Abstract
We cloned and sequenced the structural gene for Aeromonas hydrophila porin II from strain AH-3 (serogroup O:34). The genetic position of this gene, like that of ompF in Escherichia coli, is adjacent to aspC and transcribed in the same direction. However, upstream of the porin II gene no similarities with E. coli were found. We obtained defined insertion mutants in porin II gene either in A. hydrophila (O:34) or A. veronii sobria (serogroup O:11) serum-resistant or -sensitive strains. Furthermore, we complemented these mutants with a plasmid harboring only the porin II gene, which allowed us to define the role of porin II as an important surface molecule involved in serum susceptibility and C1q binding in these strains.
The complement system plays a key role in humoral defense against microbial pathogens and has been reviewed extensively (33). Its importance is clearly seen in individuals with complement deficiencies because they are at increased risk to develop severe and recurrent microbial infections (7). Resistance to complement action is thus a requisite for pathogenic microorganisms, which have developed a variety of mechanisms to ensure survival in nonimmune serum (7). Gram-negative bacteria activate complement via the classical or alternative pathway (CPC or APC, respectively) which is required for the effective elimination of serum-sensitive strains (37). In previous studies (19, 22), we focused on defining the mechanisms of complement sensitivity in mesophilic Aeromonas. Only the CPC is effective in the elimination of Aeromonas serum-sensitive strains in nonimmune serum, as we previously reported (19, 22). Activation of the CPC by these strains led to the identification of a bacterial outer membrane (OM) protein, presumably porin II (13), that binds C1q and activates this pathway in nonimmune serum and in agammaglobulinemic serum in an antibody-independent manner (24).
Mesophilic aeromonads are increasingly being reported as important pathogens of humans and lower vertebrates including amphibians, reptiles, and fish (11). Aeromonas strains have been serogrouped on the basis of the O-antigen lipopolysaccharide (LPS) (30), the polysaccharide chains in the smooth LPS, also known as the somatic antigen. Recently, a group of virulent Aeromonas hydrophila and A. veronii sobria strains isolated from humans and fish have been described (12, 15), serologically related by their O-antigen LPS (serogroup O:11) with a known chemical structure and having a surface array protein of molecular weight of ca. 52,000 (termed S-layer) (26). The S-layer-expressing (S+) strains from this serogroup are the most frequent isolates from septicemia caused by mesophilic Aeromonas sp. (12). Serogroup O:34 strains of mesophilic Aeromonas, recovered from moribund fish and from clinical specimens (21, 25), represent the single most common Aeromonas serogroup, accounting for 26.4% of all infections. Previous investigations have documented O:34 strains as an important cause of infections in humans (21, 25).
We cloned and sequenced the structural gene for A. hydrophila porin II, which allow us to obtain defined insertion mutants in this gene, and complemented these mutant strains with porin II. With all these strains, we were able to study the role of porin II in serum susceptibility and C1q binding to whole cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown on Luria-Bertani (LB) Miller broth and LB Miller agar, while Aeromonas strains were grown on tryptic soy broth or tryptic soy agar (5). Ampicillin (50 μg/ml), chloramphenicol (25 μg/ml), kanamycin (30 μg/ml), and tetracycline (20 μl/ml) were added to the different media when needed.
TABLE 1.
Bacterial strains, cosmids, and plasmids used
| Strain, cosmid, or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli | ||
| DH5α | F−endA hsdR17 (rk− mk+) supE44 thi-1 recA1 gyr-A96 φ80lacZ | 9 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F′ proAB lacIZΔM15 Tn10) | Stratagene |
| MC1061λpir | thi thr 1 leu6 proA2 his4 argE2 lacY1 galK2 ara14 xyl5 supE44 λpir | 27 |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu Kmr λpir | 27 |
| Aeromonas sp. | ||
| AH-3 | A. hydrophila wild type, serogroup O:34 | 20 |
| AH-405 | Rifampin-resistant mutant derived from AH-3 | 23 |
| AH-53 | Serum-sensitive rough mutant derived from AH-3 | 24 |
| AH-330 | Porin II insertion AH-405 mutant obtained with pFS-POR3 | This work |
| AH-336 | Porin II insertion AH-53 mutant obtained with pFS-POR3 | This work |
| AH-334 | Mutant AH-330 complemented by plasmid pLA-POR6 | This work |
| AH-338 | Mutant AH-336 complemented by plasmid pLA-POR6 | This work |
| TF7 | A. veronii sobria wild type, serogroup O:11 | 24 |
| AH-408 | Rifampin-resistant mutant derived from TF7 | This work |
| AH-26 | Rough S− serum-sensitive mutant derived from TF7 | 24 |
| AH-331 | Porin II insertion AH-408 mutant obtained with pFS-POR3 | This work |
| AH-337 | Porin II insertion AH-26 mutant obtained with pFS-POR3 | This work |
| AH-335 | Mutant AH-331 complemented by plasmid pLA-POR6 | This work |
| AH-339 | Mutant AH-337 complemented by plasmid pLA-POR6 | This work |
| Plasmids | ||
| pLA2917 | Tcr Kmr | 3 |
| COS-POR2 | pLA2917 with 20-kb chromosomal AH-3 Sau3A insert (porin II gene) | This work |
| pLA-POR3 | pLA2917 with 7.8-kb BglII insert from DNA insert of COS-POR2 (porin II gene) | This work |
| pLA-POR6 | pLA2917 with the single porin II gene | This work |
| pFS100 | pGP704 suicide plasmid, λpir-dependent, Kmr | 27 |
| pFS-POR3 | pFS100 with an internal fragment (996 bp) of porin II gene | This work |
Cell surface isolation and analysis.
Cell envelopes were prepared by lysis of whole cells in a French press at 16,000 lb/in2. Unbroken cells were removed by centrifugation at 10,000 × g for 10 min, and the envelope fraction was collected by centrifugation at 100,000 × g for 2 h. Cytoplasmic membranes were solubilized with sodium N-laurylsarcosinate, and the outer membrane (OM) fraction was collected as describe previously (24). OM proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by the Laemmli procedure (17). Protein gels were fixed and stained with Coomassie blue. LPS was purified by the method of Westphal and Jann (39). For screening purposes, LPS was obtained after proteinase K digestion of whole cells according to the procedure of Darveau and Hancock (6). SDS-PAGE was performed and LPS bands were detected by the silver staining method of Tsai and Frasch (36).
Antiserum.
Antiserum against purified porin II was obtained as previously described (24).
Western immunoblotting.
After SDS-PAGE, immunoblotting was carried out by transfer to polyvinylidine fluoride membranes (Millipore Corp., Bedford, Mass.) at 1.3 A for 1 h in the buffer of Towbin et al. (35). The membranes were then incubated sequentially with 1% bovine serum albumin, specific anti-porin II serum (1:500), alkaline phosphatase-labeled goat anti-rabbit immunoglobulin G, and 5-bromo-4-chloro-indolylphosphate disodium–nitroblue tetrazolium. Incubations were carried out for 1 h, and washing steps with 0.05% Tween 20 in phosphate-buffered saline (PBS) were included after each incubation step. Colony blotting was performed with porin II antiserum as indicated above.
Bacterial survival in human serum.
Bacterial cells (108 CFU) in the logarithmic phase were suspended in 90% serum–PBS and incubated at 37°C. Viable counts were made at different times until 3 h by dilution and plating as previously described (19, 22). A pool of nonimmune human sera (NHS) was obtained from healthy volunteers. Control experiments using heat-decomplemented NHS were also performed (19, 22).
Binding of C1q to bacterial cells.
C1q was purified from NHS and tested for purity in PAGE as previously described (2). Iodination of purified C1q was carried out with lactoperoxidase-glucose oxidase as described previously (34). Mid-logarithmic-phase bacterial cells were recovered by centrifugation, washed with PBS, and examined with radiolabeled C1q as described previously (2).
General DNA methods.
DNA manipulations were carried out essentially as previously described (28). DNA restriction endonucleases, T4 DNA ligase, E. coli DNA polymerase (Klenow fragment), and alkaline phosphatase were used as recommended by the suppliers.
Construction of an A. hydrophila AH-3 genomic library.
A. hydrophila AH-3 genomic DNA was isolated and partially digested with Sau3A as described by Sambrook et al. (28). Cosmid pLA2917 (3) was digested with BglII, dephosphorylated, and ligated to Sau3A genomic DNA fragments. DNA packaging by using Gigapack Gold III (Stratagene) and infection of E. coli DH5α were carried out as previously described (8). Recombinant clones were selected LB agar plates supplemented with tetracycline (20 μg/ml).
DNA sequencing.
Primers used for DNA sequencing were purchased from Pharmacia LKB Biotechnology. Double-stranded DNA sequencing was performed by the Sanger dideoxy-chain termination method (29) with an ABI Prism dye terminator cycle sequencing kit (Perkin-Elmer).
DNA and protein sequence analysis.
The DNA sequence was translated in all six frames, and all open reading frames (ORFs) greater than 100 bp were inspected. Deduced amino acid sequences were compared with those of DNA translated in all six frames from the nonredundant GenBank and EMBL databases using the BLAST network service at the National Center for Biotechnology Information (4). Multiple sequence alignments and determination of putative terminator sequences were done by using the PileUp and Terminator programs from the Genetics Computer Group (Madison, Wis.) package in a VAX 4300. Prediction of the secondary structure of the porin II sequence was performed using the H's program based on the prediction of beta strands of porins (31).
Construction of porin II-defined insertion mutants.
To obtain defined insertion mutants in the porin II gene, we used a method based on suicide plasmid pFS100 (27). Oligonucleotides 5′-TCTGGCTATTGCTATCCC-3′ (initial base 3283) and 5′-GCTAACACCGTTGATTTTG-3′ (initial base 4279) were used to amplify internal fragment from the porin II gene (996 bp). The amplified fragment was ligated to vector pGEM-T (Promega) and transformed into E. coli DH5α. The fragment was recovered by restriction digestion and was blunt ended with Klenow fragment; finally, it was ligated to EcoRV-digested, blunt-ended, and dephosphorylated pFS100 and transformed into E. coli MC1061(λpir), selecting for kanamycin resistance (Kmr) to generate plasmid pFS-POR3. Plasmid pFS-POR3 was isolated and transformed on E. coli SM10(λpir). Plasmid pFS-POR3 was transferred by conjugation to mesophilic Aeromonas sp. rifampin-resistant (Rifr) strains to obtain defined insertion mutants in porin II gene, selecting for Rifr and Kmr.
Nucleotide sequence accession number.
The nucleotide sequence data presented here have been assigned GenBank accession no. AF183931.
RESULTS AND DISCUSSION
Cloning of A. hydrophila AH-3 genomic region encoding porin II.
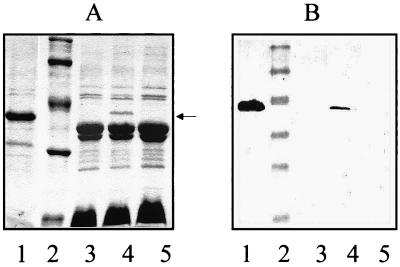
A cosmid-based genomic library of A. hydrophila AH-3 was constructed and introduced into E. coli DH5α as indicated in Materials and Methods. Tetracycline (20 μg/ml)-resistant (Tcr) clones were immunoscreened by colony blotting using specific anti-porin II serum. We found several recombinant clones, a representative one being COS-POR2. Analyses of OM proteins by SDS-PAGE with Coomassie blue stain and Western blotting of OM proteins with specific anti-porin II serum revealed that E. coli DH5α harboring COS-POR2 showed an extra band of approximately 39 kDa, which reacted with specific anti-porin II serum, in comparison with E. coli DH5α or when cured of plasmid COS-POR2 (Fig. 1).
FIG. 1.
Coomassie blue-stained gel (A) and Western immunoblot using specific anti-porin II serum (B) of OM proteins from different bacterial strains. Lanes: 1, A. hydrophila AH-3 (wild-type, serogroup O:34); 2, size standard (20, 31, 43, 62, and 97 kDa); 3, E. coli DHα5 harboring cosmid pLA2917; 4, E. coli DH5α harboring pLA-POR3 plasmid DNA (carrying the porin II gene); 5, the same strain as in lane 4 lacking the pLA-POR3 plasmid DNA. The arrow points to porin II (39 kDa).
To localize the gene responsible for the production of porin II, BglII fragments of the recombinant cosmid COS-POR2 were subcloned into the BglII site of the same cosmid vector and transformed into E. coli DH5α. The recombinant transformants were immunoscreened as previously mentioned. It is important to point out that we use as plasmid vector in order to subclone the same cosmid vector (pLA2917) were initially was cloned the gene, because none of the usual plasmid vectors (pBR328, pACYC184, or pWSK) was able to maintain the DNA insert encoding the porin gene. The initial smallest stable recombinant plasmid (pLA-POR3) exhibiting porin II production (either in SDS-PAGE or Western blotting with anti-porin II serum) was found to harbor a 7.8-kb BglII insert fragment (Fig. 1).
Sequencing of the DNA conferring porin II production.
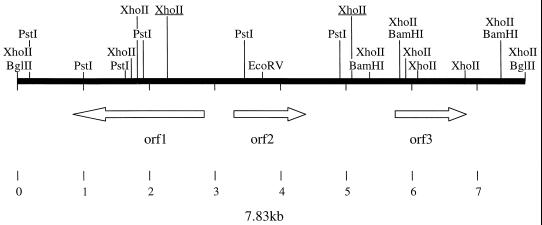
The complete nucleotide sequence of the 7.8-kb DNA insert was determined in both directions. Because we could not use the typical vectors for reasons cited above, we started the sequencing on the same cosmid vector, pLA2917, with the oligonucleotides previously described for the BglII site of this cosmid; other sequence-derived oligonucleotides were used to complete the nucleotide sequence. Analyses of the deduced sequence of pLA-POR3 showed three complete ORFs (Fig. 2). ORF1 and ORF2 are divergently transcribed; between ORF2 and ORF3, a sequence with characteristic features of Rho-independent transcription termination signals was found.
FIG. 2.
Schematic of the organization of the three ORFs in the 7.83-kb DNA insert (from A. hydrophila AH-3) contained in pLA-POR3 plasmid DNA. BglII, PstI, EcoRV, XhoII, and BamHI restriction enzyme sites are indicated. The two XhoII restriction sites underlined were used to clone the complete porin II gene to construct pLA-POR6.
Analysis of deduced amino acid sequences.
Proteins similar to each ORF gene product were analyzed to determine the levels of similarity and identity. As shown in Table 2, ORF1 was found to be similar to DING from both gram-positive and gram-negative bacteria. DING proteins have been proposed to be ATP-dependent helicases. ORF2 was found to be similar to several porin proteins of different enterobacteria and Vibrionaceae. As expected, the deduced amino acid sequence from ORF2 showed a putative signal sequence of 20 amino acid residues. On the other hand, the signal sequence characteristics and the first N-terminal amino acid of the putative mature protein strongly suggested that the AH-3 porin II is processed by a signal peptidase I. The sequence of the porin II gene predicted a protein product of 351 amino acids with a 20-residue signal peptide, whose sequence (1 to 20, MKKTILAIAIPALFASAANA) was previously confirmed by N-terminal sequencing of the purified porin (24). The mature protein deduced from the gene sequence consists of 331 amino acids with a molecular mass of 36,237 Da and has some features in common with other porins: a theoretical acidic pI (4.57), lack of cysteine residues, a regular peak and, as indicated by the Kyte-Doolittle hydropathy plot (16), a predicted structure with 16 beta strands, and a hydrophobic carboxy-terminal sequence with a final Phe that is crucial for the localization in the OM of OmpA and porin PhoE (14). Database comparisons showed that the highest scores of porin II were with porins OmpN, PhoE, and OmpF from E. coli, with porin PhoE from Enterobacter cloacae, with porins OmpK36 and OmpK37 from Klebsiella pneumoniae, and with the OmpL protein from Photobacterium sp., resulting in identity percentages of 27, 25, 24, 27, 26, 26, and 27, respectively. Detailed alignment of porin II amino acid sequence with those of enterobacterial porins with known three-dimensional structures is shown in Fig. 3. Given the low homology with other porins, many insertions and deletions were observed in the predicted loop regions, but the secondary structure of porin II could be predicted due to its beta strand content and to the presence of amino acids Lys-16, Arg-48, Glu-68, Arg-83, Asp-114, Glu-118, and Arg-129. These residues are well conserved in enterobacterial and nonenterobacterial porins (32) and in the known porin structures are distributed across the pore, resulting in a pronounced charge segregation: Asp-106, -113, -106, -114 (sequential PhoE, OmpF, OmpK36, porin II numbering); Glu-110, -117, -110, -118; main carbonyl groups from L3 on one side of the pore; and basic residues Lys-16, Arg-37, -42, -37, -48, Arg-75, -82, -75, -83, and Arg-126, -132, -125, -129 on the other side. These residues seem to be important for the function of porins, since they are also observed in the three-dimensional structure of the Rhodobacter capsulatus porin (38).
TABLE 2.
Amino acid sequence homology of ORF proteins from A. hydrophila AH-3 and other proteins
| Protein, source | No. of amino acids | % Similaritya | % Identitya | Accession no. |
|---|---|---|---|---|
| ORF1 (helicase), Aeromonas hydrophila AH-3 | 690 | AF183931 | ||
| DING (probable ATP-dependent helicase) | ||||
| Escherichia coli | 716 | 54.3 | 34.0 | P27296 |
| Mycobacterium tuberculosis | 664 | 51.2 | 29.7 | Q10640 |
| Bacillus subtilis | 931 | 49.8 | 28.5 | P54394 |
| Yom1 (putative ATP-dependent helicase in OmpH 5′ region, Photobacterium sp. | 151 | 79.5 | 49.6 | P29741 |
| ORF2 (porin II), A. hydrophila AH-3 | 352 | AF183931 | ||
| PhoE, Enterobacter cloacae | 350 | 52.7 | 30.5 | Q47490 |
| OmpF, Serratia marcescens | 374 | 51.6 | 29.9 | O33980 |
| OmpN, E. coli | 377 | 51.4 | 29.8 | P77747 |
| OmpK37, Klebsiella pneumoniae | 374 | 50.4 | 29.2 | e1325636 |
| OmpL, Photobacterium sp. | 341 | 48.7 | 28.9 | Q52581 |
| ORF3 (AspC), A. hydrophila AH-3 | 396 | AF183931 | ||
| AspC (aspartate aminotransferase) | ||||
| E. coli | 396 | 92.4 | 63.6 | P00509 |
| Haemophilus influenzae | 396 | 88.6 | 60.8 | P44425 |
| Moraxella sp. | 397 | 67.8 | 49.7 | O53137 |
Obtained from pairwise comparisons using the Gap program (gap weight, 12; length weight, 12).
FIG. 3.
Alignments of the A. hydrophila AH-3 porin II sequence with sequences of PhoE from Enterobacter aerogenes, OmpF from E. coli, and OmpK36 from K. pneumoniae. Protein sequences were derived from nucleotide sequences. Secondary structural motifs are those of OmpF structure. L1 to L8, loops 1 to 8; (in black letters); T1 to T8, turns 1 to 8; β1 to β6, beta strands (in black boxes); ∗, main basic residue conserved in enterobacterial or nonenterobacterial porin (32).
Finally, since the similarity and identity data strongly suggest that ORF3 encodes an aspartate aminotransferase, the gene was named aspC, consistent with its E. coli homolog. In E. coli the ompF gene is found between asnS and aspC, all three genes being transcribed in the same direction. The region described here showed the porin, II gene and aspC being adjacent and transcribed in the same direction (as in E. coli); however, no asnS was found upstream of the porin II gene.
Construction of defined porin II insertion mutants and complementation.
Plasmid pFS-POR3, a replication pir-dependent construct carrying an internal fragment of the porin II gene, was transferred by mating independently to Rifr mesophilic Aeromonas sp. strains AH-405 (serogroup O:34) and AH-408 (serogroup O:11), and Rifr and Kmr colonies from both matings were selected. We obtained several mutants, AH-330 and AH-331 being representative of serogroups O:34 and O:11, respectively. The insertion of plasmid pFS-POR3 in these mutants was confirmed by Southern blotting using appropriate DNA probes. As shown in Fig. 4, no porin II could be detected either in SDS-PAGE of OM proteins or in Western blot analysis using OM proteins and antibodies against porin II.
FIG. 4.
Coomassie blue-stained gel (A) and Western immunoblot using specific anti-porin II serum (B) of OM proteins from different mesophilic Aeromonas sp. strains. Lanes: 1, size standard (20, 31, 43, 62, and 97 kDa); 2, AH-331 (porin II-deficient mutant from strain AH-408); 3, AH-408 (TF7 Rifr, serogroup O:11); 4, AH-335 (AH-331 complemented with pLA-POR6 plasmid DNA [porin II gene]); 5, AH-330 (porin II-deficient mutant from strain AH-405); 6, AH-405 (AH-3 Rifr, serogroup O:34); 6, AH-334 (AH-330 complemented with pLA-POR6 plasmid DNA [porin II gene]). The arrow points to porin II; ∗∗ denotes protein S of the S-layer characteristic of strains from serogroup O:11.
Complementation of these mutants with the single gene for the porin II was accomplished as follows. pLA-POR3 was digested with XhoII; a band of 2,732 bp was recovered and ligated to pLA2917 previously digested with BglII and dephosphorylated, creating pLA-POR6. This plasmid was transferred by mating to mutants AH-330 and AH-331, selecting for Tcr and Kmr, to obtain strains AH-334 and AH-335, respectively. Strains AH-334 and AH-335 exhibited porin II, as can be observed by SDS-PAGE of OM proteins or by Western blot analysis using OM proteins and antibodies against porin II (Fig. 4).
To study the role of porin II in previously obtained serum-sensitive mutants (AH-53 and AH-26 [24]), we performed the same experiments to obtain defined insertion mutants in the porin II gene (AH-336 and AH-337, respectively) and complementation studies with pLA-POR6 on these mutants AH-336 and AH-337 (strains AH-338 and AH-339, respectively).
Serum susceptibility and C1q binding.
Porin II-defined insertion mutants (AH-330 and AH-331) from the serum-resistant wild-type strains (AH-3 and AH-1, respectively) showed similar resistance to complement-mediated killing. This result suggested that when porin II was lost but other surface molecules like O:34 antigen LPS in AH-330 (smooth strain) and O:11 antigen LPS and S-layer in AH-331 (smooth S+ strain) remained, no changes in serum susceptibility occurred (Table 3). However, strain AH-330 cultivated under conditions where no O:34 antigen LPS is expressed (rough) (37°C and low osmolarity [1, 20]) was resistant to serum, while the wild-type strain (AH-3) and the Rifr mutant (AH-405) were serum sensitive under the same growth conditions (1, 20). Complementation of AH-330 with pLA-POR6 (porin II gene) renders this strain (AH-334) serum sensitive under the growth conditions mentioned above. This finding prompted us to examine the porin II-defined insertion mutants (AH-336 and AH-337) from serum-sensitive strains (AH-53 and AH-26, respectively) which showed resistance to complement-mediated killing similar to the initial wild-type strains (AH-3 and AH-1). However, when we reintroduced the porin II gene in these strains (AH-336 and AH-337) by complementation with plasmid pLA-POR6 (strains AH-338 and AH-339, respectively), they became as serum sensitive as strains AH-53 and AH-26 (Table 3). These results indicate that when mutants lacking the O-antigen LPS or the O:34 strains not expressing this antigen (serum sensitive) are devoid of porin II, they became serum resistant, and reintroduction of the porin II gene renders the strains again serum sensitive. Because we previously showed that porin II is able to bind C1q (the initial component of the CPC pathway), we studied the C1q binding of whole cells in these strains. As shown in Table 3, porin II-defined insertion mutants (AH-336 and AH-337) from serum-sensitive strains showed a large reduction in C1q-bound molecules in comparison with AH-53 and AH-26 and became serum resistant. Complementation of these defined insertion mutants (AH-336 and AH-337) with pLA-POR6 (carrying the porin II gene), i.e., reintroduction of porin II, renders these strains (AH-338 and AH-339) serum sensitive because there are numerous of C1q molecules bound to their bacterial surface. From these results seems clear that porin II is the major C1q binding surface on these mesophilic Aeromonas sp. strains.
TABLE 3.
Serum susceptibility and binding of 125I-labeled C1q to mesophilic Aeromonas sp. whole cells
| Strain | Relevant characteristic(s) | % Survival after 3 h in NHSa | Bound C1q (molecules/bacterial cell)a |
|---|---|---|---|
| AH-3 | O:34 (smooth)b | 101 ± 4 | 89 ± 17 |
| AH-3 | O:34 (rough)c | <0.1 | 304 ± 37 |
| AH-405 | AH-3 Rifr (smooth) | 103 ± 3 | 87 ± 18 |
| AH-405 | AH-3 Rifr (rough) | <0.1 | 309 ± 40 |
| AH-53 | O− derived from AH-3 (20) | <0.1 | 378 ± 42 |
| AH-330 | Porin II mutant of AH-405 (smooth) | 108 ± 6 | <30 |
| AH-330 | Porin II mutant of AH-405 (rough) | 105 ± 4 | <30 |
| AH-334 | AH-330 + pLA-POR6 (smooth) | 99 ± 3 | 89 ± 15 |
| AH-334 | AH-330 + pLA-POR6 (rough) | <0.1 | 317 ± 33 |
| AH-336 | Porin II mutant of AH-53 | 98 ± 5 | <30 |
| AH-338 | AH-336 + pLA-POR6 | <0.1 | 369 ± 36 |
| TF7 | O:11, S+ | 145 ± 12 | <10 |
| AH-408 | TF7 Rifr | 139 ± 8 | <10 |
| AH-26 | O− S−, derived from TF7 (22) | <0.1 | 389 ± 38 |
| AH-331 | Porin II mutant of AH-408 | 141 ± 6 | <10 |
| AH-335 | AH-331 + pLA-POR6 | 143 ± 7 | <10 |
| AH-337 | Porin II mutant of AH-26 | 129 ± 8 | <30 |
| AH-339 | AH-337 + pLA-POR6 | <0.1 | 376 ± 35 |
Average of at least three independent experiments ± standard deviation. Some of the data have been previously published (24).
Strains of serogroup O:34 showed O:34 antigen LPS (smooth) when grown at 20°C (20).
Strains of serogroup O:34 are unable to show O:34 antigen LPS (rough) when grown at 37°C and in low-osmolarity medium (1).
A confirmation of this point is that porin II-defined insertion mutants from serum-resistant wild-type strains showed no difference, in either serum susceptibility or C1q-bound molecules, because of the interference of the O-antigen LPS and also the S-layer in serogroup O:11 strains. However, when porin II-defined insertion mutant AH-330 of serogroup O:34 showed a rough LPS (growing conditions of 37°C and low osmolarity [1, 20]), it was resistant to serum killing and exhibited a great reduction of C1q-bound molecules in comparison with the wild-type strains or the same mutant complemented by porin II gene (pLA-POR6) in the same growth conditions (Table 3).
It is important to point out that rough Aeromonas sp. strains (serum sensitive, easily eliminated by complement), if they lose porin II by different mechanisms (for instance, by insertion sequence interruption of the gene as happens in other gram-negative bacteria [10, 18]), could became serum resistant, develop antibiotic resistance, and emerge as a serious clinical problem.
ACKNOWLEDGMENTS
This work was supported by grants from DGICYT and Plan Nacional de I+D (Ministerio de Educación y Cultura, Spain). A.A. and M.M.N. are predoctoral fellowships from the same institution and Generalitat de Catalunya, respectively. We thank Maite Polo for technical assistance and Tilman Schirmer for help with the H's program.
REFERENCES
- 1.Aguilar A, Merino S, Rubires X, Tomás J M. The influence of osmolarity on lipopolysaccharide and virulence of Aeromonas hydrophila strains serotype O:34 grown at 37°C. Infect Immun. 1997;65:1245–1250. doi: 10.1128/iai.65.4.1245-1250.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albertí S, Marqués G, Camprubí S, Merino S, Tomás J M, Vivanco F, Benedí V J. C1q binding and activation of the complement classical pathway by Klebsiella pneumoniae outer membrane proteins. Infect Immun. 1993;61:852–860. doi: 10.1128/iai.61.3.852-860.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen L N, Hanson R S. Construction of broad-host-range cosmid cloning vector: identification of genes necessary for growth of Methylobacterium organophilum on methanol. J Bacteriol. 1985;161:955–962. doi: 10.1128/jb.161.3.955-962.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Atlas R M, Parks L C, editors. Handbook of microbiological media. Boca Raton, Fla: CRC Press Inc.; 1993. [Google Scholar]
- 6.Darveau R P, Hancock R E W. Procedure for the isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J Bacteriol. 1983;155:831–838. doi: 10.1128/jb.155.2.831-838.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figueroa J E, Densen P. Infectious diseases associated with complement deficiencies. Clin Microbiol Rev. 1991;4:359–395. doi: 10.1128/cmr.4.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guasch J F, Piqué N, Climent N, Ferrer S, Merino S, Rubirés X, Tomás J M, Regué M. Cloning and characterization of two Serratia marcescens genes involved in core lipopolysaccharide biosynthesis. J Bacteriol. 1996;178:5741–57475. doi: 10.1128/jb.178.19.5741-5747.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:577–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 10.Hernández-Allés S, Benedí V J, Martinez-Martinez L, Pascual A, Aguilar A, Tomás J M, Albertí S. Development of resistance during antimicrobial therapy caused by insertion sequence interruption of porin genes. Antimicrob Agents Chemother. 1999;43:937–939. doi: 10.1128/aac.43.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janda J M, Kokka R P. The pathogenicity of Aeromonas strains relative to genospecies and phenospecies identification. FEMS Microbiol Lett. 1991;90:29–34. doi: 10.1111/j.1574-6968.1991.tb05120.x. [DOI] [PubMed] [Google Scholar]
- 12.Janda J M, Guthertz L S, Kokka R P, Shimada T. Aeromonas species in septicemia: laboratory characteristics and clinical observations. Clin Infect Dis. 1994;19:77–83. doi: 10.1093/clinids/19.1.77. [DOI] [PubMed] [Google Scholar]
- 13.Jeanteur D, Gletsu N, Pattus F, Buckley J T. Purification of Aeromonas hydrophila major outer-membrane proteins: N-terminal sequence analysis and channel-forming properties. Mol Microbiol. 1992;6:3355–3363. doi: 10.1111/j.1365-2958.1992.tb02203.x. [DOI] [PubMed] [Google Scholar]
- 14.Jeanteur D, Lakey J H, Pattus F. The porin superfamily: diversity and common features. In: Ghuysen J M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier; 1994. [Google Scholar]
- 15.Kokka R P, Janda J M. Isolation and identification of autoagglutinating serogroup O:11 Aeromonas strains in the clinical laboratory. J Clin Microbiol. 1990;28:1297–1299. doi: 10.1128/jcm.28.6.1297-1299.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Martinez L, Hernández-Allés S, Albertí S, Tomás J M, Benedí V J, Jacoby G A. In vivo selection of porin deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum cephalosporins. Antimicrob Agents Chemother. 1996;40:342–348. doi: 10.1128/aac.40.2.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merino S, Camprubí S, Tomás J M. The role of lipopolysaccharide in complement-killing of Aeromonas hydrophila strains of serotype O:34. J Gen Microbiol. 1991;137:1583–1590. doi: 10.1099/00221287-137-7-1583. [DOI] [PubMed] [Google Scholar]
- 20.Merino S, Camprubí S, Tomás J M. Effect of the growth temperature on outer membrane components and virulence of Aeromonas hydrophila strains of serotype O:34. Infect Immun. 1992;60:4343–4349. doi: 10.1128/iai.60.10.4343-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merino S, Camprubí S, Tomás J M. Incidence of Aeromonas sp. serotypes O:34 and O:11 among clinical isolates. Med Microbiol Lett. 1993;2:48–55. [Google Scholar]
- 22.Merino S, Rubires X, Aguilar A, Albertí S, Hernández-Allés S, Benedí V J, Tomás J M. Mesophilic Aeromonas sp. serogroup O:11 resistance to complement-mediated killing. Infect Immun. 1996;64:5302–5309. doi: 10.1128/iai.64.12.5302-5309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merino S, Rubirés X, Aguilar A, Tomás J M. The role of flagella and motility in the adherence and invasion to fish cell lines by Aeromonas hydrophila serogroup O:34 strains. FEMS Microbiol Lett. 1997;151:213–217. doi: 10.1111/j.1574-6968.1997.tb12572.x. [DOI] [PubMed] [Google Scholar]
- 24.Merino S, Nogueras M M, Aguilar A, Rubires X, Albertí S, Benedí V J, Tomás J M. Activation of the complement classical pathway (C1q binding) by mesophilic Aeromonas hydrophila outer membrane protein. Infect Immun. 1998;66:3825–3831. doi: 10.1128/iai.66.8.3825-3831.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Misra S K, Shimada T, Bhadra R K, Pal S C, Nair G B. Serogroups of Aeromonas species from clinical and environmental sources in Calcutta, India. J Diarrhoeal Dis Res. 1989;7:8–12. [PubMed] [Google Scholar]
- 26.Murray R G E, Dooley J S G, Whippey P W, Trust T J. Structure of an S-layer on a pathogenic strain of Aeromonas hydrophila. J Bacteriol. 1988;170:2625–2636. doi: 10.1128/jb.170.6.2625-2630.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rubirés X, Saigi F, Piqué N, Climent N, Merino S, Albertí S, Tomás J M, Regué M. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J Bacteriol. 1997;179:7581–7586. doi: 10.1128/jb.179.23.7581-7586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakazaki R, Shimada T. O-serogrouping scheme for mesophilic Aeromonas strains. Jpn J Med Sci Biol. 1984;37:247–255. doi: 10.7883/yoken1952.37.247. [DOI] [PubMed] [Google Scholar]
- 31.Schirmer T, Cowan S. Prediction of membrane-spanning beta strands and its application to maltoporin. Protein Sci. 1993;2:1361–1363. doi: 10.1002/pro.5560020820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Struyvé M, Moons M, Tommassen J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Biol Chem. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 33.Taylor P W. Bacterial resistance to complement. In: Roth J A, editor. Virulence mechanisms of bacterial pathogens. Washington, D.C.: American Society for Microbiology; 1988. pp. 107–120. [Google Scholar]
- 34.Tenner A J, Lesavre P H, Cooper N R. Purification and radiolabeling of human C1q. J Immunol. 1981;127:648–653. [PubMed] [Google Scholar]
- 35.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai C M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharide in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 37.Vukajlovich S W, Hoffman J, Morrison D C. Activation of human serum complement by bacterial lipopolysaccharides: structural requirements for antibody independent activation of the classical and alternative pathways. Mol Immunol. 1987;24:319–331. doi: 10.1016/0161-5890(87)90173-8. [DOI] [PubMed] [Google Scholar]
- 38.Weiss M S, Kreusch A, Schiltz E, Nestel U, Welte W, Weckesser J, Schulz G E. The structure of porin from Rhodobacter capsulatus at 1.8 A resolution. FEBS Lett. 1991;280:379–382. doi: 10.1016/0014-5793(91)80336-2. [DOI] [PubMed] [Google Scholar]
- 39.Westphal O, Jann K. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure. Carbohydr Chem. 1965;5:83–91. [Google Scholar]