Abstract
Objectives:
The purpose of this study was to develop an artificial intelligence-based model to prognosticate COVID-19 patients at admission by combining clinical data and chest radiographs.
Methods:
This retrospective study used the Stony Brook University COVID-19 dataset of 1384 inpatients. After exclusions, 1356 patients were randomly divided into training (1083) and test datasets (273). We implemented three artificial intelligence models, which classified mortality, ICU admission, or ventilation risk. Each model had three submodels with different inputs: clinical data, chest radiographs, and both. We showed the importance of the variables using SHapley Additive exPlanations (SHAP) values.
Results:
The mortality prediction model was best overall with area under the curve, sensitivity, specificity, and accuracy of 0.79 (0.72–0.86), 0.74 (0.68–0.79), 0.77 (0.61–0.88), and 0.74 (0.69–0.79) for the clinical data-based model; 0.77 (0.69–0.85), 0.67 (0.61–0.73), 0.81 (0.67–0.92), 0.70 (0.64–0.75) for the image-based model, and 0.86 (0.81–0.91), 0.76 (0.70–0.81), 0.77 (0.61–0.88), 0.76 (0.70–0.81) for the mixed model. The mixed model had the best performance (p value < 0.05). The radiographs ranked fourth for prognostication overall, and first of the inpatient tests assessed.
Conclusions:
These results suggest that prognosis models become more accurate if AI-derived chest radiograph features and clinical data are used together.
Advances in knowledge:
This AI model evaluates chest radiographs together with clinical data in order to classify patients as having high or low mortality risk. This work shows that chest radiographs taken at admission have significant COVID-19 prognostic information compared to clinical data other than age and sex.
Introduction
We have all spent the last two years on the SARS-Cov-2 (COVID-19) rollercoaster, but it is not over. As the WHO weekly reports show, the global rate of new infections has been increasing since their 63rd weekly update two months ago, 1 prioritizing patients and resources remains a relevant topic.
Many prognostic models have been applied to COVID-19 patients. 2 For example, NEWS2, 3 the Brescia-COVID Respiratory Severity Scale, 4 and qSOFA 5 models are easy to use clinically, because they use only clinical data. When imaging data is used, it is often solely image findings such as the presence of opacities, pneumonia, pulmonary edema, and vascular enlargement. 6–8 These binary representations sometimes lose information which is difficult to quantify and thus have only modest improvement against simpler models. 6,8 To merge image and clinical data into a model was the next logical step, however, models which have been developed using both clinical and image data often use weighted sums from separate models, rather than creating a single model. 9 With respiratory diseases, radiographical information is as important as clinical information. So, images should be incorporated into future prognosis estimation and medical resource allocation models. 10,11
One way to incorporate images into prognostic models is to apply artificial intelligence (AI), specifically convolutional neural networks (CNNs). These can handle whole images as input and have recently found applications in medicine. 12 The potential of AI to evaluate COVID-19 on chest radiograph or CT images was explored with great success. 13–15 Results of these models indicate that image-based AI models can support radiologists in clinical practice. 14 Still, several questions about the power these models provide to support physicians remain unanswered. Specifically, are images or clinical data more useful and, to what extent are these dozens of variables effective for prognosticating COVID-19 severity? Finding the most effective variable combinations may increase model development efficiency. In this work, we will develop new models and evaluate the effect of each variable—including images—on the severity prognosis of COVID-19 patients.
Methods and materials
Study design
We conducted a retrospective model development and testing study of three AI-based prognostic models for COVID-19 mortality, ICU admission, and ventilation. We prepared three submodels for each model which used clinical data, imaging features, or a mixture of the two. Using the mixed clinical and imaging data models, we determined the relative importance of each of the variables included in the models. All models were developed and tested using a publicly available dataset. This work was prepared in accordance with the CLAIM checklist. 16 The need for review and approval from the ethics board was waived due to the open source nature of the dataset.
Patient data
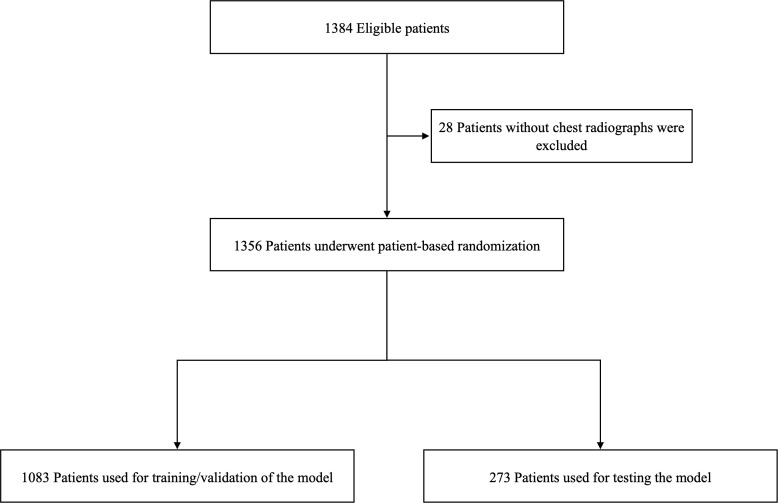
The publicly available Stony Brook University COVID-19 Positive Cases dataset 17 from The Cancer Imaging Archive 18 was used to develop the models. A total of 1356 patients were used for this study after excluding 28 patients who did not have a chest radiograph at admission. (Figure 1) Clinical data and chest radiographs at admission were extracted as explanatory variables for the model output. Patient outcomes (death, ICU admission, and ventilation requirement) were extracted as the ground truths. One chest radiograph taken closest to the date of admission was collected for each patient. All radiographs were taken in the anteroposterior view. We chose 25 variables which have been shown to be the risk factors for severe COVID-19. 19,20 Clinical data include gender, age, smoking history, BMI, and medical history (hypertension, diabetes, chronic heart disease, chronic renal failure, chronic lung disease, and malignancy), vital signs (heart rate, systolic blood pressure, respiratory rate, and blood oxygen saturation), and laboratory data (white blood cell, sodium, potassium, C-reactive protein, aspartate aminotransferase, alanine aminotransferase, urea nitrogen, creatinine, lactate, brain natriuretic peptide, and d-dimer). Detailed demographics are shown in Table 1.
Figure 1.
Eligibility flowchart.
Table 1.
Demographics
| Training / validation dataset | Test dataset | |
|---|---|---|
| Total no. of patients | 1083 | 273 |
| Male | 627 (57.8%) | 153 (56%) |
| Female | 456 (42.1%) | 120 (43.9%) |
| Age | ||
| 18–59 | 595 (54.9%) | 156 (57.1%) |
| 59–74 | 279 (25.7%) | 67 (24.5%) |
| 74–90 | 209 (19.2%) | 50 (18.3%) |
| Smoking history | 232 (21.4%) | 50 (18.3%) |
| Body mass index, mean ± std | 29.3 ± 5.8 | 29.4 ± 6.0 |
| Disease history | ||
| Hypertension | 386 (35.6%) | 103 (37.7%) |
| Diabetes | 216 (19.9%) | 58 (21.2%) |
| Chronic heart disease | 148 (13.6%) | 45 (16.4%) |
| Chronic kidney disease | 66 (6%) | 15 (5.4%) |
| Chronic lung disease | 158 (14.5%) | 46 (16.8%) |
| Malignancy | 77 (7.1%) | 16 (5.8%) |
| Outcomes | ||
| Death | 137 (12.6%) | 43 (15.7%) |
| Discharge | 946 (87.3%) | 230 (84.2%) |
| ICU admission | 166 (15.3%) | 47 (17.2%) |
| Ventilation | 200 (18.4%) | 60 (21.9%) |
Data partition
Patients were randomly divided into training and test datasets at a ratio of 4:1. (Figure 1) There was no overlap of patients among the respective datasets. Details are available in Supplementary Material 1.
Image processing
All chest radiographs were downscaled to PNG files while maintaining the aspect ratio. The shorter side was filled with black for images which were not originally square. All images were augmented using random rotation from –0.1 radians to 0.1 radians, with a random shift of 10%, a brightness range of 10%, and reflected horizontally. Further details are available in Supplementary Material 1.
Model development and evaluation
The clinical data-based AI models were developed using a multilayer perceptron (MLP), which was composed of three fully connected layers. There were 25 variables as input.
The image-based models were developed using InceptionV3, 21 ResNet50, 22 and DenseNet121 23 as the basic architectures which passed to a single fully connected layer, then connected to an MLP with three fully connected layers. The best performing basic architecture was retained.
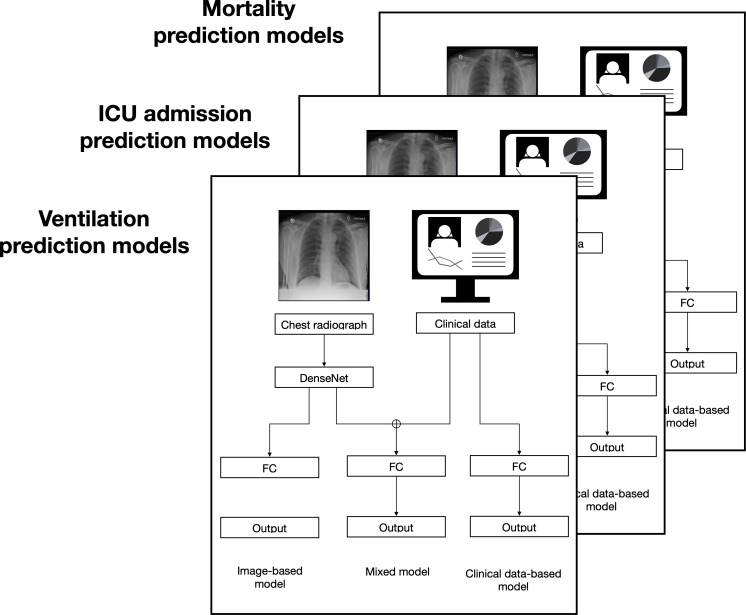
The mixed models were implemented using MLP with CNN. The images were input into the CNN as above, then this output was concatenated with clinical variables, and these continued together to an MLP with three fully connected layers. The CNN architecture was prepared with the same basic architectures as the image-based model. (Figure 2).
Figure 2.
Overview of the classification and prognostic models. The chest radiographs were input as PNG images, and the clinical data were in a CSV file. Three sets of models were created; one each with the output arranged to show if the COVID-19 patient was expected to die, require mechanical ventilation, or require ICU admission. FC = Fully connected layers
All models were developed using the PyTorch framework. 24 Each model was trained and tuned from scratch with the training dataset using five-fold cross-validation. Each output from the clinical data-based models, the image-based models, and the mixed models was obtained using cross-entropy loss. The output for all models classified if the patient was expected to experience the severe outcome targeted by that model. The performance of each model was assessed using the independent test dataset. Further model development details are available in Supplementary methods b and Supplementary Material 1.
Importance values and saliency maps
The importance of each explainable variable in the mixed model, including chest radiographs, was determined by SHapley Additive exPlanations (SHAP) values. 25 Each SHAP value was calculated 10,000 times.
A saliency map was generated for each chest radiograph evaluated by the mixed model to visualize the model’s focus as it classified patient mortality prognosis. 26 A detailed explanation of the saliency map generation model is shown in Supplementary Material 1, and the source code, including our training and testing python codes as well as our final hyperparameter selections, is available online (https://github.com/covid-cnn-mlp/).
Statistical analysis
To evaluate the models, sensitivity, specificity, accuracy, the receiver operating characteristic curve (ROC), and the area under the curve (AUC) were assessed. We used the Youden index to determine thresholds. 27 Different prediction models were compared using a binomial test to show differences in performance. The Kaplan–Meier method was used to further stratify patients into high-risk and low-risk subgroups according to the median of values from the mortality model prediction. The stratification performance was evaluated using a log-rank test based on the predicted risk scores of the stratified subgroups. Statistical inferences were performed with a two-sided 5% significance level. All analyses were performed using R, version 3.6.0 28 and Python 3.8.1.
Results
Model development
The models were each independently developed using the training dataset with five-fold cross-validation applied for 100 training epochs, then the loss value on a separate validation dataset determined the performance of the model. The final optimizer for all models was Adam (learning ratio = 0.001) with a batch size of 64. In both image-based and mixed models, the best performing models were obtained with an image size of 256 pixels; DenseNet was the best performing CNN architecture.
Model evaluation
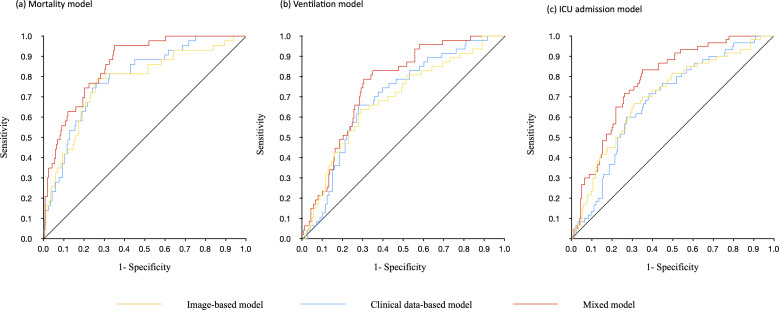
The AUC for predicting mortality was highest overall with 0.86 (0.81–0.91) for the mixed clinical and imaging data model, 0.79 (0.72–0.86) for the clinical data model, and 0.77 (0.69–0.85) for the imaging data model. The sensitivity, specificity, and accuracy, respectively, for mortality prognosis were 0.74 (0.68–0.79), 0.77 (0.61–0.88), and 0.74 (0.69–0.79) for the clinical data-based model, 0.67 (0.61–0.73), 0.81 (0.67–0.92), 0.70 (0.64–0.75) for the image-based model, and 0.76 (0.70–0.81), 0.77 (0.61–0.88), 0.76 (0.70–0.81) for the mixed model. Results for the mixed input models were consistently higher than either clinical data alone or imaging data alone for the other severity outcomes as well. (Table 2 and Figure 3)
Table 2.
Model results
| Area under the curve (95% CI) | Accuracy (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | P value | |
|---|---|---|---|---|---|
| Death | |||||
| Image-based model | 0.77 (0.69–0.85) | 0.70 (0.64–0.75) | 0.67 (0.61–0.73) | 0.81 (0.67–0.92) | 0.037 |
| Clinical data-based model | 0.79 (0.72–0.86) | 0.74 (0.69–0.79) | 0.74 (0.68–0.79) | 0.77 (0.61–0.88) | <0.001 |
| Mixed model | 0.86 (0.81–0.91) | 0.76 (0.70–0.81) | 0.76 (0.70–0.81) | 0.77 (0.61–0.88) | ref |
| Ventilation | |||||
| Image-based model | 0.68 (0.60–0.77) | 0.65 (0.59–0.70) | 0.65 (0.58–0.71) | 0.66 (0.51–0.79) | 0.047 |
| Clinical data-based model | 0.70 (0.62–0.77) | 0.67 (0.61–0.72) | 0.67 (0.60–0.73) | 0.66 (0.51–0.79) | 0.053 |
| Mixed model | 0.76 (0.69–0.83) | 0.72 (0.66–0.77) | 0.72 (0.65–0.77) | 0.72 (0.57–0.84) | ref |
| ICU admission | |||||
| Image-based model | 0.70 (0.62–0.77) | 0.66 (0.60–0.72) | 0.66 (0.59–0.72) | 0.67 (0.53–0.78) | 0.029 |
| Clinical data-based model | 0.68 (0.61–0.75) | 0.65 (0.59–0.70) | 0.65 (0.58–0.71) | 0.65 (0.52–0.77) | <0.001 |
| Mixed model | 0.78 (0.72–0.84) | 0.71 (0.66–0.77) | 0.71 (0.65–0.77) | 0.72 (0.59–0.83) | ref |
Figure 3.
Receiver operating characteristic curves. Each panel represents different outcome targets for the models. (a) Risk of death. The clinical data-based model had an AUC of 0.79 (0.72–0.86), the image-based model had an AUC of 0.77 (0.69–0.85), and the mixed model had an AUC of 0.86 (0.81–0.91). (b) Risk of mechanical ventilation. The clinical data-based model had an AUC of 0.70 (0.62–0.77), the image-based model had an AUC of 0.68 (0.60–0.77), and the mixed model had an AUC of 0.76 (0.69–0.83). (c) Risk of ICU admission. The clinical data-based model had an AUC of 0.68 (0.61–0.75), the image-based model had an AUC of 0.70 (0.62–0.77), and the mixed model had an AUC of 0.78 (0.72–0.84).
AUC = area under the curve
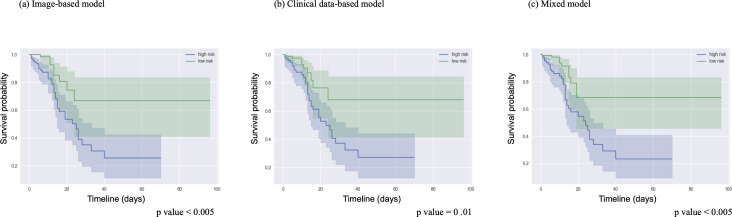
The discrimination power of the mixed model for mortality prognosis was significantly higher than that of the clinical data model (p = 0.001) and the imaging data model (p = 0.037). (Table 2) All three mortality models successfully stratified patients according to the median model score into high and low mortality risk with p values below 0.05 for each model. The accuracy of this stratification is shown by Kaplan–Meier plot. (Figure 4) Confusion matrices for the three mixed models are available in Supplementary Material 1, Figure 3.
Figure 4.
Kaplan–Meier survival plots for each model. The high-risk and low-risk patients from each mortality model were divided based on the median model output value. This plot shows the ground truth survival of these patients and the shaded area represents the accuracy of the prediction.
Importance values and saliency maps
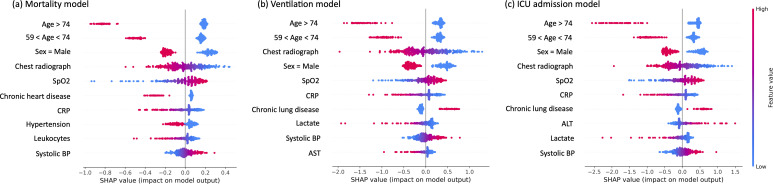
Each variable was ranked by their impact on the mixed clinical and imaging data model output using mean SHAP values. Age and sex had the largest impact on model output, followed by chest radiographs, which had the fourth highest impact. A visual representation of the impact of each variable on the model output for each severity outcome can be seen in the beeswarm plot. (Figure 5)
Figure 5.
SHAP values for each variable. Beeswarm plots of the SHAP value for each patient for the top ten variables. The plots relate to risk of death (a), risk of mechanical ventilation (b), and risk of ICU admission (c). Each dot represents one patient. The location of the dot represents if changing the value for that patient would have a positive (less likely to predict outcome) or negative (more likely to predict outcome) effect on the model, and to what extent. The color represents the value range of the variable from lowest (blue) to highest (red). Some variables only have binomial representations; for sex, red represents male. For age, red represents an age above the cutoff. When there are many patients with very similar SHAP values, the swarm expands vertically.
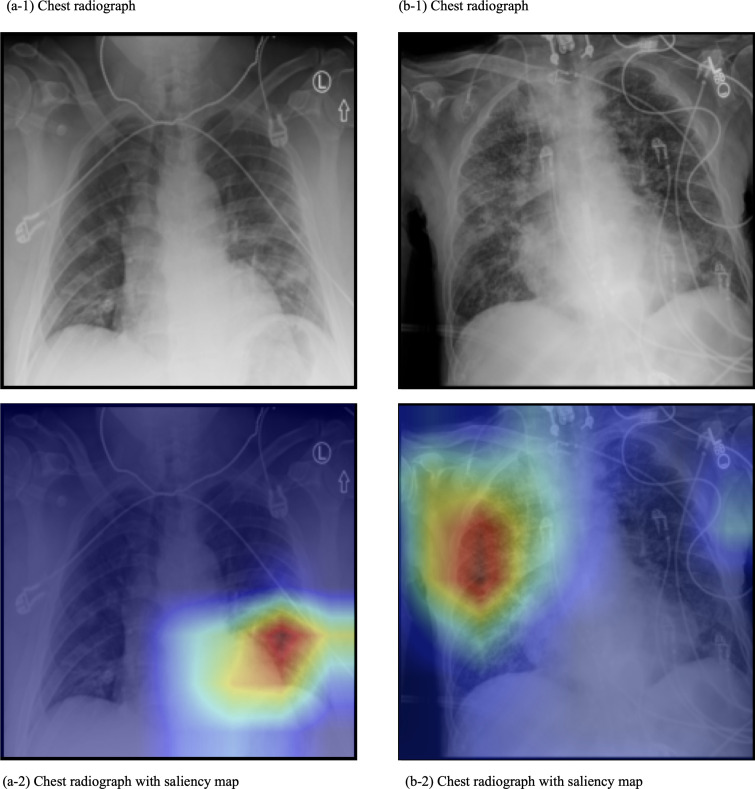
Saliency maps were produced for the chest radiographs to provide a visual representation of the focus of the model as it classified COVID-19 mortality. (Figure 6) The focus was typically on a single point on the lungs, rather than diffuse. This single point covered an area of infiltration.
Figure 6.
Representative saliency maps. These are chest radiographs and the saliency map overlay of two patients (labeled a and b) from the Stony Brook Hospital dataset. The mortality prediction model was used. In these images, the model focus was on an area of infiltration.
Discussion
In this study, we developed and compared AI-based models which predict three measures of severity of patients hospitalized with COVID-19 using clinical data, AI-derived image features, and both. The models using both performed better than models we developed using only clinical data or only image data (p value < 0.05). The area under the curve, sensitivity, specificity, and accuracy of the mortality prognosis mixed model were 0.86 (0.81–0.91), 0.76 (0.70–0.81), 0.77 (0.61–0.88), and 0.76 (0.70–0.81), respectively. We also ranked the value of image information compared to clinical data in these AI models using SHAP values. These showed that chest radiographs had the fourth largest impact on the model output, behind only age and sex in COVID-19 mortality prognosis.
AI models perform a huge number of simple calculations when extracting features from data. While it is easy to understand each calculation, the sheer volume of them makes it difficult to understand how the model made the final decision; thus, it is referred to as a black box. 29 One way to see the relative importance of each variable is by their SHAP values. 25 From previous work, we expected age, sex, and oxygen saturation to rank highly, and as radiologists, we expected the addition of images to also have a strong impact. 19,20,30 Still, we were surprised by how strongly images impacted the model. Future AI models intended for medical use can benefit from a similar quantification of variable importance in cases where it is unclear which data are valuable. The features which correlated with poor prognosis were visualized using saliency maps. The saliency maps focused on locations in the lung field showing disease features such as infiltration.
Although prognosis models using both clinical and imaging data have been reported, this technique is less common for communicable or acute diseases. Prior AI-based COVID-19 prognostication studies have shown the value of both clinical data and images separately in AI models. 14,31–36 Jiao, et al combined image data and clinical data into one model and showed the power of including chest radiographs for AI-determined prognosis. 9 Our work differs in both model design and that we evaluated the value of the image data compared to other variables in the severity classification. Gupta et al applied several models to their own dataset and found the best performance with the NEWS2 score, 3 even compared with models using imaging findings. 37 The AUC of our model surpasses that of all models evaluated in their study. Many COVID-19 prognosis studies chose various degrees of severity as the turning point, including oxygen supplementation, 38 mechanical ventilation, 9,39–44 ICU admission, 39,44,45 or mortality. 14,39,42,44,46–49 Unlike these studies, the focus of our work was not only prognosis, but also to estimate the value of using both clinical data and chest radiographs. Our model had an AUC comparable to models developed using CT but with generally higher sensitivity. CT-based models largely focused on prognosticating ICU admission or ventilation 15,36,40,50 rather than mortality. Although CT is known to be more accurate than radiographs, it has several limitations for early COVID-19 prognosis including the time required for imaging, the availability of CT-equipped facilities, and ease of cleaning between potentially contagious patients. 51 While we cannot directly compare our model to previous works because the datasets are different, it is likely that our larger cohort size and the combination of images and clinical data are reasons for our increase in AUC.
The method presented here has implications for other diseases which also currently rely on tabular clinical data to determine patient prognosis. For example, cancer staging is based on the longest diameter of the tumor(s). 52,53 This method reduces the information available from the image to a single number, while the lesion is not necessarily spherical. The inclusion of the image into a prognostic model could allow more precise information to be considered for prognosis determination. Our model can be used in a federated learning approach at centers with appropriate infrastructure. 54
There were limitations to this work. This was a single center, retrospective study and should be tested prospectively. We excluded 28 patients who were missing chest radiographs, which may have led to selection bias where patients with detailed data were those who had been determined to be at high risk at triage. Information related to the radiologist’s reports was not available in this dataset. Further study is required to determine the effect of DL-extracted information compared with using binomial scoring of image features. Although deep learning-based AI models can easily be fine-tuned to perform similar tasks, future research is needed to determine how well this technique generalizes to future COVID-19 and other respiratory disease outbreaks.
In summary, we developed and validated an AI model to determine the prognosis of patients with COVID-19 which uses both chest radiographs and clinical data. Additionally, we used an explainable AI approach to establish the value of imaging data compared to clinical data as well as to indicate the focus of the model per image. Our work is open source, and the code is available online. We hope our work leads to more accurate prognosis of the still-growing number of patients with COVID-19, as well as further research into the use of images for the prognosis of other diseases.
Footnotes
Funding: This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data statement: All data for this study was from the publicly available Stony Brook University COVID-19 dataset. Model codes are available online. (https://github.com/covid-cnn-mlp/)
Contributor Information
Shannon L Walston, Email: shannongross3@gmail.com.
Toshimasa Matsumoto, Email: matsumoto.toshimasa@gmail.com.
Yukio Miki, Email: yukio.miki@med.osaka-cu.ac.jp.
Daiju Ueda, Email: ai.labo.ocu@gmail.com.
REFERENCES
- 1. WHO . Weekly epidemiological update—16 november 2021 . Available from : https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---16-november-2021 ( accessed 16 Nov 2021 )
- 2. Wynants L, Van Calster B, Collins GS, Riley RD, Heinze G, Schuit E, et al . Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal . BMJ 2020. ; 369 : m1328 . doi: 10.1136/bmj.m1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gidari A, De Socio GV, Sabbatini S, Francisci D . Predictive value of national early warning score 2 (NEWS2) for intensive care unit admission in patients with sars-cov-2 infection . Infect Dis (Lond) 2020. ; 52 : 698 – 704 . doi: 10.1080/23744235.2020.1784457 [DOI] [PubMed] [Google Scholar]
- 4. Piva S, Filippini M, Turla F, Cattaneo S, Margola A, De Fulviis S, et al . Clinical presentation and initial management critically ill patients with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in Brescia, Italy . J Crit Care 2020. ; 58 : 29 – 33 : S0883-9441(20)30547-5 . doi: 10.1016/j.jcrc.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al . The third international consensus definitions for sepsis and septic shock (sepsis-3) . JAMA 2016. ; 315 : 801 – 10 . doi: 10.1001/jama.2016.0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Colombi D, Bodini FC, Petrini M, Maffi G, Morelli N, Milanese G, et al . Well-aerated lung on admitting chest CT to predict adverse outcome in Covid-19 pneumonia . Radiology 2020. ; 296 : E86 – 96 . doi: 10.1148/radiol.2020201433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wong HYF, Lam HYS, Fong AH-T, Leung ST, Chin TW-Y, Lo CSY, et al . Frequency and distribution of chest radiographic findings in patients positive for covid-19 . Radiology 2020; 296 : E72–78. 10.1148/radiol.2020201160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu G, Yang P, Xie Y, Woodruff HC, Rao X, Guiot J, et al . Development of a clinical decision support system for severity risk prediction and triage of covid-19 patients at hospital admission: an international multicentre study . Eur Respir J 2020. ; 56 ( 2 ): 2001104 . doi: 10.1183/13993003.01104-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jiao Z, Choi JW, Halsey K, Tran TML, Hsieh B, Wang D, et al . Prognostication of patients with covid-19 using artificial intelligence based on chest x-rays and clinical data: A retrospective study . Lancet Digit Health 2021. ; 3 : e286 – 94 : S2589-7500(21)00039-X . doi: 10.1016/S2589-7500(21)00039-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liang W, Yao J, Chen A, Lv Q, Zanin M, Liu J, et al . Early triage of critically ill covid-19 patients using deep learning . Nat Commun 2020. ; 11 ( 1 ): 3543 . doi: 10.1038/s41467-020-17280-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rodriguez VA, Bhave S, Chen R, Pang C, Hripcsak G, Sengupta S, et al . Development and validation of prediction models for mechanical ventilation, renal replacement therapy, and readmission in covid-19 patients . J Am Med Inform Assoc 2021. ; 28 : 1480 – 88 . doi: 10.1093/jamia/ocab029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hinton G . Deep learning-a technology with the potential to transform health care . JAMA 2018. ; 320 : 1101 – 2 . doi: 10.1001/jama.2018.11100 [DOI] [PubMed] [Google Scholar]
- 13. Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, et al . Using artificial intelligence to detect covid-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy . Radiology 2020. ; 296 : E65 – 71 . doi: 10.1148/radiol.2020200905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mushtaq J, Pennella R, Lavalle S, Colarieti A, Steidler S, Martinenghi CMA, et al . Initial chest radiographs and artificial intelligence (AI) predict clinical outcomes in covid-19 patients: analysis of 697 italian patients . Eur Radiol 2021. ; 31 : 1770 – 79 . doi: 10.1007/s00330-020-07269-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu Z, Li X, Sun H, Wang J, Zhao T, Chen H, et al . Rapid identification of covid-19 severity in CT scans through classification of deep features . Biomed Eng Online 2020. ; 19 ( 1 ): 63 . doi: 10.1186/s12938-020-00807-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mongan J, Moy L, Kahn CE . Checklist for artificial intelligence in medical imaging (claim): a guide for authors and reviewers . Radiol Artif Intell 2020. ; 2 : e200029 . doi: 10.1148/ryai.2020200029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. . Saltz J, Saltz M, Prasanna P, et al . Stony brook university covid-19 positive cases . The Cancer Imaging Archive ; 2021. . [Google Scholar]
- 18. Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, et al . The cancer imaging archive (TCIA): maintaining and operating a public information repository . J Digit Imaging 2013. ; 26 : 1045 – 57 . doi: 10.1007/s10278-013-9622-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barry M, Alotaibi M, Almohaya A, Aldrees A, AlHijji A, Althabit N, et al . Factors associated with poor outcomes among hospitalized patients with Covid-19: experience from a MERS-CoV referral hospital . J Infect Public Health 2021. ; 14 : 1658 – 65 : S1876-0341(21)00314-2 . doi: 10.1016/j.jiph.2021.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Izcovich A, Ragusa MA, Tortosa F, Lavena Marzio MA, Agnoletti C, Bengolea A, et al . Prognostic factors for severity and mortality in patients infected with covid-19: a systematic review . PLoS One 2020. ; 15 ( 11 ): e0241955 . doi: 10.1371/journal.pone.0241955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Szegedy C, Wei L, Yangqing J, Sermanet P, Reed S, Anguelov D, et al . Going deeper with convolutions . 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) ; Boston, MA, USA . ; 2015. . pp . 1 – 9 . doi: 10.1109/CVPR.2015.7298594 [DOI] [Google Scholar]
- 22. He K, Zhang X, Ren S, Sun J . Deep residual learning for image recognition . 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) . ; 2016. . pp . 770 – 78 . [Google Scholar]
- 23. Huang G, Liu Z, Maaten LVD, Weinberger KQ . Densely connected convolutional networks . 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) . ; 2017. . pp . 2261 – 69 . [Google Scholar]
- 24. Paszke A, Gross S, Massa F, et al . Pytorch: an imperative style, high-performance deep learning library . Adv Neural Inf Process Syst 2019. ; 32 : 8026 – 37 . [Google Scholar]
- 25. Lundberg SM, Lee S-I . A unified approach to interpreting model predictions . Proceedings of the 31st International Conference on Neural Information Processing Systems 2017. ; 4768 – 77 . [Google Scholar]
- 26. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D . Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization . In: Paper presented at the In: Paper presented at the 2017 IEEE International Conference on Computer Vision (ICCV) , Venice . doi: 10.1109/ICCV.2017.74 [DOI] [Google Scholar]
- 27. YOUDEN WJ . Index for rating diagnostic tests . Cancer 1950; 3 : 32–35. [DOI] [PubMed] [Google Scholar]
- 28. R Development Core Team . R: a language and environment for statistical computing . Vienna: : R Foundation for Statistical Computing; ; 2013. . [Google Scholar]
- 29. Reyes M, Meier R, Pereira S, Silva CA, Dahlweid F-M, von Tengg-Kobligk H, et al . On the interpretability of artificial intelligence in radiology: challenges and opportunities . Radiol Artif Intell 2020; 2 : e190043. 10.1148/ryai.2020190043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liang W, Liang H, Ou L, Chen B, Chen A, Li C, et al . Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with covid-19 . JAMA Intern Med 2020. ; 180 : 1081 – 89 . doi: 10.1001/jamainternmed.2020.2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ahmed AI, Raggi P, Al-Mallah MH . Teaching an old dog new tricks: the prognostic role of cacs in hospitalized covid-19 patients . Atherosclerosis 2021. ; 328 : 106 – 7 : S0021-9150(21)00251-3 . doi: 10.1016/j.atherosclerosis.2021.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Assaf D, Gutman Y, Neuman Y, Segal G, Amit S, Gefen-Halevi S, et al . Utilization of machine-learning models to accurately predict the risk for critical covid-19 . Intern Emerg Med 2020. ; 15 : 1435 – 43 . doi: 10.1007/s11739-020-02475-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cheng F-Y, Joshi H, Tandon P, Freeman R, Reich DL, Mazumdar M, et al . Using machine learning to predict icu transfer in hospitalized covid-19 patients . J Clin Med 2020. ; 9 ( 6 ): E1668 . doi: 10.3390/jcm9061668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luo M, Liu J, Jiang W, Yue S, Liu H, Wei S . IL-6 and cd8+ t cell counts combined are an early predictor of in-hospital mortality of patients with covid-19 . JCI Insight 2020. ; 5 : 13 : 139024 . doi: 10.1172/jci.insight.139024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma X, Ng M, Xu S, Xu Z, Qiu H, Liu Y, et al . Development and validation of prognosis model of mortality risk in patients with covid-19 . Epidemiol Infect 2020. ; 148 : e168 . doi: 10.1017/S0950268820001727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tang Z, Zhao W, Xie X, Zhong Z, Shi F, Ma T, et al . Severity assessment of covid-19 using ct image features and laboratory indices . Phys Med Biol 2021. ; 66 ( 3 ): 035015 . doi: 10.1088/1361-6560/abbf9e [DOI] [PubMed] [Google Scholar]
- 37. Gupta RK, Marks M, Samuels THA, Luintel A, Rampling T, Chowdhury H, et al . Systematic evaluation and external validation of 22 prognostic models among hospitalised adults with covid-19: an observational cohort study . Eur Respir J 2020. ; 56 ( 6 ): 2003498 . doi: 10.1183/13993003.03498-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Soda P, D’Amico NC, Tessadori J, Valbusa G, Guarrasi V, Bortolotto C, et al . AIforCOVID: predicting the clinical outcomes in patients with covid-19 applying ai to chest-x-rays. an italian multicentre study . Med Image Anal 2021. ; 74 : 102216 : S1361-8415(21)00261-9 . doi: 10.1016/j.media.2021.102216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu Q, Zhan X, Zhou Z, Li Y, Xie P, Zhang S, et al . AI-based analysis of ct images for rapid triage of covid-19 patients . NPJ Digit Med 2021. ; 4 : 75 . doi: 10.1038/s41746-021-00446-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hiremath A, Bera K, Yuan L, Vaidya P, Alilou M, Furin J, et al . Integrated clinical and CT based artificial intelligence nomogram for predicting severity and need for ventilator support in covid-19 patients: a multi-site study . IEEE J Biomed Health Inform 2021. ; 25 : 4110 – 18 . doi: 10.1109/JBHI.2021.3103389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim CK, Choi JW, Jiao Z, Wang D, Wu J, Yi TY, et al . An automated covid-19 triage pipeline using artificial intelligence based on chest radiographs and clinical data . NPJ Digit Med 2022. ; 5 : 5 . doi: 10.1038/s41746-021-00546-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kwon YJF, Toussie D, Finkelstein M, Cedillo MA, Maron SZ, Manna S, et al . Combining initial radiographs and clinical variables improves deep learning prognostication in patients with covid-19 from the emergency department . Radiol Artif Intell 2021. ; 3 : e200098 . doi: 10.1148/ryai.2020200098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shamout FE, Shen Y, Wu N, Kaku A, Park J, Makino T, et al . An artificial intelligence system for predicting the deterioration of covid-19 patients in the emergency department . NPJ Digit Med 2021. ; 4 : 80 . doi: 10.1038/s41746-021-00453-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lassau N, Ammari S, Chouzenoux E, Gortais H, Herent P, Devilder M, et al . Integrating deep learning ct-scan model, biological and clinical variables to predict severity of covid-19 patients . Nat Commun 2021. ; 12 ( 1 ): 634 . doi: 10.1038/s41467-020-20657-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Calvillo-Batllés P, Cerdá-Alberich L, Fonfría-Esparcia C, Carreres-Ortega A, Muñoz-Núñez CF, Trilles-Olaso L, et al . Development of severity and mortality prediction models for covid-19 patients at emergency department including the chest x-ray . Radiologia (Engl Ed) 2022; 64 : 214–27: S2173-5107(22)00022-2. 10.1016/j.rxeng.2021.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chamberlin JH, Aquino G, Nance S, Wortham A, Leaphart N, Paladugu N, et al . Automated diagnosis and prognosis of covid-19 pneumonia from initial er chest x-rays using deep learning . BMC Infect Dis 2022. ; 22 ( 1 ): 637 . doi: 10.1186/s12879-022-07617-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gourdeau D, Potvin O, Biem JH, Cloutier F, Abrougui L, Archambault P, et al . Deep learning of chest x-rays can predict mechanical ventilation outcome in icu-admitted covid-19 patients . Sci Rep 2022. ; 12 ( 1 ): 6193 . doi: 10.1038/s41598-022-10136-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Iori M, Di Castelnuovo C, Verzellesi L, Meglioli G, Lippolis DG, Nitrosi A, et al . Mortality prediction of covid-19 patients using radiomic and neural network features extracted from A wide chest x-ray sample size: A robust approach for different medical imbalanced scenarios . Applied Sciences 2022. ; 12 : 3903 . doi: 10.3390/app12083903 [DOI] [Google Scholar]
- 49. Matsumoto T, Walston SL, Walston M, Kabata D, Miki Y, Shiba M, et al . Deep learning-based time-to-death prediction model for covid-19 patients using clinical data and chest radiographs . J Digit Imaging 2022. . doi: 10.1007/s10278-022-00691-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang R, Jiao Z, Yang L, Choi JW, Xiong Z, Halsey K, et al . Artificial intelligence for prediction of covid-19 progression using CT imaging and clinical data . Eur Radiol 2022. ; 32 : 205 – 12 . doi: 10.1007/s00330-021-08049-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Campagnano S, Angelini F, Fonsi GB, Novelli S, Drudi FM . Diagnostic imaging in covid-19 pneumonia: A literature review . J Ultrasound 2021. ; 24 : 383 – 95 . doi: 10.1007/s40477-021-00559-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schwartz LH, Litière S, de Vries E, Ford R, Gwyther S, Mandrekar S, et al . RECIST 1.1-update and clarification: from the recist committee . Eur J Cancer 2016. ; 62 : 132 – 37 : S0959-8049(16)32043-3 . doi: 10.1016/j.ejca.2016.03.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sobin LH . TNM Online . 8th edition . Hoboken, NJ: : Wiley-Blackwell; ; 2016. . doi: 10.1002/9780471420194 [DOI] [Google Scholar]
- 54. Dayan I, Roth HR, Zhong A, Harouni A, Gentili A, Abidin AZ, et al . Federated learning for predicting clinical outcomes in patients with covid-19 . Nat Med 2021. ; 27 : 1735 – 43 . doi: 10.1038/s41591-021-01506-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.