Abstract
Infection of the mucous layer of the human stomach by Helicobacter pylori requires the bacterium to be motile and presumably chemotactic. Previous studies have shown that fully functional flagella are essential for motility and colonization, but the role of chemotaxis remains unclear. The two-component regulatory system CheA/CheY has been shown to play a major role in chemotaxis in other enteric bacteria. Scrutiny of the 26695 genome sequence suggests that H. pylori has two CheY response regulators: one a separate protein (CheY1) and the other (CheY2) fused to the histidine kinase sensor CheA. Defined deletion mutations were introduced into cheY1, cheY2, and cheA in H. pylori strains N6 and SS1. Video tracking revealed that the wild-type H. pylori strain moves in short runs with frequent direction changes, in contrast to movement of cheY2, cheAY2, and cheAY2 cheY1 mutants, whose motion was more linear. The cheY1 mutant demonstrated a different motility phenotype of rapid tumbling. All mutants had impaired swarming and greatly reduced chemotactic responses to hog gastric mucin. Neither cheY1 nor cheAY2 mutants were able to colonize mice, but they generated a significant antibody response, suggesting that despite impaired chemotaxis, these mutants were able to survive in the stomach long enough to induce an immune response before being removed by gastric flow. Additionally, we demonstrated that cheY1 failed to colonize gnotobiotic piglets. This study demonstrates the importance of the roles of cheY1, cheY2, and cheA in motility and virulence of H. pylori.
Helicobacter pylori is a human-specific gastric pathogen that colonizes the stomachs of at least half the world's population (5). H. pylori survives largely within the gastric mucous layer without attaching to host cells (5). Most infected individuals are asymptomatic; however, for a significant number, infection with H. pylori is associated with the development of duodenal and gastric ulcers, gastric adenocarcinoma, and mucosa-associated lymphoid tissue lymphoma (21). Motility is a vital adaptation for many bacterial pathogens capable of colonizing mucosal surfaces. H. pylori has been shown to be extremely motile in viscous environments, such as that encountered in the gastric lumen (15). The bacterium's sheathed flagella are composed of two proteins, FlaA and FlaB, connected to the basal body by the flagellar hook protein, which is a polymer of FlgE (34). Expression of both FlaA and FlaB is necessary for full motility and colonization of gnotobiotic piglets (9).
Chemotaxis, the purposeful movement of bacteria to and from chemical stimulants, has been studied most extensively in Escherichia coli and Salmonella enterica serovar Typhimurium, for which a model has been proposed for this important adaptation (33). Sensing of external stimulant and repellent ligands is achieved via methyl-accepting chemoreceptor proteins (MCPs), which transverse the inner membrane, possessing both a periplasmic ligand binding domain and a cytoplasmic signaling domain (24). Communication between the MCPs and the flagellar motor switch involves four proteins: CheA, CheY, CheW, and CheZ (10). CheA and CheY constitute a two-component regulatory system, although they deviate from the archetype in several ways, most notably in that CheY neither contains a DNA binding domain nor acts as a transcriptional activator (33). The effect of binding to a ligand causes a conformational change in an MCP which is recognized by an associated CheA-CheW complex, which binds to the MCP's cytoplasmic signaling domain via CheW (10). CheA has autokinase activity that is inhibited by attractant-bound receptors and is stimulated by repellent-bound or attractant-free receptors. Stimulation of CheA initiates phosphorylation of the response regulator CheY. The phosphorylated CheY (CheY-P) interacts directly with FliM in the flagellar motor switch complex to cause clockwise rotation (30). This response is terminated by the action of CheZ, which accelerates the decay of the unstable CheY-P (10). In E. coli, the flagellar rotary motor turns clockwise upon interaction with CheY-P, resulting in a tumbling motion; otherwise, it turns counterclockwise, resulting in smooth swimming of the bacterial cell (10).
In H. pylori, a CheY orthologue has been identified as part of a stress-responsive operon, but chemotaxis studies were not reported (4). The annotated genome sequence of H. pylori 26695 contains nine putative chemotaxis orthologues: a bifunctional CheAY protein (HP0392); CheW (HP0391); three CheV proteins, proteins previously identified in Bacillus subtilis which contain an amino-terminal CheW homologous domain linked to a response regulator domain of the CheY family (13) (HP0393, HP0019, and HP0616); the previously identified CheY (HP1067); (4) and three classical MCPs (HP0099, HP0082, and HP0103) (35). No CheZ orthologue was identified. Additionally, a gene (HP0599) encoding a truncated soluble MCP orthologue (with amino acid sequence similarity to the classical MCPs restricted to the highly conserved domain) has been identified and its structure has been analyzed (2). In this study, the previously identified CheY (4) is referred to as CheY1, while the CheY protein fused to CheA is termed CheY2, and the gene that encodes the bifunctional CheAY2 protein is referred to as cheAY2.
Mizote et al. have demonstrated a chemotactic response to urea and bicarbonate by H. pylori CPY3401 (26). This response is increased in a high-viscosity environment, a condition that mimics the ecological niche of H. pylori (28). It was proposed that intracellular urea hydrolyzed by cytoplasmic urease may supply the proton motive force required to drive the bacterial flagellar motor and that H. pylori chemotaxis towards urea may serve to provide urea for hydrolysis by surface urease for gastric acid neutralization (28). Mucin, the principal component of mucus which is secreted from epithelial cells of intestinal, gastric, and gall bladder tissues, has been proposed as a chemoattractant for H. pylori (37). Despite these studies, very little is understood about the mechanism of the chemotactic response in H. pylori or the role of chemotaxis genes in motility and virulence. In this study, we describe the construction and characterization of four chemotaxis mutants, cheY1, cheY2, cheAY2, and cheAY2 cheY1, in independent H. pylori strains (N6 and SS1), showing the importance of these genes in the motility and virulence of H. pylori.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. H. pylori strains were minimally passaged, aliquoted, and stored at −80°C in brain heart infusion (BHI) broth (Oxoid, Basingstoke, United Kingdom) containing 15% (vol/vol) glycerol and 10% fetal calf serum (FCS) (Sigma, Poole, United Kingdom). Strains were grown in BHI broth supplemented with 10% FCS or on Helicobacter selective agar (DENT), consisting of Blood Agar Base No. 2 (Oxoid) supplemented with 7% (vol/vol) lysed defribinated horse blood (TCS Microbiology, Botolph Claydon, United Kingdom) and DENT selective supplement (Oxoid) in a microaerophilic atmosphere at 37°C. E. coli strains were routinely grown in Luria-Bertani (LB) broth or on LB agar. The antibiotics used for selection purposes were ampicillin (100 μg/ml), kanamycin (20 μg/ml for H. pylori and 50 μg/ml for E. coli), and chloramphenicol (6 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains of H. pylori | ||
| 11637 | Typed laboratory strain | NCTC 11637 |
| N6 | Virulent wild-type strain | 11 |
| 26695 | Virulent wild-type strain | 35 |
| SS1 | Virulent wild-type strain | 23 |
| N6 cheY1 | KnrH. pylori N6 cheY1 mutant | This study |
| N6 cheY2 | KnrH. pylori N6 cheAY2 mutant | This study |
| N6 cheAY2 | KnrH. pylori N6 cheAY2 mutant | This study |
| N6 cheAY2 cheY1 | Knr CmrH. pylori N6 cheAY2 cheY1 mutant | This study |
| SS1 cheY1 | KnrH. pylori SS1 cheY1 mutant | This strain |
| SS1 cheAY2 | KnrH. pylori SS1 cheAY2 mutant | This study |
| Plasmids | ||
| pUC19 | Apr | Pharmacia |
| pJMK30 | Knr; source of CmrBamHI cassette | 36 |
| pCAT | Cmr; source of CmrBamHI cassette | C. Clayton |
| pCY | pUC19 plus 0.3-kb PCRDOP gene fragment of H. pylori cheY1 | This study |
| pCY110 | pUC19 plus 0.94-kb PCR gene fragment of cheY1 | This study |
| pCYIP | pCY110 with 20-bp deletion in cheY1 | This study |
| pCYIPk | pCYIP plus Knr | This study |
| pILLCA | pILL575 plus 6-kb H. pylori 11637 chromosomal fragment containing cheA | A. Labigne |
| pCA110 | pUC19 plus 0.71-kb PCR gene fragment of cheA | This study |
| pCAIP | pCA110 with 21-bp deletion in cheA | This study |
| pCAIPk | pCAIP plus Knr | This study |
| pCAIPc | pCAIP plus Cmr | This study |
| pCF | pUC19 plus 0.3-kb PCR gene fragment of cheY2 | This study |
| pCFIP | pCF with 5-bp deletion in cheY2 | This study |
| pCFIPk | pCFIP plus Knr | This study |
| pRS1 | pUC19 plus 0.3-kb PCR gene fragment of cheY1 | This study |
| pRS1-TI | pRS1 with 20-bp deletion in cheY1 | This study |
| pRS1-TIK | pRS1-TI plus Knr | This study |
| pSF6 | pUC19 plus 0.71-kb PCR gene fragment of cheA | This study |
| pSF6-TI | pSF6 with 21-bp deletion in cheA | This study |
| pSF6-TIK | pSF6-TI plus Knr | This study |
Apr, ampicillin resistant; Knr, kanamycin resistant; Cmr, chloramphenicol resistant.
DNA manipulations.
Unless otherwise stated, plasmid and chromosomal DNA extractions, restriction enzyme digestions, and DNA ligations were performed by standard procedures (29) using enzymes supplied by Promega (Southampton, United Kingdom). Transformations into E. coli XL2-Blue MRF′ strain (Stratagene Europe, Amsterdam, The Netherlands) were performed following the manufacturer's protocol. All chemicals were purchased from Sigma. The oligonucleotide primers used for PCRs were purchased from Genosys Biotechnologies (Europe) Ltd. (Cambridge, United Kingdom) and are summarized in Table 2. Sequencing of cloned DNA was performed by the dideoxynucleotide chain termination method with a PRISM sequencing kit (Applied Biosystems, Warrington, United Kingdom).
TABLE 2.
Oligonucleotides used for PCR
| Oligonucleotide | Method | Strand | Sequence (5′-3′)a |
|---|---|---|---|
| DOP1 | PCRDOPb | − | AAAAAGCTTNGKNGGRTTRAANGGYTT |
| DOP2 | PCRDOP | + | GCGCTGCAGTWYTWDTWGTRTAWGAT |
| SPCY1 | PCR | − | GCTTCTAATGCTGAGAT |
| SPCY2 | PCR | + | AACCATCCAATGACCCT |
| SPCY3 | PCR | + | TAAAAGGAGAAGCGC |
| SPCY4 | PCR | − | CATTGGCTTTAACACTC |
| IPCY1 | IPCRM | − | TCAGATCTGTTCATTTCAGGCAT |
| IPCY2 | IPCRM | + | CGAGATCTAGGTGCGCTCCGATAGC |
| SPCA1 | PCR | + | AGAACCCTGTGATGCTTAAA |
| SPCA2 | PCR | − | GGGATTCGTTCATCAA |
| IPCA1 | IPCRM | − | CAAGATCTAATCTACACCGTTGATG |
| IPCA2 | IPCRM | + | GCAGATCTTTGTGACTTCTTTCGCCC |
| RS1F | PCR | + | TAAAAGGAGAAGCGC |
| RS1R | PCR | − | CATTGGCTTTAACACTC |
| SF6F | PCR | + | AGAACCCTGTGATGCTTAAA |
| SF6R | PCR | − | GGGATTCGTTCATCAA |
| RS1 TIF | IPCRM | + | GCGAGATCTGGTGCGCTCCGATAG |
| RS1 TIR | IPCRM | − | GCGAGATCTGTTCATTTCAGGCAT |
| SF6 TIF | IPCRM | + | CAAGATCTAATCTACACCGTTGATG |
| SF6 TIR | IPCRM | − | GCAGATCTTTGTGACTTCTTTCGCCC |
Underlined nucleotides represent HindIII (DOP1), PstI (DOP2), and BglII (IPCY1, IPCY2, IPCA1, IPCA2, RS1 TIF, RS1 TIR, SF6 TIF, and SF6 TIR) restriction endonuclease sites.
PCRDOP, PCR with degenerate oligonucleotide primers.
Identification and cloning of H. pylori cheY1, cheA, and cheY2 gene fragments.
The H. pylori cheY1 gene was identified by PCR (34) with degenerate primers DOP1 and DOP2, designed against the conserved regions of known bacterial response regulator genes. The amplified putative cheY1 gene fragment was cloned into pUC19, sequenced, and used to probe a λZAP library NCTC 11638 to identify the entire cheY1 gene sequence. Specific primers SPCY1 and SPCY2 were used to amplify a PCR product containing the entire cheY1 gene, which was cloned into pUC19. The H. pylori cheA gene was identified by partial sequencing of plasmid pILLCA, which contained a putative cheA gene on a 6-kb fragment from H. pylori 11637. Specific primers SPCA1 and SPCA2 were used to amplify a fragment of cheA, which was cloned into pUC19. Following the release of the H. pylori 26695 genome sequence, specific primers SPF1 and SPF2 were designed to amplify the cheY2 gene from H. pylori 11637 chromosomal DNA, which was cloned into pUC19.
Construction of defined H. pylori cheY1, cheAY2, cheY2, and cheAY2 cheY1 mutants.
Defined deletions and unique BglII sites were introduced into the cloned cheY1, cheA, and cheY2 genes by inverse PCR mutagenesis (IPCRM) using the primer pairs shown in Table 2, as described previously (7, 40). A 1.4-kb BamHI restriction fragment of plasmid pJMK30, containing a gene encoding resistance to kanamycin (aph3′-III) (11), was cloned into the unique BglII sites. The constructs were introduced into H. pylori N6 or SS1 wild-type strain either by natural transformation (14) or by electroporation (31). For the construction of a double mutant, a 0.8-kb HincII restriction fragment of plasmid pCAT, containing a gene encoding resistance to chloramphenicol, was cloned into the unique BglII site in pCAIP2, which contains the mutated cheA gene fragment. The resulting construct pCAIPC was electroporated into N6 cheY1 cells, and putative double mutants were selected on DENT plates containing both kanamycin and chloramphenicol.
Motility and chemotaxis assays.
Bacterial motility was assayed on 0.27% agar plates containing Mueller-Hinton broth supplemented with 10% FCS. Plates were seeded with 10 μl of overnight broth culture, and the plates were incubated for 2 to 3 days at 37°C. Results were recorded on the basis of the swarm diameter.
To analyze free-swimming cells, a Hobson BacTracker computerized video tracking system (Hobson Tracking Systems Ltd., Sheffield, United Kingdom) was used. Motile cells grown in culture to mid-log phase (optical density at 600 nm [OD600] ≈ 0.4) were drawn into 100-μm-diameter optically flat microslides (Camlab Ltd., Cambridge, United Kingdom), and one end was sealed with vinyl plastic putty (Critoseal, Hawksley, United Kingdom) to prevent bacterial cells from drifting. Slides were observed with a Zeiss Standard 14 phase-contrast microscope at 37°C to confirm cells were motile. Free-swimming tracks were determined by motion analysis using the Hobson BacTracker system. This system provides detailed analysis of various parameters to describe motility of the bacteria, including the curvilinear velocity (CLV; the speed of the bacterium along its path) and the straight line velocity (SLV; the speed of the bacterium in a straight line from the beginning to the end of its path). The ratio of the SLV to the CLV times 100 (SLV/CLV × 100) yields a value called the track linear percentage (TL%). The more curved the route the bacterium takes, the greater will be the CLV. For a bacterium that swims in an absolute straight line, this value may approach 100%. Individual free-swimming cells were monitored for ∼2 s, and the mean values of 100 to 200 tracks were determined for duplicate samples of at least 6 replicates.
The ability of H. pylori N6 wild type and chemotaxis mutants to respond to hog gastric mucin (HGM) was compared by using Adler's capillary assay (1). Bacterial strains were grown overnight in broth to log phase. The OD600 values of the cultures were recorded, and bacterial motility was checked by microscopic analysis. The bacterial cells were harvested by centrifugation (13,000 rpm for 1 min) and resuspended in chemotaxis buffer (0.2 M Na2HPO4, 0.1 M citric acid) to 106 cells per ml (OD600 ≈ 0.1). Soluble HGM (Sigma) was prepared as 1, 0.5, and 0.1% solutions in chemotaxis buffer. The tips of 50-μl-volume capillary tubes (Sigma) were then filled with HGM or chemotaxis buffer (control), sealed at one end, and inserted vertically into 0.5-ml tubes containing 300 μl of resuspended motile cells. These were incubated horizontally under microaerophilic conditions for 45 min at 37°C. After incubation, the tubes were disassembled, and the lower 10 mm of liquid content discarded. The number of bacteria remaining in each capillary tube was then determined by performing viable counts. All assays were performed in triplicate on at least three separate occasions. The results were expressed as the chemotaxis ratio Rche ([CFU/ml in taxin capillary]/[CFU/ml in control capillary]) to normalize experimental data (27).
Colonization of H. pylori gnotobiotic piglet model.
Gnotobiotic piglet experiments were carried out essentially as described by Krakowka et al. (20). Large white hybrid piglets were delivered by cesarian section performed in a sterilized isolator unit. The piglets were maintained in sterile isolator units, and rectal swabs were cultured from the piglets to demonstrate sterility before inoculation. To suppress secretion of stomach acid, piglets were given 40 mg of cimetidine (Tagamet; SmithKline Beecham, Brentford, United Kingdom) orally 1 h before inoculation of bacterial suspensions. This was repeated 6 h after inoculation. Animals were challenged at 2 days of age with 2 ml of 109 CFU of H. pylori N6 or N6 cheY1 grown in individual broth cultures for 24 h. Seven days after infection, the mucosa from a portion of the stomach was removed, weighed, and homogenized, and the extent of the bacterial colonization was quantified by performing viable counts.
Colonization of H. pylori mouse model.
Female outbred mice (HSD/ICR strain; Harlan Ltd., Bicester, United Kingdom) with a body weight of approximately 20 g (4 to 6 weeks old) were challenged orally on successive days with SS1, SS1 cheY1, or SS1 cheAY2. Prior to challenge, all strains were pretreated with acidified 5 mM urea (pH 2) in order to boost urease activity and thus optimize colonization potential (25). Challenge inocula were 1-ml volumes of 24-h tryptose soya broth cultures containing between 1 × 107 and 1 × 108 CFU. At 2 and 8 weeks, 10 mice from each group were culled by CO2 inhalation, and the stomachs were removed and opened along the greater curvature. After washing away the stomach contents, the entire mucosal surface was spread evenly over the surface of a Columbia chocolate agar plate containing selective antibiotics (amphotericin B [50 μg/ml], vancomycin [100 μg/ml], polymyxin B [3.3 μg/ml], bacitracin [200 μg/ml], and nalidixic acid [10.7 μg/ml]) for about 10 s (25) before incubating microaerobically for 7 days at 37°C. The culture plates were then evaluated for H. pylori growth. Growth of even a single colony is sufficient to record an animal as being H. pylori positive (25). After 8 weeks, the mice were exsanguinated and the individual serum samples were stored at −20°C.
Whole-cell serum ELISA assay.
H. pylori SS1 cells were harvested from DENT agar plates, washed twice with phosphate-buffered saline (PBS), and lysed with three 30-s bursts of ultrasound (Ultrasonic Processor; Jencons Scientific Ltd., Leighton Buzzard, United Kingdom) with a 30-s cooling period on ice between each burst. The insoluble material was removed (10,000 × g for 20 min), and the soluble material was used to coat wells of an enzyme immunoassay-radioimmunoprecipitation 96-well plate (Corning Costar, High Wycombe, United Kingdom) for 18 h at 4°C (1 μg/well in 0.1 M NaHCO3, pH 9.5). The antibody levels within individual serum samples were determined by end point titration, as described previously (8). Essentially, antigen-coated wells were incubated with serum samples serially diluted twofold in PBS, and bound antibody was visualized by using a polyvalent anti-mouse immunoglobulin horseradish peroxidase conjugate (Sigma) and o-phenylenediamine as a substrate. Enzyme-linked immunosorbent assay (ELISA) titers were determined as the reciprocal of the highest serum dilution that yielded an OD490 value of 0.5 U above the background. All titers were standardized against an anti-H. pylori whole-cell antiserum. Unpaired Student's t tests were used to compare the data groups. Probabilities of P < 0.05 were taken as significant. Statistical analysis was carried out with the InStat statistical package (Sigma).
RESULTS
Cloning of chemotaxis genes.
PCR with degenerate primers was used to amplify a cheY1 gene fragment from H. pylori 11637 chromosomal DNA (39). The PCR product was cloned into pUC19 and sequenced. To isolate the entire cheY1 gene, a λZAP library NCTC 11638 was screened by radioactive colony blot hybridization using the isolated gene fragment as a probe. A positive clone was identified and partially sequenced. Specific primers were designed to amplify a 941-bp fragment containing the entire cheY1 plus some flanking DNA, which was cloned into pUC19 and termed pCY110. Searches using BLASTX software (3) revealed that the cloned fragment had a significant identity to CheY from several bacteria. Library screening and subsequent sequencing revealed that the complete CheY1 codes for a protein of 124 amino acids with the highest homology (82% identity) to Campylobacter jejuni CheY (41). The four residues found in all CheY proteins to date (Asp12, Asp13, Asp56, and Lys109) are all conserved in H. pylori CheY1 (30). Asp56 is the site of phosphorylation by CheA. Amino acids 90 to 112 are also highly conserved, an area of predicted interaction between CheY-P and FliM in E. coli (38). The EMBL accession number for the nucleotide sequence of H. pylori cheY1 is X81897.
Partial sequencing of pILLCA and subsequent searches revealed that there was extensive homology to E. coli CheA. Specific primers were designed to amplify a 707-bp fragment from the putative cheA gene, which was cloned into pUC19 and termed pCA110. The highest level of homology (50% identity) was with Thermotoga maritima CheA. Comparison of the H. pylori 11637 cheA gene fragment with H. pylori 26695 cheA (HP0392) showed that the CheA proteins from the two strains were almost identical in the region 309 to 585 (H. pylori 26695 amino acid numbering) (35). Analysis of the whole CheA amino acid sequence from 26695 revealed a high level of homology to CheA histidine protein kinases from a range of enteric bacteria. The highest level of homology (47% identity) was with Pseudomonas putida CheA. The CheA active domains are conserved, including the area surrounding His48, the site of autophosphorylation, and the four blocks of residues involved in kinase function found at the carboxyl end, which suggests that H. pylori CheA is functional as a transmitter. The main area of divergence between H. pylori CheA and E. coli CheA was between amino acids 109 and 260 (E. coli numbering). This region contains the P2 domain (124 to 257), which has CheY binding capability (33).
The cheY2 portion of cheAY2 was cloned with sequence information from the annotated H. pylori 26695 genome (35). The four conserved CheY residues are also conserved in H. pylori CheY2 (30). BLASTX searches revealed that the highest homology (40% identity) was to the N terminus of the bifunctional CheAY proteins from Streptomyces coelicolor and P. aeruginosa.
Construction of defined H. pylori cheY1, cheAY2, cheY2, and cheAY2 cheY1 mutants.
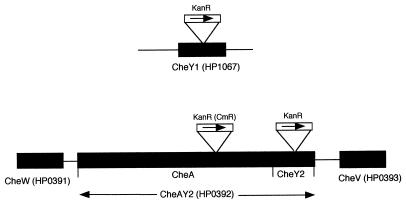
Defined deletions were introduced into the H. pylori cheY1, cheA, and cheY2 cloned gene fragments by IPCRM, followed by the insertion of a kanamycin or chloramphenicol resistance cassette (Fig. 1) (7, 40). Mutation of the cheA section of the cheAY2 gene results in a cheAY2 phenotype, as the cheY2 section is downstream of the cheA section. H. pylori cheY1, cheAY2, cheY2, and cheAY2 cheY1 mutants were constructed by allelic replacement, as described previously (12, 17). PCR using specific primer pairs and Southern hybridization analysis confirmed that double recombination events had occurred (data not shown).
FIG. 1.
Schematic representation of H. pylori chemotaxis loci. KanR and CmR are the relative insertion positions of the kanamycin and chloramphenicol resistance cassettes, respectively, with the arrows indicating the direction of transcription. HP gene designations are based on the genome sequence of H. pylori 26695 (35).
Motility assays.
Analysis of the swarm plates showed that all the H. pylori N6 and SS1 chemotaxis mutants had reduced swarming ability compared to the abilities of the respective wild-type strains. The wild-type strain formed concentric rings that increased with the period of incubation. In contrast, the mutants formed irregular growth patterns of high density limited to the area of inoculation (data not shown).
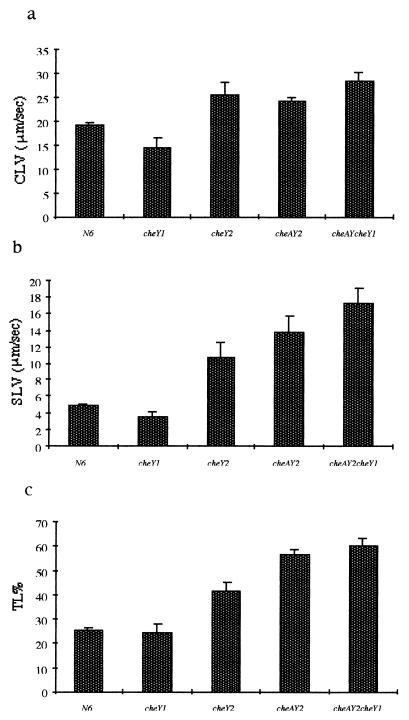
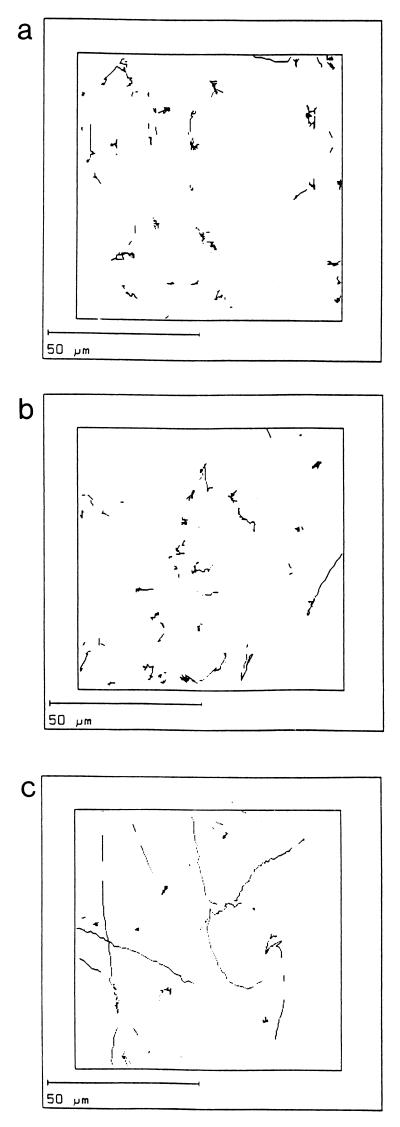
Computerized tracking showed that the H. pylori N6 wild-type strain moved with a speed of up to 20 μm/s, consistent with results reported by Karim et al. on several clinical isolates (18). In comparison, three of the deletion mutants (N6 cheY2, N6 cheAY2, and N6 cheAY2 cheY1) had significantly higher CLVs and SLVs than those observed for the wild-type strain. However, N6 cheY1 had lower CLVs and SLVs than those observed for N6 (Fig. 2a and b). The TL%s indicate that the mutant strains N6 cheY2, N6 cheAY2, and N6 cheAY2 cheY1 are significantly straighter swimming than the wild type, N6 (Fig. 2c). The linearity of these mutant strains suggests that both CheY2 and CheA contribute to tumbling motion. In contrast, the linearity of the N6 cheY1 mutant was less than that observed for the wild-type strain. Analysis of the trail draw diagrams showed that the wild-type H. pylori N6 strain moved in a random darting fashion, with frequent changes in direction and short straight runs. The mutants N6 cheY2, N6 cheAY2, and N6 cheAY2 cheY1 all moved in long straight runs or very wide circles with no sharp turns or changes in direction. In contrast, N6 cheY1 tumbled excessively, rarely moving out of the field of vision (Fig. 3a to c).
FIG. 2.
Bar charts showing data obtained from Hobson BacTracker analysis. The CLV (a), SLV (b), and TL% (c) are shown for the wild-type H. pylori N6 strain and the N6 cheY1, N6 cheY2, N6 cheAY2, and N6 cheAY2 cheY1 strains. Error bars show the standard errors.
FIG. 3.
Trail draws, showing the paths of individual bacteria tracked by computer, for the wild-type N6 (a), N6 cheY1 (b), and N6 cheAY2 cheY1 (c) strains.
Capillary tube assays.
Adler's capillary assay (1), a standard for quantitative assessment of chemotactic proficiency in enteric bacteria, was carried out to study in greater detail the phenotype of the chemotaxis deletion mutants. It has been shown that this method is applicable to H. pylori (26, 37). H. pylori N6 showed significant chemotaxis to 0.1, 0.5, and 1% (wt/vol) HGM (Table 3). By contrast, the N6 cheY1 mutant failed to show significant taxis to 0.1% HGM, but at concentrations of 0.5 and 1% HGM, chemotactic responses representing, respectively, 90 and 86% reductions in chemotaxis were observed (Table 3). The N6 cheAY2 mutant showed a significant chemotactic response only to 1% HGM (Table 3), representing an 82% reduction in chemotaxis. No response was observed with either N6 cheY2 or N6 cheAY2 cheY1, except at the highest concentration of HGM (Table 3). However, the ratios observed were too close to the minimum Rche value of 2 to be considered significant (Table 3).
TABLE 3.
Response of H. pylori N6 and chemotaxis mutant strains to HGM analyzed by capillary assaysa
| Strain | Chemotactic responses to:
|
||
|---|---|---|---|
| 0.1% HGM | 0.5% HGM | 1% HGM | |
| N6 | 16.96 ± 7.37 | 55.65 ± 6.08 | 67.69 ± 16.33 |
| N6 cheY1 | NR | 5.4 ± 0.84 | 9.33 ± 1.88 |
| N6 cheY2 | NR | NR | 2.07 ± 0.65 |
| Ne cheAY2 | NR | NR | 11.95 ± 5.41 |
| Ne cheAY2 cheY1 | NR | NR | 2.08 ± 0.4 |
Each data set represents a minimum of three experiments performed in triplicate. Results are expressed as the ratio of number of bacteria in attractant capillaries to that in the control capillary (Rche). NR, no response.
Colonization of H. pylori gnotobiotic piglet model.
Colonization of piglets by the wild-type N6 strain was approximately 105 CFU/g of gastric mucosa, consistent with previous colonization studies with this strain (9). By contrast, two independent N6 cheY1 mutants failed to colonize the gnotobiotic piglets challenged.
Colonization of H. pylori in mice and anti-H. pylori serum responses.
Three groups of 20 mice were infected with 1-ml volumes of overnight cultures of SS1 (1.05 × 108, 6.4 × 107), SS1 cheY1 (8.9 × 107, 7.0 × 107), or SS1 cheAY2 (1.1 × 108, 7.9 × 107) (the number of viable bacteria administered to each mouse on successive days is in parentheses). All mice inoculated with SS1 were colonized at 2 and 8 weeks postinfection. Using the scoring system for colonization described previously, the 2-week time point showed 58% colonization, and the 8-week time point showed 94% colonization (25). This is indicative of growth in vivo. However, mice inoculated with SS1 cheY1 or SS1 cheAY2 showed no colonization at either 2 or 8 weeks postinfection.
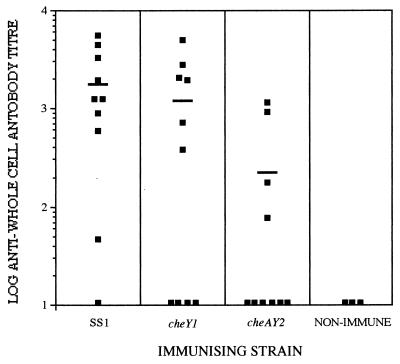
Serum harvested from individual mice 8 weeks after oral inoculation with SS1, SS1 cheY1, and SS1 cheAY2 was analyzed for the presence of anti-H. pylori antibodies by ELISA. All strains tested generated an antibody response that was significantly (P < 0.05) higher than the titer seen in the control mice (Fig. 4). Six out of the 10 mice inoculated with SS1 cheY1 generated a significant anti-H. pylori serum response (Fig. 4). This response was not significantly different (P > 0.05) than that observed for the mice challenged with the SS1 wild-type strain, of which 9 out of the 10 mice challenged generated significant anti-H. pylori serum responses. Four of the mice challenged with SS1 cheAY2 positively seroconverted. This response was significantly lower (P < 0.05) than that observed for the SS1 and SS1 cheY1 strains.
FIG. 4.
Serum anti-H. pylori SS1 whole-cell immunoglobulin responses 8 weeks postinfection. End point antibody titers from individual mice are shown. The bars denote the mean antibody responses.
DISCUSSION
H. pylori cells reside mainly in the mucous layer of the stomach or in the intestine in association with areas of gastric metaplasia. The ability to direct bacterial movement against the gastric flow towards the epithelial cell surface via chemotaxis is likely to be important in the colonization process. To determine the roles of the chemotaxis orthologues identified in the H. pylori 26695 genome sequence in motility and pathogenesis, defined cheY1, cheY2, and cheAY2 mutants were constructed in two independent strains (N6 and SS1) by using IPCRM and allelic replacement (12, 17). An H. pylori N6 cheAY2 cheY1 double mutant was also constructed.
Computerized tracking showed that H. pylori N6 moved with a speed of up to 20 μm/s, consistent with that reported by Karim et al. on several clinical isolates (18). The swimming pattern consisted of random darting movements, with frequent changes in direction and short runs. N6 cheY2, N6 cheAY2, and N6 cheAY2 cheY1 all moved in long straight runs or wide circles with no sharp turns or changes in direction. This suggests that, as in E. coli, CheY2 phosphorylated by CheA interacts with the flagellar motor switch, resulting in tumbling of H. pylori cells. This proposed system is further supported by the presence of a soluble MCP-like orthologue (HP0599) in H. pylori, which would allow the formation in the cytoplasm of a complex between CheW-CheAY2 and the truncated soluble MCP orthologue, thus allowing communication with the polar-located flagella. N6 cheY1 exhibited a tumbling phenotype closer to that of the wild type with respect to the frequency of directional changes. Swarming was not observed for the N6 cheY1 strain; therefore, the tumbling phenotype is unlikely to be due to suppression mutations in cheY1.
The recent sequencing of the C. jejuni 11168 genome has identified a similar configuration with a separate CheY and bifunctional CheAY orthologues (The Sanger Centre Campylobacter jejuni genome project [http://www.sanger.ac.uk /Project/C_jejuni/]). The H. pylori and C. jejuni CheY1 and CheY2 proteins show high sequence similarities (82 and 65%, respectively), suggesting that both proteins were derived from a common ancestral protein. The conservation of two divergent proteins suggests that these proteins have evolved vital functions. It is possible that both proteins are phosphorylated by CheA, which would explain the divergence in the P2 region between H. pylori and E. coli CheA proteins. CheY1-P and CheY2-P could then interact with a different site on the flagellar motor switch complex. Alternatively, CheY1 may act as a phosphate sink, accelerating the dephosphorylation of CheY2-P, thereby helping to terminate the clockwise tumbling response. This is consistent with the absence of an H. pylori CheZ orthologue, as CheZ accelerates the dephosphorylation of CheY-P in other bacteria (10), and also with the tumbling phenotype of N6 cheY1, which is similar to the phenotype observed for an E. coli cheZ mutant (16).
The small but significant chemotactic response observed for N6 cheAY2 reveals the importance of a functional CheY1 for the full chemotactic response of H. pylori N6 to HGM. This response in the absence of a functional CheA suggests that the CheY domains of the CheV orthologues may be phosphorylated by an alternative pathway in response to high levels of mucin. Alternatively, the CheY domains may be directly phosphorylated by small molecules linked to metabolism. In other bacterial systems, there is evidence that chemotaxis to dominant chemoattractants requires the transport into the cell and partial metabolism of these chemoattractants. In H. pylori, mucin may need to be transported into the cell in order to be recognized by the soluble MCP-like orthologue HP0599. This is consistent with the study of Nakamura et al. (28), which demonstrated that cytoplasmic urease activity was more important than external urease activity in chemotaxis. These results demonstrate that mucin is a chemoattractant for H. pylori N6 and that the chemotaxis components CheY1 and CheAY2 are involved in motility towards the mucus in the stomach.
N6 and SS1 cheY1 mutants were unable to colonize either gnotobiotic piglets or mice, respectively. Mutation of cheAY2 in H. pylori SS1 also prevented colonization of mice. Similar observations have been reported for a C. jejuni cheY (cheY1) mutant, which had a reduced ability to colonize mice and to cause disease in ferrets (41). The chemotaxis mutants of H. pylori N6 are motile, but their swimming behavior is altered; N6 cheY1 exhibits increased tumbling, whereas N6 cheAY2 swims in straight lines. In addition, their chemotactic response to mucin was significantly reduced compared with that of the N6 wild-type strain.
Mutations in the chemotaxis system would appear to affect the ability of H. pylori to move in a controlled fashion towards the gastric mucous layer in the stomach. However, serology responses observed 8 weeks after infection with SS1 cheY1 were not significantly different from the responses of mice to the wild-type strain. Significant responses were also observed in mice immunized with SS1 cheAY2. These findings suggest that chemotaxis is unnecessary for viability in vivo. It may be that significant numbers of chemotaxis-impaired bacteria remain in the mouse stomach for several days, but because they cannot maintain their position or penetrate the gastric mucus, they are eventually washed out of the stomach with the gastric flow. Studies on C. jejuni have led to the proposal that active motility combined with chemotaxis should be regarded as a potential alternative to specific attachment (22). H. pylori adherence to epithelial cells is thought to produce attachment/effacement similar to that seen in the enteropathogenic E. coli EPEC strains (32). However, it has been proposed that only a small proportion of H. pylori cells, between 1 and 5%, attach to the epithelial surface (19). Full motility is essential for H. pylori cells to colonize the gastric mucosa (6, 9). A fully functional chemotaxis system must be required for colonization by maintaining H. pylori in the mucous layer close to the epithelial cell surface, thus reducing removal of bacteria from the stomach by gastric flow.
In this study we have demonstrated that CheY1 and CheAY2 are necessary for flagellum-regulated movement and chemotaxis towards mucin. Additionally, the importance of chemotaxis in the pathogenicity of H. pylori has been demonstrated in two animal models. Chemotaxis in H. pylori appears to be distinct from the Salmonella serovar Typhimurium and E. coli paradigm. The results of this study provide the framework for the full elucidation of the complex chemotaxis system of H. pylori.
ACKNOWLEDGMENTS
We gratefully acknowledge Lynne Batty for technical assistance and Agnes Labigne, Chris Clayton, and Richard Ferrero for the generous gifts of pILLCA, pCAT, and SS1, respectively.
This work was supported by the Medical Research Council and the Joint Research Board of St. Bartholomew's Hospital.
REFERENCES
- 1.Adler J. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J Gen Microbiol. 1973;74:77–91. doi: 10.1099/00221287-74-1-77. [DOI] [PubMed] [Google Scholar]
- 2.Allan E. Identification and characterisation of potential pathogenicity determinants of Helicobacter pylori. Ph.D. thesis. London, United Kingdom: University of London; 1997. [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Beier D, Spohn G, Rappuoli R, Scarlato V. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J Bacteriol. 1997;179:4676–4683. doi: 10.1128/jb.179.15.4676-4683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaser M J. Ecology of Helicobacter pylori in the human stomach. J Clin Investig. 1997;100:759–762. doi: 10.1172/JCI119588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danon S J, Eaton K A. Isogenic flagella mutants of Helicobacter pylori: candidates for an attenuated vaccine? Gut. 1998;43(Suppl. 2):A38. [Google Scholar]
- 7.Dorrell N, Gyselman V G, Foynes S, Li S R, Wren B W. Improved efficiency of inverse PCR mutagenesis (IPCRM) BioTechniques. 1996;21:604–608. doi: 10.2144/96214bm07. [DOI] [PubMed] [Google Scholar]
- 8.Douce G, Fontana M, Pizza M, Rappuoli R, Dougan G. Intranasal immunogenicity and adjuvanticity of site-directed mutant derivatives of cholera toxin. Infect Immun. 1997;65:2821–2828. doi: 10.1128/iai.65.7.2821-2828.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eaton K A, Suerbaum S, Josenhans C, Krakowka S. Colonization of gnotobiotic piglets by Heliobacter pylori deficient in two flagellin genes. Infect Immun. 1996;64:2445–2448. doi: 10.1128/iai.64.7.2445-2448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenbach M. Control of bacterial chemotaxis. Mol Microbiol. 1996;20:903–910. doi: 10.1111/j.1365-2958.1996.tb02531.x. [DOI] [PubMed] [Google Scholar]
- 11.Ferrero R L, Cussac V, Courcoux P, Labigne A. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J Bacteriol. 1992;174:4212–4217. doi: 10.1128/jb.174.13.4212-4217.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foynes S, Dorrell N, Ward S J, Zhang Z W, McColm A A, Farthing M J G, Wren B W. Functional analysis of the roles of FliQ and FlhB in flagellar expression in Helicobacter pylori. FEMS Microbiol Lett. 1999;174:33–39. doi: 10.1111/j.1574-6968.1999.tb13546.x. [DOI] [PubMed] [Google Scholar]
- 13.Fredrick K L, Helmann J D. Dual chemotaxis signaling pathways in Bacillus subtilis: a ςD-dependent gene encodes a novel protein with both CheW and CheY homologous domains. J Bacteriol. 1994;176:2727–2735. doi: 10.1128/jb.176.9.2727-2735.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas R, Meyer T F, Vanputten J P M. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol Microbiol. 1993;8:753–760. doi: 10.1111/j.1365-2958.1993.tb01618.x. [DOI] [PubMed] [Google Scholar]
- 15.Hazell S L, Lee A, Brady L, Hennessy W. Campylobacter pyloridis and gastritis: association with intercellular spaces and adaptation to an environment of mucus as important factors in colonization of the gastric epithelium. J Infect Dis. 1986;153:658–663. doi: 10.1093/infdis/153.4.658. [DOI] [PubMed] [Google Scholar]
- 16.Hess J F, Bourret R B, Simon M I. Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature. 1988;336:139–143. doi: 10.1038/336139a0. [DOI] [PubMed] [Google Scholar]
- 17.Jenks P J, Foynes S, Ward S J, Constantinidou C, Penn C W, Wren B W. A flagellar-specific ATPase (FliI) is necessary for flagellar export in Helicobacter pylori. FEMS Microbiol Lett. 1997;152:205–211. doi: 10.1111/j.1574-6968.1997.tb10429.x. [DOI] [PubMed] [Google Scholar]
- 18.Karim Q N, Logan R P H, Puels J, Karnholz A, Worku M L. Measurement of motility of Helicobacter pylori, Campylobacter jejuni and Escherichia coli by real time computer tracking using the Hobson BacTracker. J Clin Pathol. 1998;51:623–628. doi: 10.1136/jcp.51.8.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirschner D E, Blaser M J. The dynamics of Helicobacter pylori infection of the human stomach. J Theor Biol. 1995;176:281–290. doi: 10.1006/jtbi.1995.0198. [DOI] [PubMed] [Google Scholar]
- 20.Krakowka S, Morgan D R, Kraft W G, Leunk R D. Establishment of gastric Campylobacter pylori infection in the neonatal gnotobiotic piglet. Infect Immun. 1987;55:2789–2796. doi: 10.1128/iai.55.11.2789-2796.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuipers E J. Helicobacter pylori and the risk and management of associated diseases: gastritis, ulcer disease, atrophic gastritis and gastric cancer. Aliment Pharmacol Ther. 1997;11:71–88. doi: 10.1046/j.1365-2036.11.s1.5.x. [DOI] [PubMed] [Google Scholar]
- 22.Lee A, O'Rourke J L, Barrington P J, Trust T J. Mucus colonization as a determinant of pathogenicity in intestinal infection by Campylobacter jejuni: a mouse cecal model. Infect Immun. 1986;51:536–546. doi: 10.1128/iai.51.2.536-546.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee A, O'Rourke J, DeUngria M C, Robertson B, Daskalopoulos G, Dixon M F. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 24.Maddock J R, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 25.McColm A A. Nonprimate animal models of H. pylori infection. In: Clayton C L, Mobley H L T, editors. Helicobacter pylori protocols. Totowa, N.J: Humana Press; 1997. pp. 235–251. [Google Scholar]
- 26.Mizote T, Yoshiyama H, Nakazawa T. Urease-independent chemotactic responses of Helicobacter pylori to urea, urease inhibitors, and sodium bicarbonate. Infect Immun. 1997;65:1519–1521. doi: 10.1128/iai.65.4.1519-1521.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moulton R C, Montie T C. Chemotaxis by Pseudomonas aeruginosa. J Bacteriol. 1979;137:274–280. doi: 10.1128/jb.137.1.274-280.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura H, Yoshiyama H, Takeuchi H, Mizote T, Okita K, Nakazawa T. Urease plays an important role in the chemotactic motility of Helicobacter pylori in a viscous environment. Infect Immun. 1998;66:4832–4837. doi: 10.1128/iai.66.10.4832-4837.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Sanders D A, Gillececastro B L, Stock A M, Burlingame A L, Koshland D E. Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J Biol Chem. 1989;264:21770–21778. [PubMed] [Google Scholar]
- 31.Segal E D, Tompkins L S. Transformation of Helicobacter pylori by electroporation. BioTechniques. 1993;14:225–226. [PubMed] [Google Scholar]
- 32.Segal E D, Falkow S, Tompkins L S. Helicobacter pylori attachment to gastric cells induces cytoskeletal rearrangements and tyrosine phosphorylation of host cell proteins. Proc Natl Acad Sci USA. 1996;93:1259–1264. doi: 10.1073/pnas.93.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stock J B, Surette M G. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1103–1129. [Google Scholar]
- 34.Suerbaum S. The complex flagella of gastric Helicobacter species. Trends Microbiol. 1995;3:168–170. doi: 10.1016/s0966-842x(00)88913-1. [DOI] [PubMed] [Google Scholar]
- 35.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L X, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzgerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 36.Trieu-Cuot P, Gerbaud G, Lambert T, Courvalin P. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 1985;4:3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner G A, Logan R P H, Chinnery R, Cockayne A, Hawkey C J, Borriello S P. Trefoil peptides are unique chemotaxins for Helicobacter pylori. Gastroenterology. 1997;112:A1107. [Google Scholar]
- 38.Volz K. Structural conservation in the CheY superfamily. Biochemistry. 1993;32:11741–11753. doi: 10.1021/bi00095a001. [DOI] [PubMed] [Google Scholar]
- 39.Wren B W, Colby S M, Cubberley R R, Pallen M J. Degenerate PCR primers for the amplification of fragments from genes encoding response regulators from a range of pathogenic bacteria. FEMS Microbiol Lett. 1992;99:287–291. doi: 10.1016/0378-1097(92)90042-m. [DOI] [PubMed] [Google Scholar]
- 40.Wren B W, Henderson J, Ketley J M. A PCR-based strategy for the rapid construction of defined bacterial deletion mutants. BioTechniques. 1994;16:994–996. [PubMed] [Google Scholar]
- 41.Yao R J, Burr D H, Guerry P. CheY-mediated modulation of Campylobacter jejuni virulence. Mol Microbiol. 1997;23:1021–1031. doi: 10.1046/j.1365-2958.1997.2861650.x. [DOI] [PubMed] [Google Scholar]