Abstract
A comparative analysis of the predicted protein sequences encoded in the complete genomes of Borrelia burgdorferi and Treponema pallidum provides a number of insights into evolutionary trends and adaptive strategies of the two spirochetes. A measure of orthologous relationships between gene sets, termed the orthology coefficient (OC), was developed. The overall OC value for the gene sets of the two spirochetes is about 0.43, which means that less than one-half of the genes show readily detectable orthologous relationships. This emphasizes significant divergence between the two spirochetes, apparently driven by different biological niches. Different functional categories of proteins as well as different protein families show a broad distribution of OC values, from near 1 (a perfect, one-to-one correspondence) to near 0. The proteins involved in core biological functions, such as genome replication and expression, typically show high OC values. In contrast, marked variability is seen among proteins that are involved in specific processes, such as nutrient transport, metabolism, gene-specific transcription regulation, signal transduction, and host response. Differences in the gene complements encoded in the two spirochete genomes suggest active adaptive evolution for their distinct niches. Comparative analysis of the spirochete genomes produced evidence of gene exchanges with other bacteria, archaea, and eukaryotic hosts that seem to have occurred at different points in the evolution of the spirochetes. Examples are presented of the use of sequence profile analysis to predict proteins that are likely to play a role in pathogenesis, including secreted proteins that contain specific protein-protein interaction domains, such as von Willebrand A, YWTD, TPR, and PR1, some of which hitherto have been reported only in eukaryotes. We tentatively reconstruct the likely evolutionary process that has led to the divergence of the two spirochete lineages; this reconstruction seems to point to an ancestral state resembling the symbiotic spirochetes found in insect guts.
Comparative analysis of the protein products encoded in complete genomes provides a powerful tool for evaluating evolutionary trends and adaptive strategies in pathogenic microbes. Identification of metabolic and signaling pathways that are unique to pathogens offers novel targets for therapeutic intervention. Also, such unique determinants may prove useful in developing better diagnostic and prognostic indicators of infectious diseases in humans. Conclusions from such an analysis enhance understanding of the host-parasite interactions that enable pathogens to carve out unique ecological niches in nature.
Borrelia burgdorferi is the causative agent of Lyme disease, whereas the related spirochete Treponema pallidum causes syphilis. Both organisms are fastidious in their growth requirements, have small genomes, and manifest clinically as distinct, chronic, disseminated diseases. The clinical similarities (68) include progression of disease in stages following local inoculation either through a tick in Lyme disease or through sexual contact in syphilis. Involvement of the central nervous system is seen in both infections, with sequalae ranging from neurologic deficits to neuropsychiatric abnormalities (19). Chronic, recurrent joint involvement is classically associated with Lyme disease (64). The protean clinical manifestations of these pathogens and their propensity to cause chronic infection in the human host contribute to the ongoing diagnostic and therapeutic challenges offered by these spirochetes (22, 42).
The recent determinations of the complete genome sequences of B. burgdorferi (30) and T. pallidum (31) have made a comprehensive computer-based comparison of the two spirochete genomes a feasible exercise. Recent comparative genomic studies include comparisons between two closely related mycoplasma species (36) and between bacteria that belong to related genera but encode vastly different numbers of proteins, namely, Escherichia coli and Haemophilus influenzae (72). The complete genomes of T. pallidum and B. burgdorferi give us the first chance to compare two moderately related pathogenic bacteria with approximately equivalent coding capacities. In order to systematically compare the protein sets encoded in the two spirochete genomes, we devised a measure for evaluating evolutionary relationships between families and functional classes of proteins: the orthology coefficient. With regard to the two spirochetes, it seemed likely that differences observed across families and functional groups of proteins could be indicative of the distinct evolutionary pressures of the host environment.
Completely automated computational analysis of genome sequences is often plagued by errors arising from compositional bias in protein sequences, difficulties in automatic assessment of the domain organization of proteins, and the lack of in-depth predictive power (16). Therefore, in comparative analysis of the two spirochete genomes we used a semiautomated strategy that is based on recently developed, powerful tools for sequence analysis, particularly position-specific iterating BLAST (PSI-BLAST) (2, 3), and also involves case-by-case examination of protein families and individual protein sequences. Optimization of the functional annotation of proteins accompanied by reconstruction of metabolic and signaling pathways in the two spirochetes helped uncover a number of unique features that likely play a role in host response-related functions. In this report, we compare and contrast the gene complements of the two spirochetes and provide examples of shared and unique genes and functional systems.
MATERIALS AND METHODS
Databases.
The complete protein sets of B. burgdorferi (30) and T. pallidum (31) were obtained from the genome division of the Entrez retrieval system at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Genome. All database searches were carried out against the nonredundant (NR) sequence database maintained at the National Center for Biotechnology Information (National Institutes of Health, Bethesda, Md.).
Sequence analysis: detection and interpretation of varying levels of sequence similarity.
Large-scale sequence analysis was handled using the SEALS program package (76). The first step of the analysis involved a search of the NR database of protein sequences at the National Center for Biotechnology Information using the gapped BLAST program (3), with the complete protein sets of B. burgdorferi and T. pallidum as queries. Compositionally biased regions in query sequences that tend to produce spurious hits in database searches were masked using the SEG program (81). Two different sets of parameters for SEG were used, namely, mild masking (window length, 12; trigger complexity, 2.2; extension complexity, 2.5) for routine searches and stringent masking (window length, 45; trigger complexity, 3.4; extension complexity, 3.75) for delineating particular globular domains (81). In-depth database searches were performed using the PSI-BLAST program (3) whenever specific annotation required detection of subtle relationships. Briefly, PSI-BLAST uses multiple alignments of sequences that show expectation values, or e-values, lower than a specified cutoff (in this case 0.01) in the first-pass gapped BLAST search to construct a position-specific scoring matrix (PSSM). The PSSM is then used as the input for further iterations of the database searches, which results in increased detection of subtle sequence similarities (2, 8). Statistical evaluation of the PSI-BLAST result is based on the empirical version of Karlin-Altschul statistics, modified for gapped alignments (3). The e-value indicates the number of sequences which show a certain level of similarity by chance alone for a database of the size equivalent to NR. The e-value reported on the first instance that PSI-BLAST detects the given sequence above the cutoff is a reliable indicator of statistical significance. Under the condition that properly filtered globular domains are used as queries, an e-value of less than 0.01 is generally suggestive of homology (8).
Structural features of proteins were analyzed using the FAMASK program of the SEALS package (76), which incorporates several prediction methods. Signal peptides were predicted using the SIGNALP program (49), transmembrane helices were predicted using the PHD topology program (57) or the TopPred II program (20), and coiled-coil regions were predicted using the COILS2 program with a window length of 21 (43).
The phyletic distribution of homologous proteins detected by gapped BLAST searches was evaluated using the TAX_COLLECTOR program (76) of SEALS. The hits with an e-value below 0.001 between each protein encoded in the two spirochete genomes and the NR databases were analyzed, and the best hits in the second spirochete as well as those in other bacteria, archaea, and eukarya, with their respective e-values, were tallied. This provided the starting point for identifying orthologous genes in the two genomes as well as a preliminary indication of the phyletic distribution of homologs for most of the proteins encoded in each genome. An attempt was made to delineate true orthologous relationships on the basis of symmetric best hits between the proteins from the two spirochetes as well as between the spirochete proteins and those from other bacteria, archaea, and eukaryotes. In problematic cases, phylogenetic tree analysis using multiple sequence alignments of conserved domains was carried out in order to evaluate the relationships. Phylogenetic trees were constructed using the PAUP3 or CLUSTALW programs (35) with the neighbor-joining and least squares (Fitch-Margoliash) methods, accompanied by bootstrap analysis (27).
A complementary approach was employed for case-by-case analysis of those spirochete proteins that failed to show statistically significant matches when used as queries for PSI-BLAST. These sequences were screened for the presence of known conserved domains using the previously developed libraries of PSSMs (18, 53) as well as newly developed PSSMs. Domain-family PSSMs were created by running PSI-BLAST against the NR database with the appropriate query and an e-value cutoff of 0.01 and saving the profile using the “−C” option. The resulting PSSM was rerun against the protein sequence sets of T. pallidum and B. burgdorferi using the −R option of PSI-BLAST, and hits with significant scores were detected. To test the robustness of this procedure, we prepared databases using randomized protein sequences from each of the spirochete proteomes and conducted similar searches with each of the PSSMs on them. No hits with significant e-values (<0.01) were detected in these searches, supporting the validity of the domains detected in the spirochete proteomes with the PSSMs. Additional database searches were performed with family-specific Hidden Markov models that were constructed and run using the HMMER2 program package (66). Multiple alignments of protein families were constructed using a combination of the −m4 option of PSI-BLAST and the CLUSTALW program (35).
RESULTS AND DISCUSSION
Orthology coefficient: functional classes and protein families.
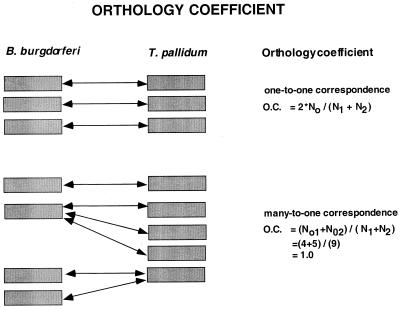
The proteins encoded by the genomes can be broadly categorized on the basis of either (predicted) cellular function (functional class) or shared conserved domains (protein families). Orthologs are genes (proteins) in different genomes that are related by vertical descent; i.e., they can be traced back to a common ancestor. Such orthologous proteins typically have similar, if not identical, biological functions in the respective organisms (71). Paralogs, in contrast, are genes related by duplication within a lineage (genome); paralogs are expected to possess generally similar biochemical functions but distinct biological roles. Given these differences in functional implications, careful delineation of orthologous and paralogous relationships between genes is of primary importance for predicting functions of proteins with an appropriate degree of precision. We adopted a quantitative measure of orthology that we termed the orthology coefficient (OC). This index can be applied to different protein categories, namely, superfamilies related by descent as opposed to functional classes of proteins, such as those involved in transcription or replication. In the simplest case of a one-to-one correspondence between genes in two genomes, OC = 2No/(N1 + N2) where No is the number of orthologs and N1 and N2 are the numbers of members of the given protein family or functional category in the two compared genomes (Fig. 1). If the gene complements in the two genomes are completely conserved, then the OC is 1.00; by contrast, in the hypothetical case where there are no orthologs, the OC is 0. If there is one or more duplications in one or both of the species that occurred after their divergence, then OC = (No1 + No2)/(N1 + N2) where No1 and No2 are the numbers of members in orthologous clusters from the two respective genomes (Fig. 1). Criteria used to identify orthologs included symmetry of retrieval in reciprocal BLAST searches, conservation of the domain organization of proteins, and, in problematic cases, phylogenetic tree analysis (71, 72).
FIG. 1.
OC as a quantitative measure of orthology between different types of protein categories. (A) In the case of a one-to-one correspondence between genes in two genomes, OC = 2No/(N1 + N2) where No is the number of orthologs and N1 and N2 are the numbers of members of the given protein family or functional category in the two compared genomes. (B) If there is a duplication (two or more members) in one or both of the species that occurred after divergence of the two species, then OC = (No1 + No2)/(N1 + N2) where No1 and No2 are the numbers of members in orthologous clusters from the two respective genomes.
Conservation and diversity of functional classes of proteins between the two spirochetes.
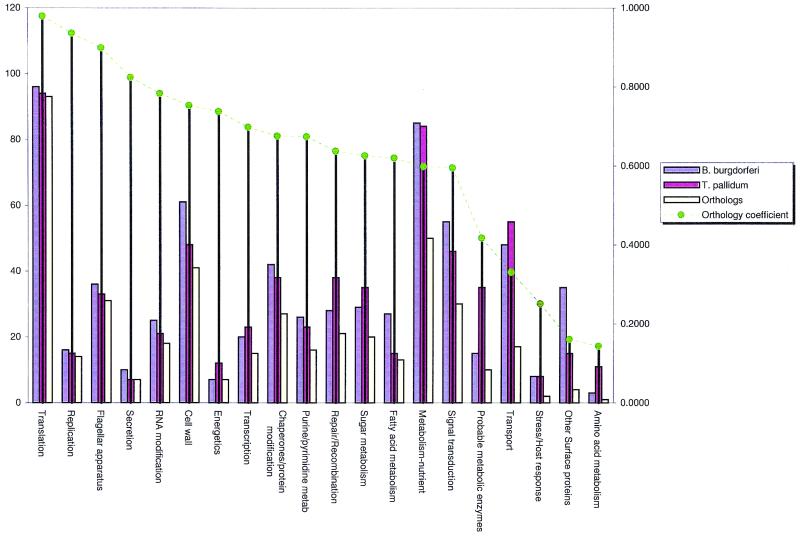
Functional classes of proteins include core functions, such as replication and cell division, repair, transcription, and translation, that are shared by all bacteria and to a large extent also by archaea and eukaryotes, and more specialized functions, such as metabolism, metabolite transport, host and environment response, and signal transduction. Figure 2 shows the representation of the functional classes in the two genomes and the level of orthology in each class measured using OC. In order to assess the effects of specific adaptations of each of the spirochetes to their distinct niches, investigation of classes with low OC values appears to be critical. Below, we discuss the distribution of functional classes in the two spirochetes along the gradient of OC values. A selection of prominent differences in the functional systems of the two spirochetes is shown in Table 1, and examples of apparent spirochete synapomorphies are listed in Table 2.
FIG. 2.
OCs for functional classes of proteins. The annotated predicted open reading frames from the proteomes of B. burgdorferi and T. pallidum were grouped by the predicted functional class of the protein. Orthologous proteins within each functional class were identified as described, and the OC values for each class were calculated as in Fig. 1. The vertical axis on the left shows the absolute number of proteins, and the one on the right shows OC values. OCs of >0.8 delineate highly conserved protein classes, OCs in the range of 0.6 to 0.8 are considered intermediate, and OCs of <0.6 represent significant divergence between the two genomes.
TABLE 1.
Selected prominent differences in the functional systems of the two spirochetesa
| B. burgdorferi | T. pallidum |
|---|---|
| Conserved systems: replication, RNA processing, and translation | |
| BB0035, BB0036; topoisomerase IV | — |
| BB0826; topo I, related to actinomycetes and cyanobacteria | TP0394; no specific affinities of Topo I to any other bacterial lineage |
| — | TP0643; DnaQ (ɛ subunit of DNA polymerase) |
| BB0552; DNA ligase with one BRCT domain | TP0634; DNA ligase with three BRCT domains |
| BB0114; one copy of single-strand binding protein | TP0062, TP0310; two copies of single-strand binding protein |
| BB0402; eukaryote-type prolyl-tRNA synthetase | TP0160; bacterial-type prolyl-tRNA synthetase |
| — | TP0559; ThiI tRNA-thiouridine biosynthesis enzyme |
| — | TP0924; Tex/SPT6 family RNA processing protein |
| Moderately conserved systems: DNA repair and transcription | |
| BB0633, BB0634, BB0632; RecB, RecC, and RecD; components of the helicase-nuclease complex involved in recombinational repair | — |
| — | TP0003, TP0442, TP0103, TP1004; RecF, RecN, RecQ, and RecR components of DNA recombination and repair |
| BB0023, BB0022; RuvA and RuvB subunits of the Holiday junction resolvase; no nuclease RuvC | TP0543, TP0162, TP0517; RuvA, RuvB, and RuvC present |
| — | TP0380; ERCC3-like DNA repair helicase |
| BB0626; small primase-like TOPRIM domain protein | — |
| BB0098, BB0797; two versions of MutS mismatch repair ATPase | TP0328; single version of MutS |
| BB0771; unique sigma factors RpoS | TP0092, TP0709, TP1012; Unique sigma factors RpoE (ς24), SigA (ς28, ς43) |
| — | TP0220, TP0233, TP0540; anti-sigma factors (RsbV) |
| — | TP0218, TP0219, TP0854; PP2C phosphatases involved in anti-sigma factor regulation |
| BBD22; MetJ/Arc family transcription regulator | — |
| BB0647; iron-related transcriptional regulation; Fur | TP0167; iron-related transcriptional regulation; TroR |
| BB0693, BB0831; XylR-type HTH + sugar kinase transcription regulators | — |
| Poorly conserved systems: metabolism, nutrient uptake, and protein modification | |
| BB0445; γ-proteobacterial-type class II aldolase | TP0662; class II aldolase shows no specific affinities to any bacterial lineage |
| BB0057; bacterial-type glyceraldehyde 3-phosphate dehydrogenase | TP0844; glyceraldehyde 3-phosphate dehydrogenase specifically related to orthologs from kinetoplastid flagellates |
| BB0015; uridine kinase; standard bacterial version | TP0667; uridine kinase with two N-terminal (probable RNA-binding) domains shared with threonyl-tRNA synthetases |
| BB0243; eukaryote-type FAD-dependent glycerol-3-phosphate dehydrogenase | — |
| BB0683, BB0684, BB0685, BB0686, BB0687, BB0688; eukaryote-type isopentenyl pyrophosphate biosynthesis operon | — |
| BB0241; glycerol kinase | — |
| — | TP0754, TP0269, TP0121, TP0068; two biotin synthase family 3 cysteine cluster enzymes |
| — | TP0053, TP1008; ribonucleotide reductase α/β subunits |
| BB0144; glycine-betaine transport system, possibly involved in osmoregulation | TP0350, TP0351, TP0797; intact proline biosynthesis operon, perhaps involved in osmoregulation |
| BB0216, BB0215, BB0042; typical bacterial phosphate transport operon including regulator PhoU | — |
| — | TP0163, TP0165, TP0166; Tro iron transport operon |
| BB0610; one FKBP-type peptidyl prolyl isomerase (Trigger factor ortholog) | TP0506, TP0349, TP0862; three distinct FKBP-like peptidyl prolyl isomerases (including the Trigger factor ortholog) |
| BB0655, BB0602, BB0517; three J-domain proteins including DnaJ ortholog | TP0098, TP0563; No DnaJ ortholog but two distinct J-domain proteins without Borrelia orthologs |
| — | TP0947; cyclophilin-like peptidyl prolyl isomerase |
| BB0296, BB0295, BB0834, BB0757, BB0612; both HlsU/HlsV and ClpP (two copies)/ClpX ATPase protease pairs present | TP0071, TP1041, TP0508, TP0507; only ClpP (two copies)/ClpX present; additionally, a SohB-like protease distantly related to ClpP present |
| — | TP0100, TP0101; thiol:disulfide interchange operon (DsbD, DsbE) |
| BB0253, BB0613; two distinct, typical ATP-dependent Lon proteases | TP0524, TP0016; typical bacterial Lon protease ortholog and archaeal-type Lon family protease-ATPase |
| — | TP0835, TP0502; two ankyrin repeat proteins (possible horizontal transfer from eukaryotes) |
| BB0030, BB0031, BB0263; three distinct signal peptidases | TP0926, TP0185; two distinct signal peptidases with only one orthologous pair |
—, absence in a given organism. Gene identification number (BB or TP) is shown in the order of the proteins described for each category.
TABLE 2.
Likely spirochete synapomorphies
| Character | Comments | |
|---|---|---|
| BB0254/TP0704; RecJ single-strand nuclease protein; ∼100-amino-acid insert | Occurs just after the DHH motif of these proteins and is absent in all other bacterial RecJ proteins | |
| BB0800/TP0892; NusA zinc ribbon at the extreme C terminus | Other bacteria lack this domain (some, e.g., E. coli, have in its place a duplicated modified HhH domain) | |
| BB0132/TP0018; GreA; has a long, unique N-terminal extension | The only other bacterial genus with this extension is Chlamydia | |
| BB0512/TP0408; large protein with a signal peptide-like sequence and C-terminal HTH domain | These proteins are so far seen only in spirochetes, although smaller paralogs without the large coiled-coil region are seen in spirochetes and some other bacteria (Fig. 3b) | |
| BB0827/TP0526; plant-type HrpA helicase | Present in many bacteria and eukaryotes, but the spirochete version is closer to eukaryotic forms | |
| BB0659/TP0644; archaeal-type lysyl-tRNA synthetase | Appears to have entered the common ancestor of these spirochetes from an archaeal lineage; among other bacteria, seen only in Rickettsia | |
| BB0513, BB0514/TP0973, TP0015; archaeal-type phenylalanyl-tRNA synthetase (subunits α and β) | Appears to have entered the common ancestor of these spirochetes from an archaeal lineage | |
| BB0819/TP0342; archaeal-type cytidylate kinase | Appears to have entered the common ancestor of these spirochetes from an archaeal lineage; shares a modified P-loop with the archaeal proteins | |
| BB0020/TP0542; eukaryote-type pyrophosphate-dependent phosphofructokinase | The spirochetes are the only bacteria with this form of PFK that is otherwise seen in plants and protists | |
| BB0670/TP0364; signaling protein that contains an N-terminal CheW domain and a C-terminal protein methyltransferase domain (Fig. 3). | Domain organization unique to spirochetes | |
| BB0236/TP0421; YWTD domain proteins | Apparent horizontal transfer from animals | |
| BB0063/TP0307; membrane protein with a fusion to penicillin-binding domains similar to those found in PKN2-like protein kinases | Domain organization unique to spirochetes | |
| BB0286/TP0567; small flagellar operon protein B | Protein unique to spirochetes | |
| BB0282/TP0726; small flagellar operon protein D | Among other bacteria, only Thermotoga encodes an ortholog of this protein |
Highly conserved functional classes with OCs close to 1.
Not surprisingly, the core functional classes, namely, DNA replication and translation, had OCs of 0.94 and 0.98, respectively, which is indicative of largely vertical inheritance (Fig. 2). The limited differences observed in these conserved functional classes could be attributed to interesting cases of horizontal gene transfer, gene loss, or duplication with divergence.
Differences in the gene complement involved in DNA replication include the lack of topoisomerase IV (parC, parE) in T. pallidum, which is of interest given the conserved role of these proteins in chromosome partitioning in other bacteria (77). Conversely, the proofreading 3′-5′ exonuclease—the ɛ subunit of DNA polymerase III (e.g., the E. coli DnaQ protein)—whose role in DNA replication seems to be conserved in all other bacteria (29), is missing in B. burgdorferi. The absence of this enzyme would result in error-prone replication, which seems to be compatible with the generation of new variants through mutation in the genes encoding variable antigens of Borrelia (58, 84). An alternative is nonorthologous displacement (E. V. Koonin, A. R. Mushegian, and P. Bork, Letter, Trends Genet. 12:334–336, 1996), that is, an unrelated or distantly related and so far unidentified exonuclease being recruited for the proofreading role. Interestingly, T. pallidum encodes a second copy of the single-stranded DNA-binding protein (TP0310), which might have a distinct function in replication or recombination.
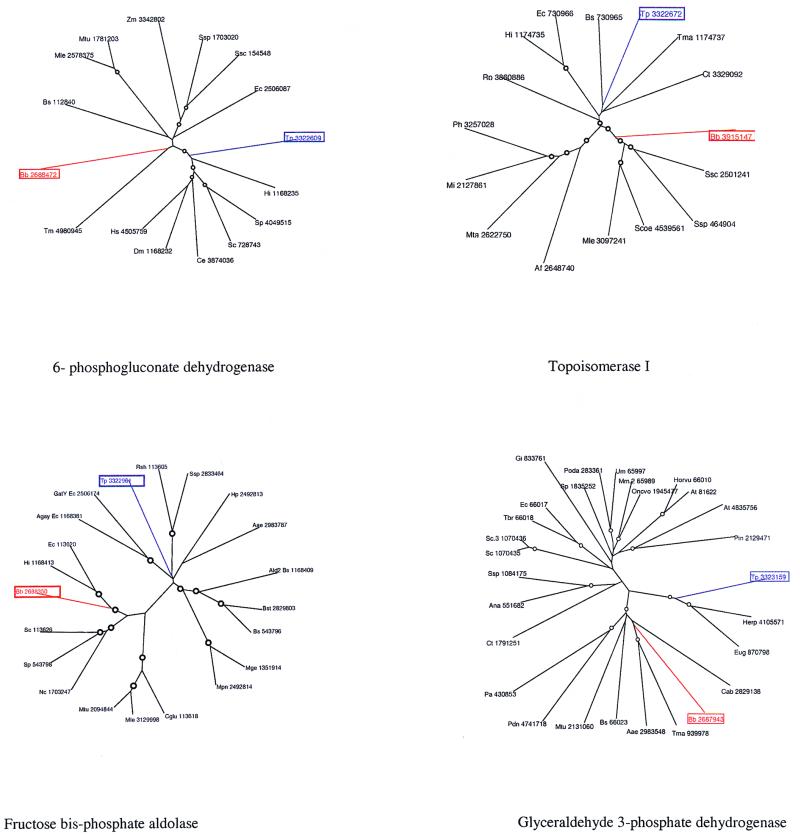
In addition to these striking differences, we detected an interesting case of apparent displacement of an existing gene by its ortholog from a distant species, probably via horizontal gene transfer. Sequence searches showed that topoisomerase I (Topo I) from B. burgdorferi shares an insert within the TOPRIM domain (10) and a C-terminal repeat domain with actinomycetes and cyanobacteria. These features are lacking in other bacterial lineages and appear to be derived, shared characters (synapomorphies) within this protein superfamily. Phylogenetic analysis using multiple alignment of the conserved region of bacterial Topo I strongly supported grouping of B. burgdorferi with actinomycetes and cyanobacteria (Fig. 3 [bootstrap value, >90%]). This phylogenetic affinity of Topo I is in stark contrast to the typical clustering of the other spirochete proteins, particularly those from the core functional classes, and suggests horizontal gene transfer into the B. burgdorferi lineage followed by loss of the ancestral form. This phenomenon, which may be termed xenologous gene displacement, that is, displacement by an orthologous gene from a phylogenetically distant source (J. P. Gogarten, Letter and comment, J. Mol. Evol. 39:541–543, 1994), is seen also in several other instances in the two spirochetes (see below).
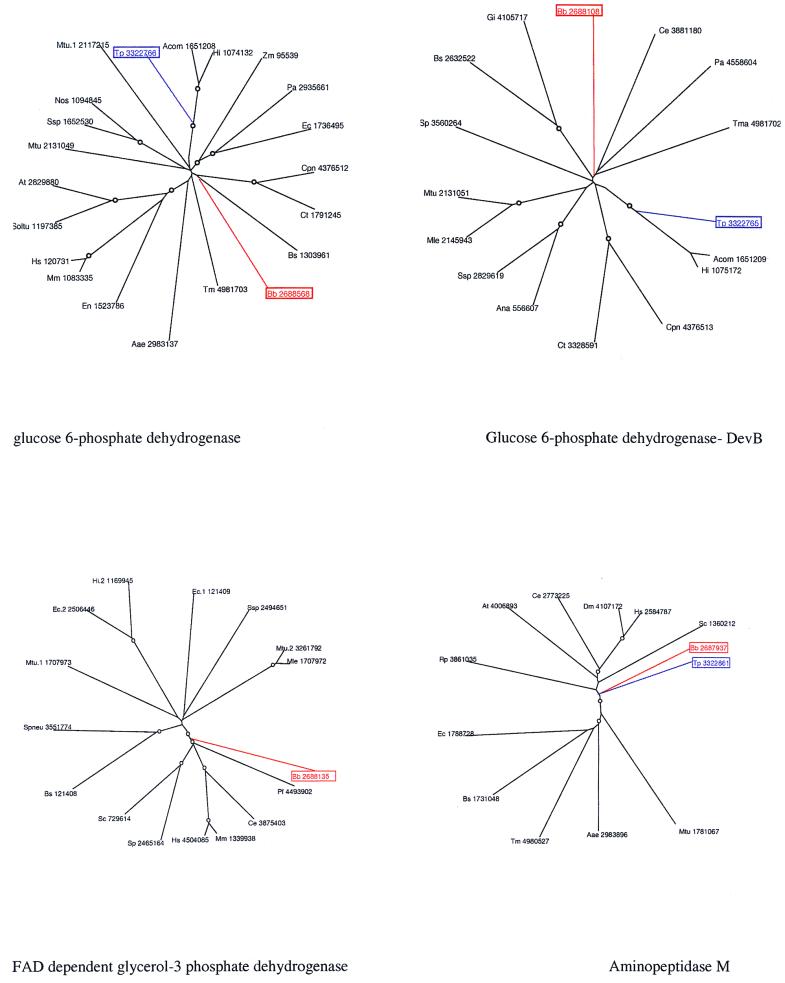
FIG. 3.
Phylogenetic trees for representative examples of xenologous gene displacement and horizontal gene transfer in the spirochetes. The trees were constructed using alignments generated with CLUSTALW, followed by manual adjustments based on PSI-BLAST outputs. The neighbor-joining (NEIGHBOR program from Phylip) and least squares (from the PAUP package) methods were used to construct the trees, and 1,000 bootstrap replications were performed. The nodes supported by bootstrap values greater than 75% are indicated with the symbol “○.” All genes are shown with the mnemonic species name along with the GenBank accession number. Species abbreviations are as follows: Tp, T. pallidum; Bb, B. burgdorferi; Bs, B. subtilis; Ec, E. coli; Hi, H. influenzae; Pa, Pseudomonas aeruginosa; Pdn, Pseudomonas denitrificans; Rp, R. prowazekii; Zm, Zymomonas mobilis; Acom, Actinobacillus actinomycetemcomitans; Mtu, Mycobacterium tuberculosis; Mle, Mycobacterium leprae; Ssp, Synechocystis sp.; Ssc, Synechococcus sp.; Ana, Anabaena sp.; Nos, Nostoc sp.; Cpn, Chlamydia pneumoniae; Ct, Chlamydia trachomatis; Tm, T. maritima; Aae, Aquifex aeolicus; Ap, Aeropyrum pernix; Af, Archaeoglobus fulgidus; Mta, Methanobacterium thermoautotrophicum; Mj, Methanococcus jannaschii; Ph, Pyrococcus horikoshi; Um, Ustilago maydis; Poda, Podospora anserina; Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae; Ce, Caenorhabditis elegans; Hs, Homo sapiens; Dm, Drosophila melanogaster; Mm, Mus musculus; Oncvo, Onchocerca volvulus; At, Arabidopsis thaliana; Horvu, Hordeum vulgare; Soltu, Solanum tuberosum; Gi, Giardia intestinalis; Tbr, Trypanosoma brucei; En, Entamoeba histolytica.
In spite of the high level of conservation in the translation apparatus, we did detect a few interesting examples of evolutionary divergence between the two species. One notable case was the prolyl-tRNA synthetase from B. burgdorferi, where phylogenetic analysis strongly suggested that the enzyme in B. burgdorferi has been acquired from a eukaryotic source by the xenologous displacement mechanism (78). The other aminoacyl-tRNA synthetases are strongly conserved between the spirochetes, but phylogenetic analysis provided evidence for likely ancient horizontal transfers into the ancestor of spirochetes accompanied by displacement of the original gene. One well-studied case in this category is class I lysyl-tRNA synthetase, which apparently was acquired from the archaea (37, 78); similar evidence of archaeal origin was obtained for the two subunits of phenylalanyl-tRNA synthetase (78). By contrast, the synthetases for arginine, glutamate, methionine, isoleucine, and serine showed evidence of likely horizontal transfer from eukaryotic sources (78).
Functional classes with intermediate OCs (0.6 to 0.8).
The gene complements involved in repair, recombination, and transcription show significant differences between the two spirochetes, which is reflected in OC values of less than 0.8 (Fig. 2). A number of important repair genes are different in the two genomes. Thus, B. burgdorferi encodes the helicase-nuclease complex RecBCD, whereas T. pallidum lacks these genes but has instead a complement of recFGNR genes. Another repair enzyme unique to T. pallidum is the ERCC3-like eukaryotic-type DNA repair helicase that is also present in mycobacteria (54). It is likely that this gene was horizontally transferred from eukaryotes into a certain bacterial lineage and subsequently disseminated amidst the bacteria. The triplication of the C-terminal BRCT domain of DNA ligase in T. pallidum is a unique feature that is in contrast to the similarly located single copy of this domain present in all other bacteria (7, 15). The RecQ helicase gene is found in T. pallidum but not in B. burgdorferi. Conversely, B. burgdorferi but not T. pallidum retains the ancient duplication of the mutS gene, which is also seen in other bacteria. Interestingly, B. burgdorferi lacks RuvC, the endonuclease subunit of the Holliday junction resolvase complex, which suggests either that the Ruv complex is nonfunctional or that a distinct nuclease complements this function. Both spirochetes possess a novel mismatched-base DNA glycosylase (TP0229/BB0013), a base excision repair enzyme, orthologs of which are conserved in a number of bacteria and in all archaea (60), but the B. burgdorferi version is highly divergent from all other bacterial orthologs. The major disparities in the repertoires of repair proteins suggest a difference in the selective forces that act on this system in the two spirochetes. The fact that B. burgdorferi undergoes antigenic variation (58, 84) that may require both active recombination and error-prone repair provides a possible explanation for the retention of only certain of its repair pathways.
The intermediate OC of the transcriptional apparatus seems to reflect a bimodal effect of selective evolutionary forces. The core transcriptional machinery, which includes RNA polymerase subunits and components of the elongation and termination complexes, is highly conserved, whereas the repertoires of specific transcriptional regulators are largely different (Fig. 2). Within the shared heritage, there are some unique features that are likely to have been derived in the common ancestor of these spirochetes and may be considered signatures of this clade. In particular, the spirochete NusA protein (a part of the transcription termination complex) contains a C-terminal Zn ribbon domain in place of the helix-hairpin-helix (HhH) domain that is found in most other bacteria (data not shown). Similarly, the transcription elongation factor GreA (40) in both spirochetes possesses a long N-terminal extension which is shared only with chlamydiae (69). An orthologous pair of spirochete proteins (TP0511 and BB0355) are homologous to the transcription factor CarD from Myxococcus (48) and share a domain with the transcription repair-coupling helicase (TRCF) (Fig. 4A). Since the corresponding region in TRCF interacts with the RNA polymerase holoenzyme (63), these proteins are likely to be novel transcription factors, characteristic of the spirochete clade, that modulate transcription by interacting directly with the RNA polymerase.
FIG. 4.
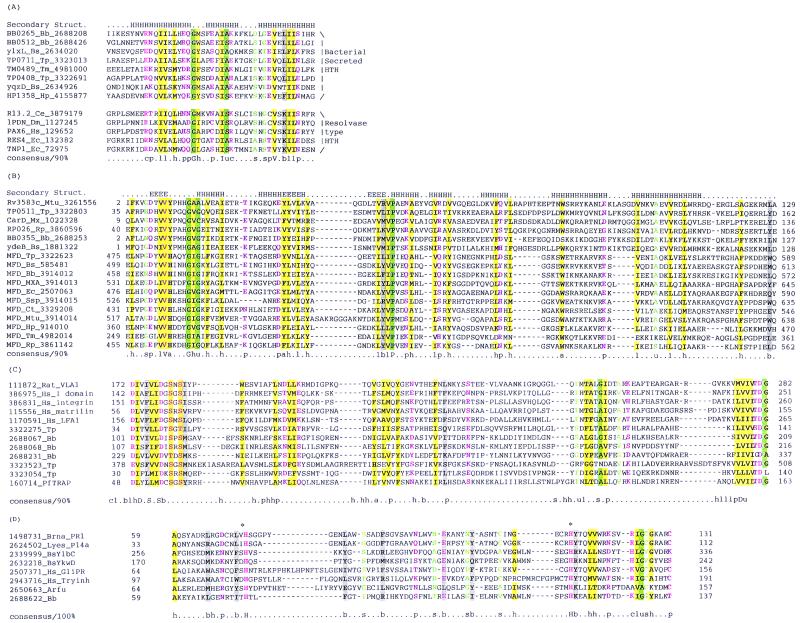
Multiple alignments of previously uncharacterized protein domains discovered in the course of comparative genome analysis of the spirochetes. (A) Resolvase-type HTH domains identified in large proteins containing an N-terminal signal peptide. (B) RNA polymerase-interacting domain found in CarD-like proteins and TRCF. (C) von Willebrand factor A domains in the spirochetes. (D) PR-1 domain in B. burgdorferi. The alignments were constructed using PSI-BLAST-derived pairwise alignments as input for the CLUSTALW program and were further adjusted based on the PSI-BLAST output. Proteins are designated with the protein name, species of origin, and GenBank accession number. Numbers preceding and following the aligned regions refer to the amino acid positions of the domain within each protein sequence. Coloring is according to the consensus shown below the alignment. Uppercase letters indicate conserved residues, by the single-letter amino acid code. Lowercase letters indicate conserved classes of amino acids that are highlighted using differential coloring or shading as follows: violet, polar residues (p) (C,D,E,H,K,N,Q,R,S,T); pink, charged residues (c) (D,E,H,K,R); green background, tiny residues (u) (A,G,S); green shading, small residues (s) (A,C,D,N,G,P,S,T,V); grey shading, bulky residues (b) (E,F,I,K,L,M,Q,R,W,Y); yellow shading, hydrophobic residues (h) (A,C,F,I,L,M,V,W,Y), aromatic residues (a) (F,H,W,Y), and aliphatic residues (l) (I,L,V). Species abbreviations are as follows: Hp, H. pylori; Mx, Myxococcus xanthus; Pf, Plasmodium falciparum. Others are as indicated in the legend to Fig. 3.
Major differences between the two spirochetes were revealed even among the sigma factors. While they share ς70 (RpoD) and ς54 (RpoN), B. burgdorferi also encodes an RpoS ortholog, whereas T. pallidum possesses ς24 (RpoE), ς28, and ς43 (SigA). The three sigma factors present in T. pallidum but not in B. burgdorferi are likely to be involved in regulating multiple gene sets specific to the former. Of particular interest in this context is ς24, whose orthologs in other bacteria regulate gene batteries associated with stress response and pathogenesis (1). Consistent with the detection of multiple sigma factors, T. pallidum encodes elements of the system of sigma factor regulators similar to that seen in Bacillus, Chlamydia, Synechocystis, and the actinomycetes. These regulators include two phosphatases of the PP2C family and an anti-sigma factor antagonist (83), which suggests that T. pallidum senses a distinct environmental signal regulating the anti-sigma factor antagonist. However, T. pallidum does not encode an ortholog of the small serine kinase of the histidine kinase class (RsbV protein), which is an essential component of the anti-sigma factor regulatory system in other bacteria (83). If this system is to be functional, it must include a kinase other than conventional histidine or serine/threonine kinases, as none of them, with the exception of CheA (which is not known to participate in this pathway), are detectable in T. pallidum.
Each of the spirochetes encodes several predicted specific transcriptional regulators of the helix-turn-helix (HTH) class of DNA-binding proteins. In addition, B. burgdorferi encodes a member of the MetJ/Arc family of β-sheet-containing transcription factors (BBD22) on its linear plasmid. With the exception of a single, novel family of HTH proteins, none of these likely transcriptional regulators are orthologous in the two spirochetes, which is consistent with the diversification of the possible target metabolic pathway operons (see below). The shared HTH proteins are an unusual group of two orthologous protein pairs (TP0711/BB0265 and TP0408/BB0512) that contain a C-terminal HTH domain similar to the PAIRED domains of resolvases (Fig. 4B). In addition, they have a hydrophobic N-terminal region resembling a signal peptide that may be responsible for an unexpected membrane association and processing associated with their function. Orthologs of these proteins with the same domain arrangement are detectable in H. pylori, Bacillus subtilis, and Thermatoga maritima. As an example of the diversified transcriptional regulation for similar functions, T. pallidum encodes a winged-HTH protein, TroR (34), whereas B. burgdorferi encodes the unrelated Fur protein (75), both of which are predicted to negatively regulate metal uptake. Two HTH-containing regulators of sugar metabolism, XylR-1 and XylR-2, that combine the HTH domain with the Hsp-70 like sugar-kinase domain are present only in B. burgdorferi.
Functional classes with low OC (<0.6) signaling.
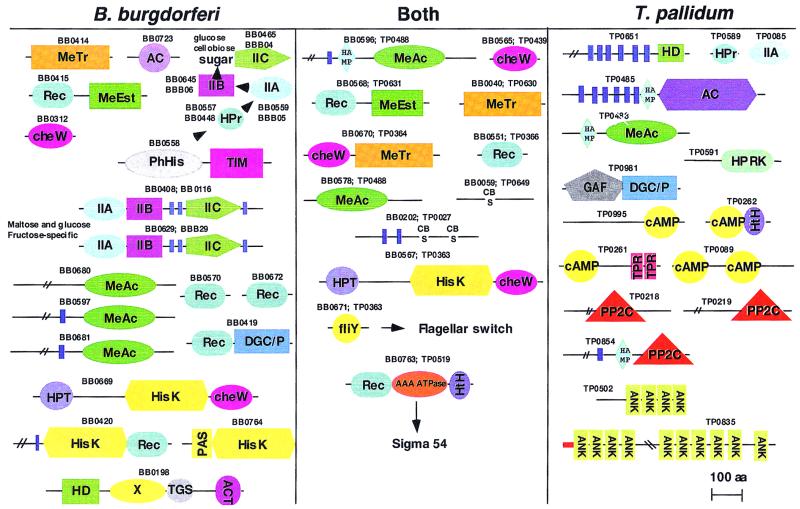
The signal transduction systems in T. pallidum and B. burgdorferi show low OC values (Fig. 2), with only a few conserved features that seem to be remnants of the signaling apparatus of the common ancestor of the spirochetes. This shared core includes components of chemotactic signaling based on the histidine kinase CheA, which contains a C-terminal CheW domain (Fig. 5), and associated downstream signaling components, such as methyl-accepting and methyltransferase chemotaxis proteins. Even within this system, however, B. burgdorferi possesses specific methylesterase, methyltransferase, and methyl-accepting proteins with no counterparts in T. pallidum (Fig. 5). Furthermore, B. burgdorferi has a larger representation of the two-component relay systems, with two additional histidine kinases other than the two CheA paralogs and multiple additional receiver domain proteins. These histidine kinases are likely to function as sensors of different stimuli, since one of them contains a PAS domain that has been implicated in redox sensing and the other one contains a large, uncharacterized extracellular (periplasmic) domain (Fig. 5). Cross-talk between these kinases and other signaling systems is suggested by the fusion of the receiver domain of the two-component system with other signaling domains, such as methylesterase and diguanylate cyclase/phosphodiesterase. Of additional interest is the presence, in both spirochetes, of CheX proteins (TP0365/BB0671) (32). These proteins are homologs of the FliM protein of E. coli, which appears to provide a link between chemotactic signaling and the flagellar motor (46).
FIG. 5.
Differences and conservation in the domain architectures of proteins involved in signaling in B. burgdorferi and T. pallidum. Proteins unique to either organism are shown in the outer panels; the middle panel represents proteins conserved in the two organisms. Domain identification/organization is based on sequence searches using PSI-BLAST-derived PSSMs and Hidden Markov models derived using the HMMER2 package and was confirmed by detailed examination of sequence alignments. The figure is drawn roughly to scale. The double slash (//) indicates that a portion of the long sequence has been omitted, blue vertical bars represent transmembrane helices predicted using the TopPred II program, and the red horizontal bar represents a signal peptide predicted using the SignalP program. Domain abbreviations are as follows: MeTr, methyl transferase; MeAc, methyl acceptor; MeEst, methyl esterase; cheW, two-component signaling adaptor domain; Rec, receiver domain; AC, adenylyl cyclase; IIA/IIB/IIC, components of phosphoenolpyruvate:sugar phosphotransferase (PTS) system; PAS, Per/Arnt/Sim domain; HisK, histidine kinase; PhHis, phosphohistidine; HPT, histidine phosphotransferase; TIM, TIM barrel domain; DGC/P, diguanylate cyclase/phosphodiesterase domain; HAMP, domain common to histidine kinase, adenylyl cyclase, methyl acceptor, and PP2C phosphatase (11); HTH, helix-turn-helix; HD, HD superfamily phosphodiesterase domain; GAF, domain present in cGMP-specific phosphodiesterase (cyclic nucleotide binding); cAMP, cyclic AMP binding domain; TPR, tetratricopeptide repeat; PP2C, PP2C family phosphatase; ANK, ankyrin repeat; CBS, cystathionine-beta synthase domain (a widespread domain implicated in signaling [13]); HPT, histidine-containing phosphotransfer domain; ACT, aspartokinase, chorismate mutase, TyrA domain (8); TGS, threonyl-tRNA synthetase, GTPase, SpoT domain (78); HPRK, HPR kinase; HPr, phosphocarrier protein; PhHis, phosphohistidine; X, conserved domain of unknown function seen in the SpoT protein; fliY, domain common to CheX and FliY/M proteins.
The phosphoenolpyruvate:sugar phosphotransferase system (PTS) (59) is fully represented in B. burgdorferi (Fig. 5). It includes multiple EII enzymes with different sugar specificities along with other PTS proteins, such as HPR and phosphoenolpyruvate-protein-phosphotransferase. By contrast, T. pallidum encodes an unusual motley of proteins from the PTS system which includes the phosphocarrier protein Hpr and its kinase, similar to the one involved in catabolite repression in gram-positive bacteria (56), and a single protein containing an EIIA domain but not the other PTS components (Fig. 5). It is possible that these remnants of the PTS system in T. pallidum are involved in a novel signal transduction mechanism.
T. pallidum possesses a well-developed cyclic AMP (cAMP) signaling system that is lacking in B. burgdorferi (Fig. 5). The presence, in T. pallidum, of four cAMP-binding domain-containing proteins and a classical “eukaryote-type” adenylyl cyclase similar to those present in some other bacteria, such as cyanobacteria, myxobacteria, and actinomycetes, indicates a prominent role for this system in signal transduction. The domain architectures of these proteins suggest functional diversity—these include duplication of the cAMP-binding domain (TP0089) and fusions to an HTH DNA-binding domain (TP0262) and to an alpha-helical domain distantly related to the tetratricopeptide repeats (TPR) (TP0261) (Fig. 5). Furthermore, diguanylate cyclase/phosphodiesterase from T. pallidum contains a GAF domain that in some instances binds cyclic nucleotides (12). This protein might participate in cross-talk between cyclic nucleotide signaling and cyclic diguanylate signaling. The adenylyl cyclase of T. pallidum (TP0485) contains a HAMP (for histidine kinases, adenylyl cyclases, methyl accepting chemotaxis receptors, and phosphatases) domain, a recently described module that is associated with the cytoplasmic face of several bacterial signaling proteins (11). This domain is predicted to transmit extracellular stimuli sensed by these receptor proteins to the respective intracellular signaling domains. All bacteria that contain this eukaryotic-type cAMP-dependent signaling system lack a counterpart to the eukaryotic cyclic nucleotide phosphodiesterase and, in addition, lack the proteobacterial-type phosphodiesterase Icc (44). However, T. pallidum encodes a predicted membrane protein with six transmembrane helices that contains a metal-dependent phosphodiesterase domain of the HD superfamily, which is distantly related to eukaryotic phosphodiesterases (9). This protein is a candidate for the cAMP phosphodiesterase function. B. burgdorferi lacks classical adenylyl cyclases and cAMP-binding proteins but encodes an unrelated adenylyl cyclase (BB0723) that belongs to a recently described class of adenylyl cyclases found in thermophilic archaea and Aeromonas hydrophila (65). The function of this protein in signal transduction remains unclear in the absence of any detectable downstream effector molecules that would recognize cAMP. It cannot be ruled out that a new class of cAMP-binding domains shared by archaea, eukaryotes, and some bacteria, including B. burgdorferi, remains to be discovered.
Most bacteria encode the SpoT protein, which is a guanosine-3′,5′-bispyrophosphate (ppGpp) 3′-pyrophosphohydrolaseguanosine 3′,5′-bispyrophosphohydrolase (62). This key enzyme is responsible for cellular (p)ppGpp degradation, which reverses ppGpp accumulation during the stringent response to amino acid deprivation. While B. burgdorferi possesses a SpoT ortholog (BB0198) (Fig. 5), with its characteristic catalytic HD domain (9) and regulatory TGS (78) and ACT (8) domains, T. pallidum surprisingly lacks this protein. The parasitic lifestyle of T. pallidum may differ from that of Borrelia, resulting in the loss of this stringent response signaling system. Of the published genomes of bacteria, only the two chlamydial species (38, 69) and T. pallidum lack this enzyme, whereas Rickettsia prowazekii encodes a degenerate and probably inactive version of the enzyme (5). This may reflect the superfluous nature of stringent response regulation in the nutrient-rich environments of these pathogens.
Functional classes with low OCs (<0.6): metabolic pathways and transport and other membrane components.
Consistent with the picture emerging from other pathogenic bacteria, the genomes of the two spirochetes are noticeably reduced in terms of genes coding for metabolic enzymes, which suggests a general degeneration of metabolic systems that most likely were represented in their common ancestor. However, even in this degenerate state, there are striking differences in the predicted metabolic pathways between T. pallidum and B. burgdorferi, suggesting different adaptation strategies. The common metabolic heritage is largely restricted to the trunk of the glycolytic Embden-Meyerhof pathway, the phosphorylation steps of nucleotide interconversion, and a system for membrane potential generation, namely, the multisubunit V-type ATPase. The latter is the principal energy-generating ATPase in archaea; in bacteria, the V-ATPase subunits are encoded in a single operon that appears to be evolutionarily mobile and shows a scattered distribution, being found, in addition to in the spirochetes, in Chlamydia, Enterococcus, Deinococcus, and Thermus (55).
As already discussed for other, more conserved pathways, even within the shared core of metabolism there are instances of likely horizontal transfer and associated xenologous and nonorthologous (E. V. Koonin et al., Letter, Trends Genet. 12:334–336, 1996) gene displacement. Apparent lateral gene transfer accompanied by displacement is illustrated by fructose-1-6-bisphosphate aldolase and the glyceraldehyde 3-phosphate dehydrogenase that belong to the central glycolytic pathway. Phylogenetic analysis of the aldolases from the two spirochetes showed that, while both of them are class II aldolases, they belong to two entirely distinct, well-supported groups, which suggests independent acquisition via lateral gene transfer from other bacteria (Fig. 3). In phylogenetic trees, glyceraldehyde 3-phosphate dehydrogenase from T. pallidum groups strongly with the orthologs of this protein from kinetoplastid eukaryotes (Fig. 3). This points to horizontal gene transfer from this eukaryotic lineage to T. pallidum after the divergence of the two spirochetes, followed by displacement of the original spirochete version. An even more interesting case is T. pallidum uridine kinase, which not only shows the unexpected grouping with the ortholog from Thermotoga in phylogenetic trees but also possesses a unique domain architecture, with two domains, including the TGS domain, apparently acquired from threonyl-tRNA synthetases (78). In the threonyl-tRNA synthetases, these domains play a role in RNA binding (61), which suggests that these unique uridine kinases could regulate their own expression by binding to mRNA. Another notable case of likely horizontal transfer followed by gene displacement involves a pair of closely linked genes in an operon, namely glucose 6-phosphate dehydrogenase (G6-PDH) and the adjacent gene that encodes the DevB protein. In T. pallidum, both of these proteins are closely related to their counterparts from Haemophilus and Actinobacillus rather than to those from B. burgdorferi. Phylogenetic analysis (Fig. 3) provides strong bootstrap support for this lineage and suggests that the entire operon comprised of G6-PDH and DevB has been exchanged between T. pallidum and the proteobacterial lineage.
Even among the genes encoding components of metabolic pathways that have been vertically inherited by the two spirochetes, there are footprints of likely ancient horizontal transfer events. Horizontal transfer from eukaryotes to spirochetes is attested by the presence of a pyrophosphate-dependent phosphofructokinase (BB0020, TP0542) that otherwise is characteristic of plants and several protists, such as Entamoeba and Giardia. This enzyme is present in the spirochetes along with the original bacterial ATP-dependent phosphofructokinase (BB0727, TP0108) in the glycolytic pathway, suggesting functional partitioning of the two versions.
Each of the spirochetes encodes two cytidylate kinases. One of these appears to have been acquired by the common ancestor of the spirochetes from the archaea. This is strongly suggested by their highly significant relationship to the archaeal proteins in phylogenetic tree analysis (Fig. 3) and the modified P-loop ATPase motif (data not shown) that is uniquely shared by these proteins with their archaeal counterparts.
Both spirochetes encode an aminopeptidase M containing an N-terminal insert sequence that is specifically shared with eukaryotes; phylogenetic analysis indicates that it might have been horizontally transferred into the ancestral spirochete from a eukaryote (Fig. 3).
Some of the drastic differences in the gene complements for metabolic processes seem to reveal adaptive features unique to each spirochete. The ability to utilize glycerol and chitin as energy sources through the glycolytic pathway, which is unique to B. burgdorferi, is consistent with the existence of an arthropod-specific stage in the life cycle of this spirochete. This is supported by the presence of genes for glycerol uptake and the FAD-dependent glycerol-3-phosphate dehydrogenase (BB0243), the latter apparently being an unusual case of horizontal transfer from a eukaryotic source (Fig. 3). Utilization of N-acetylglucosamine polymers or chitin (available in the arthropod host) as a substrate for cell wall biosynthesis (glucosamine deacetylase, NagA; BB0151) and/or as an energy source (glucosamine deaminase, NagB; BB0152) could represent an adaptation of B. burgdorferi for survival in its specific niche, in this case the arthropod host. The presence of an additional pathway for conversion of dihydroxyacetone phosphate into lactate via a methylglyoxal intermediate also could be an adaptation for growth under certain environmental conditions and carbohydrate sources (28, 74).
Another interesting metabolic feature of B. burgdorferi inferred from sequence analysis is the presence of a pathway for the biosynthesis of isopentenyl pyrophosphate (IPP). All of the enzymes necessary for the formation of IPP from 3-hydroxymethyl glutaryl-coenzyme A (HMG-CoA), along with a novel glycolate oxidase-like, TIM barrel fold oxidoreductase (BB0684) that could participate in the same process, are encoded in a single long operon. With the exception of this oxidoreductase, the remaining proteins of this pathway show eukaryotic or archaeal affinities in phylogenetic analysis (data not shown), with no closely related proteins in bacteria. Taken together with the fact that Pseudomonas mevalonii is the only other bacterium to date that encodes an orthologous HMG-CoA reductase (14), these observations suggest that this pathway has been acquired by B. burgdorferi via horizontal transfer from either an archaeal or a eukaryotic source. The enzymes encoded in this unusual operon are likely to participate in the synthesis of still uncharacterized membrane/lipoprotein components in B. burgdorferi.
The repertoires of transporters encoded by the two spirochetes are significantly different and, with an OC of 0.38, are among the most divergent functional classes revealed in our analysis; this suggests distinct substrate requirements. In this class, it is notable that, unlike B. burgdorferi, T. pallidum appears to lack a transport mechanism for proline or glycine-betaine. Given the proposed osmoprotective role of proline and glycine-betaine in bacterial stress response (4, 23), the presence of a full complement of genes for proline biosynthesis in T. pallidum likely serves as an adaptive response to osmotic stress.
Finally, the surface lipoproteins and other, uncharacterized membrane proteins of the two spirochetes show very low OC values (Fig. 2), which is compatible with the different modes of parasite-host interaction.
Individual protein families in the two spirochetes. (i) Conserved and variable families.
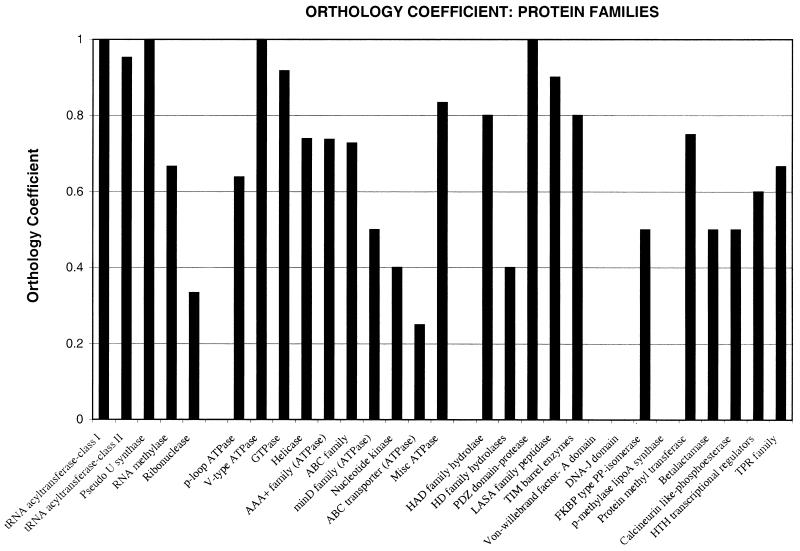
As an alternative approach to assessing the evolutionary forces acting on the spirochetes, we analyzed all of the largest families of paralogous proteins in the two genomes and calculated the OCs for each of them (Fig. 6). Like other bacteria and archaea (39, 80), the largest protein superfamily in the spirochetes includes P-loop-containing NTPases, with 86 members in B. burgdorferi and 69 in T. pallidum. Different families within this large superfamily show substantial variability in the degree of conservation. A number of ATPases, such as those involved in DNA replication, V-type ATPases, and GTPases, show a largely vertical inheritance pattern. By contrast, the ABC transporter ATPases show considerable diversity in the two spirochetes, which is consistent with the differences noted in nutrient transport and substrate specificity (see above). There is a specific expansion of the MinD family of ATPases, which are involved in chromosome partitioning in B. burgdorferi (12 members); this is consistent with the presence of several cosegregating plasmids that comprise parts of the B. burgdorferi genome (30).
FIG. 6.
OCs for protein families and superfamilies identified in the two spirochetes. Protein and domain (super)families within the two spirochete proteomes were identified using transitive BLASTP searches and family-specific PSSMs, as described in Materials and Methods. The OC was computed for each of these (super)families as described in the text.
Compared to the P-loop ATPase superfamily, all other protein families are much smaller and show widely varying degrees of orthology (Fig. 6). Some of these families, such as the pseudouridine synthetases, the PDZ domain proteins, and lysostaphin-like proteases, show OC values close to 1, which suggests functional coherence between the two organisms. The two classes of aminoacyl-tRNA synthetases and SAM-dependent methyltransferases have slightly lower OC values, which is due to the likely horizontal transfers discussed above or to relatively minor lineage-specific gene losses. Other families, such as HD superfamily hydrolases (9), metallo-β-lactamase fold hydrolases (6), calcineurin-like phosphatases (7), and peptidyl-prolyl cis-trans isomerases, show much lower OC values, which indicates major effects of lineage-specific gene loss, horizontal transfer, and rapid diversification. These families, unlike those associated with translation and replication, appear to participate in diverse roles that are under constant selection due to variations in the niches occupied by the two spirochetes. Certain protein families, such as the P-methylase/lipoate synthetase family, are seen only in T. pallidum. This raises the possibility of distinct metabolic options that are absent in B. burgdorferi.
(ii) Newly identified protein families.
We identified several previously undetected protein families in the spirochetes by using family-specific PSSMs (see Materials and Methods). These include some secreted (or possibly periplasmic) and membrane proteins that could be involved in pathogen-host interactions. The most interesting of these are the von Willebrand A factor (vWA) domain-containing proteins (52), which, until recently, had been detected only in eukaryotes. The vWA domain is a Mg2+-binding protein module with an α/β fold that participates in adhesion and protein-protein interactions in a wide range of eukaryotic proteins, such as integrins (26). Using profile searches, we identified three vWA domain-containing proteins in each of the spirochetes (Fig. 4C). However, the vWA proteins from B. burgdorferi are not highly similar to those from T. pallidum, suggesting that they either have been independently acquired or have diverged rapidly due to selective forces acting on extracellular proteins. These secreted or membrane-associated vWA domain proteins are reminiscent of similar domains present in the extracellular adhesion molecule (TRAP) of a eukaryotic pathogen, the malarial genus Plasmodium (73). By analogy to TRAP, it is likely that the spirochete vWA proteins are involved in adhesion of these bacteria to the extracellular matrix or to cells of the host connective tissues. These proteins could be potential targets for specific antispirochete therapies. The similarity between the spirochete vWA domains and that of the LFA-1 integrin is of interest given the recent report identifying LFA-1 as an autoantigen in treatment-resistant Lyme arthritis (33).
Another interesting protein of B. burgdorferi is BB0689, a secreted or periplasmic protein containing a PR1 domain (70). This domain hitherto has been detected only in eukaryotes, namely in plant pathogenesis-related proteins (PR-1) and in proteins expressed in the animal immune system such as GliPR (70). One of the human proteins in this family has been suggested to be a novel trypsin inhibitor (82), whereas other members, such as helothermine, found in the toxins of Helodermid lizards and certain snakes, act as inhibitors of potassium and calcium channels (17, 50). Another protein of this family, TPX-1, is implicated in the interaction between spermatogenic and Sertoli cells (45). All of these observations indicate that PR-1 domain-containing proteins participate in diverse extracellular protein-protein interactions, suggesting a role for BB0689 in adhesion to host extracellular proteins and, accordingly, in pathogenesis.
Conversely, T. pallidum encodes an unusual, large, secreted (periplasmic) protein (TP0544) that contains an OB-fold domain (47). This protein might interact with host cells and act as a virulence factor like the OB-fold-containing toxins from a wide range of bacteria (25).
Both spirochetes encode proteins (BB0236 and TP0421) containing a modified version of the YWTD domain (67) that is seen in intracellular animal proteins, some of which also contain the RING finger domain (L. Aravind, unpublished data). This relationship to a class of animal proteins suggests lateral transfer from an animal host into the ancestor of the two spirochetes. YWTD domains assume a β-propeller structure similar, for example, to that in low-density lipoprotein receptors and in proteins from other parasites (e.g., Trypanosoma cruzi) that mediate extracellular adhesion (51). The presence of a predicted signal peptide suggests that, like the vWA and PR-1 domain-containing proteins, the spirochete YWTD domain proteins are secreted and interact with host cell surface molecules (52).
Another interesting family that is conserved between the two spirochetes, some of whose members appear to be secreted, are the tetratricopeptide repeat (TPR) domain-containing proteins (24). These domains form an alpha-helical superstructure and mediate protein-protein interactions. The use of TPR motifs in extracellular interactions might be a novel strategy of host interaction employed by the spirochetes and is reminiscent of a family of secreted Sel-1 repeat proteins in Helicobacter pylori (52; L. Aravind, unpublished data).
To add to this repertoire of previously undetected proteins that could be important for the pathogen's interaction with host cell surfaces, T. pallidum encodes two proteins with multiple ankyrin repeat motifs (TP0502, TP0835) (Fig. 5). At least TP0835, which contains 20 ankyrin repeats, has a canonical signal peptide, indicating that, unlike the eukaryotic ankyrin repeat proteins, this Treponema protein is secreted.
Evolutionary implications of comparative analysis of the two spirochete proteomes.
The use of the OC concept, along with a case-by-case analysis of individual protein sequences, has provided considerable information that allows us to address several evolutionary issues: (i) the amount of shared heritage present in the spirochete genomes, (ii) the likely gene repertoire and lifestyle of the common ancestor, (iii) the role of horizontal transfer, and (iv) the strategies and adaptations used by these organisms in their interaction with the host, which are important for pathogenesis.
Likely orthologs comprise about 43% of the total number of proteins encoded by the two spirochetes (39.5% [495 of 1,256] of the B. burgdorferi proteins and 47% [486 of 1,031] of the T. pallidum proteins). This fairly low fraction of orthologs indicates that lineage-specific gene loss, horizontal gene transfer, and rapid divergence account for more than one-half of the gene repertoires of the two spirochetes. The relatively lower fraction of orthologs in Borrelia is to a large extent due to the fact that multiple plasmids harbored by this spirochete encode a number of highly variable proteins without counterparts in Treponema (30). Genome comparisons between two more closely related bacteria, namely H. influenzae and E. coli, have indicated that lineage-specific gene loss was a major force in the evolution of the parasitic organism, in this case H. influenzae (72). Given that both spirochetes are obligate parasites, it is likely that the two lineages have lost genes largely independently, resulting in major differences in the complement of genes eventually retained. However, the number of genes lost by the spirochetes is lower than that seen in some other obligate parasites, such as the mycoplasmas, chlamydiae, and rickettsiae (5, 69, 79).
The OC values for different functional classes of proteins (Fig. 2) illustrate the spectrum of genes that have retained identical or similar functions and were probably present in the common ancestor of the spirochetes. As expected, certain core functions, such as translation, RNA modification, and replication, are hardly affected by gene loss. By contrast, the DNA repair systems and the transcription systems show major differences, probably due to a combination of lineage-specific loss and horizontal gene transfer driven by different selective forces acting on the two organisms. The core of the glycolytic pathway and associated metabolic steps show a predominantly vertical inheritance. These systems with high OC values are very similar to their counterparts in other bacteria, which suggests a typical bacterial core physiology of the common ancestor. The presence of V-type ATPase operons in both spirochetes indicates that, like Thermus and Enterococcus (55), the common ancestor of the spirochetes used H+/Na+ exchange for ion gradient generation. The signal transduction systems in general show low OC values, but the elements involved in chemotactic response are conserved. Together with the conservation of the flagellar apparatus (Fig. 2), this suggests an actively motile chemoresponsive ancestral spirochete that used a mode of locomotion similar to that of the two extant species.
Even within the conserved systems, there are some striking cases of xenologous and nonorthologous gene displacement that are suggestive of horizontal transfer from distantly related bacteria and archaea. Some of these appear to have occurred in the common ancestor of the spirochetes, whereas others are lineage specific. Examples of apparent ancient gene transfers include the acquisition of certain aminoacyl-tRNA synthetases and cytidylate kinase from archaea. Likely xenologous displacements in one of the spirochete lineages were seen in a number of cases, such as fructose-1-6-bisphosphate aldolase, glyceraldehyde 3-phosphate dehydrogenase, glucose 6-phosphate dehydrogenase subunits, 6-phosphogluconate dehydrogenase, and topoisomerase I (Fig. 3). Other examples of likely gene exchange with distant bacteria include the small, primase-like TOPRIM domain protein (10), which is otherwise seen only in low-GC gram-positive bacteria and in archaea, and the unusual HTH proteins shared with Bacillus, Thermotoga, and Helicobacter. These observations suggest that, unlike the extant forms that lead a relatively isolated existence due to their specialized life cycles, the ancestors of B. burgdorferi and T. pallidum, both before and after their divergence from each other, existed in close proximity with other prokaryotes, which favored gene exchange.
There is considerable evidence of likely acquisition of genes from eukaryotic, and possibly animal, sources by the common ancestor of the spirochetes; on most occasions, such gene acquisitions apparently have been accompanied by displacement of the endogenous bacterial versions. Several of the aminoacyl-tRNA synthetases exemplify this phenomenon (78). Other striking cases include the YWTD domain proteins and the secreted vWA domain proteins, which until now appeared to be specific to animals, as well as predicted secreted proteins containing TPR repeats. Thus, it appears that the common ancestor of Treponema and Borrelia was already in contact with animal hosts. However, the evidence of gene exchange with other bacteria, archaea, and perhaps eukaryotic protists (see above) suggests that they were not specialized obligate parasites isolated from the rest of the microbial community. The lifestyle predicted for the common ancestor of B. burgdorferi and T. pallidum is consistent with that of the spirochetes in the microbial community in invertebrate guts (21, 41). These spirochetes are not only in contact with an animal host but also share the niche with other bacteria, methanogenic archaea, and eukaryotic protists. The alternative hypothesis, that the common ancestor of Treponema and Borrelia was a free-living spirochete and that the bacteria of these two lineages have independently established symbiosis with the eukaryotic host, cannot be ruled out but seems to be less strongly supported by the evidence. Indeed, many of the likely cases of horizontal gene transfer from eukaryotes are shared by the two spirochetes, which strongly suggests that their ancestor was already in contact with a eukaryotic host.
Subsequent to the divergence from its common ancestor, each lineage independently acquired the ability to infect vertebrate hosts. This is indicated by the presence of very different variant antigens in the two genera, a feature that might have enabled these organisms to evade the acquired immunity of the vertebrate hosts. The possibility that Borrelia has moved from arthropod parasitism to vertebrate parasitism is suggested by the presence of specific carbohydrate metabolism genes that provide for the utilization of arthropod chitin by this bacterium. The presence of commensal treponemes in vertebrates (21) suggests that the ancestors of T. pallidum were well adapted for life in a vertebrate host before the final step in the evolution of pathogenicity that might have been accompanied by the acquisition of specific variant antigens to evade the host immune system. T. pallidum lacks many of the carbohydrate metabolism genes and a functional PTS system; these systems might have been lost subsequent to its displacement from the carbohydrate-rich niches occupied by the ancestral treponeme, such as the vertebrate oral cavity.
Conclusions.
By means of local sequence similarity searches, profile searches, and analysis of individual domains and protein families, we have conducted a detailed comparative analysis of the genomes of the spirochetes B. burgdorferi and T. pallidum. The level of conservation between functional classes and paralogous families of proteins was measured using the orthology coefficient. Using this measure, it was possible to characterize, in functional terms, the nature of the divergence between the two spirochetes and the common and distinct aspects of their physiological strategies. Protein profile searches resulted in the identification of hitherto undetected components of the signal transduction machinery and novel proteins containing domains previously seen only in eukaryotes. Secreted proteins containing these domains might mediate interactions between the spirochetes and host cells or the extracellular matrix. It appears possible to tentatively reconstruct the evolutionary steps leading from a common ancestor that might have been an invertebrate gut symbiont to the divergence of the two genera of spirochetes and adaptation to their specific niches.
Availability of complete results.
The list of T. pallidum and B. burgdorferi orthologs classified by functional categories and by protein families is available by anonymous ftp at ftp://ncbi.nlm.nih.gov/pub/Koonin/Spirochetes.
REFERENCES
- 1.Akbar S, Kang C M, Gaidenko T A, Price C W. Modulator protein RsbR regulates environmental signalling in the general stress pathway of Bacillus subtilis. Mol Microbiol. 1997;24:567–578. doi: 10.1046/j.1365-2958.1997.3631732.x. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Koonin E V. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amin U S, Lash T D, Wilkinson B J. Proline betaine is a highly effective osmoprotectant for Staphylococcus aureus. Arch Microbiol. 1995;163:138–142. doi: 10.1007/BF00381788. [DOI] [PubMed] [Google Scholar]
- 5.Andersson S G, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 6.Aravind L. An evolutionary classification of the metallo-β-lactamase fold proteins. In Silico Biol. 1998;1:0008. http://www.bioinfo.de/isb/1998/01/0008 . [Online.] http://www.bioinfo.de/isb/1998/01/0008. . [PubMed] [Google Scholar]
- 7.Aravind L, Koonin E V. DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aravind L, Koonin E V. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J Mol Biol. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- 9.Aravind L, Koonin E V. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23:469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 10.Aravind L, Leipe D D, Koonin E V. Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aravind, L., and C. Ponting. HAMP: signaling domain in prokaryotes. FEMS Microbiol Lett. 176:111–116. [DOI] [PubMed]
- 12.Aravind L, Ponting C P. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 13.Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem Sci. 1997;22:12–13. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- 14.Bochar D A, Stauffacher C V, Rodwell V W. Sequence comparisons reveal two classes of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol Genet Metab. 1999;66:122–127. doi: 10.1006/mgme.1998.2786. [DOI] [PubMed] [Google Scholar]
- 15.Bork P, Hofmann K, Bucher P, Neuwald A F, Altschul S F, Koonin E V. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 16.Bork P, Koonin E V. Predicting functions from protein sequences—where are the bottlenecks? Nat Genet. 1998;18:313–318. doi: 10.1038/ng0498-313. [DOI] [PubMed] [Google Scholar]
- 17.Brown R L, Haley T L, West K A, Crabb J W. Pseudechetoxin: a peptide blocker of cyclic nucleotide-gated ion channels. Proc Natl Acad Sci USA. 1999;96:754–759. doi: 10.1073/pnas.96.2.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chervitz S A, Aravind L, Sherlock G, Ball C A, Koonin E V, Dwight S S, Harris M A, Dolinski K, Mohr S, Smith T, Weng S, Cherry J M, Botstein D. Comparison of the complete protein sets of worm and yeast: orthology and divergence. Science. 1998;282:2022–2028. doi: 10.1126/science.282.5396.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cintron R, Pachner A R. Spirochetal diseases of the nervous system. Curr Opin Neurol. 1994;7:217–222. doi: 10.1097/00019052-199406000-00007. [DOI] [PubMed] [Google Scholar]
- 20.Claros M G, von Heijne G. TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 21.Coene M, Agliano A M, Paques A T, Cattani P, Dettori G, Sanna A, Cocito C. Comparative analysis of the genomes of intestinal spirochetes of human and animal origin. Infect Immun. 1989;57:138–145. doi: 10.1128/iai.57.1.138-145.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coyle P K. Advances and pitfalls in the diagnosis of Lyme disease. FEMS Immunol Med Microbiol. 1997;19:103–109. doi: 10.1111/j.1574-695X.1997.tb01078.x. [DOI] [PubMed] [Google Scholar]
- 23.Csonka L N, Hanson A D. Prokaryotic osmoregulation: genetics and physiology. Annu Rev Microbiol. 1991;45:569–606. doi: 10.1146/annurev.mi.45.100191.003033. [DOI] [PubMed] [Google Scholar]
- 24.Das A K, Cohen P W, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dodd H N, Pemberton J M. Cloning, sequencing, and characterization of the nucH gene encoding an extracellular nuclease from Aeromonas hydrophila JMP636. J Bacteriol. 1996;178:3926–3933. doi: 10.1128/jb.178.13.3926-3933.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley J, Cruz M, Handin R, Liddington R. Crystal structure of the von Willebrand Factor A1 domain and implications for the binding of platelet glycoprotein Ib. J Biol Chem. 1998;273:10396–10401. doi: 10.1074/jbc.273.17.10396. [DOI] [PubMed] [Google Scholar]
- 27.Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 28.Ferguson G P, Totemeyer S, MacLean M J, Booth I R. Methylglyoxal production in bacteria: suicide or survival? Arch Microbiol. 1998;170:209–218. doi: 10.1007/s002030050635. [DOI] [PubMed] [Google Scholar]
- 29.Fijalkowska I J, Schaaper R M. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc Natl Acad Sci USA. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Venter J C, et al. Genomic sequence of a Lyme disease spirochete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 31.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Venter J C, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 32.Ge Y, Charon N W. Molecular characterization of a flagellar/chemotaxis operon in the spirochete Borrelia burgdorferi. FEMS Microbiol Lett. 1997;153:425–431. doi: 10.1111/j.1574-6968.1997.tb12606.x. [DOI] [PubMed] [Google Scholar]
- 33.Gross D M, Forsthuber T, Tary-Lehmann M, Etling C, Ito K, Nagy Z A, Field J A, Steere A C, Huber B T. Identification of LFA-1 as a candidate autoantigen in treatment-resistant Lyme arthritis. Science. 1998;281:703–706. doi: 10.1126/science.281.5377.703. [DOI] [PubMed] [Google Scholar]
- 34.Hardham J M, Stamm L V, Porcella S F, Frye J G, Barnes N Y, Howell J K, Mueller S L, Radolf J D, Weinstock G M, Norris S J. Identification and transcriptional analysis of a Treponema pallidum operon encoding a putative ABC transport system, an iron-activated repressor protein homolog, and a glycolytic pathway enzyme homolog. Gene. 1997;197:47–64. doi: 10.1016/s0378-1119(97)00234-5. [DOI] [PubMed] [Google Scholar]
- 35.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 36.Himmelreich R, Plagens H, Hilbert H, Reiner B, Herrmann R. Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 1997;25:701–712. doi: 10.1093/nar/25.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ibba M, Losey H C, Kawarabayasi Y, Kikuchi H, Bunjun S, Soll D. Substrate recognition by class I lysyl-tRNA synthetases: a molecular basis for gene displacement. Proc Natl Acad Sci USA. 1999;96:418–423. doi: 10.1073/pnas.96.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman R W, Olinger L, Grimwood J, Davis R W, Stephens R S. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet. 1999;21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- 39.Koonin E V, Mushegian A R, Galperin M Y, Walker D R. Comparison of archaeal and bacterial genomes: computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol Microbiol. 1997;25:619–637. doi: 10.1046/j.1365-2958.1997.4821861.x. [DOI] [PubMed] [Google Scholar]
- 40.Koulich D, Nikiforov V, Borukhov S. Distinct functions of N- and C-terminal domains of GreA, an Escherichia coli transcript cleavage factor. J Mol Biol. 1998;276:379–389. doi: 10.1006/jmbi.1997.1545. [DOI] [PubMed] [Google Scholar]
- 41.Kudo T, Ohkuma M, Moriya S, Noda S, Ohtoko K. Molecular phylogenetic identification of the intestinal anaerobic microbial community in the hindgut of the termite, Reticulitermes speratus, without cultivation. Extremophiles. 1998;2:155–161. doi: 10.1007/s007920050055. [DOI] [PubMed] [Google Scholar]
- 42.Larsen S A, Steiner B M, Rudolph A H. Laboratory diagnosis and interpretation of tests for syphilis. Clin Microbiol Rev. 1995;8:1–21. doi: 10.1128/cmr.8.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lupas A. Predicting coiled-coil regions in proteins. Curr Opin Struct Biol. 1997;7:388–393. doi: 10.1016/s0959-440x(97)80056-5. [DOI] [PubMed] [Google Scholar]
- 44.Macfadyen L P, Ma C, Redfield R J. A 3′,5′ cyclic AMP (cAMP) phosphodiesterase modulates cAMP levels and optimizes competence in Haemophilus influenzae Rd. J Bacteriol. 1998;180:4401–4405. doi: 10.1128/jb.180.17.4401-4405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maeda T, Sakashita M, Ohba Y, Nakanishi Y. Molecular cloning of the rat Tpx-1 responsible for the interaction between spermatogenic and Sertoli cells. Biochem Biophys Res Commun. 1998;248:140–146. doi: 10.1006/bbrc.1998.8918. [DOI] [PubMed] [Google Scholar]
- 46.Mathews M A, Tang H L, Blair D F. Domain analysis of the FliM protein of Escherichia coli. J Bacteriol. 1998;180:5580–5590. doi: 10.1128/jb.180.21.5580-5590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murzin A G. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicolas F J, Cayuela M L, Martinez-Argudo I M, Ruiz-Vazquez R M, Murillo F J. High mobility group I(Y)-like DNA-binding domains on a bacterial transcription factor. Proc Natl Acad Sci USA. 1996;93:6881–6885. doi: 10.1073/pnas.93.14.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 50.Nobile M, Noceti F, Prestipino G, Possani L D. Helothermine, a lizard venom toxin, inhibits calcium current in cerebellar granules. Exp Brain Res. 1996;110:15–20. doi: 10.1007/BF00241369. [DOI] [PubMed] [Google Scholar]
- 51.Pereira M E, Mejia J S, Ortega-Barria E, Matzilevich D, Prioli R P. The Trypanosoma cruzi neuraminidase contains sequences similar to bacterial neuraminidases, YWTD repeats of the low density lipoprotein receptor, and type III modules of fibronectin. J Exp Med. 1991;174:179–191. doi: 10.1084/jem.174.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ponting C P, Aravind L, Schultz J, Bork P, Koonin E V. Eukaryotic signalling domain homologues in Archaea and Bacteria. Ancient ancestry and horizontal gene transfer. J Mol Biol. 1999;289:729–745. doi: 10.1006/jmbi.1999.2827. [DOI] [PubMed] [Google Scholar]
- 53.Ponting C P, Schultz J, Milpetz F, Bork P. SMART: identification and annotation of domains from signalling and extracellular protein sequences. Nucleic Acids Res. 1999;27:229–232. doi: 10.1093/nar/27.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poterszman A, Lamour V, Egly J M, Moras D, Thierry J C, Poch O. A eukaryotic XPB/ERCC3-like helicase in Mycobacterium leprae? Trends Biochem Sci. 1997;22:418–419. doi: 10.1016/s0968-0004(97)01124-9. [DOI] [PubMed] [Google Scholar]
- 55.Radax C, Sigurdsson O, Hreggvidsson G O, Aichinger N, Gruber C, Kristjansson J K, Stan-Lotter H. F- and V-ATPases in the genus Thermus and related species. Syst Appl Microbiol. 1998;21:12–22. doi: 10.1016/S0723-2020(98)80003-9. [DOI] [PubMed] [Google Scholar]
- 56.Reizer J, Hoischen C, Titgemeyer F, Rivolta C, Rabus R, Stulke J, Karamata D, Saier M H, Jr, Hillen W. A novel protein kinase that controls carbon catabolite repression in bacteria. Mol Microbiol. 1998;27:1157–1169. doi: 10.1046/j.1365-2958.1998.00747.x. [DOI] [PubMed] [Google Scholar]
- 57.Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ryan J R, Levine J F, Apperson C S, Lubke L, Wirtz R A, Spears P A, Orndorff P E. An experimental chain of infection reveals that distinct Borrelia burgdorferi populations are selected in arthropod and mammalian hosts. Mol Microbiol. 1998;30:365–379. doi: 10.1046/j.1365-2958.1998.01071.x. [DOI] [PubMed] [Google Scholar]
- 59.Saier M H, Jr, Reizer J. The bacterial phosphotransferase system: new frontiers 30 years later. Mol Microbiol. 1994;13:755–764. doi: 10.1111/j.1365-2958.1994.tb00468.x. [DOI] [PubMed] [Google Scholar]
- 60.Sandigursky M, Franklin W A. Thermostable uracil-DNA glycosylase from Thermotoga maritima, a member of a novel class of DNA repair enzymes. Curr Biol. 1999;9:531–534. doi: 10.1016/s0960-9822(99)80237-1. [DOI] [PubMed] [Google Scholar]
- 61.Sankaranarayanan R, Dock-Bregeon A C, Romby P, Caillet J, Springer M, Rees B, Ehresmann C, Ehresmann B, Moras D. The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999;97:371–381. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 62.Sarubbi E, Rudd K E, Xiao H, Ikehara K, Kalman M, Cashel M. Characterization of the spoT gene of Escherichia coli. J Biol Chem. 1989;264:15074–15082. [PubMed] [Google Scholar]
- 63.Selby C P, Sancar A. Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J Biol Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 64.Sigal L H. Lyme disease and the Lyme disease vaccines. Bull Rheum Dis. 1999;48:1–4. [PubMed] [Google Scholar]
- 65.Sismeiro O, Trotot P, Biville F, Vivares C, Danchin A. Aeromonas hydrophila adenylyl cyclase 2: a new class of adenylyl cyclases with thermophilic properties and sequence similarities to proteins from hyperthermophilic archaebacteria. J Bacteriol. 1998;180:3339–3344. doi: 10.1128/jb.180.13.3339-3344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sonnhammer E L, Eddy S R, Birney E, Bateman A, Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Springer T A. An extracellular beta-propeller module predicted in lipoprotein and scavenger receptors, tyrosine kinases, epidermal growth factor precursor, and extracellular matrix components. J Mol Biol. 1998;6:836–862. doi: 10.1006/jmbi.1998.2115. [DOI] [PubMed] [Google Scholar]
- 68.Stechenberg B W. Lyme disease: the latest great imitator. Pediatr Infect Dis J. 1988;7:402–409. [PubMed] [Google Scholar]
- 69.Stephens R S, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov R L, Zhao Q, Koonin E V, Davis R W. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 70.Szyperski T, Fernandez C, Mumenthaler C, Wuthrich K. Structure comparison of human glioma pathogenesis-related protein GliPR and the plant pathogenesis-related protein P14a indicates a functional link between the human immune system and a plant defense system. Proc Natl Acad Sci USA. 1998;95:2262–2266. doi: 10.1073/pnas.95.5.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tatusov R L, Koonin E V, Lipman D J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 72.Tatusov R L, Mushegian A R, Bork P, Brown N P, Hayes W S, Borodovsky M, Rudd K E, Koonin E V. Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with Escherichia coli. Curr Biol. 1996;6:279–291. doi: 10.1016/s0960-9822(02)00478-5. [DOI] [PubMed] [Google Scholar]
- 73.Templeton T J, Kaslow D C. Cloning and cross-species comparison of the thrombospondin-related anonymous protein (TRAP) gene from Plasmodium knowlesi, Plasmodium vivax and Plasmodium gallinaceum. Mol Biochem Parasitol. 1997;84:13–24. doi: 10.1016/s0166-6851(96)02775-2. [DOI] [PubMed] [Google Scholar]
- 74.Totemeyer S, Booth N A, Nichols W W, Dunbar B, Booth I R. From famine to feast: the role of methylglyoxal production in Escherichia coli. Mol Microbiol. 1998;27:553–562. doi: 10.1046/j.1365-2958.1998.00700.x. [DOI] [PubMed] [Google Scholar]
- 75.Vasil M L, Ochsner U A, Johnson Z, Colmer J A, Hamood A N. The fur-regulated gene encoding the alternative sigma factor PvdS is required for iron-dependent expression of the LysR-type regulator ptxR in Pseudomonas aeruginosa. J Bacteriol. 1998;180:6784–6788. doi: 10.1128/jb.180.24.6784-6788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walker D R, Koonin E V. SEALS: a system for easy analysis of lots of sequences. ISMB. 1997;5:333–339. [PubMed] [Google Scholar]
- 77.Ward D, Newton A. Requirement of topoisomerase IV parC and parE genes for cell cycle progression and developmental regulation in Caulobacter crescentus. Mol Microbiol. 1997;26:897–910. doi: 10.1046/j.1365-2958.1997.6242005.x. [DOI] [PubMed] [Google Scholar]
- 78.Wolf, Y., L. Aravind, N. Grishin, and E. V. Koonin. Domain organization and evolutionary history of aminoacyl tRNA synthetases. Genome Res., in press. [PubMed]
- 79.Wolf Y I, Aravind L, Koonin E V. Rickettsiae and Chlamydiae: evidence of horizontal gene transfer and gene exchange. Trends Genet. 1999;15:173–175. doi: 10.1016/s0168-9525(99)01704-7. [DOI] [PubMed] [Google Scholar]
- 80.Wolf Y I, Brenner S E, Bash P A, Koonin E V. Distribution of protein folds in the three superkingdoms of life. Genome Res. 1999;9:17–26. [PubMed] [Google Scholar]
- 81.Wootton J C. Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput Chem. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- 82.Yamakawa T, Miyata S, Ogawa N, Koshikawa N, Yasumitsu H, Kanamori T, Miyazaki K. cDNA cloning of a novel trypsin inhibitor with similarity to pathogenesis-related proteins, and its frequent expression in human brain cancer cells. Biochim Biophys Acta. 1998;1395:202–208. doi: 10.1016/s0167-4781(97)00149-8. [DOI] [PubMed] [Google Scholar]
- 83.Yang X, Kang C M, Brody M S, Price C W. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996;10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]
- 84.Zhang J R, Hardham J M, Barbour A G, Norris S J. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]