Abstract
Effector proteins secreted by pathogens modulate various host cellular processes and help in bacterial pathogenesis. Some of these proteins, injected by enteric pathogens via Type Three Secretion System (T3SS) were grouped together based on a conserved signature motif (WxxxE) present in them. The presence of WxxxE motif is not limited to effectors released by enteric pathogens or the T3SS but has been detected in non-enteric pathogens, plant pathogens and in association with Type II and Type IV secretion systems. WxxxE effectors are involved in actin organization, inflammation regulation, vacuole or tubule formation, endolysosomal signaling regulation, tight junction disruption, and apoptosis. The WxxxE sequence has also been identified in TIR [Toll/interleukin-1 (IL-1) receptor] domains of bacteria and host. In the present review, we have focussed on the established and predicted functions of WxxxE effectors secreted by several pathogens, including enteric, non-enteric, and plant pathogens.
Keywords: WxxxE motif, effectors, pathogens, function, TIR domains
Introduction
Pathogenic bacteria have evolved various mechanisms to invade and survive within the host. One of these strategies is the injection of several bacterial effector proteins using diverse secretion systems (types I–IX) (Costa et al., 2015; Mak and Thurston, 2021; Rapisarda et al., 2018). These effector proteins allow the pathogen to survive and replicate within the host by disrupting various host cellular processes, rearranging the host cell morphology, modifying membrane and vesicular trafficking and evading the host immune response (Alto et al., 2006; Colonne et al., 2016; Cornelis, 2006; Galán, 2009; Weber and Faris, 2018). Many of the host cellular processes are mediated by low molecular weight GTPases, making them essential targets for bacterial effector proteins (Mattoo et al., 2007; Wennerberg et al., 2005). These small guanine nucleotide-binding G proteins are regulated alternately by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). During the presence of the extracellular stimulus, G-proteins are sequestered from the cytoplasm and localized to the membrane, where a molecular switching mechanism activates them. This process involves a conversion from inactive guanosine diphosphate (GDP) bound to active guanosine triphosphate (GTP) bound state through activation of the G protein by GEFs (Bos et al., 2007; Jaiswal et al., 2011). Once the stimulus is terminated, GAPs accelerate the GTPase reaction to hydrolyze GTP to an inactive GDP bound state (Vetter and Wittinghofer, 2001).
Some bacterial effectors target the host cytoskeleton G protein signaling cascades, and others bypass the endogenous GTPases to directly activate downstream signaling responses (Alto et al., 2006). The first group of bacterial effectors interacts directly with host GTPases, for example, RhoGTPases; where these effectors modulate the host responses either by modifying GDP/GTP cycling or acting directly on RhoGTPases, that could facilitate several processes involved in bacterial pathogenesis, such as induction of cytoskeletal rearrangements allowing entry of bacteria, modulation of membrane trafficking, and stimulation of cytokinesis (Hardt et al., 1998; Shao et al., 2002).
Previous studies had shown that pathogenic bacteria manipulate the host GTPase pathways for their own benefit by developing a unique subset of effector proteins exhibiting eukaryotic motif mimicry. This motif, first identified by Alto et al. in the effector proteins secreted by some enteric pathogens, is characterized by an invariant sequence that includes the tryptophan (W) and glutamate (E) residues known as the signature WxxxE motif (Alto et al., 2006). Following this discovery, these bacterial effectors were categorized into a single family of WxxxE effector proteins and, at the time, were believed to closely mimic low molecular weight GTPases to activate signaling cascades within host cells. Although WxxxE effectors were initially identified among enteric pathogens, recently, our group showed that the WxxxE effectors are distributed among enteric, non-enteric and plant pathogens. However, these effectors are absent in the commensals (Sayed et al., 2021). Further, studies examining the protein structure of the WxxxE containing effectors revealed GEF-like fold resembling that of Salmonella SopE effector protein despite sharing no sequence similarity (Huang et al., 2009). SopE had previously been documented as the first bacterial GEF mimic that induces membrane ruffling during bacterial invasion through activation of host GTP-binding proteins Cdc42 and Rac1 (Hardt et al., 1998). Subsequently, Huang et al. reported direct evidence of several WxxxE effector proteins such as Map, IpgB1, and IpgB2 to function as GEFs for Rho-GTPases (Huang et al., 2009). Collectively, the previous data revealed that various enteric pathogens utilize the bacterial GEF protein structure to trigger GTPase signaling cascades within host cells. Recently, we have shown the WxxxE effectors bind to host engulfment and cell motility protein 1 (ELMO1) that could modulate the host immune response and could explain how the host immune system discriminates between pathogenic and non-pathogenic bacteria (Sayed et al., 2021).
In the present review, we have focussed on bacterial effectors containing the WxxxE motif, their functions, and their roles in bacterial pathogenesis. We have also highlighted the impact of the interaction of these effectors with host proteins on the modulation of the host immune response against pathogens. Additionally, we have discussed the relevance of the WxxxE sequence identified in TIR domains of microbes.
Role of WxxxE containing effectors, secreted by enteric pathogens
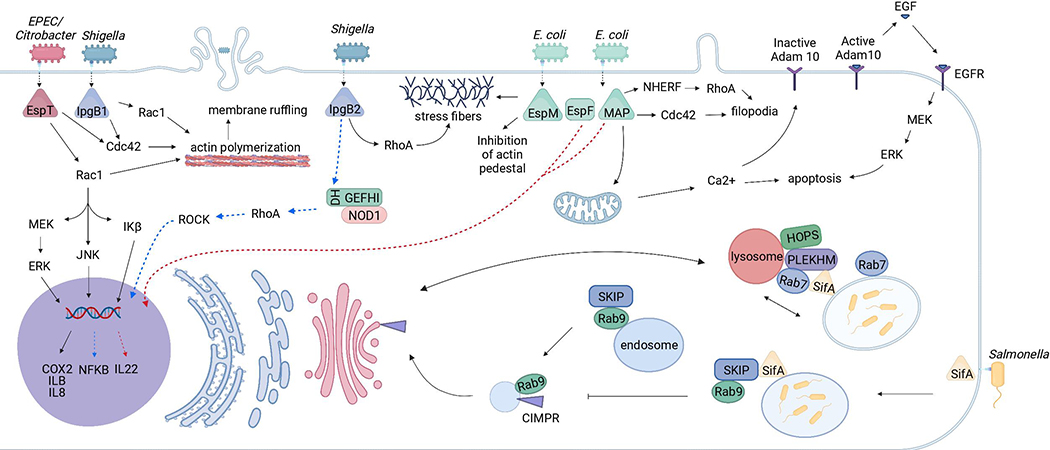
Although these effectors have a common WxxxE motif and several mimic GEF, depending upon the bacteria and their pathogenesis, these effectors exhibit different functions inside the host. The functions associated with different WxxxE effectors from various microbes are summarized in Figure 1 and Table 1.
Figure 1. Schematic representation of the effects of bacterial effectors with WxxxE motif on the host cellular pathways.
Enteric pathogens such as Shigella, EPEC/EHEC, Citrobacter rodentium and Salmonella inject WxxxE containing effectors such as IpgB1/IpgB2, EspT/Map/EspM/ SifA using the T3SS. IpgB1/IpgB2, EspT/Map/EspM/ activate Rho GTPases (either Rac1, RhoA, and or Cdc42), resulting in the formation of membrane ruffles/ lamellipodia/ filopodia/ stress fibres allowing the entry of these pathogens. Salmonella Typhimurium invades epithelial cells via SPI1- T3SS and resides inside the SCV. Effector SifA induces the formation of with Rab9 and interferes with the transport tubular structures called Sifs. SifA interacts with Rab7, HOPS, PLEKHM1, phagolysosome and LAMP1. SifA-SKIP complex interacts of MPR to the Golgi.
WxxxE containing effectors regulate the host inflammatory response; induce inflammatory responses such as COX-2, IL-8, IL-1β and PGE2 through Erk, JNK and NF-κB pathways.
Map induces mitochondrial dysfunction triggering a signalling cascade which results in apoptosis.
The same colors code indicates the effectors and the corresponding bacteria from which they are released. Dotted lines represent pathways that are not completely identified.
The pathways mentioned in the figure are adapted from (Berger et al., 2009; McGourty et al., 2012; Ramachandran et al., 2020; Raymond et al., 2011)
Table 1:
WxxxE effectors from enteric pathogens and their functions
| Bacterial effector | Bacteria species | Amino acid sequence | Function | Host target | References |
|---|---|---|---|---|---|
| SifA | Salmonella enterica serovar Typhimurium | TELRKGHLDGWKAQEKATYLAAKIQ | a) maintains the integrity of SCV. b) induces formation of SIFs c) recruits the host vesicle fusion machinery d) enhances the production of inflammatory innate cytokines |
a) binds with protein SKIP (PLEKHM2) and Rab9. b) interacts with HOPS (HOmotypic fusion and Protein Sorting), Rab7 c) interacts with ELMO1 |
(Beuzón et al., 2000; Boucrot et al., 2005; Dumont et al., 2010; Garcia-del Portillo et al., 1993; McEwan et al., 2015; Ohlson et al., 2008; Sindhwani et al., 2017) |
| Salmonella enterica serovar Typhi | SEWRKGNLDEWKTQEKATYLAAKIQ | ||||
| Salmonella enterica, strain “s3015” | SEWRKGILDEWKTQEKVTYLAAKIQ | ||||
| Yersinia frederiksenii, strain"22714/85" | NINQGDKFDMWKKEERTTYLSAVIN | ||||
| SifB | Salmonella enterica serovar Typhimurium, Salmonella enterica serovar Typhi and Salmonella enterica serovar Enteritidis | AMAEKGNLCDWKEQERKAAISSRIN | a) plays redundant function with other bacterial effectors | a) localizes to the SCV membrane LAMP1-positive structures. | (Freeman et al., 2003; Ohlson et al., 2008) |
| Map | E. coli* | RQS-TKDINGWIKDERIVYPSRVIN | a) induces actin-based filopodia b) regulates apoptosis, alters mitochondria morphology c) acts on intestinal mucosa-brush border remodeling, formation of attaching and effacing lesions, and regulation of ion channels d) mediates uptake of EPEC into non-phagocytic cells |
a) interacts with F-actin. b) interacts with GTPases (Cdc42) c) interacts with EbP50 (NHERF)1 and NHERF2 |
(Alto et al., 2006; Huang et al., 2009; Jepson et al., 2003; Martinez et al., 2010; Orchard et al., 2012; Shaw et al., 2005; Simpson et al., 2006) |
| E. coli EHEC O157:H7, strain “TW14359” | RQS-TKDIDEWIKDERIVYPSRVIN | ||||
| Citrobacter rodentium, strain“DBS100” | KQTGNGDTQQWFRQEQITFISKTVN | ||||
| E. coli EPEC, serotype“O127:H7” strain “E2348/69” | KQTGSSDTQQWFKQEQITFLSRAVN | ||||
| EHEC strain “95SF2” | KQTGSSDTQQWFKQEQITFLSRTVN | ||||
| E. coli | KQTRSGDTQQWFQQEQTTYISRTVN | ||||
|
E. coli EHEC O157:H7 strain “EC4196” |
KQTRNGDTQQWFQQEQTTYISRTVN | ||||
|
E. coli serotype“O125ac:H6” strain“aEPEC EC292/84” |
KQTRSGDTQQWFKQEQITYISRTVN | ||||
| EspT | E. coli serotype“O2:H49“ strain“FV11583”. | LKN-EGKKNEWMKEESICFVSRDVN | a) modulates the host cell cytoskeleton (formation of lamellipodia, membrane ruffles) b) plays a role in production of IL1β, IL8 and PGE2 |
a) interacts with Rac1, Cdc42 and activates Erk, JNK and NF-κB. | (Bulgin et al., 2009; Raymond et al., 2011) |
| Citrobacter rodentium | LKN-EGKMNEWMREECICFVSRDVN | ||||
| E. coli* | KQTRSGDTQQWFQQEQTTYISRTVN | ||||
| EspM | E. coli strain“700495” Citrobacter rodentium | RQS-TKDIDEWIKDERIVYPSRVIN | a) Remodels actin, b) Delocalizes tight junction (TJ). c) stabilizes focal adhesion formation |
a) interacts with RhoA and ROCK. b) phosphorylates colifin |
(Arbeloa et al., 2008; Arbeloa et al., 2010; Morita-Ishihara et al., 2013) |
| IpgB1 | Shigella flexneri serotype“5a”strain“M90T” | DSNSGNQLFCWMSQERTTYVSSMIN | a) Stimulates actin rearrangement. b) promotes bacteria uptake by non-phagocytic cells |
a) interacts with Rac1, Cdc42 b) activates ELMO1-Dock180 pathway c) interacts directly with acidic phospholipids |
(Alto et al., 2006; Costa and Lesser, 2014; Ohya et al., 2005; Weigele et al., 2017) |
| E. coli serotype“O26:H11” strain “DEC9A” | KQTRSGDTQQWFKQEQITYISRTVN | ||||
| Shigella flexneri plasmid“pINV_F6_M1382” | DSNSGNQLFCWMSQERTSYVSSMIN | ||||
| E. coli | DSNSGDQLFCWMSQERTSYVSSMIN | ||||
| IpgB2 | E. coli | RQS-TKDIHGWVSDERTVYPSRVIN | a) Stimulates actin rearrangement | a) interacts with Rac1, Cdc42 and RhoA BfpA b) activates NF-κB and requires NOD1 GEF-H1 and ROCK for activation |
(Alto et al., 2006; Fukazawa et al., 2008) |
| Shigella flexneri serotype 5a, str. M90T E. coli serotype“0144” strain “53638” | EQI-GENITDWKNDEKKVYVSRVVN | ||||
| TrcA | EPEC strain “B171-8” | RQN-TKDINGWIKDERIVYPSRVIN | a) Remodels actin | (Arbeloa et al., 2008) |
• : Bacteria serovar and/or serotype are not identified.
Organization of Actin
Actin filaments, along with intermediate filaments and microtubules, define the cellular cytoskeleton and play a key role in determining the shape as well as the organization of cell components. Hence, they serve as a target for several pathogenic bacteria. Bacterial effectors can modulate the architecture of the host cell and thus help in pathogenesis. Previous studies have shown that several WxxxE effectors such as Map, EspT, IpgB1, IpgB2, EspM1, EspM2, and EspM3 are involved in the subversion of cytoskeleton dynamics (Alto et al., 2006; Arbeloa et al., 2008; Bulgin et al., 2009). For instance, the secretion of IpgB1 by Shigella into epithelial cells induces membrane ruffles by activation of Rac1 and Cdc42 functions (Ohya et al., 2005). Handa et al. reported that IpgB1 interacts with the host engulfment protein ELMO1 and activates the ELMO–Dock180-Rac1 pathway promoting bacterial entry and invasion through induction of the membrane ruffling (Handa et al., 2007). Similarly, stimulation of Rac1 and Cdc42 by EspT secreted from Citrobacter rodentium has been shown to trigger lamellipodia formation on Swiss 3T3 cells and induce membrane ruffles on HeLa cells (Bulgin et al., 2009). Shigella also secretes IpgB2, which induces actin stress fibers in a mechanism similar to GTP-active RhoA (Alto et al., 2006). Enteropathogenic E.coli (EPEC) releases Map that triggers filopodia formation by the activation of Cdc42 (Berger et al., 2009). Map also interacts with host cell protein, NHERF (sodium/hydrogen exchanger regulatory factor-1), leading to recruitment of Ezrin and activation of RhoA-ROCK pathway, which stabilizes actin microfilaments (Berger et al., 2009). EPEC and Enterohemorrhagic E.coli (EHEC) secrete EspM effector proteins such as EspM1, EspM2 and EspM3, which induce stress fibers. EspM1 induces the formation of localized parallel stress fibers, whereas global parallel stress fibers and localized radial stress fibers are triggered by EspM2 and EspM3 respectively, through the RhoA-ROCK pathway (Arbeloa et al., 2008; Simovitch et al., 2010). EspM has also been demonstrated to inhibit the formation of actin pedestals formed by these pathogens (Arbeloa et al., 2008; Simovitch et al., 2010). Further, EspM2 has been shown to interact with another effector protein EspO1-2 which results in the regulation of EspM2 associated RhoA activity and stabilization of focal adhesion formation in EHEC infected cells (Morita-Ishihara et al., 2013).
Collectively, the previous findings showed that enteric pathogens utilize WxxxE containing bacterial effectors belonging to type III secretion system to facilitate their pathogenesis inside the host. These effectors activate different mediators (Rac1, Cdc42, RhoA, etc) and act on reorganization of actin elements to facilitate bacterial invasion and establish infection. Figure 1 summarizes the different pathways of enteric pathogens to invade host cells.
Regulation of inflammation
The effectors secreted by T3SS of pathogenic bacteria are known to modulate cell signaling pathways and disrupt host cell responses. There are very few studies that have investigated the role of WxxxE effectors with respect to inflammation signaling.
Shigella flexneri mutant strains lacking IpgB1 and IpgB2 induced pro-inflammatory response and response similar to wild-type strain, respectively, in the cornea of guinea pig (kerato-conjunctivitis model). However, a double inactivated ipgB1 ipgB2 mutant strains failed to induce inflammation and in the murine pulmonary shigellosis model, the inflammation score was low compared to the wild type (Hachani et al., 2008). However, the precise mechanism associated with the process has not been identified. Fukazawa et al demonstrated that IpgB2 activates Nuclear Factor kappa-light-chain-enhancer of activated B cells (NFκB) by a mechanism that requires guanine nucleotide exchange factor H1(GEF-H1) and NOD1, wherein RhoA mediated activation of ROCK was found to be essential for this activation (Fukazawa et al., 2008).
EspT secreted by Citrobacter rodentium has been shown to induce the production of inflammatory cytokines IL-8 and IL-1β as well as inflammatory mediators prostaglandin E2 (PGE2) and expression of cyclooxygenase-2 (COX-2) in U937 macrophages. The EspT induced cytokine release was mediated by NFκB, extracellular-signal-regulated kinase (Erk,1/2), and c-Jun N- terminus kinase (JNK) pathways (Raymond et al., 2011). Interestingly, it has also been noted that the WxxxE motif of EspT effector was essential for the release of pro-inflammatory cytokines and the substitution mutations at ‘W’ residue significantly decreased the level of IL-8 secretion (Raymond et al., 2011). Additionally, using in vivo mouse model of Citrobacter rodentium infection, Raymond and colleagues demonstrated that EspT stimulated the expression of inflammatory mediators such as CXCL-1 (also known as KC) and TNFα (Raymond et al., 2011). Moreover, a recent in vivo study using Citrobacter rodentium has revealed that T3SS effectors act as a network or in combination. WxxxE effectors such as Map have been shown to act in combination with other effectors and affect the secretion of GM-CSF and IL-6 from immune cells. Besides, Map and EspF have been shown to influence the IL-22 response (Ruano-Gallego et al., 2021).
In the case of Salmonella Typhimurium pathogenesis, the WxxxE effector SifA contributes to T3SS1 (SPI-1-encoded T3SS effectors) independent inflammation in streptomycin pretreated murine model of colitis. In the previous model, mice pretreated with streptomycin become more susceptible to oral infection with Salmonella enterica serovars Typhimurium and Enteritidis due to the elimination of commensal intestinal bacteria by the effect of the antibiotic (Barthel et al., 2003). In addition, this model can be used to study colitis (intestinal inflammation) associated with Typhimurium infection (Barthel et al., 2003). Using this model, Mastuda et al. showed thar the T3SS1 deficient strain is incapable of translocating T3SS1 effectors however they still invade the intestinal cells and induce inflammation (Matsuda et al., 2019). T3SS2 effectors (SPI-2-encoded T3SS effectors), including SifA, SpvB, SseF, SseJ and SteA collectively contribute to the T3SS1 independent inflammatory changes. Although these five effectors were required for induction of cytotoxicity in RAW macrophages in vitro, SifA was identified as most important for induction of inflammation in vivo (Matsuda et al., 2019). However, the mechanistic studies determining the role of SifA in inflammation are lacking.
Recently, we have shown SifA interacts with the C terminus end of ELMO1 and WxxxE motif of SifA is specifically required for the interaction (Sayed et al., 2021). ELMO1-SifA interaction influences the colonization and dissemination of Salmonella Typhimurium as well as the production of inflammatory cytokines such as TNFα, IL-1β, MCP-1, CXCL-1, and IL-6 in vivo. In a parallel line, ELMO1 has also been shown to interact with WxxxE effectors IpgB1, IpgB2, and Map, stimulating higher inflammatory responses upon challenge with microbes or microbial ligands (Sayed et al., 2021). Collectively, our findings show that the WxxxE containing effectors stimulate the host inflammatory responses (Sayed et al., 2021). The induction of proinflammatory cytokines could impact bacterial load and dissemination, therefore the WxxxE containing effectors affect the microbial pathogenesis and enteric diseases. The presence of WxxxE signature motif in enteric bacterial proteins but not in the commensals (Sayed et al., 2021) could explain the differential immune response of host following interaction with pathogenic vs non-pathogenic bacteria. We have previously shown that ELMO1 interacts with WxxxE effectors of enteric bacteria, generates MCP-1 in epithelial cells following infection and recruits monocytes and increases the TNF-α level (Sayed et al., 2020). This pathway could be helpful in the detection of the early biomarker of inflammatory bowel disease. Other than our published work, till date, the effect of WxxxE containing effectors on the adaptive immune response is not studied. This area of host immune responses would be useful in our understanding of the initiation of infection-associated inflammatory diseases.
Formation of vacuoles or tubules
Some pathogens, such as Salmonella, invade cells, then reside and replicate in a membrane-bound compartment called Salmonella Containing Vacuoles (SCV) (Garcia-del Portillo et al., 1993). These vacuoles initially acquire markers of early endosomes, which then mature to late SCV by dropping off early markers and then acquiring markers of late endosomes as well as lysosomes (Méresse et al., 1999; Steele-Mortimer et al., 1999). However, they have lesser lysosomal hydrolases compared to lysosomes (Drecktrah et al., 2007; McGourty et al., 2012; Méresse et al., 1999). Salmonella replicates within the SCV through the release of effectors, such as SifA, by the T3SS2. Many of these effectors are involved in the formation of tubular structures called Sifs (Salmonella-induced filaments). Sifs are tubules extended outwards from the SCV and characterized by the presence of the host lysosome-associated membrane protein-1 (LAMP1) within their membranes (Garcia-del Portillo et al., 1993; Krieger et al., 2014; Leung and Finlay, 1991). SifA promotes the Sif formation through the fusion between late endocytic compartments and the SCV and induce LAMP1-positive tubules (Brumell et al., 2001). A recent study revealed that among seven T3SS2 effectors (SifA, SopD2, PipB2, SteA, SseJ, SseF, and SseG), SifA is sufficient to induce LAMP1-positive tubule formation on its own, and multiple T3SS2 effectors are required for efficient formation and extension of LAMP1-positive tubule (Knuff-Janzen et al., 2020). The C-terminus of SifA interacts with GTPase Rab7, components of homotypic fusion and vacuole protein sorting (HOPS) complex and Pleckstrin homology domain-containing family M member 1 (PLEKHM1), which amplifies GTP loading of Rab7. This complex recruits phagolysosomal membranes and LAMP1 for SCV growth (McEwan et al., 2015; Zhao et al., 2015).
Regulation of Endolysosomal signaling
Kinesin-1, a microtubule motor protein, is involved in the transportation of cargoes towards the plus end of microtubules (Dietrich, K.A. et al., 2008). SifA interacts with the host protein SKIP (SifA and kinesin interacting protein), which controls the level of kinesin-1 on the SCV (Boucrot et al., 2005). Biochemical and crystallographic studies have revealed that the N- terminus domain of SifA interacts with the pleckstrin homology (PH) domain of SKIP, which then modulates the mobilization of kinesin-1 (Diacovich et al., 2009). The WxxxE motif of SifA is essential for maintaining the tertiary structure of SifA, which is crucial to the interaction with the SKIP protein (Diacovich et al., 2009). The tetratricopeptide repeat (TPR) domain of the kinesin interacts with the C- terminus domain of SKIP, RUN (RPIP8, UNC- 14 and NESCA) and triggers the microtubule and kinesin-1dependent anterograde movement of late endosomal/ lysosomal compartments (Dumont et al., 2010). The SifA/SKIP complex was found to be essential either for the formation or the anterograde transport of kinesin-1-enriched vesicles. (Dumont et al., 2010). SifA and SKIP interactions also influence the recruitment of LAMP1 and its distribution in the perinuclear location. SifA functions as an antagonist to Rab9 by preventing SKIP: Rab9 interaction by binding to the PH domain of SKIP. The interaction of SifA and SKIP is stronger than Rab9 interaction allowing SifA to bring SKIP to SCV and interestingly, the W197 and E201 residues of the WxxxE are required for the binding of SifA with SKIP (Jackson et al., 2008).
Salmonella containing vesicles have reduced levels of hydrolytic enzymes and Mannose Phosphate Receptors (MPR) that are responsible for the transport of these hydrolases. MPRs act as a shuttle between Trans-Golgi Network (TGN) and endosomes to deliver the new synthesized lysosomal enzymes to endosomes. Acidification of endosomes results in dissociation of the hydrolases from MPR and the empty MPR are recycled back to TGN for the next cycle of transport. Interestingly, Rab9 is involved in late endosome- TGN transport of MPRs; further, it also facilitates the tethering of MPR vesicles and its fusion at TGN by binding to the Golgi-associated protein GCC185 (McGourty et al., 2012). SifA has been shown to have a role in the subversion of this trafficking by binding to SKIP (also known as PLEKHM2) (McGourty et al., 2012). SifA forms a stable complex with SKIP and Rab9 in Salmonella-infected cells. SifA and SKIP form a sink for the sequestration of Rab9, resulting in inhibition of MPR transport and lysosome function (McGourty et al., 2012).
Disruption of tight junction
Tight junctions (TJ) are adhesion complexes by which cells adhere to each other and regulate permeability (Zihni et al., 2016). They are comprised of transmembrane proteins such as claudins, occludin, tricellulin, etc., and cytoplasmic plaque such as zonula occludens (ZO) proteins, kinases, phosphatases, GTPases, exchange factors, etc. (Singh et al., 2018). These cytoplasmic plaques are linked to the C terminal region of transmembrane proteins on one end and the other end is linked to actin (Singh et al., 2018; Zihni et al., 2016). Intestinal pathogens are known to disrupt TJs and increase the permeability of intestinal epithelium (Singh et al., 2018). Shigella interact with intestinal epithelial cells and deplete claudin1 as well as modulate the expression of ZO1, ZO2, cadherin and dephosphorylate occludin (Hachani et al., 2008). IpgB1 and IpgB2 have been predicted to have a role in disturbing TJ by unidentified signaling pathways due to their Rac and Rho mimicking ability, respectively (Hachani et al., 2008). Map secreted by EPEC affects the junctional assembly of TJ proteins by preventing their recruitment to the complex and disrupting the TJ. Map reduces the expression of claudin-1 and decreases the level of claudin-4, and occludin by lysosomal degradation (Singh et al., 2018). Map also disrupts the intestine’s barrier function and may contribute to diarrhea (Dean and Kenny, 2004). A previous study has reported that EspM2 secreted by EPEC modulates the mislocalization of ZO1 towards the basal end of polarized epithelial monolayers. However, in this case, the functioning of TJ was found to be unaffected (Simovitch et al., 2010).
Apoptosis
Map secreted from EPEC plays a role in bacterial pathogenesis through stimulation of host cell apoptosis. Map targets host mitochondria lead to disruption of the membrane and increase the Ca2+ efflux, which activates ADAM10 sheddase and release of epidermal growth factors triggering the ERK and p38 MAPK signaling cascades leading to cell apoptosis (Ramachandran et al., 2020).
WxxxE effectors/ virulence proteins identified in non-enteric pathogens
Although the WxxxE motif has been initially identified in the T3SS bacterial effector proteins belonging to a group of enteric pathogens, we have found the presence of this motif in effectors/ proteins associated with type II and type IV secretion systems of non-enteric pathogens (Table 2).
Table 2:
WxxxE effectors/ virulence proteins from non-enteric pathogens and their functions
| Bacteria Effector | Bacteria Species | Secretion System | Amino Acid Sequence | Function | Host Target | References |
|---|---|---|---|---|---|---|
| PtlH | Bordetella pertussis | Type IV | GQLWYEDRNGWNRQESGALTLDHLM | a) acts as signalling protein for a gate/channel b) provides energy for Pertussis toxin translocation |
a) interacts with proteins involved in the transport process. | (Craig-Mylius and Weiss, 1999; Farizo et al., 1996; Kotob and Burns, 1997; Verma and Burns, 2007; Williams et al., 2016) |
| Bordetella bronchiseptica | ||||||
| BepF | Bartonella henselae | Type IV | ENPALGEQLSWEVSEHPKSISKLAGKK | a) triggers F-actin rearrangements and bacterial uptake | a) interferes with Cdc42 and Rac1 signalling. | (Truttmann et al., 2011a) |
| DrrA | Legionella pneumophila | Type IV | RENEGNEVSPWQEWENGLRQIYKEM | a) helps in the formation LCV b) localizes to the LCV membrane, recruits Rab1 and regulates transport of ER-derived vesicles |
a) activates Rab1 | (Müller et al., 2010; Murata et al., 2006; Tan and Luo, 2011) |
| PulA | Klebsiella pneumoniae | Type II | AHWVDKTTLLWPGGENKPIVRLYYSH | a) plays a role in immune evasion b) helps in bacterial survival in the lung |
a) interacts with glycan on the host epithelium b) limits the activation of the TLR4-TLR2-MyD88 pathway c) interacts with alveolar macrophages and neutrophils |
(Pugsley et al., 1990; Tomás et al., 2015) |
| SGLP | Mycoplasma | GKSNINDAIKWVLGEQSSKSLRGDNM | a) plays a role in tumor cell migration and proliferation b) decreases in stress fibers and increases in filopodia and lamellipodia |
a) activates Rac1 and phosporylates Stat3 | (Hu et al., 2014) |
BepF is one of the seven effector proteins secreted through VirB/VirD4 (Type IV secretion system) by Bartonella henselae, a zoonotic pathogen (Schmid et al., 2004; Truttmann et al., 2011b). BepF along with BepC is known to trigger F-actin rearrangements leading to invasome formation and uptake of Bartonella (Truttmann et al., 2011b). BepF possesses three BID (Bartonella intracellular delivery) domains among them BIDF1 and BIDF2 was found to be sufficient for inducing invasome formation (Truttmann et al., 2011a). WxxxE motif has been identified in BIDF1 domain and mutation at ‘W’ residue was found to affect the invasome formation. Although BepF is speculated to perturb Cdc42 and Rac1 signaling, it has less similarity to the WxxxE-GEF mimics identified, and hence the mechanism of action may differ (Truttmann et al., 2011a).
DrrA (also known as SidM) is an effector protein secreted by Legionella using Dot/Icm (Type IV secretion system). DrrA has three functional domains i) C- terminus lipid phosphatidylinositol-4-phosphate binding domain (P4M) responsible for membrane attachment ii) guanine nucleotide exchange factor (GEF) domain for Rab1 activation, and iii) N- terminus adenylyltransferase (ATase) (Müller et al., 2010). Legionella replicates within specialized vacuoles called Legionella-containing vacuole (LCV). DrrA helps in the formation of this LCV by remodeling vesicles derived from the endoplasmic reticulum (ER) and prevents its fusion with lysosomes (Goody et al., 2011; Hardiman and Roy, 2014). DrrA localizes to the LCV membrane and then recruits Rab1, which is involved in the regulation of transport of ER-derived vesicles (Goody et al., 2011). Importantly, the WxxxE motif is also identified in DrrA effector (Sayed et al., 2021).
PtlH is one among the accessory proteins of Ptl (a member of the Type IV secretion system) which is required for the secretion of pertusis toxin from Bordetella pertussis (Kotob and Burns, 1997). PtlH has a nucleotide-binding motif and interacts with proteins involved in the transport process (Kotob and Burns, 1997; Verma and Burns, 2007). Although the exact role of PtlH is not known, it is speculated to provide energy for the toxin translocation by ATP hydrolysis or may act as a signaling component responsible for the opening of gate or channel due to its kinase activity (Kotob and Burns, 1997; Verma and Burns, 2007). Our recent study reported the presence of the WxxxE sequence in the PtlH proteins (Sayed et al., 2021).
PulA (Pullulanase) is a lipoprotein secreted to the cell surface by Klebsiella through the PulA secretion (Type II secretion system) (East et al., 2016). PulA plays a role in immune evasion by perturbing the TLR dependent detection of K. pneumoniae (Tomás et al., 2015). The WxxxE motif is also recorded in PulA from K. pneumoniae (Sayed et al., 2021).
The secretion systems require NTPases, mainly ATPases, as a part of its assembly to energize the transfer of substrates/molecules to the host cell. VirB 11 is an example of ATPases associated with the Type IV secretion system (Savvides et al., 2003). Interestingly, the WxxxE sequence has been identified in VirB11 present in Acinetobacter baumanni and Pseudomonas aeruginosa (Sayed et al., 2021). The functional relevance of the WxxxE motif present in the above-mentioned proteins (PtlH, DrrA, PulA, VirB 11) need further experimentation.
Small GTPase-like protein (SGLP) fragment identified in the chromosome partition protein, Smc of Mycoplasma pulmonis has been reported to possess WxxxE motif (Hu et al., 2014). SGLP has been shown to induce activation of Rac1 and phosphorylation of Stat3 and mutations at the tryptophan and glutamate residues of WxxxE motif resulted in loss of activation. SGLP and its homologues are considered as virulence factors and SGLP-transduced HeLa cells demonstrated a decrease in stress fibers and an increase in filopodia and lamellipodia. It is also involved in tumor cell migration and proliferation (Hu et al., 2014).
WxxxE effectors/ virulence proteins identified in plant pathogens
Various plant pathogens also use the secretion systems to inject effector proteins to suppress the plant immune system. AvrE is one such superfamily of effectors secreted by T3SS and present in various genera of plant pathogens (Ham et al., 2009). The presence of WxxxE motifs was recorded in several of the AvrE effectors (Table 3). Interestingly, two WxxxE motifs have been reported in AvrE effectors secreted by pathogens such as Pantoea stewartii subsp. stewartii, E. amylovora, E. pyrifoliae, P. syringae pv. tomato, P. syringae pv. phaseolicola 1448A, P. viridiflava and P. cichorii whereas only one motif has been identified in effectors of Pantoea agglomerans pv. gypsophilae, P. syringae pv. syringae B728a, and P. fluorescens SBW45 (Ham et al., 2009). Mutation of ‘W’ residue at the WxxxE motif of WtsE, Avr effector secreted by Pa. stewartii resulted in altered functioning of the protein. However, their role as GEF mimics has yet to be ascertained (Ham et al., 2009).
Table 3:
WxxxE effectors/ virulence proteins from Plant pathogens and their functions
| Bacterial Effector | Bacterial Species | Secretion System | Amino Acid Sequence | Function | Host Target | References |
|---|---|---|---|---|---|---|
| WtsE | Pantoea stewartii subsp. Stewartii | Type III | VHYFDQLTRGWTEAEAGCQQLKKGL | a) disrupts sphingolipid biosynthesis | a) mimics activated G-proteins b) targets PP2A via direct interaction with B’ regulatory subunits c) interacts with LRR-RLK proteins |
(Ham et al., 2009; Jin et al., 2016; Siamer et al., 2014) |
| AvrE1 | Pseudomonas syringae | Type III | LYQFDPISTRWKIPEGLEDTAFNSL KGLMQLKAGQWQRFEQRPVEENPRW |
a) disrupts sphingolipid biosynthesis b) promotes bacterial growth and/or suppresses callose deposition |
a) mimics activated G-proteins b) interacts with multiple PP2A B’ subunits (The same target for WtsE) |
(Ham et al,. 2009, Jin et al,. 2016) |
| DspA | Erwinia amylovora | Type III | LHYFDQLTKGWTGAESDCKQLKKGL | a) suppresses plant defence responses and promotes bacterial growth | a) de-phosphorylates Orm proteins by activating PP2A regulatory subunit CDC55 b) interacts with LRR-RLK proteins |
(Jin et al., 2016; Meng et al., 2006; Siamer et al., 2013; Siamer et al., 2014) |
| DspE | Erwinia amylovora | Type III | KNAAYATQHGWQGREGLKPLYEMQG | a) suppresses plant defence responses and promotes bacterial growth | a) de-phosphorylates Orm proteins by Activating PP2A regulatory subunit CDC55. b) interacts with LRR-RLK proteins |
(Jin et al,. 2016, Siamer et al,. 2013, Siamer et al,. 2014, Meng et al,. 2006) |
| RopE | Pseudomonas fluorescens | Type III | WQGNTAIAQSWRKVELPDRQPLESL | helps in pathogenesis | unknown | (Preston et al., 2001) |
| VirB11 | Agrobacterium tumefaciens | Type IV | GQVLTEGPGGWRTYEMPELTFEKLM | a) promotes pilus polymerization by regulating VirB4 b) modulates VirB4 pilin dislocase activity c) interacts with coupling protein VirD4 to proceed with nucleoprotein substrate transport |
a) acts as a traffic ATPase | (Atmakuri et al., 2004; Ripoll-Rozada et al., 2013) |
Jin et al., revealed that AvrE-type effectors target protein phosphatase 2A (PP2A). They demonstrated that WtsE from Pantoea stewartii subsp. stewartii and AvrE1 from Pseudomonas syringae interact with B’ regulatory subunit of (PP2A) of their respective hosts (Jin et al., 2016). AvrE-type effectors DspA/E and WtsE both hindered sphingolipid biosynthesis in yeast cells by exhausting precursor molecules in this pathway. They also found that DspA/E interacts with the PP2A B regulatory subunit, Cdc55, in yeast which disrupts the ORM(for orsomucoid like proteins)1/2 proteins involved in the rate-limiting step of sphingolipid biosynthesis (Jin et al., 2016). Importantly, the effector-mediated inhibition of sphingolipid metabolism in plants affects vesicular trafficking and may lead to cell death (Chen et al., 2006; Dietrich et al., 2008; Markham et al., 2011). AvrE- type effectors also target other molecules such as leucine-rich repeat receptor-like kinases (LRR-RLKs) involved in triggering plant defense responses (Jin et al., 2016).
VirB11, is believed to be a traffic ATPase that promotes pilus polymerization through regulation of VirB4 as it disrupts pilin subunits from the inner membrane to the periplasmic region (Ripoll-Rozada et al., 2013). It was also reported that VirB11 is involved in nucleoprotein transfer by interacting with the VirD4 coupling protein (Ripoll-Rozada et al., 2013). Ultimately, VirB11 acts as a switch between substrate transport and pilus biogenesis in which VirD4, VirB11 and VirB4 interact with one another to promote substrate transfer through an ATP-dependent and independent mechanism (Atmakuri et al., 2004; Ripoll-Rozada et al., 2013). We have identified WxxxE motif in the type IV assembly protein VirB11 from the plant pathogen A. tumefaciens.
The WxxxE motif is present in the TIR-domain of several eukaryotes and bacteria
WxxxE motif has been identified in TIR (Toll/interleukin-1 receptor (IL1R)/resistance protein) domains of several eukaryotes and prokaryotes. The common example of TIR domains in eukaryotes are TLR proteins and interestingly WxxxE is present in TLR1-10 (Felix et al., 2014; Sayed et al., 2021). TIR domain is also present in positive regulators of TIR-adaptors - TRAM (TRIF-related adaptor molecule), MyD88 (Myeloid differentiation primary response gene 88), TIRAP also known as Mal (TIR domain-containing adaptor protein), TRIF (TIR domain-containing adapter-inducing interferon-β); as well as in negative regulator of TIR adaptor protein - sterile and HEAT-Armadillo motifs containing protein SARM (O’Neill and Bowie, 2007). Among all 5 adapters, WxxxE is present only in SARM but not in TIRAP, TRIF, TRAM and MyD88 (Felix et al., 2014; Zhang et al., 2011).
Several investigators have shown that TIR domain is important as a scaffold promoting assembly of signaling complexes via protein-protein interactions. For example, TLR signaling is triggered by dimerization of the cytoplasmic Toll/interleukin receptor (TIR) domains allowing protein-protein interactions between the TLRs and signal transduction molecules (Chan et al., 2009; Jang and Park, 2014). Toshchakov et al applied Bayesian partitioning with pattern selection (BPPS) and classified TIR domains in metazoan, higher plant, archaeal, protozoan, chlorophytan, and in bacterial proteins (Toshchakov and Neuwald, 2020). Although TIR domains have been widely studied in human immune responses, these proteins in bacteria are probably involved in subversion of the vertebrate immune system (Spear et al., 2009).
An additional new role of TIR domains as catalytic enzymes has been established with the discovery of NAD+-nucleosidase (NADase) activity by several TIR domain containing proteins. The NADase activity was first detected in mammalian TIR protein SARM1, involved in axonal degeneration (Essuman et al., 2017). SARM1 plays a key role in elimination of the damaged/unhealthy axons by accelerating the depletion of NAD+ (Essuman et al., 2017; Horsefield et al., 2019). Substitution mutation in the glutamate residue to alanine in the WxxxE motif of SARM1 resulted in abolishing the NADase of SARM1 (Essuman et al., 2017; Horsefield et al., 2019). TIR domains are also present in plant nucleotide-binding leucine rich repeat immune receptors (Wan et al., 2019). Homology studies with SARM1 TIR domain identified several TIR domain in plants with catalytic glutamate and neighboring residues positionally conserved (Wan et al., 2019). These receptors are known to promote cell death and confer disease resistance. Substitution mutations of glutamate to alanine in immune receptor RBA1 (E86A), and RPS4 (E88A), RPPI (E164A) in Arabidopsis, BdTIR (E127A) in Brachypodium distachyon, L6(E135A) in Linum usitatissimum and RUN1(E100A) in Muscadinia rotundifolia resulted in the loss of NADase activity and its potential to induce cell death (Horsefield et al., 2019; Wan et al., 2019). Structural studies of TIR domain in RUN1 demonstrated that tryptophan(W96) residue is involved in a face to face stacking arrangement with adenosine group of NADP+. Mutations of W96A also resulted in loss of NAD+ cleavage activity in RUN1(Horsefield et al., 2019). A recent study by Toshchakov and Neuwald confirmed the glutamate residue responsible for NADase activity is conserved in 70% of TIR domains (Toshchakov and Neuwald, 2020).
Phylogenetic studies with the TIR domain of SARM1 found the related bacterial proteins with TIR domains (Spear et al., 2009; Zhang et al., 2011). These TIR domain containing proteins of pathogens for example Staphylococcus aureus (TirS), uropathogenic E. coli (TcpC), Brucella abortus (BtpA and BtpB) and Acinetobacter baumannii (AbTir), non-pathogens for example, Paracoccus dentrificans (PdTir) and Actinoplanes (ApTir) species, and archaea such as Theionarchaea archaeon (TcpA) and Methanobrevibacter olleyae (TcpO) have NAD hydrolase (NADase) activity. The pathogenic bacteria can reduce the levels of NAD+ and NADP+ inside the host and increases pathogenicity (Coronas-Serna et al., 2020; Essuman et al., 2018). The NADase activity of TIR domains present in non-pathogenic bacteria suggests its function in the regulation of metabolic pathways (Essuman et al., 2018). TIR domain containing bacterial proteins are also involved in a bacterial anti-phage defence system or Thoeris with ThsA and ThsB(Doron et al., 2018). Recently Ofir et al. showed that infection of phage generates cyclic ADP-ribose by the TIR-domain protein ThsB, that activates ThsA. ThsA depletes nicotinamide adenine dinucleotide and terminates phage infection(Ofir et al., 2021). We did not notice any WxxxE motif in ThsA and ThsB proteins.
Several studies have demonstrated NADase activity of TIR domains where the glutamate-E residue is conserved and mutation of glutamate can abolish the NADase activity (Coronas-Serna et al., 2020; Essuman et al., 2018; Horsefield et al., 2019; Toshchakov and Neuwald, 2020; Wan et al., 2019). Though the authors did not specify the presence of WxxxE motif but interestingly the tryptophan position is also conserved. It is possible that WxxxE motif of these TIR proteins in general have a role in nucleotide binding and NADase activity associated with the TIR domains. It needs further study. The universal presence of these TIR domains from prokaryotes to eukaryotes clearly indicates its early emergence and the functional diversity observed in these domains owes to the long evolutionary history resulting in acquired functions. Since NAD+ and NADP+ are key constituents of bioenergetics and associated with a myriad of functions, the conservation of WxxxE motif in TIR domains of bacteria, archaea, plants and mammals is justified.
The structural analysis of the TIR-domain of bacteria compared to eukaryote TLRs, exposed a conserved core protein structure with unique conformations in loop positions among the TIR domain-containing subfamilies (Jang and Park, 2014; Waldhuber et al., 2016; Xu et al., 2000). While the secondary structure of bacterial TIR domains is similar, located in either the N- or C- terminus of the protein, and these domains are highly variable in the remaining part of the protein structure (Xu et al., 2000). These findings suggest that there are structural similarities between the TIR protein domains of mammals and bacteria, which could explain the role of TIR domain in host-pathogen interactions (Zhang et al., 2011). In addition, there are strong indications that the bacterial TIR proteins disrupt the host signal transduction pathways through molecular mimicry (Chan et al., 2009; Rana et al., 2013). Bioinformatic analysis showed that TIR domains are present in pathogenic bacteria and non-pathogenic bacteria, including nonsymbiotic and commensal bacteria (Spear et al., 2009; Zhang et al., 2011), suggesting that the TIR domain-containing proteins (Tdcps) of eukaryotes and non-pathogenic bacteria could also play a crucial role between host and commensal interactions in the gut microbiota (Zhang et al., 2011). This area of Tdcps in bacteria is unexplored, and further study is required to understand their contributions in bacteria.
Several studies have explored bacterial TIR proteins and their ability to cause disease through direct manipulation of the host TLR signaling pathway. Btp1/BtpA/TcpB in Brucella and TcpC in uropathogenic Escherichia coli CFT073 target MyD88 adaptor molecule to subdue downstream TLR4 and TLR2 mediated signaling (Cirl and Miethke, 2010; Salcedo et al., 2008). TIR-like protein A (TlpA) in Salmonella enterica serovar Enteriditis diminishes NF-κB activation by interfering with TIR-domain containing protein TLR4, IL-1 receptor, and MyD88 mediated pathways (Newman et al., 2006). PumA secreted by Pseudomonas aeruginosa PA7 inhibits NF-κB and the TIR domain of PumA has been found to interact with TIRAP, MyD88, and the ubiquitin-associated protein 1 (UBAP1), leading to disruption of cytokine signaling and TLR signalingand helping the pathogen to evade host immune response (Imbert et al., 2017). In addition, other protein classes have been identified in pathogenic and non-pathogenic bacteria such as SEFIR and DUF1863 and these proteins have TIR-like domain of the unknown function (Novatchkova et al., 2003). These proteins also have WxxxE motif in the TIR-like domains. Further studies need to assess the function(s) of these proteins.
Bacterial Tdcps have been shown to downregulate inflammatory signaling pathways. However, some bacterial TIR proteins such as BaTcp from Bacillus anthracis and Brucella Btpa target and induce microtubule formation through a conserved WxxxE motif (Cirl et al., 2008; Felix et al., 2014). Essentially, the conserved WxxxE motif is present in some eukaryotic TIR domain proteins and several bacterial TIR domain proteins. The Table 4 summarizes the WxxxE motif containing TIR proteins of pathogenic bacteria. Multiple sequence alignment warrants that the WxxxE motif is highly conserved among the bacterial Tdcps such as Escherichia coli TcpC, Yersinia pestis YpTdp, Salmonella enterica TlpA, and Paracoccus denitrificans PdTir (Felix et al., 2014). The presence of this conserved WxxxE motif in the TIR domain of many bacteria suggests that this motif could have a significant structural role for the TIR domain as seen in the WxxxE GEF family proteins (Felix et al., 2014). However, the crystal structure of BtpA revealed that the WxxxE motif is positioned at different loop structures and possibly important for association with microtubules (Felix et al., 2014). While TIR proteins such as BtpA are not a part of the WxxxE GEF family despite ectopic similarities in the regulation of host actin filaments, TIR domain proteins could give us an insight into the cross-talk between the TLR and GTPase signaling pathways (Felix et al., 2014). Furthermore, ectopic expression of BtpA-TIR and BtpB-TIR in yeast and human cells has also been reported to form long filament-like structures which is independable on the WxxxE motif (Coronas-Serna et al., 2020).
Table 4:
TIR domain containing bacterial proteins with WxxxE motif
| Bacterial effector | Bacteria species | Amino acid sequence | Function | Host target | References |
|---|---|---|---|---|---|
| TcpS | Salmonella enterica serovar Enteritidis | VVVLSKSFIKKDWTEYELNGLTAREM | a) inhibits NF-κB and MAPK activation, and inhibits inflammatory responses | a) blocks MyD88- and TRIF-mediated TLR signalling | (Xiong et al., 2019) |
| BtpB | B. abortus | VVFVGDDYQRKDWCGVEFRAIREIIM | a) affects host inflammatory response, microtubule protection. b) possess NADase activity |
a) inhibits TLR signalling, probably via MyD88 | (Felix et al., 2014; Salcedo et al., 2013) |
| TcpC | uropathogenic Escherichia coli CFT073 | LSHNFLNKKWTQYELDSLINRAVYDD | a) regulates pro-inflammatory cytokines IL-6, IL-1α/β, IL-8, TNF-α NF-κB. b) increases acute mortality, bacterial persistence and tissue damage c) possess NADase activity |
a) regulates TLR and MyD88 dependent responses | (Essuman et al., 2018; Snyder et al., 2013; Yadav et al., 2010) |
| TcpB (BtpA/Btp1) | Brucella abortus, Brucella melitensis, | GIVVLSEHFFSKQWPARELDGLTAME | a) modulates maturation of dendritic cells. b) interferes with TLR2 signalling. c) inhibits host NF-κB activation, d) modulates host actin dynamics. e) possess NADase activity |
a) TcpB interacts with MAL, MyD88, and TLR4 | (Alaidarous et al., 2014; Essuman et al., 2018; Felix et al., 2014; Li et al., 2016; Salcedo et al., 2008) |
| TirS | S. aureus MSSA476 | RFVVVFLSPNFIESGWSRYEFLSFLN | a) reduces the levels of cytokines MCP-1 and G-CSF. b) possess NADase activity |
a) interacts with TLR2, MyD88 and TIRAP, NF-κB, MAPK | (Askarian et al., 2014; Essuman et al., 2018) |
| PumA | Pseudomonas aeruginosa PA7 | LNYTCWRSREDCERAWQTREDAQGPL | a) Disrupts cytokine signalling, TLR signalling, b) inhibits NF-κB |
a) interacts with TIRAP, MyD88, and the ubiquitin-associated protein 1 (UBAP1) | (Imbert et al., 2017) |
| PdTLP/PdTir | Paracoccus denitrificans | VVLSTHFFKKEWPQKELDGLFQLESS | a) Possess NADase activity | a) binds TLR4 and MyD88 | (Essuman et al., 2018; Low et al., 2007) |
| AbTir | Acinetobacter baumannii | VVLSTDFIKKDWTNYELDGLVAREMN | a) Possess NADase activity | (Essuman et al., 2018) | |
| TcpO | Methanobrevibacter olleyae | SEDFFKSKWTNYEYDNIFLDFYDEEK | a) Possess NADase activity | (Essuman et al., 2018) | |
| ApTir | Actinoplanes | ASPEAAASPWVNQEIEHWLSRHSVDR | a) Possess NADase activity | (Essuman et al., 2018) | |
| TcpA | Theionarchaea archaeon | VVLSKRFFEKEWPQKELDGLVAKEVE | a) Possess NADase activity | (Essuman et al., 2018) |
Conclusion and future perspectives
Although several studies have identified a plethora of T3SS effectors from different pathogens and their ability to hijack host cell function, there are only a few studies that have focussed on the characterization and functioning of WxxxE effectors. Most of these WxxxE effectors have been originally identified for their GEF mimicking functions, however these were additionally found to influence the host cytoskeleton structure. Besides, the effector molecules are also known to regulate cellular functions such as endolysosomal trafficking, inflammation and intestinal barrier function. However, the precise mechanisms associated with such functions and key components involved are yet to be elucidated. Recent studies have demonstrated that some WxxxE effectors function in cooperation with other effectors, and these effectors form a robust network that can sustain contraction (deletions of effector genes) without affecting the pathogenesis. Similar studies are required to explore such functions of other WxxxE effectors. The presence of the WxxxE motif in the effector proteins secreted by enteric pathogens and its absence in commensal bacteria indicates its putative role in the differentiation and recognition of pathogens by the immune system. Studies in this direction will shed light on the mechanism of homeostasis maintained by the host immune system. Further, the role of the WxxxE sequence in several of these effectors/proteins secreted/associated with the secretion system of non-enteric pathogens is yet to be explored. The presence of WxxxE motif in TIR domains of bacteria, archaea, plants and metazoans and the NADase activity of these domains signifies the role of WxxxE motif. NAD+ and NADP+ binding activity of WxxxE motif in TIR domains and the GEF as well as the ATPase activity of the WxxxE effectors/ associated proteins collectively imply the role of WxxxE motif in nucleotide binding. Although most of the studies associated with WxxxE effectors have focussed on GEF activity, the activity of these effectors on other nucleotides and possible role as NADase/ influencing NADase activity needs to be examined. This new direction may provide answers for the absence of GEF activity in some WxxxE effectors. Zhang et al indicated that the TIR family proteins had a complicated evolutionary history and probably several independent bacteria-eukaryotes lateral gene transfer events are involved (Zhang et al., 2011). Their findings suggest that bacterial TIR domain-containing proteins may play important roles in interactions between bacteria and eukaryotes where WxxxE is important. Due to the lack of physiological functions and biochemical data of prokaryotic TIR domains our understanding of these proteins are incomplete and it needs further research to determine the functional implication of WxxxE motif in these bacterial TIR proteins.
Acknowledgment:
The figure present in the manuscript is designed using www.biorender.com. We are grateful to Dr. Stella-Rita Ibeawuchi, Ms. Mahitha Shree Anandachar and Dr. Stefania Tocci for providing suggestions during the preparation of the manuscript.
Funding Details
This work was supported by the National Institutes of Health (NIH) grants DK107585 and AG069689 (to S.D.). S.D was supported by NIH/National Center for Advancing Translational Sciences (NCATS) awards UG3TR002968, UG3TR003355, and R01-AI155696 and the Leona M. and Harry B. Helmsley Charitable Trust.
Abbreviations:
- ADAM10
A Disintegrin and metalloproteinase domain-containing protein 10
- ATP
Adenosine tri phosphate
- AvrE
avirulence
- Bep
Bartonella effector proteins
- BID
Bartonella intracellular delivery
- Btp
Brucella TIR domain containing protein
- Cdc
Cell division control protein
- COX-2
cyclooxygenase-2
- CXCL
Chemokine (C-X-C motif) ligand 1
- Dock
Dedicator of cytokinesis
- DrrA
Defect in Rab1 recruitment A
- Dsp
Disease specific
- DUF
Domain of unknown function
- EHEC
Enterohemorrhagic E. coli
- ELMO1
Engulfment and cell motility protein 1
- EPEC
Enteropathogenic E. coli
- Erk,1/2
extracellular-signal-regulated kinase
- Esp
EPEC secreted protein
- G proteins
guanine nucleotide-binding proteins
- GCC185
Golgi localized coiled-coil protein
- GDP
guanosine diphosphate
- GEF
guanine nucleotide exchange factors
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
- GTP
guanosine triphosphate
- HOPS
Homotypic fusion and vacuole protein sorting
- Icm/Dot
intracellular multiplication/defect in organelle trafficking genes
- IL
Interleukin
- Ipg
Invasion plasmid gene
- JNK
c-Jun N- terminus kinase
- LAMP1
lysosome-associated membrane protein-1
- LRR-RLKs
leucine-rich repeat receptor-like kinases
- Mal
MyD88 adaptor-like protein
- Map
Mitochondrial associated protein
- MAPK
mitogen-activated protein kinases
- MCP
Monocyte chemoattractant protein-1
- MPR
Mannose Phosphate Receptors
- MyD88
Myeloid differentiation primary response gene 88
- NESCA
New molecule containing an SH3 domain at the carboxyl terminus
- NFkB
Nuclear Factor kappa-light-chain-enhancer of activated B cells
- NHERF
Na+–H+ exchanger regulatory factor-1
- NOD1
Nucleotide-binding oligomerization domain
- NTP
Nucleotide tri phosphate
- PP 2A
protein phosphatase 2A
- PGE2
Prostaglandin E2
- PH
Pleckstrin homology
- PLEKHM
Pleckstrin homology domain-containing family M member
- Ptl
Pertussis toxin liberation
- Rab
Ras-related in brain
- Rac
Ras related C3 botulinum toxin substrate
- ROCK
Rho-associated protein kinase
- RPIP8
Rap2-interactingprotein 8
- RUN
RPIP8, UNC- 14 and NESCA
- SARM
Sterile α and Armadillo motifs containing protein
- SCV
Salmonella Containing Vacuoles
- SEFIR
SEF/IL-17 receptor
- Sid
Substrate of Icm/Dot
- Sif
Salmonella-induced filament
- SKIP
SifA and kinesin interacting protein
- SopE
Salmonella Outer Proteins
- SPI
Salmonella Pathogenicity Island
- T3SS1
Salmonella pathogenicity island 1 encoded type 3 secretion system
- T3SS2
Salmonella pathogenicity island 2 encoded type 3 secretion system
- Tcp
TIR domain containing protein
- Tdp
TIR domain protein
- TGN
Trans-Golgi Network
- TIR
Toll/interleukin-1 receptor
- TIRAP
TIR domain-containing adaptor protein
- Tlp
TIR-like proteins
- TLR
Toll like Receptor
- TNF
Tumor necrosis factor
- TPR
Tetratricopeptide repeat
- TRAM
TRIF-related adaptor molecule
- TRIF
TIR domain-containing adapter-inducing interferon-β
- Vir
virulence
- Wts
Water-soaking
- ZO
zonula occludens
Footnotes
Declaration of Interest: The authors declare no conflict of interest.
References
- Alaidarous M, Ve T, Casey LW, Valkov E, Ericsson DJ, Ullah MO, Schembri MA, Mansell A, Sweet MJ, Kobe B, 2014. Mechanism of bacterial interference with TLR4 signaling by Brucella Toll/interleukin-1 receptor domain-containing protein TcpB. Journal of Biological Chemistry 289(2), 654–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, McMahon SA, Ghosh P, Hughes TR, Boone C, 2006. Identification of a bacterial type III effector family with G protein mimicry functions. Cell 124(1), 133–145. [DOI] [PubMed] [Google Scholar]
- Arbeloa A, Bulgin RR, MacKenzie G, Shaw RK, Pallen MJ, Crepin VF, Berger CN, Frankel G, 2008. Subversion of actin dynamics by EspM effectors of attaching and effacing bacterial pathogens. Cellular microbiology 10(7), 1429–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeloa A, Garnett J, Lillington J, Bulgin RR, Berger CN, Lea SM, Matthews S, Frankel G, 2010. EspM2 is a RhoA guanine nucleotide exchange factor. Cellular microbiology 12(5), 654–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askarian F, Van Sorge NM, Sangvik M, Beasley FC, Henriksen JR, Sollid JU, Van Strijp JA, Nizet V, Johannessen M, 2014. A Staphylococcus aureus TIR domain protein virulence factor blocks TLR2-mediated NF-κB signaling. Journal of innate immunity 6(4), 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Christie PJ, 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Molecular microbiology 54(5), 1199–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt W-D, 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infection and immunity 71(5), 2839–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger CN, Crepin VF, Jepson MA, Arbeloa A, Frankel G, 2009. The mechanisms used by enteropathogenic Escherichia coli to control filopodia dynamics. Cellular microbiology 11(2), 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuzón CR, Méresse S, Unsworth KE, Ruíz-Albert J, Garvis S, Waterman SR, Ryder TA, Boucrot E, Holden DW, 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. The EMBO journal 19(13), 3235–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A, 2007. GEFs and GAPs: critical elements in the control of small G proteins. Cell 129(5), 865–877. [DOI] [PubMed] [Google Scholar]
- Boucrot E, Henry T, Borg J-P, Gorvel J-P, Méresse S, 2005. The intracellular fate of Salmonella depends on the recruitment of kinesin. Science 308(5725), 1174–1178. [DOI] [PubMed] [Google Scholar]
- Brumell JH, Tang P, Mills SD, Finlay BB, 2001. Characterization of Salmonella-induced filaments (Sifs) reveals a delayed interaction between Salmonella-containing vacuoles and late endocytic compartments. Traffic 2(9), 643–653. [DOI] [PubMed] [Google Scholar]
- Bulgin RR, Arbeloa A, Chung JC, Frankel G, 2009. EspT triggers formation of lamellipodia and membrane ruffles through activation of Rac-1 and Cdc42. Cellular microbiology 11(2), 217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Low LY, Hsu S, Li S, Liu T, Santelli E, Le Negrate G, Reed JC, Woods VL, Pascual J, 2009. Molecular mimicry in innate immunity: crystal structure of a bacterial TIR domain. Journal of Biological Chemistry 284(32), 21386–21392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Han G, Dietrich CR, Dunn TM, Cahoon EB, 2006. The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. The plant cell 18(12), 3576–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirl C, Miethke T, 2010. Microbial Toll/interleukin 1 receptor proteins: a new class of virulence factors. International Journal of Medical Microbiology 300(6), 396–401. [DOI] [PubMed] [Google Scholar]
- Cirl C, Wieser A, Yadav M, Duerr S, Schubert S, Fischer H, Stappert D, Wantia N, Rodriguez N, Wagner H, 2008. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain–containing proteins. Nature medicine 14(4), 399–406. [DOI] [PubMed] [Google Scholar]
- Colonne PM, Winchell CG, Voth DE, 2016. Hijacking host cell highways: manipulation of the host actin cytoskeleton by obligate intracellular bacterial pathogens. Frontiers in cellular and infection microbiology 6, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis GR, 2006. The type III secretion injectisome. Nature Reviews Microbiology 4(11), 811–825. [DOI] [PubMed] [Google Scholar]
- Coronas-Serna JM, Louche A, Rodríguez-Escudero M, Roussin M, Imbert PR, Rodríguez-Escudero I, Terradot L, Molina M, Gorvel J-P, Cid VJ, 2020. The TIR-domain containing effectors BtpA and BtpB from Brucella abortus impact NAD metabolism. PLoS pathogens 16(4), e1007979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa SC, Lesser CF, 2014. A multifunctional region of the Shigella type 3 effector IpgB1 is important for secretion from bacteria and membrane targeting in eukaryotic cells. PLoS One 9(4), e93461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa TR, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G, 2015. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nature Reviews Microbiology 13(6), 343–359. [DOI] [PubMed] [Google Scholar]
- Craig-Mylius KA, Weiss AA, 1999. Mutants in the ptlA-H genes of Bordetella pertussis are deficient for pertussis toxin secretion. FEMS microbiology letters 179(2), 479–484. [DOI] [PubMed] [Google Scholar]
- Dean P, Kenny B, 2004. Intestinal barrier dysfunction by enteropathogenic Escherichia coli is mediated by two effector molecules and a bacterial surface protein. Molecular microbiology 54(3), 665–675. [DOI] [PubMed] [Google Scholar]
- Diacovich L, Dumont A, Lafitte D, Soprano E, Guilhon A-A, Bignon C, Gorvel J-P, Bourne Y, Méresse S, 2009. Interaction between the SifA virulence factor and its host target SKIP is essential for Salmonella pathogenesis. Journal of Biological Chemistry 284(48), 33151–33160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich Han, G., Chen M, Berg RH, Dunn TM, Cahoon EB, 2008. Loss-of-function mutations and inducible RNAi suppression of Arabidopsis LCB2 genes reveal the critical role of sphingolipids in gametophytic and sporophytic cell viability. The Plant Journal 54(2), 284–298. [DOI] [PubMed] [Google Scholar]
- Dietrich KA, Sindelar CV, Brewer PD, Downing KH, Cremo CR, Rice SE, 2008. The kinesin-1 motor protein is regulated by a direct interaction of its head and tail. Proceedings of the National Academy of Sciences 105(26), 8938–8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, Sorek R, 2018. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359(6379). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drecktrah D, Knodler LA, Howe D, Steele-Mortimer O, 2007. Salmonella trafficking is defined by continuous dynamic interactions with the endolysosomal system. Traffic 8(3), 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont A, Boucrot E, Drevensek S, Daire V, Gorvel JP, Poüs C, Holden DW, Méresse S, 2010. SKIP, the host target of the Salmonella virulence factor SifA, promotes kinesin-1-dependent vacuolar membrane exchanges. Traffic 11(7), 899–911. [DOI] [PubMed] [Google Scholar]
- East A, Mechaly AE, Huysmans GH, Bernarde C, Tello-Manigne D, Nadeau N, Pugsley AP, Buschiazzo A, Alzari PM, Bond PJ, 2016. Structural basis of pullulanase membrane binding and secretion revealed by X-ray crystallography, molecular dynamics and biochemical analysis. Structure 24(1), 92–104. [DOI] [PubMed] [Google Scholar]
- Essuman K, Summers DW, Sasaki Y, Mao X, DiAntonio A, Milbrandt J, 2017. The SARM1 toll/interleukin-1 receptor domain possesses intrinsic NAD+ cleavage activity that promotes pathological axonal degeneration. Neuron 93(6), 1334–1343. e1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essuman K, Summers DW, Sasaki Y, Mao X, Yim AKY, DiAntonio A, Milbrandt J, 2018. TIR domain proteins are an ancient family of NAD+-consuming enzymes. Current Biology 28(3), 421–430. e424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farizo KM, Cafarella TG, Burns DL, 1996. Evidence for a ninth gene, ptlI, in the locus encoding the pertussis toxin secretion system of Bordetella pertussis and formation of a PtlI-PtlF complex. Journal of Biological Chemistry 271(49), 31643–31649. [DOI] [PubMed] [Google Scholar]
- Felix C, Türköz BK, Ranaldi S, Koelblen T, Terradot L, O’Callaghan D, Vergunst AC, 2014. The Brucella TIR domain containing proteins BtpA and BtpB have a structural WxxxE motif important for protection against microtubule depolymerisation. Cell Communication and Signaling 12(1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JA, Ohl ME, Miller SI, 2003. The Salmonella enterica serovar typhimurium translocated effectors SseJ and SifB are targeted to the Salmonella-containing vacuole. Infection and immunity 71(1), 418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa A, Alonso C, Kurachi K, Gupta S, Lesser CF, McCormick BA, Reinecker H-C, 2008. GEF-H1 mediated control of NOD1 dependent NF-κB activation by Shigella effectors. PLoS pathogens 4(11), e1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán JE, 2009. Common themes in the design and function of bacterial effectors. Cell host & microbe 5(6), 571–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-del Portillo F, Zwick MB, Leung KY, Finlay BB, 1993. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proceedings of the National Academy of Sciences 90(22), 10544–10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goody RS, Müller MP, Schoebel S, Oesterlin LK, Blümer J, Peters H, Blankenfeldt W, Itzen A, 2011. The versatile Legionella effector protein DrrA. Communicative & integrative biology 4(1), 72–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachani A, Biskri L, Rossi G, Marty A, Ménard R, Sansonetti P, Parsot C, Van Nhieu GT, Bernardini ML, Allaoui A, 2008. IpgB1 and IpgB2, two homologous effectors secreted via the Mxi-Spa type III secretion apparatus, cooperate to mediate polarized cell invasion and inflammatory potential of Shigella flexenri. Microbes and infection 10(3), 260–268. [DOI] [PubMed] [Google Scholar]
- Ham JH, Majerczak DR, Nomura K, Mecey C, Uribe F, He S-Y, Mackey D, Coplin DL, 2009. Multiple activities of the plant pathogen type III effector proteins WtsE and AvrE require WxxxE motifs. Molecular plant-microbe interactions 22(6), 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa Y, Suzuki M, Ohya K, Iwai H, Ishijima N, Koleske AJ, Fukui Y, Sasakawa C, 2007. Shigella IpgB1 promotes bacterial entry through the ELMO–Dock180 machinery. Nature cell biology 9(1), 121–128. [DOI] [PubMed] [Google Scholar]
- Hardiman CA, Roy CR, 2014. AMPylation is critical for Rab1 localization to vacuoles containing Legionella pneumophila. MBio 5(1), e01035–01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardt W-D, Chen L-M, Schuebel KE, Bustelo XR, Galán JE, 1998. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93(5), 815–826. [DOI] [PubMed] [Google Scholar]
- Horsefield S, Burdett H, Zhang X, Manik MK, Shi Y, Chen J, Qi T, Gilley J, Lai J-S, Rank MX, 2019. NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 365(6455), 793–799. [DOI] [PubMed] [Google Scholar]
- Hu X, Yu J, Zhou X, Li Z, Xia Y, Luo Z, Wu Y, 2014. A small GTPase-like protein fragment of Mycoplasma promotes tumor cell migration and proliferation in vitro via interaction with Rac1 and Stat3. Molecular medicine reports 9(1), 173–179. [DOI] [PubMed] [Google Scholar]
- Huang Z, Sutton SE, Wallenfang AJ, Orchard RC, Wu X, Feng Y, Chai J, Alto NM, 2009. Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics. Nature structural & molecular biology 16(8), 853–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert PR, Louche A, Luizet JB, Grandjean T, Bigot S, Wood TE, Gagné S, Blanco A, Wunderley L, Terradot L, 2017. A Pseudomonas aeruginosa TIR effector mediates immune evasion by targeting UBAP 1 and TLR adaptors. The EMBO journal 36(13), 1869–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson LK, Nawabi P, Hentea C, Roark EA, Haldar K, 2008. The Salmonella virulence protein SifA is a G protein antagonist. Proceedings of the National Academy of Sciences 105(37), 14141–14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal M, Gremer L, Dvorsky R, Haeusler LC, Cirstea IC, Uhlenbrock K, Ahmadian MR, 2011. Mechanistic insights into specificity, activity, and regulatory elements of the regulator of G-protein signaling (RGS)-containing Rho-specific guanine nucleotide exchange factors (GEFs) p115, PDZ-RhoGEF (PRG), and leukemia-associated RhoGEF (LARG). Journal of Biological Chemistry 286(20), 18202–18212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang T. h., Park HH, 2014. Crystal structure of TIR domain of TLR6 reveals novel dimeric interface of TIR–TIR interaction for toll-like receptor signaling pathway. Journal of molecular biology 426(19), 3305–3313. [DOI] [PubMed] [Google Scholar]
- Jepson MA, Pellegrin S, Peto L, Banbury DN, Leard AD, Mellor H, Kenny B, 2003. Synergistic roles for the Map and Tir effector molecules in mediating uptake of enteropathogenic Escherichia coli (EPEC) into non-phagocytic cells. Cellular microbiology 5(11), 773–783. [DOI] [PubMed] [Google Scholar]
- Jin L, Ham JH, Hage R, Zhao W, Soto-Hernández J, Lee SY, Paek S-M, Kim MG, Boone C, Coplin DL, 2016. Direct and indirect targeting of PP2A by conserved bacterial type-III effector proteins. PLoS pathogens 12(5), e1005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Iwatsuki M, Nagai T, Matsumoto A, Takahashi Y, Shiomi K, Ōmura S, Abe A, 2011. A small-molecule inhibitor of the bacterial type III secretion system protects against in vivo infection with Citrobacter rodentium. The Journal of antibiotics 64(2), 197–203. [DOI] [PubMed] [Google Scholar]
- Knuff-Janzen K, Tupin A, Yurist-Doutsch S, Rowland JL, Finlay BB, 2020. Multiple Salmonella-pathogenicity island 2 effectors are required to facilitate bacterial establishment of its intracellular niche and virulence. Plos one 15(6), e0235020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotob SI, Burns DL, 1997. Essential role of the consensus nucleotide-binding site of PtlH in secretion of pertussis toxin from Bordetella pertussis. Journal of bacteriology 179(23), 7577–7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger V, Liebl D, Zhang Y, Rajashekar R, Chlanda P, Giesker K, Chikkaballi D, Hensel M, 2014. Reorganization of the endosomal system in Salmonella-infected cells: the ultrastructure of Salmonella-induced tubular compartments. PLoS pathogens 10(9), e1004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K, Finlay B, 1991. Intracellular replication is essential for the virulence of Salmonella typhimurium. Proceedings of the National Academy of Sciences 88(24), 11470–11474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ke Y, Wang Y, Yang M, Gao J, Zhan S, Xinying D, Huang L, Li W, Chen Z, 2016. Brucella TIR-like protein TcpB/Btp1 specifically targets the host adaptor protein MAL/TIRAP to promote infection. Biochemical and biophysical research communications 477(3), 509–514. [DOI] [PubMed] [Google Scholar]
- Low LY, Mukasa T, Reed JC, Pascual J, 2007. Characterization of a TIR-like protein from Paracoccus denitrificans. Biochemical and biophysical research communications 356(2), 481–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak H, Thurston TLM, 2021. Interesting biochemistries in the Structure and Function of Bacterial Effectors. Frontiers in Cellular and Infection Microbiology 11, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham JE, Molino D, Gissot L, Bellec Y, Hématy K, Marion J, Belcram K, Palauqui J-C, Satiat-JeuneMaître B, Faure J-D, 2011. Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. The Plant Cell 23(6), 2362–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Schroeder GN, Berger CN, Lee SF, Robinson KS, Badea L, Simpson N, Hall RA, Hartland EL, Crepin VF, 2010. Binding to Na+/H+ exchanger regulatory factor 2 (NHERF2) affects trafficking and function of the enteropathogenic Escherichia coli type III secretion system effectors Map, EspI and NleH. Cellular microbiology 12(12), 1718–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Haneda T, Saito H, Miki T, Okada N, 2019. Salmonella enterica effectors SifA, SpvB, SseF, SseJ, and SteA contribute to type III secretion system 1-independent inflammation in a streptomycin-pretreated mouse model of colitis. Infection and immunity 87(9), e00872–00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattoo S, Lee YM, Dixon JE, 2007. Interactions of bacterial effector proteins with host proteins. Current opinion in immunology 19(4), 392–401. [DOI] [PubMed] [Google Scholar]
- McEwan DG, Richter B, Claudi B, Wigge C, Wild P, Farhan H, McGourty K, Coxon FP, Franz-Wachtel M, Perdu B, 2015. PLEKHM1 regulates Salmonella-containing vacuole biogenesis and infection. Cell host & microbe 17(1), 58–71. [DOI] [PubMed] [Google Scholar]
- McGourty K, Thurston TL, Matthews SA, Pinaud L, Mota LJ, Holden DW, 2012. Salmonella inhibits retrograde trafficking of mannose-6-phosphate receptors and lysosome function. Science 338(6109), 963–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Bonasera JM, Kim JF, Nissinen RM, Beer SV, 2006. Apple proteins that interact with DspA/E, a pathogenicity effector of Erwinia amylovora, the fire blight pathogen. Molecular plant-microbe interactions 19(1), 53–61. [DOI] [PubMed] [Google Scholar]
- Méresse S, Steele-Mortimer O, Finlay BB, Gorvel JP, 1999. The rab7 GTPase controls the maturation of Salmonella typhimurium-containing vacuoles in HeLa cells. The EMBO journal 18(16), 4394–4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita-Ishihara T, Miura M, Iyoda S, Izumiya H, Watanabe H, Ohnishi M, Terajima J, 2013. EspO1-2 regulates EspM2-mediated RhoA activity to stabilize formation of focal adhesions in enterohemorrhagic Escherichia coli-infected host cells. PLoS One 8(2), e55960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller MP, Peters H, Blümer J, Blankenfeldt W, Goody RS, Itzen A, 2010. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329(5994), 946–949. [DOI] [PubMed] [Google Scholar]
- Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR, 2006. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nature cell biology 8(9), 971–977. [DOI] [PubMed] [Google Scholar]
- Newman RM, Salunkhe P, Godzik A, Reed JC, 2006. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infection and immunity 74(1), 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novatchkova M, Leibbrandt A, Werzowa J, Neubüser A, Eisenhaber F, 2003. The STIR-domain superfamily in signal transduction, development and immunity. Trends in biochemical sciences 28(5), 226–229. [DOI] [PubMed] [Google Scholar]
- O’Neill LA, Bowie AG, 2007. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nature Reviews Immunology 7(5), 353–364. [DOI] [PubMed] [Google Scholar]
- Ofir G, Herbst E, Baroz M, Cohen D, Millman A, Doron S, Tal N, Malheiro D, Malitsky S, Amitai G, 2021. Antiviral activity of bacterial TIR domains via immune signalling molecules. Nature 600(7887), 116–120. [DOI] [PubMed] [Google Scholar]
- Ohlson MB, Huang Z, Alto NM, Blanc M-P, Dixon JE, Chai J, Miller SI, 2008. Structure and function of Salmonella SifA indicate that its interactions with SKIP, SseJ, and RhoA family GTPases induce endosomal tubulation. Cell host & microbe 4(5), 434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya K, Handa Y, Ogawa M, Suzuki M, Sasakawa C, 2005. IpgB1 is a novel Shigella effector protein involved in bacterial invasion of host cells: its activity to promote membrane ruffling via RAC1 and CDC42 activation. Journal of Biological Chemistry 280(25), 24022–24034. [DOI] [PubMed] [Google Scholar]
- Orchard RC, Kittisopikul M, Altschuler SJ, Wu LF, Süel GM, Alto NM, 2012. Identification of F-actin as the dynamic hub in a microbial-induced GTPase polarity circuit. Cell 148(4), 803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston GM, Bertrand N, Rainey PB, 2001. Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25. Molecular microbiology 41(5), 999–1014. [DOI] [PubMed] [Google Scholar]
- Pugsley A, Kornacker M, Ryter A, 1990. Analysis of the subcellular location of pullulanase produced by Escherichia coli carrying the pulA gene from Klebsiella pneumoniae strain UNF5023. Molecular microbiology 4(1), 59–72. [DOI] [PubMed] [Google Scholar]
- Ramachandran RP, Spiegel C, Keren Y, Danieli T, Melamed-Book N, Pal RR, Zlotkin-Rivkin E, Rosenshine I, Aroeti B, 2020. Mitochondrial targeting of the enteropathogenic escherichia coli map triggers calcium mobilization, ADAM10-MAP kinase signaling, and host cell apoptosis. Mbio 11(5), e01397–01320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana RR, Zhang M, Spear AM, Atkins HS, Byrne B, 2013. Bacterial TIR-containing proteins and host innate immune system evasion. Medical microbiology and immunology 202(1), 1–10. [DOI] [PubMed] [Google Scholar]
- Rapisarda C, Tassinari M, Gubellini F, Fronzes R, 2018. Using Cryo-EM to investigate bacterial secretion systems. Annual review of microbiology 72, 231–254. [DOI] [PubMed] [Google Scholar]
- Raymond B, Crepin VF, Collins JW, Frankel G, 2011. The WxxxE effector EspT triggers expression of immune mediators in an Erk/JNK and NF-κB-dependent manner. Cellular microbiology 13(12), 1881–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoll-Rozada J, Zunzunegui S, de la Cruz F, Arechaga I, Cabezón E, 2013. Functional interactions of VirB11 traffic ATPases with VirB4 and VirD4 molecular motors in type IV secretion systems. Journal of bacteriology 195(18), 4195–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruano-Gallego D, Sanchez-Garrido J, Kozik Z, Núñez-Berrueco E, Cepeda-Molero M, Mullineaux-Sanders C, Clark JN-B, Slater SL, Wagner N, Glegola-Madejska I, 2021. Type III secretion system effectors form robust and flexible intracellular virulence networks. Science 371(6534). [DOI] [PubMed] [Google Scholar]
- Salcedo S, Marchesini MI, Degos C, Terwagne M, Von Bargen K, Lepidi H, Herrmann CK, Santos Lacerda TL, Imbert P, Pierre P, 2013. BtpB, a novel Brucella TIR-containing effector protein with immune modulatory functions. Frontiers in cellular and infection microbiology 3, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo SP, Marchesini MI, Lelouard H, Fugier E, Jolly G, Balor S, Muller A, Lapaque N, Demaria O, Alexopoulou L, 2008. Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS pathogens 4(2), e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, Lanka E, Buhrdorf R, Fischer W, Haas R, Waksman G, 2003. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. The EMBO journal 22(9), 1969–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed IM, Ibeawuchi S-R, Lie D, Anandachar MS, Pranadinata R, Raffatellu M, Das S, 2021. The interaction of enteric bacterial effectors with the host engulfment pathway control innate immune responses. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed IM, Suarez K, Lim E, Singh S, Pereira M, Ibeawuchi SR, Katkar G, Dunkel Y, Mittal Y, Chattopadhyay R, 2020. Host engulfment pathway controls inflammation in inflammatory bowel disease. The FEBS journal 287(18), 3967–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MC, Schulein R, Dehio M, Denecker G, Carena I, Dehio C, 2004. The VirB type IV secretion system of Bartonella henselae mediates invasion, proinflammatory activation and antiapoptotic protection of endothelial cells. Molecular microbiology 52(1), 81–92. [DOI] [PubMed] [Google Scholar]
- Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE, 2002. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109(5), 575–588. [DOI] [PubMed] [Google Scholar]
- Shaw RK, Cleary J, Murphy MS, Frankel G, Knutton S, 2005. Interaction of enteropathogenic Escherichia coli with human intestinal mucosa: role of effector proteins in brush border remodeling and formation of attaching and effacing lesions. Infection and immunity 73(2), 1243–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siamer S, Gaubert S, Boureau T, Brisset M-N, Barny M-A, 2013. Mutational analysis of a predicted double β-propeller domain of the DspA/E effector of Erwinia amylovora. FEMS microbiology letters 342(1), 54–61. [DOI] [PubMed] [Google Scholar]
- Siamer S, Guillas I, Shimobayashi M, Kunz C, Hall MN, Barny M-A, 2014. Expression of the bacterial type III effector DspA/E in Saccharomyces cerevisiae down-regulates the sphingolipid biosynthetic pathway leading to growth arrest. Journal of Biological Chemistry 289(26), 18466–18477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierocki R, Jneid B, Orsini Delgado ML, Plaisance M, Maillère B, Nozach H, Simon S, 2021. An antibody targeting type III secretion system induces broad protection against Salmonella and Shigella infections. PLoS neglected tropical diseases 15(3), e0009231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simovitch M, Sason H, Cohen S, Zahavi EE, Melamed-Book N, Weiss A, Aroeti B, Rosenshine I, 2010. EspM inhibits pedestal formation by enterohaemorrhagic Escherichia coli and enteropathogenic E. coli and disrupts the architecture of a polarized epithelial monolayer. Cellular microbiology 12(4), 489–505. [DOI] [PubMed] [Google Scholar]
- Simpson N, Shaw R, Crepin VF, Mundy R, FitzGerald AJ, Cummings N, Straatman-Iwanowska A, Connerton I, Knutton S, Frankel G, 2006. The enteropathogenic Escherichia coli type III secretion system effector Map binds EBP50/NHERF1: implication for cell signalling and diarrhoea. Molecular microbiology 60(2), 349–363. [DOI] [PubMed] [Google Scholar]
- Sindhwani A, Arya SB, Kaur H, Jagga D, Tuli A, Sharma M, 2017. Salmonella exploits the host endolysosomal tethering factor HOPS complex to promote its intravacuolar replication. PLoS pathogens 13(10), e1006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Sharma S, Pagarware K, Siraji RA, Ansari I, Mandal A, Walling P, Aijaz S, 2018. Enteropathogenic E. coli effectors EspF and Map independently disrupt tight junctions through distinct mechanisms involving transcriptional and post-transcriptional regulation. Scientific reports 8(1), 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder GA, Cirl C, Jiang J, Chen K, Waldhuber A, Smith P, Römmler F, Snyder N, Fresquez T, Dürr S, 2013. Molecular mechanisms for the subversion of MyD88 signaling by TcpC from virulent uropathogenic Escherichia coli. Proceedings of the National Academy of Sciences 110(17), 6985–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear AM, Loman NJ, Atkins HS, Pallen MJ, 2009. Microbial TIR domains: not necessarily agents of subversion? Trends in microbiology 17(9), 393–398. [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer O, Méresse S, Gorvel JP, Toh BH, Finlay BB, 1999. Biogenesis of Salmonella typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cellular microbiology 1(1), 33–49. [DOI] [PubMed] [Google Scholar]
- Tan Y, Luo Z-Q, 2011. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475(7357), 506–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomás A, Lery L, Regueiro V, Pérez-Gutiérrez C, Martínez V, Moranta D, Llobet E, González-Nicolau M, Insua JL, Tomas JM, 2015. Functional genomic screen identifies Klebsiella pneumoniae factors implicated in blocking nuclear factor κB (NF-κB) signaling. Journal of Biological Chemistry 290(27), 16678–16697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshchakov VY, Neuwald AF, 2020. A survey of TIR domain sequence and structure divergence. Immunogenetics 72(3), 181–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truttmann MC, Guye P, Dehio C, 2011a. BID-F1 and BID-F2 domains of Bartonella henselae effector protein BepF trigger together with BepC the formation of invasome structures. PLoS One 6(10), e25106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truttmann MC, Rhomberg TA, Dehio C, 2011b. Combined action of the type IV secretion effector proteins BepC and BepF promotes invasome formation of Bartonella henselae on endothelial and epithelial cells. Cellular microbiology 13(2), 284–299. [DOI] [PubMed] [Google Scholar]
- Verma A, Burns DL, 2007. Requirements for assembly of PtlH with the pertussis toxin transporter apparatus of Bordetella pertussis. Infection and immunity 75(5), 2297–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A, 2001. The guanine nucleotide-binding switch in three dimensions. Science 294(5545), 1299–1304. [DOI] [PubMed] [Google Scholar]
- Waldhuber A, Snyder GA, Römmler F, Cirl C, Müller T, Xiao TS, Svanborg C, Miethke T, 2016. A comparative analysis of the mechanism of toll-like receptor-disruption by tir-containing protein C from uropathogenic Escherichia coli. Pathogens 5(1), 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Essuman K, Anderson RG, Sasaki Y, Monteiro F, Chung E-H, Nishimura EO, DiAntonio A, Milbrandt J, Dangl JL, 2019. TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 365(6455), 799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MM, Faris R, 2018. Subversion of the endocytic and secretory pathways by bacterial effector proteins. Frontiers in cell and developmental biology 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigele BA, Orchard RC, Jimenez A, Cox GW, Alto NM, 2017. A systematic exploration of the interactions between bacterial effector proteins and host cell membranes. Nature communications 8(1), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerberg K, Rossman KL, Der CJ, 2005. The Ras superfamily at a glance. Journal of cell science 118(5), 843–846. [DOI] [PubMed] [Google Scholar]
- Williams MM, Sen K, Weigand MR, Skoff TH, Cunningham VA, Halse TA, Tondella ML, Group CPW, 2016. Bordetella pertussis strain lacking pertactin and pertussis toxin. Emerging infectious diseases 22(2), 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong D, Song L, Geng S, Jiao Y, Zhou X, Song H, Kang X, Zhou Y, Xu X, Sun J, 2019. Salmonella coiled-coil-and TIR-containing TcpS evades the innate immune system and subdues inflammation. Cell reports 28(3), 804–818. e807. [DOI] [PubMed] [Google Scholar]
- Xu Y, Tao X, Shen B, Horng T, Medzhitov R, Manley JL, Tong L, 2000. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature 408(6808), 111–115. [DOI] [PubMed] [Google Scholar]
- Yadav M, Zhang J, Fischer H, Huang W, Lutay N, Cirl C, Lum J, Miethke T, Svanborg C, 2010. Inhibition of TIR domain signaling by TcpC: MyD88-dependent and independent effects on Escherichia coli virulence. PLoS pathogens 6(9), e1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zmasek CM, Cai X, Godzik A, 2011. TIR domain-containing adaptor SARM is a late addition to the ongoing microbe–host dialog. Developmental & Comparative Immunology 35(4), 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Moest T, Zhao Y, Guilhon A-A, Buffat C, Gorvel J-P, Méresse S, 2015. The Salmonella effector protein SifA plays a dual role in virulence. Scientific reports 5(1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zihni C, Mills C, Matter K, Balda MS, 2016. Tight junctions: from simple barriers to multifunctional molecular gates. Nature reviews Molecular cell biology 17(9), 564–580. [DOI] [PubMed] [Google Scholar]