Abstract
Zinc-finger nuclease (ZFN)-based in vivo genome editing is a novel treatment that can potentially provide lifelong protein replacement with single intravenous administration. Three first-in-human open-label ascending single-dose phase 1/2 studies were performed in parallel (starting November 2017) primarily to assess safety and tolerability of ZFN in vivo editing therapy in mucopolysaccharidosis I (MPS I) (n = 3), MPS II (n = 9), and hemophilia B (n = 1). Treatment was well tolerated with no serious treatment-related adverse events. At the 1e13 vg/kg dose, evidence of genome editing was detected through albumin-transgene fusion transcripts in liver for MPS II (n = 2) and MPS I (n = 1) subjects. The MPS I subject also had a transient increase in leukocyte iduronidase activity to the lower normal range. At the 5e13 vg/kg dose, one MPS II subject had a transient increase in plasma iduronate-2-sulfatase approaching normal levels and one MPS I subject approached mid-normal levels of leukocyte iduronidase activity with no evidence of genome editing. The hemophilia B subject was not able to decrease use of factor IX concentrate; genome editing could not be assessed. Overall, ZFN in vivo editing therapy had a favorable safety profile with evidence of targeted genome editing in liver, but no long-term enzyme expression in blood.
Keywords: zinc-finger nuclease, in vivo genome editing, mucopolysaccharidosis, MPS I, MPS II, hemophilia, first-in-human
Graphical abstract

In these first-in-human studies, ZFN in vivo editing had a favorable safety profile with evidence of targeted genome editing, but no long-term sustained enzyme expression, for subjects with MPS I, MPS II, and hemophilia B at doses up to 5e13 vg/kg.
Introduction
In November 2017, the first-in-human clinical study assessing the safety and tolerability of zinc-finger nuclease (ZFN)-based in vivo genome editing therapy (ZFN in vivo editing therapy) began dosing the first subjects.1 This novel therapy offers patients the possibility of stable, long-term correction with a one-time peripheral intravenous (i.v.) treatment for genetic diseases with deficiencies in specific proteins.2,3 ZFN in vivo editing therapy consists of three recombinant adeno-associated virus (AAV) 2/6 (serotype 6 for the capsid, including AAV2 inverted terminal repeats, hereafter referred to as rAAV2/6) vectors, which are commonly used for gene therapy. AAV6 was used because it showed strong liver tropism in preclinical studies.4,5,6,7,8,9,10 Two rAAV2/6 vectors each encode ZFNs, which are engineered proteins for targeted genome editing. ZFNs are created by fusing zinc-finger (ZF) proteins with a Fok1 nuclease domain resulting in targeting a specific editing site. The ZFNs are used in pairs to create a double-strand break (DSB) at a pre-determined target site in the albumin intron 1 “safe harbor” locus with high affinity and specificity.11,12,13,14 The albumin locus can support high, consistent, and sustained transgene expression levels in the liver without disrupting neighboring genes.12 The third vector encodes a corrective promoterless transgene that can be integrated into the albumin intron 1 site after the DSB is introduced by the ZFNs. All three vectors were prepared using the baculovirus-insect (Sf9) cell system for rAAV production.
In vivo editing with custom-made ZFNs has advantages over other technologies such as transcription activator-like effector nucleases15 and clustered, regularly interspaced, short, palindromic repeats16 since the ZF DNA binding function has been extensively evolved within mammalian genomic chromatin resulting in optimal chromatin access,17 practical options for therapeutically relevant tuning of on/off target ratios,18 and a minimal ZF protein size for AAV vector compatibility.19 The ZFN in vivo editing approach creates a permanent modification at the albumin locus with the goal of providing lifelong endogenous expression of the deficient enzyme (Figure S1).
The liver is used as a target for ZFN in vivo editing therapy as it is a central organ of metabolism20 with high synthetic capacity,21 and leverages the albumin promoter to drive expression of a donor gene.4 Hepatotropic AAV serotypes, such as AAV6, are used based on their reliable liver tropism,5,6,7,8,9,10,22 which are presumed to provide long-term expression in hepatocytes.20 Inserting a normal gene into the highly transcriptionally active albumin locus of hepatocytes has been found to produce normal protein in a sustained manner in mouse models of mucopolysaccharidosis I (MPS I),23 MPS II,4 and hemophilia B13 by the process that naturally drives albumin expression. There was no effect on circulating albumin levels in these models.
All three of these diseases are strong candidates for gene therapy due to monogenic inheritance and disease pathology.24,25 In vivo genome editing may provide further benefit due to the permanent modification of the gene, which has the potential to provide lifelong treatment. Treatment options for these diseases are limited, non-curative, and require frequent administrations. All three diseases have significant morbidity, mortality, and unmet medical need for lifelong protein replacement that ZFN in vivo editing may effectively address.26,27,28,29
Here, we report the results of three first-in-human clinical studies conducted to evaluate the safety, tolerability, and efficacy of the systemic delivery of ZFN in vivo editing therapy to deliver the iduronidase enzyme to treat MPS I (SB-318-1502, Empowers, NCT02702115) and the iduronate-2-sulfatase enzyme (I2S) to treat MPS II (SB-913-1602, Champions, NCT03041324), and factor IX (FIX) to treat hemophilia B (SB-FIX-1501, NCT02695160). In the MPS studies, delivery of enzymes would be associated with a reduction in total glycosaminoglycan (GAG) levels, heparan sulfate (HS) GAG, and dermatan sulfate (DS) GAG levels in blood. In the SB-FIX-1501 study, delivery of FIX would be associated with increased FIX levels or enzyme activity intended to stop or decrease bleeding episodes.
Results
Thirteen subjects were enrolled in the three studies: nine subjects in the MPS II study, three subjects in the MPS I study, and one subject in the hemophilia B study (Table 1).
Table 1.
Baseline characteristics of MPS II subjects in SB-913-1602, MPS I subjects in SB-318-1502, and hemophilia B subjects in SB-FIX-1501 by treatment dose
| Study | No. | Dose level (vg/kg) | Age (years) | Age at onseta | Gender | Ethnicity | Weight (kg) | Disease | Genotype | Iduronidaseb(nmol/mL/h) | I2Sc(nmol/mL/h) | FIXd(IU/mL) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SB-913-1602 | 1 | 5e12 | 44 | 30 | male | white | 77 | MPSIIe | c.1037C>T (hemizygous) | N/Af | 3.66 | N/A |
| SB-913-1602 | 2 | 5e12 | 19 | unknown | male | Asian | 77 | MPSII | c.281G>A (hemizygous) | N/A | 6.2 | N/A |
| SB-913-1602 | 3 | 1e13 | 41 | 4 | male | white | 89 | MPSII | c.187A>G (hemizygous) | N/A | 1.6 | N/A |
| SB-913-1602 | 4 | 1e13 | 22 | unknown | male | white | 71 | MPSII | c.323A>C (homozygous) | N/A | 0.82 | N/A |
| SB-913-1602 | 5 | 5e13 | 61 | unknown | male | white | 56 | MPSII | c.187A>G (hemizygous) | N/A | 6.77 | N/A |
| SB-913-1602 | 6 | 5e13 | 27 | unknown | male | white | 139 | MPSII | c.253G>A (hemizygous) | N/A | 10.99 | N/A |
| SB-913-1602 | 7 | 5e13 | 49 | unknown | male | white | 60 | MPSII | c.1037C>T (hemizygous) | N/A | 2.23 | N/A |
| SB-913-1602 | 8 | 5e13 | 20 | unknown | male | white | 98 | MPSII | c.260 G>A (hemizygous) | N/A | 10.53 | N/A |
| SB-913-1602 | 9 | 5e13 | 25 | unknown | male | white | 63 | MPSII | c.1007–133 A>G (hemizygous) | N/A | 4.75 | N/A |
| SB-318-1502 | 10 | 1e13 | 23 | unknown | male | white | 61 | MPSI | c.386-2A>G/c.1898C>T (heterozygous) | 1.20 | N/A | N/A |
| SB-318-1502 | 11 | 5e13 | 37 | 31 | male | Asian, other | 79 | MPSI | c.227A>G/c.236C>T (heterozygous) | 11.92 | N/A | N/A |
| SB-318-1502 | 12 | 5e13 | 27 | 21 | female | Asian, other | 74 | MPSI | c.227A>G/c.236C>T (heterozygous) | 7.99 | N/A | N/A |
| SB-FIX-1501 | 13 | 5e13 | 60 | unknown | male | white | 75 | Hem Bg | c. 422G>A (hemizygous) | N/A | N/A | 0.011 |
Age at onset was calculated using date of onset and date of birth.
Iduronidase (normal: 6–71.4 nmol/h/mg).
I2S, iduronate-2-sulfatase (normal: 82–200 nmol/h/mL).
FIX, factor IX (normal: 60–150 IU/dL).
MPS, mucopolysaccharidosis.
N/A, not applicable.
Hem B, hemophilia B.
Safety and tolerability
ZFN in vivo editing therapy was well tolerated at doses up to 5e13 vg/kg, the highest dose tested. Thirty-three treatment-related adverse events (TRAEs) were reported in 7 of 13 (53.8%) subjects across all studies and doses. Twenty-two TRAEs were reported in 4 of 8 (50.0%) subjects receiving the 5e13 vg/kg dose. All were considered mild (grade 1 in common terminology criteria for adverse events [CTCAE]) and resolved (Table 2).
Table 2.
Treatment-related adverse events
|
System organ class Preferred term |
MPSa II |
FIXb |
ZFNcin vivo editing Overalld(N = 13) n (%) [T] |
||||
|---|---|---|---|---|---|---|---|
|
Cohort 1 Dose level 5e12 vg/kg (N = 2) n (%) [T]e |
Cohort 2 Dose level 1e13 vg/kg (N = 2) n (%) [T] |
Cohort 3 Dose level 5e13 vg/kg (N = 5) n (%) [T] |
Overall (N = 9) n (%) [T] |
Cohort 3 Dose level 5e12 vg/kg (N = 1) n (%) [T] |
Overall (N = 1) n (%) [T] | ||
| Any event | 2 (100) [5] | 1 (50.0) [6] | 4 (80.0) [22] | 7 (77.8) [33] | 1 (50.0) [5] | 1 (11.1) [5] | 8(61.5) [38] |
| Cardiac disorders | 0 (0.0) [0] | 0 (0.0) [0] | 0 (0.0) [0] | 0 (0.0) [0] | 1 (50.0) [1] | 1 (11.1) [1] | 1(7.7) [1] |
| Sinus tachycardia | 0 (0.0) [0] | 0 (0.0) [0] | 0 (0.0) [0] | 0 (0.0) [0] | 1 (50.0) [1] | 1 (11.1) [1] | 1(7.7) [1] |
| Gastrointestinal disorders | 0 (0.0) [0] | 0 (0.0) [0] | 1 (20.0) [1] | 1 (11.1) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [1] |
| Pancreatic cyst | 0 (0.0) [0] | 0 (0.0) [0] | 1 (20.0) [1] | 1 (11.1) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [1] |
| General disorders and administration site conditions | 0 (0.0) [0] | 0 (0.0) [0] | 1 (20.0) [1] | 1 (11.1) [1] | 1 (50.0) [2] | 1 (11.1) [2] | 2(15.4) [3] |
| Chills | 0 (0.0) [0] | 0 (0.0) [0] | 0 (0.0) [0] | 0 (0.0) [0] | 1 (50.0) [1] | 1 (11.1) [1] | 1(7.7) [1] |
| Pyrexia | 0 (0.0) [0] | 0 (0.0) [0] | 1 (20.0) [1] | 1 (11.1) [1] | 1 (50.0) [1] | 1 (11.1) [1] | 2(15.4) [2] |
| Hepatobiliary disorders | 0 (0.0) [0] | 1 (50.0) [1] | 2 (40.0) [3] | 3 (33.3) [4] | 0 (0.0) [0] | 0 (0.0) [0] | 3(23.1) [4] |
| Cholelithiasis | 0 (0.0) [0] | 0 (0.0) [0] | 1 (20.0) [1] | 1 (11.1) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [1] |
| Hepatic lesion | 0 (0.0) [0] | 1 (50.0) [1] | 0 (0.0) [0] | 1 (11.1) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [1] |
| Hepatomegaly | 0 (0.0) [0] | 0 (0.0) [0] | 1 (20.0) [1] | 1 (11.1) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [1] |
| Portal fibrosis | 0 (0.0) [0] | 0 (0.0) [0] | 1 (20.0) [1] | 1 (11.1) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [1] |
| Injury, poisoning, and procedural complications | 0 (0.0) [0] | 0 (0.0) [0] | 1 (20.0) [1] | 1 (11.1) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [1] |
| Infusion-related reaction | 0 (0.0) [0] | 0 (0.0) [0] | 1 (20.0) [1] | 1 (11.1) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [1] |
| Investigations | 0 (0.0) [0] | 1 (50.0) [2] | 3 (60.0) [11] | 4 (44.4) [13] | 0 (0.0) [0] | 0 (0.0) [0] | 4(30.8) [13] |
| Alanine aminotransferase increased | 0 (0.0) [0] | 1 (50.0) [1] | 2 (40.0) [6] | 3 (33.3) [7] | 0 (0.0) [0] | 0 (0.0) [0] | 3(23.1) [7] |
| Aspartate aminotransferase increased | 0 (0.0) [0] | 1 (50.0) [1] | 2 (40.0) [3] | 3 (33.3) [4] | 0 (0.0) [0] | 0 (0.0) [0] | 3(23.1) [4] |
| Blood pressure systolic increased | 0 (0.0) [0] | 0 (0.0) [0] | 1 (20.0) [2] | 1 (11.1) [2] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [2] |
| Musculoskeletal and connective tissue disorders | 1 (50.0) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1 (11.1) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [1] |
| Muscular weakness | 1 (50.0) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1 (11.1) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [1] |
| Nervous system disorders | 1 (50.0) [1] | 0 (0.0) [0] | 1 (20.0) [2] | 2 (22.2) [3] | 1 (50.0) [1] | 1 (11.1) [1] | 3(23.1) [4] |
| Dizziness | 1 (50.0) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1 (11.1) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [1] |
| Dysgeusia | 0 (0.0) [0] | 0 (0.0) [0] | 1 (20.0) [1] | 1 (11.1) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [1] |
| Headache | 0 (0.0) [0] | 0 (0.0) [0] | 1 (20.0) [1] | 1 (11.1) [1] | 1 (50.0) [1] | 1 (11.1) [1] | 2(15.4) [2] |
| Skin and subcutaneous tissue disorders | 2 (100) [3] | 1 (50.0) [2] | 1 (20.0) [1] | 4 (44.4) [6] | 0 (0.0) [0] | 0 (0.0) [0] | 4(30.8) [6] |
| Cold sweat | 1 (50.0) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1 (11.1) [1] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [1] |
| Erythema | 0 (0.0) [0] | 1 (50.0) [2] | 0 (0.0) [0] | 1 (11.1) [2] | 0 (0.0) [0] | 0 (0.0) [0] | 1(7.7) [2] |
| Pruritus | 1 (50.0) [2] | 0 (0.0) [0] | 1 (20.0) [1] | 2 (22.2) [3] | 0 (0.0) [0] | 0 (0.0) [0] | 2(15.4) [3] |
| Vascular disorders | 0 (0.0) [0] | 1 (50.0) [1] | 2 (40.0) [2] | 3 (33.3) [3] | 1 (50.0) [1] | 1 (11.1) [1] | 4(30.8) [4] |
| Flushing | 0 (0.0) [0] | 1 (50.0) [1] | 2 (40.0) [2] | 3 (33.3) [3] | 1 (50.0) [1] | 1 (11.1) [1] | 4(30.8) [4] |
MPS, mucopolysaccharidosis.
FIX, factor IX; study SB-FIX-1501 did not include the 5e12 and 1e13 vg/kg doses, so it did not have cohorts 1 or 2, only cohort 3.
ZFN, zinc-finger nuclease.
Study SB-318-1502 did not report any treatment-related adverse events.
N, total number of subjects in each group; n, number of subjects at each SOC; [T], total number of adverse events. Each subject was counted only once for each applicable specific adverse event. A subject with multiple adverse events within a system organ class was counted only once for that system organ class. The table is sorted by alphabetical order.
Twelve treatment-emergent serious adverse events (TESAEs) were reported in 7 of 13 (53.8%) subjects across all studies and doses, and 9 TESAEs were reported in 5 of 8 (62.5%) subjects receiving the 5e13 vg/kg dose (Table S1). None were assessed as related to study treatment.
See Table S2 for information on TEAEs.
Alanine aminotransferase (ALT)/aspartate aminotransferase (AST) elevations (defined as twice the upper limit of normal [ULN]) were observed in 3 of 9 subjects (33.3%) in study SB-913-1602 (MPS II) at the 1e13 and 5e13 vg/kg doses despite receiving prophylactic prednisone or a corticosteroid equivalent per the protocol (Table S3). Hepatobiliary disorders were reported in four subjects (six events), all from study SB-913-1602 (MPS II), including cholelithiasis (ongoing since day 729), hepatic lesion (day 690 to 1190), hepatocellular injury (ongoing since day 339), hepatomegaly (two events; one ongoing since day 248, the other ongoing since day 708), and portal fibrosis (days 204–344). None were classified as serious.
Genome modification testing at the molecular level
Evidence of genome editing within hepatic tissue using the albumin-transgene fusion mRNA assay was seen at both 24 and 48 weeks after infusion in MPS II subjects 3 and 4 and MPS I subject 10 dosed at 1e13 vg/kg, but not in the two MPS II subjects (subjects 6 and 8) dosed at 5e13 vg/kg with liver biopsy samples (Table 3). Subject 6 experienced an elevated ALT (84 U/L) and subject 8 had an ALT near the ULN (54 U/L). Each liver biopsy sample was tested in quadruplicate; for subject 3, two of four replicates and all four replicates tested positive for the fusion mRNA of albumin plus the iduronate-2-sulfatase gene (IDS) at weeks 24 and 48, respectively. For subject 4, two of four replicates at week 24 tested positive for albumin-IDS fusion mRNA and all four replicates tested positive at week 48. For MPS I subject 10, three of four replicate samples were positive for albumin-IDUA fusion mRNA at both weeks 24 and 48 (Table 3). All pre-treatment liver biopsy samples for these subjects tested negative for the albumin-IDS fusion mRNA.
Table 3.
Summary of genome editing in liver biopsies
| Indication (study no.) | Dose (vg/kg) | Subject no.a |
Baseline |
Week 24 |
Week 48 |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| On-target indels | Off-target indels | Fusion transcript | On-target indels | Off-target indels | Fusion transcript | On-target indels | Off-target indels | Fusion transcript | |||
| MPS II (SB-913-1602) | 5e12 | 1 | - b | – | – | – | – | – | – | – | – |
| 1e13 | 3 | – | – | – | – | – | + c | – | – | + | |
| 4 | – | – | – | – | – | + | – | – | + | ||
| 5e13 | 6 | – | – | – | – | – | – | N/Ad | |||
| 8 | – | – | – | -e | -e | -e | – | – | – | ||
| MPS I (SB-318-1502) | 1e13 | 10 | – | – | – | – | – | + | – | – | + |
| 5e13 | 12 | – | – | – | N/A | – | – | – | |||
Biopsy was not available for SB-FIX-1501, SB-913-1602 subjects 2, 5, 7, 9, or SB-318-1502 subject 11.
–, not detected.
+, detected.
N/A, not available.
Week 24 liver biopsy for subject 8 was collected at an unscheduled time point past week 24 (approximately 7 months after baseline).
No liver biopsies taken from any MPS I or MPS II subjects showed the presence of base pair insertions and/or deletions (indels) via next-generation sequencing (NGS) assay in the albumin or off-target SMCHD1 loci (Table 3). Biopsies were not collected for five subjects: two MPS II subjects (2 and 5) subsequent to the use of anticoagulation, two MPS II subjects (7 and 9) declined, and a biopsy was contraindicated for MPS I subject 11 (reason unknown). Liver biopsies were not performed in SB-FIX-1501 as the subject declined and due to risk of bleeding for the hemophilia B subject during the procedure.
Enzyme activity
In the MPS I and MPS II studies, plasma samples were collected at trough, which was initially defined as no less than 96 h (4 days) after last enzyme replacement therapy (ERT) infusion and before ERT dosing. However, variability in sample collection timing (i.e., not at true trough levels) may have resulted in the detection of variable residual enzyme activity levels from ERT even at 96 h post ERT, since low levels of residual enzyme could still be detected. To reduce interference from residual enzyme activity of ERT and to ensure consistency in sample collection, trough was later defined as 7 days after ERT administration (±1 day). For clarity, the number of days post-ERT infusion for each time point is shown on the graphs in Figures 1, 2, 3, and 4.
Figure 1.
Enzyme, substrate, and ALT levels for study SB-913-1602 (MPS II)
Notes: ALT, alanine aminotransferase; ERT, enzyme replacement therapy; GAG, glycosaminoglycan; HS, heparan sulfate; I2S, iduronate-2-sulfatase; ZFN in vivo editing, zinc-finger nuclease-based in vivo genome editing. Numbers indicate days post-ERT administration. Maximum plasma ALT level is labeled relative to study day in red.
Data on dermatan sulfate (DS) and total GAGs (T-GAG) are presented in Figure S3. The lower limit of quantitation for this quantitative method was 0.78 nmol/mL/h of I2S enzyme activity.
Figure 2.
Enzyme, substrate, and ALT levels for study SB-318-1502 (MPS I)
Notes: ALT, alanine aminotransferase; ERT, enzyme replacement therapy; GAG, glycosaminoglycan; HS, heparan sulfate; iduronidase, α-L-iduronidase; ZFN in vivo editing, zinc-finger nuclease-based in vivo genome editing. Numbers indicate days post-ERT administration. Maximum plasma ALT level is labeled relative to study day in red.
Leukocyte iduronidase activity was measured by a validated diagnostic test. Data on dermatan sulfate (DS) and total GAGs (T-GAG) are presented in Figure S4.
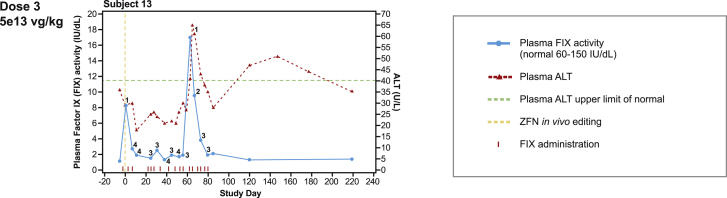
Figure 3.
Enzyme and ALT levels for study SB-FIX-1501 (hemophilia B)
Notes: ALT, alanine aminotransferase; FIX, factor IX; ZFN in vivo editing, zinc-finger nuclease-based in vivo genome editing. Numbers indicate days post-FIX administration.
Figure 4.
Enzyme, substrate, and ALT levels for subject 6 in study SB-913-1602 (MPS II)
Notes: ALT, alanine aminotransferase (normal: 7–55 U/L); ERT, enzyme replacement therapy; GAG, glycosaminoglycan; HS, heparan sulfate; I2S, iduronate-2-sulfatase; ZFN in vivo editing, zinc-finger nuclease-based in vivo genome editing. Numbers indicate days post-ERT administration. Maximum plasma ALT level is labeled relative to study day in red. Data on dermatan sulfate (DS) and total GAGs (T-GAG) are presented in Figure S3.
MPS II subjects at the lowest dose of 5e12 vg/kg had no appreciable increase in plasma I2S enzyme activity (Figure 1). Subject 1 experienced transient increases in plasma I2S activity. This was attributed to ERT administration on the day before plasma I2S measurement (versus 6–7 days post-ERT measurement of plasma I2S for the other subjects) (Figure 1). Subject 2 experienced an increase in urine HS-GAG at study day 350 despite no ERT withdrawal; levels subsequently decreased by study day 400 but were variable up to study day 1,100.
MPS II subject 6, dosed at 5e13 vg/kg, had a transient increase in plasma I2S activity approaching the normal range, peaking at 49.18 nmol/h/mL around study day 40 and declining to near baseline by study day 100 (Figure 1). The same subject also concurrently experienced liver enzyme elevation with a peak ALT level of 84 U/L (ULN, 40 U/L) (Figure 4).
MPS I subject 10 had a small transient increase in leukocyte iduronidase enzyme activity to within the lower end of the normal range (1e13 vg/kg, 18.98 nmol/h/mg), which peaked at day 138 of the study and decreased thereafter. There was a possible transient increase in leukocyte iduronidase enzyme activity in subject 11 (5e13 vg/kg) that peaked within the lower end of the normal range. Subject 12 had a transient increase in leukocyte iduronidase activity to the middle of the normal range, peaking shortly after day 0, declining rapidly, and then peaking again just after day 50 of the study. No consistent relationship between enzyme activity and urine GAG levels was observed (Figure 2). Plasma iduronidase enzyme activity was not detectable in any of the MPS I subjects. Iduronidase and I2S enzymes catalyze the breakdown of GAGs located throughout tissues, and levels of GAGs were assessed in the urine and circulation.
The FIX activity in subject 13 increased from 1 IU/dL at baseline to 17 IU/dL at day 60, which was attributed to the FIX infusion on the previous day, which the subject continued throughout the study, and not the study treatment (Figure 3). The subject’s use of recombinant FIX concentrate did not decrease after gene therapy, suggesting that the gene therapy did not produce a meaningful increment in FIX activity.
ERT withdrawal duration
Of the 12 subjects receiving ERT, 9 were withdrawn from ERT for a period of time ranging from 43 to 369 days (Figures 2, 3, and 4). Of these, 8 restarted ERT and one (subject 10) chose not to restart ERT even though he did not have an increase in iduronidase enzyme activity levels related to ZFN in vivo editing. In study SB-913-1602 (MPS II), 6 of 9 subjects were withdrawn from ERT, with off-ERT durations ranging from 43 to 131 days. All three subjects in study SB-318-1502 (MPS I) were withdrawn from ERT, with off-ERT durations of 101 to 369 days. In study SB-FIX-1501 (hemophilia B), the subject was not withdrawn from FIX treatment.
In all MPS I and MPS II subjects who underwent ERT withdrawal, enzyme activity decreased and urinary GAG levels increased independent of dose levels. In those subjects where evidence of genome editing was detected (MPS II subjects 3 and 4; MPS I subject 10) there was no evidence of sustained enzyme activity levels; increased GAG levels were also noted. Overall, urinary GAG levels followed an inverse course related to enzyme activity.
Vector shedding
The highest levels of vector genome were detected in plasma at day 1 post-treatment. By week 24, there were no measurable levels of vector genome in all tested matrices (plasma, saliva, urine, semen, and feces) in any of the subjects (Table S4).
Immunogenicity
For the SB-913-1602 (MPS II) study (data are available for all time points except for month 24 from subject 8 and subject 9), two subjects with pre-existing anti-I2S anti-drug antibodies (ADA) detected at baseline remained positive throughout the study and no treatment-boosted ADA response was observed. The remaining seven subjects had no detectable ADA at baseline and post-treatment. For the SB-318-1502 (MPS I) study, evaluation of anti-iduronidase antibodies is pending assay availability. No inhibitor was detected at baseline and post treatment for the subject treated with SB-FIX.
Other results
No clinically significant findings in either MPS I or MPS II subjects were seen in the analysis of exploratory endpoints of disease progression. The subject in the SB-FIX-1501 study experienced two bleeding episodes, which resolved during the course of the study.
Discussion
These three clinical studies were the first in vivo genome editing studies conducted in humans. They provide proof of concept for the potential for in vivo genome editing with ZFN technology and show a favorable safety profile. No subject experienced a serious TRAE. Acute reactions occurred mainly at the highest dose evaluated (5e13 vg/kg) and were generally minor and similar in nature to those seen with other AAV-based gene therapies.3,22,30,31
There was evidence of successful ZFN in vivo genome editing through the detection of albumin-IDS fusion mRNA in the liver of two MPS II subjects and albumin-IDUA fusion mRNA in the liver of one MPS I subject. At the 1e13 vg/kg dose, MPS I subject 10 had a transient increase of leukocyte iduronidase enzyme activity to the middle of normal with evidence of in vivo genome editing.
In the MPS II study, subject 6 had plasma I2S activity approaching the normal range at week 6 at the highest dose. However, this increase in I2S activity was transient with no evidence of in vivo genome editing in liver biopsies at week 24. This decline coincided with a sharp increase in ALT at week 9. Although it is possible that the ALT elevation and I2S activity reduction in the plasma for subject 6 were temporally related, their direct relationship cannot be confirmed. As well, this subject’s weight (139 kg) was double the average of other MPS II subjects dosed at 5e13 vg/kg. Therefore, subject 6 received approximately twice the total vector genome dose of other subjects in the study, which may have contributed to transient I2S plasma levels approaching the normal range in this subject. This may indicate that increasing the dose > 5e13 vg/kg has the potential to achieve I2S plasma levels reaching the normal range. As liver size increases with the weight of the subject, but not in proportion to the total body weight and liver cells may not increase proportionately,32,33 it may be worth exploring dosing per square meter rather than per kilogram.
The increased total dose that subject 6 received may also have caused or contributed to the observed ALT elevation that was seen at week 9 post-infusion. While the subject was receiving an oral prophylactic dose of prednisone (60 mg/day), it is possible that the sharp ALT elevation was caused by immune-mediated hepatocyte stress or injury due to the higher total dose of SB-913. In liver-directed genomic medicine, ALT elevations are common and usually treated with oral corticosteroids; late treatment or no treatment can lead to a decrease in transgene expression.34 The SB-FIX clinical study subject did not experience any ALT elevation or require any therapeutic corticosteroid intervention. Therefore, the relationship of prophylactic corticosteroids to reduction of ALT/AST elevation in these studies is unclear.
Despite the transient increase in plasma I2S activity, there was no evidence of genome editing in the liver biopsy for subject 6. Genome editing may have occurred but was subsequently lost between the liver enzyme elevation and the timing of the biopsy or missed due to heterogeneity in the virus distribution in the liver after i.v. delivery,35 or that the liver biopsy sample may not have been large enough to capture the area where ZFN in vivo genome editing may have occurred. Only a subset of liver biopsy samples in the MPS I and MPS II studies showed evidence of ZFN in vivo genome editing, likely due to the low incidence of DSB and subsequent transgene integration events in hepatocytes and sampling variability. The albumin-transgene fusion mRNA assay is approximately 150-fold more sensitive than the NGS assay (indels) because each molecule of albumin DNA makes hundreds of copies of mRNA, explaining why genome editing was detectable using the albumin-transgene RNA fusion assay but not the NGS assay. This indicates that in vivo genome editing occurred at a very low level in these subjects.
As with any gene therapy that targets the liver, particular attention will be given to liver surveillance over time using imaging to monitor for any possible liver mass. While ZFNs efficiently create DSBs at specific sites in the genome, on rare occasions they may bind to incorrect locations with a high level of homology to the target sequence, producing off-target mutations. Off-target mutations can be associated with chromosomal rearrangement that could activate oncogenes, potentially leading to cancer.36 Although not observed in nonclinical safety studies or in the clinical studies to date, the carcinogenic potential of ZFN off-target activity is a remote possibility that is being monitored in the long-term follow-up study.
As these three ZFN in vivo editing therapy studies were performed in a similar time frame, oversight by a Safety Monitoring Committee (SMC) was optimized as there were two (out of three) common members across the studies. The enrollment into the MPS II study was slightly ahead of the MPS I and FIX studies and, as a result, the MPS II study data were able to inform safety and dose escalation in the MPS I and hemophilia B studies. This allowed for optimization of the dose escalation within each study. After the first two subjects in the MPS II study showed an acceptable safety profile at 5e12 vg/kg, the SMC recommended dose escalating to 1e13 vg/kg. Reviewing the MPS II data allowed for a higher starting dose of 1e13 vg/kg in the MPS I study. Then, based on the safety assessment of the MPS II subjects dosed at 1e13 and 5e13 vg/kg as well as the single MPS I subject dosed at 1e13 vg/kg, the MPS I study proceeded directly to the highest dose level of 5e13 vg/kg. Based on the safety data from the MPS I and MPS II studies, the hemophilia B study’s starting dose was chosen to be the highest dose (5e13 vg/kg), as recommended by the SMC to give the highest opportunity to provide clinical benefit. The regulatory agency was thereafter informed and agreed with the SMC recommendation. This cross-study approach to dose selection increased the potential of a therapeutic benefit to subjects and minimized the number of subjects potentially exposed to a sub-therapeutic dose.
In the disease model, AAV8 was used while AAV6 was used in GLP mouse toxicology studies that supported the safe use of AAV6. These preclinical studies in mouse models demonstrated that liver-targeted ZFNs delivered in concert with a promotorless donor cDNA template could produce sustained, supraphysiological levels of circulating protein4,13,23 by the process that naturally drives albumin expression. The success rate for clinical development of new drugs has historically been low despite the use of animal models.37,38 For these liver-targeted ZFN genome editing studies, translational efficacy could have been impacted by species-specific differences in AAV transduction efficiency of the liver,39 transgene expression,40 DNA repair,41 or different proteins in human and mouse blood, which interact with AAV serotypes in distinct ways.42
Successful double-strand DNA cleavage and transgene integration requires a single hepatocyte to be simultaneously transduced by three different AAVs that individually contain the heterodimeric ZFNs and the donor cDNA template. Balancing the doses of the three AAV vectors to maximize effectiveness while minimizing potential adverse events was a critical aspect of the study design and was heavily informed by the preclinical studies.4,13,23 Those studies suggested that administering excess of donor cDNA template, relative to the amount of ZFNs administered, would lead to higher expression of the therapeutic protein, and a ratio of 1:1:8 (ZFN 1:ZFN 2:donor transgene) was selected for these clinical studies. The dose of ZFNs administered in these in vivo genome editing clinical studies was likely below this threshold for adequate transgene expression. Evidence of albumin-IDS fusion mRNA in liver and plasma I2S was found in our MPS II clinical study, and higher levels of these biomarkers may have been observed if dose escalation was continued.
These studies had certain limitations in common. A large portion of potential subjects failed screening due to single-nucleotide polymorphisms (SNPs) and pre-existing neutralizing antibodies (NAbs). SNPs at ZFN-binding sites reduce ZFN binding affinity and hence editing efficiency. In a clinical trial setting, these patients were excluded in order to provide a clear picture of potential efficacy and remove confounding variables. It is not known if these patients would be eligible for treatment in a real-world setting. The low number of available liver biopsies in 7 of the 13 subjects limits interpretability of that data. As per the MPS I and MPS II study protocols, ERT withdrawal was at the discretion of the investigator, and was not done in 3 of 12 subjects on ERT. As well, although definition of ERT infusion trough was clarified, variability in the timing of sample collection after ERT infusion may have resulted in the detection of variable levels of residual enzyme activity from the ERT infusion, as, even at 96 h post ERT, low levels of residual enzyme could still be detected. Nonetheless, the impact of ERT withdrawal was consistent in all subjects with plasma enzyme activity levels rapidly dropping and urine GAG levels rapidly increasing.
As with all AAV-based gene therapies, the presence of anti-AAV6 capsid NAbs is highly likely to elicit a strong immune response and negatively impact the potential benefit of therapy. Therefore, potential subjects with anti-AAV6 capsid NAbs were excluded from the study. This reduced the number of eligible patients for these studies and the patients that could potentially benefit from AAV-based treatments. To provide this type of potentially lifelong treatment to all patients, solutions need to be explored that allow broader access to treatment, such as higher vector doses to overcome a potential NAb response or removal of AAV NAbs by procedures such as plasmapheresis to reduce the anti-AAV response.43
These early results of first-generation ZFN in vivo genome editing studies demonstrated a favorable safety profile and evidence of low-level in vivo genome editing in the liver but no long-term sustained enzyme expression in blood. In the future, a novel in vivo genome editing therapy, potentially featuring improved ZFNs and/or an alternate delivery system, may provide therapeutic benefit in diseases similar to the ones studied here.
Materials and methods
Screening
Fifty-four patients were screened for inclusion in the studies. Of the 24 patients with MPS II screened, 9 were enrolled. Of the 20 patients with MPS I screened, 3 were enrolled. Of the 10 patients with hemophilia B screened, 1 was enrolled. The most common reason for failing screening was due to SNPs and NAbs (see Figure 5 for details).
Figure 5.
Clinical design schematics of the three clinical studies
(A) Study SB-913-1602. (B) Study SB-318-1502. (C) Study SB-FIX-1501. NAbs, neutralizing antibodies to the AAV6 capsid; SMC, safety monitoring committee; SNPs, single-nucleotide polymorphisms.
Clinical studies overview and subjects
Three phase 1/2, open-label, single-dose escalation, 36-month studies were performed in parallel to allow one study to inform on the others. Subjects in each study were dosed in ascending dose cohorts with a SMC review of data from a completed dose cohort before escalating to the next higher dose cohort. The safety data from dose cohorts in one study was also used to determine the next dose cohort in the other studies. This approach minimized the potential that subjects would get a non-efficacious dose.
Each of the three clinical studies was designed to assess primarily the safety and tolerability of ZFN in vivo editing therapy, lasting 3 months for screening and 36 months for treatment and follow-up:
-
•
Study SB-913-1602 was a phase 1/2, multicenter dose-ranging study of SB-913 (for MPS II, NCT03041324; Table S5 for inclusion/exclusion criteria)
-
•
Study SB-318-1502 was a phase 1/2, multicenter dose-ranging study of SB-318 (for MPS I, NCT02702115; Table S6 for inclusion/exclusion criteria)
-
•
Study SB-FIX-1501 was a phase 1 ascending dose study of SB-FIX (for hemophilia B, NCT02695160; Table S7 for inclusion/exclusion criteria)
Due to COVID-19, some assessments were performed remotely or not done at all.
Upon study completion, subjects were invited to participate in an ongoing long-term follow-up study (ST-IVPRP-LT01, NCT04628871) to monitor the long-term safety of ZFN in vivo editing.
Baseline assessments
Baseline assessments were performed within 21 days before ZFN in vivo editing infusion. In subjects with MPS I and MPS II receiving ERT, ERT was withheld during the week of the administration of ZFN in vivo editing to enable accurate baseline testing.
Clinical study dose escalation
Safety and tolerability data were leveraged from the three studies to determine the most appropriate dose for enrolling study cohorts (Figure 5). Studies were performed in parallel and dosing was approved by the SMC based upon dosing in the previous study. The starting dose began with SB-913-1602 at 5e12 vg/kg in MPS II subjects and was escalated to 1e13 vg/kg, then to 5e13 vg/kg (Figure 5A). Study SB-318-1502 then started at 1e13 vg/kg and escalated to 5e13 vg/kg in MPS I subjects (Figure 5B). SB-FIX-1501 started at the highest and only dose of 5e13 vg/kg in the hemophilia B subject (Figure 5C).
Investigational product and administration
The investigational therapy in each study consisted of a combination of three individual AAV 2/6 vectors with a ratio of 1:1:8 in a baculovirus-insect (Sf9) cell system encoding the left-side ZFN (ZFN 1), right-side ZFN (ZFN 2), and the promoterless donor cDNA template, which differed by product. For investigational products SB-318, SB-913, and SB-FIX, hIDUA (SB-IDUA), hI2S (SB-I2S), and human FIX donor cDNA, each with human albumin homology arms, were the cDNA templates used, respectively. Before i.v. administration of the investigational products, subjects were pre-medicated with acetaminophen 650 mg orally and diphenhydramine hydrochloride 25–50 mg orally or i.v. before the infusion. Non-steroidal anti-inflammatory drugs were used if needed to manage infusion-related adverse events, such as transient fever, chills, and/or nausea, at the discretion of the principal investigator. Subjects in studies SB-318-1502 and SB-913-1602 received pre-treatment with oral prednisone or an equivalent corticosteroid starting 2 days before the infusion. In study SB-FIX-1501, prednisone or an equivalent corticosteroid was given if subjects developed an elevation of ALT greater than twice the baseline value based on procedures from an earlier protocol for hemophilia B per guidance from key opinion leaders for the use of steroids and risks in the hemophilia patient population. ALT/AST elevations were defined as twice the ULN based on relevant literature in the patient population studied.29,43
The three investigational products were administered as a one-time infusion through an intravenous catheter using a constant rate infusion pump while monitoring the subject’s vital signs (pulse, blood pressure, respiration rate, and temperature). Dosing was based on body weight.
Primary endpoint: Safety and tolerability
The primary endpoint in each study was safety and tolerability of ZFN in vivo editing, as assessed by AE incidence. TRAEs and TESAEs were assessed by CTCAE. Investigators were responsible for monitoring and reporting all AEs. In studies SB-318-1502, SB-913-1602, and SB-FIX-1501, additional safety evaluations were performed, including routine hematology, chemistry and liver function tests, vital signs, physical exam, electrocardiogram, echocardiogram, concomitant medications, cranial nerve and muscle strength testing, and serial α-fetoprotein (AFP) testing (serial AFP testing results are reported in Tables S8–S10; AFP values fluctuated but remained within the normal range) and magnetic resonance imaging (MRI) of liver to evaluate for liver mass.
Secondary endpoints
Changes from baseline in enzyme activity in blood, GAG levels in urine for SB-318-1502 (MPS I) and SB-913-1602 (MPS II), and FIX activity in plasma for SB-FIX-1501 (hemophilia B) were measured. Changes in therapy, including frequency and dose of ERT for MPS I and MPS II, and number of FIX units infused per week for hemophilia B were recorded. Also, change from baseline in number and severity of bleeding episodes (for SB-FIX-1501) was measured. Presence and shedding of AAV2/6 vector DNA over time was measured in plasma, saliva, urine, stool, and semen by polymerase chain reaction (PCR), and changes in neutralizing antibodies to FIX.
Exploratory endpoints
Exploratory endpoints in all three studies included percentage and durability of gene modification at the albumin locus in liver biopsy tissue; changes in antibody levels to I2S (MPS II study only); and histopathological exam of liver tissue. The MPS I and MPS II studies also evaluated disease progression endpoints, such as forced vital capacity, 6-min walk test, joint range of motion, neurocognitive abilities, eye exam, liver and spleen volume, and MRI of the brain and cervical spine to evaluate soft tissue and/or bone.
GAG analysis in MPS I and MPS II subjects
Total GAG and individual disaccharides, HS and DS, were analyzed in the MPS I and MPS II clinical studies. Total GAG was measured using a validated quantitative Blyscan assay, a direct spectrophotometric determination of GAGs in centrifuged urine samples with 1,9-dimethyl-methylene blue (DMB) color reagent manufactured by Biocolor (UK). DMB formed a complex with GAGs in urine and precipitated out of solution via centrifugation. DMB was then dissociated from GAGs using a kit-specific dissociation reagent. The amount of DMB, which corresponds to urine GAG concentration, is directly proportional to absorbance of DMB at 656 nm and was quantitated using bovine tracheal chondroitin-4-sulfate as reference standard. Concentration of GAG was reported relative to the creatinine concentration in urine samples.
Urine DS and HS degraded into disaccharides by methanol HCl treatment were quantified using validated quantitative stable isotope dilution dilution-tandem mass spectrometry. Specific dimers derived from DS and HS were separated by ultra-performance liquid chromatography and analyzed by electrospray ionization tandem mass spectrometry quantified using selected reaction monitoring for each targeted GAG product (DiDS and DiHS-NS) and its corresponding internal standard (d6-DiDS and d6-DiHS-NS). Concentration of DS and HS were reported relative to the creatinine concentration in urine samples.
Plasma I2S activity
Plasma I2S activity was measured using the method adapted from published work.44 This was a two-step enzymatic activity assay using 4-methylumbelliferyl-α-L-iduronate 2-sulfate substrate and the liberated 4-methylumbelliferone (4MU) fluorescence was measured using fluorometer. The optimized assay contained two calibration curves, a recombinant I2S curve and a 4MU curve for activity calculation and assay monitoring. The lower limit of quantitation for this quantitative method was 0.78 nmol/mL/h of I2S enzyme activity.
Leukocyte iduronidase activity
α-iduronidase activity in leukocytes was measured using 4-methylumbelliferyl-α-L-iduronide substrate and the liberated 4MU fluorescence was measured using fluorometry. The optimized assay contained two calibration curves, a recombinant iduronidase curve and a 4MU curve for activity calculation and assay monitoring. The lower limit of quantitation for this quantitative method was 0.005 nmol/mL 4MU with the reported enzyme activity normalized to per mg of input leukocyte lysate concentration.
Immunogenicity
ADA against I2S in human serum were measured at baseline, weeks 4, 12, 24, 36, and 48, and months 18 and 24 using a homogenous bridging electrochemiluminescent assay. This qualitative assay utilized a commercial anti-I2S polyclonal antibody as positive control and a minimum required dilution of 1:20. The assay sensitivity was 9.73 ng/mL for the screening assay and 17.7 ng/mL for the confirmatory assay. The assay had a drug tolerance of 4 μg/mL I2S in the presence of 100 ng/mL of positive control antibody.
FIX inhibitors were measured by the FIX inhibitor Nijmegen Bethesda assay at a CLIA lab.
Complement activation was not evaluated as no toxicity was observed in animal and clinical studies at the highest clinical dose (5e13 vg/kg), and in the era that all subjects were dosed in these three studies, complement activation had not yet emerged as a distinct, potential class effect of high-dose systemic AAV administration across multiple indications.
AAV shedding
Vector genome DNA levels were measured separately for transgene and ZFN components. Evaluation for AAV2/6-transgene (FIX, IDUA, IDS) and AAV2/6-ZFN vector DNA was performed in plasma, semen, saliva, stools, and urine by qPCR at baseline, day 1 (plasma only), weeks 1, 2, and 4, and every 4 weeks after until two (SB-FIX) or three (SB-913 and SB-318) consecutive below limit of detection (LOD) samples were obtained post-treatment. The lower limit of quantitation (LLOQ) for all assays were at 10–20 copies per reaction with 100 ng of input DNA for semen, saliva, and feces and 5 μL of input if plasma or urine DNA (equivalent to DNA isolated from 10 μL of plasma or 250 μL of urine, respectively).
Liver biopsy collection
Liver biopsy samples were collected per site-specific biopsy collection procedures using a 16- or 18-gauge biopsy needle at baseline, week 24, and week 48. Collected samples were divided into three portions: ∼10 mg aliquot 1 and ∼20 mg aliquot 2 were used for indel and albumin-transgene fusion mRNA analysis, ∼30 mg aliquot 3 was used for pathology evaluation. Samples collected for indel and albumin-transgene fusion mRNA analysis were snap frozen in liquid nitrogen and stored at −70°C until they were shipped to the central lab.
Genome modification assessment at the molecular level
Two molecular assays were designed to detect ZFN-based genome editing in liver biopsy samples, including an NGS assay to detect base pair insertions and/or deletions (indels) and an albumin-transgene fusion mRNA assay (Figure S2A). ZFN-introduced DSBs can be repaired through non-homologous end joining (Figure S2B), which is an error-prone DNA repair pathway that can result in small indels at the cut site. The indel assay was developed and qualified to detect indels within the albumin on-target locus and the off-target site SMCHD1 gene locus which was the only off-target locus identified during non-biased, off-target screening of the ZFN.
To perform the NGS (MiSeq, Illumina) assay, gDNA was isolated from approximately 5 mg of liver biopsy tissues using modified a MasterPure protocol (Lucigen) and quantified using a modified Quant-iT dsDNA Assay (Thermo Fisher). Each sample was analyzed in four replicates, 150 ng gDNA each. Genetic regions corresponding to the on-target modification site in albumin and the off-target site SMCHD1 loci were amplified by multiplexed PCR reactions. These reactions were then used for a second PCR reaction that generated amplicons compatible with the MiSeq sequencing platform and sample-specific barcodes added. After the sequencing reaction was completed, paired FASTQ sequence reads were merged, filtered by read quality score, and demultiplexed for the specific amplicon (albumin or SMCHD1) and for specific samples using the two sample barcodes. Filtered sequences were then aligned to the expected sequence and the number of sequences with indels calculated. A qualified MiSeq assay was used to evaluate gene modification in liver biopsy samples. The assay qualification was performed using HepG2 cell lines that were modified by ZFN targeting the albumin locus and subsequently single cell cloned to select for a population where both albumin and SMCHD1 genes were 100% modified in the same cell. Wild-type (WT) HepG2 were used as a negative control samples (WT - no modification in either albumin or SMCHD1 genes were expected). Modified (100% indels) and WT cells (0% indels) were mixed in different ratios to prepare standard calibrators and quality control samples. The assay qualification included assessments of accuracy, precision, selectivity, the LLOQ, and LOD in liver sample matrix. The assay LLOQ was determined as 0.1% indels for the albumin gene and 0.2% indels for the SMCHD1 gene. The LOD was determined as 0.025% indels for the albumin gene and 0.05% indels for the SMCHD1 gene, allowing for indel detection in 1 modified cell mixed with 4,000 and 2,000 unmodified cells, respectively.
The presence of albumin-transgene fusion transcript was evaluated using reverse transcription (RT) qPCR. Total RNA was purified from 2 to 5 mg of liver biopsy tissues using PureLink RNA mini Kit (Thermo Fisher), the RNA concentration was measured using a NanoDrop spectrophotometer. Two μg of total RNA was evaluated in a single RT reaction. For each RT reaction, four replicate qPCR reactions were run for albumin-transgene RNA fusion detection. PCR reactions were then analyzed using LabChip GX Touch capillary electrophoresis for the detection of the specific band. The specificity and sensitivity of the integration assay were assessed using cell lines. The signal was considered positive if at least two of four replicates showed the presence of fusion transcript by both qPCR and capillary electrophoresis. The qRT-PCR LOD was determined as detection of 1 modified cell in 600,000 unmodified cells.
Statistics
These were exploratory phase 1/2 studies and thus there was limited statistical power to evaluate efficacy and related biological endpoints. Therefore, analyses were primarily descriptive and exploratory in nature.
Study approval
The studies were conducted according to the principles of the Declaration of Helsinki. All subjects provided written informed consent prior to inclusion in the study. The institutional review board (IRB)/independent ethics committee for each site and central IRB approved the protocols. Each study was overseen by independent data and safety monitoring committees. Inclusion and exclusion criteria (see Tables S4–S6) stated in the protocol were used to screen subjects for enrollment.
Trial registration: ClinicalTrials.gov NCT03041324 (MPS II); NCT02702115 (MPS I); NCT02695160 (hemophilia B).
Acknowledgments
The authors thank the study participants and their families and friends. We also thank the staff (caregivers, study support personnel, and health care professionals) at the clinical trial sites: UCSF Benioff Children’s Hospital Oakland, Ann & Robert H. Lurie Children's Hospital of Chicago, Cincinnati Children's Hospital Medical Center, NYU School of Medicine, Georgetown University, and University of North Carolina at Chapel Hill, North Carolina TraCS Clinical & Translational Research Center (UL1TR002489), and Sangamo Therapeutics Clinical Operations and Biometrics; and Yonghua Pan, Sangamo Therapeutics, for developing I2S and iduronidase enzyme activity assays. Medical writing support was provided by Patricia Rawn and Judy Wiles of Facet Communications funded by Sangamo Therapeutics. All three studies were funded by Sangamo Therapeutics who provided the study materials.
Author contributions
P.H., C.E.P., B.K.B., H.L., C.M.K., and J.M. participated in the study as investigators, reviewed and interpreted the data, and approved all drafts of the manuscript. All authors had access to and reviewed the study data, and carefully reviewed and approved all drafts of the manuscript including the final version.
Declaration of interest
Marina Falaleeva, Andres G. Villegas, Jennifer Zeitler, Kathleen Meyer, Wendy Swenson, and Lisa Shiue are employees and stockholders of Sangamo Therapeutics, Inc. Didier Rouy is a former employee of Sangamo Therapeutics, Inc. Liching Cao is an employee of Sangamo Therapeutics, Inc. and has a patent pending (16/534,483; WO 2020/05947; US 2020/0071743) for enzymatic assays supporting MPS studies. Cheryl Wong Po Foo is a former employee of Sangamo Therapeutics, Inc., and a current employee of Astellas Gene Therapies. Sagar Vaidya is a former employee of Sangamo Therapeutics, Inc., and has a patent pending for “Method for the treatment of mucopolysaccharidosis type II” and is a current employee of Travere Therapeutics.
Weston Miller is a former employee of Sangamo Therapeutics, Inc., owns stock in Sangamo Therapeutics, Inc., and is a full-time employee of Astellas Gene Therapies.
Paul Harmatz has received writing support, clinical trial support, and/or consulting fees from Sangamo Therapeutics, Inc., BioMarin Pharmaceutical Inc., Takeda/Shire, REGENXBIO, Denali Therapeutics Inc., Inventiva Pharma, QED Therapeutics, Ascendis Pharma, Orphazyme, Ultragenyx Pharmaceutical, Amicus, Aeglea BioTheraeutics, Homology, JCR Pharmaceuticals, Ltd, Paradigm, Audentes Therapeutics, and Chiesi; has received registry support from Genzyme, BioMarin Pharmaceutical Inc., and Shire (Takeda). Barbara Burton has served as a consultant for Biomarin, Shire (Takeda), Denali Therapeutics Inc., Genzyme, Ultragenyx, Moderna, Aeglea, Horizon, Alexion, Inventiva, Applied Therapeutics, SIO, and JCR Pharmaceuticals Co., Ltd. She has received clinical trial funding from Biomarin, Shire (Takeda), Ultragenyx, Sangamo Therapeutics, Inc., and Homology Medicines. Carlos Prada has served as a consultant for Sanofi-Genzyme and Shire (Takeda). Heather Lau is an employee of Ultragenyx Pharmaceutical, Inc., and reports grants and non-financial support from Sangamo Therapeutics, Inc.., during the conduct of the study. Outside the submitted work, Dr. Lau reports personal fees from Actelion, Ltd.; grants and personal fees from Amicus Therapeutics; grants and personal fees from Biomarin Pharmaceutical, Inc.; personal fees from Chiesi; grants from Denali Therapeutics, Inc.; grants from Mallinckrodt Pharmaceuticals; grants and personal fees from Orphazyme; grants from Intrabio, Ltd.; grants, personal fees, and non-financial support from Sanofi; grants, personal fees, and non-financial support from Takeda; grants, personal fees, non-financial support, and other from Ultragenyx Pharmaceutical, Inc.; grants from Pfizer, Inc.; grants and personal fees from Prevail Therapeutics, Inc.; grants, personal fees, and non-financial support from Aspa Therapeutics; grants from Protalix BioTherapeutics; personal fees and other from National Gaucher Foundation; personal fees from Taysha Therapeutics; personal fees and other from Muscular Disease Association; personal fees and other from National Fabry Disease Foundation; personal fees and other from MPS Society; grants, personal fees, and other from Adult Polyglucosan Body Disorder Disease Foundation; personal fees and other from National Fabry Disease Foundation; and grants, personal fees and other from National Tay Sachs and Allied Disease Foundation. Craig Kessler has served as an advisory board participant with an honorarium from Sangamo Therapeutics, Inc., and his university has received support from Sangamo Therapeutics, Inc., for clinical gene therapy research. He has also served as chair, DSMB for a FVIII gene therapy trial with Bayer. Joseph Muenzer has served as a principal investigator for Takeda clinical trials, received travel and speaking fees to speak at Takeda meetings, and served on Takeda advisory boards. He has participated in advisory boards and consulted for Genzyme, Bluebird Bio, Inc., Denali Therapeutics, Inc., and served as a consultant for REGENXBIO.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2022.10.010.
Supplemental information
References
- 1.Nature Editors First in vivo human genome editing trial. Nat. Biotechnol. 2018;36:5. doi: 10.1038/nbt0118-5b. [DOI] [PubMed] [Google Scholar]
- 2.Fraldi A., Serafini M., Sorrentino N.C., Gentner B., Aiuti A., Bernardo M.E. Gene therapy for mucopolysaccharidoses: in vivo and ex vivo approaches. Ital. J. Pediatr. 2018;44:130. doi: 10.1186/s13052-018-0565-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batty P., Lillicrap D. Advances and challenges for hemophilia gene therapy. Hum. Mol. Genet. 2019;28:R95–R101. doi: 10.1093/hmg/ddz157. [DOI] [PubMed] [Google Scholar]
- 4.Laoharawee K., DeKelver R.C., Podetz-Pedersen K.M., Rohde M., Sproul S., Nguyen H.O., Nguyen T., St Martin S.J., Ou L., Tom S., et al. Dose-dependent prevention of metabolic and neurologic disease in murine MPS II by ZFN-mediated in vivo genome editing. Mol. Ther. 2018;26:1127–1136. doi: 10.1016/j.ymthe.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Favaro P., Finn J.D., Siner J.I., Wright J.F., High K.A., Arruda V.R. Safety of liver gene transfer following peripheral intravascular delivery of adeno-associated virus (AAV)-5 and AAV-6 in a large animal model. Hum. Gene Ther. 2011;22:843–852. doi: 10.1089/hum.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stone D., Liu Y., Li Z.Y., Strauss R., Finn E.E., Allen J.M., Chamberlain J.S., Lieber A. Biodistribution and safety profile of recombinant adeno-associated virus serotype 6 vectors following intravenous delivery. J. Virol. 2008;82:7711–7715. doi: 10.1128/JVI.00542-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang H., Lillicrap D., Patarroyo-White S., Liu T., Qian X., Scallan C.D., Powell S., Keller T., McMurray M., Labelle A., et al. Multiyear therapeutic benefit of AAV serotypes 2, 6, and 8 delivering factor VIII to hemophilia A mice and dogs. Blood. 2006;108:107–115. doi: 10.1182/blood-2005-12-5115. [DOI] [PubMed] [Google Scholar]
- 8.Zincarelli C., Soltys S., Rengo G., Rabinowitz J.E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 9.Nathwani A.C., Gray J.T., Ng C.Y.C., Zhou J., Spence Y., Waddington S.N., Tuddenham E.G.D., Kemball-Cook G., McIntosh J., Boon-Spijker M., et al. Self-complementary adeno-associated virus vectors containing a novel liver-specific human factor IX expression cassette enable highly efficient transduction of murine and nonhuman primate liver. Blood. 2006;107:2653–2661. doi: 10.1182/blood-2005-10-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nathwani A.C., Rosales C., McIntosh J., Rastegarlari G., Nathwani D., Raj D., Nawathe S., Waddington S.N., Bronson R., Jackson S., et al. Long-term safety and efficacy following systemic administration of a self-complementary AAV vector encoding human FIX pseudotyped with serotype 5 and 8 capsid proteins. Mol. Ther. 2011;19:876–885. doi: 10.1038/mt.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papapetrou E.P., Schambach A. Gene insertion into genomic safe harbors for human gene therapy. Mol. Ther. 2016;24:678–684. doi: 10.1038/mt.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadelain M., Papapetrou E.P., Bushman F.D. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer. 2011;12:51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 13.Sharma R., Anguela X.M., Doyon Y., Wechsler T., DeKelver R.C., Sproul S., Paschon D.E., Miller J.C., Davidson R.J., Shivak D., et al. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood. 2015;126:1777–1784. doi: 10.1182/blood-2014-12-615492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeKelver R.C., Choi V.M., Moehle E.A., Paschon D.E., Hockemeyer D., Meijsing S.H., Sancak Y., Cui X., Steine E.J., Miller J.C., et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20:1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yarrington R.M., Verma S., Schwartz S., Trautman J.K., Carroll D. Nucleosomes inhibit target cleavage by CRISPR-Cas9 in vivo. Proc. Natl. Acad. Sci. USA. 2018;115:9351–9358. doi: 10.1073/pnas.1810062115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller J.C., Patil D.P., Xia D.F., Paine C.B., Fauser F., Richards H.W., Shivak D.A., Bendaña Y.R., Hinkley S.J., Scarlott N.A., et al. Enhancing gene editing specificity by attenuating DNA cleavage kinetics. Nat. Biotechnol. 2019;37:945–952. doi: 10.1038/s41587-019-0186-z. [DOI] [PubMed] [Google Scholar]
- 19.Zeitler B., Froelich S., Marlen K., Shivak D.A., Yu Q., Li D., Pearl J.R., Miller J.C., Zhang L., Paschon D.E., et al. Allele-selective transcriptional repression of mutant HTT for the treatment of Huntington's disease. Nat. Med. 2019;25:1131–1142. doi: 10.1038/s41591-019-0478-3. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs F., Gordts S.C., Muthuramu I., De Geest B. The liver as a target organ for gene therapy: state of the art, challenges, and future perspectives. Pharmaceuticals (Basel) 2012;5:1372–1392. doi: 10.3390/ph5121372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinderer C., Bell P., Gurda B.L., Wang Q., Louboutin J.P., Zhu Y., Bagel J., O'Donnell P., Sikora T., Ruane T., et al. Liver-directed gene therapy corrects cardiovascular lesions in feline mucopolysaccharidosis type I. Proc. Natl. Acad. Sci. USA. 2014;111:14894–14899. doi: 10.1073/pnas.1413645111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nathwani A.C., Tuddenham E.G.D., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C., et al. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ou L., DeKelver R.C., Rohde M., Tom S., Radeke R., St Martin S.J., Santiago Y., Sproul S., Przybilla M.J., Koniar B.L., et al. ZFN-mediated in vivo genome editing corrects murine hurler syndrome. Mol. Ther. 2019;27:178–187. doi: 10.1016/j.ymthe.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy S.L., High K.A. Gene therapy for haemophilia. Br. J. Haematol. 2008;140:479–487. doi: 10.1111/j.1365-2141.2007.06942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawamoto K., Chen H.H., Alméciga-Díaz C.J., Mason R.W., Tomatsu S. Gene therapy for mucopolysaccharidoses. Mol. Genet. Metab. 2018;123:59–68. doi: 10.1016/j.ymgme.2017.12.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muenzer J., Wraith J.E., Clarke L.A., International Consensus Panel on Management and Treatment of Mucopolysaccharidosis I mucopolysaccharidosis I: management and treatment guidelines. Pediatrics. 2009;123:19–29. doi: 10.1542/peds.2008-0416. [DOI] [PubMed] [Google Scholar]
- 27.Tomatsu S., Azario I., Sawamoto K., Pievani A.S., Biondi A., Serafini M. Neonatal cellular and gene therapies for mucopolysaccharidoses: the earlier the better? J. Inherit. Metab. Dis. 2016;39:189–202. doi: 10.1007/s10545-015-9900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giugliani R., Villarreal M.L.S., Valdez C.A.A., Hawilou A.M., Guelbert N., Garzón L.N.C., Martins A.M., Acosta A., Cabello J.F., Lemes A., et al. Guidelines for the diagnosis and treatment of Hunter Syndrome for clinicians in Latin America. Genet. Mol. Biol. 2014;37:315–329. doi: 10.1590/s1415-47572014000300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lampe C., Bosserhoff A.K., Burton B.K., Giugliani R., de Souza C.F., Bittar C., Muschol N., Olson R., Mendelsohn N.J. Long-term experience with enzyme replacement therapy (ERT) in MPS II patients with a severe phenotype: an international case series. J. Inherit. Metab. Dis. 2014;37:823–829. doi: 10.1007/s10545-014-9686-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rangarajan S., Walsh L., Lester W., Perry D., Madan B., Laffan M., Yu H., Vettermann C., Pierce G.F., Wong W.Y., Pasi K.J. AAV5-Factor VIII gene transfer in severe hemophilia A. N. Engl. J. Med. 2017;377:2519–2530. doi: 10.1056/NEJMoa1708483. [DOI] [PubMed] [Google Scholar]
- 31.Miesbach W., Meijer K., Coppens M., Kampmann P., Klamroth R., Schutgens R., Tangelder M., Castaman G., Schwäble J., Bonig H., et al. Gene therapy with adeno-associated virus vector 5-human factor IX in adults with hemophilia B. Blood. 2018;131:1022–1031. doi: 10.1182/blood-2017-09-804419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan S.C., Liu C.L., Lo C.M., Lam B.K., Lee E.W., Wong Y., Fan S.T. Estimating liver weight of adults by body weight and gender. World J. Gastroenterol. 2006;12:2217–2222. doi: 10.3748/wjg.v12.i4.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeLand F.H., North W.A. Relationship between liver size and body size. Radiology. 1968;91:1195–1198. doi: 10.1148/91.6.1195. [DOI] [PubMed] [Google Scholar]
- 34.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Rasko J., Ozelo M.C., Hoots K., Blatt P., et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 35.Bell P., Wang L., Gao G., Haskins M.E., Tarantal A.F., McCarter R.J., Zhu Y., Yu H., Wilson J.M. Inverse zonation of hepatocyte transduction with AAV vectors between mice and non-human primates. Mol. Genet. Metab. 2011;104:395–403. doi: 10.1016/j.ymgme.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koo T., Lee J., Kim J.S. Measuring and reducing off-target activities of programmable nucleases including CRISPR-cas9. Mol. Cells. 2015;38:475–481. doi: 10.14348/molcells.2015.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denayer T., Stöhr T., Roy M.V. Animal models in translational medicine: validation and prediction. Eur. J. Mol. Clin. Med. 2014;2:5. doi: 10.1016/j.nhtm.2014.08.001. [DOI] [Google Scholar]
- 38.Leenaars C.H.C., Kouwenaar C., Stafleu F.R., Bleich A., Ritskes-Hoitinga M., De Vries R.B.M., Meijboom F.L.B. Animal to human translation: a systematic scoping review of reported concordance rates. J. Transl. Med. 2019;17:223. doi: 10.1186/s12967-019-1976-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scallan C.D., Jiang H., Liu T., Patarroyo-White S., Sommer J.M., Zhou S., Couto L.B., Pierce G.F. Human immunoglobulin inhibits liver transduction by AAV vectors at low AAV2 neutralizing titers in SCID mice. Blood. 2006;107:1810–1817. doi: 10.1182/blood-2005-08-3229. [DOI] [PubMed] [Google Scholar]
- 40.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacRae S.L., Croken M.M., Calder R.B., Aliper A., Milholland B., White R.R., Zhavoronkov A., Gladyshev V.N., Seluanov A., Gorbunova V., et al. DNA repair in species with extreme lifespan differences. Aging. 2015;7:1171–1184. doi: 10.18632/aging.100866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Denard J., Beley C., Kotin R., Lai-Kuen R., Blot S., Leh H., Asokan A., Samulski R.J., Moullier P., Voit T., et al. Human galectin 3 binding protein interacts with recombinant adeno-associated virus type 6. J. Virol. 2012;86:6620–6631. doi: 10.1128/JVI.00297-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mingozzi F., High K.A. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood. 2013;122:23–36. doi: 10.1182/blood-2013-01-306647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voznyi Y.V., Keulemans J.L., van Diggelen O.P. A fluorimetric enzyme assay for the diagnosis of MPS II (Hunter disease) J. Inherit. Metab. Dis. 2001;24:675–680. doi: 10.1023/a:1012763026526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.