Abstract
The nucleus, a genome-containing organelle eponymous of eukaryotes, is enclosed by a double membrane continuous with the endoplasmic reticulum. The nuclear pore complex (NPC) is an ∼110-MDa, ∼1000-protein channel that selectively transports macromolecules across the nuclear envelope and thus plays a central role in the regulated flow of genetic information from transcription to translation. Its size, complexity, and flexibility have hindered determination of atomistic structures of intact NPCs. Recent studies have overcome these hurdles by combining biochemical reconstitution and docking of high-resolution structures of NPC subcomplexes into cryo-electron tomographic reconstructions with biochemical and physiological validation. Here, we provide an overview of the near-atomic composite structure of the human NPC, a milestone toward unlocking a molecular understanding of mRNA export, NPC-associated diseases, and viral host–pathogen interactions, serving as a paradigm for studying similarly large complexes.
The emergence of compartmentalization has provided eukaryotic cells with opportunities to spatially separate molecules and molecular processes. The nucleus segregates the regulation of gene expression and transcription from translation, while also sheltering chromatin from damaging metabolic chemistries that take place in the cytoplasm. Despite these advantages, subcellular compartmentalization poses a challenge in protein homeostasis, particularly in the delivery of proteins to their target location. Indeed, three out of four human proteins are destined to leave the cytoplasm, where nonmitochondrial ribosomes that translate the vast majority of proteins reside (Thul et al. 2017). The subcellular localization problem is solved by labeling cargo with “address tags” that are recognized by dedicated transport machineries (Blobel and Sabatini 1971). Proteins targeted to the endoplasmic reticulum (ER), mitochondria, and chloroplasts must be unfolded during their translocation across membranes (Rapoport et al. 1996), whereas vesicles shuttle cargo between compartments, including secretion across the plasma membrane (Rothman and Weiland 1996; Schekman and Orci 1996; Kirchhausen 2000).
Nucleocytoplasmic transport is distinct from these types of cargo trafficking because nuclear pores, which arise from the annular fusion of the inner and outer membranes of the nuclear envelope, are massive channels that obviate the need to perturb the tertiary and quaternary structures of cargo. This mode of transport presents advantages that are fundamental to the functions of the nucleus. Folded factors can transmit signals immediately upon translocation, enabling agile regulation of gene expression in response to intra and extracellular cues. Beyond the transmission of signals, this mode of transport allows the cell to enact quality control by spatially separating the biogenesis of large and complicated macromolecular assemblies such as pre-60S ribosomal subunits (Bataillé et al. 1990; Ho et al. 2000; Gadal et al. 2001), RNA polymerase II (Czeko et al. 2011; Gómez-Navarro et al. 2013), or mature mRNA (Stewart 2019) from the milieu in which they perform their function. For the nuclear compartment to accomplish this, nucleocytoplasmic transport must be selective. The nucleus of a typical mammalian cell presents ∼2000–5000 nuclear pores, although the number is relative to the surface area of the nuclear envelope and doubles in preparation for mitosis (Maul and Deaven 1977). Embedded in the nuclear pore is a large proteinaceous assembly, the nuclear pore complex (NPC), which establishes a diffusion barrier made up of natively unfolded phenylalanine-glycine (FG) repeat regions (Ribbeck and Görlich 2001; Denning et al. 2003; Frey and Görlich 2007; Patel et al. 2007). Historically, the diffusion barrier has been described as preventing macromolecules with a mass above ∼40 kDa from passively diffusing across the nuclear envelope (Bonner 1975; Paine et al. 1975). However, more recent research employing green fluorescent protein (GFP)-fused model cargoes indicates that the diffusion barrier represents a more gradual hurdle (Timney et al. 2016). The diffusion barrier is permeated by a dedicated set of 24 transport factors in humans, termed karyopherins (Cook et al. 2007; Wing et al. 2022), which can be divided into two main families: β-karyopherins interact directly with FG repeats and facilitate transport, whereas α-karyopherins serve as adaptors linking proteins containing nuclear localization signals (NLSs) to a β-karyopherin (Stewart 2007). The directionality of transport is established by a gradient of the small GTPase Ran, which in the nucleus is actively maintained in a GTP-bound state (Cook et al. 2007; Wing et al. 2022).
The mechanism of Ran-dependent transport of proteins and some types of non–messenger RNA is conceptually understood, though the details of specific transport events have been determined for only a small subset of cargoes. In contrast, the process by which messenger ribonucleoprotein particles (mRNPs) are remodeled and messenger RNA (mRNA) is exported from the nucleus is not generally understood (Lin and Hoelz 2019). The NPC dynamically couples transcription, splicing, and mRNA export with translation in the cytoplasm (Stewart 2019). Given this role of the NPC in the execution of the central dogma in eukaryotes, it is unsurprising that severe diseases are associated with defective NPC components, and that viruses have evolved exquisite ways to hijack the NPC and the nucleocytoplasmic transport machinery (Köhler and Hurt 2010; Yarbrough et al. 2014).
DETERMINATION OF THE NUCLEAR PORE COMPLEX STRUCTURE
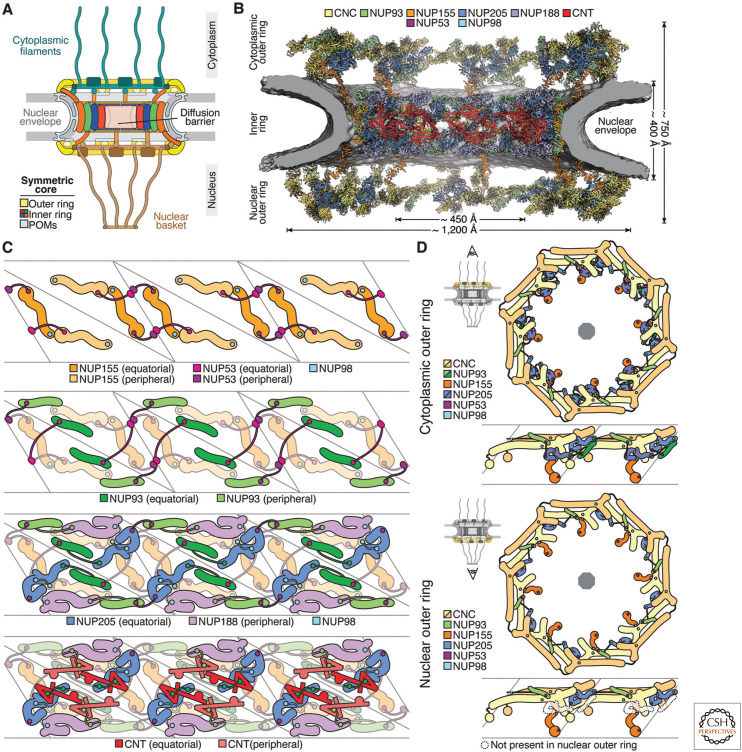
Nuclear pores were first observed more than 70 years ago (Callan and Tomlin. 1950) and structures embedded in them, corresponding to NPCs, were subsequently described (Watson 1955, 1959). Early electron microscopic (EM) studies showed that NPCs have an eightfold symmetry along the nucleocytoplasmic axis (Gall 1967) and also discovered asymmetric decorations on the cytoplasmic and nuclear sides of this symmetric core, known as the cytoplasmic filaments and nuclear basket, respectively (Fig. 1A; Franke and Scheer 1970). Over the next 40 years, EM characterizations of NPCs from various species became more refined, with cryo–electron tomography (cryo-ET) and subtomogram averaging currently representing the most prominent method for the three-dimensional reconstruction of intact NPCs (Hinshaw et al. 1992; Akey and Radermacher 1993; Akey 1995; Fahrenkrog et al. 1998; Kiseleva et al. 1998; Yang et al. 1998; Beck et al. 2004; Maimon et al. 2012).
Figure 1.
The nuclear pore complex (NPC) linker scaffold. (A) Schematic cutaway representation of the NPC architecture. (B) Near-atomic composite structure of the human NPC symmetric core generated by quantitatively docking nucleoporin and nucleoporin complex crystal and single particle cryo–electron microscopy (cryo-EM) structures into an ∼12-Å cryo–electron tomographic (cryo-ET) map of the intact human NPC (Mosalaganti et al. 2022; Petrovic et al. 2022). The nuclear envelope is shown as a gray isosurface and the nucleoporin structures are displayed in cartoon representation colored according to the legend. (C) Schematic layer-by-layer representation of a cross-section view of three spokes of the NPC inner ring showing the network of linker–scaffold interactions. (D) Schematic representation of the top views of human NPC cytoplasmic and nuclear outer rings and corresponding side views of two spokes, illustrating the reticulated head-to-tail arrangement of the coat nucleoporin complexes (CNCs) cross-linked by trans-spoke NUP93–NUP205 interactions. (Figure adapted from Petrovic et al. 2022, with permission from the authors.)
The NPC entered the molecular age with pleiotropic antibodies that were raised against nuclear envelopes (Davis and Blobel 1986) and that provided an entry point for the identification of NPC components (Davis and Fink 1990; Nehrbass et al. 1990). Over more than a decade, bootstrapping by genetic and biochemical means revealed an evolutionarily conserved set of approximately 34 different proteins termed nucleoporins, the completeness of which was confirmed by mass spectrometry (Rout et al. 2000; Cronshaw et al. 2002). To add up to a mass of ∼110 MDa in the human NPC (Reichelt et al. 1990; Ori et al. 2013), many copies of these building blocks are recruited in multiples of eight. Most nucleoporins are soluble proteins, although the set also includes integral membrane proteins of the pore membrane (POM) domain (Hallberg et al. 1993) and proteins that present membrane curvature-sensing motifs (Drin et al. 2007; Leksa et al. 2009; Lin et al. 2016; Nordeen et al. 2020).
Having inventoried the building blocks, the next challenge was to determine how they fit together in the structure of the intact NPC, which is one of the largest supramolecular assemblies in the eukaryotic cell. Although the NPC's size and complexity made it resistant to established approaches for high-resolution structure determination, progress over the last two decades has managed to overcome many previously intractable hurdles. Combining biochemical reconstitution, interaction mapping, and atomic structure determination of increasingly larger NPC subcomplexes with improving cryo-ET maps has now led to a composite near-atomic structure of an entire NPC, from which only a handful of nucleoporins remain missing (Hodel et al. 2002; Berke et al. 2004; Hsia et al. 2007; Jeudy and Schwartz 2007; Boehmer et al. 2008; Brohawn et al. 2008, Debler et al. 2008; Schrader et al. 2008; Brohawn and Schwartz 2009; Leksa et al. 2009; Nagy et al. 2009; Seo et al. 2009; Whittle and Schwartz 2009; Bilokapic and Schwartz 2012, 2013; Andersen et al. 2013; Bui et al. 2013; Sampathkumar et al. 2013; Stuwe et al. 2014; von Appen et al. 2015; Chug et al. 2015; Eibauer et al. 2015; Kelley et al. 2015, Stuwe et al. 2015a,b; Xu et al. 2015; Kosinski et al. 2016; Lin et al. 2016; Bley et al. 2022; Petrovic et al. 2022).
A pivotal event in the pursuit of the NPC structure was the elucidation of a key component of its symmetric core known as the outer ring. Determining the crystal structure of its basic building block, the Y-shaped coat nucleoporin complex (CNC; also referred to as NUP107-NUP160 complex), and docking it into an ∼32-Å cryo-ET map of the intact human NPC (Bui et al. 2013; Stuwe et al. 2015b) revealed a reticulated head-to-tail arrangement of CNCs into two concentric rings on each side of the nuclear envelope. More generally, the success of this approach demonstrated that the resolution gap between cryo-ET reconstructions of intact NPCs and high-resolution structures of their components could be bridged by quantitative docking (Bui et al. 2013; Stuwe et al. 2015b). Rapidly thereafter, biochemical reconstitution of inner ring complexes, X-ray crystallographic characterization of the constituent nucleoporins, and quantitative docking into an ∼23-Å cryo-ET map completed the structure of the symmetric core of the human NPC (Amlacher et al. 2011; Andersen et al. 2013; Sampathkumar et al. 2013; Stuwe et al. 2014, 2015a; Chug et al. 2015; Fischer et al. 2015; von Appen et al. 2015; Kosinski et al. 2016; Lin et al. 2016). The symmetric core was revealed to consist of an inner ring lining the lumen of the nuclear pore and two outer rings sitting on each side of the nuclear envelope (Fig. 1B). The eight spokes symmetrically arranged about the nucleocytoplasmic axis present additional twofold rotational symmetry along axes coplanar with the nuclear envelope (von Appen et al. 2015; Kosinski et al. 2016; Lin et al. 2016). A similar approach was subsequently employed to determine the structure of the Saccharomyces cerevisiae NPC (Kim et al. 2018; Allegretti et al. 2020), demonstrating an overall conserved architecture, with compositional differences in the outer rings of the symmetric core.
The NPC's inner ring is composed of six structured scaffold nucleoporins NUP155, NUP188, NUP205, NUP54, NUP58, and NUP62, two mostly unstructured linker nucleoporins NUP98 and NUP53, and NUP93, which presents an amino-terminal linker region and a carboxy-terminal scaffold. Whereas the CNC complex of the outer rings is held together by extensive interfaces between structured domains (Hsia et al. 2007; Boehmer et al. 2008; Brohawn et al. 2008, Debler et al. 2008; Brohawn and Schwartz 2009; Nagy et al. 2009; Bilokapic and Schwartz 2012; Kelley et al. 2015; Stuwe et al. 2015b; Nordeen et al. 2020), the integrity of the inner ring relies on multivalent linker nucleoporins simultaneously binding to multiple scaffold nucleoporins via short linear motifs (Amlacher et al. 2011; Stuwe et al. 2014, 2015a; Fischer et al. 2015; Lin et al. 2016), often potentiated by promiscuous interactions between flanking linker sequences and the scaffold surfaces (Petrovic et al. 2022). From the nuclear envelope to the central transport channel, the scaffold proteins form concentric layers that are interconnected by linker–scaffold interactions (Fig. 1C). Anchored by membrane-sensing motifs, NUP155 forms the outermost coat, followed by layers of NUP93, NUP205/NUP188, and the NUP54• NUP58•NUP62 channel nucleoporin heterotrimer (CNT) (Kosinski et al. 2016; Lin et al. 2016). The relative position of scaffold proteins in the inner ring is conserved in all known NPC structures (Kosinski et al. 2016; Lin et al. 2016; Kim et al. 2018; Mosalaganti et al. 2018, 2022; Allegretti et al. 2020; Schuller et al. 2021; Zimmerli et al. 2021; Huang et al. 2022; Petrovic et al. 2022). The recently completed structural characterization of the linker–scaffold interactions has elucidated the topology of protein interactions within and between the different scaffold layers (Stuwe et al. 2015a; Lin et al. 2016; Petrovic et al. 2022). NUP205 or NUP188 are at the center of two alternative ∼425-kDa hetero-octameric inner ring complexes, two copies of each symmetrically arranged about the NPC midplane per each of the eight inner ring spokes. Within the layered architecture of the inner ring, NUP205, NUP188, and the scaffold portion of NUP93 are anchored to the membrane-adjacent NUP155 layer by the linkers NUP53 and NUP98 (Fig. 1C). The amino-terminal linker portion of NUP93 interacts with NUP205 and NUP188, while also anchoring and positioning the CNT (Amlacher et al. 2011; Fischer et al. 2015; Stuwe et al. 2015a; Petrovic et al. 2022). Though most linker–scaffold interactions reinforce intraspoke connections, NUP53 forms flexible bridges between adjacent spokes. Parts of the linker scaffold have also been observed in a recent single particle cryo-EM structure of a detergent-extracted S. cerevisiae NPC, although the identity of all the linkers could not be assigned from the map alone (Akey et al. 2022). Despite divergent primary sequences, especially of the linker nucleoporins, the topology of the inner ring linker scaffold is conserved from fungi to humans (Petrovic et al. 2022).
The inner ring encircles the central transport channel, into which the natively unfolded amino-terminal FG-repeat regions of the CNT and NUP98 project to establish the diffusion barrier. Additionally, on either face of the NPC, asymmetric nuclear basket (NUP153, NUP50, and TPR) and cytoplasmic filament (NUP358, NUP214, and NUP42), nucleoporins contribute to the FG-repeat meshwork. FG-repeat regions can be as long as about 1000 residues with FXFG or GLFG motifs present approximately every 20 residues. Purified FG-repeat regions can undergo liquid–liquid phase separation (LLPS) (Celetti et al. 2020), which matures over time into a solid hydrogel driven by the hydrophobicity of the phenylalanine residues (Frey et al. 2006, Frey and Görlich 2007). FG-repeat peptides have also been observed to form amyloid-like structures in vitro (Ader et al. 2010; Hughes et al. 2018). β-Karyopherins or karyopherin•cargo complexes can specifically penetrate FG-repeat LLPS condensates and hydrogels with rates comparable with in vivo transit across the NPC (Frey and Görlich 2007; Celetti et al. 2020). In vivo, condensation of FG repeats is likely modulated by their persistent association with transport factors, transiting cargo, and post-translational modifications, most notably O-linked β-N-acetylglucosamine glycosylation (O-GlcNAcylation) (Labokha et al. 2013) and phosphorylation (Kosako and Imamoto 2010; Mishra et al. 2019) of serine and threonine residues. The unfolded nature of FG-repeat regions precludes their direct structural characterization, and experimental validation of FG-repeat condensation in the intact NPC's diffusion barrier has remained elusive because of a lack of adequate technology to analyze such a compartment (Celetti et al. 2020). High-speed atomic force microscopy has provided intriguing first glimpses of FG-repeat meshwork dynamics at transport-relevant timescales, capturing rapidly elongating and retracting FG-repeat regions, but has not yet been able to directly discern the condensation state of the diffusion barrier (Sakiyama et al. 2016). Whether LLPS nuclear speckles that are routinely observed in cells expressing NUP98 leukemogenic fusions are relevant to the condensation state of the diffusion barrier remains to be determined (Ahn et al. 2021).
The human NPC's symmetric core outer rings are connected to the inner ring by bridge NUP155 copies and anchored to the nuclear envelope by membrane-sensing motifs in the NUP160 and NUP133 components of the CNC (Drin et al. 2007; Bui et al. 2013; von Appen et al. 2015; Lin et al. 2016). The backbone of the outer rings is the ∼400-kDa heterononameric Y-shaped CNC, 16 copies of which are arranged in a reticulated head-to-tail fashion, giving rise to a wider distal and a narrower proximal CNC ring (Fig. 1D). On both sides of the nuclear envelope, eight copies of NUP205 and NUP93 are intercalated between the CNC double rings. The amino-terminal linker portion of NUP93 stretches to the adjacent spoke to bind to NUP205, forming a trans-spoke staple (Mosalaganti et al. 2022; Petrovic et al. 2022). This is unlike the inner ring, where the NUP93 interactions with NUP205 and NUP188 occur within the same spoke. On the cytoplasmic face, an additional eight NUP205 and NUP93 copies are present at the base of the outer ring, interacting in a cis-spoke fashion (Mosalaganti et al. 2022; Petrovic et al. 2022). The ubiquity of NUP93 in both inner and outer rings suggests it is a “linchpin” of the NPC, which is consistent with the observation that degradation-targeting of NUP93 leads to rapid dismantling of the NPC in live cells (Regmi et al. 2020). Compared to the human NPC, its fungal and algal counterparts present simpler outer ring architectures. The S. cerevisiae, Schizosaccharomyces pombe, and Chlamydomonas reinhardtii NPCs neither possess bridge NUP155 copies nor include NUP205 or NUP93 in the outer rings, and their cytoplasmic outer ring is composed of a single head-to-tail arranged CNC ring (Kim et al. 2018; Mosalaganti et al. 2018; Allegretti et al. 2020; Zimmerli et al. 2021). Interestingly, the S. pombe cytoplasmic outer ring lacks NUP107 and NUP133 and is therefore discontinuous (Asakawa et al. 2019; Zimmerli et al. 2021). On the nuclear side, S. pombe (Zimmerli et al. 2021) and C. reinhardtii (Mosalaganti et al. 2018) present a double CNC ring, whereas S. cerevisiae appears to possess NPCs with both single and double nuclear CNC rings (Kim et al. 2018; Allegretti et al. 2020; Akey et al. 2022).
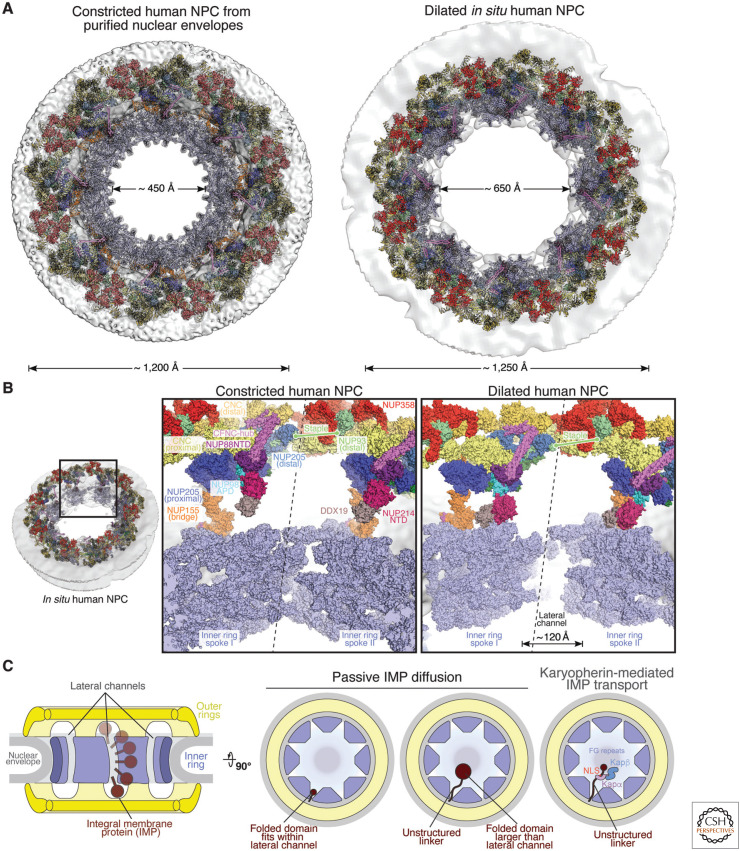
Recent advances in focused ion beam (FIB) milling-enabled in situ imaging have revealed that the central transport channel of NPCs in intact cells is dilated compared to that of NPCs from purified nuclear envelopes or detergent-extracted NPCs (Bui et al. 2013; Eibauer et al. 2015; von Appen et al. 2015; Kosinski et al. 2016; Mahamid et al. 2016; Kim et al. 2018; Mosalaganti et al. 2018, 2022; Allegretti et al. 2020; Zhang et al. 2020; Schuller et al. 2021; Zila et al. 2021; Zimmerli et al. 2021; Akey et al. 2022). The observed ∼200-Å dilation is achieved by outward displacement of the eight inner ring spokes, which the robust yet plastic linker–scaffold architecture can readily accommodate (Fig. 2A; Petrovic et al. 2022). In contrast to the inner ring, the outer rings, rigidified by large surface interfaces between CNC components and tight NUP205-NUP93 trans-spoke staples, undergo minimal rearrangement in response to dilation (Mosalaganti et al. 2022; Petrovic et al. 2022). Inner ring dilation results in the emergence of lateral channels between the eight spokes of the NPC, which can accommodate freely diffusing small pore-facing domains of inner nuclear membrane integral membrane proteins (INM-IMPs) (Fig. 2B; Ohba et al. 2004; Zuleger et al. 2011; Ungricht et al. 2015). However, INM-IMPs with larger pore-facing domains or karyopherin-binding NLSs require unstructured linkers between the soluble pore-facing and transmembrane domains to span the distance between the central transport channel and the nuclear envelope (Fig. 2C; King et al. 2006; Meinema et al. 2011; Kralt et al. 2015; Lokareddy et al. 2015; Popken et al. 2015). In S. pombe, the relaxation of nuclear envelope membrane tension following energy depletion or hypertonic osmotic shock results in constricted NPCs (Zimmerli et al. 2021). The NPC's structural responsiveness to perturbations in nuclear envelope membrane tension establishes it as the cell's largest mechanosensitive channel.
Figure 2.
Dilation of the nuclear pore complex (NPC) and lateral channel formation. (A) Cytoplasmic views of the near-atomic composite structure of constricted and dilated human NPCs (Bley et al. 2022; Mosalaganti et al. 2022). The nuclear envelope is shown as a gray isosurface and nucleoporins are shown in cartoon representation. (B) Close-up views of the interface between two spokes, separated by a dashed line, in the constricted and dilated human NPCs. Nucleoporins are shown in surface representation, with the inner ring spokes uniformly colored in pale blue. (C) Schematic representation of the NPC symmetric core cross section, illustrating inner nuclear membrane integral membrane protein (INM-IMP) passage through the lateral channels. Top views of the NPC symmetric core illustrate that lateral channels can accommodate freely diffusing small, folded pore-facing domains or unstructured linkers tethering larger folded domains or karyopherin-binding nuclear localization sequences (NLSs; classical Kapα/Kapβ-mediated import shown). (Figure adapted from Petrovic et al. 2022, with permission from the authors.)
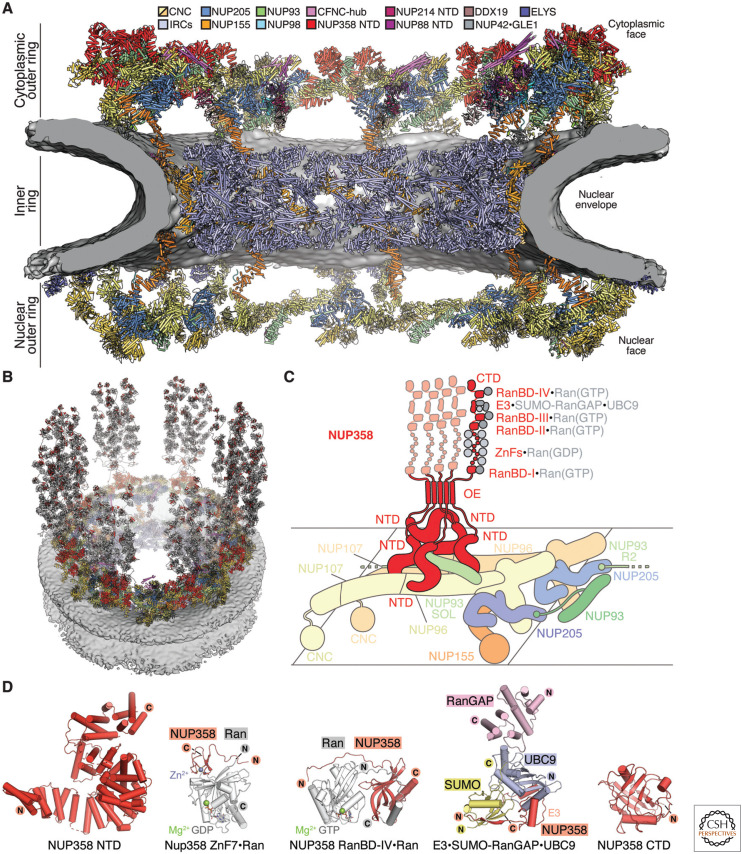
The cytoplasmic outer ring serves as a platform for the incorporation of the cytoplasmic filament nucleoporin complex (CFNC) and the metazoan-specific NUP358, whereas the bridge NUP155 provides a tentative binding site for the GLE1•NUP42 complex (Fig. 3A; Lin et al. 2018; Bley et al. 2022; Mosalaganti et al. 2022). A homo-oligomerizing pentameric bundle of NUP358 clamps down on the tandem arranged stalks of the Y-shaped CNCs in each spoke of the cytoplasmic outer ring with its amino-terminal α-helical solenoid domains (Bley et al. 2022; Mosalaganti et al. 2022). The remainder of flexibly attached NUP358 domains project as far as ∼60 nm into the cytoplasm (Walther et al. 2002), providing manifold binding sites for the small GTPase Ran (Vetter et al. 1999; Partridge and Schwartz 2009; Bley et al. 2022) and a binding site for SUMOylated Ran GTPase-activating protein (RanGAP) (Fig. 3B–D; Mahajan et al. 1998; Matunis et al. 1998; Reverter and Lima 2005). Overlooking the central transport channel, the CFNC represents a second patch of asymmetric nucleoporins attached to the cytoplasmic outer ring (Fig. 4A; Bley et al. 2022; Mosalaganti et al. 2022). The CFNC comprises NUP62, NUP88, NUP214, NUP98, RAE1, and the ATP-dependent DEAD-box RNA helicase DDX19 (Bley et al. 2022). Its modular heterohexameric architecture is conserved between fungi and humans, although the nucleoporins that anchor the CFNC coiled-coil hub to the NPC vary between species (Teimer et al. 2017; Bley et al. 2022). In the human NPC, the same NUP93 region that recruits the CNT complex to the inner ring anchors two CFNC coiled-coil hubs on the cytoplasmic face, one of which is currently unresolved in cryo-ET maps, possibly because of flexible tethering (Bley et al. 2022; Mosalaganti et al. 2022). Other folded domains can be tentatively docked nearby, including the amino-terminal β-propellers of NUP214 and NUP88, the latter bound to NUP98. The elucidated architecture of the cytoplasmic face of the NPC provides a tantalizing outlook toward the mechanistic dissection of mRNP remodeling.
Figure 3.
Architecture of the cytoplasmic face of the human nuclear pore complex (NPC). (A) Cross-sectional view of the near-atomic composite structure of the human NPC generated by docking nucleoporin and nucleoporin complex crystal and single particle cryo-EM structures into an ∼12-Å cryo-ET map of the intact human NPC (Bley et al. 2022; Mosalaganti et al. 2022). The nuclear envelope is shown as a gray isosurface, the inner ring is displayed in cartoon representation uniformly colored pale blue, and the nuclear and cytoplasmic face structures are displayed in cartoon representation colored according to the legend. (B) Architecture of the cytoplasmic face of the human NPC (Bley et al. 2022). Flexibly attached NUP358 domains protrude into the cytoplasm. (C) Schematic representation of a single cytoplasmic face protomer, illustrating the attachment of a NUP358 pentameric bundle held together by interactions between oligomerization elements (OEs). Four NUP358NTD copies wrap around the tandem arranged Y-shaped CNC stalks, generating a binding site for a fifth copy. (D) Cartoon representations of crystal structures of NUP358 complexes: NUP358NTD (PDB ID 7MNL), NUP358ZnF7•Ran(GDP) (PDB ID 7MNU), NUP358RanBD-IV•Ran(GTP) (PDB ID 7MNZ) (Bley et al. 2022), NUP358E3•SUMO-RanGAP•UBC9 (PDB ID 1Z5S) (Reverter and Lima 2005), NUP358CTD (PDB ID 4I9Y) (Lin et al. 2013). (Figure adapted from Bley et al. 2022, with permission from the authors.)
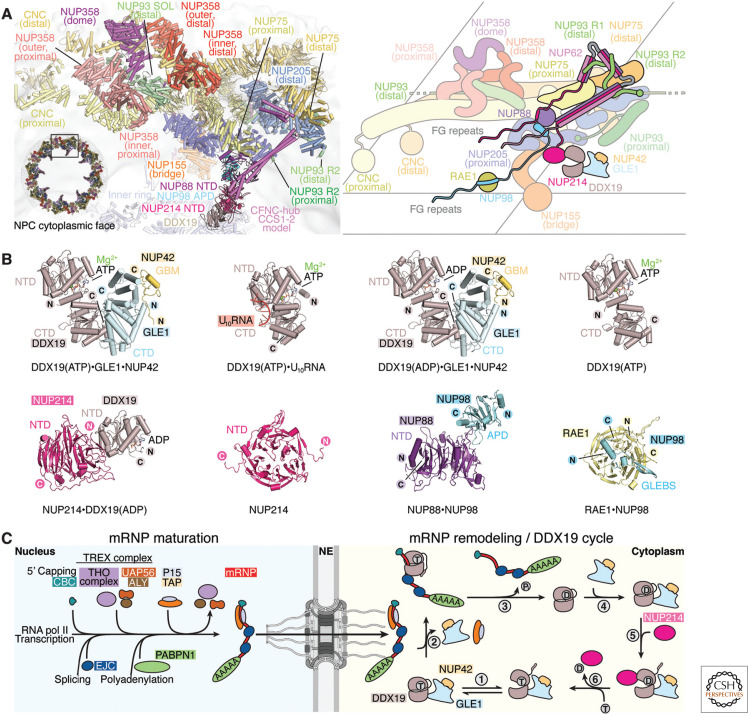
Figure 4.
mRNP export and remodeling at the cytoplasmic face of the nuclear pore complex (NPC). (A) Cartoon representation and corresponding schematic conceptualization of a spoke of the NPC cytoplasmic face illustrating the relative positions of the cytoplasmic filament nucleoporins, with the cytoplasmic filament nucleoporin complex (CFNC) anchored by NUP93R1 and a pentameric bundle of NUP358 bound to the stalk of tandem-arranged Y-shaped coat nucleoporin complexes (CNCs). (B) Cartoon representations of crystal structures of human cytoplasmic filament complexes: DDX19(ADP)•GLE1CTD•NUP42GBM (PDB ID 6B4I) (Lin et al. 2018), DDX19(ATP)• GLE1CTD•NUP42GBM (PDB ID 6B4J) (Lin et al. 2018), DDX19(ATP)•U10RNA (PDB ID 3FHT) (von Moeller et al. 2009), DDX19(ATP) (PDB ID 6B4K) (Lin et al. 2018), DDX19(ADP)•NUP214NTD (PDB ID 3FMO) (Napetschnig et al. 2009), NUP214NTD (PDB ID 2OIT) (Napetschnig et al. 2007), NUP88NTD•NUP98APD (PDB ID 7MNI) (Bley et al. 2022), RAE1•NUP98GLEBS (PDB ID 3MMY) (Ren et al. 2010). (C) Schematic representation of the maturation, export, and remodeling of mRNPs by the DDX19 helicase cycle. In the nucleus, mRNA is transcribed by RNA polymerase II, followed by 5′ capping, recruitment of the cap-binding complex (CBC), deposition of exon junction complexes (EJCs) at splice sites, loading of the transcription-export (TREX) complex in a splicing-dependent manner, 3′ polyadenylation, and deposition of polyadenylate-binding nuclear protein 1 (PABPN1). ATP-hydrolyzing DEAD-box helicase UAP56 may facilitate loading of export factors P15•TAP to produce an export-competent mRNP. The exported mRNP is remodeled at the cytoplasmic face of the NPC, where P15•TAP is removed in a DDX19-dependent manner: (1) ATP-bound DDX19 cycles between autoinhibited and closed conformations. (2) Upon binding RNA, DDX19 adopts a closed, catalytically active conformation that is incompatible with GLE1 binding but could strip P15•TAP from mRNA. (3) ATP hydrolysis by DDX19 triggers RNA release, converting DDX19 to an autoinhibited ADP-bound conformation in which the autoinhibitory α-helix binds between amino- and carboxy-terminal RecA domains. (4) GLE1 binding destabilizes the autoinhibited conformation. (5) NUP214 binding converts DDX19 to an open conformation, promoting nucleotide exchange. (6) Nucleotide exchange displaces NUP214, priming the DDX19(ATP)•GLE1•NUP42 complex to restart the cycle (Lin et al. 2018). (Figure adapted from Bley et al. 2022, with permission from the authors.)
The asymmetric nucleoporins of the nuclear face, which include the nuclear basket components NUP50, NUP153, TPR, and ELYS, are the least well-characterized parts of the NPC. The amino-terminal β-propeller and α-helical solenoid domains of ELYS are located exclusively on the nuclear face, interfacing with NUP160 copies of the outer ring (Bley et al. 2022; Mosalaganti et al. 2022). Specific localization of ELYS to the nuclear face may be mediated by its carboxy-terminal chromatin-binding AT-hook and basic RRK motifs (Gillespie et al. 2007; Rasala et al. 2008; Kobayashi et al. 2019). Interestingly, the S. pombe ELYS ortholog, which consists of only the evolutionarily conserved α-helical solenoid domain, is present on both nuclear and cytoplasmic faces (Zimmerli et al. 2021). Tubular densities atop the nuclear outer ring observed in cryo-EM reconstructions of the human (Bley et al. 2022; Mosalaganti et al. 2022) and S. cerevisiae (Akey et al. 2022) NPCs are anchoring points for the nuclear basket, likely corresponding to homo-oligomerizing coiled-coil domains of TPR (Pal et al. 2017).
Finally, POM121, POM210, and NDC1 are integral membrane proteins recruited to the NPC. The single-pass transmembrane proteins POM210 and POM121 possess soluble domains in the perinuclear space, which for POM121 is predicted to be mostly unstructured (Gerace et al. 1982; Hallberg et al. 1993; Stavru et al. 2006). Artificial intelligence (AI) models of POM210 fit into cryo-EM density surrounding the Xenopus laevis or human NPCs predict a ring-like structure circumscribing the equator of the NPC's perinuclear space, in which eight copies of POM210 per spoke align 15 immunoglobulin (Ig)-like domains into an antiparallel butterfly-like arrangement (Zhang et al. 2020; Mosalaganti et al. 2022). Imaging of NPCs from POM210-deficient human cells confirmed this prediction (Mosalaganti et al. 2022). Furthermore, modeling of an S. cerevisiae POM210 ortholog into low-resolution cryo-EM density has also delineated a luminal ring, albeit one arising from a simpler antiparallel arrangement of only two copies per spoke (Kim et al. 2018; Akey et al. 2022). Unlike POM121 and POM210, NDC1 is predicted to possess six transmembrane helices and a soluble pore-facing domain (Stavru et al. 2006). AI models of NDC1 in complex with the metazoan-specific nucleoporin ALADIN were suggested to fit into two unexplained transmembrane and membrane-adjacent cryo-ET densities per spoke, interfacing with the bridge and inner ring NUP155 copies (Mosalaganti et al. 2022).
Crystal and single-particle cryo-EM structure determination of subcomplexes combined with cryo-ET reconstruction of intact NPCs have successfully elucidated the near-atomic architecture of the human NPC, as well as NPCs from other species. A few “blind spots,” consisting primarily of the molecular details of individual binding interfaces and the nuclear basket, remain to be resolved. Although AI-based modeling only emerged subsequent to the elucidation of the near-atomic architecture of the NPC, it can now provide a first glance at regions that have yet to be structurally characterized (Mosalaganti et al. 2022). It is anticipated that AI will accelerate the approach to structural biology that conquered the NPC. However, the importance of careful experimental validation has never been greater, particularly in the interpretation of protein–protein interfaces and of structures for which no prior knowledge is available.
The divide-and-conquer approach that led to the determination of the NPC structure is a paradigm for the elucidation of similarly complex and dynamic multicomponent mega-macromolecular cellular machineries. Importantly, the structure of the NPC provides a context for predicting molecular functions, which is essential for the further study of the NPC's role in nucleocytoplasmic transport, mRNA export, nucleoporin diseases, and viral interference.
FUNCTION OF THE NUCLEAR PORE COMPLEX IN mRNA EXPORT
Eukaryotic mRNA transcription in the nucleus is closely tied to processing steps that include 5′ capping, intron removal by splicing, 3′ polyadenylation, and packaging with P15•TAP mRNA export factors to form an export-competent mRNP that can transit across the NPC (Köhler and Hurt 2007; Stewart 2019). At the cytoplasmic face of the NPC, the mRNP is remodeled to remove P15•TAP (Fig. 4B,C). Though the mechanism is relatively poorly understood, mRNP remodeling involves the CFNC components, including the DEAD-box helicase DDX19, a transient nucleoporin and RNA-dependent ATPase (Lund and Guthrie 2005). DDX19 was first identified as Dbp5/RAT8 in S. cerevisiae from a screen for genes required for poly(A+) RNA export (Snay-Hodge et al. 1998; Tseng et al. 1998). In addition to ATP and RNA, DDX19 binds to components of the NPC's cytoplasmic filaments (Collins et al. 2009; Napetschnig et al. 2009; von Moeller et al. 2009). Both GLE1•NUP42 and RNA stimulate DDX19's ATPase activity (Montpetit et al. 2011; Lin et al. 2018), ensuring that DDX19-mediated removal of P15•TAP occurs only after mRNPs have translocated through the NPC (Alcazár-Román et al. 2006; Weirich et al. 2006). By contrast, DDX19 association with NUP214, which is mutually exclusive with RNA binding, facilitates ADP release and nucleotide exchange (Schmitt et al. 1999; Napetschnig et al. 2009; von Moeller et al. 2009; Noble et al. 2011; Wong et al. 2018). DDX19 also possesses an amino-terminal autoinhibitory α-helix that binds to the same cleft as RNA (Collins et al. 2009). GLE1•NUP42 binding induces conformational changes in DDX19 to allosterically facilitate the release of the autoinhibited state (Lin et al. 2018). In fungi, but not in humans, the small molecule inositol hexaphosphate (IP6) regulates interactions at the cytoplasmic face of the NPC by promoting Gle1•Nup42's interaction with Dbp5 (Alcazár-Román et al. 2006; Weirich et al. 2006; Lin et al. 2018), while also abolishing Gle1•Nup42 binding to the outer ring CNC (Bley et al. 2022).
RAE1 is an essential RNA-binding nucleoporin of the CFNC with a prominent yet poorly understood function in mRNA export. Fungal RAE1 orthologs were discovered in genetic screens for poly(A+) RNA export defects in S. pombe (Brown et al. 1995) and S. cerevisiae (Murphy et al. 1996), whereas human RAE1 was identified by copurification of cross-linked poly(A+) RNA from UV-irradiated HeLa cells (Kraemer and Blobel 1997). RAE1 is frequently targeted by viral virulence factors, which directly compete with RNA for RAE1 binding, leading to nuclear accumulation of poly(A+) RNA (Enninga et al. 2002; Quan et al. 2014; Miorin et al. 2020).
A recent systematic screen of RNA binding by cytoplasmic filament nucleoporins identified a number of additional RNA-binding activities in the amino-terminal domains of NUP88 and NUP358, GLE1•NUP42, and the NUP358 Ran-binding domains when complexed with Ran(GTP) (Bley et al. 2022). The relevance of these binding activities needs to be determined, not only in mRNP remodeling, but potentially also in the subsequent loading of cytosolic factors and handover to the translation machinery. Coordination between mRNP remodeling at the NPC and downstream translation is hinted at by the observation that rapid depletion of NUP358 results in the reduced translation of reporter genes, but strikingly not in nuclear accumulation of RNA (Mahadevan et al. 2013; Bley et al. 2022).
Many questions regarding the coordinated action of cytoplasmic filament nucleoporins in mRNA export and mRNP remodeling remain unanswered. The recently determined near-atomic composite structure of the cytoplasmic face of the human NPC (Bley et al. 2022; Mosalaganti et al. 2022) is expected to enable the design of experiments to address these outstanding questions.
NUCLEOPORIN DISEASES
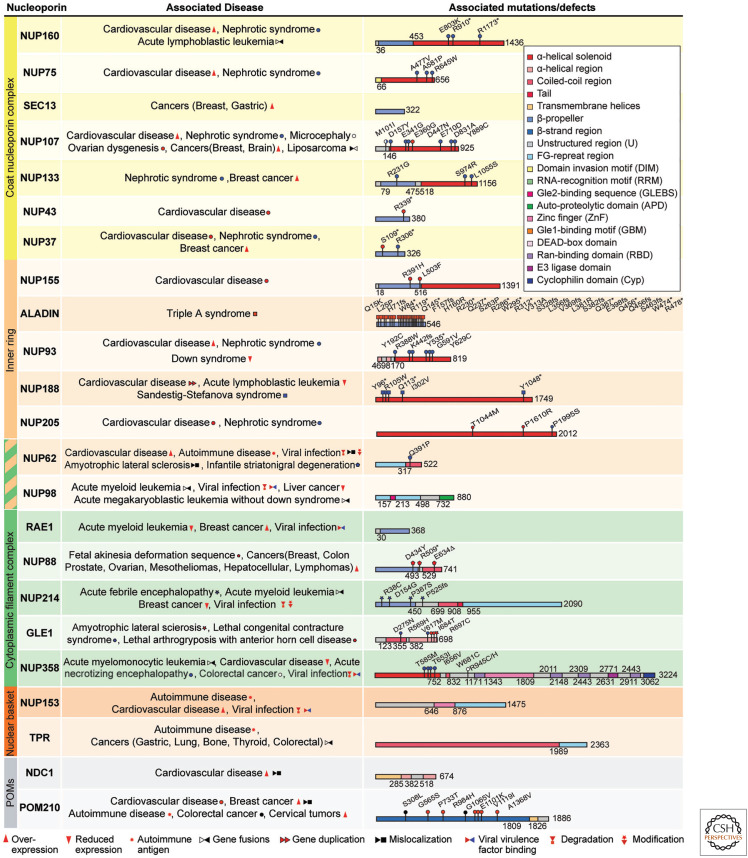
Given the central role of the NPC in controlling nuclear access and egress, it is unsurprising that nucleoporin mutations give rise to a wide variety of human diseases, and that the NPC is frequently targeted by viruses intent on entering the nucleus or altering host cell physiology in their favor. Perturbations in at least 24 nucleoporins are associated with human diseases, and at least six are targeted by viruses (Fig. 5).
Figure 5.
Nucleoporin-associated diseases. An overview of human nucleoporin diseases and viral virulence factors targeting the nuclear pore complex (NPC). Domain structures of human nucleoporins drawn as horizontal boxes with residue numbers indicated and their observed or predicted folds colored according to the legend. NUP160: dilated cardiomyopathy (Tarazón et al. 2012), steroid-resistant nephrotic syndrome (SRNS) (Braun et al. 2018; Zhao et al. 2019), B-cell acute lymphoblastic leukemia (B-ALL) (Harvey et al. 2010); NUP75: congestive heart failure (Satoh et al. 2007), SRNS (Braun et al. 2018); SEC13: breast cancer (Li and Liu 2021), gastric cancer (Mottaghi-Dastjerdi et al. 2015); NUP107: cardiac arrhythmia (Guan et al. 2019), SRNS (Braun et al. 2018; Guan et al. 2019), microcephaly (Rosti et al. 2017), ovarian dysgenesis (Weinberg-Shukron et al. 2015), breast cancer (Li and Liu 2021), brain cancer, glioblastoma multiforme (Hodgson et al. 2009), dedifferentiated liposarcoma (Wang et al. 2012); NUP133: SRNS (Braun et al. 2018), breast cancer (Hernández et al. 2007); NUP43: aortic dilatation (Haskell et al. 2017); NUP37: breast cancer (Li and Liu 2021), atrial fibrillation (Haskell et al. 2017), SRNS (Braun et al. 2018); NUP205: congenital heart disease associated with situs inversus and heterotaxy (Chen et al. 2019), SRNS (Braun et al. 2016); NUP188: congenital heart disease associated with heterotaxy (Fakhro et al. 2011), mitral valve prolapse (Haskell et al. 2017), Sandestig–Stefanova syndrome (Muir et al. 2020; Sandestig et al. 2020), B-ALL (Nowak et al. 2010); NUP93: dilated cardiomyopathy (Tarazón et al. 2012), SRNS (Braun et al. 2016; Sandokji et al. 2019), Down syndrome (Lima et al. 2011); NUP155: atrial fibrillation (Oberti et al. 2004; Zhang et al. 2008). (Continued) ALADIN: Triple A syndrome (Huebner et al. 2004); NUP62: ischemic cardiomyopathy (Chahine et al. 2015), primary biliary cirrhosis (Wesierska-Gadek et al. 1996), amyotrophic lateral sclerosis (ALS) (Aizawa et al. 2019), infantile striatonigral degeneration (Basel-Vanagaite et al. 2006), protein degradation induced by poliovirus (Gustin and Sarnow 2001) and rhinovirus (Gustin and Sarnow 2002) infections, hyperphosphorylation induced by cardiovirus infection (Porter and Palmenbert 2009), protein mislocalization induced by HIV-1 infection (Monette et al. 2011); NUP98: acute myeloid leukemia (Borrow et al. 1996; Nakamura et al. 1996; Xu and Powers 2009; Köhler and Hurt 2010; Gough et al. 2011), acute megakaryoblastic leukemia without Down syndrome (Roussy et al. 2018; Lalonde et al. 2021), protein degradation induced by poliovirus infection (Park et al. 2008), liver cancer (Singer et al. 2012); RAE1: acute myeloid leukemia (Roussy et al. 2018; Lalonde et al. 2021), breast cancer (Funasaka et al. 2011), herpesviruses (Gong et al. 2016), influenza A (Satterly et al. 2007), vesicular stomatitis virus (VSV) (Faria et al. 2005; Ren et al. 2010), and SARS-CoV2 (Oh et al. 2019) virulence factor binding; NUP88: fetal akinesia deformation sequence (Miorin et al. 2020), prostate, ovarian, breast, hepatocellular, colon, and lung cancers, and mesotheliomas and lymphomas (Bonnin et al. 2018); NUP214: acute febrile encephalopathy (Martinez et al. 1999; Gould et al. 2000), acute myeloid leukemia (von Lindern et al. 1992; Kraemer et al. 1994), breast cancer (Hernández et al. 2007), hyperphosphorylation induced by cardiovirus infection (Porter and Palmenberg 2009), protein degradation induced by rhinovirus infection (Ghildyal et al. 2009); GLE1: ALS (Kaneb et al. 2015), lethal congenital contracture syndrome (Nousiainen et al. 2008), lethal arthrogryposis with anterior horn cell disease (Paakkola et al. 2018); NUP358: acute myelomonocytic leukemia (Paakkola et al. 2018), dilated cardiomyopathy (Tarazón et al. 2012), ischemic cardiomyopathy (Lim et al. 2014), acute necrotizing encephalopathy (Neilson et al. 2009; Sell et al. 2016), colorectal cancer (Gylfe et al. 2013), protein degradation induced by rhinovirus infection (Ghildyal et al. 2009), HIV-1 virulence factor binding (Di Nunzio et al. 2012); NUP153: dilated cardiomyopathy and ischemic cardiomyopathy (Tarazón et al. 2012), cardiac arrhythmia (Nanni et al. 2016), protein degradation induced by poliovirus and rhinovirus (Gustin and Sarnow 2002) infections, hyperphosphorylation induced by cardiovirus infection (Porter and Palmenberg 2009), HIV-1 virulence factor binding (Matreyek et al. 2013), autoimmune liver disease/rheumatic disease (Enarson et al. 2004); TPR: gastric, lung, bone, thyroid, and colorectal cancers (Soman et al. 1991; Greco et al. 1997; Yu et al. 2000), autoimmune liver disease/rheumatic disease (Enarson et al. 2004); NDC1: dilated cardiomyopathy and ischemic cardiomyopathy (Tarazón et al. 2012); POM210: congenital heart disease (Chen et al. 2019), breast cancer (Curtis et al. 2012; Amin et al. 2021), primary biliary cirrhosis (Courvalin et al. 1990), colorectal cancer (Landi et al. 2012), and cervical cancer (Rajkumar et al. 2011). (fs) Frameshift, (*) nonsense mutation, (Δ) deletion.
Aberrant function of nucleoporins and NPCs has been prominently linked to autoimmune diseases, neurodegenerative diseases, cardiomyopathies, developmental disorders, and cancers. The first case of a nucleoporin implicated in human disease was found in a chromosomal rearrangement that fuses the 3′ end of NUP214 (also known as CAN) to the 5′ end of DEK, leading to acute myeloid leukemia (AML) (von Lindern et al. 1992). Gene fusions involving NUP358, NUP98, and TPR are also linked to leukemias, with more than 25 leukemogenic gene fusions involving NUP98 alone (Borrow et al. 1996; Nakamura et al. 1996; Xu and Powers 2009; Köhler and Hurt 2010; Gough et al. 2011). Leukemogenic fusions combine NUP98's natively unfolded amino-terminal FG-repeat region and part of the scaffold-binding linker region with proteins involved in transcriptional and epigenetic regulation. An attractive mechanism for NUP98 fusion protein-induced leukemogenesis involves NUP98 modulating the fusion partner's activity. For example, a NUP98-PHD finger chimera was shown to interfere with deposition of H3K27me3 marks on histones, preventing the cell from silencing developmentally significant genes, ultimately causing AML in murine models (Wang et al. 2009). NUP98's propensity to form LLPS likely contributes to this epigenetic reprogramming, as shown for NUP98 fused with the homeodomain-containing transcription factor HOXA9, which promotes HOXA9 association with genomic targets, resulting in super-enhancer-like long-distance chromatin looping that up-regulates proto-oncogenes (Ahn et al. 2021).
Aberrant nucleoporin expression levels have also been associated with several diseases. Notably, NUP88 is overexpressed in various cancers and is used as a tumor marker (Martinez et al. 1999). Increased expression of scaffold nucleoporins NUP160, NUP75, NUP107, and NUP93 has been linked to cardiovascular diseases (Satoh et al. 2007; Tarazón et al. 2012; Guan et al. 2019). Missense and nonsense mutations in scaffold nucleoporins NUP160, NUP75, NUP107, NUP133, NUP37, NUP93, and NUP205, which likely perturb the structure of the NPC and thus affect nucleocytoplasmic transport, have been associated with steroid-resistant nephrotic syndrome (SRNS) (Braun et al. 2016, 2018; Guan et al. 2019; Sandokji et al. 2019; Zhao et al. 2019). In other cases, the effect of disease-associated mutations on nucleoporin structure and function are more subtle. Point mutations in the amino-terminal domain of NUP358 linked to familial acute necrotizing encephalopathy 1 (ANE1) (Neilson et al. 2009; Sell et al. 2016) do not disturb the overall protein fold, yet reduce its thermosolubility in vitro (Bley et al. 2022). The same molecular phenotype is observed for GLE1 variants associated with neurodegenerative diseases (Nousiainen et al. 2008; Kaneb et al. 2015; Lin et al. 2018; Paakkola et al. 2018). Generally, the molecular mechanisms by which aberrations in nucleoporins and NPC structure relate to disease etiologies are not well-understood. More patient genomic sequencing and the recently determined near-atomic structure of the human NPC (Bley et al. 2022; Mosalaganti et al. 2022) are expected to provide a rich foundation for the discovery and functional characterization of nucleoporin alterations and establishing the clinical relevance of NPC function.
VIRAL TARGETING OF THE NUCLEAR PORE COMPLEX
The hijacking of the NPC and nucleocytoplasmic transport is a common event in the life cycle of a wide variety of viruses. Viruses with nuclear replication cycles have evolved various strategies to exploit the cellular transport machinery, because they have to transport viral proteins, genomes, and even intact capsids across the nuclear envelope. For example, the genomic RNA fragments of influenza A that are released from the viral particle upon its disassembly in the cytoplasm are coated with NLS-containing viral nucleoproteins, recognized by α-karyopherins, and transported into the nucleus (Neumann et al. 1997; Melén et al. 2003). In the case of herpesviruses and adenoviruses, their icosahedral capsids remain intact until they attach to the cytoplasmic face of the NPC, where partial capsid disassembly and viral genome release into the nucleus is triggered (Sodeik et al. 1997; Ojala et al. 2000; Shahin et al. 2006; Fatahzadeh and Schwartz 2007). Finally, the NPC's central transport channel is large enough for the intact NLS-displaying hepatitis B virus (HBV) or human immunodeficiency virus 1 (HIV-1) capsids to traverse the nuclear pore in a karyopherin-dependent manner before their disassembly in the nucleus (Kann et al. 1999; Zila et al. 2021).
Beyond serving as a gateway into the nucleus, the NPC represents a key target for countering the host cell's immune response and redirecting its physiology to favor viral replication. Poliovirus (PV) and human rhinovirus (HRV) encode proteases that degrade a subset of FG repeat-containing nucleoporins to disrupt the NPC's diffusion barrier (Gustin and Sarnow 2001, 2002). Human papillomavirus (HPV), severe acute respiratory syndrome coronavirus (SARS-CoV), and Ebola virus (EBOV) sequester karyopherins from their intended host cargo, thereby disrupting the nuclear import of antiviral transcription factors (Nelson et al. 2002, 2003; Frieman et al. 2007; Kopecky-Bromberg et al. 2007; Reid et al. 2007; Hussain et al. 2008; Mateo et al. 2011). Virulence factors of herpesviruses, influenza A, vesicular stomatitis virus (VSV), and SARS-CoV-2 target the nucleoporin RAE1 to inhibit mRNA export, thereby suppressing innate immune response mechanisms, decreasing the abundance of host mRNAs, and outcompeting the host for the available translation machinery and anabolic resources (Enninga et al. 2002; Satterly et al. 2007; Quan et al. 2014; Gong et al. 2016; Oh et al. 2019; Miorin et al. 2020). The near-atomic composite structure of the human NPC (Bley et al. 2022; Mosalaganti et al. 2022) puts the interactions between nucleoporins and viruses or viral virulence factors in the context of the intact NPC.
CONCLUDING REMARKS
Since the discovery of nuclear pores in 1950, the structure and function of the NPC and the nucleocytoplasmic transport machinery have been an area of active research. Over the last 20 years, efforts to solve the NPC structure have culminated in near-atomic snapshots of NPCs from several species, with only a few blind spots soon to be revealed, particularly in the nuclear basket and integral membrane portions of the NPC. The increasing detail of cryo-EM reconstructions and AI-based structural approximations are providing the first glimpses of these missing areas. Experimental structures, biochemical reconstitution, and functional studies will be vital to avoid forgoing crucial and unexpected biological insights that may arise from characterizing these missing areas. In situ imaging enabled by FIB milling of whole-cell specimens has revealed dilated states of NPCs from various species, providing glances of the NPC's membrane tension-sensing dynamics and extending the current static snapshot view of the NPC to the first frames of a molecular movie. Facing forward, the most enigmatic and challenging task for the field of nucleocytoplasmic transport is to unravel the mechanisms of mRNP remodeling at the NPC and mRNA export, along with their coupling to upstream and downstream processes. Additionally, understanding the molecular mechanisms of the myriad nucleoporin-associated diseases and viral interference mechanisms that involve nuclear transport and the NPC offers a chance for the design of novel therapies. The determination of the near-atomic composite structure of the human NPC is an important milestone that now enables tackling these problems from a molecular perspective.
ACKNOWLEDGMENTS
We apologize to the colleagues whose work could not be adequately discussed because of the length constraint of this review. We thank all current and former members of the Hoelz laboratory for the drive, tenacity, and acumen that have made the laboratory's contributions to the nucleocytoplasmic transport field possible; Ana Correia for her insight into nucleoporin diseases; and Valerie Altounian for the preparation of the NPC schematic. A.H. is a Faculty Scholar of the Howard Hughes Medical Institute and was supported by National Institutes of Health Grants R01-GM111461 and R01-GM117360.
Footnotes
Editors: Susan Ferro-Novick, Tom A. Rapoport, and Randy Schekman
Additional Perspectives on The Endoplasmic Reticulum available at www.cshperspectives.org
REFERENCES
- Ader C, Frey S, Maas W, Schmidt HB, Görlich D, Baldus M. 2010. Amyloid-like interactions within nucleoporin FG hydrogels. Proc Natl Acad Sci 107: 6281–6285. 10.1073/pnas.0910163107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JH, Davis ES, Daugird TA, Zhao S, Quiroga IY, Uryu H, Li J, Storey AJ, Tsai YH, Keeley DP, et al. 2021. Phase separation drives aberrant chromatin looping and cancer development. Nature 595: 591–595. 10.1038/s41586-021-03662-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H, Yamashita T, Kato H, Kimura T, Kwak S. 2019. Impaired nucleoporins are present in sporadic amyotrophic lateral sclerosis motor neurons that exhibit mislocalization of the 43-kDa TAR DNA-binding protein. J Clin Neurol 15: 62–67. 10.3988/jcn.2019.15.1.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey CW. 1995. Structural plasticity of the nuclear pore complex. J Mol Biol 248: 273–293. [DOI] [PubMed] [Google Scholar]
- Akey CW, Radermacher M. 1993. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J Cell Biol 122: 1–19. 10.1083/jcb.122.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey CW, Singh D, Ouch C, Echeverria I, Nudelman I, Varberg JM, Yu Z, Fang F, Shi Y, Wang J, et al. 2022. Comprehensive structure and functional adaptations of the yeast nuclear pore complex. Cell 185: 361–378.e25. 10.1016/j.cell.2021.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazár-Román AR, Tran EJ, Guo S, Wente SR. 2006. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol 8: 711–716. 10.1038/ncb1427 [DOI] [PubMed] [Google Scholar]
- Allegretti M, Zimmerli CE, Rantos V, Wilfling F, Ronchi P, Fung HKH, Lee CW, Hagen W, Turoňová B, Karius K, et al. 2020. In-cell architecture of the nuclear pore and snapshots of its turnover. Nature 586: 796–800. 10.1038/s41586-020-2670-5 [DOI] [PubMed] [Google Scholar]
- Amin R, Shukla A, Zhu JJ, Kim S, Wang P, Tian SZ, Tran AD, Paul D, Cappell SD, Burkett S, et al. 2021. Nuclear pore protein NUP210 depletion suppresses metastasis through heterochromatin-mediated disruption of tumor cell mechanical response. Nat Commun 12: 7216. 10.1038/s41467-021-27451-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. 2011. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146: 277–289. 10.1016/j.cell.2011.06.039 [DOI] [PubMed] [Google Scholar]
- Andersen KR, Onischenko E, Tang JH, Kumar P, Chen JZ, Ulrich A, Liphardt JT, Weis K, Schwartz TU. 2013. Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. eLife 2: e00745. 10.7554/eLife.00745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa H, Kojidani T, Yang HJ, Ohtsuki C, Osakada H, Matsuda A, Iwamoto M, Chikashige Y, Nagao K, Obuse C, et al. 2019. Asymmetrical localization of Nup107-160 subcomplex components within the nuclear pore complex in fission yeast. PLoS Genet 15: e1008061. 10.1371/journal.pgen.1008061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Muncher L, Straussberg R, Pasmanik-Chor M, Yahav M, Rainshtein L, Walsh CA, Magal N, Taub E, Drasinover V, et al. 2006. Mutated nup62 causes autosomal recessive infantile bilateral striatal necrosis. Ann Neurol 60: 214–222. 10.1002/ana.20902 [DOI] [PubMed] [Google Scholar]
- Bataillé N, Helser T, Fried HM. 1990. Cytoplasmic transport of ribosomal subunits microinjected into the Xenopus laevis oocyte nucleus: a generalized, facilitated process. J Cell Biol 111: 1571–1582. 10.1083/jcb.111.4.1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Förster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. 2004. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306: 1387–1390. 10.1126/science.1104808 [DOI] [PubMed] [Google Scholar]
- Berke IC, Boehmer T, Blobel G, Schwartz TU. 2004. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J Cell Biol 167: 591–597. 10.1083/jcb.200408109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S, Schwartz TU. 2012. Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex. Proc Natl Acad Sci 109: 15241–15246. 10.1073/pnas.1205151109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S, Schwartz TU. 2013. Structural and functional studies of the 252 kDa nucleoporin ELYS reveal distinct roles for its three tethered domains. Structure 21: 572–580. 10.1016/j.str.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, Mukherjee S, Harvey S, Huber FM, Lin DH, et al. 2022. Architecture of the cytoplasmic face of the nuclear pore. Science 376: eabm9129. 10.1126/science.abm9129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G, Sabatini DD. 1971. Ribosome–membrane interaction in eukaryotic cells. In Biomembranes: Volume 2 (ed. Manson LA), pp. 193–195. Springer, Boston, MA. [Google Scholar]
- Boehmer T, Jeudy S, Berke IC, Schwartz TU. 2008. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol Cell 30: 721–731. 10.1016/j.molcel.2008.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner WM. 1975. Protein migration into nuclei. I: Frog oocyte nuclei in vivo accumulate microinjected histones, allow entry to small proteins, and exclude large proteins. J Cell Biol 64: 421–430. 10.1083/jcb.64.2.421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin E, Cabochette P, Filosa A, Jühlen R, Komatsuzaki S, Hezwani M, Dickmanns A, Martinelli V, Vermeersch M, Supply L, et al. 2018. Biallelic mutations in nucleoporin NUP88 cause lethal fetal akinesia deformation sequence. PLoS Genet 14: e1007845. 10.1371/journal.pgen.1007845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow J, Shearman AM, Stanton VP Jr, Becher R, Collins T, Williams AJ, Dubé I, Katz F, Kwong YL, Morris C, et al. 1996. The t(7;11)(p15;p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin NUP96 and class I homeoprotein HOXA9. Nat Genet 12: 159–167. 10.1038/ng0296-159 [DOI] [PubMed] [Google Scholar]
- Braun DA, Sadowski CE, Kohl S, Lovric S, Astrinidis SA, Pabst WL, Gee HY, Ashraf S, Lawson JA, Shril S, et al. 2016. Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat Genet 48: 457–465. 10.1038/ng.3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Lovric S, Schapiro D, Schneider R, Marquez J, Asif M, Hussain MS, Daga A, Widmeier E, Rao J, et al. 2018. Mutations in multiple components of the nuclear pore complex cause nephrotic syndrome. J Clin Invest 128: 4313–4328. 10.1172/JCI98688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Schwartz TU. 2009. Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice. Nat Struct Mol Biol 16: 1173–1177. 10.1038/nsmb.1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. 2008. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322: 1369–1373. 10.1126/science.1165886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JA, Bharathi A, Ghosh A, Whalen W, Fitzgerald E, Dhar R. 1995. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J Biol Chem 270: 7411–7419. 10.1074/jbc.270.13.7411 [DOI] [PubMed] [Google Scholar]
- Bui KH, von Appen A, DiGuilio AL, Ori A, Sparks L, Mackmull MT, Bock T, Hagen W, Andrés-Pons A, Glavy JS, et al. 2013. Integrated structural analysis of the human nuclear pore complex scaffold. Cell 155: 1233–1243. 10.1016/j.cell.2013.10.055 [DOI] [PubMed] [Google Scholar]
- Callan HG, Tomlin SG. 1950. Experimental studies on amphibian oocyte nuclei. I: Investigation of the structure of the nuclear membrane by means of the electron microscope. Proc R Soc Lond B Biol Sci 137: 367–378. 10.1098/rspb.1950.0047 [DOI] [PubMed] [Google Scholar]
- Celetti G, Paci G, Caria J, VanDelinder V, Bachand G, Lemke EA. 2020. The liquid state of FG-nucleoporins mimics permeability barrier properties of nuclear pore complexes. J Cell Biol 219: e201907157. 10.1083/jcb.201907157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahine MN, Mioulane M, Sikkel MB, O'Gara P, Dos Remedios CG, Pierce GN, Lyon AR, Földes G, Harding SE. 2015. Nuclear pore rearrangements and nuclear trafficking in cardiomyocytes from rat and human failing hearts. Cardiovasc Res 105: 31–43. 10.1093/cvr/cvu218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zhang Y, Yang S, Shi Z, Zeng W, Lu Z, Zhou X. 2019. Bi-allelic mutations in NUP205 and NUP210 are associated with abnormal cardiac left-right patterning. Circ Genom Precis Med 12: e002492. 10.1161/CIRCGEN.119.002492 [DOI] [PubMed] [Google Scholar]
- Chug H, Trakhanov S, Hülsmann BB, Pleiner T, Görlich D. 2015. Crystal structure of the metazoan Nup62• Nup58•Nup54 nucleoporin complex. Science 350: 106–110. 10.1126/science.aac7420 [DOI] [PubMed] [Google Scholar]
- Collins R, Karlberg T, Lehtiö L, Schütz P, van den Berg S, Dahlgren LG, Hammarström M, Weigelt J, Schüler H. 2009. The DEXD/H-box RNA helicase DDX19 is regulated by an α-helical switch. J Biol Chem 284: 10296–10300. 10.1074/jbc.C900018200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. 2007. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem 76: 647–671. 10.1146/annurev.biochem.76.052705.161529 [DOI] [PubMed] [Google Scholar]
- Courvalin JC, Lassoued K, Bartnik E, Blobel G, Wozniak RW. 1990. The 210-kD nuclear envelope polypeptide recognized by human autoantibodies in primary biliary cirrhosis is the major glycoprotein of the nuclear pore. J Clin Invest 86: 279–285. 10.1172/JCI114696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. 2002. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 158: 915–927. 10.1083/jcb.200206106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486: 346–352. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeko E, Seizl M, Augsberger C, Mielke T, Cramer P. 2011. Iwr1 directs RNA polymerase II nuclear import. Mol Cell 42: 261–266. 10.1016/j.molcel.2011.02.033 [DOI] [PubMed] [Google Scholar]
- Davis LI, Blobel G. 1986. Identification and characterization of a nuclear pore complex protein. Cell 45: 699–709. 10.1016/0092-8674(86)90784-1 [DOI] [PubMed] [Google Scholar]
- Davis LI, Fink GR. 1990. The NUP1 gene encodes an essential component of the yeast nuclear pore complex. Cell 61: 965–978. 10.1016/0092-8674(90)90062-J [DOI] [PubMed] [Google Scholar]
- Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, Hoelz A. 2008. A fence-like coat for the nuclear pore membrane. Mol Cell 32: 815–826. 10.1016/j.molcel.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. 2003. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci 100: 2450–2455. 10.1073/pnas.0437902100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nunzio F, Danckaert A, Fricke T, Perez P, Fernandez J, Perret E, Roux P, Shorte S, Charneau P, Diaz-Griffero F, et al. 2012. Human nucleoporins promote HIV-1 docking at the nuclear pore, nuclear import and integration. PLoS ONE 7: e46037. 10.1371/journal.pone.0046037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Casella JF, Gautier R, Boehmer T, Schwartz TU, Antonny B. 2007. A general amphipathic α-helical motif for sensing membrane curvature. Nat Struct Mol Biol 14: 138–146. 10.1038/nsmb1194 [DOI] [PubMed] [Google Scholar]
- Eibauer M, Pellanda M, Turgay Y, Dubrovsky A, Wild A, Medalia O. 2015. Structure and gating of the nuclear pore complex. Nat Commun 6: 7532. 10.1038/ncomms8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enarson P, Rattner JB, Ou Y, Miyachi K, Horigome T, Fritzler MJ. 2004. Autoantigens of the nuclear pore complex. J Mol Med (Berl) 82: 423–433. 10.1007/s00109-004-0554-z [DOI] [PubMed] [Google Scholar]
- Enninga J, Levy DE, Blobel G, Fontoura BM. 2002. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science 295: 1523–1525. 10.1126/science.1067861 [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Hurt EC, Aebi U, Panté N. 1998. Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes. J Cell Biol 143: 577–588. 10.1083/jcb.143.3.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. 2011. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci 108: 2915–2920. 10.1073/pnas.1019645108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria PA, Chakraborty P, Levay A, Barber GN, Ezelle HJ, Enninga J, Arana C, van Deursen J, Fontoura BM. 2005. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell 17: 93–102. 10.1016/j.molcel.2004.11.023 [DOI] [PubMed] [Google Scholar]
- Fatahzadeh M, Schwartz RA. 2007. Human herpes simplex virus infections: epidemiology, pathogenesis, symptomatology, diagnosis, and management. J Am Acad Dermatol 57: 737–763; quiz 764-736. 10.1016/j.jaad.2007.06.027 [DOI] [PubMed] [Google Scholar]
- Fischer J, Teimer R, Amlacher S, Kunze R, Hurt E. 2015. Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel. Nat Struct Mol Biol 22: 774–781. 10.1038/nsmb.3084 [DOI] [PubMed] [Google Scholar]
- Franke WW, Scheer U. 1970. The ultrastructure of the nuclear envelope of amphibian oocytes: a reinvestigation. II: The immature oocyte and dynamic aspects. J Ultrastruct Res 30: 317–327. 10.1016/S0022-5320(70)80065-X [DOI] [PubMed] [Google Scholar]
- Frey S, Görlich D. 2007. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130: 512–523. 10.1016/j.cell.2007.06.024 [DOI] [PubMed] [Google Scholar]
- Frey S, Richter RP, Görlich D. 2006. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314: 815–817. 10.1126/science.1132516 [DOI] [PubMed] [Google Scholar]
- Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol 81: 9812–9824. 10.1128/JVI.01012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funasaka T, Nakano H, Wu Y, Hashizume C, Gu L, Nakamura T, Wang W, Zhou P, Moore MA, Sato H, et al. 2011. RNA export factor RAE1 contributes to NUP98-HOXA9-mediated leukemogenesis. Cell Cycle 10: 1456–1467. 10.4161/cc.10.9.15494 [DOI] [PubMed] [Google Scholar]
- Gadal O, Strauß D, Kessl J, Trumpower B, Tollervey D, Hurt E. 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol 21: 3405–3415. 10.1128/MCB.21.10.3405-3415.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. 1967. Octagonal nuclear pores. J Cell Biol 32: 391–399. 10.1083/jcb.32.2.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace L, Ottaviano Y, Kondor-Koch C. 1982. Identification of a major polypeptide of the nuclear pore complex. J Cell Biol 95: 826–837. 10.1083/jcb.95.3.826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildyal R, Jordan B, Li D, Dagher H, Bardin PG, Gern JE, Jans DA. 2009. Rhinovirus 3C protease can localize in the nucleus and alter active and passive nucleocytoplasmic transport. J Virol 83: 7349–7352. 10.1128/JVI.01748-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PJ, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ. 2007. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr Biol 17: 1657–1662. 10.1016/j.cub.2007.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Navarro N, Peiró-Chova L, Rodriguez-Navarro S, Polaina J, Estruch F. 2013. Rtp1p is a karyopherin-like protein required for RNA polymerase II biogenesis. Mol Cell Biol 33: 1756–1767. 10.1128/MCB.01449-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, Kim YH, Xiao Y, Du Y, Xie Y, Lee KK, Feng J, Farhat N, Zhao D, Shu S, et al. 2016. A herpesvirus protein selectively inhibits cellular mRNA nuclear export. Cell Host Microbe 20: 642–653. 10.1016/j.chom.2016.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough SM, Slape CI, Aplan PD. 2011. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood 118: 6247–6257. 10.1182/blood-2011-07-328880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould VE, Martinez N, Orucevic A, Schneider J, Alonso A. 2000. A novel, nuclear pore-associated, widely distributed molecule overexpressed in oncogenesis and development. Am J Pathol 157: 1605–1613. 10.1016/S0002-9440(10)64798-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco A, Miranda C, Pagliardini S, Fusetti L, Bongarzone I, Pierotti MA. 1997. Chromosome I rearrangements involving the genes TPR and NTRK1 produce structurally different thyroid-specific TRK oncogenes. Genes Chromosom Cancer 19: 112–123. [DOI] [PubMed] [Google Scholar]
- Guan Y, Gao X, Tang Q, Huang L, Gao S, Yu S, Huang J, Li J, Zhou D, Zhang Y, et al. 2019. Nucleoporin 107 facilitates the nuclear export of Scn5a mRNA to regulate cardiac bioelectricity. J Cell Mol Med 23: 1448–1457. 10.1111/jcmm.14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin KE, Sarnow P. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J 20: 240–249. 10.1093/emboj/20.1.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin KE, Sarnow P. 2002. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J Virol 76: 8787–8796. 10.1128/JVI.76.17.8787-8796.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gylfe AE, Kondelin J, Turunen M, Ristolainen H, Katainen R, Pitkänen E, Kaasinen E, Rantanen V, Tanskanen T, Varjosalo M, et al. 2013. Identification of candidate oncogenes in human colorectal cancers with microsatellite instability. Gastroenterology 145: 540–543.e22. 10.1053/j.gastro.2013.05.015 [DOI] [PubMed] [Google Scholar]
- Hallberg E, Wozniak RW, Blobel G. 1993. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J Cell Biol 122: 513–521. 10.1083/jcb.122.3.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey RC, Mullighan CG, Wang X, Dobbin KK, Davidson GS, Bedrick EJ, Chen IM, Atlas SR, Kang H, Ar K, et al. 2010. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics, and outcome. Blood 116: 4874–4884. 10.1182/blood-2009-08-239681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haskell GT, Jensen BC, Samsa LA, Marchuk D, Huang W, Skrzynia C, Tilley C, Seifert BA, Rivera-Muñoz EA, Koller B, et al. 2017. Whole exome sequencing identifies truncating variants in nuclear envelope genes in patients with cardiovascular disease. Circ Cardiovasc Genet 10: e001443. 10.1161/CIRCGENETICS.116.001443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández P, Solé X, Valls J, Moreno V, Capellá G, Urruticoechea A, Pujana MA. 2007. Integrative analysis of a cancer somatic mutome. Mol Cancer 6: 13. 10.1186/1476-4598-6-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE, Carragher BO, Milligan RA. 1992. Architecture and design of the nuclear pore complex. Cell 69: 1133–1141. 10.1016/0092-8674(92)90635-P [DOI] [PubMed] [Google Scholar]
- Ho JH, Kallstrom G, Johnson AW. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol 151: 1057–1066. 10.1083/jcb.151.5.1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel AE, Hodel MR, Griffis ER, Hennig KA, Ratner GA, Xu S, Powers MA. 2002. The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98. Mol Cell 10: 347–358. 10.1016/S1097-2765(02)00589-0 [DOI] [PubMed] [Google Scholar]
- Hodgson JG, Yeh RF, Ray A, Wang NJ, Smirnov I, Yu M, Hariono S, Silber J, Feiler HS, Gray JW, et al. 2009. Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro Oncol 11: 477–487. 10.1215/15228517-2008-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia KC, Stavropoulos P, Blobel G, Hoelz A. 2007. Architecture of a coat for the nuclear pore membrane. Cell 131: 1313–1326. 10.1016/j.cell.2007.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Zhan X, Zeng C, Liang K, Zhu X, Zhao Y, Wang P, Wang Q, Zhou Q, Tao Q, et al. 2022. Cryo-EM structure of the inner ring from Xenopus laevis nuclear pore complex. Cell Res 32: 451–460. 10.1038/s41422-022-00633-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner A, Kaindl AM, Knobeloch KP, Petzold H, Mann P, Koehler K. 2004. The triple a syndrome is due to mutations in ALADIN, a novel member of the nuclear pore complex. Endocr Res 30: 891–899. 10.1081/ERC-200044138 [DOI] [PubMed] [Google Scholar]
- Hughes MP, Sawaya MR, Boyer DR, Goldschmidt L, Rodriguez JA, Cascio D, Chong L, Gonen T, Eisenberg DS. 2018. Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science 359: 698–701. 10.1126/science.aan6398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Taft RJ, Asgari S. 2008. An insect virus-encoded microRNA regulates viral replication. J Virol 82: 9164–9170. 10.1128/JVI.01109-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeudy S, Schwartz TU. 2007. Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture. J Biol Chem 282: 34904–34912. 10.1074/jbc.M705479200 [DOI] [PubMed] [Google Scholar]
- Kaneb HM, Folkmann AW, Belzil VV, Jao LE, Leblond CS, Girard SL, Daoud H, Noreau A, Rochefort D, Hince P, et al. 2015. Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Hum Mol Genet 24: 1363–1373. 10.1093/hmg/ddu545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kann M, Sodeik B, Vlachou A, Gerlich WH, Helenius A. 1999. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J Cell Biol 145: 45–55. 10.1083/jcb.145.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley K, Knockenhauer KE, Kabachinski G, Schwartz TU. 2015. Atomic structure of the Y complex of the nuclear pore. Nat Struct Mol Biol 22: 425–431. 10.1038/nsmb.2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Raveh B, Herricks T, Slaughter BD, Hogan JA, Upla P, et al. 2018. Integrative structure and functional anatomy of a nuclear pore complex. Nature 555: 475–482. 10.1038/nature26003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Lusk CP, Blobel G. 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature 442: 1003–1007. 10.1038/nature05075 [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. 2000. Three ways to make a vesicle. Nat Rev Mol Cell Biol 1: 187–198. 10.1038/35043117 [DOI] [PubMed] [Google Scholar]
- Kiseleva E, Goldberg MW, Allen TD, Akey CW. 1998. Active nuclear pore complexes in Chironomus: visualization of transporter configurations related to mRNP export. J Cell Sci 111: 223–236. 10.1242/jcs.111.2.223 [DOI] [PubMed] [Google Scholar]
- Kobayashi W, Takizawa Y, Aihara M, Negishi L, Ishii H, Kurumizaka H. 2019. Structural and biochemical analyses of the nuclear pore complex component ELYS identify residues responsible for nucleosome binding. Commun Biol 2: 163. 10.1038/s42003-019-0385-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A, Hurt E. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8: 761–773. 10.1038/nrm2255 [DOI] [PubMed] [Google Scholar]
- Köhler A, Hurt E. 2010. Gene regulation by nucleoporins and links to cancer. Mol Cell 38: 6–15. 10.1016/j.molcel.2010.01.040 [DOI] [PubMed] [Google Scholar]
- Kopecky-Bromberg SA, Martínez-Sobrido L, Frieman M, Baric RA, Palese P. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81: 548–557. 10.1128/JVI.01782-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosako H, Imamoto N. 2010. Phosphorylation of nucleoporins: signal transduction–mediated regulation of their interaction with nuclear transport receptors. Nucleus 1: 309–313. 10.4161/nucl.1.4.11744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski J, Mosalaganti S, von Appen A, Teimer R, DiGuilio AL, Wan W, Bui KH, Hagen WJ, Briggs JA, Glavy JS, et al. 2016. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science 352: 363–365. 10.1126/science.aaf0643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer D, Blobel G. 1997. mRNA binding protein mrnp 41 localizes to both nucleus and cytoplasm. Proc Natl Acad Sci 94: 9119–9124. 10.1073/pnas.94.17.9119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer D, Wozniak RW, Blobel G, Radu A. 1994. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc Natl Acad Sci 91: 1519–1523. 10.1073/pnas.91.4.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralt A, Jagalur NB, van den Boom V, Lokareddy RK, Steen A, Cingolani G, Fornerod M, Veenhoff LM. 2015. Conservation of inner nuclear membrane targeting sequences in mammalian Pom121 and yeast Heh2 membrane proteins. Mol Biol Cell 26: 3301–3312. 10.1091/mbc.e15-03-0184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labokha AA, Gradmann S, Frey S, Hülsmann BB, Urlaub H, Baldus M, Görlich D. 2013. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J 32: 204–218. 10.1038/emboj.2012.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde E, Rentas S, Wertheim G, Cao K, Surrey LF, Lin F, Zhao X, Obstfeld A, Aplenc R, Luo M, et al. 2021. Clinical impact of genomic characterization of 15 patients with acute megakaryoblastic leukemia-related malignancies. Cold Spring Harb Mol Case Stud 7: a005975. 10.1101/mcs.a005975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi D, Gemignani F, Pardini B, Naccarati A, Garritano S, Vodicka P, Vodickova L, Canzian F, Novotny J, Barale R, et al. 2012. Identification of candidate genes carrying polymorphisms associated with the risk of colorectal cancer by analyzing the colorectal mutome and microRNAome. Cancer 118: 4670–4680. 10.1002/cncr.27435 [DOI] [PubMed] [Google Scholar]
- Leksa NC, Brohawn SG, Schwartz TU. 2009. The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture. Structure 17: 1082–1091. 10.1016/j.str.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Liu T. 2021. Evaluation of oncogene NUP37 as a potential novel biomarker in breast cancer. Front Oncol 11: 669655. 10.3389/fonc.2021.669655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JH, Jang S, Park CJ, Cho YU, Lee JH, Lee KH, Lee JO, Shin JY, Kim JI, Huh J, et al. 2014. RANBP2-ALK fusion combined with monosomy 7 in acute myelomonocytic leukemia. Cancer Genet 207: 40–45. 10.1016/j.cancergen.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Lima FA, Moreira-Filho CA, Ramos PL, Brentani H, Lima Lde A, Arrais M, Bento-de-Souza LC, Bento-de-Souza L, Duarte MI, Coutinho A, et al. 2011. Decreased AIRE expression and global thymic hypofunction in down syndrome. J Immunol 187: 3422–3430. 10.4049/jimmunol.1003053 [DOI] [PubMed] [Google Scholar]
- Lin DH, Hoelz A. 2019. The structure of the nuclear pore complex (an update). Annu Rev Biochem 88: 725–783. 10.1146/annurev-biochem-062917-011901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DH, Zimmermann S, Stuwe T, Stuwe E, Hoelz A. 2013. Structural and functional analysis of the C-terminal domain of Nup358/RanBP2. J Mol Biol 425: 1318–1329. 10.1016/j.jmb.2013.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DH, Stuwe T, Schilbach S, Rundlet EJ, Perriches T, Mobbs G, Fan Y, Thierbach K, Huber FM, Collins LN, et al. 2016. Architecture of the symmetric core of the nuclear pore. Science 352: aaf1015. 10.1126/science.aaf1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DH, Correia AR, Cai SW, Huber FM, Jette CA, Hoelz A. 2018. Structural and functional analysis of mRNA export regulation by the nuclear pore complex. Nat Commun 9: 2319. 10.1038/s41467-018-04459-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokareddy RK, Hapsari RA, van Rheenen M, Pumroy RA, Bhardwaj A, Steen A, Veenhoff LM, Cingolani G. 2015. Distinctive properties of the nuclear localization signals of inner nuclear membrane proteins Heh1 and Heh2. Structure 23: 1305–1316. 10.1016/j.str.2015.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund MK, Guthrie C. 2005. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell 20: 645–651. 10.1016/j.molcel.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Mahadevan K, Zhang H, Akef A, Cui XA, Gueroussov S, Cenik C, Roth FP, Palazzo AF. 2013. RanBP2/Nup358 potentiates the translation of a subset of mRNAs encoding secretory proteins. PLoS Biol 11: e1001545. 10.1371/journal.pbio.1001545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan R, Gerace L, Melchior F. 1998. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J Cell Biol 140: 259–270. 10.1083/jcb.140.2.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Förster F, Hyman AA, Plitzko JM, Baumeister W. 2016. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351: 969–972. 10.1126/science.aad8857 [DOI] [PubMed] [Google Scholar]
- Maimon T, Elad N, Dahan I, Medalia O. 2012. The human nuclear pore complex as revealed by cryo-electron tomography. Structure 20: 998–1006. 10.1016/j.str.2012.03.025 [DOI] [PubMed] [Google Scholar]
- Martinez N, Alonso A, Moragues MD, Ponton J, Schneider J. 1999. The nuclear pore complex protein Nup88 is overexpressed in tumor cells. Cancer Res 59: 5408–5411. [PubMed] [Google Scholar]
- Mateo M, Carbonnelle C, Reynard O, Kolesnikova L, Nemirov K, Page A, Volchkova VA, Volchkov VE. 2011. VP24 is a molecular determinant of Ebola virus virulence in guinea pigs. J Infect Dis 204: S1011–S1020. 10.1093/infdis/jir338 [DOI] [PubMed] [Google Scholar]
- Matreyek KA, Yücel SS, Li X, Engelman A. 2013. Nucleoporin NUP153 phenylalanine-glycine motifs engage a common binding pocket within the HIV-1 capsid protein to mediate lentiviral infectivity. PLoS Pathog 9: e1003693. 10.1371/journal.ppat.1003693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Wu J, Blobel G. 1998. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol 140: 499–509. 10.1083/jcb.140.3.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul GG, Deaven L. 1977. Quantitative determination of nuclear pore complexes in cycling cells with differing DNA content. J Cell Biol 73: 748–760. 10.1083/jcb.73.3.748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinema AC, Laba JK, Hapsari RA, Otten R, Mulder FA, Kralt A, van den Bogaart G, Lusk CP, Poolman B, Veenhoff LM. 2011. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science 333: 90–93. 10.1126/science.1205741 [DOI] [PubMed] [Google Scholar]
- Melén K, Fagerlund R, Franke J, Köhler M, Kinnunen L, Julkunen I. 2003. Importin α nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J Biol Chem 278: 28193–28200. 10.1074/jbc.M303571200 [DOI] [PubMed] [Google Scholar]
- Miorin L, Kehrer T, Sanchez-Aparicio MT, Zhang K, Cohen P, Patel RS, Cupic A, Makio T, Mei M, Moreno E, et al. 2020. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci 117: 28344–28354. 10.1073/pnas.2016650117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Sipma W, Veenhoff LM, Van der Giessen E, Onck PR. 2019. The effect of FG-Nup phosphorylation on NPC selectivity: a one-bead-per-amino-acid molecular dynamics study. Int J Mol Sci 20: 596. 10.3390/ijms20030596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monette A, Panté N, Mouland AJ. 2011. HIV-1 remodels the nuclear pore complex. J Cell Biol 193: 619–631. 10.1083/jcb.201008064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. 2011. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472: 238–242. 10.1038/nature09862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosalaganti S, Kosinski J, Albert S, Schaffer M, Strenkert D, Salomé PA, Merchant SS, Plitzko JM, Baumeister W, Engel BD, et al. 2018. In situ architecture of the algal nuclear pore complex. Nat Commun 9: 2361. 10.1038/s41467-018-04739-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turonova B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull M-T, et al. 2022. AI-based structure prediction empowers integrative structural analysis of human nuclear pores. Science 376: eabm9506. 10.1126/science.abm9506 [DOI] [PubMed] [Google Scholar]
- Mottaghi-Dastjerdi N, Soltany-Rezaee-Rad M, Sepehrizadeh Z, Roshandel G, Ebrahimifard F, Setayesh N. 2015. Identification of novel genes involved in gastric carcinogenesis by suppression subtractive hybridization. Hum Exp Toxicol 34: 3–11. 10.1177/0960327114532386 [DOI] [PubMed] [Google Scholar]
- Muir AM, Cohen JL, Sheppard SE, Guttipatti P, Lo TY, Weed N, Doherty D, DeMarzo D, Fagerberg CR, Kjærsgaard L, et al. 2020. Bi-allelic loss-of-function variants in NUP188 cause a recognizable syndrome characterized by neurologic, ocular, and cardiac abnormalities. Am J Hum Genet 106: 623–631. 10.1016/j.ajhg.2020.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR. 1996. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell 7: 1921–1937. 10.1091/mbc.7.12.1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy V, Hsia KC, Debler EW, Kampmann M, Davenport AM, Blobel G, Hoelz A. 2009. Structure of a trimeric nucleoporin complex reveals alternate oligomerization states. Proc Natl Acad Sci 106: 17693–17698. 10.1073/pnas.0909373106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Largaespada DA, Lee MP, Johnson LA, Ohyashiki K, Toyama K, Chen SJ, Willman CL, Chen IM, Feinberg AP, et al. 1996. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet 12: 154–158. 10.1038/ng0296-154 [DOI] [PubMed] [Google Scholar]
- Nanni S, Re A, Ripoli C, Gowran A, Nigro P, D'Amario D, Amodeo A, Crea F, Grassi C, Pontecorvi A, et al. 2016. The nuclear pore protein Nup153 associates with chromatin and regulates cardiac gene expression in dystrophic mdx hearts. Cardiovasc Res 112: 555–567. 10.1093/cvr/cvw204 [DOI] [PubMed] [Google Scholar]
- Napetschnig J, Blobel G, Hoelz A. 2007. Crystal structure of the N-terminal domain of the human protooncogene Nup214/CAN. Proc Natl Acad Sci 104: 1783–1788. 10.1073/pnas.0610828104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napetschnig J, Kassube SA, Debler EW, Wong RW, Blobel G, Hoelz A. 2009. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc Natl Acad Sci 106: 3089–3094. 10.1073/pnas.0813267106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U, Kern H, Mutvei A, Horstmann H, Marshallsay B, Hurt EC. 1990. NSP1: a yeast nuclear envelope protein localized at the nuclear pores exerts its essential function by its carboxy-terminal domain. Cell 61: 979–989. 10.1016/0092-8674(90)90063-K [DOI] [PubMed] [Google Scholar]
- Neilson DE, Adams MD, Orr CM, Schelling DK, Eiben RM, Kerr DS, Anderson J, Bassuk AG, Bye AM, Childs AM, et al. 2009. Infection-triggered familial or recurrent cases of acute necrotizing encephalopathy caused by mutations in a component of the nuclear pore, RANBP2. Am J Hum Genet 84: 44–51. 10.1016/j.ajhg.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson LM, Rose RC, Moroianu J. 2002. Nuclear import strategies of high risk HPV16 L1 major capsid protein. J Biol Chem 277: 23958–23964. 10.1074/jbc.M200724200 [DOI] [PubMed] [Google Scholar]
- Nelson LM, Rose RC, Moroianu J. 2003. The L1 major capsid protein of human papillomavirus type 11 interacts with Kap β2 and Kap β3 nuclear import receptors. Virology 306: 162–169. 10.1016/S0042-6822(02)00025-9 [DOI] [PubMed] [Google Scholar]
- Neumann G, Castrucci MR, Kawaoka Y. 1997. Nuclear import and export of influenza virus nucleoprotein. J Virol 71: 9690–9700. 10.1128/jvi.71.12.9690-9700.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble KN, Tran EJ, Alcazár-Román AR, Hodge CA, Cole CN, Wente SR. 2011. The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes Dev 25: 1065–1077. 10.1101/gad.2040611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordeen SA, Turman DL, Schwartz TU. 2020. Yeast Nup84-Nup133 complex structure details flexibility and reveals conservation of the membrane anchoring ALPS motif. Nat Commun 11: 6060. 10.1038/s41467-020-19885-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousiainen HO, Kestilä M, Pakkasjärvi N, Honkala H, Kuure S, Tallila J, Vuopala K, Ignatius J, Herva R, Peltonen L. 2008. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat Genet 40: 155–157. 10.1038/ng.2007.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak NJ, Sait SN, Zeidan A, Deeb G, Gaile D, Liu S, Ford L, Wallace PK, Wang ES, Wetzler M. 2010. Recurrent deletion of 9q34 in adult normal karyotype precursor B-cell acute lymphoblastic leukemia. Cancer Genet Cytogenet 199: 15–20. 10.1016/j.cancergencyto.2010.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberti C, Wang L, Li L, Dong J, Rao S, Du W, Wang Q. 2004. Genome-wide linkage scan identifies a novel genetic locus on chromosome 5p13 for neonatal atrial fibrillation associated with sudden death and variable cardiomyopathy. Circulation 110: 3753–3759. 10.1161/01.CIR.0000150333.87176.C7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JH, Lee JY, Yu S, Cho Y, Hur S, Nam KT, Kim MH. 2019. RAE1 mediated ZEB1 expression promotes epithelial-mesenchymal transition in breast cancer. Sci Rep 9: 2977. 10.1038/s41598-019-39574-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Schirmer EC, Nishimoto T, Gerace L. 2004. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J Cell Biol 167: 1051–1062. 10.1083/jcb.200409149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala PM, Sodeik B, Ebersold MW, Kutay U, Helenius A. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol Cell Biol 20: 4922–4931. 10.1128/MCB.20.13.4922-4931.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori A, Banterle N, Iskar M, Andrés-Pons A, Escher C, Khanh Bui H, Sparks L, Solis-Mezarino V, Rinner O, Bork P, et al. 2013. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol Syst Biol 9: 648. 10.1038/msb.2013.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paakkola T, Vuopala K, Kokkonen H, Ignatius J, Valkama M, Moilanen JS, Fahiminiya S, Majewski J, Hinttala R, Uusimaa J. 2018. A homozygous I684T in GLE1 as a novel cause of arthrogryposis and motor neuron loss. Clin Genet 93: 173–177. 10.1111/cge.13086 [DOI] [PubMed] [Google Scholar]
- Paine PL, Moore LC, Horowitz SB. 1975. Nuclear envelope permeability. Nature 254: 109–114. 10.1038/254109a0 [DOI] [PubMed] [Google Scholar]
- Pal K, Bandyopadhyay A, Zhou XE, Xu Q, Marciano DP, Brunzelle JS, Yerrum S, Griffin PR, Vande Woude G, Melcher K, et al. 2017. Structural basis of TPR-mediated oligomerization and activation of oncogenic fusion kinases. Structure 25: 867–877.e3. 10.1016/j.str.2017.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park N, Katikaneni P, Skern T, Gustin KE. 2008. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J Virol 82: 1647–1655. 10.1128/JVI.01670-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JR, Schwartz TU. 2009. Crystallographic and biochemical analysis of the Ran-binding zinc finger domain. J Mol Biol 391: 375–389. 10.1016/j.jmb.2009.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Belmont BJ, Sante JM, Rexach MF. 2007. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129: 83–96. 10.1016/j.cell.2007.01.044 [DOI] [PubMed] [Google Scholar]
- Petrovic S, Samanta D, Perriches T, Bley CJ, Thierbach K, Brown B, Nie S, Mobbs GW, Stevens TA, Liu X, et al. 2022. Architecture of the linker-scaffold in the nuclear pore. Science 376: eabm9798. 10.1126/science.abm9798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popken P, Ghavami A, Onck PR, Poolman B, Veenhoff LM. 2015. Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex. Mol Biol Cell 26: 1386–1394. 10.1091/mbc.E14-07-1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter FW, Palmenberg AC. 2009. Leader-induced phosphorylation of nucleoporins correlates with nuclear trafficking inhibition by cardioviruses. J Virol 83: 1941–1951. 10.1128/JVI.01752-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan B, Seo HS, Blobel G, Ren Y. 2014. Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1•Nup98). Proc Natl Acad Sci 111: 9127–9132. 10.1073/pnas.1409076111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkumar T, Sabitha K, Vijayalakshmi N, Shirley S, Bose MV, Gopal G, Selvaluxmy G. 2011. Identification and validation of genes involved in cervical tumourigenesis. BMC Cancer 11: 80. 10.1186/1471-2407-11-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. 1996. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem 65: 271–303. 10.1146/annurev.bi.65.070196.001415 [DOI] [PubMed] [Google Scholar]
- Rasala BA, Ramos C, Harel A, Forbes DJ. 2008. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell 19: 3982–3996. 10.1091/mbc.e08-01-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regmi SG, Lee H, Kaufhold R, Fichtman B, Chen S, Aksenova V, Turcotte E, Harel A, Arnaoutov A, Dasso M. 2020. The nuclear pore complex consists of two independent scaffolds. bioRxiv 10.1101/2020.11.13.381947 [DOI] [Google Scholar]
- Reichelt R, Holzenburg A, Buhle EL Jr, Jarnik M, Engel A, Aebi U. 1990. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol 110: 883–894. 10.1083/jcb.110.4.883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid SP, Valmas C, Martinez O, Sanchez FM, Basler CF. 2007. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin α proteins with activated STAT1. J Virol 81: 13469–13477. 10.1128/JVI.01097-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Seo HS, Blobel G, Hoelz A. 2010. Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc Natl Acad Sci 107: 10406–10411. 10.1073/pnas.1005389107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter D, Lima CD. 2005. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435: 687–692. 10.1038/nature03588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Görlich D. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J 20: 1320–1330. 10.1093/emboj/20.6.1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosti RO, Sotak BN, Bielas SL, Bhat G, Silhavy JL, Aslanger AD, Altunoglu U, Bilge I, Tasdemir M, Yzaguirrem AD, et al. 2017. Homozygous mutation in NUP107 leads to microcephaly with steroid-resistant nephrotic condition similar to Galloway–Mowat syndrome. J Med Genet 54: 399–403. 10.1136/jmedgenet-2016-104237 [DOI] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. 1996. Protein sorting by transport vesicles. Science 272: 227–234. 10.1126/science.272.5259.227 [DOI] [PubMed] [Google Scholar]
- Roussy M, Bilodeau M, Jouan L, Tibout P, Laramée L, Lemyre E, Léveillé F, Tihy F, Cardin S, Sauvageau C, et al. 2018. NUP98-BPTF gene fusion identified in primary refractory acute megakaryoblastic leukemia of infancy. Genes Chromosom Cancer 57: 311–319. 10.1002/gcc.22532 [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 148: 635–652. 10.1083/jcb.148.4.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakiyama Y, Mazur A, Kapinos LE, Lim RY. 2016. Spatiotemporal dynamics of the nuclear pore complex transport barrier resolved by high-speed atomic force microscopy. Nat Nanotechnol 11: 719–723. 10.1038/nnano.2016.62 [DOI] [PubMed] [Google Scholar]
- Sampathkumar P, Kim SJ, Upla P, Rice WJ, Phillips J, Timney BL, Pieper U, Bonanno JB, Fernandez-Martinez J, Hakhverdyan Z, et al. 2013. Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure 21: 560–571. 10.1016/j.str.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandestig A, Engström K, Pepler A, Danielsson I, Odelberg-Johnsson P, Biskup S, Holz A, Stefanova M. 2020. NUP188 biallelic loss of function may underlie a new syndrome: nucleoporin 188 insufficiency syndrome? Mol Syndromol 10: 313–319. 10.1159/000504818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandokji I, Marquez J, Ji W, Zerillo CA, Konstantino M, Lakhani SA, Khokha MK, Warejko JK. 2019. Identification of novel mutations and phenotype in the steroid resistant nephrotic syndrome gene NUP93: a case report. BMC Nephrol 20: 271. 10.1186/s12882-019-1458-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M, Akatsu T, Ishkawa Y, Minami Y, Nakamura M. 2007. A novel activator of C-C chemokine, FROUNT, is expressed with C-C chemokine receptor 2 and its ligand in failing human heart. J Card Fail 13: 114–119. 10.1016/j.cardfail.2006.11.003 [DOI] [PubMed] [Google Scholar]
- Satterly N, Tsai PL, van Deursen J, Nussenzveig DR, Wang Y, Faria PA, Levay A, Levy DE, Fontoura BM. 2007. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc Natl Acad Sci 104: 1853–1858. 10.1073/pnas.0610977104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R, Orci L. 1996. Coat proteins and vesicle budding. Science 271: 1526–1533. 10.1126/science.271.5255.1526 [DOI] [PubMed] [Google Scholar]
- Schmitt C, von Kobbe C, Bachi A, Panté N, Rodrigues JP, Boscheron C, Rigaut G, Wilm M, Séraphin B, Carmo-Fonseca M, et al. 1999. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J 18: 4332–4347. 10.1093/emboj/18.15.4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader N, Stelter P, Flemming D, Kunze R, Hurt E, Vetter IR. 2008. Structural basis of the nic96 subcomplex organization in the nuclear pore channel. Mol Cell 29: 46–55. 10.1016/j.molcel.2007.10.022 [DOI] [PubMed] [Google Scholar]
- Schuller AP, Wojtynek M, Mankus D, Tatli M, Kronenberg-Tenga R, Regmi SG, Dip PV, Lytton-Jean AKR, Brignole EJ, Dasso M, et al. 2021. The cellular environment shapes the nuclear pore complex architecture. Nature 598: 667–671. 10.1038/s41586-021-03985-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sell K, Storch K, Hahn G, Lee-Kirsch MA, Ramantani G, Jackson S, Neilson D, von der Hagen M, Hehr U, Smitka M. 2016. Variable clinical course in acute necrotizing encephalopathy and identification of a novel RANBP2 mutation. Brain Dev 38: 777–780. 10.1016/j.braindev.2016.02.007 [DOI] [PubMed] [Google Scholar]
- Seo HS, Ma Y, Debler EW, Wacker D, Kutik S, Blobel G, Hoelz A. 2009. Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex. Proc Natl Acad Sci 106: 14281–14286. 10.1073/pnas.0907453106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahin V, Hafezi W, Oberleithner H, Ludwig Y, Windoffer B, Schillers H, Kühn JE. 2006. The genome of HSV-1 translocates through the nuclear pore as a condensed rod-like structure. J Cell Sci 119: 23–30. 10.1242/jcs.02705 [DOI] [PubMed] [Google Scholar]
- Singer S, Zhao R, Barsotti AM, Ouwehand A, Fazollahi M, Coutavas E, Breuhahn K, Neumann O, Longerich T, Pusterla T, et al. 2012. Nuclear pore component Nup98 is a potential tumor suppressor and regulates posttranscriptional expression of select p53 target genes. Mol Cell 48: 799–810. 10.1016/j.molcel.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J 17: 2663–2676. 10.1093/emboj/17.9.2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodeik B, Ebersold MW, Helenius A. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol 136: 1007–1021. 10.1083/jcb.136.5.1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soman NR, Correa P, Ruiz BA, Wogan GN. 1991. The TPR-MET oncogenic rearrangement is present and expressed in human gastric carcinoma and precursor lesions. Proc Natl Acad Sci 88: 4892–4896. 10.1073/pnas.88.11.4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F, Hülsmann BB, Spang A, Hartmann E, Cordes VC, Görlich D. 2006. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J Cell Biol 173: 509–519. 10.1083/jcb.200601001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. 2007. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol 8: 195–208. 10.1038/nrm2114 [DOI] [PubMed] [Google Scholar]
- Stewart M. 2019. Polyadenylation and nuclear export of mRNAs. J Biol Chem 294: 2977–2987. 10.1074/jbc.REV118.005594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T, Lin DH, Collins LN, Hurt E, Hoelz A. 2014. Evidence for an evolutionary relationship between the large adaptor nucleoporin Nup192 and karyopherins. Proc Natl Acad Sci 111: 2530–2535. 10.1073/pnas.1311081111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T, Bley CJ, Thierbach K, Petrovic S, Schilbach S, Mayo DJ, Perriches T, Rundlet EJ, Jeon YE, Collins LN, et al. 2015a. Architecture of the fungal nuclear pore inner ring complex. Science 350: 56–64. 10.1126/science.aac9176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T, Correia AR, Lin DH, Paduch M, Lu VT, Kossiakoff AA, Hoelz A. 2015b. Nuclear pores. Architecture of the nuclear pore complex coat. Science 347: 1148–1152. 10.1126/science.aaa4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarazón E, Rivera M, Roselló-Lletí E, Molina-Navarro MM, Sánchez-Lázaro IJ, España F, Montero JA, Lago F, González-Juanatey JR, Portolés M. 2012. Heart failure induces significant changes in nuclear pore complex of human cardiomyocytes. PLoS ONE 7: e48957. 10.1371/journal.pone.0048957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teimer R, Kosinski J, von Appen A, Beck M, Hurt E. 2017. A short linear motif in scaffold Nup145C connects Y-complex with pre-assembled outer ring Nup82 complex. Nat Commun 8: 1107. 10.1038/s41467-017-01160-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thul PJ, Akesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, Alm T, Asplund A, Bjork L, Breckels LM, et al. 2017. A subcellular map of the human proteome. Science 356: eaal3321. 10.1126/science.aal3321 [DOI] [PubMed] [Google Scholar]
- Timney BL, Raveh B, Mironska R, Trivedi JM, Kim SJ, Russel D, Wente SR, Sali A, Rout MP. 2016. Simple rules for passive diffusion through the nuclear pore complex. J Cell Biol 215: 57–76. 10.1083/jcb.201601004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SS, Weaver PL, Liu Y, Hitomi M, Tartakoff AM, Chang TH. 1998. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J 17: 2651–2662. 10.1093/emboj/17.9.2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht R, Klann M, Horvath P, Kutay U. 2015. Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J Cell Biol 209: 687–704. 10.1083/jcb.201409127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. 1999. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature 398: 39–46. 10.1038/17969 [DOI] [PubMed] [Google Scholar]
- von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, et al. 2015. In situ structural analysis of the human nuclear pore complex. Nature 526: 140–143. 10.1038/nature15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs A, Grosveld G. 1992. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol 12: 1687–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moeller H, Basquin C, Conti E. 2009. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol 16: 247–254. 10.1038/nsmb.1561 [DOI] [PubMed] [Google Scholar]
- Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW, Fornerod M. 2002. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol 158: 63–77. 10.1083/jcb.200202088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. 2009. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459: 847–851. 10.1038/nature08036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Motoi T, Khanin R, Olshen A, Mertens F, Bridge J, Dal Cin P, Antonescu CR, Singer S, Hameed M, et al. 2012. Identification of a novel, recurrent HEY1-NCOA2 fusion in mesenchymal chondrosarcoma based on a genome-wide screen of exon-level expression data. Genes Chromosomes Cancer 51: 127–139. 10.1002/gcc.20937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ML. 1955. The nuclear envelope; its structure and relation to cytoplasmic membranes. J Biophys Biochem Cytol 1: 257–270. 10.1083/jcb.1.3.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ML. 1959. Further observations on the nuclear envelope of the animal cell. J Biophys Biochem Cytol 6: 147–156. 10.1083/jcb.6.2.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg-Shukron A, Renbaum P, Kalifa R, Zeligson S, Ben-Neriah Z, Dreifuss A, Abu-Rayyan A, Maatuk N, Fardian N, Rekler D, et al. 2015. A mutation in the nucleoporin-107 gene causes XX gonadal dysgenesis. J Clin Invest 125: 4295–4304. 10.1172/JCI83553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. 2006. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol 8: 668–676. 10.1038/ncb1424 [DOI] [PubMed] [Google Scholar]
- Wesierska-Gadek J, Hohenuer H, Hitchman E, Penner E. 1996. Autoantibodies against nucleoporin p62 constitute a novel marker of primary biliary cirrhosis. Gastroenterology 110: 840–847. 10.1053/gast.1996.v110.pm8608894 [DOI] [PubMed] [Google Scholar]
- Whittle JRR, Schwartz TU. 2009. Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J Biol Chem 284: 28442–28452. 10.1074/jbc.M109.023580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing CE, Fung HYJ, Chook YM. 2022. Karyopherin-mediated nucleocytoplasmic transport. Nat Rev Mol Cell Biol 23: 307–328. 10.1038/s41580-021-00446-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong EV, Gray S, Cao W, Montpetit R, Montpetit B, De La Cruz EM. 2018. Nup159 weakens Gle1 binding to Dbp5 but does not accelerate ADP release. J Mol Biol 430: 2080–2095. 10.1016/j.jmb.2018.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Powers MA. 2009. Nuclear pore proteins and cancer. Semin Cell Dev Biol 20: 620–630. 10.1016/j.semcdb.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Li Z, He H, Wernimont A, Li Y, Loppnau P, Min J. 2015. Crystal structure of human nuclear pore complex component NUP43. FEBS Lett 589: 3247–3253. 10.1016/j.febslet.2015.09.008 [DOI] [PubMed] [Google Scholar]
- Yang Q, Rout MP, Akey CW. 1998. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell 1: 223–234. 10.1016/S1097-2765(00)80023-4 [DOI] [PubMed] [Google Scholar]
- Yarbrough ML, Mata MA, Sakthivel R, Fontoura BM. 2014. Viral subversion of nucleocytoplasmic trafficking. Traffic 15: 127–140. 10.1111/tra.12137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Miehlke S, Ebert MP, Hoffmann J, Breidert M, Alpen B, Starzynska T, Stolte Prof M, Malfertheiner P, Bayerdorffer E. 2000. Frequency of TPR-MET rearrangement in patients with gastric carcinoma and in first-degree relatives. Cancer 88: 1801–1806. [PubMed] [Google Scholar]
- Zhang X, Chen S, Yoo S, Chakrabarti S, Zhang T, Ke T, Oberti C, Yong SL, Fang F, Li L, et al. 2008. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell 135: 1017–1027. 10.1016/j.cell.2008.10.022 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li S, Zeng C, Huang G, Zhu X, Wang Q, Wang K, Zhou Q, Yan C, Zhang W, et al. 2020. Molecular architecture of the luminal ring of the Xenopus laevis nuclear pore complex. Cell Res 30: 532–540. 10.1038/s41422-020-0320-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Zhu JY, Richman A, Fu Y, Huang W, Chen N, Pan X, Yi C, Ding X, Wang S, et al. 2019. Mutations in NUP160 are implicated in steroid-resistant nephrotic syndrome. J Am Soc Nephrol 30: 840–853. 10.1681/ASN.2018080786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zila V, Margiotta E, Turoňová B, Müller TG, Zimmerli CE, Mattei S, Allegretti M, Börner K, Rada J, Müller B, et al. 2021. Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 184: 1032–1046.e18. 10.1016/j.cell.2021.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli CE, Allegretti M, Rantos V, Goetz SK, Obarska-Kosinska A, Zagoriy I, Halavatyi A, Hummer G, Mahamid J, Kosinski J, et al. 2021. Nuclear pores dilate and constrict in cellulo. Science 374: eabd9776. 10.1126/science.abd9776 [DOI] [PubMed] [Google Scholar]
- Zuleger N, Kelly DA, Richardson AC, Kerr AR, Goldberg MW, Goryachev AB, Schirmer EC. 2011. System analysis shows distinct mechanisms and common principles of nuclear envelope protein dynamics. J Cell Biol 193: 109–123. 10.1083/jcb.201009068 [DOI] [PMC free article] [PubMed] [Google Scholar]