0.0030 | 0.0000 | 0.0021 | 6.21 | 0.1659 | 0.3620 | 1.66 | 0.5229 | 0.6275 |
| ROS1 | 0.0030 | 0.0012 | 0.0021 | 2.07 | 0.4740 | 0.5417 | 1.66 | 0.5226 | 0.6275 |
| GJB2 | 0.0030 | 0.0012 | 0.0025 | 2.07 | 0.4740 | 0.5417 | 1.40 | 0.6578 | 0.7518 |
| ERCC2 | 0.0030 | 0.0012 | 0.0029 | 2.07 | 0.4740 | 0.5417 | 1.22 | 0.7929 | 0.8650 |
| DHCR7 | 0.0030 | 0.0036 | 0.0037 | 0.88 | 0.8810 | 0.8810 | 0.96 | 0.9538 | 0.9538 |
| MUTYH | 0.0030 | 0.0036 | 0.0037 | 0.88 | 0.8810 | 0.8810 | 0.96 | 0.9538 | 0.9538 |
OR, Odds Ratio; P, P-value from Firth logistic regression; FDR, false discovery rate adjusted P-value. Significant results (FDR ≤ 0.05) are highlighted in bold.
P/LP carrier frequency in NB cases.
P/LP carrier frequency in controls.
P/LP carrier frequency in NIG controls.
We then compared NB P/LP variant frequencies against an independent dataset including 2424 unrelated healthy individuals of Italian origins from the NIG (http://www.nig.cineca.it/) and confirmed a significant enrichment of P/LP variants in BRCA1 gene (Firth's logistic regression FDR = 7.7 × 10−3, OR = 10.95), ALK and RAD51C (Firth's logistic regression FDR = 3.8 × 10−2, OR = 25.53, Table 2, Table S12).
In silico analysis of BRCA1, ALK, and RAD51C variants
All BRCA1 (N = 7) P/LP variants were predicted to be loss-of-function and three of them were within the RING, EIN3 and BRCT protein domains (Figure S5). Notably, the variant c.181T>G (p.Cys61Gly) was the one predicted to have the highest impact in terms of variation of unfolding free energy (ΔΔG = −3.0) among all missense variants found in the NB cohort (Table S8). All BRCA1 variants were found in single patients, except the c.5329dup variant identified in two patients.
Three different ALK mutations, all located in the tyrosine kinase domain, were detected in five different patients (Figure S5). Two patients carried the c.3383G>C (p.G1128A) variant, previously associated with familial NB,49 whereas the other two patients carried the c.3824G>A (p.R1275Q) variant, already found in germline and somatic samples.50
RAD51C was mutated in the NB cohort and in both validation cohorts. It was also significantly enriched in mutations compared with both control groups. Interestingly, all five variants showed loss-of-function features and were located in the radA domain (Figure S5).
Burden analysis in NB gene-sets
We compared the burden of a collection of biologically informed gene-sets in NB cases and controls in order to maximize the joint effect of rare pathogenic germline variants defined according to M-CAP and CADD (RPV model, see Methods) and tested a total of 56 gene-sets (Table S7) gathered from Harmonizome,43 DisGeNET,44 and literature from the following functional categories: i) DNA repair-related processes,45 to validate their involvement in NB pathogenesis with a different genetic model; ii) nervous system development, neuron and neural crest differentiation as NB arises from abnormal neuronal development and differentiation,51 and neurodevelopmental disorders (NDDs); iii) NB susceptibility genes identified through GWAS. A set of susceptibility genes associated with non-cancer disorders (e.g., Crohn's disease, psoriasis and asthma) was also selected as negative control.
Given that some gene-sets may include more than hundreds of genes, we used the burden of synonymous variants, in addition to sex, to adjust for eventual inflation (see Methods). For the gene-sets related to DNA damage repair pathways, we found a preferential enrichment of HR (logistic regression FDR = 0.035, log(OR) = 0.14) rather than other mechanisms such as direct repair or mismatch repair (logistic regression FDR = 0.68 and FDR = 0.48, respectively, Fig. 3 and Table S13). This finding was further confirmed by the higher RPV burden in genes co-expressed with BRCA1 and BRCA2 (significantly enriched in P/LP variants in the collapsing analysis) (logistic regression FDR = 0.025, log(OR) = 0.15 and FDR = 0.011, log(OR) = 0.20, Fig. 3 and Table S13).
Fig. 3.
Burden of RPV variants in selected gene-sets. The burden of RPV in the 56 gene-sets is shown on the x-axis (log-odds from logistic regression; error bars indicate the 95% confidence intervals); gene-sets are shown on the y-axis and annotated with a box indicating the respective gene-set category according to the legend. Gene-sets are ordered by increasing FDR with the dot colour proportional to the −log10(FDR). Results with FDR above the significance threshold (logistic regression, FDR > 0.05) are shown in grey.
Consistent with NB pathogenesis, we found a significant enrichment for Gene Ontology (GO) terms related to the regulation of neuronal differentiation (logistic regression FDR = 0.021; log(OR) = 0.07); however, we also observed a significant enrichment for biological processes related to neural tube (logistic regression FDR = 0.025; log(OR) = 0.23) rather than neural crest development or differentiation. We also found an enrichment of RPV in genes involved in the molecular mechanisms responsible for synapse assembly (logistic regression FDR = 0.03; log(OR) = 0.17) or ganglion development (logistic regression FDR = 0.04; log(OR) = 0.37) rather than peripheral (logistic regression FDR = 0.39; log(OR) = 0.11) or sympathetic nervous system development (logistic regression FDR = 0.32; log(OR) = 0.15). Notably, a high RPV burden was also found in genes associated with NDDs (logistic regression FDR = 0.03; log(OR) = 0.06).
Finally, we found a significant enrichment in NB samples for RPV in genes previously associated with NB from GWASdb (logistic regression FDR = 0.011; log(OR) = 0.20).52 On the contrary, sets of genes associated with diseases not related to NB, such as asthma or psoriasis, tested as negative controls, were not enriched in RPV (Fig. 3 and Table S13). In line with studies in other diseases,53 this suggests that rare germline pathogenic variants contribute to NB susceptibility in addition to common risk variants with low penetrance identified in GWAS.
Discussion
We have carried out exome sequencing in a large set of individuals diagnosed with NB and compared it with control individuals to identify the contribution of rare germline pathogenic variants to NB risk. We used two different analytic strategies: one designed to identify mutations with high clinical impact, and the other designed to discover less penetrant risk mutations that may affect non-canonical cancer gene pathways, such as those linked to the cellular origins of NB.
The first analysis allowed us to identify rare clinically-relevant pathogenic variants (P/LP), occurring in 45 out of 242 CPGs, in approximately 12% of cases. This mutational enrichment resulted statistically significant when the genes were stratified according to the inheritance pattern (dominant and recessive). We assume that heterozygous variants in autosomal recessive genes could bring on small effects, like induction of haploinsufficiency, increasing thus susceptibility to cancer without leading to an overt cancer syndrome. The 45 genes that were mutated in our Italian cohort were mutated in 14% and 17% of individuals from two separate cohorts of American European and German origins, respectively. This set of inherited rare variants, significantly more frequent in NB patients than in controls (6%), contributes to NB susceptibility. This may have relevant clinical implications and help to refine NB diagnosis with the development of a hereditary NB gene panel and the set-up of appropriate genetic consulting. Interestingly, alteration of the HR pathway seems to play a significant role in NB pathogenesis as it was the only pathway enriched when testing the P/LP mutated CPGs against the whole set of CPGs.
We performed a P/LP gene-collapsing analysis comparing cases and controls in order to increase statistical power and determine the cumulative effect on NB risk, focusing on CPGs with at least 2 carriers. Although no gene yield significant P-value after FDR correction, we detected a nominally significant P/LP enrichment in BRCA1 and BRCA2 and a suggestive enrichment in ALK and RAD51C. However, these genes were all significantly enriched (Firth's logistic regression FDR ≤ 0.05) in a validation analysis using aggregated data from a larger and independent set of controls (NIG cohort), with the exception of BRCA2. The validity of these findings was further supported from the analysis of synonymous variants whose frequency did not differ between cases and controls for the above-mentioned genes.
BRCA1 is highly expressed in neuronal progenitor cells during early development54 and mutations in this gene result in HR deficiency. A recent study has also demonstrated that it is recruited by MYCN and enhances its transcriptional activation in NB.55 ALK is the most frequently mutated gene in familial NB.12 We found germline point mutations already associated with familial NB50 that are located in the kinase domain of the protein and cause constitutive signaling.12 Notably, we did not identify any mutations in PHOX2B, the first NB susceptibility gene identified.56 Together with a significant P/LP enrichment in NB, we identified variants in both validation cohorts for RAD51C, a gene that has never been associated with NB. RAD51C, which is essential for HR, functions as a TP53-dependent tumour suppressor gene, and it has been reported to be a breast and ovarian cancer susceptibility gene.57 A homozygous RAD51C mutation has also been identified in Fanconi anaemia-like disorders.58
No significant association was found between the status of P/LP carrier and clinical factors, such as age at diagnosis, MYCN amplification status and INSS stage. Carriership of P/LP mutation was not associated with a better or worse prognosis. This may suggest that rare P/LP mutations predispose to NB initiation rather than its progression and/or severity.
We adopted a second systematic strategy (RPV model), based on two predictor scores (CADD and M-CAP)36,37 rather than clinically annotated variants used in the P/LP model, because the selection of P/LP variants in ClinVar database might be biased towards the identification of genes and mutations with high penetrance and not be suitable to investigate less penetrant pathogenic variants of non-cancer-related genes.
This analysis showed that deleterious rare variants are more frequent in neuronal differentiation and development genes when comparing NB cases to controls, suggesting their involvement in NB pathogenesis. We found mutational enrichment of gene-sets from the early phase of nervous system development (neural tube) and from later phases (e.g., synapse assembly and ganglion development). Although NB is thought to arise from improper differentiation of neural crest cells of the sympathoadrenal lineage,1 no significant association was found for gene-sets related to differentiation and development of the neural crest and sympathetic nervous system. NDD genes were enriched in rare risk variants, in line with recent data reporting that NDD genes are altered in NB at somatic and germline levels.59,60
The enrichment of rare variants in NB GWAS genes supports the convergence of rare and common variants in conferring NB risk, as demonstrated in complex diseases and cancers.53 Finally, our gene-set burden analysis confirmed the involvement of HR genes in the NB pathogenesis, and, together with the specific enriched set, we found significantly higher RPV burden in genes co-expressed with BRCA1 and BRCA2, which are known to be involved in chromosomal double strand break repair by HR.61
One possible limitation of this study is that we could not perform a validation of the gene-set burden analysis because only aggregated variant data were available for the additional control cohort (NIG). However, the reliability of the results is supported by the robustness of the model that accounted for confounders and inflation (sex and synonymous variant burden used as covariates) and by testing multiple overlapping gene-sets and negative controls from different sources to corroborate the findings that support a biological relevance. Moreover, functional studies are needed to determine the specific role of RAD51C in NB development and the penetrance of its mutations, and to ultimately confirm its role as a candidate NB susceptibility gene.
In conclusion, our study highlights a broad spectrum of germline CPGs variants in a large cohort of NB patients and indicates that inherited alterations in HR and neurodevelopmental genes contribute to NB initiation. Further analysis of the germline genome and functional validation are essential to fully understand the contribution of these variants to NB tumorigenesis and to establish an appropriate genetic risk assessment for the patients. Many of these variants hold potential for therapeutic actionability and, as precision medicine is becoming increasingly integrated into paediatric oncology, germline analysis may be additive to tumour sequence analysis for therapy selection.
Contributors
FB: Formal analysis, Data curation, Methodology, Investigation, Validation, Visualization, Writing-Review and Editing, verification of the underlying data; VAL: Formal analysis, Data curation, Validation; SC: Formal analysis, Data curation; GDA: Formal analysis, Visualization; VA: Formal analysis, Visualization; AB: Data curation, Visualization; MA: Data curation; SF: Formal analysis, Data curation; AR: Data curation, Methodology; MM: Data curation; SS: Data curation, Validation; FW: Data curation, Validation; GP: Visualization; AE: Data curation; AI: Supervision, Writing-Review and Editing, Funding acquisition, Resources; MC: Conceptualization, Supervision, Writing-Review and Editing, Funding acquisition, Resources, verification of the underlying data. All authors have read and approved the submission of this manuscript.
Data sharing statement
The aggregated variant data for the NB cohort generated in this study are available at http://nbgen.ceinge.unina.it. The code used to process WES data as described in the Methods section is available at https://github.com/nandobonf/ngspipe. WGS samples (EGA-NB) can be accessed via application at The European Genome-phenome Archive (EGA) with the ID EGAS00001004349 and EGAS00001001308. WES samples from the TARGET-NB dataset are available under license at https://ocg.cancer.gov/programs/target/data-matrix. Aggregated variant data from the NIG are available upon request at http://nigdb.cineca.it/.
Declaration of interests
All authors have no potential conflicts of interests to disclose.
Acknowledgements
This study was supported by grants from Associazione Italiana per la Ricerca sul Cancro (Grant No. 25796 to MC and Grant No. 20757 to AI), Fondazione Italiana per la Lotta al Neuroblastoma (to MC), Associazione Oncologia Pediatrica e Neuroblastoma (to MC), Regione Campania “SATIN” Grant 2018-2020 (to MC), Associazione Giulio Adelfio onlus (to MC and AI), and Italian Health Ministry (No. GR-2016-02364546 to MC). We would like to thank the BIT Biobank (Gaslini Institute, Genova, Italy) for providing neuroblastoma DNA samples and the Network for Italian Genome NIG (http://www.nig.cineca.it) for providing aggregated variant data of the Italian control population.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2022.104395.
Appendix A. Supplementary data
References
- 1.Matthay K.K., Maris J.M., Schleiermacher G., et al. Neuroblastoma. Nat Rev Dis Primers. 2016;2 doi: 10.1038/nrdp.2016.78. [DOI] [PubMed] [Google Scholar]
- 2.Capasso M., Diskin S.J. Genetics and genomics of neuroblastoma. Cancer Treat Res. 2010;155:65–84. doi: 10.1007/978-1-4419-6033-7_4. [DOI] [PubMed] [Google Scholar]
- 3.Peifer M., Hertwig F., Roels F., et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526:700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lasorsa V.A., Cimmino F., Ognibene M., et al. 19p loss is significantly enriched in older age neuroblastoma patients and correlates with poor prognosis. NPJ Genom Med. 2020;5:18. doi: 10.1038/s41525-020-0125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pugh T.J., Morozova O., Attiyeh E.F., et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lasorsa V.A., Montella A., Cantalupo S., et al. Somatic mutations enriched in cis-regulatory elements affect genes involved in embryonic development and immune system response in neuroblastoma. Cancer Res. 2022;82(7):1193–1207. doi: 10.1158/0008-5472.CAN-20-3788. [DOI] [PubMed] [Google Scholar]
- 7.Capasso M., Lasorsa V.A., Cimmino F., et al. Transcription factors involved in tumorigenesis are over-represented in mutated active DNA-binding sites in neuroblastoma. Cancer Res. 2020;80:382–393. doi: 10.1158/0008-5472.CAN-19-2883. [DOI] [PubMed] [Google Scholar]
- 8.Formicola D., Petrosino G., Lasorsa V.A., et al. An 18 gene expression-based score classifier predicts the clinical outcome in stage 4 neuroblastoma. J Transl Med. 2016;14:142. doi: 10.1186/s12967-016-0896-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barbieri E., De Preter K., Capasso M., et al. A p53 drug response signature identifies prognostic genes in high-risk neuroblastoma. PLoS One. 2013;8 doi: 10.1371/journal.pone.0079843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park J.R., Bagatell R., Cohn S.L., et al. Revisions to the international neuroblastoma response criteria: a consensus statement from the National Cancer Institute Clinical Trials Planning Meeting. J Clin Oncol. 2017;35:2580–2587. doi: 10.1200/JCO.2016.72.0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mosse Y.P., Laudenslager M., Khazi D., et al. Germline PHOX2B mutation in hereditary neuroblastoma. Am J Hum Genet. 2004;75:727–730. doi: 10.1086/424530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mossé Y.P., Laudenslager M., Longo L., et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tonini G.P., Capasso M. Genetic predisposition and chromosome instability in neuroblastoma. Cancer Metastasis Rev. 2020;39:275–285. doi: 10.1007/s10555-020-09843-4. [DOI] [PubMed] [Google Scholar]
- 14.He J., Zou Y., Wang T., et al. Genetic variations of GWAS-identified genes and neuroblastoma susceptibility: a replication study in Southern Chinese children. Transl Oncol. 2017;10:936–941. doi: 10.1016/j.tranon.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Testori A., Lasorsa V.A., Cimmino F., et al. Exploring shared susceptibility between two neural crest cells originating conditions: neuroblastoma and congenital heart disease. Genes (Basel) 2019;10:E663. doi: 10.3390/genes10090663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avitabile M., Succoio M., Testori A., et al. Neural crest-derived tumor neuroblastoma and melanoma share 1p13.2 as susceptibility locus that shows a long-range interaction with the SLC16A1 gene. Carcinogenesis. 2020;41:284–295. doi: 10.1093/carcin/bgz153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J., Walsh M.F., Wu G., et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373:2336–2346. doi: 10.1056/NEJMoa1508054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiala E.M., Jayakumaran G., Mauguen A., et al. Prospective pan-cancer germline testing using MSK-IMPACT informs clinical translation in 751 patients with pediatric solid tumors. Nat Cancer. 2021;2:357–365. doi: 10.1038/s43018-021-00172-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capasso M., Montella A., Tirelli M., Maiorino T., Cantalupo S., Iolascon A. Genetic predisposition to solid pediatric cancers. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.590033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lasorsa V.A., Formicola D., Pignataro P., et al. Exome and deep sequencing of clinically aggressive neuroblastoma reveal somatic mutations that affect key pathways involved in cancer progression. Oncotarget. 2016;7:21840–21852. doi: 10.18632/oncotarget.8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Alterio G., Lasorsa V.A., Bonfiglio F., et al. Germline rare variants of lectin pathway genes predispose to asymptomatic SARS-CoV-2 infection in elderly individuals. Genet Med. 2022;24(8):1653–1663. doi: 10.1016/j.gim.2022.04.007. S1098-3600(22)724-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.der Auwera G.A.V., O'Connor B.D. O’Reilly Media, Incorporated; 2020. Genomics in the cloud: using docker, GATK, and WDL in terra. [Google Scholar]
- 23.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poplin R., Ruano-Rubio V., DePristo M.A., et al. Scaling accurate genetic variant discovery to tens of thousands of samples. biorxiv. 2018 doi: 10.1101/201178. [DOI] [Google Scholar]
- 26.Kim S., Scheffler K., Halpern A.L., et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15:591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 27.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The 1000 Genomes Project Consortium, Auton A., Brooks L.D., et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Privé F., Aschard H., Ziyatdinov A., Blum M.G.B. Efficient analysis of large-scale genome-wide data with two R packages: bigstatsr and bigsnpr. Bioinformatics. 2018;34:2781–2787. doi: 10.1093/bioinformatics/bty185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danecek P., Bonfield J.K., Liddle J., et al. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10:giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLaren W., Gil L., Hunt S.E., et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 33.Montanucci L., Capriotti E., Birolo G., et al. DDGun: an untrained predictor of protein stability changes upon amino acid variants. Nucleic Acids Res. 2022;50(W1):W222–W227. doi: 10.1093/nar/gkac325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karczewski K.J., Francioli L.C., Tiao G., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mallick S., Li H., Lipson M., et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jagadeesh K.A., Wenger A.M., Berger M.J., et al. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat Genet. 2016;48:1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 37.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landrum M.J., Lee J.M., Benson M., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirabello L., Zhu B., Koster R., et al. Frequency of pathogenic germline variants in cancer-susceptibility genes in patients with osteosarcoma. JAMA Oncol. 2020;6:724–734. doi: 10.1001/jamaoncol.2020.0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gröbner S.N., Worst B.C., Weischenfeldt J., et al. The landscape of genomic alterations across childhood cancers. Nature. 2018;555:321–327. doi: 10.1038/nature25480. [DOI] [PubMed] [Google Scholar]
- 41.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koko M., Krause R., Sander T., et al. Distinct gene-set burden patterns underlie common generalized and focal epilepsies. eBioMedicine. 2021;72 doi: 10.1016/j.ebiom.2021.103588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rouillard A.D., Gundersen G.W., Fernandez N.F., et al. The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database. 2016;2016:baw100. doi: 10.1093/database/baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knijnenburg T.A., Wang L., Zimmermann M.T., et al. Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell Rep. 2018;23:239–254.e6. doi: 10.1016/j.celrep.2018.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Povysil G., Petrovski S., Hostyk J., Aggarwal V., Allen A.S., Goldstein D.B. Rare-variant collapsing analyses for complex traits: guidelines and applications. Nat Rev Genet. 2019;20:747–759. doi: 10.1038/s41576-019-0177-4. [DOI] [PubMed] [Google Scholar]
- 47.R Core Team . R Core Team; Vienna, Austria: 2013. R: a language and environment for statistical computing.http://www.R-project.org/ [Google Scholar]
- 48.McDaniel L.D., Conkrite K.L., Chang X., et al. Common variants upstream of MLF1 at 3q25 and within CPZ at 4p16 associated with neuroblastoma. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonatti F., Pepe C., Tancredi M., et al. RNA-based analysis of BRCA1 and BRCA2 gene alterations. Cancer Genet Cytogenet. 2006;170:93–101. doi: 10.1016/j.cancergencyto.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 50.Janoueix-Lerosey I., Lequin D., Brugières L., et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 51.Tomolonis J.A., Agarwal S., Shohet J.M. Neuroblastoma pathogenesis: deregulation of embryonic neural crest development. Cell Tissue Res. 2018;372:245–262. doi: 10.1007/s00441-017-2747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buniello A., MacArthur J.A.L., Cerezo M., et al. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Q., Dhindsa R.S., Carss K., et al. Rare variant contribution to human disease in 281,104 UK Biobank exomes. Nature. 2021;597:527–532. doi: 10.1038/s41586-021-03855-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pao G.M., Zhu Q., Perez-Garcia C.G., et al. Role of BRCA1 in brain development. Proc Natl Acad Sci U S A. 2014;111:E1240–E1248. doi: 10.1073/pnas.1400783111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herold S., Kalb J., Büchel G., et al. Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase. Nature. 2019;567:545–549. doi: 10.1038/s41586-019-1030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trochet D., Bourdeaut F., Janoueix-Lerosey I., et al. Germline mutations of the paired–like homeobox 2B (PHOX2B) gene in neuroblastoma. Am J Hum Genet. 2004;74:761–764. doi: 10.1086/383253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meindl A., Hellebrand H., Wiek C., et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42:410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 58.Vaz F., Hanenberg H., Schuster B., et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42:406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 59.Lopez G., Conkrite K.L., Doepner M., et al. Somatic structural variation targets neurodevelopmental genes and identifies SHANK2 as a tumor suppressor in neuroblastoma. Genome Res. 2020;30:1228–1242. doi: 10.1101/gr.252106.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Egolf L.E., Vaksman Z., Lopez G., et al. Germline 16p11.2 microdeletion predisposes to neuroblastoma. Am J Hum Genet. 2019;105:658–668. doi: 10.1016/j.ajhg.2019.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy R., Chun J., Powell S.N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.