Abstract
Defective genes account for ∼80% of the total of more than 7,000 diseases known to date. Gene therapy brings the promise of a one-time treatment option that will fix the errors in patient genetic coding. Recombinant viruses are highly efficient vehicles for in vivo gene delivery. Adeno-associated virus (AAV) vectors offer unique advantages, such as tissue tropism, specificity in transduction, eliciting of a relatively low immune responses, no incorporation into the host chromosome, and long-lasting delivered gene expression, making them the most popular viral gene delivery system in clinical trials, with three AAV-based gene therapy drugs already approved by the US Food and Drug Administration (FDA) or European Medicines Agency (EMA). Despite the success of AAV vectors, their usage in particular scenarios is still limited due to remaining challenges, such as poor transduction efficiency in certain tissues, low organ specificity, pre-existing humoral immunity to AAV capsids, and vector dose-dependent toxicity in patients. In the present review, we address the different approaches to improve AAV vectors for gene therapy with a focus on AAV capsid selection and engineering, strategies to overcome anti-AAV immune response, and vector genome design, ending with a glimpse at vector production methods and the current state of recombinant AAV (rAAV) at the clinical level.
Keywords: gene therapy, viral vectors, adeno-associated virus (AAV)
Graphical abstract
Pupo et al. review the different approaches to improve AAV vectors for gene therapy with a focus on AAV capsid selection and engineering, strategies to overcome anti-AAV immune response, and vector genome design, ending with a glimpse at vector production methods and the current state of rAAV at the clinical level.
Introduction
The success of gene therapy relies on an efficacious means to transfer the proper genetic material into the correct tissue. Vectors derived from adeno-associated viruses (AAVs) have become popular delivery systems for therapeutic gene transfer due to their non-pathogenic and broadly tropic nature,1,2,3 their reduced immunogenicity, and the potential to achieve efficient and long-lived gene transfer.4 AAV is a non-enveloped, single-stranded DNA virus, with a genome size of 4.7 kb and a small icosahedral capsid of ∼25 nm, that was discovered in the 1960s as a contaminant in a simian adenovirus preparation.1
The AAV capsid is composed of 60 copies in total of viral protein, VP1–3, in a population ratio of 1:1:10 on average.5,6 However, this is highly dependent on the serotype and the particular preparation.7 AAV capsid assembly is divergent and stochastic, leading to a highly heterogeneous population of capsids of variable composition, whereby even the single-most abundant VP stoichiometry represents only a small percentage of the total AAV population.7
The biology of AAV has been extensively studied.8 A defining characteristic of AAV is its dependence on a helper virus co-infection (such as adenoviruses or herpes-viruses) for productive replication.9
Replication defective AAV viral-like particles (also known as recombinant AAV [rAAV]), in which all viral open reading frames from the viral genome have been eliminated, and containing heterologous genetic information, can be assembled and packaged to high vector yields for gene transfer applications.4
AAV vectors are being explored for numerous therapeutic applications, and they are the most popular viral gene delivery system explored in clinical trials.10,11 The use of AAV vectors in early- and late-stage clinical trials for monogenic diseases such as hemophilia,12 inherited forms of blindness,12 and muscular dystrophy13 has been remarkably successful, regarding their clinical safety and efficacy. Gene transfer therapies using AAV vectors are being fast-tracked for clinical approval for congestive heart failure, hemophilia A and B, retinal disease, X-linked myotubular myopathy, glioma, glioblastoma, and spinal muscular atrophy (SMA), among others.14
Despite the fast pace of applications for AAV vectors, their usage in particular treatments is restricted due to remaining challenges, such as pre-existing humoral immunity to AAV capsids, poor transduction efficiency in certain tissues, low organ specificity, and vector dose-dependent toxicity in patients.3,8
In the present review, we address the different approaches to improve AAV vectors for gene therapy with a focus on AAV capsid, strategies to overcome anti-AAV immune response, and vector genome design, and ending with a glimpse at vector production methods and the current state of rAAV at the clinical level.
AAV capsids
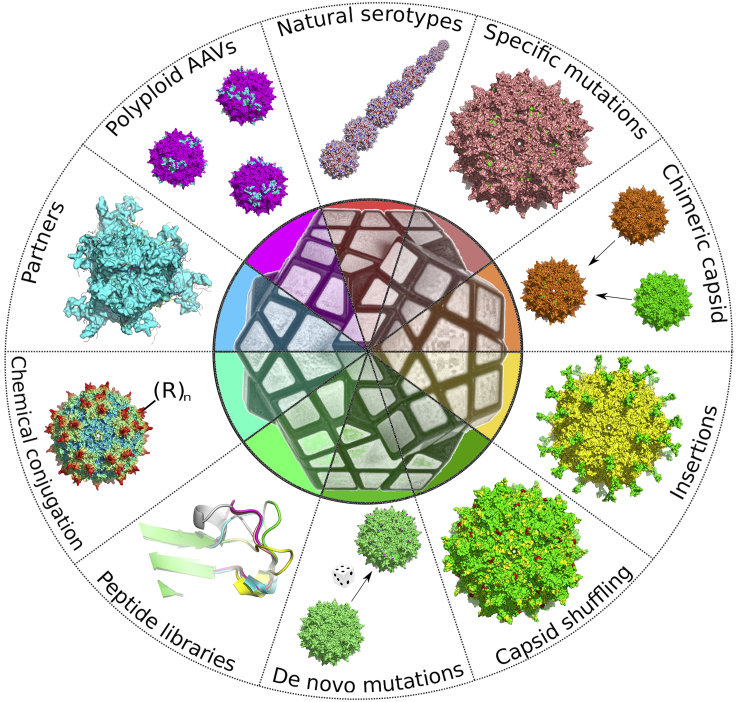
In the last 10 years, with the advances in the field of synthetic biology and our better understanding of AAV structure and biology, several new rAAVs with defined properties have been generated. These new rAAVs have displayed the potential to markedly reduce anti-capsid immune responses and vector dose administration owing to improved gene transfer efficiency and transduction of specific cells and tissues. Figure 1 summarizes the capsid-related approaches that are detailed below.
Figure 1.
Capsid-related approaches to improve AAV-based gene delivery vectors
Natural serotypes
It is frequently said that 13 AAV serotypes have been identified to date (AAV1–13),14,15 but this is just the most studied subgroup from hundreds of serotypes or distinct AAV variants that have been isolated from primates, goats, sea lions, bats, snakes, and other animals4,8,16,17 (Table 1).
Table 1.
Common serotypes/variants for AAV vectors
| Serotype | Origin | Primary receptor | Secondary receptor | Tissue tropism |
|---|---|---|---|---|
| AAV1 | primate | N-linked sialic acid | AAVR GPR108 TM9SF2 |
SMm,c,p,h, CNSm,c,p, lungm,p,h, retinam,r, pancreasm, heartm,i, liverm,p |
| AAV2 | human | heparan sulfate proteoglycan | AAVR GPR108 TM9SF2 LamR αVβ5 integrin α5β1 integrin FGFR1 CD9 HGFR |
SMm,c,h, CNSm,c,p,h, liverm,c,p,h, kidneym, retinam,c,p,h, lungp,h, jointh, ACm |
| AAV3 | human | heparan sulfate proteoglycan | AAVR GPR108 LamR FGFR1 c-MET HGFR |
Liverp,h, SMm, CNSp, cochlear inner hair cellsm |
| AAV4 | NHP | O-linked sialic acid | GPR108 | CNSm,c,p, retinam,c, lungm,p, kidneym, heartm |
| AAV5 | human | N-linked sialic acid | AAVR PDGFR TM9SF2 |
SMm, CNSm,c,p,r, lungm, retinam, liverp,h |
| AAV6 | human | heparan sulfate proteoglycan N-linked sialic acid |
AAVR GPR108 TM9SF2 EGFR |
SMm,c, heartm,s,i,c, lungc,m,p, airwaym,c,p, liverc, CNSp, retinar |
| AAV7 | rhesus macaque | – | GPR108 TM9SF2 |
SMm, retinam, CNSm, liverm |
| AAV8 | rhesus macaque | – | AAVR GPR108 TM9SF2 LamR |
Liverm,c,p,h, SMm,c, CNSm,c,p, retinam,c,p, pancreasm, heartm,c,p, kidneym, ATm |
| AAVrh.8 | rhesus macaque | – | GPR108 | SMh, liverh, CNSm |
| AAV9 | human | terminal N-linked galactose of SIA | AAVR GPR108 TM9SF2 LamR |
Liverm, heartm,i,c,p, SMm,c,p,h, lungm, pancreasm, CNSm,c,p, retinam,p, testesm, kidneym |
| AAV10 | cynomolgus monkeys | – | – | Heartm, lungm, liverm, kidneym, uterusm |
| AAVrh.10 | rhesus macaque | – | GPR108 LamR |
Liverm, heartm, SMm,c, lungm, pancreasm, CNSm,r,c, retinam, kidneym |
| AAV11 | cynomolgus monkeys | – | – | SMm, kidneym, spleenm, lungm, heartm, stomachm |
| AAV12 | NHP | – | – | SMm, salivary glandsm |
| AAVrh32.33 | rhesus macaque | – | GPR108 | – |
Some of these serotypes have been separated into clades A–F on the basis of shared serologic and functional attributes.14 From the common variants for AAV vectors, two serotypes, AAV5 and AAV4, exhibit greater differences in capsid protein sequences. AAV5 is the most phylogenetically distinct, with 58% capsid homology with AAV2 and AAV8 and 57% homology with AAV10.14 In contrast, the other serotypes commonly used in gene transfer share greater homology (∼80%).18 The variable regions in sequence correspond mainly with conformationally distinct loops in the VP structures, which are associated with receptor binding, transduction efficacy, and antigenicity, being relevant in terms of differences in tissue tropism, antigenicity, and the likelihood of cross-reactive immunogenicity between serotypes.34
Tissue tropism reflects the specific interactions between serotype-specific structures on the rAAV capsid and cellular glycans and receptors.14 The initial binding of many AAV serotypes is via primary receptors including glycans and proteoglycans such as heparan sulfate, terminal galactose (Gal), and several linkage variants of sialic acid (SA).14 AAV serotype vectors can be grouped into three categories with respect to their glycan receptor usage: Heparan Sulfate Proteoglycan (HSPG) for AAV2, AAV3, AAV6, and AAV13; sialic acid (SA) for AAV1, AAV4, AAV5, and AAV6; and Gal for AAV9.15 The primary glycan receptors, if any, are unknown for AAV7, AAV8, AAVrh10, AAV11, and AAV1215 (Table 1). We need to point out that this glycan receptor usage grouping is probably a simplification, and that actual profile of glycan recognition is more complex. An example of that is AAV2, as more than 70% of AAV2-like sequences isolated from human samples (having at least a 98% of sequence identity) lack arginine residues at position 585 and 588, being unable to bind HSPG.35 An equivalent diversity could be expected within the other serotypes, regarding their interaction with receptors. We are going to be referring to the AAV laboratory strains, cloned and with the reference genomes, in the rest of this work, unless stated otherwise.
This initial binding is followed by interactions with secondary membrane protein receptors that facilitate internalization.14 A range of secondary receptors has been reported: FGFR1,36 CD9, αVβ5, and α5β1 integrins14,37 for AAV2; hepatocyte growth factor receptor c-MET for AAV238 and AAV339; platelet-derived growth factor receptor for AAV515; EGFR for AAV640; LamR for AAV2, AAV3, AAV8, and AAV9,14 among others14 (Table 1).
AAVR, a glycoprotein with a molecular mass of 150 kDa previously known as KIAA0319L, has been identified as critical for the entry of numerous AAV serotypes, including AAV1, AAV2, AAV3B, AAV5, AAV6, AAV8, and AAV9.41,42 AAV serotypes interact differently with AAVR, while AAV2 interacts predominantly with the second immunoglobulin (Ig)-like polycystic kidney disease repeat domain (PKD2) present in the ectodomain of AAVR, AAV5 interacts primarily through the most membrane-distal PKD1,43 and other AAV serotypes, including AAV1 and AAV8, require a combination of PKD1 and PKD2 for optimal transduction.42 Different non-exclusive functions have been proposed for AAVR in AAV infection: AAVR interacts with AAV either at the surface and aids in AAV cellular uptake into an endosomal pathway, the interaction occurs in the early endosomal system and facilitates trafficking to the trans-Golgi, AAVR interacts with AAV once it reaches the Golgi and facilitates escape from the trans-Golgi network into the cytoplasm, or a combination of the three possibilities.3 Recent data support that AAVR is not a major contributor in cellular attachment and that its role is to facilitate entry at a step prior to nuclear import.19 AAVR usage is conserved among all primate AAVs except for those of the AAV4 lineage (composed of AAV4 and AAVrh32.33) that can bind and transduce cells in the absence of AAVR by an AAVR-alternate route.44
GPR108, a member of the G protein-coupled receptor superfamily, appears to mediate this AAVR-alternate route.19 Transduction was affected in GPR108 knockout for most serotypes (including AAV4 and AAVrh32.33), except for AAV5.19 GPR108 independence can be transferred to other serotypes with the VP1 unique domain of AAV5, and its usage is conserved in mice and humans, being relevant both in vitro and in vivo.19 GPR108 localized primarily to the Golgi, where it may interact with AAV and play a critical role in mediating virus escape or trafficking.20
TM9SF2, a Golgi protein involved in glyco-sphingolipid regulation and endosomal trafficking, also mediates viral transduction in at least seven different AAV serotypes (Table 1).20 TM9SF2 is important for the stabilization and localization of NDST1,45 and it has been shown to be involved in ricin activity, Shiga toxin activity and in vaccinia virus and chikungunya virus infections,46 and AIDS progression.47
The unique combination of specific glycan and protein receptors is believed to be the main factor determining AAV serotype tissue tropisms.8 In mice, AAV serotypes 1–9 show overlapping but distinct tissue expression patterns: liver (AAV 1–3 and 5–9), skeletal muscle (AAV 1–9), heart (AAV4 and 6–9), lung (AAV4 and 6–9), brain (AAV 8–9), and testes (AAV9).48 AAV9, AAVrh.8, and AAVrh.10 are special cases for central nervous system (CNS) delivery, as they are able to bypass the blood-brain barrier and efficiently target cells of the CNS when injected intravenously in different animal models.48,49,50,51,52 Although the diversity of AAVs is huge, several important cell types and tissues are not targeted by natural AAVs.3 Unfortunately, animal models may be poorly predictive of AAV tropism in humans.14,15 Given the difficulties to obtain multiple biopsies in the clinical setting, vector tropism is less characterized in humans, and tissue-specific promoters are frequently incorporated into vectors to limit the expression of the transgene to a particular target tissue.14
AAV vector efficacy depends not only on the ability of the different serotypes to enter the target cells but may also reflect the speed of uncoating and the conversion of the single-stranded vector DNA into duplex DNA that is transcriptionally active.53,54 Several reports summarize and review the transduction efficacy of AAV serotypes in different tissues and specific cells.10,55,56,57,58 A more effective vector could be administered at a lower dose, which could have potential benefits in terms of reduced immunogenicity. The final dose will also depend on other factors, such as the quality of vector manufacturing, transgene activity, and host immunity to the serotype.14
Dealing with post-translation modifications by specific mutations
A total of 52 post-translational modifications (PTMs) were detected in AAV2, AAV3, AAV4, AAV5, AAV6, AAV7, AAV8, AAV9, and AAVrh10 capsids produced in AAV-293 cells, including glycosylation (36%), phosphorylation (21%), ubiquitination (17%), SUMOylation (13%), and acetylation (11%).59 To add complexity to this matter, capsid PTMs are heterogeneous60 and differ when produced in human and baculovirus-Sf9 manufacturing platforms.61 Four AAV serotypes (AAV2, AAV5, AAV9, and AAVrh10) had more than seven PTMs in their capsid proteins.61
While no detectable PTM was found in AAV1 by Mary et al.,61 several were found in other studies,61 including N-terminal acetylation; K61, K459, and K528 methylation; and S149 and S153 phosphorylation. The fact that serine and lysine mutant AAV1 vectors (K137R, S663A, and S669A) demonstrate increased transduction both in vitro and in vivo60 is additional evidence of the presence and functional relevance of PTMs in AAV1.
AAV2 capsid proteins have eight sequence motifs that are potential sites for O- or N-linked glycosylation.62 Although no glycosylation was previously found for virus prepared in cultured HeLa cells,63 AAV2 capsid is indeed glycosylated at residues N253, N518, S537, and N551.63 The disruption of N253 N-glycosylation site (mutant N253Q) severely compromised AAV2 packaging efficiency, probably by compromising the vector assembly and stability.61
AAV2 capsids are phosphorylated at tyrosine residues by EGFR-PTK, which leads to their subsequent ubiquitination and impaired intracellular trafficking to the nucleus.61 Point mutations of specific surface-exposed single tyrosine residues (Y444F and Y730F) lead to high-efficiency transduction at lower doses.64,65 A triple-mutant (Y444F + Y500F + Y730F) vector has ∼30- to 80-fold higher level of in vivo gene transfer to murine hepatocytes66 and ∼130-fold increase in transgene expression in murine fibroblasts67 than the wild-type AAV2. These tyrosine residues are highly conserved in most AAV serotypes, with few exceptions.68
Phosphorylation sites, recognized as degradation signals by ubiquitin ligase (phosphodegrons), were predicted from AAV2 capsid sequences and the corresponding serine and threonine residues were mutated by alanine and lysine residues by arginine.66 The transduction efficiencies of 11 S/T/A and 7 K/R vectors were significantly higher than the AAV2-WT vectors.69 The AAV2 K532R mutant showed significantly reduced ubiquitination compared with AAV2-WT.69
Native levels of ubiquitinated capsids in freshly packaged AAV2 are significantly low.69 These capsids showed the presence of ubiquitination at codon K544, phosphorylation at S149, and SUMOylation by SUMO-2/3 at K258.61 The mutant vector K258Q demonstrated reduced levels of SUMO-1/2/3 proteins and negligible transduction.61
AAV2 N-terminal residue is acetylated, a PTM that is conserved in AAV1, AAV2, AAV3, AAV5, AAV7, AAV9, and AAVrh10 vectors.61
In AAV3, phosphorylation and HexNAcylation was observed in residues Y6 and S149, respectively.61,70
Only one PTM was detected in AAV4, a ubiquitination site at K479. This modification was unique and absent in all other serotypes.61 On the other hand, AAV5 has several PTMs, including glycosylation at N292; ubiquitination at K122, K451, and K639; and HexNAcylation at residues N308, T376, and N428.61 AAV5 mutant vectors neutralizing phosphorylation sites at serine (S268A, S652A, and S658A) and threonine (T107A) improve gene transfer efficiency in vitro and in vivo.61
AAV6 had a single acetylation at residue A2.62 AAV7 stands out for the glycosylation PTMs at S157, N254, N460.61
Both AAV9 and AAVrh10 had a relatively higher proportion of sumoylated residues detected in AAV9 (K84, K316, K557) compared with all other serotypes and an overall similar distribution of common PTMs.61
PTMs were distributed along the entire capsid polypeptide of AAV8.61 AAV8 has phosphorylations at Y6, S149, S153, S279, and S671; acetylation of A2 residue; ubiquitination at K137; O-GlcNAc on T494; and N-glycosylation at N499 and N521 residues.60 The point mutant vectors S279A, S671A, and K137R showed higher and preferential transduction of the liver.60,61,71,72 Spontaneous deamidation occurs on the AAV8 vector capsid at 16 positions, predominantly asparagine residues where the N + 1 residue is glycine.71 There is a correlation between vector activity loss over time and progressive deamidation.60,73 This deamidation phenomenon is not serotype specific, with experimental evidence of its presence in AAV1, AAV3B, AAV4, AAV5, AAV7, AAV9, and AAVrh32.33.73
Baculo-AAV8 capsids have more PTMs than those produced in human cells.60 These include methylation and acetylation on K530, methylation on K709, and O-GlyNAc on T663.60
New methodologies, such as an HILIC-FLR-MS protocol74 and VectorMOD,75 have been developed to ease the process of unambiguous serotype identification, stoichiometry assessment, process impurities detection, and PTM characterization.
Chimeric capsid
A common rational design approach for AAV capsid engineering is to build chimeras by the grafting of functional motives from a serotype into another. These motives are usually related with receptor binding regions that dictate tropism. Sometimes the substitution of only one amino acid is enough to achieve a significantly different phenotype. The mutation of K531 in AAV6 by the corresponding amino acid in AAV1 (E) suppresses the heparin-binding ability of AAV6. It also reduces transgene expression to levels similar to those achieved with AAV1 in HepG2 cells in vitro and in mouse. The mutant AAV1 E531K has heparin-binding ability and increased transduction efficiency.76 Intravitreally delivered heparan sulfate binding mutants AAV1-E531K and AAV8-E533K accumulate at the inner limiting membrane separating the vitreous and the retina in the eye, similar to AAV2, while wild types AAV1 and AAV8 do not.76 The single insertion of Thr residue from AAV1 into AAV2 capsid at position 265 enhanced muscle transduction and reduced neutralizing antibody (NAb) titers in mouse.77
In most cases, multiple residues or entire regions are grafted from one serotype into another to achieve the desired phenotype. Both AAV1 and AAV6 are more efficient transducing muscle than AAV2. Replacement of residues 350–736 of AAV2 VP1 with the corresponding amino acids from VP1 of AAV1 resulted in a chimeric vector that behaved very similarly to AAV1 in vitro and in vivo.78 This region can be reduced to amino acids 350–430 to obtain a chimera with similar behavior.79 Following a more comprehensive approach, a panel of synthetic AAV2 vectors was generated by replacing the hexapeptide sequence involved in heparan sulfate binding with corresponding residues from other AAV strains.80 The tropism of the generated capsid was evaluated after intravenous administration in mice. AAV2i8, an AAV2/AAV8 chimera, selectively transduced cardiac and skeletal muscle tissues with high efficiency, traversed the blood vasculature, showed markedly reduced hepatic tropism, and displayed an altered antigenic profile.81 Only four mutations (Q263A, N705A, V708A, and T716N, AAV2 numbering) and one insertion (T265, AAV1 numbering) were required in AAV2 to obtain AAV2.5, which combines the improved muscle transduction capacity of AAV1 with reduced antigenic cross-reactivity against both parental serotypes, while keeping the AAV2 receptor binding.81
Engraftment of AAV9 galactose binding footprint into AAV2 resulted in the chimera AAV2G9, which exploits both Gal and heparan sulfate receptors for infection.82 AAV2G9 remains hepatotropic, like AAV2, but mediates rapid onset and higher transgene expression in mice.82 AAV2G9 displays preferential, robust, and widespread neuronal transduction within the brain and decreased glial tropism.82 Engraftment of the same Gal footprint onto the previously mentioned AAV2i8 chimera yielded an enhanced chimera named AAV2i8G9. This chimera remains liver-detargeted like AAV2i8 and selectively transduces muscle tissues with high efficiency.83
AAV2-AAV8 chimeras, combined with site-directed mutations, were used to map the epitopes related with T cell responses to AAV capsids after intramuscular injection of vectors into mice and non-human primates (NHPs).82 The identified epitope overlaps with the heparan sulfate proteoglycan binding site, which justified the observed correlation between heparin binding, uptake into human dendritic cells, and activation of capsid-specific T cells.84
A series of chimeras termed AAV X-Vivo (AAV-XV) were obtained combining AAV12 VP1/2 sequences with the VP3 sequence of AAV6.85 These AAV variants showed enhanced infection, compared with the wild-type parental viruses, of neuronal cell lines, hematopoietic stem cells, and human primary T cells.85
Although the construction of chimeric AAV capsids is essentially a trial-and-error process, the abundance of structural information on different serotypes and mutants supports a more robust design from the beginning. In order to provide quantitative design principles for guiding site-directed recombination of AAV capsids, capsid structural perturbations predicted by the SCHEMA algorithm was correlated with experimental measurements of disruption in 17 chimeric capsid proteins.86 Protection of viral genomes and cellular transduction were inversely related to calculated disruption of the capsid structure in a chimera population created by recombining AAV2 and AAV4.86 This algorithm and equivalent analysis may be useful for delineating quantitative design principles to guide the creation of new AAV chimeras.
Peptide and protein insertions
Instead of grafting motives from one serotype into another, chimeric AAV capsids can be created by the insertion of peptides and whole proteins in specific VP regions. The main goal of these insertions has been to alter natural AAV serotype tropism. There are two choices that conform the essence of this methodology: what to insert and where.
In one of the first related reports, in order to target CD34+ hematopoietic stem cells, a chimeric AAV2 capsid was constructed where VP2 was replaced by an anti-CD34 scAb-VP2 fusion protein.87 The chimeric had a significantly increased preferential infectivity for the normally refractory CD34+ human myoleukemia cell line KG-1.87
In order to define critical components of virion assembly and infectivity in AAV2, an exhaustive linker insertional mutagenesis was performed in AAV2.88 Some insertional mutants were identified with the capability to assemble the capsid, encapsulate DNA, and infect target cells.88 A 14-amino-acid targeting peptide, including the RGD motif, was inserted into six AAV2 VP loops and the resulting mutants were evaluated by their ability to package the viral genome, A20 antibody recognition, presence of the peptide on the capsid surface, and B16F10 and RN22 target cell binding.89 Mutants with insertion in positions 447 and 587 passed all the filters.89 The mutant with the insertion in 587 efficiently transduced cells despite the presence of neutralizing antisera.89 Peptide insertions in the same loops but different positions, 449 and 588, were successful as well.90 After testing seven positions to insert a 4C-RGD peptide, only insertions following amino acids 584 or 588 were able to expand AAV2 tropism.91 In this last study, no insertion in 447–449 was tried, but in 459 instead, which failed.91 A mutant with this peptide inserted in position 453 also fails to bind its target receptor, but a structural analysis revealed a possible interaction with residues R585 and R588.92 The AAV2 capsid mutant carrying RGD-4C in position 453 and with R585A/R588A substitutions was superior to the one with the insertion in position 587 in target receptor binding and cell transduction efficiency.93 When the muscle targeting peptide MTP is inserted in positions 587 or 588 of AAV2 capsid, both viruses diminished their infectivity on non-muscle cell lines and on un-differentiated myoblast, but preserved or enhanced their infectivity on differentiated myotubes.93 MTP inserted in 587, but not in 588, abolished AAV2 heparin-binding capacity, and infected myotubes in a heparin-independent manner.94 Peptides containing a net negative charge, such as MTP, are prone to confer an HSPG non-binding phenotype when inserted in 587.95
In a comprehensive analysis of AAV2 capsid, 93 mutants at 59 different positions were constructed, including peptide insertions and alanine scanning.96 Insertion of the serpin receptor ligand in N-terminal regions of VP1 (residue 34) or VP2 (residue 138) changed the tropism of AAV2 in vitro.96 With the design of a new system of production in which expression of VP2 protein is isolated and combined with VP1 and VP3 expressed separately, proteins up to 30 kDa were successfully inserted after residue 138, within the VP1-VP2 overlap.97 Different types of proteins have been inserted in the N terminus of VP2 since then. For example, an anti-Her2/neu DARPin was inserted in the N terminus of AAV2 VP2 in order to precisely target tumor tissue and deliver a suicide gene.98 DARPins are high-affinity binding proteins of 14–17 kDa that have been developed as an alternative to antibodies. The DARPin-AAV vector substantially reduced Her2/neu+ tumor growth without causing fatal liver toxicity.98 Anthopleurin-B, a toxin specific for cardiac sodium channel Nav1.5, was inserted in VP2 after residue T138.99 The mutant with the insertion plus R585A/R588A mutations transfers genes to cardiac myocyte without hepatic and other extracardiac infectivity.99
Insertions of peptides and proteins are not exclusive to serotype 2. The tolerance of AAV5 capsid to small deletions and tandem duplication of sequence was globally addressed with the generation and characterization of a library of more than 105 mutants, revealing potential insertion sites.100 An AAV6 mutant with an RGD peptide insertion at position 585, plus Y705-731F + T492V-K531E substitutions, showed a significant increase in both specificity and efficiency in a xenograft animal model in vivo.101 A murine breast cancer tissue-specific peptide was inserted at positions 590 and 589 of AAV8 and AAV9, respectively.102 This peptide was previously selected in an AAV2-based peptide library. The insertion of this peptide augmented tumor gene delivery in both serotypes.102 Nevertheless, the insertion of a peptide derived from the same AAV2 library, with specificity for murine lung tissue, did not alter tropism of either serotype.102 This is evidence of how the context may affect the function of the inserted sequences, particularly short peptide ones, where the required functional conformations could be affected by the anchor and interacting residues in the template AAV.
In a recent approach, the seven amino acids from the GH2/GH3 loop (residues 453–459 in AAV2 VP1) were replaced by three different nanobodies, having 110–130 amino acids, with specificities for CD38, ARTC2.2, and P2X7, respectively.103 Nanobodies are single Ig variable domains from heavy-chain antibodies that naturally occur in camelids. This strategy dramatically enhanced the transduction of specific target cells by recombinant AAV2 and it was also successfully tested in AAV1, AAV8, and AAV9 serotypes.103
Enhanced AAV9 (eAAV9), a modified AAV9 with an insulin-mimetic peptide (S519) inserted into the capsid, has a significantly higher transduction efficiency of insulin receptor (IR) expressing cell lines, differentiated primary human muscle cells, and up to 18-fold enhancement over AAV9 transduction of mouse muscle in vivo.104 A new AAV variant, AAV9-Retro, was developed by inserting the 10-mer peptide into the VR-VIII loop of AAV9 VP3 domain.105 AAV9-Retro can retrogradely infect projection neurons, and can be transported across the nervous system, like AAV9, by intracranial or intravenous injections in mice.105 The insertion of bone-targeting peptide motifs (Asp)14 or (AspSerSer)6 onto the AAV9-VP2 capsid protein results in a significant reduction of transgene expression in non-bone peripheral organs in an osteoporosis model.106
In order to evaluate the capacities of different serotypes to display peptides on their capsid surfaces, 27 peptides were systematically inserted in 13 different AAV capsid variants, followed by production of reporter-encoding vectors.107 Vector performance depended not only on the combination of capsid, peptide, and cell type but also on the position of the inserted peptide and the nature of flanking residues.107 The results are available as an online database, and tools like this should accelerate the in vivo screening of new peptide insertions and the application of the resulting products as unique gene therapy vectors.
Capsid shuffling
DNA shuffling of cap proteins from different serotypes can be achieved by random recombination of homologous genes in vitro.108 These result in large chimeric AAV capsids that can be selected for a given phenotype in a classical directed evolution approach.
An adapted DNA family shuffling technology was used to create a complex library of hybrid capsids from eight different wild-type viruses.109 Selection on primary or transformed human hepatocytes and pooled human antisera led to a single type 2/8/9 chimera, named AAV-DJ, with 60 capsid mutations compared with AAV2.109 Most of these mutated residues are located in the loops extruding from the VP, some critical for natural receptor binding or for antibody recognition or escape. AAV-DJ vectors had more infectivity in vitro than eight standard AAV serotypes (AAV1–6, 8, and 9) and surpassed AAV2 in livers of naive and intravenous immunoglobulin (IVIG) immunized mice.109 AAV-DJ vector has been shown to be a useful tool for gene therapy research targeting retinal disorders, being able to transfer genes to the photoreceptor layer with intravitreal injection and having an efficient gene transfer to various cell types in the retina.110 AAV-DJ8, an AAV-DJ variant modified to specific residues of AAV8, displayed more tropism in astrocytes compared with AAV9 in the substantia nigra region than AAV9 and other vectors.17
Chimeric-1829 is an infectious clone selected from a chimeric capsid library containing randomly fragmented and reassembled capsid genomes from serotypes 1–9.111 As its name indicates, this particular clone contains genome fragments from AAV1, 2, 8, and 9, with AAV2 contributing to surface loops at the 3-fold axis of symmetry, while AAV1 and 9 contribute to 2- and 5-fold symmetry interactions, respectively. Similar to AAV2, Chimeric-1829 utilizes heparan sulfate as a primary receptor, but it shows an altered tropism in skeletal muscle, liver, and brain; transduces melanoma cells more efficiently; and has a unique immunological profile.111
A similarly obtained library was selected against a kainic acid-induced limbic seizure rat model in order to produce a novel AAV vector capable of crossing the seizure-compromised blood-brain barrier and transducing cells in the brain.112 After intravenous administration, two clones transduced the cells in areas exhibiting a seizure-compromised blood-brain barrier that were localized to the piriform cortex and ventral hippocampus.112
Rec1–4 were generated by shuffling the fragments of capsid sequences in three NHP AAV serotypes cy5, rh20, and rh39 and AAV8.113 These vectors were originally selected for retinal tissue transduction, where they are efficient, but no more so than AAV2 and AAV5.113 They have shown potential in other localizations. Rec2 vector showed high transduction efficiency in both brown and white adipose tissue, superior to AAV1, AAV8, and AAV9.114 Compared with AAV9, Rec3 improved gene delivery to mouse spinal cord, transducing a broader region of it, and displaying higher transgene expression and increased maximal transduction rates of astrocytes and neurons.115
Another vector obtained by capsid shuffling and directed evolution, named Olig001, contains a chimeric mixture of AAV1, 2, 6, 8, and 9.116 Olig001 shows a preferential oligodendrocyte tropism following intracranial injection into the striatum, as opposed to the vast majority of AAV vectors, which have shown a dominant neural tropism in the CNS. This vector has very low affinity for peripheral organs, especially the liver, after intravenous administration.116
AAV LP2-10, composed of capsids from AAV serotypes 2, 6, 8, and 9, with VP3 subunits derived from AAV8 swapped with AAV6 from residues 261 to 272, manifested a higher ability to escape NAbs in IVIG and human serum samples.117 LP2-10 transduced human hepatocytes with efficiency similar to that of AAV8.117
AAV2.5T, a capsid with lung tropism, was evolved from an AAV2/AAV5 capsid-shuffled library.118 VP2 and VP3 capsid proteins of AAV2.5T are derived from AAV5 with a single A581T mutation in VP1. VP1 of AAV2.5T is a hybrid of AAV2 and AAV5 capsids with the N-terminal unique sequence (VP1u) from the 1–128 amino acids of the AAV2 VP1, followed by the 129–725 amino acids of AAV5 capsid harboring the A581T mutation.118
De novo mutations
An alternate way to create an AAV capsid library from which to select a desired phenotype is by the generation of random mutations in capsid DNA. This has been accomplished by modified PCR protocols, including staggered extension process and error-prone PCR.119 The size of AAV libraries obtained by this approach is in the range of 106–107 mutants and they have been based in AAV2 with the main goal to escape NAbs while keeping the other functional properties of AAV2 untouched120 or modifying heparan sulfate affinity.95,121,122
To improve the liver transduction of AAV5 while retaining its low seroprevalence, a library of AAV5 mutants was constructed via random mutagenesis and screened in Huh7 cells.123 Two new AAV5 variants, MV50 and MV53, demonstrated significantly increased transduction efficiency in Huh7 cells (∼12×) and primary human hepatocytes (∼10×).123
AAV-S was isolated from a random capsid library, and it mediates highly efficient reporter gene expression in a variety of cochlear cells, including inner and outer hair cells, fibrocytes, and supporting cells in both mice and cynomolgus macaques.124
An alternative approach to random mutations is the comprehensive mutagenesis approach, where all single-codon substitution, insertion, and deletion mutants for AAV2 capsid were generated and evaluated in vitro and in vivo.125 The phenotypic characterization of this library, regarding capsid ensemble and thermal stability, tissue tropism, and delivery, allows the implementation of a machine learning (ML) guided design of new AAV capsids that outperform the one resulting from random mutagenesis.125
The combination of DNA/RNA barcoding with multiplexed next-generation sequencing enables the direct side-by-side comparison of pre-selected AAV capsids in high throughput and in the same animal.126 As a validation of the methodology, three independent libraries were created, from which a peptide-displaying AAV9 mutant called AAVMYO was discovered.126 AAVMYO exhibits superior efficiency and specificity in the musculature, including skeletal muscle, heart, and diaphragm, following peripheral delivery.126
ML models trained directly on experimental data without biophysical modeling provide one route to accessing the full potential diversity of engineered proteins. The application of deep learning allowed designing highly diverse adeno-associated virus 2 (AAV2) capsid protein variants that remain viable for packaging of a DNA payload.127
The vast structural information on AAV capsids allows the rational design of new variants. The structures of several AAV8/antibody complexes were solved by cryoelectron microscopy, and then each VP surface loop involved in the interactions was evolved by infectious cycling in the presence of a helper adenovirus.128 This process yielded a humanized AAV8 capsid (AAVhum.8) displaying non-natural surface loops that simultaneously display tropism for human hepatocytes, increased gene transfer efficiency, and NAb evasion.128
Using alanine scanning mutagenesis, S155 and the flanking residues, D154 and G158, were found to be essential for AAV2 transduction efficiency.129 Specific capsid mutants show a 5- to 9-fold decrease in viral mRNA transcripts, highlighting a potential role of this N-terminal S/T motif in transcription of the viral genome.129
Peptide libraries
The most frequent approach in direct evolution of AAV capsids consists of the insertion of random peptides (usually seven amino acids long) in a VP loop (AAV2’s VP1 587, 588, and their equivalent in other serotypes are preferred) to generate a random capsid library. This library is then used to perform selection for a desired phenotype, usually related to a given tropism or to escape NAbs. In this approach, peptides are selected in the right capsid context and, in principle, should be superior to the insertion of peptides previously selected elsewhere. In one of the first implementations of this methodology, a random AAV2 peptide library was screened on human coronary artery endothelial cells, peptide motives were selected, and their insertion enhanced transduction in coronary endothelial cells but not in control non-endothelial cells.130 New capsids targeting primary human CD34+ peripheral blood progenitor cells were selected with the methodology.131 The chimera AAV-DJ has been used to develop new capsid peptide display random libraries from which new capsids targeting mice lung alveolar cells have been selected.109 In another successful example, selected AAV2-peptide capsids target autologous human keratinocytes, transducing these cells with high efficiency and selectivity.132
Despite early successful examples and the undeniable potential of this methodology, from early on its limitations became apparent, some specific and others that extend to all directed evolution approaches (including the previously discussed capsid shuffling and de novo mutations libraries). Among the limitations are the following:
-
•
Libraries contaminated with WT.
-
•
In vitro selection results do not always translate to in vivo applications.
-
•
Collateral tropism of selected variants.
-
•
Cross-packaging and mosaicism.
-
•
The enhanced features of the selected capsids may not extend beyond the context in which the selective pressure was applied.
An improved library production system using a synthetic cap gene reliably avoids generation of wild-type AAV2.133 With this procedure, the efficiency and specificity of gene transfer to target cells was improved.133
Vectors selected for optimized transduction of primary tumor cells in vitro were not suitable for transduction of the same target cells under in vivo conditions.134 After several rounds of selections performed using tumor and lung tissue as prototype targets in living animals injected intravenously, selected clones conferred gene expression in the target tissue, while gene expression was undetectable in animals injected with control vectors.134 Nevertheless, the selected vectors also transduced a number of other tissues, particularly the heart.134 MicroRNA-regulated transgene cassettes as the combination of library-derived capsid targeting and microRNA control was proposed as an alternative to address this collateral tropism issue.135 In vivo screening of random peptide libraries by next-generation sequencing can achieve improvement in target tissue specificity.136 Using this methodology, a capsid variant was selected that specifically and very efficiently delivers genes to the endothelium of the pulmonary vasculature after intravenous administration.136
The correlation between the selected capsids and the encapsidated viral genomes is crucial to be able to obtain the correct information about the selected variants.137 AAV libraries have the potential to be affected by cross-packaging and mosaicism, in which particles are composed of genomes and capsid monomers derived from different library members.138 AAV2 libraries produced by a two-step protocol with pseudotyped library transfer shuttles, which should ensure genome-capsid correlation, display a bias in the amino acid composition. This bias confers increased heparin affinity and, thus, similarity to wild-type AAV2 tropism, which may impair the intended use of these libraries.137 A step-by-step protocol on how to obtain and screen random AAV display peptide libraries in vivo, taking into account most of these topics, is available.139 Dilution of input library DNA significantly increases capsid monomer homogeneity and increases capsid-genome correlation, reducing cross-packaging and capsid mosaic formation.138
Cre-recombination-based AAV targeted evolution (CREATE) methodology140 is one of the most interesting cases of AAV random displayed libraries, showing both their great potential and highlighting their probable main limitation. This methodology first provided the AAV-PHP.B variant, which efficiently and widely transduces the adult mouse CNS after intravenous injection.140 Two modified versions were obtained later, AAV-PHP.eB and AAV-PHP.S, that efficiently transduce the central and peripheral nervous systems, respectively.141 In adult mice, intravenous administration of a lower dose of AAV-PHP.eB transduced 69% of cortical and 55% of striatal neurons, while administration of AAV-PHP.S transduced 82% of dorsal root ganglion neurons as well as cardiac and enteric neurons.141 Being able to bypass the blood-brain barrier and efficiently target cells of the CNS after systemic injection, AAV-PHP.B and its derivatives were strong candidates for gene delivery therapies in humans. Then, it was reported that the neurotropic properties of AAV-PHP.B were not extended to NHPs or the other commonly used mouse strain, BALB/cJ.142 The same happened with AAV-PHP.B variants, such as AAV-PHP.eB, AAV-PHP.B2, and AAV-PHP.B3.143 To make matters worse, AAV-PHP.B caused an acute toxicity event, with consumption thrombocytopenia and secondary hemorrhages, leading to the euthanasia of an NHP 5 days after injection.142 Intravenous administration of PHP.B capsid also failed to upregulate transduction efficiency in the marmoset brain.144 Nevertheless, AAV-PHP.B does work in other strains of mice (C57BL/6N, SJL/J, FVB/N, and DBA/2)145 and PHP.eB works in rats.146
One of the advantages of direct evolution approaches is that there is no need to know the molecular determinants of the successful phenotypes, including the receptors and corresponding binding sites giving a new tropism, or the epitopes recognized for the avoided antibodies. The problem is that such receptors or epitopes do not necessarily have to be conserved among different species, or even strains of a given species. It was recently described that a GPI-linked protein, LY6A, is the cellular receptor for AAV-PHP.B capsids, driving their transport across the blood-brain barrier.143,147 AAV-PHP.B capsids binding and posterior transduction mediated by this receptor can occur independently of other known AAV receptors.143 The absence of LY6A in some mouse strains and primates explained the failure to reproduce the reported neurotropic effects previously observed in C57BL/6J mice.143 AAV-PHP.B and AAV-PHP.eB are now valuable and widely used vectors for mouse neuroscience studies, but they cannot be used in their current form as vectors to be delivered systemically to target CNS in human gene therapy.
These results suggest that the libraries should be selected in their intended final target. As we have seen that there is not a good translation between libraries selected in vitro with the in vivo results, the challenge to select good vectors from AAV libraries for gene delivery in humans is high.
Alternative approaches have been developed. A new RNA-driven screen platform, termed Tropism Redirection of AAV by Cell-type-specific Expression of RNA (TRACER) was developed as a directed evolution approach based on recovery of capsid library RNA transcribed from CNS-restricted promoters.148 The platform was tested in mice with AAV9 peptide display libraries, and 10 individual variants were characterized and showed up to 400-fold higher brain transduction over AAV9 following systemic administration in mice.148 It remains to be seen whether this or equivalent methodologies will allow the selection of better vectors for human use.
Chemical conjugation and unnatural rAAV vectors
Chemical conjugation is an alternative approach to disrupt receptor binding motifs, or introduce new ones, and to mask epitopes and escape NAbs. PEGylation moderately protects AAV2 against NAbs with ∼2-fold less neutralization over unmodified vector.149 The PEGylated AAV2 conserves the infectivity up to a given PEG:lysine conjugation ratio, which depends on the polymer chain size.149 PEGylation of AAV vectors with succinimidyl succinate (SSPEG) and tresyl chloride (TMPEG) has been reported as a more effective approach, protecting the vector from NAbs without compromising transduction efficiency to the liver and muscle, and improving gene expression in the lung.149
Masking of arginine residues on AAV capsids can be achieved by an exogenous glycation reaction, in which surface-exposed guanidinium side chains are modified into charge neutral hydroimidazolones.150 This reaction disrupted the cluster of basic amino acid residues implicated in heparan sulfate binding in AAV2.150 Glycated AAV2 is then unable to bind heparan sulfate but retained the ability to infect neurons in the mouse brain and was redirected from liver to skeletal and cardiac muscle following systemic administration in mice, while also showing a 2-fold decrease in binding to the A20 monoclonal antibody.150
Due to the abundance of lysine and arginine residues in AAV capsids, the previous reactions intrinsically lack site specificity, making it difficult to avoid the unwanted modification of functionally critical residues. Two approaches have emerged to address the site-specific chemical modification of AAV capsids. The first strategy involves the genetic insertion of a peptide with a consensus sequence into the cap gene. The insertion of a 15-amino-acid biotin acceptor peptide allows the site-specific ligation of a ketone analog of biotin with the enzyme biotin ligase.151 This ketone group can be specifically conjugated to a hydrazide- or hydroxylamine-functionalized molecules, such as fluorophores, and a synthetic cyclic RGD peptide, targeting the virus to the αvβ3 integrin receptors.151 Another 13-amino-acid peptide, with a consensus sequence that is recognized by a cellular formylglycine-generating enzyme, was inserted into the cap gene at amino acid position 587.152 This enzyme converts the cysteine residue of the peptide to aldehyde-bearing formylglycine (FGly) residue. This aldehyde group allows the attachment of functionalized elements, such as peptides, antibodies, and fluorophores, without significant loss of viral function and infectivity.152 The conjugation of an anti-human leukocyte antigen (HLA) mouse monoclonal antibody increased transduction in 293T and HepG2 cells, while the conjugation of an anti-CD20 MAb enhanced transduction in CD20-expressing 293T (293T.CD20).152 Conjugation of a chemically modified cyclic RGD peptide displayed enhanced transduction in HeLa cells.152
The second strategy involves the incorporation of unnatural amino acids (UAAs) into specific sites on the virus capsid.153 This technology makes use of an engineered tRNA/aminoacyl-tRNA synthetase pair to deliver the UAA of interest in response to a repurposed nonsense codon, and over 150 UAAs have been genetically encoded.154,155 The unnatural amino acid AzK was chosen (by its azido functionality, which enables a highly specific bioconjugation reaction) to be introduced in distinct surface-exposed sites of AAV2.153 AzK residue was well tolerated in multiple regions of the capsid, which significantly expands the selection of sites that can be evaluated for attaching new elements.153 When a cyclic RGD peptide was attached to AzK in T454 and R588 sites, the R588AzK virus, which is non-infective due to the lack of R588 residue, regained significant infectivity toward SK-OV-3 cells, but not HEK293T cells.153
With the goal to protect AAV2 from pre-existing NAbs and increase its serum stability, a site-specific PEGylation was achieved using a lysine mimic called Nε-2-azideoethyloxycarbonyl-L-lysine (NAEK), an azide moiety.156 AAVs conjugated with 20-kDa PEG at sites Q325 + 1, S452 + 1, and R585 + 1 showed improved stability in pooled human serum, a nearly 2-fold reduction in antibody recognition, and a 20% reduction in antibody inducement in Sprague-Dawley rats.156
Building on the UAAs insertion approach, oligonucleotides were coupled to AAV2 and AAV-DJ capsids.157 Oligonucleotide-pseudotyped AAVs were then incubated with lipofectamine, which interacts with the negative charges of the oligonucleotides and forms a complex with them, creating a cloak around the capsids. The cloaked AAVs are resistant to serum-based NAbs and retained full functionality, keeping their ability to transduce a range of cell types and also enabling a robust delivery of CRISPR-Cas9 effectors.157 Tethering of oligonucleotides will enable the coating of AAV capsids with proteins, nucleotides, and small molecules through the use of specific DNA aptamers.157
These results demonstrate the feasibility of chemical alteration of rAAV vector tropism and/or antigenicity, becoming a practical orthogonal strategy to engineer AAV vectors for gene therapy applications.
Partners
AAV capsids can be associated non-covalently with partner biomolecules in order to alter their tropism or protect them from NAbs. The first successful example involved a bispecific F(ab’)2 antibody with one arm that recognizes cell-surface αIIβ3 (receptor expressed on human megakaryocytes), and the other, the AAV2 capsid.158 Targeted AAV vectors were able to selectively transduce megakaryocyte cell lines, which are not permissible to AAV2, while the endogenous tropism of AAV2 was significantly reduced.158 The binding of the bispecific antibody did not affect other steps required to successfully transduce target cells, such as escape from endosomes, migration to nucleus, or uncoating.158 Despite the potential of this particular approach, the use of bispecific antibodies to alter AAVs tropism has been practically abandoned since then.
Peptide affinity reagents have been used as alternative partners to antibodies in AAV capsid targeting. Pre-incubation of AAV2 vector with vascular endothelial cell membrane-specific peptides markedly increased AAV2 transduction of human umbilical vein vascular endothelial cells, without affecting AAV2 expression in other cell types.159 Through panning of a commercially available phage display peptide library, a heptapeptide specific for AAV8 serotype was obtained.160 This peptide blocked AAV8 vector gene transduction in vitro and in vivo and could be used as a reagent for affinity column chromatography of recombinant AAV8 vectors.160 This peptide could potentially be used as an anchor to conjugate other molecules to modify the properties of the AAV8 capsid.
Tat-Y1068, a small, cell permeable peptide consisting of epidermal growth factor receptor tyrosine kinase inhibitor peptide and the HIV-Tat sequence, was designed to improve the transduction efficiency of AAV2.161 Pre- or co-treatment of CYNOM-K1 cells (from cynomolgus monkey embryo skin) and rat fibroblast cell line RAT-1 with Tat-Y1068 increased the transduction efficiencies of AAV2 in a dose-dependent manner.161
Cell-penetrating peptides can significantly enhance the in vitro transduction efficiency of AAV9, the best of which was the LAH4 peptide.162 The enhancement of AAV9 transduction by LAH4 relied on binding of the AAV9 capsid to the peptide. LAH4 peptide increased the AAV9 transduction in the CNS in vitro and in vivo after systemic administration.162
Several potential blood-brain barrier shuttle peptides, that significantly enhance AAV8 transduction in the brain after a systemic administration, have been identified.163 The best results were obtained with a peptide named THR. The enhancement of AAV8 brain transduction by THR is dose dependent, with neurons as primary targets. THR directly binds to the AAV8 virion, increasing its ability to cross the endothelial cell barrier, without interfering with AAV8 infection biology.163
During production, a fraction of AAV vectors are associated with microvesicles/exosomes, termed vexosomes (vector exosomes) or, alternatively, exosome-associated AAV (exo-AAV).164 In exo-AAV, capsids are associated with the surface and in the interior of microvesicles.164 Purified exo-AAV outperformed conventionally purified AAV vectors in transduction efficiency and were more resistant to a neutralizing anti-AAV antibody.164 Exo-AAV bound to magnetic beads can be attracted to a magnetized area in cultured cells,164 which could be of use for their specific targeting.
Exo-AAV9 was up to 136-fold more resistant over a range of NAb concentrations relative to standard AAV2 vector in vitro.165 While passively transferred human antibodies decreased intravenously administered AAV9 transduction of brain by 80% in mice, transduction of exo-AAV9 was 4,000-fold higher.165 Simple pelleting of exo-AAV9 from media via ultracentrifugation results in high-titer vector preparations, capable of a more efficient transduction of CNS cells than AAV9 after systemic injection in mice.166
A significant enhancement of transduction efficiency in liver was observed in a hemophilia B mouse model treated with exo-AAV8 expressing human coagulation factor IX, compared with AAV8.167 Exo-AAV8 gene delivery allowed for efficient liver transduction even in the presence of moderate NAb titers.167
Gene delivery to sensory hair cells of the inner ear is inefficient with available vectors.168 Exo-AAV1 is more efficient than conventional AAV1, both in mouse cochlear explants in vitro and with direct cochlear injection in vivo.168 Exo-AAV1 gene therapy partially rescues hearing in a mouse model of hereditary deafness and shows no toxicity in vivo.168
Exo-AAV2 vector mediates robust gene delivery into the murine retina upon intravitreal injection, efficiently reaching the inner nuclear and outer plexiform, and, to a lesser extent, the outer nuclear layer.169 The simplicity of its production and isolation should make it widely applicable to basic research of the eye.169
An intratumoral administration of AAV6 vexosomes carrying a suicide gene in a murine xenograft model revealed a 2.3-fold increase in hepatocellular carcinoma regression compared with untreated animals.170
Given the positive results obtained so far with exo-AAVs and its applicability to any existing AAV vector, clinical applications based on exo-AAVs are expected. Before that, an exhaustive characterization of other biomolecules contained in such exosomes, besides the capsids, will be required. Other issues, such as the homogeneity and reproducibility of the exo-AAVs, will have to be addressed.
In an innovative approach, a hybrid viral vector composed of a bacteriophage T4 and more than one “piggy-backed” AAV was recently developed.171 The AAV capsid acts as a driver by virtue of its natural ability to enter human cells. The delivery payload capacity of the whole hybrid vector is up to 170 kb.171 T4-AAV particles packaged with influenza virus hemagglutinin DNA, and displayed with plague F1mutV antigen, elicited durable immune responses against both flu and plague pathogens, providing complete protection to mice against pneumonic plague.171 This hybrid vector holds a great potential for applications when delivery of large genes is required.
As larger insertions often have unpredictable deleterious impacts on capsid formation and gene delivery, Velcro-AAV, a coiled-coil-based platform for non-covalent attachment of proteins/peptides to the surface of the AAV capsid, was developed.172 AAV capsids were decorated with leucine-zipper coiled-coil binding motifs that exhibit specific non-covalent heterodimerization.172 This protein display platform may facilitate the incorporation of biological moieties on the AAV surface, expanding possibilities for vector enhancement and engineering.
Polyploid AAV capsid
Polyploid or transcapsidation approach consisted of the transfection of combinations of AAV serotype helper plasmids to produce mosaic capsid recombinant AAV.173,174 To combine the advantages of AAV1 and AAV2 vectors, a mixture of the corresponding helper plasmids was used in the transfection process, resulting in packaged virions with capsid proteins from both serotypes.173 The resulting chimeric vectors could be purified by heparin column, and they showed expression levels similar to those of AAV1 in muscle or AAV2 in liver, in vivo.173 Surprisingly, the polyploids vectors did not escape NAbs, being inhibited by both AAV1 or AAV2 antiserum.173
To systematically evaluate how these mosaic capsid recombinant AAV can be formed, and their functionality, AAV1–5 helper plasmids were mixed at five ratios.174 This revealed the existence of functional subgroups among the serotypes, which correspond to pairwise VP sequence identities: subgroup A (AAV1–3), B (AAV4), and C (AAV5). The rAAV titer depends on the serotype’s subgroup: it is high with mixtures within subgroup A, intermediate from serotype 5 mixtures, while mixtures containing the AAV4 capsid exhibited reduced packaging capacity.174 This suggests a reduced compatibility of subunit interaction between serotypes of different subgroups. The binding profiles of the mixed-virus preparations to heparin sulfate or mucin agarose revealed either an abrupt shift or a gradual alteration in the binding profile to the respective ligand upon increase of a capsid component that conferred ligand specificity, only AAV3–AAV5 mixtures at the 3:1 ratio exhibited duality in binding. The transduction profiles either do not change with the ratio of the mixtures, increase gradually with titration of a second capsid component, or increase abruptly given a threshold. A synergistic effect in transduction and an unexpected new tropism were observed in some combinations of AAV1 helper constructs with type 2 or type 3 recipient helpers.174
Transduction efficiencies in the rat hypothalamus of polyploid AAV1/2 and AAV2/8 vectors carrying short hairpin RNAs were compared versus AAV1, AAV2, and AAV8.175 AAV1 vector was more efficient than the polyploids and the other serotypes in neuronal transduction of the rat lateral hypothalamic nucleus.175
Polyploid viruses might potentially acquire advantages from parental serotypes for enhancement of AAV transduction and evasion of NAb recognition without increasing capsid antigen presentation in target cells.176 rAAV resulting from the co-transfection of AAV2 and AAV8 helper plasmids at different ratios were evaluated for both their transduction efficiency and NAb escape activity.176 All the polyploid viruses induced higher transduction than their parental AAV vectors after muscular injection. After systemic administration, a 4-fold higher transduction in the liver was observed with AAV2/8 1:3 than that with AAV8.176 This same AAV2/8 1:3 virus was able to escape AAV2 neutralization and did not increase capsid antigen presentation capacity. NAb evasion ability was improved with the triploid AAV2/8/9 (ratio 1:1:1), which was able to escape NAb activity from mouse sera immunized with parental serotypes.176
Chai et al.177 explored the transduction efficiency of several haploid viruses, which were made from the VP1/VP2 of one serotype and VP3 of another compatible serotype. The haploid AAV vectors, composed of VP1/VP2 from serotypes 8 or 9, and VP3 from AAV2, displayed an increased liver transduction compared with those of AAV2 vectors, while those with VP1/VP2 from serotypes 8 or 9 and VP3 from AAV3 achieved higher transductions in multiple tissue types compared with those of AAV3 vectors.
Polyploid AAV vectors can potentially be generated from any compatible AAV, whether a natural serotype, rational designed, library derived, or any combination thereof, providing a novel strategy that should be explored in future clinical trials.
Strategies to overcome anti-AAV immune response
One reason AAV is the vector of choice for gene therapy is its relatively low immunogenicity. Paradoxically, finding efficient strategies to circumvent pre-existing anti-AAV immunity and to prevent de novo humoral and cellular responses to the vector is a necessity for the advancement of the field of gene therapy.
The ability of pre-existing NAbs to reduce or abrogate AAV-mediated therapy has been well documented.178,179,180 AAV2 has the highest seroprevalence of NAbs in the general population,181 which makes this serotype less suitable for systemic administration. The serotypes with the lowest reported seroprevalence of NAbs in humans are AAV8 and the phylogenetically distinct AAV5.178,181 Unlike other serotypes, AAV5 has proved to be more resistant to NAbs, as it is able to successfully transduce liver in humans with relatively high titers of NAbs.182,183 Based on these results, participants with titers of NAbs were included in trials in hemophilia A and B using AAV5 vector.14 NAb prevalence is moderate at birth, decreases markedly from 7 to 11 months, and then progressively increases through childhood and adolescence.184 The seroprevalence of NAbs to AAV vector serotypes also varies by geographic region.178 Although all four IgG subclasses have been detected in seropositive individuals or patients receiving AAV-mediated gene therapy, IgG1 constitutes the predominant subclass.59,185,186 Additionally, non-neutralizing AAV-specific antibodies have been observed that can potentially increase transduction in certain tissues.187 Complement activation by AAV-specific antibodies is currently being investigated as a possible source of tissue toxicity.188
Based on rates of pre-existing immunity, it makes sense to choose a vector serotype with the lowest prevalence in the general population, such as AAV5 or AAV8.14 However, anti-AAV antibody cross-reactivity caused by the high degree of conservation in the amino acid sequence among AAVs is another element to consider,181 as well as differences in immunogenicity between AAV serotypes.189 Due to this complex scenario, an important effort is being made in the design of novel AAV capsids or capsid modification strategies directed to avoid Nabs, as discussed elsewhere in this review.
Other strategies have focused on eliminating circulating NAbs or antibody-producing B cells. In this sense, plasmapheresis has been used to eliminate AAV-specific NAbs from patients’ sera with a degree of success in the context of low NAb titers.190 B cell depletion before AAV administration caused a reduction of AAV-specific humoral response in preclinical studies,191 and combination of rituximab with rapamycin is under study in a clinical setting (clinical trial NCT02240407). A promising approach based on IgG cleavage with the streptococcal endopeptidase IdeS (Imlifidase) was described last year by Leborgne et al.192 The authors demonstrated that IdeS degraded anti-AAV8 NAbs both in vitro and in vivo in a setting of passive transference of human IVIG followed by AAV8 administration in mice.192 Furthermore, elimination of anti-AAVs NAbs by IdeS allowed efficient AAV8-mediated liver transduction in mice infused with IVIG. These results were extended to NHPs naturally carrying anti-AAV8 NAbs, where IdeS treatment significantly increased transduction with AAV8-hFIX vector in the presence of NAbs and during a re-dosing schedule using AAV-LK03.192 Similar results were obtained by Elmore et al. using IdeZ endopeptidase, an IdeS analog from Streptococcus zooepidemicus.193 IdeZ rescued AAV8- and AAV9-mediated liver transduction in mice passively immunized with human IVIG and individual human sera, as well as in seropositive NHPs injected with AAV9.193
Despite the growing research effort, AAV re-dosing continues to be an unanswered need. Strategies such as IgG cleavage with IdeS or IdeZ can transiently eliminate anti-AAV NAbs before vector injections, which could be a solution to the major issue of pre-existing NAbs. However, the generation of capsid-specific CD8+ T cells with the ability to eliminate transduced target cells has also been described.194,195,196,197 Capsid immunodominant peptides are presented in major histocompatibility complex (MHC) classes I and II by antigen-presenting cells (APCs),198,199,200,201 which constitute the first signal for the activation of AAV-specific CD8+ and CD4+ T cells, respectively. Co-stimulation and cytokine signals provided by APCs after sensing of pathogen-associated molecular patterns (PAMPs) on AAV capsid and viral genome are also required for priming naive AAV-specific T cells.200,201,202 Evidence indicates that capsid proteins are sensed by Toll-like receptor 2 (TLR2) in the surface of APCs,202 whereas unmethylated CpG motifs, present in the viral genome, are recognized by endosomal TLR9.200,203 Interestingly, Rogers et al. demonstrated that activation of TLR9-MyD88 pathway in plasmacytoid dendritic cells (pDC) is essential for the cross-priming of capsid-specific CD8+ T cells, in a complex process that also requires a crosstalk with conventional DC (cDC) and type I interferon (IFN I).200 Early evidence also suggests that MDA5 sensor could have a role in AAV innate sensing through recognition of dsRNA molecules generated during vector transduction, which contributes to IFN I production.204
Based on the important role of TLR9 signaling for anti-AVV T cell responses, depletion of stimulatory CpG motifs or incorporation of TLR9 inhibitory oligonucleotides in the transgene sequence have been studied as potential solutions to avoid T cell activation in the context of gene therapy.205,206,207 Faust et al. showed that CpG-depleted AAVrh32.33 vectors allowed a stable transgene expression in mouse muscle tissue, which was associated with a significant reduction of the frequency and activation of CD8+ T cells specific for the capsid and the transgene.205 More recently, Chang et al. demonstrated that engineered AAV2, AAV8, and AAVrh32.33 vectors containing short DNA oligonucleotides that antagonize TLR9 activation elicited impaired T cell responses and enhanced transgene expression in different animal models.207 Nonetheless, Xiang et al.206 showed that CpG depletion does not always lead to reduced T cell responses and pointed out the importance of properly interpreting the results obtained in available animal models. In fact, most of the work commented on here evaluated the effect of CpG depletion on naive T cells directed against the capsid or the transgene, where the consensus is a reduction of their expansion and functionality. Conversely, Xiang et al. observed an enhanced in vivo proliferation of capsid-specific memory CD8+ T cells in response to CpG-depleted AAV vectors compared with vectors containing CpG motifs.206 Therefore, a deeper understanding of the potential outcome of CpG depletion in AAV vectors is needed for proper clinical application. Some evidence in this regard starts to accumulate in the clinical setting. In a phase I/II clinical trial, transient transgene expression was observed in hemophilia B patients treated with AAV8-based FIX Padua (BAX 335) gene therapy, which was associated with an increased content of CpG motifs introduced into the transgene coding sequence by codon optimization.208 This link between CpG content, immune system activation, and transgene elimination was corroborated in other gene therapy clinical trials for hemophilia B.209 Therefore, finding the right balance between the potential increase of transgene expression and the number of immunostimulatory CpG motifs can be critical when codon optimization strategies are applied to AAV gene therapy vectors for clinical use.
Experience from clinical trials has shown immune-related toxicity in patients receiving AAV-based gene therapy, particularly at high vector doses.12,194,198,210 The most common course of treatment has been corticosteroid administration,12,196,210 which has not always been successful to control anti-AAV immune responses and has known side effects.211,212 This highlights the importance of using more selective and efficient drugs to target AAV-specific T cell responses, which could potentially be used in patients in combination with strategies focused on avoiding NAbs, such as IdeS enzyme treatment, to enable re-dosing efforts. Particularly desirable are the strategies promoting specific immune tolerance mechanisms, such as expansion of regulatory T cells (Tregs). In this sense, ImmTOR nanoparticles, containing the ImmTOR inhibitor rapamycin, can promote a tolerogenic response characterized by the induction of tolerogenic DCs and antigen-specific Tregs when co-administered with antigens in animal models.213,214,215 Meliani et al. demonstrated that ImmTOR co-administered in mice with AAV vectors reduced both NAb titers and T cell responses against capsid proteins, including memory CD4+ T cell reactivation, with concomitant Treg expansion.216 Therefore, re-dosing was possible in both mice and NHP within the experimental conditions tested.216 More recently, Ilyinskii et al. showed that co-administration of ImmTOR with AAV vectors enhanced transgene expression in the liver through a mechanism unrelated to the immunoregulatory effects of ImmTOR and independent of the AAV receptor.217 The efficacy of co-administration of ImmTOR with AAV vectors to control anti-AAV immune responses in patients, as well as potential side effects, will be crucial points for future use in the clinical setting.
Vector genome design
Promoters “in the small work” of rAAV
Along with the selection of new capsids, design of the transgene sequence has also evolved during the past years. Given the limited cargo capacity of AAV and the broad tropism of most of the capsids, many efforts have been made for the identification of short and strong promoters.218 Although ubiquitous promoters are currently in use in the clinic and can still be used depending on the indication, e.g., when multiple cell types or tissues need to be treated, current efforts are more focused on the selection of tissue-specific promoter sequences. When selecting such sequences, promoter strength must also be considered in parallel to tissue specificity, since use of a strong promoter would allow increased vector potency and, thus, reduced doses. One strategy simply consists of identifying the minimal/core promoter sequence through testing different shortened sequences of natural gene promoters, as illustrated, for example, by the photoreceptor-specific rhodopsin kinase core promoter.219 Another strategy is to rationally assemble hybrid promoters using elements from different known enhancers and promoters, leading to the generation of short and strong promoters, either ubiquitous or tissue specific, suitable for delivery of large coding sequences such as s.p. Cas9 or coagulation factor VIII.218,220,221
This strategy can be further refined using computational approaches, by which transcription factor binding sites and other cis-acting sequences involved in tissue specificity and promoter strength can be identified from microarrays and genome-wide functional analyses.222 By assembling different combinations of such cis-regulatory motifs, it has been possible to select short tissue-specific enhancers able to greatly increase tissue-specific gene expression when added upstream of different promoters, as first demonstrated in mice and NHPs to direct coagulation factor IX expression in the liver from AAV9 vectors.223 The same strategy was also used to enhance specific gene expression in the heart or in skeletal muscles.224,225 Computational approaches have also been used to assemble libraries of completely synthetic promoters that are active in specific cell types.226 This technology has already allowed identification of new tissue-specific promoters of short length and high strength compared with natural promoter sequences that would be useful for AAV-mediated gene therapy in different tissues, including liver, muscle, retina, and brain.227,228,229 In addition to tissue specificity, such synthetic promoters can be selected to be constitutive, or inducible or repressible by chemical stimuli, as recently reported using liver as the target tissue 230.
AAV terminal repeats
In the process of screening promoter specificity and strength, it is critical to select the final candidates in an AAV vector context; i.e., in the presence of AAV terminal repeats in cis. Indeed, it is well known that AAV-2 inverted terminal repeat (ITR) sequence displays some promoter activity,231 which was shown to interfere with drug-inducible promoter regulation and was mapped within the A/D sequence element.232 Several putative transcription factor binding motifs have been found within the AAV-2 ITR sequence through bioinformatic analysis, and, substituting the D sequence in one, the ITR was reported to change transgene expression levels from the same promoter, both in vitro and in vivo.233,234 Substitution of AAV-2 terminal repeats with ITR from other serotypes was also shown to modify gene transfer efficiency, as shown using vectors with serotype 2 or serotype 3 ITRs packaged into an AAV-3 capsid.235 It is theoretically possible to encapsidate AAV vector genomes having ITR from any serotype into any AAV capsid serotype provided that the appropriate Rep proteins are used,236,237 so changing the ITR sequence depending on the promoter and the target tissue might have to be considered in the future.
RNA processing elements
Beyond promoter choice, other aspects of the transgene expression cassette design need to be considered to improve expression and, thus, increase vector potency.
First, it is now obvious that an intronic sequence must be included for optimal transgene RNA processing and mRNA export. A number of intronic sequences of varying length have been described and used in combination with different promoters (hybrid beta-actin/beta-globin,238 hybrid adenovirus/immunoglobin,239 hybrid beta-globin/Ig heavy chain or minute virus of mice (MVM)240). The addition of processing sequences at the 3′ end of the transgene has also been shown to improve transgene expression, such as the Woodchuck hepatitis virus posttranscriptional regulatory element (WPRE), which can promote mRNA export to the cytoplasm,241,242,243 or short UC-rich elements, which can improve mRNA stability through PCPB1 protein binding.244 The addition of an S/MAR sequence can also improve transgene expression, and it has been shown to allow episomal vector persistence in proliferating cells with integration-deficient lentiviral vectors (IDLV),245,246 and, more recently, with AAV,247 which could be an advantage in differentiating or renewing tissues, such as the liver or the damaged muscle.
Selection of the polyadenylation signal can also affect transgene expression levels. The choice of a very short poly(A) signal is sometimes dictated by transgene size but can have a negative impact on expression levels. For example, short synthetic or shortened versions of the SV40 poly(A) signals were shown to decrease transgene expression from AAV vectors compared with bGH or SV40 larger sequences.242
In addition to the poly(A) signal required for transgene expression, a second poly(A) signal placed in the reverse orientation relative to the transgene sequence should be considered. Indeed, a recent study has shown that transcription of antisense RNA from the 3′ ITR can induce an innate immune response triggered by cellular double-stranded RNA sensors, resulting in IFN-β expression and lower transgene levels, and this was prevented by the addition of a reverse poly(A) signal close to the 3′ ITR to stop antisense transcription.204
Apart from sequences that act to increase transgene expression, other elements acting at the RNA level can also be used to modulate or restrict expression. Indeed, the addition of microRNA target sequences within the 3′ untranslated region can be used to inhibit transgene expression in some cell types where it is undesirable. This can be particularly useful in the case of systemic administration, for example to limit expression in the liver (where large amounts of vector are often found), through addition of miR122a target sequences.248,249 Similarly, the addition of miR142-3b target sequences has been shown to prevent immune response against the transgene by inhibiting expression in hematopoietic APCs from an AAV vector,250 as initially shown with lentivectors.251 Another interesting example is the addition of miR208a target sequences to prevent expression in the heart in case of transgene-induced cardiac toxicity, as shown with an AAV vector expressing calpain 3 under a muscle-specific promoter.252 The addition of miRNA target sequences was also shown to allow cell-type-specific transgene expression in the retina while using AAV vectors with ubiquitous cytomegalovirus (CMV) promoter.253
Coding sequence optimization
Optimization of the transgene coding sequence in AAV vectors can cover several aspects, including enhanced gene expression and reduced transgene immunogenicity. One aspect involves optimization of codon usage to improve translation efficiency based on codon adaptation index or, more recently, on tRNA adaptation index (tAI), which relies on codons/tRNA pool co-adaptation. Indeed, tAI has been correlated to both translation efficiency and importance of the gene function.254 Reduction of secondary structures is also another approach to improve ribosome processivity on transgene.255 Codon optimization was shown to improve expression of engineered microdystrophin transgene from an AAV8 vector in muscular dystrophy X-linked (mdx) mice256 and has been widely used to enhance gene transfer efficiency in the liver for various indications.195,221,257,258 An interesting example is the optimization of the complex RPGR coding sequence through codon adaptation index, together with reduction of repeated sequences and secondary structures, which was shown to improve both transgene expression and vector stability following AAV-mediated retinal gene transfer in mice and dog models of X-linked retinitis pigmentosa (XLRP).259,260
In addition, a suitable codon optimization algorithm needs to avoid the addition of unwanted sequences such as alternative translation initiation codons, splicing donor and acceptor, or polyadenylation signals. Hence, the design of an ideal coding sequence to achieve both high expression and low immunogenicity appears difficult, and, in most cases, several sequences optimized with different algorithms need to be tested to select the optimal one.
Vector genome size
Beyond vector genome optimization for improved transgene expression, another parameter must be considered when designing an AAV vector. Genome size of wild-type AAV is around 4.7–4.8 kb, and the packaging limit of the AAV capsids has been shown to be around 5.0–5.1 kb. Oversized vectors exceeding this packaging capacity result in the encapsidation of fragmented genomes. These fragmented vectors are still able to mediate transgene expression through reconstitution of full-length vectors by homologous recombination between overlapping fragments in the target cells, but the vector preparations contain more heterogenous particle populations with a higher ratio of empty capsids, which is undesirable for a clinical product.261,262,263,264,265,266
Several strategies have been explored to overcome this packaging limitation and to allow gene transfer of large coding sequences by splitting the transgene sequence into two or even three AAV vectors. The trans-splicing vectors is the first strategy and was described 20 years ago by different groups.267,268,269,270 This strategy consists of one AAV vector containing the 5′ half of the transgene cassette ending with an intronic splicing donor sequence and a second vector carrying the 3′ half of the transgene starting with an intronic splicing acceptor site. The full-length transgene is reconstituted through intermolecular recombination between the ITR of both vectors in the target cell, and the ITR-ITR junction is removed from the mature mRNA by intron splicing. This allows, for example, delivery of a split minidystrophin gene in mdx mice.271
The second described strategy makes use of overlapping AAV vectors, which share a common sequence from the middle of the transgene, allowing full-length transgene reconstitution through homologous recombination in the target cells.272,273 This has been used, for example, to deliver the 6.7-kb MYO7A cDNA in the mouse retina,274 or a 6.2-kb cDNA coding for minidystrophin or for Dysferlin in mouse muscles.275,276
More recently, hybrid dual vectors combining both the trans-splicing and overlapping strategies have been developed and have been shown to be more efficient and potentially adaptable to any transgene sequence.277,278 This strategy further evolved and was used in dual AAV vectors carrying cDNA for MYO7A or ABCA4, which were shown to be effective in both the mouse and pig retina.279,280,281 This dual vector strategy allows delivery of transgene sequences of about 9 kb, and a similar design was used to develop triple AAV vectors able to deliver larger transgenes of up to 14 kb. Feasibility of the triple vector strategy was demonstrated through expression of a full-length dystrophin protein in the mdx mouse muscle,282 and through delivery of CDH23 and ALMS1 transgenes in the mouse retina.283
An alternative strategy to trans-splicing and overlapping vectors has also been described, which makes use of split inteins to reconstitute the full-length transgene product at the protein level, through protein trans-splicing (see Aranko et al.284 for review). This was first tested in dual AAV vectors for expression of full-length coagulation factor VIII protein in hemophilic mice285and, very recently, for ABCA4 and CEP290 transgene delivery in the retina of STGD1 and LCA10 mouse models and in human iPSC-derived retinal organoids.286
While many efforts have been made to adapt AAV vectors for delivery of large transgenes, little is known about strategies to adapt vector size in the case of small transgenes. Indeed, it has been shown that empty particles are impaired in the ability to expose the VP1 N-terminal domain outside, a critical step for vector infectivity.287,288 Similarly, AAV particles containing small genomes of 3.4 and 4.1 kb have been found to be more stable than particles containing wild-type size genomes, which was correlated with reduced ability to release vector DNA.289 Thus, lower size vectors may have lower potency than wild-type size vectors independently of transgene cassette design due to reduced transgene delivery in the target cell nucleus.
In addition, some experimental data have highlighted that sequences from the vector backbone flanking the ITRs outside the AAV cassette could be co-packaged during vector assembly in the producer cells. This was shown by the detection of ITR-backbone junctions in purified AAV particles by both PCR and next-generation sequencing,290 and both length and relative quantities of these co-packaged sequences were found to be inversely related to AAV vector size.291 Taken together, all these findings indicate that vector genome size should be ideally adjusted to 4.7 ± 0.1 kb for a single-stranded AAV to match wild-type viral genome size. Several strategies could be considered to achieve this in case of small transgenes. The first obvious strategy is to select transgene regulatory elements that reach the target size when added together. However, this can be challenging given the limited number of available and characterized regulatory elements, and the wide range of coding sequence sizes. To better adjust the vector size once all regulatory elements have been selected, the most straightforward strategy would be to use stuffer (or spacer) DNA sequences, which could theoretically be located upstream or downstream of the transgene expression cassette, and/or between some of the regulatory elements. The addition of stuffer DNA sequence has been investigated for the helper-dependent adenoviral vectors, for which it is mandatory to match the wild-type viral genome size to achieve packaging. First generation HD-Ad vectors used bacteriophage lambda DNA as a stuffer, but this was shown to induce an innate immune response upon mice injection,292 which was not seen when replacing lambda DNA with non-coding DNA sequences from human origin.292,293 Thus, choice of the stuffer DNA sequence must be carefully considered since it can dramatically influence transgene expression, and an ideal sequence would be biologically neutral. To achieve this, the following features should be avoided in the stuffer sequence design:
-
•
Transcription factor binding sites that could interfere with transgene expression or promote transcription of antisense RNA.
-
•
Target sequences for microRNA expressed in the target cells or tissues that would promote transgene extinction.
-
•
Splice acceptor and donor sites that may interfere with accurate transgene splicing.
-
•
Strong secondary structures that would impede vector DNA replication and packaging.
-
•
ATG or alternative translation initiation codon not followed by an in-frame stop codon, to prevent expression of any unwanted peptide.
-
•
CpG motifs that could promote immune response.
Besides these, the ideal stuffer sequence should not share any homology with prokaryotic or viral DNA, or with human DNA, since presence of non-therapeutic human sequences in gene therapy vectors could potentially raise some safety concerns. To design such a perfect stuffer DNA, a dedicated bioinformatic pipeline would be welcome.
Production methods for rAAV
The following methods have been established for the production of rAAV vectors: transient plasmid DNA transfection in mammalian cells, adenovirus infection of stable mammalian cell lines, infection of insect cells with recombinant baculoviruses, and infection of mammalian cells with recombinant herpes simplex viruses (rHSVs).294
Transient transfection of mammalian cells with plasmids carrying the necessary components for the assembly of the vector is one of the most popular and straightforward methods to produce AAV.295 HEK293 cells constitutively expressing adenovirus E1a and E1b genes are transfected with a helper plasmid containing other Adenovirus (AdV) genes (required for replication, mRNA processing, and translation), a trans-plasmid expressing rep and cap gene, and a cis-plasmid to be packaged in the AAV capsid (which usually contains an expression cassette with the gene of interest flanked by two ITRs).296 A GMP-compatible two-plasmid cotransfection system, in which rep and cap genes and Ad helper genes are on the same plasmid, is also available.297 These HEK293 cells have been adapted for growth in suspension to allow for scale-up and increased production yields.298 Advantages of transient transfection are that new rAAV constructs can be produced and tested quickly, as well as the absence of residual helper virus contaminants.296
One concern associated with such a process is the packaging of plasmid backbone sequences, which can lead to activation of the innate immune system, and to dissemination of antibiotic resistance genes.299,300 To avoid this problem, alternative DNA production systems have been developed.
One alternative to standard plasmids is the use of minicircles, for which the sequence of interest is separated from the plasmid backbone through inducible expression of a recombinase in the bacterial cells,301,302,303 and, indeed, this system has been shown to improve vector purity.304 However, both the fermentation process and the purification methods used for minicircle production are quite complex to set up.
Another possible alternative is using plasmids devoid of an antibiotic resistance gene and contain minimal bacterial sequences. Such a system, called nanoplasmid, has been developed using a sacB-targeted antisense RNA that has the selection marker and an R6K origin for replication.305 These plasmids are propagated in a bacterial host strain expressing the sacB gene for sucrose selection, and the pir Rep protein for replication. Thus, they are devoid of antibiotic selection and the R6K origin restricts replication to bacterial host expressing the pir protein. In addition, the resulting short bacterial backbone has been shown to prevent negative effect on transgene expression seen with standard plasmids, even when inserted within an AAV transgene cassette.306 However, this technology still requires fermentation for DNA production.
A third alternative is now also available, which involves production of DNA completely devoid of bacterial plasmid backbone through an entirely in vitro synthesis approach.307 Briefly, the starting DNA is a plasmid with the sequence of interest flanked by the target sequences for the bacteriophage N15 protelomerase TelN.308,309 This plasmid is used as a template for DNA amplification by the phi29 DNA polymerase through rolling circle replication. The plasmid backbone is then separated from the sequence of interest by the TelN enzyme, and the final product, called doggybone DNA, is linear double-stranded DNA with covalently closed ends. In addition to providing DNA free of bacterial sequence, except the remaining short prototelomer sequences, such a process does not require fermentation, which results in reduced quality controls and production timelines, and it has been shown to be effective for AAV production.307
Mammalian HeLa-based stable cell lines infected with Ad were actually the first system to provide a scalable upstream process for manufacturing of purified rAAV particles.295 This methodology is based on HeLa cells stably transfected with copies of the AAV rep-cap genes and the rAAV vector genome with flanking ITRs, where production is initiated by the introduction of AdV.310 A process for a 250-L production has been designed.311 This approach has two main limitations: a new cell line has to be generated for each combination of capsid and gene of interest to be delivered, and the need to deal with the introduced AdV.295 The establishment and screening of stable and high-producer clones is an arduous and time-consuming step which has limited the rate of adoption of this technology.294
Infection of insect cells with recombinant baculovirus (BEV) has been developed as an alternative to the rAAV production in mammalian cells.312 The baculovirus-Sf9 platform has been notably established as a GMP-compatible and scalable system.313,314 AAV production has been scaled-up to 200 L in bioreactors, resulting in a yield exceeding 1016 viral genomes (VG).315 In terms of specific yields, mammalian cell systems are comparable with the baculovirus expression vector system (BEVS), with 103 to 106 AAV particles/cell.316 This is about to change, as a new method to improve the baculovirus-based AAV production using Thermo Fisher Scientific’s ExpiSf Baculovirus Expression System was recently developed.317 Using a chemically defined medium for suspension culture of high-density ExpiSf9 cells, baculovirus-infected ExpiSf9 produced up to 5 × 1011 genome copies of highly purified AAV vectors per 1 mL of suspension culture, which is up to a 19-fold higher yield than the titers obtained from the conventional Sf9 cell-based systems.317 This methodology will increase the overall appeal of baculovirus-based AAV production. Sf9 cells do not require additional helper viral genes beyond those provided by the recombinant baculovirus.318 BEV-Sf9 systems exhibit reduced encapsidation of contaminating DNAs.295 Baculovirus-Sf9 and human manufacturing platforms can produce vector lots exhibiting chemical and functional differences,60 which include different capsid PTMs, insect and baculoviral impurities, and potentially poorer packaging percentages than human-produced vector as human-produced rAAVs can be more potent than baculovirus-Sf9 vectors in vitro and in vivo.60
AAV can also be produced by infecting mammalian cells (HEK293 cells or BHK cells) with replication-deficient herpes simplex virus (HSV).319,320 Usually two HSV are used; one carries the expression cassette for the gene of interest flanked by the two ITRs and the other virus carries the rep and cap genes regulated by their endogenous promoter.296,320 This methodology appears to produce rAAV stocks of improved viral potency.320,321 The main limitation of the HSV system is the presence of contaminating HSV and its derived products.296
Other rAAV production methodologies have been proposed. A method based on dual infection of HeLa-S3 cells with one vaccinia vector and one Ad-AAV hybrid vector consistently produce ∼1 × 1015 genome containing rAAV vectors per liter of suspension cells.322 The capsids obtained by this method differ from those obtained by traditional systems because VP1 is produced more abundantly than VP2 (19:1 ratio), which appears to improve the transduction levels in vitro and in vivo.322 As the vaccinia virus replicates in the cytoplasm, the rep-cap sequence and rAAV genome are in different sub-cellular compartments, which eliminates the contamination with replication-competent AAV.294 rAAV2 infectious particles can be generated in Saccharomyces cerevisiae after transformation with four plasmids.323,324,325 These plasmids allow the expression of rep78/52, VP1–3, and assembly activating protein (AAP) under the control of galactose-inducible promoters. The vector loading efficiency is lower than in the other methods,294,323,324,325 but yeast offer great possibilities for scale-up and reduced cost, so it is very probable that we will see more development on this methodology in the near future.
Despite these potential alternatives and the significant improvements made with the transient production systems, development of stable mammalian AAV producer cell lines is still a relevant objective, in particular when thinking about commercial production of high-quality vectors. Although production through adenovirus infection of stable cell lines containing the AAV rep-cap genes and the recombinant vector genomes was shown to be effective, the ultimate goal is to establish cell lines that also contain the helper virus components; i.e., adenovirus E1, E4, E2A, and VA RNA genes. To achieve this, a tightly regulated and strongly inducible expression system must be implemented. Such stable producer cells have been recently developed using E1-immortalized amniocytes,326,327 into which AAV2 Rep, adenovirus E2A, E4, and VA RNA, and finally AAV Cap sequences, have been added, under control of a doxycycline-inducible expression system.328 These cells are reported to efficiently produce AAV once the recombinant vector genome has also been integrated, but, in addition to AAV and adenovirus components, such a system also requires expression of an inducer or a repressor into the cells. The use of promoters that can be induced by small molecules or by simply modulating the cell culture conditions would be an advantage. For example, temperature-inducible promoters have been proposed to control AAV rep and cap expression.329 Using computational approaches, the design of synthetic promoters that can be controlled by chemical compounds or cell culture parameters to mimic expression kinetics of the natural AAV life cycle could be considered for optimal vector production.230
rAAV at clinical
Recombinant adeno-associated viral vectors have been used in more than 200 ongoing clinical studies with a positive safety profile and significant clinical benefit in a variety of genetic diseases. Based on its increasingly promising clinical trial results, AAV gene therapy is proving to be a potent treatment option with the potential to achieve long-term correction addressing unmet medical needs.
In 2012, the first AAV1-based drug, Glybera, was approved by the European Medicines Agency (EMA) to treat lipoprotein lipase deficiency,330 a rare monogenic genetic disorder characterized by accumulation of triglycerides in plasma due to mutations in lipoprotein lipase.331 However, while promising to be a viable treatment option, the drug ended up a commercial failure 332due to a combination of the low demand for this treatment and its high cost. Glybera was subsequently withdrawn from the market, highlighting that a major challenge for rare disease treatments is the availability of appropriate funding and reimbursement mechanisms within the healthcare system.333
Luxturna, an AAV2 vector for the treatment of blindness has become the first viral-based drug approved by the Food and Drug Administration (FDA) in 2017. Luxturna is a one-time gene therapy treatment used to improve vision in patients with established genetic vision loss due to Leber congenital amaurosis or retinitis pigmentosa. Patients with confirmed biallelic RPE65 mutation-associated retinal dystrophy will be treated with Luxturna and are expected to have their vision restored within several months.334 After the approval of Luxturna, another rare genetic eye disease AAV-based drug is expected to be released in the near future as a treatment for choroideremia. This X-linked inherited retinal dystrophy is caused by mutations in Rab escort protein 1 (REP1). REP1 is a protein involved in lipid modification of Rab proteins and has been implicated as a cause of choroideremia disease. Patients that received AAV-REP1 therapy showed a significant increase in their visual acuity.335
Recently, Zolgensma, also known as AVXS-101, was approved by the FDA to treat SMA. SMA is caused by a mutation in the survival motor neuron 1 (SMN1) gene,336,337 occurs in approximately 1 in 10,000 live births, and is the leading genetic cause of infant mortality.338,339 This AAV9-based drug consists of double-stranded DNA and is used to treat pediatric patients less than 2 years of age with bi-allelic mutations in the SMN1 gene.338 A clinical trial, START, enrolled 15 infants (six male and nine female) with infantile-onset SMA, three in a low-dose cohort and 12 in a high-dose cohort. The 15 patients were treated with Zolgensma and all survived at 24 months post-treatment without any events recorded. None required permanent ventilation. Out of the 12 children in the high-dose cohort, 11 achieved a Children's Hospital of Philadelphia infant test of neuromuscular disorders (CHOP-INTEND) score of 40 or greater, 10 reached 50 or greater, and two of the children were able to sit without assistance for at least 30 s. All 15 children are currently part of a long-term study with the goal to further observe their development annually for another 15 years.340
Conclusions
There are still many questions unanswered regarding AAV vector receptors in different tissues, the mechanism of endosomal escape and nucleus delivery, rAAV-mediated immune responses, and what regulates the duration and efficacy of delivered gene expression. Results from in vivo experiments in small animals does not easily translate to humans. Required rAAV doses are still high and production is extremely expensive. A significant percentage of patients are being excluded from current clinical trials due to pre-existing NAbs. In this work, we reviewed different approaches aimed at overcoming AAV’s limitations as a gene delivery system. We are confident in the success of gene therapy and believe we are witnessing the birth of a new kind of medicine, with rAAVs as protagonists.
Acknowledgments
We would like to thank Cheryl Everette for the revision and editing of the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Atchison R.W., Casto B.C., Hammon W.M.D. Adenovirus-associated defective virus particles. Science. 1965;149:754–756. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- 2.Hastie E., Samulski R.J. Adeno-associated virus at 50: a golden anniversary of discovery, research, and gene therapy success—a personal perspective. Hum. Gene Ther. 2015;26:257–265. doi: 10.1089/hum.2015.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pillay S., Carette J.E. Host determinants of adeno-associated viral vector entry. Curr. Opin. Virol. 2017;24:124–131. doi: 10.1016/j.coviro.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zinn E., Vandenberghe L.H. Adeno-associated virus: fit to serve. Curr. Opin. Virol. 2014;8:90–97. doi: 10.1016/j.coviro.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rose J.A., Maizel J.V., Inman J.K., Shatkin A.J. Structural proteins of adenovirus-associated viruses. J. Virol. 1971;8:766–770. doi: 10.1128/jvi.8.5.766-770.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snijder J., van de Waterbeemd M., Damoc E., Denisov E., Grinfeld D., Bennett A., Agbandje-McKenna M., Makarov A., Heck A.J.R. Defining the stoichiometry and cargo load of viral and bacterial nanoparticles by orbitrap mass spectrometry. J. Am. Chem. Soc. 2014;136:7295–7299. doi: 10.1021/ja502616y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wörner T.P., Bennett A., Habka S., Snijder J., Friese O., Powers T., Agbandje-McKenna M., Heck A.J.R. Adeno-associated virus capsid assembly is divergent and stochastic. Nat. Commun. 2021;12:1642. doi: 10.1038/s41467-021-21935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asokan A., Schaffer D.V., Samulski R.J. The AAV vector toolkit: poised at the clinical crossroads. Mol. Ther. 2012;20:699–708. doi: 10.1038/mt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geoffroy M.-C., Salvetti A. Helper functions required for wild type and recombinant adeno- associated virus growth. Curr. Gene Ther. 2005;5:265–271. doi: 10.2174/1566523054064977. [DOI] [PubMed] [Google Scholar]
- 10.Chen S.H., Haam J., Walker M., Scappini E., Naughton J., Martin N.P. Recombinant viral vectors as neuroscience tools. Curr. Protoc. Neurosci. 2019;87:e67. doi: 10.1002/cpns.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabinowitz J., Chan Y.K., Samulski R.J. Adeno-associated Virus (AAV) versus immune response. Viruses. 2019;11:E102–E111. doi: 10.3390/v11020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nathwani A.C., Reiss U.M., Tuddenham E.G.D., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D., et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowles D.E., McPhee S.W.J., Li C., Gray S.J., Samulski J.J., Camp A.S., Li J., Wang B., Monahan P.E., Rabinowitz J.E., et al. Phase 1 gene therapy for duchenne muscular dystrophy using a translational optimized AAV vector. Mol. Ther. 2012;20:443–455. doi: 10.1038/mt.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pipe S., Leebeek F.W.G., Ferreira V., Sawyer E.K., Pasi J. Clinical considerations for capsid choice in the development of liver-targeted AAV-based gene transfer. Mol. Ther. Methods Clin. Dev. 2019;15:170–178. doi: 10.1016/j.omtm.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivastava A. In vivo tissue-tropism of adeno-associated viral vectors. Curr. Opin. Virol. 2016;21:75–80. doi: 10.1016/j.coviro.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao G., Vandenberghe L.H., Wilson J.M. New recombinant serotypes of AAV vectors. Curr. Gene Ther. 2005;5:285–297. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- 17.Hammond S.L., Leek A.N., Richman E.H., Tjalkens R.B. Cellular selectivity of AAV serotypes for gene delivery in neurons and astrocytes by neonatal intracerebroventricular injection. PLoS ONE. 2017;12:01888300–e188922. doi: 10.1371/journal.pone.0188830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vance M.A., Mitchell A., Samulski R.J. Gene Therapy - Principles and Challenges. InTech; 2015. AAV biology, infectivity and therapeutic use from bench to clinic. [Google Scholar]
- 19.Dudek A.M., Zabaleta N., Zinn E., Pillay S., Zengel J., Porter C., Franceschini J.S., Estelien R., Carette J.E., Zhou G.L., Vandenberghe L.H. GPR108 is a highly conserved AAV entry factor. Mol. Ther. 2020;28:367–381. doi: 10.1016/j.ymthe.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meisen W.H., Nejad Z.B., Hardy M., Zhao H., Oliverio O., Wang S., Hale C., Ollmann M.M., Collins P.J. Pooled screens identify GPR108 and TM9SF2 as host cell factors critical for AAV transduction. Mol. Ther. Methods Clin. Dev. 2020;17:601–611. doi: 10.1016/j.omtm.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agbandje-McKenna M., Kleinschmidt J. Gene Therapy. 2012. AAV capsid structure and cell interactions; pp. 47–92. [DOI] [PubMed] [Google Scholar]
- 22.Xiao P.J., Lentz T.B., Samulski R.J. Recombinant adeno-associated virus: clinical application and development as a gene-therapy vector. Ther. Deliv. 2012;3:835–856. doi: 10.4155/tde.12.63. [DOI] [PubMed] [Google Scholar]
- 23.Lisowski L., Tay S.S., Alexander I.E. Adeno-associated virus serotypes for gene therapeutics. Curr. Opin. Pharmacol. 2015;24:59–67. doi: 10.1016/j.coph.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Salganik M., Hirsch M.L., Samulski R.J. Adeno-associated virus as a mammalian DNA vector. Microbiol. Spectr. 2015;3:829–851. doi: 10.1128/microbiolspec.MDNA3-0052-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albright B.H., Storey C.M., Murlidharan G., Castellanos Rivera R.M., Berry G.E., Madigan V.J., Asokan A. Mapping the structural determinants required for AAVrh.10 transport across the blood-brain barrier. Mol. Ther. 2018;26:510–523. doi: 10.1016/j.ymthe.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bates R., Huang W., Cao L. Adipose tissue: an emerging target for adeno-associated viral vectors. Mol. Ther. Methods Clin. Dev. 2020;19:236–249. doi: 10.1016/j.omtm.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown H.C., Doering C.B., Herzog R.W., Ling C., Markusic D.M., Spencer H.T., Srivastava A., Srivastava A. Development of a clinical candidate AAV3 vector for gene therapy of hemophilia B. Hum. Gene Ther. 2020;31:1114–1123. doi: 10.1089/hum.2020.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Q., Luo H., Zhou C., Yu H., Yao S., Fu F., Seeley R., Ji X., Yang Y., Chen P., et al. Comparative intra-articular gene transfer of seven adeno-associated virus serotypes reveals that AAV2 mediates the most efficient transduction to mouse arthritic chondrocytes. PLoS ONE. 2020;15:e0243359. doi: 10.1371/journal.pone.0243359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colon-Cortes Y., Hasan M.A., Aslanidi G. Intra-tracheal delivery of AAV6 vectors results in sustained transduction in murine lungs without genomic integration. Gene. 2020;763S:100037. doi: 10.1016/j.gene.2020.100037. [DOI] [PubMed] [Google Scholar]
- 30.Han I.C., Cheng J.L., Burnight E.R., Ralston C.L., Fick J.L., Thomsen G.J., Tovar E.F., Russell S.R., Sohn E.H., Mullins R.F., et al. Retinal tropism and transduction of adeno-associated virus varies by serotype and route of delivery (intravitreal, subretinal, or suprachoroidal) in rats. Hum. Gene Ther. 2020;31:1288–1299. doi: 10.1089/hum.2020.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang M.H., Hu J., Pratt R.E., Hodgkinson C.P., Asokan A., Dzau V.J. Optimizing delivery for efficient cardiac reprogramming. Biochem. Biophys. Res. Commun. 2020;533:9–16. doi: 10.1016/j.bbrc.2020.08.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z., Klose K., Neuber S., Jiang M., Gossen M., Stamm C. Comparative analysis of adeno-associated virus serotypes for gene transfer in organotypic heart slices. J. Transl. Med. 2020;18:437. doi: 10.1186/s12967-020-02605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pietersz K.L., Martier R.M., Baatje M.S., Liefhebber J.M., Brouwers C.C., Pouw S.M., Fokkert L., Lubelski J., Petry H., Martens G.J.M., et al. Transduction patterns in the CNS following various routes of AAV-5-mediated gene delivery. Gene Ther. 2021;28:435–446. doi: 10.1038/s41434-020-0178-0. [DOI] [PubMed] [Google Scholar]
- 34.Cearley C.N., Vandenberghe L.H., Parente M.K., Carnish E.R., Wilson J.M., Wolfe J.H. Expanded repertoire of AAV vector serotypes mediate unique patterns of transduction in mouse brain. Mol. Ther. 2008;16:1710–1718. doi: 10.1038/mt.2008.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C.-L., Jensen R.L., Schnepp B.C., Connell M.J., Shell R., Sferra T.J., Bartlett J.S., Clark K.R., Johnson P.R. Molecular characterization of adeno-associated viruses infecting children. J. Virol. 2005;79:14781–14792. doi: 10.1128/JVI.79.23.14781-14792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qing K., Mah C., Hansen J., Zhou S., Dwarki V., Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 37.Akache B., Grimm D., Pandey K., Yant S.R., Xu H., Kay M.A. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J. Virol. 2006;80:9831–9836. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori S., Wang L., Takeuchi T., Kanda T. Two novel adeno-associated viruses from cynomolgus monkey: pseudotyping characterization of capsid protein. Virology. 2004;330:375–383. doi: 10.1016/j.virol.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Ling C., Lu Y., Kalsi J.K., Jayandharan G.R., Li B., Ma W., Cheng B., Gee S.W.Y., McGoogan K.E., Govindasamy L., et al. Human hepatocyte growth factor receptor is a cellular coreceptor for adeno-associated virus serotype 3. Hum. Gene Ther. 2010;21:1741–1747. doi: 10.1089/hum.2010.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weller M.L., Amornphimoltham P., Schmidt M., Wilson P.A., Gutkind J.S., Chiorini J.A. Epidermal growth factor receptor is a co-receptor for adeno-associated virus serotype 6. Nat. Med. 2010;16:662–664. doi: 10.1038/nm.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murlidharan G., Corriher T., Ghashghaei H.T., Asokan A. Unique glycan signatures regulate adeno-associated virus tropism in the developing brain. J. Virol. 2015;89:3976–3987. doi: 10.1128/JVI.02951-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pillay S., Meyer N.L., Puschnik A.S., Davulcu O., Diep J., Ishikawa Y., Jae L.T., Wosen J.E., Nagamine C.M., Chapman M.S., Carette J.E. An essential receptor for adeno-associated virus infection. Nature. 2016;530:108–112. doi: 10.1038/nature16465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silveria M.A., Large E.E., Zane G.M., White T.A., Chapman M.S. The structure of an AAV5-AAVR complex at 2.5 Å resolution: implications for cellular entry and immune neutralization of AAV gene therapy vectors. Viruses. 2020;12:1326. doi: 10.3390/v12111326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pillay S., Zou W., Cheng F., Puschnik A.S., Meyer N.L., Ganaie S.S., Deng X., Wosen J.E., Davulcu O., Yan Z., et al. Adeno-associated virus (AAV) serotypes have distinctive interactions with domains of the cellular AAV receptor. J. Virol. 2017;91 doi: 10.1128/JVI.00391-17. e003911-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka A., Tumkosit U., Nakamura S., Motooka D., Kishishita N., Priengprom T., Sa-ngasang A., Kinoshita T., Takeda N., Maeda Y. Genome-wide screening uncovers the significance of N-sulfation of heparan sulfate as a host cell factor for chikungunya virus infection. J. Virol. 2017;91:e517. doi: 10.1128/JVI.00432-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luteijn R.D., van Diemen F., Blomen V.A., Boer I.G.J., Manikam Sadasivam S., van Kuppevelt T.H., Drexler I., Brummelkamp T.R., Lebbink R.J., Wiertz E.J. A genome-wide haploid genetic screen identifies heparan sulfate-associated genes and the macropinocytosis modulator TMED10 as factors supporting vaccinia virus infection. J. Virol. 2019;93 doi: 10.1128/JVI.02160-18. e021600-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chinn L.W., Tang M., Kessing B.D., Lautenberger J.A., Troyer J.L., Malasky M.J., McIntosh C., Kirk G.D., Wolinsky S.M., Buchbinder S.P., et al. Genetic associations of variants in genes encoding HIV-dependency factors required for HIV-1 infection. J. Infect. Dis. 2010;202:1836–1845. doi: 10.1086/657322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zincarelli C., Soltys S., Rengo G., Rabinowitz J.E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H., Yang B., Mu X., Ahmed S.S., Su Q., He R., Wang H., Mueller C., Sena-Esteves M., Brown R., et al. Several rAAV vectors efficiently cross the blood–brain barrier and transduce neurons and astrocytes in the neonatal mouse central nervous system. Mol. Ther. 2011;19:1440–1448. doi: 10.1038/mt.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gray S.J., Matagne V., Bachaboina L., Yadav S., Ojeda S.R., Samulski R.J. Preclinical differences of intravascular AAV9 delivery to neurons and glia: a comparative study of adult mice and nonhuman primates. Mol. Ther. 2011;19:1058–1069. doi: 10.1038/mt.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenberg J.B., Sondhi D., Rubin D.G., Monette S., Chen A., Cram S., De B.P., Kaminsky S.M., Sevin C., Aubourg P., Crystal R.G. Comparative efficacy and safety of multiple routes of direct CNS administration of adeno-associated virus gene transfer vector serotype rh.10 expressing the human arylsulfatase A cDNA to nonhuman primates. Hum. Gene Ther. Clin. Dev. 2014;25:164–177. doi: 10.1089/humc.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang B., Li S., Wang H., Guo Y., Gessler D.J., Cao C., Su Q., Kramer J., Zhong L., Ahmed S.S., et al. Global CNS transduction of adult mice by intravenously delivered rAAVrh.8 and rAAVrh.10 and nonhuman primates by rAAVrh.10. Mol. Ther. 2014;22:1299–1309. doi: 10.1038/mt.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas C.E., Storm T.A., Huang Z., Kay M.A. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J. Virol. 2004;78:3110–3122. doi: 10.1128/JVI.78.6.3110-3122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davidoff A.M., Gray J.T., Ng C.Y.C., Zhang Y., Zhou J., Spence Y., Bakar Y., Nathwani A.C. Comparison of the ability of adeno-associated viral vectors pseudotyped with serotype 2, 5, and 8 capsid proteins to mediate efficient transduction of the liver in murine and nonhuman primate models. Mol. Ther. 2005;11:875–888. doi: 10.1016/j.ymthe.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 55.Lebherz C., Maguire A., Tang W., Bennett J., Wilson J.M. Novel AAV serotypes for improved ocular gene transfer. J. Gene Med. 2008;10:375–382. doi: 10.1002/jgm.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe S., Sanuki R., Ueno S., Koyasu T., Hasegawa T., Furukawa T. Tropisms of AAV for subretinal delivery to the neonatal mouse retina and its application for in vivo rescue of developmental photoreceptor disorders. PLoS ONE. 2013;8:e54146. doi: 10.1371/journal.pone.0054146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colella P., Ronzitti G., Mingozzi F. Emerging issues in AAV-mediated in vivo gene therapy. Mol. Ther. Methods Clin. Dev. 2018;8:87–104. doi: 10.1016/j.omtm.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hardcastle N., Boulis N.M., Federici T. AAV gene delivery to the spinal cord: serotypes, methods, candidate diseases, and clinical trials. Expert Opin. Biol. Ther. 2018;18:293–307. doi: 10.1080/14712598.2018.1416089. [DOI] [PubMed] [Google Scholar]
- 59.Kruzik A., Fetahagic D., Hartlieb B., Dorn S., Koppensteiner H., Horling F.M., Scheiflinger F., Reipert B.M., de la Rosa M. Prevalence of anti-adeno-associated virus immune responses in international cohorts of healthy donors. Mol. Ther. Methods Clin. Dev. 2019;14:126–133. doi: 10.1016/j.omtm.2019.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rumachik N.G., Malaker S.A., Poweleit N., Maynard L.H., Adams C.M., Leib R.D., Cirolia G., Thomas D., Stamnes S., Holt K., et al. Methods matter: standard production platforms for recombinant AAV produce chemically and functionally distinct vectors. Mol. Ther. Methods Clin. Dev. 2020;18:98–118. doi: 10.1016/j.omtm.2020.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mary B., Maurya S., Arumugam S., Kumar V., Jayandharan G.R. Post-translational modifications in capsid proteins of recombinant adeno-associated virus ( AAV ) 1-rh10 serotypes. FEBS J. 2019;286:4964–4981. doi: 10.1111/febs.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sen D., Balakrishnan B., Gabriel N., Agrawal P., Roshini V., Samuel R., Srivastava A., Jayandharan G.R. Improved adeno-associated virus (AAV) serotype 1 and 5 vectors for gene therapy. Sci. Rep. 2013;3:1832–1836. doi: 10.1038/srep01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murray S., Nilsson C.L., Hare J.T., Emmett M.R., Korostelev A., Ongley H., Marshall A.G., Chapman M.S. Characterization of the capsid protein glycosylation of adeno-associated virus type 2 by high-resolution mass spectrometry. J. Virol. 2006;80:6171–6176. doi: 10.1128/JVI.02417-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong L., Zhao W., Wu J., Li B., Zolotukhin S., Govindasamy L., Agbandje-McKenna M., Srivastava A. A dual role of EGFR protein tyrosine kinase signaling in ubiquitination of AAV2 capsids and viral second-strand DNA synthesis. Mol. Ther. 2007;15:1323–1330. doi: 10.1038/sj.mt.6300170. [DOI] [PubMed] [Google Scholar]
- 65.Zhong L., Li B., Jayandharan G., Mah C.S., Govindasamy L., Agbandje-McKenna M., Herzog R.W., Weigel-Van Aken K.A., Hobbs J.A., Zolotukhin S., et al. Tyrosine-phosphorylation of AAV2 vectors and its consequences on viral intracellular trafficking and transgene expression. Virology. 2008;381:194–202. doi: 10.1016/j.virol.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhong L., Li B., Mah C.S., Govindasamy L., Agbandje-McKenna M., Cooper M., Herzog R.W., Zolotukhin I., Warrington K.H., Weigel-Van Aken K.A., et al. Next generation of adenoassociated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. USA. 2008;105:7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Markusic D.M., Herzog R.W., Aslanidi G.V., Hoffman B.E., Li B., Li M., Jayandharan G.R., Ling C., Zolotukhin I., Ma W., et al. High-efficiency transduction and correction of murine hemophilia B using AAV2 vectors devoid of multiple surface-exposed tyrosines. Mol. Ther. 2010;18:2048–2056. doi: 10.1038/mt.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li M., Jayandharan G.R., Li B., Ling C., Ma W., Srivastava A., Zhong L. High-efficiency transduction of fibroblasts and mesenchymal stem cells by tyrosine-mutant AAV2 vectors for their potential use in cellular therapy. Hum. Gene Ther. 2010;21:1527–1543. doi: 10.1089/hum.2010.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gabriel N., Hareendran S., Sen D., Gadkari R.A., Sudha G., Selot R., Hussain M., Dhaksnamoorthy R., Samuel R., Srinivasan N., et al. Bioengineering of AAV2 capsid at specific serine, threonine, or lysine residues improves its transduction efficiency in vitro and in vivo. Hum. Gene Ther. Methods. 2013;24:80–93. doi: 10.1089/hgtb.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jin X., Liu L., Nass S., O’Riordan C., Pastor E., Zhang X.K. Direct liquid chromatography/mass spectrometry analysis for complete characterization of recombinant adeno-associated virus capsid proteins. Hum. Gene Ther. Methods. 2017;28:255–267. doi: 10.1089/hgtb.2016.178. [DOI] [PubMed] [Google Scholar]
- 71.Sen D., Gadkari R.A., Sudha G., Gabriel N., Kumar Y.S., Selot R., Samuel R., Rajalingam S., Ramya V., Nair S.C., et al. Targeted modifications in adeno-associated virus serotype 8 capsid improves its hepatic gene transfer efficiency in vivo. Hum. Gene Ther. Methods. 2013;24:104–116. doi: 10.1089/hgtb.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Aloor A., Zhang J., Gashash E.A., Parameswaran A., Chrzanowski M., Ma C., Diao Y., Wang P.G., Xiao W. Site-specific N-glycosylation on the AAV8 capsid protein. Viruses. 2018;10:644. doi: 10.3390/v10110644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giles A.R., Sims J.J., Turner K.B., Govindasamy L., Alvira M.R., Lock M., Wilson J.M. Deamidation of amino acids on the surface of adeno-associated virus capsids leads to charge heterogeneity and altered vector function. Mol. Ther. 2018;26:2848–2862. doi: 10.1016/j.ymthe.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu A.P., Patel S.K., Xing T., Yan Y., Wang S., Li N. Characterization of adeno-associated virus capsid proteins using hydrophilic interaction chromatography coupled with mass spectrometry. J. Pharm. Biomed. Anal. 2020;189:113481. doi: 10.1016/j.jpba.2020.113481. [DOI] [PubMed] [Google Scholar]
- 75.Rumachik N.G., Malaker S.A., Paulk N.K. VectorMOD: method for bottom-up proteomic characterization of rAAV capsid post-translational modifications and vector impurities. Front. Immunol. 2021;12:657795. doi: 10.3389/fimmu.2021.657795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu Z., Asokan A., Grieger J.C., Govindasamy L., Agbandje-McKenna M., Samulski R.J. Single amino acid changes can influence titer, heparin binding, and tissue tropism in different adeno-associated virus serotypes. J. Virol. 2006;80:11393–11397. doi: 10.1128/JVI.01288-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Woodard K.T., Liang K.J., Bennett W.C., Samulski R.J. Heparan sulfate binding promotes accumulation of intravitreally delivered adeno-associated viral vectors at the retina for enhanced transduction but weakly influences tropism. J. Virol. 2016;90:9878–9888. doi: 10.1128/JVI.01568-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li C., Diprimio N., Bowles D.E., Hirsch M.L., Monahan P.E., Asokan A., Rabinowitz J., Agbandje-McKenna M., Samulski R.J. Single amino acid modification of adeno-associated virus capsid changes transduction and humoral immune profiles. J. Virol. 2012;86:7752–7759. doi: 10.1128/JVI.00675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hauck B., Xiao W. Characterization of tissue tropism determinants of adeno-associated virus type 1. J. Virol. 2003;77:2768–2774. doi: 10.1128/JVI.77.4.2768-2774.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hauck B., Xu R.R., Xie J., Wu W., Ding Q., Sipler M., Wang H., Chen L., Wright J.F., Xiao W. Efficient AAV1-AAV2 hybrid vector for gene therapy of hemophilia. Hum. Gene Ther. 2006;17:46–54. doi: 10.1089/hum.2006.17.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Asokan A., Conway J.C., Phillips J.L., Li C., Hegge J., Sinnott R., Yadav S., DiPrimio N., Nam H.-J., Agbandje-McKenna M., et al. Reengineering a receptor footprint of adeno-associated virus enables selective and systemic gene transfer to muscle. Nat. Biotechnol. 2010;28:79–82. doi: 10.1038/nbt.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shen S., Horowitz E.D., Troupes A.N., Brown S.M., Pulicherla N., Samulski R.J., Agbandje-McKenna M., Asokan A. Engraftment of a galactose receptor footprint onto adeno-associated viral capsids improves transduction efficiency. J. Biol. Chem. 2013;288:28814–28823. doi: 10.1074/jbc.M113.482380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murlidharan G., Sakamoto K., Rao L., Corriher T., Wang D., Gao G., Sullivan P., Asokan A. CNS-Restricted transduction and CRISPR/Cas9-mediated gene deletion with an engineered AAV vector. Mol. Ther. Nucleic Acids. 2016;5:e338. doi: 10.1038/mtna.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vandenberghe L.H., Wang L., Somanathan S., Zhi Y., Figueredo J., Calcedo R., Sanmiguel J., Desai R.A., Chen C.S., Johnston J., et al. Heparin binding directs activation of T cells against adeno-associated virus serotype 2 capsid. Nat. Med. 2006;12:967–971. doi: 10.1038/nm1445. [DOI] [PubMed] [Google Scholar]
- 85.Viney L., Bürckstümmer T., Eddington C., Mietzsch M., Choudhry M., Henley T., Agbandje-McKenna M. Adeno-associated virus (AAV) capsid chimeras with enhanced infectivity reveal a core element in the AAV genome critical for both cell transduction and capsid assembly. J. Virol. 2021;95 doi: 10.1128/JVI.02023-20. JVI.02023-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ho M.L., Adler B.A., Torre M.L., Silberg J.J., Suh J. SCHEMA computational design of virus capsid chimeras: calibrating how genome packaging, protection, and transduction correlate with calculated structural disruption. ACS Synth. Biol. 2013;2:724–733. doi: 10.1021/sb400076r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Q., Mamounas M., Yu G., Kennedy S., Leaker B., Merson J., Wong-Staal F., Yu M., Barber J.R. Development of novel cell surface CD34-targeted recombinant adenoassociated virus vectors for gene therapy. Hum. Gene Ther. 1998;9:1929–1937. doi: 10.1089/hum.1998.9.13-1929. [DOI] [PubMed] [Google Scholar]
- 88.Rabinowitz J.E., Xiao W., Samulski R.J. Insertional mutagenesis of AAV2 capsid and the production of recombinant virus. Virology. 1999;265:274–285. doi: 10.1006/viro.1999.0045. [DOI] [PubMed] [Google Scholar]
- 89.Girod A., Ried M., Wobus C., Lahm H., Leike K., Kleinschmidt J., Deléage G., Hallek M. Genetic capsid modifications allow efficient re-targeting of adeno- associated virus type 2. Nat. Med. 1999;5:1438. doi: 10.1038/71021. [DOI] [PubMed] [Google Scholar]
- 90.Huttner N.A., Girod A., Perabo L., Edbauer D., Kleinschmidt J.A., Büning H., Hallek M. Genetic modifications of the adeno-associated virus type 2 capsid reduce the affinity and the neutralizing effects of human serum antibodies. Gene Ther. 2003;10:2139–2147. doi: 10.1038/sj.gt.3302123. [DOI] [PubMed] [Google Scholar]
- 91.Grifman M., Trepel M., Speece P., Gilbert L.B., Arap W., Pasqualini R., Weitzman M.D. Incorporation of tumor-targeting peptides into recombinant adeno-associated virus capsids. Mol. Ther. 2001;3:964–975. doi: 10.1006/mthe.2001.0345. [DOI] [PubMed] [Google Scholar]
- 92.Shi W., Bartlett J.S. RGD inclusion in VP3 provides adeno-associated virus type 2 (AAV2)-based vectors with a heparan sulfate-independent cell entry mechanism. Mol. Ther. 2003;7:515–525. doi: 10.1016/s1525-0016(03)00042-x. [DOI] [PubMed] [Google Scholar]
- 93.Boucas J., Lux K., Huber A., Schievenbusch S., von Freyend M.J., Perabo L., Quadt-Humme S., Odenthal M., Hallek M., Büning H. Engineering adeno-associated virus serotype 2-based targeting vectors using a new insertion site-position 453-and single point mutations. J. Gene Med. 2009;11:1103–1113. doi: 10.1002/jgm.1392. [DOI] [PubMed] [Google Scholar]
- 94.Yu C.Y., Yuan Z., Cao Z., Wang B., Qiao C., Li J., Xiao X. A muscle-targeting peptide displayed on AAV2 improves muscle tropism on systemic delivery. Gene Ther. 2009;16:953–962. doi: 10.1038/gt.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Perabo L., Goldnau D., White K., Endell J., Boucas J., Humme S., Work L.M., Janicki H., Hallek M., Baker A.H., Büning H. Heparan sulfate proteoglycan binding properties of adeno-associated virus retargeting mutants and consequences for their in vivo tropism. J. Virol. 2006;80:7265–7269. doi: 10.1128/JVI.00076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu P., Xiao W., Conlon T., Hughes J., Agbandje-McKenna M., Ferkol T., Flotte T., Muzyczka N. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 2000;74:8635–8647. doi: 10.1128/jvi.74.18.8635-8647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Warrington K.H., Gorbatyuk O.S., Harrison J.K., Opie S.R., Zolotukhin S., Muzyczka N. Adeno-associated virus type 2 VP2 capsid protein is nonessential and can tolerate large peptide insertions at its N terminus. J. Virol. 2004;78:6595–6609. doi: 10.1128/JVI.78.12.6595-6609.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Münch R.C., Janicki H., Völker I., Rasbach A., Hallek M., Büning H., Buchholz C.J. Displaying high-affinity ligands on adeno-associated viral vectors enables tumor cell-specific and safe gene transfer. Mol. Ther. 2013;21:109–118. doi: 10.1038/mt.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Finet J.E., Wan X., Donahue J.K. Fusion of Anthopleurin-B to AAV2 increases specificity of cardiac gene transfer. Virology. 2018;513:43–51. doi: 10.1016/j.virol.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hida K., Won S.Y., Di Pasquale G., Hanes J., Chiorini J.A., Ostermeier M. Sites in the AAV5 capsid tolerant to deletions and tandem duplications. Arch. Biochem. Biophys. 2010;496:1–8. doi: 10.1016/j.abb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sayroo R., Nolasco D., Yin Z., Colon-Cortes Y., Pandya M., Ling C., Aslanidi G. Development of novel AAV serotype 6 based vectors with selective tropism for human cancer cells. Gene Ther. 2016;23:18–25. doi: 10.1038/gt.2015.89. [DOI] [PubMed] [Google Scholar]
- 102.Michelfelder S., Varadi K., Raupp C., Hunger A., Körbelin J., Pahrmann C., Schrepfer S., Müller O.J., Kleinschmidt J.A., Trepel M. Peptide ligands incorporated into the threefold spike capsid domain to re-direct gene transduction of AAV8 and AAV9 in vivo. PLoS ONE. 2011;6:e23101. doi: 10.1371/journal.pone.0023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Eichhoff A.M., Börner K., Albrecht B., Schäfer W., Baum N., Haag F., Körbelin J., Trepel M., Braren I., Grimm D., et al. Nanobody-enhanced targeting of AAV gene therapy vectors. Mol. Ther. Methods Clin. Dev. 2019;15:211–220. doi: 10.1016/j.omtm.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jackson C.B., Richard A.S., Ojha A., Conkright K.A., Trimarchi J.M., Bailey C.C., Alpert M.D., Kay M.A., Farzan M., Choe H. AAV vectors engineered to target insulin receptor greatly enhance intramuscular gene delivery. Mol. Ther. Methods Clin. Dev. 2020;19:496–506. doi: 10.1016/j.omtm.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lin K., Zhong X., Li L., Ying M., Yang T., Zhang Z., He X., Xu F. AAV9-Retro mediates efficient transduction with axon terminal absorption and blood–brain barrier transportation. Mol. Brain. 2020;13:138. doi: 10.1186/s13041-020-00679-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang Y.-S., Xie J., Chaugule S., Wang D., Kim J.-M., Kim J., Tai P.W.L., Seo S.K., Gravallese E., Gao G., Shim J.H. Bone-targeting AAV-mediated gene silencing in osteoclasts for osteoporosis therapy. Mol. Ther. Methods Clin. Dev. 2020;17:922–935. doi: 10.1016/j.omtm.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Börner K., Kienle E., Huang L.-Y., Weinmann J., Sacher A., Bayer P., Stüllein C., Fakhiri J., Zimmermann L., Westhaus A., et al. Pre-arrayed pan-AAV peptide display libraries for rapid single-round screening. Mol. Ther. 2020;28:1016–1032. doi: 10.1016/j.ymthe.2020.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao H., Arnold F.H. Optimization of DNA shuffling for high fidelity recombination. Nucleic Acids Res. 1997;25:1307–1308. doi: 10.1093/nar/25.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Grimm D., Lee J.S., Wang L., Desai T., Akache B., Storm T.A., Kay M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Katada Y., Kobayashi K., Tsubota K., Kurihara T. Evaluation of AAV-DJ vector for retinal gene therapy. PeerJ. 2019;7:e6317. doi: 10.7717/peerj.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li W., Asokan A., Wu Z., Van Dyke T., DiPrimio N., Johnson J.S., Govindaswamy L., Agbandje-McKenna M., Leichtle S., Eugene Redmond D., Jr., et al. Engineering and selection of shuffled AAV genomes: a new strategy for producing targeted biological nanoparticles. Mol. Ther. 2008;16:1252–1260. doi: 10.1038/mt.2008.100. [DOI] [PubMed] [Google Scholar]
- 112.Gray S.J., Blake B.L., Criswell H.E., Nicolson S.C., Samulski R.J., McCown T.J., Li W. Directed evolution of a novel adeno-associated virus (AAV) vector that crosses the seizure-compromised blood-brain barrier (BBB) Mol. Ther. 2010;18:570–578. doi: 10.1038/mt.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Charbel Issa P., de Silva S.R., Lipinski D.M., Singh M.S., Mouravlev A., You Q., Barnard A.R., Hankins M.W., During M.J., MacLaren R.E. Assessment of tropism and effectiveness of new primate-derived hybrid recombinant AAV serotypes in the mouse and primate retina. PLoS One. 2013;8:603611. doi: 10.1371/journal.pone.0060361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu X., Magee D., Wang C., McMurphy T., Slater A., During M., Cao L. Adipose tissue insulin receptor knockdown via a new primate-derived hybrid recombinant AAV serotype. Mol. Ther. - Methods Clin. Develop. 2014;1:8. doi: 10.1038/mtm.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Siu J.J., Queen N.J., Huang W., Yin F.Q., Liu X., Wang C., McTigue D.M., Cao L. Improved gene delivery to adult mouse spinal cord through the use of engineered hybrid adeno-associated viral serotypes. Gene Ther. 2017;24:361–369. doi: 10.1038/gt.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Powell S.K., Khan N., Parker C.L., Samulski R.J., Matsushima G., Gray S.J., McCown T.J. Characterization of a novel adeno-associated viral vector with preferential oligodendrocyte tropism. Gene Ther. 2016;23:807–814. doi: 10.1038/gt.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pei X., Shao W., Xing A., Askew C., Chen X., Cui C., Abajas Y.L., Gerber D.A., Merricks E.P., Nichols T.C., et al. Development of AAV variants with human hepatocyte tropism and neutralizing antibody escape capacity. Mol. Ther. Methods Clin. Dev. 2020;18:259–268. doi: 10.1016/j.omtm.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tang Y., Yan Z., Lin S., Huntemann E.D., Feng Z., Park S.-Y., Sun X., Yuen E., Engelhardt J.F. Repeat dosing of AAV2.5T to ferret lungs elicits an antibody response that diminishes transduction in an age-dependent manner. Mol. Ther. Methods Clin. Dev. 2020;19:186–200. doi: 10.1016/j.omtm.2020.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhao H., Giver L., Shao Z., Affholter J.A., Arnold F.H. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat. Biotechnol. 1998;16:258–261. doi: 10.1038/nbt0398-258. [DOI] [PubMed] [Google Scholar]
- 120.Perabo L., Endell J., King S., Lux K., Goldnau D., Hallek M., Büning H. Combinatorial engineering of a gene therapy vector: directed evolution of adeno-associated virus. J. Gene Med. 2006;8:155–162. doi: 10.1002/jgm.849. [DOI] [PubMed] [Google Scholar]
- 121.Koerber J.T., Maheshri N., Kaspar B.K., Schaffer D.V. Construction of diverse adeno-associated viral libraries for directed evolution of enhanced gene delivery vehicles. Nat. Protoc. 2006;1:701–706. doi: 10.1038/nprot.2006.93. [DOI] [PubMed] [Google Scholar]
- 122.Maheshri N., Koerber J.T., Kaspar B.K., Schaffer D.V. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat. Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- 123.Qian R., Xiao B., Li J., Xiao X. Directed evolution of AAV serotype 5 for increased hepatocyte transduction and retained low humoral seroreactivity. Mol. Ther. Methods Clin. Dev. 2021;20:122–132. doi: 10.1016/j.omtm.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ivanchenko M.V., Hanlon K.S., Hathaway D.M., Klein A.J., Peters C.W., Li Y., Tamvakologos P.I., Nammour J., Maguire C.A., Corey D.P. AAV-S: a versatile capsid variant for transduction of mouse and primate inner ear. Mol. Ther. Methods Clin. Dev. 2021;21:382–398. doi: 10.1016/j.omtm.2021.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ogden P.J., Kelsic E.D., Sinai S., Church G.M. Comprehensive AAV capsid fitness landscape reveals a viral gene and enables machine-guided design. Science. 2019;366:1139–1143. doi: 10.1126/science.aaw2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Weinmann J., Weis S., Sippel J., Tulalamba W., Remes A., El Andari J., Herrmann A.-K., Pham Q.H., Borowski C., Hille S., et al. Identification of a myotropic AAV by massively parallel in vivo evaluation of barcoded capsid variants. Nat. Commun. 2020;11:5432. doi: 10.1038/s41467-020-19230-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bryant D.H., Bashir A., Sinai S., Jain N.K., Ogden P.J., Riley P.F., Church G.M., Colwell L.J., Kelsic E.D. Deep diversification of an AAV capsid protein by machine learning. Nat. Biotechnol. 2021;39:691–696. doi: 10.1038/s41587-020-00793-4. [DOI] [PubMed] [Google Scholar]
- 128.Havlik L.P., Simon K.E., Smith J.K., Klinc K.A., Tse L.V., Oh D.K., Fanous M.M., Meganck R.M., Mietzsch M., Kleinschmidt J., et al. Coevolution of adeno-associated virus capsid antigenicity and tropism through a structure-guided approach. J. Virol. 2020;94 doi: 10.1128/JVI.00976-20. e009766-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Robinson T.M., Ho M.L., Wahlig B., Gough V., Banta A., Reyes Gamas K., Kang B., Lee E., Chen W., Suh J. An essential N-terminal serine-rich motif in the AAV VP1 and VP2 subunits that may play a role in viral transcription. Virology. 2020;546:127–132. doi: 10.1016/j.virol.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Müller O.J., Kaul F., Weitzman M.D., Pasqualini R., Arap W., Kleinschmidt J.A., Trepel M. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat. Biotechnol. 2003;21:1040–1046. doi: 10.1038/nbt856. [DOI] [PubMed] [Google Scholar]
- 131.Sellner L., Stiefelhagen M., Kleinschmidt J.A., Laufs S., Wenz F., Fruehauf S., Zeller W.J., Veldwijk M.R. Generation of efficient human blood progenitor-targeted recombinant adeno-associated viral vectors (AAV) by applying an AAV random peptide library on primary human hematopoietic progenitor cells. Exp. Hematol. 2008;36:957–964. doi: 10.1016/j.exphem.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 132.Sallach J., Di Pasquale G., Larcher F., Niehoff N., Rübsam M., Huber A., Chiorini J., Almarza D., Eming S.A., Ulus H., et al. Tropism-modified AAV vectors overcome barriers to successful cutaneous therapy. Mol. Ther. 2014;22:929–939. doi: 10.1038/mt.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Waterkamp D.A., Müller O.J., Ying Y., Trepel M., Kleinschmidt J.A. Isolation of targeted AAV2 vectors from novel virus display libraries. J. Gene Med. 2006;8:1307–1319. doi: 10.1002/jgm.967. [DOI] [PubMed] [Google Scholar]
- 134.Michelfelder S., Kohlschütter J., Skorupa A., Pfennings S., Müller O., Kleinschmidt J.A., Trepel M. Successful expansion but not complete restriction of tropism of adeno-associated virus by in vivo biopanning of random virus display peptide libraries. PLoS ONE. 2009;4:e5122. doi: 10.1371/journal.pone.0005122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Trepel M., Körbelin J., Spies E., Heckmann M.B., Hunger A., Fehse B., Katus H.A., Kleinschmidt J.A., Müller O.J., Michelfelder S. Treatment of multifocal breast cancer by systemic delivery of dual-targeted adeno-associated viral vectors. Gene Ther. 2015;22:848. doi: 10.1038/gt.2015.76. [DOI] [PubMed] [Google Scholar]
- 136.Körbelin J., Sieber T., Michelfelder S., Lunding L., Spies E., Hunger A., Alawi M., Rapti K., Indenbirken D., Müller O.J., et al. Pulmonary targeting of adeno-associated viral vectors by next-generation sequencing-guided screening of random capsid displayed peptide libraries. Mol. Ther. 2016;24:1050–1061. doi: 10.1038/mt.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Körbelin J., Hunger A., Alawi M., Sieber T., Binder M., Trepel M. Optimization of design and production strategies for novel adeno-associated viral display peptide libraries. Gene Ther. 2017;24:470–481. doi: 10.1038/gt.2017.51. [DOI] [PubMed] [Google Scholar]
- 138.Schmit P.F., Pacouret S., Zinn E., Telford E., Nicolaou F., Broucque F., Andres-mateos E., Xiao R., Penaud-budloo M., Bouzelha M., et al. Cross-packaging and capsid mosaic formation in multiplexed AAV libraries. Mol. Ther. Methods Clin. Dev. 2020;17:107–121. doi: 10.1016/j.omtm.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Körbelin J., Trepel M. How to successfully screen random adeno-associated virus display peptide libraries in vivo. Hum. Gene Ther. Methods. 2017;28:109–123. doi: 10.1089/hgtb.2016.177. [DOI] [PubMed] [Google Scholar]
- 140.Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.L., Yang B., Huber N., Pasca S.P., Gradinaru V. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chan K.Y., Jang M.J., Yoo B.B., Greenbaum A., Ravi N., Wu W.L., Sánchez-Guardado L., Lois C., Mazmanian S.K., Deverman B.E., Gradinaru V. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017;20:1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hordeaux J., Wang Q., Katz N., Buza E.L., Bell P., Wilson J.M. The neurotropic properties of AAV-PHP.B are limited to C57BL/6J mice. Mol. Ther. 2018;26:664–668. doi: 10.1016/j.ymthe.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Huang Q., Chan K.Y., Tobey I.G., Chan Y.A., Poterba T., Boutros C.L., Balazs A.B., Daneman R., Bloom J.M., Seed C., et al. Delivering genes across the blood-brain barrier: LY6A, a novel cellular receptor for AAV-PHP.B capsids. bioRxiv. 2019 doi: 10.1101/538421. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Matsuzaki Y., Konno A., Mochizuki R., Shinohara Y., Nitta K., Okada Y., Hirai H. Intravenous administration of the adeno-associated virus-PHP.B capsid fails to upregulate transduction efficiency in the marmoset brain. Neurosci. Lett. 2018;665:182–188. doi: 10.1016/j.neulet.2017.11.049. [DOI] [PubMed] [Google Scholar]
- 145.Matsuzaki Y., Tanaka M., Hakoda S., Masuda T., Miyata R., Konno A., Hirai H. Neurotropic properties of AAV-PHP.B are shared among diverse inbred strains of mice. Mol. Ther. 2019;27:700–704. doi: 10.1016/j.ymthe.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dayton R.D., Grames M.S., Klein R.L. More expansive gene transfer to the rat CNS: AAV PHP.EB vector dose–response and comparison to AAV PHP. Gene Ther. 2018;25:392–400. doi: 10.1038/s41434-018-0028-5. [DOI] [PubMed] [Google Scholar]
- 147.Hordeaux J., Yuan Y., Clark P.M., Wang Q., Martino R.A., Sims J.J., Bell P., Raymond A., Stanford W.L., Wilson J.M. The GPI-linked protein LY6A drives AAV-PHP.B transport across the blood-brain barrier. Mol. Ther. 2019;27:912–921. doi: 10.1016/j.ymthe.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nonnenmacher M., Wang W., Child M.A., Ren X.-Q., Huang C., Ren A.Z., Tocci J., Chen Q., Bittner K., Tyson K., et al. Rapid evolution of blood-brain-barrier-penetrating AAV capsids by RNA-driven biopanning. Mol. Ther. Methods Clin. Dev. 2021;20:366–378. doi: 10.1016/j.omtm.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee G.K., Maheshri N., Kaspar B., Schaffer D.V. PEG conjugation moderately protects adeno-associated viral vectors against antibody neutralization. Biotechnol. Bioeng. 2005;92:24–34. doi: 10.1002/bit.20562. [DOI] [PubMed] [Google Scholar]
- 150.Horowitz E.D., Weinberg M.S., Asokan A. Glycated AAV vectors: chemical redirection of viral tissue tropism. Bioconjug. Chem. 2011;22:529–532. doi: 10.1021/bc100477g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stachler M.D., Chen I., Ting A.Y., Bartlett J.S. Site-specific modification of AAV vector particles with biophysical probes and targeting ligands using biotin ligase. Mol. Ther. 2008;16:1467–1473. doi: 10.1038/mt.2008.129. [DOI] [PubMed] [Google Scholar]
- 152.Liu Y., Fang Y., Zhou Y., Zandi E., Lee C.L., Joo K.I., Wang P. Site-specific modification of adeno-associated viruses via a genetically engineered aldehyde tag. Small. 2013;9:421–429. doi: 10.1002/smll.201201661. [DOI] [PubMed] [Google Scholar]
- 153.Kelemen R.E., Mukherjee R., Cao X., Erickson S.B., Zheng Y., Chatterjee A. A precise chemical strategy to alter the receptor specificity of the adeno-associated virus. Angew. Chem. Int. Ed. Engl. 2016;55:10645–10649. doi: 10.1002/anie.201604067. [DOI] [PubMed] [Google Scholar]
- 154.Liu C.C., Schultz P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 155.Dumas A., Lercher L., Spicer C.D., Davis B.G. Designing logical codon reassignment - expanding the chemistry in biology. Chem. Sci. 2015;6:50–69. doi: 10.1039/c4sc01534g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yao T., Zhou X., Zhang C., Yu X., Tian Z., Zhang L., Zhou D. Site-specific PEGylated adeno-associated viruses with increased serum stability and reduced immunogenicity. Molecules. 2017;22:1155. doi: 10.3390/molecules22071155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Katrekar D., Moreno A.M., Chen G., Worlikar A., Mali P. Oligonucleotide conjugated multi-functional adeno-associated viruses. Sci. Rep. 2018;8:3589–3598. doi: 10.1038/s41598-018-21742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bartlett J.S., Kleinschmidt J., Boucher R.C., Samulski R.J. Targeted adeno-associated virus vector transduction of nonpermissive cells mediated by a bispecific f(ab’γ)2 antibody. Nat. Biotechnol. 1999;17:181–186. doi: 10.1038/6185. [DOI] [PubMed] [Google Scholar]
- 159.Merchan J.A., Dean J., Azpurua F., Sen S., Zhu Y., Aikawa R. Utility of vascular endothelial specific peptides for enhancement of adeno-associated virus-mediated gene transfer. Int. J. Biomed. Sci. 2008;4:217–220. [PMC free article] [PubMed] [Google Scholar]
- 160.Pulicherla N., Asokan A. Peptide affinity reagents for AAV capsid recognition and purification. Gene Ther. 2011;18:1020–1024. doi: 10.1038/gt.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tabata K., Sugano E., Murakami F., Yamashita T., Ozaki T., Tomita H. Improved transduction efficiencies of adeno-associated virus vectors by synthetic cell-permeable peptides. Biochem. Biophys. Res. Commun. 2016;478:1732–1738. doi: 10.1016/j.bbrc.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 162.Meng Y., Sun D., Qin Y., Dong X., Luo G., Liu Y. Cell-penetrating peptides enhance the transduction of adeno-associated virus serotype 9 in the central nervous system. Mol. Ther. Methods Clin. Dev. 2021;21:28–41. doi: 10.1016/j.omtm.2021.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang X., He T., Chai Z., Samulski R.J., Li C. Blood-brain barrier shuttle peptides enhance AAV transduction in the brain after systemic administration. Biomaterials. 2018;176:71–83. doi: 10.1016/j.biomaterials.2018.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Maguire C.A., Balaj L., Sivaraman S., Crommentuijn M.H.W., Ericsson M., Mincheva-Nilsson L., Baranov V., Gianni D., Tannous B.A., Sena-Esteves M., et al. Microvesicle-associated AAV vector as a novel gene delivery system. Mol. Ther. 2012;20:960–971. doi: 10.1038/mt.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.György B., Fitzpatrick Z., Crommentuijn M.H.W., Mu D., Maguire C.A. Naturally enveloped AAV vectors for shielding neutralizing antibodies and robust gene delivery invivo. Biomaterials. 2014;35:7598–7609. doi: 10.1016/j.biomaterials.2014.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hudry E., Martin C., Gandhi S., György B., Scheffer D.I., Mu D., Merkel S.F., Mingozzi F., Fitzpatrick Z., Dimant H., et al. Exosome-associated AAV vector as a robust and convenient neuroscience tool HHS Public Access Author manuscript. Gene Ther. 2016;23:380–392. doi: 10.1038/gt.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Meliani A., Boisgerault F., Fitzpatrick Z., Marmier S., Leborgne C., Collaud F., Simon Sola M., Charles S., Ronzitti G., Vignaud A., et al. Enhanced liver gene transfer and evasion of preexisting humoral immunity with exosome-enveloped AAV vectors. Blood Adv. 2017;1:2019–2031. doi: 10.1182/bloodadvances.2017010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.György B., Sage C., Indzhykulian A.A., Scheffer D.I., Brisson A.R., Tan S., Wu X., Volak A., Mu D., Tamvakologos P.I., et al. Rescue of hearing by gene delivery to inner-ear hair cells using exosome-associated AAV. Mol. Ther. 2017;25:379–391. doi: 10.1016/j.ymthe.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wassmer S.J., Carvalho L.S., György B., Vandenberghe L.H., Maguire C.A. Exosome-associated AAV2 vector mediates robust gene delivery into the murine retina upon intravitreal injection. Sci. Rep. 2017;7:45329–45410. doi: 10.1038/srep45329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Khan N., Maurya S., Bammidi S., Jayandharan G.R. AAV6 vexosomes mediate robust suicide gene delivery in a murine model of hepatocellular carcinoma. Mol. Ther. Methods Clin. Dev. 2020;17:497–504. doi: 10.1016/j.omtm.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhu J., Tao P., Mahalingam M., Sha J., Kilgore P., Chopra A.K., Rao V. A prokaryotic-eukaryotic hybrid viral vector for delivery of large cargos of genes and proteins into human cells. Sci. Adv. 2019;5:eaax0064. doi: 10.1126/sciadv.aax0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Thadani N.N., Yang J., Moyo B., Lee C.M., Chen M.Y., Bao G., Suh J. Site-specific post-translational surface modification of adeno-associated virus vectors using leucine zippers. ACS Synth. Biol. 2020;9:461–467. doi: 10.1021/acssynbio.9b00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hauck B., Chen L., Xiao W. Generation and characterization of chimeric recombinant AAV vectors. Mol. Ther. 2003;7:419–425. doi: 10.1016/s1525-0016(03)00012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rabinowitz J.E., Bowles D.E., Faust S.M., Ledford J.G., Cunningham S.E., Samulski R.J. Cross-dressing the virion the transcapsidation of adeno-associated. J. Virol. 2004;78:4421–4432. doi: 10.1128/JVI.78.9.4421-4432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.de Backer M.W.A., Brans M.A.D., Luijendijk M.C., Garner K.M., Adan R.A.H. Optimization of adeno-associated viral vector-mediated gene delivery to the hypothalamus. Hum. Gene Ther. 2010;21:673–682. doi: 10.1089/hum.2009.169. [DOI] [PubMed] [Google Scholar]
- 176.Chai Z., Sun J., Rigsbee K.M., Wang M., Samulski R.J., Li C. Application of polyploid adeno-associated virus vectors for transduction enhancement and neutralizing antibody evasion. J. Control Release. 2017;262:348–356. doi: 10.1016/j.jconrel.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Chai Z., Zhang X., Dobbins A.L., Frost E.A., Samulski R.J., Li C. Chimeric capsid proteins impact transduction efficiency of haploid adeno-associated virus vectors. Viruses. 2019;11:1138. doi: 10.3390/v11121138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Calcedo R., Vandenberghe L.H., Gao G., Lin J., Wilson J.M. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J. Infect. Dis. 2009;199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wang L., Calcedo R., Wang H., Bell P., Grant R., Vandenberghe L.H., Sanmiguel J., Morizono H., Batshaw M.L., Wilson J.M. The pleiotropic effects of natural AAV infections on liver-directed gene transfer in macaques. Mol. Ther. 2010;18:126–134. doi: 10.1038/mt.2009.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Perocheau D.P., Cunningham S., Lee J., Antinao Diaz J., Waddington S.N., Gilmour K., Eaglestone S., Lisowski L., Thrasher A.J., Alexander I.E., et al. Age-related seroprevalence of antibodies against AAV-LK03 in a UK population cohort. Hum. Gene Ther. 2019;30:79–87. doi: 10.1089/hum.2018.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M.F., Masurier C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010;21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 182.Miesbach W., Meijer K., Coppens M., Kampmann P., Klamroth R., Schutgens R., Tangelder M., Castaman G., Schwäble J., Bonig H., et al. Gene therapy with adeno-associated virus vector 5-human factor IX in adults with hemophilia B. Blood. 2018;131:1022–1031. doi: 10.1182/blood-2017-09-804419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Majowicz A., Nijmeijer B., Lampen M.H., Spronck L., de Haan M., Petry H., van Deventer S.J., Meyer C., Tangelder M., Ferreira V. Therapeutic hFIX activity achieved after single AAV5-hFIX treatment in hemophilia B patients and NHPs with pre-existing anti-AAV5 NABs. Mol. Ther. Methods Clin. Dev. 2019;14:27–36. doi: 10.1016/j.omtm.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mays L.E., Wang L., Tenney R., Bell P., Nam H.-J., Lin J., Gurda B., Van Vliet K., Mikals K., Agbandje-McKenna M., Wilson J.M. Mapping the structural determinants responsible for enhanced T cell activation to the immunogenic adeno-associated virus capsid from isolate rhesus 32.33. J. Virol. 2013;87:9473–9485. doi: 10.1128/JVI.00596-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Murphy S.L., Li H., Mingozzi F., Sabatino D.E., Hui D.J., Edmonson S.A., High K.A. Diverse IgG subclass responses to adeno-associated virus infection and vector administration: IgG Subclass Responses to AAV Capsid. J. Med. Virol. 2009;81:65–74. doi: 10.1002/jmv.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Leborgne C., Latournerie V., Boutin S., Desgue D., Quéré A., Pignot E., Collaud F., Charles S., Simon Sola M., Masat E., et al. Prevalence and long-term monitoring of humoral immunity against adeno-associated virus in Duchenne Muscular Dystrophy patients. Cell. Immunol. 2019;342:103780. doi: 10.1016/j.cellimm.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 187.Fitzpatrick Z., Leborgne C., Barbon E., Masat E., Ronzitti G., van Wittenberghe L., Vignaud A., Collaud F., Charles S., Simon Sola M., et al. Influence of pre-existing anti-capsid neutralizing and binding antibodies on AAV vector transduction. Mol. Ther. Methods Clin. Dev. 2018;9:119–129. doi: 10.1016/j.omtm.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ronzitti G., Gross D.-A., Mingozzi F. Human immune responses to adeno-associated virus (AAV) vectors. Front. Immunol. 2020;11:670. doi: 10.3389/fimmu.2020.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Thwaite R., Pagès G., Chillón M., Bosch A. AAVrh.10 immunogenicity in mice and humans. Relevance of antibody cross-reactivity in human gene therapy. Gene Ther. 2015;22:196–201. doi: 10.1038/gt.2014.103. [DOI] [PubMed] [Google Scholar]
- 190.Monteilhet V., Saheb S., Boutin S., Leborgne C., Veron P., Montus M.-F., Moullier P., Benveniste O., Masurier C. A 10 patient case report on the impact of plasmapheresis upon neutralizing factors against adeno-associated virus (AAV) types 1, 2, 6, and 8. Mol. Ther. 2011;19:2084–2091. doi: 10.1038/mt.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Corti M., Elder M., Falk D., Lawson L., Smith B., Nayak S., Conlon T., Clément N., Erger K., Lavassani E., et al. B-cell depletion is protective against anti-AAV capsid immune response: a human subject case study. Mol. Ther. Methods Clin. Dev. 2014;1:14033. doi: 10.1038/mtm.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Leborgne C., Barbon E., Alexander J.M., Hanby H., Delignat S., Cohen D.M., Collaud F., Muraleetharan S., Lupo D., Silverberg J., et al. IgG-cleaving endopeptidase enables in vivo gene therapy in the presence of anti-AAV neutralizing antibodies. Nat. Med. 2020;26:1096–1101. doi: 10.1038/s41591-020-0911-7. [DOI] [PubMed] [Google Scholar]
- 193.Elmore Z.C., Oh D.K., Simon K.E., Fanous M.M., Asokan A. Rescuing AAV gene transfer from neutralizing antibodies with an IgG-degrading enzyme. JCI Insight. 2020;5:e139881. doi: 10.1172/jci.insight.139881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Rasko J., Ozelo M.C., Hoots K., Blatt P., et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 195.Nathwani A.C., Rosales C., McIntosh J., Rastegarlari G., Nathwani D., Raj D., Nawathe S., Waddington S.N., Bronson R., Jackson S., et al. Long-term safety and efficacy following systemic administration of a self-complementary AAV vector encoding human FIX pseudotyped with serotype 5 and 8 capsid proteins. Mol. Ther. 2011;19:876–885. doi: 10.1038/mt.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.George L.A., Sullivan S.K., Giermasz A., Rasko J.E.J., Samelson-Jones B.J., Ducore J., Cuker A., Sullivan L.M., Majumdar S., Teitel J., et al. Hemophilia B gene therapy with a high-specific-activity factor IX variant. N. Engl. J. Med. 2017;377:2215–2227. doi: 10.1056/NEJMoa1708538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Palaschak B., Marsic D., Herzog R.W., Zolotukhin S., Markusic D.M. An immune-competent murine model to study elimination of AAV-transduced hepatocytes by capsid-specific CD8+ T cells. Mol. Ther. Methods Clin. Dev. 2017;5:142–152. doi: 10.1016/j.omtm.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Mingozzi F., Maus M.V., Hui D.J., Sabatino D.E., Murphy S.L., Rasko J.E.J., Ragni M.V., Manno C.S., Sommer J., Jiang H., et al. CD8(+) T-cell responses to adeno-associated virus capsid in humans. Nat. Med. 2007;13:419–422. doi: 10.1038/nm1549. [DOI] [PubMed] [Google Scholar]
- 199.Hui D.J., Edmonson S.C., Podsakoff G.M., Pien G.C., Ivanciu L., Camire R.M., Ertl H., Mingozzi F., High K.A., Basner-Tschakarjan E. AAV capsid CD8+ T-cell epitopes are highly conserved across AAV serotypes. Mol. Ther. Methods Clin. Dev. 2015;2:15029. doi: 10.1038/mtm.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Rogers G.L., Shirley J.L., Zolotukhin I., Kumar S.R.P., Sherman A., Perrin G.Q., Hoffman B.E., Srivastava A., Basner-Tschakarjan E., Wallet M.A., et al. Plasmacytoid and conventional dendritic cells cooperate in crosspriming AAV capsid-specific CD8+ T cells. Blood. 2017;129:3184–3195. doi: 10.1182/blood-2016-11-751040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Shirley J.L., Keeler G.D., Sherman A., Zolotukhin I., Markusic D.M., Hoffman B.E., Morel L.M., Wallet M.A., Terhorst C., Herzog R.W. Type I IFN sensing by cDCs and CD4+ T cell help are both requisite for cross-priming of AAV capsid-specific CD8+ T cells. Mol. Ther. 2020;28:758–770. doi: 10.1016/j.ymthe.2019.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hösel M., Broxtermann M., Janicki H., Esser K., Arzberger S., Hartmann P., Gillen S., Kleeff J., Stabenow D., Odenthal M., et al. Toll-like receptor 2-mediated innate immune response in human nonparenchymal liver cells toward adeno-associated viral vectors. Hepatology. 2012;55:287–297. doi: 10.1002/hep.24625. [DOI] [PubMed] [Google Scholar]
- 203.Zhu J., Huang X., Yang Y. The TLR9-MyD88 pathway is critical for adaptive immune responses to adenoassociated virus gene therapy vectors in mice. J. Clin. Invest. 2009;119:2388–2398. doi: 10.1172/JCI37607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Shao W., Earley L.F., Chai Z., Chen X., Sun J., He T., Deng M., Hirsch M.L., Ting J., Samulski R.J., Li C. Double-stranded RNA innate immune response activation from long-term adeno-associated virus vector transduction. JCI insight. 2018;3:120474. doi: 10.1172/jci.insight.120474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ashley S.N., Bell P., Cutler B.J., Faust S.M., Zhu Y., Rabinowitz J.E., Wilson J.M., Zhu Y., Rabinowitz J.E., Wilson J.M. CpG-depleted adeno-associated virus vectors evade immune detection Find the latest version : CpG-depleted adeno-associated virus vectors evade immune detection. J. Clin. Invest. 2013;123:2994–3001. doi: 10.1172/JCI68205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Xiang Z., Kurupati R.K., Li Y., Kuranda K., Zhou X., Mingozzi F., High K.A., Ertl H.C.J. The effect of CpG sequences on capsid-specific CD8+ T cell responses to AAV vector gene transfer. Mol. Ther. 2020;28:771–783. doi: 10.1016/j.ymthe.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chan Y.K., Wang S.K., Chu C.J., Copland D.A., Letizia A.J., Costa Verdera H., Chiang J.J., Sethi M., Wang M.K., Neidermyer W.J., et al. Engineering adeno-associated viral vectors to evade innate immune and inflammatory responses. Sci. Transl. Med. 2021;13:eabd3438. doi: 10.1126/scitranslmed.abd3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Konkle B.A., Walsh C.E., Escobar M.A., Josephson N.C., Young G., von Drygalski A., McPhee S.W.J., Samulski R.J., Bilic I., de la Rosa M., et al. BAX 335 hemophilia B gene therapy clinical trial results: potential impact of CpG sequences on gene expression. Blood. 2021;137:763–774. doi: 10.1182/blood.2019004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wright J.F. Codon modification and PAMPs in clinical AAV vectors: the tortoise or the hare? Mol. Ther. 2020;28:701–703. doi: 10.1016/j.ymthe.2020.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Nathwani A.C., Tuddenham E.G.D., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C., et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Monahan P.E., Sun J., Gui T., Hu G., Hannah W.B., Wichlan D.G., Wu Z., Grieger J.C., Li C., Suwanmanee T., et al. Employing a gain-of-function factor IX variant R338L to advance the efficacy and safety of hemophilia B human gene therapy: preclinical evaluation supporting an ongoing adeno-associated virus clinical trial. Hum. Gene Ther. 2015;26:69–81. doi: 10.1089/hum.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Pipe S., Stine K., Rajasekhar A., Everington T., Poma A., Crombez E., Hay C.R. 101HEMB01 is a phase 1/2 open-label, single ascending dose-finding trial of DTX101 (AAVrh10FIX) in patients with moderate/severe hemophilia B that demonstrated meaningful but transient expression of human factor IX (hFIX) Blood. 2017;130:3331. [Google Scholar]
- 213.Kishimoto T.K., Ferrari J.D., LaMothe R.A., Kolte P.N., Griset A.P., O’Neil C., Chan V., Browning E., Chalishazar A., Kuhlman W., et al. Improving the efficacy and safety of biologic drugs with tolerogenic nanoparticles. Nat. Nanotechnol. 2016;11:890–899. doi: 10.1038/nnano.2016.135. [DOI] [PubMed] [Google Scholar]
- 214.Zhang A.-H., Rossi R.J., Yoon J., Wang H., Scott D.W. Tolerogenic nanoparticles to induce immunologic tolerance: prevention and reversal of FVIII inhibitor formation. Cell. Immunol. 2016;301:74–81. doi: 10.1016/j.cellimm.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 215.Mazor R., King E.M., Onda M., Cuburu N., Addissie S., Crown D., Liu X.-F., Kishimoto T.K., Pastan I. Tolerogenic nanoparticles restore the antitumor activity of recombinant immunotoxins by mitigating immunogenicity. Proc. Natl. Acad. Sci. USA. 2018;115:E733–E742. doi: 10.1073/pnas.1717063115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Meliani A., Boisgerault F., Hardet R., Marmier S., Collaud F., Ronzitti G., Leborgne C., Costa Verdera H., Simon Sola M., Charles S., et al. Antigen-selective modulation of AAV immunogenicity with tolerogenic rapamycin nanoparticles enables successful vector re-administration. Nat. Commun. 2018;9:4098. doi: 10.1038/s41467-018-06621-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ilyinskii P.O., Michaud A.M., Roy C.J., Rizzo G.L., Elkins S.L., Capela T., Chowdhury A.C., Leung S.S., Kishimoto T.K. Enhancement of liver-directed transgene expression at initial and repeat doses of AAV vectors admixed with ImmTOR nanoparticles. Sci. Adv. 2021;7:eabd0321. doi: 10.1126/sciadv.abd0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Senís E., Fatouros C., Große S., Wiedtke E., Niopek D., Mueller A.K., Börner K., Grimm D. CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol. J. 2014;9:1402–1412. doi: 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- 219.Khani S.C., Pawlyk B.S., Bulgakov O.V., Kasperek E., Young J.E., Adamian M., Sun X., Smith A.J., Ali R.R., Li T. AAV-mediated expression targeting of rod and cone photoreceptors with a human rhodopsin kinase promoter. Invest. Ophthalmol. Vis. Sci. 2007;48:3954–3961. doi: 10.1167/iovs.07-0257. [DOI] [PubMed] [Google Scholar]
- 220.Juven-Gershon T., Cheng S., Kadonaga J.T. Rational design of a super core promoter that enhances gene expression. Nat. Methods. 2006;3:917–922. doi: 10.1038/nmeth937. [DOI] [PubMed] [Google Scholar]
- 221.Greig J.A., Wang Q., Reicherter A.L., Chen S.J., Hanlon A.L., Tipper C.H., Clark K.R., Wadsworth S., Wang L., Wilson J.M. Characterization of adeno-associated viral vector-mediated human factor VIII gene therapy in hemophilia a mice. Hum. Gene Ther. 2017;28:392–402. doi: 10.1089/hum.2016.128. [DOI] [PubMed] [Google Scholar]
- 222.Roberts M.L. Computational Biology and Applied Bioinformatics. InTech); 2011. The use of functional genomics in synthetic promoter design; p. 13. [Google Scholar]
- 223.Chuah M.K., Petrus I., De Bleser P., Le Guiner C., Gernoux G., Adjali O., Nair N., Willems J., Evens H., Rincon M.Y., et al. Liver-specific transcriptional modules identified by genome-wide in silico analysis enable efficient gene therapy in mice and non-human primates. Mol. Ther. 2014;22:1605–1613. doi: 10.1038/mt.2014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Rincon M.Y., VandenDriessche T., Chuah M.K. Gene therapy for cardiovascular disease: advances in vector development, targeting, and delivery for clinical translation. Cardiovasc. Res. 2015;108:4–20. doi: 10.1093/cvr/cvv205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sarcar S., Tulalamba W., Rincon M.Y., Tipanee J., Pham H.Q., Evens H., Boon D., Samara-Kuko E., Keyaerts M., Loperfido M., et al. Next-generation muscle-directed gene therapy by in silico vector design. Nat. Commun. 2019;10:492–516. doi: 10.1038/s41467-018-08283-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Roberts B., Haupt A., Tucker A., Grancharova T., Arakaki J., Fuqua M.A., Nelson A., Hookway C., Ludmann S.A., Mueller I.A., et al. Systematic gene tagging using CRISPR/Cas9 in human stem cells to illuminate cell organization. Mol. Biol. Cell. 2017;28:2854–2874. doi: 10.1091/mbc.E17-03-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Iglesias J.M., Fraser R.M., Moran S., Rio P.D., Baker K., Cooper S., Whyteside G., Kippen N., Koutsoupi P., Rajan R., et al. A novel genomics-based platform for the creation of environmental-responsive gene promoters. Mol. Ther. 2018;26:5S1. [Google Scholar]
- 228.Green J.C.G., Willacy T.J., Yanez J., Iglesias J.M., Macnamara K., Schneider J. Identification of novel muscle specific promoters for AAV gene expression in skeletal and cardiac muscles. Mol. Ther. 2019;27:4S1. [Google Scholar]
- 229.Tijani M., Privolizzi R., Oosterveen T., Iglesias J.M., Cooper S., Waddington S.N., Roberts M.L. Development of novel promoters for neurological gene therapy. Mol. Ther. 2019;27:4S1. [Google Scholar]
- 230.Whyteside G., Braae A., Torranc V., Kippen N., Iglesias J.M., Karda R., Waddington S., Fraser R.M., Cuna J.O.Y., Baker K., et al. Controlled gene expression in the liver; design of constitutive, inducible and repressible synthetic promoters for use in gene medicine and their in vivo validation. Mol. Ther. 2019;27:4S1. [Google Scholar]
- 231.Flotte T.R., Afione S.A., Solow R., Drumm M.L., Markakis D., Guggino W.B., Zeitlin P.L., Carter B.J. Expression of the cystic fibrosis transmembrane conductance regulator from a novel adeno-associated virus promoter. J. Biol. Chem. 1993;268:3781–3790. [PubMed] [Google Scholar]
- 232.Haberman R.P., McCown T.J., Samulski R.J. Novel transcriptional regulatory signals in the adeno-associated virus terminal repeat A/D junction element. J. Virol. 2000;74:8732–8739. doi: 10.1128/jvi.74.18.8732-8739.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ling C., Wang Y., Lu Y., Wang L., Jayandharan G.R., Aslanidi G.V., Li B., Cheng B., Ma W., Lentz T., et al. Enhanced transgene expression from recombinant single-stranded D-sequence-substituted adeno-associated virus vectors in human cell lines in vitro and in murine hepatocytes in vivo. J. Virol. 2015;89:952–961. doi: 10.1128/JVI.02581-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Brown N., Song L., Kollu N.R., Hirsch M.L. Adeno-associated virus vectors and stem cells: friends or foes? Hum. Gene Ther. 2017;28:450–463. doi: 10.1089/hum.2017.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Ling C., Yin Z., Li J., Zhang D., Aslanidi G., Srivastava A. Strategies to generate high-titer, high-potency recombinant AAV3 serotype vectors. Mol. Ther. Methods Clin. Dev. 2016;3:16029. doi: 10.1038/mtm.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Grimm D., Pandey K., Nakai H., Storm T.A., Kay M.A. Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J. Virol. 2006;80:426–439. doi: 10.1128/JVI.80.1.426-439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Hewitt F.C., Li C., Gray S.J., Cockrell S., Washburn M., Samulski R.J. Reducing the risk of adeno-associated virus (AAV) vector mobilization with AAV type 5 vectors. J. Virol. 2009;83:3919–3929. doi: 10.1128/JVI.02466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Miyazaki J., Takaki S., Araki K., Tashiro F., Tominaga A., Takatsu K., Yamamura K. Expression vector system based on the chicken β-actin promoter directs efficient production of interleukin-5. Gene. 1989;79:269–277. doi: 10.1016/0378-1119(89)90209-6. [DOI] [PubMed] [Google Scholar]
- 239.Choi T., Huang M., Gorman C., Jaenisch R. A generic intron increases gene expression in transgenic mice. Mol. Cell. Biol. 1991;11:3070–3074. doi: 10.1128/mcb.11.6.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wu Z., Sun J., Zhang T., Yin C., Yin F., Van Dyke T., Samulski R.J., Monahan P.E. Optimization of self-complementary AAV vectors for liver-directed expression results in sustained correction of hemophilia B at low vector dose. Mol. Ther. 2008;16:280–289. doi: 10.1038/sj.mt.6300355. [DOI] [PubMed] [Google Scholar]
- 241.Schambach A., Mueller D., Galla M., Verstegen M.M.A., Wagemaker G., Loew R., Baum C., Bohne J. Overcoming promoter competition in packaging cells improves production of self-inactivating retroviral vectors. Gene Ther. 2006;13:1524–1533. doi: 10.1038/sj.gt.3302807. [DOI] [PubMed] [Google Scholar]
- 242.Choi J.-H., Yu N.-K., Baek G.-C., Bakes J., Seo D., Nam H.J., Baek S.H., Lim C.-S., Lee Y.-S., Kaang B.-K. Optimization of AAV expression cassettes to improve packaging capacity and transgene expression in neurons. Mol. Brain. 2014;7:17. doi: 10.1186/1756-6606-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Patrício M.I., Barnard A.R., Orlans H.O., McClements M.E., MacLaren R.E. Inclusion of the woodchuck hepatitis virus posttranscriptional regulatory element enhances AAV2-driven transduction of mouse and human retina. Mol. Ther. Nucleic Acids. 2017;6:198–208. doi: 10.1016/j.omtn.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wang D., Zhong L., Li M., Li J., Tran K., Ren L., He R., Xie J., Moser R.P., Fraser C., et al. Adeno-associated virus neutralizing antibodies in large animals and their impact on brain intraparenchymal gene transfer. Mol. Ther. Methods Clin. Dev. 2018;11:65–72. doi: 10.1016/j.omtm.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Verghese S.C., Goloviznina N.A., Skinner A.M., Lipps H.J., Kurre P. S/MAR sequence confers long-term mitotic stability on non-integrating lentiviral vector episomes without selection. Nucleic Acids Res. 2014;42:e53. doi: 10.1093/nar/gku082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Jin C., Fotaki G., Ramachandran M., Nilsson B., Essand M., Yu D. Safe engineering of CAR T cells for adoptive cell therapy of cancer using long-term episomal gene transfer. EMBO Mol. Med. 2016;8:702–711. doi: 10.15252/emmm.201505869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Hagedorn C., Schnödt-Fuchs M., Boehme P., Abdelrazik H., Lipps H.J., Büning H. S/MAR element facilitates episomal long-term persistence of adeno-associated virus vector genomes in proliferating cells. Hum. Gene Ther. 2017;28:1169–1179. doi: 10.1089/hum.2017.025. [DOI] [PubMed] [Google Scholar]
- 248.Xie Q., Lerch T.F., Meyer N.L., Chapman M.S. Structure-function analysis of receptor-binding in adeno-associated virus serotype 6 (AAV-6) Virology. 2011;420:10–19. doi: 10.1016/j.virol.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Reid C.A., Boye S.L., Hauswirth W.W., Lipinski D.M. MiRNA-mediated post-transcriptional silencing of transgenes leads to increased adeno-associated viral vector yield and targeting specificity. Gene Ther. 2017;24:462–469. doi: 10.1038/gt.2017.50. [DOI] [PubMed] [Google Scholar]
- 250.Majowicz A., Maczuga P., Kwikkers K.L., van der Marel S., van Logtenstein R., Petry H., van Deventer S.J., Konstantinova P., Ferreira V. Mir-142-3p target sequences reduce transgene-directed immunogenicity following intramuscular adeno-associated virus 1 vector-mediated gene delivery. J. Gene Med. 2013;15:219–232. doi: 10.1002/jgm.2712. [DOI] [PubMed] [Google Scholar]
- 251.Brown B.D., Venneri M.A., Zingale A., Sergi Sergi L., Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat. Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 252.Roudaut C., Le Roy F., Suel L., Poupiot J., Charton K., Bartoli M., Richard I. Restriction of calpain3 expression to the skeletal muscle prevents cardiac toxicity and corrects pathology in a murine model of limb-girdle muscular dystrophy. Circulation. 2013;128:1094–1104. doi: 10.1161/CIRCULATIONAHA.113.001340. [DOI] [PubMed] [Google Scholar]
- 253.Karali M., Manfredi A., Puppo A., Marrocco E., Gargiulo A., Allocca M., Corte M.D., Rossi S., Giunti M., Bacci M.L., et al. MicroRNA-Restricted transgene expression in the retina. PLoS ONE. 2011;6:e22166. doi: 10.1371/journal.pone.0022166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Waldman Y.Y., Tuller T., Shlomi T., Sharan R., Ruppin E. Translation efficiency in humans: tissue specificity, global optimization and differences between developmental stages. Nucleic Acids Res. 2010;38:2964–2974. doi: 10.1093/nar/gkq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Gaspar P., Moura G., Santos M.A.S., Oliveira J.L. MRNA secondary structure optimization using a correlated stem-loop prediction. Nucleic Acids Res. 2013;41:e73–e77. doi: 10.1093/nar/gks1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Foster H., Sharp P.S., Athanasopoulos T., Trollet C., Graham I.R., Foster K., Wells D.J., Dickson G. Codon and mRNA sequence optimization of microdystrophin transgenes improves expression and physiological outcome in dystrophic mdx mice following AAV2/8 gene transfer. Mol. Ther. 2008;16:1825–1832. doi: 10.1038/mt.2008.186. [DOI] [PubMed] [Google Scholar]
- 257.Bell P., Wang L., Chen S.J., Yu H., Zhu Y., Nayal M., He Z., White J., Lebel-Hagan D., Wilson J.M. Effects of self-complementarity, codon optimization, transgene, and dose on liver transduction with AAV8. Hum. Gene Ther. Methods. 2016;27:228–237. doi: 10.1089/hgtb.2016.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Ronzitti G., Bortolussi G., van Dijk R., Collaud F., Charles S., Leborgne C., Vidal P., Martin S., Gjata B., Sola M.S., et al. A translationally optimized AAV-UGT1A1 vector drives safe and long-lasting correction of Crigler-Najjar syndrome. Mol. Ther. Methods Clin. Dev. 2016;3:16049. doi: 10.1038/mtm.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Beltran W.A., Cideciyan A.V., Boye S.E., Ye G.J., Iwabe S., Dufour V.L., Marinho L.F., Swider M., Kosyk M.S., Sha J., et al. Optimization of retinal gene therapy for X-linked retinitis pigmentosa due to RPGR mutations. Mol. Ther. 2017;25:1866–1880. doi: 10.1016/j.ymthe.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Fischer M.D., McClements M.E., Martinez-Fernandez de la Camara C., Bellingrath J.S., Dauletbekov D., Ramsden S.C., Hickey D.G., Barnard A.R., MacLaren R.E. Codon-optimized RPGR improves stability and efficacy of AAV8 gene therapy in two mouse models of X-linked retinitis pigmentosa. Mol. Ther. 2017;25:1854–1865. doi: 10.1016/j.ymthe.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Grieger J.C., Samulski R.J. Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J. Virol. 2005;79:9933–9944. doi: 10.1128/JVI.79.15.9933-9944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Dong B., Nakai H., Xiao W. Characterization of genome integrity for oversized recombinant AAV vector. Mol. Ther. 2010;18:87–92. doi: 10.1038/mt.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wu Z., Yang H., Colosi P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Grose W.E., Clark K.R., Griffin D., Malik V., Shontz K.M., Montgomery C.L., Lewis S., Brown R.H., Janssen P.M.L., Mendell J.R., Rodino-Klapac L.R. Homologous recombination mediates functional recovery of dysferlin deficiency following AAV5 gene transfer. PLoS ONE. 2012;7:39233–39310. doi: 10.1371/journal.pone.0039233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Hirsch M.L., Li C., Bellon I., Yin C., Chavala S., Pryadkina M., Richard I., Samulski R.J. Oversized AAV transductifon is mediated via a DNA-PKcs-independent, Rad51C-dependent repair pathway. Mol. Ther. 2013;21:2205–2216. doi: 10.1038/mt.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kyostio-Moore S., Berthelette P., Piraino S., Sookdeo C., Nambiar B., Jackson R., Burnham B., O’Riordan C.R., Cheng S.H., Armentano D. The impact of minimally oversized adeno-associated viral vectors encoding human factor VIII on vector potency in vivo. Mol. Ther. Methods Clin. Dev. 2016;3:16006. doi: 10.1038/mtm.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Duan D., Yue Y., Yan Z., Engelhardt J.F. A new dual-vector approach to enhance recombinant adeno-associated virus-mediated gene expression through intermolecular cis activation. Nat. Med. 2000;6:595–598. doi: 10.1038/75080. [DOI] [PubMed] [Google Scholar]
- 268.Nakai H., Storm T.A., Kay M.A. Increasing the size of rAAV-mediated expression cassettes in vivo by intermolecular joining of two complementary vectors. Nat. Biotechnol. 2000;18:527–532. doi: 10.1038/75390. [DOI] [PubMed] [Google Scholar]
- 269.Sun L., Li J., Xiao X. Overcoming adeno-associated virus vector size limitation through viral DNA heterodimerization. Nat. Med. 2000;6:599–602. doi: 10.1038/75087. [DOI] [PubMed] [Google Scholar]
- 270.Yan Z., Zhang Y., Duan D., Engelhardt J.F. Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proc. Natl. Acad. Sci. USA. 2000;97:6716–6721. doi: 10.1073/pnas.97.12.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Lai Y., Yue Y., Liu M., Ghosh A., Engelhardt J.F., Chamberlain J.S., Duan D. Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nat. Biotechnol. 2005;23:1435–1439. doi: 10.1038/nbt1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Duan D., Yue Y., Engelhardt J.F. Expanding AAV packaging capacity with Trans-splicing or overlapping vectors: a quantitative comparison. Mol. Ther. 2001;4:383–391. doi: 10.1006/mthe.2001.0456. [DOI] [PubMed] [Google Scholar]
- 273.Halbert C.L., Allen J.M., Miller A.D. Efficient mouse airway transduction following recombination between AAV vectors carrying parts of a larger gene. Nat. Biotechnol. 2002;20:697–701. doi: 10.1038/nbt0702-697. [DOI] [PubMed] [Google Scholar]
- 274.Lopes V.S., Boye S.E., Louie C.M., Boye S., Dyka F., Chiodo V., Fofo H., Hauswirth W.W., Williams D.S. Retinal gene therapy with a large MYO7A cDNA using adeno-associated virus. Gene Ther. 2013;20:824–833. doi: 10.1038/gt.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Odom G.L., Gregorevic P., Allen J.M., Chamberlain J.S. Gene therapy of mdx mice with large truncated dystrophins generated by recombination using rAAV6. Mol. Ther. 2011;19:36–45. doi: 10.1038/mt.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Pryadkina M., Lostal W., Bourg N., Charton K., Roudaut C., Hirsch M.L., Richard I. A comparison of AAV strategies distinguishes overlapping vectors for efficient systemic delivery of the 6.2 kb Dysferlin coding sequence. Mol. Ther. Methods Clin. Dev. 2015;2:15009. doi: 10.1038/mtm.2015.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ghosh A., Yue Y., Lai Y., Duan D. A hybrid vector system expands adeno-associated viral vector packaging capacity in a transgene-independent manner. Mol. Ther. 2008;16:124–130. doi: 10.1038/sj.mt.6300322. [DOI] [PubMed] [Google Scholar]
- 278.Ghosh A., Yue Y., Duan D. Efficient transgene reconstitution with hybrid dual AAV vectors carrying the minimized bridging sequences. Hum. Gene Ther. 2011;22:77–83. doi: 10.1089/hum.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Colella P., Trapani I., Cesi G., Sommella A., Manfredi A., Puppo A., Iodice C., Rossi S., Simonelli F., Giunti M., et al. Efficient gene delivery to the cone-enriched pig retina by dual AAV vectors. Gene Ther. 2014;21:450–456. doi: 10.1038/gt.2014.8. [DOI] [PubMed] [Google Scholar]
- 280.Trapani I., Colella P., Sommella A., Iodice C., Cesi G., de Simone S., Marrocco E., Rossi S., Giunti M., Palfi A., et al. Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol. Med. 2014;6:194–211. doi: 10.1002/emmm.201302948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Trapani I., Toriello E., De Simone S., Colella P., Iodice C., Polishchuk E.V., Sommella A., Colecchi L., Rossi S., Simonelli F., et al. Improved dual AAV vectors with reduced expression of truncated proteins are safe and effective in the retina of a mouse model of Stargardt disease. Hum. Mol. Genet. 2015;24:6811–6825. doi: 10.1093/hmg/ddv386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Lostal W., Kodippili K., Yue Y., Duan D. Full-length dystrophin reconstitution with adeno-associated viral vectors. Hum. Gene Ther. 2014;25:552–562. doi: 10.1089/hum.2013.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Maddalena A., Tornabene P., Tiberi P., Minopoli R., Manfredi A., Mutarelli M., Rossi S., Simonelli F., Naggert J.K., Cacchiarelli D., Auricchio A. Triple vectors expand AAV transfer capacity in the retina. Mol. Ther. 2018;26:524–541. doi: 10.1016/j.ymthe.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Aranko A.S., Wlodawer A., Iwaï H. Nature’s recipe for splitting inteins. Protein Eng. Des. Sel. 2014;27:263–271. doi: 10.1093/protein/gzu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Chen L., Zhu F., Li J., Lu H., Jiang H., Sarkar R., Arruda V.R., Wang J., Zhao J., Pierce G.F., et al. The enhancing effects of the light chain on heavy chain secretion in split delivery of factor VIII gene. Mol. Ther. 2007;15:1856–1862. doi: 10.1038/sj.mt.6300268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Tornabene P., Trapani I., Minopoli R., Centrulo M., Lupo M., De Simone S., Tiberi P., Dell’Aquila F., Marrocco E., Iodice C., et al. Intein-mediated protein trans-splicing expands adeno-associated virus transfer capacity in the retina. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aav4523. eaav4523–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Kronenberg S., Böttcher B., von der Lieth C.W., Bleker S., Kleinschmidt J.A. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 2005;79:5296–5303. doi: 10.1128/JVI.79.9.5296-5303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Johnson J.S., Li C., DiPrimio N., Weinberg M.S., McCown T.J., Samulski R.J. Mutagenesis of adeno-associated virus type 2 capsid protein VP1 uncovers new roles for basic amino acids in trafficking and cell-specific transduction. J. Virol. 2010;84:8888–8902. doi: 10.1128/JVI.00687-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Horowitz E.D., Rahman K.S., Bower B.D., Dismuke D.J., Falvo M.R., Griffith J.D., Harvey S.C., Asokan A. Biophysical and ultrastructural characterization of adeno-associated virus capsid uncoating and genome release. J. Virol. 2013;87:2994–3002. doi: 10.1128/JVI.03017-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Lecomte E., Tournaire B., Cogné B., Dupont J.B., Lindenbaum P., Martin-Fontaine M., Broucque F., Robin C., Hebben M., Merten O.W., et al. Advanced characterization of DNA molecules in rAAV vector preparations by single-stranded virus next-generation sequencing. Mol. Ther. Nucleic Acids. 2015;4:e260. doi: 10.1038/mtna.2015.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Penaud-Budloo M., Lecomte E., Guy-Duché A., Saleun S., Roulet A., Lopez-Roques C., Tournaire B., Cogné B., Léger A., Blouin V., et al. Accurate identification and quantification of DNA species by next-generation sequencing in adeno-associated viral vectors produced in insect cells. Hum. Gene Ther. Methods. 2017;28:148–162. doi: 10.1089/hgtb.2016.185. [DOI] [PubMed] [Google Scholar]
- 292.Parks R.J., Bramson J.L., Wan Y., Addison C.L., Graham F.L. Effects of stuffer DNA on transgene expression from helper-dependent adenovirus vectors. J. Virol. 1999;73:8027–8034. doi: 10.1128/jvi.73.10.8027-8034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Sandig V., Youil R., Bett A.J., Franlin L.L., Oshima M., Maione D., Wang F., Metzker M.L., Savino R., Caskey C.T. Optimization of the helper-dependent adenovirus system for production and potency in vivo. Proc. Natl. Acad. Sci. USA. 2000;97:1002–1007. doi: 10.1073/pnas.97.3.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Penaud-Budloo M., François A., Clément N., Ayuso E. Pharmacology of recombinant adeno-associated virus production. Mol. Ther. Methods Clin. Dev. 2018;8:166–180. doi: 10.1016/j.omtm.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Robert M.A., Chahal P.S., Audy A., Kamen A., Gilbert R., Gaillet B. Manufacturing of recombinant adeno-associated viruses using mammalian expression platforms. Biotechnol. J. 2017;12:1600193–1600216. doi: 10.1002/biot.201600193. [DOI] [PubMed] [Google Scholar]
- 297.Tang Q., Keeler A.M., Zhang S., Su Q., Lyu Z., Cheng Y., Gao G., Flotte T.R. Two-plasmid packaging system for recombinant adeno-associated virus. Biores. Open Access. 2020;9:219–228. doi: 10.1089/biores.2020.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Grieger J.C., Soltys S.M., Samulski R.J. Production of recombinant adeno-associated virus vectors using suspension HEK293 cells and continuous harvest of vector from the culture media for GMP FIX and FLT1 clinical vector. Mol. Ther. 2016;24:287–297. doi: 10.1038/mt.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Chadeuf G., Ciron C., Moullier P., Salvetti A. Evidence for encapsidation of prokaryotic sequences during recombinant adeno-associated virus production and their in vivo persistence after vector delivery. Mol. Ther. 2005;12:744–753. doi: 10.1016/j.ymthe.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 300.Hauck B., Murphy S.L., Smith P.H., Qu G., Liu X., Zelenaia O., Mingozzi F., Sommer J.M., High K.A., Wright J.F. Undetectable transcription of cap in a clinical AAV vector: implications for preformed capsid in immune responses. Mol. Ther. 2009;17:144–152. doi: 10.1038/mt.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Bigger B.W., Tolmachov O., Collombet J.M., Fragkos M., Palaszewski I., Coutelle C. An araC-controlled bacterial cre expression system to produce DNA minicircle vectors for nuclear and mitochondrial gene therapy. J. Biol. Chem. 2001;276:23018–23027. doi: 10.1074/jbc.M010873200. [DOI] [PubMed] [Google Scholar]
- 302.Chen Z.-Y., He C.-Y., Kay M.A. Improved production and purification of minicircle DNA vector free of plasmid bacterial sequences and capable of persistent transgene expression in vivo. Hum. Gene Ther. 2005;16:126–131. doi: 10.1089/hum.2005.16.126. [DOI] [PubMed] [Google Scholar]
- 303.Mayrhofer P., Blaesen M., Schleef M., Jechlinger W. Minicircle-DNA production by site specific recombination and protein-DNA interaction chromatography. J. Gene Med. 2008;10:1253–1269. doi: 10.1002/jgm.1243. [DOI] [PubMed] [Google Scholar]
- 304.Schnödt M., Schmeer M., Kracher B., Krüsemann C., Espinosa L.E., Grünert A., Fuchsluger T., Rischmüller A., Schleef M., Büning H. DNA minicircle technology improves purity of adeno-associated viral vector preparations. Mol. Ther. Nucleic Acids. 2016;5:e355. doi: 10.1038/mtna.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Hodgson C.P., Carnes A.E., Williams J.A. Recent advances in non-viral vectors for gene therapy & vaccination. Cell Gene Ther. Insights. 2017;3:95–101. [Google Scholar]
- 306.Lu J., Williams J.A., Luke J., Zhang F., Chu K., Kay M.A. A 5′ noncoding exon containing engineered intron enhances transgene expression from recombinant AAV vectors in vivo. Hum. Gene Ther. 2017;28:125–134. doi: 10.1089/hum.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Karbowniczek K., Rothwell P., Extance J., Milsom S., Lukashchuk V., Bowes K., Smith D., Caproni L. Doggybone™ DNA: an advanced platform for AAV production. Cell Gene Ther. Insights. 2017;3:731–738. [Google Scholar]
- 308.Deneke J., Ziegelin G., Lurz R., Lanka E. Phage N15 telomere resolution. Target requirements for recognition and processing by the protelomerase. J. Biol. Chem. 2002;277:10410–10419. doi: 10.1074/jbc.M111769200. [DOI] [PubMed] [Google Scholar]
- 309.Deneke J., Ziegelin G., Lurz R., Lanka E. The protelomerase of temperate Escherichia coli phage N15 has cleaving-joining activity. Proc. Natl. Acad. Sci. USA. 2000;97:7721–7726. doi: 10.1073/pnas.97.14.7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Clark K.R., Voulgaropoulou F., Fraley D.M., Johnson P.R. Cell lines for the production of recombinant adeno-associated virus. Hum. Gene Ther. 1995;6:1329–1341. doi: 10.1089/hum.1995.6.10-1329. [DOI] [PubMed] [Google Scholar]
- 311.Thorne B.A., Takeya R.K., Peluso R.W. Manufacturing recombinant adeno-associated viral vectors from producer cell clones. Hum. Gene Ther. 2009;20:707–714. doi: 10.1089/hum.2009.070. [DOI] [PubMed] [Google Scholar]
- 312.Urabe M., Ding C., Kotin R.M. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum. Gene Ther. 2002;13:1935–1943. doi: 10.1089/10430340260355347. [DOI] [PubMed] [Google Scholar]
- 313.Virag T., Cecchini S., Kotin R.M. Producing recombinant adeno-associated virus in foster cells: overcoming production limitations using a baculovirus-insect cell expression strategy. Hum. Gene Ther. 2009;20:807–817. doi: 10.1089/hum.2009.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Kotin R.M., Snyder R.O. Manufacturing clinical grade recombinant adeno-associated virus using invertebrate cell lines. Hum. Gene Ther. 2017;28:350–360. doi: 10.1089/hum.2017.042. [DOI] [PubMed] [Google Scholar]
- 315.Cecchini S., Virag T., Kotin R.M. Reproducible high yields of recombinant adeno-associated virus produced using invertebrate cells in 0.02- to 200-liter cultures. Hum. Gene Ther. 2011;22:1021–1030. doi: 10.1089/hum.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Aucoin M.G., Perrier M., Kamen A.A. Critical assessment of current adeno-associated viral vector production and quantification methods. Biotechnol. Adv. 2008;26:73–88. doi: 10.1016/j.biotechadv.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 317.Kurasawa J.H., Park A., Sowers C.R., Halpin R.A., Tovchigrechko A., Dobson C.L., Schmelzer A.E., Gao C., Wilson S.D., Ikeda Y. Chemically defined, high-density insect cell-based expression system for scalable AAV vector production. Mol. Ther. Methods Clin. Dev. 2020;19:330–340. doi: 10.1016/j.omtm.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Kotin R.M. Large-scale recombinant adeno-associated virus production. Hum. Mol. Genet. 2011;20:2–6. doi: 10.1093/hmg/ddr141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Clément N., Knop D.R., Byrne B.J. Large-scale adeno-associated viral vector production using a herpesvirus-based system enables manufacturing for clinical studies. Hum. Gene Ther. 2009;20:796–806. doi: 10.1089/hum.2009.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Thomas D.L., Wang L., Niamke J., Liu J., Kang W., Scotti M.M., Ye G.j., Veres G., Knop D.R. Scalable recombinant adeno-associated virus production using recombinant herpes simplex virus type 1 coinfection of suspension-adapted mammalian cells. Hum. Gene Ther. 2009;20:861–870. doi: 10.1089/hum.2009.004. [DOI] [PubMed] [Google Scholar]
- 321.Adamson-Small L., Potter M., Byrne B.J., Clément N. Sodium chloride enhances recombinant adeno-associated virus production in a serum-free suspension manufacturing platform using the herpes simplex virus system. Hum. Gene Ther. Methods. 2017;28:1–14. doi: 10.1089/hgtb.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Wang Q., Wu Z., Zhang J., Firrman J., Wei H., Zhuang Z., Liu L., Miao L., Hu Y., Li D., et al. A robust system for production of superabundant VP1 recombinant AAV vectors. Mol. Ther. Methods Clin. Dev. 2017;7:146–156. doi: 10.1016/j.omtm.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Backovic A., Cervelli T., Salvetti A., Zentilin L., Giacca M., Galli A. Capsid protein expression and adeno-associated virus like particles assembly in Saccharomyces cerevisiae. Microb. Cell Fact. 2012;11:124. doi: 10.1186/1475-2859-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Barajas D., Aponte-Ubillus J.J., Akeefe H., Cinek T., Peltier J., Gold D. Generation of infectious recombinant Adeno-associated virus in Saccharomyces cerevisiae. PLOS ONE. 2017;12:e0173010. doi: 10.1371/journal.pone.0173010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Galli A., Della Latta V., Bologna C., Pucciarelli D., Cipriani F., Backovic A., Cervelli T. Strategies to optimize capsid protein expression and single-stranded DNA formation of adeno-associated virus in Saccharomyces cerevisiae. J. Appl. Microbiol. 2017;123:414–428. doi: 10.1111/jam.13511. [DOI] [PubMed] [Google Scholar]
- 326.Schiedner G., Hertel S., Bialek C., Kewes H., Waschütza G., Volpers C. Efficient and reproducible generation of high-expressing, stable human cell lines without need for antibiotic selection. BMC Biotechnol. 2008;8:13. doi: 10.1186/1472-6750-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Schiedner G., Hertel S., Kochanek S. Efficient transformation of primary human amniocytes by E1 functions of Ad5: generation of new cell lines for adenoviral vector production. Hum. Gene Ther. 2000;11:2105–2116. doi: 10.1089/104303400750001417. [DOI] [PubMed] [Google Scholar]
- 328.Hein K., Goncalves I., Hudjetz B., Strempel N., Riebesehl N., Kewes H., Bauer T., Bialek C., Schmidt-Hertel S., Patil A., et al. Scalable AAV production using stable helper virus-free AAV producer cell lines based on CEVEC’s CAP-GT cells. Hum. Gene Ther. 2019;30:A222–A241. [Google Scholar]
- 329.Emmerling V.V., Holzmann K., Lanz K., Kochanek S., Hörer M. Novel approaches to render stable producer cell lines viable for the commercial manufacturing of rAAV-based gene therapy vectors. BMC Proc. 2013;7:P12. [Google Scholar]
- 330.Mullard A. Gene therapies advance towards finish line. Nat. Rev. Drug Discov. 2011;10:719–720. doi: 10.1038/nrd3572. [DOI] [PubMed] [Google Scholar]
- 331.Salmon F., Grosios K., Petry H. Safety profile of recombinant adeno-associated viral vectors: focus on alipogene tiparvovec (Glybera®) Expert Rev. Clin. Pharmacol. 2014;7:53–65. doi: 10.1586/17512433.2014.852065. [DOI] [PubMed] [Google Scholar]
- 332.Senior M. After Glybera’s withdrawal, what’s next for gene therapy? Nat. Biotechnol. 2017;35:491–492. doi: 10.1038/nbt0617-491. [DOI] [PubMed] [Google Scholar]
- 333.Touchot N., Flume M. Early insights from commercialization of gene therapies in Europe. Genes. 2017;8:78. doi: 10.3390/genes8020078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Goswami R., Subramanian G., Silayeva L., Newkirk I., Doctor D., Chawla K., Chattopadhyay S., Chandra D., Chilukuri N., Betapudi V. Gene therapy leaves a vicious cycle. Front. Oncol. 2019;9:297. doi: 10.3389/fonc.2019.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Edwards T.L., Jolly J.K., Groppe M., Barnard A.R., Cottriall C.L., Tolmachova T., Black G.C., Webster A.R., Lotery A.J., Holder G.E., et al. Visual acuity after retinal gene therapy for choroideremia. N. Engl. J. Med. 2016;374:1996–1998. doi: 10.1056/NEJMc1509501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 337.Anderton R.S., Mastaglia F.L. Advances and challenges in developing a therapy for spinal muscular atrophy. Expert Rev. Neurother. 2015;15:895–908. doi: 10.1586/14737175.2015.1059757. [DOI] [PubMed] [Google Scholar]
- 338.Ogino S., Leonard D.G.B., Rennert H., Ewens W.J., Wilson R.B. Genetic risk assessment in carrier testing for spinal muscular atrophy. Am. J. Med. Genet. 2002;110:301–307. doi: 10.1002/ajmg.10425. [DOI] [PubMed] [Google Scholar]
- 339.Lunn M.R., Wang C.H. Spinal muscular atrophy. Lancet. 2008;371:2120–2133. doi: 10.1016/S0140-6736(08)60921-6. [DOI] [PubMed] [Google Scholar]
- 340.Al-Zaidy S.A., Kolb S.J., Lowes L., Alfano L.N., Shell R., Church K.R., Nagendran S., Sproule D.M., Feltner D.E., Wells C., et al. AVXS-101 (onasemnogene abeparvovec) for SMA1: comparative study with a prospective natural history cohort. J. Neuromuscul. Dis. 2019;6:307–317. doi: 10.3233/JND-190403. [DOI] [PubMed] [Google Scholar]