Abstract
The pathogenic species Neisseria meningitidis and Neisseria gonorrhoeae cause dramatically different diseases despite strong relatedness at the genetic and biochemical levels. N. meningitidis can cross the blood-brain barrier to cause meningitis and has a propensity for toxic septicemia unlike N. gonorrhoeae. We previously used subtractive hybridization to identify DNA sequences which might encode functions specific to bacteremia and invasion of the meninges because they are specific to N. meningitidis and absent from N. gonorrhoeae. In this report we show that these sequences mark eight genetic islands that range in size from 1.8 to 40 kb and whose chromosomal location is constant. Five of these genetic islands were conserved within a representative set of strains and/or carried genes with homologies to known virulence factors in other species. These were deleted, and the mutants were tested for correlates of virulence in vitro and in vivo. This strategy identified one island, region 8, which is needed to induce bacteremia in an infant rat model of meningococcal infection. Region 8 encodes a putative siderophore receptor and a disulfide oxidoreductase. None of the deleted mutants was modified in its resistance to the bactericidal effect of serum. Neither were the mutant strains altered in their ability to interact with endothelial cells, suggesting that such interactions are not encoded by large genetic islands in N. meningitidis.
Neisseria meningitidis colonizes the nasopharynx, from which it can seed the bloodstream before crossing the blood-brain barrier (BBB) to cause meningitis. In contrast, Neisseria gonorrhoeae colonizes and invades the epithelium of the genitourinary tract, where it can cause a localized inflammation; bacteremia, though frequent, is asymptomatic and dissemination is rare. Thus, both species are capable of crossing a cellular barrier at their port-of-entry but they differ in their abilities to subsequently disseminate in the blood. The ability to induce intense and prolonged bacteremia is one of the prerequisites for a bacterial pathogen to cross the BBB. In contrast, the details of specific interactions with the cellular components of the BBB remain unclear. Therefore, in order to understand the mechanisms that allow N. meningitidis to cross the BBB, it will be necessary to identify the genes that are involved in bloodstream dissemination and/or specific interaction with the cellular components of the BBB. Such genes might be present in both N. meningitidis and N. gonorrhoeae but differ subtly in sequence or regulation, or they might be present in only one of the two species.
Results from in vitro models have shown that most of the mechanisms mediating cellular interactions are common to both N. meningitidis and N. gonorrhoeae. On the other hand, several determinants have been identified that are specific to N. meningitidis: the polysaccharide capsule (8), the enzyme rotamase (26), the RTX toxin-like Frp proteins (29, 30), and a glutathione peroxidase (20). Of these, the capsule locus is required for systemic dissemination and bloodstream survival (34), whereas a role in virulence has not been demonstrated for the other genes.
We have recently created a bank of N. meningitidis-specific sequences after subtractive hybridization between N. meningitidis and N. gonorrhoeae in order to identify genes which are present only in N. meningitidis and might therefore account for its differential pathogenesis (32). Some of the clones mapped closely together, suggesting that they may have been derived from larger regions of N. meningitidis-specific DNA. One region containing such clones (region 1) corresponds to the locus of capsule synthesis which had previously been well characterized (8, 12, 13). However, the significance of the other regions was unknown. We have now investigated the other regions of N. meningitidis-specific DNA in order to obtain details on the differences between N. meningitidis and N. gonorrhoeae and to possibly identify mechanisms responsible for the specificity of N. meningitidis pathogenesis. Our data identify eight novel DNA islands that are specifically present in N. meningitidis and absent from N. gonorrhoeae. Those islands that were conserved among a representative set of meningococcal strains and/or showed homologies with known virulence factors were deleted, and the resulting strains were tested for phenotypes that are associated with crossing the BBB. The results show that one of the eight islands is required for high levels of bacteremia.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains of N. meningitidis were tested that represent the genetic diversity of this species according to multilocus sequence typing (MLST) (18). Their MLST assignments were: ST1 (subgroup I, strain B40), ST2 (subgroup VI, Z6835), ST4 (subgroup IV-1, Z5463, Z2491 [27]), ST5 (subgroup III, Z3524), ST8 (A4 cluster, BZ 10), ST11 (ET-37 complex, serogroup C: FAM18; serogroup W135: ROU [24]), ST25 (NG G40), ST30 (NG 4/88), ST32 (ET-5 complex, 44/76), ST41 (lineage 3, BZ 198); ST48 (BZ147), ST49 (297-0), ST60 (subgroup IX, 890592), and ST74 (ET-5 complex, MC58 [33]). Additional strains were N. meningitidis 8013, N. gonorrhoeae FA1090 and two strains of Neisseria lactamica (Z6793 and Z6784). N. meningitidis strains were grown on GC agar (GCB; Difco), with the addition of Kellogg's defined supplement plus ferric nitrate (14) for 12 to 20 h at 37°C in a moist atmosphere containing 5% CO2. Liquid media were GC-PO4 (1.5% Proteose Peptone number 3 [Difco], 0.5% NaCl, 30 mM potassium phosphate; pH 7.5) and GC-HEPES (like GC-PO4 but the potassium phosphate was replaced by 30 mM HEPES [pH 7.5]), both supplemented as for the solid medium. Escherichia coli were grown on Luria-Bertani (LB) agar or in LB liquid medium. Antibiotics used were: ampicillin, 50 μg/ml; kanamycin, 50 μg/ml (N. meningitidis) or 100 μg/ml (E. coli); nalidixic acid, 20 μg/ml; and spectinomycin, 40 μg/ml.
Oligonucleotide primers and PCR conditions.
The sequences of the primers used to amplify the individual regions and to construct deletions are listed in Table 1; the other primer sequences are available on request. Template chromosomal DNA was isolated as described elsewhere (27).
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′–3′)a |
|---|---|
| Primers to amplify each region from flanking sequences | |
| Reg2-for | TTGGCGAAGAGCAACGAACCTGTTTATCTCGTG |
| Reg2-rev | GGCGCAATGTTCGATTTTTCCTCTATCCGGTTC |
| Reg3-for | GACAAACTGATGACTTGGGGCTTCTGGCTGCTC |
| Reg3-rev | ATATGCCCGACATCGACGGCGGCATTTTGTTCC |
| Reg4-for | GCAAATGGTTGGAAGTCGGCGGTTGCGGTATGG |
| Reg4-rev | GCAATGCCCTTAACACTCAAGCAATCAGCGCGG |
| Reg5-for | CGAAGCCATGTACCTTGCCAATACTGCGGCCGG |
| Reg5-rev | ACCGCCGCACTGGAGGCATAAAGGAAGGGGATG |
| Reg6-for | GATGGCGAAAGTGATGAGTTTCGGCGTGTGGTG |
| Reg6-rev | AACTGCCGTGCCTCATACGATGCGGCGCAAATC |
| Reg7-for | CCAAAATGGACGCGACCGTATAGGACGTCTTCC |
| Reg7-rev | AAAAGTACTGGACGCAAGTTGGGCGGCGTTGCA |
| Reg8-for | AGCAGTTCCCTGCTTATGCCGCCAACTTTCCCG |
| Reg8-rev | GAAACAAATCCCCGAAGGTACGGCTGAAAAAGC |
| Reg9-for | ATTTGAAAGGAAGACGGCAGGCCTGCGTCGCGG |
| Reg9-rev | CAGTATCCGCAACTCTTGGACAACTACGGCAAC |
| Primers from Fig. 1A | |
| T1 | GATCCGAAAAGCAGCCGTCTGAAACAGATCTGCA |
| T2 | GATCTGTTTCAGACGGCTGCTTTTCG (5′ PO4) |
| T3 | TGTTTCAGACGGCTGCTTTTG (5′ PO4) |
| T4 | GATCCAAAAGCAGCCGTCTGAAACATGCA |
| BE1 | GATCGCAGGG(5′ PO4) |
| BE2 | AATTCCCTGC |
| Primers to amplify flanking sequences as in Fig. 1B | |
| R3001 | GTCGGCAGATCTTACGCGGGCAACTTCTT |
| R3002 | CGTTTGGAATTCCAAACGCCGTTCAATTCAA |
| R3003 | TTTACCGAATTCCTCGTCAACCGCGACGGCGA |
| R3004 | GTTACGGGTACCTCGATTTGGGACGTTTCT |
| R6001 | CCGTCTGAATTCTTTCAGACGGCATTTTTGCCGA |
| R6002 | TGCAAAGGATCCAAAGGCCCTCACAACTGTTTT |
| R6003 | TTTTGCGGATCCCGTCTGAAACAGGGTATGTTT |
| R6004 | TTCGGCTCTAGATGCCCCACGCCGATACCGA |
| R7001 | TCCCACGGATCCTCGCCAGCCTCGGCGACGTCA |
| R7002 | TAAAGCAGATCTGAAACGGTTATGAAATTCCCAA |
| R7003 | CGTGTAAGATCTTTTGGAAGACGAAGATTTTAT |
| R7004 | TTTTTTGGATCCTTGTGTGATTAAACGTCTTT |
| R8001 | CGTCGAAGATCTTCGCCTTCCCCATTGAAATG |
| R8002 | ACAACCGAATTCTATTGCCTCACGGAGGAAATGA |
| R8003 | AAATAGGAATTCAGACGGCCTTTTGTATTTAGGCT |
| R8004 | CGACGGGTACCGCCCTGCCGCGATTGAA |
| R9001 | CTGCGCGAATTCCAGCACGCCCAAGTCTTCCGT |
| R9002 | ATTCTTAGATCTGTTCCAACCACTAATACACTA |
| R9003 | GATCTCAGATCTCATTTGTTGTTCATTTTGGTT |
| R9004 | GGGCGCGTCGATGTGGAATTCT |
Restriction sites are is indicated in boldface.
The PCR conditions used depended on the length of the desired product. For products up to 3 kb, the reaction mixture contained template DNA (1 μg ml−1); reaction buffer (10 mM Tris-Cl, pH 8.0; 50 mM KCl; 1.5 mM MgCl2; 0.01% gelatin); dATP, dCTP, dGTP, and dTTP (200 μM concentrations of each); dimethyl sulfoxide (5%); forward and reverse primers (100 nM concentrations of each); and Taq polymerase. The PCR reactions were incubated 1 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1.5 min at 5°C below the Tm of the oligonucleotide primers, and 2 min at 72°C, followed by incubation for 5 min at 72°C. For PCR products between 3 and 8 kb, semi-long-range PCR was performed by using the Expand Long Template PCR System (Boehringer Mannheim) under the same conditions except that the mixture contained Buffer 1 and the polymerase mix (0.75 μl) supplied with the kit. The thermocycling conditions were 1 min at 94°C, 30 cycles of 45 s at 94°C, 1 min at 65°C, and 3 min at 68°C, and a final incubation for 5 min at 68°C. Template DNA longer than 8 kb was amplified by using the same kit and conditions except that higher concentrations of dATP, dCTP, dGTP, and dTTP (350 μM concentrations of each) and oligonucleotide primers (300 nM) and 2 μl of the polymerase mix were used. Incubation was for 1 min at 94°C, 30 cycles of 10 s at 94°C, 30 s at 65°C, and 20 min at 68°C, followed by 7 min at 68°C.
Sequencing of the eight regions.
Chromosomal DNA of N. meningitidis Z2491 was restricted by partial Sau3AI digestion and fragments of 12- to 23-kb size fractionated by gel electrophoresis were cloned into the BamHI site of the Lambda DASH II (Stratagene) phage vector by using E. coli XL1-Blue MRA (Stratagene). Details of the following steps were according to the DIG System Users Guide (Boehringer Mannheim). Plaques were transferred to nylon membranes (Hybond N; Amersham). N. meningitidis-specific clones (32) were digoxigenin labeled during PCR amplification and used as probes for plaque hybridization under stringent conditions. Phages containing hybridizing sequences were detected colorimetrically, and single plaques were purified before lysates were prepared. Two microliters of each lysate was used as a template for long-range PCR with primers in the phage vector immediately flanking the inserts. Then, 15 μg of PCR product was randomly sheared by nebulization (no. 4100 Nebuliser; Inhalation Plastics) for 20 min at 0.7 atm as described elsewhere (http://bric.postech.ac.kr/resources/rprotocol/rprotocol_partii.html). The sheared fragments were precipitated, end repaired with T4 DNA polymerase and Klenow DNA polymerase (New England Biolabs), and size fractionated on a 0.8% agarose gel. Fragments of between 0.4 and 0.6 kb and between 0.8 and 1 kb were separately eluted from the gel by using the Qiaquick Gel Extraction Kit (Qiagen). dATP overhangs were added to the fragments and ligated with the TA cloning vector pCR2.1 (Invitrogen). These preparations were transformed into E. coli XL1-Blue by electroporation. A total of 96 recombinant colonies were picked per transformation and grown in LB medium with ampicillin, and their inserts were amplified by PCR by using primers complementary to the flanking vector sequences. The PCR products were purified and sequenced by using the M13 reverse primer, a dRhodamine terminator cycle sequencing kit, and ABI Prism 377 DNA sequencers (Perkin-Elmer Applied Biosystems). Raw data from the ABI sequencer were prepared for assembly by using the ASP program (http://www.sanger.ac.uk/Software/Sequencing/ASD/asp/MODULES.shtml), and sequences were assembled with GAP4 from the Staden sequence analysis package (28).
Sequences that were 100% identical to those available in the public domain (Sanger Center; http://www.sanger.ac.uk/Projects/N_meningitidis/) at that time were accepted as correct, whereas all discrepancies were resequenced as follows using PCR products from the chromosomal DNA of strain Z2491. Fragments of approximately 5 kb were amplified by semi-long-range PCR by using primers designed from the sequences of the phage inserts. The PCR products were purified using the Qiaquick PCR Purification Kit (Qiagen) and sequenced from both strands with appropriate primers as described above. Additional smaller PCR products from bacterial chromosomes were sequenced from other strains of N. meningitidis using the same strategy.
Region 2 was sequenced from both strands by using a different strategy than for the other regions. Primers were designed according to the sequences of the clones isolated by Tinsley and Nassif (32), and products were obtained from chromosomal DNA by semi-long-range PCR. Sequence walking was used to complete the sequences of each of these products.
Analysis of nucleotide sequences.
Open reading frames (ORFs) were recognized by using the Codon Use program written by Conrad Halling, which supplies a graphical output for a sliding window of the codon adaptation index in all six frames. The permitted start codons were ATG and GTG, and the permitted stop codons were TAA, TAG, and TGA. Homology searches at the nucleotide level were performed by using BLASTN (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-blast?) and at the amino acid level by using PSI-BLAST (http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST/nph-psi_blast). Repetitive nucleotide sequences were detected by using Miropeats (22).
DNA dot blot and Southern hybridization.
DNA dot blot hybridization was performed according to the DIG System Users Guide (Boehringer) by spotting 1 μl containing 100 ng of denatured chromosomal DNA from each strain onto nylon membranes (Hybond N; Amersham). For Southern hybridization analysis, chromosomal DNA was digested with restriction endonucleases and separated by conventional electrophoresis or by pulsed-field gel electrophoresis (PFGE) and then transferred to nylon membranes. The DNA dot blots were hybridized with digoxigenin-labeled probes obtained by PCR amplification of each ORF, and Southern blots were hybridized with labeled probes corresponding to ORFs or entire regions. For the analysis of the distribution of the regions among meningococcal strains, hybridizations were performed at 37°C in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 50 mM sodium phosphate, 7% sodium dodecyl sulfate (SDS), 2% Blocking Reagent (Boehringer-Mannheim), 0.1% N-lauroylsarcosine, and 50% formamide, and the last washing step was with 0.5× SSC–0.1% SDS at 50°C in order to allow approximately 30% mismatch. Positive hybridization signals were detected by chemiluminescence. For verification of the mutants by Southern blotting, probes were labeled with [α-32P]dCTP. Hybridization in 500 mM sodium phosphate (pH 7.2) containing 7% SDS and 1 mM EDTA and washing in 40 mM sodium phosphate (pH 7.2), containing 1% SDS and 1 mM EDTA were performed at 65°C.
PCR analysis of DNA islands in different N. meningitidis strains.
The sizes of the eight islands were determined by PCR amplification with primers Reg2-for to Reg9-rev (Table 1) that are complementary to the 5′ and 3′ flanking sequences, respectively, in both N. meningitidis and N. gonorrhoeae. Semi-long-range PCR was used for all regions. Region 3 containing Pnm1 was amplified by eight sets of long-range PCRs. The locations of each island were confirmed by additional PCR reactions by using each forward primer (Reg2-for, Reg3-for, etc.) with a reverse primer in the leftmost ORF and each reverse primer (Reg2-rev, Reg3-rev, etc.) with a forward primer in the rightmost ORF of the corresponding island. Additionally, each ORF was separately amplified from strains that reacted in dot blot hybridization by primers specific to its 5′ and 3′ ends, and the sizes were confirmed by gel electrophoresis.
Production of deletion mutants in the N. meningitidis-specific islands.
Briefly, deletions were produced by transforming N. meningitidis ROU, MC58, and Z5463 with plasmid DNA into which sequences flanking an island had been cloned, such that they were separated by an antibiotic resistance cassette in place of the N. meningitidis-specific island. After transformation of N. meningitidis, DNA from a single colony expressing the selected antibiotic resistance was used to retransform the same strain, and a mixture of several hundred transformants was pooled and used for biological assays.
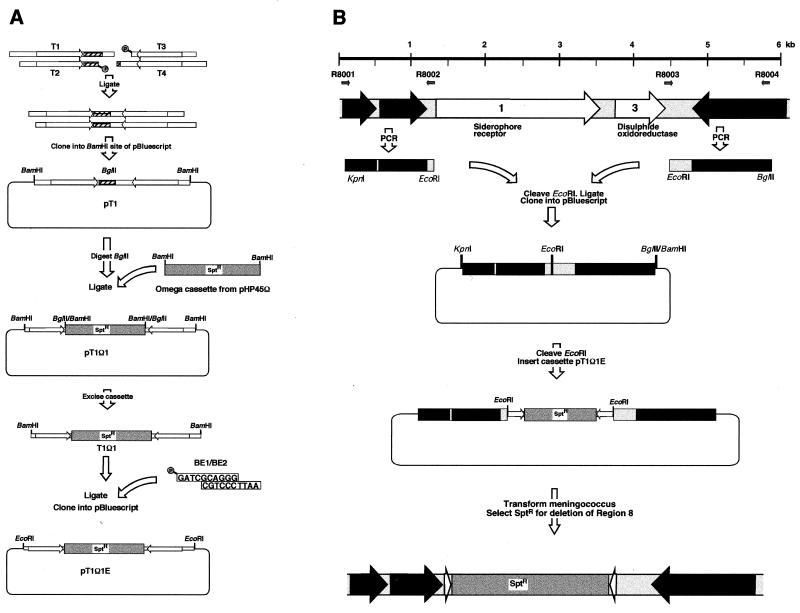
Figure 1B outlines the procedure for the production of the plasmids used for the deletions, taking region 8 as its example. The PCR products of the flanking regions (approximately 1 kb) were amplified using the primers shown in Table 1. The primers were designed so that both PCR products contained either a BamHI or a EcoRI restriction site at the internal end and one of a variety of different restriction sites at the external end. The PCR products were digested with BamHI or EcoRI, purified, and ligated. The ligation products were reamplified by using the “external” primers, cleaved at the ends with the appropriate enzyme(s) and cloned into pBluescript KS(+) (Stratagene). A resistance cassette (see below) flanked by two neisserial uptake sequences was then cloned into the BamHI or the EcoRI site.
FIG. 1.
Production of a cassette, flanked by neisserial uptake sequences, and deletion of the N. meningitidis-specific islands. (A) For production of the cassette, an inverted repeat containing two neisserial uptake sequences (arrows) and an internal BglII site (shaded) was constructed by ligating synthetic oligonucleotides T1 to T4 (Table 1). This molecule was cloned into pBluescript, and an Ω spectinomycin resistance cassette was cloned into the BglII site to yield a cassette that may be excised with BamHI. (B) Deletion of region 8. For replacement of the N. meningitidis-specific islands with the cassette, the flanking regions of the islands were PCR amplified by using the oligonucleotides in Table 1. These were ligated together at an internal restriction enzyme site (EcoRI), reamplified, and cloned into pBluescript. The construct was reopened at the EcoRI site, and the cassette was inserted between the flanking sequences. This plasmid was then used to transform N. meningitidis and replace the chromosomal island with the resistance cassette by homologous recombination.
Construction of the resistance cassette.
The resistance cassette contains the omega fragment (23), which encodes resistance to spectinomycin due to aadA (aminoglycoside adenyltransferase) and interrupts both translation and transcription, flanked on each side in inverted orientation by the neisserial uptake sequence GCCGTCTGAA. Due to the degree of secondary structure in the omega fragment, the construct was made without using PCR technology, as outlined in Fig. 1A. An artificial inverted repeat of uptake sequences was produced as follows. The oligonucleotides T1 and T2 (Table 1) were mixed (25 μM, 30 μl in T4 DNA ligase buffer), heated at 75°C for 10 min, and allowed to cool to room temperature for 1 h. This results in a double-stranded DNA containing one copy of the neisserial uptake sequence, with a 5′ overhang of GATC (compatible with BamHI) and a 3′ overhang of TGCA. A similar hybridization of oligonucleotides T3 and T4 also produced a DNA with the same overhangs. After mixing and ligation, only the desired internal join (TGCA) can be ligated due to the 5′ phosphate groups. The resulting mixture was cloned into the BamHI site in pBluescript KS(+), and the correct construct (plasmid pT1) was detected by its possession of a BglII site (present in oligonucleotide T1).
The omega fragment was excised from plasmid pHP45Ω by using BamHI and inserted into the unique BglII site of pT1, resulting in plasmid pT1Ω1, where the omega cassette is flanked by neisserial uptake sequences and can be excised by digestion with BamHI. We also constructed plasmid pT1Ω1E, where the fragment can be excised by digestion with EcoRI. To this end, the BamHI fragment of pT1Ω1 was ligated to the adaptors BE1 and BE2 and cloned into the EcoRI site of pBluescript KS(+).
Transformation of N. meningitidis.
After overnight growth on GC agar plates, bacteria were resuspended in GC-HEPES containing 1 mM K2HPO4 and Kellogg supplement 1 (14) to an optical density at 600 nm (OD600) of 0.1. MgCl2 was added to a final concentration of 10 mM, and transforming DNA (0.5 to 5 μg of linearized plasmid or 0.5 μg of chromosome) was added to 500 μl of the suspension. After incubation without shaking at 37°C for 30 min, 4.5 ml of GC-PO4 was added, and the mixture was incubated with shaking for a further 2 h. The bacteria were plated onto GC agar containing 40 μg of spectinomycin per ml for selection of transformants.
Complement-dependent serum bactericidal assay.
Antiserum against strains ROU and MC58 was obtained by immunizing rabbits with a mixture of paraformaldehyde-treated and sonicated bacteria with Freund's adjuvant and boosting three times using Freund's incomplete adjuvant. After overnight growth on GC agar plates, bacteria were resuspended to a final concentration of 106 CFU/ml in phosphate-buffered saline (PBS) containing 5 mM MgCl2 and 0.25 mM CaCl2 (PBSB). Then, 10 μl of bacterial suspension was mixed with 360 μl of antiserum (decomplemented by heating at 56°C for 30 min) diluted in PBSB and 40 μl of freshly thawed guinea pig serum (Gibco-BRL) as a complement source. Killing was measured by determining colony counts after 45 min incubation at 37°C, and the survival was compared to bacteria incubated in the absence of antiserum. In each experiment, a positive killing control used a serum-sensitive polyphosphate kinase mutant (ppk) (31), while as a negative control the ppk bacteria were incubated with decomplemented guinea pig serum in the presence of opsonizing antiserum.
Adhesion and invasion assays.
Bacteria from GC agar plates were grown in RPMI (Gibco-BRL) containing 10% heat-inactivated fetal calf serum (FCS; Gibco-BRL) with gentle shaking for 2 h to an OD600 of 0.1. Then, 1 ml of a 100-fold dilution was added to a confluent monolayer of human umbilical vein endothelial cells (HUVEC) in 2-cm2 tissue culture wells (Costar). One well was used per N. meningitidis strain to test adherence, and two wells were used to assay invasion.
Adhesion.
After incubation for 1 h at 37°C in 5% CO2, the supernatant was removed (nonadherent bacteria) and the monolayer was washed three times with RPMI. The adherent bacteria were released by adding 1 ml of PBS–1% saponin and scraping the bottom of the wells with a micropipette tip. The numbers of adherent and nonadherent bacteria were determined after plating them on supplemented GC agar. Adherence was calculated as the number of adherent bacteria divided by the total number of adherent plus nonadherent bacteria.
Invasion.
The wells were washed every hour as described above for 6 h and then filled with RPMI-FCS containing 150 μg of gentamicin per ml. After 1 h of incubation to kill external bacteria, internalized bacteria were harvested and enumerated as described above. Invasion was calculated as the number of internalized bacteria divided by the total bacteria at 1 h after infection.
Infant rat model of meningococcal infection.
Bacteria grown on GC agar plates for 14 h were resuspended in pyrogen-free 0.9% NaCl to an OD600 of 0.06. Four- to five-day-old Lewis rats (IFFA Credo, L'Arbresle, France) anesthetised with diethyl ether were injected intraperitoneally with 100 μl of bacterial suspension. Half of each litter (usually 10 animals) was injected with the parental strain and half was injected with the mutant strain. Samples of blood (5 μl) were taken from an incision in the tail after 1, 3, 6, 9, and 24 h. The blood samples were diluted in GC-PO4 and plated on GC agar for enumeration.
Nucleotide sequence accession numbers. The DNA sequences described in this work have been deposited in the EMBL database under the following accession numbers: for region 2 of N. meningitidis Z2491 (fhaB and fhaC homologues and genes of unknown function) and flanking genes, AJ391255; for region 3 (prophage Pnm1 and gpx/A) and flanking genes, AJ391256; for region 4 (genes of unknown function) and flanking genes, AJ391257; for region 5 (restriction/modification system) and flanking gene, AJ391258; for region 6 (pseudogene with homology to siderophore receptor genes) and flanking genes, AJ391259; for region 7 (homology to type I secretion system) and flanking genes, AJ391260; for region 8 (fhuA and dsbA homologues) and flanking genes, AJ391261; for region 9 (cluster of putative ORFs and insertion element IS4351N2) and flanking genes, AJ391262; for strain FAM18 hlyD gene (putative component of type I secretion system), AJ391263; for strain FAM18 tolC gene (putative component of type I secretion system), AJ391264; for strain NG 4/88 tolC gene (putative component of type I secretion system), AJ391265; for strain 297-0 tolC pseudogene (putative component of type I secretion system), AJ391266; for strain FAM18 fhuA gene for putative siderophore receptor, AJ391267; for strain NG 4/88 fhuA gene for putative siderophore receptor, AJ391268; for strain MC58 fhuA gene for putative siderophore receptor, AJ391269; for strain ROU fhuA gene for putative siderophore receptor, AJ391270; for strain BZ 10 fhuA pseudogene for putative siderophore receptor, AJ391271; for strain BZ 147 fhuA pseudogene for putative siderophore receptor, AJ391272; for strain BZ 198 fhuA pseudogene for putative siderophore receptor, AJ391273; for strain B40 fhuA pseudogene for putative siderophore receptor, AJ391274; for strain Z3524 fhuA pseudogene for putative siderophore receptor, AJ391275; for strain 297-0 fhuA pseudogene for putative siderophore receptor, AJ391276; for strain 44/76 fhuA pseudogene, AJ391277; for strain FAM18 dsbA gene for putative disulfide oxidoreductase, AJ391278; for strain BZ10 dsbA gene for putative disulfide oxidoreductase, AJ391279; for strain NG 4/88 dsbA gene for putative disulfide oxidoreductase, AJ391280; for strain 44/76 DNA for region 6 (rsi1 pseudogene) and flanking fnr and dinP genes (partial), AJ391281; for strain BZ 198 DNA for region 6, insertion sequence, partial rsi1 pseudogene and flanking fnr and dinP genes (partial), AJ391282; for strain 297-0 partial fnr gene for putative ferredoxin-NADP+ reductase and partial dinP gene for putative DNA-damage inducible protein P, AJ391283; and for strain FAM18 DNA for region 2 (fhaB and fhaC homologues and genes of unknown function) and flanking genes, AJ391284.
RESULTS
Identification of N. meningitidis-specific regions in strain Z2491.
Twenty-eight subtractive clones from N. meningitidis serogroup A, subgroup IV-1 strain Z2491 have been described that have no homologies in N. gonorrhoeae (32). DNA probes corresponding to these clones were used to screen a Lambda Dash II library containing 12- to 23-kb DNA fragments of strain Z2491. Other clones comprising “region 1” were not investigated further because they correspond to the well-characterized locus of capsule production.
A minimal set of Lambda Dash II recombinant phages were identified whose inserts hybridized with 25 of the probes (all except B305, B333, and E103), and these inserts were sequenced. Comparison of these N. meningitidis sequences with that of N. gonorrhoeae strain FA1090 (University of Oklahoma; http://dna1.chem.ou.edu/gono.html) identified the flanking ends that were homologous in both species and, hence, the extent of eight regions of N. meningitidis-specific sequences. The eight regions were designated regions 2 to 9 and are shown in Fig. 2, 3, and 4. They contain 85 ORFs, most with codon usage typical of N. meningitidis, of which 43 are homologous to previously described ORFs in other species (Table 2). ORFs with no significant homology to genes of known function were named according to the region (rtw [region two], rth [region three], rfo [region four], rfi [region five], rsi [region six], rse [region seven], rei [region eight], and rni [region nine]) plus a sequential number corresponding to the position of the ORF (rtw1, rth52, etc.).
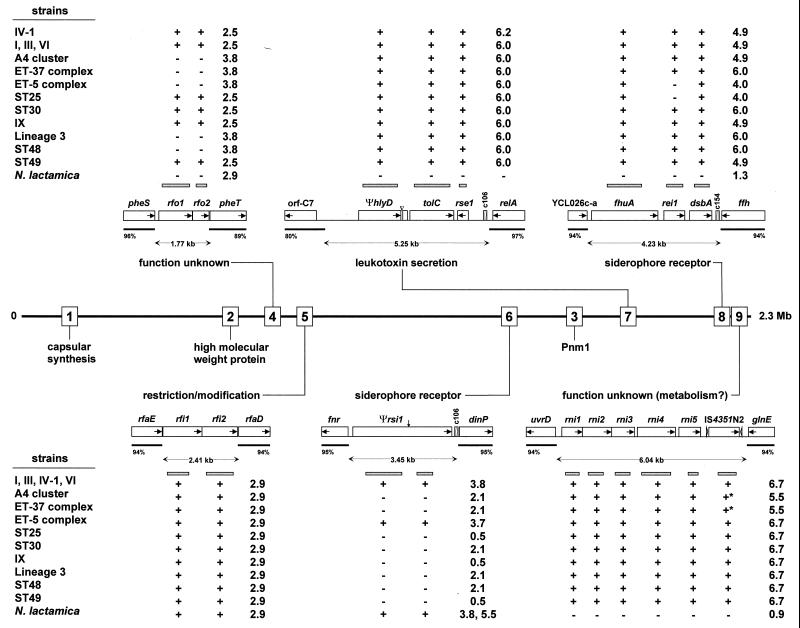
FIG. 2.
Relative genetic locations of N. meningitidis-specific regions 1 to 9 in the serogroup A subgroup IV-1 strain Z2491. For the six smaller regions, insets indicate the size of ORFs (open rectangles) and the orientation of transcription (internal arrows). Flanking sequences that are homologous in N. meningitidis and N. gonorrhoeae are indicated by black bars, and the numbers show the percent homology between N. meningitidis and N. gonorrhoeae. c106 and c154 in regions 6, 7, and 8 refer to 106- and 154-bp Correia elements, respectively, whereas IS4351N2 in region 9 is an IS element with flanking inverted repeats (open arrowheads). The PCR probes used for hybridization are indicated by gray bars above and below the map. The results from dot blot hybridization and PCR analyses with diverse strains of N. meningitidis and N. lactamica are summarized at the top and bottom of the figure by “+” and “−”. ∗, positive hybridization due to other copies of IS4351N2 present at other locations in the chromosome. The numbers indicate the sizes (in kilobases) of the PCR products obtained after amplification of the regions with primers complementary to the 5′ and 3′ flanking sequences. An explanation of the genetic designations is in Table 2. ▿, duplication in hlyD of region 7; ↓, frameshift in Ψrsi1 of region 6.
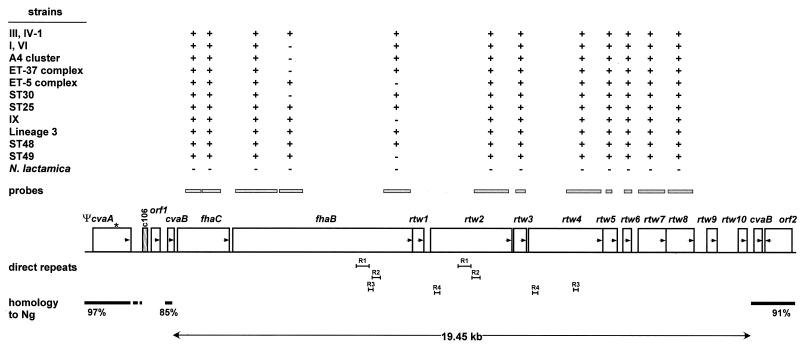
FIG. 3.
Contents of region 2. Region 2 is located within a homologue of the E. coli cvaB gene that is a pseudogene in N. gonorrhoeae inactivated by the insertion of a Correia element (4). In N. meningitidis, region 2 has replaced the Correia element plus 693 bp of the cvaB gene. Symbols are as described for Fig. 2. Repetitive stretches within the region from fhaB to rtw4 are indicated by R1 to R4. A stop codon in the ΨcvaA gene is indicated by an asterisk. c106, a 106-bp Correia element.
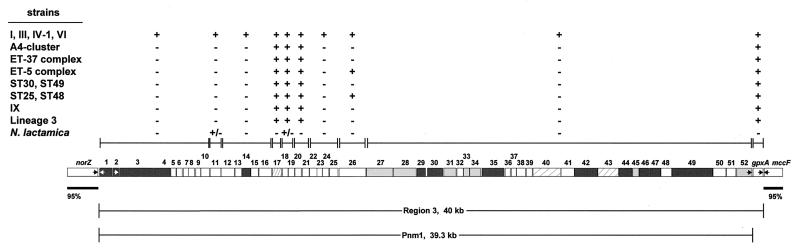
FIG. 4.
Region 3 and the Pnm1 prophage. Details are as given in Fig. 2. The numbers 1 to 52 above the ORFs indicate genes rth1 to rth52. The transcription orientation is from left to right for rth2 to rth52. Dark gray ORFs are homologous to both phage genes and to clustered genes in H. influenzae. ORFs shaded in light gray are only homologous to clustered genes in H. influenzae. ORFs in hatched boxes are only homologous to phage proteins (rth17, rth40, and rth43). Blocks of ORFs that yielded identical results in dot blot analysis are delineated by paired vertical lines above the map. +/−, positive result with one of two strains of N. lactamica.
TABLE 2.
ORFs in the eight DNA islands and their homologies to other bacterial proteinsa
| ORF (length; probes) | Length (aa) | Homologous protein
|
|||||
|---|---|---|---|---|---|---|---|
| Function | Species | Length (aa) | Statistical significance (P) | % Identity/% similarity | Accession no. | ||
| Region 2 (19.45 kb; B322, B220, B132, E139, B233, E145, B328, B108, and B101) | |||||||
| Region 2, 5′ flanking | |||||||
| ΨcvaA pseudogene | 411 | Colicin V secretion protein | E. coli | 413 | 7e-31 | 25/46 | P25519 |
| orf1 | 100 | Unknown | |||||
| cvaB left end | 79 | Colicin V secretion ATP-binding protein | E. coli | 698 | 4e-05 | 48/68 | P22520 |
| Region 2b | |||||||
| fhaC | 580 | Hemolysin activator-like protein precursor | B. pertussis | 584 | 5e-27 | 25/41 | P35077 |
| fhaB | 2,015 | FHA B precursor | B. pertussis | 3,591 | 1e-50 | 25/42 | P12255 |
| Region 2, 3′ flanking cvaB right end | 93 | Colicin V secretion ATP-binding protein | E. coli | 698 | 8e-23 | 54/73 | P22520 |
| orf2 (fragment) | 303 | Unknown protein | Vitreoscilla sp. | 376 | 2e-47 | 39/53 | AF067083 |
| Region 3 (40.0 kb; E142, E137, E107, E120, E146, E115, E114, B306, and E124) | |||||||
| Region 3, 5′ flanking norZ (fragment) | 677 | Nitric oxide reductase | R. eutropha | 762 | 0.0 | 58/69 | AF002217 |
| Region 3c | |||||||
| rth1 | 249 | Repressor protein | Phage D3112 | 240 | 2e-15 | 36/56 | S13498 |
| Transcriptional regulatory protein HI1476 | H. influenzae | 239 | 5e-06 | 25/45 | P44207 | ||
| rth2 | 87 | DNA-binding protein Ner | Phage Mu | 75 | 9e-14 | 50/66 | P06020 |
| Ner protein homolog HI1477 | H. influenzae | 89 | 2e-17 | 53/70 | P46496 | ||
| rth3 | 681 | Transposase | Phage D3112 | 690 | 2e-64 | 30/46 | S62728 |
| Transposase A (MuA)-homolog HI1478 | H. influenzae | 687 | 4e-07 | 21/36 | B64126 | ||
| rth4 | 304 | MuB (DNA transposition protein) | Phage Mu | 312 | 9e-16 | 26/44 | P03763 |
| MuB protein homolog HI1481 | H. influenzae | 287 | 2e-25 | 29/52 | C64126 | ||
| rth13 | 91 | DNA-binding protein HU-beta | S. typhimurium | 90 | 5e-24 | 58/75 | P05515 |
| rth14 | 144 | muE16 | Phage Mu | 195 | 4e-06 | 40/61 | M64097 |
| E16 homolog HI1488 | H. influenzae | 185 | 0.002 | 35/53 | E64126 | ||
| rth17 | 181 | Gene 25 | Phage SPP1 | 271 | 4e-04 | 28/39 | X97918 |
| rth27 | 519 | HI1501 | H. influenzae | 520 | 1e-118 | 47/63 | P44225 |
| rth28 | 448 | HI1502 | H. influenzae | 414 | 7e-22 | 31/46 | P44226 |
| rth29 | 165 | MuG (virion morphogenesis) | Phage Mu | 156 | 0.004 | 29/45 | Q01261 |
| MuG homolog HI1568 | H. influenzae | 138 | 2e-19 | 37/50 | P45255 | ||
| rth30 | 354 | MuI homolog HI1504 | H. influenzae | 355 | 4e-19 | 26/38 | I64126 |
| rth31 | 300 | HI1505 | H. influenzae | 308 | 3e-83 | 52/65 | P44227 |
| rth33 | 141 | HI1508 | H. influenzae | 141 | 3e-13 | 32/51 | P44230 |
| rth34 | 222 | HI1509 | H. influenzae | 194 | 0.81 | 22/41 | P44231 |
| rth35 | 475 | Sheath protein gpL | Phage Mu | 495 | 2e-23 | 26/37 | AB000833 |
| gpL (MuL) homolog HI1511 | H. influenzae | 487 | 6e-27 | 25/38 | P44233 | ||
| rth40 | 559 | ORF15 | Phage phi PVL | 694 | 5e-07 | 22/38 | AB009866 |
| rth42 | 455 | 64-kDa virion protein muN | Phage Mu | 491 | 0.011 | 21/38 | P08557 |
| MuN homolog HI1515 | H. influenzae | 457 | 7e-05 | 22/41 | A64127 | ||
| rth43 | 379 | 43-kDa tail protein | Phage Mu | 379 | 5e-16 | 23/40 | P08558 |
| rth44 | 209 | Baseplate assembly protein V (gpV) | Phage P2 | 211 | 3e-06 | 25/40 | P31340 |
| HI1518 | H. influenzae | 182 | 6e-09 | 26/44 | P44238 | ||
| rth45 | 117 | HI1519 | H. influenzae | 135 | 2e-04 | 37/46 | P44239 |
| rth46 | 161 | HI1520 (N terminus) | H. influenzae | 355 | 4e-06 | 29/44 | P44240 |
| rth47 | 191 | Protein xkdT (C terminus) | PBSX prophage | 348 | 1e-07 | 33/48 | P54339 |
| HI1520 (C terminus) | H. influenzae | 355 | 8e-10 | 28/44 | P44240 | ||
| rth48 | 188 | Hypothetical protein YmfQ (27% identical to HI1521) | E. coli | 194 | 6e-08 | 25/43 | P75982 |
| rth49 | 760 | HI1522 | H. influenzae | 623 | 3e-14 | 36/50 | P44242 |
| rth52 | 279 | HI1523 | H. influenzae | 296 | 3e-71 | 47/65 | P44243 |
| gpxA | 177 | Glutathione peroxidase | N. meningitidis | 177 | 0.0 | 100 | P52036 |
| Region 3, 3′ flanking | |||||||
| mccF | 394 | Microcin immunity | E. coli | 344 | 1e-16 | 27/42 | X57583 |
| Region 4 (1.77 kb; B342) | |||||||
| Region 4, 5′ flanking pheS | 330 | Phenylalanine-tRNA synthetase alpha chain | E. coli | 327 | 1e-111 | 59/73 | P08312 |
| Region 4 | |||||||
| rfo2 | 172 | No homologies | |||||
| rfo1 | 354 | No homologies | |||||
| Region 4, 3′ flanking | |||||||
| pheT (fragment) | 693 | Phenylalanine-tRNA synthetase beta chain | E. coli | 795 | 1e-148 | 41/59 | P07395 |
| Region 5 (2.41 kb; E136) | |||||||
| Region 5, 5′ flanking | |||||||
| rfaE | 323 | ADP-heptose synthase | H. influenzae | 342 | 6e-77 | 49/64 | U17642 |
| Region 5 | |||||||
| rfi1 | 411 | scrFI-a, modification methylase | L. lactis | 389 | 1e-46 | 31/48 | P34877 |
| rfi2 | 376 | dcrH, type II restriction enzyme | N. gonorrhoeae | 374 | 6e-08 | 35/55 | AF001598 |
| Region 5, 3′ flanking | |||||||
| rfaD | 334 | ADP-l-glycero-d-mannoheptose epimerase | N. gonorrhoeae | 334 | 0.0 | 97/99 | L07845 |
| Region 6 (3.45 kb; B208) | |||||||
| Region 6, 5′ flanking | |||||||
| fnr | 259 | ferredoxin-NADP+ reductase | A. vinelandii | 258 | 9e-32 | 32/53 | A57432 |
| Region 6 | |||||||
| Ψrsi1 pseudogene | 1045 | fpvA, ferripyoveridine receptor precursor | P. aeruginosa | 813 | 5e-38 | 33/51 | P48632 |
| Region 6, 3′ flanking | |||||||
| dinP | 352 | DNA-damage-inducible protein P | E. coli | 351 | 2e-65 | 41/58 | Q47155 |
| Region 7 (5.25 kb; E116) | |||||||
| Region 7, 5′ flanking | |||||||
| orf-C7 | 333 | ORF on plasmid pJTPS1 | R. solanacearum | 444 | 8e-20 | 32/47 | AB015669 |
| Region 7 | |||||||
| ΨhlyD pseudogene | 454 (+54) | Hemolysin secretion protein D | E. coli | 478 | 3e-77 | 35/56 | P09986 |
| tolC | 467 | Outer membrane protein | E. coli | 495 | 5e-20 | 23/40 | P02930 |
| rse1 | 111 | sll0201, putative transposase | Synechocystis sp. | 164 | 4e-12 | 42/64 | D64000 |
| Region 7, 3′ flanking | |||||||
| relA (fragment) | 345 | GTP-pyrophosphokinase | E. coli | 744 | 5e-58 | 36/57 | P11585 |
| Region 8 (4.23 kb; B313 and B341) | |||||||
| Region 8, 5′ flanking | |||||||
| YCL026c-a | 201 | Hypothetical protein on chromosome III | S. cerevisiae | 192 | 9e-20 | 31/53 | P37261 |
| Region 8 | |||||||
| fhuA | 703 | Ferrichrome iron receptor | E. coli | 747 | 5e-26 | 23/40 | P06971 |
| rei1 | 219 | HP1334, hypothetical protein | H. pylori | 224 | 2e-46 | 47/61 | AE000635 |
| dsbA | 231 | Disulfide oxidoreductase | P. syringae | 214 | 3e-18 | 28/47 | AF036929 |
| Region 8, 3′ flanking | |||||||
| ffh | 456 | Signal recognition particle protein | E. coli | 453 | 1e-174 | 64/76 | P07019 |
| Region 9 (6.04 kb; E102) | |||||||
| Region 9, 5′ flanking | |||||||
| uvrD | 735 | DNA helicase II | E. coli | 720 | 0.0 | 47/65 | P03018 |
| Region 9 | |||||||
| rni1 | 215 | HI1731, urea amidolyase homolog | H. influenzae | 213 | 6e-69 | 58/72 | P44299 |
| rni2 | 309 | HI1730, urea amidolyase homolog | H. influenzae | 309 | 1e-101 | 59/71 | P44298 |
| rni3 | 245 | HI1729, lactam utilization protein homolog | H. influenzae | 257 | 3e-80 | 59/77 | P45347 |
| rni4 | 397 | HI1728, braB homolog | H. influenzae | 397 | 1e-162 | 72/84 | G64138 |
| rni5 | 230 | MTH939, unknown function | Methanobacterium | 188 | 1e-7 | 27/47 | AE000868 |
| rni6 | 321 | tra4, transposase for IS element IS4351 | B. fragilis | 326 | 3e-62 | 41/60 | P37247 |
| Region 9, 5′ flanking | |||||||
| glnE (fragment) | 719 | Glutamine-synthetase adenyltransferase | E. coli | 946 | 1e-130 | 37/54 | P30870 |
aa, amino acids. Flanking genes designated as fragments were only partially sequenced. Genes indicated as pseudogenes contain stop codons compared with their homologues in other bacteria.
Region 2 ORFs with no homologies (length in aa): rtw1 (127), rtw2 (893), rtw3 (143), rtw4 (833), rtw5 (162), rtw6 (90), rtw7 (313), rtw8 (311), rtw9 (116), and rtw10 (98).
Region 3 ORFs with no homologies (length in aa): rth5 (80), rth6 (99), rth7 (71), rth8 (122), rth9 (97), rth10 (151), rth11 (203), rth12 (261), rth15 (148), rth16 (225), rth18 (78), rth19 (155), rth20 (101), rth21 (115), rth22 (113), rth23 (93), rth24 (71), rth25 (168), rth26 (539), rth32 (157), rth36 (124), rth37 (128), rth38 (155), rth39 (129), rth41 (238), rth50 (207), and rth51 (162).
The N. meningitidis-specific regions are imported DNA islands in N. gonorrhoeae and N. meningitidis.
The GC contents of most of the ORFs in regions 2 to 3 and 6 to 9 were close to the 51% value typical of N. meningitidis. However, rei1 of region 8 has a GC content of 40%, and regions 4 and 5 have average GC contents of only 33%. The ends of the eight regions are defined by longer stretches of flanking DNA that are 80 to 96% homologous between the two species. Except for region 2 (Fig. 3), the regions begin and end in intergenic stretches. No repeat structures similar to those flanking pathogenicity islands in Enterobacteriaceae were found, except possibly for region 9. In region 9, 39 bp at the 5′ end are repeated 180 bp to the left of the 3′ end of the N. meningitidis-specific sequence. Furthermore, the right end of region 9 includes a putative IS element, IS4351N2, containing a transposase that might have been involved in the insertion of this region.
The comparison between N. meningitidis and N. gonorrhoeae shows that the regions do not correspond to simple insertions but rather replace alternative sequences, 44 bp to over 12 kb in size, in N. gonorrhoeae FA1090 (Table 3). Thus, the N. meningitidis specific regions correspond to DNA islands that are species specific. The following data suggest that some or all of these islands may have arisen by import from other species via recombination in the homologous flanking DNA, similar to the mechanism deduced for three small islands described elsewhere (39). First, DNA uptake sequences (DUS) (GCCGTCTGAA) (9, 10) were found in all the N. meningitidis islands except regions 4 and 5, where they were found in their immediate borders. The presence of a DUS has been associated with import of DNA into N. meningitidis from Haemophilus influenzae (15) and would facilitate the import of such islands from unrelated bacteria. Second, regions of 70 to 88% homology over 75 to 1,100 bp were found at one of the two borders in regions 4 and 6 to 8, whereas the homology between N. meningitidis and N. gonorrhoeae is normally at least 90%. These borders of low homology might represent the remnants of the recombination with foreign DNA (39). Taken together, all of these results are compatible with the import of these specific sequences from different foreign species into N. meningitidis and/or N. gonorrhoeae and justify using the term DNA island for the eight regions.
TABLE 3.
Sizes of the eight islands in N. meningitidis and N. gonorrhoeaea
| Region | Flanking genes
|
Size of
region (kb)
|
Contents of N. gonorrhoeae region (homologies) | ||
|---|---|---|---|---|---|
| Left | Right | N. meningitidis | N. gonorrhoeae | ||
| 2 | cvaBΔ1 | cvaBΔ2 | 19.5 | 0.746 | cvaBΔ3, Correia element |
| 3 | norZ | mccF | 40.0 | 0.047 | |
| 4 | pheS | pheT | 1.8 | 1.841 | hpaIIV, hpaIIM, hphIR |
| 5 | rfaE | rfaD | 2.4 | 0.044 | |
| 6 | fnr | dinP | 3.5 | 0.755 | ORF98 |
| 7 | orf-C7 | relA | 5.3 | >12 | Unknown |
| 8 | YCL026c-a | ffh | 4.2 | 0.640 | ORF151 |
| 9 | uvrD | glnE | 6.0 | 0.328 | ORF58 |
hpaIIV, gene for very-short-patch-repair enzyme from H. parainfluenzae; hpaIIM, gene for modification enzyme from Haemophilus parainfluenzae; hphIR, gene for restriction enzyme from Haemophilus parahaemolyticus. ORF98, ORF151, and ORF58 are ORFs of unknown function whose numbers indicate the length of the putatively encoded protein. The genes flanking region 7 in N. meningitidis are on separate contigs in N. gonorrhoeae. Those N. gonorrhoeae contigs contain a total of 12 kb of DNA at the position where region 7 is located in N. meningitidis, as confirmed by PCR analysis.
Conservation of the eight islands among diverse N. meningitidis.
Twelve variant capsular polysaccharides are expressed by different N. meningitidis, suggesting the existence of at least as many variants of region 1. The data presented above for regions 2 to 9 were based on the sequence analysis of one serogroup A strain of N. meningitidis and do not indicate whether the eight islands are generally present at the same location in diverse strains of N. meningitidis or whether their genetic content is constant. To address this issue, three kinds of experiments were performed with 13 representative N. meningitidis strains, N. gonorrhoeae FA1090, and two commensal N. lactamica strains. The 13 N. meningitidis strains were chosen to represent the genetic diversity of this species according to MLST (18) and include members of the eight hypervirulent clonal groupings called subgroups I, III, IV-1, and VI, ET-5 and ET-37 complex, the A4 cluster, and lineage 3, as well as unrelated endemic strains (serogroup A subgroup IX, STs 25, 30, 48, and 49). (i) To confirm the localization of each island, PCR amplifications were performed by using one primer in each flanking sequence plus a matching reverse primer in the neighboring gene within the island. (ii) The length of each region was determined after PCR amplification by using primers located in the flanking sequences. (iii) Dot blot and Southern hybridizations against chromosomal DNA were performed using PCR products amplified from each of the 85 ORFs as probes. Representative examples of the Southern and dot blot analyses are shown in Fig. 5. Finally, the regions were sequenced at least in part from some of the 13 N. meningitidis strains.
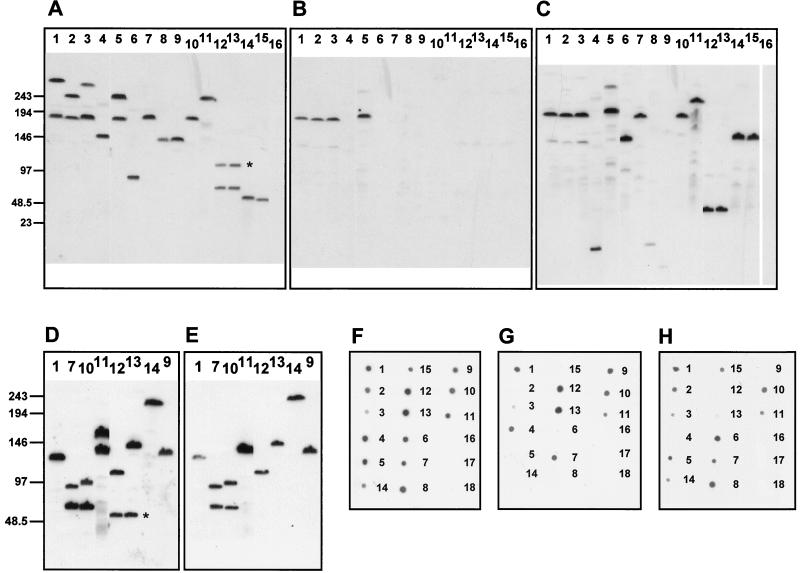
FIG. 5.
Southern analysis of pulsed-field gels of SpeI-digested chromosomal DNA (A to E) and DNA dot blot analysis (F to H) of regions 2 and 3. The numeration of bacterial strains is the same in all panels: 1, subgroup IV-1; 2, subgroup I; 3, subgroup III; 4, subgroup IX; 5, subgroup VI; 6, A4 cluster; 7, lineage 3; 8, ST30; 9, ST49; 10, ST48; 11, ST25; 12, ET-5 complex (44/76); 13, ET-5 complex (MC58); 14, ET-37 complex (FAM18); 15, ET-37 complex (ROU); 16, N. gonorrhoeae FA1090; 17, N. lactamica Z6793; 18, N. lactamica Z6784. (A to C) Southern analysis of region 3 with probes corresponding to rth18 (A), rth33 (B), and gpxA (C). Results identical to those shown in panel A were obtained with probes for rth17 and rth19. Hybridization with probes for rth20 and rth21 resulted in a similar pattern except for lanes 12 and 13, where the upper signal (marked by an asterisk) disappeared. The same results as in panel B were obtained with probes for rth1, rth4, rth28, and rth30. The other ORFs of Pnm1 were not tested by Southern analysis, but similar results can be expected according to dot blot analysis except for rth26, which is also present in strains of the ET-5 complex, ST25, and ST48. (D and E) Southern analysis of region 2 with probes for fhaC (D) and rtw7 (E). The same results as in panel D were obtained with probes for fhaB (5′-part), rtw2, and rtw4. With a probe for rtw5, the lower signals in lanes 12 and 13 (marked by an asterisk) disappeared. The result of hybridization with a probe for rtw8 was the same as with rtw7 (E). Positions of the molecular size markers are shown in kilobases. (F to H) DNA dot blot analysis with probes for the 5′ end (F), the central part (G), and the 3′ end (H) of fhaB.
As expected, none of the probes hybridized with the N. gonorrhoeae strain in dot blots. The other results are summarized in Fig. 2, 3, and 4. They show that, when present, the islands are flanked by the same sequences in all N. meningitidis strains tested and are therefore located at the same chromosomal locations. Furthermore, regions 2, 5, 7, 8, and 9 were present in all representative N. meningitidis strains (Fig. 2 and 3), with little apparent variation in content, except that the central and 3′ end of fhaB in region 2 and rei1 in region 8 did not hybridize with some strains. On the other hand, region 4 was absent in about half of the strains tested and region 6 showed considerable interstrain variation (Fig. 2) due to replacement by an IS element and/or deletions (data not shown). Probes from regions 5 and 6 and two probes from region 3 hybridized with the N. lactamica strains, showing that these genes are not totally specific to N. meningitidis.
Data presented below show that region 3 consists of the gpxA gene encoding glutathione peroxidase (19, 20) plus an integrated 39.3-kb prophage that we have called Pnm1 (prophage in N. meningitidis 1). Pnm1 was present only in serogroup A strains of the epidemic subgroups I, III, IV-1, and VI (Fig. 4) and was lacking in serogroup A subgroup IX, which is unrelated to the epidemic subgroups of serogroup A (1). Furthermore, Pnm1 was also lacking in all other N. meningitidis strains tested. From N. meningitidis strains outside the epidemic serogroup A subgroups, primers specific for the flanking norZ and mccF genes amplified a 1.4-kb product whose sequence contained only gpxA plus small, flanking intergenic regions. Within Pnm1, only probes from rth17 to rth21 hybridized with N. meningitidis strains outside the epidemic serogroup A subgroups. Homology searches in the Sanger Centre genome project for serogroup A strain Z2491 (http://www.sanger.ac.uk/Projects/N_meningitidis/) revealed that these five genes had homologues within a second prophage containing at least 22 ORFs. Furthermore, Southern blots revealed that rth17 to rth21 hybridized with two pulsed-field DNA fragments in strains possessing Pnm1 (Fig. 5). Thus, the hybridization of rth17 to rth21 with all strains of N. meningitidis probably reflects the universal presence of a second prophage.
As with rth17 to rth21, some of the region 2 genes (fhaC, fhaB, rtw2, and rtw4 to rtw8) also hybridized with two pulsed-field DNA fragments in Southern blots from strains of the ET-5 complex, lineage 3, ST25, and ST48 and are therefore present twice in these bacteria (Fig. 5).
Together, these data show that the N. meningitidis-specific islands 2, 5, 7, 8, and 9 are fairly well conserved among representative strains of N. meningitidis and that Pnm1 is conserved among epidemic serogroup A strains.
Sequence analysis of the N. meningitidis-specific islands.
The possible functions of the ORFs in the eight islands were investigated by protein homology searches and computer analysis of the sequences (Table 2). The following description concentrates on those homologies that might be relevant to the virulence potential of N. meningitidis.
The two ORFs at the left of region 2 are homologous to and approximately the same size as genes in Bordetella pertussis encoding the filamentous hemagglutinin (FHA) precursor, FhaB, and the accessory protein, FhaC, involved in the secretion of FHA (17). In B. pertussis, FhaB (367 kDa) is processed during secretion, yielding the 220 kDa FHA protein (5, 6). FHA is thought to be one of the major adhesins of B. pertussis and is also a component of several vaccines against whooping cough. The homology between FhaB in N. meningitidis and B. pertussis is largely restricted to the N-terminal 900 amino acids, including the NPNGI(S/T) sequence involved in secretion and the signal peptide cleavage site (HA↓Q), at the end of an unusually long signal peptide. The central and 3′ end of the fhaB gene are variable between strains according to the dot blot hybridization results (Fig. 3). In addition, the sequencing of region 2 was complicated by the presence of direct repeats (>95% similarity) within fhaB, rtw2, and rtw4 that were designated R1 (431 bp), R2 (271 bp), R3 (160 bp), and R4 (185 bp) (Fig. 3). An interesting hypothesis is that recombination between these repeats could provide for variation in the C-terminal portion of the FhaB protein, a feature that is common among virulence factors in pathogenic neisseriae.
Integrated prophages are known to encode virulence factors in other bacteria (3, 35). Prophage Pnm1 in region 3 contains 52 ORFs, of which 16 are homologous to Mu and related phages (including genes for the transposase and repressor protein). A total of 13 of these 16 ORFs, as well as 7 others, also show homologies to a group of genes from H. influenzae in section 140 to 143 of the genome (7) (Table 2), suggesting that homologous prophages have integrated into both serogroup A N. meningitidis and H. influenzae. In N. meningitidis Z2491, Pnm1 is flanked by direct repeats of AACT (21 bp downstream of norZ and 153 bp upstream of gpxA) which are present only once in nonepidemic serogroup A strains. Pnm1 is the first example of a neisserial prophage, although N. meningitidis bacteriophages have previously been reported (2).
Region 7 contains the pseudogene ΨhlyD and the tolC gene (Fig. 2) in N. meningitidis Z2491. There is a 61-bp insertion containing a stop codon 1,311 bp after the ATG start codon of the ΨhlyD pseudogene in N. meningitidis strain Z2491, followed by a direct duplication of the 73 bp preceding the insertion. On the other hand, sequencing region 7 from the ET-37 complex strain revealed that this strain contains a complete hlyD ORF. In the other N. meningitidis strains, region 7 was the same size as from the ET-37 complex (6.0 kb) (Fig. 2). Furthermore, sequences of the 3′ end of hlyD revealed that none of these other strains contained the insertion from strain Z2491. Thus, with the exception of Z2491, hlyD is normally not a pseudogene. Considering the homologies of hlyD and tolC with genes of the type 1 secretion apparatus in other bacteria, it seems possible that region 7 might be involved in the virulence of most N. meningitidis strains.
Region 8 contains the fhuA, rei1, and dsbA genes in Z2491 (Fig. 2). PCR analysis of region 8 revealed size variations in half of the representative N. meningitidis strains. This region was subsequently sequenced from several strains to determine the basis for this variability. The data revealed that rei1 is deleted in the ET-5 complex, whereas in four other strains (ET-37 complex, lineage 3, ST30, and ST48) an IS element, IS4351N1 (1,076 bp), has been inserted between dsbA and the Correia element, c154 (4). However, fhuA and dsbA were present in all strains. The fhuA gene is a homologue of an E. coli ferrichrome-iron receptor that is involved in the uptake of siderophore-bound iron. fhuA was a pseudogene in some of the strains (subgroup I, subgroup III, A4 cluster, lineage 3, ST48, and ST49) due to stop codons introduced by base changes, insertions, or deletions, but the reading frame was intact in strains belonging to the ET-37 and ET-5 complex, subgroup IV-1, and in ST30. dsbA is homologous to genes in E. coli and Pseudomonas syringae that encode a disulfide oxidoreductase. The predicted N. meningitidis DsbA protein contains the motif C-X-X-C (residues 76 to 79), characteristically present at the active sites of DsbA enzymes. The N. meningitidis DsbA protein also contains a typical lipoprotein signal peptide and cleavage site between A18 and C19 (25) within the motif L-X-A-C. The amino acid following the LXAC motif is S, indicating that the N. meningitidis DsbA is sorted to the outer membrane.
Region 9 contains five ORFs of unknown function plus an IS element, IS4351N2, homologous to that inserted in region 8 in some strains. Sequence analysis revealed that the size differences in region 9 (Fig. 2) were due to the lack of IS4351N2 in strains of the A4 cluster and ET37 complex. The other five genes in region 9 were present in all bacteria, and therefore region 9 was retained for further study of its possible role in virulence.
Regions 4 and 5 are probably not important in N. meningitidis virulence. The former region contains two genes of unknown function and is absent from a large proportion of N. meningitidis strains, and the latter region encodes a restriction-modification system and is also present in the commensal N. lactamica. Region 6 encodes a pseudogene having homologies with a siderophore receptor in Z2491 but is deleted in most of the strains. Regions 4, 5, and 6 were not investigated further.
Virulence analysis of the N. meningitidis-specific islands.
Based on the results described above, regions 2, 3, 7, 8, and 9 were chosen for subsequent investigation of their possible roles in virulence. The entire regions were inactivated by deletion (Fig. 1) and confirmed by Southern blot (Fig. 6). These regions had been sequenced from N. meningitidis strain Z2491, but this strain is not competent for DNA transformation and could not be used to construct the deletion mutants. Therefore, regions 7, 8, and 9 were deleted within N. meningitidis strains MC58 (ET-5 complex) and ROU (ET-37 complex). These strains were chosen to give consistent results in the tests for the different biological phenomena and have been tested previously by several models related to bacterial infection. Neither of these strains contained pseudogenes in any of these regions. Region 2 was deleted only within strain ROU because the chromosome of MC58 contains at least two copies at distinct locations of several of the ORFs from region 2 (see above). The deletions were constructed by using cloned PCR products from the chromosome of strain MC58, except that region 2 was deleted using cloned PCR products from strain ROU. The existence of the desired deletion mutations was confirmed by PCR and Southern blotting. All of these mutants grew well on GC agar and were as transformable as are their parents, indicating that they are piliated.
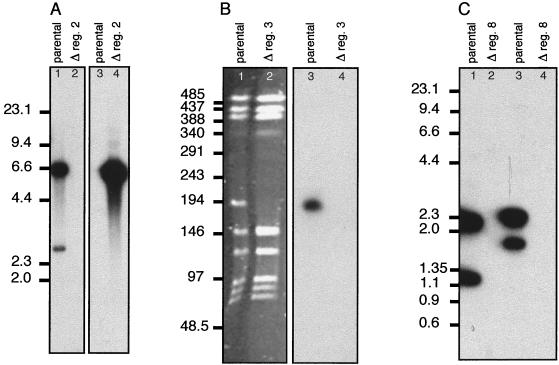
FIG. 6.
Verification of the deletion mutants. Examples are shown of the methods used to verify the deletion of the regions. Positions of the molecular size markers are shown in kilobases for each gel. (A) Deletion of region 2 from strain ROU. Chromosomal DNA from the parental strain (lanes 1 and 3) and the region 2 deletion mutant (lanes 2 and 4) was digested with ClaI. Lanes 1 and 2 were probed with a PCR product corresponding to the gene fhaC; lanes 3 and 4 were probed with the cassette “omega” used to replace the region. Due to its size (about 25 kb) region 2 encompassed several ClaI fragments; similar results were obtained after probing with PCR products corresponding to several of the other ORFs. (B) Deletion of region 3 from strain Z5463. Chromosomal DNA from the parental strain (lanes 1 and 3) and the region 2 deletion mutant (lanes 2 and 4) was digested with SgfI. Lanes 1 and 2 are the pulsed-field gel electrophoresis analysis of the deletion. Note the disappearance of a band at about 194 kb in the mutant and the new band appearing with a size of about 146 kb; this corresponds to the deletion of about 50 kb (region 3). Lanes 3 and 4 were probed with a PCR product corresponding to a portion of the phage transposase gene. (C) Deletion of region 8 from strains MC58 and ROU. Lane 1, MC58 parental strain; lane 2, MC58 region 8 deletion; lane 3, ROU; lane 4, ROU region 8 deletion. Chromosomal DNA was digested with ClaI, and Southern blots were probed with a PCR product corresponding to the entire region 8 from strain Z2491.
First, the deletion mutants of strain ROU were assessed for their ability to adhere to and to invade HUVEC, a model for interactions with the endothelial cells of the BBB. No differences were detected between the parental strain and the deletion mutants (Table 4), indicating that regions 2, 7, 8, and 9 do not affect interactions with endothelial cells.
TABLE 4.
Percent adherence and invasion by deletion mutants of strain ROUa
| Strain | % Adherence | % Invasion |
|---|---|---|
| Parental | 2 | 0.24 |
| ΔRegion 2 | 2 | 0.21 |
| ΔRegion 7 | 2.6 | 0.29 |
| ΔRegion 8 | 2.7 | 0.29 |
| ΔRegion 9 | 2.9 | 0.27 |
Percent adherence is the number of bacteria adherent to HUVEC × 100, divided by the total number of bacteria present after 1 h of infection. Percent invasion is the number of internalized bacteria after 6 h × 100, divided by the number of bacteria present 1 h after infection.
Second, the deletion mutants were tested for sensitivity to the bactericidal activity of complement (Table 5). For these bactericidal assays, meningococci were incubated with 10% guinea pig serum as a complement source supplemented with different dilutions of antiserum raised against the homologous parental strain as an antibody source. The deletion mutants of region 2, 7, 8, or 9 were as resistant to the bactericidal action of the immune serum as their parents in contrast to the serum-sensitive ppk mutant control. Thus, these regions do not seem to be involved in serum resistance.
TABLE 5.
Complement-dependent percent killing of deletion mutantsa
| Strain | % Killing of strain at (serum
dilution):
|
|||
|---|---|---|---|---|
| MC58
|
ROU
|
|||
| 1:3,000 | 1:10,000 | 1:3,000 | 1:10,000 | |
| Parent | 85 | 9 | 93 | 28 |
| ΔRegion 2 | 77 | 7 | 88 | 34 |
| ΔRegion 7 | 80 | −7 | 81 | 18 |
| ΔRegion 8 | 86 | 12 | 86 | 44 |
| ΔRegion 9 | 88 | 8 | 85 | 31 |
| Ppk− control | 100 | 79 | 100 | 98 |
| Ppk− without complement | 6 | 3 | 33 | 13 |
The data show the average percent killing from two experiments with two dilutions of homologous sera against strains MC58 and ROU. As a positive killing control, a serum-sensitive polyphosphate kinase mutant (Ppk−) was tested while, as a negative control, decomplemented guinea pig serum was tested with the Ppk− mutant rather than normal guinea pig serum.
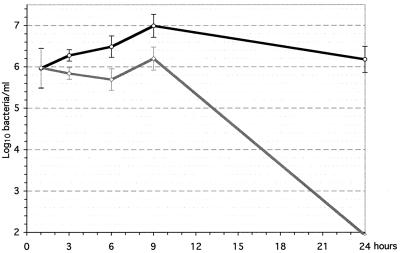
Finally, the mutants were tested for their ability to multiply in the bloodstream in an infant rat model. These tests were performed with the mutants deleted for regions 7, 8, and 9 of strain MC58, which causes high levels of bacteremia in this model. Region 2 could not be tested in this strain because ORFs in region 2 are present at two distinct locations in MC58, the flanking sequences are not known in this strain, and the sequence of region 2 is highly variable between strains. For each experiment, half of the animals in each litter were injected intraperitoneally with the parental strain, and the other half were injected with the deletion mutant; the numbers of bacteria in the blood were then quantitated for 24 h. No differences were detected with deletion mutants of regions 7 or 9 (data not shown), showing that these regions are not important for causing bacteremia. On the other hand, lower numbers of bacteria in blood samples than with the parental control were consistently found with the MC58 deletion mutant lacking region 8 (Fig. 7). Furthermore, only 2 of 15 infant rats died within 48 h after injection of the region 8 deletion mutant, whereas 11 of 14 rats died after injection of the parental control. These data demonstrate that region 8 is important for bacterial survival in the bloodstream.
FIG. 7.
Bacterial concentrations in sequential blood samples after the intraperitoneal infection of infant rats. One typical experiment is shown with average counts and standard deviations. ○, MC58 parental strain; ◊, MC58 Δregion 8.
Virulence determinants may be carried on bacterial prophages (3, 35). Though region 3, including Pnm1, is only present in epidemic serogroup A strains, we deleted this region in order to investigate its possible role in the pathogenesis of these strains. We identified a subgroup IV-1 strain, Z5463, that is transformable and then constructed a region 3 deletion of this strain. Z5463 does not yield high levels of bacteremia in infant rats, and we could not carry out this assay. Because of this strain's sensitivity to complement, bactericidal assays were performed with guinea pig complement in the absence of opsonizing antiserum. The deletion of region 3 did not affect the resistance to complement killing; both the wild-type and mutant strains were killed at concentrations of serum of >1%. Neither did deletion of region 3 affect the interaction of Z5463 with monolayers of HUVEC in the adhesion and invasion assay (data not shown).
DISCUSSION
We investigated genes within eight DNA islands that comprise approximately 5% of the chromosome (100 kb) of N. meningitidis and are lacking in N. gonorrhoeae (Table 6). Many of the genes in these islands seem to be irrelevant to the differences in pathogenicity between these species. However, the islands designated as regions 2, 7, 8, and 9 contained genes homologous to genes from other pathogenic bacteria that are involved in virulence and were present in diverse, representative N. meningitidis strains. Deletion mutants lacking region 8 were impaired in their ability to cause bacteremia in an infant rat model. A ninth island containing genes encoding the synthesis of capsular polysaccharide is also known to be necessary for dissemination in the bloodstream. Thus, two of the nine known islands in N. meningitidis are important for bloodstream dissemination, a prerequisite for causing meningitis. It remains possible that the other seven islands may play a role in virulence that was not detected by the assays used here.
TABLE 6.
Features of the eight islands in N. meningitidis
| Region | Size (kb) | Important homologies | Distribution | Comments |
|---|---|---|---|---|
| 2 | 19.5 | fhaB (FHA of B. pertussis); fhaC (FHA activator protein) | Present in all N. meningitidis strains; high variability of fhaB | Some region 2 genes are present twice in some serogroup B strains |
| 3 (Pnm1) | 40 | Prophage in H. influenzae gpxA (glutathione peroxidase) | Pnm1 in clonally related serogroup A strains only; rth17–rth21 and gpxA in all N. meningitidis strains | Southern blot data suggests a second prophage present in all N. meningitidis strains |
| 4 | 1.8 | None | Present in only 60% of the N. meningitidis strains | Replaced by restriction/modification system in N. gonorrhoeae |
| 5 | 2.4 | Restriction-modification system | Present in all N. meningitidis strains | Also present in N. lactamica |
| 6 | 3.5 | Siderophore receptor (Ψrsi1) | Pseudogene, replaced by IS element or deleted in all N. meningitidis strains | |
| 7 | 5.3 | Leukotoxin secretion (hlyD, tolC) | Present in all N. meningitidis strains | hlyD is a pseudogene in serogroup A, subgroup IV-1 |
| 8 | 4.2 | Siderophore receptor (fhuA), disulfide oxidoreductase (dsbA) | Present in all N. meningitidis strains, but fhuA is a pseudogene in half of the strains | Deletion of region 8 results in reduced bacteremia in the infant rat |
| 9 | 6.0 | Four proteins of H. influenzae with putative metabolic functions | Present in all N. meningitidis strains |
Region 8 contains homologues of fhuA and dsbA, as well as a new gene called rei1. However, rei1 is deleted within MC58, the N. meningitidis strain that was used in the infant rat model, leaving only fhuA and dsbA as candidates for genes involved in bacteremia. FhuA is a siderophore-iron receptor in E. coli. The fhuA gene was defective in several of the N. meningitidis strains, suggesting that it is not important for virulence. The DsbA homologue is present in diverse N. meningitidis strains and is probably sorted to the outer membrane. DsbA proteins in other pathogenic bacteria have been shown to play an indirect role in virulence. They are involved in the correct folding of proteins responsible for both the secretion of invasion proteins by S. flexneri (36) and for its intracellular survival (37), as well as for the formation of type IV fimbriae in enteropathogenic E. coli (38). The DsbA homologue in region 8 does not affect piliation because mutants deleted for region 8 were normally transformable and adhered to endothelial cells in a pilus-dependent assay as efficiently as their parent.
In several bacterial species (for example, E. coli, Helicobacter pylori, Salmonella enterica, and Yersinia enterocolitica), genes encoding increased pathogenicity are clustered in so-called pathogenicity islands (PAIs) (11). PAIs are usually large (50 to 200 kb) and often have a different GC-content from that of the host chromosome. PAIs can be genetically unstable due to flanking repetitive sequences or IS elements and, within the Enterobacteriaceae, tRNA loci often serve as targets for their integration and excision. None of the N. meningitidis-specific islands in regions 2 to 9 fulfills all of these criteria, and most fulfill none of them. Thus, the generic term “DNA island” seems more appropriate for these sequences in the neisseriae than the term PAI. We note that some of the differences between these islands and PAIs may reflect the fact that the neisseriae are naturally transformable and readily undergo genetic exchange via homologous recombination after DNA transformation, whereas mobile genetic elements are often more important for the Enterobacteriaceae.
N. meningitidis probably contains more DNA islands than the eight described here. For example, upstream of region 2 in N. meningitidis is another small island containing orf1 plus a 106-bp long Correia element (Fig. 3), whereas two small ORFs are present at that location in N. gonorrhoeae. Similarly, three other islands in the opcA and ΨopcB regions were described elsewhere (39), and the regions corresponding to three N. meningitidis-specific clones (32) were also not identified here. Finally, region 1 encodes genes involved in capsular biosynthesis and was not investigated further in this report. Thus, a considerable proportion of the differences between N. meningitidis and N. gonorrhoeae may be encoded by species-specific DNA islands, only some of which have been analyzed here.
The eight islands were present in most of the strains studied, and their overall organization was conserved, despite the presence of a number of pseudogenes. The conservation of region 5 (restriction-modification system) can be explained, since functional restriction-modification systems act as selfish genetic units (21). Similarly, region 4 of N. meningitidis is replaced by a restriction-modification system in N. gonorrhoeae FA1090 (see Table 3, hpaIIM and hphIR), which has no homologue in N. meningitidis and whose corresponding genes exhibit a low GC content. The presence of some of the other islands might be accounted for by the selfish operon theory (16), whereby the integrity of a group of genes linked by a common function is favored by stochastic mechanisms. On the other hand, conserved, functional genes are classically the result of natural selection by the environment, and those in the N. meningitidis-specific islands should therefore be beneficial to N. meningitidis in those environments which it specifically inhabits. N. meningitidis and N. gonorrhoeae differ not only in their characteristic pathogenicities but also in the anatomical sites they colonize. Thus, these islands might be relevant to multiplication under the particular biochemical conditions and in the presence of the microbial competition experienced by the meningococcus in its natural habitat. Region 2 encodes fhaB and fhaC homologues, as well as a number of ORFs with no obvious homologies, and is present in all N. meningitidis strains analyzed. The FHA protein is an adhesin of B. pertussis. Although the N. meningitidis fhaB homologue is considerably shorter than that in B. pertussis, it could produce a 224-kDa protein after cleavage of the signal peptide that is of comparable size to the processed FHA. The conservation of an amino-terminal asparagine-rich domain plus the conserved NPNGI motif might reflect a similar secretion mechanism. However, the C termini differ extensively between B. pertussis and N. meningitidis; such sequence variability might reflect functional differences or diversifying selection due to the human immune system. Serum sensitivity and adhesion were not affected by deletion of region 2, indicating that it is not essential for these phenotypes in N. meningitidis. It was not possible to test whether region 2 is important for blood dissemination because MC58, the strain used for these tests, possesses two copies of ORFs from region 2 at distinct locations, and ROU, the other strain in which a deletion of region 2 was introduced, does not replicate in the infant rat for unknown reasons, although it is a clinical isolate from a case of meningitis with septicemia. Notwithstanding the lack of sequence information, we were able to produce a double mutation in the gene fhaB in the two region 2 loci of another strain, 8013, by the deletion of one region 2 followed by the insertional inactivation of the fhaB gene in the other. The double mutant had no alteration with respect to the in vitro or in vivo assays (data not shown); however, it is not possible to extrapolate this result to consider the whole of region 2.
In summary, we describe the characteristics and distribution of eight DNA islands that are specific to N. meningitidis and show that genes on one of these islands are important for virulence. These results will form the basis of additional experiments to develop new protein vaccine candidates and to define factors that are important for bacterial invasion of the bloodstream.
ACKNOWLEDGMENTS
This work was supported by a grant from SmithKline Beecham Biologicals s.a., Rixensart, Belgium.
We gratefully acknowledge the helpful advice of Martin Schenker.
ADDENDUM IN PROOF
During the course of this work, sequences corresponding to two of the regions have been submitted to GenBank by other groups. Region 5 with its flanking rfaE and rfaD genes has been sequenced in the N. meningitidis serogroup B strain CDC 8201085 (C. M. Kahler and D. S. Stephens, GenBank accession number AF125564), where it is 98.6% identical to region 5 of Z2491. The rfi1 and rfi2 genes are designated nmgII and nmgI, respectively. Region 7 was sequenced in the N. meningitidis serogroup C strain IR1075 (I. Stojiljkovic, GenBank accession number AF121772), where it contains natC, which is 99.8% identical to tolC of Z2491, and natD, with 99% identity to hlyD of Z2491 over the first 1,310 bases.
REFERENCES
- 1.Bart A, Schuurman I G A, Achtman M, Caugant D A, Dankert J, van der Ende A. Random amplified polymorphic DNA (RAPD) genotyping of serogroup A meningococci yields similar results to multilocus enzyme electrophoresis (MEE) and reveals new genotypes. J Clin Microbiol. 1998;36:1746–1749. doi: 10.1128/jcm.36.6.1746-1749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cary S G, Hunter D H. Isolation of bacteriophages active against Neisseria meningitidis. J Virol. 1967;1:538–542. doi: 10.1128/jvi.1.3.538-542.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheetham B F, Katz M E. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 4.Correia F F, Inouye S, Inouye M. A family of small repeated elements with some transposon-like properties in the genome of Neisseria gonorrhoeae. J Biol Chem. 1988;263:12194–12198. [PubMed] [Google Scholar]
- 5.Delisse-Gathoye A M, Locht C, Jacob F, Raaschou-Nielsen M, Heron I X R J, de Wilde M, Cabezon T. Cloning, partial sequence, expression, and antigenic analysis of the filamentous hemagglutinin gene of Bordetella pertussis. Infect Immun. 1990;58:2895–2905. doi: 10.1128/iai.58.9.2895-2905.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domenighini M, Relman D, Capiau C, Falkow S, Prugnola A, Scarlato V, Rappuoli R. Genetic characterization of Bordetella pertussisfilamentous haemagglutinin: a protein processed from an unusually large precursor. Mol Microbiol. 1990;4:787–800. doi: 10.1111/j.1365-2958.1990.tb00649.x. [DOI] [PubMed] [Google Scholar]
- 7.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, Mckenney K, Sutton G, Fitzhugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzaeRd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 8.Frosch M, Weisgerber C, Meyer T F. Molecular characterization and expression in Escherichia coli of the gene complex encoding the polysaccharide capsule of Neisseria meningitidisgroup B. Proc Natl Acad Sci USA. 1989;86:1669–1673. doi: 10.1073/pnas.86.5.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodman S D, Scocca J J. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1988;85:6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodman S D, Scocca J J. Factors influencing the specific interaction of Neisseria gonorrhoeaewith transforming DNA. J Bacteriol. 1991;173:5921–5923. doi: 10.1128/jb.173.18.5921-5923.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 12.Hammerschmidt S, Birkholz C, Zähringer U, Robertson B D, Van Putten J, Ebeling O, Frosch M. Contribution of genes from the capsule gene complex (cps) to lipooligosaccharide biosynthesis and serum resistance in Neisseria meningitidis. Mol Microbiol. 1994;11:885–896. doi: 10.1111/j.1365-2958.1994.tb00367.x. [DOI] [PubMed] [Google Scholar]
- 13.Hammerschmidt S, Müller A, Sillmann H, Müllenhoff M, Borrow R, Fox A, Van Putten J, Zollinger W, Gerardy-Schahn R, Frosch M. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand misspairing in the polysialyltransferase gene (siaD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol Microbiol. 1996;20:1211–1220. doi: 10.1111/j.1365-2958.1996.tb02641.x. [DOI] [PubMed] [Google Scholar]
- 14.Kellogg D S, Peacock W L, Deacon W E, Brown L, Pirkle C I. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J Bacteriol. 1963;85:1274–1279. doi: 10.1128/jb.85.6.1274-1279.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroll J S, Wilks K E, Farrant J L, Langford P R. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc Natl Acad Sci USA. 1998;95:12381–12385. doi: 10.1073/pnas.95.21.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence J G, Roth J R. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics. 1996;143:1843–1860. doi: 10.1093/genetics/143.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Locht C, Bertin P, Menozzi F D, Renauld G. The filamentous haemagglutinin, a multifaceted adhesion produced by virulent Bordetellaspp. Mol Microbiol. 1993;9:653–660. doi: 10.1111/j.1365-2958.1993.tb01725.x. [DOI] [PubMed] [Google Scholar]
- 18.Maiden M C J, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore T D, Sparling P F. Interruption of the gpxA gene increases the sensitivity of Neisseria meningitidisto paraquat. J Bacteriol. 1996;178:4301–4305. doi: 10.1128/jb.178.14.4301-4305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore T D E, Sparling P F. Isolation and identification of a glutathione peroxidase homolog gene, gpxA, present in Neisseria meningitidis but absent in Neisseria gonorrhoeae. Infect Immun. 1995;63:1603–1607. doi: 10.1128/iai.63.4.1603-1607.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naito T, Kusano K, Kobayashi I. Selfish behavior of restriction-modification systems. Science. 1995;267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 22.Parsons J D. Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- 23.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 24.Pron B, Taha M K, Rambaud C, Fournet J C, Pattey N, Monnet J P, Musilek M, Beretti J L, Nassif X. Interaction of Neisseria meningitidiswith the components of the blood-brain barrier correlates with an increased expression of PilC. J Infect Dis. 1997;176:1285–1292. doi: 10.1086/514124. [DOI] [PubMed] [Google Scholar]
- 25.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sampson B A, Gotschlich E C. Neisseria meningitidisencodes an FK506-inhibitable rotamase. Proc Natl Acad Sci USA. 1992;89:1164–1168. doi: 10.1073/pnas.89.4.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkari J, Pandit N, Moxon E R, Achtman M. Variable expression of the Opc outer membrane protein in Neisseria meningitidisis caused by size variation of a promoter containing polycytidine. Mol Microbiol. 1994;13:207–217. doi: 10.1111/j.1365-2958.1994.tb00416.x. [DOI] [PubMed] [Google Scholar]
- 28.Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 29.Thompson S A, Sparling P F. The RTX cytotoxin-related FrpA protein of Neisseria meningitidis is secreted extracellularly by meningococci and by HlyBD+Escherichia coli. Infect Immun. 1993;61:2906–2911. doi: 10.1128/iai.61.7.2906-2911.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson S A, Wang L L, Sparling P F. Cloning and nucleotide sequence of frpC, a second gene from Neisseria meningitidisencoding a protein similar to RTX cytotoxins. Mol Microbiol. 1993;9:85–96. doi: 10.1111/j.1365-2958.1993.tb01671.x. [DOI] [PubMed] [Google Scholar]
- 31.Tinsley C R, Gotschlich E C. Cloning and characterization of the meningococcal polyphosphate kinase gene: production of polyphosphate synthesis mutants. Infect Immun. 1995;63:1624–1630. doi: 10.1128/iai.63.5.1624-1630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tinsley C R, Nassif X. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae: Two closely related bacteria expressing two different pathogenicities. Proc Natl Acad Sci USA. 1996;93:11109–11114. doi: 10.1073/pnas.93.20.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Virji M, Makepeace K, Peak I, Ferguson D J P, Jennings M P, Moxon E R, Peak I R A. Opc- and pilus-dependent interactions of meningococci with human endothelial cells: molecular mechanisms and modulation by surface polysaccharides. Mol Microbiol. 1995;18:741–754. doi: 10.1111/j.1365-2958.1995.mmi_18040741.x. [DOI] [PubMed] [Google Scholar]
- 34.Vogel U, Hammerschmidt S, Frosch M. Sialic acids of both the capsule and the sialylated lipooligosaccharide of Neisseria meningitidisserogroup B are prerequisites for virulence of meningococci in the infant rat. Med Microbiol Immunol. 1996;185:81–87. doi: 10.1007/s004300050018. [DOI] [PubMed] [Google Scholar]
- 35.Waldor M K. Bacteriophage biology and bacterial virulence. Trends Microbiol. 1998;6:295–297. doi: 10.1016/s0966-842x(98)01320-1. [DOI] [PubMed] [Google Scholar]
- 36.Watarai M, Tobe T, Yoshikawa M, Sasakawa C. Disulfide oxidoreductase activity of Shigella flexneriis required for release of Ipa proteins and invasion of epithelial cells. Proc Natl Acad Sci USA. 1995;92:4927–4931. doi: 10.1073/pnas.92.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu J. Inactivation of DsbA, but not DsbC and DsbD, affects the intracellular survival and virulence of Shigella flexneri. Infect Immun. 1998;66:3909–3917. doi: 10.1128/iai.66.8.3909-3917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H Z, Donnenberg M S. DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:787–797. doi: 10.1046/j.1365-2958.1996.431403.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhu P, Morelli G, Achtman M. The opcA and ΨopcB regions in Neisseria: genes, pseudogenes, deletions, insertion elements and DNA islands. Mol Microbiol. 1999;33:635–650. doi: 10.1046/j.1365-2958.1999.01514.x. [DOI] [PubMed] [Google Scholar]