Abstract
Legionella pneumophila, the agent of Legionnaires' disease, is an intracellular pathogen of protozoa and macrophages. Previously, we had determined that the Legionella pilD gene is involved in type IV pilus biogenesis, type II protein secretion, intracellular infection, and virulence. Since the loss of pili and a protease do not account for the infection defect exhibited by a pilD-deficient strain, we sought to define other secreted proteins absent in the mutant. Based upon the release of p-nitrophenol (pNP) from p-nitrophenyl phosphate, acid phosphatase activity was detected in wild-type but not in pilD mutant supernatants. Mutant supernatants also did not release either pNP from p-nitrophenyl caprylate and palmitate or free fatty acid from 1-monopalmitoylglycerol, suggesting that they lack a lipase-like activity. However, since wild-type samples failed to release free fatty acids from 1,2-dipalmitoylglycerol or to cleave a triglyceride derivative, this secreted activity should be viewed as an esterase-monoacylglycerol lipase. The mutant supernatants were defective for both release of free fatty acids from phosphatidylcholine and degradation of RNA, indicating that PilD-negative bacteria lack a secreted phospholipase A (PLA) and nuclease. Finally, wild-type but not mutant supernatants liberated pNP from p-nitrophenylphosphorylcholine (pNPPC). Characterization of a new set of mutants defective for pNPPC-hydrolysis indicated that this wild-type activity is due to a novel enzyme, as opposed to a PLC or another known enzyme. Some, but not all, of these mutants were greatly impaired for intracellular infection, suggesting that a second regulator or processor of the pNPPC hydrolase is critical for L. pneumophila virulence.
Legionella pneumophila is the agent of Legionnaires' disease, a potentially fatal form of pneumonia (76). L. pneumophila, a gram-negative inhabitant of fresh water, enters the respiratory tract following either the inhalation of contaminated aerosols generated by air conditioners and other devices or the aspiration of contaminated potable water (22, 38, 76). Once in the alveoli, the bacterium invades and replicates to high numbers within macrophages (32, 76). Ultimately, host cell death and lysis as well as bacterial degradative enzymes result in damage to lung tissue. We recently discovered that L. pneumophila possesses a gene (pilD) whose analogs, in other gram-negative bacteria, promote both pilus biogenesis and protein secretion (42). The inner membrane PilD-related proteins facilitate type IV pilus formation in two ways (15, 28, 35, 40, 43, 44, 51, 56, 77). First, they cleave the signal sequence from type IV prepilin and methylate the amino terminus of the resultant mature pilin (52, 71). Second, they cleave and methylate six prepilin-like proteins that, once processed, help form the scaffold through which pilin is assembled into a pilus (1, 2, 23, 61). The PilD peptidases also facilitate the passage of proteins through the main terminal branch of the general secretory pathway, a form of protein export that is commonly known as type II secretion (9, 23, 25, 45, 58, 60, 70). PilD and its analogs promote secretion by processing another set of pseudopilins, which constitute part of the type II secretion apparatus (8, 10, 53). Factors whose secretion is PilD dependent include the aerolysin and protease of Aeromonas hydrophila, the exotoxin A, lipase, and phospholipase C (PLC) of Pseudomonas aeruginosa, and the cholera toxin of Vibrio cholerae (24, 44, 56, 58, 63, 70).
Given the pivotal role that PilD-like proteins can have in virulence, we recently began an analysis of an L. pneumophila pilD mutant (41). It was first observed that the mutant was nonpiliated, confirming a role for PilD in the biogenesis of L. pneumophila type IV pili (41, 42, 69). Examination of the mutant's supernatants revealed the loss of several protein species, confirming that PilD is required for Legionella protein secretion (41). This latter observation indicated that L. pneumophila possesses a type II secretion system, a hypothesis that was later confirmed by the identification of genes encoding components of the secretion apparatus, including the pseudopilins (29). Most importantly, the pilD mutant was defective for intracellular infection and virulence (41). The strain was ca. 1,000-fold impaired in its ability to infect a human macrophage (U937) cell line and a Hartmannella strain of fresh water amoebae. In addition, it did not replicate within the lungs of guinea pigs, displaying a 50% lethal dose that was at least 100-fold greater than wild type. Since the Legionella type IV pilus is not critical for intracellular growth (69), we reasoned that the mutant's attenuation was due to the loss of PilD-dependent, secreted proteins (41). However, the only exoprotein known to be lacking in the PilD-negative strain was a metalloprotease, an enzyme that is not required for intracellular infection and has only a minor role in pulmonary disease (41, 48, 72). Shortly after its discovery, L. pneumophila was found to exhibit phosphatase, lipase, nuclease, and PLC-like activities (7, 16, 49, 50, 73). Because some of these activities are linked to type II secretion in other bacteria, they served as a starting point in our search for new PilD-dependent exoproteins. Here, we report that the L. pneumophila pilD mutant is defective for the secretion of an acid phosphatase, monoacylglycerol lipase, RNase, PLA, and p-nitrophenylphosphorylcholine (pNPPC)-hydrolase.
(Portions of this work were presented at the 99th General Meeting of the American Society for Microbiology [abstr. D/B-71, p. 223] in 1999.)
MATERIALS AND METHODS
Bacterial strains and media.
The wild-type L. pneumophila used in this study was serogroup 1 strain 130b (Wadsworth), a virulent clinical isolate (21). Mutant NU243, a direct derivative of 130b, contains a stable mini-Tn10 (kanamycin resistance) insertion in the Legionella pilD gene (41). The strains NU243 (pMRL13), NU243 (pBBR1MCS), and 130b (pBBR1MCS) that were used for trans-complementation analysis were also previously described (41). To ultimately screen for mutants deficient in specific secretion activities, strain 130b was mutagenized with mini-Tn10phoA, as previously described (46, 57). After mini-Tn10 mutagenesis, at least 96% of L. pneumophila mutants contain single DNA insertions (57). Bacteria were generally cultured on buffered charcoal yeast extract agar for 3 days at 37°C (18). However, to facilitate the detection of certain lytic enzymes, legionellae were also cultured on buffered starch yeast extract agar containing 5% egg yolk (7, 73). Finally, in preparation for assessing secreted enzymatic activities, bacteria were grown in buffered yeast extract (BYE) broth, the standard liquid medium for culturing L. pneumophila. Growth was assessed by measuring the optical density of the culture at 660 nm (OD660) (41).
Preparation of supernatants and cell lysates.
Supernatants from L. pneumophila cultures to be tested for secreted enzymes were prepared in the following manner. First, bacteria from buffered charcoal yeast extract agar were suspended in 25 ml of BYE broth, contained within 125-ml flasks, at an OD660 of approximately 0.1. Then, after overnight growth at 37°C, the broth-adapted legionellae were subcultured into 25 ml of fresh medium, and the cultures were returned to the 37°C shaking incubator. At various times postinoculation, a 1.5-ml portion of the culture was removed and centrifuged for 5 min at 12,000 × g at 4°C. Finally, after careful removal from the centrifuge tube, the supernatant was sterilized by passage through a 0.2-μm (pore-size) filter and either assayed immediately or stored at −20°C. Frozen samples retained all activities tested for up to at least 6 months. In order to detect some activities, ca. 200 ml of chilled supernatants were concentrated 40-fold by passage through Millipore YM10 ultrafiltration cells (41). To assay for cell-associated activities, the pellet obtained from centrifugation of the culture sample was lysed by resuspension in 300 μl of phosphate-buffered saline containing 0.1% Triton X-100 and 0.2 mg of lysozyme per ml. After repeated passage through a 26-gauge needle, the lysate was tested immediately or stored at −20°C.
Enzymatic assays.
To detect phosphatase activity, samples were assayed, as is routinely done, for their ability to release p-nitrophenol (pNP) from p-nitrophenylphosphate (Sigma Chemical, St. Louis, Mo.) (24, 37, 67, 70, 73). Briefly, 10 μl of sample was added to 100 μl of a 50 mM citric acid buffer containing 7.6 mM p-nitrophenylphosphate, and then after 1 (for concentrated supernatants) or 5 h of incubation at 37°C, the production of pNP was monitored at 410 nm. To distinguish between acid and alkaline phosphatase activities, the reactions were performed at pH 5 and pH 10. The alkaline phosphatase (type III-L) of Escherichia coli and the acid phosphatase (type IV-S) of potato, both obtained from Sigma, served as standards in this assay. One unit of enzyme activity was defined as that which yields 1 nM pNP in 1 min.
Secreted protease activity, which was previously documented to be deficient in the pilD mutant, was quantitated by using hide powder azure and azocasein assays (17, 41, 73).
Lipase activity was monitored in three different ways. First, supernatants were assayed for the release of pNP from p-nitrophenyl caprylate and p-nitrophenyl palmitate (Sigma) (3, 20, 24, 34, 73, 75). Briefly, 100 μl of sample was added to 1 ml of buffer (i.e., 100 mM Tris [pH 8] and 0.2% Triton X-100) containing 1 mM of substrate, and then the increase in absorbance at 410 nm was measured after 5 and 15 min of incubation at 37°C. One unit of enzyme was defined as that which yields 1 nM pNP in 1 min. Second, the samples were tested for their ability to release free fatty acid from 1-monopalmitoylglycerol (1-MG) and 1,2-dipalmitoylglycerol (1,2-DG) (34). Thus, supernatants were incubated for 15 h at 37°C in 20 mM Tris-HCl containing 3 mM sodium azide, 0.5% Triton X-100, and either 2 mg of 1-MG (Sigma) or 1.6 mg of 1,2-DG (Sigma) per ml. After this incubation, free fatty acid levels were determined by the NEFA-C-Kit obtained from Wako Chemicals (Neuss, Germany) (31). Finally, culture supernatants were examined for their ability to release resorufin from 1,2-O-dilauryl-rac-glycero-3-glutaric acid resorufin ester (Boehringer, Indianapolis, Ind.) (34). Toward that end, 50 μl of sample was added to a tube containing 0.85 ml of 100 mM KH2PO4 (pH 6.8) and 0.1 ml of a 1-mg/ml ester solution. After incubation at room temperature, the release of the red resorufin was monitored spectrophotometrically at 572 nm. Standards in these assays were the lipases of Rhizopus arrhizus (Boehringer) and Pseudomonas sp. (Sigma).
PLA activity was determined by assaying for the release of free fatty acid from a dipalmitoylphosphatidylcholine (DPPC) (26, 31). Thus, unconcentrated supernatants were incubated for 15 h at 37°C in 20 mM Tris-HCl containing 3 mM sodium azide, 0.5% Triton X-100, and 5 mg of DPPC (Sigma) per ml. Then, free fatty acid levels were determined by the NEFA-C-Kit and visualized by thin-layer chromatography after staining with copper sulfate phosphoric acid reagent (26, 74). The standard for this series of experiments was the PLA2 of Streptomyces violaceoruber, obtained from Sigma.
To detect nuclease activity, supernatants were examined for their ability to hydrolyze RNA and DNA contained within Noble agar (64, 73). Toward that end, a thin layer of agar containing either 0.15% RNA or 0.1% DNA was placed onto a glass slide, and then 40 μl of concentrated supernatant was placed within a well that had been centered in the overlay. After overnight incubation at room temperature, the gel was stained with ethidium bromide and observed for clearing. RNA (type II-C) of Torula yeast and DNA (type I) of calf thymus from Sigma served as the substrates for these experiments, while RNase ONE and RQ1 DNAse from Promega (Madison, Wis.) were used as standards.
The release of pNP from pNPPC, a reaction most often ascribed to PLC enzymes, was monitored as previously described (6, 24, 39, 68, 70). Briefly, 100 μl of supernatant samples was added to 1 ml of 50 mM HEPES (pH 7.5) buffer containing 5 mM CaCl2, 5 mM MnCl2, 3 mM sodium azide, 0.5% Triton X-100, and 2.5 mM pNPPC, and then after overnight incubation at 37°C the amount of pNP was recorded as above. The PLC (type IV) of Bacillus cereus (Sigma) served as a control in the pNPPC hydrolysis tests, with one unit of enzyme activity being defined as that which yields 1 nM pNP in 1 min.
To assay for the possible presence of a Legionella glycerophosphorylcholine-phosphocholine (GPC)-phosphodiesterase, culture supernatants were incubated for 4 h at 37°C in 20 mM Tris-HCl (pH 7.2) containing 3 mM sodium azide, 0.5% Triton X-100, and 5 mg of GPC (Sigma) per ml. Subsequently, inorganic phosphate levels were estimated as previously described (19). When no activity was detected, 0.4 U of both the acid and alkaline phosphatase standards (see above) were added to the mixture. After an additional 19 h of incubation, a second inorganic phosphate determination was done.
Intracellular infection of U937 cells and Hartmannella amoebae.
U937, a human cell line that differentiates into macrophage-like cells after treatment with phorbol esters, served as a host for in vitro infection by L. pneumophila (12). The cell line was maintained and infected as previously described (12, 41). To assess the relative infectivity of L. pneumophila strains, 50% infective doses (ID50) were determined after a 3-day incubation period (41). In a number of studies, the ID50 value has proven to be a reliable predictor of a given strain's intracellular growth capacity (12, 30, 54). To quantitate intracellular growth, monolayers containing 105 macrophages were inoculated with approximately 105 CFU, incubated for 0, 24, 48, 72, or 96 h, and then lysed. Serial dilutions of the lysates were plated on BCYE agar to determine the corresponding numbers of bacteria per monolayer (41). To ascertain the cytopathic effect of L. pneumophila on U937 cells, infected monolayers were treated with alamar blue (Biosource International, Vacaville, Calif.) or MTT (Sigma), as previously described (12, 41). Subsequent assessments were performed by using neutral red, as follows (4). The dye (Sigma) was added to infected cells to give a final concentration of 40 μg/ml and then, after a 4-h incubation at 37°C, the wells were washed with 1% CaCl2–0.5% formaldehyde. Finally, after 100 μl of 1% acetic acid in ethanol was added to extract the dye, the plates were examined at 540 nm. To examine the ability of legionellae to grow within a protozoan host, Hartmannella vermiformis was infected and characterized as previously indicated (13, 41). Thus, approximately 103 CFU were added to wells containing 105 amoebae and then, at 0, 24, 48, 72, or 96 h postinoculation, the numbers of bacteria within the coculture were determined by plating.
RESULTS
For the following experiments, the pilD mutant NU243 and its wild-type parent 130b were grown in BYE broth, and then supernatants were analyzed for enzymatic activities. Since NU243 and 130b have identical growth patterns in BYE, except for slightly reduced viability and/or recoverability in very late stationary phase (41), culture supernatants could be directly compared.
Acid phosphatase secretion by L. pneumophila strains.
Early studies reported acid and alkaline phosphatase activities for 10 different strains of L. pneumophila (49, 50, 73). Although the alkaline phosphatase is now known to be a periplasmic enzyme that is not critical for intracellular infection (37), the location and significance of the acid phosphatase has remained unclear. Thus, we began our study by determining whether filter-sterilized supernatants from wild-type cultures effectively released pNP from p-nitrophenylphosphate at pH 5. The initial experiment indicated that an acid phosphatase activity was detectable at any time after mid-log phase (Fig. 1A and B). Supernatants from late-log-phase 130b cultures also had acid phosphatase activity (Fig. 2). For three reasons, we believe that this activity is reflective of a secreted enzyme rather than a cell-associated protein that had simply leaked into the medium. First, the supernatants containing acid phosphatase lacked appreciable alkaline phosphatase activity (data not shown). Second, the acid phosphatase was easily detected by using unconcentrated supernatants. Third, it was apparent in mid log phase, a stage of growth lacking significant cell lysis.
FIG. 1.
Acid phosphatase, protease, and pNPPC-hydrolase activity within L. pneumophila wild-type and pilD mutant supernatants. (A) Wild-type 130b (●) and mutant NU243 (□) were inoculated into BYE broth, and then bacterial growth was monitored spectrophotometrically. After various times of incubation, filtered culture supernatants were examined for phosphatase activity at pH 5 (B), protease activity against azocasein (C), and hydrolysis of pNPPC (D). The values given represent the mean and standard deviations from triplicate cultures. Significant differences in enzyme activity were most evident during late-log and early-stationary phases, i.e., P < 0.001 and 0.005 for the 10- and 13-h time points, respectively, in panels B and C, and P < 0.001 for the 13 h time point in panel D (Student's t test). The increasing levels of enzymatic activity in the older mutant cultures likely reflect inevitable cell lysis (see Fig. 2). Comparable results were obtained in an additional, identical experiment. Single readings of late-log cultures were also obtained on at least five additional occasions (see Fig. 2 and data not shown).
FIG. 2.
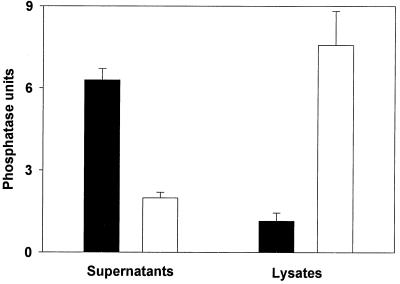
Acid phosphatase secretion by and accumulation within L. pneumophila wild-type and pilD mutant bacteria. Wild-type 130b (black columns) and mutant NU243 (white columns) were grown in BYE broth, and then, at late log phase, culture supernatants (left) and cell lysates (right) were examined for phosphatase activity at pH 5. The values presented are the mean and standard deviations from triplicate cultures. For both the supernatant and lysate comparisons, the differences in strain activity were significant (P < 0.001, Student's t test). Similar results were obtained on six additional occasions.
With the realization that the L. pneumophila acid phosphatase is secreted, we next examined the supernatants from NU243 cultures for loss of activity. The pilD mutant's supernatants exhibited a level of phosphatase activity that was significantly less than that of wild type (Fig. 1B and 2). The loss of acid phosphatase was most apparent when mid- to late-log-phase culture supernatants were compared. In a similar way, the general protein secretion defect of NU243 was most clearly seen by sodium dodecyl sulfate-polyacrylamide gel electrophoresis when log-phase cultures were analyzed (41). Since type II secreted proteins generally accumulate within pilD mutants (24, 56, 70), we compared mutant and wild-type cell lysates for differences in acid phosphatase activity. Upon examination of late-log-phase lysates, NU243 contained fivefold more acid phosphatase activity than did strain 130b (Fig. 2). Taken together, these data indicate that the L. pneumophila acid phosphatase is a secreted enzyme whose export is dependent upon the prepilin peptidase.
Protease secretion by L. pneumophila strains.
During these experiments, we took the opportunity to assess the kinetics of metalloprotease secretion. Although earlier work had determined the protease to be secreted and pilD dependent (17, 41), these studies apparently did not monitor when, in the course of growth, the enzyme was released. As presented in Fig. 1C, the kinetics of protease production paralleled that of acid phosphatase secretion.
Esterase-lipase secretion by L. pneumophila strains.
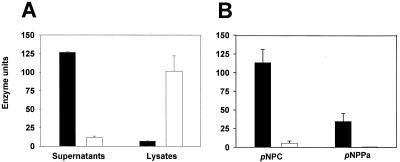
All 13 strains of L. pneumophila previously tested possessed lipase-like activities (5, 7, 49, 50, 73). Since there has been some debate as to whether these activities reflect a true lipase or a simple esterase (49, 50), we examined wild-type legionellae in three different lipase assays. First, we assessed their supernatants' ability to release pNP from p-nitrophenyl caprylate. Strain 130b supernatants, but not cell lysates, contained significant levels of reactivity (Fig. 3A). As was noted for the phosphatase and protease activities, the lipase-like activity first appeared in mid-log phase and peaked by late-log phase (data not shown). For the same reasons cited above, we believe that this activity also represents a bona fide secreted enzyme. With the confirmation of the lipase-like activity in Legionella supernatants, we reinvestigated the capacity of the enzyme to process long-chain fatty acids by using p-nitrophenyl palmitate as the substrate (73, 75). Indeed, supernatants from strain 130b split the palmitate substrate, albeit not as well as the caprylate substrate (Fig. 3B), suggesting the existence of a lipase. To test this hypothesis, we determined whether the 130b product could release free fatty acid from 1-MG and 1,2-DG substrates. The Legionella sample cleaved 1-monoacylglycerol (Fig. 4) but not the diacylglycerol substrate (data not shown), suggesting that L. pneumophila only possesses a monoacylglycerol lipase. Finally, we employed a new spectrophotometric assay that uses an artificial triglyceride with long-chain fatty acids in the sn-1 and sn-2 positions, i.e., measurement of the release of resorufin from 1,2-O-dilauryl-rac-glycero-3-glutaric acid resorufin ester by lipases (35). Neither concentrated nor unconcentrated 130b supernatants released resorufin from the triglyceride derivative, demonstrating that L. pneumophila indeed lacks a triacylglycerol lipase.
FIG. 3.
Secretion, accumulation, and substrate specificity of a L. pneumophila esterase-lipase. (A) 130b (black columns) and NU243 (white columns) were grown in BYE broth, and then, at late log phase, culture supernatants (left) and cell lysates (right) were examined for their ability to cleave p-nitrophenyl caprylate. (B) In a separate experiment, late-log supernatants from wild-type and pilD mutant cultures were compared for their ability to cleave p-nitrophenyl caprylate (pNPC) (left) versus p-nitrophenyl palmitate (pNPPa) (right). The values presented are the mean and standard deviations from triplicate cultures. For both the supernatant and lysate comparisons, the differences between 130b and NU243 activity were significant (P < 0.001, Student's t test). The results obtained in panel A were observed on six other occasions, while those in panel B were seen in two other experiments.
FIG. 4.
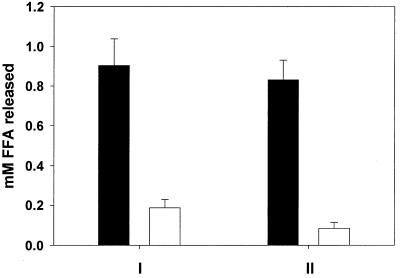
Activity of the L. pneumophila esterase upon monoacylglycerol. On two occasions (i.e., I and II), supernatants from 130b (black columns) and NU243 (white columns) late-log-phase cultures were examined for their ability to release free fatty acid (FFA) from monoacylglycerol as measured by the NEFA-C-Kit. The values presented are the mean and standard deviation from two cultures. Differences between 130b and NU243 activity were significant (P < 0.001, Student's t test).
With the clarification of an esterase-lipase activity in wild-type supernatants, we could examine the PilD mutant for another secretion defect. NU243 was >10-fold impaired for esterase secretion, regardless of whether the substrate was p-nitrophenyl caprylate or p-nitrophenyl palmitate (Fig. 3). Similarly, the strain's supernatants had a diminished ability to release free fatty acid from 1-MG (Fig. 4). However, esterase activity was apparent within the NU243 cell, again indicating that the strain is a secretion mutant (Fig. 3A). In sum, these data indicate that L. pneumophila secretes an esterase-monoacylglycerol lipase whose export is dependent upon PilD.
PLA secretion by L. pneumophila strains.
Using methods such as thin-layer chromatography and mass spectrometry, Flieger et al. recently found an L. pneumophila PLA (26). Interestingly, the PLA was evident in log-phase cultures and peaked during late-log to early-stationary phase (26), suggesting that its expression is controlled in a manner similar to that of the acid phosphatase, protease, and esterase-lipase activities. Thus, we examined supernatants from strain 130b and its pilD-negative derivative for the presence of PLA (Fig. 5). Strain 130b, like other wild-type legionellae, secreted an enzyme that was capable of releasing free fatty acid from phosphatidylcholine. In contrast, NU243 was defective for PLA secretion, indicating that the processing of this newly described enzyme is also influenced by the prepilin peptidase.
FIG. 5.
PLA secretion by wild-type and pilD mutant L. pneumophila. In two trials (I and II), supernatants from 130b (black column) and NU243 (white column) late-log cultures were tested for their ability to release free fatty acid (FFA) from phosphatidylcholine as measured by the NEFA-C-Kit. The values presented are the mean and standard deviations from duplicate cultures. The differences between 130b and NU243 activity were significant (P < 0.01 and < 0.001 for I and II, respectively; Student's t test). Similar conclusions were obtained by using thin-layer chromatography.
Nuclease secretion by L. pneumophila strains.
It had been reported that L. pneumophila secretes nuclease activities (73). Thus, we compared supernatants from late-log-phase cultures of 130b and NU243 for their ability to clear agar matrices impregnated with either RNA or DNA (64). On three occasions, the wild-type samples completely cleared the RNA-containing agar, yielding hydrolysis zones that were approximately 30 mm in diameter. The 130b supernatants also exhibited a DNase activity, yielding zones that were ca. 20 mm in diameter and were nearly clear. The supernatants from strain NU243 showed a consistent, albeit modest, reduction in RNase activity, i.e., their hydrolysis zones were about 30% smaller and less clear compared to those of the wild type. On the other hand, the mutant was not impaired for DNA-degrading activity, as evidenced by the normal size and the clarity of its hydrolysis zones. In sum, the secretion of only one of the L. pneumophila nucleases was notably diminished by the loss of PilD.
Secretion of a pNPPC-hydrolase by L. pneumophila strains.
Since its colonies produce a zone of opacity on egg yolk plates and its supernatants release pNP from pNPPC, L. pneumophila has long been believed to possess a PLC (6–8). Indeed, strain 130b secreted a factor during log phase that cleaves pNPPC (Fig. 1D). Importantly for us, supernatants from pilD mutant cultures were lacking in pNPPC hydrolysis (Fig. 1D), while mutant lysates showed elevated pNPPC-hydrolase activity (data not shown). Although our interest in the pNPPC-hydrolase activity had been piqued, new data raised doubts about its molecular basis. First, acid and alkaline phosphatases and GPC-phosphodiesterases can also release pNP from pNPPC (27, 66, 68). Second, a very recent study concluded that L. pneumophila does not produce a PLC (26). Since we found that strain 130b did not express a GPC-phosphodiesterase activity (data not shown), it now seemed plausible that the pilD-dependent, pNPPC-hydrolyzing activity was another manifestation of the secreted acid phosphatase and/or esterase.
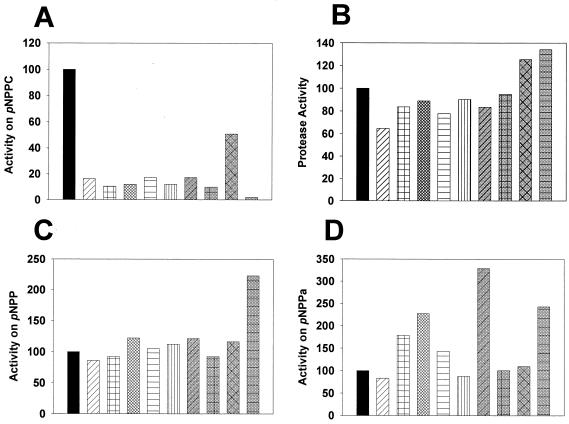
As one approach to determining whether the pNPPC-hydrolyzing activity reflects a known Legionella enzyme, we sought a 130b mutant that is specifically defective for one of the secreted activities. Toward that end, transposon-mutagenized legionellae were first screened for alterations on egg yolk plates. Nine mutants were obtained that had diminished iridescence. Interestingly, these mutants, designated as strains NU245 through NU253, produced supernatants that had reduced pNPPC-hydrolyzing activity (Fig. 6A). However, none of the mutants had altered growth in BYE or significantly reduced levels of the secreted zinc metalloprotease, as measured by supernatant hydrolysis of hide powder azure (Fig. 6B) and azocasein (data not shown), indicating that they are not generally impaired for growth or protein secretion. Importantly, none of the mutants had a loss of acid phosphatase activity (Fig. 6C), and all retained full esterase activity, as measured by cleavage of either p-nitrophenyl palmitate (Fig. 6D) or p-nitrophenyl caprylate (data not shown). These results signal that the hydrolysis of pNPPC by wild-type legionellae is not another manifestation of the identified acid phosphatase or esterase. In addition, they further document why iridescence on egg yolk plates should not be ascribed to only lipase-like activities. Although there was no reason to suspect that the Legionella PLA was responsible for pNPPC hydrolysis, we nonetheless tested two of the new mutants for loss of ability to release free fatty acid from phosphatidylcholine. Both NU247 and NU253 had normal PLA activity (data not shown). In summary, mutant analysis indicates that the pNPPC-hydrolase activity of L. pneumophila is promoted partly, if not completely, by a distinct, pilD-dependent factor.
FIG. 6.
Secreted activities of L. pneumophila pNPPC-hydrolase mutants. Wild-type (black columns) and mutant bacteria (hatched columns) were grown in BYE broth, and then, at late log phase, culture supernatants were examined for their relative abilities to cleave pNPPC (A), hide powder azure (B), p-nitrophenylphosphate (pNPP) (C), and p-nitrophenyl palmitate (pNPPa) (D). The activities for wild type are set at 100%. In each figure, the mutants' results are arranged in the same order, beginning with NU245 on the left and ending with NU253 on the right. For NU247 and NU253, similar results were obtained in one additional experiment. For all others, similar results were obtained on two other occasions.
Complementation of the L. pneumophila pilD mutation.
To confirm that the secretion defects of NU243 were caused by the loss of pilD and not a second site mutation, we examined the supernatant activities from NU243 harboring a plasmid (i.e., pMRL13) that contains as its Legionella DNA component only pilD. For all activities tested, NU243(pMRL13) exhibited, as expected, a level of activity that was comparable to that of 130b but greater than that of mutant bacteria containing only the pBBR1MCS vector (Table 1). Thus, we believe that the altered secretion phenotype displayed by the pilD mutant is indeed due to the loss of prepilin peptidase.
TABLE 1.
Secreted activities of wild-type, pilD mutant, and complemented mutant bacteria
| Activity assayed | Level of enzymatic activity within strain supernatants (mean ± SD)a
|
||
|---|---|---|---|
| 130b and 130b(pBBR1MCS) | NU243 and NU243(pBBR1MCS) | NU243(pMRL13) | |
| Acid phosphatase | + (5.14 ± 0.61)b | − (1.41 ± 0.06)b | + (4.49 ± 0.12)b |
| Proteasec | + (0.45 ± 0.02) | − (0.05 ± 0.01) | + (0.44 ± 0.05) |
| Esterase-lipased | + (37.46 ± 2.85) | − (2.31 ± 0.67) | + (38.50 ± 1.83) |
| PLA | + | − | NDe |
| DNase | + | + | ND |
| RNase | + | − | ND |
| pNPPC-hydrolase | + (0.28 ± 0.04) | − (0.00 ± 0.00) | + (0.31 ± 0.05) |
+, Wild-type level of activity; −, a significantly reduced or undetectable level of activity.
The values in parentheses represent the mean and standard deviations from triplicate late-log cultures of either 130b(pBBR1MCS), NU243(pBBR1MCS), or NU243(pMRL13) and are representative of two additional complementation experiments. In all trials, the secreted activities exhibited by the uncomplemented mutant were significantly less than those of the wild type and the complemented strain (P < 0.0001, Student's t test). The complemented mutant behaved as did the wild type (P > 0.5).
As measured by the azocasein assay.
As measured by the release of pNP from p-nitrophenyl caprylate.
ND, not determined.
Intracellular infection by L. pneumophila strains.
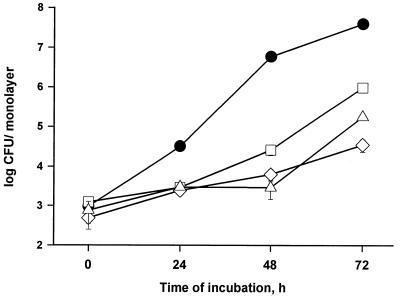
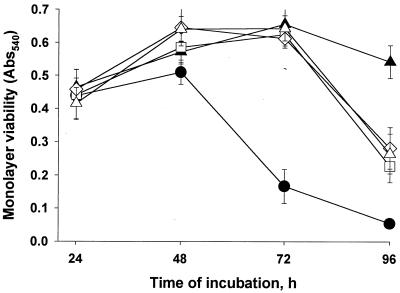
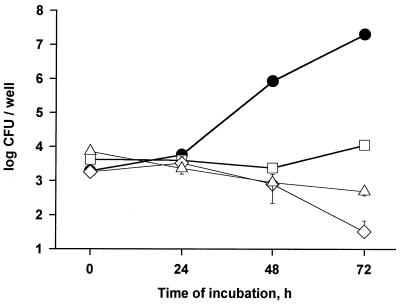
As a first step to ultimately determining which, if any, of the newly defined exoproteins promote intracellular infection and virulence, we assayed the nine pNPPC-hydrolase mutants for their ability to infect U937 cells. Based upon ID50 analysis, seven of the mutants did not show a significant defect in macrophage infection (data not shown), indicating that the pNPPC-hydrolase activity is not required for intracellular infection. Interestingly, however, two of the mutants, NU247 and NU253, exhibited ID50s that were at least 100-fold greater than wild type, suggesting that they are notably impaired for macrophage infection. To confirm this hypothesis, U937 cells were infected with equal amounts of wild-type and mutant bacteria and then, at various times, the bacteria within the monolayers were quantitated. Both NU247 and NU253 displayed a dramatic intracellular growth defect, which was slightly greater in magnitude to that of the pilD mutant (Fig. 7). Following an apparently normal uptake period, the numbers of mutant bacteria did not significantly increase for 2 days. Although replication was evident by the third day, the mutants ultimately produced 1,000-fold fewer progeny than did strain 130b. Inoculation of U937 cell monolayers with a low multiplicity of infection of L. pneumophila generally results in death and lysis of host cells (12). To determine whether these pNPPC-hydrolase mutants were also defective for cytopathic effect, we examined the viability of the infected monolayers with vital stains (Fig. 8). Within the first 72 h of incubation, strain 130b destroyed 75% of host cells, while inoculation with at least fourfold-greater numbers of NU243, NU247, and NU253 failed to reduce monolayer viability. By 96 h postinoculation, the mutants did elicit a significant cytopathic effect, albeit one that was still less than that of the wild type. Given the strong similarities that exist between Legionella macrophage and protozoan infection, we finally assessed the ability of NU247 and NU253 to infect Hartmannella amoebae (Fig. 9). The two pNPPC-hydrolase mutants were greatly impaired for protozoan infection, even more so than the pilD mutant, i.e., the numbers of NU247 and NU253 never increased during the 72-h incubation. In summary, NU247 and NU253 lack a factor that is necessary for optimal intracellular infection. Since the pNPPC-hydrolase activity is not required for intracellular infection, we suspect that this factor is a regulator of pNPPC-hydrolase expression or secretion.
FIG. 7.
Macrophage infection by wild-type and mutant L. pneumophila. U937 cell monolayers were infected with approximately 5 × 105 CFU of wild-type 130b (●), pilD mutant NU243 (□), and pNPPC-hydrolase mutants NU247 (◊) and NU253 (▵). CFU per well were quantitated at 0, 24, 48, and 72 h. Each datum point represents the mean and standard deviation for three monolayers. Significant differences in recovery between 130b and its mutant derivatives were evident at 24 h (P < 0.05) and beyond (P < 0.001). These differences were seen in three additional experiments (data not shown).
FIG. 8.
Cytopathic effect of L. pneumophila strains on U937 cells. Replicate monolayers (n = 6) were either not infected (▴) or were infected with 103 CFU of strain 130b (●), 5 × 104 CFU of NU243 (□), 8 × 104 CFU of NU247 (◊), or 6.5 × 104 CFU of NU253 (▵). After various periods of incubation, the viability of the host cells was measured by neutral red uptake. Since the pilD mutant does not elicit any cytopathic effect within the typical 72-h infection assay (41), the monolayers were monitored for 96 h and were purposely infected with greater numbers of mutant relative to wild-type bacteria. Datum points represent the mean OD540, and vertical bars indicate the standard deviations. Differences in cytopathic effect between 130b and its mutant derivatives were significant at 72 and 96 h after inoculation (P < 0.001, Student's t test). Similar conclusions were obtained from two additional experiments with neutral red and a third trial with alamar blue (data not shown).
FIG. 9.
Infection of H. vermiformis by L. pneumophila strains. Wells containing Hartmannella amoebae were infected with approximately 5 × 103 CFU of wild-type 130b (●), pilD mutant NU243 (□), and pNPPC-hydrolase mutants NU247 (◊) and NU253 (▵). Bacterial CFU per well were quantitated at 0, 24, 48, and 72 h after inoculation. Each datum point represents the mean and standard deviations for three wells. Significant differences in recovery between 130b and its mutant derivatives were evident at 48 and 72 h (P < 0.005, Student's t test). These differences were observed in two additional experiments (data not shown).
DISCUSSION
The present study provides seven basic conclusions about L. pneumophila secretion (Table 1). First, L. pneumophila does indeed secrete an acid phosphatase, PLA, DNase, and RNase. Second, the organism secretes an esterase-monoacylglycerol lipase. Third, the pNPPC-hydrolase activity of Legionella, originally ascribed to a PLC, is not, as yet, accounted for by known major enzymatic activities. Fourth, the export of the acid phosphatase, zinc metalloprotease, esterase-monoacylglycerol lipase, and pNPPC-hydrolase was detectable during log phase. A companion study demonstrated that the PLA is similarly expressed (26). Although nuclease release was not monitored over the entire growth cycle, it was apparent in late-log-phase cultures. Fifth, secretion of the acid phosphatase, esterase-monoacylglycerol lipase, PLA, RNase, and pNPPC-hydrolase, like that of the protease, is deficient in a L. pneumophila pilD mutant (Table 1). Sixth, as in other bacteria, the mutation in pilD results in the intracellular accumulation of the exoenzymes (24, 56, 70). Seventh, the effect of the pilD mutation may only be evident when the comparisons between wild type and mutant utilize mid- to late-log-phase bacteria. Thus, we suspect that our earlier study overlooked the effect of PilD on phosphatase expression because stationary-phase cultures had been examined (41). Based upon analyses of other gram-negative bacteria, the changes in secretion activity in the L. pneumophila pilD mutant are likely due, at least in part, to the absence of a type II secretion apparatus, some of whose components are substrates for the PilD (23, 60).
The finding of up to six pilD-dependent exoproteins in L. pneumophila adds considerably to an expanding appreciation for PilD and type II secretion in bacterial physiology. The exoenzymes previously found lacking from pilD or other type II secretion mutants include the following: the esterase and lipase of Acinetobacter calcoaceticus; the acyltransferase, aerolysin, and protease of A. hydrophila; the lipases of Burkholderia sp.; the pectate lyase and cellulase of Erwinia chrysanthemi; the pullulanase of Klebsiella oxytoca; the alkaline phosphatase, lipase, elastase, exotoxin A, LasA protease, and PLC of P. aeruginosa; the cholera toxin, protease, and endochitinase of V. cholerae; and the amylase, cellulase, endoglucanase, and protease of Xanthomonas campestris (14, 20, 24, 33, 36, 44, 55, 56, 58, 63, 70). Thus, there is precedent for a linkage between pilD and an esterase-lipase and protease. However, this study is the first to document how the loss of pilD is associated with changes in acid phosphatase, PLA, and RNase activity. Furthermore, we believe that L. pneumophila has other PilD-dependent activities, including the factor responsible for pNPPC hydrolysis. Indeed, visualization of supernatant proteins by Coomassie staining of polyacrylamide gels suggested that the pilD mutant is missing at least eight exoproteins (41). In addition, the L. pneumophila pilD mutant displays an altered colony morphology that is not simply due to the loss of pili, suggesting that PilD influences the expression of surface components (41). We do not believe, however, that all Legionella exoproteins are controlled by PilD and type II secretion. For example, our current data suggest that the L. pneumophila DNAse is not dependent upon PilD, a finding that has a precedent in the V. cholerae system (63).
Along with the increased understanding of the gram-negative secretion machinery, recent attention has been directed toward defining the role of PilD- and type II secretion-dependent exoproteins in pathogenesis. For example, our previous study was the first to implicate PilD-dependent secretion in intracellular infection (41), and the present study signifies an initial step toward identifying those secreted proteins that potentiate L. pneumophila macrophage infection and overall virulence. Although the characterization of a new panel of mutants indicated that the pNPPC-hydrolase activity is not required for U937 cell infection, the acid phosphatase, esterase-monoacylglycerol lipase, PLA, and RNase constitute potential cell infectivity determinants. Alternatively, the newly defined activities, including that of pNPPC-hydrolase, might promote virulence by fostering extracellular survival as opposed to or in addition to intracellular infection. Interestingly, acid phosphatases, due to their ability to inhibit superoxide anion production, have been implicated in the intracellular survival of Francisella tularensis, L. micdadei, and Leishmania donovanii (59, 62). In addition, PLAs promote the pathogenesis of A. hydrophila and Yersinia enterocolitica, and an RNase is required for the virulence of Shigella flexneri (11, 47, 65). The characterization of additional mutants that are defective for single exoenzymes will show if and how the various secreted products promote L. pneumophila pathogenesis.
Unlike other pNPPC-hydrolase mutants, NU247 and NU253 were greatly impaired for intracellular infection. Although other scenarios exist, we hypothesize that these two strains represent regulatory or processing mutants, i.e., they lack a factor(s) that, in addition to affecting the expression of the pNPPC-hydrolase, influences the production of molecules that do promote intracellular growth. Since NU247 and NU253 were not deficient in the acid phosphatase, esterase, PLA, and protease activities, this factor would have to be acting with a certain degree of specificity. However, there are many examples of transcriptional regulators that coordinately control the expression of some but not all secreted activities, including PilD- or type II-dependent exoproteins, e.g., ToxR/S/T in Vibrio sp. and Fur in Pseudomonas sp. (25). Similarly, a periplasmic chaperone can promote the secretion of some but not all exoproteins, e.g., Lif specifically influences lipase secretion in Pseudomonas sp. (23, 34). Thus, further examination of these mutants should provide yet additional new insights into L. pneumophila regulation, secretion, and pathogenesis.
ACKNOWLEDGMENTS
We thank Tracy Aber Scheel, Mark Liles, Ombeline Rossier, V. K. Viswanathan, and Jamie Borensztajn for helpful discussions.
This work was supported by NIH grant AI43987 awarded to N.P.C.
REFERENCES
- 1.Alm R A, Hallinan J P, Watson A A, Mattick J S. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol Microbiol. 1996;22:161–173. doi: 10.1111/j.1365-2958.1996.tb02665.x. [DOI] [PubMed] [Google Scholar]
- 2.Alm R A, Mattick J S. Identification of two genes with prepilin-like leader sequences involved in type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1996;178:3809–3817. doi: 10.1128/jb.178.13.3809-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anguita J, Rodriguez Aparicio L B, Naharro G. Purification, gene cloning, amino acid sequence analysis, and expression of an extracellular lipase from an Aeromonas hydrophila human isolate. Appl Environ Microbiol. 1993;59:2411–2417. doi: 10.1128/aem.59.8.2411-2417.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babich H, Borenfreund E, Stern A. Comparative cytotoxicities of selected minor dietary non-nutrients with chemopreventive properties. Cancer Lett. 1993;73:127–133. doi: 10.1016/0304-3835(93)90254-7. [DOI] [PubMed] [Google Scholar]
- 5.Baine W B. Cytolytic and phospholipase C activity in Legionella species. J Gen Microbiol. 1985;131:1383–1391. doi: 10.1099/00221287-131-6-1383. [DOI] [PubMed] [Google Scholar]
- 6.Baine W B. A phospholipase C from the Dallas 1E strain of Legionella pneumophila serogroup 5: purification and characterization of conditions for optimal activity with an artificial substrate. J Gen Microbiol. 1988;134:489–498. doi: 10.1099/00221287-134-2-489. [DOI] [PubMed] [Google Scholar]
- 7.Baine W B, Rasheed J K, Mackel D C, Bopp C A, Wells J G, Kaufmann A F. Exotoxin activity associated with the Legionnaires' disease bacterium. J Clin Microbiol. 1979;9:453–456. doi: 10.1128/jcm.9.3.453-456.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bally M, Filloux A, Akrim M, Ball G, Lazdunski A, Tommassen J. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol Microbiol. 1992;6:1121–1131. doi: 10.1111/j.1365-2958.1992.tb01550.x. [DOI] [PubMed] [Google Scholar]
- 9.Bally M, Wretlind B, Lazdunski A. Protein secretion in Pseudomonas aeruginosa: Molecular cloning and characterization of the xcp-1 gene. J Bacteriol. 1989;171:4342–4348. doi: 10.1128/jb.171.8.4342-4348.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bleves S, Voulhoux R, Michel G, Lazdunski A, Tommassen J, Filloux A. The secretion apparatus of Pseudomonas aeruginosa: identification of a fifth pseudopilin, XcpX (GspK family) Mol Microbiol. 1998;27:31–40. doi: 10.1046/j.1365-2958.1998.00653.x. [DOI] [PubMed] [Google Scholar]
- 11.Cheng Z F, Zuo Y, Li Z, Rudd K E, Deutscher M P. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J Biol Chem. 1998;273:14077–14080. doi: 10.1074/jbc.273.23.14077. [DOI] [PubMed] [Google Scholar]
- 12.Cianciotto N P, Eisenstein B I, Mody C H, Toews G B, Engleberg N C. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect Immun. 1989;57:1255–1262. doi: 10.1128/iai.57.4.1255-1262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cianciotto N P, Fields B S. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc Natl Acad Sci USA. 1992;89:5188–5191. doi: 10.1073/pnas.89.11.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connell T D, Metzger D J, Lynch J, Folster J P. Endochitinase is transported to the extracellular milieu by the eps-encoded general secretory pathway of Vibrio cholerae. J Bacteriol. 1998;180:5591–5600. doi: 10.1128/jb.180.21.5591-5600.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donnenberg M S, Zhang H-Z, Stone K D. Biogenesis of the bundle-forming pilus of enteropathogenic Escherichia coli: reconstitution of fimbriae in recombinant E. coli and role of DsbA in pilin stability—a review. Gene. 1997;192:33–38. doi: 10.1016/s0378-1119(96)00826-8. [DOI] [PubMed] [Google Scholar]
- 16.Dowling J N, Saha A K, Glew R H. Virulence factors of the family Legionellaceae. Microbiol Rev. 1992;56:32–60. doi: 10.1128/mr.56.1.32-60.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreyfus L A, Iglewski B. Purification and characterization of an extracellular protease of Legionella pneumophila. Infect Immun. 1986;51:736–743. doi: 10.1128/iai.51.3.736-743.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edelstein P H. Improved semiselective medium for isolation of Legionella pneumophila from contaminated clinical and environmental specimens. J Clin Microbiol. 1981;14:298–303. doi: 10.1128/jcm.14.3.298-303.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eibl H, Lands W E. A new, sensitive determination of phosphate. Anal Biochem. 1969;30:51–57. doi: 10.1016/0003-2697(69)90372-8. [DOI] [PubMed] [Google Scholar]
- 20.El Khattabi M, Ockhuijsen C, Bitter W, Jaeger K-E, Tommassen J. Specificity of the lipase-specific foldases of gram-negative bacteria and the role of the membrane anchor. Mol Gen Genet. 1999;261:770–776. doi: 10.1007/s004380050020. [DOI] [PubMed] [Google Scholar]
- 21.Engleberg N C, Drutz D J, Eisenstein B I. Cloning and expression of Legionella pneumophila antigens in Escherichia coli. Infect Immun. 1984;44:222–227. doi: 10.1128/iai.44.2.222-227.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fields B S. The molecular ecology of legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 23.Filloux A, Michel G, Bally M. GSP-dependent protein secretion in Gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. Vol. 22. 1998. pp. 177–198. [DOI] [PubMed] [Google Scholar]
- 24.Filloux A, Murgier M, Wretlind B, Lazdunski A. Characterization of two Pseudomonas aeruginosa mutants with defective secretion of extracellular proteins and comparison with other mutants. FEMS Microbiol Lett. 1987;40:159–163. [Google Scholar]
- 25.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flieger, A., S. Gong, M. Faigle, M. Deeg, P. Bartmann, and B. Neumeister. Characterization of a novel phospholipase A (PLA) activity secreted by Legionella species. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 27.Flieger, A., S. Gong, M. Faigle, and B. Neumeister. Critical evaluation of p-nitrophenylphosphorylcholine (p-NPPC) as artificial substrate for the detection of phospholipase C. Enzyme Microb. Technol., in press. [DOI] [PubMed]
- 28.Fullner K J, Mekalanos J J. Genetic characterization of a new type IV-A pilus gene cluster found in both Classical and El Tor biotypes of Vibrio cholerae. Infect Immun. 1999;67:1393–1404. doi: 10.1128/iai.67.3.1393-1404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hales L M, Shuman H A. Legionella pneumophila contains a type II general secretion pathway required for growth in amoebae as well as for secretion of the Msp protease. Infect Immun. 1999;67:3662–3666. doi: 10.1128/iai.67.7.3662-3666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hickey E K, Cianciotto N P. An iron- and fur-repressed Legionella pneumophila gene that promotes intracellular infection and encodes a protein with similarity to the Escherichia coli aerobactin synthetases. Infect Immun. 1997;65:133–143. doi: 10.1128/iai.65.1.133-143.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann G E, Schmidt D, Bastian B, Guder W G. Photometric determination of phospholipase A. J Clin Chem Clin Biochem. 1986;24:871–875. doi: 10.1515/cclm.1986.24.11.871. [DOI] [PubMed] [Google Scholar]
- 32.Horwitz M A. Interactions between macrophages and Legionella pneumophila. Curr Top Microbiol Immunol. 1992;181:265–282. doi: 10.1007/978-3-642-77377-8_10. [DOI] [PubMed] [Google Scholar]
- 33.Hu N-T, Hung M-N, Chen D C, Tsai R-T. Insertion mutagenesis of XpsD, an outer membrane protein involved in extracellular protein secretion in Xanthomonas campestris pv. campestris. Microbiology. 1998;144:1479–1486. doi: 10.1099/00221287-144-6-1479. [DOI] [PubMed] [Google Scholar]
- 34.Jaeger K-E, Dijkstra B W, Reetz M T. Bacterial biocatalysts: molecular biology, three dimensional structures, and biotechnological applications of lipases. Annu Rev Microbiol. 1999;53:315–351. doi: 10.1146/annurev.micro.53.1.315. [DOI] [PubMed] [Google Scholar]
- 35.Johnston J L, Billington S J, Haring V, Rood J I. Identification of fimbrial assembly genes from Dichelobacter nodosus: evidence that fimP encodes the type-IV prepilin peptidase. Gene. 1995;161:21–26. doi: 10.1016/0378-1119(95)00264-7. [DOI] [PubMed] [Google Scholar]
- 36.Kessler E, Safrin M, Gustin J K, Ohman D E. Elastase and the LasA protease of Pseudomonas aeruginosa are secreted with their propeptides. J Biol Chem. 1998;273:30225–30231. doi: 10.1074/jbc.273.46.30225. [DOI] [PubMed] [Google Scholar]
- 37.Kim M J, Rogers J E, Hurley M C, Engleberg N C. Phosphatase-negative mutants of Legionella pneumophila and their behavior in mammalian cell infection. Microb Pathog. 1994;17:51–62. doi: 10.1006/mpat.1994.1051. [DOI] [PubMed] [Google Scholar]
- 38.Kramer M H, Ford T E. Legionellosis: ecological factors of an environmentally ‘new’ disease. Zentbl Hyg. 1994;195:470–482. [PubMed] [Google Scholar]
- 39.Kurioka S, Matsuda M. Phospholipase C assay using p-nitrophenylphosphorylcholine together with sorbitol and its application to studying the metal and detergent requirement of the enzyme. Anal Biochem. 1976;75:281–289. doi: 10.1016/0003-2697(76)90078-6. [DOI] [PubMed] [Google Scholar]
- 40.Lauer P, Albertson N H, Koomey M. Conservation of genes encoding components of a type IV pilus assembly/two-step protein export pathway in Neisseria gonorrhoeae. Mol Microbiol. 1993;8:357–368. doi: 10.1111/j.1365-2958.1993.tb01579.x. [DOI] [PubMed] [Google Scholar]
- 41.Liles M R, Edelstein P H, Cianciotto N P. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol Microbiol. 1999;31:959–970. doi: 10.1046/j.1365-2958.1999.01239.x. [DOI] [PubMed] [Google Scholar]
- 42.Liles M R, Viswanathan V K, Cianciotto N P. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II secretion. Infect Immun. 1998;66:1776–1782. doi: 10.1128/iai.66.4.1776-1782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lory S, Strom M S. Structure-function relationship of type-IV prepilin peptidase of Pseudomonas aeruginosa—a review. Gene. 1997;192:117–121. doi: 10.1016/s0378-1119(96)00830-x. [DOI] [PubMed] [Google Scholar]
- 44.Marsh J W, Taylor R K. Identification of the Vibrio cholerae type 4 prepilin peptidase required for cholera toxin secretion and pilus formation. Mol Microbiol. 1998;29:1481–1492. doi: 10.1046/j.1365-2958.1998.01031.x. [DOI] [PubMed] [Google Scholar]
- 45.Martinez A, Ostrovsky P, Nunn D N. Identification of an additional member of the secretin superfamily of proteins in Pseudomonas aeruginosa that is able to function in type II secretion. Mol Microbiol. 1998;28:1235–1246. doi: 10.1046/j.1365-2958.1998.00888.x. [DOI] [PubMed] [Google Scholar]
- 46.McClain M S, Engleberg N C. Construction of an alkaline phosphatase fusion-generating transposon, mTn10phoA. Gene. 1996;170:147–148. doi: 10.1016/0378-1119(95)00856-x. [DOI] [PubMed] [Google Scholar]
- 47.Merino S, Aguilar A, Nogueras M M, Regue M, Swift S, Tomas J M. Cloning, sequencing, and role in virulence of two phospholipases (A1 and C) from mesophilic Aeromonas sp. serogroup O:34. Infect Immun. 1999;67:4008–4013. doi: 10.1128/iai.67.8.4008-4013.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moffat J F, Edelstein P H, Regula D P, Jr, Cirillo J D, Tompkins L S. Effects of an isogenic Zn-metalloprotease-deficient mutant of Legionella pneumophila in a guinea-pig pneumonia model. Mol Microbiol. 1994;12:693–705. doi: 10.1111/j.1365-2958.1994.tb01057.x. [DOI] [PubMed] [Google Scholar]
- 49.Muller H E. Enzymatic profile of Legionella pneumophila. J Clin Microbiol. 1981;13:423–426. doi: 10.1128/jcm.13.3.423-426.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nolte F S, Hollick G E, Robertson R G. Enzymatic activities of Legionella pneumophila and Legionella-like organisms. J Clin Microbiol. 1982;15:175–177. doi: 10.1128/jcm.15.1.175-177.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nunn D, Bergman S, Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990;172:2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nunn D N, Lory S. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc Natl Acad Sci USA. 1991;88:3281–3285. doi: 10.1073/pnas.88.8.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nunn D N, Lory S. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J Bacteriol. 1993;175:4375–4382. doi: 10.1128/jb.175.14.4375-4382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Connell W A, Dhand L, Cianciotto N P. Infection of macrophage-like cells by Legionella species that have not been associated with disease. Infect Immun. 1996;64:4381–4384. doi: 10.1128/iai.64.10.4381-4384.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parche S, Geissdorfer W, Hillen W. Identification and characterization of xcpR encoding a subunit of the general secretory pathway necessary for dodecane degradation in Acinetobacter calcoaceticus ADP1. J Bacteriol. 1997;179:4631–4634. doi: 10.1128/jb.179.14.4631-4634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pepe C M, Eklund M W, Strom M S. Cloning of an Aeromonas hydrophila type IV pilus biogenesis gene cluster: complementation of pilus assembly functions and characterization of a type IV leader peptidase/N-methyltransferase required for extracellular protein secretion. Mol Microbiol. 1996;19:857–869. doi: 10.1046/j.1365-2958.1996.431958.x. [DOI] [PubMed] [Google Scholar]
- 57.Pope C D, Dhand L, Cianciotto N P. Random mutagenesis of Legionella pneumophila with mini-Tn10. FEMS Microbiol Lett. 1994;124:107–111. doi: 10.1111/j.1574-6968.1994.tb07269.x. [DOI] [PubMed] [Google Scholar]
- 58.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reilly T J, Baron G S, Nano F E, Kuhlenschmidt M S. Characterization and sequencing of a respiratory burst-inhibiting acid phosphatase from Francisella tularensis. J Biol Chem. 1996;271:10973–10983. doi: 10.1074/jbc.271.18.10973. [DOI] [PubMed] [Google Scholar]
- 60.Russel M. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J Mol Biol. 1998;279:485–499. doi: 10.1006/jmbi.1998.1791. [DOI] [PubMed] [Google Scholar]
- 61.Russell M A, Darzins A. The pilE gene product of Pseudomonas aeruginosa, required for pilus biogenesis, shares amino acid sequence identity with the N-termini of type 4 prepilin proteins. Mol Microbiol. 1994;13:973–985. doi: 10.1111/j.1365-2958.1994.tb00489.x. [DOI] [PubMed] [Google Scholar]
- 62.Saha A K, Dowling J N, Pasculle A W, Glew R H. Legionella micdadei phosphatase catalyzes the hydrolysis of phosphatidylinositol 4,5-bisphosphate in human neutrophils. Arch Biochem Biophys. 1988;265:94–104. doi: 10.1016/0003-9861(88)90375-x. [DOI] [PubMed] [Google Scholar]
- 63.Sandkvist M, Overbye Michel L, Hough L P, Morales V R, Bagdasarian M, Koomey M, DiRita V J, Bagdasarian M. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J Bacteriol. 1997;179:6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schill W-B, Schumacher G F B. Radial diffusion in gel for microdetermination of enzymes. Anal Biochem. 1972;46:502–533. doi: 10.1016/0003-2697(72)90324-7. [DOI] [PubMed] [Google Scholar]
- 65.Schmiel D H, Wagar E, Karamanou L, Weeks D, Miller V L. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect Immun. 1998;66:3941–3951. doi: 10.1128/iai.66.8.3941-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sok D E, Kim M R. A spectrophotometric assay of Zn2+-glycerophosphorylcholine phosphocholine phosphodiesterase using p-nitrophenylphosphorylcholine. Anal Biochem. 1992;203:201–205. doi: 10.1016/0003-2697(92)90303-o. [DOI] [PubMed] [Google Scholar]
- 67.Sommer A J. The determination of acid and alkaline phosphatase using p-nitrophenylphosphate as substrate. Am J Med Technol. 1954;20:244–253. [PubMed] [Google Scholar]
- 68.Srivastava P N, Brewer J M, White R A., Jr Hydrolysis of p-nitrophenylphosphorylcholine by alkaline phosphatase and phospholipase C from rabbit sperm-acrosome. Biochem Biophys Res Commun. 1982;108:1120–1125. doi: 10.1016/0006-291x(82)92116-7. [DOI] [PubMed] [Google Scholar]
- 69.Stone B, Abu Kwaik Y. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to and intracellular replication within mammalian and protozoan cells. Infect Immun. 1998;66:1768–1775. doi: 10.1128/iai.66.4.1768-1775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Strom M S, Nunn D, Lory S. Multiple roles of the pilus biogenesis protein PilD: involvement of PilD in excretion of enzymes from Pseudomonas aeruginosa. J Bacteriol. 1991;173:1175–1180. doi: 10.1128/jb.173.3.1175-1180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Strom M S, Nunn D N, Lory S. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci USA. 1993;90:2404–2408. doi: 10.1073/pnas.90.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szeto L, Shuman H A. The Legionella pneumophila major secretory protein, a protease, is not required for intracellular growth or cell killing. Infect Immun. 1990;58:2585–2592. doi: 10.1128/iai.58.8.2585-2592.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thorpe T C, Miller R D. Extracellular enzymes of Legionella pneumophila. Infect Immun. 1981;33:632–635. doi: 10.1128/iai.33.2.632-635.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Touchstone J C, Levin S S, Dobbins M F, Matthews L, Beers P C, Gabbe S G. (3-sn-Phosphatidyl)cholines (lecithins) in amniotic fluid. Clin Chem. 1983;29:1951–1954. [PubMed] [Google Scholar]
- 75.Winkler U K, Stuckmann M. Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J Bacteriol. 1979;138:663–670. doi: 10.1128/jb.138.3.663-670.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Winn W C., Jr Legionnaires disease: historical perspective. Clin Microbiol Rev. 1988;1:60–81. doi: 10.1128/cmr.1.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu S S, Wu J, Cheng Y L, Kaiser D. The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus. Mol Microbiol. 1998;29:1249–1261. doi: 10.1046/j.1365-2958.1998.01013.x. [DOI] [PubMed] [Google Scholar]