Abstract
Lorlatinib is currently the most advanced, potent and selective ALK tyrosine kinase inhibitor (TKI) for the treatment of ALK-positive non-small cell lung cancer (NSCLC) in the clinic. However, diverse compound ALK mutations driving therapy resistance emerge. Here, we determine the spectrum of lorlatinib-resistant compound ALK mutations in patients, following treatment with lorlatinib, the majority of which involve ALK G1202R or I1171N/S/T. We further identify structurally diverse lorlatinib analogs that harbor differential selective profiles against G1202R- versus I1171N/S/T- compound ALK mutations. Structural analysis revealed increased potency against compound mutations through improved inhibition of either G1202R- or I1171N/S/T-mutant kinases. Overall, we propose a classification of heterogenous ALK compound mutations enabling the development of distinct therapeutic strategies for precision targeting following sequential TKIs.
Keywords: ALK, compound mutation, lorlatinib resistance, lorlatinib analog, NSCLC
INTRODUCTION
Anaplastic lymphoma kinase (ALK) gene fusions are the driver oncogene identified across diverse tumor types, including in 3–5% of lung adenocarcinoma (1–3). In these tumors, aberrant expression of a constitutively active ALK fusion protein provides a potent oncogenic stimulus that promotes dysregulation and hyperactivation of downstream growth and survival pathways (1). Over the past decade, the treatment strategies for metastatic ALK fusion-positive (ALK-positive) non-small cell lung cancer (NSCLC) have rapidly evolved with the development of successive generations of increasingly potent and selective ALK tyrosine kinase inhibitors (TKIs) (4–9). Multiple randomized studies have demonstrated the superiority of next-generation, highly potent ALK TKIs as initial therapy for metastatic ALK-positive NSCLC (7–9). The significant strides made towards optimizing therapeutic targeting of ALK in this short timeframe have resulted in dramatic improvements in patient survival that are unprecedented in metastatic lung cancer (10–12) and yielded lessons on targeted therapy applicable across cancers.
Despite marked initial responses to ALK TKIs in most patients, tumors inevitably develop drug resistance leading to disease progression. In 50% to 60% of patients treated with second-generation ALK TKIs (e.g., ceritinib, alectinib, brigatinib), resistance results from the acquisition of secondary mutations in the ALK kinase domain (13). These secondary mutations restore ALK activity in the presence of an ALK inhibitor—so-called “on-target” resistance—and the tumors retain dependency on ALK signaling. In particular, the ALK G1202R mutation in the solvent front accounts for approximately one-half of on-target resistance across all second-generation ALK TKIs, and I1171T/N/S has been found in 10–15% of patients progressing on alectinib (13). Lorlatinib is a third-generation ALK TKI that has demonstrated preclinical potency and clinical efficacy against almost all single ALK mutations, including ALK G1202R and I1171N/S/T (13–17). Of all currently FDA-approved ALK TKIs, lorlatinib is considered the most effective treatment option for ALK-positive NSCLCs refractory to a second-generation ALK TKI (16). However, resistance to lorlatinib also emerges. Recent work by our group and others has demonstrated that sequential therapy with a second-generation ALK TKI followed by lorlatinib can drive the development of compound ALK resistance mutations (18–20). As these tumors are expected to retain dependency on ALK signaling, the development of novel, fourth-generation ALK TKIs that can overcome compound mutation-mediated lorlatinib resistance are urgently needed for patients with ALK-positive NSCLC.
Here, we describe the spectrum of compound ALK mutations detected in biopsies from patients with ALK-positive NSCLC treated with lorlatinib. Using Ba/F3 and patient-derived models harboring compound mutant EML4-ALK, we identify lorlatinib analogs with activity against recurrent ALK compound mutants involving either G1202R or I1171N/S. These results provide new insights into the clinical and biological significance of compound ALK mutations and provide a conceptual framework to guide the development of next-generation inhibitors that can overcome heterogeneous ALK compound mutations.
RESULTS
Lorlatinib resistance cohort
In our initial description of the molecular landscape of resistance to lorlatinib, approximately one-third of tissue biopsies harbored double or triple ALK resistance mutations (18). Here, we analyzed a larger updated dataset comprised of 47 patients with lorlatinib-resistant tissue biopsies to determine the prevalence of ALK-positive tumors with ≥2 ALK mutations. Demographic characteristics of this group of patients are summarized in Supplementary Table 1.
The majority of patients (85%) received lorlatinib in the third- or later-line setting. All patients had received at least one prior second-generation ALK TKI, and 77% had received prior crizotinib followed by at least one second-generation ALK TKI (Supplementary Table 1). The median time to treatment discontinuation (TTD) of lorlatinib was 8.5 months (95% CI, 6.4–16.1) and the median time to progression (TTP) on lorlatinib was 6.8 months (95% CI, 5.5–10.0), consistent with the known efficacy of lorlatinib in this patient population in the phase 2 trial (16). Among 21 patients who had known baseline ALK mutation status with a pre-lorlatinib biopsy, both the TTP and TTD were significantly longer in patients with known baseline ALK mutations in the tumor (pre-lorlatinib) as compared to those without known baseline ALK mutations (TTP: 7.8 months vs 2.8 months, HR 0.363, p<0.0001; TTD: 13.5 months vs 4.1 months, HR 0.128, p=0.0002) (Extended Data Fig. 1), again concordant with the phase 2 trial results (17).
Landscape of compound ALK resistance mutations in tumors
A total of 48 lorlatinib-resistant tissue biopsies from 47 patients were assessed for resistance mechanisms. Two (4%) demonstrated evidence of histologic transformation: one case from adenocarcinoma to small cell, and another from adenocarcinoma to squamous cell. One case was known to have neuroendocrine histology at initial diagnosis, and this was retained at resistance to lorlatinib. All remaining cases showed non-small cell histology without evidence of histologic transformation.
ALK kinase domain mutations were identified in 23 biopsies (48%), of which 14 (29%) were found to harbor double or triple ALK resistance mutations (Fig. 1A–B, Supplementary Table 2) including the previously unreported ALK C1156Y/G1269A (isolated in an ENU mutagenesis screen by our group (18) but not previously identified in clinic) and ALK G1202R/C1156Y. Using a variety of sequencing techniques as annotated in Supplementary Table 2, we were able to confirm that 9 of the 14 compound mutations were in cis (Fig. 1C, Supplementary Table 2); for the remainder, it was not feasible to confirm whether the mutations occurred in cis versus in trans configuration due to the distance between the mutated nucleotides. While no single predominant compound ALK mutation was identified, among the 14 cases with ≥2 ALK mutations, 8 (57%) harbored ALK G1202R—the most common ALK resistance mutation after a second-generation ALK TKI—and 3 (21%) harbored ALK I1171N—the second most common ALK mutation after alectinib (13).
Figure 1. Spectrum of compound ALK mutations identified in lorlatinib-resistant biopsies.
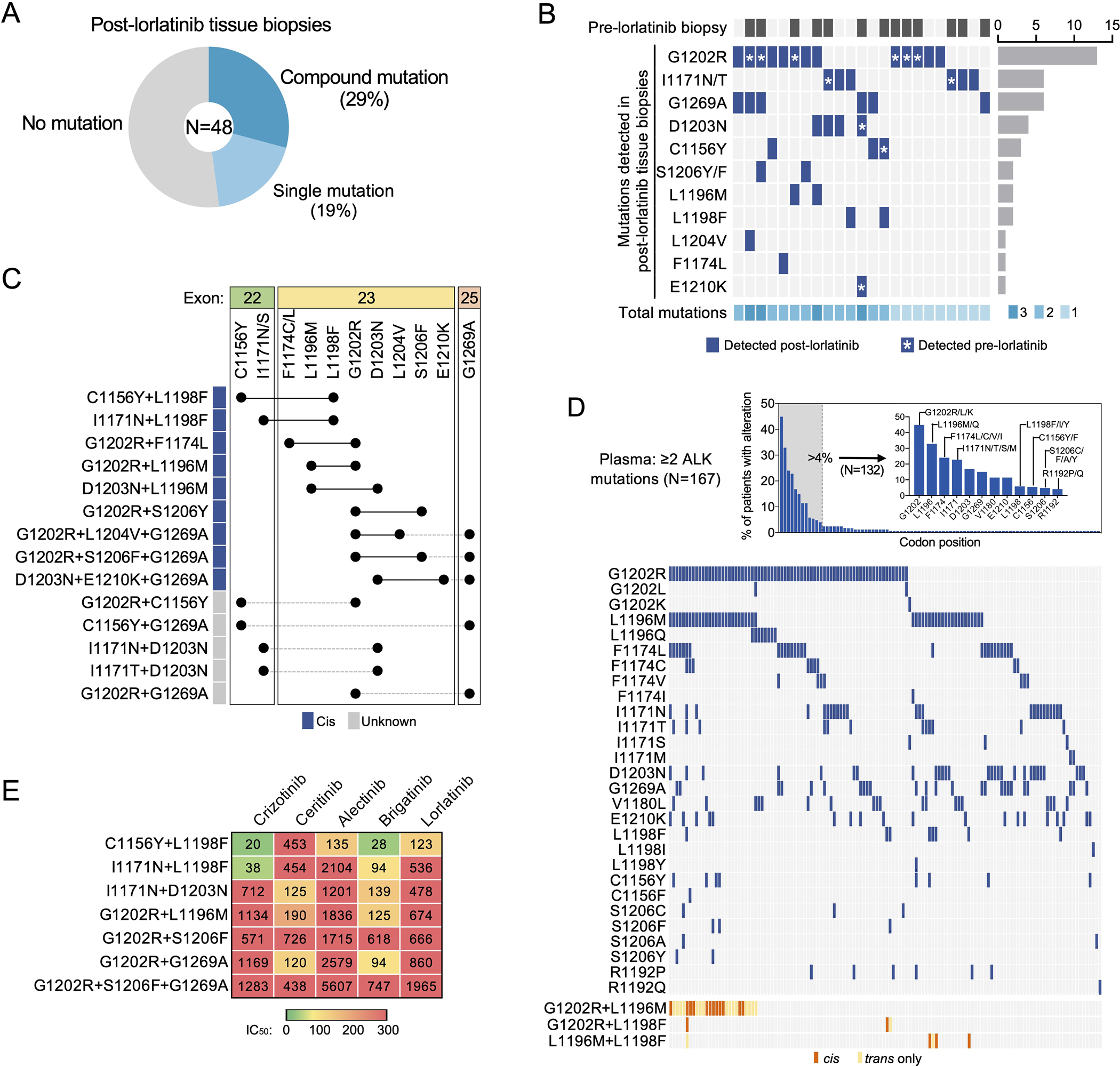
(A) The frequencies of compound, single, vs no ALK mutation detected in lorlatinib-resistant tissue biopsies (N = 48 biopsies). (B) Heatmap demonstrating the distribution of ALK mutations (dark blue) detected in post-lorlatinib tissue biopsies with at least one ALK mutation. Those cases with paired pre-lorlatinib biopsy available are indicated by dark gray (top row). The total number of ALK mutations identified are shown by shades of cyan (bottom row). Asterisk indicates the ALK mutation was present at baseline (i.e., detected in pre-lorlatinib biopsy). (C) Configuration (cis vs trans) of ALK mutations in tissue biopsies harboring ≥2 ALK mutations. (D) ALK mutations in an independent cohort of de-identified ALK TKI-resistant plasma samples harboring ≥2 ALK mutations (N = 194 plasma specimens). Top panel shows frequency of alterations per codon position. Bottom panel shows co-occurrence of individual mutations at positions altered in greater than 4% of patients. Co-occurring mutations involving closely spaced L1196, L1198 and G1202 positions could be assessed for cis (orange) versus trans (yellow) configuration. (E) Cellular IC50 values of FDA-approved ALK TKIs against clinical compound ALK mutations. Data are mean of three independent experiments.
The frequency of compound ALK mutations was higher in the tumors with acquired vs primary resistance (41% vs 6%, p=0.018). Of the 7 cases with primary lorlatinib resistance (defined as primary progression or disease stabilization for less than 6 months) and paired pre-lorlatinib specimen available, only 1 had harbored a baseline ALK resistance mutation, indicative of a pre-existing ALK-independent mechanism in most of these cases (Supplementary Table 2). In contrast, of the 15 cases with acquired lorlatinib resistance and paired pre-lorlatinib tissue, 12 harbored a baseline ALK mutation. Of these 12 cases with a baseline ALK mutation, an ALK mutation was no longer identified in 3 cases at lorlatinib resistance (suggestive of acquired ALK-independent resistance on lorlatinib), and another ALK mutation was identified in addition to the pre-existing baseline ALK mutation in 5 cases (suggestive of stepwise accumulation of ALK mutations on lorlatinib) (Supplementary Table 2, Fig. 1B). Additionally, in one patient with an ALK I1171T compound mutation for whom a pre-treatment tissue biopsy was not available, the I1171T mutation was detected in plasma prior to initiation of lorlatinib. Of note, not all paired pre- and post-lorlatinib tumor specimens were analyzed using the same genotyping method as this was a retrospective analysis (Supplementary Table 2); however, all of these genotyped specimens were assayed for the presence of known ALK kinase domain mutations.
Recurrent compound ALK resistance mutations in plasma
To further assess whether particular ALK mutations co-occurred in TKI-resistant ALK-positive NSCLC, we reviewed an independent de-identified Guardant Health dataset comprised of plasma specimens with ≥2 ALK mutations. This Guardant cohort included 194 plasma specimens from 167 patients with ALK-positive NSCLC (no treatment history available). The plasma specimens contained between 2 to 10 ALK mutations, with the majority (n=127, 65%) harboring 2 mutations. The most frequent ALK mutations detected in plasma overall were: G1202R (44%), L1196M (29%), F1174C/V/L (23%), I1171N/S/T (22%), D1203N (17%), and G1269A (15%) (Fig. 1D). ALK L1198F was identified in 5% of specimens, of which half also harbored ALK G1202R. Because these specimens represent patients treated with different generations of ALK inhibitors, we focused our subsequent analysis on specimens containing ALK mutations most commonly associated with alectinib resistance, G1202R (n=73) and I1171N/S/T (n=36). Of the 73 plasma samples with ≥2 ALK mutations including G1202R, the ALK co-mutations detected in ≥15% of specimens included L1196M (n=27, 37%), F1174C/V/L (n=23, 32%) and I1171N/T (n=15, 21%). Among 36 specimens with ≥2 ALK mutations including I1171N/T/S, the ALK co-mutations detected in ≥15% of specimens were: G1202R (n=15, 42%), L1196M (n=11, 31%), and V1180L (n=9, 25%), E1210K (n=7, 19%), and D1203N (n=7, 19%). To distinguish whether co-occurring mutations represent compound mutations in the same cell versus independent clones harboring single resistance mutations, we specifically examined the 27 cases with evidence of both G1202R and L1196M, 3 cases with both G1202R and L1198F, and 5 cases with both L1196M and L1198F, all of which lie in close proximity on exon 23 sufficient to be captured on single ctDNA fragments. Collectively, in 17 of 35 (48.6%) cases of co-occurrence, we found evidence that these mutations were in cis (12/27 G1202R+L1196M, 2/3 G1202R+L1198F, 3/5 L1196M+L1198F), confirming the recurrent nature of these compound ALK mutations in TKI-resistant cases. As the I1171 residue resides on exon 22 whereas the 1180–1210 positions are on exon 23, the allelic relationship between these mutations could not be determined.
Potency of approved ALK TKIs against compound ALK mutations
Our prior work demonstrated that the lorlatinib-resistant ALK C1156Y+L1198F double mutation re-sensitized tumor cells to the first-generation inhibitor crizotinib (21). In order to assess the sensitivity of additional lorlatinib-resistant compound ALK mutations to the currently available, FDA-approved first- and second-generation ALK TKIs, we generated Ba/F3 cell lines harboring distinct ALK compound mutations, including the newly discovered G1202R+S1206F+G1269A clinical mutation. Among them, we found distinct patterns of sensitivity vs resistance (Fig. 1E, Extended Data Fig. 2). Those harboring ALK L1198F were sensitive to crizotinib, consistent with prior findings (21). On the other hand, compound lorlatinib-resistant ALK mutations harboring either G1202R or I1171N without L1198F that were examined here were largely refractory to first- and second-generation ALK inhibitors tested, underscoring the need to discover novel ALK TKIs able to overcome these compound mutations.
Lorlatinib analogs targeting compound ALK mutations
To identify novel compounds potent against compound ALK mutations, we evaluated a panel of 20 lorlatinib analogs (Las) (14, 22) (Extended Data Fig. 3), which were selected based on three criteria: 1) structural diversity (acyclic vs cyclic), 2) cellular phosphorylation IC50 against ALK L1196M mutation in the NIH3T3 model of <100 nM, and 3) reduced bulk of features predicted to orient toward solvent front residues (to reduce potential steric clash with ALK G1202R). These 20 LAs were assessed in a three-step series of functional screens, using the Ba/F3 models generated above (Fig. 2A). In the initial screen, we sought to identify compounds with higher potency than lorlatinib against single ALK mutations. This aim was based on our previous observation that the effect of some compound mutations may result from the additive decremental effects of each single mutation on ALK binding and activity (18). We then performed a counter screen using PC9 (EGFR-mutant NSCLC) and Ba/F3 parental cells to identify compounds with non-ALK-specific cell toxicity. Treatment of Ba/F3 cells harboring single ALK mutations with LAs revealed a wide range of effects on cell viability (Fig. 2B, Extended Data Fig. 4). While all compounds were highly potent against nonmutant EML4-ALK, five of the LAs (LA1, 14, 16, 17, 18) exhibited IC50 values higher than 100 nM in one or more single ALK mutant models. Additionally, three LAs (LA10, 13, 15) reduced viability of Ba/F3 parental cells with IC50’s lower than 500 nM with non-ALK-specific cell toxicity (Fig. 2B). On the basis of these results, we selected 12 LAs for further testing.
Figure 2. Drug screening of Ba/F3 ALK mutation models identifies lorlatinib analogs with increased potency against compound ALK mutations.

(A) The schema summarizes the 3-step functional screening of 20 lorlatinib analogs (LAs). (B) Heatmap of cellular IC50 values of each LA against single ALK mutations (top) and parental Ba/F3 and PC9 cells (bottom). Asterisks indicate the 12 LAs selected for further assessment. (C) Potency of 12 LAs against clinical ALK compound mutations. Asterisks indicate the 6 LAs selected for additional validation. (D) Potency of 6 LAs against an expanded set of clinical and putative compound ALK mutations. Relative mutation positions within the ALK kinase domain are shown. Red, ALK G1202R; blue, ALK I1171N/S.
Next, we tested LAs against Ba/F3 models harboring seven distinct compound ALK mutations that were identified in post-lorlatinib patients. In general, compound mutation models exhibited higher cellular viability IC50 values compared to single mutation models (Fig. 2C, Extended Data Fig. 4). Among the 12 LAs tested, three (LA9, 11, 12) suppressed cell viability with IC50’s lower than 100 nM in five compound mutation models, demonstrating higher potency compared to lorlatinib especially against G1202R-containing compound mutations (Extended Data Fig. 4B). LA4 and LA7 were selected based on their improved sensitivity profile against ALK I1171N single and compound mutations. Finally, we selected LA19, a cyclic molecule similar to lorlatinib, to ensure structural diversity. Using these six candidate LAs, we assessed cell survival of an expanded panel of 17 Ba/F3 cell models harboring clinical and putative ALK compound mutations. Similar to our prior results, LAs displayed greater potency with generally lower IC50 values against compound mutants than lorlatinib (Fig. 2D, Extended Data Fig. 4C, 5). Among these six LAs, LA7 exhibited the broadest potency, suppressing cell proliferation of 13 compound mutation models with IC50’s lower than 100 nM; however, LA7 was not the most potent against all compound mutations and was notably less effective at inhibiting cells harboring G1202R-containing compound mutations. Indeed, by categorizing the sensitivity of compound mutations against LAs, we identified distinct patterns of LA drug efficacy (Fig. 2D, Extended Data Fig. 4). I1171N/S-containing compound mutations were most sensitive to LA7, whereas LA9/11/12 were most active against G1202R-containing compound mutations. Non-I1171 and G1202 compound mutations—including those containing L1198F—were generally sensitive to all six LAs. Together, these results suggest that distinct compounds may be required to target different categories of ALK compound mutations.
LA7 and LA9 target I1171N or G1202R compound ALK mutations
Based on the Ba/F3 results demonstrating the differential activity of LAs against G1202R and I1171N/S compound mutations, we selected LA7 and LA9 for further validation. Direct comparison of IC50 values of LA7 and LA9 (and vs lorlatinib) revealed increased selectivity against I1171N and G1202R single and compound mutants, respectively (Fig. 3A, B, C). When we directly compared the relative potency of LA7 and LA9 against Ba/F3 models classified according to G1202R-based, I1171N/S-based, vs other compound mutants, we confirmed that LA7 suppressed cell viability of I1171N/S and other compound mutation models with greater efficacy, whereas LA9 inhibited G1202R compound mutation models more effectively (Fig. 3D), supporting the classification of compound mutations according to the dominant pre-lorlatinib resistance mutation. Next, since LA7 and LA9 have off-target effects on several other kinases (Supplementary Table 3), we performed western blotting on G1202R and I1171N compound mutant Ba/F3 models after treatment with LA7 and LA9 to confirm the on-target effects on ALK phosphorylation and the effects on downstream signaling pathways. While lorlatinib failed to suppress pALK in both compound mutation models, LA7 and LA9 suppressed pALK and downstream pAKT, pERK and pS6 signaling (Fig. 3E, Extended Data Fig. 6A). Consistent with the cell survival assay results, we again observed a distinct pattern of differential LA activity. LA7 suppressed pALK in I1171N+D1203N and I1171N+L1198F models more effectively than LA9, and conversely, LA9 more potently suppressed G1202R+L1196M, G1202R+G1269A and G1202R+S1206F+G1269A models. Finally, we compared the activity of LA7 and LA9 to clinically available ALK inhibitors. Overall, LA7 and LA9 were the most potent against the seven compound mutations that we identified in post-lorlatinib patients (Extended Data Fig. 6B).
Figure 3. Selectivity of LA7 and LA9 against I1171N and G1202R single and compound mutants, respectively.

(A) Relative selectivity of LA7 and LA9 for single ALK mutants, calculated by the ratio IC50(LA7)/IC50(LA9). (B)-(C) Comparison of relative fold increase in potency of LA7 vs LA9 (over lorlatinib) against single and compound ALK mutants. LA9 has a greater increase in potency relative to lorlatinib against G1202R single and compound mutations compared to LA7 (B). Conversely, LA7 has greater increase in potency relative to lorlatinib against I1171N single and compound mutations compared to LA9 (C). (D) Selectivity of LA7 and LA9 against Ba/F3 models harboring G1202R (G1202R+L1198F, G1202R+L1196M, G1202R+S1206F, G1202R+G1269A, G1202R+S1206F+G1269A), I1171N/S (I1171S+L1198F, I1171N+L1198F, I1171N+D1203N, I1171N+C1156Y, I1171S+C1156Y), or other compound ALK mutations (F1174C+D1203N, D1203N+E1210K, C1156Y+L1198F, F1174C+L1198F, L1196M+L1198F, L1198F+G1269A) calculated by the ratio IC50(LA7)/IC50(LA9). Circles indicate clinical mutations, and diamonds indicate putative mutations. (E) Western blot analysis performed in Ba/F3 cells expressing ALK G1202R+L1196M or ALK I1171N+D1203N after treatment with DMSO, LA7, LA9, or lorlatinib for 6 hours. The images shown are representative of at least two repeats with similar results.
Next, we compared the efficacy of LA7 and LA9 against patient-derived cell lines generated from tumors harboring G1202R single mutation (MGH953-4), G1202R+L1196M compound mutation (MGH953-7) and I1171N+D1203N compound mutation (MGH990-2). Concordant with the results in Ba/F3 models, LA9 inhibited the growth of MGH953-7 harboring G1202R+L1196M most effectively, whereas LA7 was most potent against MGH990-2 harboring I1171N+D1203N (Fig. 4A, Extended Data Fig. 6C, D). Direct comparison of the selectivity profiles of LA7 and LA9 in the patient-derived cells showed close concordance with the Ba/F3 models harboring the same mutations (Fig. 4B). Consistent with the cell viability results, LA7 and LA9 more effectively suppressed pALK signaling in MGH990-2 and MGH953-7 cells, respectively (Fig. 4C). To test the ability for LA7 and LA9 to suppress the growth of ALK compound mutants over time, we treated MGH953-7 and MGH990-2 cells with concentrations of LA7, LA9 or lorlatinib predicted to discriminate between compound mutations for 14 days and monitored the effects on cell growth. Consistent with the signaling and short-term viability results, LA9 suppressed cell growth of MGH953-7 (G1202R+L1196M) to a greater extent than LA7 or lorlatinib, and conversely, LA7 inhibited cell growth of MGH990-2 (I1171N+D1203N) more effectively than LA9 or lorlatinib (Fig. 4D, Extended Data Fig. 6E).
Figure 4. Differential selectivity of LA7 and LA9 in patient-derived ALK-positive NSCLC cell lines.

(A) MGH953-7 (G1202R+L1196M) and MGH990-2 (I1171N+D1203N) patient-derived cell lines treated with the indicated drugs for 5 days, with viability measured using CellTiter-Glo assay. Data are mean and SEM of three independent experiments. The treatment histories of the MGH953 and MGH990 patients are summarized (top left), and cellular IC50 values in MGH953-7, MGH990-2, and MGH953-4 (G1202R) are shown (top right). (B) Selectivity of LA7 and LA9 against patient-derived and Ba/F3 models harboring I1171N vs G1202R mutations, respectively. (C) Western blot analysis performed in MGH990-2 and MGH953-7 cells after treatment with indicated drugs for 6 hours. The images shown are representative of at least two repeats with similar results. (D) Cell growth of MGH953-7 and MGH990-2 cells treated with the indicated drugs for 14 days. 300 nM of each drug was used for MGH953-7 and 100 nM of each drug was used for MGH990-2. Data are mean and SEM of three independent experiments.
Finally, we investigated whether LA7 and LA9 are able to inhibit the growth of ALK-positive NSCLC tumors harboring compound mutations in vivo. As expected, MGH953-7 primary patient-derived xenograft (PDX) tumors harboring G1202R+L1196M were resistant to clinically relevant doses of lorlatinib (Fig. 5A). To account for differences in plasma protein binding, we estimated the unbound plasma concentrations of lorlatinib, LA7 and LA9 to select comparable doses using a once-daily (QD) dosing regimen (Supplementary Table 4). All doses were well tolerated (Extended Data Fig. 7A, B) and both LA7 and LA9 initially slowed tumor growth; however, upon extended treatment, LA7-treated tumors regrew, while the LA9-treated tumors continued to be largely suppressed (p=0.0076 for tumor size difference on day 32; Fig. 5A). Consistent with these results, immunohistochemical analysis of MGH953-7 xenograft tumors demonstrated that ALK phosphorylation was effectively inhibited in LA9-treated tumors, but not lorlatinib or LA7-treated tumors (Fig. 5B, Extended Data Fig. 8). Despite multiple attempts, we were unable to establish xenograft tumors from the MGH990-2 (I1171N+D1203N) cell line; therefore, in order to test the efficacy of LA7 in vivo, we generated NIH3T3 cells expressing ALK I1171N+D1203N. Consistent with the I1171N+D1203N Ba/F3 and MGH990-2 cell lines, we confirmed that LA7 more potently suppressed ALK phosphorylation compared to LA9 or lorlatinib in vitro (Fig. 5C). Treatment of 3T3 I1171N+D1203N xenograft tumors in immunocompromised mice with lorlatinib, LA7 or LA9 at the same doses as those used for the MGH953-7 (G1202R+L1196M) demonstrated significantly greater tumor growth inhibition by LA7 compared to LA9 or lorlatinib (Fig. 5D). In contrast, only LA9 achieved significant suppression of tumor growth of 3T3 G1202R+L1196M xenograft tumors (Extended Data Fig. 7C–E), further demonstrating greater efficacy of LA7 against I1171N+D1203N and LA9 against G1202R+L1196M (Extended Data Fig. 7F). To investigate the unbound systemic exposures of each drug, we performed pharmacokinetic analysis of mouse blood sampled at steady state (Extended Data Fig. 7G, Supplementary Table 5, 6). The unbound plasma concentrations of lorlatinib and LA9 were similar (483 nM and 499 nM, respectively), confirming the superior activity of LA9 over lorlatinib in overcoming G1202R+L1196M in vivo. The concentration of LA7 achieved in vivo was comparatively lower (79.7 nM); yet despite this difference in exposure, it was capable of suppressing I1171N+D1203N xenograft tumors to a greater extent than LA9 or lorlatinib. Together, these results confirm our in vitro findings that LA9 and LA7 differentially target G1202R and I1171N-based compound mutations, respectively.
Figure 5. LA7 and LA9 inhibit tumor growth in xenograft tumors bearing ALK compound mutations.
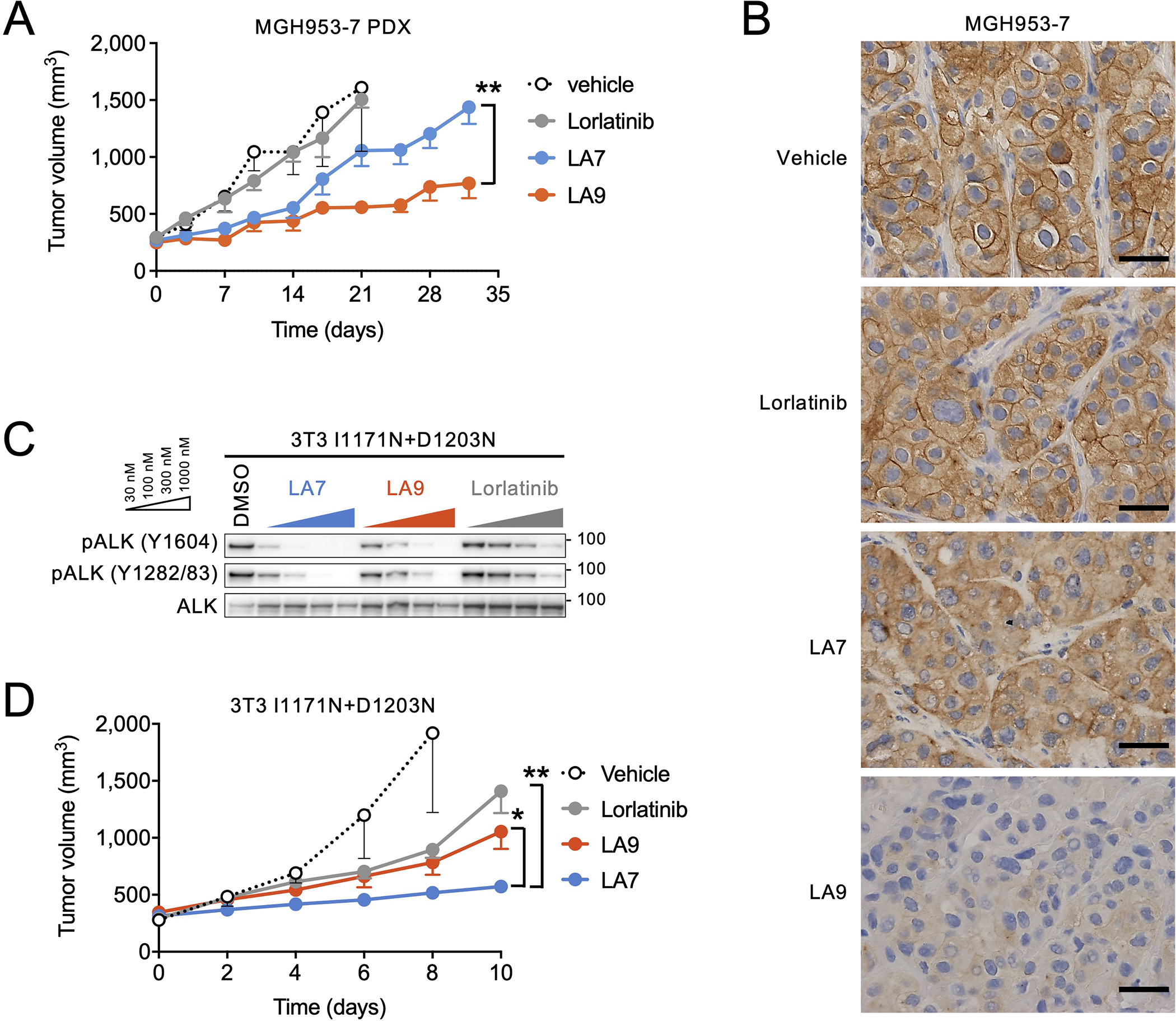
(A) Change in tumor volume of MGH953-7 (G1202R+L1196M) PDX tumors. Mice were treated with vehicle (N = 6 mice), lorlatinib 6 mpk QD (N = 5 mice), LA7 20 mpk QD (N = 6 mice), or LA9 40 mpk QD (N = 6 mice), 5 days per week. Tumor sizes on day 32 were significantly different between LA7 and LA9 (p=0.0076, two-tailed t test). Error bars indicate SEM. (B) Immunohistochemistry for pALK performed on MGH953-7 xenograft tumors harvested after 3-day treatment with the indicated drugs. The experiments were performed once after antibody validation. The images are representative of three tumors in each treatment group shown in Extended Data Fig. 8A. (C) Western blot analysis of NIH3T3 cells expressing ALK I1171N+D1203N and treated with the indicated drugs for 1 hour. The images shown are representative of at least two repeats with similar results. (D) Change in tumor volume of NIH3T3 I1171N+D1203N xenograft tumors. Mice were treated with lorlatinib 6 mpk QD, LA7 20 mpk QD, or LA9 40 mpk QD (N = 6 mice per group). Tumors treated with LA7 were significantly smaller on day 10 compared to those treated with LA9 (p=0.0133) or lorlatinib (p=0.0018, two-tailed t test). Error bars indicate SEM.
Structural basis for differential selectivity of LA7 and LA9
To better understand the mechanistic basis for the selectivity of LA9 and LA7 against G1202R- and I1171N/S/T compound mutants, respectively, we first compared the cellular IC50 values of lorlatinib for nonmutant, single and compound mutant ALK. Consistent with previous reports (13), both single mutations G1202R and I1171N resulted in relative increases in lorlatinib IC50 values (~88-fold and ~43-fold, respectively) compared to nonmutant ALK; however, the absolute IC50 values remained within the range of clinical exposures (~200 nM) (15) (Fig. 6A, B, Extended Data Fig. 9A). Compound mutations resulted in a further relative increase in lorlatinib IC50 of approximately 10-fold (range, ~9–26 fold), increasing the absolute IC50 values above the upper range of plasma concentrations that are clinically achievable. By comparison, LA7 and LA9 exhibited IC50 values against I1171N- and G1202R-containing compound mutations that ranged from ~6 to 139-fold lower than lorlatinib, respectively (Fig. 6C, D, Extended Data Fig. 9B). Notably, LA7 and LA9 exhibited a similar relative improvement in potency against I1171N and G1202R single mutations, respectively (~11 and ~18 fold, respectively), suggesting that the improved drug efficacy against compound mutations is predominantly attributable to improved ability to overcome either the I1171N or G1202R single site substitutions. Consistent with this notion, neither drug was capable of overcoming the putative ALK I1171N+G1202R compound mutation (Fig. 2D).
Fig. 6. Cellular IC50 changes with single and compound ALK mutations.

(A)-(B) Comparison of absolute and fold shift in cellular IC50 values for nonmutant, single and compound ALK mutants (corresponding to data shown in Extended Data Fig. 9A). G1202R (A) and I1171N (B) single point mutations result in large shift in IC50 for lorlatinib; however, this remains within range of clinical exposure (<200nM, indicated by dashed line). Compound mutations result in comparable fold shift in IC50 values of lorlatinib, LA7 and LA9 relative to single mutations; however, the improved potency of LA7 and LA9 against single mutations results in substantial improvement in IC50 against compound mutations. (C)-(D) Fold improvement of LA7 (C) or LA9 (D) compared to lorlatinib against single and compound mutations (calculated by dividing the lorlatinib IC50 by the LA IC50, corresponding to data shown in Extended Data Fig. 9B).
Next, we examined the structural basis for the improved potency of LA9 and LA7 against G1202R- and I1171N-compound mutants, respectively. Comparison of the co-crystal structures of lorlatinib, LA7 and LA9 bound to nonmutant ALK confirmed similar binding modes of all three ligands, with the open conformation LA7 and LA9 adopting a conformation similar to the constrained macrocyclic lorlatinib (Fig. 7A). Previously, we reported that the introduction of the G1202R solvent front mutation increases steric bulk near the pyrazole ring of lorlatinib, leading to partial destabilization of ligand binding and an altered P-loop conformation (18). Modeling the G1202R mutation based on the LA9/ALK co-crystal structure revealed that the orientation of the unsubstituted phenyl ring near G1202 enables accommodation of the G1202R substitution without significant steric clash or disruption of the ligand position (Fig. 7B). Furthermore, the G1202R side chain amino group makes a new productive interaction with the lactam carbonyl of LA9. This interaction was preserved upon modeling the G1202R+L1196M compound mutation, as were key interactions with hinge residues (Extended Data Fig. 10A), consistent with our experimental observation that LA9 exhibits improved potency against both G1202R and G1202R-containing compound mutations.
Figure 7. Structural basis for selectivity of LA7 and LA9 against ALK compound mutations.

(A) Co-crystal structures of lorlatinib (4CLI, 2.05 Å), LA7 (7R7R, 1.93 Å) and LA9 (7R7K, 1.83 Å) in the wild-type (WT) ALK kinase domain. Left panels highlight residues mutated in resistant patients. Right panels show structure of the ligand binding pocket. (B) Co-crystal structure of LA9 bound to WT ALK (ligand and protein colored gray) superimposed with energy-minimized model of G1202R (ligand and protein colored blue). Solvent front G1202/G1202R and D1203 residues are bolded for emphasis. Productive interactions between both LA9/WT and LA9/G1202R are shown by dotted lines, including a novel interaction between G1202R side chain –NH2 and the lactam carbonyl of LA9 (distances are reported in Angstroms; red spheres represent structural water molecules). Note the minimal movement of both ligand (LA9) and protein (G1202R) relative to LA9/WT co-crystal structure. (C) Co-crystal structure of LA7 bound to WT ALK (ligand and protein colored gray) superimposed with energy-minimized model of I1171N (ligand and protein colored pink), highlighting nearly identical productive interactions, including a novel network of hydrogen bonds between thiazole ring hydroxyl groups and solvent front residues (1201–1203, bolded). Distances are I1171N (bold) and WT (in parentheses). (D) Co-crystal structure of LA7 in WT ALK (ligand and protein colored gray) superimposed with energy-minimized model of G1202R (ligand and protein colored blue) showing significant movement of both ligand and protein relative to LA7/WT co-crystal structure. This movement disrupts key protein ligand interactions, including the two hydroxyl groups with G1201 and G1203 shown in panel D. Distances of LA7/G1202R interactions are shown. (E) Overlap of binding modes of lorlatinib, LA7 and LA9 (in WT ALK) revealing orientation of key structural features conferring mutant selectivity.
In contrast to the predominantly steric constraints introduced by the solvent front G1202R substitution, I1171N lies within the hydrophobic core and confers ALK TKI resistance by increasing the catalytic activity of ALK (23–25). Interestingly, LA7 exhibited improved potency against nonmutant ALK as well as I1171N (Extended Data Fig. 9C), suggesting favorable interactions between the ligand and wild-type residues. Examination of the co-crystal structure of LA7 bound to nonmutant ALK revealed stabilizing interactions between the two hydroxyl groups of the ligand thiazole ring and the side chain carboxylate of D1203, as well as a hydrogen bond network involving the backbone carbonyl of G1201, the amino nitrogen of D1203 and a structural water (Fig. 7C, Extended Data Fig. 10B), that are not present with LA9 or lorlatinib. Modeling the I1171N mutation, which is distant from the ligand binding pocket, had minimal impact on LA7 binding, as shown by the nearly superimposable structures of LA7 bound to nonmutant and I1171N ALK (Fig. 7C, Extended Data Fig. 10B). Furthermore, the solvent front interactions were largely retained upon modeling the I1171N+D1203N compound mutation (Extended Data Fig. 10B), consistent with our experimental observation that LA7 retains potency against I1171N-containing compound mutations. While not involving interactions with G1202 directly, the position of substitutions on the thiazole ring of LA7 adjacent to solvent front residues results in a significant steric clash upon G1202R substitution (both single G1202R and compound G1202R+L1196M substitutions). This disrupts key protein-ligand interactions (including hinge binding and the two hydroxyl groups with G1201 and D1203) and causes significant movement of ligand relative to the LA7/ALK co-crystal structure (Fig. 7D, Extended Data Fig. 10C). Thus, mutually exclusive molecular features of LA7 and LA9 enable improved targeting of I1171N/S/T- and G1202R-class mutations, respectively.
Finally, we examined the structural basis for overcoming L1198F-containing compound mutations. Originally, the lorlatinib nitrile group was installed to improve ALK-selectivity based on the observation that most kinases contain either a phenylalanine or tyrosine corresponding to the position at L1198 (14). The L1198F mutation coupled with the rigid nature of the lorlatinib macrocycle causes a steric clash between the phenyl group and nitrile group of the inhibitor, reducing affinity (21). Five out of the six selected LAs (Fig. 2D), including LA7 and LA9, lack the nitrile and consistently exhibited high potency against L1198F compound mutations (Extended Data Fig. 4C, 5). Consistent with this result, structural modeling of L1198F on the LA7 and LA9/ALK co-crystal structures revealed that the mutation had minimal impact on ligand binding (Extended Data Fig. 10D–F). LA4, on the other hand, contains a nitrile but is otherwise identical to LA7 (Extended Data Fig. 3). Comparison of cellular IC50 values of LA4 vs LA7 across compound mutants revealed a significant decrease in potency of LA4 compared to LA7 specifically against L1198F-containing compound mutations (Extended Data Fig. 10H). Modeling of L1198F on the LA4/ALK co-crystal structure revealed orientation of the nitrile toward L1198F (Extended Data Fig. 10G), albeit with less steric clash compared to lorlatinib (owing to the more flexible LA4 structure), consistent with the decreased activity against L1198F mutants compared to LA7 (Extended Data Fig. 10I). Collectively, these findings reveal the structural basis for a rational approach to targeting diverse compound resistance mutations classified according G1202R and I1171N (Fig. 7E).
DISCUSSION
Currently, most patients with advanced ALK-positive lung cancer are treated using a sequential therapy paradigm with successive generations of ALK TKIs, often culminating with the third-generation and most potent TKI, lorlatinib. While this sequential targeted therapy approach has enabled prolonged survival and quality of life, acquired resistance to lorlatinib is inevitable, and treatment options following lorlatinib remain an area of critical unmet need. Our current study extends prior work by our group and others demonstrating that in a subset of these patients, acquired resistance is caused by recurrent lorlatinib-refractory ALK compound mutations that sequentially evolve from ALK single mutant clones arising after prior treatment with first- or second-generation ALK TKIs (18–20). Efforts to develop fourth-generation TKIs that can overcome ALK compound mutations are underway (26, 27); however, the diverse spectrum of ALK compound mutations observed in patients after lorlatinib suggests that one approach is unlikely to be effective for all patients. Indeed, our prior finding that the acquisition of a compound ALK mutation containing L1198F confers resensitization to crizotinib (21) is not always recapitulated with other compound mutations, many of which remain largely refractory to currently available ALK TKIs. By analyzing a structurally diverse set of lorlatinib analogs, we identified molecules with distinct sensitivity patterns against classes of ALK compound mutations grouped according to the predominant resistance mutation that exists pre-lorlatinib, G1202R or I1171N/S/T. As these represent the two most common resistance mutations observed in patients progressing on alectinib (the preferred first-line ALK TKI in the recent years), our findings offer a roadmap for rational development of ALK inhibitors targeting subgroups of ALK-positive lung cancers with resistance to lorlatinib.
Comparison of potencies of lorlatinib and lorlatinib analogs against diverse compound mutations, integrated with our analysis of co-crystal structures of lorlatinib analogs bound to ALK, reveals the structural basis for overcoming ALK compound mutations. While single residue substitutions in the kinase domain are sufficient to cause resistance to first- and second-generation ALK TKIs, lorlatinib binds more potently and is able to inhibit G1202R and I1171N/S/T resistance mutations at clinically achievable drug exposures (~200 nM) (15), However, the relative potency of lorlatinib against these mutations is reduced compared to nonmutant ALK, and our analysis reveals this to be a major factor contributing to clinical resistance conferred by subsequent compound mutations. In virtually all cases, the introduction of a second mutation in ALK that already harbors a G1202R or I1171N single point mutation leads to a further ~10-fold decrease in the potency of both lorlatinib and the lorlatinib analogs. For lorlatinib, this decreased potency translates to a requirement for drug concentrations that are not clinically achievable. On the other hand, for the lorlatinib analogs tested, we identified multiple compounds including LA9 and LA7 with markedly improved potency (≥10-fold) against ALK G1202R or I1171N/S/T single mutations. Thus, even with a similar relative reduction in potency incurred upon the introduction of a second mutation, LA9 and LA7 exhibited >10-fold increased potency relative to lorlatinib against diverse G1202R or I1171N/S/T compound mutations, respectively, leading to improved activity in vitro and in vivo.
From a structural perspective, LA7 and LA9 achieve improved targeting of I1171N/S/T and G1202R substitutions by distinct mechanisms. Kinase domain substitutions may contribute to resistance in complex ways, including by reducing the effectiveness of the ligand’s ability to bind and block ATP, or alternatively by increasing the catalytic efficiency of the kinase (which in turn may derive from changes in substrate binding, ATP binding, or conformational changes in the kinase that favor the active form). Our prior work revealed that the G1202R solvent front mutation in ALK increases steric bulk near the pyrazole ring of lorlatinib, leading to destabilization of ligand binding and an altered P-loop conformation (18). Consistent with this, we observed that the lorlatinib-like macrocyclic analogs LA2 and LA19, which lack the N-methyl pyrazole substitution, exhibited improved potency against G1202R and G1202R compound mutations compared to lorlatinib (Fig. 2B, C), further highlighting the importance of decreasing ligand bulk adjacent to G1202. Modeling the G1202R substitution based on the co-crystal structure of LA9 bound to wild-type ALK revealed that LA9 can maintain identical binding without significant steric clash, consistent with the nearly identical cellular IC50 values of LA9 against nonmutant and G1202R ALK. In contrast to G1202 positioned in the ALK solvent front, I1171 is buried within the ALK hydrophobic core, and I1171N/S/T mutations confer resistance by increasing catalytic activity of the kinase rather than steric hindrance of ligand binding (23, 24). Analysis of the co-crystal structure of LA7 bound to wild-type ALK revealed several interactions between the hydroxyl groups of the ligand and the D1203 side chain carboxylate and amino nitrogen, the G1201 backbone carbonyl and a structural water – all together forming a network of optimal interactions that strengthen ligand binding. Notably, the features of LA9 and LA7 that lead to improved selectivity against G1202R vs I1171N/S/T appear to be mutually exclusive. The increased functionality of the thiazole ring of LA7 that interacts with G1201 and D1203 becomes a liability in the presence of the G1202R mutation due to increased steric clash. Conversely, the lack of functionality on the phenyl ring of LA9 allows accommodation of the sterically bulky G1202R substitution, but prevents LA9 from forming similar favorable interactions with adjacent G1201 and D1203. It is also worth noting that in contrast to the closed and constrained macrocyclic structure of lorlatinib, both LA7 and LA9 are open structures with greater molecular flexibility. This allows them to adopt a similar pose to lorlatinib within the ATP binding pocket of nonmutant ALK (Fig. 7A, E), while likely also allowing them to better accommodate subtle changes to the overall protein structure that accompany the resistance mutations. Collectively, these findings provide a structure-based conceptual framework for different approaches to target distinct classes of ALK compound mutations that emerge following treatment with sequential second- and third-generation ALK inhibitors.
Given the current dearth of therapeutic options for patients after lorlatinib, several efforts to target lorlatinib-resistant compound ALK mutations are underway. Most notably, fourth-generation ALK inhibitors to target G1202R compound resistance mutations are being developed. As an example, TPX-0131 has demonstrated excellent potency against G1202R compound mutations in preclinical models (26). Reminiscent of our results, TPX-0131 achieves improved activity through increased potency against the G1202R single mutation and does not target I1171 compound mutations, owing to a compact molecular structure that positions an amide group near the solvent front residues that is presumably capable of avoiding a steric clash with the G1202R substitution (in contrast to the N-methyl pyrazole ring of lorlatinib) (26). In a similar fashion, NVL-655 harbors preclinical potency against G1202R compound mutations (27). Other studies have reported repurposing of available kinase inhibitors to overcome specific compound mutations. Okada et al. found that AG-957, initially developed as a BCR-ABL inhibitor, suppressed the viability of cells harboring G1202R+L1196M (IC50: 87 nM) (20), while Mizuta et al. found that gilteritinib, a FLT3 inhibitor approved for the treatment of FLT3-mutant AML, could overcome I1171N compound mutations but not G1202R compound mutations (28). These reports together with our findings support that distinct compounds may be required in clinic to overcome G1202R- vs I1171N/S/T-based (vs other) ALK compound mutations.
Importantly, these results further underscore the challenges inherent to sequential targeted therapies and subsequent evolution of heterogeneous compound mutations. While our findings point to the G1202R- and I1171N/S/T-containing ALK compound mutations as the predominant lorlatinib-resistant mutations and therefore, the most urgent and salient target for drug development, we anticipate that not all of the broad array of ALK compound mutations that emerge in patients after lorlatinib will ultimately be surmountable, even with fourth-generation ALK inhibitors in development. Furthermore, tumor heterogeneity and polyclonal resistance can emerge (19), including with different resistant clones harboring distinct compound ALK resistance mutations; and although in these cases, combinations of ALK inhibitors with distinct sensitivity profiles may conceptually offer a more comprehensive coverage of the compound mutations, combinations are often challenging to pursue in clinic due to augmented toxicities. Therefore, a more effective strategy may be to suppress the emergence of ALK mutations upfront. Our group previously showed in accelerated mutagenesis screening of cells expressing nonmutant EML4-ALK (modeling treatment-naïve tumors) that no single ALK mutation could confer resistance to lorlatinib, whereas in cells harboring pre-existing single ALK mutations (modeling tumors resistant to prior ALK TKIs), compound ALK mutations could emerge to confer lorlatinib resistance (18). Thus, upfront lorlatinib therapy may suppress, or at least delay, the development of ALK mutations. Recently, the interim analysis of the phase III CROWN study comparing front-line lorlatinib to crizotinib was published, demonstrating an impressive prolongation in the progression-free survival with the hazard ratio for disease progression or death of 0.28 (95% CI, 0.19–0.41) that was numerically the lowest among all phase III trials that evaluated next-generation ALK TKIs in the first-line setting (7–9, 29). On the basis of these results, the FDA approval of lorlatinib was expanded to include the first-line indication. While numerous considerations will ultimately factor into which next-generation ALK inhibitor is selected for first-line use in patients, one potential benefit of upfront lorlatinib may be in delaying or suppressing on-target resistance and circumventing the emergence of refractory compound ALK mutations, at least some of which may not be able to be overcome in patients. On the other hand, such an approach may result in the selection for ALK-independent resistance requiring combination strategies. Deeper understanding of resistance mechanisms after the various next-generation ALK TKIs in the first-line setting, integrated with the identification of potential biomarkers that predict whether certain tumors are more likely to develop ALK-dependent versus ALK-independent resistance, may additionally help optimize first-line treatment selection for an individual patient.
In summary, we have provided here an expanded catalogue of compound ALK mutations observed in patients treated with lorlatinib, highlighting G1202R- and I1171N/S/T-containing mutations as occurring most commonly after sequential therapy. We have shown through functional and structural analysis of a diverse set of lorlatinib analogs that distinct compounds may be required to target selective subsets of mutant ALK kinases, including G1202R- vs I1171N/S/T-harboring compound mutants. Our data offer a framework for the functional classification of heterogeneous compound mutations to guide the development of novel therapeutic strategies addressing on-target resistance after sequential targeted therapy.
METHODS
Patients and Tumor Genotyping
Patients were included if they had the diagnosis of advanced ALK-positive NSCLC, were treated at Massachusetts General Hospital (MGH), had disease progression on lorlatinib and underwent resistant tissue biopsies with genotyping after providing informed consent between November 2014 and October 2020. All cases underwent histopathology review on a clinical basis. Genotyping was performed using: the MGH SNaPshot DNA-based genotyping panel and a separate RNA-based NGS assay (Solid Fusion Assay) as previously described (30), FoundationOne (Foundation Medicine, Inc.; Cambridge, MA), Caris NGS (Caris Life Sciences; Irving, TX), Tempus xT panel (Tempus; Chicago, IL), Oncomine Comprehensive Assay (Cornell, New York, NY), MD Anderson Solid Tumor Genomic Assay (MD Anderson Cancer Center; Houston, TX), UW-Oncoplex (University of Washington; Seattle, WA), 556-gene panel NGS (China), NeoGenomics ALK Analysis (NeoGenomics; Fort Myers, FL), and Oncopanel (Brigham and Women’s Hospital; Boston, MA).
Electronic medical records were retrospectively reviewed to extract data on clinical, pathologic, and molecular features. Data were updated as of January 2021. Time to progression (TTP) was measured from the time of therapy initiation to clinical/radiologic disease progression. Time to treatment discontinuation (TTD) was measured from the time of lorlatinib initiation to lorlatinib discontinuation. Patients continuing on treatment at the time of data cut-off were censored at their last follow-up. The study was performed under an Institutional Review Board–approved protocol at Massachusetts General Hospital. This study was conducted in accordance with the Belmont Report and the U.S. Common Rule.
Plasma Biopsy Dataset
We queried a de-identified dataset at Guardant Health (Redwood City, CA) comprised of plasma specimens analyzed between October 2015 and December 2020 in order to identify specimens harboring ≥2 ALK kinase domain mutations. All plasma specimens were genotyped using the Guardant360 cell-free DNA (cfDNA) assay, as previously described (31). The Guardant360 assay detects ALK fusions in addition to mutations in critical exons of ALK, including exons 22–25 which encode the ALK kinase domain. The allelic configuration of ALK mutations (i.e., cis vs trans) was determined for a subset of specimens with multiple ALK mutations arising in the same exon.
Cell Lines and Reagents
Ba/F3 immortalized murine bone marrow-derived pro-B cells were obtained from the RIKEN BRC Cell Bank (RCB0805, RIKEN BioResource Center) in 2010 and cultured in DMEM with 10% FBS with (parental) or without (EML4-ALK) IL3 (0.5 ng/mL). cDNAs encoding EML4-ALK variant 1 (E13; A20) or variant 3a (E6a; A20) containing different point mutations were cloned into retroviral expression vectors, and Ba/F3 cells were infected with the virus as described previously (24). After retroviral infection, Ba/F3 cells were selected in puromycin (0.7 μg/mL) for 2 weeks. IL3 was withdrawn from the culture medium for more than 2 weeks before experiments. NIH/3T3 murine fibroblast cells (CRL-1658) were purchased from ATCC in 2014 and cultured in DMEM with 10% FBS. The 3T3 cells were infected with the vectors of EML4-ALK variant 3a containing I1171N+D1203N or G1202R+L1196M mutations and selected in puromycin (2 μg/mL). The MGH953-4 cell line was developed from a malignant pleural effusion from a patient with ALK-positive NSCLC who had progressed on alectinib, and the MGH953-7 cell line was subsequently developed from a patient-derived xenograft (PDX) model established from a lorlatinib-resistant malignant pleural effusion from the same patient. The MGH990-2 cell line was developed from an adrenal biopsy from a patient with lorlatinib-resistant ALK-positive NSCLC. Prior to cell line generation, the patients signed informed consent to participate in a Dana Farber/Harvard Cancer Center Institutional Review Board-approved protocol giving permission for research to be performed on their sample. MGH953-4, MGH953-7, and MGH990-2 were cultured in RPMI1640 with 10% FBS. Cell lines were sequenced to confirm the presence of ALK rearrangement and ALK mutations. Additional authentication was performed by SNP fingerprinting. Crizotinib and ceritinib were purchased from Selleck Chemicals. Alectinib and brigatinib were purchased from MedChem Express. Lorlatinib and lorlatinib analogs were provided by Pfizer, Inc.
Mutagenesis
QuickChange XL Site-Directed mutagenesis kit (Agilent Technologies) was used to generate point mutations in the expression vectors of EML4-ALK in accordance with the manufacturer’s protocol.
Survival Assays
Ba/F3 cells (2,000) or patient derived cells (5,000) were plated in triplicate into 96-well plates. Cells were incubated with CellTiter-Glo (CTG; Promega) after 2 days or 5 days after drug treatment, and luminescence was measured with a SpectraMax M5 Multi-Mode Microplate Reader (Molecular Devices, LLC). GraphPad Prism (GraphPad Software) was used to graphically display data and determine IC50 values by a nonlinear regression model utilizing a four-parameter analytic method. The screening method is summarized in Supplementary Table 7.
14-Day Cell Viability Assay
MGH953-7 and MGH990-2 cells were seeded at 1,000 cells/well in replicate 96-well plates. One plate was immediately frozen at –80°C (“Day 0”). The next day, cells on the remaining plates were drugged with LA7, LA9 or lorlatinib (300 nM of each drug for MGH953-7, 100 nM of each drug for MGH990-2). Media and drug were replaced every 3–4 days. Plates were frozen on days 3, 7, 10 and 14. CTG assay was performed on all frozen plates simultaneously. CTG values were normalized to corresponding “Day 0” values and are displayed on the y-axis in Fig. 4D as relative cell proliferation.
Western Blot Analysis
Ba/F3 cells and patient derived cell lines were treated for 6 hours with lorlatinib, LA7 or LA9 unless otherwise noted. Total protein lysates were analyzed by western blotting with the following antibodies and dilutions. (all from Cell Signaling Technology): phospho-ALK Y1604 (3341, 1:1000), phospho-ALK Y1282/83 (9687, 1:1000), ALK (3633, 1:1000), phospho-AKT S473 (4060, 1:1000), AKT (4691, 1:1000), phospho-ERK1/2 T202/Y204 (9101, 1:1000), ERK1/2 (9102, 1:1000), phospho-S6 S240/244 (5364, 1:10,000), S6 (2217, 1:10,000), and β-Actin (4970, 1:1000).
Development of Xenografts and in vivo Pharmacological Studies
All animal studies were conducted in accordance with the guidelines as published in the Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of MGH (Protocol number: 2008N000095). Female NSG mice and nude (Nu/Nu) mice aged 6 to 8 weeks were obtained from Jackson Laboratory and MGH Cox-7 facility, respectively. Mice were maintained in laminar flow units in sterile filter-top cages with Alpha-Dri bedding in a 12/12 dark/light cycle, at controlled temperature (20–26°C) and humidity conditions (30–70%). MGH953-7 cells were implanted subcutaneously into the flank of NSG mice. Similarly, 2 × 106 3T3-ALK I1171N+D1203N or G1202R+L11196M cells in 0.2 mL 50% BD Matrigel Basement Membrane Matrix in PBS were subcutaneously injected into the flank of nude mice. Mice were randomized into groups once the tumors had attained a volume of 200 mm3. The treatment groups were treated 5 days (MGH953-7) or 7 days (NIH3T3) per week with drug solution dissolved in acid water (lorlatinib and LA9) or water (LA7) by oral gavage. Tumor volumes were measured twice weekly and calculated using the formula: mm3 = 0.52 × L × W2. For pharmacodynamics studies, tumor-bearing mice were administered drugs daily for 3 days, and tumors were harvested 3 hours after the last treatment for western blot and immunohistochemistry. The mice were euthanized when the tumor size exceeded 20 mm in any one dimension according to the IACUC-approved protocol.
Immunohistochemistry
Tissue sections 4 μm thick were cut from 10% buffered formalin-fixed and paraffin-embedded (FFPE) mice tumor tissue blocks. The deparaffinized and rehydrated sections were autoclaved in Bond TM Epitope Retrieval 2-1L EDTA (pH 9.0) at 100°C for 20 minutes for antigen retrieval. They were then incubated with rabbit polyclonal anti-phospho ALK (Y1507) antibody diluted 1:50 (ABCAM™) for 15 minutes at room temperature. Immunoactivity was detected with mixed DAB refine after 10 minutes (Leica Biosystems). The sections were counterstained with hematoxylin for 5 minutes and dehydrated prior to coverslipping. The staining was tested in EGFR-mutant NSCLC PDXs (negative control) and ALK-positive NSCLC PDXs (positive control) (Extended Data Fig. 8B).
Pharmacokinetic Analysis
During the in vivo pharmacology studies, serial whole blood samples (~8 uL/time point) were collected from the tail vein of mice (n=3/group) at 0 (pre-dose),1, 4, 7, and 24 hours after the last dose of a 5-day or longer treatment period. The concentration of lorlatinib, LA7 or LA9 in whole blood was determined using quantitative LC-MS/MS methods. Pharmacokinetic parameters for each analyte were calculated from individual animal concentration-time profiles using standard non-compartmental methods. The unbound fraction (fu,p) of each analyte in NOD-scid mouse plasma was determined in vitro using an equilibrium dialysis method as described previously (32). The blood-to-plasma concentration ratio (BPR) of each analyte was determined in vitro in fresh whole mouse (CD-1) blood as described previously (32). The unbound fraction in blood (fu,b) for each analyte was calculated from their respective measured fu,p and BPR values as follows: fu,b = fu,p / BPR.
Structural Modeling of Lorlatinib Analogs Bound to mutant ALK
Co-crystal structures of wild-type ALK were used as the starting point for modelling mutations. Mutations were introduced and side chains minimized using the OPLS2003 forcefield. Protein and ligand minimizations were also performed with flexible protein using OPLS2003 forcefield. Original and protein-ligand minimized structures were overlapped to visualize extent of protein movement, which inversely correlates with fit from original. The structural and refinement statistics are summarized in Supplementary Table 8.
Statistics and Reproducibility
For the clinical outcome data, TTP and TTD curves were estimated using the Kaplan-Meier method with the differences between groups expressed as a hazard ratio (HR) and compared using the log-rank test. The Cox proportional hazards model was used to estimate the HR for TTP, while the HR for TTD was estimated as a ratio of the observed group medians under the assumption of an exponential distribution as none of the patients with known baseline ALK mutations had discontinued treatment earlier than any of those without known baseline ALK mutations. All statistical analyses of clinical outcomes were performed using Stata version 14.2. All Western blots were performed at least twice, and representative images are shown. Immunohistochemistry was performed once after antibody validation with repeated controls. In animal studies, the sample size was calculated by one-way Analysis of Variance (ANOVA) with the assumption that the standard deviation of the tumor volumes at the end of treatments will be 350 mm3 and the treatment effect will be 2.5–3 fold of the standard deviation. For a study of four treatment groups with a significance level of 0.05, 80% power is attained with 4–5 animals per group. Five or six mice per group were used in all studies. Mice were assigned randomly to each treatment group and blinding was not used in this study. No data were excluded from the analyses. Data distribution was assumed to be normal but this was not formally tested.
Extended Data
Extended Data Fig. 1. Clinical outcomes on lorlatinib.

(A) Time to progression on lorlatinib according to presence (N = 13 patients) or absence (N = 8 patients) of a baseline ALK mutation. (B) Time to treatment discontinuation from lorlatinib according to the presence (N = 13 patients) or absence (N = 8 patients) of a baseline ALK mutation. Only those patients with available pre-lorlatinib tissue biopsy and known status of baseline ALK mutations are included in these analyses. Both time to progression and time to treatment discontinuation curves were estimated using the Kaplan-Meier method.
Extended Data Fig. 2. Sensitivity of compound ALK mutations to currently FDA-approved ALK tyrosine kinase inhibitors.
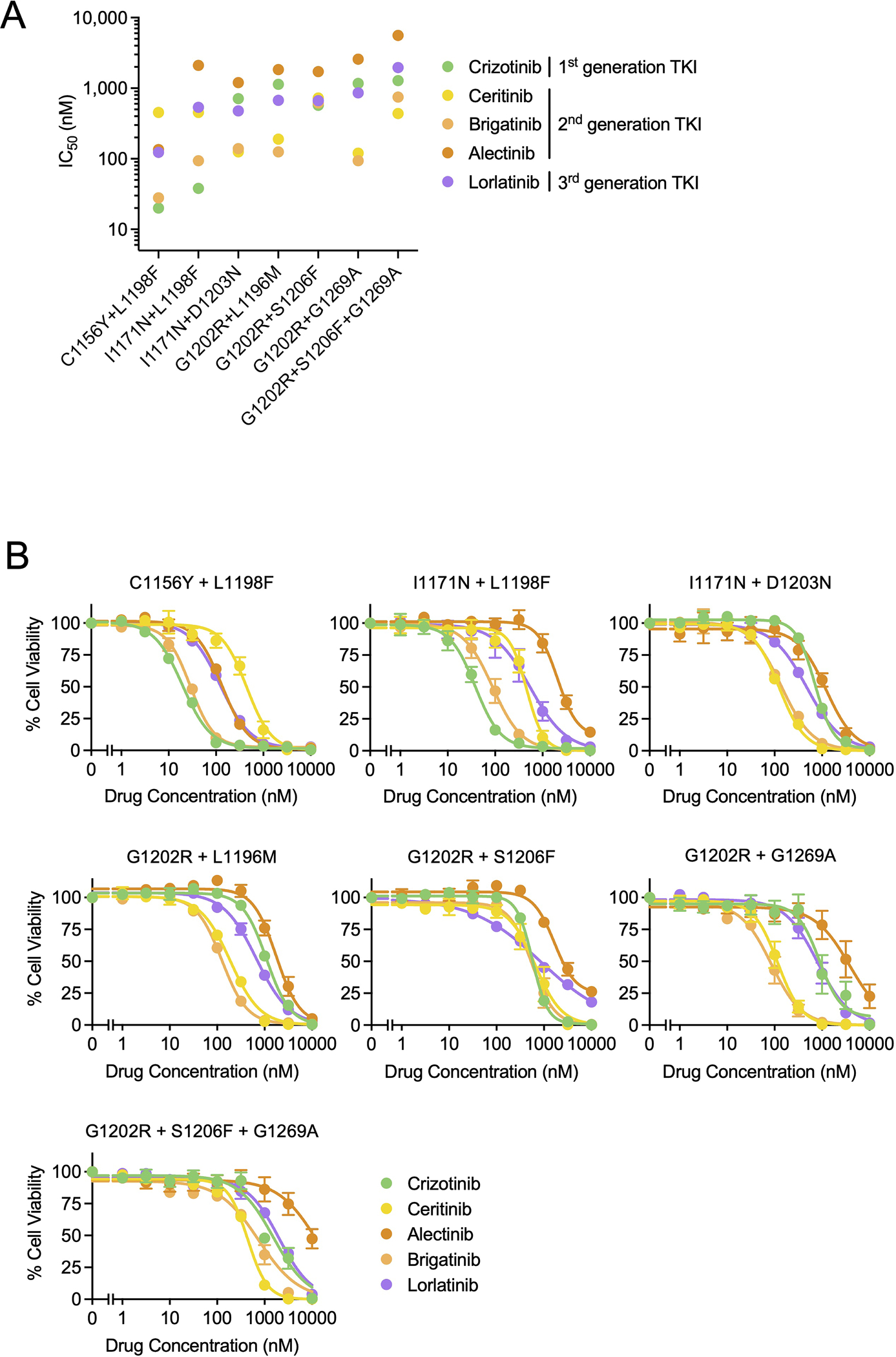
Cellular IC50 values (A) and dose response curves (B) of Ba/F3 cells expressing clinical ALK compound mutations to crizotinib, ceritinib, brigatinib, alectinib, and lorlatinib. Data correspond to the heatmap shown in Fig. 1E. Data are mean and SEM of three independent experiments.
Extended Data Fig. 3. Molecular structures of lorlatinib analogs (LAs) 1–20.

Extended Data Fig. 4. Drug screening with lorlatinib analogs using Ba/F3 ALK mutation models.

(A) Cellular IC50 values of each LA against single ALK mutations (top) and Ba/F3 parental and PC9 cells (bottom). This data corresponds to the heatmap in Fig. 2B (N = 1). When the calculated IC50 exceeded the highest concentration used in the experiment (10μM), the value was plotted as >10000. (B) Cellular IC50 values of 12 selected LAs against clinical ALK compound mutations. This data corresponds to the heatmap in Fig. 2C (N = 1). (C) Cellular IC50 values of 6 selected LAs against an expanded set of clinical and putative compound ALK mutations. This data corresponds to the heatmap in Fig. 2D (N = 1).
Extended Data Fig. 5. Drug dose response curves of lorlatinib analogs against Ba/F3 ALK mutation models.

Cell viability assays performed with Ba/ F3 cells expressing EML4-ALK with the indicated compound mutations treated with 6 LAs or lorlatinib for 48 hours. The viabilities were measured with CellTiter-Glo assay. These data correspond to the heatmap in Fig. 2D and dot plots in Extended Data Fig. 4C. The experiments were performed once.
Extended Data Fig. 6. Distinct activity of LA7 and LA9 in Ba/F3 models and the patient-derived models.

A) Western blot analysis performed on compound mutation Ba/F3 models. Ba/F3 cells expressing nonmutant EML4-ALK, EML4-ALK G1202R + G1269A, G1202R + S1206F + G1269A or I1171N + L1198F were treated with DMSO, LA7, LA9, or lorlatinib for 6 hours, and total cell lysates were analyzed by western blotting. The images are representative of at least two repeats with similar results. (B) Comparison of LA7 and LA9 with currently approved ALK inhibitors. Cellular IC50 values of LA7, LA9 and currently approved ALK inhibitors including lorlatinib against clinical ALK compound mutations. IC50 values for approved ALK inhibitors are replotted from Extended Data Fig. 2A for comparison purposes. Data are mean of three independent experiments. (C) MGH953-4 (G1202R) cells were treated with the indicated drugs for 5 days and cell viability assessed with CellTiter-Glo assay. Data are mean and SEM of three independent experiments. (D) Cellular IC50 values of LA7 and LA9 compared to approved ALK inhibitors in MGH953-4, MGH953-7 (G1202R + L1196M), and MGH990-2 (I1171N + D1203N) cells. Data are mean of three independent experiments. (E) The ratio of cell viability with LA7 or LA9 in MGH953-7 (G1202R + L1196M) at 300 nM or MGH990-2 (I1171N + D1203N) at 100 nM, corresponding to Fig. 4D. Data are mean and SEM of three independent experiments.
Extended Data Fig. 7. In vivo activity of LA7 and LA9.
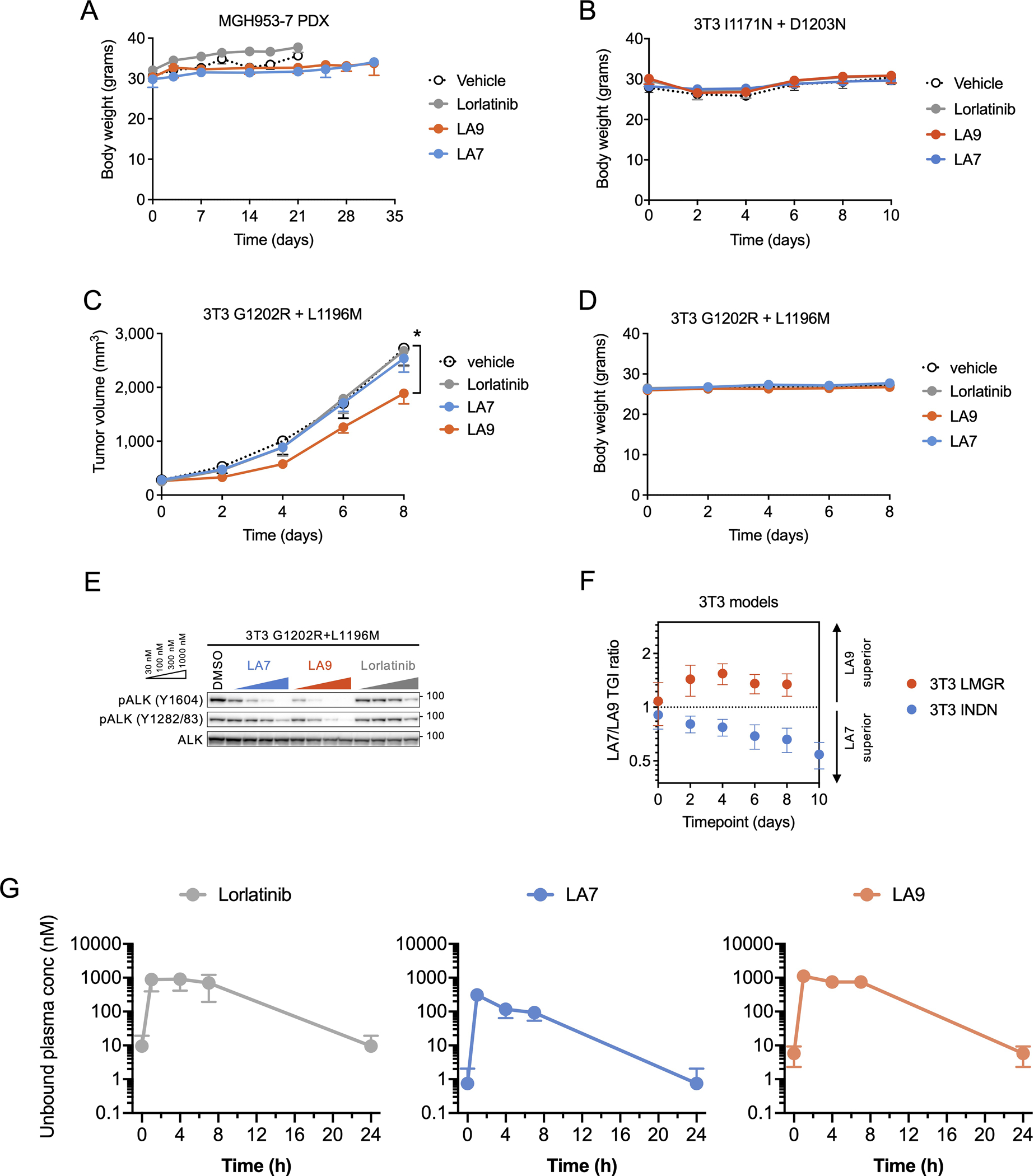
(A) Body weight of mice bearing MGH953-7 during the course of treatment (N = 6 mice per group). (B) Body weight of mice bearing NIH3T3 EML4-ALK I1171N + D1203N xenograft tumors during the course of treatment (N = 6 per group). (C) Change in tumor volume of NIH3T3 G1202R + L1196M xenograft tumors. Mice were treated with lorlatinib 6 mpk QD, LA7 20 mpk QD, or LA9 40 mpk QD (N = 6 mice per group). Tumors treated with LA9 were significantly smaller on day 8 compared to those with vehicle (p = 0.048, two-tailed t-test). (D) Body weight of mice bearing NIH3T3 EML4-ALK I1171N + D1203N xenograft tumors during the course of treatment (N = 6 mice per group). (E) Western blot analysis of NIH3T3 cells expressing ALK G1202R + L1196M and treated with the indicated drugs for 1 hour. (F) The ratio of tumor growth inhibition (TGI) with LA7 and LA9 in NIH3T3 EML4-ALK I1171N + D1203N or G1202R + L1196M xenograft tumors, corresponding to Fig. 5D and Extended Data Fig. 7C. Error bars indicate SEM for 6 mice per group. (G) Unbound systemic concentrations of lorlatinib, LA7 and LA9 were calculated based on the total drug concentrations in whole blood measured in vivo and the unbound fraction in blood (fu,b; calculated as fu,p / BPR). Data are mean and SEM of three mice.
Extended Data Fig. 8. Phospho-ALK immunohistochemistry of MGH953-7 PDX tumors and control specimens.

(A) MGH953-7 PDX tumors treated with lorlatinib, LA7 or LA9 were harvested after 3-day treatment and FFPE sections were stained with anti-phospho-ALK antibody. Scale bar in low magnification images: 300μm. Scale bar in high magnification images: 50μm. (B) Patient-derived xenografts (PDXs) of two EGFR-mutated NSCLC were used as negative controls and PDXs of two ALK-positive NSCLC were used as positive controls. FFPE sections were stained with anti-phospho-ALK antibody. Phospho-ALK is localized in the cytoplasm and exhibits a diffuse staining pattern in tumor cells. Scale bar in low magnification images: 300μm. Scale bar in high magnification images: 20μm.
Extended Data Fig. 9. Relative changes in IC50 values for lorlatinib, LA7 and LA9 against single and compound ALK mutations.
(A) Lorlatinib, LA7 and LA9 cellular IC50 values (upper left) and IC50 ratios (lower right) of single mutant vs nonmutant ALK (left panel) or compound mutant vs single mutant ALK (middle and right panels). IC50 values correspond to data shown in the Source Data. (B) Fold improvement of LA7 or LA9 compared to lorlatinib against single and compound mutations (calculated by dividing the lorlatinib IC50 by the LA IC50). (C) Cell viability assays performed with Ba/F3 cells expressing nonmutant EML4-ALK with 0.01–100 nM lorlatinib, LA7 or LA9 for 48 hours. The viabilities were measured with CellTiter-Glo assay. Data are mean and SEM of three independent experiments.
Extended Data Fig. 10. Structural basis for selectivity of lorlatinib analogs against ALK compound mutations and the L1198F mutation.

(A) Energy-minimized model of LA9 bound to G1202R/L1196M. Solvent front G1202R and D1203 residues are bolded for emphasis. Productive interactions shown by dotted lines are similar to G1202R single mutant (see Fig. 7B). (B) Co-crystal structure of LA7 bound to wild-type ALK (upper) and energy minimized models of LA7 bound to I1171N (middle) and I1171N/D1203N (bottom) superimposed with energy-minimized model of I1171N (ligand and protein colored pink) showing that the network of hydrogen bonds between thiazole ring hydroxyl groups and solvent front residues are largely preserved. (C) Energy minimized model of LA7 bound to G1202R/L1196M showing disruption of solvent front hydrogen bond network, similar to G1202R single mutant (see Fig. 7D). (D)-(G) The L1198F mutation was modeled onto co-crystal structures of lorlatinib (D), LA7 (E), LA9 (F) or LA4 (G) bound to WT ALK. The L1198F substitution results in steric clash with the selectivity nitrile of lorlatinib. LA7 and LA9 lack the selectivity nitrile and can easily accommodate the L1198F substitution, whereas the nitrile of LA4 is positioned toward L1198F but exhibits reduced steric clash compared to lorlatinib due to the more flexible ligand structure. (H) Relative fold potency decrease of LA4 compared with LA7 (calculated by dividing the cellular IC50 values of LA4 by the that of LA7) against Ba/F3 models harboring compound ALK mutants. IC50 values correspond to data shown in the Source Data. (I) Superimposition of LA7 onto the LA4/L1198F model shown in Panel G comparing the position of the corresponding thiazole methyl or nitrile groups near L1198F.
Supplementary Material
ACKNOWLEDGEMENTS
We thank patients and their families, as well as members of the Hata lab and MGH Thoracic Oncology Group for helpful discussion and support. This study was supported by a JSPS Overseas Research Fellowships (to AS), a National Cancer Institute Career Development Award (K12CA087723–16 to IDJ), NIH/NCI R01CA164273 (to JJL), a Lung Cancer Research Foundation grant (to SY), by Be a Piece of the Solution, and by Targeting a Cure for Lung Cancer Research Fund at MGH.
Footnotes
COMPETING INTERESTS STATEMENT
TWJ, TRJ, PW, SLW and MAM are employees of Pfizer, Inc. IDJ has received honoraria from Foundation Medicine, Creative Education Concepts, and American Lung Association, consulting fees from Guidepost, AstraZeneca, Boehringer Ingelheim, BostonGene, Catalyst, Novocure, Pfizer, Syros, and Xcovery, research support from Array, Genentech, Novartis, Pfizer, and Guardant Health, and travel support from Array and Pfizer. LAK is an employee and shareholder of Guardant Health. JFG has served as a compensated consultant or received honoraria from Bristol-Myers Squibb, Genentech, Ariad/Takeda, Loxo, Pfizer, Incyte, Novartis, Merck, Agios, Amgen, Jounce, Karyopharm, GlydeBio, Regeneron, Oncorus, Helsinn, Jounce, Array, and Clovis Oncology, has an immediate family member who is an employee with equity in Ironwood Pharmaceuticals, has received research funding from Novartis, Genentech/Roche, and Ariad/Takeda, and institutional research support from Tesaro, Moderna, Blueprint, BMS, Jounce, Array, Adaptimmune, Novartis, Genentech/Roche, Alexo and Merck. JJL has served as a compensated consultant for Genentech, C4 Therapeutics, Blueprint Medicines, Nuvalent, Turning Point Therapeutics, Mirati Therapeutics, Bayer, and Elevation Oncology; received honorarium and travel support from Pfizer; received institutional research funds from Hengrui Therapeutics, Turning Point Therapeutics, Neon Therapeutics, Relay Therapeutics, Bayer, Elevation Oncology, Roche/Genentech, Pfizer, Nuvalent, Linnaeus Therapeutics, and Novartis; received CME funding from OncLive, MedStar Health, and Northwell Health. SY is an employee of Tango Therapeutics and has received a consulting fee from Pfizer Japan and Nuvalent. ANH has received research support from Pfizer, Nuvalent Inc., Roche/Genentech, Amgen, Blueprint Medicines, Eli Lilly, Scorpion Therapeutics, Bristol-Myers Squibb, BridgeBio and Relay Therapeutics; has served as a compensated consultant for Nuvalent, Tolremo Therapeutics, Engine Biosciences and TigaTx. The remaining authors declare no competing interests.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
The crystal structural data and methods have been deposited to the Protein Data Bank (PDB) and have been assigned ID codes: lorlatinib, PDB 4CLI (14); LA7, PDB 7R7R (1.93 Å); LA9, PDB 7R7K (1.83 Å). Source data have been provided as Source Data files. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Soda M et al. , Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 448, 561–566 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Shaw AT et al. , Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 27, 4247–4253 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jordan EJ et al. , Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer discovery 7, 596–609 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwak EL et al. , Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. The New England journal of medicine 363, 1693–1703 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solomon BJM, T.; Kim D; Wu Y; Nakagawa K; Mekhail T; Felip E; Cappuzzo F; Paolini J; Usari T; Iyer S; Reisman A; Wilner KD; Tursi J; Blackhall F, First-Line Crizotinib versus Chemotherapy in ALK-Positive Lung Cancer. N Engl J Med 373, 1582 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Soria JC et al. , First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): a randomised, open-label, phase 3 study. Lancet 389, 917–929 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Peters S et al. , Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. The New England journal of medicine, (2017). [DOI] [PubMed] [Google Scholar]
- 8.Camidge DR et al. , Brigatinib versus Crizotinib in ALK-Positive Non-Small-Cell Lung Cancer. The New England journal of medicine 379, 2027–2039 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Shaw AT et al. , First-Line Lorlatinib or Crizotinib in Advanced ALK-Positive Lung Cancer. The New England journal of medicine 383, 2018–2029 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Perol M et al. , Patient-reported outcomes from the randomized phase III ALEX study of alectinib versus crizotinib in patients with ALK-positive non-small-cell lung cancer. Lung cancer 138, 79–87 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Mok T et al. , Updated overall survival and final progression-free survival data for patients with treatment-naive advanced ALK-positive non-small-cell lung cancer in the ALEX study. Annals of oncology : official journal of the European Society for Medical Oncology 31, 1056–1064 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Camidge DR et al. , Brigatinib Versus Crizotinib in Advanced ALK Inhibitor-Naive ALK-Positive Non-Small Cell Lung Cancer: Second Interim Analysis of the Phase III ALTA-1L Trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 38, 3592–3603 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gainor JF et al. , Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer discovery 6, 1118–1133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson TW et al. , Discovery of (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(m etheno)pyrazolo[4,3-h][2,5,11]-benzoxadiazacyclotetradecine-3-carbonitrile (PF-06463922), a macrocyclic inhibitor of anaplastic lymphoma kinase (ALK) and c-ros oncogene 1 (ROS1) with preclinical brain exposure and broad-spectrum potency against ALK-resistant mutations. Journal of medicinal chemistry 57, 4720–4744 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Shaw AT et al. , Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. The Lancet. Oncology 18, 1590–1599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solomon BJ et al. , Lorlatinib in patients with ALK-positive non-small-cell lung cancer: results from a global phase 2 study. The Lancet. Oncology 19, 1654–1667 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Shaw AT et al. , ALK Resistance Mutations and Efficacy of Lorlatinib in Advanced Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 37, 1370–1379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoda S et al. , Sequential ALK Inhibitors Can Select for Lorlatinib-Resistant Compound ALK Mutations in ALK-Positive Lung Cancer. Cancer discovery 8, 714–729 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dagogo-Jack I et al. , Treatment with Next-Generation ALK Inhibitors Fuels Plasma ALK Mutation Diversity. Clinical cancer research : an official journal of the American Association for Cancer Research 25, 6662–6670 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada K et al. , Prediction of ALK mutations mediating ALK-TKIs resistance and drug re-purposing to overcome the resistance. EBioMedicine 41, 105–119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaw AT et al. , Resensitization to Crizotinib by the Lorlatinib ALK Resistance Mutation L1198F. The New England journal of medicine 374, 54–61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Q et al. , Design of potent and selective inhibitors to overcome clinical anaplastic lymphoma kinase mutations resistant to crizotinib. Journal of medicinal chemistry 57, 1170–1187 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Lee CC et al. , Crystal structure of the ALK (anaplastic lymphoma kinase) catalytic domain. The Biochemical journal 430, 425–437 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Bresler SC et al. , ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer cell 26, 682–694 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson TW et al. , Reviving B-Factors: Activating ALK Mutations Increase Protein Dynamics of the Unphosphorylated Kinase. ACS medicinal chemistry letters 9, 872–877 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray BW et al. , TPX-0131, a Potent CNS-penetrant, Next-generation Inhibitor of Wild-type ALK and ALK-resistant Mutations. Molecular cancer therapeutics 20, 1499–1507 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelish EH et al. , Abstract 1468: NUV-655 is a selective, brain-penetrant ALK inhibitor with antitumor activity against the lorlatinib-resistant G1202R/L1196M compound mutation. In: Proceedings of the Annual Meeting of the American Association for Cancer Research 2021, (2021). [Google Scholar]
- 28.Mizuta H et al. , Gilteritinib overcomes lorlatinib resistance in ALK-rearranged cancer. Nature communications 12, 1261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selvaggi G et al. , ID:1882 Phase III Randomized Study of Ensartinib vs Crizotinib in Anaplastic Lymphoma Kinase (ALK) POSITIVE NSCLC Patients: eXalt3. Journal of Thoracic Oncology 15, e41–e42 (2020). [Google Scholar]
- 30.Zheng Z et al. , Anchored multiplex PCR for targeted next-generation sequencing. Nature medicine 20, 1479–1484 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Odegaard JI et al. , Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clinical cancer research : an official journal of the American Association for Cancer Research 24, 3539–3549 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Leung L et al. , Clearance Prediction of Targeted Covalent Inhibitors by In Vitro-In Vivo Extrapolation of Hepatic and Extrahepatic Clearance Mechanisms. Drug metabolism and disposition: the biological fate of chemicals 45, 1–7 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The crystal structural data and methods have been deposited to the Protein Data Bank (PDB) and have been assigned ID codes: lorlatinib, PDB 4CLI (14); LA7, PDB 7R7R (1.93 Å); LA9, PDB 7R7K (1.83 Å). Source data have been provided as Source Data files. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.


