Abstract
Staphylococcus aureus is isolated from a substantial number of patients with infective endocarditis who are not known to have predisposing heart abnormalities. It has been suggested that the infection is initiated by the direct binding of S. aureus to human vascular endothelium. To determine the mutual response of the endothelial cells and the bacteria, we studied the interaction between S. aureus and human vascular endothelium. Scanning electron microscopic analyses showed that binding of S. aureus to human umbilical vein endothelial cells (HUVEC) mainly occurred via thread-like protrusions extending from the cell surface. Bound bacteria appeared to be internalized via retraction of the protrusions into newly formed invaginations of the endothelial cell surface. The growth phase of S. aureus had a major impact on the interaction with HUVEC. Logarithmically growing bacteria showed increased binding to, and were more readily internalized by, HUVEC compared to stationary-phase bacteria. To assess the bacterial response to the cellular environment, an expression library of S. aureus was used to identify genes whose expression was induced after 4 h of exposure to HUVEC. The identified genes could be divided into different categories based on the functions of the encoded proteins (transport, catabolism, biosynthesis, and DNA repair). Further analyses of five of the S. aureus transposon clones showed that HUVEC as well as human serum are stimuli for triggering gene expression in S. aureus.
Infective endocarditis (IE) due to Staphylococcus aureus is an acute infection of the heart. S. aureus IE frequently has a fulminant course, and mortality is up to 40%. Approximately half of patients with S. aureus IE have no known history of heart disease or heart damage (12, 20, 30, 32). It is assumed that these patients develop IE due to the ability of S. aureus to directly interact with the undamaged endocardial lining.
S. aureus has a tropism for endocardial tissue (19) and adheres much more readily to vascular endothelial cells (EC) than other bacterial species (23). Following initial binding, adherent S. aureus cells are actively internalized by the EC. Intracellular S. aureus resides and persists in phagosome-like vacuoles (2, 10, 19, 23). Although in EC the phagosomes were found to fuse with lysosomes, no bacterial degradation could be observed (19). The intracellular presence of S. aureus eventually leads to cell destruction (6, 19, 33), due to the direct action of bacterial toxins (34) or through induction of apoptosis of the EC (21). Damage of the vascular endothelial lining exposes the subendothelial matrix to the bloodstream, causing deposition of platelets and fibrin, and can thus result in the onset of IE.
A recent study indicated that S. aureus responds to the complex in vivo environment by altering gene expression (17). However, the in vivo stimuli to which S. aureus responds remained undetermined. Contact with eukaryotic host cells has recently been identified as a signal for bacterial pathogens, resulting in the expression of genes that are specifically required for survival or virulence (7, 26, 37). As interaction of S. aureus with vascular EC might be a primary step in the pathogenesis of S. aureus IE, the present study focuses on the initial process of colonization and invasion of the endothelial lining. In particular, we aimed to identify S. aureus genes whose expression is specifically induced in the presence of EC in order to understand the mechanism by which these pathogens enhance their pathogenicity. This was done by studying the response of S. aureus to human umbilical vein endothelial cells (HUVEC) and analyzing S. aureus genes whose expression was induced upon exposure to HUVEC.
MATERIALS AND METHODS
Reagents and media.
M199 medium was purchased from Life Technologies (Grand Island, N.Y.). Human serum (HS) was prepared from blood collected from healthy donors and was heat inactivated at 56°C for 30 min (HSi). Lysostaphin was from Sigma Chemical Co. (St. Louis, Mo.), gelatin was from Difco Laboratories (Detroit, Mich.), l-glutamine was from Flow Laboratories (Irvine, United Kingdom), penicillin G was from Brocades Pharma B.V. (Leiderdorp, The Netherlands), streptomycin was from Gist-Brocades N.V. (Delft, The Netherlands), and amphotericin B was from Squibb B.V. (Rijswijk, The Netherlands). EC growth factor was prepared from bovine hypothalamus as described previously (3).
HUVEC.
HUVEC were isolated from the human umbilical cord vein by digestion with 0.1% collagenase as described previously (3). The cells were cultured to confluency in M199 cell culture medium, i.e., M199 medium supplemented with 10% HSi, 1 mM l-glutamine, 0.1 mg of streptomycin/ml, 100 U of penicillin G/ml, 100 U of amphotericin B/ml, 0.1 mg of EC growth factor/ml, and 5 U of heparin/ml in plastic tissue culture dishes (Falcon no. 3080; Becton Dickinson, Lincoln Park, N.J.) in a 5% CO2 atmosphere at 37°C.
S. aureus inoculum preparation.
S. aureus strain RN4220, a restriction-negative mutant used for general cloning purposes (16), was used in these experiments. Inocula for infection of HUVEC were prepared from either overnight or early-exponential (3 h of growth) cultures grown at 37°C in nutrient broth no. 2 (Oxoid Ltd., London, United Kingdom). The bacteria were harvested by centrifugation at 1,500 × g for 10 min, washed twice in 0.9% NaCl, and resuspended in M199 medium supplemented with 0.1% (wt/vol) gelatin (gelatin-M199). S. aureus was opsonized by incubation with 20% (vol/vol) fresh HS in gelatin-M199 at 37°C under rotation for 30 min. The bacteria were washed once with gelatin-M199 and resuspended in M199 plus 10% HSi. The numbers of CFU were determined by plating serial dilutions in phosphate-buffered saline (pH 7.4).
Infection of HUVEC with S. aureus.
HUVEC cultures grown to confluency on gelatin-coated glass coverslips in 24-well tissue culture plates were washed with M199 cell culture medium without antibiotics. When confluent, each well contained about 2 × 105 EC. Subsequently, 1 ml of opsonized S. aureus in M199 plus 10% HSi was added, and HUVEC cultures were incubated at 37°C in a 5% CO2 atmosphere. Infection was allowed to proceed for 1 h. The cell monolayers were then washed twice with prewarmed M199 at 37°C to remove extracellular bacteria, incubated with 2 U of lysostaphin/ml for 5 min at room temperature to lyse the remaining cell-bound S. aureus organisms, and then washed twice more with M199 cell culture medium at 37°C. Lysostaphin does not induce EC activation (2) or cell damage or affect monolayer integrity (unpublished results). Determination of the percentage of infected HUVEC and assessment of the number of intracellular bacteria were done by light microscopy and by plating of HUVEC lysed by the addition of 1 ml H2O, respectively. For light microscopical counting, EC monolayers on 0.5% gelatin-coated glass coverslips were fixed by incubation in methanol for 15 min and stained with Giemsa stain for 15 min (2). EC were scored positive for infection when intracellular bacteria could be distinguished. Intracellular bacteria stained dark purple and could easily be distinguished from extracellular bacteria, which stained light pink, apparently damaged by the lysostaphin treatment. On three individual glass coverslips from three independent HUVEC infections, bacteria were counted in 100 microscopic fields of vision, containing 15 to 20 EC per field. The results are the averages of these counts in two infection assays performed on different days.
Scanning electron microscopy (SEM).
HUVEC grown to confluency on gelatin-coated glass coverslips in culture dishes were infected for 1 h with S. aureus grown overnight. After being washed as described above, the EC were fixed overnight in a cacodylate-buffered (pH 7.4) gluteraldehyde-formaldehyde mixture (25). Subsequently, the coverslips were thoroughly rinsed with cacodylate buffer (pH 7.4), dehydrated in a graded series of ethanol, and dried with hexamethyl disilasane. Samples were examined in an XL20 scanning electron microscope (Philips, Eindhoven, The Netherlands).
Construction of a Tn917-lacZ transposon bank.
A genomic expression library of S. aureus RN4220 was constructed using the transposon Tn917-lacZ containing vector pLTV1 (13), essentially as described by Camilli et al. (5). Vector pLTV1 was introduced into S. aureus RN4220 by electroporation (29). Final freezer stocks from this S. aureus bank were stored at −70°C. Over 99% of the bacteria from these stocks were tetracycline sensitive and erythromycin resistant, indicating loss of pLTV1 and insertion of the transposon into the chromosome. The randomness of insertions was checked by Southern blotting (A. Lammers, E. Kruijt, C. van de Kuijt, P. J. M. Nuijten, and H. E. Smith, personal communication).
Selection of HUVEC-inducible S. aureus genes.
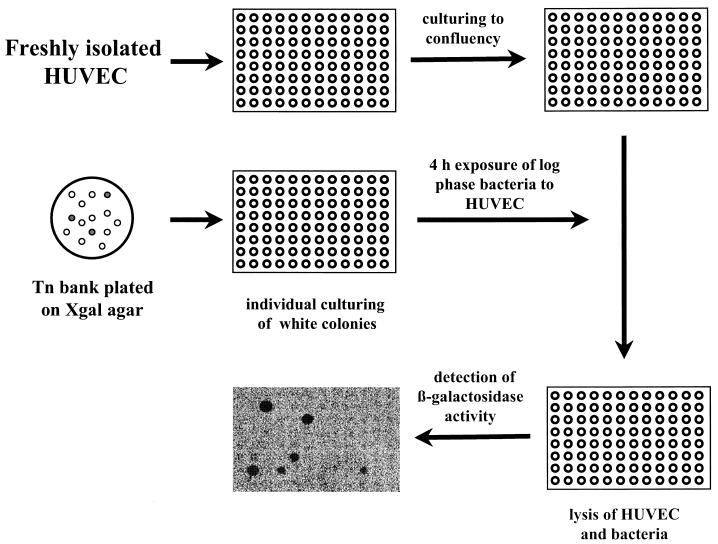
The approach for the selection of inducible S. aureus genes is schematically depicted in Fig. 1. The staphylococcal expression library was plated onto Luria-Bertani (LB) agar plates containing 250 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) (Boehringer Mannheim) per ml. After overnight incubation at 37°C, white colonies were transferred to single wells of a 96-well microtiter plate containing 100 μl of LB broth supplemented with erythromycin (masterplate). Early-log-phase cultures, obtained by subculturing overnight cultures from the wells of the masterplates in LB broth with erythromycin for 3 h, were used to infect HUVEC cultured in the same format (107 CFU/well and 3 × 104 to 4 × 104 HUVEC/well, respectively). The bacteria were allowed to interact with the HUVEC during a 4-h incubation period. After this incubation period, the HUVEC were lysed by the addition of lysis reagent (Boehringer Mannheim), and all of remaining bacteria were washed once in 0.9% NaCl, resuspended in 100 μl of lysis reagent, and lysed by the addition of 2.5 U of lysostaphin and incubation at 37°C for 30 min. Bacterial β-galactosidase activity was determined using a chemiluminescent β-galactosidase reporter gene assay (Boehringer Mannheim). The assays were performed in opaque 96-well microtiter plates (Boehringer Mannheim) to reduce background, and chemiluminescent signals were detected in a Lumi-Imager (Boehringer Mannheim). Clones were only selected when the signals were at least twofold higher than background levels (i.e., signal of bacteria not expressing β-galactosidase). To obtain higher signals, several of the selected S. aureus clones were retested by culturing HUVEC in 24-well cell culture plates (2 × 105 HUVEC/well) and using higher start inocula (5 × 107 CFU/well). As controls, bacteria were cultured in M199 medium with or without HSi or in LB medium. To correct for background signal, S. aureus not expressing β-galactosidase were included, and bacterial counts were performed. Experiments were repeated at least three times.
FIG. 1.
Schematic representation of the selection strategy for the identification of S. aureus gene expression, as described in Materials and Methods. The black dots on the final photographic image represent those S. aureus clones positive for β-galactosidase activity after exposure to HUVEC, as detected with a chemiluminescent β-galactosidase gene assay and a Lumi-Imager.
DNA manipulations, transformation, and sequencing.
DNA manipulations were done by standard techniques (28). Chromosomal DNA was isolated from S. aureus using the Puregene chromosomal DNA isolation kit for gram-positive bacteria and yeast (Gentra Systems Inc., Minneapolis, Minn.) with lysostaphin at a final concentration of 5 U/ml. Chromosomal DNA fragments were self-ligated using T4 DNA ligase (Boehringer Mannheim). Plasmid DNA was introduced into Escherichia coli BHB2600 (11) or DH5α (Gibco-BRL, Life Technologies, Breda, The Netherlands) by electroporation (9). Plasmid DNA was isolated from E. coli using the Wizard Plus minipreps kit from Promega Corporation (Madison, Wis.). DNA sequencing was performed using the PCR-mediated Taq dye deoxy terminator cycle-sequencing kit (Perkin-Elmer, Foster City, Calif.) and primer AV33 (5′-CAC AAT AGA GAG ATG TCA GCG-3′). Reactions were analyzed on an Applied Biosystems (San Jose, Calif.) model 373 DNA sequencer. The obtained sequences were compared to entries in the GenBank database using the BLAST program (1). Homology scores were considered significant when the sum probability was higher than e−10. Lower homologies (e−3 to e−9) to previously described genes were indicated. Sequences with homology scores lower than e−3 were classified as having no significant database match (8).
RESULTS
Influence of bacterial growth phase on HUVEC infection.
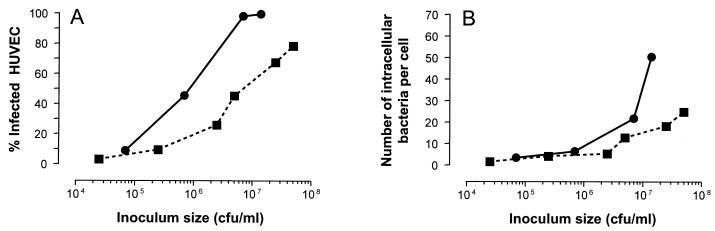
Expression of the various adhesive structures of S. aureus, such as protein A, clumping factor, and adhesins for fibronectin, fibrinogen, and collagen, is maximal in the early and mid-logarithmic growth phase and is down-regulated in the stationary phase (18). We therefore infected HUVEC with either early-logarithmic-phase or stationary-phase S. aureus RN4220 using various inocula. The percentage of infected cells and the numbers of intracellular bacteria per cell increased with increasing inoculum size, irrespective of the bacterial growth phase. Logarithmic-phase S. aureus organisms were much more infective than stationary-phase bacteria, since inocula of logarithmic-phase bacteria required to obtain the same percentage of HUVEC infection were much smaller than those of stationary-phase bacteria (Fig. 2). The number of bacteria per cell was also higher with the logarithmic-phase bacteria than with the stationary-phase bacteria. This was most pronounced at higher inocula (Fig. 2).
FIG. 2.
Infection with different inocula of either logarithmic-phase (circles) or stationary-phase (squares) bacteria by determination of the percentage of infected cells (A) and the bacterial numbers per cell (B). The values are the averages of counts in 100 microscopic fields of vision from three independent HUVEC infections and are representative of duplicate experiments performed on different days.
SEM analysis of S. aureus-HUVEC interaction.
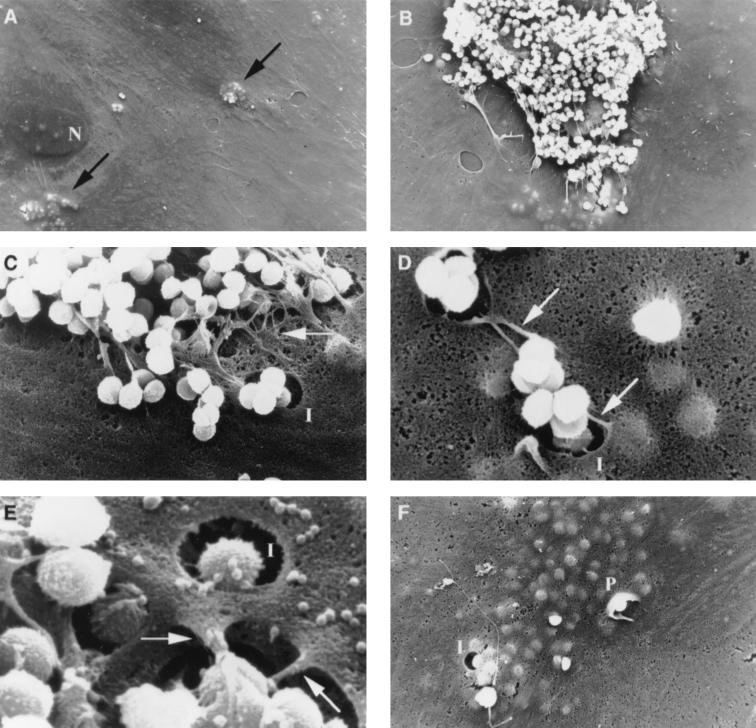
The primary contact of S. aureus with HUVEC and the subsequent bacterial internalization was studied by SEM. After 1 h of infection with 107 bacteria and removal of nonadherent S. aureus organisms, internalized bacteria were observed within HUVEC (Fig. 3A). Most cell-bound S. aureus organisms were present as clusters of various sizes (Fig. 3B), and bacteria were attached to thread-like protrusions extending from the EC (Fig. 3C to E). Uninfected EC did not show such protrusions (not shown). Higher magnification revealed close contact between bacteria and cells via these protrusions (Fig. 3D and E). The protrusions with adherent bacteria seemed to be internalized or possibly retracted into already opened invaginations of the endothelial cell surface (Fig. 3C to E). Although occasionally bacteria were internalized by HUVEC through cup-shaped uptake processes with pseudopod-like structures (Fig. 3F), this was only in a minority of the phagocytotic processes (not shown).
FIG. 3.
SEM analysis of the interaction between HUVEC and S. aureus RN4220 1 h after infection. (A) Bacteria were internalized by HUVEC and could be observed within EC, as indicated by the black arrows. (B) Most cell surface-bound S. aureus were present as clusters of various sizes. (C to E) S. aureus cells were mainly bound to thread-like protrusions (white arrows) extending from the cell surface, which appeared to be internalized or retracted into opened invaginations of the cellular surface. (F) Occasionally, pseudopod-like structures were observed during uptake of the bacteria by HUVEC. N, cell nucleus with several nuclei; I, invagination of the cellular surface; P, pseudopod-like membrane structure. Magnifications: ×1,500 (A), ×3,000 (B), ×10,000 (C), ×15,000 (D), ×30,000 (E), and ×5,000 (F).
Isolation of inducible S. aureus genes upon exposure to HUVEC.
As the bacterial response to a new environment is, at least in part, accomplished by altering gene expression profiles, we determined the induced gene expression of S. aureus RN4220 in the presence of HUVEC (see Materials and Methods). Since the number of intracellular S. aureus organisms in this experimental setup was too low to assess bacterial β-galactosidase activity, the β-galactosidase activity of the entire bacterial population after 4 h of HUVEC exposure was determined. From a total of approximately 800 bacterial clones that were white on X-Gal-containing agar plates, 41 were identified that showed β-galactosidase activity when exposed to HUVEC. This indicated that transposon integration in the selected clones had occurred in a gene whose expression was induced under the conditions used. To determine the site of transposon integration, genomic DNA was isolated from these clones, digested with EcoRI, and self-ligated. Because of the presence of only one EcoRI site within the integrated vector pLTV1, self-ligation resulted in plasmids that contained vector sequences necessary for plasmid replication, as well as chromosomal DNA of S. aureus upstream of the promoterless lacZ gene up to a chromosomally located EcoRI site. After transformation of E. coli BHB2600 with these ligation mixtures, erythromycin-resistant colonies were selected on erythromycin-containing LB agar. Plasmid DNA was isolated from these colonies and digested with EcoRI to remove possible multiple chromosomal inserts, and the inserts upstream of the promoterless lacZ were sequenced.
A total of 33 different insertions were identified within the 41 isolated clones, and in 19 of these clones the identified S. aureus sequence showed homology at the protein level to entries in the database (Table 1). These sequences could be divided into four categories according to their functions. Four were sequences of transporter proteins, involved in amino acid (BrnQ), peptide (OppD and OppF), or sugar (FruA) transport. Homology to two different peptidases was found, and N-acyl-l-amino acid amidohydrolase was identified in six of the isolated sequences with three different transposon insertion positions within the gene. Homologs of six proteins known to be involved in different biosynthetic routes, e.g., amino acid and nucleotide biosynthesis, were found. Strikingly, four of these were enzymes involved in the synthesis of lysine from aspartate (Fig. 4). One of the insertions was in a gene encoding a UV damage repair protein. Finally, 3 of the identified sequences showed homology to unknown or hypothetical proteins from Bacillus subtilis, and 14 did not give any significant database match.
TABLE 1.
Characterization of isolated S. aureus sequences
| Classification | Strain | No.a | Blast descriptionb |
|---|---|---|---|
| Transporters | 4-C1/5-H9c | 3 | (U87144) branched-chain amino acid carrier protein; BrnQ (S. aureus) |
| 4-H11 | 1 | (Z99111) PTS fructose-specific enzyme IIBC component; FruA (B. subtilis) | |
| 5-C6 | 2 | (X89237) oligopeptidepermease; OppF (Streptococcus pyogenes) | |
| 8-C2 | 1 | (AE001293) oligopeptide transporter ATPase; OppD (Chlamydia trachomatis) | |
| Peptidases | 5-G3/3-D11/9-D10cd | 6 | (D90917) N-acyl-l-amino acid amidohydrolase; Ama Synechocystis sp.) |
| 5-H3 | 1 | (Z99110) similar to oligoendopeptidase; YjbG (B. subtilis) | |
| Biosynthetic routes | 8-C1 | 2 | (P39593) hydroxyethylthiazole kinase (thiamin biosynthesis); ThiM (B. subtilis) |
| 9-B12 | 1 | (B26532) prephenate dehydrogenase (tyrosine biosynthesis); TyrA (B. subtilis) | |
| 5-B6 | 2 | (P23630) diaminopimelate decarboxylase (DAP-lysine biosynthesis); LysA (B. subtilis) | |
| 6-C6 | 1 | (Q57865) dihydrodipicolinate reductase (DAP-lysine biosynthesis); DapB (Methanococcus jannaschii) | |
| 8-H10d | 1 | (P08495) aspartokinase (DAP-lysine biosynthesis); Ask2 (B. subtilis) | |
| 7-H6 | 1 | (Q59291) aspartate-semialdehyde dehydrogenase (DAP-lysine biosynthesis); Asd (Campylobacter jejuni) | |
| DNA repair | 7-B11 | 1 | (Z99115) UV-damage repair protein; UvrX (B. subtilis) |
| Unknown/hypothetical | 5-H6 | 1 | (Z99121) similar to hypothetical protein; YvgP (B. subtilis) |
| 6-G10 | 1 | (Z99114) unknown; Yoj0 (B. subtilis) | |
| 10-G2 | 2 | (P37572) putative DNA repair protein; RadA (B. subtilis) | |
| No database matchc | 14 |
Number of recovered strains with transposon integrated within the same gene.
Determined by comparison of the sequences and coding regions with the EMBL GenBank DNA database, using the BLASTX and N determinations (>e−10). PTS, phosphoenolpyruvate: sugar phosphotransferase system; DAP, diaminopimelic acid.
Strains with transposon integration at different positions within the same gene.
Low homology scores (e−3 to e−9).
Homology scores with no significant database match (<e−3).
FIG. 4.
Successive steps in the biosynthesis pathway of l-lysine from l-aspartate. The intermediates ll-2,6-diaminopimelate and meso-2,6-diaminopimelate are incorporated into bacterial cell walls. Expression of the genes encoding the enzymes printed in boldface was found to be up-regulated in S. aureus exposed to HUVEC.
Determination of inducing stimuli.
The stimuli resulting in the induced bacterial gene expression were investigated in more detail. From each of the four categories (see above) (Table 1) one clone was chosen randomly. The four clones were 7-B11, 8-C2, 8-H10, and 9-D10. In addition, clone 4-G8, with no database match, was used. We first assessed the β-galactosidase activities in these clones cultured in M199 medium or in M199 medium supplemented with 10% HSi and compared them to the activity in LB broth. Clones 7-B11, 8-C2, 8-H10, and 9-D10 had no detectable β-galactosidase activity when grown in LB broth, whereas clone 4-G8 had very limited activity (data not shown). All five clones showed strongly induced β-galactosidase expression when cultured in plain M199 medium. Although the exact induction levels could not be determined due to the detection limit of the assay, the levels were more than 100-fold increased in most cases. In addition to the strong induction by the M199 medium, four of the tested clones showed even higher levels of β-galactosidase activity due to the presence of HSi in the M199 medium (Table 2). This indicates that components from serum can be stimuli for gene induction in S. aureus.
TABLE 2.
Induction levels of gene expression in five of the isolated S. aureus RN4220 clones when cultured in the presence of HSi
| Clone | Homolog | Fold induction in M199 + HSia |
|---|---|---|
| 8-C2 | OppD | 2.1 |
| 9-D10 | Ama | 1.0 |
| 8-H10 | Ask2 | 1.3 |
| 7-B11 | UvrX | 3.0 |
| 4-G8 | Unknown | 3.1 |
Induction is expressed as fold increase in β-galactosidase activity of bacteria cultured for 4 h in M199 supplemented with 10% HSi compared to that of bacteria cultured for 4 h in M199. All clones were retested three times, and the fold induction is representative of the individual tests.
Next, we determined if monolayers of HUVEC presented a signal to S. aureus resulting in induction of bacterial gene expression. The β-galactosidase activity of bacteria exposed to HUVEC in M199 cell culture medium supplemented with 10% HSi was determined and compared to the β-galactosidase activity of S. aureus incubated in the same medium in the absence of HUVEC. Four clones showed increased β-galactosidase activities in the presence of HUVEC (1.4- to 4.0-fold), whereas clone 8-C2 showed no difference under these conditions (Table 3). This indicates that S. aureus responds to signals from HUVEC, resulting in the expression of a specific subset of S. aureus genes.
TABLE 3.
Induction levels of gene expression in five of the isolated S. aureus RN4220 clones when cultured in the presence of HUVEC
| Clone | Homolog | Fold induction in presence of HUVECa |
|---|---|---|
| 8-C2 | OppD | 1.0 |
| 9-D10 | Ama | 2.1 |
| 8-H10 | Ask2 | 2.7 |
| 7-B11 | UvrX | 4.0 |
| 4-G8 | Unknown | 1.4 |
Induction is expressed as fold increase in β-galactosidase activity of bacteria cultured for 4 h in the presence of HUVEC compared to that of bacteria cultured for 4 h in the absence of HUVEC. All clones were retested at least three times, and the fold induction is representative of the individual tests.
DISCUSSION
In patients without predisposing underlying heart disease, development of S. aureus IE is believed to be due to a direct interaction of the bacteria with EC of the endocardium. EC actively internalize S. aureus (23), but the precise nature of this interaction is largely unknown.
S. aureus expresses several different adhesins which might play a role in the interaction with EC. Expression of these adhesins is maximal during logarithmic growth and is down-regulated at the late exponential and stationary phases (18). The growth phase of S. aureus also influenced the interaction with HUVEC. Both the number of infected cells and the total numbers of intracellular bacteria per EC were higher with logarithmic-phase S. aureus than with stationary-phase S. aureus. Expression of adhesins and toxins in S. aureus is largely regulated by the agr locus (14, 18), which is partially absent in the restriction-negative mutant S. aureus RN4220 (22). The observed differences in infectivity between early-log-phase and stationary-phase S. aureus indicate that regulation of the expression of bacterial structures involved in EC binding may not depend solely on agr. In addition, RN4220 induced killing of most EC within 24 h of infection, as was also found for the virulent peritonitis isolate S. aureus CAPD but not for the avirulent S. aureus strain 42D (31). It thus seems that, although S. aureus RN4220 lacks part of one of its regulatory loci, this strain still behaves much like a virulent S. aureus strain in HUVEC infections.
One hour after exposure of HUVEC to S. aureus in vitro, SEM showed both surface-bound and intracellular bacteria. Surface-bound S. aureus organisms were mostly observed in clusters, resembling the patchy manner of binding of S. aureus to cultured human valvular EC, rabbit endovascular tissue, and human aortic tissue (19) and suggesting a nonuniform distribution of cellular receptors on the HUVEC surface. Thread-like protrusions extending from the surfaces of the HUVEC, to which most of the surface-bound bacteria were attached, appear to be specifically induced by the bacteria, since such structures were never observed on HUVEC which were not exposed to S. aureus. As the protrusions of the HUVEC show similarity to structures identified on the surfaces of endothelial cells from human aortic walls (19), formation of such structures might be a general response of vascular EC to the presence of S. aureus. The specific bacterial stimulus responsible for the induction of the thread-like protrusions is unknown. Internalization of bound bacteria appeared to occur mostly via retraction of the thread-like protrusions into opened invaginations of the EC membrane. Cup-shaped internalization processes (10, 23) were observed, but very infrequently. Although it is known that internalization of S. aureus by HUVEC involves cytoskeletal rearrangements (2, 10, 19), the exact uptake mechanism, including the formation of the observed protrusions, remains to be elucidated.
Studies of the interaction of bacteria with vascular EC have mainly focused on the cellular responses, including surface receptor and cytokine expression (2, 4, 35, 36), and induction of apoptosis (21), whereas little attention has been paid to the bacterial responses in the interaction with EC. This study is the first report describing the identification of S. aureus genes whose expression was induced upon exposure of the bacteria to HUVEC, using a transposon-based expression library of S. aureus RN4220. There are some limitations to using a transposon-based system, including the possibility of disruption of a single gene or of an entire operon or bias for transposition. However, except for genes or operons that are essential for bacterial maintenance, induction of disrupted genes will still result in detectable β-galactosidase activity in the EC interaction model due to the selective activation of the promoter of the identified gene or its operon. Additionally, as Southern blotting has indicated randomness of transposon integration of Tn917-lacZ in S. aureus (Lammers et al., personal communication), this approach is suitable for the identification of inducible S. aureus genes and can give new insights into the bacterial response to the environment.
Most of the genes identified by our screening method are homologous to genes involved in amino acid and cell wall synthesis, transport of a number of (macro) molecules, and DNA repair and recombination. It is striking that none of the sequences found in our study was identified in a recent study of induced gene expression of S. aureus in an experimental murine abscess model using in vivo expression technology (IVET) (17). This suggests a specific response of S. aureus to stimuli from the in vitro HUVEC model. The similarity between our study and the IVET study was the inability to detect genes encoding classical virulence factors (e.g., adhesins or toxins). As expression of such factors is regulated (18), the corresponding genes must already be expressed when S. aureus is cultured on laboratory media.
The identified S. aureus genes were induced either in extracellular, cell-associated, or internalized bacteria. HUVEC and HS could act as stimuli for the up-regulation of expression of several S. aureus genes. The presence of HUVEC increased expression of an ask2 homolog, encoding an aspartate kinase involved in the conversion of l-aspartate to l-lysine. Interestingly, genes encoding three other enzymes of this biosynthesis route were also identified. This was not due to a lysine deficiency, as M199 contains sufficient lysine. Activation of the l-lysine biosynthetic pathway may have another function, as it can cause alterations in the bacterial cell wall. Such alterations may be beneficial or even required for bacterial survival in the presence of HUVEC, as is the case in in vivo infection models (8, 17). Expression of a putative ama gene, encoding an N-acyl-l-amino acid amidohydrolase and catalyzing the hydrolysis of N-alpha-acylated amino acids, was also increased due to the presence of HUVEC. The specific function of this protein in relation to the response to HUVEC is unknown.
Expression of oppD (clone 8-C2) and of the unknown sequence (clone 4-G8) was increased in the presence of HS but not of HUVEC. OppD is part of an oligopeptide ATP binding cassette transporter operon of S. aureus and encodes one of the ATP binding proteins of the transporter complex. Mutations in this operon strongly decreased the viability of S. aureus in different experimental infection models, including the rabbit endocarditis model (8), possibly due to defective import of peptides used by S. aureus for growth. The observed upregulation of OppD expression could be the result of the presence of such peptides in HS. Oligopeptide transporters are also involved in adherence to host cells, resistance to host defensins, and production of toxins (15, 24, 27). Further research is required to establish the possible role of OppD in the interaction of S. aureus with HUVEC.
Finally, expression of a gene encoding a putative DNA repair protein was increased in the presence of both serum and HUVEC, suggesting that in the bloodstream S. aureus is subject to increased environmental stress.
In conclusion, we have assessed the cross talk between HUVEC and S. aureus. The EC react to the bacteria by extending large, thread-like protrusions from their surfaces. We have shown that S. aureus reacts to HUVEC and to HS by activating genes potentially involved in survival and adaptation to the host. Further studies of the exact nature of the inducing signals and the role of the induced genes will elucidate the mechanism of S. aureus survival and growth in interaction with EC. These insights may contribute to an understanding of the pathogenesis of S. aureus IE in patients without prior heart disease.
ACKNOWLEDGMENTS
We thank Willem van Wamel for S. aureus RN4220 and Carla C. T. Hopman for technical assistance. We also thank Eelco Roos, Wim van Est, and Ton But for excellent photographical work and Martine J. van Vugt for critical reading of the manuscript.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E F, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Beekhuizen H, van de Gevel J S, Olsson B, van Benten I J, van Furth R. Infection of human vascular endothelial cells with Staphylococcus aureus induces hyperadhesiveness for human monocytes and granulocytes. J Immunol. 1997;158:774–782. [PubMed] [Google Scholar]
- 3.Beekhuizen H, van Furth R. Growth characteristics of cultured human macrovascular venous and arterial and microvascular endothelial cells. J Vasc Res. 1994;31:230–239. doi: 10.1159/000159048. [DOI] [PubMed] [Google Scholar]
- 4.Bengualid V, Hatcher V B, Diamond B, Blumberg E A, Lowy F D. Staphylococcus aureus infection of human endothelial cells potentiates Fc receptor expression. J Immunol. 1990;145:4279–4283. [PubMed] [Google Scholar]
- 5.Camilli A, Portnoy D A, Youngman P. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J Bacteriol. 1990;172:3738–3744. doi: 10.1128/jb.172.7.3738-3744.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper M D, Jeffery C, Gall D L, Anderson A S. Scanning electron microscopy studies of staphylococcal adherence to heart valve endothelial cells in organ culture: an in vitro model of acute endocarditis. Scan Electron Microsc. 1985;3:1231–1237. [PubMed] [Google Scholar]
- 7.Cornelis G R. Contact with eukaryotic cells: a new signal triggering bacterial gene expression. Trends Microbiol. 1997;5:43–45. doi: 10.1016/S0966-842X(96)30040-1. [DOI] [PubMed] [Google Scholar]
- 8.Coulter S N, Schwan W R, Ng E Y W, Langhorne M H, Ritchie H D, Westbrock-Wadman S, Hufnagle W O, Folger K R, Bayer A S, Stover C K. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol. 1998;30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 9.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamill R J, Vann J M, Proctor R A. Phagocytosis of Staphylococcus aureus by cultured bovine aortic endothelial cells: model for postadherence events in endovascular infections. Infect Immun. 1986;54:833–836. doi: 10.1128/iai.54.3.833-836.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hohn B. In vitro packaging of λ and cosmid DNA. Methods Enzymol. 1979;68:299–309. doi: 10.1016/0076-6879(79)68021-7. [DOI] [PubMed] [Google Scholar]
- 12.Johnson C M. Adherence events in the pathogenesis of infective endocarditis. Infect Dis Clin N Am. 1993;7:21–36. [PubMed] [Google Scholar]
- 13.Johnson C M, Helgeson S C. Platelet adherence to cardiac and noncardiac endothelial cells in culture: lack of a prostacyclin effect. J Lab Clin Med. 1988;112:372–379. [PubMed] [Google Scholar]
- 14.Kornblum J, Kreiswirth B N, Projan S J, Ross H, Novick R P. agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers, Inc.; 1990. pp. 373–402. [Google Scholar]
- 15.LeDeaux J R, Solomon J M, Grossman A D. Analysis of non-polar deletion mutations in the genes of the spo0K (opp) operon of Bacillus subtilis. FEMS Microbiol Lett. 1997;153:63–69. doi: 10.1111/j.1574-6968.1997.tb10464.x. [DOI] [PubMed] [Google Scholar]
- 16.Lee J C. Electrotransformation of staphylococci. In: Nickoloff J A, editor. Methods in molecular microbiology. Totowa, N.J: Humana Press Inc.; 1995. pp. 209–216. [DOI] [PubMed] [Google Scholar]
- 17.Lowe A M, Beattie D T, Deresiewicz R L. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol Microbiol. 1998;27:967–976. doi: 10.1046/j.1365-2958.1998.00741.x. [DOI] [PubMed] [Google Scholar]
- 18.Lowy F D. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 19.Lowy F D, Fant J, Higgins L L, Ogawa S K, Hatcher V B. Staphylococcus aureus-human endothelial cell interactions. J Ultrastruct Mol Struct Res. 1988;98:137–146. doi: 10.1016/s0889-1605(88)80906-6. [DOI] [PubMed] [Google Scholar]
- 20.McKinsey D S, Ratts T E, Bisno A L. Underlying cardiac lesions in adults with infective endocarditis. Am J Med. 1987;82:681–688. doi: 10.1016/0002-9343(87)90001-5. [DOI] [PubMed] [Google Scholar]
- 21.Menzies B E, Kourteva I. Internalization of Staphylococcus aureus by endothelial cells induces apoptosis. Infect Immun. 1998;66:5994–5998. doi: 10.1128/iai.66.12.5994-5998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B N, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogawa S K, Yurberg E R, Hatcher V B, Levitt M A, Lowy F D. Bacterial adherence to human endothelial cells in vitro. Infect Immun. 1985;50:218–224. doi: 10.1128/iai.50.1.218-224.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parra-Lopez C, Baer M T, Groisman E A. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 1993;12:4053–4062. doi: 10.1002/j.1460-2075.1993.tb06089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peters A. The fixation of central nervous tissue and analysis of electron micrographs, with special reference to the cerebral cortex. In: Nauta W J H, Ebbeson S O E, editors. Contemporary research methods in neuroanatomy. New York, N.Y: Springer; 1970. pp. 56–76. [Google Scholar]
- 26.Petterson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson K E, Wolf-Watz H. Modulation of virulence factor expression by pathogen target cell contact. Science. 1996;273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- 27.Podbielski A, Pohl B, Woischnik M, Korner C, Schmidt K H, Rozdzinski E. Molecular characterization of group A streptococcal (GAS) oligopeptide permease (opp) and its effect on cysteine protease production. Mol Microbiol. 1996;21:1087–1099. doi: 10.1046/j.1365-2958.1996.661421.x. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Schenk S, Laddaga R A. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;94:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 30.Selton-Suty C, Hoen B, Grentzinger A, Houplon P, Maignan M, Juilliere Y, Danchin N, Canton P, Cherrier F. Clinical and bacteriological characteristics of infective endocarditis in the elderly. Heart. 1997;77:260–263. doi: 10.1136/hrt.77.3.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tekstra J, Beekhuizen H, van de Gevel J S, van Benten I J, Tuk C W, Beelen R H J. Infection of human endothelial cells with Staphylococcus aureus induces the production of monocyte chemotactic protein-1 and monocyte chemotaxis. Clin Exp Immunol. 1999;117:489–495. doi: 10.1046/j.1365-2249.1999.01002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson R L. Staphylococcal infective endocarditis. Mayo Clin Proc. 1982;57:106–114. [PubMed] [Google Scholar]
- 33.Vann J M, Proctor R A. Ingestion of Staphylococcus aureus by bovine endothelial cells results in time- and inoculum-dependent damage to endothelial cell monolayers. Infect Immun. 1987;55:2155–2163. doi: 10.1128/iai.55.9.2155-2163.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vann J M, Proctor R A. Cytotoxic effects of ingested Staphylococcus aureus on bovine endothelial cells: role of S. aureus α-hemolysin. Microb Pathog. 1988;4:443–453. doi: 10.1016/0882-4010(88)90029-0. [DOI] [PubMed] [Google Scholar]
- 35.Yao L, Bengualid V, Lowy F D, Gibbons J J, Hatcher V B, Berman J W. Internalization of Staphylococcus aureus by endothelial cells induces cytokine gene expression. Infect Immun. 1995;63:1835–1839. doi: 10.1128/iai.63.5.1835-1839.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao L, Lowy F D, Berman J W. Interleukin-8 gene expression in Staphylococcus aureus-infected endothelial cells. Infect Immun. 1996;64:3407–3409. doi: 10.1128/iai.64.8.3407-3409.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J P, Normark S. Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science. 1996;273:1234–1236. doi: 10.1126/science.273.5279.1234. [DOI] [PubMed] [Google Scholar]