Abstract
Specific virulence factors associated with the pathogenesis of Escherichia coli strains causing neonatal meningitis (ECNM), such as the K1 capsular polysaccharide, the S fimbriae, and the Ibe10 protein, have been previously identified. However, some other yet unidentified factors are likely to be involved in the pathogenesis of ECNM. To identify specialized unique DNA regions associated with ECNM virulence, we used the representational difference analysis technique. The genomes of two strains belonging to nonpathogenic phylogenetic group A of the ECOR reference collection were subtracted from E. coli strain C5, isolated from a case of neonatal meningitis. Strain C5 belongs to the phylogenetic group B2 as do the majority of ECNM. We have isolated and mapped 64 DNA fragments which are specific for strain C5 and not found in nonpathogenic strains. Of these clones, 44 were clustered in six distinct regions on the chromosome. The sfa and ibe10 genes were located in regions 2 and 6, respectively. A group of genes (cnf1, hra, hly, and prs) known to be present in a pathogenicity island of the uropathogenic strain E. coli J96 colocalized with region 6. The occurrence of these DNA regions was tested in a set of meningitis-associated strains and in a control group composed of non-meningitis-associated strains belonging to the same B2 group. Regions 1, 3, and 4 were present in 91, 82, and 81%, respectively, of the meningitis strains and in 40, 13, and 47% of the control strains. Together, these data suggest that regions 1, 3, and 4 code for factors associated with the ability of E. coli to invade the meninges of neonates.
Escherichia coli is responsible for a third of the cases of neonatal meningitis (NM), with an incidence of 0.1 per 1,000 live births (8). Case fatality rates are still very high and range from 25 to 40%. Furthermore, the occurrence of long-term neurologic sequelae in nonfatal cases is 33 to 50% of neonates with E. coli meningitis (8, 9, 32). Understanding the pathogenesis of this disease and characterizing these pathogenic strains are prerequisites to the development of new treatments.
Few specific pathogenic determinants have been described for E. coli strains causing NM (ECNM). Both expression of the K1 capsular polysaccharide (17) and production of aerobactin (21) are believed to be important for bloodstream dissemination. On the other hand, S fimbrial adhesin (sfa) (11, 17, 23) and Ibe10 protein (13), involved in the adhesion and invasion of brain microvascular endothelial cells, likely promote the crossing of the blood-brain barrier.
Phylogenetic approaches have helped to characterize the pathogenic strains. The E. coli species has been divided into four main phylogenetic groups designated A, B1, B2, and D (12, 28). Previous studies have shown that ECNM has a clonal structure (3, 27) and that strains mostly belong to the B2 group (5). Considering that only 38% of ECNM have both sfa and ibe10, it is likely that other determinants remain to be identified (5). In favor of this hypothesis is the fact that 10 pathogen-specific chromosomal segments have recently been detected by comparative macrorestriction mapping of the chromosomes of neonatal meningitis-associated E. coli RS218 and the laboratory strain E. coli K-12 (26). Two of these segments colocalized with the sfa and ibe10 genes. However, no functional or epidemiological studies have confirmed the role of these 10 DNA segments in the pathogenesis of ECNM.
In this work, we compared the chromosome of the C5 strain, which belongs to the B2 group, with those of two nonpathogenic E. coli strains by using representational difference analysis (18, 30). We obtained a library of sequences which are present only in the virulent strain. These sequences clustered onto the chromosome in six distinct regions. Comparing the presence of these regions among meningitis and non-meningitis-associated strains of the B2 group allowed us to identify three regions as being specific for the ECNM.
MATERIALS AND METHODS
Bacterial strains.
Strains used for subtractive hybridization were E. coli C5 (serotype O18:K1:H7), isolated from the cerebrospinal fluid (CSF) of a newborn (15) and obtained from R. Bertolussi (Dhalousie University, Halifax, Nova Scotia, Canada), and two nonpathogenic E. coli strains, ECOR4 and ECOR15, belonging to the ECOR collection (22). E. coli C5 harbors several virulence factors, such as the capsular antigen K1, an sfa operon, the ibe10 gene, Pap pili, and the hemolysin gene (hly) (5, 13, 16). This strain belongs to the phylogenetic B2 group (5). On the other hand, the ECOR4 and ECOR15 strains, which belong to the phylogenetic A group, do not express any identified virulence factors, a property of almost all strains of this group (5, 7, 10, 24). Other E. coli strains were RS218 (serotype O18:K1:H7), an isolate from the CSF of a newborn, kindly provided by K. Kim (Childrens Hospital, Los Angeles, Calif.) (13), which harbored the same virulence factors as the C5 strain, and the E. coli laboratory strain K-12 MG1655, the genome of which has recently been sequenced (6).
In addition we used a set of 54 ECNM obtained from the CSF of newborns with meningitis (age range, 1 to 28 days) and belonging to the phylogenetic B2 group (5). This population was compared to the 15 non-meningitis-associated E. coli strains from phylogenetic group B2 of the 72 strains of the ECOR collection (22). This collection was obtained from R. Selander (Department of Biology, University of Rochester, Rochester, N.Y.). These reference strains, isolated from a variety of hosts and geographical locations, are representative of the range of genotypic variation in the species and are divided in four main phylogenetic groups (A, B1, B2, and D) (12, 28). Bacteria were grown at 37°C in Luria-Bertani broth or on Luria-Bertani agar. When necessary, ampicillin was used at a concentration of 100 μg per ml.
Southern blotting.
Southern blotting was performed by capillary transfer onto positively charged nylon membranes. Hybridizations were performed at 65°C in 1% sodium dodecyl sulfate–1 M NaCl–50 mM Tris HCl (pH 7.5)–1% blocking reagent (Boehringer Mannheim, Mannheim, Germany). The membranes were washed first in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 min at room temperature, then in 2× SSC–0.1 sodium dodecyl sulfate for 30 min at 65°C, and finally in 0.1× SSC for 5 min at room temperature. Detection by chemiluminescence was performed with the DIG luminescence detection kit for nucleic acid (Boehringer Mannheim) according to the manufacturer's instructions. The sfa and ibe10 probes were produced by PCR, using primers and an amplification procedure previously described (4).
Representational difference analysis.
The procedure was that of Tinsley and Nassif (30). Chromosomal DNA from the ECOR strains was sheared by repeated passage through a hypodermic needle to obtain fragments ranging between 3 and 10 kb long. This digested DNA was purified by phenol extraction. Chromosomal DNA from E. coli C5 was cleaved with the restriction endonuclease Sau3AI or Tsp509I. This DNA (20 μg) was ligated with 10 nmol of the annealed oligonucleotides RBam12 (5′-GATCCTCGGTGA-3′) and RBam24 (5′-AGCACTCTCCAGCCTCTCACCGAG-3′) or REco12 (5′-AATTCTCGGTGA-3′) and REco24 (5′-AGCACTCTCCAGCCTCTCACCGAG-3′) when the restriction was done by SauAI or Tsp509I, respectively. DNA was separated from the excess primers by electrophoresis through a low-melting-point 2% agarose gel. The portion of the gel containing fragments of more than 200 bp was excised and digested with β-agarase. This DNA was purified by phenol extraction.
For the subtractive hybridization (first round), 0.2 μg of E. coli C5 DNA, ligated to the oligonucleotides, was mixed with 40 μg of sheared of ECOR4 or ECOR15 DNA in a total volume of 8 μl of 3× EE buffer [1× EE buffer is 10 mM N-(2-hydroxyethyl) piperazine-N′-(3-propanesulfonic acid)–1 mM EDTA (pH 8.0)]. The solution was overlaid with mineral oil, and the DNA was denatured by heating at 100°C for 2 min; 2 μl of 5 M NaCl was added, and the mixture was allowed to hybridize at 55°C for 48 h. The reaction mixture was diluted 10-fold with preheated 3× EE buffer–1 M NaCl and immediately placed on ice. A portion of the dilution (10 μl) was added to 400 μl of PCR mixture (10 mM Tris HCl [pH 9.0], 50 mM KCl, 1.5 mM MgCl2, 0.1% Triton X-100, a 0.25 mM concentration of each deoxynucleoside triphosphate, 50 U of Taq polymerase per ml) and incubated for 3 min at 70°C to fill in the ends of the reannealed E. coli C5 fragments. After denaturation at 94°C for 5 min and addition of the oligonucleotide RBam24 or REco24 (0.1 nmol per 100 μl), the hybridizations were amplified by PCR (30 cycles of 1 min at 94°C, 1 min at 70°C, and 3 min at 72°C, followed by 1 min at 94°C and 10 min at 72°C in a GeneAmp 9600 thermal cycler [Perkin-Elmer]). The PCR products were gel purified to separate amplified E. coli C5 fragments from the primer and high-molecular-weight subtracting ECOR DNA. A second round of subtractive hybridization was performed, using 40 μg of sheared E. coli ECOR4 or ECOR15 and 25 ng of RBam24- or REco24-ligated DNA obtained from the first round. These second-round difference products were radiolabeled en masse and used as the probe in Southern hybridization experiments to ensure that the amplified fragments were unique to the pathogenic DNA and not present in the nonpathogenic strains. Thus, four subtractive libraries were obtained.
Analysis of clones from the subtractive libraries.
DNA from the subtractive libraries was cloned into the BamHI (Sau3AI libraries) or EcoRI (Tsp509I libraries) site of pUC19 (New England Biolabs, Beverly, Mass.) and then transformed into Epicurian coli XL2-blue ultracompetent cells (Stratagene, La Jolla, Calif.). The inserts were amplified by PCRs performed on transformant colonies, using the following primers: P1 (5′-CATGCCTGCAGGTCGACTCT-3′) and P2 (5′-CGTTGTAAAACGACGGCCAG-3′). Clones were designated by the following (in order): the restriction enzyme used (“Tsp” or “Sau”), the strain used for the subtraction (E4 or E15), and an alphanumeric designation.
(i) DNA sequencing.
After purification of PCR products by solid-phase reversible immobilization, purified PCR fragments were sequenced using the BigDye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq DNA polymerase FS (Perkin-Elmer) on an automated ABI PRISM 377 XL DNA Sequencer (Perkin-Elmer) by following the manufacturer's instructions. When problems in obtaining a good-quality sequence were encountered with a particular primer, a sequencing reaction was performed with the dGTP BigDye Terminator Ready Reaction kit (Perkin-Elmer). Sequences were screened for homologies with previously published sequences using the computer programs BLASTN and BLASTX at the National Center for Biotechnology Information (2).
(ii) Southern blot hybridization.
To check for their specificity, the PCR products obtained by using primers P1 and P2 from the colonies of transformants were labeled by incorporation of digoxigenin-11-dUTP (Boehringer Mannheim) and used as probes for Southern blot analysis of DraI-digested chromosomal DNA from strains E. coli C5, ECOR4, and ECOR15 and E. coli K-12 MG1655.
(iii) Pulsed-field gel electrophoresis and mapping of the clones on the chromosomes of strains RS218 and C5.
The position of the DNA sequences corresponding to the cloned difference products was determined relative to the map of E. coli RS218 (26) by probing Southern blots of pulsed-field agarose gels. DNA of strain RS218 was digested with BlnI, NotI, and XbaI and subjected to pulsed-field gel electrophoresis, as was DNA of strain C5 that had been digested by BlnI and NotI. Gels were 1% agarose in 0.5× Tris-borate-EDTA buffer, and were subjected to electrophoresis at 6 V/cm for 27 h, with pulse times varying linearly between 2 and 49 s. The positions on the RS218 chromosome of sequences reactive with each of the clones were determined by comparison of the recognized BlnI and NotI restriction fragments with the published macrorestriction map (26).
Nucleotide sequence accession numbers.
The sequences of the subtractive DNA fragments have been assigned GenBank accession numbers AF222070 to AF222222 for sequences showing no homology and numbers AF222223 to AF222307 for sequences with significant homology.
RESULTS
Production of libraries of DNA fragments of pathogenetic strain C5 not found in the genome of nonpathogenic E. coli.
Using the technique of representational difference analysis, we subtracted the chromosomes of the two nonpathogenic (ECOR4 and ECOR15) strains from the chromosome of the C5 pathogenic strain. Four libraries were produced and designated SauE4, SauE15, TspE4, and TspE15, according to the enzyme used to digest the chromosome of the C5 strain and the strain used for subtraction. In each case, the amplified difference product from the second round of subtraction was labeled and used as a probe against DraI-digested DNA from C5, RS218, ECOR4, and ECOR15. A strong reactivity with the chromosome of pathogenic strains was observed. On the other hand, little or no signal was present in the lanes corresponding to the subtractive nonpathogenic strains (data not shown). Altogether, 494 clones were subsequently isolated and sequenced. Of these, 140 had significant homology with sequences of E. coli K-12 and were subsequently discarded. Among the 354 remaining fragments, 259 sequences were unique. Table 1 shows all the clones which had a significant homology with previously described genes (except bacteriophages). Some of these clones corresponded to genes already known to be present in the C5 strain, such as pap, hly, and kps. None of these clones were found to be homologous with sfa or ibe10. On the other hand, sequences corresponding to virulence factors found in strains not responsible for NM were present, as follows: (i) prs, cnf, and hra, all part of a pathogenicity island (PAI) in the uropathogenic strain E. coli J96 (29); (ii) chuA, a gene involved in an iron transport system and found in enterohemorrhagic E. coli O157:H7 (31); and (iii) senB, a gene encoding an enterotoxin on the virulence plasmid of Shigella and enteroinvasive E. coli (20). Finally, 153 fragments showed no significant homology with any published sequence.
TABLE 1.
Summary of BLAST search of clones that were specific for E. coli strain C5 and had significant homologiesa
| Cloneb | Insert size (bp) | Sequence(s) with similarityc | Score | Probability | GenBank accession no. |
|---|---|---|---|---|---|
| SauE15.A1 | 163 | IS629 (N), plasmid pO157, E. coli O157:H7 | 315 | e−84 | AB011549 |
| SauE15.A7 | 190 | iroC (N), ATP cassette transporter (iron-regulated locus), Salmonella enterica serovar Typhi | 157 | e−37 | U6129 |
| SauE15.B2 | 294 | repB (N); replication protein, plasmid pCD1; Yersinia pestis | 123 | e−26 | AF053946 |
| SauE15.B6 | 157 | kps (N), promoter region of polysialic acid gene cluster region 3, E. coli | 242 | e−62 | U05251 |
| SauE15.B9 | 107 | traD (N), F sex factor plasmid, E. coli | 198 | e−50 | M29254 |
| SauE15.B10 | 240 | ORF 34 and 35 (P), 102-kb unstable region, Y. pestis | 69 | e−12 | CAA21357 |
| SauE15.B12 | 479 | unknown protein (P), E. coli ec11 | 102 | e−21 | AF044503 |
| SauE15.C1 | 100 | r6 (N), transposase, pathogenicity island of E. coli CFT073 | 198 | e−49 | AF081285 |
| SauE15.C6 | 119 | IS100 (N), Y. pestis | 228 | e−58 | L19030 |
| SauE15.C7 | 155 | TonB-dependent receptor HI1217 precursor (P), Haemophilus influenzae | 52 | e−7 | P45114 |
| SauE15.C9 | 273 | rhuM (N), pathogenicity island of Salmonella enterica serovar Typhimurium (SPI3) | 311 | e−83 | AF106566 |
| SauE15.C11 | 77 | orfE (N); promoter-distal region of the tra operon, plasmid R100; S. flexneri | 129 | e−29 | X55815 |
| SauE15.D4 | 153 | IS100 (N), Y. pestis | 287 | e−76 | L19030 |
| SauE15.D8 | 347 | r3 (N), beta-cystathionase, pathogenicity island of E. coli CFT073 | 615 | e−174 | AF081286 |
| SauE15.E4 | 281 | senB (N), enterotoxin, E. coli | 541 | e−152 | Z54195 |
| SauE15.E11 | 314 | traJ, Y (N), plasmid R1–19, E. coli | 523 | e−147 | M19710 |
| SauE15.F3 | 422 | chuA (P), heme utilization gene, E. coli O157:H7 | 98 | e−20 | U67920 |
| SauE15.F9 | 137 | Thioesterase (P), Bacillus sp. | 48 | e−6 | AB016427 |
| SauE15.F10 | 210 | r3 (N), beta-cystathionase, pathogenicity island of E. coli CFT073 | 408 | e−112 | AF081286 |
| SauE15.G3 | 206 | traG (N), plasmid R100, S. flexneri | 165 | e−39 | U01159 |
| SauE15.G6 | 328 | IS100 (N), Y. pestis | 480 | e−134 | L19030 |
| SauE15.H5 | 200 | HMWP1 protein (P), Yersinia enterocolitica | 80 | e−15 | CAA73127 |
| SauE15.H7 | 150 | Oxydoreductase (P), Thermotoga maritima | 160 | e−11 | AE001762 |
| SauE15.H10 | 141 | traT (N), plasmid R100, E. coli | 280 | e−74 | J01769 |
| SauE15.H11 | 160 | Hemoglobin protease (P), E. coli EB1 | 50 | e−6 | CAA11507 |
| SauE15.I3 | 176 | asst (N), arylsulfate sulfotransferase, Klebsiella sp. | 341 | e−92 | U32616 |
| SauE15.I11 | 162 | chuA (N), heme utilization gene, E. coli O157:H7 | 305 | e−82 | U67920 |
| SauE15.J7 | 118 | iroB (N), glucosyl transferase homolog, Salmonella serovar Typhi | 74 | e−12 | U62129 |
| SauE15.J9 | 96 | IS100 (N), Y. pestis | 174 | e−42 | L19030 |
| SauE15.M4 | 193 | r3 and malX (N), pathogenicity island of E. coli CFT073 | 383 | e−104 | AF081286 |
| SauE15.M8 | 149 | Delta-(l-α-aminoadipyl)-l-cyteinyl-d-valine synthetase (P), Penicillium sp. | 65 | e−11 | P26046 |
| SauE15.M12 | 119 | senB (N), enterotoxin of enteroinvasive E. coli | 228 | e−58 | Z54195 |
| SauE15.N7 | 188 | Plasmid pColBM-C1139 (N), E. coli | 208 | e−52 | M35683 |
| SauE4.A2 | 321 | ORF 36 (N), 102-kb unstable region, Y. pestis | 135 | e−30 | AL031866 |
| SauE4.A5 | 249 | r3 (N), beta-cystathionase, pathogenicity island of E. coli CTF073 | 355 | e−96 | AF081286 |
| SauE4.B4 | 360 | IS200 (N), E. coli | 523 | e−147 | L25845 |
| SauE4.C7 | 275 | Hippurate hydrolase (P), Campylobacter jejuni | 54 | e−7 | P45493 |
| SauE4.C11 | 255 | Pristinamycine I synthase (P), Streptomyces spp. | 51 | e−6 | CAA67248 |
| SauE4.D3 | 239 | hlyB (N), hemolysine, E. coli | 474 | e−132 | M81823 |
| SauE4.E3 | 263 | shuX genes (N), heme utilization genes, Shigella dysenteriae | 387 | e−106 | U64516 |
| SauE4.E11 | 242 | IS66 (N), E. coli | 329 | e−88 | AF119170 |
| SauE4.F8 | 188 | sorC genes (N), sor operon for l-sorbose utilization, Klebsiella pneumoniae | 139 | e−31 | X66059 |
| SauE4.F9 | 439 | YfkN (P), Bacillus subtilis | 57 | e−8 | BAA23404 |
| SauE4.F12 | 324 | kpsM (N), polysialic acid gene cluster region 3, E. coli K1 | 642 | 0 | M57382 |
| SauE4.H2 | 85 | sorM (N), sor operon for l-sorbose utilization, K. pneumoniae | 105 | e−22 | X66059 |
| SauE4.I2 | 431 | yihA (N), plasmid R100, S. flexneri | 829 | 0 | AP000342 |
| TspE4.A5 | 271 | pap and prsK (N), P-pili protein, E. coli | 498 | e−139 | X61239 |
| TspE4.A8 | 216 | ORF 17 kD of prs pili operon (N), cytoplasmic protein, E. coli | 387 | e−106 | X61238 |
| TspE4.A9 | 179 | kpsT (N), polysialic acid gene cluster region 3, E. coli K1 | 347 | e−94 | M57381 |
| TspE4.A10 | 212 | HecB (P), putative hemolysin activator transporter, Erwinia chrysanthemi | 73 | e−13 | AAC31980 |
| TspE4.B1 | 229 | r1 (N), pathogenicity island of E. coli CFT073 | 430 | e−119 | AF081286 |
| TspE4.B5 | 215 | Sensory transduction histidine kinase (P), Synechocystis sp. | 52 | e−7 | BAA18223 |
| TspE4.B9 | 319 | senB (N), enterotoxin of enteroinvasive E. coli | 617 | e−175 | Z54195 |
| TspE4.B12 | 430 | IS100 (N), Y. pestis | 698 | 0 | L19030 |
| TspE4.C10 | 267 | Intergenic K42 capsule cluster (N), E. coli | 466 | e−129 | AF118251 |
| TspE4.D2 | 232 | waaL (N), lipid A core-to-surface polymer ligase, E. coli | 404 | e−111 | AF019746 |
| TspE4.D4 | 245 | ORF 169 (N), plasmid F, E. coli | 456 | e−126 | X17539 |
| TspE4.D10 | 222 | cnf1 (N), cytotoxic necrotizing factor, E. coli | 440 | e−122 | X70670 |
| TspE4.D11 | 217 | hlyB (N), hemolysin, E. coli | 422 | e−117 | M81823 |
| TspE4.E3 | 298 | hlyD (N), hemolysin, E. coli | 553 | e−156 | M10133 |
| TspE4.E4 | 267 | ORF 95 (N), plasmid F, E. coli | 482 | e−134 | X17539 |
| TspE4.E6 | 190 | l-sorbose P reductase (P), K. pneumoniae | 112 | e−25 | P37084 |
| TspE4.E8 | 285 | hlyB (N), hemolysin, E. coli | 541 | e−152 | M81823 |
| TspE4.G7 | 238 | tra (N), plasmid F, E. coli | 448 | e−124 | X61575 |
| TspE4.G8 | 323 | Transmembrane protein (P), E. coli | 82 | e−15 | AAA92620 |
| TspE4.H1 | 283 | Arginine deiminase (P), Pseudomonas aeruginosa | 63 | e−10 | P13981 |
| TspE4.H9 | 179 | traT (N), plasmid R100, E. coli | 353 | e−96 | J01769 |
| TspE4.H10 | 223 | prf and papI (N), adhesin regulatory gene, E. coli | 418 | e−115 | X76613 |
| TspE4.H11 | 279 | ORF 9 (N), plasmid F, E. coli | 456 | e−127 | X17539 |
| TspE4.I10 | 269 | neuC (N), capsule gene cluster, E. coli | 492 | e−137 | M84026 |
| TspE4.J1 | 327 | yhtA (N), plasmid R100, E. coli | 521 | e−146 | AP000342 |
| TspE4.J6 | 221 | chuA (N), heme utilization gene, E. coli O157:H7 | 375 | e−102 | U67920 |
| TspE4.K3 | 180 | iss (N), serum survival, E. coli | 270 | e−70 | AF042279 |
| TspE4.K8 | 184 | IS100 (N), E. coli | 190 | e−47 | L19030 |
| prf and papB (N), E. coli | 143 | e−32 | X76613 | ||
| TspE15.A1 | 332 | Na+ H+ antiporter (P), H. influenzae | 96 | e−20 | Q57007 |
| TspE15.C1 | 299 | hra (N), heat-resistant agglutinin, E. coli 99 | 537 | e−151 | U07174 |
| TspE15.C3 | 386 | hcp (N), E. coli | 81 | e−14 | AF044503 |
| TspE15.D7 | 239 | Protein STBA (P), plasmid NR1, E. coli | 87 | e−17 | P11904 |
| TspE15.D9 | 230 | chuA (P), heme utilization gene, E. coli O157:H7 | 89 | e−18 | AAC44857 |
| TspE15.E7 | 360 | kpsS (N), capsule gene cluster region 1, E. coli K5 | 531 | e−149 | X74567 |
| TspE15.G12 | 287 | Putative amino transferase (P), B. subtilis | 72 | e−12 | Q08432 |
| TspE15.H2 | 258 | Pyruvate formate lyase-activating enzyme (P), Streptococcus mutans | 51 | e−6 | AAB89799 |
| TspE15.H5 | 310 | cnf1 (N), cytotoxic necrotizing factor, E. coli | 601 | e−170 | U42629 |
| TspE15.H9 | 273 | Major fimbrial subunit of F17-like fimbriae (P), E. coli | 48 | e−5 | I41206 |
| TspE15.I2 | 112 | prs and papE (N), P-pili protein, E. coli | 222 | e−57 | X62158 |
Only homologies with at least probability of e−5 were retained. Homologies to bacteriophages (n = 21) are not shown.
Clones are designated by the name of the enzyme (Sau or Tsp), followed by the name of the strain used for the subtraction (E4 or E15) and a code composed of a letter and a number.
The name of the gene sequence is given (with similarity type in parentheses), followed by the product or function that is encoded by the gene and/or the gene's location, as well as the organism name. N, similarity at the nucleotide level; P, similarity at the protein level; ORF, open reading frame.
Mapping of the ECNM-specific sequences on the E. coli chromosome.
The availability of a physical map of the chromosome of E. coli RS218 (26) made possible the investigation of the distribution of some of the above-mentioned sequences. Of 64 clones that were chosen, 7 showed homology to known virulence factors (kps, hly, prs, hra, cnf1, chuA, and senB) and 57 showed no known homology. These latter clones were chosen at random from the TspE4 and SauE15 libraries. These two libraries were chosen because they contained most of the expected pathogen-associated genes (e.g., pap, hly, and kps) and thus were considered the most comprehensive. All the clones showed homology by Southern hybridization to the chromosome of strain RS218. PCR products from these clones were labeled and used to probe Southern blots of DNA from RS218 digested with the infrequently cutting enzymes BlnI, NotI, and XbaI. The location of both sfa and ibe10 was also determined. To confirm that each one of these clones was specific for ECNM, they were all used to probe DraI-digested DNA from strains ECOR4, ECOR15, and MG1655 and shown to be nonreactive against these strains.
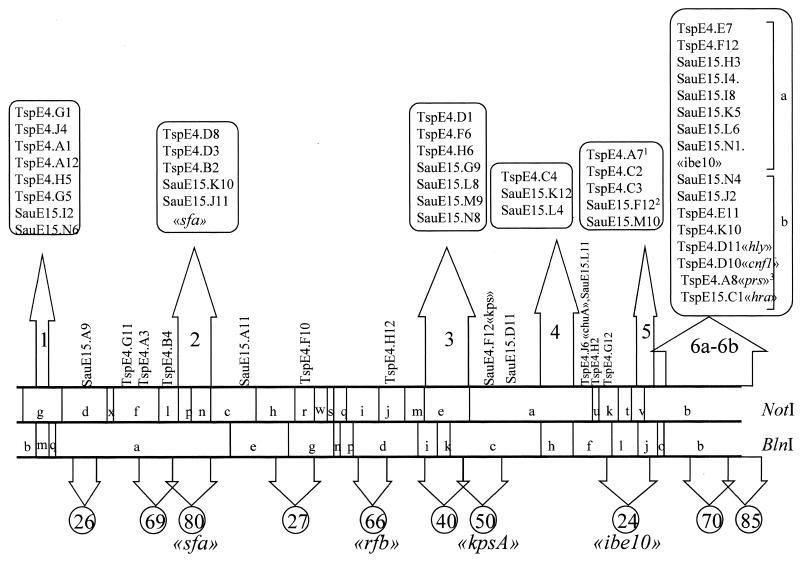
Mapping of these clones revealed a nonrandom distribution of the ECNM-specific sequences (Fig. 1). Forty-four of the clones clustered in six distinct groups on the chromosome. One clone encoding part of the chuA gene found in enterohemorragic E. coli was not associated with any of these clusters and remained isolated. Region 1 is contained within the BlnI fragment m (85 kb). The clones of region 2 mapped on NotI fragments p and n (∼240 kb). Region 3 is contained within NotI fragment e (∼310 kb), and region 4 is contained within BlnI fragment h (∼210 kb). Region 5 maps to BlnI fragment j (∼135 kb). Region 6, which overlaps the NotI fragment b and BlnI fragment b (∼550 kb), has been divided into two subregions by the enzyme XbaI (data not shown), regions 6a and 6b. The latter subregion contained clones with homologies to cnf1, hly, prs, and hra. The sfa and ibe10 probes hybridized in regions 2 and 6a, respectively. The capsule-encoding genes were not linked to any of these regions. Six clones presenting no homology with known sequences and the senB gene all mapped on a large plasmid present in the RS218 strain (1).
FIG. 1.
Distribution of specific sequences on the chromosome of E. coli strain RS218. E. coli strain C5-specific clones were used as probes on Southern blots of DNA from strain RS218 digested with the infrequently cutting enzymes BlnI and NotI and were positioned on the linearly represented restriction map (26). The upper arrows indicate the six regions found in this study and delineated by NotI and BlnI fragments with a high density of C5-specific clones. Clones with known homologies are indicated, as are the positions of the probes sfa and ibe10. Region 6 was divided into two subregions according to the mapping of the clones on different XbaI fragments. The lower arrows indicate the 10 segments of the RS218 chromosome in which Rode et al. found additions relative to the E. coli K-12 chromosome. The length (in kilobases) of each addition is indicated in the respective circle. The positions of various virulence factors are also indicated (26). Superscripts to clone names designate the following: 1, TspE4.A7 was positioned by overlapping SauE15.F12; 2, SauE15.F12 also shows reactivity on the plasmid; and 3, TspE4.A8 also shows a weak reactivity on the NotI fragment p.
Distribution of E. coli C5-specific genomic regions among two collections of E. coli strains.
To assess the relevance of these regions in terms of pathogenesis, we determined the frequency of occurrence of clones located on regions 1, 3, 4, 5, and 6b as well as of the clone containing part of the chuA gene in our collection of 54 ECNM belonging to the phylogenetic group B2. We excluded from this study regions 2 and 6a because they contained the sfa and ibe10 genes, the distribution of which among group B2 E. coli strains is well established (5). For each region, two to four clones were used independently to probe Southern blots of genomic DNA of ECNM isolates. The control group corresponded to the 15 B2 strains of the ECOR collection which contains no meningitis strains. Results are shown in Table 2. All the above-mentioned regions are widely present among ECNM except for region 6b which is underrepresented in meningitis-associated strains. In addition, regions 1, 3, and 4 appeared with a significantly higher frequency (P < 0.05) among ECNM than the other B2 strains, thus suggesting that these regions may encode factors specific to ECNM.
TABLE 2.
Strains isolated from cases of NM and from the ECOR collection that hybridized with subtractive clones used as probes
| Strain sourcee | No. of strains | % of isolates positive by Southern blot hybridizationd
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region 1
|
Region 3
|
Region 4
|
Region 5
|
Region 6b
|
TspE4J6 | ||||||||
| TspE4.J4 | TspE4.H5 | SauE15M9 | TspE4H6 | SauE15.L4 | SauE15K12 | TspE4.C4 | SauE15.M10 | TspE4C2 | SauE15.N4 | PAI Va | |||
| NM | 54 | 91b | 91b | 80b | 84b | 81b | 81b | 81b | 100 | 98c | 17b | 17b | 100 |
| ECOR collection | 15 | 40 | 40 | 13 | 13 | 47 | 47 | 47 | 100 | 87 | 47 | 47 | 100 |
The prevalence of PAI V was assessed using the clones TspE4.D11, TspE4.D10, and TspE15.C1, homologous to the hly, cnf1, and hra genes, respectively.
P < 0.05, compared with strains of the ECOR collection (existence of a difference in the distribution of the studied clones was tested by the χ2 test).
Not significant, compared with strains of the ECOR collection.
TspE4.J6 is homologous to chuA. The other clones are representative of their respective regions.
All strains were in the phylogenetic B2 group.
DISCUSSION
The pathogenesis of ECNM is incompletely understood. Until now, only four specific virulence factors have been described: the K1 capsular polysaccharide, the aerobactin, the S fimbrial adhesin, and the Ibe10 protein (11, 13, 17, 21, 23). However, E. coli isolates that were obtained from the CSF of neonates with meningitis and which do not express these virulence factors have been described, thus suggesting that some other yet unidentified bacterial attributes may be involved (5). Very recently, a new gene (ibeB) required for penetration of brain microvascular endothelial cells was reported (14). In this work, we performed subtractive hybridization to identify regions of the chromosome likely to encode attributes responsible for specific aspects of the pathogenesis of ECNM. We performed two rounds of subtractive hybridization according to a previously described method (30) and obtained libraries of clones specific for ECNM containing inserts with sizes ranging from 100 to 500 bp. The specificity of the subtractive libraries was assessed (i) by Southern blotting with the nonpathogenic strains and (ii) by sequence analysis which showed that 72% of the clones had no homology with the published sequence of E. coli K-12. Some clones corresponded to expected virulence-associated genes (kps, pap, and hly). On the other hand, we did not isolate clones corresponding to sfa and ibe10 genes. However, clones derived from regions containing both of these genes were obtained. Together these data confirm the comprehensiveness of these libraries.
Very recently, Rode et al. carried out comparative macrorestriction mapping using rare-restriction-site alleles between the chromosomes of the laboratory strain K-12 MG1655 and E. coli RS218 (26). They identified 10 large chromosomal additions in the RS218 strain but not the MG1655 strain (Fig. 1). Most of these segments are in perfect accordance with our results. However, four segments undetected by our approach were reported by Rode et al. On the other hand, our regions 1 and 4 were not detected in this previous work. The most likely explanation for not having isolated clones in some of the regions reported by Rode et al. is, first, that these may correspond to sequences deleted during the course of laboratory isolation of E. coli K-12 and, second, that these deletions are unlikely to have occurred in the group A ECOR strains. In favor of this hypothesis is the fact that segments 66 and 69 (Fig. 1), which we did not obtain, correspond to rfb genes and the λ bacteriophage, respectively, which are deleted in strain MG1655 (19, 25).
An epidemiological approach was undertaken to shed light on the role of these regions in the infective process of ECNM. Considering that most ECNM belong to the phylogenetic group B2, we determined the prevalence of each region as well as of chuA among ECNM of group B2 and among non-meningitis-associated group B2 strains from the ECOR collection. Although small, this control group was chosen because it is composed of reference strains that belong to the ECOR collection, which is considered representative of the range of genotypic variations of the species. We used two to four clones from each region and the clone TspE4.J6, homologous to chuA, as probes against Southern blots of genomic DNA prepared from isolates belonging to both groups. The occurrence of these clones among the meningitis isolates indicates that all these regions, except the region 6b, are widely represented among ECNM, thus suggesting the involvement of genes encoded by these regions in the pathogenesis of these strains. On the other hand and surprisingly, region 6b which resembles PAI V, has a low prevalence in ECNM (17%) but was widely represented in the B2 group strains from the ECOR collection (47%). Region 5 had a high but a similar prevalence in both collections and thus may correspond to segments characteristic of phylogenetic group B2. The same distribution was observed for chuA, but interestingly, this gene was present in all B2 group strains tested without exception. Considering that regions 1, 3, and 4, which are specific for ECNM, do not contain known virulence factors, one may speculate that these regions correspond to islands of DNA associated with the invasion of meninges by E. coli. Further work is in progress to characterize the contents of these regions.
ACKNOWLEDGMENTS
This work was supported in part by the Programme de Recherche Fondamentale en Microbiologie, Maladies Infectieuses et Parasitaires (Appel d'offre 1998) (Recherche de déterminants génétiques de pathogénicité chez E. coli K1 responsable de méningite néonatale) and by the Programme Hospitalier de Recherche Clinique (grant AOM 96069). The laboratory of X.N. is supported by the INSERM, the Université Paris 5 and the Fondation pour le Recherche Médicale. S.P.P.B. was the recipient of a fellowship obtained from Hoescht Marion Roussel and from the Société de Pathologies Infectieuses de Langue Française.
We thank P. Berche for his help, A. Perrin for her constant support, and H. Dabernat, M. Thibaut, H. Chardon, J. C. Philippe, J. Freney, T. Lambert, J. Raymond, M. Weber, M. Benbachir, K. Kim, J. Hacker, R. Bertolussi, H. Vu Thien, and R. Selander for providing some of the strains used in this study.
Footnotes
Editor: E. I. Tuomanen
REFERENCES
- 1.Achtman M, Mercer A, Kusecek B, Pohl A, Heuzenroeder M, Aaronson W, Sutton A, Silver R P. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect Immun. 1983;39:315–335. doi: 10.1128/iai.39.1.315-335.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bingen E, Denamur E, Brahimi N, Elion J. Genotyping may provide rapid identification of Escherichia coli K1 organisms that cause neonatal meningitis. Clin Infect Dis. 1996;22:152–156. doi: 10.1093/clinids/22.1.152. [DOI] [PubMed] [Google Scholar]
- 4.Bingen E, Bonacorsi S, Brahimi N, Denamur E, Elion J. Virulence patterns of Escherichia coli strains associated with neonatal meningitis. J Clin Microbiol. 1997;35:2981–2982. doi: 10.1128/jcm.35.11.2981-2982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bingen E, Picard B, Brahimi N, Mathy S, Desjardins P, Elion J, Denamur E. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J Infect Dis. 1998;177:642–650. doi: 10.1086/514217. [DOI] [PubMed] [Google Scholar]
- 6.Blattner F R, Plunkett G I, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1461. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Boyd E F, Hartl D L. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J Bacteriol. 1998;180:1159–1165. doi: 10.1128/jb.180.5.1159-1165.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Louvois J. Acute bacterial meningitis in the newborn. J Antimicrob Chemother. 1994;34(Suppl. A):61–73. doi: 10.1093/jac/34.suppl_a.61. [DOI] [PubMed] [Google Scholar]
- 9.Franco S M, Cornelius V E, Andrews B F. Long-term outcome of neonatal meningitis. Am J Dis Child. 1992;146:567–571. doi: 10.1001/archpedi.1992.02160170047014. [DOI] [PubMed] [Google Scholar]
- 10.Goulet P, Picard B. Comparative electrophoretic polymorphism of esterases and other enzymes in Escherichia coli. J Gen Microbiol. 1989;135:135–143. doi: 10.1099/00221287-135-1-135. [DOI] [PubMed] [Google Scholar]
- 11.Hacker J, Kestler H, Hoschutzky H, Jann K, Lottspeich F, Korhonen T. Cloning and characterization of the S fimbrial adhesin II complex of an Escherichia coli O18-K1 meningitis isolate. Infect Immun. 1993;61:544–550. doi: 10.1128/iai.61.2.544-550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herzer P J, Inouye S, Inouye M, Whittman T S. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang S H, Wass C, Fu Q, Prasadarao N V, Stins M, Kim K S. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect Immun. 1995;63:4470–4475. doi: 10.1128/iai.63.11.4470-4475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang S, Chen Y, Fu Q, Stins M, Wang Y, Wass C, Kim K S. Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infect Immun. 1999;67:2103–2109. doi: 10.1128/iai.67.5.2103-2109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim K S, Manocchio M, Anthony B F. Efficacy of trimethoprim/sulfamethoxazole in experimental Escherichia coli bacteremia and meningitis. Chemotherapy. 1983;29:428–435. doi: 10.1159/000238231. [DOI] [PubMed] [Google Scholar]
- 16.Kim K S, Itabashi H, Gemski P, Sadoff J, Warren R L, Cross A S. The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J Clin Investig. 1992;90:897–905. doi: 10.1172/JCI115965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korhonen T K, Valtonen M V, Parkkinen J, Vaïsänen-Rhen V, Finne J, Ørskov F, Ørskov I, Svenson S B, Mäkelä P H. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect Immun. 1985;48:486–491. doi: 10.1128/iai.48.2.486-491.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lisistsyn N A, Lisistsyn N M, Wigler M. Cloning the difference between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 19.Liu D, Reeves P R. Escherichia coli K12 regains its O antigen. Microbiology. 1994;140:49–57. doi: 10.1099/13500872-140-1-49. [DOI] [PubMed] [Google Scholar]
- 20.Nataro J P, Seriwatana J, Fasano A, Maneval D R, Guers L D, Noriega F, Dubovsky F, Levine M M, Morris J G., Jr Identification and cloning of a novel plasmid-encoded enterotoxin of enteroinvasive Escherichia coli and Shigella strains. Infect Immun. 1995;63:4721–4728. doi: 10.1128/iai.63.12.4721-4728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neilands J B, Bindereif A, Montgomerie J Z. Genetic basis of iron assimilation in pathogenic Escherichia coli. Curr Top Microbiol Immunol. 1985;118:179–196. doi: 10.1007/978-3-642-70586-1_10. [DOI] [PubMed] [Google Scholar]
- 22.Ochman H, Selander R K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984;157:690–692. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parkkinen J, Korhonen T K, Pere A, Hacker J, Soinila S. Binding sites in the rat brain for Escherichia coli S fimbriae associated with neonatal meningitis. J Clin Investig. 1988;81:860–865. doi: 10.1172/JCI113395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Picard B, Garcia J S, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun. 1999;67:546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rode C K, Obreque V H, Bloch C A. New tools for integrated genetic and physical analysis of the Escherichia coli chromosome. Gene. 1995;166:1–9. doi: 10.1016/0378-1119(95)00630-5. [DOI] [PubMed] [Google Scholar]
- 26.Rode C K, Melkerson-Watson L, Johnson A T, Bloch C A. Type-specific contributions to chromosome size differences in Escherichia coli. Infect Immun. 1999;67:230–236. doi: 10.1128/iai.67.1.230-236.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selander R, Korhonen T, Vaisanen-Rhen V, Williams P, Pattison P, Caugant D. Genetic relationships and clonal structure of strains of Escherichia coli causing neonatal septicemia and meningitis. Infect Immun. 1986;52:213–222. doi: 10.1128/iai.52.1.213-222.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selander R K, Caugant D A, Whittman T S. Genetic structure and variation in natural populations of Escherichia coli. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C.: American Society for Microbiology; 1987. pp. 1625–1648. [Google Scholar]
- 29.Swenson D L, Bukanov N O, Berg D E, Welch R A. Two pathogenicity islands in uropathogenic Escherichia coli J96: cosmid cloning and sample sequencing. Infect Immun. 1996;64:3736–3743. doi: 10.1128/iai.64.9.3736-3743.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tinsley C R, Nassif X. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc Natl Acad Sci USA. 1996;93:11109–11114. doi: 10.1073/pnas.93.20.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torres A, Payne S. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 1997;23:825–833. doi: 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- 32.Unhanand M, Mustafa M M, McCracken G H, Jr, Nelson J D. Gram-negative enteric bacillary meningitis: a twenty-year experience. J Pediatr. 1993;122:15–21. doi: 10.1016/s0022-3476(05)83480-8. [DOI] [PubMed] [Google Scholar]