Abstract
Hypoxia is a key characteristic of the breast cancer microenvironment that promotes expression of the transcriptional activator hypoxia-inducible factor 1 (HIF-1) and is associated with poor patient outcome. HIF-1 increases the expression or activity of stem cell pluripotency factors, which control breast cancer stem cell (BCSC) specification and are required for cancer metastasis. Here, we identify nuclear prelamin A recognition factor (NARF) as a hypoxia-inducible, HIF-1 target gene in human breast cancer cells. NARF functions as an essential coactivator by recruiting the histone demethylase KDM6A to OCT4 bound to genes encoding the pluripotency factors NANOG, KLF4, and SOX2, leading to demethylation of histone H3 trimethylated at lysine-27 (H3K27me3), thereby increasing the expression of NANOG, KLF4, and SOX2, which, together with OCT4, mediate BCSC specification. Knockdown of NARF significantly decreased the BCSC population in vitro and markedly impaired tumor initiation capacity and lung metastasis in orthotopic mouse models.
NARF recruits KDM6A to OCT4, leading to histone H3K27 demethylation and licensing of pluripotency factor gene transcription.
INTRODUCTION
As of 2020, breast cancer has surpassed lung cancer as the most common cancer type in women worldwide, and the 2.3 million new cases diagnosed in 2020 accounted for 11.7% of all cancer diagnoses (1). Breast cancer is the leading cause of cancer death for women globally, and the vast majority of breast cancer–related deaths are due to metastasis (2, 3). Advanced breast cancers are characterized by intratumoral hypoxia, which is associated with metastasis, treatment failure, and patient mortality (4–6). Measurements of the partial pressure of oxygen in locally advanced breast cancers revealed a median partial pressure of oxygen (PO2) of 10 mmHg (~1.4% O2), with one quarter of all tumor regions having a PO2 < 2.5 mmHg, as compared to normal breast tissue, which had a median PO2 of 65 mmHg (~9.3% O2) and no regions with PO2 < 15 mmHg (6).
Intratumoral hypoxia leads to expression of hypoxia-inducible factor 1 (HIF-1) in many advanced cancers (7, 8). HIF-1, which is a heterodimer composed of HIF-1α and HIF-1β subunits (9), binds to hypoxia response elements (HREs) to activate transcription of hundreds of target genes that control cellular responses to decreased O2 availability (10). Under normoxic conditions, HIF-1α is subject to O2-dependent hydroxylation, ubiquitination, and proteasomal degradation, whereas under hypoxic conditions, HIF-1α is stabilized, dimerizes with HIF-1β, and activates transcription of target genes (8). HIF-1α expression is increased in many breast cancers and is associated with increased patient mortality (11, 12) by mediating expression of genes that play critical roles in tumor vascularization, metabolic reprogramming, invasion, cancer stem cell specification, and immune evasion leading to metastasis and patient mortality (8, 13, 14). HIF-2α, which dimerizes with HIF-1β and binds to HREs to activate transcription, is also overexpressed in many cancers (15).
A large and growing body of work suggests that breast cancer stem cells (BCSCs) are a subpopulation of cancer cells that (i) express pluripotency factors including Kruppel-like factor 4 (KLF4),Nanog homeobox transcription factor (NANOG), octamer-binding transcription factor 4 (OCT4), and SRY-box transcription factor 2 (SOX2); (ii) are characterized by their ability to self-renew; and (iii) are required for the formation of secondary (recurrent and/or metastatic) tumors (16, 17). Exposure of breast cancer cells to hypoxia enriches the BCSC population (17–19). HIF-1 activates the transcription of multiple genes encoding proteins that drive the BCSC phenotype by increasing the expression of the pluripotency factors NANOG, SOX2, and KLF4 (17); by enabling transcription mediated by the master pluripotency factor OCT4 (20); or by activating TERT gene transcription to maintain replicative capacity (21).
Nuclear prelamin A recognition factor (NARF) is a relatively uncharacterized protein that was previously found to associate with the nuclear lamina and potentially maintain the structural integrity of the nucleus (22, 23). NARF has also been identified as a mitochondrial protein, and increased NARF expression was associated with aberrant iron deposition in multiple sclerosis brains (24). Last, NARF mRNA was overexpressed in retinal cells of early growth response protein 1 knockout mouse embryos, suggesting a potential role in retinal development (25). However, no studies have reported a role for NARF in cancer or transcriptional regulation.
In this study, we identify NARF as a HIF-1 target gene and demonstrate a critical role for NARF in BCSC specification. Our studies show that exposure of breast cancer cells to hypoxia induces NARF mRNA and protein expression as a result of direct binding of HIF-1 to the NARF gene promoter. Exposure of breast cancer cells to hypoxia enriches the BCSC population, and knockdown (KD) of NARF abrogates this effect. Consistent with these findings, NARF KD inhibited tumor initiation and the metastatic potential of breast cancer cells in an orthotopic mouse model. Investigation of the molecular mechanisms underlying these effects revealed that NARF recruits histone lysine demethylase 6A (KDM6A) to OCT4 binding sites, thereby licensing OCT4-mediated transcriptional activation of the NANOG, SOX2, and KLF4 genes.
RESULTS
Hypoxia induces NARF expression in a HIF-1–dependent manner
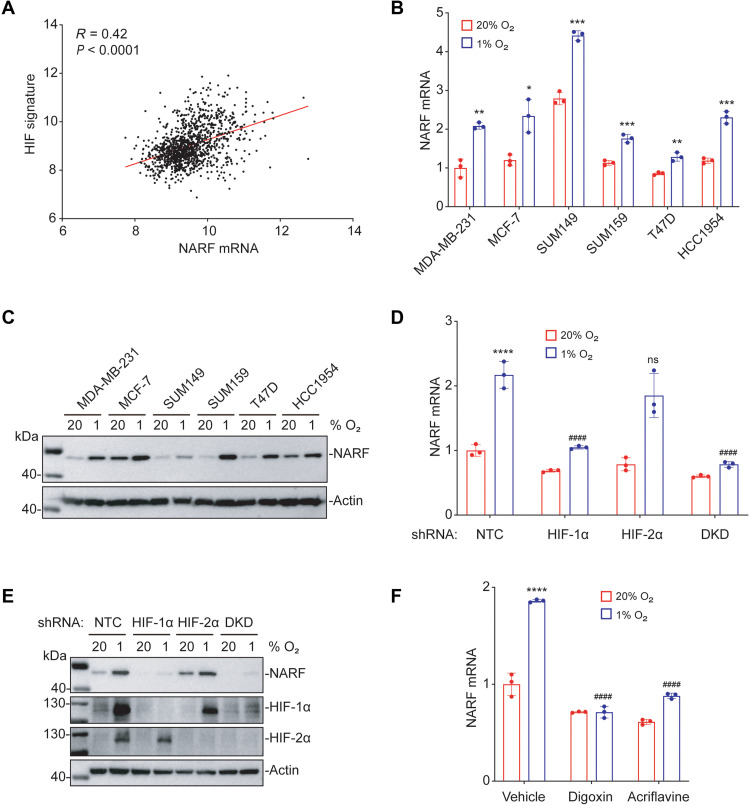
Recently published RNA sequencing data from our laboratory revealed that NARF mRNA expression was significantly induced by hypoxia in SUM159 human breast cancer cells (26). To investigate whether NARF expression is correlated with a HIF signature in breast cancers, we analyzed mRNA expression data from 1218 breast cancer specimens in The Cancer Genome Atlas (TCGA) breast cancer database using Pearson’s correlation (R) test. NARF gene expression was significantly correlated (R = 0.42) with a HIF signature consisting of 10 HIF-regulated genes (ANGPTL4, BNIP3, CA9, CXCR3, L1CAM, LDHA, P4HA1, P4HA2, PGK1, and PLOD1; Fig. 1A), suggesting that NARF expression is hypoxia-induced and HIF-dependent in human breast cancers. To test this hypothesis, we exposed six human breast cancer cell lines (estrogen/progesterone receptor–positive MCF-7 and T47D cells; HER2-positive HCC1954 cells; and triple-negative MDA-MB-231, SUM149, and SUM159 cells) to either nonhypoxic (20% O2) or hypoxic (1% O2) culture conditions for 24 hours, and NARF mRNA expression was quantified using reverse transcription (RT) and quantitative real-time polymerase chain reaction (qPCR). Hypoxia induced significantly increased NARF expression in all six breast cancer cell lines (Fig. 1B). Immunoblot assays using lysates of cells exposed to 20 or 1% O2 for 48 hours revealed hypoxic induction of NARF protein expression in all six lines as well (Fig. 1C).
Fig. 1. Hypoxia induces NARF expression in a HIF-1–dependent manner.
(A) NARF mRNA levels in 1218 human breast cancers from TCGA database were compared with the HIF signature using Pearson’s correlation test. (B and C) Six human breast cancer cell lines were exposed to 20 or 1% O2 for 24 hours (B) or 48 hours (C), and the expression of NARF mRNA (B) or protein (C) was analyzed by RT-qPCR (B) and immunoblot (C) assays, respectively. For each cell line, the expression of NARF mRNA was quantified relative to 18S rRNA and then normalized to the result obtained from MDA-MB-231 cells at 20% O2 (means ± SD; n = 3). *P < 0.05, **P < 0.01, and ***P < 0.001 versus 20% O2 [one-way analysis of variance (ANOVA)]. (D and E) MDA-MB-231 subclones, which stably expressed an NTC shRNA or shRNA targeting HIF-1α, HIF-2α, or both HIF-1α and HIF-2α (DKD), were exposed to 20 or 1% O2 for 24 hours, followed by analysis of NARF mRNA levels by RT-qPCR (means ± SD; n = 3) (D), or for 48 hours, followed by immunoblot assay (E). ****P < 0.0001 versus NTC at 20% O2; ####P < 0.0001 versus NTC at 1% O2; ns, not significant (two-way ANOVA with Tukey’s multiple comparisons test). (F) MDA-MB-231 cells were exposed to 20 or 1% O2 for 24 hours in the presence of vehicle, digoxin (100 nM), or acriflavine (5 μM), and expression of NARF mRNA was assayed by RT-qPCR (means ± SD; n = 3). ****P < 0.0001 versus vehicle at 20% O2; ####P < 0.0001 versus vehicle at 1% O2 (two-way ANOVA with Tukey’s multiple comparisons test).
To determine the HIF dependence of NARF expression, we established MDA-MB-231 subclones stably transduced with an expression vector encoding either a nontargeting control (NTC) short-hairpin RNA (shRNA) or a vector encoding shRNA targeting HIF-1α, HIF-2α, or both [double KD (DKD)] (27). RT-qPCR results revealed that KD of HIF-1α, but not HIF-2α, abrogated the induction of NARF mRNA expression in cells exposed to 1% O2 (Fig. 1D). Hypoxia-induced NARF protein expression was also abrogated by HIF-1α KD (Fig. 1E). HIF-1α–dependent, hypoxia-induced NARF expression was also observed in MCF-7 cells (fig. S1, A and B). To confirm by an orthogonal approach that hypoxia induces NARF expression in a HIF–dependent manner, we treated MDA-MB-231 or MCF-7 cells with either of two known pharmacologic HIF inhibitors, digoxin and acriflavine (27–29), during exposure to 20 or 1% O2 for 24 hours. RT-qPCR assays revealed that treatment with either digoxin or acriflavine blocked hypoxia-induced NARF mRNA expression (Fig. 1F and fig. S1C). Together, these data demonstrate that NARF is a hypoxia-inducible gene whose expression in breast cancer cells depends on HIF-1α but not HIF-2α.
NARF is a direct HIF-1 target gene
A search of the NARF human genome sequence revealed a match to the HIF-1 consensus binding sequence, 5′-(A/G)CGTG-3′ (30), which was located 95 base pairs (bp) upstream of the transcription start site (Fig. 2A). To test whether HIF-1 binds at this position on the NARF promoter, MDA-MB-231 and MCF-7 cells exposed to 20 or 1% O2 for 24 hours were analyzed by chromatin immunoprecipitation (ChIP) and qPCR. Chromatin fragments were precipitated using antibody against HIF-1α, HIF-1β, or HIF-2α, and fragments containing the putative HIF binding site were quantified by qPCR. HIF-1α and HIF-1β, but not HIF-2α, were significantly enriched at the NARF promoter of hypoxic cells, demonstrating that NARF is bound by HIF-1 but not HIF-2 (Fig. 2B and fig. S2).
Fig. 2. NARF is a direct HIF-1 target gene.
(A) A match to the HIF consensus binding site 5′-(A/G)CGTG-3′ was identified in the human NARF gene at −95 bp relative to the transcription start site. (B) MDA-MB-231 cells were exposed to 20 or 1% O2 for 16 hours, and ChIP assays were performed using antibodies against HIF-1α, HIF-1β, or HIF-2α. Primers flanking the candidate HIF binding site were used for qPCR, and results were normalized to the mean result at 20% O2 (means ± SEM; n = 3). ***P < 0.001; ****P < 0.0001 versus 20% O2; ns, not significant (two-tailed Student’s t test). (C) The following reporter plasmids were generated: pNARF-HRE, containing a 55-bp candidate HRE, which was either wild type [WT; nucleotide sequence shown in (A)] or mutant (MUT), downstream of an SV40 promoter and FLuc coding sequences (top), and pSV-RL, a control plasmid containing RLuc coding sequences downstream of the SV40 promoter (bottom). (D) MDA-MB-231 cells were cotransfected with pSV-RL and pNARF-HRE (WT or MUT) and then exposed to 20 or 1% O2 for 24 hours. The FLuc/RLuc ratio was determined and normalized to WT at 20% O2 (means ± SEM; n = 3). ***P < 0.001 versus WT at 20% O2; ###P < 0.001 versus WT at 1% O2 (two-way ANOVA with Tukey’s multiple comparisons test).
Next, we performed reporter assays to demonstrate that the NARF gene HIF-1 binding site was present within a functional HRE. The reporter plasmid pNARF-HRE was generated by inserting a 55-bp oligonucleotide, encompassing the HIF-1 binding site, downstream of SV40 promoter and firefly luciferase (FLuc) coding sequences (Fig. 2C, top). pNARF-HRE was cotransfected with pSV-RL, which encodes Renilla luciferase (RLuc) driven by the SV40 promoter alone (Fig. 2C, bottom). The FLuc/RLuc ratio serves as a measure of HIF-dependent transcriptional activity. In cells cotransfected with wild-type pNARF-HRE and pSV-Renilla, exposure to hypoxia significantly increased the FLuc/RLuc ratio. This hypoxic induction of FLuc was abrogated in cells cotransfected with mutant pNARF-HRE, which contains a 3-bp mutation (Fig. 2D) that abrogates HIF-1 binding (30). Together, the results presented in Fig. 2 demonstrate that NARF is a direct HIF-1 target gene whose expression is induced by hypoxia in breast cancer cells.
NARF KD inhibits hypoxia-induced BCSC enrichment
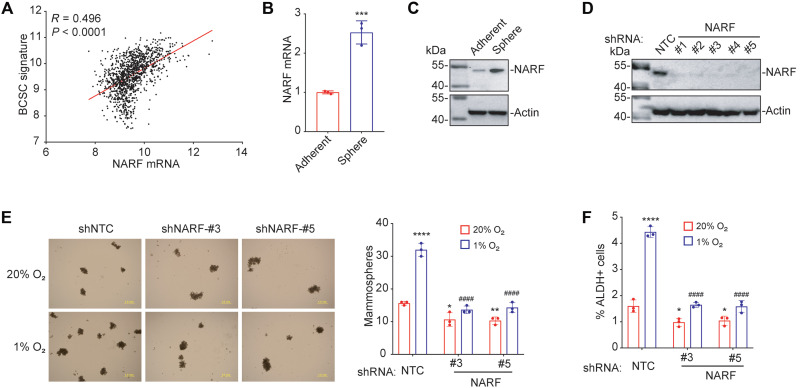
Many genes that contribute to the maintenance and specification of BCSCs and tumor metastasis are hypoxia-inducible HIF target genes (8, 13, 17). To explore the relevance of NARF for the BCSC phenotype, Pearson’s correlation test was performed to compare NARF gene expression in 1218 breast cancer patient samples from the TCGA database with a 20-gene BCSC signature (MMP1, SNF, MIEN1, PHLDA2, EPB41L5, RACGAP1, H2AFZ, H2AFJ, APOBEC3B, CENPW, TOP2A, CDK1, MRPS23, NDUFB10, PHB, ALYREF, EXOSC4, NOL3, LY6E, and EIF4EBP1) (31). NARF expression was significantly correlated (R = 0.496) with expression of genes associated with the BCSC phenotype (Fig. 3A).
Fig. 3. NARF KD blocks hypoxia-induced BCSC enrichment.
(A) Expression of NARF mRNA and a BCSC signature composed of 20 mRNAs expressed in primary breast cancer samples were accessed from TCGA database, and the correlation was analyzed by Pearson’s test. (B and C) MDA-MB-231 cells were cultured on standard polystyrene tissue culture plates (Adherent) or ultralow attachment plates (Sphere) for 7 days and harvested for analysis of NARF mRNA (B) and protein (C) expression. Results were normalized to Adherent (means ± SD; n = 3). ***P < 0.001 versus Adherent (two-tailed Student’s t test). (D) MDA-MB-231 cells were stably transfected with NTC or a NARF shRNA vector (#1 to #5), and immunoblot assays were performed. (E) MDA-MB-231 subclones transduced with NTC or a NARF shRNA (#3 or #5) were exposed to 20 or 1% O2 for 72 hours; then, cells were cultured on ultralow attachment plates for 7 days, and the number of mammospheres per 1000 cells seeded was calculated (means ± SD; n = 3). *P < 0.05; **P < 0.01; ****P < 0.0001 versus NTC at 20% O2; ####P < 0.0001 versus NTC at 1% O2 (two-way ANOVA with Tukey’s multiple comparisons test). Scale bar, 1 mm. (F) MDA-MB-231 subclones were exposed to 20 or 1% O2 for 72 hours, and the percentage of aldehyde dehydrogenase-expressing (ALDH+) cells was determined (means ± SD; n = 3). *P < 0.05; ****P < 0.0001 versus NTC at 20% O2; ####P < 0.0001 versus NTC at 1% O2 (two-way ANOVA with Tukey’s multiple comparisons test).
To determine whether NARF contributes to BCSC enrichment induced by hypoxia, MDA-MB-231 and MCF-7 cells were cultured for 7 days on either standard polystyrene tissue culture plates as an adherent monolayer of bulk breast cancer cells or on ultralow attachment plates, which enable sphere formation by BCSCs (32). NARF expression was significantly increased in the sphere-forming cells compared to adherent monolayer cells at both the mRNA and protein levels (Fig. 3, B and C, and fig. S3, A and B). To further investigate the role of NARF in BCSC enrichment, we generated MDA-MB-231 and MCF-7 subclones by stable transduction with lentiviral vectors encoding one of five different shRNAs targeting NARF or the NTC vector, and immunoblot assays were performed to confirm KD efficiency (Fig. 3D and fig. S3C). NTC and NARF-KD subclones were exposed to 20 or 1% O2 for 72 hours. Mammosphere-forming cells were significantly increased in response to hypoxia in the NTC subclones but not in the NARF-KD subclones (Fig. 3E and fig. S3D). NARF silencing also decreased the percentage of cells expressing the BCSC marker aldehyde dehydrogenase (33) after hypoxic exposure of MDA-MB-231 and MCF-7 cells (Fig. 3F and fig. S3E).
NARF KD impairs tumor initiation and lung metastasis
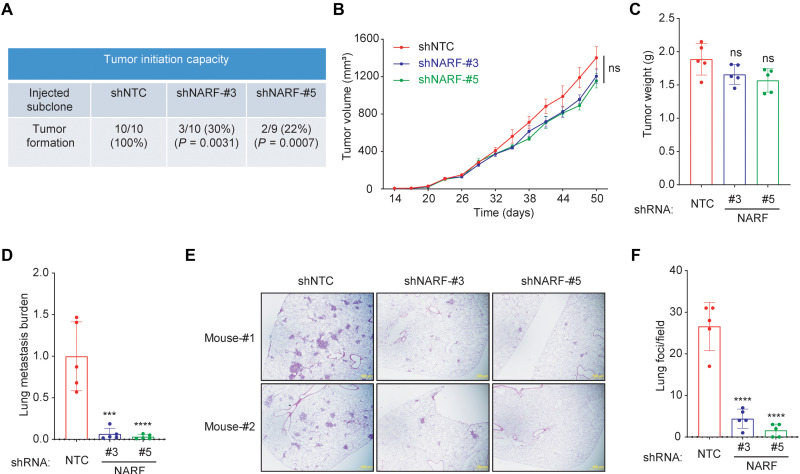
To test tumor initiation capacity, limiting numbers of MDA-MB-231 NTC or NARF-KD (shNARF#3 or shNARF#5) cells (1 × 103) were transplanted into the mammary fat pad (MFP) of female severe combined immunodeficiency (SCID) mice. All of the mice transplanted with NTC cells developed palpable tumors, whereas tumor formation was significantly decreased in mice injected with NARF-KD cells (Fig. 4A). To determine whether NARF contributes to primary tumor growth, 2 × 106 MDA-MB-231 subclone cells were injected into the MFP of SCID mice, such that BCSCs were not limiting for tumor initiation. Tumor growth curves and final tumor weight revealed no significant difference between NTC and NARF-KD subclones (Fig. 4, B and C). The lungs were harvested; genomic DNA was extracted from one lung, while the other lung was used for histological staining. qPCR using human DNA-specific primers was performed as a measure of human cancer cells that had metastasized to the lungs. Metastatic burden was significantly lower in mice injected with NARF-KD as compared to NTC cells (Fig. 4D). Hematoxylin and eosin staining of contralateral lung sections revealed that compared to NTC, NARF-KD subclones generated significantly fewer lung metastases (Fig. 4, E and F). Together, these studies revealed that NARF is required for breast cancer initiation and metastasis but not bulk tumor growth, which is consistent with an essential role for NARF in BCSC specification and/or maintenance.
Fig. 4. NARF KD impairs tumor formation and metastasis.
(A) MDA-MB-231 subclones (1 × 103 cells) transduced with NTC or a NARF shRNA vector (#3 or #5) were implanted into the MFP of 7-week-old female SCID mice. The proportion of mice with tumors after 10 weeks and P value (versus NTC; Fisher’s exact test) are shown. (B and C) MDA-MB-231 subclones (2 × 106 cells) were implanted into the MFP. Primary tumor volume (B) was determined starting at day 14. On day 50, the primary tumor was harvested and weighed (C). (D) To analyze lung metastasis, genomic DNA was purified from one lung for qPCR using human-specific HK2 primers, and the results were normalized to the NTC group. (E and F) The contralateral lung was fixed under inflation and paraffin-embedded, and sections were stained with hematoxylin and eosin (E) (scale bar, 0.5 mm) to count the number of metastatic foci per field (F). For all graphs, means ± SEM (n = 5) are shown; ***P < 0.001; ****P < 0.0001 versus NTC; ns, not significant (one-way ANOVA).
NARF is required for OCT4-mediated transcription
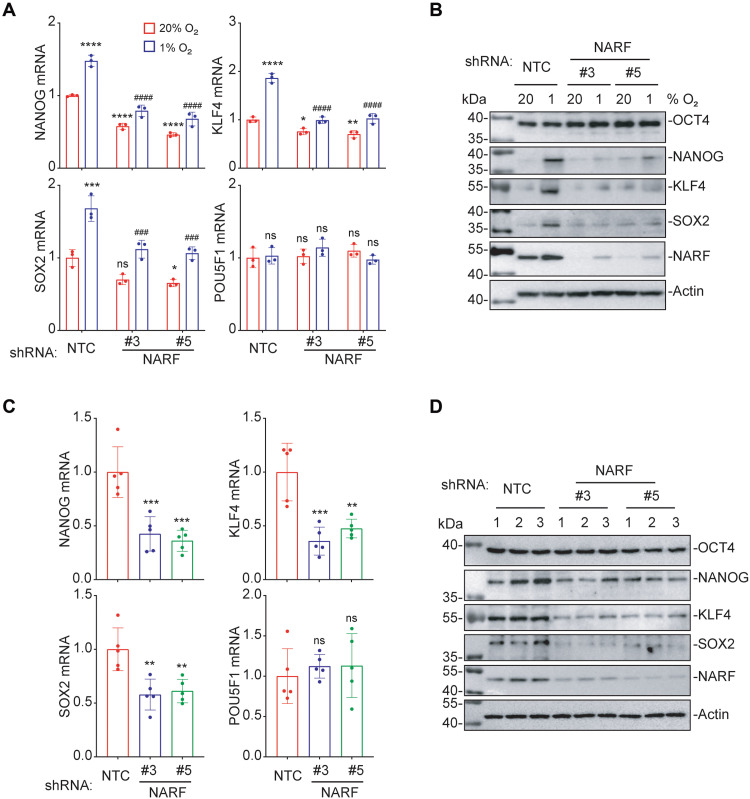
NANOG, KLF4, SOX2, and OCT4 are core pluripotency factors and stem cell master regulators that are critical for BCSC specification and maintenance (17). To explore the mechanism by which hypoxia-inducible NARF mediates BCSC specification, we analyzed the expression of these four pluripotency factors. RT-qPCR and immunoblot assays revealed that hypoxia-induced NANOG, SOX2, and KLF4 mRNA and protein expression were significantly decreased in NARF-KD subclones of MDA-MB-231 or MCF-7 cells (Fig. 5, A and B, and fig. S4, A and B). By contrast, neither POU class 5 homeobox 1 (POU5F1) mRNA (encoding OCT4) expression nor OCT4 protein expression was hypoxia induced, and POU5F1 mRNA and OCT4 protein levels did not change significantly in NARF-KD subclones. Similar to the effects observed in vitro, the expression of NANOG, SOX2, and KLF4 mRNA and protein was significantly decreased in NARF-KD tumors compared to NTC tumors (Fig. 5, C and D). OCT4 participates in a complex gene regulatory network with other pluripotency factors, and OCT4-mediated transactivation of the NANOG, KLF4, and SOX2 genes is an essential feature of BCSCs (17, 20). The observed pluripotency factor gene expression patterns suggested that NARF might be required for OCT4-mediated NANOG, SOX2, and KLF4 transcription.
Fig. 5. NARF is required for OCT4-mediated pluripotency factor expression in vitro and in vivo.
(A) MDA-MB-231 subclones were exposed to 20 or 1% O2 for 24 hours, and RT-qPCR assays were performed to analyze expression of NANOG, KLF4, SOX2, and POU5F1 (OCT4) mRNA. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 versus NTC at 20% O2; ###P < 0.001; ####P < 0.0001 versus NTC at 1% O2; ns, not significant (two-way ANOVA with Tukey’s multiple comparisons test). (B) MDA-MB-231 subclones were exposed to 20 or 1% O2 for 48 hours, and immunoblot assays were performed. (C and D) MDA-MB-231 subclones (2 × 106 cells) were implanted into the MFP. On day 50, tumors were harvested for RT-qPCR (C) and immunoblot assays (D). **P < 0.01; ***P < 0.001 versus NTC; ns, not significant (one-way ANOVA).
NARF facilitates OCT4 binding to pluripotency factor genes
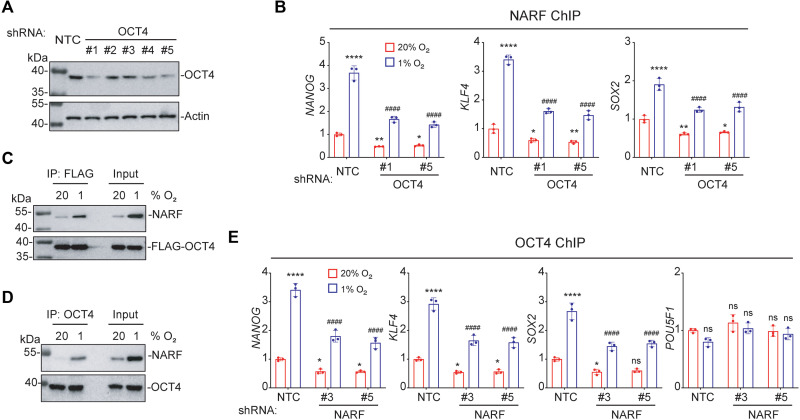
To investigate the role of NARF in OCT4-mediated transcription, we generated OCT4-KD subclones of MDA-MB-231 cells by stable transduction with lentiviral vectors encoding one of five different shRNAs (Fig. 6A). Cells were exposed to 20 or 1% O2 for 48 hours, and ChIP was performed using antibody against NARF, followed by qPCR. Hypoxia-induced NARF was recruited to the OCT4 binding sites in the NANOG, SOX2, and KLF4 genes in the NTC cells. However, the recruitment of NARF was decreased in OCT4 KD cells, indicating that NARF binding was OCT4 dependent (Fig. 6B). Next, lysates of MDA-MB-231 cells that overexpressed FLAG-tagged OCT4 and were exposed to either 20 or 1% O2 for 48 hours were subjected to IP assays, which revealed that NARF binds to OCT4 in a hypoxia-induced manner consistent with the increase in NARF protein levels (Fig. 6C). IP of endogenous proteins also showed that OCT4 interacts with NARF in hypoxic cells (Fig. 6D). ChIP-qPCR assays using antibody against OCT4 revealed that NARF is required for stable OCT4 binding to the NANOG, SOX2, and KLF4 genes (but not the POU5F1 gene) under hypoxic conditions (Fig. 6E). Together, these data suggest OCT4 recruits NARF to OCT4 binding sites and NARF stabilizes OCT4 binding to pluripotency factor gene promoters.
Fig. 6. OCT4 recruits NARF to pluripotency factor genes.
(A) MDA-MB-231 cells were stably transfected with NTC or an OCT4 shRNA vector (#1 to #5), and immunoblot assays were performed. (B) MDA-MB-231 subclones were exposed to 20 or 1% O2 for 48 hours, and ChIP assays were performed using antibody against NARF. Primers flanking OCT4 binding sites in the indicated genes were used for qPCR. For each primer pair, the qPCR data were normalized to the mean result at 20% O2 (means ± SEM; n = 3); *P < 0.05; **P < 0.01; ****P < 0.0001 versus NTC at 20% O2; ####P < 0.0001 versus NTC 1% O2 (two-way ANOVA with Tukey’s multiple comparisons test). (C) MDA-MB-231 cells were transfected with FLAG-OCT4 expression vector and exposed to 20 or 1% O2 for 48 hours. IP was performed using FLAG antibody followed by immunoblot assays with NARF antibody. (D) MDA-MB-231 cells were exposed to 20 or 1% O2 for 48 hours. IP was performed using OCT4 antibody, followed by immunoblot assays. (E) MDA-MB-231 subclones were exposed to 20 or 1% O2 for 48 hours, and ChIP-qPCR assays were performed using antibody against OCT4 and primers flanking OCT4 binding sites in the indicated genes. The qPCR data were normalized to the mean result at 20% O2 (means ± SEM; n = 3); *P < 0.05; ****P < 0.0001 versus NTC at 20% O2; ####P < 0.0001 versus NTC 1% O2 (two-way ANOVA with Tukey’s multiple comparisons test).
OCT4 recruits KDM6A to erase H3K27me3 chromatin marks in hypoxic cells
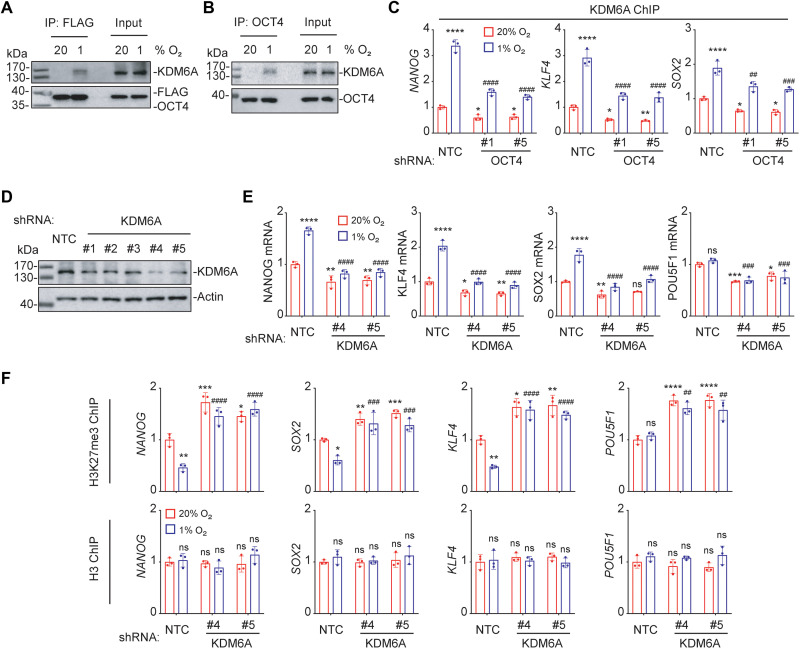
The histone demethylase KDM6A was identified as a component of the OCT4 transcriptional complex that is critical for OCT4-mediated breast cancer stemness (20). In response to chemotherapy, OCT4 recruits KDM6A to erase H3K27me3 marks (20), which are associated with gene silencing at transcriptional regulatory regions (34). Hypoxia increased interaction of OCT4 and KDM6A as determined by IP assays in MDA-MB-231 cells (Fig. 7, A and B). ChIP-qPCR assays showed that KDM6A was recruited to OCT4 binding sites in the NANOG, KLF4, and SOX2 genes in response to hypoxia (Fig. 7C). KD of KDM6A in MDA-MB-231 cells (Fig. 7D) abrogated hypoxia-induced expression of NANOG, KLF4, and SOX2 mRNA (Fig. 7E). ChIP-qPCR assays revealed that hypoxia significantly decreased H3K27me3 marks at the OCT4 binding sites of the NANOG, SOX2, and KLF4 genes, but not at the POU5F1 gene, in a KDM6A-dependent manner, whereas total H3 levels were unchanged (Fig. 7F). Together, these results indicate that KDM6A is recruited by OCT4 to erase H3K27me3 marks and stimulate OCT4-mediated pluripotency factor gene transcription in response to hypoxia.
Fig. 7. OCT4 recruits KDM6A to erase H3K27me3 chromatin marks.
(A) MDA-MB-231 cells were transfected with FLAG-OCT4 expression vector and exposed to 20 or 1% O2 for 48 hours. IP was performed using FLAG antibody followed by immunoblot assays. (B) MDA-MB-231 cells were exposed to 20 or 1% O2 for 48 hours. IP was performed using OCT4 antibody, followed by immunoblot assays. (C) MDA-MB-231 subclones were exposed to 20 or 1% O2 for 48 hours, and ChIP-qPCR assays were performed using antibody against KDM6A and primers flanking OCT4 binding sites. The qPCR data were normalized to the mean result at 20% O2 (means ± SEM; n = 3); *P < 0.05; **P < 0.01; ****P < 0.0001 versus NTC at 20% O2; ##P < 0.01; ###P < 0.001; ####P < 0.0001 versus NTC 1% O2 (two-way ANOVA with Tukey’s multiple comparisons test). (D) MDA-MB-231 cells were stably transfected with NTC or a KDM6A shRNA vector (#1 to #5), and immunoblot assays were performed. (E) MDA-MB-231subclones were exposed to 20 or 1% O2 for 24 hours, and RT-qPCR assays were performed. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 versus NTC at 20% O2; ###P < 0.001; ####P < 0.0001 versus NTC at 1% O2; ns, not significant (two-way ANOVA with Tukey’s multiple comparisons test). (F) MDA-MB-231 subclones were exposed to 20 or 1% O2, and ChIP-qPCR was performed (means ± SEM; n = 3); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 versus NTC at 20% O2; ##P < 0.01; ###P < 0.001; ####P < 0.0001 versus NTC at 1% O2; ns, not significant (two-way ANOVA with Tukey’s multiple comparisons test).
NARF is required for KDM6A-mediated H3K27me3 demethylation at OCT4 binding sites
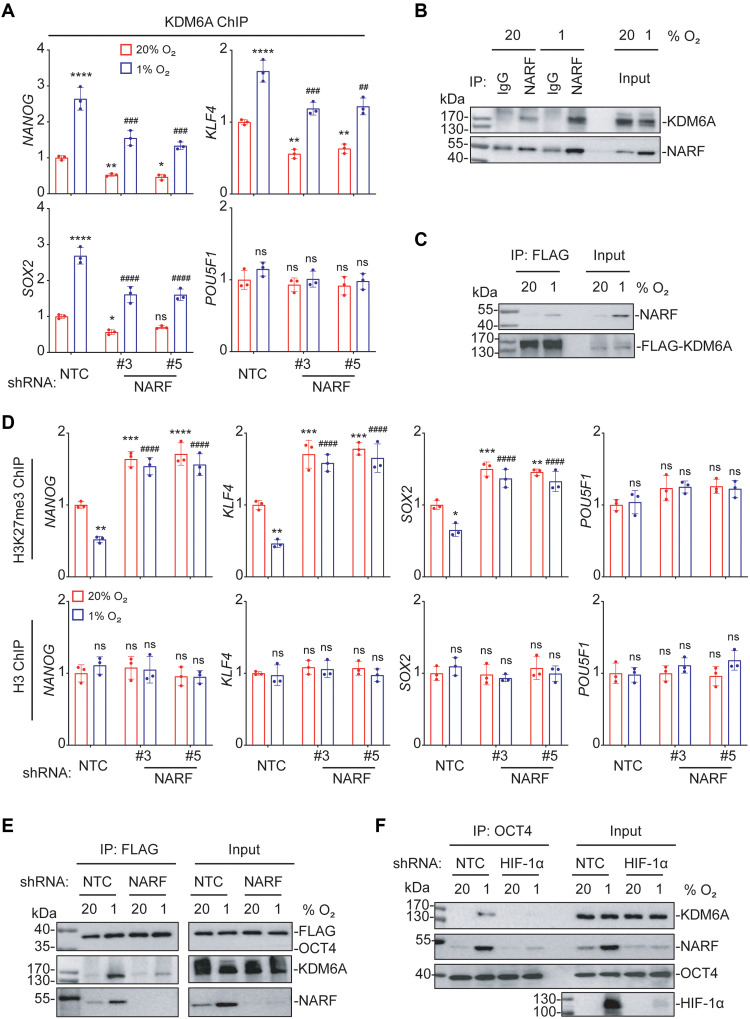
To investigate whether NARF is required for OCT4-KDM6A complex formation, KDM6A occupancy of OCT4 binding sites in the NANOG, KLF4, SOX2, and POU5F1 promoters was measured by ChIP-qPCR in NTC and NARF-KD subclones of MDA-MB-231 cells exposed to 20 or 1% O2. KDM6A was recruited in a hypoxia-dependent manner to the NANOG, KLF4, and SOX2 promoters, but not to the POU5F1 promoter, in NTC cells, whereas hypoxia-induced KDM6A recruitment was decreased in NARF-KD cells (Fig. 8A). IP assays revealed interaction of NARF and KDM6A, especially under hypoxic conditions (Fig. 8, B and C). H3K27me3 marks were depleted at the NANOG, KLF4, and SOX2 promoters in hypoxic NTC cells but not in NARF-KD cells; by contrast, H3K27me3 marks at the POU5F1 promoter did not change significantly as a result of hypoxia in NTC or NARF-KD cells (Fig. 8D). The results of immunofluorescence assays showed that NARF protein expression was increased in hypoxic cells and that OCT4, NARF, and KDM6A colocalized in the nuclei of hypoxic cells, particularly in the vicinity of the nuclear lamina (fig. S5). IP assays further demonstrated that the interaction of KDM6A and OCT4 is NARF dependent (Fig. 8E) and that the formation of the OCT4-NARF-KDM6A complex is HIF-1α dependent (Fig. 8F). Together, these results demonstrate that HIF-1–dependent expression of NARF is required for hypoxia-induced KDM6A recruitment and KDM6A-mediated H3K27 demethylation at OCT4 binding sites.
Fig. 8. NARF is required for KDM6A-mediated H3K27me3 demethylation at OCT4 binding sites.
(A) MDA-MB-231 subclones were exposed to 20 or 1% O2 for 48 hours, and ChIP-qPCR assays were performed using antibody against KDM6A and primers flanking OCT4 binding sites in the indicated genes. The qPCR data were normalized to the mean result at 20% O2 (means ± SEM; n = 3); *P < 0.05; **P < 0.01; ****P < 0.0001 versus NTC at 20% O2; ##P < 0.01; ###P < 0.001; ####P < 0.0001 versus NTC 1% O2 (two-way ANOVA Tukey’s multiple comparisons). (B) MDA-MB-231 cells were exposed to 20 or 1% O2 for 48 hours. IP was performed using immunoglobulin G or NARF antibody, followed by immunoblot assays. (C) MDA-MB-231 subclones were transfected with FLAG-KDM6A expression vector and exposed to 20 or 1% O2 for 48 hours. IP was performed using FLAG antibody, followed by immunoblot assays. (D) MDA-MB-231 subclones were exposed to 20 or 1% O2, and ChIP-qPCR assays were performed using H3K27me3 or H3 antibody and primers flanking OCT4 binding sites in the indicated genes (means ± SEM; n = 3); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 versus NTC at 20% O2; ####P < 0.0001 versus NTC at 1% O2; ns, not significant (two-way ANOVA Tukey’s multiple comparisons). (E) MDA-MB-231 subclones were transfected with FLAG-OCT4 expression vector and exposed to 20 or 1% O2 for 48 hours. IP was performed using FLAG antibody, followed by immunoblot assays. (F) NTC or HIF-1α-KD subclones of MDA-MB-231 cells were exposed to 20 or 1% O2 for 48 hours. IP was performed using OCT4 antibody, followed by immunoblot assays.
NARF is highly expressed in human breast cancers and is correlated with patient mortality
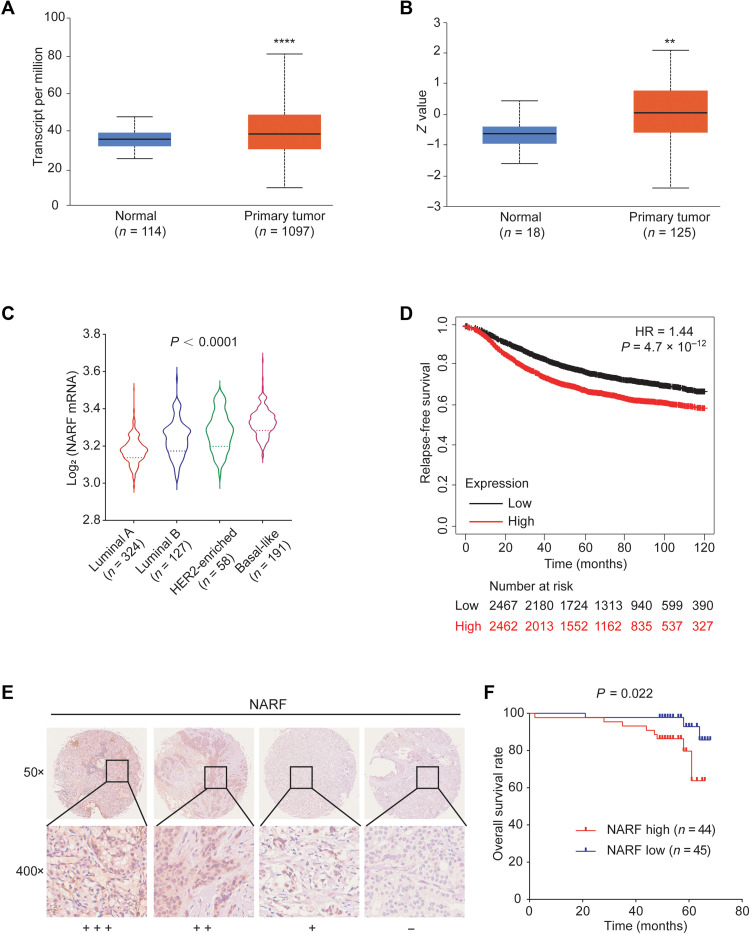
Comparison of NARF mRNA expression in breast cancer samples and normal breast tissue revealed that NARF is overexpressed in tumor as compared to normal tissue (Fig. 9, A and B). As observed for other HIF target genes (35), NARF expression is highest in basal-like breast cancers (Fig. 9C), which have the poorest prognosis of the breast cancer molecular subtypes. Last, Kaplan-Meier survival analysis revealed that high NARF expression (greater than the median) is significantly associated (P = 4.7 × 10–12) with decreased relapse-free survival over 10 years (Fig. 9D). We analyzed two additional smaller datasets, which revealed that high levels of NARF expression in breast cancer biopsies were correlated with decreased disease-free and overall patient survival (fig. S6, A to C). Immunohistochemical analysis of a tissue microarray containing 89 matched tumor and surrounding nontumor breast tissues revealed that NARF protein was more highly expressed in breast cancer compared to normal tissue (Fig. 9E). Moreover, Kaplan-Meier analysis demonstrated that high NARF protein expression was associated with decreased overall survival in these patients with breast cancer (Fig. 9F). Furthermore, treatment with chemotherapy is known to induce HIF-1–dependent BCSC enrichment by the induction of OCT4-mediated pluripotency factor expression (20). To investigate whether hypoxia-inducible NARF may play a role in chemoresistance in patients with breast cancer, we analyzed publicly available gene expression data. NARF mRNA expression is higher in tumors from patients exhibiting residual disease after neoadjuvant chemotherapy, compared to those with pathological complete response (fig. S7). These data suggest that NARF promotes chemoresistance and offer a direction for future investigation.
Fig. 9. NARF is highly expressed in human breast cancers and is correlated with patient mortality.
(A) NARF mRNA expression in primary breast cancer samples (n = 1097) relative to adjacent normal tissue (n = 114) from TCGA database is shown. ****P < 0.0001 versus normal (two-tailed Student’s t test). (B) NARF protein expression in primary breast cancer samples (n = 125) relative to normal breast tissue (n = 18) from Clinical Proteomic Tumor Analysis Consortium database is shown. **P < 0.01 versus normal (two-tailed Student’s t test). (C) The relative log2 expression of NARF mRNA from 700 human breast cancer specimens that were stratified according to molecular subtype is shown. Statistical analysis was performed by one-way ANOVA with Bonferroni’s post hoc testing; P < 0.0001 for basal-like versus other groups. (D) Kaplan-Meier analysis of relapse-free survival over 10 years was performed on the basis of clinical and molecular data from 4929 breast cancer patients. The patients were stratified by NARF mRNA levels in the primary tumor, which were greater (red) or less (black) than the median. The hazard ratio (HR) and P value (log-rank test) are shown. (E and F) Immunohistochemical staining for NARF was performed in tumor tissue from 89 patients with breast cancer (E). Kaplan-Meier analysis for overall survival (F) was performed according to scoring of NARF expression.
DISCUSSION
The results of this study have delineated a molecular mechanism by which hypoxia induces BCSC specification and promotes breast cancer metastasis. Binding of HIF-1 to the NARF gene in MCF-7 cells was previously identified in ChIPsequencing studies (36, 37). Here, we have provided a detailed analysis of the regulation of NARF gene expression in multiple breast cancer cell lines. Furthermore, our studies have revealed that NARF protein is recruited to OCT4 binding sites at the NANOG, SOX2, and KLF4 genes in hypoxic breast cancer cells. NARF, in turn, recruits KDM6A, which erases repressive H3K27me3 marks, enabling OCT4-mediated transcription of the NANOG, SOX2, and KLF4 genes, which is required for specification of BCSCs that are essential for tumor initiation and metastasis (Fig. 10). Future studies will investigate whether NARF is recruited to OCT4 binding sites at other locations in the genome and determine what other OCT4 target genes are induced by hypoxia in a NARF- and KDM6A-dependent manner.
Fig. 10. HIF-1–dependent NARF expression promotes OCT4-mediated breast cancer stem cell specification.
(A and B) Hypoxia induces HIF-1–mediated expression of NARF (A), which enables KDM6A recruitment to OCT4 binding sites to erase inhibitory H3K27me3 chromatin marks, thereby licensing transcription of OCT4 target genes encoding pluripotency factors (NANOG, KLF4, and SOX2) that specify the BCSC phenotype (B).
NARF is also known as iron-only hydrogenase-like protein 2 (IOP2) because of the presence of a protein domain with homology to bacterial iron hydrogenases (22). NARFL, which is also known as IOP1, has been shown to increase the activity of cytosolic aconitase and xanthine oxidase, two Fe-S proteins, suggesting that IOP1 may play a role in Fe-S protein biogenesis, whereas IOP2 does not have this effect (38, 39). IOP1 has been reported to negatively regulate HIF-1α mRNA expression and HIF-1 transcriptional activity (40).
We previously reported that another HIF target gene, S100A10, plays an important role in BCSC enrichment through OCT4-mediated pluripotency factor gene transcription. In response to exposure of breast cancer cells to cytotoxic chemotherapy, S100A10 expression is increased in a HIF–dependent manner and S100A10 protein forms a complex with ANXA2 and SPT6 to facilitate interaction of KDM6A with OCT4, thereby licensing transcription of NANOG, SOX2, and KLF4 genes for BCSC specification (20). It appears that in hypoxic cells NARF plays a role that is functionally similar to that of the S100A10-ANXA2-SPT6 complex in chemotherapy-treated cells.
In addition to the role of NARF and S100A10 in facilitating OCT4-mediated transactivation, protein products of other HIF target genes increase NANOG gene transcription (A2BR, CD73, DUSP9, GCLM, GSTO1, and SLC7A11) or increase NANOG mRNA stability (ALKBH5, REST, and ZNF217) (17). Furthermore, NANOG functions as a coactivator for HIF-1–dependent TERT gene transactivation (21). All of the HIF target genes involved in hypoxia-induced BCSC specification are transactivated by HIF-1 and not by HIF-2. Thus, HIF-1 functions as a master regulator of hypoxia-induced BCSC specification, and inhibitors that target HIF-1 may prevent breast cancer recurrence and metastasis by eliminating BCSCs.
MATERIALS AND METHODS
Cell culture
MDA-MB-231 and MCF-7 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Corning, catalog no. 10-013-CV). SUM149 cells were maintained in Ham’s F-12 medium (Corning, catalog no. 10-080-CV) supplemented with hydrocortisone (MilliporeSigma) and insulin (Thermo Fisher Scientific). SUM159 cells were maintained in DMEM/F12 (50:50) medium (Corning, catalog no. 10-092-CV). HCC1954 and T47D cells were maintained in RPMI 1640 medium (Corning, catalog no. 10-040-CV). The culture media were supplemented with 10% (v/v) fetal bovine serum (GeminiBio, catalog no. 100-106) and 1% (v/v) penicillin-streptomycin. Cells were cultured at 37°C in a 5% CO2 and 95% air incubator. Cells were subjected to hypoxia in a modular incubator chamber (Billups-Rothenberg) that was flushed with a gas mixture of 1% O2, 5% CO2, and 94% N2. Breast cancer cell lines were mycoplasma-free and authenticated by short tandem repeat DNA profiling analysis by the Johns Hopkins Genetics Resources Core Facility. Digoxin and acriflavine were obtained from MilliporeSigma and dissolved in DMSO at 1000x relative to their final concentration in tissue culture media.
RT-qPCR
RT-qPCR assays were performed as described previously (26). Briefly, total RNA was isolated using TRIzol (Invitrogen) and reverse-transcribed using the High-Capacity RNA-to-cDNA Kit (Applied Biosystems). qPCR analysis was performed using SYBR Green and the CFX96 Real-Time PCR Detection System (Bio-Rad). The expression (E) of each target mRNA relative to 18S rRNA was calculated on the basis of the cycle threshold (Ct) as E = 2-Δ(ΔCt), in which ΔCt = Ct (target) − Ct (18S) and Δ (ΔCt) = ΔCt (test sample) − ΔCt (control sample). PCR primer sequences are shown in table S1.
Immunoblot assays
Immunoblot assays were performed as described previously (26). Briefly, cultured cells were lysed in radioimmunoprecipitation assay buffer (Thermo Fisher Scientific), and proteins were fractionated by SDS–polyacrylamide gel electrophoresis, blotted onto nitrocellulose membranes, which were blocked with 5% (w/v) nonfat milk in phosphate-buffered saline (PBS) with Tween 20 [PBST; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, and 0.1% Tween 20 (w/v)] for 1 hour at room temperature, probed with primary antibody (table S2) in blocking solution (5% nonfat milk in PBST) overnight at 4°C, and incubated with horseradish peroxidase–conjugated secondary antibody (GE Healthcare) for 1 hour at room temperature; the chemiluminescent signal was detected using ECL Plus (GE Healthcare).
ChIP-qPCR
ChIP-qPCR assays were performed as described previously (26). Briefly, MDA-MB-231 and MCF-7 cells were cross-linked in 1% formaldehyde for 10 min, quenched in 0.125 M glycine for 5 min, and lysed with SDS lysis buffer [1% SDS, 10 mM EDTA, and 50 mM tris (pH 8.1)]. Chromatin was sheared by sonication to fragments of 200 to 1000 bp. Lysates were diluted 1:10 with ChIP dilution buffer [0.01% SDS, 1.1% Triton X100, 1.2 mM EDTA, 16.7 mM tris-HCl (pH 8.1), and 167 mM NaCl], precleared with salmon sperm DNA/protein A–agarose slurry (MilliporeSigma) at 4°C for 1 hour, and incubated with antibody (table S3) in the presence of protein A–agarose beads overnight. After sequential washes with low-salt [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM tris-HCl (pH 8.1), and 150 mM NaCl], high-salt [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM tris-HCl (pH 8.1), and 500 mM NaCl], LiCl [0.25 M LiCl, 1% IGEPAL-CA630, 1% deoxycholic acid, 1 mM EDTA, and 10 mM tris (pH 8.1)], and tris-EDTA [10 mM tris-HCl (pH 8.0) and 1 mM EDTA] buffers, DNA was recovered in elution buffer (1% SDS and 0.1 M NaHCO3), and cross-links were reversed by addition of 0.2 M NaCl at 65°C for 4 hours. DNA was purified by phenol-chloroform extraction and ethanol precipitation and analyzed by qPCR (primer sequences are shown in table S4).
IP assays
IP assays were performed as described previously (26). Briefly, cells were lysed in IP lysis buffer [150 mM NaCl, 50 mM Hepes (pH 7.9), 1 mM EDTA, 10% glycerol, 1% IGEPAL, 1 mM phenylmethylsulfonyl fluoride, and 1× cOmplete protease inhibitor cocktail (Roche)] and incubated for 20 min on ice, and debris was pelleted by centrifugation at 13,000 rpm at 4°C. Lysates were precleared by incubating with Protein G–Sepharose (Amersham Biosciences) for 1 hour with rotation. A 50-μg aliquot was removed for input analysis, and 1-mg aliquots of lysates were incubated overnight with rotation at 4°C with 2 μg of the indicated antibody (table S5). The next day, Protein G–Sepharose (20-μl bed volume) was added to the samples and incubated for 4 hour at 4°C with rotation. The resulting immunoprecipitate was washed with IP lysis buffer four times for 5 min at 4°C with rotation. Immunoprecipitates were subject to immunoblot assay using the indicated antibody (table S2).
Lentiviral transduction
Vectors encoding shRNA targeting HIF-1α and HIF-2α were described previously (27). Briefly, pLKO.1-puro lentiviral shuttle vectors encoding shRNA targeting NARF, KDM6A, and OCT4 (MilliporeSigma; table S6) were transfected into human embryonic kidney 293T cells for packaging (27). MDA-MB-231 and MCF-7 cells were transduced with viral supernatant for 24 hours, and stably transfected clones were selected with puromycin (0.5 μg/ml; MilliporeSigma). Cells were maintained in puromycin-containing medium.
Luciferase reporter assays
Complementary oligonucleotides (55 bp; table S7) were annealed and inserted into the Bam HI and Sal I sites of pGL2-Promoter (Promega), which contains a basal SV40 promoter upstream of FLuc coding sequences. For reporter assays, MDA-MB-231 cells were seeded onto six-well plates, transfected with the indicated plasmids, and exposed to 20 or 1% O2 for 24 hours. The FLuc/RLuc activities were measured using the Dual-Luciferase Reporter Assay System (Promega).
Mammosphere assay
Cells were counted after trypsinization, and single-cell suspensions were seeded in six-well ultralow attachment plates (Corning) at a density of 5000 cells/ml in complete MammoCult Medium (STEMCELL Technologies) (41). Mammosphere cultures were photographed 7 days later using a phase contrast microscope (Olympus), and mammospheres ≥50 μm in diameter were counted using ImageJ software [National Institutes of Health (NIH)].
Aldehyde dehydrogenase assay
The ALDEFLUOR assay (STEMCELL Technologies) was performed according to the manufacturer’s instructions (33). Cells were counted after trypsinization, and 1 × 106 live cells were suspended in assay buffer containing 1 mM BODIPY-aminoacetaldehyde and incubated at 37°C for 45 min. An aliquot of cells from each sample was treated with 50 mM diethylaminobenzaldehyde, an ALDH inhibitor, as a negative control for gating. Samples were analyzed by flow cytometry using FACSCalibur (BD Biosciences).
Animal studies
For tumorigenicity assays, 1 × 103 MDA-MB-231 subclone cells were injected into the MFP of 6- to 8-week-old female SCID mice (Charles River Laboratories, strain 561) in a 1:1 (v/v) suspension of Matrigel (BD Biosciences) in PBS. All mice were monitored weekly for tumor formation by gentle palpation. Ten weeks after injection, the mice were euthanized, and the number of mice that developed palpable tumors was recorded. For all other assays with SCID mice, 2 × 106 MDA-MB-231 subclone cells were injected into the MFP. Primary tumors were measured with calipers for length (L) and width (W) by a blinded observer, and tumor volume (V) was calculated as V = L × W2 × 0.524. The mice were euthanized on day 50 after injection. The wet weight of resected tumors was measured.
Immunofluorescence assays
Cells were fixed in chilled methanol for 5 min, rehydrated three times with PBS, and blocked in 3% bovine serum albumin/PBS for 30 min. The cells were stained with primary antibody in blocking buffer overnight at 4°C, rinsed with PBS, and stained with secondary antibody for 1 hour at room temperature. The cells were rinsed with PBS three times and mounted (ProLong Glass Antifade Mountant; Thermo Fisher Scientific, catalog no. P36980). The cells were observed under an Olympus Station 1 immunofluorescence microscope using a 60× oil (numerical aperture 1.49) objective lens. Antibodies used in the assay are shown in table S8.
Immunohistochemistry
A tissue microarray containing 89 breast cancer specimens was obtained from Shanghai Zhuoli Company. Formalin-fixed and paraffin-embedded 4-μm sections mounted on glass slides were incubated at 60°C for 60 min. The slides were deparaffinized with xylene and rehydrated in decreasing concentrations of ethanol (100, 95, and 80%). For antigen retrieval, the slides were boiled in 10 mM sodium citrate buffer (pH 6.0) for 10 min. After inhibition of endogenous peroxidase activities by heat-induced recovery with methanol containing 3% H2O2 for 10 min, the sections were blocked with tris-buffered saline with Tween 20 (Sigma-Aldrich) containing 5% normal goat serum for 30 min and incubated overnight at 4°C with primary antibody against NARF (1:50 dilution; Novus Biologicals, NBP2-55660) followed by incubation with a biotin-labeled secondary antibody (Jackson ImmunoResearch, catalog no. 111-035-003) for 30 min. Color was developed with 3,3′-diaminobenzidine, and instant hematoxylin (Thermo Fisher Scientific, catalog no. 7211) was used for counterstaining. Staining was independently assessed by two senior pathologists without knowledge of patient characteristics. The intensity of staining was scored as 0 (−), 1 (+), 2 (++), or 3 (+++), denoting no staining, light staining, intermediate staining, or dark staining, respectively, by visual assessment. The H-score was calculated on the basis of the following formula: H = (1 × percentage of cells of light staining) + (2 × percentage of cells of intermediate staining) + (3 × percentage of cells of dark staining). For Kaplan-Meier analysis of overall survival, samples were divided into high and low groups according to the median H-score for NARF expression.
Bioinformatics and statistics
TCGA breast cancer data were obtained from an online data portal (https://xenabrowser.net/) (42). Kaplan-Meier curves were generated from a dataset containing gene expression and survival data from 4929 breast cancer patients, and the log-rank test was performed (https://kmplot.com/analysis/) (43). Kaplan-Meier survival analysis for GSE4922 was performed for the Uppsala cohort patients (44, 45). Log-rank and Cox proportional hazard regression tests were performed with the “survival” package in base R (v4.0.5) (46). Kaplan-Meier plots were generated using the survminer (v0.4.9) package (47) with ggplot2 (v3.3.6) (48). The UALCAN database was used to analyze the mRNA and protein expression of NARF in tumor and normal samples based on data from TCGA and the Clinical Proteomic Tumor Analysis Consortium dataset (49, 50). Pearson’s correlation test was used to compare NARF expression with the HIF signature or BCSC signature, based on mRNA levels from TCGA Breast Invasive Carcinoma dataset of 1218 patients. For the tumorigenicity assay, Fisher’s exact test was performed. For all other assays, values are expressed as means ± SEM, two-tailed Student’s t test was used to test differences between two groups, differences between multiple groups were analyzed by two-way analysis of variance, and P values < 0.05 were considered significant for all analyses.
Public data analysis
NARF expression in primary patient tumors and corresponding patient survival data were analyzed using datasets from the Gene Expression Omnibus (GEO) database. The data are publicly available and can be accessed using GEO accession numbers GSE4922, GSE12276, and GSE163882. Normalized series matrix files were downloaded directly from GEO. Patient survival data and expression data corresponding to the NARF gene probe(s) were extracted. Expression data from multiple probes corresponding to the NARF gene (if applicable) were averaged before analysis. Data sorting and statistical analyses were performed in base R (v4.0.5). Plotting was performed with the ggplot2 (v3.3.6) package in R.
Study approval
Mouse protocols were approved by the Johns Hopkins University Animal Care and Use Committee and were in accordance with the NIH Guide for the Care and Use of Laboratory Animals (51).
Supplementary Material
Acknowledgments
We thank R. Geisler, S. Garcia, and N. Sardo (Novus Biologicals Inc.) for providing antibodies listed in tables S2, S3, S5, and S8. G.L.S. is an American Cancer Society Research Professor and the C. Michael Armstrong Professor at the Johns Hopkins University School of Medicine.
Funding: This work was supported by grants from the American Cancer Society (122473-RP-13-090-001-COUN), Armstrong Family Foundation, and the Cindy Rosencrans Fund for Triple-Negative Breast Cancer.
Author contributions: Conceptualization: Y.Y. and G.L.S. Investigation: Y.Y., C.C., Q.Z., T.Y.-T.H., E.E.W., W.J., E.D., R.W., Y.W., N.L., Y.Z., and W.Q. Methodology: H.L., S.S., and Y.L. Visualization: Y.Y. and C.C. Writing (original draft): Y.Y. and C.C. Writing (review and editing): G.L.S. Supervision: G.L.S.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S8
Correction (21 June 2023):
The original version of table S6 in the Supplementary Materials included a duplicated nucleotide sequence. The table has been corrected in the Supplementary Materials PDF. The original version is available here:
REFERENCES AND NOTES
- 1.H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal, F. Bray,Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71,209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.G. P. Gupta, J. Massagué,Cancer metastasis: Building a framework. Cell 127,679–695 (2006). [DOI] [PubMed] [Google Scholar]
- 3.P. S. Steeg,Tumor metastasis: Mechanistic insights and clinical challenges. Nat. Med. 12,895–904 (2006). [DOI] [PubMed] [Google Scholar]
- 4.R. Bos, H. Zhong, C. F. Hanrahan, F. C. Mommers, G. L. Semenza, H. M. Pinedo, M. D. Abeloff, J. W. Simons, P. J. van Diest, E. van der Wall,Levels of hypoxia-inducible factor-1α during breast carcinogenesis. J. Natl. Cancer Inst. 93,309–314 (2001). [DOI] [PubMed] [Google Scholar]
- 5.A. L. Harris,Hypoxia—A key regulatory factor in tumour growth. Nat. Rev. Cancer 2,38–47 (2002). [DOI] [PubMed] [Google Scholar]
- 6.P. Vaupel, M. Höckel, A. Mayer,Detection and characterization of tumor hypoxia using pO2 histography. Antioxid. Redox Signal. 9,1221–1236 (2007). [DOI] [PubMed] [Google Scholar]
- 7.H. Zhong, A. M. De Marzo, E. Laughner, M. Lim, D. A. Hilton, D. Zagzag, P. Buechler, W. B. Isaacs, G. L. Semenza, J. W. Simons,Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 59,5830–5835 (1999). [PubMed] [Google Scholar]
- 8.L. Schito, G. L. Semenza,Hypoxia-inducible factors: Master regulators of cancer progression. Trends Cancer 2,758–770 (2016). [DOI] [PubMed] [Google Scholar]
- 9.G. L. Wang, B. H. Jiang, E. A. Rue, G. L. Semenza,Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 92,5510–5514 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Z. Andrysik, H. Bender, M. D. Galbraith, J. M. Espinosa,Multi-omics analysis reveals contextual tumor suppressive and oncogenic gene modules within the acute hypoxic response. Nat. Commun. 12,1375 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.R. Bos, P. van der Groep, A. E. Greijer, A. Shvarts, S. Meijer, H. M. Pinedo, G. L. Semenza, P. J. van Diest, E. van der Wall,Levels of hypoxia-inducible factor-1α independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer 97,1573–1581 (2003). [DOI] [PubMed] [Google Scholar]
- 12.M. Schindl, S. F. Schoppmann, H. Samonigg, H. Hausmaninger, W. Kwasny, M. Gnant, R. Jakesz, E. Kubista, P. Birner, G. Oberhuber; Austrian Breast and Colorectal Cancer Study Group ,Overexpression of hypoxia-inducible factor 1α is associated with an unfavorable prognosis in lymph node-positive breast cancer. Clin. Cancer Res. 8,1831–1837 (2002). [PubMed] [Google Scholar]
- 13.G. L. Semenza,The hypoxic tumor microenvironment: A driving force for breast cancer progression. Biochim. Biophys. Acta 1863,382–391 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.G. L. Semenza,Intratumoral hypoxia and mechanisms of immune evasion mediated by hypoxia-inducible factors. Phys. Ther. 36,73–83 (2021). [DOI] [PubMed] [Google Scholar]
- 15.K. L. Talks, H. Turley, K. C. Gatter, P. H. Maxwell, C. W. Pugh, P. J. Ratcliffe, A. L. Harris,The expression and distribution of the hypoxia-inducible factors HIF-1α and HIF-2α in normal human tissues, cancers, and tumor-associated macrophages. Am. J. Pathol. 157,411–421 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.V. Plaks, N. Kong, Z. Werb,The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 16,225–238 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.L. Xiang, G. L. Semenza,Hypoxia-inducible factors promote breast cancer stem cell specification and maintenance in response to hypoxia or cytotoxic chemotherapy. Adv. Cancer Res. 141,175–212 (2019). [DOI] [PubMed] [Google Scholar]
- 18.S. J. Conley, E. Gheordunescu, P. Kakarala, B. Newman, H. Korkaya, A. N. Heath, S. G. Clouthier, M. S. Wicha,Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc. Natl. Acad. Sci. U.S.A. 109,2784–2789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L. P. Schwab, D. L. Peacock, D. Majumdar, J. F. Ingels, L. C. Jensen, K. D. Smith, R. C. Cushing, T. N. Seagroves,Hypoxia-inducible factor 1α promotes primary tumor growth and tumor-initiating cell activity in breast cancer. Breast Cancer Res. 14,R6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.H. Lu, Y. Xie, L. Tran, J. Lan, Y. Yang, N. L. Murugan, R. Wang, Y. J. Wang, G. L. Semenza,Chemotherapy-induced S100A10 recruits KDM6A to facilitate OCT4-mediated breast cancer stemness. J. Clin. Invest. 130,4607–4623 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.H. Lu, Y. Lyu, L. Tran, J. Lan, Y. Xie, Y. Yang, N. L. Murugan, Y. J. Wang, G. L. Semenza,HIF-1 recruits NANOG as a coactivator for TERT gene transcription in hypoxic breast cancer stem cells. Cell Rep. 36,109757 (2021). [DOI] [PubMed] [Google Scholar]
- 22.R. M. Barton, H. J. Worman,Prenylated prelamin A interacts with NARF, a novel nuclear protein. J. Biol. Chem. 274,30008–30018 (1999). [DOI] [PubMed] [Google Scholar]
- 23.T. Kiel, A. Busch, A. Meyer-Rachner, S. Hübner,Laminopathy-inducing mutations reduce nuclear import of expressed prelamin A. Int. J. Biochem. Cell Biol. 53,271–280 (2014). [DOI] [PubMed] [Google Scholar]
- 24.D. Ding, A. O. Valdivia, S. K. Bhattacharya,Nuclear prelamin A recognition factor and iron dysregulation in multiple sclerosis. Metab. Brain Dis. 35,275–282 (2020). [DOI] [PubMed] [Google Scholar]
- 25.R. Schippert, F. Schaeffel, M. P. Feldkaemper,Microarray analysis of retinal gene expression in Egr-1 knockout mice. Mol. Vis. 15,2720–2739 (2009). [PMC free article] [PubMed] [Google Scholar]
- 26.Y. Yang, H. Lu, C. Chen, Y. Lyu, R. N. Cole, G. L. Semenza,HIF-1 interacts with TRIM28 and DNA-PK to release paused RNA polymerase II and activate target gene transcription in response to hypoxia. Nat. Commun. 13,316 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.H. Zhang, C. C. L. Wong, H. Wei, D. M. Gilkes, P. Korangath, P. Chaturvedi, L. Schito, J. Chen, B. Krishnamachary, P. T. Winnard Jr., V. Raman, L. Zhen, W. A. Mitzner, S. Sukumar, G. L. Semenza,HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene 31,1757–1770 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.H. Zhang, D. Z. Qian, Y. S. Tan, K. Lee, P. Gao, Y. R. Ren, S. Rey, H. Hammers, D. Chang, R. Pili, C. V. Dang, J. O. Liu, G. L. Semenza,Digoxin and other cardiac glycosides inhibit HIF-1α synthesis and block tumor growth. Proc. Natl. Acad. Sci. U.S.A. 105,19579–19586 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.C. C.-L. Wong, H. Zhang, D. M. Gilkes, J. Chen, H. Wei, P. Chaturvedi, M. E. Hubbi, G. L. Semenza,Inhibitors of hypoxia-inducible factor 1 block breast cancer metastatic niche formation and lung metastasis. J. Mol. Med. 90,803–815 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.G. L. Semenza, B. H. Jiang, S. W. Leung, R. Passantino, J. P. Concordet, P. Maire, A. Giallongo,Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271,32529–32537 (1996). [DOI] [PubMed] [Google Scholar]
- 31.S. Pece, D. Disalvatore, D. Tosoni, M. Vecchi, S. Confalonieri, G. Bertalot, G. Viale, M. Colleoni, P. Veronesi, V. Galimberti, P. P. Di Fiore,Identification and clinical validation of a multigene assay that interrogates the biology of cancer stem cells and predicts metastasis in breast cancer: A retrospective consecutive study. EBioMedicine 42,352–362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D. Ponti, A. Costa, N. Zaffaroni, G. Pratesi, G. Petrangolini, D. Coradini, S. Pilotti, M. A. Pierotti, M. G. Daidone,Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 65,5506–5511 (2005). [DOI] [PubMed] [Google Scholar]
- 33.C. Ginestier, M. H. Hur, E. Charafe-Jauffret, F. Monville, J. Dutcher, M. Brown, J. Jacquemier, P. Viens, C. G. Kleer, S. Liu, A. Schott, D. Hayes, D. Birnbaum, M. S. Wicha, G. Dontu,ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1,555–567 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.E. T. Wiles, E. U. Selker,H3K27 methylation: A promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 43,31–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The Cancer Genome Atlas Network ,Comprehensive molecular portraits of human breast tumors. Nature 490,61–70 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D. R. Mole, C. Blancher, R. R. Copley, P. J. Pollard, J. M. Gleadle, J. Ragoussis, P. J. Ratcliffe,Genome-wide association of hypoxia-inducible factor (HIF)-1α and HIF-2α DNA binding with expression profiling of hypoxia-inducible transcripts. J. Biol. Chem. 284,16767–16775 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J. Schödel, S. Oikonomopoulos, J. Ragoussis, C. W. Pugh, P. J. Ratcliffe, D. R. Mole,High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood 117,e207–e217 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D. Song, F. S. Lee,A role for IOP1 in mammalian cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 283,9231–9238 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D. Song, F. S. Lee,Mouse knock-out of IOP1 protein reveals its essential role in mammalian cytosolic iron-sulfur protein biogenesis. J. Biol. Chem. 286,15797–15805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.J. Huang, D. Song, A. Flores, Q. Zhao, S. M. Mooney, L. M. Shaw, F. S. Lee,IOP1, a novel hydrogenase-like protein that modulates hypoxia-inducible factor-1α activity. Biochem. J. 401,341–352 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D. Samanta, G. L. Semenza,In vitro assays of breast cancer stem cells. Methods Mol. Biol. 1742,237–246 (2018). [DOI] [PubMed] [Google Scholar]
- 42.M. J. Goldman, B. Craft, M. Hastie, K. Repečka, F. McDade, A. Kamath, A. Banerjee, Y. Luo, D. Rogers, A. N. Brooks, J. Zhu, D. Haussler,Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38,675–678 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.A. Lánczky, B. Győrffy,Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J. Med. Internet Res. 23,e27633 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.J. Bergh, T. Norberg, S. Sjögren, A. Lindgren, L. Holmberg,Complete sequencing of the p53 gene provides prognostic information in breast cancer patients, particularly in relation to adjuvant systemic therapy and radiotherapy. Nat. Med. 1,1029–1034 (1995). [DOI] [PubMed] [Google Scholar]
- 45.A. V. Ivshina, J. George, O. Senko, B. Mow, T. C. Putti, J. Smeds, T. Lindahl, Y. Pawitan, P. Hall, H. Nordgren, J. E. Wong, E. T. Liu, J. Bergh, V. A. Kuznetsov, L. D. Miller,Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 66,10292–10301 (2006). [DOI] [PubMed] [Google Scholar]
- 46.R. C. Team, R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2021).
- 47.H. Wickham, ggplot2: Elegant Graphics for Data Analysis. (Springer, 2016). [Google Scholar]
- 48.A. Kassambara, M. Kosinski, P. Biecek, survminer: Drawing Survival Curves using ‘ggplot2’. R package version 0.4.9 (2021).
- 49.D. S. Chandrashekar, B. Bashel, S. A. H. Balasubramanya, C. J. Creighton, I. Ponce-Rodriguez, B. V. S. K. Chakravarthi, S. Varambally,UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19,649–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.F. Chen, D. S. Chandrashekar, S. Varambally, C. J. Creighton,Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 10,5679 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.National Research Council, Guide for the Care and Use of Laboratory Animals, Eighth Edition. (The National Academies Press, 2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S8
Correction (21 June 2023):
The original version of table S6 in the Supplementary Materials included a duplicated nucleotide sequence. The table has been corrected in the Supplementary Materials PDF. The original version is available here: