Abstract
The vls (variable major protein [VMP]-like sequence) locus of Borrelia burgdorferi encodes an antigenic variation system that closely resembles the VMP system of relapsing fever borreliae. To determine whether vls sequences are present consistently in low-passage, infectious isolates of B. burgdorferi, 22 blood and erythema migrans biopsy isolates from Lyme disease patients in Westchester County, New York, were examined by Southern blot and PCR analysis. Each of the strains contained a single plasmid varying in size from 21 to 38 kb that hybridized strongly with a vlsE probe based on the B. burgdorferi B31 sequence. In contrast, PCR products were obtained with only 10 of the 22 strains when primers corresponding to the 5′ and 3′ regions of the B31 vlsE sequence outside the variable cassette region were used. Only 2 of 16 B. burgdorferi-infected tick specimens yielded detectable PCR product. Eight of 10 strains that yielded a PCR product under these conditions were type 1 (a genotype with a high rate of dissemination), according to PCR-restriction fragment length polymorphism analysis of intergenic rDNA sequences, whereas the isolates that did not yield vlsE PCR products were either type 2 or type 3. Comparison of the sequences of cloned PCR products from the patient isolates indicated a high degree of identity to the B31 sequence, with most of the differences restricted to the hypervariable regions known to undergo sequence variation. Taken together, these results both reinforce previous evidence that vls sequences are present consistently in low-passage Lyme disease spirochetes and indicate that both highly conserved and heterogeneous subgroups exist with regard to vlsE sequences.
Lyme disease, a multisystem disorder with possible cutaneous, neurologic, and rheumatologic manifestations, is the most common arthropod-borne disease in the United States (4). The etiologic agent of the disease, Borrelia burgdorferi, is transmitted to humans by the bite of an infected tick of the Ixodes ricinus complex (Ixodes scapularis or Ixodes pacificus in the United States) (1). In untreated humans and other mammalian hosts, spirochetal infection can persist in certain tissues for extended periods, even in the presence of an active immune response (11, 18). Mechanisms underlying this long-term persistence have not been elucidated.
The vls locus of B. burgdorferi strain B31 consists of a single expressed gene (vlsE), which encodes a surface-exposed lipoprotein, and 15 silent (unexpressed) cassettes, which have >90% sequence identity to the central cassette region of vlsE (23). Most of the sequence differences between the cassette regions are concentrated in six short DNA segments termed variable regions (VR-I through VR-VI). Unidirectional recombination of segments of the vls silent cassettes into the cassette region of vlsE results in extensive antigenic variation in the expressed VlsE protein (23, 24). In strain B31, the vls locus is present on the linear plasmid lp28-1 (23), which was detected by hybridization in high-infectivity clones of B. burgdorferi B31 and Sh-2-82 but not in most low-infectivity clones (23; J. E. Purser and S. J. Norris, unpublished data). Therefore, it has been postulated that VlsE represents a virulence factor, as well as an antigenic variation protein (23).
It is important to establish whether vls systems are consistently present in Lyme disease borreliae. In the present study, the presence of vls sequences in multiple isolates from Lyme disease patients and in ticks was examined.
Detection of vls sequences in clinical isolates of B. burgdorferi.
Twenty-two B. burgdorferi clinical isolates were analyzed for the presence of vls sequences by Southern blot analysis of pulsed-field gel electrophoresis (PFGE)-separated plasmid DNA. B. burgdorferi isolates were obtained from either skin biopsies of erythema migrans (EM) lesions or blood of Lyme disease patients with EM attending the Lyme Disease Diagnostic Center of Westchester Medical Center as previously described (16, 21). B. burgdorferi isolates were grown in BSK-H medium (Sigma Chemicals, St. Louis, Mo.) supplemented with 6% rabbit serum for 2 to 4 weeks at 33°C. Spirochetes were harvested and lysed in agarose blocks (1.8% low-gelling agarose) as previously described (19), and plasmids were resolved by PFGE on a Bio-Rad contour-clamped homogeneous electric field DR II apparatus at constant voltage (6 V/cm) for 19 h with a switch time of 0.9 to 2.5 s in 0.5× Tris-borate-EDTA buffer (pH 8.3) at 14°C. DNA was transferred to a positively charged nylon membrane and hybridized with a vlsE probe which was prepared by PCR amplification of B. burgdorferi B31 DNA with the vlsE-specific primers F4120 and R4066 as previously described (23).
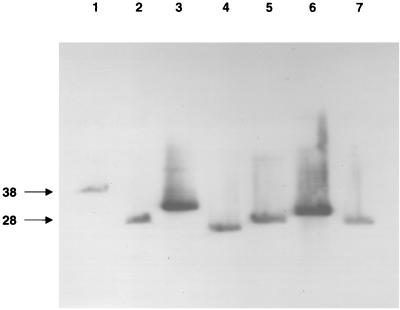
A representative experiment is shown in Fig. 1, and the data for all isolates are presented in Table 1. All isolates, regardless of the source (skin biopsy or blood), contained a single band that hybridized with the vls probe (Fig. 1), indicating that each isolate contained a single plasmid harboring vls sequences. All but 3 of the 22 isolates had been passaged less than two times following the initial culture and none was passaged more than four times. Thus, their plasmid content is likely to be quite similar to that of the organisms infecting the patients. The availability of such low-passage isolates for studies of plasmid content is important in that many plasmids, including lp28-1, are rapidly lost during in vitro culture (2, 12, 14, 17, 22, 23; J. E. Purser and S. J. Norris, unpublished data).
FIG. 1.
Localization of vls sequence region to various plasmids by Southern hybridization in representative clinical isolates. Plasmids were separated by PFGE and transferred to nylon membranes, and the blots were hybridized with a vlsE probe. Lane 1, BL36; lane 2, BL146; lane 3, B57; lane 4, B282; lane 5, B287; lane 6, B294; lane 7, B311. The migration positions of lp28-1 and lp38 are indicated on the left, based on the migration of contour-clamped homogeneous electric field DNA size standards (Bio-Rad) and hybridization to the ospD probe.
TABLE 1.
vlsE plasmid hybridization and PCR results in clinical isolates of B. burgdorferi
| Isolatea | Passage no. | Size (kb) of plasmid hybridizing with vlsE probeb | vlsE PCR resultc | RFLP typed |
|---|---|---|---|---|
| B14 | 1 | 36 | + | 1 |
| B16 | 1 | 28 | − | 2 |
| B23 | 1 | 32 | − | 3 |
| B57 | 2 | 32 | + | 1 |
| B247 | 4 | 28 | + | 2 |
| B282 | 2 | 25 | − | 2 |
| B287 | 2 | 28 | − | 2 |
| B294 | 2 | 32 | + | 1, 2 |
| B296 | 2 | 28 | + | 1 |
| B305 | 2 | 28 | + | 1 |
| B310 | 2 | 32 | + | 1 |
| B311 | 2 | 28 | − | 2 |
| B322 | 1 | 32 | − | 3 |
| B327 | 2 | 21 | + | 3 |
| B329 | 1 | 28 | − | 2 |
| B332 | 3 | 32 | − | 2 |
| B333 | 2 | 32 | − | 3 |
| BL36 | 2 | 38 | − | 3 |
| BL146 | 2 | 28 | − | 2 |
| BL152 | 2 | 28 | + | 1 |
| BL154 | 4 | 28 | − | 2 |
| CD160 | 2 | 28 | + | 1 |
Isolates with a B or CD designation were cultured from EM lesion biopsies, and those with a BL designation were cultivated from blood.
Molecular size of plasmid to which B31-5A3 vlsE probe hybridized, as determined by Southern blot analysis of PFGE-separated plasmids.
Presence (+) or absence (−) of a band detectable by ethidium bromide staining following PCR amplification using B31 vlsE-based primers F4120 and R4066.
Based on PCR-RFLP analysis of the 16S-23S rDNA spacer (8).
Plasmids hybridizing with B31-based vls probes are also present in B. burgdorferi strains Sh-2-82 and N40, in Borrelia afzelii ACA-1, and in Borrelia garinii Ip90 (23). In addition, vls sequences were identified in B. burgdorferi 297 by Kawabata et al. (6), and a survey of GenBank entries revealed B. burgdorferi cN40 (S. Feng and S. Barthold, GenBank entries AF005053 to AF005057 and AF011454) and B. garinii N34 (M.-C. Misonne and P. P. Hoet, GenBank entry AAB58528) sequences with high predicted amino acid similarity to vls. Most of these sequences appear to represent short segments of vls silent cassettes. Thus, vls sequences are present in all Lyme disease borreliae with a high-infectivity phenotype (13, 23) examined to date. Further study is needed to determine the organization, expression, and genetic variation of the vls loci in infectious strains other than B31.
For 11 of the 22 clinical isolates, the vls probe hybridized to a 28-kb plasmid, as is the situation for strain B31. Significantly, positive hybridization with the remaining isolates was observed for plasmids ranging in size from 21 to 38 kb (Fig. 1 and Table 1). This variation in migration could be due to the different sizes of linear plasmids or to the presence of vls sequences in circular plasmids. It is also not known at this point whether these plasmids represent size variants of lp28-1 or whether the vls sequences are present in plasmids that otherwise have a different genetic content. Size variation could also be due to expansion or shrinkage of the vls silent cassette locus, which spans 8.5 kb in B. burgdorferi B31-5A3 (23). Comparisons of the plasmid gene content of different strains indicate a high incidence of interplasmid recombination during borrelial evolution (3). In any event, it is of interest that within an individual isolate, vls sequences were limited to a single plasmid species.
PCR amplification of vlsE.
Given the finding that vls sequences exist in all clinical isolates analyzed, it was of interest to determine the relative sequence variation of vlsE in these isolates. As already stated, the vls region of strain B31 consists of 15 silent cassettes and 1 expression cassette within vlsE (23). The expression cassette is flanked by unique sequences, which allows specific amplification of this region by PCR. DNA was extracted from 1-ml cultures of B. burgdorferi clinical isolates using the Isoquick nucleic acid extraction kit (9, 10, 15). PCR amplification was performed using the primers F4120 and R4066 described previously (23). Amplification was carried out for 35 cycles with denaturation at 94°C for 30 s, annealing at 52°C for 30 s, and extension at 72°C for 30 s, and PCR products were analyzed by electrophoresis on 1% agarose gels.
Although all isolates contained sequences that hybridized to the vls probe, only 10 of 22 yielded a 660-bp, vlsE-specific PCR product (Table 1). The most likely explanation for this finding is that the sequences in the region of the primers (based on the B31-5A3 vlsE sequence upstream and downstream of the central cassette) were divergent in the strains where amplification was not obtained. Indeed, the vls sequences reported for the North American B. burgdorferi isolates 297 (6) and cN40 (S. Feng and S. W. Barthold, unpublished data) are only ∼40% identical and ∼60% similar to the B31-5A3 sequence; many of these sequence differences occur in the invariant sequences outside the variable regions. Attempts to amplify vlsE from the cN40 strain using a variety of primers based on the B31-5A3 sequence have been unsuccessful (M. B. Lawrenz and S. J. Norris, unpublished data). Preliminary sequence data from B. garinii vlsE indicate a moderate degree of sequence divergence both within and outside the central cassette (D. Wang and S. J. Norris, unpublished data). Another possibility is that an intact vlsE gene is not present in all strains and the vls sequences that are present represent an inactive, vestigial locus. This explanation seems unlikely, however, in that a large proportion of culture-positive EM patients and nearly all Lyme disease patients beyond this stage express antibodies against VlsE (7).
There is a remarkable correlation between vlsE PCR positivity and ribosomal DNA (rDNA) spacer restriction fragment length polymorphism (RFLP) type. Previous studies have demonstrated that clinical isolates of B. burgdorferi consist of several different genotypes, based on PCR-RFLP typing of the 16S-23S rDNA spacer (8, 9). Furthermore, patients infected with type 1 organisms had a greater likelihood of disseminated infection than those infected with other genotypes (20). It is interesting that 8 of 10 isolates which were vlsE positive by PCR are classified as type 1 organisms, whereas none of the 12 vlsE PCR-negative isolates were type 1 (P = 0.0001) (Table 1). This intriguing result could mean that a certain vlsE subgroup correlates with dissemination, or simply that there is a predominant clonal line in the New York area that contains both RFLP type 1 and B31-like vlsE genotypes.
Similar PCR analysis was carried out for I. scapularis collected by the drag-cloth method in Westchester County, New York (5). DNA was isolated from individual ticks as previously described (15) and employed directly for PCR. Of 16 B. burgdorferi-infected ticks (based on PCR amplification of the 16S-23S rDNA spacer [8, 9]), only 2 yielded PCR product with vlsE-specific primers. Southern hybridization with the vlsE probe was not attempted with tick extracts due to the small amounts of material available. However, it is reasonable to assume that tick isolates generally contain vls sequences, since partial sequences of vls silent cassettes or vlsE have been identified in the tick-derived strains examined to date (B31, N40, and Ip90). Thus, this result suggests that only a small subset of infected ticks contain B. burgdorferi with a vlsE sequence that could be amplified with a primer pair based on the B31 sequence. The reason for the relatively low frequency of vlsE amplification in tick-derived organisms relative to the clinical isolates in this study is not clear. It is possible that mammalian infection may “select” for strains containing the vls locus or vls sequences in an expression cassette. lp28-1 is rapidly lost during in vitro passage of B. burgdorferi B31 and Sh-2-82 (23), yet in vitro growth rates are apparently unaffected by this loss (12). Survival and growth of B. burgdorferi in ticks may also be unaffected by lp28-1 loss, permitting the accumulation of vls-deficient strains. There are other possible explanations, including a positive selection for a B. burgdorferi B31 vlsE subgroup in Lyme disease patients, as suggested above.
Sequence analysis of individual vlsE clones.
The availability of vlsE PCR products from some of the clinical and tick specimens permitted comparison of these sequences with the well-characterized B31-5A3 vlsE sequence. Preliminary direct sequence analysis of the PCR products revealed that some of the samples contained a mixture of B. burgdorferi clones expressing different vlsE variants. It was thus necessary to clone the PCR products into a plasmid vector prior to sequence analysis. After amplification, the 3′ ends of the PCR products were adenylated with Taq polymerase, and the resultant products were cloned into the pCR2.1 vector (Invitrogen, Carlsbad, Calif.) and transformed into E. coli SURE2 cells (Stratagene, La Jolla, Calif.).
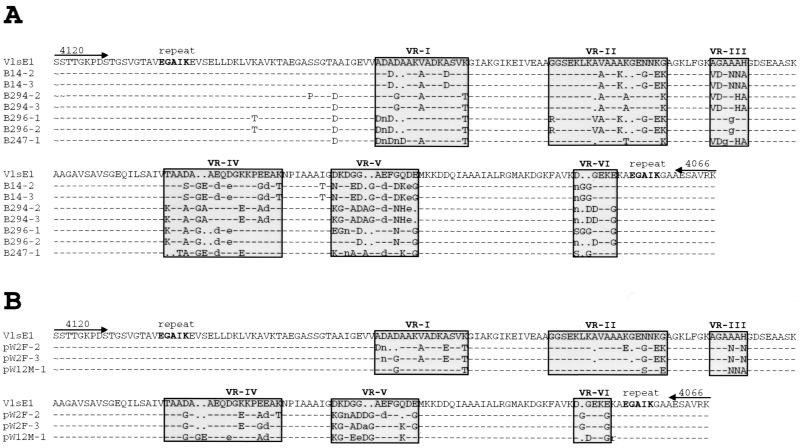
Sequences obtained with representative PCR product clones from Lyme disease patient isolates are shown in Fig. 2A. A total of seven PCR product clones were analyzed: two each from patient isolates B14, B247, and B296 and one from B294. When the deduced amino acid sequences were aligned with the corresponding VlsE sequence from B31-5A3, a high degree of sequence identity was observed. This high homology was also present at the DNA sequence level (data not shown). Nearly all of the differences were restricted to the six variable regions (VR-I through VR-VI), which contain most of the sequence differences among the vls silent cassettes of B. burgdorferi B31-5A3 (23). Indeed, all but six of the deduced amino acid sequence differences could be attributed to recombination with silent cassette sequences that had been identified in B31-5A3 (data not shown). Based on these results, each of these four clinical isolates contain vls loci nearly identical to that of B31-5A3. As mentioned above, two PCR product clones were sequenced from each of the human isolates B14, B247, and B296. The pairs of PCR sequences were identical for strains B14 and B247. However, the B296 PCR product sequences had differences in VR-V and VR-VI (Fig. 2A). Thus, strain B296 contains at least two vlsE variants.
FIG. 2.
Comparison of sequences from representative clinical isolate and tick vlsE PCR products with the B. burgdorferi B31-5A3 vlsE sequence. PCR products were cloned into the plasmid pCR2.1 and sequenced as described in the text. Deduced amino acid sequences were aligned with the B31-5A3 vlsE sequence by using ClustalW and formatted by using Boxshade. Dashes indicate identity with the B31-5A3 sequence, lowercase letters identify similar residues, and capital letters represent dissimilar amino acids. Variable regions (VR) I through VI previously identified by alignment of B31-5A3 vls silent cassette sequences with vlsE (23) are shown. (A) Alignment of sequences from Lyme disease clinical isolates B14, B294, B296, and B247. Deduced sequences from two different PCR product clones (e.g., B14-2 and B14-3) are shown for each of the isolates except B247, where only one PCR product clone was sequenced. (B) Sequences obtained from PCR product clones obtained from tick W2F (clones W2F-2 and W2F-3) and tick W12M (clone W12M-1).
The PCR products obtained from tick midgut extracts were also cloned and sequenced (Fig. 2B). As with the clinical isolates, the nucleotide and deduced amino acid sequences were nearly identical to those of vlsE of B. burgdorferi B31-5A3, except for differences in the variable regions. The two PCR product clones obtained from tick W2F were different in VR-I, -II, -IV, and -V, indicating the coexistence of vlsE variants in the midgut of the tick. In this case, all but one of the variable-region sequence differences could be attributed to vls silent cassette recombinations.
Previous studies have shown that multiple vlsE variants arise within days after experimental infection of mice with B. burgdorferi B31-5A3 or other B31-derived strains (25). In this report, one of the human isolates (B296) and one tick extract (W2F) were each shown to contain at least two different vlsE cassette region sequences (Fig. 2). These results confirm that Lyme disease patients and ticks can be infected with more than one vlsE variant. Alternatively, variation may be generated during host infection.
Most of the sequence differences in the patient and tick specimens shown in Fig. 2 could be explained by the occurrence of vls silent cassette-vlsE recombinations relative to B31-5A3. This is perhaps not surprising, since all sequences were obtained from PCR-amplified products primed with B31-derived sequences. If variants with divergent sequences exist in the population, they would probably not be represented in the isolates which were sequenced in this study. These results argue strongly for the existence of a subgroup of B. burgdorferi strains in the New York area that has nearly identical vls loci.
In conclusion, this study demonstrates that B. burgdorferi clinical isolates from Lyme disease patients in the lower Hudson Valley of New York consistently contain vls sequences and that a significant proportion of these isolates have a high degree of similarity to the B31 vlsE genotype. In addition, a vlsE expression cassette is found predominantly in isolates with a high dissemination genotype (rDNA spacer type 1). The B31 vlsE genotype was present in a smaller proportion of B. burgdorferi-infected ticks from this same region, but further analysis is necessary to interpret this finding. Previous mouse infection studies show that the presence of lp28-1 correlates with a high-infectivity phenotype (23). The consistent presence of vls sequences in Lyme disease isolates provides further evidence supporting the view that the vls locus or associated sequences may be required for infection of mammalian hosts.
Acknowledgments
We thank Jerrilyn K. Howell for technical support for this project and Denis Liveris for the RFLP typing.
This work was supported by grants AR41511 (to I.S.) and AI37277 (to S.J.N.) from the National Institutes of Health.
REFERENCES
- 1.Barbour A G, Fish D. The biological and social phenomenon of Lyme disease. Science. 1993;260:1610–1616. doi: 10.1126/science.8503006. [DOI] [PubMed] [Google Scholar]
- 2.Barbour A G, Garon C F. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987;237:409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- 3.Casjens, S., W. Huang, G. Sutton, J. Peterson, N. Palmer, R. van Vugt, B. Stevenson, P. Rosa, R. Lathigra, and C. Fraser. A genome in flux: the twelve linear and nine circular extrachromosomal DNAs of an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol., in press. [DOI] [PubMed]
- 4.Centers for Disease Control and Prevention. Lyme disease—United States, 1996. Morbid Mortal Weekly Rep. 1997;46:531–535. [PubMed] [Google Scholar]
- 5.Daniels T J, Boccia T M, Varde S, Marcus J, Le J, Bucher D J, Falco R C, Schwartz I. Geographic risk for Lyme disease and human granulocytic ehrlichiosis in southern New York state. Appl Environ Microbiol. 1998;64:4663–4669. doi: 10.1128/aem.64.12.4663-4669.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawabata H, Myouga F, Inagaki Y, Murai N, Watanabe H. Genetic and immunological analyses of Vls (VMP-like sequences) of Borrelia burgdorferi. Microb Pathog. 1998;24:155–166. doi: 10.1006/mpat.1997.0183. [DOI] [PubMed] [Google Scholar]
- 7.Lawrenz M B, Hardham J M, Owens R T, Nowakowski J, Steere A C, Wormser G P, Norris S J. Human antibody responses to VlsE antigenic variation protein of Borrelia burgdorferi. J Clin Microbiol. 1999;37:3997–4004. doi: 10.1128/jcm.37.12.3997-4004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liveris D, Gazumyan A, Schwartz I. Molecular typing of Borrelia burgdorferi sensu lato by PCR-restriction fragment length polymorphism analysis. J Clin Microbiol. 1995;33:589–595. doi: 10.1128/jcm.33.3.589-595.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liveris D, Varde S, Iyer R, Koenig S, Bittker S, Cooper D, McKenna D, Nowakowski J, Nadelman R B, Wormser G P, Schwartz I. Genetic diversity of Borrelia burgdorferi in Lyme disease patients as determined by culture versus direct PCR with clinical specimens. J Clin Microbiol. 1999;37:565–569. doi: 10.1128/jcm.37.3.565-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liveris D, Wormser G P, Nowakowski J, Nadelman R, Bittker S, Cooper D, Varde S, Moy F H, Forseter G, Pavia C S, Schwartz I. Molecular typing of Borrelia burgdorferi from Lyme disease patients by PCR-restriction fragment length polymorphism analysis. J Clin Microbiol. 1996;34:1306–1309. doi: 10.1128/jcm.34.5.1306-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nocton J J, Steere A C. Lyme disease. Adv Intern Med. 1995;40:69–117. [PubMed] [Google Scholar]
- 12.Norris S J, Carter C J, Howell J K, Barbour A G. Low-passage-associated proteins of Borrelia burgdorferi B31: characterization and molecular cloning of OspD, a surface-exposed, plasmid-encoded lipoprotein. Infect Immun. 1992;60:4662–4672. doi: 10.1128/iai.60.11.4662-4672.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norris S J, Howell J K, Garza S A, Ferdows M S, Barbour A G. High- and low-infectivity phenotypes of clonal populations of in vitro-cultured Borrelia burgdorferi. Infect Immun. 1995;63:2206–2212. doi: 10.1128/iai.63.6.2206-2212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwan T G, Burgdorferi W, Garon C F. Changes in infectivity and plasmid profile of the Lyme disease spirochete, Borrelia burgdorferi, as a result of in vitro cultivation. Infect Immun. 1988;56:1831–1836. doi: 10.1128/iai.56.8.1831-1836.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz I, Varde S, Nadelman R B, Wormser G P, Fish D. Inhibition of efficient polymerase chain reaction amplification of Borrelia burgdorferi DNA in blood-fed ticks. Am J Trop Med Hyg. 1997;56:339–342. doi: 10.4269/ajtmh.1997.56.339. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz I, Wormser G P, Schwartz J J, Cooper D, Weissensee P, Gazumyan A, Zimmermann E, Goldberg N S, Bittker S, Campbell G L, Pavia C S. Diagnosis of early Lyme disease by polymerase chain reaction amplification and culture of skin biopsies from erythema migrans lesions. J Clin Microbiol. 1992;30:3082–3088. doi: 10.1128/jcm.30.12.3082-3088.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simpson W J, Garon C F, Schwan T G. Analysis of supercoiled circular plasmids in infectious and non-infectious Borrelia burgdorferi. Microb Pathog. 1990;8:109–118. doi: 10.1016/0882-4010(90)90075-2. [DOI] [PubMed] [Google Scholar]
- 18.Steere A C. Lyme disease. N Engl J Med. 1989;321:586–596. doi: 10.1056/NEJM198908313210906. [DOI] [PubMed] [Google Scholar]
- 19.Walker E M, Howell J K, You Y, Hoffmaster A R, Heath J D, Weinstock G M, Norris S J. Physical map of the genome of Treponema pallidum subsp. pallidum (Nichols) J Bacteriol. 1995;177:1797–1804. doi: 10.1128/jb.177.7.1797-1804.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wormser G P, Liveris D, Nowakowski J, Nadelman R B, Cavaliere L F, McKenna D, Holmgren D, Schwartz I. Association of specific subtypes of Borrelia burgdorferi with hematogenous dissemination in early Lyme disease. J Infect Dis. 1999;180:720–725. doi: 10.1086/314922. [DOI] [PubMed] [Google Scholar]
- 21.Wormser G P, Nowakowski J, Nadelman R B, Bittker S, Cooper D, Pavia C. Improving the yield of blood cultures for patients with early Lyme disease. J Clin Microbiol. 1998;36:296–298. doi: 10.1128/jcm.36.1.296-298.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Kodner C, Coleman L, Johnson R C. Correlation of plasmids with infectivity of Borrelia burgdorferi sensu stricto type strain B31. Infect Immun. 1996;64:3870–3876. doi: 10.1128/iai.64.9.3870-3876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J R, Hardham J M, Barbour A G, Norris S J. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell. 1997;89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J-R, Norris S J. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect Immun. 1998;66:3698–3704. doi: 10.1128/iai.66.8.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J-R, Norris S J. Kinetics and in vivo induction of genetic variation of vlsE in Borrelia burgdorferi. Infect Immun. 1998;66:3689–3697. doi: 10.1128/iai.66.8.3689-3697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]