Abstract
Understanding the genomic basis of memory processes may help in combating neurodegenerative disorders. Hence, we examined the associations of common genetic variants with verbal short-term memory and verbal learning in adults without dementia or stroke (N = 53,637). We identified novel loci in the intronic region of CDH18, and at 13q21 and 3p21.1, as well as an expected signal in the APOE/APOC1/TOMM40 region. These results replicated in an independent sample. Functional and bioinformatic analyses supported many of these loci and further implicated POC1. We showed that polygenic score for verbal learning associated with brain activation in right parieto-occipital region during working memory task. Finally, we showed genetic correlations of these memory traits with several neurocognitive and health outcomes. Our findings suggest a role of several genomic loci in verbal memory processes.
Subject terms: Genetics, Neuroscience
Introduction
The ability to focus attention and to encode, store, and recall information are not only imperative for survival but these memory-related cognitive processes also reflect healthy brain aging [1, 2]. Cognitive decline, especially episodic memory impairment, is a clinical hallmark and genetic endophenotype of several types of dementia, especially Alzheimer’s disease (AD) [3]. Understanding the genetic and molecular basis of inter-individual variation in normal memory function could improve precision in screening for dementias, and identify novel drug targets to support cognitive reserve, and to prevent and treat dementia.
Both episodic memory in cognitively normal individuals [3, 4] and AD [5] show moderate to high heritability in twin studies. Large-scale genome-wide association meta-analyses (GWAMAs) across several cohorts have identified over 30 genomic loci for AD [6], but GWAMAs for episodic memory among dementia-free adults have shown less consistent findings [7–17]. In the largest GWAMA of episodic memory, Davies et al. [17] did not find any significant genomic variants for visuo-spatial memory in the UK Biobank sample of 112,067 persons. As visuo-spatial encoding of information involves partially different brain networks compared to verbal encoding [18], genomic architecture of visuo-spatial memory and verbal memory may differ. Indeed, an earlier GWAMA from the CHARGE consortium showed that rs4420638 at 19q13.3 near the APOE-APOC1-TOMM40 locus, that shows the largest known effects on AD [6], was associated with verbal long-term memory (delayed recall) in a sample of 29,076 persons [7]. There is ample evidence for differences in brain networks and thus, genetic networks, that are involved in long-term and short-term episodic memory processes [19]. A relatively small (N = 7486) genome-wide association study of immediate recall scores in tests of verbal episodic memory (verbal short-term memory; VSTM), however, detected the same APOE-APOC1-TOMM40 locus [16]. GWAMAs with considerably larger sample sizes are needed to find novel loci beyond this locus.
Therefore, we examined if common genetic variants were associated with verbal episodic memory in adults of European ancestry without dementia or stroke in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. We operationalized VSTM as immediate recall scores in tests of verbal episodic memory and conducted a GWAMA in a sample of 53,637persons (32 cohorts). As verbal learning (VL) tasks may constitute a more sensitive marker of cognitive deficits than tests of VSTM without a learning component [20] and to our knowledge, only one small (N = 700) GWAMA for VL exists [15], we also examined genetic underpinnings of VL in 32,762 persons (19 cohorts). To assess the functional role of the identified variants, we analyzed fMRI activations during working memory performance and computed genomic associations.
Results
The characteristics of the study cohorts, details of memory tests administered, genotyping quality control and genomic inflation factors are shown in Supplementary information S1–S4 and Supplement 1.
Due to differences in verbal memory tests used in the different cohorts, we performed sample-size based meta-analyses using METAL [21]. All models were adjusted for age, sex, and population substructure. Table 1 shows results for the lead SNPs and Figs. 1–3 shows regional plots of genome-wide significant associations. Supplementary Figs. 1–7 show Manhattan plots of all genomic associations and Supplementary Table S1 shows all genome-wide significant (p < 5 × 10−8) and suggestive (5 × 10−8 ≥ p < 5 × 10−6) associations in the discovery sample.
Table 1.
Meta-analyses results of lead SNPs for verbal short-term memory, paragraph recall, and verbal learning in discovery, replication, and combined samples (N = 53,637).
| Discovery cohorts | Replication cohorts | Discovery + Replication cohorts | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pheno-type | SNP | Chr | Genomic position | EA/OA | EAF | Nearby gene | Predicted function | N | Z-score | P-val | Direction | Het P-val | N | Z-score | P-val | Direction | Het P-val | Na | Z-score | P-val |
| VL | rs4687625 | 3 | 52538758 | t/c | 0.38 | NT5DC2 | Intronic | 28579 | 5.70 | 1.2E−08 | +?+++++++++++−++++++++ | 0.97 | 3853 | 2.74 | 0.006 | ++ | 0.28 | 32432 | 6.3 | 3.1E−10 |
| VL | rs2015971 | 3 | 52521860 | t/c | 0.38 | STAB1 | Intronic | 28127 | 5.64 | 1.7E−08 | +?−+++?++++++−++++++++ | 0.96 | 3853 | 2.70 | 0.007 | ++ | 0.23 | 31980 | 6.2 | 4.8E−10 |
| VL | rs11711421 | 3 | 52536819 | t/c | 0.39 | NT5DC2 | Intronic | 28579 | 5.53 | 3.3E−08 | +?−++++++++++−++++++++ | 0.96 | 3853 | 2.55 | 0.01 | ++ | 0.29 | 32432 | 6.1 | 1.3E−9 |
| VL | rs2276816 | 3 | 52835856 | t/c | 0.12 | ITIH4 | Synon | 27896 | 5.53 | 3.3E−08 | +++++++++−+−+?++++++++ | 0.53 | 3067 | 0.61 | 0.54 | ?+ | 1 | 30963 | 5.4 | 5.4E−8 |
| VSTM | rs425724 | 5 | 19518050 | c/t | 0.12 | CDH18 | Intronic | 44668 | 5.56 | 2.7E−08 | ?++++++++−−+++++++++++++++− | 0.67 | 8763 | 2.04 | 0.04 | +++++ | 0.84 | 53431 | 5.91 | 3.4E−09 |
| VSTM: Paragraph | rs9528369 | 13 | 62216927 | a/c | 0.32 | Intergenic | 18098 | −6.0 | 2.0E−09 | ?−−−−−−??−−−+ | 0.62 | 4293 | −0.79 | 0.43 | −+− | 0.66 | 22391 | −5.74 | 9.4E−9 | |
| VSTM | rs4420638 | 19 | 45422946 | g/a | 0.18 | APOC1 | 3’down-stream | 37556 | −7.23 | 4.9E−13 | ?+−−−−−?−−−−−−??−?−−−−+−?−− | 5.5E−05 | 8164 | −2.89 | 0.004 | −−?−+ | 1.4E−04 | 44317 | −7.83 | 4.9E−15 |
| VL | −“− | −“− | −“− | −“− | −“− | −“− | −“− | 23575 | −7.05 | 1.8E−12 | −?+−−−−?−−−?−?−−−−−−+? | 0.14 | 3067 | 0.56 | 0.58 | ?+ | 1 | 26642 | −6.4 | 1.2E−10 |
| VSTM: Paragraph | −“− | −“− | −“− | −“− | −“− | −“− | −“− | 16216 | −6.93 | 4.2E−12 | ?−−−−?−?−−−−− | 4.4E−05 | 3694 | −5.14 | 2.78E−07 | −?− | 0.37 | 19910 | −8.47 | 2.5E−17 |
| VL: Visual | −“− | −“− | −“− | −“− | −“− | −“− | −“− | 15096 | −6.0 | 3.1E−09 | −−?−−−−−−−+ | |||||||||
| VL | rs6857 | 19 | 45392254 | t/c | 0.16 | PVRL2 | 3’utr | 26849 | −6.00 | 2.0E−09 | −?−−−−−?−−−+−?−−−−−−++ | 2.2E−05 | 3067 | 0.63 | 0.53 | ?+ | 1 | 29916 | −5.5 | 4.2E−8 |
All models were adjusted for sex, age, and population substructure and results reflect analyses in participants of European ancestry without dementia or stroke.
VSTM Verbal short-term memory, VL Verbal learning, VSTM Paragraph VSTM meta-analyses restricted to those cohorts with paragraph recall test, VL Visual meta-analyses restricted to those cohorts with VL test using visual presentation of the words, EA Effect allele, OA Other allele, EAF Effect allele frequency.
aN varies due to missing data.
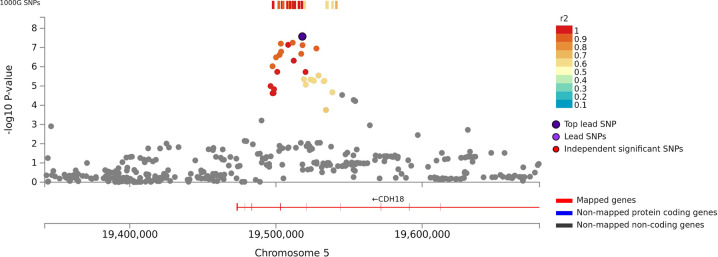
Fig. 1. Regional plot of associations of SNPs at the 5p14.3 region with verbal short-term memory in the discovery sample (N = 44,874).
Dots indicate p-values of SNPs and rs425724 in an intron of CDH18 is marked in violet.
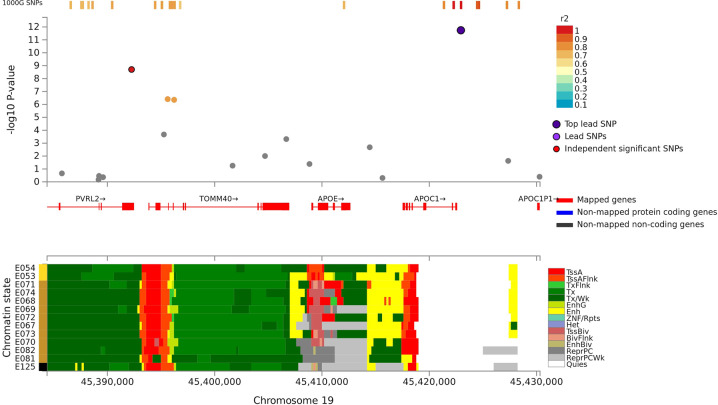
Fig. 3.
Regional plot of associations of SNPs at the 19q13.3 region with verbal learning (Upper panel). Dots indicate p values of SNPs and the top lead SNP rs4420638 is marked in violet and another independent and significant SNP rs6857 is marked in red. Lower panel indicates 15-core chromatin state in Roadmap brain-related tissues (E053-E082) and E125 ENCODE NH-A Astrocytes primary cells and shows that both significant SNPs are in or flanking transcriptionally active region.
For VSTM, we observed two significant associations in the discovery sample (N = 44,874): rs425724 (p = 2.7 × 10−8) within an intron of CDH18 and rs4420638 (p = 4.9 × 10−13) downstream of APOC1 at 19q13.3. Associations of both SNPs with VSTM were replicated in an independent sample at nominal significance (p values < 0.04; N = 8763).
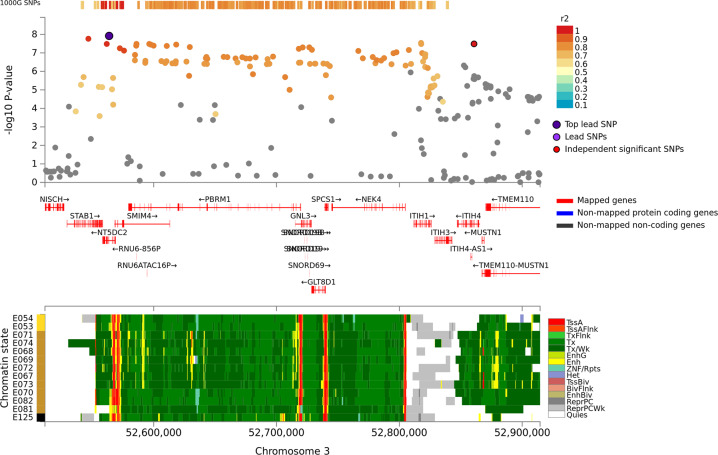
For VL, we observed significant associations at the same 19q13.3 locus and at 3p21 in the discovery sample (N = 28,909). At the 19q13.3 locus the strongest associations were observed with rs4420638 (p = 1.8 × 10−12) and rs6857 (p = 2.0 × 10−9) that are in linkage disequilibrium (LD; r2: 0.45) with each other. The 3p21 locus harbors a large LD block in/near NT5DC2, STAB1, ITIH1, ITIH4, and PBRM1. Out of 14 SNPs showing a significant association at this locus, rs4687625, within an intron of NT5DC2, and a synonymous ITIH4 variant rs2276816 were independently significant SNPs (r2: 0.12, distance: 297 kb). Three of the significant 3p21 SNPs (rs4687625, rs2015971, and rs11711421; all intronic to or near NT5DC2) showed nominally significant association with VL scores in an independent replication sample (p values < 0.01; N = 3853).
Despite some heterogeneity between the cohorts in 19q13.3 SNPs (rs4420638 and rs6857), no single cohort drove the results (Supplementary Figs. 7–13). We further examined with meta-regression if cohort-level characteristics influenced estimates of the association between these SNPs and memory test scores. Larger effect estimates in both 19q13.3 SNPs associated with smaller proportion of women in the cohort and rs4420638 effect estimates for VL associated with younger mean age of the cohort (Supplementary Table S16).
There were no other significant signals in the analyses combining discovery and replication cohorts (Supplementary Table S8).
Analyses stratified by the type of the memory test
As in Debette et al. [7], we further meta-analyzed cohorts based on the specific type of memory test applied. In the analyses of VSTM, cohorts were classified into those with paragraph recall test data (13 cohorts, N = 19,420) and those with word list recall test data (14 cohorts, N = 25,454). In the analyses of VL, cohorts were classified into those with orally presented words (11 cohorts, N = 12,593) and those with visually presented words (11 cohorts, N = 16,191).
In the analyses restricted to cohorts with the VSTM paragraph recall tests, we observed a novel locus in an intergenic region at 13q21 (lead SNP rs9528369, p = 2.0 × 10−9) and a second locus at 19q13.3 (lead SNP rs4420638, p = 4.2 × 10−12). Additionally, rs4420638 showed a significant association with VL in those cohorts with visually presented words (p = 3.1 × 10−9). Of these results, we were able to replicate the association of rs4420638 with paragraph recall (p = 1.4 × 10−4) in an independent replication sample (N = 4293). There were no significant associations in the other stratified meta-analyses.
Analyses adjusting for educational attainment
Following Debette et al. [7], we ran secondary analyses to test if associations were independent of education. All associations in the significant lead SNPs remained significant after further adjusting the models for educational attainment except that the associations of rs4687625 (p = 8.8 × 10−7) and rs2276816 (p = 5.3 × 10−6) at 3p21 with VL became only suggestively significant.
Gene-based, gene-set, and gene property analysis results with MAGMA
Gene-based association analyses with MAGMA identified one gene for VSTM (APOC1 at 19q13.3), 15 genes for VL (SMIM4, STAB1, PBRM1, NEK4, NT5DC2, ITIH4, GNL3, ITIH1, MUSTN1, GLT8D1, and ITIH3 at 3p21; CALN1 at 7q11; TOMM40 and APOC1 at 19q13.3; and AGXT2 at 5p13), and two genes for paragraph recall (APOC1 and TOMM40 at 19q13.3) after Bonferroni correction for multiple testing (Supplementary Table S2 and Supplementary Fig. 21). We found no significant enrichment in gene-set analyses (Supplementary Table S3).
Gene-property analysis tests if tissue-specific expression is predictive of the association of the gene with the phenotype. These analyses indicate that genes with the highest expression levels in the pituitary and all available brain regions, except for the rostral intracranial portion of the spinal cord, were the same genes showing significant associations with VSTM and with paragraph recall, but not with VL (Supplementary Table S4).
Functional analyses and colocalization
We identified potential functionality of SNPs showing significant associations with FUMA [22] (Supplementary Tables S5 and S6). Fourteen SNPs at the 3p21 locus that associated with VL are significant eQTLs for POC1A, GNL3, GLYCTK, DUSP7, ITIH4, PPM1M, and GLT8D1 in putamen, cerebellum, frontal cortex, and/or hippocampus in the Genotype-Tissue Expression (GTeX) and in putamen, white matter, and/or hippocampus in the Brain eQTL Almanac (Braineac) database. Of these, rs2276816 is also a synonymous exonic SNP with a Combined Annotation Dependent Depletion (CADD) score indicating a potential functionally deleterious effect (CADD > 12.37) [23]. Additionally, rs1961958, that associated with VL, and rs11148561, that associated with paragraph recall, have high CADD scores. Moreover, 3p21 locus SNPs rs4687625, rs1961959, rs6798246, and rs3774355, that associated with VL, also may influence gene regulation as indicated by both eQTL data and transcription factor binding data (regulomeDB category 1f [24]). Roadmap 15-core chromatin states show that 3p21 and 19q13.3 loci are situated in transcriptionally active regions and rs6798246 flanks an active transcription start site in brain tissues (Figs. 2 and 3). Additionally, our methylation QTL (mQTL) and amyloid/tau accumulation PET analyses corroborate the functional role of the 3p21, 13q21 and 19q13.3 loci in the brain tissues. In the dorsolateral prefrontal cortex (DLPFC) samples of the Religious Orders Study and Rush Memory and Aging Project (ROSMAP) (N = 322), the top 3p21 SNPs associated with methylation levels of CpGs corresponding to ITIH4, ITIH1, STAB1, NEK4, MUSTN1, DNAH1, TLR9, GNL3, SNORD69, TMEM110, and NT5DC2 (p(Benjamini-Hochberg false discovery rate [FDR]) < 0.01). Moreover, rs9528369 associated with a cg09367879 located in the open sea region in chromosome 13, and rs6857 associated with a CpG in the APOE (Supplementary Table S13). Both 19q13.3 SNPs marginally associated with tau accumulation in the precuneus, and rs4420638 also associated with overall amyloid accumulation in a Framingham Heart Study (FHS) sample of young adults with PET imaging (N = 183) (Supplementary Table S15). Chromatin-chromatin interaction analyses show that all genomic regions implicated in VSTM, VL, and paragraph recall showed significant interactions with other intra-chromosomal regions (Supplementary Figs. 14–19 and Supplementary Table S6). For example, the intronic CDH18 region implicated in VSTM analyses interacts with the CDH18 promoter region in the Roadmap Epigenomics Project brain tissue samples. In these same brain samples, the intergenic 13q21 region implicated in the paragraph recall analyses interacts with the promoter region of TDRD3. This same region also interacted with the PCDH20 gene region in non-brain tissue samples.
Fig. 2.
Regional plot of associations of SNPs at the 3q21 region with verbal learning in the discovery sample (N = 28,909; Upper panel). Dots indicate p-values of SNPs and the top lead SNP rs4687625 is marked in violet and another independent and significant SNP rs2276816 is marked in red. Lower panel indicates 15-core chromatin state in Roadmap brain-related tissues (E053-E082) and E125 ENCODE NH-A Astrocytes primary cells and shows that both significant SNPs are in transcriptionally active region.
Using S-PrediXcan [25], after Bonferroni correction for multiple testing we identified a single gene (POC1A) whose expression in the putamen was negatively associated with VL (Z = −5.02; p = 5.04 × 10−7) whereas no significant associations were observed for VSTM (Supplementary Table S7 and Supplementary Fig. 20).
Finally, we tested with polygenic scores (PGSs) the overall association of VSTM (PGSVSTM) and VL (PGSVL) with brain activation assessed via fMRI during a working memory task in 435 healthy participants in the Clinical Brain Disorders Branch Sibling Study. The intermediate PGSVL (SNP inclusion p value < 10−4) correlated negatively with activity in a right parieto-occipital cluster with a peak in BA19 (peak Z = 4.73; pFWE = 0.016; 55 voxels; MNI coordinates x = 45; y = −64; z = 10; Fig. 4). At a lower p < 0.001 (uncorrected) threshold, a symmetric cluster was significant on the left with a peak in BA39 (peak Z = 3.55; 24 voxels; MNI coordinates x = −45; y = −58; z = 13; Fig. 4). No results survived correction for multiple comparisons using the PGSVSTM.
Fig. 4. Associations of the polygenic score for verbal learning (PGSVL) with activation in the right BA19 during the 2-back working memory task in a sample of N = 435 healthy adults (upper panel left view and lower panel right view).
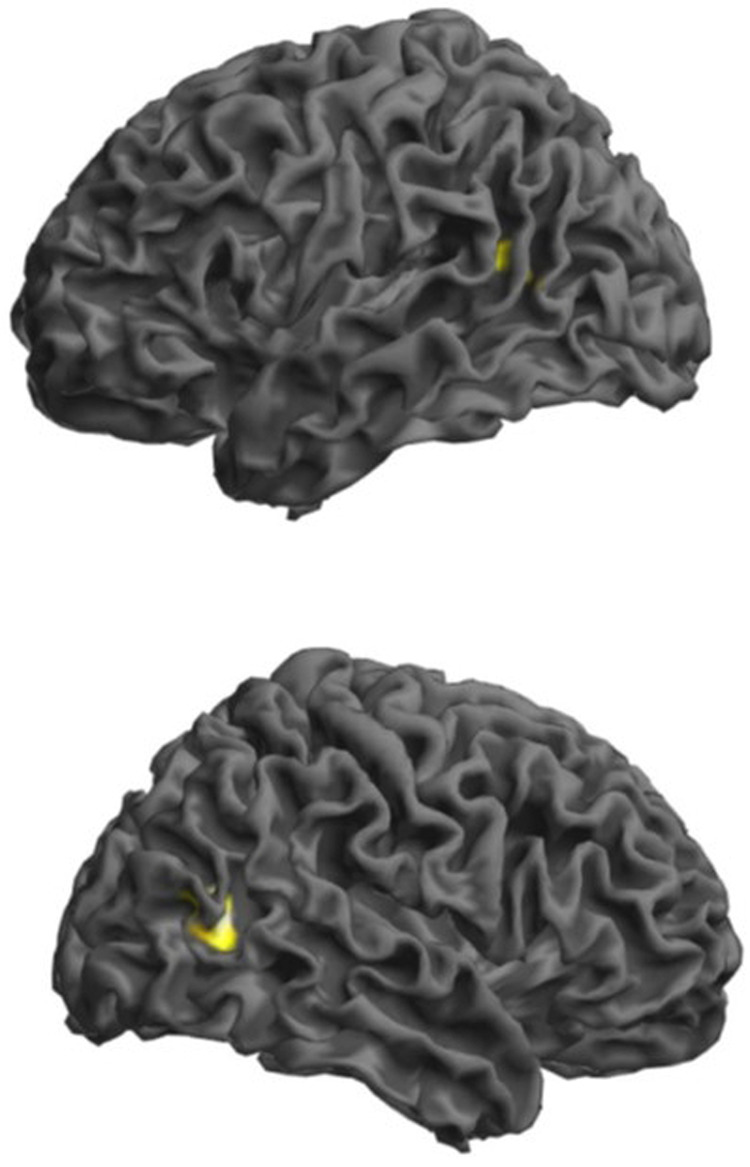
Results are thresholded at peak-level p < 0.001 and masked for significantly increased activity during 2-back relative to 0-back. Rendered image illustrates clusters in which activity is negatively correlated with the PGSVL (the right cluster survives correction for multiple comparisons at BA19; MNI coordinates x = 45, y = −64, Z = 10; FWE corrected p = 0.016). Left in the figure is left in the brain.
Protein-protein interactions
We investigated protein-protein interactions with DAPPLE [26] and results are presented in Supplementary Table 12. Fourteen, 30, and 11 proteins were included in the network construction for VSTM, VL, and paragraph recall, respectively, but six, 16, and two proteins were present in direct or indirect networks, respectively. None of the network parameters were significant. In the analyses of single proteins, SYT9 and NRXN1 were significant for VSTM (p = 0.006), ZFAND5, GRIK2, and ZC3H18 were nominally significant for VL (p = 0.018–0.05), and PRLHR was nominally significant for paragraph recall (p = 0.044).
Genetic correlation analyses
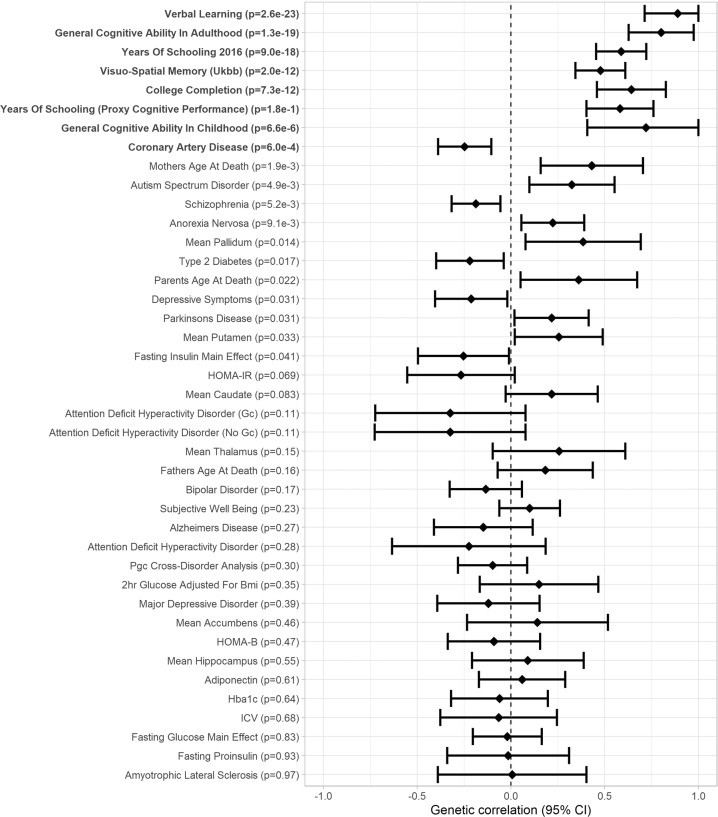
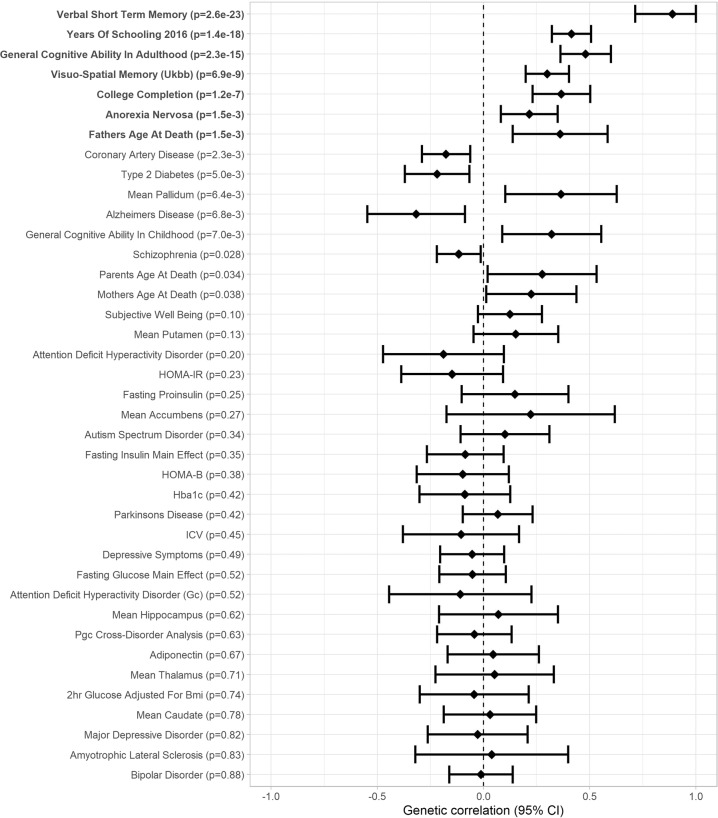
We used LDHub [27] for analyses of SNP-based heritability and genetic correlations. In the cohorts that could pool individual participant data (16 cohorts N = 26,977 in VSTM and 15 cohorts N = 25,180 in VL), SNP-based heritability was 0.06 (SE: 0.02) and 0.18 (SE: 0.02) for VSTM and VL, respectively. Genetic correlations between VSTM, VL, and health-related phenotypes are presented in Figs. 5, 6 and in Supplementary Table S11. After FDR correction, VSTM and VL showed positive genetic correlation with each other (rg = 0.89, p = 2.6 × 10−23) and with general cognitive ability (GCA; rg > 0.44, p < 2.3 × 10−16) in adults (and VSTM also with GCA in childhood, rg > 0.72, p < 7.3 × 10−6), visuo-spatial memory in the UK Biobank (rg > 0.30, p < 6.9 × 10−9), years of schooling (rg > 0.41, p < 1.4 × 10−18), and college completion (rg > 0.37, p < 1.2 × 10−7). In addition, VSTM showed negative genetic correlation with coronary artery disease (rg = −0.25, p = 6.0 × 10−4), and VL showed positive genetic correlation with anorexia nervosa (rg = 0.37, p = 1.2 × 10−7) and father’s age at death (rg = 0.36, p = 1.5 × 10−8).
Fig. 5.
Forest plot of genetic correlations between verbal short-term memory and 46 traits related to cognitive abilities or health (Genetic correlation [95% confidence interval]; significant genetic correlations after FDR correction in boldface).
Fig. 6.
Forest plot of genetic correlations between verbal learning and 46 traits related to cognitive abilities or health (Genetic correlation [95% confidence interval]; significant genetic correlations after FDR correction in boldface).
Consistency of findings with earlier studies
As our results might reflect genetic effects on more general cognitive abilities, we also show the GWAS results for visuo-spatial memory test scores in the UK Biobank sample (N = 336,881; http://www.nealelab.is/uk-biobank) and Davies et al (2018) [28] GWAMA results for GCA in the Supplemental Table 1. Only SNPs in 3p21 showed significant association with GCA implying that associations between CDH18, 13q21, and 19q13.3 SNPs with VSTM and VL are not secondary to the effect of this loci on GCA or general memory processes, but may show specificity to verbal episodic memory. However, as the UK Biobank memory test has showed low test-retest reliability, these results need to be interpreted with caution [29]. Further, we examined if the top SNPs of this study also linked with brain structure [30–32] and function [33] in previous GWA studies (Supplementary Table S14). We noticed that all our 3p21 top SNPs were associated with smaller intracranial volume and larger alpha oscillation during rest and both 19q13.3 (APOE-TOMM40-APOC1) SNPs linked with smaller volumes of hippocampus, amygdala, and nucleus accumbens.
Finally, in Supplementary Table S9 we show that of the top candidate SNPs for episodic verbal memory phenotypes (e.g., in KIBRA [10], CTNNBL1 [9], SCN1A [8], and FASTKD2 [11]) [7, 9–16], our meta-analyses showed at least suggestive signals only at the APOE-TOMM40-APOC1 complex (rs4420638, rs2075650, rs6857, and rs157582).
Discussion
We studied if common genetic variants associated with VSTM and VL in 53,637 adults without history of stroke or dementia within the CHARGE consortium. We identified four novel loci for VSTM/VL. The top SNPs showed wide range of functional properties in the brain tissues: Some were eQTLs, meQTLs, or associated with tau or amyloid accumulation in the brain, and an aggregate polygenic score for VL associated with working memory activity in the right parieto-occipital cortex.
The first novel peak for VSTM locates at 5p14.3 and encompasses rs425724, an intronic SNP within CDH18 (aka CDH14 and CDH24) as the lead SNP. Functional effects of rs425724 remain poorly known, but Hi-C chromosomal interaction tests suggest that it may influence regulation of CDH18 expression. CDH18 is specifically expressed in the brain [34] and it belongs to the Type II classic cadherin family, which is involved in neuronal cell-adhesion [35]. Cadherins are critically important in the development of cells and synapses early in life, and in maintaining neuronal and synaptic structure in mature synapses [36]. Cadherins are also suggested to play a central role in synaptic plasticity in general, and in long-term potentiation (LTP), the molecular basis of learning and memory, in particular [37, 38]. Cadherin-related alterations in LTP have been demonstrated in pharmacological, gene knockout, and RNAi experiments [39, 40], but little is known about the role of genomic variation in cadherin genes in memory processes in humans. We report that rs425724 may affect specifically processing of verbal information. Interestingly, a variant in CDH13 associated with verbal but not spatial working memory in patients with ADHD [41], pointing again towards modality specificity. Some studies exist linking cadherin genes with neurodevelopmental outcomes (Supplement 1).
We also discovered a new locus for VL in 3p21 containing 14 SNPs in high LD in a ~300 kb region that showed significant associations with VL. Of these variants, we replicated rs4687625 and rs2015971, both intronic to NT5DC2, and rs2015971, which is intronic to STAB1. This locus harbors several genes and gene-based analyses implicated 11 genes (NT5DC2, STAB1, ITIH1, ITIH4, PBRM1, SMIM4, NEK4, GLT8D1, ITIH3, MUSTN1, and GNL3). We identified several potentially functional variants at this locus. All significant 3p21 SNPs are either intronic or exonic, are significant eQTLs and mQTLs in brain tissues, and link with brain intracranial volume [30] and alpha oscillation [33] in the previous studies. Some are also considered deleterious or regulatory. Moreover, the locus is in a transcriptionally active region and, finally, SNP associations of 3p21 variants with VL colocalized with imputed expression of POC1A in the putamen. The putamen is part of a cortico-striatal loop and it receives input from different parts of the cortex and projects back to the cortex via the globus pallidus and thalamus. Traditionally it has been linked with motor control functions, but recently both neuroimaging studies [42, 43] and studies on effects of focal lesions [44] have suggested an additional role in memory functions. Prior studies have associated SNPs at 3p21 locus with various neurodevelopmental outcomes, such as GCA [28] and schizophrenia [45], but causal variant(s) are not known and in the studies with functional analyses, no specific gene has been conclusively shown to account for the many association findings at this locus (Supplement 1). Interestingly, a recent study reported an association between GLT8D1-variant rs6795646 and working memory in healthy Chinese persons [46].
We observed a third novel locus in the intergenic region in 13q21 in meta-analyses of discovery sample cohorts with paragraph recall tests to measure VSTM. The lead SNP was rs9528369 and the locus harbors 36 other significant SNPs. Again, the causal SNP or gene underlying this association is not known, but earlier studies point towards influences of this locus on language processing [47] and educational attainment [48] (Supplement 1). In line with this, rs9528369 showed no association with visuo-spatial memory test performance in the UK Biobank sample (http://www.nealelab.is/uk-biobank). Functional influences of this locus remain poorly understood, but rs9528369 was a mQTL in the dorsolateral prefrontal cortex and Hi-C analyses of this study showed chromatin-chromatin interactions with the promoter region of TDRD3 in brain tissue and PCDH20 in other tissues. TDRD3 is part of the TOP3beta-TDRD3-FMRP complex, and TOP3beta deletion was recently linked with schizophrenia, cognitive impairment, and learning difficulties [49], while lack of FMRP causes the Fragile X syndrome characterized by severe learning deficits and mental retardation.
In line with Debette et al. [7] in the GWAS for long-term verbal memory, we showed that rs4420638 in the APOE-TOMM40-APOC1 locus at 19q13.3 is associated consistently with VSTM, especially paragraph recall, and overall VL and visually presented VL test scores. Also, rs6857 associated with VL. It is near PVRL2 and locates ~30 kb downstream from rs4420638 and is in LD with rs4420638. Both significant SNPs are located near transcriptionally active region, associate with tau accumulation, and with the size of the memory-relevant regions (e.g., hippocampus) [31, 32]. Prior studies have linked many SNPs in this locus with a variety of cognitive outcomes and dementias although not previously with VSTM or VL in cognitively normal adults (Supplement 1) [6, 7, 16]. These various signals may merely reflect an impact of genetic variation at the APOE locus or suggest that additional genes in this region are involved in episodic memory, but this distinction requires functional studies; the strong LD in this region precludes further conclusions based solely on genetic association studies.
We also showed gene-level associations and significant enrichment with genes expressed widely in the brain, especially in the cerebellum and the frontal cortex for VSTM, and the cerebellum and striatal nuclei for paragraph recall - a pattern that parallels one shown recently for the GCA [28]. Gene-based analyses implicated AGXT2 and CALN1 for VL, while analyses of protein-protein interactions implicated synaptic proteins previously associated with Alzheimer disease biology, SYT9 and NRXN1 for VSTM; ZFAND5, GRIK2, and ZC3H18 for VL; and PRLHR for paragraph recall. There is some evidence that AGXT2, CALN1, NRXN, and GRIK2 may influence neurodevelopmental outcomes (Supplement 1).
Previous fMRI studies on short-term word list recall associated performance with a network of brain regions including the medial temporal lobe, superior temporal gyrus, medial and inferior parietal cortex, and dorsolateral prefrontal cortex [50, 51]. Within this network, joint analysis of episodic and working memory tasks observed the involvement of the prefrontal cortex, supplementary motor area, and bilateral ventral posterior parietal cortex spanning into the extrastriate cortex [52]. Consistently, here we show that a polygenic score for VL associated with activity in the posterior parietal and extrastriate cortex during the N-back fMRI task. This association was not due to years of education. This visual association area is active during recognition memory [53, 54]. The association had a negative direction, consistent with N-back performance data which correlate negatively with frontoparietal network activity in healthy individuals. [55, 56]
The heritability estimates of ~6% for VSTM and 18% for VL are in line with a recent phenome-wide study that showed SNP-based estimates between 6% and 11% for visuo-spatial memory in the UK Biobank [57]. Moreover, our estimates are in line with a twin study showing lower estimates for VSTM than for VL [4]. In our study, VSTM and VL showed strong positive genetic correlations with each other and with GCA in adulthood, completion of college, and years of schooling, consistent with recent findings from the UK Biobank [58]; and VSTM with childhood GCA and VL with anorexia nervosa and father’s age at death. VSTM also showed negative genetic correlation with coronary artery disease, in agreement with a previous study showing a negative association between a polygenic risk score for cardiovascular disease and verbal short-term memory [59]. To our knowledge, no previous studies have suggested a shared genetic background between verbal episodic memory and anorexia nervosa. However, anorexia nervosa shows positive genetic correlation with years of education and attending college [60] and children born to mothers with anorexia nervosa have shown increased working memory capacity [61].
There are limitations to our study. Heterogeneity in the testing methods and phenotypes across cohorts may have hindered our ability to find associations. Since majority of the samples (91.2% for VSTM and 93.3% for VL) were imputed against the HapMap2 reference panel resulting in ~2.5 Million SNPs in the meta-analyses, re-analyses with higher resolution genotyping is warranted. Moreover, despite reporting GWAMA results of the largest sample with VSTM and VL, our study is still underpowered to detect all genomic variation related to verbal episodic memory and larger studies are needed. Finally, as VSTM and VL showed strong genetic correlation with GCA, it is possible that our results reflect genomic influences on GCA. However, there are several lines of evidence against this: of several cognitive abilities, memory has shown largest unique genetic variance [62], adjusting for educational attainment only marginally altered our results, and finally, of our lead SNPs only those in a highly pleiotropic region at 3q21 were implicated in the recent GWAS for GCA [28].
To sum up, we report the results of the largest GWAMA of verbal episodic memory. We show novel genome-wide significant associations between common SNPs in four loci, CDH18, 3p21, 13q21, and 19q13.3, and VSTM and VL, and link combined polygenic variation for VL with brain activity during working memory task in the parieto-occipital cortex. Whereas many SNPs in these loci, especially in 3p21 and in 19q13.3, have been linked to other neurocognitive outcomes and show functional significance and associations with brain structure and function, their exact biological role needs to be studied further. We also show moderate SNP-based heritability and high genetic correlation of these memory traits and GCA, as well as coronary artery disease and anorexia nervosa suggesting some shared biology. These results improve our understanding of the biology underlying learning and memory and could lead to improved risk stratification scores and new drug targets for preserving memory, and preventing or treating dementias.
Online methods
Participants
This study comprised 37 cohorts and 53,637 adult participants (age > 18 years) of European descent brought together by the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. Exclusion criteria included clinical stroke and any form of prevalent dementia.
The discovery sample comprised 44,874 participants from 27 cohorts for VSTM and 28,909 participants from 22 cohorts for VL. Replication samples comprised 8763 participants (five cohorts) and 3853 participants (two cohorts) for VSTM and VL, respectively. All studies were approved by their institutional ethics review committees and all participants provided written informed consent. Characteristics of the study cohorts are shown in Supplementary information Table 1 and Supplement 1.
Phenotypes
All verbal memory tests are standardized and validated and have shown psychometrically adequate properties. Cognitive tests were administered by trained personnel following standardized protocols and blind to genetic information. To assess VSTM, cohorts administered either word list tests, e.g., the California Verbal Learning Test (CVLT), or paragraph tests, e.g., the Paragraph/Story recall test in the Wechsler Memory Scale (WMS) test battery, with immediate recall (Supplement 1 and Supplementary information Table 3). In all tests, participants were asked to recall as many words or story elements as possible immediately after their presentation.
In addition, some of the word list tests, e.g., CVLT, RAVLT, and CERAD, included assessment of VL. In these tests, the recalled material was presented, either orally or visually, and recalled more than once, hence the tests are tapping into the ability to learn across trials. In these tests, the first round of recall was also used in the VSTM analyses. Thus, these cohorts contributed both to the VL meta-analyses and to the VSTM meta-analyses.
We decided a priori to run meta-analyses combining all cohorts with verbal episodic memory tests with immediate recall (VSTM) and another meta-analyses across cohorts that administered tests of verbal learning with immediate recall (VL). Following Debette et al. [7] we also ran additional meta-analyses combining only the cohorts that administered similar tests. In these meta-analyses, we combined cohorts with word list tests with immediate recall (VSTM word list), paragraph tests with immediate recall (VSTM paragraph recall), verbal learning tests with orally presented material (VL orally presented words), and finally, verbal learning tests with visually presented words (VL visually presented words).
Genotyping, QC, and imputation
Genome-wide genotyping was conducted in each cohort on several platforms following manufacturer protocols. Quality control was performed independently for each study. In addition, each group performed genotype imputation with appropriate software using the HapMap Phase II release 22 reference panel (70% of the cohorts) or 1000 Genomes, Phase 1, Release v3 panel. To harmonize the datasets, we updated the SNP IDs in those cohorts with HapMap Phase II imputation to match 1000 genomes, phase 1, release v3 panel (hg 19) by using LiftOver tool. Imputation quality scores for each SNP were obtained from IMPUTE (“proper_info”) or MACH (“rsq_hat”). Details on the genotyping are presented in Supplementary Information Table 2.
Cohort-level genome-wide association analyses
Each cohort applied multiple linear regressions with additive genetic effect models to test for phenotype-genotype association using ~2.5 million genotyped and/or imputed autosomal SNPs (cohorts with HapMap II imputation) and 10–12 million SNPs in cohorts with 1000 genomes, phase 1 imputation. In our primary model, we adjusted for sex, age, population substructure, and study-specific covariates if deemed appropriate such as clinical center for multi-center cohorts. Furthermore, in family-based studies we fitted familial relationships, if necessary. In the secondary model, we adjusted for primary model covariates and educational attainment.
Meta-analyses and detection of genomic risk loci
We performed quality control of the cohort-level summary statistics before the meta-analyses with the QCGWAS R package, version 1.0–8 [63], in the cohorts with HapMap II imputed data and EasyQC version 9.0 [64] in the cohorts with 1000 Genomes imputed data. We conducted the meta-analyses using METAL software [21]. We used the sample-size weighting and fixed effect model approach. We ran meta-analyses first separately in the discovery and replication samples and then in the combined sample including both discovery and replication cohorts. At the meta-analysis stage, we filtered out SNPs with low minor allele frequency (MAF <1%), poor imputation quality (proper_info <0.4 for IMPUTE and rsq_hat <0.3), or small sample size in the meta-analyses (N < 4000). We applied genomic control correction. A threshold of p < 5 × 10−8 was pre-specified as genome-wide significant, while a threshold of p < 1 × 10−6 was considered suggestive genome-wide significant. We used lambda values and quantile–quantile (Q-Q) plots of observed versus expected –log10(P value) to examine the genome-wide distribution of P values for signs of excessive false positive results. Genomic inflation factors are shown in Supplementary Information Table 4.
We applied FUnctional Mapping and Annotation of genetic associations (FUMA) [22] with default values to detect individual significant SNPs (p < 5 × 10−8 and independent of other genome wide significant SNPs at r2 < 0.6) and corresponding genomic risk loci (independent significant SNPs with r2 ≥ 0.1 and distance <250 kb are assigned to the same genomic risk locus) based on the meta-analysis results.
We also report associations on visuo-spatial memory test scores (variable #399, “Number of incorrect matches in round”) in the UKBiobank sample (N = 336,881; http://www.nealelab.is/uk-biobank) and on GCA in the Davies et al. [28] for those SNPs showing at least suggestively significant results (p < 5 × 10−6) in our discovery cohort.
Functional annotation
For each of the SNPs showing a significant genome-wide signal, we derived several indices suggesting functionality using FUMA [22]: a) annotations with ANNOVAR [65] and the Ensembl genes build 85; b) CADD (http://cadd.gs.washington.edu/) scores that reflect deleteriousness of variants computed by integrating 63 functional annotations and applying a cut-off score of 12.37 as previously suggested (in general the higher the CADD score the more deleterious the variant is likely to be) [23]; c) regulome DB scores indicating the level of evidence for a variant to be a regulatory element, with lower scores indicating stronger evidence [24]; d) 15-core chromatin states for 127 epigenomes as characterized by ChromHMM v1.10 derived from 5 chromatin markers (H3K4me3, H3K4me1, H3K36me3, H3K27me3, H3K9me3) [66]; e) significant brain-related eQTLs defined as FDR (gene q-value) ≤ 0.05, using eQTL information on gene expression in 13 brain tissues obtained from GTEx v7 (http://www.gtexportal.org/home/) [67, 68] and 10 brain tissues obtained from Braineac (http://www.braineac.org/) [69] databases; f) chromatin-chromatin interactions (using pre-processed significant loops filtered at FDR 0.05 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE87112) [70] between independent significant SNPs and gene promoter regions (predicted using DNase peaks and core 15-state chromatin state model (http://egg2.wustl.edu/roadmap/web_portal/DNase_reg.html#delieation) in Roadmap Epigenomics Project brain tissues (E007, E009, E010, E053, E054, E067, E068, E069, E070, E071, E072, E073, E074, E081, E082) [71].
Additionally, we tested if the top SNPs reaching genome-wide significance associated with i) methylation levels in the dorsolateral prefrontal cortex (DLPFC) in the participants of the ROSMAP cohort (N = 322) and ii) brain amyloid and tau burden in a sample of 183 persons from the Framingham Heart Study (FHS) Third Generation cohort (mean age 46 ± 8years, 44% women) who underwent positron emission tomography (PET) imaging (Please see Supplement 1 for methods).
Gene-based, gene-set, and gene property analyses
We performed gene-based association analysis with MAGMA (v1.6) [72] with default settings as implemented in FUMA [22]. SNPs were assigned to protein coding genes obtained from Ensembl build 85. We applied Bonferroni correction and genome-wide significance was set at 2.777 × 10−6 (0.05/18,007).
We also performed MAGMA (v1.6) [72] competitive gene-set analysis, using the results of the gene-based analyses, to examine whether genes in a gene-set are more strongly associated with VSTM and VL than other genes. A total of 10,655 gene sets (curated gene sets: N = 4738, GO terms: N = 5917) from MsigDB v6.1 [73] were used. We applied Bonferroni correction and genome-wide significance was set at 4.69 × 10−6 (0.05/10,655).
In addition, we performed MAGMA tissue expression analysis as implemented in FUMA with default settings and GTEx v7 gene expression data. This test examines the (positive) relationship between highly expressed genes in a specific tissue and genetic associations with those phenotypes showing significant genes (VSTM, VL, and VSTM tests with paragraph recall).
S-PrediXscan analyses
We used S-PrediXcan [25] to integrate eQTL information with GWAS summary statistics to identify genes for which genetically predicted expression levels are associated with VSTM and VL. We used expression weights derived from 13 brain tissues in the GTEx v7 database and LD information from the 1000 Genomes Project Phase 3 [74]. These data were processed with beta values and standard errors from the VSTM and VL GWAS to estimate the expression-GWAS association statistic. We used a transcriptome-wide significance threshold of p < 1.10 × 10−6, which is the Bonferroni-corrected threshold when adjusting for all brain tissues and genes and visualized the colocalization (if any) with locus compare plot (http://locuscompare.com/ /accessed 17.5.2019).
PGSVSTM, PGSVL, and brain activity during 2-Back working memory task
To compute the short-term memory (PGSVSTM) and verbal learning (PGSVL) polygenic scores, we obtained betas associating allele dose with performance for 115,414 and 57,689, respectively, linkage disequilibrium-independent (R2 < 0.1) index SNPs. We then computed a weighted sum of the cumulative SNP effects by summing the imputation probability for the reference allele of the index SNP, weighted by the effect size of association with performance, at each independent locus across the genome, as described elsewhere [75]. We analyzed fMRI data of 435 healthy adult (≥18 years) volunteers of Caucasian ancestry who participated in the Clinical Brain Disorders Branch Sibling Study of schizophrenia (Supplement 1). Participants were genotyped according to standard procedures. In the PGS, we included SNPs at whole-genome (p = 5 × 10−8), intermediate (p = 10−4), and nominal significance levels (p = 0.05). Participants performed the N-back task during fMRI (block design version: 2-Back vs. 0-Back, lasting 240 s) working memory (WM) task. This task is widely used in imaging genetics studies [76–78]. fMRI data collection, pre-processing, and analysis followed standard procedures (Supplement 1) [79]. We used SPM12 to perform multiple regression analyses using PGSs as predictors. We report results surviving pFWE < 0.05 threshold at whole brain level masked by task activity with a minimum cluster extent of 10 voxels (Supplement 1). Results are illustrated at p < 0.001 (uncorrected) in Fig. 4.
Protein-protein interactions with DAPPLE
We investigated a possible causal role for genes at the loci associated with VSTM and VL by searching for physical connections between proteins encoded by genes within these loci. The hypothesis is that causal genetic variants are likely to affect common mechanisms and these mechanisms may be revealed by these protein-protein interaction (PPI) networks. We performed the analyses using Disease Association Protein-Protein Link Evaluator (DAPPLE) [26] in GenePattern. DAPPLE searches for PPI in the InWeb database and assigned a probabilistic score. The InWeb database collects PPI data reported in the literature from numerous sources including IntAct, Reactome, the Molecular Interaction Database (MINT), the Biomolecular Interaction Network Database (BIND) and the Kyoto Encyclopaedia of Genes and Genomes (KEGG). DAPPLE constructs PPI networks where proteins are nodes and interactions in the InWeb databases are edges connecting the nodes. Input SNPs are those associated with memory phenotypes at p value < 0.10 and minor allele frequency >0.05. Genes harboring any of the input SNPs or those in LD (r2 > 0.5) with the input SNPs, or located within the closest recombination hotspots plus 50 kb are identified. Proteins coded by these genes are used to construct an interaction network. Four parameters are estimated for the observed network: (1) number of edges in the direct network; (2) the average number of proteins with which each seed protein directly interacts; (3) the average number of proteins with which each seed protein indirectly interacts; (4) the average number of seed proteins bound by common interactor (CI) proteins. The distributions of these estimates are then enumerated via 20,000 permutations by randomly reassigning proteins of the same binding degree (i.e., the total number of interactions a protein has in the InWeb database) as the proteins in the observed network to each node. Individual seed proteins are then scored based on their presence in direct and indirect networks. The significance of these scores are evaluated in the same permutation procedure and Bonferroni-corrected for the number of possible candidate proteins from each locus to prioritize genes (pcorr < 0.05).
Genetic correlation analyses
We used LDscore (LDSC) regression as implemented in LD Hub [27] to estimate the degree of overlap between the polygenic architecture of the traits. We estimated genetic correlations between verbal episodic memory traits and traits that may be phenotypically linked with memory (categories: Neurological, Psychiatric, Brain volume, Aging, Cognitive, Education, Cardiometabolic, and Glycemic). In these analyses, we excluded the American cohorts as their consent precluded the use of their data to examine an association with education. Therefore, sample size was 26,977 in the analyses of genetic correlation with VSTM and 25,180 in the analyses of genetic correlation with the VL. We used FDR correction to account for multiple comparisons. Heritability z-scores were 4.9 and 7.4 for VSTM and VL, respectively, suggesting that the datasets for both traits are suitable for LDSC analyses.
Supplementary information
Supplementary cohort information tables S1-S4
Author contributions
JL wrote the first draft and prepared all drafts for submission; JL and STT, meta-analyzed the results; STT, QY, JP, ZG, and GP provided secondary analyses; JL, GP, SA, NA, NJA, AB, KB, EB, AC, HC, QC, JC, SRC, GD, PLJ, JDF, ALF, AEF, IF, MF, HJG, FG, VG, ES, EK, JK, LK, NAK, JBK, ML, ML, TL, AL, TL, KAM, KN, AP, SR, SLR, RJR, VS, CS, PS, RS, JAS, DJS, ST, STT, AV, UV, RSW, KW, EV, KY, HZ, WZ, DA, JA, DAB, HB, DIC, ALG, CH, MI, JJ, SLK, TL, ML, VSM, AP, BMP, AR, PMR, SGR, PSS, AJS, MS, PSS, SS, JMS, JT, WU, MW, DRW, JFW, MJW, DRW, JGE, THM, LJL, CMD, IJD, SS, and KR facilitated data collection in the cohorts and provided critical phenotypic and/or genotypic analytic feedback for individual cohorts; JL, STT, QY, GP, SA, NA, NJA, AB, KB, JCB, EB, JB, AC, QC, GD, ED, JDF, AEF, IF, MF, ES, EGH, PKJ, PK, LK, MJK, ML, ML, SL, REM, KAM, SM, SM, MAN, KN, AO, CO, JP, VS, MS, KES, JS, AVS, JAS, IS, AT, AT, ST, SL, EV, RX, LY, HZ, WZ, DIC, ALG, VSM, WU, DRW, SD, and THM prepared individual datasets and conducted analyses and provided summary statistics or results; SS, SD, JGE, THM, LJL, CMD, IJD, and KR provided critical support in planning the study and the manuscript; JL, SS, and KR supervised all analyses; All authors reviewed the manuscript and approved it for submission.
Funding
Open Access funding provided by University of Helsinki including Helsinki University Central Hospital.
Code availability
Code of the primary statistical analyses can be obtained from the corresponding author.
Competing interests
HB is in an Advisory Board member in Nutricia; HJG reported Fresenius Medical Care, BMP serves on the DSMB of a clinical trial funded by Zoll LifeCor and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Deceased: John M. Starr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41380-022-01710-8.
References
- 1.Nyberg L, Lovden M, Riklund K, Lindenberger U, Backman L. Memory aging and brain maintenance. Trends Cogn Sci. 2012;16:292–305. doi: 10.1016/j.tics.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Stern Y. Cognitive reserve in ageing and Alzheimer’s disease. Lancet Neurol. 2012;11:1006–12. doi: 10.1016/S1474-4422(12)70191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindgren N, Kaprio J, Rinne JO, Vuoksimaa E. Immediate verbal recall and familial dementia risk: population-based study of over 4000 twins. J Neurol Neurosurg Psychiatry. 2019;90:90–97. doi: 10.1136/jnnp-2018-319122. [DOI] [PubMed] [Google Scholar]
- 4.Panizzon MS, Lyons MJ, Jacobson KC, Franz CE, Grant MD, Eisen SA, et al. Genetic architecture of learning and delayed recall: a twin study of episodic memory. Neuropsychology. 2011;25:488–98. doi: 10.1037/a0022569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen psychiatry. 2006;63:168–74. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 6.Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51:414–30. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Debette S, Verbaas CAI, Bressler J, Schuur M, Smith A, Bis JC, et al. Genome-wide studies of verbal declarative memory in nondemented older people: The Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. Biol Psychiatry. 2015;77:749–63. doi: 10.1016/j.biopsych.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papassotiropoulos A, Henke K, Stefanova E, Aerni A, Muller A, Demougin P, et al. A genome-wide survey of human short-term memory. Mol Psychiatry. 2011;16:184–92. doi: 10.1038/mp.2009.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papassotiropoulos A, Stefanova E, Vogler C, Gschwind L, Ackermann S, Spalek K, et al. A genome-wide survey and functional brain imaging study identify CTNNBL1 as a memory-related gene. Mol Psychiatry. 2013;18:255–63. doi: 10.1038/mp.2011.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV, et al. Common Kibra alleles are associated with human memory performance. Science. 2006;314:475–8. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- 11.Ramanan VK, Nho K, Shen L, Risacher SL, Kim S, McDonald BC, et al. FASTKD2 is associated with memory and hippocampal structure in older adults. Mol Psychiatry. 2015;20:1197–204. doi: 10.1038/mp.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KL, Hunt P, et al. A genome-wide study of common SNPs and CNVs in cognitive performance in the CANTAB. Hum Mol Genet. 2009;18:4650–61. doi: 10.1093/hmg/ddp413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seshadri S, DeStefano AL, Au R, Massaro JM, Beiser AS, Kelly-Hayes M, et al. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham Study. BMC Med Genet. 2007;8:S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huentelman MJ, Papassotiropoulos A, Craig DW, Hoerndli FJ, Pearson JV, Huynh KD, et al. Calmodulin-binding transcription activator 1 (CAMTA1) alleles predispose human episodic memory performance. Hum Mol Genet. 2007;16:1469–77. doi: 10.1093/hmg/ddm097. [DOI] [PubMed] [Google Scholar]
- 15.LeBlanc M, Kulle B, Sundet K, Agartz I, Melle I, Djurovic S, et al. Genome-wide study identifies PTPRO and WDR72 and FOXQ1-SUMO1P1 interaction associated with neurocognitive function. J Psychiatr Res. 2012;46:271–8. doi: 10.1016/j.jpsychires.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Arpawong TE, Pendleton N, Mekli K, McArdle JJ, Gatz M, Armoskus C, et al. Genetic variants specific to aging-related verbal memory: Insights from GWASs in a population-based cohort. PLoS One. 2017;12:e0182448. doi: 10.1371/journal.pone.0182448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies G, Marioni RE, Liewald DC, Hill WD, Hagenaars SP, Harris SE, et al. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151) Mol Psychiatry. 2016;21:758–67. doi: 10.1038/mp.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirchhoff BA, Buckner RL. Functional-anatomic correlates of individual differences in memory. Neuron. 2006;51:263–74. doi: 10.1016/j.neuron.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Norris D. Short-term memory and long-term memory are still different. Psychol Bull. 2017;143:992–1009. doi: 10.1037/bul0000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabin LA, Pare N, Saykin AJ, Brown MJ, Wishart HA, Flashman LA, et al. Differential memory test sensitivity for diagnosing amnestic mild cognitive impairment and predicting conversion to Alzheimer’s disease. Neuropsychol Dev Cogn B Aging Neuropsychol Cogn. 2009;16:357–76. doi: 10.1080/13825580902825220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 2018;9:1825. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–9. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies G, Lam M, Harris SE, Trampush JW, Luciano M, Hill WD, et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat Commun. 2018;9:2098. doi: 10.1038/s41467-018-04362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyall DM, Cullen B, Allerhand M, Smith DJ, Mackay D, Evans J, et al. Cognitive test scores in UK Biobank: data reduction in 480,416 participants and longitudinal stability in 20,346 participants. PLoS One. 2016;11:e0154222. doi: 10.1371/journal.pone.0154222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams HH, Hibar DP, Chouraki V, Stein JL, Nyquist PA, Renteria ME, et al. Novel genetic loci underlying human intracranial volume identified through genome-wide association. Nat Neurosci. 2016;19:1569–82. doi: 10.1038/nn.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hibar DP, Adams HHH, Jahanshad N, Chauhan G, Stein JL, Hofer E, et al. Novel genetic loci associated with hippocampal volume. Nat Commun. 2017;8:13624. doi: 10.1038/ncomms13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satizabal CL, Adams HHH, Hibar DP, White CC, Knol MJ, Stein JL, et al. Genetic architecture of subcortical brain structures in 38,851 individuals. Nat Genet. 2019;51:1624–36. doi: 10.1038/s41588-019-0511-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smit DJA, Wright MJ, Meyers JL, Martin NG, Ho YYW, Malone SM, et al. Genome-wide association analysis links multiple psychiatric liability genes to oscillatory brain activity. Hum Brain Mapp. 2018;39:4183–95. doi: 10.1002/hbm.24238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibata T, Shimoyama Y, Gotoh M, Hirohashi S. Identification of human cadherin-14, a novel neurally specific type II cadherin, by protein interaction cloning. J Biol Chem. 1997;272:5236–40. doi: 10.1074/jbc.272.8.5236. [DOI] [PubMed] [Google Scholar]
- 35.Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat Rev Neurosci. 2007;8:11–20. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- 36.Friedman LG, Benson DL, Huntley GW. Cadherin-based transsynaptic networks in establishing and modifying neural connectivity. Curr Top Dev Biol. 2015;112:415–65. doi: 10.1016/bs.ctdb.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murase S, Schuman EM. The role of cell adhesion molecules in synaptic plasticity and memory. Curr Opin Cell Biol. 1999;11:549–53. doi: 10.1016/s0955-0674(99)00019-8. [DOI] [PubMed] [Google Scholar]
- 38.Tai CY, Kim SA, Schuman EM. Cadherins and synaptic plasticity. Curr Opin Cell Biol. 2008;20:567–75. doi: 10.1016/j.ceb.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 39.Bozdagi O, Shan W, Tanaka H, Benson DL, Huntley GW. Increasing numbers of synaptic puncta during late-phase LTP: N-cadherin is synthesized, recruited to synaptic sites, and required for potentiation. Neuron. 2000;28:245–59. doi: 10.1016/s0896-6273(00)00100-8. [DOI] [PubMed] [Google Scholar]
- 40.Bozdagi O, Wang XB, Nikitczuk JS, Anderson TR, Bloss EB, Radice GL, et al. Persistence of coordinated long-term potentiation and dendritic spine enlargement at mature hippocampal CA1 synapses requires N-cadherin. J Neurosci Off J Soc Neurosci. 2010;30:9984–9. doi: 10.1523/JNEUROSCI.1223-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arias-Vasquez A, Altink ME, Rommelse NN, Slaats-Willemse DI, Buschgens CJ, Fliers EA, et al. CDH13 is associated with working memory performance in attention deficit/hyperactivity disorder. Genes Brain Behav. 2011;10:844–51. doi: 10.1111/j.1601-183X.2011.00724.x. [DOI] [PubMed] [Google Scholar]
- 42.Ystad M, Eichele T, Lundervold AJ, Lundervold A. Subcortical functional connectivity and verbal episodic memory in healthy elderly-a resting state fMRI study. Neuroimage. 2010;52:379–88. doi: 10.1016/j.neuroimage.2010.03.062. [DOI] [PubMed] [Google Scholar]
- 43.Seger CA. The basal ganglia in human learning. Neuroscientis: Rev J bringing Neurobiol Neurol Psychiatry. 2006;12:285–90. doi: 10.1177/1073858405285632. [DOI] [PubMed] [Google Scholar]
- 44.Ell SW, Marchant NL, Ivry RB. Focal putamen lesions impair learning in rule-based, but not information-integration categorization tasks. Neuropsychologia. 2006;44:1737–51. doi: 10.1016/j.neuropsychologia.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 45.Ripke SBM, Neale A, Corvin JT, Walters K, Farh PA, Holmans P, et al. Biological Insights From 108 Schizophrenia-Associated Genetic Loci. Nature. 2014; 511:421–7. [DOI] [PMC free article] [PubMed]
- 46.Yang CP, Li X, Wu Y, Shen Q, Zeng Y, Xiong Q, et al. Comprehensive integrative analyses identify GLT8D1 and CSNK2B as schizophrenia risk genes. Nat Commun. 2018;9:838. doi: 10.1038/s41467-018-03247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Truong DT, Shriberg LD, Smith SD, Chapman KL, Scheer-Cohen AR, DeMille MM, et al. Multipoint genome-wide linkage scan for nonword repetition in a multigenerational family further supports chromosome 13q as a locus for verbal trait disorders. Hum Genet. 2016;135:1329–41. doi: 10.1007/s00439-016-1717-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50:1112–21. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoll G, Pietilainen OPH, Linder B, Suvisaari J, Brosi C, Hennah W, et al. Deletion of TOP3beta, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat Neurosci. 2013;16:1228–37. doi: 10.1038/nn.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saykin AJ, Johnson SC, Flashman LA, McAllister TW, Sparling M, Darcey TM, et al. Functional differentiation of medial temporal and frontal regions involved in processing novel and familiar words: an fMRI study. Brain. 1999;122:1963–71. doi: 10.1093/brain/122.10.1963. [DOI] [PubMed] [Google Scholar]
- 51.Kircher T, Weis S, Leube D, Freymann K, Erb M, Jessen F, et al. Anterior hippocampus orchestrates successful encoding and retrieval of non-relational memory: an event-related fMRI study. Eur Arch Psychiatry Clin Neurosci. 2008;258:363–72. doi: 10.1007/s00406-008-0805-z. [DOI] [PubMed] [Google Scholar]
- 52.Sambataro F, Safrin M, Lemaitre HS, Steele SU, Das SB, Callicott JH, et al. Normal aging modulates prefrontoparietal networks underlying multiple memory processes. Eur J Neurosci. 2012;36:3559–67. doi: 10.1111/j.1460-9568.2012.08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slotnick SD, Schacter DL. A sensory signature that distinguishes true from false memories. Nat Neurosci. 2004;7:664–72. doi: 10.1038/nn1252. [DOI] [PubMed] [Google Scholar]
- 54.Hofer A, Weiss EM, Golaszewski SM, Siedentopf CM, Brinkhoff C, Kremser C, et al. An FMRI study of episodic encoding and recognition of words in patients with schizophrenia in remission. Am J Psychiatry. 2003;160:911–8. doi: 10.1176/appi.ajp.160.5.911. [DOI] [PubMed] [Google Scholar]
- 55.Pergola G, Di Carlo P, Andriola I, Gelao B, Torretta S, Attrotto MT, et al. Combined effect of genetic variants in the GluN2B coding gene (GRIN2B) on prefrontal function during working memory performance. Psychol Med. 2016;46:1135–50. doi: 10.1017/S0033291715002639. [DOI] [PubMed] [Google Scholar]
- 56.Callicott JH, Mattay VS, Bertolino A, Finn K, Coppola R, Frank JA, et al. Physiological characteristics of capacity constraints in working memory as revealed by functional MRI. Cereb Cortex. 1999;9:20–26. doi: 10.1093/cercor/9.1.20. [DOI] [PubMed] [Google Scholar]
- 57.Ge T, Chen CY, Neale BM, Sabuncu MR, Smoller JW. Phenome-wide heritability analysis of the UK Biobank. PLoS Genet. 2017;13:e1006711. doi: 10.1371/journal.pgen.1006711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hagenaars SP, Harris SE, Davies G, Hill WD, Liewald DC, Ritchie SJ, et al. Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N=112 151) and 24 GWAS consortia. Mol Psychiatry. 2016;21:1624–32. doi: 10.1038/mp.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hagenaars SP, Harris SE, Clarke TK, Hall L, Luciano M, Fernandez-Pujals AM, et al. Polygenic risk for coronary artery disease is associated with cognitive ability in older adults. Int J Epidemiol. 2016;45:433–40. doi: 10.1093/ije/dyv354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brainstorm C, Anttila V, Bulik-Sullivan B, Finucane HK, Walters RK, Bras J et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360:eaap8757. [DOI] [PMC free article] [PubMed]
- 61.Kothari R, Solmi F, Treasure J, Micali N. The neuropsychological profile of children at high risk of developing an eating disorder. Psychol Med. 2013;43:1543–54. doi: 10.1017/S0033291712002188. [DOI] [PubMed] [Google Scholar]
- 62.Petrill SA, Plomin R, Berg S, Johansson B, Pedersen NL, Ahern F, et al. The genetic and environmental relationship between general and specific cognitive abilities in twins age 80 and older. Psychological Sci. 1998;9:183–9. [Google Scholar]
- 63.van der Most PJ, Vaez A, Prins BP, Munoz ML, Snieder H, Alizadeh BZ, et al. QCGWAS: A flexible R package for automated quality control of genome-wide association results. Bioinformatics. 2014;30:1185–6. doi: 10.1093/bioinformatics/btt745. [DOI] [PubMed] [Google Scholar]
- 64.Winkler TW, Day FR, Croteau-Chonka DC, Wood AR, Locke AE, Magi R, et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9:1192–212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–6. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Battle A, Brown CD, Engelhardt BE, Montgomery SB. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–13. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–28. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 2016;17:2042–59. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen Q, Ursini G, Romer AL, Knodt AR, Mezeivtch K, Xiao E, et al. Schizophrenia polygenic risk score predicts mnemonic hippocampal activity. Brain. 2018;141:1218–28. doi: 10.1093/brain/awy004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fazio L, Pergola G, Papalino M, Di Carlo P, Monda A, Gelao B, et al. Transcriptomic context of DRD1 is associated with prefrontal activity and behavior during working memory. Proc Natl Acad Sci USA. 2018;115:5582–7. doi: 10.1073/pnas.1717135115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Selvaggi P, Pergola G, Gelao B, Di Carlo P, Nettis MA, Amico G, et al. Genetic variation of a DRD2 co-expression network is associated with changes in prefrontal function after D2 receptors stimulation. Cereb Cortex. 2019;29:1162–73. doi: 10.1093/cercor/bhy022. [DOI] [PubMed] [Google Scholar]
- 78.Taurisano P, Pergola G, Monda A, Antonucci LA, Carlo PD, Piarulli F et al. The interaction between cannabis use and a CB1-related polygenic co-expression index modulates dorsolateral prefrontal activity during working memory processing. Brain Imaging Behav. 2021;15:288–299. [DOI] [PubMed]
- 79.Pergola G, Di Carlo P, D’Ambrosio E, Gelao B, Fazio L, Papalino M, et al. DRD2 co-expression network and a related polygenic index predict imaging, behavioral and clinical phenotypes linked to schizophrenia. Transl Psychiatry. 2017;7:e1006. doi: 10.1038/tp.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary cohort information tables S1-S4
Data Availability Statement
Code of the primary statistical analyses can be obtained from the corresponding author.