In this Resource/Methodology, Farrell et al. designed a macrocyclic bispecific degrader targeting BET proteins and validated CUL3KLHL20 as an E3 ligase system suitable for TPD. This work thus contributes to the expansion of usable E3 ligases for potential drug development.
Keywords: targeted protein degradation, CUL3, KLHL20, macrocycles, PROTAC
Abstract
Targeted protein degradation (TPD) has risen as a promising therapeutic modality. Leveraging the catalytic nature of the ubiquitin–proteasome enzymatic machinery, TPD exhibits higher potency to eliminate disease-causing target proteins such as oncogenic transcription factors that may otherwise be difficult to abrogate by conventional inhibitors. However, there are challenges that remain. Currently, nearly all degraders engage CUL4CRBN or CUL2VHL as the E3 ligase for target ubiquitination. While their immediate efficacies are evident, the narrowed E3 ligase options make TPD vulnerable to potential drug resistance. In addition, E3 ligases show differential tissue expression and have intrinsic limitations in accessing varying types of disease-relevant targets. As the success of TPD is closely associated with the ability of E3 ligases to efficiently polyubiquitinate the target of interest, the long-term outlook of TPD drug development will depend on whether E3 ligases such as CUL4CRBN and CUL2VHL are accessible to the targets of interest. To overcome these potential caveats, a broad collection of actionable E3 ligases is required. Here, we designed a macrocyclic degrader engaging CUL3KLHL20 for targeting BET proteins and validated CUL3KLHL20 as an E3 ligase system suitable for TPD. This work thus contributes to the expansion of usable E3 ligases for potential drug development.
Ablating proteins to abrogate pathological progression has been a desired therapeutic pursuit. Over the past decades, progress has been made by using gene silencing strategies such as RNAi or antisense oligo approaches. However, in recent years, the targeted protein degradation (TPD) approach, which leverages the ubiquitin–proteasome machinery for direct protein degradation, has been proven a paradigm-shifting modality for efficient target depletion (Bekes et al. 2022). The general principle of TPD is based on the observations that most of the ubiquitin E3 ligases function in a proximity-driven manner and have significant flexibility for substrate acceptance (Sakamoto et al. 2001; Chen and Hellmann 2013; Verma et al. 2020) such that, once recruited in proximity to the E3 ligase, a targeted protein can potentially be degraded even though it is not naturally a cognate substrate for a particular E3 ligase. This artificial, targeted substrate recruitment can be achieved by creating a bifunctional PROTAC (proteolysis targeting chimera) molecule that has one chemical moiety with specificity for a target protein and another moiety with specificity for the desired E3 ligase. Some examples include degraders for EGFR (Jang et al. 2020), AKT (You et al. 2020), cyclin-dependent kinases (CDKs) (Olson et al. 2018; Teng et al. 2020), STAT3 (Bai et al. 2019), and BET family bromodomain proteins (Raina et al. 2016; Winter et al. 2017). With these promising outcomes endorsing the potential of TPD, it is also noteworthy that the majority of the demonstrated protein degraders were based on engaging CUL4CRBN and CUL2VHL as their executing E3 ligases. Some other E3 ligases such as MDM2 (Hines et al. 2019) and cIAP1 (Schiemer et al. 2021) have been explored as well, but not extensively. Part of the reason that CUL4CRBN and CUL2VHL prevail in protein degrader design could stem from the limited availability of synthetic E3 ligase binders that have been reported and validated. Consequently, all protein degraders that have entered the clinical stage use the CUL4CRBN E3 ligase for targeted degradation (Bekes et al. 2022). Nevertheless, in the past few years, TPD has advanced from an experimental concept to the clinic.
Despite the convincing results of many TPDs as experimental therapies, some issues remain to be addressed. First, the differential expression and localization of E3 ligases in different cell types and target tissues could limit the therapeutic potential in treating certain types of cancers if only a narrow set of E3 ligases is available to be used. Another concerning aspect is whether drug resistance and therapy-escaping mechanisms would occur, to what extent, and by which mechanism. Similar to what has been found with many targeted therapies, TPD drug resistance could arise from acquired mutations in the cancer cells, rendering the loss of E3 ligase engagement and therefore allowing the target protein to escape from ubiquitination (Zhang et al. 2019; Gooding et al. 2021). Consequently, if solely focused on CUL4CRBN and CUL2VHL E3 ligases, TPD modality may suffer from drug resistance once cancer cells acquire such mutations. To circumvent potential drug resistance, we set out to identify new E3 ligases as alternatives to the currently explored ligases to broaden the targeting options. Hence, widening the available E3 ligases should allow for targeting a more diverse range of proteins in a broader range of tissues.
To search for a new E3 ligase capable of targeting a broad range of proteins, our selection criteria required the E3 ligase to be ubiquitously expressed and have a wide subcellular distribution so that colocalization with potential targets can occur. In addition, we required the E3 ligase to have sufficient catalytic activity toward all substrates and not be overly selective in substrate acceptance or require stringent 3D orientation for substrate engagement. With these criteria in mind, we chose the CUL3 E3 ligase system. CUL3 has been shown to have a broad subcellular localization pattern with high expression levels in many cell types (Jang et al. 2018) and is therefore an ideal candidate E3 ligase system to exploit. Like CUL2 and CUL4, CUL3 also belongs to the Cullen-Ring ubiquitin ligase (CRL) family (Furukawa et al. 2003; Wimuttisuk and Singer 2007), which requires an adaptor protein to anchor the substrate protein to be ubiquitinated (Geyer et al. 2003; Xu et al. 2003). There are >60 member proteins of the BTB-Kelch family that serve as the adaptor proteins for CUL3 (Canning et al. 2013), and among the BTB-Kelch adaptor proteins, KLHL20 has been discovered as an oncoprotein mediating the degradation of the death-activated protein kinase 1 (DAPK1) and promyelocytic leukemia protein (PML) in cancer cells (Lee et al. 2010; Yuan et al. 2011). Furthermore, KLHL20 is expressed in a wide range of cancer cell lines (Chen et al. 2015; Chen and Chen 2016) and was found to be up-regulated by HIF1-a, a crucial antihypoxic transcription factor abundant in many cancer cells (Lee et al. 2010; Yuan et al. 2011). Altogether, the evidence suggested that CUL3KLHL20 should be a suitable E3 ligase for abrogating challenging drug targets such as transcription factors. Here, we describe our discovery of a synthetic KLHL20 ligand for developing CUL3KLHL20-based protein degraders (Fig. 1). To our knowledge, this is the first CUL3KLHL20-based protein degrader, thus putting CUL3KLHL20 into the list of actionable E3 ligases for TPD applications and thereby providing more opportunities for the TPD modality.
Figure 1.
Developing the CULKLHL20 E3 ligase TPD system. The CULKLHL20-based TPD molecules are comprised of three parts: (1) the synthetic KLHL20 ligand for E3 ligase engagement, (2) the target protein-engaging ligand for specific target protein recruitment, and (3) a linker moiety that connects both ligand types. The design of target-selective degraders is highly modular, with the synthetic KLHL20 ligand serving as the constant part to engage the E3 ligase and an exchangeable part to target the protein of choice (e.g., a transcription factor in this study).
Results
Design and synthesis of the KLHL20 ligand molecule BTR2000
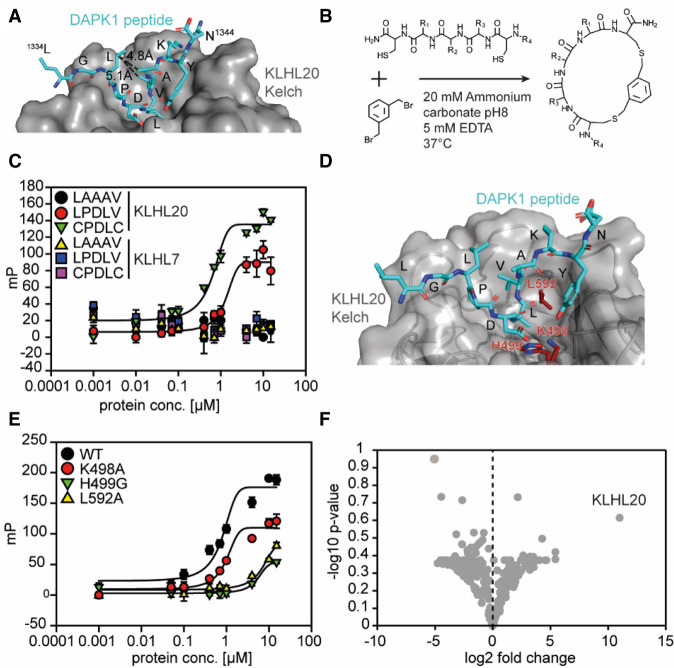
To develop CUL3KLHL20-based TPD molecules, we first aimed to create a KLHL20-engaging ligand that can eventually serve as the template for a target-specific degrader design (Fig. 1). To this end, structure-guided design was carried out based on the crystal structure of a DAPK1 peptide fragment (residues 1334–1344) in complex with the KLHL20 Kelch domain (PDB: 6GY5; Chen et al. 2019). DAPK1 has been identified as a KLHL20 substrate for polyubiquitination (Lee et al. 2010). In the crystal structure, the nine-residue DAPK peptide fragment 1334LGLPDLVAK1342 formed direct contact with KLHL20 (Fig. 2A); notably, DAPK1 D1338 with KLHL20 K498 and H499, as well as DAPK1 L1339 with KLHL20 L592. Furthermore, in the center of this nine-residue fragment is a five-residue motif (LPDLV) with a hairpin-like conformation, which has been reported as the minimum core motif required for KLHL20 binding (Chen et al. 2019). Although the LPDLV peptide was shown to be capable of binding to KLHL20, the affinity was low (Chen et al. 2019). However, when evaluating the hairpin-like structure of the peptide bound to KLHL20, we envisioned that the binding affinity can be significantly improved if the linear peptide motif is engineered into a macrocyclic molecule to constrain the hairpin structure, thereby reducing the entropic cost of adopting the favorable binding conformation. The LPDLV motif thus served as the template for our ligand design.
Figure 2.
Cyclization of a linear DAPK1-derived peptide motif provides increased affinity for the KLHL20 Kelch binding pocket. (A) Crystal structure of the KLHL20 Kelch domain bound to the DAPK-derived peptide fragment 1334LGLPDLVAK1342 (PDB 6GY5). (B) Cyclization scheme. Free sulfhydryls from cysteine residues react with 1,3-bis(bromomethyl)benzene to form two thioether bonds, yielding macrocycles. (C) Fluorescence polarization (FP) binding curves of FITC-labeled peptides incubated with serial dilutions of the KLHL20 Kelch domain. Peptide LAAAV was used as a negative control. Error bars show standard deviation (SD; N = 3). Data are background-corrected, and curves were fitted using a three-parameter dose response model. (D) Crystal structure of the KLHL20 Kelch domain, as in A. Residues mutated for BTR2000 binding studies are highlighted in red. (E) FP curves, as in C, showing binding of FITC-labeled BTR2000 to KLHL20 Kelch wild type (WT) or mutants. (F) Volcano plot showing proteomics data of the pull-down experiment comparing BTR2000 with cyclized CPGAC. The −log10 P-value (N = 3) is plotted against the log2 fold change BTR2000/CPGAC. KLHL20 showed the highest enrichment, with a log2 value of 11.
Our general strategy to form a macrocycle entailed the introduction of two cysteine residues to flank this KLHL20 binding motif, followed by cyclization via thioether bond formation between the two flanking cysteine residues and the benzylic carbons of 1,3-bis(bromomethyl)benzene (Fig. 2B). When examining the crystal structure of the 1334LGLPDLVAK1342 fragment, we noticed that L1336 and A1341 were not in direct contact with KLHL20 and could be substituted by cysteine for cyclization. The distance measured between residues L1336 and A1341 was ∼4.8 Å (Fig. 2A), a distance feasible for allowing the 1,3 disubstituted benzyl group to be introduced. This strategy yielded macrocycle CPDLVC. To reduce the molecular weight of the macrocycle, a similar approach was applied to generate the macrocycle CPDLC by substituting L1336 and V1340 (by 5.1-Å separation) with cysteines. We synthesized FITC-labeled macrocycles CPDLVC and CPDLC and then measured their binding affinity to the KLHL20 Kelch domain by fluorescence polarization (FP) (Fig. 2C; Supplemental Fig. S1A). The FP assay indicated that the approximate KD of the linear LPDLV motif was >5 μM. In contrast, both macrocycle compounds CPDLVC and CPDLC exhibited improved affinity, with an approximate KD of ∼600 nM. Hence, the cyclization approach was successful in improving the affinity of the KLHL20 ligand. Binding of the macrocycle compounds was also KLHL20 Kelch domain-specific, without any detectable binding to KLHL7 Kelch domain. Interestingly, although the crystal structure revealed that V1340 was in direct contact with KLHL20, we did not observe deterred binding affinity with macrocycle CPDLC. This could be due to the new binding contact offered by the cysteine introduced to replace the valine binding contact. For its smaller molecular weight, the macrocycle CPDLC (referred to here as BTR2000) was therefore chosen as our KLHL20 anchor molecule to further exploit KLHL20-based TPD molecules.
BTR2000 binds to KLHL20 in a mode similar to that of the wild-type ligand and with high specificity
While the linear LPDLV motif was found to be essential for KLHL20 binding (Chen et al. 2019), it is intriguing that BTR2000 is capable of binding to the KLHL20 Kelch domain with higher affinity than linear LPDLV while only containing core residues PDL. To confirm that BTR2000 is indeed binding to the KLHL20 ligand pocket in a manner analogous to the LPDLV motif, we mutagenized the key LPDLV-contacting residues on KLHL20 based on the X-ray crystal structure. In this structure, the KLHL20 residues K498, H499, and L592 form crucial contacts with the DAPK peptide with both the aspartic and leucine residues (Fig. 2D; Chen et al. 2019). We therefore generated three KLHL20 mutants (KLHL20K498A, KLHL20H499G, and KLHL20L592G) to measure their affinity with BTR2000 and the linear LPDLV peptide. We observed that these three residues are indeed important for BTR2000 binding, analogous to the binding mode suggested by the crystal structure of the DAPK1 peptide in complex with KLHL20, with H499A and L592G showing more prominent loss in binding, indicating that BTR2000 binds to KLHL20 in a mode similar to that of the wild-type ligand (Fig. 2E). Likewise, mutating the aspartic acid (cyclized CPGLC) and leucine residues (cyclized CPDAC) on BTR2000 showed a similar loss in KLHL20 binding, indicating that these residues are the interacting residues corresponding to K498, H499, and L592 of KLHL20 (Supplemental Fig. S1B). Since the interaction of BTR2000 with KLHL20 is only based on a three-residue peptide motif, low target selectivity and resulting off-target effects are possible risks. To rule out this possibility, we performed pull-down experiments from PC3 lysates with immobilized compounds. BTR2000 successfully enriched KLHL20 but not KLHL12 from lysates, while cyclized CPGLC, cyclized CPDAC, and a cyclized double-mutant CPGAC showed no binding to KLHL20 (Supplemental Fig. S1C). In order to analyze the BTR2000 interactome in more depth, the eluates from the pull-down experiment were subjected to MS-based proteomics. KLHL20 was the only E3 ubiquitin ligase component enriched in the experiment and showed the highest binding preference for BTR2000 over the CPGAC (Fig. 2F) or empty bead control (Supplemental Fig. S1D). Together, these results demonstrate a high KLHL20 specificity of BTR2000, which makes it a suitable building block for a KLHL20-based PROTAC molecule.
BTR2000-derived PROTAC molecule BTR2003 facilitates KLHL20–bromodomain interaction
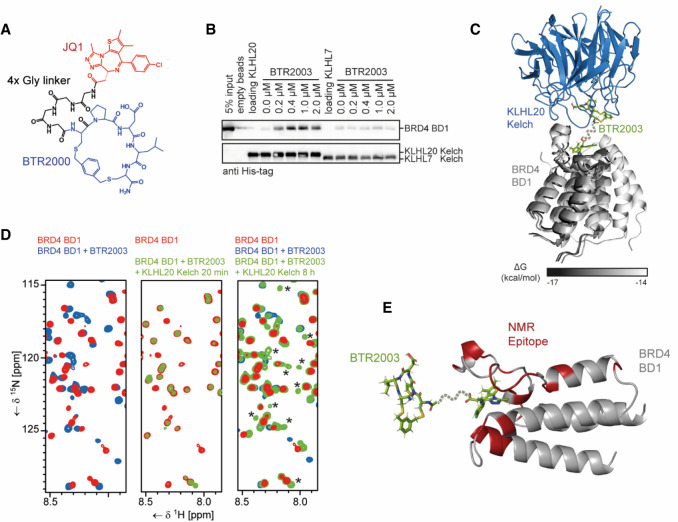
After confirming that BTR2000 binds to the canonical KLHL20 substrate binding site with high specificity, we envisioned multiple options for generating BTR2000-based TPD molecules to recruit target proteins to KLHL20. As a proof of concept, we selected JQ1 as a model molecule to build such bifunctional molecules. The BET bromodomain inhibitor JQ1 has been widely used in generating a number of BET family bromodomain protein degraders that use different E3 ligase systems such as CUL2VHL and CUL4CRBN (Zengerle et al. 2015; Raina et al. 2016; Winter et al. 2017) and therefore serves as a suitable compound to demonstrate the utility of BTR2000 and the CUL3KLHL20 E3 ligase system for ablating oncogenic transcriptional regulators. We first synthesized a bifunctional JQ1–BTR2000 linked compound (namely, BTR2003) by coupling JQ1 carboxylic acid to BTR2000 with a four-glycine (4× Gly) linker (Fig. 3A) to afford flexibility between BTR2000 and JQ1. To test whether BTR2003 can initiate ternary complex formation upon KLHL20 and BET protein colocalization, we performed a pull-down experiment using the biotinylated KLHL20 Kelch domain as the bait to capture purified BRD4 bromodomain 1 (BD1) in the presence or absence of BTR2003 (Fig. 3B). The BTR2003 concentration-dependent BD1 pull-down indicated that BTR2003 does promote KLHL20–BTR2003–BD1 ternary complex formation. Based on ELISA, we quantitated an EC50 value of 8 nM for the ternary complex (Supplemental Fig. S2). Overall, we confirmed that BTR2003 is capable of promoting ternary complex formation at low nanomolar concentration.
Figure 3.
BTR2003 promotes ternary complex formation between KLHL20 and BRD4 BD1. (A) Structure of the bifunctional molecule BTR2003. The BET bromodomain inhibitor JQ1 (red) was conjugated to BTR2000 (blue) via a four-glycine linker (black). (B) BRD4 BD1 pull-down experiment with the KLHL20 or KLHL7 Kelch domains immobilized to beads in the presence of different BTR2003 concentrations. Bound His-BRD4 BD1 was detected by Western blotting using an anti-His antibody. (C) Binding model of the KLHL20 Kelch domain to the BRD4 BD1 in the presence of BTR2003. Shown are the five best results of a docking experiment by Rosetta docking as judged by the free energy. (D) NMR TROSY spectra of 15N-labeled BRD4 BD1. Shown excerpts of full spectral overlays are of the domain alone (red) or in the presence of BTR2003 (blue) or BTR2003 and KLHL20 Kelch domain (green). The asterisks indicate signals exclusive to the trimeric complex. (E) The KLHL20 Kelch binding epitope on the BTR2003-bound BRD4 BD1 based on the NMR experiment. Residues highlighted in red showed significant reduction in signal intensities relative to the BTR2003-bound domain after addition of the KLHL20 Kelch domain. BTR2003's position is based on the docking experiment.
Neo-protein–protein interaction mediated by BTR2003
Most of the reported ternary complex structures mediated by PROTAC molecules revealed the existence of additional protein–protein interactions (neo-protein–protein interactions) upon degrader binding (Winter et al. 2015; Gadd et al. 2017; Schiemer et al. 2021; Yu et al. 2021). To better understand the BTR2003-mediated ternary complex formation and the possible neo-protein–protein interaction on a structural level, we used the Rosetta protein modeling suite to evaluate the likelihood of protein–protein interface formation. Rosetta docking (Maguire et al. 2021) was used to simulate KLHL20 Kelch domain, BTR2000, and JQ1-bound BRD4 BD1 ternary complex formation. Since BTR2000 and JQ1 moieties will be tethered by the linker, we performed local docking of KLHL20–BTR2000 and BRD4 BD1-JQ1 to limit the search space so that docking on the irrelevant protein surface (for example, the opposite site of the KLHL20) can be avoided. Our search results mostly predicted that BRD4 BD1 and KLHL20 can form direct protein–protein interactions permitted by BTR2000 linked to JQ1 (Fig. 3C). In this model, JQ1 and BTR2000 molecules are within 5-Å distance, which can be easily tethered through a short linker.
To further verify the existence of direct protein–protein contacts between the KLHL20 Kelch domain and BRD4 BD1, we performed two-dimensional NMR experiments. TROSY spectra were recorded of 15N-labeled BRD4 BD1 alone, with the presence of BTR2003, or with BTR2003 and the unlabeled KLHL20 Kelch domain. Incubation of BD1 with an excess of BTR2003 led to extensive chemical shift changes (Fig. 3D, left panel). However, when adding equimolar amounts of BTR2003 and KLHL20 Kelch to BD1 to result in a 1:1:1 complex, only weak signals representative of a small fraction of unbound BD1 remained. This phenomenon was only observed if BTR2003 was present in the sample (Supplemental Fig. S3A), and indicated that the majority of the 15N-labeled BRD4 BD1 protein had become part of a higher-molecular-weight species that was not observable under the chosen measurement conditions, likely representing the trimeric BD1–BTR2003–KLHL20 Kelch complex (Fig. 3D, middle panel). Significantly increasing the measurement time then unveiled an additional set of peaks that was partly identical to the spectrum of the BTR2003-bound BD1 but also featured new signals that are exclusive to the BD1–BTR2003–KLHL20 Kelch complex (Fig. 3D, right panel). Those signals could indicate direct contacts between the two domains induced by BTR2003-mediated recruitment of KLHL20 Kelch into close proximity to BRD4 BD1, verifying our earlier hypothesis that additional protein–protein contacts were formed upon BTR2003 engagement. Closer analysis of the signal intensities of BRD4 BD1 in the trimeric complex in comparison with its BTR2003-bound form (Supplemental Fig. S3B) allowed us to map the complex-specific signal changes onto the crystal structure of the domain (Fig. 3E). The resulting epitope was largely in agreement with the results of the docking experiment.
Characterizing the effect of linker design on ternary complex formation
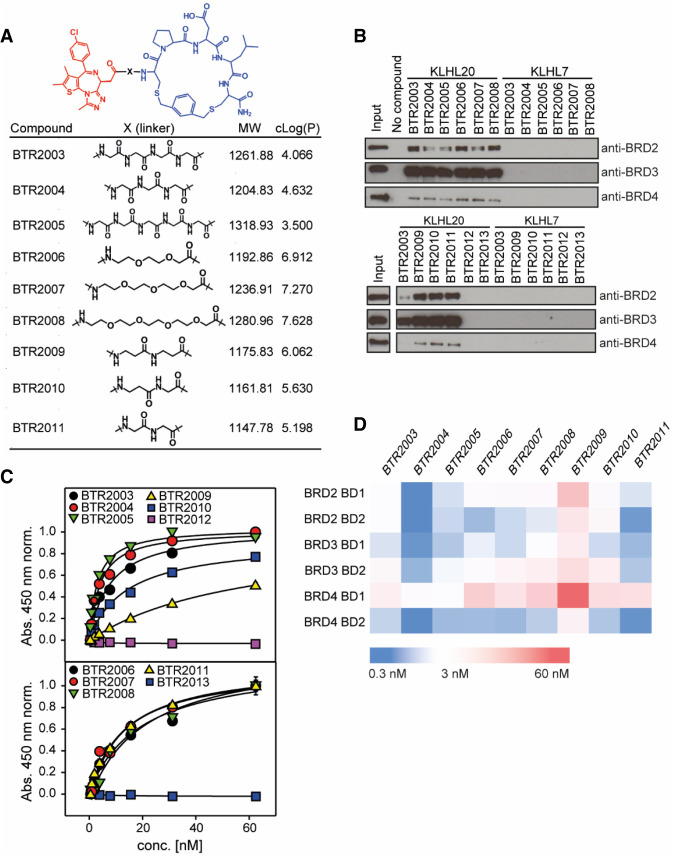
Linker properties such as length or physiochemical features (lipophilicity, for example) have been shown to be crucial for TPD potency (Nowak et al. 2018; Klein et al. 2021). We therefore created a panel of BTR2003 linker variants—implementing commonly used linker building blocks—to evaluate the activity of these variant molecules (Fig. 4A). In addition, for negative control, two small molecules that do not bind to BET family proteins acetylsalicylic acid and indomethacin were conjugated to BTR2000 to yield to BTR2012 (acetylsalicylic acid) and BTR2013 (indomethacin) (Supplemental Fig. S4). In pull-down experiments using PC3 whole-cell lysates, we observed that all JQ1-conjugated molecules (BTR2003–BTR2011), but not the negative controls BTR2012 and BTR2013, could recruit endogenous full-length BRD2, BRD3, and BRD4 to the KLHL20 Kelch domain (Fig. 4B), indicating their potential as novel BRD2, BRD3, or BRD4 protein degraders. Interestingly, the BTR2003 analogs showed a higher propensity for BRD2/3 engagement over BRD4 to KLHL20.
Figure 4.
Linker length and composition have effects on trifunctional complex formation and affinity. (A) Structure of 10 different linker variants of BTR2003 derivatives (BTR2004–BTR2013). (B) Pull-down experiment of full-length BRD2, BRD3, and BRD4 from PC3 lysates using immobilized KLHL20 and KLHL7 Kelch in the presence of different BTR2003 derivatives or negative controls. Bound protein was detected by Western blotting using antibodies against the respective BET proteins. (C) ELISA binding curves of T7-BRD4 BD1 to the immobilized KLHL20 Kelch domain in the presence of serial dilutions of BTR2003–BTR2013. Error bars show SD (N = 2). Data were normalized over the full concentration range of 0–1 µM, and data below the hook point were fitted to a three-parameter hyperbolic model. Plots of the full concentration range are shown in Supplemental Figure S5A. (D) Heat map illustrating the ELISA EC50 values of six different BET bromodomains binding to the KLHL20 Kelch domain in the presence of BTR compounds.
In order to quantitatively compare the BTR2003 variants in ternary complex formation, we used ELISA to quantitate the differential EC50 of BRD4 BD1–BTR2003 analog–KLHL20 ternary complex formation (Fig. 4C; Supplemental Fig. S5A). Although all of the tested compounds were capable of promoting ternary complex formation, variability among the analogs was observed. To further investigate whether the trend of differential ternary complex affinity would be conserved with other BET family bromodomains, we included all six bromodomains from BRD2, BRD3, and BRD4 for comparison. We observed that not only did the linkers affect the ternary complex affinity with a particular bromodomain, they also varied affinities across individual bromodomains (Fig. 4D). Even though the six bromodomains exhibited high levels of sequence conservation (Supplemental Fig. S5B), their individual ternary complex-forming propensity also differed. For instance, BRD4 BD1 has weaker interaction among all the bromodomains across all BTR analogs tested, while BRD4 BD2 and BRD3 BD1 have higher affinities across the panel (Fig. 4D), consistent with the observation from the pull-down experiment of the full-length proteins (Fig. 4B). The variation in affinity could possibly originate from the differential protein–protein interface fitness between the bromodomain and KLHL20 afforded by individual binding modes of the varying linkers.
Although linker spacers are generally flexible, subtle differences can contribute to additional favorable molecular interaction of the ternary complex or unfavorable steric hinderance. In our example with JQ1 and BTR2000 linkage, the 2× β-alanine (β-Ala) linker (BTR2009) exhibited the weakest affinity for the ternary complex, while substituting the 2× β-Ala with the β-Ala–Gly linker (BTR2010) had improved affinity with all six bromodomains (Fig. 4D), suggesting subtle steric interactions can play a large role in ternary complex formation. In general, we observed that 2×–5× Gly linkers (BTR2003, BTR2004, BTR2005, and BTR2011) seemed to perform better than the PEG linkers (BTR2006, BTR2007, and BTR2008), with 3× Gly (BTR2004) exhibiting the highest affinity across the six bromodomains.
BTR2003 analogs degrade BET family BRD proteins in multiple cancer cell lines
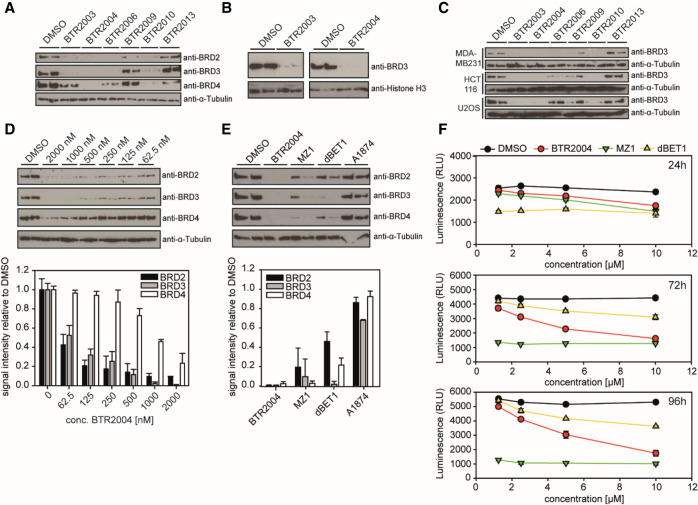
We next examined whether the BTR2003 analogs can degrade the target BET proteins in cancer cells. Treating PC3 cells (a human prostate cancer cell line) with various BTR compounds, we observed significant protein degradation of BRD2, BRD3, and BRD4 within 4 h by BTR2003 analogs but not the negative control compounds BTR2013–BTR2015 (Fig. 5A; Supplemental Fig. S6), indicating that BTR compounds are cell-permeable and capable of catalyzing targeted protein degradation. We also noticed that among various BTR compounds, BTR2009 showed nearly no degradation activity (Fig. 5A). This was consistent with our in vitro binding experiment, where BTR2009 showed the least ternary complex-forming activity. Following the trend of differential BD binding, most of the BTR compounds showed higher efficacy toward BRD2 and BRD3 degradation over BRD4, whereas BTR2004 showed complete depletion of all three proteins. In addition, since BRD2, BRD3, and BRD4 are primarily nuclear-localized, we collected nuclear fractions of PC3 cells to confirm the protein depletion by BTR2003 and BTR2004 in the nucleus (Fig. 5B). KLHL20 has been reported to be present in the cell body, with enriched localization at the trans-Golgi network (TGN) (Yuan et al. 2014), the autophagosome (Liu et al. 2016), and the nuclear plasma (Lee et al. 2010; Yuan et al. 2011). The presence of potent BRD protein degradation indicated that there was indeed sufficient nuclear activity of CUL3KLHL20 for BTR compound-facilitated protein degradation, suggesting that CUL3KLHL20 is a promising new E3 ligase system for the TPD of transcriptional regulators.
Figure 5.
Degradation of BET family proteins in cancer cells. (A) PC3 cells treated with 10 μM BTR compounds for 4 h. Cell lysates from biological duplicates were loaded and probed by Western blotting using the respective BET protein antibodies. α-Tubulin was used as a loading control. (B) BRD3 protein level in PC3 nuclear fraction. Histone-H3 was used as a nuclear loading control. (C) Experiments were as in A using MDA-MB-231, HCT116, and U-2 OS cell lines treated with 10 μM BTR compounds. BRD3 was detected by Western blotting. (D) Quantitation of BRD2, BRD3, and BRD4 protein degradation in PC3 cells treated with varying concentrations of BTR2004 for 4 h. Values are Western blot signal intensities relative to the DMSO negative control. (E) Protein levels of BRD2, BRD3, and BRD4 after 4-h treatment of PC3 cells with 5 μM BTR2004, A1874, MZ-1, and dBET1. DMSO negative control was used as a reference. (F) PC3 cells treated with different concentrations of BTR2004, MZ-1, and dBET1 for 24, 72, and 96 h. DMSO was used as a negative control. Cell titer after treatment was quantitated via luminescence measurement by using CellTiter-Glo 2.0. Error bars show standard deviation (SD; N = 3).
In addition to the PC3 prostate cancer cell line, we also verified the efficacy of BTR compounds in several human cell lines of different cancer types (Fig. 5C). These results illustrated both the potency of BTR compounds and the robust activity of CUL3KLHL20 E3 ligase in a wide variety of cancer cells. Further dose curve treatment of BTR2004 in PC3 cells confirmed that BTR2004 exhibited potency for BRD2, BRD3, and BRD4 at EC50 of ∼46, 87, and 777 nM, respectively (Fig. 5D; Supplemental Fig. S7A–C), again supporting specificity for BRD2/3 over BRD4.
So far, CUL4CRBN and CUL2VHL have been the most frequently used Cullin-Ring-type E3 ligases for designing TPD molecules, including the widely used JQ1-based BET family BRD degraders dBET1 analogs (via CUL4CRBN) (Winter et al. 2017) and MZ1 analogs (via CUL2VHL) (Gadd et al. 2017). Other types of E3 ligases such as human MDM2 (hDM2) have also been explored (Hines et al. 2019); for example, A1874 (Hines et al. 2019), a JQ1-based degrader that engages hDM2 via the idasanutlin moiety. To examine whether there are any differential degradation effects through different E3 ligases, we treated cancer cells with BTR2003, BTR2004, MZ1, dBET1, and A1874 (Fig. 5E; Supplemental Fig. S7D–F). Interestingly, in the PC3 cell line, while BTR2003, BTR2004, MZ1, and dBET1 showed efficacy on BET protein degradation, A1874 was not effective (Fig. 5E). This finding suggested that E3 ligase activities can vary among different cell types and that the capability of engaging various suitable E3 ligases is important to achieve potent target protein degradation. With the BRD protein degradation efficacy validated, BTR2004 treatment also led to a significant cell growth inhibition in PC3 (Fig. 5F) as well as MDA-MB-231 and HepG2 cells (Supplemental Fig. S8A,B). We also tested MZ1, dBET1, and BTR2004 in normal human cell line hTERT-BJ1 and observed some degree of cytotoxicity with all three TPD molecules. However, while MZ1 showed higher efficiency than BTR2004 in PC3 and MDA-MB-231 cancer cells at lower concentrations, BTR2004 demonstrated a lower normal cell toxicity, as judged by the treatment of hTERT-BJ1 cells (Supplemental Fig. S8C). Altogether, as a proof of concept, these data demonstrate the potency of the CUL3KLHL20 E3 ligase for TPD and the potential of the CUL3KLHL20-engaging ligand BTR2000 for future development of therapeutics.
Discussion
Leveraging the plasticity of E3 ubiquitin ligases, targeted protein degradation (TPD) has immense potential to target and degrade a wide range of proteins in the cell by creating target proximity to the E3 ligase assembly. Unlike many conventional inhibitors, which have stringent requirements for their target engagement sites (active sites or functional allosteric sites), TPD molecules have greater flexibility in target engagement if the binding mode can afford efficient polyubiquitination by the E3 ligase. Furthermore, since protein ubiquitination and the subsequent proteasome degradation processes are catalytic, efficient TPD does not necessarily require high affinity or a large amount of drug molecule to be present inside the cell. In contrast, conventional inhibitors achieve their activities by stoichiometry rather than by catalytic mechanisms and hence require high affinity or high dose for efficient stoichiometric target abrogation. The catalytic nature has provided opportunities for targeting proteins conventionally considered to be difficult, such as transcription factors, by the TPD approach. However, the TPD field is still in its early phase in demonstrating the therapeutic efficacy of protein degraders in the clinic. It is anticipated that the next few years for the TPD field will be a phase of broadening target protein coverage by the creation of a wide variety of E3-based degraders to be tested in the clinic.
In the present work, we have designed a KLHL20 macrocycle ligand, BTR2000, to demonstrate the utility of CUL3KLHL20 as a new modality for TPD. Our characterization of BTR2000-based protein degraders described here has validated that CUL3KLHL20 is capable of degrading nuclear target proteins such as the BET family BRD proteins. In addition to nuclear targets, preliminary data from our laboratory suggest that CUL3KLHL20 can also be directed to degrade cytosolic target proteins using another BTR2000-based target-specific degrader. BTR2000 thus can form the basis for building degrader molecules for further functional exploration of target accessibility and degradability by engaging CUL3KLHL20. A potential drawback of BTR2000 is the relatively large size of the macrocyclic molecule, which might limit its cell permeability. In our cell-based experiments, BTR2004 already showed activity comparable with or even better than other JQ1-based protein degraders at low micromolar concentrations, demonstrating the general ability of the molecules to enter cultured cells. As recent studies have identified key parameters to improve cell permeability for macrocycles with molecular weight >1000 Da (Furukawa et al. 2020), high membrane permeability of BTR2000 potentially can be achieved through further optimization to increase pharmacological potency.
It is worth noting that TPD molecules in general can be classified into two categories: the “molecular glue” (Schreiber 2021) and the PROTAC (Bekes et al. 2022). Although functionally both categories can deliver similar outcomes, at the molecular level, they present different features. Molecular glues are strictly defined by the formation of neo-protein–protein interaction to bridge together two proteins that otherwise do not interact (Schreiber 2021). Examples are the phthalimide derivative IMiD molecules (Schreiber 2021). In contrast, PROTAC molecules originate from the concept of connecting two binding moieties through a linker to create a heterofunctional molecule capable of fostering ternary complex formation regardless of whether a neo-protein–protein interaction is formed (Bekes et al. 2022). As the BTR2000 core structure has the potential to be derivatized into both classes of TPD molecules, here we used a more generalized term, “TPD molecules,” to refer to all the degrader molecules, even though the BTR2003 series molecules described here fit better to the PROTAC class. Technically speaking, it takes more molecular crafting to create a molecular glue that has defined binding modes, whereas the design process is more modular for making a PROTAC. Future work can be done to create BTR2000-based molecular glues and PROTACs for a side-by-side comparison to better understand whether their therapeutic efficacies and outcomes can differ.
One interesting phenomenon revealed from most PROTAC ternary complex crystal structures is the existence of neo-protein–protein interaction upon degrader binding (Winter et al. 2015; Gadd et al. 2017; Schiemer et al. 2021; Yu et al. 2021). Given the paradigm that protein–protein interaction in the proteome has been shaped to maintain high specificity for function, the emergence of degrader-facilitated neo-protein–protein interfaces indicates that the proteome does contain a high potential to be rewired to engage artificial interaction. These interactions can confer the plastic nature of neo-protein–protein interaction for gaining high binding affinity upon ternary complex formation and hence the protein degradation activity.
Besides the biophysical properties that contribute to ternary complex formation and degradability, factors such as E3 expression level may account for the efficacy of a chosen E3 ligase. The fact that hDM2-based degrader A1874 was ineffective in degrading the target BRD proteins in the PC3 cancer cell line highlighted the importance of choosing the appropriate E3 ligase for a specific type of cancer cell as part of the targeting strategy design. In addition, while existing CUL2- and CUL4-based BRD protein degraders have been demonstrated to show selectivity for BRD4 over BRD2 and BRD3 (Zengerle et al. 2015), the CUL3KLHL20 -based degraders presented in this study show potency on all three BRD proteins, in particular with higher selectivity for BRD2 and BRD3 over BRD4. Although JQ1 is a panbromodomain inhibitor, coupling this molecule to BTR2000 was able to confer selectivity for BRD2/3 over BRD4 by several-fold. Whether this will make BTR2003 analogs less ideal for targeting cancer is yet to be investigated further. However, our results clearly show that target selectivity can be reshaped by the choice of the CRL E3 ligases, probably via the differences in neo-protein–protein interface. On top of the E3 abundance, which can affect substrate degradation efficiency, protein colocalization can also play an essential role in the success of protein degradation. CUL3 has both higher cellular abundance and a wider range of subcellular localization patterns among the CRL E3 ligases (Uhlén et al. 2015; Jang et al. 2018). Therefore, the capacity of engaging additional CUL3 E3 ligase systems, such as CUL3KLHL20, should provide advantages to the TPD field in accessing a wide variety of cellular target proteins. The BTR2000 derivative molecules therefore lay the groundwork for future efforts toward a wide variety of TPD molecules for a broad range of disease targets and for broader options of TPD modality.
Materials and methods
Protein constructs
Human KLHL20 Kelch (317–603), KLHL7 Kelch (294–579), KLHL12 Kelch (272–568), BRD2 BD1 (74–194), BRD2 BD2 (348–455), BRD3 BD1 (24–144), BRD3 BD2 (306–413), BRD4 BD1 (44–168), and BRD4 BD2 (350–457) were cloned in pETM11 (omitting the MAD tag) or pET21a for E. coli expression. Eventually, an N-terminal T7 tag for antibody detection or a C-terminal AviTag for in vivo biotinylation was added in the cloning process. All mutants were generated from WT constructs by quick change PCR using suitable mutagenesis primers.
Protein expression and purification
All Kelch-like and BET protein constructs were expressed in E. coli Rosetta (DE3). Culture volumes of 1–4 L of 2× YT medium were grown to an OD600 of 0.7–0.9 at 37°C, and protein expression was induced by adding 0.3 mM IPTG and carried out overnight at 18°C. For in vivo biotinylation, constructs containing a C-terminal Avitag were coexpressed with BirA in the presence of 50 µM biotin in the culture medium. After cell lysis by sonication, His-tagged proteins were batch-purified using 1 mL of Ni-NTA beads (IMAC buffer: 20 mM TRIS at pH 8, 150 mM NaCl, 20 mM imidazole, 2% glycerol, 1 mM DTT, Roche cOmplete EDTA-free protease inhibitor cocktail) and a salt gradient of 500–150 mM NaCl in lysis buffer for washing. Elution was carried out by 200 mM imidazole in lysis buffer. Purity of the eluted proteins was checked by SDS-PAGE, and eventually proteins were further purified by MonoQ column chromatography if the purity was not sufficient or if His or T7 tags were removed by TEV cleavage after IMAC. Pure proteins were dialyzed into TBS (pH 7.5) and 1 mM DTT overnight at 4°C and concentrated to up to 2 mg/mL. The biotinylation level of proteins coexpressed with BirA was assessed by Western blotting using an HRP-conjugated antibiotin antibody (Cell Signaling).
Solid-phase peptide synthesis
All reagents were purchased through Sigma Aldrich and VWR. Solid-phase peptide synthesis was performed on Rink amide MBHA resin at 0.65 mmol/g loading (Novabiochem 8.55003.005). All peptides were synthesized at 5-μmol scale using standard Fmoc solid-phase chemistry. Three 0.5-mL NMP and four 0.5-mL DCM washes were performed between each step. Fmoc deprotection was performed using 0.5 mL of 20% 4-methylpiperidine in NMP for 20 min with nitrogen purging. Three equivalents of 0.2 M amino acid was activated with three equivalents of 0.2 M HCTU and 5.7 equivalents of 2.0 M DIPEA for 1 min before adding to deprotected resin. JQ1-COOH and all carboxylic acid-containing warheads were coupled in the same fashion. Couplings were allowed to proceed for 20 min to 1 h. All couplings were checked by Kaiser test for any remaining unreacted amines. Where necessary, resin was capped with 50 equivalents of acetic anhydride and 50 equivalents of pyridine in 0.2 mL of NMP. All cysteine residues were Mmt- or Trt-protected. The Mmt group was removed on resin with 2% TFA in DCM. On-resin cyclization was performed using four equivalents of 1,3-bis(bromomethyl)benzene and four equivalents of DIPEA, both at 25 mM, in NMP for 3 h at room temperature under inert atmosphere. FITC was conjugated to free amine by dissolving three equivalents of the isothiocyanate in 200 μL of NMP, adding three equivalents of DIPEA, and adding this mixture to the deprotected resin. Coupling was allowed to proceed overnight under inert atmosphere. Biotinylated compounds were prepared by coupling PEG3 followed by Fmoc-Lys(Biotin) to the N terminus. Peptides were cleaved using 95:2.5:2.5 TFA:TIS:H2O for 2 h with nitrogen purging. Solvent volume was reduced to 50 μL with nitrogen flow. Peptides were precipitated with −70°C diethyl ether. Crude peptides were dissolved in 1:1 acetonitrile:water and subsequently purified using RP-HPLC. Flow rate was 5 mL/min at 95% B for 30 min. The correct fraction was identified by LC-MS and lyophilized to dryness. The representative yield of at least three reactions was equal to 60%. Examples of LC-MS data for linear and cyclized BTR2003 and BTR2004 (crude and purified) are shown in Supplemental Figures S9 and S10, respectively.
Fluorescence polarization (FP) assay
Varying concentrations of protein from 0 to 15 μM were incubated with 20 nM peptide for 30 min at 4°C. Plates were then read for fluorescence polarization with a Cytation 5 plate reader (BioTek) using a cube with excitation wavelength of 485/20 nm, emission wavelength of 528/20 nm, and M = 510 nm. All dilutions were performed with 1× TBS and 1 mM DTT.
General pull-down procedure
Twenty micrograms of NeutrAvidin-conjugated M-280 tosylactivated Dynabeads (Invitrogen 14204) was blocked with 0.5 mL of 0.1% BSA (Amresco 0332-500G) for 2 h at 4°C. Beads were subsequently washed with 0.5 mL of 0.1% BSA in 1× TBS and then incubated with 110 μL of 2 μM KLHL20 or KLHL7 or 430 nM biotinylated compound for 1 h at 4°C. Proteins were diluted with 1× TBS, 1 mM DTT, and 0.1% BSA. Lysate was prepared by incubating cells with 1.5 mL of 10 mM Tris (pH 7.5), 100 mM NaCl, 0.5% deoxycholate, 10% glycerol, 1% Triton X-100, and protease and phosphatase inhibitors (Thermo Scientific A32961). The total protein concentration was determined by Bradford assay. Cell lysate was diluted 1:7 (total final protein concentration 1.6 mg/mL) with 1× TBS, 1 mM DTT, 0.1% BSA, and protease and phosphatase inhibitors. KLHL20/7 loaded beads were then incubated with either 0.5 mL of lysate or 110 μL of 2 μM BRD4 BD1, depending on the experiment, and varying concentrations of peptide (2 μM if not listed) for 1 h at 4°C. Beads were then washed three times with 0.5 mL of 1× TBS, 1 mM DTT, and 0.1% BSA, and proteins were eluted from the beads with 20 μL of 1× Laemmli buffer and 5% BME. For the proteomics experiments, eluates were in-gel-digested with trypsin, peptides were analyzed by LC-MS/MS, and label-free quantification was used to measure protein differential abundance across samples. Detailed values for all proteins detected in these experiments are in Supplemental Table S1.
Molecular modeling of ternary complex interaction
Docking simulation was performed using RosettaCommons v3 (2021.16). The structure of the BTR2000 was first simulated with Avogadro software and was exported as a PDB file. The PDB files of both BTR2000 and the KLHL20 Kelch domain (adopted from PDB: 6GY5) were processed with Rosetta Relax (Khatib et al. 2011) followed by RosettaLigandDocking to simulate the BTR2000–KLHL20 complex. The best 50 simulated outputs, ranked by the overall score and the interface score, were used to model the ternary complex. We imported the BRD4 BD1–JQ1 structure (adopted from PDB: 3MXF1) into the simulated BTR2000–KLHL20 complex and aligned the structure as described (Maguire et al. 2021). RosettaRelax was performed to generate 100 relaxed structures, and the top 10 structures were then simulated by RosettaDock. Thirty-thousand structures were generated and ranked.
NMR chemical shift
All NMR data were recorded on a Bruker 800-MHz Avance III spectrometer equipped with a 5-mm TCI cryoprobe. Spectra were processed using TopSpin 3 (Bruker), and the spectral data were evaluated in CCPNmr analysis (Vranken et al. 2005). Transverse relaxation optimized spectroscopy (TROSY) spectra of uniformly 15N-labeled BRD4 BD1 in PBS (pH 7.5) and 10% (v/v) D2O were measured at 298 K with a protein concentration of 80 μM. In total, 1024 × 256 complex data points were acquired with eight scans, or 128 scans in the case of the BRD4 BD1–BTR2003–KLHL20 Kelch complex. The backbone assignment of the BRD4 BD1 was downloaded from the Biological Magnetic Resonance Data Bank (BMRB entry 50145; Patel et al. 2021). To determine the epitope of the KLHL20 Kelch domain on BRD4 BD1 in the trimeric complex, and normalized signal intensities of 15N-BRD4 BD1 measured in the presence of equimolar amounts of BTR2003 alone or BTR2003 and KLHL20 Kelch were compared. A reduction of the normalized signal intensity ratio to lower than the average intensity ratio minus standard deviation was considered significant, and respective assigned residues were taken into account for the epitope.
ELISA
For the ELISA analysis of BTR compound-mediated complex formation between BRD bromodomains and the KLHL20 Kelch domain, ELISA plates were coated with 150 ng of NeutrAvidin per well. After blocking with 0.5% BSA in PBS for 2 h at 4°C, the plates were washed three times with PBS-T and subsequently incubated with 90 ng of biotinylated KLHL20 or KLHL12 Kelch domain (WT or mutant) per well for 1 h at room temperature. Plates were washed again three times with PBS-T and subsequently incubated with 50 ng of T7-tagged BRD2, BRD3, or BRD4 BD1 or BD2 per well and different concentrations of BTR compound (1 µM to 0.5 nM dilution series) for 1 h at room temperature. Plates were again washed three times with PBS-T and then incubated with an HRP-conjugated anti-T7 antibody (100 ng/mL) for 0.5 h at room temperature. After a final washing step, signals of bound T7 bromodomain were detected using 1-Step Ultra TMB-ELISA mix (Thermo Scientific) on a Cytation 5 imaging reader (BioTek). All proteins and compounds were diluted into blocking buffer supplemented with 1 mM DTT to reduce background binding.
Cell culture and compound treatment experiments
All cells were cultured in DMEM with 10% FBS. For protein degradation Western blots, cells were seeded in 24-well plates 24 h prior to compound treatments. At the time of treatment, cells were changed to fresh culture medium containing the compounds or equal volume of DMSO control. At the end of treatment, cells were washed once with TBS/0.05% Tween 20, followed by cell lysis. Cell lysis buffer (120 µL) (see “General Pull-Down Procedure”) was added to each well. For collecting the nuclear fraction, cells were seeded in 10-cm culture plates according to the nuclear fractionation protocol (https://www.abcam.com/ps/pdf/protocols/Nuclear%20fractionation%20protocol.pdf)
For the cell proliferation experiment, cells were seeded in 96-well plates at a density of 50% confluency 24 h prior to compound treatments. At the time of treatment, cells were changed to fresh culture medium containing the compounds or an equal volume of DMSO control. At the end of treatment, cell titers were measured by using CellTiter-Glo 2.0 (Promega) according to the manufacturer's instruction.
Supplementary Material
Acknowledgments
We thank Dr. Eszter Boros, Darek Smilowicz, Grace Kim, and Nan Wang at Stony Brook University for access to instrumentation; Dr. Yousef Al-Abed, Dr. Mingzhu He, and Dr. Kai F. Cheng at Feinstein Institute for Medical Research for access to prep-HPLC; Dr. Michael Goger and Dr. Shibani Bhattacharya at New York Structural Biology Consortium for assisting NMR instrumentation; Dr. Paolo Cifani at Cold Spring Harbor Laboratory Proteomics Shared Resource for assisting with mass spectrometry proteomics; Michelle Yang, Carmelita Bautista, Megan Chamberland, and Anna Catherine Unser for assistance with experiments; Cold Spring Harbor Laboratory Antibody and Phage Display Shared Resource for anti-T7 tag and anti-α tubulin (DM1A) monoclonal antibodies; and Dr. Ruey-Hwa Chen at Academia Sinica for kindly providing the full-length human KLHL20 plasmid. This work was supported in part by Development Funds and the Cold Spring Harbor Laboratory Shared Resources funded by Cancer Center Support 5P30CA045508, and the Cold Spring Harbor Laboratory and Northwell Health affiliation. NMR data were collected using the 800-MHz Avance III spectrometer at the New York Structural Biology Consortium NMR Facility, which is supported by National Institutes of Health grant S10OD016432.
Author contributions: J.T.-H.Y. conceived and designed the study, designed the compounds, performed the experiments and data analysis, and wrote the manuscript. B.M.F. designed the compounds, performed the experiments and data analysis, and wrote the manuscript. F.G. performed the experiments and data analysis and wrote the manuscript. C.-H.R.Y. performed the experiments and data analysis and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.349717.122.
Competing interest statement
The authors declare no competing interests.
References
- Bai L, Zhou H, Xu R, Zhao Y, Chinnaswamy K, McEachern D, Chen J, Yang CY, Liu Z, Wang M, et al. 2019. A potent and selective small-molecule degrader of STAT3 achieves complete tumor regression in vivo. Cancer Cell 36: 498–511.e17. 10.1016/j.ccell.2019.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekes M, Langley DR, Crews CM. 2022. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov 21: 181–200. 10.1038/s41573-021-00371-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canning P, Cooper CDO, Krojer T, Murray JW, Pike ACW, Chaikuad A, Keates T, Thangaratnarajah C, Hojzan V, Marsden BD, et al. 2013. Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases. J Biol Chem 288: 7803–7814. 10.1074/jbc.M112.437996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HY, Chen RH. 2016. Cullin 3 ubiquitin ligases in cancer biology: functions and therapeutic implications. Front Oncol 6: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Hellmann H. 2013. Plant E3 ligases: flexible enzymes in a sessile world. Mol Plant 6: 1388–1404. 10.1093/mp/sst005 [DOI] [PubMed] [Google Scholar]
- Chen HY, Hu JY, Chen TH, Lin YC, Liu X, Lin MY, Lang YD, Yen Y, Chen RH. 2015. KLHL39 suppresses colon cancer metastasis by blocking KLHL20-mediated PML and DAPK ubiquitination. Oncogene 34: 5141–5151. 10.1038/onc.2014.435 [DOI] [PubMed] [Google Scholar]
- Chen Z, Picaud S, Filippakopoulos P, D'Angiolella V, Bullock AN. 2019. Structural basis for recruitment of DAPK1 to the KLHL20 E3 ligase. Structure 27: 1395–1404.e4. 10.1016/j.str.2019.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M, He YJ, Borchers C, Xiong Y. 2003. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol 5: 1001–1007. 10.1038/ncb1056 [DOI] [PubMed] [Google Scholar]
- Furukawa A, Schwochert J, Pye CR, Asano D, Edmondson QD, Turmon AC, Klein VG, Ono S, Okada O, Lokey RS. 2020. Drug-like properties in macrocycles above MW 1000: backbone rigidity versus side-chain lipophilicity. Angew Chem Int Ed Engl 59: 21571–21577. 10.1002/anie.202004550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadd MS, Testa A, Lucas X, Chan KH, Chen W, Lamont DJ, Zengerle M, Ciulli A. 2017. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol 13: 514–521. 10.1038/nchembio.2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer R, Wee S, Anderson S, Yates J, Wolf DA. 2003. BTB/POZ domain proteins are putative substrate adaptors for cullin 3 ubiquitin ligases. Mol Cell 12: 783–790. 10.1016/S1097-2765(03)00341-1 [DOI] [PubMed] [Google Scholar]
- Gooding S, Ansari-Pour N, Towfic F, Ortiz Estévez M, Chamberlain PP, Tsai KT, Flynt E, Hirst M, Rozelle D, Dhiman P, et al. 2021. Multiple cereblon genetic changes are associated with acquired resistance to lenalidomide or pomalidomide in multiple myeloma. Blood 137: 232–237. 10.1182/blood.2020007081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines J, Lartigue S, Dong H, Qian Y, Crews CM. 2019. MDM2-Recruiting PROTAC offers superior, synergistic antiproliferative activity via simultaneous degradation of BRD4 and stabilization of p53. Cancer Res 79: 251–262. 10.1158/0008-5472.CAN-18-2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SM, Redon CE, Aladjem MI. 2018. Chromatin-bound cullin-ring ligases: regulatory roles in DNA replication and potential targeting for cancer therapy. Front Mol Biosci 5: 19. 10.3389/fmolb.2018.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J, To C, De Clercq DJH, Park E, Ponthier CM, Shin BH, Mushajiang M, Nowak RP, Fischer ES, Eck MJ, et al. 2020. Mutant-selective allosteric EGFR degraders are effective against a broad range of drug-resistant mutations. Angew Chem Int Ed Engl 59: 14481–14489. 10.1002/anie.202003500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatib F, Cooper S, Tyka MD, Xu K, Makedon I, Popović Z, Baker D, Players F. 2011. Algorithm discovery by protein folding game players. Proc Natl Acad Sci 108: 18949–18953. 10.1073/pnas.1115898108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein VG, Bond AG, Craigon C, Lokey RS, Ciulli A. 2021. Amide-to-ester substitution as a strategy for optimizing PROTAC permeability and cellular activity. J Med Chem 64: 18082–18101. 10.1021/acs.jmedchem.1c01496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YR, Yuan WC, Ho HC, Chen CH, Shih HM, Chen RH. 2010. The cullin 3 substrate adaptor KLHL20 mediates DAPK ubiquitination to control interferon responses. EMBO J 29: 1748–1761. 10.1038/emboj.2010.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Lin YC, Chen YH, Chen CM, Pang LY, Chen HA, Wu PR, Lin MY, Jiang ST, Tsai TF, et al. 2016. Cul3-KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Mol Cell 61: 84–97. 10.1016/j.molcel.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Maguire JB, Haddox HK, Strickland D, Halabiya SF, Coventry B, Griffin JR, Pulavarti S, Cummins M, Thieker DF, Klavins E, et al. 2021. Perturbing the energy landscape for improved packing during computational protein design. Proteins 89: 436–449. 10.1002/prot.26030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak RP, DeAngelo SL, Buckley D, He Z, Donovan KA, An J, Safaee N, Jedrychowski MP, Ponthier CM, Ishoey M, et al. 2018. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat Chem Biol 14: 706–714. 10.1038/s41589-018-0055-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson CM, Jiang B, Erb MA, Liang Y, Doctor ZM, Zhang Z, Zhang T, Kwiatkowski N, Boukhali M, Green JL, et al. 2018. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol 14: 163–170. 10.1038/nchembio.2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K, Solomon PD, Walshe JL, Ford DJ, Wilkinson-White L, Payne RJ, Low JKK, Mackay JP. 2021. BET-family bromodomains can recognize diacetylated sequences from transcription factors using a conserved mechanism. Biochemistry 60: 648–662. 10.1021/acs.biochem.0c00816 [DOI] [PubMed] [Google Scholar]
- Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AM, Wang J, Chen X, Dong H, Siu K, et al. 2016. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci 113: 7124–7129. 10.1073/pnas.1521738113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. 2001. Protacs: chimeric molecules that target proteins to the Skp1–Cullin–F box complex for ubiquitination and degradation. Proc Natl Acad Sci 98: 8554–8559. 10.1073/pnas.141230798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiemer J, Horst R, Meng Y, Montgomery JI, Xu Y, Feng X, Borzilleri K, Uccello DP, Leverett C, Brown S, et al. 2021. Snapshots and ensembles of BTK and cIAP1 protein degrader ternary complexes. Nat Chem Biol 17: 152–160. 10.1038/s41589-020-00686-2 [DOI] [PubMed] [Google Scholar]
- Schreiber SL. 2021. The rise of molecular glues. Cell 184: 3–9. 10.1016/j.cell.2020.12.020 [DOI] [PubMed] [Google Scholar]
- Teng M, Jiang J, He Z, Kwiatkowski NP, Donovan KA, Mills CE, Victor C, Hatcher JM, Fischer ES, Sorger PK, et al. 2020. Development of CDK2 and CDK5 dual degrader TMX-2172. Angew Chem Int Ed Engl 59: 13865–13870. 10.1002/anie.202004087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjöstedt E, Asplund A, et al. 2015. Proteomics. Tissue-based map of the human proteome. Science 347: 1260419. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Verma R, Mohl D, Deshaies RJ. 2020. Harnessing the power of proteolysis for targeted protein inactivation. Mol Cell 77: 446–460. 10.1016/j.molcel.2020.01.010 [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. 2005. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59: 687–696. 10.1002/prot.20449 [DOI] [PubMed] [Google Scholar]
- Wimuttisuk W, Singer JD. 2007. The Cullin3 ubiquitin ligase functions as a Nedd8-bound heterodimer. Mol Biol Cell 18: 899–909. 10.1091/mbc.e06-06-0542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, Bradner JE. 2015. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348: 1376–1381. 10.1126/science.aab1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter GE, Mayer A, Buckley DL, Erb MA, Roderick JE, Vittori S, Reyes JM, di Iulio J, Souza A, Ott CJ, et al. 2017. BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol Cell 67: 5–18.e19. 10.1016/j.molcel.2017.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Wei Y, Reboul J, Vaglio P, Shin TH, Vidal M, Elledge SJ, Harper JW. 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425: 316–321. 10.1038/nature01985 [DOI] [PubMed] [Google Scholar]
- You I, Erickson EC, Donovan KA, Eleuteri NA, Fischer ES, Gray NS, Toker A. 2020. Discovery of an AKT degrader with prolonged inhibition of downstream signaling. Cell Chem Biol 27: 66–73.e7. 10.1016/j.chembiol.2019.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Li D, Kottur J, Shen Y, Kim HS, Park KS, Tsai YH, Gong W, Wang J, Suzuki K, et al. 2021. A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models. Sci Transl Med 13: eabj1578. 10.1126/scitranslmed.abj1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan WC, Lee YR, Huang SF, Lin YM, Chen TY, Chung HC, Tsai CH, Chen HY, Chiang CT, Lai CK, et al. 2011. A Cullin3-KLHL20 ubiquitin ligase-dependent pathway targets PML to potentiate HIF-1 signaling and prostate cancer progression. Cancer Cell 20: 214–228. 10.1016/j.ccr.2011.07.008 [DOI] [PubMed] [Google Scholar]
- Yuan WC, Lee YR, Lin SY, Chang LY, Tan YP, Hung CC, Kuo JC, Liu CH, Lin MY, Xu M, et al. 2014. K33-linked polyubiquitination of coronin 7 by Cul3-KLHL20 ubiquitin E3 ligase regulates protein trafficking. Mol Cell 54: 586–600. 10.1016/j.molcel.2014.03.035 [DOI] [PubMed] [Google Scholar]
- Zengerle M, Chan KH, Ciulli A. 2015. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem Biol 10: 1770–1777. 10.1021/acschembio.5b00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Riley-Gillis B, Vijay P, Shen Y. 2019. Acquired resistance to BET-PROTACs (proteolysis-targeting chimeras) caused by genomic alterations in core components of E3 ligase complexes. Mol Cancer Ther 18: 1302–1311. 10.1158/1535-7163.MCT-18-1129 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.