Summary
Single-cell combinatorial indexing RNA sequencing (sci-RNA-seq3) enables high-throughput single-nucleus transcriptomic profiling of multiple samples in one experiment. Here, we describe an optimized protocol of mouse kidney nuclei isolation and sci-RNA-seq3 library preparation. The use of a dounce tissue homogenizer enables nuclei extraction with high yield. Fixed nuclei are processed for sci-RNA-seq3, and self-loaded transposome Tn5 is used for tagmentation in library generation. The step-by-step protocol allows researchers to generate scalable single-cell transcriptomic data with common laboratory supplies at low cost.
For complete details on the use and execution of this protocol, please refer to Li et al. (2022).1
Subject areas: Cell Biology, Cell isolation, Model Organisms, Molecular Biology, RNAseq, Single Cell
Graphical abstract

Highlights
-
•
Optimized nuclei isolation protocol for mouse kidneys with high efficiency
-
•
Tn5 assembly with annealed oligonucleotides to make functional transposome
-
•
Transposome activity titration test to determine optimal working concentration
-
•
Performing a small-scale sci-RNA-seq3 experiment as a proof of principle
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Single-cell combinatorial indexing RNA sequencing (sci-RNA-seq3) enables high-throughput single-nucleus transcriptomic profiling of multiple samples in one experiment. Here, we describe an optimized protocol of mouse kidney nuclei isolation and sci-RNA-seq3 library preparation. The use of a dounce tissue homogenizer enables nuclei extraction with high yield. Fixed nuclei are processed for sci-RNA-seq3, and self-loaded transposome Tn5 is used for tagmentation in library generation. The step-by-step protocol allows researchers to generate scalable single-cell transcriptomic data with common laboratory supplies at low cost.
Before you begin
In sci-RNA-seq3,2,3,4 each nucleus is indexed with a unique combination of three oligonucleotide barcodes, introduced by reverse transcription, hairpin ligation and indexed PCR reactions, respectively. Currently popular droplet microfluidics platforms such as the one offered by 10X Genomics5 requires a chromium controller to physically isolate individual cells, whereas sci-RNA-seq3 can be performed solely with commonly available lab supplies. Challenges in executing the original sci-RNA-seq3 protocol2 on adult mouse kidney tissues include low nuclei extraction yield, reduced library quality due to non-uniform transposase activity in tagmentation, incomplete purification and lack of a workflow for performing small-scale pilot experiments as a proof-of-principle.
The protocol below describes the specific steps for nuclei isolation from adult mouse kidneys and profiling of multiple samples simultaneously with sci-RNA-seq3. The protocol is composed of three major sections: (I) nuclei isolation from mouse kidneys with fixation, (II) sci-RNA-seq3 on fixed nuclei and (III) sub-library generation. Section I includes an optimized nuclei isolation method based on dounce tissue homogenizer which enables extraction of >15 million fresh nuclei from mouse kidney tissues that are as small as 0.1 grams. The backbone of Sections II and III is adapted from the original publication2 and includes optimizations in nuclei permeabilization, transposome assembly (with addition of a transposome activity titration assay) and library purification. Finally, we describe a workflow for performing pilot experiments at reduced scale to enable researchers to adapt this protocol in their own laboratories.
Institutional permissions
This protocol involves experimental procedures on mice. All mouse experiments of this study were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee at Washington University in St. Louis.
Mouse sacrifice and sample preservation
Timing: 15–30 min per sample
CRITICAL: Please refer to Institutional Permissions for all experiments conducted on mice.
-
1.
After the mouse is completely euthanized (not responsive to painful stimuli such as paw pitch), cut the rib cage to expose the heart.
-
2.
Clear red blood cells by cardiac perfusion with ice-cold phosphate-buffered saline (PBS) using a perfusion pump.
Note: Perfusion is performed by inserting the needle into the apex of the left ventricle with procedures described before.6 Mouse sacrifice techniques can vary across laboratories and should not affect the performance of this protocol.
Note: This protocol works for both healthy kidneys as well as those that may be inflamed or fibrotic, for example as a consequence of ischemia reperfusion injury or unilateral ureteral obstruction.
-
3.
Perfusion typically takes 2–5 min and can be terminated when the dark red kidneys turn into light brown color.
-
4.
Carefully dissect the kidney and remove the renal capsule.
-
5.
Flash freeze the tissue in a cryogenic tube in liquid nitrogen. The tissue can be stored at −80°C for over a year.
Note: We recommend dissecting at least 100 mg mouse kidney tissues for flash freezing to ensure sufficient number of nuclei can be extracted and processed for sci-RNA-seq3.
CRITICAL: The tissue should be frozen immediately after dissection to reduce RNA degradation.
-
6.
Repeat steps 1–5 to collect all samples in the study cohort.
Prepare split-pool barcoding oligos and plates
Timing: 1 day
Note: The sequences of all poly-T oligonucleotides (RT oligos), hairpin ligation oligonucleotides and PCR P5/P7 oligonucleotides have been described in the original publication2 or are available through its GitHub resource page. Figure 1 presents structures of these uniquely indexed oligos including certain oligo modifications. Resuspend all oligos to 100 μM with nuclease-free water.
Note: We recommend use nuclease-free laboratory supplies throughout this protocol. We recommend researchers clean the laboratory bench with RNaseZap Decontamination Solution before conducting this experiment.
-
7.Prepare four 96-well plates for reverse transcription by adding 2 μL uniquely indexed RT oligos (100 μM) into each well.
-
a.Seal the plates.
-
b.The plates can be stored at −20°C for over a month.
-
a.
CRITICAL: The position of each RT oligo in the 96-well plate must be recorded since the sequence of each oligo will be used for sample demultiplexing in downstream data analysis. Table 1 presents an example of depositing the 384 RT oligos into desired well positions.
-
8.Prepare four 96-well plates for hairpin ligation by adding 8 μL uniquely indexed ligation oligonucleotides (100 μM) into each well.
-
a.Seal the plates.
-
b.The plates can be stored at −20°C for over a month.
-
a.
Figure 1.
Structures of oligonucleotides used in sci-RNA-seq3
An RT oligo contains a UMI sequence (8 base pairs in length), 10-bp RT barcode sequence, 30-bp poly-T sequence and a phosphorylation modification at the 5′ end. A hairpin ligation oligo contains two ligation barcode sequences (9 or 10 base pairs in length) that are reverse complemented with each other and an internal deoxyuridine modification. Both P5 and P7 oligos contain a 10-bp PCR barcode sequence. The total number of barcode combinations can be calculated with the equation presented. Then, an estimated cell collision rate can be calculated as previous described.7,8
Table 1.
An example of oligo deposition locations in four RT plates
| Plate#1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | RT#1 | RT#2 | RT#3 | RT#4 | RT#5 | RT#6 | RT#7 | RT#8 | RT#9 | RT#10 | RT#11 | RT#12 |
| B | RT#13 | RT#14 | RT#15 | RT#16 | RT#17 | RT#18 | RT#19 | RT#20 | RT#21 | RT#22 | RT#23 | RT#24 |
| … | …... | |||||||||||
| H | RT#85 | RT#86 | RT#87 | RT#88 | RT#89 | RT#90 | RT#91 | RT#92 | RT#93 | RT#94 | RT#95 | RT#96 |
| Plate#2 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | RT#97 | RT#98 | RT#99 | RT#100 | RT#101 | RT#102 | RT#103 | RT#104 | RT#105 | RT#106 | RT#107 | RT#108 |
| B | RT#109 | RT#110 | RT#111 | RT#112 | RT#113 | RT#114 | RT#115 | RT#116 | RT#117 | RT#118 | RT#119 | RT#120 |
| … | …... | |||||||||||
| H | RT#181 | RT#182 | RT#183 | RT#184 | RT#185 | RT#186 | RT#187 | RT#188 | RT#189 | RT#190 | RT#191 | RT#192 |
| Plate#3 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | RT#193 | RT#194 | RT#195 | RT#196 | RT#197 | RT#198 | RT#199 | RT#200 | RT#201 | RT#202 | RT#203 | RT#204 |
| B | RT#205 | RT#206 | RT#207 | RT#208 | RT#209 | RT#210 | RT#211 | RT#212 | RT#213 | RT#214 | RT#215 | RT#216 |
| … | …... | |||||||||||
| H | RT#277 | RT#278 | RT#279 | RT#280 | RT#281 | RT#282 | RT#283 | RT#284 | RT#285 | RT#286 | RT#287 | RT#288 |
| Plate#4 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | RT#289 | RT#290 | RT#291 | RT#292 | RT#293 | RT#294 | RT#295 | RT#296 | RT#297 | RT#298 | RT#299 | RT#300 |
| B | RT#301 | RT#302 | RT#303 | RT#304 | RT#305 | RT#306 | RT#307 | RT#308 | RT#309 | RT#310 | RT#311 | RT#312 |
| … | …... | |||||||||||
| H | RT#373 | RT#374 | RT#375 | RT#376 | RT#377 | RT#378 | RT#379 | RT#380 | RT#381 | RT#382 | RT#383 | RT#384 |
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| NaCl 5 M | Invitrogen | AM9759 |
| MgCl2 1 M | Invitrogen | AM9530G |
| Tris-HCl pH 7.5 1 M | Invitrogen | Cat# 15567-027 |
| Tris-HCl pH 8.0 1 M | Invitrogen | Cat# 15568-025 |
| Triton X-100 | Sigma | T8787 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat# 15713 |
| Nuclei EZ lysis buffer | Sigma | NUC101 |
| EDTA-free protease inhibitor tablets | Roche | Cat# 5892791001 |
| RNasin Plus Ribonuclease inhibitor | Promega | N2615 |
| SUPERase·In RNase inhibitor | Thermo Scientific | AM2696 |
| RNaseOUT RNase inhibitor | Thermo Scientific | Cat# 10777019 |
| Bovine serum albumin (BSA) (20 mg/mL) | NEB | B9000S |
| dNTP | Clontech | Cat# 639125 |
| SuperScript IV reverse transcriptase and buffer | Thermo Scientific | Cat# 18090050 |
| Quick Ligase and buffer | NEB | M2200L |
| Second-strand synthesis enzyme | NEB | E6111L |
| Dimethylformamide | Thermo Scientific | Cat# 20673 |
| USER enzyme | NEB | M5505L |
| NEBNext High-Fidelity 2× PCR Master Mix | NEB | M0541L |
| DNA binding buffer | Zymo | D4004-1-L |
| Nuclease-free H2O | Invitrogen | AM9932 |
| Unloaded Tn5 transposase | Diagenode | C01070002 |
| Glycerol | Sigma | G5516-1L |
| Elution buffer | Qiagen | Cat# 19086 |
| AMPure XP Reagent | Beckman Coulter | A63880 |
| Select-a-Size DNA Clean & Concentrator MagBead | Zymo | D4084 |
| RNaseZap Decontamination Solution | Invitrogen | AM9780 |
| Deposited data | ||
| Mendeley Data | This study | https://data.mendeley.com/datasets/59z97k52x7 |
| Experimental models: Organisms/strains | ||
| C57BL/6J mice (8- to 9-week-old male mice) | The Jackson Lab | Cat# 000664 |
| Oligonucleotides | ||
| Uniquely indexed oligos for reverse transcription, hairpin ligation and indexed PCR | IDT | Sequences available at https://github.com/JunyueC/sci-RNA-seq3_pipeline/blob/master/sci3_primer_sequences_plate.xls |
| ME_REV: [PHO]CTGTCTCTTATACACATCT | IDT | N/A |
| ME_A: GTCTCGTGGGCTCG GAGATGTGTATAAGAGACAG |
IDT | N/A |
| Other | ||
| 2 mL Dounce All-Glass Tissue Grinders and pestles | Kimble | Cat# 885300-0002 |
| 200-μm cell strainer | pluriSelect | Cat# 43-50200-03 |
| 40-μm cell strainer | pluriSelect | Cat# 43-50040-51 |
| Flowmi 40-μm cell strainer | Bel-Art | H13680-0040 |
| 96 Well LoBind PCR plates | Fisher Scientific | Cat# 0030129512 |
| Microseal PCR plate sealing film | Bio-Rad | MSB1001 |
Materials and equipment
Nuclei Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 7.5 (1 M) | 10 mM | 2,000 μL |
| NaCl (5 M) | 10 mM | 400 μL |
| MgCl2 (1 M) | 3 mM | 600 μL |
| Nuclease-free H2O | N/A | 197 mL |
| Total | N/A | 200 mL |
Nuclei Buffer can be stored at 4°C for at least 6 months.
Annealing Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 8.0 (1 M) | 40 mM | 400 μL |
| NaCl (5 M) | 50 mM | 100 μL |
| Nuclease-free H2O | N/A | 9.5 mL |
| Total | N/A | 10 mL |
Annealing Buffer can be stored at 4°C for at least 6 months.
Tagmentation Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 7.5 (1 M) | 20 mM | 200 μL |
| MgCl2 (1 M) | 10 mM | 100 μL |
| Dimethylformamide | 20% (v/v) | 2 mL |
| Nuclease-free H2O | N/A | 7.7 mL |
| Total | N/A | 10 mL |
Tagmentation Buffer can be stored at −20°C for at least 12 months.
CRITICAL: Dimethylformamide can be absorbed through the skin and cause adverse health effects. Avoid skin contact and prolonged exposure.
10% (v/v) Triton X-100
| Reagent | Final concentration | Amount |
|---|---|---|
| Triton X-100 | 10% | 1 mL |
| Nuclease-free H2O | N/A | 9 mL |
| Total | N/A | 10 mL |
10% Triton X-100 can be stored at 20°C–25°C for at least 12 months.
Step-by-step method details
Nuclei extraction from mouse kidneys and fixation
Timing: 1–2 h per sample
Timing: 30 min (for step 1)
Timing: ∼ 1 h (for step 2)
Timing: 30 min (for step 3)
Here we generate nuclear suspensions by performing tissue homogenization on mouse kidney with a dounce homogenizer and fix the extracted nuclei, which will then be frozen for future sci-RNA-seq3 processing. The nuclei isolation protocol is adapted from a previous study9 and optimized specifically for the purpose of extracting a large number of high-quality nuclei in a single preparation to satisfy the requirement of sci-RNA-seq3.
-
1.Prepare materials and equipment for nuclei extraction.
-
a.Prepare NLB1 buffer with recipe provided below. Place the buffer on ice.
-
i.Add one tablet of cOmplete Mini Protease Inhibitor Cocktail (EDTA-free) to 10 mL Nuclei EZ Lysis Buffer.
-
ii.Ensure the tablet is completely dissolved by gentle vortex at room temperature (20°C–25°C) for 10–15 min.NLB1
Reagent Amount Nuclei EZ Lysis Buffer supplemented with protease inhibitor 4 mL RNasin Plus Ribonuclease Inhibitor 20 μL SUPERase·In RNase Inhibitor 20 μL Note: Extra protease inhibitor-supplemented Nuclei EZ Lysis Buffer can be saved at −20°C for future use.
-
i.
-
b.Prepare NLB2 buffer with recipe provided below. Place the buffer on ice.NLB2
Reagent Amount Nuclei EZ Lysis Buffer (without protease inhibitor) 4 mL RNasin Plus Ribonuclease Inhibitor 4 μL SUPERase·In RNase Inhibitor 4 μL -
c.Clean the dounce tissue grinder, as well as the large and small grinder pestles, with RNaseZap Decontamination Solution and wash the grinder with RNase-free water.
-
d.Prepare Nuclei Isolation Buffer (NIB) with recipe provided below. Place the buffer on ice.Nuclei Isolation Buffer (NIB)
Reagent Amount Nuclei Buffer (see materials and equipment) 5 mL SUPERase·In RNase Inhibitor 50 μL BSA 100 μL Note: We recommend NIB to be freshly prepared at each time of use, though Nuclei Buffer can be prepared ahead. -
e.Prepare 4% paraformaldehyde (PFA) solution and place it on ice.
-
i.PFA should be diluted with Nuclei Buffer if the stock concentration is over 4%.
 CRITICAL: PFA reacts violently with strong oxidizers. Avoid inhalation and skin contact. Dispose unused PFA following proper hazardous waste procedures.
CRITICAL: PFA reacts violently with strong oxidizers. Avoid inhalation and skin contact. Dispose unused PFA following proper hazardous waste procedures.
-
i.
-
f.Precool a 6-cm dish, a razor blade, a 200-μm cell strainer, a 40-μm cell strainer and several 15-mL and 50-mL centrifuge tubes in the 4°C cold room.
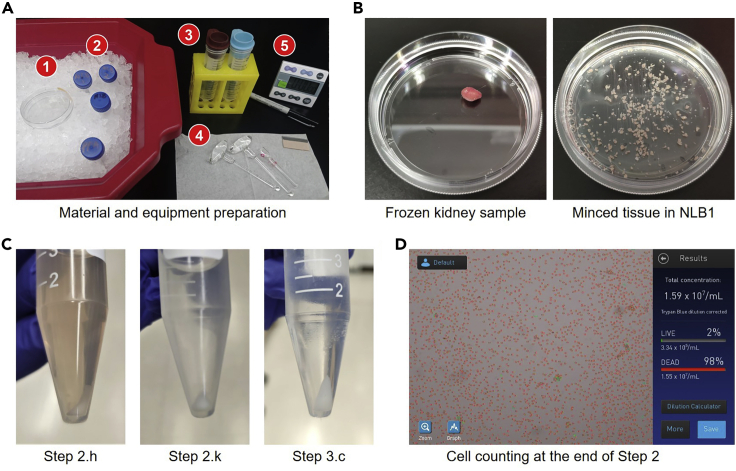
 CRITICAL: Kidney is a RNase-rich tissue and reduced library quality can be observed if RNA degradation occurs. Thus, the nuclei isolation experiment should be conducted in a 4°C cold room and it is important to ensure all reagents and equipment are pre-prepared to reduce the amount of time needed for tissue homogenization after the tissue is thawed. Figure 2A presents reagents and equipment that should be cleaned, pre-prepared and precooled in the 4°C cold room.
CRITICAL: Kidney is a RNase-rich tissue and reduced library quality can be observed if RNA degradation occurs. Thus, the nuclei isolation experiment should be conducted in a 4°C cold room and it is important to ensure all reagents and equipment are pre-prepared to reduce the amount of time needed for tissue homogenization after the tissue is thawed. Figure 2A presents reagents and equipment that should be cleaned, pre-prepared and precooled in the 4°C cold room.
-
a.
-
2.Tissue homogenization and cell lysis (all procedures at 4°C).
-
a.Thaw the kidney tissue (> 100 mg) stored at −80°C and immediately transfer the tissue onto the 6-cm dish (Figure 2B).
-
b.Add 2 mL NLB1 to the dish and keep the tissue exposed to NLB1.
-
c.Mince the tissue thoroughly with a razor blade and avoid big chunks of tissue (Figure 2B).
-
i.This step can take 2–5 min.
-
i.
-
d.Transfer 1–1.5 mL tissue suspension into the dounce grinder.
-
i.Move the small grinder pestle (loose pestle) up and down for 15 times to induce cell dissociation.
-
i.
-
e.Transfer homogenates in the dounce grinder onto a 200-μm cell strainer which is laid on a 50-mL centrifuge tube (Figure 2A).
-
i.Fibrotic kidney tissues, such as tissues harvested from the unilateral ureteral obstruction model, can require more times of grinding (20–25 times).
-
ii.Repeat this step until all tissue suspension in the 6-cm dish generated in step 2.c is processed for the dounce grinder.
-
i.
-
f.Transfer 1–1.5 mL filtered homogenates in the 50-mL tube (obtained in step 2.e) back into the dounce grinder.
-
i.Slowly press the large grinder pestle (tight pestle) up and down for 5 times to induce further nuclear dissociation.
-
ii.Transfer the nuclei suspension to a 15-mL centrifuge tube.
-
iii.Add another 1 mL NLB1. Incubate for 5 min.
-
iv.Repeat this step until all homogenates generated in step 2.e are processed for the dounce grinder.
-
i.
-
g.Filter the nuclei suspension with the 40-μm cell strainer.
-
i.Transfer the filtered nuclei suspension into a 15-mL centrifuge tube.
-
i.
-
h.Spin down the nuclei by centrifugation at 500 g for 4 min at 4°C. A representative picture of the nuclei pellet is shown in Figure 2C.Note: A refrigerated centrifuge with a swinging bucket should be used to increase the nuclei yield.
-
i.Remove supernatant and resuspend the nuclei pellet with 1.5 mL NLB2 by pipetting 10 times.
-
j.Mix the nuclei suspension with the rest of NLB2 and incubate for 5 min.
-
k.Spin down the nuclei by centrifugation at 500 g for 4 min at 4°C. A representative picture of the nuclei pellet is shown in Figure 2C.
-
l.Then, remove supernatant thoroughly and resuspend the nuclei with 2 mL NIB.Note: The number of fresh nuclei can be counted at this point. At least 15 million nuclei can be extracted from a kidney tissue of 100 mg.Note: Count the number of nuclei with either an automated cell counter or a hemacytometer. We usually use an automated cell counter (e.g., Countess II Automated Cell Counter) because it is more efficient in processing multiple samples. A representative snapshot of nuclei counting with the Countess Cell Counter at this step is shown in Figure 2D, where nuclei are stained with Trypan Blue following the manufacture’s instruction and the majority of nuclei are marked as “Dead” cells with this instrument.
-
a.
-
3.Nuclei fixation and preservation.
-
a.Add 3 mL ice-cold 4% PFA to the 2 mL nuclei suspension for a final concentration of 2.4%.
-
i.Mix thoroughly by inverting the tube for 3 times.
-
i.
-
b.Fix the nuclei on ice for 10 min.
-
c.Spin down the fixed nuclei by centrifugation at 500 g for 5 min at 4°C.
-
i.We may observe the nuclei pellet with a larger size (compared with the nuclei pellet obtained in previous steps) due to reduced nuclei density after fixation (Figure 2C).
-
ii.Remove supernatant and wash with 2 mL NIB.
-
i.
-
d.Resuspend the nuclei pellet with 300 μL NIB.
-
e.Count the number of nuclei.Note: PFA fixation in step 3.b and buffer washes in step 3.c can cause 10%–20% nuclei loss at each step. Thus, we may observe over 40% of cell loss at step 3.e compared to the fresh nuclei obtained at step 2.l. With a kidney tissue over 100 mg, we typically obtain at least 8 million fixed nuclei at this point.
-
f.Dilute the nuclei with NIB to a final concentration of 5–10 million/mL.
-
g.Aliquot the nuclei suspension to several cryogenic tubes with 300 μL materials per tube.
 Pause point: Flash freeze the nuclei suspension with liquid nitrogen. Store the sample in a liquid nitrogen tank for future sci-RNA-seq3 processing.
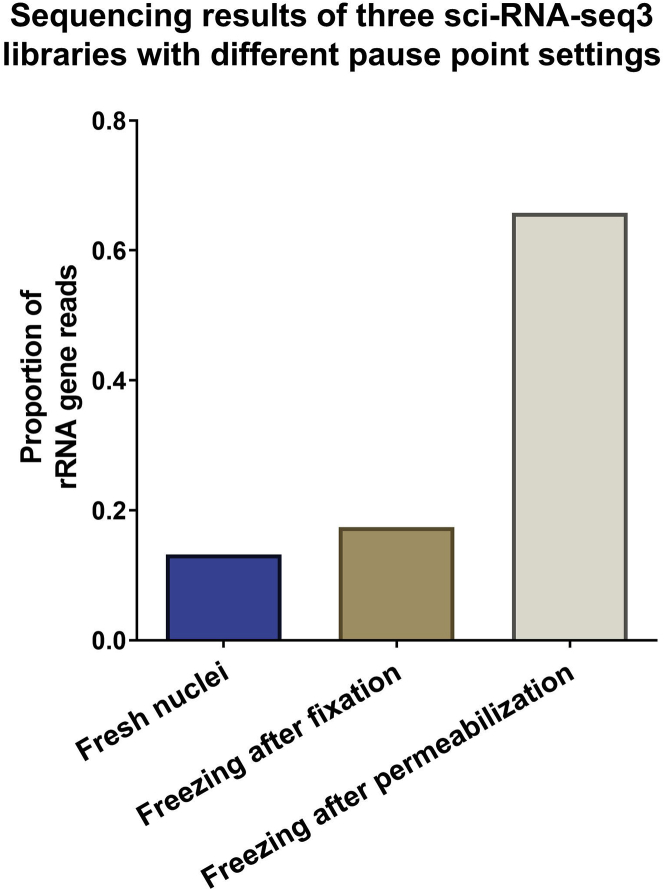
Pause point: Flash freeze the nuclei suspension with liquid nitrogen. Store the sample in a liquid nitrogen tank for future sci-RNA-seq3 processing. CRITICAL: A pause point is needed to collect samples from multiple mouse kidneys and process them together in sci-RNA-seq3 of the next section. This is the only pause point (i.e., after fixation and before permeabilization in step 5) we can include before starting the sci-RNA-seq3 experiment. Compared to processing fresh and unfixed nuclei, freezing nuclei after PFA fixation can offer comparable library quality. On the other hand, we found that freezing nuclei after permeabilization caused significant ribosomal RNA contamination and reduced library quality, as presented in Figure 3.
CRITICAL: A pause point is needed to collect samples from multiple mouse kidneys and process them together in sci-RNA-seq3 of the next section. This is the only pause point (i.e., after fixation and before permeabilization in step 5) we can include before starting the sci-RNA-seq3 experiment. Compared to processing fresh and unfixed nuclei, freezing nuclei after PFA fixation can offer comparable library quality. On the other hand, we found that freezing nuclei after permeabilization caused significant ribosomal RNA contamination and reduced library quality, as presented in Figure 3.
-
a.
Figure 2.
Overview of mouse kidney nuclei isolation and fixation procedures
(A) Before performing nuclei isolation on the mouse kidney tissue, all reagents and equipment should be prepared ahead and precooled in a 4°C cold room. This includes: (1) a 6-cm dish, (2) buffers of NLB1, NLB2, NIB and 4% PFA, (3) a 200-μm cell strainer and a 40-μm cell strainer, assembled on 50-mL tubes, (4) a tissue dounce homogenizer and the large and small pestles, and (5) supplies such as a razor blade, a timer and a marker pen. We also assume that common laboratory supplies, including a P1000 pipette, 1,250-μL pipet tips and tube racks, are available at the 4°C cold room.
(B) A frozen mouse kidney tissue is transferred onto a 6-cm dish (left panel) and minced with a razor blade in the NLB1 buffer.
(C) Representative pictures of nuclei pellets at specific steps, in the same experiment processing the tissue presented in (B). At step 2.h, the supernatant contains tissue lysates. At step 2.k, the supernatant becomes clear. Fixation can reduce the density of nuclei, and therefore, a large size of nuclei pellet may be observed at step 3.c.
(D) A representative Countess II Automated Cell Counter profile which counts the concentration of fresh nuclei obtained the end of step 2, in the same experiment processing the tissue presented in (B). In this case, we obtained 2 mL fresh nuclei suspension with concentration over 15 million/mL, and therefore, over 30 million nuclei were isolated.
Figure 3.
Quality of sci-RNA-seq3 libraries with different pause point settings
The proportion of ribosomal RNA (rRNA) reads of three sci-RNA-seq3 libraries. In the first library, nuclei were not fixed and processed directly for subsequent reactions (steps 4–14). In the second library, nuclei were fixed and then frozen in liquid nitrogen following procedures of step 3. In the third library, nuclei were fixed and permeabilized right after fixation (steps 3 and 5) and then frozen in liquid nitrogen. The three libraries were generated from pilot experiments as proposed in step 15.
Reverse transcription and hairpin ligation of sci-RNA-seq3
Timing: 1 day
Here we will process the fixed and permeabilized nuclei from multiple mouse kidney samples for reverse transcription and hairpin ligation of sci-RNA-seq3. This protocol is adapted from the original sci-RNA-seq publication2 and includes modifications in nuclei permeabilization and sonication and described in more detail.
Note: Before you start this section, make sure you have prepared the oligo plates as mentioned in ‘before you begin’ and obtained fixed nuclei from all samples in your study cohort following steps 1–3 in ‘nuclei extraction from mouse kidneys and fixation’.
Note: A large number of samples may be processed simultaneously in this section, and therefore, previewing this protocol and labeling tubes with sample identifications ahead of the experiment are recommended.
-
4.Buffer preparation.
-
a.Prepare 35 mL NIB with recipe provided below. Place the buffer on ice.Nuclei Isolation Buffer (NIB)
Reagent Amount Nuclei Buffer (see materials and equipment) 35 mL SUPERase·In RNase Inhibitor 350 μL BSA 700 μL -
b.Prepare Nuclei Buffer with BSA (NBB) with recipe provided below. Place the buffer on ice.Nuclei Buffer with BSA (NBB)
Reagent Amount Nuclei Buffer (see materials and equipment) 95 mL BSA (final concentration 2% (v/v)) 1.9 mL -
c.Prepare permeabilization buffer with recipe provided below. Place the buffer on ice.Permeabilization buffer
Reagent Amount 10% Triton X-100 (see materials and equipment) 300 μL NIB 11.7 mL
-
a.
-
5.Nuclei permeabilization.
-
a.For each sample, take one vial of fixed nuclei in a cryogenic tube (300 μL; generated in step 3.g) out of the liquid nitrogen tank.Note: All samples are thawed in 37°C water bath for 30–60 s and immediately placed on ice.
-
b.For each sample, transfer the nuclei suspension into a 15-mL centrifuge tube.
-
c.Spin down the nuclei of all samples by centrifugation at 500 g for 5 min at 4°C.
-
d.Remove supernatant, resuspend the nuclei thoroughly with 100 μL NIB and add 400 μL permeabilization buffer to each sample.
-
e.Incubate the mix on ice for 5 min.
-
f.Spin down the nuclei by centrifugation at 500 g for 5 min at 4°C.
-
g.Carefully remove supernatant and resuspend the pellet with 250 μL NIB and transfer the nuclei suspension to a 1.5 mL tube for each sample.
-
h.Place the 1.5 mL tubes in a Bioruptor Pico Sonication device and perform sonication for 10 s.
-
i.For each sample, filter the nuclei suspension through a 40-μm Flowmi cell strainer and place the sample on ice. Please refer to the manufacturer’s instructions for proper use of a Flowmi cell strainer: https://www.belart.com/media/catalogstudio/Instructions/913680015.pdf
 CRITICAL: Permeabilization of fixed nuclei can cause significant nuclei clumping. Performing light sonication at step 5.h is critical to break up these nuclei aggregates and reduce nuclei loss in the subsequent filtration step 5.i.
CRITICAL: Permeabilization of fixed nuclei can cause significant nuclei clumping. Performing light sonication at step 5.h is critical to break up these nuclei aggregates and reduce nuclei loss in the subsequent filtration step 5.i. -
j.Measure and record the concentration of nuclei of each sample. Dilute each sample to around 3.6 × 106 nuclei/mL with NIB.Note: This will help us to aliquot 80,000 nuclei (in 22 μL suspension) in the next step.
-
a.
-
6.384-well reverse transcription.
-
a.Thaw the four 96-well plates (Table 1) for reverse transcription prepared ahead of time.
-
i.Each well contains 2 μL uniquely indexed RT oligos (100 μM). Briefly centrifuge the plates.
-
i.
-
b.Add 2 μL dNTP (10 mM) into each of the 384 wells. Briefly centrifuge the plates.Note: A multichannel pipette and pipette reservoirs can be used to accelerate reagent deposition.
-
c.Add around 80,000 nuclei (in 22 μL suspension) into each of the 384 wells.
 CRITICAL: The well positions where nuclei from each sample are deposited must be recorded since nuclei from different samples will be indexed with different sets of RT oligos for the purpose of sample hashing. Table 2 presents an example of depositing nuclei from a total of 16 samples into each of the 384 wells (24 wells per sample). Table 2 can be combined with Table 1 to create a “Sample-RT oligo” look-up table for downstream sample demultiplexing in data analysis.Note: Add the 80,000 nuclei to the bottom of each well since the plates may not be further centrifuged.
CRITICAL: The well positions where nuclei from each sample are deposited must be recorded since nuclei from different samples will be indexed with different sets of RT oligos for the purpose of sample hashing. Table 2 presents an example of depositing nuclei from a total of 16 samples into each of the 384 wells (24 wells per sample). Table 2 can be combined with Table 1 to create a “Sample-RT oligo” look-up table for downstream sample demultiplexing in data analysis.Note: Add the 80,000 nuclei to the bottom of each well since the plates may not be further centrifuged. -
d.Incubate the four plates at 55°C for 5 min and immediately place them on ice after incubation.
-
e.Prepare reverse transcription reaction mix with recipe provided below.Reverse transcription reaction mix
Reagent Amount 5× Superscript IV First-Strand Buffer 3225.6 μL DTT (100 mM) 806.4 μL SuperScript IV reverse transcriptase 806.4 μL RNaseOUT Recombinant Ribonuclease Inhibitor 806.4 μL -
f.Distribute 14 μL reaction mix into each well with a multichannel pipette.
-
i.The final volume per well is 40 μL.
-
i.
-
g.Start the reverse transcription reaction with the following program.Reverse transcription thermocycling conditions
Temperature Time 4°C 2 min 10°C 2 min 20°C 2 min 30°C 2 min 40°C 2 min 50°C 2 min 55°C 15 min 4°C forever -
h.Place the plates on ice and add 60 μL NBB into each well with a multichannel pipette to dilute the nuclei suspension.
-
i.Pool the nuclei suspension from all 384 wells together into a reservoir.
-
j.Split the nuclei suspension into two 50-mL tubes.
-
k.Centrifuge at 500 g for 10 min at 4°C and remove supernatant carefully.
-
a.
-
7.384-well hairpin ligation.
-
a.Thaw the four 96-well plates for hairpin ligation prepared ahead of time. Briefly centrifuge the plates.
-
b.Prepare ligation reaction mix with recipe provided below.Ligation reaction mix
Reagent Amount Quick ligase buffer 8,064 μL Quick ligase 806.4 μL -
c.Distribute 22 μL reaction mix into each of the 384 wells with a multichannel pipette.
-
d.Briefly centrifuge the plates.
-
e.Resuspend the nuclei pellet obtain in step 6.k with 4.3 mL NIB thoroughly.
-
f.Distribute 10 μL nuclei suspension into each of the 384 wells with a multichannel pipette.Note: Add the nuclei to the bottom of each well since the plates may not be further centrifuged.
-
g.Perform ligation reaction by incubating the plates at 25°C for 10 min.
-
h.After the reaction, add 60 μL NBB into each well with a multichannel pipette to dilute the nuclei suspension.
-
i.Pool the nuclei suspension from all 384 wells together into a reservoir.
-
j.Split the nuclei suspension into two 50-mL tubes and add another 20 mL NBB to each tube.
-
k.Centrifuge at 600 g for 10 min and remove supernatant.
-
l.For each 50-mL tube, resuspend the nuclei pellet with 2.5 mL NBB and combine them into one 15-mL tube.
-
m.Centrifuge at 600 g for 10 min and remove supernatant.
-
n.Resuspend the nuclei pellet with 4 mL NBB.
-
o.Filter the nuclei suspension with 40-μm Flowmi cell strainers. Then determine the concentration of nuclei with either an automated cell counter or a hemacytometer.Note: Please refer to the manufacturer’s instructions for proper use of a Flowmi cell strainer.
-
p.Dilute the nuclei suspension to 600–800 nuclei/μL with NBB.
-
q.Distribute 5 μL nuclei suspension (3,000–4,000 nuclei) into each well of several 96-well plates.
-
r.Seal the plates and briefly centrifuge the plates.
 Pause point: The plates can be stored at −80°C for at least a month.Note: We typically obtained a total of 8–12 plates at this point.Note: One plate is used to generate one sub-library in the next section.
Pause point: The plates can be stored at −80°C for at least a month.Note: We typically obtained a total of 8–12 plates at this point.Note: One plate is used to generate one sub-library in the next section.
-
a.
Table 2.
An example of sample deposition locations in four RT plates
| Plate#1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 |
| B | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 | Kidney#1 |
| … | …... | |||||||||||
| H | Kidney#4 | Kidney#4 | Kidney#4 | Kidney#4 | Kidney#4 | Kidney#4 | Kidney#4 | Kidney#4 | Kidney#4 | Kidney#4 | Kidney#4 | Kidney#4 |
| Plate#2 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 |
| B | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 | Kidney#5 |
| … | …... | |||||||||||
| H | Kidney#8 | Kidney#8 | Kidney#8 | Kidney#8 | Kidney#8 | Kidney#8 | Kidney#8 | Kidney#8 | Kidney#8 | Kidney#8 | Kidney#8 | Kidney#8 |
| Plate#3 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 |
| B | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 | Kidney#9 |
| … | …... | |||||||||||
| H | Kidney#12 | Kidney#12 | Kidney#12 | Kidney#12 | Kidney#12 | Kidney#12 | Kidney#12 | Kidney#12 | Kidney#12 | Kidney#12 | Kidney#12 | Kidney#12 |
| Plate#4 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 |
| B | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 | Kidney#13 |
| … | …... | |||||||||||
| H | Kidney#16 | Kidney#16 | Kidney#16 | Kidney#16 | Kidney#16 | Kidney#16 | Kidney#16 | Kidney#16 | Kidney#16 | Kidney#16 | Kidney#16 | Kidney#16 |
sci-RNA-seq3 sub-library generation
Timing: 1 day per sub-library
In the above section, two levels of combinatorial indexing have been introduced through reverse transcription (384 barcodes) and hairpin ligation (384 barcodes). A third-level combinatorial indexing will be introduced by indexed PCR in this sub-library generation step. In this section, each sub-library is generated by processing one 96-well plate obtained in step 7.r and introduces 96 unique PCR primer combinations. Therefore, if a number of N sub-libraries are generated in this experiment, the total number of barcode combinations will be: 384×384×96×N.
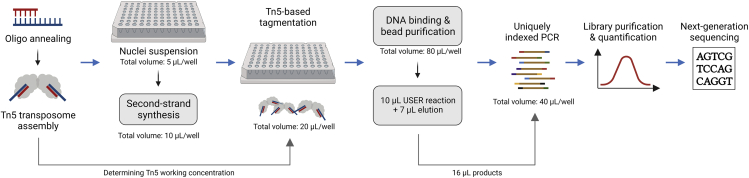
The sub-library generation protocol includes second-strand synthesis, transposase Tn5-based tagmentation, USER (Uracil-Specific Excision Reagent) reaction, indexed PCR and library purification (Figure 4).
Figure 4.
Scheme of sci-RNA-seq3 sub-library generation workflow
A workflow of sci-RNA-seq3 sub-library generation. All experiments are performed on a 96-well plate before library purification and quantification. The volume per well at each step is labeled.
In the tagmentation step, the Tn5 must be pre-loaded with specific oligonucleotides to make functional transposase. To the best of our knowledge, the Tn5 transposome specifically used in the original sci-RNA-seq3 paper is not commercially available, and therefore, we will present a protocol for generating this construct with commercially available naked Tn5 (Figure 5).
-
8.Tn5 transposome assembly (Figure 5).
 Timing: 2–3 h
Note: The Tn5 transposome can be prepared once and used for generation of all sub-libraries.Note: Both unmodified Tn5 and protein A-fused Tn5 can be used for tagmentation. We have validated this protocol on two different sources of naked Tn5 (Diagenode C01070002 and Lucigen TNP92110).
Timing: 2–3 h
Note: The Tn5 transposome can be prepared once and used for generation of all sub-libraries.Note: Both unmodified Tn5 and protein A-fused Tn5 can be used for tagmentation. We have validated this protocol on two different sources of naked Tn5 (Diagenode C01070002 and Lucigen TNP92110).-
a.Resuspend lyophilized ME_REV and ME_A oligos with Annealing Buffer (see materials and equipment) to a stock concentration of 100 μM.
 CRITICAL: Using HPLC-purified oligos for transposome assembly is highly recommended.
CRITICAL: Using HPLC-purified oligos for transposome assembly is highly recommended. -
b.Mix 5 μL ME_REV (100 μM) and 10 μL ME_A (100 μM) in a PCR tube.
-
c.Briefly spin down and start the oligo annealing reaction with the following program.Oligo annealing thermocycling conditions
Temperature Time 95°C 5 min Cool to 65°C −0.1°C/s 65°C 5 min Cool to 4°C −0.1°C/s 4°C forever -
d.Add 10 μL naked Tn5 transposase to 12.5 μL annealed oligos generated above.
-
e.Mix well and briefly centrifuge the tube.
-
f.Incubate the Tn5-oligo mix at 23°C for 30–40 min.Optional: Performing the reaction with gentle shaking (50–300 rpm) on a thermomixer may moderately improve the efficiency of transposome assembly.
-
g.After the reaction, supplement the suspension with 12.5 μL glycerol and mix well.Note: The loaded transposome can be stored at −20°C for at least a month.
-
a.
-
9.Second-strand synthesis.
-
a.Thaw one 96-well plate generated in step 7.s at room temperature (20°C–25°C).
-
i.Each well contains 5 μL nuclei suspension. Briefly centrifuge the plate.
-
i.
-
b.Prepare second-strand synthesis reaction mix with recipe provided below.Second-strand synthesis reaction mix
Reagent Amount Elution buffer 297 μL Second-strand synthesis buffer 132 μL Second-strand synthesis enzyme 66 μL -
c.Distribute 5 μL second-strand synthesis reaction mix into each of the 96 wells with a multichannel pipette.
-
i.The final volume per well is 10 μL.
-
i.
-
d.Vortex and briefly centrifuge the plate.
-
e.Seal the plate and perform second-strand synthesis at 16°C for 3 h.
-
a.
-
10.Tn5-based tagmentation.
-
a.Dilute 2 μL oligo-loaded Tn5 transposome generated in step 8.g with Tagmentation Buffer (see materials and equipment) to a final concentration of 100 nM.Note: Contact the provider of naked Tn5 for its stock concentration and calculate the concentration of assembled transposome with an example presented below.Tn5 dilution ratio
Description Calculation Example Stock concentration ε μM 50 μM Transposome concentration (step 8.g) 0.286ε μM 14.3 μM Volume added to make a final 100 nM concentration (step 10.a) (5.71ε – 2) μL 283.5 μL -
b.Further dilute the 100 nM transposome with Tagmentation Buffer to the working concentration.
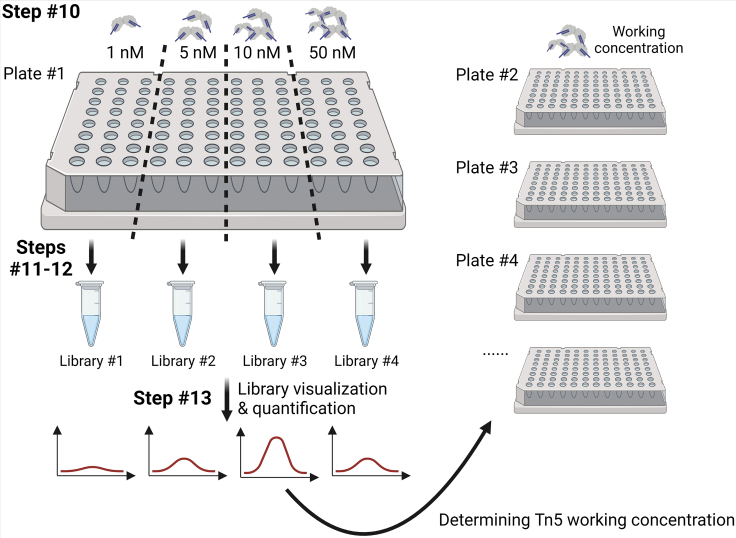
 CRITICAL: The baseline activity of Tn5 varies substantially across different manufacturers or even across different batches from the same manufacturer, and different amounts of DNA inputs may also require different Tn5 doses for tagmentation. Therefore, it is critical to determine the optimal Tn5 concentration of use (working concentration) when generating the first sci-RNA-seq3 sub-library with an activity titration test. An example of implementing the titration test is presented in Figure 6. After a working concentration is determined, keep using Tn5 at this concentration when generating the other sub-libraries.
CRITICAL: The baseline activity of Tn5 varies substantially across different manufacturers or even across different batches from the same manufacturer, and different amounts of DNA inputs may also require different Tn5 doses for tagmentation. Therefore, it is critical to determine the optimal Tn5 concentration of use (working concentration) when generating the first sci-RNA-seq3 sub-library with an activity titration test. An example of implementing the titration test is presented in Figure 6. After a working concentration is determined, keep using Tn5 at this concentration when generating the other sub-libraries. -
c.Vortex and briefly centrifuge the plate after the reaction in step 9.e.
-
i.Now each of the 96 wells contains 10 μL products post second-strand synthesis.
-
ii.Place the plate on ice and add 10 μL Tn5 transposome at the working concentration into each of the 96 wells with a multichannel pipette.
-
i.
-
d.Incubate the plate at 55°C for 5 min.
-
e.Add 20 μL DNA binding buffer to each well with a multichannel pipette.
-
i.The final volume per well is approximately 40 μL.
-
ii.Vortex and briefly centrifuge the plate.
-
i.
-
f.Incubate the plate at room temperature (20°C–25°C) for 5 min and then briefly centrifuge the plate.
-
a.
-
11.Bead purification and USER reaction.
-
a.Perform AMPure XP bead purification.
-
i.Add 40 μL AMPure XP beads (i.e., 1× beads) to each well with a multichannel pipette and mix well.
-
ii.Incubate at room temperature (20°C–25°C) for 5 min and then briefly centrifuge the plate.
-
iii.Place the plate on a magnet for 5 min and then carefully remove supernatant.
-
iv.Wash each well with 100 μL 80% ethanol with a multichannel pipette while keeping the plate on the magnet.
-
v.Incubate for 30 s and then remove supernatant.
-
vi.Repeat steps 11.a.iv–11.a.v for the second wash.
-
vii.Remove the plate from the magnet and place the plate at room temperature (20°C–25°C) for 5 min to let residual ethanol evaporate.
-
i.
-
b.Prepare USER reaction mix with recipe provided below.USER reaction mix
Reagent Amount Nuclease-free water 864 μL 10× USER CutSmart buffer 108 μL USER enzyme 108 μL -
c.Add 10 μL USER reaction mix to each well (containing beads only) with a multichannel pipette.
-
d.Mix the beads with USER reaction mix by pipetting for 10–20 times.
-
e.Seal the plate and incubate the plate at 37°C for 15 min.
-
f.Vortex and briefly centrifuge the plate.
-
g.Add 7 μL Elution buffer to each well with a multichannel pipette.
-
h.Vortex and briefly centrifuge the plate.
-
i.Place the plate on the magnet for 5 min.
-
j.Transfer 16 μL supernatant of each well to a new 96-well plate.
-
k.Briefly centrifuge the new plate and place it on ice.
-
a.
-
12.Indexed PCR.
-
a.Incubate the plate obtained in step 11.k at 80°C for 10 min.
-
b.Add 2 μL uniquely indexed P5 primers (10 μM) and 2 μL uniquely indexed P7 primers (10 μM) into each well.
-
i.At this point, the volume per well is 20 μL. The final volume for PCR is 40 μL per well, so the final concentration of P5/P7 primers is 0.5 μM.
 CRITICAL: The combination of indexed P5 and P7 primers in a well must be different from the combination of any other wells, including wells of the other plates for generation of the other sub-libraries. For example, if the combination of P5_#1 and P7_#1 is used in generating the first sub-library, this combination cannot not be used for downstream steps when generating the other sub-libraries. A feasible example is to add the 96 uniquely indexed P7 primers into the 96-well plate when generating all sub-libraries, but choose different P5 primers for different sub-libraries, as presented in Table 3.Optional: Primers with a working concentration (10 μM) can be pre-prepared from stock primers (100 μM).
CRITICAL: The combination of indexed P5 and P7 primers in a well must be different from the combination of any other wells, including wells of the other plates for generation of the other sub-libraries. For example, if the combination of P5_#1 and P7_#1 is used in generating the first sub-library, this combination cannot not be used for downstream steps when generating the other sub-libraries. A feasible example is to add the 96 uniquely indexed P7 primers into the 96-well plate when generating all sub-libraries, but choose different P5 primers for different sub-libraries, as presented in Table 3.Optional: Primers with a working concentration (10 μM) can be pre-prepared from stock primers (100 μM).
-
i.
-
c.Add 20 μL NEBNext High-Fidelity PCR Master Mix into each well. The recipe of indexed PCR reaction mix is presented below.Indexed PCR reaction mix
Reagent Amount NEBNext High-Fidelity PCR Master Mix 20 μL Indexed P5 primer (10 μM) 2 μL Indexed P7 primer (10 μM) 2 μL DNA products (step 11.k) 16 μL -
d.Briefly centrifuge the plate and start the PCR reaction with the following program.Indexed PCR cycling conditions
Steps Temperature Time Cycles Initial extension 72°C 5 min 1 Initial Denaturation 98°C 30 s 1 Denaturation 98°C 10 s 13–15 cycles Annealing 66°C 30 s Extension 72°C 30 s Final extension 72°C 5 min 1 Hold 4°C Forever Note: A 14-cycle PCR reaction can be used in the Tn5 activity titration test when generating the first sub-library. The cycle number can be adjusted according to the library quantification result in the next section. Pause point: Pool PCR products from all 96 wells together in a 15-mL tube.Note: The PCR products can be stored at 4°C for 1–2 days or at −20°C for at least a month.Note: For the Tn5 activity titration test, only pool PCR products generated from the same condition (Figure 6). In the example of Figure 6, a total of 900–960 μL PCR products can be obtained in each condition. For subsequent sub-library generation, a total of 3,600–3,840 μL PCR products can be obtained.
Pause point: Pool PCR products from all 96 wells together in a 15-mL tube.Note: The PCR products can be stored at 4°C for 1–2 days or at −20°C for at least a month.Note: For the Tn5 activity titration test, only pool PCR products generated from the same condition (Figure 6). In the example of Figure 6, a total of 900–960 μL PCR products can be obtained in each condition. For subsequent sub-library generation, a total of 3,600–3,840 μL PCR products can be obtained.
-
a.
-
13.Library purification and quantification.
-
a.Thaw pooled PCR products generated in step 12 and transfer 900 μL to a 2-mL tube.Note: The remaining PCR products can be stored at −20°C for troubleshooting.
-
b.Perform library purification with 0.8× Select-a-Size MagBeads.
-
i.Add 720 μL beads and mix thoroughly. Incubate at room temperature (20°C–25°C) for 5 min.
-
ii.Separate the beads by placing the tube against a magnet for 5 min.
-
iii.Remove supernatant and wash the beads twice with 200 μL 80% ethanol while keeping the tube on the magnet.
-
iv.Carefully remove all ethanol and air dry for 3–5 min.
-
v.Remove the tube from the magnet. Add 100 μL Elution buffer and mix with the beads thoroughly.
-
vi.Incubate at room temperature (20°C–25°C) for 3–5 min.
-
vii.Separate the beads by placing the tube on the magnet for 3 min.
-
viii.Transfer all supernatant carefully to a new 1.5-mL tube.
-
i.
-
c.Perform a second-round 0.7× Select-a-Size MagBead purification by adding 70 μL beads and repeat procedures of step 13.b.
-
d.Elute the sub-library with 20 μL Elution buffer.
 Pause point: The sub-library can be stored at −20°C for several months.Visualize the sub-library on a Bioanalyzer or Tapestation instrument (see expected outcomes). Determine the concentration of the sub-library. The library typically has an average insert at 300–600 bp, with concentration varied between 5–50 nM.
Pause point: The sub-library can be stored at −20°C for several months.Visualize the sub-library on a Bioanalyzer or Tapestation instrument (see expected outcomes). Determine the concentration of the sub-library. The library typically has an average insert at 300–600 bp, with concentration varied between 5–50 nM.
-
a.
-
14.Next-generation sequencing.
-
a.Balance all sub-libraries to the same molarity to ensure that sequencing power is distributed evenly to each sub-library.
-
b.Pool all sub-libraries together and sequence on a NovaSeq 6000 platform (Read1-Index1-Index2-Read2: 34-10-10-100 bp). Table 4 presents an example of pooling 8 sub-libraries with different concentrations.Note: A higher number of sub-libraries pooled will result in higher number of cells and will require a higher sequencing power. We typically sequence at least 8 sub-libraries on one flow cell of NovaSeq S4 to increase data throughput.Optional: One sub-library can be sequenced on a NextSeq platform (Read1-Index1-Read2: 34-10-46 bp) if we want to briefly check the quality of library before proceeding to a full sequencing depth. Such data can usually be used for standard downstream analysis such as cell clustering. An even lower sequencing depth (e.g., sequencing a sub-library on a MiSeq platform) may also be used if we just want to check the quality of captured reads and whether oligo barcodes are successfully incorporated.Note: Please contact the sequencing service center for a recommended final volume and concentration of the pooled library.
-
c.Raw undemultiplexed fastq files should be expected.
-
a.
Figure 5.
Scheme of Tn5 transposome assembly for sci-RNA-seq3
Two oligos (ME_REV and ME_A) are annealed and assembled with naked Tn5 transposase to make functional transposome for tagmentation.
Figure 6.
An example of Tn5 activity titration test to determine the working concentration
When processing the first 96-well plate for sci-RNA-seq sub-library generation, four different concentrations of Tn5 transposome are tested, with 24 wells per condition. The working concentration of Tn5 transposome usually varies between 1–50 nM. After steps 10–12, PCR products from 24 wells are pooled together and processed for sub-library generation in step 13. The four sub-libraries are visualized and quantified. The sub-library with highest concentration is chosen and the working concentration of Tn5 transposome is determined and can be used for generation of the other sub-libraries. Figure created with biorender.com.
Table 3.
An example of depositing indexed P5 and P7 PCR primers in sub-library generation
| P7 primers (a total of 96) used for all sub-libraries | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plate | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| A | P7_#1 | P7_#2 | P7_#3 | P7_#4 | P7_#5 | P7_#6 | P7_#7 | P7_#8 | P7_#9 | P7_#10 | P7_#11 | P7_#12 |
| B | P7_#13 | P7_#14 | P7_#15 | P7_#16 | P7_#17 | P7_#18 | P7_#19 | P7_#20 | P7_#21 | P7_#22 | P7_#23 | P7_#24 |
| … | …... | |||||||||||
| H | P7_#85 | P7_#86 | P7_#87 | P7_#88 | P7_#89 | P7_#90 | P7_#91 | P7_#92 | P7_#93 | P7_#94 | P7_#95 | P7_#96 |
| One unique P5 primer added to all 96 wells of each sub-library | ||||||||||||
| Sub-library #1 | P5_#1 | |||||||||||
| Sub-library #2 | P5_#2 | |||||||||||
| Sub-library #3 | P5_#3 | |||||||||||
| Sub-library #4 | P5_#4 | |||||||||||
| … | …... | |||||||||||
Table 4.
An example of balancing and pooling sub-libraries
| Sublibrary | #1 | #2 | #3 | #4 | #5 | #6 | #7 | #8 |
|---|---|---|---|---|---|---|---|---|
| Concentration (nM) | 40 | 40 | 40 | 30 | 30 | 20 | 20 | 20 |
| Volume used (μL) | 5 | 5 | 5 | 6.67 | 6.67 | 10 | 10 | 10 |
| H2O added for dilution (μL) | 5 | 5 | 5 | 3.33 | 3.33 | 0 | 0 | 0 |
| Final concentration | 20 nM | |||||||
Performing a pilot small-scale sci-RNA-seq3 experiment
Timing: 3–5 days
Following the above procedures, researchers should be able to generate a high-throughput and a highly multiplexed sci-RNA-seq3 library which includes a total of 384×384×96×N barcode combinations, where N is the number of sub-libraries. However, generating such a large-scale library requires preparation of large amount of experimental materials (e.g., mouse kidney tissues and oligonucleotides) and is not financially efficient for pilot tests. Therefore, in this section, we describe how to generate a small-scale sci-RNA-seq3 library with a total of 10×10×20 barcode combinations to help researchers to reproduce this protocol.
-
15.Nuclei isolation from one mouse kidney and sci-RNA-seq3 with reduced scale.
-
a.Perform nuclei isolation and fixation from one mouse kidney tissue following steps 1–3.
-
b.Prepare 4 mL NIB, 4 mL NBB and 400 μL permeabilization buffer at step 4.
-
c.Perform nuclei permeabilization on fixed nuclei generated from the tissue of use following step 5. At least 800,000 nuclei should be retained at the end of this step.
-
d.Distribute the nuclei suspension into 10 wells of a 96-well plate for reverse transcription. Add a uniquely indexed RT oligo into each of the 10 wells. Prepare 154 μL reverse transcription reaction mix and follow the other procedures of step 6 accordingly.
-
e.Distribute the nuclei suspension into 10 wells of a 96-well plate for hairpin ligation.
-
i.Add a uniquely ligation oligo into each of the 10 wells.
-
ii.Prepare 231 μL ligation reaction mix and follow the other procedures of step 7 accordingly.
-
iii.The nuclei suspension can be distributed to at least 20 wells of a 96-well plate (3,000–4,000 nuclei in 5 μL per well) at the end of this step.
-
i.
-
f.Follow procedures mentioned in steps 8–11 accordingly. The Tn5 activity titration test can be omitted since the aim of this pilot experiment is to reproduce its chemistry instead of achieving optimal library complexity. The Tn5 transposome can be used as a final concentration of 10 nM for tagmentation.
-
g.Perform indexed PCR by adding 20 different P5/P7 primer combinations to the 20 wells. The PCR cycling number can be set as 16 to ensure that the library is concentrated enough for next-generation sequencing. Follow the other procedures of step 12 accordingly.
-
h.Process approximately 800 μL PCR products and follow the other procedures of step 13 accordingly.
-
i.We can sequence the library on a MiSeq platform or with a spike-in approach (Read1-Index1-Index2-Read2: 34-10-10-100 bp) to obtain a relatively small number of reads for quality check.
-
a.
Note: The sequencing result (fastq files) can be analyzed to examine whether all 10×10×20 barcode combinations have been successfully incorporated, whether reads can be mapped to the reference genome and read quality, etc., but may not be used for downstream analysis such as cell clustering.
As a summary, we present the major optimizations made in this protocol in Table 5.
Table 5.
Optimized procedures made in this protocol and comparison with the original protocol
| Steps | Procedures of the original protocol | Optimizations made in this protocol |
|---|---|---|
| Tissue homogenization | With the rubber tip of a syringe plunger | With a dounce homogenizer |
| Lysis buffer | Homemade buffer supplemented with RNase inhibitor | Nuclei EZ Lysis Buffer supplemented with both RNase and protease inhibitors |
| Nuclei filtration | 40-μm cell strainer | First 200-μm and then 40-μm cell strainers |
| Nuclei fixation | 4% PFA; 15 min | 2.4% PFA; 10 min |
| Nuclei permeabilization | 100 μL material; 3 min | 300 μL material; 5 min |
| Tn5 of use | N7-Tn5 (commercially unavailable) | We describe a protocol of transposome assembly with commercially available Tn5. |
| Tn5 Tagmentation | 1:400 diluted N7-Tn5 | We describe a protocol of performing Tn5 activity titration test on a sub-library plate to determine the best Tn5 dilution ratio. |
| Size-select purification | Column-based purification | Bead-based purification |
| Performing a small-scale experiment for proof-of-principle | Not mentioned | Described and validated in this protocol |
Expected outcomes
Each sub-library can be visualized and quantified as mentioned in step 13. The library should have fragments with an average insert at 300–600 bp. The library concentration is typically between 5–50 nM. Figure 7 presents an example of the Bioanalyzer profile of a sci-RNA-seq3 sub-library.
Note: For the Tn5 activity titration test (Figure 6), the library with highest concentration is chosen and the working concentration of Tn5 transposome is determined.
Figure 7.
The Bioanalyzer profile of a typical sci-RNA-seq3 sub-library
(A) A Bioanalyzer trace (electropherogram) showing an average fragment insert at around 500 bp. The region of most library fragment inserts is selected and quantified.
(B) Bioanalyzer gel images of the ladder and the sub-library presented in Figure 7A.
With one flow cell of Novaseq 6000 S4 sequencing platform, 8–10 billion paired raw reads are expected. A data preprocessing pipeline has been described in the previous study2 with codes available at https://github.com/JunyueC/sci-RNA-seq3_pipeline. An illustration is presented in Figure 8. Briefly, starting from undemultiplexed fastq files, we will perform index1/2 demultiplexing, read filtering based on RT and ligation barcodes, adapter trimming, reference genome mapping, UMI error correction, duplicate removal and gene counting, which ultimately generates a cell-by-gene count matrix. We expect to identify more reads mapped to intronic regions than genome exons. An example of downstream analysis such as data quality control, cell artefact identification and cell clustering can be found in Li et al.1 with codes available at https://github.com/TheHumphreysLab/sci-RNA-seq-kidney. The data throughput may vary from 100,000 to millions of cells depending on sample type, library complexity and the number of sub-libraries pooled at step 14.b.
Figure 8.
sci-RNA-seq3 data preprocessing workflow
The computational pipeline described in Cao et al.2 can be used for preprocessing raw, undemultiplexed sci-RNA-seq3 sequencing data. Software required for this analysis and package versions used in Li et al.1 are presented. ED, edit distance.
Limitations
sci-RNA-seq3 is a single-nucleus RNA sequencing method and we have not validated this protocol on isolated kidney cells. The protocol requires a relatively large number of extracted nuclei and may not work efficiently on small tissues where nuclei input is limited. In addition, one key experimental material, the Tn5 transposome loaded with specific oligos, is not commercially available, and therefore, titrating the Tn5 activity by our suggested titration test proposed in step 10 is highly recommended. Compared to 10X Genomics technologies, sci-RNA-seq3 can profile cells at very high throughput (100,000 to millions of cells) but may not be practical if researchers only want to profile fewer than 10,000 cells. In addition, the sci-RNA-seq3 library typically shows a reduced gene detection rate compared with 10X Genomics data.
Troubleshooting
Problem 1
Tissue homogenates cannot pass through cell strainers in steps 2.e–2.g.
Potential solution
Large tissue chunks may clog the strainer mesh. Please ensure complete tissue homogenization in steps 2.c–2.d. Nuclei clumps can also be avoided by pipetting the homogenates against the strainer’s wall, instead of the strainer mesh.
Problem 2
Lots of nuclei aggregates or significant nuclei debris are observed when performing cell counting in step 2.l.
Potential solution
Resuspend the nuclei thoroughly by pipetting for 10 times. A second-round filtration with a 40-μm cell strainer may also be performed.
Problem 3
Pellet size is very small when spinning down nuclei after fixation in step 3.c or the number of nuclei is insufficient when executing step 3.f.
Potential solution
Please use a centrifuge with a swinging bucket to reduce nuclei loss during centrifugation. PFA should be diluted with Nuclei Buffer if the stock concentration is not 4%. The centrifugation speed can be increased to 1,000 g if significant nuclei loss still exists.
Problem 4
Pellet size is very small when spinning down nuclei after reverse transcription in step 6.k.
Potential solution
Please make sure the nuclei concentration is counted accurately in step 5.j. In addition, completely resuspend the nuclei in step 6.c before distributing nuclei into the 96-well plate because nuclei may sink to the bottom due to gravity.
Problem 5
Fewer than 8 96-well plates are obtained in step 7.
Potential solution
We typically obtain 8–12 96-well plates at this point. This may not be a critical problem if > 5 sub-libraries can be generated. Reasons and solutions include: (1) Insufficient number of nuclei are distributed to each well in step 6.c. Please refer to problem 4. (2) Cell loss during reverse transcription and hairpin ligation (steps 6–7). Make sure the pipet tips are tightly attached to the multichannel pipette so that reagents can be distributed with an accurate volume. The centrifugation speed can be increased to 1,000 g if significant nuclei loss still exists. (3) Cell loss in the use of the Flowmi cell strainer in step 7.o. Please refer to the manufacturer’s instructions for proper use.
Problem 6
The library concentration is very low (< 5 nM) in step 13.
Potential solution
We typically obtain sub-libraries with a concentration between 5–50 nM. PCR cycling number in step 12 may be increased to 16–17 to increase the final concentration although this may reflect a reduced library complexity. Reasons and solutions include: (1) RNA degradation. Try to perform kidney sample preservation and nuclei isolation as quickly as possible and avoid prolonged exposure to room temperature (20°C–25°C). Use nuclease-free water in buffer preparation and use nuclease-free laboratory supplies all the time. (2) Insufficient Tn5-based tagmentation. All libraries generated in the Tn5 activity titration test (Figure 6) could be lowly concentrated if Tn5 is inefficiently assembled with annealed oligos in step 8. Please use HPLC-purified oligos for transposome assembly and avoid prolonged incubation (> 1 h) at step 8.f. The tagmentation reaction may also be extended to 10 min in step 10.d. (3) Product loss in library purification. Although the library can be purified with either column-based or bead-based approaches, we typically observe a higher library yield with the bead-based method as proposed in this protocol.
Problem 7
The library contains many small fragments (inserts < 200 bp) as measured in step 13.
Potential solution
The issue is caused by insufficient size selection and purification. We have experienced this problem with column-based size selection, and therefore, we have validated the bead-based size selection method as proposed in this protocol. In addition, perform Select-a-Size MagBead purification for two times (steps 13.b–13.c).
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Benjamin D. Humphreys (humphreysbd@wustl.edu).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
These experiments were funded by NIH grants DK103740 and UC2DK126024 to B.D.H. The authors also acknowledge the Washington University Genome Technology Access Center and Center for Genome Sciences & Systems Biology for sequencing support.
Author contributions
H.L. and B.D.H. conceived, coordinated, and designed the study. H.L. performed experiments. H.L. and B.D.H. analyzed data. H.L. and B.D.H. wrote and approved the final manuscript.
Declaration of interests
B.D.H. is a consultant for Janssen Research & Development, LLC, Pfizer, and Chinook Therapeutics and holds equity in Chinook Therapeutics and grant funding from Chinook Therapeutics, Janssen Research & Development, LLC, and Pfizer; all interests are unrelated to the current work.
Contributor Information
Haikuo Li, Email: haikuolibio@gmail.com.
Benjamin D. Humphreys, Email: humphreysbd@wustl.edu.
Data and code availability
Source data of Figures 3 and 7 and raw sequencing files required for generating Figure 3 are available at Mendeley Data (https://data.mendeley.com/datasets/59z97k52x7).
References
- 1.Li H., Dixon E.E., Wu H., Humphreys B.D. Comprehensive single-cell transcriptional profiling defines shared and unique epithelial injury responses during kidney fibrosis. Cell Metab. 2022 doi: 10.1016/J.CMET.2022.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao J., O’Day D.R., Pliner H.A., Kingsley P.D., Deng M., Daza R.M., Zager M.A., Aldinger K.A., Blecher-Gonen R., Zhang F., et al. A human cell atlas of fetal gene expression. Science. 2020;370:eaba7721. doi: 10.1126/SCIENCE.ABA7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin B.K., Qiu C., Nichols E., Phung M., Green-Gladden R., Srivatsan S., Blecher-Gonen R., Beliveau B.J., Trapnell C., Cao J., Shendure J. Optimized single-nucleus transcriptional profiling by combinatorial indexing. Nat. Protoc. 2022;2022:1–20. doi: 10.1038/s41596-022-00752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049–14112. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gage G.J., Kipke D.R., Shain W. Whole animal perfusion fixation for rodents. J. Vis. Exp. 2012:e3564. doi: 10.3791/3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma S., Zhang B., LaFave L.M., Earl A.S., Chiang Z., Hu Y., Ding J., Brack A., Kartha V.K., Tay T., et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell. 2020;183:1103–1116.e20. doi: 10.1016/j.cell.2020.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H., Humphreys B.D. Single cell technologies: beyond microfluidics. Kidney360. 2021;2:1196–1204. doi: 10.34067/KID.0001822021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirita Y., Wu H., Uchimura K., Wilson P.C., Humphreys B.D. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc. Natl. Acad. Sci. USA. 2020;117:15874–15883. doi: 10.1073/pnas.2005477117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Picelli S., Björklund A.K., Reinius B., Sagasser S., Winberg G., Sandberg R. Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res. 2014;24:2033–2040. doi: 10.1101/GR.177881.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Priore I., Ma S., Strecker J., Jacks T., LaFave L.M., Buenrostro J.D. Protocol for single-cell ATAC sequencing using combinatorial indexing in mouse lung adenocarcinoma. STAR Protoc. 2021;2:100583. doi: 10.1016/J.XPRO.2021.100583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Source data of Figures 3 and 7 and raw sequencing files required for generating Figure 3 are available at Mendeley Data (https://data.mendeley.com/datasets/59z97k52x7).