Abstract
Pseudomonas aeruginosa produces siderophores, pyoverdin and pyochelin, for high-affinity iron uptake. To investigate their contribution to P. aeruginosa infections, we constructed allelic exchange mutants from strain PAO1 which were deficient in producing one or both of the siderophores. When inoculated into the calf muscles of immunosuppressed mice, pyochelin-deficient and pyoverdin-deficient mutants grew and killed the animals as efficiently as PAO1. In contrast, the pyochelin- and pyoverdin-deficient (double) mutant did not show lethal virulence, although it did infect the muscles. On the other hand, when inoculated intranasally, all mutants grew in the lungs and killed immunosuppressed mice. Compared with PAO1, however, the pyoverdin-deficient mutant and the double mutant grew poorly in the lungs, and the latter was significantly attenuated for virulence. Irrespective of the inoculation route, the pyoverdin-deficient and doubly deficient mutants detected in the blood were significantly less numerous than PAO1. Additionally, in vitro examination demonstrated that the growth of the double mutant was extremely reduced under a free-iron-restricted condition with apotransferrin but that the growth reduction was completely canceled by supplementation with hemoglobin as a heme source. These results suggest that both pyoverdin and pyochelin are required for efficient bacterial growth and full expression of virulence in P. aeruginosa infection, although pyoverdin may be comparatively more important for bacterial growth and dissemination. However, the siderophores were not always required for infection. It is possible that non-siderophore-mediated iron acquisition, such as via heme uptake, might also play an important role in P. aeruginosa infections.
Iron is essential for almost all living organisms, including bacteria. The ability of pathogenic bacteria to acquire iron in hosts is absolutely essential for bacterial growth and infection (7, 24). In animal hosts, iron is usually bound to proteins such as transferrin and lactoferrin in extracellular fluid and to ferritin, hemoglobin, and heme-containing enzymes in cells (26, 45). To utilize such complexes as iron sources, bacteria generally possess some sophisticated mechanisms, which include an iron uptake system mediated by high-affinity iron chelators called siderophores and a system for heme uptake via specific receptors (18, 26, 45).
Pseudomonas aeruginosa, a ubiquitous gram-negative rod, is considered an important opportunistic pathogen. It is highly pathogenic for individuals with compromised immunity; some infections involving this organism have a poor prognosis despite recent advances in antimicrobial chemotherapy (5, 14). P. aeruginosa is known to produce two chemically distinct siderophores, pyoverdin (Pvd) and pyochelin (Pch), for high-affinity iron uptake (10, 11, 28).
Pvd is a fluorescent dihydroxyquinoline derivative connected to a small peptide and contains hydroxamate and catecholate residues to chelate ferric ion, Fe (III) (42). Pch is a structurally unique siderophore possessing phenolate, but neither a hydroxamate nor a catecholate moiety (12). Pvd chelates Fe (III) in a 1:1 stoichiometry with high affinity (stability constant, 1032) (42), whereas Pch binds Fe (III) in a 2:1 stoichiometry with comparatively low affinity (stability constant, 105) (11). The genes pvdA (40), pvdD (20), and pvdE (19) are considered to encode enzymes responsible for Pvd synthesis, and similarly a pchDCBA operon (34) and pchEF genes (31) are responsible for Pch synthesis. The outer membrane proteins FpvA (29) and FptA (3) are characterized as specific receptors for Fe (III)-Pvd and Fe (III)-Pch complexes, respectively. Fe (III)-siderophore complexes bound to receptors are thought, generally in gram-negative bacteria, to be internalized into the periplasm across the outer membrane with energy transduced by the TonB protein from the cytoplasmic membrane (6, 30).
Both Pvd and Pch have been shown to remove iron from transferrin and lactoferrin and to promote P. aeruginosa growth in media containing these iron-binding proteins or human serum (2, 36, 46). Pvd is more active than Pch in this regard in vitro, leading to speculation of the primacy of Pvd over Pch for bacterial iron acquisition in vivo. The relationship between these siderophores and P. aeruginosa pathogenicity or virulence has also been studied. Pvd production was shown to be required for bacterial colonization of the lung in a rat infection model (28) and to correlate with lethal virulence in a burned-mouse infection model (23). In these previous studies, Pvd production mutants obtained by mutagenesis with UV or chemical treatment or transposon insertion were used successfully. On the other hand, Pch was shown to promote bacterial growth and lethality when injected into the peritoneal cavities of mice simultaneously with a virulent mutant of P. aeruginosa strain PAO1, which was isolated by repetitive passage in mice (8). These experimental facts have undoubtedly led to advanced understanding about the importance of Pvd and Pch and iron acquisition with them for P. aeruginosa infections. However, the comparative roles or importance of these siderophores in vivo are still unclear and have never been investigated systematically.
In this study, in order to address the comparative importance of Pvd and Pch production for P. aeruginosa infection, we constructed Pch-deficient, Pvd-deficient, and doubly deficient mutants from a wild-type strain of P. aeruginosa, PAO1, by a DNA allelic exchange method and examined them both in vivo and in vitro.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. The P. aeruginosa mutants PAD02, PAD06, and PAD07 and the pHT series of plasmids were generated in this study as described below. Escherichia coli strains DH5α (TOYOBO, Tokyo, Japan) and S17-1 (35) were utilized as a host for plasmid multiplication and a donor for conjugal transfer and mobilization of plasmids, respectively. Competent cells of E. coli were prepared as described elsewhere (32). Media used were Luria broth (LB); Vogel-Bonner minimal medium (VB) (41), which is selective for P. aeruginosa; and succinate minimal medium (22) containing 0.2% Casamino Acids (SMMCA; the concentration of contaminated iron in this medium, measured with Fe-750 reagents [Eiken Chemical Co., Ltd., Tokyo, Japan], was less than 1 μM). Solid media were prepared by addition of agar (1.5%). Selective agents included in the media were as follows: ampicillin, 100 μg/ml for E. coli; tetracycline, 10 μg/ml for E. coli and 50 μg/ml for P. aeruginosa; and streptomycin, 25 μg/ml for E. coli and 500 μg/ml for P. aeruginosa. Unless otherwise stated, bacteria were cultured at 37°C. For conjugal transfer and mobilization of plasmids from E. coli to P. aeruginosa, the recipient cells were grown overnight at 43°C (38).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Prototroph | |
| PAD02 | PAO1 ΔpchD::Tc | This study |
| PAD06 | PAO1 ΔpvdA::ΩSm/Sp | This study |
| PAD07 | PAO1 ΔpchD::Tc ΔpvdA::ΩSm/Sp | This study |
| E. coli | ||
| DH5α | recA1 endA1 gylA96 thi-1 hsdR17 supE44 Δ(lac)U169 (φ80dlac ΔM15) | TOYOBO |
| S17-1 | pro thi recA hsdR Tpr Smr; chromosomally integrated RP4-2-Tc::Mu-Km::Tn7; mobilizer of plasmids carrying the R68-derived Mob region | 35 |
| Plasmids | ||
| pMT5059 | Apr; pBR322 derivative carrying multiple cloning sites and a NotI site | 38 |
| pMT5071 | Kmr Cmr; pMOB3 derivative carrying NotI-flanked Mob cassette with ΩCm gene cartridge | 37 |
| pMT5056 | Apr Tcr; pBend2 derivative carrying Tc gene cartridge flanked with multiple restriction enzyme sites | 38 |
| pHRP316 | Apr Smr/Spr; pSL301 derivative carrying ΩSm/Sp gene cartridge flanked with superpolylinkers | 27 |
| pHT004 | Apr; pMT5059 carrying pchD in EcoRI-BamHI site | This study |
| pHT005 | Apr Tcr; pMT5059::ΔpchD::Tc | This study |
| pHT001 | Apr; pMT5059 carrying pvdA in EcoRI-BamHI site | This study |
| pHT010 | Apr Smr/Spr; pMT5059::ΔpvdA::ΩSm/Sp | This study |
Tpr, trimethoprim resistant; Smr, streptomycin resistant; Mob, plasmid mobilization; Apr, ampicillin resistant; Kmr, kanamycin resistant; Cmr, chloramphenicol resistant; Tcr, tetracycline resistant; Spr, spectinomycin resistant.
Recombinant DNA techniques.
Established procedures were used for preparation of plasmids, DNA manipulation, agarose gel electrophoresis, and transformation of E. coli (17). Plasmids and DNA fragments were purified with a kit (Prep-A-Gene DNA Purification Systems; Bio-Rad).
PCR and gene cloning.
Bacterial chromosomal DNA was extracted with TRIzol LS Reagent (Life Technologies) as described by the manufacturer. The genes pchD (on a DNA fragment corresponding to base positions 115 to 2253 of GenBank sequence X82644) and pvdA (on a DNA fragment corresponding to base positions 753 to 2772 of GenBank sequence Z25465) were amplified from the chromosomal DNA of P. aeruginosa strain PAO1 by PCR with the following synthesized primers: 5′-CGGAATTCAGATGGACAAAGCGCCCTGC-3′ (sense) and 5′-CGGGATCCGATGGGCGGAGACGAACAGG-3′ (antisense) for pchD and 5′-CGGAATTCCATCGTCAACGCGATGAAGC-3′ (sense) and 5′-CGGGATCCGCCTTCGAGCAACTGGCGTA-3′ (antisense) for pvdA. The 5′ regions of the sense primers were artificially flanked with 2 additional bases, CG, plus EcoRI sequence (GAATTC), and those of the antisense primers were artificially flanked with the 2 additional bases plus BamHI sequence (GGATCC). The reaction mixture (50 μl) consisted of 2.5 U of Taq polymerase, 0.2 μM paired primers, 0.2 mM deoxynucleoside triphosphate mixture, 1.5 mM MgCl2, and 200 ng of chromosomal DNA in PCR buffer (Life Technologies). The reaction mixtures were treated for 5 min at 95°C followed by 30 cycles of 30 s at 95°C, 1 min at 60°C, and 1 min at 72°C, before finishing with 5 min at 72°C. Each of the amplified pchD and pvdA genes was separated by agarose gel electrophoresis, purified from the gel, digested with EcoRI and BamHI, and cloned into the EcoRI-BamHI site of pMT5059 (38).
Construction of mutants.
Allelic exchange mutagenesis of the P. aeruginosa chromosome was carried out with a system established by Schweizer (33) except that the ColE1 type vector and the source of the mobilization cassette used were pMT5059 and pMT5071 (37), respectively. The pMT5059 derivative carrying the pchD gene, pHT004, was digested with StuI for deletion of an internal part (0.83 kb) of the gene; into this plasmid was inserted a StuI-flanked Tc gene cartridge (1.6 kb) excised from pMT5056 (38), resulting in pHT005. An 8.5-kb NotI fragment containing the mobilization cassette of pMT5071 was subsequently inserted into the NotI site of pHT005 carrying ΔpchD::Tc. The plasmid thus constructed (pHT006) was conjugally mobilized from E. coli strain S17-1 to P. aeruginosa strain PAO1, and then P. aeruginosa transconjugants were selected on VB agar plates containing tetracycline. A colony of tetracycline-resistant transconjugants was next spread onto LB agar plates containing 5% sucrose and tetracycline for selection of an allelic exchange mutant, PAD02. The pMT5059 derivative carrying the pvdA gene, pHT001, was digested with BglII and StuI for deletion of an internal part (0.47 kb) of the gene and treated with the Klenow fragment of DNA polymerase I; into this plasmid was inserted a blunt-end ΩSm gene cartridge (2.0 kb) prepared from pHRP316 (27), resulting in pHT010. The mobilization cassette was subsequently inserted into the NotI site of pHT010 carrying ΔpvdA::ΩSm. The plasmid generated (pHT012) was introduced into PAO1 and PAD02 as described above, and then transconjugants were selected on VB agar plates containing streptomycin. Each colony of streptomycin-resistant transconjugants, derived from PAO1 and PAD02, was next spread onto LB agar plates containing 5% sucrose and streptomycin for selection of allelic exchange mutants PAD06 and PAD07, respectively. When chromosomal DNAs of the mutants were subjected to PCRs under the same conditions as those described for amplification of normal pchD and pvdA genes (2.1 and 2.0 kb, respectively), size changes were observed in PCR products as expected (2.9 kb for pchD in PAD02 and PAD07; 3.5 kb for pvdA in PAD06 and PAD07), indicating that the expected allelic exchanges had successfully occurred in the mutants obtained. In addition, the mutants, as well as PAO1, were all sensitive to carbenicillin and chloramphenicol (the MICs of carbenicillin and chloramphenicol against the mutants in SMMCA supplemented with 1 μM FeCl3 were 64 and 16 μg/ml, respectively), indicating that the vector plasmid which mediated the allelic exchange did not remain in the mutants.
Siderophore assays.
P. aeruginosa strain PAO1 and its mutants were inoculated at approximately 106 CFU/ml into, and cultured in, 5 ml of SMMCA containing 1 μM FeCl3. After 20 h, Pvd secreted was assayed by measuring the optical density (OD) of the culture supernatant at 405 nm (OD405). The Pch assay was performed as described previously (2) with minor modifications. Three milliliters of the supernatant of the P. aeruginosa culture was made acidic with 0.3 ml of acetic acid and extracted with 1.5 ml of dichloromethane. The extracts were concentrated by evaporation and applied onto silica thin-layer plates for chromatography in chloroform-acetic acid-ethanol (90:5:2.5). Spots corresponding to Rf 0.35 were scraped from the plates, eluted with 3 ml of ethanol, and measured fluorometrically for Pch (excitation at 350 nm and emission at 440 nm). The values for amounts of Pvd and Pch were corrected by dividing by the values for bacterial growth (OD590s of cultures).
Animal experiments.
Male ddY mice (Japan SLC Ltd., Hamamatsu, Japan), 5 to 6 weeks old, were used. They were housed in cages in an experimental room at 22 ± 2°C and 55 ± 15% humidity and were provided commercial food and drinking water ad libitum. The mice received intraperitoneal injections of 3 mg of cyclophosphamide (Endoxane; Shionogi & Co., Ltd., Osaka, Japan) for immunosuppression 3 days before the bacterial inoculation. P. aeruginosa strains for inocula were first fully grown in SMMCA containing 40 μM FeSO4 for 16 h; then bacterial cells collected by centrifugation were suspended in SMMCA containing 1 μM FeCl3, an iron-poor medium, and were cultured for an additional 4 h. Bacteria were harvested by centrifugation and were suspended and diluted appropriately in saline. Bacterial suspensions were inoculated into mice by injection into the calf muscles of both legs (25 μl/muscle) under ether anesthesia or by instillation into the nostrils (40 μl) under anesthesia with a mixture of ketamine and xyladine. At various times, the animals were sacrificed by collection of blood from the axillary vein under ether anesthesia, and then the calf muscles, in the case of intramuscular inoculation, or the lungs, in the case of intranasal inoculation, were removed. Tissue samples were homogenized in 2 ml of potassium phosphate buffer (33 mM; pH 7.0). The blood and tissue homogenates were appropriately diluted with the phosphate buffer and plated onto LB agar plates for assaying counts of viable bacteria. In these animal studies, we followed the animal experimentation guidelines of the Daiichi Pharmaceutical Co., Ltd., Animal Care and Use Committee.
In vitro growth assays.
Assays were performed in 96-well round-bottom plates. For the examination of the effect of apotransferrin (apoTsf, from bovines; Life Technologies) on bacterial growth, apoTsf was dissolved and diluted twofold serially with SMMCA in the wells (40 μl/well), to which was added SMMCA containing 20 μM FeCl3 and 40 mM sodium bicarbonate (50 μl/well). The plates were placed at 37°C for 1 h under 5% CO2 and 95% air for promoting iron chelation by apoTsf. Then each well was inoculated with P. aeruginosa strain PAO1 or one of its mutants (10 μl of bacterial suspension in SMMCA) at approximately 105 CFU/ml and incubated further for 20 h under the same conditions. Bacterial growth was measured as the OD590 of the culture in a microplate spectrophotometer (MultiScan Plus MKII; Flow Laboratories). When FeSO4 or hemoglobin (bovine; Sigma) was used as a further supplement in the apoTsf-containing medium, their twofold serial dilutions in SMMCA (40 μl/well) were mixed with SMMCA containing 20 μM FeCl3, 40 mM sodium bicarbonate, and 50 μM apoTsf (50 μl/well).
Statistics.
Significant differences between two groups were analyzed by Student's t test (for means) or by Fisher's exact probability test (for frequencies). Probability values below 0.05 were considered significant.
RESULTS
Characteristics of P. aeruginosa mutants constructed.
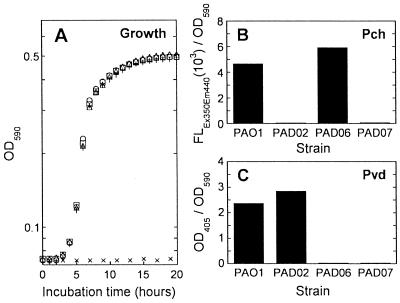
We constructed P. aeruginosa mutants PAD02, PAD06, and PAD07, in which pchD, pvdA, and both genes of strain PAO1, respectively, were inactivated. It is known that pchD encodes a putative adenylate-forming enzyme responsible for Pch synthesis (34) and that pvdA encodes an l-ornithine N5-oxygenase responsible for Pvd synthesis (40). To examine siderophore production by the mutants, we cultured them in SMMCA supplemented with 1 μM FeCl3 and assayed Pch and Pvd in culture supernatants. In this medium, in which the iron concentration was kept low for promoting siderophore production, the mutants were indistinguishable from PAO1 in their growth (Fig. 1A). Whereas PAO1 as the control was capable of producing Pch and Pvd, mutants PAD02, PAD06, and PAD07 were defective in production of Pch, Pvd, and both siderophores, respectively (Fig. 1B and C).
FIG. 1.
Growth (A) and production of siderophores Pch (B) and Pvd (C) by P. aeruginosa strain PAO1 and its allelic exchange mutants in SMMCA supplemented with 1 μM FeCl3. A bacterial growth assay was performed in a 96-well plate (0.1 ml of culture/well), and the growth was measured as OD590 in a SPECTRAmax (Molecular Devices, Sunnyvale, Calif.), which is a microplate spectrophotometer with incubation and mixing functions, after bacterial inoculations at approximately 106 CFU/ml. ○, strain PAO1; ▵, the pchD-inactivated mutant, PAD02; □, the pvdA-inactivated mutant, PAD06; +, the pchD- and pvdA-inactivated mutant, PAD07; ×, no inoculum (medium control). Pch (B) and Pvd (C) secreted into the culture supernatant were fluorometrically or photometrically measured as described in Materials and Methods; the values for Pch and Pvd amounts (fluorometric measurement with excitation at 350 nm and emission at 440 nm and OD405, respectively) were corrected by dividing by the values for bacterial growth (OD590).
In vivo growth and virulence of P. aeruginosa strain PAO1 and its mutants after intramuscular inoculation.
To examine the infectivity of strain PAO1 and its siderophore production mutants, we first inoculated them intramuscularly into immunosuppressed mice. Viable bacteria in inoculation sites (the calf muscles) and the blood were assayed at 16 h postinoculation (hpi). Bacterial growth was evaluated as the increase in the number of viable cells in the muscles over that in the inoculum (approximately 106 CFU). The virulence of the bacteria was assessed as lethality in the mice.
Like the parental strain, PAO1, the Pch-deficient mutant, PAD02, and the Pvd-deficient mutant, PAD06, grew very efficiently in the muscles (increase in viable bacteria, approximately 3 orders of magnitude [Table 2]). They were disseminated and detectable at 16 hpi in the blood (103 to 105 CFU/ml), but the mean count of PAD06 was significantly smaller than that of PAO1 (P < 0.05 [Table 2]). These mutants and PAO1 showed similar virulence in the mice by 24 hpi (Table 2).
TABLE 2.
Growth and virulence of P. aeruginosa PAO1 and its siderophore production mutants inoculated intramuscularly into immunosuppressed mice
| Inoculum
|
Count of viable bacteriaa at 16 hpi in:
|
Lethality in miceb at:
|
||||
|---|---|---|---|---|---|---|
| Strain | Log10 CFU | Muscles | Blood | 16 hpi | 24 hpi | 48 hpi |
| PAO1 | 6.32 | 9.28 ± 0.13 (2.96) | 4.46 ± 0.21 | 2/5 | 5/5 | 5/5 |
| PAD02 | 6.30 | 9.08 ± 0.09 (2.78) | 4.39 ± 0.44 | 0/5 | 5/5 | 5/5 |
| PAD06 | 6.42 | 9.19 ± 0.21 (2.77) | 3.84 ± 0.23c | 0/5 | 4/5 | 5/5 |
| PAD07 | 6.24 | 7.09 ± 0.06d (0.89)d | <2.00d | 0/5 | 0/5d | 0/5d |
Samples were collected from three mice per group at 16 hpi and were assayed for viable bacteria. Values are mean log10 CFU ± SDs in muscles or in 1 ml of blood. Values in parentheses represent bacterial growth, expressed as the mean of the increases in the count (each count in the group at 16 hpi minus the inoculum), in log10 CFU. SDs for mean increases in the counts are theoretically identical to those for mean counts at 16 hpi.
Number of dead mice/number of mice inoculated for observation of bacterial virulence.
Significantly different (P < 0.05) from value for PAO1-inoculated group as calculated by Student's t test (for means) or Fisher's exact probability test (for frequencies).
Significantly different (P < 0.01) from value for PAO1-inoculated group as calculated by Student's t test (for means) or Fisher's exact probability test (for frequencies).
In contrast, the growth of the Pch- and Pvd-deficient mutant, PAD07, in the muscles was very poor (increase in viable bacteria, less than 1 order of magnitude), and the mutant was not detected in the blood (<102 CFU/ml) (Table 2). These results were significantly different from those for PAO1 and the other mutants (P < 0.01 [Table 2]). Although the double mutant PAD07 in the inoculation sites decreased gradually in terms of viable counts (mean log10 CFU ± standard deviation [SD] per sample, 6.84 ± 0.87 at 40 hpi and 5.66 ± 1.71 at 64 hpi), this mutant apparently persisted in vivo. Surprisingly, the mutant was detectable at 7 days postinoculation in the muscles (mean log10 CFU ± SD per sample, 4.96 ± 2.13), showing establishment of a chronic local infection. No mice died from such an infection (Table 2).
In vivo growth and virulence of P. aeruginosa strain PAO1 and its mutants after intranasal inoculation.
Next, to examine further the infectivity of strain PAO1 and its siderophore production mutants, we inoculated them intranasally into immunosuppressed mice. Bacterial growth was evaluated as the increase in the number of viable cells at 12 hpi over that at 1 hpi (indicated as amounts of bacteria introduced; approximately 106 CFU expected) in the lungs. Also in this experiment, viable bacteria in the blood were assayed, and bacterial lethality in the mice was observed.
The Pch-deficient mutant, PAD02, grew efficiently in the lung (increase in viable bacteria, more than 2 orders of magnitude), as did the parental strain, PAO1 (Table 3). This mutant and PAO1 were detected in substantial amounts (104 to more than 105 CFU/ml) in the blood and similarly killed almost all of the mice by 24 hpi (Table 3).
TABLE 3.
Growth and virulence of P. aeruginosa PAO1 and its siderophore production mutants inoculated intranasally into immunosuppressed mice
| Strain inoculated | Count of viable bacteriaa detected in:
|
Lethality in miceb at:
|
||||
|---|---|---|---|---|---|---|
| Lung
|
Blood (12 hpi) | |||||
| 1 hpi | 12 hpi | 12 hpi | 24 hpi | 48 hpi | ||
| PAO1 | 5.73 ± 0.37 | 8.01 ± 0.41 (2.28) | >4.21 ± 0.65 | 3/7 | 6/7 | 6/7 |
| PAD02 | 6.03 ± 0.18 | 8.24 ± 0.32 (2.22) | >4.66 ± 0.58 | 3/7 | 6/7 | 7/7 |
| PAD06 | 6.31 ± 0.07 | 7.61 ± 0.46 (1.30)d | 3.11 ± 0.73c | 0/7 | 4/7 | 5/7 |
| PAD07 | 6.33 ± 0.10 | 7.22 ± 0.58c (0.89)d | <2.36 ± 0.39d | 0/7 | 0/7d | 7/7 |
a Samples were collected from three or five mice a group at 1 and 12 hpi, respectively, and assayed for viable bacteria. Values are mean log10 CFU ± SDs in the lung or in 1 ml of blood. The detection limit of bacteria in the blood was 2.00 to 5.00; the limit values were included for calculation of the mean, if data were below the limit. Values in parentheses represent bacterial growth, expressed as the mean of the increases in the count (each count in the group at 12 hpi minus the mean count at 1 hpi), in log10 CFU. SDs for the mean increases in the counts are theoretically identical to those for mean counts at 12 hpi.
Number of dead mice/number of mice inoculated for observation of bacterial virulence.
c Significantly different (P < 0.05) from value for PAO1-inoculated group as calculated by Student's t test (for means) or Fisher's exact probability test (for frequencies).
Significantly different (P < 0.01) from value for PAO1-inoculated group as calculated by Student's t test (for means) or Fisher's exact probability test (for frequencies).
The Pvd-deficient mutant, PAD06, also grew in the lungs (increasing by more than 1 order of magnitude) and was detectable in the blood (102 to 104 CFU/ml) (Table 3). However, the levels of growth in the lungs and detection in the blood of this mutant were significantly lower than those of PAO1 (P < 0.01 and P < 0.05, respectively [Table 3]), in contrast to the result in the intramuscular infection model. The virulence of PAD06 appeared to be a little attenuated, because this mutant took somewhat longer to kill the mice than did PAO1 (Table 3).
The growth level of the doubly deficient mutant, PAD07, in the lungs (increase, less than 1 order of magnitude) was also significantly lower than that of PAO1 (P < 0.01 [Table 3]). In contrast with the results in the intramuscular infection model, PAD07 was detectable in the blood (102 to 103 CFU/ml), although the counts were significantly lower than those of PAO1 (P < 0.01), and showed lethal virulence within 24 to 48 hpi (Table 3). However, the virulence of PAD07 was also certainly attenuated in this lung infection model, because no mice died from its inoculation by 24 hpi, when almost all mice were dead from the PAO1 inoculation (the difference was significant: P < 0.01 [Table 3]).
In vitro growth of P. aeruginosa strain PAO1 and its mutants under a free-iron-restricted condition and the effect of hemoglobin.
As described above, mutant PAD07, in spite of its inability to produce Pch and Pvd, resulted in infections of immunosuppressed mice, suggesting the contribution of non-siderophore-mediated iron acquisition to P. aeruginosa growth in vivo. We speculated on the possible participation of heme uptake and so conducted in vitro experiments to examine it.
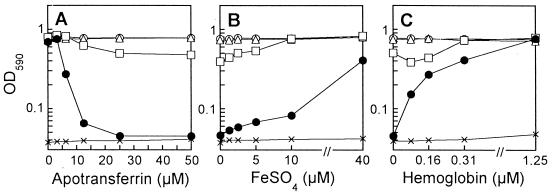
To constitute a free-iron-restricted condition in vitro, similar to the condition in vivo, we used a host iron chelator protein, apoTsf. We first tested the effect of apoTsf on the growth of strain PAO1 and its mutants in SMMCA containing 10 μM FeCl3 and 20 mM sodium bicarbonate. As shown in Fig. 2A, the growth of the double mutant, PAD07, was reduced in a manner dependent on increased concentrations of apoTsf and was almost completely abolished at 25 μM (2 mg/ml). The concentration of apoTsf was very similar to those of transferrin (including apo-form) in the sera of mice and humans (43). The growth of the Pvd-deficient mutant, PAD06, was reduced partially by the addition of apoTsf at more than 12.5 μM, but the growth of the Pch-deficient mutant and the parental strain was not affected at all (Fig. 2A). The reduction in the growth of PAD07 and PAD06 in the medium including 25 μM apoTsf was canceled by supplementation with FeSO4 at 10 to 40 μM (Fig. 2B), indicating that the growth reduction was based on the restriction of available free iron. In this free-iron-restricted medium prepared by addition of 25 μM apoTsf, where iron might be present as transferrin, we examined the effect of hemoglobin as a heme source on bacterial growth. Interestingly, the growth of mutants PAD07 and PAD06 was completely restored by supplementation with hemoglobin at 0.31 to 1.25 μM (Fig. 2C), concentrations much lower than those required for FeSO4. These results demonstrate that P. aeruginosa certainly possesses a high-affinity heme uptake system, as described recently (16, 25).
FIG. 2.
Effects of apoTsf on the growth of P. aeruginosa strain PAO1 and its siderophore production mutants in SMMCA containing 10 μM FeCl3 and 20 mM sodium bicarbonate (A), and effects of supplementation with FeSO4 (B) or hemoglobin (C) on their growth in the medium including 25 μM apoTsf. Growth was measured as the end point OD590 of culture 20 h after bacterial inoculation. ○, strain PAO1; ▵, the Pch-deficient mutant, PAD02; □, the Pvd-deficient mutant, PAD06; ●, the Pch- and Pvd-deficient mutant, PAD07; ×, no inoculum (medium control).
DISCUSSION
In this study, we approached the impacts of siderophores on P. aeruginosa infections by using a set of allelic exchange mutants of strain PAO1 that were unable to produce Pch, Pvd, or both siderophores. Two kinds of infection models in immunosuppressed mice were used. The results obtained in the present study included novel findings and increased our understanding of the contribution of these siderophores to P. aeruginosa infections, since the mutants possessed the same genetic background except for mutations in the genes responsible for Pch and Pvd production.
Compared with the parental wild-type strain, PAO1, the Pvd-deficient mutant (PAD06) grew poorly in the lung (Table 3) and was detected as lower counts in the blood after both intramuscular and intranasal inoculations (Tables 2 and 3), but this was not the case for the Pch-deficient mutant (PAD02). These results indicate that Pvd production is more important than Pch production for P. aeruginosa growth in vivo and probably for its dissemination. The primacy of Pvd in vivo that we observed may be consistent with previous findings in animal infection models (23, 28). Furthermore, it is well supported by previous in vitro findings that showed superior activities of Pvd over Pch in removing iron from transferrin and lactoferrin and promoting growth in media supplemented with serum or host iron-binding proteins (2, 36).
On the other hand, a certain importance of Pch production for P. aeruginosa infection was also demonstrated in this study, because the Pch- and Pvd-deficient mutant (PAD07) showed clearly lower growth ability and virulence than the Pvd-deficient mutant (PAD06) in the intramuscular inoculation model (Table 2; P < 0.01 for all the differences in viable counts in the muscles and blood and in lethality) and a similar tendency also in the intranasal inoculation model (Table 3). Together with the primacy of Pvd, our data indicate that both Pvd and Pch are required for efficient bacterial growth and full expression of virulence in P. aeruginosa infections.
Thus, we succeeded in showing experimentally the impact of siderophore production on P. aeruginosa infections. At the same time, however, we should mention unequivocally that neither Pch nor Pvd nor both siderophores were always required for P. aeruginosa infection and virulence, because the doubly deficient mutant (PAD07) was certainly able to persist or grow in the muscles and lungs (Tables 2 and 3) and showed substantial lethality after intranasal inoculation (Table 3) in immunosuppressed mice.
Before this study, Pvd of P. aeruginosa had been believed to be essential for bacterial colonization of the lungs in a rat infection model (28) and for virulence in a burned-mouse infection model (23). Poole et al. showed that Pvd-deficient mutants could not colonize the lungs of rats when the bacteria (104 CFU) were introduced incorporated into agar beads (28). The mutants used in the previous study, IA130 and IAJ40M1, had been generated from a PAO1 auxotroph, PAO25 (argF10 leu-10), by chemical treatment and from PAO1 by transposon insertion, respectively (1). Meyer et al. demonstrated that the Pvd-deficient mutants PAO6606 and PAO6609 had significantly reduced lethal virulence when inoculated (102 CFU) into burned mice (23); the mutants had been obtained from a methionine auxotroph of PAO1 (PAO6049) by UV mutagenesis (15). Discrepancies between these previous results and ours might be due to differences in infection models, including immunological states of the animals, characters of the mutants used, and sizes of bacterial inocula. In our study, the artificial immunosuppression of the mice by cyclophosphamide treatment might be related to considerable virulence of the mutants, including the Pvd-deficient mutant (PAD06), but the immunosuppression was unavoidable because even the wild-type strain PAO1 did not cause infections without it (data not shown). In addition, although the inoculum size used by us (approximately 106 CFU) was larger than that in the previous studies, it was required for determining constantly the lethal virulence of PAO1 as the control in our models (data not shown). On the other hand, it is also of concern whether mutations not responsible for siderophore production, defined or undefined, might be implicated in the previous results, since the previous mutants used were generated by fundamentally unspecific mutagenesis and actually included some other mutations associated with the auxotrophy in some strains.
Our animal experiments posed the question of what could support iron acquisition in vivo and infections by the Pch- and Pvd-deficient mutant, PAD07. To gain insight into this question, we established a free-iron-restricted condition by using apoTsf, which was thought to partly reflect the host conditions, and performed in vitro examinations. As a result, it was clarified that the double mutant was able to utilize a heme source during its growth under such a condition (Fig. 2C). We consider this experimental fact to be strong evidence for a possibility that heme uptake may be also involved in iron acquisition in vivo and infection by P. aeruginosa.
As another possibility, we cannot ignore the effect of pyocyanin in supporting the infectivity of the doubly deficient mutant by its activity in the reductive release of soluble iron from transferrin (9). In addition, a role for iron reductases excreted by P. aeruginosa (39) could be a matter of speculation. It is also possible that, in place of Pch and Pvd, some metabolites might participate as siderophores in iron acquisition. For example, salicylate, which is a precursor of Pch and may be produced after pchD inactivation with non-Ω gene cartridge insertion (4, 34) as in our mutants (PAD02 and PAD07), could be a candidate (21). However, further studies are needed to evaluate these possibilities.
We would propose, nevertheless, that the heme uptake system might be comparably important to siderophore-mediated systems for in vivo iron acquisition, because heme-containing proteins are very abundant in animal hosts and seem available as sources for efficient iron uptake (26). In concert with the heme uptake system, it is likely that some pathogenic factors, including hemolysins and proteases, might work to damage cells and host proteins and to facilitate the release of heme, a mechanism similar to that by which proteases were supposed to promote siderophore-mediated iron acquisition from transferrin (13, 44). As a heme uptake system in P. aeruginosa, the HasA (heme-binding protein)-mediated system has been partially characterized (16). Moreover, an additional system that would involve a putative heme receptor, PhuR, might be present as suggested through the analysis of the P. aeruginosa genome (R. E. W. Hancock laboratory website [http://www.cmdr.ubc.ca/bobh/TonBfamily.html]; Pseudomonas Genome Project website [http://www.pseudomonas.com/]). Our proposal, the contribution of the heme uptake system to P. aeruginosa growth in vivo and virulence, could be supported by further studies using some genetically defined heme utilization mutants in the future, when the systems would be further characterized at the molecular and functional levels. In addition, recent advances in the analysis of the genome suggest that putative heme receptors (PhuR and HasR) of P. aeruginosa might be TonB dependent, as are receptors for Fe (III)-siderophore complexes (R. E. W. Hancock laboratory and Pseudomonas Genome Project websites). It is of interest to determine whether TonB protein (30) is essential for heme utilization in addition to siderophore-mediated iron acquisition, and also for infections by P. aeruginosa.
ACKNOWLEDGMENTS
We thank N. Gotoh and M. Tsuda for their gifts of bacterial strains and plasmids.
REFERENCES
- 1.Ankenbauer R, Hanne L F, Cox C D. Mapping of mutations in Pseudomonas aeruginosa defective in pyoverdin production. J Bacteriol. 1986;167:7–11. doi: 10.1128/jb.167.1.7-11.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ankenbauer R, Sriyosachati S, Cox C D. Effects of siderophores on the growth of Pseudomonas aeruginosa in human serum and transferrin. Infect Immun. 1985;49:132–140. doi: 10.1128/iai.49.1.132-140.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ankenbauer R G, Quan H N. FptA, the Fe (III)-pyochelin receptor of Pseudomonas aeruginosa: a phenolate siderophore receptor homologous to hydroxamate siderophore receptors. J Bacteriol. 1994;176:307–319. doi: 10.1128/jb.176.2.307-319.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ankenbauer R G, Toyokuni T, Staley A, Rinehart K, Jr, Cox C D. Synthesis and biological activity of pyochelin, a siderophore of Pseudomonas aeruginosa. J Bacteriol. 1988;170:5344–5351. doi: 10.1128/jb.170.11.5344-5351.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodey G P, Bolivar R, Fainstein V, Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983;5:279–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- 6.Braun V. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol Rev. 1995;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 7.Bullen J J. The significance of iron in infection. Rev Infect Dis. 1981;3:1127–1138. doi: 10.1093/clinids/3.6.1127. [DOI] [PubMed] [Google Scholar]
- 8.Cox C D. Effect of pyochelin on the virulence of Pseudomonas aeruginosa. Infect Immun. 1982;36:17–23. doi: 10.1128/iai.36.1.17-23.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox C D. Role of pyocyanin in the acquisition of iron from transferrin. Infect Immun. 1986;52:263–270. doi: 10.1128/iai.52.1.263-270.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox C D, Adams P. Siderophore activity of pyoverdin for Pseudomonas aeruginosa. Infect Immun. 1985;48:130–138. doi: 10.1128/iai.48.1.130-138.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox C D, Graham R. Isolation of an iron-binding compound from Pseudomonas aeruginosa. J Bacteriol. 1979;137:357–364. doi: 10.1128/jb.137.1.357-364.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox C D, Rinehart K, Jr, Moore M L, Cook J., Jr Pyochelin: novel structure of an iron-chelating growth promoter for Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1981;78:4256–4260. doi: 10.1073/pnas.78.7.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doring G, Pfestorf M, Botzenhart K, Abdallah M A. Impact of proteases on iron uptake of Pseudomonas aeruginosa pyoverdin from transferrin and lactoferrin. Infect Immun. 1988;56:291–293. doi: 10.1128/iai.56.1.291-293.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hohnadel D, Haas D, Meyer J M. Mapping of mutations affecting pyoverdine production in Pseudomonas aeruginosa. FEMS Microbiol Lett. 1986;36:195–199. [Google Scholar]
- 16.Létoffé S, Redeker V, Wandersman C. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence similarities with the Serratia marcescens HasA haemophore. Mol Microbiol. 1998;28:1223–1234. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 18.Marinez J L, Delgado-Iribarren A, Baquero F. Mechanisms of iron acquisition and bacterial virulence. FEMS Microbiol Rev. 1990;75:45–56. doi: 10.1111/j.1574-6968.1990.tb04085.x. [DOI] [PubMed] [Google Scholar]
- 19.McMorran B J, Merriman M E, Rombel I T, Lamont I L. Characterisation of the pvdE gene which is required for pyoverdine synthesis in Pseudomonas aeruginosa. Gene. 1996;176:55–59. doi: 10.1016/0378-1119(96)00209-0. [DOI] [PubMed] [Google Scholar]
- 20.Merriman T R, Merriman M E, Lamont I L. Nucleotide sequence of pvdD, a pyoverdine biosynthetic gene from Pseudomonas aeruginosa: PvdD has similarity to peptide synthetases. J Bacteriol. 1995;177:252–258. doi: 10.1128/jb.177.1.252-258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer J M. Exogenous siderophore-mediated iron uptake in Pseudomonas aeruginosa: possible involvement of porin OprF in iron translocation. J Gen Microbiol. 1992;138:951–958. doi: 10.1099/00221287-138-5-951. [DOI] [PubMed] [Google Scholar]
- 22.Meyer J M, Abdallah M A. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. J Gen Microbiol. 1978;107:319–328. [Google Scholar]
- 23.Meyer J M, Neely A, Stintzi A, Georges C, Holder I A. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect Immun. 1996;64:518–523. doi: 10.1128/iai.64.2.518-523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neilands J B. Iron absorption and transport in microorganisms. Annu Rev Nutr. 1981;1:27–46. doi: 10.1146/annurev.nu.01.070181.000331. [DOI] [PubMed] [Google Scholar]
- 25.Noya F, Arias A, Fabiano E. Heme compounds as iron sources for nonpathogenic Rhizobium bacteria. J Bacteriol. 1997;179:3076–3078. doi: 10.1128/jb.179.9.3076-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otto B R, Verweij-van Vudht A M J J, MacLaren D M. Transferrin and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–233. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 27.Parales R E, Harwood C S. Construction and use of a new broad-host range lacZ transcriptional fusion vector, pHRP309, for Gram− bacteria. Gene. 1993;133:23–30. doi: 10.1016/0378-1119(93)90220-w. [DOI] [PubMed] [Google Scholar]
- 28.Poole K, Dean C, Heinrichs D, Neshat S, Krebs K, Young L, Kilburn L. Siderophore-mediated iron transport in Pseudomonas aeruginosa. In: Nakazawa T, editor. Molecular biology of Pseudomonas. Washington, D.C.: American Society for Microbiology; 1996. pp. 371–383. [Google Scholar]
- 29.Poole K, Neshat S, Krebes K, Heinrichs D E. Cloning and nucleotide sequence analysis of the ferripyoverdine receptor gene fpvA of Pseudomonas aeruginosa. J Bacteriol. 1993;175:4597–4604. doi: 10.1128/jb.175.15.4597-4604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poole K, Zhao Q, Neshat S, Heinrichs D E, Dean C R. The Pseudomonas aeruginosa tonB gene encodes a novel TonB protein. Microbiology. 1996;142:1449–1458. doi: 10.1099/13500872-142-6-1449. [DOI] [PubMed] [Google Scholar]
- 31.Reimann C, Serino L, Beyeler M, Haas D. Dihydroaeruginoic acid synthetase and pyochelin synthetase, products of the pchEF genes, are induced by extracellular pyochelin in Pseudomonas aeruginosa. Microbiology. 1998;144:3135–3148. doi: 10.1099/00221287-144-11-3135. [DOI] [PubMed] [Google Scholar]
- 32.Schweizer H. The agmR gene, an environmentally responsive gene, complements defective glpR, which encodes the putative activator for glycerol metabolism in Pseudomonas aeruginosa. J Bacteriol. 1991;173:6798–6806. doi: 10.1128/jb.173.21.6798-6806.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 34.Serino L, Reimmann C, Visca P, Beyeler M, Chiesa V D, Haas D. Biosynthesis of pyochelin and dihydroaeruginoic acid requires the iron-regulated pchDCBA operon in Pseudomonas aeruginosa. J Bacteriol. 1997;179:248–257. doi: 10.1128/jb.179.1.248-257.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon R, O'Connell M, Labe M, Puhler A. Plasmid vectors for the genetic analysis and manipulation of rhizobia and other gram-negative bacteria. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 36.Sriyosachati S, Cox C D. Siderophore-mediated iron acquisition from transferrin by Pseudomonas aeruginosa. Infect Immun. 1986;52:885–891. doi: 10.1128/iai.52.3.885-891.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsuda M. Use of a transposon-encoded site-specific resolution system for construction of large and defined deletion mutations in bacterial chromosome. Gene. 1998;207:33–41. doi: 10.1016/s0378-1119(97)00601-x. [DOI] [PubMed] [Google Scholar]
- 38.Tsuda M, Miyazaki H, Nakazawa T. Genetic and physical mapping of genes involved in pyoverdin production in Pseudomonas aeruginosa PAO. J Bacteriol. 1995;177:423–431. doi: 10.1128/jb.177.2.423-431.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vartivarian S E, Cowart R E. Extracellular iron reductases: identification of a new class of enzymes by siderophore-producing microorganisms. Arch Biochem Biophys. 1999;364:75–82. doi: 10.1006/abbi.1999.1109. [DOI] [PubMed] [Google Scholar]
- 40.Visca P, Ciervo A, Orsi N. Cloning and nucleotide sequence of the pvdA gene encoding the pyoverdin biosynthetic enzyme l-ornithine N5-oxygenase in Pseudomonas aeruginosa. J Bacteriol. 1994;176:1128–1140. doi: 10.1128/jb.176.4.1128-1140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel H J, Bonner D M. Acetylornithase of E. coli: partial purification and properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 42.Wendenbaum S, Demange P, Dell A, Meyer J M, Abdallah M A. The structure of pyoverdinePa, the siderophore of Pseudomonas aeruginosa. Tetrahedron Lett. 1983;24:4877–4880. [Google Scholar]
- 43.Werner R G, Erne A S, Stecher A, Goeth H. Quantitative determination of transferrins in serum of different species. FEMS Microbiol Lett. 1983;17:131–136. [Google Scholar]
- 44.Wolz C, Hohloch K, Ocaktan A, Poole K, Evans R W, Rochel N, Albrecht-Gary A M, Abdallah M A, Doring G. Iron release from transferrin by pyoverdin and elastase from Pseudomonas aeruginosa. Infect Immun. 1994;62:4021–4207. doi: 10.1128/iai.62.9.4021-4027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wooldridge K G, Williams P H. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol Rev. 1993;12:325–348. doi: 10.1111/j.1574-6976.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 46.Xiao R, Kisaalita W S. Iron acquisition from transferrin and lactoferrin by Pseudomonas aeruginosa pyoverdin. Microbiology. 1997;143:2509–2515. doi: 10.1099/00221287-143-7-2509. [DOI] [PubMed] [Google Scholar]