Abstract
Microtubule-associated proteins (MAPs) play essential roles in cancer development. This study aimed to identify transcriptomic biomarkers among MAP genes for the diagnosis and prognosis of lung cancer by analyzing differential gene expressions and correlations with tumor progression. Gene expression data of patients with lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) from the Cancer Genome Atlas (TCGA) database were used to identify differentially expressed MAP genes (DEMGs). Their prognostic value was evaluated by Kaplan–Meier and Cox regression analysis. Moreover, the relationships between alterations in lung cancer hallmark genes and the expression levels of DEMGs were investigated. The candidate biomarker genes were validated using three independent datasets from the Gene Expression Omnibus (GEO) database and by quantitative reverse transcription polymerase chain reaction (qRT-PCR) on clinical samples. A total of 88 DEMGs were identified from TCGA data. The 20 that showed the highest differential expression were subjected to association analysis with hallmark genes. Genetic alterations in TP53, EGFR, PTEN, NTRK1, and PIK3CA correlated with the expression of most of these DEMGs. Of these, six candidates—NUF2, KIF4A, KIF18B, DLGAP5, NEK2, and LRRK2—were significantly differentially expressed and correlated with the overall survival (OS) of the patients. The mRNA expression profiles of these candidates were consistently verified using three GEO datasets and qRT-PCR on patient lung tissues. The expression levels of NUF2, KIF4A, KIF18B, DLGAP5, NEK2, and LRRK2 can serve as diagnostic biomarkers for LUAD and LUSC. Moreover, the first five can serve as prognostic biomarkers for LUAD, while LRRK2 can be a prognostic biomarker for LUSC. Our research describes the novel role and potential application of MAP-encoding genes in clinical practice.
Keywords: diagnosis, gene expression, lung cancer, microtubule-associated proteins, prognosis
1. Introduction
Based on histology, most patients suffering from non-small cell lung cancer (NSCLC) are diagnosed with two main subtypes, namely lung adenocarcinoma (LUAD, 40%) and lung squamous cell carcinoma (LUSC, 25%) [1]. The survival rate of lung cancer remains low due to its high metastatic potential and chemotherapeutic resistance, making it the major cause of cancer-related death [2]. Moreover, the specific symptoms of lung cancer are unclear; about 70% of patients are diagnosed only at an advanced stage, which highly correlates with low 5-year survival rates [1,3]. Treatment of lung cancer depends on the type of cancer and diagnosis stage. For the NSCLC, surgical resection combined with chemotherapy or chemoradiation is recommended for the early stage; however, most of the patients undergo disease relapse, recurrence, and metastasis. The guidelines have recommended targeted therapy for the advanced stage treatment, in which molecular biomarker testing is required for appropriate drug selection [4,5]. As a drug target, the genetic alteration status of several genes, such as EGFR, ALK, KRAS, ROS1, BRAF, NTRK1/2/3, MET, and RET, was indicated as predictive biomarkers, but approximately 30% of patients do not carry these kinds of genetic alterations [4,5,6]. In daily clinical practice, only a few diagnosis biomarkers have been used, including thyroid transcription factor 1 (TTF-1) and p40 for immunohistochemistry (IHC) and cytokeratin 19 fragment (CYFRA 21-1) and carcinoembryonic antigen (CEA) for blood/serum testing. The addition of novel markers is required to improve specificity and sensitivity [7]. Prognostic prediction is important to classify the patient’s risk and decide the treatment strategy. Until now, numerous molecular biomarkers have been reported, but there is still no effective prognostic biomarker for clinical use [7]. Therefore, identifying diagnostic and prognostic biomarkers is urgently needed and may provide precise diagnostic tools and therapeutic targets to improve the clinical outcome.
Microtubule dynamics play an essential role in several cellular processes, including cell division, cell motility, cell morphology maintenance, cell signaling, and intracellular trafficking [8]. Perturbations in microtubule dynamics are tightly associated with cancer cell behaviors [9,10]. Microtubules reorganize to facilitate cancer-related activities, such as mitosis and migration, contributing to tumor growth and metastasis, respectively [9,10,11]. In addition, microtubule instability participates in signaling related to cancer cell survival, cell death, and stress response [9,12]. The expression levels and functional changes in microtubule-associated proteins (MAPs), key regulators of microtubule dynamics, were shown to correlate with the prognosis and clinical outcomes of several cancers [13,14,15]. For example, MAPs, such as tau, MAP2, and MAP4, are biomarkers for responsiveness to microtubule-targeting drugs [16,17,18]. Based on these data, MAPs are crucial for tumor development and aggressiveness. However, their roles and clinical application in lung cancer are still poorly understood.
Several MAPs have reported significantly altered expression in various cancers. However, there are a number of MAPs that differently express in each cancer type; for example, MAP2 upregulation was found in several types of carcinoma and myeloma, especially, and it was indicated as a potential tumor marker among neuronal epithelial tumor subtypes due to its limited expression in neurons. Contrastingly, MAP2 was rarely expressed in metastatic melanoma [19,20]. Upregulation of MAP4 was distinctly found in esophageal squamous cell carcinoma, prostate cancer, and lung adenocarcinoma, but its downregulation was observed in oral squamous cell carcinomas, which was associated with poor differentiation and proliferation [18,19]. Likewise, kinesins were elevated in cancers, and KIFAP3 and KIF3A were highly expressed in breast cancer, while these genes were unchanged in lung cancer as to whether there are several KIFs reported from our results [19,20]. These data have pointed out the cancer-type specific expression of MAPs and, therefore, we extensively performed transcriptomic analysis to identify their expressions relevant to lung cancer prognosis.
Bioinformatic analyses of gene expression and genetic alteration profiles obtained from international public databases, such as the TCGA program and Gene Expression Omnibus (GEO), have revealed potential biomarkers and drug targets in various cancers [21,22,23]. The differential expression of several genes has been associated with lung cancers and their specific subtypes, including LUAD and LUSC [24,25,26,27,28]. However, specific targets still need to be identified and validated, and their functions and prognostic relevance need to be examined.
The aim of this study was to investigate the differential expression of MAP genes in LUAD and LUSC by analyzing TCGA data. The association between DEMGs and lung cancer hallmark genes was evaluated, and the prognostic and predictive value of DEMGs was assessed. In addition, the potential biomarker genes were validated using three GEO datasets and quantitative reverse transcription polymerase chain reaction (qRT-PCR) tests on patient lung specimens.
2. Results
2.1. Identification of DEMGs between Tumor and Normal Lung Tissues
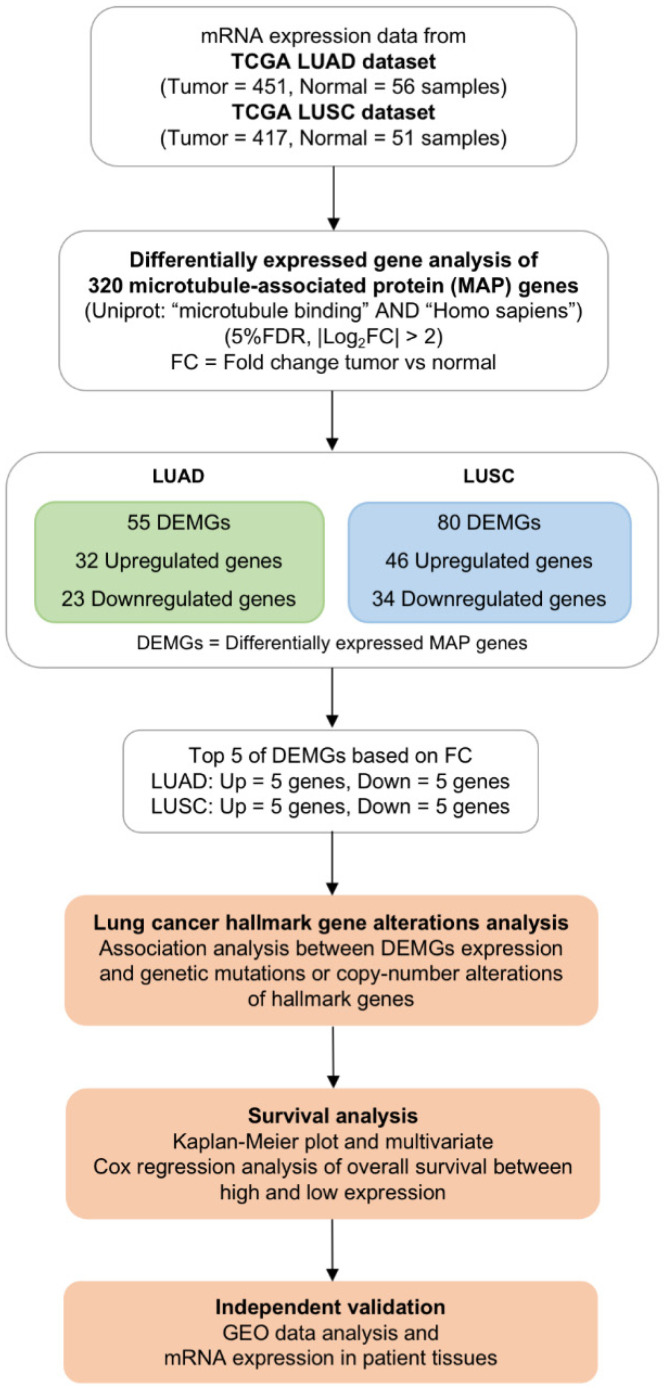
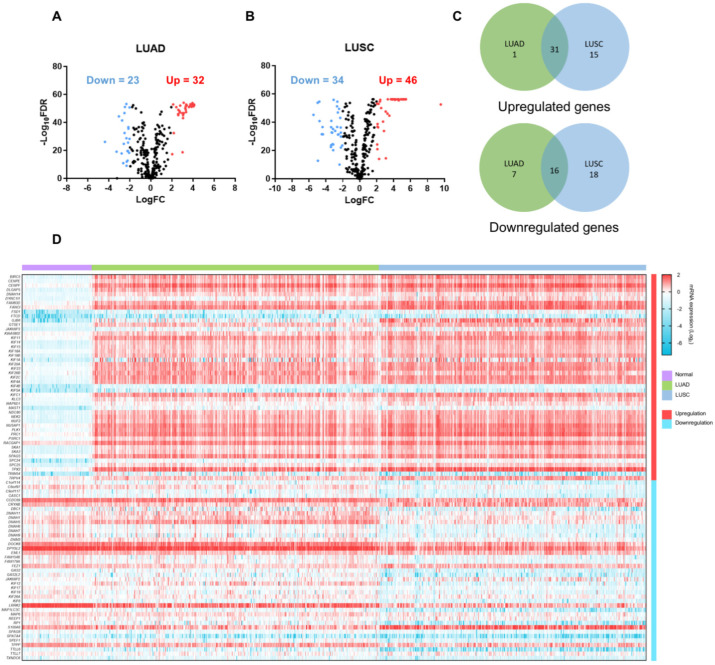
To screen for transcriptomic biomarkers, 320 MAP genes were identified from the UniProt database using the search terms “microtubule binding” AND “Homo sapiens” [29]. The workflow of this study is shown in Figure 1. Gene expression levels retrieved from the TCGA database were compared between tumor and normal lung tissues. The clinical characteristics of patients from the TCGA database are shown in Table 1. Sex, age, and race of patients were comparable between the tumor (n = 868) and normal (n = 107) groups. There were 451 LUAD and 417 LUSC tumor tissues. Volcano plots were used to display the significance of and fold change in DEMGs (FDR < 0.05, |log2FC| > 2) identified in each subtype (Figure 2A,B). Differential expression analysis of TCGA data demonstrated 32 upregulated and 23 downregulated genes in LUAD, and 46 upregulated and 34 downregulated genes in LUSC. Among DEMGs, 31 upregulated and 16 downregulated genes overlapped between the 2 subtypes (Figure 2C). The heatmap represents the expression level of each DEMG in normal, LUAD, and LUSC samples (Figure 2D, Supplementary Figure S1). The top five most upregulated genes in the LUAD dataset included NUF2, KIF4A, KIF18B, NEK2, and DLGAP5, while those in the LUSC dataset included NUF2, KIF4A, KIF18B, GJB6, and BIRC5. The top five most downregulated genes in the LUAD dataset were DNAH9, SPATA4, LRRK2, FAM154B, and REEP1, while those in the LUSC dataset were RP1, MAP1LC3C, DNAH11, LRRK2, and TTLL6. These 20 DEMGs were selected for subsequent analyses. Here, NUF2, KIF4A, and KIF18B were commonly upregulated, and LRRK2 was commonly downregulated in both lung cancer subtypes.
Figure 1.
Schematic workflow for screening microtubule-associated protein (MAP)-encoding gene biomarkers.
Table 1.
Clinical characteristics of the Cancer Genome Atlas dataset.
| Characteristic | Tumor (n = 868) |
Normal (n = 107) |
p-Value * |
|---|---|---|---|
| Sex―no. (%) | |||
| Female | 351 (40.44) | 46 (42.99) | 0.612 |
| Male | 517 (59.56) | 61 (57.01) | |
| Age―years | |||
| Median (Min, Max) | 67 (38, 88) | 67 (42, 86) | 0.330 |
| Overall Survival―no. (%) | |||
| Alive | 576 (66.36) | ||
| Deceased | 292 (33.64) | ||
| Race―no. (%) | |||
| White | 613 (87.20) | 93 (92.08) | 0.360 |
| Black or African American | 73 (10.38) | 8 (7.92) | |
| Asian | 16 (2.28) | 0 (0) | |
| American Indian or Alaskan Native | 1 (0.14) | 0 (0) | |
| Cancer subtype―no. (%) | |||
| LUAD | 451 (51.96) | ||
| LUSC | 417 (48.04) | ||
| Stage―no. (%) | |||
| I | 457 (52.65) | ||
| II | 247 (28.46) | ||
| III | 134 (15.44) | ||
| IV | 30 (3.46) | ||
* Calculated based on Mann–Whitney U test and chi-square test. Here, LUAD is lung adenocarcinoma; LUSC is lung squamous cell carcinoma.
Figure 2.
Differentially expressed MAP genes (DEMGs). Volcano plots depict the DEMGs in (A) lung adenocarcinoma (LUAD) and (B) lung squamous cell carcinoma (LUSC). Red indicates upregulated genes with log fold change (log2FC) > 2; blue indicates downregulated genes with log2FC < −2, false discovery rate (FDR) < 0.05. (C) Venn diagrams represent the intersection of upregulated or downregulated genes between LUAD and LUSC. (D) Heatmap represents the mRNA expression (log2) of the DEMGs in normal, LUAD, and LUSC samples.
2.2. Association between DEMG Expressions and Lung Cancer Hallmark Gene Alterations
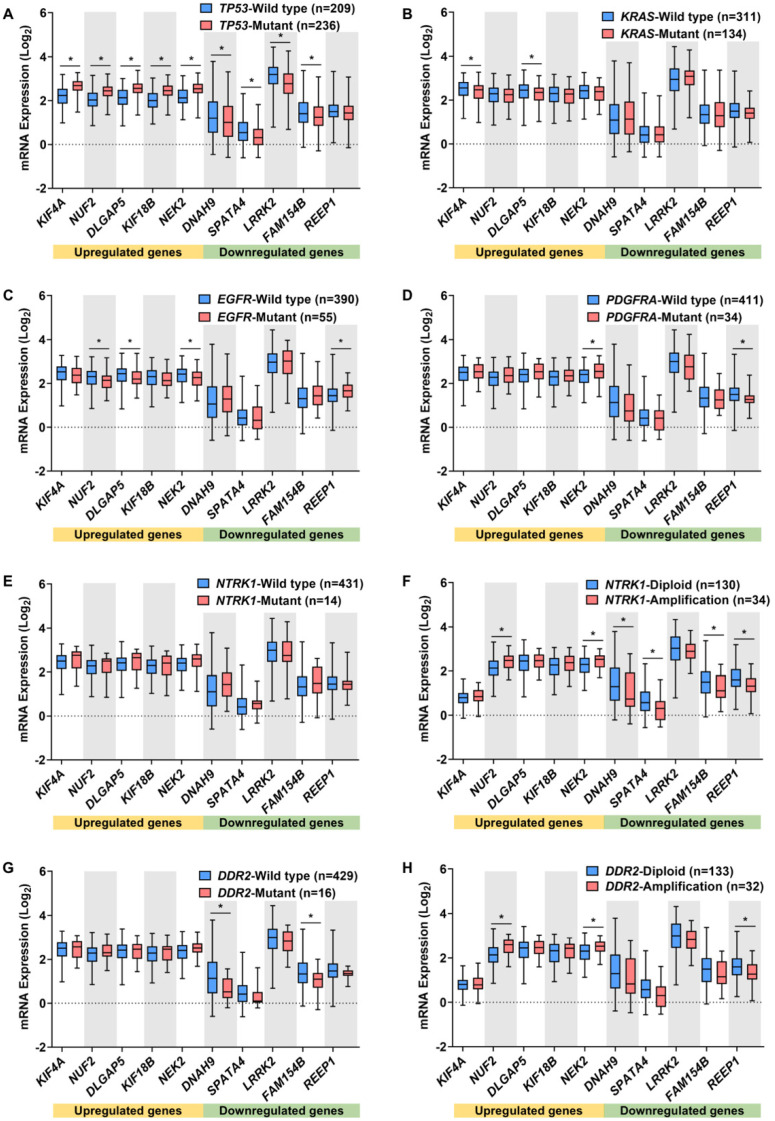
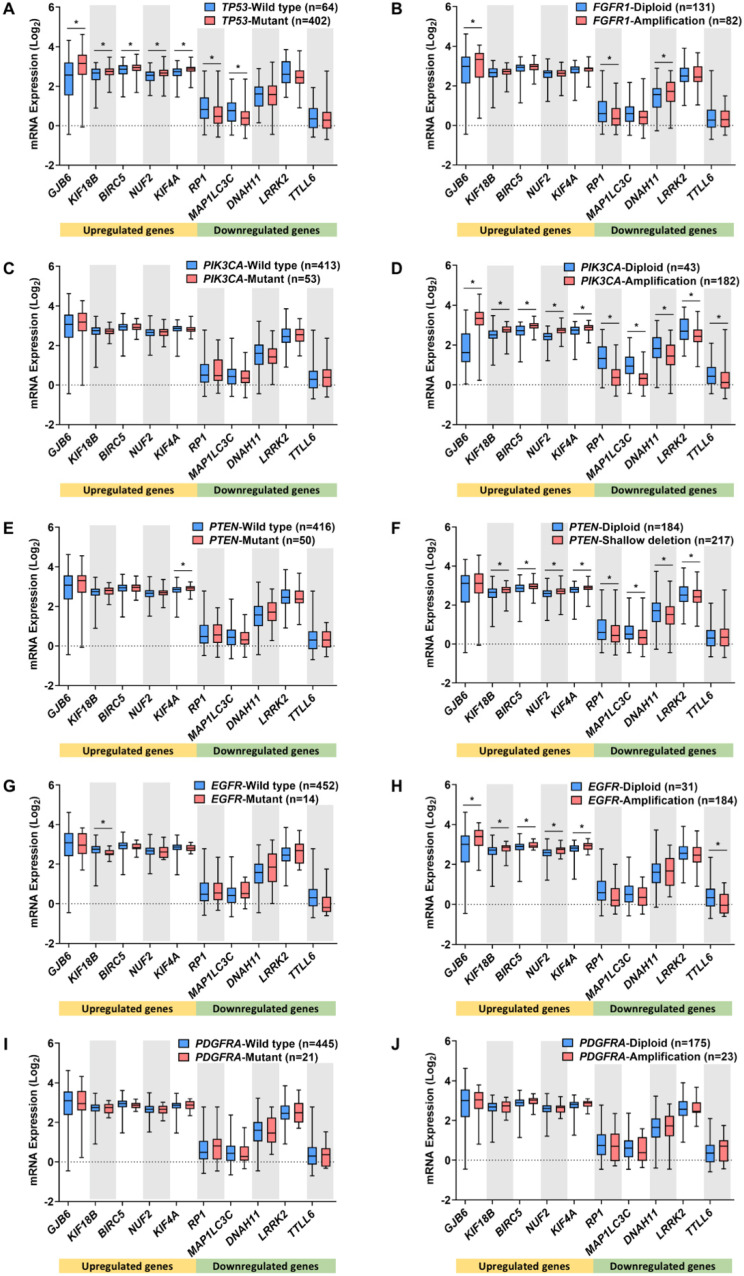
The regulatory roles of DEMGs in oncogenesis can be discerned by evaluating the association between their expression and the genetic alterations in hallmark genes. The hallmark genes included oncogenes and tumor suppressor genes, whose mutation, amplification, or deletion was strongly associated with the pathology and aggressiveness of lung cancer [1,30,31,32,33,34,35]. The alteration frequencies of hallmark genes in LUAD and LUSC tumor samples were confirmed by cBioPortal [36] and the top five most altered genes were included in this study, as shown in Supplementary Table S1. The mRNA expression levels of the 20 DEMGs were investigated in different alteration profiles of the hallmark genes (Figure 3 and Figure 4).
Figure 3.
Relationships between the top five DEMG expressions in LUAD and its lung cancer hallmark gene alterations. Box plots demonstrate the association between DEMG expressions and mutation of TP53 (A), KRAS (B), EGFR (C), PDGFRA (D), NTRK1 (E), and DDR2 (G), and amplification of NTRK1 (F), and DDR2 (H). * p < 0.05 vs. wild-type or diploid group.
Figure 4.
Relationships between the top five DEMG expressions in LUSC and its lung cancer hallmark gene alterations. Box plots demonstrate the association between DEMG expressions and mutation of TP53 (A), PIK3CA (C), PTEN (E), EGFR (G), and PDGFRA (I), amplification of FGFR1 (B), PIK3CA (D), EGFR (H), and PDGFRA (J), and shallow deletion of PTEN (F). * p < 0.05 vs. wild-type or diploid group.
In LUAD, the mutation of TP53, the most frequently altered gene, was significantly related to the expression levels of nine DEMGs (Figure 3A). Compared with TP53 wild-type, tumors with TP53 mutations showed significantly higher levels of the upregulated genes KIF4A, NUF2, DLGAP5, KIF18B, and NEK2, and lower levels of the downregulated genes DNAH9, SPATA4, LRRK2, and FAM154B, potentially implicating these genes in oncogenesis. Furthermore, KRAS mutation status was associated with decreased levels of KIF4A and DLGAP5 (Figure 3B), while patients with an EGFR mutation exhibited a decrease in NUF2, DLGAP5, and NEK2 expression and an increase in REEP1 expression (Figure 3C). Additionally, NEK2 and REEP1 expressions were associated with PDGFRA mutation (Figure 3D), NUF2, NEK2, DNAH9, SPATA4, FAM154B, and REEP1 expression with NTRK1 amplification (Figure 3F), DNAH9 and FAM154B expression with DDR2 mutations (Figure 3G), and NUF2, NEK2, and REEP1 expression with DDR2 amplification (Figure 3H). The NTRK1 mutation was not significantly related to the expression of DEMGs (Figure 3E).
In the LUSC cohort, TP53 mutations were associated with expression changes in the following seven DEMGs: GJB6, KIF18B, BIRC5, NUF2, and KIF4A were upregulated, while RP1 and MAP1L3C3 were downregulated (Figure 4A). Compared with FGFR1 diploid patients, those with FGFR1 amplification displayed significantly altered levels of GJB6, RP1, and DNAH11 (Figure 4B). Remarkably, while PIK3CA mutations were not associated with fluctuating levels of DEMGs, PIK3CA amplification was significantly related to the levels of all DEMGs, as follows: the upregulated DEMGs showed an increase, while the downregulated ones showed a decrease (Figure 4C,D). Shallow deletions and mutations are commonly found in PTEN alterations. These PTEN shallow deletions were associated with an increase in the expressions of KIF18B, BIRC5, NUF2, and KIF4A, and a decrease in the expressions of RP1, MAP1L3C3, DNAH11, and LRRK2 (Figure 4F). The PTEN mutations were related to the high expression of KIF4A (Figure 4E). Moreover, only the low level of KIF18B expression was associated with EGFR mutations (Figure 4G), while the high expressions of GJB6, KIF18B, BIRC5, NUF2, and KIF4A, and the low expression of TTLL6, were found in patients with EGFR amplification (Figure 4H). The expression of DEMGs did not significantly alter with PDGFRA mutation and amplification in LUSC tissues (Figure 4I,J).
2.3. Prognostic Value of DEMGs
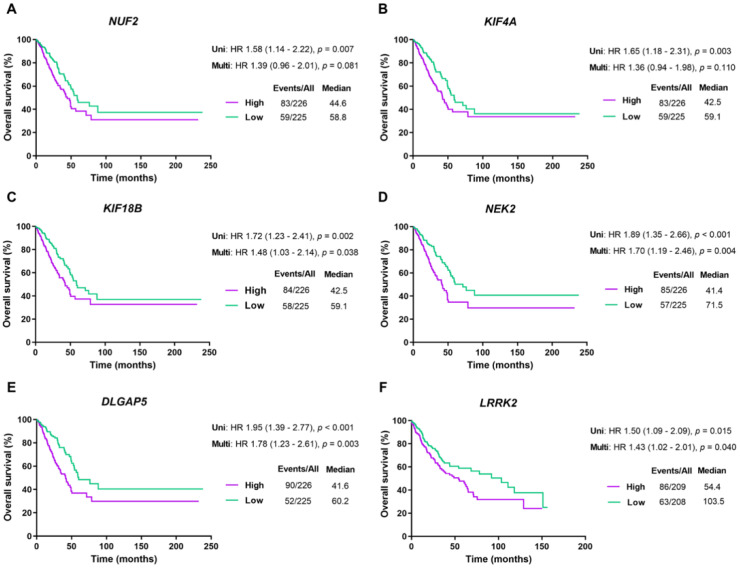
The prognostic value of the 20 DEMGs was evaluated via the Kaplan–Meier method [37]. Based on the median expression level of each gene, patients from the TCGA dataset were stratified into either low- or high-expression groups. Statistical analysis using a logrank test suggested that the high expressions of NUF2, KIF4A, KIF18B, NEK2, and DLGAP5 in LUAD (logrank p = 0.007, 0.003, 0.002, <0.001, <0.001, respectively) and LRRK2 in LUSC (logrank p = 0.015) were significantly related to worse OS for the patients (Figure 5).
Figure 5.
Survival analysis of the top five DEMGs in LUAD and LUSC patients. Kaplan–Meier curves present the overall survival rate of NUF2 (A), KIF4A (B), KIF18B (C), NEK2 (D), and DLGAP5 (E) in LUAD, and LRRK2 (F) in LUSC. Abbreviations are as follows: Uni, univariate analysis; Multi, multivariate analysis; HR, hazard ratio; Events/All, No. of events/No. of patients; Median (months).
Furthermore, univariate and multivariate Cox regression analyses were applied to assess the independent predictive value of each gene for the survival outcomes of lung cancer patients, including OS, disease-specific survival (DSS), and progression-free survival (PFS). Since TP53 mutation status was related to the expression of most DEMGs and significantly correlated with survival outcomes in both LUAD and LUSC patients (Supplementary Tables S2 and S3), it was included as a covariate in the multivariate analysis of the identified DEMGs.
In LUAD patients, the clinical stage and the expression of NUF2, NEK2, and DLGAP5 were significantly correlated with all types of survival outcomes, while the expression of KIF4A and KIF18B and TP53 mutation status were significantly correlated with OS and DSS in univariate analyses. In the multivariate analysis, the expression of NEK2 and DLGAP5 was related to all types of survival outcomes, that of NUF2 was associated with DSS and PFS, and that of KIF18B was correlated with OS (Supplementary Table S2).
In the LUSC dataset, the clinical stage significantly correlated with all types of survival outcomes, TP53 mutation status correlated with OS and DSS in univariate analyses, and LRRK2 expression was associated with OS in both univariate and multivariate analyses (Supplementary Table S3). Taken together, the high expression of six genes was remarkably associated with poor survival outcomes and independently correlated with the prognosis of lung cancer patients.
2.4. Gene Set Enrichment Analysis (GSEA) of DEMGs
To further evaluate the associated pathway of the DEMGs, we perform the GSEA using gene expression data from the TCGA, and hallmark gene sets from the Molecular Signatures Database (MsigDB). We compared the datasets for high- and low-expression of each gene. Considering the most significantly enriched signaling pathways based on normalized enrichment score (NES), high expression of NUF2, KIF4A, KIF18B, DLGAP5, and NEK2 and low expression of LRRK2 were mostly associated with G2/M checkpoint, E2F targets, MTORC1 signaling, and MYC targets. In addition, KIF4A, KIF18B, and DLGAP5 upregulation was associated with an increment in the mitotic spindle regulation, and elevation of NEK2 was linked to an increase in unfolded protein response (Supplementary Figure S2). As the roles of the G2/M checkpoint, E2F targets, MTORC1 signaling, MYC targets, and mitotic spindle regulation were mainly implicated in the cell cycle progression, this finding supported the significant role of candidate genes in tumor growth, and suggests that their function might participate in these pathways.
2.5. Validation of the Six Candidate Biomarkers Using Patient Samples and GEO Databases
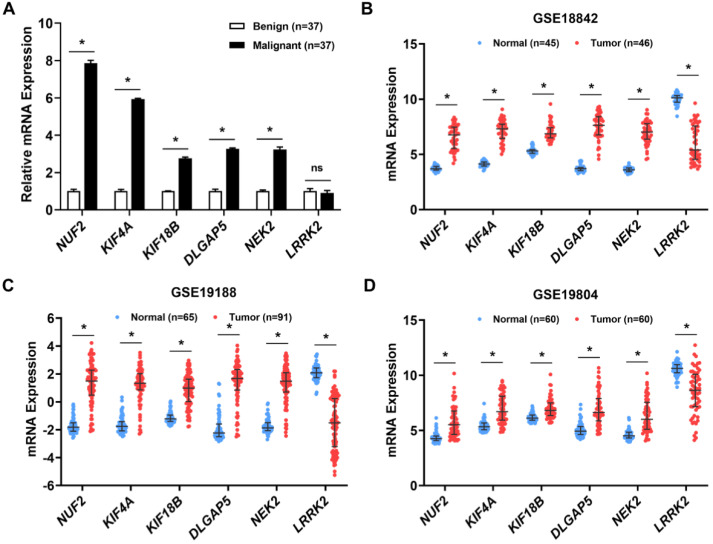
The six candidate biomarkers, associated with hallmark gene alterations and poor survival outcomes of patients, were verified for their differential expression using patient lung tissues and GEO databases. These candidates included five upregulated genes—NUF2, KIF4A, KIF18B, NEK2, and DLGAP5—and one downregulated gene—LRRK2—in the tumor tissues of TCGA databases. The expression levels of these biomarkers were measured by qRT-PCR in patient lung biopsy tissues that had pathologically been classified as benign (n = 37) or malignant (n = 37). The baseline clinical characteristics of patients in the two groups were not significantly different, except for age, which was lower in the benign group (p-value = 0.030) (Table 2). The mRNA levels of NUF2, KIF4A, KIF18B, DLGAP5, and NEK2 were significantly more than two-fold higher in malignant tissues than in benign tissues. However, LRRK2 expression levels did not differ significantly between the two groups (Figure 6A). Although age was significantly different between the two groups (Table 2), the expressions of these candidate genes were age-independent (Supplementary Figure S3). To further validate these biomarkers, the expression of the six candidate genes was compared between normal and tumor lung tissues from three independent GEO datasets, namely GSE18842, GSE19188, and GSE19804. Consistently, NUF2, KIF4A, KIF18B, DLGAP5, and NEK2 were highly overexpressed, whereas LRRK2 was significantly downregulated in the tumor tissues of all datasets (Figure 6B–D). These results corroborate the evidence indicating the involvement of these six candidate genes in lung cancer pathogenesis.
Table 2.
Clinical characteristics of patients for clinical validation.
| Characteristic | Malignancy (n = 37) |
Benign (n = 37) |
p-Value * |
|---|---|---|---|
| Sex―no. (%) | |||
| Female | 17 (45.95) | 19 (51.35) | 0.816 |
| Male | 20 (54.05) | 18 (48.65) | |
| Age―years | |||
| Median (Min, Max) | 65 (42, 87) | 63 (17, 81) | 0.030 |
| Smoking―no. (%) | |||
| Yes | 18 (48.65) | 15 (40.54) | 0.640 |
| No | 19 (51.35) | 22 (59.46) | |
| Carcinogen exposure―no. (%) | |||
| Yes | 7 (18.92) | 5 (13.51) | 0.754 |
| No | 30 (81.08) | 32 (74.42) | |
| Family history―no. (%) | |||
| Yes | 8 (21.62) | 2 (5.41) | 0.085 |
| No | 29 (78.38) | 35 (94.60) | |
| Stage―no. (%) | |||
| I | 4 (10.81) | ||
| II | 9 (24.32) | ||
| III | 11 (29.73) | ||
| IV | 13 (35.14) | ||
* Calculated based on a Mann–Whitney U test and chi-square test.
Figure 6.
Validation of the candidate DEMG expressions. Clinical validation in benign (n = 37) and malignant (n = 37) lung tissues was analyzed by quantitative reverse transcription polymerase chain reaction (A). * p < 0.05 vs. benign tissue, ns = not significant. The DEMG expressions were examined in the following three Gene Expression Omnibus datasets: GSE18842 (B), GSE19188 (C), and GSE19804 (D), compared between normal and tumor tissues. * p < 0.05 vs. normal tissue.
The relevance of DEMGs in lung cancer was further confirmed by the in vitro experiments. We first determined the proliferation profile of several lung cancer cell lines. Then, the cells were classified into a high and low proliferation based on their relative cell proliferation values. The mRNA expression of candidate genes was compared between the high proliferative lung cancer cell lines (A549 and H460 cells) and the low proliferative lung cancer cell lines (H292 and H23 cells). The results demonstrated that NUF2, KIF4A, KIF18B, DLGAP5, and NEK2 were higher expressed, while LRRK2 levels were lower in the cells that have a greater proliferative activity (Supplementary Figure S4). These data, at least, provided the oncogenic and tumor suppressive effect of the upregulated and downregulated candidate genes in lung cancer, respectively, supporting our findings from the bioinformatic and clinical sample analyses.
3. Discussion
Lung cancers display aggressive characteristics with metastasis and chemotherapeutic resistance [38,39]. Most patients are only diagnosed at an advanced stage due to non-specific symptoms, leading to the high progression and high death rate of lung cancer [1,3]. Identifying novel biomarkers for earlier diagnosis or prognosis could improve the clinical outcomes of patients. Multiple studies have reported that MAPs contribute to several cancer-related processes, including tumor growth, metastasis, and chemoresistance [13,14,15,20]. Several mitosis-associated genes, including AURKA, DLGAP5, TPX2, KIF11, and CKAP5, were overexpressed in tumor tissues, and associated with cell proliferation and poor OS [40]. Despite much evidence implicating MAPs in the pathogenesis of lung cancer, over 300 MAPs are still unexplored. In the present study, the transcriptomics of 320 MAPs were investigated, and the highly differentially expressed ones that were associated with alterations in hallmark genes and lung cancer prognosis were characterized. For the first time, we propose NUF2, KIF4A, KIF18B, DLGAP5, NEK2, and LRRK2 as biomarkers for lung cancer progression.
The NUF2 biomarker is a component of the nuclear division cycle 80 complex, which serves to segregate chromosomes during cell division [41]. Pan-cancer analysis in 31 distinct tumor types has revealed the overexpression of NUF2 in 23 cancer types. In contrast, it showed no significant alteration in kidney, prostate, and thyroid cancers, while its downregulation was found in leukemia and testicular cancers [42]. Silencing of NUF2 suppresses an in vitro cell proliferation and inhibits tumor growth in pancreatic and liver cancers [43,44]. We demonstrated the consistent upregulation of NUF2 in LUAD and LUSC datasets, patient tumor tissues, and GEO datasets, supporting the muti-omics analysis that its protein level was upregulated in clinical tissues [45,46]. Furthermore, NUF2 knockdown was shown to induce cell death and cell invasion in lung cancer cell lines [47], indicating the potential role of NUF2 in lung cancer progression. Furthermore, functional enrichment analysis revealed roles of NUF2 in the cell cycle and p53 signaling, mutations which resulted in the overexpression of NUF2 [45,46]. Similarly, we found that TP53 mutation was strongly related to the upregulation of NUF2 in both LUAD and LUSC, suggesting that TP53 negatively regulates NUF2.
The gene encoding EGFR plays an oncogenic role in lung cancer, and its mutation and amplification have been commonly recorded [31,48]. The expressions of EGFR and NUF2 were found to be positively correlated [46]. We further demonstrated that EGFR mutation was associated with decreasing expression of NUF2 in LUAD and, interestingly, EGFR amplification, mostly identified in LUSC, was highly related to the increased expression of NUF2 in this subtype. These data highlight the distinct influence of EGFR in regulating NUF2 in lung cancer subtypes, but their interaction at the molecular level remains to be elucidated.
Mutations in NTRK1 and DDR2, which encode tyrosine kinase receptors, have been reported in lung cancer [32]. The rearrangement of NTRK1 constitutively activates the receptor and its downstream signaling, which mediates tumor growth [33]. When DDR2 binds to extracellular collagen, it triggers SHP-2, SRC, and mitogen-activated protein kinase signaling; hence, DDR2 mutations induce cancer cell proliferation, differentiation, and metastasis [49,50]. In the present study, amplifications of NTRK1 and DDR2 were associated with elevated expression of NUF2 only in the LUAD subtype. However, the regulatory roles of NTRK1 and DDR2 amplification on lung cancer cell behaviors have not yet been characterized.
We found that PIK3CA amplification was associated with the upregulation of NUF2 in LUSC. Here, PIK3CA encodes phosphatidylinositol-3-kinase, which plays an important role in activating the Akt signaling pathway, the regulator of several cancer-related activities [51]. Mutation or amplification of PIK3CA has been reported and associated with lung cancer progression [52]. Consistent with our study, a positive correlation between PIK3CA and NUF2 expression has been reported [46].
In addition, we showed that shallow deletion of PTEN, a well-known tumor suppressor, was tightly correlated with increased expression of NUF2 in LUSC. Indeed, PTEN, a phosphatase enzyme, suppresses cancer cell growth through its downstream transcriptional activity [53]. Our data suggest that PTEN negatively regulates NUF2 expression, but the exact regulatory mechanism needs to be further elucidated. Overall, our findings provide a more detailed picture of the regulation of NUF2 in lung cancer and indicate that its expression can independently predict the DSS and PFS outcomes in LUAD patients.
Furthermore, KIF4A and KIF18B, members of the kinesin superfamily, execute essential functions in microtubule trafficking [54]. Overexpression of KIF4A and KIF18B was observed in various cancers [55,56]. The previous study revealed that KIF4A promotes cell proliferation by inducing p21-mediated cell cycle progression in colorectal cancer [57]. In contrast, KIF4A was downregulated in gastric cancer, and its elevation inhibits the proliferation of human gastric carcinoma cells [58]. Additionally, KIF18B participates in the Wnt/beta-catenin signaling pathway that induces cell proliferation, migration, and invasion in cervical and breast cancers [59,60]. In cases of lung cancer, KIF4A was reported to promote cell proliferation and migration and inhibit apoptosis in LUAD cell lines, whereas KIF18B was suggested to promote LUAD cell proliferation, migration, and invasion via the Rac1/Akt/mammalian target of rapamycin (mTOR) signaling pathway [61,62]. Roles of KIF4A and KIF18B have been substantially reported in LUAD [61,62,63], but the present study provides new evidence suggesting their potential roles in LUSC. The overexpression and clinical relevance of KIF4A and KIF18B have also been previously established through cross-validation between the public databases and clinical samples [61,62,63].
We discovered that TP53 mutation correlated with increased KIF4A and KIF18B expression in LUAD and LUSC. Supporting this finding, elevated levels of KIF4A were encountered in TP53-mutant compared with wild-type lung cancer cell lines, indicating the negative regulation of this candidate gene by TP53 [64]. Tumor suppressor p53, a transcription factor, participates in microtubule organization by regulating the expression of tubulins and MAPs [9]. Therefore, p53 might govern KIF4A and KIF18B expression through its transcriptional activity.
Furthermore, KRAS, which encodes a GTPase downstream of the tyrosine kinase receptor, is an essential mediator for cancer cell growth, differentiation, and apoptosis [65]. In this study, KRAS mutation was related to decreased expression of KIF4A in LUAD. In contrast, an analysis of cBioPortal data revealed that KIF4A was upregulated in KRAS-mutant pancreatic ductal adenocarcinoma patient tissues [66]. It is possible that KRAS might regulate KIF4A in a cell type-specific manner, and this particular regulation in the context of lung cancer requires further elucidation.
Additionally, EGFR amplification was found to be correlated with increased expression of KIF4A and KIF18B, while EGFR mutation was related to the decreased expression of KIF18B in LUSC. Since EGFR initiates signal transduction through a network of downstream pathways that activate transcription of target genes [67], such as signal transducer and activator of transcription 3 (STAT3), extracellular signal-regulated kinase (ERK), and mTOR, it might indirectly regulate KIF4A and KIF18B transcription via these pathways. In addition, the multivariate analysis suggested that KIF18B was an independent predictor of OS in LUAD. Taken together, these findings demonstrate the impact of these MAP genes in the oncogenic regulation of lung cancer hallmark genes.
Furthermore, DLGAP5, a microtubule stabilizer, plays a crucial role in the formation of tubulin polymers [40]. Cohort and bioinformatic studies have revealed high expression of DLGAP5 and its association with poor OS in numerous cancers [40,68]. Thus, DLGAP5 knockdown could attenuate cell growth and induce apoptosis via cyclin-dependent kinase 1/cyclin D1/Bcl-2 signaling in ovarian cancer cells [69]. Consistent with these findings, we found that DLGAP5 was upregulated in LUAD, positively associated with TP53 mutation, and negatively associated with KRAS and EGFR mutation, indicating the tumor oncogenic activity of DLGAP5 in relation to the lung cancer hallmark genes. Moreover, it showed the potential to predict OS, DSS, and PFS in LUAD, making it a promising prognostic biomarker.
Likewise, the serine/threonine protein kinase NEK2 was upregulated in multiple cancers [70]. Targeting NEK2 inhibits tumorigenesis through the Wnt1/beta-catenin signaling pathway in cervical cancer [71]. Furthermore, overexpression of NEK2 in triple-negative breast cancer cells promotes cell migration and invasion [72]. Additionally, NEK2 has also been found to be overexpressed and possess significant prognostic value in lung cancer [68,73]. It is known that NEK2 plays an important role in stabilizing microtubules during mitotic processes. Elevated levels of NEK2 induce cell proliferation and chromosome instability in NSCLC cells [74]. We provided consistent evidence for the upregulation and prognostic value of NEK2 in LUAD and found that TP53 and PDGFRA mutations were correlated with its elevated expression. The TP53 genetic lesion was previously found to be correlated with the amplification/overexpression of NEK2 in multiple myeloma, suggesting NEK2 as a promising target in TP53-mutant myeloma [75]. Interestingly, EGFR mutation was reported to induce NEK2 expression via the ERK signaling pathway, promoting in vitro NSCLC cell proliferation, and this NEK2 upregulation was found to impair the sensitivity of EGFR-targeting drugs [76]. We also found increased levels of NEK2 in clinical specimens with EGFR mutation. We further indicated that amplification of the hallmark genes NTRK1 and DDR2 may be related to the upregulation of NEK2, but further clarification is needed to accurately define their association.
Furthermore, LRRK2, a well-known regulator in Parkinson’s disease, was recently reported to be downregulated and associated with lung cancer progression [77,78], consistent with our findings. This LRRK2 was shown to participate in host immune responses involving the recruitment of macrophages in the tumor microenvironment, and its overexpression was reported in kidney and thyroid cancers [79]. The depletion of LRRK2 inhibited cell proliferation and migration and induced thyroid cancer cell apoptosis by inhibiting the c-Jun N-terminal kinase signaling pathway [80]. However, its role in lung cancer was largely unknown. Here, we have reported its involvement in lung cancer type for the first time and identified the relationship between TP53 mutation and decreased expression of LRRK2 in LUAD. In contrast, PIK3CA amplification and PTEN shallow deletion were correlated with the upregulation of LRRK2. However, we found no difference in the expression level of LRRK2 in patient tissues from our cohort. Therefore, the differential expression of LRRK2 in tumor versus normal tissues must be validated with a larger sample size to accurately discern its regulatory role in lung cancer.
These identified molecules also exhibit potential drug targets; however, there is no inhibitor approved for all candidate genes. Even some inhibitors are in the processes of in vitro, in vivo, and clinical investigation. Indeed, NUF2 and DLGAP5 inhibitors have not been reported. This is similar to KIF4A and KIF18B, although several KIF-inhibitors are undergoing clinical trials. For example, Ispinesib, a KIF11 inhibitor, was evaluated in a phase I clinical trial in breast cancer, although the specific inhibitors for KIF4A and KIF18B are still limited [81]. While NEK2 inhibitors have been widely established, and the efficacy was tested in both in vitro and in vivo in several types of cancer, including multiple myeloma, leukemia, gastric, colorectal, glioma, breast, and liver cancers, their effect on lung cancer requires further investigation [82]. As a key target for Parkinson’s disease, inhibitors for LRRK2 have been developed [83], but the anticancer effect of this inhibitor has not yet been evaluated. In cancer research, a recent study has demonstrated the role of LRRK2 only in thyroid cancer [80]. Likewise, our study has highlighted the significance of LRRK2 in lung cancer; however, the effect of LRRK2 inhibitors in the cancer context requires further clarification. Taken together, our study revealed the significant alterations of these genes in lung cancer in a subtype-specific manner. Further in vitro and in vivo investigations could strengthen the impact of these molecules in lung cancer cell biology and provide an advantage for clinical application.
4. Materials and Methods
4.1. Gene Expression Datasets
The mRNA expression data and clinicopathological data of LUAD and LUSC patients from the TCGA database were obtained from cBioPortal (https://www.cbioportal.org/ accessed on 4 August 2021) [36]. The LUAD dataset included 507 samples (451 tumors and 56 normal tissues), while the LUSC dataset included 468 samples (417 tumors and 51 normal tissues). Samples lacking survival and clinical data were excluded from the study. The clinical characteristics of patients are shown in Table 1.
4.2. Differential Expression Analysis
The differential expression of MAP genes between tumor and normal samples was analyzed using the R program version 2022.02.0+443 [84]. The false discovery rate (FDR) was calculated by Benjamini–Hochberg adjustment of the p-value. Fold change (FC) was used to quantify the difference in mRNA expression levels. The MAP genes with FDR < 0.05 and |log2FC| > 2 were determined as DEMGs. The DEMGs were presented in a volcano plot, Venn diagram, and heatmap, which were all generated using GraphPad Prism 9. From each dataset, the top five DEMGs exhibiting the highest fold changes were selected for subsequent analyses.
4.3. Lung Cancer Hallmark Gene Alteration Analysis
Genetic alteration profiles of lung cancer hallmark genes in LUAD and LUSC, including mutations and copy-number variations from the TCGA database, were obtained from cBioPortal (https://www.cbioportal.org/, accessed on 4 August 2021) [36]. The top five altered hallmark genes from each dataset, shown in Supplementary Table S1, were included in the analysis. Gene expression levels of DEMGs were compared between patients with wild-type and mutant genes or among patients with diploid, amplified, or deleted hallmark genes.
4.4. Survival Analysis
Kaplan–Meier plots of the OS of patients, categorized by high or low mRNA expression determined by the median expression of DEMGs [27,85], were generated using GraphPad Prism 9. The p-value was calculated by the logrank test [37]. Patients in the high expression group were characterized by their expression levels being above the median, while those in the low expression group showed expression levels below the median. Univariate and multivariate analyses were performed by the Cox’s proportional hazards regression model using a stepwise selection of variables. Clinicopathological variables, including age, sex, clinical stage, and TP53 mutation status, were selected for the analyses. A p-value less than 0.05 was considered statistically significant.
4.5. Patients and Tissue Samples
A total of 74 lung cancer patients from the Central Chest Institute of Thailand were enrolled in this study after obtaining written informed consent from them. All experiments were approved by the Central Research Ethics Committee of the Central Chest Institute of Thailand (approval number 086/2563) and were performed in accordance with the Helsinki Declaration of 1975. Lung biopsy samples were collected and defined as benign or malignant by the pathologist. The clinical information of patients is summarized in Table 2. Fresh lung tissues were frozen in RNA stabilizing solution (Invitrogen, Carlsbad, CA, USA) and stored at −20 °C for subsequent biochemical assays.
4.6. qRT-PCR
Total RNA was extracted from the patient’s lung tissues using GENEzolTM reagent (Geneaid, New Taipei City, Taiwan) and reverse-transcribed to cDNA using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, CA, USA), following the manufacturer’s instructions. Then, qRT-PCR was performed using SensiFAST™ SYBR Green Supermix (Meridian Bioscience, OH, USA). The expression levels were normalized to the internal control, glyceraldehyde-3-phosphate dehydrogenase, and relative expressions were calculated by the 2−∆∆Ct method. Primers used in this study are shown in Supplementary Table S4.
4.7. GEO Data Validation
To validate the associations of candidate biomarkers, three lung cancer datasets (i.e., GSE18842, GSE19188, GSE19804), containing gene expression data on tumor and normal tissues, were used for an independent analysis. The gene expression profile from the microarray and corresponding clinical information were obtained from GEO (https://www.ncbi.nlm.nih.gov/gds, accessed on 7 September 2022) [22]. The GSE18842 included dataset 46 tumors and 45 adjacent normal lung tissues. The GSE19188 dataset included 91 tumors and 65 adjacent normal lung tissues. The GSE19804 dataset included 60 tumors and 60 adjacent normal lung tissues.
4.8. Statistical Analysis
Statistical analysis was performed using GraphPad Prism 9. Student’s t-test and Mann–Whitney U test were used for comparisons between two groups of continuous variables, as appropriate. The chi-square test was applied for comparisons between two groups of categorical variables. A p-value less than 0.05 was considered statistically significant.
5. Conclusions
The present study identified and validated NUF2, KIF4A, KIF18B, DLGAP5, NEK2, and LRRK2 as promising diagnostic biomarkers for lung cancer. The increased expressions of NUF2, KIF4A, KIF18B, DLGAP5, and NEK2 were prognostic biomarkers in LUAD, and the reduced expression of LRRK2 was a prognostic biomarker in LUSC. Their gene expression levels were strongly related to alterations in lung cancer hallmark genes, including TP53, KRAS, EGFR, PDGFRA, NTRK1, DDR2, PIK3CA, and PTEN. Further in vitro and in vivo investigations of the molecular mechanisms underlying these associations might support their clinical application as biomarkers for lung cancer.
Supplementary Materials
The supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms232314724/s1.
Author Contributions
Data curation, formal analysis, investigation, writing—original draft, N.S.; conceptualization, supervision, writing—review and editing, V.Y.; formal analysis, writing—review and editing, S.S.; investigation, writing-review and editing, S.K.; investigation, writing—review, and editing, S.P.; investigation, writing—review, and editing, N.M.; investigation, writing—review and editing, W.P.; investigation, writing—review and editing, W.D.; conceptualization, data curation, investigation, methodology, project administration, supervision, resources, writing—original draft, writing—review and editing, V.P. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Central Research Ethics Committee of the Central Chest Institute of Thailand (approval number 086/2563).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data supporting the findings of this study are available within the article and its Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was supported by the National Research Council of Thailand (N41A640133, to V.P.) and the Second Century Fund (C2F, to N.S.), Chulalongkorn University, Thailand.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schabath M.B., Cote M.L. Cancer Progress and Priorities: Lung Cancer. Cancer Epidemiol. Biomark. Prev. 2019;28:1563–1579. doi: 10.1158/1055-9965.EPI-19-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 4.Aisner D.L., Akerley W., Bauman J.R., Bruno D.S., Chang J.Y., Chirieac L.R., Center Thomas D.C.A., DeCamp M., Dilling T.J., Dowell J., et al. NCCN Guidelines Version 5.2022 Non-Small Cell Lung Cancer Continue NCCN Guidelines Panel Disclosures. 2022. [(accessed on 11 November 2022)]. Available online: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf.
- 5.Planchard D., Popat S., Kerr K., Novello S., Smit E.F., Faivre-Finn C., Mok T.S., Reck M., Van Schil P.E., Hellmann M.D., et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018;29((Suppl. 4)):iv192–iv237. doi: 10.1093/annonc/mdy275. [DOI] [PubMed] [Google Scholar]
- 6.Suh J.H., Johnson A., Albacker L., Wang K., Chmielecki J., Frampton G., Gay L., Elvin J.A., Vergilio J.-A., Ali S., et al. Comprehensive Genomic Profiling Facilitates Implementation of the National Comprehensive Cancer Network Guidelines for Lung Cancer Biomarker Testing and Identifies Patients Who May Benefit from Enrollment in Mechanism-Driven Clinical Trials. Oncologist. 2016;21:684–691. doi: 10.1634/theoncologist.2016-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Šutić M., Vukić A., Baranašić J., Försti A., Džubur F., Samaržija M., Jakopović M., Brčić L., Knežević J. Diagnostic, Predictive, and Prognostic Biomarkers in Non-Small Cell Lung Cancer (NSCLC) Management. J. Pers. Med. 2021;11:1102. doi: 10.3390/jpm11111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Forges H., Bouissou A., Perez F. Interplay between microtubule dynamics and intracellular organization. Int. J. Biochem. Cell Biol. 2012;44:266–274. doi: 10.1016/j.biocel.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 9.Parker A.L., Kavallaris M., McCarroll J.A. Microtubules and Their Role in Cellular Stress in Cancer. Front. Oncol. 2014;4:153. doi: 10.3389/fonc.2014.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wattanathamsan O., Pongrakhananon V. Post-translational modifications of tubulin: Their role in cancers and the regulation of signaling molecules. Cancer Gene Ther. 2021:1–8. doi: 10.1038/s41417-021-00396-4. [DOI] [PubMed] [Google Scholar]
- 11.Etienne-Manneville S. Microtubules in Cell Migration. Annu. Rev. Cell Dev. Biol. 2013;29:471–499. doi: 10.1146/annurev-cellbio-101011-155711. [DOI] [PubMed] [Google Scholar]
- 12.Lopes D., Maiato H. The Tubulin Code in Mitosis and Cancer. Cells. 2020;9:2356. doi: 10.3390/cells9112356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo J., Hu Q., Gou M., Liu X., Qin Y., Zhu J., Cai C., Tian T., Tu Z., Du Y., et al. Expression of Microtubule-Associated Proteins in Relation to Prognosis and Efficacy of Immunotherapy in Non-Small Cell Lung Cancer. Front. Oncol. 2021;11:680402. doi: 10.3389/fonc.2021.680402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schiewek J., Schumacher U., Lange T., Joosse S.A., Wikman H., Pantel K., Mikhaylova M., Kneussel M., Linder S., Schmalfeldt B., et al. Clinical relevance of cytoskeleton associated proteins for ovarian cancer. J. Cancer Res. Clin. Oncol. 2018;144:2195–2205. doi: 10.1007/s00432-018-2710-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baquero M.T., Lostritto K., Gustavson M.D., ABassi K., Appia F., Camp R.L., Molinaro A.M., Harris L.N., Rimm D.L. Evaluation of prognostic and predictive value of microtubule associated protein tau in two independent cohorts. Breast Cancer Res. 2011;13:R85. doi: 10.1186/bcr2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J., Yu Y., Liu W., Li Z., Wei Z., Jiang R. Microtubule-associated protein tau is associated with the resistance to docetaxel in prostate cancer cell lines. Res. Rep. Urol. 2017;9:71–77. doi: 10.2147/RRU.S118966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng S., Shi L., Zhang Y., He T. Expression of SNCG, MAP2, SDF-1 and CXCR4 in gastric adenocarcinoma and their clinical significance. Int. J. Clin. Exp. Pathol. 2014;7:6606–6615. [PMC free article] [PubMed] [Google Scholar]
- 18.Xia X., He C., Wu A., Zhou J., Wu J. Microtubule-Associated Protein 4 Is a Prognostic Factor and Promotes Tumor Progression in Lung Adenocarcinoma. Dis. Markers. 2018;2018:8956072. doi: 10.1155/2018/8956072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khwaja S., Kumar K., Das R., Negi A.S. Microtubule associated proteins as targets for anticancer drug development. Bioorgan. Chem. 2021;116:105320. doi: 10.1016/j.bioorg.2021.105320. [DOI] [PubMed] [Google Scholar]
- 20.Wattanathamsan O., Pongrakhananon V. Emerging role of microtubule-associated proteins on cancer metastasis. Front. Pharmacol. 2022;13:935493. doi: 10.3389/fphar.2022.935493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoadley K.A., Yau C., Hinoue T., Wolf D.M., Lazar A.J., Drill E., Shen R., Taylor A.M., Cherniack A.D., Thorsson V., et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell. 2018;173:291–304.e6. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X., Wang J., Chen M., Liu S., Yu X., Wen F. Combining data from TCGA and GEO databases and reverse transcription quantitative PCR validation to identify gene prognostic markers in lung cancer. OncoTargets Ther. 2019;12:709–720. doi: 10.2147/OTT.S183944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zengin T., Önal-Süzek T. Comprehnsive Profiling of Genomic and Transcriptomic Differences between Risk Groups of Lung Adenocarcinoma and Lung Squamous Cell Carcinoma. J. Pers. Med. 2021;11:154. doi: 10.3390/jpm11020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Y., Tian X. Analysis of genes associated with prognosis of lung adenocarcinoma based on GEO and TCGA databases. Medicine. 2020;99:e20183. doi: 10.1097/MD.0000000000020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Q., Min S., Zhou Q. Identification of potential diagnostic and prognostic biomarkers for LUAD based on TCGA and GEO databases. Biosci. Rep. 2021;41:BSR20204370. doi: 10.1042/BSR20204370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao J., Guo C., Ma Z., Liu H., Yang C., Li S. Identification of a novel gene expression signature associated with overall survival in patients with lung adenocarcinoma: A comprehensive analysis based on TCGA and GEO databases. Lung Cancer. 2020;149:90–96. doi: 10.1016/j.lungcan.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Li Y., Gu J., Xu F., Zhu Q., Ge D., Lu C. Transcriptomic and functional network features of lung squamous cell carcinoma through integrative analysis of GEO and TCGA data. Sci. Rep. 2018;8:15834. doi: 10.1038/s41598-018-34160-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bateman A., Martin M.J., Orchard S., Magrane M., Agivetova R., Ahmad S., Alpi E., Bowler-Barnett E.H., Britto R., Bursteinas B., et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/NAR/GKAA1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibbons D.L., Byers L.A., Kurie J.M. Smoking, p53 Mutation, and Lung Cancer. Mol. Cancer Res. 2014;12:3–13. doi: 10.1158/1541-7786.MCR-13-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz-Ceja K.A., Chirino Y.I. Current FDA-approved treatments for non-small cell lung cancer and potential biomarkers for its detection. Biomed. Pharmacother. 2017;90:24–37. doi: 10.1016/j.biopha.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 32.Hirsch F.R., Suda K., Wiens J., Bunn P.A., Jr. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet. 2016;388:1012–1024. doi: 10.1016/S0140-6736(16)31473-8. [DOI] [PubMed] [Google Scholar]
- 33.Vaishnavi A., Capelletti M., Le A.T., Kako S., Butaney M., Ercan D., Mahale S., Davies K.D., Aisner D.L., Pilling A.B., et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat. Med. 2013;19:1469–1472. doi: 10.1038/nm.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oxnard G.R., Binder A., Jänne P.A. New Targetable Oncogenes in Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2013;31:1097–1104. doi: 10.1200/JCO.2012.42.9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Álvarez-Garcia V., Tawil Y., Wise H.M., Leslie N.R. Mechanisms of PTEN loss in cancer: It’s all about diversity. Semin. Cancer Biol. 2019;59:66–79. doi: 10.1016/j.semcancer.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bland J.M., Altman D.G. The logrank test. BMJ. 2004;328:1073. doi: 10.1136/bmj.328.7447.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhuyan J., Chen M., Zhu T., Bao X., Zhen T., Xing K., Wang Q., Zhu S. Critical steps to tumor metastasis: Alterations of tumor microenvironment and extracellular matrix in the formation of pre-metastatic and metastatic niche. Cell Biosci. 2020;10:89. doi: 10.1186/s13578-020-00453-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim E.S. Chemotherapy Resistance in Lung Cancer. Adv. Exp. Med. Biol. 2015;893:189–209. doi: 10.1007/978-3-319-24223-1_10/TABLES/1. [DOI] [PubMed] [Google Scholar]
- 40.Schneider M.A., Christopoulos P., Muley T., Warth A., Klingmueller U., Thomas M., Herth F.J., Dienemann H., Mueller N.S., Theis F., et al. AURKA, DLGAP5, TPX2, KIF11 and CKAP5: Five specific mitosis-associated genes correlate with poor prognosis for non-small cell lung cancer patients. Int. J. Oncol. 2017;50:365–372. doi: 10.3892/ijo.2017.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang T., Zhou Y., Qi S., Wang Z.-B., Qian W.-P., Ouyang Y.-C., Shen W., Schatten H., Sun Q.-Y. Nuf2 is required for chromosome segregation during mouse oocyte meiotic maturation. Cell Cycle. 2015;14:2701–2710. doi: 10.1080/15384101.2015.1058677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang X., Jiang Y., Luo S., Sekar K., Koh C.K.T., Deivasigamani A., Dong Q., Zhang N., Li S., Hao F., et al. Correlation of NUF2 Overexpression with Poorer Patient Survival in Multiple Cancers. Cancer Res. Treat. 2021;53:944–961. doi: 10.4143/crt.2020.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q., Dai S.-J., Li H., Dong L., Peng Y.-P. Silencing of NUF2 Inhibits Tumor Growth and Induces Apoptosis in Human Hepatocellular Carcinomas. Asian Pac. J. Cancer Prev. 2014;15:8623–8629. doi: 10.7314/APJCP.2014.15.20.8623. [DOI] [PubMed] [Google Scholar]
- 44.Hu P., Chen X., Sun J., Bie P., Zhang L.D. SiRNA-Mediated Knockdown against NUF2 Suppresses Pancreatic Cancer Proliferation in Vitro and in Vivo. Biosci. Rep. 2015;35:170. doi: 10.1042/BSR20140124. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Li X., Zhang L., Yi Z., Zhou J., Song W., Zhao P., Wu J., Song J., Ni Q. NUF2 Is a Potential Immunological and Prognostic Marker for Non-Small-Cell Lung Cancer. J. Immunol. Res. 2022;2022:1161931. doi: 10.1155/2022/1161931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen M., Li S., Liang Y., Zhang Y., Luo D., Wang W. Integrative Multi-Omics Analysis of Identified NUF2 as a Candidate Oncogene Correlates with Poor Prognosis and Immune Infiltration in Non-Small Cell Lung Cancer. Front. Oncol. 2021;11:656509. doi: 10.3389/fonc.2021.656509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.You X., Ren H., Wen L. MicroRNA-34c-5p Inhibition of NUF2 Suppresses Lung Adenocarcinoma Cell Viability and Invasion. J. Nanomater. 2021;2021:9152985. doi: 10.1155/2021/9152985. [DOI] [Google Scholar]
- 48.Nukaga S., Yasuda H., Tsuchihara K., Hamamoto J., Masuzawa K., Kawada I., Naoki K., Matsumoto S., Mimaki S., Ikemura S., et al. Amplification of EGFR Wild-Type Alleles in Non–Small Cell Lung Cancer Cells Confers Acquired Resistance to Mutation-Selective EGFR Tyrosine Kinase Inhibitors. Cancer Res. 2017;77:2078–2089. doi: 10.1158/0008-5472.CAN-16-2359. [DOI] [PubMed] [Google Scholar]
- 49.Terai H., Tan L., Beauchamp E.M., Hatcher J.M., Liu Q., Meyerson M., Gray N.S., Hammerman P.S. Characterization of DDR2 Inhibitors for the Treatment of DDR2 Mutated Nonsmall Cell Lung Cancer. ACS Chem. Biol. 2015;10:2687–2696. doi: 10.1021/acschembio.5b00655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hammerman P.S., Sos M.L., Ramos A.H., Xu C., Dutt A., Zhou W., Brace L.E., Woods B.A., Lin W., Zhang J., et al. Mutations in the DDR2 Kinase Gene Identify a Novel Therapeutic Target in Squamous Cell Lung Cancer. Cancer Discov. 2011;1:78–89. doi: 10.1158/2159-8274.CD-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Wang Y., Li J., Li J., Che G. Clinical Significance of PIK3CA Gene in Non-Small-Cell Lung Cancer: A Systematic Review and Meta-Analysis. BioMed Res. Int. 2020;2020:3608241. doi: 10.1155/2020/3608241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan A.C. Targeting the PI3K/Akt/mTOR pathway in non-small cell lung cancer (NSCLC) Thorac. Cancer. 2020;11:511–518. doi: 10.1111/1759-7714.13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brito M.B., Goulielmaki E., Papakonstanti E.A. Focus on PTEN regulation. Front. Oncol. 2015;5:166. doi: 10.3389/fonc.2015.00166/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hirokawa N., Tanaka Y. Kinesin superfamily proteins (KIFs): Various functions and their relevance for important phenomena in life and diseases. Exp. Cell Res. 2015;334:16–25. doi: 10.1016/j.yexcr.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 55.Wu J., Li L., Zhong H., Zhang H.-H., Li J., Zhang H.-B., Zhao Y.-Q., Xu B., Song Q.-B. Bioinformatic and Experimental Analyses Reveal That KIF4A Is a Biomarker of Therapeutic Sensitivity and Predicts Prognosis in Cervical Cancer Patients. Curr. Med. Sci. 2022:1–12. doi: 10.1007/s11596-022-2636-y. [DOI] [PubMed] [Google Scholar]
- 56.Qiu M.-J., Wang Q.-S., Li Q.-T., Zhu L.-S., Li Y.-N., Yang S.-L., Xiong Z.-F. KIF18B is a Prognostic Biomarker and Correlates with Immune Infiltrates in Pan-Cancer. Front. Mol. Biosci. 2021;8:559800. doi: 10.3389/fmolb.2021.559800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hou P.-F., Jiang T., Chen F., Shi P.-C., Li H.-Q., Bai J., Song J. KIF4A facilitates cell proliferation via induction of p21-mediated cell cycle progression and promotes metastasis in colorectal cancer. Cell Death Dis. 2018;9:477. doi: 10.1038/s41419-018-0550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao J., Sai N., Wang C., Sheng X., Shao Q., Zhou C., Shi Y., Sun S., Qu X., Zhu C. Overexpression of chromokinesin KIF4 inhibits proliferation of human gastric carcinoma cells both in vitro and in vivo. Tumor Biol. 2010;32:53–61. doi: 10.1007/s13277-010-0090-0. [DOI] [PubMed] [Google Scholar]
- 59.Wu Y., Wang A., Zhu B., Huang J., Lu E., Xu H., Xia W., Dong G., Jiang F., Xu L. KIF18B promotes tumor progression through activating the Wnt/β-catenin pathway in cervical cancer. OncoTargets Ther. 2018;11:1707–1720. doi: 10.2147/OTT.S157440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu L., Zhang Z., Xia X., Lei J. KIF18B Promotes Breast Cancer Cell Proliferation, Migration and Invasion by Targeting TRIP13 and Activating the Wnt/β-Catenin Signaling Pathway. Oncol. Lett. 2022;23:112. doi: 10.3892/ol.2022.13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song Y., Tang W., Li H. Identification of KIF4A and its effect on the progression of lung adenocarcinoma based on the bioinformatics analysis. Biosci. Rep. 2021;41:BSR20203973. doi: 10.1042/BSR20203973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji Z., Pan X., Shang Y., Ni D.-T., Wu F.-L. KIF18B as a regulator in microtubule movement accelerates tumor progression and triggers poor outcome in lung adenocarcinoma. Tissue Cell. 2019;61:44–50. doi: 10.1016/j.tice.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 63.Zhong Y., Jiang L., Long X., Zhou Y., Deng S., Lin H., Li X. Clinical Significance and Integrative Analysis of Kinesin Family Member 18B In Lung Adenocarcinoma. OncoTargets Ther. 2019;12:9249–9264. doi: 10.2147/OTT.S227438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hanselmann S., Wolter P., Malkmus J., Gaubatz S. The microtubule-associated protein PRC1 is a potential therapeutic target for lung cancer. Oncotarget. 2017;9:4985–4997. doi: 10.18632/oncotarget.23577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fathi Z., Mousavi S.A.J., Roudi R., Ghazi F. Distribution of KRAS, DDR2, and TP53 gene mutations in lung cancer: An analysis of Iranian patients. PLoS ONE. 2018;13:e0200633. doi: 10.1371/journal.pone.0200633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Y., Gao L., Weng N.-N., Li J.-J., Liu J.L., Zhou Y., Liao R., Xiong Q.-L., Xu Y.-F., Varela-Ramirez A., et al. Identification of Novel Molecular Therapeutic Targets and Their Potential Prognostic Biomarkers Among Kinesin Superfamily of Proteins in Pancreatic Ductal Adenocarcinoma. Front. Oncol. 2021;11:3260. doi: 10.3389/fonc.2021.708900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosenkranz A.A., Slastnikova T.A. Epidermal Growth Factor Receptor: Key to Selective Intracellular Delivery. Biochemistry. 2020;85:967–993. doi: 10.1134/S0006297920090011. [DOI] [PubMed] [Google Scholar]
- 68.Shi Y.-X., Yin J.-Y., Shen Y., Zhang W., Zhou H.-H., Liu Z. Genome-scale analysis identifies NEK2, DLGAP5 and ECT2 as promising diagnostic and prognostic biomarkers in human lung cancer. Sci. Rep. 2017;7:8072. doi: 10.1038/s41598-017-08615-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang H., Liu Y., Tang S., Qin X., Li L., Zhou J., Zhang J., Liu B. Knockdown of DLGAP5 suppresses cell proliferation, induces G2/M phase arrest and apoptosis in ovarian cancer. Exp. Ther. Med. 2021;22:1245. doi: 10.3892/etm.2021.10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fang Y., Zhang X. Targeting NEK2 as a promising therapeutic approach for cancer treatment. Cell Cycle. 2016;15:895–907. doi: 10.1080/15384101.2016.1152430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu T., Zeng Y., Shi L., Yang Q., Chen Y., Wu G., Li G., Xu S. Targeting NEK2 impairs oncogenesis and radioresistance via inhibiting the Wnt1/β-catenin signaling pathway in cervical cancer. J. Exp. Clin. Cancer Res. 2020;39:183. doi: 10.1186/s13046-020-01659-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rivera-Rivera Y., Marina M., Jusino S., Lee M., Velázquez J.V., Chardón-Colón C., Vargas G., Padmanabhan J., Chellappan S.P., Saavedra H.I. The Nek2 centrosome-mitotic kinase contributes to the mesenchymal state, cell invasion, and migration of triple-negative breast cancer cells. Sci. Rep. 2021;11:9016. doi: 10.1038/s41598-021-88512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhong X., Guan X., Liu W., Zhang L. Aberrant expression of NEK2 and its clinical significance in non-small cell lung cancer. Oncol. Lett. 2014;8:1470–1476. doi: 10.3892/ol.2014.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shah D., Joshi M., Patel B.M. Role of NIMA-related kinase 2 in lung cancer: Mechanisms and therapeutic prospects. Fundam. Clin. Pharmacol. 2022;36:766–776. doi: 10.1111/fcp.12777. [DOI] [PubMed] [Google Scholar]
- 75.Cusan M., Wang L. NEK2, a promising target in TP53 mutant cancer. Blood Sci. 2022;4:97–98. doi: 10.1097/BS9.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen C., Peng S., Li P., Ma L., Gan X. High expression of NEK2 promotes lung cancer progression and drug resistance and is regulated by mutant EGFR. Mol. Cell. Biochem. 2020;475:15–25. doi: 10.1007/s11010-020-03854-z. [DOI] [PubMed] [Google Scholar]
- 77.Ma Q., Xu Y., Liao H., Cai Y., Xu L., Xiao D., Liu C., Pu W., Zhong X., Guo X. Identification and validation of key genes associated with non-small-cell lung cancer. J. Cell. Physiol. 2019;234:22742–22752. doi: 10.1002/jcp.28839. [DOI] [PubMed] [Google Scholar]
- 78.Lebovitz C., Wretham N., Osooly M., Milne K., Dash T., Thornton S., Tessier-Cloutier B., Sathiyaseelan P., Bortnik S., Go N.E., et al. Loss of Parkinson’s susceptibility gene LRRK2 promotes carcinogen-induced lung tumorigenesis. Sci. Rep. 2021;11:2097. doi: 10.1038/s41598-021-81639-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan J., Zhao W., Yu W., Cheng H., Zhu B. LRRK2 correlates with macrophage infiltration in pan-cancer. Genomics. 2021;114:316–327. doi: 10.1016/j.ygeno.2021.11.037. [DOI] [PubMed] [Google Scholar]
- 80.Jiang Z.-C., Chen X.-J., Zhou Q., Gong X.-H., Chen X., Wu W.-J. Downregulated LRRK2 gene expression inhibits proliferation and migration while promoting the apoptosis of thyroid cancer cells by inhibiting activation of the JNK signaling pathway. Int. J. Oncol. 2019;55:21–34. doi: 10.3892/ijo.2019.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li T.-F., Zeng H.-J., Shan Z., Ye R.-Y., Cheang T.-Y., Zhang Y.-J., Lu S.-H., Zhang Q., Shao N., Lin Y. Overexpression of kinesin superfamily members as prognostic biomarkers of breast cancer. Cancer Cell Int. 2020;20:123. doi: 10.1186/s12935-020-01191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dana D., Das T., Choi A., Bhuiyan A.I., Das T.K., Talele T.T., Pathak S.K. Nek2 Kinase Signaling in Malaria, Bone, Immune and Kidney Disorders to Metastatic Cancers and Drug Resistance: Progress on Nek2 Inhibitor Development. Molecules. 2022;27:347. doi: 10.3390/molecules27020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thakur G., Kumar V., Lee K.W., Won C. Structural Insights and Development of LRRK2 Inhibitors for Parkinson’s Disease in the Last Decade. Genes. 2022;13:1426. doi: 10.3390/genes13081426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Therneau T.M. Survival Analysis [R Package Survival Version 3.4-0] 2022. [(accessed on 19 August 2022)]. Available online: https://cran.r-project.org/web/packages/survival/index.html.
- 85.Ji Q., Cai G., Liu X., Zhang Y., Wang Y., Zhou L., Sui H., Li Q. MALAT1 regulates the transcriptional and translational levels of proto-oncogene RUNX2 in colorectal cancer metastasis. Cell Death Dis. 2019;10:378. doi: 10.1038/s41419-019-1598-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the article and its Supplementary Materials.