Abstract
Background
The coronavirus disease 2019 (COVID‐19) pandemic has had an unprecedented impact on the healthcare system, economy, and society. Studies have reported that COVID‐19 may cause various neurologic symptoms, including cognitive impairment. We aimed to assess the causal effect of COVID‐19 on neurodegenerative diseases using two‐sample Mendelian randomization (MR) study.
Methods
Genetic variants were obtained from genome‐wide association studies (GWAS) summary‐level data and meta‐analyses. We used the inverse variance–weighted (IVW) method as the primary analysis to estimate causal effects. Sensitivity analyses were performed to make the conclusions more robust and reliable.
Results
We found that the COVID‐19 infection phenotype was associated with a higher risk of AD and inverse associated with the risk of ALS and MS. The hospitalized COVID‐19 phenotype was associated with the risk of AD and wasn't associated with ALS and MS. We also found that the severe COVID‐19 (main analysis) phenotype was associated with the AD outcome from UK biobank datasets but was not associated with other outcomes. The severe COVID‐19 infection phenotype, the severe COVID‐19 (subtype analysis) phenotype and the hospitalization risk of COVID‐19 were not associated with each outcome.
Conclusion
This MR study suggests a potential association between genetically predicted COVID‐19 and a higher risk of AD and a reduced risk of ALS and MS. Further elucidations of this association and underlying mechanisms may inform public health messages to prevent COVID‐19 and AD.
Background
The coronavirus disease 2019 (COVID‐19), caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has been a global pandemic and resulted in substantial morbidity and mortality, especially among the elderly. 1 , 2 Due to its rapid spread and lack of effective therapy methods, COVID‐19 has brought an enormous economic burden globally. Although the typical symptoms of COVID‐19 are respiratory complications, there is growing evidence that SARS‐CoV‐2 infection involves central nervous system (CNS) damage. 3 Recent studies have revealed that patients infected with SARS‐CoV‐2 showed shrinkage in brain size, cognitive decline, and damage to brain regions related to smell. 4 In addition, studies have observed the CNS invasion of SARS‐CoV‐2 in the postmortem brain of COVID‐19 patients and animal models. 5 , 6 However, the long‐term impact on neurodegenerative diseases during this pandemic is unclear. Alzheimer's disease (AD), a common neurodegenerative disease mainly affecting the elderly, is characterized by cognitive decline and brain degeneration. 7 , 8 The pathogenesis of AD is still unclear. Prevalent theories include the amyloid hypothesis, the tau hypothesis, and the neuroinflammation hypothesis. Additionally, previous studies have shown that specific viral infections could impact the neuropathology of AD. 9 AD patients depend solely on caregivers and family members in the middle‐late stage. 10 As the COVID‐19 pandemic requires isolation and quarantine management, it adds an extra burden on AD patients, caregivers, and families. Amyotrophic lateral sclerosis (ALS) is an idiopathic and fatal neurodegenerative disease characterized by the degeneration of both upper and lower motor neurons. 11 The typical symptoms of ALS are related to motor dysfunction (such as muscle weakness, spasticity, respiratory failure, and dysphagia). 12 , 13 Also, cognitive and behavioral impairment is a vital feature of ALS. 14 During the COVID‐19 pandemic, isolation management has challenged the diagnosis, clinical care, and outpatient follow‐up visits for ALS patients. 15 Moreover, a recent study indicates COVID‐19 may accelerate the disease progression in ALS patients. 16 Multiple sclerosis (MS) is a chronic autoimmune neurodegenerative disease characterized by selective primary demyelination, axonal damage, and reactive astrocytic gliosis. 17 Clinically, MS displays muscle weakness, sensory loss, cognitive impairment, and fatigue. 18 As the COVID‐19 pandemic continues, SARS‐CoV‐2 infection poses a particularly concerning threat to MS patients, especially those with disease‐modifying therapies (DMTs), which may increase the risk of infections. Furthermore, recent studies have assessed the safety and response of the COVID‐19 vaccine in MS patients. 19 , 20
To date, researchers have been concerned about the correlation between COVID‐19 and neurodegenerative diseases, 21 , 22 but accurate data on SARS‐CoV‐2 infection in neurodegenerative disease patients are unavailable. Also, it is limited in observational studies to investigate causality because of the possible bias due to confounding and reverse causation. Mendelian randomization (MR) study can minimize these biases by using genetic variants as instrumental variables (IVs) to evaluate the causal relationship between exposure (COVID‐19) and outcome (AD, ALS, and MS). 23 , 24 Here, we used the MR method to assess the causal effect of COVID‐19 on the risk of AD, ALS, and MS (Fig. 1).
Figure 1.
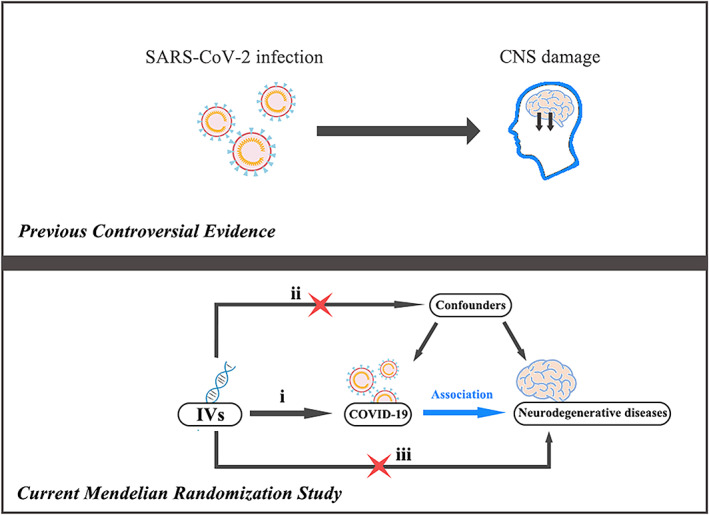
(Top) Previous observational studies have found that patients infected with SARS‐CoV‐2 may experience central nervous system (CNS) damage. (Bottom) Our 2‐sample MR analyses show a causal role of COVID‐19 on the neurodegenerative diseases. MR study relies on three assumptions: (i) the instrumental variables (IVs) should be associated with the exposure (COVID‐19). (ii) the IVs should not be related to confounders. (iii) the IVs should influence the outcome (NDs) risk via the exposure, not through other pathways.
Methods
Selection of instrumental variables
IVs from summary data
Summary data of COVID‐19 phenotypes were obtained from recent Genome‐Wide Association Studies (GWAS) by the COVID‐19 host genetics initiative (RELEASE 5). 25 We used genetic variants associated with three different COVID‐19 phenotypes in individuals of European ancestry (1) COVID‐19 infection, (2) hospitalized COVID‐19 (hospitalized vs. population), and (3) severe COVID‐19 infection (very severe respiratory confirmed vs. population). As genetic IVs, single‐nucleotide polymorphisms (SNPs) were selected with genome‐wide significance (5 × 10−8). We also calculated the proportion of variance explained by each of the genetic instruments using the following formula: R 2 = 2 × EAF × (1‐EAF) × (Beta2)/SD, where Beta is the beta coefficient for the association of the IVs with the exposure, SD is the variance of the exposure, and EAF is the effect allele frequency of the IVs. Then, SNPs were not in linkage disequilibrium (LD, r 2 > 0.001) and had an F statistic greater than 10. Finally, ambiguous and palindromic SNPs were removed in the harmonizing process.
IVs from meta‐analysis
We also obtained genetic IVs associated with three COVID‐19 phenotypes from two genome‐wide association meta‐analyses 26 , 27 of European ancestry: (1) severe COVID‐19 (main analysis, hospitalization with respiratory support), (2) severe COVID‐19 (subtype analysis, hospitalization with mechanical ventilation), and (3) hospitalization risk with COVID‐19. Information on SNPs used as IVs is presented in Table S1 and S2.
Outcome sources
Summary‐level data for AD were obtained from a large genome‐wide meta‐analysis contributed by the Alzheimer's disease working group of the Psychiatric Genomics Consortium (PGC‐ALZ), the International Genomics of Alzheimer's Project (IGAP), the Alzheimer's Disease Sequencing Project (ADSP), and UK biobank, including clinically diagnosed AD and AD‐by‐proxy of European ancestry. 28 Then, we validated our results using recently published GWAS summary‐level data, including 954 AD cases and 487,331 controls from the UK Biobank. 29 The details of the data were described in the original study. Additionally, summary‐level data for ALS were obtained from a recent GWAS, including 20,806 ALS patients diagnosed with ALS and 59,804 neurologically normal control individuals of European ancestry. 30 Furthermore, GWAS summary‐level data for MS outcome was obtained from the International Multiple Sclerosis Genetics Consortium with a sample size of 47,429 multiple cases and 68,374 control subjects. 31
MR analysis
We used multiplicative random‐effects inverse‐variance weighted (IVW‐RE) as the primary analysis to estimate the causal effect. A Bonferroni‐corrected significance p‐value threshold of 0.05/9 = 0.0056 was used to correct multiple comparisons of exposures. A significant association was suggested for p < 0.0056, and a suggestive association for p < 0.05. As sensitivity analyses, we also used MR Egger, 32 weighted median (WM), 33 and the MR‐Robust Adjusted Profile Score (MR‐RAPS) 34 methods to strengthen causal evidence. In addition, Mendelian Randomization Pleiotropy RESidual Sum and Outlier (MR‐PRESSO) 35 and Cochrane's Q test 36 were conducted to assess heterogeneity, and the MR‐Egger intercept was used to evaluate directional pleiotropy. Then, we performed MR‐Steiger to determine the direction of the causal effect. 37 Power calculations were done for the MR study using online tools at http://cnsgenomics.com/shiny/mRnd/. 38
All analyses were performed using R Version 4.1.2 with the R package “TwosampleMR,” “MendelianRandomization,” and “MRPRESSO.” 35 , 39 , 40 All the data used for the current study are publicly available.
Results
The MR estimates from different methods of assessing the causal effect of COVID‐19 on neurodegenerative diseases are presented in Table S3 and Figures 2, 3, 4. The results of power calculations are presented in Table S4.
Figure 2.
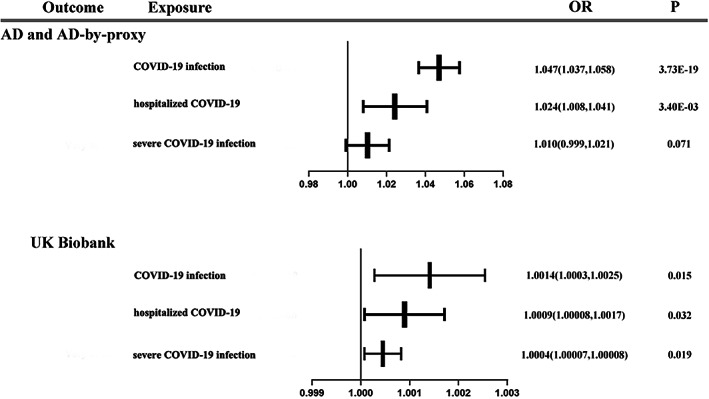
Forest plot of primary analysis for association between each phenotype of COVID‐19 and AD.
Figure 3.
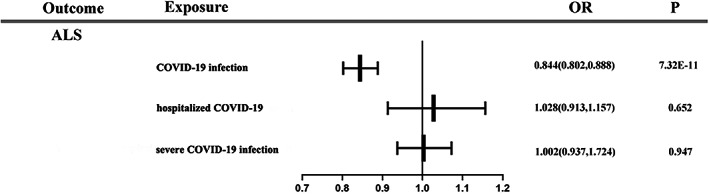
Forest plot of primary analysis for association between each phenotype of COVID‐19 and ALS.
Figure 4.
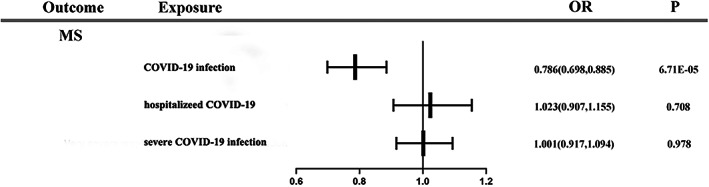
Forest plot of primary analysis for association between each phenotype of COVID‐19 and MS.
COVID‐19 and AD
For the AD and AD‐by‐proxy outcome, we found a significant association between the COVID‐19 infection phenotype (ORIVW‐RE = 1.047 [95%CI: 1.037, 1.058]; P = 3.73 E‐19) and a higher risk of AD. The association was consistent in fixed‐effects IVW (IVW‐FE), WM, MR‐PRAS, and MR‐PRESSO but inconsistent in MR‐Egger. We also found that the hospitalized COVID‐19 phenotype (OR IVW‐RE = 1.024 [95%CI: 1.008, 1.041]; p = 0.0034) was associated with the risk of AD. A consistent association was observed in IVW‐FE and MR‐RAPS but inconsistent in WM, MR‐Egger, and MR‐PRESSO. There was no association between the severe COVID‐19 infection phenotype (OR IVW‐RE = 1.011 [95%CI: 0.999, 1.021]; p = 0.071) and the risk of AD. The null association was consistent in sensitivity analyses. Additionally, using IVs from meta‐analyses, no significant association was found between the severe COVID‐19 (main analysis and subtype analysis) phenotypes (OR IVW‐RE (main analysis) = 1.012 [95%CI: 0.998, 1.025]; p = 0.088, OR IVW‐RE (subtype analysis) = 1.012 [95%CI: 0.998, 1.025]; p = 0.088) and the hospitalization risk with COVID‐19 (OR IVW‐RE = 1.006 [95%CI: 0.998, 1.014]; p = 0.152) and the risk of AD.
For the UK Biobank database outcome, we found that the COVID‐19 infection phenotype (OR IVW‐RE = 1.0014 [95%CI: 1.0003, 1.0025]; p = 0.015) was associated with the risk of AD. The association was consistent in IVW‐FE, WM, and MR‐RAPS but inconsistent in MR‐Egger and MR‐PRESSO. Then, suggestive evidence was observed in the hospitalized COVID‐19 phenotype (OR IVW‐RE = 1.0009 [95%CI: 1.00008, 1.0017]; p = 0.032). The association was consistent in IVW‐FE, WM and MR‐RAPS but inconsistent in MR‐Egger and MR‐PRESSO. We also found suggestive evidence that the severe COVID‐19 infection phenotype (OR IVW‐RE = 1.0004 [95%CI: 1.00007, 1.00082]; p = 0.019) was associated with the risk of AD. However, the association was inconsistent in sensitivity analyses. The result of IVW‐RE showed a suggestive association between the severe COVID‐19 (main analysis) phenotype (OR IVW‐RE = 1.0005 [95%CI: 1.0001, 1.0008]; p = 0.010) and higher risk of AD. Estimates were similar in IVW‐FE and MR‐RAPS analyses. Using IVW‐RE method, we found a suggestive association between the severe COVID‐19 (subtype analysis) phenotype (OR IVW‐RE = 1.0003 [95%CI: 1.00002, 1.0006]; p = 0.037) and the risk of AD. However, the association was inconsistent in sensitivity analyses. The result of IVW‐RE showed an inverse association between the hospitalization risk with COVID‐19 (OR IVW‐RE = 0.9996 [95%CI: 0.9994, 0.9998]; p = 9.15 E‐06) and the risk of AD. But the result was inconsistent in sensitivity analyses.
COVID‐19 and ALS
An inversely association was observed between the COVID‐19 infection phenotype (ORIVW‐RE = 0.844 [95%CI: 0.802, 0.888]; p = 7.23 E‐11) and the risk of ALS. The result was consistent in IVW‐FE, WM, and MR‐PRESSO analyses but inconsistent in MR‐RAPS and MR‐Egger methods. We found that the hospitalized COVID‐19 phenotype (OR IVW‐RE = 1.028 [95%CI: 0.913, 1.157]; p = 0.652), the severe COVID‐19 infection phenotype (OR IVW‐RE = 1.002 [95%CI: 0.937, 1.072]; p = 0.947) and the hospitalization risk with COVID‐19 (OR IVW‐RE = 0.972 [95%CI: 0.861, 1.097]; p = 0.644) were not associated with the risk of ALS. The null association was consistent in sensitivity analyses. Using IVW‐RE method, we found that the severe COVID‐19 (main analysis and subtype analysis) phenotypes (OR IVW‐RE (main analysis) = 0.943 [95%CI: 0.935, 0.951]; p = 1.13 E‐39, OR IVW‐RE (subtype analysis) = 0.963 [95%CI: 0.944, 0.981]; p = 1.00 E‐04) were associated with the risk of ALS. However, the association was inconsistent in supplementary sensitivity analyses.
COVID‐19 and MS
An inversely association was observed between the COVID‐19 infection phenotype (ORIVW‐RE = 0.786 [95%CI: 0.698, 0.885]; p = 6.71 E‐05) and the risk of MS. The association was consistent in IVW‐FE, MR‐RAPS, and MR‐PRESSO methods but inconsistent in WM and MR‐Egger. We did not find that the hospitalized COVID‐19 phenotype (OR IVW‐RE = 1.023 [95%CI: 0.907, 1.155]; p = 0.708), the severe COVID‐19 infection phenotype (OR IVW‐RE = 1.001 [95%CI: 0.917, 1.094]; p = 0.978), the severe COVID‐19 (subtype analysis) phenotype (OR IVW‐RE = 0.923 [95%CI: 0.818, 1.042]; p = 0.194) and the hospitalization risk with COVID‐19 (OR IVW‐RE = 1.010 [95%CI: 0.943, 1.083]; p = 0.769) were associated with the risk of MS. The null association was consistent in the sensitivity analyses. There was a suggestive association between the severe COVID‐19 (main analysis) phenotype (OR IVW‐RE = 0.863 [95%CI: 0.753, 0.989]; p = 0.034) and the risk of MS. Estimates were consistent in the IVW‐FE, WM, and MR‐RAPS analyses, but inconsistent in MR‐Egger. However, the result of the Q test suggested heterogeneity across instrument SNP effects (p = 0.039).
No directional pleiotropy or heterogeneity was detected in the current study by Cochrane's Q test, MR‐Egger intercept test, and MR‐PRESSO global test.
Discussion
To the best of our knowledge, this is the first MR study to assess the causal relationship between COVID‐19 and neurodegenerative diseases. Our findings indicate that genetically predicted COVID‐19 may contribute to the higher risk of AD and may be inversely associated with the risk of ALS and MS. The sensitivity analyses were overall consistent with the primary analysis. However, there was no evidence that very severe respiratory confirmed COVID‐19 may be associated with AD, ALS, and MS risk.
MR design rests on three key assumptions: (1) the genetic variants are robustly associated with the exposure; (2) the genetic variants are not associated with other confounders of the exposure‐outcome association; and (3) the genetic variants are associated with the outcome only through the investigated exposure. 41 For the first assumption, we have selected SNPs with genome‐wide significance (5 × 10−8) used as IVs from summary data. Then, F statistics for every IV were greater than 10, indicating the small possibility of weak instrumental variable bias. The second and third assumptions might be violated if a genetic variant used as an IV is also associated with other outcome risk factors. Pleiotropy refers to genetic variants associated with multiple risk factors, which may lead to biased effect estimates for MR analysis. However, the MR‐Egger regression and MR‐PRESSO global test showed no evidence of pleiotropic effects in the current analysis. Furthermore, according to the existing knowledge, there is no obvious evidence that SNPs in our study affect each outcome through other pathways, demonstrating the validity of our MR analysis. Besides, violation of MR assumptions may also occur in population stratification that the population under investigation is a mixture of individuals of different ethnic origins. Thus, we restricted the study population to European ancestry to alleviate the bias from population stratification.
Studies have shown the negative influence of SARS‐CoV‐2 infection on cognitive function. A retrospective observational study in China that included 214 hospitalized patients with COVID‐19 found that 14.8% had impaired consciousness. 42 Data from 50 hospitalized patients with COVID‐19 from a retrospective study in Chicago, United States, showed that 24% had short‐term memory loss. 43 In a cross‐specialty surveillance study, 6 of 23 patients with altered mental status showed a neurocognitive (dementia‐like) syndrome. 44 In an observational study in Strasbourg, France, 26 of 40 patients were shown to have confusion according to the Confusion Assessment Method for the intensive care unit (ICU). 45 A recently published study showed that risks of cognitive deficit and dementia were still increased after a 2‐years follow‐up period. 46 Given this evidence, we speculate that SARS‐CoV‐2 infection might contribute to the initiation or acceleration of neurodegenerative diseases.
We found evidence that COVID‐19 was associated with a higher risk of AD. Studies have demonstrated that the COVID‐19 virus may access the CNS via the indirect hematogenous or direct neural route, 47 which could contribute to neurological complications. A recent 3D microfluidic model study has found that the SARS‐CoV‐2 protein destabilizes the blood–brain barrier (BBB) and triggers a pro‐inflammatory response in brain endothelial cells. 48 Moreover, patients with COVID‐19 have shown an inflammatory response and a rise in systemic cytokine levels. Inflammation has been proven to impact cognitive function and contribute to neurodegenerative progression. 49 Furthermore, SARS‐CoV‐2 can directly activate the NOD‐, LRR‐, and pyrin domain‐containing protein 3 (NLRP3) inflammasome, 50 which is involved in the changes of amyloid‐beta (Aβ) deposition in the brain. 51 In addition, a recent genotyping analysis revealed an SNP in oligoadenylate synthetase 1 (OAS1) linked to AD in the same locus that predisposes to COVID‐19‐related critical illness. 52 The biological mechanisms underlying the association between COVID‐19 and the risk of AD remain to be disclosed in longitudinal and multifaceted studies. SARS‐CoV‐2 infections might be involved in AD pathogenesis in complex ways. We speculated that CNS damage might have ongoing activity well past the acute infection despite the short duration of the COVID‐19 infection. Moreover, it is possible that there are shared genetic factors that affect the diseases independently.
Interestingly, we found evidence that COVID‐19 was inversely associated with the risk of ALS. A recent study showed that the COVID‐19 pandemic might accelerate the early progression of ALS. 15 However, studies on the impact of COVID‐19 on ALS are still limited. The underlying mechanisms of COVID‐19 in ALS are unclear. We speculated that establishing protective immunity after SARS‐CoV‐2 infection might reduce the risk of ALS. However, this remains an area of future study to investigate the exact impacts of COVID‐19 on ALS.
Similarly, we found evidence that COVID‐19 was inversely associated with the risk of MS. COVID‐19 pandemic poses a challenge to MS patients concerning their risk for SARS‐CoV‐2 infection and guiding disease‐modifying treatment (DMT). Studies have proved the safety of the COVID‐19 vaccine for MS patients, including patients with DMT. 19 , 20 Also, the underlying effects of COVID‐19 on MS are worth further studies.
Due to the COVID‐19 pandemic having been ongoing for <4 years, it is difficult to identify the long‐term sequela of SARS‐CoV‐2 infection. Although previous studies have shown that SARS‐CoV‐2 infection impaired cognitive function, indicating that COVID‐19 may play a potential role in the risk of neurodegenerative diseases, there is still insufficient evidence to support the causal association. Using genetic variants as IVs, the MR approach could overcome the limitations of traditional observational studies. Our MR analysis suggests that COVID‐19 may increase the risk of AD and may be inversely associated with the risk of ALS and MS. These findings may provide evidence for the long‐term consequence of COVID‐19 and pivotal information for public health and prevention guidelines. However, further studies still need to be conducted to clarify this association in the future.
Strength and Limitation
The main strength of this study is the MR design using genetic variants to proxy COVID‐19 exposure, which could reduce typical bias for observational research. Moreover, selection bias could occur when considering genetic associations with a disease outcome in an elderly population, as a participant can be recruited if they have survived to old age. 53 The current MR design uses the largest GWAS data, including clinically diagnosed AD and AD‐by‐proxy participants could avoid selection bias, as the diagnosed cause of death is unlikely to influence whether proxy data are available for analysis. In addition, we used 2 separate sets of AD outcome data to validate our findings. However, our study also has several limitations. First, the SARS‐CoV‐2 virus identified at the end of 2019 has evolved and emerged with various variants. There are no available published GWAS data for these variants, so we were not able to explore the association between COVID‐19 variants and neurodegenerative diseases. Second, horizontal pleiotropy cannot be completely expelled, although we have conducted various sensitivity analyses to minimize the impact of pleiotropy. Third, the summary GWAS data used in the current study was merely obtained from European population ancestry, so our results should be interpreted with caution when generalizing to the whole population. Fourth, we could not estimate the overlap of participants between exposure and outcome studies. However, the use of strong instruments can minimize bias from sample overlap. 54 Fifth, sensitivity analysis based on different assumptions may increase the possibility of getting inconsistent or contrary results, leading to obscured conclusions. Nonetheless, each method has its strengths. The IVW method assumes that all SNPs are valid IVs or are invalid in such a way that the overall bias is zero, and this method has been regarded as the most efficient analysis method. 55 Therefore, we used the IVW as the primary analysis method.
Conclusion
In conclusion, our MR results suggest a positive association between genetically predicted COVID‐19 and a higher risk of AD and an inverse association between genetically predicted COVID‐19 and the risk of ALS and MS. During the COVID‐19 pandemic, our research may contribute to predictions for the prevention, diagnosis, and treatment of neurodegenerative diseases. Further studies are needed to provide more evidence of our findings and explore the underlying biological processes.
Conflict of Interest
The authors declare no competing interests.
Authors' Contributions
Both authors designed the study, analyzed the data, interpreted the results, and wrote the manuscript. Both authors read and approved the final manuscript.
Consent for Publication
Not applicable.
Supporting information
Table S1. IVs from summary data.
Table S2. IVs from meta‐analysis.
Table S3. Results of primary analysis and sensitivity analyses.
Table S4. Power calculations in the current MR study.
Acknowledgements
We gratefully thank all the participants and researchers who contributed to the GWAS summary statistics and meta‐analyses in the original studies.
Funding InformationThis research did not receive any specific grant from the public, commercial, or nonprofit funding agencies.
References
- 1. Li H, Liu S‐M, Yu X‐H, Tang S‐L, Tang C‐K. Coronavirus disease 2019 (COVID‐19): current status and future perspectives. Int J Antimicrob Agents. 2020;55(5):105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O'Driscoll M, Ribeiro Dos Santos G, Wang L, et al. Age‐specific mortality and immunity patterns of SARS‐CoV‐2. Nature. 2021;590(7844):140‐145. [DOI] [PubMed] [Google Scholar]
- 3. Paterson RW, Brown RL, Benjamin L, et al. The emerging spectrum of COVID‐19 neurology: clinical, radiological and laboratory findings. Brain. 2020;143(10):3104‐3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Douaud G, Lee S, Alfaro‐Almagro F, et al. SARS‐CoV‐2 is associated with changes in brain structure in UK biobank. Nature. 2022;604(7907):697‐707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matschke J, Lütgehetmann M, Hagel C, et al. Neuropathology of patients with COVID‐19 in Germany: a post‐mortem case series. Lancet Neurol. 2020;19(11):919‐929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Song E, Zhang C, Israelow B, et al. Neuroinvasion of SARS‐CoV‐2 in human and mouse brain. J Exp Med. 2021;218(3):e20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Desikan RS, Cabral HJ, Hess CP, et al. Automated MRI measures identify individuals with mild cognitive impairment and Alzheimer's disease. Brain. 2009;132(Pt 8):2048‐2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wenk GL. Neuropathologic changes in Alzheimer's disease. J Clin Psychiatry. 2003;64(Suppl):9. [PubMed] [Google Scholar]
- 9. Readhead B, Haure‐Mirande J‐V, Funk CC, et al. Multiscale analysis of independent Alzheimer's cohorts finds disruption of molecular, genetic, and clinical networks by human herpesvirus. Neuron. 2018;99(1):64‐82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hardy JA, Higgins GA. Alzheimer's disease: the amyloid cascade hypothesis. Science. 1992;256(5054):184‐185. [DOI] [PubMed] [Google Scholar]
- 11. Hardiman O, Al‐Chalabi A, Chio A, et al. Amyotrophic lateral sclerosis. Nat Rev Dis Primers. 2017;3(1):17071. [DOI] [PubMed] [Google Scholar]
- 12. Kiernan MC, Vucic S, Cheah BC, et al. Amyotrophic lateral sclerosis. Lancet. 2011;377(9769):942‐955. [DOI] [PubMed] [Google Scholar]
- 13. Brown RH, Al‐Chalabi A. Amyotrophic lateral sclerosis. N Engl J Med. 2017;377(2):162‐172. [DOI] [PubMed] [Google Scholar]
- 14. Pender N, Pinto‐Grau M, Hardiman O. Cognitive and behavioural impairment in amyotrophic lateral sclerosis. Curr Opin Neurol. 2020;33(5):649‐654. [DOI] [PubMed] [Google Scholar]
- 15. De Marchi F, Gallo C, Sarnelli MF, et al. Accelerated early progression of amyotrophic lateral sclerosis over the COVID‐19 pandemic. Brain Sci. 2021;11(10):1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li X, Bedlack R. COVID‐19‐accelerated disease progression in two patients with amyotrophic lateral sclerosis. Muscle Nerve. 2021;64(3):E13‐e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lassmann H. Multiple sclerosis pathology and its reflection by imaging technologies: introduction. Brain Pathol. 2018;28(5):721‐722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Multiple sclerosis. Nat Rev Dis Primers 2018;4(1):44. [DOI] [PubMed] [Google Scholar]
- 19. Tallantyre EC, Vickaryous N, Anderson V, et al. COVID‐19 vaccine response in people with multiple sclerosis. Ann Neurol. 2022;91(1):89‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Achiron A, Dolev M, Menascu S, et al. COVID‐19 vaccination in patients with multiple sclerosis: what we have learnt by February 2021. Mult Scler. 2021;27(6):864‐870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xia X, Wang Y, Zheng J. COVID‐19 and Alzheimer's disease: how one crisis worsens the other. Transl Neurodegener. 2021;10(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu C, Chen C, Dong XP. Impact of COVID‐19 pandemic on patients with neurodegenerative diseases. Front Aging Neurosci. 2021;13:664965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Davies NM, Holmes MV, Davey SG. Reading mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ. 2018;362:k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burgess S, Timpson NJ, Ebrahim S, Davey SG. Mendelian randomization: where are we now and where are we going? Int J Epidemiol. 2015;44(2):379‐388. [DOI] [PubMed] [Google Scholar]
- 25. The COVID‐19 host genetics initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS‐CoV‐2 virus pandemic. Eur J Hum Genet. 2020;28(6):715‐718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Degenhardt F, Ellinghaus D, Juzenas S, et al. Detailed stratified GWAS analysis for severe COVID‐19 in four European populations. Hum Mol Genet. 2022. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cruz R, Almeida SD‐D, Heredia ML, et al. Novel genes and sex differences in COVID‐19 severity. Hum Mol Genet. 2022. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jansen IE, Savage JE, Watanabe K, et al. Genome‐wide meta‐analysis identifies new loci and functional pathways influencing Alzheimer's disease risk. Nat Genet. 2019;51(3):404‐413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Larsson SC, Woolf B, Gill D. Plasma caffeine levels and risk of Alzheimer's disease and Parkinson's disease: mendelian randomization study. Nutrients. 2022;14(9):1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nicolas A, Kenna KP, Renton AE, et al. Genome‐wide analyses identify KIF5A as a novel ALS gene. Neuron. 2018;97(6):1268‐83.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 2019;365(6460):eaav7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int J Epidemiol. 2015;44(2):512‐525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304‐314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao Q, Wang J, Hemani G, Bowden J, Small DS. Statistical inference in two‐sample summary‐data mendelian randomization using robust adjusted profile score. Ann Stat. 2020;48(3):1742‐1769. [Google Scholar]
- 35. Verbanck M, Chen C‐Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693‐698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Greco MFD, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in mendelian randomisation studies with summary data and a continuous outcome. Stat Med. 2015;34(21):2926‐2940. [DOI] [PubMed] [Google Scholar]
- 37. Lutz SM, Voorhies K, Wu AC, Hokanson J, Vansteelandt S, Lange C. The influence of unmeasured confounding on the MR Steiger approach. Genet Epidemiol. 2022;46(2):139‐141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in mendelian randomization studies. Int J Epidemiol. 2013;42(5):1497‐1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yavorska OO, Burgess S. MendelianRandomization: an R package for performing mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734‐1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hemani G, Zheng J, Elsworth B, et al. The MR‐base platform supports systematic causal inference across the human phenome. Elife. 2018;30:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA. 2017;318(19):1925‐1926. [DOI] [PubMed] [Google Scholar]
- 42. Mao L, Jin H, Wang M, et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77(6):683‐690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pinna P, Grewal P, Hall JP, et al. Neurological manifestations and COVID‐19: experiences from a tertiary care center at the frontline. J Neurol Sci. 2020;415:116969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Varatharaj A, Thomas N, Ellul MA, et al. Neurological and neuropsychiatric complications of COVID‐19 in 153 patients: a UK‐wide surveillance study. Lancet Psychiatry. 2020;7(10):875‐882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Helms J, Kremer S, Merdji H, et al. Neurologic features in severe SARS‐CoV‐2 infection. N Engl J Med. 2020;382(23):2268‐2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taquet M, Sillett R, Zhu L, et al. Neurological and psychiatric risk trajectories after SARS‐CoV‐2 infection: an analysis of 2‐year retrospective cohort studies including 1 284 437 patients. Lancet Psychiatry. 2022;9(10):815‐827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Baig AM, Khaleeq A, Ali U, Syeda H. Evidence of the COVID‐19 virus targeting the CNS: tissue distribution, host‐virus interaction, and proposed neurotropic mechanisms. ACS Chem Nerosci. 2020;11(7):995‐998. [DOI] [PubMed] [Google Scholar]
- 48. Buzhdygan TP, DeOre BJ, Baldwin‐Leclair A, et al. The SARS‐CoV‐2 spike protein alters barrier function in 2D static and 3D microfluidic in‐vitro models of the human blood‐brain barrier. Neurobiol Dis. 2020;146:105131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heneka MT, Carson MJ, El Khoury J, et al. Neuroinflammation in Alzheimer's disease. Lancet Neurol. 2015. Apr;14(4):388‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van den Berg DF, Te Velde AA. Severe COVID‐19: NLRP3 inflammasome dysregulated. Front Immunol. 2020;11:1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tejera D, Mercan D, Sanchez‐Caro JM, et al. Systemic inflammation impairs microglial Aβ clearance through NLRP3 inflammasome. EMBO J. 2019;38(17):e101064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Magusali N, Graham AC, Piers TM, et al. A genetic link between risk for Alzheimer's disease and severe COVID‐19 outcomes via the OAS1 gene. Brain. 2021;144(12):3727‐3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schooling CM, Lopez PM, Yang Z, Zhao JV, Au Yeung SL, Huang JV. Use of multivariable mendelian randomization to address biases due to competing risk before recruitment. Front Genet. 2020;11:610852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pierce BL, Burgess S. Efficient design for mendelian randomization studies: subsample and 2‐sample instrumental variable estimators. Am J Epidemiol. 2013;178(7):1177‐1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658‐665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. IVs from summary data.
Table S2. IVs from meta‐analysis.
Table S3. Results of primary analysis and sensitivity analyses.
Table S4. Power calculations in the current MR study.


