Abstract
Systemic autoimmune rheumatic diseases (SARDs) are a heterogeneous group of chronic multisystem inflammatory disorders that are thought to have a complex pathophysiology, which is not yet fully understood. Recently, the role of non-coding RNAs, including long non-coding RNA (lncRNA), has been of particular interest in the pathogenesis of SARDs. We aimed to summarize the potential roles of lncRNA in SARDs affecting the skin including, systemic sclerosis (SSc), dermatomyositis (DM) and cutaneous lupus erythematosus (CLE). We conducted a narrative review summarizing original articles published until July 19, 2021, regarding lncRNA associated with SSc, DM, and CLE. Several lncRNAs were hypothesized to play an important role in disease pathogenesis of SSc, DM and CLE. In SSc, Negative Regulator of IFN Response (NRIR) was thought to modulate Interferon (IFN) response in monocytes, anti-sense gene to X-inactivation specific transcript (TSIX) to regulate increased collagen stability, HOX transcript antisense RNA (HOTAIR) to increase numbers of myofibroblasts, OTUD6B-Anti-Sense RNA 1 to decrease fibroblast apoptosis, ncRNA00201 to regulate pathways in SSc pathogenesis and carcinogenesis, H19X potentiating TGF-β-driven extracellular matrix production, and finally PSMB8-AS1 potentiates IFN response. In DM, linc-DGCR6-1 expression was hypothesized to target the USP18 protein, a type 1 IFN-inducible protein that is considered a key regulator of IFN signaling. Additionally, AL136018.1 is suggested to regulate the expression Cathepsin G, which increases the permeability of vascular endothelial cells and the chemotaxis of inflammatory cells in peripheral blood and muscle tissue in DM. Lastly, lnc-MIPOL1-6 and lnc-DDX47-3 in discoid CLE were thought to be associated with the expression of chemokines, which are significant in Th1 mediated disease. In this review, we summarize the key lncRNAs that may drive pathogenesis of these connective tissue diseases and could potentially serve as therapeutic targets in the future.
Keywords: LncRNA, Systemic sclerosis, Cutaneous lupus erythematosus, Dermatomyositis
Background
Only 1.2% of RNA encodes messenger RNA (mRNA) that is further translated into proteins (Jarroux et al. 2017). Remaining RNA belongs to a class of non-coding RNA (ncRNA) and includes regulatory RNAs such as microRNAs (miRNAs), small interfering RNAs (siRNAs) and long (> 200 nucleotides) non-coding RNA (lncRNA) (Jarroux et al. 2017; Mariotti et al. 2019; Le et al. 2020; Zhang et al. 2019). There are ~ 30,000 lncRNAs detected in the human genome. They are further classified according to their location into intergenic, intronic, bidirectional, enhancer, sense, and antisense (AS) (Devaux et al. 2015). They exert important biologic effects including remodeling chromatin and regulating gene expression at transcriptional and post-transcriptional levels. Previous studies suggested that the transcription process of the lncRNA appears to regulate the corresponding coding DNA expression (Salviano-Silva et al. 2018). Antisense-lncRNAs (AS-lncRNAs) are transcribed from the opposite strand to protein-coding genes and overlap in one or several exons and introns with the sense strand. High-throughput RNA sequencing analysis showed that for most AS-lncRNAs, their expression was ~ tenfold lower than their corresponding coding genes (Derrien et al. 2012; Ozsolak et al. 2010). Interestingly, the expression of AS-lncRNAs was found to be more tissue specific than those of protein coding genes. AS-lncRNAs can also regulate the expression of transcription sites on the same (cis) or other chromosomes (trans) (Kornienko et al. 2013).
To date, lncRNAs have been implicated in carcinogenesis and autoimmune disease pathways. Particularly, the function of lncRNAs in systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA) have been well-documented (Gao et al. 2018; Chen et al. 2019; Wang et al. 2020). However, the role of lncRNA in Cutaneous Lupus Erythematosus (CLE) and other Systemic Autoimmune Rheumatic Diseases (SARDs) affecting the skin such as systemic sclerosis (SSc) and dermatomyositis (DM) remains poorly understood. In this manuscript, we comprehensively review published literature regarding the role of lncRNAs in the pathogenesis of SSc, DM, and CLE and present the key lncRNAs, discovered over the last 40 years, that are thought to be involved with the pathogenesis of these conditions (Nor Muhammad 2019; Jarroux et al. 2017).
Methods
We searched PubMed, Medline and EMBASE up to July 19th, 2021 and included original studies. Review papers were excluded. Studies must have reported one or more lncRNA as was identified through their experiments. Studies on SLE were excluded given there are recent review papers summarizing the key roles of ncRNAs including lncRNA in disease pathogenesis (Cai et al. 2021; Tsai et al. 2020; Taheri et al. 2020).
Systemic sclerosis
SSc is a rare, autoimmune disease resulting in fibrosis of the skin and multiple internal organs (Denton and Khanna 2017; Gabrielli et al. 2009), which leads to increased morbidity and mortality (Ouchene et al. 2020). Diagnosis is usually confirmed according to the 2013 American College of Rheumatology (ACR)/European Alliance of Associations for Rheumatology (EULAR) criteria (except one study which used Subcommittee for scleroderma criteria 1980 (Messemaker et al. 2018)) and patients are classified into limited cutaneous SSc (lcSSc) and diffuse cutaneous SSc (dcSSc) (van den Hoogen et al. 2013; LeRoy et al. 1988). Subtypes include overlap SSc and SSc sine scleroderma (ssSc) (also known as non-cutaneous SSc) (Diab et al. 2014). Similar to other autoimmune diseases, SSc is thought to be environmentally triggered in individuals with genetic susceptibility (Kassamali et al. 2021). In this respect ncRNAs may be central to the pathogenesis of this disease since they often modulate this gene-environment interaction (Bergmann and Distler 2017; Aslani et al. 2018).
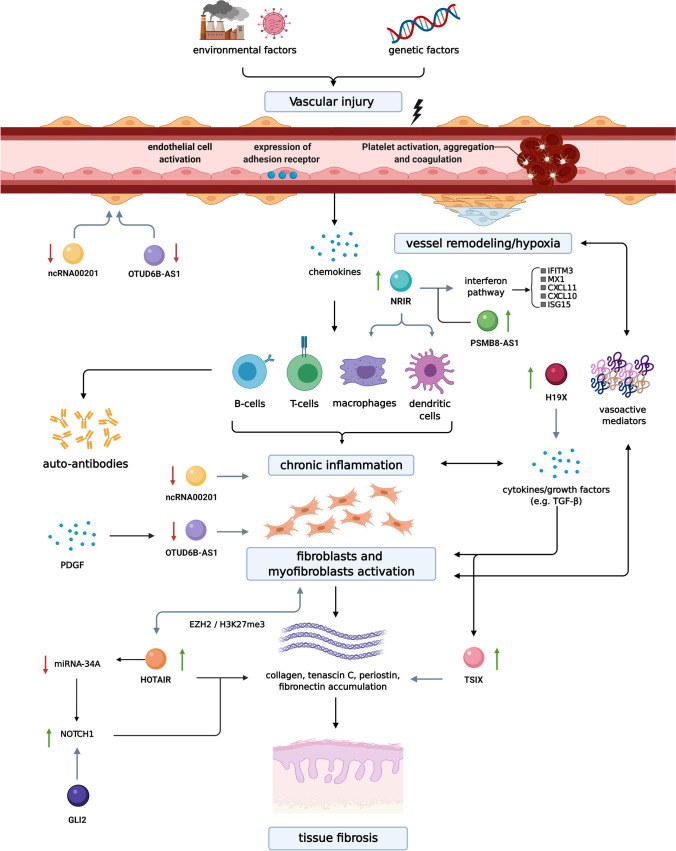
The SSc pathogenesis and signaling pathways have been described in detail elsewhere (Pattanaik et al. 2015; Katsumoto et al. 2011; Sierra-Sepúlveda et al. 2019; Boleto et al. 2018). Briefly, immune dysregulation, endothelial dysfunction, and subsequent fibrosis are the key features (Fig. 1). The initial damage to the small/medium vessel walls in the skin and internal organs triggers subsequent inflammation (Matucci-Cerinic et al. 2013). Interferon (IFN) signaling pathway has been initially described based on observations of patients receiving IFN-α injections leading to new onset and/or worsening of SSc-related features (Solans et al. 2004; Black et al. 1999). Increased expression of type I IFN-regulated genes, is a hallmark of SSc, which is present both in the fibrotic skin, peripheral blood cells (Eloranta et al. 2010; Tan et al. 2006), and in monocytes of SSc patients from the earliest phases of the disease, even prior to the evident skin fibrosis (Brkic et al. 2016). The role for adaptive immunity involving autoantibodies as well as Th2 and Th17 pathways has been demonstrated (Ihn et al. 1995; Badea et al. 2009) with the Th2 pathway gaining more importance in the late fibrotic stages (Saracino et al. 2017). IL-4 and IL-13 are thought to stimulate fibrosis through their ability to activate type 2 macrophages and fibroblasts and via upregulation of the critical profibrotic cytokines such as the transforming growth factor beta (TGF-β) (Saracino et al. 2017; Huang et al. 2015; Kawakami et al. 1998; Varga and Pasche 2009; Zehender et al. 2018) and platelet-derived growth factor (PDGF) (Iwayama and Olson 2013; Liakouli et al. 2018; Klareskog et al. 1990). While vasculopathy and immune dysregulation play a crucial role in SSc pathogenesis, fibroblasts, that are capable of excess matrix production and deposition, are thought to be the effectors cells and hallmark of this disease. Once removed from affected tissues, SSc fibroblasts maintain their profibrotic phenotype in vitro, with elevated production of collagen and extracellular matrix proteins (ECM) (mainly type 1 collagen which consists of α1(I) and α2(I) collagen) and higher frequency of α-smooth muscle actin (SMA) with subsequent differentiation into myofibroblasts (Garrett et al. 2017; Hinz et al. 2012; Kähäri et al. 1988). Activation of fibroblasts, differentiation into apoptosis resistant myofibroblasts, and subsequent sustained production of ECM components leading to tissue fibrosis is thought to occur through TGF-β signaling (Pachera et al. 2020). Therefore, its deletion by genetic modification in animal models was found to decrease fibrosis and scar formation (Hoyles et al. 2011).
Fig. 1.
Pathogenesis of systemic sclerosis (SSc). The first step in the pathogenesis is thought to involve vascular injury leading to vessel remodeling and hypoxia on one hand and inflammation on the other. Chronic inflammation is important for subsequent activation of fibroblasts/myofibroblasts and tissue fibrosis, it also contributes to further vascular remodeling and hypoxia. The long non-coding RNAs (lncRNAs) at their proposed sites of action are shown. Upregulation of Negative Regulator of IFN Response (NRIR) modulates Interferon (IFN) response in monocytes, the overexpression of anti-sense gene to X-inactivation specific transcript (TSIX) regulates increased collagen stability, higher levels of HOX transcript antisense RNA (HOTAIR) leads to increased numbers of myofibroblasts, downregulation of OTUD6B-Anti-Sense RNA 1 (OTUD6B-AS1) leads to decreased fibroblast/endothelial smooth muscle cell apoptosis, decrease in ncRNA00201 regulates pathways of all three parts of SSc pathogenesis and is also involved in carcinogenesis, H19X potentiating TGF-β–driven extracellular matrix production, and finally PSMB8-AS1 potentiating the IFN response. Created with Biorender.com
In total there were 11 studies (Table 1) which identified 14 important lncRNAs that were differentially expressed in SSc patients and had a plausible role in disease pathogenesis (Table 2). Since fibrosis, the last step in proposed SSc pathogenesis, clinically leads to complications and poor outcomes, the majority of studies focused on evaluating lncRNAs role in tissue fibrosis. This includes Negative Regulator of IFN Response (NRIR) modulating IFN response in monocytes, the anti-sense gene to X-inactivation specific transcript (TSIX) regulating increased collagen stability, HOX transcript antisense RNA (HOTAIR) leading to increased numbers of myofibroblasts, OTUD6B-Anti-Sense RNA 1 (OTUD6B-AS1) leading to decreased fibroblast apoptosis, ncRNA00201 which regulates pathways of all three parts of SSc pathogenesis and is also involved in carcinogenesis, H19X potentiating TGF-β–driven ECM production, and finally PSMB8-AS1 is implicated in cell activation, response to viral and external stimuli, and pro-fibrotic cytokine production.
Table 1.
Summary of the studies identified for SSc, DM and DLE
| Study | Patient number | Patients additional characteristics | Controls type and number | Specimen source |
|---|---|---|---|---|
|
Mariotti et al NRIR (Mariotti et al. 2019) |
Definite SSc cohort ncSSc (7) lcSSc (11) dcSSc (7) Non-fibrotic SSc cohort: eaSSc (11) ncSSc (10) SSc cohort 3: eaSSc (15) ncSSc (27) lcSSc (23) dcSSc (19) |
Age (range); female % Definite SSc cohort ncSSc 45 (26–63); 86% lcSSc 59 (45–70); 73% dcSSc 58 (34–72); 43% Non-fibrotic SSc cohort eaSSc 57 (40–77); 100% ncSSc 52 (25–70); 100% SSc cohort 3 eaSSc 62 (25–81); 100% ncSSc 59 (29–80); 100% lcSSc 60 (41–80); 96% dcSSc 52 (27–80); 79% |
All sex and age-matched Definite SSc cohort HCs (9) Non-fibrotic SSc cohort HCs (9) SSc cohort 3 (21) |
Blood |
| Takata et al. OTUDB-AS1(Takata et al. 2019) |
dcSSc (4) lcSSc (5) |
dcSSc average disease duration 5.34 years NA for lcSSc |
HCs (14) |
Skin biopsy dcSSc: 1/3 had fibrosis at biopsy site lcSSc: 0/5 had fibrosis at biopsy site |
| Dolcino et al. ncRNA00201 (Dolcino et al. 2019) |
dcSSc (10) lcSSc (10) |
Mean age ± standard deviation; female/male number dcSSc: 55 ± 10; 9/1 lcSSc: 57 ± 13; 8/2 HCs: 55 ± 11; 17/3 |
Sex and age matched HCs (20) | Blood |
|
Wang et al |
dcSSc (13) lcSSc (23) |
Females/males number: 28/8 Mean age SSc: 60.9 years |
Sex and age matched SLE (8) SSD (10) HCs (12) |
Blood Skin biopsy of lesional skin |
|
Messemaker et al CTBP1-AS2, OTUD6B-AS1, AGAP2-AS1(Messemaker et al. 2018) |
dcSSc (10) lcSSc (4) |
Mean disease duration of 1.5 years 43% received systemic drug (methotrexate ± prednisone) |
Sex and age matched HCs (6) | Skin biopsy from proximal part of the lower arm, distal from the elbow. In 10 patients from affected area and 4 from unaffected area |
|
1) Wasson et al 2) Wasson et.al HOTAIR |
(1) SSc (12) (2) SSc (3) |
(1) Early onset SSc (2) Recent onset SSc (< 18 months from clinical skin induration) |
(1) HCs fibroblasts (2) NA |
1,2)Skin biopsy forearm of affected skin |
| (Pachera et al. 2020) H19X |
SSc: cohort 1: 5 cohort 2: 48 cohort 3: 14 cohort 4: 6 SSc – ILD: 11 patients (lung tissue in patients for lung transplant) |
Cohort 2: early dcSSc enrolled in Prospective Registry of Early Systemic Sclerosis (PRESS) Cohort 3 and 4: more diverse disease duration and severity |
HCs for each cohorts: cohort 1: 5 cohort 2: 33 cohort 3: 6 cohort 4: 6 HC—lung tissue: 11 |
Skin biopsy Lung tissue |
| (Servaas et al. 2021)PSMB8-AS1 |
Definite Cohort ncSSc (7) lcSSc (11) dcSSc (7) |
Female/male number; mean age 6/1; 45 8/3; 59 3/4; 58 |
HC (9) Females (5) Males (4) Mean age: 52 |
Blood |
| MGC12916 | SSc (15) | African American patients only included | HC (age-matched within 5 years) (15) | |
|
(Abd-Elmawla et al. 2020) ANCR TINCR HOTTIP SPRY4-IT1 |
dcSSc (20) lcSSc (43) |
Female/male number; age mean (range) SSc—47/16; 34.5 (20–60 years old) HC—24/11; 36.1 (19–55 years old) |
Age and sex matched HC (35) | Blood |
|
Peng et al. linc-DGCR6-1 (Peng et al. 2016) |
DM (15) | Not reported | HCs (5) | Muscle biopsy |
|
Liang et al. (Liang et al. 2021) |
DM (6) | Not reported | Paracancerous muscle tissue (3) | Muscle biopsy |
|
Xuan et al. lnc-MIPOL1-6 lnc-DDX47-3 (Xuan et al. 2019) |
DLE (3) | Not reported | None | Skin biopsy |
The value in brackets indicated the number of patients. The treatment regimen the patients were using is described when reported
HC healthy controls, ncSSc non-cutaneous SSc defined as patients satisfying the classification criteria without skin fibrosis, eaSSc-early SSc defined as presence of Raynaud’s and SSc autoantibodies and/or typical nailfold videocapillaroscopy abnormalities (LeRoy and Medsger 2001), SSD scleroderma spectrum disorder, lcSSc localized cutaneous SSc, dcSSc diffuse cutaneous SSc, SLE systemic lupus erythematosus, NA not available, ILD interstitial lung disease, DM dermatomyositis, DLE discoid lupus erythematosus
Table 2.
Summary of important lncRNAs in systemic autoimmune diseases affecting the skin
| lncRNA | Expression level | Tissue or cell type | Target mRNA | Role |
|---|---|---|---|---|
| Systemic sclerosis | ||||
| TSIX (Wang, et al. 2016a, b) |
↑ in fibroblasts in vivo and in vitro ↑ in serum |
Skin tissue | mRNA expression of a1(I) and a2(I) collagen | High TSIX levels led to significantly elevated mRNA expression of α1(I) and α2(I) collagen, playing a role in tissue fibrosis |
| OTUD6B-AS1 (Wang et al. 2019; Takata et al. 2019) |
↓ Skin ↓ Human Pulmonary Artery Smooth Muscle Cells |
Skin tissue Human pulmonary artery smooth muscle cells |
Cyclin D1 | Regulates proliferation and apoptosis of fibroblasts and vascular endothelial smooth muscle cells via cyclin D1 expression in a sense gene independent manner |
| NRIR (Mariotti et al. 2019) | ↑ | Monocytes | CXCL10, CXCL11, MX1, IFITM3, ISG15 |
Drives IFN-Response in monocytes Correlates strongly with the IFN score of SSc patients |
| AGAP2-AS1 (Messemaker et al. 2018) | ↑ | Skin | Not directly assessed | Not evaluated in this study. Previously shown to be involved in cell migration and can repress transcription by interacting with histone methyltransferase (EZH2) and LSD1 in cancer cells. (Messemaker et al. 2018) |
| CTBP1-AS2 (Messemaker et al. 2018) | ↑ | Skin | Not directly assessed | Not evaluated in this study |
| ncRNA00201 (HNRNPU-AS1)(Dolcino et al. 2019) | ↓ | Monocytes |
hnRNPC (Heterogeneous nuclear ribonucleoproteins C) 56 miRNAs 31 genes |
Regulates genes involved in the vasculopathy, immune dysregulation, and fibrosis seen in SSc and genes in tumor-associated pathways |
| HOTAIR (Wasson, et al. 2020a, b; Wasson, et al. 2020a, b) | ↑ | Skin |
Direct the EZH2 to induce H3K27me3 in specific target genes Repression of miRNA-34A |
Associated with increase in collagen and α-SMA expression in vitro Represses miRNA-34a activating NOTCH signaling This leads to expression of Hedgehog pathway transcription factor GLI2 which mediates expression of pro-fibrotic markers downstream |
| H19X (Pachera et al. 2020) | ↑ |
Skin Lungs |
DDIT4L gene expression, changing chromatin accessibility of DDIT4L enhancer upstream | Upregulated in the skin and lungs of patients with SSc. H19X is an obligatory factor for TGF-β-induced synthesis of extracellular matrix and differentiation and survival of myofibroblasts |
| PSMB8-AS1(Servaas et al. 2021) | ↑ | Monocytes |
IL-6 TNF-α |
May be implicated in cell activation and response to viral and external stimuli IL-6 and TNF-α may be implicated in disease pathogenesis and fibrosis |
| MGC12916 (Baker Frost et al. 2021) |
↑ methylation ↓ expression |
Skin | Not specified | Not specified |
|
ANCR TINCR HOTTIP SPRY4-IT1 (Abd-Elmawla et al. 2020) |
↓ ↑ ↑ ↑ |
Plasma | Not specified | Serve as possible biomarkers for SSc and help distinguish subtypes |
| Dermatomyositis | ||||
| linc-DGCR6-1 (Peng et al. 2016) | ↑ | Muscle tissue | USP18 |
USP18 is a known gene encoding type 1 IFN-inducible protein, which is considered to be a key regulator of interferon signaling |
|
(Liang and Peng 2021) |
↑ | Muscle tissue | Cathepsin G (CTSG) | CTSG is thought to play an important role in muscle inflammatory cells infiltration by increasing the permeability of vascular endothelial cells |
| Discoid lupus erythematosus | ||||
|
lnc-MIPOL1-6 (Xuan et al. 2019) |
↑ | Skin tissue | IL19, CXCL1, CXCL11, and TNFSF15 | IL19, CXCL11 and TNSF15, have each been shown to be associated with Th1 dominant diseases (Azuma et al. 2011), promote Th1 cell recruitment (Kumar and Herbert 2017) and stimulate Th1 cytokine production (Zhang and Li 2012; Zhang and Zhang 2015) |
|
lnc-DDX47-3 (Xuan et al. 2019) |
↓ | Skin tissue | IL19, CXCL1, CXCL11, and TNFSF15 | IL19, CXCL11 and TNSF15, have each been shown to be associated with Th1 dominant diseases (Azuma et al. 2011), promote Th1 cell recruitment (Kumar and Herbert 2017) and stimulate Th1 cytokine production (Zhang and Li 2012; Zhang and Zhang 2015) |
NRIR
NRIR is currently thought to be an important regulator of IFN response, a mechanism implicated in SSc. Activation of type 1 IFN pathway, described above, may occur by the action of Toll-like receptor 4 (TLR4). Initially, TLR4 was thought to bind to lipopolysaccharides (LPS), but overtime, endogenous ligand binding has also been described such as the damage-associated molecular patterns (DAMPs), released upon cell damage or stress (Farrugia and Baron 2017; Bhattacharyya and Varga 2015). In SSc increased levels of DAMPs have been reported and their binding to TLR4 contributes to fibrosis (O'Reilly and van Laar 2018) through type 1 IFN pathway and downstream TGF-β production (Bhattacharyya et al. 2013; Bhattacharyya and Varga 2015; Ciechomska et al. 2013; Lafyatis and Farina 2012; Ivashkiv and Donlin 2014).
The study by Mariotti et al. (2019) evaluated the role and function of lncRNAs in regulating the type 1 IFN pathway in peripheral blood monocytes. Their study revealed that NRIR was highly expressed and was implicated in biological processes related to immune response and the IFN/antiviral response. It also demonstrated that this IFN-dependent lncRNA was a positive regulator of the LPS-induced IFN response in human monocytes and suggested that abnormal expression of NRIR can be involved in the dysregulation of immune system seen in SSc. Monocytes from lcSSc and ssSSc patients showed NRIR overexpression which correlated with the IFN signature, thus, confirming the implication of NRIR in the IFN response. The highest levels of NRIR were consistently seen in ssSSc alongside the strongest IFN-signature (Brkic et al. 2016).
Upregulation of NRIR was seen in early SSc patients suggesting a potential role of NRIR in disease initiation and progression. NRIR-silencing reduced the LPS-induced expression of type 1 IFN target genes, such as C-X-C motif chemokine ligand 10 (CXCL10), also known as Interferon gamma-induced protein 10 (IP-10), MX dynamin like GTPase 1 (MX1), IFN Induced Transmembrane Protein 3 (IFITM3), IFN Induced Protein 44 (IFI44) and Ubiquitin Like Modifier (ISG15). CXCL10 and CXCL11 are pro-inflammatory chemokines involved in multiple processes including chemotaxis, differentiation, and activation of peripheral immune cells, regulation of cell growth, apoptosis and modulation of angiogenesis. They were found to be upregulated and clinically are associated with more severe disease including lung and kidney involvement (Eloranta et al. 2010). These two chemokines have been proposed to act as biomarkers for identification of early and non-fibrotic subset of SSc (Cossu et al. 2017). Anifrolumab, an agent which inhibits IFN receptor signaling was found to lead to a reduction in CXCL10 levels and in transcripts related to fibrosis (e.g., CXCL10 and CD40 Ligand (CD40LG)) (Guo et al. 2015) and could be explored as a therapeutic target in the future. IFI44 correlated with modified Rodnan skin score (Merkel et al. 2003) and MX1 with ischemic skin ulcers and reduced forced vital capacity of the lungs, therefore predicting more severe disease. (Christmann et al. 2011; Mariotti et al. 2019). Thus, NRIR was found to control the expression of certain IFN stimulated genes, and overexpression of NRIR in SSc monocytes could explain the type 1 IFN signature in SSc. NRIR is thought to contribute to eventual development of fibrosis seen in SSc through regulation of the IFN pathway in monocytes (Fig. 1).
TSIX
TSIX is the anti-sense gene to XIST (X-inactivation specific transcript, a key gene regulating X-chromosome inactivation in females) (Brooks and Renaudineau 2015) and may help explain the female predisposition in a number of autoimmune diseases. Both TSIX and XIST are expressed on the X-chromosome, which is marked for inactivation. There is a balance in their action, and overexpression of TSIX prevents increases in XIST expression and blocks inactivation of that X-chromosome. Numerous studies have shown that some genes on the inactive-X (Xi) escape inactivation and become expressed. One study looking at mature naïve T and B cells, found dispersed patterns of XIST/XIST RNA, and they lacked the typical heterochromatic modifications of Xi (Wang, et al. 2016a, b). Of note, there was evidence of biallelic expression of immunity-related genes in SLE.
Increased expression of TSIX observed in SSc serum in vivo and in vitro fibroblasts from affected patients, which was thought to be due to intrinsic TGF-β activation seen in disease pathogenesis (Wang, et al. 2016a, b). Increased TSIX expression correlated to mRNA expression of α1(I) and α2(I) collagen in dermal fibroblasts and was thought to have a direct regulatory effect on collagen production through regulating collagen mRNA stability (Wang, et al. 2016a, b). Increased collagen production may explain the fibrosis observed in SSc pathogenesis (Fig. 1). Elevated levels of TSIX in the serum of dcSSc patients appeared to correlate with the level of skin fibrosis, and hence were proposed as a novel disease biomarker (Wang, et al. 2016a, b). A trial of inhibition of TSIX in vivo led to subsequent decreased collagen production which could be a possible treatment target in SSc.
HOTAIR
HOTAIR is an important factor in the epigenetic differentiation of skin and has previously been implicated in various cancers (Tang and Hann 2018). HOTAIR also appears to have a role in tissue fibrosis through priming of myofibroblasts (Wasson, et al. 2020a, b; Wasson, et al. 2020a, b). Myofibroblasts are defined by α-SMA expression (Gilbane et al. 2013). In the study by Wasson et al. increased expression of HOTAIR was observed in SSc skin fibroblasts derived from cutaneous biopsies, and this overexpression resulted in myofibroblast activation through EZH2 activation and subsequent histone H3K27me3 methylation (Fig. 1) (Wasson, et al. 2020a, b). EZH2 is an enzymatic subunit of the polycomb repressor complex involved in silencing target genes. In fact, HOTAIR expression was sufficient to induce profibrotic activation of dermal fibroblasts in vitro. As a result of HOTAIR overexpression, there was reduced expression of miRNA-34 which leads to increased NOTCH signaling and increased downstream collagen, ECM secretion, and α-SMA positive cells – all leading to the fibrosis phenotype. Importantly, the number of myofibroblasts was previously found in vivo to correlate with disease severity (Farina et al. 2009). Thus, when the fibroblasts were treated with a HOTAIR inhibitor, reductions in type 1 collagen and α-SMA protein levels were noted. Therefore, it was hypothesized that a population of cells can be epigenetically ‘primed’ to differentiate into myofibroblasts. HOTAIR was identified as one such epigenetic factor. Interestingly, the expression of HOTAIR is higher in dermal fibroblasts in the hands and feet which may explain why those are typically the first areas of disease manifestation (Rinn et al. 2007; Wasson, et al. 2020a, b).
A subsequent study revealed that HOTAIR-resultant Notch pathway activation stimulated expression of the Hedgehog pathway transcription factor GLI2 (Wasson, et al. 2020a, b). This observation was further strengthened by inhibiting H3K27 methylation and Notch signaling which led to decreased expression of GLI2 in HOTAIR expressing fibroblasts and SSc fibroblasts. Additionally, the inhibition of GLI2 reduced the pro-fibrotic phenotype induced by HOTAIR. Thus, GLI2 expression is upregulated in SSc myofibroblasts through HOTAIR expression and GLI2 mediates the expression of pro-fibrotic markers downstream of Notch and may serve as a more specific therapeutic target in the future.
H19X
H19 X-linked co-expressed lncRNA (H19X), also referred to as MIR503HG, is an intergenic lncRNA with its gene located on chromosome X. The study by Pachara et al. identified it as a direct potent mediator of prolonged myofibroblast survival, potentiating TGF-β–driven ECM production and progressive tissue fibrosis where upregulation of H19X was dose dependent across biologically relevant TGF-β concentrations (Pachera et al. 2020). Other pro-fibrotic cytokines such as IL-1β, IL-4, IL-13, and IL-17a did not induce H19X expression. H19X was found to be upregulated in the skin of patients with SSc and lung tissues in patients with severe SSc related interstitial lung disease undergoing lung transplantation compared to controls. When tested in other fibrotic diseases including idiopathic pulmonary fibrosis, fibrotic ileum in Crohn’s disease, liver tissue in primary sclerosing cholangitis, and dermal wounds compared to normal tissue, expression was also significantly elevated. Specifically, of all the cell types explored, fibroblasts of different origins and fibroblast-like cells showed the strongest induction of H19X by TGF-β.
Functional experiments following H19X silencing revealed that H19X regulates DDIT4L gene expression, specifically interacting with a region upstream of the DDIT4L gene and changing the chromatin accessibility of a DDIT4L enhancer. DDIT4L is possibly implicated in mediating the profibrotic effects of TGF-β–induced H19X. The resulting transcriptional repression of DDIT4L leads to increased collagen expression and fibrosis. Hence, H19X is an obligatory factor for TGF-β–induced ECM synthesis, differentiation and survival of ECM-producing myofibroblasts. Silencing of H19X and therefore inducing apoptosis of myofibroblasts could be a therapeutic strategy which will allow selective removal of ECM-overproducing cells from fibrotic tissues (Lagares et al. 2017).
OTUD6B-AS1
OTUD6B-AS1 has a role in regulating apoptosis resistance in fibroblasts and vascular smooth muscle cells (Fig. 1) (Takata et al. 2019). Regulation of proliferation and apoptosis is thought to be mediated by a cell cycle regulator, cyclin D1, in a sense gene (OTUD6B) independent manner.
Significantly reduced expression of OTUD6B-AS1 and its sense gene expression was observed in skin biopsies of SSc patients compared to healthy controls and non-affected skin biopsies in SSc individuals. This expression could be regulated by PDGF, as stimulation with PDGF in this study led to time-dependent down regulation of OTUD6B-AS1 and OTUD6B in fibroblasts of healthy controls. OTUD6B-AS1 did not appear to have an impact on expression of fibrotic genes such as type 1 collagen, fibronectin-1 (FN1), and α-SMA. Inhibition of OTUD6B-AS1, using antisense oligonucleotide (ASO)-mediated transcripts, showed suppression of apoptosis in dermal fibroblasts and human pulmonary artery smooth muscle cells (HPASMC) (Takata et al. 2019). Although, inhibition of proliferation was observed, this was thought to be a compensatory mechanism and further larger experiments are required to characterize this effect further. Expression of Cyclin D1 was significantly increased at both mRNA level and protein levels in both cell types which provides evidence for involvement of this lncRNA in proliferation and apoptosis. The study by Messemaker et al. also found this lncRNA to be dysregulated in patients with SSc, although no hypothesis as to function was presented (Messemaker et al. 2018).
Although most studies focus on expression in fibroblasts, the study by Takata et al. also studied HPASMC to further elucidate the pathogenesis of SSc in the lungs. Proliferation, and resistance to apoptosis of HPASMC in SSc is thought to be important in the development of microvascular lesions, which leads to proliferation of vascular smooth muscle cells and consequent vessel wall thickening and an occlusion of small arteries, which clinically translates to pulmonary arterial hypertension (Allanore et al. 2015; Distler et al. 2017). Thus, the regulation of Cyclin D1 by downregulation of OTUD6B-AS1 is a possible novel mechanism to explain apoptosis resistance observed in SSc.
ncRNA00201
The ncRNA00201 was found to control many pathways related to the 3 steps of SSc pathogenesis (Fig. 1), as well as tumor development (Dolcino et al. 2019). This could serve as the link between SSc and increased rates of cancer observed in these patients. Transcriptional profiles of SSc patients compared to controls, including more than 50,000 lncRNAs, found only one lncRNA (HNRNPU-AS1, also referred to as ncRNA00201) to be significantly decreased (Dolcino et al. 2019). There were 56 known miRNA and 31 gene targets of ncRNA00201 identified, many of which were involved in known SSc pathways. One of these is hnRNPC, which encodes a known autoantigen in SSc, the ribonucleoprotein complex (RNP) (Stanek et al. 1997). lncRNA00201 was found to also be an important player in disease pathways of immune and inflammatory response, vasculitis, and fibrosis, which are seen in SSc as well as cancer proliferation (Sutaria et al. 2017; Dolcino et al. 2018). This provides insight into disease pathogenesis and opens avenues for the design of novel therapeutic strategies.
PSMB8-AS1
Proteasome 20S Subunit Beta 8 Anti-Sense 1 (PSMB8-AS1) was identified as a top-ranking hub gene in co-expression modules implicated in cell activation and response to viral and external stimuli (Servaas et al. 2021). In the study by Servaas et al., transcriptomic data from two independent SSc patient cohorts revealed 886 lncRNAs with differential expression in monocytes. Then, the identified lncRNAs were associated with neighboring protein coding genes that are involved in regulating of IFN responses and apoptosis in monocytes. Further characterization of function of increased PSMB8-AS1 expression in monocytes demonstrated that this lncRNA is involved in the secretion of IL-6 and TNFα both of which are important cytokines in SSc pathogenesis and associated with fibrosis (De Lauretis et al. 2013; Schmidt et al. 2009). Thus, PSMB8-AS1 lncRNA is linked to monocyte dysregulation in SSc patients and may contribute to the pathogenesis.
MGC12916
DNA methylation levels in dermal fibroblasts from African American patients with SSc revealed hypermethylation of Uncharacterized Protein MGC12916 (MGC12916) lncRNA which was associated with its downregulation (Baker Frost et al. 2021). In general, dermal fibroblasts from African American patients showed widespread reduced DNA methylation. Differential methylation was identified in 17 genes and 11 promoters, mostly in ncRNA genes and pseudogenes. Gene set enrichment analysis and gene ontology analyses showed enhancement of pathways related to IFN signaling and mesenchymal differentiation. This is the first report of this lncRNAs of differential gene methylation in African American patients.
CTBP1-AS2, AGAP2-AS1
One study (Messemaker et al. 2018), evaluating 15,941 known annotated, lncRNAs found that 676 were dysregulated (122 decreased and 554 increased) in tissues of SSc patients compared to healthy controls. The top 3 deregulated anti-sense lncRNAs were CTBP1-AS2, AGAP2-AS1, and this study also identified OTUD6B-AS1. These were found to have a strong correlation with their paired sense genes. CTBP1 and CTBP1-AS2 levels had a positive correlation across cell types studied, especially immune cells. Interestingly, AGAP2 was only expressed in immune cells, while AGAP2-AS1 was only expressed in dermal cell types, which in keeping with other studies that showed this discordance. Thus, several AS-lncRNAs were found to be differentially expressed in SSc and levels were found to be altered in a disease-specific manner. However, specific functions of these were not elucidated in this study.
ANCR, TINCR, HOTTIP, SPRY4-IT1
The expression of these four lncRNAs, which have a role in skin biology, were explored in the plasma of SSc patients and correlated with presence of autoantibodies and subtype (Abd-Elmawla et al. 2020). Anti-differentiation ncRNA (ANCR) is a prototypical epidermal lncRNA which is expressed on undifferentiated keratinocytes and helps to maintain them in the basal layer (Kretz et al. 2012). Terminal differentiation–induced ncRNA (TINCR) is elevated in keratinocytes undergoing differentiation and contributes to regulation of gene expression involved with differentiation (Kretz et al. 2013). SPRY4 intronic transcript 1 (SPRY4-IT1) which arises from the intron region of Sprouty4 (SPRY4) gene, is found to be expressed in melanocytes, keratinocytes, and has been found to block apoptosis and promote viability and motility of melanoma cells (Khaitan et al. 2011). Finally, the HOXA transcript at the distal tip (HOTTIP) controls cellular growth, proliferation, and apoptosis (Wang et al. 2011).
Plasma levels of TINCR, HOTTIP, and SPRY4-IT1 in SSc patients were found to be elevated and ANCR-downregulated compared to controls (Abd-Elmawla et al. 2020). SPRY4-IT1 was found to be a strong diagnostic indicator and was higher in patients with dcSSc compared to lcSSc. Both SPRY4-IT1 and HOTTIP correlated positively with the modified Rodnan skin score depicting the degree of fibrosis, and the latter also with antinuclear antibody profile. Both SPRY4-IT1 and ANCR were positively associated with pulmonary hypertension. Hence, plasma SPRY4-IT1, HOTTIP, ANCR and TINCR may represent novel candidate biomarkers for SSc, where SPRY4-IT1 could also help with subtype determination.
Dermatomyositis
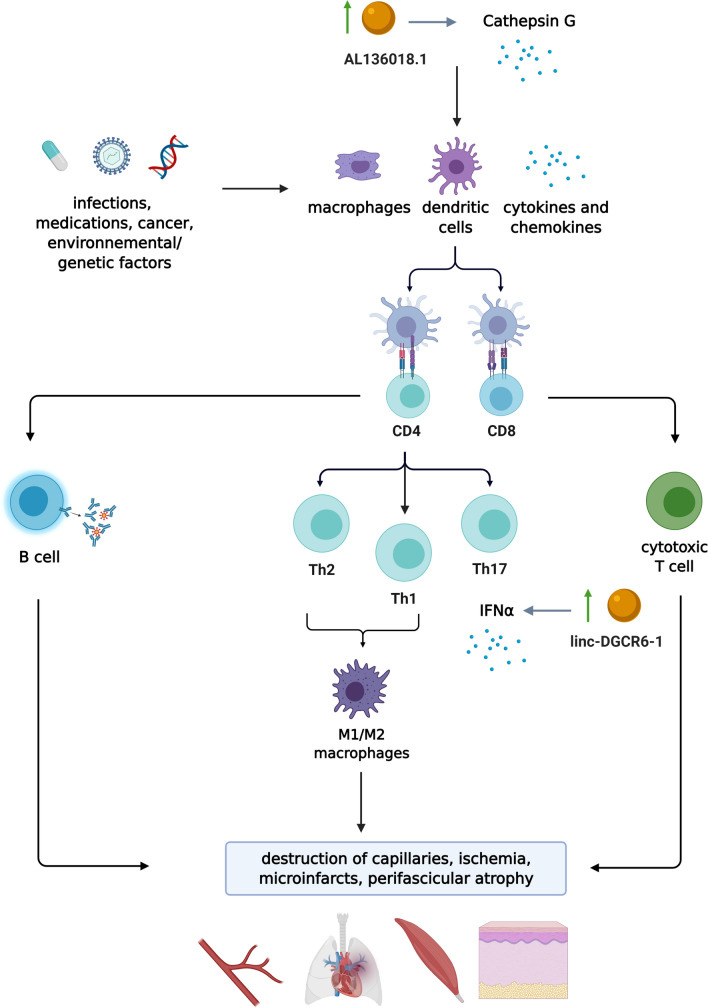
DM is an autoimmune disease primarily affecting skeletal muscle and skin (Waldman, DeWane, and Lu 2020). Clinically, DM is characterized by symmetrical skeletal muscle weakness and cutaneous manifestations including, heliotrope rash, Gottron papules, and Gottron sign (Lundberg et al. 2017; Sharma et al. 2017). A subset of DM may present as cutaneous stigmata of solid organ malignancy (Qiang et al. 2017; Yang et al. 2015), whereas all forms may be associated with systemic complications including interstitial lung disease (ILD), oesophageal dysmotility and cardiac disease (Kurasawa et al. 2018; Chen et al. 2013). While genetic associations and environmental factors, including infections, ultraviolet radiation, vitamin D deficiency and drugs, have been linked to DM, the overall molecular mechanism of DM remains largely unknown (Thompson et al. 2018). However, similar to other SARDs, the role innate immunity, humoral and cellular immune responses are of importance (Dalakas 2015). Studies have shown that the characteristic muscle pathology of DM demonstrates capillary abnormalities and small, abnormal appearing muscle fibers bordering the perimysial connective tissue, a lesion called perifascicular atrophy (PFA) (Salajegheh et al. 2010). Microarray data have pointed towards a mechanism of tissue injury in DM associated with the overexpression of type 1 IFN-inducible genes (Salajegheh et al. 2010). Compared to normal muscle and other inflammatory myopathies, over 85% of the highest expressed transcripts in muscle from DM are from genes known to be induced by type 1 IFNs (Greenberg et al. 2005a, b).
linc-DGCR6-1
The role of lncRNA in DM was first studied by Peng et al. (2016) (Fig. 2). Of the differentially expressed lncRNAs, lncRNAs ENST00000428205.1 and ENST00000450016.1 displayed the most significant increase and decrease, respectively. Based on the results of correlation analyses of the differentially expressed lncRNAs and mRNAs, 12 mRNA-lncRNA co-expression networks were identified. This suggests the possibility that these lncRNAs are regulators of corresponding mRNA expression. Although the function of the target mRNA has not yet been elucidated in 11 lncRNA-mRNA pairs, linc-DGCR6-1 was thought to target the USP18 protein, a type 1 IFN-inducible protein. This is in keeping with findings by Salajegheh et al., which showed marked USP18 transcript elevation (68-fold increase) in skeletal muscle biopsies from patients with DM compared to control subjects (Salajegheh et al. 2010). It is thought that USP18 plays a critical role in the enzymatic pathway for IFN-stimulated gene 15 (ISG15) conjugation to a target protein. ISG15 is known as a type 1 IFN-inducible protein and is the single most overexpressed gene in DM muscle compared to both normal muscle and muscle from patients with other inflammatory myopathies (Salajegheh et al. 2010; Greenberg, et al. 2005a, b). Specifically, USP18 deconjugates ISG15 from target proteins, although the role of ISG15 in the mediation of myofiber injury in DM/ PM remains to elucidated (Basters et al. 2018; Salajegheh et al. 2010). Besides being an active enzyme, USP18 has also been found to negatively regulate type 1 IFN signalling independent of its protease activity (Malakhova et al. 2006; Basters et al. 2018). Although, USP18 is currently hypothesized to play a critical role in the enzymatic pathway for ISG15 conjugation to a target protein, more studies are necessary to further elucidate the role of USP18 in the mediation of myofiber injury in DM/ PM.
Fig. 2.
Pathogenesis of dermatomyositis (DM). While the pathogenesis of DM is poorly understood, it is believed to be triggered by an environmental factor, medication or cancer in a genetically susceptible host. Complex interplay of aberrant innate and adaptive immune response leads to downstream capillary damage/ischemia in end organs and characteristic muscle disease (i.e. perifascicular atrophy). While several lncRNAs have been found to be differentially expressed and several were co-expressed along with mRNA, linc-DGCR6-1 and AL136018.1 have a proposed role in DM’s pathogenesis through targeting the USP18 protein involved in the type 1 IFN pathway, and regulating CTSG expression involved in increasing vasopermeability and chemotaxis of inflammatory cells, respectively. Created with Biorender.com
AL136018.1
In a recent study by Liang and Peng (2021), the antisense lncRNA, AL136018.1, was discovered to be highly expressed in skeletal muscle tissues of patients with DM (Liang and Peng 2021). Interestingly, AL136018.1 expression positively correlated with transcription level and DNA methylation of its contiguous CTSG gene, which encodes Cathepsin G. A member of the serine protease family, Cathepsin G plays an important role in increasing the permeability of vascular endothelial cells and the chemotaxis of inflammatory cells. Previous studies have demonstrated increased expression and activity of Cathepsin G in peripheral blood and muscle tissue in patients with DM (Gao et al. 2017). It is suggested that CTSG may play an important role in facilitating CD4 + T cell and B cell infiltration in perivascular and muscle tissue by increasing the permeability of vascular endothelial cells in DM (Gao et al. 2017).
Further studies investigating the function of AL136018.1 transcripts may reveal their role in regulating CTSG expression and their overall importance in the pathogenesis of DM.
Discoid CLE
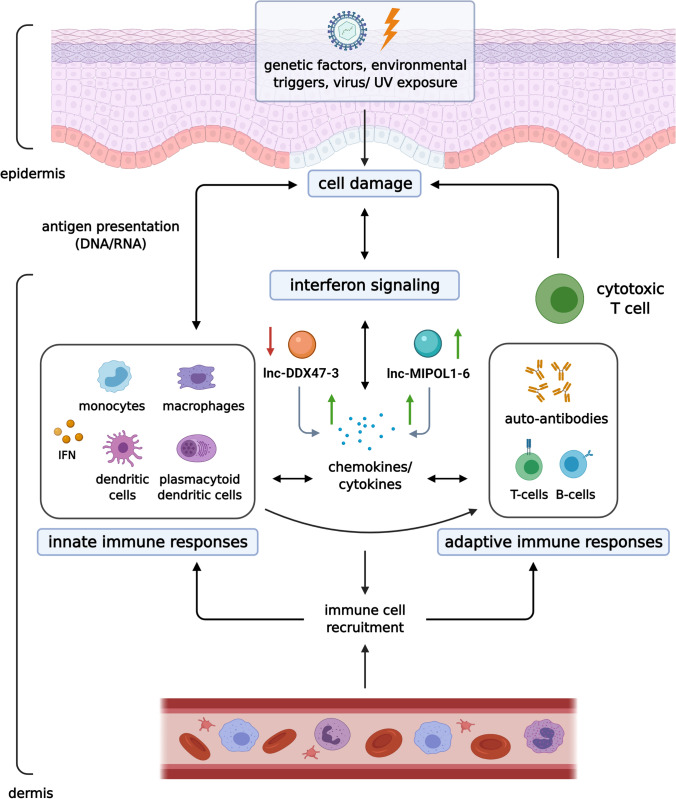
CLE encompasses a wide range of dermatologic manifestations that may or may not be associated with the development of systemic disease (Okon and Werth 2013). In fact, the pathophysiology of CLE and SLE has been hypothesized to be closely comparable where elevations in cytokine expression, such as IFN-α, have-been identified in both diseases (Lee and Sinha 2006). CLE is divided into three main subtypes: acute CLE (ACLE), subacute CLE (SCLE) and chronic CLE (CCLE) (Okon and Werth 2013; Grönhagen and Nyberg 2014). ACLE typically presents as a malar rash (Kuhn, Landmann, and Bonsmann 2016) while SCLE presents as with widespread, non-scarring, photosensitive erythematous annular or psoriasiform plaques (Walling and Sontheimer 2009). The most common form of CCLE is discoid CLE (DLE), which is characterized by indurated discoid (coin-shaped) plaques that commonly involve the head and neck, including the scalp and ears (Walling and Sontheimer 2009). Unless diagnosed and treated in a timely fashion, DLE can lead to disfiguring scarring (Ghauri et al. 2012; Kuhn et al. 2016). Detailed molecular mechanisms of the pathogenesis of DLE are not well understood and the role of lncRNA in CLE has not yet been extensively explored. Current pathogenic concept focuses on epidermal DNA/RNA damage revealing neo-antigens and eliciting complex innate and adaptive immune responses (Fig. 3) (Wenzel 2019).
Fig. 3.
Pathogenesis of discoid lupus (DLE). It is believed that epidermal cell damage revealing nuclear DNA initiate the inflammatory process in DLE, featuring complex innate and adaptive immune responses. The role of Th1 response and interferon signaling has been demonstrated. In regards to lncRNAs, upregulation of lnc-MIPOL1-6 and downregulation of lnc-DDX47-3 in lesional lip mucosa was associated with increased expression of Th1 cytokines and chemokines (IL19, CXCL1, CXCL11, and TNFSF15). Created with Biorender.com
lnc-MIPOL1-6 and lnc-DDX47-3
The role of lncRNAs in DLE pathogenesis was first investigated by Xuan et al., who discovered the dysregulation of two lncRNAs, lnc-MIPOL1-6 and lnc-DDX47-3 (Xuan et al. 2019) (Fig. 3). In total, 507 lncRNAs were found to be differentially expressed in lesional lip in comparison to non-lesional mucosa. Using 37 significantly expressed coding genes, authors constructed a coding-noncoding co-expression (CNC) network whereby a significant correlation was identified between lncRNA and their potential nearby genes. Up-regulation of lnc-MIPOL1-6 was associated with increased IL-19, CXCL1, CXCL11, and TNFSF15 chemokine expression. On the other hand, downregulation of lnc-DDX47 was also associated with the increased expression of the aforementioned chemokines (Fig. 3). Specifically, IL-19, has been shown to be associated with Th1 dominant diseases (Azuma et al. 2011; Wenzel 2019); CXCL11 has been found to promote Th1 cell recruitment (Kumar and Herbert 2017); and TNSF15 has a role in stimulating Th1 cytokine production (Zhang and Li 2012; Zhang and Zhang 2015). A recent study by Jabbari et al. correspondingly demonstrated a predominance of IFN-γ-producing Th1 cells from the skin of DLE patients, suggesting the important role of Th1 cells in the pathogenesis of DLE (Jabbari et al. 2014). Results also showed that Transcription Factors (TF), STAT4, ETV6, and ZNF597, regulated most of the pathways that involved the differentially expressed lncRNAs, which suggested the importance of these TFs in the pathogenesis of this disease. STAT4 has been found to regulate the IL-12 response and is important in Th1 cell differentiation (Jordan et al. 2014). ETV6 has been linked to a number of hematologic neoplasms and was found to be an important TF for blood cell development (Lambert 2019). The role of ZNF597 has not been discovered (Schulze et al. 2019). Taken together, the role of Th1 pathway in DLE pathogenesis and the role of elicited lncRNAs warrants further investigation in larger studies.
Limitations and call for future studies
The main limitation for all studies included in this review was the small sample size. Thus, the interpreted results can only be applied to select individuals given that a diversity of participants is excluded. Additionally, a small sample size increases the risk of type II statistical error, and it is important to note that the predicted correlations of lncRNA-mRNA networks does not signify causation. Selection bias in choosing certain lncRNA and mRNA to create lncRNA-mRNA networks also limits the scope of studying the differential lncRNAs and their role in the studied diseases. Furthermore, in many instances, studies identifying significant lncRNAs are unique and have not been replicated or verified by external sources.
Future studies with a larger sample sizes designed to investigate the differential expression of lncRNA in patients with autoimmune and connective tissue diseases are warranted. With larger studies, the clinical application of these results could yield lncRNAs as biomarkers for disease development, targets for genetic testing, or for following the progression of the disease in patients with autoimmune skin conditions (Galeotti and Bayry 2020). Knockout lncRNA in vivo animal models or in vitro cell lines using CRISPR/Cas9 technologies for genomic engineering may be helpful in determining essential lncRNAs in disease development (Fu 2014). Further, these models may help in determining suitable lncRNAs as specific drug targets in autoimmune diseases. Given the strong family history among autoimmune diseases, it would also be of interest to investigate if lncRNA profiles suggest a familial inheritance pattern (Galeotti and Bayry 2020).
Overall, our understanding of the role of the differential expression of lncRNAs in connective tissue diseases is expanding, although there is still a large knowledge gap that must be addressed before making definitive conclusions regarding the significance of lncRNA in disease pathophysiology. Although lncRNAs have up to tenfold lower expression than mRNAs, these serve important regulatory functions including activation of IFN pathways in SSc, DM and DLE, increased collagen production and fibroblast survival in SSc, and Th1 shift in DLE, important in the pathogenesis of these cutaneous connective tissue conditions. The key lncRNAs that may drive pathogenesis of these connective tissue diseases could potentially serve as therapeutic targets in the future.
Funding
None.
Availability of data and materials
Not applicable.
Code availability
Not applicable.
Declarations
Conflict of interest
None.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Anastasiya Muntyanu and Michelle Le have contributed equally to this work.
Contributor Information
Anastasiya Muntyanu, Email: anastasiya.muntyanu@mail.mcgill.ca.
Michelle Le, Email: michelle.le@mail.mcgill.ca.
Zainab Ridha, Email: zainab.ridha.1@ulaval.ca.
Elizabeth O’Brien, Email: elizabeth.o'brien@mcgill.ca.
Ivan V. Litvinov, Email: ivan.litvinov@mcgill.ca
Philippe Lefrançois, Email: philippe.lefrancois2@mail.mcgill.ca.
Elena Netchiporouk, Email: elena.netchiporouk@mail.mcgill.ca.
References
- Abd-Elmawla MA, Hassan M, Elsabagh YA, Alnaggar A, Senousy MA. Deregulation of long noncoding RNAs ANCR, TINCR, HOTTIP and SPRY4-IT1 in plasma of systemic sclerosis patients: SPRY4-IT1 as a novel biomarker of scleroderma and its subtypes. Cytokine. 2020;133:155124. doi: 10.1016/j.cyto.2020.155124. [DOI] [PubMed] [Google Scholar]
- Allanore Y, Simms R, Distler O, Trojanowska M, Pope J, Denton CP, Varga J. Systemic sclerosis. Nat Rev Dis Primers. 2015;1:15002. doi: 10.1038/nrdp.2015.2. [DOI] [PubMed] [Google Scholar]
- Aslani S, Sobhani S, Gharibdoost F, Jamshidi A, Mahmoudi M. Epigenetics and pathogenesis of systemic sclerosis; the ins and outs. Hum Immunol. 2018;79:178–187. doi: 10.1016/j.humimm.2018.01.003. [DOI] [PubMed] [Google Scholar]
- Azuma YT, Nakajima H, Takeuchi T. IL-19 as a potential therapeutic in autoimmune and inflammatory diseases. Curr Pharm Des. 2011;17:3776–3780. doi: 10.2174/138161211798357845. [DOI] [PubMed] [Google Scholar]
- Badea I, Taylor M, Rosenberg A, Foldvari M. Pathogenesis and therapeutic approaches for improved topical treatment in localized scleroderma and systemic sclerosis. Rheumatology (oxford) 2009;48:213–221. doi: 10.1093/rheumatology/ken405. [DOI] [PubMed] [Google Scholar]
- Baker Frost D, da Silveira W, Hazard ES, Atanelishvili I, Wilson RC, Flume J, Day KL, Oates JC, Bogatkevich GS, Feghali-Bostwick C, Hardiman G, Ramos PS. Differential DNA methylation landscape in skin fibroblasts from african americans with systemic sclerosis. Genes (basel) 2021;12:129. doi: 10.3390/genes12020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basters A, Knobeloch KP, Fritz G. USP18—a multifunctional component in the interferon response. Biosci Rep. 2018;38:BSR20180250. doi: 10.1042/BSR20180250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C, Distler JH. Epigenetic factors as drivers of fibrosis in systemic sclerosis. Epigenomics. 2017;9:463–477. doi: 10.2217/epi-2016-0150. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Varga J. Emerging roles of innate immune signaling and toll-like receptors in fibrosis and systemic sclerosis. Curr Rheumatol Rep. 2015;17:474. doi: 10.1007/s11926-014-0474-z. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Kelley K, Melichian DS, Tamaki Z, Fang F, Su Y, Feng G, Pope RM, Budinger GR, Mutlu GM, Lafyatis R, Radstake T, Feghali-Bostwick C, Varga J. Toll-like receptor 4 signaling augments transforming growth factor-beta responses: a novel mechanism for maintaining and amplifying fibrosis in scleroderma. Am J Pathol. 2013;182:192–205. doi: 10.1016/j.ajpath.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black CM, Silman AJ, Herrick AI, Denton CP, Wilson H, Newman J, Pompon L, Shi-Wen X. Interferon-alpha does not improve outcome at one year in patients with diffuse cutaneous scleroderma: results of a randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 1999;42:299–305. doi: 10.1002/1529-0131(199902)42:2<299::AID-ANR12>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Boleto G, Allanore Y, Avouac J. Targeting costimulatory pathways in systemic sclerosis. Front Immunol. 2018;9:2998. doi: 10.3389/fimmu.2018.02998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brkic Z, van Bon L, Cossu M, van Helden-Meeuwsen CG, Vonk MC, Knaapen H, van den Berg W, Dalm VA, Van Daele PL, Severino A, Maria NI, Guillen S, Dik WA, Beretta L, Versnel MA, Radstake T. The interferon type I signature is present in systemic sclerosis before overt fibrosis and might contribute to its pathogenesis through high BAFF gene expression and high collagen synthesis. Ann Rheum Dis. 2016;75:1567–1573. doi: 10.1136/annrheumdis-2015-207392. [DOI] [PubMed] [Google Scholar]
- Brooks WH, Renaudineau Y. Epigenetics and autoimmune diseases: the X chromosome-nucleolus nexus. Front Genet. 2015;6:22–22. doi: 10.3389/fgene.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B, Cai J, Yin Z, Jiang X, Yao C, Ma J, Xue Z, Miao P, Xiao Q, Cheng Y, Qin J, Guo Q, Shen N, Ye Z, Qu B, Ding H. Long non-coding RNA expression profiles in neutrophils revealed potential biomarker for prediction of renal involvement in SLE patients. Rheumatology (oxford) 2021;60:1734–1746. doi: 10.1093/rheumatology/keaa575. [DOI] [PubMed] [Google Scholar]
- Chen Z, Cao M, Plana MN, Liang J, Cai H, Kuwana M, Sun L. Utility of anti-melanoma differentiation–associated gene 5 antibody measurement in identifying patients with dermatomyositis and a high risk for developing rapidly progressive interstitial lung disease: a review of the literature and a meta-analysis. Arthritis Care Res. 2013;65:1316–1324. doi: 10.1002/acr.21985. [DOI] [PubMed] [Google Scholar]
- Chen W, Liu Di, Li Q-Z, Zhu H. The function of ncRNAs in rheumatic diseases. Epigenomics. 2019;11:821–833. doi: 10.2217/epi-2018-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmann RB, Hayes E, Pendergrass S, Padilla C, Farina G, Affandi AJ, Whitfield ML, Farber HW, Lafyatis R. Interferon and alternative activation of monocyte/macrophages in systemic sclerosis-associated pulmonary arterial hypertension. Arthritis Rheum. 2011;63:1718–1728. doi: 10.1002/art.30318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechomska M, Cant R, Finnigan J, van Laar JM, O'Reilly S. Role of toll-like receptors in systemic sclerosis. Expert Rev Mol Med. 2013;15:e9. doi: 10.1017/erm.2013.10. [DOI] [PubMed] [Google Scholar]
- Cossu M, van Bon L, Preti C, Rossato M, Beretta L, Radstake T. Earliest phase of systemic sclerosis typified by increased levels of inflammatory proteins in the serum. Arthritis Rheumatol. 2017;69:2359–2369. doi: 10.1002/art.40243. [DOI] [PubMed] [Google Scholar]
- Dalakas MC. Inflammatory muscle diseases. N Engl J Med. 2015;373:393–394. doi: 10.1056/NEJMc1506827. [DOI] [PubMed] [Google Scholar]
- De Lauretis A, Sestini P, Pantelidis P, Hoyles R, Hansell DM, Goh NS, Zappala CJ, Visca D, Maher TM, Denton CP, Ong VH, Abraham DJ, Kelleher P, Hector L, Wells AU, Renzoni EA. Serum interleukin 6 is predictive of early functional decline and mortality in interstitial lung disease associated with systemic sclerosis. J Rheumatol. 2013;40:435–446. doi: 10.3899/jrheum.120725. [DOI] [PubMed] [Google Scholar]
- Denton CP, Khanna D. Systemic sclerosis. Lancet. 2017;390:1685–1699. doi: 10.1016/S0140-6736(17)30933-9. [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux Y, Zangrando J, Schroen B, Creemers EE, Pedrazzini T, Chang CP, Dorn GW, 2nd, Thum T, Heymans S. Long noncoding RNAs in cardiac development and ageing. Nat Rev Cardiol. 2015;12:415–425. doi: 10.1038/nrcardio.2015.55. [DOI] [PubMed] [Google Scholar]
- Diab S, Dostrovsky N, Hudson M, Tatibouet S, Fritzler MJ, Baron M, Khalidi N. Systemic sclerosis sine scleroderma: a multicenter study of 1417 subjects. J Rheumatol. 2014;41:2179–2185. doi: 10.3899/jrheum.140236. [DOI] [PubMed] [Google Scholar]
- Distler JH, Feghali-Bostwick C, Soare A, Asano Y, Distler O, Abraham DJ. Review: frontiers of antifibrotic therapy in systemic sclerosis. Arthritis Rheumatol. 2017;69:257–267. doi: 10.1002/art.39865. [DOI] [PubMed] [Google Scholar]
- Dolcino M, Pelosi A, Fiore PF, Patuzzo G, Tinazzi E, Lunardi C, Puccetti A. Gene profiling in patients with systemic sclerosis reveals the presence of oncogenic gene signatures. Front Immunol. 2018;9:449. doi: 10.3389/fimmu.2018.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolcino M, Tinazzi E, Puccetti A, Lunardi C. In systemic sclerosis, a unique long non coding rna regulates genes and pathways involved in the three main features of the disease (vasculopathy, fibrosis and autoimmunity) and in carcinogenesis. J Clin Med. 2019;8:320. doi: 10.3390/jcm8030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloranta ML, Franck-Larsson K, Lovgren T, Kalamajski S, Ronnblom A, Rubin K, Alm GV, Ronnblom L. Type I interferon system activation and association with disease manifestations in systemic sclerosis. Ann Rheum Dis. 2010;69:1396–1402. doi: 10.1136/ard.2009.121400. [DOI] [PubMed] [Google Scholar]
- Farina G, Lemaire R, Pancari P, Bayle J, Widom RL, Lafyatis R. Cartilage oligomeric matrix protein expression in systemic sclerosis reveals heterogeneity of dermal fibroblast responses to transforming growth factor beta. Ann Rheum Dis. 2009;68:435–441. doi: 10.1136/ard.2007.086850. [DOI] [PubMed] [Google Scholar]
- Farrugia M, Baron B. The role of toll-like receptors in autoimmune diseases through failure of the self-recognition mechanism. Int J Inflam. 2017;2017:8391230. doi: 10.1155/2017/8391230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D. Non-coding RNA: a new frontier in regulatory biology. Natl Sci Rev. 2014;1:190–204. doi: 10.1093/nsr/nwu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielli A, Avvedimento EV, Krieg T. Scleroderma. N Engl J Med. 2009;360:1989–2003. doi: 10.1056/NEJMra0806188. [DOI] [PubMed] [Google Scholar]
- Galeotti C, Bayry J. Autoimmune and inflammatory diseases following COVID-19. Nat Rev Rheumatol. 2020;16:413–414. doi: 10.1038/s41584-020-0448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Zhu H, Yang H, Zhang H, Li Q, Luo H. The role and mechanism of cathepsin G in dermatomyositis. Biomed Pharmacother. 2017;94:697–704. doi: 10.1016/j.biopha.2017.07.088. [DOI] [PubMed] [Google Scholar]
- Gao Y, Li S, Zhang Z, Xinhua Yu, Zheng J. The role of long non-coding RNAs in the pathogenesis of RA SLE, and SS. Front Med. 2018;5:193–293. doi: 10.3389/fmed.2018.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett SM, Baker Frost D, Feghali-Bostwick C. The mighty fibroblast and its utility in scleroderma research. J Scleroderma Relat Disord. 2017;2:69–134. doi: 10.5301/jsrd.5000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghauri A-J, Valenzuela AA, O'Donnell B, Selva D, Madge SN. Periorbital discoid lupus erythematosus. Ophthalmology. 2012;119:2193.e11–2194.e11. doi: 10.1016/j.ophtha.2012.05.041. [DOI] [PubMed] [Google Scholar]
- Gilbane AJ, Denton CP, Holmes AM. Scleroderma pathogenesis: a pivotal role for fibroblasts as effector cells. Arthritis Res Ther. 2013;15:215. doi: 10.1186/ar4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg SA, Pinkus JL, Pinkus GS, Burleson T, Sanoudou D, Tawil R, Barohn RJ, Saperstein DS, Briemberg HR, Ericsson M, Park P, Amato AA. Interferon-alpha/beta-mediated innate immune mechanisms in dermatomyositis. Ann Neurol. 2005;57:664–678. doi: 10.1002/ana.20464. [DOI] [PubMed] [Google Scholar]
- Greenberg SA, Pinkus JL, Pinkus GS, Burleson T, Sanoudou D, Tawil R, Barohn RJ, Saperstein DS, Briemberg HR, Ericsson M. Interferon-α/β-mediated innate immune mechanisms in dermatomyositis. Ann Neurol. 2005;57:664–678. doi: 10.1002/ana.20464. [DOI] [PubMed] [Google Scholar]
- Grönhagen CM, Nyberg F. Cutaneous lupus erythematosus: an update. Indian Dermatol Online J. 2014;5:7–13. doi: 10.4103/2229-5178.126020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Higgs BW, Bay-Jensen AC, Karsdal MA, Yao Y, Roskos LK, White WI. Suppression of T cell activation and collagen accumulation by an anti-IFNAR1 mAb, anifrolumab, in adult patients with systemic sclerosis. J Investig Dermatol. 2015;135:2402–2409. doi: 10.1038/jid.2015.188. [DOI] [PubMed] [Google Scholar]
- Hinz B, Phan SH, Thannickal VJ, Prunotto M, Desmouliere A, Varga J, De Wever O, Mareel M, Gabbiani G. Recent developments in myofibroblast biology: paradigms for connective tissue remodeling. Am J Pathol. 2012;180:1340–1355. doi: 10.1016/j.ajpath.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyles RK, Derrett-Smith EC, Khan K, Shiwen X, Howat SL, Wells AU, Abraham DJ, Denton CP. An essential role for resident fibroblasts in experimental lung fibrosis is defined by lineage-specific deletion of high-affinity type II transforming growth factor β receptor. Am J Respir Crit Care Med. 2011;183:249–261. doi: 10.1164/rccm.201002-0279OC. [DOI] [PubMed] [Google Scholar]
- Huang XL, Wang YJ, Yan JW, Wan YN, Chen B, Li BZ, Yang GJ, Wang J. Role of anti-inflammatory cytokines IL-4 and IL-13 in systemic sclerosis. Inflamm Res. 2015;64:151–159. doi: 10.1007/s00011-015-0806-0. [DOI] [PubMed] [Google Scholar]
- Ihn H, Sato S, Fujimoto M, Kikuchi K, Takehara K. Demonstration of interleukin-2, interleukin-4 and interleukin-6 in sera from patients with localized scleroderma. Arch Dermatol Res. 1995;287:193–197. doi: 10.1007/BF01262331. [DOI] [PubMed] [Google Scholar]
- Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwayama T, Olson LE. Involvement of PDGF in fibrosis and scleroderma: recent insights from animal models and potential therapeutic opportunities. Curr Rheumatol Rep. 2013;15:304. doi: 10.1007/s11926-012-0304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbari A, Suárez-Fariñas M, Fuentes-Duculan J, Gonzalez J, Cueto I, Franks AG, Jr, Krueger JG. Dominant Th1 and minimal Th17 skewing in discoid lupus revealed by transcriptomic comparison with psoriasis. J Investig Dermatol. 2014;134:87–95. doi: 10.1038/jid.2013.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. Adv Exp Med Biol. 2017;1008:1–46. doi: 10.1007/978-981-10-5203-3_1. [DOI] [PubMed] [Google Scholar]
- Jordan MA, Field J, Butzkueven H, Baxter AG. Genetic predisposition, humans. In: Rose NR, Mackay IR, editors. The autoimmune diseases. 5. Boston: Academic Press; 2014. [Google Scholar]
- Kähäri VM, Sandberg M, Kalimo H, Vuorio T, Vuorio E. Identification of fibroblasts responsible for increased collagen production in localized scleroderma by in situ hybridization. J Investig Dermatol. 1988;90:664–670. doi: 10.1111/1523-1747.ep12560826. [DOI] [PubMed] [Google Scholar]
- Kassamali B, Kassamali AA, Muntyanu A, Netchiporouk E, Vleugels RA, LaChance A. Geographic distribution and environmental triggers of systemic sclerosis cases from two large academic tertiary centers in massachusetts. J Am Acad Dermatol. 2021;17:351. doi: 10.1016/j.jaad.2021.03.055. [DOI] [PubMed] [Google Scholar]
- Katsumoto TR, Whitfield ML, Kari Connolly M. The pathogenesis of systemic sclerosis. Annu Rev Pathol. 2011;6:509–537. doi: 10.1146/annurev-pathol-011110-130312. [DOI] [PubMed] [Google Scholar]
- Kawakami T, Ihn H, Xu W, Smith E, LeRoy C, Trojanowska M. Increased expression of TGF-beta receptors by scleroderma fibroblasts: evidence for contribution of autocrine TGF-beta signaling to scleroderma phenotype. J Investig Dermatol. 1998;110:47–51. doi: 10.1046/j.1523-1747.1998.00073.x. [DOI] [PubMed] [Google Scholar]
- Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- Klareskog L, Gustafsson R, Scheynius A, Hallgren R. Increased expression of platelet-derived growth factor type B receptors in the skin of patients with systemic sclerosis. Arthritis Rheum. 1990;33:1534–1541. doi: 10.1002/art.1780331011. [DOI] [PubMed] [Google Scholar]
- Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59–59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, Qu K, Zheng GX, Chow J, Kim GE, Rinn JL, Chang HY, Siprashvili Z, Khavari PA. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012;26:338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, Johnston D, Kim GE, Spitale RC, Flynn RA, Zheng GX, Aiyer S, Raj A, Rinn JL, Chang HY, Khavari PA. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Landmann A, Bonsmann G. Cutaneous lupus erythematosus. In: Tsokos GC, editor. Systemic lupus erythematosus. Boston: Academic Press; 2016. [Google Scholar]
- Kumar RK, Herbert C. Airway epithelial cytokines in asthma and chronic obstructive pulmonary disease. In: Foti M, Locati M, editors. Cytokine effector functions in tissues. Cambridge: Academic Press; 2017. [Google Scholar]
- Kurasawa K, Arai S, Namiki Y, Tanaka A, Takamura Y, Owada T, Arima M, Maezawa R. Tofacitinib for refractory interstitial lung diseases in anti-melanoma differentiation-associated 5 gene antibody-positive dermatomyositis. Rheumatology. 2018;57:2114–2119. doi: 10.1093/rheumatology/key188. [DOI] [PubMed] [Google Scholar]
- Lafyatis R, Farina A. New insights into the mechanisms of innate immune receptor signalling in fibrosis. Open Rheumatol J. 2012;6:72–79. doi: 10.2174/1874312901206010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagares D, Santos A, Grasberger PE, Liu F, Probst CK, Rahimi RA, Sakai N, Kuehl T, Ryan J, Bhola P, Montero J, Kapoor M, Baron M, Varelas X, Tschumperlin DJ, Letai A, Tager AM. Targeted apoptosis of myofibroblasts with the BH3 mimetic ABT-263 reverses established fibrosis. Sci Transl Med. 2017;9:eaal3765. doi: 10.1126/scitranslmed.aal3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert MP. Congenital thrombocytopenia'. In: Shaz BH, Hillyer CD, Reyes Gil M, editors. Transfusion medicine and hemostasis. 3. New York: Elsevier; 2019. [Google Scholar]
- Le M, Muntyanu A, Netchiporouk E. IncRNAs and circRNAs provide insight into discoid lupus pathogenesis and progression. Ann Transl Med. 2020;8:260. doi: 10.21037/atm.2020.03.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Sinha AA. Cutaneous lupus erythematosus: understanding of clinical features, genetic basis, and pathobiology of disease guides therapeutic strategies. Autoimmunity. 2006;39:433–444. doi: 10.1080/08916930600886851. [DOI] [PubMed] [Google Scholar]
- LeRoy EC, Medsger TA., Jr Criteria for the classification of early systemic sclerosis. J Rheumatol. 2001;28:1573–1576. [PubMed] [Google Scholar]
- LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, Medsger TA, Jr, Rowell N, Wollheim F. Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. J Rheumatol. 1988;15:202–205. [PubMed] [Google Scholar]
- Liakouli V, Elies J, El-Sherbiny YM, Scarcia M, Grant G, Abignano G, Derrett-Smith EC, Esteves F, Cipriani P, Emery P, Denton CP, Giacomelli R, Mavria G, Del Galdo F. Scleroderma fibroblasts suppress angiogenesis via TGF-beta/caveolin-1 dependent secretion of pigment epithelium-derived factor. Ann Rheum Dis. 2018;77:431–440. doi: 10.1136/annrheumdis-2017-212120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Peng Y. Gene body methylation facilitates the transcription of CTSG via antisense lncRNA AL136018.1 in dermatomyositic myoideum. Cell Biol Int. 2021;45:456–462. doi: 10.1002/cbin.11508. [DOI] [PubMed] [Google Scholar]
- Lundberg IE, Tjärnlund A, Bottai M, Werth VP, Pilkington C, de Visser M, Alfredsson L, Amato AA, Barohn RJ, Liang MH. 2017 European League Against Rheumatism/American College of Rheumatology classification criteria for adult and juvenile idiopathic inflammatory myopathies and their major subgroups. Arthritis Rheumatol. 2017;69:2271–2282. doi: 10.1002/art.40320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, Fuchs SY, Shuai K, Zhang DE. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. Embo j. 2006;25:2358–2367. doi: 10.1038/sj.emboj.7601149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti B, Servaas NH, Rossato M, Tamassia N, Cassatella MA, Cossu M, Beretta L, van der Kroef M, Radstake T, Bazzoni F. The long non-coding RNA NRIR drives IFN-response in monocytes: implication for systemic sclerosis. Front Immunol. 2019;10:100. doi: 10.3389/fimmu.2019.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matucci-Cerinic M, Kahaleh B, Wigley FM. Review: evidence that systemic sclerosis is a vascular disease. Arthritis Rheum. 2013;65:1953–1962. doi: 10.1002/art.37988. [DOI] [PubMed] [Google Scholar]
- Merkel PA, Clements PJ, Reveille JD, Suarez-Almazor ME, Valentini G, Furst DE. Current status of outcome measure development for clinical trials in systemic sclerosis. Report from OMERACT 6. J Rheumatol. 2003;30:1630–1647. [PubMed] [Google Scholar]
- Messemaker TC, Chadli L, Cai G, Goelela VS, Boonstra M, Dorjee AL, Andersen SN, Mikkers HMM, van Hof P, Mei H, Distler O, Draisma HHM, Johnson ME, Orzechowski NM, Simms RW, Toes REM, Aarbiou J, Huizinga TW, Whitfield ML, DeGroot J, de Vries-Bouwstra J, Kurreeman F. Antisense long non-coding rnas are deregulated in skin tissue of patients with systemic sclerosis. J Investig Dermatol. 2018;138:826–835. doi: 10.1016/j.jid.2017.09.053. [DOI] [PubMed] [Google Scholar]
- Muhammad N, Nor A. Biological database searching. In: Ranganathan S, Gribskov M, Nakai K, Schönbach C, editors. Encyclopedia of bioinformatics and computational biology. Oxford: Academic Press; 2019. [Google Scholar]
- Wang J, Yan S, Yang J, Lu H, Xu D, Wang Z. Non-coding RNAs in rheumatoid arthritis: from bench to bedside. Front Immunol. 2020;10:1986. doi: 10.3389/fimmu.2019.03129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okon LG, Werth VP. Cutaneous lupus erythematosus: diagnosis and treatment. Best Pract Res Clin Rheumatol. 2013;27:391–404. doi: 10.1016/j.berh.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly S, van Laar JM. Targeting the TLR4-MD2 axis in systemic sclerosis. Nat Rev Rheumatol. 2018;14:564–566. doi: 10.1038/s41584-018-0077-6. [DOI] [PubMed] [Google Scholar]
- Ouchene L, Muntyanu A, Lavoué J, Baron M, Litvinov IV, Netchiporouk E. Towards understanding of environmental risk factors in Systemic Sclerosis. J Cutan Med Surg. 2020;25:188–204. doi: 10.1177/1203475420957950. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachera E, Assassi S, Salazar GA, Stellato M, Renoux F, Wunderlin A, Blyszczuk P, Lafyatis R, Kurreeman F, de Vries-Bouwstra J, Messemaker T, Feghali-Bostwick CA, Rogler G, van Haaften WT, Dijkstra G, Oakley F, Calcagni M, Schniering J, Maurer B, Distler JH, Kania G, Frank-Bertoncelj M, Distler O. Long noncoding RNA H19X is a key mediator of TGF-β-driven fibrosis. J Clin Investig. 2020;130:4888–4905. doi: 10.1172/JCI135439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanaik D, Brown M, Postlethwaite BC, Postlethwaite AE. Pathogenesis of systemic sclerosis. Front Immunol. 2015;6:272. doi: 10.3389/fimmu.2015.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q-L, Zhang Y-M, Yang H-B, Shu X-M, Xin Lu, Wang G-C. Transcriptomic profiling of long non-coding RNAs in dermatomyositis by microarray analysis. Sci Rep. 2016;6:32818–32918. doi: 10.1038/srep32818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang JK, Kim WB, Baibergenova A, Alhusayen R. Risk of malignancy in dermatomyositis and polymyositis: a systematic review and meta-analysis. J Cutan Med Surg. 2017;21:131–136. doi: 10.1177/1203475416665601. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salajegheh M, Kong SW, Pinkus JL, Walsh RJ, Liao A, Nazareno R, Amato AA, Krastins B, Morehouse C, Higgs BW, Jallal B, Yao Y, Sarracino DA, Parker KC, Greenberg SA. Interferon-stimulated gene 15 (ISG15) conjugates proteins in dermatomyositis muscle with perifascicular atrophy. Ann Neurol. 2010;67:53–63. doi: 10.1002/ana.21805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salviano-Silva A, Lobo-Alves SC, Almeida RC, Malheiros D, Petzl-Erler ML. Besides pathology: long non-coding RNA in cell and tissue homeostasis. Noncoding RNA. 2018;4:3. doi: 10.3390/ncrna4010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracino AM, Denton CP, Orteu CH. The molecular pathogenesis of morphoea: from genetics to future treatment targets. Br J Dermatol. 2017;177:34–46. doi: 10.1111/bjd.15001. [DOI] [PubMed] [Google Scholar]
- Schmidt K, Martinez-Gamboa L, Meier S, Witt C, Meisel C, Hanitsch LG, Becker MO, Huscher D, Burmester GR, Riemekasten G. Bronchoalveoloar lavage fluid cytokines and chemokines as markers and predictors for the outcome of interstitial lung disease in systemic sclerosis patients. Arthritis Res Ther. 2009;11:R111. doi: 10.1186/ar2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze KV, Szafranski P, Lesmana H, Hopkin RJ, Hamvas A, Wambach JA, Shinawi M, Zapata G, Carvalho CMB, Liu Q, Karolak JA, Lupski JR, Hanchard NA, Stankiewicz P. Novel parent-of-origin-specific differentially methylated loci on chromosome 16. Clin Epigenet. 2019;11:60. doi: 10.1186/s13148-019-0655-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servaas NH, Mariotti B, van der Kroef M, Wichers CGK, Pandit A, Bazzoni F, Radstake T, Rossato M. Characterization of long non-coding RNAs in systemic sclerosis monocytes: a potential role for PSMB8-AS1 in altered cytokine secretion. Int J Mol Sci. 2021;22:4365. doi: 10.3390/ijms22094365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Hudson M, Watters K, Billick R, Fallavolita S, Netchiporouk E. Rapidly progressive melanoma differentiation-associated protein 5-positive amyopathic dermatomyositis in an HIV-positive patient. JAAD Case Rep. 2017;3:158–161. doi: 10.1016/j.jdcr.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra-Sepúlveda A, Esquinca-González A, Benavides-Suárez SA, Sordo-Lima DE, Caballero-Islas AE, Cabral-Castañeda AR, Rodríguez-Reyna TS. Systemic sclerosis pathogenesis and emerging therapies, beyond the fibroblast. Biomed Res Int. 2019;2019:4569826–4569926. doi: 10.1155/2019/4569826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solans R, Bosch JA, Esteban I, Vilardell M. Systemic sclerosis developing in association with the use of interferon alpha therapy for chronic viral hepatitis. Clin Exp Rheumatol. 2004;22:625–628. [PubMed] [Google Scholar]
- Stanek D, Vencovsky J, Kafkova J, Raska I. Heterogenous nuclear RNP C1 and C2 core proteins are targets for an autoantibody found in the serum of a patient with systemic sclerosis and psoriatic arthritis. Arthritis Rheum. 1997;40:2172–2177. doi: 10.1002/art.1780401211. [DOI] [PubMed] [Google Scholar]
- Sutaria DS, Jiang J, Azevedo-Pouly ACP, Lee EJ, Lerner MR, Brackett DJ, Vandesompele J, Mestdagh P, Schmittgen TD. Expression profiling identifies the noncoding processed transcript of HNRNPU with proliferative properties in pancreatic ductal adenocarcinoma. Noncoding RNA. 2017;3:24. doi: 10.3390/ncrna3030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri M, Eghtedarian R, Dinger ME, Ghafouri-Fard S. Exploring the role of non-coding RNAs in the pathophysiology of systemic lupus erythematosus. Biomolecules. 2020;10:937. doi: 10.3390/biom10060937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Pachera E, Frank-Bertoncelj M, Kozlova A, Jungel A, Whitfield ML, Assassi S, Calcagni M, de Vries-Bouwstra J, Huizinga TW, Kurreeman F, Kania G, Distler O. OTUD6B-AS1 might be a novel regulator of apoptosis in systemic sclerosis. Front Immunol. 2019;10:1100. doi: 10.3389/fimmu.2019.01100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan FK, Zhou X, Mayes MD, Gourh P, Guo X, Marcum C, Jin L, Arnett FC., Jr Signatures of differentially regulated interferon gene expression and vasculotrophism in the peripheral blood cells of systemic sclerosis patients. Rheumatology (oxford) 2006;45:694–702. doi: 10.1093/rheumatology/kei244. [DOI] [PubMed] [Google Scholar]
- Tang Q, Hann SS. HOTAIR: an oncogenic long non-coding RNA in human cancer. Cell Physiol Biochem. 2018;47:893–913. doi: 10.1159/000490131. [DOI] [PubMed] [Google Scholar]
- Thompson C, Piguet V, Choy E. The pathogenesis of dermatomyositis. Br J Dermatol. 2018;179:1256–1262. doi: 10.1111/bjd.15607. [DOI] [PubMed] [Google Scholar]
- Tsai CY, Shen CY, Liu CW, Hsieh SC, Liao HT, Li KJ, Lu CS, Lee HT, Lin CS, Wu CH, Kuo YM, Yu CL. Aberrant non-coding RNA expression in patients with systemic lupus erythematosus: consequences for immune dysfunctions and tissue damage. Biomolecules. 2020;10:1641. doi: 10.3390/biom10121641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen F, Khanna D, Fransen J, Johnson SR, Baron M, Tyndall A, Matucci-Cerinic M, Naden RP, Medsger TA, Jr, Carreira PE, Riemekasten G, Clements PJ, Denton CP, Distler O, Allanore Y, Furst DE, Gabrielli A, Mayes MD, van Laar JM, Seibold JR, Czirjak L, Steen VD, Inanc M, Kowal-Bielecka O, Muller-Ladner U, Valentini G, Veale DJ, Vonk MC, Walker UA, Chung L, Collier DH, Ellen Csuka M, Fessler BJ, Guiducci S, Herrick A, Hsu VM, Jimenez S, Kahaleh B, Merkel PA, Sierakowski S, Silver RM, Simms RW, Varga J, Pope JE. 2013 classification criteria for systemic sclerosis: an American college of rheumatology/European league against rheumatism collaborative initiative. Ann Rheum Dis. 2013;72:1747–1755. doi: 10.1136/annrheumdis-2013-204424. [DOI] [PubMed] [Google Scholar]
- Varga J, Pasche B. Transforming growth factor beta as a therapeutic target in systemic sclerosis. Nat Rev Rheumatol. 2009;5:200–206. doi: 10.1038/nrrheum.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman R, DeWane ME, Jun Lu. Dermatomyositis: Diagnosis and treatment. J Am Acad Dermatol. 2020;82:283–296. doi: 10.1016/j.jaad.2019.05.105. [DOI] [PubMed] [Google Scholar]
- Walling HW, Sontheimer RD. Cutaneous lupus erythematosus: issues in diagnosis and treatment. Am J Clin Dermatol. 2009;10:365–381. doi: 10.2165/11310780-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Syrett CM, Kramer MC, Basu A, Atchison ML, Anguera MC. Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc Natl Acad Sci USA. 2016;113:E2029–E2038. doi: 10.1073/pnas.1520113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Jinnin M, Nakamura K, Harada M, Kudo H, Nakayama W, Inoue K, Nakashima T, Honda N, Fukushima S, Ihn H. Long non-coding RNA TSIX is upregulated in scleroderma dermal fibroblasts and controls collagen mRNA stabilization. Exp Dermatol. 2016;25:131–136. doi: 10.1111/exd.12900. [DOI] [PubMed] [Google Scholar]
- Wang G, Zhang ZJ, Jian WG, Liu PH, Xue W, Wang TD, Meng YY, Yuan C, Li HM, Yu YP, Liu ZX, Wu Q, Zhang DM, Zhang C. Novel long noncoding RNA OTUD6B-AS1 indicates poor prognosis and inhibits clear cell renal cell carcinoma proliferation via the Wnt/beta-catenin signaling pathway. Mol Cancer. 2019;18:15. doi: 10.1186/s12943-019-0942-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson CW, Abignano G, Hermes H, Malaab M, Ross RL, Jimenez SA, Chang HY, Feghali-Bostwick CA, Del Galdo F. Long non-coding RNA HOTAIR drives EZH2-dependent myofibroblast activation in systemic sclerosis through miRNA 34a-dependent activation of NOTCH. Ann Rheum Dis. 2020;79:507–517. doi: 10.1136/annrheumdis-2019-216542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson CW, Ross RL, Wells R, Corinaldesi C, Georgiou IC, Riobo-Del Galdo NA, Del Galdo F. Long non-coding RNA HOTAIR induces GLI2 expression through Notch signalling in systemic sclerosis dermal fibroblasts. Arthritis Res Ther. 2020;22:286. doi: 10.1186/s13075-020-02376-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel J. Cutaneous lupus erythematosus: new insights into pathogenesis and therapeutic strategies. Nat Rev Rheumatol. 2019;15:519–532. doi: 10.1038/s41584-019-0272-0. [DOI] [PubMed] [Google Scholar]
- Xuan J, Xiong Y, Shi L, Aramini B, Wang H. Do lncRNAs and circRNAs expression profiles influence discoid lupus erythematosus progression?-a comprehensive analysis. Ann Transl Med. 2019;7:728–828. doi: 10.21037/atm.2019.12.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Lin F, Qin B, Liang Y, Zhong R. Polymyositis/dermatomyositis and malignancy risk: a metaanalysis study. J Rheumatol. 2015;42:282–291. doi: 10.3899/jrheum.140566. [DOI] [PubMed] [Google Scholar]
- Zehender A, Huang J, Gyorfi AH, Matei AE, Trinh-Minh T, Xu X, Li YN, Chen CW, Lin J, Dees C, Beyer C, Gelse K, Zhang ZY, Bergmann C, Ramming A, Birchmeier W, Distler O, Schett G, Distler JHW. The tyrosine phosphatase SHP2 controls TGFbeta-induced STAT3 signaling to regulate fibroblast activation and fibrosis. Nat Commun. 2018;9:3259. doi: 10.1038/s41467-018-05768-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li L-Y. TNFSF15 modulates neovascularization and inflammation. Cancer Microenviron. 2012;5:237–247. doi: 10.1007/s12307-012-0117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Zhang R. Epigenetics in autoimmune diseases: pathogenesis and prospects for therapy. Autoimmun Rev. 2015;14:854–863. doi: 10.1016/j.autrev.2015.05.008. [DOI] [PubMed] [Google Scholar]
- Zhang P, Wenyi Wu, Chen Qi, Chen M. Non-coding RNAs and their integrated networks. J Integr Bioinform. 2019;16:20190027. doi: 10.1515/jib-2019-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.
Not applicable.