Abstract
Background
Autoantibodies against type I IFNs occur in approximately 10% of adults with life-threatening coronavirus disease 2019 (COVID-19). The frequency of anti-IFN autoantibodies in children with severe sequelae of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is unknown.
Objective
We quantified anti–type I IFN autoantibodies in a multicenter cohort of children with severe COVID-19, multisystem inflammatory syndrome in children (MIS-C), and mild SARS-CoV-2 infections.
Methods
Circulating anti–IFN-α2 antibodies were measured by a radioligand binding assay. Whole-exome sequencing, RNA sequencing, and functional studies of peripheral blood mononuclear cells were used to study any patients with levels of anti–IFN-α2 autoantibodies exceeding the assay’s positive control.
Results
Among 168 patients with severe COVID-19, 199 with MIS-C, and 45 with mild SARS-CoV-2 infections, only 1 had high levels of anti–IFN-α2 antibodies. Anti–IFN-α2 autoantibodies were not detected in patients treated with intravenous immunoglobulin before sample collection. Whole-exome sequencing identified a missense variant in the ankyrin domain of NFKB2, encoding the p100 subunit of nuclear factor kappa–light-chain enhancer of activated B cells, aka NF-κB, essential for noncanonical NF-κB signaling. The patient’s peripheral blood mononuclear cells exhibited impaired cleavage of p100 characteristic of NFKB2 haploinsufficiency, an inborn error of immunity with a high prevalence of autoimmunity.
Conclusions
High levels of anti–IFN-α2 autoantibodies in children and adolescents with MIS-C, severe COVID-19, and mild SARS-CoV-2 infections are rare but can occur in patients with inborn errors of immunity.
Key words: Anti-interferon autoantibody, COVID-19, MIS-C, NFKB2, inborn errors of immunity
Introduction
Neutralizing autoantibodies against type I IFNs occur in approximately 10% of adults with life-threatening coronavirus disease 2019 (COVID-19).1 , 2 Less is known about levels of anti-IFN antibodies in pediatric populations. Small studies of 7 to 59 children identified autoantibodies to several tissue antigens in patients with multisystem inflammatory syndrome in children (MIS-C), a postinfectious inflammatory disorder typically occurring within 2 to 6 weeks of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection.3, 4, 5, 6 Although inborn errors of immunity can be associated with autoantibodies,7 it is unknown if anti-IFN autoantibodies are associated with severe COVID-19 or MIS-C in the general pediatric population. Treatment of patients with MIS-C with intravenous immunoglobulin (IVIG), which can contain autoantibodies to tissue antigens, may confound the measurement of endogenous autoantibodies.8
Here we present results from a multicenter study investigating anti–IFN-α2 autoantibodies in a large cohort of children and adolescents with MIS-C, severe COVID-19, or mild SARS-CoV-2 infections.
Results and discussion
This study included 412 patients: 199 patients with MIS-C, 168 patients hospitalized for COVID-19 in an intensive care or step-down unit (henceforth referred to as severe COVID-19), and 45 with SARS-CoV-2 infections managed as outpatients. All but 2 patients were under 21 years of age (Table I ). IVIG was administered to 137 patients (68.8%) with MIS-C and to 9 patients (5%) with severe COVID-19 before specimen collection. Critical care was required for 85.4% of patients with MIS-C and 69.6% of patients with severe COVID-19 (Table I). Five patients (3%) who were hospitalized for COVID-19 died; all with MIS-C survived.
Table I.
Patient characteristics of children with MIS-C, severe COVID-19 requiring intensive care unit or step-down unit hospital care, and outpatients with mild SARS-CoV-2 infections evaluated for anti–IFN-α2 autoantibodies
| Characteristic | MIS-C (n = 199) | Severe COVID-19 (n = 168) | Mild SARS-CoV-2 infection (n = 45) |
|---|---|---|---|
| Male sex | 118 (59.3) | 83 (49.4) | 24 (53.3) |
| Age (years), median (interquartile range) | 10.9 (7.5-14.7) | 13.6 (6.3-17.2) | 5.5 (2.2-11.1) |
| Race/ethnicity | |||
| White, non-Hispanic | 63 (31.7) | 62 (36.9) | 14 (31.1) |
| Black, non-Hispanic | 74 (37.2) | 41 (24.4) | 5 (11.1) |
| Hispanic or Latino | 47 (23.6) | 46 (27.4) | 15 (33.3) |
| Other race, non-Hispanic | 8 (4.0) | 14 (8.3) | 2 (4.4) |
| Unknown | 7 (3.5) | 5 (3.0) | 9 (20.0) |
| Previously healthy | 134 (67.3) | 48 (28.6) | 42 (93)∗ |
| Preexisting condition | |||
| Obesity† | 29 (14.6) | 50 (29.8) | 8 (17.8) |
| Asthma | 22 (11.1) | 34 (20.2) | 1 (2.2) |
| Cardiovascular | 4 (2.0)‡ | 15 (8.9)§ | 0 |
| Interventions | |||
| Intensive care unit admission | 170 (85.4) | 117 (69.6) | 0 |
| Shock requiring vasopressors | 90 (45.2) | 16 (9.5) | 0 |
| Mechanical ventilation | 38 (19.1) | 60 (35.7) | 0 |
Data are presented as nos. (%) unless otherwise indicated.
Among outpatients, 1 had asthma requiring inhaled steroids, 1 had sickle cell trait, and 1 had a seizure disorder.
Because height was not available for many outpatients, weight for age of >95% percentile for age was used as a proxy to assign potential obesity status in lieu of body mass index.
Congenital heart disease (n = 4).
Congenital heart disease (n = 7), systemic hypertension (n = 6), acquired heart disease (n = 2).
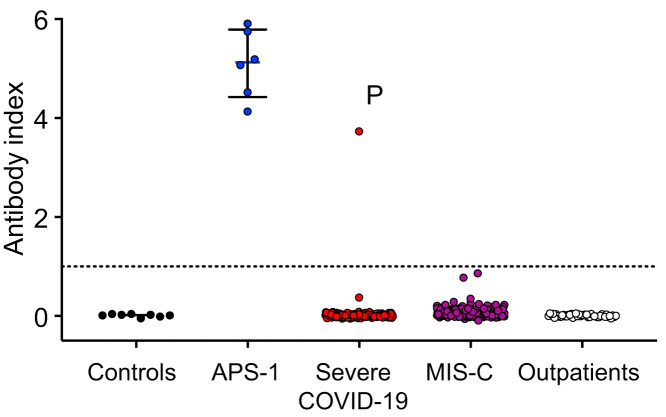
Anti–IFN-α2 autoantibodies were measured using an established radioligand binding assay (see the Methods in this article’s Online Repository at www.jacionline.org). The antibody index indicates the ratio of IFN-α2 protein precipitated by patient plasma normalized to the assay’s positive control.2 Plasma from 8 healthy adults served as negative controls (mean antibody index of 0.011). Positive disease controls included 6 individuals with anti–IFN-α2 autoantibodies due to autoimmune polyglandular syndrome type 1 (APS-1), a disease known to cause neutralizing anti–type I IFN autoantibodies.9 , 10 In our cohort, only 1 patient had an antibody index (3.73) exceeding that of the assay’s positive control and the levels previously found in adults with severe COVID-19 due to neutralizing anti–IFN-α2 antibodies2 (Fig 1 ). This patient’s sample was obtained before treatment with IVIG. Additionally, none of the other 137 patients who received IVIG before specimen collection had high levels of anti–IFN-α2 autoantibody, thus excluding IVIG as a confounding source of autoantibodies. Our study was not designed to quantify the incidence of anti–IFN-α2 autoantibodies in children with COVID-19 or MIS-C, as not all eligible children consented to enrollment. However, the largest study of anti-IFN autoantibodies in adults identified anti–IFN-α2 autoantibodies in 88 (8.9%) of 987 patients with severe COVID-19,1 which is higher than the 0.5% (n = 1) with autoantibodies and severe COVID-19 in our cohort.
Fig 1.
High levels of anti–IFN-α2 autoantibodies in children and adolescents with MIS-C, severe COVID-19, and mild SARS-CoV-2 infections are rare. Levels of anti–IFN-α2 were measured by radioligand binding assay. The dotted line represents the antibody index of the anti-Myc assay–positive control. P denotes the patient with high levels of anti–IFN-α2 autoantibodies. APS-1 was used as positive disease control. Medians and interquartile ranges are shown for the control (n = 8) and APS-1 (n = 6) cohorts.
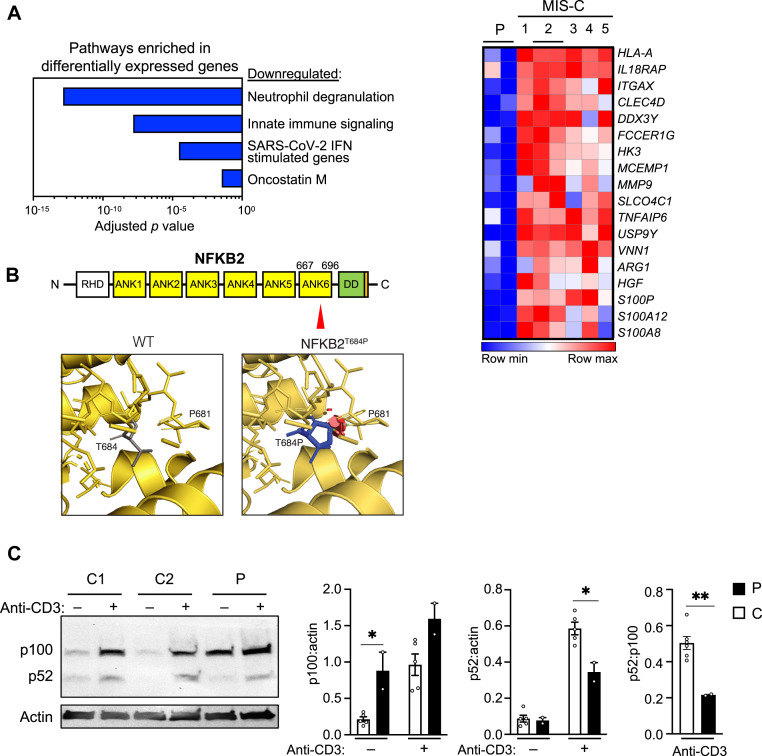
The patient with high levels of anti–IFN-α2 autoantibodies was an adolescent female subject with acute hypoxemic respiratory failure due to severe COVID-19. Neutralizing antibodies to SARS-CoV-2 were not detected until the fifth day of hospitalization, rising to levels comparable to that of other patients with severe COVID-19 by day 12.11 Her respiratory failure resolved after 2 weeks of hospitalization, but she subsequently developed left ventricular dysfunction, which was treated with milrinone in the setting of persistently elevated inflammatory markers, prompting diagnostic consideration of MIS-C. Compared to other patients similarly treated for MIS-C, the patient’s peripheral blood mononuclear cells (PBMCs) exhibited reduced expression of differentially expressed genes in pathways of neutrophil degranulation, innate immune signaling, SARS-CoV-2 IFN-stimulated genes, and oncostatin M, an enhancer of type I IFN signaling (Fig 2 , A). Reduced IFN signaling is found in adults with acute COVID-19, particularly those with anti–type I IFN autoantibodies.1 , 2 However, the reduced neutrophil degranulation and innate immune signaling in our patient contrasts with that of neutrophil and monocyte activation found in adults with severe COVID-196 , 12 , 13 and children with MIS-C,4 , 14 suggesting that additional factors beyond anti–IFN-α2 autoantibodies contributed to her immune dysregulation.
Fig 2.
A, Differentially expressed genes (≥2-fold change, false discovery rate < 0.05) determined via bulk RNA sequencing of whole blood from the single patient with increased levels of anti–IFN-α2 autoantibodies, compared to 5 disease controls (patients with MIS-C of comparable age, disease severity, and treatment). The patient and one of the disease controls (labeled 2) had 2 samples available at the midpoint of their hospitalizations. B,Top, Schematic of NFKB2 with the patient’s variant indicated with a red triangle in ankyrin repeat domain 6 (ANK6). DD, Death domain, with the degron domain needed for p100 processing noted in orange; RHD, Rel homology domain. Bottom, Steric clash (red disks) is predicted to arise from the patient’s substitution of a bulky proline residue for threonine 684. C, Representative immunoblot of full-length p100 and processed p52 in PBMCs from the patient (P) and 2 healthy controls (C1 and C2) with and without anti-CD3 stimulation for 2 days. Bar graphs show densitometric quantitation of indicated proteins pooled from 2 experiments in PBMCs from the patient and 5 controls, with and without 2 days of anti-CD3 stimulation. ∗P < .05, ∗∗P < .01 by Student t test.
In addition to prolonged hospitalization for COVID-19 followed by MIS-C, this patient had a history of influenza A pandemic H1N1/09 viral pneumonia requiring noninvasive ventilation. Immunologic evaluation after recovery from that hospitalization was notable only for reduced levels of IgG and IgA, although titers to tetanus and pneumococcus were normal (see Table E1 in the Online Repository available at www.jacionline.org). Genetic testing and immunoglobulin replacement were not initiated then, as she had no prior significant infections. Immunologic evaluation after recovery from COVID-19 and MIS-C in 2020 revealed panhypogammaglobulinemia, with reduced titers to pneumococcal subtypes. Whole-exome sequencing identified a heterozygous variant in the ankyrin domain of NFKB2 (p.Thr684Pro), encoding the p100 subunit of nuclear factor kappa–light-chain enhancer of activated B cells (aka NF-κB) essential for noncanonical NF-κB signaling. This variant is absent from the gnomAD database (gnomad.broadinstitute.org) and is predicted to be pathogenic, with a CADD score of 27.6. Structural modeling indicates that the Thr684Pro variant causes steric clash (Fig 2, B). After stimulation with anti-CD3, the patient’s PBMCs exhibited increased p100 levels and reduced p52 levels, indicative of impaired cleavage of p100 into its active form (Fig 2, C). This finding is consistent with the importance of the ankyrin domain for p100 ubiquitinylation and cleavage.15 All previously published missense mutations affect the protein’s C-terminal degron domain essential for p100 processing.16 , 17 The patient’s pan-hypogammaglobulinemia, anti–IFN-α2 autoantibodies, and susceptibility to severe viral infections indicate the deleterious effect of her NFKB2Thr683Pro variant. Her residual levels of p52 likely contributed to the sporadic nature of her infections, which emerged only when exposed to newly emerged pathogens during 2 pandemics. Similarly, all 3 previously reported patients with NFKB2 haploinsufficiency and COVID-19 required critical care.7 , 18 , 19
Table E1.
Immunologic evaluations in the patient with NFKB2Thr684Pro
| Characteristic | Patient’s approximate age group at laboratory testing |
|
|---|---|---|
| Young childhood after recovery from influenza A(H1N1)pdm09 virus pneumonia (reference range) | Older adolescence after recovery from severe COVID-19 (reference range) | |
| Hemogram | ||
| White blood cells, 103 cells/μL | 11.28 (5.41-9.7) | 7.78 (5.52-9.29) |
| Neutrophils, 103 cells/μL | 5.31 (2.58-5.95) | 3.51 (3.04-6.06) |
| Lymphocytes, 103 cells/μL | 4.92 (1.23-2.76) | 3.09 (1.17-3.10) |
| Platelets, 103 cells/μL | 406 (187-376) | 384 (189-342) |
| Lymphocyte subsets∗ | ||
| CD3+, 103 cells/μL | 3879 (1000-2600) | 3166 (1000-2600) |
| CD3+CD4+, 103 cells/μL | 2070 (225-1100) | 1649 (530-1500) |
| CD3+CD8+, 103 cells/μL | 1616 (3330-1100) | 1367 (330-1100) |
| CD19+, 103 cells/μL | 705 (270-860) | 251 (110-570) |
| Naive, % CD19+ | 74.0 (48.4-79.7) | 72.3 (48.4-79.7) |
| Unswitched memory, % CD19+ | 10.4 (7.0-23.8) | 6.6 (7.0-23.8)† |
| Switched memory, % CD19+ | 12.4 (8.30-27.8) | 16.7 (8.30-27.8) |
| Plasmablasts | Not done | 0.3 (0.1-2.4) |
| Marginal zone–like B cells, % CD19+ | Not done | 19.6 (11.8-59.7) |
| CD3−CD56+, 103 cells/μL | 591 (70-480) | 62 (70-480)† |
| Immunoglobulins | ||
| IgG, mg/dL | 501 (639-1434)† | 369 (639-1344)† |
| IgM, mg/dL | 182 (40-240) | 28 (40-240)† |
| IgA, mg/dL | 61 (70-312)† | 24 (70-312)† |
| Vaccine titers | ||
| Positive titers to pneumococcal subtypes | 13/14 (>7) | 5/23 (>14) |
| Tetanus | >7.0 (>0.15) | 0.44 (>0.15) |
This patient had 2 immunologic evaluations: after recovery from influenza A(H1N1)pdm09 virus pneumonia in early childhood, and after recovery from SARS-CoV-2 during adolescence. The parenthetical reference ranges are matched to the patient’s age group and are derived from healthy controls, as determined by the clinical immunology laboratory at Boston Children’s Hospital (Boston, Mass).
Naive, CD19+CD27−IgD+, unswitched memory CD19+CD27+IgD+, switched memory CD19+CD27+IgD−, plasmablast CD19+CD24loCD38hi.
Clinically relevant values outside the parenthetical reference ranges.
Autoimmunity, including anticytokine antibodies, occurs in up to 80% of patients with NFKB2 haploinsufficiency.17 Although we did not measure the neutralizing capacity of our patient’s anti–IFN-α2 autoantibodies, her antibody index exceeds that of autoantibodies with neutralizing capacity in published studies using the same assay in independent cohorts.2 , 20 Additionally, differentially expressed genes from her whole-blood transcriptome were reduced in pathways downstream of type I IFN signaling compared to other patients with MIS-C, further supporting a neutralizing effect of her anti–IFN-α2 autoantibodies. At the time of this study, she had no clinical evidence of autoimmunity beyond the anti–IFN-α2 autoantibodies. Because our study is limited by the identification of only a single patient with NFKB2 haploinsufficiency, future studies with larger cohorts are needed to determine the prevalence and levels of anti-IFN autoantibodies in patients with this disease.
Our study underscores the rarity of high levels of anti–IFN-α2 antibodies in most children. While the majority of adults with severe COVID-19 and autoantibodies to type 1 IFNs have anti–IFN-α2 autoantibodies, Bastard et al1 have shown that a subset of individuals have only anti–IFN-ω antibodies, for which we did not screen. In samples collected before the COVID-19 pandemic, neutralizing autoantibodies to IFN-α were identified in less than 0.3% of individuals younger than 69 years,1 , 21 compared to 1.1% of adults aged 70 through 79 years and 3.4% in those over 80 years.21 Neutralizing autoantibodies to IFN occurred in 10% of individuals with severe COVID-19, the majority of whom were over 75 years of age. Other than APS-1, no inborn errors of immunity, including defects in the NFKB2 pathway, have been reported in adults with severe COVID-19 and autoantibodies to type I IFNs.
Autoantibodies occur more frequently in the elderly as a result of progressive B-cell dysfunction, differentiation of age-associated B cells into autoantibody-producing plasma cells, and the release of self-antigens from tissue damage. In children, anti-IFN autoantibodies may instead reflect early onset B-cell dysfunction with underlying immune dysfunction. In support of this, a study of 31 individuals with known inborn errors of immunity identified neutralizing autoantibodies against IFN-α2 and IFN-ω in one child with a combined immunodeficiency and another with immune dysregulation.7 This previously published cohort had 3 additional pediatric patients with antinuclear antibodies, reflecting the spectrum of autoantibodies in patients with inborn errors of immunity. Thus, diagnostic studies for genetic causes of immune dysregulation are merited in children with anticytokine antibodies.
Clinical implications.
Anti–IFN-α2 autoantibodies should prompt diagnostic evaluation for inborn errors of immunity if identified in children or adolescents.
Acknowledgments
We thank the patients and their families for participation in this study. We thank Michail S. Lionakis (National Institute of Allergy and Infectious Diseases) for providing positive control APS-1 serum, and the New York Blood Center for providing pre–COVID-19 control plasma. Support for document creation and format was provided by AuthorArranger (analysistools.cancer.gov/), a tool developed at the National Cancer Institute.
The following members of the Overcoming COVID-19 Network Study Group Investigators (presented alphabetically by state) were all closely involved with the design, implementation, and oversight of the Overcoming COVID-19 study as well as collecting patient samples and data. Alabama: Children’s of Alabama, Birmingham: Michele Kong, MD. Arkansas: Arkansas Children’s Hospital, Little Rock: Katherine Irby, MD; Ronald C. Sanders Jr, MD, MS; Masson Yates; Chelsea Smith. California: UCSF Benioff Children’s Hospital Oakland, Oakland: Natalie Z. Cvijanovich, MD; UCSF Benioff Children’s Hospital, San Francisco: Matt S. Zinter, MD. Florida: Holtz Children’s Hospital, Miami: Brandon Chatani, MD; Gwenn McLaughlin, MD, MSPH. Georgia: Children’s Healthcare of Atlanta at Egleston, Atlanta: Keiko M. Tarquinio, MD. Illinois: Ann & Robert H. Lurie Children’s Hospital of Chicago, Chicago: Bria M. Coates, MD. Indiana: Riley Children’s Hospital, Indianapolis: Courtney M. Rowan, MD, MScr. Massachusetts: Boston Children’s Hospital, Boston: Adrienne G. Randolph, MD; Margaret M. Newhams, MPH; Suden Kucukak, MD; Tanya Novak, PhD; Hye Kyung Moon, MA; Takuma Kobayashi BS; Jeni Melo, BS; Cameron Young, BS; Sabrina R. Chen, BS; Janet Chou, MD. Michigan: University of Michigan C. S. Mott Children’s Hospital, Ann Arbor: Heidi R. Flori, MD, FAAP; Mary K. Dahmer, PhD. Minnesota: Mayo Clinic, Rochester: Emily R. Levy, MD, FAAP; Supriya Behl, MSc; Noelle M. Drapeau, BA. Missouri: Children’s Mercy Hospital, Kansas City: Jennifer E. Schuster, MD. Nebraska: Children’s Hospital & Medical Center, Omaha: Melissa L. Cullimore, MD, PhD; Russell J. McCulloh, MD. New Jersey: Cooperman Barnabas Medical Center, Livingston: Shira J. Gertz, MD. North Carolina: University of North Carolina, Chapel Hill: Stephanie P. Schwartz, MD; Tracie C. Walker, MD. Ohio: Akron Children’s Hospital, Akron: Ryan A. Nofziger, MD; Cincinnati Children’s Hospital, Cincinnati: Mary Allen Staat, MD, MPH; Chelsea C. Rohlfs, BS, MBA. Pennsylvania: Children’s Hospital of Philadelphia, Philadelphia: Julie C. Fitzgerald, MD, PhD, MSCE. South Carolina: MUSC Shawn Jenkins Children’s Hospital, Charleston: Elizabeth H. Mack, MD, MS; Nelson Reed, MD. Tennessee: Monroe Carell Jr Children’s Hospital at Vanderbilt, Nashville: Natasha B. Halasa, MD, MPH. Texas: Texas Children’s Hospital and Baylor College of Medicine, Houston: Laura L. Loftis, MD. Utah: Primary Children’s Hospital and University of Utah, Salt Lake City: Hillary Crandall, MD, PhD.
Members of the US Centers for Disease Control and Prevention COVID-19 Response Team on Overcoming COVID-19 were Laura D. Zambrano, PhD, MPH, Manish M. Patel, MD, MPH, and Angela P. Campbell, MD, MPH.
Footnotes
Supported by contracts 75D30120C07725 and 75D30121C10297 from the Centers for Disease Control and Prevention. Additional support was from the US National Institute of Allergy and Infectious Diseases (R01AI154470 to A.G.R., 5P01AI118688 for M. Anderson, and R01AI139633-04S1 to R.S.G. and J.C.), National Institute of Diabetes and Digestive and Kidney Diseases (1F30DK123915 to S. Vazquez; R01DK130465 to J.C.), Pediatric Scientist Development Program and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (K12-HD000850 to A. Bodansky), Chan Zuckerberg Biohub for J. DeRisi. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the US Centers for Disease Control and Prevention.
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Contributor Information
Overcoming COVID-19 Network Study Group Investigators,:
Michele Kong, Ronald C. Sanders, Jr., Masson Yates, Chelsea Smith, Natalie Z. Cvijanovich, MattS. Zinter, Gwenn McLaughlin, Keiko M. Tarquinio, Bria M. Coates, Courtney M. Rowan, Adrienne G. Randolph, Margaret M. Newhams, Suden Kucukak, Tanya Novak, Hye Kyung Moon, Takuma Kobayashi, Jeni Melo, Cameron Young, Sabrina R. Chen, Janet Chou, Heidi R. Flori, Mary K. Dahmer, Emily R. Levy, Supriya Behl, Noelle M. Drapeau, Jennifer E. Schuster, Melissa L. Cullimore, Russell J. McCulloh, Shira J. Gertz, Stephanie P. Schwartz, Tracie C. Walker, Ryan A. Nofziger, Mary Allen Staat, Chelsea C. Rohlfs, Julie C. Fitzgerald, Elizabeth H. Mack, Nelson Reed, Natasha B. Halasa, Laura L. Loftis, and Hillary Crandall
Methods
Study design and subjects
Patients were recruited through the prospectively enrolling multicenter Overcoming COVID-19 study in the United States.E1 , E2 A total of 412 patients were enrolled onto 1 of the following independent cohorts: 199 patients hospitalized with MIS-C, 168 patients hospitalized for COVID-19 in either an intensive care or step-down unit (referred to as severe COVID-19 in this study), and 45 outpatients with SARS-CoV-2 infections associated with mild or no symptoms. The demographic and clinical data are summarized in Table I. US Centers for Disease Control and Prevention case definitions were used to define MIS-CE3; those with acute COVID-19 had a positive antigen test or nucleic acid amplification test.E4 All patients with MIS-C had positive SARS-CoV-2 serology results and/or positive SARS-CoV-2 test results by reverse transcriptase quantitative PCR. All patients with severe COVID-19 or outpatient SARS-CoV-2 infections tested positive for SARS-CoV-2. For outpatients, samples were collected from 36 to 190 days after the positive test (median, 70 days after positive test; interquartile range, 56-81 days).
To maintain deidentification of clinical data from the patient with the NFKB2Thr684Pro variant, approximate age, rather than exact age, is provided. Informed consent was provided by participants or legal guardians. All protocols were approved by the institutional review board at Boston Children’s Hospital, which served as the single IRB for the study (IRB-P00033157). Data of APS-1–positive control samples were previously published and collected as described in Ferre et al.E5 All patients with APS-1 were enrolled onto research study protocols approved by the US National Institute of Allergy and Infectious Diseases, National Institutes of Health Clinical Center, and National Cancer Institute institutional review board committees, and all provided written informed consent for study participation. All patients recruited at the National Institutes of Health gave passive consent for use of their medical record for research purposes, thus allowing eligible participants to opt out of study inclusion (protocol 11-I-0187). Healthy, pre–COVID-19 control plasma samples were obtained from the New York Blood Center, where they were collected under informed consent, including use for research.
Anti–IFN-α2 antibody radioligand binding assay
A sequence-verified plasmid encoding the IFN-Α2 cDNA sequence with a Flag-Myc tag (OriGene Technologies, Rockville, Md; catalog RC221091) was used as template in T7-promoter-based in vitro transcription/translation reactions (Promega, Madison, Wis; L1170) with 35S-methionine (PerkinElmer, Waltham, Mass; NEG709A). IFNΑ2 protein was purified via Nap-5 columns (GE Healthcare, Chicago, Ill; 17-0853-01) incubated with 2.5 μL of study participant plasma or 1 μL of anti–myc-positive control antibody (Cell Signaling Technology, Danvers, Mass; 2272), followed by immunoprecipitation with Sephadex protein A/G beads (Sigma-Aldrich, St Louis, Mo; GE17-5280-02 and GE17-0618-05, 4:1 ratio) in 96-well polyvinylidene difluoride filtration plates (Corning, Corning, NY; EK-680860). A Microbeta Trilux liquid scintillation plate reader (PerkinElmer) was used to measure the radioactive counts (cpm) of immunoprecipitated protein samples. The antibody index was calculated as follows: (sample cpm value − mean blank cpm value)/(positive control antibody cpm value − mean blank cpm value).
Whole-exome sequencing
Whole-exome sequencing and candidate variant analysis was performed as previously described.E6
Bulk RNA sequencing
Whole blood was collected in PAXgene tubes (Qiagen, Germantown, Md). Messenger RNA was extracted using the PAXgene blood RNA Kit (Qiagen), followed by globin messenger RNA depletion and polyA capture. Barcoded nondirectional libraries were sequenced using the Illumina NovaSeq platform (Illumina, San Diego, Calif), generating paired-end 150 bp reads. Differential gene expression analysis was performed by Partek Flow software (Partek, Chesterfield, Mo).
Assessment of NFKB2 activation
A total of 1 × 106 PBMCs, corresponding to 120 μg of protein, were stimulated with anti-CD3 (clone OKT3, Thermo Fisher Scientific, Waltham, Mass) for 2 days, then lysed, transferred to nitrocellulose membranes, and immunoblotted with antibodies against p100/p52 (Cell Signaling Technology; 4882) and β-actin (clone 13E5, Cell Signaling Technology), both used at 1:1000 dilution in 5% BSA. Images were acquired by an iBright Imager device (Thermo Fisher Scientific) and quantified by ImageJ2 software (imagej.net/software/imagej2/).E7
Statistical analysis
Pathway analysis was performed by Ingenuity Pathway Analysis (Qiagen) on differentially expressed genes with a fold change of at least 2-fold and a false discovery rate of <0.05.
References
- 1.Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Wijst M.G.P., Vazquez S.E., Hartoularos G.C., Bastard P., Grant T., Bueno R., et al. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci Transl Med. 2021 doi: 10.1126/scitranslmed.abh2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consiglio C.R., Cotugno N., Sardh F., Pou C., Amodio D., Rodriguez L., et al. The immunology of multisystem inflammatory syndrome in children with COVID-19. Cell. 2020 doi: 10.1016/j.cell.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porritt R.A., Binek A., Paschold L., Rivas M.N., McArdle A., Yonker L.M., et al. The autoimmune signature of hyperinflammatory multisystem inflammatory syndrome in children. J Clin Invest. 2021;131 doi: 10.1172/JCI151520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruber C.N., Patel R.S., Trachtman R., Lepow L., Amanat F., Krammer F., et al. Mapping systemic inflammation and antibody responses in multisystem inflammatory syndrome in children (MIS-C) Cell. 2020;183:982–995.e14. doi: 10.1016/j.cell.2020.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Cevins C., Luka M., Smith N., Meynier S., Magérus A., Carbone F., et al. A monocyte/dendritic cell molecular signature of SARS-CoV-2–related multisystem inflammatory syndrome in children with severe myocarditis. Med (N Y) 2021;2:1072–1092.e7. doi: 10.1016/j.medj.2021.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abolhassani H., Delavari S., Landegren N., Shokri S., Bastard P., Du L., et al. Genetic and immunologic evaluation of children with inborn errors of immunity and severe or critical COVID-19. J Allergy Clin Immunol. 2022;150:1059–1073. doi: 10.1016/j.jaci.2022.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burbelo P.D., Castagnoli R., Shimizu C., Delmonte O.M., Dobbs K., Discepolo V., et al. Autoantibodies against proteins previously associated with autoimmunity in adult and pediatric patients with COVID-19 and children with MIS-C. Front Immunol. 2022 doi: 10.3389/fimmu.2022.841126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bastard P., Orlova E., Sozaeva L., Lévy R., James A., Schmitt M.M., et al. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. J Exp Med. 2021 doi: 10.1084/jem.20210554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyer S., Woodward M., Hertel C., Vlaicu P., Haque Y., Kärner J., et al. AIRE-deficient patients harbor unique high-affinity disease-ameliorating autoantibodies. Cell. 2016;166:582–595. doi: 10.1016/j.cell.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang J., Novak T., Hecker J., Grubbs G., Zahra F.T., Bellusci L., et al. Cross-reactive immunity against the SARS-CoV-2 Omicron variant is low in pediatric patients with prior COVID-19 or MIS-C. Nat Commun. 2022;13:2979. doi: 10.1038/s41467-022-30649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mann E.R., Menon M., Knight S.B., Konkel J.E., Jagger C., Shaw T.N., et al. Longitudinal immune profiling reveals key myeloid signatures associated with COVID-19. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abd6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramaswamy A., Brodsky N.N., Sumida T.S., Comi M., Asashima H., Hoehn K.B., et al. Immune dysregulation and autoreactivity correlate with disease severity in SARS-CoV-2–associated multisystem inflammatory syndrome in children. Immunity. 2021;54:1083–1095.e7. doi: 10.1016/j.immuni.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vatsyayan J., Qing G., Xiao G., Hu J. SUMO1 modification of NF-kappaB2/p100 is essential for stimuli-induced p100 phosphorylation and processing. EMBO Rep. 2008;9:885–890. doi: 10.1038/embor.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wirasinha R.C., Davies A.R., Srivastava M., Sheridan J.M., Sng X.Y.X., Delmonte O.M., et al. Nfkb2 variants reveal a p100-degradation threshold that defines autoimmune susceptibility. J Exp Med. 2021;218 doi: 10.1084/jem.20200476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klemann C., Camacho-Ordonez N., Yang L., Eskandarian Z., Rojas-Restrepo J.L., Frede N., et al. Clinical and immunological phenotype of patients with primary immunodeficiency due to damaging mutations in NFKB2. Front Immunol. 2019;10:297. doi: 10.3389/fimmu.2019.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyts I., Bucciol G., Quinti I., Neven B., Fischer A., Seoane E., et al. Coronavirus disease 2019 in patients with inborn errors of immunity: an international study. J Allergy Clin Immunol. 2021;147:520–531. doi: 10.1016/j.jaci.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abraham R.S., Marshall J.M., Kuehn H.S., Rueda C.M., Gibbs A., Guider W., et al. Severe SARS-CoV-2 disease in the context of a NF-κB2 loss-of-function pathogenic variant. J Allergy Clin Immunol. 2021;147:532–544.e1. doi: 10.1016/j.jaci.2020.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vazquez S.E., Bastard P., Kelly K., Gervais A., Norris P.J., Dumont L.J., et al. Neutralizing autoantibodies to type i interferons in COVID-19 convalescent donor plasma. J Clin Immunol. 2021;41:1169–1171. doi: 10.1007/s10875-021-01060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bastard P., Gervais A., Le Voyer T., Rosain J., Philippot Q., Manry J., et al. Autoantibodies neutralizing type I IFNs are present in ∼4% of uninfected individuals over 70 years old and account for ∼20% of COVID-19 deaths. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- Tang J., Novak T., Hecker J., Grubbs G., Zahra F.T., Bellusci L., et al. Cross-reactive immunity against the SARS-CoV-2 Omicron variant is low in pediatric patients with prior COVID-19 or MIS-C. Nat Commun. 2022;13:2979. doi: 10.1038/s41467-022-30649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal G.B., Novak T., Mathew A., Chou J., Zhang Y., Manjula N., et al. Measurement of SARS-CoV-2 antigens in plasma of pediatric patients with acute COVID-19 or MIS-C using an ultrasensitive and quantitative immunoassay. Clin Infect Dis. 2022;75:1351–1358. doi: 10.1093/cid/ciac160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Centers for Disease Control and Prevention (CDC). For parents: multisystem inflammatory syndrome in children (MIS-C) associated with COVID-19. Last reviewed September 20, 2021. Available at: https://www.cdc.gov/mis/mis-c.html.
- US Centers for Disease Control and Prevention (CDC); National Notifiable Diseases Surveillance System (NNDSS). Coronavirus disease 2019 (COVID-19) 2021 case definition. Last reviewed August 24, 2021. Available at: https://ndc.services.cdc.gov/case-definitions/coronavirus-disease-2019-2021/.
- Ferre E.M.N., Rose S.R., Rosenzweig S.D., Burbelo P.D., Romito K.R., Niemela J.E., et al. Redefined clinical features and diagnostic criteria in autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy. JCI Insight. 2016;1 doi: 10.1172/jci.insight.88782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt C.D., Zaman F., Bainter W., Stafstrom K., Almutairi A., Reigle M., et al. Efficacy and economics of targeted panel versus whole exome sequencing in 878 patients with suspected primary immunodeficiency. J Allergy Clin Immunol. 2021;147:723–726. doi: 10.1016/j.jaci.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden C.T., Schindelin J., Hiner M.C., DeZonia B.E., Walter A.E., Arena E.T., et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics. 2017;18:529. doi: 10.1186/s12859-017-1934-z. [DOI] [PMC free article] [PubMed] [Google Scholar]