Abstract
Detecting breast cancer (BC) at the initial stages of progression has always been regarded as a lifesaving intervention. With modern technology, extensive studies have unraveled the complexity of BC, but the current standard practice of early breast cancer screening and clinical management of cancer progression is still heavily dependent on tissue biopsies, which are invasive and limited in capturing definitive cancer signatures for more comprehensive applications to improve outcomes in BC care and treatments. In recent years, reviews and studies have shown that liquid biopsies in the form of blood, containing free circulating and exosomal microRNAs (miRNAs), have become increasingly evident as a potential minimally invasive alternative to tissue biopsy or as a complement to biomarkers in assessing and classifying BC. As such, in this review, the potential of miRNAs as the key BC signatures in liquid biopsy are addressed, including the role of artificial intelligence (AI) and machine learning platforms (ML), in capitalizing on the big data of miRNA for a more comprehensive assessment of the cancer, leading to practical clinical utility in BC management.
Keywords: liquid biopsy, circulating miRNA, breast cancer, AI, ML
1. Introduction
Globally, breast cancer (BC) remains the most common cancer and the leading cause of cancer death for women [1]. Despite the advancement in BC screening, diagnostics and therapy, the overall rates of BC incidence and mortality around the world have been on an increasing trend. Although BC mortality rates have declined over time in most high-income countries (HICs) [2], they remain high and are increasing in many low-middle-income and low-income countries [3,4], partly due to poor awareness and perception of early BC detection, leading to delays in diagnosis and treatment [1]. Recent studies show that the mortality rate is also compounded by disparities in BC screening between rural and urban rural/urban areas [5] as well as among those from different socio-economic and ethnic backgrounds of HICs [2]. Nevertheless, there has been a more established understanding of the heterogenous nature of BC and its application in the development of personalized medicine and targeted therapy [6]. However, the complex interaction within the tumor microenvironment (TME) and the influence of cancer stem cells (CSCs) from cancer recurrence to drug resistance [7] as well as population-based variation in terms of immunological response [8] continue to pose challenges in deciphering feasible approaches in managing BC diagnosis, monitoring and administrating effective treatment options. In the past decade, the evolution of BC diagnosis and classification has resulted in greater supporting tools, ranging from classical mammography and histopathology [9] to molecular-based markers and multigene prognosticators [10], which ultimately guided the overall management of BC (Table 1) [11,12,13,14]. Still, many if not all of these existing tools are heavily reliant on invasive tissue biopsies as the starting point for screening and monitoring BC progression, evaluating cancer prognosis and deciding on the best therapeutic options. Moreover, studies have shown that the BC signatures are often not clearly manifested and well represented in the current screening methods, namely mammography [15] and tissue biopsies [16]—more so in the diagnosis of metastatic cancers [17]. Liquid biopsy (LB), on the other hand, has emerged as a potential feasible approach in overcoming these shortfalls, from obtaining samples in a noninvasive manner to early detection of cancer and more comprehensive monitoring [18,19], thereby offering patients less stressful experiences and a better sense of worthiness in managing cancer treatments. Among the commonly known cancer biomarkers in LB [19], circulating microRNAs (miRNAs) stand out as a feasible and practical option [20]. Hence, this review will address the role of miRNA as a feasible candidate for liquid biopsy, not only in personalized BC management and targeted therapy but also in consolidating the big data of miRNA with artificial intelligence (AI) and machine learning platforms (ML) for a more comprehensive and inclusive approach in promoting effective BC patient care and outcome.
Table 1.
Clinical and molecular classification of BC.
| Cancer Type | Benign | Pre-Malignant/ In-Situ (20–25%) [13] |
Malignant/Invasive [IDC (80%), ILC (20%)] [13] | |||||
|---|---|---|---|---|---|---|---|---|
| Categories | Fibroadenoma Intraductal papilloma Lipoma |
Early Breast Cancer Detection |
Molecular Subtypes (St Gallen) | Recurrence/ Metastatic |
||||
| Lubular Carcinoma In-situ (LCIS) | Ductal Carcinoma In-situ (DCIS) | Luminal A | Luminal B (HER2-) |
Luminal B (HER2+) | HER2+ Enriched | TNBC | ||
| Cancer/Bio markers [11,12] |
ER, PR, HER2 & Ki67 (low < 10%); Germline test BRCA1 & 2 (High risk group) |
ER+; PR+; HER2−; Ki67 low (<10–14%); Germline test BRCA1 & 2 (High Risk Group) |
ER+; PR−; HER2−; Ki67 high (>14–30%); Germline test BRCA1 & 2 (High Risk Group) |
ER+; PR+/−; HER2+; Ki67 high/low; Germline test BRCA1 & 2 (High Risk Group) |
ER−; PR−; HER2+; Ki67 high; Germline test BRCA1 & 2 (High Risk Group) |
ER−; PR−; HER2−; Ki67 high; (CK 5/6+; EGFR+); Germline test BRCA1 & BRCA2 (High Risk Group) |
Metastatic Site: Bone, liver, lungs, brain ESCAT score: I = Good prognosis II = Poor Prognosis |
|
| Frequency of cases [14] | 20–25% | 40–50% | 20–30% | 20–30% | 15–20% | 10–20% | 4% | |
| Histological grade (Majority) | Well differentiated (G1) |
Moderately differentiated (G2) |
Moderately differentiated (G2) |
Poorly differentiated (G3) |
Poorly differentiated (G3) |
Poorly differentiated (G4) |
||
| TNM Stage | NR | I-III | I-III | I-III | I-III | I-III | IV | |
| Prognosis | NR | Good | Intermediate | Intermediate | Poor | Poor | Poor | |
| Response to therapies [11,12,14] | Surgery Breast-conserving surgery (BCS) Radiotherapy Lumpectomy Mastectomy |
Endocrine | Endocrine Chemotherapy |
Endocrine Chemotherapy Targeted Therapy |
Chemotherapy Targeted Therapy |
Chemotherapy PARP inhibitors |
Chemotherapy CKD4/6 Inhibitor Fulvestrant |
|
Abbreviations: BRCA 1 & 2: Breast Cancer gene 1 and 2; TNM: tumor, node, and metastasis; DCIS: Ductal Carcinoma In-situ; ESCAT: ESMO Scale for Clinical Actionability of molecular Targets; ER: Estrogen receptor; HER2: Human epidermal growth factor receptor; IDC: Invasive Ductal Carcinoma; ILC: Invasive Lobular Carcinoma; LCIS: Lubular Carcinoma In-situ; PALB2: Partner and localizer of BRCA2; PDL1: Program death-ligand 1; PR: Progesterone receptor, 2; MSI: Microsatellite instability.
2. miRNA as Liquid Biopsy in Personalized Breast Cancer Management and Targeted Therapy
The liquid biopsy (LB) approach enables the securing of essential information on cancer progression and tumor through simple body-fluid-based samples, mainly through routine blood sampling. As gene-regulatory molecules in the body, circulating microRNAs (miRNAs) may be readily detected in plasma or serum of blood samples, enabling measurable changes in their levels, which are associated with the various conditions of the body, including cancers. Moreover, with the rapid advancement in bioinformatics in molecular data analysis, the inference of miRNAs with oncogene targets, cancer signaling pathways, survival analysis, prognostic values and drug targets is commonly obtainable [21] with cross-validation from clinical cancer-associated databases [22,23]. Although the application of miRNA as LBs for BC in the clinical setting is fairly new, with only two clinical studies recorded in www.clinicaltrials.gov as of October 2022 (Table 2), its utility may complement well with the existing standard clinical approach (Table 1), enhance diagnosis and monitoring of BC progression, as well as the response to treatments.
Table 2.
Clinical trials that involved circulating miRNAs as liquid biopsy of human breast cancer (n = 2).
| Study Name | Year Launched | Study ID | Location | Status |
|---|---|---|---|---|
| Onco-liq: Kit for Breast Cancer Diagnosis. | 2021 | NCT04906330 | Argentina | On-going |
| Prospective Breast Cancer Biobanking (PBCB) | 2020 | NCT04488614 | Norway | On-going |
3. Current Trends and Research Outcomes of Circulating miRNA as Liquid Biopsy
Circulating miRNAs are extracellular miRNAs that are present in body fluids, such as blood, serum, plasma, milk, saliva and urine, either in the form of free-circulating miRNAs or encapsulated within extracellular vesicles (EVs), such as exosomes [24,25,26]. The advancement in molecular biology techniques has allowed scientists to employ different methods, such as real-time polymerase chain reaction (qPCR) [27], miRNA-sequencing (miRNA-seq) [28] and microarray [29], to detect the levels of circulating miRNAs among BC patients. These techniques have been reported to play essential roles in the diagnosis, classification and prognosis of BC [30,31]. We hope to provide an updated overview of the research findings that reported the association of circulating miRNAs to the diagnosis and prognosis of BC. Notably, EV-derived and exosomal miRNAs have attracted remarkable interest due to their superior stability to those of free-circulating miRNAs. As such, a separate cluster of studies that researched specifically miRNAs derived from EVs is also highlighted in this review.
3.1. Diagnostic Significance of Circulating miRNAs in Human Breast Cancer
To date, more than 50 original research articles have reported the roles of circulating miRNAs as a minimally invasive tool in diagnosing BC, either as free-circulating miRNA BC in plasma or serum (Table 3) [32,33] or transported in EVs and exosomes (Table 4) [34,35]. Many circulating miRNAs were shown to be elevated in BC patients as compared to healthy controls and these include miR-21 [36], miR-24 [32], miR-34 [37], miR-155 [38], miR-140-5p [39] and many other miRNAs [40]. Similarly, the downregulation of specific circulating miRNAs, such as miR-363-5p [25] and miR-4488 [41], was demonstrated to be useful to distinguish between BC patients and healthy controls. In addition, the elevation of serum exosomal miR-21-5p and miR-23a-3p levels [33] and plasma miR-133a, miR-148b and miR-200 levels [42,43] was found to be sensitive to distinguish early and advanced BC and this is particularly useful to detect early BC. Other miRNAs that had been reported to be useful in detecting early BC include miR-23b, miR-26b-5p, miR-106b-5p, miR-127-3p, miR-142-3p, miR-142-5p, miR-148b, miR-185-5p, miR-362-5p, miR-409-3p, miR-652 and miR-801 [44,45,46].
Table 3.
Potential free-circulating miRNAs in diagnosing and predicting the prognosis or treatment responses among breast cancer patients. ↑ increase; ↓ reduce.
| miRNAs | miRNAs Source | Diagnostic Significance | Significance in Grading/Classification | Prognostic Significance | Ref. | ||
|---|---|---|---|---|---|---|---|
| Response to Treatment | Overall Survival | Relapse/Recurrence | |||||
| miR-21 | Serum | ↑ miR-21 in BC | ↑ miR-21 in advanced BC | ↑ miR-21 linked to ↑ radioresistance | ↑ miR-21 in BC ↓ survival | NIA | [36] |
| miR-125b | Serum | NIA | ↑ miR-125b linked to ↑ disease staging | ↑ miR-125b linked to ↑ chemoresistance | NIA | NIA | [61] |
| miR-140-5p | Plasma | ↓ miR-140-5p in BC as compared to CT | ↓ miR-140-5p linked to worst disease prognosis | ↓ miR-140-5p linked to ↑ chemoresistance | ↓ miR-140-5p linked to ↓ EFS | ↓ miR-140-5p linked to ↑ relapse/recurrence | [39] |
| miR-335 | Serum | ↓ miR-335 in BC as compared to CT | ↓ miR-335 in TNBC | NIA | ↓ miR-335 linked to ↓ OS | ↓ miR-335 linked to ↑ relapse/recurrence | [54] |
| miR-34a/b/c | Plasma | ↓ In the 3 miRNAs levels in BC as compared to CT | ↓ miR-34a levels linked to advanced clinical staging & histopathological grading | NIA | ↓ miR-34a levels linked to ↓ survival | NIA | [37] |
| miR-21, miR-23b, miR-200c, miR-190 |
Plasma | ↑ miR-21, miR-23b & miR-200c levels & ↓ miR-190 in BC |
The 4 miRNAs distinguished relapsed & non-relapsed BC cases | NIA | ↑ miR-21 & miR-200c linked to short DFS | ↑ miR-21, miR-23b & miR-200c & ↓ miR-190 in relapsed as compared to non-relapsed case | [42] |
| miR-16-5p, miR-17-3p, miR-451a, miR-940 | Serum | No significant difference in the 4 miRNAs levels between BC & CT cases | The 4 miRNAs distinguished metastatic & non-metastatic BC cases | ↓ In the 4 miRNAs in trastuzumab-resistant BC | ↑ In the 4 miRNAs in improved BC survival | ↑ In the 4 miRNAs in reduced incidence of relapse/recurrence | [50] |
| miR-18b, miR-103, miR-107, miR-652 | Serum | ↑ In the 4 miRNAs levels in TNBC | ↑ In the 4 miRNAs levels linked to advanced clinical staging & histopathological grading | NIA | ↑ In the 4 miRNAs ↓ RFS & OS | ↑ In the 4 miRNAs in relapse group | [66] |
| Let-7a, miR-10b, miR-21, miR-145, miR-181a | Plasma | ↑ miR-10b, miR-21 & miR-181a & ↓ let-7a & miR-145 in BC | ↑ miR-10b, miR-21 & miR-181a & ↓ let-7a & miR-145 in locally advanced BC | NIA | ↑ miR-10b & ↓ miR-21 linked to survival | ↑ miR-10b & miR-21 linked to ↑ relapse | [57] |
| miR-26b-5p, miR-106b-5p, miR-142-3p, miR-142-5p, miR-185-5p, miR-362-5p | Whole blood | ↑ In the 6 miRNAs levels in BC | ↑ In the 6 miRNAs levels in early BC | NIA | ↑ In the 6 miRNAs levels linked to ↓ OS/DFS | NIA | [45] |
| miR-19a, miR-19b-3p, miR-22-3p, miR-25-3p, miR-93-5p, miR-199a-3p, miR-210-3p | Plasma | ↑ In the 7 miRNAs levels in BC | These miRNAs predicted BC patient survival & relapse | The 7 miRNAs regulate chemotherapy & targeted therapy resistance | ↑ miR-19a, miR-19b, miR-93 & miR-201 linked to poor OS in TNBC patients | NIA | [30] |
| miR-296-3p, miR-575, miR-3610-5p, miR-4483, miR-4710, miR-4755-3p, miR-5698, miR-8089 | Serum | ↑ In the 8 miRNAs levels in BC | The 8 miRNAs distinguished metastatic & non-metastatic BC cases | NIA | ↓ miR-5698 & miR-8089 linked to ↑ improved survival | The 8 miRNAs predicted distant metastases | [31] |
BC: breast cancer; DCIS: ductal carcinoma in situ; DFS: disease-free survival; EFS: event-free survival; ER: estrogen receptor; LN: lymph node; OS: overall survival; NACT: neoadjuvant chemotherapy; TNBC: triple-negative breast cancer; PFS: progression-free survival; PR: progesterone receptor; NIA: no information available. The complete list of miRNA studies surveyed in this review is provided as Supplementary Tables S1 and S2.
Table 4.
Potential exosomal and extracellular-vesicle miRNAs in diagnosing and predicting the prognosis or treatment responses among breast cancer patients. ↑ increase; ↓ reduce.
| miRNAs | miRNAs Source | Diagnostic Significance | Significance in Grading/Classification | Prognostic Significance | Ref. | ||
|---|---|---|---|---|---|---|---|
| Response to Treatment | Overall Survival | Relapse/Recurrence | |||||
| miR-24-3p | Plasma | ↑ miR-24-3p in BC | ↑ miR-24-3p linked to advanced clinical & histopathological grading | NIA | ↓ miR-24-3p linked to improved survival | NIA | [47] |
| miR-363-5p | Plasma | ↓ miR-363-5p in BC | ↓ miR-363-5p in LN+ve BC cases as compared to LN –ve BC cases | NIA | ↑ miR-363-5p linked to ↑ survival | NIA | [25] |
| miR-141, miR-200c |
Plasma | ↑ miR-141 & miR-200c in BC | ↑ miR-141 in invasive BC; ↑ miR-141 & miR-200c in metastatic BC | NIA | ↑ miR-200c linked to short OS | NIA | [64] |
| miR-155, miR-1246 |
Plasma | ↑ Both miRNAs in trastuzumab-resistant BC | ↑ Both miRNAs advanced BC as compared to non-advanced BC | ↑ Both miRNAs in trastuzumab-resistant BC | ↑ Both miRNAs linked to poor survival | ↑ Both miRNAs linked to relapse & poor EFS | [38] |
| miR-21, miR-105, miR-222 |
Serum | ↑ In the 3 miRNAs levels linked to presence of circulating BC cells | ↑ miR-222 linked to advanced clinical staging & histopathological grading; ↑ miR-21 & miR-105 in metastatic than non-metastatic BC | ↑ miR-21 reduced NACT response | NIA | NIA | [58] |
| miR-150-5p, miR-576-3p, miR-4665-5p |
Plasma | ↑ In the 3 miRNAs levels in BC | The 3 miRNAs distinguished recurrence & non-recurrence in BC cases | NIA | NIA | ↑ In the 3 miRNAs levels in recurrent BC as compared to non-recurrent BC | [34] |
| miR-16, miR-30b, miR-93 |
Plasma | ↑ miR-16 in BC & ↑ miR-93 in DCIS | ↑ miR-93 in ER & PR +ve BC | NIA | NIA | ↓ miR-30b linked to recurrence | [63] |
| miR-195-5p, miR-548ab, miR-2392, miR-2467-3p, miR-4448, miR-4800-3p |
Serum | ↑ miR-2392, miR-2467-3p, miR-4448 & miR-4800-3p levels in BC | The 6 miRNAs distinguished recurrence & non-recurrence in BC cases | ↑ miR-2392, miR-2467-3p, miR-4448 & miR-4800-3p levels in BC with complete NACT response | ↑ miR-2392, miR-2467-3p, miR-4448 & miR-4800-3p levels linked to ↑ OS in BC | ↑ In miR-195-5p & ↓ miR-548ab in recurrent BC cases | [60] |
| miR-30b, miR-34a, miR-127, miR-141, miR-182, miR-183, miR-328, miR-423 |
Plasma | Dysregulation in the 8 miRNA levels in BC | ↓ miR-30b, miR-127 & miR-328 in invasive BC | ↑ miR-127 & miR-141 linked to complete NACT response; ↑ miR-34a, miR-182 & miR-183 linked to poor NACT response |
↓ miR-141, miR-34a, miR-423, miR-182 & miR-183 linked to ↑ OS | NIA | [59] |
BC: breast cancer; DCIS: ductal carcinoma in situ; DFS: disease-free survival; EFS: event-free survival; ER: estrogen receptor; LN: lymph node; OS: overall survival; NACT: neoadjuvant chemotherapy; TNBC: triple-negative breast cancer; PFS: progression-free survival; PR: progesterone receptor; NIA: no information available. The complete list of miRNA studies surveyed in this review is provided as Supplementary Tables S1 and S2.
On the contrary, the level of circulating miRNAs is helpful to classify BC based on the clinical and histopathological grading [47,48]. For instance, the increased levels of exosomal miR-363-5p [25] and circulating miR-106a levels [49] were useful to differentiate BC with and without lymph node involvement. A clinical study reported that the elevation of four serum miRNAs, including miR-16-5p, miR-17-3p, miR-451a and miR-940, was observed in metastatic BC as compared to local, non-metastatic BC [50] and downregulation of plasma miR-195 level was also observed in metastatic BC [51]. The combined findings from these studies [50,51] highlighted the significance of circulating miRNA levels in BC staging. In addition, upregulation of plasma exosomal miR-181-5p and miR-222-3p is reported to link to advanced inflammatory BC as compared to non-inflammatory BC [35]. In terms of molecular grading, circulating miR-182 and miR-200c were shown to be downregulated and upregulated, respectively, in estrogen receptor (ER)- and progesterone receptor (PR)-positive patients. Upregulation of miR-10b and miR-21 was reported in ER-negative BC according to an Irish study [52], whereas suppression of miR-17 was shown to be observed in ER-positive BC [24]. For human epidermal growth factor receptor 2 (HER2)-positive patients, circulating miR-373 was reported to be upregulated [24] and, in another Chinese study [53], it was shown that the circulating level of miR-106a-5p and miR-20b-5p was increased in HER2-negative BC patients. In triple-negative BC (TNBC), serum miR-335 was reported to be downregulated [54], whereas serum miR-200c was upregulated [55]. Other circulating miRNAs that were upregulated in TNBC include miR-188-5p, miR-642b-3p, miR-1202, miR-1207-5p, miR-1225-5p, miR-1290, miR-3141, miR-4270 and miR-4281 [56].
3.2. Prognostic Significance of Circulating miRNAs in Human Breast Cancer
Apart from being employed as minimally invasive biomarkers in diagnosing and classifying different stages or types of BC (Table 3 and Table 4) [57], circulating miRNAs are also important in predicting the prognosis and treatment responses of BC patients [38,58]. For instance, the expression of miR-155 and miR-1246 was elevated in the plasma exosomes isolated from BC patients, and the upregulation of both miRNA levels was linked to poor survival, recurrence and trastuzumab resistance among BC patients [38]. In another study [39], downregulation of plasma miR-140-5p was correlated to increased chemoresistance, reduced event-free survival (EFS) and increased recurrence among BC patients. On the other hand, upregulation of exosomal miR-21 [58], exosomal miR-34a, miR-182 and miR-183 levels [59] was shown to contribute to poor chemotherapy response among BC patients, while the elevation of blood exosomal miR-2392, miR-4448 and miR-4800-3p was demonstrated to correlate to good response after neoadjuvant chemotherapy [60]. In terms of response towards targeted therapy, such as trastuzumab, decreased serum levels of miR-16-5p, miR-17-3p, miR-451a and miR-940 were observed in trastuzumab-resistant BC and the increased expression of these miRNAs was shown to promote treatment response to trastuzumab and improve survival among BC patients [50]. In another Arabic study [30], dysregulations of seven circulating miRNAs that include miR-19a, miR-19b-3p, miR-22-3p, miR-25-3p, miR-93-5p, miR-199a-3p and miR-210-3p were shown to be related to resistance to both chemotherapy and targeted therapy. On the contrary, the upregulation of circulating miR-21 correlated tightly to radio resistance among BC patients [36]. Notably, all the circulating miRNAs that were reported to modulate treatment responses in BC patients appeared to share a common function in promoting uncontrolled proliferation and apoptosis evasion among the BC cells [61,62]. For example, miR-373 was reported to upregulate the expression of vascular endothelial growth factor (VEGF) in BC cells, which would lead to enhanced proliferation and angiogenesis [62].
By studying the relationships between circulating miRNA levels, clinical conditions and treatment responses, clinicians could predict the survival and likelihood of disease recurrence among BC patients [63]. Elevation of several circulating exosomal miRNAs was shown to be associated with good survival and it was suggested that the upregulation of these miRNAs may improve patient survival by enhancing patient response to chemotherapy [60]. On the other hand, the upregulation of exosomal miR-200c [64] and miR-24-3p [47] was shown to correlate to poor overall survival (OS) among BC patients, as the increased expressions of these miRNAs were hypothesized to directly correlate to advanced disease staging [47,64]. Similarly, the downregulation of several circulating miRNAs, such as miR-34a [37] and miR-335 [54], was shown to reduce BC patient survival and this would contribute to an increased likelihood of relapse and recurrence. Metastasis is one of the important factors causing BC recurrence and the dysregulations. Eight miRNAs, including miR-296-3p, miR-575, miR-3610-5p, miR-4483, miR-4710, miR-4755-3p, miR-5698 and miR-8089, were demonstrated to be able to predict the likelihood of recurrence secondary to BC metastasis [31]. Other circulating miRNAs that are reported to play vital roles in influencing patient survival include miR-17, miR-18b, miR-103, miR-107, miR-652, miR-26b-5p, miR-106b-5p, miR-142-3p, miR-142-5p, miR-185-5p and miR-362-5p [45,65,66]. Patients with dysregulated circulating levels of these miRNAs were found to have more advanced disease staging and were more prone to face disease relapse and recurrence with reduced survival rate [45,65,66]. In two other studies [29,31], at least 20 circulating or exosomal miRNAs were reported to be sensitive and useful in distinguishing between recurrent and non-recurrent BC cases and this is helpful to predict patient prognosis and survival.
3.3. Multifunctional Roles of Circulating miRNAs as Potential Biomarker for Human Breast Cancer
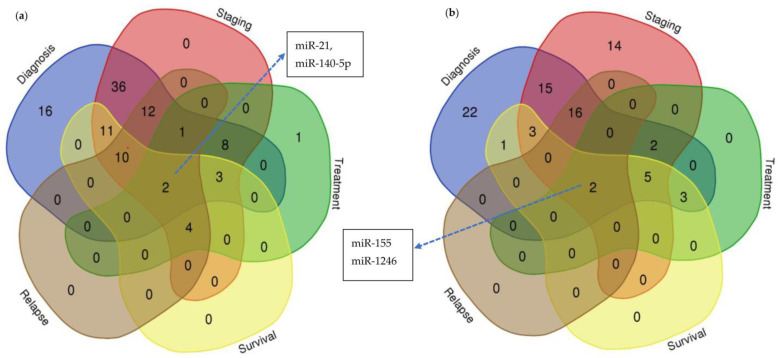
Evidently, circulating miRNAs have great potential to be employed as minimally invasive biomarkers in diagnosing BC at an early stage and complementary to the distinguishment of BC based on its clinical and histopathological grading [62,67]. In addition, circulating miRNAs are also helpful to predict the likelihood of relapse, recurrence and treatment responses among BC patients and this is particularly useful in guiding clinicians in planning a personalized treatment approach for different BC patients [30,68]. Given the challenge in identifying a miRNA panel useful for these functions within the growing number of related studies, we hope to provide an overview of the current status of the reported multifunctional roles of circulating miRNAs. Based on our findings from Table 3 and Table 4 as well as the Supplementary Tables S1 and S2, the miRNAs were further classified based on their reported roles in the diagnosis, staging classification and the prediction for relapse, treatment outcome and survival prognosis for BC patients (Figure 1a,b). We found that most of the reported free-circulating miRNAs were suitable in achieving the purpose of diagnosis only or concurrently in diagnosis and staging classification (Figure 1a). On the other hand, more reported exosomal and EV miRNAs were classified with either diagnosis only or staging only (Figure 1b), suggesting that exosomal and EV miRNAs may have more specific targets than free-circulating miRNA to be translated into clinical validation for diagnostic and staging purposes. Interestingly, there are two free-circulating (miR-21, miR-140-5p) and exosomal and EV (miR-155, miR-1246) miRNAs that were reported with the multifunctional roles, i.e., diagnosis, staging, treatment, survival and relapse.
Figure 1.
Number of (a) circulating and (b) exosomal and extracellular-vesicle (EV) miRNAs that play key roles in diagnosing and predicting prognosis or treatment responses among breast cancer patients. Number of (a) free-circulating (non exosomal) and (b) exosomal and extracellular-vesicle (EV) miRNAs. Numbers in the chart indicating the amount of miRNAs with the dedicated roles.
3.4. Sensitivity and Specificity Levels of miRNA Detection in BC Patients
Several potential miRNA biomarkers have been identified in BC patients’ serum or plasma. With a receiver operating characteristic (ROC) curve analysis, miR-21-5p was shown to have greater potential in discriminating between BC patients and the control group than that of miR-221-3p [69]. Additionally, a recent meta-analysis on miR-21-5p and BC that comprises six publications, consisting of Asian and Caucasian study cohorts, further confirmed the potential early diagnostic role of miR-21-5p in BC patients due to its high pooled AUC and diagnostic odds ratio [70]. Further, another recent meta-analysis conducted with the aim of determining the overall diagnostic performance of 56 eligible studies involving circulating miRNAs via qPCR revealed a pooled sensitivity and specificity of 0.85 and 0.83, respectively [71]. Moreover, multiple miRNA panels with sensitivity and specificity scores of 0.90 and 0.86, respectively, were significantly higher compared to that of the single miRNA panels with corresponding sensitivity and specificity scores of 0.82 and 0.83, respectively. With regard to specimen type, pool sensitivity and specificity of plasma were 0.83 and 0.85, respectively, and the pool sensitivity and specificity of serum were 0.87 and 0.83, respectively, indicating little difference in the diagnostic performance between serum and plasma samples. These studies revealed that cell-free circuiting miRNA could function as a promising early diagnostic biomarker for the detection of BC [71].
4. Current Challenges and Issues in Circulating miRNAs as a Common Candidate for Liquid Biopsy in BC Management
While establishing significant dysregulated miRNA expression of significance blood samples, studies have shown contrary findings between the use of serum or plasma as starting materials for obtaining miRNAs and in multicentered heterogenous patient samples, as well as in the use of various statistical models. Nevertheless, the need for standardization of pre-analytical variables, namely sample processing, storage and handling, as well as the data normalization strategy for the quantification of miRNA, are often highlighted as possible causes for the discordant outcomes of the identified miRNA as diagnostic, predictive or prognostic markers [72].
4.1. Biological Parameters
The expression profiles between miRNAs obtained from human BC serum versus tumors using RNA-sequencing revealed a total of 109 significant differentially expressed miRNAs between the patient’s serum and healthy individuals’ serum. Furthermore, 174 significant differentially expressed miRNAs between normal tissues and tumors were observed, of which only 10 common miRNAs were differentially expressed in serum and tumor biopsy [73]. Furthermore, an in-depth analysis of data obtained from the HMDD v3.0 database and individual papers showed circulating miRNAs as BC diagnostic biomarkers lack specificity due to different expressions in tissues and blood of cancer patients and even miR-21-5p being cited as the most commonly dysregulated miRNA in BC studies was shown to be highly expressed in other cancer types and diseases [74]. On the other hand, a study on circulating miRNA among BC tumors, serum and normal tissues using microarray and qPCR was able to show that miRNA profiles between tumors matched to that of their corresponding serums, indicating the possible selective release of miRNAs from the tumor site to the blood [75]. In another study, in which the influence of the heterogeneous population setting on miRNA profile was obtained from two different geographical populations, one from Belgium (n = 110; primary BC = 55, healthy individuals = 55) and another from Rwanda (n = 110; primary BC = 55, healthy individuals = 55), using qPCR from plasma samples, revealed two distinct pools of circulating miRNA corresponding to each of the studied populations [76]. However, a multicenter study comprising Caucasian and Asian ethnicities from five different geographical locations obtained a common pool of miRNA, with AUC values ranging from 0.88 to 0.97 in the detection of early BC [15].
4.2. Statistical Models
The detection of dysregulated miRNA expression level methods requires normalization to remove variations across samples and different normalization methods were shown to have likely contributed to differences in the miRNA profiles obtained. In one study, qPCR was validated using the >2-fold change method from plasma circulating miRNAs and obtained 24 significant upregulated miRNAs and 16 significant downregulated miRNAs in BC patients compared to controls; however, only three miRNAs (miR-22-5p, miR-27b-3p, miR-423-5p) were able to distinguish cancer patients from healthy individuals [77]. Another study also used qPCR for the detection of possible circulating miRNA biomarkers in BC by creating a panel from an unbiased exploration among all expressed miRNAs via the two-fold cross-validation consolidating logistic regression and feature selection algorithm in the discovery cohort. Results from the study revealed the identification of six miRNA potential biomarker panels with an AUC of 0.78 and 0.77 in the discovery and validation cohorts, respectively, using the global geometric mean normalization method [15].
It is obvious that numerous studies (Table 3 and Table 4) have identified dysregulated miRNA profiles, which significantly represent early cancer detection, molecular subtype classification status and monitoring signatures of recurrence and metastatic BC progression. However, these studies often reveal a diverse pool of miRNA, with roles typically associated with the various hallmarks of cancer [78]. Therefore, by consolidating the big data of miRNA with artificial intelligence (AI) and machine learning platforms (ML), a more comprehensive and inclusive approach may be established for complementing clinical-based decisions in promoting effective BC patient care and recovery outcome.
5. Machine Learning and Deep Learning Approaches in BC Research
The field of healthcare has been transformed by technological advancements, such as the generation of large digital datasets. Over the past few decades, many researchers have put their efforts in exploring the application of machine learning (ML) in various healthcare applications, including cancer detection, diagnoses, prognoses, treatment and recurrence prediction [79,80,81,82]. Basically, there are two main common types of ML techniques, i.e., (i) supervised learning and (ii) unsupervised learning. The main difference between these two types of learning is the need for labelled training data. Supervised learning relies on labelled input and output training data. Based on the input and output data, the model will first identify their relationship before it can be used to classify new and unseen datasets and predict outcomes. On the contrary, unsupervised learning processes unlabeled or raw data. It is often used to identify the trends or patterns in raw datasets and perform initial data analysis.
5.1. Machine Learning and Deep Learning for Detection and Diagnosis
Detection and diagnosis of BC at the early stage helps in reducing the fatality rate to a greater extent. Rana et al. [83] conducted a comparative experiment with four different supervised algorithms, including the support vector machine (SVM), logistic regression (LR), k-nearest neighbor (KNN) and Naïve Bayes (NB) on the Wisconsin Breast Cancer Diagnostic dataset (WBCD) in predicting and diagnosing BC. Based on their analysis, the KNN technique provided the best results. NB and LR have also performed well in BC diagnosis. Nonetheless, they highlighted that SVM is a strong predictive and sophisticated machine learning algorithm, especially when it comes to predictive analysis; thus, this technique is also the most suited technique for recurrence or non-recurrence prediction of BC. Similar findings were also found by many researchers [84,85], whereby SVM has demonstrated its efficiency in BC prediction and diagnosis and achieved the best performance in terms of accuracy and precision.
In addition to BC detection and diagnosis, machine learning techniques were also used for subtypes classification. Three different ML techniques, including fuzzy SVM, Bayesian classifier and random forest (RF), were compared in categorizing the types of cancer from a sequence of mammography images in the MIAS database. They found that fuzzy SVM has the best performance compared to other ML techniques, with over 90% accuracy, sensitivity, specificity, precision and recall [86]. Similar findings were also obtained by Wu and Hicks [87], whereby the SVM technique was effective in discriminating the existence of triple-negative breast cancer (TNBC) based on the RNA sequencing datasets. The clinical significance of this investigation is that ML algorithms could be used not only to improve diagnostic accuracy, but also for identifying women who are at high risk of developing TNBC, which could be prioritized for treatment.
ML techniques have been widely utilized for cancer prognosis and survival prediction purposes too. For instance, in a study [88] involving eight different ML techniques to develop models for identifying and visualizing relevant prognostic indications of BC survival rates, based on 5 years’ BC patient database of the National Cancer Institute’s SEER Program from 2006 to 2010, the RF technique was found to be the best technique, with an accuracy level of 94.64%. A study focusing on the analyses of the impact of chemotherapy and establishment of prediction model of prognosis in early elderly TNBC was conducted by using machine learning, with 4696 patients in the SEER Database who were 70 years or older, diagnosed with primary early TNBC, from 2010 to 2016 [89]. The propensity-score-matched method was utilized to reduce covariable imbalance. Univariable and multivariable analyses were used to compare BC-specific survival (BCSS) and overall survival (OS). Nine models were developed by ML to predict the 5-year OS and BCSS for patients who received chemotherapy. The multivariate analyses showed a better survival in the chemotherapy group and the Light Gradient Boosting Machine (LightGBM) is a practical model for predicting survival and providing precious systemic treatment for patients who received chemotherapy [90].
Researchers have also used deep learning (DL), a subset of ML, in cancer applications. Based on the literature review, it was noted that some of the DL applications were found to have better performance compared to the conventional ML techniques. For instance, an ensemble deep learning approach for the definite classification of non-carcinoma and carcinoma BC histopathology images was able show a sensitivity of 97.73% for carcinoma classification, with an overall accuracy of 95.29% [91]. On the other hand, particle-swarm-optimized wavelet neural network (PSOWNN) was found relatively superior compared to other conventional ML techniques, such as CNN, KNN and SVM [92]. Meanwhile, the deep-learning-assisted efficient AdaBoost algorithm (DLA-EABA), a combined ML approach with AdaBoost algorithm as the base, for early BC detection showed a high accuracy level of 97.2%, sensitivity at 98.3% and specificity at 96.5% [93]. Apart from that, this method was reported to increase the patient survival rate.
5.2. Studies with miRNAs as Breast Cancer Biomarkers with ML/DL Approaches
MicroRNAs (miRNAs) have been suggested as the biomarkers or therapeutic targets in BCs [94,95,96]. However, there are not many BC studies utilizing the ML or DL approaches on miRNA biomarkers. Table 5 summarizes some of the related work for BC that adopted ML or DL approaches on miRNA biomarkers.
Table 5.
Related work for BC studies with miRNA as biomarkers.
| Reference | Function/Purpose | Methods | Accuracy of Model |
|---|---|---|---|
| [97] | Cancer Classification | Gradient Boosting | Accuracy 93.59% |
| RF | Accuracy 93.24% | ||
| LR | Accuracy 92.37% | ||
| Passive Aggressive | Accuracy 88.31% | ||
| SGD | Accuracy 90.35% | ||
| SVM | Accuracy 91.54% | ||
| Ridge | Accuracy 83.05% | ||
| Bagging | Accuracy 91.1% | ||
| [98] | Cancer Classification | NB | Accuracy 94.9% |
| [99] | Cancer detection | RF | AUC 99.5–99.9% |
| SVM | AUC 93.8–99.6% | ||
| ANN | Accuracy 97.3% | ||
| KNN | Accuracy 99.2% | ||
| SVM | Accuracy 96.3% | ||
| LR | Accuracy 95.8% | ||
| [100] | Cancer Classification | Tree-based model | NIA |
| [101] | Cancer Classification | DT | Accuracy 99.12% |
| NB | Accuracy 93.86% | ||
| ANN | Accuracy 100% | ||
| DL | Accuracy 100% |
Abbreviations: BC: breast cancer; SVM: Support Vector Machine; LR: Logistic Regression; KNN: k-Nearest Neighbor; NB: Naïve Bayes; WBCD: Wisconsin Breast Cancer Diagnostic dataset; RF: Random Forest; SGD: Stochastic Gradient Descent; ANN: Artificial Neural Network; DT: Decision Tree; DL: Deep Learning; NIA: no information available.
As the amount of miRNA expression data in the Genomic Data Commons (GDC) Data Portal increased dramatically, several researchers proposed feature-selection methods to reduce the size of datasets, before they proceed with their analysis. An ensemble feature-selection methodology for miRNA signatures was proposed to identify the most robust and reliable miRNAs to be used in clinically relevant prediction tasks [97]. In their research, including over 8000 samples from TCGA, 100 miRNA signatures were identified and distinguished between tumor and normal tissues. The proposed approach provided better accuracy after 10-fold cross-validation with different ML classifiers, showing over 90% classification accuracy. In another study, the ensemble methodology was performed to identify the important biomarkers for BC and then classified by different ML techniques, such as NB, LR, KNN, SVM and multilayer perceptron [98]. In their preliminary analysis, default parameters were changed only when experimentation showed that classifier performance generally improved significantly across all datasets. Rehman et al. [99] performed four different feature-selection methods, including the Information Gain (IG), Chi-Squared (CHI2) and Least Absolute Shrinkage and Selection Operation (LASSO), to identify the most specific and effective miRNAs in discriminating normal and cancerous tissues. After feature selection, they applied the RF and SVM algorithms to identify the cancerous cell. The study demonstrated that the miRNAs ranked higher by their analysis had higher classifier performance. Performance becomes lower as the rank of the miRNA decreases, which shows that these miRNAs had different degrees of importance as biomarkers.
The tree-based ML models were normally applied on specific miRNAs for classifying the upregulated and downregulated BC cells [100]. In addition, several supervised methods, such as DT, NB, neural network and DL, were adopted, to classify cancer cells based on the expression of the microRNA gene to obtain the best method that can be used for gene analysis [101]. It was found that the DL method, which was developed based on a multilayer feed-forward ANN trained with stochastic gradient descent using backpropagation outperformed other conventional ML methods [101].
6. Conclusions
Key cancer signatures are represented by the dysregulated expression of miRNAs detected in liquid biopsy and shown to be associated with BC diagnosis, subtype classification and recurrence, as well as metastatic spread of the cancerous cells. Therefore, using miRNA-based liquid biopsy may be much more feasible in BC management, which also may come with greater patient acceptance as it could be conducted as simple routine blood collection, but the amount of information contained in the miRNA profile is immense. Using bioinformatics and with current and emerging AI and ML platforms, this huge amount of miRNA data may be able to be analyzed in ways that provide cancer progression indicators that complement standard clinical practices. However, translation of the miRNA targets selected by ML requires clinical validation to achieve the number of biomarkers that can accurately perform the expected roles cost effectively. This will eventually result in better treatment outcomes and supportive BC management from early detection to personalized therapy, ultimately improving the quality of life among BC patients.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232315382/s1.
Author Contributions
L.L., A.F.A., Z.X.C., R.J.C., E.Z.X.S. and J.F.K. conducted the data reviews and analysis and wrote the original draft preparation; W.Y.H., S.K.Y., Y.L.Y., J.L.L. and N.S.Y. participated in critically reviewing and proofreading the manuscript. A.H.K.O. conceived, developed and led the overall study; All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable for this study.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by funding from UTARF Cycle 1 2020 (IPSR/RMC/UTARRF/2020-C1/A02) (Vot number: 6200/A46).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Giaquinto A.N., Sung H., Miller K.D., Kramer J.L., Newman L.A., Minihan A., Jemal A., Siegel R.L. Breast Cancer Statistics, 2022. CA Cancer J. Clin. 2022;72:524–541. doi: 10.3322/caac.21754. [DOI] [PubMed] [Google Scholar]
- 3.Lima S.M., Kehm R.D., Terry M.B. Global Breast Cancer Incidence and Mortality Trends by Region, Age-Groups, and Fertility Patterns. EClinicalMedicine. 2021;38:100985. doi: 10.1016/j.eclinm.2021.100985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mubarik S., Malik S.S., Wang Z., Li C., Fawad M., Yu C. Recent Insights into Breast Cancer Incidence Trends among Four Asian Countries Using Age-Period-Cohort Model. Cancer Manag. Res. 2019;11:8145–8155. doi: 10.2147/CMAR.S208323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schliemann D., Hoe W.M.K., Mohan D., Allotey P., Reidpath D.D., Tan M.M., Taib N.A.M., Donnelly M., Su T.T. Challenges and Opportunities for Breast Cancer Early Detection among Rural Dwelling Women in Segamat District, Malaysia: A Qualitative Study. PLoS ONE. 2022;17:267308. doi: 10.1371/journal.pone.0267308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuasha N., Petros B. Heterogeneity of Tumors in Breast Cancer: Implications and Prospects for Prognosis and Therapeutics. Scientifica (Cairo) 2020;2020:4736091. doi: 10.1155/2020/4736091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zambelli A., Tondini C., Munkácsy G., Santarpia L., Gyorffy B. Gene Expression Profiling in Early Breast Cancer-Patient Stratification Based on Molecular and Tumor Microenvironment Features. Biomedicines. 2022;10:248. doi: 10.3390/biomedicines10020248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan J.W., Zabidi M.M.A., Ng P.S., Meng M.Y., Hasan S.N., Sandey B., Sammut S.J., Yip C.H., Rajadurai P., Rueda O.M., et al. The Molecular Landscape of Asian Breast Cancers Reveals Clinically Relevant Population-Specific Differences. Nat. Commun. 2020;11:6433. doi: 10.1038/s41467-020-20173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eliyatkin N., Yalcin E., Zengel B., Aktaş S., Vardar E. Molecular Classification of Breast Carcinoma: From Traditional, Old-Fashioned Way to A New Age, and A New Way. J. Breast Health. 2015;11:59–66. doi: 10.5152/tjbh.2015.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zubair M., Wang S., Ali N. Advanced Approaches to Breast Cancer Classification and Diagnosis. Front. Pharmacol. 2021;11:632079. doi: 10.3389/fphar.2020.632079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gennari A., André F., Barrios C.H., Cortés J., de Azambuja E., DeMichele A., Dent R., Fenlon D., Gligorov J., Hurvitz S.A., et al. ESMO Clinical Practice Guideline for the Diagnosis, Staging and Treatment of Patients with Metastatic Breast Cancer. Ann. Oncol. 2021;32:1475–1495. doi: 10.1016/j.annonc.2021.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Gradishar W.J., Moran M.S., Abraham J., Aft R., Agnese D., Allison K.H., Anderson B., Burstein H.J., Chew H., Dang C., et al. Breast Cancer, Version 3.2022. JNCCN J. Natl. Compr. Cancer Netw. 2022;20:691–722. doi: 10.6004/jnccn.2022.0030. [DOI] [PubMed] [Google Scholar]
- 13.Saha T., Lukong K.E. Breast Cancer Stem-Like Cells in Drug Resistance: A Review of Mechanisms and Novel Therapeutic Strategies to Overcome Drug Resistance. Front. Oncol. 2022;12:961. doi: 10.3389/fonc.2022.856974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Do Nascimento R.G., Otoni K.M. Histological and Molecular Classification of Breast Cancer: What Do We Know? Mastology. 2020;30:e20200024. doi: 10.29289/25945394202020200024. [DOI] [Google Scholar]
- 15.Zou R., Loke S.Y., Tan V.K.M., Quek S.T., Jagmohan P., Tang Y.C., Madhukumar P., Tan B.K.T., Yong W.S., Sim Y., et al. Development of a Microrna Panel for Classification of Abnormal Mammograms for Breast Cancer. Cancers (Basel) 2021;13:2130. doi: 10.3390/cancers13092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oluwaseyi Temilola D., Wium M., Herve Coulidiati T., Ademola Adeola H., Maria Carbone G., Vittorio Catapano C., Fernando Zerbini L. The Prospect and Challenges to the Flow of Liquid Biopsy in Africa. Cells. 2019;8:862. doi: 10.3390/cells8080862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hashimoto K., Nishimura S., Ito T., Oka N., Akagi M. Limitations and Usefulness of Biopsy Techniques for the Diagnosis of Metastatic Bone and Soft Tissue Tumors. Ann. Med. Surg. 2021;68:102581. doi: 10.1016/j.amsu.2021.102581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodríguez J., Avila J., Rolfo C., Ruíz-Patiño A., Russo A., Ricaurte L., Ordóñez-Reyes C., Arrieta O., Zatarain-Barrón Z.L., Recondo G., et al. When Tissue Is an Issue the Liquid Biopsy Is Nonissue: A Review. Oncol. Ther. 2021;9:89–110. doi: 10.1007/s40487-021-00144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirahata T., ul Quraish R., ul Quraish A., ul Quraish S., Naz M., Razzaq M.A. Liquid Biopsy: A Distinctive Approach to the Diagnosis and Prognosis of Cancer. Cancer Inform. 2022;21:11769351221076062. doi: 10.1177/11769351221076062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X., Papukashvili D., Wang Z., Liu Y., Chen X., Li J., Li Z., Hu L., Li Z., Rcheulishvili N., et al. Potential Utility of MiRNAs for Liquid Biopsy in Breast Cancer. Front. Oncol. 2022;12:940314. doi: 10.3389/fonc.2022.940314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng X., Shi G., He Q., Zhu P. Screening and Predicted Value of Potential Biomarkers for Breast Cancer Using Bioinformatics Analysis. Sci. Rep. 2021;11:20799. doi: 10.1038/s41598-021-00268-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li R., Qu H., Wang S., Chater J.M., Wang X., Cui Y., Yu L., Zhou R., Jia Q., Traband R., et al. CancerMIRNome: An Interactive Analysis and Visualization Database for MiRNome Profiles of Human Cancer. Nucleic Acids Res. 2022;50:D1139–D1146. doi: 10.1093/nar/gkab784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lánczky A., Nagy Á., Bottai G., Munkácsy G., Szabó A., Santarpia L., Győrffy B. MiRpower: A Web-Tool to Validate Survival-Associated MiRNAs Utilizing Expression Data from 2178 Breast Cancer Patients. Breast Cancer Res. Treat. 2016;160:439–446. doi: 10.1007/s10549-016-4013-7. [DOI] [PubMed] [Google Scholar]
- 24.Eichelser C., Flesch-Janys D., Chang-Claude J., Pantel K., Schwarzenbach H. Deregulated Serum Concentrations of Circulating Cell-Free MicroRNAs MiR-17, MiR-34a, MiR-155, and MiR-373 in Human Breast Cancer Development and Progression. Clin. Chem. 2013;59:1489–1496. doi: 10.1373/clinchem.2013.205161. [DOI] [PubMed] [Google Scholar]
- 25.Wang X., Qian T., Bao S., Zhao H., Chen H., Xing Z., Li Y., Zhang M., Meng X., Wang C., et al. Circulating Exosomal MiR-363-5p Inhibits Lymph Node Metastasis by Downregulating PDGFB and Serves as a Potential Noninvasive Biomarker for Breast Cancer. Mol. Oncol. 2021;15:2466–2479. doi: 10.1002/1878-0261.13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moloney B.M., Gilligan K.E., Joyce D.P., O’Neill C.P., O’Brien K.P., Khan S., Glynn C.L., Waldron R.M., Maguire C.M., Holian E., et al. Investigating the Potential and Pitfalls of EV-Encapsulated MicroRNAs as Circulating Biomarkers of Breast Cancer. Cells. 2020;9:141. doi: 10.3390/cells9010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M., Zou X., Xia T., Wang T., Liu P., Zhou X., Wang S., Zhu W. A Five-MiRNA Panel in Plasma Was Identified for Breast Cancer Diagnosis. Cancer Med. 2019;8:7006–7017. doi: 10.1002/cam4.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen W., Cao R., Su W., Zhang X., Xu Y., Wang P., Gan Z., Xie Y., Li H., Qin J. Simple and Fast Isolation of Circulating Exosomes with a Chitosan Modified Shuttle Flow Microchip for Breast Cancer Diagnosis. Lab Chip. 2021;21:1759–1770. doi: 10.1039/D0LC01311K. [DOI] [PubMed] [Google Scholar]
- 29.Sueta A., Yamamoto Y., Tomiguchi M., Takeshita T., Yamamoto-Ibusuki M., Iwase H. Differential Expression of Exosomal MiRNAs between Breast Cancer Patients with and without Recurrence. Oncotarget. 2017;8:69934–69944. doi: 10.18632/oncotarget.19482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qattan A., Al-Tweigeri T., Alkhayal W., Suleman K., Tulbah A., Amer S. Clinical Identification of Dysregulated Circulating MicroRNAs and Their Implication in Drug Response in Triple Negative Breast Cancer (TNBC) by Target Gene Network and Meta-Analysis. Genes. 2021;12:549. doi: 10.3390/genes12040549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satomi-Tsushita N., Shimomura A., Matsuzaki J., Yamamoto Y., Kawauchi J., Takizawa S., Aoki Y., Sakamoto H., Kato K., Shimizu C., et al. Serum MicroRNA-Based Prediction of Responsiveness to Eribulin in Metastatic Breast Cancer. PLoS ONE. 2019;14:e0222024. doi: 10.1371/journal.pone.0222024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jang J.Y., Kim Y.S., Kang K.N., Kim K.H., Park Y.J., Kim C.W. Multiple MicroRNAs as Biomarkers for Early Breast Cancer Diagnosis. Mol. Clin. Oncol. 2021;14:31. doi: 10.3892/mco.2020.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koi Y., Tsutani Y., Nishiyama Y., Ueda D., Ibuki Y., Sasada S., Akita T., Masumoto N., Kadoya T., Yamamoto Y., et al. Predicting the Presence of Breast Cancer Using Circulating Small RNAs, Including Those in the Extracellular Vesicles. Cancer Sci. 2020;111:2104–2115. doi: 10.1111/cas.14393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu H., Wang Q., Zhong H., Li L., Zhang Q., Huang Q., Yu Z. Differentially Expressed MicroRNAs in Exosomes of Patients with Breast Cancer Revealed by Next-Generation Sequencing. Oncol. Rep. 2020;43:240–250. doi: 10.3892/or.2019.7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed S.H., Espinoza-Sánchez N.A., El-Damen A., Fahim S.A., Badawy M.A., Greve B., El-Shinawi M., Götte M., Ibrahim S.A. Small Extracellular Vesicle-Encapsulated MiR-181b-5p, MiR-222-3p and Let-7a-5p: Next Generation Plasma Biopsy-Based Diagnostic Biomarkers for Inflammatory Breast Cancer. PLoS ONE. 2021;16:e0250642. doi: 10.1371/journal.pone.0250642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dan T., Shastri A.A., Palagani A., Buraschi S., Neill T., Savage J.E., Kapoor A., Deangelis T., Addya S., Camphausen K., et al. MiR-21 Plays a Dual Role in Tumor Formation and Cytotoxic Response in Breast Tumors. Cancers. 2021;13:888. doi: 10.3390/cancers13040888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng Z., Chen X., Zhu D., Luo Z., Yang M. Low Expression of Circulating MicroRNA-34c Is Associated with Poor Prognosis in Triple-Negative Breast Cancer. Yonsei Med. J. 2017;58:697–702. doi: 10.3349/ymj.2017.58.4.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z., Zhang L., Yu G., Sun Z., Wang T., Tian X., Duan X., Zhang C. Exosomal MiR-1246 and MiR-155 as Predictive and Prognostic Biomarkers for Trastuzumab-Based Therapy Resistance in HER2-Positive Breast Cancer. Cancer Chemother. Pharmacol. 2020;86:761–772. doi: 10.1007/s00280-020-04168-z. [DOI] [PubMed] [Google Scholar]
- 39.Di Cosimo S., Appierto V., Pizzamiglio S., Tiberio P., Iorio M.V., Hilbers F., De Azambuja E., De La Peña L., Izquierdo M., Huober J., et al. Plasma MiRNA Levels for Predicting Therapeutic Response to Neoadjuvant Treatment in HER2-Positive Breast Cancer: Results from the NeoALTTO Trial. Clin. Cancer Res. 2019;25:3887–3895. doi: 10.1158/1078-0432.CCR-18-2507. [DOI] [PubMed] [Google Scholar]
- 40.Stevic I., Müller V., Weber K., Fasching P.A., Karn T., Marmé F., Schem C., Stickeler E., Denkert C., Van Mackelenbergh M., et al. Specific MicroRNA Signatures in Exosomes of Triple-Negative and HER2-Positive Breast Cancer Patients Undergoing Neoadjuvant Therapy within the GeparSixto Trial. BMC Med. 2018;16:179. doi: 10.1186/s12916-018-1163-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng X., Lu S., He Z., Huang H., Yao Z., Miao Y., Cai C., Zou F. MCU-Dependent Negative Sorting of MiR-4488 to Extracellular Vesicles Enhances Angiogenesis and Promotes Breast Cancer Metastatic Colonization. Oncogene. 2020;39:6975–6989. doi: 10.1038/s41388-020-01514-6. [DOI] [PubMed] [Google Scholar]
- 42.Papadaki C., Stratigos M., Markakis G., Spiliotaki M., Mastrostamatis G., Nikolaou C., Mavroudis D., Agelaki S. Circulating MicroRNAs in the Early Prediction of Disease Recurrence in Primary Breast Cancer. Breast Cancer Res. 2018;20:72. doi: 10.1186/s13058-018-1001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen J., Hu Q., Schrauder M., Yan L., Wang D., Medico L., Guo Y., Yao S., Zhu Q., Liu B., et al. Circulating MiR-148b and MiR-133a as Biomarkers for Breast Cancer Detection. Oncotarget. 2014;5:5284–5294. doi: 10.18632/oncotarget.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papadaki C., Stoupis G., Tsalikis L., Monastirioti A., Papadaki M., Maliotis N., Stratigos M., Mastrostamatis G., Mavroudis D., Agelaki S. Circulating MiRNAs as a Marker of Metastatic Disease and Prognostic Factor in Metastatic Breast Cancer. Oncotarget. 2019;10:966–981. doi: 10.18632/oncotarget.26629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang K., Wang Y.Y., Xu Y., Zhang L., Zhu J., Si P.C., Wang Y.W., Ma R. A Two-MiRNA Signature of Upregulated MiR-185-5p and MiR-362-5p as a Blood Biomarker for Breast Cancer. Pathol. Res. Pract. 2021;222:153458. doi: 10.1016/j.prp.2021.153458. [DOI] [PubMed] [Google Scholar]
- 46.Cuk K., Zucknick M., Madhavan D., Schott S., Golatta M., Heil J., Marmé F., Turchinovich A., Sinn P., Sohn C., et al. Plasma MicroRNA Panel for Minimally Invasive Detection of Breast Cancer. PLoS ONE. 2013;8:e76729. doi: 10.1371/journal.pone.0076729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khodadadi-Jamayran A., Akgol-Oksuz B., Afanasyeva Y., Heguy A., Thompson M., Ray K., Giro-Perafita A., Sánchez I., Wu X., Tripathy D., et al. Prognostic Role of Elevated Mir-24-3p in Breast Cancer and Its Association with the Metastatic Process. Oncotarget. 2018;9:12868–12878. doi: 10.18632/oncotarget.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshikawa M., Iinuma H., Umemoto Y., Yanagisawa T., Matsumoto A., Jinno H. Exosome-Encapsulated MicroRNA-223-3p as a Minimally Invasive Biomarker for the Early Detection of Invasive Breast Cancer. Oncol. Lett. 2018;15:9584–9592. doi: 10.3892/ol.2018.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohmmed E.A., Shousha W.G., EL-Saiid A.S., Ramadan S.S. A Clinical Evaluation of Circulating MiR-106a and Raf-1 as Breast Cancer Diagnostic and Prognostic Markers. Asian Pac. J. Cancer Prev. 2021;22:3513–3520. doi: 10.31557/APJCP.2021.22.11.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H., Liu J., Chen J., Wang H., Yang L., Chen F., Fan S., Wang J., Shao B., Yin D., et al. A Serum MicroRNA Signature Predicts Trastuzumab Benefit in HER2-Positive Metastatic Breast Cancer Patients. Nat. Commun. 2018;9:1614. doi: 10.1038/s41467-018-03537-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McAnena P., Tanriverdi K., Curran C., Gilligan K., Freedman J.E., Brown J.A.L., Kerin M.J. Circulating MicroRNAs MiR-331 and MiR-195 Differentiate Local Luminal a from Metastatic Breast Cancer. BMC Cancer. 2019;19:436. doi: 10.1186/s12885-019-5636-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heneghan H.M., Miller N., Lowery A.J., Sweeney K.J., Newell J., Kerin M.J. Circulating MicroRNAs as Novel Minimally Invasive Biomarkers for Breast Cancer. Ann. Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 53.Li M., Zhou Y., Xia T., Zhou X., Huang Z., Zhang H., Zhu W., Ding Q., Wang S. Circulating MicroRNAs from the MiR-106a–363 Cluster on Chromosome X as Novel Diagnostic Biomarkers for Breast Cancer. Breast Cancer Res. Treat. 2018;170:257–270. doi: 10.1007/s10549-018-4757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swellam M., Mahmoud M.S., Hashim M., Hassan N.M., Sobeih M.E., Nageeb A.M. Clinical Aspects of Circulating MiRNA-335 in Breast Cancer Patients: A Prospective Study. J. Cell. Biochem. 2018;120:8975–8982. doi: 10.1002/jcb.28168. [DOI] [PubMed] [Google Scholar]
- 55.Niedźwiecki S., Piekarski J., Szymańska B., Pawłowska Z., Jeziorski A. Serum Levels of Circulating MiRNA-21, MiRNA-10b and MiRNA-200c in Triple-Negative Breast Cancer Patients. Ginekol. Pol. 2018;89:414–419. doi: 10.5603/GP.a2018.0071. [DOI] [PubMed] [Google Scholar]
- 56.Hamam R., Ali A.M., Alsaleh K.A., Kassem M., Alfayez M., Aldahmash A., Alajez N.M. MicroRNA Expression Profiling on Individual Breast Cancer Patients Identifies Novel Panel of Circulating MicroRNA for Early Detection. Sci. Rep. 2016;6:25997. doi: 10.1038/srep25997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ibrahim A.M., Said M.M., Hilal A.M., Medhat A.M., Abd Elsalam I.M. Candidate Circulating MicroRNAs as Potential Diagnostic and Predictive Biomarkers for the Monitoring of Locally Advanced Breast Cancer Patients. Tumor Biol. 2020;42:1010428320963811. doi: 10.1177/1010428320963811. [DOI] [PubMed] [Google Scholar]
- 58.Rodríguez-Martínez A., De Miguel-Pérez D., Ortega F.G., García-Puche J.L., Robles-Fernández I., Exposito J., Martorell-Marugan J., Carmona-Sáez P., Garrido-Navas M.D.C., Rolfo C., et al. Exosomal MiRNA Profile as Complementary Tool in the Diagnostic and Prediction of Treatment Response in Localized Breast Cancer under Neoadjuvant Chemotherapy. Breast Cancer Res. 2019;21:21. doi: 10.1186/s13058-019-1109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Todorova V.K., Byrum S.D., Gies A.J., Haynie C., Smith H., Reyna N.S., Makhoul I. Circulating Exosomal MicroRNAs as Predictive Biomarkers of Neoadjuvant Chemotherapy Response in Breast Cancer. Current Oncology. 2022;29:613–630. doi: 10.3390/curroncol29020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sueta A., Fujiki Y., Goto Yamaguchi L., Tomiguchi M., Yamamoto Ibusuki M., Iwase H., Yamamoto Y. Exosomal MiRNA Profiles of Triple negative Breast Cancer in Neoadjuvant Treatment. Oncol. Lett. 2021;22:819. doi: 10.3892/ol.2021.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang H., Tan G., Dong L., Cheng L., Li K., Wang Z., Luo H. Circulating Mir-125b as a Marker Predicting Chemoresistance in Breast Cancer. PLoS ONE. 2012;7:e34210. doi: 10.1371/journal.pone.0034210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bakr N.M., Mahmoud M.S., Nabil R., Boushnak H., Swellam M. Impact of Circulating MiRNA-373 on Breast Cancer Diagnosis through Targeting VEGF and Cyclin D1 Genes. J. Genet. Eng. Biotechnol. 2021;19:84. doi: 10.1186/s43141-021-00174-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ni Q., Stevic I., Pan C., Müller V., Oliviera-Ferrer L., Pantel K., Schwarzenbach H. Different Signatures of MiR-16, MiR-30b and MiR-93 in Exosomes from Breast Cancer and DCIS Patients. Sci. Rep. 2018;8:12974. doi: 10.1038/s41598-018-31108-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang G., Zhang W., Li B., Stringer-Reasor E., Chu C., Sun L., Bae S., Chen D., Wei S., Jiao K., et al. MicroRNA-200c and MicroRNA-141 Are Regulated by a FOXP3-KAT2B Axis and Associated with Tumor Metastasis in Breast Cancer. Breast Cancer Res. 2017;19:73. doi: 10.1186/s13058-017-0858-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hesari A.R., Azizian M., Darabi H., Nesaei A., Hosseini S.A., Salarinia R., Motaghi A.A., Ghasemi F. Expression of Circulating MiR-17, MiR-25, and MiR-133 in Breast Cancer Patients. J. Cell. Biochem. 2019;120:7109–7114. doi: 10.1002/jcb.27984. [DOI] [PubMed] [Google Scholar]
- 66.Sahlberg K.K., Bottai G., Naume B., Burwinkel B., Calin G.A., Børresen-Dale A.-L., Santarpia L. A Serum MicroRNA Signature Predicts Tumor Relapse and Survival in Triple-Negative Breast Cancer Patients. Clin. Cancer Res. 2015;21:1207–1214. doi: 10.1158/1078-0432.CCR-14-2011. [DOI] [PubMed] [Google Scholar]
- 67.Diansyah M.N., Prayogo A.A., Sedana M.P., Savitri M., Romadhon P.Z., Amrita P.N.A., Wijaya A.Y., Hendrata W.M., Bintoro U.Y. Early Detection Breast Cancer: Role of Circulating Plasma MiRNA-21 Expression as a Potential Screening Biomarker. Turk. J. Med. Sci. 2021;51:562–569. doi: 10.3906/sag-2005-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lopes B.C., Braga C.Z., Ventura F.V., de Oliveira J.G., Kato-Junior E.M., Bordin-Junior N.A., Zuccari D.A.P.C. MiR-210 and MiR-152 as Biomarkers by Liquid Biopsy in Invasive Ductal Carcinoma. J. Pers. Med. 2021;11:31. doi: 10.3390/jpm11010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Motawi T.M.K., Sadik N.A.H., Shaker O.G., el Masry M.R., Mohareb F. Study of MicroRNAs-21/221 as Potential Breast Cancer Biomarkers in Egyptian Women. Gene. 2016;590:210–219. doi: 10.1016/j.gene.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 70.Li S., Yang X., Yang J., Zhen J., Zhang D. Serum MicroRNA-21 as a Potential Diagnostic Biomarker for Breast Cancer: A Systematic Review and Meta-Analysis. Clin. Exp. Med. 2016;16:29–35. doi: 10.1007/s10238-014-0332-3. [DOI] [PubMed] [Google Scholar]
- 71.Sehovic E., Urru S., Chiorino G., Doebler P. Meta-Analysis of Diagnostic Cell-Free Circulating MicroRNAs for Breast Cancer Detection. BMC Cancer. 2022;22:634. doi: 10.1186/s12885-022-09698-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hamam R., Hamam D., Alsaleh K.A., Kassem M., Zaher W., Alfayez M., Aldahmash A., Alajez N.M. Circulating MicroRNAs in Breast Cancer: Novel Diagnostic and Prognostic Biomarkers. Cell Death Dis. 2017;8:e3045. doi: 10.1038/cddis.2017.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu J., Zheng Z., Wang J., Sun J., Wang P., Cheng X., Fu L., Zhang L., Wang Z., Li Z. Different MiRNA Expression Profiles between Human Breast Cancer Tumors and Serum. Front. Genet. 2014;5:149. doi: 10.3389/fgene.2014.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grimaldi A.M., Incoronato M. Clinical Translatability of “Identified” Circulating Mirnas for Diagnosing Breast Cancer: Overview and Update. Cancers. 2019;11:901. doi: 10.3390/cancers11070901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chan M., Liaw C.S., Ji S.M., Tan H.H., Wong C.Y., Thike A.A., Tan P.H., Ho G.H., Lee A.S.G. Identification of Circulating MicroRNA Signatures for Breast Cancer Detection. Clin. Cancer Res. 2013;19:4477–4487. doi: 10.1158/1078-0432.CCR-12-3401. [DOI] [PubMed] [Google Scholar]
- 76.Uyisenga J.P., Debit A., Poulet C., Frères P., Poncin A., Thiry J., Mutesa L., Jerusalem G., Bours V., Josse C. Differences in Plasma MicroRNA Content Impair MicroRNA-Based Signature for Breast Cancer Diagnosis in Cohorts Recruited from Heterogeneous Environmental Sites. Sci. Rep. 2021;11:11698. doi: 10.1038/s41598-021-91278-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jusoh A.R., Mohan S.V., Ping T.L., Din T.A.D.A.A.B.T., Haron J., Romli R.C., Jaafar H., Nafi S.N., Salwani T.I.T., Yahya M.M. Plasma Circulating Mirnas Profiling for Identification of Potential Breast Cancer Early Detection Biomarkers. Asian Pac. J. Cancer Prev. 2021;22:1375–1381. doi: 10.31557/APJCP.2021.22.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chong Z.X., Yeap S.K., Ho W.Y. Roles of Circulating MicroRNA(s) in Human Breast Cancer. Arch. Biochem. Biophys. 2020;695:108583. doi: 10.1016/j.abb.2020.108583. [DOI] [PubMed] [Google Scholar]
- 79.Jiang F., Jiang Y., Zhi H., Dong Y., Li H., Ma S., Wang Y., Dong Q., Shen H., Wang Y. Artificial Intelligence in Healthcare: Past, Present and Future. Stroke Vasc. Neurol. 2017;2:230–243. doi: 10.1136/svn-2017-000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Davenport T., Kalakota R. The Potential for Artificial Intelligence in Healthcare. Future Healthc. J. 2019;6:94–98. doi: 10.7861/futurehosp.6-2-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dlamini Z., Francies F.Z., Hull R., Marima R. Artificial Intelligence (AI) and Big Data in Cancer and Precision Oncology. Comput. Struct. Biotechnol. J. 2020;18:2300–2311. doi: 10.1016/j.csbj.2020.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang H.Y., Chen C.H., Shi S., Chung C.R., Wen Y.H., Wu M.H., Lebowitz M.S., Zhou J., Lu J.J. Improving Multi-Tumor Biomarker Health Check-up Tests with Machine Learning Algorithms. Cancers. 2020;12:1442. doi: 10.3390/cancers12061442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rana M., Chandorkar P., Dsouza A., Kazi N. Breast Cancer Diagnosis And Recurrence Prediction Using Machine Learning Techniques. Int. J. Res. Eng. Technol. 2015;4:372–376. [Google Scholar]
- 84.Shravya C., Pravalika K., Subhani S. Prediction of Breast Cancer Using Supervised Machine Learning Techniques. Int. J. Innov. Technol. Explor. Eng. (IJITEE) 2019;8:1106–1110. [Google Scholar]
- 85.Naji M.A., el Filali S., Aarika K., Benlahmar E.H., Abdelouhahid R.A., Debauche O. Machine Learning Algorithms for Breast Cancer Prediction and Diagnosis. Procedia Comput. Sci. 2021;191:487–492. doi: 10.1016/j.procs.2021.07.062. [DOI] [Google Scholar]
- 86.Chaudhury S., Krishna A.N., Gupta S., Sankaran K.S., Khan S., Sau K., Raghuvanshi A., Sammy F. Effective Image Processing and Segmentation-Based Machine Learning Techniques for Diagnosis of Breast Cancer. Comput. Math. Methods Med. 2022;2022:6841334. doi: 10.1155/2022/6841334. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87.Wu J., Hicks C. Breast Cancer Type Classification Using Machine Learning. J. Pers. Med. 2021;11:61. doi: 10.3390/jpm11020061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haque M.N., Tazin T., Khan M.M., Faisal S., Ibraheem S.M., Algethami H., Almalki F.A. Predicting Characteristics Associated with Breast Cancer Survival Using Multiple Machine Learning Approaches. Comput. Math. Methods Med. 2022;2022:1249692. doi: 10.1155/2022/1249692. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89.Huang K., Zhang J., Yu Y., Lin Y., Song C. The Impact of Chemotherapy and Survival Prediction by Machine Learning in Early Elderly Triple Negative Breast Cancer (ETNBC): A Population Based Study from the SEER Database. BMC Geriatr. 2022;22:268. doi: 10.1186/s12877-022-02936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boughorbel S., Al-Ali R., Elkum N. Model Comparison for Breast Cancer Prognosis Based on Clinical Data. PLoS ONE. 2016;11:146413. doi: 10.1371/journal.pone.0146413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hameed Z., Zahia S., Garcia-Zapirain B., Aguirre J.J., Vanegas A.M. Breast Cancer Histopathology Image Classification Using an Ensemble of Deep Learning Models. Sensors. 2020;20:4373. doi: 10.3390/s20164373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nomani A., Ansari Y., Nasirpour M.H., Masoumian A., Pour E.S., Valizadeh A. PSOWNNs-CNN: A Computational Radiology for Breast Cancer Diagnosis Improvement Based on Image Processing Using Machine Learning Methods. Comput. Intell. Neurosci. 2022;2022:5667264. doi: 10.1155/2022/5667264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zheng J., Lin D., Gao Z., Wang S., He M., Fan J. Deep Learning Assisted Efficient AdaBoost Algorithm for Breast Cancer Detection and Early Diagnosis. IEEE Access. 2020;8:96946–96954. doi: 10.1109/ACCESS.2020.2993536. [DOI] [Google Scholar]
- 94.Song T., Liang Y., Cao Z., Du W., Li Y. Computational Analysis of Specific MicroRNA Biomarkers for Noninvasive Early Cancer Detection. Biomed. Res. Int. 2017;2017:4680650. doi: 10.1155/2017/4680650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lopez-Rincon A., Mendoza-Maldonado L., Martinez-Archundia M., Schönhuth A., Kraneveld A.D., Garssen J., Tonda A. Machine Learning-Based Ensemble Recursive Feature Selection of Circulating Mirnas for Cancer Tumor Classification. Cancers. 2020;12:1785. doi: 10.3390/cancers12071785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robertus Fujii Y. The Quantum Language of the MicroRNA Gene and Anti-Cancer: With a Dynamic Computer Simulation of Human Breast Cancer Drug Resistance. Integr. Mol. Med. 2018;5 doi: 10.15761/IMM.1000346. [DOI] [Google Scholar]
- 97.Lopez-Rincon A., Martinez-Archundia M., Martinez-Ruiz G.U., Schoenhuth A., Tonda A. Automatic Discovery of 100-MiRNA Signature for Cancer Classification Using Ensemble Feature Selection. BMC Bioinform. 2019;20:480. doi: 10.1186/s12859-019-3050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kotlarchyk A., Khoshgoftaar T., Pavlovic M., Zhuang H., Pandya A.S. Identification of MicroRNA Biomarkers for Cancer by Combining Multiple Feature Selection Techniques. J. Comput. Methods Sci. Eng. 2011;11:283–298. doi: 10.3233/JCM-2011-0395. [DOI] [Google Scholar]
- 99.Rehman O., Zhuang H., Ali A.M., Ibrahim A., Li Z. Validation of MiRNAs as Breast Cancer Biomarkers with a Machine Learning Approach. Cancers. 2019;11:431. doi: 10.3390/cancers11030431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sherafatian M. Tree-Based Machine Learning Algorithms Identified Minimal Set of MiRNA Biomarkers for Breast Cancer Diagnosis and Molecular Subtyping. Gene. 2018;677:111–118. doi: 10.1016/j.gene.2018.07.057. [DOI] [PubMed] [Google Scholar]
- 101.Waspada I., Wibowo A., Meraz N.S. Supervised Machine Learning Model For Microrna Expression Data In Cancer. J. Ilmu Komput. Dan Inf. 2017;10:108. doi: 10.21609/jiki.v10i2.481. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable for this study.