In this Outlook, Glousker and Lingner highlight a study in this issue of Genes & Development from Yang et al., who discover that TFIIH, a basic component of the PolII transcription initiation and nucleotide excision repair machinery, facilitates telomere replication.
Keywords: telomere, shelterin, replication, TRF1, TFIIH, fragile telomere
Abstract
Although telomeres are essential for chromosome stability, they represent fragile structures in our genome. Telomere shortening occurs during aging in cells lacking telomerase due to the end replication problem. In addition, recent work uncovered that the bulk of telomeric DNA poses severe hurdles for the semiconservative DNA replication machinery, requiring the assistance of an increasing number of specialized factors that prevent accidental telomere loss or damage events. In this issue of Genes & Development, Yang and colleagues (pp. 956–969) discover that TFIIH, a basic component of the PolII transcription initiation and nucleotide excision repair machinery, facilitates telomere replication. TFIIH is recruited to telomeres by the shelterin component TRF1, taking on at telomeres a moonlighting function.
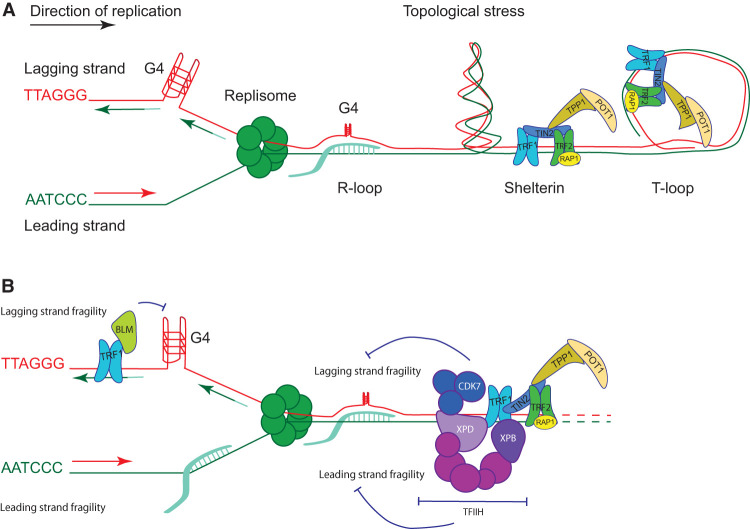
The end replication problem, which was described half a century ago, mainly stems from the nucleolytic processing of telomeres after replication to yield 3′ overhangs of telomeric DNA at chromosome ends. The end replication problem can be counteracted by telomerase in human germ and stem cells that express this enzyme. The absence of telomerase in differentiated cells, however, limits their proliferative potential and counteracts tumor formation (Maciejowski and de Lange 2017). The bulk telomere replication problem refers to the difficulties of the semiconservative DNA replication machinery to replicate through telomeric DNA (Glousker and Lingner 2021). Vertebrate telomeric DNA consists of thousands of 5′-TTAGGG-3′/5′-CCCTAA-3′ repeats with the G-rich strand containing the 3′ end. The G-rich strand is replicated by lagging strand synthesis, whereas the C-rich strand serves as the template for leading strand synthesis (Fig. 1). Multiple difficulties may occur during telomere replication of either strand (Fig. 1A) including G-quadruplexes forming on the lagging strand template and R-loops forming with the long noncoding RNA TERRA on the leading strand template. Topological stress, T-loops, and protein roadblocks may also interfere with replication fork progression.
Figure 1.
The bulk telomere replication and its suppression by TRF1-recruited factors. (A) The G strand, which provides the template for lagging strand synthesis, may form highly stable secondary structures when separated from the C-rich strand during replication. The guanines can assemble into four-stranded G-quadruplex structures and other nonstandard base interactions and entanglements that impair their function as templates (Vannier et al. 2012; Yang et al. 2020). Telomeres are transcribed into the TERRA long noncoding RNAs that associate with telomeres, forming DNA:RNA hybrid structures known as R-loops. If TERRA R-loops are not removed in S phase, they interfere with replication (Feretzaki et al. 2020). In addition, TERRA R-loops may favor formation of G-quadruplexes on the displaced G-rich strand. Telomeres form T-loop structures in which the 3′ overhang invades and base-pairs with the double-stranded part of the telomere. T-loop structures must also be unwound during replication (Vannier et al. 2012; Margalef et al. 2018). Topological stress at telomeres during replication requires mechanisms of relief. Telomere binding proteins have been observed to counteract fork progression in vitro and may need to dissociate from telomeres for replication (Douglas and Diffley 2021). Replication at telomeres proceeds mostly unidirectionally from replication forks, moving toward the ends of chromosomes. Therefore, stalled forks may not be rescued from converging forks coming from the end of the chromosome. (B) TRF1 counteracts telomere fragility, recruiting TFIIH and BLM to chromosome ends. While BLM sustains lagging strand replication, TFIIH has very pronounced roles at leading and lagging strand telomeres. The exact targets of TFIIH at telomeres remain to be explored. It is also not known whether TFIIH acts ahead of the replication fork as depicted or behind as arbitrarily illustrated for BLM. (Dark blue) The CAK subcomplex of TFIIH, (magenta) GTF2H1–5 subunits, (lilac) XPD, (purple) XPB.
Several factors have been identified to facilitate telomere replication. For example, the BLM and RTEL1 helicases have been implicated in unwinding G-quadruplex secondary structures for replication (Vannier et al. 2012; Yang et al. 2020). RTEL1 can also unwind T-loops. Several factors counteract TERRA expression or TERRA R-loops in S phase. These include RNase H, which degrades RNA in DNA:RNA hybrid structures; RNA surveillance factors, which are present at telomeres and counteract TERRA accumulation at chromosome ends; and others (Glousker and Lingner 2021). In addition, proteomic analysis of the telomeric replisome identified a large number of components that are specific to the telomeric replisome to facilitate telomere replication (Lin et al. 2021).
The shelterin component TRF1 binds to double-stranded telomeric DNA and has been known to facilitate telomere replication (Sfeir et al. 2009). TRF1 loss gives rise to so-called fragile telomeres, in which the telomeres display an abnormal appearance in metaphase chromosomes, manifested by multiple or smeared telomeric signals at individual chromosome ends. Telomere fragility reflects replication stress, though the exact nature of these structures remains to be elucidated. Consistently, TRF1 depletion also leads to impairment of fork progression (Sfeir et al. 2009). The new study by Yang et al. (2022) develops like a detective story. They first undertake domain swap experiments between TRF1 and its paralog, TRF2, in mouse cells to identify the domains, and finally amino acids in two distant regions of TRF1 (helix 2 of the TRFH domain and helix 1 of the Myb domain) that are critical for telomere replication. In a parallel investigation, they screened DNA repair factors for their involvement in fragility suppression. The two approaches converge on the transcription factor TFIIH. The 10-subunit complex physically interacts with TRF1 through both of the above-mentioned domains in TRF1. Epistasis analysis further demonstrates that TRF1 and TFIIH act in the same pathway to suppress telomere fragility. Together, these experiments convincingly demonstrate that TRF1 most prominently promotes telomere replication through the recruitment of TFIIH.
How then does TFIIH facilitate telomere replication? TFIIH is a basic component of the PolII transcription initiation machinery, commonly known as the preinitiation complex (PIC). TFIIH enables ATP-dependent opening of the promoter DNA at transcription start sites. Thus, loss of this factor could have impaired transcription of the TRF1 gene. However, several experiments refute this possibility. Most convincingly, inhibition of the CDK7 protein kinase of TFIIH induced fragility while not impairing transcription initiation. TFIIH also has crucial tasks for DNA opening during nucleotide excision repair (NER), which removes UV-induced and other bulky lesions in DNA. However, the NER pathway is not involved, as the targeting of other essential components of NER did not induce telomere fragility.
Of note, three of the 10 subunits of TFIIH possess catalytic activity. XPB is an ATPase and translocase that enables ATP-dependent opening of the promoter DNA at the transcription start site. XPD is a 5′–3′ helicase. CDK7 is a Ser/Thr kinase. It phosphorylates Ser5 and Ser7 in the C-terminal domain (CTD) heptad repeats of RNA PolII, allowing promoter escape. CDK7 also activates cyclin-dependent kinases during the cell cycle. It will be interesting to determine which of the three catalytic activities of TFIIH are critical at telomeres to facilitate telomere replication. It is conceivable that TFIIH facilitates fork progression, contributing to DNA unwinding upon encountering DNA secondary structures, damaged DNA, or protein blocks. It is also possible that XPB and XPD help resolve TERRA R-loops. Finally, CDK7 may phosphorylate critical proteins at telomeres to mediate their catalytic activation or release from telomeres. Consistent with the latter, Yang et al. (2022) demonstrate that chemical inhibition of CDK7 induces telomere fragility.
Another interesting question relates to the mechanism of fragility induction and its structure. Previous work demonstrated that in BLM-deficient cells, a repair pathway termed break-induced replication (BIR) leads to fragility of telomeres replicated by lagging strand synthesis (Yang et al. 2020). In the absence of BLM, broken telomeres generated by SLX4/SLX1 nuclease-mediated cleavage invade other chromosome ends, using them as a template for DNA synthesis. This pathway depends on, among others, POLD3, an accessory subunit of DNA Polδ (Dilley et al. 2016). However, in TRF1 deficiency, telomere fragility was observed at both telomeres replicated by leading and lagging strand synthesis. Furthermore, POLD3 was not involved in the generation of telomere fragility of TRF1-deficient telomeres and neither was fork reversal, as seen in RTEL1 deficiency (Margalef et al. 2018). Thus, another repair pathway downstream from impairment with replication appears to be involved (Yang et al. 2022). Finally, the deficiency in TFIIH subunits has been implicated in rare autosomal recessive diseases, including xeroderma pigmentosum (XP) and trichothiodystrophy (TTD). The work of Yang et al. (2022) opens up the possibility that telomeres may be affected in a subset of these patients.
Acknowledgments
The laboratory is supported by the Swiss National Science Foundation (SNSF; 310030_184718), the SNSF-funded National Centre of Competence in Research RNA and Disease Network (205601), and an Innovative Training Network (ITN; aDDRess) from the European Commission's Seventh Framework Programme (812829).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.350140.122.
References
- Dilley RL, Verma P, Cho NW, Winters HD, Wondisford AR, Greenberg RA. 2016. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 539: 54–58. 10.1038/nature20099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas ME, Diffley JFX. 2021. Budding yeast Rap1, but not telomeric DNA, is inhibitory for multiple stages of DNA replication in vitro. Nucleic Acids Res 49: 5671–5683. 10.1093/nar/gkab416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feretzaki M, Pospisilova M, Valador Fernandes R, Lunardi T, Krejci L, Lingner J. 2020. RAD51-dependent recruitment of TERRA lncRNA to telomeres through R-loops. Nature 587: 303–308. 10.1038/s41586-020-2815-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glousker G, Lingner J. 2021. Challenging endings: how telomeres prevent fragility. Bioessays 43: e2100157. 10.1002/bies.202100157 [DOI] [PubMed] [Google Scholar]
- Lin C-YG, Näger AC, Lunardi T, Vančevska A, Lossaint G, Lingner J. 2021. The human telomeric proteome during telomere replication. Nucleic Acids Res 49: 12119–12135. 10.1093/nar/gkab1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejowski J, de Lange T. 2017. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol 18: 175–186. 10.1038/nrm.2016.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalef P, Kotsantis P, Borel V, Bellelli R, Panier S, Boulton SJ. 2018. Stabilization of reversed replication forks by telomerase drives telomere catastrophe. Cell 172: 439–453.e14. 10.1016/j.cell.2017.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. 2009. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 138: 90–103. 10.1016/j.cell.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannier J-B, Pavicic-Kaltenbrunner V, Petalcorin MIR, Ding H, Boulton SJ. 2012. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149: 795–806. 10.1016/j.cell.2012.03.030 [DOI] [PubMed] [Google Scholar]
- Yang Z, Takai KK, Lovejoy CA, de Lange T. 2020. Break-induced replication promotes fragile telomere formation. Genes Dev 34: 1392–1405. 10.1101/gad.328575.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Sharma K, de Lange T. 2022. TRF1 uses a noncanonical function of TFIIH to promote telomere replication. Genes Dev (this issue). 10.1101/gad.349975.122 [DOI] [PMC free article] [PubMed] [Google Scholar]