SUMMARY
The ATP-dependent ring-shaped chaperonin TRiC/CCT is essential for cellular proteostasis. To uncover why some eukaryotic proteins can only fold with TRiC assistance, we reconstituted the folding of β-tubulin using human Prefoldin and TRiC. We find unstructured β-tubulin is delivered by Prefoldin to the open TRiC chamber followed by ATP-dependent chamber closure. CryoEM resolves four near-atomic resolution structures containing progressively folded β-tubulin intermediates within the closed TRiC chamber, culminating in native tubulin. This substrate folding pathway appears closely guided by site-specific interactions with conserved regions in the TRiC chamber. Initial electrostatic interactions between the TRiC interior wall and both the folded tubulin N-domain and its C-terminal E-hook tail establish the native substrate topology, thus enabling C-domain folding. Intrinsically disordered CCT C-termini within the chamber promote subsequent folding of Tubulin’s Core and Middle domains and GTP-binding. Thus, TRiC’s chamber provides chemical and topological directives that shape the folding landscape of its obligate substrates.
Keywords: Chaperonin, Chaperone, TRiC/CCT, Prefoldin, Tubulin, CryoEM, XL-MS
Graphical Abstract
In brief eTOC
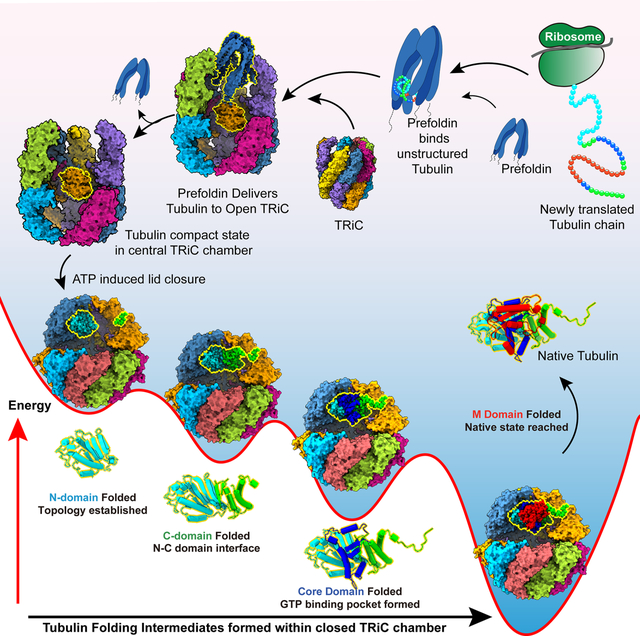
Starting from unstructured tubulin bound to the chaperone Prefoldin, the steps of TriC-guided tubulin folding are revealed in structural detail illuminating the folding mechanism.
INTRODUCTION
How proteins fold in vivo is a central question in biology, highly relevant to human health, as many diseases ranging from cancer to neurodegeneration, are linked to protein misfolding1–5. Several classes of molecular chaperones play central roles in the folding of newly translated polypeptides6–8. While some chaperones, such as NAC or Hsp70 facilitate folding of a wide range of proteins, others, such as the ring-shaped chaperonin TRiC/CCT facilitate folding of a subset of proteins comprising ~10% of the eukaryotic proteome9. The proteins that require TRiC for folding, many of them essential for viability, are distinguished by their aggregation propensity and topological complexity9,10. In vivo, TRiC functions in cooperation with upstream chaperones, including Hsp70 and Prefoldin (PFD, also called GIMc). Prefoldin binds directly to TRiC through an electrostatic pivot, suggesting a direct mechanism of substrate transfer11.
How TRiC/CCT assists folding is intriguing because several eukaryotic proteins can only fold with the assistance of TRiC. These proteins cannot fold either spontaneously nor with any other chaperone, under any known condition12,13. An explanation for this obligate requirement is lacking. An intriguing hypothesis is that the information present in their amino acid sequence does not suffice to reach the native state. In this hypothesis, TRiC may contribute the chemical and topological environment necessary to guide their folding, suggesting a co-evolution between TRiC and some eukaryotic proteins.
TRiC is a ~1 MDa chaperonin, consisting of two identical rings stacked back-to-back14. Unlike bacterial or archaeal chaperonins, each TRiC ring is assembled from eight different subunits, called CCT1-CCT8, in an evolutionarily conserved arrangement15. Access to the central chamber of each ring is regulated in an ATP-dependent manner by a built-in lid, formed by flexible loops protruding from each subunit. The lid is open in the unliganded apo-state of TRiC, allowing substrates to bind through subunit-specific contacts in the apical domains12,16. ATP hydrolysis leads to lid closure, encapsulating the substrate within the central chamber, where folding occurs17,18.
The hetero-oligomeric nature of TRiC is likely key to its unique ability to fold obligate substrates. Subunit divergence likely contributes to the folding process in various ways. The specific subunit arrangement segregates substrate and ATP binding properties within the ring and produces an asymmetric charge distribution within the closed chamber15,18. Unfolded substrates such as actin, AML or VHL bind open TRiC through subunit specific contacts with CCT apical domains12,19–21. This could bias the initial topology of the TRiC-bound substrate to a conformation that is productive for folding. Analysis of TRiC-mediated actin folding indicates that the gradient of ATP affinities within the ring promotes stepwise release during chamber closure to facilitate folding12,18. However, how folding occurs inside the closed TRiC folding chamber is not understood. Is the bound substrate released into the ATP-closed chaperonin chamber, as shown for bacterial chaperonin GroEL-ES? How do the properties of the TRiC chamber affect the folding pathway of its substrates?
To address these questions, we reconstituted and analysed the Prefoldin-TRiC mediated folding pathway of the archetype TRiC obligate substrates α- and β-tubulin. Prefoldin maintains tubulin in a highly unstructured state and directly transfers substrate into the TRiC chamber. ATP addition then leads to lid closure and tubulin encapsulation and folding within the closed TRiC chamber. Biochemical and cryogenic electron microscopy (cryoEM) structural analyses of the chaperone-mediated folding reaction reveal highly specific substrate contacts with the inner chamber of TRiC. These electrostatic and polar tubulin contacts with individual CCT subunits direct the tubulin folding trajectory by promoting domain-wise formation of four progressively folded intermediates culminating in the native state.
RESULTS
Reconstituting the human Prefoldin-TRiC pathway for efficient tubulin folding
α- or β-tubulin monomers are folded in vivo by a chaperone pathway consisting of Prefoldin and TRiC/CCT22,23 and subsequently steered into tubulin dimers by a complex network of poorly understood assembly factors24,25. We reconstituted the Prefoldin-TRiC-tubulin monomer folding pathway using purified human components (Figure 1A, S1A–C). Folding reactions were performed starting from Prefoldin-bound α- and β-tubulin complexes (Figure S1C) that were incubated with purified human TRiC in the presence and absence of ATP. Folding reactions were analyzed using Native PAGE (N-PAGE) to assess formation of tubulin chaperone complexes during the folding reaction while limited-proteolysis assays followed by gel electrophoresis (LIP-PAGE) were used to examine the conformation of chaperone-bound tubulin (Figure 1B, C). N-PAGE revealed that TRiC addition to Prefoldin:α-tubulin or Prefoldin:β-tubulin complexes led to formation of a ternary complex with TRiC, reflected in a characteristic mobility shift (Figure 1B for β-tubulin and S1D for α-tubulin)11. Subsequent ATP addition led to production of folded protease-resistant tubulin monomer (Figure 1C, note GTP is added to stabilized folded tubulin). To further assess whether TRiC promotes ATP-dependent tubulin folding, we incubated these reactions with two factors known to bind to either α- or β-tubulin monomers, called TBCB and TBCA respectively22–25. N-PAGE revealed that, in the presence of ATP, TRiC yielded tubulin competent to form a characteristic complex with these monomer-binding factors (Figure 1B; S1D).
Figure 1: Reconstitution of the Tubulin folding pathway from purified components.
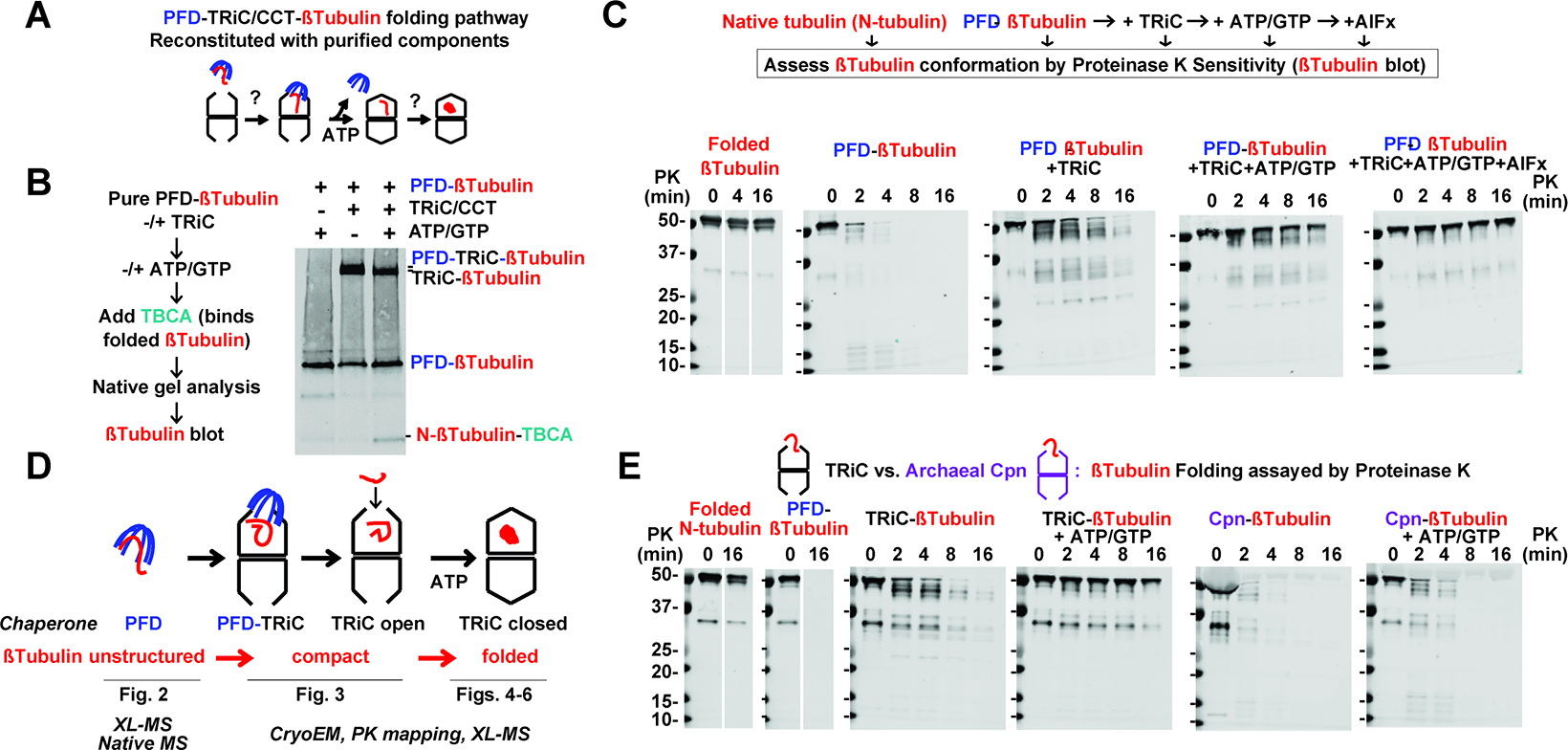
(A) Prefoldin (PFD) and TRiC mediated Tubulin folding pathway reconstituted from purified components. (B) N-PAGE analysis of β-tubulin interactions with Prefoldin and TRiC. Addition of nucleotide (ATP/GTP) drives folding and partial binding of β-tubulin to TBCA. (C) Proteinase K sensitivity assay (PK) of Tubulin conformation along the Prefoldin-TRiC folding pathway. (D) Tubulin conformation along the chaperone folding pathway was further analysed as indicated. (E) PK assay of β-tubulin copurified with either TRiC or group II archaeal chaperonin CPN with or without ATP/GTP addition.
Susceptibility to low concentrations of non-specific protease Proteinase K (PK) is a hallmark of unstructured polypeptides, while folded domains tend to exhibit enhanced PK resistance26,27. We used LIP-PAGE with PK to characterize the conformation of α- and β-tubulin folding intermediates along the Prefoldin:TRiC chaperone pathway (LIP-PAGE, Figure 1C, S1E). Similar results were obtained for α- and β-tubulins suggesting a shared mechanism of chaperone action for these proteins (Figure S1D, E). As expected, folded tubulin in α/β dimers is nearly fully PK resistant, except for some degradation of flexible C-terminal tails. In contrast, both α- and β-tubulin bound to Prefoldin were highly PK sensitive, indicating the bound substrate adopts an unstructured protease-sensitive conformation. Addition of TRiC and ensuing formation of a ternary Prefoldin:tubulin:TRiC complex, led to enhanced α- and β-tubulin protection against PK, suggesting tubulin becomes compacted upon binding to TRiC. We reached similar conclusions using an antibody directed against the N-terminal domain of tubulin or one directed against the C-terminus of tubulin (Figure S1G–I). Interestingly, probing with the N-terminal directed antibody but not the C-terminal directed antibody also detected smaller protected protease fragments, suggesting TRiC promoted formation of partially folded N-terminal domain structures. We next incubated the Prefoldin:tubulin:TRiC complex with ATP, which promotes TRiC chamber closure and Prefoldin dissociation11. ATP incubation led to formation of full-length protease protected tubulin, with a similar level of protection as that observed for folded α- and β-tubulins in the native dimer. Since in the presence of ATP, the TRiC lid alternates between open and closed states, we examined if the increased protease protection was due to tubulin folding or to obstruction by the closed lid. First, we extended the time course of PK incubation to 16 min, which should give ample time to PK to access the substrate during the open states28,29. Second, TRiC-tubulin was incubated with ATP to promote folding, followed by subsequent addition of apyrase for 30 or 90 min to hydrolyze all ATP, causing the TRiC lid to reopen11. Under these conditions, tubulin remained fully protease protected, confirming the ATP-induced protease-protection arises from tubulin reaching the folded conformation (Figure S1G, I).
To confirm the TRiC dependence of β-tubulin folding, we incubated Prefoldin:β-tubulin with archaeal M. maripaludis chaperonin, Mm-Cpn29. Despite its structural and mechanistic similarity to TRiC29, incubating the tubulin-Mm-Cpn complex with ATP/GTP does not yield PK-protection for tubulin, but instead produces a digestion pattern similar to that of Prefoldin:β-tubulin alone (Figure 1E). This experiment indicates tubulin cannot fold when encapsulated by a non-TRiC group II chaperonin. It also confirmed tubulin protease protection does not derive from a lack of PK accessibility upon substrate enclosure in a folding chamber. These experiments suggest Prefoldin and TRiC differentially affect the conformation of the substrate polypeptide during ATP-dependent tubulin folding (Figure 1D). Prefoldin maintains tubulin in a protease-sensitive unstructured state, with no obvious protease-protected fragments observed. Upon Prefoldin transfer to TRiC, tubulin becomes compacted, acquiring some protease-resistance, including formation of N-terminal domain structures. Finally, ATP-driven encapsulation leads to a fully protease-resistant tubulin that can bind to tubulin monomer binding factors. Since locking the TRiC chamber into a closed conformation by the addition of ATP-AlFx, also led to strong protection from proteolysis, we conclude the substrate is fully encapsulated within the central chamber.
We next examined folding when tubulin is directly bound to TRiC and the Prefold-indelivery step is bypassed (Figure 1E). The LIP-PAGE assay indicated tubulin adopts the same compact intermediate upon TRiC binding, regardless whether tubulin is copurified with TRiC or delivered via Prefoldin; furthermore ATP/GTP addition to TRiC-tubulin also yielded a fully protease protected conformation (Figure 1E). Thus, TRiC is primarily responsible for tubulin folding, while Prefoldin functions to maintain tubulin in an unstructured state, likely to prevent formation of off-pathway trapped intermediates. We set out to study each stage of the reconstituted Prefoldin:TRiC mediated tubulin folding pathway using a hybrid structural and biochemical approach (Figure 1D).
Defining Prefoldin interactions with non-native tubulin
Prefoldin is a jellyfish-shaped complex with six different subunits forming a double beta-sheet barrel from which coiled-coils extend forming “tentacles” (Figure 2A). The flexible nature and relatively small size of Prefoldin:tubulin hinders its structural characterization by cryoEM. Accordingly, we mapped the Prefoldin:tubulin contacts using cross-linking-mass spectrometry (XL-MS), with similar results obtained for α- and β-tubulins (Figure 2A, B; Figure S2A). Substrate crosslinks (XL) mapped primarily to the coiled coil tentacles and the terminal disordered tails of Prefoldin, indicating non-native tubulin binds inside the Prefoldin “chamber” (Figure 2B; S2B). Structural mapping of the Prefoldin:tubulin XL contact information suggests substrate binds Prefoldin through highly heterogeneous interactions within the chamber (Figure 2C, D). The N-terminal domain of tubulin made numerous contacts throughout the Prefoldin inner chamber, with most XL mapping to contiguous Prefoldin 1/5/6 coiled coils. In contrast, the tubulin core domain was crosslinked primarily to one specific region in Prefoldin 6. The C-terminal domain of tubulin also crosslinked to a continuous patch on Prefoldin 3/5/6, while the M domain crosslinked to several points at the bottom of the Prefoldin chamber. These analyses, together with the high protease sensitivity of Prefoldin-bound tubulin, suggest tubulin populates a highly dynamic conformational ensemble, even though some regions of tubulin bind more stably to specific regions within the Prefoldin chamber.
Figure 2: Defining Prefoldin interactions with non-native tubulin.
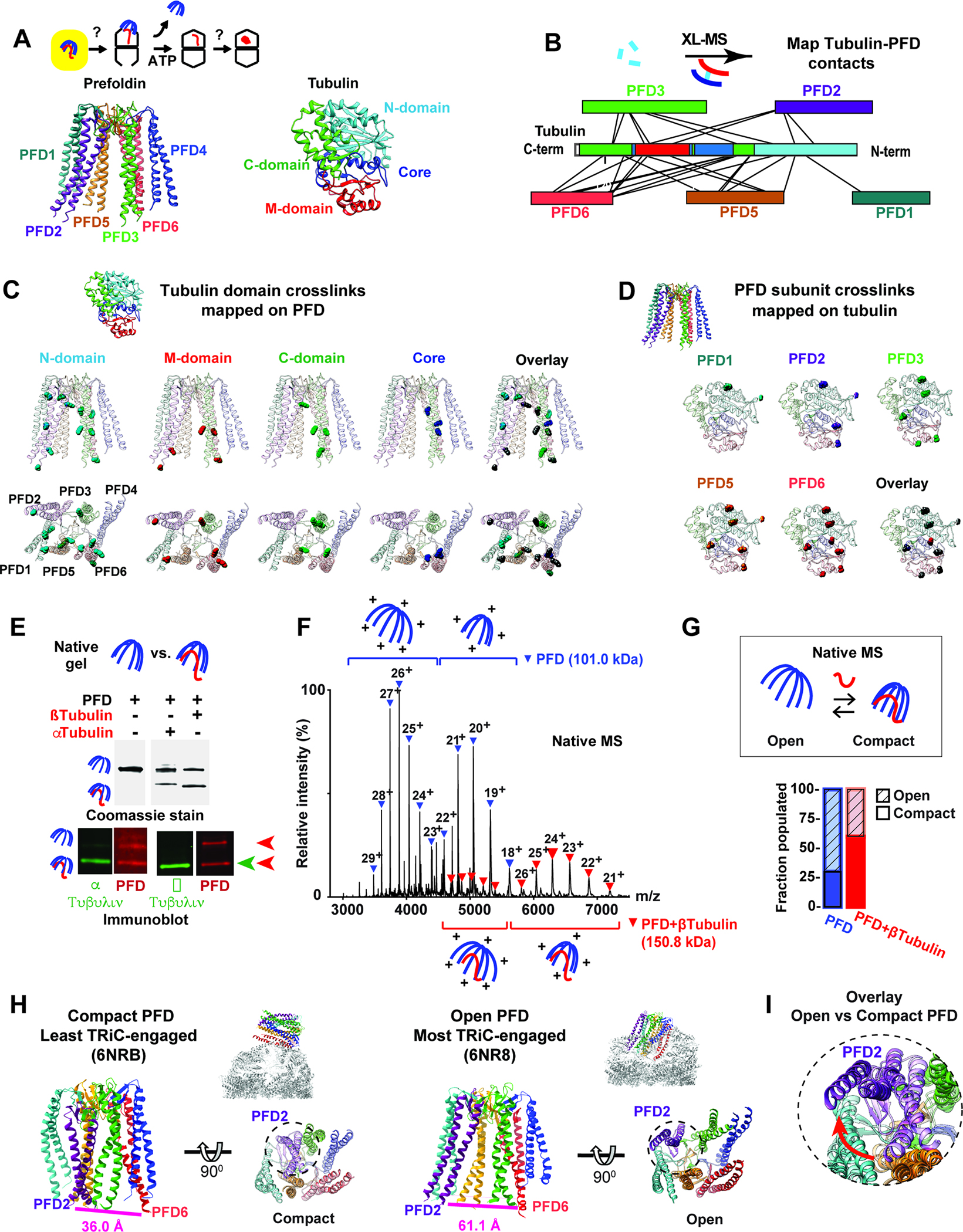
(A) Role of Prefoldin:tubulin within folding pathway highlighting color scheme for Prefoldin subunits and tubulin domains. (B) XL-MS sites for Prefoldin-bound α- and β-tubulin mapped onto linear bar diagrams of Prefoldin subunits and tubulin domain. (C) All tubulin XL sites mapped onto Prefoldin structure, note XL to unstructured regions of Prefoldin tentacles are mapped to the last resolved residue. (D) XL sites mapped onto the Tubulin structure. (E) N-PAGE followed by immunoblot analyses for Prefoldin with or without bound Tubulin. (F) Representative native MS of Prefoldin with or without bound β-tubulin. Charge state distributions are inferred to “open” or “compact” conformations. (G) Quantification of the fractions of Prefoldin and Prefoldin:β-tubulin populating open and compact conformations by native MS. (H) Structures of Prefoldin from the least engaged (PDB:6NRB11) and most engaged (PDB:6NR811) states with TRiC with distances between the terminal residues of PFD2 (Ile 124) and PFD6 (Glu 114) indicated. (I) Overlay of Prefoldin structures focusing on PFD2 (purple) movement in least engaged (light color) and most engaged (dark color) states.
Prefoldin is an ATP-independent chaperone, raising the question of how substrate binding and release are regulated. Surprisingly, while binding either α- or β-tubulin to Prefoldin adds an additional molecular mass of ~50 kDa to the 101 kDa Prefoldin complex, the Prefoldin-substrate complexes migrated faster on N-PAGE than Prefoldin alone (Figure 2E). We hypothesized substrate binding to Prefoldin induces a conformational compaction. Native mass spectrometry analyses support this idea (Figure 2F, G, S2C, D). Prefoldin alone produced a bimodal charge distribution for the single expected Prefoldin mass (101 kDa) (Figure S2C, D). Because the charge states produced during gentle ionization are strongly dependent on the exposed surface area30–33, these data indicate pure Prefoldin adopts two distinct structural states: one more compact and one more extended. Analysis of Prefoldin:β-tubulin complexes by native mass spectrometry also yielded a bimodal charge series consistent with the expected mass of the binary Prefoldin:β-tubulin complex (151 kDa) (Figure 2F, S2E). Of note, the relative intensity, and therefore abundance, of the compact species was higher in Prefoldin:β-tubulin peaks compared to Prefoldin peaks (Figure 2E–G, S2E). The conclusion that tubulin binding induces a shift to the more compact Prefoldin conformation supports the N-PAGE analyses as well as our finding of extensive XL between tubulin and the tentacles of PFD2 and PFD6, located at opposite sides of the Prefoldin chamber. A possible function for this compaction arises from cryoEM structural analyses of the Prefoldin-TRiC interaction, which found a flexible contact point between the base of Prefoldin and the apical domain of CCT4 in TRiC that allows Prefoldin to pivot into the chamber11. Prefoldin adopts a compact conformation in the less engaged TRiC contact, where the two substrate binding chambers are not aligned, but shifts to an extended conformation when the Prefoldin-TRiC chambers become aligned allowing for substrate transfer (Figure 2H, I; S2F, G). This suggests Prefoldin initially binds TRiC in the compact, substrate-bound conformation but then TRiC interactions promote the extended Prefoldin state to facilitate substrate release into the chaperonin chamber.
The ternary complex of Prefoldin:β-tubulin:TRiC in the open conformation
To visualize the substrate handoff from Prefoldin to TRiC, we added Prefoldin:β-tubulin to purified TRiC and studied the ternary complex by cryoEM (Figure 3A; S3A–D; Table S1). The Prefoldin:β-tubulin:TRiC complex showed the chamber of Prefoldin fully aligned with that of TRiC through subunit specific contacts (Figure 3B). We observed a significant extra density inside the inter-ring space formed at the TRiC equatorial region not observed in the absence of substrate (Figure 3B–D)11. The Prefoldin tentacles extend into the TRiC chamber. Strikingly, the PFD6 coiled-coil made a clear connection to the substrate-dependent density in the inter-ring space (Figure 3C). The Prefoldin in this TRiC complex is in the open extended conformation, suggesting TRiC binding reduces its affinity for the bound substrate. Indeed, no significant substrate density is observed inside the Prefoldin chamber, despite the substantial extra density observed between the two TRiC rings. The oval-shaped substrate-induced extra-density is asymmetrically located within the inter-ring TRiC chamber, close to subunits CCT8, 6, and 3. Of note, we also observe clear connections between this density and the N-terminal tails of CCT5 and 7, which are normally not resolved in open TRiC structures (Figure 3D).
Figure 3: Ternary Prefoldin:β-tubulin:TRiC complex in the open conformation.
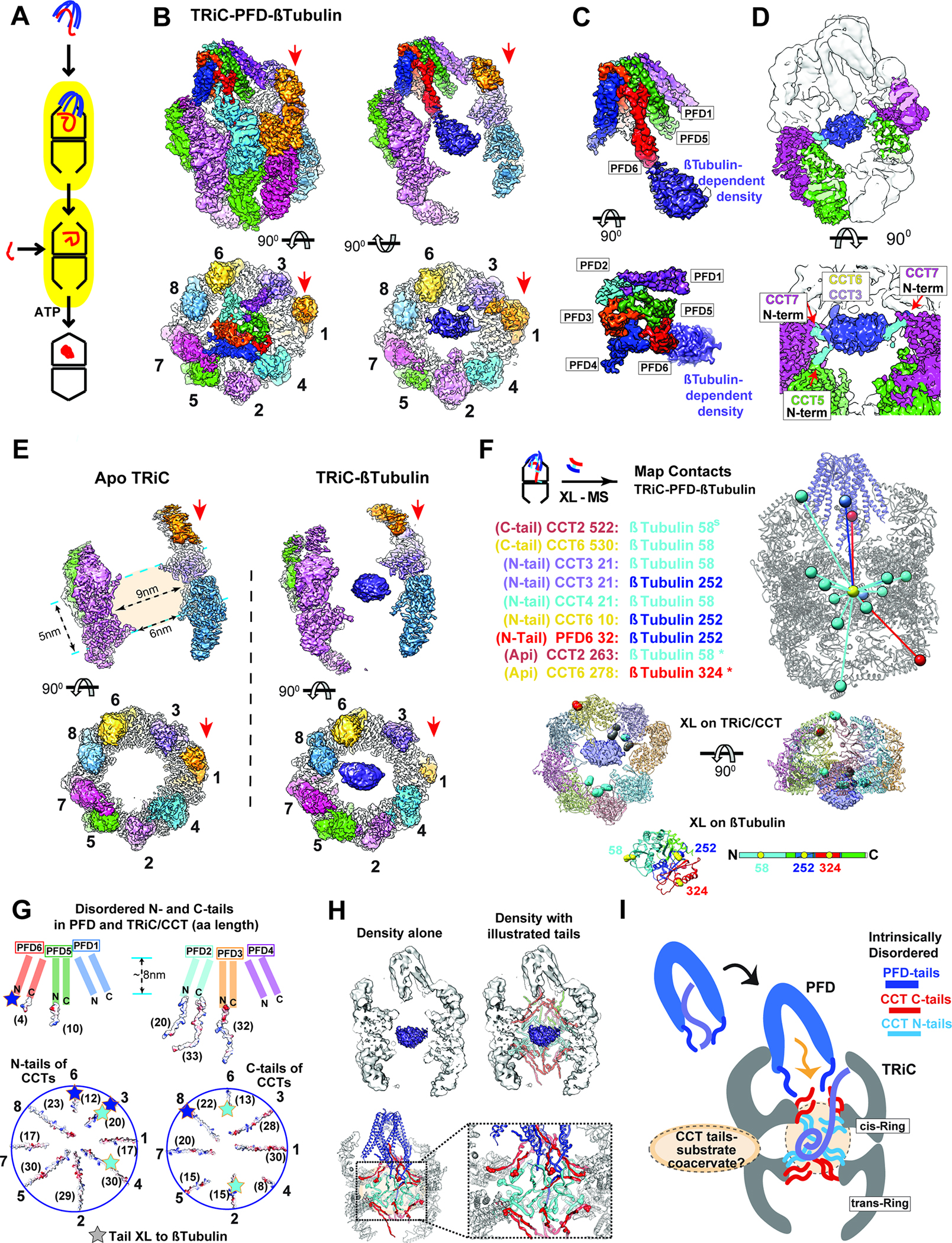
(A) Model of substrate loading to TRiC. (B-D) 3D-reconstruction of TRiC-Prefoldin:β-tubulin ternary complex, (B) Side and end-on view of the ternary complex, and slice views showing β-tubulin density bound to TRiC. Red arrows indicate CCT1. (C) Side and end-on views segmented Prefoldin:β-tubulin density. (D) Side and top view at lower threshold to illustrate N-terminal tail contacts from CCT5 and CCT7. (E) Side and top views from 3D-reconstruction of apo-TRiC and TRiC:β-tubulin highlighting the β-tubulin density is absent from apo-TRiC. Red arrows indicate CCT1. (F) TRiC:β-tubulin inter-molecular XL mapped onto the model, note XL are mapped to the center of mass of the density and colored by the domain of Tubulin they XL to. Residues that XL to β-tubulin are highlighted on a single ring of TRiC and colored by the domain of β-tubulin they XL to, residues that XL to multiple domains are colored grey. XL between β-tubulin and TRiC apical domains (*) from previous studies16,58. (G) Schematic of disordered tail residues from the N and C termini of Prefoldin (top) and TRiC (bottom) highlighting their electrostatic surface. Brackets denote the length (as number of residues) of each subunit’s disordered tail. Star: indicates residues within the disordered tails that XL to β-tubulin domains. (H) Schematic of disordered tails in the TRiC inter-chamber space with map (top) and model (bottom). Spheres in the model indicates the location of the first resolved N-terminal (cyan) and the last resolved C-terminal residues (red). (I) Cartoon of substrate delivery by the interplay of TRiC and Prefoldin’s disordered tails. The dotted circle in the TRiC chamber represents a hypothetical coacervate formed by substrate polypeptide and disordered CCT tails.
To further assess the origin of the extra density, we prepared a TRiC:β-tubulin complex generated without Prefoldin and carried out cryoEM analyses of both substrate-free apo-TRiC and TRiC:β-tubulin. Only the β-tubulin-containing TRiC has density in the central inter-ring region, while no notable density is observed in substrate-free apo-TRiC (Figure 3E). The substrate-dependent density is located within an inter-ring chamber created by the equatorial domains of CCTs protruding into the central axis, forming a ~6 nm open cap covering a ~9×5 nm globular inner space (Figure 3E). However, extensive focused 3D classification on the substrate-dependent extra-density failed to discern any folded domain or secondary structure. While the presence of substrate-dependent density indicates the formation of a compact state, this structural uncertainty suggests heterogeneity in either the folding state or the positioning of β-tubulin within this region of the open TriC chamber.
We used XL-MS to further characterize the contacts of β-tubulin with Prefoldin and TRiC within the ternary complex and in the binary TRiC:β-tubulin complex (Figure 3E; S2A and Table S2). TRiC addition to Prefoldin:β-tubulin led to a near complete loss of β-tubulin’s XL to Prefoldin and to the appearance of extensive XL to TRiC, consistent with substrate transfer between these chaperones (Figure 3F). The ternary complex only had one β-tubulin XL to PFD1 and one β-tubulin XL to the N-terminus of PFD6 (Figure 3E). We also observed multiple PFD6 XL to the tails of CCT1 and CCT2 in the equatorial region of TRiC (Figure S3E, F). Together, these XL-MS observations support the structural data showing PFD6 inserting into the TRiC chamber placing the tip of the PFD6 tentacle proximal to the substrate-dependent density. Importantly, XL-MS also indicated extensive contacts between β-tubulin and the N- and C-terminal tails extending from the equatorial domains of CCT subunits into the chamber (Figure 3F, G). These XL-MS findings also agree with the cryoEM maps showing the substrate localizes to the inter-ring chamber region in close proximity to the disordered N- and C-terminal CCT tails. XL between β-tubulin and the CCT apical domains were observed in the TRiC β-tubulin complex generated without Prefoldin but not in the Prefoldin:β-tubulin:TRiC ternary sample16,34. We did not observe notable substrate density at the similar contour level in the apical domain regions of TRiC. Perhaps the substrate interactions with apical domains are too dynamic to be clearly observed, or alternatively apical contacts serve in the initial capture of substrate for subsequent engagement with the CCT tails. The ternary complex with Prefoldin may thus reflect the substrate state after transfer to the inter-ring chamber.
While TRiC has been considered as a two-chamber folding system, our data suggests the existence of a third chamber that holds the substrate in open TRiC. Theoretically, the third inter-ring chamber provides enough space for ~50 kDa protein, but it is most likely that both the substrate and the CCT-tails contribute to the observed substrate-dependent density in our maps. Both TRiC and Prefoldin contain long intrinsically disordered N- and C-termini tails; the CCT tails projecting into this chamber comprise ~35.5 kDa of protein mass (schematized in Figure 3G). The observed substrate-dependent density within TRiC, together with the increased substrate protection from PK, both indicate the substrate becomes compacted upon TRiC binding, without leading to any stable domain formation. The unstructured CCT tails contacting β-tubulin in this region of the open inter-ring TRiC chamber contain both polar and hydrophobic character (Figure 3G–H). We speculate that these disordered tails function as a tethered solvent that maintains the unfolded polypeptide in a conformationally dynamic coacervate state, thus preventing the formation of trapped intermediates. This may stabilize β-tubulin folding intermediates in a folding-competent state by interactions with CCT tails in the open chaperonin conformation (Figure 3H, I).
CryoEM identifies β-tubulin folding intermediates within the ATP-closed TRiC chamber
To examine the next step of the β-tubulin folding cycle (Figure 4A), we incubated the TRiC:β-tubulin complex with ATP-AlFx to stabilize the closed conformation and examined β-tubulin folding in the chamber by cryoEM. Following incubation with ATP-AlFx, we observed open and closed TRiC states containing β-tubulin dependent density (Figure 4A). As expected, the 3.8 Å open TRiC state contains the β-tubulin-dependent density in the inter-ring space (Figure 4; S4A, B; Table S1). In contrast, the closed TRiC conformation contains the substrate density in the apical region within one of the chambers (Figure 4A right panel). The repositioned β-tubulin in the 2.8 Å resolution closed TRiC density map had well-defined secondary structural element features (Figure 4A). Additionally, most of the CCT N- and C-terminal tails could be traced in the closed conformation but not in the open conformation.
Figure 4: CryoEM identifies β-tubulin folding intermediates in ATP-closed TRiC chamber.
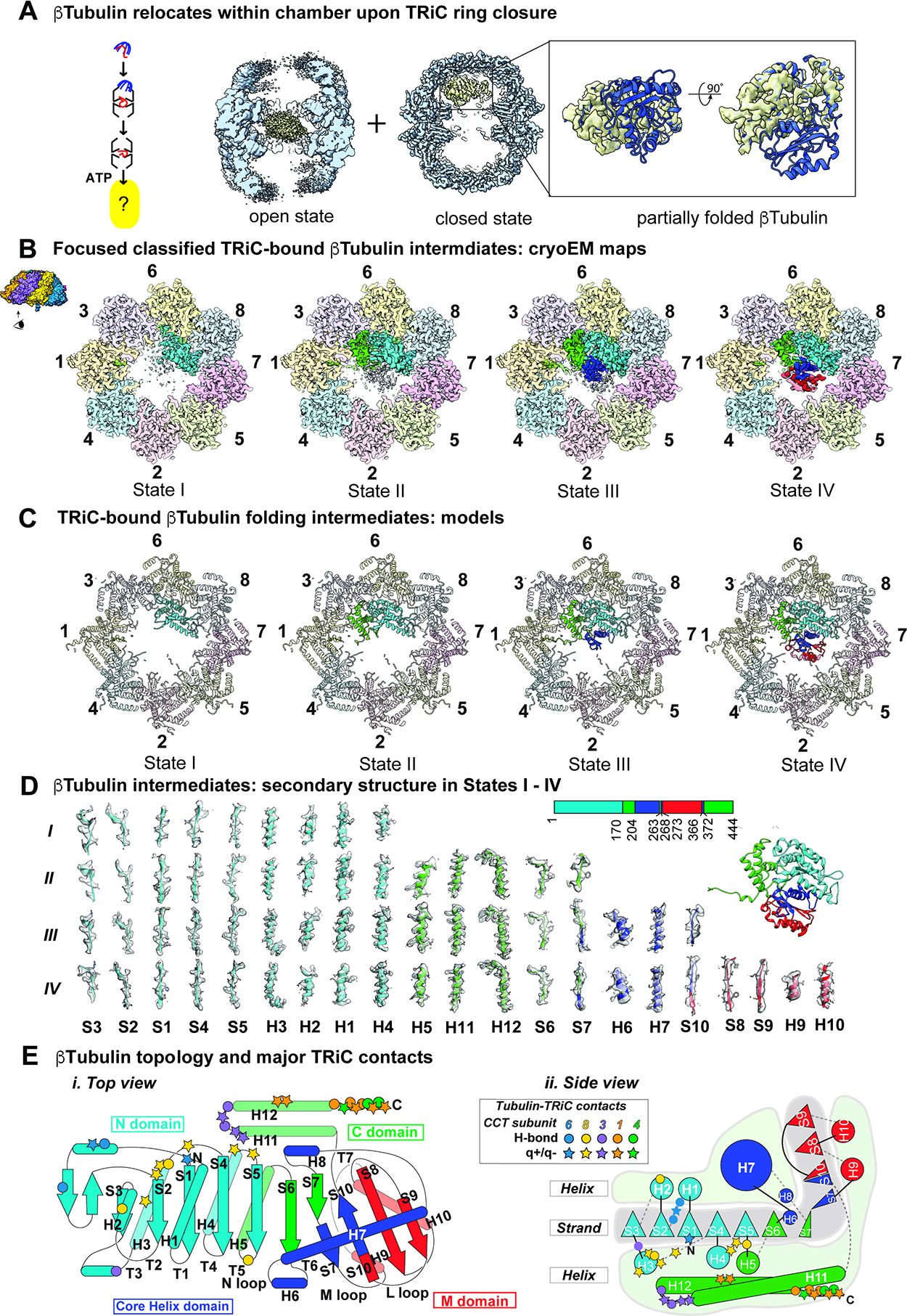
(A) CryoEM consensus maps of open and closed TRiC:β-tubulin indicates dramatic repositioning of β-tubulin density. (B) Focused classification on β-tubulin in the closed state identified four distinct states I-IV that could be separated by the amount of progressively folded β-tubulin relative to the final state of β-tubulin. (C) Atomic models of TRiC bound β-tubulin folding intermediates. (D) Local resolvability of secondary structure elements in β-tubulin folding intermediates reveals progressive formation of folded domains. (E) Cartoon illustration of β-tubulin topology highlighting major contacts with TRiC subunits.
In the initial overall map of closed TRiC:β-tubulin we visualized partially folded β-tubulin with varying resolution, suggesting compositional and conformational heterogeneity. 3D classification focusing on the β-tubulin density revealed four distinct conformations of β-tubulin with structurally resolvable domains. The TRiC:β-tubulin conformations were reconstructed to resolutions ranging from 2.9 Å to 3.6 Å (Figure 4B, S4C–E) allowing us to build models of each TRiC bound state (Figure 4C). In all four maps, the resolvable densities corresponded to native tubulin domains with well resolved α-helices and β-strands (Figure 4B, C; Movie S1). The four β-tubulin structures are aligned along an axis of increased domain formation, suggesting β-tubulin is progressively folded in the TRiC chamber. We also observed clear density for the negatively charged C-terminal β-tubulin “E-hook” tail (Figure 4D, S5A), which is flexible in native tubulin. To assess the resolvability of our densities, we used a per-residue Q-score profile in a sliding window of 10 residues35 (Figure S4F). In agreement with their estimated resolution, we obtained consistently high Q-scores for the folded regions demonstrating the high resolvability of structural elements. In contrast, the poorly resolved segments have low Q-scores, suggestive of unfolded or conformationally dynamic regions. These conformations, designated States I–IV, reveal progressively folded β-tubulin inside closed TRiC, with State IV corresponding to fully folded β-tubulin (Movie S1).
The well-resolved residues in states I-IV map to specific domains formed by discontinuous sequences suggestive of domain-wise folding of β-tubulin (Figure 4D, 4E). The least folded intermediate, State I, contains the folded N-terminal β-tubulin domain (residues 1–170) (Fig. 4E) with the exposed surface contacting the inner wall of CCT6-CCT8 (Fig. 4C) and the C-terminal β-tubulin E-Hook (residue 438–444) bound to a pocket between CCT1 and CCT4 (Fig. S5A). State II contains the resolved regions of State I as well as resolvable discontinuous sequence elements corresponding to the folded C-terminal domain (C-domain) of β-tubulin, spanning residues 171–203; 263–267 and C-terminal helices 372–426 (Fig. 4D, 4E). The folded C-terminal domain also binds the inner wall of TRiC, contacting residues of subunits CCT1 and CCT3 (Fig. 4C). State III also contained the folded N- and C-terminal domains observed in State II, as well as the discontinuous resolvable sequence elements corresponding to the folded helical Core-domain, spanning residues 204–262, 268–272 and 366–371 (Fig. 4D, 4E). Finally, State IV resembles State III but also contains the folded middle M-domain, composed of residues 273–365. Folding of this last M-domain completes formation of a fully native β-tubulin monomer (Fig. 4D, 4E). Though the TRiC-bound β-tubulin states exhibit C-alpha backbone variation in certain loop regions, the folded secondary structural elements in all four states are almost identical to the structure of native β-tubulin (PDB 6I2I36) (Figure S4G). The C-terminal E-hook tail of β-tubulin was clearly traceable in all four states nestled in a pocket formed by CCT1 and CCT4 (Figure S5A). These four structural states of β-tubulin within the chamber of TRiC suggest a sequential folding pathway where discontinuous sequence elements fold into specific native-like domains in close association with the inner chaperonin chamber, culminating in the native state (Figure 4E).
Domain specific CCT contacts spatially orient and restrain β-tubulin within the TRiC chamber
Upon ATP-driven encapsulation the inner TRiC wall engages in domain specific contacts with all four β-tubulin intermediates. The extent of electrostatic interactions with the inner TRiC chamber increases from the less folded to more folded β-tubulin intermediates (Figure 5B, Movie S2). This suggests a different mechanism to bacterial chaperonin GroEL-ES, which promotes substrate folding by releasing encapsulated polypeptides into their central hydrophilic chamber2, often termed an “Anfinsen cage”37.
Figure 5: TRiC and β-tubulin interaction in a domain specific manner.
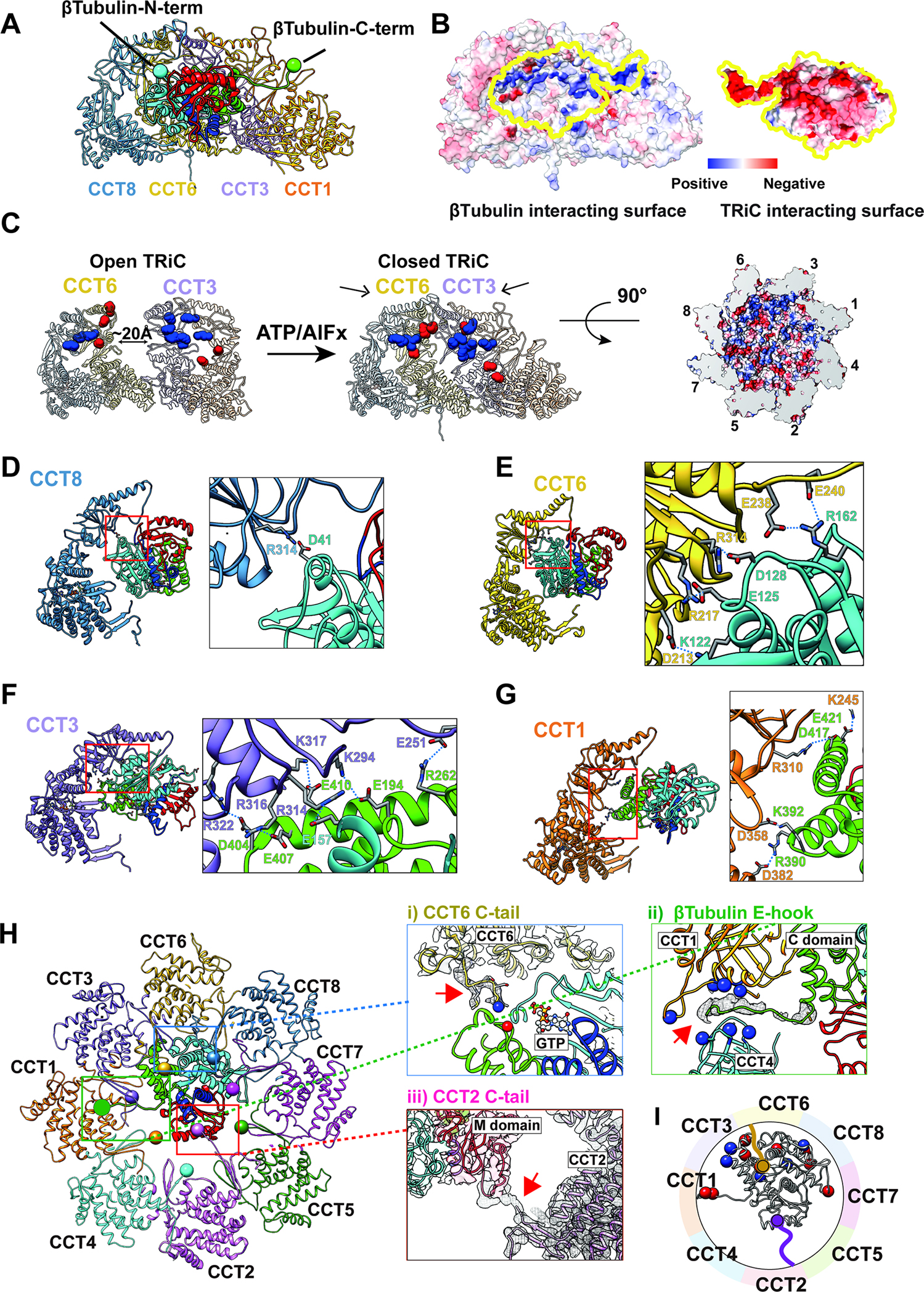
(A) Side view of a ribbon diagram of β-tubulin and the CCT subunits (1,3,6,8) that have a large contact interface. The β-tubulin N- and C-termini are indicated as a cyan and green ball, respectively. (B) The surface electrostatic distribution of the interacting surfaces of CCT (1,3,6,8) and β-tubulin. (C) The formation of the electrostatic patch inner chamber of CCT (1,3,6,8) induced by the ring closure and the asymmetric charge distribution. (D-G) Side view of CCT (1,3,6,8) and β-tubulin. β-tubulin is colored by progressively folded domains. Residues making salt bridges are labeled and side chains are displayed with contact shown as a dotted line. (H) Specific tail contacts between TRiC and β-tubulin. Magnified views are displayed for (i) the CCT6 C-tail and β-tubulin GTP binding pocket, (ii) β-tubulin CCT1/4 binding pocket and β-tubulin C-terminal E-hook tail, and (iii) CCT2 C-tail and β-tubulin M-domain. (I) Schematic of β-tubulin, negatively charged residues (red) and positively charged residues (blue) that contact TRiC are indicated by spheres, the CCT2 (purple) and CCT6 (gold) C-terminal tails are also displayed with spheres on the terminal residue.
We next examined in detail the interactions between β-tubulin and the TRiC chamber in each state. In all four β-tubulin intermediates, negatively charged residues exposed by folded N-terminal domain contacts specific positively charged residues in CCT6 and CCT8 while the negatively charged, normally unstructured, C- terminal E-hook tail of β tubulin contacts a positively charged pocket at the interface of CCT1 and CCT4 (Figure 5B, S5A, Movie S2). The folded C-terminal β-tubulin domain in states II-IV also engages in electrostatic contacts with positively charged residues in CCT1 and CCT3. These electrostatic contacts between the folded N- and C-terminal domains and TRiC persist even once β-tubulin reaches the fully folded state (Figure 5A).
Previous analysis recognized that the TRiC inner chamber contains an asymmetric charge distribution, with a strong positively charged inner surface contributed by subunits CCT1/3/6/8, and a strong negatively charged inner surface contributed by CCT2/4/5/715 (Figure 5C). Of note, these charged surface patches are only formed upon lid closure following ATP hydrolysis (Figure 5C). The β-tubulin electrostatic interactions with the positively charged surface of TRiC segregate β-tubulin to one side of the TRiC chamber via contacts present in all folding intermediates, while the polypeptide regions folded only in States III and IV extend into the chamber cavity (Figure 4B, S5B), suggesting additional TRiC regions contribute to β-tubulin folding.
We next examined the specific contacts between the CCT subunits and each β-tubulin folding intermediate (Figure 5D–I). In the folded β-tubulin N domain (blue in Figure 5D, E), residues on the N terminus, H2, H3, H4, and loops H1-S2, H3-S4 and H4-S5 form salt bridges and hydrogen bonds with residues on CCT6 and CCT8 (Figure 5D, E). Within the folded C-domain (green in Figure 5F, G), H11 and H12 are anchored to CCT1 and CCT3 by additional salt bridges and hydrogen bonds (Figure 4E). Our analysis also revealed specific contacts between β-tubulin and tails of CCT6 and CCT2, in addition to the previously highlighted E-hook tail of β-tubulin (Figure 5H, S5B). Remarkably, both the E-hook and the CCT tails are normally disordered, indicating that formation of the TRiC:β-tubulin complex promotes stabilization of these flexible regions. The negatively charged E-hook of β-tubulin is nestled into a positively charged pocket at the interface of CCT1 and CCT4 (Figure 5H, S5A). The CCT6 C-terminal tail is proximal to the β-tubulin T5 loop in the C domain, which is part of the GTP binding pocket (Figure 5Hi). Since the T5 loop forms a possible hydrogen bond to GTP, it is possible that the CCT6 tail facilitates formation of the tubulin GTP binding pocket. Indeed, we observe nucleotide density in State III of β-tubulin (Fig. 5H).
The regions corresponding to the Core and M- domains extend into the center of the TRiC chamber without any appreciable interactions with the inner wall. Closer examination reveals that TRiC contacts the Core and M-domains via the C-terminal tail of CCT2 in all the observed β-tubulin folding states (Figure 5Hiii; S5B). In State I and II, the C-terminus of CCT2 interacts with unfolded Core/M domain density, while in States III and IV the CCT2 tail engages the N and M loops in the taxol binding site38 as the Core and M domains form sequentially during folding (Figure S5B). This suggests the C-terminal tail of CCT2 interacts with and assists the folding of the Core and M-domains of β-tubulin within the lumen of the chamber. Interestingly, although the flexible CCT2 C-tail is poorly resolved in substrate-free closed TRiC chamber, we observe at a lower threshold an association between the vicinal C-terminal tails of CCT1 and CCT2 which extend together into the chamber (Figure S5E). Consistent with this observation, the C-terminal tail of CCT1 is negatively charged and that of CCT2 is positively charged and possesses several prolines that may rigidify this flexible region. It is interesting to speculate that the C-termini of CCT1-CCT2 jointly act as a fulcrum that, upon lid closure, facilitates the repositioning of β-tubulin to the vicinity of the CCT1-CCT3-CCT6-CCT8 hemisphere.
These structures indicate that all the β-tubulin folding intermediates interact with closed TRiC through a combination of more rigid and complementary electrostatic interactions with the chamber wall and flexible interactions with the CCTs tails (Figure 5I). Importantly, the negatively charged surface character of the N- and C-terminal domains is conserved across α-, β- and γ−tubulins (Figure S5F), suggesting a similar mode of TRiC-facilitated folding. Emphasizing the importance of these contacts for tubulin folding, the residues within the TRiC chamber making both electrostatic inner chamber contacts and tail contacts with β-tubulin are highly conserved (Figure S5G). It thus appears that TRiC does not release the β-tubulin polypeptide into the closed chamber for folding. Instead, the TRiC chamber binds β-tubulin throughout the folding process. The TRiC contacts directly restrain and orient the N-domain and C terminal tail of β-tubulin to one hemisphere in the chamber, thus limiting the folding polypeptide’s degree of freedom and preventing its free rotation in the chamber.
TRiC interactions shaping the tubulin folding landscape
Our four cryoEM structures of progressively folded β-tubulin (Figure 5) demonstrate the involvement of different CCT subunits at different folding intermediates via specific chemical interactions and the unexpected roles of the intrinsically disordered CCT tails guiding the unstructured domains of β-tubulin towards the fully folded structure (Figure 6A). A pathway invoking early folding of the N-terminal β-tubulin domain, which is well resolved in all four TRiC-bound folding intermediates, resonates with the PK-sensitivity assays of the folding reaction, which indicate early protection of an N-terminal domain, but rapid digestion of the C-terminus until completion of full-length β-tubulin folding (Figure S1G–I).
Figure 6: The directed folding of β-tubulin intermediates by specific CCT interactions.
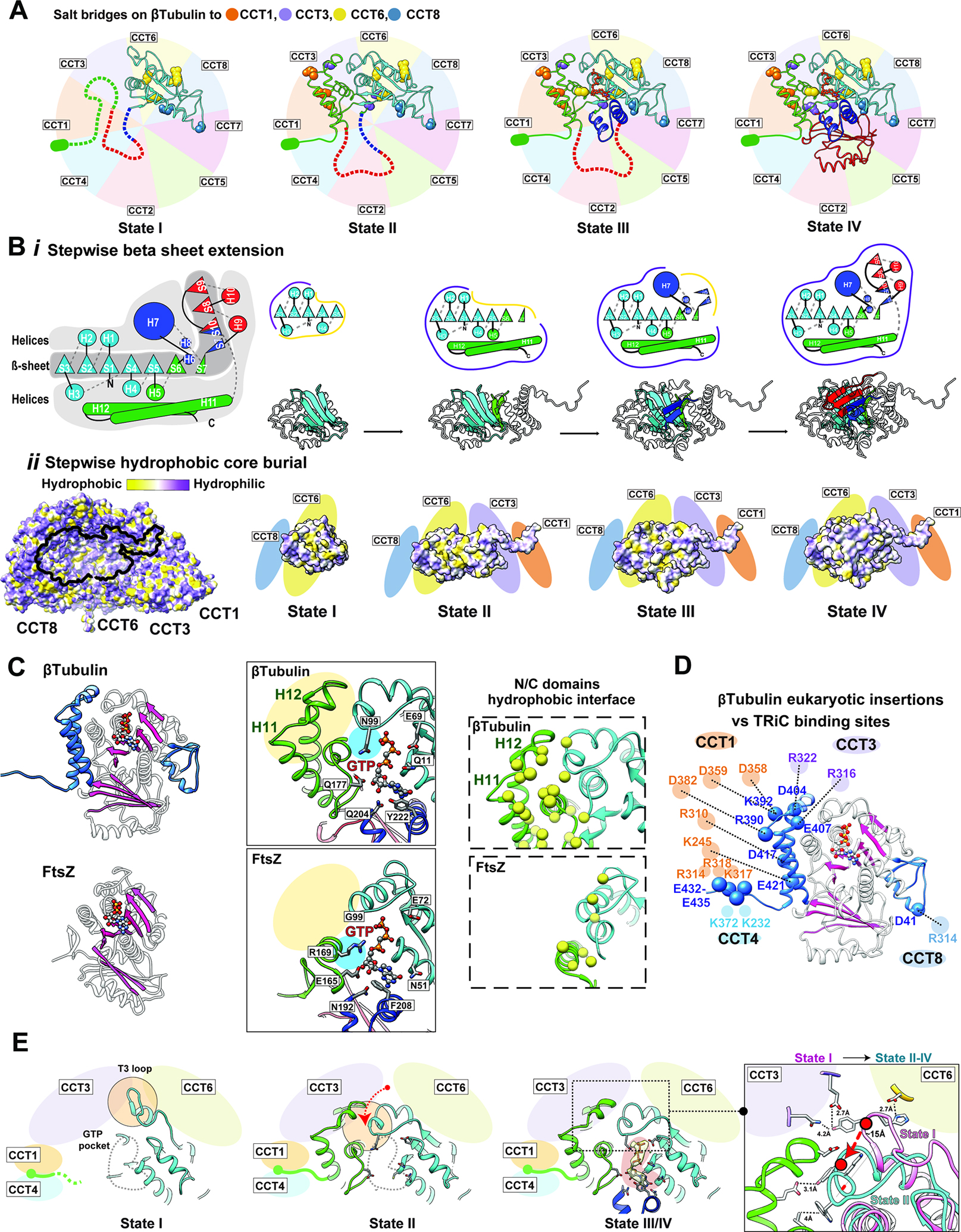
(A) Specific contacts between β-tubulin domains and CCT subunits at each intermediate state; salt bridges shown as balls colored according to the interacting CCT subunit. Not yet folded domains of each state shown as dotted lines. The negatively charged E-hook annotated as a green ellipse. (B) (i) Cartoon of β-tubulin emphasizes the hydrophobic β-sheet running the length of the β-tubulin structure that is surrounded by α−helices, and individual cartoons for the sequential formation of this β-sheet core through each folding state. (ii) Hydrophobicity distribution on TRiC of the β-tubulin interacting surface, and the hydrophobic core burial of β-tubulin as it extends towards the interior of the chamber through the folding states. (C) Ribbon diagram of β-tubulin (PDB 6I2I36, top panel) and FtsZ (PDB 1FSZ48; bottom panel); additions unique to β-tubulin structure shown in blue. The overall β-strand folds in β-tubulin and FtsZ are colored in pink. Zoom-in views of the GTP binding pocket (contacting residues indicated) and N/C-domain interface (hydrophobic residues yellow) are shown for comparison. (D) The contacts between β-tubulin specific insertions and CCT subunits are shown with potential salt bridges. (E) Formation of the β-tubulin GTP binding pocket through β-tubulin intermediates. GTP pocket is indicated as a gray dotted line in states I and II, density map corresponding to the nucleotide displayed as a yellow density with pink background in state III/IV. T3 loop makes a large conformational change between state I and the other states indicated by the red arrow and displayed in the focused overlay between states I and II-IV.
Our results suggest how TRiC-directed β-tubulin folding circumvents the challenges posed by its complex topology and aggregation-prone β-sheets. The progressively folded intermediates observed in the chamber form domains that are discontinuous in sequence but orchestrate the continuous formation of a hydrophobic β-sheet running the length of the β-tubulin structure (Figure 6Bi, Movie S3). Anchoring the surface of the folded N- and C-domains to the TRiC chamber wall not only establishes the native topology but also positions the growing folding intermediates so that their hydrophobic core extends towards the interior of the chamber (Figure 6Bii). Comparing State I through State IV, each increasingly folded intermediate adds another β-strand layer to the exposed hydrophobic core, likely priming it for the next folding step. Importantly, each intermediate also incorporates the helices above and below the growing β-sheet thus protecting its edges and reducing the potential for off-pathway interactions. In State I, the folded N-domain exposes the hydrophobic strands S4 and S5. Completion of the Rossman fold in State II allows helices H5, H11 and H12 to cover the newly incorporated strands S6 and S739. Formation of the Core domain in State III requires a change in direction between strands S7 and S10, mediated by helix H7. The CCT2 tail may participate in this step, as it interacts with this region of the Core domain. The last folding step to reach native β-tubulin, involved completion of the β-sheet by addition of sheets S8-S10 and positioning of helices H7, H9 and H10 to cover the hydrophobic core of tubulin. We propose that these TRiC interactions shape the tubulin folding landscape to establish the native topology and preclude the formation of trapped folding intermediates, thereby reducing the frustration of the folding landscape.
TRiC action in folding pathway in the context of tubulin evolution
While eukaryotic tubulin folding exhibits an obligate requirement for TRiC, its prokaryotic homolog FtsZ can spontaneously fold without chaperones40,41. Both proteins share a similar overall topology with other superfamily members in prokaryotes and archaea, including the overall domain organization and the extended core beta sheet (Figure 6C; S6A–B)40,42–44. However, eukaryotic tubulins are distinguished by additional features integral to their unique function and regulation, including remodeling of GTP affinity and hydrolysis kinetics; ability to engage in microtubule lateral contacts and binding microtubule-associated proteins (MAPs)40,45–47. These tubulin functions are accompanied by insertions in the H1-S2 loop and two negatively charged helices, H11 and H12; by the presence of the E-hook tail; by the remodeling of the residues contacting GTP, and by extensive changes to the C-domain leading to an increase in hydrophobic interdomain interfaces40,42–44 (highlighted in Figure 6C for 6I2I36 vs 1FSZ48, S6C). Eukaryotic tubulins are also distinguished by the strongly negative surface charge on the folded N- and C-domains as well as a negatively charged C-terminal E-hook tail. We hypothesize these features of eukaryotic tubulins hinder their ability to fold spontaneously or with simpler chaperone systems. Remarkably, the tubulin-specific insertions correspond to the sites of TRiC binding during folding in the closed chamber, suggesting their incorporation into ancestral tubulin was accompanied by a need for specialized folding assistance (Figure 6D).
This hypothesis is supported by close examination of how TRiC mediates formation of the unique N-C-domain interface in β-tubulin (Figure 6E, S6D). The T3 loop, which participates in the folded β-tubulin N-C interdomain interface, shifts between two alternative conformations in the TRiC-bound β-tubulin folding intermediates, with 13 Å C-alpha variation (Fig. S4G, S5C, Movie S4). Our maps’ good Q score resolve the bulky side chains in the T3 loop, such as H105 and Y106, allowing us to trace the density of T3 in the folding intermediates (Figure S6D). The H1-S2 loop forms charged and hydrogen bonded interactions with the TRiC chamber suggesting a stabilizing effect on the inserted β-tubulin loops by the TRiC chamber (Figure 4E, 6D). In State I, the T3 loop, resolved at residues 100–109 adopts an extended conformation directed by contacts with E202 of CCT3 and D86 of CCT6 (Fig. S5C). In State II, a decrease in resolvability of residues 96–106 in T3 (Fig. S5C), suggests the loop changes conformation upon folding of the C-domain. In States III and IV, the T3 loop becomes well resolved due to its stabilization by interaction with the folded C-domain, adopting the native β-tubulin conformation (PDB: 6I2I36) (Figure S4G, S5C). Accordingly, N99 of T3 serves in GTP γ-phosphate sensing while hydrophobic T3 residues including W101 are embedded in the hydrophobic interface between the N and C-domains (Figure 6C). Folding of the core helix domain completes the GTP binding pocket by contributing the H6-H7, consistent with the nucleotide density in the GTP binding pocket observed in States III and IV (Fig. S5C,5D, Figure 6E). Thus, TRiC contacts keep the T3 loop in an extended conformation when the C-domain is not folded, and that these contacts are released to allow the native conformation at the interdomain interface upon C-domain folding.
DISCUSSION
Why some proteins have an obligate requirement for TRiC assistance has long been enigmatic. We combined XL-MS and cryoEM to elucidate the pathway delivering unstructured β-tubulin from PFD to the TRiC chamber where it folds through specific contacts between the substrate and the CCT subunits (Figure 7A). We determined near atomic resolution structures of multiple stable folding intermediates of β-tubulin engaged in specific interactions with various CCT subunits including the intrinsically disordered termini. Our data suggest that these interactions guide tubulin to the native state through stepwise formation of partially folded intermediates (Figure 7B). Throughout this pathway the substrate is not released into the central TRiC chamber, unlike what is observed with GroEL-ES, but instead remains anchored through specific contacts with the CCT wall and the C-tails. These findings establish that, for tubulin, and perhaps other TRiC substrates, the folding pathway is guided by explicit interactions with the inner TRiC chamber.
Figure 7: Mechanism of TRiC directed folding pathway for β-tubulin.
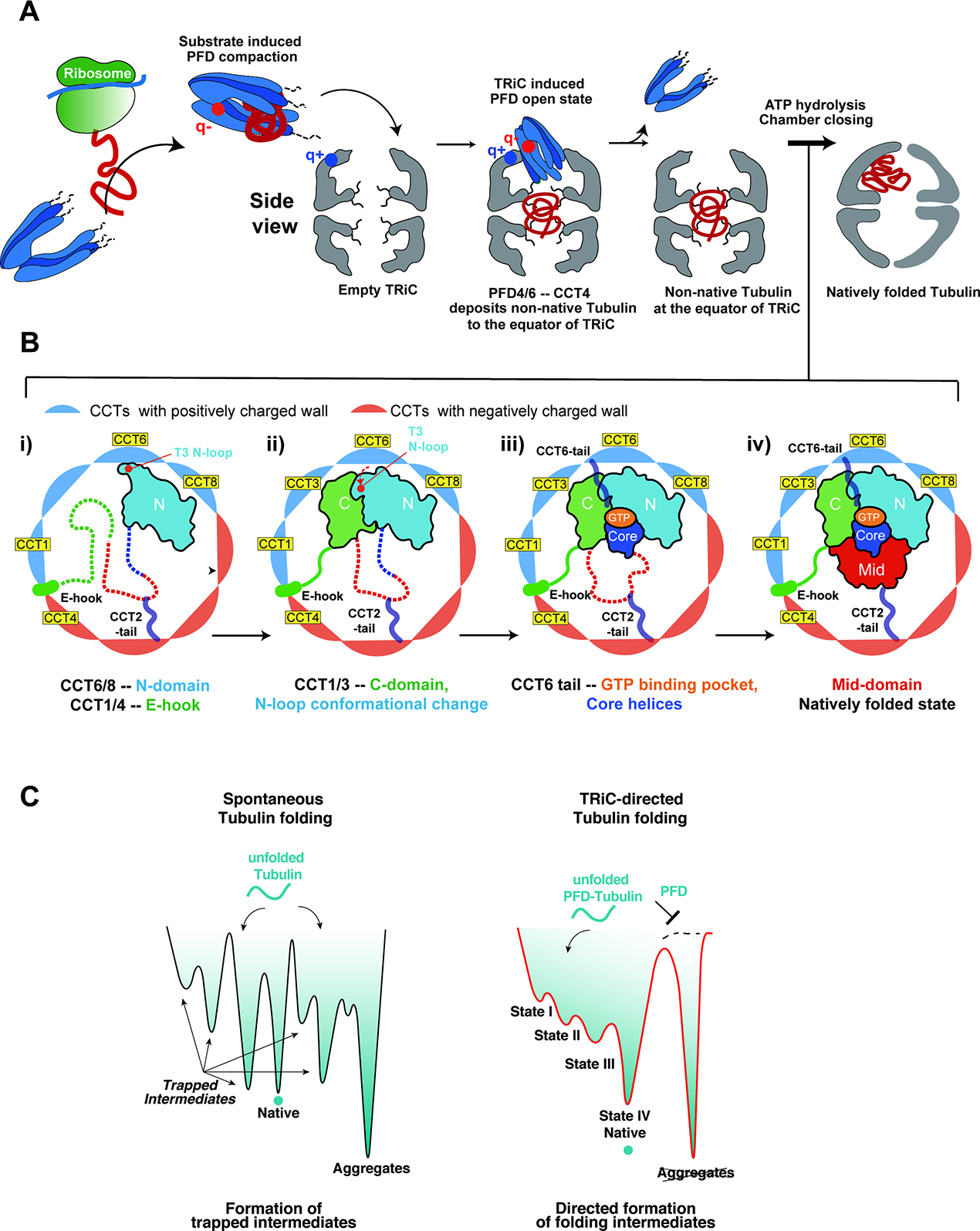
(A) Model of β-tubulin folding pathway; Nascent chain β-tubulin is captured by Prefoldin in an unfolded state. Red and blue dots on Prefoldin and TRiC show key electrostatic points mediating Prefoldin binding and repositioning into the chamber. See text for additional details. (B) TRiC directed folding pathway of β-tubulin through distinct intermediates. Unfolded domains shown in dotted lines; CCT subunits colored according to their charged state. See text for details. (C) Schematic folding landscape of tubulin: (left) Spontaneous tubulin folding leads to trapped intermediates and/or in aggregates. (right) Prefoldin maintains tubulin unfolded and prevents aggregation. TRiC directs the formation of progressively folded intermediates and prevents off-pathway trapped intermediates.
Beyond Anfinsen: the TRiC chamber shapes folding intermediates
Prefoldin bound β-tubulin is delivered to the open TRiC chamber, where interactions with the disordered CCT tails generates a compact entity without discernible folded elements. We speculate the disordered tails confined within the TRiC chamber act as a tethered solvent to maintain the fluidity of the substrate promoting a dynamic yet compacted state. The liquid-like nature of such a coacervate may prevent the formation of non-productive or trapped intermediates, priming the substrate for productive folding upon chamber closure (Figure 7A). Upon ATP-dependent chamber closure, the substrate repositions from the equatorial chamber into one of the ring-enclosed folding chambers (Figure 7B). The ATP-closed TRiC chamber directs the stepwise formation of β-tubulin folding intermediates that remain bound to specific regions in the chamber through electrostatic and H-bond interactions between β-tubulin and the TRiC chamber. These contacts establish the necessary native topology for β-tubulin and help orchestrate a hydrophobic collapse to then drive sequential folding of the β-tubulin domains. One feature of the discontinuous formation of β-tubulin domains in this folding pathway is the unidirectional assembly of the β-sheet spanning the entire hydrophobic core in folded β-tubulin. These TRiC contacts appear to direct β-tubulin folding through sequential domain formation without releasing the substrate into the chamber. This contrasts to the mechanism of chaperonin GroEL-ES, which releases polypeptides into an inner chamber which provides a unique confined environment for folding49–53. The fact that contacts with the closed TRiC chamber guide multiple steps in β-tubulin folding raises doubts on the universality of Anfinsen’s Principle.
The concept of TRiC directing the folding pathway is illustrated by the notable structural change in the T3-loop of β-tubulin between States I and II. TRiC keeps the T3-loop in an extended conformation through contacts with the inner wall until the C-terminal domain folds through association with the TRiC wall, then the T3-loop is released from the chamber wall to become nestled at the N-C domain interface. The T3 loop acts as a folding switch to control sequential folding of the GTP binding pocket after completion of the N-C domain interface (Figure 6E). Thus, TRiC binding to the T3-loop ensures the proper folding of the C-domain, establishing the overall fold topology of β-tubulin and allowing the hydrophobic core of the M- and Core domains to fold. Perhaps GTP binding to the incipient nucleotide pocket drives these subsequent steps, as observed for bacterial Tubulin FtsZ (Figure 6C), where GTP binding is a limiting step during spontaneous refolding54. The TRiC: β-tubulin contacts directly orchestrate the progression of events needed to form the GTP binding pocket by sequential positioning of the nucleotide sensing T3 loop to form the N-C domain interface, which are not conserved in FtsZ (Figure 6C–E). Consistent with this, tubulin mutations in the N/C-domain interface affect β-tubulin folding and release from TRiC55 (Figure S6E). While TRiC appears to directs the folding landscape through formation of specific intermediates (Figure 7B), the tubulin-CCT contacts may also disfavor formation of kinetically trapped states (Figure 7C).
Insights into evolution of eukaryotic proteins that cannot fold without TRiC
It is intriguing to speculate why and how this complex TRiC-tubulin requirement evolved from simpler ancestral proteins like FtsZ, which can refold without assistance. Compared with FtsZ, tubulin contains a series of structural changes which serve to enable functional interactions at the lateral interface of protofilaments and binding of microtubules associated proteins (MAPs) and Kinesin46,47,56. While these additional elements support the novel properties of eukaryotic tubulin, they appear to impair spontaneous folding56, perhaps leading to off-pathway or kinetically trapped intermediates (Figure 7C). Strikingly, the major sites of β-tubulin interaction with the TRiC-chamber are in these eukaryote-specific regions. We propose these eukaryotic-specific changes in tubulin evolved to exploit the hetero-oligomeric and asymmetric nature of TRiC to circumvent the inherent challenges they pose to the folding pathway.
The finding that tubulin folding is closely directed by TRiC chamber interactions opens new perspectives to understand the role of TRiC in eukaryotic protein evolution. The striking expansion of TRiC subunits directs the subunit-specific diversification of substrate binding sites, ATP affinity and chamber properties all of which contribute to its unique ability to fold some eukaryotic proteins. In turn, these may underlie their obligate requirement for TRiC to fold. Given that TRiC facilitates folding of ~10% of the proteome, it is likely that some proteins employ TRiC like an Anfinsen Cage, similar to GroEL:ES, to prevent aggregation and smooth the folding landscape57. Indeed, the now more limited space present in the chamber upon ATP-induced lid closure may restrain the possible orientations or conformations of the encapsulated polypeptide, significantly reducing the structural ensembles available to the substrate within the chamber of closed TRiC. Other proteins may additionally exploit other evolved features of the TRiC hetero-oligomer for which the diverse apical domain sequence and structure can provide substrate interaction specificity to promote a specific polypeptide binding topology12,16,19,20. Also, some substrates may interact distinctly with specific regions in the chamber58. Our finding that the eukaryote-specific tubulin insertions evolved surface-exposed motifs that bind specific regions in the TRiC chamber to shape the folding pathway provides an intriguing clue as to how some eukaryotic proteins evolved to exploit the unique properties of TRiC to acquire new functionalities that would otherwise impede their correct folding.
Limitations of the study
A TRiC directed β-tubulin folding pathway that progresses from less folded to more folded states is here derived from snapshot images at single equilibrium condition. Time-resolved kinetic analyses of tubulin folding, such as time-resolved H-D exchange and spectroscopic approaches are needed to validate and extend this pathway. The observation that the CCT intrinsically disordered tails promote TRiC-mediated folding resonates with findings for other chaperones that also use disordered domains for function59,60. Extending the idea that regions of intrinsic disorder promote folding will open new perspectives on what constitutes a chaperone and revise the classic notion that chaperones only function through domains that recognize non-native proteins through hydrophobic surfaces.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Judith Frydman (jfrydman@stanford.edu).
Materials availability
Reagents generated in this study are available from the lead contact upon request without restriction.
Data and code availability
The 3D cryoEM density maps have been deposited in the Electron Microscopy Data Bank under the accession number EMD: EMD-32822, 32823, 26089, 26120, 26123, 26131. Coordinates have been deposited in the Protein Data Bank under the accession number PDB: 7WU7, 7TRG, 7TTN, 7TTT, 7TUB. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD030590. This paper does not report the original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Initial bacmid DNA was generated in DH10Bac cells (Thermo Fisher Scientific) according to manufacturer protocols. Generation of baculovirus was performed in SF9 cells (Thermo Fisher Scientific), SF9 cells were maintained in SF900 iii SFM at 27 C and baculovirus was generated at a cell concentration of ~ 1×10^6 cells/ml. Protein expression was performed in High Five cells (Thermo Fisher Scientific) adapted to and maintained in ESF 921 media (Expression Systems) at 27 C. Viral infections for protein expression were performed at a cell concentration of ~ 2×10^6 cells/ml.
METHOD DETAILS
Cloning
pFB-α-tubulin and pFB-β-tubulin:
Human β-tubulin (TBB5) and α-tubulin (TUBA1B) were cloned into the pFB-Dual vector using conventional methods. Briefly, βTubulin was amplified using 5’ primer TTT TAG CGG CCG CCA AAA CAT GAG GGA AAT CGT GCA CAT CC and 3’ primer ATT TTT CTA GAT TAT CAG GCC TCC TCT TCG GCC TCC, and αTubulin was amplified using 5’ primer TTT TAG CGG CCG CCA AAA CAT GCG TGA GTG CAT CTC CAT CC and 3’ primer ATT TTT CTA GAT TAT CAG TAT TCC TCT CCT TCT TCC TCA CC from the plasmids pTS3067 and pTS4119 respectively. Amplified products and pFB-Dual vector were combined after digestion with NotI and XbaI, and sequence of isolated clones was verified.
TBCA and TBCB:
TBCA was cloned into pST39 and TBCB was cloned into pFB-Dual vectors using conventional methods. Briefly, TBCA was amplified using 5’ primer AGT TTC TAG ATT TGT TTA ACT TTA AGA AGG AGA TAT ACA TAT GGC CGA TCC TCG CGT GAG and 3’ primer AAT TAG GAT CCT CAT TAG TGA TGG TGA TGA TGG TGG GCT TCT AAC TTC ACT G and cloned into pST39 using XbaI and BamHI. TBCB was first amplified using 5’ primer AGT TTC TAG ATT TGT TTA ACT TTA AGA AGG AGA TAT ACA TAT GGA GGT GAC GGG GGT GTC and 3’ primer (with 6XHis) AAT TAG GAT CCT CAT TAG TGA TGG TGA TGA TGG TGA CTG CCT ATC TCG TCC AAC CCG TAG TCC and cloned into pST39 with XbaI and BamHI. To move TBCB into the pFB-Dual vector for insect cell expression TBCB was PCR amplified out of pST39 using 5’ TAA TTC TCG AGC TAT AAA TAT GGA GGT GAC GGG GGT GTC G and 3’ TAA TTG GTA CCT CAT TAG TGA TGG TGA TGA TGG TGA CTG CC and inserted into pFB-Dual using XhoI and KpnI.
Protein expression and purification
TRiC alone.
High Five insect cells (1 L) were co-infected with rBVs encoding his-tagged TRiC with CCT1 tagged with GFP. Cells were incubated at 27°C for ~72 h, pelleted by centrifugation at 500 × g for 10 min, and resuspended in TRiC lysis buffer (100 mM HEPES pH 7.4, 50 mM NaCl, 20 mM imidazole, 10 % glycerol, 5 mM PMSF) supplemented with benzonase (Sigma-Aldrich-Aldrich, E1014) (1,000 units) and a protease inhibitor cocktail [Roche]). Cells were lysed using dounce homogenization and debris was cleared by ultracentrifugation at 50,000 × g at 4 °C for 40 min. Cleared supernatant was passed over nickel resin and washed with column wash buffer (50 mM HEPES pH 7.4, 50 mM NaCl, 5 mM MgCl2 20 mM imidazole, 10 % glycerol) with an additional 250 mM NaCl, column wash buffer + 1 mM ATP, column wash buffer + an additional 500 mM NaCl, and finally column wash buffer alone. Nickel-bound protein was eluted with elution buffer (50 mM HEPES pH 7.4, 50 mM NaCl, 5 mM MgCl2 400 mM imidazole, 10 % glycerol). Protein-containing fractions were pooled and passed over a heparin column equilibrated with MQA buffer (50 mM HEPES pH 7.4, 50 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 10 % glycerol). Protein was eluted with a linear gradient of 20 % to 100 % MQB buffer (50 mM HEPES pH 7.4, 1 M NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 10 % glycerol). TRiC-containing fractions were pooled and diluted with MQ buffer (50 mM HEPES pH 7.4, 5 mM MgCl2, 1 mM DTT, 10 % glycerol) to remove excess NaCl. Pooled TRiC was loaded onto a MonoQ ion exchange column and eluted with a 200 ml linear gradient of 0 % to 100 % MQB. TRiC containing fractions were pooled, concentrated with a 100 kDa MWCO Centricon device, and passed over a Superose 6 size exclusion column equilibrated with MQA. TRiC containing fractions were pooled, concentrated, and snap-frozen in liquid nitrogen for long-term storage.
TRiC:β-tubulin purification:
High Five insect cells (1 L) were co-infected with rBVs encoding his-tagged TRiC and the Human β-tubulin protein. Cells were incubated at 27°C for ~72 h, pelleted by centrifugation at 500 × g for 10 min, and resuspended in TRiC lysis buffer (100 mM HEPES pH 7.4, 50 mM NaCl, 20 mM imidazole, 10 % glycerol, 5 mM PMSF) supplemented with benzonase (Sigma-Aldrich-Aldrich, E1014) (1,000 units) and a protease inhibitor cocktail [Roche]). Cells were lysed using dounce homogenization and debris was cleared by ultracentrifugation at 50,000 × g at 4 °C for 40 min. Cleared supernatant was passed over nickel resin and washed with column wash buffer (50 mM HEPES pH 7.4, 50 mM NaCl, 20 mM imidazole, 10 % glycerol). Nickel-bound protein was eluted with column wash buffer containing 400 mM imidazole. Protein-containing fractions were pooled and passed over a heparin column equilibrated with MQA buffer (50 mM HEPES pH 7.4, 50 mM NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 10 % glycerol). Protein was eluted with a linear gradient of 20 % to 100 % MQB buffer (50 mM HEPES pH 7.4, 1 M NaCl, 5 mM MgCl2, 0.5 mM EDTA, 1 mM DTT, 10 % glycerol). TRiC-containing fractions were pooled and diluted with MQ buffer (50 mM HEPES pH 7.4, 5 mM MgCl2, 1 mM DTT, 10 % glycerol) to remove excess NaCl. Pooled TRiC was loaded onto a MonoQ ion exchange column and eluted with a 200 ml linear gradient of 0 % to 100 % MQB. Fractions were assayed for the presence of β-tubulin and TRiC, pooled, concentrated with a 100 kDa MWCO Centricon device, and passed over a Superose 6 size exclusion column equilibrated with MQA. TRiC containing fractions were pooled, concentrated, and snap-frozen in liquid nitrogen for long-term storage. The integrity of TRiC and β-tubulin occupancy were confirmed by SDS-PAGE, native-PAGE, Coomassie staining, and immunoblotting with a rabbit CCT5-specific antibody (Abcam ab129016) and a mouse β-tubulin specific monoclonal antibody (Proteintech 66240-1-Ig).
Human Prefoldin:
All Human Prefoldin subunits were co-expressed using baculovirus in High Five insect cells. The Prefoldin complex was isolated by affinity (Nickel-NTA), anion exchange (MonoQ 10/100), and gel filtration (Superdex 200) chromatography. At each step, fractions containing Prefoldin were identified by SDS-PAGE. Final protein was concentrated to ~100 μM using an Amicon Ultra 30 kDa MWCO, and 50 % glycerol was added to yield a final concentration of 10 %. Protein was aliquoted, snap-frozen, and stored at −80 °C11.
Human Prefoldin + αTubulin, βTubulin:
Human Prefoldin was co-expressed with human α-tubulin or β-tubulin in High Five insect cells and purified as described above without the anion exchange step. All subunits were coexpressed using baculovirus in High Five insect cells. The Prefoldin complex was isolated by affinity (NickelNTA) and gel filtration (Superdex 200) chromatography. At each step, fractions containing Prefoldin and Tubulin were identified by SDS-PAGE. Final protein was concentrated to ~20 μM using an Amicon Ultra 30 kDa MWCO, and 50 % glycerol was added to yield a final concentration of 10 %. Protein was aliquoted, snap-frozen, and stored at −80 °C.
TBCA:
TBCA was expressed in 4 L of BL21 Rosetta2 pLysS cells O/N at 16 °C. Cells were lysed in 50 ml of column buffer (300 mM NaCl, 50 mM HEPES pH 7.4) with benzonase, fresh PMSF, 10 mM imidazole and Complete protease inhibtiors, EDTA free. Cells were lysed 2X using an emulsiflex (Avestin), and lysate was cleared at 20,000 × g for 30 min. Protein was affinity purified in a 10 ml Ni column equilibrated with column buffer and eluted with column buffer + 400 mM imidazole after a wash of ~300 ml column buffer + 10 mM imidazole. Protein was concentrated using an Amicon ultra 3 kD MWCO device to less than 2 ml and run over an Superdex 75 SEC column equilibrated with column buffer + 1 mM DTT. Protein was concentrated using an Amicon ultra 3 kD MWCO device, glycerol added to 5 %, aliquoted, snap-frozen, and stored at −80 °C.
TBCB:
TBCB was expressed in Hi-5 cells adapted to ESF media by infection with 15 ml of P2 virus into 1 L of cells at ~2 × 106 cells/ml. Cells were harvested 3 days after infection. Cells were lysed in column buffer (300 mM NaCl, 50 mM HEPES pH 7.4) with Benzonase, fresh PMSF, 20 mM imidazole and Complete protease inhibitors, EDTA free. Cell pellet was resuspended in ~90 ml lysis buffer and lysed using dounce homogenization. Lysate was cleared at 40,000 × g for 45 min. Protein was affinity purified in a 25 ml Ni column equilibrated with column buffer and eluted with column buffer + 400 mM imidazole after washes of ~75 ml column buffer + 15 mM imidazole, 50 ml column buffer + 500 mM NaCl + 15 mM imidazole, 50 ml column buffer + 15 mM imidazole. Protein was concentrated using an Amicon ultra 3 kD MWCO to less than 2 ml and run over an Superdex 200 SEC column equilibrated with 200 mM NaCl, 50 mM HEPES pH 7.4, 1 mM TCEP. Protein was concentrated using an Amicon ultra 3 kD MWCO device, glycerol added to 5 %, aliquoted, snap-frozen, and stored at −80 °C.
Native and SDS PAGE analysis
Native and SDS gel analysis for Prefoldin-tubulin:
Prefoldin samples were diluted to 5 μM. For native gels 7.5 μl of sample was run out on 4–16 % Native-PAGE gels. For SDS-PAGE, 8 μl of 4XPSB (200 mM Tris pH 6.8, 8% SDS, 40% glycerol, 20% β-mercaptoethanol, bromophenol blue to color) was added to 24 μl of sample, and 10 μl of each sample was separated in 15 % SDS-PAGE gels. Both gels were run in duplicate with one used for Coomassie staining and the other for immunoblot.
Native and SDS gel analysis for TRiC:tubulin:
TRiC samples were diluted to 0.5 μg/μl and bovine Tubulin to ~0.05 μg/μl for each Tubulin. For native gels, 5 μl of TRiC samples was separated. SDS-PAGE samples were made by adding 4XPSB to 1X final concentration, 6 μl of TRiC samples and 1, 2, 4 and 8 μl of Tubulin were separated.
Tubulin folding assays
β-tubulin folding assay was performed in folding buffer (20 mM Tris-HCl pH 7.4, 50 mM KCl, 10 mM MgCl2 and 0.2 mM CaCl2)12 with fresh TCEP added to 1 mM. Prefoldin: β-tubulin was prepared to a final concentration of 1 μM, TRiC to 0.1 μM, and TBCA to 1 μM. Samples were incubated in the presence/absence of 0.9 mM ATP, 1 mM GTP as indicated for 1 h. Folding assay samples were separated by native page and β-tubulin was detected by immunoblot with a monoclonal mouse antibody (Proteintech 66240-1-lg). α-tubulin folding was performed in folding buffer with an additional 50 mM KCl and fresh TCEP added to 1 mM. Prefoldin: α-tubulin was prepared to a final concentration of 1 μM, TRiC to 0.25 μM, and TBCB to 1 μM. Samples were incubated in the presence/absence of 0.9 mM ATP, 1 mM GTP as indicated for 1 h at 37 C. Folding assay samples were separated by native page and α-tubulin was detected by immunoblot with a monoclonal mouse antibody (Sigma-Aldrich T5168).
Proteinase K assays
Prefoldin:tubulin, Bovine tubulin, TRiC, and mmCPN were brought to a final concentration of 1 μM in 1XATPase buffer (50 mM HEPES, 50 mM KCl, 5 mM MgCl2, 10 % glycerol, 1 mM DTT). Samples were brought to 37 °C and 1 mM ATP/GTP was added to the Tubulin dimer control, Prefoldin:tubulin sample, TRiC:Prefoldin:tubulin sample (+ATP/GTP), and TRiC:Prefoldin:tubulin sample (+ATP/GTP-AlFx). After 5 min, NaF to 1 mM, and AlNO3 to 6 mM was added to the TRiC:Prefoldin:tubulin sample (+ATP/GTP-AlFx). After 30 min, samples were brought to room temperature. 15 μl was removed from each sample for zero-minute time points, and digestion was initiated by addition of proteinase K to a final concentration of 2 ng/μl. Timepoints were taken at 2, 4, 8 and 16 min by adding 15 μl sample to 2 μl of PMSF, followed by 6 μl 4XPSB. Samples were analyzed by SDS-PAGE followed by immunoblot with indicated antibodies. For assays with denatured Tubulin, chaperones were diluted to 0.25 μM. Bovine Tubulin at a concentration of 12.5 μM was denatured in 6 M GuHCl, 100 mM HEPES pH 7.4, 1 mM DTT, and chaperone mixes were used to dilute denatured Tubulin 1:100 for a final concentration of 0.125 μM. Aggregated Tubulin was pelleted by centrifugation at 21 K*G for 10 min. Supernatant was removed and PK assay was carried out as described as above with 14 μl timepoints. Tubulin binding by chaperones was assayed by running 10 μl samples in Native PAGE. For western blots, β-tubulin was probed for with a monoclonal mouse antibody towards the N-terminus (Proteintech 66240-1-lg), a polyclonal rabbit antibody (abcam ab6046) or a C-terminal epitope specific AA2 antibody (Millipore sigma 05–661), and α-tubulin with a mouse monoclonal antibody (Sigma-Aldrich T5168).
Crosslinking-Mass Spectrometry
For hPrefoldin:β-tubulin with hTRiC samples, TRiC was brought to 1 μM, and Prefoldin to 2 μM, hPrefoldin:α/β-tubulin alone was prepared at 5 μM, hTRiC:β-tubulin alone was prepared at 1 μM. All samples were prepared in 50 mM HEPES pH 7.4, 50 mM NaCl and 1 mM DTT. TRiC samples were incubated at 37 °C for 30 min with nucleotide to reach equilibrium/closed state. Fresh DSS crosslinker dissolved in DMSO was added to a final concentration of 1 mM, TRiC samples were incubated at 37 °C for 1 h with crosslinker, hPrefoldin:α/β-tubulin alone was incubated at room temperature for 1 h at room temperature. Crosslinking was quenched by incubation for 30 min after addition of 1 M Tris pH 7.4 to a final concentration of 100 mM. Crosslinked samples processing steps included reduction and alkylation of cysteine residues with tris(2-carboxyethyl)phosphine and iodoacetamide, respectively, sequential digestion with endoprotease Lys-C (Wako) and trypsin (Promega), clean-up using solid-phase extraction (Waters Sep-Pak tC18 cartridges) and fractionation of the purified digests by size exclusion chromatography (SEC; GE Superdex Peptide PC 3.2/300)66.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed on an Easy nLC-1200 HPLC system coupled to an Orbitrap Fusion Lumos mass spectrometer (both ThermoFisher Scientific). Peptides were separated by reversed-phase chromatography on an Acclaim PepMap RLSC C18 column (250 mm × 75 μm, ThermoFisher Scientific) at a flow rate of 300 nl/min. The mobile phase gradient was 11 to 40% B in 60 min, with mobile phases A = water/acetonitrile/formic acid (98:2:0.15, v/v/v) and B = acetonitrile/water/formic acid (80:20:0.15, v/v/v). MS/MS data were acquired in the data-dependent acquisition top speed mode with a cycle time of 3 s. Precursor and fragment ion spectra were acquired in the Orbitrap at 120000 and 30000 resolution, respectively. Precursor ions with a charge state of +3 to +7 were fragmented in the linear ion trap at a normalized collision energy of 35%. Dynamic exclusion was enabled for 30 s after one sequencing event.
MS/MS spectra were analyzed using xQuest, version 2.1.5 (available from https://gitlab.ethz.ch/leitner_lab/xquest_xprophet). The database contained the entries of all human TRiC and prefoldin subunits, human tubulin beta (TBB5_HUMAN), and insect tubulins as possible contaminants. No other contaminant proteins were observed at relevant levels. Search parameters for xQuest included: enzyme = trypsin, maximum number of missed cleavages = 2, carbamidomethylation of Cys as fixed modification, oxidation of Met as variable modification, Lys and protein N terminus as cross-linking sites, mass error tolerances of ±15 ppm at the MS1 level and ±20 ppm at the MS2 level. Search results were further filtered with stricter mass tolerances depending on the dataset, and identifications were required to have an xQuest delta score <0.9, a TIC score >0.1, and a minimum of four bond cleavages overall or three consecutive bond cleavages per peptide. A parallel search against the reversed and shuffled sequences of the database entries revealed no decoy hits fulfilling the search and filter criteria, suggesting the false discovery rate is close to 0% for the selected score thresholds. XLs are shown in Table S2. Mapping of XLs was performed using xiVIEW (Graham, M., Combe, C. W., Kolbowski, L. & Rappsilber, J. xiView: A common platform for the downstream analysis of Crosslinking Mass Spectrometry data. doi: 10.1101/561829)
Native Mass Spectrometry
Prefoldin and Prefoldin:β-tubulin were diluted 20-fold from storage buffer into 250 mM ammonium acetate, pH 7.0, before completing buffer exchange using 10 kDa MWCO Amicon centrifugal filters (Millipore Sigma). Final concentrations were estimated by UV absorbance to be between 10 and 20 μM Prefoldin. Native mass spectra were then recorded within several hours, during which proteins were kept on ice. Samples were injected using borosilicate emitters (ThermoFisher Scientific ES380) into a QExactive Plus EMR orbitrap mass spectrometer (ThermoFisher Scientific). The quadrupole was set to transmit m/z range 1,000 to 10,000. Resolution was 17,500 at 200 m/z for a transient time of 64 ms. Injection time was 50 ms and automatic gain control was off. The capillary voltage was 1.2 kV and S-lens RF was 100. Microscans were grouped sequentially in sets of 10. In-source activation was 50 V and activation in the higher-energy collisional dissociation (HCD) cell was 20 V, based on values found to sharpen peaks without discernable complex dissociation. Pressure in the HCD cell (nitrogen) was 5e-10 mbar. For transmission of Prefoldin, the injection flatapole was set to 20 V, the inter flatapole lens to 12 V, and the bent flatapole to 6 V; for Prefoldin:β-tubulin transmission, these values were respectively 12, 10, and 3 V. Raw data were summed and visualized in XCalibur v4.1.31.9 (ThermoFisher Scientific). Initial deconvolution was performed using UniDec v3.1.0(Marty et al. 2015). Masses were measured manually by minimizing error over different charge state assignments. Each charge state arising from Prefoldin and Prefoldin:β-tubulin was assigned to the open or compact conformation based on Gaussian peak envelopes. Raw signal intensities of each peak were recorded from XCalibur, summed within assigned conformations, and used to derive fractional occupancies. Conclusions based on native MS data were cross-validated by computing changes in solvent-accessible surface area for Prefoldin models.
Cryo-EM specimen preparation and data collection
Apo-TRiC:
3 μl of apo-TRiC sample was vitrified at 2 mg/ml and applied to 200-mesh R1.2/1.3 holey-carbon grids (Quantifoil) coated with Poly-L-lysine and vitrified using Vitrobot Mark IV (Thermo Fisher Scientific, CMCI in Seoul). 6,084 Movies were collected on a Titan Krios (Thermo Fisher Scientific) equipped with a Falcon 4 (Thermo Fisher Scientific) detector. The detailed imaging conditions are shown in Table S1.
Prefoldin:β-tubulin:TRiC:
0.5 μM TRiC sample was mixed with 2 μM Prefoldin:β-tubulin complex at RT. CryoEM grids (Quantifoil R1.2/1.3 200 Cu) were glow discharged (PELCO easiGlow) for 45 s. 0.07 % Octyl-beta-glucoside was mixed with the sample prior to vitrification. Sample was vitrified using Vitrobot Mark IV, where 2.7 μl sample was applied on the cryoEM grids and blotted for 3 s. 11,796 movies were collected on a Titan Krios (Thermo Fisher Scientific) equipped with the K2 Summit (Gatan) detector. The detailed imaging conditions are shown in Table S1.
TRiC:β-tubulin with ATP-AlFx:
1 μM co-purified TRiC:β-tubulin sample was incubated with ATP-AlFx for 1 hr at 37 °C. CryoEM grids (Quantifoil R1.2/1.3 200 Cu) were glow discharged (PELCO easiGlow) for 45 s. Sample was vitrified using a Gatan Leica GP plunger, where 2.7 μl sample was applied on the cryoEM grids and blotted from the back for 3~5 s. 31,204 movies were collected on Titan Krios (Thermo Fisher Scientific) equipped with the K2 Summit (Gatan) detector. The detailed imaging conditions are shown in Table S1.
Data processing and 3D refinement
All image processing was done in RELION 3.167 and cryoSPARC v3.268. Computing resources were utilized in the S2C2 SLAC national facility and at CMCI at Seoul National University. The detailed processing parameters are summarized in Table S1.
Apo-TRiC:
Movies were aligned in 5 × 5 patches in MotionCor269 and CTF parameters were estimated with GCTF70. Utilizing template-based autopicking in cryoSPARC v3.2, 2,100,259 particles were initially picked. After 2D classification and removing bad particles, 945,248 particles were subjected to 3D heterogeneous refinement using Ab initio model in cryoSPARC. After further 3D classification and CTF refinements, non-uniform refinement was performed using 662,744 particles yielding a 3.11 Å map of apo-TRiC based on the gold-standard Fourier shell correlation (FSC) at 0.143. This analysis workflow is illustrated in Fig. S3.
Prefoldin:β-tubulin:TRiC:
Movies were aligned in 5 × 5 patches in MotionCor2, and CTF parameters were estimated with GCTF. After initial template-based picking of 1,621,636 particles, 2D classification was performed. After selection, 443,858 particles were used for 3D classification yielding 2 major classes, Prefoldin bound TRiC with 194,013 particles and non-bound TRiC with 249,845 particles. Using non-uniform refinement, prefoldin bound and non-bound TRiC was reconstructed at a resolution of 3.9 Å and 4.2 Å, respectively. This analysis workflow is illustrated in Fig. S3.
TRiC:β-tubulin with ATP-AlFx:
21,486 movies were aligned in 5 × 5 patches by MotionCor2 (v), and CTF parameters were estimated with CTFFIND (v 4.1). Particles picked with a template matching method were subject to multiple rounds of 2D classification followed by 3D classification. 819,553 particles in open state were used to reconstruct an open state TRIC:β-tubulin map to 3.8 Å, and 529,181 particles were used to obtain a 2.7 Å C1 symmetry map. This analysis workflow is illustrated in Fig. S4A.
Compositional heterogeneity analysis of TRiC:β-tubulin with ATP-AlFx:
529,181 particles of closed state are aligned to the C2 (D1) symmetry axis to obtain a 2.5 Å consensus map of closed state TRIC-tubulin. For compositional analysis of tubulin in the TRiC chamber, the star file of the related particles was extended 2 times by relion_particle_symmetry_expand with D1 symmetry to cover the orientation for each of two rings in each particle. 3D focused classification of the tubulins was performed with a tubulin mask but without symmetry and orientational search. The identified tubulin occupied rings were thus traced back to the original double-ring TRiC particles for the tubulin occupancy analysis. This analysis workflow is illustrated in Fig. S4A.
Conformational heterogeneity analysis of TRiC:β-tubulin with ATP-AlFx:
The tubulin occupied rings were further subject to focused classification for conformational heterogeneity analysis. In brief, a tubulin mask was applied to the related rings in multiple 3D classifications without orientational search, four tubulin intermediate conformations were identified and each conformation was further locally refined to high resolution. This analysis workflow is illustrated in Fig. S4B.
Model building
Prefoldin:β-tubulin:TRiC:
Model building started from previous Prefoldin:TRiC model (PDB: 6NR811). The initial model was fitted into the density map and manually refined in COOT71. Then, the model was refined on the Namdinator server using MDFF72 and Phenix real space refinement default options72. After few rounds, the model was further corrected using Phenix and COOT (Afonine et al., 2018).
Copurified TRiC:β-tubulin under ATP-AlFx condition:
The model of TRiC in the closed form (PDB ID: 7LUM58) reference was rigidly fit into the closed state TRiC density by rigid body fitting with Fit in Map tool from Chimera v1.14. This fitted model was further refined with phenix.real_space_refine in Phenix v1.18.1, ISOLDE v1.1.0. And refined models were inspected and adjusted in COOT. The model of tubulin (PDB: 6I2I36) was rigidly fitted to each tubulin intermediate density, and manually adjusted by COOT and ISOLDE v1.1.0. All adjusted models were then refined using phenix.real_space_refine in Phenix.
All models were validated by Q-score35 and phenix.validation_cryoem73. Difference maps showing the nucleotide density in TRiC-tubulin closed state were calculated between the complex density map and the map calculated from the model of protein only, generated by phenix.real_diff_map. The figures of the difference map were generated by Chimera with the same contour level 3 sigma. All other figures were generated by Chimera and ChimeraX74.
PISA analysis
The interactions between TRiC and β-tubulin model in each state were calculated using the PISA server (https://www.ebi.ac.uk/pdbe/pisa/).
Logo Plot generation
Residue conservation was compared across 393 mainly eukaryotic with some archaeal species (group II chaperonins) for residues predicted to make interactions by PISA. Plots were generated using EDlogo plot75.
Sequence alignment and phylogenetic tree building
Multiple sequence alignments for tubulin and tubulin homologues (Data S1) were generated using the multiple sequence alignment tool T-coffee Espresso and visualized76. A phylogenetic tree of tubulin and tubulin homologs was made in MEGA X software77.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical tests were carried out in Prism. Pie, bar charts, heat maps and scatter plots were produced using R and Prism. Gel and radiograph densitometry were done in ImageJ. ATPase data was plotted in Prism. Average XL distance for PFD-TRiC interXLs was calculated by mapping all interXLs in which both XL residues are resolved onto PDB 6NR8 and averaging the distance. SEM was calculated by dividing the standard deviation by the square root of the number of XLs.
Supplementary Material
(A) Purification scheme for Prefoldin:tubulin and TRiC:β-tubulin (B) Purified TRiC:β-tubulin or α-tubulin with SDS-PAGE and Native-PAGE analysis, immunoblot confirms β-tubulin or α-tubulin co-migrates with TRiC in native gel. (C) Purified Prefoldin:tubulin (β or α-tubulin) with SDS-PAGE and Native-PAGE analysis, note faster migration of Prefoldin:tubulin relative to Prefoldin alone in native gel. (D) α-tubulin folding assay. α-tubulin co-expressed and co-purified with Prefoldin comigrates on a native gel. In the absence of nucleotide α-tubulin comigrates with TRiC, and addition of nucleotide (ATP/GTP) drives folding and partial release of α-tubulin in the presence of TBCB. (E) Protease sensitivity assay (Proteinase K) of Prefoldin:tubulin samples; Fully folded α/β Tubulin dimer is insensitive to PK digestion, while Prefoldin:tubulin samples show rapid degradation by SDS-PAGE visualized by Coomassie stain (i) or immunoblot (ii). (F) Protease sensitivity assay of Prefoldin:α-tubulin folding by TRiC. Like β-tubulin, α-tubulin bound to Prefoldin is protease sensitive, transfer to TRiC from Prefoldin causes a decrease in sensitivity, and addition of nucleotide drives folding resulting in limited protease sensitivity equivalent to α-tubulin in a native α/β Tubulin dimer. (G-I) Protease sensitivity assay to test if TRiC closure is providing protease protection with degradation assayed by both an N-terminal region polyclonal antibody, and a C-terminal (AA2) monoclonal antibody. After completion of β-tubulin folding by TRiC as assayed in (H), apyrase is added to reopen the lid TRiC complex, followed by incubation at the indicated times. PK protease sensitivity is assayed at indicated time points (I). No increase in protease sensitivity is observed immediately after 30 min apyrase addition, the 90 min incubation leads to a mild increase in sensitivity likely due to slow unfolding.
(A) (i) Scheme of XL-MS analysis pipeline. XL of Prefoldin:tubulin (ii), Prefoldin:tubulin:TRiC (ii), and tubulin:TRiC (iii) samples were carried out in the indicated nucleotide condition and analysed by Native PAGE and SDS-PAGE as indicated, prior to Mass Spectrometry analysis. Native PAGE analysis shows XLed samples run as single complexes, while SDS-PAGE shows XLed samples migrate as high molecular weight multi-protein species due to inter-subunit XLing. (B) Bottom view of XLs mapped onto Prefoldin, note extensive XLs between PFD2 and PFD6. (C) Representative native mass spectrum of Prefoldin without β-tubulin. Two charge state distributions can be inferred to correspond to conformations that are relatively open and compact, represented by cartoons with more or fewer charges (+), the calculated solvent exposed surface area is listed next to the cartoons for open (PDB:6NRB11) and compact (PDB:6NR811) Prefoldin. The most abundant charge state of each conformation is labeled. (D) Mass to charge plot of deconvolved data in (C). A single mass, Prefoldin, gives rise to both peak distributions of charge. (E) Alternative representation of the native mass spectrometry data in main figure 2F, with charge plotted on the vertical axis and intensity as a color scale (right). Open and compact states of Prefoldin and Prefoldin:β-tubulin are outlined in blue and red respectively, with outline widths proportional to relative abundances. These data show a shift in conformational equilibrium upon β-tubulin binding. (F,G) Prefoldin structures alone (i) and with the top ring of TRiC (ii) from the least engaged (F) PDB:6NRB11 and most engaged (G) PDB:6NR811 Prefoldin:TRiC structures. The distance between the terminally resolved residues of PFD2 (Ile 124) and PFD6 (Glu 114) are indicated in magenta.
(A) Representative cryoEM micrograph and 2D averages of apo-TRiC and the complex of Prefoldin:β-tubulin-TRiC in the open conformation. (B) Image processing workflow of apo-TRiC and Prefoldin:β-tubulin-TRiC and Gold standard FSC curve (C). (D) Local resolution estimation of three reconstructed maps: apo-TRiC, TRiC:β-tubulin (Prefoldin unbound), Prefoldin:TRiC:β-tubulin. (E) Crosslink mapping on Prefoldin:β-tubulin bound to TRiC (F) Histogram of shortest measured intra/interring Cα -Cα distances for identified XLs between Prefoldin and TRiC.
(A) Representative micrograph and 2D class averages of TRiC:β-tubulin with ATP-AlFx. (B) Processing workflow of TRiC:β-tubulin with ATP-AlFx and (C) 3D classification strategy for focusing the density inner chamber of TRiC. Our compositional heterogeneity analysis identifies different β-tubulin occupancies for TRiC, including no substrate bound (apo state), and single or double ring occupancy. Further analysis identified four progressively folded tubulin conformations inside the chamber: State I (41% of the rings); State II (16% of the rings); State III (34% of the rings) and State IV (9% of the rings) corresponding to folded tubulin. (D) Local resolution map estimation of β-tubulin intermediate states. (E) FSC curve of reconstructed maps showing resolution according to the gold standard FSC (0.143 criterion). (F) Q-score plots of the β-tubulin intermediate states shown with the expected Q-score according to 3.55 and 3.00 Å. (G) C-alpha backbone RMSD per residue between intermediate state and native β-tubulin (PDB: 6I2I36). Large variation indicated at T3 loop (red star) and N loop (yellow hexagon)
(A) cryoEM density of β-tubulin E hook tail in the CCT1/4 pocket for the different folding states (I-IV). (B) cryoEM density of CCT2 C terminal tail connected to β-tubulin for the different folding states (I-IV). (C) cryoEM density of the β-tubulin at the GTP binding pocket: T3 loop (red star), N loop (yellow hexagon), nucleotide (magenta eclipse). (D) Difference map analysis of the nucleotide occupancy in β-tubulin GTP binding pocket in comparison to CCT ATP binding pockets. The difference maps are displayed with +3σ. (E) Slice side and top view of CCT1/2 C terminal tail contacts in apo closed TRiC. (F) Electrostatic surface display of human α-tubulin (PDB: 6I2I36), β-tubulin (PDB: this study) and γ-tubulin (1Z5V61), respectively. (I) Logo plot of the conservation of CCT residues. The residues making direct contact with β-tubulin are indicated by red arrows
(A) Sequence alignment for selected areas of human α-, β-, γ-tubulins and archaeal/prokaryotic homologues (CetZ1: H. Volcanii CetZ2: H. Volcanii, FtsZ: M. jannaschii and TubZ: B.cereus). T3 loop, GTP binding motif, N/C-domain interface and C-domain including H12 and E-hook are represented. Red box indicates conserved N99 and L189 in T3 loop and N/C-domain interface in human tubulins. (B) Phylogenetic tree of tubulins and their structures 36,48,61–63. Structures are colored in domain (N-domain: cyan, C-domain: green, Core domain: blue, M-domain: red). The scale bar represents the number of amino acids substitutions per site. (C) The hydrophobic core and domain interfaces of β-tubulin and FtsZ with hydrophobic residues (Trp, Phe, Ile, Leu) highlighted with yellow (β-tubulin) and gray (FtsZ) balls. Red arrow indicates the N/C-domain interface. Blue arrows and dashed line indicate the C/Core-domain interface. Colored circles represent hydrophobic core of each domain. The progressive assembly of the hydrophobic cores for N-, C- and Core domain of β-tubulin (bottom panels). (D) Model B factor distribution of each state highlights the N-domain with T3 loop in red dashed box (i) and corresponding cryo-EM density maps with fitted model are presented (ii). Per residue RMSD plots of T3 loop among four states (iii) and Q-score plot of T3 loop (iv). Dashed black line indicates average Q score expected at 3.5 Å resolution. (E) β-tubulin model with point mutations at the GTP binding pocket indicated as balls on residues that have folding defects. Their locations and effects are summarized55,64,65.
This movie demonstrates the local density of three β-tubulin intermediates and fully folded states. The model of the folded β-tubulin domain is fitted into each density map and displayed. GTP is fitted into the GTP binding pocket as an example to illustrate the observed extra density in State III and IV.
This movie illustrates β-tubulin N-, C-domain interaction with the CCT1/3/6/8 by complementary charged surface.
The first part of the movie illustrates that TRiC orients hydrophobic surface of β-tubulin N-, C-domain to face the chamber center and allows the subsequent hydrophobic burial with Core helix and M-domain folding. The second part of the movie demonstrates the progressive β-sheet building along with continuously significant hydrophobic framing from helices as β-tubulin folds.
The movie shows the model trajectory from state I to the other states highlighting a conformational change of T3 loop in β-tubulin.
List of inter/intra-molecular crosslinks between PFD, α/β-tubulin and TRiC in indicated conditions.
fileFASTA file with alignment of tubulin homologues across evolution. Related to Figure 6.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti β-tubulin (Mouse) | ProteinTech | Cat# 66240-1-Ig, RRID: AB 2881629 |
| anti β-tubulin (Rabbit) Polyclonal | Abcam | Cat# AB6046, RRID: AB_2210370 |
| anti β-tubulin (Mouse) Monoclonal C-term AA2 epitope | Millipore Sigma | Cat# 05-661, RRID: AB_309885 |
| anti α-tubulin (Mouse) Monoclonal | Sigma-Aldrich | Cat# T5168, RRID: AB_477579 |
| Custom anti human PFD (Rabbit) | Cocalico Biologicals | N/A |
| IRDye® 680RD Goat anti-mouse IgG | LiCOr | Cat# 926-68070, RRID: AB_10956588 |
| IRDye® 800RD Donkey anti-mouse IgG | LiCOr | Cat# 926-32212, RRID: AB_621847 |
| anti -aTubulin (Mouse) | DSHB Univ. of Iowa | Cat#12G10, RRID: AB 1157911 |
| IRDye® 680RD Donkey anti-rabbit IgG | LiCOr | Cat# 926-68073, RRID: AB_10954442 |
| IRDye® 800RD Goat anti-rabbit IgG | LiCOr | Cat# 926-32211, RRID: AB_621843 |
| NativePAGE™ Novex® 4–16% Bis-Tris Protein Gels, 1.0 mm, 15 well | Life Technologies | Cat# BN1004BOX |
| NativePAGE™ Novex® 4–16% Bis-Tris Protein Gels, 1.0 mm, 10 well | Life Technologies | Cat# BN1002BOX |
| Bacterial and virus strains | ||
| Rosetta™(DE3)pLysS Competent Cells | EMD Millipore (Novagen) | Cat# 71403 |
| Max Efficiency DH10Bac Cells | Thermo Fisher Scientific | Cat# 10361-012 |
| Biological samples | ||
| N/A | N/A | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| hTRiC | This study | N/A |
| hTRiC C1GFP | This study | N/A |
| hTRiC:β-tubulin | This study | N/A |
| hPFD:β-tubulin | This study | N/A |
| hPFD:α-tubulin | This study | N/A |
| TBCA | This study | N/A |
| TBCB | This study | N/A |
| Benzonase nuclease | Sigma-Aldrich | Cat# E1014-25KU |
| DSS crosslinker H12/D12 | Creative Molecules Inc. | Cat# 001S |
| PMSF | Sigma-Aldrich | Cat# P7626-5G |
| CNBr-Activated Sepharose 4 Fast Flow | GE Healthcare | Cat# 17098101 |
| Apyrase ATPase | Sigma-Aldrich | Cat# A6410-500UN |
| GTP | Thermo Fisher scientific | Cat# R0461 |
| ATP (magnesium salt) | Sigma-Aldrich | Cat# A9187-1G |
| Proteinase K | Sigma-Aldrich | Cat# P2308-10MG |
| Sequencing grade Trypsin | Promega | Cat# V5117 |
| Critical commercial assays | ||
| N/A | N/A | N/A |
| Deposited data | ||
| Apo-TRiC map | This study | EMD-32822 |
| Prefoldin:β-tubulin:TRiC complex map | This study | EMD-32823 |
| Prefoldin:β-tubulin:TRiC complex model | This study | 7WU7 |
| TRiC:β-tubulin intermediate I model | This study | 7TRG |
| TRiC:β-tubulin intermediate II model | This study | 7TTN |
| TRiC:β-tubulin intermediate III model | This study | 7TTT |
| TRiC:β-tubulin intermediate IV model | This study | 7TUB |
| TRiC:β-tubulin intermediate I map | This study | EMD-26089 |
| TRiC:β-tubulin intermediate II map | This study | EMD-26120 |
| TRiC:β-tubulin intermediate III map | This study | EMD-26123 |
| TRiC:β-tubulin intermediate IV map | This study | EMD-26131 |
| Cross-linking mass spectrometry proteomics data | This study | PXD030590 |
| Human α-tubulin | Adib, R. et al, 201936 | 6I2I |
| Human β-tubulin | Adib, R. et al, 201936 | 6I2I |
| Human γ-tubulin | Aldaz. et al, 200561 | 1Z5V |
| Tubulin homologue: FtsZ from M. jannaschii | Lowe, et al, 199848 | 1FSZ |
| Experimental models: Cell lines | ||
| High Five™ Cells (BTI-TN-5B1-4) | Thermo Fisher scientific | Cat# B85502 |
| Sf9 cells | Thermo Fisher scientific | Cat# 11496015 |
| Experimental models: Organisms/strains | ||
| N/A | N/A | N/A |
| Oligonucleotides | ||
| 5’ pFB-β-tubulin TTT TAG CGG CCG CCA AAA CAT GAG GGA AAT CGT GCA CAT CC | This study | N/A |
| 3’ pFB-β-tubulin ATT TTT CTA GAT TAT CAG GCC TCC TCT TCG GCC TCC | This study | N/A |
| 5’ pFB-α-tubulin TTT TAG CGG CCG CCA AAA CAT GCG TGA GTG CAT CTC CAT CC | This study | N/A |
| 5’ pFB-α-tubulin ATT TTT CTA GAT TAT CAG TAT TCC TCT CCT TCT TCC TCA CC | This study | N/A |
| 5’ TBCA pST39 AGT TTC TAG ATT TGT TTA ACT TTA AGA AGG AGA TAT ACA TAT GGC CGA TCC TCG CGT GAG | This study | N/A |
| 3’ TBCA pST39 AAT TAG GAT CCT CAT TAG TGA TGG TGA TGA TGG TGG GCT TCT AAC TTC ACT G | This study | N/A |
| 5’ TBCB pST39 AGT TTC TAG ATT TGT TTA ACT TTA AGA AGG AGA TAT ACA TAT GGA GGT GAC GGG GGT GTC | This study | N/A |
| 3’ TBCB pST39 AAT TAG GAT CCT CAT TAG TGA TGG TGA TGA TGG TGA CTG CCT ATC TCG TCC AAC CCG TAG TCC | This study | N/A |
| 5’ TBCB pFB TAA TTC TCG AGC TAT AAA TAT GGA GGT GAC GGG GGT GTC G | This study | N/A |
| 3’ TBCB pFB TAA TTG GTA CCT CAT TAG TGA TGG TGA TGA TGG TGA CTG CC | This study | N/A |
| Recombinant DNA | ||
| pFB-α-tubulin | This study | N/A |
| pFB-β-tubulin | This study | N/A |
| pFB-TRiC | Knowlton et al, 202158 | N/A |
| pFB-TBCB | This study | N/A |
| pST39-TBCA | This study | N/A |
| pTS-4119 α-tubulin template | Tim Stearns lab | N/A |
| pTS-3067 β-tubulin template | Tim Stearns lab | N/A |
| pFB-PFD1,2 | Gestaut et al, 201911 | N/A |
| pFB-PFD3,5 | Gestaut et al, 201911 | N/A |
| pFB-PFD4,6 | Gestaut et al, 201911 | N/A |
| pFB-CCT1-GFP,CCT8 | Gestaut et al, 201911 | N/A |
| pFB-CCT2, CCT4 | Gestaut et al, 201911 | N/A |
| pFB-CCT3, CCT6 | Gestaut et al, 201911 | N/A |
| pFB-CCT5, CCT7-6XHis | Gestaut et al, 201911 | N/A |
| Software and algorithms | ||
| RELION v3.1 | Zivanov, J., 201867 | http://www2.mrc-lmb.cam.ac.uk/relion |
| PHENIX | Afonine et al., 201873 | https://www.phenix-online.org |
| ChimeraX | Pettersen et al., 202174 | https://www.cgl.ucsf.edu/chimera |
| ProMod | Pintilie et al., 202035 | http://cryoem.bcm.edu/cryoem/downloads/segger |
| Igor Pro 6.22A | Wavemetrics Inc. | https://www.wavemetrics.com |
| Imagequant V 5.2 and 8.1 | GE Healthcare Lifecsciences | https://www.gelifesciences.com |
| Pymol 1.8.6.2 | Schrödinger | https://pymol.org |
| CryoSPARC v3.2 | Punjani et al., 201768 | https://cryosparc.com/ |
| Namdinator | Kidmose et al., 201972 | https://namdinator.au.dk/ |
| T-coffee | Di Tommaso et al., 201176 | https://tcoffee.crg.eu/ |
| Mega X | Hall, 201377 | https://www.megasoftware.net/ |
| Other | ||
| Quantifoil R1.2/1.3 200-mesh holey carbon-coated grids | Quantifoil | https://www.emsdiasum.com/microscopy/products/grids/quantifoil.aspx |
Highlights.
Reconstitution of chaperone-mediated tubulin folding with human Prefoldin and TRiC/CCT
Prefoldin delivers unstructured tubulin directly into the TRiC chamber
TRiC orchestrates ATP-driven tubulin folding via progressively formed intermediates
Specific contacts within the TRiC chamber shape the tubulin folding landssssssscape
ACKNOWLEDGMENTS.
Supported by NIH grants R01GM074074 to JF; P41GM103832, S10OD021600 and R01GM079429 to WC and the Korean National Research Foundation (2019R1C1C1004598, 2020R1A5A1018081, 2021M3A9I4021220, 2019M3E5D6063871) and SUHF foundation to SHR. CryoEM data collected at the Stanford-SLAC CryoEM facilities. We thank Dr. P. Picotti and the UCSF Mass Spectrometry Facility for access to instrumentation.
Footnotes
DECLARATION OF INTERESTS.
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Buchner J (2019). Molecular chaperones and protein quality control: an introduction to the JBC Reviews thematic series. J Biol Chem 294, 2074–2075. 10.1074/jbc.REV118.006739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartl FU, Bracher A, and Hayer-Hartl M (2011). Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332. 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 3.Labbadia J, and Morimoto RI (2015). The biology of proteostasis in aging and disease. Annu Rev Biochem 84, 435–464. 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mogk A, Bukau B, and Kampinga HH (2018). Cellular Handling of Protein Aggregates by Disaggregation Machines. Mol Cell 69, 214–226. 10.1016/j.molcel.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Plate L, Paxman RJ, Wiseman RL, and Kelly JW (2016). Modulating protein quality control. Elife 5. 10.7554/eLife.18431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deuerling E, and Bukau B (2004). Chaperone-assisted folding of newly synthesized proteins in the cytosol. Crit Rev Biochem Mol Biol 39, 261–277. 10.1080/10409230490892496. [DOI] [PubMed] [Google Scholar]
- 7.Deuerling E, Gamerdinger M, and Kreft SG (2019). Chaperone Interactions at the Ribosome. Cold Spring Harb Perspect Biol 11. 10.1101/cshperspect.a033977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willmund F, del Alamo M, Pechmann S, Chen T, Albanese V, Dammer EB, Peng J, and Frydman J (2013). The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152, 196–209. 10.1016/j.cell.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yam AY, Xia Y, Lin HT, Burlingame A, Gerstein M, and Frydman J (2008). Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol 15, 1255–1262. 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stein KC, Kriel A, and Frydman J (2019). Nascent Polypeptide Domain Topology and Elongation Rate Direct the Cotranslational Hierarchy of Hsp70 and TRiC/CCT. Mol Cell 75, 1117–1130 e1115. 10.1016/j.molcel.2019.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gestaut D, Roh SH, Ma B, Pintilie G, Joachimiak LA, Leitner A, Walzthoeni T, Aebersold R, Chiu W, and Frydman J (2019). The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis. Cell 177, 751–765 e715. 10.1016/j.cell.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balchin D, Milicic G, Strauss M, Hayer-Hartl M, and Hartl FU (2018). Pathway of Actin Folding Directed by the Eukaryotic Chaperonin TRiC. Cell 174, 1507–1521 e1516. 10.1016/j.cell.2018.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Balchin D, Hayer-Hartl M, and Hartl FU (2016). In vivo aspects of protein folding and quality control. Science 353, aac4354. 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 14.Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, et al. (2010). 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc Natl Acad Sci U S A 107, 4967–4972. 10.1073/pnas.0913774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leitner A, Joachimiak LA, Bracher A, Monkemeyer L, Walzthoeni T, Chen B, Pechmann S, Holmes S, Cong Y, Ma B, et al. (2012). The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 20, 814–825. 10.1016/j.str.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joachimiak LA, Walzthoeni T, Liu CW, Aebersold R, and Frydman J (2014). The structural basis of substrate recognition by the eukaryotic chaperonin TRiC/CCT. Cell 159, 1042–1055. 10.1016/j.cell.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cong Y, Schroder GF, Meyer AS, Jakana J, Ma B, Dougherty MT, Schmid MF, Reissmann S, Levitt M, Ludtke SL, et al. (2012). Symmetry-free cryo-EM structures of the chaperonin TRiC along its ATPase-driven conformational cycle. EMBO J 31, 720–730. 10.1038/emboj.2011.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reissmann S, Joachimiak LA, Chen B, Meyer AS, Nguyen A, and Frydman J (2012). A gradient of ATP affinities generates an asymmetric power stroke driving the chaperonin TRIC/CCT folding cycle. Cell Rep 2, 866–877. 10.1016/j.celrep.2012.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roh SH, Kasembeli M, Galaz-Montoya JG, Trnka M, Lau WC, Burlingame A, Chiu W, and Tweardy DJ (2016). Chaperonin TRiC/CCT Modulates the Folding and Activity of Leukemogenic Fusion Oncoprotein AML1-ETO. J Biol Chem 291, 4732–4741. 10.1074/jbc.M115.684878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spiess C, Miller EJ, McClellan AJ, and Frydman J (2006). Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol Cell 24, 25–37. 10.1016/j.molcel.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tam S, Spiess C, Auyeung W, Joachimiak L, Chen B, Poirier MA, and Frydman J (2009). The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol 16, 1279–1285. 10.1038/nsmb.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geissler S, Siegers K, and Schiebel E (1998). A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J 17, 952–966. 10.1093/emboj/17.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, and Cowan NJ (1998). Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 93, 863–873. [DOI] [PubMed] [Google Scholar]
- 24.Lewis SA, Tian G, and Cowan NJ (1997). The alpha- and beta-tubulin folding pathways. Trends Cell Biol 7, 479–484. 10.1016/S0962-8924(97)01168-9. [DOI] [PubMed] [Google Scholar]
- 25.Nithianantham S, Le S, Seto E, Jia W, Leary J, Corbett KD, Moore JK, and Al-Bassam J (2015). Tubulin cofactors and Arl2 are cage-like chaperones that regulate the soluble alphabeta-tubulin pool for microtubule dynamics. Elife 4. 10.7554/eLife.08811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frydman J, Nimmesgern E, Ohtsuka K, and Hartl FU (1994). Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature 370, 111–117. 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- 27.Thulasiraman V, Ferreyra RG, and Frydman J (2000). Folding assays. Assessing the native conformation of proteins. Methods Mol Biol 140, 169–177. 10.1385/1-59259-061-6:169. [DOI] [PubMed] [Google Scholar]
- 28.Meyer AS, Gillespie JR, Walther D, Millet IS, Doniach S, and Frydman J (2003). Closing the folding chamber of the eukaryotic chaperonin requires the transition state of ATP hydrolysis. Cell 113, 369–381. 10.1016/s0092-8674(03)00307-6. [DOI] [PubMed] [Google Scholar]
- 29.Reissmann S, Parnot C, Booth CR, Chiu W, and Frydman J (2007). Essential function of the built-in lid in the allosteric regulation of eukaryotic and archaeal chaperonins. Nat Struct Mol Biol 14, 432–440. 10.1038/nsmb1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collier MP, Alderson TR, de Villiers CP, Nicholls D, Gastall HY, Allison TM, Degiacomi MT, Jiang H, Mlynek G, Furst DO, et al. (2019). HspB1 phosphorylation regulates its intramolecular dynamics and mechanosensitive molecular chaperone interaction with filamin C. Sci Adv 5, eaav8421. 10.1126/sciadv.aav8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaltashov IA, and Mohimen A (2005). Estimates of protein surface areas in solution by electrospray ionization mass spectrometry. Anal Chem 77, 5370–5379. 10.1021/ac050511+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santambrogio C, Frana AM, Natalello A, Papaleo E, Regonesi ME, Doglia SM, Tortora P, Invernizzi G, and Grandori R (2012). The role of the central flexible region on the aggregation and conformational properties of human ataxin-3. FEBS J 279, 451–463. 10.1111/j.1742-4658.2011.08438.x. [DOI] [PubMed] [Google Scholar]
- 33.Scott D, Layfield R, and Oldham NJ (2015). Ion mobility-mass spectrometry reveals conformational flexibility in the deubiquitinating enzyme USP5. Proteomics 15, 2835–2841. 10.1002/pmic.201400457. [DOI] [PubMed] [Google Scholar]
- 34.Cuellar J, Ludlam WG, Tensmeyer NC, Aoba T, Dhavale M, Santiago C, Bueno-Carrasco MT, Mann MJ, Plimpton RL, Makaju A, et al. (2019). Structural and functional analysis of the role of the chaperonin CCT in mTOR complex assembly. Nat Commun 10, 2865. 10.1038/s41467-019-10781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pintilie G, Zhang K, Su Z, Li S, Schmid MF, and Chiu W (2020). Measurement of atom resolvability in cryo-EM maps with Q-scores. Nat Methods 17, 328–334. 10.1038/s41592-020-0731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adib R, Montgomery JM, Atherton J, O’Regan L, Richards MW, Straatman KR, Roth D, Straube A, Bayliss R, Moores CA, and Fry AM (2019). Mitotic phosphorylation by NEK6 and NEK7 reduces the microtubule affinity of EML4 to promote chromosome congression. Sci Signal 12. 10.1126/scisignal.aaw2939. [DOI] [PubMed] [Google Scholar]
- 37.Ellis RJ (1996). Revisiting the Anfinsen cage. Fold Des 1, R9–15. 10.1016/S1359-0278(96)00004-1. [DOI] [PubMed] [Google Scholar]
- 38.Orr GA, Verdier-Pinard P, McDaid H, and Horwitz SB (2003). Mechanisms of Taxol resistance related to microtubules. Oncogene 22, 7280–7295. 10.1038/sj.onc.1206934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rossmann MG, Moras D, and Olsen KW (1974). Chemical and biological evolution of nucleotide-binding protein. Nature 250, 194–199. 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 40.Andreu JM, Oliva MA, and Monasterio O (2002). Reversible unfolding of FtsZ cell division proteins from archaea and bacteria. Comparison with eukaryotic tubulin folding and assembly. J Biol Chem 277, 43262–43270. 10.1074/jbc.M206723200. [DOI] [PubMed] [Google Scholar]
- 41.Erickson HP, Anderson DE, and Osawa M (2010). FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol Biol Rev 74, 504–528. 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aylett CHS, and Duggin IG (2017). The Tubulin Superfamily in Archaea. Subcell Biochem 84, 393–417. 10.1007/978-3-319-53047-5_14. [DOI] [PubMed] [Google Scholar]
- 43.Nogales E, Downing KH, Amos LA, and Lowe J (1998). Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol 5, 451–458. 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 44.Yutin N, and Koonin EV (2012). Archaeal origin of tubulin. Biol Direct 7, 10. 10.1186/1745-6150-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janke C, and Bulinski JC (2011). Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol 12, 773–786. 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- 46.Nogales E, Wolf SG, and Downing KH (1998). Structure of the alpha beta tubulin dimer by electron crystallography. Nature 391, 199–203. 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 47.Skiniotis G, Cochran JC, Muller J, Mandelkow E, Gilbert SP, and Hoenger A (2004). Modulation of kinesin binding by the C-termini of tubulin. EMBO J 23, 989–999. 10.1038/sj.emboj.7600118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lowe J, and Amos LA (1998). Crystal structure of the bacterial cell-division protein FtsZ. Nature 391, 203–206. 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 49.Chen DH, Madan D, Weaver J, Lin Z, Schroder GF, Chiu W, and Rye HS (2013). Visualizing GroEL/ES in the act of encapsulating a folding protein. Cell 153, 1354–1365. 10.1016/j.cell.2013.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fenton WA, and Horwich AL (1997). GroEL-mediated protein folding. Protein Sci 6, 743–760. 10.1002/pro.5560060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta AJ, Haldar S, Milicic G, Hartl FU, and Hayer-Hartl M (2014). Active cage mechanism of chaperonin-assisted protein folding demonstrated at single-molecule level. J Mol Biol 426, 2739–2754. 10.1016/j.jmb.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 52.Motojima F (2015). How do chaperonins fold protein? Biophysics (Nagoya-shi) 11, 93–102. 10.2142/biophysics.11.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma S, Chakraborty K, Muller BK, Astola N, Tang YC, Lamb DC, Hayer-Hartl M, and Hartl FU (2008). Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell 133, 142–153. 10.1016/j.cell.2008.01.048. [DOI] [PubMed] [Google Scholar]
- 54.Huecas S, Canosa-Valls AJ, Araujo-Bazan L, Ruiz FM, Laurents DV, Fernandez-Tornero C, and Andreu JM (2020). Nucleotide-induced folding of cell division protein FtsZ from Staphylococcus aureus. FEBS J 287, 4048–4067. 10.1111/febs.15235. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Tian G, Cowan NJ, and Cabral F (2006). Mutations affecting beta-tubulin folding and degradation. J Biol Chem 281, 13628–13635. 10.1074/jbc.M513730200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bertrand S, Barthelemy I, Oliva MA, Carrascosa JL, Andreu JM, and Valpuesta JM (2005). Folding, stability and polymerization properties of FtsZ chimeras with inserted tubulin loops involved in the interaction with the cytosolic chaperonin CCT and in microtubule formation. J Mol Biol 346, 319–330. 10.1016/j.jmb.2004.11.054. [DOI] [PubMed] [Google Scholar]
- 57.Georgescauld F, Popova K, Gupta AJ, Bracher A, Engen JR, Hayer-Hartl M, and Hartl FU (2014). GroEL/ES chaperonin modulates the mechanism and accelerates the rate of TIM-barrel domain folding. Cell 157, 922–934. 10.1016/j.cell.2014.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Knowlton JJ, Gestaut D, Ma B, Taylor G, Seven AB, Leitner A, Wilson GJ, Shanker S, Yates NA, Prasad BVV, et al. (2021). Structural and functional dissection of reovirus capsid folding and assembly by the prefoldin-TRiC/CCT chaperone network. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2018127118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quan S, Wang L, Petrotchenko EV, Makepeace KA, Horowitz S, Yang J, Zhang Y, Borchers CH, and Bardwell JC (2014). Super Spy variants implicate flexibility in chaperone action. Elife 3, e01584. 10.7554/eLife.01584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reichmann D, Xu Y, Cremers CM, Ilbert M, Mittelman R, Fitzgerald MC, and Jakob U (2012). Order out of disorder: working cycle of an intrinsically unfolded chaperone. Cell 148, 947–957. 10.1016/j.cell.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aldaz H, Rice LM, Stearns T, and Agard DA (2005). Insights into microtubule nucleation from the crystal structure of human gamma-tubulin. Nature 435, 523–527. 10.1038/nature03586. [DOI] [PubMed] [Google Scholar]
- 62.Hoshino S, and Hayashi I (2012). Filament formation of the FtsZ/tubulin-like protein TubZ from the Bacillus cereus pXO1 plasmid. J Biol Chem 287, 32103–32112. 10.1074/jbc.M112.373803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duggin IG, Aylett CH, Walsh JC, Michie KA, Wang Q, Turnbull L, Dawson EM, Harry EJ, Whitchurch CB, Amos LA, and Lowe J (2015). CetZ tubulin-like proteins control archaeal cell shape. Nature 519, 362–365. 10.1038/nature13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zabala JC, Fontalba A, and Avila J (1996). Tubulin folding is altered by mutations in a putative GTP binding motif. J Cell Sci 109 (Pt 6), 1471–1478. 10.1242/jcs.109.6.1471. [DOI] [PubMed] [Google Scholar]
- 65.Farr GW, and Sternlicht H (1992). Site-directed mutagenesis of the GTP-binding domain of beta-tubulin. J Mol Biol 227, 307–321. 10.1016/0022-2836(92)90700-t. [DOI] [PubMed] [Google Scholar]
- 66.Leitner A, Walzthoeni T, and Aebersold R (2014). Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat Protoc 9, 120–137. 10.1038/nprot.2013.168. [DOI] [PubMed] [Google Scholar]
- 67.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7. 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 69.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang K (2016). Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12. 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kidmose RT, Juhl J, Nissen P, Boesen T, Karlsen JL, and Pedersen BP (2019). Namdinator - automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ 6, 526–531. 10.1107/S2052252519007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Afonine PV, Klaholz BP, Moriarty NW, Poon BK, Sobolev OV, Terwilliger TC, Adams PD, and Urzhumtsev A (2018). New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr D Struct Biol 74, 814–840. 10.1107/S2059798318009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82. 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dey KK, Xie D, and Stephens M (2018). A new sequence logo plot to highlight enrichment and depletion. BMC Bioinformatics 19, 473. 10.1186/s12859-018-2489-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, and Notredame C (2011). T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res 39, W13–17. 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hall BG (2013). Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30, 1229–1235. 10.1093/molbev/mst012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Purification scheme for Prefoldin:tubulin and TRiC:β-tubulin (B) Purified TRiC:β-tubulin or α-tubulin with SDS-PAGE and Native-PAGE analysis, immunoblot confirms β-tubulin or α-tubulin co-migrates with TRiC in native gel. (C) Purified Prefoldin:tubulin (β or α-tubulin) with SDS-PAGE and Native-PAGE analysis, note faster migration of Prefoldin:tubulin relative to Prefoldin alone in native gel. (D) α-tubulin folding assay. α-tubulin co-expressed and co-purified with Prefoldin comigrates on a native gel. In the absence of nucleotide α-tubulin comigrates with TRiC, and addition of nucleotide (ATP/GTP) drives folding and partial release of α-tubulin in the presence of TBCB. (E) Protease sensitivity assay (Proteinase K) of Prefoldin:tubulin samples; Fully folded α/β Tubulin dimer is insensitive to PK digestion, while Prefoldin:tubulin samples show rapid degradation by SDS-PAGE visualized by Coomassie stain (i) or immunoblot (ii). (F) Protease sensitivity assay of Prefoldin:α-tubulin folding by TRiC. Like β-tubulin, α-tubulin bound to Prefoldin is protease sensitive, transfer to TRiC from Prefoldin causes a decrease in sensitivity, and addition of nucleotide drives folding resulting in limited protease sensitivity equivalent to α-tubulin in a native α/β Tubulin dimer. (G-I) Protease sensitivity assay to test if TRiC closure is providing protease protection with degradation assayed by both an N-terminal region polyclonal antibody, and a C-terminal (AA2) monoclonal antibody. After completion of β-tubulin folding by TRiC as assayed in (H), apyrase is added to reopen the lid TRiC complex, followed by incubation at the indicated times. PK protease sensitivity is assayed at indicated time points (I). No increase in protease sensitivity is observed immediately after 30 min apyrase addition, the 90 min incubation leads to a mild increase in sensitivity likely due to slow unfolding.
(A) (i) Scheme of XL-MS analysis pipeline. XL of Prefoldin:tubulin (ii), Prefoldin:tubulin:TRiC (ii), and tubulin:TRiC (iii) samples were carried out in the indicated nucleotide condition and analysed by Native PAGE and SDS-PAGE as indicated, prior to Mass Spectrometry analysis. Native PAGE analysis shows XLed samples run as single complexes, while SDS-PAGE shows XLed samples migrate as high molecular weight multi-protein species due to inter-subunit XLing. (B) Bottom view of XLs mapped onto Prefoldin, note extensive XLs between PFD2 and PFD6. (C) Representative native mass spectrum of Prefoldin without β-tubulin. Two charge state distributions can be inferred to correspond to conformations that are relatively open and compact, represented by cartoons with more or fewer charges (+), the calculated solvent exposed surface area is listed next to the cartoons for open (PDB:6NRB11) and compact (PDB:6NR811) Prefoldin. The most abundant charge state of each conformation is labeled. (D) Mass to charge plot of deconvolved data in (C). A single mass, Prefoldin, gives rise to both peak distributions of charge. (E) Alternative representation of the native mass spectrometry data in main figure 2F, with charge plotted on the vertical axis and intensity as a color scale (right). Open and compact states of Prefoldin and Prefoldin:β-tubulin are outlined in blue and red respectively, with outline widths proportional to relative abundances. These data show a shift in conformational equilibrium upon β-tubulin binding. (F,G) Prefoldin structures alone (i) and with the top ring of TRiC (ii) from the least engaged (F) PDB:6NRB11 and most engaged (G) PDB:6NR811 Prefoldin:TRiC structures. The distance between the terminally resolved residues of PFD2 (Ile 124) and PFD6 (Glu 114) are indicated in magenta.
(A) Representative cryoEM micrograph and 2D averages of apo-TRiC and the complex of Prefoldin:β-tubulin-TRiC in the open conformation. (B) Image processing workflow of apo-TRiC and Prefoldin:β-tubulin-TRiC and Gold standard FSC curve (C). (D) Local resolution estimation of three reconstructed maps: apo-TRiC, TRiC:β-tubulin (Prefoldin unbound), Prefoldin:TRiC:β-tubulin. (E) Crosslink mapping on Prefoldin:β-tubulin bound to TRiC (F) Histogram of shortest measured intra/interring Cα -Cα distances for identified XLs between Prefoldin and TRiC.
(A) Representative micrograph and 2D class averages of TRiC:β-tubulin with ATP-AlFx. (B) Processing workflow of TRiC:β-tubulin with ATP-AlFx and (C) 3D classification strategy for focusing the density inner chamber of TRiC. Our compositional heterogeneity analysis identifies different β-tubulin occupancies for TRiC, including no substrate bound (apo state), and single or double ring occupancy. Further analysis identified four progressively folded tubulin conformations inside the chamber: State I (41% of the rings); State II (16% of the rings); State III (34% of the rings) and State IV (9% of the rings) corresponding to folded tubulin. (D) Local resolution map estimation of β-tubulin intermediate states. (E) FSC curve of reconstructed maps showing resolution according to the gold standard FSC (0.143 criterion). (F) Q-score plots of the β-tubulin intermediate states shown with the expected Q-score according to 3.55 and 3.00 Å. (G) C-alpha backbone RMSD per residue between intermediate state and native β-tubulin (PDB: 6I2I36). Large variation indicated at T3 loop (red star) and N loop (yellow hexagon)
(A) cryoEM density of β-tubulin E hook tail in the CCT1/4 pocket for the different folding states (I-IV). (B) cryoEM density of CCT2 C terminal tail connected to β-tubulin for the different folding states (I-IV). (C) cryoEM density of the β-tubulin at the GTP binding pocket: T3 loop (red star), N loop (yellow hexagon), nucleotide (magenta eclipse). (D) Difference map analysis of the nucleotide occupancy in β-tubulin GTP binding pocket in comparison to CCT ATP binding pockets. The difference maps are displayed with +3σ. (E) Slice side and top view of CCT1/2 C terminal tail contacts in apo closed TRiC. (F) Electrostatic surface display of human α-tubulin (PDB: 6I2I36), β-tubulin (PDB: this study) and γ-tubulin (1Z5V61), respectively. (I) Logo plot of the conservation of CCT residues. The residues making direct contact with β-tubulin are indicated by red arrows
(A) Sequence alignment for selected areas of human α-, β-, γ-tubulins and archaeal/prokaryotic homologues (CetZ1: H. Volcanii CetZ2: H. Volcanii, FtsZ: M. jannaschii and TubZ: B.cereus). T3 loop, GTP binding motif, N/C-domain interface and C-domain including H12 and E-hook are represented. Red box indicates conserved N99 and L189 in T3 loop and N/C-domain interface in human tubulins. (B) Phylogenetic tree of tubulins and their structures 36,48,61–63. Structures are colored in domain (N-domain: cyan, C-domain: green, Core domain: blue, M-domain: red). The scale bar represents the number of amino acids substitutions per site. (C) The hydrophobic core and domain interfaces of β-tubulin and FtsZ with hydrophobic residues (Trp, Phe, Ile, Leu) highlighted with yellow (β-tubulin) and gray (FtsZ) balls. Red arrow indicates the N/C-domain interface. Blue arrows and dashed line indicate the C/Core-domain interface. Colored circles represent hydrophobic core of each domain. The progressive assembly of the hydrophobic cores for N-, C- and Core domain of β-tubulin (bottom panels). (D) Model B factor distribution of each state highlights the N-domain with T3 loop in red dashed box (i) and corresponding cryo-EM density maps with fitted model are presented (ii). Per residue RMSD plots of T3 loop among four states (iii) and Q-score plot of T3 loop (iv). Dashed black line indicates average Q score expected at 3.5 Å resolution. (E) β-tubulin model with point mutations at the GTP binding pocket indicated as balls on residues that have folding defects. Their locations and effects are summarized55,64,65.
This movie demonstrates the local density of three β-tubulin intermediates and fully folded states. The model of the folded β-tubulin domain is fitted into each density map and displayed. GTP is fitted into the GTP binding pocket as an example to illustrate the observed extra density in State III and IV.
This movie illustrates β-tubulin N-, C-domain interaction with the CCT1/3/6/8 by complementary charged surface.
The first part of the movie illustrates that TRiC orients hydrophobic surface of β-tubulin N-, C-domain to face the chamber center and allows the subsequent hydrophobic burial with Core helix and M-domain folding. The second part of the movie demonstrates the progressive β-sheet building along with continuously significant hydrophobic framing from helices as β-tubulin folds.
The movie shows the model trajectory from state I to the other states highlighting a conformational change of T3 loop in β-tubulin.
List of inter/intra-molecular crosslinks between PFD, α/β-tubulin and TRiC in indicated conditions.
fileFASTA file with alignment of tubulin homologues across evolution. Related to Figure 6.
Data Availability Statement
The 3D cryoEM density maps have been deposited in the Electron Microscopy Data Bank under the accession number EMD: EMD-32822, 32823, 26089, 26120, 26123, 26131. Coordinates have been deposited in the Protein Data Bank under the accession number PDB: 7WU7, 7TRG, 7TTN, 7TTT, 7TUB. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD030590. This paper does not report the original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


