Summary
The gut microbiota influences host brain function, but the underlying gut-brain axis connections and molecular processes remain unclear. One pathway along this bidirectional communication system involves circulating microbially derived metabolites, such as short-chain fatty acids (SCFAs), which include butyrate and propionate. Brain endothelium is the main interface of communication between circulating signals and the brain, and it constitutes the main component of the blood-brain barrier (BBB). Here, we used a well-established in vitro BBB model treated with physiologically relevant concentrations of butyrate and propionate with and without lipopolysaccharide (LPS) to examine the effects of SCFAs on the actin cytoskeleton and tight junction protein structure. Both SCFAs induced distinct alterations to filamentous actin directionality. SCFAs also increased tight junction protein spikes and protected from LPS-induced tight-junction mis-localization, improved BBB integrity, and modulated mitochondrial network dynamics. These findings identify the actin cytoskeletal dynamics as another target further illuminating how SCFAs can influence BBB physiology.
Subject areas: Biological sciences, cell biology, functional aspects of cell biology, organizational aspects of cell biology
Graphical abstract

Highlights
-
•
Short-chain fatty acids (SCFAs), butyrate and propionate, alter actin directionality
-
•
SCFAs increase tight junction protein spikes and colocalization with actin
-
•
SCFAs protect from LPS-induced decrease in tight junction proteins
-
•
SCFAs protect against LPS-induced barrier and mitochondrial footprint disruption
Biological sciences; Cell biology; Functional aspects of cell biology; Organizational aspects of cell biology
Introduction
A community of microbes have co-evolved with the human host over thousands of years, influencing host health.1,2 Most of these microbes reside in the gut and this gut microbiota is known to communicate with and influence a variety of host health aspects, including key brain processes and behaviors.3,4 Gut microbiota composition changes throughout the lifespan under steady-state conditions. Moreover, changes in the composition have also been associated with neurological disorders including autism spectrum disorder, major depressive disorder, obesity, Parkinson’s disease, Alzheimer’s disease, and Multiple Sclerosis.3 Communication pathways between the gut microbiota and the brain include the vagus nerve, immune system, microbial metabolites, and endocrine pathways.5,6 The brain vasculature and its associated blood-brain barrier (BBB) play an important role in regulating what enters and leaves the brain, thus constituting a communication interface between peripheral signals, such as microbial metabolites, and the brain. This highly selective structural and biochemical barrier between the circulation and the brain is essential for maintaining an optimal microenvironment for brain homeostasis7,8,9
In the last years, the BBB has been identified as a target of microbial influence.10,11,12 This link was first identified in germ-free mice which lack a gut-microbiota and have increased BBB permeability from embryonic development through adulthood.10 There are major implications if the integrity or cellular physiology of the BBB is altered, potentially causing detrimental effects in the brain. BBB disruption can be triggered by stress,13,14,15 depression,16 aging,17,18 and notably is a hallmark of neurodegenerative disorders.18 Interestingly, detrimental changes in gut microbiota composition have been described in all these pathological phenomena.18,19,20
BBB function is primarily maintained by highly specialized endothelial cytoarchitecture and junctional complexes which largely reduces paracellular and transcellular diffusion.21 Endothelial cells are squamous cells that form a thin monolayer that lines the interior of the brain’s blood vessels in direct contact with the circulation responsible for exchange between blood and surrounding brain.22 Tight junction proteins, a specialized feature of brain endothelial cells, help to greatly restrict paracellular permeability and are localized to the cell-cell junctions.23 The tight junction proteins include the transmembrane proteins claudin-5 and occludin, as well as the peripherally associated protein ZO-1.18,23 ZO-1 helps anchor the claudins and occludins to the cytoskeleton,24 greatly contributing to the formation and function of cell-cell junctions in addition to maintaining cell morphology, stabilization of the cells, and transport mechanisms.25
The gut microbiota produces and influences a diverse array of metabolites which reach the host circulation to act at sites distal from the gut.26 Several of these microbial metabolites can alter BBB integrity including butyrate, propionate, urolithin A, trimethylamine (TMA), trimethylamine N-oxide (TMAO), deoxycholic acid, and chenodeoxycholic acid.18 Butyrate and propionate, are short-chain fatty acids (SCFAs) which have received major attention for their broad impact on host physiology.27 SCFAs are the main metabolites produced by bacterial fermentation of dietary fiber.28,29 Butyrate has been identified to protect BBB function12 and improve BBB integrity in germ free mice treated with either the SCFA or colonized with SCFA producing bacteria.10 Propionate protects BBB integrity from a post-inflammatory insult (via lipopolysaccharide (LPS)) in an in vitro BBB model.12 Although butyrate and propionate protect BBB function, the involvement of various cellular structures specifically at the level of the cytoskeleton, remain unknown.
In this study, we examined the effects of physiologically relevant concentrations of butyrate and propionate on the structure and function of brain endothelial cells. In particular, we focus on the impact of SCFAs on cytoskeletal structure in the context of filamentous actin directionality, tight junction protein localization, and barrier integrity.
Results
Butyrate and propionate alter actin filamentous directionality
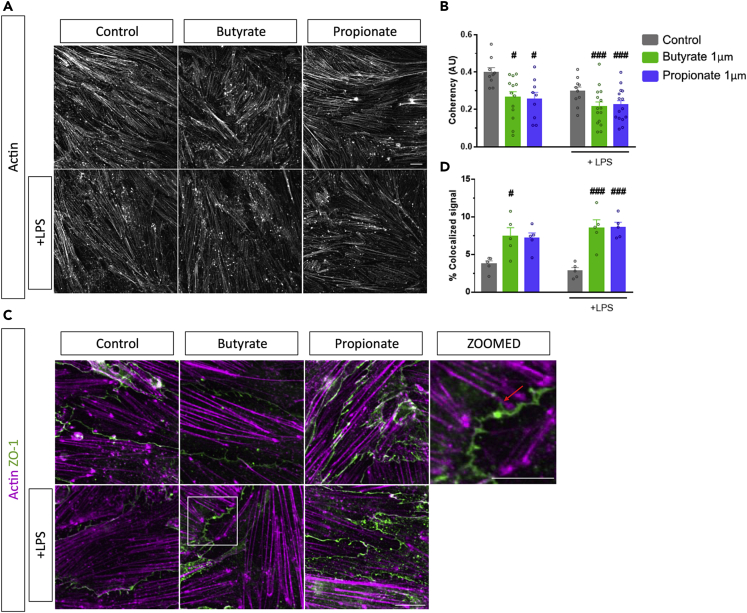
Filamentous actin was stained and visualized using phalloidin following butyrate and propionate treatment with and without LPS in bEnd.3 endothelial cells. Confocal microscopy revealed changes in the organization of actin filaments following butyrate and propionate exposure (Figure 1A). Particularly, the coherency of the directionality of the filamentous actin was significantly altered upon treatment with both SCFAs independent of LPS since there was no interaction [SCFAs; F (2,65) = 8.813, p < 0.001, LPS; F (1,65) = 6.810, p = 0.011, interaction; F (2,65) = 0.722, p = 0.490]. Butyrate or propionate significantly altered the directionality with and without the addition of LPS (Figure 1A).
Figure 1.
Short-chain fatty acids (SCFAs) alter filamentous actin arrangement and increase actin-ZO-1 colocalization.
(A) Representative 60x confocal images of phalloidin-stained actin following butyrate or propionate treatment with and without LPS.
(B) Coherency coefficients of the filamentous actin in the 60x confocal image following butyrate or propionate treatment with and without LPS.
(C) Representative 300x confocal images of actin (magenta) and ZO-1 (green) following butyrate or propionate treatment with and without LPS.
(D) Percentage of actin-ZO-1 fluorescent signal that is co-localized. Mean ± S.E.M. two-way ANOVA followed by Tukey’s post hoc. #p < 0.05, ###p < 0.001 compared to control group. Scale bars 20μm (60x), 10μm (100x zoom3).
Next, we wanted to examine the interactions between actin filaments and ZO-1 tight junction protein, as these are known to be crucial for the proper coordination of barrier function with cytoskeletal dynamics. Confocal imaging following incubation with phalloidin and an anti-ZO-1 antibody revealed the interaction between cytoskeletal actin and tight junction proteins at the endothelial cell-cell junctions (Figure 1C). The percentage of co-localized signal between ZO-1 and actin was increased with SCFAs treatment [SCFAs; F (2,24) = 22.189, p < 0.001]. Butyrate with or without LPS or propionate with LPS treatment significantly increased the percentage of co-localized signal (Figure 1C), indicating the modulation of actin-ZO-1 interactions.
Butyrate and propionate protect tight junction protein localization and induce tight junction protein spikes
As previously described, tight junction protein localization can be influenced by interactions with the actin cytoskeleton.25 Confocal microscopy of the tight junction proteins ZO-1 and claudin-5 revealed differences in the localization upon treatment with LPS, reflected in a reduced relative intensity of these proteins at the cell-cell junctions (Figures 2A and 2B). There was an effect of SCFAs treatment, LPS, and an interaction [SCFA; F (2,86) = 12.650, p < 0.001, LPS; F (1,86) = 46.502, p < 0.001, interaction; F (2,86) = 0.722, p = 0.038]. LPS significantly reduced the relative intensity of both ZO-1 and claudin-5 at the cell-cell junctions. Remarkably, butyrate or propionate rescued the observed reduction in a relative intensity of ZO-1 and claudin-5 signal in the presence of LPS at the cell-cell junctions (Figures 2C and 2E). Of interest, butyrate and propionate treatment also induced expression of ZO-1 and claudin-5 spike-like structures (from now on “spikes”), which are perpendicular to the direction of cell-cell junctions (Figures 2A and 2B highlighted with red arrows). There was an effect of SCFAs on both ZO-1 and claudin-5 spikes and an effect of LPS on ZO-1 spikes, but no interaction effect with either proteins spikes [ZO-1: SCFAs; F (2,80) = 10.155, p < 0.001, LPS; F (1,80) = 17.456, p < 0.001, interaction; F (2,80) = 2.152, p = 0.123], [Claudin-5: SCFAs; F (2,101) = 8.889, p < 0.001, LPS; F (1,101) = 0.721, p = 0.389, interaction; F (2,101) = 2.158, p = 0.121]. The percentage of cells with claudin-5 and ZO-1 spikes significantly increased after butyrate and propionate treatments (Figures 2D and 2F). This suggests that butyrate and propionate modulate the localization and dynamics of tight junction proteins.
Figure 2.
Short-chain fatty acids increase tight junction protein spikes and protect expression and localization.
(A-F) Representative 60x confocal images following butyrate and propionate treatment with and without LPS stained with claudin-5 (A) and ZO-1 (B) antibodies. Relative intensity of claudin-5 (C) and ZO-1 (E) at cell-cell junctions following butyrate or propionate treatment with or without LPS. Percentage of cells containing cells with spikes normalized to the percentage of the control for claudin-5 (D) and ZO-1 (F). Mean ± SEM two-way ANOVA followed by Tukey’s post hoc. ∗∗p < 0.01, ∗∗∗p < 0.001 SCFAs group compared to LPS control group; ##p < 0.01, ###p < 0.001 compared to control. Scale bar 20μm.
Butyrate and propionate protect mitochondrial networks and function
Actin filaments play a large role in modulating mitochondrial dynamics, trafficking, biogenesis, and metabolism.30 Additionally, mitochondrial dysfunction and a decrease in tight junction protein expression can be induced by LPS.31 Given these previous observations, we proceeded to visualize the mitochondrial networks by treating endothelial cell monolayers with Mitotracker™ Red CMXRos (Figure 3A). Using mitochondrial network image analysis tools,32 networks were analyzed with the three major readouts: i) mitochondrial footprint or landscape of the mitochondria in the cells; ii) the number of branches each network made, and iii) average branch length made by each network. There was an effect of LPS and SCFAs treatment on the mitochondrial footprint as well as an interaction effect [SCFAs; F (2,142) = 4.280, p = 0.016, LPS; F (1,142) = 7.650, p = 0.007, interaction; F (2,142) = 4.421, p = 0.014]. LPS exposure significantly decreased the mitochondrial footprint compared to the control (Figure 3B). The decrease in mitochondrial footprint was restored with both butyrate or propionate pre-treatments (Figure 3B). There was no effect of LPS or SCFAs on the number of branches or branch length of the mitochondrial networks; however, there was an interaction effect on the average number of branches [SCFAs; F (2,141) = 1.563, p = 0.213, LPS; F (1,141) = 0.299, p = 3.659, interaction; F (2,141) = 3.659, p = 0.028]. Propionate treatment in the presence of LPS increased the number of branches in mitochondrial networks compared to LPS alone (Figure 3D).
Figure 3.
Short-chain fatty acids protect the mitochondrial network visualization and quantification.
(A) Representative 60x confocal immunostaining images of Mitotracker staining following butyrate or propionate treatment with and without LPS. Quantification of the mitochondria footprint (B), average number of branches each mitochondrial networks has (C), and average branch length of each branch within the mitochondrial networks (D) in brain endothelial cells. Mean ± SEM two-way ANOVA followed by Tukey’s post hoc. ∗p < 0.05, ∗∗p < 0.01 SCFAs group compared to LPS control group; #p < 0.05, ##p < 0.01, compared to control group. Scale bar 20μm.
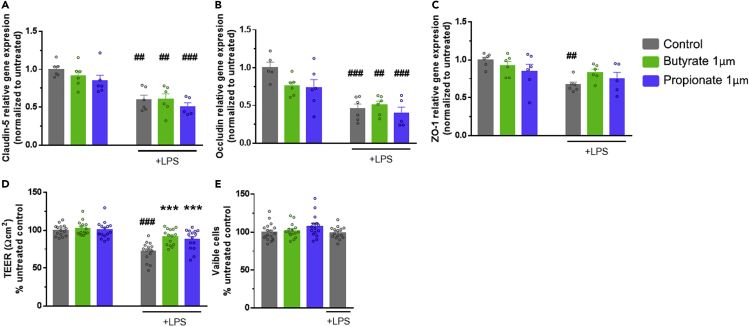
Butyrate and propionate do not protect against lipopolysaccharide-mediated reduction in tight junction protein mRNA expression levels
To determine if the observed protection to tight junction protein intensity upon treatment with butyrate or propionate reflects modulation at a mRNA expression level, real-time PCR was used to assess mRNA expression. There was an effect of LPS exposure on all three tight junction proteins mRNA expression; however, there was no SCFA effect or interaction effect [Claudin-5: LPS; F (1,29) = 46.164, p < 0.001, Occludin: LPS; F (1,28) = 37.538, p < 0.001, ZO-1: LPS; F (1,29) = 11.259, p < 0.001, interaction; F (2,47) = 1.237, p = 0.002]. As expected, the mRNA expression of the three tight junction proteins was decreased with LPS exposure compared to the controls. Interestingly, however, neither butyrate nor propionate pre-treatment protected against LPS-mediated decreased expression (Figures 4A, 4B, and 4C). These data indicate that the butyrate and propionate-mediated protection of the localization of the tight junction proteins was not at the transcriptional level through mRNA expression of the tight junctions.
Figure 4.
Short-chain fatty acids protect LPS-induced barrier disruption.
(A-C) mRNA levels of claudin-5 (A-C), occludin (B), and ZO-1 (C) in bEnd.3 cells measured following butyrate, propionate, and LPS treatment by TaqMan qPCR and normalized to the actin beta housekeeping gene.
(D).Trans-endothelial electrical resistance of bEnd.3 cellular monolayer following 24h butyrate or propionate exposure with or without LPS (1ug/mL) for the last 12hrs of incubation.
(E).Viability of bEnd.3 cells following butyrate, propionate, or LPS treatment determined using the MTT assay. Data are Mean ± SEM two-way ANOVA followed by Tukey’s post hoc. ∗∗∗p < 0.001 SCFA group compared to LPS control group; ##p < 0.01, ###p < 0.001 compared to control group.
Butyrate and propionate protect against lipopolysaccharide-induced disruption of blood-brain barrier function
Next, we wanted to determine the functional consequences of the observed alterations in actin filament coherency, tight junction protein spikes and localization and mitochondrial networks on barrier properties in the cultured endothelial cells. There was an effect of LPS and SCFA treatment on TEER as well as an interaction effect between LPS and SCFAs treatments [SCFAs; F (2,114) = 10.826, p < 0.001, LPS; F (1,114) = 48.691, p < 0.001, interaction; F (2,114) = 8.962, p < 0.001]. Following LPS exposure, TEER in endothelial cells was significantly reduced compared to the controls (Figure 4D). This reduction in TEER was protected against pre-treatments of either butyrate or propionate (Figure 4D). Moreover, an MTT assay was performed to assess cell viability. Neither butyrate, nor propionate, nor LPS exposure had an impact on cell viability compared to the control cells (Figure 4E).
Discussion
As research grows in the microbiome-gut-brain axis field, mechanistic studies to understand how the gut microbiota influence CNS function in health and disease are urgently required.33,34,35 In the current study, we show that the microbial metabolites butyrate and propionate may act on brain endothelial cells by modulating cytoskeletal arrangement/architecture. Butyrate and propionate also increase tight junction protein spike formation, protect cell-cell tight junction protein expression, and regulate mitochondrial network dynamics.
Our understanding of the effects of microbial metabolites such as SCFAs on individual host cell types is rudimentary and requires further elaboration27,28,36 to expedite translation and move toward therapeutic applications.37 For this study, the concentrations of butyrate and propionate were chosen based on the range of circulating concentrations found in the blood of heathy adults,27 further emphasizing the impact these metabolites have on brain endothelial cells. Due to their proximity to the gut microbiota, there are more studies investigating the effects of gut microbiota products on the intestinal epithelial barrier cells than on brain barrier cells per se. SCFAs have been shown to affect cytoskeletal proteins in the intestinal epithelial barrier.38,39 However, the effect of butyrate and propionate on the actin cytoskeleton within brain endothelial cells has remained under-investigated until our current findings.
Formation of tight junction protein spikes has been noted in several endothelial and epithelial cell types.40 The functional implications of increased tight junction protein spikes remain unknown, but perpendicular spikes are associated with characteristic differences in the organization of the associated actin filaments which form a complex with ZO-1 bound to claudins.40 To our knowledge, this is the first study to describe an increase in the percentage of cells with tight junction protein spikes following exposure to butyrate or propionate in brain endothelial cells. The increase in the percentage of cells with these spikes may reflect the altered directionality of the actin cytoskeleton. Furthermore, the localized expression of the tight junction proteins indicated the protected function of the tight junctions even though the decrease in mRNA expression induced following LPS exposure appears to be non-recoverable at this concentration of butyrate and propionate.
Mitochondrial content in endothelial cells is low (ca. 2-6% of total cytoplasm volume) compared with other cell types.41 In fact, endothelial cells rely on glycolysis for producing more than 80% of their ATP.41 This low mitochondrial content in endothelial cells in general, and brain endothelial cells in particular, points to endothelial cells being possibly more involved in regulating signaling responses to environmental cues rather than in energy production. Indeed, the activity of endothelial cell mitochondria is influenced by a variety of peripheral circulating factors such as oxygen and nutrients by modifying their dynamics.41 Here, we propose that circulating microbial factors can also act as modulators of brain endothelial mitochondrial dynamics thereby influencing endothelial cell signaling and BBB function. The mitochondrial footprint is one of the readouts of mitochondrial network, and a decrease in mitochondrial network complexity reflects an increase in mitochondrial fragmentation or fission,32 so the decrease in mitochondrial footprint with LPS possibly reflects an increase in mitochondrial fission. Previous studies show actin plays a role in mediating fission and fusion balance in mitochondrial networks.42,43
The observed alterations to the actin arrangement following butyrate and propionate exposure, are accompanied by the functional protection in barrier integrity upon LPS-mediated disruption, in line with previous literature.12 Importantly, other microbial metabolites such as TMA and TMAO, have been also shown to influence BBB integrity as well as alter the expression of actin cytoskeleton regulation genes.12 The protection of BBB integrity we observed was not mediated by cellular mechanisms involving cell viability per se, which is not surprising at the physiologically relevant concentrations of butyrate and propionate.27 Caution, however, should be taken when interpreting the translatability of the LPS effects since the LPS concentration used in this study is much higher than what is reported to be in the circulation of patients with sepsis.44 The LPS concentration used in this study was strong enough to significantly reduce TEER while having no effect on cell viability. At higher doses, both LPS and butyrate are reported to cause toxicity in cells and are detrimental to cell viability.45,46 What is currently unclear is whether the cytoskeletal effects of SCFAs contribute to their ability to impart barrier integrity protection and through which mechanisms. Based on previous studies, future work should focus on understanding the crucial roles of histone deacetylases (HDACs),47,48 CD14,12 FFAR2/3,48,49,50 G-protein coupled receptor 109a (GPR109a),51 monocarboxylate transporter (MCT1),52 sodium-coupled monocarboxylate transporter (SMCT1),52 and aryl hydrocarbon receptor (AhR)53,54,55 in mediating the physiological effects of butyrate and propionate. Additionally, these findings are intriguing and should be followed up incase SCFAs protect against barrier disruption upon LPS exposure in vivo (e.g. due to bacterial infection).
This study confirms the protective effects of both SCFAs, butyrate, and propionate, on an in vitro BBB model integrity. Further, we found that these two metabolites alter actin cytoskeletal arrangement in brain endothelial cells and increase tight junction protein spikes. Additionally, the metabolites protect cell-cell tight junction protein localized expression and mitochondrial networks. Our data provide insight into the effects microbial metabolites have on the cytoskeleton of brain endothelial cells.
Limitations of the study
While this study further elucidates the role of microbial factors (SCFAs) in modulating brain barriers, more studies are needed to uncover the exact molecular mechanisms behind SCFA-mediated changes in cytoskeletal architecture and their link to the barrier function and mitochondrial dynamics. Moreover, leveraging the advantages of this simple but robust and widely used in vitro BBB model will inform future in vivo studies to confirm our current observations.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ZO-1 Polyclonal Antibody | Thermo Fisher Scientific | #61-7300 |
| Claudin-5 Polyclonal Antibody | Thermo Fisher Scientific | #34-1600 |
| Alexa Fluor™ 647 Phalloidin | Thermo Fisher Scientific | #A22287 |
| MitotrackerTM Red CMXRos | Thermo Fisher Scientific | #M7512 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | #A-21206, RRID:AB_2535792 |
| Chemicals, peptides, and recombinant proteins | ||
| Sodium butyrate | Sigma Merk | #B5887-250MG |
| Sodium propionate | Sigma Merk | #P1880-100G |
| Lipopolysaccharides (LPS) from Escherichia coli 026:B6, ≥10,000 EU/mg, purified by phenol extraction | Sigma Merk | #L8274-100MG |
| MTT (3-(4,5dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) | Thermo Fisher Scientific | #M6494 |
| Dulbecco’s modified eagle medium F12 Nutrient Mixture + L-glutamine | Thermo Fisher Scientific | #11320-074 |
| Trichloracetic acid | Sigma Merk | #T6399-500G |
| Critical commercial assays | ||
| GenElute™ Mammalian Total RNA Miniprep Kit | Sigma-Aldrich | #RTN70 |
| High-capacity cDNA reverse transcription kit | Thermo Fisher Scientific | #4368814 |
| Deposited data | ||
| Raw and analyzed | This paper | ISCIENCE-D-22-02,918 |
| Experimental models: Cell lines | ||
| bEnd.3 [BEND3] | ATCC | CRL-2299, RRID:CVCL_0170 |
| Oligonucleotides | ||
| Primer: Hypoxanthine-guanine phosphoribosyltransferase (HPRT) | Integrated DNA Technologies | Mm.PT.39a.22214828, Ref# NM_013556 |
| Primer: Claudin-5 | Integrated DNA Technologies | Mm.PT.58.33394738.g, Ref# NM_013805 |
| Primer: ZO-1 | Integrated DNA Technologies | Mm.PT.58.12952721, Ref# NM_009386 |
| Primer: Occludin | Integrated DNA Technologies | Mm.PT.58.30118962, Ref#NM_008756 |
| Software and algorithms | ||
| FIJI/ImageJ (Java 1.8.0_172 (64-bit) | Wayne Rasband and contributors, National Institute of Health, USA (Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012; 9(7):676-82.) | https://imagej.net/software/fiji/downloads |
| OrientationJ plugin for FIJI/ImageJ | Barroso A, Mahler JV, Fonseca-Castro PH, Quintana FJ. The aryl hydrocarbon receptor and the gut–brain axis. Cellular & Molecular Immunology. 2021; 18(2):259-68. | http://bigwww.epfl.ch/demo/orientation/ |
| Coloc2 plugin for FIJI/ImageJ | license GPLv3+, release 3.0.5 | https://imagej.net/plugins/coloc-2 |
| Other | ||
| Thincert cell culture insert for 24 well plates | Greiner Bio-one | #662641 |
| μ-Slide 8 Well | Ibidi | #80826 |
| Confocal laser scanning microscope (Olympus FV1000) | Olympus | https://www.olympus-lifescience.com/en/technology/museum/micro/2004/ |
| Epithelial volt/Ohm (TEER) Meter | World Precision Instrument | https://www.wpiinc.com/var-2754-epithelial-volt-ohm-teer-meter |
Resource availability
Lead contact
Further information or requests for resources and reagents should be directed to the lead author, John F. Cryan (j.cryan@ucc.ie).
Materials availability
This study did not generate any unique reagents or materials.
Experimental model and subject details
Brain endothelial cells
Murine brain endothelial cell line bEnd.3 (ATCC CRL-2299, Middlesex, UK) were cultured in Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F12) containing 10% heat-inactivated fetal bovine serum and 1% penicillin/streptomycin. Cells were maintained in an incubator at 37°C and 5% of CO2. Cells under passage 20 were used for all experimentation and were seeded at a density of 1.2∗104 cells/cm2 and grown to confluence. Similar to another study,12 cells were treated with sodium butyrate (1 μM) or sodium propionate (1 μM) in media 24 hrs before beginning terminal assays. Control cells were treated with media absent of SCFAs to control for the media change required for SCFA treatment. These concentrations of butyrate and propionate were chosen based on the concentrations of found in blood of healthy adults (butyrate 1.0 (0.3–1.5) μM; propionate 0.9 ± 1.2μM).27 Cells were challenged with lipopolysaccharide (LPS) (Escherichia coli O 26:B6; 1 μg/mL) 12 hrs12 after the start of butyrate or propionate treatments with no wash out period.
Method details
Immunofluorescence assays
Cells were cultured on Ibidi μ-slide 8-well (Ibidi #80826). Following treatments, cells were fixed with ice-cold 10% trichloroacetic acid (TCA) or 4% PFA, washed with 1xPBS, permeabilized with 0.1% Triton X-, and blocked with 10% normal donkey serum. Alexa Fluor 647 Phalloidin (Thermo Fisher Scientific #A22287) was used to stain the actin cytoskeleton. Primary antibodies were used against claudin-5 (1:150, Thermo Fisher Scientific #34-1600) and ZO-1 (1:150, Thermo Fisher Scientific #61-7300). Mitotracker™ Red CMXRos (Thermo Fisher Scientific #M7512) was used in live cells before fixation according to the manufacturer’s protocol. Images were captured using a confocal laser scanning microscope (Olympus FV1000) fitted with 488, 545, 633 nm lasers and 60x or 100× oil immersion objective lenses.
Image analysis
The coherence of the actin filamentous directionality was measured using OrientationJ56 (http://bigwww.epfl.ch/demo/orientation/), a plugin for Fiji/ImageJ57 (Java 1.8.0_172 (64-bit); Wayne Rasband and contributors, National Institute of Health, USA; https://imagej.net/software/fiji/downloads), with the region of interest (ROI) set to the entirety of each image. The scale of coherency is 1 to 0 with 1 being the dominant orientation and 0 being completely isotropic.56
The tight junction mean fluorescence of claudin-5 and ZO-1 was used to measure pixel intensity at the cell – cell junctions as adapted from previous literature58. A linear ROI was placed along the maximum fluorescent plane between the cells and the mean pixel intensity was measured then a linear ROI identical to the first ROI used was placed within the cell body in close proximity to the cell – cell junction. The first measurement at the cell – cell junction was divided by the second measurement to express the cell – cell fluorescent intensity ratio. Five tight junction fluorescent measurements were taken per image and averaged. The tight junction mean fluorescent results are expressed as the percent average of the control.
The colocalization signal was calculated using Pearson’s correlation coefficient in the Fiji/ImageJ2 plugin Coloc 2 (license GPLv3+, release 3.0.5) (https://imagej.net/plugins/coloc-2) and expressed as a percentage of the control.
Similar to other studies,59 the percentage of cells with tight junction protein spikes was quantified by counting the number of cells containing projections perpendicular to the cell – cell junction divided by the total number of cells in the image.
Mitochondrial networks were analyzed using the Fiji/ImageJ macro tool described for analyzing mitochondrial network morphology in mammalian cell culture.32 Briefly, the images were preprocessed in Fiji/ImageJ to uniformly optimize quality and contrast then processed into a binary image. The binary image was used to measure the mitochondrial footprint, or area occupied by mitochondria. The branch length and number of branches in each image were calculated by skeletonizing the binary image and then analyzing the skeleton.
Evaluation of relative gene expression
Cells were seeded on 24 well plates. Following treatment, total RNA was purified with GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich #RTN70). RNA concentration was quantified using the ND-1000 spectrophotometer (NanoDrop). Equal amounts of total RNA were reverse transcribed to cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Life Technologies, Carlsbad, CA). Gene expression was analyzed using real-time PCR on the AB7300 system (Applied Biosystems, Thermo Fisher Scientific). At least three technical replicates were averaged and normalized to a stable housekeeping gene, Hypoxanthine-guanine phosphoribosyltransferase (HPRT). Changes in gene expression levels were calculated using the ΔΔCt method.60
Evaluation of endothelial cell barrier integrity
Cells were grown on 24-well plate polyethylene terephthalate (PET) transwell inserts (surface area 0.33 cm2, pore size 0.4 μm; Grenier Bio-one #662641). Trans-endothelial electrical resistance performed using a World Precision Instrument Epithelial volt/Ohm (TEER) Meter on 100% confluent cells. Values obtained from cell-free inserts were subtracted from the total values and expressed as percentage of control. The average of the control group is 32.95 Ω cm2 in accordance with previous literature.61
Cell viability assay
Cell viability was assessed by MTT (3-(4,5dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (Thermo Fisher Scientific #M6494) assay.62 Cells were grown on 96 well plates until confluent. 24 h before the assay, cells were treated for 24 h with butyrate (1 μM) or propionate (1 μM) or 12 h with LPS (1 μg/mL). Media was removed and replaced with 50μL of 0.5 mg/mL MTT solution and incubated at 37°C for 3hrs. Once crystals had formed, the MTT was removed, and crystals were solubilized with dimethyl sulfide. Absorbance was read at 590nm. The expressed cell viability is the percent dehydrogenase activity relative to control cells.
Quantification and statistical analysis
In vitro experimental data was expressed as mean ± SEM, n = biological replicates. At least three independent experiments were performed with at least three biological replicates in each experiment. Grubs test was used to remove any outliers from datasets. Data was analyzed by two-way ANOVA for metabolite treatment with and without LPS insult (reported in results) followed by Tukey-adjusted post hoc testing (reported in figures/figure legends) using SPSS Statistics software (IBM, Armonk, NY, US). A p value of less than 0.05 was considered significant.
Acknowledgments
The research was conducted in the APC Microbiome Ireland which is funded by Science Foundation Ireland (SFI/12/RC/2273-P2). G.C. and J.F.C. are funded by the European Office of Aerospace Research and Development, Air Force Office of Scientific Research, and 711 Human Performance Wing, Air Force Research Laboratory (FA9550-17-1-0016). M.R.A. Is funded by the European Research Council under Horizon 2020 Work research and innovation program (grant agreement No. 101040951) and by the Science Foundation Ireland and Irish Research Council under SFI-IRC Pathway program (21/PATH-S/9424)
Author contributions
E.G.K. and M.R.A. contributed experimentally. E.G.K., M.R.A., J.N., C.T., G.C., C.M.O., and J.F.C. critically analyzed the data. E.G.K., M.R.A., G.C., C.M.O., and J.F.C. conceived the project, contributed to the experimental design, and wrote the article.
Declaration of interests
G.C. has received honoraria from Janssen, Probi, and Apsen as an invited speaker; is in receipt of research funding from Pharmavite, Fonterra, Tate and Lyle, and Reckitt; and is a paid consultant for Yakult, Zentiva and Heel pharmaceuticals. This support neither influenced nor constrained the content of this article.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: December 22, 2022
Contributor Information
Maria R. Aburto, Email: maria.rodriguezaburto@ucc.ie.
John F. Cryan, Email: j.cryan@ucc.ie.
Data and code availability
-
•
All data reported in this paper will be shared by the corresponding authors upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Groussin M., Mazel F., Alm E.J. Co-evolution and co-speciation of host-gut bacteria systems. Cell Host Microbe. 2020;28:12–22. doi: 10.1016/j.chom.2020.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Gomaa E.Z. Human gut microbiota/microbiome in health and diseases: a review. Antonie Leeuwenhoek. 2020;113:2019–2040. doi: 10.1007/s10482-020-01474-7. [DOI] [PubMed] [Google Scholar]
- 3.Cryan J.F., O'Riordan K.J., Cowan C.S.M., Sandhu K.V., Bastiaanssen T.F.S., Boehme M., Codagnone M.G., Cussotto S., Fulling C., Golubeva A.V., et al. The microbiota-gut-brain axis. Physiol. Rev. 2019;99:1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- 4.Morais L.H., Schreiber H.L., Mazmanian S.K. The gut microbiota–brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 2021;19:241–255. doi: 10.1038/s41579-020-00460-0. [DOI] [PubMed] [Google Scholar]
- 5.Sherwin E., Sandhu K.V., Dinan T.G., Cryan J.F. May the force be with you: the light and dark sides of the microbiota-gut-brain axis in neuropsychiatry. CNS Drugs. 2016;30:1019–1041. doi: 10.1007/s40263-016-0370-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cryan J.F., Dinan T.G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 7.Mentor S., Makhathini K.B., Fisher D. The role of cytoskeletal proteins in the formation of a functional in vitro blood-brain barrier model. Int. J. Mol. Sci. 2022;23:742. doi: 10.3390/ijms23020742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daneman R., Prat A. The blood-brain barrier. Cold Spring Harbor Perspect. Biol. 2015;7:a020412. doi: 10.1101/cshperspect.a020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segarra M., Aburto M.R., Acker-Palmer A. Blood-brain barrier dynamics to maintain brain homeostasis. Trends Neurosci. 2021;44:393–405. doi: 10.1016/j.tins.2020.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Braniste V., Al-Asmakh M., Kowal C., Anuar F., Abbaspour A., Tóth M., Korecka A., Bakocevic N., Ng L.G., Guan N.L., et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 2014;6:263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoyles L., Pontifex M.G., Rodriguez-Ramiro I., Anis-Alavi M.A., Snelling T., Solito E., Fonseca S., Carvalho A.L., Carding S.R., Müller M., et al. Regulation of blood–brain barrier integrity and cognition by the microbiome-associated methylamines trimethylamine-N-oxide and trimethylamine. bioRxiv. 2021 doi: 10.1101/2021.01.28.428430. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoyles L., Snelling T., Umlai U.-K., Nicholson J.K., Carding S.R., Glen R.C., McArthur S. Microbiome-host systems interactions: protective effects of propionate upon the blood-brain barrier. Microbiome. 2018;6:55. doi: 10.1186/s40168-018-0439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dion-Albert L., Cadoret A., Doney E., Kaufmann F.N., Dudek K.A., Daigle B., Parise L.F., Cathomas F., Samba N., Hudson N., et al. Vascular and blood-brain barrier-related changes underlie stress responses and resilience in female mice and depression in human tissue. Nat. Commun. 2022;13:164. doi: 10.1038/s41467-021-27604-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Menard C., Pfau M.L., Hodes G.E., Kana V., Wang V.X., Bouchard S., Takahashi A., Flanigan M.E., Aleyasin H., LeClair K.B., et al. Social stress induces neurovascular pathology promoting depression. Nat. Neurosci. 2017;20:1752–1760. doi: 10.1038/s41593-017-0010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welcome M.O., Mastorakis N.E. Stress-induced blood brain barrier disruption: molecular mechanisms and signaling pathways. Pharmacol. Res. 2020;157:104769. doi: 10.1016/j.phrs.2020.104769. [DOI] [PubMed] [Google Scholar]
- 16.Wu S., Yin Y., Du L. Blood–brain barrier dysfunction in the pathogenesis of major depressive disorder. Cell. Mol. Neurobiol. 2022;42:2571–2591. doi: 10.1007/s10571-021-01153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banks W.A., Reed M.J., Logsdon A.F., Rhea E.M., Erickson M.A. Healthy aging and the blood–brain barrier. Nat. Aging. 2021;1:243–254. doi: 10.1038/s43587-021-00043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knox E.G., Aburto M.R., Clarke G., Cryan J.F., O’Driscoll C.M. The blood-brain barrier in aging and neurodegeneration. Mol. Psychiatr. 2022;27:2659–2673. doi: 10.1038/s41380-022-01511-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simpson C.A., Diaz-Arteche C., Eliby D., Schwartz O.S., Simmons J.G., Cowan C.S.M. The gut microbiota in anxiety and depression - a systematic review. Clin. Psychol. Rev. 2021;83:101943. doi: 10.1016/j.cpr.2020.101943. [DOI] [PubMed] [Google Scholar]
- 20.Foster J.A., Rinaman L., Cryan J.F. Stress & the gut-brain axis: regulation by the microbiome. Neurobiol. Stress. 2017;7:124–136. doi: 10.1016/j.ynstr.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Z., Nelson A.R., Betsholtz C., Zlokovic B.V. Establishment and dysfunction of the blood-brain barrier. Cell. 2015;163:1064–1078. doi: 10.1016/j.cell.2015.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dugina V.B., Shagieva G.S., Shakhov A.S., Alieva I.B. The cytoplasmic actins in the regulation of endothelial cell function. Int. J. Mol. Sci. 2021;22:7836. doi: 10.3390/ijms22157836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sweeney M.D., Zhao Z., Montagne A., Nelson A.R., Zlokovic B.V. Blood-brain barrier: from physiology to disease and back. Physiol. Rev. 2019;99:21–78. doi: 10.1152/physrev.00050.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stamatovic S.M., Johnson A.M., Keep R.F., Andjelkovic A.V. Junctional proteins of the blood-brain barrier: new insights into function and dysfunction. Tissue Barriers. 2016;4:e1154641. doi: 10.1080/21688370.2016.1154641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bayir E., Sendemir A. Role of intermediate filaments in blood-brain barrier in health and disease. Cells. 2021;10:1400. doi: 10.3390/cells10061400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caspani G., Swann J. Small talk: microbial metabolites involved in the signaling from microbiota to brain. Curr. Opin. Pharmacol. 2019;48:99–106. doi: 10.1016/j.coph.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 27.O'Riordan K.J., Collins M.K., Moloney G.M., Knox E.G., Aburto M.R., Fülling C., Morley S.J., Clarke G., Schellekens H., Cryan J.F. Short chain fatty acids: microbial metabolites for gut-brain axis signalling. Mol. Cell. Endocrinol. 2022;546:111572. doi: 10.1016/j.mce.2022.111572. [DOI] [PubMed] [Google Scholar]
- 28.Dalile B., Van Oudenhove L., Vervliet B., Verbeke K. The role of short-chain fatty acids in microbiota–gut–brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019;16:461–478. doi: 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- 29.Silva Y.P., Bernardi A., Frozza R.L. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. 2020;11:25. doi: 10.3389/fendo.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Illescas M., Peñas A., Arenas J., Martín M.A., Ugalde C. Regulation of mitochondrial function by the actin cytoskeleton. Front. Cell Dev. Biol. 2021;9:795838. doi: 10.3389/fcell.2021.795838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haileselassie B., Joshi A.U., Minhas P.S., Mukherjee R., Andreasson K.I., Mochly-Rosen D. Mitochondrial dysfunction mediated through dynamin-related protein 1 (Drp1) propagates impairment in blood brain barrier in septic encephalopathy. J. Neuroinflammation. 2020;17:36. doi: 10.1186/s12974-019-1689-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valente A.J., Maddalena L.A., Robb E.L., Moradi F., Stuart J.A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017;119:315–326. doi: 10.1016/j.acthis.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Martin C.R., Osadchiy V., Kalani A., Mayer E.A. The brain-gut-microbiome Axis. Cell. Mol. Gastroenterol. Hepatol. 2018;6:133–148. doi: 10.1016/j.jcmgh.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rutsch A., Kantsjö J.B., Ronchi F. The gut-brain Axis: how microbiota and host inflammasome influence brain physiology and pathology. Front. Immunol. 2020;11:604179. doi: 10.3389/fimmu.2020.604179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cryan J.F., Mazmanian S.K. Microbiota-brain axis: context and causality. Science. 2022;376:938–939. doi: 10.1126/science.abo4442. [DOI] [PubMed] [Google Scholar]
- 36.Spichak S., Donoso F., Moloney G.M., Gunnigle E., Brown J.M., Codagnone M., Dinan T.G., Cryan J.F. Microbially-derived short-chain fatty acids impact astrocyte gene expression in a sex-specific manner. Brain Behav. Immun. Health. 2021;16:100318. doi: 10.1016/j.bbih.2021.100318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gill P.A., van Zelm M.C., Muir J.G., Gibson P.R. Review article: short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment. Pharmacol. Ther. 2018;48:15–34. doi: 10.1111/apt.14689. [DOI] [PubMed] [Google Scholar]
- 38.Wang R.X., Lee J.S., Campbell E.L., Colgan S.P. Microbiota-derived butyrate dynamically regulates intestinal homeostasis through regulation of actin-associated protein synaptopodin. Proc. Natl. Acad. Sci. USA. 2020;117:11648–11657. doi: 10.1073/pnas.1917597117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qualtrough D., Smallwood K., Littlejohns D., Pignatelli M. The actin-bundling protein Fascin is overexpressed in inflammatory bowel disease and may be important in tissue repair. BMC Gastroenterol. 2011;11:14. doi: 10.1186/1471-230X-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lynn K.S., Peterson R.J., Koval M. Ruffles and spikes: control of tight junction morphology and permeability by claudins. Biochim. Biophys. Acta Biomembr. 2020;1862:183339. doi: 10.1016/j.bbamem.2020.183339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caja S., Enríquez J.A. Mitochondria in endothelial cells: sensors and integrators of environmental cues. Redox Biol. 2017;12:821–827. doi: 10.1016/j.redox.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rehklau K., Hoffmann L., Gurniak C.B., Ott M., Witke W., Scorrano L., Culmsee C., Rust M.B. Cofilin1-dependent actin dynamics control DRP1-mediated mitochondrial fission. Cell Death Dis. 2017;8:e3063. doi: 10.1038/cddis.2017.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore A.S., Wong Y.C., Simpson C.L., Holzbaur E.L.F. Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission–fusion balance within mitochondrial networks. Nat. Commun. 2016;7:12886. doi: 10.1038/ncomms12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Opal S.M., Scannon P.J., Vincent J.L., White M., Carroll S.F., Palardy J.E., Parejo N.A., Pribble J.P., Lemke J.H. Relationship between plasma levels of lipopolysaccharide (LPS) and LPS-binding protein in patients with severe sepsis and septic shock. J. Infect. Dis. 1999;180:1584–1589. doi: 10.1086/315093. [DOI] [PubMed] [Google Scholar]
- 45.Cao M., Zhang Z., Han S., Lu X. Butyrate inhibits the proliferation and induces the apoptosis of colorectal cancer HCT116 cells via the deactivation of mTOR/S6K1 signaling mediated partly by SIRT1 downregulation. Mol. Med. Rep. 2019;19:3941–3947. doi: 10.3892/mmr.2019.10002. [DOI] [PubMed] [Google Scholar]
- 46.Hosseini N., Jose S., Vidyadaran S., Nordin S.A. Optimization of cell density and LPS concentration for the evaluation of nitric oxide production on BV-2 cells in a griess assay. Malays. J. Med. Health Sci. 2014:1–8. [Google Scholar]
- 47.Silva L.G., Ferguson B.S., Avila A.S., Faciola A.P. Sodium propionate and sodium butyrate effects on histone deacetylase (HDAC) activity, histone acetylation, and inflammatory gene expression in bovine mammary epithelial cells. J. Anim. Sci. 2018;96:5244–5252. doi: 10.1093/jas/sky373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li M., van Esch B.C.A.M., Henricks P.A.J., Folkerts G., Garssen J. The anti-inflammatory effects of short chain fatty acids on lipopolysaccharide- or tumor necrosis factor α-stimulated endothelial cells via activation of GPR41/43 and inhibition of HDACs. Front. Pharmacol. 2018;9:533. doi: 10.3389/fphar.2018.00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexander S.P., Davenport A.P., Kelly E., Marrion N., Peters J.A., Benson H.E., Faccenda E., Pawson A.J., Sharman J.L., Southan C., Davies J.A., CGTP Collaborators The Concise Guide to PHARMACOLOGY 2015/16: G protein-coupled receptors. Br. J. Pharmacol. 2015;172:5744–5869. doi: 10.1111/bph.13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mishra S.P., Karunakar P., Taraphder S., Yadav H. Free fatty acid receptors 2 and 3 as microbial metabolite sensors to shape host health: pharmacophysiological view. Biomedicines. 2020;8:154. doi: 10.3390/biomedicines8060154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gong Y., Jin X., Yuan B., Lv Y., Yan G., Liu M., Xie C., Liu J., Tang Y., Gao H., et al. G protein-coupled receptor 109A maintains the intestinal integrity and protects against ETEC mucosal infection by promoting IgA secretion. Front. Immunol. 2020;11:583652. doi: 10.3389/fimmu.2020.583652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vijay N., Morris M.E. Role of monocarboxylate transporters in drug delivery to the brain. Curr. Pharmaceut. Des. 2014;20:1487–1498. doi: 10.2174/13816128113199990462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marinelli L., Martin-Gallausiaux C., Bourhis J.-M., Béguet-Crespel F., Blottière H.M., Lapaque N. Identification of the novel role of butyrate as AhR ligand in human intestinal epithelial cells. Sci. Rep. 2019;9:643. doi: 10.1038/s41598-018-37019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korecka A., Dona A., Lahiri S., Tett A.J., Al-Asmakh M., Braniste V., D'Arienzo R., Abbaspour A., Reichardt N., Fujii-Kuriyama Y., et al. Bidirectional communication between the Aryl hydrocarbon Receptor (AhR) and the microbiome tunes host metabolism. NPJ Biofilms Microbiomes. 2016;2:16014. doi: 10.1038/npjbiofilms.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barroso A., Mahler J.V., Fonseca-Castro P.H., Quintana F.J. The aryl hydrocarbon receptor and the gut–brain axis. Cell. Mol. Immunol. 2021;18:259–268. doi: 10.1038/s41423-020-00585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fonck E., Feigl G.G., Fasel J., Sage D., Unser M., Rüfenacht D.A., Stergiopulos N. Effect of aging on elastin functionality in human cerebral arteries. Stroke. 2009;40:2552–2556. doi: 10.1161/strokeaha.108.528091. [DOI] [PubMed] [Google Scholar]
- 57.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hatte Guillaume, Prigent Claude, Tassan Jean-Pierre. Tight junctions negatively regulate mechanical forces applied to adherens junctions in vertebrate epithelial tissue. Journal of Cell Science. 2018;131(3) doi: 10.1242/jcs.208736. [DOI] [PubMed] [Google Scholar]
- 59.Schlingmann B., Overgaard C.E., Molina S.A., Lynn K.S., Mitchell L.A., Dorsainvil White S., Mattheyses A.L., Guidot D.M., Capaldo C.T., Koval M. Regulation of claudin/zonula occludens-1 complexes by hetero-claudin interactions. Nat. Commun. 2016;7:12276. doi: 10.1038/ncomms12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 61.Mc Carthy D.J., Malhotra M., O’Mahony A.M., Cryan J.F., O’Driscoll C.M. Nanoparticles and the blood-brain barrier: advancing from in-vitro models towards therapeutic significance. Pharm. Res. 2015;32:1161–1185. doi: 10.1007/s11095-014-1545-6. [DOI] [PubMed] [Google Scholar]
- 62.O’Neill M.J., O’Mahony A.M., Byrne C., Darcy R., O’Driscoll C.M. Gastrointestinal gene delivery by cyclodextrins – in vitro quantification of extracellular barriers. Int. J. Pharm. 2013;456:390–399. doi: 10.1016/j.ijpharm.2013.08.073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
-
•
All data reported in this paper will be shared by the corresponding authors upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.