Summary
In vitro assembly of amyloid fibrils that recapitulate those in human brains is very useful for fundamental and applied research on the amyloid formation, pathology, and clinical detection. Recent success in the assembly of Tau fibrils in vitro enables the recapitulation of the paired helical filament (PHF) of Tau extracted from brains of patients with Alzheimer’s disease (AD). However, following the protocol, we observed that Tau constructs including 297-391 and a mixture of 266-391 (3R)/297-391, which are expected to predominantly form PHF-like fibrils, form highly heterogeneous fibrils instead. Moreover, the seemingly PHF-like fibril formed by Tau 297-391 exhibits a distinctive atomic structure with a spindle-like fold, that is neither PHF-like or similar to any known Tau fibril structures revealed by cryo-electron microscopy (cryo-EM). Our work highlights the high sensitivity of amyloid fibril formation to subtle conditional changes and suggests high-resolution structural characterization to in vitro assembled fibrils prior to further laboratory use.
Subject areas: Neuroscience, cell biology, structural biology
Graphical abstract
Highlights
-
•
Slightly tuning fibrillation condition largely changes the composition of Tau fibrils
-
•
Preparation of PHF-like Tau fibril unexpectedly results in a new polymorph
-
•
High-resolution structural characterization ensures the production of desired Tau fibril
Neuroscience; Cell biology; Structural biology
Introduction
Tau protein forms amyloid fibrils which serve as a pathological entity in AD and other Tauopathies such as chronic traumatic encephalopathy (CTE), corticobasal degeneration (CBD), and Pick’s disease (PiD).1,2,3,4 Tau fibrils extracted from the patients’ brains of different Tauopathies exhibit distinct pathologic features in cellular and animal models.5 Moreover, cryo-EM studies reveal that these ex vivo Tau fibrils have distinct structures with different folding of Tau, so-called polymorphs.3,6,7,8,9 Accumulating evidence have shown that polymorphism is a common feature of amyloid fibrils.1,10,11,12,13,14,15 Accordingly, many important questions remain to be answered. For example, what determines fibril polymorphs; how polymorphic fibril structures determine their neuropathological activity; can we develop a specific binder to recognize certain disease-related fibril polymorph for clinical diagnosis and therapeutical treatment of the disease? To investigate these issues, a large amount of fibrils are required. However, the extraction of amyloid fibrils from diseased human brains can hardly match the needs, not even mention the inaccessibility of human brain tissues for most labs. Therefore, in vitro preparation of recombinant fibrils provides an important strategy to supply adequate amounts of fibrils for further investigation in fundamental and applied research.
However, many studies have shown that in vitro prepared recombinant proteins such as Tau and α-synuclein (α-syn) form fibrils with structural polymorphs largely distinct from those extracted from diseased brains,6,13,16,17,18,19,20,21,22 which leads to a concern of the biological relevance of in vitro prepared fibrils. Thus, is it possible to assemble fibrils in vitro that recapitulate those in brains? In 2017, Serpell and colleagues reported that Tau 297-391 can assemble into the AD PHF-like fibrils in vitro in the presence of reductant, and the disulfide bond formation by Cys322 prevents the AD PHF-like fibril formation.23,24 Later, Scheres and colleagues screened various truncations of Tau under different conditions, and obtained several Tau truncations that can form fibrils under reducing conditions featuring structures similar to the fibril polymorphs found in AD and CTE.25 This result is promising for in vitro preparation of disease-related fibril polymorphs for further studies of fibril pathology and potential pharmaceuticals.
In this work, we originally sought to produce Tau PHF-like polymorph in vitro by following the published protocol for further study. Unexpectedly, we obtained highly heterogeneous fibrils with either Tau 297-391 or a mixture of Tau 266-391 (3R)/297-391 (equal molar), which was reported to predominantly form the PHF-like fibril of AD.25 By optimizing the fibrillation condition, we managed to populate the PHF-like fibril with both Tau samples. However, by using cryo-EM, we found that Tau 297-391 surprisingly folds into a previously unobserved structure, although the fibril is morphologically similar to the PHF-like fibril under low-resolution EM imaging. Our work demonstrates that the in vitro assembly of Tau fibrils is highly sensitive to subtle changes in fibrillation conditions. Even following a similar protocol, different labs may generate fibrils morphologically similar, but totally different in atomic structures.
Results
Purification and fibrillation of recombinant Tau 297-391 and Tau 266-391 (3R)
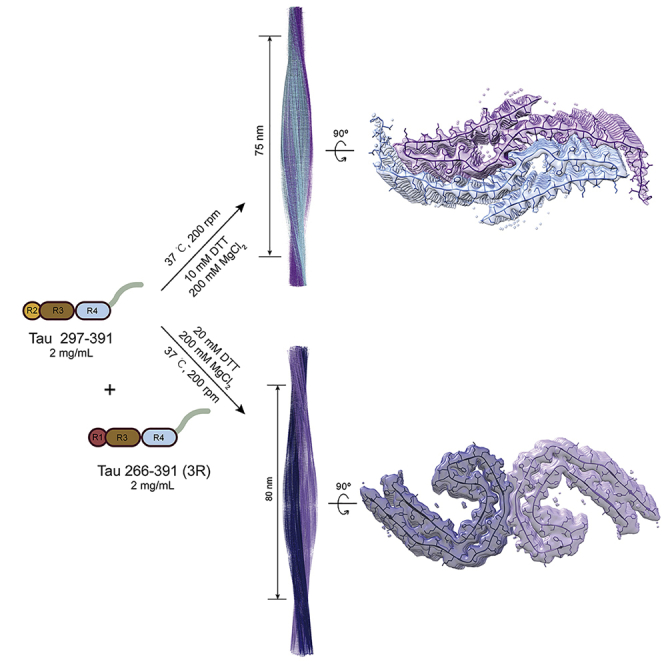
To assemble the PHF-like fibril in vitro, we followed the previously published protocol to the most extent in our lab to prepare Tau 297-391 and Tau 266-391 (3R) proteins25 (Figures 1A and S1; Table S1). We obtained monomeric recombinant Tau 297-391 and Tau 266-391 (3R) with a high purity measured by size exclusion chromatography (SEC), SDS-PAGE, and MALDI-TOF mass spectrometry (Figures S1B, S1D, and S1E). We next assembled Tau fibrils using a mixture of Tau 266-391 (3R) and Tau 297-391 at an equal molar or Tau 297-391 alone in a PB buffer (10 mM phosphate buffer, pH 7.4) with the addition of 10 mM dithiothreitol (DTT) and 200 mM MgCl2. Of note, the addition of MgCl2 into the PB buffer can change the buffer pH to different levels depending on the concentration of MgCl2 (Table S2), which may also play a role in regulating fibril formation. We firmly followed the reported incubation time (48 h) and shaking conditions (orbital shaking using FLUOstar Omega Microplate Reader (BMG LABTECH) at 200 rpm in the Corning 96 Well Black Polystyrene Microplate (Thermo Fisher Scientific)25 (Table S1).
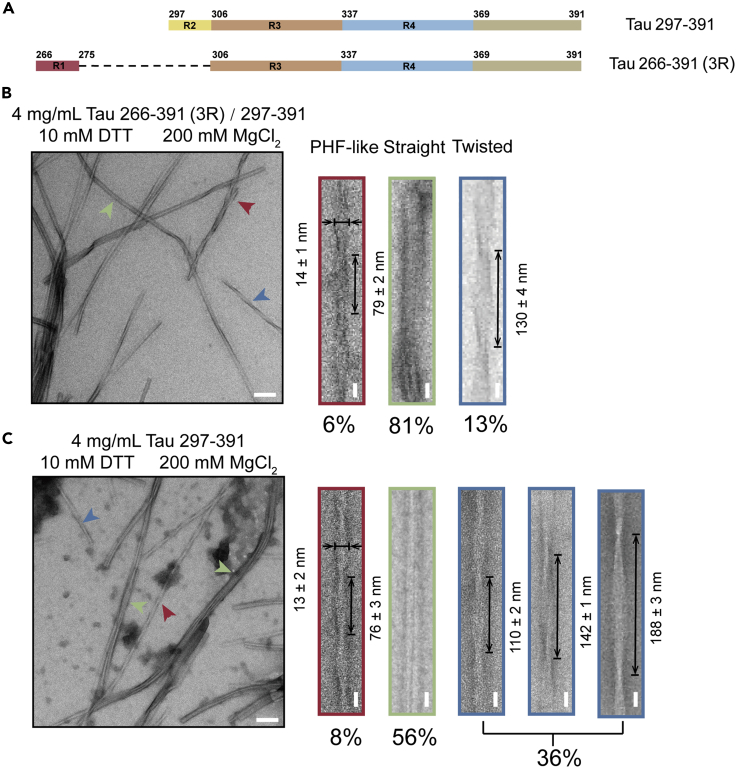
Figure 1.
Characterization of the fibrils formed with Tau 297-391 and 266-391 (3R) under the previously reported fibrillation conditions
(A) Domain organization of the two Tau truncated constructs used in this study. Tau 266-391 excluding R2 only contains R1, R3, and R4, and thus is referred to as Tau 266-391 (3R).
(B and C) TEM micrographs of fibrils formed by 1:1 ratio of Tau 266-391 (3R)/297-391 (B) and Tau 297-391 (C). The fibrillation conditions are indicated. The different morphological fibrils are shown on the right. The half pitch and width of the PHF-like fibril and the half pitch of the twisted fibril are shown. These parameters are shown as mean ± SD with n > 100 for each sample. The percentage of each fibril species that was calculated from NS-TEM images is indicated. The horizontal scale bar represents 100 nm, and the vertical scale bar represents 20 nm.
Next, we examined the fibril morphology by using negative-staining transmission electron microscopy (NS-TEM). Unexpectedly, we obtained highly heterogeneous fibrils. As for fibrils formed by the Tau 266-391 (3R)/297-391 sample, the main proportion (∼81% calculated from NS-TEM images) of the fibrils exhibited a ribbon-like straight morphology (Figure 1B). In the rest twisted fibrils, we observed a small proportion (∼6%) of fibrils featuring a half pitch of 79 ± 2 nm and a width of 14 ± 1 nm (Figure 1B), which is very close to that of the AD PHF (half pitch: 65-80 nm; width: ∼15 nm) (3). The remaining twisted fibrils (∼13%) exhibited an elongated half pitch of ∼130 nm (Figure 1B).
As for fibrils formed by Tau 297-391, the main proportion (∼56%) was also ribbon-like straight fibrils (Figure 1C). In the twisted fibrils, ∼8% fibrils appeared similar to the PHF fibril with a half pitch of 76 ± 3 nm and a width of 13 ± 2 nm (Figure 1C). The remaining twisted fibrils (∼36%) featured a longer half-pitch ranging from ∼110 nm to ∼188 nm (Figure 1C). Taken together, in our hands, both Tau 266-391 (3R)/297-391 and Tau 297-391 formed a mixture of fibrils with distinct morphologies, in which only a small proportion exhibited helical parameters similar to those of the AD PHF fibril.
Optimization of fibrillation conditions to populate the paired helical filament-like fibrils
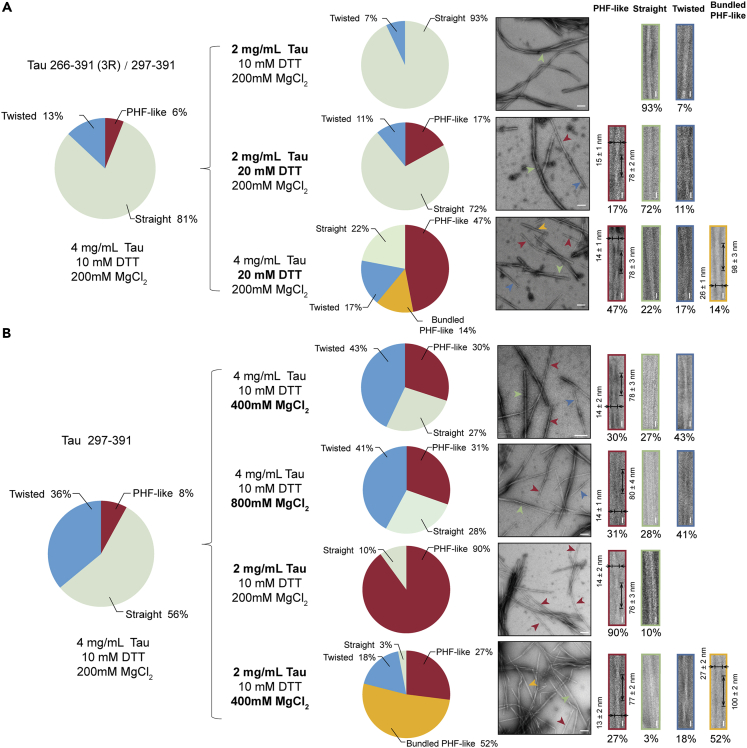
Next, we sought to examine whether we could obtain a higher proportion of PHF-like fibrils by adjusting the fibrillation conditions. As for Tau 266-391 (3R)/297-391, we changed the protein and DTT concentrations, respectively. The result showed that as the DTT concentration increased from 10 mM to 20 mM, Tau 266-391 (3R)/297-391 formed fibrils with the proportion of the PHF-like fibril increasing from ∼6% to ∼47% (Figure 2A). Meanwhile, an additional fibril species (∼14%) appeared with a half pitch of 98 ± 3 nm and a width of 26 ± 1 nm (Figure 2A), which as we later characterized, represents a bundled PHF-like fibril. As for Tau 297-391, the increase of DTT concentration showed no promising effect the population of PHF-like fibril (Figure S2A). We then changed the protein and MgCl2 concentrations, respectively. The result showed that as we decreased the protein concentration from 4 mg/mL to 2 mg/mL, the proportion of PHF-like fibril drastically increased from ∼8% to ∼90% (Figure 2B). While, if the protein concentration was further decreased, fibril formation was inefficient and only short filaments were observed (Figure S2B). In addition, we also tested the influences of solution volumes and shaking conditions, which did not help on the population of PHF-like fibril (Figures S2C and S2D). Collectively, by slightly adjusting the fibrillation condition, the PHF-like fibril can be significantly populated.
Figure 2.
Optimization of fibrillation conditions to populate the PHF-like fibrils
(A and B) The percentage of different morphological fibrils is shown in the pie chart on the left. The NS-TEM images were used to calculate the percentage of each fibril polymorph. The optimized conditions, the pie chart, and the representative TEM images of fibrils formed by Tau 266-391 (3R)/297-391 are shown in (A). Those of the fibrils formed by Tau 297-391 are shown in (B). The half pitch and width of the PHF-like and bundled PHF-like fibrils are shown as mean ± SD with n > 100 for each sample. The horizontal scale bars represent 100 nm, and the vertical scale bars represent 20 nm.
In fact, other fibril species can also be populated under certain conditions. For example, as we decreased the protein concentration of Tau 266-391 (3R)/297-391, the population of straight fibril can be further increased to ∼93% (Figure 2A). As we decreased the concentration of Tau 297-391 and increased the MgCl2 concentration, the bundled PHF-like fibril, which was not observed in the original condition, can be populated to ∼52% (Figure 2B). Taken together, these results indicate that Tau fibril formation is highly sensitive to conditions. Small changes in fibrillation condition may largely tune the population, or even change the species, of the fibrils.
Tau 266-391 (3R)/297-391 fibril recapitulates the Alzheimer’s disease paired helical filament-like fold
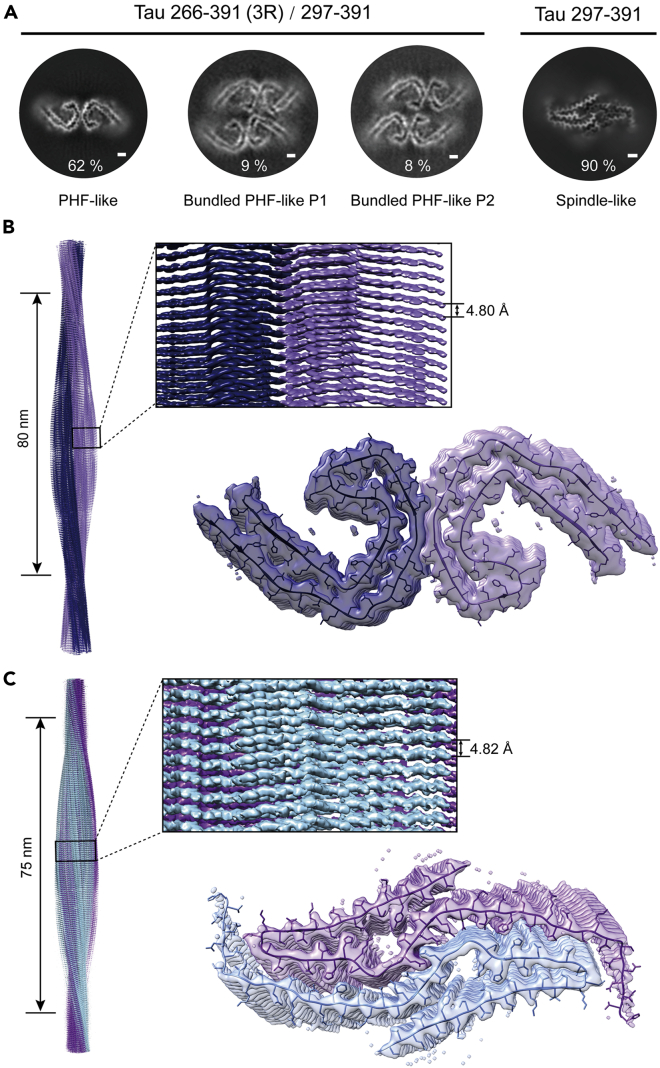
We further used cryo-EM to determine the structures of the in vitro prepared PHF-like fibrils. As for the fibrils formed by Tau 266-391 (3R)/297-391 under the optimized condition (2 mg/mL Tau 266-391 (3R)/297-391 in 10 mM PB buffer, pH 7.4 with the addition of 20 mM DTT and 200 mM MgCl2) (Figure 2A). 5,766 fibrils (straight fibrils were excluded) from 3,732 micrographs were manually selected for the reconstruction of both the PHF-like and the bundled PHF-like fibrils (Table 1). By two-dimensional (2D) classification and three-dimensional (3D) reconstructions, we obtained three different fibril polymorphs including one major specie (the PHF-like fibril) composed of two protofilaments and the other two minor species (the bundled PHF-like fibril) composed of four protofilaments (Figures 3A and S3). Based on the 3D density maps, the structures of Tau monomers in each protofilament of the three polymorphs are similar to each other and to that of the AD fold Tau (Figure 3A).3 The 3D density map of the PHF-like fibril was reconstructed to an overall resolution of 3.46 Å at 0.143 cutoff of Fourier shell correlation (FSC) by performing helical reconstruction using RELION3.126 (Figures 3B and S4), which confirms that the in vitro prepared fibril formed by Tau 266-391 (3R)/297-391 indeed well resembles that of the AD PHF (3). Both the in vitro PHF-like fibril and the AD PHF fibril share the same left-handed twist with a similar fibril pitch (Figure 3B) (3). Meanwhile, the in vitro PHF-like fibril can further pack via different protofilamental interfaces to form two different polymorphs of the bundled PHF-like fibrils (Figure 3A). We next built the structural model based on the cryo-EM density of the PHF-like fibril. The result further confirmed that the dimer structure of Tau from the two neighboring protofilaments of the PHF-like fibril is nearly identical to that of the paired C-shaped fold of Tau in AD PHF with an all-atom RMSD of 0.604 Å (Figure S5).
Table 1.
Statistics of cryo-EM data collection and refinement
| Name | PHF-like fibril formed by Tau 266–391 (3R)/297–391 | Spindle-like fibril formed by Tau 297–391 |
|---|---|---|
| PDB ID | 7YMN | 7YPG |
| EMDB ID | EMD-33934 | EMD-33999 |
| Data collection | ||
| Magnification | 105,000 | 105,000 |
| Pixel size (Å) | 0.83 | 0.83 |
| Defocus Range (μm) | −1.4 to −2.2 | −1.4 to −2.2 |
| Voltage (kV) | 300 | 300 |
| Camera | BioContinuum K3 | BioContinuum K3 |
| Microscope | Krios G4 | Krios G4 |
| Exposure time (s/frame) | 0.05 | 0.05 |
| Number of frames | 40 | 40 |
| Total dose (e−/Å2) | 55 | 55 |
| Reconstruction | ||
| Micrographs | 3,732 | 1,268 |
| Manually picked fibrils | 5,766 | 11,170 |
| Box size (pixel) | 288 | 288 |
| Inter-box distance (Å) | 24 | 24 |
| Segments extracted (no.) | 156,320 | 723,243 |
| Segments after Class2D (no.) | 142,335 | 395,966 |
| Segments after Class3D (no.) | 92,332 | 354,966 |
| Resolution (Å) | 3.46 | 2.54 |
| Map sharpening B-factor (Å2) | −184.211 | −90.56 |
| Helical rise (Å) | 4.80 | 4.82 |
| Helical twist (°) | 179.46 | 179.43 |
| Atomic model | ||
| Non-hydrogen atoms | 3,468 | 2,676 |
| Protein residues | 456 | 354 |
| Ligands | 0 | 0 |
| r.m.s.d. Bond lengths | 0.007 | 0.008 |
| r.m.s.d. Bond angles | 0.722 | 0.793 |
| All-atom clash score | 11.6 | 9.02 |
| Rotamer outliers | 0% | 0% |
| Ramachandran Outliers | 0% | 0% |
| Ramachandran Allowed | 17.57% | 12.28% |
| Ramachandran Favored | 82.43% | 87.72% |
Figure 3.
Cryo-EM structures of the in vitro fibrils formed by Tau 266-391 (3R)/297-391 and Tau 297-391
(A) Central slices of the 3D maps of the in vitro PHF-like and bundled fibrils formed by Tau 266-391 (3R)/297-391 and the spindle-like fibril formed by Tau 297-391. The proportions of each polymorph in the picked fibrils are indicated. Scale bar: 1 nm.
(B and C) Cryo-EM density maps of the PHF-like fibril formed by Tau 266-391 (3R)/297-391 (B) and the spindle-like fibril formed by Tau 297-391. Fibril parameters including half pitch, twist angle, and rise are labeled. Two protofilaments in one fibril are colored differently. Cross-section views of the structural model fitted in the density map are shown.
The “PHF-like” fibril of Tau 297-391 exhibits a distinctive spindle-like fold
We next performed cryo-EM to solve the structure of the seemingly PHF-like fibril formed by Tau 297-391. We prepared Tau 297-391 fibrils under the optimized condition with most populated “PHF-like” fibril (2 mg/mL Tau 297-391 in 10 mM PB buffer, pH 7.4 with the addition of 10 mM DTT and 200 mM MgCl2) (Figure 2B). 11,170 fibrils (straight fibrils were excluded) from 1,268 micrographs were selected for reconstruction (Table 1). Consistent with the NS-TEM result, only one dominant species of the “PHF-like” fibril with a population of about 90% was identified in the 2D classification with an 864-pixel box size (Figure S3). We reconstructed a 3D density map of Tau 297-391 fibril to an overall resolution of 2.54 Å at 0.143 cutoff of Fourier shell correlation (FSC) by performing helical reconstruction using RELION3.126 (Figures 3C and S6). Surprisingly, the 3D map showed a spindle-like structure that is distinctive from either the AD PHF fibril or any known Tau fibrils (Figure 3A). The fibril consists of two intertwining protofilaments featuring a half pitch of ∼75 nm (Figure 3C and Table 1).
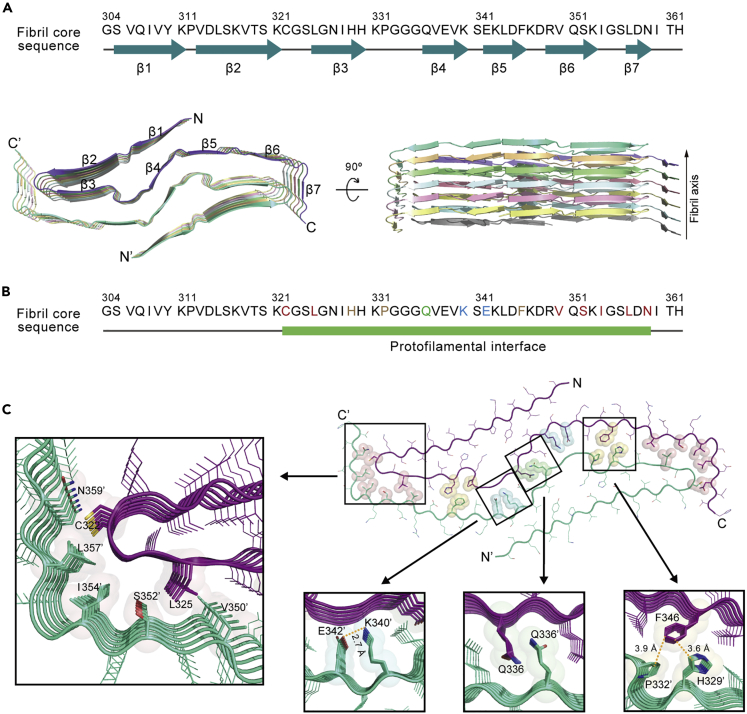
According to the high-quality density map, we were able to unambiguously build an atomic model for the Tau 297-391 spindle-like fibril (Figure 3C). The fibril core is composed of residues 304-362 which forms 7 β-strands (Figure 4A). Within each Tau subunit, β1-β4 constitute a β-hairpin structure, which is followed by an extended conformation of β5-β7 (Figure 4A). Remarkably, the two protofilaments are paired together via an extraordinarily large interface formed by residues 322-359 involving β3-β7 (Figures 4A and 4B). The protofilamental interface is stabilized by a variety of different interactions. The central Q-zipper is formed by the Q336 pair from opposing subunits via van der Waals forces (Figure 4C). K340 and E342 form two salt bridges adjacent to the Q-zipper (Figure 4C). H329, P332, and the opposing F346 interact via π-π stacking (Figure 4C). On each edge of the interface, C322, L325 and the opposing V350, S352, I354, L357, N359 form hydrophobic interactions (Figure 4C). Taken together, our structural study reveals that rather than forming PHF-like fibril, Tau 297-391 actually formed a new Tau fibril polymorph featuring a spindle-like fold of a tight dimer.
Figure 4.
Overall structure and the interface of the Tau 297-391 spindle-like fibril
(A) The primary and secondary sequences of the spindle-like fibril core are shown on the top. The β-strands are indicated below the amino acid sequence. Top and side views of six layers of the spindle-like fibril are shown in cartoon at the bottom. The structure is colored by the chain. The fibril axis is indicated. The β-strands are numbered and labeled accordingly.
(B) The primary sequence of the fibril core with the protofilamental interface highlighted by a green bar. Residues forming the Q-zipper in the interface are green. Residues forming salt bridge are in blue. Residues forming π-π stacking are brown. Residues forming hydrophobic interaction are in red.
(C) Zoom-in views of the interactions formed within the protofilamental interface of the spindle-like fibril structure.
Structural comparison of the spindle-like Tau fibril with known Tau fibril polymorphs
In the comparison of the spindle-like Tau fibril with the structures of Tau fibrils extracted from diseased human brains, we found that although the spindle-like Tau fibril exhibits a distinctive structure, its secondary structure composition is in line with the known polymorphs (Figure S7A). Especially, the topology of β1-β4 of the spindle-like fibril is similar to that of the PiD and CBD (Figure S7B). However, distinctive from the brain-derived fibrils, significantly less residues are involved in the formation of fibril core of the spindle-like fibril. The C-terminal residues 363-380, which commonly form a β-strand in the brain-derived fibrils and involve to compose the fibril core, are flexible and absent in the fibril core of the spindle-like fibril (Figure S7A). Thus, the spindle-like fibril has less intramolecular interactions within Tau monomer. Instead, a remarkably large interface is formed between Tau dimers, which plays an essential role in stabilizing the overall fibril structure as shown in cryo-EM structure and calculated by FoldX27 (Figures 4C and S8; Table S3). In contrast, in the brain-derived Tau fibrils or other in vitro prepared Tau fibrils, interactions between protofilaments are often unnecessary. Taken together, the spindle-like Tau fibril represents a new polymorph of Tau fibril that is distinctive from known polymorphs.
Discussion
Structural polymorphism has been widely observed as a common characteristic of amyloid fibrils.1,8,10,11,28,29,30 Structural polymorphs of Tau and α-syn fibrils are most intensively studied, which demonstrates that even the same protein can form fibrils with distinct structures under different diseased or in vitro conditions.6,8,9,13,16,17,25,31,32 Notably, fibrils prepared in vitro (both Tau and α-syn) feature different structures compared with the ex vivo fibrils purified from the brains of patients.6,12,16 The ex vivo fibrils contain prevalent post-translational modifications (PTMs) and non-covalently bound co-factors in their structures,3,33 absence of which usually generates smaller fibril cores with structural variations.12 On the other hand, in vitro fibril assembly can hardly mimic these chemical modifications, especially as we don’t even know the identities of the co-factors and many of the PTMs. Nevertheless, proteins are the building blocks of amyloid fibrils after all. Is it possible to reproduce the ex vivo fibril structures under certain in vitro conditions? Many efforts have been made toward this aim, and in vitro assembly of Tau fibrils recapitulating the structure of the AD and CTE fibrils have been reported.23,24,25
However, given the high sensitivity of in vitro fibril assembly to conditions, it has been recognized that different labs may obtain different amyloid polymorphs using nominally the same preparation protocols. It seems that slight differences in fibril preparation procedure that usually might not be noticed or regarded as influencing factors in other experiments actually make the discrepancy of in vitro preparation of amyloid fibrils in different laboratories; however, evidence to support this hypothesis is lacking. In this study, we followed the protocol of previous studies to reproduce AD Tau fibrils. Although we tried our best to follow the reported conditions for fibril preparation (Table S1), the reproducibility is still unsatisfying. Our work provides structural evidence to support that in vitro fibril formation is highly sensitive to fibrillation condition and warns that by following fibril preparation protocols, one may not obtain the expected fibrils: neither the purity nor the polymorphs of fibrils. More disturbingly, totally different polymorphic fibrils with undistinguishable appearances by low-resolution microscopic imaging may be generated. Trivial discrepancy during the in vitro preparation of amyloid fibrils among different laboratories may cause significant polymorphism of fibril formation. Therefore, we strongly suggest high-resolution structural characterization for in vitro prepared fibrils prior to application in further investigation.
Limitation of the study
We have listed our experimental conditions for in vitro Tau fibril assembly in detail in Table S1; however, production of the same Tau fibril polymorphs by other researchers following such a condition cannot be fully guaranteed. Exactly as we conclude in this study, subtle changes in conditions would alter fibril conformations. Unfortunately, the effecting factor(s) is unknown and in most cases not even recognized. Therefore, in vitro preparation of specific fibril polymorphs by tuning conditions might not be a robust method, especially for amyloid proteins such as Tau that exhibit high polymorphism.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| BL21 (DE3) E. coli | TransGenBiotech | Cat#CD601-03 |
| Chemicals, peptides, and recombinant proteins | ||
| Tau 297–391 | This study | N/A |
| Tau 266–391(3R) | This study | N/A |
| Deposited data | ||
| PHF-like fibril core, coordinate | This paper | PDB: 7YMN |
| PHF-like fibril core, cryo-EM map | This paper | EMD: 33934 |
| Spindle-like fibril, coordinate | This paper | PDB: 7YPG |
| Spindle-like fibril core, cryo-EM map | This paper | PDB: 33999 |
| PHF fibril from Alzheimer’s disease, coordinate | Shi et al.34 | PDB: 7NRV |
| PHF fibril from Alzheimer’s disease, coordinate | Fitzpatrick et al.3 | PDB: 5O3O |
| Narrow Pick fibril from Pick’s disease, coordinate | Falcon et al.9 | PDB: 6GX5 |
| CTE typeⅡfibril, coordinate | Falcon et al.7 | PDB: 6NWQ |
| CBD typeⅡfibril, coordinate | Zhang et al.31 | PDB: 6TJX |
| See data and code availability section | N/A | N/A |
| Software and algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| GraphPad Prism Software | GraphPad | https://www.graphpad.com/scientificsoftware/prism/ |
| MotionCor2 | Zheng et al.35 | https://emcore.ucsf.edu/ucsf-software |
| CTFFIND | Rohou et al.36 | https://grigoriefflab.umassmed.edu/ctffind4 |
| RELION | Scheres et al.26 | https://www3.mrc-lmb.cam.ac.uk/relion |
| COOT | Emsley et al.37 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PHENIX | Adams et al.38 | https://phenix-online.org/documentation/reference/real_space_refine.html |
| UCSF Chimera | Pettersen et al.39 | https://www.cgl.ucsf.edu/chimera/ |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact Dan Li (lidan2017@sjtu.edu.cn).
Materials availability
Reagents generated in this study will be available upon request.
Method details
Plasmids
The gene encoding Tau 297–391 and Tau 266–391 (3R) were amplified using pRK172 0N4R human tau (gift from Dr. Zhuohao He). Reverse and forward primers were designed to share 15–20 nucleotides of homologous region and 15–30 nucleotides for annealing to the template with melting temperatures ranging from 58°C to 67°C. The resulting construct was verified by DNA sequencing (Azenta, Inc Suzhou, China).
Expression and purification of recombinant truncated Tau
Tau 297–391 and Tau 266–391 (3R) were over-expressed in E. coli BL21 (DE3) cells. Cells were grown in LB medium at 37°C to OD 600 nm of 0.8–1.2. Then, protein over-expression was induced by addition of 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG), and the cells were further incubated at 24°C for 16 hours. Cells were then harvested by centrifugation at 4,000× g for 20 minutes and lysed in 50 mL washing buffer (WB: 50 mM MES at pH 6.0; 10 mM EDTA; 10 mM DTT, supplemented with 0.1 mM PMSF and complete EDTA-free protease cocktail inhibitors, at 10 mL/g of pellet) by a high-pressure homogenizer (800–1,000 bar, 10 min). Lysed cells were centrifuged at 20,000× g for 35 min at 4°C, filtered through 0.45 μm cut-off filters. Tau proteins were purified by using a HighTraP HP SP column (5 mL, GE Healthcare), Fractions were collected and analyzed by SDS-PAGE. Protein-containing fractions were pooled and precipitated using 0.3 g/mL ammonium sulphate and left on a rocker for 30 min at 4°C. Precipitated proteins were then centrifuged at 20,000× g for 35 min at 4°C, and resuspended in 2 mL of 10 mM phosphate buffer (PB), pH 7.2–7.4, with 10 mM DTT, and loaded onto a Superdex 75 gel filtration column (120 mL, GE Healthcare). Protein-containing fractions pooled and concentrated using molecular weight concentrators with a cut-off filter of 3 kDa. Size exclusion fractions were analyzed by SDS-PAGE. Protein concentration was determined by NANODROP 2000c spectrophotometer (Thermo Fisher). The purified proteins were stored in a buffer of 10 mM PB, pH 7.4, with 10 mM DTT, flash frozen in liquid nitrogen, and stocked at −80°C.
Fibril formation
Purified protein samples were thawed on ice and diluted with 10 mM PB and 10 mM DTT to different concentrations (4 mg/mL, 2 mg/mL, 1 mg/mL, respectively). Different concentrations of MgCl2 were added as indicated. Fibrils were assembled in a FLUOstar Omega Microplate Reader (BMG LABTECH) using Corning 96 Well Black Polystyrene Microplate (Thermo Fisher Scientific) with orbital shaking at 37°C for 48 h. The presence of filaments was assessed by using NS-TEM.
Negative-staining transmission electron microscopy (NS-TEM)
After fibril growth, a 5 μL aliquots of fibrillization reactions were adsorbed onto a freshly glow-discharged grid with 200 mesh carbon support film (Beijing Zhongjingkeyi Technology Co., Ltd.) for 45 s. Then, the grid was washed with 5 μL double-distilled water and followed by another wash of 5 μL 3% w/v uranyl acetate. The grid was further stained with 3% (w/v) uranyl acetate for 45 s. After removing the excess buffer by filter paper, the grid was dried by infrared lamp. TEM images were measured on a Tecnai T12 microscope (FEI Company) operated at 120 kV.
As for the measurement of fibril parameters, for each fibril sample, 3 to 4 different areas on the EM grid were used for quantification The dimensions of the fibrils were sampled manually using the built-in measurement tool of ImageJ.40 The fibril was selected and quantified once it can be clearly traced from start to end. Over 100 well-traced fibrils from 9 different TEM images were sampled for each fibril sample formed under different conditions.
Cryo-EM data collection
Samples with confirmed fibrils were centrifuged at 2,000× g for 1 min to remove precipitation components, and a 4 μL aliquot was applied onto a glow-discharged holey copper grid (Quantifoil R2/1, 300 mesh), followed by plunge-frozen in liquid ethane using Vitrobot Mark IV (FEI, Thermo). Cryo-EM micrographs (40 frames per micrograph) were collected on Thermo Fisher Titan Krios G4 cryo transmission electron microscope, operated at 300 kV with a BioContinuum K3 direct detector (Gatan), and a GIF Quantum energy filter (Gatan) that was used with a slit width of 20 eV. All images were automated recorded at a total dose of ∼56 electrons per Å2, using EPU software (Thermo Fisher Scientific).
Imaging processing and helical reconstruction
All 40 frames were aligned, summed and dose-weighted by MotionCorr2 and further binned to a pixel size of 0.83 Å. The defocus values of dose-weighted micrographs were estimated by CTFFIND4.1.836. Helical reconstruction was performed in RELION 3.1. For Tau 266–391 (3R)/297–391 dataset, 5766 fibrils were manually picked from 3732 micrographs. For Tau 297–391 dataset, 11,170 fibrils were picked manually from 1268 micrographs. The extracted segments (box size: 864 pixels, inter-box distance of 71.7 Å) were classified by 2D classification with suitable segments selected for further processing. Initial models were generated de novo from 2D class average images using “relion_helix_inimodel2d” program. Subsequently, 3D classification was used to optimize helical twist and rise, and 3D auto-refinement was used. In order to improve the resolution of reconstruction map, the contrast transfer function (CTF) refinement was performed. Finally, the maps were sharpened with a soft-edge solvent mask using the standard “post-processing” program in RELION 3.1. Overall resolution estimates were calculated based on the gold-standard 0.143 Fourier shell correlation (FSC) between the two independently refined half-maps. Local resolution was estimated using the Local resolution procedure in RELION 3.1 with the same mask and B-factor in post-processing.
Model building and refinement
Based on the density map after post-processing, the atomic models were built and modified by COOT37 using Tau structure (PDB entry code 6GX5) as an initial model. The model with three adjacent layers (six promoters) was refined using the real-space refinement program in PHENIX.38 The subunit in the middle of three layers was extracted and used as the final model.
Protein structure analysis
The PDB files were extended to six layers via UCSF Chimera V1.16.39 Next, we used FoldX “Stability” command to predict the thermodynamic stability of the protein structure.27 The function computes and returns the DG (kcal/mol) of the folding process of the requested protein. The Residue-specific stability was predicted by using “SequenceDetail” command, one layer spindle-like fibril structure PDB files was analyzed.
Quantification and statistical analysis
One-way ANOVA followed by Tukey HSD post hoc test was performed in the GraphPad Prism software following standard procedures.
Acknowledgments
We thank the cryo-EM Microscopy center at the Interdisciplinary Research Center on Biology and Chemistry, Shanghai Institute of Organic Chemistry for help with data collection. This work was supported by the National Natural Science Foundation (NSF) of China (82188101, 32171236, and 31872716 to C.L.; 32170683 to D.L.), the Major State Basic Research Development Program (2019YFE0120600 to C.L.), the Science and Technology Commission of Shanghai Municipality (STCSM) (Grant No. 20XD1425000, 2019SHZDZX02 and 22JC1410400 to C.L.), the Shanghai Pilot Program for Basic Research – Chinese Academy of Science, Shanghai Branch (Grant No. CYJ-SHFY-2022-005 to C.L.) and Shanghai Municipal Commission of Science and Technology Program (21dz2210100 to D.L.).
Author contributions
X.L. and D.L. designed the project. Z.L., S.Z., and X.L. purified recombinant Tau protein and prepared cryo-EM samples. S.Z., X.L., Y.T., W.X., and B.S. collected and processed the cryo-EM data. All the authors are involved in analyzing the structure, and contributed to article editing. X.L., S.Z., C.L., W. L., and D.L. wrote the article.
Declaration of interests
The authors declare no competing interests.
Published: December 22, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105645.
Supplemental information
Data and code availability
-
•
The cryo-EM density map and corresponding atomic model generated in this work have been deposited at the EMDB database and PDB database and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Li D., Liu C. Conformational strains of pathogenic amyloid proteins in neurodegenerative diseases. Nat. Rev. Neurosci. 2022;23:523–534. doi: 10.1038/s41583-022-00603-7. [DOI] [PubMed] [Google Scholar]
- 2.Spillantini M.G., Goedert M. Tau pathology and neurodegeneration. Lancet Neurol. 2013;12:609–622. doi: 10.1016/s1474-4422(13)70090-5. [DOI] [PubMed] [Google Scholar]
- 3.Fitzpatrick A.W.P., Falcon B., He S., Murzin A.G., Murshudov G., Garringer H.J., Crowther R.A., Ghetti B., Goedert M., Scheres S.H.W. Cryo-EM structures of tau filaments from Alzheimer's disease. Nature. 2017;547:185–190. doi: 10.1038/nature23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goedert M. Tau filaments in neurodegenerative diseases. FEBS Lett. 2018;592:2383–2391. doi: 10.1002/1873-3468.13108. [DOI] [PubMed] [Google Scholar]
- 5.Xu H., O'Reilly M., Gibbons G.S., Changolkar L., McBride J.D., Riddle D.M., Zhang B., Stieber A., Nirschl J., Kim S.J., et al. In vitro amplification of pathogenic tau conserves disease-specific bioactive characteristics. Acta Neuropathol. 2021;141:193–215. doi: 10.1007/s00401-020-02253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W., Falcon B., Murzin A.G., Fan J., Crowther R.A., Goedert M., Scheres S.H. Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer's and Pick's diseases. Elife. 2019;8:e43584. doi: 10.7554/eLife.43584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falcon B., Zivanov J., Zhang W., Murzin A.G., Garringer H.J., Vidal R., Crowther R.A., Newell K.L., Ghetti B., Goedert M., Scheres S.H.W. Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature. 2019;568:420–423. doi: 10.1038/s41586-019-1026-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi Y., Zhang W., Yang Y., Murzin A.G., Falcon B., Kotecha A., van Beers M., Tarutani A., Kametani F., Garringer H.J., et al. Structure-based classification of tauopathies. Nature. 2021;598:359–363. doi: 10.1038/s41586-021-03911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falcon B., Zhang W., Murzin A.G., Murshudov G., Garringer H.J., Vidal R., Crowther R.A., Ghetti B., Scheres S.H.W., Goedert M. Structures of filaments from Pick's disease reveal a novel tau protein fold. Nature. 2018;561:137–140. doi: 10.1038/s41586-018-0454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiang W., Yau W.M., Lu J.X., Collinge J., Tycko R. Structural variation in amyloid-β fibrils from Alzheimer's disease clinical subtypes. Nature. 2017;541:217–221. doi: 10.1038/nature20814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riek R., Eisenberg D.S. The activities of amyloids from a structural perspective. Nature. 2016;539:227–235. doi: 10.1038/nature20416. [DOI] [PubMed] [Google Scholar]
- 12.Li D., Liu C. Hierarchical chemical determination of amyloid polymorphs in neurodegenerative disease. Nat. Chem. Biol. 2021;17:237–245. doi: 10.1038/s41589-020-00708-z. [DOI] [PubMed] [Google Scholar]
- 13.Guerrero-Ferreira R., Taylor N.M., Arteni A.A., Kumari P., Mona D., Ringler P., Britschgi M., Lauer M.E., Makky A., Verasdonck J., et al. Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy. Elife. 2019;8:e48907. doi: 10.7554/eLife.48907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Y., Long H., Xia W., Wang K., Zhang X., Sun B., Cao Q., Zhang Y., Dai B., Li D., Liu C. The hereditary mutation G51D unlocks a distinct fibril strain transmissible to wild-type α-synuclein. Nat. Commun. 2021;12:6252. doi: 10.1038/s41467-021-26433-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X., Ma Y., Zhao K., Zhang J., Sun Y., Li Y., Dong X., Hu H., Liu J., Wang J., et al. The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2022933118. e2022933118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Zhao C., Luo F., Liu Z., Gui X., Luo Z., Zhang X., Li D., Liu C., Li X. Amyloid fibril structure of α-synuclein determined by cryo-electron microscopy. Cell Res. 2018;28:897–903. doi: 10.1038/s41422-018-0075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schweighauser M., Shi Y., Tarutani A., Kametani F., Murzin A.G., Ghetti B., Matsubara T., Tomita T., Ando T., Hasegawa K., et al. Structures of α-synuclein filaments from multiple system atrophy. Nature. 2020;585:464–469. doi: 10.1038/s41586-020-2317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lövestam S., Schweighauser M., Matsubara T., Murayama S., Tomita T., Ando T., Hasegawa K., Yoshida M., Tarutani A., Hasegawa M., et al. Seeded assembly in vitro does not replicate the structures of α-synuclein filaments from multiple system atrophy. FEBS Open Bio. 2021;11:999–1013. doi: 10.1002/2211-5463.13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao K., Lim Y.J., Liu Z., Long H., Sun Y., Hu J.J., Zhao C., Tao Y., Zhang X., Li D., et al. Parkinson's disease-related phosphorylation at Tyr39 rearranges α-synuclein amyloid fibril structure revealed by cryo-EM. Proc. Natl. Acad. Sci. USA. 2020;117:20305–20315. doi: 10.1073/pnas.1922741117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Y., Hou S., Zhao K., Long H., Liu Z., Gao J., Zhang Y., Su X.D., Li D., Liu C. Cryo-EM structure of full-length α-synuclein amyloid fibril with Parkinson's disease familial A53T mutation. Cell Res. 2020;30:360–362. doi: 10.1038/s41422-020-0299-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyer D.R., Li B., Sun C., Fan W., Zhou K., Hughes M.P., Sawaya M.R., Jiang L., Eisenberg D.S. The α-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure. Proc. Natl. Acad. Sci. USA. 2020;117:3592–3602. doi: 10.1073/pnas.1917914117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyer D.R., Li B., Sun C., Fan W., Sawaya M.R., Jiang L., Eisenberg D.S. Structures of fibrils formed by α-synuclein hereditary disease mutant H50Q reveal new polymorphs. Nat. Struct. Mol. Biol. 2019;26:1044–1052. doi: 10.1038/s41594-019-0322-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Hilaly Y.K., Pollack S.J., Vadukul D.M., Citossi F., Rickard J.E., Simpson M., Storey J.M.D., Harrington C.R., Wischik C.M., Serpell L.C. Alzheimer's disease-like paired helical filament assembly from truncated Tau protein is independent of disulfide crosslinking. J. Mol. Biol. 2017;429:3650–3665. doi: 10.1016/j.jmb.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Al-Hilaly Y.K., Foster B.E., Biasetti L., Lutter L., Pollack S.J., Rickard J.E., Storey J.M.D., Harrington C.R., Xue W.F., Wischik C.M., Serpell L.C. Tau (297-391) forms filaments that structurally mimic the core of paired helical filaments in Alzheimer's disease brain. FEBS Lett. 2020;594:944–950. doi: 10.1002/1873-3468.13675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lövestam S., Koh F.A., van Knippenberg B., Kotecha A., Murzin A.G., Goedert M., Scheres S.H.W. Assembly of recombinant tau into filaments identical to those of Alzheimer's disease and chronic traumatic encephalopathy. Elife. 2022;11:e76494. doi: 10.7554/eLife.76494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheres S.H.W. Amyloid structure determination in RELION-3.1. Acta Crystallogr. D Struct. Biol. 2020;76:94–101. doi: 10.1107/s2059798319016577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tycko R. Amyloid polymorphism: structural basis and neurobiological relevance. Neuron. 2015;86:632–645. doi: 10.1016/j.neuron.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petkova A.T., Leapman R.D., Guo Z., Yau W.M., Mattson M.P., Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 30.Lu J.X., Qiang W., Yau W.M., Schwieters C.D., Meredith S.C., Tycko R. Molecular structure of β-amyloid fibrils in Alzheimer's disease brain tissue. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W., Tarutani A., Newell K.L., Murzin A.G., Matsubara T., Falcon B., Vidal R., Garringer H.J., Shi Y., Ikeuchi T., et al. Novel tau filament fold in corticobasal degeneration. Nature. 2020;580:283–287. doi: 10.1038/s41586-020-2043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guerrero-Ferreira R., Taylor N.M., Mona D., Ringler P., Lauer M.E., Riek R., Britschgi M., Stahlberg H. Cryo-EM structure of alpha-synuclein fibrils. Elife. 2018;7:e36402. doi: 10.7554/eLife.36402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wesseling H., Mair W., Kumar M., Schlaffner C.N., Tang S., Beerepoot P., Fatou B., Guise A.J., Cheng L., Takeda S., et al. Tau PTM profiles identify patient heterogeneity and stages of Alzheimer's disease. Cell. 2020;183:1699–1713.e13. doi: 10.1016/j.cell.2020.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi Y., Murzin A.G., Falcon B., Epstein A., Machin J., Tempest P., Newell K.L., Vidal R., Garringer H.J., Sahara N., et al. Cryo-EM structures of tau filaments from Alzheimer's disease with PET ligand APN-1607. Acta Neuropathol. 2021;141:697–708. doi: 10.1007/s00401-021-02294-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohou A., Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/s0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/s0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 40.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The cryo-EM density map and corresponding atomic model generated in this work have been deposited at the EMDB database and PDB database and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.