Abstract
Cardiovascular diseases represent a major cause of morbidity and mortality, necessitating research to improve diagnostics, and to discover and test novel preventive and curative therapies, all of which warrant experimental models that recapitulate human disease. The translation of basic science results to clinical practice is a challenging task, in particular for complex conditions such as cardiovascular diseases, which often result from multiple risk factors and comorbidities. This difficulty might lead some individuals to question the value of animal research, citing the translational ‘valley of death’, which largely reflects the fact that studies in rodents are difficult to translate to humans. This is also influenced by the fact that new, human-derived in vitro models can recapitulate aspects of disease processes. However, it would be a mistake to think that animal models do not represent a vital step in the translational pathway as they do provide important pathophysiological insights into disease mechanisms particularly on an organ and systemic level. While stem cell-derived human models have the potential to become key in testing toxicity and effectiveness of new drugs, we need to be realistic, and carefully validate all new human-like disease models. In this position paper, we highlight recent advances in trying to reduce the number of animals for cardiovascular research ranging from stem cell-derived models to in situ modelling of heart properties, bioinformatic models based on large datasets, and state-of-the-art animal models, which show clinically relevant characteristics observed in patients with a cardiovascular disease. We aim to provide a guide to help researchers in their experimental design to translate bench findings to clinical routine taking the replacement, reduction, and refinement (3R) as a guiding concept.
Keywords: iPSC, Tissue engineering, Multiomics, Network medicine, Bioinformatics, Big data, Comorbidities, Cardiovascular disease
This manuscript was handled by a Consulting Editor, Prof. Ajay M. Shah.
1. Introduction
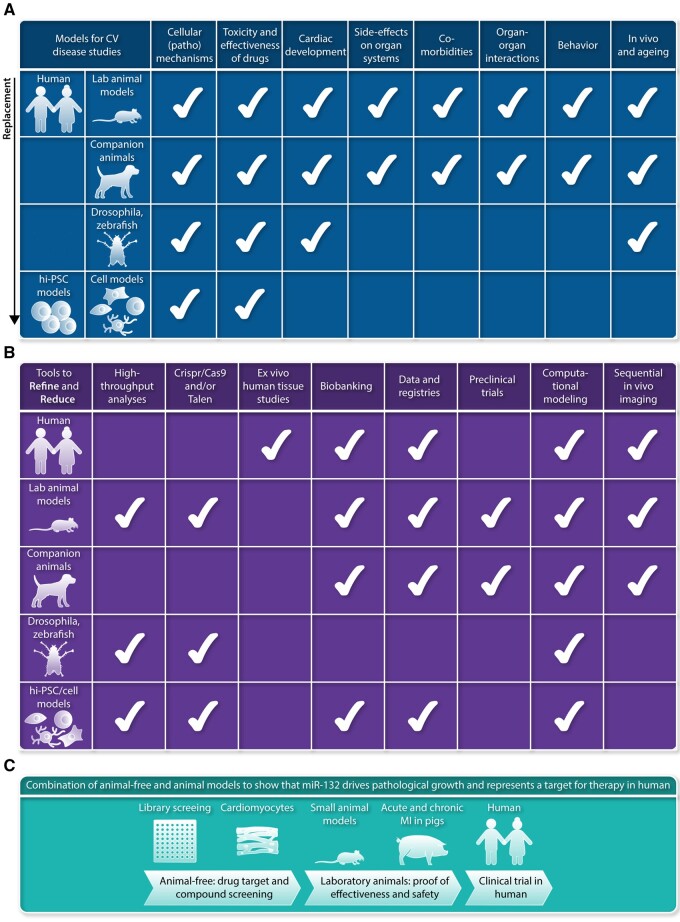
The chronic and progressive nature of cardiovascular disease represents an enormous economical and societal challenge.1 Economic consequences are largely due to high healthcare expenses and loss of healthy years and ability to work of affected individuals. Moreover, the burden of cardiovascular disease is high not only for affected individuals but also for their relatives. This justifies research models that resemble human cardiovascular pathology and strategies to make optimal use of obtained data. In past years, many new potential drug targets turned out to be ineffective in the treatment of ischaemic heart disease and heart failure (HF). This is principally due to a lack of reproducibility and limited translation from rodent models to large animal models and subsequently to humans. Reproducibility and validation of key research findings in experimental models that represent human cardiovascular disease characteristics is essential for the implementation of new diagnostics and therapies in a routine clinical setting. The design of models for studies on cardiac pathophysiology is challenging, as cardiovascular disease is complex and involves multiple causes and comorbidities, resulting in a multiple-organ disease in an ageing population. In this position paper, we focus on replacement, reduction, and refinement of animal experiments, also known as the 3Rs. This concept had already been introduced in 1959 by Russel and Burch2 (Table 1). The objective of this consensus document is to provide an overview of current state-of-the-art in animal models, studies in human and stem cell-derived models (Figure 1A), and highlight how tools have been developed to advance our knowledge of cardiac muscle, vascular and valve diseases (VDs) based on the 3R principles (Figure 1B).
Table 1.
Definitions of the 3Rs2
| Standard | Scientific approach | |
|---|---|---|
| Replacement | Methods which avoid or replace the use of animals | Accelerating the development and use of models and tools, based on the latest science and technologies, to address important scientific questions without the use of animals |
| Reduction | Methods which minimize the number of animals used per experiment | Appropriately designed and analysed animal experiments that are robust and reproducible, and truly add to the knowledge base |
| Refinement | Methods which minimize animal suffering and improve welfare | Advancing animal welfare by exploiting the latest in vivo technologies and by improving understanding of the impact of welfare on scientific outcomes |
Figure 1.
(A) Models that are available for studies on cardiovascular disease, ranging from human and laboratory animals to stem cell-derived models. Aspects that can be measured currently in the different models are indicated with the white check mark. This overview shows that several models allow to reduce the number of studies in laboratory animals, as many initial steps in identification of pathomechansisms, testing drug toxicity and drug effectiveness can be studied in cell-based models. Clearly, studies in human itself offers multiple opportunities to reduce the work in laboratory animals. (B) Multiple tools have been developed in past years to refine and replace studies in the models used for cardiovascular research, and range from tools and expertise to characterize human tissue samples obtained during surgery to models derived from hiPSCs (human induced pluripotent stem cells). (C) Example of an experimental design making use of available complementary research models based on the 3R principles.2
2. Cardiovascular diseases and current experimental models
2.1 Epidemiology of acquired and inherited forms of cardiovascular disease
HF has a high prevalence, is often lethal and patient care is expensive. This condition is now estimated to affect ∼38 million people worldwide and represents the main cause of death and disability.3 Despite the remarkable progress in clinical management of patients and the use of devices assisting the failing myocardium,4 the prognosis of HF remains poor, with mortality rates ranging from 6% to 7% at 1 year in patients with stable HF to ≥25% in patients hospitalized with acute HF,5 and with an overall mortality rate estimated at 40% at 4 years from diagnosis.6 HF is also tremendously expensive, accounting for 2–3% of national health expenditures in high-income countries,7 and is projected to more than double in the next 20 years as a result of the ageing population.8 The most common progressive cardiac rhythm disorder, atrial fibrillation (AF), is associated with HF, stroke and increased mortality. AF affects 2–3% of the Western population, and this percentage will increase in the ageing population.9 Inherited cardiomyopathies caused by pathogenic variants in genes encoding regulatory and structural cardiomyocyte (CM) proteins, and channelopathies, caused primarily by pathogenic variants in genes encoding ion channels are a major cause of sudden cardiac death and morbidity in the young.10,11 In addition to acquired and inherited forms of heart disease and rhythm disorders, pathologies such as aortic aneurysms and valvular disease affect many individuals. Abdominal aortic aneurysms (AAAs) occur in 4–7% of men and up to 2% of women over the age of 55 and are the 10th leading cause of death worldwide.12 Heart VD is highly prevalent, with a mortality risk ratio of 1.36 in developed countries. VD is a progressive disease that increases with the ageing of the population and up to 30% of patients undergo surgical or percutaneous interventions. Valvular dysfunction can be congenital or acquired, and in each case may lead to either stenosis or regurgitation.13 Below we describe the main pathological features of cardiovascular diseases, animal models that mimic disease features observed in humans and the availability of animal-free models.
2.2 Heart failure with reduced ejection fraction
HF is a haemodynamic concept, and failure of the pump to deliver blood (i.e. systolic failure) is often quantified as a reduced left ventricular ejection fraction (LVEF). HF with an LVEF <40% is termed heart failure with reduced ejection fraction (HFrEF). Failure of the heart to properly relax and fill (i.e. diastolic failure) may produce similar symptoms as HFrEF, although with a preserved ejection fraction of >50% (HFpEF; Section 2.3). HF with an LVEF between 40% and 50% is termed HF with mildly reduced EF. At least half of all HF patients present with reduced systolic function.14 Loss of contractile capacity of the heart in HFrEF is due to loss of myocytes and to adverse remodelling of the surviving myocytes, reducing their contractile function (Table 2). The most common cause is myocardial infarction (MI), and subsequent post-MI remodelling, due to coronary artery disease and all its underlying causes (hypertension, hypercholesterolaemia, diabetes, and obesity).15 Other common causes of HFrEF are exposure to cardiotoxic agents, including cancer chemotherapy,16 viral myocarditis,17 peripartum cardiomyopathy (PPCM) (Section 6.1),18 and genetic defects (Section 2.5).19
Table 2.
Comorbidities, causes and cellular, structural and functional remodelling of the heart in HFrEF and HFpEF patients
| Co-morbidities and causes | Vascular changes | Cellular changes in the heart | Structural remodelling | Cardiac dysfunction |
|---|---|---|---|---|
| HFrEF | ||||
|
|
|
|
|
|
||||
| HFpEF | ||||
| Multiple comorbidities: hypertension, obesity, diabetes mellitus, coronary artery disease, sleep apnoea, and lung disease | Proposed: Systemic inflammation-mediated endothelial dysfunction |
|
|
|
Current standard of care includes first-generation drugs: angiotensin-converting enzyme inhibitors, angiotensin receptor blockers (ARBs), β-blockers, mineralocorticoid receptor antagonists, ivabradine and, more recently, combined ARB-neprilysin inhibitors (ARNIs-sacubitril/valsartan).20 These drugs were developed decades ago to target both myocardium and vasculature to improve haemodynamics, and they may also mitigate the adverse remodelling of CMs. Hope has been raised by the unexpected discovery of the remarkable effect on HF of gliflozins (i.e. inhibitors of the sodium–glucose cotransporter 2). However, this effect is still awaiting a molecular explanation.21 Recently, an oral soluble guanylate cyclase stimulator, vericiguat, has been shown to reduce cardiovascular deaths or hospitalization in patients with high-risk HF.22 The fact that not a single biological drug (protein, peptide, antibody, and nucleic acid) exists for a condition that is as prevalent as HF23 is explained by the complex multifactorial nature of this disease.
The stalling of molecular therapeutic innovation24 is in stark contrast to the significant progress in the understanding of HFrEF pathophysiology. Cardiac injury and coincident reduced strain results in increased myocardial stress and determines a common endpoint, largely independent from the original cause of damage and diverse response and pathways triggered by the initial cardiac injury. This includes CM remodelling and alteration of metabolism, followed by progressive LV dilatation (eccentric remodelling), associated with extensive remodelling of the extracellular matrix (ECM), fibrosis and significant changes in viscoelastic properties.25 This, in turn, reduces contraction efficiency and increases oxygen consumption, leading to the activation of the sympathetic nervous system and the renin–angiotensin–aldosterone system, which are initially adaptive but eventually worsen the condition.26,27 The main features of adverse remodelling in HFrEF patients are summarized in Table 2. Various aspects of HFrEF pathophysiology can be mimicked in cellular or tissue models in vitro by applying stress factors (Table 3). Correlates of molecular causes of HFrEF in CMs include de-regulation of β-adrenergic receptor signalling, transition from compensatory to pathological hypertrophy, switch to a fetal type of gene expression and metabolism, changes in post-translational modification profiles, alterations in the calcium cycle and dysfunction of the sarcomere. Virtually all these cellular events can be experimentally mimicked to a significant extent in cell-based model systems where the molecular events involved can be dissected. Analogous considerations can be made for the other cell types that are involved in the myocardial response to injury, namely cardiac fibroblasts and endothelial cells.
Table 3.
Examples of animal models with reduced contractile function and animal-free alternatives
| Species | Experimental animal model and pathological features | Applications | Limitations animal model | Animal-free alternatives | Limitations animal-free alternatives |
|---|---|---|---|---|---|
| Mouse, rat, pig, dog |
|
|
|
Mimicking ischaemia–reperfusion in primary CMs, hiPSC-CMs, EHT, cardiac organoids32,33 |
|
| Mouse, rat, pig, dog, sheep, non-human primate |
|
Mimicking acute and chronic ischaemia in cell-based models32,33 | |||
| Mouse, pig | Spontaneous myocardial infarction in genetic mouse models, large animals on special diets. Spontaneous plaque rupture with thrombotic occlusion, MI40,41 | High heterogeneity and unpredictability | None | ||
| Mouse, Pig | Cancer chemotherapy cardiotoxicity. CM death, vascular injury, contractile dysfunction42–44 | Some commonly used models do not recapitulate the dosing regime used in humans, and typically use healthy (not tumour-bearing) animals |
|
||
| Mouse | Non-physiological methods of administration (e.g. intravenous) | hiPSC-CM, EHT47,48 | Can only assess direct effects since no inflammatory cells are present |
Abbreviations: MI, myocardial infarction; CM, cardiomyocyte; LV, left ventricle; hiPSC-CMs, human induced pluripotent stem cell-derived cardiomyocytes; EHT, engineered heart tissue.
Nevertheless, to address the wide gap in translation, and to reproduce the complex sequential events that occur in HFrEF, small and large animal models are complementary and still required.28 Such models are essential for proof of concept of treatment strategies and for evaluation of systemic effects of cardiac insults and therapies at different stages of the disease. Table 3 illustrates animal models showing reduced cardiac function upon acute and chronic cardiac insults, and animal-free models, including primary CMs, induced pluripotent stem cell (iPSC)-derived CMs, engineered heart tissue (EHT), and organoids.29–48
2.3 Heart failure with preserved ejection fraction
HFpEF prevalence is continuously increasing but many large clinical trials have failed to improve outcomes.49 The lack of improved outcomes is due to the absence of a specific therapy because of incomplete understanding of the pathophysiology of the disease, and the recognition that the more cardio-centric view of HFrEF does not fit HFpEF. Furthermore, there is a large heterogeneity in the patient population as HFpEF is a complex syndrome with varying contribution of the pathophysiological substrate.50,51 HFpEF is more common among the elderly and is associated with multiple comorbidities, such as hypertension, obesity, diabetes mellitus, coronary artery disease, sleep apnoea, lung disease, and remarkable sex-related differences.52 Classic common features include abnormal LV compliance and relaxation, with resultant elevations in LV filling pressure, abnormal systemic and pulmonary vasorelaxation, and neurohumoral activation.50,51,53 Recent principles in HFpEF management rely on the fact that the underlying mechanisms of this syndrome are not the same in all affected patients. This highlights the need to identify the specific causes that can lead to HFpEF and the different HFpEF phenotypes.52 Recent implementation of phenomapping54 has enabled identification of phenotypically distinct HFpEF categories to better classify pathophysiologically similar individuals who may respond in a more homogeneous and predictable way to interventions, regardless of the associated comorbidities.
An important limitation in understanding the HFpEF pathomechanisms and developing new pharmaceutical substances is the scarcity of proper animal models for this complex syndrome, leading to failure in the translation of basic research to the clinical setting. In fact, most animal models suggested to be ‘HFpEF’ present with elevated diastolic pressure but rarely demonstrate the development of HF, which is an essential condition to recapitulate the human situation. Excellent, in-depth reviews on this subject are available.55–60 A true animal model of HFpEF should present with all of the following: an ejection fraction in the normal range for that animal model of at least 50%; diastolic dysfunction; exercise intolerance and pulmonary oedema (Table 2).58 Concentric cardiac hypertrophy can be observed depending on the studied pathomechanism. The challenge is to reliably and reproducibly trigger these characteristic changes in small or large animal models. Several diabetes and obesity rodent models show HFpEF disease features (Table 4).61–66 Unfortunately, pure gene-knockout animal models, so successful in other fields when studying a pathomechanism, are unlikely to generate the complex HFpEF phenotype, although aspects of the disease may appear. Typical examples are the db/db and ob/ob mice, two common models of type-2 diabetes mellitus that lack the leptin receptor or functional leptin, respectively and do show HFpEF characteristics. However, potentially confounding adverse effects arise from altered leptin signalling.58,59 Table 4 provides an overview of the different models which are used to mimic HFpEF disease characteristics based on the different comorbidities and various ways to induce cardiac remodelling.61–85 We also indicate how well the model reflects the HFpEF phenotype observed in patients and the strengths and limitations of specific models. Questionable HFpEF models that incompletely mimic the phenotype include the classical transverse aortic constriction approach, as well as various other interventions predominantly causing hypertension and cardiac hypertrophy.67–70,73–78 Altogether, it is unlikely that there will be a single animal model that can combine all HFpEF sub-phenotypes. This caveat notwithstanding, a good animal model of a common form of HFpEF has emerged as one that is both metabolically and mechanically stressed, similar to what is observed in patients. A recently proposed and interesting concept is that HFpEF presents as a multisystem inflammatory metabolic disease86 driven mainly by excess adiposity linked with imbalance of nitric oxide (NO) levels.84,87,88 An additional, commonly observed risk factor is hypertension, which is also associated with generalized imbalance in NO metabolism and bioavailability. In light of these findings, HFpEF models that recapitulate the metabolic inflammatory phenotype are warranted.
Table 4.
Examples of animal models that mimic disease characteristics of HFpEF patients
| Experimental model | Species | Pathological features | Strengths and limitations of model | Score as HFpEF model (− to +++) |
|---|---|---|---|---|
| Diabetes and obesity model | db/db (leptin deficient) and ob/ob (leptin receptor-deficient) mice61–63 | Hypertrophy, diastolic dysfunction |
|
+ |
| Obese Zucker rats64 | Hypertrophy, fibrosis, diastolic dysfunction |
|
+/++ | |
| ZDF (Zucker Diabetic Fatty) rats65 | Hypertrophy, diastolic dysfunction | +/++ | ||
| Otsuka Long-Evans Tokushima Fatty rats66 | Hypertrophy, diastolic dysfunction |
|
++ | |
| Hypertension models | Deocycoticosterone acetate-salt hypertensive mice67 | Hypertension, diastolic dysfunction |
|
−/+ |
| Dahl Salt-sensitive rats68 | Hypertension, eccentric or concentric hypertrophy, and systolic and/or diastolic dysfunction dependening on age-dependent timing of high-salt diet | −/+ | ||
| Bilateral renal wrapping in dogs69 | Hypertension, hypertrophy, fibrosis, diastolic dysfunction | −/+ | ||
| Deocycoticosterone acetate combined with a Western diet in pigs70 | Hypertension, hypertrophy, impaired relaxation |
|
+ | |
| Hormones | Low dose angiotensin II in mice71 | Diastolic dysfunction |
|
− |
| Hypertrophy | Inbred Hypertrophic Heart in rats72 | Hypertrophy, diastolic dysfunction |
|
−/+ |
| Aortic constriction or banding | Mice with mild and severe transverse aortic constriction73 | Hypertrophy, fibrosis, diastolic and systolic dysfunction |
|
−/+ |
| Rats with aortic banding74 | Hypertrophy, diastolic dysfunction | |||
| LV pressure overload by an implantable stent or inflatable aortic cuff in pigs75,76 or cats77 | Hypertrophy, fibrosis, impaired relaxation, symptoms of heart failure | |||
| Dogs with aortic banding78 | Hypertrophy | |||
| Ageing models | Physiologic or accelerated ageing in mice79,80 | Hypertrophy, fibrosis, diastolic dysfunction |
|
+ |
| Fischer F344 rats81 | ||||
| Cardiometabolic syndrome models | Dahl Salt-sensitive-Obese rats65 | Diabetes, hypertension, hypertrophy, fibrosis, diastolic dysfunction |
|
++ |
| ZSF1: ZDFxSHHF (spontaneously hypertensive heart failure)-hybrid rats82,83 | Diabetes, hypertension, obese at older age, hypertrophy, fibrosis, diastolic dysfunction |
|
++/+++ | |
| L-NAME plus high-fat diet in mice84 | Hypertrophy, fibrosis, diastolic dysfunction |
|
++/+++ | |
| Pigs with streptozotocin-induced diabetes, high-fat diet, and hypertension caused by renal artery embolization85 | Hypertrophy, fibrosis, diastolic dysfunction |
|
++ |
Abbreviations: VICs, valvular interstitial cells; CAVD, calcification aortic valve disease.
One of these rare HFpEF-mimicking models is the obese Zucker diabetic, spontaneously hypertensive Fatty (ZSF1) rat that presents with hypertension, type 2 diabetes, hyperlipidaemia, obesity, and nephropathy. This hybrid rat is a Charles River Laboratories cross between a Zucker Diabetic Fatty female rat and a Spontaneously Hypertensive Heart Failure male rat. Unlike the lean ZSF1 rat that can serve as a convenient control, the obese ZSF1 rat shows multiple HFpEF characteristics known in patients and typical cardiac hallmarks of the disease including modest fibrosis, titin modifications, and CM stiffening.83,87 Furthermore, a large animal model of metabolic inflammatory disease has been generated, which clearly supports the concept of mechanical and metabolic hits as triggers of the disease. Manifestation of ‘patient-like’ HFpEF was evident in pigs with hypertension, diabetes, and hypercholesterolaemia.85 A robust small-animal model of HFpEF was recently made by combining meta-inflammation induced by adiposity (high-fat diet) and hypertension induced by disruption of NO signalling (suppression of constitutive NO synthases) in wild-type mice.84 Importantly, the individual insults alone did not recapitulate HFpEF pathology. A remarkable finding in this two-hit insult mouse model is the disruption of the unfolded protein response that is also linked to autophagy in various diseases.89 Autophagy activators such as caloric restriction mimetics are pleiotropic agents that are beneficial for diastolic heart function in rodent models of ageing and hypertensive heart disease.88
The few available patient-mimicking animal models of HFpEF, driven by metabolic and mechanical stress, represent useful platforms for testing novel treatments in common HFpEF subtypes. The overview provided in Table 4 highlights the progress that has been made in refinement of HFpEF animal studies. However, there remains a need to generate additional models that also represent other HFpEF phenotypes and allow for testing of specific treatments. Whether animal-free models of HFpEF can be successfully developed is questionable due to the complexity of the HFpEF pathophenotypes. iPSC-CMs may be of potential use as they can also be cultured as 3D cardiac tissues. These systems have the advantage of being derived from humans (including patients). This would be useful given the scarcity of cardiac biopsies from the HFpEF patient population. Human iPSC-CMs (hiPSC-CMs) could be used to model specific parameters of cardiac function, such as relaxation, for drug testing, and in co-culture studies to define the effect of endothelial cell dysfunction on CM performance.90 However, with very few exceptions,91 the application of hiPSC-CMs as well as other cell culture types has not really been explored in HFpEF research.
2.4 Atrial fibrillation
Atrial fibrillation is more than just an irregular rhythm on an ECG. It is a condition that requires a multifaceted approach and a variety of research. Known risk factors associated with AF include ageing, common cardiovascular diseases, cardiomyopathies, and channelopathies.92,93 Furthermore, genetic studies have demonstrated an appreciable genetic component in the determination of risk for AF, and genome-wide association studies have identified ∼100 risk loci.94,95 This combination of inherited risk factors, acquired risk and DNA damage96 makes research into AF both especially interesting and challenging. Experimental models to study AF are shown in Table 5. Various research groups discovered that AF perpetuates itself, ‘AF begets AF’, as a landmark paper put it.97 The signalling pathways, structural, and functional alterations of this self-perpetuation have been dissected in large animal models and in patients with AF.92 The interaction between genomic factors leading to AF and other stressors is less well understood. Small animal models like murine models, fish and Drosophila are useful for studying genetic and genomic modifications, and due to their shorter lifespan provide an opportunity to include research on ageing (Figure 1A).96,98,99
Table 5.
Examples of animal models of atrial fibrillation and animal-free innovations
| Species | Pathological features | Applications | Animal-free alternatives |
|---|---|---|---|
| Dog, pig, sheep, goat | Pacing induced tachycardia97,104,105 | Understanding mechanisms of tachycardia-induced ion channel remodelling, therapeutic interventions to prevent electrical remodelling | Paced cell systems, immortalized myocytes |
| Dog, pig, sheep, goat | Electrically induced AF |
|
Cell based models are not available, but in-depth phenotyping of patients with AF may offer solutions: electrical mapping, imaging, blood/tissue biomarkers, genetics |
| Rodents, zebrafish, Drosophila | Mono-causal AF |
|
Animal-free innovations like human cell models, immortalized CM cell lines, and EHT will be instrumental in exploring these interactions and the underlying transcriptional and pathophysiological adaptations in detail.100 Different forms of AF (paroxysmal, persistent, and chronic) are very difficult to mimic in animal or non-animal models. To date, there is no model for paroxysmal AF. Moreover, as AF is often a result of long-term exposure to risk factors partly on top of a genetic vulnerability it is especially difficult to copy a chronic disease like AF in cells. While experiments studying cellular adaptive processes and intracellular signalling require experiments in cells and cell-colonies allowing for genetic and pharmacological interventions, there are challenges with the use of such models for studying human chronic conditions like AF. Human iPSCs have already been differentiated into atrial CMs,101 and atrial CMs have been generated from fetal immortalized CMs.102 An important limitation is that such cells do not mimic all aspects of the adult CM phenotype, such as cell–cell coupling between cells (myocyte–myocyte or myocyte–fibroblast), making studies on the pathophysiology of, for example, conduction disturbances challenging. 3D formats facilitate in vitro maturation, and these 3D cell arrangements including EHT and bioprinting have overcome many of the previous limitations of cellular-based solutions and have been specifically adapted for AF research.103
As in other disease models, validation in more complex systems, occasionally large animals but ideally in patients with AF98, will be required for successful translation of new findings into better diagnostics or therapies.9,98,104–106 For this purpose, data collection in human cohorts should be improved and intensified by for example: analysing algorithms in smartphones and wearables, machine learning and artificial intelligence analysis, phenotyping of patients at risk of AF and with AF. This should be done not only with electrophysiological studies like high-density electrical mapping, but also imaging, biomarkers, proteomics, metabolomics, genetics, and genomics.
2.5 Inherited cardiac diseases—cardiomyopathies, channelopathies, and ventricular arrhythmias
The clinical classification of genetic cardiomyopathies considers structural, functional, and arrhythmogenic alterations. Genetic cardiomyopathies mainly consist of dilated, hypertrophic, and arrhythmogenic phenotypes (i.e. DCM, HCM and AC).10,107–109 Many pathogenic genetic variants in over hundred different genes encoding for sarcomeric (HCM, DCM), desmosomal (AC), nuclear (DCM), mitochondrial (DCM, HCM), and ion channel (AC, DCM) proteins have been identified. Inherited channelopathies, caused by mutations in ion channel genes and their interacting/modulating proteins, lead to a wide range of clinical phenotypes, including conduction disorders, AF and familial syndromes associated with life-threatening arrhythmias and a high risk of sudden cardiac death (e.g. long QT syndrome, Brugada syndrome, catecholaminergic polymorphic ventricular tachycardia). The clinical variability in the expression of the phenotype, in part due to environmental factors,110 and the genetic and phenotypic overlap among different cardiomyopathies and channelopathies,111,112 have challenged the proper evaluation of the clinical, therapeutic, and prognostic impact of genotyping. Animal models, iPSC-CMs, and human cardiac samples (Section 4.1) are currently used to study the consequences of specific genetic variants. Table 6 illustrates animal models and animal-free cell models that are commonly used for cardiomyopathy studies, and highlights how these models relate to the 3Rs.
Table 6.
Examples of animal models of inherited cardiac diseases and animal-free innovations
| Species | Pathological features | Applications | Animal-free alternatives |
|---|---|---|---|
| Mouse, zebrafish, Drosophila |
|
|
|
| Mouse, zebrafish, rabbit, pig |
|
|
|
| Mouse, zebrafish, pig |
|
Mimicking heterozygous and homozygous mutations as present in cardiomyopathy patients |
|
| Rat, cat, dog |
|
|
|
All animal models enable in vivo/ex vivo/in vitro analysis of (electro)physiology, histology, and molecular biology. Abbreviations: GWAS, genome-wide association studies; hiPSC-CMs, human induced pluripotent stem cell-derived cardiomyocytes.
Animal models of cardiomyopathies, such as mice and occasionally rats, have been obtained through genetic engineering.113 These transgenic or knock-in models carrying human pathogenic gene variants (mutations) are the most widely used models of cardiomyopathies. Transgenic mouse models were the most often used method to show pathogenicity of mutant proteins in vivo. In this approach, a large number of copies of the mutant gene are introduced on top of the wild-type gene, which may lead to artificially high expression levels. Gene targeting approaches such as CRISPR/Cas9 in which a mutation is introduced in one or both alleles of the endogenous gene reflect the genetic state of cardiomyopathy patients better. Still, due to important biological and physiological differences between mice and humans, these models may not always recapitulate the human phenotypes. Recent technologies, including CRISPR/Cas9 have advanced the field helping to extend manipulation of genes to large mammals such as pigs, whose hearts are physiologically closer to humans.114 Alternative animal models for studying genetic cardiomyopathies include Caenorhabditis elegans, animals with naturally occurring cardiomyopathy (Section 3.3), Drosophila melanogaster (Section 3.4), and zebrafish (Section 3.5). Similarities at the level of embryonic development, structure, function, and high conservation of gene function, combined with their ease of maintenance, short lifespan, and easy access to approaches for genetic manipulation, make these organisms attractive models for identifying mutations affecting proteins, signalling pathways and biological processes implicated in cardiomyopathies. They allow high-throughput screening (HTS) of gene function as well as druggable targets that can be further validated in larger animal models.
Research into inherited channelopathies traditionally employed heterologous expression systems, such as Chinese hamster ovary cells, human embryonic kidney (HEK293) cells, and Xenopus oocytes, for functional investigations of the consequences and putative pathogenicity of mutations. While these cell systems are inexpensive and easy to maintain and transfect, they are limited by their dissimilarities to CMs environments. Similarly, neonatal cells from rat, mouse or rabbit allow for overexpression or knock-down of genes followed by electrophysiological assessment. However, their immaturity makes them less well suited because of inherent differences in, for example, ion channel isoform expression and (t-tubule) structure. These limitations can be partly overcome by the use of transgenic animal models such as mice, rats, rabbits, and pigs. Although mice differ in certain ion current characteristics, most notably, potassium channels, heart rate, and autonomic regulation, they are easy to breed and to genetically modify by either overexpression or deletion of genes of interest, and it is easier to introduce genetic variants. More recently, rabbits have been successfully used in transgenic studies, which more closely resemble human electrophysiology. Overall, transgenic animals allow for in-depth electrophysiological studies in vivo (ECG, echocardiography), in the whole heart ex vivo (optical mapping, arrhythmia inducibility), on the CM level (patch clamp analysis, calcium fluorescence) and in combination with histological and molecular analyses as well as long-term therapeutic studies. Advances in gene editing resulted in step-wise refinement of animal models, moving from deletion or overexpression of genes of interest to transgenic overexpression of specific gene mutations, and CRISPR/Cas9 models that mimic the heterozygous gene mutations present in most cardiomyopathy patients (Table 6).
Human iPSC-CMs provide an unlimited source of CMs from healthy controls and patients with inherited conditions, and thereby represent an important animal-free method for replacing animal cell studies and reducing the number of animal experiments. They maintain the patient’s genotype as cells are derived from the affected patient skin biopsy or circulating cells. In addition, gene editing with CRISPR/Cas9 enables the generation of isogenic controls that allow for the characterization of the consequences of the genetic defect and rule out the confounding effect of the genetic background.115 However, reprogramming and differentiation remains time-consuming (up to 3 months) and costly. Furthermore, hiPSC-CMs remain immature compared to human adult CMs at the metabolic, structural, and functional level (Section 4.2). For instance, hiPSC-CMs typically lack T-tubules, form only precursory intercalated disks, and their sarcomeres are relatively disorganized. Moreover, hiPSC-CMs have depolarized resting membrane potentials as a result of a lack of inward rectifier potassium current, with potential consequences for electrophysiological analyses. Human iPSC-CMs also lack the multicellular cardiac composition and neurohumoral control. Their integration into EHT with fibroblasts and/or endothelial cells has, nevertheless, been shown to increase their structural and functional maturation, as have various hormonal factors and mechanical activity.115,116 Both hiPSC-CMs and EHTs allow molecular, functional, and electrophysiological phenotyping, facilitating research aimed at developing strategies for personalized risk stratification and therapy in inherited cardiomyopathies.117
Overall, there are important advantages and disadvantages of the different models. The selection of which model to use might be guided by the type of research that is being conducted. Frequently, a combination of models enabling both in vivo and in vitro studies may be required to define the molecular and functional consequences of mutations.
2.6 Valve diseases
For a long time, pathology of cardiac VD has remained elusive. Research on this subject has been limited to observational studies in small animals, such as mice, where genetic manipulation allows for a relatively rapid screening of phenotypes describing valve malformations (e.g. the development of the bicuspid aortic valve) or the evolution of valves towards a stenotic-like condition.13 On the other hand, the lack of consistent larger animals models of valve calcification, except for sheep, has prevented an in-depth investigation of the molecular pathways underlying valve pathophysiology.
Valves contain two major cell types: valvular endothelial cells (VECs), which prevent thromboembolic events by covering the surface of the aortic and ventricular side of the aortic valve producing NO, and valvular interstitial cells (VICs), the most prevalent cell type and crucial for calcification aortic VD (CAVD) pathogenesis.118 VICs are responsible for the homeostasis of the ECM proteins, including collagen, elastin, and glycosaminoglycans, which assure mechanical stability and elasticity of the aortic valve119 and respond to inflammatory cues by inducing a robust calcification response.120 Therefore, VIC functions have prompted new investigations on paracrine pathways involved in CAVD [e.g. transforming growth factor-β (TGF-β) signalling]. The human aortic valve opens and closes over three billion times over an average human lifespan and is thereby subjected to major mechanical forces. These forces include: axial stress during diastole upon valvular closure, mainly sensed by VICs, laminar shear stress on the ventricular side during systole, and oscillatory shear stress on the aortic side of the cusps during diastole both sensed by VECs.121 Both excessive axial stress and lack of laminar shear promote the phenotype switch of VICs towards myofibroblasts, which acting as ‘mechanosensors’, promote valve pathologic ECM remodelling, including fibrosis and valvular sclerosis.120 With further progression of CAVD, increased valvular stiffness, myofibroblasts differentiate into osteoblasts.122
Individuals with increased mechanical strain on the aortic cusps, due to the congenital malformation of bicuspid aortic valves show increased prevalence at a younger age for the development of CAVD.123 Moreover, calcification of the aortic valve predominantly starts at areas subjected to the highest mechanical strain and the lowest laminar shear stress, namely the non-coronary cusp.124 It is the mechanically challenged aortic side of the valve leaflet that calcifies in contrast to the ventricular side of the leaflet. Patients with increased blood pressure, and thus valve overload, show higher risks for the development of CAVD, highlighting that therapeutic strategies should aim to reduce biomechanical forces on the valve.
Until now, no pharmacological agent was able to prevent valvular calcification or promote valve repair, as valve tissue is unable to regenerate spontaneously. Thus, heart valve replacement/repair is currently the only available treatment to prevent HF in VD. The research focuses on two approaches: animal models (mostly large animal models) and animal-free strategies. Animal models have been critical for the development of devices or innovative valve repairing/replacing techniques. Sheep is currently accepted as the gold standard model for valve replacement using defined survival surgeries that meet FDA requirements.125 Normal cardiovascular physiological parameters of sheep approximate those of humans in blood pressure, heart rate, cardiac output, and intracardiac pressures. Also, the valve orifice diameters are similar to humans. Animal-free strategies have become exciting alternatives to promote the development of matrix-guided regenerated or bioengineered valves and studies on the cardiac impact of VD. Considering the highly controlled in vitro conditions, the potential of these animal-free strategies to uncover the pathophysiologic mechanisms underlying VD may even surpass the potential of animal studies. Nevertheless, animal models are still indispensable for studying specific aspects of VD. Table 7 depicts the most commonly used animal models of VD, their potential applications and currently available animal-free alternatives.126–135
Table 7.
Examples of animal models of valve disease and animal-free alternatives
| Species | Experimental animal model and pathological features | Applications | Animal-free alternatives | Refs |
|---|---|---|---|---|
| Calcific aortic valve disease | ||||
| Mouse |
|
To study valve sclerosis early during valve disease progression | Notch-signalling can be studied in cultured aortic VICs as a model of cell-autonomous valve calcificationReplacement and reduction | 126 |
|
|
Not available | 127 | |
| Rabbit |
|
To investigate the mechanisms underlying the association between hypertension and aortic stenosis and the efficacy of different medical treatments to delay, or even hinder, the disease progression | Not available | 128 |
|
To study the link between atherosclerosis and aortic valve stenosis; results are similar to changes reported in human sclerotic aortic valves, suggesting the suitability of this model of atherosclerosis as a model for CAVD |
|
129,130 | |
| Watanabe heritable hyperlipidaemic (WHHL) rabbits fed with a high-fat/high carbohydrate diet display a spontaneous LDLR mutation; the valve does not show significant haemodynamic stenosis but presents lipid deposition, fibrosis, calcification, and inflammatory cell infiltrations | To study early-stage of CAVD and the impact of dietary cholesterol on valve disease | Not available | 130 | |
| White rabbits fed with a standard diet supplemented with 0.5% cholesterol and 50,000 IU/day vitamin D3; non-invasive echocardiographic and invasive measurements confirmed the increase in transvalvular pressure gradient and development of valvular aortic stenosis; histology showed severe calcified and thickened aortic valve | To evaluate the haemodynamic and transvalvular gradient measurements after percutaneous balloon dilation of the valve, for translational research | Not available | 131 | |
| Pig |
|
|
|
132 |
| Valve insufficiency or stenosis | ||||
| Dog, pig | Severing the chorda tendinae, ischaemic injury of the posterior papillary muscle | Mitral valve regurgitation | Not available | 133 |
| Sheep | Pacing-induced heart failure with tricuspidal insufficiency | Tricuspidal valve insufficiency | Not available | 134 |
| Cat, dog, sheep, pig | Supravalvular aortic stenosis by surgical banding of the aorta | Aortic stenosis | Not available | 135 |
2.7 Vascular pathology—atherosclerosis
Atherosclerosis, the underlying process of the majority of cardiovascular diseases, is a lipid driven chronic inflammatory disease. The disease is characterized by the accumulation of lipids and immune cells in the arterial wall: the atherosclerotic plaque. Atherosclerotic plaques can cause stenosis by gradually reducing the arterial lumen or cause acute arterial occlusion by plaque erosion or rupture. These processes result in ischaemia and, depending on the arterial bed affected, result in cardiovascular events including angina pectoris, MI, stroke, or peripheral arterial disease.136 The pathogenesis of atherosclerosis is complex and years of research in patients and experimental animal models have taught us that a combination of systemic environmental factors (e.g. flow, shear stress, oxidative stress, inflammation, endocrine factors and hyperlipidaemia) and plaque intrinsic factors [e.g. cellular lipid uptake, endothelial cell activation, vascular smooth muscle cell (SMC) migration, ECM production, immune cell recruitment and activation] and most importantly cell-cell interactions between immune cells and between immune cells and non-immune cells all drive atherogenesis.137
For decades, most groundbreaking insights into this complex disease have been obtained by studies in laboratory animals (Table 8).40,41,138–151 Until the 1990s, the most widely used animal models for atherosclerosis were cholesterol-fed rabbits, pigs, and non-human primates. These models, especially the pig and non-human primate, have a very similar cardiovascular physiology to humans, but need a long time (>1 year) for developing minimal disease and even longer to develop advanced atherosclerosis (see Section 3.2).147 The design of transgenic mice that lack genes important in lipid metabolism, such as the LDL-receptor and apolipoprotein E, was a major step forward and further refined animal models for investigation of atherosclerosis. Not only do these mouse models develop widespread atherosclerotic lesions in a reproducible way within a few months, but the development, progression, and growth of lesions show features reminiscent of human atherogenesis.148,149 A major advantage of these mouse models is that they can easily be backcrossed to other cell-type specific genetically modified mice in order to not only study the role of specific genes on plaque development, progression, and composition but also the effects of systemic alterations caused by these respective genes on atherosclerosis.148 One of the major drawbacks of animal models of atherosclerosis is the lack of end-stage atherosclerosis with spontaneous plaque rupture.149 Although very old ApoE−/− mice do develop intraplaque haemorrhages, spontaneous rupture of the fibrous cap whereby the thrombus is in continuity with the necrotic core, or spontaneous plaque erosions have only rarely been observed.149 For studying the process of atherosclerotic plaque rupture or the post-rupture healing process, models in which acute plaque rupture is induced mechanically or by vasoconstriction have been developed. For example, in atherosclerotic mice, mechanical plaque rupture was induced by gently squeezing the plaque-bearing aortic segment of the abdominal aorta between blunt forceps.150 Other models of plaque rupture include models in which a plastic cuff is placed around the carotid artery, followed by ligation of the artery.151 A few genetic models, including SRBI−/−/ApoE−/− mice40 and Fb1−/−ApoE−/− mice41 show spontaneous plaque rupture with end-organ damage including stroke and MI.
Table 8.
Examples of animal models that mimic human atherosclerosis
| Species | Model | Main changes in the heart and vasculature | Animal-free alternatives | Refs |
|---|---|---|---|---|
| Pig | Familial hypercholesterolaemia | Atherosclerotic lesions of all vessels | 138,139 | |
| Yucatan and Sinclair miniature pigs fed with Alloxan resulting in diabetes | Human-like atherosclerotic lesions and microvascular diseases | 140,141 | ||
| Ossabaw pigs | Obesity and metabolic syndrome like humans | 142 | ||
| PCSK9 gain-of-function mutant | Hypertension, diabetes, kidney disease, endothelial dysfunction | 143,144 | ||
| Non-human primate | High-fat, high-cholesterol diet in Rhesus and cynomolgous macaques | Slow development of atherosclerosis | 146 | |
| Novel gene-modification technologies, e.g. CRISPR/Cas9 | Accelerated atherosclerosis | 146 | ||
| Mouse | Transgenic mice with lack of genes involved in lipid metabolism (LDL-receptor, apolipoprotein E) | Accelerated atherosclerosis; spontaneous plaque rupture is rare | 148,149 | |
| Refinement: Induction of plaque rupture | 150,151 | |||
|
Spontaneous plaque rupture with end-organ damage including stroke and MI | 40,41 |
Many alternative cell- and model-based efforts are currently being developed and the first results are quite promising. However, atherosclerosis is a complex, multifactorial disease which cannot be mimicked using such a ‘lab on a chip’ approach. As the interactions between many different immune cell types, flow, shear stress, hyperlipidaemia, and endocrine factors all affect its pathogenesis, we still need to make use of living organisms, especially mice. Noteworthy, in atherosclerosis research, we are reducing the number of laboratory animals used by carefully designing our experiments and testing aspects of the disease as much as possible in in vitro systems. Recent developments in single-cell technologies (transcriptomics and mass cytometry)152–154 and the design of novel computational tools has enabled us to more carefully select our candidates and targets, thereby reducing the number of laboratory animals being used. Aspects of the disease, including endothelial cell biology, lipid uptake, leucocyte recruitment, and immune cell activation can be studied in 2D in vitro systems, using cell-lines or iPSCs, thereby limiting research in laboratory animals. Advanced 3D in vitro models are being developed. Furthermore, new and improved animal models of vascular disease (i.e. humanized mouse models) are currently under development.
2.8 Vascular pathology—aneurysms
Aortic aneurysms (AAs) are a complex cardiovascular disease, most likely to develop in the abdominal area. It is associated with risk factors such as advanced age, male gender, genetic predisposition, smoking, and other cardiovascular comorbidities. Currently, the only available treatment for AAA is surgical repair or efforts to improve general cardiovascular health. There are no other effective therapies or drugs because the process leading to AA is ambiguous.155 Previous studies implicate defects in SMCs, ECM remodelling, inflammation, and oxidative stress as key factors in the pathogenesis.156 However, treatment strategies to intervene in the oxidative stress pathway or inflammation have all failed in clinical practice. The underlying pathophysiological processes behind the long-term chronic development of AAA have to be unravelled.
Extensive studies and models have been developed to understand AAA (Table 9).157–162 Research started with in vivo animal models. Murine models are the gold standard of experimental in vivo AAA research. Various different models, each with individual limitations, are capable of providing partial simulation of human pathology. One common feature of all AAA models are the required external stimuli to initiate aortic dilatation. The most common ones are angiotensin II (AngII), porcine pancreatic elastase (PPE), and CaCl2 instillation.157 Experimental AngII-induced AAAs require mice with an atherosclerosis-prone background, such as Apolipoprotein E/ApoE or Low-density lipoprotein receptor (Ldlr) deficiency. AngII-AAAs display suprarenal aortic aneurysms and are commonly associated with covered ruptures or dissections.158 The murine PPE model presents many histo-morphological features associated with human AAA disease.159 A promising modification of the model that utilizes external peri-adventitial elastase application in combination with β-aminopropionitrile (BAPN) to provoke acute rupture and intraluminal thrombus formation has been reported.161 In addition to small animal models, several studies report AAA formation in large animals (mainly pigs) that have the advantage of exploiting similar anatomical and physiological dimensions to humans, allowing the application of devices and surgical techniques.162–164 It appears evident that further advancements in small animal models as well as refinement of large animal models (e.g. using Ldlr-deficient mini-pigs) will enhance studies of unmet translational research questions. However, today no available model closely resembles human AAA characteristics. Recent studies are conducted on the first steps towards the development of an in vitro pre-clinical disease model for AAA (Section 4.4).
Table 9.
Examples of animal models of aneurysms and animal-free alternatives
| Species | Pathological features | Applications | Animal-free alternatives | Refs |
|---|---|---|---|---|
|
Dilation of suprarenal aorta, dissection, covered ruptures, intraluminal thrombus formation | Therapeutic intervention studies | Not available | 158 |
|
Dilation of infrarenal aorta, elastic layer fragmentation, smooth muscle cell apoptosis, increased immune cell infiltration | Therapeutic intervention studies | Not available | 159 |
|
Dilation of infrarenal aorta, enhanced inflammation, smooth muscle cell apoptosis | Therapeutic intervention studies | Not available | 160 |
|
Chronic, advanced-stage AAA with persistent growth, thrombus formation, spontaneous rupture |
|
|
161 |
|
Dilation of infrarenal aorta, elastic layer fragmentation, smooth muscle cell apoptosis, increased immune cell infiltration |
|
|
162–164 |
Abbreviations: ANGII, angiotensin II; PPE, porcine pancreatic elastase; BAPN, β-aminopropionitrile.
3. State-of-the-art in animal models
Animal models allow for in vivo and ex vivo functional and electrophysiological studies at various disease stages in correlation with molecular and histological findings, as well as for research into the impact of stressors such as exercise and comorbidities, ageing and chronic effects of pharmacological interventions. The latter aspects are not easily mimicked in animal-free cell and tissue models (Figure 1A). The following paragraphs describe limitations and opportunities of current animal models.
3.1 Rodent models
Rodent models are widely exploited as they provide biological insight at the organ and cell level, are hypothesis-generating in pathophysiological processes and provide the opportunity for body dose-response testing. The major advantages of these models are relatively easy genetic manipulation, availability of biomedical tools with rodent specificity and their relatively low cost. Below we review some of the major limitations of rodent models and provide promising perspectives to refine and improve their research use.
Rodent models are often used to study the function of a specific protein or mutation. This was initially analysed using pharmacological inhibitors and/or activators, but pharmacological treatments were increasingly criticized for their unspecific effects. Nowadays, genetically engineered mice are the standard in cardiovascular biology, because they permit the modification of a single gene or specific mutation and to examine their function in an integrated physiological system. Two genetic technologies exist, insertional transgenesis (transgenic animals in which additional copies of a gene are inserted) and gene targeting (knock-out to functionally remove a gene, or knock-in to introduce a mutation in a gene). Inducible tissue-specific gene-targeting systems based on the Cre-loxP technology are preferred, to overcome the limitations of global gene targeting which include: embryonic lethality, compensatory changes over time and effects related to gene deletion in organs not under investigation. However, numerous pitfalls have to be considered when interpreting data obtained from genetically modified animals.165 For example, both the Cre protein and Tamoxifen, used to activate the Cre, can have cardiotoxic effects.166,167 While overexpression of any protein might induce undesired effects, its knock-out might also affect the whole proteome.168 Both pharmacological and genetic approaches have potential limitations and may be combined to strengthen the understanding of protein–function relationship.
Additional limitations are the difficulty in translating results generated in rodents to humans, with particular reference to novel therapeutic strategies. Firstly, rodent models are usually developed in healthy and young animals. While some models consider comorbidities, they fail to reproduce the complexity of cardiovascular disorders in humans and lack routinely used medication or other disease-influencing effectors thereby oversimplifying human disease. A second issue to consider is genetic background of mice, as phenotypes may differ significantly between different strains which may confound results. However, combining phenotypic analysis, expression data in cardiac tissue and genetics offers the unique opportunity to identify new disease-related genes and pathways.169,170 Thirdly, rodent hearts poorly mimic the human heart, particularly in terms of heart rate and collaterals. Fourthly, while systematic reviews/meta-analyses are commonly performed to improve clinical practice,171 they are underused in experimental research. Most rodent studies are conducted in a single research facility as a proof-of-concept study. Just like clinical trials, large multi-centre preclinical studies should be initiated to validate findings and to ensure their reproducibility (see Section 5.1), although sustainability may be challenging and require the support of large funding schemes. Societies, funding agencies, and journals should agree on common standards for experimental animal studies with regard to randomization, blinding, and information on age, sex, and comorbidities, to at least be made available as supplemental data. Standardization would allow increasing data robustness and quality, extracting new data from previous studies, reducing the number of animals, and be in compliance with the 3R policy.172 Along the same line, an additional step forward would be establishing repositories of samples from rodent models, with biobanks maximizing tissue usage from euthanized animals. While a particular organ might be the target of a specific study, the remaining tissues could serve the goal of research groups focusing on other organs and systems, thereby reducing the number of research animals and replacing living animals with stored samples. Again, the critical aspect here is assuring that organs, tissue or cells are collected and preserved according to established protocols, to ensure high-quality samples, paired with controls and accurately linked to comprehensive databases providing relevant information. Finally, assessment of cardiovascular function in rodents should privilege methods that avoid invasive or terminal procedures, such as echocardiography, magnetic resonance imaging (MRI), and telemetry. Both echocardiography and MRI allow for complete, repeated and non-invasive assessment of systolic and diastolic function. MRI shows the advantage of providing information regarding cardiac metabolism. However, its use is limited due to its high costs. In contrast, echocardiography is widely used and standard procedures for echocardiographic assessment have been recently published aiming to increase accuracy and reproducibility of the data.173 Telemetry systems involve surgically implanting small devices (telemeters) into the animal. These telemeters assess and emit wireless signals from conscious, non-restrained animals, to a receiver outside the cage. Progress in device miniaturization and battery duration allow for continuous recording of data and for the merging of several cardiovascular parameters in the same telemeter (ECG, blood, and intraventricular pressure) with minimal human-animal contact.174
3.2 Large animal models
While ‘refine’ and ‘reduce’ of the 3R principles (Table 1)2 can be considered in many animal experiments, the ‘replace’ is difficult and is often questioned. Large animal models are mandatory for translational research before entering into clinical trials in most of the drug and class III medical device development projects. The translational value of large animal models, including dogs, pigs, sheep, and non-human primates is high, due to their similar cardiovascular physiology and cellular biology to humans.175–178 An additional advantage of large animal models is their size, allowing for the study of clinical imaging modalities, device implantations, and mechanical interventions. Another advantage, as compared to small rodents, is that per animal many simultaneous or serial tissue and blood samples can be taken, avoiding the need for a separate group of animals for each measurement. Despite their non-disputable advantages, large animal models are costly, require specific infrastructure and handling and lifespan and gestation times are longer. Genetic manipulation of these animals is difficult and may raise ethical questions, but if successful, genetic pig models are extremely helpful in the design of new therapies.114 Below is a brief, non-exhaustive overview of available large animal models.
HFrEF or ischaemic–reperfusion injury without infarction mimic human ischaemic heart diseases very closely (Table 3).30,35–39 In contrast to dogs, pigs (like humans) have sparse coronary collaterals. Therefore, pig or mini-pig ischaemic/reperfusion/infarction models were introduced. The porcine closed-chest reperfused MI model mimics the primary percutaneous coronary intervention in ST-segment MI, and just as in humans, cardiac function can be comprehensively investigated with cardiac MRI.35 Such models successfully mimicked the neutral or minimal cardioprotective effect of ischaemic conditioning seen in clinical trials.31 The size and shape of MIs in pigs are also more like those in humans as compared with infarctions in rats and mice, where infarct size often amounts >50% of LV mass, which is lethal in large animals and in humans. Therefore, results from studies on infarction in pigs are better compatible with those in humans than rodent studies. Atherosclerosis-induced vessel lesions, a major cause of HFrEF, can be simulated in large animal models with high translational power (Table 8).138–146 Whereas dogs are more resistant to the development of atherosclerosis, spontaneous atherosclerosis occurs with ageing in pigs and non-human primates, as it does in humans, which can be accelerated with a Western diet.142,146 Currently, there are four atherosclerotic pig models available: diabetic (type 1 or type 2) and/or diet-induced hypercholesterolaemic pigs; the Rapacz familial hypercholesterolaemic (LDL receptor mutant) pig; and Ossabaw pigs and PCSK9 gain of function pigs.138,140,142–146 These porcine models produce human-like atherosclerotic plaques and importantly diagnostic and treatment studies in these models have corroborated observations in humans. Interestingly, these models also display marked coronary microvascular dysfunction and as such are excellent models for investigating microvascular disease.140,144 Non-human primates, including rhesus and cynomolgous macaques, also recapitulate human-like hypercholesterolaemia when put on a high-fat/high-cholesterol diet, which after several years results in fibrofatty plaques.146 This slow development of atherosclerosis, together with societal concerns, has resulted in restricted use of the non-human primate model for atherosclerosis studies. Perhaps with the advancement of genetic manipulation, accelerated atherosclerosis of primate models will be possible.146
Structural cardiac remodelling, such as hypertrophy or fibrosis, can be induced in pigs by implantation of stents or an inflatable aortic cuff, which results in a gradual pressure overload of the LV thereby causing hypertrophy, impairment of relaxation and HF symptoms.75,76 The latter models may be used to model HFpEF-related structural concentric remodelling and coincident diastolic dysfunction (Table 4). Subcutaneous implantation of deoxycorticosteroneacetate (DOCA) pellets in combination with a Western diet resulted in chronic hypertension-induced myocardial hypertrophy with impaired relaxation and preserved LVEF in pigs,70 while treatment with cardiotoxic cancer drugs such as doxorubicin cause remodelling of the pig heart, including fibrosis and reduced systolic function.44 As described in Section 2.3, mimicking HFpEF in a large animal model represents a challenge, and thus far most models incompletely mimic the clinical phenotype and may show hypertrophy and diastolic dysfunction without clinical HF characteristics. The addition of relevant interventions or comorbidities is essential to trigger the microvascular dysfunction associated with systemic metabolic stress.85,179
An area where experiments on dogs have been indispensable for developments in understanding of disease and development of new therapy is dyssynchrony, induced by intrinsic conduction block in one of the bundle branches or by pacemaker therapy for bradycardia purposes. Dog experiments showed how abnormal conduction of the electrical impulse through the ventricles creates different contraction patterns and loading conditions in opposing ventricular wall segments, thereby lowering ventricular pump function, followed by adverse remodelling over time, with very diverse molecular abnormalities.180 These experiments also showed how cardiac resynchronization could cure all these abnormalities.181 Other animal species turned out to reflect the human situation less well.182 Atrial and ventricular arrhythmias and sudden cardiac death can occur during the development of myocardial disease, or during pacing-induced rhythm disturbances in several large animal models.97,104,105,183,184 In large animals AC, DCM and HCM are diagnosed and represent an interesting alternative model to study arrhythmias and cardiac dysfunction in genetic heart disease (described in Section 3.3). In addition, valve insufficiency and stenosis are mimicked in several large animal models133–135 and are used to study pathomechanisms as well as to test novel therapeutic interventions. For the development and testing of heart valve prostheses large animal models became indispensable (see Section4.4). Sheep were extensively used to test prostheses based on biological materials especially as sheep had a very sensitive reaction with calcification if there were impaired graft conditions. As a result, heart valve prostheses based on decellularized allogenic valve matrices were directly introduced into clinical application after successful testing in sheep.185,186 The pig has become a common transgenic animal model, and genetically modified porcine tissues and organs are gaining the attention of xenogeneic transplantation medicine. Furthermore, whole animals may also serve as ‘humanized’ recipients. Baboon, an old world monkey, lacking the prominent xenoantigen alpha-Gal is considered to be the large animal for testing immunological aspects. Therefore, genetically modified porcine tissues (e.g. decellularized heart valves, and organs) are tested in baboons.185
An example of the complexity and paradox of the cardiovascular system research is tissue-engineered heart valves (TEHVs), any other vascular conduits, or organic patches that can be constructed without using animals. However, to prove the safety and efficacy of the medicinal product, they must first be implanted in animals before human use. Additional comorbidities, such as diabetes and/or hypertonia-induced chronic kidney disease and related alterations in organ function would be possible to mimic in large animal models, but due to their complexity and cost, such models are rarely applied.
3.3 Companion animals
Naturally occurring large animal models have mostly been found in companion animals or livestock, as these animals ubiquitous in our society because of their emotional and economic value.187 The most prevalent non-ischaemic cardiomyopathies in humans are commonly diagnosed in companion animals. HCM is the most common feline cardiac disease affecting around 15% of all cats.188 Mutations have been reported in MYH7189 and MYBPC3.190,191 DCM is more common in dogs and affects mainly large breeds, including Doberman, in which its prevalence reaches 58% and predominantly affects males.192–197 The two main histological findings described in canine cardiomyopathies include attenuated wavy fibres, occurring in various breeds, and fibro‐fatty infiltration of the myocardium, mainly observed in Boxers and Doberman Pinschers.
As in humans, canine DCM has a strong genetic basis with marked familial transmission. Human DCM-associated mutations have been reported in dogs in PDK4, TTN, DMD, and PLN gene.194,195 Finally, AC is commonly diagnosed in Boxers and as in humans, it is characterized by fibrofatty replacement, ventricular premature complexes and ventricular tachycardia.196,197 Being large animals, companion animals have weight, metabolism, and pharmacokinetics that are closer to humans than rodents, allowing therapeutics to be tested for efficacy and toxicity using a relevant regimen. Coupled with the fact that they are relatively outbred, share our environment, are often aged and affected by multiple comorbidities, companion animals make ideal models for testing novel therapeutic interventions (i.e. gene therapy).198,199
3.4 Drosophila
For several years, the Drosophila heart has been used as a tool to study various aspects of the heart, including development, mechanisms of cardiac diseases, and drug screening. The Drosophila heart is a linear tube, reminiscent of the primitive vertebrate embryonic heart tube. Although the final heart structure in Drosophila is very different compared with that in vertebrates, the basic elements for heart development, function, and ageing are conserved.200 In addition, Drosophila offers the opportunity to manipulate gene expression in a highly precise spatial and temporal fashion, using the UAS/GAL4 system.201 This system was successfully utilized to identify genes causing cardiac diseases, including AF and cardiomyopathies.201 New techniques, such as optical coherence tomography, allow accurate phenotyping of cardiac diseases, including HF, HCM, DCM and AC as well as cardiac arrhythmias, such as AF, in flies.96
Because of its simplicity, ease of culturing, and genetic interventions, the Drosophila heart has also been successfully used for drug and genome-wide screening assays, for example, to screen for novel drugs directed at conservation of the proteostasis pathway, which underlies AF.202 Finally, the Drosophila heart has been exploited to verify the outcomes of a human genome-wide association study (GWAS) on genes related to heart rate.203 In this GWAS, 21 loci associated with the heart rate were identified. Experimental down-regulation of gene expression in Drosophila confirmed the relevance of 20 genes at 11 loci for heart rate regulation and highlighted a role for the involved signal transduction routes, embryonic cardiac development and the pathophysiology of DCM, congenital HF, and/or sudden cardiac death.
3.5 Zebrafish
Since their introduction into the biomedical research arena in the 1970s, zebrafish (Danio rerio) have become widely used to study cardiac function and disease due to their tractable genetics.204 Sequencing the zebrafish genome in 2013 revealed that >80% of human disease-related genes have an orthologous gene in zebrafish.205 Together with new developments in genome editing techniques, such as Talens and CRISPR/Cas9, efficient protocols were generated for gene knock-outs, knock-ins, and ‘humanized’ fish carrying human-specific disease alleles.206 A promising feature is that the larvae are small, completely transparent, display similar cardiac electrophysiology to humans and readily take up chemicals from the water, so that they can be grown in a 96-well plate and used for drug screenings.207 Several compounds that have been identified in zebrafish-based assays, are now being tested in clinical trials.
Despite clear anatomical differences, as the two-chambered zebrafish heart consists of an atrium and a ventricle, all major cardiac cell types are present, this allows for the study of their origin, regulation and function. Thus, the zebrafish has proven useful for studying numerous cardiac pathologies. Due to its regenerative capacities, cardiac regeneration remains the most frequently studied process. Upon injury, CMs are able to de-differentiate, proliferate and re-differentiate into mature CMs recapitulating embryonic development of the myocardium.208 In addition to cardiac regeneration, inhibition or genetic deletion of pathways can be very helpful for identifying mechanisms of congenital malformations.204
What the zebrafish community currently lacks is a reliable method to create conditional knock-outs, allowing for the investigation of gene functions in a tissue-specific manner. Hopefully, new developments using CRISPR/Cas9 will resolve these.
4. Models and tools to reduce, refine, and replace research in laboratory animals
4.1 Human tissue samples
Research tools to study cardiovascular (patho)physiologic properties in adult myocardium and blood vessels require careful tissue sampling and storage. A pioneer in setting up a cardiac tissue bank is Prof. dos Remedios, who initiated The Sydney Heart Bank in 1989. Cardiac samples in the Sydney Heart Bank have been collected in a highly routine manner, assuring high quality of tissue samples that have been key in advancing cardiovascular science in many areas ranging from genetics to functional muscle studies.209 RNA deep sequencing of human samples (e.g. cardiac muscle biopsies, vessels) that are obtained during cardiac catheterization or surgery from patients at different disease stages allows molecular profiling, pathway analysis and therapeutic target discovery in relation to different cardiac disease phenotypes.210 Adult human tissue, either as membrane-permeabilized myofibrils, CMs and muscle strips, or intact CMs and SMCs, allow studying myofilament kinetics, myofilament calcium sensitivity, ATP consumption, metabolism and mitochondrial function, electrophysiology and response to different pharmacological agents.211–217 As the preparations are derived from adult hearts, the physiological relevance and pharmacological predictivity are high. Adult CMs are relatively delicate cells, difficult to maintain in culture and have a limited lifespan and potential for expansion. Myocardial tissue slices of human samples represent a new opportunity for studying human tissue over a longer time span in culture. The methodological and technological progress associated with living myocardial slices (LMS) preparations and in vitro culture have increased the interest in this research platform. LMS are 200–400 μm thick sections of living myocardium where structure, function and biochemical properties of the in situ heart are largely preserved.218,219 As such, LMS can be used to study the connections, networks and interplay between the different cardiac cells in a more controlled, comprehensive and realistic manner. LMS thinness allows for oxygen and nutrients diffusion which is critical during experimentation and chronic culture. A high-precision vibratome is required to produce LMS, the slicing is very precise and automated, this is a prerequisite for higher throughput. Between 2 and 9 LMS can be prepared from mouse or rat hearts. However, this number can increase to hundreds when large portions of myocardium are available from large animals or human samples. The LMS technology may significantly reduce the number of animals needed for experimental studies. The preparation of LMS from human specimens is also crucial for translational research.220 A large variety of assays can be applied to interrogate LMS. Functional parameters include, but is not limited to: contractility, conduction velocity, Ca2+ transients, action potentials and metabolism.218,221 Structural assessment provides analysis of cellular and ECM organization, In addition, specific biomolecules can easily be labelled and visualized. Biochemical assessment can also be used to assess LMS genomic and proteomic signatures.222,223
Novel biomimetic technologies allow LMS to be maintained in vitro in a highly functional state and cultured in stable conditions for extended periods,224,225 this allows for novel areas of cardiovascular research to be unravelled. Unique therapeutic research applications may utilize long-term efficacy prediction, RNA-based target evaluation, cell-based regeneration, and high-content analysis by RNA-seq. With standard couriers being used for tissue specimens or LMS movement, it is likely that laboratory networks will soon be formed to share human material that will reduce waste of tissue and increase data collection.
Like any other research model, LMS have limitations that should be carefully considered. Tissue damage occurs during cutting which is likely to trigger inflammatory responses and tissue remodelling. In addition, LMS are disconnected from the circulatory system and neuro-hormonal stimulation. The heterogeneity among LMS obtained from the same heart, as a result of the region that is sliced, should also be considered.226 Furthermore, the lack of standardization across laboratories may result in variable readouts. Biomimetic approaches have enormously improved LMS in vitro culture, however, the preparations progressively adapt to the new in vitro environment that over time results in an alternative phenotype. This adaptation could potentially be controlled by culture conditions and improved biomimetic technologies. It might even level out the variability among samples from diseased individuals. Even though LMS have a bright future several challenges remain that have to be tackled. The standardization of LMS preparation and culture requiring refinement, education and validation of research readouts and applications, are a priority.
Isolated segments of human blood vessels (e.g. human mammary arteries, human coronary arteries, renal arteries, organ-specific vessels or aneurysm samples) can provide unique insights into disease pathology in patients, through western blotting, RNA studies as well as functional vasomotor studies.227,228 Moreover, 24–48 h orgainoid culture can provide valuable pharmacological and mechanistic information. Human mammary arteries (IMAs) are most readily available as a model of systemic vascular function regulation and vascular oxidative stress. While IMA does not develop atherosclerosis, it is sensitive to local pro-atherosclerotic insults eliciting endothelial dysfunction and oxidative stress.229 This approach may be most effectively used in combination with other methodologies described here to identify key novel mechanisms in a translational fashion.
While the demanding logistics represent a challenge, and sample availability is relatively limited, human cardiac, vascular and valvular tissue samples have proven an essential tool to uncover mechanisms of human disease and sex differences. Moreover, human tissue samples provide an excellent basis for validation of the hiPSC-derived models described below.
4.2 Human stem cell-derived cardiovascular cells and their 3D derivatives
The advent of methods to reprogramme somatic cells (e.g. from skin, adipose tissue, peripheral blood and urine) to human iPSC as well as the derivation of bona fide CMs and other cardiovascular cell types at principally unlimited scale, has boosted research in this area by complementing, and occasionally replacing animal experimentation. Recent advances in differentiation protocols230 and mimicking organ-like function in vitro will further enhance this trend.
The human biology of hiPSC-derivatives principally increases the validity and translatability of experimental results when compared with cells from animal species, particularly rodents. Cultures of hiPSC-derivatives are generally more stable and produce more robust data than freshly isolated primary cells, tissues or organs (e.g. Langendorff-perfused hearts), which represent dying-cell-models. Human iPSC-derivatives represent a biological basis that is more physiologically relevant for mechanistic studies than the available immortalized cell lines. The genetic background of patient-derived hiPSC allows for modelling of individual disease mechanisms and susceptibility. Furthermore, direct access to pharmacological and genetic manipulation in vitro (e.g. by gene editing) facilitates studying direct drug/gene cause–effect relationships under controlled conditions. Moreover, cellular models can be exploited to identify both cardioprotective and pro-proliferative therapies and are particularly amenable to HTSs (Section 4.5). Co-cultures of various hiPSC-derived cell types can decipher some cell–cell interactions in a forward manner, which can be combined with tissue engineering to provide organoid-shaped and biomechanical-modelled platforms.
Human iPSC-derivatives exhibit a fetal rather than adult phenotype with only partially canonical function.231 Human iPSC-CM, such as foetal, neonatal and immortalized cells, have poorly developed mitochondria and rely on glycolysis rather than substrate oxidation.232 Consequently, they exhibit a high basal glucose catabolism with poor insulin responsiveness (i.e. only at supra-physiological insulin concentration).233 Whereas differentiation protocols introduce batch-to-batch variation, reprogramming and long-term culture can induce artefacts such as karyotype abnormalities and epigenetic alterations that are difficult to control.234 In vitro assays only partially capture disease-relevant whole organ functions (e.g. arrhythmias and diastolic heart function). Of the most common human pathology ischaemic damage by blood vessel occlusion, only the earliest stage of ischaemia can be modelled in vitro (Table 3). Cell–cell-based mechanisms (e.g. through the dynamic influx of inflammatory and immune cells) are difficult to explore in vitro. In models of iPSC-derived cardiac tissue, vascularization and ultimately perfusion are key challenges that are often underestimated in their influence on cell behaviour and in their relevance for rebuilding more physiological tissue. Moreover, the limited time lines of in vitro experiments impede assessment of cardiovascular disease mechanisms that often act over many years. This limitation also applies to the most common animal models, but multicellular responses could, in principle, be better assessed in animals. Major cardiovascular risk factors and comorbidities such as ageing and metabolic diseases, including hyperlipidemia and diabetes, can only partially be addressed in vitro. Organ–organ interactions (e.g. effects of the liver, gut or brain on heart function) cannot be captured in current in vitro hiPSC cultures.
Solutions to increase the applicability of hiPSC-derived cell systems for cardiovascular studies are described below:
Reduce experimental variation: Employing established quality standards, such as: the obligatory use of standard operating procedures, master and working cell banks, defined passage number, proven normal karyotype, high pluripotency marker expression, isogenic controls (e.g. by CRIPSR/Cas9 gene editing), minimum repetition of experiments in three batches from three lines, and standardizing circadian time will reduce variability.235,236 Worldwide hiPSC banking initiatives such a hPSCreg (http://hpscreg.eu) add to this standardization. Furthermore, automation has the potential to reduce experimental variation237 and will likely become more common in high-throughput facilities (e.g. for drug screening). The high costs for initial investment and maintenance limit a more widespread application in academia.
Improve maturity: Refinement of culture media composition (e.g. energy substrates, hormones and growth factors)238,239 as well as culturing of hiPSC-CM on matrices with tunable stiffness,240,241 Matrigel mattresses,242 or micropatterned surfaces203,243 have been shown to improve the maturity. Consistently, lowering glucose and adding fatty acids have been shown to improve the metabolic maturity of hiPSC-CM, reflecting the fact that the use of glucose is inhibited by fatty acid oxidation in a fasting state and is stimulated by insulin in a fed state.244 3D Multicellular constructs, mechanical loading, and electrical pacing (e.g. in EHT) are some of the most effective means to improve the structural, metabolic, electrophysiological, and contractile maturity of hiPSC-CM and the spectrum of functional readouts.245,246 Further improvements are expected from co-cultures of hiPSC-derived CMs, fibroblasts, endothelial cells, neurons, immune cells, and others.247 So far, several differentiation protocols for the respective cell types are available,248 but it is still not known how well these cells resemble the organ-specific cells in their respective environment (e.g. cardiac endothelial cells). More work is needed to achieve truly adult-like CMs/heart tissue from hiPSC.
Improve the functional readout: Simultaneous measurements of force, calcium transients, and membrane voltage by fluorescent dyes (e.g. Fluo-4, FURA-2, Arclight, Fluovolt,249,250 or genetically encoded calcium sensors such as GCaMP6f116) improve the depth of phenotypic characterization of hiPSC-CM/EHT and allow analysis, including arrhythmias, in intact preparations.251 Sharp microelectrode action potential recordings reduce confounding influences of cell isolation and the small size of hiPSC-CM compared to patch clamp recordings.252 However, tissue damage and localized ischaemia may occur, and patch clamp recordings in isolated hiPSC-CMs with or without dynamic clamp may be considered for certain studies.
Study hiPSC phenotypes under disease-provoking conditions: Experimental setups that allow the manipulation of matrix stiffness or afterload in 3D constructs can provoke phenotypes masked under basal condition.241,253 Influences of common comorbidities on disease phenotypes in patient-derived hiPSC-CM or the effect of simulated ischaemia may be studied by applying hyperglycaemic and hypercholesterolaemic culture conditions as shown in fetal rat myocytes.254 In vitro vascularization may allow for the study of mechanisms of thrombosis and ischaemia in vitro.255
Study organ–organ interactions: Organ-on-chip approaches (i.e. microfluidic culture systems in which organotypic cell types are cultured in one or multiple compartments connected by circulating medium) offer the opportunity to study organ-like function or complex interactions between organs of the human body, for example, between the drug-metabolizing liver and the heart (multi-organs-on-chips).256 Even though perfusable tissue surrogates are available, but they are still far from replicating a vascularized organ with chambers, conduction system, and physiological function, and would therefore only enable partial replacement of animal experiments. The potential of these new approaches has to be weighed against their technical complexity. Moreover, the necessary simplification of culture conditions may interfere with the desired maturity of the respective ‘mini-organs’.
Alternatives: The necessary level of maturity and complexity depends on the question being asked. For some high-throughput screens, a simple and cheap cell line might be appropriate as a first choice (e.g. the rodent cardiomyoblastic cell line H9C2). These cells have primarily skeletal muscle characteristics and lack cardiac contractility. HL-1 cells, derived from a mouse atrial tumour, exhibit several cardiac-specific phenotypes but proliferate possibly involving more genetic alterations than the initial SV40 antigen expression.257 More recently, rat atrial CMs were transduced with a doxycycline-dependent SV40 LT antigen that could be easily expanded and differentiated into excitable and contractile atrial CMs upon removal of doxycycline.258 The rodent background of these CM-like cells has, however, a considerable limitation. More recently, a similar approach was used for generation of a human atrial immortalized cell line.102
As indicated above, further fine-tuning of differentiating iPSC-derived cell types and generation of multi-cellular models is ongoing. Promising developments that may be able to reduce the use of animal models, include the generation of simple 3D microtissues or organoids containing iPS-derived cardiac endothelial cells, fibroblasts and CMs,247 most likely applying matrix-like substances,259 containing a vascular network,260 or using printed scaffold materials to tailor microstructural mechanical design and mimic cardiac stiffness.261
4.3 Animal-free strategies to mimic valve disease and vascular pathology
In recent years, animal-free strategies have been introduced to uncover the pathophysiologic mechanisms underlying VD, atherosclerosis and AAA.
For VD several studies focused on decrypting the cellular pro-calcific phenotype by evolving 3D pathology modelling involving substrates with defined chemical and mechanical characteristics using an integrated vision of ‘mechano-paracrine’ signalling controlling the physiological versus the pathological phenotype of VICs. The stiffness sensitivity of VICs was demonstrated, for example, in studies performed with hydrogels with tuneable mechanical characteristics,262 as well as in the presence of paracrine signalling by TGF-β.263 More recently, investigations have allowed for the characterization of the molecular signalling underlying the activation of VICs towards the pro-fibrotic phenotype. In particular, for describing the relevance of the mechanically activated Hippo transcriptional machinery264 for porcine265 and human266 aortic VICs pro-fibrotic activation. In aortic VICs, this pathway was more active close to the calcified areas.267 Another option relies on complex fabrication processes of valve microenvironments combining different ratios of matrix components (e.g. glycosaminoglycans, GAG) with hydrogels (e.g. Gelatin-Methacrylate) mimicking mechanical features of structural valve components such as collagen.268 In addition to mechanical valves and valve prostheses made from fixed biological materials like porcine heart valves or bovine pericardia, prostheses made from decellularized heart valve matrices may become the gold standard as these display fundamental beneficial characteristics.269 With these approaches, it is more feasible to investigate the complex response of valve cells to pathophysiologic stimuli in the context of valve tissue-mimicking architecture and essential biophysical characteristics (Figure 2).
Figure 2.
In situ valve engineering.
AA for atherosclerosis, flow chambers coated with human atherosclerotic plaque lysates are being applied to study the dynamics of platelet and leucocyte plaque interactions under flow conditions. Tissue-engineered vascular grafts, composed of polymers, and implanted in bioreactors or animal models for vascular tissue regeneration, have been successfully created.270,271 Chip-based microfluidics systems containing 3D structures with an arterial geometry build, containing iPSC-derived pericytes, vascular SMCs and endothelial cells, can be subjected to flow and shear stress. These are useful for studying the effects of flow and shear stress on endothelial cell biology, as well as arterial thrombosis.272,273 These novel 3D tissue-engineered arteries can be considered a prelude to the 3D in vitro generation of atherosclerotic plaques. However, engineering an artery that contains the arterial geometry, is subjected to flow conditions, contains a plaque in which all cells are represented, immune cells are recruited, and lipids are processed, is still not possible and poses a future challenge.
AAA studies in aortic tissues or models developed with patient cells from biobanks studying the SMC contractility and AA pathophysiology (Section 4.1),217,274 as well as novel in vitro 3D models to study SMC-ECM interactions are forthcoming. Advancements are made to integrate mechanical components into these models to mimic shear stress, which can activate inflammatory pathways, atherosclerosis, intima hyperplasia, and aneurysm formation.275,276 The evolution of imaging-based models of intravascular flow dynamics has revealed that pathological programming of the vessel wall may also occur with the crucial contribution of the wall stress.276 Recently, the concept of cell mechanosensation has come to connect the transmission of mechanical forces to cells from the ECM or vice-versa and to discrete gene regulation patterns affecting the cellular homeostasis within the cardiovascular system.277 This has confirmed the existence of novel mechano-dependent pathologic pathways. For example, through an in vitro model of circumferential wall strain associated with coronary flow dynamics occurring in arterialized saphenous veins, involvement of Thrombospondin-1 (TSP-1) in pathological activation of resident myofibroblasts in the wall was revealed for the first time, with consequences for neointima accumulation and vein graft failure.278 Since TSP-1 has a role in the formation of ascending aneurysm through a mechanism involving changes in mechanical characteristics of the vessel wall,279 it could be a key factor connecting alterations in tissue biophysical features, modifications in cellular composition and signal transduction.
Molecular modelling with ‘vasculature-on-a-chip’ devices mimicking the architecture, mechanics and cell setup of arteries and veins has finally become a novel way to investigate vascular pathology programming (Figure 3).280 These models have the advantage of being easily manufactured with biocompatible materials, are miniaturized and reproduce the haemodynamic patterns typical of pathologic vasculature. This is expected to allow an unprecedented multiplex analysis power with cells that can be directly derived from patient biopsies without involving animals, providing immediate translational and personalized therapeutic perspectives.
Figure 3.
Aneurysm-on-a-chip manufactured with a 3D printing-based microfluidic channel patterned inside a polydimethylsiloxane (PDMS) block. The heatmap represents the distribution of the flow velocity reproducing the hemodynamic conditions occurring into aneurysms. Figure 3 is original and contains unpublished modelling and manufacturing images.
4.4 Production and testing of heart valves
Given the limited number and sizes available from human donor material, current research focuses on the development of non-immunogenic xenogeneic heart valves matrices.281 Developed in the sheep model, orthotopically implanted acellular allogeneic pulmonary and aortic heart valve matrices get repopulated with autologous interstitial cells, whereas the lumen gets re-endothelialized by autologous endothelial cells.269 With this, the grafts are non-thrombogenic and regain the ability to adapt to the growth of the recipient. Therefore, these animal-free based strategies are easily translated into the clinical setting as they provide the possibility to create new transplantable valves which are of utmost importance, for instance, for paediatric patients.282
The principle of the tissue engineered heart valve (TEHV) is based on the construction of a biodegradable heart valve-figured scaffold that develops into living valve-formed tissue by autologous cell invasion after resolving the scaffold. The basic requirements of TEHVs are: biocompatibility, non-immunogenicity, non-thrombogenicity, capacity to mimic function and structure of the heart valves, and adaptability to physiological and pathophysiological conditions.283
The strategies of TEHV fabrications include molded or sutured scaffolds with using: natural or synthetic polymers, decellularization, electrospinning, 3D printing, in vivo bioengineering, and combination of these techniques (hybrid TEHVs).284 The majority of the TEVHs are constructed by molding of polymeric substances into a valve-like shape, or attaching to an appropriately formed stent.285 For the engineered tissue, either natural biopolymers, such as collagen or fibrin or synthetic polymers (e.g. polyglycolic acid, polylactic acid, polyε-caprolactone, poly4-hydroxybutyrate) are used. The stent-polymeric scaffolds are then populated with different types of cells (e.g. marrow stromal or endothelial cells, or mesenchymal stem cells) in bioreactors to avoid foreign body reaction. The second most frequently used TEHV fabrication is the decellularization of animal heart valves by using detergents, immersion, or perfusion approaches.286 Currently, two TEHVs have been approved for human use: the Cryolife’s SynerGraft® in Europe and the USA, and AutoTissue GmbH's Matrix P plus N™ in Europe. Unfortunately, the safety and efficacy of these products are currently rather insufficient, showing controversial results in clinical applications.287,288
Electrospinning is less frequently used due to its complexity. This technique is based on creating a solid controlled fibre structure of TEHV. The construction of which fits better to the anisotropic mechanical characteristics of the natural valve, simulating the microarchitecture of the valve better than the other technologies.289 To enable a 3D Bioprinting of a TEHV, a 3D imaging (computed tomography or magnetic resonance) is first applied, and converted to a stereolithography computed file of the 3D printer, followed by bioprinting of the TEHVs (inkjet, extrusion or laser-assisted) by using bioinks of cell-free or cell-encapsulated biomaterial.290 The hybrid technique to construct TEHV combines decellularization, and cell seeding technologies, as well as tubular fibrin gels, encapsulating cells followed by decellularization or the electrospinning method recombining with gelatin hydrogels, or others. The in vivo tissue engineering of a valve requires its implantation in an animal species chosen for the experiment (in vivo ‘bioreactor or cell culture’), and cellularization in vivo, followed by orthotopic implantation.291 Each TEHV construction technology has its advantages and disadvantageous, and a great deal more scientific and technological development is needed for human translation of the TEHVs.
4.5 High-throughput screenings
Over the last decade, there has been an explosion of studies based on HTSs of both small molecules and small nucleic acids in cultured CMs for drug and gene discovery. This was rendered possible by the development of biological assays amenable to miniaturization and automation, and by the availability of technologies for processive high content (HC) microscopy imaging, determination of mechanical forces, and electrophysiology measurements. The use of cultured cell lines of cardiac derivation, primary fibroblasts or neonatal CMs or human embryonic stem cell (hESC)/hiPSC-derived CMs has been instrumental in the possibility of identifying active compounds through large library screenings.
A number of cellular, molecular, and functional assays can be adapted to 96- or 384-well plates and thus rendered amenable to HTS analyses. To search for small molecules or nucleic acids regulating these processes at the cellular level in primary CMs or CMs derived from hESC/hiPSC lines the following has been implemented: the incorporation of thymidine analogue to measure CM proliferation,292–294 assessment of CM cross-sectional area,295–297 inhibition of pathologic aggregate formation,298 protection from cardiotoxic treatments,299–301 or regulation of Ca2+ handling.302 The development of HTS assays aimed at assessing two fundamental parameters of CM function, namely electrical activity and contraction force, is definitely more demanding in terms of instrumentation and complicated by the immature nature of hESC/hiPSC-CMs. Electrophysiology assays, such as patch clamping recording, are too low throughput for HTS, although automated patch clamp technology is advancing. Nevertheless, this limitation can be overcome by using optical recording of fluorescent sensor probes of transmembrane voltage, current transients using dedicated devices or by HC microscopy.303,304 Mechanical force exerted by CMs can be measured, in an HTS format, by culturing cells on thin films of materials that can be bent by systolic contraction,305 or by measuring contraction and relaxation of substrates embedded with fluorescent microspheres.306 In addition to studies in CMs, a recent HTS in primary human cardiac fibroblasts identified drug candidates to target cardiac fibrosis and diastolic dysfunction.307
As indicated in Section 4.2, a major limitation remains the embryonic nature of hESC/hiPSC-CMs. As some embryonic characteristics can mature in vitro CM maturation itself can become the read-out of specific HTS with small molecules or microRNAs. In addition to the cell studies which replace animal studies, recent advances in HTS measurements in enzymatically isolated intact single CMs from rodent hearts reduce the number of animals required for high-throughput testing of compounds and stressors.308,309
Finally, the possibility of growing CMs, either alone or in various combinations with cardiac fibroblasts or other cells offers the opportunity of conducting screenings in conditions of load and CM maturation closer to those of the heart in vivo.310
5. The power of data
5.1 Registration of preclinical trials: data repository for animal research
Preclinical research is pivotal to understand basic mechanisms of diseases and to provide information about the safety and efficacy of new strategies. The ultimate final goal is to make advances in medical science and to improve patient healthcare. Currently, only a relatively small number of the products from translational research finds application in the clinical setting.311 One of the main issues with preclinical studies is publication bias. Positive and/or significant results are more likely to be published than negative study results. This leads to an overestimation of the effects of therapies and unjustified transition of interventions to clinical trials. Moreover, the lack of sharing both negative and positive results contributes to the repetition of research, and failure to comply with the 3R principles.
The development and use of an animal registry and/or preclinical network represent a possible solution for minimizing publication bias. To this end, two platforms (www.preclinicaltrials.eu312 and www.animalstudyregistry.org313) were recently launched for preregistration of animal studies to increase transparency and reproducibility of bioscience research and to promote animal welfare. The registration form helps scientists plan their study thoroughly by asking detailed questions concerning study design, methods, and statistics. Although most researchers are in favour of more transparency, major disadvantages of preregistration exist, especially intellectual property (IP) issues and administrative burden. At present, these are the most likely reasons why there are only a limited number of preregistered studies. Several solutions are currently being incorporated to circumvent these obstacles. One example is when registering a study, it automatically receives a digital object identifier (DOI) that marks it as the original research idea of the investigator. In addition to this, the users can decide to restrict the visibility of their registered studies for up to 5 years. The Consortium for Preclinical Assessment of Cardioprotective Therapies (CAESAR)314 and Mouse Phenome Database (https://phenome.jax.org/) are examples of networks in which experienced laboratories work together and share data on rodent models. The implementation of an independent and prospective animal registry and preclinical network can, therefore, support the researcher in enhancing the quality of the study, as it requires addressing blinding, randomization, sample size calculation, and power. Furthermore, they will lead to standardized protocols, and a reduction of unnecessarily repeated studies, animal use, and costs. A data repository for animal research could be exploited for advanced analysis through artificial intelligence and data mining, which could help to establish rules or formulas for predicting adverse and/or therapeutic responses.
5.2 Patient registries, biobanking, -omics studies and imaging
Further acceleration of clinical cardiovascular research will only be possible if networks are created across institutes and countries to facilitate collaborative data science. In particular, the implementation of (trans)national networks across institutes using similar data models and harmonized clinical care pathways will facilitate patient recruitment in targeted clinical trials and enable genotype–phenotype association studies with appropriate statistical power, for example, in cardiomyopathy patient groups. Furthermore, it would provide a framework for a learning healthcare system through benchmarking, cross-validation of novel strategies and artificial intelligence algorithms in both research and routine care. Unsupervised learning allows for the clustering, structuring and compressing of the information content for a high-dimensional dataset of important features or main components. Common methods are principal component analysis, spectral clustering315 or deep autoencoders.316–318 A well-known extension to autoencoders are variational autoencoders that allow efficient inference and learning in directed probabilistic models.319 Autoencoders are neural networks used to learn an efficient representation in an unsupervised manner. They contain a bottle-neck layer that then generates the latent space of compressed variables. Understanding the underlying data distribution and the effect of involved parameters with such a deep autoencoder, generates predictive models320 and simulates the effect of different parameters, such as drug responses.321
Great steps in creating collaborative networks for human data exchange have been made through the creation of large biobanks, for example the, UK Biobank (https://www.ukbiobank.ac.uk/about-biobank-uk/) and Generation Scotland project (https://www.ed.ac.uk/generation-scotland). Both are resources of demographic, clinical information, biological samples and in some cases imaging data from thousands of volunteers from the South of England and Scotland, respectively. Both biobanks have established multi-disciplinary skills networks in health informatics, epidemiology, genetics, health economics, and focused data analyses from cross-sectional whole-body imaging and specific cardiac imaging. Significant ethical, legal and social issues need to be addressed to allow such complex biobanks to operate safely. The fundamental aim of such large biorepository resources is to improve the prevention, diagnosis, and treatment of a wide range of serious and life-threatening illnesses. Scotland in particular has a unique electronic health record system with data linkage dating back to its creation in 1986, the information available from the Biobankscan be data-linked with clinical outcomes and long-term follow-up, as well as genetic analysis of its participants. Whilst these Biobanks have only recently been established in the past decade, there are much older and implicitly extremely valuable long-term follow-up registries. For example, the Aberdeen Children of the 1950’s, which comprises 12150 participants born between 1950 and 1956 who were subsequently deeply phenotyped every decade with state-of-the-art investigations contemporaneously available at each such time point.
An example of utilizing the maximal potential of data obtained within the different disciplines is Network Medicine. It originated from the fact that conventional scientific reductionism is inadequate for understanding complex diseases and developing precise therapies. Moreover, it views health and disease as an interplay among molecular and environmental determinants that must be fully considered in precision medicine. Network Medicine, therefore, uses big data to create an integrated set of principles and discoveries that can fully capture these inherent dependencies. Focusing on the interaction of biological components, such as proteins, mRNAs, microRNAs, or metabolites, allows us to understand molecular pathways that underlie the pathogenesis of diseases. In addition, Network Medicine has expanded to integrate molecular data with phenotypic features to clarify mechanisms driving clinical disorders.322 The strategy used in Network Medicine to address a clinical question (i.e. absence of a priori hypotheses on the molecular mechanisms causing diseases or a priori molecular target selection) and the technologies used in network analysis are, by definition, unbiased, and do not affect how networks are defined in different data sets or network layers. Therefore, the network medicine approach can lead to a significant reduction of the number of animal experiments designed in the classical reductionist way. As a simple example, the miRNA expression fingerprint of the hypercholesterolaemic myocardium, allows to build the miRNA–mRNA target networks and predict key molecular targets in an unbiased way, thus remarkably reducing the necessary in vivo experiments for validation of predicted targets.323
The cardiovascular community should provide guidelines to establish a framework according to FAIR principles to: enhance findability using metadata catalogues of patients with clinical, genetic, imaging and -omics data; create transparency about accessibility protocols of existing data sources for external researchers and other third parties; stimulate interoperability across institutes to enable collaborative science and federated learning and promote reuse of data in spirit of open science and improve durability of financial and non-financial public investment.324 Instead of manual curation of clinical care data, the cardiovascular community should aim to standardize clinical care pathways and harmonize phenotypes and outcomes within electronic health records to minimize the burden of data collection, and access the wealth of data available within our hospital systems including clinical notes, imaging and -omics data. To facilitate collaborative analyses a common data model should be adopted, like the one developed by the Observational Health Data Sciences and Informatics programme (https://ohdsi.org). A common data model will also enable distributed learning. Currently, collaboration across institutes is limited by privacy and security concerns of data sharing. However, with the development of federated learning, these restrictions could be resolved.325 Instead of sharing data within a huge central data storage (data-to-code), the algorithms will be distributed across centres (code-to-data) without any actual data sharing. The created statistical models and its parameters can subsequently be validated across different clinical settings, patient characteristics (e.g. age, sex and ethnicity), and countries to ensure that those algorithms are generalizable or calibrated to the individual patient in front of us. The importance of such an infrastructure is clearly illustrated by the COVID-19 pandemic. Already existing networks such as REMAP-CAP (Randomized, Embedded, Multifactorial Adaptive Platform Trial for Community-Acquired Pneumonia, www.remapcap.org) and newly founded networks like CAPACITY-COVID (www.capacity-covid.eu) initiated by the cross-institutional Dutch CardioVascular Alliance (www.dcvalliance.nl) have accelerated clinical research to inform patients and caregivers about risk assessment and potential therapies for COVID-19 in a relatively short period. Further development and expansion of networks across countries are needed to collect real-time clinical information to perform point of care pragmatic trials across different groups of patients and healthcare systems.
Lastly, the cardiovascular scientific committee should not forget to involve the main group of interest, the patients.
Quote from a patient: ‘I have given permission to take blood and tissue for scientific research but I have never heard again about the results or outcome of the research’.
Too often, scientists forget to correspond about the results obtained with patient’s data/tissues once a publication is accepted. Participation of patients and their family members is key for successful translational research, in particular in chronic cardiovascular diseases, where follow-up studies in patients and their families are central for improving our knowledge of disease pathomechanisms and effectiveness of treatments. The fact that the questions of cardiovascular biomedical research are scientifically relevant does not necessarily mean that they are relevant from the patient’s perspective. Most research questions are posed from a medical or regulatory perspective and are often based on a laboratory point of view and is focused on basic science that is often removed from the true needs of patients.326 Patient participation in research is thus crucial for identifying patient-relevant questions and outcomes.
5.3 Computational modelling of cardiovascular function
Over the last two decades, there has been rapid development in cardiovascular research methodologies (e.g. advanced methods for quantification of cellular function, better understanding of intercellular communication, new methods for genetic targeting of selected pathways and advanced high-resolution medical imaging), which has increased the quality and quantity of available data on the complex and dynamic function of the cardiovascular system. The availability and the level of details of data have enabled the development of thoroughly validated computational models of heart and vessels.327,328 These models capture the complex non-linear dynamics of the cardiovascular system across different scales, from genetic mutations to subcellular protein function and cellular electrophysiology, to tissue-scale myocardial and vascular mechanics, to organ-scale cardiac pump function and system-scale blood flow dynamics. Computational models provide a unique alternative research platform for integration of experimental data and for performing in silico experiments to better understand cardiovascular physiology and pathophysiology, support clinical decision making and improve safety and efficacy of drug and biomedical device therapies.328
The application of computational models for both fundamental, preclinical and clinical research in biomedicine is rapidly increasing329 and this has led to many examples showing that in silico experiments can lead to refinement, reduction, and in some cases even replacement of animal experiments. For example, research has demonstrated that computational models of cellular cardiac electrophysiology can predict adverse drug effects (e.g. life-threatening arrhythmias) with higher accuracy than animal models330 showing that human computational models can help to reduce the use of animal experiments in early stages of drug testing. This research is part of the Comprehensive in vitro Proarrhythmia Assay initiative (https://cipaproject.org/about-cipa/) that aims to integrate predictions by in vitro, in silico and hiPSC-CM models with clinical evaluation for drug safety testing and is promoted by regulatory bodies.
In fundamental cardiovascular research, in silico cardiovascular models have mainly been used to translate changes in cellular physiology observed in vitro or in animal models to cellular changes in human cells and whole-organ human clinical phenotypes. For example, in the context of cardiac myocyte Ca2+ handling, where in vivo measurements are not available, simulation studies have shown how in silico models can be used to extrapolate changes observed in vitro or in animal models into an in vivo human context.331
In a more clinical setting, multi-scale computational models of heart and vessels are being personalized using the rapidly growing wealth of patient-specific diagnostic data available in the clinic. The resulting virtual representation of the individual patient, also referred to as ‘Digital Twin’,332 can be used to gain better insights into the patient’s cardiovascular pathology, underlying symptoms and to predict the individual’s response to therapy. Studies have demonstrated successful applications of personalized computational models, including prediction of arrhythmia risk in post-MI patients,333 non-invasive measurement of fractional flow reserve from computed tomographic images of patients with coronary artery disease,334 and non-invasive electrocardiographic imaging.335
In conclusion, computational modelling and simulation, sometimes called the third paradigm of science, already established a prominent role in the quest to refine and reduce the use of animal experiments for cardiovascular research. However, computational modelling is not likely to fully replace animal experiments in the foreseeable future. Animal models continue to provide novel insights into pathophysiological processes which have not yet been implemented in computational models. Moreover, animal experimental data are required for validation of computational models when human data are unavailable. What all aforementioned successful applications of computational models have in common is that they are the result of decades of basic research and multidisciplinary collaborations between researchers, computer scientists, and clinicians.
6. Moving from bench to clinic
Our paper highlights the evolution in the design of cardiovascular disease models that has taken place in a relatively brief time-span. Multiple animal-free models and tools to increase power of studies became available, and animal models have been refined in the past ∼20 years. Translation of basic and clinical research to actual implementation in the clinic represents a major challence, and warrants a careful experimental design making use of available complimentary research models ranging from in vitro experiments in cells and iPSC-derived models to studies in rodents, large animals and patients. Recent examples, described below, illustrate the potential of such an approach to move from bench to clinic.
6.1 Peripartum cardiomyopathy
PPCM is a potentially life-threatening heart disease that emerges with acute or with slow progression of LV systolic dysfunction (LVEF < 45%) late in pregnancy, during delivery, or in the first postpartum months, in women with no other known causes of HF.336 Risk factor profiles (i.e. higher risk for PPCM in women with African ancestry) for women with pregnancy-associated hypertensive complications, such as older women or women with twin pregnancies, suggests that PPCM consists of multiple pathomechanisms pointing to a syndrome and not a single defined disease.336,337 This notion is further supported by the prevalence of cardiomyopathy-causing mutations in about 15% of patients338,339 Experimental data confirm that different factors can induce and drive PPCM, including inflammation and immunity, pregnancy hormone impairment, catecholamine stress, defective cAMP-protein kinase A, and G-protein-coupled-receptor signalling genetic variants336 and aberrant cardiac metabolism. Under physiological circumstances, maternal lipid metabolism is increased during the last trimester of pregnancy and normalizes after delivery. Recently, it has been shown that lipid metabolism is widely affected in hiPSC from patients with PPCM, findings that were replicated in a PPCM mouse model.340 Evidence is accumulating that several of these mechanisms may merge into a common major pathway, which includes unbalanced oxidative stress and the cleavage of the nursing hormone prolactin (PRL) into an angiostatic, pro-apoptotic and pro-inflammatory 16 kDa-PRL fragment, resulting in subsequent vascular damage and HF.336 Based on this common pathway, potential disease-specific biomarkers and therapies have emerged that are currently tested in a bench to bedside approach. One therapy concept has been developed in mice where HF medication is combined with the PRL blocker bromocriptine and had already been introduced into 2018 European Society of Cardiology (ESC) Guidelines for the management of cardiovascular diseases during pregnancy.341
6.2 microRNAs - route to the clinic
Based on initial miRNA library screens miR-132 was identified as driver of pathological growth of CMs in vitro and next in vivo (Figure 1C).342 In a number of mouse studies it was shown that oligonucleotide-based inhibiton of miR-132 halted and reverted pathological cardiac remodelling.343 Following this, the therapeutic efficacy was tested in an acute343 and a chronic344 model of MI in pigs. These activities were recently translated to chronic HF patients where the miR-132 inhibitor drug showed a good safety profile and indicative therapeutic efficacy based on improvement of several parameters, such as reduction of N-terminal pro-B-type natriuretic peptide, paving the way for further clinical development of this new generation of HF medication.345
7. Conclusion and future challenges
Globally, there is a mounting belief that biomedical sciences can progress without animal research by replacing in vivo experiments with tests performed in human-derived in vitro models. While this is in part justified as multiple research questions can be answered without the use of animals, the use of animal pathological modelling is still necessary for several applications such as, implantation of medical devices (e.g. stents, new catheter-guided endoscopy systems, implant devices), in vivo drug testing, and for identifying mechanisms underlying cardiovascular disease as outlined in the current paper. Stem cell-based human pathology models have the potential to become key in testing toxicity and effectiveness of new drugs at a cellular or organ-like levels, but lack the complexity present in multiple forms of cardiovascular disease. As cardiovascular disease is a complex, multifactorial disorder, and the current knowledge is limited, we will have to continue to rely on laboratory animals, enabling thorough studies in a well-controlled in vivo setting.
In coming years, animal models will be further refined and made more ‘human-like’ on the basis of big data sets obtained in human studies. As pathomechanisms and treatment response differ between male and female cardiovascular patients, the effect of sex should be taken into account in the design of animal studies. Novel 2D and 3D in vitro technologies, and advanced computational analyses will certainly result in a more refined experimental design reducing the number of laboratory animals currently required to perform studies and test drugs. A major challenge in the refinement of iPSC-derived models is their validation, i.e. do models capture human pathophysiology? The iPSC-derived models may ultimately be used for precision medicine, however, currently, a gap exists between iPSC-derived heart models and the clinical phenotype of patients, as human cardiac muscle systems have not been validated (i.e. not compared to individual patient characteristics and human cardiac tissue samples). This limits their applicability for studies on pathomechanisms and use in the clinical setting. In addition, mimicking sex differences in stem cell-derived heart models is a largely unexplored area and warrants further research and development. Successful translation of cardiovascular research warrants integration of results obtained in animals, animal-free models and patients.
Acknowledgements
We thank Dr Marianna Barbuto and Stefano Rizzi from the Unità di Ingegneria Tissutale Cardiovascolare, Centro cardiologico Monzino, IRCCS in Milan, Italy, for the conception of the Figures 2 and 3.
Contributor Information
Jolanda van der Velden, Amsterdam UMC, Vrije Universiteit, Physiology, Amsterdam Cardiovascular Science, Amsterdam, The Netherlands; Netherlands Heart Institute, Utrecht, The Netherlands.
Folkert W Asselbergs, Division Heart & Lungs, Department of Cardiology, University Medical Center Utrecht, Utrecht University, Utrecht, the Netherlands; Faculty of Population Health Sciences, Institute of Cardiovascular Science and Institute of Health Informatics, University College London, London, UK.
Jeroen Bakkers, Hubrecht Institute-KNAW and University Medical Centre Utrecht, Utrecht, The Netherlands.
Sandor Batkai, Hannover Medical School, Institute of Molecular and Translational Therapeutic Strategies, Hannover, Germany.
Luc Bertrand, Hannover Medical School, Institute of Molecular and Translational Therapeutic Strategies, Hannover, Germany.
Connie R Bezzina, Université catholique de Louvain, Institut de Recherche Expérimentale et Clinique, Pole of Cardiovascular Research, Brussels, Belgium.
Ilze Bot, Heart Center, Department of Experimental Cardiology, Amsterdam UMC, Location Academic Medical Center, Amsterdam Cardiovascular Sciences, University of Amsterdam, Amsterdam, The Netherlands; Division of BioTherapeutics, Leiden Academic Centre for Drug Research, Leiden University, Leiden, The Netherlands.
Bianca J J M Brundel, Amsterdam UMC, Vrije Universiteit, Physiology, Amsterdam Cardiovascular Science, Amsterdam, The Netherlands.
Lucie Carrier, Institute of Experimental Pharmacology and Toxicology, University Medical Center Hamburg Eppendorf, Hamburg, Germany; DZHK (German Centre for Cardiovascular Research), Partner Site Hamburg/Kiel/Lübeck, Hamburg, Germany.
Steven Chamuleau, Amsterdam UMC, Heart Center, Cardiology, Amsterdam Cardiovascular Science, Amsterdam, The Netherlands.
Michele Ciccarelli, Department of Medicine, Surgery and Odontology, University of Salerno, Fisciano (SA), Italy.
Dana Dawson, Department of Cardiology, Aberdeen Cardiovascular and Diabetes Centre, Aberdeen Royal Infirmary and University of Aberdeen, Aberdeen, UK.
Sean M Davidson, The Hatter Cardiovascular Institute, University College London, 67 Chenies Mews, London WC1E 6HX, UK.
Andreas Dendorfer, Walter-Brendel-Centre of Experimental Medicine, University Hospital, Ludwig-Maximilians-University, Munich, Germany.
Dirk J Duncker, Division of Experimental Cardiology, Department of Cardiology, Thoraxcenter, Erasmus MC, University Medical Center Rotterdam, Rotterdam, The Netherlands.
Thomas Eschenhagen, Institute of Experimental Pharmacology and Toxicology, University Medical Center Hamburg Eppendorf, Hamburg, Germany; DZHK (German Centre for Cardiovascular Research), Partner Site Hamburg/Kiel/Lübeck, Hamburg, Germany.
Larissa Fabritz, DZHK (German Centre for Cardiovascular Research), Partner Site Hamburg/Kiel/Lübeck, Hamburg, Germany; University Center of Cardiovascular Sciences and Department of Cardiology, University Heart Center Hamburg, Germany and Institute of Cardiovascular Sciences, University of Birmingham, UK.
Ines Falcão-Pires, UnIC - Cardiovascular Research and Development Centre, Department of Surgery and Physiology, Faculty of Medicine, University of Porto, Portugal.
Péter Ferdinandy, Cardiometabolic Research Group and MTA-SE System Pharmacology Research Group, Department of Pharmacology and Pharmacotherapy, Semmelweis University, Budapest, Hungary; Pharmahungary Group, Szeged, Hungary.
Mauro Giacca, Department of Medicine, Surgery and Health Sciences and Cardiovascular Department, Centre for Translational Cardiology, Azienda Sanitaria Universitaria Integrata Trieste, Trieste, Italy; International Center for Genetic Engineering and Biotechnology (ICGEB), Trieste, Italy; King’s British Heart Foundation Centre, King’s College London, London, UK.
Henrique Girao, Univ Coimbra, Center for Innovative Biomedicine and Biotechnology, Faculty of Medicine, Coimbra, Portugal; Clinical Academic Centre of Coimbra, Coimbra, Portugal.
Can Gollmann-Tepeköylü, Department of Cardiac Surgery, Medical University of Innsbruck, Innsbruck, Austria.
Mariann Gyongyosi, Division of Cardiology, Department of Internal Medicine II, Medical University of Vienna, Vienna, Austria.
Tomasz J Guzik, Instutute of Cardiovascular and Medical Sciences, University of Glasgow, Glasgow, UK; Jagiellonian University, Collegium Medicum, Kraków, Poland.
Nazha Hamdani, Division Cardiology, Molecular and Experimental Cardiology, Ruhr University Bochum, Bochum, Germany; Institute of Physiology, Ruhr University Bochum, Bochum, Germany.
Stephane Heymans, Department of Cardiology, Cardiovascular Research Institute Maastricht (CARIM), Maastricht University Medical Centre, Maastricht University, Maastricht, The Netherlands; Department of Cardiovascular Sciences, University of Leuven, Leuven, Belgium.
Andres Hilfiker, Department for Cardiothoracic, Transplant, and Vascular Surgery, Hannover Medical School, Hannover, Germany.
Denise Hilfiker-Kleiner, Department for Cardiology and Angiology, Hannover Medical School, Hannover, Germany; Department of Cardiovascular Complications in Pregnancy and in Oncologic Therapies, Comprehensive Cancer Centre, Philipps-Universität Marburg, Germany.
Alfons G Hoekstra, Computational Science Lab, Informatics Institute, Faculty of Science, University of Amsterdam, Amsterdam, the Netherlands.
Jean-Sébastien Hulot, Université de Paris, INSERM, PARCC, F-75015 Paris, France; CIC1418 and DMU CARTE, AP-HP, Hôpital Européen Georges-Pompidou, F-75015 Paris, France.
Diederik W D Kuster, Amsterdam UMC, Vrije Universiteit, Physiology, Amsterdam Cardiovascular Science, Amsterdam, The Netherlands.
Linda W van Laake, Division Heart & Lungs, Department of Cardiology, University Medical Center Utrecht, Utrecht University, Utrecht, the Netherlands.
Sandrine Lecour, Department of Medicine, Hatter Institute for Cardiovascular Research in Africa and Cape Heart Institute, University of Cape Town, Cape Town, South Africa.
Tim Leiner, Department of Radiology, Utrecht University Medical Center, Utrecht, the Netherlands.
Wolfgang A Linke, Institute of Physiology II, University of Muenster, Robert-Koch-Str. 27B, 48149 Muenster, Germany.
Joost Lumens, Department of Biomedical Engineering, Cardiovascular Research Institute Maastricht (CARIM), Maastricht University, Maastricht, the Netherlands.
Esther Lutgens, Experimental Vascular Biology Division, Department of Medical Biochemistry, University of Amsterdam, Amsterdam Cardiovascular Sciences, Amsterdam University Medical Centers, Amsterdam, The Netherlands; Institute for Cardiovascular Prevention, Ludwig-Maximilians-Universität München (LMU), Munich, Germany; DZHK, Partner Site Munich Heart Alliance, Munich, Germany.
Rosalinda Madonna, Department of Pathology, Cardiology Division, University of Pisa, 56124 Pisa, Italy; Department of Internal Medicine, Cardiology Division, University of Texas Medical School in Houston, Houston, TX, USA.
Lars Maegdefessel, DZHK, Partner Site Munich Heart Alliance, Munich, Germany; Department for Vascular and Endovascular Surgery, Klinikum rechts der Isar, Technical University Munich, Munich, Germany; Department of Medicine, Karolinska Institutet, Stockholm, Sweden.
Manuel Mayr, King’s British Heart Foundation Centre, King’s College London, London, UK.
Peter van der Meer, Department of Cardiology, University Medical Center Groningen, University of Groningen, Groningen, the Netherlands.
Robert Passier, Department of Applied Stem Cell Technologies, TechMed Centre, University of Twente, 7500AE Enschede, The Netherlands; Department of Anatomy and Embryology, Leiden University Medical Centre, 2300 RC Leiden, The Netherlands.
Filippo Perbellini, Hannover Medical School, Institute of Molecular and Translational Therapeutic Strategies, Hannover, Germany.
Cinzia Perrino, Department of Advanced Biomedical Sciences, Federico II University, Naples, Italy.
Maurizio Pesce, Unità di Ingegneria Tissutale Cardiovascolare, Centro cardiologico Monzino, IRCCS, Milan, Italy.
Silvia Priori, Molecular Cardiology, Istituti Clinici Scientifici Maugeri, Pavia, Italy; University of Pavia, Pavia, Italy.
Carol Ann Remme, Université catholique de Louvain, Institut de Recherche Expérimentale et Clinique, Pole of Cardiovascular Research, Brussels, Belgium.
Bodo Rosenhahn, Institute for information Processing, Leibniz University of Hanover, 30167 Hannover, Germany.
Ulrich Schotten, Department of Physiology, Cardiovascular Research Institute Maastricht, Maastricht University, Maastricht, the Netherlands.
Rainer Schulz, Institute of Physiology, Justus Liebig University Giessen, Giessen, Germany.
Karin R Sipido, Department of Cardiovascular Sciences, KU Leuven, 3000 Leuven, Belgium.
Joost P G Sluijter, Experimental Cardiology Laboratory, Department of Cardiology, Regenerative Medicine Center Utrecht, Circulatory Health Laboratory, Utrecht University, University Medical Center Utrecht, Utrecht, The Netherlands.
Frank van Steenbeek, Division Heart & Lungs, Department of Cardiology, University Medical Center Utrecht, Utrecht University, Utrecht, the Netherlands; Department of Clinical Sciences, Faculty of Veterinary Medicine, Utrecht University, Utrecht, The Netherlands.
Sabine Steffens, Institute for Cardiovascular Prevention, Ludwig-Maximilians-Universität München (LMU), Munich, Germany; DZHK, Partner Site Munich Heart Alliance, Munich, Germany.
Cesare M Terracciano, National Heart & Lung Institute, Imperial College London, London, UK.
Carlo Gabriele Tocchetti, Cardio-Oncology Unit, Department of Translational Medical Sciences, Center for Basic and Clinical Immunology Research (CISI), Interdepartmental Center for Clinical and Translational Research (CIRCET), Interdepartmental Hypertension Research Center (CIRIAPA), Federico II University, Naples, Italy.
Patricia Vlasman, Amsterdam UMC, Vrije Universiteit, Physiology, Amsterdam Cardiovascular Science, Amsterdam, The Netherlands.
Kak Khee Yeung, Amsterdam UMC, Vrije Universiteit, Surgery, Amsterdam Cardiovascular Science, Amsterdam, The Netherlands.
Serena Zacchigna, Department of Medicine, Surgery and Health Sciences and Cardiovascular Department, Centre for Translational Cardiology, Azienda Sanitaria Universitaria Integrata Trieste, Trieste, Italy; International Center for Genetic Engineering and Biotechnology (ICGEB), Trieste, Italy.
Dayenne Zwaagman, Amsterdam UMC, Heart Center, Cardiology, Amsterdam Cardiovascular Science, Amsterdam, The Netherlands.
Thomas Thum, Hannover Medical School, Institute of Molecular and Translational Therapeutic Strategies, Hannover, Germany; Fraunhofer Institute for Toxicology and Experimental Medicine, Hannover, Germany.
Authors’ contributions
All authors contributed to the design of the consensus document, and drafted and approved the final version of the manuscript. All authors agree to be accountable for all aspects of the work, and have confidence in the integrity of the contributions of their co-authors.
Funding
J.v.d.V. acknowledges support from NWO-ZonMW (91818602 VICI grant), ZonMW and Heart Foundation for the translational research program, project 95105003; the Dutch Cardiovascular Alliance (DCVA) grant Double Dose 2021; the Leducq Foundation grant number 20CVD01; and Proper Therapy project funded by the Dutch Research Council, domain Applied and Engineering Sciences (NWO-AES), the Association of Collaborating Health Foundations (SGF), and ZonMW within the Human models 2.0 call. F.A. is supported by UCL Hospitals NIHR Biomedical Research Centre, and the DCVA grant Double Dose 2021. J.B. is supported by the Netherlands CardioVascular Research Initiative CVON (CVON2014-18, CVON2018-30, and CVON2019-002), Stichting Hartekind and the Dutch Research Counsel (NWO) (OCENW.GROOT.2019.029). L.B. is supported by National Fund for Scientific Research, Belgium and Action de Recherche Concertée de la Communauté Wallonie-Bruxelles, Belgium. C.R.B. acknowledges support from NWO-ZonMW (016.150.610 VICI grant), the Netherlands CardioVascular Research Initiative CVON (PREDICT2 and CONCOR-genes projects), the Leducq Foundation (project 17CVD02), and ERA PerMed (PROCEED study). B.B. acknowledges support from the Netherlands Cardiovascular Research Initiative: an initiative with support of the Dutch Heart Foundation, CVON2014-40 DOSIS, CVON-STW2016-14728 and the Medical Delta. L.C. is supported by the German Centre of Cardiovascular Research (DZHH); and the Leducq Foundation grant number 20CVD01. D.D. is supported by the British Heart Foundation (FS/RTF/20/30009, NH/19/1/34595, PG/18/35/33786, CS/17/4/32960, PG/15/88/31780, and PG/17/64/33205), Chest Heart and Stroke Scotland (19/53), Tenovus Scotland (G.18.01), Friends of Anchor and Grampian NHS-Endowments. S.D. was supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre (BRC233/CM/SD/101320) from the British Heart Foundation (PG/18/44/33790). A.D. is supported by the German Centre for Cardiovascular Research (DZHK, 81X2600253 and 81X2600257). D.J.D. was supported by the Netherlands CardioVascular Research Initiative CVON (CVON2014 RECONNECT and CVON2016 ARENA-PRIME). The work of T.E. was supported by the European Research Council (ERC-AG IndivuHeart), the Deutsche Forschungsgemeinschaft (DFG Es 88/12-1), the European Union Horizon 2020 (REANIMA and TRAINHEART), the German Ministry of Education and Research (BMBF), and the Centre for Cardiovascular Research (DZHK). L.F. was supported by European Union Horizon 2020 [grant agreement No 633196 (CATCH ME) and 965286 (MAESTRIA)]; British Heart Foundation (FS/13/43/30324; PG/17/30/32961; PG/20/22/35093; and AA/18/2/34218); DFG FA413. The Institute of Cardiovascular Sciences, University of Birmingham is a recipient of a BHF Accelerator Award (AA/18/2/34218). P.F. was supported by the National Research, Development and Innovation Office of Hungary (Research Excellence Program—TKP; National Heart Program NVKP 16-1-2016-0017); by the Higher Education Institutional Excellence Program of the Ministry of Human Capacities in Hungary, within the framework of the Therapeutic Development thematic program of the Semmelweis University; and by the European Union Horizon 2020 (COVIRNA, CRYTAL). H.G. is supported by PAC ‘NETDIAMOND’ POCI‐01‐0145‐FEDER‐016385; HealthyAging2020 CENTRO‐01‐0145‐FEDER‐000012‐N2323; POCI‐01‐0145‐FEDER‐007440, CENTRO‐01‐0145‐FEDER‐032179, CENTRO‐01‐0145‐FEDER‐032414, POCI-01-0145-FEDER-022122, UID/NEU/04539/2019, UIDB/04539/2020, and UIDP/04539/2020. C.G.-T. was supported by the Austrian Science Fund (P 32821). S.H. acknowledges the European Union Commission’s Seventh Framework programme under grant agreement N° 305507 [HOMAGE0, IMI2-CARDIATEAM (N° 821508)] and support from the Netherlands Cardiovascular Research Initiative, an initiative with support of the Dutch Heart Foundation, CVON2016-Early HFPEF, 2015-10, CVON She-PREDICTS, grant 2017-21, CVON Arena-PRIME, 2017-18, CVON Double Dosis, and support of FWO G091018N and FWO G0B5930N. A.G.H. acknowledges support from the INSIST project (www.insist-h2020.eu) and the CompBioMed2 project (https://www.compbiomed.eu) that both received funding from the European Union’s Horizon 2020 research and innovation programme under respectively grant agreement No 777072 and No 823712. D.H. was supported by the Deutsche Forschungsgemeinschaft (DFG, Hi 842/4-3; 842/10-2;) and the Leducq Foundation (transatlantic network of excellence: Targeted Approaches for Prevention and Treatment of Anthracycline-Induced Cardiotoxicity) and Volkswagenstiftung (A128871). A.H. was/is supported by the Deutsche Forschungsgemeinschaft (DFG) via the Cluster of Excellence ‘From regenerative biology to reconstructive therapy’ (REBIRTH), via the project C7 of TRR127 (Biology of xeno-geneic cell and organ transplantation—from bench to bedside), and via the Project HA 13 06/9-1, the BMBF Project ‘AUREKA’, the project B4 of R2N by the Federal State of Lower Saxony, the Fördergemeinschaft ‘Deutsche Kinderherzzentren e.V.’ and the ‘Cortiss’ foundation. J.-S.H. is supported by AP-HP, INSERM, the French National Research Agency (NADHeart ANR-17-CE17-0015-02, PACIFIC ANR-18-CE14-0032-01, CORRECT_LMNA ANR-19-CE17-0013-02), the ERA-Net-CVD (ANR-16-ECVD-0011-03, Clarify project), Fédération Française de Cardiologie, the Fondation pour la Recherche Médicale (EQU201903007852), and by a grant from the Leducq Foundation (18CVD05) and is coordinating a French PIA Project (2018-PSPC-07, PACIFIC-preserved, BPIFrance) and a University Research Federation against heart failure (FHU2019, PREVENT Heart Failure). D.K. acknowledges the PPP Allowance made available by Health_Holland, Top Sector Life Sciences & Health, to stimulate public–private partnerships. L.W.v.L. is supported by the Netherlands Heart Foundation [Dekker Senior Clinical Scientist (2019T056), Health Holland TKI-LSH (LSHM19035), and TUe/UMCU/UU Alliance Fund]. S.L. is supported by grants from the south African National Foundation, the Cancer Association of South Africa and Winetech. T.L. is supported by the Netherlands Heart Foundation/Applied & Engineering Sciences grant number 14741 and Institutional research grant by Dutch Technology Foundation (P15-26) with participation of Pie Medical Imaging and Philips Healthcare; Institutional research grant by Dutch Technology Foundation (12726) with participation of Pie Medical Imaging; institutional research grant by The Netherlands Organisation for Health Research and Development with participation of Pie Medical Imaging; Industrial research grant by Pie Medical Imaging. J.L. was supported by the Netherlands Organisation for Scientific Research (NWO-ZonMw, grant 016.176.340) and the Dutch Heart Foundation (ERA-CVD JTC2018 grant 2018T094, EMPATHY project; Dr. Dekker Program grant 2015T082). E.L. acknowledges the support from the Netherlands CardioVascular Research Initiative: the Dutch Heart Foundation, Dutch Federation of University Medical Centres, the Netherlands Organization for Health Research and Development and the Royal Netherlands Academy of Sciences for the GENIUS-II project ‘Generating the best evidence-based pharmaceutical targets for atherosclerosis’ (CVON2017-20), the Deutsche Forschungsgemeinschaft (CRC 1123), the Netherlands Organization for Scientific Research (NWO) (VICI grant); the European Research Council (ERC consolidator grant 681493). R.M. is supported by grants from Incyte s.r.l. and from Ministero dell’Istruzione, Università e Ricerca Scientifica (549901_2020). L.M. is supported by the German Center for Cardiovascular Research (Junior Research Group & Translational Research Project), the European Research Council (ERC Starting Grant NORVAS), the SFB1123 and TRR267 of the German Research Council (DFG), the Swedish Heart-Lung-Foundation (20180680), the Swedish Research Council (Vetenkapsrådet 2019-01577), the National Institutes of Health (NIH; 1R011HL150359-01), and the Bavarian State Ministry of Health and Care through the research project DigiMed Bayern. P.v.d.M. is supported by the ERC (StG 715732). R.P. is supported by ERA-CVD 2016T092, Health Holland TKI-LSH (LSHM19004), the Dutch Heart Foundation, ZonMw and by the NWO Gravitation project (024.003.001). C.P. was supported by Ministero dell'Istruzione, Università e Ricerca Scientifica grant (2015583WMX) and Programma STAR grant by Federico II University and Compagnia di San Paolo. M.P. is supported by grants of the Italian Ministry of Health (Ricerca Corrente, 5 per 1000) and from Regione Lombardia. C.A.R. is supported by the Netherlands CardioVascular Research Initiative CVON (CVON2018-30 and CVON2015-12) and the Netherlands Organisation for Health Research and Development (ZonMw 91714371). U.S. is supported by grants of the Netherlands Heart Foundation (CVON2014-09, RACE V Reappraisal of Atrial Fibrillation: Interaction between hyperCoagulability, Electrical remodelling, and Vascular Destabilisation in the Progression of AF) and the European Union (ITN Network Personalize AF: Personalized Therapies for Atrial Fibrillation: a translational network, grant number 860974; MAESTRIA: Machine Learning Artificial Intelligence Early Detection Stroke Atrial Fibrillation, grant number 965286; REPAIR: Restoring cardiac mechanical function by polymeric artificial muscular tissue, grant number 952166). R.S. was supported by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) (Project number 268555672—SFB 1213, Project B05). J.S. was supported by European Union H2020 program to the project TECHNOBEAT (grant number 66724), EVICARE (grant number 725229) and BRAV3 (grant number 874827), and ZonMw program No. 116006102. S.S. is supported by the Deutsche Forschungsgemeinschaft (DFG CRC 1123) and the German Centre for Cardiovascular Research (DZHK). C.T. is supported by the British Heart Foundation Centre for Cardiac Regeneration RM/17/1/33377, British Heart Foundation studentship FS/18/37/33642, NC3Rs grant NC/T001488/1. S.Z. is supported by the Interreg ITA-AUS project InCARDIO (B56J19000210005) and by the Italian Association for Cancer Research (AIRC IG 2020 ID 24529). T.T. acknowledges funding from the Deutsche Forschungsgemeinschaft (KFO311, TRR267 and SFB1470).
Data availability
No new data were generated or analysed in support of this consensus document.
References
- 1. Pearson J, Sipido KR, Musialek P, van Gilst WH. The Cardiovascular Research community calls for action to address the growing burden of cardiovascular disease. Cardiovasc Res 2019;115:e96–e98. [DOI] [PubMed] [Google Scholar]
- 2. Russell WMS, Burch RL. The Principles of Humane Experimental Technique. Maryland: Johns Hopkins Bloomberg School of Public Health; 1959. [Google Scholar]
- 3. Roth GA, Johnson C, Abajobir A, Abd-Allah F, Abera SF, Abyu G, Ahmed M, Aksut B, Alam T, Alam K, Alla F, Alvis-Guzman N, Amrock S, Ansari H, Arnlov J, Asayesh H, Atey TM, Avila-Burgos L, Awasthi A, Banerjee A, Barac A, Barnighausen T, Barregard L, Bedi N, Belay Ketema E, Bennett D, Berhe G, Bhutta Z, Bitew S, Carapetis J, Carrero JJ, Malta DC, Castaneda-Orjuela CA, Castillo-Rivas J, Catala-Lopez F, Choi JY, Christensen H, Cirillo M, Cooper LJr, Criqui M, Cundiff D, Damasceno A, Dandona L, Dandona R, Davletov K, Dharmaratne S, Dorairaj P, Dubey M, Ehrenkranz R, El Sayed Zaki M, Faraon EJA, Esteghamati A, Farid T, Farvid M, Feigin V, Ding EL, Fowkes G, Gebrehiwot T, Gillum R, Gold A, Gona P, Gupta R, Habtewold TD, Hafezi-Nejad N, Hailu T, Hailu GB, Hankey G, Hassen HY, Abate KH, Havmoeller R, Hay SI, Horino M, Hotez PJ, Jacobsen K, James S, Javanbakht M, Jeemon P, John D, Jonas J, Kalkonde Y, Karimkhani C, Kasaeian A, Khader Y, Khan A, Khang YH, Khera S, Khoja AT, Khubchandani J, Kim D, Kolte D, Kosen S, Krohn KJ, Kumar GA, Kwan GF, Lal DK, Larsson A, Linn S, Lopez A, Lotufo PA, El Razek HMA, Malekzadeh R, Mazidi M, Meier T, Meles KG, Mensah G, Meretoja A, Mezgebe H, Miller T, Mirrakhimov E, Mohammed S, Moran AE, Musa KI, Narula J, Neal B, Ngalesoni F, Nguyen G, Obermeyer CM, Owolabi M, Patton G, Pedro J, Qato D, Qorbani M, Rahimi K, Rai RK, Rawaf S, Ribeiro A, Safiri S, Salomon JA, Santos I, Santric Milicevic M, Sartorius B, Schutte A, Sepanlou S, Shaikh MA, Shin MJ, Shishehbor M, Shore H, Silva DAS, Sobngwi E, Stranges S, Swaminathan S, Tabares-Seisdedos R, Tadele Atnafu N, Tesfay F, Thakur JS, Thrift A, Topor-Madry R, Truelsen T, Tyrovolas S, Ukwaja KN, Uthman O, Vasankari T, Vlassov V, Vollset SE, Wakayo T, Watkins D, Weintraub R, Werdecker A, Westerman R, Wiysonge CS, Wolfe C, Workicho A, Xu G, Yano Y, Yip P, Yonemoto N, Younis M, Yu C, Vos T, Naghavi M, Murray C. Global, Regional, and National Burden of Cardiovascular Diseases for 10 Causes, 1990 to 2015. J Am Coll Cardiol 2017;70:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Birks EJ. Molecular changes after left ventricular assist device support for heart failure. Circ Res 2013;113:777–791. [DOI] [PubMed] [Google Scholar]
- 5. Crespo-Leiro MG, Anker SD, Maggioni AP, Coats AJ, Filippatos G, Ruschitzka F, Ferrari R, Piepoli MF, Delgado Jimenez JF, Metra M, Fonseca C, Hradec J, Amir O, Logeart D, Dahlstrom U, Merkely B, Drozdz J, Goncalvesova E, Hassanein M, Chioncel O, Lainscak M, Seferovic PM, Tousoulis D, Kavoliuniene A, Fruhwald F, Fazlibegovic E, Temizhan A, Gatzov P, Erglis A, Laroche C, Mebazaa A; Heart Failure Association (HFA) of the European Society of Cardiology (ESC) . European Society of Cardiology Heart Failure Long-Term Registry (ESC-HF-LT): 1-year follow-up outcomes and differences across regions. Eur J Heart Fail 2016;18:613–625. [DOI] [PubMed] [Google Scholar]
- 6. Roger VL. Epidemiology of heart failure. Circ Res 2013;113:646–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cook C, Cole G, Asaria P, Jabbour R, Francis DP. The annual global economic burden of heart failure. Int J Cardiol 2014;171:368–376. [DOI] [PubMed] [Google Scholar]
- 8. Heidenreich PA, Albert NM, Allen LA, Bluemke DA, Butler J, Fonarow GC, Ikonomidis JS, Khavjou O, Konstam MA, Maddox TM, Nichol G, Pham M, Pina IL, Trogdon JG; American Heart Association Advocacy Coordinating Committee; Council on Arteriosclerosis, Thrombosis and Vascular Biology; Council on Cardiovascular Radiology and Intervention; Council on Clinical Cardiology; Council on Epidemiology and Prevention; Stroke Council . Forecasting the impact of heart failure in the United States: a policy statement from the American Heart Association. Circ Heart Fail 2013;6:606–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fabritz L, Crijns HJGM, Guasch E, Goette A, Häusler KG, Kotecha D, Lewalter T, Meyer C, Potpara TS, Rienstra M, Schnabel RB, Willems S, Breithardt G, Camm AJ, Chan A, Chua W, de Melis M, Dimopoulou C, Dobrev D, Easter C, Eckardt L, Haase D, Hatem S, Healey JS, Heijman J, Hohnloser SH, Huebner T, Ilyas BS, Isaacs A, Kutschka I, Leclercq C, Lip GYH, Marinelli EA, Merino JL, Mont L, Nabauer M, Oldgren J, Pürerfellner H, Ravens U, Savelieva I, Sinner MF, Sitch A, Smolnik R, Steffel J, Stein K, Stoll M, Svennberg E, Thomas D, Van Gelder IC, Vardar B, Wakili R, Wieloch M, Zeemering S, Ziegler PD, Heidbuchel H, Hindricks G, Schotten U, Kirchhof P. Dynamic risk assessment to improve quality of care in patients with atrial fibrillation: the 7th AFNET/EHRA Consensus Conference. Europace 2021;23:329–344. [DOI] [PubMed] [Google Scholar]
- 10. Authors/Task Force members; Elliott PM, Anastasakis A, Borger MA, Borggrefe M, Cecchi F, Charron P, Hagege AA, Lafont A, Limongelli G, Mahrholdt H, McKenna WJ, Mogensen J, Nihoyannopoulos P, Nistri S, Pieper PG, Pieske B, Rapezzi C, Rutten FH, Tillmanns C, Watkins H. 2014 ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC). Eur Heart J 2014;35:2733–2779. [DOI] [PubMed] [Google Scholar]
- 11. Bagnall RD, Weintraub RG, Ingles J, Duflou J, Yeates L, Lam L, Davis AM, Thompson T, Connell V, Wallace J, Naylor C, Crawford J, Love DR, Hallam L, White J, Lawrence C, Lynch M, Morgan N, James P, Du Sart D, Puranik R, Langlois N, Vohra J, Winship I, Atherton J, McGaughran J, Skinner JR, Semsarian C. A prospective study of sudden cardiac death among children and young adults. N Engl J Med 2016;374:2441–2452. [DOI] [PubMed] [Google Scholar]
- 12. Sakalihasan N, Michel J-B, Katsargyris A, Kuivaniemi H, Defraigne J-O, Nchimi A, Powell JT, Yoshimura K, Hultgren R. Abdominal aortic aneurysms. Nat Rev Dis Primers 2018;4:35. [DOI] [PubMed] [Google Scholar]
- 13. Kheradvar A, Zareian R, Kawauchi S, Goodwin RL, Rugonyi S. Animal models for heart valve research and development. Drug Discov Today Dis Models 2017;24:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McMurray JJ. Clinical practice. Systolic heart failure. N Engl J Med 2010;362:228–238. [DOI] [PubMed] [Google Scholar]
- 15. Lloyd-Jones DM, Larson MG, Leip EP, Beiser A, D’Agostino RB, Kannel WB, Murabito JM, Vasan RS, Benjamin EJ, Levy D; Framingham Heart Study . Lifetime risk for developing congestive heart failure: the Framingham Heart Study. Circulation 2002;106:3068–3072. [DOI] [PubMed] [Google Scholar]
- 16. Tocchetti CG, Ameri P, de Boer RA, D'Alessandra Y, Russo M, Sorriento D, Ciccarelli M, Kiss B, Bertrand L, Dawson D, Falcao-Pires I, Giacca M, Hamdani N, Linke WA, Mayr M, van der Velden J, Zacchigna S, Ghigo A, Hirsch E, Lyon AR, Görbe A, Ferdinandy P, Madonna R, Heymans S, Thum T. Cardiac dysfunction in cancer patients: beyond direct cardiomyocyte damage of anticancer drugs. Novel cardio-oncology insights from the joint 2019 meeting of the ESC Working Groups of Myocardial Function and Cellular Biology of the Heart. Cardiovasc Res 2020;116:1820–1834. [DOI] [PubMed] [Google Scholar]
- 17. Sagar S, Liu PP, Cooper LTJr. Myocarditis. Lancet 2012;379:738–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davis MB, Arany Z, McNamara DM, Goland S, Elkayam U. Peripartum cardiomyopathy: JACC State-of-the-Art Review. J Am Coll Cardiol 2020;75:207–221. [DOI] [PubMed] [Google Scholar]
- 19. Rosenbaum AN, Agre KE, Pereira NL. Genetics of dilated cardiomyopathy: practical implications for heart failure management. Nat Rev Cardiol 2020;17:286–297. [DOI] [PubMed] [Google Scholar]
- 20. Ponikowski P, Voors AA, Anker SD, Bueno H, Cleland JGF, Coats AJS, Falk V, Gonzalez-Juanatey JR, Harjola VP, Jankowska EA, Jessup M, Linde C, Nihoyannopoulos P, Parissis JT, Pieske B, Riley JP, Rosano GMC, Ruilope LM, Ruschitzka F, Rutten FH, van der Meer P; ESC Scientific Document Group . 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC)Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur Heart J 2016;37:2129–2200. [DOI] [PubMed] [Google Scholar]
- 21. Cowie MR, Fisher M. SGLT2 inhibitors: mechanisms of cardiovascular benefit beyond glycaemic control. Nat Rev Cardiol 2020;17:761–772. [DOI] [PubMed] [Google Scholar]
- 22. Armstrong PW, Pieske B, Anstrom KJ, Ezekowitz J, Hernandez AF, Butler J, Lam CSP, Ponikowski P, Voors AA, Jia G, McNulty SE, Patel MJ, Roessig L, Koglin J, O'Connor CM; VICTORIA Study Group . Vericiguat in patients with heart failure and reduced ejection fraction. N Engl J Med 2020;382:1883–1893. [DOI] [PubMed] [Google Scholar]
- 23. Packer M. The future treatment of heart failure? Eur Heart J 2018;39:5–7. [DOI] [PubMed] [Google Scholar]
- 24. McClellan M, Brown N, Califf RM, Warner JJ. Call to Action: urgent challenges in cardiovascular disease: a presidential advisory from the american heart association. Circulation 2019;139:e44–e54. [DOI] [PubMed] [Google Scholar]
- 25. Katz AM, Rolett EL. Heart failure: when form fails to follow function. Eur Heart J 2016;37:449–454. [DOI] [PubMed] [Google Scholar]
- 26. Metra M, Teerlink JR. Heart failure. Lancet 2017;390:1981–1995. [DOI] [PubMed] [Google Scholar]
- 27. Bloom MW, Greenberg B, Jaarsma T, Januzzi JL, Lam CSP, Maggioni AP, Trochu JN, Butler J. Heart failure with reduced ejection fraction. Nat Rev Dis Primers 2017;3:17058. [DOI] [PubMed] [Google Scholar]
- 28. Riehle C, Bauersachs J. Small animal models of heart failure. Cardiovasc Res 2019;115:1838–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bøtker HE, Hausenloy D, Andreadou I, Antonucci S, Boengler K, Davidson SM, Deshwal S, Devaux Y, Di Lisa F, Di Sante M, Efentakis P, Femminò S, García-Dorado D, Giricz Z, Ibanez B, Iliodromitis E, Kaludercic N, Kleinbongard P, Neuhäuser M, Ovize M, Pagliaro P, Rahbek-Schmidt M, Ruiz-Meana M, Schlüter KD, Schulz R, Skyschally A, Wilder C, Yellon DM, Ferdinandy P, Heusch G. Practical guidelines for rigor and reproducibility in preclinical and clinical studies on cardioprotection. Basic Res Cardiol 2018;113:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pavo N, Lukovic D, Zlabinger K, Zimba A, Lorant D, Goliasch G, Winkler J, Pils D, Auer K, Jan Ankersmit H, Giricz Z, Baranyai T, Sarkozy M, Jakab A, Garamvölgyi R, Emmert MY, Hoerstrup SP, Hausenloy DJ, Ferdinandy P, Maurer G, Gyöngyösi M. Sequential activation of different pathway networks in ischemia-affected and non-affected myocardium, inducing intrinsic remote conditioning to prevent left ventricular remodeling. Sci Rep 2017;7:43958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baranyai T, Giricz Z, Varga ZV, Koncsos G, Lukovic D, Makkos A, Sárközy M, Pávó N, Jakab A, Czimbalmos C, Vágó H, Ruzsa Z, Tóth L, Garamvölgyi R, Merkely B, Schulz R, Gyöngyösi M, Ferdinandy P. In vivo MRI and ex vivo histological assessment of the cardioprotection induced by ischemic preconditioning, postconditioning and remote conditioning in a closed-chest porcine model of reperfused acute myocardial infarction: importance of microvasculature. J Transl Med 2017;15:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fiedler LR, Chapman K, Xie M, Maifoshie E, Jenkins M, Golforoush PA, Bellahcene M, Noseda M, Faust D, Jarvis A, Newton G, Paiva MA, Harada M, Stuckey DJ, Song W, Habib J, Narasimhan P, Aqil R, Sanmugalingam D, Yan R, Pavanello L, Sano M, Wang SC, Sampson RD, Kanayaganam S, Taffet GE, Michael LH, Entman ML, Tan TH, Harding SE, Low CMR, Tralau-Stewart C, Perrior T, Schneider MD. MAP4K4 inhibition promotes survival of human stem cell-derived cardiomyocytes and reduces infarct size in vivo. Cell Stem Cell 2019;24:579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Richards DJ, Li Y, Kerr CM, Yao J, Beeson GC, Coyle RC, Chen X, Jia J, Damon B, Wilson R, Starr Hazard E, Hardiman G, Menick DR, Beeson CC, Yao H, Ye T, Mei Y. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat Biomed Eng 2020;4:446–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Waard MC, van der Velden J, Bito V, Ozdemir S, Biesmans L, Boontje NM, Dekkers DH, Schoonderwoerd K, Schuurbiers HC, de Crom R, Stienen GJ, Sipido KR, Lamers JM, Duncker DJ. Early exercise training normalizes myofilament function and attenuates left ventricular pump dysfunction in mice with a large myocardial infarction. Circ Res 2007;100:1079–1088. [DOI] [PubMed] [Google Scholar]
- 35. Pavo IJ, Pavo N, Kastner N, Traxler D, Lukovic D, Zlabinger K, Spannbauer A, Riesenhuber M, Lorant D, Bartko PE, Goliasch G, Hülsmann M, Winkler J, Gyöngyösi M. Heart failure with reduced ejection fraction is characterized by systemic NEP downregulation. JACC Basic Transl Sci 2020;5:715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nasi-Er BG, Lou X, Zhang Y, Sun H, Zhou X, Li Y, Zhou Q, Zhang J, Tang B, Lu Y. Renal sympathetic denervation improves outcomes in a canine myocardial infarction model. Med Sci Monit 2019;25:3887–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rienzo M, Imbault J, El Boustani Y, Beurton A, Carlos Sampedrano C, Pasdois P, Pernot M, Bernus O, Haïssaguerre M, Couffinhal T, Ouattara A. A total closed chest sheep model of cardiogenic shock by percutaneous intracoronary ethanol injection. Sci Rep 2020;10:12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Contamin H, Rioufol G, Bettinger T, Helbert A, Portier KG, Lepage OM, Thomas R, Broillet A, Tranquart F, Schneider M. A minimally-invasive closed chest myocardial occlusion-reperfusion model in rhesus monkeys (Macaca mulatta): monitoring by contrast-enhanced ultrasound imaging. Int J Cardiovasc Imaging 2012;28:531–554. [DOI] [PubMed] [Google Scholar]
- 39. van der Velden J, Merkus D, Klarenbeek BR, James AT, Boontje NM, Dekkers DH, Stienen GJ, Lamers JM, Duncker DJ. Alterations in myofilament function contribute to left ventricular dysfunction in pigs early after myocardial infarction. Circ Res 2004;95:e85–e95. [DOI] [PubMed] [Google Scholar]
- 40. Zhang S, Picard MH, Vasile E, Zhu Y, Raffai RL, Weisgraber KH, Krieger M. Diet-induced occlusive coronary atherosclerosis, myocardial infarction, cardiac dysfunction, and premature death in scavenger receptor class B type I-deficient, hypomorphic apolipoprotein ER61 mice. Circulation 2005;111:3457–3464. [DOI] [PubMed] [Google Scholar]
- 41. Van der Donckt C, Van Herck JL, Schrijvers DM, Vanhoutte G, Verhoye M, Blockx I, Van Der Linden A, Bauters D, Lijnen HR, Sluimer JC, Roth L, Van Hove CE, Fransen P, Knaapen MW, Hervent AS, De Keulenaer GW, Bult H, Martinet W, Herman AG, De Meyer GR. Elastin fragmentation in atherosclerotic mice leads to intraplaque neovascularization, plaque rupture, myocardial infarction, stroke, and sudden death. Eur Heart J 2015;36:1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Efentakis P, Varela A, Chavdoula E, Sigala F, Sanoudou D, Tenta R, Gioti K, Kostomitsopoulos N, Papapetropoulos A, Tasouli A, Farmakis D, Davos CH, Klinakis A, Suter T, Cokkinos DV, Iliodromitis EK, Wenzel P, Andreadou I. Levosimendan prevents doxorubicin-induced cardiotoxicity in time- and dose-dependent manner: implications for inotropy. Cardiovasc Res 2020;116:576–591. [DOI] [PubMed] [Google Scholar]
- 43. Galán-Arriola C, Lobo M, Vílchez-Tschischke JP, López GJ, de Molina-Iracheta A, Pérez-Martínez C, Agüero J, Fernández-Jiménez R, Martín-García A, Oliver E, Villena-Gutierrez R, Pizarro G, Sánchez PL, Fuster V, Sánchez-González J, Ibanez B. Serial magnetic resonance imaging to identify early stages of anthracycline-induced cardiotoxicity. J Am Coll Cardiol 2019;73:779–791. [DOI] [PubMed] [Google Scholar]
- 44. Gyöngyösi M, Lukovic D, Zlabinger K, Spannbauer A, Gugerell A, Pavo N, Traxler D, Pils D, Maurer G, Jakab A, Riesenhuber M, Pircher A, Winkler J, Bergler-Klein J. Liposomal doxorubicin attenuates cardiotoxicity via induction of interferon-related DNA damage resistance. Cardiovasc Res 2020;116:970–982. [DOI] [PubMed] [Google Scholar]
- 45. Kishimoto C, Hiraoka Y, Takamatsu N, Takada H, Kamiya H, Ochiai H. An in vivo model of autoimmune post-coxsackievirus B3 myocarditis in severe combined immunodeficiency mouse. Cardiovasc Res 2003;60:397–403. [DOI] [PubMed] [Google Scholar]
- 46. Pinkert S, Pryshliak M, Pappritz K, Knoch K, Hazini A, Dieringer B, Schaar K, Dong F, Hinze L, Lin J, Lassner D, Klopfleisch R, Solimena M, Tschöpe C, Kaya Z, El-Shafeey M, Beling A, Kurreck J, Van Linthout S, Klingel K, Fechner H. Development of a new mouse model for coxsackievirus-induced myocarditis by attenuating coxsackievirus B3 virulence in the pancreas. Cardiovasc Res 2020;116:1756–1766. [DOI] [PubMed] [Google Scholar]
- 47. Sharma A, Marceau C, Hamaguchi R, Burridge PW, Rajarajan K, Churko JM, Wu H, Sallam KI, Matsa E, Sturzu AC, Che Y, Ebert A, Diecke S, Liang P, Red-Horse K, Carette JE, Wu SM, Wu JC. Human induced pluripotent stem cell-derived cardiomyocytes as an in vitro model for coxsackievirus B3-induced myocarditis and antiviral drug screening platform. Circ Res 2014;115:556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bailey AL, Dmytrenko O, Greenberg L, Bredemeyer AL, Ma P, Liu J, Penna V, Winkler ES, Sviben S, Brooks E, Nair AP, Heck KA, Rali AS, Simpson L, Saririan M, Hobohm D, Stump WT, Fitzpatrick JA, Xie X, Zhang X, Shi PY, Hinson JT, Gi WT, Schmidt C, Leuschner F, Lin CY, Diamond MS, Greenberg MJ, Lavine KJ. SARS-CoV-2 infects human engineered heart tissues and models COVID-19 myocarditis. JACC Basic Transl Sci 2021;6:331–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, de Ferranti SD, Floyd J, Fornage M, Gillespie C, Isasi CR, Jiménez MC, Jordan LC, Judd SE, Lackland D, Lichtman JH, Lisabeth L, Liu S, Longenecker CT, Mackey RH, Matsushita K, Mozaffarian D, Mussolino ME, Nasir K, Neumar RW, Palaniappan L, Pandey DK, Thiagarajan RR, Reeves MJ, Ritchey M, Rodriguez CJ, Roth GA, Rosamond WD, Sasson C, Towfighi A, Tsao CW, Turner MB, Virani SS, Voeks JH, Willey JZ, Wilkins JT, Wu JH, Alger HM, Wong SS, Muntner P; American Heart Association Statistics Committee and Stroke Statistics Subcommittee . Heart disease and stroke statistics-2017 update: a report from the American Heart Association. Circulation 2017;135:e146–e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Argulian E, Chandrashekhar Y, Shah SJ, Huttin O, Pitt B, Zannad F, Bonow RO, Narula J. Teasing apart heart failure with preserved ejection fraction phenotypes with echocardiographic imaging: potential approach to research and clinical Practice. Circ Res 2018;122:23–25. [DOI] [PubMed] [Google Scholar]
- 51. Senni M, Paulus WJ, Gavazzi A, Fraser AG, Díez J, Solomon SD, Smiseth OA, Guazzi M, Lam CS, Maggioni AP, Tschöpe C, Metra M, Hummel SL, Edelmann F, Ambrosio G, Stewart Coats AJ, Filippatos GS, Gheorghiade M, Anker SD, Levy D, Pfeffer MA, Stough WG, Pieske BM. New strategies for heart failure with preserved ejection fraction: the importance of targeted therapies for heart failure phenotypes. Eur Heart J 2014;35:2797–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shah SJ, Kitzman DW, Borlaug BA, van Heerebeek L, Zile MR, Kass DA, Paulus WJ. Phenotype-specific treatment of heart failure with preserved ejection fraction: a multiorgan roadmap. Circulation 2016;134:73–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Udelson JE. Heart failure with preserved ejection fraction. Circulation 2011;124:e540–e543. [DOI] [PubMed] [Google Scholar]
- 54. Pieske B, Tschöpe C, de Boer RA, Fraser AG, Anker SD, Donal E, Edelmann F, Fu M, Guazzi M, Lam CSP, Lancellotti P, Melenovsky V, Morris DA, Nagel E, Pieske-Kraigher E, Ponikowski P, Solomon SD, Vasan RS, Rutten FH, Voors AA, Ruschitzka F, Paulus WJ, Seferovic P, Filippatos G. How to diagnose heart failure with preserved ejection fraction: the HFA-PEFF diagnostic algorithm: a consensus recommendation from the Heart Failure Association of the ESC. Eur J Heart Fail 2020;22:391–412. [DOI] [PubMed] [Google Scholar]
- 55. Dubi S, Arbel Y. Large animal models for diastolic dysfunction and diastolic heart failure-a review of the literature. Cardiovasc Pathol 2010;19:147–152. [DOI] [PubMed] [Google Scholar]
- 56. Horgan S, Watson C, Glezeva N, Baugh J. Murine models of diastolic dysfunction and heart failure with preserved ejection fraction. J Card Fail 2014;20:984–995. [DOI] [PubMed] [Google Scholar]
- 57. Conceição G, Heinonen I, Lourenço AP, Duncker DJ, Falcão-Pires I. Animal models of heart failure with preserved ejection fraction. Neth Heart J 2016;24:275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Valero-Muñoz M, Backman W, Sam F. Murine models of heart failure with preserved ejection fraction: a "Fishing Expedition". JACC Basic Transl Sci 2017;2:770–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Noll NA, Lal H, Merryman WD. Mouse models of heart failure with preserved or reduced ejection fraction. Am J Pathol 2020;190:1596–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mishra S, Kass DA. Cellular and molecular pathobiology of heart failure with preserved ejection fraction. Nat Rev Cardiol 2021;18:400–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mori J, Patel VB, Abo Alrob O, Basu R, Altamimi T, Desaulniers J, Wagg CS, Kassiri Z, Lopaschuk GD, Oudit GY. Angiotensin 1-7 ameliorates diabetic cardiomyopathy and diastolic dysfunction in db/db mice by reducing lipotoxicity and inflammation. Circ Heart Fail 2014;7:327–339. [DOI] [PubMed] [Google Scholar]
- 62. van Bilsen M, Daniels A, Brouwers O, Janssen BJ, Derks WJ, Brouns AE, Munts C, Schalkwijk CG, van der Vusse GJ, van Nieuwenhoven FA. Hypertension is a conditional factor for the development of cardiac hypertrophy in type 2 diabetic mice. PLoS One 2014;9:e85078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hammoudi N, Jeong D, Singh R, Farhat A, Komajda M, Mayoux E, Hajjar R, Lebeche D. Empagliflozin improves left ventricular diastolic dysfunction in a genetic model of type 2 diabetes. Cardiovasc Drugs Ther 2017;31:233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Conti M, Renaud IM, Poirier B, Michel O, Belair MF, Mandet C, Bruneval P, Myara I, Chevalier J. High levels of myocardial antioxidant defense in aging nondiabetic normotensive Zucker obese rats. Am J Physiol Regul Integr Comp Physiol 2004;286:R793–R800. [DOI] [PubMed] [Google Scholar]
- 65. Murase T, Hattori T, Ohtake M, Abe M, Amakusa Y, Takatsu M, Murohara T, Nagata K. Cardiac remodeling and diastolic dysfunction in DahlS.Z-Lepr(fa)/Lepr(fa) rats: a new animal model of metabolic syndrome. Hypertens Res 2012;35:186–193. [DOI] [PubMed] [Google Scholar]
- 66. Yu Y, Ohmori K, Chen Y, Sato C, Kiyomoto H, Shinomiya K, Takeuchi H, Mizushige K, Kohno M. Effects of pravastatin on progression of glucose intolerance and cardiovascular remodeling in a type II diabetes model. J Am Coll Cardiol 2004;44:904–913. [DOI] [PubMed] [Google Scholar]
- 67. Silberman GA, Fan TH, Liu H, Jiao Z, Xiao HD, Lovelock JD, Boulden BM, Widder J, Fredd S, Bernstein KE, Wolska BM, Dikalov S, Harrison DG, Dudley SCJr. Uncoupled cardiac nitric oxide synthase mediates diastolic dysfunction. Circulation 2010;121:519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Doi R, Masuyama T, Yamamoto K, Doi Y, Mano T, Sakata Y, Ono K, Kuzuya T, Hirota S, Koyama T, Miwa T, Hori M. Development of different phenotypes of hypertensive heart failure: systolic versus diastolic failure in Dahl salt-sensitive rats. J Hypertens 2000;18:111–120. [DOI] [PubMed] [Google Scholar]
- 69. Hart CY, Meyer DM, Tazelaar HD, Grande JP, Burnett JCJr, Housmans PR, Redfield MM. Load versus humoral activation in the genesis of early hypertensive heart disease. Circulation 2001;104:215–220. [DOI] [PubMed] [Google Scholar]
- 70. Schwarzl M, Hamdani N, Seiler S, Alogna A, Manninger M, Reilly S, Zirngast B, Kirsch A, Steendijk P, Verderber J, Zweiker D, Eller P, Höfler G, Schauer S, Eller K, Maechler H, Pieske BM, Linke WA, Casadei B, Post H. A porcine model of hypertensive cardiomyopathy: implications for heart failure with preserved ejection fraction. Am J Physiol Heart Circ Physiol 2015;309:H1407–H1418. [DOI] [PubMed] [Google Scholar]
- 71. Regan JA, Mauro AG, Carbone S, Marchetti C, Gill R, Mezzaroma E, Valle Raleigh J, Salloum FN, Van Tassell BW, Abbate A, Toldo S. A mouse model of heart failure with preserved ejection fraction due to chronic infusion of a low subpressor dose of angiotensin II. Am J Physiol Heart Circ Physiol 2015;309:H771–H778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Curl CL, Danes VR, Bell JR, Raaijmakers AJA, Ip WTK, Chandramouli C, Harding TW, Porrello ER, Erickson JR, Charchar FJ, Kompa AR, Edgley AJ, Crossman DJ, Soeller C, Mellor KM, Kalman JM, Harrap SB, Delbridge LMD. Cardiomyocyte functional etiology in heart failure with preserved ejection fraction is distinctive—a new preclinical model. J Am Heart Assoc 2018;7:e007451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. van Deel ED, de Boer M, Kuster DW, Boontje NM, Holemans P, Sipido KR, van der Velden J, Duncker DJ. Exercise training does not improve cardiac function in compensated or decompensated left ventricular hypertrophy induced by aortic stenosis. J Mol Cell Cardiol 2011;50:1017–1025. [DOI] [PubMed] [Google Scholar]
- 74. Miranda-Silva DG, Rodrigues P, Alves E, Rizo D, Fonseca ACRG, Lima T, Baganha F, Conceição G, Sousa C, Gonçalves A, Miranda I, Vasques-Nóvoa F, Magalhães J, Leite-Moreira A, Falcão-Pires I. Mitochondrial reversible changes determine diastolic function adaptations during myocardial (reverse) remodeling. Circ Heart Fail 2020;13:e006170. [DOI] [PubMed] [Google Scholar]
- 75. Gyöngyösi M, Pavo N, Lukovic D, Zlabinger K, Spannbauer A, Traxler D, Goliasch G, Mandic L, Bergler-Klein J, Gugerell A, Jakab A, Szankai Z, Toth L, Garamvölgyi R, Maurer G, Jaisser F, Zannad F, Thum T, Bátkai S, Winkler J. Porcine model of progressive cardiac hypertrophy and fibrosis with secondary postcapillary pulmonary hypertension. J Transl Med 2017;15:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Charles CJ, Lee P, Li RR, Yeung T, Ibraham Mazlan SM, Tay ZW, Abdurrachim D, Teo XQ, Wang WH, de Kleijn DPV, Cozzone PJ, Lam CSP, Richards AM. A porcine model of heart failure with preserved ejection fraction: magnetic resonance imaging and metabolic energetics. ESC Heart Fail 2020;7:92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wallner M, Eaton DM, Berretta RM, Borghetti G, Wu J, Baker ST, Feldsott EA, Sharp TE, Mohsin S, Oyama MA, von Lewinski D, Post H, Wolfson MR, Houser SR. A Feline HFpEF model with pulmonary hypertension and compromised pulmonary function. Sci Rep 2017;7:16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hittinger L, Mirsky I, Shen YT, Patrick TA, Bishop SP, Vatner SF. Hemodynamic mechanisms responsible for reduced subendocardial coronary reserve in dogs with severe left ventricular hypertrophy. Circulation 1995;92:978–986. [DOI] [PubMed] [Google Scholar]
- 79. Gevaert AB, Shakeri H, Leloup AJ, Van Hove CE, De Meyer GRY, Vrints CJ, Lemmens K, Van Craenenbroeck EM. Endothelial senescence contributes to heart failure with preserved ejection fraction in an aging mouse model. Circ Heart Fail 2017;10:e003806. [DOI] [PubMed] [Google Scholar]
- 80. Koch SE, Haworth KJ, Robbins N, Smith MA, Lather N, Anjak A, Jiang M, Varma P, Jones WK, Rubinstein J. Age- and gender-related changes in ventricular performance in wild-type FVB/N mice as evaluated by conventional and vector velocity echocardiography imaging: a retrospective study. Ultrasound Med Biol 2013;39:2034–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Forman DE, Cittadini A, Azhar G, Douglas PS, Wei JY. Cardiac morphology and function in senescent rats: gender-related differences. J Am Coll Cardiol 1997;30:1872–1877. [DOI] [PubMed] [Google Scholar]
- 82. Tofovic SP, Kusaka H, Kost CKJr, Bastacky S. Renal function and structure in diabetic, hypertensive, obese ZDFxSHHF-hybrid rats. Ren Fail 2000;22:387–406. [DOI] [PubMed] [Google Scholar]
- 83. Hamdani N, Franssen C, Lourenço A, Falcão-Pires I, Fontoura D, Leite S, Plettig L, López B, Ottenheijm CA, Becher PM, González A, Tschöpe C, Díez J, Linke WA, Leite-Moreira AF, Paulus WJ. Myocardial titin hypophosphorylation importantly contributes to heart failure with preserved ejection fraction in a rat metabolic risk model. Circ Heart Fail 2013;6:1239–1249. [DOI] [PubMed] [Google Scholar]
- 84. Schiattarella GG, Altamirano F, Tong D, French KM, Villalobos E, Kim SY, Luo X, Jiang N, May HI, Wang ZV, Hill TM, Mammen PPA, Huang J, Lee DI, Hahn VS, Sharma K, Kass DA, Lavandero S, Gillette TG, Hill JA. Nitrosative stress drives heart failure with preserved ejection fraction. Nature 2019;568:351–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sorop O, Heinonen I, van Kranenburg M, van de Wouw J, de Beer VJ, Nguyen ITN, Octavia Y, van Duin RWB, Stam K, van Geuns RJ, Wielopolski PA, Krestin GP, van den Meiracker AH, Verjans R, van Bilsen M, Danser AHJ, Paulus WJ, Cheng C, Linke WA, Joles JA, Verhaar MC, van der Velden J, Merkus D, Duncker DJ. Multiple common comorbidities produce left ventricular diastolic dysfunction associated with coronary microvascular dysfunction, oxidative stress, and myocardial stiffening. Cardiovasc Res 2018;114:954–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Paulus WJ, Tschöpe C. A novel paradigm for heart failure with preserved ejection fraction: comorbidities drive myocardial dysfunction and remodeling through coronary microvascular endothelial inflammation. J Am Coll Cardiol 2013;62:263–271. [DOI] [PubMed] [Google Scholar]
- 87. Franssen C, Chen S, Unger A, Korkmaz HI, De Keulenaer GW, Tschöpe C, Leite-Moreira AF, Musters R, Niessen HW, Linke WA, Paulus WJ, Hamdani N. Myocardial microvascular inflammatory endothelial activation in heart failure with preserved ejection fraction. JACC Heart Fail 2016;4:312–324. [DOI] [PubMed] [Google Scholar]
- 88. Eisenberg T, Abdellatif M, Schroeder S, Primessnig U, Stekovic S, Pendl T, Harger A, Schipke J, Zimmermann A, Schmidt A, Tong M, Ruckenstuhl C, Dammbrueck C, Gross AS, Herbst V, Magnes C, Trausinger G, Narath S, Meinitzer A, Hu Z, Kirsch A, Eller K, Carmona-Gutierrez D, Büttner S, Pietrocola F, Knittelfelder O, Schrepfer E, Rockenfeller P, Simonini C, Rahn A, Horsch M, Moreth K, Beckers J, Fuchs H, Gailus-Durner V, Neff F, Janik D, Rathkolb B, Rozman J, de Angelis MH, Moustafa T, Haemmerle G, Mayr M, Willeit P, von Frieling-Salewsky M, Pieske B, Scorrano L, Pieber T, Pechlaner R, Willeit J, Sigrist SJ, Linke WA, Mühlfeld C, Sadoshima J, Dengjel J, Kiechl S, Kroemer G, Sedej S, Madeo F. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med 2016;22:1428–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Porrello ER, Delbridge LMD. HFpEF-Time to explore the role of genetic heterogeneity in phenotypic variability: new mechanistic insights offer promise for personalized therapies. Circulation 2019;140:1607–1609. [DOI] [PubMed] [Google Scholar]
- 90. Juni RP, Kuster DWD, Goebel M, Helmes M, Musters RJP, van der Velden J, Koolwijk P, Paulus WJ, van Hinsbergh VWM. Cardiac microvascular endothelial enhancement of cardiomyocyte function is impaired by inflammation and restored by empagliflozin. JACC Basic Transl Sci 2019;4:575–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kriegel AJ, Gartz M, Afzal MZ, de Lange WJ, Ralphe JC, Strande JL. Molecular approaches in HFpEF: microRNAs and iPSC-derived cardiomyocytes. J Cardiovasc Transl Res 2017;10:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Schotten U, Verheule S, Kirchhof P, Goette A. Pathophysiological mechanisms of atrial fibrillation: a translational appraisal. Physiol Rev 2011;91:265–325. [DOI] [PubMed] [Google Scholar]
- 93. Fabritz L, Guasch E, Antoniades C, Bardinet I, Benninger G, Betts TR, Brand E, Breithardt G, Bucklar-Suchankova G, Camm AJ, Cartlidge D, Casadei B, Chua WWL, Crijns HJGM, Deeks J, Hatem S, Hidden-Lucet F, Kääb S, Maniadakis N, Martin S, Mont L, Reinecke H, Sinner MF, Schotten U, Southwood T, Stoll M, Vardas P, Wakili R, West A, Ziegler A, Kirchhof P. Expert consensus document: defining the major health modifiers causing atrial fibrillation: a roadmap to underpin personalized prevention and treatment. Nat Rev Cardiol 2016;13:230–237. [DOI] [PubMed] [Google Scholar]
- 94. Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K, Palsson A, Blondal T, Sulem P, Backman VM, Hardarson GA, Palsdottir E, Helgason A, Sigurjonsdottir R, Sverrisson JT, Kostulas K, Ng MC, Baum L, So WY, Wong KS, Chan JC, Furie KL, Greenberg SM, Sale M, Kelly P, MacRae CA, Smith EE, Rosand J, Hillert J, Ma RC, Ellinor PT, Thorgeirsson G, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 2007;448:353–357. [DOI] [PubMed] [Google Scholar]
- 95. Roselli C, Chaffin MD, Weng L-C, Aeschbacher S, Ahlberg G, Albert CM, Almgren P, Alonso A, Anderson CD, Aragam KG, Arking DE, Barnard J, Bartz TM, Benjamin EJ, Bihlmeyer NA, Bis JC, Bloom HL, Boerwinkle E, Bottinger EB, Brody JA, Calkins H, Campbell A, Cappola TP, Carlquist J, Chasman DI, Chen LY, Chen Y-DI, Choi E-K, Choi SH, Christophersen IE, Chung MK, Cole JW, Conen D, Cook J, Crijns HJ, Cutler MJ, Damrauer SM, Daniels BR, Darbar D, Delgado G, Denny JC, Dichgans M, Dörr M, Dudink EA, Dudley SC, Esa N, Esko T, Eskola M, Fatkin D, Felix SB, Ford I, Franco OH, Geelhoed B, Grewal RP, Gudnason V, Guo X, Gupta N, Gustafsson S, Gutmann R, Hamsten A, Harris TB, Hayward C, Heckbert SR, Hernesniemi J, Hocking LJ, Hofman A, Horimoto ARVR, Huang J, Huang PL, Huffman J, Ingelsson E, Ipek EG, Ito K, Jimenez-Conde J, Johnson R, Jukema JW, Kääb S, Kähönen M, Kamatani Y, Kane JP, Kastrati A, Kathiresan S, Katschnig-Winter P, Kavousi M, Kessler T, Kietselaer BL, Kirchhof P, Kleber ME, Knight S, Krieger JE, Kubo M, Launer LJ, Laurikka J, Lehtimäki T, Leineweber K, Lemaitre RN, Li M, Lim HE, Lin HJ, Lin H, Lind L, Lindgren CM, Lokki M-L, London B, Loos RJF, Low S-K, Lu Y, Lyytikäinen L-P, Macfarlane PW, Magnusson PK, Mahajan A, Malik R, Mansur AJ, Marcus GM, Margolin L, Margulies KB, März W, McManus DD, Melander O, Mohanty S, Montgomery JA, Morley MP, Morris AP, Müller-Nurasyid M, Natale A, Nazarian S, Neumann B, Newton-Cheh C, Niemeijer MN, Nikus K, Nilsson P, Noordam R, Oellers H, Olesen MS, Orho-Melander M, Padmanabhan S, Pak H-N, Paré G, Pedersen NL, Pera J, Pereira A, Porteous D, Psaty BM, Pulit SL, Pullinger CR, Rader DJ, Refsgaard L, Ribasés M, Ridker PM, Rienstra M, Risch L, Roden DM, Rosand J, Rosenberg MA, Rost N, Rotter JI, Saba S, Sandhu RK, Schnabel RB, Schramm K, Schunkert H, Schurman C, Scott SA, Seppälä I, Shaffer C, Shah S, Shalaby AA, Shim J, Shoemaker MB, Siland JE, Sinisalo J, Sinner MF, Slowik A, Smith AV, Smith BH, Smith JG, Smith JD, Smith NL, Soliman EZ, Sotoodehnia N, Stricker BH, Sun A, Sun H, Svendsen JH, Tanaka T, Tanriverdi K, Taylor KD, Teder-Laving M, Teumer A, Thériault S, Trompet S, Tucker NR, Tveit A, Uitterlinden AG, Van Der Harst P, Van Gelder IC, Van Wagoner DR, Verweij N, Vlachopoulou E, Völker U, Wang B, Weeke PE, Weijs B, Weiss R, Weiss S, Wells QS, Wiggins KL, Wong JA, Woo D, Worrall BB, Yang P-S, Yao J, Yoneda ZT, Zeller T, Zeng L, Lubitz SA, Lunetta KL, Ellinor PT. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet 2018;50:1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Zhang D, Hu X, Li J, Liu J, Baks-Te Bulte L, Wiersma M, Malik NU, van Marion DMS, Tolouee M, Hoogstra-Berends F, Lanters EAH, van Roon AM, de Vries AAF, Pijnappels DA, de Groot NMS, Henning RH, Brundel BJJM. DNA damage-induced PARP1 activation confers cardiomyocyte dysfunction through NAD+ depletion in experimental atrial fibrillation. Nat Commun 2019;10:1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wijffels MC, Kirchhof CJ, Dorland R, Allessie MA. Atrial fibrillation begets atrial fibrillation. A study in awake chronically instrumented goats. Circulation 1995;92:1954–1968. [DOI] [PubMed] [Google Scholar]
- 98. Reyat JS, Chua W, Cardoso VR, Witten A, Kastner PM, Kabir SN, Sinner MF, Wesselink R, Holmes AP, Pavlovic D, Stoll M, Kääb S, Gkoutos GV, de Groot JR, Kirchhof P, Fabritz L. Reduced left atrial cardiomyocyte PITX2 and elevated circulating BMP10 predict atrial fibrillation after ablation. JCI Insight 2020;5:e139179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Parahuleva MS, Kockskämper J, Heger J, Grimm W, Scherer A, Bühler S, Kreutz J, Schulz R, Euler G. Structural, Pro-Inflammatory and calcium handling remodeling underlies spontaneous onset of paroxysmal atrial fibrillation in JDP2-overexpressing mice. Int J Mol Sci 2020;21:9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. van Ouwerkerk AF, Hall AW, Kadow ZA, Lazarevic S, Reyat JS, Tucker NR, Nadadur RD, Bosada FM, Bianchi V, Ellinor PT, Fabritz L, Martin JF, de Laat W, Kirchhof P, Moskowitz IP, Christoffels VM. Epigenetic and transcriptional networks underlying atrial fibrillation. Circ Res 2020;127:34–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Devalla HD, Schwach V, Ford JW, Milnes JT, El-Haou S, Jackson C, Gkatzis K, Elliott DA, Chuva de Sousa Lopes SM, Mummery CL, Verkerk AO, Passier R. Atrial-like cardiomyocytes from human pluripotent stem cells are a robust preclinical model for assessing atrial-selective pharmacology. EMBO Mol Med 2015;7:394–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Harlaar N, Liu J, Volkers L, Ramkisoensing AA, Schalij MJ, Klautz RJM, van Brakel TJ, Pijnappels DA, de Vries AAF. Massive expansion of native human atrial cardiomyocytes through immortogenetics: generation of the hiAM cell lines. Eur Heart J 2019;40:P1229. [Google Scholar]
- 103. Lemme M, Ulmer BM, Lemoine MD, Zech ATL, Flenner F, Ravens U, Reichenspurner H, Rol-Garcia M, Smith G, Hansen A, Christ T, Eschenhagen T. Atrial-like engineered heart tissue: an in vitro model of the human atrium. Stem Cell Reports 2018;11:1378–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Yamashita K, Silvernagel J, Kwan E, Kamali R, Ghafoori E, MacLeod R, Dosdall DJ, Ranjan R. Changes in atrial electrophysiological and structural substrate and their relationship to histology in a long-term chronic canine atrial fibrillation model. Pacing Clin Electrophysiol 2019;42:930–936. [DOI] [PubMed] [Google Scholar]
- 105. Frydrychowski P, Michałek M, Sławuta A, Noszczyk-Nowak A. Large animals as models of atrial fibrillation. Adv Clin Exp Med 2020;29:757–767. [DOI] [PubMed] [Google Scholar]
- 106. Wiersma M, van Marion DMS, Bouman EJ, Li J, Zhang D, Ramos KS, Lanters EAH, de Groot NMS, Brundel BJJM. Cell-free circulating mitochondrial DNA: a potential blood-based marker for atrial fibrillation. Cells 2020;9:1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Pinto YM, Elliott PM, Arbustini E, Adler Y, Anastasakis A, Böhm M, Duboc D, Gimeno J, de Groote P, Imazio M, Heymans S, Klingel K, Komajda M, Limongelli G, Linhart A, Mogensen J, Moon J, Pieper PG, Seferovic PM, Schueler S, Zamorano JL, Caforio AL, Charron P. Proposal for a revised definition of dilated cardiomyopathy, hypokinetic non-dilated cardiomyopathy, and its implications for clinical practice: a position statement of the ESC working group on myocardial and pericardial diseases. Eur Heart J 2016;37:1850–1858. [DOI] [PubMed] [Google Scholar]
- 108. James CA, Syrris P, van Tintelen JP, Calkins H. The role of genetics in cardiovascular disease: arrhythmogenic cardiomyopathy. Eur Heart J 2020;41:1393–1400. [DOI] [PubMed] [Google Scholar]
- 109. Corrado D, Basso C, Thiene G. Is it time to include ion channel diseases among cardiomyopathies? J Electrocardiol 2005;38:81–87. [DOI] [PubMed] [Google Scholar]
- 110. Bondue A, Arbustini E, Bianco A, Ciccarelli M, Dawson D, De Rosa M, Hamdani N, Hilfiker-Kleiner D, Meder B, Leite-Moreira AF, Thum T, Tocchetti CG, Varricchi G, Van der Velden J, Walsh R, Heymans S. Complex roads from genotype to phenotype in dilated cardiomyopathy: scientific update from the Working Group of Myocardial Function of the ESC. Cardiovasc Res 2018;114:1287–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Haas J, Frese KS, Peil B, Kloos W, Keller A, Nietsch R, Feng Z, Müller S, Kayvanpour E, Vogel B, Sedaghat-Hamedani F, Lim WK, Zhao X, Fradkin D, Köhler D, Fischer S, Franke J, Marquart S, Barb I, Li DT, Amr A, Ehlermann P, Mereles D, Weis T, Hassel S, Kremer A, King V, Wirsz E, Isnard R, Komajda M, Serio A, Grasso M, Syrris P, Wicks E, Plagnol V, Lopes L, Gadgaard T, Eiskjær H, Jørgensen M, Garcia-Giustiniani D, Ortiz-Genga M, Crespo-Leiro MG, Deprez RH, Christiaans I, van Rijsingen IA, Wilde AA, Waldenstrom A, Bolognesi M, Bellazzi R, Mörner S, Bermejo JL, Monserrat L, Villard E, Mogensen J, Pinto YM, Charron P, Elliott P, Arbustini E, Katus HA, Meder B. Atlas of the clinical genetics of human dilated cardiomyopathy. Eur Heart J 2015;36:1123–1135a. [DOI] [PubMed] [Google Scholar]
- 112. Verdonschot JAJ, Merlo M, Dominguez F, Wang P, Henkens MTHM, Adriaens ME, Hazebroek MR, Masè M, Escobar LE, Cobas-Paz R, Derks KWJ, van den Wijngaard A, Krapels IPC, Brunner HG, Sinagra G, Garcia-Pavia P, Heymans SRB. Phenotypic clustering of dilated cardiomyopathy patients highlights important pathophysiological differences. Eur Heart J 2021;42:162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Duncker DJ, Bakkers J, Brundel BJ, Robbins J, Tardiff JC, Carrier L. Animal and in silico models for the study of sarcomeric cardiomyopathies. Cardiovasc Res 2015;105:439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Moretti A, Fonteyne L, Giesert F, Hoppmann P, Meier AB, Bozoglu T, Baehr A, Schneider CM, Sinnecker D, Klett K, Fröhlich T, Rahman FA, Haufe T, Sun S, Jurisch V, Kessler B, Hinkel R, Dirschinger R, Martens E, Jilek C, Graf A, Krebs S, Santamaria G, Kurome M, Zakhartchenko V, Campbell B, Voelse K, Wolf A, Ziegler T, Reichert S, Lee S, Flenkenthaler F, Dorn T, Jeremias I, Blum H, Dendorfer A, Schnieke A, Krause S, Walter MC, Klymiuk N, Laugwitz KL, Wolf E, Wurst W, Kupatt C. Somatic gene editing ameliorates skeletal and cardiac muscle failure in pig and human models of Duchenne muscular dystrophy. Nat Med 2020;26:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wijnker PJM, van der Velden J. Mutation-specific pathology and treatment of hypertrophic cardiomyopathy in patients, mouse models and human engineered heart tissue. Biochim Biophys Acta Mol Basis Dis 2020;1866:165774. [DOI] [PubMed] [Google Scholar]
- 116. Saleem U, Mannhardt I, Braren I, Denning C, Eschenhagen T, Hansen A. Force and calcium transients analysis in human engineered heart tissues reveals positive force-frequency relation at physiological frequency. Stem Cell Reports 2020;14:312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Prondzynski M, Lemoine MD, Zech AT, Horváth A, Di Mauro V, Koivumäki JT, Kresin N, Busch J, Krause T, Krämer E, Schlossarek S, Spohn M, Friedrich FW, Münch J, Laufer SD, Redwood C, Volk AE, Hansen A, Mearini G, Catalucci D, Meyer C, Christ T, Patten M, Eschenhagen T, Carrier L. Disease modeling of a mutation in α-actinin 2 guides clinical therapy in hypertrophic cardiomyopathy. EMBO Mol Med 2019;11:e11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Taylor PM, Batten P, Brand NJ, Thomas PS, Yacoub MH. The cardiac valve interstitial cell. Int J Biochem Cell Biol 2003;35:113–118. [DOI] [PubMed] [Google Scholar]
- 119. Dweck MR, Boon NA, Newby DE. Calcific aortic stenosis: a disease of the valve and the myocardium. J Am Coll Cardiol 2012;60:1854–1863. [DOI] [PubMed] [Google Scholar]
- 120. Wang H, Leinwand LA, Anseth KS. Cardiac valve cells and their microenvironment—insights from in vitro studies. Nat Rev Cardiol 2014;11:715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Back M, Gasser TC, Michel JB, Caligiuri G. Biomechanical factors in the biology of aortic wall and aortic valve diseases. Cardiovasc Res 2013;99:232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Leopold JA. Cellular mechanisms of aortic valve calcification. Circ Cardiovasc Interv 2012;5:605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Katayama S, Umetani N, Hisada T, Sugiura S. Bicuspid aortic valves undergo excessive strain during opening: a simulation study. J Thorac Cardiovasc Surg 2013;145:1570–1576. [DOI] [PubMed] [Google Scholar]
- 124. Cujec B, Pollick C. Isolated thickening of one aortic cusp: preferential thickening of the noncoronary cusp. J Am Soc Echocardiogr 1988;1:430–432. [DOI] [PubMed] [Google Scholar]
- 125. Robinson N, Souslian L, Gallegos RP, Rivard AL, Dalmasso AP, Bianco RW. Animal models for cardiac research. In: Iaizzo PA (ed). Handbook of Cardiac Anatomy, Physiology, and Devices. Cham: Springer International Publishing, 2015. pp. 469–491. [Google Scholar]
- 126. Nigam V, Srivastava D. Notch1 represses osteogenic pathways in aortic valve cells. J Mol Cell Cardiol 2009;47:828–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Tanaka K, Sata M, Fukuda D, Suematsu Y, Motomura N, Takamoto S, Hirata Y, Nagai R. Age-associated aortic stenosis in apolipoprotein E-deficient mice. J Am Coll Cardiol 2005;46:134–141. [DOI] [PubMed] [Google Scholar]
- 128. Cuniberti LA, Stutzbach PG, Guevara E, Yannarelli GG, Laguens RP, Favaloro RR. Development of mild aortic valve stenosis in a rabbit model of hypertension. J Am Coll Cardiol 2006;47:2303–2309. [DOI] [PubMed] [Google Scholar]
- 129. Cimini M, Boughner DR, Ronald JA, Aldington L, Rogers KA. Development of aortic valve sclerosis in a rabbit model of atherosclerosis: an immunohistochemical and histological study. J Heart Valve Dis 2005;14:365–375. [PubMed] [Google Scholar]
- 130. Rajamannan NM, Subramaniam M, Caira F, Stock SR, Spelsberg TC. Atorvastatin inhibits hypercholesterolemia-induced calcification in the aortic valves via the Lrp5 receptor pathway. Circulation 2005;112:I229–I234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Spargias K, Gyongyosi M, Hemetsberger R, Posa A, Pavo N, Pavo IJ, Huber K, Petrasi Z, Petnehazy O, von Strandmann RP, Park J, Glogar D, Maurer G, Rajamannan NM. Valvuloplasty with a paclitaxel-eluting balloon prevents restenosis in an experimental animal model of aortic stenosis. J Heart Valve Dis 2014;23:484–491. [PubMed] [Google Scholar]
- 132. Sider KL, Zhu C, Kwong AV, Mirzaei Z, de Lange CF, Simmons CA. Evaluation of a porcine model of early aortic valve sclerosis. Cardiovasc Pathol 2014;23:289–297. [DOI] [PubMed] [Google Scholar]
- 133. Perry GJ, Wei CC, Hankes GH, Dillon SR, Rynders P, Mukherjee R, Spinale FG, Dell’Italia LJ. Angiotensin II receptor blockade does not improve left ventricular function and remodeling in subacute mitral regurgitation in the dog. J Am Coll Cardiol 2002;39:1374–1379. [DOI] [PubMed] [Google Scholar]
- 134. Malinowski M, Proudfoot AG, Langholz D, Eberhart L, Brown M, Schubert H, Wodarek J, Timek TA. Large animal model of functional tricuspid regurgitation in pacing induced end-stage heart failure. Interact Cardiovasc Thorac Surg 2017;24:905–910. [DOI] [PubMed] [Google Scholar]
- 135. Chien SF, Diana JN, Brum JM, Bove AAA. simple technique for producing supravalvular aortic stenosis in animals. Cardiovasc Res 1988;22:739–745. [DOI] [PubMed] [Google Scholar]
- 136. Libby P, Buring JE, Badimón L, Hansson GK, Deanfield J, Bittencourt MS, Tokgözoğlu L, Lewis EF. Atherosclerosis. Nat Rev Dis Primers 2019;5:56–18. [DOI] [PubMed] [Google Scholar]
- 137. Libby P, Lichtman AH, Hansson GK. Immune effector mechanisms implicated in atherosclerosis: from mice to humans. Immunity 2013;38:1092–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Hasler-Rapacz J, Prescott MF, Von Linden-Reed J, Rapacz JMJr, Hu Z, Rapacz J. Elevated concentrations of plasma lipids and apolipoproteins B, C-III, and E are associated with the progression of coronary artery disease in familial hypercholesterolemic swine. Arterioscler Thromb Vasc Biol 1995;15:583–592. [DOI] [PubMed] [Google Scholar]
- 139. Vilahur G, Padro T, Badimon L. Atherosclerosis and thrombosis: insights from large animal models. J Biomed Biotechnol 2011;2011:907575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Sorop O, van de Wouw J, Chandler S, Ohanyan V, Tune JD, Chilian WM, Merkus D, Bender SB, Duncker DJ. Experimental animal models of coronary microvascular dysfunction. Cardiovasc Res 2020;116:756–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Lee YT, Laxton V, Lin HY, Chan YWF, Fitzgerald-Smith S, To TLO, Yan BP, Liu T, Tse G. Animal models of atherosclerosis. Biomed Rep 2017;6:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Li ZL, Woollard JR, Ebrahimi B, Crane JA, Jordan KL, Lerman A, Wang SM, Lerman LO. Transition from obesity to metabolic syndrome is associated with altered myocardial autophagy and apoptosis. Arterioscler Thromb Vasc Biol 2012;32:1132–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Hedayat AF, Park KH, Kwon TG, Woollard JR, Jiang K, Carlson DF, Lerman A, Lerman LO. Peripheral vascular atherosclerosis in a novel PCSK9 gain-of-function mutant Ossabaw miniature pig model. Transl Res 2018;192:30–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Hamamdzic D, Wilensky RL. Porcine models of accelerated coronary atherosclerosis: role of diabetes mellitus and hypercholesterolemia. J Diabetes Res 2013;2013:761415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Schwartz RS, Edelman E, Virmani R, Carter A, Granada JF, Kaluza GL, Chronos NA, Robinson KA, Waksman R, Weinberger J, Wilson GJ, Wilensky RL. Drug-eluting stents in preclinical studies: updated consensus recommendations for preclinical evaluation. Circ Cardiovasc Interv 2008;1:143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Shim J, Al-Mashhadi RH, Sørensen CB, Bentzon JF. Large animal models of atherosclerosis—new tools for persistent problems in cardiovascular medicine. J Pathol 2016;238:257–266. [DOI] [PubMed] [Google Scholar]
- 147. Hoogendoorn A, Hoedt den S, Hartman EMJ, Krabbendam-Peters IL, Hekkert Te M, van der Zee L, van Gaalen K, Witberg KT, Dorst K, Ligthart JMR, Drouet L, Van der Heiden K, van Lennep JR, van der Steen AFW, Duncker DJ, Mulder MT, Wentzel JJ Variation in coronary atherosclerosis severity related to a distinct LDL (low-density lipoprotein) profile: findings from a familial hypercholesterolemia pig model. Arterioscler Thromb Vasc Biol 2019;39:2338–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Scheidt von M, Zhao Y, Kurt Z, Pan C, Zeng L, Yang X, Schunkert H, Lusis AJ. Applications and limitations of mouse models for understanding human atherosclerosis. Cell Metab 2017;25:248–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Lutgens E, van Suylen R-J, Faber BC, Gijbels MJ, Eurlings PM, Bijnens AP, Cleutjens KB, Heeneman S, Daemen MJAP. Atherosclerotic plaque rupture: local or systemic process? Arterioscler Thromb Vasc Biol 2003;23:2123–2130. [DOI] [PubMed] [Google Scholar]
- 150. Reddick RL, Zhang SH, Maeda N. Aortic atherosclerotic plaque injury in apolipoprotein E deficient mice. Atherosclerosis 1998;140:297–305. [DOI] [PubMed] [Google Scholar]
- 151. Hartwig H, Silvestre-Roig C, Hendrikse J, Beckers L, Paulin N, Van der Heiden K, Braster Q, Drechsler M, Daemen MJ, Lutgens E, Soehnlein O. Atherosclerotic plaque destabilization in mice: a comparative study. PLoS One 2015;10:e0141019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Fernandez DM, Rahman AH, Fernandez NF, Chudnovskiy A, Amir E-AD, Amadori L, Khan NS, Wong CK, Shamailova R, Hill CA, Wang Z, Remark R, Li JR, Pina C, Faries C, Awad AJ, Moss N, Bjorkegren JLM, Kim-Schulze S, Gnjatic S, Ma'ayan A, Mocco J, Faries P, Merad M, Giannarelli C. Single-cell immune landscape of human atherosclerotic plaques. Nat Med 2019;25:1576–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Cole JE, Park I, Ahern D, Kassiteridi C, Danso Abeam D, Goddard M, Green P, Maffia P, Monaco C. Immune cell census in murine atherosclerosis: cytometry by time of flight illuminates vascular myeloid cell diversity. Cardiovasc Res 2018;390:1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Depuydt MAC, Prange KHM, Slenders L, Örd T, Elbersen D, Boltjes A, de Jager SCA, Asselbergs FW, de Borst GJ, Aavik E, Lönnberg T, Lutgens E, Glass CK, den Ruijter HM, Kaikkonen MU, Bot I, Slütter B, van der Laan SW, Yla-Herttuala S, Mokry M, Kuiper J, de Winther MPJ, Pasterkamp G. Microanatomy of the human atherosclerotic plaque by single-cell transcriptomics. Circ Res 2020;127:1437–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Groeneveld ME, Meekel JP, Rubinstein SM, Merkestein LR, Tangelder GJ, Wisselink W, Truijers M, Yeung KK. Systematic review of circulating, biomechanical, and genetic markers for the prediction of abdominal aortic aneurysm growth and rupture. J Am Heart Assoc 2018;7:e007791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Bogunovic N, Meekel JP, Micha D, Blankensteijn JD, Hordijk PL, Yeung KK. Impaired smooth muscle cell contractility as a novel concept of abdominal aortic aneurysm pathophysiology. Sci Rep 2019;9:6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Lysgaard Poulsen J, Stubbe J, Lindholt JS. Animal models used to explore abdominal aortic aneurysms: a systematic review. Eur J Vasc Endovasc Surg 2016;52:487–499. [DOI] [PubMed] [Google Scholar]
- 158. Daugherty A, Manning MW, Cassis LA. Angiotensin II promotes atherosclerotic lesions and aneurysms in apolipoprotein E-deficient mice. J Clin Invest 2000;105:1605–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Busch A, Chernogubova E, Jin H, Meurer F, Eckstein HH, Kim M, Maegdefessel L. Four surgical modifications to the classic elastase perfusion aneurysm model enable haemodynamic alterations and extended elastase perfusion. Eur J Vasc Endovasc Surg 2018;56:102–109. [DOI] [PubMed] [Google Scholar]
- 160. Wang Y, Krishna S, Golledge J. The calcium chloride-induced rodent model of abdominal aortic aneurysm. Atherosclerosis 2013;226:29–39. [DOI] [PubMed] [Google Scholar]
- 161. Lu G, Su G, Davis JP, Schaheen B, Downs E, Roy RJ, Ailawadi G, Upchurch GRJr. A novel chronic advanced stage abdominal aortic aneurysm murine model. J Vasc Surg 2017;66:232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Li DY, Busch A, Jin H, Chernogubova E, Pelisek J, Karlsson J, Sennblad B, Liu S, Lao S, Hofmann P, Bäcklund A, Eken SM, Roy J, Eriksson P, Dacken B, Ramanujam D, Dueck A, Engelhardt S, Boon RA, Eckstein HH, Spin JM, Tsao PS, Maegdefessel L. H19 induces abdominal aortic aneurysm development and progression. Circulation 2018;138:1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Kloster BO, Lund L, Lindholt JS. Induction of continuous expanding infrarenal aortic aneurysms in a large porcine animal model. Ann Med Surg (Lond) 2015;4:30–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Busch A, Pauli J, Winski G, Bleichert S, Chernogubova E, Metschl S, Winter H, Trenner M, Wiegering A, Otto C, Fischer J, Reiser J, Werner J, Roy J, Brostjan C, Knappich C, Eckstein HH, Paloschi V, Maegdefessel L. Lenvatinib halts aortic aneurysm growth by restoring smooth muscle cell contractility. JCI Insight 2021;6:140364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Davis J, Maillet M, Miano JM, Molkentin JD. Lost in transgenesis: a user's guide for genetically manipulating the mouse in cardiac research. Circ Res 2012;111:761–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Ved N, Curran A, Ashcroft FM, Sparrow DB. Tamoxifen administration in pregnant mice can be deleterious to both mother and embryo. Lab Anim 2019;53:630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Rehmani T, Salih M, Tuana BS. Cardiac-specific Cre induces age-dependent dilated cardiomyopathy (DCM) in mice. Molecules 2019;24:1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Kotini M, Barriga EH, Leslie J, Gentzel M, Rauschenberger V, Schambony A, Mayor R. Gap junction protein Connexin-43 is a direct transcriptional regulator of N-cadherin in vivo. Nat Commun 2018;9:3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Nicod J, Davies RW, Cai N, Hassett C, Goodstadt L, Cosgrove C, Yee BK, Lionikaite V, McIntyre RE, Remme CA, Lodder EM, Gregory JS, Hough T, Joynson R, Phelps H, Nell B, Rowe C, Wood J, Walling A, Bopp N, Bhomra A, Hernandez-Pliego P, Callebert J, Aspden RM, Talbot NP, Robbins PA, Harrison M, Fray M, Launay JM, Pinto YM, Blizard DA, Bezzina CR, Adams DJ, Franken P, Weaver T, Wells S, Brown SD, Potter PK, Klenerman P, Lionikas A, Mott R, Flint J. Genome-wide association of multiple complex traits in outbred mice by ultra-low-coverage sequencing. Nat Genet 2016;48:912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Podliesna S, Bezzina CR, Lodder EM. Complex genetics of cardiovascular traits in mice: F2-mapping of QTLs and their underlying genes. Methods Mol Biol 2017;1488:431–454. [DOI] [PubMed] [Google Scholar]
- 171. Mullen PD, Ramirez G. The promise and pitfalls of systematic reviews. Annu Rev Public Health 2006;27:81–102. [DOI] [PubMed] [Google Scholar]
- 172. Hooijmans CR, IntHout J, Ritskes-Hoitinga M, Rovers MM. Meta-analyses of animal studies: an introduction of a valuable instrument to further improve healthcare. ILAR J 2014;55:418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Zacchigna S, Paldino A, Falcao-Pires I, Daskalopoulos EP, Dal Ferro M, Vodret S, Lesizza P, Cannata A, Miranda-Silva D, Lourenco AP, Pinamonti B, Sinagra G, Weinberger F, Eschenhagen T, Carrier L, Kehat I, Tocchetti CG, Russo M, Ghigo A, Cimino J, Hirsch E, Dawson D, Ciccarelli M, Oliveti M, Linke WA, Cuijpers I, Heymans S, Hamdani N, de Boer M, Duncker D, Kuster D, van der Velden J, Beauloye C, Bertrand L, Mayr M, Giacca M, Leuschner F, Backs J, Thum T. Towards standardization of echocardiography for the evaluation of left ventricular function in adult rodents: a position paper of the ESC Working Group on Myocardial Function. Cardiovasc Res 2021;117:43–59. [DOI] [PubMed] [Google Scholar]
- 174. Niemeyer JE. Telemetry for small animal physiology. Lab Anim (NY) 2016;45:255–257. [DOI] [PubMed] [Google Scholar]
- 175. Tsang HG, Rashdan NA, Whitelaw CB, Corcoran BM, Summers KM, MacRae VE. Large animal models of cardiovascular disease. Cell Biochem Funct 2016;34:113–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Camacho P, Fan H, Liu Z, He JQ. Large mammalian animal models of heart disease. J Cardiovasc Dev Dis 2016;3:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Houser SR, Margulies KB, Murphy AM, Spinale FG, Francis GS, Prabhu SD, Rockman HA, Kass DA, Molkentin JD, Sussman MA, Koch WJ. Animal models of heart failure: a scientific statement from the American Heart Association. Circ Res 2012;111:131–150. [DOI] [PubMed] [Google Scholar]
- 178. Vernooy K, Verbeek XA, Peschar M, Crijns HJ, Arts T, Cornelussen RN, Prinzen FW. Left bundle branch block induces ventricular remodelling and functional septal hypoperfusion. Eur Heart J 2005;26:91–98. [DOI] [PubMed] [Google Scholar]
- 179. Heinonen I, Sorop O, van Dalen BM, Wüst RCI, van de Wouw J, de Beer VJ, Octavia Y, van Duin RWB, Hoogstrate Y, Blonden L, Alkio M, Anttila K, Stubbs A, van der Velden J, Merkus D, Duncker DJ. Cellular, mitochondrial and molecular alterations associate with early left ventricular diastolic dysfunction in a porcine model of diabetic metabolic derangement. Sci Rep 2020;10:13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Nguyên UC, Verzaal NJ, van Nieuwenhoven FA, Vernooy K, Prinzen FW. Pathobiology of cardiac dyssynchrony and resynchronization therapy. Europace 2018;20:1898–1909. [DOI] [PubMed] [Google Scholar]
- 181. Vernooy K, Cornelussen RN, Verbeek XA, Vanagt WY, van Hunnik A, Kuiper M, Arts T, Crijns HJ, Prinzen FW. Cardiac resynchronization therapy cures dyssynchronopathy in canine left bundle-branch block hearts. Eur Heart J 2007;28:2148–2155. [DOI] [PubMed] [Google Scholar]
- 182. Strik M, van Middendorp LB, Vernooy K. Animal models of dyssynchrony. J Cardiovasc Transl Res 2012;5:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Zhou M, Liu Y, He Y, Xie K, Quan D, Tang Y, Huang H, Huang C. Selective chemical ablation of transient receptor potential vanilloid 1 expressing neurons in the left stellate ganglion protects against ischemia-induced ventricular arrhythmias in dogs. Biomed Pharmacother 2019;120:109500. [DOI] [PubMed] [Google Scholar]
- 184. Killingsworth CR, Walcott GP, Gamblin TL, Girouard SD, Smith WM, Ideker RE. Chronic myocardial infarction is a substrate for bradycardia-induced spontaneous tachyarrhythmias and sudden death in conscious animals. J Cardiovasc Electrophysiol 2006;17:189–197. [DOI] [PubMed] [Google Scholar]
- 185. Lichtenberg A, Tudorache I, Cebotari S, Suprunov M, Tudorache G, Goerler H, Park JK, Hilfiker-Kleiner D, Ringes-Lichtenberg S, Karck M, Brandes G, Hilfiker A, Haverich A. Preclinical testing of tissue-engineered heart valves re-endothelialized under simulated physiological conditions. Circulation 2006;114:I559–I565. [DOI] [PubMed] [Google Scholar]
- 186. Längin M, Mayr T, Reichart B, Michel S, Buchholz S, Guethoff S, Dashkevich A, Baehr A, Egerer S, Bauer A, Mihalj M, Panelli A, Issl L, Ying J, Fresch AK, Buttgereit I, Mokelke M, Radan J, Werner F, Lutzmann I, Steen S, Sjöberg T, Paskevicius A, Qiuming L, Sfriso R, Rieben R, Dahlhoff M, Kessler B, Kemter E, Kurome M, Zakhartchenko V, Klett K, Hinkel R, Kupatt C, Falkenau A, Reu S, Ellgass R, Herzog R, Binder U, Wich G, Skerra A, Ayares D, Kind A, Schönmann U, Kaup FJ, Hagl C, Wolf E, Klymiuk N, Brenner P, Abicht JM. Consistent success in life-supporting porcine cardiac xenotransplantation. Nature 2018;564:430–433. [DOI] [PubMed] [Google Scholar]
- 187. van Steenbeek FG, Hytonen MK, Leegwater PA, Lohi H. The canine era: the rise of a biomedical model. Anim Genet 2016;47:519–527. [DOI] [PubMed] [Google Scholar]
- 188. Payne JR, Brodbelt DC, Luis Fuentes V. Cardiomyopathy prevalence in 780 apparently healthy cats in rehoming centres (the CatScan study). J Vet Cardiol 2015;17(Suppl 1):S244–S257. [DOI] [PubMed] [Google Scholar]
- 189. Schipper T, Van Poucke M, Sonck L, Smets P, Ducatelle R, Broeckx BJG, Peelman LJ. A feline orthologue of the human MYH7 c.5647G>A (p.(Glu1883Lys)) variant causes hypertrophic cardiomyopathy in a Domestic Shorthair cat. Eur J Hum Genet 2019;27:1724–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Meurs KM, Norgard MM, Ederer MM, Hendrix KP, Kittleson MD. A substitution mutation in the myosin binding protein C gene in ragdoll hypertrophic cardiomyopathy. Genomics 2007;90:261–264. [DOI] [PubMed] [Google Scholar]
- 191. Meurs KM, Sanchez X, David RM, Bowles NE, Towbin JA, Reiser PJ, Kittleson JA, Munro MJ, Dryburgh K, Macdonald KA, Kittleson MD. A cardiac myosin binding protein C mutation in the Maine Coon cat with familial hypertrophic cardiomyopathy. Hum Mol Genet 2005;14:3587–3593. [DOI] [PubMed] [Google Scholar]
- 192. Gasparini S, Fonfara S, Kitz S, Hetzel U, Kipar A. Canine dilated cardiomyopathy: diffuse remodeling, focal lesions, and the involvement of macrophages and new vessel formation. Vet Pathol 2020;57:397–408. [DOI] [PubMed] [Google Scholar]
- 193. Meurs KM, Lahmers S, Keene BW, White SN, Oyama MA, Mauceli E, Lindblad-Toh K. A splice site mutation in a gene encoding for PDK4, a mitochondrial protein, is associated with the development of dilated cardiomyopathy in the Doberman pinscher. Hum Genet 2012;131:1319–1325. [DOI] [PubMed] [Google Scholar]
- 194. Simpson S, Edwards J, Ferguson-Mignan TF, Cobb M, Mongan NP, Rutland CS. Genetics of human and canine dilated cardiomyopathy. Int J Genomics 2015;2015:204823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Meurs KM, Friedenberg SG, Kolb J, Saripalli C, Tonino P, Woodruff K, Olby NJ, Keene BW, Adin DB, Yost OL, DeFrancesco TC, Lahmers S, Tou S, Shelton GD, Granzier H. A missense variant in the titin gene in Doberman pinscher dogs with familial dilated cardiomyopathy and sudden cardiac death. Hum Genet 2019;138:515–524. [DOI] [PubMed] [Google Scholar]
- 196. Basso C, Fox PR, Meurs KM, Towbin JA, Spier AW, Calabrese F, Maron BJ, Thiene G. Arrhythmogenic right ventricular cardiomyopathy causing sudden cardiac death in boxer dogs: a new animal model of human disease. Circulation 2004;109:1180–1185. [DOI] [PubMed] [Google Scholar]
- 197. Yamada N, Kitamori T, Kitamori F, Ishigami K, Iwanaga K, Itou T, Kobayashi R, Kumabe S, Doi T, Sato J, Wako Y, Tsuchitani M. Arrhythmogenic right ventricular cardiomyopathy coincided with the cardiac fibrosis in the inner muscle layer of the left ventricular wall in a boxer dog. J Vet Med Sci 2015;77:1299–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Paradies P, Carlucci L, Woitek F, Staffieri F, Lacitignola L, Ceci L, Romano D, Sasanelli M, Zentilin L, Giacca M, Salvadori S, Crovace A, Recchia FA. Intracoronary gene delivery of the cytoprotective factor vascular endothelial growth factor-B167 in canine patients with dilated cardiomyopathy: a short-term feasibility study. Vet Sci 2019;6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Sleeper MM. Status of therapeutic gene transfer to treat cardiovascular disease in dogs and cats. Vet Clin North Am Small Anim Pract 2017;47:1113–1121. [DOI] [PubMed] [Google Scholar]
- 200. Olson EN. Gene regulatory networks in the evolution and development of the heart. Science 2006;313:1922–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Wolf MJ, Amrein H, Izatt JA, Choma MA, Reedy MC, Rockman HA. Drosophila as a model for the identification of genes causing adult human heart disease. Proc Natl Acad Sci USA 2006;103:1394–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Hoogstra-Berends F, Meijering RA, Zhang D, Heeres A, Loen L, Seerden JP, Kuipers I, Kampinga HH, Henning RH, Brundel BJ. Heat shock protein-inducing compounds as therapeutics to restore proteostasis in atrial fibrillation. Trends Cardiovasc Med 2012;22:62–68. [DOI] [PubMed] [Google Scholar]
- 203. den Hoed M, Eijgelsheim M, Esko T, Brundel BJ, Peal DS, Evans DM, Nolte IM, Segre AV, Holm H, Handsaker RE, Westra HJ, Johnson T, Isaacs A, Yang J, Lundby A, Zhao JH, Kim YJ, Go MJ, Almgren P, Bochud M, Boucher G, Cornelis MC, Gudbjartsson D, Hadley D, van der Harst P, Hayward C, den Heijer M, Igl W, Jackson AU, Kutalik Z, Luan J, Kemp JP, Kristiansson K, Ladenvall C, Lorentzon M, Montasser ME, Njajou OT, O'Reilly PF, Padmanabhan S, St Pourcain B, Rankinen T, Salo P, Tanaka T, Timpson NJ, Vitart V, Waite L, Wheeler W, Zhang W, Draisma HH, Feitosa MF, Kerr KF, Lind PA, Mihailov E, Onland-Moret NC, Song C, Weedon MN, Xie W, Yengo L, Absher D, Albert CM, Alonso A, Arking DE, de Bakker PI, Balkau B, Barlassina C, Benaglio P, Bis JC, Bouatia-Naji N, Brage S, Chanock SJ, Chines PS, Chung M, Darbar D, Dina C, Dorr M, Elliott P, Felix SB, Fischer K, Fuchsberger C, de Geus EJ, Goyette P, Gudnason V, Harris TB, Hartikainen AL, Havulinna AS, Heckbert SR, Hicks AA, Hofman A, Holewijn S, Hoogstra-Berends F, Hottenga JJ, Jensen MK, Johansson A, Junttila J, Kaab S, Kanon B, Ketkar S, Khaw KT, Knowles JW, Kooner AS, Kors JA, Kumari M, Milani L, Laiho P, Lakatta EG, Langenberg C, Leusink M, Liu Y, Luben RN, Lunetta KL, Lynch SN, Markus MR, Marques-Vidal P, Mateo Leach I, McArdle WL, McCarroll SA, Medland SE, Miller KA, Montgomery GW, Morrison AC, Muller-Nurasyid M, Navarro P, Nelis M, O'Connell JR, O'Donnell CJ, Ong KK, Newman AB, Peters A, Polasek O, Pouta A, Pramstaller PP, Psaty BM, Rao DC, Ring SM, Rossin EJ, Rudan D, Sanna S, Scott RA, Sehmi JS, Sharp S, Shin JT, Singleton AB, Smith AV, Soranzo N, Spector TD, Stewart C, Stringham HM, Tarasov KV, Uitterlinden AG, Vandenput L, Hwang SJ, Whitfield JB, Wijmenga C, Wild SH, Willemsen G, Wilson JF, Witteman JC, Wong A, Wong Q, Jamshidi Y, Zitting P, Boer JM, Boomsma DI, Borecki IB, van Duijn CM, Ekelund U, Forouhi NG, Froguel P, Hingorani A, Ingelsson E, Kivimaki M, Kronmal RA, Kuh D, Lind L, Martin NG, Oostra BA, Pedersen NL, Quertermous T, Rotter JI, van der Schouw YT, Verschuren WM, Walker M, Albanes D, Arnar DO, Assimes TL, Bandinelli S, Boehnke M, de Boer RA, Bouchard C, Caulfield WL, Chambers JC, Curhan G, Cusi D, Eriksson J, Ferrucci L, van Gilst WH, Glorioso N, de Graaf J, Groop L, Gyllensten U, Hsueh WC, Hu FB, Huikuri HV, Hunter DJ, Iribarren C, Isomaa B, Jarvelin MR, Jula A, Kahonen M, Kiemeney LA, van der Klauw MM, Kooner JS, Kraft P, Iacoviello L, Lehtimaki T, Lokki ML, Mitchell BD, Navis G, Nieminen MS, Ohlsson C, Poulter NR, Qi L, Raitakari OT, Rimm EB, Rioux JD, Rizzi F, Rudan I, Salomaa V, Sever PS, Shields DC, Shuldiner AR, Sinisalo J, Stanton AV, Stolk RP, Strachan DP, Tardif JC, Thorsteinsdottir U, Tuomilehto J, van Veldhuisen DJ, Virtamo J, Viikari J, Vollenweider P, Waeber G, Widen E, Cho YS, Olsen JV, Visscher PM, Willer C, Franke L, Global BC, Consortium CA, Erdmann J, Thompson JR, Consortium PG, Pfeufer A, Consortium QG, Sotoodehnia N, Consortium Q-I, Newton-Cheh C, Consortium C-A, Ellinor PT, Stricker BH, Metspalu A, Perola M, Beckmann JS, Smith GD, Stefansson K, Wareham NJ, Munroe PB, Sibon OC, Milan DJ, Snieder H, Samani NJ, Loos RJ; CHARGE-AF Consortium . Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet 2013;45:621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Bakkers J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc Res 2011;91:279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Kettleborough RN, Busch-Nentwich EM, Harvey SA, Dooley CM, de Bruijn E, van Eeden F, Sealy I, White RJ, Herd C, Nijman IJ, Fenyes F, Mehroke S, Scahill C, Gibbons R, Wali N, Carruthers S, Hall A, Yen J, Cuppen E, Stemple DL. A systematic genome-wide analysis of zebrafish protein-coding gene function. Nature 2013;496:494–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Wierson WA, Welker JM, Almeida MP, Mann CM, Webster DA, Torrie ME, Weiss TJ, Kambakam S, Vollbrecht MK, Lan M, McKeighan KC, Levey J, Ming Z, Wehmeier A, Mikelson CS, Haltom JA, Kwan KM, Chien CB, Balciunas D, Ekker SC, Clark KJ, Webber BR, Moriarity BS, Solin SL, Carlson DF, Dobbs DL, McGrail M, Essner J. Efficient targeted integration directed by short homology in zebrafish and mammalian cells. Elife 2020;9:e53968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. MacRae CA, Peterson RT. Zebrafish as tools for drug discovery. Nat Rev Drug Discov 2015;14:721–731. [DOI] [PubMed] [Google Scholar]
- 208. Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science 2002;298:2188–2190. [DOI] [PubMed] [Google Scholar]
- 209. dos Remedios CG, Lal SP, Li A, McNamara J, Keogh A, Macdonald PS, Cooke R, Ehler E, Knöll R, Marston SB, Stelzer J, Granzier H, Bezzina C, van Dijk S, De Man F, Stienen GJM, Odeberg J, Pontén F, Linke WA, Linke W, van der Velden J. The Sydney Heart Bank: improving translational research while eliminating or reducing the use of animal models of human heart disease. Biophys Rev 2017;9:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Verdonschot JAJ, Derks KWJ, Hazebroek MR, Wang P, Robinson EL, Adriaens ME, Krapels IPC, van den Wijngaard A, Brunner HG, Heymans SRB. Distinct cardiac transcriptomic clustering in titin and lamin A/C-associated dilated cardiomyopathy patients. Circulation 2020;142:1230–1232. [DOI] [PubMed] [Google Scholar]
- 211. Borbély A, van der Velden J, Papp Z, Bronzwaer JGF, Edes I, Stienen GJM, Paulus WJ. Cardiomyocyte stiffness in diastolic heart failure. Circulation 2005;111:774–781. [DOI] [PubMed] [Google Scholar]
- 212. Coppini R, Ferrantini C, Yao L, Fan P, Del Lungo M, Stillitano F, Sartiani L, Tosi B, Suffredini S, Tesi C, Yacoub M, Olivotto I, Belardinelli L, Poggesi C, Cerbai E, Mugelli A. Late sodium current inhibition reverses electromechanical dysfunction in human hypertrophic cardiomyopathy. Circulation 2013;127:575–584. [DOI] [PubMed] [Google Scholar]
- 213. Chen CY, Caporizzo MA, Bedi K, Vite A, Bogush AI, Robison P, Heffler JG, Salomon AK, Kelly NA, Babu A, Morley MP, Margulies KB, Prosser BL. Suppression of detyrosinated microtubules improves cardiomyocyte function in human heart failure. Nat Med 2018;24:1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Piroddi N, Witjas-Paalberends ER, Ferrara C, Ferrantini C, Vitale G, Scellini B, Wijnker PJM, Sequiera V, Dooijes D, Dos Remedios C, Schlossarek S, Leung MC, Messer A, Ward DG, Biggeri A, Tesi C, Carrier L, Redwood CS, Marston SB, van der Velden J, Poggesi C. The homozygous K280N troponin T mutation alters cross-bridge kinetics and energetics in human HCM. J Gen Physiol 2019;151:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Nijenkamp LLAM, Bollen IAE, Niessen HWM, Dos Remedios CG, Michels M, Poggesi C, Ho CY, Kuster DWD, van der Velden J. Sex-specific cardiac remodeling in early and advanced stages of hypertrophic cardiomyopathy. PLoS One 2020;15:e0232427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Bogunovic N, Meekel JP, Majolée J, Hekhuis M, Pyszkowski J, Jockenhövel S, Kruse M, Riesebos E, Micha D, Blankensteijn JD, Hordijk PL, Ghazanfari S, Yeung KK. Patient-specific 3-dimensional model of smooth muscle cell and extracellular matrix dysfunction for the study of aortic aneurysms. J Endovasc Ther 2021;28:604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Meekel JP, Mattei G, Costache VS, Balm R, Blankensteijn JD, Yeung KK. A multilayer micromechanical elastic modulus measuring method in ex vivo human aneurysmal abdominal aortas. Acta Biomater 2019;96:345–353. [DOI] [PubMed] [Google Scholar]
- 218. Watson SA, Scigliano M, Bardi I, Ascione R, Terracciano CM, Perbellini F. Preparation of viable adult ventricular myocardial slices from large and small mammals. Nat Protoc 2017;12:2623–2639. [DOI] [PubMed] [Google Scholar]
- 219. Pitoulis FG, Watson SA, Perbellini F, Terracciano CM. Myocardial slices come to age: an intermediate complexity in vitro cardiac model for translational research. Cardiovasc Res 2020;116:1275–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Perbellini F, Thum T. Living myocardial slices: a novel multicellular model for cardiac translational research. Eur Heart J 2020;41:2405–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Wang K, Lee P, Mirams GR, Sarathchandra P, Borg TK, Gavaghan DJ, Kohl P, Bollensdorff C. Cardiac tissue slices: preparation, handling, and successful optical papping. Am J Physiol Heart Circ Physiol 2015;308:H1112–H1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Thomas RC, Singh A, Cowley PM, Myagmar B-E, Montgomery MD, Swigart PM, Marco T, De Baker AJ, Simpson PC. A myocardial slice culture model reveals alpha-1A-adrenergic receptor signaling in the human heart. JACC Basic Transl Sci 2016;1:155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Watson SA, Dendorfer A, Thum T, Perbellini F. A practical guide for investigating cardiac physiology using living myocardial slices. Basic Res Cardiol 2020;115:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Fischer C, Milting H, Fein E, Reiser E, Lu K, Seidel T, Schinner C, Schwarzmayr T, Schramm R, Tomasi R, Husse B, Cao-Ehlker X, Pohl U, Dendorfer A. Long-term functional and structural preservation of precision-cut human myocardium under continuous electromechanical stimulation in vitro. Nat Commun 2019;10:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Watson SA, Duff J, Bardi I, Zabielska M, Atanur SS, Jabbour RJ, Simon A, Tomas A, Smolenski RT, Harding SE, Perbellini F, Terracciano CM. Biomimetic electromechanical stimulation to maintain adult myocardial slices in vitro. Nat Commun 2019;10:2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Pitoulis FG, Hasan W, Papadaki M, Clavere NG, Perbellini F, Harding SE, Kirk JA, Boateng SY, Tombe PP, de Terracciano CM. Intact myocardial preparations reveal intrinsic transmural heterogeneity in cardiac mechanics. J Mol Cell Cardiol 2020;141:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Guzik TJ, Mussa S, Gastaldi D, Sadowski J, Ratnatunga C, Pillai R, Channon KM. Mechanisms of increased vascular superoxide production in human diabetes mellitus: role of NAD(P)H oxidase and endothelial nitric oxide synthase. Circulation 2002;105:1656–1662. [DOI] [PubMed] [Google Scholar]
- 228. Guzik TJ, Sadowski J, Guzik B, Jopek A, Kapelak B, Przybylowski P, Wierzbicki K, Korbut R, Harrison DG, Channon KM. Coronary artery superoxide production and nox isoform expression in human coronary artery disease. Arterioscler Thromb Vasc Biol 2006;26:333–339. [DOI] [PubMed] [Google Scholar]
- 229. Guzik TJ, West NE, Black E, McDonald D, Ratnatunga C, Pillai R, Channon KM. UltraRapid communications: vascular superoxide production by NAD(P)H OxidaseAssociation with endothelial dysfunction and clinical risk factors. Circ Res 2000;86:1008. [DOI] [PubMed] [Google Scholar]
- 230. Buikema JW, Lee S, Goodyer WR, Maas RG, Chirikian O, Li G, Miao Y, Paige SL, Lee D, Wu H, Paik DT, Rhee S, Tian L, Galdos FX, Puluca N, Beyersdorf B, Hu J, Beck A, Venkamatran S, Swami S, Wijnker P, Schuldt M, Dorsch LM, van Mil A, Red-Horse K, Wu JY, Geisen C, Hesse M, Serpooshan V, Jovinge S, Fleischmann BK, Doevendans PA, van der Velden J, Garcia KC, Wu JC, Sluijter JPG, Wu SM. Wnt activation and reduced cell-cell contact synergistically induce massive expansion of functional human iPSC-derived cardiomyocytes. Cell Stem Cell 2020;27:50–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Cai W, Zhang J, de Lange WJ, Gregorich ZR, Karp H, Farrell ET, Mitchell SD, Tucholski T, Lin Z, Biermann M, McIlwain SJ, Ralphe JC, Kamp TJ, Ge Y. An unbiased proteomics method to assess the maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res 2019;125:936–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Madonna R, Gorbe A, Ferdinandy P, de Caterina R. Glucose metabolism, hyperosmotic stress, and reprogramming of somatic cells. Mol Biotechnol 2013;55:169–178. [DOI] [PubMed] [Google Scholar]
- 233. Bowman PRT, Smith GL, Gould GW. GLUT4 expression and glucose transport in human induced pluripotent stem cell-derived cardiomyocytes. PLoS One 2019;14:e0217885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Rohani L, Johnson AA, Naghsh P, Rancourt DE, Ulrich H, Holland H. Concise review: molecular cytogenetics and quality control: clinical guardians for pluripotent stem cells. Stem Cells Transl Med 2018;7:867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Dierickx P, Vermunt MW, Muraro MJ, Creyghton MP, Doevendans PA, van Oudenaarden A, Geijsen N, van Laake LW. Circadian networks in human embryonic stem cell-derived cardiomyocytes. EMBO Rep 2017;18:1199–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Pamies D, Bal-Price A, Chesné C, Coecke S, Dinnyes A, Eskes C, Grillari R, Gstraunthaler G, Hartung T, Jennings P, Leist M, Martin U, Passier R, Schwamborn JC, Stacey GN, Ellinger-Ziegelbauer H, Daneshian M. Advanced Good Cell Culture Practice for human primary, stem cell-derived and organoid models as well as microphysiological systems. Altex 2018;35:353–378. [DOI] [PubMed] [Google Scholar]
- 237. Denning C, Borgdorff V, Crutchley J, Firth KS, George V, Kalra S, Kondrashov A, Hoang MD, Mosqueira D, Patel A, Prodanov L, Rajamohan D, Skarnes WC, Smith JG, Young LE. Cardiomyocytes from human pluripotent stem cells: from laboratory curiosity to industrial biomedical platform. Biochim Biophys Acta 2016;1863:1728–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Hu D, Linders A, Yamak A, Correia C, Kijlstra JD, Garakani A, Xiao L, Milan DJ, van der Meer P, Serra M, Alves PM, Domian IJ. Metabolic maturation of human pluripotent stem cell-derived cardiomyocytes by inhibition of HIF1alpha and LDHA. Circ Res 2018;123:1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Horikoshi Y, Yan Y, Terashvili M, Wells C, Horikoshi H, Fujita S, Bosnjak ZJ, Bai X. Fatty acid-treated induced pluripotent stem cell-derived human cardiomyocytes exhibit adult cardiomyocyte-like energy metabolism phenotypes. Cells 2019;8:1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Heras-Bautista CO, Katsen-Globa A, Schloerer NE, Dieluweit S, Abd El Aziz OM, Peinkofer G, Attia WA, Khalil M, Brockmeier K, Hescheler J, Pfannkuche K. The influence of physiological matrix conditions on permanent culture of induced pluripotent stem cell-derived cardiomyocytes. Biomaterials 2014;35:7374–7385. [DOI] [PubMed] [Google Scholar]
- 241. Corbin EA, Vite A, Peyster EG, Bhoopalam M, Brandimarto J, Wang X, Bennett AI, Clark AT, Cheng X, Turner KT, Musunuru K, Margulies KB. Tunable and reversible substrate stiffness reveals a dynamic mechanosensitivity of cardiomyocytes. ACS Appl Mater Interfaces 2019;11:20603–20614. [DOI] [PubMed] [Google Scholar]
- 242. Feaster TK, Cadar AG, Wang L, Williams CH, Chun YW, Hempel JE, Bloodworth N, Merryman WD, Lim CC, Wu JC, Knollmann BC, Hong CC. Matrigel mattress: a method for the generation of single contracting human-induced pluripotent stem cell-derived cardiomyocytes. Circ Res 2015;117:995–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Salick MR, Napiwocki BN, Sha J, Knight GT, Chindhy SA, Kamp TJ, Ashton RS, Crone WC. Micropattern width dependent sarcomere development in human ESC-derived cardiomyocytes. Biomaterials 2014;35:4454–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Bertrand L, Horman S, Beauloye C, Vanoverschelde JL. Insulin signalling in the heart. Cardiovasc Res 2008;79:238–248. [DOI] [PubMed] [Google Scholar]
- 245. Weinberger F, Mannhardt I, Eschenhagen T. Engineering cardiac muscle tissue: a maturating field of research. Circ Res 2017;120:1487–1500. [DOI] [PubMed] [Google Scholar]
- 246. Leonard A, Bertero A, Powers JD, Beussman KM, Bhandari S, Regnier M, Murry CE, Sniadecki NJ. Afterload promotes maturation of human induced pluripotent stem cell derived cardiomyocytes in engineered heart tissues. J Mol Cell Cardiol 2018;118:147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Giacomelli E, Meraviglia V, Campostrini G, Cochrane A, Cao X, van Helden RWJ, Krotenberg Garcia A, Mircea M, Kostidis S, Davis RP, van Meer BJ, Jost CR, Koster AJ, Mei H, Miguez DG, Mulder AA, Ledesma-Terron M, Pompilio G, Sala L, Salvatori DCF, Slieker RC, Sommariva E, de Vries AAF, Giera M, Semrau S, Tertoolen LGJ, Orlova VV, Bellin M, Mummery CL. Human-iPSC-derived cardiac stromal cells enhance maturation in 3D cardiac microtissues and reveal non-cardiomyocyte contributions to heart disease. Cell Stem Cell 2020;26:862–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Olmer R, Engels L, Usman A, Menke S, Malik MNH, Pessler F, Gohring G, Bornhorst D, Bolten S, Abdelilah-Seyfried S, Scheper T, Kempf H, Zweigerdt R, Martin U. Differentiation of human pluripotent stem cells into functional endothelial cells in scalable suspension culture. Stem Cell Reports 2018;10:1657–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Shinnawi R, Huber I, Maizels L, Shaheen N, Gepstein A, Arbel G, Tijsen AJ, Gepstein L. Monitoring human-induced pluripotent stem cell-derived cardiomyocytes with genetically encoded calcium and voltage fluorescent reporters. Stem Cell Reports 2015;5:582–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. de Korte T, van Meer B, Garcia AK, Tertoolen L, Clements PJ, Bahinski A, Rossman EI, Xu X, Turner S, Denning C, Vlaming M, Braam S, Mummery C. Simultaneous measurement of contraction, voltage and calcium in HIPSC-CMS for the detection of inotropic effects under blinded conditions. J Pharmacol Toxicol Methods 2019;99:106595. [Google Scholar]
- 251. Park SJ, Zhang D, Qi Y, Li Y, Lee KY, Bezzerides VJ, Yang P, Xia S, Kim SL, Liu X, Lu F, Pasqualini FS, Campbell PH, Geva J, Roberts AE, Kleber AG, Abrams DJ, Pu WT, Parker KK. Insights into the pathogenesis of catecholaminergic polymorphic ventricular tachycardia from engineered human heart tissue. Circulation 2019;140:390–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Lemoine MD, Mannhardt I, Breckwoldt K, Prondzynski M, Flenner F, Ulmer B, Hirt MN, Neuber C, Horvath A, Kloth B, Reichenspurner H, Willems S, Hansen A, Eschenhagen T, Christ T. Human iPSC-derived cardiomyocytes cultured in 3D engineered heart tissue show physiological upstroke velocity and sodium current density. Sci Rep 2017;7:5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Ma Z, Huebsch N, Koo S, Mandegar MA, Siemons B, Boggess S, Conklin BR, Grigoropoulos CP, Healy KE. Contractile deficits in engineered cardiac microtissues as a result of MYBPC3 deficiency and mechanical overload. Nat Biomed Eng 2018;2:955–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Makkos A, Szantai A, Paloczi J, Pipis J, Kiss B, Poggi P, Ferdinandy P, Chatgilialoglu A, Gorbe A. A comorbidity model of myocardial ischemia/reperfusion injury and hypercholesterolemia in rat cardiac myocyte cultures. Front Physiol 2019;10:1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Zhang YS, Davoudi F, Walch P, Manbachi A, Luo X, Dell'Erba V, Miri AK, Albadawi H, Arneri A, Li X, Wang X, Dokmeci MR, Khademhosseini A, Oklu R. Bioprinted thrombosis-on-a-chip. Lab Chip 2016;16:4097–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. van den Berg A, Mummery CL, Passier R, van der Meer AD. Personalised organs-on-chips: functional testing for precision medicine. Lab Chip 2019;19:198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle regulation. Circ Res 2002;90:1044–1054. [DOI] [PubMed] [Google Scholar]
- 258. Liu J, Volkers L, Jangsangthong W, Bart CI, Engels MC, Zhou G, Schalij MJ, Ypey DL, Pijnappels DA, de Vries AAF. Generation and primary characterization of iAM-1, a versatile new line of conditionally immortalized atrial myocytes with preserved cardiomyogenic differentiation capacity. Cardiovasc Res 2018;114:1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Drakhlis L, Biswanath S, Farr CM, Lupanow V, Teske J, Ritzenhoff K, Franke A, Manstein F, Bolesani E, Kempf H, Liebscher S, Schenke-Layland K, Hegermann J, Nolte L, Meyer H, de la Roche J, Thiemann S, Wahl-Schott C, Martin U, Zweigerdt R. Human heart-forming organoids recapitulate early heart and foregut development. Nat Biotechnol 2021;39:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Lai BFL, Lu RXZ, Davenport Huyer L, Kakinoki S, Yazbeck J, Wang EY, Wu Q, Zhang B, Radisic M. A well plate-based multiplexed platform for incorporation of organoids into an organ-on-a-chip system with a perfusable vasculature. Nat Protoc 2021;16:2158–2189. [DOI] [PubMed] [Google Scholar]
- 261. Castilho M, van Mil A, Maher M, Metz CHG, Hochleitner G, Groll J, Doevendans PA, Ito K, Sluijter JPG, Malda J. Melt electrowriting allows tailored microstructural and mechanical design of scaffolds to advance functional human myocardial tissue. Adv Funct Mater 2018;28:1803151. [Google Scholar]
- 262. Wang H, Tibbitt MW, Langer SJ, Leinwand LA, Anseth KS. Hydrogels preserve native phenotypes of valvular fibroblasts through an elasticity-regulated PI3K/AKT pathway. Proc Natl Acad Sci USA 2013;110:19336–19341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Kural MH, Billiar KL. Mechanoregulation of valvular interstitial cell phenotype in the third dimension. Biomaterials 2014;35:1128–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature 2011;474:179–183. [DOI] [PubMed] [Google Scholar]
- 265. Ma H, Killaars AR, DelRio FW, Yang C, Anseth KS. Myofibroblastic activation of valvular interstitial cells is modulated by spatial variations in matrix elasticity and its organization. Biomaterials 2017;131:131–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Santoro R, Scaini D, Severino LU, Amadeo F, Ferrari S, Bernava G, Garoffolo G, Agrifoglio M, Casalis L, Pesce M. Activation of human aortic valve interstitial cells by local stiffness involves YAP-dependent transcriptional signaling. Biomaterials 2018;181:268–279. [DOI] [PubMed] [Google Scholar]
- 267. Elosegui-Artola A, Andreu I, Beedle AEM, Lezamiz A, Uroz M, Kosmalska AJ, Oria R, Kechagia JZ, Rico-Lastres P, Le Roux AL, Shanahan CM, Trepat X, Navajas D, Garcia-Manyes S, Roca-Cusachs P. Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell 2017;171:1397–1410. [DOI] [PubMed] [Google Scholar]
- 268. Porras AM, Westlund JA, Evans AD, Masters KS. Creation of disease-inspired biomaterial environments to mimic pathological events in early calcific aortic valve disease. Proc Natl Acad Sci USA 2018;115:E363–E371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Amadeo F, Boschetti F, Polvani G, Banfi C, Pesce M, Santoro R. Aortic valve cell seeding into decellularized animal pericardium by perfusion-assisted bioreactor. J Tissue Eng Regen Med 2018;12:1481–1493. [DOI] [PubMed] [Google Scholar]
- 270. Wissing TB, van Haaften EE, Koch SE, Ippel BD, Kurniawan NA, Bouten CVC, Smits AIPM. Hemodynamic loads distinctively impact the secretory profile of biomaterial-activated macrophages—implications for in situ vascular tissue engineering. Biomater Sci 2019;8:132–147. [DOI] [PubMed] [Google Scholar]
- 271. Fu J, Ding X, Stowell CET, Wu Y-L, Wang Y. Slow degrading poly(glycerol sebacate) derivatives improve vascular graft remodeling in a rat carotid artery interposition model. Biomaterials 2020;257:120251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Manz XD, Albers HJ, Symersky P, Aman J, van der Meer AD, Bogaard HJ, Szulcek R. In vitro microfluidic disease model to study whole blood-endothelial interactions and blood clot dynamics in real-time. J Vis Exp 2020;159:e61068. [DOI] [PubMed] [Google Scholar]
- 273. Cochrane A, Albers HJ, Passier R, Mummery CL, van den Berg A, Orlova VV, van der Meer AD. Advanced in vitro models of vascular biology: human induced pluripotent stem cells and organ-on-chip technology. Adv Drug Deliv Rev 2019;140:68–77. [DOI] [PubMed] [Google Scholar]
- 274. Jalalzadeh H, Indrakusuma R, Blankensteijn JD, Wisselink W, Yeung KK, Lindeman JHN, Hamming JF, Koelemay MJW, Legemate DA, Balm R. Design and protocol of a comprehensive multicentre biobank for abdominal aortic aneurysms. BMJ Open 2019;9:e028858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Min E, Schwartz MA. Translocating transcription factors in fluid shear stress-mediated vascular remodeling and disease. Exp Cell Res 2019;376:92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Lipp SN, Niedert EE, Cebull HL, Diorio TC, Ma JL, Rothenberger SM, Boster KA, Goergen CJ Computational hemodynamic modeling of arterial aneurysms: a mini-review. Front Physiol 2020;11:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Pesce M, Santoro R. Feeling the right force: how to contextualize the cell mechanical behavior in physiologic turnover and pathologic evolution of the cardiovascular system. Pharmacol Ther 2017;171:75–82. [DOI] [PubMed] [Google Scholar]
- 278. Garoffolo G, Ruiter MS, Piola M, Brioschi M, Thomas AC, Agrifoglio M, Polvani G, Coppadoro L, Zoli S, Saccu C, Spinetti G, Banfi C, Fiore GB, Madeddu P, Soncini M, Pesce M. Coronary artery mechanics induces human saphenous vein remodelling via recruitment of adventitial myofibroblast-like cells mediated by thrombospondin-1. Theranostics 2020;10:2597–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Yamashiro Y, Thang BQ, Shin SJ, Lino CA, Nakamura T, Kim J, Sugiyama K, Tokunaga C, Sakamoto H, Osaka M, Davis EC, Wagenseil JE, Hiramatsu Y, Yanagisawa H. Role of thrombospondin-1 in mechanotransduction and development of thoracic aortic aneurysm in mouse and humans. Circ Res 2018;123:660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280. Kim S, Kim W, Lim S, Jeon JS. Vasculature-on-a-chip for in vitro disease models. Bioengineering (Basel) 2017;4:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Findeisen K, Morticelli L, Goecke T, Kolbeck L, Ramm R, Höffler HK, Brandes G, Korossis S, Haverich A, Hilfiker A. Toward acellular xenogeneic heart valve prostheses: histological and biomechanical characterization of decellularized and enzymatically deglycosylated porcine pulmonary heart valve matrices. Xenotransplantation 2020;27:e12617. [DOI] [PubMed] [Google Scholar]
- 282. Sarikouch S, Theodoridis K, Hilfiker A, Boethig D, Laufer G, Andreas M, Cebotari S, Tudorache I, Bobylev D, Neubert L, Teiken K, Robertus JL, Jonigk D, Beerbaum P, Haverich A, Horke A. Early insight into in vivo recellularization of cell-free allogenic heart valves. Ann Thorac Surg 2019;108:581–589. [DOI] [PubMed] [Google Scholar]
- 283. Cheung DY, Duan B, Butcher JT. Current progress in tissue engineering of heart valves: multiscale problems, multiscale solutions. Expert Opin Biol Ther 2015;15:1155–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Pina S, Ribeiro VP, Marques CF, Maia FR, Silva TH, Reis RL, Oliveira JM. Scaffolding strategies for tissue engineering and regenerative medicine applications. Materials (Basel) 2019;12:1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Claiborne TE, Slepian MJ, Hossainy S, Bluestein D. Polymeric trileaflet prosthetic heart valves: evolution and path to clinical reality. Expert Rev Med Devices 2012;9:577–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Emmert MY, Schmitt BA, Loerakker S, Sanders B, Spriestersbach H, Fioretta ES, Bruder L, Brakmann K, Motta SE, Lintas V, Dijkman PE, Frese L, Berger F, Baaijens FPT, Hoerstrup SP. Computational modeling guides tissue-engineered heart valve design for long-term in vivo performance in a translational sheep model. Sci Transl Med 2018;10:eaan4587. [DOI] [PubMed] [Google Scholar]
- 287. Simon P, Kasimir MT, Seebacher G, Weigel G, Ullrich R, Salzer-Muhar U, Rieder E, Wolner E. Early failure of the tissue engineered porcine heart valve SYNERGRAFT in pediatric patients. Eur J Cardiothorac Surg 2003;23:1002–1006. [DOI] [PubMed] [Google Scholar]
- 288. Brown JW, Ruzmetov M, Eltayeb O, Rodefeld MD, Turrentine MW. Performance of SynerGraft decellularized pulmonary homograft in patients undergoing a Ross procedure. Ann Thorac Surg 2011;91:416–422. [DOI] [PubMed] [Google Scholar]
- 289. Fallahiarezoudar E, Ahmadipourroudposht M, Idris A, Mohd Yusof N. A review of: application of synthetic scaffold in tissue engineering heart valves. Mater Sci Eng C Mater Biol Appl 2015;48:556–565. [DOI] [PubMed] [Google Scholar]
- 290. Vashistha R, Kumar P, Dangi AK, Sharma N, Chhabra D, Shukla P. Quest for cardiovascular interventions: precise modeling and 3D printing of heart valves. J Biol Eng 2019;13:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Cohn D, Sloutski A, Elyashiv A, Varma VB, Ramanujan R. In situ generated medical devices. Adv Healthc Mater 2019;8:e1801066. [DOI] [PubMed] [Google Scholar]
- 292. Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 2012;492:376–381. [DOI] [PubMed] [Google Scholar]
- 293. Diez-Cunado M, Wei K, Bushway PJ, Maurya MR, Perera R, Subramaniam S, Ruiz-Lozano P, Mercola M. miRNAs that induce human cardiomyocyte proliferation converge on the Hippo Pathway. Cell Rep 2018;23:2168–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Klimas A, Ambrosi CM, Yu J, Williams JC, Bien H, Entcheva E. OptoDyCE as an automated system for high-throughput all-optical dynamic cardiac electrophysiology. Nat Commun 2016;7:11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Jentzsch C, Leierseder S, Loyer X, Flohrschutz I, Sassi Y, Hartmann D, Thum T, Laggerbauer B, Engelhardt S. A phenotypic screen to identify hypertrophy-modulating microRNAs in primary cardiomyocytes. J Mol Cell Cardiol 2012;52:13–20. [DOI] [PubMed] [Google Scholar]
- 296. Carlson C, Koonce C, Aoyama N, Einhorn S, Fiene S, Thompson A, Swanson B, Anson B, Kattman S. Phenotypic screening with human iPS cell-derived cardiomyocytes: HTS-compatible assays for interrogating cardiac hypertrophy. J Biomol Screen 2013;18:1203–1211. [DOI] [PubMed] [Google Scholar]
- 297. Reid BG, Stratton MS, Bowers S, Cavasin MA, Demos-Davies KM, Susano I, McKinsey TA. Discovery of novel small molecule inhibitors of cardiac hypertrophy using high throughput, high content imaging. J Mol Cell Cardiol 2016;97:106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. McLendon PM, Davis G, Gulick J, Singh SR, Xu N, Salomonis N, Molkentin JD, Robbins J. An unbiased high-throughput screen to identify novel effectors that impact on cardiomyocyte aggregate levels. Circ Res 2017;121:604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. da Rocha AM, Campbell K, Mironov S, Jiang J, Mundada L, Guerrero-Serna G, Jalife J, Herron TJ. hiPSC-CM monolayer maturation state determines drug responsiveness in high throughput pro-arrhythmia screen. Sci Rep 2017;7:13834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Doherty KR, Talbert DR, Trusk PB, Moran DM, Shell SA, Bacus S. Structural and functional screening in human induced-pluripotent stem cell-derived cardiomyocytes accurately identifies cardiotoxicity of multiple drug types. Toxicol Appl Pharmacol 2015;285:51–60. [DOI] [PubMed] [Google Scholar]
- 301. Sharma A, Burridge PW, McKeithan WL, Serrano R, Shukla P, Sayed N, Churko JM, Kitani T, Wu H, Holmstrom A, Matsa E, Zhang Y, Kumar A, Fan AC, Alamo D, Wu JC, Moslehi SM, Mercola JJ, Wu Jc M. High-throughput screening of tyrosine kinase inhibitor cardiotoxicity with human induced pluripotent stem cells. Sci Transl Med 2017;9:eaaf2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Wahlquist C, Jeong D, Rojas-Munoz A, Kho C, Lee A, Mitsuyama S, van Mil A, Park WJ, Sluijter JP, Doevendans PA, Hajjar RJ, Mercola M. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature 2014;508:531–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Del Alamo JC, Lemons D, Serrano R, Savchenko A, Cerignoli F, Bodmer R, Mercola M. High throughput physiological screening of iPSC-derived cardiomyocytes for drug development. Biochim Biophys Acta 2016;1863:1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Wells SP, Waddell HM, Sim CB, Lim SY, Bernasochi GB, Pavlovic D, Kirchhof P, Porrello ER, Delbridge LMD, Bell JR. Cardiomyocyte functional screening: interrogating comparative electrophysiology of high-throughput model cell systems. Am J Physiol Cell Physiol 2019;317:C1256–C1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Feinberg AW, Feigel A, Shevkoplyas SS, Sheehy S, Whitesides GM, Parker KK. Muscular thin films for building actuators and powering devices. Science 2007;317:1366–1370. [DOI] [PubMed] [Google Scholar]
- 306. Jacot JG, McCulloch AD, Omens JH. Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes. Biophys J 2008;95:3479–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Schimmel K, Jung M, Foinquinos A, José GS, Beaumont J, Bock K, Grote-Levi L, Xiao K, Bär C, Pfanne A, Just A, Zimmer K, Ngoy S, López B, Ravassa S, Samolovac S, Janssen-Peters H, Remke J, Scherf K, Dangwal S, Piccoli MT, Kleemiss F, Kreutzer FP, Kenneweg F, Leonardy J, Hobuß L, Santer L, Do QT, Geffers R, Braesen JH, Schmitz J, Brandenberger C, Müller DN, Wilck N, Kaever V, Bähre H, Batkai S, Fiedler J, Alexander KM, Wertheim BM, Fisch S, Liao R, Diez J, González A, Thum T. Natural compound library screening identifies new molecules for the treatment of cardiac fibrosis and diastolic dysfunction. Circulation 2020;141:751–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Nollet EE, Manders EM, Goebel M, Jansen V, Brockmann C, Osinga J, van der Velden J, Helmes M, Kuster DWD. Large-scale contractility measurements reveal large atrioventricular and subtle interventricular differences in cultured unloaded rat cardiomyocytes. Front Physiol 2020;11:815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Schuldt M, Pei J, Harakalova M, Dorsch LM, Schlossarek S, Mokry M, PhD Knol JC, Pham TV, Schelfhorst T, Piersma SR, Dos Remedios C, Dalinghaus M, Michels M, Asselbergs FW, Moutin MJ, Carrier L, Jimenez CJ, Van Der Velden J, Kuster DWD. Proteomic and functional studies reveal detyrosinated tubulin as treatment target in sarcomere mutation-induced hypertrophic cardiomyopathy. Circ Heart Fail 2021;14:e007022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Mills RJ, Parker BL, Quaife-Ryan GA, Voges HK, Needham EJ, Bornot A, Ding M, Andersson H, Polla M, Elliott DA, Drowley L, Clausen M, Plowright AT, Barrett IP, Wang QD, James DE, Porrello ER, Hudson JE. Drug screening in human PSC-cardiac organoids identifies pro-proliferative compounds acting via the mevalonate pathway. Cell Stem Cell 2019;24:895–907. e896. [DOI] [PubMed] [Google Scholar]
- 311. van Hout GP, Jansen Of Lorkeers SJ, Wever KE, Sena ES, Kouwenberg LH, van Solinge WW, Macleod MR, Doevendans PA, Pasterkamp G, Chamuleau SA, Hoefer IE. Translational failure of anti-inflammatory compounds for myocardial infarction: a meta-analysis of large animal models. Cardiovasc Res 2016;109:240–248. [DOI] [PubMed] [Google Scholar]
- 312. Jansen Of Lorkeers SJ, Doevendans PA, Chamuleau SA. All preclinical trials should be registered in advance in an online registry. Eur J Clin Invest 2014;44:891–892. [DOI] [PubMed] [Google Scholar]
- 313. Bert B, Heinl C, Chmielewska J, Schwarz F, Grune B, Hensel A, Greiner M, Schönfelder G. Refining animal research: the Animal Study Registry. PLoS Biol 2019;17:e3000463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Jones SP, Tang XL, Guo Y, Steenbergen C, Lefer DJ, Kukreja RC, Kong M, Li Q, Bhushan S, Zhu X, Du J, Nong Y, Stowers HL, Kondo K, Hunt GN, Goodchild TT, Orr A, Chang CC, Ockaili R, Salloum FN, Bolli R. The NHLBI-sponsored Consortium for preclinicAl assESsment of cARdioprotective therapies (CAESAR): a new paradigm for rigorous, accurate, and reproducible evaluation of putative infarct-sparing interventions in mice, rabbits, and pigs. Circ Res 2015;116:572–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Zare H, Shooshtari P, Gupta A, Brinkman RR. Data reduction for spectral clustering to analyze high throughput flow cytometry data. BMC Bioinformatics 2010;11:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Dwivedi SK, Tjärnberg A, Tegnér J, Gustafsson M. Deriving disease modules from the compressed transcriptional space embedded in a deep autoencoder. Nat Commun 2020;11:856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Kiarashinejad Y, Abdollahramezani S, Adibi A. Deep learning approach based on dimensionality reduction for designing electromagnetic nanostructures. NPJ Comput Mater 2020;6:12. [Google Scholar]
- 318. Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science 2006;313:504–507. [DOI] [PubMed] [Google Scholar]
- 319. Kingma DP, Welling M. Auto-encoding variational bayes; Mathematics, Computer Science 2013. arXiv:1312.6114. [Google Scholar]
- 320. Choi H, Kang H, Lee DS. Alzheimer's disease neuroimaging initiative. Predicting aging of brain metabolic topography using variational autoencoder. Front Aging Neurosci 2018;10:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Rampášek L, Hidru D, Smirnov P, Haibe-Kains B, Goldenberg A. Dr.VAE: improving drug response prediction via modeling of drug perturbation effects. Bioinformatics 2019;35:3743–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Parini P, Altucci L, Balligand JL, Baumbach J, Ferdinandy P, Filetti S, Maron BA, Petrillo E, Silverman EK, Barabasi AL, Loscalzo J; International Network Medicine Consortium . The Network medicine imperative and the need for an International Network Medicine Consortium. Am J Med 2020;133:e451–e454. [DOI] [PubMed] [Google Scholar]
- 323. Ágg B, Baranyai T, Makkos A, Vető B, Faragó N, Zvara Á, Giricz Z, Veres DV, Csermely P, Arányi T, Puskás LG, Varga ZV, Ferdinandy P. MicroRNA interactome analysis predicts post-transcriptional regulation of ADRB2 and PPP3R1 in the hypercholesterolemic myocardium. Sci Rep 2018;8:10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJ, Groth P, Goble C, Grethe JS, Heringa J, 't Hoen PA, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone SA, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016;3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Rieke N, Hancox J, Li W, Milletarì F, Roth HR, Albarqouni S, Bakas S, Galtier MN, Landman BA, Maier-Hein K, Ourselin S, Sheller M, Summers RM, Trask A, Xu D, Baust M, Cardoso MJ. The future of digital health with federated learning. NPJ Digit Med 2020;3:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Sacristán JA, Aguarón A, Avendaño-Solá C, Garrido P, Carrión J, Gutiérrez A, Kroes R, Flores A. Patient involvement in clinical research: why, when, and how. Patient Prefer Adherence 2016;10:631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Hoekstra AG, Alowayyed S, Lorenz E, Melnikova N, Mountrakis L, van Rooij B. Towards the virtual artery: a multiscale model for vascular physiology at the physics-chemistry-biology interface. Philos Trans A Math Phys Eng Sci 2016;374:20160146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Niederer SA, Lumens J, Trayanova NA. Computational models in cardiology. Nat Rev Cardiol 2019;16:100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Viceconti M, Hunter P. The virtual physiological human: ten years after. Annu Rev Biomed Eng 2016;18:103–123. [DOI] [PubMed] [Google Scholar]
- 330. Passini E, Britton OJ, Lu HR, Rohrbacher J, Hermans AN, Gallacher DJ, Greig RJH, Bueno-Orovio A, Rodriguez B. Human in silico drug trials demonstrate higher accuracy than animal models in predicting clinical pro-arrhythmic cardiotoxicity. Front Physiol 2017;8:668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Sutanto H, Lyon A, Lumens J, Schotten U, Dobrev D, Heijman J. Cardiomyocyte calcium handling in health and disease: insights from in vitro and in silico studies. Prog Biophys Mol Biol 2020;157:54–75. [DOI] [PubMed] [Google Scholar]
- 332. Corral-Acero J, Margara F, Marciniak M, Rodero C, Loncaric F, Feng Y, Gilbert A, Fernandes JF, Bukhari HA, Wajdan A, Martinez MV, Santos MS, Shamohammdi M, Luo H, Westphal P, Leeson P, DiAchille P, Gurev V, Mayr M, Geris L, Pathmanathan P, Morrison T, Cornelussen R, Prinzen F, Delhaas T, Doltra A, Sitges M V, Igmond EJ, Zacur E, Grau V, Rodriguez B, Remme EW, Niederer S, Mortier P, McLeod K, Potse M, Pueyo E, Bueno-Orovio A, Lamata P. The ‘Digital Twin’ to enable the vision of precision cardiology. Eur Heart J 2020;41:4556–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Arevalo HJ, Vadakkumpadan F, Guallar E, Jebb A, Malamas P, Wu KC, Trayanova NA. Arrhythmia risk stratification of patients after myocardial infarction using personalized heart models. Nat Commun 2016;7:11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Taylor CA, Fonte TA, Min JK. Computational fluid dynamics applied to cardiac computed tomography for noninvasive quantification of fractional flow reserve: scientific basis. J Am Coll Cardiol 2013;61:2233–2241. [DOI] [PubMed] [Google Scholar]
- 335. Ramanathan C, Ghanem RN, Jia P, Ryu K, Rudy Y. Noninvasive electrocardiographic imaging for cardiac electrophysiology and arrhythmia. Nat Med 2004;10:422–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336. Ricke-Hoch M, Pfeffer TJ, Hilfiker-Kleiner D. Peripartum cardiomyopathy: basic mechanisms and hope for new therapies. Cardiovasc Res 2020;116:520–531. [DOI] [PubMed] [Google Scholar]
- 337. Hilfiker-Kleiner D, Sliwa K. Pathophysiology and epidemiology of peripartum cardiomyopathy. Nat Rev Cardiol 2014;11:364–370. [DOI] [PubMed] [Google Scholar]
- 338. van Spaendonck-Zwarts KY, Posafalvi A, van den Berg MP, Hilfiker-Kleiner D, Bollen IAE, Sliwa K, Alders M, Almomani R, van Langen IM, van der Meer P, Sinke RJ, van der Velden J, van Veldhuisen DJ, van Tintelen JP, Jongbloed JDH. Titin gene mutations are common in families with both peripartum cardiomyopathy and dilated cardiomyopathy. Eur Heart J 2014;35:2165–2173. [DOI] [PubMed] [Google Scholar]
- 339. Ware JS, Li J, Mazaika E, Yasso CM, DeSouza T, Cappola TP, Tsai EJ, Hilfiker-Kleiner D, Kamiya CA, Mazzarotto F, Cook SA, Halder I, Prasad SK, Pisarcik J, Hanley-Yanez K, Alharethi R, Damp J, Hsich E, Elkayam U, Sheppard R, Kealey A, Alexis J, Ramani G, Safirstein J, Boehmer J, Pauly DF, Wittstein IS, Thohan V, Zucker MJ, Liu P, Gorcsan J3rd, McNamara DM, Seidman CE, Seidman JG, Arany Z Imac Investigators I. Shared IMAC-2; and IPAC Investigators . Shared genetic predisposition in peripartum and dilated cardiomyopathies. N Engl J Med 2016;374:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Hoes MF, Bomer N, Ricke-Hoch M, de Jong TV, Arevalo Gomez KF, Pietzsch S, Hilfiker-Kleiner D, van der Meer P. Human iPSC-derived cardiomyocytes of peripartum patients with cardiomyopathy reveal aberrant regulation of lipid metabolism. Circulation 2020;142:2288–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341. Regitz-Zagrosek V, Roos-Hesselink JW, Bauersachs J, Blomstrom-Lundqvist C, Cifkova R, De Bonis M, Iung B, Johnson MR, Kintscher U, Kranke P, Lang IM, Morais J, Pieper PG, Presbitero P, Price S, Rosano GMC, Seeland U, Simoncini T, Swan L, Warnes CA; ESC Scientific Document Group . 2018 ESC Guidelines for the management of cardiovascular diseases during pregnancy. Eur Heart J 2018;39:3165–3241. [DOI] [PubMed] [Google Scholar]
- 342. Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, Dangwal S, Kumarswamy R, Bang C, Holzmann A, Remke J, Caprio M, Jentzsch C, Engelhardt S, Geisendorf S, Glas C, Hofmann TG, Nessling M, Richter K, Schiffer M, Carrier L, Napp LC, Bauersachs J, Chowdhury K, Thum T. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun 2012;3:1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343. Foinquinos A, Batkai S, Genschel C, Viereck J, Rump S, Gyöngyösi M, Traxler D, Riesenhuber M, Spannbauer A, Lukovic D, Weber N, Zlabinger K, Hasimbegovic E, Winkler J, Fiedler J, Dangwal S, Fischer M, Roche J, Wojciechowski D, Kraft T, Garamvölgyi R, Neitzel S, Chatterjee S, Yin X, Bär C, Mayr M, Xiao K, Thum T. Preclinical development of a miR-132 inhibitor for heart failure treatment. Nat Commun 2020;11:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Batkai S, Genschel C, Viereck J, Rump S, Bär C, Borchert T, Traxler D, Riesenhuber M, Spannbauer A, Lukovic D, Zlabinger K, Hašimbegović E, Winkler J, Garamvölgyi R, Neitzel S, Gyöngyösi M, Thum T. CDR132L improves systolic and diastolic function in a large animal model of chronic heart failure. Eur Heart J 2020;42:192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345. Täubel J, Hauke W, Rump S, Viereck J, Batkai S, Poetzsch J, Rode L, Weigt H, Genschel C, Lorch U, Theek C, Levin AA, Bauersachs J, Solomon SD, Thum T. Novel antisense therapy targeting microRNA-132 in patients with heart failure: results of a first-in-human Phase 1b randomized, double-blind, placebo-controlled study. Eur Heart J 2020;42:178–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analysed in support of this consensus document.