Text
Although vaccinations and SARS-CoV-2 infections have resulted in a high level of antibody-positive individuals SARS-CoV-2 omicron variants continue to spread, including with high infection rates in triple-vaccinated individuals.1 The first waves of SARS-CoV-2 infections were caused by distinct variants of concern (VOC), each dominating until being replaced by a novel VOC. Since the emergence of omicron in late 2021, new subvariants exhibiting increased capacity for immune escape have emerged.2,3 Currently, various BA.2/4/5 sublineages circulate in the population, strongly dominated by BA.5.
Recently, sequencing data has revealed the emergence of new circulating omicron variants in e.g. the United Kingdom,4 with indication of accelerated convergent evolution4,5 resulting in shared phenotypes between variants. This marks a possible shift in the transmission landscape; rather than one specific sublineage dominating, several distinct subvariants with a specific phenotype may together form a dominating cluster. One of such important mutations is R346X, in particular R346T, situated in the receptor binding domain (RBD) of the spike protein, with an enhanced capacity to escape neutralizing antibodies.5 Current sequencing data is largely generated from group-specific testing, possibly delaying detection of novel variants and transmission patterns in society.4
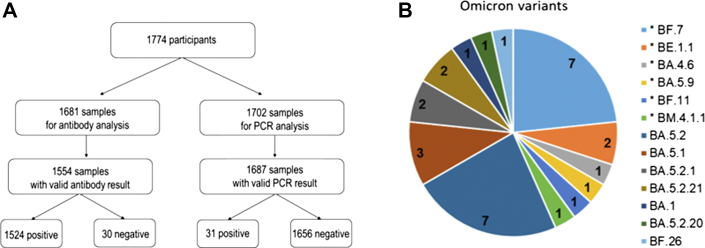
To estimate the prevalence of SARS-CoV-2 infection and to identify variants causing them, we invited individuals being part of a Swedish nationwide probability-based web panel to participate in a survey taking place September 26–29, 2022. Enrolled participants (n = 1774) received material for self-sampling of upper respiratory tract for PCR-analysis for presence of SARS-CoV-2 and of blood for serological analysis (Fig. 1A, Supplementary Appendix for methods).
Fig. 1.
High frequency of circulating R346X mutated omicron-variants in circulation late September, 2022, Sweden. (A) Overview of enrolled study participants, collected and analyzed samples. (B) Variants detected (pangolin classification) in the PCR-screening. Of the 31 PCR-positive samples, 30 could be sequenced. In total 13 different omicron sublineages was detected. Variants with R346X-mutations are marked with an asterix.
In total, 1524 of 1554 participants were positive for anti-spike IgG. Ongoing SARS-CoV-2-infection was detected in 31 out of 1687 participants, resulting in an age-, sex- and region-adjusted estimated overall point-prevalence of 1·5% (95% CI: 0·9–2·5%). Of the 31 positive samples, 30 were successfully sequenced. In total, 13 different omicron sublineages were identified, among which BA.5.2 (n = 7) and BF.7 (n = 7) were the most common ones (Fig. 1B, Table S1). Strikingly, 40% of infections were caused by R346X-mutated variants represented by six different sublineages. Moreover, five of these sublineages showed an identical pattern of RBD mutations (Table S2).
To conclude, these results show a rapid emergence of several R346T expressing sublineages with identical or very similar RBD, indicating that the next wave of SARS-CoV-2 infections may be caused by a group of sublineages sharing a phenotype, rather than by one specific sublineage.
Contributors
Literature search: R.G., I.G., J.K., K.B.
Figures: R.G., I.G., J.K., K.B.
Study design: R.G., I.G., P.B., M.R., A.B., J.K., K.B.
Data collection: R.G., I.G., K.S., M.S., E.M., P.B., T.E., L.P., M.R., V.S., S.S., K.V.A., S.Z., M.L.K., J.K., K.B.
Data analysis: R.G., I.G., K.V.A, J.K., K.B.
Data interpretation R.G., I.G., J.K., K.B.
Writing – original draft: R.G., I.G., J.K., K.B.
Writing – review & editing: All authors.
Data sharing statement
The datasets that support the findings of this study are not publicly available, but are available from the corresponding author upon reasonable request.
Declaration of interests
All authors have no conflicts of interest related to this work.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lanepe.2022.100564.
Appendix A. Supplementary data
References
- 1.Blom K., Havervall S., Marking S., et al. Infection rate of SARS-CoV-2 in asymptomatic healthcare workers, Sweden, June 2022. Emerg Infect Dis. 2022;28:2119–2121. doi: 10.3201/eid2810.221093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Viana R., Moyo S., Amoako D.G., et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hachmann N.P., Miller J., Collier A.-R.Y., et al. Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N Engl J Med. 2022;387:86–88. doi: 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1109820/Technical-Briefing-46.pdf. 2022.
- 5.Cao Y., Jian F., Wang J., et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. bioRxiv. 2022 doi: 10.1101/2022.09.15.507787. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.