Abstract
Lactic acid bacteria were reported as a promising alternative to antibiotics against pathogens. Among them, Lactobacillus rhamnosus could be used as probiotics and inhibit several pathogens, but its antibacterial mechanisms are still less known. Here, L. rhamnosus SCB0119 isolated from fermented pickles could inhibit bacterial growth or even cause cell death in Escherichia coli ATCC25922 and Staphylococcus aureus ATCC6538, which was mainly attributed to the cell-free culture supernatant (CFS). Moreover, CFS induced the accumulation of reactive oxygen species and destroyed the structure of the cell wall and membrane, including the deformation in cell shape and cell wall, the impairment of the integrity of the cell wall and inner membrane, and the increases in outer membrane permeability, the membrane potential, and pH gradient in E. coli and S. aureus. Furthermore, the transcriptomic analysis demonstrated that CFS altered the transcripts of several genes involved in fatty acid degradation, ion transport, and the biosynthesis of amino acids in E. coli, and fatty acid degradation, protein synthesis, DNA replication, and ATP hydrolysis in S. aureus, which are important for bacterial survival and growth. In conclusion, L. rhamnosus SCB0119 and its CFS could be used as a biocontrol agent against E. coli and S. aureus.
Keywords: Lactobacillus rhamnosus, cell-free culture supernatant, Escherichia coli, Staphylococcus aureus, antimicrobial activity, transcriptomic analysis
1. Introduction
Foodborne pathogens, including Escherichia coli and Staphylococcus aureus, cause foodborne diseases and bring serious harm to human health [1,2,3]. Approximately 2.8 billion people worldwide suffer from diarrhea due to foodborne illness, and about 3000 people die each year from illness by ingesting food contaminated with foodborne pathogens in the United States [4]. Existing widely in food, water, food-producing animals, and so on [5], E. coli can cause various human diseases ranging from gastroenteritis, such as diarrhea, to extra-intestinal infections, including those of the urinary tract, bloodstream, and central nervous system [6,7]. For example, Shiga toxin-producing E. coli caused 2,801,000 acute illnesses annually, 3890 cases of hemolytic uremic syndrome, 270 cases of permanent end-stage renal disease, and 230 deaths, according to databases from 21 countries published between 1 January 1990 and 30 April 2012 [8]. As one of the main foodborne pathogens contaminating ready-to-eat products, meat, egg, dairy products, cream-filled pastries, and cakes [9] and existing in food production animals [10], S. aureus could produce exotoxins and cause food poisoning with the symptoms including nausea, vomiting, and abdominal cramps with or without diarrhea [11]. Additionally, S. aureus is a leading cause of bacteremia, which harbors high rates in the first year of life, a low incidence through young adulthood, and a gradual rise in incidence with advancing age, and the population incidence of S. aureus bacteremia is 10–30 per 100,000 person-years in industrialized countries [12]. To prevent the contamination of foodborne pathogens, antibiotics and/or chemical preservatives are widely used in animal husbandry and food manufacturing, but the excessive and unreasonable use of antibiotics and chemical preservatives resulted in new hazards to human health, such as the accumulation of chemical residues in food and feed chain [13], the development of antibiotic resistance in bacteria, and environmental pollution [14]. Thus, it is necessary to find an effective and alternative way to replace antibiotics and chemical preservatives.
Lactic acid bacteria are regarded as a promising alternative to antibiotics against the contamination of pathogens due to high-effective antibacterial activities, various antimicrobial substances, beneficial effects on human and animal health, and so on [15,16]. Among them, Lactobacillus rhamnosus is generally considered to be beneficial to human health and commonly used as a probiotic [17]. Previous studies showed that L. rhamnosus could inhibit the growth of several pathogens, such as S. aureus, E. coli, Candida albicans, Shigella sonnei, Salmonella enteritidis, Pseudomonas fluorescens, Pseudomonas putida, and Listeria monocytogene, and its antibacterial activities differed among different L. rhamnosus strains [18,19]. However, it was still little known about the antibacterial mechanisms of L. rhamnosus, though it was reported to inhibit the growth of S. enteritidis by elevating the reactive oxygen species level and destructing bacterial membrane [20] and inhibiting the growth of S. aureus by increasing cell permeability [21]. In this study, we evaluated the potential antibacterial effects of L. rhamnosus SCB0119 isolated from fermented pickles against E. coli and S. aureus by investigating the changes in bacterial growth and cell structures. Moreover, transcriptomic analysis was used to unveil the underlying mechanism of actions. The findings will be helpful in preventing the contamination of E. coli and S. aureus and developing a novel preventative strategy against pathogens not limited to E. coli and S. aureus.
2. Results
2.1. Reduction in Bacterial Growth of E. coli and S. aureus Caused by L. rhamnosus SCB0119
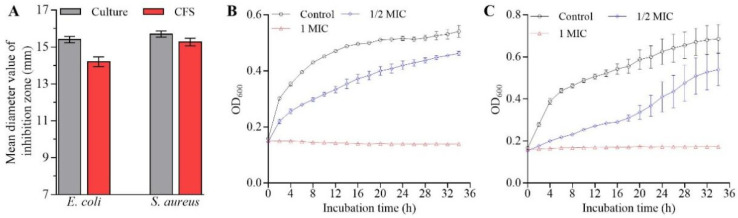
The L. rhamnosus SCB0119 culture solution and cell-free culture supernatant (CFS) showed antimicrobial activity against E. coli and S. aureus with mean inhibition zone diameters of 15.4 and 14.2 mm for E. coli ATCC25922, and 15.7 and 15.27 mm for S. aureus ATCC6538, respectively (Figure 1A). However, resuspended bacterial cells displayed no antagonistic activity (no inhibition zone) against E. coli ATCC25922 and S. aureus ATCC6538. These results demonstrated the antimicrobial activity of L. rhamnosus SCB0119 was mainly attributed to its metabolites.
Figure 1.
Antimicrobial activity of L. rhamnosus SCB0119 against E. coli ATCC25922 and S. aureus ATCC6538. (A) The mean diameters of inhibition zones after E. coli ATCC25922 and S. aureus ATCC6538 were incubated with L. rhamnosus SCB0119 culture solution and CFS for 12 h, respectively. (B,C) Effects of CFS on the growth of E. coli ATCC25922 and S. aureus ATCC6538, respectively. Error bars: SD of the mean from three repeated assays.
To further investigate the effects of L. rhamnosus SCB0119 on bacterial growth and its potential mechanisms, CFS was chosen to conduct the following studies. The minimum inhibitory concentrations (MICs) of CFS against E. coli ATCC25922 and S. aureus ATCC6538 were 3.91 and 7.81 mg/mL, respectively. Obviously, 1 MIC of CFS completely inhibited the growth of E. coli ATCC25922 (Figure 1B) and S. aureus ATCC6538 (Figure 1C). Moreover, the reduction in bacterial growth of E. coli ATCC25922 and S. aureus ATCC6538 started at the early stationary phase, and the time needed to reach a plateau was delayed by 6 h for E. coli ATCC25922 (Figure 1B) and 4 h for S. aureus ATCC6538 (Figure 1C), when 1/2 MIC of CFS was added into Luria Bertani (LB) medium.
2.2. Effects of CFS on the Production of Intracellular ATP and Reactive Oxygen Species in E. coli and S. aureus
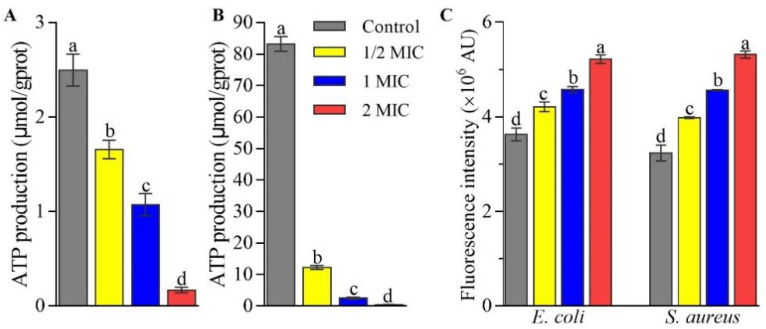
CFS dose-dependently inhibited the production of intracellular ATP in E. coli ATCC25922 and S. aureus ATCC6538. In comparison to 2.49 and 83.22 nmol/mg protein on average for E. coli ATCC25922 (Figure 2A) and S. aureus ATCC6538 (Figure 2B), respectively, in the control group, the ATP production decreased by 33.68% and 85.27% in 1/2 MIC group, 57.01% and 96.95% in 1 MIC group, and 93.31% and 99.49% in 2 MIC group, respectively, suggesting CFS inhibited E. coli ATCC25922 and S. aureus ATCC6538 growth or even caused death because the decreases in intracellular ATP levels mean that cells undergo apoptosis, necrosis, or are in a toxic state [22].
Figure 2.
Effects of CFS from L. rhamnosus SCB0119 on the production of ATP (A,B) and ROS (C) in E. coli ATCC25922 (A,C) and S. aureus ATCC6538 (B,C). ROS production was detected using DCFH-DA staining. Different lowercase letters indicate significant differences among the treatments (p < 0.05). Error bars: SD of the mean from three repeated assays.
Moreover, CFS induced oxidative stress in E. coli ATCC25922 and S. aureus ATCC6538 cells. As shown in Figure 2C, the fluorescence intensities using DCFH-DA staining in E. coli ATCC25922 and S. aureus ATCC6538 cells increased by 16.10% and 23.09% in 1/2 MIC group, 26.12% and 41.03% in 1 MIC group, and 43.90% and 64.36% in 2 MIC group, respectively, compared with those in the control group, suggesting CFS increased the reactive oxygen species (ROS) levels in a dose-dependent manner.
2.3. Changes in Cell Morphology and Structure of E. coli and S. aureus Caused by CFS
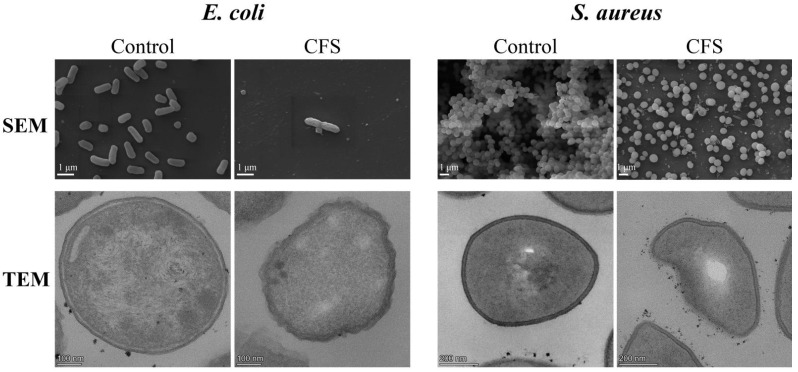
Cell morphology and structure of E. coli ATCC25922 and S. aureus ATCC6538 were affected by CFS. Firstly, scanning electron microscopy (SEM) images showed that fewer cells and more shrunk cells were observed in the CFS group than that in the control group (Figure 3). Moreover, S. aureus ATCC6538 cells became dispersed rather than clustered after being treated with CFS (Figure 3). Additionally, transmission electron microscopy (TEM) images displayed that E. coli ATCC25922 and S. aureus ATCC6538 cells treated with CFS were deformed, ruptured, and less distinctly outlined compared with untreated cells (Figure 3).
Figure 3.
SEM and TEM images after E. coli ATCC25922 and S. aureus ATCC6538 cells were treated with 1/2 MIC of CFS from L. rhamnosus SCB0119.
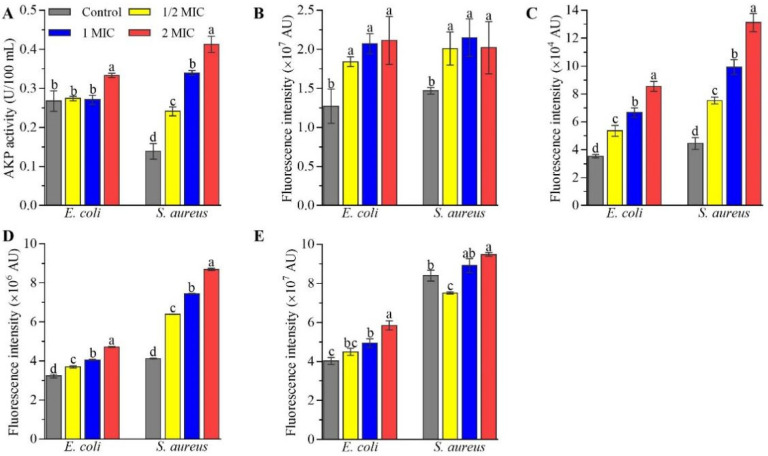
Secondly, CFS impaired cell wall integrity based on alkaline phosphatase (AKP) activity. As shown in Figure 4A, the AKP contents in S. aureus ATCC6538 cells increased by 73.81% in the 1/2 MIC group, 144.76% in the 1 MIC group, and 197.62% in the 2 MIC group, and those in E. coli ATCC25922 cells increased 24.69% in 2 MIC group, compared with those in the control group, though CFS at 1/2 and 1 MIC did not change the AKP contents in E. coli ATCC25922 cells.
Figure 4.
Effects of CFS from L. rhamnosus SCB0119 on AKP contents (A), outer membrane permeability (B), inner membrane integrity (C), membrane potential (D) and pH gradients (E) in E. coli ATCC25922 and S. aureus ATCC6538. Changes in the outer membrane permeability, inner membrane integrity, membrane potential, and pH gradients were assayed using NPN, PI, DiBAC4(3), and BCECF-AM staining, respectively. Different lowercase letters indicate significant differences among the treatments (p < 0.05). Error bars: SD of the mean from three repeated assays.
Moreover, CFS increased outer membrane permeability and impaired inner membrane integrity. The fluorescence intensities using 1-N-phenylnaphthylamine (NPN) staining, which has become a common fluorescent detection signal to evaluate the extent of bacterial outer membrane damage, increased by 36.92–66.18% in E. coli ATCC25922 and S. aureus ATCC6538 cells treated with CFS at different concentrations, compared with those in untreated cells (Figure 4B). Additionally, the fluorescence intensities using propidium iodide (PI) staining in E. coli ATCC25922 and S. aureus ATCC6538 cells increased by 51.39% and 69.13% in the 1/2 MIC group, 88.44% and 123.12% in the 1 MIC group, and 141.69% and 194.72% in the 2 MIC group (Figure 4C), respectively, compared with those in the control group, suggesting CFS impaired inner membrane integrity in a dose-dependent manner.
Accompanied by the destruction of cell membrane integrity, CFS increased the membrane potential (ΔΨ) and pH gradient (ΔpH). As shown in Figure 4D, the fluorescence intensities using DiBAC4(3) staining in E. coli ATCC25922 and S. aureus ATCC6538 cells increased by 14.55% and 55.03% in the 1/2 MIC group, 25.83% and 80.51% in the 1 MIC group, and 46.30% and 110.69% in the 2 MIC group, respectively, compared with those in the control group, suggesting CFS caused depolarization of E. coli ATCC25922 and S. aureus ATCC6538 cells. Similarly, the fluorescence intensities using BCECF-AM staining in E. coli ATCC25922 cells increased by 11.73% in the 1/2 MIC group, 22.73% in the 1 MIC group, and 45.31% in the 2 MIC group, and those in S. aureus ATCC6538 cells increased by 6.12% and 12.92 in the 1 MIC and 2 MIC groups, respectively, but decreased by 10.57% in the 1/2 MIC group (Figure 4E), compared with those in the control group.
2.4. Transcriptomic Analysis of E. coli and S. aureus Cells in Response to CFS
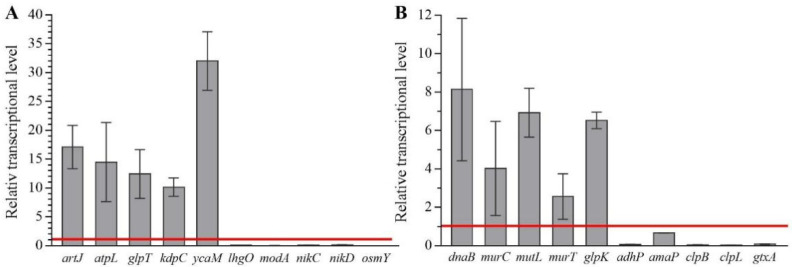
In E. coli ATCC25922, the RNA sequencing results showed that the raw sequence outputs were 32823244, 35152464, 35876796, and 33148174 reads with 88.70%, 90.15%, 88.54%, and 88.21% total matches for the two CFS-free experiments and two CFS experiments, respectively (Table S2). Compared with those in CFS-free experiments, 1/2 MIC of CFS differentially upregulated the expression of 327 genes and downregulated the expression of 70 genes (Table S3). For S. aureus ATCC6538, the raw sequence outputs were 25524088, 21597232, 26838638, and 24129642 reads with 91.41%, 86.25%, 90.26%, and 80.69% total matches for the two CFS-free experiments and two CFS experiments, respectively (Table S2). When treated with 1/2 MIC of CFS, 814 genes were significantly up-regulated, and 233 genes were differentially down-regulated (Table S4). Moreover, all five upregulated and five downregulated genes selected for evaluation in E. coli ATCC25922 and S. aureus ATCC6538 were differentially expressed in response to CFS (Figure 5), confirming the reliability of the transcriptome data.
Figure 5.
Fold change of selected genes by qRT-PCR after bacterial cells were treated with CFS from L. rhamnosus SCB0119 in E. coli ATCC25922 (A) and S. aureus ATCC6538 (B). The bacterial 16S rRNA was used as an internal standard. The red line at value 1 represented the expression of chosen genes in the control groups. Error bars: SD of the mean from three repeated assays.
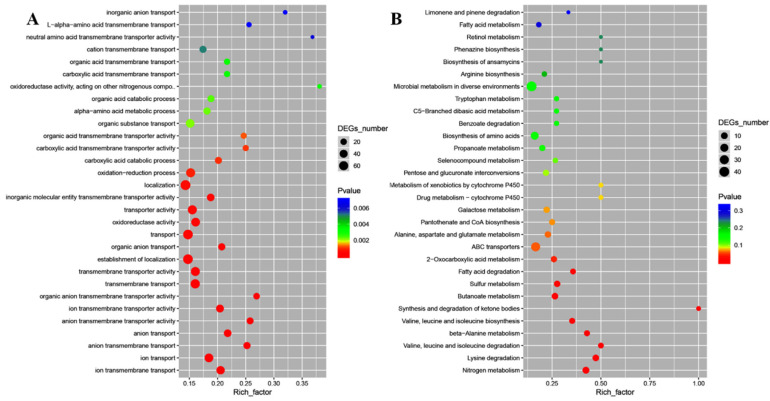
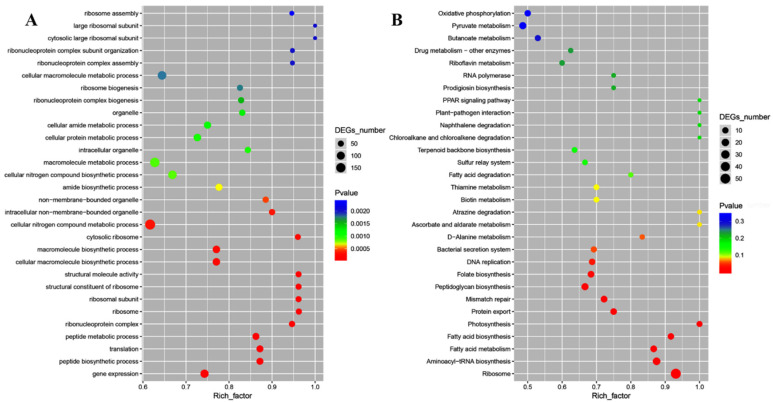
Moreover, gene ontology (GO) enrichment analysis classified the functions of the differentially expressed genes (DEGs) into biological process, cellular component, and molecular function, which included 57, 1, and 23 for E. coli ATCC25922, and 44, 15, and 4 for S. aureus ATCC6538, respectively. In E. coli ATCC25922, ion transmembrane transport and ion transport, outer membrane-bounded periplasmic space, and anion transmembrane transporter activity and ion transmembrane transporter activity were dominant for biological process, cellular component, and molecular function, respectively (Figure 6A). Kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis demonstrated that DEGs were mapped into nitrogen metabolism, amino acid metabolism, synthesis and degradation of ketone bodies, sulfur metabolism, ABC transporters, carbohydrate metabolism, metabolism of cofactors and vitamins, benzoate degradation, microbial metabolism in diverse environments, drug metabolism-cytochrome P450, and fatty acid metabolism (Figure 6B), which were crucial for lipid metabolism, amino acid metabolism, cellular structure and processes, and membrane transport. For S. aureus ATCC6538, GO enrichment analysis showed that gene expression and peptide biosynthetic process, ribonucleoprotein complex and ribosome, and structural molecule activity and structural constituent of ribosome were dominant for biological process, cellular component, and molecular function, respectively (Figure 7A). KEGG enrichment analysis demonstrated that DEGs were mapped into the ribosome, aminoacyl-tRNA biosynthesis, peptidoglycan biosynthesis, protein export, mismatch repair, DNA replication, fatty acid metabolism, bacterial secretion system, metabolism of cofactors and vitamins, the PPAR signaling pathway, RNA polymerase, carbohydrate metabolism, and oxidative phosphorylation (Figure 7B), which are crucial for lipid metabolism, energy metabolism, translation, and replication and repair. These data implied that the targeted genes of CFS in E. coli might differ from those in S. aureus.
Figure 6.
The GO enrichment scatter plot (A) and KEGG enrichment scatter plot (B) of differentially expressed genes treated with CFS from L. rhamnosus SCB0119 in E. coli ATCC25922.
Figure 7.
The GO enrichment scatter plot (A) and KEGG enrichment scatter plot (B) of differentially expressed genes treated with CFS from L. rhamnosus SCB0119 in S. aureus ATCC6538.
To illuminate the antimicrobial mechanisms of CFS against E. coli and S. aureus, we further analyzed the DEGs involved in the biosynthesis of amino acids, ABC transporters, and fatty acid degradation in E. coli ATCC25922, and ribosome, aminoacyl-tRNA biosynthesis, DNA replication, oxidative phosphorylation, and fatty acid degradation in S. aureus ATCC6538. In E. coli ATCC25922, CFS increased the expressions of ilvN, ilvI, leuA, leuB, and lysC involved in the biosynthesis of amino acids by 2.08–8.75-fold and decreased the expressions of ABC transporter nikE, nikD, nikC, nikB, nikA, and fepD by 52.89–81.45% and the expression of adhP involved in fatty acid degradation by 27.94% (Table 1). Moreover, the expression of osmY encoding one periplasmic protein was down-regulated by 18.14% (Table S3). In S. aureus ATCC6538, CFS upregulated the expression of genes encoding 30S small subunits, 50S large subunits, and aminoacyl-tRNA by 3.66–104.47 folds and the expressions of atpD, atpA, atpE, and atpB related to oxidative phosphorylation by 5.48–16.79 folds, but decreased the expressions of adhP and EKM74_RS04795 related to DNA replication by 38.05% and 53.22%, respectively (Table 2). Moreover, the expressions of frr and rbfA related to ribosome cofactor were up-regulated by 3.19- and 12.50-fold (Table S4), respectively.
Table 1.
Gene annotation and KEGG pathway of the differentially expressed genes in E. coli ATCC25922 in response to CFS from L. rhamnosus SCB0119.
| Gene ID | RPKM Control | RPKM CFS | log2 Ratio | Gene Annotation |
|---|---|---|---|---|
| Fatty acid degradation | ||||
| C7A06_RS17240 | 9.40 ± 0.16 | 2.62 ± 0.05 | −1.83961 | adhP; alcohol dehydrogenase AdhP |
| Biosynthesis of amino acids | ||||
| C7A06_RS27640 | 4.88 ± 1.53 | 21.23 ± 2.32 | 2.119947 | ilvN; acetolactate synthase small subunit |
| C7A06_RS27645 | 5.18 ± 0.68 | 24.79 ± 0.65 | 2.259582 | ilvI; acetolactate synthase 3 large subunit |
| C7A06_RS27660 | 17.62 ± 0.97 | 58.19 ± 0.51 | 1.723499 | leuA; 2-isopropylmalate synthase |
| C7A06_RS27665 | 15.63 ± 2.17 | 48.10 ± 7.45 | 1.621654 | leuB; 3-isopropylmalate dehydrogenase |
| C7A06_RS30700 | 15.96 ± 0.02 | 155.56 ± 1.24 | 3.284926 | lysC; lysine-sensitive aspartokinase 3 |
| ABC transporters | ||||
| C7A06_RS01665 | 14.27 ± 0.12 | 6.44 ± 0.43 | −1.14856 | nikE; nickel import ATP-binding protein NikE |
| C7A06_RS01670 | 39.57 ± 0.39 | 15.70 ± 1.38 | −1.33379 | nikD; nickel import ATP-binding protein NikD |
| C7A06_RS01675 | 43.38 ± 0.05 | 11.55 ± 1.74 | −1.90906 | nikC; nickel ABC transporter permease subunit NikC |
| C7A06_RS01680 | 39.74 ± 2.21 | 10.25 ± 2.12 | −1.95437 | nikB; nickel ABC transporter permease subunit NikB |
| C7A06_RS01685 | 10.05 ± 1.12 | 1.86 ± 0.26 | −2.43055 | nikA; nickel ABC transporter substrate-binding protein |
| C7A06_RS01780 | 0.15 ± 0.08 | 1.01 ± 0.42 | 2.788886 | livH; ABC transporter permease LivH |
| C7A06_RS01790 | 4.29 ± 0.27 | 14.90 ± 1.90 | 1.794947 | livG;ABC transporter ATP-binding protein LivG |
| C7A06_RS24375 | 3.74 ± 0.98 | 1.53 ± 0.65 | −1.29111 | fepD; Fe3+ -siderophore ABC transporter permease |
Table 2.
Gene annotation and KEGG pathway of the differentially expressed genes in S. aureus ATCC6538 in response to CFS from L. rhamnosus SCB0119.
| Gene ID | RPKM Control | RPKM CFS | log2 Ratio | Gene Annotation |
|---|---|---|---|---|
| Fatty acid degradation | ||||
| EKM74_RS11915 | 1832.07 ± 490.15 | 697.14 ± 121.09 | −1.39396 | adhP; alcohol dehydrogenase |
| DNA replication | ||||
| EKM74_RS04795 | 85.2 ± 44.94 | 39.85 ± 1.91 | −1.09611 | single-stranded DNA-binding protein |
| Ribosome | ||||
| EKM74_RS01000 | 77.06 ± 19.54 | 474.46 ± 30.77 | 2.622215 | accC; acetyl-CoA carboxylase biotin carboxylase subunit |
| EKM74_RS01125 | 36.7 ± 12.69 | 642.43 ± 184.29 | 4.129847 | rpmG; 50S ribosomal protein L33 |
| EKM74_RS01310 | 21.83 ± 0.96 | 168.95 ± 53.18 | 2.951911 | rpsT; 30S ribosomal protein S20 |
| EKM74_RS01660 | 458.35 ± 97.45 | 2756.92 ± 263.1 | 2.588527 | rpmA; 50S ribosomal protein L27 |
| EKM74_RS01670 | 102.05 ± 29.64 | 4057 ± 488.97 | 5.313122 | rplU; 50S ribosomal protein L21 |
| EKM74_RS01855 | 19.42 ± 10.05 | 621.08 ± 94.23 | 4.999194 | rplT; 50S ribosomal protein L20 |
| EKM74_RS01860 | 51.94 ± 21.24 | 5478.49 ± 45.03 | 6.720704 | rpmI; 50S ribosomal protein L35 |
| EKM74_RS02080 | 80.66 ± 17.27 | 3451.94 ± 642.32 | 5.419314 | rpsD; 30S ribosomal protein S4 |
| EKM74_RS05465 | 190.13 ± 11.18 | 3317.01 ± 500.37 | 4.124828 | rpsI; 30S ribosomal protein S9 |
| EKM74_RS05470 | 150.17 ± 31.96 | 5880.76 ± 17.12 | 5.291357 | rplM; 50S ribosomal protein L13 |
| EKM74_RS05495 | 122.74 ± 4.32 | 3341.82 ± 309.69 | 4.766903 | rplQ; 50S ribosomal protein L17 |
| EKM74_RS05505 | 287.56 ± 78.6 | 2321.16 ± 704.2 | 3.01292 | rpsK; 30S ribosomal protein S11 |
| EKM74_RS05510 | 582.25 ± 5.82 | 10,572.69 ± 1079.55 | 4.182553 | rpsM; 30S ribosomal protein S13 |
| EKM74_RS05515 | 259.55 ± 26.92 | 8724.69 ± 798.78 | 5.071017 | rpmJ; 50S ribosomal protein L36 |
| EKM74_RS05535 | 328.44 ± 51.85 | 2770.35 ± 239.31 | 3.076378 | rplO; 50S ribosomal protein L15 |
| EKM74_RS05540 | 370.08 ± 105.31 | 13,564.79 ± 1089.74 | 5.195877 | rpmD; 50S ribosomal protein L30 |
| EKM74_RS05545 | 183.9 ± 22.05 | 4254.61 ± 754.45 | 4.532035 | rpsE; 30S ribosomal protein S5 |
| EKM74_RS05550 | 205.01 ± 26.43 | 9035.82 ± 328.78 | 5.461917 | rplR; 50S ribosomal protein L18 |
| EKM74_RS05555 | 81.16 ± 4.22 | 1809.37 ± 428.25 | 4.478643 | rplF; 50S ribosomal protein L6 |
| EKM74_RS05560 | 98.44 ± 3.27 | 3014.88 ± 247.03 | 4.93666 | rpsH; 30S ribosomal protein S8 |
| EKM74_RS05570 | 126.29 ± 2.95 | 4279.22 ± 391.93 | 5.082565 | rplE; 50S ribosomal protein L5 |
| EKM74_RS05575 | 138.6 ± 11.64 | 4600.35 ± 70.63 | 5.052719 | rplX; 50S ribosomal protein L24 |
| EKM74_RS05580 | 181.27 ± 11.93 | 4136.6 ± 837.23 | 4.512211 | rplN; 50S ribosomal protein L14 |
| EKM74_RS05585 | 140.46 ± 16.11 | 4981.9 ± 113.47 | 5.148408 | rpsQ; 30S ribosomal protein S17 |
| EKM74_RS05590 | 154.79 ± 34.38 | 4723.29 ± 418.72 | 4.931395 | rpmC; 50S ribosomal protein L29 |
| EKM74_RS05595 | 194.99 ± 8.39 | 3442.61 ± 618.09 | 4.142009 | rplP; 50S ribosomal protein L16 |
| EKM74_RS05600 | 333.68 ± 100.38 | 6017.46 ± 493.68 | 4.172617 | rpsC; 30S ribosomal protein S3 |
| EKM74_RS05605 | 267.84 ± 59.12 | 9325.05 ± 0.95 | 5.121682 | rplV; 50S ribosomal protein L22 |
| EKM74_RS05610 | 210.47 ± 4.59 | 7349.55 ± 191.55 | 5.125963 | rpsS; 30S ribosomal protein S19 |
| EKM74_RS05615 | 231.04 ± 32.27 | 4313.16 ± 576.59 | 4.222508 | rplB; 50S ribosomal protein L2 |
| EKM74_RS05620 | 87.68 ± 7.24 | 1918.56 ± 543.34 | 4.451662 | rplW; 50S ribosomal protein L23 |
| EKM74_RS05625 | 145.16 ± 12.67 | 3427.73 ± 563.02 | 4.561522 | rplD; 50S ribosomal protein L4 |
| EKM74_RS05630 | 188.98 ± 15.87 | 7819.97 ± 221.75 | 5.37082 | rplC; 50S ribosomal protein L3 |
| EKM74_RS05635 | 294.61 ± 86.56 | 9259.91 ± 978.26 | 4.97412 | rpsJ; 30S ribosomal protein S10 |
| EKM74_RS07510 | 151.39 ± 9.3 | 705.28 ± 120.89 | 2.219912 | rplI; 50S ribosomal protein L9 |
| EKM74_RS07590 | 114.58 ± 17.68 | 1144.54 ± 138.16 | 3.320336 | rpmH; 50S ribosomal protein L34 |
| EKM74_RS10450 | 236.72 ± 37.3 | 7542.84 ± 237.11 | 4.993827 | rpsF; 30S ribosomal protein S6 |
| EKM74_RS10460 | 55.84 ± 4.69 | 531.48 ± 99.45 | 3.250636 | rpsR; 30S ribosomal protein S18 |
| EKM74_RS11530 | 51.86 ± 16.74 | 493.86 ± 62.28 | 3.251456 | rpmG; 50S ribosomal protein L33 |
| EKM74_RS11545 | 230.05 ± 33.59 | 6745.81 ± 496 | 4.873994 | rplK; 50S ribosomal protein L11 |
| EKM74_RS11550 | 89.54 ± 9.77 | 1345.42 ± 304.26 | 3.909295 | rplA; 50S ribosomal protein L1 |
| EKM74_RS11560 | 75.58 ± 8 | 6432.15 ± 34.02 | 6.411134 | rplJ; 50S ribosomal protein L10 |
| EKM74_RS11565 | 38.15 ± 13.06 | 2351.81 ± 364.44 | 5.945945 | rplL; 50S ribosomal protein L7/L12 |
| EKM74_RS11590 | 185.09 ± 16.78 | 6759.64 ± 1120.81 | 5.190643 | rpsL; 30S ribosomal protein S12 |
| EKM74_RS11595 | 156.22 ± 21.95 | 3889.22 ± 770.69 | 4.637864 | rpsG; 30S ribosomal protein S7 |
| EKM74_RS14380 | 74.47 ± 6.98 | 956.98 ± 89.75 | 3.683755 | rpmF; 50S ribosomal protein L32 |
| EKM74_RS15340 | 44.89 ± 6.07 | 387.7 ± 59.81 | 3.110434 | rpsP; 30S ribosomal protein S16 |
| EKM74_RS15355 | 56.77 ± 3.55 | 839.03 ± 212.16 | 3.885462 | rplS; 50S ribosomal protein L19 |
| EKM74_RS15435 | 112.72 ± 30.02 | 3718.42 ± 314.52 | 5.043867 | rpsB; 30S ribosomal protein S2 |
| EKM74_RS15530 | 178.88 ± 2.05 | 2219.27 ± 90.62 | 3.633007 | rpsO; 30S ribosomal protein S15 |
| Aminoacyl-tRNA biosynthesis | ||||
| EKM74_RS02305 | 35.37 ± 21.65 | 401.03 ± 43.46 | 3.503254 | leuS; leucine-tRNA ligase |
| EKM74_RS03605 | 63.4 ± 16.82 | 997.64 ± 109.83 | 3.975903 | gatB; Asp-tRNA (Asn)/Glu-tRNA (Gln) amidotransferase subunit |
| EKM74_RS11370 | 90.84 ± 8.58 | 509.92 ± 53.53 | 2.488778 | lysS; lysine-tRNA ligase |
| EKM74_RS11490 | 107.77 ± 37.9 | 605.02 ± 32.1 | 2.489052 | gltX; glutamate-tRNA ligase |
| EKM74_RS14775 | 34.9 ± 14.25 | 239.53 ± 20.28 | 2.77887 | pheT; phenylalanine-tRNA ligase subunit beta |
| EKM74_RS15105 | 30.17 ± 17.78 | 273.57 ± 29.78 | 3.180715 | ileS; isoleucine-tRNA ligase |
| Oxidative phosphorylation | ||||
| EKM74_RS04830 | 170.47 ± 3.15 | 3033.08 ± 240.81 | 4.153207183 | atpD; F0-F1 ATP synthase subunit beta |
| EKM74_RS04840 | 97.18 ± 1.86 | 1251.21 ± 170.89 | 3.686521282 | atpA; F0-F1 ATP synthase subunit alpha |
| EKM74_RS04855 | 241.14 ± 56.65 | 1679.25 ± 147.57 | 2.799862175 | atpE; F0-F1 ATP synthase subunit C |
| EKM74_RS04860 | 239.15 ± 52.69 | 1550.63 ± 98.56 | 2.696892312 | atpB; F0-F1 ATP synthase subunit A |
3. Discussion
Lactic acid bacteria were reported as a promising alternative to antibiotics against several pathogens [18]. Here, L. rhamnosus SCB0119 could inhibit the growth of E. coli ATCC25922 and S. aureus ATCC6538. Moreover, its CFS, the main component for antibacterial activity, inhibited bacterial growth or even caused death, induced oxidative stress, and impaired cell shape and structure, as discussed below.
CFS from L. rhamnosus SCB0119 reduced bacterial growth of E. coli ATCC25922 and S. aureus ATCC6538. According to the diameter of the inhibition zone, CFS from L. rhamnosus SCB0119 exhibited greater antibacterial activity against E. coli ATCC25922 and S. aureus ATCC6538 than that from L. rhamnosus XN2 (7.2 ± 0.5 mm for E. coli CICC10302 and 15.1 ± 1.2 mm for S. aureus CICC10384) and LMG 23,522 (8.9 ± 0.2 mm for E. coli O157:H7 and 11.3 ± 0.4 mm for S. aureus) [21,23]. Additionally, the decreases in intracellular ATP levels suggested that CFS from L. rhamnosus SCB0119 might cause the death of E. coli and S. aureus cells [22]. The cell death of pathogens caused by L. rhamnosus was also reported in S. typhimurium treated with L. rhamnosus SQ511 [20].
Moreover, ROS production plays an important role in commencing cessation of the cell during the initiation of cell death [24]. Here, CFS from L. rhamnosus SCB0119 accumulated the ROS levels in E. coli ATCC25922 and S. aureus ATCC6538, which was similar to the increases of ROS production in S. typhimurium caused by CFS from L. rhamnosus SQ511 [20]. The high level of ROS production causes oxidative stress, which is closely associated with adverse effects on cellular components and causes cell death [25,26].
Furthermore, CFS from L. rhamnosus SCB0119 destroyed the cell wall, outer membrane, and plasma membrane. First, the cell wall is the first line to protect cells, including the cell membrane and organelles [27,28]. When the cell wall is damaged, AKP existing between the cell wall and the membrane of bacteria leaks into the extracellular milieu, causing an increase in extracellular AKP activity [29]. In this study, CFS increased AKP contents in E. coli ATCC25922 and S. aureus ATCC6538, suggesting the cell wall of E. coli and S. aureus was impaired by CFS from L. rhamnosus SCB0119. The impairment of the cell wall might affect the cell membrane. Generally, effective antibacterial compounds must penetrate or destroy the plasma membrane of the pathogen [30]. The increases in fluorescence intensity of PI staining, ΔΨ, and ΔpH indicated that CFS from L. rhamnosus SCB0119 destroyed the integrity of the cell membrane and improved the cell membrane permeability in E. coli ATCC25922 and S. aureus ATCC6538, which resulted in the dissipation of membrane ΔΨ and ΔpH and depolarization of the cytoplasmic membrane [31,32]. Changes in PI staining, ΔΨ, and ΔpH were consistent with that in E. coli cells treated with a bacteriocin produced by L. rhamnosus 1.0320 [32]. Additionally, changes in PI staining coincided with that in S. aureus cells conditioned with CFS from L. rhamnosus XN2 [21], and the depolarization of cell membrane was in agreement with that in S. enteritidis caused by CFS from L. rhamnosus SQ511 [20]. Besides, disruption of membrane integrity leads to intracellular ATP leakage, which is also responsible for the decreased intracellular ATP concentration [33]. The elimination of the transmembrane electrochemical gradient might also lead to the loss of the ability of bacterial cells to synthesize ATP [34]. Moreover, the outer membrane is important for barrier function and nutrient flow [35]. The increases in the amount of NPN staining demonstrated that CFS from L. rhamnosus SCB0119 increased the permeability of the outer membrane in E. coli ATCC25922 and S. aureus ATCC6538, which concurred with the findings found in E. coli cells treated with a bacteriocin produced by L. rhamnosus 1.0320 [32]. These data indicated that CFS from L. rhamnosus SCB0119 destroyed the integrity of the cell wall and membrane, increased the permeability of the cell membrane, released the cell contents, and induced cell death in E. coli and S. aureus.
Additionally, CFS from L. rhamnosus SCB0119 affected the transcriptional expression of genes involved in fatty acid degradation, biosynthesis of amino acids, ABC transporters, ribosome, aminoacyl-tRNA biosynthesis, DNA replication, and/or oxidative phosphorylation in E. coli ATCC25922 and S. aureus ATCC6538. Firstly, adhP encoding an alcohol dehydrogenase contributes to the reduction of fatty aldehydes to fatty alcohols [36], and its deletion resulted in the accumulation of fatty aldehydes, which caused damage to the cell or the inhibition of enzymes activities by metabolic disorders or potential toxicity [37]. Here, CFS inhibited the expression of adhP in E. coli ATCC25922 and S. aureus ATCC6538, which might be an explanation for the inhibition of bacterial growth caused by CFS from L. rhamnosus SCB0119. Secondly, nickel is an essential catalytic cofactor of enzymes allowing organisms to inhabit diverse environmental niches, and its pumping through the cytoplasmic membrane is regulated by NikABCDE [38]. Iron intake positively correlated with the synthesis of some ferritin proteins, such as catalases, whose reduction will cause the intracellular accumulation of H2O2, induce the oxidative stress response inside the thallus, and bring adverse effects on cellular components such as DNA, proteins, and membrane lipids [26,39,40]. The deletion of E. coli fepD caused a reduction in iron intake [41]. Therefore, the downregulation of nikA, nikB, nikC, nikD, nikE, and fepD might result in the damage of nickel and iron homeostasis and then induce the inhibition of bacterial growth in E. coli caused by CFS from L. rhamnosus SCB0119. Thirdly, the accumulation of amino acids could be attributed to related to protein denaturation and blockage of protein synthesis [42]. The overexpression of lysC, ilvI, ilvN, leuA, and leuB might increase the production of lysine, leucine, isoleucine, and valine [43], suggesting the presence of CFS from L. rhamnosus SCB0119 might promote the biosynthesis of lysine, leucine, isoleucine, and valine and then cause the damage to cell survival in E. coli. In addition, the decrease in the expression of osmY might be another explanation for the antibacterial activities of CFS from L. rhamnosus SCB0119 against E. coli because OsmY is a molecular chaperone through a genetic selection that forces cells to optimize unstable protein folding in vivo [44] and positively regulates the integrity of the outer membrane [45]. In S. aureus ATCC6538, the inhibition of bacterial growth caused by CFS from L. rhamnosus SCB0119 might be due to the changes in protein synthesis by mediating the expression of the ribosome cofactor genes (frr and rbfA), genes encoding 30S ribosomal proteins (rpsT, rpsD, rpsI, rpsK, rpsM, rpsE, rpsH, rpsJ, rpsQ, rpsC, and rpsS), genes encoding 50S ribosomal proteins (rpmG, rpmA, rplU, rpmI, rpmJ, rplO, rplR, rplF, rplE, rplN, and rplP), and genes in the pathway of the aminoacyl-tRNA biosynthesis (leuS, gatB, lysS, gltX, pheT, and ileS), because they were involved in the initiation, elongation, and termination of the assembly of the ribosome [46] and important for protein synthesis [47,48]. Moreover, gene EKM74-RS04795 encodes a single-stranded DNA-binding protein, which is crucial in DNA replication, repair, and recombination [49], was down-regulated by CFS in S. aureus ATCC6538, suggesting CFS from L. rhamnosus SCB0119 might cause cell damage or death in S. aureus by interfering with DNA repair pathways and leading to genomic instability. Besides, the expression of four genes (atpA, atpB, atpD, and atpE) encoding ATP synthases hydrolyzing ATP [50] were up-regulated by CFS from L. rhamnosus SCB0119 in S. aureus ATCC6538, which might result in the decreases of ATP content and alive cells.
Lastly, previous studies demonstrated that the consumption of L. rhamnosus strains gave positive results in preventing diarrhea and bacterial vaginosis, restoring normal gastrointestinal microflora, and so on [17]. In addition, when directly mixed with beef, CFS from Lactobacillus with stable antimicrobial activity could effectively reduce the microbial load of inoculated pathogens and enhance the quality and safety of beef products [51], and the addition of L. rhamnosus into milk could inhibit the growth of P. fluorescens and P. putida from raw milk and improve the raw milk quality [52]. Therefore, effectiveness in reducing the growth of E. coli ATCC25922 and S. aureus ATCC6538 suggested that L. rhamnosus SCB0119 and its CFS might be directly consumed by humans and animals or mixed with various foods to prevent the infection or contamination of E. coli and S. aureus, although the findings need further validation of a clinical, practical, and/or industrial scale. In the future, the safety of L. rhamnosus SCB0119 and its CFS, active components from CFS, and the evaluation of their application potential in preventing the infection and contamination of E. coli and S. aureus should be further investigated.
4. Material and Methods
4.1. Bacterial Strains and Culture Condition
Lactobacillus rhamnosus SCB0119 (CGMCC NO: 17618) was isolated from fermented pickles by ourselves and cultivated in De Man Rogosa Sharpe (MRS) medium at 37 °C under anaerobic conditions. Escherichia coli ATCC25922 and Staphylococcus aureus ATCC6538 were obtained from the American Type Culture Collection (ATCC) and cultivated at 37 °C in LB medium under constant shaking at 180 rpm.
4.2. Preparation of Cell-Free Culture Supernatants and Cells of L. rhamnosus SCB0119
Briefly, L. rhamnosus SCB0119 was inoculated into MRS medium at a ratio of 3:100 (v/v) and statically cultured at 37 °C for 60 h. The supernatants were collected, filtered with a 0.22 µm pore filter (Merck-Millipore, Darmstadt, Germany), and served as CFS. Bacterial cells were washed with phosphate buffer saline (PBS) and then resuspended in an equal volume of PBS.
4.3. Determination of the Antibacterial Activity of L. rhamnosus SCB0119
The antimicrobial activity of L. rhamnosus SCB0119 was evaluated using the agar well diffusion method [53] with some modifications. Briefly, the dilutions of E. coli ATCC25922 and S. aureus ATCC6538 cells (107 CFU/mL) were added into LB medium with 0.75% agar, respectively. Then, the mixture was poured onto LB agar plates (1.5% agar) with several Oxford cups (6 × 7.8 mm), which were removed when the agar solidified. Aliquots of 100 μL of L. rhamnosus SCB0119 culture, CFS, resuspended bacterial cells, and MRS medium were added to the wells and incubated at 37 °C. After 12 h of incubation, the mean diameters of the inhibition zones were recorded as antibacterial activity of L. rhamnosus SCB0119 against E. coli ATCC25922 and S. aureus ATCC6538. The experiments were conducted in triplicate.
4.4. Minimum Inhibitory Concentrations of CFS against E. coli and S. aureus
MICs were determined via the microdilution method in 96-well polystyrene plates (Corning Inc., Corning, NY, USA) [54]. Briefly, CFS was evaporated, dissolved using ultrapure water to 500 mg/mL and stored at −20 °C. Wells containing 100 μL of E. coli ATCC25922/S. aureus ATCC6538 dilutions (107 CFU/mL) in LB medium were supplemented with different concentrations (0.49–250 mg/mL) of CFS and incubated at 37 °C for 12 h. Optical densities at 600 nm (OD600) were read using a Multiskan GO Reader (Thermo Scientific, Waltham, MA, USA). The experiments were conducted in triplicate.
4.5. Growth Curve of E. coli and S. aureus after Treatment with CFS
The bacterial growth curve was assayed using the method reported by Yan et al. [55]. Briefly, E. coli ATCC25922/S. aureus ATCC6538 dilutions (107 CFU/mL) in LB medium were supplemented with 0, 1/2 MIC, and 1 MIC of CFS and incubated in Bioscreen C (Oy Growthcurves Ab Ltd., Turku, Finland), where the temperature was set at 37 °C and the rotating speed was set at 180 rpm. During a 34-h incubation, OD600s were read at 2-h intervals. The experiments were conducted in triplicate.
4.6. Scanning Electron Microscopy and Transmission Electron Microscopy
The potential effects of CFS on cell morphology and structure of E. coli ATCC25922 and S. aureus ATCC6538 were determined by SEM and TEM [56,57]. Briefly, bacterial dilutions (107 CFU/mL) in LB medium were supplemented with 1/2 MIC of CFS and incubated for 10 h at 37 °C by shaking. After centrifugation at 4 °C for 5 min, bacterial cells were collected, washed with PBS (0.1 mM, pH 7.2) three times, fixed overnight with 4% glutaraldehyde, washed with PBS once, and post-fixed in 1% OsO4 for 30 min. Some post-fixed cells were dehydrated in a graded ethanol series (50%, 70%, 80%, 90%, and 100%), dried by CO2 for 4 h, coated with gold, and observed under a field emission scanning electron microscope (FEI/Talos L120C, Thermo Fisher Scientific, New York, NY, USA). Other post-fixed cells were dyed with uranium acetate overnight, dehydrated in a graded ethanol series (30%, 50%, 70%, 85%, 95%, and 100%), embedded in Spurr resin, polymerized at 55 °C for 48 h, and observed under a transmission electron microscope (ZEISS, Oberkochen, Germany).
4.7. Concentrations of Alkaline Phosphatase and Intracellular ATP
Bacterial dilutions of E. coli ATCC25922/S. aureus ATCC6538 (108 CFU/mL) in PBS for the activity of AKP or 5 mM HEPES buffer for the concentration of intracellular ATP were supplemented with 0, 1/2 MIC, 1 MIC, and 2 MIC of CFS and incubated at 37 °C by shaking. After 1 h, the supernatants or bacteria were collected and used to measure the AKP activity using an AKP kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) and ATP content using an ATP detection kit (Beyotime Biotechnology, Shanghai, China) [58] according to the method described by the manufacturer. The concentration of ATP was determined using external calibration curves of the standard ATP solution at different concentrations. Moreover, the protein concentration in the sample was measured using the total protein quantitative determination kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China), and the concentration of ATP was converted into nmol/mg protein. All of the experiments were conducted in triplicate.
4.8. Assay for Outer Membrane Permeability and Inner Membrane Integrity
For the outer membrane permeability, E. coli ATCC25922/S. aureus ATCC6538 dilutions (108 CFU/mL) in 5 mM HEPES buffer containing 5 mM glucose (pH 7.0) were incubated with 10 µM NPN for 30 min at 25 °C and then supplemented with 0, 1/2 MIC, 1 MIC, and 2 MIC of CFS and incubated at 37 °C by shaking [59]. After 30 min, the fluorescence intensities were recorded using a Varioskan Flash (spectraMax i3x, Molecular Devices, San Francisco, CA, USA) at the excitation/emission wavelengths of 350/420 nm. The experiments were conducted in triplicate.
For the inner membrane integrity, E. coli ATCC25922/S. aureus ATCC6538 dilutions (108 CFU/mL) in PBS were incubated with 10 µM PI for 30 min at 25 °C and then supplemented with 0, 1/2 MIC, 1 MIC, and 2 MIC of CFS and incubated at 37 °C by shaking [60]. After 1 h, the fluorescence intensities were recorded using the Varioskan Flash at the excitation/emission wavelengths of 535/615 nm. The experiments were conducted in triplicate.
4.9. Assays for Membrane Potential, pH Gradient, and Reactive Oxygen Species
ΔΨ, ΔpH, and ROS were assayed with the DiBAC4(3) (Solarbio, Beijing, China) fluorescence probe [61], the BCECF-AM (Yuanye, Shanghai, China) fluorescence probe [62], and a ROS assay kit (Beyotime Biotechnology, Shanghai, China), respectively. Briefly, E. coli ATCC25922/S. aureus ATCC6538 dilutions (108 CFU/mL) in 5 mM HEPES buffer were incubated with 5 µM DiBAC4(3) dilution, 5 µM BCECF-AM dilution, or 10 µM DCFH-DA dilution incubator for 30 min at 37 °C and then supplemented with 0, 1/2 MIC, 1 MIC, and 2 MIC of CFS and incubated at 37 °C by shaking. After 1 h, the fluorescence intensities were recorded using the Varioskan Flash at the excitation/emission wavelengths of 470/500 nm, 480/535 nm, and 488/525 nm, respectively. All the experiments were conducted in triplicate.
4.10. Transcriptome Sequencing and Analysis
Total RNA was extracted from E. coli ATCC25922 and S. aureus ATCC6538 cells grown in LB medium with 1/2 MIC of CFS for 10 h at 37 °C using TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and genomic DNA was removed using DNase I (TaKaRa, Kyoto, Japan). Then RNA quality was determined using 2100 Bioanalyser (Agilent, Palo Alto, CA, USA) and quantified using the ND-2000 (NanoDrop Technologies, Thermo Fisher Scientific, New York, NY, USA). RNA-seq strand-specific libraries were created using 5 g of total RNA and the TruSeq RNA sample preparation Kit (Illumina, San Diego, CA, USA). rRNA was removed by RiboZero rRNA removal kit (Epicenter, Stockholm, Sweden) and fragmented using fragmentation buffer. The Illumina-indexed adaptors were ligated after cDNA synthesis, end repair, A-base addition, and ligation according to Illumina’s protocol. cDNA target fragments of 200–300 bp were selected on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. Paired-end libraries were sequenced by Illumina NovaSeq 6000 sequencing (150 bp × 2, Shanghai BIOZERON Co., Ltd, Shanghai, China) after being quantified by TBS380. The sequencing data were deposited in NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra) under the accession numbers SRP395161 for E. coli ATCC25922 and SRP394988 for S. aureus ATCC6538.
Clean data were mapped to the data from the E. coli genome (NCBI accession: ASM301845v1) and the S. aureus genome (NCBI accession: ASM394542v1), respectively. The expression level of each gene was evaluated by calculating the fragments per kilobase per million fragments mapped (FPKM), and edgeR (https://bioconductor.org/packages/release/bioc/html/edgeR.html) was used for differential expression analysis. To compare the differential gene expression among the libraries, the false discovery rate (FDR) ≤0.05 and the absolute value of the log2 ratio ≥1 were used as the threshold to screen the DEGs. Functional classification and pathway analysis of DEGs were performed using GO functional enrichment (Goatools, https://github.com/tanghaibao/Goatools) and KEGG pathway analysis (KOBAS, http://kobas.cbi.pku.edu.cn/kobas3), respectively. The experiments were conducted in twice.
4.11. qRT-PCR
Total RNA was extracted from E. coli ATCC25922 and S. aureus ATCC6538 cells grown in LB medium with 1/2 MIC of CFS for 10 h at 37 °C using TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA), detected by agarose gel electrophoresis, quantified using the ND-2000 (NanoDrop Technologies, Thermo Fisher Scientific, New York, NY, USA), and transcribed reversely into cDNA with PrimeScript RT reagent kit (TaKaRa, Kyoto, Japan). Tenfold dilutions of each cDNA were used as templates to analyze the expression of tested genes via qRT-PCR with paired primers (Table S1) using SYBR® Premix Ex Taq™ (TaKaRa, Kyoto, Japan). The reaction cycling consisted of initial denaturation at 95 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing and extension at 60 °C for 34 s, and ended up with a dissolution curve. The analysis of the dissolution curve evaluated the specificity of the primers. The 16 s rRNA was used as an internal standard. The relative transcription level of each gene was defined as the ratio of its transcript in cells treated with CFS over that without CFS using the 2−ΔΔCt method, where Ct means the threshold cycle, ΔCt is equal to the difference in threshold cycles for target and reference (Ctx − Ctr), and ΔΔCt equals (ΔCtin CFS group − ΔCtin control group) [63]. The experiments were conducted in triplicate.
4.12. Data Analysis
Results from replicates were expressed as mean ± standard deviation (SD), and the data analysis was subjected to a one-factor analysis of variance, followed by Tukey’s HSD test. p < 0.05 was considered a significant difference in all experiments.
5. Conclusions
Lactobacillus rhamnosus SCB0119 could inhibit the growth and induce cell death of E. coli and S. aureus, and its antibacterial activity was mainly attributed to the CFS. Its CFS could induce oxidative stress involving ROS and cause damage to the integrity and permeability of the cell wall and membrane, which resulted in the release of the cell contents. Moreover, the transcriptomic analysis showed that CFS aimed at the expression of genes involved in fatty acid degradation, ion transport, and the biosynthesis of amino acids in E. coli and affected the expression of genes involved in fatty acid degradation, protein synthesis, DNA replication, and ATP hydrolysis in S. aureus. These findings demonstrated L. rhamnosus SCB0119 and its CFS have the potential for use as a biocontrol agent against pathogens, including E. coli and S. aureus.
Abbreviations
| CFS | cell-free culture supernatant |
| LB | Luria Bertani |
| MICs | minimum inhibitory concentrations |
| SEM | scanning electron microscopy |
| TEM | transmission electron microscopy |
| AKP | alkaline phosphatase |
| NPN | 1-N-phenylnaphthylamine |
| PI | propidium iodide |
| ΔΨ | membrane potential |
| ΔpH | pH gradient |
| ROS | reactive oxygen species |
| DEGs | differentially expressed genes |
| GO | gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| MRS | De Man Rogosa Sharpe |
| PBS | phosphate buffer saline |
| FPKM | fragments per kilobase per million fragments mapped |
| ATCC | American Type Culture Collection |
| SD | standard deviation |
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232315159/s1.
Author Contributions
Conceptualization, J.W., H.P. and G.Z.; Methodology, X.-M.Y.; Validation, G.-J.C.; Formal Analysis, G.Z.; Investigation, J.W., H.P. and H.-B.C.; Resources, X.F. and J.W.; Data Curation, H.P.; Writing-Original Draft Preparation, H.P.; Writing-Review & Editing, J.W., Q.-P.Z., L.W. and X.F.; Visualization, Z.-L.L.; Supervision, J.W.; Project Administration, J.W.; Funding Acquisition, J.W. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study does not involve any experiments with human participants or animals.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data supporting the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare that there was no conflict of interest.
Funding Statement
This work was financially supported by Guangzhou Science and Technology Research Projects (201904010274), the opening foundation from State Key Laboratory of Applied Microbiology Southern China, Guangdong Institute of Microbiology (SKLAM003-2018), and Guangdong Province Science and Technology Innovation Strategy Special Fund (2020B020226008 and 2022B0202040002).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kirk M.D., Pires S.M., Black R.E., Caipo M., Crump J.A., Devleesschauwer B., Dopfer D., Fazil A., Fischer-Walker C.L., Hald T., et al. World health organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: A data synthesis. PLos Med. 2015;12:e1001921. doi: 10.1371/journal.pmed.1001921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mangal M., Bansal S., Sharma S.K., Gupta R.K. Molecular detection of foodborne pathogens: A rapid and accurate answer to food safety. Crit. Rev. Food Sci. Nutr. 2016;56:1568–1584. doi: 10.1080/10408398.2013.782483. [DOI] [PubMed] [Google Scholar]
- 3.Scallan E., Hoekstra R.M., Angulo F.J., Tauxe R.V., Widdowson M.A., Roy S.L., Jones J.L., Griffin P.M. Foodborne illness acquired in the United States-major pathogens. Emerg. Infect. Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson J.R. Foodborne illness acquired in the United States. Emerg. Infect. Dis. 2011;17:1338–1339. doi: 10.3201/eid1707.110256. [DOI] [PubMed] [Google Scholar]
- 5.Croxen M.A., Law R.J., Scholz R., Keeney K.M., Wlodarska M., Finlay B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013;26:822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Croxen M.A., Finlay B.B. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 2010;8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 7.Muller A., Stephan R., Nuesch-Inderbinen M. Distribution of virulence factors in ESBL-producing Escherichia coli isolated from the environment, livestock, food and humans. Sci. Total Environ. 2016;541:667–672. doi: 10.1016/j.scitotenv.2015.09.135. [DOI] [PubMed] [Google Scholar]
- 8.Majowicz S.E., Scallan E., Jones-Bitton A., Sargeant J.M., Stapleton J., Angulo F.J., Yeung D.H., Kirk M.D. Global incidence of human Shiga toxin-producing Escherichia coli infections and deaths: A systematic review and knowledge synthesis. Foodborne Pathog. Dis. 2014;11:447–455. doi: 10.1089/fpd.2013.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Argudin M.A., Mendoza M.C., Rodicio M.R. Food poisoning and Staphylococcus aureus enterotoxins. Toxins. 2010;2:1342–1751. doi: 10.3390/toxins2071751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mourenza A., Gil J.A., Mateos L.M., Letek M. Novel treatments and preventative strategies against food-poisoning caused by staphylococcal species. Pathogens. 2021;10:91. doi: 10.3390/pathogens10020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadariya J., Smith T.C., Thapaliya D. Staphylococcus aureus and staphylococcal food-borne disease: An ongoing challenge in public health. Biomed Res. Int. 2014;2014:827965. doi: 10.1155/2014/827965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong S., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mostafa A.A., Al-Askar A.A., Almaary K.S., Dawoud T.M., Sholkamy E.N., Bakri M.M. Antimicrobial activity of some plant extracts against bacterial strains causing food poisoning diseases. Saudi J. Biol. Sci. 2018;25:361–366. doi: 10.1016/j.sjbs.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies J., Davies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010;74:417. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reis J.A., Paula A.T., Casarotti S.N., Penna A. Lactic acid bacteria antimicrobial compounds: Characteristics and applications. Food Eng. Rev. 2012;4:124–140. doi: 10.1007/s12393-012-9051-2. [DOI] [Google Scholar]
- 16.Vieco-Saiz N., Belguesmia Y., Raspoet R., Auclair E., Gancel F., Kempf I., Drider D. Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front. Microbiol. 2019;10:57. doi: 10.3389/fmicb.2019.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zamberlin S., Spehar I.D., Kelava N., Samarzija D. Probiotic bacterium Lactobacillus rhamnosus: Beneficial and a dverse effects on human health. Milchwissenschaft-Milk Sci. Int. 2012;67:30–33. [Google Scholar]
- 18.Ozogul F., Hamed I. The importance of lactic acid bacteria for the prevention of bacterial growth and their biogenic amines formation: A review. Crit. Rev. Food Sci. 2018;10:1660–1670. doi: 10.1080/10408398.2016.1277972. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y.C., Zhang L.W., Du M., Yi H.X., Guo C.F., Tuo Y.F., Han X., Li J.Y., Zhang L.L., Yang L. Antimicrobial activity against Shigella sonnei and probiotic properties of wild Lactobacilli from fermented food. Microbiol. Res. 2011;167:27–31. doi: 10.1016/j.micres.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Shi S.Q., Gong L., Yu H., He G.Y., Zhang J.J., Han Y., Liu Y.N., Hu J., Dong J.S., Liu J., et al. Antagonistic activity and mechanism of Lactobacillus rhamnosus SQ511 against Salmonella enteritidis. 3 Biotech. 2022;12:126. doi: 10.1007/s13205-022-03176-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei Y.H., Wang J.Z., Liu Z., Pei J.J., Brennan C., Abd El-Aty A.M. Isolation and characterization of bacteriocin-producing Lacticaseibacillus rhamnosus XN2 from yak yoghurt and its bacteriocin. Molecules. 2022;27:2066. doi: 10.3390/molecules27072066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zorova L.D., Popkov V.A., Plotnikov E.Y., Silachev D.N., Pevzner I.B., Jankauskas S.S., Babenko V.A., Zorov S.D., Balakireva A.V., Juhaszova M., et al. Mitochondrial membrane potential. Anal. Biochem. 2018;552:50–59. doi: 10.1016/j.ab.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamal R.M., Alnakip M.E., Abd El Aal S.F., Bayoumi M.A. Bio-controlling capability of probiotic strain Lactobacillus rhamnosus against some common foodborne pathogens in yoghurt. Int. Dairy J. 2018;85:1–7. doi: 10.1016/j.idairyj.2018.04.007. [DOI] [Google Scholar]
- 24.Khan I., Bahuguna A., Kumar P., Bajpai V.K., Kang S.C. Antimicrobial potential of carvacrol against uropathogenic Escherichia coli via membrane disruption, depolarization, and reactive oxygen species generation. Front. Microbiol. 2017;8:2421. doi: 10.3389/fmicb.2017.02421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ezraty B., Gennaris A., Barras F., Collet J.F. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017;15:385–396. doi: 10.1038/nrmicro.2017.26. [DOI] [PubMed] [Google Scholar]
- 26.Warnes S.L., Caves V., Keevil C.W. Mechanism of copper surface toxicity in Escherichia coli O157:H7 and Salmonella involves immediate membrane depolarization followed by slower rate of DNA destruction which differs from that observed for Gram-positive bacteria. Environ. Microbiol. 2012;14:1730–1743. doi: 10.1111/j.1462-2920.2011.02677.x. [DOI] [PubMed] [Google Scholar]
- 27.Sun D.D., Zhang W.W., Mou Z.P., Chen Y., Guo F., Yang E.D., Wang W.Y. Transcriptome analysis reveals silver nanoparticle-decorated quercetin antibacterial molecular mechanism. ACS Appl. Mater. Interfaces. 2017;9:10047–10060. doi: 10.1021/acsami.7b02380. [DOI] [PubMed] [Google Scholar]
- 28.Tang H., Chen W.X., Dou Z.M., Chen R.H., Hu Y.Y., Chen W.J., Chen H.M. Antimicrobial effect of black pepper petroleum ether extract for the morphology of Listeria monocytogenes and Salmonella typhimurium. J. Food Sci. Tech. Mys. 2017;54:2067–2076. doi: 10.1007/s13197-017-2644-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diao M.M., Qi D.P., Xu M.M., Lu Z.X., Lv F.X., Bie X.M., Zhang C., Zhao H.Z. Antibacterial activity and mechanism of monolauroyl-galactosylglycerol against Bacillus cereus. Food Control. 2018;85:339–344. doi: 10.1016/j.foodcont.2017.10.019. [DOI] [Google Scholar]
- 30.Ibrahim H.R., Sugimoto Y., Aoki T. Ovotransferrin antimicrobial peptide (OTAP-92) kills bacteria through a membrane damage mechanism. Biochim. Biophys. Acta. 2000;1523:196–205. doi: 10.1016/S0304-4165(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 31.Cotter P.D., Ross R.P., Hill C. Bacteriocins—A viable alternative to antibiotics? Nat. Rev. Microbiol. 2013;11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 32.Xu C., Fu Y.Y., Liu F., Liu Z.J., Ma J.G., Jiang R., Song C.N., Jiang Z.M., Hou J.C. Purification and antimicrobial mechanism of a novel bacteriocin produced by Lactobacillus rhamnosus 1.0320. LWT-Food Sci. Technol. 2021;137:110338. doi: 10.1016/j.lwt.2020.110338. [DOI] [Google Scholar]
- 33.Burt S. Essential oils: Their antibacterial properties and potential applications in foods—A review. Int. J. Food Microbiol. 2004;94:223–253. doi: 10.1016/j.ijfoodmicro.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 34.Gong H.S., Meng X.C., Wang H. Mode of action of plantaricin MG, a bacteriocin active against Salmonella typhimurium. J. Basic Microbiol. 2010;50:S37–S45. doi: 10.1002/jobm.201000130. [DOI] [PubMed] [Google Scholar]
- 35.Lou Z.X., Wang H.X., Zhu S., Ma C.Y., Wang Z.P. Antibacterial activity and mechanism of action of chlorogenic acid. J. Food Sci. 2011;76:M398–M403. doi: 10.1111/j.1750-3841.2011.02213.x. [DOI] [PubMed] [Google Scholar]
- 36.Liu R., Zhu F., Lu L., Fu A., Lu J., Deng Z., Liu T. Metabolic engineering of fatty acyl-ACP reductase-dependent pathway to improve fatty alcohol production in Escherichia coli. Metab. Eng. 2014;22:10–21. doi: 10.1016/j.ymben.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Cao Y., Xiao W., Zhang J., Xie Z., Ding M., Yuan Y. Heterologous biosynthesis and manipulation of alkanes in Escherichia coli. Metab. Eng. 2016;38:19–28. doi: 10.1016/j.ymben.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Li Y., Zamble D.B. Nickel homeostasis and nickel regulation: An overview. Chem. Rev. 2009;109:4617–4643. doi: 10.1021/cr900010n. [DOI] [PubMed] [Google Scholar]
- 39.Dixon S.J., Stockwell B.R. The role of iron and reactive oxygen species in cell death. Nat. Chem. Biol. 2014;10:9–17. doi: 10.1038/nchembio.1416. [DOI] [PubMed] [Google Scholar]
- 40.Kohanski M.A., Dwyer D.J., Wierzbowski J., Cottarel G., Collins J.J. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chenault S.S., Earhart C.F. Organization of genes encoding membrane proteins of the Escherichia coli ferrienterobactin permease. Mol. Microbiol. 1991;5:1405–1413. doi: 10.1111/j.1365-2958.1991.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 42.Mousavi F., Bojko B., Bessonneau V., Pawliszyn J. Cinnamaldehyde characterization as an antibacterial agent toward E. coli metabolic profile using 96-blade solid-phase microextraction coupled to liquid chromatography–mass spectrometry. J. Proteome Res. 2016;15:963–975. doi: 10.1021/acs.jproteome.5b00992. [DOI] [PubMed] [Google Scholar]
- 43.Miyanoiri Y., Ishida Y., Takeda M., Terauchi T., Inouye M., Kainosho M. Highly efficient residue-selective labeling with isotope-labeled Ile, Leu, and Val using a new auxotrophic E. coli strain. J. Biomol. NMR. 2016;65:109–119. doi: 10.1007/s10858-016-0042-0. [DOI] [PubMed] [Google Scholar]
- 44.Lennon C.W., Thamsen M., Friman E.T., Cacciaglia A., Sachsenhauser V., Sorgenfrei F.A., Wasik M.A., Bardwell J.C.A. Folding optimization in vivo uncovers new chaperones. J. Mol. Biol. 2015;427:2983–2994. doi: 10.1016/j.jmb.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan Z., Hussain S., Wang X., Bernstein H.D., Bardwell J.C.A. Chaperone OsmY facilitates the biogenesis of a major family of autotransporters. Mol. Microbiol. 2019;112:1373–1387. doi: 10.1111/mmi.14358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCoy L.S., Xie Y., Tor Y. Antibiotics that target protein synthesis. WIREs RNA. 2011;2:209–232. doi: 10.1002/wrna.60. [DOI] [PubMed] [Google Scholar]
- 47.Bai J., Wu Y., Bu Q., Zhong K., Gao H. Comparative study on antibacterial mechanism of shikimic acid and quinic acid against Staphylococcus aureus through transcriptomic and metabolomic approaches. LWT-Food Sci. Technol. 2022;153:112441. doi: 10.1016/j.lwt.2021.112441. [DOI] [Google Scholar]
- 48.Peacock J.R., Walvoord R.R., Chang A.Y., Kozlowski M.C., Gamper H., Hou Y. Amino acid-dependent stability of the acyl linkage in aminoacyl-tRNA. RNA. 2014;20:758–764. doi: 10.1261/rna.044123.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin E., Huang C. Crystal structure of the single-stranded DNA-binding protein SsbB in complex with the anticancer drug 5-fluorouracil: Extension of the 5-fluorouracil interactome to include the oligonucleotide/oligosaccharide-binding fold protein. Biochem. Biophys. Res. Commun. 2021;534:41–46. doi: 10.1016/j.bbrc.2020.11.125. [DOI] [PubMed] [Google Scholar]
- 50.Grosser M.R., Paluscio E., Thurlow L.R., Dillon M.M., Cooper V.S., Kawula T.H., Richardson A.R. Genetic requirements for Staphylococcus aureus nitric oxide resistance and virulence. PLoS Pathog. 2018;14:e1006907. doi: 10.1371/journal.ppat.1006907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arrioja-Breton D., Mani-Lopez E., Palou E., Lopez-Malo A. Antimicrobial activity and storage stability of cell-free supernatants from lactic acid bacteria and their applications with fresh beef. Food Control. 2020;115:107286. doi: 10.1016/j.foodcont.2020.107286. [DOI] [Google Scholar]
- 52.De Alcntara A., Bruzaroski S.R., Luiz L.L., de Souza C., Poli-Frederico R.C., Fagnani R., de Santana E. Antimicrobial activity of Lactobacillus rhamnosus against Pseudomonas fluorescens and Pseudomonas putida from raw milk. J. Food Process Preserv. 2019;43:e14082. doi: 10.1111/jfpp.14082. [DOI] [Google Scholar]
- 53.Benavides A.B., Ulcuango M., Yepez L., Tenea G.N. Assessment of the in vitro bioactive properties of lactic acid bacteria isolated from native ecological niches of Ecuador. Rev. Argent. Microbiol. 2016;48:236–244. doi: 10.1016/j.ram.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 54.Wang R.X., Li J.H., Qu G.R., Guo D., Yang Y.P., Ma X.Y., Wang M.X., Xu Y.F., Wang Y.T., Xia X.D., et al. Antibacterial activity and mechanism of coenzyme Q (0) against Escherichia coli. Foodborne Pathog. Dis. 2021;18:398–404. doi: 10.1089/fpd.2020.2884. [DOI] [PubMed] [Google Scholar]
- 55.Yan H., Lu Y.Y., Li X., Yi Y.L., Wang X., Shan Y.Y., Liu B.F., Zhou Y., Lu X. Action mode of bacteriocin BM1829 against Escherichia coli and Staphylococcus aureus. Food Biosci. 2021;39:100794. doi: 10.1016/j.fbio.2020.100794. [DOI] [Google Scholar]
- 56.Ali Z.I., Saudi A.M., Albrecht R., Talaat A.M. The inhibitory effect of nisin on Mycobacterium avium ssp. paratuberculosis and its effect on mycobacterial cell wall. J. Dairy Sci. 2019;102:4935–4944. doi: 10.3168/jds.2018-16106. [DOI] [PubMed] [Google Scholar]
- 57.Modiri S., Kermanshahi R.K., Soudi M.R., Arab S.S., Khammari A., Cousineau B., Vali H., Zahiri H.S., Noghabi K.A. Multifunctional acidocin 4356 combats Pseudomonas aeruginosa through membrane perturbation and virulence attenuation: Experimental results confirm molecular dynamics simulation. Appl. Environ. Microbiol. 2020;86:e00367-20. doi: 10.1128/AEM.00367-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuroff A.S., Sabat G., Hickey W.J. Transporter-mediated uptake of 2-chloro- and 2-hydroxybenzoate by Pseudomonas huttiensis strain D1. Appl. Environ. Microbiol. 2003;69:7401–7408. doi: 10.1128/AEM.69.12.7401-7408.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ma B., Fang C., Lu L.S., Wang M.Z., Xue X.Y., Zhou Y., Li M.K., Hu Y., Luo X.X., Hou Z. The antimicrobial peptide thanatin disrupts the bacterial outer membrane and inactivates the NDM-1 metallo-beta-lactamase. Nat. Commun. 2019;10:3517. doi: 10.1038/s41467-019-11503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yarlagadda V., Akkapeddi P., Manjunath G.B., Haldar J. Membrane active vancomycin analogues: A strategy to combat bacterial resistance. J. Med. Chem. 2014;57:4558–4568. doi: 10.1021/jm500270w. [DOI] [PubMed] [Google Scholar]
- 61.Fei P., Ali M.A., Gong S.Y., Sun Q., Bi X., Liu S.F., Guo L. Antimicrobial activity and mechanism of action of olive oil polyphenols extract against Cronobacter sakazakii. Food Control. 2018;94:289–294. doi: 10.1016/j.foodcont.2018.07.022. [DOI] [Google Scholar]
- 62.Molenaar D., Abee T., Konings W.N. Continuous measurement of the cytoplasmic pH in Lactococcus lactis with a fluorescent pH indicator. Biochim. Biophys. Acta. 1991;1115:75–83. doi: 10.1016/0304-4165(91)90014-8. [DOI] [PubMed] [Google Scholar]
- 63.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available from the corresponding author upon reasonable request.