Abstract
Proteins secreted or exported by Treponema denticola have been implicated as mediators of specific interactions between the spirochete and subgingival tissues in periodontal diseases. However, limited information is available on the ability of this peptidolytic organism to bind or transport soluble peptides present in the subgingival environment. A prominent 70-kDa protein was isolated from surface extracts of T. denticola ATCC 35405. A clone expressing a portion of the protein was identified in an Escherichia coli expression library of T. denticola DNA. DNA sequence analysis showed that the cloned gene encoded a peptide homologous to OppA, the solute binding protein of an ATP-binding cassette-type peptide transporter involved in peptide uptake and environmental signaling in a wide range of bacteria. Genes encoding OppB, -C, -D, and -F were identified directly downstream of oppA in T. denticola. OppA was present in representative strains of T. denticola and in Treponema vincentii but was not detected in Treponema pectinovorum or Treponema socranskii. Immunogold electron microscopy suggested that OppA was accessible to proteins at the surface of the spirochete. Native OppA bound soluble plasminogen and fibronectin but did not bind to immobilized substrates or epithelial cells. A T. denticola oppA mutant bound reduced amounts of soluble plasminogen, and plasminogen binding to the parent strain was inhibited by the lysine analog ɛ-aminocaproic acid. Binding of soluble host proteins by OppA may be important both for spirochete-host interactions in the subgingival environment and for uptake of peptide nutrients.
Treponema denticola is recognized as one of several potential pathogens in acute and chronic forms of human periodontal disease (50, 55, 62), and closely related spirochetes have been identified in bovine digital dermatitis lesions (10). Likely virulence factors of oral spirochetes include the ability to attach to host tissue and other microorganisms, motility and chemotaxis, immunomodulation, production of toxic metabolic byproducts, and direct cytopathogenicity (reviewed in reference 22). In the case of periodontal diseases, bacterial factors that contribute to the overgrowth of subgingival microflora must also be considered as potential virulence factors. These could include, for instance, uptake systems for peptide nutrients present in a high concentration in the inflamed gingival sulcus. Characterization of these processes will aid in understanding the biology of this organism and may suggest targets for treatment or prophylaxis.
T. denticola derives energy primarily from anaerobic degradation of peptides and amino acids (63). Nutrient requirements of this organism are complex (71), and the mechanisms of nutrient uptake are not well understood (12, 27, 28, 61). Peptide uptake requires specific systems for the binding and transport of substrates across the bacterial cell envelope. Oligopeptide uptake systems, members of a superfamily of highly conserved ATP-binding cassette (ABC) transporters, have been described for many bacteria (41, 65). In gram negative bacteria, the transporter includes a periplasmic solute-binding protein and an inner membrane complex consisting of an integral membrane protein(s) and membrane-bound cytoplasmic ATP-binding protein(s). In gram positive bacteria, SBPs are lipoproteins anchored to the cell membrane by their N-terminal lipid moiety (65). Oligopeptide uptake systems may be used for nutrient acquisition or turnover, though in organisms with multiple peptide uptake systems, one or more of these may function in environmental sensing, sporulation, or uptake of pheromones (59). While mechanisms for peptide uptake are likely to be important for T. denticola metabolism and chemotaxis, no studies of the molecular mechanisms of peptide uptake in oral spirochetes have been reported. In other spirochetes, including Treponema pallidum and Borrelia burgdorferi, genes encoding putative nutrient uptake systems have been cloned from genomic libraries (18, 40, 44, 58) or identified in the genomic sequences of these organisms (32, 33), but none of the proposed uptake activities have been demonstrated.
Secreted and exported proteins of T. denticola mediate specific interactions between the spirochete and the subgingival epithelium in periodontal diseases (reviewed in reference 22). Previous studies focused on potential adhesins (37, 47) and on spirochete surface proteins (21, 52, 66) or other cellular components (11, 35) cytotoxic to eukaryotic cells. Studies of membrane-associated proteins of two distinct strains of T. denticola identified a 70-kDa protein having fibronectin (FN)-binding (67) or FN-, laminin-, and fibrinogen-binding (37) activity. This protein was distinct from the 53-kDa Msp pore-forming adhesin in these strains, which also bound FN (23, 37, 67). We set out to identify and characterize the 70-kDa protein as a possible mediator of spirochete interaction with host tissue components. The present study describes initial molecular and functional characterization of a treponemal membrane-associated protein that is the product of a conserved genetic locus homologous to those encoding oligopeptide uptake systems in a wide range of bacteria. We propose that the binding of soluble host components by this protein may contribute to the survival and proliferation of the spirochete in the subgingival environment.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Oral Treponema strains used in this study are listed in Table 1. Cultures were grown and maintained in NOS broth medium as previously described (38) or in NOS broth supplemented with 0.3% pectin (69). For allelic replacement, mutants were selected on NOS/GN plates (9) containing erythromycin (40 μg ml−1) as described previously (24, 48). For some studies of T. denticola mutant strains, the growth medium was supplemented with triornithine (400 μM), trilysine (400 μM), aminopterin (1 μM), or bialafos (80 μg ml−1; gift of J. Davies). Cultures were examined by phase-contrast microscopy for purity and typical strain morphology before use. Four-day-old cultures were harvested by centrifugation at 10,000 × g (10 min, 4°C), washed in phosphate-buffered saline (PBS; 10 mM Na2HPO4, 150 mM NaCl, 2.5 mM KCl, 1.5 mM KH2PO4 [pH 7.2]), and then suspended in PBS to an optical density at 600 nm (OD600) of 0.2 (5 × 109 cells per ml) for use in assays.
TABLE 1.
Oral Treponema strains used in this study
| Species description | Strain | Source or reference |
|---|---|---|
| T. denticola serovar a | ATCC 35405 | ATCCa |
| T. denticola serovar c | ATCC 35404 | ATCC |
| T. denticola W | ATCC 33520 | ATCC |
| T. vincentii LA-1 | ATCC 35580 | ATCC |
| T. denticola isolate | OTK | R. Johnson |
| T. denticola isolate | GM-1 | A. Weinberg |
| T. socranskii subsp. socranskii | ATCC 35536 | ATCC |
| T. pectinovorum | ATCC 33768 | ATCC |
| T. denticola isogenic oppA mutant | OHE | This study |
| T. denticola isogenic oppF mutant | OXE | This study |
| T. denticola isogenic msp mutant | MHE | 24 |
ATCC, American Type Culture Collection.
Escherichia coli XL1-Blue (Stratagene Cloning Systems, La Jolla, Calif.) was used for plasmid preparations. Plasmid vector pTZ19R (U.S. Biochemicals, Cleveland, Ohio) was used for the library construction, subcloning, and preparation of sequencing templates. E. coli XL1-Blue was grown in Luria-Bertani broth or agar medium supplemented with ampicillin (50 μg ml−1) and erythromycin (200 μg ml−1) as appropriate.
Chemicals.
Unless otherwise noted, chemicals were purchased at the highest available purity from Sigma Chemical Co. (St. Louis, Mo.) or Fisher Scientific (Ottawa, Ontario, Canada).
Gel electrophoresis and immunoblotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western immunoblotting were done as described previously (23). Proteins in gels were detected by silver staining or Coomassie brilliant blue staining. Proteins blotted to nitrocellulose membranes were probed with rabbit polyclonal primary antibodies and alkaline phosphatase-conjugated anti-rabbit secondary antibodies, and the membranes were developed with 5-bromo-4-chloro-3-indolylphosphate (BCIP) and nitroblue tetrazolium.
Purification of the 70-kDa protein.
Extraction of treponemal outer membranes and associated proteins with Triton X-114 (Calbiochem, La Jolla, Calif.) and partitioning of the extracts into aqueous and detergent phases were performed as described for T. pallidum (17), with slight modifications as described previously (21). Phenylmethylsulfonyl fluoride (100 μM) was included in the Triton X-114 extract throughout the procedure. The detergent phase of Triton X-114 extracts from 1-liter batch cultures of T. denticola ATCC 35405 was subjected to preparative SDS-PAGE using a Model 491 Prep Cell (Bio-Rad Laboratories, Richmond, Calif.) as described previously (21). Fractions containing the protein of interest were concentrated by ultrafiltration, precipitated in acetone to remove detergent (4), resuspended in PBS at concentrations of 100 to 500 μg ml−1, and stored in aliquots at −70°C. Preparative electrophoresis of E. coli overproducing recombinant protein was performed similarly, except that E. coli cultures were lysed by suspension in SDS-PAGE sample buffer and heated prior to electrophoresis.
Preparation of antisera.
Polyclonal antisera to purified native and recombinant proteins were raised in New Zealand White rabbits as described previously by intramuscular injections with approximately 1 mg of purified protein in complete Freund's adjuvant (37). The titer of the antiserum was determined by enzyme-linked immunosorbent assay (ELISA), using alkaline phosphatase-conjugated goat anti-rabbit antibody (1:5,000; Life Technologies, Gaithersburg, Md.). The specificity of the antiserum was determined by Western immunoassay of T. denticola parent and oppA mutant strains.
Electron microscopy.
Preparation of T. denticola cells for transmission electron microscopy and immunogold labeling were done as described previously (20, 37), with some modifications designed to minimize disruption of intact cells. Briefly, 3- to 4-day-old cultures were harvested by centrifugation at 10,000 × g for 10 min at 4°C. Cells were washed twice in PBS at 4°C and applied to Parlodion-coated and carbon-stabilized copper grids at a density of approximately 107 cells per ml. The grid was first treated with 0.5% bovine serum albumin (BSA)–PBS for 30 min and then applied onto drops of primary antibodies in 0.5% BSA–PBS for 1 h. The grids were washed in 0.05% Tween 20–PBS (30 s) and PBS (30 s) after each incubation with antibodies. Grids were then incubated with 10-nm gold-bead-conjugated goat anti-rabbit antibodies for 1 h. After being washed, the grids were negatively stained with freshly prepared 4% uranyl acetate (pH 4.5) and studied by transmission electron microscopy with a Phillips Model 300 operating at 60 or 80 kV.
Recombinant DNA methods.
Unless stated otherwise, standard methods found in studies by Ausubel et al. (4) or Sambrook et al. (60) were followed. Southern and Northern blots were hybridized with biotin-labeled DNA probes and detected by development in streptavidin-alkaline phosphatase followed by BCIP and nitroblue tetrazolium, as described previously (23, 25). DNA fragments were eluted from agarose gels with the Gene Clean kit (Bio101, La Jolla, Calif.). Inverse PCR was performed as described by Ochman et al. (56). T. denticola genomic DNA was digested to completion with KpnI and ligated under conditions favoring circularization. The ligation products were subjected to PCR using oligonucleotide primers MX23 (5′ dCGG TAA TTT ATC TTA CCG AC 3′) and MX24 (5′ dGTC GGT AAG ATA AAT TAC CG 3′).
Preparation and screening of a genomic library of T. denticola.
T. denticola genomic DNA, prepared as described previously (23), was partially digested with Sau3AI and fractionated on a sucrose density gradient to yield fragments of 3 to 10 kb. A plasmid library of these fragments was constructed with BamHI-digested pTZ19R. Following induction with isopropyl-1-thio-β-d-galactopyranoside (IPTG), the library was screened for clones reactive with antiserum raised against the purified 70-kDa protein (anti-70).
DNA sequence analysis.
Sequencing reactions were performed with a Taq Cycle Sequencing Kit with fluorescent-labeled dideoxynucleoside triphosphates (Applied Biosystems Inc., Foster City, Calif.) using standard M13 sequencing primers or sequence-derived internal primers, electrophoresed and recorded by an Applied Biosystems model 373A automated DNA sequencer in the Nucleic Acids and Protein Sequencing Unit at the University of British Columbia. Both strands of the DNA sequence reported here were sequenced in their entirety. Analysis of DNA sequence data was performed with SeqEd 1.0 (Applied Biosystems Inc.), DNA Strider (Service de Biochimie, Department de Biologie, Institut de Recherche Fondamentale Commissariat a l'Energie Atomique, Cedex, France), and University of Wisconsin Genetics Computer Group software (19). The nonredundant SWISS-PROT, PIR, EMBL, and GenBank databases were searched for homologous peptide and nucleotide sequences by using the BLAST (3) network service at the National Center for Biotechnology Information, National Institutes of Health.
Construction of allelic replacement opp mutants.
To construct mutations in the opp locus, an ermF/AM cassette (31) was cloned into unique restriction sites in oppA or oppF sequences carried on an E. coli plasmid vector. Subsequent to the digestion of each construct with appropriate restriction enzymes to remove vector DNA, the fragments of interest were isolated after agarose gel electrophoresis. T. denticola was electroporated with linear DNA consisting of the ermF/AM cassette flanked by the desired target sequence, and erythromycin-resistant colonies were selected for further analysis (24, 48).
Adherence and binding assays.
Binding of T. denticola cells and purified components to soluble and immobilized substrates was assayed by ELISA or by ligand blotting as described previously (23, 37) and detected with antibodies to specific T. denticola proteins or with commercial antibodies to the ligand of interest. For ligand-blotting experiments, T. denticola cells or proteins bound to nitrocellulose membranes were probed with the ligand of interest (10 μg per ml in Tris-buffered saline) as described previously (37). Ligand binding was detected with specific antibodies directed against the ligand, as described for Western immunoblotting. For some experiments assaying plasminogen binding, plasminogen was pretreated with ɛ-aminocaproic acid (EACA; 100 mM, 30 min, 4°C) as described by Coleman et al. (14). Porcine periodontal ligament epithelial cells (7) and human gingival fibroblasts were isolated, cultured, and fixed in 0.25% glutaraldehyde as described previously (7, 21). Adherence and cytotoxicity of T. denticola cells and purified proteins to cultured cell monolayers was assayed as described previously (21).
Nucleotide sequence accession number.
The nucleotide sequence of the T. denticola oppA-F region has been assigned GenBank accession no. AF042861.
RESULTS
Isolation of the 70-kDa protein.
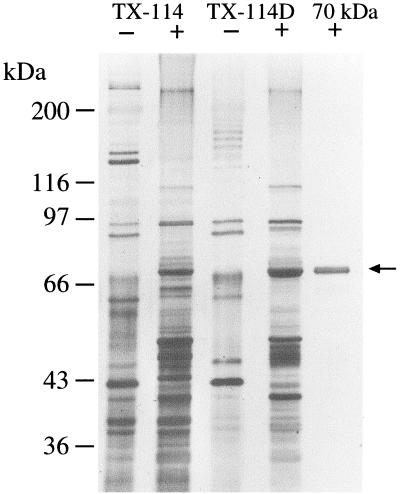
Previous studies of T. denticola surface-associated proteins had noted that, in addition to the pore-forming adhesin Msp, a prominent protein with a mass of approximately 70 kDa that was present in surface extracts also bound soluble FN (37, 67). Triton X-114 selectively solubilizes treponemal outer membranes and releases associated proteins of a likely outer membrane and periplasmic location (17). As shown in Fig. 1, the 70-kDa protein was released from T. denticola ATCC 35405 cells by mild treatment with Triton X-114 and, upon sequential phase partitioning at 37°C, localized to the hydrophobic detergent phase of the extracted material. The 70-kDa protein band was not seen in the aqueous phase of the Triton X-114 extract (data not shown). The 70-kDa protein was visible as a diffuse band when samples were not heated prior to electrophoresis. The protein was further purified by preparative electrophoresis, and the homogeneity of the protein was assessed by silver-stained PAGE (Fig. 1).
FIG. 1.
Purification of the 70-kDa protein from the detergent phase of a Triton X-114 extract of the T. denticola cell surface. Shown is a silver stained SDS-PAGE gel of samples which were heated at 100°C (+) or unheated (−) before electrophoresis. Lanes: TX-114, Triton X-114 extract; TX-114D, detergent phase of Triton X-114 extract; 70 kDa, purified 70-kDa protein.
Cloning and recombinant expression in E. coli.
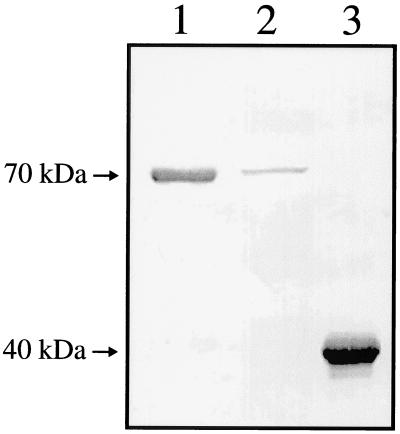
A plasmid library of T. denticola ATCC 35405 genomic DNA was probed for the expression of peptides recognized by antibodies raised against the purified 70-kDa protein (anti-70), and a clone expressing high levels of an immunoreactive 40-kDa peptide was identified. The recombinant protein, subsequently designated rOppA40, was purified by preparative electrophoresis from the E. coli strain carrying pMT1 and was used to immunize rabbits. As shown in Fig. 2 (lanes 1 and 2), antibodies raised against rOppA40 recognized the native T. denticola 70-kDa protein.
FIG. 2.
Western immunoblot probed with antibodies raised against recombinant protein rOppA40 (anti-40). Samples were heated at 100°C prior to electrophoresis. Arrows indicate the location of native OppA and rOppA40. Lanes: 1, purified 70-kDa protein; 2, T. denticola whole-cell extract; 3, E. coli pMT1 whole-cell extract.
Sequence analysis and identification of adjacent related genes.
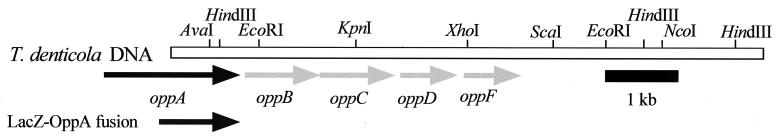
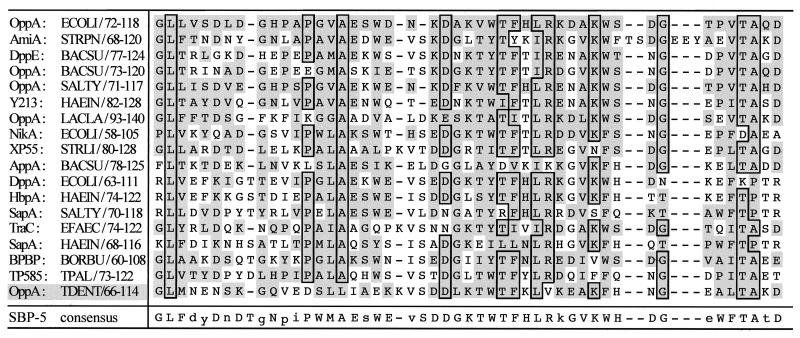
DNA encoding rOppA40 was localized to one end of the 8-kb fragment of T. denticola DNA carried on pMT1 (Fig. 3). Preliminary sequencing indicated that the immunoreactive peptide was a fusion with the vector-encoded amino terminus of LacZ (data not shown). Since no clones carrying the 5′ end of the gene were detected in the genomic library, this region was identified by inverse PCR. Self-ligated T. denticola genomic DNA was amplified with oligonucleotide primers derived from the T. denticola DNA insert carried on pMT1, and a single PCR product containing the 5′ end of the gene was obtained. The DNA sequence of the PCR product was combined with that obtained from pMT1 to yield the complete open reading frame (data not shown). The deduced peptide exhibited significant homology with the solute binding proteins of Cluster 5 of the superfamily of bacterial ABC transporters, which primarily includes oligopeptide uptake systems (65). The deduced T. denticola peptide, designated OppA, consists of 591 residues and contains the consensus sequence for lipid modification (L-X-X-C) at the predicted signal peptidase II cleavage site (residues 17 to 20). The predicted molecular mass of the mature peptide is 64,485 Da, and its hydropathy profile did not indicate significant hydrophobicity (data not shown). OppA residues 66 to 114, corresponding to the highly conserved signature sequence of this group, are shown aligned with other Cluster 5 SBPs in Fig. 4. Sequence analysis directly downstream of oppA revealed four open reading frames (depicted in Fig. 3) whose predicted peptide products exhibited high homology with OppB, -C, -D, and -F proteins involved in oligopeptide uptake in a wide range of bacteria (42). The proteins encoded by oppB and oppC appear to be integral membrane proteins, while those encoded by oppD and oppF corresponded to the ATP-hydrolyzing subunits of the deduced ABC transporter. Potential ribosome binding sites are located fewer than 10 bp upstream of each gene of oppA through oppF (data not shown). Northern blot analysis of T. denticola mRNA revealed a 1.9-kb band hybridizing with an internal oppA probe, which corresponded with the size of the oppA gene (data not shown). A promoter and transcription start site remain to be identified in the AT-rich DNA directly upstream of oppA.
FIG. 3.
Map of the T. denticola opp locus. The 8-kb fragment of T. denticola DNA carried on pMT1 is shown as an open bar, with the locations of some identified restriction enzyme sites shown. Immediately below are arrows representing locations of the genes identified in the opp locus. The solid arrow extending beyond the 8-kb insert represents oppA, while the shaded arrows downstream represent oppB, -C, -D, and -F. Below oppA, the solid arrow represents DNA encoding the 40-kDa LacZ-OppA fusion.
FIG. 4.
Optimal alignment of the conserved signature sequence of members of the OppA family (Cluster 5) of bacterial SBPs (65). Residues identical for >75% of the sequences are outlined. Residues conserved as compared with those of E. coli OppA are shaded. The sequences are presented as follows: protein name:organism/residue numbers. ECOLI, E. coli; STRPN, Streptococcus pneumoniae; BACSU, Bacillus subtilis; SALTY, Salmonella typhimurium; HAEIN, H. influenzae; LACLA, Lactococcus lactis; STRLI, Streptomyces lividans; EFAEC, Enterococcus faecalis; BORBU, B. burgdorferi; TPAL, T. pallidum; TDENT, T. denticola.
Conservation and expression of oppA in oral treponemes.
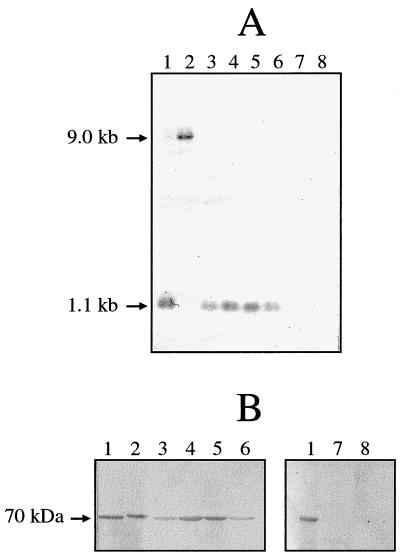
Strains representing the major serovars of T. denticola, as well as Treponema vincentii, Treponema pectinovorum, and Treponema socranskii were screened for oppA gene homologues and for proteins reactive with antibodies to OppA. The oppA gene was conserved in all T. denticola strains tested and in T. vincentii, but was not detected in T. pectinovorum and T. socranskii (Fig. 5A). In the strains in which oppA was detected, the oppA probe hybridized with a 1.1-kb HindIII fragment, except that in T. denticola ATCC 35404 the oppA band was 9 kb (Fig. 5A, lane 2). The OppA peptide was detected in all strains in which the gene was detected (Fig. 5B) and had a similar apparent molecular mass in all strains of T. denticola, as well as in T. vincentii. The OppA peptide of T. denticola ATCC 35404 had a slightly higher apparent molecular mass than that of the other OppA peptides (Fig. 5B, lane 2). No immunoreactive proteins were detected in either T. pectinovorum or T. socranskii.
FIG. 5.
Conservation of oppA in oral Treponema strains. (A) Southern blot of HindIII-digested chromosomal DNA probed with a 0.9-kb internal fragment of oppA. (B) Western blot of whole-cell extracts probed with anti-40 antibodies. Lanes: 1, T. denticola ATCC 35405; 2, T. denticola ATCC 35404; 3, T. denticola ATCC 33520; 4, T. vincentii LA-1; 5, T. denticola OTK; 6, T. denticola GM-1; 7, T. socranskii ATCC 35536; 8, T. pectinovorum ATCC 33768.
Mutagenesis of the opp locus.
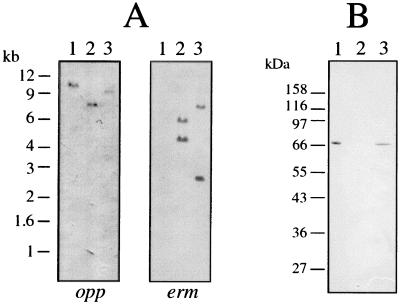
To gain information on the biological function of OppA, genetically defined allelic replacement mutations were constructed at two points in the opp locus and selected by resistance to erythromycin. Southern blot analysis, shown in Fig. 6A, confirmed the presence of the ermF/AM cassette within a single mutated copy of the target sequence in each strain. In strain OHE, the ermF/AM cassette was inserted at the HindIII site within oppA, while strain OXE was mutated at the XhoI site located in the 5′ end of oppF (Fig. 3). As shown in Western blots probed with anti-70 antibodies (Fig. 6B), strain OHE produced no OppA, while strain OXE appeared to produce normal levels of OppA. No differences were observed between parent and mutant strains when compared for ability to grow in NOS broth medium or in NOS supplemented with toxic peptide antibiotics, including triornithine, trilysine, bialafos, or aminopterin (data not shown). Other than erythromycin resistance in both mutant strains and the loss of OppA expression in strain OHE, no significant phenotypic differences between the parent and mutant strains were observed when assayed for surface-associated proteolytic activity, expression and assembly of Msp oligomers and CTLP protease complexes, or adherence and cytotoxicity toward epithelial cells (data not shown).
FIG. 6.
Confirmation of construction of allelic replacement mutants in the opp locus. (A) Southern blots of ScaI-digested T. denticola genomic DNA were probed with a biotinylated internal oppA fragment (opp) and the 2.1-kb ermF/AM cassette (erm). The ermF/AM cassette contains a single ScaI site. (B) Western immunoblot of T. denticola lysates probed with anti-70 antibodies. Lanes: 1, parent strain ATCC 35405; 2, oppA mutant strain OHE; 3, oppF mutant strain OXE.
Surface association of OppA.
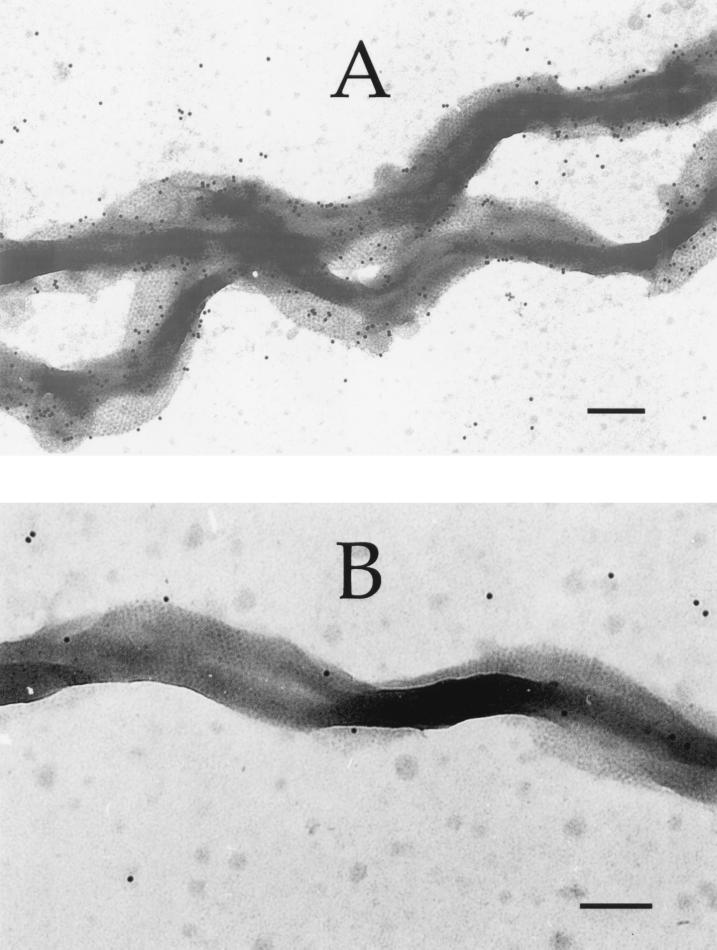
The deduced OppA peptide sequence is not predicted to be significantly hydrophobic (data not shown). However, the partitioning of OppA to the hydrophobic detergent phase of the Triton X-114 extract suggested that the mature protein is membrane associated, probably due to its predicted N-terminal lipid modification. While the nonacylated SBPs of gram-negative bacteria are localized to the periplasmic space, the acylated SBPs of gram-positive bacteria are anchored in the cell membrane. The cellular location of acylated spirochete SBPs has not been directly addressed. To study the cellular location and surface accessibility of OppA, whole spirochete cells were probed with either antibodies raised against the purified OppA (anti-70) or with preimmune rabbit antibodies and were visualized by immunogold electron microscopy (Fig. 7). The outer membrane hexagonal array formed by the Msp protein (20) is clearly visible in both micrographs, indicating that the cells were essentially intact. As shown in Fig. 7A, the anti-70 antibodies recognized epitopes in the spirochete outer membrane, while antibodies present in the preimmune rabbit serum did not bind to the cells (Fig. 7B). This suggested that OppA is accessible to extracellular proteins and could interact with host tissue components.
FIG. 7.
Immunogold electron micrograph showing localization of OppA to the cell surface of T. denticola organisms. Spirochete cells were probed with anti-70 antibodies (A) or preimmune rabbit serum (B), and bound antibodies were detected with gold conjugated anti-rabbit immunoglobulins. Bars, 300 nm.
Binding activity of OppA toward host proteins and cells.
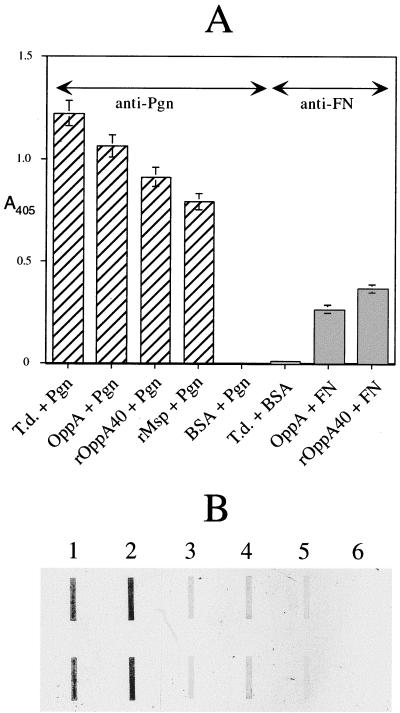
Since the 70-kDa protein was originally described as a FN-binding protein, we first assayed the ability of OppA to bind FN in an ELISA format. As shown in Fig. 8A, both native OppA and a 40-kDa LacZ fusion protein (rOppA40) immobilized in microtiter plate wells bound soluble FN. Anti-FN antibodies did not recognize T. denticola cells in this assay. In parallel assays shown in the same figure, soluble plasminogen bound to immobilized T. denticola cells, native OppA, rOppA40, and recombinant Msp (rMsp) (23). While there can be no direct comparison between the levels of FN binding and plasminogen binding from these experiments, these data suggest that both OppA and Msp are involved in the accretion of plasminogen to T. denticola cells. No binding of OppA to immobilized FN or plasminogen was detected (data not shown). In ligand blotting experiments, isogenic T. denticola strains OHE and MHE, deficient in OppA and Msp, respectively, bound less plasminogen than did the parent strain (Fig. 8B, lanes 2 to 4). Preincubation of plasminogen with the lysine analog EACA inhibited the binding of plasminogen to the parent strain (Fig. 8B, lanes 2 and 5) and to purified OppA (data not shown). Plasminogen binding to parent and mutant strains was only partially inhibited by specific antibodies against OppA and Msp (data not shown), suggesting that soluble host proteins bind to multiple receptors on the spirochete.
FIG. 8.
Binding of soluble host proteins to T. denticola cells and proteins. (A) ELISA format. FN, plasminogen (Pgn), or BSA in PBS were added to wells containing immobilized T. denticola ATCC 35405 cells (T.d.), native OppA (OppA), recombinant LacZ-OppA fusion protein (rOppA40), recombinant Msp (rMsp), or BSA. Bound FN and plasminogen were detected with anti-FN (shaded bars) and anti-plasminogen (hatched bars) antibodies, respectively. The y axis shows absorbance at 405 nm. Data shown represent the means ± standard deviations of at least two experiments using triplicate samples. (B) Ligand blot format. Duplicate samples of protein (1 μg) or cells (0.1 ml, OD600 = 0.2) applied to nitrocellulose membranes were probed with plasminogen (samples 2 to 4) or plasminogen plus EACA (sample 5). Bound plasminogen was detected with anti-plasminogen immunoglobulin G. Lanes: 1, Plasminogen; 2, 5, and 6, T. denticola ATCC 35405; 3, T. denticola OHE; 4, T. denticola MHE.
Previously characterized T. denticola outer membrane proteins with demonstrated binding activity were cytotoxic to cultured cells (21, 37, 47). In contrast, OppA was not cytotoxic to epithelial cells or fibroblasts, nor did it bind to glutaraldehyde-fixed epithelial cell monolayers, and anti-OppA antibodies did not significantly affect these processes (data not shown). The abilities of certain bacterial proteins to bind either (but not both) soluble or immobilized forms of host proteins is well documented (46, 54, 70) and appears to be due to conformation differences between soluble and immobilized polypeptides. Taken together, these suggest that OppA has a role in binding soluble peptides or proteins, but does not participate in the adherence of the spirochete to cell-bound receptors or the extracellular matrix.
DISCUSSION
Previous studies independently described a 70- or 72-kDa T. denticola surface-associated protein as a possible adhesin, based on its FN-binding activity (37, 67). We hypothesized that this protein might be involved in the interaction between the spirochete and host tissues in periodontal diseases. In the present study, we purified this protein from surface extracts of T. denticola ATCC 35405 cells and identified the genetic locus encoding it as a homologue of those encoding SBPs of oligopeptide transporters. Characterization of its binding activity suggested that T. denticola OppA has a role in binding soluble peptides or proteins, but does not participate in bacterial adherence to cell-bound receptors.
DNA encoding all but the amino portion of the 70-kDa protein was cloned and expressed in E. coli as a LacZ fusion protein, and the DNA sequence of the 5′ end of the gene was obtained by inverse PCR. The deduced peptide, designated OppA, is homologous to the substrate binding protein of oligopeptide permease systems found in many bacteria. Homologues of oppB, -C, -D, and -F are located directly downstream of oppA, suggesting that the complete permease system is present in T. denticola. Both native OppA and rOppA40 fusion protein bound soluble FN and plasminogen, which are abundant in gingival crevicular fluid (49), but neither OppA species bound to immobilized forms of these proteins. In contrast to other T. denticola surface-associated proteins that bind host proteins present in the subgingival environment (21, 37, 47), OppA was not cytotoxic to epithelial cells or fibroblasts nor did it adhere to glutaraldehyde-fixed epithelial monolayers. Similarly, antibodies specific for OppA did not significantly inhibit adherence or cytotoxicity of whole T. denticola to epithelial cells, suggesting that OppA does not interact significantly with cell-bound receptors.
In contrast to the heterogeneous Msp proteins of T. denticola strains (25), there is little evidence of variability in OppA or in the oppA gene. The oppA gene and the OppA peptide are highly conserved in representative strains of T. denticola and T. vincentii. Neither the OppA peptide nor the gene encoding it were detected in T. pectinovorum or T. socranskii. Interestingly, metabolic requirements of T. pectinovorum and T. socranskii are somewhat distinct from those of T. denticola and T. vincentii (26, 28, 53, 69) and neither possesses the high levels of cell surface peptidase activity characteristic of T. denticola and T. vincentii (53, 64). This suggests that OppA may be important for survival of T. denticola in the subgingival environment, perhaps functioning in the acquisition of peptide nutrients.
T. denticola requires peptide and amino acid nutrients (63) and is likely to possess several mechanisms for their acquisition. In other bacterial systems, oligopeptide uptake systems have been characterized by the ability to grow on minimal media supplemented with specific peptide nutrients. Alternatively, mutants with alterations in opp-related loci have been selected and characterized by their resistance to specific naturally occurring or synthetic toxic peptides that are transported by one or more oligopeptide permeases (2, 41, 57). While a complex defined growth medium for T. denticola has been described (71), in our hands, T. denticola required a medium containing yeast extract and serum. The viability and growth of oppA mutant strain OHE and oppF mutant OXE were not impaired under these conditions. Similarly, no differences in viability were observed between the parent strain and oppA mutant strain OHE grown in the presence of triornithine, trilysine, bialafos, or aminopterin. Taken together, these data suggest that, if OppA is involved in oligopeptide binding for nutrient uptake, it is part of a redundant system. These results may also be reflective of significant differences between in vitro culture conditions and the in vivo environment.
Native OppA preferentially segregates to the detergent phase of partitioned Triton X-114 extracts, indicating that the native protein is quite hydrophobic. While the hydropathy profile of the deduced OppA peptide did not indicate significant hydrophobicity (data not shown), sequence data support the inference that the mature protein is acylated at the amino terminus. This would account for the segregation of OppA to the hydrophobic phase of the extract and suggests that OppA is membrane bound rather than free in the periplasmic space. Studies to characterize the predicted acylation of OppA are in progress. While SBPs of gram-negative bacteria are generally characterized as nonacylated periplasmic proteins, T. denticola OppA and several other spirochete SBPs appear unique in this regard. With the exception of a Haemophilus influenzae hemin binding protein (39) that is not genetically linked to an ABC transporter (30), the only acylated SBPs identified in gram-negative bacteria are in spirochetes. The 38-kDa MglB lipoprotein of T. pallidum appears to be the SBP of an ABC transporter specific for glucose and galactose (6, 58). BPBP, a 70-kDa plasminogen binding lipoprotein of B. burgdorferi (44), has significant homology with T. denticola OppA (Fig. 4) as well as with at least two other Borrelia OppA lipoprotein homologues (45). The acylation of T. denticola OppA and its high homology with Bacillus SBPs (Fig. 4 and data not shown) are consistent with reports of other spirochete proteins whose closest homologues are in gram-positive bacteria and archaebacteria (34, 40) and lend support to earlier speculations on membrane transport architecture in spirochetes (6).
In immunogold electron microscopy of whole T. denticola cells, OppA appeared to be exposed on the surface of T. denticola cells. Anti-OppA gold beads were associated with the typically prominent outer membrane hexagonal array of T. denticola cells rather than with the protoplasmic cylinder. The hexagonal array ultrastructure is believed to be primarily comprised of the Msp protein (20, 25, 43, 51), and this feature is absent in isogenic msp mutants (24). A recent study questioning the outer membrane localization of Msp has been published, but the authors failed to detect the hexagonal array in the wild-type strain and the specificity of the anti-Msp antibodies used was not documented (8). Notwithstanding this single report, the present data suggest that OppA is bound to the outer membrane by its lipid moiety. These results are supportive of a recent model of spirochete outer membrane architecture that proposed that spirochete outer membrane-associated lipoproteins might have both transient periplasmic and cell surface localization (15). Both of the proposed functions of OppA (peptide binding for uptake and soluble host protein binding) are consistent with this model. Our cell localization results are also supported by the data on plasminogen binding by OppA. While further studies are required to determine the precise membrane localization of OppA and the extent to which OppA is exposed to the extracellular environment in vivo, the plasminogen binding activity of OppA is similar to that reported for BPBP, an OppA homologue in B. burgdorferi (44). In addition to their apparent surface localization and similar molecular weights, the deduced T. denticola OppA and B. burgdorferi BPBP peptides have identical predicted signal peptidase cleavage and acylation sites, possess similar signature sequences for Cluster 5 SBPs (Fig. 4), have the same carboxyl terminus dilysine motif, and are involved in binding plasminogen to the bacterial surface (44). Taken together, these observations may reflect the unique cell envelope architecture of spirochetes and are likely to be significant for the study of substrate binding and transport in these organisms, as compared with the classical model for gram-negative bacteria.
While no direct role for OppA in host tissue cytopathology is indicated, further studies are required to rule out a more subtle role in the interaction of the spirochete with host cells (29). For instance, data presented here could support a role for OppA in the evasion of the host immune response by mediating the accretion of soluble host proteins to the bacterial surface. While there is presently no consensus on this scenario as a virulence mechanism of pathogenic spirochetes (1, 5, 16, 36, 68), data presented here are consistent with recent studies of the role of borrelial plasminogen binding proteins. In B. burgdorferi, at least one surface-accessible OppA homologue binds plasminogen (44). Plasminogen bound to the spirochete surface appears to become activated to plasmin and is implicated in borrelial tissue invasion and extracellular matrix degradation (13, 14). Binding of soluble FN and plasminogen by T. denticola OppA, surface accessibility of OppA, and in vitro viability of the oppA mutant are consistent with a scenario that suggests a significant role for OppA in interaction with host tissues. Studies are in progress to further characterize host protein binding by T. denticola parent and oppA mutant strains in order to define more clearly the biological role of this molecule.
ACKNOWLEDGMENTS
We thank Andre Wong of the Faculty of Dentistry, University of British Columbia, for assistance in electron microscopy.
This study was supported by the Medical Research Council of Canada.
REFERENCES
- 1.Alderete J F, Baseman J B. Serum lipoprotein binding by Treponema pallidum: possible role for proteoglycans. Genitourin Med. 1989;65:177–182. doi: 10.1136/sti.65.3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alloing G, de Philip P, Claverys J P. Three highly homologous membrane-bound lipoproteins participate in oligopeptide transport by the Ami system of the gram-positive Streptococcus pneumoniae. J Mol Biol. 1994;241:44–58. doi: 10.1006/jmbi.1994.1472. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Wiley-Interscience; 1995. [Google Scholar]
- 5.Baughn R E. Role of fibronectin in the pathogenesis of syphilis. Rev Infect Dis. 1987;9(Suppl. 4):S372–S385. doi: 10.1093/clinids/9.supplement_4.s372. [DOI] [PubMed] [Google Scholar]
- 6.Becker P S, Akins D R, Radolf J D, Norgard M V. Similarity between the 38-kilodalton lipoprotein of Treponema pallidum and the glucose/galactose-binding (MglB) protein of Escherichia coli. Infect Immun. 1994;62:1381–1391. doi: 10.1128/iai.62.4.1381-1391.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunette D M, Melcher A H, Moe H K. Culture and origin of epithelium-like and fibroblast-like cells from porcine periodontal ligament explants and cell suspensions. Arch Oral Biol. 1976;21:393–400. doi: 10.1016/0003-9969(76)90001-7. [DOI] [PubMed] [Google Scholar]
- 8.Caimano M J, Bourell K W, Bannister T D, Cox D L, Radolf J D. The Treponema denticola major sheath protein is predominantly periplasmic and has only limited surface exposure. Infect Immun. 1999;67:4072–4083. doi: 10.1128/iai.67.8.4072-4083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan E C S, DeCiccio A, McLaughlin R, Klitorinos A, Siboo R. An inexpensive solid medium for obtaining colony-forming units of oral spirochetes. Oral Microbiol Immunol. 1997;12:372–376. doi: 10.1111/j.1399-302x.1997.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 10.Choi B K, Nattermann H, Grund S, Haider W, Gobel U B. Spirochetes from digital dermatitis lesions in cattle are closely related to treponemes associated with human periodontitis. Int J Syst Bacteriol. 1997;47:175–181. doi: 10.1099/00207713-47-1-175. [DOI] [PubMed] [Google Scholar]
- 11.Chu L, Ebersole J L, Kurzban G P, Holt S C. Cystalysin, a 46-kilodalton cysteine desulfhydrase from Treponema denticola, with hemolytic and hemoxidative activities. Infect Immun. 1997;65:3231–3238. doi: 10.1128/iai.65.8.3231-3238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu L, Song M, Holt S C. Effect of iron regulation on expression and hemin-binding function of outer-sheath proteins from Treponema denticola. Microb Pathog. 1994;16:321–335. doi: 10.1006/mpat.1994.1033. [DOI] [PubMed] [Google Scholar]
- 13.Coleman J L, Roemer E J, Benach J L. Plasmin-coated Borrelia burgdorferi degrades soluble and insoluble components of the mammalian extracellular matrix. Infect Immun. 1999;67:3929–3936. doi: 10.1128/iai.67.8.3929-3936.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman J L, Sellati T J, Testa J E, Kew R R, Furie M B, Benach J L. Borrelia burgdorferi binds plasminogen, resulting in enhanced penetration of endothelial monolayers. Infect Immun. 1995;63:2478–2484. doi: 10.1128/iai.63.7.2478-2484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox D L, Akins D R, Bourell K W, Lahdenne P, Norgard M V, Radolf J D. Limited surface exposure of Borrelia burgdorferi outer surface lipoproteins. Proc Natl Acad Sci USA. 1996;93:7973–7978. doi: 10.1073/pnas.93.15.7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox D L, Chang P, McDowall A W, Radolf J D. The outer membrane, not a coat of host proteins, limits antigenicity of virulent Treponema pallidum. Infect Immun. 1992;60:1076–1083. doi: 10.1128/iai.60.3.1076-1083.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham T M, Walker E M, Miller J N, Lovett M A. Selective release of the Treponema pallidum outer membrane and associated polypeptides with Triton X-114. J Bacteriol. 1988;170:5789–5796. doi: 10.1128/jb.170.12.5789-5796.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das S, Shraga D, Gannon C, Lam T T, Feng S, Brunet L R, Telford S R, Barthold S W, Flavell R A, Fikrig E. Characterization of a 30-kDa Borrelia burgdorferi substrate-binding protein homologue. Res Microbiol. 1996;147:739–751. doi: 10.1016/s0923-2508(97)85121-2. [DOI] [PubMed] [Google Scholar]
- 19.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egli C, Leung W K, Müller K H, Hancock R E, McBride B C. Pore-forming properties of the major 53-kilodalton surface antigen from the outer sheath of Treponema denticola. Infect Immun. 1993;61:1694–1699. doi: 10.1128/iai.61.5.1694-1699.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenno J C, Hannam P M, Leung W K, Tamura M, Uitto V-J, McBride B C. Cytopathic effects of the major surface protein (Msp) and the chymotrypsinlike protease (CTLP) of Treponema denticola. Infect Immun. 1998;66:1869–1877. doi: 10.1128/iai.66.5.1869-1877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenno J C, McBride B C. Virulence factors of oral treponemes. Anaerobe. 1998;4:1–17. doi: 10.1006/anae.1997.0131. [DOI] [PubMed] [Google Scholar]
- 23.Fenno J C, Müller K-H, McBride B C. Sequence analysis, expression and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola. J Bacteriol. 1996;178:2489–2496. doi: 10.1128/jb.178.9.2489-2497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fenno J C, Wong G W K, Hannam P M, McBride B C. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola. FEMS Microbiol Lett. 1998;163:209–215. doi: 10.1111/j.1574-6968.1998.tb13047.x. [DOI] [PubMed] [Google Scholar]
- 25.Fenno J C, Wong G W K, Hannam P M, Müller K-H, Leung W K, McBride B C. Conservation of msp, the gene encoding the major outer membrane protein of oral Treponema spp. J Bacteriol. 1997;179:1082–1089. doi: 10.1128/jb.179.4.1082-1089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiehn N E. Nutrient and environmental growth factors for nine oral small-sized spirochete strains containing one endoflagellum from each cell end. Acta Pathol Microbiol Immunol Scand Sect B. 1989;97:287–296. doi: 10.1111/j.1699-0463.1989.tb00790.x. [DOI] [PubMed] [Google Scholar]
- 27.Fiehn N E, Frandsen A. Evaluation of serum-containing substrates for cultivation of oral spirochetes. J Periodontal Res. 1984;19:61–66. doi: 10.1111/j.1600-0765.1984.tb01196.x. [DOI] [PubMed] [Google Scholar]
- 28.Fiehn N E, Westergaard J. Nutrient and environmental growth factors for eight small-sized oral spirochetes. Scand J Dent Res. 1986;94:208–218. doi: 10.1111/j.1600-0722.1986.tb01755.x. [DOI] [PubMed] [Google Scholar]
- 29.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L, Glodek A, Kelley J, Weidman J, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 31.Fletcher H M, Schenkein H A, Morgan R M, Bailey K A, Berry C R, Macrina F L. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun. 1995;63:1521–1528. doi: 10.1128/iai.63.4.1521-1528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature (London) 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 33.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Sandusky M, Weidman J, Smith H O, Venter J C. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 34.Ge Y, Charon N W. Molecular characterization of a flagellar/chemotaxis operon in the spirochete Borrelia burgdorferi. FEMS Microbiol Lett. 1997;153:425–431. doi: 10.1111/j.1574-6968.1997.tb12606.x. [DOI] [PubMed] [Google Scholar]
- 35.Grenier D, Uitto V-J. Cytotoxic effect of peptidoglycan from Treponema denticola. Microb Pathog. 1993;15:389–397. doi: 10.1006/mpat.1993.1088. [DOI] [PubMed] [Google Scholar]
- 36.Guner E S. Complement evasion by the Lyme disease spirochete Borrelia burgdorferi grown in host-derived tissue co-cultures: role of fibronectin in complement-resistance. Experientia. 1996;52:364–372. doi: 10.1007/BF01919542. [DOI] [PubMed] [Google Scholar]
- 37.Haapasalo M, Müller K-H, Uitto V-J, Leung W K, McBride B C. Characterization, cloning, and binding properties of the major 53-kilodalton Treponema denticola surface antigen. Infect Immun. 1992;60:2058–2065. doi: 10.1128/iai.60.5.2058-2065.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haapasalo M, Singh U, McBride B C, Uitto V-J. Sulfhydryl-dependent attachment of Treponema denticola to laminin and other proteins. Infect Immun. 1991;59:4230–4237. doi: 10.1128/iai.59.11.4230-4237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanson M S, Hansen E J. Molecular cloning, partial purification, and characterization of a haemin-binding lipoprotein from Haemophilus influenzae type b. Mol Microbiol. 1991;5:267–278. doi: 10.1111/j.1365-2958.1991.tb02107.x. [DOI] [PubMed] [Google Scholar]
- 40.Hardham J M, Stamm L V, Porcella S F, Frye J G, Barnes N Y, Howell J K, Mueller S L, Radolf J D, Weinstock G M, Norris S J. Identification and transcriptional analysis of a Treponema pallidum operon encoding a putative ABC transport system, an iron-activated repressor protein homolog, and a glycolytic pathway enzyme homolog. Gene. 1997;197:47–64. doi: 10.1016/s0378-1119(97)00234-5. [DOI] [PubMed] [Google Scholar]
- 41.Higgins C F, Gibson M M. Peptide transport in bacteria. Methods Enzymol. 1986;125:365–377. doi: 10.1016/s0076-6879(86)25031-4. [DOI] [PubMed] [Google Scholar]
- 42.Higgins C F, Hyde S C, Mimmack M M, Gileadi U, Gill D R, Gallagher M P. Binding protein-dependent transport systems. J Bioenerg Biomembr. 1990;22:571–592. doi: 10.1007/BF00762962. [DOI] [PubMed] [Google Scholar]
- 43.Holt S C, Bramanti T E. Factors in virulence expression and their role in periodontal disease pathogenesis. Crit Rev Oral Biol Med. 1991;2:177–281. doi: 10.1177/10454411910020020301. [DOI] [PubMed] [Google Scholar]
- 44.Hu L T, Pratt S D, Perides G, Katz L, Rogers R A, Klempner M S. Isolation, cloning, and expression of a 70-kilodalton plasminogen binding protein of Borrelia burgdorferi. Infect Immun. 1997;65:4989–4995. doi: 10.1128/iai.65.12.4989-4995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kornacki J A, Oliver D B. Lyme disease-causing Borrelia species encode multiple lipoproteins homologous to peptide-binding proteins of ABC-type transporters. Infect Immun. 1998;66:4115–4122. doi: 10.1128/iai.66.9.4115-4122.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuusela P, Vartio T, Vuento M, Myhre E B. Attachment of staphylococci and streptococci on fibronectin, fibronectin fragments, and fibrinogen bound to a solid phase. Infect Immun. 1985;50:77–81. doi: 10.1128/iai.50.1.77-81.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leung W K, Haapasalo M, Uitto V-J, Hannam P M, McBride B C. The surface proteinase of Treponema denticola may mediate attachment of the bacteria to epithelial cells. Anaerobe. 1996;2:39–46. [Google Scholar]
- 48.Li H, Ruby J, Charon N, Kuramitsu H. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J Bacteriol. 1996;178:3664–3667. doi: 10.1128/jb.178.12.3664-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loesche W J. Bacterial mediators in periodontal disease. Clin Infect Dis. 1993;16(Suppl. 4):S203–S210. doi: 10.1093/clinids/16.supplement_4.s203. [DOI] [PubMed] [Google Scholar]
- 50.Loesche W J, Syed S A, Laughon B E, Stoll J. The bacteriology of acute necrotizing ulcerative gingivitis. J Periodontol. 1982;53:223–230. doi: 10.1902/jop.1982.53.4.223. [DOI] [PubMed] [Google Scholar]
- 51.Masuda K, Kawata T. Isolation, properties, and reassembly of outer sheath carrying a polygonal array from an oral treponeme. J Bacteriol. 1982;150:1405–1413. doi: 10.1128/jb.150.3.1405-1413.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathers D A, Leung W K, Fenno J C, Hong Y, McBride B C. Major surface protein complex of Treponema denticola depolarizes and induces ion channels in HeLa cell membranes. Infect Immun. 1996;64:2904–2910. doi: 10.1128/iai.64.8.2904-2910.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mikx F H. Comparison of peptidase, glycosidase and esterase activities of oral and non-oral Treponema species. J Gen Microbiol. 1991;137(Pt. 1):63–68. doi: 10.1099/00221287-137-1-63. [DOI] [PubMed] [Google Scholar]
- 54.Mintz K P, Fives-Taylor P M. Binding of the periodontal pathogen Actinobacillus actinomycetyemcomitans to extracellular matrix proteins. Oral Microbiol Immunol. 1999;14:109–116. doi: 10.1034/j.1399-302x.1999.140206.x. [DOI] [PubMed] [Google Scholar]
- 55.Moore W E, Moore L H, Ranney R R, Smibert R M, Burmeister J A, Schenkein H A. The microflora of periodontal sites showing active destructive progression. J Clin Periodontol. 1991;18:729–739. doi: 10.1111/j.1600-051x.1991.tb00064.x. [DOI] [PubMed] [Google Scholar]
- 56.Ochman H, Medhora M M, Garza D, Hartl D L. Amplification of flanking sequences by inverse PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 219–227. [Google Scholar]
- 57.Perego M, Higgins C F, Pearce S R, Gallagher M P, Hoch J A. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol Microbiol. 1991;5:173–185. doi: 10.1111/j.1365-2958.1991.tb01838.x. [DOI] [PubMed] [Google Scholar]
- 58.Porcella S F, Popova T G, Hagman K E, Penn C W, Radolf J D, Norgard M V. A mgl-like operon in Treponema pallidum, the syphilis spirochete. Gene. 1996;177:115–121. doi: 10.1016/0378-1119(96)00286-7. [DOI] [PubMed] [Google Scholar]
- 59.Saier M H., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58:71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 61.Scott D, Chan E C, Siboo R. Iron acquisition by oral hemolytic spirochetes: isolation of a hemin-binding protein and identification of iron reductase activity. Can J Microbiol. 1996;42:1072–1079. doi: 10.1139/m96-137. [DOI] [PubMed] [Google Scholar]
- 62.Simonson L G, Goodman C H, Bial J J, Morton H E. Quantitative relationship of Treponema denticola to severity of periodontal disease. Infect Immun. 1988;56:726–728. doi: 10.1128/iai.56.4.726-728.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smibert R M. Order I. Spirochetales. In: Krieg N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 49–57. [Google Scholar]
- 64.Syed S A, Salvador S L, Loesche W J. Enzyme profiles of oral spirochetes in RapID-ANA system. J Clin Microbiol. 1988;26:2226–2228. doi: 10.1128/jcm.26.10.2226-2228.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tam R, Saier M H., Jr Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uitto V-J, Pan Y M, Leung W K, Larjava H, Ellen R P, Finlay B B, McBride B C. Cytopathic effects of Treponema denticola chymotrypsin-like proteinase on migrating and stratified epithelial cells. Infect Immun. 1995;63:3401–3410. doi: 10.1128/iai.63.9.3401-3410.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Umemoto T, Nakatani Y, Nakamura Y, Namikawa I. Fibronectin-binding proteins of a human oral spirochete Treponema denticola. Microbiol Immunol. 1993;37:75–78. doi: 10.1111/j.1348-0421.1993.tb03182.x. [DOI] [PubMed] [Google Scholar]
- 68.van der Sluis J J, Kant M, Onvlee P C, Stolz E. The inaccessibility of the outer membrane of adherent Treponema pallidum (Nichols strain) to anti-treponemal antibodies, a possible role of serum proteins. Genitourin Med. 1990;66:165–170. doi: 10.1136/sti.66.3.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weber F H, Canale-Parola E. Pectinolytic enzymes of oral spirochetes from humans. Appl Environ Microbiol. 1984;48:61–67. doi: 10.1128/aem.48.1.61-67.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Westerlund B, Korhonen T K. Bacterial proteins binding to the mammalian extracellular matrix. Mol Microbiol. 1993;9:687–694. doi: 10.1111/j.1365-2958.1993.tb01729.x. [DOI] [PubMed] [Google Scholar]
- 71.Wyss C. Growth of Porphyromonas gingivalis, Treponema denticola, T. pectinovorum, T. socranskii, and T. vincentii in a chemically defined medium. J Clin Microbiol. 1992;30:2225–2229. doi: 10.1128/jcm.30.9.2225-2229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]