Abstract
Coelimycin P1 and argimycins P are two types of polyketide alkaloids produced by Streptomyces coelicolor and Streptomyces argillaceus, respectively. Their biosynthesis pathways share some early steps that render very similar aminated polyketide chains, diverging the pathways afterwards. By expressing the putative isomerase cpkE and/or the putative epoxidase/dehydrogenase cpkD from the coelimycin P1 gene cluster into S. argillaceus wild type and in argimycin mutant strains, five novel hybrid argimycins were generated. Chemical characterization of those compounds revealed that four of them show unprecedented scaffolds (quinolizidine and pyranopyridine) never found before in the argimycin family of compounds. One of these compounds (argimycin DM104) shows improved antibiotic activity. Noticeable, biosynthesis of these quinolizidine argimycins results from a hybrid pathway created by combining enzymes from two different pathways, which utilizes an aminated polyketide chain as precursor instead of lysine as it occurs for other quinolizidines.
Coelimycin P1 and argimycins P are two polyketide alkaloids produced by Streptomyces coelicolor and Streptomyces argillaceus respectively, whose biosynthesis pathways share some early steps. By expressing cpkD or cpkDE genes from later biosynthetic steps of coelimycin P1 into S. argillaceus strains, five novel hybrid argimycins were generated with unprecedented scaffolds, one of which with improved antibiotic activity.
INTRODUCTION
Alkaloids are a group of complex nitrogen‐containing natural products derived from a variety of sources, including bacteria, fungi, insects, plants, and animals (Cushnie et al., 2014; Rathbone & Bruce, 2002; Sigrist et al., 2015). They have attracted a lot of interest due to their multiple biological activities, such as analgesics, antibiotic, antibacterial‐enhancing, antivirulence, antifungal, anticancer, anti‐inflammatory, or antiplatelet (Ain et al., 2016; Cushnie et al., 2014; Khan et al., 2017; Mondal et al., 2019; Peng et al., 2019; Rathbone & Bruce, 2002). Currently, more than 27,000 alkaloids have been identified from natural sources that according to their structures can be divided into non‐heterocyclic (or atypical/protoalkaloids) and heterocyclic (or typical) alkaloids (Cushnie et al., 2014). Despite the abundant diversity of typical alkaloids, their basic units are limited and only around 20 different rings have been identified so far, being the two most frequently found the indolizidine and piperidine rings (Cushnie et al., 2014; Peng et al., 2016). Most piperidine‐containing alkaloids or true alkaloids derive from amino acids, but the so‐called pseudoalkaloids have a polyketide origin and acquire their nitrogen atoms via transamination reactions (Awodi et al., 2017; Sigrist et al., 2015) (Figure 1).
FIGURE 1.
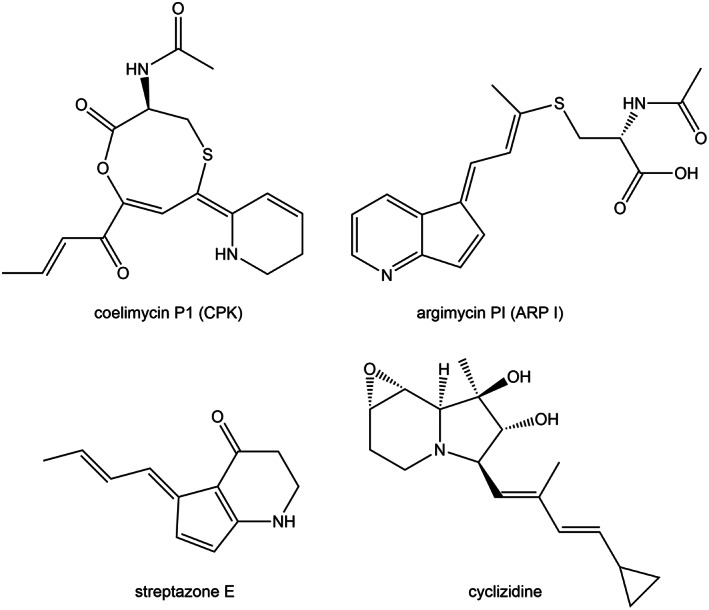
Structures of some piperidine‐containing polyketide alkaloids from Streptomyces.
These polyketides are synthesized by Type I polyketide synthases (PKSs), which are multifunctional enzymes organized into modules. Each module is responsible for one elongation step and contains different domains in which specific enzymatic reactions occur. They contain three essential domains that collaborate to produce a β‐keto ester intermediate: a β‐ketoacyl synthase (KS), an acyltransferase (AT), and an acyl carrier protein (ACP). In addition, those modules may contain optional domains such as a β‐ketoreductase (KR), a dehydratase (DH), and an enoyl reductase (ER), which can act on the resultant β‐keto group originating hydroxy groups, double bonds or methylene groups at different locations of the molecule, thus generating a variety of polyketide chains (Risdian et al., 2019).
Actinobacteria are the most prolific microbial source of natural products, which show diverse chemical structures and complexity (Katz & Baltz, 2016). Several indolizidine and piperidine polyketide alkaloids have been isolated from actinomycetes such as osmanicin (Samy et al., 2019), strepchazolins (Yang et al., 2017), chartrenoline (Liu et al., 2019), cyclizidines (Freer et al., 1982; Jiang et al., 2018), streptopiridines (Groenhagen et al., 2014), coelimycin P1 (Gomez‐Escribano et al., 2012), streptazones (Liu et al., 2013; Puder et al., 2000), or argimycins P (Ye et al., 2017). However, to date, only the biosynthesis pathway for cyclizidine (Huang et al., 2015; Peng et al., 2016), coelimycin P1 (Awodi et al., 2017; Gomez‐Escribano et al., 2012), streptazone E (Ohno et al., 2015) and argimycins P (Ye et al., 2017, 2018) have been partially or totally characterized. These biosynthesis pathways share some common initial steps. They start by synthesizing a polyketide chain by a type I PKS. This chain is released from the PKS as an aldehyde by its thioester reductase (TR) domain, and subsequently reductively aminated by a ω‐transaminase (Awodi et al., 2017; Peng et al., 2016). The resultant intermediate may further proceed through different steps such as cyclization and/or modifications by other tailoring enzymes.
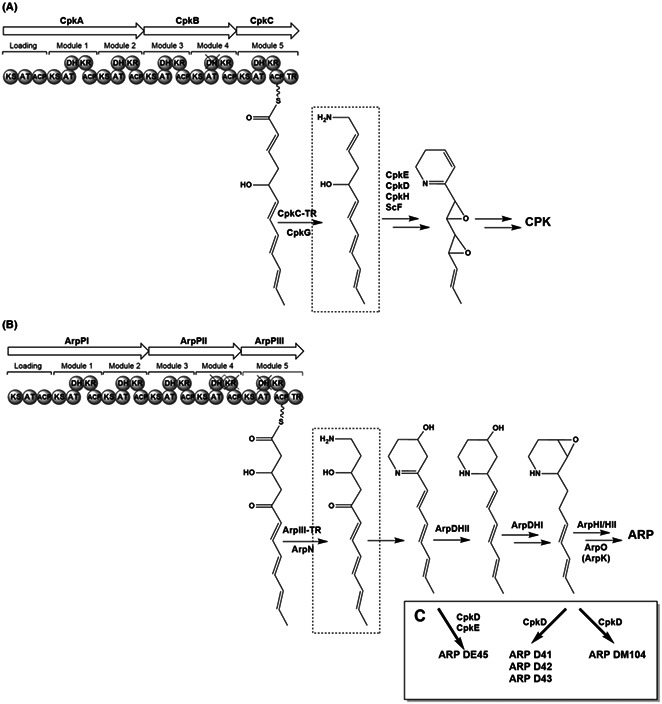
Coelimycin P1 (CPK) and argimycins P (ARP) (Figure 1) are two polyketide alkaloids encoded by the two cryptic biosynthesis gene clusters (BGCs) cpk and arp from S. coelicolor and S. argillaceus, respectively (Gomez‐Escribano et al., 2012; Ye et al., 2017). They contain genes encoding a type I PKS, which synthesize polyketide chains of the same length. After being released, these carbon chains suffer a transamination by CpkG and ArpN respectively, as it has been shown in vitro for CPK, and in vivo for ARP (Awodi et al., 2017; Ye et al., 2017) (Figure 2). Afterwards both pathways diverge. It has been proposed that the biosynthesis pathway for CPK might proceed through a series of oxidations, a cyclodehydration, and the addition of N‐acetylcysteine to give rise the final product coelimycin P1 (Gomez‐Escribano et al., 2012). On the other hand, by characterizing the metabolite profiles of mutants in different arp genes it was shown that the biosynthesis of ARP proceeds through the formation of a piperideine ring and reduction of its imine group, cyclization to give rise to the five‐membered ring, aminoxidation and attachment of a N‐acetylcysteine residue (Ye et al., 2017, 2018).
FIGURE 2.
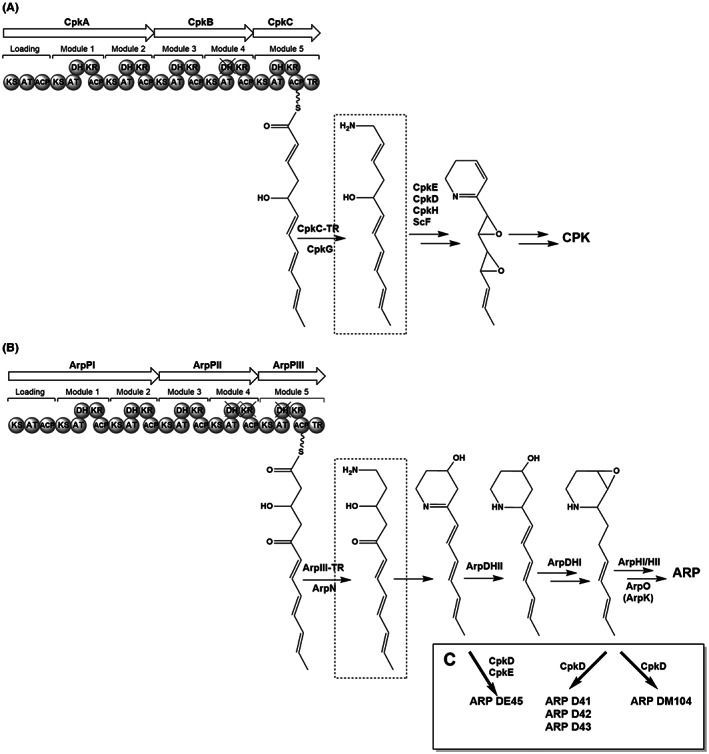
Analysis of PKS modules and biosynthesis steps of coelimycin P1 and argimycins P. (A) coelimycin P1, (B) argimycins P and (C) proposed biosynthesis of hybrid argimycin compounds. ARP, argimycins P; CPK, coelimycin P1.
In this work, we show that by expressing selected cpk genes from S. coelicolor involved in tailoring modifications of CPK, into S. argillaceus wild type and arp mutant strains, novel hybrid ARP compounds were generated, some of which constitute novel scaffolds with improved antibiotic activity.
EXPERIMENTAL PROCEDURES
Strains, culture conditions, plasmids and DNA manipulations
Streptomyces coelicolor M145 (Kieser et al., 2000) was used as source of DNA to amplify cpk genes. S. argillaceus ATCC 12956 and S. argillaceus MARPPIII, MARPDHI, MARPHI, MARPHII and MARPO mutant strains (Ye et al., 2017, 2018) were used as hosts to express cpk genes. Escherichia coli DH10B (Invitrogen) and E. coli ET12567/pUB307 (Kieser et al., 2000) were used as cloning hosts for plasmid propagation and for conjugation experiments, respectively. MA and SM10 media (Fernández et al., 1998; Ye et al., 2017) were used for sporulation and ARP production, respectively. When required, antibiotics were added to media at the following final concentrations: kanamycin (50 μg/ml), nalidixic acid (25 μg/ml), apramycin (25 μg/ml), and thiostrepton (50 μg/ml). Plasmid pCR‐Blunt (Invitrogen) was used for subcloning. Plasmid pSETEc and pSETETc (Cano‐Prieto et al., 2015) were used to express cpk genes in S. argillaceus strains. DNA manipulations, intergeneric conjugations and transformations were carried out according to standard procedures for Streptomyces (Kieser et al., 2000) and for E. coli (Sambrook & Russell, 2001). PCR amplifications were carried out using Herculase II (Stratagene) and 5% dimethyl‐sulphoxide (DMSO). Purified amplicons were sequenced, and sequences were compared with others in databases. Sequence analysis was carried out using BLAST (Altschul et al., 1997). Bioassays were performed as previously described (Vilches et al., 1990).
Plasmid constructs for expressing cpk genes
Several plasmids were constructed to express cpk genes from S. coelicolor into S. argillaceus (Table 1). To this aim, genes were amplified and subcloned under the control of an erythromycin resistance promoter (see Supporting Information).
TABLE 1.
Plasmids and Streptomyces argillaceus recombinant strains generated in this work
| Plasmid | Gene | Recombinant strain |
|---|---|---|
| pSETEc | – |
WT‐pSETEc MARPPIII‐pSETEc |
| pSETEcScF | scF | WT‐scF |
| pSETEcCpkH | cpkH | WT‐cpkH |
| pSETEcCpkD | cpkD |
WT‐cpkD MARPPIII‐cpkD |
| pSETEcCpkE | cpkE |
WT‐cpkE MARPPIII‐cpkE |
| pSETEcCpkDE | cpkD + cpkE |
WT‐cpkDE MARPPIII‐cpkDE |
| pSETETc | – |
MARPDHI‐pSETETc MARPHI‐pSETETc MARPO‐pSETETc |
| pSETETcCpkD | cpkD |
MARPDHI‐cpkD MARPHI‐cpkD MARPO‐cpkD |
| pSETETcCpkDE | cpkD + cpkE |
MARPDHI‐cpkDE MARPHI‐cpkDE MARPO‐cpkDE |
UPLC analysis and purification of argimycin derivatives
Argimycins P derivatives were extracted with n‐butanol and preliminary analyses were carried out as previously reported (Ye et al., 2017). Further analysis of cultures of cpkD, cpkE and cpkDE expressing strains were carried out using a different column, an HSS T3 column (1.8 μm, 2.1 × 100 mm; Waters), with mixtures of acetonitrile and 0.1% trifluoroacetic acid as mobile phase. Samples were eluted with pure 0.1% trifluoroacetic acid for 1 min, followed by a linear gradient from 0% to 60% acetonitrile in 7 min, at a flow rate of 0.5 ml/min and a column temperature of 35°C. Detection and spectral characterization was performed by photodiode array detection and Empower software (Waters). Chromatograms were extracted at 300 and 280 nm.
For purification purposes, strains were grown by a two‐step culture method, as previously described (Fernández et al., 1998), using forty 250‐mililitre Erlenmeyer flasks in the production step. Purification of ARP novel compounds was carried out as previously described (Ye et al., 2017), but using an Atlantis T3 column (3 μm, 2.1 × 150 mm; Waters) and using isocratic chromatography conditions optimized for each compound.
Structural elucidation
Structural elucidation of each compound was carried out using a combination of ESI‐TOF mass spectrometry and NMR spectroscopy (see Supporting Information). HRMS spectra were collected from LC‐DAD‐MS analyses using an Agilent 1200 Rapid Resolution HPLC system equipped with a SB‐C8 column (2.1 × 30 mm, Zorbax) and coupled to a Bruker maXis mass spectrometer. Chromatographic and ionization conditions were identical to those previously described (Martín et al., 2014; Pérez‐Victoria et al., 2016). UV/vis (DAD) spectra were also collected in the same chromatographic analyses. NMR spectra were recorded in CD3OD at 24°C on a Bruker AVANCE III‐500 (500 MHz and 125 MHz for 1H and 13C NMR, respectively) equipped with a 1.7 mm TCI MicroCryoProbe™, using the residual solvent signal as internal reference (δH 3.32 and δC 47.5). The molecular formula obtained from the experimental accurate mass of each compound alongside the analysis of the 1D and 2D NMR spectra rendered the full connectivity and relative stereochemistry of the compounds.
RESULTS
Comparative analysis between coelimycin P1 and argimycins P biosynthesis pathways
A comparative analysis between cpk and arp reveals only five structural genes in common: the ones encoding a type I PKS (cpkA, cpkB, cpkC and arpPI, arpPII, arpPIII), the aminotransferase (cpkG and arpN), and a type II thioesterase (scoT and arpT) (Figure S1). Both PKSs are quite similar (Figure 2); they consist of three subunits with a similar module and domain organization. They only differ at modules 4 and 5: module 4 from CpkB contains a KR domain that is inactive in ArpPII; and module 5 in CpkC contains a DH domain that is inactive in ArpPIII (Figure 2) (Pawlik et al., 2007; Ye et al., 2017). Therefore, these PKSs synthesize polyketide chains of the same length that differ at C‐2, C‐3 and C‐5, which are aminated after being released from those PKSs (Figure 2). However, afterwards both pathways follow different routes that will involve different and specific enzymatic reactions in each pathway. Thus, the cpk BGC contains three genes (scF, cpkH and cpkD) that encode proteins similar to known Flavin‐dependent epoxidases/dehydrogenases, which have been proposed as candidates to catalyse the two epoxidations and/or the oxidation of a hydroxy group of CPK biosynthesis intermediates (Gomez‐Escribano et al., 2012). Since those gene functions are absent in the arp BGC, we hypothesized that expression of some of those genes in S. argillaceus could generate hybrid ARP compounds. Therefore, we selected those genes (as well as cpkE that encodes a possible isomerase) for being expressed into S. argillaceus. The reason why cpkE was included was that its coding region overlaps with that of cpkD, which suggests a functional relationship between them.
Heterologous expression of cpk genes in S. argillaceus wild type strain
scF, cpkH, cpkD, cpkE and cpkDE genes were PCR amplified from S. coelicolor M145 and independently cloned under the control of the erythromycin resistance promoter (ermEp) in the integrative plasmid pSETEc, as described in Supporting Information (Table 1).
The respective constructs (as well as the empty vector) were independently introduced into S. argillaceus wild type strain (WT), and the metabolite profiles of the resultant recombinant strains analysed by UPLC. As control, the different constructs were also expressed into MARPPIII, a mutant in the arpPIII PKS gene that is blocked in the biosynthesis of the ARP polyketide chain (Ye et al., 2017). Metabolite profiles produced by MARPPIII recombinant strains were used to discriminate differential peaks not related to ARP.
Expression of either scF or cpkH into S. argillaceus WT did not result in production of any detectable new compound (data not shown). However, when either cpkD or cpkDE was expressed in S. argillaceus WT, some differential peaks were detected at 300 nm that were absent in the control strain containing the empty vector (Figure 3A). Peak c was detected in both strains WT‐cpkD and WT‐cpkDE, while peak a was only observed in WT‐cpkDE. Expression of cpkE alone did not result in production of neither of these peaks (Figure 3A). These two peaks were not produced when either cpkD or cpkDE were expressed in MARPPIII mutant (MARPPIII‐cpkD and MARPPIII‐cpkDE; Figure 3B), which indicates that compounds in peaks a and c were ARP derivatives. These results suggest formation of novel hybrid ARP compounds, and show that formation of compounds in peak c only requires expression of cpkD, while both CpkD and CpkE are necessary for production of compounds in peak a, indicating that CpkD would act before than CpkE. In addition, a careful analysis of compounds in peak b from WT‐cpkD and WT‐cpkDE revealed they contain a new compound with a similar absorption spectrum to those compounds in peak c. This newly identified compound was absent in the control strain (WT‐pSETEc), in WT‐cpkE, and in all MARPPIII recombinant strains (Figure 3), indicating it was a hybrid ARP compound.
FIGURE 3.
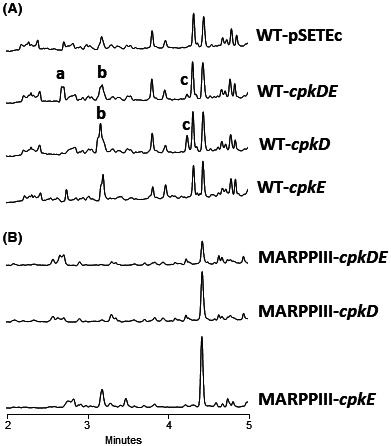
UPLC chromatograms at 300 nm of butanol extracts of S. argillaceus strains expressing cpk genes. (A) S. argillaceus wild type strain (WT) and (B) S. argillaceus MARPPIII. Peaks a to c contain novel argimycins (ARP) compounds identified in this work: ARP DE45 (peak a), ARP D43 and ARP D44 (peak b) and ARP D41 and ARP D42 (peak c).
Heterologous expression of cpkD , cpkE and cpkDE in S. argillaceus mutant strains
In light of the positive results obtained above by expressing cpkD and cpkDE in S. argillaceus WT strain, and taking advantage of the existence of a collection of knockout mutant strains in different structural genes of arp BGC (Ye et al., 2018), we attempted to express those genes in some of these mutant strains. This had two purposes: (i) to generate new ARP derivatives resulting from the enzymatic activity of Cpk proteins on compounds accumulated by those mutant strains and (ii) to determine the contribution of arp gene products to the formation of the different new hybrid compounds. We selected S. argillaceus MARPDHI, S. argillaceus MARPHI, S. argillaceus MARPHII and S. argillaceus MARPO, which are mutants in specific steps of ARP biosynthesis that accumulate different biosynthesis intermediates (Ye et al., 2018). To express cpkD and cpkDE in these mutants, it was necessary to generate two new constructs (pSETETcCpkD and pSETETcCpkDE) using as a vector pSETETc that confers resistance to thiostrepton, since the constructs used before (pSETEcCpkD and pSETEcCpkDE) confer resistance to apramycin and the S. argillaceus mutants already display resistance to this antibiotic (Ye et al., 2018).
MARPDHI is a knockout mutant strain in arpDHI. This gene encodes a putative acyl‐CoA dehydrogenase, proposed to oxidize a biosynthesis intermediate at an early step of ARP biosynthesis, before the five‐membered ring formation (Ye et al., 2018). When cpkD or cpkDE were expressed into MARPDHI, the resultant strains MARPDHI‐cpkD and MARPDHI‐cpkDE did not produce any of those ARP derivatives contained in peaks b and c (Figure 4A). This indicates that production of hybrid ARPs in those peaks requires expression of arpDHI. However, peak a could be detected in chromatograms from MARPDHI‐cpkDE (Figure 4A), which indicates that production of this peak is independent of expression of arpDHI.
FIGURE 4.
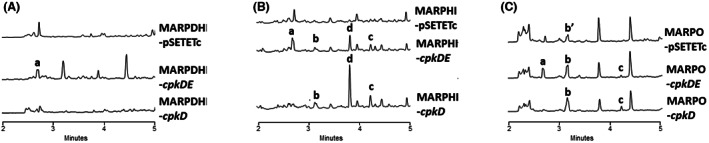
UPLC chromatograms at 300 nm of butanol extracts of S. argillaceus mutant strains expressing cpk genes. (A) S. argillaceus MARPDHI, (B) S. argillaceus MARPHI and (C) S. argillaceus MARPO. Peak b’ corresponds to ARP D44 and peak d to ARP DM104.
MARPHI and MARPHII are knockout S. argillaceus mutants in arpHI and arpHII, respectively. These genes encode putative cyclases that are essential for the formation of the five‐membered ring of bicyclic ARP (Ye et al., 2018). Previous studies have shown that both mutant strains produce the same metabolite profiles and accumulate the same ARP compounds (Ye et al., 2018). Likewise, expression of cpkD and cpkDE genes in MARPHI and MAPRHII led to the production of the same compounds in both strains. Here for simplification we only show chromatograms corresponding to MARPHI recombinant strains. All peaks previously identified in S. argillaceus WT either expressing cpkD or cpkDE (peaks a, b and c; Figure 3A) were also detected in the corresponding MARPHI‐cpkD and MARPHI‐cpkDE strains (Figure 4B), indicating that neither ArpHI nor ArpHII enzymatic activities were essential for their biosynthesis. Additionally, a new peak (peak d) was identified at 280 nm UV/vis spectrum in MARPHI‐cpkD and MARPHI‐cpkDE, which was absent in the control strain (Figure 4B).
MARPO is a knockout mutant in arpO, which encodes a putative oxygenase necessary for the biosynthesis of ARP PI and ARP PII (Ye et al., 2018). All compounds identified in peaks a to c were also identified in the MARPO strains expressing cpkD and cpkDE (Figure 4C), indicating that ArpO enzymatic activity is likewise not necessary for their biosynthesis. No other additional ARP compound was identified. Interestingly, MARPO containing the empty vector shows a peak (peak b’ in Figure 4C) with a UV/vis spectrum with a maximum at 302 nm. A mass spectrometry analysis of this peak revealed the same protonated adduct (192 [M + H]+) as ARP D44 (Figure 5; see below), suggesting that albeit being a new ARP derivative identified in this work, ARP D44 does not result from the enzymatic activity of CpkD on ARP biosynthesis pathway, and therefore is not a hybrid compound.
FIGURE 5.
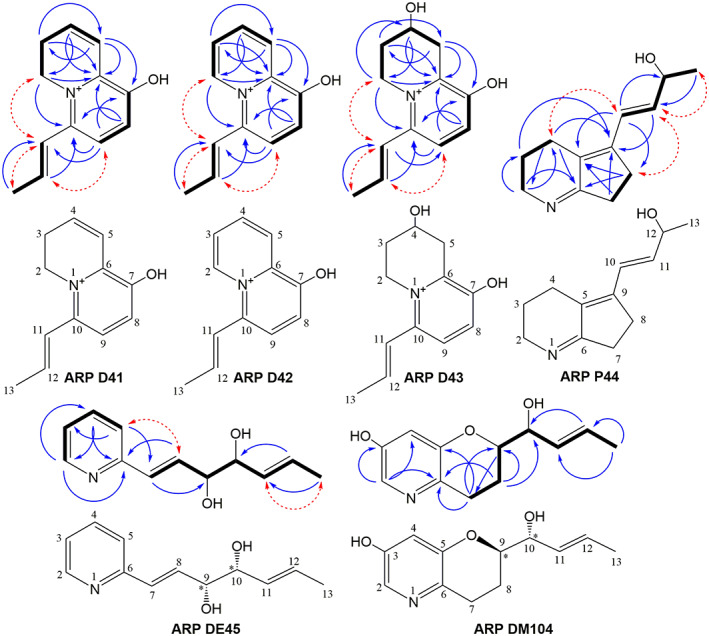
Structural elucidation of novel argimycin compounds. Structures and key COSY (bold bonds), HMBC (blue arrows) and NOESY (dashed red arrows) correlations used to determine the chemical structure of the novel argimycin P (ARP) derivatives identified in this work.
Purification, structural elucidation and bioactivity of novel argimycins
Compounds from peaks a to d were purified and chemically characterized by MS and NMR (see Supporting Information). Compound from peak a was purified from S. argillaceus WT‐cpkDE and was named ARP DE45. Compounds from peak c were purified from S. argillaceus WT‐cpkD. This peak contained two different compounds, ARP D41 and ARP D42, which were purified by HPLC and independently characterized. Peak b contains one major compound, ARP D43, and two minor ones, whose separation proved to be impossible in all conditions tested. The structure of one of the minor compounds, ARP D44, could also be determined. These compounds were purified from two strains, S. argillaceus WT‐cpkD and S. argillaceus WT‐cpkDE, what confirmed that compound composition of both peaks b was similar in both strains. The compound contained in peak d was purified from MARPHI‐cpkD and was named ARP DM104. The purification procedure afforded ARP D41 (1.5 mg), ARP D42 (1.3 mg), ARP D43 (4.1 mg), ARP D44 (1.9 mg), ARP DE45 (0.9 mg) and ARP DM104 (0.8 mg).
ARP D41 was assigned the molecular formula C12H14NO+ based on the observed M+ ion at m/z = 188.1076 (calcd. For C12H14NO+ = 188.1070, Δ = 3.2 ppm) which is in the range of that observed for argimycins P encoded by the cryptic gene cluster arp of S. argillaceus (Ye et al., 2017, 2018) and indicated seven degrees of unsaturation. Analysis of the 1H and HSQC NMR spectra revealed a total of 13 non‐exchangeable hydrogens. A further experiment was carried out to unequivocally confirm the number of exchangeable hydrogens by HRMS analysis. The NMR sample, prepared in CD3OD and thus displaying D/H exchange of the exchangeable hydrogens in the molecule, was diluted with the same solvent, and analysed by HRMS employing direct infusion. The new M+ ion was observed at m/z = 189.1145 corresponding to C12H13DNO+ (calcd. For C12H13DNO+ = 189.1133, Δ = 6.3 ppm) confirming the presence of only one exchangeable hydrogen in the compound. The HSQC spectrum revealed the presence of five sp 2 methines, two aliphatic methylenes (one likely bound to nitrogen), and one aliphatic methyl group. The key correlations observed in the COSY spectrum identified the different spin systems, which were connected via the long‐range correlations observed in the HMBC spectrum (Figure 5), which likewise was essential to unambiguously determine the position of the hydroxy group to finally determine the connectivity of the compound. Key NOESY correlations (Figure 5) provided further evidence of the structure and, alongside the coupling constants observed (J H11‐H12 = 15.4 Hz), allowed determining the E stereochemistry of the double bond in the exocyclic chain. The compound is positively charged due to the presence of a quaternized nitrogen and displays the expected chemical shifts for such a dihydroquinozilinium structure. The trifluoracetate counter ion (derived from the use of trifluoroacetic acid in the chromatographic purification) must be present in the isolated compound but it has no observable signals by 1H NMR. The similarity of the UV–vis (DAD) spectrum with that reported for 3,4‐dihydroquinozilinium iodide (Boekerheide & Gall, 1954) further corroborated the determined structure.
The UV–vis (DAD) spectrum of ARP D42 was very similar to that obtained for ARP D41, suggesting their structural relationship and a similar pattern of conjugation. Assuming a cationic form of the molecule with a quaternized nitrogen, the compound was assigned the molecular formula C12H12NO+ based on the observed M+ ion at m/z = 186.0920 (calcd. For C12H12NO+ = 186.0913, Δ = 3.7 ppm), indicating eight degrees of unsaturation. A deuterium exchange experiment again confirmed that the molecule contained just one exchangeable hydrogen due to the M+ ion observed at m/z = 187.0989 (calcd. For C12H11DNO+ = 187.0976, Δ = 6.9 ppm). Comparing its 1H and HSQC NMR spectra with those of ARP D41, it was clear the disappearance of the two sp 3 methylenes, replaced by signals corresponding to two aromatic hydrogens, suggesting the presence of a quinozilinium core rather than the dihydroquinozilinium present in ARP D41. Key correlations observed in its COSY and HMBC spectra (Figure 5) confirmed that, as expected, the compound was identical to ARP D41 but carrying an extra double bond that turns aromatic the second ring. Likewise, again key NOESY correlations (Figure 5) provided further evidence of the structure and, alongside the J H11‐H12 coupling constant of 15.4 Hz observed, allowed determining the configuration of the double bond in the exocyclic chain as E. The compound is positively charged and displays the expected NMR chemical shifts and UV–vis (DAD) spectrum for such a quinozilinium structure (Boekerheide & Gall, 1954).
The UV–vis (DAD) spectrum of ARP D43 was similar to that obtained for ARP D41 and ARP D42, suggesting their structural relationship and a similar pattern of conjugation. Again, assuming a cationic form of the molecule with a quaternized nitrogen, the compound was assigned the molecular formula C12H16NO2 + based on the observed M+ ion at m/z = 206.1181 (calcd. For C12H16NO2 + = 206.1176, Δ = 2.4 ppm), indicating six degrees of unsaturation. Its 1H NMR spectrum showed various signals very similar to those observed in ARP D41 and ARP D42, confirming their structural relationship. Integration of the spectrum revealed 14 non‐exchangeable hydrogens, indicating that—according to the molecular formula—two hydrogens were exchangeable and likely corresponded to hydroxy or phenolic groups. Additional analysis of the HSQC spectrum indicated a total of four sp 2 protons (two of them of aromatic nature), three aliphatic methylene groups (one of them bound to nitrogen), one aliphatic oxygenated methine and an allylic methyl. This information, together with the determined molecular formula and comparisons with ARP D41 were in agreement with a common backbone but losing a double bond of the non‐aromatic ring and gaining a hydroxy substituent in such cycle. The observed key COSY and HMBC correlations (Figure 5) confirmed the backbone connectivity and unambiguously determined the position of the new hydroxy group. Likewise, the key NOESY correlations provided further evidence on the structure and, alongside the J H11‐H12 coupling constant of 15.4 Hz observed, again stablished the E configuration of the double bond in the exocyclic chain. The absolute configuration of the single hydroxylated methine chiral center was not determined. The compound is positively charged and displays the expected chemical shifts and UV–vis (DAD) spectrum for such a tetrahydroquinozilinium structure (Boekerheide & Gall, 1954).
ARP D44 appeared as a minor component in a preparative HPLC fraction containing ARP D43 as main component. Its molecular formula could be stablished as C12H17NO+ based on the observed ion [M + H]+ at m/z = 192.1383 (calcd. For C12H18NO+ = 192.1383, Δm = 0 ppm), indicating five degrees of unsaturation. Fortunately, the NMR signals of this minor component did not overlap with the signals of ARP D43 (main component in the sample) and were intense enough for enabling structural elucidation by 2D NMR. The 1H and HSQC NMR spectra revealed that this minor component contained two olefinic hydrogens, one oxygenated methine, five aliphatic methylenes (one bound to nitrogen) and an aliphatic methyl instead of the vinyl methyl found in the previously described compounds. The different spin systems were determined from the cross‐peaks observed in its COSY spectrum and the long‐range correlations observed in the HMBC spectrum were employed to connect those and establish its final connectivity (Figure 5). The E configuration of the double bond in the lateral chain is based on the J H10‐H11 large coupling constant observed between the two olefinic hydrogens (15.7 Hz). Key NOESY correlations (Figure 5) provided further evidence for the structure proposed. The absolute configuration of the single hydroxylated methine chiral center was not determined.
ARP DE45 was assigned a molecular formula of C12H15NO2 based on the observed ion [M + H]+ at m/z = 206.1177 (calcd. For C12H16NO2 + = 206.1176, Δ = 0.5 ppm), indicating six degrees of unsaturation. Its 1H and HSQC NMR spectra revealed four aromatic, pyridine‐like, hydrogens plus four olefinic hydrogens. Two oxygenated methine groups were observed and also an aliphatic methyl (in vinylic position). Two spins systems were identified in the COSY spectrum (Figure 5). One corresponds to a monosubstituted pyridine (in the carbon contiguous to the nitrogen) and the other to the hydroxylated aliphatic chain substituent. The key long‐range correlations observed in the HMBC spectrum (Figure 5) confirmed this connectivity. The configuration of the double bonds was established based on the observed coupling constants (JH7‐H8 = 16 Hz, JH11‐H12 = 15.3 Hz) and key NOESY correlations (Figure 5). The target compound thus shares the same backbone as nigrifactin (Terashima et al., 1969). Not surprisingly, its UV–vis (DAD) spectrum also showed strong similarity with that of 2‐vinylpyridine in acidic methanol (Akito et al., 1993). The relative configuration of the vicinal diol moiety was stablished as syn (9R*, 10R*) by comparison of the coupling constants of H9 with those reported for the equivalent hydrogens in the syn and anti diastereomers of the analogous synthetic (1 E)‐1‐phenyl‐hexa‐1,5‐dien‐3,4‐diol (Lombardo et al., 2001). The absolute configuration of the diol motif remained undetermined.
ARP DM104 was assigned a molecular formula of C12H15NO3 based on the observed ion [M + H]+ at 222.1135 (calcd. For C12H16NO3 + = 222.1125, Δm = 4.5 ppm), indicating six degrees of unsaturation. Its UV–vis (DAD) spectrum differed from that of the compounds described above. Analysis of its 1H and HSQC NMR spectra revealed two aromatic and two olefinic hydrogens, two oxygenated methine groups, two aliphatic methylenes and finally an aliphatic methyl in vinylic position. Two spin systems were identified in the COSY spectrum, one belonging to an aromatic ring and the second corresponding to an aliphatic substituent of the former (Figure 5). The COSY correlation of the two aromatic hydrogens was weak, indicating a small J coupling typical of their meta positioning. The observed key HMBC correlations together with the molecular formula and the 13C NMR spectrum allowed stablishing the connectivity of the compound and secured the ring closure between C5 and C9 via and ether bridge (Figure 5). Since the two olefinic hydrogens are essentially isochronous, the configuration of this double bond could not be established from NOE or J coupling data. Nevertheless, it is safe to assume an E configuration for such double bond as for the argimycins described above, assuming a common biosynthesis route for that moiety. The absence of NOE correlation between H10 and methyl H13 further supports this proposal. The relative configuration of the two vicinal oxymethines could not be established due to the lack of measurable coupling constants (because signal broadening and overlap with the water signal). However, it can be assumed to be the same syn relative configuration (9R*, 10R*) indicated before for ARP DE45, again based on biosynthetic arguments. The absolute configuration was not determined but should match that of ARP DE45.
The antibiotic activity of all novel ARP compounds was tested against M. luteus. It is worth mentioning that ARP D43 sample contains a mixture of three compounds, being the major one ARP D43. In this assay, we also included ARP PIII (also named as nigrifactin) as control, since it had shown the highest antibiotic activity among all ARP tested before (Ye et al., 2017). Among all compounds assayed, ARP DM104 was the only one that showed a clear antibiotic activity at the highest amount of compound tested (100 μg), while the same amount of ARP PIII did not show antibiotic activity (Figure 6).
FIGURE 6.
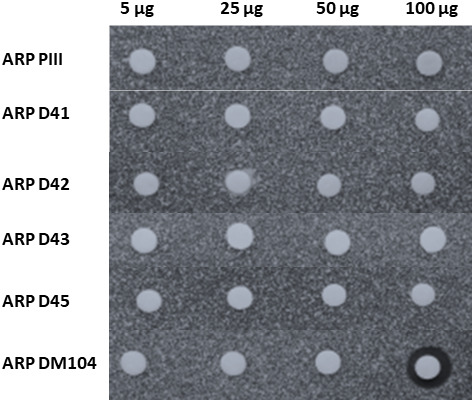
Bioassay of novel argimycin compounds. Bioassay against Micrococcus luteus. ARP, argimycins P.
DISCUSSION
The ARP family of compounds includes piperidine and pyridine‐containing alkaloids, some of which contain one heterocycle attached to a polyene chain, while others also contain a five‐membered fused ring with a shorter polyene chain. To date, 13 ARP compounds have been identified and chemically characterized from cultures of S. argillaceus WT and knockout mutants in arp genes (Ye et al., 2017, 2018). In this work, we have been able to expand the number and scaffold diversity of ARP family through combinatorial biosynthesis. To do that, we took advantage of the fact that the ARP and CPK biosynthesis pathways have similar initial steps that lead to structurally similar aminated polyketide chains, diverting their pathways afterwards (Figure 2). Therefore, by expressing cpkD or cpkDE genes involved in later steps of CPK biosynthesis into S. argillaceus WT novel ARP derivatives were generated, either containing a quinolizidine backbone (ARP D41, ARP D42 and ARP D43) or a pyridine ring (ARP DE45) (Figure 5). In addition, by expressing cpkD into S. argillaceus MARPHI mutant strain the novel compound ARP DM104 with a pyranopyridine scaffold was produced. Although the precise steps for the biosynthesis of these novel ARP remain unclear, the heterologous expression of cpkD and cpkDE genes in S. argillaceus knockout mutants have revealed some hints about the involvement of some Arp proteins in the biosynthesis of these novel compounds (Figure 2C). Thus, ARP DE45 would derive from an early dehydrated ARP biosynthesis intermediate synthesized by the ArpP PKS and the ArpN ϖ‐transaminase; and the quinolizidine and pyranopyridine ARP compounds would do from an ArpDHI product. Also, it is discarded a role of cyclases ArpHI/HII in the cyclization events leading to the formation of quinolizidine and pyranopyridine scaffolds, pointing out that processes leading to formation of their second ring would occur spontaneously and not by a dedicated Arp enzyme.
Based on the structure of these novel ARP derivatives, it is proposed that CpkD would introduce an epoxide group at the C9‐C10 double bond either on an ArpDHI product (ARP D41‐ARP 43 and ARP DM104) or on an earlier intermediate (ARP DE45). CpkE might be responsible for the subsequent hydrolysation of the epoxide on that generated intermediate to produce ARP DE45 (Figure 2C). CpkE has been proposed to be a putative isomerase whose action would facilitate formation of the heterocycle in CPK (Gomez‐Escribano et al., 2012). However, a BlastP search against the Protein Data Bank (PDB) reveals similarity with epoxide hydrolases, activity that fits better to its proposed role in the formation of ARP DE45. Moreover, individual expression of cpkE does not lead to any new compound, suggesting that its coding enzyme would act after CpkD, what would support a coordinated activity between CpkD and CpkE. Since the other two flavin‐dependent epoxidases/dehydrogenases (CpkH and ScF) encoded by the cpk BGC do not modify the ARP biosynthetic pathway when expressed alone in S. argillaceus, it is proposed that in CPK biosynthesis, CpkD would be involved in epoxidation of the C9‐C10 double bond in the corresponding CPK intermediate, which would occur before epoxidation of C7‐C8 double bond by either CpkH or ScF.
Four of the novel ARP derivatives are bicyclic showing unprecedented scaffolds never found before in the ARP family of compounds. Thus, ARP D41 to ARP D43 show a bicyclic quinolizidine scaffold with an unusual iminium ion, in which the six‐membered fused ring is formed by an unnatural cyclization between C10 and the nitrogen atom in the heterocycle, instead of the natural cyclization between C9 and C5 that leads to the formation of five‐membered fused ring of bicyclic ARPs. All these three quinolizidine ARPs share a hydroxy group at C7 but differ at the heterocycle. Quinolizidines constitute a major class of alkaloid compounds that are mainly produced by plants, in which the biosynthesis pathway starts with the amino acid lysine (Bunsupa et al., 2012; Michael, 2008). Noticeable, in this work we have set up an alternative pathway for the biosynthesis of quinolizidine compounds using as precursor an aminated polyketide chain (Figure 2C), by creating a hybrid pathway combining genes from two different but related BGCs each of which unable to direct the biosynthesis of a quinolizidine (Gomez‐Escribano et al., 2012; Ye et al., 2017). Another novel scaffold in ARPs corresponds to ARP DM104, which contains a pyranopyridine backbone. Most probably this scaffold would derive from a biosynthesis intermediate accumulated by mutant MARPHI and produced as result of ArpDHI activity, which after being modified by CpkD would suffer dehydration of hydroxy groups at C5 and C9 (Figure 2C).
We have previously reported that most ARP do not show antibiotic activity, except ARP PIII that shows a very weak antibiotic activity against M. luteus (Ye et al., 2017). Now we report that ARP DM104, apart from bearing an unprecedented scaffold in ARP compounds, shows improved activity against M. luteus. Thus, combinatorial biosynthesis applied to polyketide alkaloids has proven as a successful strategy for the generation of structural diversity and enhanced bioactivity in ARP, paving the way for the exploitation of an unexplored source of new compounds.
AUTHOR CONTRIBUTIONS
Suhui Ye: Conceptualization (equal); investigation (equal); writing – original draft (equal). Giovanni Ballin: Investigation (equal). Ignacio Pérez‐Victoria: Investigation (equal). Alfredo Férnandez Braña: Investigation (supporting). Jesús Martín: Investigation (supporting). Fernando Reyes: Investigation (supporting). Jose A Salas: Conceptualization (equal); funding acquisition (equal). Carmen Méndez: Conceptualization (equal); formal analysis (lead); funding acquisition (lead); supervision (lead); writing – original draft (lead); writing – review and editing (lead).
FUNDING INFORMATION
This work was supported by grants to CM from the Spanish Ministry of Economy and Competitiveness, MINECO (Grants BIO2014‐56752‐R and PIM2010EEI‐00752) and by the grant “Apoyo a Grupos de Excelencia”, Principado de Asturias‐FEDER (FC‐15‐GRUPIN14‐014).
CONFLICT OF INTEREST
The authors declare no competing financial interest.
Supporting information
Appendix S1
ACKNOWLEDGMENTS
This work was supported by grants to CM from the Spanish Ministry of Economy and Competitiveness, MINECO (Grants BIO2014‐56752‐R and PIM2010EEI‐00752) and by the grant “Apoyo a Grupos de Excelencia”, Principado de Asturias‐FEDER (FC‐15‐GRUPIN14‐014).
Ye, S. , Ballin, G. , Pérez‐Victoria, I. , Braña, A.F. , Martín, J. & Reyes, F. et al. (2022) Combinatorial biosynthesis yields novel hybrid argimycin P alkaloids with diverse scaffolds in Streptomyces argillaceus . Microbial Biotechnology, 15, 2905–2916. Available from: 10.1111/1751-7915.14167
REFERENCES
- Ain, Q.U. , Khan, H. , Mubarak, M.S. & Pervaiz, A. (2016) Plant alkaloids as antiplatelet agent: drugs of the future in the light of recent developments. Frontiers in Pharmacology, 7, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akito, I. , Tatsumi, U. & Setsuo, T. (1993) Photoinduced alkoxylation of 2‐vinylpyridinium ion. Bulletin of the Chemical Society of Japan, 66, 1580–1582. [Google Scholar]
- Altschul, S.F. , Madden, T.L. , Schäffer, A.A. , Zhang, J. , Zhang, Z. , Miller, W. et al. (1997) Gapped BLAST and PSI‐BLAST: a new generation of protein database search programs. Nucleic Acids Research, 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awodi, U.R. , Ronan, J.L. , Masschelein, J. , de los Santos, E.L.C. & Challis, G.L. (2017) Thioester reduction and aldehyde transamination are universal steps in actinobacterial polyketide alkaloid biosynthesis. Chemical Science, 8, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekerheide, V. & Gall, W.G. (1954) A synthesis of quinolizinium and dehydroquinolizinium derivatives. Journal of the American Chemical Society, 76, 1832–1836. [Google Scholar]
- Bunsupa, S. , Yamazaki, M. & Saito, K. (2012) Quinolizidine alkaloid biosynthesis: recent advances and future prospects. Frontiers in Plant Science, 3, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano‐Prieto, C. , García‐Salcedo, R. , Sánchez‐Hidalgo, M. , Braña, A.F. , Fiedler, H.P. , Méndez, C. et al. (2015) Genome mining of Streptomyces sp. Tü 6176: characterization of the nataxazole biosynthesis pathway. Chembiochem, 16, 1461–1473. [DOI] [PubMed] [Google Scholar]
- Cushnie, T.P. , Cushnie, B. & Lamb, A.J. (2014) Alkaloids: an overview of their antibacterial, antibiotic‐enhancing and antivirulence activities. International Journal of Antimicrobial Agents, 44, 377–386. [DOI] [PubMed] [Google Scholar]
- Fernández, E. , Weibbach, U. , Sánchez Reillo, C. , Braña, A.F. , Méndez, C. & Salas, J.A. (1998) Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. Journal of Bacteriology, 180, 4929–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freer, A.A. , Gardner, D. , Greatbanks, D. , Poyser, J.P. & Sim, G.A. (1982) Structure of cyclizidine (antibiotic M146791): X‐ray crystal structure of an indolizidinediol metabolite bearing a unique cyclopropyl side‐chain. Journal of the Chemical Society, Chemical Communications, 20, 1160–1162. [Google Scholar]
- Gomez‐Escribano, J.P. , Song, L. , Fox, D.J. , Yeo, V. , Bibb, M.J. & Challis, G.L. (2012) Structure and biosynthesis of the unusual polyketide alkaloid coelimycin P1, a metabolic product of the cpk gene cluster of Streptomyces coelicolor M145. Chemical Science, 3, 2716–2720. [Google Scholar]
- Groenhagen, U. , Maczka, M. , Dickschat, J.S. & Schulz, S. (2014) Streptopyridines, volatile pyridine alkaloids produced by Streptomyces sp. FORM5. Beilstein Journal of Organic Chemistry, 10, 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W. , Kim, S.J. , Liu, J. & Zhang, W. (2015) Identification of the polyketide biosynthetic machinery for the indolizidine alkaloid cyclizidine. Organic Letters, 17, 5344–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y.J. , Li, J.Q. , Zhang, H.J. , Ding, W.J. & Ma, Z.J. (2018) Cyclizidine‐type alkaloids from Streptomyces sp. HNA39. Journal of Natural Products, 81, 394–399. [DOI] [PubMed] [Google Scholar]
- Katz, L. & Baltz, R.H. (2016) Natural products discovery: past, present, and future. Journal of Industrial Microbiology & Biotechnology, 43, 155–176. [DOI] [PubMed] [Google Scholar]
- Khan, H. , Mubarak, M.S. & Amin, S. (2017) Antifungal potential of alkaloids as an emerging therapeutic target. Current Drug Targets, 18, 1825–1835. [DOI] [PubMed] [Google Scholar]
- Kieser, T. , Bibb, M.J. , Buttner, M.J. , Chater, K.F. & Hopwood, D.A. (2000) Practical Streptomyces genetics. Norwich: The John Innes Foundation. [Google Scholar]
- Liu, C. , Yang, C. , Zeng, Y. , Shi, J. , Li, L. , Li, W. et al. (2019) Chartrenoline, a novel alkaloid isolated from a marine Streptomyces chartreusis NA02069. Chinese Chemical Letters, 30, 44–46. [Google Scholar]
- Liu, Q.F. , Wang, J.D. , Wang, X.J. , Liu, C.X. , Zhang, J. , Pang, Y.W. et al. (2013) Two new piperidine alkaloids from Streptomyces sp. NEAU‐Z4. Journal of Asian Natural Products Research, 15, 221–224. [DOI] [PubMed] [Google Scholar]
- Lombardo, M. , Girotti, R. , Morganti, S. & Trombini, C. (2001) A new protocol for the acetoxyallylation of aldehydes mediated by indium in THF. Organic Letters, 3, 2981–2983. [DOI] [PubMed] [Google Scholar]
- Martín, J. , Crespo, G. , González‐Menéndez, V. , Pérez‐Moreno, G. , Sánchez‐Carrasco, P. , Pérez‐Victoria, I. et al. (2014) MDN‐0104, an antiplasmodial betaine lipid from Heterospora chenopodii . Journal of Natural Products, 77, 2118–2123. [DOI] [PubMed] [Google Scholar]
- Michael, J.P. (2008) Indolizidine and quinolizidine alkaloids. Natural Product Reports, 25, 139–165. [DOI] [PubMed] [Google Scholar]
- Mondal, A. , Gandhi, A. , Fimognari, C. , Atanasov, A.G. & Bishayee, A. (2019) Alkaloids for cancer prevention and therapy: current progress and future perspectives. European Journal of Pharmacology, 858, 172472. [DOI] [PubMed] [Google Scholar]
- Ohno, S. , Katsuyama, Y. , Tajima, Y. , Izumikawa, M. , Takagi, M. , Fujie, M. et al. (2015) Identification and characterization of the streptazone E biosynthetic gene cluster in Streptomyces sp. MSC090213JE08. Chembiochem, 16, 2385–2391. [DOI] [PubMed] [Google Scholar]
- Pawlik, K. , Kotowska, M. , Chater, K.F. , Kuczek, K. & Takano, E. (2007) A cryptic type I polyketide synthase (cpk) gene cluster in Streptomyces coelicolor A3(2). Archives of Microbiology, 187, 87–99. [DOI] [PubMed] [Google Scholar]
- Peng, H. , Wei, E. , Wang, J. , Zhang, Y. , Cheng, L. , Ma, H. et al. (2016) Deciphering piperidine formation in polyketide‐derived indolizidines reveals a thioester reduction, transamination, and unusual imine reduction process. ACS Chemical Biology, 11, 3278–3283. [DOI] [PubMed] [Google Scholar]
- Peng, J. , Zheng, T.T. , Li, X. , Liang, Y. , Wang, L.J. , Huang, Y.C. et al. (2019) Plant‐derived alkaloids: the promising disease‐modifying agents for inflammatory bowel disease. Frontiers in Pharmacology, 10, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Victoria, I. , Martín, J. & Reyes, F. (2016) Combined LC/UV/MS and NMR strategies for the dereplication of marine natural products. Planta Medica, 82, 857–861. [DOI] [PubMed] [Google Scholar]
- Puder, C. , Krastel, P. & Zeeck, A. (2000) Streptazones A, B(1), B(2), C, and D: new piperidine alkaloids from streptomycetes. Journal of Natural Products, 63, 1258–1260. [DOI] [PubMed] [Google Scholar]
- Rathbone, D.A. & Bruce, N.C. (2002) Microbial transformation of alkaloids. Current Opinion in Microbiology, 5, 274–281. [DOI] [PubMed] [Google Scholar]
- Risdian, C. , Mozef, T. & Wink, J. (2019) Biosynthesis of polyketides in streptomyces. Microorganisms, 7, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. & Russell, D.W. (2001) Molecular cloning: a laboratory manual, 3rd edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Samy, M.N. , Le Goff, G. , Lopes, P. , Gergousaki, K. , Gumeni, S. , Almeida, C. et al. (2019) Osmanicin, a polyketide akaloid isolated from Streptomyces osmaniensis CA‐244599 inhibits elastase in human fibroblasts. Molecules, 24, 2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist, R. , da Costa, B.Z. , Marsaioli, A.J. & de Oliveira, L.G. (2015) Nature‐inspired enzymatic cascades to build valuable compounds. Biotechnology Advances, 33, 394–411. [DOI] [PubMed] [Google Scholar]
- Terashima, T. , Kuroda, Y. & Kaneko, Y. (1969) Studies on a new alkaloid of Streptomyces structure of nigrifactin. Tetrahedron Letters, 10, 2535–2537. [DOI] [PubMed] [Google Scholar]
- Vilches, C. , Méndez, C. , Hardisson, C. & Salas, J.A. (1990) Biosynthesis of oleandomycin by Streptomyces antibioticus: influence of nutritional conditions and development of resistance. Journal of General Microbiology, 136, 1447–1454. [DOI] [PubMed] [Google Scholar]
- Yang, C.L. , Wang, Y.S. , Liu, C.L. , Zeng, Y.J. , Cheng, P. , Jiao, R.H. et al. (2017) Strepchazolins A and B: two new alkaloids from a marine Streptomyces chartreusis NA02069. Marine Drugs, 15, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, S. , Braña, A.F. , González‐Sabín, J. , Morís, F. , Olano, C. , Salas, J.A. et al. (2018) New insights into the biosynthesis pathway of polyketide alkaloid argimycins P in Streptomyces argillaceus . Frontiers in Microbiology, 9, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, S. , Molloy, B. , Braña, A.F. , Zabala, D. , Olano, C. , Cortés, J. et al. (2017) Identification by genome mining of a type I polyketide gene cluster from Streptomyces argillaceus involved in the biosynthesis of pyridine and piperidine alkaoids argimycins P. Frontiers in Microbiology, 8, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1


