Abstract
African wild suids consist of several endemic species that represent ancient members of the family Suidae and have colonized diverse habitats on the African continent. However, limited genomic resources for African wild suids hinder our understanding of their evolution and genetic diversity. In this study, we assembled high-quality genomes of a common warthog (Phacochoerus africanus), a red river hog (Potamochoerus porcus), as well as an East Asian Diannan small-ear pig (Sus scrofa). Phylogenetic analysis showed that common warthog and red river hog diverged from their common ancestor around the Miocene/Pliocene boundary, putatively predating their entry into Africa. We detected species-specific selective signals associated with sensory perception and interferon signaling pathways in common warthog and red river hog, respectively, which contributed to their local adaptation to savannah and tropical rainforest environments, respectively. The structural variation and evolving signals in genes involved in T-cell immunity, viral infection, and lymphoid development were identified in their ancestral lineage. Our results provide new insights into the evolutionary histories and divergent genetic adaptations of African suids.
Keywords: African suids, common warthog, red river hog, pig, genome, local adaptation
Introduction
The family Suidae includes many widely distributed species known as pigs and hogs which show large morphological differences and have adapted to diverse habitats. There are at least 15 extant species of suids, which are currently grouped into six genera, including Babyrousa, Porcula, and Sus distributed in Eurasia, and Hylochoerus, Potamochoerus, and Phacochoerus inhabiting sub-Saharan Africa (Grubb 2005; Groves and Grubb 2011; Ruvinsky et al. 2011; Gongora et al. 2017). The majority are classified into the subfamily Suinae (Grubb 2005), whereas Babyrousa is frequently proposed as a distinct sister clade Babyrousinae (Gongora et al. 2011; Ruvinsky et al. 2011; Orliac 2013). The well-known suid species Sus scrofa gave rise to domestic pigs which have contributed greatly to the agricultural development of human civilization since ∼9,000 BP (Ruvinsky et al. 2011; Frantz et al. 2016). The origin of Suidae can be traced back to Early Oligocene in Eurasia, followed by diversification into multiple genera (Ruvinsky et al. 2011; Frantz et al. 2016; Gongora et al. 2017). Subsequent evolution and multiple dispersals have resulted in diverse ranges of extant Suidae species that have successfully occupied Africa, Asia, and Europe (Ruvinsky et al. 2011; Frantz et al. 2013, 2016; Gongora et al. 2017; Liu et al. 2019). The Suidae colonized Africa from Eurasia several times, but only members of the subfamily Suinae survived, ultimately leading to the suid species now present in sub-Saharan Africa (Ruvinsky et al. 2011; Frantz et al. 2016; Gongora et al. 2017). Nowadays, while wild boars comprise a single species across all over Eurasia (Liu et al. 2019), sub-Saharan Africa is renowned for the diversity of indigenous wild Suidae species in multiple niches, indicating the adaptation of these suids to their local environments, distinct from Eurasia.
A high level of species diversity and successful colonization of diverse habitats have been described for the African suids. Five extant suid species, including common warthog (Phacochoerus africanus), desert warthog (Ph. aethiopicus), bushpig (Potamochoerus larvatus), red river hog (P. porcus), and giant forest hog (Hylochoerus meinertzhageni), are found exclusively in sub-Saharan Africa, and inhabit different ecosystems, including savannah, tropical rainforest, and grassland (fig. 1A) (Grubb 2005; Ruvinsky et al. 2011; Frantz et al. 2016; Gongora et al. 2017). Among them, common warthog typically occurs in the vast open areas of savannah with long dry seasons (Butynski and de Jong 2017) while red river hog is restricted primarily to the Guinean and Congo Basin rainforests with dense cover and considerable annual rainfall (Leslie and Huffman 2015; Melletti et al. 2017), representing two of the major ecosystems within sub-Saharan Africa. Huge differences between the African savannahs and rainforests are linked to the genetic divergence and adaptative traits of their associated fauna. Some mammals of the same lineage or even the same species, including African savannah (Loxodonta africana) and forest (L. cyclotis) elephants (Grubb et al. 2000; Rohland et al. 2010; Roca et al. 2015), African buffalo (Syncerus caffer) (Smitz et al. 2013), and chimpanzee (Pan troglodytes) (Wessling et al. 2018; Schmidt et al. 2019), displayed both distinct genotypes and adaptive phenotypic divergence between the savannah and rainforest environments. It seems likely that similar adaptive evolution has also occurred in the African suids. For instance, the African suids have developed resistance to African swine fever virus (ASFV), showing asymptomatic infections as natural reservoirs (Jori and Bastos 2009), while Sus scrofa suffers from a high morbidity and mortality (Oura et al. 1998; Dixon et al. 2019).
Fig. 1.
Phylogeny and demographic history of African suids. (A) Habitat range of wild suids in Africa. Sampling sites are indicated by swine symbols in blue and red for common warthog and red river hog, respectively. Data of the spatial range were accessed from IUCN Red List (https://www.iucnredlist.org). (B) Species phylogeny with divergence time estimation. The maximum-likelihood tree was reconstructed with RAxML v8.2.12 (Stamatakis 2014). The divergence times were estimated from the concatenated CDSs with MCMCTREE (Yang 2007). The 95% HPD confidence intervals of estimates are indicated by horizontal bars at nodes. The nodes in dots were calibrated by information in Supplementary material. (C) Demographic history inferred with pairwise sequentially Markovian coalescence (Li and Durbin 2011). The generation times of 6.0, 8.6, and 7.3 years were used for common warthog, red river hog, and Sus scrofa (including Diannan small-ear pig, southern East Asian wild boar, and European wild boar), respectively (Pacifici et al. 2013). The mutation rate is 2.5 × 10−8 per site per generation (Groenen et al. 2012; Frantz et al. 2013). The Early-Middle Pleistocene transition (EMPT) and Last Glacial Maximum (LGM) are highlighted in purple and light blue, respectively. The geological time boundaries of the early/middle and the middle/late Pleistocene are indicated by vertical solid lines.
The success of extant African suids in various ecological niches indicates that they are excellent models to study the genetics of mammalian adaptive evolution in sub-Saharan Africa, which can provide additional valuable information on the spread of the family Suidae in Africa. Nevertheless, most of the previous studies associated with the extant African suids focused largely on phylogeny (Gongora et al. 2011; Liu et al. 2019), phylogeography (Muwanika et al. 2003; Garcia-Erill et al. 2022), social organization (Muwanika et al. 2007; White et al. 2010), conservation biology (Adeola et al. 2021; Codjia et al. 2021), feeding and reproductive biology (Boshe 1981; Berger et al. 2006; Edossa et al. 2021), as well as infectious and parasitic diseases (Luther et al. 2007; Everett et al. 2011; Blomstrom et al. 2012; Apanaskevich et al. 2013; Ebhodaghe et al. 2021; Friant et al. 2022). These have laid foundations for the preliminary understanding on their phylogeny, behavior, and adaptation. With the accumulation of genomic data in recent years for the family Suidae (Groenen et al. 2012; Frantz et al. 2013, 2016; Groenen 2016; Liu et al. 2019, 2022; Warr et al. 2020; Garcia-Erill et al. 2022), this provides a baseline that will underpin further studies on the potential molecular mechanisms of the evolution and phenotypic adaptations of the African suids with genomic approaches.
Herein, we applied long-read sequencing and de novo assembly of the genomes from two African suids (i.e., common warthog and red river hog) and one East Asian domestic pig (i.e., Diannan small-ear pig). Based on the newly generated high-quality genomes, we provided a phylogenomic framework for African suids. We further identified species-specific genomic signatures potentially associated with the local adaptations of common warthog and red river hog to their very distinct habitats. Additionally, we defined common evolutionary features in their ancestral lineage, as compared to their Eurasian relatives. These results extend and deepen our understanding of the evolution and local adaptations of the African suids.
Results
Genome Assembly and Annotation of African Suids
We performed long-read sequencing and de novo genome assembly for a male common warthog and a female red river hog, both sampled from Nigeria (Materials and Methods, fig. 1A). A total of 286.71 Gb (N50 read length 31.83 Kb) and 291.93 Gb (N50 read length 13.85 Kb) Oxford Nanopore long-read sequencing data were generated, accounting for 109-fold and 113-fold coverage of the common warthog and red river hog genomes, respectively, based on sizes estimated by a K-mer strategy. A total of 263.27 Gb and 330.34 Gb Illumina reads were also generated from the same two individuals, respectively. Contigs were assembled using the Nanopore reads and polished using the Illumina reads (supplementary table S1, Supplementary Material online). The contig N50 were 45.73 and 29.43 Mb for the common warthog and red river hog, respectively (table 1). However, their genomic DNA failed to meet criteria for the Bionano and Hi-C assembly technologies, so we used the latest Duroc assembly (Sscrofa11.1) (Warr et al. 2020) as the reference for scaffolding. In total, 96.81% and 97.73% of contigs from common warthog and red river hog, respectively, were assigned to pseudo-chromosomes (supplementary table S2, Supplementary Material online).
Table 1.
Comparison of Assembly Statistics and Assessment of the Four Suidae Genomes.
| Common warthog | Red river hog | Diannan small-ear pig | Duroc pig (Sscrofa11.1)a |
|
|---|---|---|---|---|
| Assembly size (bp) | 2,453,766,712 | 2,457,697,124 | 2,648,650,142 | 2,501,912,388 |
| Sequencing technology | Nanopore | Nanopore | Pacbio | Pacbio |
| Coverage depth (X) | 109.0 | 113.2 | 81.1 | 65.0 |
| Gap length (bp) | 197,800 | 172,501 | 10,364,137 | 29,864,641 |
| Read N50 (bp) | 31,826 | 13,850 | 14,500 | 19,786 |
| Number of contigs | 1,968 | 1,711 | 3,651 | 1,118 |
| Contig N50 (bp) | 45,725,369 | 29,428,946 | 14,072,921 | 48,231,277 |
| Longest contig (bp) | 138,745,353 | 104,680,965 | 70,343,318 | - |
| Placed contig length (bp) | 2,375,606,863 | 2,406,887,618 | 2,408,896,715 | 2,436,858,178 |
| Scaffold N50 (bp) | 138,295,081 | 139,403,444 | 137,346,382 | 88,231,837 |
| Repeat rate | 39.9% | 39.5% | 42.0% | 45.1% |
| Predicted protein-coding gene number | 21,300 | 21,711 | 21,463 | 20,790 |
| Average gene length (bp) | 27,351 | 27,200 | 38,871 | 50,936 |
| Average CDS length (bp) | 1,469 | 1,450 | 1,531 | 2,085 |
| Average exons per gene | 8.10 | 8.00 | 8.65 | 10.41 |
| Average exon length (bp) | 181 | 181 | 177 | 439 |
| Average intron length (bp) | 3,647 | 3,677 | 4,884 | 7,604 |
| Complete BUSCOs | 93.2% | 93.4% | 94.5% | 93.8% |
| Complete & single-copy BUSCOs | 92.6% | 92.9% | 93.7% | 93.3% |
| Complete & duplicated BUSCOs | 0.6% | 0.5% | 0.8% | 0.5% |
| Fragmented BUSCOs | 3.2% | 3.4% | 3.2% | 3.5% |
| Missing BUSCOs | 3.6% | 3.2% | 2.3% | 2.7% |
Summary statistics for the Duroc pig assembly (Sscrofa11.1) were accessed from Warr et al. (2020) and NCBI (https://www.ncbi.nlm.nih.gov/assembly/GCA_000003025.6).
For the common warthog, 98.61% of Illumina reads were mapped to the long-read assembly, of which 97.33% had at least 20-fold coverage. Similarly, 98.73% of Illumina reads were mapped to the red river hog assembly, of which 98.26% exhibited at least 20-fold coverage. Benchmarking Universal Single-Copy Orthologs (BUSCO) (Simão et al. 2015) assessment showed that 93.2% and 93.4% of the 4,104 orthologs in the mammalia_odb9 database were identified in the common warthog and red river hog genomes, respectively (table 1). We further checked 248 house-keeping genes in six eukaryotes and found that 229 and 231 genes were present in the common warthog and red river hog genomes, respectively. These results indicated a high level of completeness of the two newly assembled African suid genomes. Based on homology search and ab initio prediction, we annotated 21,300 and 21,711 protein-coding genes in the common warthog and red river hog genomes, respectively (table 1; supplementary figs. S1, S2, and tables S3–S5, Supplementary Material online). Screening of the predicted genes in seven protein annotation databases (Supplementary material online) showed that 20,107 common warthog (94.40%) and 20,545 red river hog (94.63%) genes were represented in these databases (supplementary table S3, Supplementary Material online). Additionally, the annotated genes on the unplaced contigs (accounting for 2.37–3.19% of the total assemblies, supplementary table S2, Supplementary Material online) of the two African suids were both primarily involved in olfactory receptors and cytokine activity (supplementary tables S4 and S5, Supplementary Material online). We further identified a total of 978.14 and 970.95 Mb of repetitive elements, accounting for 39.87% and 39.51% of the total genome size of the common warthog and red river hog, respectively (table 1; supplementary figs. S3, S4, and tables S6 and S7, Supplementary Material online).
Genome Assembly and Annotation of East Asian Pig
A de novo genome assembly of a male Diannan small-ear pig, which was an indigenous breed in southwestern China locating at one of the basal maternal lineages of East Asian domestic pigs (Wu et al. 2007), was used for comparison. A total of 243.3 Gb PacBio Sequel reads (read N50 of 14.5 Kb), 171.0 Gb Illumina reads, 147.3 Gb Bionano Genomic Mapping data, and 333.9 Gb Hi-C reads were generated from the same individual (supplementary table S1, Supplementary Material online). Based on the PacBio reads, 3,651 contigs were assembled with a contig N50 of 14.07 Mb (table 1). The scaffolding was performed with the Bionano technology. A total of 3,290 scaffolds were obtained with the longest scaffold of 284.85 Mb and a scaffold N50 of 137.35 Mb. After polishing with the Illumina reads, the original scaffolds were assigned to the chromosome level based on the Hi-C data (supplementary fig. S5, Supplementary Material online). The chromosome-level scaffolds were ordered based on the alignment with the Duroc reference chromosomes (Sscrofa11.1). Despite 361 gaps in this new Diannan small-ear pig genome, the assembly showed a closure of 244 (sum length of 75.6 Mb) out of the 506 gaps in the Duroc reference. According to the whole-genome alignment, the Diannan small-ear pig genome and Duroc reference showed a synteny conservation across all chromosomes except for chromosome 11 possibly with a large structural variation (supplementary fig. S6, Supplementary Material online) that needs further confirmation. BUSCO analysis showed that 94.5% of the 4,104 orthologs in the mammalia_odb9 database were complete in the Diannan small-ear pig assembly (table 1). We annotated 21,463 protein-coding genes in the Diannan small-ear pig assembly by integrating the results from the PacBio ISO-seq data of transcripts of six tissues from the same individual as well as sequence homology-based and ab initio predictions (table 1; supplementary figs. S1 and S2, Supplementary Material online), of which 21,090 (98.26%) were represented in seven known protein annotation databases (supplementary tables S3 and S8, Supplementary Material online). We also annotated its unplaced scaffolds and found many of the genes related to olfactory transduction and pathogen infection (supplementary table S8, Supplementary Material online). We further identified a total of 1,111.16 Mb of repetitive elements, accounting for 41.95% of the total genome of Diannan small-ear pig (table 1; supplementary fig. S7, tables S6 and S7, Supplementary Material online).
Phylogeny, Divergence, and Demographic History
Based on orthologous genes identified from the ten-genome alignment (supplementary table S9, Supplementary Material online), the species phylogenetic trees were reconstructed with a concatenated 4D-site matrix (four-fold degenerate sites, fig. 1B) and a coalescent method (constructed from gene trees with ASTRAL-III, supplementary fig. S8AandB, Supplementary Material online). The phylogeny clearly revealed that common warthog and red river hog shared a more recent common ancestor than with Eurasian pigs. Additionally, we reconstructed the phylogeny for the family Suidae using supermatrix and supertree approaches (Supplementary material online) based on the newly and previously re-sequenced genomes of 42 wild samples representing ten extant Suidae species from five genera (including ten common warthogs and two red river hogs newly sequenced in this study as well as previously released for other wild suids, supplementary tables S10 and S11, Supplementary Material online). Both methods revealed a similar main topology that supported a basal divergence of African suids within the family Suidae (supplementary figs. S9 and S10, Supplementary Material online).
A molecular clock analysis using the CDS matrix of orthologous genes from the ten-genome alignment indicated a 6.9 Mya (95% highest posterior density, HPD = 9.9–4.1) divergence between African suids and Eurasian pigs as well as a divergence at ∼4.2 Mya (95% HPD = 6.1–2.5) between common warthog and red river hog (supplementaryfig. 1B, Supplementary material online). To check the robustness of divergence estimates within the family Suidae, we also incorporated additional re-sequenced Suidae samples to date the nodes in the phylogeny (supplementary tables S10 and S11, Supplementary Material online). The results showed that African suids diverged from their Eurasian relatives (except for Babyrousa) at ∼9.8 Mya (95% HPD = 12.9–6.6) while the split between the common warthog and red river hog lineages occurred at ∼5.5 Mya (95% HPD = 7.8–3.4, supplementary fig. S11, Supplementary Material online).
We employed a pairwise sequential Markovian coalescent (PSMC) model (Li and Durbin 2011) to examine the dynamics in effective population size (Ne) of the ancestral populations for common warthog, red river hog, and Diannan small-ear pig, in combination with one European (SAMN02904855) and one southern East Asian (SAMN02298082) wild boars for comparison (fig. 1C and supplementary table S10, Supplementary Material online). Common warthog exhibited a larger Ne than red river hog throughout the history (from ∼1 Mya to ∼0.01 Mya, fig. 1C), consistent with its greater habitat range (fig. 1A). Further, a decline in Ne of red river hog and Diannan small-ear pig from start of the Late Pleistocene coincided with the Last Glacial period (c. 115,000 to c. 11,700 years ago), while the Ne of common warthog still increased. Diannan small-ear pig and the southern East Asian wild boar showed a similar Ne trend during most of their histories in line with the close geographical origins. All suids showed a population decline before/during the Last Glacial Maximum (LGM, c. 20,000 years ago) (Clark et al. 2009), but the Ne of common warthog, Diannan small-ear pig, and the European wild boar reached a minor peak shortly after LGM in a contrast to red river hog and the southern East Asian wild boar (fig. 1C).
Selective Genes in Adaptive Evolution of African Suids
To assess selective pressures on the African suid genomes, we first estimated dN/dS ratios (ω) of a total of 8,726 orthologous genes characterized from the ten-genome alignment for each animal (supplementary table S9 and fig. S12, Supplementary Material online). We then identified 406 and 452 rapidly evolving genes (REGs) in common warthog and red river hog, respectively, as well as 250 REGs in their ancestral lineage, of which 333, 384, and 192 were lineage-specific REGs, namely not detected in other suid lineages (including the Diannan small-ear and Duroc pigs) (supplementary tables S12–S17, Supplementary Material online). Likewise, we observed 85 and 80 positively selected genes (PSGs) in common warthog and red river hog, respectively as well as 56 PSGs in their ancestral lineage, with 79, 73, and 54 to be lineage-specific PSGs in the African suids (supplementary tables S18–S23, Supplementary Material online). Some overlaps were observed between their respective lineage-specific REGs and PSGs (17, 21, and 10 for common warthog, red river hog, and their ancestral lineage; respectively, supplementary tables S12–S14, tables S18–S20, Supplementary Material online).
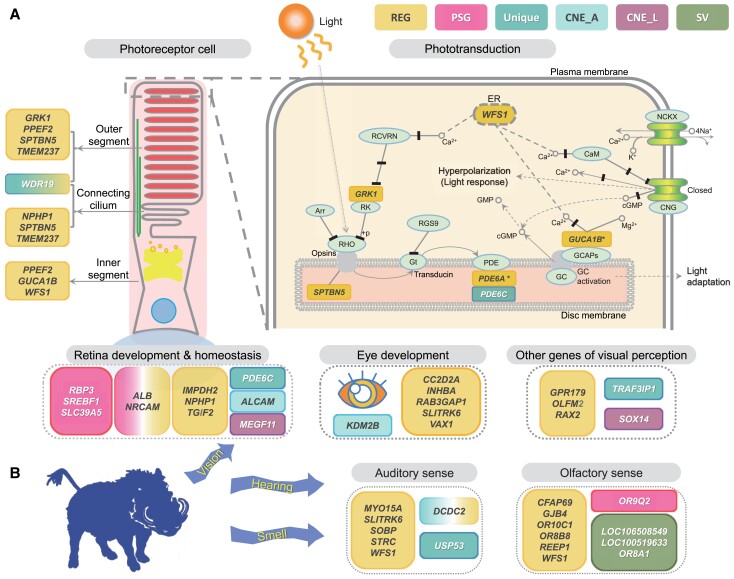
With the aid of functional enrichment by KOBAS-i (Bu et al. 2021) for the lineage-specific REGs and PSGs separately, we found that common warthog showed selective signals on sensory perception (including vision, olfaction, and audition) (fig. 2AandB), immune system, metabolism, and central nervous system (supplementary tables S24 and S25, Supplementary Material online), while the red river hog had private REGs and PSGs primarily related to immune system, viral infection, metabolism, and nervous system development (supplementary tables S26 and S27, Supplementary Material online). In-depth inspection revealed that 19 REGs of common warthog were related to several aspects of visual perception, including retina development (ALB, NRCAM, NPHP1, TGIF2, and IMPDH2), photoreceptor maintenance (GPR179, GRK1, NPHP1, OLFM2, PPEF2, RAX2, SPTBN5, TMEM237, WDR19, and WFS1), and eye development (CC2D2A, INHBA, RAB3GAP1, SLITRK6, and VAX1), along with four of its PSGs related to retina and eye development (ALB, NRCAM, RBP3, and SLC39A5) (fig. 2A). Other aspects of sensory perception were also represented by REGs and PSGs of common warthog, including audition and ear development (DCDC2, MYO15A, SLITRK6, SOBP, STRC, and WFS1) as well as olfaction (CFAP69, GJB4, OR10C1, OR8B8, REEP1, WFS1, and OR9Q2) (fig. 2B). Although a total of 12 sensory REGs and PSGs were also identified in red river hog, but statistically more of the sensory genes were detected in common warthog (31 REGs and PSGs in total; P = 7.2×10−4, Fisher's exact test). When the REGs and PSGs were considered separately, the REGs relevant to sensory perception were still enriched in common warthog (P = 2.6×10−4; supplementary table S28, Supplementary Material online), suggesting a stronger adaptation in sensory perception in common warthog.
Fig. 2.
Genes involved in adaptation of sensory perception in common warthog. (A) Genetic changes specific to common warthog related to multiple aspects of the visual sense. Graph of the phototransduction pathway (adapted from KEGG:04744) shows the response in the photoreceptor outer segment when light stimulus comes in, in which SPTBN5 binding opsins and WFS1 regulating calcium balance on endoplasmic reticulum (ER) of photoreceptor are added onto the graph. One of the noteworthy sensory REGs, WFS1, encodes a protein wolframin, of which many mutations have been reported to cause a Wolfram syndrome in humans leading to defects in vision, hearing, and olfaction (Rigoli et al. 2011). Other genes related to the function of different sub-structures of photoreceptor cell, retina development, and homeostasis, eye development, and visual perception are also listed. Asterisk marks (*) indicate genes also detected as REG in red river hog. (B) Genes showing evolving signals unique in common warthog and associated with the senses of hearing and smell. Genes identified with genetic variations or under selection related to sensory systems for common warthog are in italics and listed inside round rectangles: rapidly evolving genes (REG), positively selected genes (PSG), genes with unique amino acid substitutions (Unique), genes with structural variations (SV), genes with accelerated conserved noncoding elements (CNE_A), and genes with CNE lost (CNE_L).
Many of the species-specific REGs and PSGs were found to be associated with multiple immune processes in the two African suids. The most significantly enriched immune categories indicated that they both had REGs/PSGs related to thymus development and T-cell differentiation (e.g., PRR7, RAG1, RAG2, and ZFP36L2 in common warthog together with ABL1 and BCL11B in red river hog) (supplementary tables S24–S27, Supplementary Material online). A discrepancy between their respective immune genes under selection were observed as well, represented by the detection of more REGs/PSGs relevant to viral infection in red river hog (P = 0.010 for REGs and PSGs combined and P = 0.044 for REGs only, Fisher's exact test; supplementary table S28, Supplementary Material online). These evolving viral infection-related genes in red river hog were primarily associated with cytokine production (e.g., HERC5, ISG15, TREX1, TRIM21, and YTHDF3) and defense response to virus (e.g., AP1M2, FDPS, HERC5, HSPG2, IPO7, ISG15, NOS2, and POLR3C) (fig. 3A–C and supplementary tables S26 and S27, Supplementary Material online). In addition, in the ancestral lineage of African suids, we found some of the REGs and PSGs were relevant to RNA transport and nuclear pore complex assembly (NDC1, NUP93, and SMG1), which also participated in the viral transcription, transport, and infection processes, together with four genes (BCL3, DFFA, NKX2-3, and SPEN) involved in lymphoid organ development (e.g., spleen development; supplementary tables S29 and S30, Supplementary Material online).
Fig. 3.
Genes associated with modifications in interferon signaling pathways of red river hog. (A) Genes with red river hog-specific variations involved in the type I/II interferon (IFN) signaling in antiviral responses. Genes listed inside round rectangles in italics are IFN-stimulated genes (ISGs). Notably among them, ISG15, which is identified as both REG and PSG in red river hog, functions as a crucial role in type I/II interferon (IFN) signaling pathways and responses to viral infection (Perng and Lenschow 2018). ISG15 activates ISGylation, a reversible process with ubiquitylation aided by HERC5 and mediated by nitrosylation of NOS2 (Sadler and Williams 2008; MacMicking 2012; Perng and Lenschow 2018), to modify protein substrates in the IFN pathways and to regulate immune responses. This further suggests the noticeable antiviral adaptation of red river hog. (B) Mechanisms of TRIM21 and POLR3C to trigger intracellular viral nucleic acid sensing by cytosolic pathogen-recognition receptors RIG-I and cGAS and subsequent transcription of antiviral IFNs and cytokines. (C) Mechanisms of inhibition of viral RNA transcription and protein translation by NOS2 as well as inhibition of viral protein stability by ISGylation of ISG15 with the aid of HERC5. Genes identified with genetic variations or evolving signals related to type I/II IFN signaling pathways for red river hog are in italics and listed inside round rectangles: rapidly evolving genes (REG), positively selected genes (PSG), genes with unique amino acid substitutions (Unique). The dashed and solid lines with arrow/bar at the ends between molecules indicate molecular regulation (activate/inhibit).
Further permutation tests revealed that REGs exclusive to common warthog were enriched in sensory perception (including vision, audition, and olfaction) (P < 0.05) and PSGs in vision (P < 0.05), while REGs exclusive to red river hog were enriched in viral process and Type I IFN pathway (P < 0.05) together with PSGs in Type II/III IFN, lymph, and RIG-I-like receptor pathways (P < 0.05). The ancestral lineage of African suids had more-than-expected unique REGs and PSGs in lymphoid development (P = 0.0029 and P = 0.0159, respectively; supplementary table S31, Supplementary Material online). Besides, we found that sensory genes (especially for vision and olfaction) in common warthog showed much higher dN/dS than the background lineages (P < 0.05, Student's t-test; supplementary fig. S13, Supplementary Material online) and elevated dN/dS compared with the whole-genomic level (P < 0.01, Wilcoxon rank-sum test; supplementary table S32, Supplementary Material online). Viral process/Type I IFN-related genes and viral process/lymph genes showed similarly elevated trends of dN/dS for the red river hog and African ancestral lineages, respectively (supplementary fig. S14 and table S32, Supplementary Material online).
Lineage-Specific Substitutions in Adaptive Genes of African Suids
Given that nonsynonymous mutations have played important roles in phenotype variation and adaptive evolution (Eyre-Walker 2006; Andolfatto 2007; Halligan et al. 2010), we investigated the lineage-specific amino acid substitutions in African suids by applying a multiple-genome alignment to the two African suids (i.e., common warthog and red river hog) and the two Eurasian pigs (i.e., Diannan small-ear and Duroc pigs) (Supplementary material). A total of 39,441 and 38,315 resulted in amino acid substitutions specific in common warthog and red river hog, respectively, while their ancestral lineage had 60,266 such nonsynonymous mutations in comparison with the Eurasian pigs (supplementary table S33, Supplementary Material online). We further used FST ≥ 0.9 calculated from a re-sequencing genome dataset of 42 suid individuals (supplementary table S10, Supplementary Material online) to validate the lineage-specific nonsynonymous mutations highly differentiated between each of the African lineages and the other suids in the dataset. We then employed SIFT (Kumar et al. 2009) to identify deleterious variants and pCADD (Groß et al. 2020) with scores ≥30 to evaluate the most functionally significant substitutions, leading to an identification of 47, 60, and 46 nonsynonymous mutations specific to common warthog, red river hog, and their ancestral lineages, affecting 42, 56, and 41 protein-coding genes, respectively (supplementary tables S34–S36, Supplementary Material online). With a functional enrichment by KOBAS-i, we found that some of the most significantly enriched signals in these genes were mainly related to eye development (PDE6C, TRAF3IP1, and WDR19) in common warthog, immune responses in red river hog (e.g., AXL, CXCR6, HDAC4, HDAC7, HNMT, IKZF3), and viral entry in their ancestral lineage (CDHR3 and NECTIN1) (supplementary tables S37–S39, Supplementary Material online).
Conserved Noncoding Elements (CNEs) in African Suids
In view of the functional importance of CNEs as cis-regulatory elements and their association with transcription factor binding sites (TFBS), we analyzed CNEs in the genomes of common warthog and red river hog. Following the pipeline in Chen et al. (2019), we characterized 72, 75, and 45 lineage-specific accelerated CNEs with phyloP (Pollard et al. 2010) in common warthog, red river hog, and their ancestral lineage, respectively, with an overall mean length of ∼284 bp (supplementary tables S40–S42, Supplementary Material online), which all intersected with recognizing motifs for TFBS and highly differentiated lineage-specific SNVs (Supplementary material). After the equivalent analysis for the Eurasian pigs (supplementary table S43, Supplementary Material online), we extracted 94, 100, and 52 genes associated with these accelerated CNEs specific in the African suid lineages accordingly (supplementary tables S40–S42, Supplementary Material online). Additionally, we found that both common warthog and red river hog had lost seven CNEs, which were validated by re-sequencing data (Supplementary material and supplementary table S44, Supplementary Material online).
We found that the functional enrichment (P < 0.05) of the lineage-specific genes associated with accelerated CNEs in common warthog included nervous activities and development, immunity, body growth, and development, etc., in which anterior/posterior pattern specification was nearly most significantly enriched by four genes (HOXC9, HOXC10, MLLT3, and MSX2, supplementary table S45, Supplementary Material online). While in red river hog, similar functions were also enriched, especially by around a dozen of genes (ACVR2A, ALX1, CCN1, FBN1, GAB2, HOXD12, HOXD13, PKDCC, RGMB, SBNO2, SMAD3, STAG1, and THBS3) involved in body growth such as skeletal system development and anatomical structure morphogenesis (supplementary table S46, Supplementary Material online). In the ancestral lineage, pathways relevant to developmental processes such as male gonad development were enriched (supplementary table S47, Supplementary Material online). It was noted that two genes of common warthog relevant to visual perception and nervous system development (MEGF11 and SOX14) lost the associated CNEs (supplementary table S44, Supplementary Material online). In red river hog, one CNE was lost in an intergenic region (between the genes ENSSSCG00000049953 and ENSSSCG00000051043, supplementary table S44, Supplementary Material online), a locus reported to be associated with the immune response to a swine pneumonia etiologic agent Mycoplasma hyopneumoniae (Raaphorst 2020).
Structural Variations in African Suids
We integrated smartie-sv (Kronenberg et al. 2018) and LAST v982 (Kielbasa et al. 2011) to screen structural variations (SVs, ≥50 bp) in the newly assembled genomes based on the alignment with the Duroc reference genome (Sscrofa11.1) (supplementary table S48, Supplementary Material online). We focused on the non-repetitive SVs (supplementary table S49, Supplementary Material online) that intersected with protein-coding regions to evaluate their loss-of-function effects in the African suids. The non-repetitive SVs included 2635, 2512, and 1581 insertions along with 2669, 2916, and 1060 deletions unique in each of the common warthog, red river hog, and their common ancestral lineage, respectively, of which 0.048–0.283% were found to have open reading frames disrupted or shifted (supplementary table S50, Supplementary Material online) and thus interrupt the coding sequences (CDS) of 11, 20, and 11 genes (supplementary table S51, Supplementary Material online). In common warthog, these SVs produced effects primarily on the CDSs of genes related to olfactory receptors (LOC106508549, LOC100519633, and ENSSSCG00000015193) and pathogen defense (IFI44L, NBR1, RAMP1, and TIMD4). The protein-coding genes influenced by the SVs in red river hog were associated with different categories, such as carbohydrate metabolism (PFKFB4 and UGT2A3), nervous system development (DCLK2), and especially, immune processes (including antiviral activity with LTN1, USP24, and ZNF550, chemokine activity with ENSSSCG00000037962, lymphocyte proliferation with RXRA, and a tumor suppressor PDGFRL) (supplementary table S51, Supplementary Material online). In the ancestral lineage of African suids, we found a remarkable gene TRBV27 with a 284-bp deletion causing frameshift and spanning its two coding regions (fig. 4A), which encodes a partial T-cell receptor (TCR) beta chain. Additional whole-genome re-sequencing data (supplementary tables S10, Supplementary Material online) confirmed the fixation of this deletion in the two African suids (fig. 4A). The complementarity-determining region 1 domain and the adjacent conserved motif of TRBV27 were lost due to this deletion (fig. 4B), possibly altering the antigen presenting process between the associated TCRs and antigen presenting cells in both common warthog and red river hog (fig. 4CandD) (Turner et al. 2006). The recombination signal sequences recognized by RAG1/RAG2 for VDJ somatic recombination (Oettinger et al. 1990) remained intact in the common warthog and red river hog genomes (supplementary fig. S15, Supplementary Material online), implying that VDJ recombination of TRBV27 was still functional during T-cell differentiation. In addition, two identical 11-bp motifs (5′-GGACCCAGCCC-3′) in the same orientation (supplementary fig. S15, Supplementary Material online) might have mediated the deletion of this in-between 284-bp sequence by non-allelic homologous recombination (Parks et al. 2015).
Fig. 4.
The 284-bp deletion in TRBV27 and the binding model proposed for TCR and MHC in the African suids. (A) Schematic diagram of the canonical gene structure of TRBV27 in the Duroc pig reference genome and the structural variation revealed by the sequencing depth of SNPs. The 284-bp deletion in TRBV27 is indicated between the vertical dashed lines. The critical binding domains (CDR1 and CDR2) and RSS of TRBV27 are indicated on the gene structure diagram. The deletion was validated by the SNP depth calculated based on the genomic re-sequencing data of 320 suid individuals (including 61 common warthogs and six desert warthogs of Phacochoerus, four red river hogs and one bushpig of Potamochoerus, 20 suid individuals of South and Southeast Asia, 140 East Asian domestic pigs, 20 East Asian wild boars, 61 European domestic pigs, and seven European wild boars), in which ten common warthogs and two red river hogs were newly generated in this study (supplementary table S10, Supplementary Material online). (B) Multiple-species alignment of the partial TRBV27 protein with the deleted amino acids denoted in dashes “-”. The motifs of CDR1 and CDR2 are indicated by horizontal bars. (C, D) The proposed normal and mutant models for MHC and TRBV27-used TCR binding in Eurasian pigs and African wild suids, respectively. MHC: major histocompatibility complex; TCR: T-cell receptor; CDR1: complementarity-determining region 1; CDR2: complementarity-determining region 2; CDR3: complementarity-determining region 3; RSS: recombination signal sequence; α, β, γ, δ, ε, and ζ are TCR sub-units.
Discussion
In this study, we sequenced, assembled, and annotated genomes of common warthog, red river hog, and East Asian Diannan small-ear pig. The three new genomes were primarily generated using long-read sequencing technologies and exhibited a high level of completeness (table 1 and supplementary table S3, Supplementary Material online). The Diannan small-ear pig genome assembly filled gaps and improved the completeness of the current pig reference genome (Sscrofa11.1). The African suids genome assemblies, together with re-sequenced genomic data newly generated in this study, provide valuable resources for various research on the evolution of the family Suidae as well as porcine genomics.
Based on the genomic data, we depicted the phylogenomic context for studying the evolution of Suidae species (fig. 1B and supplementary fig. S8–S11, Supplementary Material online). The phylogenetic framework corroborate previous results based on a limited number of mitochondrial DNA and nuclear markers (Wu et al. 2006; Gongora et al. 2011) as well as the re-sequencing data (Frantz et al. 2013; Liu et al. 2019), suggesting that the de novo assemblies and reconstructed phylogeny are robust for evolutionary and comparative genomics analyses. The emergence of the most recent common ancestor (MRCA) of extant sub-Saharan African suids (except for Babyrousa), compared with the Eurasian suids, was estimated in this study at approximately 9.8–6.9 Mya, which coincided with the rapid expansion of Suinae during the Late Miocene (∼11.6–5.3 Mya) in Eurasia (Gongora et al. 2011; Frantz et al. 2016; Liu et al. 2019). Interestingly, our estimates of the divergence between the two major lineages of extant African suids (∼5.5–4.2 Mya) roughly coincided with the first Suinae fossil in Africa around the Miocene/Pliocene boundary at ∼5.5 Mya (Brunet and MPFT 2000; Brunet and White 2001). The ancestors of the major African Suinae lineages may have appeared in Africa at different times from fossil records, in which Kolpochoerus and Metridiochoerus, representing the ancestral Hylochoerus and Phacochoerus, respectively, occurred in Africa during the Early Pliocene whereas the earliest African representative of Potamochoerus was absent at that time (Cooke 1978; Harris and White 1979; Pickford 2006, 2012). Instead, the Potamochoerus fossils were found from the Early Pliocene in India and the Late Pliocene in Spain, implying its origin in eastern Eurasia followed by westward dispersal conceivably to Europe and finally to Africa in the Early Pleistocene (Harris and White 1979; Arribas and Garrido 2008; Pickford 2012; Kumar and Gaur 2013). Although our current results cannot completely rule out the possibility of their derivations from an unknown common ancestor rooted in Africa and the following migrations back to Eurasia (Pickford 2006, 2012; Frantz et al. 2016; Gongora et al. 2017; Liu et al. 2019), our divergence time estimates, together with current paleontological insights, tend to provide an inference about the split of the red river hog and common warthog lineages outside Africa.
The demographic history (PSMC) suggested a much larger population size (Ne) of common warthog than that of red river hog, especially during the Late Pleistocene (fig. 1C) which coincided with the Last Glacial period (c. 115,000 to c. 11,700 years ago). This may reflect the vast and diverse habitats of common warthog, including sub-Saharan savannah, woodland, and even semi-desert (fig. 1A) (Butynski and de Jong 2017), in comparison with the relatively restricted and fragmented range of red river hog primarily confined to the tropical rainforest belt along the western and central Africa (fig. 1A) (Leslie and Huffman 2015; Melletti et al. 2017). The Last Glaciation has led to a similar trend in Ne decline for almost all suids, apart from common warthog (fig. 1C). This corresponds to the expansion of western populations of common warthog into eastern and then southern Africa from the start of the Late Pleistocene (Garcia-Erill et al. 2022). During the Last Glaciation, a cold arid climate prevailed with the fragmented forests as well as the expanded savannah-grassland areas in the tropical African belt (Hamilton and Taylor 1991), whereas vegetation (shrubs and grassland) in the LGM covered markedly more areas on the land than today in the Sahara (Shao et al. 2018). Altogether, the pronounced fluctuation such as the increase of common warthog as well as the decline of red river hog in Ne could be attributed to a putative expansion of the savannah-like habitats in contrast with a contracted tropical forest during the time. Although all suids have their populations shrunk before or during the LGM, but the rapid recovery of common warthog shortly after that could benefit from its large range (fig. 1AandC). This emphasizes the diverging evolutionary trajectories experienced by common warthog and red river hog after the entry of their ancestors into Africa. It is therefore expected that the deep divergence time and distinct demographic histories of common warthog and red river hog (fig. 1BandC; supplementary fig. S11, Supplementary Material online), together with the different habitats, would have left distinguishing adaptive signatures in their genomes in the face of diverse survival requirements.
The comparative genomics analyses of the African suid genomes in this study provide new insights into the genetic basis of local adaptations to diverse environments on the African continent. Our analyses suggest that common warthog, well adapted to the savannah conditions, may have evolved adaptive signals in several aspects of sensory perception including vision, audition, and olfaction (fig. 2AandB, supplementary fig. S13, tables S31 and S32, Supplementary Material online). The evolution of visual perception in common warthog is mainly reflected by many of the evolving genes, which are associated with some of the specific cells and tissues in the eyes, such as photoreceptor and retina (fig. 2A). Some of these visual genes play crucial roles in the phototransduction, maintenance of photoreceptor sub-structures (fig. 2A), contributing to the detection and conversion of light stimulus in the first step of vision (Molday and Moritz 2015). Other visual genes participate in the development and homeostasis of retina and eye (fig. 2A), maintaining the normal functional structures in eyes. The extra two visual genes (MEGF11 and SOX14) of common warthog have lost their associated CNEs (supplementary table S44, Supplementary Material online), of which MEGF11 contributes to the normal arrangement of retinal layer (Kay et al. 2012), and SOX14 is associated with two syndromes of abnormal craniofacial development possibly causing vision and hearing problems (Arsic et al. 1998) and circadian mediation in the subcortical visual shell (Delogu et al. 2012; LeGates et al. 2014). This hints at a contribution of regulatory elements to the adaptive change in the vision of common warthog. Notably, one of the noteworthy REGs in common warthog, WFS1, regulates calcium flux in the endoplasmic reticulum and is therefore important for the intracellular calcium homeostasis, such as in the hyperpolarization of photoreceptors in response to photon stimuli (fig. 2A) (Rigoli et al. 2011). Many mutations in WFS1 have been reported to be of potential relevance to a human disease known as Wolfram syndrome with serious defects in vision, audition, and olfaction (Rigoli et al. 2011). As a typical example, it has thus provided the insight that along with the visual perception, the detection of adaptive genes and variants in the hearing and smell of common warthog (fig. 2B) highlighted its significant co-adaptations in multiple senses for the savannah lifestyle, which is mainly characterized by open areas with risks posed by a range of various carnivores (fig. 1A) (Butynski and de Jong 2017). Adaptive evolution in multiple sensory systems and some of the relevant genes has been identified in other African savannah fauna, such as giraffe with a related suite of sensory co-adaptations in vision, olfaction, hearing, and circadian rhythm (Liu et al. 2021). Although it has been proposed that common warthog might not have excellent vision, its senses of hearing and smell are indeed acute, by which it could flee rapidly in collaboration with other species, such as in response to warning calls from African green monkeys, when facing predators in open areas (Butynski and de Jong 2017). All these suggest an adaptive sensory evolution in common warthog that may also be shared by many African savannah animals.
Both the red river hog and common warthog have shown substantial species-specific evolving signals on genes relevant to the immune system, which agrees with the knowledge that immune genes often evolve rapidly in the genome due to their involvement in dynamic co-evolutions with pathogens (Jiggins and Kim 2007; Nielsen et al. 2007; Obbard et al. 2009). Especially, red river hog has significantly more viral infection-associated genes showing detectable adaptive evolutionary signatures, particularly in REGs and PSGs (supplementary fig. S14, tables S26, S27, S31, and S32, Supplementary Material online), suggesting its prominent antiviral adaptation. Among the antiviral responses, IFN signaling pathways (especially in Type I IFN) are strongly represented by evolving genes identified in red river hog (fig. 3A–C; supplementary fig. S14, Supplementary Material online). Many of them are IFN-stimulated genes (ISGs) or involved in the IFN pathways in response mainly to viral infection, among which a prominent antiviral effector, ISG15, has been well characterized. ISG15 acts as an early essential effector in regulation of antiviral defense by mobilizing a type I IFN response and inducing IFN-γ secretion (fig. 3A), which can activate a ubiquitin-like protein response called ISGylation by targeting many important substrates in the IFN responses, including pattern-recognition receptor RIG-I (fig. 3B) and cytosolic viral components (fig. 3C) (Sadler and Williams 2008; MacMicking 2012; Perng and Lenschow 2018). The central role of ISG15 in host responses to viral infection is further highlighted by the involvement of three REGs encoding E3 ubiquitin ligase (HERC5 and TRIM21) in the regulation of type I IFN and inflammatory responses together with an IFN-induced nitric oxide synthase (NOS2) in virus restriction, of which HERC5 and NOS2 are crucial for ISG15's ISGylation (fig. 3A–C) (Sadler and Williams 2008; MacMicking 2012; Perng and Lenschow 2018) while TRIM21 is important for sensing RNA and DNA viruses via the interactions with cytosolic nucleic acid sensors RIG-I and cGAS, respectively (fig. 3B) (Watkinson et al. 2015). Furthermore, two REGs sensing cytosolic DNA (TREX1 and POLR3C) and one gene with unique amino acid mutations (AXL) as a membrane receptor can regulate type I IFN response by induction or suppression (fig. 3AandB) (Stetson et al. 2008; Bhattacharyya et al. 2013; Cai et al. 2021). The IFN signaling pathways are at frontlines in combat with viral invasion to initiate signaling cascades and subsequently to induce hundreds of cytokines, functioning as crucial antiviral responses (Sadler and Williams 2008; Perng and Lenschow 2018). Therefore, the adaptive evolution in this respect is of particular importance to confer antiviral activities on red river hog. The red river hog is naturally confined to the tropical rainforest belt along the west and central Africa (fig. 1A), which is primarily featured with a constant damp and dense vegetation canopy (Leslie and Huffman 2015; Melletti et al. 2017). Quite a few studies on the selective history or local adaptation of mammals distributed in the African tropical rainforests, such as chimpanzees (Leffler et al. 2013; Schmidt et al. 2019), gorillas (McManus et al. 2015), and guenons (Ayoola et al. 2021), and even humans (Lopez et al. 2019), have revealed remarkable adaptive signals on host–pathogen interactions such as virus defense and parasite resistance. The comparable adaptive patterns among red river hog and other rainforest animals in immune responses to pathogens, especially in antiviral activity, may indicate that nowhere is a greater adaptive challenge from miscellaneous pathogens than to the rainforest residents in the tropical Africa (Fan et al. 2016).
As the natural mammalian hosts of African swine fever virus (ASFV), common warthog, and red river hog exhibit tolerance/resistance to ASFV infection (Jori and Bastos 2009; Butynski and de Jong 2017; Melletti et al. 2017). Our screening of CD163 and RELA, namely, two proposed candidate genes involved in tolerance to ASFV (Sánchez-Torres et al. 2003; Palgrave et al. 2011), found no specific evolving signals in the genomes of the two African suids. Otherwise, our results reveal that common warthog and red river hog share certain common genetic features. In their ancestral lineage, genes with evolving signals relevant to lymphoid organ development as well as viral RNA transcription, transport, and infection are identified from our comparative genomics analyses. Especially, the structural variations shared in the genomes of the African suids may have occurred in their ancestral lineage after divergence from the ancestor of Eurasian pigs (fig. 1B and supplementary fig. S8–S11, Supplementary Material online). The fixation of structural variation in the T-cell receptor gene (TRBV27, fig. 4A) that was likely under some selective pressure, together with many different evolving signals in the immune system, suggested underlying mechanisms in the extant African suids in adaptation to various pathogens across the diverse sub-Saharan African environments (Darfour-Oduro et al. 2015). Based on these genomic resources, the host–pathogen coevolution in African suids will be eventually revealed by leveraging multiple-omic approaches (Forth et al. 2020; Hu et al. 2021), together with functional assays and animal models (Popescu et al. 2017; McCleary et al. 2020).
Materials and Methods
Sample Collection and Genome Sequencing
All experiments and sample collection were approved by the Internal Review Board of Kunming Institute of Zoology, Chinese Academy of Sciences (SMKX-20191213-03). The sampling in Nigeria was approved by Nigeria National Park Service (NPH/GEN/121/XXV/368). The genomic DNA isolated from blood samples of a female red river hog (sample accession: SAMC146518) and a male common warthog (SAMC146517) from Gashaka-Gumti National Park, Nigeria, were subjected to fragmentation and adapter ligation to construct libraries, and they were long-read de novo sequenced on Oxford Nanopore PromethION. The blood samples of a male Diannan small-ear pig (SAMC146519) from the farm of Kunming Institute of Zoology were subjected to genomic DNA isolation, and then a PacBio SMRT library with a length of 20–25 Kb was de novo sequenced on PacBio Sequel. Additionally, the samples of a male red river hog (SAMC146529) from Cross River National Park, Nigeria, and further nine common warthogs (SAMC146520–SAMC146528) from the central Kenyan highlands supplied by The International Livestock Research Institute (ILRI), Kenya, in collaboration with The Kenya Wildlife Service (KWS) were further collected for 150-bp paired-end re-sequencing on Illumina NovaSeq 6000 with libraries of 350-bp insert size.
De Novo Genome Assembly
The raw Nanopore reads of the common warthog and red river hog were assembled with Wtdbg2 v2.3 (Ruan and Li 2020). The assemblies were polished by the Nanopore long reads with Racon v1.3.1 (Vaser et al. 2017) and Illumina short reads with Pilon v1.22 (Walker et al. 2014) of the same individuals. The resultant contigs were mapped to the pig reference (Sscrofa11.1) for scaffolding and constructing pseudo-chromosomes. The contigs of the Diannan small-ear pig were assembled with FALCON v1.8.7 (Chin et al. 2016) and then error-corrected with Quiver v2.3.2 (Chin et al. 2013) based on the PacBio Sequel long reads. Contigs were error-corrected and anchored into scaffolds with a physical map produced by Bionano system. Scaffolds were gap-filled by Blasr v4.0 (Chaisson and Tesler 2012) and polished by PBJelly v15.8.24 (English et al. 2012) and Pilon v1.22 with the PacBio long reads and Illumina short reads of the same individual. Hi-C was employed to assist in constructing a chromosome-scale assembly of Diannan small-ear pig. BUSCO v3.0.2 (Simão et al. 2015) was used to evaluate the completeness of the three new assembled genomes with library “mammalia_odb9”.
Genome Annotation
Repeat elements were annotated by combining RepeatModeler v1.0.11 (Smit et al. 2008), RepeatMasker v4.0.7 (Smit et al. 2015), RepeatProteinMask v1.23 (Tarailo-Graovac and Chen 2009), RepeatScout v1.0.5 (Price et al. 2005), LTR_FINDER v1.0.7 (Xu and Wang 2007), and Tandem Repeat Finder v4.07b (Benson 1999). Genes were annotated by homology-based prediction with the latest references of pig, human, mouse, cow, horse, goat, and dog (access date: March, 2019) in combination with de novo predictions of Augustus v3.3.2 (Stanke et al. 2006), GlimmerHMM v3.0.3 (Majoros et al. 2004), SNAP (Korf 2004), GeneID (Blanco and Abril 2009), and GenScan v1.0 (Burge and Karlin 1997). Gene functions were predicted via GO (Ashburner et al. 2000), InterPro (Mitchell et al. 2018), KEGG (Kanehisa et al. 2017), Nr (Yu and Zhang 2013), Pfam (El-Gebali et al. 2018), Swiss-Prot (O’Donovan et al. 2002), and TrEMBL (O’Donovan et al. 2002) databases (access date: March, 2019).
Phylogeny, Molecular Clock, and Demographic History Analyses
Orthologous genes identified from the ten-genome alignment (common warthog, red river hog, Diannan small-ear pig, and Duroc pig (Warr et al. 2020), taurine cattle (Rosen et al. 2020), goat (Bickhart et al. 2017), reindeer (Li et al. 2017), Arabian camel (Elbers et al. 2019), sperm whale (Fan et al. 2019), and horse (Kalbfleisch et al. 2018)) with LAST v982 (Kielbasa et al. 2011) and Multiz v11.2 (Blanchette et al. 2004), and their CDSs were used to infer ML gene trees with RAxML v8.2.12 (Stamatakis 2006, 2014) under GTR+Γ+I model determined by jModelTest v2.1.10 (Guindon and Gascuel 2003; Darriba et al. 2012). A coalescent species tree was constructed with ASTRAL-III v5.6.3 (Zhang et al. 2018) from the gene trees. A concatenated four-fold degenerate (4D) site matrix from the CDSs was also used to infer ML species tree. In addition, the CDSs of autosomal pig-to-cow orthologous genes extracted from the re-sequencing data of 42 Suidae individuals were concatenated to infer a Suidae phylogeny with the ML approach. They were also used for constructing individual ML gene trees, from which a coalescent consensus tree was produced. The concatenated CDSs from the above ten-genome alignment and re-sequencing data were separately used to estimate divergence times with MCMCTree of PAML v4.9j (Yang 2007). A GTR model and a strict clock (clock = 1) were used for mutation rate estimation first. The fossil calibration was detailed in Supplementary material. Demographic history inference was performed for the three newly assembled genomes in combination with a European and an East Asian wild boars for comparison with PSMC v0.6.5-r67 (Li and Durbin 2011), and the results were scaled to real time and population size with different suid generation times (g) from Pacifici et al. (2013) and the sanme mutation rate (μ = 2.5 × 10−8) as in Groenen et al. (2012) and Frantz et al. (2013).
Detection of Rapidly Evolving and Positively Selected Genes
The dN/dS ratios of the genes for each animal were first estimated with the free ratio model in Codeml of PAML v4.9i (Yang 2007). Genes putatively under selection were identified with the optimized branch and branch-site models to detect rapidly evolving genes (REGs) with an elevated dN/dS and positively selected genes (PSGs) with codons under positive selection (dN/dS > 1), respectively, in a pre-specified branch. Genes with CDS < 150 bp, premature stop codon/frameshift mutations, and dS > 2 were precluded. The likelihood ratio test (LRT) based on χ2 statistics (P < 0.05) was employed to define the significant REGs/PSGs.
Detection of Unique Amino Acid Substitutions
The lineage-specific single-nucleotide variations (SNVs) were identified from the genome alignment with LAST v982 and in-house programs against the pig reference (Sscrofa11.1). The resultant SNVs were annotated by SnpEff v4.3t (Cingolani et al. 2012) to recognize nonsynonymous mutations (i.e., amino acid substitutions), which were further evaluated for their differentiation between lineages by FST computed using SAMTools v1.10 (Li et al. 2009) with re-sequencing data and the potential functional impact with pCADD (Groß et al. 2020) and SIFT v5.2.2 (Kumar et al. 2009).
Detection of Structural Variations
Structural variations including deletions and insertions (≥50 bp) were identified by a default pipeline of smartie-sv (Kronenberg et al. 2018), cross-checked with LAST v982 genome alignment and in-house programs against the pig reference (Sscrofa11.1), and finally validated by re-sequencing reads mapping with BWA-MEM v0.7.17 (Li 2014) and SAMtools v1.10.
Conserved Noncoding Element Analysis
Several following tools implemented in PHAST v1.4 (Hubisz et al. 2011) and the pipeline in Chen et al. (2019) were utilized for CNEs identification. Based on the ten-genome alignment, a nonconserved model was estimated by phyloFit (Siepel and Haussler 2004) from the 4D sites. Then a conserved model was estimated by phastCons (Siepel et al. 2005) with the nonconserved model. The highly conserved elements predicted by phastCons that intersected with noncoding regions were identified as CNEs. The CNEs of accelerated evolution were defined as those with LRT P < 0.05 computed by phyloP (Pollard et al. 2010) for each lineage. The lost CNEs in the African suids were validated in the same way as in SV analysis.
Statistical Analysis
The statistical tests were indicated throughout the article wherever necessary. The one-tailed Fisher's exact test compared the differences in numbers of viral/sensory REGs/PSGs between common warthog and red river hog. The permutation test evaluated if a particular category of REGs/PSGs were observed in focal lineages more than the expected, by random sampling the equal number of genes to REGs/PSGs in each specified lineage 10,000 times without replacement and calculating the P value by the percentage of the sampling distribution higher than or equal to the observed gene number of a certain category (supplementary table S31, Supplementary Material online). The one-tailed Student's t-test was conducted to compare if dN/dS of a particular category of genes in specified foreground lineages were higher than those in background branches (supplementary figs. S13 and S14, Supplementary Material online). The one-tailed Wilcoxon rank-sum test was performed for comparison of dN/dS in specified lineages between a particular category of genes and the whole-genomic level (supplementary table S32, Supplementary Material online). Above statistical tests were performed in R v3.6.3 (R Core Team 2020).
More details about methods and settings are available in Supplementary material.
Supplementary Material
Acknowledgements
We thank the management of Nigerian National Park Service for the permits and provision of technical assistance for the field survey, particularly Gashaka-Gumti National Park, Taraba and Cross River National Park, Cross River in Nigeria. We also thank the Kenya Wildlife Services for providing warthog resources for this study. This work was supported by the African Swine Fever Research Emergency Program of the Chinese Academy of Sciences (KJZD-SW-L06), National Key Research and Development Program of China (2021YFF1000602), the Sino-Africa Joint Research Center, Chinese Academy of Sciences (SAJC202103), the Talents Team Construction Fund of Northwestern Polytechnical University (NWPU), the Southwest Research Center for Pig Molecular Breeding and Translational Medicine, National Natural Science Foundation of China (31750110480), the Spring City Plan: the High-level Talent Promotion and Training Project of Kunming (2022SCP001), and the Animal Branch of the Germplasm Bank of Wild Species, Chinese Academy of Sciences (the Large Research Infrastructure Funding). A.C.A. is supported by the Chinese Academy of Sciences President's International Fellowship Initiative (2018FYB0003 and 2021FYB0006). M.-S.P. is supported by the Yunnan Revitalization Talent Support Program. The Chinese Government contribution to CAAS-ILRI Joint Laboratory on Livestock and Forage Genetic Resources in Beijing is also appreciated. The paper contributes to the CGIAR Research Program on Livestock.
Contributor Information
Hai-Bing Xie, State Key Laboratory of Genetic Resources and Evolution & Yunnan Laboratory of Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650201, China.
Chen Yan, State Key Laboratory of Genetic Resources and Evolution & Yunnan Laboratory of Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650201, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming 650204, China.
Adeniyi C Adeola, State Key Laboratory of Genetic Resources and Evolution & Yunnan Laboratory of Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650201, China; Sino-Africa Joint Research Center, Chinese Academy of Sciences, Kunming 650204, China; Centre for Biotechnology Research, Bayero University, Kano 700006, Nigeria.
Kun Wang, School of Ecology and Environment, Northwestern Polytechnical University, Xi’an 710129, China.
Cui-Ping Huang, State Key Laboratory of Genetic Resources and Evolution & Yunnan Laboratory of Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650201, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming 650204, China.
Ming-Min Xu, State Key Laboratory of Genetic Resources and Evolution & Yunnan Laboratory of Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650201, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming 650204, China.
Qiang Qiu, School of Ecology and Environment, Northwestern Polytechnical University, Xi’an 710129, China.
Xue Yin, State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, School of Life Sciences, Yunnan University, Kunming 650091, China.
Chen-Yu Fan, State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, School of Life Sciences, Yunnan University, Kunming 650091, China.
Yun-Fei Ma, State Key Laboratory of Genetic Resources and Evolution & Yunnan Laboratory of Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650201, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming 650204, China.
Ting-Ting Yin, State Key Laboratory of Genetic Resources and Evolution & Yunnan Laboratory of Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650201, China.
Yun Gao, State Key Laboratory of Genetic Resources and Evolution & Yunnan Laboratory of Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650201, China.
Jia-Kun Deng, State Key Laboratory of Genetic Resources and Evolution & Yunnan Laboratory of Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650201, China.
Agboola O Okeyoyin, National Park Service Headquarter, Federal Capital Territory, Abuja 900108, Nigeria.
Olufunke O Oluwole, Institute of Agricultural Research and Training, Obafemi Awolowo University, Ibadan, Nigeria.
Oladipo Omotosho, Department of Veterinary Medicine, University of Ibadan, Ibadan 200005, Nigeria.
Victor M O Okoro, Department of Animal Science and Technology, School of Agriculture and Agricultural Technology, Federal University of Technology, Owerri 460114, Nigeria.
Ofelia G Omitogun, Department of Animal Sciences, Obafemi Awolowo University, Ile-Ife 220282, Nigeria.
Philip M Dawuda, Department of Veterinary Surgery and Theriogenology, College of Veterinary Medicine, University of Agriculture Makurdi, Makurdi 970001, Nigeria.
Sunday C Olaogun, Department of Veterinary Medicine, University of Ibadan, Ibadan 200005, Nigeria.
Lotanna M Nneji, State Key Laboratory of Genetic Resources and Evolution & Yunnan Laboratory of Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650201, China; Sino-Africa Joint Research Center, Chinese Academy of Sciences, Kunming 650204, China.
Adeola O Ayoola, State Key Laboratory of Genetic Resources and Evolution & Yunnan Laboratory of Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650201, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming 650204, China; Sino-Africa Joint Research Center, Chinese Academy of Sciences, Kunming 650204, China.
Oscar J Sanke, Taraba State Ministry of Agriculture and Natural Resources, Jalingo 660213, Nigeria.
Pam D Luka, National Veterinary Research Institute, Vom 930103, Nigeria.
Edward Okoth, International Livestock Research Institute (ILRI), Nairobi 00100, Kenya.
Isaac Lekolool, Kenya Wildlife Services, Nairobi 00100, Kenya.
Dominic Mijele, Kenya Wildlife Services, Nairobi 00100, Kenya.
Richard P Bishop, International Livestock Research Institute (ILRI), Nairobi 00100, Kenya.
Jianlin Han, International Livestock Research Institute (ILRI), Nairobi 00100, Kenya; CAAS-ILRI Joint Laboratory on Livestock and Forage Genetic Resources, Institute of Animal Science, Chinese Academy of Agricultural Sciences (CAAS), Beijing 100094, China.
Wen Wang, State Key Laboratory of Genetic Resources and Evolution & Yunnan Laboratory of Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650201, China; School of Ecology and Environment, Northwestern Polytechnical University, Xi’an 710129, China.
Min-Sheng Peng, State Key Laboratory of Genetic Resources and Evolution & Yunnan Laboratory of Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650201, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming 650204, China; Sino-Africa Joint Research Center, Chinese Academy of Sciences, Kunming 650204, China.
Ya-Ping Zhang, State Key Laboratory of Genetic Resources and Evolution & Yunnan Laboratory of Molecular Biology of Domestic Animals, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650201, China; Kunming College of Life Science, University of Chinese Academy of Sciences, Kunming 650204, China; Sino-Africa Joint Research Center, Chinese Academy of Sciences, Kunming 650204, China; State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, School of Life Sciences, Yunnan University, Kunming 650091, China.
Supplementary material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
Y.-P.Z., M.-S.P., W.W., and J.H. conceived the project. H.-B.X., C.Y., C.-P.H., Y.-F.M. performed genome sequencing and assembly. C.Y., H.-B.X., K.W., M.-M.X., Q.Q. performed comparative genomics analysis. A.C.A., M.-S.P., J.H., T.-T.Y., C.-P.H., Y.-F.M., Y.G., J.-K.D., A.O.O., O.O.O., O.O., V.M.O.O., O.G.O., P.M.D., S.C.O., L.M.N., A.O.A., O.J.S., P.D.L., E.O., I.L., D.M., R.P.B., X.Y., and C.-Y.F. organized and collected the samples. C.Y., H.-B.X., and A.C.A. wrote the manuscript. M.-S.P., Y.-P.Z., W.W., J.H., and R.P.B. revised the manuscript. All authors have approved the final manuscript.
Data Availability
The three newly assembled genomes have been deposited in the Genome Warehouse and the Illumina reads generated in this study have been submitted to the Genome Sequence Archive in Big Data Center (CNCB-NGDC Members and Partners 2022) under the project accession PRJCA002378, publicly accessible at https://bigd.big.ac.cn/bioproject/browse/PRJCA002378. The whole-genome alignment of the ten genomes, the CDS alignments of orthologous genes, the resulted gene trees, in-house scripts/tools have been deposited in the OMIX database available at https://ngdc.cncb.ac.cn/omix/release/OMIX938.
References
- Adeola AJ, Adeyemo AI, Ejidike BN, Olaniyi OE, Akande OA, Ajayi SR, Azeez OK. 2021. Conservation status and habitat preferences of common warthog (Phacochoerus africanus) in Old Oyo National Park, Nigeria. J Appl Sci Environ Manage. 25(1):87–92. [Google Scholar]
- Andolfatto P. 2007. Hitchhiking effects of recurrent beneficial amino acid substitutions in the Drosophila melanogaster genome. Genome Res. 17(12):1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apanaskevich DA, Horak IG, Mulumba-Mfumu LK. 2013. A new species of Rhipicephalus (Acari: Ixodidae), a parasite of red river hogs and domestic pigs in the Democratic Republic of Congo. J Med Entomol. 50(3):479–484. [DOI] [PubMed] [Google Scholar]
- Arribas A, Garrido G. 2008. A new wild boar belonging to the genus Potamochoerus (Suidae, Artiodactyla, Mammalia) from the Eurasian Late Upper Pliocene (Fonelas P-1, Cuenca de Guadix, Granada). Quad. Mus. Geominero. 10:337–364. [Google Scholar]
- Arsic N, Rajic T, Stanojcic S, Goodfellow PN, Stevanovic M. 1998. Characterisation and mapping of the human SOX14 gene. Cytogenet Cell Genet. 83(1-2):139–146. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoola AO, Zhang BL, Meisel RP, Nneji LM, Shao Y, Morenikeji OB, Adeola AC, Ng’ang’a SI, Ogunjemite BG, Okeyoyin AO, et al. 2021. Population genomics reveals incipient speciation, introgression, and adaptation in the African mona monkey (Cercopithecus mona). Mol Biol Evol. 38(3):876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger EM, Leus K, Vercammen P, Schwarzenberger F. 2006. Faecal steroid metabolites for non-invasive assessment of reproduction in common warthogs (Phacochoerus africanus), red river hogs (Potamochoerus porcus) and babirusa (Babyrousa babyrussa). Anim Reprod Sci. 91(1-2):155–171. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Zagorska A, Lew ED, Shrestha B, Rothlin CV, Naughton J, Diamond MS, Lemke G, Young JA. 2013. Enveloped viruses disable innate immune responses in dendritic cells by direct activation of TAM receptors. Cell Host Microbe. 14(2):136–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickhart DM, Rosen BD, Koren S, Sayre BL, Hastie AR, Chan S, Lee J, Lam ET, Liachko I, Sullivan ST, et al. 2017. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet. 49(4):643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14(4):708–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco E, Abril JF. 2009. Computational gene annotation in new genome assemblies using GeneID. In: Posada D, editors. Bioinformatics for DNA sequence analysis. Totowa: (NJ: ): Humana Press. p. 243–261. [DOI] [PubMed] [Google Scholar]
- Blomstrom AL, Stahl K, Masembe C, Okoth E, Okurut AR, Atmnedi P, Kemp S, Bishop R, Belak S, Berg M. 2012. Viral metagenomic analysis of bushpigs (Potamochoerus larvatus) in Uganda identifies novel variants of Porcine parvovirus 4 and Torque teno sus virus 1 and 2. Virol J. 9(1):192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshe JI. 1981. Reproductive ecology of the warthog Phacochoerus aethiopicus and its significance for management in the Eastern Selous Game Reserve, Tanzania. Biol Conserv. 20(1):37–44. [Google Scholar]
- Brunet M, MPFT . 2000. Chad: discovery of a vertebrate fauna close to the Mio-Pliocene boundary. J Vert Paleontol. 20(1):205–209. [Google Scholar]
- Brunet M, White T. 2001. Two new suin species (Mammalia, Suidae) from Africa (Chad, Ethiopia). C R Acad Sci Ser II Fasc A Sci Terre Planètes. 332:51–57. [Google Scholar]
- Bu D, Luo H, Huo P, Wang Z, Zhang S, He Z, Wu Y, Zhao L, Liu J, Guo J, et al. 2021. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 49(W1):W317–W325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge C, Karlin S. 1997. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 268(1):78–94. [DOI] [PubMed] [Google Scholar]
- Butynski TM, de Jong YA. 2017. Common warthog Phacochoerus africanus (Gmelin, 1788). In: Meijaard E, Melletti M, editors. Ecology, conservation and management of wild pigs and peccaries. Cambridge: Cambridge University Press. p. 85–100. [Google Scholar]
- Cai C, Tang YD, Xu G, Zheng C. 2021. The crosstalk between viral RNA- and DNA-sensing mechanisms. Cell Mol Life Sci. 78(23):7427–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, Tesler G. 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinf. 13(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Qiu Q, Jiang Y, Wang K, Lin Z, Li Z, Bibi F, Yang Y, Wang J, Nie W, et al. 2019. Large-scale ruminant genome sequencing provides insights into their evolution and distinct traits. Science. 364(6446):eaav6202. [DOI] [PubMed] [Google Scholar]
- Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, et al. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 10(6):563–569. [DOI] [PubMed] [Google Scholar]
- Chin CS, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O’Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. 2016. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 13(12):1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark PU, Dyke AS, Shakun JD, Carlson AE, Clark J, Wohlfarth B, Mitrovica JX, Hostetler SW, McCabe AM. 2009. The last glacial maximum. Science. 325(5941):710–714. [DOI] [PubMed] [Google Scholar]
- CNCB-NGDC Members and Partners . 2022. Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 50(D1):D27–D38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codjia FG, Lougbegnon TO, SdF L, Mensah GA. 2021. Conservation facing the uses knowledge of Phacochoerus africanus (Gmelin 1788), common warthog and Potamochoerus porcus (Linnaeus 1758), red river hog in south of Benin. Research Square. 10.21203/rs.3.rs-156828/v1 [DOI] [Google Scholar]
- Cooke HB. 1978. Suid evolution and correlation of African hominid localities: an alternative taxonomy. Science. 201(4354):460–463. [DOI] [PubMed] [Google Scholar]
- Darfour-Oduro KA, Megens HJ, Roca AL, Groenen MA, Schook LB. 2015. Adaptive evolution of Toll-Like receptors (TLRs) in the family Suidae. PLoS One. 10(4):e0124069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. Jmodeltest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delogu A, Sellers K, Zagoraiou L, Bocianowska-Zbrog A, Mandal S, Guimera J, Rubenstein JL, Sugden D, Jessell T, Lumsden A. 2012. Subcortical visual shell nuclei targeted by ipRGCs develop from a Sox14+-GABAergic progenitor and require Sox14 to regulate daily activity rhythms. Neuron. 75(4):648–662. [DOI] [PubMed] [Google Scholar]
- Dixon LK, Islam M, Nash R, Reis AL. 2019. African swine fever virus evasion of host defences. Virus Res. 266:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebhodaghe FI, Okal MN, Kalayou S, Bastos ADS, Masiga DK. 2021. Tsetse bloodmeal analyses incriminate the common warthog Phacochoerus africanus as an important cryptic host of animal Trypanosomes in smallholder cattle farming communities in Shimba Hills, Kenya. Pathogens. 10(11):1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edossa A, Bekele A, Debella HJ. 2021. Diet preferences of common warthogs (Phacochoerus africanus) in Gassi and Haro Aba Diko controlled hunting areas, Western Ethiopia. Glob Ecol Conserv. 29:e01722. [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. 2018. The Pfam protein families database in 2019. Nucleic Acids Res. 47(D1):D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbers JP, Rogers MF, Perelman PL, Proskuryakova AA, Serdyukova NA, Johnson WE, Horin P, Corander J, Murphy D, Burger PA. 2019. Improving illumina assemblies with Hi-C and long reads: An example with the North African dromedary. Mol Ecol Resour. 19(4):1015–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, et al. 2012. Mind the gap: upgrading genomes with Pacific biosciences RS long-read sequencing technology. PLoS One. 7(11):e47768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett H, Crooke H, Gurrala R, Dwarka R, Kim J, Botha B, Lubisi A, Pardini A, Gers S, Vosloo W, et al. 2011. Experimental infection of common warthogs (Phacochoerus africanus) and bushpigs (Potamochoerus larvatus) with classical swine fever virus. I: susceptibility and transmission. Transbound Emerg Dis. 58(2):128–134. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A. 2006. The genomic rate of adaptive evolution. Trends Ecol Evol. 21(10):569–575. [DOI] [PubMed] [Google Scholar]
- Fan S, Hansen ME, Lo Y, Tishkoff SA. 2016. Going global by adapting local: a review of recent human adaptation. Science. 354(6308):54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Zhang Y, Liu X, Wang J, Sun Z, Sun S, Zhang H, Chen J, Lv M, Han K, et al. 2019. The first chromosome-level genome for a marine mammal as a resource to study ecology and evolution. Mol Ecol Resour. 19(4):944–956. [DOI] [PubMed] [Google Scholar]
- Forth JH, Forth LF, Lycett S, Bell-Sakyi L, Keil GM, Blome S, Calvignac-Spencer S, Wissgott A, Krause J, Hoper D, et al. 2020. Identification of African swine fever virus-like elements in the soft tick genome provides insights into the virus’ evolution. BMC Biol. 18(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz L, Meijaard E, Gongora J, Haile J, Groenen MA, Larson G. 2016. The evolution of Suidae. Annu Rev Anim Biosci. 4(1):61–85. [DOI] [PubMed] [Google Scholar]
- Frantz LA, Schraiber JG, Madsen O, Megens HJ, Bosse M, Paudel Y, Semiadi G, Meijaard E, Li N, Crooijmans RP, et al. 2013. Genome sequencing reveals fine scale diversification and reticulation history during speciation in Sus. Genome Biol. 14(9):R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friant S, Young DK, Goldberg TL. 2022. Typical intracranial myiasis in Nigerian red river hogs (Potamochoerus porcus) caused by an unknown bot fly (Diptera: Oestridae). Int J Parasitol Parasites Wildl. 17:14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Erill G, Jorgensen CHF, Muwanika VB, Wang X, Rasmussen MS, de Jong YA, Gaubert P, Olayemi A, Salmona J, Butynski TM, et al. 2022. Warthog genomes resolve an evolutionary conundrum and reveal introgression of disease resistance genes. Mol Biol Evol. 39(7):msac13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gongora J, Cuddahee RE, do Nascimento FF, Palgrave CJ, Lowden S, Ho SYW, Simond D, Damayanti CS, White DJ, Tay WT, et al. 2011. Rethinking the evolution of extant sub-Saharan African suids (Suidae. Artiodactyla). Zool Scr. 40(4):327–335. [Google Scholar]
- Gongora J, Groves C, Meijaard E. 2017. Evolutionary relationships and taxonomy of Suidae and Tayassuidae. In: Meijaard E and Melletti M, editors. Ecology, conservation and management of wild pigs and peccaries. Cambridge: Cambridge University Press. p. 1–19. [Google Scholar]
- Groenen MA. 2016. A decade of pig genome sequencing: a window on pig domestication and evolution. Genet Sel Evol. 48:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen MA, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, Rogel-Gaillard C, Park C, Milan D, Megens HJ, et al. 2012. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 491(7424):393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groß C, Derks M, Megens HJ, Bosse M, Groenen MAM, Reinders M, de Ridder D. 2020. pCADD: SNV prioritisation in Sus scrofa. Genet Sel Evol. 52(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves CP, Grubb P. 2011. Ungulate taxonomy. Baltimore: (MD: ): Johns Hopkins University Press. [Google Scholar]
- Grubb P. 2005. Order artiodactyla. In: Wilson DE and Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference guide. Baltimore: (MD: ): Johns Hopkins University Press. p. 637–722. [Google Scholar]
- Grubb P, Groves CP, Dudley JP, Shoshani J. 2000. Living African elephants belong to two species: Loxodonta africana (Blumenbach, 1797) and Loxodonta cyclotis (Matschie,1900). Elephant. 2(4):1–4. [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704. [DOI] [PubMed] [Google Scholar]
- Halligan DL, Oliver F, Eyre-Walker A, Harr B, Keightley PD. 2010. Evidence for pervasive adaptive protein evolution in wild mice. PLoS Genet. 6(1):e1000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AC, Taylor D. 1991. History of climate and forests in tropical Africa during the last 8 million years. Clim Change. 19(1-2):65–78. [Google Scholar]
- Harris JM, White TD. 1979. Evolution of the Plio-Pleistocene African Suidae. Trans Am Philos Soc. 69(2):1–128. [Google Scholar]
- Hu Y, Yu L, Fan H, Huang G, Wu Q, Nie Y, Liu S, Yan L, Wei F. 2021. Genomic signatures of coevolution between nonmodel mammals and parasitic roundworms. Mol Biol Evol. 38(2):531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Pollard KS, Siepel A. 2011. PHAST And RPHAST: phylogenetic analysis with space/time models. Brief Bioinform. 12(1):41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins FM, Kim KW. 2007. A screen for immunity genes evolving under positive selection in Drosophila. J Evol Biol. 20(3):965–970. [DOI] [PubMed] [Google Scholar]
- Jori F, Bastos ADS. 2009. Role of wild suids in the epidemiology of African swine fever. EcoHealth. 6(2):296–310. [DOI] [PubMed] [Google Scholar]
- Kalbfleisch TS, Rice ES, DePriest MS jr, Walenz BP, Hestand MS, Vermeesch JR, Connell BLO, Fiddes IT, Vershinina AO, Saremi NF, et al. 2018. Improved reference genome for the domestic horse increases assembly contiguity and composition. Commun Biol. 1(1):197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45(D1):D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, Chu MW, Sanes JR. 2012. MEGF10 And MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature. 483(7390):465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielbasa SM, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res. 21(3):487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinf. 5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg ZN, Fiddes IT, Gordon D, Murali S, Cantsilieris S, Meyerson OS, Underwood JG, Nelson BJ, Chaisson MJP, Dougherty ML, et al. 2018. High-resolution comparative analysis of great ape genomes. Science. 360(6393):eaar6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Gaur R. 2013. First record of maxillary dentition of Potamochoerus theobaldi (Suidae, Mammalia) from the Upper Siwaliks of India. Riv Ital Paleontol S. 119:57–63. [Google Scholar]
- Kumar P, Henikoff S, Ng PC. 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 4(7):1073–1081. [DOI] [PubMed] [Google Scholar]
- Leffler EM, Gao Z, Pfeifer S, Segurel L, Auton A, Venn O, Bowden R, Bontrop R, Wall JD, Sella G, et al. 2013. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science. 339(6127):1578–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGates TA, Fernandez DC, Hattar S. 2014. Light as a central modulator of circadian rhythms, sleep and affect. Nat Rev Neurosci. 15(7):443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie DM, Huffman BA. 2015. Potamochoerus porcus (Artiodactyla: Suidae). Mamm Species. 47(919):15–31. [Google Scholar]
- Li H. 2014. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 30(20):2843–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2011. Inference of human population history from individual whole-genome sequences. Nature. 475(7357):493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lin Z, Ba H, Chen L, Yang Y, Wang K, Qiu Q, Wang W, Li G. 2017. Draft genome of the reindeer (Rangifer tarandus). Gigascience. 6(12):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Bosse M, Megens HJ, Frantz LAF, Lee YL, Irving-Pease EK, Narayan G, Groenen MAM, Madsen O. 2019. Genomic analysis on pygmy hog reveals extensive interbreeding during wild boar expansion. Nat Commun. 10(1):1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Gao J, Cui X, Li Z, Chen L, Yuan Y, Zhang Y, Mei L, Zhao L, Cai D, et al. 2021. A towering genome: experimentally validated adaptations to high blood pressure and extreme stature in the giraffe. Sci Adv. 7(12):eabe9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Megens HJ, Crooijmans R, Bosse M, Huang Q, van Sonsbeek L, Groenen MAM, Madsen O. 2022. The Visayan warty pig (Sus cebifrons) genome provides insight into chromosome evolution and sensory adaptation in pigs. Mol Biol Evol. 39(6):msac110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez M, Choin J, Sikora M, Siddle K, Harmant C, Costa HA, Silvert M, Mouguiama-Daouda P, Hombert JM, Froment A, et al. 2019. Genomic evidence for local adaptation of hunter-gatherers to the African rainforest. Curr Biol. 29(17):2926–2935.e4. [DOI] [PubMed] [Google Scholar]
- Luther NJ, Majiyagbe KA, Shamaki D, Lombin LH, Antiabong JF, Bitrus Y, Owolodun O. 2007. Detection of African swine fever virus genomic DNA in a Nigerian red river hog (Potamochoerus porcus). Vet Rec. 160(2):58–59. [DOI] [PubMed] [Google Scholar]
- MacMicking JD. 2012. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat Rev Immunol. 12(5):367–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majoros WH, Pertea M, Salzberg SL. 2004. Tigrscan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 20(16):2878–2879. [DOI] [PubMed] [Google Scholar]
- McCleary S, Strong R, McCarthy RR, Edwards JC, Howes EL, Stevens LM, Sanchez-Cordon PJ, Nunez A, Watson S, Mileham AJ, et al. 2020. Substitution of warthog NF-κB motifs into RELA of domestic pigs is not sufficient to confer resilience to African swine fever virus. Sci Rep. 10(1):8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus KF, Kelley JL, Song S, Veeramah KR, Woerner AE, Stevison LS, Ryder OA, Great Ape Genome Project, Kidd JM, Wall JD, et al. 2015. Inference of gorilla demographic and selective history from whole-genome sequence data. Mol Biol Evol. 32(3):600–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melletti M, Breuer T, Huffman BA, Turkalo AK, Mirabile M, Maisels F. 2017. Red river hog Potamochoerus porcus (Linnaeus, 1758). In: Meijaard E and Melletti M, editors. Ecology, conservation and management of wild pigs and peccaries. Cambridge: Cambridge University Press. p. 134–149. [Google Scholar]
- Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang H-Y, El-Gebali S, Fraser MI, et al. 2018. Interpro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47(D1):D351–D360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molday RS, Moritz OL. 2015. Photoreceptors at a glance. J Cell Sci. 128(22):4039–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muwanika VB, Nyakaana S, Siegismund HR, Arctander P. 2003. Phylogeography and population structure of the common warthog (Phacochoerus africanus) inferred from variation in mitochondrial DNA sequences and microsatellite loci. Heredity (Edinb). 91(4):361–372. [DOI] [PubMed] [Google Scholar]
- Muwanika VB, Nyakaana S, Siegismund HR, Arctander P. 2007. Population genetic structure of the common warthog (Phacochoerus africanus) in Uganda: evidence for a strong philopatry among warthogs and social structure breakdown in a disturbed population. Afr J Ecol. 45(1):22–30. [Google Scholar]
- Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. 2007. Recent and ongoing selection in the human genome. Nat Rev Genet. 8(11):857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Welch JJ, Kim KW, Jiggins FM. 2009. Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 5(10):e1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan C, Martin MJ, Gattiker A, Gasteiger E, Bairoch A, Apweiler R. 2002. High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Brief Bioinform. 3(3):275–284. [DOI] [PubMed] [Google Scholar]
- Oettinger MA, Schatz DG, Gorka C, Baltimore D. 1990. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 248(4962):1517–1523. [DOI] [PubMed] [Google Scholar]
- Orliac MJ. 2013. The petrosal bone of extinct Suoidea (Mammalia, Artiodactyla). J Syst Palaeontol. 11(8):925–945. [Google Scholar]
- Oura CA, Powell PP, Anderson E, Parkhouse RM. 1998. The pathogenesis of African swine fever in the resistant bushpig. J Gen Virol. 79(6):1439–1443. [DOI] [PubMed] [Google Scholar]
- Pacifici M, Santini L, Di Marco M, Baisero D, Francucci L, Marasini GG, Visconti P, Rondinini C. 2013. Generation length for mammals. Nat Conserv-Bulgaria. 5(5):87–94. [Google Scholar]
- Palgrave CJ, Gilmour L, Lowden CS, Lillico SG, Mellencamp MA, Whitelaw CB. 2011. Species-specific variation in RELA underlies differences in NF-κB activity: a potential role in African swine fever pathogenesis. J Virol. 85(12):6008–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks MM, Lawrence CE, Raphael BJ. 2015. Detecting non-allelic homologous recombination from high-throughput sequencing data. Genome Biol. 16(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perng YC, Lenschow DJ. 2018. ISG15 In antiviral immunity and beyond. Nat Rev Microbiol. 16(7):423–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickford M. 2006. Synopsis of the biochronology of African Neogene and Quaternary Suiformes. Trans R Soc S Afr. 61(2):51–62. [Google Scholar]
- Pickford M. 2012. Ancestors of Broom's Pigs. Trans R Soc South Africa. 67(1):17–35. [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. 2010. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20(1):110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu L, Gaudreault NN, Whitworth KM, Murgia MV, Nietfeld JC, Mileham A, Samuel M, Wells KD, Prather RS, Rowland RRR. 2017. Genetically edited pigs lacking CD163 show no resistance following infection with the African swine fever virus isolate, Georgia 2007/1. Virology. 501:102–106. [DOI] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA. 2005. De novo identification of repeat families in large genomes. Bioinformatics. 21(Suppl 1):i351–i358. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2020. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Raaphorst E. 2020. Antibody responses to porcine reproductive and respiratory syndrome virus, influenza A virus, and Mycoplasma hyopneumoniae and their association with single-nucleotide variants. [Thesis]. Guelph: (Ontario, Canada: ): The University of Guelph. [Google Scholar]
- Rigoli L, Lombardo F, Di Bella C. 2011. Wolfram syndrome and WFS1 gene. Clin Genet. 79(2):103–117. [DOI] [PubMed] [Google Scholar]
- Roca AL, Ishida Y, Brandt AL, Benjamin NR, Zhao K, Georgiadis NJ. 2015. Elephant natural history: a genomic perspective. Annu Rev Anim Biosci. 3(1):139–167. [DOI] [PubMed] [Google Scholar]
- Rohland N, Reich D, Mallick S, Meyer M, Green RE, Georgiadis NJ, Roca AL, Hofreiter M. 2010. Genomic DNA sequences from mastodon and woolly mammoth reveal deep speciation of forest and savanna elephants. PLoS Biol. 8(12):e1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen BD, Bickhart DM, Schnabel RD, Koren S, Elsik CG, Tseng E, Rowan TN, Low WY, Zimin A, Couldrey C, et al. 2020. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience. 9(3):giaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Li H. 2020. Fast and accurate long-read assembly with wtdbg2. Nat Methods. 17(2):155–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky A, Rothschild MF, Larson G, Gongora J. 2011. Systematics and evolution of the pig. In: Rothschild MF and Ruvinsky A, editors. The genetics of the pig. Wallingford: CAB International. p. 1–13. [Google Scholar]
- Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol. 8(7):559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Torres C, Gómez-Puertas P, Gómez-del-Moral M, Alonso F, Escribano JM, Ezquerra A, Domínguez J. 2003. Expression of porcine CD163 on monocytes/macrophages correlates with permissiveness to African swine fever infection. Arch Virol. 148(12):2307–2323. [DOI] [PubMed] [Google Scholar]
- Schmidt JM, de Manuel M, Marques-Bonet T, Castellano S, Andres AM. 2019. The impact of genetic adaptation on chimpanzee subspecies differentiation. PLoS Genet. 15(11):e1008485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Anhäuser A, Ludwig P, Schlüter P, Williams E. 2018. Statistical reconstruction of global vegetation for the last glacial maximum. Glob Planet Change. 168:67–77. [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15(8):1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Haussler D. 2004. Phylogenetic estimation of context-dependent substitution rates by maximum likelihood. Mol Biol Evol. 21(3):468–488. [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P.. 2008. RepeatModeler Open-1.0. Available from: http://www.repeatmasker.org/RepeatModeler.html.
- Smit AFA, Hubley R, Green P.. 2015. RepeatMasker Open-4.0. Available from: http://www.repeatmasker.org
- Smitz N, Berthouly C, Cornelis D, Heller R, Van Hooft P, Chardonnet P, Caron A, Prins H, van Vuuren BJ, De Iongh H, et al. 2013. Pan-African genetic structure in the African buffalo (Syncerus caffer): investigating intraspecific divergence. PLoS One. 8(2):e56235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. 2006. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34(Web Server issue):W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 134(4):587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 25(1):1–14. [DOI] [PubMed] [Google Scholar]
- Turner SJ, Doherty PC, McCluskey J, Rossjohn J. 2006. Structural determinants of T-cell receptor bias in immunity. Nat Rev Immunol. 6(12):883–894. [DOI] [PubMed] [Google Scholar]
- Vaser R, Sović I, Nagarajan N, Šikić M. 2017. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 27(5):737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr A, Affara N, Aken B, Beiki H, Bickhart DM, Billis K, Chow W, Eory L, Finlayson HA, Flicek P, et al. 2020. An improved pig reference genome sequence to enable pig genetics and genomics research. Gigascience. 9(6):giaa051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkinson RE, McEwan WA, Tam JC, Vaysburd M, James LC. 2015. TRIM21 Promotes cGAS and RIG-I sensing of viral genomes during infection by antibody-opsonized virus. PLoS Pathog. 11(10):e1005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessling EG, Kuhl HS, Mundry R, Deschner T, Pruetz JD. 2018. The costs of living at the edge: seasonal stress in wild savanna-dwelling chimpanzees. J Hum Evol. 121:1–11. [DOI] [PubMed] [Google Scholar]
- White AM, Cameron EZ, Peacock MM. 2010. Grouping patterns in warthogs, Phacochoerus africanus: is communal care of young enough to explain sociality? Behaviour. 147(1):1–18. [Google Scholar]
- Wu G-S, Pang J-F, Zhang Y-P. 2006. Molecular phylogeny and phylogeography of Suidae. Zool Res. 27(2):197–201. [Google Scholar]
- Wu GS, Yao YG, Qu KX, Ding ZL, Li H, Palanichamy MG, Duan ZY, Li N, Chen YS, Zhang YP. 2007. Population phylogenomic analysis of mitochondrial DNA in wild boars and domestic pigs revealed multiple domestication events in East Asia. Genome Biol. 8(11):R245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wang H. 2007. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35(Web Server issue):W265–W268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yu K, Zhang T. 2013. Construction of customized sub-databases from NCBI-nr database for rapid annotation of huge metagenomic datasets using a combined BLAST and MEGAN approach. PLoS One. 8(4):e59831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Rabiee M, Sayyari E, Mirarab S. 2018. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinf. 19(Suppl 6):153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The three newly assembled genomes have been deposited in the Genome Warehouse and the Illumina reads generated in this study have been submitted to the Genome Sequence Archive in Big Data Center (CNCB-NGDC Members and Partners 2022) under the project accession PRJCA002378, publicly accessible at https://bigd.big.ac.cn/bioproject/browse/PRJCA002378. The whole-genome alignment of the ten genomes, the CDS alignments of orthologous genes, the resulted gene trees, in-house scripts/tools have been deposited in the OMIX database available at https://ngdc.cncb.ac.cn/omix/release/OMIX938.