Abstract
The genetic background of each person might affect the severity of radiotherapy (RT)-induced normal tissue toxicity. The aim of study was to evaluate the influence of TGFB1 C-509T and Leu10Pro, XRCC1 Arg280His and XRCC3 Thr241Met polymorphisms as well as the level of radiation-induced CD8 T-lymphocyte apoptosis (RILA) on adverse effects of RT for prostate cancer (PCa). The study included 88 patients with localized or locally advanced PCa who were treated with RT. The polymorphisms were determined by PCR–RFLP analysis on DNA from peripheral blood mononuclear cells. RILA values were measured by flow cytometry. We found that CT genotype of TGFB1 C-509T could be protective biomarker for acute genitourinary (GU) and gastrointestinal (GI) radiotoxicity, while Thr variant of XRCC3 Thr241Met could predict the risk for acute GU radiotoxicity. Correlation between RILA values and toxicity was not detected. Univariate logistic regression analysis showed that Gleason score and risk group were risk factors for late GU, while for late GI radiotoxicity it was diabetes mellitus type 2. However, in multivariate model those were not proven to be significant and independent risk factors. Identification of assays combination predicting individual radiosensitivity is a crucial step towards personalized RT approach.
Subject terms: Cancer, Molecular biology, Biomarkers, Molecular medicine, Oncology
Introduction
About half of all prostate cancer (PCa) patients receive radiotherapy (RT) at some stage of the disease, either as a single curative treatment or as adjuvant/salvage treatment after radical prostatectomy1. However, RT is associated with a wide spectrum of side effects (toxicity) in the surrounding malignantly non-transformed tissues. Acute toxicity occurs during the treatment, or shortly after the end of therapy. Usually, it is transient, and reversible, and affects highly dividing tissues, such as the skin and mucosa of the bladder and intestine, resulting in dermatitis, urinary incontinence, cystitis or diarrhea, respectively2, whereas late toxicity effects, such as fibrosis, rectal bleeding, a bladder and erectile dysfunction, or even secondary malignancies, occur months to years after treatment and can persist for lifetime3. Radiotherapy toxicity may have significant impact on the quality of life of PCa patients because most of them have clinically localized and indolent tumors at diagnosis with a very successful 5-year survival rate of nearly 100%4.
The aim of radiogenomics is to find the genetic variants associated with adverse reactions to RT such as single nucleotide polymorphisms (SNPs), gene alteration, copy number variations etc. SNPs make up 90% of germinal variations in DNA sequence5. These polymorphic variations may underlie the basis of individual patients' radiosensitivity because they change the thermodynamic stability of mRNA molecules, transcription and translation rates, and/or protein–protein interactions, which may reflect on oxidative balance, contributing to altered DNA damage signaling and cell cycle control. SNPs or gene variants can also change the structure and conformation of DNA repair proteins, members of inflammatory response, and can influence cytokine activity related to fibrosis as well as general metabolism and homeostasis5.
Transforming growth factor b1 (TGFβ1), the protein encoded by TGFB1 gene, is a multifunctional cytokine produced primarily by endothelial, hematopoietic, and connective tissue cells. TGFβ1 regulates various cell functions such as proliferation, differentiation, embryonic development, immune response, wound healing, and angiogenesis6. In irradiated cells, TGFβ1 is a mediator of inflammatory response involved in proliferation, differentiation, the production of extracellular matrix proteins and fibrosis7.
Previous studies have hypothesized that the TGFB1 promoter phenotype affects gene transcriptional activity and TGFβ1 plasma levels. The presence of the T allele at − 509 bp of the promoter region (C>T, rs1800469) is associated with higher concentrations of TGFβ1 than C allele8. The T version increases the amount of TGFβ1 produced by preventing AP1 from binding to this region whereas in the C version it would normally downregulate production9.
The T>C transition at codon 10 in exon 1 of TGFB results in leucine to proline substitution (Leu10Pro) (rs1800470). This transition results in increased levels of TGFB1 mRNA and protein in individuals with the proline allele compared to those with the leucine allele10.
XRCC1 (X-ray repair cross complementing group 1) and XRCC3 (X-ray repair cross complementing group 3) genes encode for proteins that are involved in single-strand DNA breaks and base excision repair and homologous recombination (HR) repair of radiation-induced DNA double-strand breaks (DSBs), respectively11. Disruption of these pathways has the potential to affect the normal tissue response to RT. One of the most extensively studied SNP in XRCC1 is Arg280His in exon 9 (G>A, rs25489). The 280His protein variant is associated with increased repair activity and could be potential biomarker of acute and/or late radiotoxicity12. One of the most common SNP investigated in XRCC3 is Thr241Met in exon 7 (C>T, rs861539). The 241Met protein variant was reported to be associated with elevated levels of DNA adducts13, chromosomal deletions14, sensitivity to ionizing radiation, and cross-linking agents15.
Radiation-Induced Lymphocyte Apoptosis (RILA) is defined as the percentage of apoptosis of peripheral blood lymphocytes (PBLs) irradiated with 8 Gy minus the percentage of apoptosis of control non-irradiated PBLs (0 Gy). No association was found between early toxicity and T-lymphocyte apoptosis levels while radiation-induced T-lymphocyte apoptosis can significantly predict differences in late toxicity between individuals. A negative predictive value was found in the case of high RILA value and a late toxicity grade < 2. On the contrary, all severe side-effects (grade ≥ 2) were observed in patients with low values of RILA16. Further studies confirmed that RILA levels can significantly predict the risk for development late radiation-induced toxicity17,18.
According to what was previously mentioned, the aim of this study was to examine the impact of polymorphisms TGFB1 C-509T and Leu10Pro, XRCC1 Arg280His, XRCC3 Thr241Met as well as the radiation-induced CD8 T-lymphocyte apoptosis rate on the RT-induced normal tissue toxicity in Serbian PCa patients.
Results
Patient', clinical and treatment characteristics
The characteristics of the study cohort and RT treatment are shown in Table 1.
Table 1.
Patient's, clinical and treatment characteristics.
| Mean age ± standard deviation | 69.5 years ± 6.6 |
| Initial PSA level | 3.9–70 ng/mL (median 12.1 ng/mL) |
| Diabetes mellitus type 2 | |
| Yes | 16 (18.2%) |
| No | 72 (81.8%) |
| Smoking status | |
| Active/former | 62 (70.5%) |
| Non-smokers | 26 (29.5%) |
| Chronic hypertension | |
| Yes | 54 (61.4%) |
| No | 34 (38.6%) |
| Gleason score, median (min- max) | 7 (6–9) |
| Risk group | |
| Low | 9 (10.2%) |
| Intermediate | 47 (53.4%) |
| High | 32 (36.4%) |
| Type of radiotherapy | |
| Radical | 50 (56.8%) |
| Postoperative |
38 (43.2%) Adjuvant: 22/38 Salvage: 16/38 |
PSA- prostate specific antigen.
Radiation‑induced toxicity description
All 88 patients included in the study were evaluated for toxicity throughout follow-up time (acute and late). Among them, 83 patients (94.3%) experienced acute genitourinary (GU) toxicity (grade 1, 2, and 3: 65.1%, 25.3%, and 9.6% patients, respectively. Eighty (90.9%) patients had acute gastrointestinal (GI) toxicity (grade 1 and 2 in 75% and 25% patients).
Late GU toxicity was developed in 62 (70.5%) patients. Among them, grade 1, 2, and 3 had: 62.9%, 32.3%, and 4.8% patients, respectively. Late GI toxicity appeared in 24 of 88 patients (27.3%) (grade 1 and 2 in 83.3% and 16.7% patients). The follow up period for late GU and GI was 3 to 54 months after RT cessation (median 42 and 39 months, respectively).
TGFB1, XRCC1 and XRCC3 polymorphisms
The distributions of genotypes of TGFB1 C-509T, TGFB1 Leu10Pro, XRCC1 Arg280His, and XRCC3 Thr241Met were given in Table 2. The frequencies of alleles were: C (69.8%) and T (30.2%) for TGFB1 C-509T, Leu (56.7%) and Pro (43.3%) for TGFB1 Leu10Pro, Arg (98.8%) and His (1.2%), Thr (63.6%) and Met (36.4%). The study group was in Hardy–Weinberg equilibrium in relation to the investigated gene loci.
Table 2.
The distributions of genotypes and alleles of investigated SNPs in prostate cancer patients.
| Genotype | n (%) | Alelle (%) | Hardy–Weinberg equilibrium (p value) |
|---|---|---|---|
| TGFβ1 C-509T | |||
| CC | 45 (52.3%) | C (69.8%) | 0.108 |
| CT | 30 (34.9%) | T (30.2%) | |
| TT | 11 (12.8%) | ||
| TGFβ1 Leu10Pro | |||
| LeuLeu | 25 (30.5%) | Leu (56.7%) | 0.538 |
| LeuPro | 43 (52.4%) | Pro (43.3%) | |
| ProPro | 14 (17.1%) | ||
| XRCC1 Arg280His | |||
| ArgArg | 81 (97,6%) | Arg (98.8%) | 0.911 |
| ArgHis | 2 (2.4%) | His (1.2%) | |
| HisHis | 0 (0%) | ||
| XRCC3 Thr241Met | |||
| ThrThr | 33 (40.7%) | Thr (63.6%) | 0.902 |
| ThrMet | 37 (45.7%) | Met (36.4%) | |
| MetMet | 11 (13.6%) | ||
n- Number of patients.
The differences in the distribution of genotypes of TGFB1 C-509T, TGFB1 Leu10Pro, and XRCC3 Thr241Met between patients with or without acute or late RT-induced GU or GI toxicity as well as different grades of toxicity were given in Table 3.
Table 3.
Distribution of acute and late RT-induced GU or GI toxicity between patients with genotypes of TGFB1 C-509T, TGFB1 Leu10Pro and XRCC3 Thr241Met.
| TGFB1 C-509T | TGFB1 Leu10Pro | XRCC3 Thr241Met | |
|---|---|---|---|
|
Acute GU toxicity |
CC: 97.8% CT: 86.7% TT: 100% |
LeuLeu: 100% LeuPro: 90.7% ProPro: 100% |
ThrThr: 100% ThrMet: 94.6% MetMet: 81.8% |
|
Acute GU toxicity (grade 1 vs. Grade 2 plus 3) |
Grade 1: CC: 70.5% CT: 61.5% TT: 54.5% Grade 2 plus 3: CC: 29.5% CT: 38.5% TT: 45.5% |
Grade 1: LeuLeu: 76.0% LeuPro: 59.0% ProPro: 64.3% Grade 2 plus 3: LeuLeu: 24.0% LeuPro: 41% ProPro: 35.7% |
Grade 1: ThrThr: 54.5% ThrMet: 71.4% MetMet: 77.8% Grade 2 plus 3: ThrThr: 45.5% ThrMet: 28.6% MetMet: 22.2% |
|
Acute GI toxicity |
CC: 95.6% CT: 83.3% TT: 100% |
LeuLeu: 92.0% LeuPro: 88.4% ProPro: 100% |
ThrThr: 90.9% ThrMet: 91.9% MetMet: 90.9% |
|
Acute GI toxicity (grade 1 vs. Grade 2 plus 3) |
Grade 1: CC: 76.7% CT: 68% TT: 81.8% Grade 2: CC: 23.3% CT: 32% TT: 18.2% |
Grade 1: LeuLeu: 78.3% LeuPro: 73.7% ProPro: 78.6% Grade 2: LeuLeu: 21.7% LeuPro: 26.3% ProPro: 21.4% |
Grade 1: ThrThr: 76.7% ThrMet: 76.5% MetMet: 60% Grade 2: ThrThr: 23.3% ThrMet: 23.5% MetMet: 40% |
|
Late GU toxicity |
Yes: CC: 71.1% CT: 70.0% TT: 63.6% |
Yes: LeuLeu: 72.0% LeuPro: 72.1% ProPro: 64.3% |
Yes: ThrThr: 78.8% ThrMet: 64.9% MetMet: 63.6% |
|
Late GU toxicity (grade 1 vs. Grade 2 plus 3) |
Grade 1: CC: 71.9% CT: 47.6% TT: 71.4% Grade 2 plus 3: CC: 28.1% CT: 52.4% TT: 28.6% |
Grade 1: LeuLeu: 72.2% LeuPro: 58.1% ProPro: 66.7% Grade 2 plus 3: LeuLeu: 27,8% LeuPro: 41.9% ProPro: 33.3% |
Grade 1: ThrThr: 61.5% ThrMet: 70.8% MetMet: 42.9% Grade 2 plus 3: ThrThr: 38.5% ThrMet: 29.1% MetMet: 57.1% |
|
Late GI toxicity |
Yes: CC: 28.9% CT: 23.3% TT: 36.4% |
Yes: LeuLeu: 28.0% LeuPro: 27.9% ProPro: 28.6% |
Yes: ThrThr: 24.2% ThrMet: 32.4% MetMet: 18.2% |
|
Late GI toxicity (grade 1 vs. Grade 2 plus 3) |
Grade 1: CC: 76.9% CT: 100% TT: 75.0% Grade 2: CC: 23.1% CT: 0% TT: 25.0% |
Grade 1: LeuLeu: 71.4% LeuPro: 91.7% ProPro: 75.0% Grade 2: LeuLeu: 28.6% LeuPro: 8.3% ProPro: 25.0% |
Grade 1: ThrThr: 62.5% ThrMet: 91.7% MetMet: 100% Grade 2: ThrThr: 37.5% ThrMet: 8.3% MetMet: 0% |
GU- genitourinary; GI- gastrointestinal.
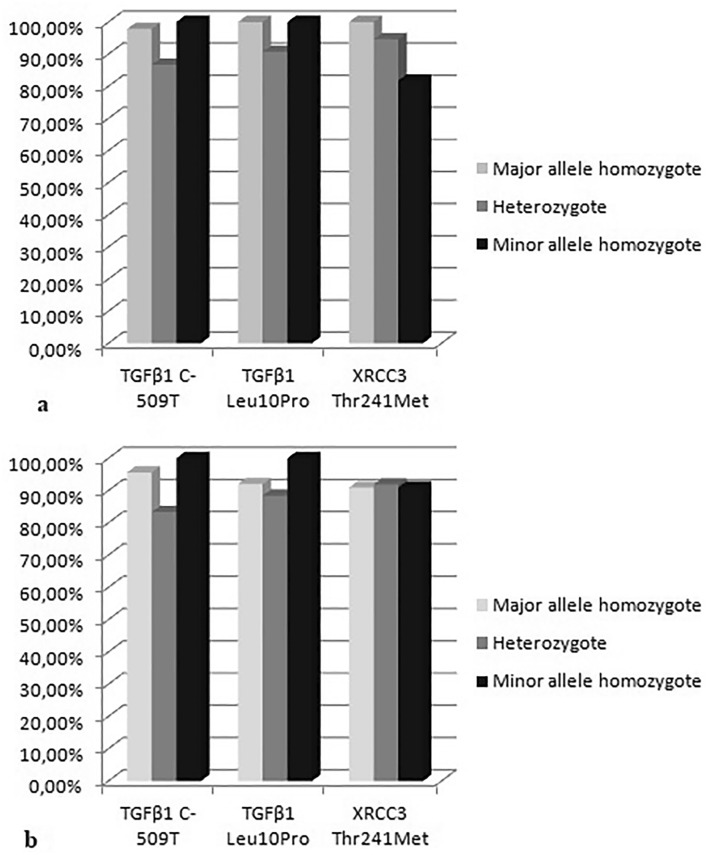
Heterozygote carriers of TGFB1 C-509T (CT) had lower rate of acute GU as well as acute GI toxicity than homozygotes (CC plus TT) (Fig. 1a,b) and this difference was statistically significant (p = 0.048 and p = 0.047, Fisher exact test). Additionally, the OR (95% CI) in the over-dominant model (0.12 (0.01–1.11) for acute GU and 0.19 (0.03–1.02) for acute GI) indicated a lower risk for the development of acute RT-induced toxicity in heterozygote patients. The frequencies of acute GU toxicity grade ≥ 2 arose from CC and CT to TT genotype while in acute GI toxicity the heterozygote patients had higher frequency of grade 2 toxicity than CC and TT genotype but without statistically significant difference in any of the examined genetic models. In late GU toxicity, the CC and CT carriers had slightly higher rate than TT but without statistical significance. However, heterozygote carriers had nearly double higher rate of grade ≥ 2 toxicity than homozygotes. Contrary to late GU toxicity, individuals with TT genotype had higher rate of overall late GI toxicity, and the heterozygotes had lower rate of grade ≥ 2 than homozygotes but these differences did not show statistical significance.
Figure 1.
The differences in the distribution of genotypes of TGFB1 C-509T, TGFB1 Leu10Pro, and XRCC3 Thr241Met between patients with (a) acute RT-induced GU toxicity and (b) acute RT-induced GI toxicity.
Heterozygote Pca patients of TGFB1 Leu10Pro (LeuPro) had lower rate of acute GU as well as acute GI toxicity than homozygotes (LeuLeu plus ProPro) (Fig. 1a,b). The frequency of acute GU and GI toxicity grade ≥ 2 were higher in LeuPro than homozygote carriers but without statistically significant difference. In late GU toxicity, LeuLeu and LeuPro carriers had slightly higher rate than ProPro. For late GI toxicity, there was a similar frequency of all genotypes of TGFB1 Leu10Pro. However, heterozygote carriers had higher rate of grade ≥ 2 GU but lower rate of grade ≥ 2 GI toxicity than homozygotes but without statistically significant difference.
We did not perform statistical analysis for XRCC1 Arg280His polymorphism because only two of 83 analyzed patients had ArgHis genotype and everyone else ArgArg. Both of ArgHis patients had acute GU and GI toxicity grade 1. One of them experienced late GU toxicity grade 1 and the other late GI toxicity grade 1.
There was the statistical trend for higher acute GU toxicity in carriers of Thr variant of XRCC3 (ThrThr plus ThrMet) compared to MetMet (Fig. 1a) (p = 0.087, Fisher exact test) as well as ThrThr vs. MetMet (p = 0.058, Fisher exact test). Additionally, PCa patients with ThrThr genotype had higher rate of acute GU toxicity grade ≥ 2. For acute GI toxicity, there was a similar frequency of genotypes of XRCC3 Thr241Met (Fig. 1b) but contrary to acute GU in GI toxicity MetMet had higher rate of grade ≥ 2. Late GU adverse RT-induced effects were appeared more frequent in patients with ThrThr variant of XRCC3 while late GI RT-induced effects were appeared more frequent in patients with ThrMet variant. Grade ≥ 2 GU toxicity was the frequent in MetMet while grade ≥ 2 GI toxicity in ThrThr carriers but without statistical significance.
Radiation-induced CD8 T-lymphocyte apoptosis
RILA levels were measured for 67 out of 88 patients. Median RILA value was 30.24 (mean, 30.65; SD, 13.49; range, 7.96 to 69.86). Kolmogorov–Smirnov test showed that the values of RILA assay have a normal distribution (K–S Dist. = 0.092; p = 0.164).
The median value of RILA levels in patients with grade 1 acute GU and GI toxicity was higher than in patients with grade 2 plus 3 group but without statistical significance. Also, in patients with late GU toxicity RILA levels were higher in grade 1 compared to grade 2 plus 3 group (Table 4). In patients who were analyzed, only four patients experienced late GI toxicity grade 2 and none experienced grade 3. Because of that we did not perform statistical analysis for late GI toxicity and RILA.
Table 4.
RILA and RT-induced toxicity.
| RILA number of patient (median; mean; range) | Test; p value | |
|---|---|---|
|
Acute GU toxicity |
Grade 1: 41 (31.1; 31.6; 8.0 to 69.9) Grade 2 plus 3: 23 (22.8; 29.2; 8.1 to 66.9) |
MWU = 524.0; p = 0.467* |
|
Acute GI toxicity |
Grade 1:50 (30.3; 30.9; 8.0 to 69.9) Grade 2: 10 (27.7; 30.7; 15.8 to 47.5) |
t (58) = 0.040; p = 0.969** |
|
Late GU toxicity |
Grade 1: 29 (33.2; 33.3; 12.2 to 66.9) Grade 2 plus 3: 21 (30.1; 31.1; 8.0 to 69.9) |
t (48) = 0.527; p = 0.600** |
|
First tertile: Grade 1:9 Grade 2 plus 3: 6 Second and third tertile: Grade 1: 20 Grade 2 plus 3: 15 |
χ21 = 0.016; p = 0.900** | |
|
Late GI toxicity |
Grade 1: 15 (30.1; 32.3; 13.0 to 69.9) Grade 2: 4 (43.3; 44.2; 40.0–50.3) |
t (17) = − 1.485; p = 0.156** |
|
First tertile: Grade 1: 5 Grade 2: 0 Second and third tertile: Grade 1: 10 Grade 2: 4 |
Fisher exact test p = 0.530 |
*Wilcoxon rank sum test; **t test.
MWU- Mann–Whitney U Statistic.
NA- not applicable.
GU- genitourinary; GI- gastrointestinal.
According to the REQUITE project report, tertile cut-off points could be used to divide the patients into three equal groups of low, mid and high radiation-induced cell death. In our case the first tertile cut-off value was 21.44% and 36.18%. We did not observe a statistically significant difference in the distribution of patients with different radiotoxicity grade among tertiles (Table 4).
Association of TGFB1 C-509T, TGFB1 Leu10Pro, XRCC1 Arg280His and XRCC3 Thr241Met with RILA values
In the light of incidence for late GU and GI radiotoxicity, patients with the highest mean value of RILA had TT genotypes for C-509T and ProPro for Leu10Pro TGFB1 polymorphism. These data reached statistical trend for GU and GI toxicity for C-509T and statistical significance for GI for Leu10Pro polymorphism. For XRCC3 Thr241Met polymorphism, heterozygotes patients had the highest mean value of RILA but without statistical significance (Table 5).
Table 5.
Association of polymorphic variants with RILA values.
| Polymorphism | Genotype (No of patients): Mean value late GU toxicity RILA | ANOVA | Genotype (No of patients): Mean value late GI toxicity RILA | ANOVA |
|---|---|---|---|---|
|
TGFB1 C-509T |
CC (26): 33.8 CT (17): 27.7 TT (5): 43.0 |
p = 0.080 |
CC (12): 32.4 CT (5): 31.4 TT (2): 57.8 |
p = 0.055 |
| TGFB1 Leu10Pro |
LeuLeu (14): 34.4 LeuPro (24): 29.9 ProPro (8): 35.4 |
p = 0.522 |
LeuLeu (6): 33.5 LeuPro (10): 30.3 ProPro (2): 57.8 |
p = 0.050 |
| XRCC1 Arg280His | NA | NA | NA | NA |
| XRCC3 Thr241Met |
ThrThr (21): 32.5 ThrMet (18): 35.1 MetMet (7): 27.1 |
p = 0.438 |
ThrThr (7): 32.7 ThrMet (9): 40.2 MetMet (2): 27.5 |
p = 0.422 |
NA- not applicable.
Bolded values indicate statistically significant data or statistical trend.
GU- genitourinary; GI- gastrointestinal.
Logistic regression analysis
According to the concept that minimum ten event for variable is acceptable for logistic regression analysis19 and the small number of least frequent outcome in our study (absence of acute GU and GI toxicity- in five and eight patients respectively), we performed logistic regression analysis only for late toxicity.
The logistic regression analysis data were shown in Table 6. Gleason score and risk group were risk factors for late GU while for late GI RT-induced toxicity it was diabetes mellitus type 2. However, in multivariate model Gleason score and risk group (p = 0.192 and p = 0.367) did not prove to be significant and independent risk factors for late GU toxicity.
Table 6.
Prediction of late GU and GI radiotoxicity.
| Variable | Occurrence of late GU toxicity | Occurrence of late GI toxicity | ||
|---|---|---|---|---|
| p value | OR (95% CI) | p value | OR (95% CI) | |
| Age | 0.385 | 1.031 (0.962–1.106) | 0.299 | 1.041 (0.965–1.122) |
| Initial PSA value | 0.610 | 1.011 (0.970–1.053) | 0.076 | 1.035 (0.996–1.076) |
| Diabetes mellitus type 2 | 0.869 | 1.104 (0.342–3.567) | 0.001 | 0.145 (0.045–0.466) |
| Smoking status | 0.871 | 0.920 (0.340–2.495) | 0.634 | 1.278 (0.466–3.503) |
| Chronic hypertension | 0.983 | 1.011 (0.394–2.589) | 0.721 | 1.190 (0.457–3.098) |
| Type of radiotherapy | 0.716 | 0.843 (0.335–2.117) | 0.511 | 0.724 (0.277–1.895) |
| Gleason score | 0.030 | 2.440 (1.092–5.449) | 0.663 | 1.158 (0.598–2.244) |
| Risk group | 0.037 | 2.256 (1.052–4.837) | 0.303 | 1.497 (0.695–3.227) |
| RILA value | 0.073 | 1.045 (0.996–1.096) | 0.118 | 1.033 (0.992–1.075) |
| TGFB1 C-509T genotypes | 0.670 | 0.869 (0.455–1.659) | 0.867 | 1.058 (0.544–2.060) |
| TGFB1 Leu10Pro genotypes | 0.662 | 1.170 (0.579–2.362) | 0.975 | 0.989 (0.485–2.015) |
| XRCC3 Thr241Met genotypes | 0.215 | 0.644 (0.321–1.292) | 0.993 | 0.997 (0.488–2.037) |
Bolded values indicate statistically significant data or statistical trend.
GU- genitourinary; GI- gastrointestinal.
Discussion
A combination of different approaches should be developed to predict patients' individual radiosensitivity, and to reduce side effects of radiation treatment. This will allow clinicians to modify treatment regiments, to improve success and efficacy of the therapy1.
Symptoms of GI toxicity can range from a mild increase in bowel movement frequency to more severe complications such as rectal bleeding, pain, or fistula. The acute phase of RT injury is characterized by inflammation in response to therapy, while the late phase is characterized by fibrosis and sclerosis within the GI tract20. Urinary toxicity includes the following symptoms: changes in urinary frequency, bladder obstruction, hematuria, urinary incontinence or dysuria21.
Irradiation causes direct DNA damage and increases production of free radicals which interact with DNA, thus inducing single-strand or double-strand breaks, and DNA crosslinks 22. Cells respond to irradiation by cell cycle arrest thus inducing DNA damage repair. Unrepaired damage causes cell cycle arrest or cell death. Tumors are more susceptible to the DNA damage caused by radiation because of their higher proliferation rates, thus having less time to repair damage than normal tissue23. Free radicals induce stress responses, inflammation, and release of cytokines, growth factors, and chemokines. Deficiency in DNA repair genes, cells' ability to undergo apoptosis or impaired cell cycle arrest, as well as changes in genes encoding enzymes involved in free-radical metabolism, and immune response contribute to increased radiosensitivity23.
Individual genetic background might be crucial for prediction of normal tissue reaction to RT. In light of that, we tried to find an association between SNPs TGFB1 C-509T, TGFB1 Leu10Pro, XRCC1 Arg280His, and XRCC3 Thr241Met and RT-induced toxicity of normal tissue.
XRCC1 gene encodes protein that is involved in single-strand DNA break and base excision repair of radiation-induced damage11. XRCC1 fixes base damage and DNA single-strand breaks caused by ionizing radiation interacting with polymerase-beta, DNA ligase III, and poly (ADP-ribose) polymerase12. One of candidate biomarker of radiotoxicity could be XRCC1 Arg280His SNP because of fact that the 280His protein variant is associated with increased repair activity12. It has been shown that ArgArg individuals have substantially more chromosome breaks compared to those carrying at least one 280His allele24. These findings were supported by the observation that workers exposed to organic solvents carrying the wild type 280 ArgArg had significantly higher levels of chromosomal aberrations than those with one or two variant His alleles25.
It has been shown in PCa patients that presence of 280His allele was associated with decreased risk of late toxicity12. Other authors have not found any association between late toxicity after RT for treatment of PCa and XRCC1 Arg280His SNP26. Also, in breast cancer, the authors did not observe association of this SNP with fibrosis as late effect of RT or acute skin reaction27,28. However, the meta-analysis on 2199 breast cancer patients showed that 280His allele had protective against radiation-induced toxicity29. Regarding unexplained role of XRCC1 Arg280His SNP in normal tissue response to RT, it will require further research. In our study, only 2.4% of patients had ArgHis genotype and everyone else ArgArg. Because of that, we did not perform statistical analysis for XRCC1 Arg280His polymorphism and RT-induced toxicity. Also, it is important to mention that ArgHis and HisHis were rare variants in some other studies on PCa as well breast cancer patients and were in range 5.1–8.0% and 0–0.9%, respectively11,26,27.
XRCC3 gene encodes protein involved in HR repair of radiation-induced DSBs. During HR repair of DSBs, XRCC3 interacts with RAD51 to promote the initiation of HR and stabilize DNA heteroduplex30. One of the most common SNP investigated in XRCC3 is Thr241Met. The 241Met variant was reported to be associated with elevated levels of DNA adducts13, chromosomal deletions14, sensitivity to ionizing radiation and cross-linking agents15. Thr241Met amino acid substitution may affect the structure of XRCC3 protein and lead to a deficiency in the HR pathway. Consequently, the repair mechanism of DSBs could be moved toward non-homologous end joining, which may result in chromosome instability and the cell’s ability to repair DNA damage lessions30. The XRCC3 Thr241Met was found to be significantly associated with radiation-induced acute skin toxicity and mucositis. Furthermore, XRCC3 Thr241Met was significantly associated with radiation-induced fibrosis. The presence of XRCC3 Thr241Met variant was significantly correlated with a higher risk of developing normal tissue reactions to RT after head and neck area and breast irradiation. The absence of significant correlations between this variant and side normal tissue effects was observed after lung or pelvic irradiation31.
Contrary to expectations, in our study, there was the statistical trend towards higher acute GU toxicity in carriers of Thr variant. For acute GI toxicity, there was a similar distribution in Thr241Met SNP genotypes. PCa patients with ThrThr genotype had higher rate of acute GU toxicity grade ≥ 2 while in GI toxicity MetMet had higher rate of grade ≥ 2. The late GU toxicity was more common in ThrThr while late GI toxicity in ThrMet patients. The grade ≥ 2 late GU toxicity was more common in MetMet while the grade ≥ 2 late GI toxicity in ThrThr individuals. The possible explanation of these findings is that a combination of unknown genotypes may confer RT-induce toxicity. For example, in PCa patients with the combination ValAla of the antioxidant SOD2 (superoxide dismutase 2) Val16Ala SNP and ThrMet of XRCC3 Thr241Met SNP experienced a significant increase in grade 2 late rectal bleeding compared to patients without this particular genotypic arrangement11.
It is also interesting to mention that the same polymorphic variant was related to GU but not GI toxicity and vice versa. It indicates that probably exist cell and tissue-specific characteristics that lead to distinct phenotypes upon radiation treatment. Additionally, for different tissue higher grade of toxicity was related to different polymorphic variants. Further study on larger group is necessary to confirm this date and to clarify mechanism underlying this observation.
Fibrosis, as a late toxicity effect of radiation, represents an inflammatory-mediated proliferation response of surviving fibrocytes to growth factors, cytokines, and chemokines released following irradiation23. TGFβ1 is a key cytokine associated with inflammation and fibrosis. The presence of the T allele at − 509 bp of the promoter region of TGFB1 is associated with higher concentrations of TGFβ1 than the C allele8. The T>C transition at codon 10 of TGFB1 results in leucine to proline substitution (Leu10Pro) and in increased levels of TGFβ1 protein10.
Initial results from a relatively small Danish patient’s cohort indicated significant associations between the TGFB1 − 509T and codon 10 Pro allele and increased risk of radiation-induced breast fibrosis32,33 but the authors were unable to confirm this association in a subsequent larger study27. Data from two British cohorts showed a significant association of the − 509T allele with breast fibrosis following RT34,35 as well as later study on early-stage breast cancer patients from MD Anderson Cancer Center (Houston, Texas)36. Contrary to previous finding, the studies on nasopharyngeal and breast cancer patients showed an association between 10 Pro allele and lower risk of fibrosis37,38. However, a large study on 778 breast cancer patients failed to confirm any association of C-509T and Leu10Pro TGFB1 and radiation-induced toxicity39 as well as further meta-analysis on 2782 participants who received adjuvant breast RT40. In PCa, study on 413 patients did not find any association of mentioned TGFB1 SNPs and late GU and GI toxicity41. Damaraju et al. did not find association of C-509T SNP and late rectal and bladder toxicity after RT for prostate26 cancer while Peters et al. found that patients with the -509TT genotype had a significantly increased risk of developing late rectal bleeding7.
In our study, for late GU toxicity, the CC and CT carriers of C-509T had slightly higher rate than TT. However, heterozygote carriers had nearly double higher rate of grade ≥ 2 toxicity than homozygotes. Contrary to late GU toxicity, individuals with TT genotype had the higher rate of overall late GI toxicity and the heterozygotes had lower rate of grade ≥ 2 than homozygotes. For GI toxicity it is in accordance with some previous data7 but for GU toxicity data are missing.
For Leu10Pro SNP in late GU toxicity, LeuLeu and LeuPro carriers had slightly higher rate than ProPro while in late GI toxicity there was a similar frequency of all genotypes of TGFB1 Leu10Pro. Similar to C-509T for Leu10Pro SNP, heterozygote carriers had higher rate of grade ≥ 2 GU but lower rate of grade ≥ 2 GI toxicity than homozygotes. Our results indicate that different level of TGFβ1 protein is included in late GU in regard to late GI toxicity and the different mechanism that lies behind it. Also, probably different mechanism is at the base of higher grade of toxicity of genitourinary and gastrointestinal tissues.
Although TGFβ1 is a key proinflammatory and profibrotic cytokine, its role in acute toxicity after RT is still unclear. We found that heterozygote carriers of TGFB1 C-509T had statistically significant lower rate of acute GU and GI toxicity than homozygotes (CC plus TT). Additionally, the OR indicated lower risk for acute toxicity development in heterozygote than homozygote PCa patients. The obtained data indicate that CT genotype of TGFB1 C-509T could be potential biomarkers of lower risk for acute RT-induced toxicity. Additionally, heterozygote PCa patients for TGFB1 Leu10Pro had lower rate of acute GU and GI toxicity than homozygotes. This is to be expected because of the strong linkage disequilibrium between C-509T and Leu10Pro polymorphisms39,42. The obtained results indicated that median dose of TGFβ1 is protective for the development of acute toxicity. The exact mechanism underlining this should be further investigated.
Interestingly, the frequency of acute GU toxicity in grade ≥ 2 arose from CC and CT to TT C-509T genotype while in acute GI toxicity the heterozygote patients had higher frequency of grade ≥ 2 toxicity than CC and TT genotype. For Leu10Pro SNP, the frequency of acute GU and GI toxicity grade ≥ 2 were higher in LeuPro than in homozygote carriers. These data may be explained by different mechanisms of development of RT-induced injury in different tissue or relatively small sample size of PCa patients with grade ≥ 2 toxicity.
PCa clinical variables may contribute to RT-induced toxicity. Because of that, we performed the logistic regression analysis to elucidate these effects. We found that Gleason score and risk group were risk factors for late GU while for late GI RT-induced toxicity it was diabetes mellitus type 2 in univariante model. None of the genotypes of investigated SNPs was shown to be a risk factor for RT-induced toxicity of normal tissue.
Radiosensitivity is typically measured in research studies using patient-derived cells (mainly PBLs or fibroblasts). Cells are irradiated and radiosensitivity can be measured using clonogenic assay, detection of chromosome and DNA damage, cell cycle delay as well as RILA23,43. The molecular mechanism linking compromised RILA and late toxicity is still unclear. One could hypothesize that a slow apoptotic response in irradiated lymphocytes promotes cytokine production and attracts inflammatory immune cells that could be at the origin of late toxicities43.
According to the REQUITE project report, tertile cut-off points could be used to divide the patients into three equal groups of low, mid and high RILA. Although, the mean value of RILA in grade 1 was slightly higher than in grade 2 plus 3 group for acute GU and GI toxicity as well as late GU toxicity, we did not observe statistically significant correlation. For late GI toxicity the mean value of RILA in patients with grade 1 was lower than in patients with grade 2 plus 3. This could be explained by fact that we only had four patients in group ≥ 2 toxicity and according to the extremely small sample size we did not perform statistical analysis. Additionally, our tertile cut-off was slightly higher than those in REQUITE project study.
Previous multicenter studies showed a good negative predictive value in patients with high RILA values and low-grade late toxicity following RT16,17. However, the first study of Ozsahin et al. (2005) included 399 patients from different cancer cite and there were only 36 of PCa patients among them. We hypothesize that CD4 and CD8 lymphocytes from blood of patients with different cancer should react differently to radiation and the chemotherapy and hormone therapy maybe have some impact on radiation response. The other study of Azria et al. related RILA to RT-induced fibrosis17. One of recent study that included 245 patients with PCa showed that RILA does not correlate with the inter-individual variation in maximal late GU or GI toxicity after radiation44. In the same manner, the REQUITE study showed that the RILA assay is a very robust marker for fibrosis (breast or head and neck fibrosis) after RT. The assay seemed less robust with other types of toxicity like vascular damage lesions, which could explain why the correlation is not as good as in our study. In the REQUITE project, the toxicity endpoints were changes in breast appearance at 2 years measured using photographs, pneumonitis at 1 year and rectal bleeding at 2 years. These primary endpoints were selected because they show radiation dose–response relationships.
We found correlation of TGFB1 variants (TT for C-509T and ProPro for Leu10Pro) and highest mean value RILA for late GU and GI radiotoxicity as statistical trend in case of C-509T and significance in Leu10Pro for GI radiotoxicity. For XRCC3 Thr241Met, heterozygotes patients had the highest but not statistically significant mean value of RILA. But these data should be taken with caution for GI toxicity given the small number of patients with GI toxicity, particularly for TT and ProPro TGFB1 and MetMet XRCC3 with only two patients each. Furthermore, the study by Azria et al. showed that high risk of late radiotoxicity was associated with a low RILA values and the presence of four or more SNPs in selected genes (ATM, SOD2, TGFB1, XRCC1, XRCC3, and RAD2)45,46. Combining determination of RILA and profiling of a specific set of SNPs in genes implicated in radiosensitivity might represent a promising strategy for identification of cancer patients at high risk of late normal tissue reactions after RT, individual RT optimization, and improvement of patients’ quality of life. Complex genetic and biological factors underlying associations between RILA and late radiotoxicity in patients with cancer should be further explored18. The large multicentric studies for better defining cut-off RILA values for each cancer type are also needed.
Conclusions
We found that CT genotype of TGFB1 C-509T SNP could be potential protective biomarker for acute GU as well as GI RT-induced toxicity while Thr variant of XRCC3 Thr241Met SNP could identify patients at risk for acute GU toxicity. For other investigated polymorphisms we did not observe any association with acute or late adverse effects of RT. Also, we did not show correlation of RILA value with RT-induced normal tissues injury. Further studies on larger study groups are needed to confirm our results.
The finding combination of assays that are characterized by a high level of sensitivity and specificity to predict susceptibility to the development of adverse effect of RT will serve as a powerful tool in precision medicine approach. This approach might contribute to the development of tests for the prediction of individual radiosensitivity, and even RT-induced secondary malignancies, not only for RT for prostate cancer but also for the prediction of other radiation-induced adverse effects in other malignancies treated with RT, such as breast cancer, brain malignancies, lung cancer, etc.
Methods
Study population
Eighty-eight patients who had a histologically confirmed localized or locally advanced PC were included in the study. Patients were treated with 3D conformal RT (3DCRT) (n = 76) or Volumetric Modulated Arc Therapy (VMAT) (n = 12) at the Institute for Oncology and Radiology of Serbia from January 2016 to August 2019 with radical (72 Gy in 36 fractions) (in 50 patients) or postoperative/salvage (66 Gy in 33 fractions) (38 patients) RT without previous hormonal therapy. The details of the study were explained to the patients and informed consent forms were signed by the participants. The study protocol was approved by the Ethics Committee of the Institute of Oncology and Radiology of Serbia (approval No 3348/1–01). The study was carried out according to the principles of the Declaration of Helsinki.
Acute and late radiotoxicity were evaluated according to RTOG/EORTC Radiation Morbidity Scoring Criteria modified by Peeters47. Side-effects occurring within 120 days from the start of RT were considered as acute radiation morbidity. The radiation oncologist recorded baseline symptoms and acute symptoms during RT and at first control (30 days after the end of RT). Late toxicity was scored from 120 days after the start of treatment. All clinical data and blood samples were prospectively collected. Exclusion criteria were as follows: chronic infective diseases, neoadjuvant or concomitant hormonal therapy, the presence of enlarged lymph nodes (N1 stage) at the time of diagnosis, the presence of distant metastasis (M1 stage), Karnofsky index < 80, and previous pelvic irradiation.
Peripheral blood mononuclear cell and DNA extraction
Peripheral blood mononuclear cells (PBMC) were extracted with Histopaque-1077 (Sigma-Aldrich, USA) solution according to the manufacturer’s instructions. DNA from PBMC was extracted by salting out method48.
Genotyping of TGFB1, XRCC1 and XRCC3 polymorphisms
The polymorphic variants of promoter C-509T (rs1800469) and Leu10Pro (rs1800470) of TGFB1, Arg280His (rs25489) of XRCC1, and Thr241Met (rs861539) of XRCC3 were determined by polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP) analysis. The DNA regions around mentioned SNPs in TGFB1, XRCC1, and XRCC3 genes were amplified, separately, in 25 μL PCR reaction volume contained 12.5 μL DreamTaq PCR Master Mix 2x (Thermo Fisher Scientific, USA), 200 ng of genomic DNA, and 0.4 µM of each sense and antisense primers (Invitrogene, USA). Primers sequences and PCR conditions were given in Table 749–51. PCR products of TGFB1, XRCC1, and XRCC3 were visualized by electrophoresis on 2% agarose gel and concentrations were measured on NanoDrop (Shimadzu, Japan). PCR products were digested by BspTI, PvuII, RsaI and FatI fast digest restriction enzyme (Thermo Fisher Scientific, USA), respectively. After digestion, the fragments were separated on 2100 Bioanalyzer (Agilent, USA) using DNA 1000 kit according to the manufacturer’s instructions. The length of PCR products and fragments were given in Table 8. Figure 2a–d show electropherograms from 2100 Bioanalyzer which contain DNA fragments obtained by PCR–RFLP method.
Table 7.
Primers’ sequences and PCR conditions.
| Gene | SNP | Primers (5′–3′) | PCR conditions |
|---|---|---|---|
| TGFB1 | C-509T |
F:GTCGCAGGGTGTTGAGTGACAG R:AGGGGGCAACAGGACACCTTA |
95 °C, 5 min- initial denaturation, 95 °C, 30 s; 62 °C, 45 s; 72 °C, 1 min- 35 cycles, 72 °C, 10 min- final elongation |
| TGFB1 | Leu10Pro |
F:CTCCGGGCTGCGGCTGCAGC R:GGCCTCGATGCGCTTCCGCTTCA |
95 °C, 5 min- initial denaturation, 95 °C, 30 s; 62 °C, 45 s; 72 °C, 1 min- 35 cycles, 72 °C, 10 min- final elongation |
| XRCC1 | Arg280His |
F:TGGGGCCTGGATTGCTGGGTCTG R:CAGCACCACTACCACACCCTGAAGG |
94 °C, 5 min- initial denaturation, 94 °C, 40 s; 55 °C to 69 °C, 40 s; 72 °C, 30 s- 35 cycles, 72 °C, 10 min- final elongation |
| XRCC3 | Thr241Met |
F: GGTCGAGTGACAGTCCAAAC R: TGCAACGGCTGAGGGTCTT |
95 °C, 5 min- initial denaturation, 95 °C, 1 min; 60 °C, 1 min; 72 °C, 1 min- 40 cycles, 72 °C, 5 min- final elongation |
Table 8.
The length of PCR products, restriction enzymes and fragments.
| Gene | SNP | Restriction enzymes | PCR products (bp) | Fragments size (bp) |
|---|---|---|---|---|
| TGFB1 | C-509T | BspTI | 123 |
CC: 123 CT: 123, 101,22 TT: 101, 22 |
| TGFB1 | Leu10Pro | PvuII | 136 |
CC (ProPro): 136 CT (LeuPro):136, 117, 19 TT (LeuLeu): 117, 19 |
| XRCC1 | Arg280His | RsaI | 280 |
GG (ArgArg): 140 GA (ArgHis): 280, 140 AA (HisHis): 280 |
| XRCC3 | Thr241Met | FatI | 456 |
CC (ThrThr): 315, 141 CT (ThrMet): 315, 210, 141, 105 TT (MetMet):210, 141, 105 |
Figure 2.
Detection of polymorphisms by PCR–RFLP method. (a) TGFB1 C-509TC: column L- DNA Ladder; column 1, 2, 8, 10- CC genotype; column 5, 7, 9, 11, 12- CT genotype; columns 4, 6- TT genotype. (b) TGFB1 Leu10Pro: column L- DNA Ladder; columns 1, 4- ProPro genotype; column 2, 3, 5, 7, 9, 11- LeuPro genotype; columns 6, 8, 10- LeuLeu genotype, column 12- PCR product. (c) XRCC1 Arg280His: column L- DNA Ladder; columns 1, 2, 3, 4, 5, 6, 7, 9, 10, 11- ArgArg genotype; column 8- ArgHis genotype; column 12- PCR product. (d) XRCC3 Thr241Met: column L- DNA Ladder; columns 1, 5, 6, 7, 8- ThrMet genotype; columns 3, 4, 11- ThrThr genotype; columns 9, 10- MetMet genotype.
RILA assay
Before RT a single blood sample was collected from each patient in a 4-ml BD Vacutainer lithium heparin tubes. The whole blood samples were diluted 1:10 in RPMI 1640 containing 20% fetal bovine serum, irradiated at 0 and 8 Gy, and incubated for 48 h. The cells were then labeled with FITC-conjugated anti-CD8 monoclonal antibodies, red blood cells were lysed, and the DNA of the remaining cells stained with propidium iodide. Samples were measured using a FACScan flow cytometer, and data analysis was done using Kazula software (Beckman Coulter, USA). Apoptotic CD8 T-lymphocytes were defined as those cells staining positively for their cell type-specific antibodies and displaying reduced propidium iodide fluorescence and cell size. Data from at least 10,000 cells per sample were acquired45. The addition of CD4 into the model did not contribute significantly in separating the two groups16. Therefore, we took into account only CD8 results for the present study.
Statistical analysis
Deviations of the genotype frequencies of TGFB1 C-509T and Leu10Pro, XRCC1 Arg280His, and XRCC3 Thr241Met polymorphisms from those expected under Hardy–Weinberg equilibrium in the studied groups were assessed using the χ2 test.
The differences in the distribution of genotypes of TGFB1 C-509T and Leu10Pro, XRCC1 Arg280His, and XRCC3 Thr241Met between patients with or without acute or late RT-induced GU or GI toxicity, as well as different grade of toxicity and RILA were tested by χ2 and Fisher’s exact test. Kolmogorov–Smirnov test was used to test normality of RILA assay date. To test the differences in RILA value between the groups with or without toxicity as well as different grade of toxicity t-test or Wilcoxon rank sum test were used depending on the distribution of data in groups. The RILA data in different polymorphic variants of TGFB1, XRCC1, and XRCC3 were compared by one way ANOVA test. p Values ≤ 0.05 were considered statistically significant, while p values between 0.1 and 0.05 were pointed out as a statistical trend. The genotype-specific risks were estimated as odds ratios (OR) with associated 95% confidence intervals (CIs) for dominant, recessive, codominant, over-dominant genetic model in case of statistically significant distribution between groups were observed52. Positive OR (OR > 1) with 95% CI that does not overlap the null value of OR = 1 was considered statistically significant.
For evaluation of the relationship of the outcome dependent variable (RT-induced toxicity) with potential predictors (clinical-pathological characteristics, polymorphisms and RILA) logistic regression analysis was used. The models were computed both as univariate for all predictors and multivariate for selected predictors that reached p < 0.05 in univariate analysis. Results of statistical evaluations were accompanied by OR with 95% CIs. Data were analyzed by SigmaStat 3.5 and MedCalc.
Acknowledgements
This study was financially supported by the Ministry of Education, Science and Technological Development, Republic of Serbia (451-03-68/2022-14/200043) and SIRIC Montpellier Cancer Grant (INCa-DGOS-Inserm 12553). The authors would like to thank Center for Leadership Development – Programme “Move for Science”. Also, the authors would like to thank Tatjana Petrović for her excellent technical assistance for preparation of PBMC for SNP analyses and blood sample for RILA as well as Laura Bourillon and Tiphany Gouveia for their technical assistance with the RILA assay.
Author contributions
E.M. designed the study, performed experiments, analyzed and interpreted obtained experimental data, performed the statistical analysis, and wrote the main part of the manuscript. N.P. designed the study, performed experiments, analyzed and interpreted obtained experimental data, and participated in writing the manuscript. M.B. performed experiments, participate in the interpretation of data, and critically revised the manuscript. I.M. participated in writing the manuscript and critically revised the manuscript. I.S. performed part of the experiments. K.K. enrolled patients in the study and interpreted obtained clinical data. T.S., M.N., and D.A. were responsible for the execution of the entire project, participated in study design, and critically revised the manuscript. All authors read and approved the final manuscript.
Data availability
The data that support the findings of this study are not openly available due to [reasons of sensitivity e.g. human data] and are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Massi, M. C. et al. A deep learning approach validates genetic risk factors for late toxicity after prostate cancer radiotherapy in a REQUITE multi-national cohort. Front. Oncol.10, (2020). [DOI] [PMC free article] [PubMed]
- 2.Berkey, F. Managing the adverse effects of radiation therapy. Am. Fam. Physician.82, 381–8, 394 https://pubmed.ncbi.nlm.nih.gov/20704169/ (2010). [PubMed]
- 3.Rattay, T. & Talbot, C. J. Finding the genetic determinants of adverse reactions to radiotherapy. Clin. Oncol. (R. Coll. Radiol).26, 301–308 (2014). [DOI] [PubMed]
- 4.Klusa, D. et al. Metastatic spread in prostate cancer patients influencing radiotherapy response. Front. Oncol.10, (2021). [DOI] [PMC free article] [PubMed]
- 5.Andreassen CN, Alsner J, Overgaard J. Does variability in normal tissue reactions after radiotherapy have a genetic basis—Where and how to look for it? Radiother. Oncol. 2002;64:131–140. doi: 10.1016/S0167-8140(02)00154-8. [DOI] [PubMed] [Google Scholar]
- 6.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N. Engl. J. Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 7.Peters CA, et al. TGFB1 single nucleotide polymorphisms are associated with adverse quality of life in prostate cancer patients treated with radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2008;70:752–759. doi: 10.1016/j.ijrobp.2007.05.023. [DOI] [PubMed] [Google Scholar]
- 8.Grainger DJ, et al. Genetic control of the circulating concentration of transforming growth factor type beta1. Hum. Mol. Genet. 1999;8:93–97. doi: 10.1093/hmg/8.1.93. [DOI] [PubMed] [Google Scholar]
- 9.Shah R, Hurley CK, Posch PE. A molecular mechanism for the differential regulation of TGF-beta1 expression due to the common SNP -509C-T (c. -1347C>T) Hum. Genet. 2006;120:461–469. doi: 10.1007/s00439-006-0194-1. [DOI] [PubMed] [Google Scholar]
- 10.Dunning AM, et al. A transforming growth factorβ1 signal peptide variant increases secretion in vitro and is associated with increased incidence of invasive breast cancer. Cancer Res. 2003;63:2610–2615. [PubMed] [Google Scholar]
- 11.Burri RJ, et al. Association of single nucleotide polymorphisms in SOD2, XRCC1 and XRCC3 with susceptibility for the development of adverse effects resulting from radiotherapy for prostate cancer. Radiat. Res. 2008;170:49–59. doi: 10.1667/RR1219.1. [DOI] [PubMed] [Google Scholar]
- 12.Langsenlehner T, et al. Association between single nucleotide polymorphisms in the gene for XRCC1 and radiation-induced late toxicity in prostate cancer patients. Radiother. Oncol. 2011;98:387–393. doi: 10.1016/j.radonc.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 13.Matullo G, et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22:1437–1445. doi: 10.1093/carcin/22.9.1437. [DOI] [PubMed] [Google Scholar]
- 14.Au WW, Salama SA, Sierra-Torres CH. Functional characterization of polymorphisms in DNA repair genes using cytogenetic challenge assays. Environ. Health Perspect. 2003;111:1843–1850. doi: 10.1289/ehp.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su CH, et al. Contribution of DNA double-strand break repair gene XRCC3 genotypes to triple-negative breast cancer risk. Cancer Genomics Proteomics. 2015;12:359–368. [PubMed] [Google Scholar]
- 16.Ozsahin M, et al. CD4 and CD8 T-lymphocyte apoptosis can predict radiation-induced late toxicity: A prospective study in 399 patients. Clin. Cancer Res. 2005;11:7426–7433. doi: 10.1158/1078-0432.CCR-04-2634. [DOI] [PubMed] [Google Scholar]
- 17.Azria D, et al. Radiation-induced CD8 T-lymphocyte apoptosis as a predictor of breast fibrosis after radiotherapy: Results of the prospective multicenter French trial. EBioMedicine. 2015;2:1965–1973. doi: 10.1016/j.ebiom.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talbot CJ, et al. Multi-centre technical evaluation of the radiation-induced lymphocyte apoptosis assay as a predictive test for radiotherapy toxicity. Clin. Transl. Radiat. Oncol. 2019;18:1–8. doi: 10.1016/j.ctro.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peduzzi P, Concato J, Kemper E, Holford TR, Feinstem AR. A simulation study of the number of events per variable in logistic regression analysis. J. Clin. Epidemiol. 1996;49:1373–1379. doi: 10.1016/S0895-4356(96)00236-3. [DOI] [PubMed] [Google Scholar]
- 20.Peach MS, Showalter TN, Ohri N. Systematic review of the relationship between acute and late gastrointestinal toxicity after radiotherapy for prostate cancer. Prostate Cancer. 2015;2015:1. doi: 10.1155/2015/624736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rancati T, Palorini F, Cozzarini C, Fiorino C, Valdagni R. Understanding urinary toxicity after radiotherapy for prostate cancer: First steps forward. Tumori. 2017;103:395–404. doi: 10.5301/tj.5000681. [DOI] [PubMed] [Google Scholar]
- 22.Baskar R, Lee KA, Yeo R, Yeoh KW. Cancer and radiation therapy: current advances and future directions. Int. J. Med. Sci. 2012;9:193–199. doi: 10.7150/ijms.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West CM, Barnett GC. Genetics and genomics of radiotherapy toxicity: Towards prediction. Genome Med. 2011;3:52. doi: 10.1186/gm268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuimala J, et al. Genetic polymorphisms of DNA repair and xenobiotic-metabolizing enzymes: Effects on levels of sister chromatid exchanges and chromosomal aberrations. Mutat. Res. 2004;554:319–333. doi: 10.1016/j.mrfmmm.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Hoyos-Giraldo LS, Carvajal S, Cajas-Salazar N, Ruíz M, Sánchez-Gómez A. Chromosome aberrations in workers exposed to organic solvents: Influence of polymorphisms in xenobiotic-metabolism and DNA repair genes. Mutat. Res. 2009;666:8–15. doi: 10.1016/j.mrfmmm.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Damaraju S, et al. Association of DNA repair and steroid metabolism gene polymorphisms with clinical late toxicity in patients treated with conformal radiotherapy for prostate cancer. Clin. Cancer Res. 2006;12:2545–2554. doi: 10.1158/1078-0432.CCR-05-2703. [DOI] [PubMed] [Google Scholar]
- 27.Andreassen CN, Alsner J, Overgaard M, Sørensen FB, Overgaard J. Risk of radiation-induced subcutaneous fibrosis in relation to single nucleotide polymorphisms in TGFB1, SOD2, XRCC1, XRCC3, APEX and ATM–a study based on DNA from formalin fixed paraffin embedded tissue samples. Int. J. Radiat. Biol. 2006;82:577–586. doi: 10.1080/09553000600876637. [DOI] [PubMed] [Google Scholar]
- 28.Zhou L, Xia J, Li H, Dai J, Hu Y. Association of XRCC1 variants with acute skin reaction after radiotherapy in breast cancer patients. Cancer Biother. Radiopharm. 2010;25:681–685. doi: 10.1089/cbr.2010.0811. [DOI] [PubMed] [Google Scholar]
- 29.Xie XX, et al. Predictive value of Xrcc1 gene polymorphisms for side effects in patients undergoing whole breast radiotherapy: A meta-analysis. Asian Pac. J. Cancer Prev. 2012;13:6121–6128. doi: 10.7314/APJCP.2012.13.12.6121. [DOI] [PubMed] [Google Scholar]
- 30.Brenneman MA, Wagener BM, Miller CA, Allen C, Nickoloff JA. XRCC3 controls the fidelity of homologous recombination: Roles for XRCC3 in late stages of recombination. Mol. Cell. 2002;10:387–395. doi: 10.1016/S1097-2765(02)00595-6. [DOI] [PubMed] [Google Scholar]
- 31.Song YZ, et al. Association between single nucleotide polymorphisms in XRCC3 and radiation-induced adverse effects on normal tissue: A meta-analysis. PLoS ONE. 2015;10:e130388. doi: 10.1371/journal.pone.0130388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andreassen CN, Alsner J, Overgaard M, Overgaard J. Prediction of normal tissue radiosensitivity from polymorphisms in candidate genes. Radiother. Oncol. 2003;69:127–135. doi: 10.1016/j.radonc.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 33.Andreassen CN, et al. TGFB1 polymorphisms are associated with risk of late normal tissue complications in the breast after radiotherapy for early breast cancer. Radiother. Oncol. 2005;75:18–21. doi: 10.1016/j.radonc.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Quarmby S, Fakhoury H, Levine E, Barber J, Wylie J, Hajeer AH. Association of transforming growth factor beta-1 single nucleotide polymorphisms with radiation-induced damage to normal tissues in breast cancer patients—PubMed. Int J Radiat Biol. 2003;79:137–143. doi: 10.1080/0955300021000045673. [DOI] [PubMed] [Google Scholar]
- 35.Giotopoulos G, et al. The late radiotherapy normal tissue injury phenotypes of telangiectasia, fibrosis and atrophy in breast cancer patients have distinct genotype-dependent causes. Br. J. Cancer. 2007;96:1001–1007. doi: 10.1038/sj.bjc.6603637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grossberg AJ, et al. Association of transforming growth factor β polymorphism C-509T with radiation-induced fibrosis among patients with early-stage breast cancer: A secondary analysis of a randomized clinical trial. JAMA Oncol. 2018;4:1751–1757. doi: 10.1001/jamaoncol.2018.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alsbeih G, Al-Harbi N, Al-Hadyan K, El-Sebaie M, Al-Rajhi N. Association between normal tissue complications after radiotherapy and polymorphic variations in TGFB1 and XRCC1 genes. Radiat. Res. 2010;173:505–511. doi: 10.1667/RR1769.1. [DOI] [PubMed] [Google Scholar]
- 38.Terrazzino S, et al. Common variants of GSTP1, GSTA1, and TGFβ1 are associated with the risk of radiation-induced fibrosis in breast cancer patients. Int. J. Radiat. Oncol. Biol. Phys. 2012;83:504–511. doi: 10.1016/j.ijrobp.2011.06.2012. [DOI] [PubMed] [Google Scholar]
- 39.Barnett GC, et al. No association between SNPs regulating TGF-β1 secretion and late radiotherapy toxicity to the breast: Results from the RAPPER study. Radiother. Oncol. 2010;97:9–14. doi: 10.1016/j.radonc.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnett GC, et al. Individual patient data meta-analysis shows no association between the SNP rs1800469 in TGFB and late radiotherapy toxicity. Radiother. Oncol. 2012;105:289–295. doi: 10.1016/j.radonc.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fachal L, et al. TGFβ1 SNPs and radio-induced toxicity in prostate cancer patients. Radiother. Oncol. 2012;103:206–209. doi: 10.1016/j.radonc.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 42.De Langhe S, et al. Acute radiation-induced nocturia in prostate cancer patients is associated with pretreatment symptoms, radical prostatectomy, and genetic markers in the TGFβ1 gene. Int. J. Radiat. Oncol. Biol. Phys. 2013;85:393–399. doi: 10.1016/j.ijrobp.2012.02.061. [DOI] [PubMed] [Google Scholar]
- 43.Brengues M, et al. T lymphocytes to predict radiation-induced late effects in normal tissues. Expert Rev. Mol. Diagn. 2017;17:119–127. doi: 10.1080/14737159.2017.1271715. [DOI] [PubMed] [Google Scholar]
- 44.Vogin G, et al. Absence of correlation between radiation-induced CD8 T-lymphocyte apoptosis and sequelae in patients with prostate cancer accidentally overexposed to radiation. Oncotarget. 2018;9:32680–32689. doi: 10.18632/oncotarget.26001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Azria D, et al. Single nucleotide polymorphisms, apoptosis, and the development of severe late adverse effects after radiotherapy. Clin. Cancer Res. 2008;14:6284–6288. doi: 10.1158/1078-0432.CCR-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Azria D, Betz M, Bourgier C, Sozzi WJ, Ozsahin M. Identifying patients at risk for late radiation-induced toxicity. Crit. Rev. Oncol. Hematol. 2012;84(Suppl 1):e35–e41. doi: 10.1016/j.critrevonc.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 47.Peeters STH, et al. Acute and late complications after radiotherapy for prostate cancer: Results of a multicenter randomized trial comparing 68 Gy to 78 Gy. Int. J. Radiat. Oncol. Biol. Phys. 2005;61:1019–1034. doi: 10.1016/j.ijrobp.2004.07.715. [DOI] [PubMed] [Google Scholar]
- 48.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan X, et al. Single nucleotide polymorphism at rs1982073:T869C of the TGFbeta 1 gene is associated with the risk of radiation pneumonitis in patients with non-small-cell lung cancer treated with definitive radiotherapy. J. Clin. Oncol. 2009;27:3370–3378. doi: 10.1200/JCO.2008.20.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiy F, et al. Prevalence and persistence of chromosomal damage and susceptible genotypes of metabolic and DNA repair genes in Chinese vinyl chloride-exposed workers. Carcinogenesis. 2010;31:648–653. doi: 10.1093/carcin/bgq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mutlu P, et al. Identification of XRCC1 Arg399Gln and XRCC3 Thr241Met polymorphisms in a Turkish population and their association with the risk of chronic lymphocytic leukemia. Indian J. Hematol. Blood Transfus. 2015;31:332–338. doi: 10.1007/s12288-014-0482-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horita N, Kaneko T. Genetic model selection for a case-control study and a meta-analysis. Meta Gene. 2015;5:1–8. doi: 10.1016/j.mgene.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are not openly available due to [reasons of sensitivity e.g. human data] and are available from the corresponding author upon reasonable request.