Abstract
Platelet-rich plasma (PRP) has great potential in regenerative medicine. In addition to the well-known regenerative potential of secreted growth factors, extracellular vesicles (EVs) are emerging as potential key players in the regulation of tissue repair. However, little is known about their therapeutic potential as regenerative agents. In this study, we have identified and subtyped circulating EVs (platelet-, endothelial-, and leukocyte-derived EVs) in the peripheral blood of athletes recovering from recent muscular injuries and undergoing a submaximal strength rehabilitation program. We found a significant increase in circulating platelet-derived EVs at the end of the rehabilitation program. Moreover, EVs from PRP samples were isolated by fluorescence-activated cell sorting and analyzed by label-free proteomics. The proteomic analysis of PRP-EVs revealed that 32% of the identified proteins were associated to “defense and immunity”, and altogether these proteins were involved in vesicle-mediated transport (GO: 0016192; FDR = 3.132 × 10−19), as well as in wound healing (GO: 0042060; FDR = 4.252 × 10−13) and in the events regulating such a process (GO: 0061041; FDR = 2.812 × 10−12). Altogether, these data suggest that platelet-derived EVs may significantly contribute to the regeneration potential of PRP preparations.
Keywords: platelet-derived extracellular vesicles, regenerative medicine, PRP, proteomics
1. Introduction
Platelet-rich plasma (PRP) is a peripheral blood-derived preparation, containing higher concentrations of platelets than whole blood [1]. PRP has a great potential in regenerative medicine, given that its therapeutic properties were demonstrated in a wide range of clinical fields, including cardiac [2], maxillofacial [3], and plastic surgery, as well as orthopedics [4], dermatology [5], and sports medicine [6]. In addition to PRP’s well-known safety, convenience, and clinical potential, PRP’s mechanisms involved in regenerative medicine are not completely understood. Recently, it has been hypothesized that, in addition to secreted growth factors, extracellular vesicles (EVs) could have a role as key players in the PRP regulation of tissue repair [7,8,9]. EVs are cell-derived nanosized particles, surrounded by a lipid bilayer, and are similar, in terms of lipid composition (i.e., desatured lipids, sphingomyelinase and gangliosides), to that of cell plasma membranes [10]. EVs carry active cargoes, consisting of proteins, lipids, mRNAs, long/short noncoding RNAs, DNA fragments, and even organelles (i.e., mitochondria) [11]. Traditionally, three different EV subtypes, namely exosomes, microvesicles (MVs), and apoptotic bodies, were identified based on their biogenesis and size. Exosomes, the smallest EVs (with diameters ranging from 30 to 150 nm), originate within the lumens of multivesicular bodies (MVBs), and are released by exocytosis [12,13]. MVs generally range from 100 to 1000 nm in diameter and are released by blebbing or budding, therefore retaining the parental phenotype [14,15]. Apoptotic bodies, the larger EVs (~0.1 to ~5 µm), are released by apoptotic cells [16]. However, it has been underlined that the above reported classification does not fit the heterogeneity of the EV subtypes populations and their overlaps in size, cargoes, functions and biodistribution [17]. For this reason, the International Society of Extracellular Vesicles endorsed the use of the term “extracellular vesicle” for all EV subtypes, with a generic subclassification as small, if within 100 nm, and medium/large, if above 100–200 nm [18]. In any case, EVs have been identified in many body fluids, such as peripheral blood, cerebrospinal fluid, and tears [19,20,21,22,23,24,25]. EVs also express surface antigens that allow them to reach target cells [26]. Once attached the related recipient cell, EVs activate specific signaling responses interacting with their ligands; EVs can be also internalized by endocytosis and/or phagocytosis or can fuse with the membrane of the target cell, therefore delivering their cargo into the target cell cytosol, modifying the recipient cell biology [27]. In this case, EVs can directly activate the target cells by acting as signaling complexes, or can transfer genetic information, inducing transient or persistent phenotypic changes in recipient cells [28]. The involvement of EVs in the events regulating tissue, and in particular muscle, concerns the mechanisms of regeneration and repair [29]. The regeneration of damaged skeletal muscle largely relies on the presence of a population of cells, called satellite cells, that are mononuclear and myogenic elements that retain the capacity to proliferate and further differentiate to generate new fibers [30]. More recently, it has been also demonstrated that muscle tissue regeneration is a process characterized by a complex and coordinated interaction between muscles and the immune system [31]. In this context, it has been shown that after an acute injury, M1 cells infiltrate early during the first stages to promote the clearance of necrotic debris, whereas M2 macrophages develop later and sustain tissue healing [32]. More interestingly, as active messengers, leukocyte-derived, and more specifically macrophage-derived EVs have emerged as vital mediators in the mechanisms associated with tissue repair [33]. Recent data shows that platelet-derived EVs have also been involved in different healing responses [34].
Furthermore, even if a role for the EVs contained in PRP preparations (PRP-EVs) has been hypothesized [1], little is known about their therapeutic potential.
Here, we have analyzed leukocyte-, platelet- and endothelial-derived EV levels in a cohort of subjects recovering from muscle injuries. We showed that the concentrations of platelet-derived EVs was increased in the peripheral blood during the recovering phase. We have, therefore, purified PRP-EVs by fluorescence-activated cell sorting, to study their protein cargos with the final aim to understand their role as regenerative agents.
2. Results
2.1. Peripheral Blood EV Identification and Count in Athletes Recovering from Muscle Injuries
To study the possible physiological involvement of EVs in the processes associated with regeneration, a cohort of athletes recovering from muscle injuries (at T0) and undergoing submaximal strength rehabilitation was analyzed for the presence of EV of different phenotypes, as shown in the gating strategy depicted in Figure 1. As shown in Figure S1, no differences in terms of EV counts between athletes recovering from muscle injuries (at T0) and control subjects were evidenced.
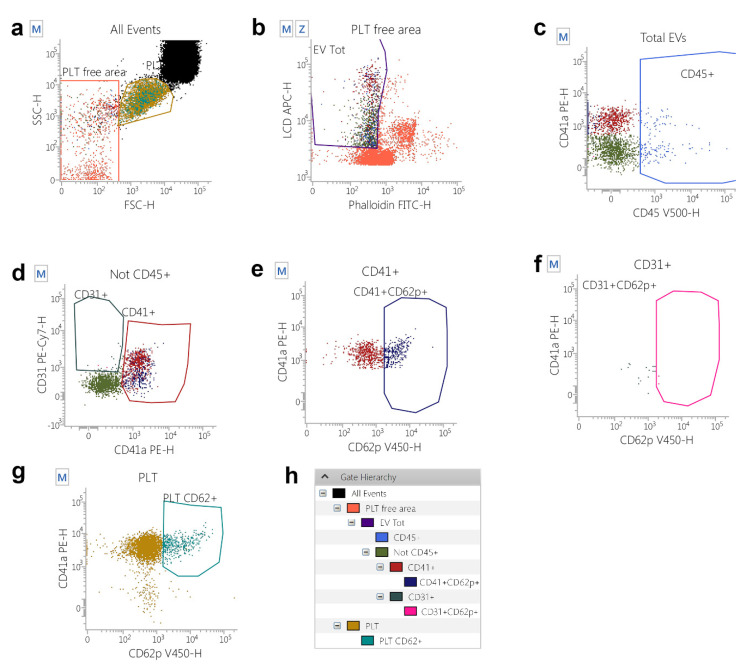
Figure 1.
Gating Strategy for extracellular vesicles (EVs) identification and subtyping. (a) A platelet-free area region was defined on a Forward Scatter-H/Side Scatter-H dot-plot and, by using platelets (PLT) as a reference population. (b) The “Platelet-free area” was shown on a Phalloidin-H/Lipophilic Cationic Dye (LCD)-H dot-plot and EVs were identified as LCD positive/phalloidin negative events. (c) EVs (LCD+/Phalloidin- events) were analysed on a CD45-H/CD41a-H dot-plot and CD45+ events were identified as leukocyte-derived EVs. (d) A logical gate excluding all the CD45+ events was then obtained, and the resulting population was plotted on a CD31-H/CD41a-H dot-plot. Events showing the CD31+/CD41a+ phenotype were identified as platelet-derived EVs, whereas the CD31+/CD41a- compartment represented endothelium-derived EVs. (e) CD31+/CD41a+ platelet-derived EVs, CD31+/CD41a- endothelial-derived EVs (f) and whole platelets (g) were analysed for activation marker CD62P. (h) The applied gating hierarchy is shown as a scheme.
More interestingly, when athletes were measured before starting the rehabilitation (T0) and at the end of the program (T1), a significant increase of platelet-derived EVs was observed (p = 0.0095, Figure 2), whereas the other analyzed EV phenotypes did not change in terms of concentrations (Figure S2).
Figure 2.

CD41+ circulating EVs in the peripheral blood of athletes recovering from muscle injuries. The graph shows absolute counts a of CD41+ circulating EVs, analyzed before (T0, blue dots) and after (T1, green dots) the rehabilitation program.
2.2. Phenotypes of PRP-EVs
The EVs from six PRP samples, isolated by fluorescence-activated cell sorting, were subtyped using the panel described in Table S1 and the gating strategy depicted in Figure 1. Figure 3 shows the Box and Whiskers Plots, representing the distribution of the EV subtypes that we have analyzed. Leukocyte-derived EVs (Leuko EVs) are the most represented (mean = 2677.27 + 1706.11) EV subpopulation, followed by platelet-derived EVs (PLT EVs, mean = 1163.4 + 1193.52) and EVs stemming from the endothelium (Endo EVs, mean = 232.4 + 269.70).
Figure 3.
EV subtypes from PRP samples. The whole EV compartment, as well as EVs derived from the endothelium, leukocytes, and platelets were analyzed from the PRP of six subjects.
2.3. Proteomic Characterization of the PRP-EV Cargo
1 × 106 PRP-EVs isolated by fluorescence-activated cell sorting (FACS) were analyzed by shotgun proteomics from four different patients. Table 1 shows the list of the quantified proteins common in at least two of the analyzed samples.
Table 1.
The represented list has been obtained by identifying the proteins displayed by at least two PRP-EV isolated samples. The Uniprot code and the Gene name, Protein full name, Protein Classification, and Functional Network interaction are reported. WH means Wound Healing, VH means Vesicle-mediated Transport.
| Protein UniProt Code | Gene Name | Protein Name | Protein Class | Functional Network Interaction |
|---|---|---|---|---|
| P01834 | IGKC | Immunoglobulin kappa constant | defense/immunity protein | - |
| P01860 | IGHG3 | Immunoglobulin heavy constant gamma 3 | defense/immunity protein | - |
| A0A0C4DH42 | IGHV3–66 | Immunoglobulin heavy variable 3–66 | defense/immunity protein | - |
| P0DOY3 | IGLC3 | Immunoglobulin lambda constant 3 | defense/immunity protein | - |
| P0DP04 | IGHV3–43D | Immunoglobulin heavy variable 3–43D | defense/immunity protein | VT |
| P01857 | IGHG1 | Immunoglobulin heavy constant gamma 1 | defense/immunity protein | - |
| P02749 | APOH | Beta-2-glycoprotein 1 | defense/immunity protein | VT |
| P01876 | IGHA1 | Immunoglobulin heavy constant alpha 1 | defense/immunity protein | - |
| P0DP01 | IGHV1–8 | Immunoglobulin heavy variable 1–8 | defense/immunity protein | - |
| P25311 | AZGP1 | Zinc-alpha-2-glycoprotein | defense/immunity protein | - |
| A0A075B7B8 | IGHV3OR16–12 | Immunoglobulin heavy variable 3/OR16–12 (non-functional) | defense/immunity protein | - |
| P01871 | IGHM | Immunoglobulin heavy constant mu | defense/immunity protein | - |
| A0A0C4DH31 | HV1–18 | Immunoglobulin heavy variable 1–18 | defense/immunity protein | - |
| P08603 | CFH | Complement factor H | defense/immunity protein | - |
| P0DP08 | IGHV4–38–2 | Immunoglobulin heavy variable 4–38–2 | defense/immunity protein | VT |
| P0DP03 | HV3–30–5 | Immunoglobulin heavy variable 3–30–5 | defense/immunity protein | - |
| P01591 | IGJ | Immunoglobulin J chain | defense/immunity protein | VT |
| P04003 | C4BPA | C4b-binding protein alpha chain | defense/immunity protein | VT |
| A2NJV5 | KV2–29 | Immunoglobulin kappa variable 2–29 | defense/immunity protein | - |
| P0CG04 | IGLL5 | Immunoglobulin lambda like polypeptide 5 | defense/immunity protein | VT |
| P01861 | IGHG4 | Immunoglobulin heavy constant gamma 4 | defense/immunity protein | - |
| P01619 | KV320 | Immunoglobulin kappa variable 3–20 | defense/immunity protein | - |
| A0A0B4J1V6 | HV373 | Immunoglobulin heavy variable 3–73 | defense/immunity protein | - |
| P07357 | C8A | Complement component C8 alpha chain | defense/immunity protein | - |
| Q96PD5 | PGLYRP2 | N-acetylmuramoyl-L-alanine amidase | defense/immunity protein | - |
| P01877 | IGHA2 | Immunoglobulin heavy constant alpha 2 | defense/immunity protein | - |
| P01859 | IGHG2 | Immunoglobulin heavy constant gamma 2 | defense/immunity protein | - |
| Q0VDD8 | DNAH14 | Dynein axonemal heavy chain 14 | cytoskeletal protein | - |
| Q8TF72 | SHROOM3 | Protein Shroom3 | cytoskeletal protein | - |
| Q13835 | PKP1 | Plakophilin-1 | cytoskeletal protein | VT |
| P15924 | DSP | Desmoplakin | cytoskeletal protein | WH, VT |
| P63261 | ACTG1 | Actin, cytoplasmic 2 | cytoskeletal protein | WH, VT |
| P69905 | HBA2 | Hemoglobin subunit alpha 2 | transfer/carrier protein | VT |
| P68871 | HBB | Hemoglobin subunit beta | transfer/carrier protein | WH, VT |
| P02787 | TF | Serotransferrin | transfer/carrier protein | VT |
| P02042 | HBD | Hemoglobin subunit delta | transfer/carrier protein | WH |
| P02656 | APOC3 | Apolipoprotein C-III | transfer/carrier protein | - |
| P02774 | GC | Vitamin D-binding protein | transfer/carrier protein | - |
| P04114 | APOB | Apolipoprotein B-100 | transfer/carrier protein | VT |
| P05090 | APOD | Apolipoprotein D | transfer/carrier protein | - |
| P43652 | AFM | Afamin | transfer/carrier protein | - |
| P06727 | APOA4 | Apolipoprotein A-IV | transfer/carrier protein | - |
| P02649 | APOE | Apolipoprotein E | transfer/carrier protein | WH, VT |
| P02652 | APOA2 | Apolipoprotein A-II | transfer/carrier protein | - |
| P02647 | APOA1 | Apolipoprotein A-I | transfer/carrier protein | VT |
| P00738 | HP | Haptoglobin | protein-modifying enzyme | VT |
| P00734 | F2 | Prothrombin | protein-modifying enzyme | WH, VT |
| P00747 | PLG | Plasminogen | protein-modifying enzyme | WH, VT |
| P31944 | CASP14 | Caspase-14 | protein-modifying enzyme | - |
| P02790 | HPX | Hemopexin | protein-modifying enzyme | VT |
| P02675 | FGB | Fibrinogen beta chain | intercellular signal molecules | WH, VT |
| P02679 | FGG | Fibrinogen gamma chain | intercellular signal molecules | WH, VT |
| P01024 | C3 | Complement C3 | protein-binding activity modulator | VT |
| P01009 | SERPINA1 | Alpha-1-antitrypsin | protein-binding activity modulator | WH, VT |
| P19827 | ITIH1 | Inter-alpha-trypsin inhibitor heavy chain H1 | protein-binding activity modulator | - |
| P01011 | GIG25 | Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | protein-binding activity modulator | VT |
| P01042 | KNG1 | Kininogen-1 | protein-binding activity modulator | WH, VT |
| P01008 | SERPINC1 | Antithrombin-III | protein-binding activity modulator | WH |
| P19823 | ITIH2 | Inter-alpha-trypsin inhibitor heavy chain H2 | protein-binding activity modulator | - |
| P01019 | AGT | Angiotensinogen | protein-binding activity modulator | - |
| P05155 | SERPING1 | Plasma protease C1 inhibitor | protein-binding activity modulator | WH, VT |
| P0C0L5 | C4B | Complement C4-B | protein-binding activity modulator | - |
| P01031 | C5 | Complement C5 | protein-binding activity modulator | - |
| Q14624 | ITIH4 | Inter-alpha-trypsin inhibitor heavy chain H4 | protein-binding activity modulator | VT |
| P05546 | SERPIND1 | Heparin cofactor 2 | protein-binding activity modulator | WH |
| P08185 | SERPINA6 | Corticosteroid-binding globulin | protein-binding activity modulator | - |
| P01023 | A2M | Alpha-2-macroglobulin | protein-binding activity modulator | WH, VT |
| P02452 | COL1A1 | Collagen alpha-1(I) chain | extracellular matrix protein | WH, VT |
| P02751 | FN1 | Fibronectin type III domain containing | extracellular matrix protein | VT |
| P08123 | COL1A2 | Collagen alpha-2(I) chain | extracellular matrix protein | WH, VT |
| P06702 | S100A9 | Protein S100-A9 | calcium-binding protein | VT |
| P07355 | ANXA2 | Annexin A2 | calcium-binding protein | WH, VT |
| P05109 | S100A8 | Protein S100-A8 | calcium-binding protein | WH, VT |
| P00915 | CA1 | Carbonic anhydrase 1 | metabolite interconversion enzyme | - |
| P00918 | CA2 | Carbonic anhydrase 2 | metabolite interconversion enzyme | - |
| P02766 | TTR | Transthyretin | metabolite interconversion enzyme | VT |
| P32119 | PRDX2 | Peroxiredoxin-2 | metabolite interconversion enzyme | WH |
| P04406 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | metabolite interconversion enzyme | - |
| P04040 | CAT | Catalase | metabolite interconversion enzyme | VT |
| Q08188 | TGM3 | Protein-glutamine gamma-glutamyltransferase E | metabolite interconversion enzyme | - |
| P00450 | CP | Ceruloplasmin | metabolite interconversion enzyme | - |
| P02747 | C1QC | Complement C1q subcomponent subunit C | scaffold/adaptor protein | - |
| P31947 | SFN | 14–3–3 protein sigma | scaffold/adaptor protein | VT |
| P63104 | YWHAZ | 14–3–3 protein zeta/delta | scaffold/adaptor protein | WH, VT |
| O75882 | ATRN | Attractin | gene-specific transcriptional regulator | - |
| Q08554 | DSC1 | Desmocollin-1 | cell adhesion molecule | VT |
| Q02413 | DSG1 | Desmoglein-1 | cell adhesion molecule | VT |
| P12273 | PIP | Prolactin induced protein | - | - |
| P02763 | ORM1 | Alpha-1-acid glycoprotein 1 | - | VT |
| P02671 | FGA | Fibrinogen alpha chain | - | WH, VT |
| S4R460 | ENSG00000259680 | Uncharacterized protein | - | - |
| P81605 | DCD | Dermcidin | - | - |
| P10909 | CLU | Clusterin | - | VT |
| P02760 | AMBP | Protein AMBP | - | VT |
| P04217 | A1BG | Alpha-1B-glycoprotein | - | VT |
| P27169 | PON1 | Serum paraoxonase/arylesterase 1 | - | - |
| O43866 | CD5L | CD5 antigen-like | - | VT |
| P04004 | VTN | Vitronectin | - | WH, VT |
| P62987 | UBA52 | Ubiquitin-60S ribosomal protein L40 | - | VT |
| P00751 | CFB | Complement factor B | - | - |
| O75223 | GGCT | Gamma-glutamylcyclotransferase | - | - |
| P14923 | JUP | Junction plakoglobin | - | VT |
| Q32MH5 | FAM214A | Protein FAM214A | - | - |
| Q5T749 | KPRP | Keratinocyte proline rich protein | - | - |
| Q5TA81 | LCE2C | Late cornified envelope protein 2C | - | - |
Panther protein classification analysis was performed and uploading PRP-EV proteins are listed in Table 1 with proteins grouped according to their biological functions. As shown in Figure 4, more than 32% of PRP-EV proteins are involved in the “defense and immunity” biological function. The String analysis (PPI enrichment p-value < 10−16) of PRP-EV proteins listed in Table 1 was carried out and the results, shown in Figure 5, demonstrated that they were mainly involved in vesicle-mediated transport (GO: 0016192; FDR = 3.132 × 10−19) (Figure 5A), as well as in wound healing (GO: 0042060; FDR = 4.252 × 10−13) and in the events regulating such a process (GO: 0061041; FDR = 2.812 × 10−12, Figure 5B).
Figure 4.
Protein classification viewed in pie charts of the 105 PRP-EV proteins reported in Table 1.
Figure 5.
Network representing the interactions existing among the 105 PRP-EV proteins reported in Table 1. (A) Red dots represent the proteins involved in the “vesicle-mediated transport”. (B) Blue dots represent the proteins associated with “wound healing”, whereas green dots are related to the proteins regulating the wound healing process.
3. Discussion
Platelet-derived EVs represent most of the peripheral blood circulating EVs, and, for this reason, they have a long history of discovery [35,36]. Traditionally, platelet-derived EVs have been described as procoagulant agents [37,38]. On the other hand, it is well known that PRP has great potential in promoting tissue repair and regeneration [39,40]. PRP is enriched in platelets and contains extracellular vesicles (EVs) [41], which have also attracted great interest in regenerative medicine [7,8,9].
It is known that an injury-responsive production of EVs exists, which carry specific proteins, lipids, RNAs, and DNA fragments that facilitate tissue repair and regeneration. This EV cargo appears to be selectively packaged, depending on the context, the type of injury, and the cellular targets. Therefore, EVs produced upon muscle injuries participate in the orchestration of responses from myofiber repair and regeneration [29]. In detail, EVs from young serum play a key role in the rejuvenating effects exerted on aged skeletal muscles [42]. Several other studies demonstrated that mesenchymal-derived EVs (MSC-EVs) promoted myogenesis and angiogenesis in vitro, and muscle regeneration in in vivo models of muscle injury [43]. Such a regeneration process may be mediated, at least in part by miRNAs (i.e., mir-494) [44]. In this context, EVs are involved in a complex intercellular crosstalk process, and MSC-EVs, circulating in the body fluids, passing across the biological barriers, home to damaged tissues, participate in the repair of injured skeletal muscles [45]. Furthermore, adipose stem cell-derived EVs display skeletal muscle protective properties, associated with their cargo, which is enriched in Neuregulin-1/mRNA [46]. It has been also demonstrated that platelet-derived EVs carry many growth factors, such as VEGF, bFGF, TGF-β1, and PDGF-BB, as well as cytokines [7,47]. More specifically, it has been shown that PRP-EVs induce immunomodulatory effects and accelerate muscle recovery after injury in rat models [47,48]. A summary of the most important EVs in muscle regeneration is reported in Figure 6.
Figure 6.
Scheme of EV interaction in the muscle repair process.
Therefore, we have analyzed the concentrations of EVs in the peripheral blood samples of professional athletes recovering from muscle injuries. Platelet-, leukocyte- and endothelial-derived EVs, already reported to be released during exercise [49], were characterized in subjects undergoing a recovering program including submaximal strength rehabilitation after the completion of such a program. Our data show that, during the recovery, the compartment of platelet-derived EVs significantly increased, suggesting that platelet-derived EVs may be involved in the processes driving muscle repair and regeneration. It can therefore be hypothesized that platelet-derived EVs may have a role in the muscle repair/regeneration during this phase, possibly also exerting anti-inflammatory effects by reshaping the inflammatory environment, as it was also demonstrated for rheumatoid arthritis-affected mice [50].
We have therefore carried out, for the first time to our knowledge, an in-depth analysis of the PRP-EV phenotypes and cargoes. We observed that the PRP-EVs have heterogeneous phenotypes, including leukocyte- and endothelial-derived EVs, even if platelet-derived EVs are well represented, as it was already observed [51].
To deeply study the protein cargo of PRP EVs, we have carried out a shotgun proteomics experiment. From the related molecular and functional characterization, 105 proteins were identified, mostly classified in the “defense and immunity” biological function. This data underlines that PRP EVs actively participate in the well-known interactions between muscle and immune system, orchestrating skeletal muscle regeneration [31]. Unfortunately, there are not accepted specific markers for the immune EV subtyping [18], but we can hypothesize that the leukocyte-derived EV compartment circulating in the peripheral blood of athletes affected by muscle injuries and therefore present in PRP preparations stems from the wide variety of innate and adaptive immune cells that have been involved in muscle repair processes [52]. These data are in line with previously reported evidence demonstrating that EVs convey immunomodulatory messengers that have been already involved in tissue repair and regeneration. Interestingly, PRP EVs that we have isolated by fluorescence-activated cell sorting showed a protein cargo associated with the vesicle-mediated transport. We observed that 49 out of 105 proteins were implicated in the “vesicle-mediated transport” function, demonstrating how the here-applied integrated EV sort-omics approach brings out typical proteins of EVs, potentially implicated in conveying biological information [22,25].
Therefore, those EVs may mediate the intercellular crosstalk, passing across the biological barriers, and reaching remote target cells and tissues [11]. Indeed, our data demonstrate that the cargo of PRP EVs plays a main role in conveying information related to regeneration processes. Furthermore, among the functional networks, “wound healing” emerged as a significant biological function contextually related to “vesicular-mediated transport”, given that more than 80% of wound healing-related proteins were also associated to the vesicle-mediated transport function.
Therefore, PRP EVs may have great potential in promoting tissue repair and regeneration. Some disadvantages have been associated with the use of whole PRP-based therapies. It has been demonstrated that many PRP-derived active biomolecules are damaged by lytic enzymes from the extracellular milieu, not being protected by the plasma membranes, and losing their activities [1]. Additionally, even if platelets lack an integral cell structure, cases of immunological rejection between allogeneic individuals have been registered [1]. On the other hand, several advantages have been demonstrated to be associated with the use of EVs, including their scarce immunogenicity and their ability to be locally released [1]. Interestingly, the use of purified EVs instead of whole PRP preparations as regenerative agents may, therefore, circumvent all these limitations.
Here, we combined, for the first time to our knowledge, the analysis of circulating EVs in untouched peripheral blood samples of athletes recovering from recent muscular injuries with the analysis of the cargo of PRP preparations. The major limitation of this study is the small number of enrolled subjects, even if both flow cytometry and proteomics data underline a possible relevant role of platelet-derived EVs in the processes regulating both repair and regeneration of muscle tissue. In this context, EV-based subcellular therapies have great potential for clinical applications in regenerative medicine, overcoming the challenges of cell-based therapies. Therefore, even if further enlarged studies are needed, our data offer exciting new insights into the complex process of muscle repair, underlying that the use of EVs can be optimized for therapeutic purposes in disease as well as in sport.
4. Materials and Methods
4.1. Patients
The study, carried out according to the ethical principles laid down by the latest version of the Declaration of Helsinki, was approved by the Ethical Committee of the University “G. d’Annunzio”, Chieti-Pescara, Italy (approval code: 29012020). Written informed consent was obtained from all study participants. All patients were enrolled from January to November 2021. We enrolled 4 athletes recovering from recent muscular injuries (all males, mean age: 25.2 + 3.8) and 3 healthy volunteers who did not reported any injury during the last 5 years, and matched them for age and sex with recently injured athletes. We also enrolled 6 athletes recovering from injuries whose peripheral blood samples were used to produce PRP samples. To identify, count, and subtype circulating EVs, peripheral blood samples were collected from all control subjects and from recently injured athletes before (T0) starting a recovering program including submaximal strength rehabilitation and strength development and after (T1) the rehabilitation program [53,54].
4.2. Flow Cytometry Identification, Count, and Subtyping of EVs from Peripheral Blood Samples
Peripheral blood samples were drawn using sodium citrate tubes (Becton Dickinson Biosciences-BD, San Jose, CA, USA, Ref 454387) and analyzed within 4 h from venipuncture. The EV staining was carried out as previously described [21,55,56,57]. Briefly, the reagent mix summarized in Table S1 was added to 195 μL of PBS 1X; then, 5 μL of whole blood were added to the mix. After 45 min of staining (RT, in the dark), 500 μL of PBS 1X were added to each tube and 1 × 106 events/sample were acquired by flow cytometry (FACSVerse, BD Biosciences, San Jose, CA, USA), placing the trigger threshold on the lipophilic cationic dye channel (LCD) [58]. To avoid immune complex formation and antibody aggregation, each reagent stock solution was centrifuged before its use (21,000× g, 12 min). For all measured parameters, the height (H) signals are shown.
Instrument performances, data reproducibility, and fluorescence calibrations were monitored by the Cytometer Setup and Tracking Module (BD Biosciences). The evaluation of non-specific fluorescence was obtained by acquiring FMO combined with the respective isotype control [59,60]. Data were analyzed using FACSuite v 1.0.6.5230 (BD Biosciences) and FlowJo X v 10.0.7 (BD Biosciences) software. EVs concentrations were calculated based on the volumetric count function.
4.3. Flow Cytometry Gating Strategy
Figure 1A shows the gating strategy used to identify, count, and subtype circulating EVs. In detail, a forward scatter height (FSC-H)/ Side Scatter-H (SSC-H) dot-plot was used to establish a region, defined as “platelet free area”, containing the events with the EV scatter features (Figure 1A) [55,56]. Those events were represented on an LCD-H/Phalloidin-H dot-plot and EVs were identified as LCD positive/phalloidin negative dots (Figure 1B). Therefore, EVs (LCD+/Phalloidin- events) were analyzed on a CD45-H/CD41a-H dot-plot and CD45+ events were identified as leukocyte-derived EVs (Figure 1C) [18]. A CD45 negative logical gate was obtained, and the resulting population was represented on a CD31-H/CD41a-H dot-plot. Events showing the CD31+/CD41a+ phenotype were identified as platelet-derived EVs (Platelet EVs) (Figure 1D) [18], whereas the compartment was identified as endothelial-derived EVs (Figure 1D) [18]. Platelet-derived EVs (Figure 1E) and endothelial-derived EVs (Figure 1F) and all platelets (Figure 1G) were further analyzed for the activation marker CD62P. In Figure 1H, the used gating hierarchy is shown as a scheme.
4.4. Platelet-Enriched Plasma Preparation
The preparation of PRP was carried out as already reported [61]. Briefly, peripheral blood samples (60 mL) were drawn in 10% citrate dextrose A (ACD) solution and centrifuged (600× g, 10 min). The plasma fraction was then centrifuged at 4000× g for 15 min. Supernatants, representing the platelet poor plasma (PPP), were removed, and the remaining suspension, the PRP, was collected and mixed with the platelets present in the PPP, previously trapped by filtration. The mixture was passed through a WBC filter (Terumo Imuguard, CO, USA) to remove the leukocyte fraction. Platelet and leukocyte counts were carried out, and platelet concentrations were adjusted to 1 × 1010 in 8 mL.
4.5. EV Separation by Fluorescence-Activated Cell Sorting
PRP-EVs were isolated by fluorescence activated cell sorting, as previously reported [55]. Briefly, PRP samples were stained by 0.5 µL of FITC-conjugated phalloidin and LCD (BD Biosciences–Catalog, #626267, Custom Kit), as described above. After 45 min of staining (RT, in the dark), at least 500 µL of PBS 1X was added to each tube. Such a dilution allowed us to maintain the correct event rate recommended for 100 μm nozzle, which we have used. Total EVs (LCD+/Phalloidin- events) were separated using a FACSAria III cell sorter (BD Biosciences San Jose, CA, USA). The trigger threshold was placed on the APC channel and, for all parameters, the height (H) signals, as well as bi-exponential or logarithmic modes were selected. The post-sorting purity was assessed by using the same instrument (FACSAria III) and the same setting applied for EV separation. Instrument performances, data reproducibility, and fluorescence calibrations were sustained by the Cytometer Setup and Tracking Module (BD Biosciences).
4.6. EV Protein Cargo Detection by Label-Free Proteomics
Proteomics analyses were normalized by using the number of purified EVs. As already described, 1 × 106 purified PRP-EVs were used for each proteomic detection [22,25]. Digested proteins were acquired in triplicate by LC-MS/MS using the UltiMateTM 3000 UPLC (Thermo Fisher Scientific, Waltham, MA, USA) chromatographic system coupled to the Orbitrap FusionTM TribridTM (Thermo Fisher Scientific, Waltham, MA, USA) mass spectrometer. Briefly, the flow rate was set at 300 nL/min with a total run of 65 min by using an EASY-spray AcclaimTM PepMapTM C18 (75 μm ID, 25 cm L, 2 μm PS, Thermo Fisher Scientific) nanoscale chromatographic column. Details of LC-MS/MS parameters are reported in our previous work [62]. Proteomics MS/MS raw data were processed using the Andromeda peptide search engine through MaxQuant version 1.6.10.50 (Max-Planck Institute for Biochemistry, Martinsried, Germany) matching spectra against the UniProt database (released 2020_06, taxonomy Homo Sapiens, 20,588 entries) supplemented with frequently observed contaminants and containing forward and reverse sequences. iBAQ (intensity-based absolute quantification) values were used to quantify protein abundance in each sample whenever the protein was quantified in at least two analytical replicates. STRING version 11.5 database was used to perform Protein–Protein Interaction (PPI) networks.
4.7. Statistical Analysis
Data were analyzed using the XLSTAT 2022 (Addinsoft, Paris, France) and GraphPad Prism 9 (GraphPad Software Inc., La Jolla, CA, USA). Two-sided Student’s t-test or paired t-test were used as indicated. Statistical significance was accepted for p < 0.05.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232314913/s1.
Author Contributions
Conceptualization, P.D.B., D.P. and P.L.; methodology, G.C., M.C.C., D.D.B., S.V., F.T., G.M. and L.S.; software, M.C.C., I.C. and P.S.; validation, G.C., M.C.C., I.C. and P.S.; formal analysis, G.C. and M.C.C.; investigation, G.C., M.C.C. and P.S.; resources, V.D.L. and P.L.; data curation, G.C., M.C.C., D.D.B. and I.C.; writing—original draft preparation, G.C. and M.C.C.; writing—review and editing, P.S. and P.L.; visualization, P.D.B. and D.P.; supervision, V.D.L. and P.L.; project administration, P.D.B., D.P., V.D.L. and P.L.; funding acquisition, P.S. and V.D.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the ethical Committee of the University “G. d’Annunzio”, Chieti-Pescara, Italy. (protocol code: Report N. 26 of 29 December 2020).
Informed Consent Statement
Written informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data will be available under reasonable requests.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
Domenico De Bellis has a PhD fellowship (code: n. 1353889) in the framework of PON RI 2014/2020, I.1-“Innovative PhDs with industrial characterization”, funded by the Italian Ministry of University and Research (MUR), Italy, FSE-FESR.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wu J., Piao Y., Liu Q., Yang X. Platelet-rich plasma-derived extracellular vesicles: A superior alternative in regenerative medicine? Cell Prolif. 2021;54:e13123. doi: 10.1111/cpr.13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel A.N., Selzman C.H., Kumpati G.S., McKellar S.H., Bull D.A. Evaluation of autologous platelet rich plasma for cardiac surgery: Outcome analysis of 2000 patients. J. Cardiothorac. Surg. 2016;11:62. doi: 10.1186/s13019-016-0452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anitua E., Fernández-de-Retana S., Alkhraisat M.H. Platelet rich plasma in oral and maxillofacial surgery from the perspective of composition. Platelets. 2021;32:174–182. doi: 10.1080/09537104.2020.1856361. [DOI] [PubMed] [Google Scholar]
- 4.Muchedzi T.A., Roberts S.B. A systematic review of the effects of platelet rich plasma on outcomes for patients with knee osteoarthritis and following total knee arthroplasty. Surgeon. 2018;16:250–258. doi: 10.1016/j.surge.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Emer J. Platelet-Rich Plasma (PRP): Current Applications in Dermatology. Skin Ther. Lett. 2019;24:1–6. [PubMed] [Google Scholar]
- 6.Dohan Ehrenfest D.M., Andia I., Zumstein M.A., Zhang C.-Q., Pinto N.R., Bielecki T. Classification of platelet concentrates (Platelet-Rich Plasma-PRP, Platelet-Rich Fibrin-PRF) for topical and infiltrative use in orthopedic and sports medicine: Current consensus, clinical implications and perspectives. Muscles. Ligaments Tendons J. 2014;4:3–9. doi: 10.32098/mltj.01.2014.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torreggiani E., Perut F., Roncuzzi L., Zini N., Baglìo S.R., Baldini N. Exosomes: Novel effectors of human platelet lysate activity. Eur. Cells Mater. 2014;28 doi: 10.22203/eCM.v028a11. [DOI] [PubMed] [Google Scholar]
- 8.Tao S.-C., Yuan T., Rui B.-Y., Zhu Z.-Z., Guo S.-C., Zhang C.-Q. Exosomes derived from human platelet-rich plasma prevent apoptosis induced by glucocorticoid-associated endoplasmic reticulum stress in rat osteonecrosis of the femoral head via the Akt/Bad/Bcl-2 signal pathway. Theranostics. 2017;7:733–750. doi: 10.7150/thno.17450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo S.-C., Tao S.-C., Yin W.-J., Qi X., Yuan T., Zhang C.-Q. Exosomes derived from platelet-rich plasma promote the re-epithelization of chronic cutaneous wounds via activation of YAP in a diabetic rat model. Theranostics. 2017;7:81–96. doi: 10.7150/thno.16803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaborowski M.P., Balaj L., Breakefield X.O., Lai C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience. 2015;65:783–797. doi: 10.1093/biosci/biv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simeone P., Bologna G., Lanuti P., Pierdomenico L., Guagnano M.T., Pieragostino D., Del Boccio P., Vergara D., Marchisio M., Miscia S., et al. Extracellular Vesicles as Signaling Mediators and Disease Biomarkers across Biological Barriers. Int. J. Mol. Sci. 2020;21:2514. doi: 10.3390/ijms21072514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.György B., Szabó T.G., Pásztói M., Pál Z., Misják P., Aradi B., László V., Pállinger E., Pap E., Kittel A., et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colombo M., Raposo G., Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 14.Cufaro M.C., Pieragostino D., Lanuti P., Rossi C., Cicalini I., Federici L., De Laurenzi V., Del Boccio P. Extracellular Vesicles and Their Potential Use in Monitoring Cancer Progression and Therapy: The Contribution of Proteomics. J. Oncol. 2019;2019:1639854. doi: 10.1155/2019/1639854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akers J.C., Gonda D., Kim R., Carter B.S., Chen C.C. Biogenesis of extracellular vesicles (EV): Exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies. J. Neurooncol. 2013;113:1–11. doi: 10.1007/s11060-013-1084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poon I.K.H., Parkes M.A.F., Jiang L., Atkin-Smith G.K., Tixeira R., Gregory C.D., Ozkocak D.C., Rutter S.F., Caruso S., Santavanond J.P., et al. Moving beyond size and phosphatidylserine exposure: Evidence for a diversity of apoptotic cell-derived extracellular vesicles in vitro. J. Extracell. Vesicles. 2019;8:1608786. doi: 10.1080/20013078.2019.1608786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coumans F.A.W., Brisson A.R., Buzas E.I., Dignat-George F., Drees E.E.E., El-Andaloussi S., Emanueli C., Gasecka A., Hendrix A., Hill A.F., et al. Methodological Guidelines to Study Extracellular Vesicles. Circ. Res. 2017;120:1632–1648. doi: 10.1161/CIRCRESAHA.117.309417. [DOI] [PubMed] [Google Scholar]
- 18.Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossi C., Cicalini I., Cufaro M.C., Agnifili L., Mastropasqua L., Lanuti P., Marchisio M., De Laurenzi V., Del Boccio P., Pieragostino D. Multi-Omics Approach for Studying Tears in Treatment-Naïve Glaucoma Patients. Int. J. Mol. Sci. 2019;20:4029. doi: 10.3390/ijms20164029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falasca K., Lanuti P., Ucciferri C., Pieragostino D., Cufaro M.C., Bologna G., Federici L., Miscia S., Pontolillo M., Auricchio A., et al. Circulating extracellular vesicles as new inflammation marker in HIV infection. AIDS. 2021;35:595–604. doi: 10.1097/QAD.0000000000002794. [DOI] [PubMed] [Google Scholar]
- 21.Brocco D., Lanuti P., Pieragostino D., Cufaro M.C., Simeone P., Bologna G., Di Marino P., De Tursi M., Grassadonia A., Irtelli L., et al. Phenotypic and Proteomic Analysis Identifies Hallmarks of Blood Circulating Extracellular Vesicles in NSCLC Responders to Immune Checkpoint Inhibitors. Cancers. 2021;13:585. doi: 10.3390/cancers13040585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pieragostino D., Cicalini I., Lanuti P., Ercolino E., di Ioia M., Zucchelli M., Zappacosta R., Miscia S., Marchisio M., Sacchetta P., et al. Enhanced release of acid sphingomyelinase-enriched exosomes generates a lipidomics signature in CSF of Multiple Sclerosis patients. Sci. Rep. 2018;8:3071. doi: 10.1038/s41598-018-21497-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brocco D., Lanuti P., Simeone P., Bologna G., Pieragostino D., Cufaro M.C., Graziano V., Peri M., Di Marino P., De Tursi M., et al. Circulating Cancer Stem Cell-Derived Extracellular Vesicles as a Novel Biomarker for Clinical Outcome Evaluation. J. Oncol. 2019;2019:5879616. doi: 10.1155/2019/5879616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cela I., Cufaro M.C., Fucito M., Pieragostino D., Lanuti P., Sallese M., Del Boccio P., Di Matteo A., Allocati N., De Laurenzi V., et al. Proteomic Investigation of the Role of Nucleostemin in Nucleophosmin-Mutated OCI-AML 3 Cell Line. Int. J. Mol. Sci. 2022;23:7655. doi: 10.3390/ijms23147655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pieragostino D., Lanuti P., Cicalini I., Cufaro M.C., Ciccocioppo F., Ronci M., Simeone P., Onofrj M., van der Pol E., Fontana A., et al. Proteomics characterization of extracellular vesicles sorted by flow cytometry reveals a disease-specific molecular cross-talk from cerebrospinal fluid and tears in multiple sclerosis. J. Proteom. 2019;204:103403. doi: 10.1016/j.jprot.2019.103403. [DOI] [PubMed] [Google Scholar]
- 26.Buca D., D’Antonio F., Buca D., Di Sebastiano F., Simeone P., Di Girolamo R., Bologna G., Vespa S., Catitti G., Liberati M., et al. Extracellular Vesicles in pregnancy: Their potential role as a liquid biopsy. J. Reprod. Immunol. 2022;154:103734. doi: 10.1016/j.jri.2022.103734. [DOI] [PubMed] [Google Scholar]
- 27.Tkach M., Théry C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 28.Tetta C., Ghigo E., Silengo L., Deregibus M.C., Camussi G. Extracellular vesicles as an emerging mechanism of cell-to-cell communication. Endocrine. 2013;44:11–19. doi: 10.1007/s12020-012-9839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bittel D.C., Jaiswal J.K. Contribution of Extracellular Vesicles in Rebuilding Injured Muscles. Front. Physiol. 2019;10:828. doi: 10.3389/fphys.2019.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tidball J.G., Villalta S.A. Regulatory interactions between muscle and the immune system during muscle regeneration. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010;298:R1173–R1187. doi: 10.1152/ajpregu.00735.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tidball J.G. Regulation of muscle growth and regeneration by the immune system. Nat. Rev. Immunol. 2017;17:165–178. doi: 10.1038/nri.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rigamonti E., Zordan P., Sciorati C., Rovere-Querini P., Brunelli S. Macrophage plasticity in skeletal muscle repair. Biomed Res. Int. 2014;2014:560629. doi: 10.1155/2014/560629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Zhao M., Liu S., Guo J., Lu Y., Cheng J., Liu J. Macrophage-derived extracellular vesicles: Diverse mediators of pathology and therapeutics in multiple diseases. Cell Death Dis. 2020;11:924. doi: 10.1038/s41419-020-03127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antich-Rosselló M., Forteza-Genestra M.A., Monjo M., Ramis J.M. Platelet-Derived Extracellular Vesicles for Regenerative Medicine. Int. J. Mol. Sci. 2021;22:8580. doi: 10.3390/ijms22168580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vajen T., Mause S.F., Koenen R.R. Microvesicles from platelets: Novel drivers of vascular inflammation. Thromb. Haemost. 2015;114:228–236. doi: 10.1160/TH14-11-0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berckmans R.J., Lacroix R., Hau C.M., Sturk A., Nieuwland R. Extracellular vesicles and coagulation in blood from healthy humans revisited. J. Extracell. Vesicles. 2019;8:1688936. doi: 10.1080/20013078.2019.1688936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melki I., Tessandier N., Zufferey A., Boilard E. Platelet microvesicles in health and disease. Platelets. 2017;28:214–221. doi: 10.1080/09537104.2016.1265924. [DOI] [PubMed] [Google Scholar]
- 38.Santilli F., Marchisio M., Lanuti P., Boccatonda A., Miscia S., Davì G. Microparticles as new markers of cardiovascular risk in diabetes and beyond. Thromb. Haemost. 2016;116:220–234. doi: 10.1160/TH16-03-0176. [DOI] [PubMed] [Google Scholar]
- 39.Gentile P., Garcovich S. Systematic Review-The Potential Implications of Different Platelet-Rich Plasma (PRP) Concentrations in Regenerative Medicine for Tissue Repair. Int. J. Mol. Sci. 2020;21:5702. doi: 10.3390/ijms21165702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marques L.F., Stessuk T., Camargo I.C.C., Sabeh Junior N., dos Santos L., Ribeiro-Paes J.T. Platelet-rich plasma (PRP): Methodological aspects and clinical applications. Platelets. 2015;26:101–113. doi: 10.3109/09537104.2014.881991. [DOI] [PubMed] [Google Scholar]
- 41.Spakova T., Janockova J., Rosocha J. Characterization and Therapeutic Use of Extracellular Vesicles Derived from Platelets. Int. J. Mol. Sci. 2021;22:9701. doi: 10.3390/ijms22189701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahu A., Clemens Z.J., Shinde S.N., Sivakumar S., Pius A., Bhatia A., Picciolini S., Carlomagno C., Gualerzi A., Bedoni M., et al. Regulation of aged skeletal muscle regeneration by circulating extracellular vesicles. Nat. Aging. 2021;1:1148–1161. doi: 10.1038/s43587-021-00143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H., Wang B. Extracellular vesicle microRNAs mediate skeletal muscle myogenesis and disease. Biomed. Rep. 2016;5:296–300. doi: 10.3892/br.2016.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakamura Y., Miyaki S., Ishitobi H., Matsuyama S., Nakasa T., Kamei N., Akimoto T., Higashi Y., Ochi M. Mesenchymal-stem-cell-derived exosomes accelerate skeletal muscle regeneration. FEBS Lett. 2015;589:1257–1265. doi: 10.1016/j.febslet.2015.03.031. [DOI] [PubMed] [Google Scholar]
- 45.Zhu Y.-G., Feng X.-M., Abbott J., Fang X.-H., Hao Q., Monsel A., Qu J.-M., Matthay M.A., Lee J.W. Human mesenchymal stem cell microvesicles for treatment of Escherichia coli endotoxin-induced acute lung injury in mice. Stem Cells. 2014;32:116–125. doi: 10.1002/stem.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Figliolini F., Ranghino A., Grange C., Cedrino M., Tapparo M., Cavallari C., Rossi A., Togliatto G., Femminò S., Gugliuzza M.V., et al. Extracellular Vesicles From Adipose Stem Cells Prevent Muscle Damage and Inflammation in a Mouse Model of Hind Limb Ischemia: Role of Neuregulin-1. Arterioscler. Thromb. Vasc. Biol. 2020;40:239–254. doi: 10.1161/ATVBAHA.119.313506. [DOI] [PubMed] [Google Scholar]
- 47.Vajen T., Benedikter B.J., Heinzmann A.C.A., Vasina E.M., Henskens Y., Parsons M., Maguire P.B., Stassen F.R., Heemskerk J.W.M., Schurgers L.J., et al. Platelet extracellular vesicles induce a pro-inflammatory smooth muscle cell phenotype. J. Extracell. Vesicles. 2017;6:1322454. doi: 10.1080/20013078.2017.1322454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iyer S.R., Scheiber A.L., Yarowsky P., Henn R.F., Otsuru S., Lovering R.M. Exosomes Isolated From Platelet-Rich Plasma and Mesenchymal Stem Cells Promote Recovery of Function after Muscle Injury. Am. J. Sports Med. 2020;48:2277–2286. doi: 10.1177/0363546520926462. [DOI] [PubMed] [Google Scholar]
- 49.Brahmer A., Neuberger E., Esch-Heisser L., Haller N., Jorgensen M.M., Baek R., Möbius W., Simon P., Krämer-Albers E.-M. Platelets, endothelial cells and leukocytes contribute to the exercise-triggered release of extracellular vesicles into the circulation. J. Extracell. Vesicles. 2019;8:1615820. doi: 10.1080/20013078.2019.1615820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma Q., Bai J., Xu J., Dai H., Fan Q., Fei Z., Chu J., Yao C., Shi H., Zhou X., et al. Reshaping the Inflammatory Environment in Rheumatoid Arthritis Joints by Targeting Delivery of Berberine with Platelet-Derived Extracellular Vesicles. Adv. NanoBiomed Res. 2021;1:2100071. doi: 10.1002/anbr.202100071. [DOI] [Google Scholar]
- 51.Chandler W.L. Microparticle counts in platelet-rich and platelet-free plasma, effect of centrifugation and sample-processing protocols. Blood Coagul. Fibrinolysis. 2013;24:125–132. doi: 10.1097/MBC.0b013e32835a0824. [DOI] [PubMed] [Google Scholar]
- 52.Ziemkiewicz N., Hilliard G., Pullen N.A., Garg K. The Role of Innate and Adaptive Immune Cells in Skeletal Muscle Regeneration. Int. J. Mol. Sci. 2021;22:3265. doi: 10.3390/ijms22063265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casolo A., Del Vecchio A., Balshaw T.G., Maeo S., Lanza M.B., Felici F., Folland J.P., Farina D. Behavior of motor units during submaximal isometric contractions in chronically strength-trained individuals. J. Appl. Physiol. 2021;131:1584–1598. doi: 10.1152/japplphysiol.00192.2021. [DOI] [PubMed] [Google Scholar]
- 54.Casolo A., Farina D., Falla D., Bazzucchi I., Felici F., Del Vecchio A. Strength Training Increases Conduction Velocity of High-Threshold Motor Units. Med. Sci. Sports Exerc. 2020;52:955–967. doi: 10.1249/MSS.0000000000002196. [DOI] [PubMed] [Google Scholar]
- 55.Marchisio M., Simeone P., Bologna G., Ercolino E., Pierdomenico L., Pieragostino D., Ventrella A., Antonini F., Del Zotto G., Vergara D., et al. Flow Cytometry Analysis of Circulating Extracellular Vesicle Subtypes from Fresh Peripheral Blood Samples. Int. J. Mol. Sci. 2020;22:48. doi: 10.3390/ijms22010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brocco D., Simeone P., Buca D., Di Marino P., De Tursi M., Grassadonia A., De Lellis L., Martino M.T., Veschi S., Iezzi M., et al. Blood Circulating CD133+ Extracellular Vesicles Predict Clinical Outcomes in Patients with Metastatic Colorectal Cancer. Cancers. 2022;14:1357. doi: 10.3390/cancers14051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buca D., Bologna G., D’Amico A., Cugini S., Musca F., Febbo M., D’Arcangelo D., Buca D., Simeone P., Liberati M., et al. Extracellular Vesicles in Feto-Maternal Crosstalk and Pregnancy Disorders. Int. J. Mol. Sci. 2020;21:2120. doi: 10.3390/ijms21062120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simeone P., Celia C., Bologna G., Ercolino E., Pierdomenico L., Cilurzo F., Grande R., Diomede F., Vespa S., Canonico B., et al. Diameters and Fluorescence Calibration for Extracellular Vesicle Analyses by Flow Cytometry. Int. J. Mol. Sci. 2020;21:7885. doi: 10.3390/ijms21217885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lanuti P., Ciccocioppo F., Bonanni L., Marchisio M., Lachmann R., Tabet N., Pierdomenico L., Santavenere E., Catinella V., Iacone A., et al. Amyloid-specific T-cells differentiate Alzheimer’s disease from Lewy body dementia. Neurobiol. Aging. 2012;33:2599–2611. doi: 10.1016/j.neurobiolaging.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 60.Lanuti P., Simeone P., Rotta G., Almici C., Avvisati G., Azzaro R., Bologna G., Budillon A., Di Cerbo M., Di Gennaro E., et al. A standardized flow cytometry network study for the assessment of circulating endothelial cell physiological ranges. Sci. Rep. 2018;8:5823. doi: 10.1038/s41598-018-24234-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bansal H., Leon J., Pont J.L., Wilson D.A., Bansal A., Agarwal D., Preoteasa I. Platelet-rich plasma (PRP) in osteoarthritis (OA) knee: Correct dose critical for long term clinical efficacy. Sci. Rep. 2021;11:3971. doi: 10.1038/s41598-021-83025-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Potenza F., Cufaro M.C., Di Biase L., Panella V., Di Campli A., Ruggieri A.G., Dufrusine B., Restelli E., Pietrangelo L., Protasi F., et al. Proteomic Analysis of Marinesco-Sjogren Syndrome Fibroblasts Indicates Pro-Survival Metabolic Adaptation to SIL1 Loss. Int. J. Mol. Sci. 2021;22:12449. doi: 10.3390/ijms222212449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be available under reasonable requests.