Abstract
Basal cell carcinoma (BCC) is the most common human cancer, especially in individuals with light skin phototypes (i.e., Fitzpatrick I-II skin type). Many affected develop multiple BCCs during their lifetime. It is not uncommon to observe elderly patients with >5 BCCs. In this study, we explored whether for patients diagnosed with multiple BCCs, analyzing the genomic mutations in one tumor could be sufficient to derive meaningful molecular/genetic conclusions regarding the other BCC tumors. Following the Genome Analysis Toolkit (GATK) best practices we have completed the study of 6 BCCs that occurred in an 83-year-old Caucasian male due to sun exposure. We have analyzed exome sequencing data of each BCC tumor and matched normal skin samples. We identified that BCCs from the same patient shared some of the key driver mutations, but they also displayed significant intertumoral heterogeneity. This finding may in part explain the different clinical progression/evolution of BCCs observed in the same patient. This work also highlights the value of characterizing multiple BCCs in one individual to identify patient-specific genetic events with a potential link to other malignancies and implications for personalized medicine.
To the editor
Basal Cell Carcinoma (BCC) is the most common human cancer (Lefrancois et al. 2020; Litvinov et al. 2021). BCC driver mutations are typically found in the Sonic Hedgehog pathway genes, such as PTCH1, SMO and SUFU, and in TP53. Genome-wide profiling has expanded the implicated pathways in BCC tumorigenesis, including N-Myc and Hippo-YAP signaling (Bonilla et al. 2016). In this study, we explored whether for a patient with multiple BCCs, studying a single BCC tumor may be sufficient to derive meaningful molecular/genetic conclusions regarding the other tumors.
We analyzed exome sequencing data on matched tumor-normal skin pairs from 40 BCCs, and focused on one individual, who was diagnosed with 6 BCCs (Bonilla et al. 2016). This 83-year-old Caucasian male with extensive history of sun exposure developed 3 superficial BCCs on the legs, and 3 nodular BCCs in the head and neck region. He did not have a prior diagnosis of the nevoid basal cell carcinoma syndrome based on the lack of clinical or radiologic features (e.g., pits in the palms and soles of the feet, macrocephaly etc.) He had no known innate, acquired or iatrogenic immunodeficiency/immunosuppression.
Following the Genome Analysis Toolkit (GATK) best practices (DePristo et al. 2011), we identified 41,946 somatic cancer-associated mutations when comparing 6 BCC samples to normal control skin from this individual. The mutational signatures varied among BCCs (Fig. 1). As expected, the Catalogue of Somatic Mutations in Cancer (COSMIC) mutational signature 7 (UV radiation signature; highlighted in yellow; 27–62%) was the dominant one in all BCCs analyzed. However, there was a significant variation (p < 0.01; ANOVA) in the importance of secondary mutational signatures, mostly in COSMIC signature 3 (failure of DNA double-stranded break repair; highlighted in blue; 0–19%) and in COSMIC signature 11 (alkylating agents; highlighted in red; 10–21%).
Fig. 1.
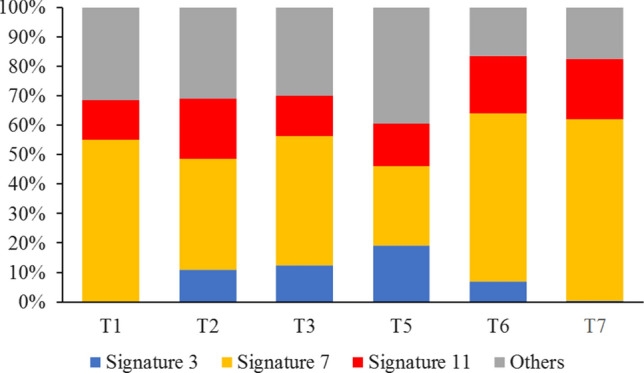
Predominant COSMIC mutational signatures display significant heterogeneity in 6 BCC tumors from the same patient. Percentages of COSMIC mutational signatures 3 (failure of DNA double-stranded break repair; blue), 7 (UV radiation; yellow), and 11 (alkylating agents; red) are shown, along with the remaining 27 signatures (grouped in “others”; grey). Different tumors are referred by T1–T3, and T5–T7
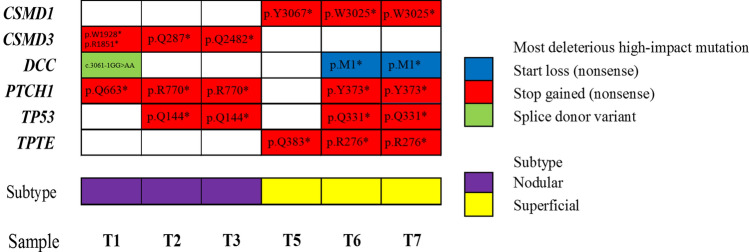
Focusing on high-impact variants including nonsense mutations (start codon loss/stop codon gain/frameshift) and important splice site mutations (splice donor/acceptor variants), we identified recurrent (i.e., present in ≥ 3/6 BCCs) putative driver mutations in 6 genes (Fig. 2). Not surprisingly, PTCH1 was the most frequently mutated gene (seen in 5/6 samples), with TP53 being a close second (4/6 samples). Interestingly, we observed mutually exclusive mutations either in CSMD1 (3/6 samples; in 3 nodular BCCs) or in CSMD3 (the remaining 3/6 samples; in 3 superficial BCCs), two paralogs whose loss-of-function results in epithelial proliferation in several cancers (Kamal et al. 2017). Furthermore, we uncovered two patient-specific mutations. The first one was in TPTE/PTEN2 (3/6 samples; p = 0.008; Fisher’s Exact Test using the remaining 34 BCC tumors as controls), a cancer testis antigen signal transduction phosphatase. The second one was in DCC (3/6 samples; p = 0.03), a netrin receptor, whose loss-of-function is a feature in colorectal cancer development/metastasis (Kroigard et al. 2018). Finding patient-specific recurrent mutations may explain a propensity for certain genomic alterations in a patient, with potential implications for personalized medicine, cancer risk and screening for other malignancies/diseases using the same predisposition genes. Indeed, based on a previous report, in patients with ≥ 6 BCCs, ~ 20% of them represented germline mutations in DNA repair genes (Cho et al. 2018). These individuals were also ~ 3 times more likely to develop another malignancy (Cho et al. 2018).
Fig. 2.
Recurrent high-impact putative driver mutations can be BCC-characteristic or patient-specific. A diagram showing genes encompassing high-impact mutations, in at least 3/6 BCC tumors (T1–T3; T5–T7) originating from 1 patient. If two deleterious high-impact mutations are present in the same gene in one tumor, the most deleterious (start loss > stop gained > splice donor variant) is plotted. Histopathological subtype information is overlaid
Our analysis suggests that while different BCCs from the same individual share some of the key driver mutations, they also display significant intertumoral molecular heterogeneity, which together with the heterogeneity due to subclones, may in part explain drug resistance, metastasis and adverse prognosis in a subset of BCCs (Oh et al. 2019). This study further highlights the value of characterizing multiple BCCs in one individual since collecting a single clinical specimen in an individual with multiple BCCs may miss important genetic/clinical information on other BCCs.
Acknowledgements
We thank Dr. Sergey Nikolaev Ph.D (Institute Gustave Roussy, Villejuif, France) for generously sharing the exome sequencing data used in this study and providing valuable suggestions for the analyses.
Funding
This work was further supported by the Cancer Research Society (CRS)-CIHR Partnership Grant #25343 to Dr. Litvinov, CIHR Project Scheme Grant #426655 to Dr. Litvinov, and by the Fonds de la recherche du Québec—Santé to Dr. Litvinov (#296643).
Declarations
Conflict of interest
None declared.
Ethical approval
The data for the analyses was obtained from open source repository and prior studeis.
Footnotes
Pingxing Xie and Philippe Lefrançois have contributed equally to this publication.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bonilla X, Parmentier L, King B, Bezrukov F, Kaya G, Zoete V, Seplyarskiy VB, Sharpe HJ, McKee T, Letourneau A, et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet. 2016;48:398–406. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- Cho HG, Kuo KY, Li S, Bailey I, Aasi S, Chang ALS, Oro AE, Tang JY, Sarin KY (2018) Frequent basal cell cancer development is a clinical marker for inherited cancer susceptibility. JCI Insight 3(15):e122744 [DOI] [PMC free article] [PubMed]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal M, Holliday DL, Morrison EE, Speirs V, Toomes C, Bell SM. Loss of CSMD1 expression disrupts mammary duct formation while enhancing proliferation, migration and invasion. Oncol Rep. 2017;38:283–292. doi: 10.3892/or.2017.5656. [DOI] [PubMed] [Google Scholar]
- Kroigard AB, Larsen MJ, Laenkholm AV, Knoop AS, Jensen JD, Bak M, Mollenhauer J, Thomassen M, Kruse TA. Identification of metastasis driver genes by massive parallel sequencing of successive steps of breast cancer progression. PLoS ONE. 2018;13:e0189887. doi: 10.1371/journal.pone.0189887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois P, Xie P, Gunn S, Gantchev J, Villarreal AM, Sasseville D, Litvinov IV. In silico analyses of the tumor microenvironment highlight tumoral inflammation, a Th2 cytokine shift and a mesenchymal stem cell-like phenotype in advanced in basal cell carcinomas. J Cell Commun Signal. 2020;14:245–254. doi: 10.1007/s12079-020-00563-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvinov IV, Xie P, Gunn S, Sasseville D, Lefrancois P (2021) The transcriptional landscape analysis of basal cell carcinomas reveals novel signalling pathways and actionable targets. Life Sci Alliance 4(7):e202000651 [DOI] [PMC free article] [PubMed]
- Oh BY, Shin HT, Yun JW, Kim KT, Kim J, Bae JS, Cho YB, Lee WY, Yun SH, Park YA, et al. Intratumor heterogeneity inferred from targeted deep sequencing as a prognostic indicator. Sci Rep. 2019;9:4542. doi: 10.1038/s41598-019-41098-0. [DOI] [PMC free article] [PubMed] [Google Scholar]