Abstract
With the advent of chromatin-interaction maps, chromosome-level genome assemblies have become a reality for a wide range of organisms. Scaffolding quality is, however, difficult to judge. To explore this gap, we generated multiple chromosome-scale genome assemblies of an emerging wild animal model for carcinogenesis, the California sea lion (Zalophus californianus). Short-read assemblies were scaffolded with two independent chromatin interaction mapping data sets (Hi-C and Chicago), and long-read assemblies with three data types (Hi-C, optical maps and 10X linked reads) following the “Vertebrate Genomes Project (VGP)” pipeline. In both approaches, 18 major scaffolds recovered the karyotype (2n = 36), with scaffold N50s of 138 and 147 Mb, respectively. Synteny relationships at the chromosome level with other pinniped genomes (2n = 32–36), ferret (2n = 34), red panda (2n = 36) and domestic dog (2n = 78) were consistent across approaches and recovered known fissions and fusions. Comparative chromosome painting and multicolour chromosome tiling with a panel of 264 genome-integrated single-locus canine bacterial artificial chromosome probes provided independent evaluation of genome organization. Broad-scale discrepancies between the approaches were observed within chromosomes, most commonly in translocations centred around centromeres and telomeres, which were better resolved in the VGP assembly. Genomic and cytological approaches agreed on near-perfect synteny of the X chromosome, and in combination allowed detailed investigation of autosomal rearrangements between dog and sea lion. This study presents high-quality genomes of an emerging cancer model and highlights that even highly fragmented short-read assemblies scaffolded with Hi-C can yield reliable chromosome-level scaffolds suitable for comparative genomic analyses.
Keywords: California sea lion (Zalophus californianus), cancer, Carnivora, chromatin interaction mapping, genome assembly, genome evolution, Hi-C
1 ∣. INTRODUCTION
Chromosomes vary among taxa in number, content and linear organization. Chromosomal organization is subject to evolutionary change induced by structural mutations causing inter- and intrachromosomal rearrangements (Tusso et al., 2019; Weissensteiner et al., 2020; Wellenreuther et al., 2019). Rearrangements can be of relevance to fitness and contribute to evolution (Avelar et al., 2013): they unite or disrupt co-adapted gene complexes (Schwander et al., 2014), modify the recombination landscape affecting the efficiency of selection (Peñalba & Wolf, 2020; Stapley et al., 2017), interact with the epigenetic background (Feng & Riddle, 2020; Shiao, 2015) and, in the case of gene movement between sex chromosomes and autosomes, alter sex-specific gene expression (Emerson et al., 2004). When passed on vertically through the germline, these changes can accumulate over time and shape genome evolution. Yet, structural mutations can also accrue in a subset of somatic cells during the course of a single lifetime, often with deleterious effects to individual fitness. Such deleterious affects were among the first to be seen over 60 years ago, with the discovery of the Philadelphia chromosome (Nowell & Hungerford, 1960), where cancers were associated with numerous somatic genome alterations. Evaluation of over 70,000 human cases across over 75 different types of cancer has identified over 16,900 and 7,100 structural and numerical chromosome abnormalities, respectively (https://mitelmandata-base.isb-cgc.org). Accurate identification of structural recurrent chromosome aberrations in cancers offers a means to advance diagnosis, subclassification, prognosis and even guide treatment selection. Moreover, embracing the One Health concept, a comparative approach to cancers shared across numerous species should provide opportunities to identify genome changes suggestive of an ancestral mechanism of pathogenesis. There is an accumulating body of work identifying shared numerical and/or structural genome changes detected in comparable cancers across species, which suggest that such events may reflect ancestral mechanisms of pathogenesis. This work is most advanced for the domestic dog (e.g., Megquier et al., 2019; Schiffman & Breen, 2015; Shapiro et al., 2015; Thomas et al., 2009).
One group of species that will allow understanding structural genome evolution and structural changes associated with disease in a wild setting are the Carnivora. For example, high rates of chromosome evolution are seen in members of the Canidae (Duke Becker et al., 2011; Yang et al., 1999), Ursidae (Nash et al., 1998) and Mephitidae (Perelman et al., 2008), whereas Feliformes in general show substantial chromosome conservation (Perelman et al., 2012; Rettenberger et al., 1995). To unravel the principles behind structural genomic changes requires the reliable characterization of genomic rearrangements unfolding across evolutionary time, as well as during ontogenetic trajectories of aberrant somatic cells. This goal is greatly facilitated by the generation of new, high-quality chromosome-scale genome assemblies.
With the introduction of recent scaffolding technologies utilizing in vivo chromosome conformation capture (“Hi-C”) (Lieberman-Aiden et al., 2009) or in vitro reconstituted chromatin interaction maps (the “Chicago” method) (Putnam et al., 2016), it is now possible to construct chromosome-scale genome assemblies in essentially any organism of choice—without reliance on difficult-to-obtain linkage maps or the costly and time-consuming generation of bacterial artificial (BAC) libraries (Ekblom & Wolf, 2014; Peichel et al., 2017; Waterhouse et al., 2020). Originally conceived to investigate the three-dimensional architecture of genomes (Burton et al., 2013; Kaplan & Dekker, 2013; Marie-Nelly et al., 2014), Hi-C mapping uses chromosome interactions to gain information on long-range contiguity. It builds on the principle that even at distances of several hundred megabases (Mb), intrachromosome interactions are more common than interactions between different chromosomes (Lieberman-Aiden et al., 2009). The related “Chicago” method uses in vitro reconstituted chromatin outside of its native, cellular context (Putnam et al., 2016). Both approaches open the opportunity to investigate the mechanisms underlying genome rearrangements across large evolutionary timescales (e.g. Gemmell et al., 2020; Strijk et al., 2019). As we move towards de novo assembly of genomes at a population scale, as opposed to read mapping (Chaisson et al., 2015; Tusso et al., 2019), individual-level, highly contiguous genome assemblies could soon be the norm. Despite this potential, information on the accuracy and reproducibility of chromosome reconstruction based on chromatin interaction is essentially lacking.
The goals of this study are two-fold. First, we present two annotated, chromosome-level genome assemblies of the California sea lion, Zalophus californianus (for taxonomic considerations see Lopes et al., 2021; Wolf et al., 2007). The California sea lion is a carnivoran species that is attracting increasing interest as a wild animal model to understand the genetic and environmental interactions involved in carcinogenesis (Browning et al., 2015; Buckles et al., 2007) and other diseases (Neely et al., ,2015, 2018). The lack of a high-quality genome assembly has impeded progress in this area. Second, we use these two assemblies to compare the robustness of syntenic inference between Illumina short-read vs. Pacbio long-read primary assemblies and different scaffolding technologies (Hi-C, Chicago, 10X Genomics and Bionano optical mapping data) combining bioinformatic and cytogenetic methods. We then expand genome comparisons to chromosome-level Hi-C scaffolded assemblies from three additional pinniped species (2n = 32–36), and several outgroup species from other families within the Caniformia including ferret (2n = 34), red panda (2n = 36) and domestic dog (2n = 78), comprising 45 million years of evolution. In summary, this study adds annotated high-quality genomes of the California sea lion, reveals technical aspects of syntenic inference and provides biological insight into chromomsome evolution within Carnivora.
2 ∣. MATERIAL AND METHODS
2.1 ∣. Assembly and annotation of the California sea lion (Zalophus californianus) genome
We constructed two different types of assemblies: (a) based on short-read (SR) shotgun sequencing with Illumina technology (SRassembly); and (b) based on long reads (SMRT sequencing, Pacific Biosciences) and independent scaffolding data (10X genomics, BioNano optical maps and Hi-C) following the pipeline of the Vertebrate Genome Project (VGPassembly).
2.1.1. ∣. SRassembly and annotation
The primary SRassembly (SRassembly.v0, ZalCal_v1_BIUU-GCA_004024565.1) was constructed as described in Zoonomia Consortium (2020), from 250-bp paired-end shotgun sequencing data and assembled using discovar de novo version 52488. Subsequently, this primary assembly was scaffolded to high contiguity using Dovetail Chicago in vitro proximity ligation (SRassembly.v1), with two Chicago libraries prepared from the same sample (SAMN07678053) as described previously (Putnam et al., 2016). Briefly, for each library, ~500 ng of high-molecular-weight genomic DNA was reconstituted into chromatin in vitro and fixed with formaldehyde. Fixed chromatin was digested with DpnII, and the 5′ overhangs were filled in with biotinylated nucleotides. After ligation of the free blunt ends, crosslinks were reversed, the DNA was purified from protein and terminal biotinylated nucleotides were removed with exonucleases. After shearing to a mean fragment size of ~350 bp, the DNA was subjected to library preparation using NEBNext Ultra enzymes and Illumina-compatible adapters. Biotin-containing fragments were isolated using streptavidin beads before PCR enrichment of each library. The libraries were sequenced on an Illumina HiSeq X machine (rapid run mode). The number and length of read pairs produced for each library was: 191 million, 2 × 101 bp for library 1; and 127 million, 2 × 101 bp for library 2. Together, these Chicago library reads provided 74.67× physical coverage of the genome with insert sizes of 1–100 kb. The input de novo assembly, shotgun reads generated both for the primary assembly and the Chicago libraries were used as input data for hirise version 1.3.0-72-gcd4fb8a, a software pipeline designed specifically for using proximity ligation data to scaffold genome assemblies (Putnam et al., 2016). Both the shotgun and Chicago library sequences were aligned to the draft input assembly using a modified snap read mapper (http://snap.cs.berkeley.edu). The location of Chicago read pairs mapped within draft scaffolds was then analysed by hirise estimating genomic distance between read pairs in a likelihood framework. This likelihood model was used to identify and break putative misjoins of the primary assembly and to establish new joins. After scaffolding, shotgun sequences were used to close gaps between contigs resulting in the final version of SRassembly.v1 (as used in Peart et al., 2020; 10.5281/zenodo.3741488).
The SRassembly.v1 assembly was then further scaffolded to chromosomal level with Hi-C. To do so, three Hi-C libraries were prepared from blood by dovetail in a similar manner as described previously (Lieberman-Aiden et al., 2009). Due to lack of material from the genome individual the blood sample was from a different female individual recovered from the same geographical area (ZCA 13399, Biosample SAMEA5145493). For each library, chromatin was cross-linked with formaldehyde in the nucleus prior to DNA extraction. The remaining steps, from DNA extraction, chromatin digestion to sequencing, were identical to the preparation of the Chicago libraries described above. The number and length of read pairs produced for each library was: 151 million, 2 × 151 bp for library 1; 126 million, 2 × 151 bp for library 2; and 151 million, 2 × 151 bp for library 3. Together, these Hi-C library reads provided 17,218.17×physical coverage of the genome with insert sizes of 10–10,000 kb. Hi-C libraries were then used for scaffolding following an iterative approach. The draft SRassembly.v1 assembly was used as input for alignment of read-pairs generated from the Hi-C libraries, and scaffolded with hirise version 2.1.5-a028029ddb34; shotgun sequences generated for the primary assembly were used to close gaps between contigs. To guarantee equal coverage between autosomes and sex chromosomes, both individuals used for the assembly were female, representing the homogametic sex in pinnipeds. The final, scaffolded assembly (SRassembly.v2, NCBI acronym zalCal 2.2) has been deposited in NCBI's Database at https://www.ncbi.nlm.nih.gov/ under GenBank accession no. GCF_900631625.1.
SRassembly.v2 was annotated using the NCBI Eukaryotic Genome Annotation Pipeline (https://www.ncbi.nlm.nih.gov/genome/annotation_euk/process/). The annotation included evidence-based information from RNAseq data that were generated from 10 tissues plus a pool from six different brain regions (biosamples SAMN10285328–SAMN10285338) from a single juvenile male (named Ensign; Marine Mammal Center CSL-13825). Accession numbers and further details on sequencing are provided in Table S1. The number of tissue-specific transcripts was subsequently assessed by mapping to the reference genome using star with default parameters (Dobin et al., 2013). The annotation was released in NCBI Zalophus californianus Annotation Release 100.
To test for the effect of possible limitation of Hi-C data or library complexity, we also compared SRassembly.v2 to an additional assembly of the California sea lion constructed by the DNAzoo team using SRassembly.v2 as input and another round of Hi-C scaffolding with independent Hi-C libraries using the 3D-DNA pipeline (Dudchenko et al., 2017) and juicebox Assembly Tools (Dudchenko et al., 2018). This assembly (SRassembly.v3) is available at https://www.dnazoo.org/assemblies.
2.1.2 ∣. VGPassembly
We constructed a second high-quality Zalophus californianus genome assembly following the version 1.6 pipeline of the Vertebrate Genome Project for which we generated four data sets: Pacbio single-molecule continuous long reads (CLR), 10X Genomics linked-read sequencing, Bionano optical mapping and Arima Hi-C. In brief, DNA was extracted from 400 μl of whole blood sample from a single sea lion male (biosample SAMN12368149) using the Bionano SP kit (#80030) yielding a total of 17.88 μg of ultra-high-molecular weight (uHMW) DNA. We then sheared the DNA using a 26G blunt end needle (Pacbio protocol PN 101-181-000 Version 05) to 20- to 50-kb fragments. We used 8 μg of fragmented DNA to prepare a large-insert Pacbio library using the Pacific Biosciences Express Template Prep Kit version 2.0 (#100-938–900) following the manufacturer's protocol and subjected it to size selection (>20 kb) using the Sage Science BluePippin Size-Selection System. The final PacBio Library was sequenced on three PacBio 8 M (#101-820–200) smrtcells on the Sequel instrument with the sequencing kit 2.0 (#101-820–200) using the Binding Kit 2.0 (#101-842–900) and 15 h movie. A total of 253.67 Gb of raw PacBio data were generated (insert N50 ~36 kb). Unfragmented uHMW DNA was also used to generate a linked-reads library on the 10X Genomics Chromium (Genome Library Kit & Gel Bead Kit version 2 PN-120258, Genome Chip Kit version 2 PN-120257, i7 Multiplex Kit PN-120262). We sequenced this 10X library on an Illumina Novaseq S4 150-bp paired-end lane (~60× coverage). The same uHMW DNA were labelled for Bionano Genomics optical mapping using the Bionano Prep Direct Label and Stain (DLS) Protocol (30206E) and run on one Saphyr instrument chip flow-cell. We generated 323.37 Gb of data with read length ≥150 kb to 0.3787 Mb (read length N50 = 378.7 kb). These optical reads were assembled into a Consensus Map (CMAP) and we obtained a total of 82 maps (N50 = 110.8 Mb) and 2.5 Gb total length. Hi-C data were generated by Arima Genomics (https://arimagenomics.com/) using an Arima-HiC kit (P/N: A510008). Proximally ligated DNA was sheared and size-selected at ~200–600 bp using SPRI beads. Enriched biotin-labelled proximity-ligated DNA was prepared into an Illumina library (KAPA Hyper Prep kit; P/N: 51KK8504). The final library was sequenced on an Illumina HiSeq X at ~60× coverage following the manufacturer's protocols.
Pacific Biosciences CLR data were used to generate haplotype-phased contigs, and 10X linked-read sequencing, Bioano optical mapping and Arima Hi-C scaffolding were sequentially used to scaffold the contigs (Rhie et al., 2020). The resulting assembly was manually curated. This included removing microbial contaminations, analysing the concordance of the raw data and assembly in geval (Chow et al., 2016) and correcting the encountered errors (process described in Howe et al., 2020; Rhie et al., 2020), and using the Hi-C juicer maps to the assembly and prior karyotyping to assign chromosomes. The curation involved breaking 18 misjoins, adding 26 missed joins and removing three instances of false duplications (4.2 Mb sequence total), increasing the scaffold N50 from 141 to 147 Mb and assigning 99.93% of the assembled sequence to 17 autosomes plus X and Y.
The primary pseudohaplotype of the diploid assembly is available at https://www.ncbi.nlm.nih.gov/ under accession no. GCA_009762305.1 and forms the basis of the between-species alignments in this study. A subsequent assembly with improved consensus quality is available as GCA_009762305.2 and was used for the remainder of the analyses. The assembly of the alternative pseudohaplotype contigs are available under accession nos. GCA_009762295.1 and GCA_009762295.2.
busco version 3 was used to benchmark assembly quality in the protein coding regions using the universal single-copy orthologue set from mammals (odb9) (Simão et al., 2015). The annotation was generated using the same procedure as above for SRassembly.v2 and was released in NCBI Zalophus californianus as Annotation Release 101.
2.2 ∣. Synteny between genomes
Synteny comparisons were made between our California sea lion assemblies and chromosomal-level genome assemblies of species within the mammalian order Carnivora, suborder Caniformia, of different phylogenetic distance from the California sea lion. Outside of the pinnipeds we included the sole representative of the Ailuridae (red panda, Ailurus fulgens available at DNAzoo https://www.dnazoo.org/assemblies/Ailurus_fulgens; Hu et al., 2017), a representative of the Mustelidae (ferret, Mustela putorius furo, available at DNAzoo https://www.dnazoo.org/assemblies/Mustela_putorius_furo; Peng et al., 2014) and the Canidae (domestic dog, Canis lupus familiaris, canFam3: GenBank accession no. GCF_000002285.3).
Within pinnipeds, genomes scaffolded to (pseudo)chromosome level using Hi-C were available for at least one member from the three major families: Odobenidae, Phocidae and Otariidae. This included the only extant member of the Odobenidae, the walrus Odobenus rosmarus (available at DNAzoo https://www.dnazoo.org/assemblies/Odobenus_rosmarus; Foote et al., 2015), and the northern elephant seal Mirounga angustirostris (available at DNAzoo https://www.dnazoo.org/assemblies/Mirounga_angustirostris), a member of the Phocidae. The family Otariidae was represented by the California sea lion of this study and an improved assembly of the Antarctic fur seal (Arctocephalus gazella) (Humble et al., 2018). To increase contiguity, we scaffolded the Antarctic fur seal genome with Hi-C data using assembly version 1.4 (Humble et al., 2018) as the input assembly; the original assembly was constructed from a combination of short read, mate-pair sequencing using the Illumina platform improved with Pacific Biosciences CLR reads. Three Hi-C libraries were prepared from Biosample (SAMEA4666125) and sequenced by dovetail in the same manner as described above for the California sea lion resulting in 180 million, 179 million and 136 million reads, respectively. Together, these dovetail Hi-C library reads provided 2,619× physical coverage of the genome with insert sizes of 10–10,000-kb. All Hi-C libraries were aligned and scaffolded with hirise (Putnam et al., 2016) using the same procedure as that for the California sea lion described above. After scaffolding, shotgun sequences were used to close gaps between contigs. The resulting scaffolded genome assembly is available at https://www.ncbi.nlm.nih.gov/ under accession no. GCA_900642305.1.
To assess synteny, we performed pairwise alignment of the above-mentioned genomes using the pipeline implemented in the UCSC genome browser (Kent et al., 2002). Scripts and utilities for performing genome alignments were downloaded from http://genomewiki.cse.ucsc.edu/index.php/DoBlastzChainNet.pl and locally installed. The pipeline from UCSC uses lastz (Harris, 2007) as an aligner, and downstream netting and chaining of alignments are performed by various scripts in the pipeline. The pipeline uses different parameter sets depending on the evolutionary distance. For all within-pinniped alignments we used the parameter set “human to other primates,” and for the outgroups “human to other mammals.” Prior to alignment, each assembly was repeat-masked by first performing de novo prediction of repeats specific to the California sea lion using repeatmodeler (version 1.0.8 http://www.repeatmasker.org/RepeatModeler.html). These repeats were combined with repeats present in the Carnivora library, and masking was performed using repeatmasker (version 4.0.6 http://www.repeatmasker.org/). The alignments were visualized using circos plots (Krzywinski et al., 2009), and only alignments where both template and query comprised at least 1,000 bp were used. Contigs of <1 Mb were excluded from subsequent analysis. Circos also provides a set of tools which convert the chain file coordinates to links which can be plotted. The bundlelinks utility from circos tools was used with the parameter -min_bundle_membership 3. The BAC coordinates were lifted over using the liftOver utility (Hinrichs, 2006). The chain files needed for liftover were also generated by the DoBlastzChainNet.pl pipeline (http://genomewiki.cse.ucsc.edu/index.php/DoBlastzChainNet.pl) and were filtered to remove spans in query and template which were <100 kb avoiding spurious matches. The Rbest chains were also produced and then filtered using the bundlelinks utility from circos tools with the parameters -max_gap 500000 -min_bundle_size 250000 -min_bundle_membership 3. SRassembly.v2 and SRassembly.v3 were both aligned separately to the VGPassembly using mummer 4.0 (Kurtz et al., 2004) using the nucmer pipeline with a variety of different parameters, which produced concordant results.
2.3 ∣. Chromosome painting, validation and cross-referencing to the genome of the domestic dog
As an initial “synteny guide” we developed whole chromosome paint probes for the California sea lion and used these to identify large evolutionarily conserved chromosome segments (ECCS) shared with the domestic dog. To refine and orient each ECCS and additional unpainted segments in the karyotype of the California sea lion, we then used a panel of over 250 canine bacterial artificial chromosome (BAC) clones, each with defined physical locations in the canine genome assembly, to perform multicolour single-locus probe tiling as described below.
Cells were collected from a female California sea lion after its death while in veterinary care at “The Marine Mammal Center,” under MMPA permit no. 18786. A primary cell culture of female sea lion kidney cells was established and cultured in MEM (Mediatech) supplemented with L-glutamine. Cells were subcultured to 50% confluency, grown for 24 h prior to addition of 50 ng ml−1 demecolcine (Sigma) for 16 h, and then harvested. Following mitotic shake-off, cells were recovered, centrifuged at 289 g for 5 min, resuspended in hypotonic solution (75 mM KCl, 10 mM MgSO4, 0.2 mM spermine, 0.5 mM spermidine, pH 8.0), and incubated at room temperature for 30 min. The suspension of swollen cells was centrifuged at 289 g for 5 min, the cell pellet resuspended in 1.5 ml of ice-cold polyamine isolation buffer (PAB, containing 15 mM Tris, 2 mM EDTA, 0.5 mM EGTA, 80 mM KCl, 3 mM dithiothreitol, 0.25% Triton X-100, 0.2 mM spermine, 0.5 mM spermidine, pH 7.50), and vortexed for 30 s to disrupt cell membranes. Five microlitres of the resulting chromosome suspension was stained with propidium iodide to assess the extent of cell lysis. The chromosome suspension was centrifuged gently (201 g, 2 min) to pellet large material/debris, and the supernatant containing single chromosomes was filtered through a 20-μm mesh filter (Celltrics, Partec). Chromosomes were stained overnight with 5 μg ml−1 of Hoechst (Sigma), 40 μg ml−1 of Chromomycin A3 (Sigma), and 10 mM MgSO4. In addition, 10 mM sodium citrate and 25 mM sodium sulphite were added to the stained suspension and left overnight before flow analysis and sorting. Subsequently, the stained chromosome suspension was flow sorted on a MoFlo cell sorter (Beckman Coulter) with lasers and optics set up as described previously (Ng & Carter, 2006; Ng et al., 2007). Chromosomes were isolated on a bivariate plot of Hoechst fluorescence vs. Chromomycin fluorescence. For each peak, 500 chromosomes were collected into sterile 500-μl Eppendorf tubes containing 33 μl of sterile UV-treated distilled water.
Each of the 15 discrete sorted pools was subject to routine DNA isolation, amplification of the DNA using the GenomiPhi DNA Amplification kit (GE Healthcare), and then labelling with one of five spectrally resolvable fluorophore-conjugated dNTPs. The resulting probes were hybridized in groups to metaphase preparations of several California sea lions to verify their chromosomal content. To obtain positional information of the ECCS between species, we proceeded as follows. In a previous study, a panel of 264 canine BAC clones from the CHORI-82 library (https://bacpacresources.org/library.php?id=253) had been integrated into the canine genome (CanFam2.0) at 10-Mb intervals and their cytogenetic location was determined by multicolour chromosome tiling (Thomas et al., 2007). DNA was isolated from each of these canine BAC clones using routine methods, grouped into sets of five adjacent clones (spaced at 10-Mb intervals in the canine genome) and labelled with one of five spectrally resolvable fluorophores, as described previously (Thomas et al., 2007, 2008). Each group of five BAC clones was then hybridized to metaphase preparations of the California sea lion and imaged as described previously (Thomas et al., 2007, 2008) to determine their physical chromosomal location and to orient each ECCS. All multicolour fluorescence in situ hybridization (FISH) images were acquired using an Olympus BX61 semi-automated microscope equipped with series of zero-shift, narrow pass fluorescence filters, driven by smartcapture version 3.0 (Digital Scientific).
3 ∣. RESULTS AND DISCUSSION
3.1 ∣. Genome assemblies
The primary short-read assembly of a female California sea lion (CSL) sample resulted in 47,532 contigs totaling 2.520 Gb in length (SRassembly.v0, Table 1). With a contig N50 of 132 kb it was highly fragmented, but contained the vast majority of single-copy genes (3,547) in the mammalian busco core gene set (Table 2). Scaffolding with the Chicago method (SRassembly.v1) and additional Hi-C chromosome conformation capture (SRassembly.v2) improved gene model completeness only slightly from 3,843 to 3,854 genes. As expected, scaffold N50 contiguity increased from 12.6 to 138.14 Mb, respectively. Summary statistics were only marginally affected when SRassembly.v2 was compared to the DNAzoo assembly (SRassembly. v3, Tables 1 and 2) suggesting that a standard, single Hi-C library was sufficient with regard to scaffold sizes and gene model completeness. Comparable to other recently published pinniped genome assemblies ranging from ~2.3 to 2.5 Gb (Mohr et al., 2017; Park et al., 2018) the final assembly (SRassembly.v2) had a total ungapped length of 2.37 Gb, contained in 10,421 scaffolds larger than 1 kb (thus excluding contigs that had not been scaffolded at all). The number of scaffolds spanning more than 90% of the genome (L90) was 17, which compares well with the 18 chromosomes expected from the karyotype (Figure 1a; Figure S1). Functional annotation including primary evidence of RNA-sequencing (RNA-seq) data from 11 tissues (Table S1)resulted in gene models for 19,617 protein-coding genes (Table S2). Testis provided the largest number of transcripts overall and the greatest number of tissue-specific transcripts (Table S3).
TABLE 1.
Assembly statistics for successive improvements of the Zalophus californianus genome assembly
| Assembly versiona | SRassembly.v0 | SRassembly.v1 | SRassembly.v2 | SRassembly.v3 | VGPassembly (RefSeq) |
|---|---|---|---|---|---|
| GenBank accession no. | GCA_004024565.1 | — | GCA_900631625.1 | Available at https://www.dnazoo.org/assemblies) | GCA_009762305.2 |
| GenBank annotation | Release 100c | Release 101d | |||
| Number of scaffolds >1 kb | 47,532 | 11,428 | 10,421 | 10,444 | 42 |
| Scaffold N90b | 0.017 (22,718) | 0.848 (301) | 59.71 (17) | 91.40 (15) | 95.46 (14) |
| Scaffold N50b | 0.132 (5284) | 12.6 (49) | 138.14 (8) | 143.40 (7) | 146.93 (7) |
| Longest scaffold (Mb) | 1.48 | 72.15 | 212.60 | 212.59 | 216.12 |
| Total (gapped) size (Mb) | 2,519.86 | 2,524.24 | 2,524.65 | 2,372.42 | 2,408.66 |
| Total size scaffolds ≥1 kb (Mb) | 2,368.57 | 2,371.92 | 2,372.34 | 2,372.42 | 2,408.66 |
| Number of gaps | 10,284 | 40,168 | 47,448 | 47,547 | 122 |
SR: short-read; LR: long-read.
Size in Mb (number of scaffolds).
TABLE 2.
Counts of Benchmarking Universal Single-Copy Ortholog (BUSCO) genes using the mammalia odb9 data set for the different assemblies of California sea lion (Zalophus californianus) and the species used as outgroups in Figure 2
| Species | Assembly version | Single | Duplicated | Fragmented | Missing | Total |
|---|---|---|---|---|---|---|
| Zalophus californianus | SRassembly.v0 | 3,547 | 69 | 318 | 170 | 4,104 |
| Zalophus californianus | SRassembly.v1 | 3,843 | 68 | 95 | 98 | 4,104 |
| Zalophus californianus | SRassembly.v2 | 3,854 | 62 | 91 | 97 | 4,104 |
| Zalophus californianus | SRassembly.v3 | 3,836 | 63 | 101 | 104 | 4,104 |
| Zalophus californianus | VGPassembly | 3,828 | 65 | 99 | 112 | 4,104 |
| Arctocephalus gazella | ArcGazvl.5 | 3,605 | 28 | 274 | 197 | 4,104 |
| Odobenus rosmarus | 3,855 | 59 | 88 | 102 | 4,104 | |
| Mirounga angustirostris | 3,844 | 40 | 118 | 102 | 4,104 | |
| Ailurus fulgens | 3,885 | 34 | 94 | 91 | 4,104 | |
| Mustela putorius furo | 3,877 | 27 | 108 | 92 | 4,104 |
FIGURE 1.
Chromosomal painting and identification of chromosomes of the California sea lion (CSL). (a) DAPI-banded ideogram of the chromosomes of a male California sea lion, diplaying 17 pairs of autosomes and the sex chromosomes. (b) Bivariate flow karyotype of a female sea lion, showing 15 distinct peaks (labelled A–O), with 13 peaks (labelled A–G, I–K, M–O) representing single autosomes and two peaks (H and L) each representing two autosomes. (c) Example of a five-colour FISH analysis of DNA purified from CSL chromosome sort peaks J (yellow), L (green), M (aqua), N (red) and O (magenta) hybridized to DAPI stained CSL chromosomes. (d) Data from C with DAPI stain inverted to reveal the DAPI banding used for chromosome identification. (e) Six painted chromosome pairs from (d) aligned and identified as ZCA 12 (peak J), ZCA 15 + 16 (peak L), ZCA 13 (peak M), ZCA 14 (peak N) and ZCA 17 (peak O) using DAPI banding
Consistent with other recent genomes generated with the Vertebrate Genome Project pipeline (Rhie et al., 2020), the California sea lion VGPassembly resulted in a less fragmented primary assembly with a contig N50 of 24.59 Mb, and scaffold N50 of 129.41 Mb. The ungapped assembly size was 2.39 Gb, comparable to the SRassemblies (see above), but busco gene content statistics were slightly worse (3,828 single-copy genes; Tables 1 and 2), presumably due to low-level frameshift errors. With 41.40%, the assembly had similar repeat content to the SRassembly.v2. (43.26%) of which 21.80% was identified as long interspersed nuclear elements (26.69% in SRassembly.v2; Table S4). The VGPassembly was constructed from a male individual allowing for assembly of the Y-chromosome with an ungapped length of 4,004,775 bp, of which 2,200,796 bp was identified as repetitive. In total, 42% of the Y assembly was covered by uniquely mapping RNA-seq reads (from Table S1) at a depth above 10. Even though the busco identified genes were fewer, annotation of the VGPassembly resulted in 21,397 protein-coding genes (Table S4), 1,780 more than in the SRassembly.v2 (Table S3).
3.2 ∣. Comparison of California sea lion assemblies
Despite strongly differing primary assemblies, the final scaffolds (representing chromosomes 1-17 and X) of both SRassembly.v2 and VGPassembly had a similar total length scaffold size distribution (Table 1), and both assemblies showed near-complete collinearity (Figure S2 also including SRassembly.v3). Hi-C-based scaffolding may thus be suitable for broad-scale syntenic comparisons even when the primary assembly is highly fragmented. However, at a local level, many differences were present between the assemblies (Figure S3). A small number of regions were assembled on different scaffolds representing different chromosomes in both SRassembly. v2 and SRassembly.v3 compared to VGPassembly, and all were located towards the ends of chromosomes, where telomeres would be expected. Smaller scaffolds from the short-read assemblies that could be placed within the longer scaffolds of the VGPassembly (matching >10,000 bp) differed only slightly among assemblies with six matches in SRassembly.v2 and four in SRassembly.v3 (Figure S3) When SRassembly.v2 and SRassembly.v3 are compared to the VGPassembly, SRassembly.v2 has the most differences in both contig ordering and orientation with the total length of misassembled blocks also larger in SRassembly.v2. These inconsistencies between the SRassemblies are probably due to a combination of differences in the Hi-C library and the different bioinformatic approaches. Overall, however, differences between all three Hi-C scaffolded assemblies (SRassembly.v2, SRassembly.v3; VGPassembly) were minor.
3.3 ∣. In silico inference of synteny between Hi-C scaffolded assemblies across pinnipeds
Summary statistics on genome contiguity, gene and repeat content were comparable between the California sea lion assemblies, the additional pinniped Hi-C scaffolded assemblies (Antarctic fur seal, walrus and northern elephant seal) and the additional carnivore outgroups (ferret and red panda) (see Tables 1 and 2; Tables S2 and S4-S10).
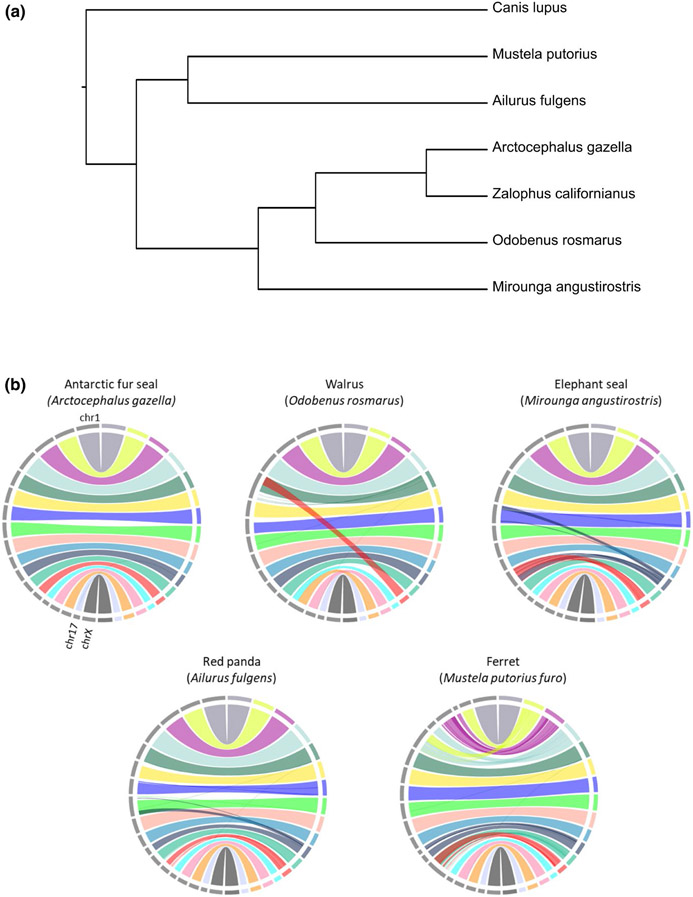
Consistent with expectations, synteny between the California sea lion and Antarctic fur seal, which have an identical karyotype (2n = 36) and diverged only ~5.4 million years ago (Nyakatura & Bininda-Emonds, 2012), was near-complete with the exception of two small interchromosome translocations (Figure 2). Whole genome alignments to the remaining four species were largely concordant and corresponded to the expected karyotypic shifts from cytological studies (Árnason, 1974; Cavagna et al., 2000; Nie et al., 2002) (VGPassembly, Figure 2; SRassembly.v2, Figure S4). The walrus alignments (Figure 2; Figures S4 and S5) recapitulated the known reduction in the number of chromosomes from 2n = 36 in otariids to 2n = 32 (Fay et al., 1967). Based on an ancestral carnivore karyotype, inferred to be 2n = 38, and thus more similar to the otariids (Beklemisheva et al., 2016), this is consistent with two fusion events in the lineage leading to the walrus. A fusion event different from transition to walrus was seen in the lineage leading to the elephant seal with a karyotype of 2n = 34 (Árnason, 1974). This may be accompanied by further translocation events from sea lion chromosome 11 (ZCA 11) to chromosome 7 of the elephant seal, although these remain to be investigated. There was also a large degree of chromosome-level conservation between the otariids and other members of the Caniformia, red panda and ferret (Figure 2). This includes near total synteny to the red panda with the potential translocation of similar regions to those seen in the elephant seal. The differences between the sea lion and ferret were characterized by multiple fission/fusion events rather than large-scale translocations. These results stand in contrast to the high level of chromosomal rearrangement seen in the dog.
FIGURE 2.
Chromosomal synteny. (a) Phylogeny showing the relationships between the taxa that were aligned to the California sea lion (edited from timetree. org; Kumar et al., 2017). (b) Circos plots showing the alignment of the California sea lion genome (VGPassembly, left side) to other species (right side). California sea lion chromosomes (ZCA 1–17, ZCA X) are shown in colour, while chromosomes of the other species are depicted with grey bars
Overall, these results attest to the suitability of chromatin interaction mapping for inferring chromosomal organization from even highly fragmented primary assemblies. Interchromosomal rearrangements were only inferred between distantly related species and were consistent with known karyotypic transitions. Intrachromosomal rearrangements were rare and sensitive to filtering both between different assemblies of the California sea lions and the Antarctic fur seal. Note, however, that our design did notallow differentiation between the effect of individual and technical variation, a topic that warrants further study.
3.4 ∣. Comparison of in silico inference and comparative cytogenetic map to dog
For validation and guarding against possible systematic biases of the scaffolding technology permeating all Hi-C-based assemblies (Bickhart et al., 2017), we cross-validated the alignment-based syntenic inference with cytogenetic evidence using the domestic dog for comparison. While otariids (2n = 36) closely resemble the ancestral carnivore karyotype, the dog has a highly rearranged karyotype with the highest diploid chromosome number in the Carnivora (2n = 78) (e.g., Breen et al., 1999; Selden et al., 1975).
Flow sorting allowed us to isolate most chromosomes and develop chromosome-specific probes for the California sea lion (Figure 1a). Probes were then hybridized in groups to metaphase preparations of California sea lions to verify their chromosomal content (Figure 1c-e). The sea lion paint probes were then hybridized to metaphase chromosomes of the domestic dog to identify the ECCS to within 5- to 10-Mb resolution. To refine and orient each ECCS in the karyotype of the California sea lion, we then hybridized a panel of 255 canine BAC clones, each with defined physical locations in the canine genome assembly (CanFam3.1.), to the chromosomes of the California sea lion by multicolour FISH. The sequences of 228 (86%), 225 (85%) and 229 (87%) clones could be lifted over into SRassembly.v2, SRassembly.v3 and VGPassembly, respectively (Table 3). Comparative FISH analysis indicated that the BACs (n = 14) from dog chromosome X (CFA X) were found solely on California sea lion chromosome X (ZCA X) in all assemblies. Moreover, judging by the subset of four (SRassembly.v2/v3) and five clones (VGPassembly) that could be lifted over with high confidence, probe order from the dog was maintained in the sea lion. This high level of synteny suggests that the X chromosome can be accurately aligned across large evolutionary distances in Carnivora (e.g., Liu et al., 2019; Ross et al., 2005) and corroborates previous findings of a highly conserved gene order on the X chromosome across a range of placental mammals (Murphy et al., ,1999, 2005; Raudsepp et al., 2004; Rodriguez Delgado et al., 2009); with notable deviations in some groups such as rodents (Romanenko et al., 2020) or cetartiodactyla (Proskuryakova et al., 2017). This provides a promising outlook for studies on the collocation of genomic elements on the X chromosome, small-scale rearrangements and the effect of different species-specific traits on its evolution (Emerson et al., 2004).
TABLE 3.
Comparison of the locations of the BACs in chromosome-scale assemblies of Zalophus californianus compared to the FISH analysis
| Assembly version | SRassembly. v2 |
SRassembly. v3 |
VGPassembly |
|---|---|---|---|
| Total BACs lifted | 204 | 202 | 205 |
| BACs deleted in assembly | 1 | 1 | 1 |
| BACs split in assembly | 9 | 9 | 9 |
| BACs partial in assembly | 24 | 26 | 23 |
| Assembled on another chromosome | 6 | 5 | 5 |
| FISH centromere-assembly telomere | 3 | 1 | 1 |
| FISH telomere-moved to centromere | 0 | 0 | 0 |
| Location moved within chromosome arm | 5 | 3 | 3 |
Note: Congruence among assemblies probably suggests errors in cytogenetic inference or systematic errors common to all.
In contrast, autosomes showed a substantial degree of rearrangement between sea lion and dog including both inter- as well as intrachromosomal rearrangements. Results from comparative FISH analyses were broadly concordant with inference from whole genome alignments (Figures S6-S8). To exemplify the procedure and complexity of comparing the whole genome alignments to the comparative FISH analyses, we consider here in detail chromosome 6 of the California sea lion. FISH-based chromosome painting of ZCA 6 (Figure 3a, chromosome model to the right) supports synteny blocks with three dog chromosomes, CFA 1, CFA 12 and CFA 35. These were also the only dog chromosomes found to align with the scaffold corresponding to this chromosome (VGPassembly: Figure 3a, circos plot and chromosome model to the left; SRassembly.v2 &.v3: Figures S6-S8). Within these blocks (alignment chains that bundle together), chromosomal synteny was less well resolved, as alignments overlapped in all assemblies in a similar fashion (Figures S6-S8). For example, the sequence from CFA 35 aligned to both ends of the chromosome rich in repetitive sequences. The sequence from CFA 1 had a small alignment where expected from the FISH, but also a larger alignment overlapping with that from CFA 35, highlighting the challenge of syntenic inference by whole genome alignment alone. No reciprocal best chains passed filtering/bundling, which would allow confident inference of orthology for a large section of the q-arm (Figure 3a, overlapping colours in left chromosome model). In addition, there were differences at the resolution level of single BACs (using liftover locations). For instance, BAC 122I05 (from CFA 12) found on the p-arm of ZCA 6 was lifted over to the q-arm of ZCA 6 between two BACs from CFA 1, 283L04 and 326P14, in all three assembly versions. These type of inconsistencies have been seen before with VGP assemblies (Rhie et al., 2020), where more often multiple long-read and scaffolding data sets support the VGP assembly, possibly indicating erroneous placement during cytological inference, or point at alignment errors.
FIGURE 3.
Schematic showing the synteny between the California sea lion and domestic dog (CanFam3.1). (a) Left: Circos plot showing alignment-based syntenic relationships of ZCA 6 (grey bar; VGPassembly) with all 39 dog chromosomes (numbered clockwise) depicted in colour. Regions with vertical patterns show alignment overlaps. Synteny blocks are depicted in the same colour scheme as the chromosome model. Right: four inverted DAPI-banded images of sea lion chromosome 6 (ZCA 6), showing the physical location of 14 clones from the CH82 canine BAC library (which map to dog chromosomes CFA 35, 12 and 1) ordered from the distal end of ZCA 6p to the distal end of ZCA 6q. Raw data underlying synthetic reconstruction from FISH analyses are shown on sea lion metaphase chromosomes with independent colours. (b) Synteny inferred from alignment (left chromosome model) and reciprocal chromosome painting corresponding to the FISH analyses (right chromosome model) depicted with the same colour scheme as the chromosome models in (a) for the entire California karyotype with the VGPassembly alignment generated from reciprocal best chains and bundle size of 250 kb, allowing gaps up to 500 kb and at least three links in a bundle. (c) Similar results as in (b) for SRassembly.v2
3.4.1 ∣. Chromosomal synteny
Considering the classic definition of synteny restricted to chromosomal identity, but not sequence order, we found full chromosome-level synteny for eight of the 17 chromosomes (using the reciprocal best chains). For ZCA 1, 3, 4, 6, 10, 11, 12 and 14 the same dog chromosomes were identified as syntenic between the FISH analysis and the whole genome alignments of dog with all three California sea lion assemblies. An additional five chromosomes (ZCA 2, 8, 9, 13 and 17) showed near-perfect synteny including all chromosomes from the FISH analysis plus small alignments to a further chromosome. These additional alignments were supported by all assemblies except for ZCA 17 where alignments differed slightly (Figures S6-S8). These additional chromosomes may either represent spurious matches or real inclusion of small fragments not captured at the level of BAC resolution.
For the remaining four chromosomes (ZCA 5, 7, 15, 16), synteny to dog was difficult to establish. The p-arm of ZCA 5 showed a repetitive signal in the FISH analyses and was not assembled in the scaffold that contained the q-arm in SRassembly.v2 and v3 (Figure 3c; Figure S9). One BAC attributed to ZCA 5p was assembled on ZCA 15 in SRassembly.v2 (Table 3). In the VGPassembly, however, the p-arm was at least partially assembled (Figure 3b). Within the q-arm of ZCA SRassembly.v2 was syntenic whereas SRassembly.v3 and VGPassembly also contained additional matches to further chromosomes. Overall, among all chromosomes there was consistency in the California sea lion short- (SRassembly.v2) and long-read (VGPassembly) based assemblies with somewhat more complete, contiguous regions assembled in the latter (Figure 3b,c).
The largest disconnect between the FISH and assembly results was for ZCA 15 and ZCA 16. FISH analysis showed that two BACs from CFA 9 hybridized to ZCA 15 (506N23 and 332E15), whereas these were assembled in the scaffold representing ZCA 16 in the sea lion assemblies. This difference may be related to the fact that these two chromosomes could not be separated by bivariate chromosome sorting (Figure 1e), which may influence the accuracy of the FISH analyses, and perhaps explains the large gap in chromosome painting on ZCA 15. BACs from CFA 7, which were found in the FISH analysis on ZCA 7 (Figure S10), were assembled in this gap in all the alignments.
3.4.2 ∣. Sequence order
The order of BACs, as inferred by FISH, matched the probe order predicted from the alignment (BAC liftover coordinates) between dog and the most contiguous sea lion assemblies (VGPassembly, SRassembly.v3) for 13 of the 17 autosomes (ZCA 1–5, 8–14, 17; Table S11). The BAC order for SRassembly.v2 matched the FISH results for the same chromosomes with the exception of ZCA 5, 8 and 9. Inconsistencies were most commonly seen in repetitive chromosomal regions, such as centromeres, that are known to be notoriously difficult to assemble (Miga, 2015, 2019). These types of misassemblies were more common in SRassembly.v2, where three BACs aligned towards the telomeres despite being found in the FISH analyses towards the centromeric region. In contrast, in both the SRassembly.v3 and VGPassembly (Figure S11; Table 3) only a single BAC showed this behaviour. Local misassemblies were only seen in SRassembly.v2 with three BACs assembled at different locations within the same chromosome arm predicted by FISH (Table 3). The reduction in all types of misassemblies coupled with the slightly higher liftover of the BACs in the VGPassembly speaks in favour of the long reads used to construct it, increasing contig lengths known to benefit the scaffolding process (Bickhart et al., 2017; Rhie et al., 2020). The VGPassembly also benefited from the use of optical maps, which further improves structural accuracy (Rhie et al., 2020; Udall & Dawe, 2018). However, SRassembly.v3, based on the same simple short-read primary assembly as SRassembly.v2, but constructed with a different assembly pipeline and independent Hi-C libraries, also resolved discrepancies specific to SRassembly.v2. Some of the intrachromosomal misassemblies seen in SRassembly. v2 were also resolved in the Antarctic fur seal assembly, constructed with long reads for gap filling, but no further long-range technology. Dedicated experiments are needed to uncover the contribution of the primary assembly, Hi-C libraries and scaffolding pipeline to correct syntenic inference.
4 ∣. CONCLUSIONS
This study resulted in two well-annotated assemblies for the California sea lion (Zalophus californianus) and a synteny map to dog (Canis lupus familiaris) anchored to sequence data. Chromosome-scale assemblies of the quality presented here allow the study of syntenic regions across individuals as well as across large evolutionary distances. They are readily generated and will contribute to debates surrounding the role of genome structure in evolution and can readily be combined with the raw Hi-C reads in assessing three-dimensional structure of the genome (Oluwadare et al., 2020). Assembly quality is an important consideration when engaging in comparative analyses either assessing individual variation or variation between evolutionarily distant clades (Fan et al., 2019). A central, yet hitherto poorly explored, result of this study is the general suitability of assemblies derived from simple short-read data combined with Hi-C scaffolding to infer long-range synteny across large evolutionary time spans (here 45 million years of evolution). Results indicate that quality for broad-scale chromosomal inference was comparable to the high-quality genome integrating long-read data and additional scaffolding information (BioNano, 10X genomics): at the 10-Mb resolution level of the BACs the SRassembly.v3 performed very similarly to the VGPassembly. Yet, on a local scale there are differences between the assemblies, and the high-quality VGPassembly in combination with the raw long-read data will be essential to resolve locally restricted structural variation (Rhie et al., 2020; Weissensteiner et al., 2020).
Supplementary Material
ACKNOWLEDGEMENTS
California sea lion samples were collected under Marine Mammal Protection Act (MMPA) permit no. 18786. The sequencing and scaffolding of the CSL genome was supported by a Deutsche Forschungsgemeinschaft (DFG) standard grant to J.B.W.W. and J.I.H. (HO 5122/4-1) and by LMU Munich to J.B.W.W. The scaffolding of the Antarctic genome was funded by the DFG in the framework of a Sonderforschungsbereich (project nos. 316099922 and 396774617–TRR 212) and the priority programme “Antarctic Research with Comparative Investigations in Arctic Ice Areas” SPP 1158 (project no. 424119118) to J.I.H. The molecular cytogenetic components of this study were funded in part by a Morris Animal Foundation award to M.B. and F.M.D.G. (Grant no. D10ZO-003). Additional funding for this project at NC State University was provided by the NCSU Cancer Genomics Funds (M.B.) and HHMI (E.D.J.). Identification of certain commercial equipment, instruments, software or materials does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the products identified are necessarily the best available for the purpose. We thank the DNAzoo team for making their California sea lion assembly available before release. Open Access funding enabled and organized by Projekt DEAL.
Funding information
Morris Animal Foundation, Grant/Award Number: D10ZO-003; Deutsche Forschungsgemeinschaft, Grant/Award Number: 424119118 and HO 5122/4-1; NCSU Cancer Genomics Funds; LUDWIG-MAXIMILIANS-UNIVERSITAET MUNCHEN
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
DATA AVAILABILITY STATEMENT
All data are publicly available at the National Center for Biotechnology Information (NCBI). Accession numbers for genome assemblies and annotations of the California sea lion are provided in Table 1. Accessory raw data and publicly available genome information for other species are specified in the Material and Methods section. The improved genome assembly of the Antarctic fur seal (Arctocephalus gazeiia) is available at NCBI under accession no. GCA_900642305.1. Source code used to generate the VGP assembly (which forms part of the VGP BioProject ID PRJNA489243) to run locally or on the DNAnexus platform is publicly available on github (https://github.com/VGP/vgp-assembly). The scaffolding pipeline is also available to run on generic architecture and Docker containers. Intermediate assemblies and raw data are available to download on Genome Ark (https://vgp.github.io) until archived. All code for comparative analyses is available at https://github.com/EvoBioWolf/2021_ZalophusCalifornianus_genome.
REFERENCES
- Árnason Ú. (1974). Comparative chromosome studies in Pinnipedia. Hereditas, 76(2), 179–226. 10.1111/j.1601-5223.1974.tb01340.x [DOI] [PubMed] [Google Scholar]
- Avelar AT, Perfeito L, Gordo I, & Ferreira MG (2013). Genome architecture is a selectable trait that can be maintained by antagonistic pleiotropy. Nature Communications, 4(1), 2235. 10.1038/ncomms3235 [DOI] [PubMed] [Google Scholar]
- Beklemisheva VR, Perelman PL, Lemskaya NA, Kulemzina AI, Proskuryakova AA, Burkanov VN, & Graphodatsky AS (2016). The ancestral carnivore karyotype as substantiated by comparative chromosome painting of three pinnipeds, the walrus, the steller sea lion and the baikal seal (Pinnipedia, Carnivora). PLoS One, 11(1), e0147647. 10.1371/journal.pone.0147647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickhart DM, Rosen BD, Koren S, Sayre BL, Hastie AR, Chan S, Lee J, Lam ET, Liachko I, Sullivan ST, Burton JN, Huson HJ, Nystrom JC, Kelley CM, Hutchison JL, Zhou Y, Sun J, Crisà A, Ponce de León FA, … Smith TPL (2017). Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nature Genetics, 49(4), 643–650. 10.1038/ng.3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen M, Bullerdiek J, & Langford CF (1999). The DAPI banded karyotype of the domestic dog (Canis familiaris) generated using chromosome-specific paint probes. Chromosome Research, 7(5), 401–406. 10.1023/A:1009224232134 [DOI] [PubMed] [Google Scholar]
- Browning HM, Gulland FMD, Hammond JA, Colegrove KM, & Hall AJ (2015). Common cancer in a wild animal: The California sea lion (Zalophus californianus) as an emerging model for carcinogenesis. Philosophical Transactions of the Royal Society B: Biological Sciences, 370(1673), 20140228. 10.1098/rstb.2014.0228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckles EL, Lowenstine LJ, DeLong RL, Melin SR, Vittore RK, Wong H-N, Ross GL, St Leger JA, Greig DJ, Duerr RS, Gulland FMD, & Stott JL (2007). Age-prevalence of Otarine Herpesvirus-1, a tumor-associated virus, and possibility of its sexual transmission in California sea lions. Veterinary Microbiology, 120(1–2), 1–8. 10.1016/j.vetmic.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Burton JN, Adey A, Patwardhan RP, Qiu R, Kitzman JO, & Shendure J (2013). Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nature Biotechnology, 31(12), 1119–1125. 10.1038/nbt.2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavagna P, Menotti A, & Stanyon R (2000). Genomic homology of the domestic ferret with cats and humans. Mammalian Genome, 11(10), 866–870. 10.1007/s003350010172 [DOI] [PubMed] [Google Scholar]
- Chaisson MJP, Wilson RK, & Eichler EE (2015). Genetic variation and the de novo assembly of human genomes. Nature Reviews Genetics, 16(11), 627–640. 10.1038/nrg3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow W, Brugger K, Caccamo M, Sealy I, Torrance J, & Howe K. (2016). GEVAL — A web-based browser for evaluating genome assemblies. Bioinformatics, 32(16), 2508–2510. 10.1093/bioinformatics/btw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, & Gingeras TR (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics, 29(1), 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, & Aiden EL (2017). De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science, 356(6333), 92–95. 10.1126/science.aal3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Shamim MS, Batra SS, Durand NC, Musial NT, Mostofa R, & Aiden EL (2018). The Juicebox Assembly Tools module facilitates de novo assembly of mammalian genomes with chromosome-length scaffolds for under $1000. BioRxiv. 10.1101/254797 [DOI] [Google Scholar]
- Duke Becker SE, Thomas R, Trifonov VA, Wayne RK, Graphodatsky AS, & Breen M (2011). Anchoring the dog to its relatives reveals new evolutionary breakpoints across 11 species of the Canidae and provides new clues for the role of B chromosomes. Chromosome Research, 19(6), 685–708. 10.1007/S10577-011-9233-4 [DOI] [PubMed] [Google Scholar]
- Ekblom R, & Wolf JBW (2014). A field guide to whole-genome sequencing, assembly and annotation. Evolutionary Applications, 7(9), 1026–1042. 10.1111/eva.12178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, Kaessmann H, Betrán E, & Long M (2004). Extensive gene traffic on the mammalian X chromosome. Science, 303(5657), 537. 10.1126/science.1090042 [DOI] [PubMed] [Google Scholar]
- Fan H, Wu Q, Wei F, Yang F, Ng BL, & Hu Y (2019). Chromosome-level genome assembly for giant panda provides novel insights into Carnivora chromosome evolution. Genome Biology, 20(1), 267. 10.1186/s13059-019-1889-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay FH, Rausch VR, & Feltz ET (1967). Cytogenetic comparison of some pinnipeds (mammalia: Eutheria). Canadian Journal of Zoology, 45(5), 773–778. 10.1139/z67-088 [DOI] [Google Scholar]
- Feng JX, & Riddle NC (2020). Epigenetics and genome stability. Mammalian Genome, 10.1007/s00335-020-09836-2 [DOI] [PubMed] [Google Scholar]
- Foote AD, Liu Y, Thomas GWC, Vinař T, Alföldi J, Deng J, Dugan S, van Elk CE, Hunter ME, Joshi V, Khan Z, Kovar C, Lee S, Lindblad-Toh K, Mancia A, Nielsen R, Qin X, Qu J, Raney BJ, … Gibbs RA (2015). Convergent evolution of the genomes of marine mammals. Nature Genetics, 47(3), 272–275. 10.1038/ng.3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell NJ, Rutherford K, Prost S, Tollis M, Winter D, Macey JR, Adelson DL, Suh A, Bertozzi T, Grau JH, Organ C, Gardner PP, Muffato M, Patricio M, Billis K, Martin FJ, Flicek P, Petersen B, Kang L, … Stone C (2020). The tuatara genome reveals ancient features of amniote evolution. Nature, 584(7821), 403–409. 10.1038/s41586-020-2561-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS (2007). Improved pairwise alignment of genomic DNA. The Pennsylvania State University. (PhD thesis). [Google Scholar]
- Hinrichs AS (2006). The UCSC genome browser database: Update 2006. Nucleic Acids Research, 34(90001), D590–D598. 10.1093/nar/gkj144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Chow W, Collins J, Pelan S, Pointon D-L, Sims Y, Wood J(2020). Significantly improving the quality of genome assemblies through curation. BioRxiv. 10.1101/2020.08.12.247734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Wu QI, Ma S, Ma T, Shan L, Wang X, Nie Y, Ning Z, Yan LI, Xiu Y, & Wei F (2017). Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proceedings of the National Academy of Sciences, 114(5), 1081–1086. 10.1073/pnas.1613870114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humble E, Dasmahapatra KK, Martinez-Barrio A, Gregório I, Forcada J, Polikeit A-C, Goldsworthy SD, Goebel ME, Kalinowski J, Wolf JBW, & Hoffman JI (2018). RAD sequencing and a hybrid Antarctic fur seal genome assembly reveal rapidly decaying linkage disequilibrium, global population structure and evidence for inbreeding. G3: Genes, Genomes, Genetics, 8(8), 2709–2722. 10.1534/g3.118.200171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, & Dekker J (2013). High-throughput genome scaffolding from in vivo DNA interaction frequency. Nature Biotechnology, 31(12), 1143–1147. 10.1038/nbt.2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, & Haussler AD (2002). The human genome browser at UCSC. Genome Research, 12(6), 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, & Marra MA (2009). Circos: An information aesthetic for comparative genomics. Genome Research, 19(9), 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, & Hedges SB (2017). TimeTree: A resource for timelines, timetrees, and divergence times. Molecular Biology and Evolution, 34(7), 1812–1819. 10.1093/molbev/msx116 [DOI] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, & Salzberg SL (2004). Versatile and open software for comparing large genomes. Genome Biology, 5, R12. 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, & Dekker J (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science, 326(5950), 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Low WY, Tearle R, Koren S, Ghurye J, Rhie A, Phillippy AM, Rosen BD, Bickhart DM, Smith TPL, Hiendleder S, & Williams JL (2019). New insights into mammalian sex chromosome structure and evolution using high-quality sequences from bovine X and Y chromosomes. BMC Genomics, 20(1), 1000. 10.1186/s12864-019-6364-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes F, Oliveira LR, Kessler A, Beux Y, Crespo E, Cárdenas-Alayza S, Majluf P, Sepúlveda M, Brownell RL, Franco-Trecu V, Páez-Rosas D, Chaves J, Loch C, Robertson BC, Acevedo-Whitehouse K, Elorriaga-Verplancken FR, Kirkman SP, Peart R, Wolf JBW, … Bonatto SL (2021). Phylogenomic discordance in the eared seals is best explained by incomplete lineage sorting following explosive radiation in the southern hemisphere. Systematic Biology, 70(4), 786–802, (syaa099). 10.1093/sysbio/syaa099 [DOI] [PubMed] [Google Scholar]
- Marie-Nelly H, Marbouty M, Cournac A, Flot J-F, Liti G, Parodi DP, Syan S, Guillén N, Margeot A, Zimmer C, & Koszul R (2014). High-quality genome (re)assembly using chromosomal contact data. Nature Communications, 5(1), 5695. 10.1038/ncomms6695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megquier K, Turner-Maier J, Swofford R, Kim J-H, Sarver AL, Wang C, Sakthikumar S, Johnson J, Koltookian M, Lewellen M, Scott MC, Schulte AJ, Borst L, Tonomura N, Alfoldi J, Painter C, Thomas R, Karlsson EK, Breen M, … Lindblad-Toh K (2019). Comparative genomics reveals shared mutational landscape in canine hemangiosarcoma and human angiosarcoma. Molecular Cancer Research, 17(12), 2410–2421. 10.1158/1541-7786. MCR-19-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH (2015). Completing the human genome: The progress and challenge of satellite DNA assembly. Chromosome Research, 23(3), 421–426. 10.1007/s10577-015-9488-2 [DOI] [PubMed] [Google Scholar]
- Miga KH (2019). Centromeric satellite DNAs: Hidden sequence variation in the human population. Genes, 10(5), 352. 10.3390/genes10050352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr DW, Naguib A, Weisenfeld N, Kumar V, Shah P, Church DM, & Scott AF (2017). Improved de novo genome assembly: Linked-read sequencing combined with optical mapping produce a high quality mammalian genome at relatively low cost. BioRxiv. 10.1101/128348 [DOI] [Google Scholar]
- Murphy WJ, Larkin DM, der Wind AE, Bourque G, Tesler G, Auvil L, & Lewin HA (2005). Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science, 309(5734), 613–617. 10.1126/science.1111387 [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Sun S, Chen Z-Q, Pecon-Slattery J, O’Brien SJ (1999). Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Research, 9(12), 1223–1230. 10.1101/gr.9.12.1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash WG, Wienberg J, Ferguson-Smith MA, Menninger JC, & O’Brien SJ (1998). Comparative genomics: Tracking chromosome evolution in the family Ursidae using reciprocal chromosome painting. Cytogenetic and Genome Research, 83(3–4), 182–192. 10.1159/000015176 [DOI] [PubMed] [Google Scholar]
- Neely BA, Prager KC, Bland AM, Fontaine C, Gulland FM, & Janech MG (2018). Proteomic analysis of urine from California sea lions (Zalophus californianus): A resource for urinary biomarker discovery. Journal of Proteome Research, 17(9), 3281–3291. 10.1021/acs.jproteome.8b00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely BA, Soper JL, Gulland FMD, Bell PD, Kindy M, Arthur JM, & Janech MG (2015). Proteomic analysis of cerebrospinal fluid in California sea lions (Zalophus californianus) with domoic acid toxicosis identifies proteins associated with neurodegeneration. Proteomics, 15(23–24), 4051–4063. 10.1002/pmic.201500167 [DOI] [PubMed] [Google Scholar]
- Ng BL, & Carter NP (2006). Factors affecting flow karyotype resolution. Cytometry Part A, 69A(9), 1028–1036. 10.1002/cyto.a.20330 [DOI] [PubMed] [Google Scholar]
- Ng BL, Yang F, & Carter NP (2007). Flow analysis and sorting of microchromosomes (<3 Mb). Cytometry Part A, 71A(6), 410–413. 10.1002/cyto.a.20394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie W, Wang J, O’Brien PCM, Fu B, Ying T, Ferguson-Smith MA, & Yang F (2002). The genome phylogeny of domestic cat, red panda and five mustelid species revealed by comparative chromosome painting and G-banding. Chromosome Research, 10, 209–222. 10.1023/A:1015292005631 [DOI] [PubMed] [Google Scholar]
- Nowell PC, & Hungerford DA (1960). A minute chromosome in human chronic granulocytic leukemia. Science, 132(1497). [DOI] [PubMed] [Google Scholar]
- Nyakatura K, & Bininda-Emonds OR (2012). Updating the evolutionary history of Carnivora (Mammalia): A new species-level supertree complete with divergence time estimates. BMC Biology, 10, 12. 10.1186/1741-7007-10-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oluwadare O, Highsmith M, Turner D, Lieberman Aiden E, & Cheng J (2020). GSDB: A database of 3D chromosome and genome structures reconstructed from Hi-C data. BMC Molecular and Cell Biology, 21(1), 60. 10.1186/s12860-020-00304-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Kim K, Sohn H, Kim HW, An Y-R, Kang J-H, Kim E-M , Kwak W, Lee C, Yoo DA, Jung J, Sung S, Yoon J, & Kim H (2018). Deciphering the evolutionary signatures of pinnipeds using novel genome sequences: The first genomes of Phoca largha, Callorhinus ursinus, and Eumetopias jubatus. Scientific Reports, 8(1), 16877. 10.1038/s41598-018-34758-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart CR, Tusso S, Pophaly SD, Botero-Castro F, Wu C-C, Aurioles-Gamboa D, Baird AB, Bickham JW, Forcada J, Galimberti F, Gemmell NJ, Hoffman JI, Kovacs KM, Kunnasranta M, Lydersen C, Nyman T, de Oliveira LR, Orr AJ, Sanvito S, … Wolf JBW (2020). Determinants of genetic variation across eco-evolutionary scales in pinnipeds. Nature Ecology & Evolution, 4(8), 1095–1104. 10.1038/s41559-020-1215-5 [DOI] [PubMed] [Google Scholar]
- Peichel CL, Sullivan ST, Liachko I, & White MA (2017). Improvement of the threespine stickleback genome using a hi-c-based proximity-guided assembly. Journal of Heredity, 108(6), 693–700. 10.1093/jhered/esx058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalba JV, & Wolf JBW (2020). From molecules to populations: Appreciating and estimating recombination rate variation. Nature Reviews Genetics, 21, 476–492. 10.1038/s41576-020-0240-1 [DOI] [PubMed] [Google Scholar]
- Peng X, Alföldi J, Gori K, Eisfeld AJ, Tyler SR, Tisoncik-Go J, Brawand D, Law GL, Skunca N, Hatta M, Gasper DJ, Kelly SM, Chang J, Thomas MJ, Johnson J, Berlin AM, Lara M, Russell P, Swofford R, … Katze MG (2014). The draft genome sequence of the ferret (Mustela putorius furo) facilitates study of human respiratory disease. Nature Biotechnology, 32(12), 1250–1255. 10.1038/nbt.3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelman PL, Beklemisheva VR, Yudkin DV, Petrina TN, Rozhnov V, Nie W, & Graphodatsky AS (2012). Comparative chromosome painting in carnivora and pholidota. Cytogenetic and Genome Research, 137(2–4), 174–193. 10.1159/000341389 [DOI] [PubMed] [Google Scholar]
- Perelman PL, Graphodatsky AS, Dragoo JW, Serdyukova NA, Stone G, Cavagna P, Menotti A, Nie W, O’Brien PCM, Wang J, Burkett S, Yuki K, Roelke ME, O’Brien SJ, Yang F, & Stanyon R (2008). Chromosome painting shows that skunks (Mephitidae, Carnivora) have highly rearranged karyotypes. Chromosome Research, 16(8), 1215–1231. 10.1007/S10577-008-1270-2 [DOI] [PubMed] [Google Scholar]
- Proskuryakova A, Kulemzina A, Perelman P, Makunin A, Larkin D, Farré M, Kukekova A, Lynn Johnson J, Lemskaya N, Beklemisheva V, Roelke-Parker M, Bellizzi J, Ryder O, O’Brien S, & Graphodatsky A (2017). X chromosome evolution in cetartiodactyla. Genes, 8(9), 216. 10.3390/genes8090216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, O'Connell BL, Stites JC, Rice BJ, Blanchette M, Calef R, Troll CJ, Fields A, Hartley PD, Sugnet CW, Haussler D, Rokhsar DS, & Green RE (2016). Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Research, 26(3), 342–350. 10.1101/gr.193474.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudsepp T, Lee E-J, Kata SR, Brinkmeyer C, Mickelson JR, Skow LC, Womack JE, & Chowdhary BP (2004). Exceptional conservation of horse-human gene order on X chromosome revealed by high-resolution radiation hybrid mapping. Proceedings of the National Academy of Sciences, 101(8), 2386–2391. 10.1073/pnas.0308513100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettenberger G, Klett CH, Zechner U, Bruch J, Just W, Vogel W, & Hameister H (1995). ZOO-FISH analysis: Cat and human karyotypes closely resemble the putative ancestral mammalian karyotype. Chromosome Research, 3(8), 479–486. 10.1007/BF00713962 [DOI] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, & Jarvis ED (2020). Towards complete and error-free genome assemblies of all vertebrate species. BioRxiv. 10.1101/2020.05.22.110833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez Delgado CL, Waters PD, Gilbert C, Robinson TJ, & Graves JAM (2009). Physical mapping of the elephant X chromosome: Conservation of gene order over 105 million years. Chromosome Research, 17(7), 917–926. 10.1007/S10577-009-9079-1 [DOI] [PubMed] [Google Scholar]
- Romanenko SA, Fedorova YE, Serdyukova NA, Zaccaroni M, Stanyon R, & Graphodatsky AS (2020). Evolutionary rearrangements of X chromosomes in voles (Arvicolinae, Rodentia). Scientific Reports, 10(1), 13235. 10.1038/s41598-020-70226-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, Frankish A, Lovell FL, Howe KL, Ashurst JL, Fulton RS, Sudbrak R, Wen G, Jones MC, Hurles ME, … Bentley DR (2005). The DNA sequence of the human X chromosome. Nature, 434(7031), 325–337. 10.1038/nature03440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffman JD, & Breen M (2015). Comparative oncology: What dogs and other species can teach us about humans with cancer. Philosophical Transactions of the Royal Society B: Biological Sciences, 370(1673), 20140231. 10.1098/rstb.2014.0231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander T, Libbrecht R, & Keller L (2014). Supergenes and complex phenotypes. Current Biology, 24(7), R288–R294. 10.1016/j.cub.2014.01.056 [DOI] [PubMed] [Google Scholar]
- Selden JR, Moorhead PS, Oehlert ML, & Patterson DF (1975). The Giemsa banding pattern of the canine karyotype. Cytogenetic and Genome Research, 15(6), 380–387. 10.1159/000130537 [DOI] [PubMed] [Google Scholar]
- Shapiro SG, Raghunath S, Williams C, Motsinger-Reif AA, Cullen JM, Liu T, Albertson D, Ruvolo M, Bergstrom Lucas A, Jin J, Knapp DW, Schiffman JD, & Breen M (2015). Canine urothelial carcinoma: Genomically aberrant and comparatively relevant. Chromosome Research, 23(2), 311–331. 10.1007/s10577-015-9471-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao Y-H (2015). Interplay of epigenetics, genome rearrangement, and environment during development. In Su LJ, & Chiang T (Eds.), Environmental epigenetics (pp. 281–294). Springer. 10.1007/978-1-4471-6678-8_12 [DOI] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, & Zdobnov EM (2015). BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics, 31(19), 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Stapley J, Feulner PGD, Johnston SE, Santure AW, & Smadja CM (2017). Variation in recombination frequency and distribution across eukaryotes: Patterns and processes. Philosophical Transactions of the Royal Society B: Biological Sciences, 372(1736), 20160455. 10.1098/rstb.2016.0455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strijk JS, Hinsinger DD, Zhang F, & Cao K. (2019). Trochodendronaralioides, the first chromosome-level draft genome in Trochodendrales and a valuable resource for basal eudicot research. GigaScience, 8(11). 10.1093/gigascience/giz136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Duke SE, Bloom SK, Breen TE, Young AC, Feiste E, Seiser EL, Tsai P-C, Langford CF, Ellis P, Karlsson EK, Lindblad-Toh K, & Breen M (2007). A cytogenetically characterized, genome-anchored 10-Mb BAC set and CGH array for the domestic dog. Journal of Heredity, 98(5), 474–484. 10.1093/jhered/esm053 [DOI] [PubMed] [Google Scholar]
- Thomas R, Duke SE, Karlsson EK, Evans A, Ellis P, Lindblad-Toh K, & Breen M (2008). A genome assembly-integrated dog 1 Mb BAC microarray: A cytogenetic resource for canine cancer studies and comparative genomic analysis. Cytogenetic and Genome Research, 122(2), 110–121. 10.1159/000163088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Duke SE, Wang HJ, Breen TE, Higgins RJ, Linder KE, Ellis P, Langford CF, Dickinson PJ, Olby NJ, & Breen M (2009). ‘Putting our heads together’: Insights into genomic conservation between human and canine intracranial tumors. Journal of Neuro-Oncology, 94(3), 333–349. 10.1007/s11060-009-9877-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusso S, Nieuwenhuis BPS, Sedlazeck FJ, Davey JW, Jeffares DC, & Wolf JBW (2019). Ancestral admixture is the main determinant of global biodiversity in fission yeast. Molecular Biology and Evolution, 36(9), 1975–1989. 10.1093/molbev/msz126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udall JA, & Dawe RK (2018). Is it ordered correctly? Validating genome assemblies by optical mapping. The Plant Cell, 30(1), 7–14. 10.1105/tpc.17.00514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, Aganezov S, Anselmetti Y, Lee J, Ruzzante L, Reijnders MJMF, Feron R, Bérard S, George P, Hahn MW, Howell PI, Kamali M, Koren S, Lawson D, Maslen G, Peery A, Phillippy AM, Sharakhova MV, Tannier E, … Sharakhov IV (2020). Evolutionary superscaffolding and chromosome anchoring to improve Anopheles genome assemblies. BMC Biology, 18. 10.1186/s12915-019-0728-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissensteiner MH, Bunikis I, Catalán A, Francoijs K-J, Knief U, Heim W, Peona V, Pophaly SD, Sedlazeck FJ, Suh A, Warmuth VM, & Wolf JBW (2020). Discovery and population genomics of structural variation in a songbird genus. Nature Communications, 11(1), 3403. 10.1038/s41467-020-17195-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellenreuther M, Mérot C, Berdan E, & Bernatchez L (2019). Going beyond SNPs: The role of structural genomic variants in adaptive evolution and species diversification. Molecular Ecology, 28(6), 1203–1209. 10.1111/mec.15066 [DOI] [PubMed] [Google Scholar]
- Wolf JBW, Tautz D, & Trillmich F (2007). Galápagos and Californian sea lions are separate species: Genetic analysis of the genus Zalophus and its implications for conservation management. Frontiers in Zoology, 4(20), 1–13. 10.1186/1742-9994-4-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, O'Brien P, Milne BS, Graphodatsky AS, Solanky N, Trifonov V, Rens W, Sargan D, & Ferguson-Smith MA (1999). A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics, 62(2), 189–202. 10.1006/geno.1999.5989 [DOI] [PubMed] [Google Scholar]
- Zoonomia Consortium (2020). A comparative genomics multitool for scientific discovery and conservation. Nature, 587(7833), 240–245. 10.1038/s41586-020-2876-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are publicly available at the National Center for Biotechnology Information (NCBI). Accession numbers for genome assemblies and annotations of the California sea lion are provided in Table 1. Accessory raw data and publicly available genome information for other species are specified in the Material and Methods section. The improved genome assembly of the Antarctic fur seal (Arctocephalus gazeiia) is available at NCBI under accession no. GCA_900642305.1. Source code used to generate the VGP assembly (which forms part of the VGP BioProject ID PRJNA489243) to run locally or on the DNAnexus platform is publicly available on github (https://github.com/VGP/vgp-assembly). The scaffolding pipeline is also available to run on generic architecture and Docker containers. Intermediate assemblies and raw data are available to download on Genome Ark (https://vgp.github.io) until archived. All code for comparative analyses is available at https://github.com/EvoBioWolf/2021_ZalophusCalifornianus_genome.