Abstract
Influenza viruses cause acute respiratory infections, especially in the autumn–winter period. They are characterized by a high mutation frequency and cause annual seasonal epidemics. The detection of antibodies that neutralize the virus is an important criterion in the assessment of population immunity and the influenza vaccine effectiveness. In this study, a method for determining the titer of virus-neutralizing antibodies in blood serum has been developed. A new test called the luciferase neutralization assay uses a bioluminescent signal for detection. The assay is based on engineered influenza reporter viruses with various surface antigens and a nanoluciferase reporter protein in the NS1 reading frame. Using the developed method, we studied paired sera of volunteers obtained before and after vaccination. The proposed assay was compared with the conventional antibody assessment methods (microneutralization and hemagglutination inhibition assay); a high degree of correlation was observed. At the same time, the use of the luciferase neutralization assay made it possible to reduce the time required for the analysis and to simplify the detection procedure.
Supplementary Information
The online version contains supplementary material available at 10.1134/S0003683822070067.
Keywords: microneutralization, reporter virus, influenza virus, bioluminescence, nanoluciferase
INTRODUCTION
According to the World Health Organization, influenza virus poses a serious threat to public health, causing yearly from 3 million to 5 million severe infections with 290 000–650 000 deaths worldwide.1
Serological methods play a critical role in various aspects of influenza epidemic surveillance and in the assessment of immunogenicity of influenza vaccines. The hemagglutination inhibition assay (HI) and microneutralization assay (MNA) are used for the assessment of the individual immune status, for instance, before and after vaccination, and for retrospective studies of paired sera at the beginning and end of the epidemic season.
Hemagglutination inhibition assay developed in the 1940s remains the most widespread among serological methods. It is rather easy to perform and can use both live and inactivated viruses, which makes it safer and more accessible to a wide range of laboratories. However, HI only detects antibodies that prevent the influenza virus from binding to receptors on the surface of erythrocytes. The antibody titer in HI correlates with the degree of protection from influenza [2, 3], which underlies the requirements for registration of inactivated human influenza vaccines.2
MNA evaluates the ability of antibodies to block viral reproduction in a sensitive system and, unlike HI, detects not only antibodies to the receptor-binding site of the hemagglutinin (HA) molecule, but also to other neutralizing epitopes among the viral surface antigens [4]. MNA has become critical for the detection of the A(H3N2) viruses, since beginning from 2005, due to mutations in the receptor-binding site, these viruses gradually lost the ability to agglutinate chicken red blood cells and are characterized by differences in agglutination of turkey, guinea pig, and human erythrocytes, up to a complete lack of binding [5].
Historically, MNA appeared earlier than HI and at first test animals were used as a sensitive system [6]. The modern MNA version, which was first described in 1980, is based on the use of the Madin–Darby canine kidney (MDCK) cell culture and is carried out in 96-well plate format [7]. The classic MNA variant measures viral reproduction by the cytopathic effect and in the hemagglutination reaction 3–7 days after the infection of a sensitive culture.3 The test time can be reduced by using ELISA, which determines the viral reproduction as soon as 1–2 days after infection.4
MNA can be simplified and accelerated by using, for example, reporter strains, which allows virus detection by the direct measurement of a fluorescent or bioluminescent signal. By now, a great number of recombinant influenza viruses have been created that encode reporter fluorescent or luciferase proteins [8]. As an example, an influenza reporter virus encoding green fluorescent protein (GFP) in the neuraminidase (NA) reading frame was used to detect neutralizing antibodies to high-pathogenic influenza A(H5N1) viruses. However, the incubation time of infected cells before detection is 2–4 days due to the low fluorescence level of the GFP reporter and high background cell autofluorescence, which is practically not shorter than the analysis time in MNA [9]. In addition, the low genetic stability of these reporter constructs has been shown [8].
The goal of this work was to create recombinant influenza A viruses encoding a small reporter protein nanoluciferase (NanoLuc) in the NS1 protein open reading frame and to develop an express method for evaluation of virus-neutralizing antibodies on its basis (the luciferase neutralization assay, LNA). This method, as well as the classic HI and MNA, were used to examine the blood sera of volunteers before and after immunization with the seasonal inactivated influenza vaccine.
MATERIALS AND METHODS
Plasmids
The nucleotide sequence of the chimeric NS gene segment encoding the NS1 protein truncated to 124 amino acids (NS124) of the A/PR/8/1934 (H1N1) influenza virus and the NanoLuc protein (Promega, United States) was synthesized de novo and cloned into the pHW2006 vector by Evrogen (Russia). The pHW-PR8-NS124-Luc plasmid was designed so that the nucleotide sequence for the NanoLuc protein was optimized to correspond to the genomic sequence of the A/PR/8/1934 influenza virus in the codon frequency and GC content.
Plasmids encoding internal and surface proteins of the A/PR/8/1934 (H1N1) influenza virus, as well as the surface antigens of the epidemic influenza viruses A/Mississipi/10/2013 (H1N1)pdm09, A/St. Petersburg/01/2019 (H1N1)pdm09, A/Switzerland/9715293/2013 (H3N2) and A/Kansas/14/2017 (H3N2) were received from the Laboratory of Vector Vaccines of the Smorodintsev Research Institute of Influenza.
Cell Cultures
Vero cells (ATCC CCL81, United States) adapted for growing in a serum-free OptiPro medium (Gibco, United States) with the addition of 2% GlutaMax (Gibco) were used in this work. MDCK cell culture, (IRR FR-58, United States), was grown in AlphaMEM medium (BioloT, Russia) with 10% SC-biol fetal serum (BioloT). A primary chicken embryo kidney culture (CEK) was obtained by extraction of the kidney tissue from 19-day-old developing chicken embryos (СE), tissue decomposition in trypsin-EDTA solution (Sigma) for 30 min at 37°C in a water bath and filtration through the syringe nozzle with a 70 μm mesh diameter (pluriSelect Life Sciences UG&Co) followed by inoculation into DMEM/F12 medium (PanEko, Russia), containing 10% SC-biol fetal serum, 1% GlutaMax, 1% sodium pyruvate (Gibco) and 1% antibiotic-antimycotic (Gibco).
Viruses
Epidemic influenza viruses A/Mississipi/10/2013 (H1N1)pdm09 and А/Kansas/14/2017 (H3N2) were received from the Centers for Disease Control and Prevention (CDC, United States). The influenza virus A/Switzerland/9715293/2013 (H3N2) was received from the Worldwide Influenza Center (Great Britain). The A/St. Petersburg/01/2019 (H1N1)pdm09 influenza virus was isolated in the Laboratory of Evolutionary Variability of Influenza Viruses, at the Smorodintsev Research Institute of Influenza. The viruses were cultured in 10–12-day-old CE.
Recombinant influenza viruses based on the A/PR/8/1934 strain with the chimeric NS124-Luc protein and various surface antigens were obtained by reverse genetics [10]. Vero cells were transfected with a set of bidirectional plasmids encoding eight gene segments of influenza virus using a Nucleofector II electroporator (Lonza, Switzerland). After the transfection and attachment, the cells were incubated in medium supplemented with TPCK trypsin (1 μg/mL, Sigma) until the development of a specific cytopathic effect. Viruses resulting from transfection were cultivated in CE. The reporter strains with surface antigens of epidemic viruses were deposited in the Collection of Influenza and ARVI Viruses (Smorodintsev Research Institute of Influenza),5 and the A/PR8/NS124-Luc strain was deposited in the State Collection of Viruses of II–IV pathogenicity groups (no. 2906).6
Virus Infectivity Analysis
The virus infectious activity was determined by the limiting dilution method. For this purpose, a series of 10-fold dilutions of a virus-containing liquid was prepared. CE infection was carried out using dilutions in Dulbecco’s phosphate-buffered saline (DPBS, BioloT) with the addition of an antibiotic–antimycotic. Embryos were infected by injecting this solution (0.2 mL) into the allantoic cavity followed by incubation at 34°С for 48 h using 3 CE for each dilution. When Vero cell culture was used for the analysis of virus infectious titer, the dilutions were prepared using OptiPro medium containing an antibiotic-antimycotic and TPCK trypsin (1 μg/mL); for MDCK cell culture, AlphaMEM medium with antibiotic-antimycotic and TPCK trypsin (2.5 μg/mL) was used; CEK was infected in full DMEM/F12 medium without serum and trypsin. Ninety six-well plates (TPP, Switzerland) with a cell monolayer were infected with virus dilutions (0.1 mL per well) using four wells for each dilution. The plates were incubated at 37°С in 5% СО2 for 72 h. At the end of the incubation, the presence of the virus was determined by hemagglutination in allantoic or culture fluid. The infection activity was calculated by the Reed and Muench method [11] and was expressed in decimal logarithms of 50% embryo/tissue infectious dose (EID50/TID50).
Analysis of Luciferase Activity
The activity was measured in infected cells and in allantoic fluid of infected CE. To this end, the culture media was removed, and cells were washed twice with DPBS. The substrate and the buffer from the reagent kit of the Nano-Glo Luciferase Assay System (Promega) were then added to the washed cells; the mixture was incubated for 2 min and transferred to dark-walled plates. The luminescent signal (in relative luminescent units, RLU) was evaluated using a CLARIOstar multiphotometer (BMG LABTECH, ЕС).
Clinical Materials
Pre- and post-vaccination sera (day 0 and day 21) from 20 volunteers over 18 years old were used. The volunteers were vaccinated once before the epidemic season 2018/2019 with a trivalent inactivated influenza vaccine that contained A/Michigan/45/2015 (H1N1)pdm09, A/Singapore/INFIMH-16-0019/2016 (H3N2) and B/Colorado/06/2017-like strains.7
Blood serum samples were obtained within the framework of the State Task for Evaluation of the Epidemiological Effectiveness of Influenza Vaccines. The serum samples were stored at –20°C.
Luciferase Neutralization Assay
The sera were treated with the RDE enzyme (Denka Seiken Co, Ltd, Japan) in a ratio of 1 : 3 and incubated for 16–19 h at 37°C followed by 30 min at 56°C. Serial two-fold dilutions beginning from 1 : 10 (AlphaMEM medium + 1% antibiotic–antimycotic) were prepared in a 96-well U-bottom plate. An equal volume of pre-titrated reporter virus was then added to the wells, which provided a signal detection at the level of 200–300 RLU/50 μL. The mixture of serum and virus was incubated for 1 h at 37°C, after which it was transferred to a 96-well plate (100 μL per well) with a monolayer of MDCK cells. Luciferase activity in cells was measured after 6 h of incubation.
The luminescence threshold value (RLUcut_off) was determined by the formula: RLUcut_off = RLUVC/2, where RLUVC is the luciferase activity in the well of the viral control (without serum). The reciprocal of the maximal serum dilution, at which the luciferase activity was less or equal to RLUcut_off, was considered the serum titer. If the luminescent signal of the 1 : 10 dilution exceeded the threshold value, the titer was taken equal to 5.
Microneutralization Assay
Sera treated with RDE were used in MNA. Two-fold dilutions (1 : 10—1 : 1280) in AlphaMEM medium with a 1% antibiotic were added to an equal volume of the pre-titrated virus antigen containing 100 TID50/50 μL. After a 1-h incubation at room temperature, an aliquot (100 μL) of each mixture was transferred to 96-well plates with a pre-washed monolayer of MDCK cells. The plates were incubated at 37°C in an atmosphere of 5% CO2 for 24 h. Then medium was removed, and the cells were washed and fixed with 80% acetone for 15 min at room temperature. The wells were blocked with a 5% suspension of skimmed milk powder (TF DITOL, Russia) in PBS supplemented with 0.1% Tween-20 (PBS-T, Serva, EU) at room temperature for 2 h.
The virus reproduction was detected using the murine monoclonal antibodies 6D11 to influenza A virus nucleoprotein, labeled with horse radish peroxidase (courtesy of Dr. V. Z. Krivitskaya). The antibodies in a dilution of 1 : 2000 in blocking buffer were added into plate wells (100 μL per well) and incubated for 1 h at room temperature. After washing with PBS-T, a TMB solution (100 μL, Dako, EU), which is the peroxidase substrate, was added, the plate was incubated for 15 min, and the reaction was stopped by the addition of 2 N H2SO4 (100 μL per well, VECTON, Russia). Optical density was measured at a wavelength of 450 nm (OD450) on a CLARIOstar multiphotometer. The optical density threshold (ODcut_off) was calculated by the formula: ODcut_off = (ODVC + ODCC)/2, where ODVC is the optical density of infected cells without serum, and ODСC is the optical density of uninfected cells. The final serum titer was expressed as the reciprocal of the maximal dilution, at which OD was less than or equal to ODcut_off. If OD of the 1 : 10 dilution exceeded the threshold value, the serum titer was taken equal to 5.
Hemagglutination Inhibition Assay
HI was carried out as described in Methodical Guidelines MU 3.3.2.1758-03.8 Epidemic influenza viruses, sera treated with RDE and 0.5% suspension of human erythrocytes of blood group I(0) were used for the analysis. Sera were additionally treated with concentrated erythrocytes (incubation for 1 h at 4°C) to prevent nonspecific binding. The HI titer of antibodies was expressed as the reciprocal of the highest serum dilution, at which agglutination inhibition was observed. If hemagglutination inhibition was not detected at the 1 : 10 dilution, the titer was taken equal to 5.
Statistical Data Analysis
The MS Office Excel 2016, GraphPad Prizm 6.07 and RStudio Desktop 1.0.153 software packages were used for visualization and statistical processing of the data. The results were described using such indicators as arithmetic mean, standard deviation (SD), and geometric mean. The antibody titer increase was calculated as the ratio of the antibody titer on day 21 (after vaccination) to the titer on day 0 (before vaccination). Correlation of the antibody titers and the titer increase values obtained using different methods was assessed using the Pearson test applied to the logarithms.
RESULTS AND DISCUSSION
Obtaining Recombinant Nanoluciferase-Encoding Influenza Viruses
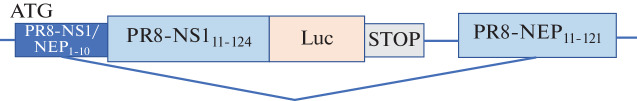
Recombinant viruses were obtained using the NanoLuc reporter gene. This enzyme is a genetically engineered protein obtained by directed evolution from the small subunit of luciferase from deep sea shrimp Oplophorus gracilirostris [12]. The nucleotide sequence encoding Nanoluc was optimized and inserted into the open reading frame of the influenza virus NS1 protein, which was truncated to 124 amino acids (Fig. 1).
Fig. 1.
A map of the chimeric A/PR/8/1934 (H1N1) NS genomic segment coding for the truncated NS1 protein, fused with the NanoLuc insert, and a nuclear export protein (NEP).
The A/PR/8/1934 (H1N1) strain characterized by high reproduction in various biological systems was used as a source of genes for internal and nonstructural proteins of reporter viruses. A/PR/8/1934 (H1N1) virus and reference epidemic strains of the A(H1N1)pdm09 and A(H3N2) subtypes served as a source of surface antigens. Reverse genetics was used to obtain a set of recombinant viruses encoding the NanoLuc reporter protein (Table 1). The resulting reporter viruses were characterized by high genetic stability of the NS chimeric gene segment with the heterologous insert throughout 4 consecutive passages in CE. High luciferase activity (about 104–106 RLU) was observed in various cell cultures infected with the reporter viruses. In their level of luciferase activity the resulting reporter viruses were similar to the viruses encoding NanoLuc in the PA gene segment [13] or other small luciferase (Gaussia luciferase) in the full-length NS gene segment [14].
Table 1.
Recombinant influenza viruses with a luciferase reporter and different surface antigens
| Reporter influenza virus* | Wild type virus, a source of surface antigens |
Titer, logEID50/mL/logTID50/mL* | |||
|---|---|---|---|---|---|
| CE | MDCK | Vero | CEK | ||
| A/PR8‑NS124‑Luc | A/PR/8/1934 (H1N1) | 8.28 | 7.75 ± 0.43 | 7.59 ± 0.63 | 8.24 ± 0.25 |
| A/Miss/PR8‑NS124‑Luc | A/Mississippi/10/2013 (H1N1)pdm09 | 6.2 | <1.5 | 2.5 | 2.5 |
| A/SPb/PR8‑NS124‑Luc | A/St.‑Petersburg/01/2019 (H1N1)pdm09 | 6.7 | 6.44 ± 0.10 | 2.5 ± 0.00 | 4.4 ± 0.09 |
| A/Switz/PR8‑NS124‑Luc | A/Switzerland/9715293/2013 (H3N2) | 7.2 | 6.53 ± 0.22 | 5.80 ± 0.38 | 8.17 ± 0.44 |
| A/Kans/PR8‑NS124‑Luc | А/Kansas/14/2017 (H3N2) | 7.45 | 6.5 ± 0.00 | 3.11 ± 0.54 | <1.5 |
* Virus titer is expressed in logEID50/mL for CE and in logTID50/mL for cell cultures.
The obtained reporter viruses were characterized by high infectious activity in the CE system, regardless of the subtype of surface antigens; however, they differed in replication in different cell cultures (Table 1). The A/PR8-NS124-Luc (H1N1) strain, as was expected, replicated actively in all tested systems. The A/SPb/PR8-NS124-Luc (H1N1)pdm09 and A/Kans/PR8-NS124-Luc (H3N2) strains had lower reproduction activity in Vero and CEK cells. The A/Miss/PR8-NS124-Luc (H1N1)pdm09 strain showed a low replication level in all used cell cultures.
The Kinetics of Luciferase Accumulation in Cells Infected with Reporter Viruses
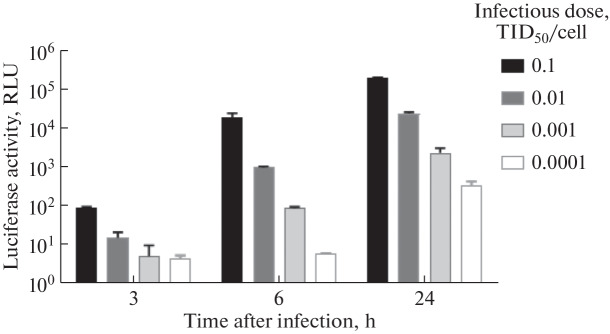
The luciferase activity kinetics was studied using the A/PR8-NS124-Luc influenza strain. Vero cells were infected with the virus at various doses (0.0001–0.1 TID50/cell) and luciferase activity of the expressed NanoLuc protein was evaluated after 3–24 h.
The luminescent signal in the infected cells increased gradually over 24 h (Fig. 2). Using an infectious dose of 0.001 TID50/cell (about 40 TID50 per well), the luminescence was observed at the level of 102 RLU (in the absence of a background signal) as early as 6 h after infection. Thus, the bioluminescent reporter significantly reduced the time from cell infection to detection.
Fig. 2.
The increase in the luciferase activity over time in Vero cells infected with the recombinant influenza A/PR8-NS124-Luc virus at various infectious doses, from 0.1 TID50/cell to 0.0001 TID50/cell.
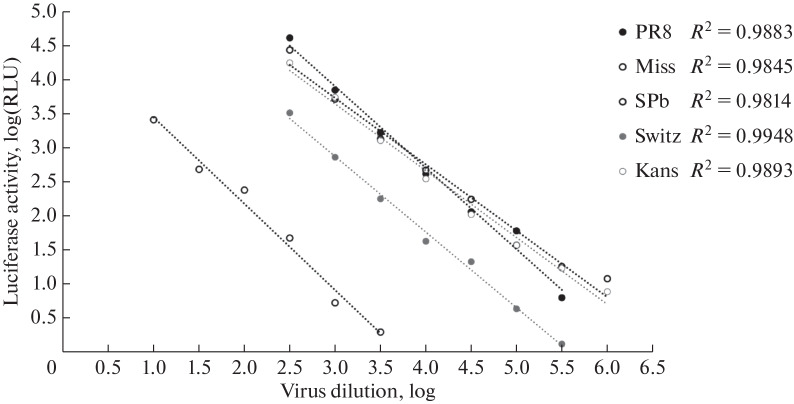
In addition, there is a direct correlation between the luminescent signal in the cells and the infectious dose of the virus, which makes the parameter of luciferase activity applicable not only for virus detection, but also for assessment of its quantity. In order to clarify the nature of this correlation, the curves of the dependence of the luminescent signal in MDCK cells on the virus infectious dose were plotted for each engineered recombinant virus (Fig. 3). The graphs were linear in the RLU range from 101 to 105, where the approximation accuracy (R2) was 0.98–0.99.
Fig. 3.
The dependence of luciferase activity in MDCK cells 6 h after infection on the infectious dose of recombinant reporter viruses. The linear fit results in MSOffice16 Excel (R2) are shown on the diagram.
The Titer of Virus-Neutralizing Antibodies in the Sera of Volunteers
The engineered recombinant viruses were used for the assessment of the titer of virus-neutralizing antibodies in blood serum samples of volunteers taken before and after immunization with an inactivated vaccine against seasonal influenza virus. The neutralizing antibody assessment method was developed based on the classical MNA; however, for detection, a direct measurement of the luciferase activity of the reporter viruses in cells was used. Based on the results of the luciferase activity kinetics in infected cells, the following parameters were selected for LNA performance: incubation time, 6 h, and control signal level, 200–300 RLU. The selected parameters were validated by testing the sera of laboratory animals (rats) immunized with the reporter viruses or their combinations (the results are presented in Supplementary Materials, Table S1).
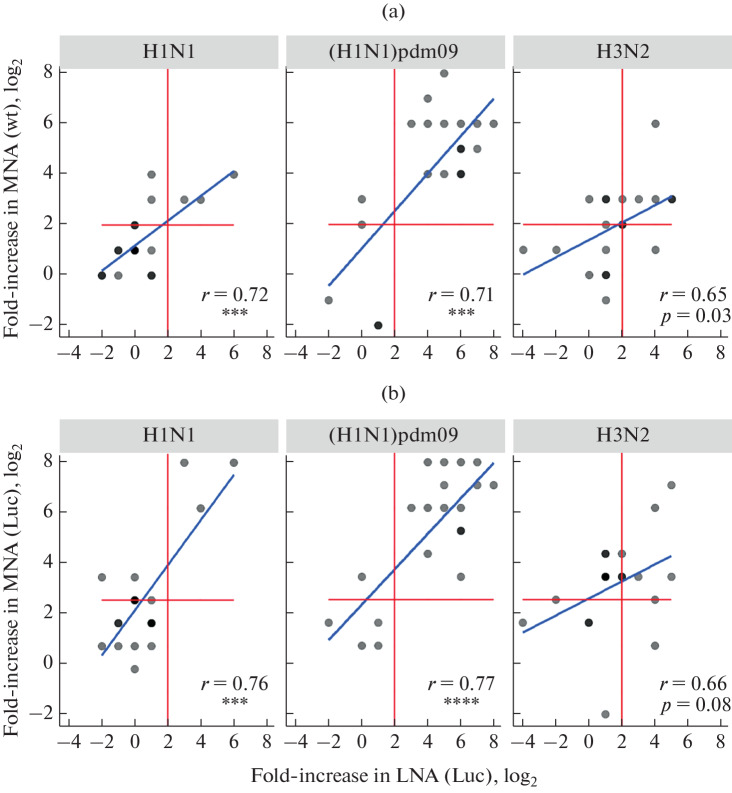
The LNA antibody titers of the volunteer sera were compared with the levels of neutralizing antibodies obtained in classical MNA, as well as with the amounts of anti-hemagglutinating antibodies detected in HI. In the comparative assay, the following reporter strains were used: A/PR8-NS124-Luc (H1N1), A/SPb/PR8-NS124-Luc (H1N1)pdm09 and A/Kans/PR8-NS124-Luc (H3N2), as well as the related epidemic viruses. The results of the correlation analysis of neutralizing antibody titers are shown in Fig. 4, while the data on the comparison of LNA and HI are presented in Supplementary Materials, Fig. S1. The results demonstrate a high correlation degree between the LNA and MNA/HI data for the influenza virus of the A(H1N1)pdm09 subtype (r > 0.7) and a moderate correlation level for the A(H1N1) and A(H3N2) viruses (r > 0.5).
Fig. 4.
Comparative analysis of neutralizing antibody titers measured in volunteer sera by LNA (X axis) and MNA (Y axis) using (a) wild-type virus (wt) and (b) reporter virus (Luc). The graphs show the individual values (log2) of antibody titers; a darker color means the coincidence of several values at one point. The blue line represents the result of the correlation analysis; Pearson’s correlation coefficients and p values are shown on graphs: (***), p < 0.001 or (****), p < 0.0001.
A four-fold or more increase in the antibody titer on day 21 is an important characteristic of the immune response to vaccination (included in the three European criteria for influenza vaccines).9 In this regard, we also compared the antibody titer increase measured using LNA and MNA/HI. The results of these two methods correlated well for the A(H1N1)pdm09 and A(H1N1) influenza viruses (r > 0.7) and moderately for the A(H3N2) virus (r > 0.5) (Fig. 5).
Fig. 5.
Comparative analysis of the antibody titer increase in sera of vaccinated volunteers measured by LNA (X axis) and MNA (Y axis). Viruses used: (a), wild-type (wt) and (b), reporter (Luc). A darker gray color means the coincidence of several values at one point. Red lines mark the threshold of a significant increase in antibody titer (4 times or more). Blue line represents the results of the correlation analysis; Pearson’s correlation coefficients and p values are shown on the graphs: (***), p < 0.001, and (****), p < 0.0001.
In general, the antibody titer increase to the A(H1N1)pdm09 influenza virus after vaccination measured by both LNA and MNA was higher than that to the A(H1N1) and A(H3N2) viruses (Fig. 5). A similar dependence was observed when comparing the results of LNA and HI (Figure S2). This may be due to the fact that the A(H1N1)pdm09 influenza viruses are subject to very slow antigenic drift. According to HI with post-infectious ferret sera, the A/St.-Petersburg/01/2019 (H1N1)pdm09 virus used in the experiment does not differ antigenically from the A/Michigan/45/2015 (H1N1)pdm09 virus, which is part of the vaccine composition of the season 2018/2019.10 A(H3N2)viruses are more variable and the antigenic activity of the А/Kansas/14/2017 (H3N2) strain used in this experiment differs significantly from that of the A/Singapore/INFIMH-16-0019/2016 vaccine virus: the difference in the titers of post-infectious ferret sera in HI was 64 or more times11. It should be noted that the A/PR/8/1934 (H1N1) strain was not a component of the vaccine composition at all. Thus, we can conclude that LNA is a sensitive test for identification of antibodies to antigenically close and distant influenza viruses.
In this study, recombinant influenza strains that stably expressed the luciferase reporter throughout a series of passages on CE were obtained. NanoLuc has a molecular weight of 19 kDa and is much smaller in size than fluorescent proteins, the most widespread firefly luciferase (61 kDa) and coral (Renilla) enzyme (36 kDa). In the case of the influenza vector, the smaller size provides a higher genetic stability for the reporter gene. In addition, NanoLuc differs from other luciferases in its higher activity and independence from ATP, which increases its sensitivity and facilitates the detection of the bioluminescent signal.
We created reporter influenza A viruses based on the high-yield A/PR/8/1934 strain and surface antigens (HA and NA) of the epidemic A(H1N1)pdm09 and A(H3N2) influenza viruses. NanoLuc in the reporter viruses was linked to the truncated C terminus of the NS1 protein, which is actively expressed during the first hours after infection [15]; this provided the registration of the bioluminescent signal as rapidly as 6 h after the cell infection. This advantage of the created reporter viruses was used for the development of an express method for evaluating virus-neutralizing antibodies.
The developed LNA method was used to assess antibody titers in blood serum samples obtained from volunteers before and after the immunization with inactivated vaccine against seasonal influenza. The LNA results correlated with the MNA results, as well as with the HI results. At the same time, the use of luciferase vectors significantly reduced the time of the neutralization test due to the higher sensitivity of bioluminescent detection and skipping the additional stages of immunoenzyme staining carried out in the classical MNA.
A rapid neutralization test based on the concept of fluorescence and luminescence detection has been developed to identify antibodies to metapneumovirus, rabies virus, and several animal and avian viruses [16]. The approach based on the use of pseudoviruses with a luminescent reporter is widely used, for instance, for the assessment of neutralizing antibodies to coronavirus SARS-CoV-2 [17] and papillomovirus [18]. The method for identification of neutralizing antibodies with influenza pseudoviruses [19], in particular, its version with two reporters [20], was also described. The latter is characterized by the disadvantage of complicated process of obtaining pseudoviruses, which requires a specialized MDCK-HA cell culture expressing a specific hemagglutinin subtype. In addition, pseudoviruses only partially mimic the infection process, reducing it to the stage of virus penetration into the cell. The use of recombinant viruses with an inserted reporter gene is free from these disadvantages.
CONCLUSIONS
Overall within the framework of this study, we have developed a neutralization test based on influenza A reporter viruses with bioluminescent activity, and have shown that luciferase neutralization assay can be successfully used for the analysis of post-vaccination immunity to influenza A viruses.
Supplementary Information
FUNDING
The study was supported by the grant of the President of the Russian Federation for young PhDs (no. 075-15-2019-226) and the grant of the Government of Saint-Petersburg for PhD students (25.09.2018, No. 124).
COMPLIANCE WITH ETHICAL STANDARDS
The authors declare that they have no conflicts of interest.
This article does not contain any studies involving animals performed by any of the authors.
The blood samples from volunteers were taken according to the research protocol PEV-2018/2019, version 01 of September 14, 2018 approved by the Local Ethic Committee of the Smorodintsev Research Institute of Influenza (LEC meeting no. 131 dated October 10, 2018).
Footnotes
WHO Influenza FactSheet, https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal).
European Medicines Agency. Guideline on Influenza Vaccines, https://www.ema.europa.eu/en/documents/scientific-guideline/influenza-vaccines-non-clinical-clinical-module_en.pdf.
WHO Manual on Animal Influenza Diagnosis and Surveillance, https://www.who.int/csr/resources/publications/influenza/whocdscsrncs20025rev.pdf.
WHO Manual for the laboratory diagnosis and virological surveillance of influenza, https://www.who.int/influenza/gisrs_ laboratory/manual_diagnosis_surveillance_influenza/en/.
Russian Biobank of Influenza at the Smorodintsev Research Institute of Influenza, https://rubin.influenza.spb.ru/
State Collection of Viruses of II–IV Pathogenicity Groups on the basis of the D.I. Ivanovsky Institute of Virology of the FSBI N.F. Gamaleya National Research Center of the Ministry of Health of Russia, https://virology.gamaleya.org/structure/otdel-gosudarstvennoy-kollektsii-virusov/.
WHO Recommended composition of influenza virus vaccines for use in the 2018–2019 northern hemisphere influenza season, https://www.who.int/influenza/vaccines/virus/recommendations/2018_19_north/en/.
Methods for determining the quality indicators of immunobiological preparations for the prevention and diagnosis of influenza: Methodical instructions MU 3.3.2.1758-03 (approved by the Chief State Sanitary Doctor of the Russian Federation on September 28, 2003).
European Medicines Agency. Guideline on Influenza Vaccines. https://www.ema.europa.eu/en/documents/scientific-guideline/influenza-vaccines-non-clinical-clinical-module_en.pdf.
Worldwide Influenza Center. Report prepared for the WHO annual consultation on the composition of influenza vaccine for the Northern Hemisphere 2019–2020. https://www.crick.ac.uk/sites/default/files/2019-04/Crick%20-VCMFeb2019%20report_toPost.pdf (Table 5–11).
WHO. Addendum to the recommended composition of influenza virus vaccines for use in the 2019–2020 northern hemisphere influenza season. (Table 1). https://www.who.int/influenza/vaccines/virus/recommendations/201902_recommendation_addendum.pdf?ua=1.
Abbreviations: CEK, primary culture of chicken embryo kidney; CE, developing chicken embryos; DPBS, Dulbecco’s phosphate-buffered saline; EID50, 50% embryo infectious dose; ELISA, enzyme-linked immonosorbent assay; HA, hemagglutinin; HI, hemagglutination inhibition assay; LNA, luciferase neutralization assay; MNA, microneutralization assay; NA, neuraminidase; NanoLuc, nanoluciferase; NS1, influenza virus non-structural protein 1; RLU, relative luminescence units; TID50, 50% tissue culture infectious dose.
Translated by I. Gordon
REFERENCES
- 1.Hirst G.K. The quantitative determination of influenza virus and antibodies by means of red cell agglutination. J. Exp. Med. 1942;75:49–64. doi: 10.1084/jem.75.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potter C.W., Oxford J.S. Determinants of immunity to influenza infection in man. Br. Med. Bull. 1979;35:69–75. doi: 10.1093/oxfordjournals.bmb.a071545. [DOI] [PubMed] [Google Scholar]
- 3.Black S., Nicolay U., Vesikari T. Hemagglutination inhibition antibody titers as a correlate of protection for inactivated influenza vaccines in children. Pediatr. Infect. Dis. J. 2011;30:1081–1085. doi: 10.1097/INF.0b013e3182367662. [DOI] [PubMed] [Google Scholar]
- 4.Krivitskaya V.Z., Kuznetsova E.V., Mayorova V.G. Microneutralization reaction versus hemagglutination inhibition reaction in assessing the immunogenicity of influenza vaccines and diagnosing influenza. Infekts. Immun. 2019;9:763–772. doi: 10.15789/2220-7619-2019-5-6-763-772. [DOI] [Google Scholar]
- 5.Lin Y.P., Xiong X., Wharton S.A. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc. Natl. Acad. Sci. U. S. A. 2012;109:21474–21479. doi: 10.1073/pnas.1218841110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis T., Shope R.E. Neutralization tests with sera of convalescent or immunized animals and the viruses of swine and human influenza. J. Exp. Med. 1936;63:645–653. doi: 10.1084/jem.63.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank A.L., Puck J., Hughes B.J. Microneutralization test for influenza A and B and parainfluenza 1 and 2 viruses that uses continuous cell lines and fresh serum enhancement. J. Clin. Microbiol. 1980;12:426–432. doi: 10.1128/JCM.12.3.426-432.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breen M., Nogales A., Baker S.F. Replication-competent influenza A viruses expressing reporter genes. Viruses. 2016;8:179. doi: 10.3390/v8070179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rimmelzwaan G.F., Verburgh R.J., Nieuwkoop N.J. Use of GFP-expressing influenza viruses for the detection of influenza virus A/H5N1 neutralizing antibodies. Vaccine. 2011;29:3424–3430. doi: 10.1016/j.vaccine.2011.02.082. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann E., Neumann G., Hobom G. “Ambisense” approach for the generation of influenza A virus: vRNA and mRNA synthesis from one template. Virology. 2000;267:310–307. doi: 10.1006/viro.1999.0140. [DOI] [PubMed] [Google Scholar]
- 11.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 12.Hall M.P., Unch J., Binkowski B.F. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran V., Poole D.S., Jeffery J.J. Multi-modal imaging with a toolbox of influenza A reporter viruses. Viruses. 2015;7:5319–5327. doi: 10.3390/v7102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckert N., Wrensch F., Gärtner S. Influenza A virus encoding secreted Gaussia luciferase as useful tool to analyze viral replication and its inhibition by antiviral compounds and cellular proteins. PLoS One. 2014;9:e97695. doi: 10.1371/journal.pone.0097695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young J.F., Desselberger U., Palese P. Efficient expression of influenza virus NS1 nonstructural proteins in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1983;80:6105–6109. doi: 10.1073/pnas.80.19.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y., Li L.F., Yu S. Applications of replicating-competent reporter-expressing viruses in diagnostic and molecular virology. Viruses. 2016;8:127. doi: 10.3390/v8050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nie J., Li Q., Wu J. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nie J., Huang W., Wu X. Optimization and validation of a high throughput method for detecting neutralizing antibodies against human papillomavirus (HPV) based on pseudovirons. J. Med. Virol. 2014;86:1542–1555. doi: 10.1002/jmv.23995. [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Sobrido L., Cadagan R., Steel J. Hemagglutinin-pseudotyped green fluorescent proteinexpressing influenza viruses for the detection of influenza virus neutralizing antibodies. J. Virol. 2010;84:2157–2163. doi: 10.1128/JVI.01433-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker S.F., Nogales A., Santiago F.W. Competitive detection of influenza neutralizing antibodies using a novel bivalent fluorescence-based microneutralization assay (BiFMA) Vaccine. 2015;33:3562–3570. doi: 10.1016/j.vaccine.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.