Abstract
Recent clinical development of KRAS inhibitors has heightened interest in the genomic landscape of KRAS-altered cancers. We performed a pan-cancer analysis of KRAS-altered samples from 426,706 adult patients with solid or hematologic malignancies using comprehensive genomic profiling; additional analyses included 62,369 liquid biopsy and 7241 pediatric samples. 23% of adult pan-cancer samples had KRAS alterations; 88% were mutations, most commonly G12D/G12V/G12C/G13D/G12R, and prevalence was similar in liquid biopsies. Co-alteration landscapes were largely similar across KRAS mutations but distinct from KRAS wild-type, though differences were observed in some tumor types for tumor mutational burden, PD-L1 expression, microsatellite instability, and other mutational signatures. Prognosis of KRAS-mutant versus other genomic cohorts of lung, pancreatic, and colorectal cancer were assessed using a real-world clinicogenomic database. As specific KRAS inhibitors and combination therapeutic strategies are being developed, genomic profiling to understand co-alterations and other biomarkers that may modulate response to targeted or immunotherapies will be imperative.
Subject terms: Molecular medicine, Predictive markers, Cancer genomics, Outcomes research
Introduction
The Kirsten rat sarcoma viral oncogene homolog (KRAS) gene belongs to the rat sarcoma (RAS) family of oncogenes that also includes Harvey rat sarcoma (HRAS) and neuroblastoma rat sarcoma (NRAS) viral oncogene homologs and, when mutated, can initiate or promote cancer growth1–5. Activating mutations in KRAS are among the most prevalent oncogenic driver mutations in human cancers and are associated with tumorigenesis as well as aggressive tumor growth. Despite decades of research, KRAS had been an “undruggable” target until the landmark discovery of covalent inhibitors specific for KRAS G12C6. Clinical trials of mutant-specific KRAS G12C inhibitors have shown promising activity7–12. Sotorasib and adagrasib each received FDA breakthrough designation for the treatment of advanced or metastatic non-small cell lung cancer (NSCLC) harboring a KRAS G12C mutation, and sotorasib has now received marketing authorization in the US and other countries for the treatment of certain patients with KRAS G12C NSCLC13. Besides the clinical development of direct covalent KRAS G12C inhibitors, there are significant efforts underway to develop other mutant-specific and pan-KRAS inhibitors, and inhibitors that target upstream of the RAS pathway (SOS1, SHP2)14–16.
Here we performed a comprehensive pan-cancer genomic analysis to identify the incidence of KRAS alterations across 24 tumor types, the distribution of KRAS alterations inclusive of and beyond G12C. We evaluated the genomic co-alteration landscapes and immune biomarker patterns in association with different KRAS mutations in terms of tumor mutational burden (TMB), PD-L1 expression, co-alterations, and mutational signatures that may modulate response to KRAS inhibitors, immune checkpoint inhibitors (ICI) or other therapies. We also interrogated a real-world clinicogenomic database (CGDB) to assess prognostic implications for KRAS mutated subsets compared to other genomically defined cohorts of NSCLC, colorectal cancer (CRC), and pancreatic ductal adenocarcinoma (PDAC).
Results
Prevalence of KRAS alterations in adult and pediatric cancers
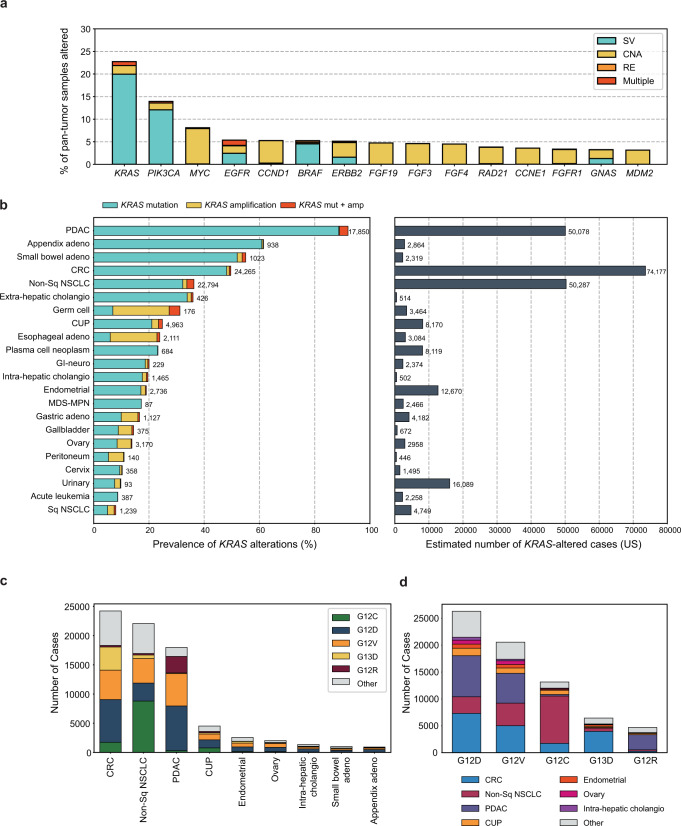
A total of 426,706 unique tissue or hematologic samples from adult patients with cancer were submitted for testing during routine clinical care from December 2013 to December 2021. KRAS was the most frequently altered oncogene with alterations identified in 97,062 (23%) pan-tumor tissue samples. The vast majority (88%) of the alterations were mutations (99.7% substitutions and 0.2% insertion or deletions [indel]), 8.4% were KRAS amplifications and 3.8% a combination of mutation with amplification (Fig. 1a). The estimated incidence for KRAS altered cancers in the US based on this prevalence data is highest in CRC with almost 75,000 cases followed by PDAC and non-squamous (non-Sq) NSCLC (>50,000 cases each; Fig. 1b and Supplemental Table 1). KRAS G12D (29%), G12V (23%), G12C (15%), G13D (7%), and G12R (5%) were the five most common KRAS mutant isoforms together accounting for ~80% of all KRAS alterations. The tumor types with the highest prevalence of KRAS mutations (KRASm) were PDAC (92%), appendiceal adenocarcinoma (61%), small bowel adenocarcinoma (SBA, 53%), CRC (49%), and non-squamous (non-Sq) NSCLC (35%) (Supplemental Table 2); CRC, non-Sq NSCLC, PDAC together represented 71% (63,480/88,907) of the KRASm pan-tumor population (Fig. 1c, d). Additionally, several other tumor types harbored KRASm including most notably: extrahepatic cholangiocarcinoma (35%), carcinoma of unknown primary (CUP, 22%), intrahepatic cholangiocarcinoma (ICC, 18%), endometrial (17%), gastric (11%), as well as breast (2.1%) and prostate (1.3%) carcinomas (Supplemental Table 2). KRAS amplification (median 11 copies, range 5-421) was rare across most tumor types but common in germ cell tumors (24%) and esophageal adenocarcinoma (18%).
Fig. 1. Prevalence of KRAS alterations among adult patients with cancer.
a In the Foundation Medicine (FM) dataset of tissue or hematologic samples from 426,706 adult patients with cancer, KRAS was the most frequently altered oncogene with alterations in 23% of samples. A longtail of other top frequently altered oncogenes is shown; bar colors indicate alteration classes, SV: short variant mutation (e.g., substitutions, indels), CNA: copy number alteration, RE: rearrangement. b Prevalence in the FM dataset (left) and incidence estimates in the United States (right) of KRAS alterations in common adult tumor types (Supplemental Table 1). KRAS alterations are most prevalent in PDAC, appendix adenocarcinoma, small bowel and CRC tumor types. The highest incidence of KRAS alterations is estimated in CRC, non-Sq NSCLC and PDAC. c, d Number of cases in the FM database with KRAS mutations among (c) the 8 top indications with highest incidence of KRAS alterations and carcinoma of unknown primary (CUP) and d the 5 most common KRAS mutant isoforms.
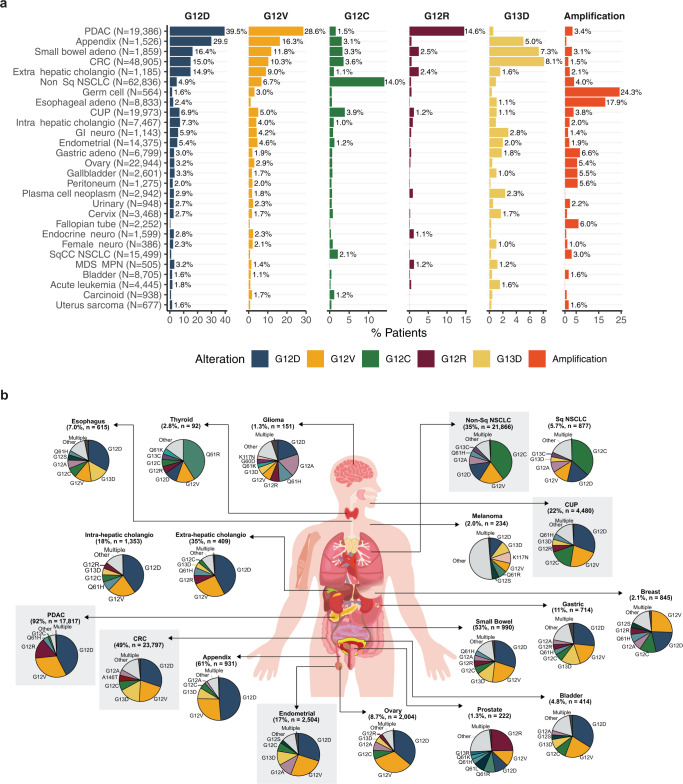
Although the KRASm distribution varied across adult tumor types, similar patterns of KRASm were observed among tumors originating from similar tissue types. KRAS G12C was the most common variant among NSCLC (40% and 36% of KRASm non-Sq and Sq, respectively). Gastrointestinal cancers (including CRC and cancers of the esophagus, stomach, small bowel, and appendix) also share a similar profile where KRAS G12D and G12V were the top two common variants. KRAS G12D was also most common among many other tumor types including PDAC (43%), and endometrial (30%), and KRAS G12V was the second most common in the majority of tumor types studied and the most common KRASm in breast (26%) (Supplemental Table 2 and Fig. 2a, b). KRASm isoforms were all largely clonal indicating that these mutations are likely truncal in all four major tumor types studied (Supplemental Fig. 1 and Supplemental Table 3). For comparative analysis, 62,369 samples underwent liquid genotyping from May 2016 to December 2021. KRAS alterations were detected in 15% of all liquid biopsies, and in 19% of liquid biopsies with elevated tumor fraction, including mutations (95.5%), amplification (3.2%) and a combination of mutation with amplification (1.3%; Supplemental Fig. 2A). KRAS G12D (26%), G12V (20%), G12C (17%), G13D (6%), and G12A (4%) were the five most common KRASm detected by liquid genotyping, which was similar to the pan-tumor distribution seen in tissue (Supplemental Fig. 2B).
Fig. 2. Prevalence of KRAS alteration subtypes by tumor type.
a Bar graph showing the prevalence of the most common types of KRAS alterations across indications. Disease subtypes are ordered based on the cumulative prevalence across the six highlighted KRAS alteration subtypes. Only tumor types with at least 250 total samples and a total KRAS alteration prevalence of at least 5% are shown. b Anatomic visualization of the prevalence of KRAS mutant isoforms. Percentage of total cases with KRAS mutations is shown for each tumor type and the corresponding ‘n’ for number of KRAS mutated cases. The four major tumor types assessed in this study as well as carcinoma of unknown primary (CUP) are highlighted in gray and represent the largest KRAS mutant populations. Major tumor types with relatively low prevalence of KRAS alterations such as breast, prostate, glioma, and melanoma are also shown. The body part background was custom designed by www.slideteam.net [slideteam.net].
Available clinicogenomic characteristics of the four major KRASm tumor types (NSCLC, CRC, PDAC, endometrial) selected based on prevalence, incidence, and the landscape of KRAS inhibitor development, stratified by major KRAS isoforms, are shown in Table 1, and additional tumor types as well as pan-cancer analysis are described in Supplemental Table 4. The pan-tumor KRAS G12C subset was notably distinct from cases with non-G12C mutations or KRAS WT, with patients carrying G12C mutant tumors being older (median age 66 vs 64 vs 64 years), more often female (58% vs 52% vs 54%), having higher TMB (32% vs 12% vs 16% with TMB ≥ 10 mutations/Mb), and a greater fraction with high PD-L1 expression (37% vs 14% vs 14%) (all comparisons p < 0.001).
Table 1.
Clinico-pathologic characteristics of patients with four major KRAS mutated tumors by KRAS mutation subtype.
| CRC (N = 23,797 KRAS mutated cases) | ||||||||
|---|---|---|---|---|---|---|---|---|
| KRAS G12C | KRAS G12D | KRAS G12V | KRAS G13D | KRAS WT | KRAS non-G12C | G12C vs non-G12C | G12C vs. WT | |
| # cases | 1739 | 7312 | 5026 | 3963 | 24,640 | 22,058 | – | – |
| Median age (range) | 60 (22–89+) | 60 (18–89+) | 61 (20–89+) | 61 (19–89+) | 60 (18–89+) | 60 (18–89+) | 0.21 | 0.27 |
| ≥65 years old | 35% (608) | 38% (2761) | 38% (1902) | 38% (1518) | 37% (9194) | 38% (8349) | 0.04 | 0.10 |
| Sex M:F % | 52:48 | 53:47 | 53:47 | 53:47 | 57:43 | 53:47 | 0.78 | P < 0.001 |
| Median TMB (range) | 3.8 (0–90) | 3.8 (0–219) | 3.5 (0–373) | 3.8 (0–608) | 3.8 (0–865) | 3.8 (0–608) | 0.59 | 0.07 |
| TMB ≥ 10 mutations/Mb (n) | 4.7% (81) | 6.6% (482) | 3.2% (163) | 9.5% (377) | 11% (2670) | 6.8% (1510) | 0.002 | P < 0.001 |
| MSI-H % (n) | 0.65% (11/1691) | 3.4% (237/7048) | 0.57% (28/4870) | 5.6% (217/3860) | 7.5% (1784/23,929) | 3.2% (690/21,347) | P < 0.001 | P < 0.001 |
| PD-L1 1–49% | 15% (68/445) | 17% (315/1832) | 13% (173/1282) | 16% (166/1022) | 13% (836/6552) | 16% (880/5595) | 0.92 | 0.21 |
| PD-L1 ≥ 50% | 1.6% (7/445) | 1.4% (26/1832) | 0.47% (6/1282) | 1.2% (12/1022) | 2.0% (134/6552) | 1.0% (58/5595) | 0.50 | 0.72 |
| Non-Sq NSCLC (N = 21,866 KRAS mutated cases) | ||||||||
|---|---|---|---|---|---|---|---|---|
| KRAS G12C | KRAS G12D | KRAS G12V | KRAS G12A | KRAS WT | KRAS non-G12C | G12C vs non-G12C | G12C vs. WT | |
| # cases | 8790 | 3085 | 4197 | 1548 | 40,042 | 13,076 | – | – |
| Median age (range) | 67 (24–89+) | 69 (23–89+) | 68 (24–89+) | 69 (30–89+) | 67 (18–89+) | 68 (23–89+) | P < 0.001 | P < 0.001 |
| ≥65 years old | 60% (5282) | 64% (1986) | 62% (2584) | 65% (1000) | 59% (23,651) | 62% (8130) | 0.003 | 0.08 |
| Sex M:F % | 40:60 | 44:56 | 42:58 | 42:58 | 49:51 | 43:57 | P < 0.001 | P < 0.001 |
| Median TMB (range) | 7.8 (0–104) | 5.0 (0–164) | 6.3 (0–937) | 6.3 (0–68) | 5.2 (0–1765) | 6.3 (0–1255) | P < 0.001 | P < 0.001 |
| TMB ≥ 10 mutations/Mb (n) | 40% (3546) | 24% (754) | 34% (1412) | 31% (485) | 32% (12,842) | 33% (4260) | P < 0.001 | P < 0.001 |
| MSI-H % (n) | 0.20% (17/8473) | 0.30% (9/2974) | 0.20% (8/4054) | 0.13% (2/1491) | 0.35% (136/38,548) | 0.26% (33/12,605) | 0.59 | 0.03 |
| PD-L1 1–49% | 28% (1067/3879) | 29% (377/1285) | 28% (506/1820) | 29% (189/655) | 30% (4937/16,351) | 28% (1578/5633) | 0.72 | 0.001 |
| PD-L1 ≥ 50% | 44% (1690/3879) | 37% (477/1285) | 39% (705/1820) | 37% (244/655) | 29% (4769/16,351) | 38% (2147/5633) | P < 0.001 | P < 0.001 |
| PDAC (N = 17,817 KRAS mutated cases) | ||||||||
|---|---|---|---|---|---|---|---|---|
| KRAS G12C | KRAS G12D | KRAS G12V | KRAS G12R | KRAS WT | KRAS non-G12C | G12C vs non-G12C | G12C vs. WT | |
| # cases | 300 | 7654 | 5542 | 2825 | 1536 | 17,517 | – | – |
| Median age (range) | 67 (37–87) | 66 (21–89+) | 66 (28–89+) | 66 (29–89+) | 64 (23–89+) | 66 (21–89+) | 0.42 | P < 0.001 |
| ≥65 years old | 59% (176) | 56% (4253) | 55% (3065) | 59% (1668) | 49% (757) | 56% (9888) | 0.64 | 0.02 |
| Sex M:F % | 59:41 | 53:47 | 52:48 | 49:51 | 59:41 | 52:48 | 0.06 | 0.95 |
| Median TMB (range) | 1.7 (0–21) | 1.3 (0–344) | 1.3 (0–166) | 1.3 (0–211) | 1.7 (0–6906) | 1.3 (0–355) | 0.001 | 0.55 |
| TMB ≥ 10 mutations/Mb (n) | 2.0% (6) | 1.0% (78) | 0.79% (44) | 0.85% (24) | 5.5% (85) | 1.0% (177) | 0.32 | 0.03 |
| MSI-H % (n) | 0.34% (1/292) | 0.37% (28/7472) | 0.20% (11/5387) | 0.15% (4/2759) | 1.5% (23/1487) | 0.32% (55/17,065) | 0.74 | 0.37 |
| PD-L1 1%-49% | 35% (29/84) | 33% (706/2118) | 30% (457/1528) | 25% (187/755) | 24% (98/410) | 31% (1484/4821) | 0.64 | 0.16 |
| PD-L1 ≥50% | 2.4% (2/84) | 7.7% (164/2118) | 4.6% (71/1528) | 6.0% (45/755) | 6.6% (27/410) | 6.3% (302/4821) | 0.35 | 0.37 |
| Endometrial (N = 2504 KRAS mutated cases) | ||||||||
|---|---|---|---|---|---|---|---|---|
| KRAS G12C | KRAS G12D | KRAS G12V | KRAS G12A | KRAS WT | KRAS non-G12C | G12C vs non-G12C | G12C vs. WT | |
| # cases | 175 | 771 | 657 | 291 | 11,639 | 2329 | – | – |
| Median age (range) | 64 (35–89+) | 63 (27–89+) | 63 (24–89+) | 64 (33–89+) | 66 (23–89+) | 64 (24–89+) | 0.95 | 0.01 |
| ≥65 years old | 46% (81) | 44% (340) | 44% (286) | 49% (144) | 56% (6484) | 46% (1082) | 1.0 | 0.03 |
| Sex M:F % | 0.57:99 | 0:100 | 0:100 | 0:100 | 0:100 | 0:100 | 0.21 | 0.03 |
| Median TMB (range) | 3.8 (0–54) | 3.8 (0–509) | 2.6 (0–384) | 3.8 (0–393) | 2.6 (0–908) | 3.8 (0–733) | 0.95 | 0.01 |
| TMB ≥ 10 mutations/Mb (n) | 30% (52) | 34% (264) | 21% (141) | 31% (89) | 16% (1919) | 33% (759) | 0.78 | P < 0.001 |
| MSI-H % (n) | 26% (43/166) | 31% (225/718) | 19% (120/621) | 27% (72/270) | 13% (1459) | 29% (619) | 0.80 | P < 0.001 |
| PD-L1 1–49% | 39% (25/64) | 26% (66/258) | 24% (54/224) | 38% (39/104) | 28% (1135/4005) | 30% (245/814) | 0.32 | 0.09 |
| PD-L1 ≥50% | 4.7% (3/64) | 1.2% (3/258) | 1.3% (3/224) | 2.9% (3/104) | 1.8% (73/4005) | 2.0% (16/814) | 0.32 | 0.14 |
Ordinal relationships were examined using the Mann–Whitney U test; categorical relationships were examined using Fisher’s exact with correction for multiple comparisons. PD-L1 expression was only available for a subset of cases.
KRAS WT includes samples WT for KRAS mutations and amplifications.
Analysis of a pediatric cohort was also performed. Among tissue or hematologic samples from pediatric patients (n = 7241 unique patients), KRAS alterations were present in 5.5% of samples. The pediatric group with the largest number of KRASm cases was acute leukemia where diverse KRASm were represented with KRAS G13D and G12D being the most common. KRASm were most prevalent in CRC (28%), germ cell tumors (20%), and myelodysplastic-myeloproliferative neoplasms (MDS-MPN, 20%); however, the total number of patients with KRASm are small due to their rarity (Supplemental Fig. 3). Based on the estimated incidence rates of different pediatric tumors in the US and prevalence of KRAS alterations observed in our cohort, we estimate the highest incidence of KRAS altered cases to be in acute leukemia (n = 553), followed by colorectal (n = 88), ovary (n = 71), and glioma (n = 64; Supplemental Table 1).
Co-alteration landscapes, mutational signatures, and immunotherapy biomarkers
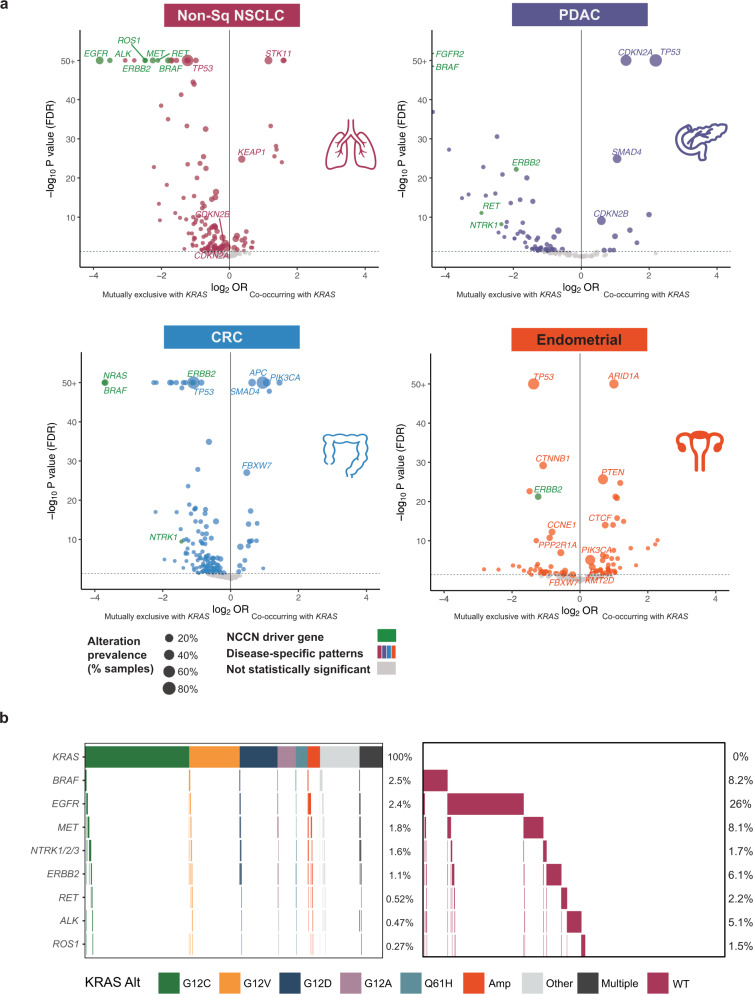
Among four major KRAS altered tumor types, volcano plots of co-occurring or mutually exclusive alterations with KRAS are shown in Fig. 3a (see also Supplemental Tables 5–8). In non-Sq NSCLC, CRC and endometrial cancer TP53 was the most frequently altered gene and tended to be mutually exclusive from KRAS, whereas TP53 alterations tended to co-occur with KRAS in PDAC. Consistent with KRAS being a key oncogenic driver, alterations in KRAS were highly mutually exclusive with other driver alterations in the RTK/MAPK pathway, including EGFR, ALK, MET, ERBB2, BRAF, RET, and ROS1 in non-Sq NSCLC (Fig. 3b); BRAF, FGFR2, ERBB2, RET and NTRK1 in PDAC; NRAS, BRAF, and ERBB2 in CRC; and ERBB2 in endometrial. Genes with co-alterations notably enriched in KRASm subsets included STK11 in non-Sq NSCLC, PIK3CA and APC in CRC, and ARID1A, PTEN, and PIK3CA in endometrial tumors. Of note, largely similar patterns of co-mutations and mutual exclusivity were observed when limiting to a subset of microsatellite stable (MSS) CRC and endometrial tumors (Supplemental Fig. 4 and Supplemental Tables 6 and 8). Volcano plots for four additional tumor types (Sq NSCLC, SBA, ICC, and appendix adenocarcinoma) also support mutual exclusivity of KRAS with other driver alterations (Supplemental Fig. 5 and Supplemental Tables 9–12).
Fig. 3. Co-occurrence of gene alterations among KRAS altered non-Sq NSCLC, PDAC, CRC and endometrial cancer.
a The prevalence of alterations was compared for KRAS altered and KRAS wild type (WT) non-Sq NSCLC (N = 62,836), PDAC (N = 19,386), CRC (N = 48,905), and endometrial (N = 14,375) tumor samples. For each tumor cohort, only genes altered in at least 50 cases and targeted across all the assay versions were included. For each gene, substitutions, short insertions/deletions, rearrangements, and copy number changes of known or likely functional significance detected using our assay were included. Driver genes highlighted in the National Comprehensive Cancer Network (NCCN) Guidelines as well as genes altered at a high prevalence (≥10%) are labeled for each volcano plot. Alterations in known driver oncogenes (labeled in green) tend to be mutually exclusive with KRAS alterations (left side of plots) in all four major tumor types studied (p ≤ 0.05; Odd’s ratio <1). b Oncoprints showing the frequency and mutual exclusivity of NCCN driver genes in KRAS altered (N = 22,794) vs KRAS WT (N = 40,042) non-Sq NSCLC. Fisher’s exact test was applied to assess patterns of co-occurrence and mutual exclusivity between KRAS and other genes alterations. P values were corrected with the Benjamini–Hochberg FDR method.
For the top differentially occurring genes with KRAS, the co-alteration landscape was largely similar across KRASm isoforms, but distinct for KRAS WT within a given tumor type (Supplemental Fig. 6). Statistical analysis of the three most common KRASm isoforms revealed additional allele-specific differences, especially in relatively rare gene alterations, within specific tumor types (Supplemental Table 13). For example, alterations in GNAS were more common in KRAS G12D compared to KRAS G12C mutated non-Sq NSCLC (5.7% vs. 2.0% respectively; FDR p < 10−4). Co-mutations in other MAPK/PI3K pathway genes tended to be more frequent in KRAS G13D compared to KRAS G12D mutated CRCs (NF1: 3.0% vs. 1.2%, AKT1: 2.0% vs. 0.8%, BRAF: 1.7% vs. 0.7% respectively; all FDR p < 10−4). Similarly, in endometrial tumors, KRAS G13D mutated tumors showed a higher prevalence of NF1 and PTEN alterations compared to KRAS G12D (NF1: 14.0% vs. 6.1%, FDR p = 0.005; PTEN: 70.6% vs. 55.1%, p = 0.0006, respectively). In PDAC, ARID1A and ERBB2 alterations were detected more commonly in KRAS G12D mutated tumors compared to KRAS G12R mutated tumors (ARID1A: 9.7% vs. 4.6%, FDR p < 10−4; ERBB2: 2.2% vs. 1.0%, FDR p = 0.0003).
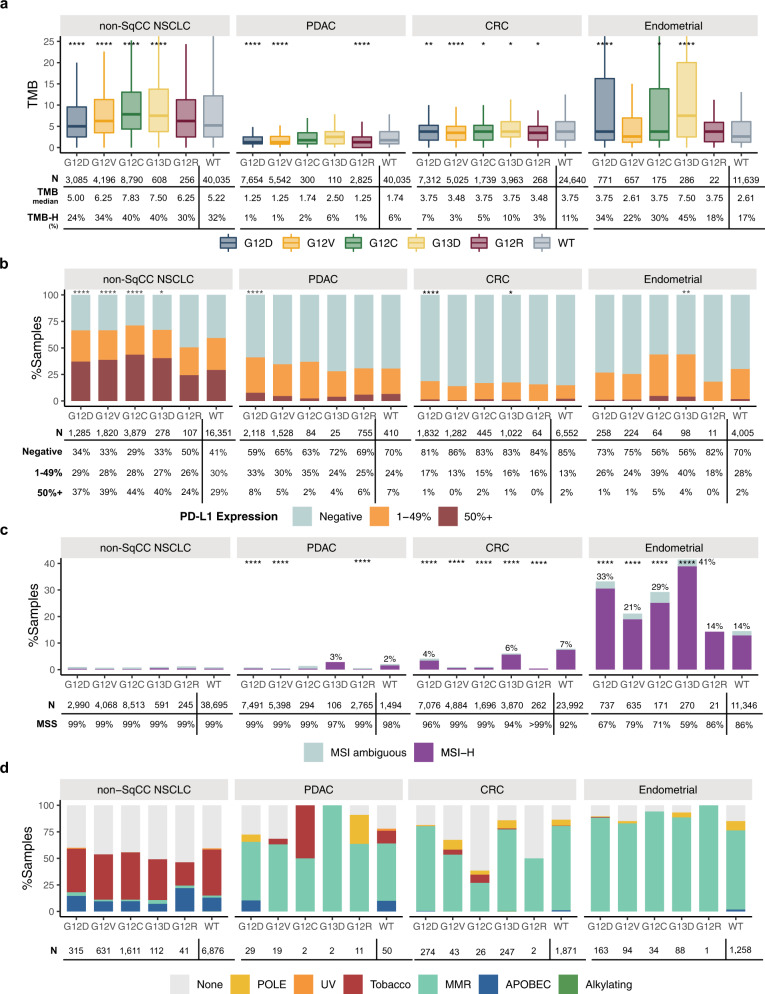
In non-Sq NSCLC, KRAS G12C mutated tumors were enriched for high TMB ≥ 10 mutations/Mb (40% vs 33% vs 32% for KRAS non-G12C and WT, both p < 0.001) and for high PD-L1 expression (44% vs 38% for KRAS non-G12C and 29% for WT, both p < 0.001), whereas KRAS G12D mutated tumors had lower incidence of elevated TMB (24% with TMB ≥ 10 mutations/Mb) relative to other KRASm isoforms assessed (Fig. 4a, b and Table 1). In Sq NSCLC the same trends were observed, but most differences were not statistically significant (Supplemental Table 4). Relative to non-Sq NSCLC, the fraction of samples positive for high PD-L1 expression in KRASm CRC, PDAC, and endometrial was low (Fig. 4b), and relatively few ICC, SBA and appendiceal samples were tested for PD-L1 (Supplemental Table 4). High TMB was also less frequent in KRASm CRC, PDAC, ICC, SBA, and appendiceal compared to NSCLC; however, endometrial was similar to NSCLC with 21–34% of samples having TMB ≥ 10. In endometrial cancers, high TMB was enriched in KRAS G13D, G12D, G12C, and G12A (45%, 34%, 30%, and 31%) compared to G12V and WT (21% and 16%, p < 0.05 for all comparisons; Table 1 and Fig. 4a). High MSI was rare (<1%) across non-Sq NSCLC subtypes, and infrequent in PDAC and CRC relative to endometrial cancer where 14–39% of KRASm isoforms were MSI high. Across tumor types, KRAS G13D and G12D were associated with the highest levels of MSI high relative to other KRASm isoforms (Fig. 4c). We also investigated KRAS allele-specific patterns of the loss of heterozygosity in the human leukocyte antigen class I locus (HLA LOH) in the four major disease subtypes and identified largely similar patterns of HLA LOH across the different KRASm isoforms (Supplemental Fig. 7). In non-Sq NSCLC, 22% of G12C-mutated cases exhibited HLA LOH; in comparison 20% to 23% of the other isoforms and 21% of WT cases had HLA LOH. In PDAC, G12D mutated cases showed a higher prevalence of HLA LOH (28%) compared to G12V (23%, p = 0.03) and G12R mutated cases (22%, p = 0.04). Rate of HLA LOH was also slightly elevated in KRAS G12D mutated CRC (18%) compared to KRAS-WT CRC (15%, p = 0.008).
Fig. 4. Immunotherapy biomarkers and mutational signatures associated with KRASm isoforms in non-Sq NSCLC, PDAC, CRC, and endometrial cancers.
a Box plots showing the distribution of tumor mutational burden (TMB) in KRASm and KRAS WT tumors. TMB is higher in KRASm vs KRAS wild-type (WT) non-Sq NSCLC and endometrial cancer, and in particular for G12C and G12D subsets of non-Sq NSCLC and G13D in endometrial cancer. Each box plot displays the interquartile range (IQR), with the lower boundary representing the 25th percentile and the upper boundary representing 75th percentile. The line within the box displays the median and the whiskers extend to ±1.5 x IQR. b PD-L1 expression was relatively consistent across KRASm and WT subsets for the four major tumor types. In non-Sq NSCLC PD-L1 high expression was enriched in G12D/V/C and G13D subsets relative to WT and in endometrial tumors, any PD-L1 expression was enriched in G12C and G13D relative to WT. c Microsatellite instability (MSI) was low across non-Sq NSCLC and PDAC. In CRC, MSI-high was enriched in KRAS WT compared to KRASm subsets, whereas in endometrial tumors, G12D/V/C and G13D subsets had elevated MSI-high compared to WT. Each KRAS mutation isoform was compared against WT with p value thresholds: 0.0001: ****, 0.001: ***, 0.01: **, 0.05: *. d Six mutational signatures were assessed for KRASm isoforms. Tobacco signature was common across KRASm and WT non-Sq NSCLC and mismatch repair (MMR) was common across PDAC, CRC and endometrial tumors. Only a subset of cases were able to be assessed for mutational signatures and number of cases is shown below each bar.
We also looked at mutational signatures across KRASm isoforms of the four major disease subtypes (Fig. 4d and Supplemental Table 14). Tobacco signature was detected in 38–45% of different KRASm and KRAS WT subsets of non-Sq NSCLC but was less frequent in G12R non-Sq NSCLC (22%) and rare in other tumor subtypes. Mismatch repair (MMR) signature was rare in non-Sq NSCLC but common in KRASm and KRAS WT PDAC, CRC and endometrial cancers, although relatively less frequent in KRAS G12C CRC relative to other subgroups. POLE signature was detected in small subsets of PDAC, CRC, and endometrial.
We further assessed the co-occurrence of KRASm isoforms and co-altered genes with implications for immunotherapy response including TMB ≥ 10 mutations/Mb, and PD-L1 expression in non-Sq NSCLC and Sq NSCLC for comparison. In non-Sq NSCLC, TMB and PD-L1 were independent biomarkers; across KRASm isoforms 49–62% had TMB ≥ 10 mutations/Mb or high PD-L1, but only 15–20% had both. With KRAS G12C mutated non-Sq NSCLC samples, co-mutations in STK11 and KEAP1 were more commonly associated with low (28% and 16%) or negative (53% and 27%) PD-L1 expression vs high (9.4% and 8.3%), and these associations were largely consistent across KRASm subsets. Across KRASm isoforms of non-Sq NSCLC, 46–51% harbored TP53, 5.1–9.2% TP53/STK11 and 1.3–2.6% TP53/STK11/KEAP1 co-alterations (Supplemental Fig. 8). In non-Sq NSCLC, there were 1487 distinct mutations throughout the TP53 gene in KRASm tumors, where 45% resulted in single amino acid changes. Co-mutations occurred throughout both STK11 and KEAP1 genes in KRASm tumors; NEF2L2 co-mutations were generally uncommon and clustered around G31 and G81 positions (Supplemental Fig. 9).
Clinico-genomic database outcomes analysis
A total of 16,357 patients with advanced NSCLC (aNSCLC), 10,430 patients with metastatic CRC (mCRC), and 3323 patients with mPDAC were included in the Flatiron Health-Foundation Medicine CGDB with 5938, 3838, and 1398 patients, respectively, meeting eligibility criteria, having started first line (1 L) of therapy after tissue biopsy CGP, and having available clinical characteristics for outcomes assessment (Supplemental Fig. 10).
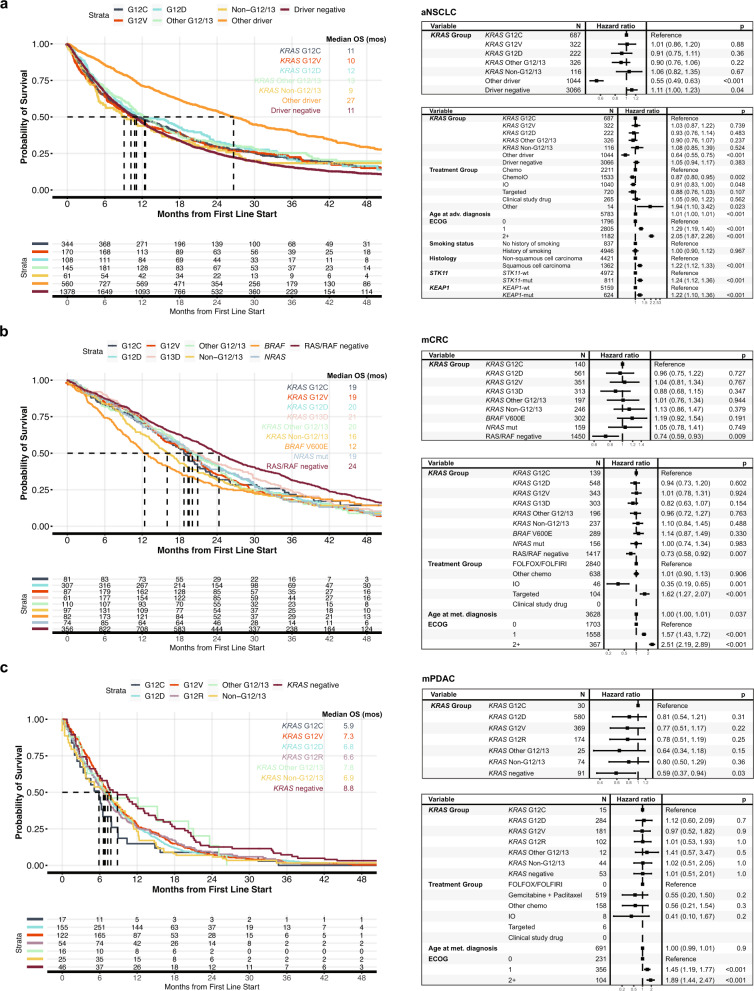
Among aNSCLC patients, KRASm were identified in 29% of cases including 93% at codons G12 or G13 (G12/13) and 6.8% at non-G12/13 codons. Patients with KRASm predominantly had non-Sq histology and most frequently were treated with 1 L chemotherapy or chemotherapy with immune checkpoint inhibitor (ICI), although over 20% of KRASm patients received ICI monotherapy (Supplemental Table 15). Patients with KRAS G12C mutation had similar overall survival (OS) to patients with other common KRAS mutations including G12V (11 vs. 10 mos, HR 1.0, 95% CI 0.86–1.20, p = 0.88) and G12D (11 vs. 12 mos, HR 0.91, 95% CI 0.75–1.11, p = 0.36) and also similar OS with rarer non-G12/13 KRASm (11 vs. 9 mos, HR 1.1, 95% CI 0.82–1.35, p = 0.67). OS for NSCLC oncodriver negative patients, although statistically different, there was very minimal clinical difference (11 vs 11 mos, HR 1.1, 95% CI 1.00–1.23, p = 0.04). Patients with aNSCLC without a KRAS alteration but exhibiting other oncogenic drivers with available approved targeted therapies, had significantly better OS than patients with NSCLC harboring KRAS G12C mutations (27 vs. 11 mos, HR 0.55, 95% CI 0.49–0.63, p < 0.001). These trends remained similar in a multivariable model incorporating treatment group, age, performance status, smoking history, histology, and concurrent STK11 and KEAP1 alterations (Fig. 5a).
Fig. 5. Real-world outcomes for patients with NSCLC, PDAC, and CRC carrying different oncodriver alterations.
Kaplan Meier curves for real world overall survival (rwOS) are shown with univariate (top) and multivariate (bottom) analysis tables for each disease subtype. Analysis was performed using the Flatiron Health-Foundation Medicine real-world clinicogenomic database. Patients with multiple driver alterations spanning >1 category were excluded. a In patients with advanced NSCLC harboring KRAS G12C mutant tumors have similar rwOS to other KRAS G12/13, KRAS non-G12/G13C, and driver negative patients. b In metastatic CRC, patients with KRAS G12C had similar rwOS compared to other KRAS mutant isoforms, BRAF V600E, and NRAS mutations, but worse rwOS compared to patients negative for KRAS and NRAS mutations and BRAF V600E (RAS/RAF negative). c In metastatic PDAC, rwOS was marginally inferior for KRAS G12C vs KRAS WT, although the differences were small and were not observed in the multivariable model.
Among mCRC patients, 49% had a KRASm, 8.1% had BRAF V600E, 4.2% had an NRASm, and 39% of cases were negative for KRASm, NRASm, and BRAF V600E (Supplemental Table 16).
Patients with KRAS G12C mutated tumors had similar OS to patients with other common KRAS mutations including G12D (19 vs. 20 mos, HR 0.96, 95% CI 0.75-1.22, p = 0.73) and G12V (19 vs. 19 mos, HR 1.04, 95% CI 0.81-1.34, p = 0.77). Patients with KRAS G12C mutated tumors had slightly elevated OS compared to patients with BRAF V600E (19 vs. 12 mos, HR 1.19, 95% CI 0.92-1.54, p = 0.19) and patients with non-G12/13 mutations (19 vs 16 mos, HR 1.13, 95% CI 0.86-1.47, p = 0.38), although not statistically significant. KRAS/NRAS/BRAF V600E negative (RAS/RAF negative) patients had more favorable OS than the KRAS G12C subgroup (24 vs. 19 mos, HR 0.74, 95% CI 0.59–0.93, p = 0.009), which was consistent in a multivariable model (Fig. 5b).
Finally, KRASm were identified in 93% of patients with metastatic PDAC, 94% of which occurred within codons G12/G13 (Supplemental Table 17). Patients with tumors harboring KRAS G12C mutation had statistically worse, though clinically similar, OS compared those with KRAS negative tumors (5.9 vs. 8.8 mos, HR 0.59, 95% CI 0.37-0.94), p = 0.03) and similar OS to patients with non-G12/13 KRASm tumors (5.9 vs. 6.9 mos, HR 0.80, 95% CI 0.50–1.29, p = 0.36). However, in a multivariable model, patients with KRAS G12C mutated tumors were not significantly associated with worse OS compared to patients with KRAS negative tumors, although this may be limited by the small number of KRAS negative cases (Fig. 5c).
Discussion
We performed the largest pan-cancer survey to date of KRAS alterations in patients with solid tumors and hematologic malignancies. KRAS is a pan-tumor oncodriver with alterations identified in 23% of over 400,000 adult cases, including 21% with mutations, similar to smaller published studies17–19. In common tumor types including NSCLC, CRC and PDAC, KRAS mutation frequencies and hotspot mutation clusters were confirmatory of published literature20,21. Taking a disease focused approach, we observed that non-Sq NSCLC, CRC, and PDAC represented 71% of all patients with KRASm cancers; the most common alterations pan-cancer were G12D (30%), G12V (23%) and G12C (15%), totaling 68% of KRASm cases, which represent potential therapeutic targets. However, we also explored less well characterized KRAS landscapes such as in endometrial cancer, where KRAS G12V and G12D are predominant, as well as small bowel and appendiceal cancers, cholangiocarcinoma, and others. Across 24 major tumor types studied, KRASm were detected at frequencies from 1.3% to 92%. We also observed KRAS alterations in 5.5% of over 7000 pediatric cases assessed, with G13D being the most common variant. In a sub analysis of CGP of liquid biopsies, we observed a similar frequency and distribution of KRAS alteration and mutation isoforms pan-tumor. KRAS amplification is also a potential therapeutic target being explored, and we saw notable enrichment of this alteration in esophageal adenocarcinomas.
In our analysis, we assessed the presence of co-alterations in over 300 genes across KRASm isoforms for eight disease types and observed some degree of similarity across KRASm isoforms, which tended to be distinct from the KRAS WT landscape. On the other hand, we did observe some KRASm specific differences in the prevalence of co-occurring gene alterations, immunotherapy-associated biomarkers and across six mutational signatures. In NSCLC, there is clear evidence that the co-alteration landscape in KRASm cancers is an important determinant of outcomes. Previously published exploratory analysis of KRAS G12C inhibitor trials suggests that co-alterations in KEAP1 and STK11 may modulate outcomes to KRAS targeted therapies, though additional studies assessing these biomarkers are warranted7,8,22. STK11 and KEAP1/NEF2L2 have also been identified as negative predictors of outcomes to chemotherapy and immunotherapy, which remain important factors to consider when treating KRASm NSCLC23–25. Our results confirm prior findings showing STK11 and KEAP1 alterations each occur more frequently in KRAS altered vs WT tumors, whereas driver alterations across disease types tend to be mutually exclusive with KRAS. Of note, our findings of elevated co-alterations in MAPK/PI3K pathway genes among KRAS G13D mutated CRCs and endometrial cancers compared to G12D mutated tumors, further supports that KRAS G13D mutated tumors may present unique biochemical and clinical mechanisms26–28. Overall, while we observed statistically significant differences in prevalence of some co-alterations, additional studies are warranted to examine the biological implications of these findings.
It is well established in NSCLC, CRC and other solid tumors that responses to KRAS inhibitors are variable; combination strategies in development will depend on characterization of the diverse genomic landscape of KRAS mutant cancers both pre-treatment and upon acquired resistance to therapies29–31. The significance of co-alterations, PD-L1 expression and mutational signatures in NSCLC and other KRASm tumor types as prognostic and predictive markers for targeted therapies and immunotherapies, warrant future investigations. Numerous combination trials with KRAS and other MAPK pathway (SHP2/MEK/ERK) inhibitors are in progress aimed at improving outcomes in patients with KRASm cancers and exploiting the presence of targetable co-alterations. Notably, in NSCLC and other solid tumors, several KRAS G12C inhibitors are being combined with immune checkpoint inhibitors (ICI), SHP2 inhibitors, EGFR inhibitors, and bevacizumab in clinical trials22,29. In CRC, trials of KRAS G12C inhibitors in combination with anti-EGFR therapies have shown promising initial results with 100% disease control rate reported for the combination of adagrasib and cetuximab32. With recent evidence for tumors employing loss of heterozygosity of the HLA locus as a common mechanism of immune evasion33–35, our study also presented the incidence of HLA LOH across cancers with frequent KRAS alterations. Investigating the role of tumor HLA status in the context of the clinical development of vaccines targeting KRAS mutations will play an important role in addressing biomarkers associated with clinical benefit from such therapies.
This study is limited by lack of available clinical and treatment information for patients in the Foundation Medicine genomic database not included in the CGDB. In the CGDB, clinical data were derived from EHR and may be incomplete or missing, particularly for events occurring outside of the Flatiron Health network. Finally, all patients in this study received CGP, which likely introduces selection bias. Our genomic analysis employs tumor-only sequencing (without matched normals) and subsequent filtering is relied on to select variants known or likely to be pathogenic for inclusion in analysis.
Notwithstanding these limitations, the findings from this study have significant implications for the development of KRAS inhibitors targeting G12C, G12D, G12V and beyond. Genomic profiling to detect co-alterations and mutational signatures, and trials to understand the clinical importance of these biomarkers as predictors of response to targeted therapies and immunotherapies in patients with a wide range of KRAS altered tumor types will be imperative to improve therapy selection and outcomes.
Methods
Foundation medicine comprehensive genomic profiling
We interrogated pan-cancer cases submitted for comprehensive genomic profiling (CGP) during routine clinical care (Foundation Medicine Inc., Cambridge, MA). For tissue biopsy samples, DNA was extracted from 40 microns of FFPE sections, and CGP was performed on hybridization-captured, adapter ligation based libraries to a mean coverage depth of >550X for 315 (n = 143,020), 324 (n = 250,197) or 405 (n = 40,730) cancer-related genes and selected introns from 28, 31, or 36 genes frequently rearranged in cancer36 (Supplemental Table 18). TMB was calculated by counting the number of non-driver synonymous and non-synonymous mutations across a 0.8–1.2 megabase (Mb) region, with computational germline status filtering, and reporting as mutations/Mb. This method has been previously validated for accuracy against whole exome sequencing37. Microsatellite instability (MSI) was determined by analyzing intronic homopolymer repeat loci for length variability and compiled into an overall MSI score via principal component analysis38. Results were analyzed for substitutions, short insertions/deletions and rearrangements, and copy number changes. A statistical copy number model was generated and fitted to each sample to determine gene copy numbers. KRAS amplifications as described here include amplifications ≥4 copies above the overall ploidy of the specimen. In most cases, this represents amplifications ≥6 copies. The detection of gene alterations followed a multi-step approach, as described previously36,39,40. Briefly, commonly occurring germline variants were excluded based on their presence in public databases such as dbSNP, 1000 Genomes Project and ExAC41–43; however, known pathogenic germline variants (e.g., in BRCA1, BRCA2) were considered as reportable. Genomic alterations were designated as known or likely pathogenic using annotations such as presence in the COSMIC database, additional knowledge about the gene affected (e.g., truncations and deletions in known tumor suppressor genes), or mutations that have been characterized as pathogenic in the scientific literature; all other uncharacterized short variant alterations were denoted as variants of unknown significance (VUS) (Supplemental Fig. 11). Detected copy number alterations (amplifications of oncogenes and homozygous deletions of tumor suppressors) recurrent fusions or rearrangements (predicted to activate oncogenes or inactivate tumor suppressors) were also designed as known or likely to be pathogenic. For the analysis outlined in this study, only genomic alterations known or likely to be pathogenic were included and VUS were excluded.
Analysis of co-occurring and mutually exclusive gene alterations with KRAS alterations was limited to genes altered in at least 50 cases and targeted across all the assay versions. For each gene, substitutions, short insertions/deletions, rearrangements, and copy number changes of known or likely functional significance detected using our assay were included. A Fisher’s exact test with FDR-based correction for multiple testing was applied for this co-mutation analysis.
Liquid CGP
For blood samples, cell free DNA (cfDNA) was extracted from blood plasma to create adapted sequencing libraries before hybrid capture and sample-multiplexed sequencing to a median unique exon coverage depth of >6000x for 62, 70, or 324 genes (Supplemental Table 18)40. Testing was performed in a CLIA-certified/CAP-accredited laboratory (Foundation Medicine Inc., Cambridge, MA).
The levels of ctDNA shed for each specimen was quantified by calculating an investigational composite tumor fraction (TF)44, which merges two methods for estimation of TF45. When TF is elevated (generally above 10%), an estimate is returned based on measure of tumor aneuploidy that incorporates observed deviations in coverage across the genome46. This aneuploidy-based approach avoids erroneously inferring elevated TF due to the presence of germline variants detected at high variant allele frequency. When lack of tumor aneuploidy limits the ability to estimate TF (generally at lower TF) a variant-based calculation is made by identifying the highest allele fraction non-germline variant, excluding specific clonal hematopoiesis (CH) associated alterations.
PD-L1 expression
PD-L1 expression was determined by immunohistochemistry (IHC) performed on FFPE tissue sections in a CLIA certified/CAP-accredited laboratory using the Dako 22C3 PD-L1 antibody. A pathologist determined the percentage of tumor cells with expression (0–100%) and the intensity of expression (0, 1+, 2+). PD-L1 expression was reported as a continuous variable with the percentage of tumor cells staining with ≥1+ intensity. PD-L1 expression was summarized as negative (<1%), low positive (1–49%), or high positive (≥50% of tumor cells staining with ≥1+ intensity). IHC staining for PD-L1 was performed with Dako 22C3 antibody (catalog number SK006) according to the manufacturer’s instructions.
Foundation Medicine-Flatiron Health Clinico-Genomic Database
We leveraged real-world data from the Flatiron Health (FH)-Foundation Medicine (FM) Clinico-Genomic Database (CGDB), a nationwide de-identified electronic health record (EHR)-derived database which includes patients sequenced at FM who received care within the FH network. The de-identified data originated from approximately 280 US cancer clinics (~800 sites of care). The FH-FM CGDB includes 16,357 patients with a diagnosis of advanced NSCLC, 10,430 with metastatic CRC, and 4438 with metastatic PDAC who received care within the FH network between 01/2011-09/2021. Cohorts included in our analysis were limited to those who had tissue CGP (FoundationOne® or FoundationOne®CDx). Patients who were diagnosed with metastatic disease greater than 90 days prior to their first visit within the FH network or received their FMI report greater than 60 days after their last FH visit date were excluded to ensure all therapies received prior to CGP were captured. Additionally, patients who had their biopsy collected after starting first line (1 L) treatment were excluded to ensure all therapies received prior to CGP were captured and that the tumor genomics were accurate prior to 1 L treatment initiation. Retrospective longitudinal clinical data were derived from EHR data, comprising patient-level structured and unstructured data, curated via technology-enabled abstraction of clinical notes and radiology/pathology reports and linked to CGP data by de-identified, deterministic matching47.
Mutational signatures
Mutational signatures for each sample were determined by examining the distribution of point mutations in the following six substitution classes: C > A, C > G, C > T, T > A, T > C, T > G, and their trinucleotide context from the bases flanking the mutated base, producing 96 possible combinations48. All point mutations were included in the analysis after excluding known oncogenic driver mutations and predicted germline mutations. In samples with at least 20 assessable mutations, the composition of the following six major mutational signatures was determined: Alkylating, APOBEC, MMR, POLE, Tobacco, and UV. A sample was deemed to have a dominant signature if a mutational class harbored a score of 0.4 or greater. Cases where a single dominant signature could not be identified were annotated as ‘None’.
Estimation of clonality
The somatic-germline-zygosity (SGZ) algorithm was applied to each sample to distinguish somatic49. For each predicted somatic alteration observed in a sample, a tumor fraction was estimated from the variant allele fraction (AF), mutant copies (mc) and wild-type copies (wc), using the following formula: 2AF/(mc−AF(wc + mc-2)). The highest estimated tumor fraction from all the somatic alterations in a sample was used as an approximate for the tumor fraction of the sample. Clonal fraction of each alteration is obtained as the ratio of the variant and sample estimated tumor fractions, with ≥50% considered clonal. Samples that failed pipeline quality control thresholds and for which SGZ could not be run were excluded from this analysis.
Determination of HLA class I loss of heterozygosity
Determination of HLA loss of heterozygosity was performed for samples profiled using FoundationOne® or FoundationOne® Heme assays34. Briefly, the minor allele frequency (MAF) of each HLA-I gene (HLA-A, HLA-B, and HLA-C) was calculated separately. HLA-I genotyping was performed using OptiType to a four-digit resolution50 and HLA-I reference sequences that matched the germline alleles for each sample were obtained. Only germline heterozygous alleles were assessed for LOH using the SGZ algorithm49.
Statistical considerations
For analysis of clinical outcomes, real world overall survival (rwOS) was defined as time from first therapy administration to date of death51. Patients without a death event were censored at their date of last known activity. To account for left truncation and to reflect the process of cohort eligibility, a patient’s entry date into the clinicogenomic database (CGDB) was considered the later of the date of a patient’s second visit within the FH network or their first eligible FM CGP report. Risk set adjustment was used to ensure patients treated prior to entry date were not included in the at-risk population in OS analysis until they reached their entry date.
IRB approval
For Foundation Medicine (FMI) genomic analysis, approval for this study, including a waiver of informed consent and a HIPAA waiver of authorization, was obtained from the WCG Institutional Review Board (IRB; Protocol No. 20152817). The IRB granted a waiver of informed consent under 45 CFR § 46.116 based on review and determination that this research meets the following requirements: (i) the research involves no more than minimal risk to the subjects; (ii) the research could not practicably be carried out without the requested waiver; (iii) the waiver will not adversely affect the rights and welfare of the subjects. The CGDB is a de-identified database, where data from Foundation Medicine and Flatiron are linked by an independent third party and there is no route to identify the included patients. For the Flatiron Health-Foundation Medicine CGDB analysis, IRB approval with waiver of informed consent based on the observational, non-interventional nature of the study (WCG IRB, Protocol No. 420180044) was also obtained prior to study conduct.
Supplementary information
Acknowledgements
This analysis is supported by Foundation Medicine Inc, Cambridge, MA, USA.
Previous presentation: a subset of the included data were presented at the annual meeting of European Society of Medical Oncology (ESMO) in 2019 and 2020 and AACR 2021. 1433: Ou et al. Annals of Oncology (2019) 30 (suppl_5): v25-v54. 10.1093/annonc/mdz239. 99P: Lee et al. Annals of Oncology (2020) 31 (suppl_4): S274-S302. 10.1016/annonc/annonc266. 2233: Sivakumar et al. Cancer Res (2021) 81 (13_Supplement): 10.1158/1538-7445.AM2021-2233.
Author contributions
J.K.L. formal analysis, data curation, visualization, writing, and review and editing; S.S. formal analysis, data curation, visualization, writing, and review and editing; A.B.S. supervision, conceptualization, writing-original draft, writing, and review and editing. R.M. formal analysis, data curation, visualization, writing, and review and editing; D.F. supervision, conceptualization, writing, and review and editing; O.G., project administration, writing, and review and editing; J.S.R., conceptualization, writing, and review and editing; G.M.F., conceptualization, writing, and review and editing; P.N., data curation, writing-review, and editing; M.M., data curation, visualization, and writing-review and editing; J.L.S., writing-review and editing; X.Y., writing-review and editing; P.S.H., supervision, conceptualization, writing, and review and editing; M.N., conceptualization, writing, and review and editing; G.R.O., supervision, conceptualization, writing, and review and editing; E.S.S., supervision, conceptualization, writing-original draft, writing, and review and editing, S.H.I.O., supervision, conceptualization, writing, and review and editing; Z.S., supervision, conceptualization, writing, and review and editing. J.K.L., S.S., and A.B.S. are co-first authors. S.H.I.O. and Z.S. are co-corresponding authors.
Data availability
The authors declare that all relevant aggregate data supporting the findings of this study are available within the article and its supplementary information files. In accordance with the Health Insurance Portability and Accountability Act, we do not have IRB approval or patient consent to share individualized patient genomic data, which contains potentially identifying or sensitive patient information and cannot be reported in a public data repository. Foundation Medicine is committed to collaborative data analysis and has well established and widely used mechanisms by which qualified researchers can query our core genomic database of >500,000 de-identified sequenced cancers. More information and mechanisms for data access can be obtained by contacting the corresponding author or the Foundation Medicine Data Governance Council at data.governance.council@foundationmedicine.com.
Code availability
This study made use of publicly available packages with R version 4.0.3 and Python 2.7. Code used to generate figures are available upon request.
Competing interests
J.K.L., S.S., A.B.S., R.M., D.F., O.G., J.S.R., G.M.F., M.M., G.R.O., P.S.H., and E.S.S. are employees of Foundation Medicine Inc, a wholly owned subsidiary of Roche, and have stock ownership in Roche. J.S.R. is consultant and stock owner in Celsius TX and Tango Tx. S.H.I.O. has received honorarium from Pfizer, Roche/Genentech, Takeda/ARIAD, Astra Zeneca, Daiichi Sankyo, BeiGene, Lilly, Caris Life Sciences Inc, has stock ownership in Turning Point Therapeutics and Elevation Oncology, and has been a member of scientific advisory board for Elevation Oncology. M.N. is on the advisory board for AstraZeneca, Daiichi Sankyo, Takeda, Novartis, EMD Serono, Janssen, Pfizer, Eli Lilly, and Company and Genentech; consultant for Caris Life Sciences (virtual tumor board); speaker for Blueprint Medicines and Takeda; and reports travel support from AnHeart Therapeutics. P.N., J.L.S., X.Y., and Z.S. are employees of Genentech, Inc. and have equity in Roche.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jessica K. Lee, Smruthy Sivakumar, Alexa B. Schrock.
Contributor Information
Sai-Hong Ignatius Ou, Email: Ignatius.ou@uci.edu.
Zhen Shi, Email: shi.zhen@gene.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41698-022-00334-z.
References
- 1.Der, C. J., Krontiris, T. G. & Cooper, G. M. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses (human tumors/transfection/retroviruses). Proc. Natl Acad. Sci. USA79, 3637–3640 (1982). [DOI] [PMC free article] [PubMed]
- 2.Parada, L. F., Tabin, C. J., Shih, C. & Weinberg, R. A. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature10.1038/297474a0 (1982). [DOI] [PubMed]
- 3.Santos, E., Tronick, S. R., Aaronson, S. A., Pulciani, S. & Barbacid, M. T24 human bladder carcinoma oncogene is an activated form of the normal human homologue of BALB- and Harvey-MSV transforming genes. Nature10.1038/298343a0 (1982). [DOI] [PubMed]
- 4.Taparowsky, E. et al. Activation of the T24 bladder carcinoma transforming gene is linked to a single amino acid change. Nature10.1038/300762a0 (1982). [DOI] [PubMed]
- 5.Capon, D. J. et al. Activation of Ki-ras2 gene in human colon and lung carcinomas by two different point mutations. Nature10.1038/304507a0 (1983). [DOI] [PubMed]
- 6.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jänne PA, et al. Adagrasib in non–small-cell lung cancer harboring a KRASG12C mutation. N. Engl. J. Med. 2022;387:120–131. doi: 10.1056/NEJMoa2204619. [DOI] [PubMed] [Google Scholar]
- 8.Skoulidis F, et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N. Engl. J. Med. 2021;384:2371–2381. doi: 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong DS, et al. KRASG12C inhibition with sotorasib in advanced solid tumors. N. Engl. J. Med. 2020;383:1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ou, S.-H. I. et al. First-in-human phase I/IB dose-finding study of adagrasib (MRTX849) in patients with advanced KRASG12C solid tumors (KRYSTAL-1). JCO JCO.21.02752 10.1200/JCO.21.02752 (2022). [DOI] [PMC free article] [PubMed]
- 11.Hallin J, et al. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Disco. 2020;10:54–71. doi: 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fakih MG, et al. Sotorasib for previously treated colorectal cancers with KRASG12C mutation (CodeBreaK100): a prespecified analysis of a single-arm, phase 2 trial. Lancet Oncol. 2022;23:115–124. doi: 10.1016/S1470-2045(21)00605-7. [DOI] [PubMed] [Google Scholar]
- 13.Nakajima, E. C. et al. FDA approval summary: sotorasib for KRAS G12C-mutated metastatic NSCLC. Clin. Cancer Res.10.1158/1078-0432.CCR-21-3074 (2021). [DOI] [PMC free article] [PubMed]
- 14.Koltun E, et al. Abstract 1260: first-in-class, orally bioavailable KRASG12V(ON) tri-complex inhibitors, as single agents and in combinations, drive profound anti-tumor activity in preclinical models of KRASG12V mutant cancers. Cancer Res. 2021;81:1260. doi: 10.1158/1538-7445.AM2021-1260. [DOI] [Google Scholar]
- 15.Wang X, et al. Identification of MRTX1133, a noncovalent, potent, and selective KRASG12D Inhibitor. J. Med. Chem. 2022;65:3123–3133. doi: 10.1021/acs.jmedchem.1c01688. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann MH, et al. BI-3406, a potent and selective SOS1-KRAS interaction inhibitor, is effective in KRAS-driven cancers through combined MEK inhibition. Cancer Discov. 2021;11:142–157. doi: 10.1158/2159-8290.CD-20-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salem, M. E. et al. Landscape of KRASG12C, associated genomic alterations, and interrelation with immuno-oncology biomarkers in kras-mutated cancers. JCO Precis. Oncol.10.1200/PO.21.00245 (2022). [DOI] [PMC free article] [PubMed]
- 18.Prior IA, Hood FE, Hartley JL. The frequency of Ras mutations in cancer. Cancer Res. 2020;80:2969–2974. doi: 10.1158/0008-5472.CAN-19-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burge RA, Hobbs GA. Not all RAS mutations are equal: a detailed review of the functional diversity of RAS hot spot mutations. Adv. Cancer Res. 2022;153:29–61. doi: 10.1016/bs.acr.2021.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Waters AM, Der CJ. KRAS: the critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb. Perspect. Med. 2018;8:a031435. doi: 10.1101/cshperspect.a031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez-Jiménez F, et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer. 2020;20:555–572. doi: 10.1038/s41568-020-0290-x. [DOI] [PubMed] [Google Scholar]
- 22.Palma G, Khurshid F, Lu K, Woodward B, Husain H. Selective KRAS G12C inhibitors in non-small cell lung cancer: chemistry, concurrent pathway alterations, and clinical outcomes. NPJ Precis. Oncol. 2021;5:98. doi: 10.1038/s41698-021-00237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arbour KC, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non–small cell lung cancer. Clin. Cancer Res. 2018;24:334–340. doi: 10.1158/1078-0432.CCR-17-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skoulidis F, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ricciuti B, et al. Diminished efficacy of programmed death-(ligand)1 inhibition in STK11- and KEAP1-mutant lung adenocarcinoma is affected by KRAS mutation status. J. Thorac. Oncol. 2022;17:399–410. doi: 10.1016/j.jtho.2021.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson CW, et al. Isoform-specific destabilization of the active site reveals a molecular mechanism of intrinsic activation of KRas G13D. Cell Rep. 2019;28:1538–1550.e7. doi: 10.1016/j.celrep.2019.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunter JC, et al. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol. Cancer Res. 2015;13:1325–1335. doi: 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- 28.McFall T, et al. A systems mechanism for KRAS mutant allele-specific responses to targeted therapy. Sci. Signal. 2019;12:eaaw8288. doi: 10.1126/scisignal.aaw8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim D, Xue J, Lito P. Targeting KRAS G12C: from inhibitory mechanism to modulation of antitumor effect in patients. Cell. 2020;183:850–859. doi: 10.1016/j.cell.2020.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, et al. Diverse alterations associated with resistance to KRAS(G12C) inhibition. Nature. 2021;599:679–683. doi: 10.1038/s41586-021-04065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Awad MM, et al. Acquired resistance to KRASG12C inhibition in cancer. N. Engl. J. Med. 2021;384:2382–2393. doi: 10.1056/NEJMoa2105281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiss J, et al. LBA6 - KRYSTAL-1: adagrasib (MRTX849) as monotherapy or combined with cetuximab (Cetux) in patients (Pts) with colorectal cancer (CRC) harboring a KRASG12C mutation. Ann. Oncol. 2021;32:S1283–S1346. doi: 10.1016/j.annonc.2021.08.2093. [DOI] [Google Scholar]
- 33.Tran E, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 2016;375:2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Montesion M, et al. Somatic HLA class I loss is a widespread mechanism of immune evasion which refines the use of tumor mutational burden as a biomarker of checkpoint inhibitor response. Cancer Discov. 2021;11:282–292. doi: 10.1158/2159-8290.CD-20-0672. [DOI] [PubMed] [Google Scholar]
- 35.McGranahan N, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;171:1259–1271.e11. doi: 10.1016/j.cell.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frampton GM, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chalmers ZR, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trabucco SE, et al. A novel next-generation sequencing approach to detecting microsatellite instability and pan-tumor characterization of 1000 microsatellite instability-high cases in 67,000 patient samples. J. Mol. Diagn. 2019;21:1053–1066. doi: 10.1016/j.jmoldx.2019.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milbury CA, et al. Clinical and analytical validation of FoundationOne®CDx, a comprehensive genomic profiling assay for solid tumors. PLoS ONE. 2022;17:e0264138. doi: 10.1371/journal.pone.0264138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark TA, et al. Analytical validation of a hybrid capture-based next-generation sequencing clinical assay for genomic profiling of cell-free circulating tumor DNA. J. Mol. Diagn. 2018;20:686–702. doi: 10.1016/j.jmoldx.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–679. doi: 10.1101/gr.9.8.677. [DOI] [PubMed] [Google Scholar]
- 42.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tukachinsky H, et al. Genomic analysis of circulating tumor DNA in 3,334 patients with advanced prostate cancer identifies targetable BRCA alterations and AR resistance mechanisms. Clin. Cancer Res. 2021;27:3094–3105. doi: 10.1158/1078-0432.CCR-20-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li G, et al. Genomic profiling of cell-free circulating tumor DNA in patients with colorectal cancer and its fidelity to the genomics of the tumor biopsy. J. Gastrointest. Oncol. 2019;10:831–840. doi: 10.21037/jgo.2019.05.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li M, et al. Abstract 2231: utility of plasma tumor fraction (TF) to inform sensitivity of FoundationOne Liquid CDx (F1LCDx) Cancer Res. 2021;81:2231. doi: 10.1158/1538-7445.AM2021-2231. [DOI] [Google Scholar]
- 47.Singal G, et al. Association of patient characteristics and tumor genomics with clinical outcomes among patients with non-small cell lung cancer using a clinicogenomic database. JAMA. 2019;321:1391–1399. doi: 10.1001/jama.2019.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zehir A, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun JX, et al. A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal. PLoS Comput. Biol. 2018;14:e1005965. doi: 10.1371/journal.pcbi.1005965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szolek A, et al. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30:3310–3316. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Q, Gossai A, Monroe S, Nussbaum NC, Parrinello CM. Validation analysis of a composite real-world mortality endpoint for patients with cancer in the United States. Health Serv. Res. 2021;56:1281–1287. doi: 10.1111/1475-6773.13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all relevant aggregate data supporting the findings of this study are available within the article and its supplementary information files. In accordance with the Health Insurance Portability and Accountability Act, we do not have IRB approval or patient consent to share individualized patient genomic data, which contains potentially identifying or sensitive patient information and cannot be reported in a public data repository. Foundation Medicine is committed to collaborative data analysis and has well established and widely used mechanisms by which qualified researchers can query our core genomic database of >500,000 de-identified sequenced cancers. More information and mechanisms for data access can be obtained by contacting the corresponding author or the Foundation Medicine Data Governance Council at data.governance.council@foundationmedicine.com.
This study made use of publicly available packages with R version 4.0.3 and Python 2.7. Code used to generate figures are available upon request.